- Department of Molecular Biology and Biophysics, University of Connecticut School of Medicine, Farmington, CT, United States
The rules of the genetic code are implemented by the unique features that define the amino acid identity of each transfer RNA (tRNA). These features, known as “identity elements,” mark tRNAs for recognition by aminoacyl-tRNA synthetases (ARSs), the enzymes responsible for ligating amino acids to tRNAs. While tRNA identity elements enable stringent substrate selectivity of ARSs, these enzymes are prone to errors during amino acid selection, leading to the synthesis of incorrect aminoacyl-tRNAs that jeopardize the fidelity of protein synthesis. Many error-prone ARSs have evolved specialized domains that hydrolyze incorrectly synthesized aminoacyl-tRNAs. These domains, known as editing domains, also exist as free-standing enzymes and, together with ARSs, safeguard protein synthesis fidelity. Here, we discuss how the same identity elements that define tRNA aminoacylation play an integral role in aminoacyl-tRNA editing, synergistically ensuring the correct translation of genetic information into proteins. Moreover, we review the distinct strategies of tRNA selection used by editing enzymes and ARSs to avoid undesired hydrolysis of correctly aminoacylated tRNAs.
1 Introduction
Accurate translation of mRNAs into proteins requires the correct synthesis of aminoacyl-tRNAs (aa-tRNAs). This reaction, known as tRNA aminoacylation or charging, is catalyzed by aminoacyl-tRNA synthetases (ARSs), which ligate amino acids to tRNAs (Ibba and Soll, 2000). Errors in amino acid or tRNA selection by ARSs lead to incorrectly synthesized aa-tRNAs (Figure 1A). Generally, ARSs display a more robust specificity for their tRNA substrates than for amino acids. The relatively weaker amino acid specificity is mainly due to the structural and chemical similarities shared by many proteinogenic and non-proteinogenic amino acids (Ling et al., 2009a; Bullwinkle et al., 2014a; Hoffman et al., 2017; Mohler and Ibba, 2017). As a result, many ARSs do not effectively discern between cognate and near-cognate amino acid substrates. Prominent examples of tRNA mischarging include threonyl-tRNA synthetase (ThrRS), which confuses Ser for Thr (Dock-Bregeon et al., 2000), and isoleucyl-tRNA synthetase (IleRS), which mistakes Val for Ile (Berg et al., 1961). If uncorrected, tRNA aminoacylation errors lead to the translation of codons with the wrong amino acid (mistranslation), which can cause cellular dysregulation, growth defects, and death (Lee et al., 2006; Nangle et al., 2006; Ling and Soll, 2010; Bullwinkle et al., 2014b; Cvetesic et al., 2014; Liu et al., 2014, 2015; Lu et al., 2014; Kelly et al., 2019; Lant et al., 2019; Zhang et al., 2021; Schuntermann et al., 2023).
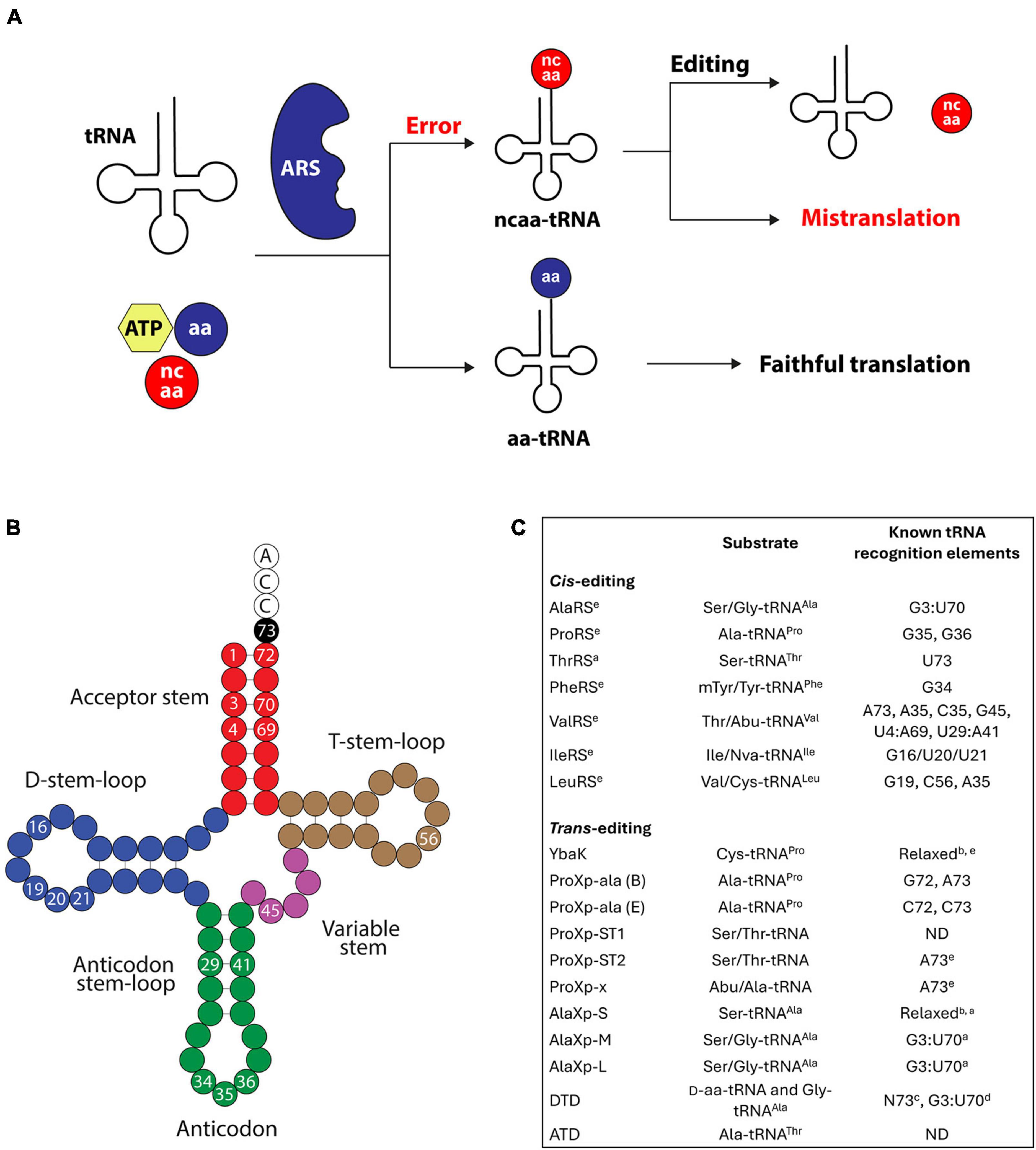
Figure 1. (A) Steps in tRNA aminoacylation and editing. tRNAs are aminoacylated by ARSs producing aa-tRNAs. If the ARS uses a non-cognate amino acid (ncaa), the resulting ncaa-tRNA can be hydrolyzed by the editing enzymes. In the absence of editing checkpoints, the ncaa is incorporated into proteins in response to the wrong codon, causing mistranslation. (B) Representative secondary structures of tRNAs. As discussed in the main text, the numbered bases indicate the various positions important for editing. (C) Summary of the trans- and cis-editing domains with characterized functions and their known tRNA recognition elements. aArchaeal origin; bindicates weak or no tRNA specificity; cthe specificity of N73 depends on the DTD’s origin; din the context of tRNAAla; eBacterial origin; “ND” indicates not determined. B and E for ProXp-Ala indicate bacterial and eukaryotic, respectively.
Due to their propensity to charge tRNAs with the wrong amino acid, ARSs acquired specialized hydrolytic domains to “edit” their aa-tRNA products. These domains, known as “editing” domains, catalyze the hydrolysis of mischarged tRNAs, ensuring that only correctly aminoacylated tRNAs accumulate in the cell (Figure 1A). In addition to the editing domains embedded in ARSs (known as cis-editing domains), aa-tRNA hydrolysis is catalyzed by standalone deacylases (known as trans-editing domains) (Kuzmishin Nagy et al., 2020; Jani and Pappachan, 2022). Cis- and trans-editing domains act as essential quality control checkpoints to maintain the integrity of the genetic code. The importance of aa-tRNA editing is underscored by the negative phenotypes associated with defects in editing domains (Lee et al., 2006; Nangle et al., 2006; Ling and Soll, 2010; Bullwinkle et al., 2014b; Cvetesic et al., 2014; Liu et al., 2014, 2015; Lu et al., 2014; Kelly et al., 2019; Lant et al., 2019; Zhang et al., 2021).
In contrast to amino acids, ARSs identify their tRNA substrates through an intricate set of structural and sequence features unique to each tRNA (Schimmel et al., 1993; Giege et al., 1998; Giege and Eriani, 2023). These tRNA features, collectively known as identity elements, promote faithful interactions between tRNAs and ARSs, preventing ARSs from cross-reacting with non-cognate tRNAs. Notably, growing evidence indicates that many editing domains rely on the same tRNA elements to gain aa-tRNA specificity and avoid hydrolysis of correctly aminoacylated tRNAs. This tRNA specificity is crucial to elude unintended energy loss due to the depletion of correctly aminoacylated tRNAs and to maintain adequate aa-tRNA supply for protein synthesis. More importantly, the role of tRNA identity elements in aa-tRNA editing highlights how identity elements secure the accurate translation of the genetic code.
2 tRNA identities
The elements that define the identity of tRNAs for a particular amino acid primarily reside in the tRNA acceptor stem and the anticodon loop (Figure 1B; Giege et al., 1998; Beuning and Musier-Forsyth, 1999; Giege and Eriani, 2023). Positions 1, 72, and 73 in the acceptor stem, and 35 and 36 in the anticodon are major contributors to tRNA selection. These elements act as an operational code to mark tRNAs for aminoacylation by a specific ARS (Schimmel et al., 1993; Ribas de Pouplana and Schimmel, 2001). Identity elements in the acceptor stem are generally recognized in the aminoacylation site of ARSs, whereas dedicated anticodon binding domains mediate the recognition of tRNA anticodon elements. tRNA identity elements are typically conserved within a single domain of life. However, with few exceptions, they diverge across domains of life (Lin et al., 2019). For example, the operational code for aminoacylation of tRNAPro diverged during evolution from G72 and A73 in bacteria to C72/A73 and C72/C73 in archaea and eukaryotes, respectively (Liu et al., 1995; Stehlin et al., 1998; Burke et al., 2001). These changes in tRNAPro were accompanied by changes in the selection mechanism of prolyl-tRNA synthetase (ProRS), preventing cross-reaction between ProRS and tRNAPro from different domains of life (Stehlin et al., 1998; Burke et al., 2001). Similar changes in the operational code of other tRNAs are known (Giege and Eriani, 2023).
3 The diversity of editing
Seven ARS families have editing domains to proofread aa-tRNA synthesis, whereas five families and superfamilies of trans-editing domains are currently known (Figure 2; Kuzmishin Nagy et al., 2020; Jani and Pappachan, 2022). In most cases, trans-editing domains are evolutionarily related to the editing domains of ARSs, sharing structural homology and, sometimes, substrate specificity. Trans- and cis-editing domains employ diverse mechanisms of substrate selection, which can involve unique characteristics of the amino acid side chain or tRNA features. Most editing domains use steric exclusion and/or chemical mechanisms to differentiate aminoacyl moieties of aa-tRNAs. Consequently, they tend to display relaxed amino acid specificities. For example, bacterial ProXp-ala, a trans-editing domain, hydrolyzes Ala- and Ser-tRNA with similar efficiency (Danhart et al., 2017). In contrast to their aminoacyl moiety selectivity, both trans- and cis-editing domains, with some exceptions, exhibit more robust tRNA specificities. The tRNA selectivity of editing enzymes can be mediated via direct or indirect interactions. These mechanisms of tRNA recognition are discussed in the following section.
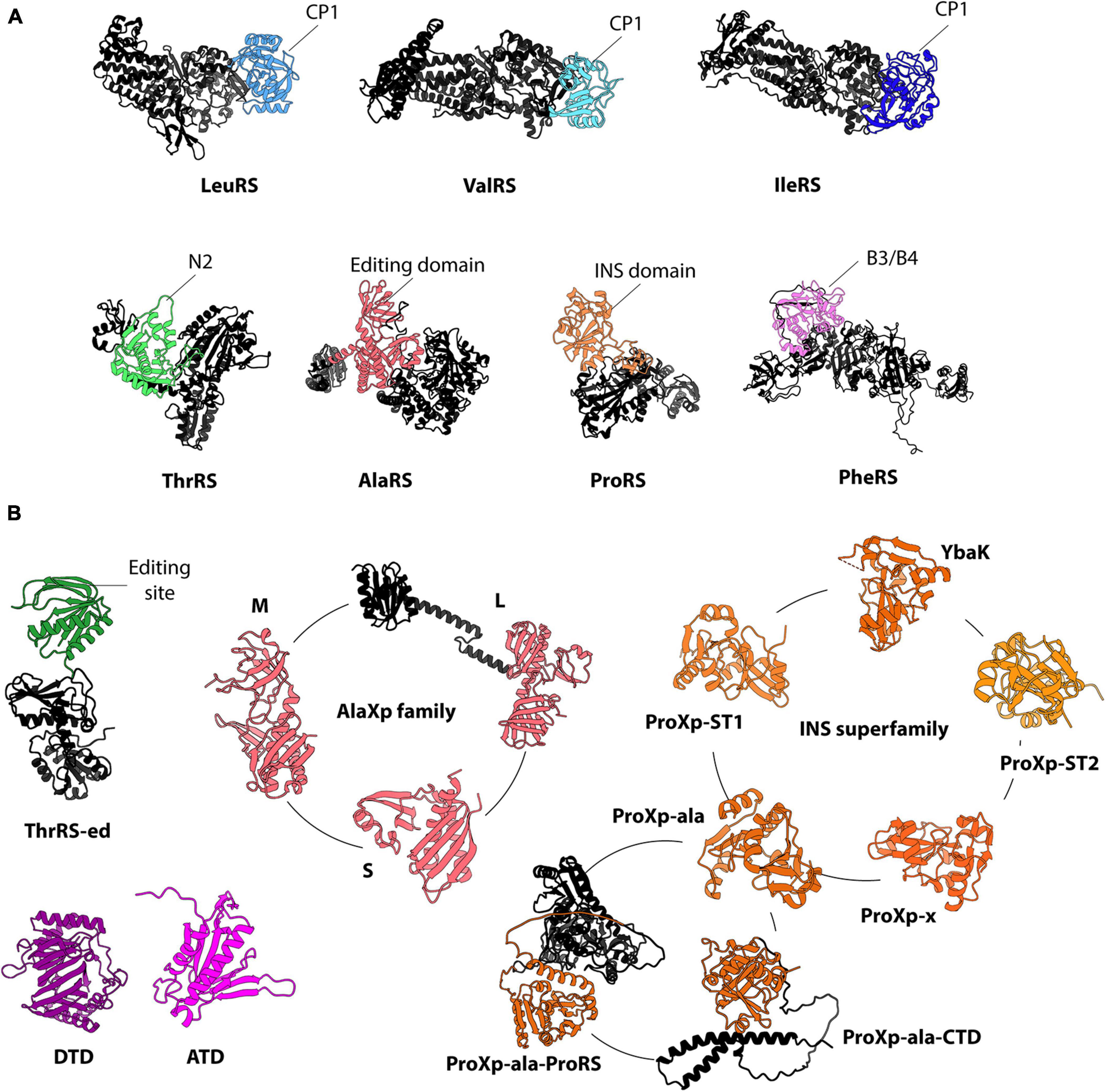
Figure 2. Representative structures of ARSs’ editing domains (A) and free-standing editing enzymes (B). The CP1 domains of LeuRS (PDB 3ZJU), ValRS (PDB 1IVS), and IleRS (PDB 1FFY) are colored in light blue, teal, and navy blue, respectively. The editing domains of ThrRS (PDB 1NYQ), AlaRS (PDB 3WQY), ProRS (PDB 2J3L), and PheRS (PDB 3PCO) are shown in green, pink, orange, and purple, respectively. The other domains (e.g., aminoacylation and anticodon binding domains) are in black. For ThrRS-ed (an AlphaFold model of S. solfataricus), the hydrolytic active domain is shown in green, while the anticodon binding domain is in black. The structure of E. coli DTD (PDB 1JKE) and the AlphaFold model of human ATD are shown. The INS superfamily is represented by ProRS and five single-domain families: YbaK (PDB 1DBU), ProXp-ST1 (an AlphaFold model of E. coli), ProXp-ST2 (an AlphaFold model of Bordetella parapertussis), ProXp-ala (PDB 5VXB), ProXp-ala-CTD (an AlphaFold model of Arabidopsis thaliana), ProXp-ala-ProRS (an AlphaFold model of Plasmodium falciparum) and ProXp-x (PDB 2CX5). ProXp-7, ProXp-8, and ProXp-9 were omitted from the INS superfamily because their activities are unknown. The three known isoforms of AlaXp are represented by the structures of AlaXp-S (PDB 1WXO), AlaXp-M (PDB 2E1B), and AlaXp-L (an AlphaFold model of Pyrococcus horikoshii). For simplicity, all structures are displayed in monomeric form.
4 Identity elements in aminoacyl-tRNA editing
Accurate recognition of mischarged tRNAs by editing enzymes is essential to avoid deacylation of correctly aminoacylated tRNAs. Because aa-tRNA synthesis requires an ATP molecule, indiscriminate hydrolysis of correctly charged tRNA by editing enzymes would be energetically costly and could impact cell growth and homeostasis by decreasing the available pool of aa-tRNAs for protein synthesis. As discussed in the following subsections, editing domains have evolved distinct mechanisms of substrate selection that ensure hydrolysis of the incorrect aa-tRNAs. Notably, in many cases, the same tRNA identity elements that define aminoacylation are used to gain specificity during editing (Figure 1C). However, lacking tRNA specificity in other cases may offer a functional advantage in acting on diverse mischarged tRNA substrates emerging from different ARSs.
4.1 ARS editing domains
4.1.1 Alanyl-tRNA synthetase (AlaRS)
AlaRS erroneously synthesizes Ser- and Gly-tRNAAla. The appended editing domain of AlaRS is responsible for clearing these mischarged products (Figure 2A; Beebe et al., 2003). The editing domain relies on the almost universally conserved wobble base pair G3:U70 to recognize tRNAAla (Beebe et al., 2008). G3:U70 is also indispensable for tRNA aminoacylation by AlaRS (Hou and Schimmel, 1988; McClain and Foss, 1988). Thus, a single base pair defines tRNAAla aminoacylation and editing. How the aa-tRNAAla substrate is transferred from the aminoacylation site to the editing domain remains unknown. Channeling the aa-tRNAAla between the two active sites would require substantial structural rearrangement of AlaRS to bring the editing domain closer to the aminoacylation domain and prevent complete dissociation of the tRNA (Naganuma et al., 2014). The C-Ala domain could facilitate the movement of the tRNA between the two domains (Guo et al., 2009). Alternatively, the editing domain could bind the tRNA after being released from the aminoacylation domain. Biochemical and biophysical characterization and structural studies are needed to determine the molecular mechanism of aa-tRNA selection by the editing domain of AlaRS.
4.1.2 ThrRS
Most ThrRSs encode a dedicated editing domain that deacylates Ser-tRNAThr produced in the aminoacylation domain (Dock-Bregeon et al., 2000; Beebe et al., 2004; Korencic et al., 2004). The editing domain is located at the N-terminus of ThrRS and exhibits evolutionary differences. Eukaryotic and bacterial ThrRS have a structurally similar editing domain known as the N2 (Figure 2A). In contrast, the archaeal ThrRS possesses an editing domain structurally homologous to D-aminoacyl-tRNA deacylases (DTD) (Dwivedi et al., 2005; Hussain et al., 2006). Notably, while the N2 and DTD-like domains effectively hydrolyze Ser-tRNAThr, they display distinct tRNA selectivity. For example, the N2 editing domain of E. coli ThrRS indiscriminately deacylates bacterial and archaeal Ser-tRNAThr. In contrast, the DTD-like domain of ThrRS from the archaeon Methanosarcina mazei only hydrolyzes archaeal Ser-tRNAThr (Beebe et al., 2004). Similarly, the editing domain of Pyrococcus abyssi ThrRS was shown to recognize Ser-tRNAThr while discriminating against other Ser-tRNA substrates (Novoa et al., 2015). These observations suggest that the tRNA specificity of the archaeal ThrRS editing domain may rely on the identity of position 73 (Beebe et al., 2004; Novoa et al., 2015), a conserved U73 in archaeal tRNAThr. In contrast, the same position is variable in bacterial and eukaryotic tRNAThr, consisting of A73 or U73 (Lin et al., 2019). Therefore, the N2 domain may have evolved a relaxed specificity that enables deacylation of tRNAThr with U73 and A73. This relaxed specificity toward N73 is also observed in the aminoacylation of bacterial and eukaryotic tRNAThr (Hasegawa et al., 1992; Nameki, 1995). In archaea, the role of N73 in aminoacylation is species-specific, with some species lacking N73 specificity (e.g., Haloferax volcanii) and others (e.g., Aeropyrum pernix) strongly depending on U73 (Ishikura et al., 2000; Nagaoka et al., 2002). Consequently, a weak correlation exists between editing and aminoacylation of tRNAThr in the context of N73. In contrast to N73, the anticodon bases play a more important and conserved role in tRNAThr aminoacylation (Giege and Eriani, 2023). Although direct evidence of the importance of the anticodon bases in editing is not available, a model based on E. coli ThrRS suggests that tRNAThr is held by the ThrRS anticodon binding domain, facilitating the CCA-end repositioning from the aminoacylation site to the editing domain (Dock-Bregeon et al., 2004). Whether the DTD-like editing domain of archaeal ThrRS uses a similar mechanism and how it recognizes the U73 is unknown.
4.1.3 Phenylalanyl-tRNA synthetase (PheRS)
The editing activity of PheRS resides in the B3/B4 domain of the β-subunit of the enzyme’s heterodimer (Figure 2A). The B3/B4 domain clears aminoacylation errors involving Tyr and meta-Tyr (Roy et al., 2004; Bullwinkle et al., 2014b). This activity of PheRS is essential for preventing mistranslation of Phe codons and maintaining cellular homeostasis. While a detailed investigation of its tRNA specificity is missing, the activity of the PheRS editing domain is affected by changes in the anticodon, as demonstrated by the lack of deacylation of a tRNAPhe G34A mutant (Ling et al., 2009b). Because G34 is an essential element for aminoacylation (Peterson and Uhlenbeck, 1992; Ling et al., 2009b), this result supports a 3′-end translocation model similar to ThrRS N2 editing, in which the anticodon binding domain provides indirect specificity to the editing by holding the tRNA and enabling the transfer of the 3′-end from the aminoacylation site to the editing site (Roy et al., 2004). Whether elements in the acceptor stem or other tRNA regions are directly recognized by the B3/B4 domain of PheRS requires further investigation.
4.1.4 ProRS
ProRS exists in different structural isoforms. In bacteria, the predominant ProRS isoform encodes an editing domain known as the insertion (INS) domain (Figure 2A). The INS domain catalyzes the deacylation of Ala-tRNAPro, which is incorrectly synthesized in the aminoacylation domain of ProRS. To avoid deacylation of cognate Ala-tRNAAla, the INS domain relies on the anticodon binding domain (ABD) of ProRS. The ABD offers specificity by interacting with the unique tRNAPro anticodon bases G35 and G36 (Das et al., 2014). These bases also serve as identity elements for aminoacylation (Liu et al., 1995; Stehlin et al., 1998). Changes in the identity of these bases prevent the binding of ProRS to the tRNA, impeding tRNA aminoacylation and deacylation. In contrast, mutations in the acceptor stem of tRNAPro are inconsequential for the catalysis of the INS domain. The role of the anticodon sequence in ProRS editing is further supported by the deacylation of Ala-tRNAAla mutants with a Pro UGG anticodon (Das et al., 2014). The dependency of the INS domain on the anticodon bases suggests that the ProRS ABD anchors the tRNA, enabling the translocation of the tRNA’s 3′-CCA end for editing. However, the molecular basis of this process remains poorly understood.
4.1.5 IleRS, LeuRS, and ValRS
IleRS, leucyl-tRNA synthetase (LeuRS), and valyl-tRNA synthetase (ValRS) share an evolutionarily related editing domain called CP1 (connecting peptide 1) (Figure 2A). However, the aa-tRNA specificity of each CP1 corresponds to the amino acid(s) mischarged by each ARS. IleRS’s CP1 catalyzes Val- and Cys-tRNA deacylation, whereas LeuRS’s editing domain hydrolyzes Ile- and Nva (norvaline)-tRNA, and ValRS edits Thr- and Abu (α-aminobutyrate)-tRNA (Baldwin and Berg, 1966; Englisch et al., 1986; Lin et al., 1996; Döring et al., 2001; Mursinna et al., 2004; Cvetesic et al., 2014). In addition to their different CP1 substrate specificities, these ARSs use distinct selection strategies for tRNA aminoacylation. IleRS and ValRS rely on anticodon bases and position 73, while LeuRS uses A73 and the unique long variable stem-loop of tRNALeu (Giege and Eriani, 2023).
For editing by ValRS’s CP1, A73, A35, and C36 are crucial, while other elements like the U4:A69, the anticodon stem U29:A41 base pair, and the core nucleotide G45 moderately contribute to editing (Tardif and Horowitz, 2002). The ValRS CP1’s reliance on the anticodon bases suggests that the ABD facilitates the CCA-end translocation between the aminoacylation and editing sites. The ValRS-tRNA complex supports this model (Fukai et al., 2000). Similarly, some overlap between elements for aminoacylation and editing has been established for LeuRS, albeit with antagonistic evidence emerging from two bacterial LeuRS models. For E. coli LeuRS, the interaction between G19 in the D-loop and C56 in the T-loop serves as a critical element for aminoacylation and editing (Du and Wang, 2003). However, LeuRS from Aquifex aeolicus, a deep-branching bacterium, may lack robust tRNA specificity for editing as it effectively edits Thr, Val, and Ile from different tRNA substrates (Zhu et al., 2007). Nonetheless, the anticodon stem-loop may contribute to transferring the tRNA acceptor stem from the aminoacylation to the editing site, as a mutation of A35 in tRNALeu mildly decreases editing (Yao et al., 2008). Structural evidence of LeuRS suggests that the anticodon binding domain holds the tRNA in place while the CCA-end moves from the aminoacylation state to the CP1 domain (Tukalo et al., 2005; Palencia et al., 2012). However, how changes in the tRNA anticodon influence LeuRS editing activity remains unclear.
Unlike ValRS and LeuRS, IleRS editing requires nucleotides that are different from those needed for aminoacylation. Nucleotides 16, 20, and 21 in the D-loop are the principal features that facilitate editing by E. coli IleRS CP1 (Hale et al., 1997). However, a mutant tRNAIle G16C/Δ20/U21G tRNAIle is deacylated with similar efficiency as wild-type (Farrow et al., 1999). These discrepancies suggest that D-loop bases influence the transfer of the tRNA but not the chemical step of deacylation (Farrow et al., 1999; Nomanbhoy et al., 1999). Notably, the crystal structure of IleRS bound to the tRNA in an editing conformation did not reveal direct interactions between IleRS and the tRNA D-loop (Silvian et al., 1999). Thus, additional biochemical and structural insights are needed to clarify the tRNA specificity of the IleRS CP1 domain, and how the aa-tRNAIle traffics between the two IleRS active sites is unknown. This could explain if a direct role of identity elements in editing exists.
4.2 Trans- editing domains
In contrast to ARSs, trans-editing domains generally lack dedicated RNA binding domains (Figure 2B). Nonetheless, several of these enzyme families have developed tRNA specificities based on recognizing tRNA acceptor stem elements. This recognition may be mediated in the same catalytic domain.
4.2.1 INS superfamily
In addition to the INS domain of ProRS, the INS superfamily groups eight families of trans-editing domains, YbaK, ProXp-ala, ProXp-x, ProXp-ST1, ProXp-ST2, ProXp-7, ProXp-8, and ProXp-9 (Vargas-Rodriguez and Musier-Forsyth, 2013; Kuzmishin Nagy et al., 2020). Most INS superfamily members are found in bacteria, but each family’s phylogenetic distribution pattern is unique. For example, ProXp-ala is found in all domains of life, whereas YbaK is present only in bacteria. Except for the INS domain, INS superfamily members are single-domain proteins. Interestingly, while these enzymes share high structural homologies and active site features, they display a wider range of aa-tRNA specificities catalyzed by several aaRSs. These deacylases also display distinct mechanisms of substrate selection, including tRNA recognition. In the following subsections, each family’s activities and tRNA specificities are described, except for ProXp-7, ProXp-8, and ProXp-9, whose functions remain unknown (Kuzmishin Nagy et al., 2020).
4.2.1.1 YbaK
YbaK is responsible for the deacylation of Cys-tRNAPro produced by ProRS (Ahel et al., 2002; An and Musier-Forsyth, 2004; Ruan and Söll, 2005). YbaK uses thiol-specific chemistry for Cys recognition and catalysis (Kumar et al., 2013). However, YbaK lacks robust tRNA selectivity, which results in the deacylation of Cys-tRNACys in vitro (An and Musier-Forsyth, 2005; Ruan and Söll, 2005; Das et al., 2014; Chen et al., 2019). In a cellular context, YbaK may gain indirect substrate specificity by forming a YbaK-tRNA-ProRS ternary complex that allows shuttling of Cys-tRNAPro from ProRS to YbaK, avoiding interaction with Cys-tRNACys (An and Musier-Forsyth, 2005; Chen et al., 2019). Additionally, the elongation factor Tu protects Cys-tRNACys from YbaK but not Cys-tRNAPro. How Cys-tRNAPro transitions from ProRS to YbaK is unknown.
4.2.1.2 ProXp-ala
ProXp-ala shares the same activity with the ProRS INS domain (Ahel et al., 2003; Vargas-Rodriguez and Musier-Forsyth, 2013). However, unlike the INS domain, ProXp-ala has a robust selectivity for tRNAPro based on the acceptor stem bases N72 and N73, which corresponds to G72 and A73 in bacteria and C72 and C73 in eukaryotes (Vargas-Rodriguez and Musier-Forsyth, 2013; Das et al., 2014; Ma et al., 2023). ProXp-ala’s specificity prevents cross-reaction with Ala-tRNAAla. Remarkably, ProXp-ala retained its tRNAPro specificity during evolution from bacteria to eukaryotes, adapting to changes in the identity of the N72 and N73 bases (Vargas-Rodriguez et al., 2020). ProXp-ala is also found fused to the N-terminus of ProRS (lacking an INS domain) in lower eukaryotes from the Stramenopila, Aveolates, and Rhizaria supergroups and the Leishmania and Trypanosoma genera (Ahel et al., 2003; Vargas-Rodriguez et al., 2020; Parrot et al., 2021). Evidence suggests that the ProRS-fused ProXp-ala can discriminate against Ala-tRNAAla (Figure 2B; Ahel et al., 2003). In plants, ProXp-ala contains a unique C-terminal domain (CTD) that contributes to the enzyme’s tRNA binding affinity (Figure 2B; Byun et al., 2022). However, the mechanism of substrate selection still needs to be determined for the ProXp-ala-ProRS fusion and plant ProXp-ala.
4.2.1.3 ProXp-x
ProXp-x deacylates tRNAs charged with the non-proteinogenic amino acid Abu, and to a lesser extent, Ala-tRNAPro (Bacusmo et al., 2018). ProXp-x prefers tRNA substrates carrying an A73, allowing it to recognize different Abu-tRNA substrates. This characteristic of ProXp-x is critical because ProRS, ValRS, LeuRS, and IleRS mischarge Abu (Döring et al., 2001; Nangle et al., 2002; Cvetesic et al., 2014; Bacusmo et al., 2018). Therefore, ProXp-x prevents broad mistranslation of the genetic code with Abu.
4.2.1.4 ProXp-ST1 and ProXp-ST2
ProXp-ST1 and ProXp-ST2 are homologous deacylases that catalyze the hydrolysis of Ser- and Thr-tRNAs (Liu et al., 2015). Both enzymes display broad tRNA specificity, recognizing diverse tRNAs, including tRNAVal, tRNAIle, tRNAThr, tRNAAla, and tRNALys, all of which are mischarged with either Ser or Thr by the corresponding ARS (Jakubowski, 2012; Liu et al., 2015). Thus, the broad tRNA specificity of ProXp-ST1 and ProXp-ST2 prevents mistranslation caused by Ser and Thr mischarging. Despite their overlapping substrate specificities, only ProXp-ST2 has developed direct tRNA recognition based on A73. This bias for tRNAs with A73 prevents hydrolysis of Ser-tRNASer due to the G73 of tRNASer (Liu et al., 2015). ProXp-ST1 is indifferent to the identity of N73, but whether it hydrolyzes Ser-tRNASer is unknown. Because tRNAThr has an A73, ProXp-ST1 and ProXp-ST2 can efficiently hydrolyze Thr-tRNAThr in vitro. However, ThrRS effectively prevents Thr-tRNAThr from both enzymes, offering a mechanism that protects correctly aminoacylated tRNAThr (Liu et al., 2015). A ProXp-ST1-related deacylase, FthB, that hydrolyzes fluorothreonyl-tRNAThr also exists, but little is known about its tRNA specificity (McMurry and Chang, 2017).
4.2.2 AlaXp
Like the AlaRS editing domain, AlaXp hydrolyzes Ser- and Gly-tRNAAla (Ahel et al., 2003; Sokabe et al., 2005; Fukunaga and Yokoyama, 2007; Beebe et al., 2008; Chong et al., 2008). AlaXp and the editing domain of AlaRS share high structural and sequence homology and possibly emerged from a common ancestor (Sokabe et al., 2005; Fukunaga and Yokoyama, 2007; Guo et al., 2009). AlaXp exists in three distinct isoforms classified based on their sequence length (Beebe et al., 2008; Novoa et al., 2015). While AlaXp-L and AlaXp-M are functionally identical, AlaXp-S only hydrolyzes Ser-tRNAAla (Sokabe et al., 2005). Moreover, AlaXp-L and AlaXp-M exhibit tRNA selectivity, achieved via recognition of the G3:U70 base pair that defines the identity of tRNAAla (Beebe et al., 2008). In contrast, AlaXp-S lacks tRNA specificity (Novoa et al., 2015). AlaXp-S is considered an ancestral version of the AlaXp family. Thus, AlaXp may have been a general aa-tRNA deacylase that gradually evolved tRNA specificity. A single Arg residue may determine the tRNA specificity of AlaXp (Novoa et al., 2015).
4.2.3 D-aminoacyl-tRNA deacylase (DTD)
DTDs prevent the cellular accumulation of D-aa-tRNAs stemming from several ARSs (Calendar and Berg, 1967; Soutourina et al., 2000). Three distinct DTD isoforms are found in organisms from all domains of life: DTD1 in most bacteria and eukaryotes, DTD2 in plants and archaea, and DTD3 in cyanobacteria (Kumar et al., 2022). Bacterial DTD requires a purine (A/G) in position 73 for effective aa-tRNA deacylation (Kuncha et al., 2018b). The specificity of bacterial DTD enables deacylation of several tRNA substrates while preventing deacylation of Gly-tRNAGly, which has a conserved U73 in bacteria (Routh et al., 2016). Interestingly, N73 evolved from U to A73 in cytosolic tRNAGly. This change in the identity of N73 prompted a switch in the tRNA specificity of eukaryotic DTD1, which prefers pyrimidine instead of purine (Gogoi et al., 2022). Whether the identity of N73 plays a role in the deacylation of D-aa-tRNAs is yet to be determined.
In addition to D-aa-tRNAs, bacterial DTD can inherently deacylate the achiral Gly from tRNAAla (Pawar et al., 2017). Bacterial DTD selects tRNAAla based on the G3:U70 and A73, which are essential for tRNAAla aminoacylation by AlaRS (Hou and Schimmel, 1988; McClain and Foss, 1988; Pawar et al., 2017). Finally, the Animalia-specific tRNA deacylase (ATD), a DTD paralog that hydrolyzes Ala-tRNAThr synthesized by AlaRS, may use G4:U69 and U73 for tRNA selection. The G4:U69 of tRNAThr enables mischarging by AlaRS (Sun et al., 2016; Kuncha et al., 2018a).
5 Outlook
Despite the strong correlation between the role of identity elements in tRNA editing and aminoacylation, our overall knowledge is limited. The tRNA specificities of several editing enzymes are unknown or poorly understood. For example, whether the B3/B4 domain of PheRS relies on tRNA acceptor stem is still unknown. The lack of molecular tools to prepare aa-tRNA substrates has significantly contributed to our poor understanding of the relationship between identity elements and editing. Producing mischarged tRNA variants using ARSs is challenging because mutating identity elements results in poor aminoacylation. Most available data for the tRNA specificity determination of CP1 domains are based on ATP consumption assays (Farrow et al., 1999; Tardif and Horowitz, 2002; Du and Wang, 2003; Zhu et al., 2007). This method integrates the effect of tRNA mutations in aminoacylation and editing. Thus, establishing the direct contribution of tRNA elements to editing can be intricate because the same elements can impact aminoacylation. The development of flexizyme technology now offers a powerful tool to investigate the role of identity elements in aa-tRNA editing (Murakami et al., 2006). This catalytic RNA ligates virtually any amino acid to tRNAs regardless of their sequence. Thus, it enables the preparation of diverse aa-tRNA mutant substrates to examine identity elements in the context of editing comprehensively (Das et al., 2014; Liu et al., 2015; Novoa et al., 2015; Danhart et al., 2017; Vargas-Rodriguez et al., 2020; Watkins et al., 2024). Adopting flexizyme can help establish and clarify the substrate specificities of many cis- and trans-editing enzymes from diverse species and across domains of life. Ultimately, this will expand our understanding of the dual role of identity elements in editing and aminoacylation, which, in turn, can provide novel insights into the contribution of editing enzymes to the establishment of the genetic code (Beebe et al., 2003).
Author contributions
EC: Writing – review and editing, Writing – original draft. OV-R: Writing – review and editing, Writing – original draft.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. Work in the Vargas-Rodriguez laboratory is partly supported by the National Science Foundation Grant (IOS-2304710 to OV-R).
Acknowledgments
We thank members of the Vargas-Rodriguez lab for critically reading the manuscript.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Ahel, I., Korencic, D., Ibba, M., and Söll, D. (2003). Trans-editing of mischarged tRNAs. Proc. Natl. Acad. Sci. U.S.A. 100, 15422–15427. doi: 10.1073/pnas.2136934100
Ahel, I., Stathopoulos, C., Ambrogelly, A., Sauerwald, A., Toogood, H., Hartsch, T., et al. (2002). Cysteine activation is an inherent in vitro property of prolyl-tRNA synthetases. J. Biol. Chem. 277, 34743–34748. doi: 10.1074/jbc.M206928200
An, S., and Musier-Forsyth, K. (2004). Trans-editing of Cys-tRNAPro by Haemophilus influenzae YbaK protein. J. Biol. Chem. 279, 42359–42362. doi: 10.1074/jbc.C400304200
An, S., and Musier-Forsyth, K. (2005). Cys-tRNA(Pro) editing by Haemophilus influenzae YbaK via a novel synthetase.YbaK.tRNA ternary complex. J. Biol. Chem. 280, 34465–34472. doi: 10.1074/jbc.M507550200
Bacusmo, J. M., Kuzmishin, A. B., Cantara, W. A., Goto, Y., Suga, H., and Musier-Forsyth, K. (2018). Quality control by trans-editing factor prevents global mistranslation of non-protein amino acid α-aminobutyrate. RNA Biol. 15, 576–585. doi: 10.1080/15476286.2017.1353846
Baldwin, A. N., and Berg, P. (1966). Transfer ribonucleic acid-induced hydrolysis of valyladenylate bound to isoleucyl ribonucleic acid synthetase. J. Biol. Chem. 241, 839–845.
Beebe, K., Merriman, E., Ribas De Pouplana, L., and Schimmel, P. (2004). A domain for editing by an archaebacterial tRNA synthetase. Proc. Natl. Acad. Sci. U.S.A. 101, 5958–5963. doi: 10.1073/pnas.0401530101
Beebe, K., Mock, M., Merriman, E., and Schimmel, P. (2008). Distinct domains of tRNA synthetase recognize the same base pair. Nature 451, 90–93. doi: 10.1038/nature06454
Beebe, K., Ribas De Pouplana, L., and Schimmel, P. (2003). Elucidation of tRNA-dependent editing by a class II tRNA synthetase and significance for cell viability. Embo J 22, 668–675. doi: 10.1093/emboj/cdg065
Berg, P., Bergmann, F. H., Ofengand, E., and Dieckmann, M. (1961). The enzymic synthesis of amino acyl derivatives of ribonucleic acid: I. The mechanism of leucyl-, valyl-, isoleucyl-, and methionyl ribonucleic acid formation. J. Biol. Chem. 236, 1726–1734.
Beuning, P. J., and Musier-Forsyth, K. (1999). Transfer RNA recognition by aminoacyl-tRNA synthetases. Biopolymers 52, 1–28. doi: 10.1002/(sici)1097-0282199952:1<1::Aid-bip1<3.0.Co;2-w
Bullwinkle, T. J., Reynolds, N. M., Raina, M., Moghal, A., Matsa, E., Rajkovic, A., et al. (2014b). Oxidation of cellular amino acid pools leads to cytotoxic mistranslation of the genetic code. eLife 3:e02501. doi: 10.7554/eLife.02501
Bullwinkle, T., Lazazzera, B., and Ibba, M. (2014a). Quality control and infiltration of translation by amino acids outside of the genetic code. Annu. Rev. Genet. 48, 149–166. doi: 10.1146/annurev-genet-120213-092101
Burke, B., Lipman, R. S., Shiba, K., Musier-Forsyth, K., and Hou, Y. M. (2001). Divergent adaptation of tRNA recognition by Methanococcus jannaschii prolyl-tRNA synthetase. J. Biol. Chem. 276, 20286–20291. doi: 10.1074/jbc.m100456200
Byun, J. K., Vu, J. A., He, S. L., Jang, J. C., and Musier-Forsyth, K. (2022). Plant-exclusive domain of trans-editing enzyme ProXp-ala confers dimerization and enhanced tRNA binding. J Biol Chem 298:102255. doi: 10.1016/j.jbc.2022.102255
Calendar, R., and Berg, P. (1967). D-Tyrosyl RNA: Formation, hydrolysis and utilization for protein synthesis. J. Mol. Biol. 26, 39–54. doi: 10.1016/0022-2836(67)90259-8
Chen, L., Tanimoto, A., So, B. R., Bakhtina, M., Magliery, T. J., Wysocki, V. H., et al. (2019). Stoichiometry of triple-sieve tRNA editing complex ensures fidelity of aminoacyl-tRNA formation. Nucleic Acids Res. 47, 929–940. doi: 10.1093/nar/gky1153
Chong, Y. E., Yang, X. L., and Schimmel, P. (2008). Natural homolog of tRNA synthetase editing domain rescues conditional lethality caused by mistranslation. J. Biol. Chem. 283, 30073–30078. doi: 10.1074/jbc.M805943200
Cvetesic, N., Palencia, A., Halasz, I., Cusack, S., and Gruic-Sovulj, I. (2014). The physiological target for LeuRS translational quality control is norvaline. EMBO J. 33, 1639–1653. doi: 10.15252/embj.201488199
Danhart, E. M., Bakhtina, M., Cantara, W. A., Kuzmishin, A. B., Ma, X., Sanford, B. L., et al. (2017). Conformational and chemical selection by a trans-acting editing domain. Proc. Natl. Acad. Sci. U.S.A. 114, E6774–E6783. doi: 10.1073/pnas.1703925114
Das, M., Vargas-Rodriguez, O., Goto, Y., Suga, H., and Musier-Forsyth, K. (2014). Distinct tRNA recognition strategies used by a homologous family of editing domains prevent mistranslation. Nucleic Acids Res. 42, 3943–3953. doi: 10.1093/nar/gkt1332
Dock-Bregeon, A. C., Rees, B., Torres-Larios, A., Bey, G., Caillet, J., and Moras, D. (2004). Achieving error-free translation; the mechanism of proofreading of threonyl-tRNA synthetase at atomic resolution. Mol. Cell 16, 375–386. doi: 10.1016/j.molcel.2004.10.002
Dock-Bregeon, A., Sankaranarayanan, R., Romby, P., Caillet, J., Springer, M., Rees, B., et al. (2000). Transfer RNA-mediated editing in threonyl-tRNA synthetase. The class II solution to the double discrimination problem. Cell 103, 877–884. doi: 10.1016/s0092-8674(00)00191-4
Döring, V., Mootz, H. D., Nangle, L. A., Hendrickson, T. L., de Crécy-Lagard, V., Schimmel, P., et al. (2001). Enlarging the amino acid set of Escherichia coli by infiltration of the valine coding pathway. Science 292, 501–504. doi: 10.1126/science.1057718
Du, X., and Wang, E. D. (2003). Tertiary structure base pairs between D- and TψC-loops of Escherichia coli tRNALeu play important roles in both aminoacylation and editing. Nucleic Acids Res. 31, 2865–2872. doi: 10.1093/nar/gkg382
Dwivedi, S., Kruparani, S. P., and Sankaranarayanan, R. (2005). A D-amino acid editing module coupled to the translational apparatus in archaea. Nat. Struct. Mol. Biol. 12, 556–557. doi: 10.1038/nsmb943
Englisch, S., Englisch, U., von der Haar, F., and Cramer, F. (1986). The proofreading of hydroxy analogues of leucine and isoleucine by leucyl-tRNA synthetases from E. coli and yeast. Nucleic Acids Res. 14, 7529–7539. doi: 10.1093/nar/14.19.7529
Farrow, M. A., Nordin, B. E., and Schimmel, P. (1999). Nucleotide determinants for tRNA-dependent amino acid discrimination by a class I tRNA synthetase. Biochemistry 38, 16898–16903. doi: 10.1021/bi9920782
Fukai, S., Nureki, O., Sekine, S., Shimada, A., Tao, J., Vassylyev, D. G., et al. (2000). Structural basis for double-sieve discrimination of L-valine from L-isoleucine and L-threonine by the complex of tRNA(Val) and valyl-tRNA synthetase. Cell 103, 793–803. doi: 10.1016/s0092-8674(00)00182-3
Fukunaga, R., and Yokoyama, S. (2007). Structure of the AlaX-M trans-editing enzyme from Pyrococcus horikoshii. Acta Crystallogr. D Biol. Crystallogr. 63(Pt 3), 390–400. doi: 10.1107/s090744490605640x
Giege, R., and Eriani, G. (2023). The tRNA identity landscape for aminoacylation and beyond. Nucleic Acids Res. 51, 1528–1570. doi: 10.1093/nar/gkad007
Giege, R., Sissler, M., and Florentz, C. (1998). Universal rules and idiosyncratic features in tRNA identity. Nucleic Acids Res. 26, 5017–5035. doi: 10.1093/nar/26.22.5017
Gogoi, J., Bhatnagar, A., Ann, K. J., Pottabathini, S., Singh, R., Mazeed, M., et al. (2022). Switching a conflicted bacterial DTD-tRNA code is essential for the emergence of mitochondria. Sci. Adv. 8:eabj7307. doi: 10.1126/sciadv.abj7307
Guo, M., Chong, Y. E., Beebe, K., Shapiro, R., Yang, X. L., and Schimmel, P. (2009). The C-Ala domain brings together editing and aminoacylation functions on one tRNA. Science 325, 744–747. doi: 10.1126/science.1174343
Hale, S. P., Auld, D. S., Schmidt, E., and Schimmel, P. (1997). Discrete determinants in transfer RNA for editing and aminoacylation. Science 276, 1250–1252. doi: 10.1126/science.276.5316.1250
Hasegawa, T., Miyano, M., Himeno, H., Sano, Y., Kimura, K., and Shimizu, M. (1992). Identity determinants of E. coli threonine tRNA. Biochem. Biophys. Res. Commun. 184, 478–484. doi: 10.1016/0006-291x(92)91219-g
Hoffman, K. S., O’Donoghue, P., and Brandl, C. J. (2017). Mistranslation: From adaptations to applications. Biochim. Biophys. Acta Gen. Subj. 1861(11 Pt B), 3070–3080. doi: 10.1016/j.bbagen.2017.01.031
Hou, Y. M., and Schimmel, P. (1988). A simple structural feature is a major determinant of the identity of a transfer RNA. Nature 333, 140–145. doi: 10.1038/333140a0
Hussain, T., Kruparani, S. P., Pal, B., Dock-Bregeon, A. C., Dwivedi, S., Shekar, M. R., et al. (2006). Post-transfer editing mechanism of a D-aminoacyl-tRNA deacylase-like domain in threonyl-tRNA synthetase from archaea. Embo j 25, 4152–4162. doi: 10.1038/sj.emboj.7601278
Ibba, M., and Soll, D. (2000). Aminoacyl-tRNA synthesis. Annu. Rev. Biochem. 69, 617–650. doi: 10.1146/annurev.biochem.69.1.617
Ishikura, H., Nagaoka, Y., Yokozawa, J., Umehara, T., Kuno, A., and Hasegawa, T. (2000). Threonyl-tRNA synthetase of archaea: Importance of the discriminator base in the aminoacylation of threonine tRNA. Nucleic Acids Symp. Ser. 44, 83–84. doi: 10.1093/nass/44.1.83
Jakubowski, H. (2012). Quality control in tRNA charging. Wiley Interdiscip. Rev. RNA 3, 295–310. doi: 10.1002/wrna.122
Jani, J., and Pappachan, A. (2022). A review on quality control agents of protein translation - The role of Trans-editing proteins. Int. J. Biol. Macromol. 199, 252–263. doi: 10.1016/j.ijbiomac.2021.12.176
Kelly, P., Backes, N., Mohler, K., Buser, C., Kavoor, A., Rinehart, J., et al. (2019). Alanyl-tRNA synthetase quality control prevents global dysregulation of the Escherichia coli proteome. mBio 10, e2921–e2919. doi: 10.1128/mBio.02921-19
Korencic, D., Ahel, I., Schelert, J., Sacher, M., Ruan, B., Stathopoulos, C., et al. (2004). A freestanding proofreading domain is required for protein synthesis quality control in Archaea. Proc. Natl. Acad. Sci. U.S.A. 101, 10260–10265. doi: 10.1073/pnas.0403926101
Kumar, P., Bhatnagar, A., and Sankaranarayanan, R. (2022). Chiral proofreading during protein biosynthesis and its evolutionary implications. FEBS Lett. 596, 1615–1627. doi: 10.1002/1873-3468.14419
Kumar, S., Das, M., Hadad, C. M., and Musier-Forsyth, K. (2013). Aminoacyl-tRNA substrate and enzyme backbone atoms contribute to translational quality control by YbaK. J. Phys. Chem. B 117, 4521–4527. doi: 10.1021/jp308628y
Kuncha, S. K., Mazeed, M., Singh, R., Kattula, B., Routh, S. B., and Sankaranarayanan, R. (2018a). A chiral selectivity relaxed paralog of DTD for proofreading tRNA mischarging in Animalia. Nat. Commun. 9:511. doi: 10.1038/s41467-017-02204-w
Kuncha, S. K., Suma, K., Pawar, K. I., Gogoi, J., Routh, S. B., Pottabathini, S., et al. (2018b). A discriminator code-based DTD surveillance ensures faithful glycine delivery for protein biosynthesis in bacteria. eLife 7:e38232. doi: 10.7554/eLife.38232
Kuzmishin Nagy, A. B., Bakhtina, M., and Musier-Forsyth, K. (2020). Trans-editing by aminoacyl-tRNA synthetase-like editing domains. Enzymes 48, 69–115. doi: 10.1016/bs.enz.2020.07.002
Lant, J. T., Berg, M. D., Heinemann, I. U., Brandl, C. J., and O’Donoghue, P. (2019). Pathways to disease from natural variations in human cytoplasmic tRNAs. J. Biol. Chem. 294, 5294–5308. doi: 10.1074/jbc.REV118.002982
Lee, J. W., Beebe, K., Nangle, L. A., Jang, J., Longo-Guess, C. M., Cook, S. A., et al. (2006). Editing-defective tRNA synthetase causes protein misfolding and neurodegeneration. Nature 443, 50–55. doi: 10.1038/nature05096
Lin, B. Y., Chan, P. P., and Lowe, T. M. (2019). tRNAviz: Explore and visualize tRNA sequence features. Nucleic Acids Res. 47, W542–W547. doi: 10.1093/nar/gkz438
Lin, L., Hale, S. P., and Schimmel, P. (1996). Aminoacylation error correction. Nature 384, 33–34. doi: 10.1038/384033b0
Ling, J., and Soll, D. (2010). Severe oxidative stress induces protein mistranslation through impairment of an aminoacyl-tRNA synthetase editing site. Proc. Natl. Acad. Sci. U.S.A. 107, 4028–4033. doi: 10.1073/pnas.1000315107
Ling, J., Reynolds, N., and Ibba, M. (2009a). Aminoacyl-tRNA synthesis and translational quality control. Annu. Rev. Microbiol. 63, 61–78. doi: 10.1146/annurev.micro.091208.073210
Ling, J., So, B. R., Yadavalli, S. S., Roy, H., Shoji, S., Fredrick, K., et al. (2009b). Resampling and editing of mischarged tRNA prior to translation elongation. Mol. Cell 33, 654–660. doi: 10.1016/j.molcel.2009.01.031
Liu, H., Peterson, R., Kessler, J., and Musier-Forsyth, K. (1995). Molecular recognition of tRNAPro by Escherichia coli proline tRNA synthetase in vitro. Nucleic Acids Res. 23, 165–169. doi: 10.1093/nar/23.1.165
Liu, Y., Satz, J. S., Vo, M. N., Nangle, L. A., Schimmel, P., and Ackerman, S. L. (2014). Deficiencies in tRNA synthetase editing activity cause cardioproteinopathy. Proc. Natl. Acad. Sci. U.S.A. 111, 17570–17575. doi: 10.1073/pnas.1420196111
Liu, Z., Vargas-Rodriguez, O., Goto, Y., Novoa, E. M., Ribas de Pouplana, L., Suga, H., et al. (2015). Homologous trans-editing factors with broad tRNA specificity prevent mistranslation caused by serine/threonine misactivation. Proc. Natl. Acad. Sci. U.S.A. 112, 6027–6032. doi: 10.1073/pnas.1423664112
Lu, J., Bergert, M., Walther, A., and Suter, B. (2014). Double-sieving-defective aminoacyl-tRNA synthetase causes protein mistranslation and affects cellular physiology and development. Nat. Commun. 5:5650. doi: 10.1038/ncomms6650
Ma, X., Bakhtina, M., Shulgina, I., Cantara, W. A., Kuzmishin Nagy, A. B., Goto, Y., et al. (2023). Structural basis of tRNAPro acceptor stem recognition by a bacterial trans-editing domain. Nucleic Acids Res. 51, 3988–3999. doi: 10.1093/nar/gkad192
McClain, W. H., and Foss, K. (1988). Changing the identity of a tRNA by introducing a G-U wobble pair near the 3′ acceptor end. Science 240, 793–796. doi: 10.1126/science.2452483
McMurry, J. L., and Chang, M. C. Y. (2017). Fluorothreonyl-tRNA deacylase prevents mistranslation in the organofluorine producer Streptomyces cattleya. Proc. Natl. Acad. Sci. U.S.A. 114, 11920–11925. doi: 10.1073/pnas.1711482114
Mohler, K., and Ibba, M. (2017). Translational fidelity and mistranslation in the cellular response to stress. Nat. Microbiol. 2:17117. doi: 10.1038/nmicrobiol.2017.117
Murakami, H., Ohta, A., Ashigai, H., and Suga, H. (2006). A highly flexible tRNA acylation method for non-natural polypeptide synthesis. Nat. Methods 3, 357–359. doi: 10.1038/nmeth877
Mursinna, R. S., Lee, K. W., Briggs, J. M., and Martinis, S. A. (2004). Molecular dissection of a critical specificity determinant within the amino acid editing domain of leucyl-tRNA synthetase. Biochemistry 43, 155–165. doi: 10.1021/bi034919h
Naganuma, M., Sekine, S., Chong, Y. E., Guo, M., Yang, X. L., Gamper, H., et al. (2014). The selective tRNA aminoacylation mechanism based on a single GU pair. Nature 510, 507–511. doi: 10.1038/nature13440
Nagaoka, Y., Yokozawa, J., Umehara, T., Iwaki, J., Okamoto, K., Kawarabayasi, Y., et al. (2002). Molecular recognition of threonine tRNA by threonyl-tRNA synthetase from an extreme thermophilic archaeon, Aeropyrum pernix K1. Nucleic Acids Res. 2, 81–82. doi: 10.1093/nass/2.1.81
Nameki, N. (1995). Identity elements of tRNA(Thr) towards Saccharomyces cerevisiae threonyl-tRNA synthetase. Nucleic Acids Res. 23, 2831–2836. doi: 10.1093/nar/23.15.2831
Nangle, L. A., De Crecy Lagard, V., Doring, V., and Schimmel, P. (2002). Genetic code ambiguity. Cell viability related to the severity of editing defects in mutant tRNA synthetases. J. Biol. Chem. 277, 45729–45733. doi: 10.1074/jbc.M208093200
Nangle, L. A., Motta, C. M., and Schimmel, P. (2006). Global effects of mistranslation from an editing defect in mammalian cells. Chem. Biol. 13, 1091–1100. doi: 10.1016/j.chembiol.2006.08.011
Nomanbhoy, T. K., Hendrickson, T. L., and Schimmel, P. (1999). Transfer RNA-dependent translocation of misactivated amino acids to prevent errors in protein synthesis. Mol. Cell 4, 519–528. doi: 10.1016/s1097-2765(00)80203-8
Novoa, E. M., Vargas-Rodriguez, O., Lange, S., Goto, Y., Suga, H., Musier-Forsyth, K., et al. (2015). Ancestral AlaX editing enzymes for control of genetic code fidelity are not tRNA-specific. J. Biol. Chem. 290, 10495–10503. doi: 10.1074/jbc.M115.640060
Palencia, A., Crépin, T., Vu, M. T., Lincecum, T. L. Jr., Martinis, S. A., and Cusack, S. (2012). Structural dynamics of the aminoacylation and proofreading functional cycle of bacterial leucyl-tRNA synthetase. Nat. Struct. Mol. Biol. 19, 677–684. doi: 10.1038/nsmb.2317
Parrot, C., Moulinier, L., Bernard, F., Hashem, Y., Dupuy, D., and Sissler, M. (2021). Peculiarities of aminoacyl-tRNA synthetases from trypanosomatids. J. Biol. Chem. 297:100913. doi: 10.1016/j.jbc.2021.100913
Pawar, K. I., Suma, K., Seenivasan, A., Kuncha, S. K., Routh, S. B., Kruparani, S. P., et al. (2017). Role of D-aminoacyl-tRNA deacylase beyond chiral proofreading as a cellular defense against glycine mischarging by AlaRS. eLife 6:e24001. doi: 10.7554/eLife.24001
Peterson, E. T., and Uhlenbeck, O. C. (1992). Determination of recognition nucleotides for Escherichia coli phenylalanyl-tRNA synthetase. Biochemistry 31, 10380–10389. doi: 10.1021/bi00157a028
Ribas de Pouplana, L., and Schimmel, P. (2001). Operational RNA code for amino acids in relation to genetic code in evolution. J. Biol. Chem. 276, 6881–6884. doi: 10.1074/jbc.R000032200
Routh, S. B., Pawar, K. I., Ahmad, S., Singh, S., Suma, K., Kumar, M., et al. (2016). Elongation factor Tu prevents misediting of Gly-tRNA(Gly) caused by the design behind the chiral proofreading site of D-aminoacyl-tRNA deacylase. PLoS Biol. 14:e1002465. doi: 10.1371/journal.pbio.1002465
Roy, H., Ling, J., Irnov, M., and Ibba, M. (2004). Post-transfer editing in vitro and in vivo by the beta subunit of phenylalanyl-tRNA synthetase. EMBO J. 23, 4639–4648. doi: 10.1038/sj.emboj.7600474
Ruan, B., and Söll, D. (2005). The bacterial YbaK protein is a Cys-tRNAPro and Cys-tRNA Cys deacylase. J. Biol. Chem. 280, 25887–25891. doi: 10.1074/jbc.M502174200
Schimmel, P., Giegé, R., Moras, D., and Yokoyama, S. (1993). An operational RNA code for amino acids and possible relationship to genetic code. Proc. Natl. Acad. Sci. U.S.A. 90, 8763–8768. doi: 10.1073/pnas.90.19.8763
Schuntermann, D. B., Fischer, J. T., Bile, J., Gaier, S. A., Shelley, B. A., Awawdeh, A., et al. (2023). Mistranslation of the genetic code by a new family of bacterial transfer RNAs. J. Biol. Chem. 299:104852. doi: 10.1016/j.jbc.2023.104852
Silvian, L. F., Wang, J., and Steitz, T. A. (1999). Insights into editing from an Ile-tRNA synthetase structure with tRNAile and mupirocin. Science 285, 1074–1077.
Sokabe, M., Okada, A., Yao, M., Nakashima, T., and Tanaka, I. (2005). Molecular basis of alanine discrimination in editing site. Proc. Natl. Acad. Sci. U.S.A. 102, 11669–11674. doi: 10.1073/pnas.0502119102
Soutourina, J., Plateau, P., and Blanquet, S. (2000). Metabolism of D-aminoacyl-tRNAs in Escherichia coli and Saccharomyces cerevisiae cells. J. Biol. Chem. 275, 32535–32542. doi: 10.1074/jbc.M005166200
Stehlin, C., Burke, B., Yang, F., Liu, H., Shiba, K., and Musier-Forsyth, K. (1998). Species-specific differences in the operational RNA code for aminoacylation of tRNAPro. Biochemistry 37, 8605–8613. doi: 10.1021/bi980364s
Sun, L., Gomes, A. C., He, W., Zhou, H., Wang, X., Pan, D. W., et al. (2016). Evolutionary gain of alanine mischarging to noncognate tRNAs with a G4:U69 base pair. J. Am. Chem. Soc. 138, 12948–12955. doi: 10.1021/jacs.6b07121
Tardif, K. D., and Horowitz, J. (2002). Transfer RNA determinants for translational editing by Escherichia coli valyl-tRNA synthetase. Nucleic Acids Res. 30, 2538–2545. doi: 10.1093/nar/30.11.2538
Tukalo, M., Yaremchuk, A., Fukunaga, R., Yokoyama, S., and Cusack, S. (2005). The crystal structure of leucyl-tRNA synthetase complexed with tRNALeu in the post-transfer-editing conformation. Nat. Struct. Mol. Biol. 12, 923–930. doi: 10.1038/nsmb986
Vargas-Rodriguez, O., and Musier-Forsyth, K. (2013). Exclusive use of trans-editing domains prevents proline mistranslation. J. Biol. Chem. 288, 14391–14399. doi: 10.1074/jbc.M113.467795
Vargas-Rodriguez, O., Bakhtina, M., McGowan, D., Abid, J., Goto, Y., Suga, H., et al. (2020). Human trans-editing enzyme displays tRNA acceptor-stem specificity and relaxed amino acid selectivity. J. Biol. Chem. 295, 16180–16190. doi: 10.1074/jbc.RA120.015981
Watkins, R. R., Kavoor, A., and Musier-Forsyth, K. (2024). Strategies for detecting aminoacylation and aminoacyl-tRNA editing In Vitro and in cells. Isr. J. Chem. e202400009. doi: 10.1002/ijch.202400009
Yao, P., Zhu, B., Jaeger, S., Eriani, G., and Wang, E. D. (2008). Recognition of tRNALeu by Aquifex aeolicus leucyl-tRNA synthetase during the aminoacylation and editing steps. Nucleic Acids Res. 36, 2728–2738. doi: 10.1093/nar/gkn028
Zhang, H., Wu, J., Lyu, Z., and Ling, J. (2021). Impact of alanyl-tRNA synthetase editing deficiency in yeast. Nucleic Acids Res. 49, 9953–9964. doi: 10.1093/nar/gkab766
Keywords: tRNA, mistranslation, translational fidelity, protein synthesis, aminoacyl-tRNA synthetases, editing
Citation: Cruz E and Vargas-Rodriguez O (2024) The role of tRNA identity elements in aminoacyl-tRNA editing. Front. Microbiol. 15:1437528. doi: 10.3389/fmicb.2024.1437528
Received: 23 May 2024; Accepted: 18 June 2024;
Published: 18 July 2024.
Edited by:
Jiqiang Ling, University of Maryland, College Park, United StatesReviewed by:
Karin Musier-Forsyth, The Ohio State University, United StatesRajan Sankaranarayanan, Centre for Cellular and Molecular Biology (CCMB), India
Copyright © 2024 Cruz and Vargas-Rodriguez. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Oscar Vargas-Rodriguez, dmFyZ2Fzcm9kcmlndWV6QHVjaGMuZWR1
 Edwin Cruz
Edwin Cruz Oscar Vargas-Rodriguez
Oscar Vargas-Rodriguez