- 1School of Life Sciences, Yunnan University, Kunming, China
- 2State Key Laboratory of Microbial Resources, Institute of Microbiology, Chinese Academy of Sciences, Beijing, China
- 3College of Life Sciences, University of Chinese Academy of Sciences, Beijing, China
- 4Kulun Banner Agricultural Technology Extension Center, Tongliao, China
Promoting the availability of silage with a high protein content on farms can lead to profitable and sustainable ruminant production systems. Whole plant soybean (Glycine max L. Merrill, WPS) is a promising high-protein forage material for silage production. In this study, we investigated the fermentation quality, amino acids profile and microbial communities of WPS silage in response to inoculation of lactic acid bacteria (LAB) alone or in combination with non-LAB agents. Before preparing the treatments, the chopped WPS was homogenized thoroughly with 0.3% molasses (0.3 g molasses per 100 g fresh matter). The treatments included CK (sterilized water), LP (Lactiplantibacillus plantarum B90), LPBS (LP combined with Bacillus subtilis C5B1), and LPSC (LP combined with Saccharomyces cerevisiae LO-1), followed by 60 days of fermentation. The inoculants significantly decreased the bacterial diversity and increased the fungal diversity of WPS silage after ensiling. As a result, the contents of lactic acid and acetic acid increased, while the pH value and propionic acid content decreased in the inoculated silages. The amino acids profile was not influenced by inoculants except phenylalanine amino acid, but LP and LPSC silages had substantial greater (p < 0.05) relative feed values of 177.89 and 172.77, respectively, compared with other silages. Taken together, the inoculation of LP alone or in combination with BS was more effective in preserving the nutrients of WPS silage and improve fermentation quality.
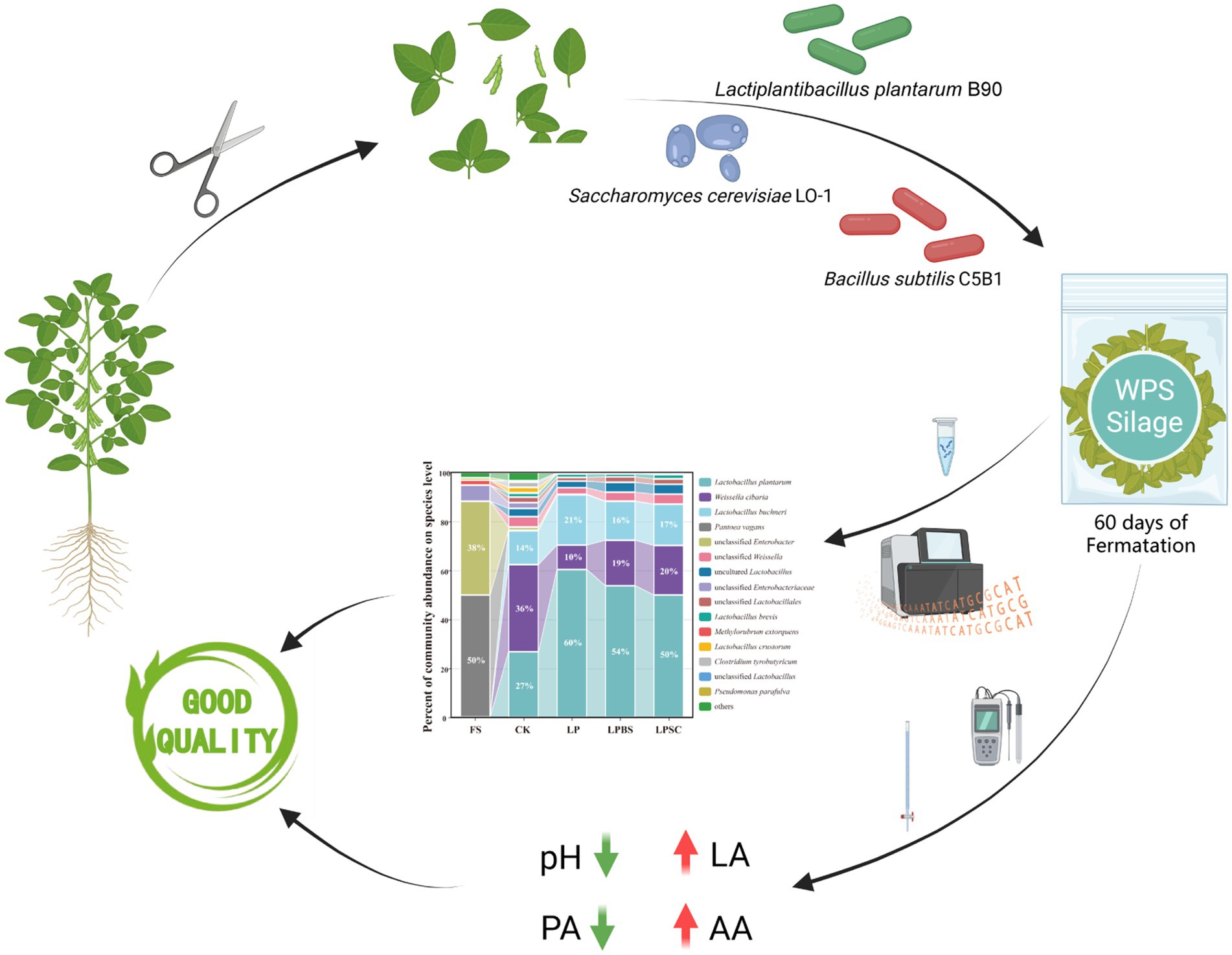
Graphical Abstract. This study investigated the quality of whole plant soybean (WPS) silage and its microbial communities when inoculated with Lactiplantibacillus plantarum B90 (LP) alone or in combination with Bacillus subtilis C5B1 (BS) and Saccharomyces cerevisiae LO-1 (SC) agents. The results demonstrate that the inoculation of LP alone or in combination with BS is more effective in preserving the nutrients of WPS silage. Moreover, it significantly influences the composition of bacterial and fungal communities, resulting in decreased bacterial diversity and increased fungal diversity. These findings offer valuable insights for the production of protein-rich silage in the future.
1 Introduction
The animal husbandry industry in China has emerged significantly in recent years, owing to the country’s swift economic expansion (Wang et al., 2023a). Despite rapid development of animal husbandry, there is an imbalance between feed demand and supply provided by feed industry. The inadequate availability of high-quality green fodder is the primary limiting factor for further development of animal husbandry (Li et al., 2022a). Recently, ensiling has attracted attention as a forage preservation method to help livestock survive in winters and dry seasons (Yang et al., 2022). Ensiling is a method of storing and processing forage through crushing, compacting, and sealing, which facilitates anaerobic fermentation to produce organic acids, extending the forage’s shelf life and ensuring a year-round supply. Under anaerobic conditions, fermentation drive by lactic acid bacteria (LAB) converts water-soluble carbohydrates (WSC) into organic acids, which are used to lower the pH of silage to inhibit microbial activity and nutrient depletion (Liu et al., 2019). The acidification process is beneficial for the preservation of silages for extended periods with a reduced risk of spoilage. However, unfavorable ensiling conditions can promote clostridial activity, leading to excessive production of ammonia nitrogen (NH3-N) and butyric acid (BA), which can result in poor-quality silage.
Promoting the availability of high-protein forages on farms is a profitable and sustainable strategy for ruminant production systems. Legume forages are becoming more popular as a source of protein in dairy farms as an alternative to home-grown protein due to their increased agricultural sustainability benefits such as carbon sequestration and nitrogen fixation, and reduced greenhouse gas emissions by ruminal fermentation and crop N fertilization (Ghizzi et al., 2023). Soybean (Glycine max L. Merrill), one of the most valuable oil seed crops, is a promising source of protein for both human and animal diets worldwide (Ni et al., 2017; Dubey et al., 2019). Whole plant soybean (Glycine max L. Merrill, WPS) is nutrient rich with high protein and fat contents due to the harvesting of stems, leaves, pods with seeds. The use of WPS as silage for animal feed could be advantageous due to its high protein content, which can help decrease dependence on fluctuating protein prices in the market (Wang et al., 2021). Meanwhile, WPS has a high buffering capacity and low WSC content (Ni et al., 2017), which impairs its silage fermentation profile by increasing the BA content and producing an unpleasant smell, creating a bottleneck problem for its preservation (Carpici, 2016). Therefore, it is of great importance to explore more effective strategies for preservation of WPS in order to fully utilize its potential as a feed source for ruminants.
Various biological additives are applied to enhance fermentation quality of silages, including LAB and molasses. The molasses inoculation generally increases the substrate for LAB proliferation, while LAB generally produce the acids, inhibit protein degradation, and reduce nutrients depletion and improve the fermentation quality by reducing the pH of silage (Ni et al., 2017; Ren et al., 2020). A study has established that 3% addition of molasses substantially improved the fermentation quality (pH < 4.5) and taste of alfalfa silage with increased abundance of Lactobacillus genus (Luo et al., 2021). Our previous studies have proved that the application of Lactiplantibacillus plantarum B90 (formerly Lactobacillus plantarum B90) could dominate the silage microbial community, and obviously improve the fermentation quality in many forages, such as soybean silage (Ni et al., 2017), sugarcane top silage (Wang et al., 2020), Sesbania cannabina and sweet sorghum mixed silage (Wang et al., 2022). Moreover, some non-LAB agents are also used as additives to perform specific functions during ensiling (Muck et al., 2018). Bacillus subtilis could potentially improve the nutritional quality of silages by producing cellulase enzyme, which increase the release of plant cell carbohydrates (Bonaldi et al., 2021). Bacillus subtilis can also produce antibiotics, mainly peptides, and antifungal compounds, such as bacillomycin, mycobacillin, and fungistatin (Tabbene et al., 2009), which inhibit the proliferation of undesirable microorganisms, preventing silage from deterioration and mildew (Lara et al., 2016). Saccharomyces cerevisiae, a member of yeast family, can help to modulate the immune system of young animals, improve the rumen fermentation, and enhance the nutrient degradability of roughage in the hindgut (Zhou et al., 2019). Studies have reported that population of Saccharomyces cerevisiae in corn silage survived during ensiling and increased after feed-out without affecting the silage quality and aerobic stability, when Saccharomyces cerevisiae was inoculated at a dose of 103–105 colony forming unit (CFU)/g fresh matter (FM) (Duniere et al., 2015; Xu et al., 2019). Our research group has screened two functional non-LAB strains: Bacillus subtilis C5B1 (BS, producing antimicrobial substance), and Saccharomyces cerevisiae LO-1 (SC, improving growth performance of cattle, unpublished data). However, how these strains interacts with Lactiplantibacillus plantarum B90 (LP) to influence the microbiome structure and fermentation quality of WPS silage is still unknown.
Therefore, current study evaluated the silage quality and microbial communities of WPS silage in response to inoculation of Lactiplantibacillus plantarum B90 alone or in combination with BS or SC. We hypothesized that additives synergistically improved silage quality through modulating microbial communities of WPS silage. The results may provide new insights into the regulation mechanism of novel microbial inoculant in silage fermentation, and theoretical support and guidance for future protein-rich silage production.
2 Materials and methods
2.1 Materials and silage preparation
The WPS (at filling period) was collected from Tongliao City, Inner Mongolia Autonomous Region, China (122°24 E′, 43°65 N′) on September 23, 2020. The WPS was chopped into a particle size of 2 cm using a crop chopper. The chopped WPS was thoroughly homogenized with 0.3% molasses (0.3 g molasses per 100 g fresh matter), then following treatments were applied: CK, sterilized water; LP (Lactiplantibacillus plantarum B90); LPBS, LP combined with BS (Bacillus subtilis C5B1); and LPSC, LP combined with SC (Saccharomyces cerevisiae LO-1). The LP and BS inoculants were applied at theoretical levels of 106 CFU/g fresh weight (FW), while SC inoculant was applied at a theoretical level of 5 × 104 CFU/g FW. An equal amount of sterilized water according to microbial inoculants was prepared for CK group. After spraying of prepared inoculants onto the chopped WPS, about 500 g well-mixed WPS was packed into polyethylene bags and vacuum sealed. The silage bags were kept at temperature around 20–30°C for 60 days (d) to evaluate the fermentation quality, amino acid profile, and microbial communities of WPS silage.
2.2 Analysis of fermentation quality and chemical composition
The silage bags were opened after 60 d of ensiling, and 10 g of silage samples were homogenized with 90 mL of sterilized water for 20 min. The resulting mixture was filtered through a 0.22 μm filter, and the filtrate was used to measure pH, organic acids such as lactic acid (LA), acetic acid (AA), propionic acid (PA), BA, and NH3-N content. The pH was determined using a glass electrode pH meter (pH 213; HANNA; Italy). Organic acid content was analyzed using high-performance liquid chromatography (HPLC) with an ICSep COREGEL-87H column, and a 210 nm UV detector at a temperature of 55°C. The mobile stage was composed of 0.005 M H2SO4 with a flow rate of 0.6 mL/min. NH3-N concentration was analyzed using the ninhydrin colorimetric and phenol-hypochlorite protocols (Broderick and Kang, 1980).
The post-ensiling samples were dried at 65°C for 48 h in a forced-air oven until a constant weight was achieved to determine the dry matter (DM). To analyze the nutritional components, the dried silage samples were ground into a 1.0 mm particle diameter. The WSC content was determined using the anthrone colorimetric method (Owens et al., 1999), while the measurement of crude protein (CP) was performed using the method described by the Association of Official Analytical Chemists (Helrich, 1990). The levels of neutral detergent fiber (NDF) and acid detergent fiber (ADF) were examined according to the method previously described (van Soest et al., 1991).
2.3 Cultivable microbial count
The 20 g of silage samples were blended thoroughly with 180 mL of sterilized saline solution (0.85% NaCl) to homogenize the solution. The homogenized solution was filtered with a single-layer sterilized gauze and then subjected to continuous dilution from 100 to 10−6. The filtrate was subsequently inoculated on MRS and PDA (Land Bridge, Beijing, China) under sterile conditions to determine the population of LAB and yeasts, respectively. The MRS plates were incubated under anaerobic conditions at 37°C for 48 h, while the PDA plates were incubated at 25°C for 4 d to estimate the colony count. The microbial populations were expressed as CFU/g of FM and then transformed logarithmically.
2.4 Relative feed value
The relative feed value (RFV) was estimated by digestible dry matter (DDM) and dry matter intake (DMI) according to following formula:
2.5 Amino acids profile analysis
The amino acids contents of samples were analyzed by following previously reported method (Tiwari and Jha, 2017). Briefly, the HPLC system (UltiMate 3,000; Thermo Scientific, Waltham, MA, USA) coupled with a fluorescence detector with precolumn derivatization was used to separate and quantify amino acids in silage samples using fluoraldehyde as the reagent (Sedgwick et al., 1991). Prior to injection, the samples were hydrolyzed with 6 mol/L HCl for 24 h at 110°C, except for cysteine, methionine, and tryptophan. An internal standard of β-amino-n-butyric acid and ethanol amine mixture was used for analysis. Cysteine content was determined as cysteic acid, methionine content as methionine sulfone after oxidation with performic acid before hydrolysis with 6 mol/L HCl.
2.6 DNA extraction, amplification, and sequencing analysis
The DNA extraction was performed according to previously reported method (Li et al., 2022a). The full-length bacterial 16S rRNA genes were amplified by PCR with primers 817F (5′-TTAGCATGGAATAATRRAATAGGA-3′) and 1196R (5′-TCTGGACCTGGTGAGTTTCC-3′), and the fungal internal transcribed spacer (ITS) was amplified with primers ITS1F (5′-CTTGGTCATTTAGAGGAAGTAA-3′) and ITS2R (5′-GCTGCGTTCTTCATCGATGC-3′) primers. The PCR products were analyzed on a 2% agarose gel by electrophoresis. Qualified PCR products were purified using magnetic beads and quantified by enzyme labeling. The purified samples were then mixed equally based on PCR product concentration and loaded onto a 2% agarose gel for detection using glycogel electrophoresis. Target bands were recovered using a gel recovery kit. To ensure PCR accuracy, each sample was set up in three groups for the reaction. The PCR products were sequenced using the Illumina MiSeq platform (Shanghai Majorbio Biopharm Technology Co. Ltd.) with paired terminal read (2 × 300 bp) and standard protocols. Barcodes and primers were removed to obtain high-quality sequencing. Sequences less than 200 bp with a maxhomop value greater than 10 were filtered using Mothur (v.1.34.4). Chimeras were checked in de novo mode by USEARCH 8.0 (Edgar, 2013) and remaining sequences were used for downstream analysis. The operational taxonomic units (OTUs) at a 97% similarity level were clustered using QIIME (v1.8.0). Microbial species annotation of OTUs was performed using the SILVA 138 database and UNITE 8.0 database. We have updated the names of Lactobacillus plantarum, Lactobacillus buchneri, Lactobacillus brevis to Lactiplantibacillus plantarum, Lentilactobacillus buchneri and Levilactobacillus brevis, respectively, according to the new taxonomic system in the text. The resulting OTUs file was used for calculating rarefaction [R (v.22)] and alpha diversity [Mothur (v1.34.4)]. We also used linear discriminant analysis effect size (LEfSe) to identify significant associations between bacterial and fungi taxa in the treatments. LEfSe was performed using the OmicStudio tools at: https://www.omicstudio.cn/tool/.
2.7 Statistical analysis
The reported results represent the mean of three replicates. Statistical analysis was conducted using GraphPad Prism (version 8.0.0, San Diego, California, USA). One-way analysis of variance was conducted for multiple groups followed by Duncan’s multiple range test. The spearman correlation coefficients were calculated to determine the relationships between the microbiome and silage quality variables. The correlation coefficients were plotted using the “pheatmap” libraries in R. Statistical significance was declared at a threshold of p < 0.05.
3 Results
3.1 Fermentation profile of WPS silage
The fermentation profile of WPS silage is presented in Figure 1. The inoculated silages had substantial lower pH values compared with CK silage (p < 0.001); specifically, LP silage exhibited lower pH value of 4.83 compared with CK silage (5.60), but it was not substantially different than other inoculated silages (Figure 1A). The inoculated silages had substantial greater LA and AA contents compared with CK silage (p < 0.001), but neither LA content nor AA content was substantially different among inoculated groups and were ranged from 76.05–78.94 g/kg DM and 38.66–44.59 g/kg DM, respectively (Figures 1B,C). The PA content was only found in CK silage with value of 22.78 g/kg DM, while it was not found in inoculated silages (Figure 1D). The LA/AA ratios were significantly higher in inoculated silages compared with CK silage (p < 0.001), but there was no substantial difference among inoculated silages (Figure 1E). However, NH3-N content was not significantly different among silages, but LP silage had numerically lower NH3-N content of 0.09 %DM compared with other groups (Figure 1F). Taken together, results presented here highlighted that LP inoculation alone or in combination with BS or SC has similar fermentation quality of WPS silage.
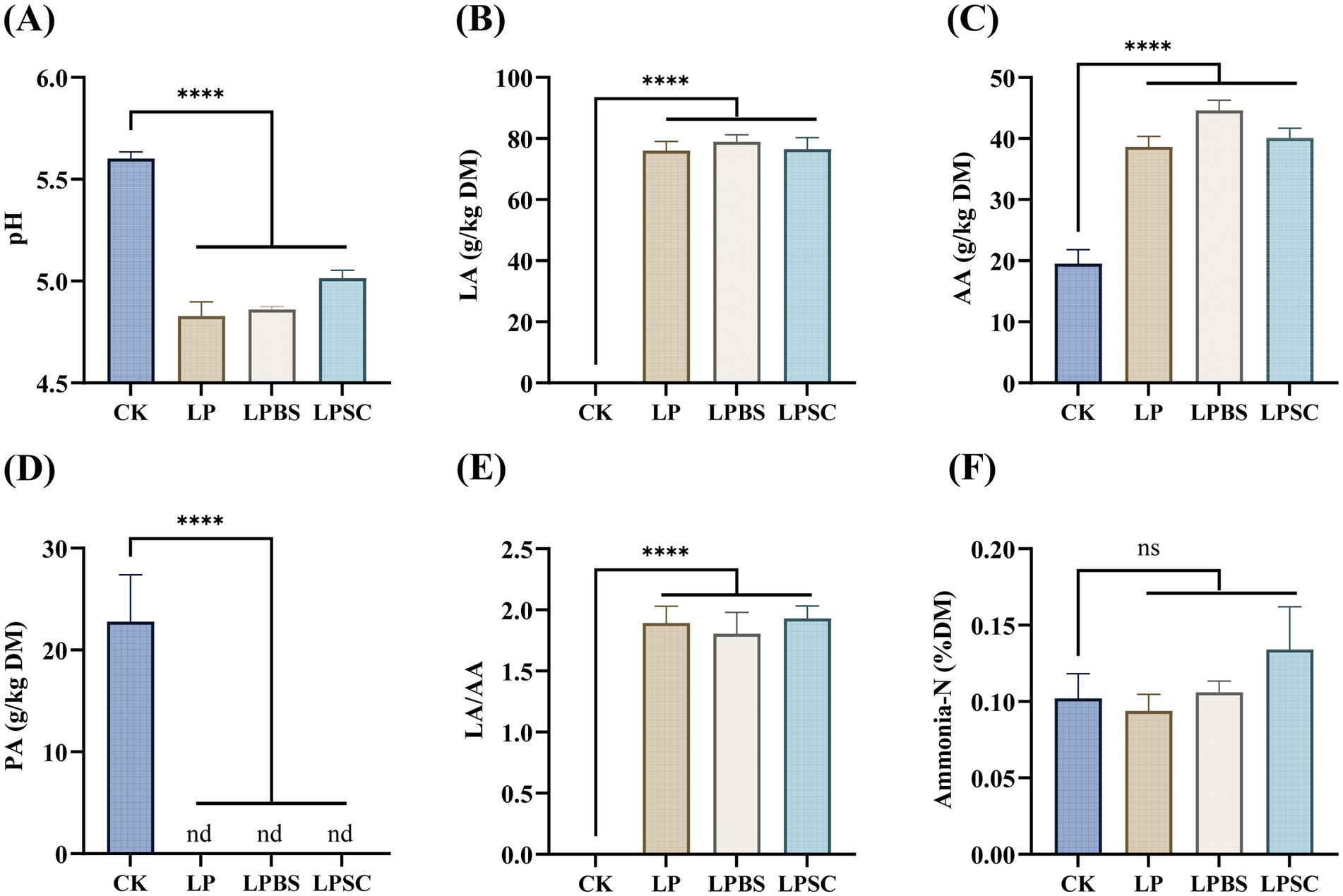
Figure 1. Fermentation characteristics of whole plant soybean silages after 60 days of ensiling. (A) For pH; (B) for LA; (C) for AA; (D) for PA; (E) for LA/AA; and (F) for ammonia-N. CK, sterilized water; LP, Lactiplantibacillus plantarum B90; LPBS, LP combined with Bacillus subtilis C5B1; LPSC, LP combined with Saccharomyces cerevisiae LO-1; ns, non-significance; nd, no detection; ****p < 0.001.
3.2 Chemical composition and cultivable microbial counts of WPS silage
The chemical composition and cultivable microbial counts of WPS silage are depicted in Table 1. The inoculated silages had significant greater DM contents and lower DM losses compared with CK silage (p < 0.001), but neither DM content nor DM loss was substantially different among inoculated groups and were ranged from 29.60 to 29.93% and 1.80 to 2.44%, respectively. The contents of CP were not influenced by inoculants and were between 22.82 and 23.94 %DM, the contents of WSC decreased with the addition of inoculant and ranged from 0.52 to 0.60 %DM (p < 0.05), respectively. The NDF and ADF contents of the LP and LPSC silages were significantly lower by 4.07 and 3.35%, and 4.35 and 3.58%, respectively, compared to the CK silage (p < 0.001). The LP and LPSC silages had substantial higher RFV of 177.89 and 172.77, respectively, compared with other silages. Taken together, results suggested that all inoculants were effective in conserving the nutrients of WPS silage, but inoculation of LP alone or in combination with SC may perform better to degrade the digestible fiber. The inoculated silages had significant higher LAB and yeast counts than CK silage (p < 0.05), but neither LAB count nor yeast count was substantially different among inoculated silages and were ranged from 9.02 to 9.15 and 8.76 to 8.83 log10 CFU/g FM, respectively.
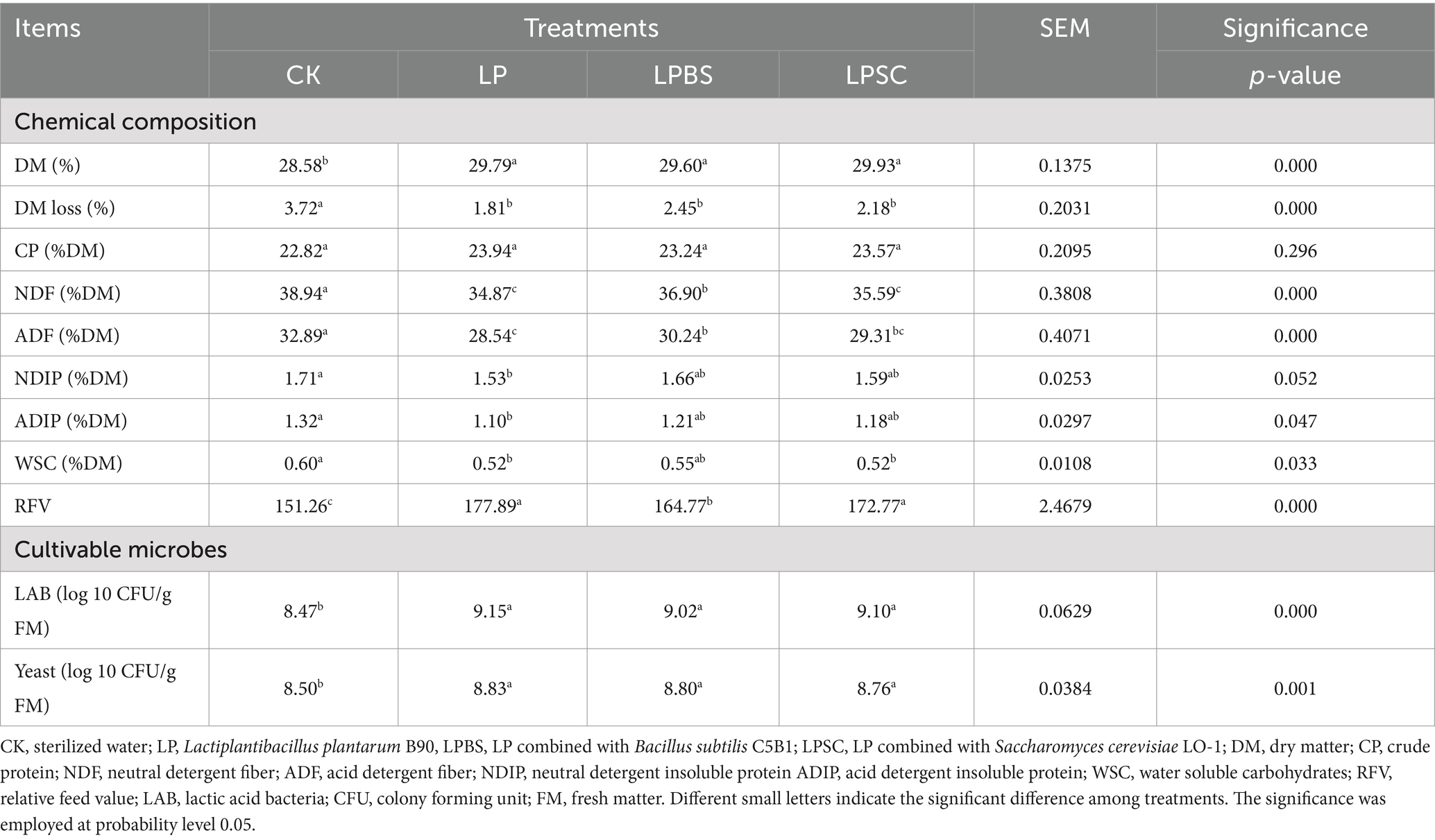
Table 1. Chemical characteristics and cultivable microbial count of whole plant soybean silages after 60 days of ensiling.
3.3 Amino acids profile of WPS silage
The amino acids composition of WPS silage is presented in Table 2. There were no substantial differences for amino acids contents except phenylalanine among all silages, and were ranged from 0.12 to 2.02 %DM. However, the content of phenylalanine amino acid was substantial higher in LPBS silage with quantity of 0.94 %DM compared with other silages (p < 0.01). Taken together, the inoculants did not influence the amino acids composition except phenylalanine of WPS silage.
3.4 Bacterial community of fresh and fermented WPS
The alpha diversity analysis revealed a decrease of the bacterial biodiversity in WPS silage when it was inoculated with LP alone or with in combination with BS or LC compared with CK silage (Supplementary Figures S1A,B). More specifically, the greater decrease in bacterial biodiversity was seen in LP silage compared with other inoculated silages. The principal coordinate’s analysis (PCoA) based on OTU level was applied to identify the factors that influence the variations in microbiome of WPS silage (beta diversity). The results showed a substantial bacterial species succession dynamic when inoculants were applied, while they were indistinguishable among silage inoculated with LP alone or with in combination with BS or LC (Supplementary Figure S1C).
The relative abundance of bacterial communities in WPS before and after ensiling are shown in Figures 2A,B. At genus level (Figure 2A), epiphytic microflora of fresh WPS was mainly comprised of unwanted bacteria for ensiling such as Pantoea (50%) and Enterobacter (38%). After 60 d of fermentation, Lactobacillus and Weissella were dominant genera in CK silage, accounting relative abundances of 47 and 43%, respectively. The inoculants substantially increased the relative abundance of Lactobacillus and decreased the relative abundance of Weissella in WPS silage. The LP silage had the greater relative abundance of Lactobacillus (83%) and lower relative abundance of Weissella (15%) compared with other inoculated silages. The relative abundances of Lactobacillus and Weissella in LPBS and LPSC silages were 71 and 26%, and 68 and 28%, respectively. At specie level (Figure 2B), the Pantoea vagans (50%) and unclassified Enterobacter (38%) were dominant species in fresh WPS. After 60 d of fermentation, the Weissella cibaria, Lactiplantibacillus plantarum and Lentilactobacillus buchneri were dominant species in CK silage comprising the relative abundances of 36, 27, and 14%, respectively. The inoculants substantially increased the relative abundance of Lactiplantibacillus plantarum and decreased the relative abundance of Weissella cibaria in WPS silage (Supplementary Figure S3). The relative abundances of Lactiplantibacillus plantarum and Weissella cibaria in LP, LPBS and LPSC silages were 60, 54, and 50%, and 10, 19, and 20%, respectively (Figure 2B). Taken together, inoculants significantly influenced the bacterial community of WPS silage, particularly increased the relative abundance of beneficial Lactiplantibacillus plantarum in WPS silage.
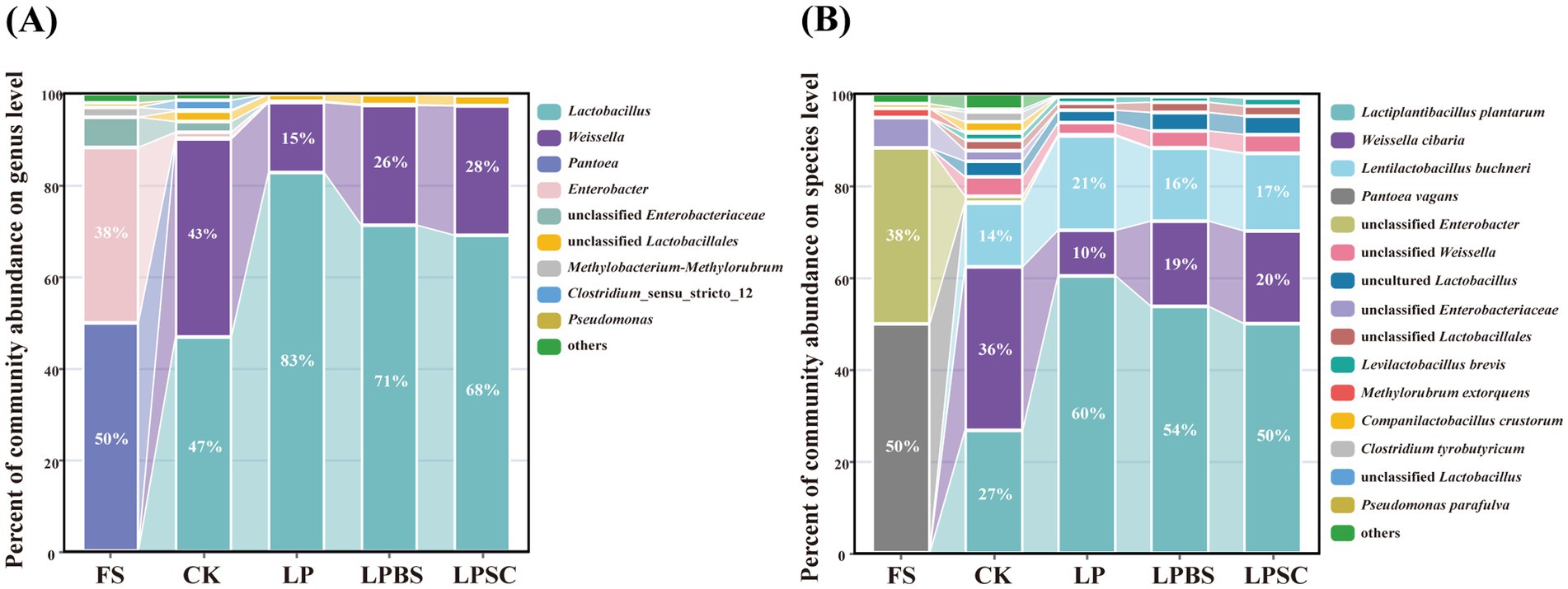
Figure 2. Microbial community of whole plant soybean silages at genus (A) and species (B) levels after 60 days of ensiling. CK, sterilized water; LP, Lactiplantibacillus plantarum B90; LPBS, LP combined with Bacillus subtilis C5B1; LPSC, LP combined with Saccharomyces cerevisiae LO-1.
The LEfSe analysis revealed that the LP, LPBS, and LPSC groups had higher Lactiplantibacillus plantarum enrichment, while the CK group had higher enrichment of Leuconostocaceae, Weissella and Clostridiaceae suggesting that this different microbial composition contributed to the difference in fermentation quality (Supplementary Figures S5A–C). The LP group had higher Lactobacillus enrichment compared to the LPBS and LPSC groups, while the LPBS and LPSC groups had higher Weissella enrichment, probably because the activity of the added BS and SC in the early fermentation phase which affected the microbial composition of the silage, as well as the growth of the LP (Supplementary Figures S5D,E). The LPBS and LPSC groups analyses showed that there were also differences in microbial species between them, with LPBS having a higher enrichment of Enterobacterales, Gammaproteobacteria and LPSC having a higher enrichment of Sphingomonadales (Supplementary Figure S5F).
3.5 Fungal community in fresh and fermented WPS
The alpha diversity analysis indicated an increase in fungal biodiversity of WPS silage when it was inoculated with LP alone or in combination with BS or LC compared with CK silage (Supplementary Figures S2A,B). The PCoA based on OTU level was applied to identify the factors that that influence the variations in fungal community of WPS silage (beta diversity). The results suggested a substantial fungal species succession change when inoculants were applied, while they were indistinguishable among inoculated silages (Supplementary Figure S2C).
The relative abundance of fungal communities in WPS before and after ensiling are shown in Figures 3A,B. At genus level (Figure 3A), epiphytic fungal microflora of fresh WPS was mainly comprised of Cladosporium, Derxomyces, and Bipolaris, accounting relative abundances of 68, 5, and 5%, respectively. After 60 d of fermentation, Pichia, Kazachstania-Candida_clade, and unclassified Agaricomycetes were most prevalent genera in CK silage comprising the relative abundances of 49, 30, and 8%, respectively. The inoculants substantially influenced the diversity of fungal microflora in WPS silage after 60 d of fermentation. The Cladosporium, norank Agaricales, and unclassified Agaricomycetes were dominant genera in LP and LPBS silages accounting the relative abundances of 27 and 21%, 29 and 26%, and 13 and 24%, respectively. In LPSC silage, the main fungal genera were comprised of Kazachstania-Candida_clade, norank Agaricales, Cladosporium and unclassified Agaricomycetes accounting the relative abundances of 37, 19, 16, and 10%, respectively. At specie level (Figure 3B), epiphytic fungal microflora of fresh WPS was mainly comprised of Cladosporium herbarum (68%). After 60 d of fermentation, Pichia kudriavzevii (49%), unclassified Kazachstania-Candida_clade (30%), and unclassified Agaricomycetes (8%) were most abundant fungal species in CK silage. The inoculants substantially influenced the fungal species and decreased the abundance of Pichia kudriavzevii (Supplementary Figure S4); and Cladosporium herbarum, unclassified Agaricales, and unclassified Agaricomycetes were most dominant species in LP and LPBS silages comprising the relative abundances of 27 and 21%, 29 and 26%, and 13 and 24%, respectively (Figure 3B). However, unclassified Kazachstania-Candida_clade (37%), unclassified Agaricales (19%), Cladosporium herbarum (16%), and unclassified Agaricomycetes (10%) were most prevalent fungal species in LPSC silage. Taken together, inoculants substantially increased the fungal diversity and substantially decreased the relative abundance of Pichia kudriavzevii in WPS silage.
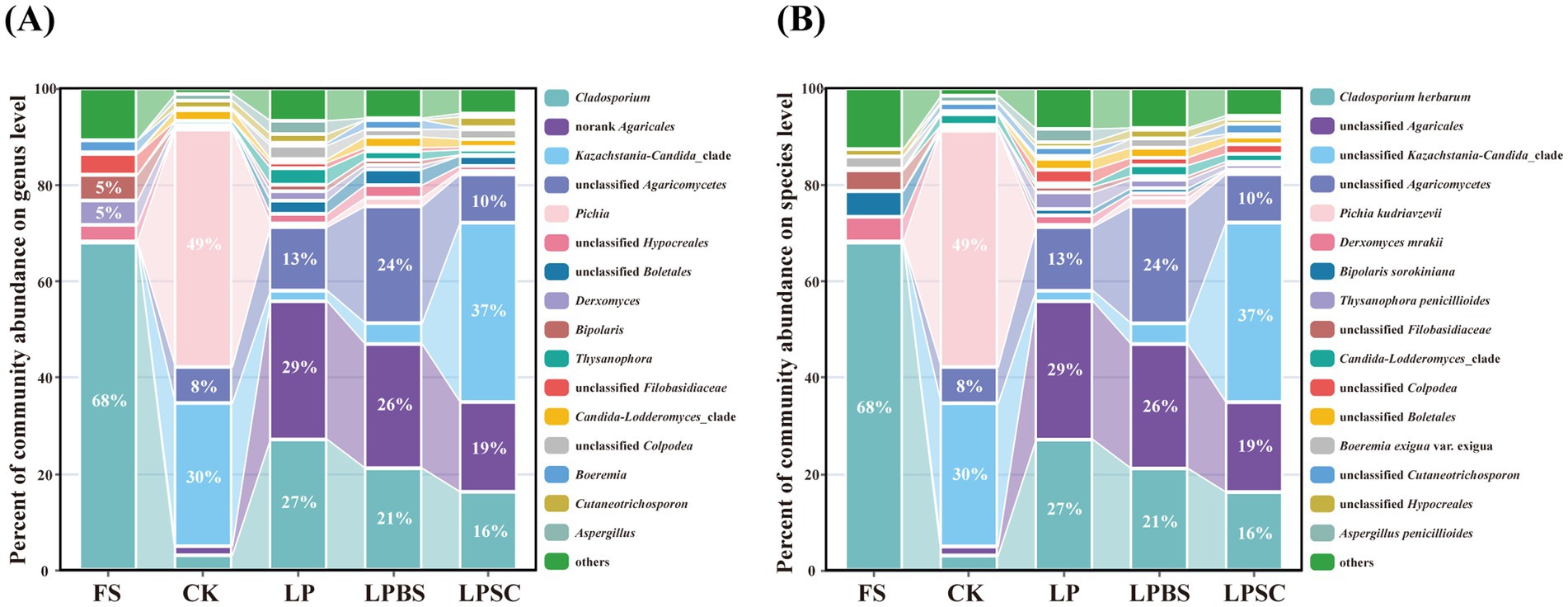
Figure 3. Fungal community of whole plant soybean silages at genus (A) and species (B) levels after 60 days of ensiling. CK, sterilized water; LP, Lactiplantibacillus plantarum B90; LPBS, LP combined with Bacillus subtilis C5B1; LPSC, LP combined with Saccharomyces cerevisiae LO-1.
The LEfSe analysis revealed that both the LP and LPBS groups had higher enrichment of Basidiomycota and Agaricomycetes compared to the CK group. In contrast, the LPSC group exhibited only an increased enrichment of Agaricomycetes. Compared to other groups, the CK group showed higher levels of Saccharomycetes and Pichia kudriavzevii (Supplementary Figures S6A–C), indicating a distinct difference in fungal composition that may influence the aerobic stability of WPS silage. The LPBS group demonstrated a greater enrichment of Agaricomycetes, while the LPSC group exhibited higher levels of Dothideomycetes and Trichomeriaceae enrichment compared to LP group, suggesting that the addition of SC significantly altered the fungal community composition in WPS silage (Supplementary Figures S6D,E).
3.6 Correlations between microbiome and fermentation products of WPS silage
To further characterize the effects of bacterial and fungal species on fermentation quality, the correlation analysis between bacterial and fungal genera and fermentation quality of silages was investigated (Figure 4). The results indicated that Lactobacillus was positively correlated with RFV, LA and LA/AA and negatively correlated with ADF and NDF, whereas Weissella, Enterobacter, Clostridium_sensu_stricto_12, unclassified Enterobacteriaceae and Lactococcus were positively correlated with ADF and NDF and negatively correlated with LA (Figure 4A). The Cladosporium and norank Agaricales were negatively correlated with NDF, ADF, pH and PA, and positively correlated with DM, RFV, LA, AA and LA/AA, whereas Kazachstania-Candida_clade was positively correlated with pH (Figure 4B).
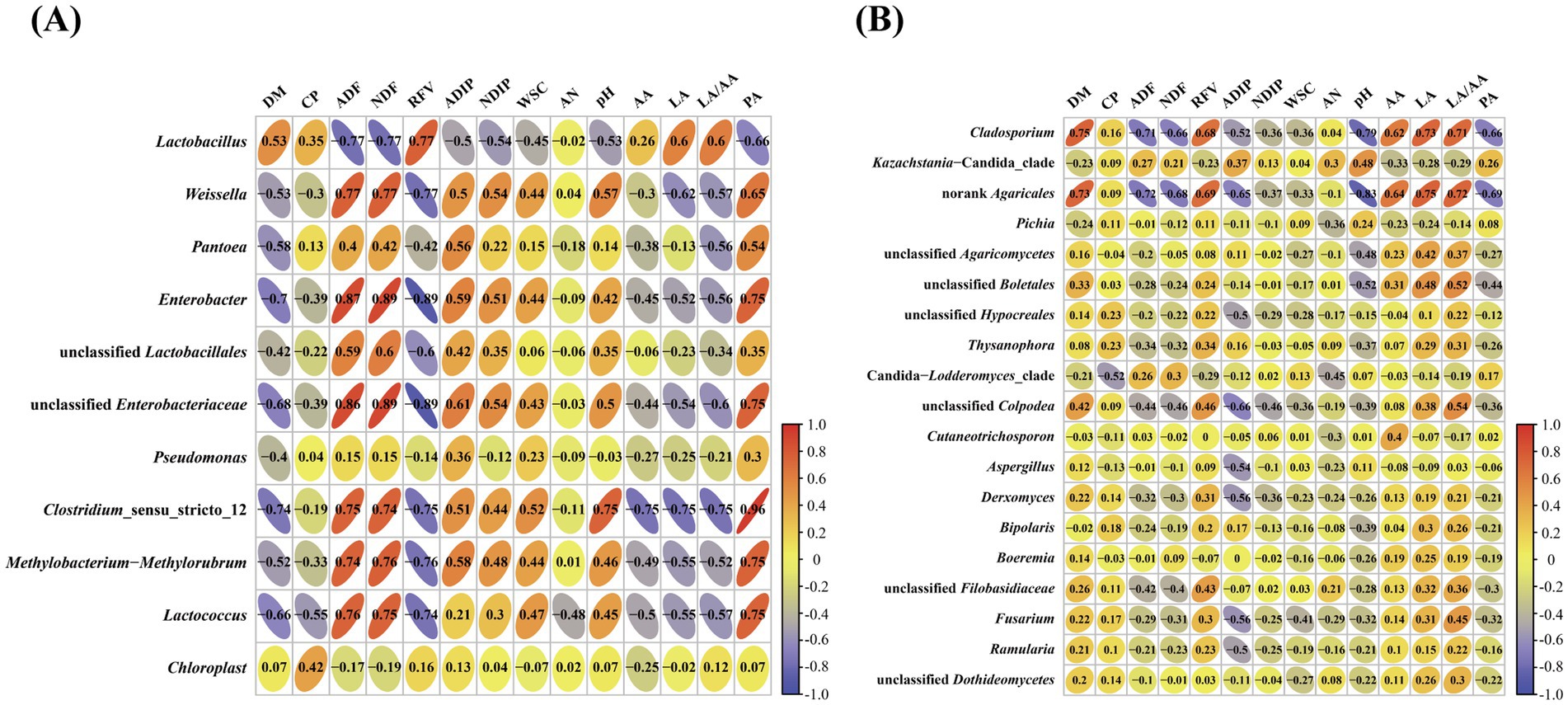
Figure 4. Associations between fermentation products and bacterial (A) and fungal (B) communities. DM, dry matter; CP, crude protein; NDF, neutral detergent fiber; ADF, acid detergent fiber; WSC, water soluble carbohydrates; RFV, relative feed value; AN, ammonia-N; LA, lactic acid; AA, acetic acid; PA, propionic acid.
4 Discussion
4.1 LAB and functional strains enhance WPS silage fermentation and nutrient preservation
Soybean is widely planted worldwide as for both human and ruminant’s feed. However, its low WSC content and high buffer capacity made it difficult to be ensiled. Inoculating silage with LAB and molasses was found to ensure good fermentation quality because LAB can efficiently transform WSC into LA and reduce silage pH to inhibit the growth of undesirable microorganisms at the initial stage of ensiling (Li et al., 2022b). Moreover, some non-LAB inoculants may have the ability to secrete cellulose-related enzymes that help in hydrolyzing the fiber structure of forages and release more fermentable sugar during ensiling (Bai et al., 2020), whereas some species have potential to improve the rumen fermentation and can be applied as a direct-fed microbial strain at ensiling (Zhu et al., 2017). Therefore, in this study, we explored the addition of lactic acid-producing bacteria (LP) with or without non-LAB agents (namely BS and SC) and molasses as additives before ensiling. The goal was to enhance the fermentation of WPS silage and tackle the challenges related to the challenging ensiling of WPS.
The silages treated with LP, LPBS, and LPSC had higher LA and AA contents compared with CK silage, whereas no significance difference was found among inoculated silages. Previous studies have shown that LAB strains can decline pH value by accelerating WSC transformation into lactic and acetic acids (Ding et al., 2019; Zhang et al., 2021). Silages treated with inoculants were found to have the lowest pH values compared to CK silage, whereas no substantial differences in pH values were found between LP, LPBS, and LPSC silages after 60 d of ensiling. The most reasonable account was that Lactiplantibacillus plantarum dominated the silage bacterial community during the fermentation (Keshri et al., 2018). In addition, a study has shown that combined inoculation of LP and BS raised the LA content and decreased the pH of corn silage (Lara et al., 2018), whereas some studies have reported that SC inoculation at a dose of 103–105 CFU/g of fresh forage did not alter the nutritional quality, fermentation traits, and Lactobacillus populations of corn silage and survived during ensiling (Duniere et al., 2015; Xu et al., 2019). Therefore, it can be speculated that the antimicrobial peptide producing ability of BS that could accelerate the growth of the homofermentative LAB and accurate dose of SC inoculation with LP did not restrict each other’s effect but benefited fermentation by increasing LA and AA concentrations compared to those in the CK silage. Generally, PA is produced from secondary fermentation of Clostridia by consuming LA. The PA was not found in inoculated silages compared to CK silage, indicating that application of microbial inoculants inhibited secondary fermentation thereby preserving more nutrients excellently (Li et al., 2020).
The lower DM losses in inoculated silages compared with CK silage further proved that additives are favorable to WPS ensilage in the present study. After 60 d of fermentation, inoculated silages LP, LPBS, and LPSC had remarkably higher DM concentrations compared with CK silage, indicating that LAB inhibited the spoilage microbes and their fermentation activities by reducing pH of silage which agrees with previous study (Zhang et al., 2023a). The consumption rate of WSC is associated with the extent of fermentation (Long et al., 2022). However, the WSC content was not affected by inoculants and there was no substantial difference among inoculated silages and CK silage. This could be attributed to the 0.3% molasses addition in this study which provided sufficient fermentable substrates to LAB to carry on the fermentation. Proteolysis mainly resulted from plant proteases, but it could be inhibited when the pH has declined (Tahir et al., 2023). The addition of inoculants in the silages did not result in a reduction of NH3-N content after 60 d of ensiling, which can be further confirmed by the CP content in all silages. This result might be attributed to the presence of aerobic bacteria and clostridia in all silages (Kung and Shaver, 2001). The substantial degradation of NDF and ADF contents was found in LP and LPSC silages compared with CK silage which could be the co-action of various enzymes produced by the LAB to hydrolyze more digestible cell wall fractions. Numerous studies have found that inoculation with LAB promoted the degradation of lignocellulose (Li et al., 2022a; Tahir et al., 2023). In this study, the degradation of NDF and ADF contents in LPBS silage was lower than other inoculated silages suggesting that combined inoculation of LP and BS restricted each other effect on cell wall degradation. Moreover, the inoculants did not influence the amino acids profile of WPS silage suggesting that inoculants were not able to efficiently break down proteins into amino acids, leading to minimal impact on the amino acid composition.
4.2 Inoculants influence bacterial community and enhance WPS silage quality
The fermentation quality of silages with or without additives depends on the bacterial composition and changes during the ensiling time (Xu et al., 2021). Bacterial alpha diversity of WPS decreased after fermentation, and silage with inoculants had lower bacterial alpha diversity after fermentation, which could be due to the dramatic decrease in pH value. The acidic anaerobic environment after ensiling led to the modification of the bacterial community where most of the epiphytic bacteria disappeared due to their inability to adapt to low pH (Zhang et al., 2023b). The presence of Pantoea in silage is generally considered undesirable because it competes with LAB for sugars (Jiang et al., 2020), while Enterobacter has the potential to ferment LA into AA and other products, leading to significant nutrition loss (Ni et al., 2017). In the present study, the inoculants substantially decreased the relative abundance of Enterobacter, Pantoea and other epiphytic bacteria, and increased the relative abundance of Lactobacillus after ensiling. This could be attributed to the adaptability as well as the rapid growth and multiplication of Lactobacillus species which produces higher LA that swiftly declined the pH to inhibit the growth of spoilage microorganisms during the fermentation (Yang et al., 2019; Bai et al., 2020). The Lactobacillus genus is known to regulate the fermentation process in anaerobic conditions and could grow vigorously during fermentation owing to its high acid resistance (Muck et al., 2018). In the present study, the relative abundance of Lactobacillus was greater, while the relative abundance of Weissella was lower in LP silage; however, the relative abundance of Lactobacillus decreased and the relative abundance of Weissella increased when WPS was inoculated with LP in combination with BS or SC in agreement with the results of pH and LA. This highlights that combined inoculation of BS or SC with LP slightly weakened the growth of Lactobacillus, which might be related to their different metabolic functions leading to competition among each other. The Weissella is typically considered an early colonizer, but is eventually surpassed by acid-resistant Lactobacillus due to the pH decline during fermentation (Graf et al., 2016), which explains its higher relative abundances in silages with greater pH value in the present study. Moreover, the Weissella cibaria was enriched bacterial specie in the CK silage; and most possible explanation lies in the high pH value after 60 d of silage fermentation. As expected, inoculants increased the relative abundance of Lactiplantibacillus plantarum specie in the treated silages compared with the control silage. This result suggested that inoculation of LP alone or in combination with BS or SC could accelerate the growth of Lactobacillus species by providing conducive environment for their proliferation resulting in better silage quality.
4.3 Inoculants altered the fungal community which may potentially enhance the aerobic stability of WPS silage
The fungal community in silage is complex, comprising various species, including yeasts, molds and filamentous fungi. In the present study, fungal alpha diversity of WPS increased after ensiling, and silage with inoculants had higher fungal alpha diversity compared with control silage after fermentation. Likewise, a study has found that the fungal richness and diversity in barley silage inoculated with LAB was higher than that of untreated silage during ensiling (Liu et al., 2019). The Cladosporium was the main fungal genus in the fresh WPS, but its growth was substantially inhibited after ensiling, suggesting the need for oxygen by latter. Cladosporium produces mycotoxins during their growth by infections of forages, posing a significant risk to livestock health during ensiling and grazing (Huang et al., 2022). In the present study, the fungal community was mainly represented by Pichia, Kazachstania-Candida_clade, unclassified Agaricomycetes, Cladosporium, and norank Agaricales genera after ensiling, supported by previous studies (Xu et al., 2019; Wang et al., 2020). The Pichia and Kazachstania-Candida_clade were most prevalent fungal genera in CK silage and generally these considered as spoilage microbes during aerobic exposure of silage (Zhang et al., 2019). The presence of these spoilage microbes in CK silage might be related to its greater pH value which was conducive for the growth of these fungi (Wang et al., 2022). Moreover, the addition of inoculants substantially changed the fugal community and Cladosporium, unclassified Agaricomycetes, and norank Agaricales became the dominant genera in inoculated silages, suggesting that these fungi could proliferate in greater acidic environment. Meanwhile, it was quite fascinating to found the substantial higher relative abundance of Kazachstania-Candida_clade genus in LPSC silage, which might be related to its lower acidic environment compared to other treated silages and inoculation of SC which provided stable environment for its growth. This result suggests that addition of LP with SC may not reduce aerobic deterioration caused by the post-spoilage fungal growth such as Kazachstania-Candida_clade and Cladosporium (Wang et al., 2020).
The main fungal specie observed in fresh WPS was Cladosporium herbarum. The Cladosporium herbarum could grow at low temperatures, and usually associated with foods spoilage and discoloration (Bullerman, 2003). The Pichia kudriavzevii and unclassified Kazachstania-Candida_clade were dominant fungal species in CK silage which might be related to its higher pH value; and these species have been involved in silage corruption during aerobic exposure (Santos, 2011; Carvalho et al., 2017). Meanwhile, the addition of LP alone or in combination with BS substantially inhibited the growth of Pichia kudriavzevii and unclassified Kazachstania-Candida_clade, and increased the relative abundances of unclassified Agaricales, Cladosporium herbarum, and unclassified Agaricomycetes fungal species after ensiling which could be attributed to lower pH values of LP and LPBS silages. This result suggests that addition of LP alone or in combination with BS could improve the aerobic stability of WPS silage by inhibiting the growth of spoilage fungal species. However, unclassified Kazachstania-Candida_clade and unclassified Agaricales were the most abundant fungal species in LPSC silage that could be directly related to the addition of SC which provided conducive environment for their growth. Moreover, the population of SC did not survive after ensiling of WPS when LPSC was inoculated and the reason for this is unknown. This phenomenon may be related to the aerobic conditions required for SC survival. At the beginning of fermentation, the residual oxygen in the silage likely provided favorable conditions for SC, allowing it to survive temporarily. During this early stage, SC may have contributed to a rise in the silage temperature, influencing the microbial composition. As the residual oxygen was consumed, LAB and other fast-growing anaerobic microbes began to dominate the fermentation. Due to the lack of a suitable growth environment, SC was eventually decomposed. However, this hypothesis requires further validation through subsequent experiments. This result highlights that addition of LP with SC may not improve the aerobic stability of WPS after exposure to air likewise to previous study (Xu et al., 2019), as unclassified Kazachstania-Candida_clade was abundant. However, it has been reported that the relative quantitative analyzes cannot reflect the true absolute abundance of microorganism groups in a sample. For instance, the increase in the relative abundance of a certain group of microorganisms may not reflect the increase of its absolute abundance. Instead, it may be linked to the decrease in the absolute abundance of other microorganisms (Yang et al., 2021).
4.4 Interaction of multiple microorganisms and metabolites influences fermentation quality of WPS silage
Silage fermentation is a very complex biological process involving a variety of microorganisms and biochemical reactions, and produces many different metabolites during ensiling (Wang et al., 2023b; Xin et al., 2023). Understanding the relationship between metabolites with fermentation bacteria and fungi can provide better insight into the fermentation mechanism of silage. Lactobacillus is the core bacteria to produce LA during ensiling process and determine the silage quality. Like previous studies (Guan et al., 2018; Xia et al., 2022; Tahir et al., 2023), the current study also reported that concentration of LA is positively correlated with Lactobacillus. The unclassified Enterobacteriaceae and Lactococcus had positive associations with ADF, NDF and PA, while negative correlations with DM, RFV, and LA, highlighting that these bacteria are involved in silage corruption. The Cladosporium and norank Agaricales were positively correlated with LA and negatively correlated with pH, highlighting that these species are beneficial to preserve the silage nutrients. However, a study has reported that Cladosporium can produce mycotoxins posing a significant health to livestock (Huang et al., 2022), therefore, it is important to manage and minimize the presence of Cladosporium in silage through proper harvesting, storage, and management practices. Kazachstania might have a strong tolerance to LA and be a yeast species that is crucially involved in initiating the aerobic deterioration of silage with a relatively low pH and AA content which is in line with the result of present study as a positive association between Kazachstania-Candida_clade and pH was observed.
5 Conclusion
The addition of LP alone or in combination with BS or SC substantially improved the WPS silage quality by increasing the LA and AA contents and decreasing the pH value and the PA content. The inoculants had no impact on the amino acid profile of WPS silage. However, it is also substantially influenced the bacterial and fungal communities, resulting in reduced bacterial diversity and increased fungal diversity. The Lactobacillus was the most prevalent bacterial genus in the inoculated silages, while Cladosporium, norank Agaricales, and unclassified Agaricomycetes were dominant fungal genera after 60 d of ensiling. Moreover, the bacterial and fungal communities were substantially correlated with fermentation characteristics. Taken together, the inoculation of LP alone or in combination with BS was more effective in preserving the nutrients of WPS silage which may provide theoretical support and guidance for future protein rich silage production.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found at: https://www.ncbi.nlm.nih.gov/, PRJNA1068694.
Author contributions
SJ: Writing – review & editing, Data curation, Formal analysis, Visualization, Writing – original draft. MT: Data curation, Formal analysis, Visualization, Writing – original draft, Writing – review & editing. FH: Data curation, Writing – review & editing, Methodology. TW: Writing – review & editing. HL: Writing – review & editing, Conceptualization, Formal analysis. WS: Conceptualization, Formal analysis, Writing – review & editing. YL: Conceptualization, Formal analysis, Writing – review & editing. WL: Writing – review & editing, Resources. JZ: Writing – review & editing, Funding acquisition, Project administration.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This research was funded by the Strategic Priority Research Program of the Chinese Academy of Sciences (XDA26040201), and Science and Technology Assistance Project of the Chinese Academy of Sciences (KFJ-FP-202209).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2024.1458287/full#supplementary-material
References
Bai, J., Xu, D., Xie, D., Wang, M., Li, Z., and Guo, X. (2020). Effects of antibacterial peptide-producing Bacillus subtilis and Lactobacillus buchneri on fermentation, aerobic stability, and microbial community of alfalfa silage. Bioresour. Technol. 315:123881. doi: 10.1016/j.biortech.2020.123881
Bonaldi, D., Carvalho, B., Ávila, C., and Silva, C. (2021). Effects of Bacillus subtilis and its metabolites on corn silage quality. Lett. Appl. Microbiol. 73, 46–53. doi: 10.1111/lam.13474
Broderick, G., and Kang, J. (1980). Automated simultaneous determination of ammonia and total amino acids in ruminal fluid and in vitro media. J. Dairy Sci. 63, 64–75. doi: 10.3168/jds.s0022-0302(80)82888-8
Bullerman, L. (2003). Spoilage: Fungi in food–An overview in encyclopedia of food sciences and nutrition. New York, NY: Academic Press, 5511–5522.
Carpici, E. B. (2016). Nutritive values of soybean silages ensiled with maize at different rates. Legume Res. Int. J. 39, 810–813. doi: 10.18805/lr.v0iOF.3772
Carvalho, B., Ávila, C., Pereira, M., and Schwan, R. (2017). Methylotrophic yeast, lactic acid bacteria and glycerine as additives for sugarcane silage. Grass Forage Sci. 72, 355–368. doi: 10.1111/gfs.12248
Ding, Z., Xu, D., Bai, J., Li, F., Adesogan, A., Zhang, P., et al. (2019). Characterization and identification of ferulic acid esterase-producing Lactobacillus species isolated from Elymus nutans silage and their application in ensiled alfalfa. J. Appl. Microbiol. 127, 985–995. doi: 10.1111/jam.14374
Dubey, A., Kumar, A., Abd_Allah, E. F., Hashem, A., and Khan, M. L. (2019). Growing more with less: breeding and developing drought resilient soybean to improve food security. Ecol. Indic. 105, 425–437. doi: 10.1016/j.ecolind.2018.03.003
Duniere, L., Jin, L., Smiley, B., Qi, M., Rutherford, W., Wang, Y., et al. (2015). Impact of adding Saccharomyces strains on fermentation, aerobic stability, nutritive value, and select lactobacilli populations in corn silage. J. Anim. Sci. 93, 2322–2335. doi: 10.2527/jas.2014-8287
Edgar, R. C. (2013). UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 10, 996–998. doi: 10.1038/nmeth.2604
Ghizzi, L. G., del Valle, T. A., Zilio, E. M. C., Dias, M. S. S., Nunes, A. T., Gheller, L. S., et al. (2023). Effects of homofermentative lactic acid bacteria and powdered molasses on fermentative losses, chemical composition and aerobic stability in whole-plant soybean silage. N. Z. J. Agric. Res. 66, 651–664. doi: 10.1080/00288233.2022.2108851
Graf, K., Ulrich, A., Idler, C., and Klocke, M. (2016). Bacterial community dynamics during ensiling of perennial ryegrass at two compaction levels monitored by terminal restriction fragment length polymorphism. J. Appl. Microbiol. 120, 1479–1491. doi: 10.1111/jam.13114
Guan, H., Yan, Y., Li, X., Li, X., Shuai, Y., Feng, G., et al. (2018). Microbial communities and natural fermentation of corn silages prepared with farm bunker-silo in Southwest China. Bioresour. Technol. 265, 282–290. doi: 10.1016/j.biortech.2018.06.018
Huang, R., Zhang, F., Wang, T., Zhang, Y., Li, X., Chen, Y., et al. (2022). Effect of intrinsic tannins on the fermentation quality and associated with the bacterial and fungal community of sainfoin silage. Microorganisms 10:844. doi: 10.3390/microorganisms10050844
Jiang, F. G., Cheng, H. J., Liu, D., Wei, C., An, W. J., Wang, Y. F., et al. (2020). Treatment of whole-plant corn silage with lactic acid bacteria and organic acid enhances quality by elevating acid content, reducing pH, and inhibiting undesirable microorganisms. Front. Microbiol. 11:593088. doi: 10.3389/fmicb.2020.593088
Keshri, J., Chen, Y., Pinto, R., Kroupitski, Y., Weinberg, Z. G., and Sela, S. (2018). Microbiome dynamics during ensiling of corn with and without Lactobacillus plantarum inoculant. Appl. Microbiol. Biotechnol. 102, 4025–4037. doi: 10.1007/s00253-018-8903-y
Kung, L., and Shaver, R. (2001). Interpretation and use of silage fermentation analysis reports. Focus Forage 3, 1–5.
Lara, E. C., Basso, F. C., de Assis, F. B., Souza, F. A., Berchielli, T. T., and Reis, R. A. (2016). Changes in the nutritive value and aerobic stability of corn silages inoculated with Bacillus subtilis alone or combined with Lactobacillus plantarum. Anim. Prod. Sci. 56, 1867–1874. doi: 10.1071/AN14686
Lara, E. C., Bragiato, U. C., Rabelo, C. H., Messana, J. D., and Reis, R. A. (2018). Inoculation of corn silage with Lactobacillus plantarum and Bacillus subtilis associated with amylolytic enzyme supply at feeding. 1. Feed intake, apparent digestibility, and microbial protein synthesis in wethers. Anim. Feed Sci. Technol. 243, 22–34. doi: 10.1016/j.anifeedsci.2018.07.004
Li, Y., Du, S., Sun, L., Cheng, Q., Hao, J., Lu, Q., et al. (2022b). Effects of lactic acid bacteria and molasses additives on dynamic fermentation quality and microbial community of native grass silage. Front. Microbiol. 13:830121. doi: 10.3389/fmicb.2022.830121
Li, F., Ke, W., Ding, Z., Bai, J., Zhang, Y., Xu, D., et al. (2020). Pretreatment of Pennisetum sinese silages with ferulic acid esterase-producing lactic acid bacteria and cellulase at two dry matter contents: fermentation characteristics, carbohydrates composition and enzymatic saccharification. Bioresour. Technol. 295:122261. doi: 10.1016/j.biortech.2019.122261
Li, H., Wang, T., Tahir, M., Zhang, J., Sun, J., Xia, T., et al. (2022a). Influence of Lactobacillus plantarum inoculation on the silage quality of intercropped Lablab purpureus and sweet sorghum grown in saline-alkaline region. Front. Microbiol. 13:1059551. doi: 10.3389/fmicb.2022.1059551
Liu, B., Huan, H., Gu, H., Xu, N., Shen, Q., and Ding, C. (2019). Dynamics of a microbial community during ensiling and upon aerobic exposure in lactic acid bacteria inoculation-treated and untreated barley silages. Bioresour. Technol. 273, 212–219. doi: 10.1016/j.biortech.2018.10.041
Long, S., Li, X., Yuan, X., Su, R., Pan, J., Chang, Y., et al. (2022). The effect of early and delayed harvest on dynamics of fermentation profile, chemical composition, and bacterial community of king grass silage. Front. Microbiol. 13:864649. doi: 10.3389/fmicb.2022.864649
Luo, R., Zhang, Y., Wang, F., Liu, K., Huang, G., Zheng, N., et al. (2021). Effects of sugar cane molasses addition on the fermentation quality, microbial community, and tastes of alfalfa silage. Animals 11:355. doi: 10.3390/ani11020355
Muck, R., Nadeau, E., McAllister, T., Contreras-Govea, F., Santos, M., and Kung, L. Jr. (2018). Silage review: recent advances and future uses of silage additives. J. Dairy Sci. 101, 3980–4000. doi: 10.3168/jds.2017-13839
Ni, K., Wang, F., Zhu, B., Yang, J., Zhou, G., Pan, Y., et al. (2017). Effects of lactic acid bacteria and molasses additives on the microbial community and fermentation quality of soybean silage. Bioresour. Technol. 238, 706–715. doi: 10.1016/j.biortech.2017.04.055
Owens, V., Albrecht, K., Muck, R., and Duke, S. (1999). Protein degradation and fermentation characteristics of red clover and alfalfa silage harvested with varying levels of total nonstructural carbohydrates. Crop Sci. 39, 1873–1880. doi: 10.2135/cropsci1999.3961873x
Ren, H., Feng, Y., Pei, J., Li, J., Wang, Z., Fu, S., et al. (2020). Effects of Lactobacillus plantarum additive and temperature on the ensiling quality and microbial community dynamics of cauliflower leaf silages. Bioresour. Technol. 307:123238. doi: 10.1016/j.biortech.2020.123238
Santos, M. C. (2011). Identification and characterization of yeasts from high moisture corn and corn silages and their potential impact on in vitro rumen fermentation. Newark: University of Delaware.
Sedgwick, G., Fenton, T., and Thompson, J. (1991). Effect of protein precipitating agents on the recovery of plasma free amino acids. Can. J. Anim. Sci. 71, 953–957. doi: 10.4141/cjas91-116
Tabbene, O., Ben Slimene, I., Bouabdallah, F., Mangoni, M.-L., Urdaci, M.-C., and Limam, F. (2009). Production of anti-methicillin-resistant Staphylococcus activity from Bacillus subtilis sp. strain B38 newly isolated from soil. Appl. Biochem. Biotechnol. 157, 407–419. doi: 10.1007/s12010-008-8277-1
Tahir, M., Li, J., Xin, Y., Wang, T., Chen, C., Zhong, Y., et al. (2023). Response of fermentation quality and microbial community of oat silage to homofermentative lactic acid bacteria inoculation. Front. Microbiol. 13:1091394. doi: 10.3389/fmicb.2022.1091394
Tiwari, U. P., and Jha, R. (2017). Nutrients, amino acid, fatty acid and non-starch polysaccharide profile and in vitro digestibility of macadamia nut cake in swine. Anim. Sci. J. 88, 1093–1099. doi: 10.1111/asj.12750
van Soest, P. J., Robertson, J. B., and Lewis, B. A. (1991). Methods for dietary fiber, neutral detergent fiber, and nonstarch polysaccharides in relation to animal nutrition. J. Dairy Sci. 74, 3583–3597. doi: 10.3168/jds.s0022-0302(91)78551-2
Wang, Y., Ke, W., Lu, Q., and Zhang, G. (2023b). Effects of Bacillus coagulans and Lactobacillus plantarum on the fermentation characteristics, microbial community, and functional shifts during alfalfa silage fermentation. Animals 13:932. doi: 10.3390/ani13050932
Wang, T., Teng, K., Cao, Y., Shi, W., Xuan, Z., Zhou, J., et al. (2020). Effects of Lactobacillus hilgardii 60TS-2, with or without homofermentative Lactobacillus plantarum B90, on the aerobic stability, fermentation quality and microbial community dynamics in sugarcane top silage. Bioresour. Technol. 312:123600. doi: 10.1016/j.biortech.2020.123600
Wang, T., Zhang, J., Shi, W., Sun, J., Xia, T., Huang, F., et al. (2022). Dynamic changes in fermentation quality and structure and function of the microbiome during mixed silage of Sesbania cannabina and sweet sorghum grown on saline-alkaline land. Microbiol. Spectr. 10, e02483–e02422. doi: 10.1128/spectrum.02483-22
Wang, C., Zhang, J., and Zhou, W. (2023a). The effect of animal husbandry on economic growth: evidence from 13 provinces of North China. Front. Environ. Sci. 10:1085219. doi: 10.3389/fenvs.2022.1085219
Wang, C., Zheng, M., Wu, S., Zou, X., Chen, X., Ge, L., et al. (2021). Effects of gallic acid on fermentation parameters, protein fraction, and bacterial community of whole plant soybean silage. Front. Microbiol. 12:662966. doi: 10.3389/fmicb.2021.662966
Xia, T., Wang, T., Sun, J., Shi, W., Liu, Y., Huang, F., et al. (2022). Modulation of fermentation quality and metabolome in co-ensiling of Sesbania cannabina and sweet sorghum by lactic acid bacterial inoculants. Front. Microbiol. 13:851271. doi: 10.3389/fmicb.2022.851271
Xin, Y., Chen, C., Zhong, Y., Bu, X., Huang, S., Tahir, M., et al. (2023). Effect of storage time on the silage quality and microbial community of mixed maize and faba bean in the Qinghai-Tibet plateau. Front. Microbiol. 13:1090401. doi: 10.3389/fmicb.2022.1090401
Xu, H., Sun, L., Na, N., Wang, C., Yin, G., Liu, S., et al. (2021). Dynamics of bacterial community and fermentation quality in Leymus chinensis silage treated with lactic acid bacteria and/or water. Front. Microbiol. 12:717120. doi: 10.3389/fmicb.2021.717120
Xu, S., Yang, J., Qi, M., Smiley, B., Rutherford, W., Wang, Y., et al. (2019). Impact of Saccharomyces cerevisiae and Lactobacillus buchneri on microbial communities during ensiling and aerobic spoilage of corn silage1. J. Anim. Sci. 97, 1273–1285. doi: 10.1093/jas/skz021
Yang, F., Wang, Y., Zhao, S., Feng, C., and Fan, X. (2022). Dynamics of the fermentation products, residual non-structural carbohydrates, and bacterial communities of wilted and non-wilted alfalfa silage with and without Lactobacillus plantarum inoculation. Front. Microbiol. 12:824229. doi: 10.3389/fmicb.2021.824229
Yang, L., Yuan, X., Li, J., Dong, Z., and Shao, T. (2019). Dynamics of microbial community and fermentation quality during ensiling of sterile and nonsterile alfalfa with or without Lactobacillus plantarum inoculant. Bioresour. Technol. 275, 280–287. doi: 10.1016/j.biortech.2018.12.067
Yang, F., Zhao, S., Wang, Y., Fan, X., Wang, Y., and Feng, C. (2021). Assessment of bacterial community composition and dynamics in alfalfa silages with and without Lactobacillus plantarum inoculation using absolute quantification 16S rRNA sequencing. Front. Microbiol. 11:629894. doi: 10.3389/fmicb.2020.629894
Zhang, Y., Ke, W., Vyas, D., Adesogan, A., Franco, M., Li, F., et al. (2021). Antioxidant status, chemical composition and fermentation profile of alfalfa silage ensiled at two dry matter contents with a novel Lactobacillus plantarum strain with high-antioxidant activity. Anim. Feed Sci. Technol. 272:114751. doi: 10.1016/j.anifeedsci.2020.114751
Zhang, J., Liu, Y., Wang, Z., Bao, J., Zhao, M., Si, Q., et al. (2023a). Effects of different types of LAB on dynamic fermentation quality and microbial community of native grass silage during anaerobic fermentation and aerobic exposure. Microorganisms 11:513. doi: 10.3390/microorganisms11020513
Zhang, Y., Wang, M., Usman, S., Li, F., Bai, J., Zhang, J., et al. (2023b). Lignocellulose conversion of ensiled Caragana korshinskii Kom. Facilitated by Pediococcus acidilactici and cellulases. Microb. Biotechnol. 16, 432–447. doi: 10.1111/1751-7915.14130
Zhang, L., Zhou, X., Gu, Q., Liang, M., Mu, S., Zhou, B., et al. (2019). Analysis of the correlation between bacteria and fungi in sugarcane tops silage prior to and after aerobic exposure. Bioresour. Technol. 291:121835. doi: 10.1016/j.biortech.2019.121835
Zhou, X., Ouyang, Z., Zhang, X., Wei, Y., Tang, S., Ma, Z., et al. (2019). Sweet corn stalk treated with Saccharomyces cerevisiae alone or in combination with Lactobacillus plantarum: nutritional composition, fermentation traits and aerobic stability. Animals 9:598. doi: 10.3390/ani9090598
Zhu, W., Wei, Z., Xu, N., Yang, F., Yoon, I., Chung, Y., et al. (2017). Effects of Saccharomyces cerevisiae fermentation products on performance and rumen fermentation and microbiota in dairy cows fed a diet containing low quality forage. J. Anim. Sci. Biotechnol. 8, 36–39. doi: 10.1186/s40104-017-0167-3
Keywords: whole plant soybean silage, microbial communities, Lactiplantibacillus plantarum, Bacillus subtilis, Saccharomyces cerevisiae
Citation: Jin S, Tahir M, Huang F, Wang T, Li H, Shi W, Liu Y, Liu W and Zhong J (2024) Fermentation quality, amino acids profile, and microbial communities of whole-plant soybean silage in response to Lactiplantibacillus plantarum B90 alone or in combination with functional microbes. Front. Microbiol. 15:1458287. doi: 10.3389/fmicb.2024.1458287
Edited by:
Jie Zhao, Nanjing Agricultural University, ChinaCopyright © 2024 Jin, Tahir, Huang, Wang, Li, Shi, Liu, Liu and Zhong. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Tianwei Wang, d2FuZ3R3QGltLmFjLmNu; Jin Zhong, emhvbmdqQGltLmFjLmNu
†These authors have contributed equally to this work and share first authorship
 Sijie Jin1†
Sijie Jin1† Muhammad Tahir
Muhammad Tahir Fuqing Huang
Fuqing Huang Tianwei Wang
Tianwei Wang Jin Zhong
Jin Zhong