- 1University Diagnostic Laboratory, Institute of Microbiology, University of Veterinary and Animal Science, Lahore, Pakistan
- 2Department of Microbiology and Molecular Genetics, University of California, Irvine, Irvine, CA, United States
- 3Department of Pharmacology and Toxicology, University of Veterinary and Animal Sciences, Lahore, Pakistan
This study involves the development and molecular characterization of the isogenic markerless knockout mutant SG ΔpurB, a genetically engineered live attenuated strain aimed at controlling Salmonella Gallinarum (SG) infection in poultry. The mutant was generated by deleting the purB gene using λ-Red recombination technology, impairing adenylosuccinate lyase, necessary for purine biosynthesis. An 1,180 bp deletion was engineered within the purB gene, leaving a residual 298 bp genomic scar resulting in a purine auxotrophic mutant. Phenotypically, SG ΔpurB showed a 66.5% reduction in growth in LB broth compared to the wild-type strain and failed to grow in minimal media without adenosine. Growth was restored to near wild-type levels with 0.3 mM adenosine supplementation, demonstrating the strain’s conditional attenuation. In vivo pathogenicity assessments revealed that oral inoculation of SG ΔpurB into 3-day-old chickens at a dose of 2 × 108 CFU resulted in zero mortality, compared to an 80% mortality rate in chickens challenged with the wild-type strain. The SG ΔpurB strain exhibited significantly reduced clinical signs and lesion scores, with clinical sign scores dropping from 2.5/3 with the wild-type to 0.4/3 with the ΔpurB mutant, and lesion scores decreasing from 2.9/3 to 0.3/3. Additionally, the mutant was efficiently cleared from liver and spleen tissues by 14 days post-inoculation, unlike the wild-type strain, which persisted until the experiment’s end on day 21. The SG ΔpurB mutant shows potential as a safe alternative for preventing fowl typhoid, highlighting the promise of targeted genetic attenuation in developing effective vaccines for poultry diseases.
1 Introduction
Salmonellosis continues to be a prevalent infectious disease within the poultry sector globally (Calnek, 1991; Chappell et al., 2009). Salmonella enterica serovar Gallinarum (SG), a host-specific serovar., induces fowl typhoid (FT), a systemic condition characterized by septicemia affecting domestic poultry of all ages, including week-old chicks (Calnek, 1991; Kwon et al., 2010). Fowl typhoid persists as a significant concern in various regions experiencing elevated ambient temperatures, which complicate environmental hygiene, leading to considerable economic losses due to mortality, morbidity, and reduced egg production (Zhang-Barber et al., 1998; Berchieri et al., 2001; Kwon et al., 2010). Vaccination represents an effective approach for the prevention of Salmonella infections (Mastroeni et al., 2001). Live attenuated Salmonella strains are more effective as vaccines against salmonellosis in various animal species compared to inactivated vaccines (Smith, 1956). Inactivated vaccines stimulate antibody production but do not significantly enhance the cellular immunity (Gast et al., 1993). In contrast, live vaccines generate strong humoral and cellular immune responses, particularly when the vaccine strain is invasive (Babu et al., 2003). Despite not having full accreditation, the widely recognized live vaccine strain 9R has substantially contributed to reducing the prevalence of the disease (Smith, 1956; Bouzoubaa et al., 1989; Kwon et al., 2010), However, the persistence of residual virulence, incomplete immunity, and the undefined genotype of the 9R vaccine strain have prompted researchers to pursue the development of improved vaccine strains (Griffin and Barrow, 1993; Shah et al., 2007; Matsuda et al., 2010; Penha Filho et al., 2010). To improve vaccine safety, it is recommended to explore attenuated strains with well-characterized genotypes. Previously various genes have been mutated in SG toward developing a safe vaccine candidate including aroA (Griffin and Barrow, 1993), SPI-2 (Jones et al., 1998), nuoG (Zhang-Barber et al., 1998), aroA-serC (Barrow et al., 2000), metC (Shah et al., 2007), ΔlonΔcpxR (Matsuda et al., 2011a), fur (Łaniewski et al., 2014), ΔcobSΔcbiA (Penha Filho et al., 2010; Penha Filho et al., 2016) among others. In the early 1950s, Bacon and his colleagues observed that mutation in gene involved in purine production results in reduced virulence (Bacon et al., 1950a,b). Another experiment was conducted to evaluate the impact of various purine auxotrophic mutations on the virulence of a Vi-positive strain of Salmonella dublin and two strains of Salmonella typhimurium in mice (McFarland and Stocker, 1987). To our knowledge, the generation and safety assessment of purine-deficient Salmonella Gallinarum have not been previously documented. This study investigates the impact of purB deletion in SG on virulence, lesion scoring, and bacterial persistence, offering insights into its potential role in pathogenicity modulation. Specifically, we describe the creation of a single isogenic, markerless knockout mutant, denoted as SG ΔpurB, engineered using λ-Red recombination. This mutant strain, lacking purB, disrupts adenylosuccinate lyase function, impacting purine biosynthesis and resulting in slower growth. The observed reduction in virulence in certain auxotrophic strains is attributed to compromised growth, impeding evasion of host defenses in contexts where essential metabolites are deficient (McFarland and Stocker, 1987). Further investigations are necessary to evaluate its immunogenicity and protective efficacy. Strategies like co-administration with adjuvants or combination with additional attenuating mutations may enhance its immunogenicity and effectiveness against Salmonella Gallinarum infection, addressing poultry health concerns.
2 Materials and methods
2.1 Bacterial strains and plasmids
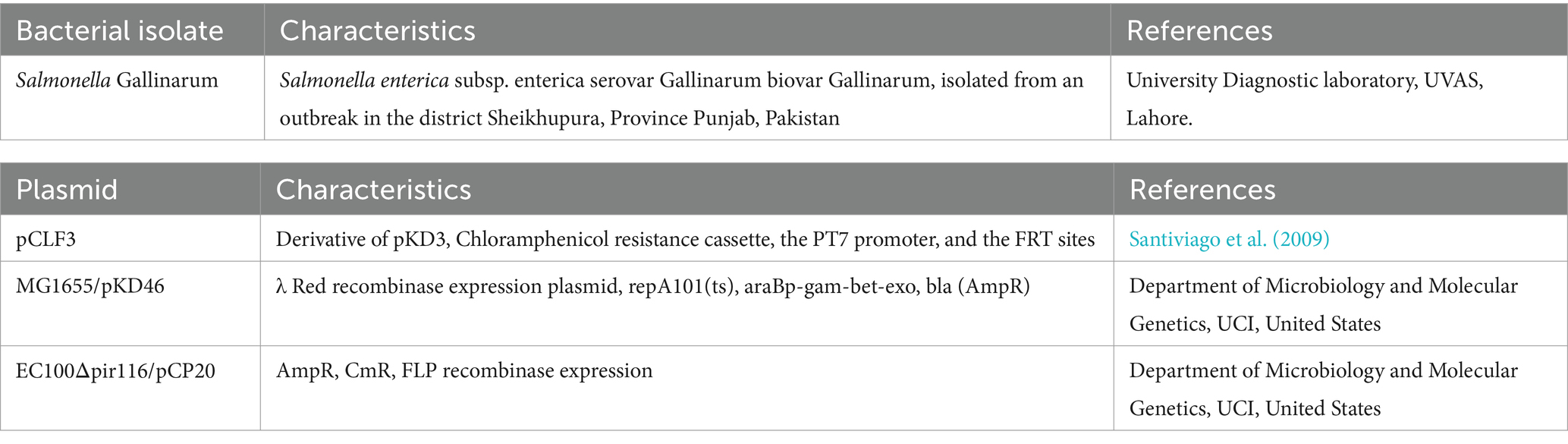
Bacterial strains and plasmids employed in this study are delineated in Table 1. Local isolates of Salmonella enterica subsp. enterica serovar Gallinarum biovar Gallinarum (SG) (Accession no. CP150644) were sourced from the University Diagnostic Laboratory (UDL) of UVAS, Lahore. Plasmids utilized in the study, denoted as pCLF3 (EU629213), pKD46 and pCP20 (Datsenko and Wanner, 2000) were acquired from the McClelland laboratory, UCI, United States.
2.2 Construction of single mutant ΔpurB of Salmonella Gallinarum
The markerless isogenic single mutant ΔpurB:ΩCm of SG was constructed by λ-Red-mediated recombination system (Datsenko and Wanner, 2000). The primer sequences employed for the generation and verification of mutant are detailed in Table 2.
2.2.1 Amplification of purB-F50: CmR: purB-R50
For targeted deletion of the purB, specific 70-base pair primers purB-F and purB-R were utilized to amplify segments containing the Chloramphenicol resistance marker along with Promoter T7 and FLP recombination target (FRT) sites from pCLF3. The resulting PCR products, flanked by 50-base homologous sequences at their ends, were designed to match regions proximal to the 5′ and 3′ ends of the purB gene. Cycling parameters included initial denaturation at 98°C for 1 min 30 s, followed by denaturation at 98°C for 15 s, annealing at 54°C for 20 s, and elongation at 72°C for 1 min 10 s for five cycles. Subsequently, there were 30 cycles of denaturation at 98°C for 15 s and elongation at 72°C for 1 min 30 s, with a final elongation step at 72°C for 3 min. After confirmation via gel electrophoresis, the amplified selection cassette was purified from the PCR mixture using the QIAquick PCR purification kit (QIAGEN, Germany).
2.2.2 Electro-transformation of Salmonella Gallinarum with pKD46 (λ red plasmid)
The Lambda red plasmid pKD46 was extracted from MG1655 Escherichia coli using the commercially available plasmid extraction kit QIAprep® Spin Miniprep Kit (QIAGEN, United States). S. gallinarum was prepared for electroporation using the subsequent protocol with some modifications (Cox et al., 2007) Datsenko and Wanner. The prepared electrocompetent cells were electroporated with the pKD46 plasmid at 1.8 kV in 1 mm cuvette (Bio-Rad, United States) for 5.7 milliseconds (ms) using EC1 on MicroPulser electroporator (Bio-Rad, United States), followed by recovery in SOC media and plating on LB medium supplemented with ampicillin (100 μg/mL). All resulting Salmonella Gallinarum transformants carrying the pKD46 plasmid were stored at-80°C for further analysis.
2.2.3 Electro-transformation of purB-F50: CmR: purB-R50 into SG: pKD46
SG cells harboring the pKD46 plasmid (46 μL) were inoculated into 23 mL of LB/Amp broth and incubated at 30°C with agitation for 1 h, followed by induction with 0.02% L-arabinose. Cells were grown to an OD600 of 0.47–0.48, and electrocompetent cells were prepared as described. Purified PCR product purB-F50: CmR: purB-R50 having DNA concentration 1.5 μg (Serra-Moreno et al., 2006) was electroporated into electrocompetent cells of SG:pKD46 at 1.8 kV in a 1 mm cuvette (Bio-Rad, United States), followed by recovery in SOC media for 50 min at 37°C with shaking. Both, the cells harboring the purB (CmR) marker and the cells subjected to the water control (IDT Nuclease-free water), were spread onto LB/Cm (15 μg/mL) plates and incubated overnight at 37°C (Karlinsey, 2007). Confirmation of SG ΔpurB:ΩCm colonies was achieved through PCR amplification using purB flank-F, purB flank-R, and Cassette-R primers.
2.2.4 Excision of resistance marker and curing of pCP20
To generate an isogenic markerless mutant, the Cm resistance marker was deleted following specific modifications (Czarniak and Hensel, 2015). A volume of 50 μL of competent cells of SG ΔpurB:ΩCm was mixed with 3 μL of plasmid pCP20 in a pre-chilled electroporation cuvette and electroporated at 1.8 kV, followed by recovery in SOC media for 50 min at 30°C with shaking. Cells harboring the pCP20 plasmid were then spread onto LB/Amp (100 μg/mL) plates and incubated at room temperature. After 48 h, the cells were scraped, suspended in LB/20% glycerol, and diluted for overnight culture at 43°C. The culture was then streaked and spread on LB plates and incubated at 37°C. Eight colonies were selected and tested on LB/Cm (15 μg/mL) and LB/Amp (100 μg/mL) plates. The resulting SG ΔpurB mutants were further verified by PCR and stored in 20% glycerol at-80°C.
2.3 Phenotypic characteristics of SG ΔpurB
For the auxotrophic experiment, a single colony of SG ΔpurB was cultured in 3 mL LB broth overnight at 37°C. Subsequently, 200ul of the overnight culture was inoculated into 20 mL of M9 minimal media, with one set of tubes supplemented with 0.3 mM adenosine and the other set without adenosine, incubated overnight at 37°C (Park et al., 2007; Jelsbak et al., 2014). To assess the in vitro growth pattern, a confirmed colony of SG ΔpurB was inoculated into 3 mL LB broth and incubated overnight at 37°C. The following day, 200 μL of the overnight culture was transferred into 20 mL of LB broth, and optical density (OD600) was measured every hour for a duration of 10 h. The growth pattern of the wild-type SG was similarly observed as a control under the same conditions (Kang et al., 2022).
2.4 Animal ethics and husbandry conditions
The chicken experiments in this study were approved by the Animal Ethics Committee of UVAS and conducted in accordance with institutional ethical guidelines. Disease-free broiler chickens were obtained as day-old chicks from a commercial hatchery and housed in pre-sterilized pens within an environmentally controlled room, provided with water and antibiotic-free food ad lib.
2.5 Assessment of bacterial virulence
This experiment involved the evaluation of SG wild-type strain and SG ΔpurB virulence by administering different doses of each strain orally to chickens. One hundred and ten day-old chickens were divided into three major groups (Group A, B, and C). Groups A and B, each comprising fifty chickens, were further subdivided into subsequent sub-groups, each containing ten birds. Group C consisted of 10 birds. On day 3, each sub-group of Group A (n = 10) received a 10-fold dilution (ranging from 1 × 105 to 1 × 109) dose of SG ΔpurB, while each sub-group of Group B was inoculated with a 10-fold dilution (1 × 105 to 1 × 109) dose of SG wild-type via oral route in 100 μL PBS. Ten birds in group C were inoculated with 100 μL PBS as a control group. Bird mortality was monitored for duration of 2 weeks. The Lethal dose (LD50) was calculated by Probit analysis (Finney, 1971).
2.5.1 General condition, mortality and gross lesion observations
In this experiment, ninety Salmonella-negative day-old chickens were allocated into three groups. On day 3, Group A and Group B were orally inoculated with the SG ΔpurB mutant and SG wild-type strains, respectively, at a dosage of 2 × 108 CFU per bird in 100 μL PBS, while Control Group C received 100 μL PBS via the same route. Body weights of the birds were recorded at 0, 7, 14, 21, and 28 days post-infection (DPI) (Kang et al., 2022). Throughout the 28-day observation period, clinical symptoms and lesion scores were monitored. Gross lesions in the spleen and liver were assessed through post-mortem examination of 5 randomly selected birds from each group on 7 DPI, with the remaining chickens being humanely slaughtered at the trial’s conclusion. Clinical symptoms, such as depression and diarrhea, were monitored daily from 5DPI to 10DPI. The scoring criteria for both clinical symptoms and gross lesions were based on methods described in a previous study (Matsuda et al., 2011b).
2.5.2 Persistence of bacteria in liver and spleen
Bacteriological examination of the organs was conducted to assess bacterial persistence. Three birds from each group were slaughtered at 3, 7, 10, 14 and 21 DPI, and samples of the liver and spleen were collected aseptically in sterile zip bags. One gram samples of the liver and spleen were minced and homogenized in 1 mL of PBS. Subsequently, 10-fold serially diluted samples (100 μL) were inoculated onto BGA and XLD agar plates, which were then incubated at 37°C for 24 h. Additionally, homogenized samples in PBS were inoculated into Rappaport-Vassiliadis broth at 42°C for 24 to 48 h for enrichment, followed by inoculation of 100 μL onto BGA agar plates, which were incubated at 37°C for 24 h. Confirmation of the mutant strain (SG ΔpurB) and SG wild-type from the samples was achieved via PCR using specific primers purB flank-F/purB flank-R. The persistence of SG ΔpurB and SG wild-type was quantified and expressed in log10 CFU/g. Samples that tested positive only after enrichment were considered as 1 CFU/g, while those that remained negative after enrichment were considered as 0 CFU/g for data analysis (Matsuda et al., 2010).
3 Results
3.1 Molecular validation of purB deletion in Salmonella Gallinarum
The amplification of target sequences with purB-F and purB-R primers resulted in 1180 bp PCR product, designated as purB-F50:CmR:purB-R50. This purified PCR product (1.5 μg) when transformed into SG:pKD46, produced 15 colonies on LB/Cm (15 μg/mL) plates, whereas, no colonies were observed in the water control. PCR screening of SG ΔpurB:ΩCm mutants demonstrated a 1,222 bp product in the mutant strain and a 1,421 bp product in the wild-type strain using purB flank-F/purB flank-R primers, confirming the inactivation of purB (Supplementary Figure S1). Additionally, amplification with purB flank-F/Cassette-R primers generated a 114 bp product in the mutant strain, verifying the insertion of the chloramphenicol resistance cassette at the target site (Supplementary Figure S2). The antibiotic cassette inserted into the inactivated purB gene was removed using FLP recombinase, resulting in the intended 298 bp genomic scars, as confirmed by PCR using specific flanking primers purB flank-F/purB flank-R (Supplementary Figure S3).
3.2 Auxotrophic evaluation and growth dynamics of SG ΔpurB
The growth dynamics of SG ΔpurB mutant and SG wild-type were monitored over a 10 h period in LB broth. Following the specified duration, the optical density at 600 nm (OD600) for SG ΔpurB reached 0.47, reflecting a notable 66.5% reduction compared to the robust OD600 of 1.40 observed for the SG wild-type (p = 0.02; Figure 1). The growth dynamics of the SG ΔpurB mutant in M9 minimal media, with or without supplemented adenosine, were evaluated. At the time of inoculation, the SG ΔpurB mutant exhibited an initial OD600 of 0.03. After ten hours of incubation, the SG ΔpurB mutant showed no significant growth, maintaining an OD600 of 0.02. In contrast, the SG wild-type reached an OD600 of 1.40 under the same conditions (p = 0.7). Notably, when the M9 minimal media was supplemented with 0.3 mM adenosine, the OD600 of the SG ΔpurB mutant increased significantly to 1.30, closely approaching the OD600 of 1.40 observed for the SG wild-type (Figure 2).
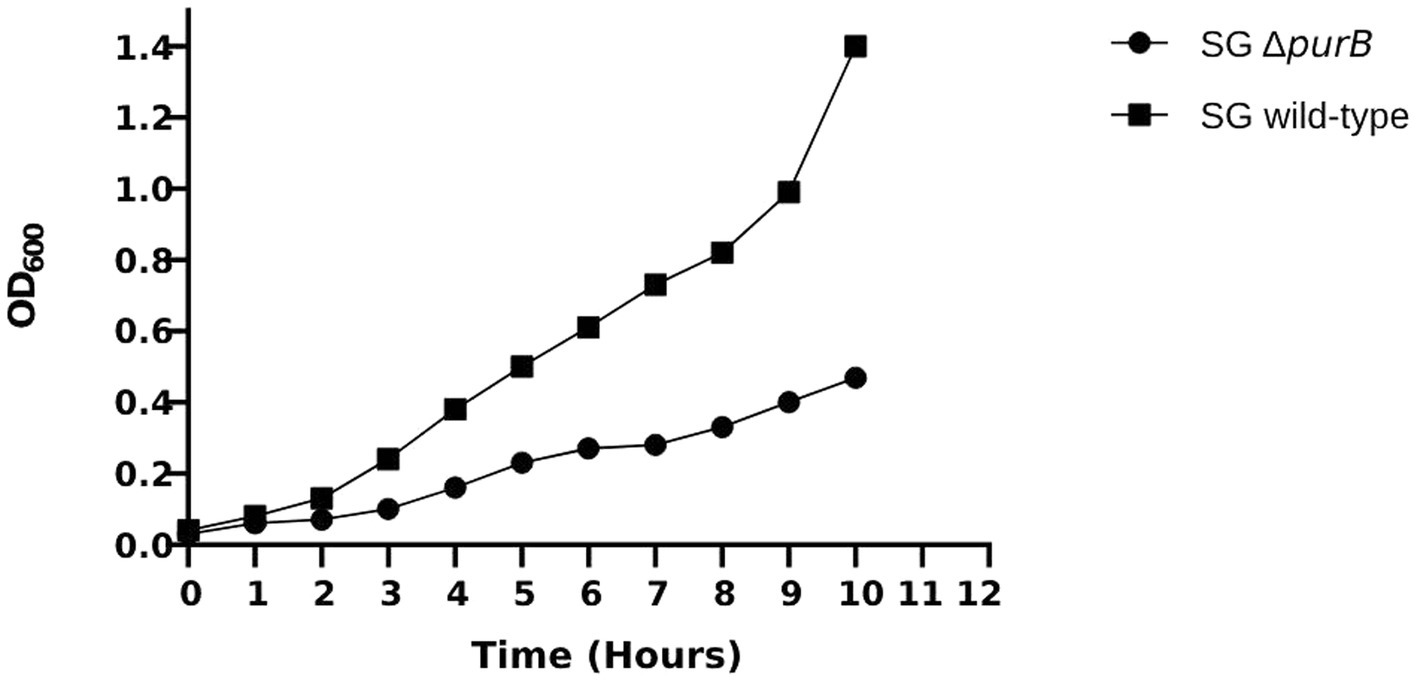
Figure 1. Growth patterns of the SG ΔpurB mutant and the SG wild-type strains in LB broth over a 10 h period. A significant difference in growth between the SG ΔpurB mutant and the SG wild-type was observed at the 10 h mark, as determined by a two-tailed Student’s t-test with equal variance (p = 0.02).
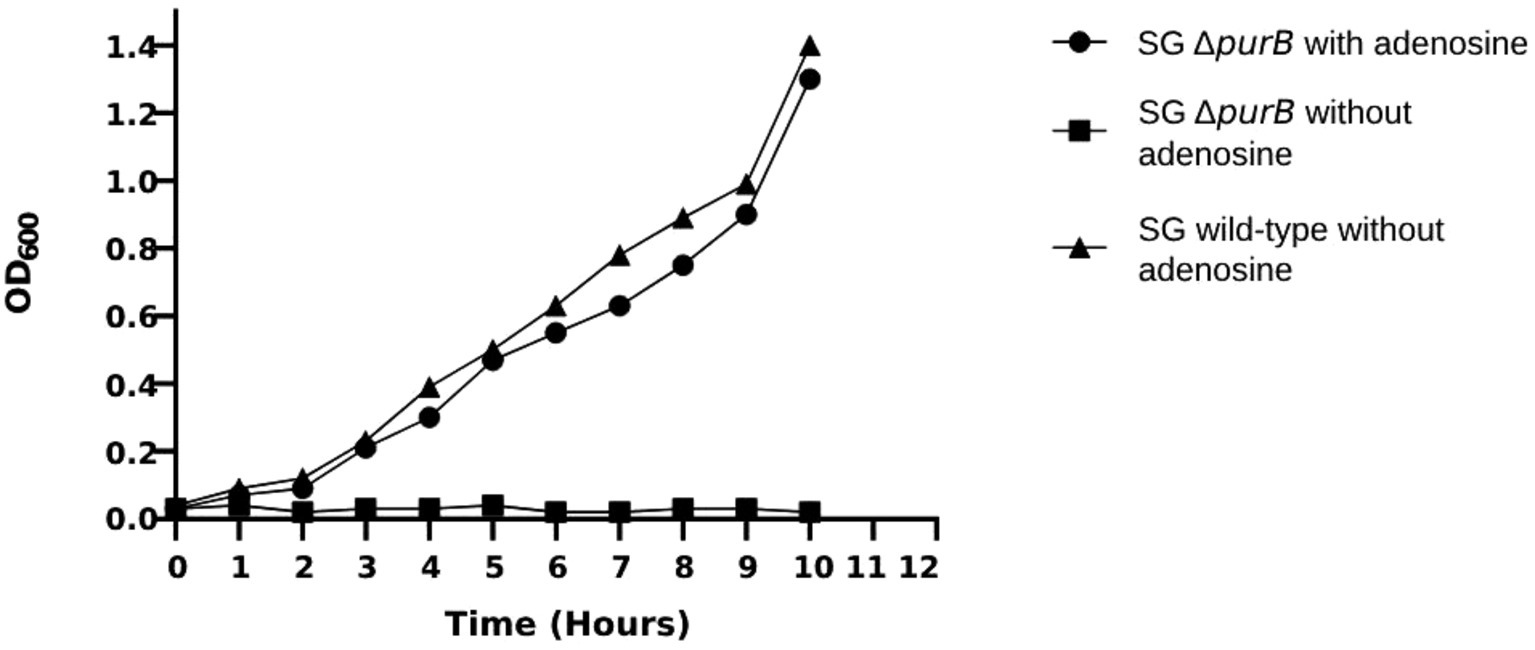
Figure 2. Growth dynamics of the SG ΔpurB mutant and SG wild-type strains in M9 minimal media, with and without adenosine supplementation. The growth curve of SG ΔpurB supplemented with adenosine showed no significant difference compared to SG wild-type without adenosine (p = 0.7). However, a significant difference was observed between the growth curve of SG ΔpurB and SG wild-type without adenosine supplementation (p < 0.05), as determined by a t-test.
3.3 Virulence assessment of SG ΔpurB mutant vs. SG wild-type
Virulence was evaluated in 3-day old chickens by calculating LD50 of SG ΔpurB and SG wild-type. LD50 of SG wild-type was 2 × 108 CFU/mL whereas no chickens died in the group challenged by SG ΔpurB.
Following the oral administration of 2 × 108 CFU in 100 μL PBS of SG ΔpurB and SG wild-type to 3-day old chickens, body weight changes were monitored weekly, with values expressed as Mean ± SEM and analyzed using GraphPad Prism 10 software. The data, presented in Table 3, showed that Group B, inoculated with SG wild-type, demonstrated significantly lower body weights than control of 84.4 ± 0.53 g, 221.9 ± 0.60 g, 365.9 ± 0.84 g, 724.5 ± 0.70 g, and 1137.3 ± 0.57 g at 0, 7, 14, 21, and 28 DPI, respectively (p = 0.01). Group A, inoculated with SG ΔpurB, exhibited body weights of 84.6 ± 0.30 g, 255.1 ± 0.80 g, 430.5 ± 0.84 g, 851.2 ± 0.72 g, and 1318.6 ± 1.19 g at 0, 7, 14, 21, and 28 DPI, respectively. These values were similar to those of the control group (Group C), which received 100 μL PBS and showed weights of 84.3 ± 0.36 g, 262 ± 0.89 g, 442.5 ± 0.45 g, 873.5 ± 0.833 g, and 1,340 ± 1.11 g at the same intervals (p = 0.7), which indicate that SG ΔpurB has significantly reduced its ability to cause detrimental effect on chicken growth (Figure 3). Mortality was also recorded throughout the study period, as illustrated in Figure 4. Both Group C (PBS control) and Group A (SG ΔpurB) exhibited no mortality, whereas Group B (SG wild-type) experienced a high mortality rate of 80%. These findings underscore the marked attenuation of virulence in the SG ΔpurB mutant relative to the wild-type strain, as well as its non-lethal effects on the growth performance of the host.
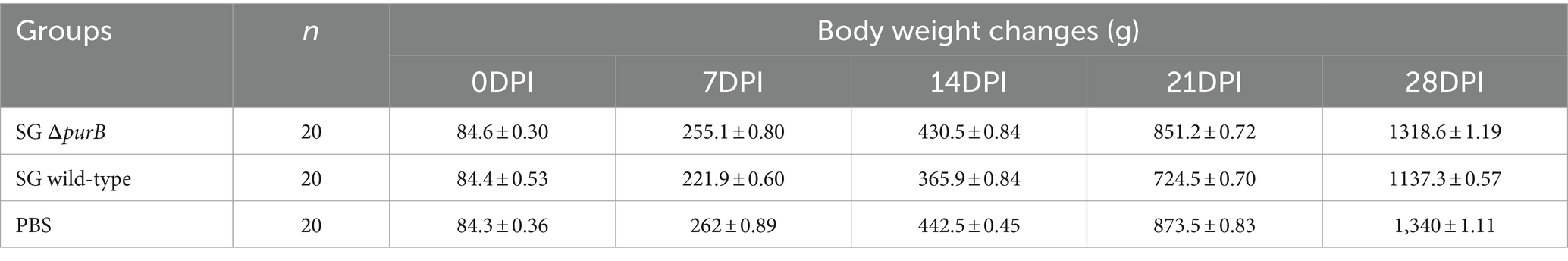
Table 3. Comparison of body weight changes in chickens following infection with SG ΔpurB and SG wild-type.
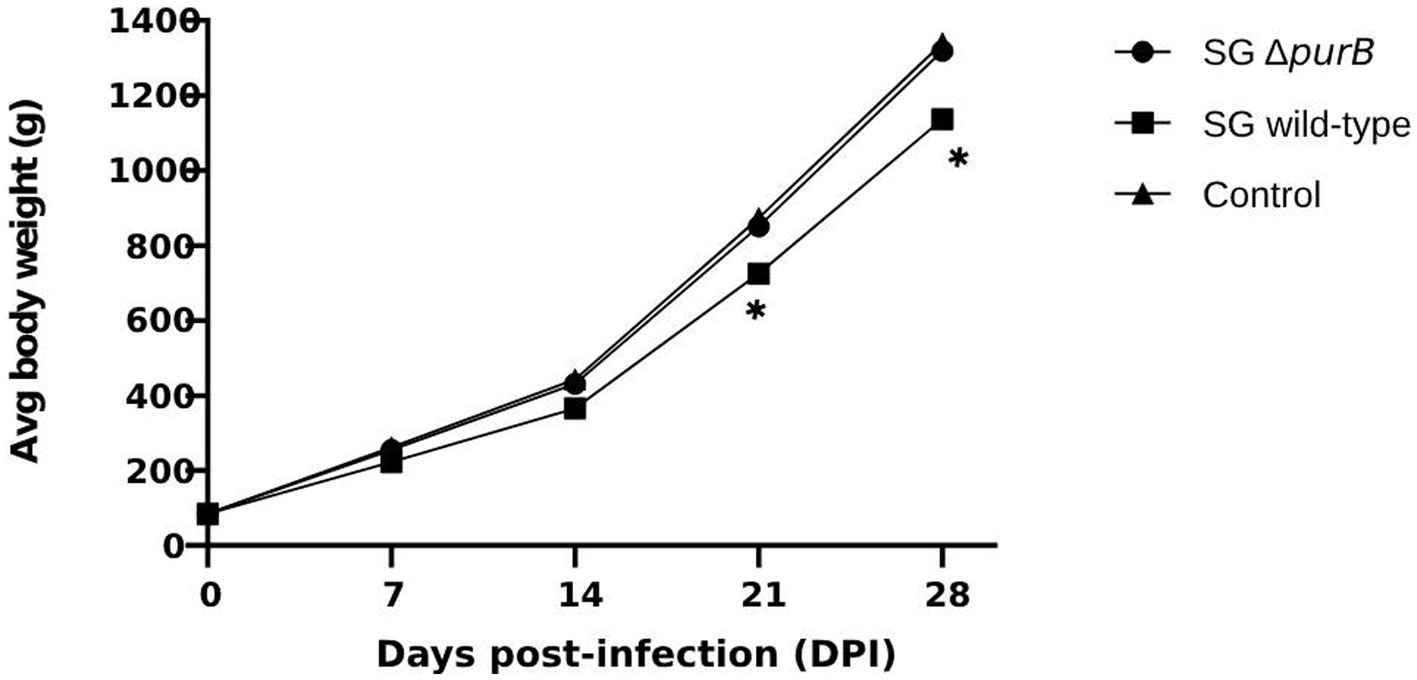
Figure 3. Body weight changes of chickens following infection with SG ΔpurB, SG wild-type, and a control group administered 100 μL of PBS. Body weights were measured at 0, 7, 14, 21, and 28 days post-infection (DPI). A significant difference was observed in the group infected with SG wild-type on 21 and 28 DPI compared to the group infected with SG ΔpurB and the uninfected control group (* p < 0.05), as determined by two-way ANOVA followed by Bonferroni’s multiple comparison tests. Data are presented as mean ± SEM.
Figure 4. Mortality of chickens observed after infection with SGΔpurB, SG wild-type, and control groups administered PBS during the experimental period. The group infected with SG wild-type exhibited 80% mortality, whereas no mortality was observed in the group infected with SGΔpurB and the uninfected control group.
3.4 Clinical manifestations and macroscopic lesions
Clinical observations were conducted bi-daily across all experimental groups (Groups A, B, and C) as delineated in Table 4, with findings presented as mean ± SEM. Notably, chickens in Group B (inoculated with SG wild-type) exhibited significantly heightened depression scores from 5 DPI to 10 DPI compared to Group A, as determined by the Mann- Whitney U-test (p = 0.001). The peak depression scores recorded during the study period for Groups A, B, and C were 0.2 ± 0.1, 2.5 ± 0.3, and 0.0 ± 0.0, respectively (Figure 5).
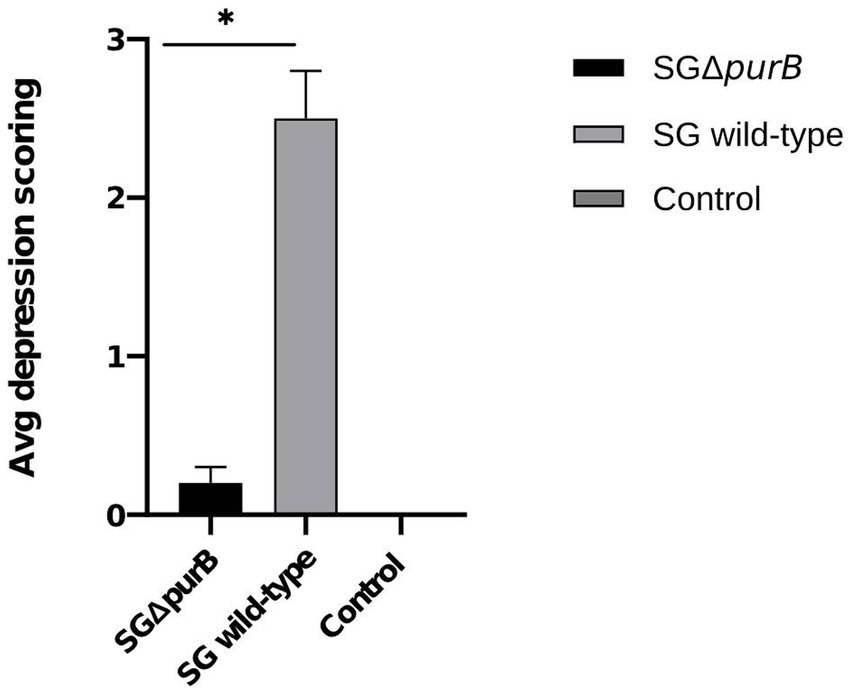
Figure 5. Maximum depression scores of all groups from 5 DPI to 10 DPI, represented as Mean ± SEM. The group infected with SG wild-type (grey bar) showed significantly severe depression scores from 5 DPI compared to the group infected with SG ΔpurB (black bar) (Mann–Whitney U-test, * p < 0.05). The uninfected control group showed no clinical signs.
Similarly, Group B also demonstrated significantly more severe diarrheal symptoms compared to Group A, as evidenced by Mann–Whitney U-test results (p = 0.001). The maximum diarrheal scores observed for Groups A, B, and C throughout the experiment was 0.4 ± 0.2, 2.7 ± 0.2, and 0.0 ± 0.0, respectively, as depicted in Figure 6. To observe the gross lesions, spleen and liver was collected from 5 randomly chickens each group on the 7 DPI. Group B showed significantly severe systemic infection when compared with Group A according to Mann–Whitney U-test (p = 0.001) and data presented as Mean ± SEM as shown in Table 5. Mean score for liver enlargement in group A (SG ΔpurB) and group B (SG wild-type) was 0.6 ± 0.3 and 2.5 ± 0.2, respectively. Whereas mean score for liver necrotic foci in the group A and group B was 0.2 ± 0.1 and 2.7 ± 0.3, respectively. Similarly, the mean score for spleen enlargement in the group A and group B was 0.4 ± 0.1 and 2.7 ± 0.4, respectively. Whereas the mean score for spleen necrotic foci in group A and group B was 0.5 ± 0.3 and 2.5 ± 0.4, respectively. Group C inoculated with PBS, a control group was negative for all gross lesions as shown in Table 5. Graphical presentation also showed in Figures 7, 8.
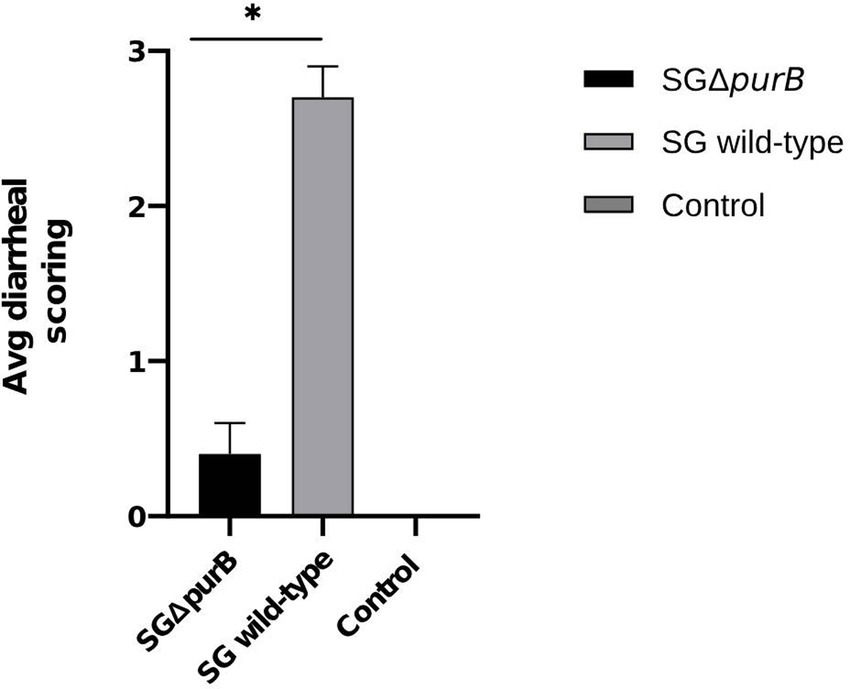
Figure 6. Maximum diarrheal scores of all groups from 5 DPI to 10 DPI, represented as Mean ± SEM. The group infected with SG wild-type (grey bar) showed significantly severe diarrheal scores compared to the group infected with SG ΔpurB (black bar) (Mann–Whitney U-test, * p < 0.05). The uninfected control group showed no clinical signs.
Figure 7. Gross liver lesions were observed at 7 DPI. (A) Mean liver enlargement scores in the SG wild-type group (grey bar) were significantly higher than in the SG ΔpurB group (black bar), as measured by the Mann–Whitney U-test, * p < 0.05 (LE scoring: 0- Clear ridge; 1- Soft tissue; 2- Large without covering gizzard; 3- Large covering gizzard). (B) Mean liver necrotic foci scores in the SG wild-type group (grey bar) were significantly higher than in the SG ΔpurB group (black bar), as measured by the Mann–Whitney U-test, * p < 0.05 (LN scoring: 0- No foci; 1- < 5 foci; 2- < 20 foci; 3- > 20 foci). Data are presented as mean ± SEM. The uninfected control group showed no gross lesions.
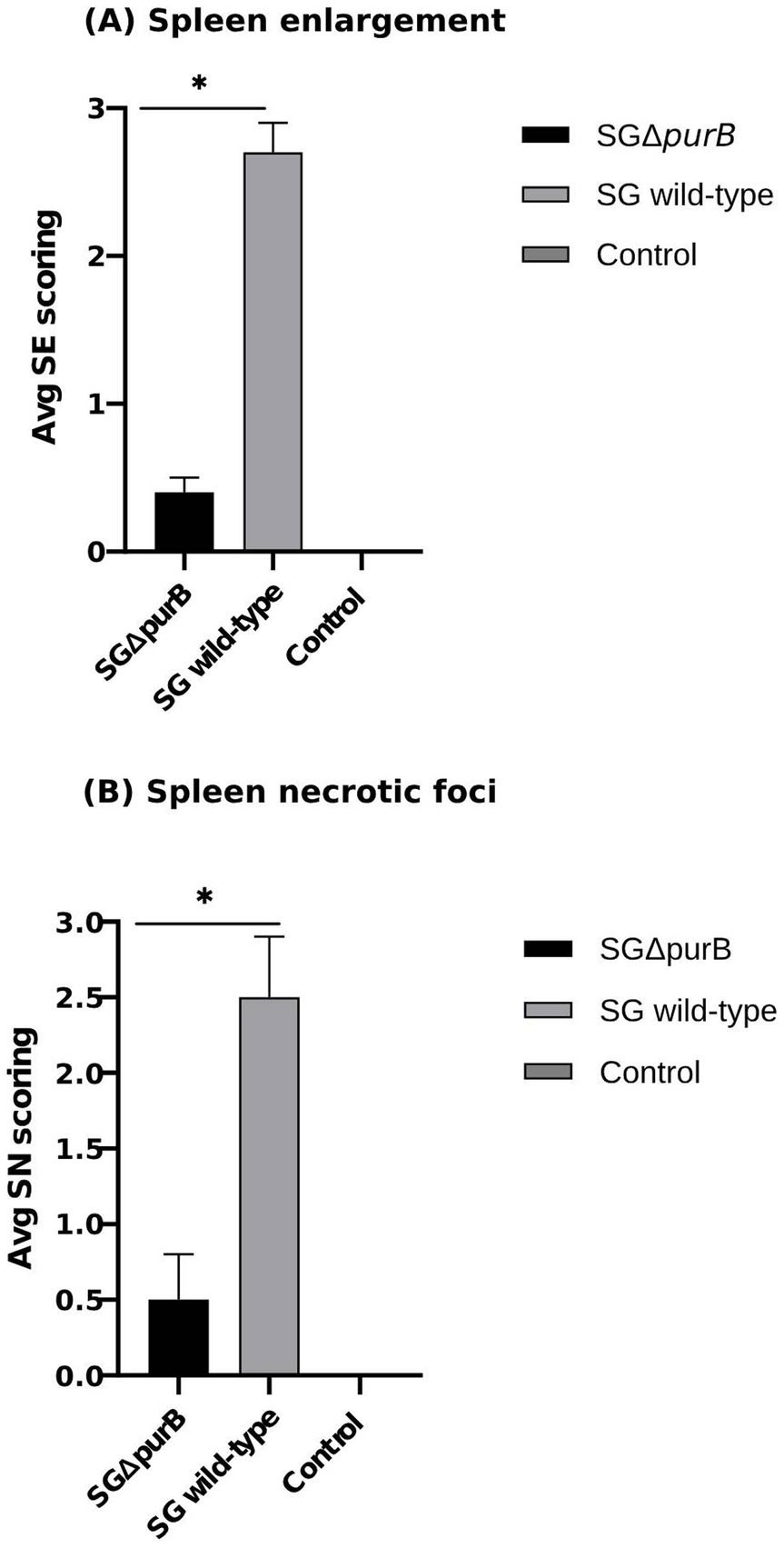
Figure 8. Gross lesions on the spleen were observed on 7 DPI. (A) Mean spleen enlargement scores in SG wild-type group (grey bar) was significantly higher than SG ΔpurB group (black bar) as measured by Mann–Whitney U-test, * p < 0.05 (SE scoring, 0-not soft; 1-soft without enlargement; 2-large with <1.5 dm; 3-large with >1.5 dm) (B) Mean spleen necrotic foci scores in SG wild-type group (grey bar) was significantly higher than SG ΔpurB group (black bar) as measured by Mann–Whitney U-test, * p < 0.05 (SN scoring, 0- No foci; 1- if <3 foci; 2- if <10 foci; 3- if >10 foci), data presented as (Mean ± SEM). The uninfected control group had no gross lesions.
3.5 Duration of SG retention in liver and spleen
Bacterial colonization and persistence within the liver and spleen were systematically evaluated at 3, 7, 10, 14 and 21 days post-infection (DPI). The control group showed no Salmonella recovery from either organ following enrichment in Rappaport Vassiliadis broth, confirming the specificity of the infection model. In contrast, colonization by SG ΔpurB was markedly reduced in both the liver and spleen throughout the study period, demonstrating significantly lower bacterial counts (p < 0.05) compared to the SG wild-type. Quantitative analysis revealed that in Group A (SG ΔpurB), the bacterial load in the liver measured 3.11 ± 0.23, 2.19 ± 0.19, 0.98 ± 0.20, 0.0 ± 0.0, and 0.0 ± 0.0 CFU/g, and in the spleen 3.21 ± 0.19, 2.58 ± 0.25, 1.34 ± 0.21, 0.0 ± 0.0, and 0.0 ± 0.0 CFU/g at 3, 7, 10, 14, and 21 DPI, respectively (Figure 9; Table 6). Conversely, Group B (SG wild-type) exhibited consistently higher CFU/g in the liver (5.50 ± 0.17, 6.34 ± 0.32, 5.76 ± 0.22, 4.54 ± 0.21, 3.49 ± 0.16) and spleen (5.12 ± 0.21, 5.98 ± 0.24, 4.89 ± 0.22, 3.88 ± 0.21, 2.89 ± 0.29) at corresponding time points (Figure 10; Table 6).
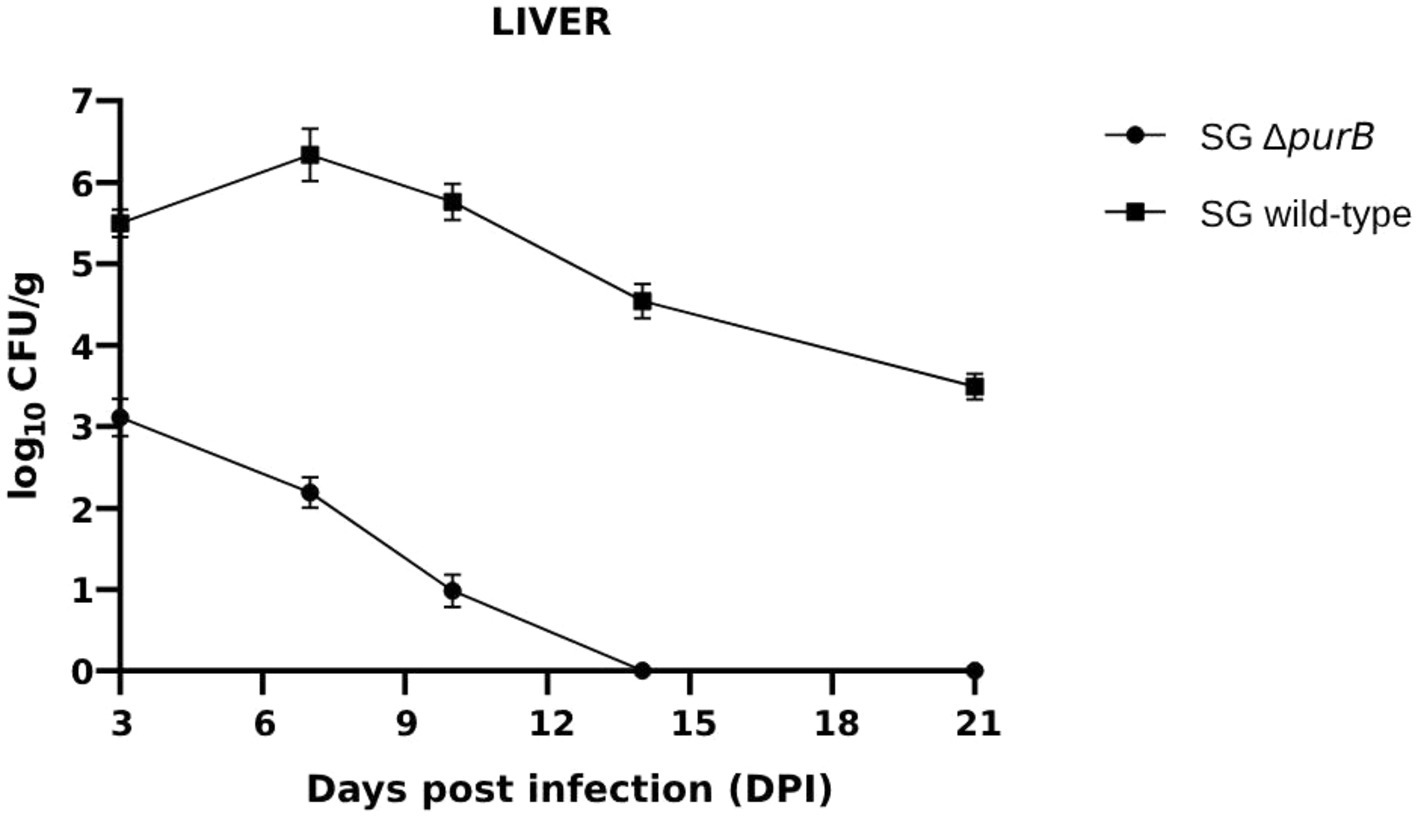
Figure 9. Bacterial colonization in the liver was assessed at 3, 7, 10, 14, and 21 days post-infection (DPI). Bacterial counts are expressed as the log10 of CFU/g. Each data point represents the mean of five samples, with data presented as mean ± SEM. Bacterial persistence in the group infected with the SG wild-type strain was significantly higher than in the group infected with the SG ΔpurB strain, as determined by two-tailed t-tests p < 0.05. The SG ΔpurB strain was cleared from the liver by 14 DPI.
Figure 10. Bacterial colonization in the spleen was assessed at 3, 7, 10, 14, and 21 days post-infection (DPI). Bacterial counts are expressed as the log10 of CFU/g. Each data point represents the mean of five samples, with data presented as mean ± SEM. Bacterial persistence in the group infected with the SG wild-type strain was significantly higher than in the group infected with the SG ΔpurB strain, as determined by two-tailed t-tests p < 0.05. The SG ΔpurB strain was cleared from the spleen by 14 DPI.
4 Discussion
Fowl Typhoid (FT), a septicemic disease induced by Salmonella Gallinarum (SG), causes substantial economic setbacks to the poultry industry worldwide (Matsuda et al., 2011b). Previous studies demonstrated that SG is transmitted both vertically and horizontally (Berchieri et al., 2001). To control and prevent Salmonella infections within poultry flocks, vaccination of chickens is a helpful method in addition to adequate management, good agricultural practices, and stringent biosecurity measures (Revolledo and Ferreira, 2012). When it comes to eliciting an immune response against Salmonella infection, live attenuated vaccines work better than inactivated or subunit vaccines (Barrow, 2007). In the current investigation, we endeavored to develop a single markerless isogenic mutant denoted as SG ΔpurB and subsequently assess its safety, virulence, lesion scoring, and bacterial colonization as a potential live attenuated vaccine candidate. To the best of our knowledge, this is the first example of purine biosynthesis deficient mutant in SG and use of chicken as an infection model for purB mutant.
The current study demonstrated the attenuation of SG through the deletion of the purB gene, which is responsible for encoding adenylosuccinate lyase—a key enzyme in the final step of the purine biosynthesis pathway that converts adenylosuccinate to AMP. Previously, studies have been conducted on mutations in the purine biosynthesis pathway in the purE and purH, purD genes of Brucella melitensis and Shigella flexneri, respectively. These auxotrophic mutants were unable to grow in minimal media without purine supplements (Drazek et al., 1995; Crawford et al., 1996; Cersini et al., 2003). It was also demonstrated that auxotrophic mutants exhibit slow growth due to insufficient nutrient availability (McFarland and Stocker, 1987). In the present study, deletion of purB in SG resulted in 66.5% reduction in growth in LB broth when compared to wild-type. In addition, SG ΔpurB could not maintain its growth in minimal media but restored its growth when supplemented with 0.3 mM adenosine in minimal media. This phenotypic characteristic confirms that purB mutant growth was impaired due to inability of the mutant to produce its endogenous adenosine and requires exogenous adenosine for normal growth which might be due to deficiency of purine biosynthesis as studied previously (Park et al., 2007).
Previous studies in a mouse model showed that purine biosynthesis is crucial for Listeria monocytogenes virulence, as purB mutants exhibited reduced virulence due to impaired multiplication within intestinal epithelial cells (Faith et al., 2012). To study virulence of SG in chicken model, a previously developed lesion scoring method was adopted (Matsuda et al., 2011b). The lesions in the liver and spleen, such as necrotic foci and hepatosplenomegaly caused by lymphocyte infiltration, are characteristic of SG infection. However, their role in protective immunity remains unclear, likely due to the acute pathogenicity of SG in chickens (Permin and Bisgaard, 2013). In this study, clinical signs and lesion scores were significantly higher in chickens infected with the SG wild-type compared to those infected with the SG ΔpurB mutant, indicating significant attenuation of pathogenicity due to the mutation. Oral inoculation with SG ΔpurB resulted in minimal adverse effects, with only a few small necrotic foci observed on the liver and spleen within a few days post-inoculation. Additionally, by 7 days post-inoculation (DPI), there was moderate enlargement of the spleen and liver in the SG ΔpurB-infected group, with recovery observed by 14 DPI. These mild lesions are likely due to a cellular immune response rather than significant tissue and functional disturbances. The significant attenuation observed in the SG ΔpurB mutant aligns with previous findings that mutations introducing new auxotrophic requirements can reduce virulence by hindering bacterial growth in host tissues (Lan et al., 2010). According to previous study (Christensen et al., 1996), the presence of gross lesions and clinical symptoms may be related to the quantity of completely virulent SG in the spleen. However, in cases when the host is resistant or the SG is attenuated, the spleen can remove the SG without causing severe clinical symptoms that would indicate the peak of the immune response (Wigley et al., 2005). To determine if the attenuation of virulence in the purB mutant was due to reduced invasiveness or a lower microbial burden in reticuloendothelial organs, we compared the purB mutant persistence in liver and spleen, with that of the wild-type strain. The purB mutant bacteria were present in the livers and spleens from day 3 post-inoculation, but their numbers were significantly lower (p < 0.01) than those of the WT strain. This indicates that the deletion of the purB gene reduced SG virulence by decreasing the bacterial burden in the liver and spleen of chickens. Importantly, the mutant bacteria were undetectable in the spleen and in the liver from day 14 onwards, suggesting a significant reduction in in vivo growth rate. This reduction is likely due to inadequate adenosine availability in the chicken intestinal tract and/or more effective elimination by the host’s immune defense mechanisms as mentioned in a previous study (Shah et al., 2007). Similarly, in our study, the body weights of chicks infected with SG ΔpurB closely paralleled those of the control group, whereas the weights of the wild-type infected group were significantly lower at 21 and 28 days post-infection (DPI). This weight reduction in the wild-type infected group was associated with high fever, reduced feed intake as described in previous study (Audisio and Terzolo, 2002) and severe clinical signs starting from 5 days post-inoculation, underscoring that purB deletion effectively diminishes SG virulence.
Our study shows that SG ΔpurB caused minimal adverse effects, with only a few minor necrotic foci in the liver and spleen, and are rapidly cleared from these organs within 14 DPI. Infected birds recovered swiftly, indicating a markedly attenuated phenotype which ensures its safety. Further studies would be required to determine the immunogenicity of this strain when used as a vaccine candidate. These findings underscore the promise of SG ΔpurB as a foundation for developing optimized attenuated strains for vaccine design, against Salmonella Gallinarum, addressing key poultry health concerns.
5 Conclusion
Our investigation demonstrates a significant reduction in virulence of the purB-deleted SG strain in chickens following oral inoculation. This strain exhibited early organ clearance, reduced lesion severity, enhanced safety, and no adverse impact on chicken growth rate or weight gain, poses it as a promising vaccine candidate.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary material, further inquiries can be directed to the corresponding author.
Ethics statement
The animal study was approved by Animal Ethics Committee of University of Veterinary and Animal Sciences, Lahore, Pakistan. The study was conducted in accordance with the local legislation and institutional requirements.
Author contributions
MaM: Formal analysis, Methodology, Writing – original draft. AG: Conceptualization, Data curation, Project administration, Resources, Writing – review & editing. MiM: Conceptualization, Data curation, Writing – review & editing. FA: Writing – review & editing. MR: Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. A small travel grant to USA was partially funded by IRSIP HEC. The project was funded by the personal funds of Aamir Ghafoor, Professor at University of Veterinary and Animal Sciences, Lahore.
Acknowledgments
We would also like to express our gratitude to Weiping Chu and Steffen Porwollik in the McClelland lab for their invaluable assistance in lab work. Additionally, we are thankful to Umar Bin Zahoor, Umar Farooq and Shakil Abbas for managing birds in experiments.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2024.1467230/full#supplementary-material
References
Audisio, M. C., and Terzolo, H. R. (2002). Virulence analysis of a Salmonella Gallinarum strain by oral inoculation of 20-day-old chickens. Avian Dis. 46, 186–191. doi: 10.1637/0005-2086(2002)046[0186:VAOASG]2.0.CO;2
Babu, U., Scott, M., Myers, M., Okamura, M., Gaines, D., Yancy, H., et al. (2003). Effects of live attenuated and killed Salmonella vaccine on T-lymphocyte mediated immunity in laying hens. Vet. Immunol. Immunopathol. 91, 39–44. doi: 10.1016/S0165-2427(02)00265-9
Bacon, G., Burrows, T., and Yates, M. (1950a). The effects of biochemical mutation on the virulence of bacterium typhosum: the induction and isolation of mutants. Br. J. Exp. Pathol. 31, 703–713.
Bacon, G., Burrows, T., and Yates, M. (1950b). The effects of biochemical mutation on the virulence of bacterium typhosum: the virulence of mutants. Br. J. Exp. Pathol. 31, 714–724.
Barrow, P. (2007). Salmonella infections: immune and non-immune protection with vaccines. Avian Pathol. 36, 1–13. doi: 10.1080/03079450601113167
Barrow, P., Lovell, M., and Stocker, B. (2000). Protection against experimental fowl typhoid by parenteral administration of live SL5828, an aroA-serC (aromatic dependent) mutant of a wild-type Salmonella Gallinarum strain made lysogenic for P22 sie. Avian Pathol. 29, 423–431. doi: 10.1080/030794500750047171
Berchieri, A. Jr., Murphy, C., Marston, K., and Barrow, P. (2001). Observations on the persistence and vertical transmission of Salmonella enterica serovars Pullorum and Gallinarum in chickens: effect of bacterial and host genetic background. Avian Pathol. 30, 221–231. doi: 10.1080/03079450120054631
Bouzoubaa, K., Nagaraja, K., Kabbai, F., Newman, J., and Pomeroy, B. (1989). Feasibility of using proteins from Salmonella gallinarum vs. 9R live vaccine for the prevention of fowl typhoid in chickens. Avian Dis. 33, 385–391. doi: 10.2307/1591094
Calnek, B. (1991). JOHN BARNES H., BEARD CW, REID WM, YODER HW diseases of poultry, Iowa, USA: University Press Ames.
Cersini, A., Martino, M. C., Martini, I., Rossi, G., and Bernardini, M. L. (2003). Analysis of virulence and inflammatory potential of Shigella flexneri purine biosynthesis mutants. Infect. Immun. 71, 7002–7013. doi: 10.1128/iai.71.12.7002-7013.2003
Chappell, L., Kaiser, P., Barrow, P., Jones, M. A., Johnston, C., and Wigley, P. (2009). The immunobiology of avian systemic salmonellosis. Vet. Immunol. Immunopathol. 128, 53–59. doi: 10.1016/j.vetimm.2008.10.295
Christensen, J., Barrow, P., Olsen, J., Poulsen, J., and Bisgaard, M. (1996). Correlation between viable counts of Salmonella Gallinarum in spleen and liver and the development of anaemia in chickens as seen in experimental fowl typhoid. Avian Pathol. 25, 769–783. doi: 10.1080/03079459608419180
Cox, M. M., Layton, S. L., Jiang, T., Cole, K., Hargis, B. M., Berghman, L. R., et al. (2007). Scarless and site-directed mutagenesis in Salmonella enteritidischromosome. BMC Biotechnol. 7, 1–10. doi: 10.1186/1472-6750-7-59
Crawford, R. M., Van De Verg, L., Yuan, L., Hadfield, T. L., Warren, R. L., Drazek, E. S., et al. (1996). Deletion of purE attenuates Brucella melitensis infection in mice. Infect. Immun. 64, 2188–2192. doi: 10.1128/iai.64.6.2188-2192.1996
Czarniak, F., and Hensel, M. (2015). Red-mediated recombineering of Salmonella enterica genomes. Methods Mol Biol 1225, 63–79. doi: 10.1007/978-1-4939-1625-2_4
Datsenko, K. A., and Wanner, B. L. (2000). One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. 97, 6640–6645. doi: 10.1073/pnas.120163297
Drazek, E. S., Houng, H., Crawford, R. M., Hadfield, T. L., Hoover, D. L., and Warren, R. L. (1995). Deletion of purE attenuates Brucella melitensis 16M for growth in human monocyte-derived macrophages. Infect. Immun. 63, 3297–3301. doi: 10.1128/iai.63.9.3297-3301.1995
Faith, N. G., Kim, J.-W., Azizoglu, R., Kathariou, S., and Czuprynski, C. (2012). Purine biosynthesis mutants (purA and purB) of serotype 4b Listeria monocytogenes are severely attenuated for systemic infection in intragastrically inoculated a/J mice. Foodborne Pathog. Dis. 9, 480–486. doi: 10.1089/fpd.2011.1013
Gast, R. K., Stone, H. D., and Holt, P. S. (1993). Evaluation of the efficacy of oil-emulsion bacterins for reducing fecal shedding of Salmonella enteritidis by laying hens. Avian Dis. 37:1085. doi: 10.2307/1591918
Griffin, H., and Barrow, P. (1993). Construction of an aroA mutant of Salmonella serotype Gallinarum: its effectiveness in immunization against experimental fowl typhoid. Vaccine 11, 457–462. doi: 10.1016/0264-410X(93)90288-9
Jelsbak, L., Hartman, H., Schroll, C., Rosenkrantz, J. T., Lemire, S., Wallrodt, I., et al. (2014). Identification of metabolic pathways essential for fitness of Salmonella Typhimurium in vivo. PLoS One 9:e101869. doi: 10.1371/journal.pone.0101869
Jones, M. A., Wood, M. W., Mullan, P. B., Watson, P. R., Wallis, T. S., and Galyov, E. E. (1998). Secreted effector proteins of Salmonella Dublin act in concert to induce enteritis. Infect. Immun. 66, 5799–5804. doi: 10.1128/iai.66.12.5799-5804.1998
Kang, X., Yang, Y., Meng, C., Wang, X., Liu, B., Geng, S., et al. (2022). Safety and protective efficacy of Salmonella Pullorum spiC and rfaH deletion rough mutant as a live attenuated DIVA vaccine candidate. Poult. Sci. 101:101655. doi: 10.1016/j.psj.2021.101655
Karlinsey, J. E. (2007). λ-Red genetic engineering in Salmonella enterica serovar typhimurium. Methods Enzymol. 421, 199–209. doi: 10.1016/S0076-6879(06)21016-4
Kwon, Y.-K., Kim, A., Kang, M.-S., Her, M., Jung, B.-Y., Lee, K.-M., et al. (2010). Prevalence and characterization of Salmonella Gallinarum in the chicken in Korea during 2000 to 2008. Poult. Sci. 89, 236–242. doi: 10.3382/ps.2009-00420
Lan, L., Cheng, A., Dunman, P. M., Missiakas, D., and He, C. (2010). Golden pigment production and virulence gene expression are affected by metabolisms in Staphylococcus aureus. J. Bacteriol. 192, 3068–3077. doi: 10.1128/jb.00928-09
Łaniewski, P., Mitra, A., Karaca, K., Khan, A., Prasad, R., Curtiss Iii, R., et al. (2014). Evaluation of protective efficacy of live attenuated Salmonella enterica serovar Gallinarum vaccine strains against fowl typhoid in chickens. Clin. Vaccine Immunol. 21, 1267–1276. doi: 10.1128/CVI.00310-14
Mastroeni, P., Chabalgoity, J., Dunstan, S., Maskell, D., and Dougan, G. (2001). Salmonella: immune responses and vaccines. Vet. J. 161, 132–164. doi: 10.1053/tvjl.2000.0502
Matsuda, K., Chaudhari, A. A., Kim, S. W., Lee, K. M., and Lee, J. H. (2010). Physiology, pathogenicity and immunogenicity of lon and/or cpxR deleted mutants of Salmonella Gallinarum as vaccine candidates for fowl typhoid. Vet. Res. 41:59. doi: 10.1051/vetres/2010031
Matsuda, K., Chaudhari, A. A., and Lee, J. H. (2011a). Comparison of the safety and efficacy of a new live Salmonella Gallinarum vaccine candidate, JOL916, with the SG9R vaccine in chickens. Avian Dis. 55, 407–412. doi: 10.1637/9680-020611-Reg.1
Matsuda, K., Chaudhari, A. A., and Lee, J. H. (2011b). Evaluation of safety and protection efficacy on cpxR and lon deleted mutant of Salmonella Gallinarum as a live vaccine candidate for fowl typhoid. Vaccine 29, 668–674. doi: 10.1016/j.vaccine.2010.11.039
Mcfarland, W. C., and Stocker, B. A. (1987). Effect of different purine auxotrophic mutations on mouse-virulence of a vi-positive strain of Salmonella Dublin and of two strains of Salmonella typhimurium. Microb. Pathog. 3, 129–141. doi: 10.1016/0882-4010(87)90071-4
Park, Y.-J., Song, E.-S., Kim, Y.-T., Noh, T.-H., Kang, H.-W., and Lee, B.-M. (2007). Analysis of virulence and growth of a purine auxotrophic mutant of Xanthomonas oryzae pathovar oryzae. FEMS Microbiol. Lett. 276, 55–59. doi: 10.1111/j.1574-6968.2007.00909.x
Penha Filho, R. A. C., de Paiva, J. B., da Silva, M. D., de Almeida, A. M., and Berchieri Junior, A. (2010). Control of Salmonella Enteritidis and Salmonella Gallinarum in birds by using live vaccine candidate containing attenuated Salmonella Gallinarum mutant strain. Vaccine 28, 2853–2859. doi: 10.1016/j.vaccine.2010.01.058
Penha Filho, R. A. C., Diaz, S. J. A., Medina, T. D. S., Chang, Y. F., da Silva, J. S., and Berchieri, A. Jr. (2016). Evaluation of protective immune response against fowl typhoid in chickens vaccinated with the attenuated strain Salmonella Gallinarum ΔcobSΔcbiA. Res. Vet. Sci. 107, 220–227. doi: 10.1016/j.rvsc.2016.06.011
Permin, A., and Bisgaard, M. (2013). “A general review on some important diseases in free-range chickens” in Food & Agriculture Organization, the scope and effect of family poultry research and development. First INFPD/FAO electronic conference on family poultry, Ed. E. F. Gueye (Food and Agriculture Organization of the United Nations (FAO)) 163–167.
Revolledo, L., and Ferreira, A. J. P. (2012). Current perspectives in avian salmonellosis: vaccines and immune mechanisms of protection. J. Appl. Poult. Res. 21, 418–431. doi: 10.3382/japr.2011-00409
Santiviago, C. A., Reynolds, M. M., Porwollik, S., Choi, S.-H., Long, F., Andrews-Polymenis, H. L., et al. (2009). Analysis of pools of targeted Salmonella deletion mutants identifies novel genes affecting fitness during competitive infection in mice. PLoS Pathog. 5:e1000477. doi: 10.1371/journal.ppat.1000477
Serra-Moreno, R., Acosta, S., Hernalsteens, J. P., Jofre, J., and Muniesa, M. (2006). Use of the lambda red recombinase system to produce recombinant prophages carrying antibiotic resistance genes. BMC Mol. Biol. 7, 1–12. doi: 10.1186/1471-2199-7-31
Shah, D. H., Shringi, S., Desai, A. R., Heo, E.-J., Park, J.-H., and Chae, J.-S. (2007). Effect of metC mutation on Salmonella Gallinarum virulence and invasiveness in 1-day-old white Leghorn chickens. Vet. Microbiol. 119, 352–357. doi: 10.1016/j.vetmic.2006.09.002
Smith, H. W. (1956). The use of live vaccines in experimental Salmonella gallinarum infection in chickens with observations on their interference effect. Epidemiol. Infect. 54, 419–432. doi: 10.1017/S0022172400044685
Wigley, P., Hulme, S., Powers, C., Beal, R., Smith, A., and Barrow, P. (2005). Oral infection with the Salmonella enterica serovar Gallinarum 9R attenuated live vaccine as a model to characterise immunity to fowl typhoid in the chicken. BMC Vet. Res. 1, 1–6. doi: 10.1186/1746-6148-1-2
Keywords: Salmonella enterica serovar Gallinarum, purine biosynthesis, purB gene, virulence, safety
Citation: Mukhtar M, Ghafoor A, McClelland M, Akhtar F and Rasheed MA (2024) Construction, molecular characterization, and safety assessment of purB mutant of Salmonella Gallinarum. Front. Microbiol. 15:1467230. doi: 10.3389/fmicb.2024.1467230
Edited by:
Agapi Doulgeraki, Aristotle University of Thessaloniki, GreeceReviewed by:
Young Min Kwon, University of Arkansas, United StatesSagar Mal Goyal, University of Minnesota, United States
Copyright © 2024 Mukhtar, Ghafoor, McClelland, Akhtar and Rasheed. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Aamir Ghafoor, YWFtaXIuZ2hhZm9vckB1dmFzLmVkdS5waw==
 Masham Mukhtar
Masham Mukhtar Aamir Ghafoor
Aamir Ghafoor Michael McClelland
Michael McClelland Fareeha Akhtar
Fareeha Akhtar Muhammad Adil Rasheed
Muhammad Adil Rasheed