
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Microbiol. , 18 June 2024
Sec. Virology
Volume 15 - 2024 | https://doi.org/10.3389/fmicb.2024.1401997
Influenza A virus (IAV) is a negative-sense single-stranded RNA virus that causes acute lung injury and acute respiratory distress syndrome, posing a serious threat to both animal and human health. N6-methyladenosine (m6A), a prevalent and abundant post-transcriptional methylation of RNA in eukaryotes, plays a crucial regulatory role in IAV infection by altering viral RNA and cellular transcripts to affect viral infection and the host immune response. This review focuses on the molecular mechanisms underlying m6A modification and its regulatory function in the context of IAV infection and the host immune response. This will provide a better understanding of virus–host interactions and offer insights into potential anti-IAV strategies.
Influenza A virus (IAV) is a pathogen that primarily affects the respiratory system, often leading to seasonal influenza outbreaks and global pandemics. In severe cases, the virus can cause acute respiratory distress syndrome (ARDS), septic shock, and multiple organ failure (MOF), which can be life-threatening (Taubenberger and Kash, 2010; Worobey et al., 2014; Herold et al., 2015). The World Health Organization (WHO) estimates that influenza is responsible for ~1 billion infections worldwide each year, resulting in 3–5 million severe cases and up to 650,000 deaths from respiratory illness (World Health Organization, 2023). IAV belongs to the Orthomyxoviridae family and is an enveloped virus with a single-stranded negative-sense RNA genome (Chou et al., 2012). Its genome is composed of eight fragments that encode three viral polymerase proteins (PB1, PB2, and PA), hemagglutinin (HA), nucleoprotein (NP), neuraminidase (NA), matrix protein (M), and non-structural (NS) protein (McGeoch et al., 1976; Cho et al., 2020). The HA protein recognizes sialic acid receptors on the host cell surface, facilitating virus attachment and entry through endocytosis (Skehel and Wiley, 2000; Fan et al., 2019). Within the cytoplasm, viral proteins and RNAs are synthesized by viral polymerase catalysis and assembled into mature viral particles using various host factors that enhance polymerase activity (Te Velthuis and Fodor, 2016; Peacock et al., 2019; Carrique et al., 2020). Subsequently, the NA protein cleaves the glycosidic bonds between viral particles and the host cell membrane, initiating virion budding and completing the replication cycle of IAV (Mora et al., 2002; Yang et al., 2016). During the process of viral replication, both double-stranded and single-stranded RNAs are generated. These RNAs act as pathogen-associated pattern molecules (PAMPs) that recognize and bind to pattern recognition receptors (PRRs) in the host, activating innate immune responses (Brennan and Bowie, 2010; Yoo et al., 2013; Downey et al., 2017). This leads to the production of various cytokines and antiviral molecules that inhibit viral replication and help control infection.
The pathogenesis of IAV involves two main processes: direct damage caused by viral replication within host cells and indirect damage caused by a virus-induced cytokine storm (Liu et al., 2016; Zhang H. et al., 2022). Existing strains of the virus have developed resistance to anti-IAV drugs, and current IAV vaccines often do not provide sufficient protection against new and emerging strains during local and global pandemics (Gubareva et al., 2000; Ann et al., 2012). Therefore, there is an urgent need to identify new targets and develop effective drugs for the control of influenza infections.
The discovery of various chemical modifications to RNA, such as the methylation and acetylation of bases such as adenine, guanine, and cytosine, has increased in recent years. Among these modifications, methylation is considered to be the most significant (Saletore et al., 2012; Jia et al., 2013). N6-methyladenosine (m6A) modification, which involves methylation of the N6 position of adenosine in RNA, is a common epigenetic transcriptional modification in eukaryotes. m6A is involved in multiple aspects of RNA biology and metabolism, including variable splicing, nuclear export, stability, and translation, with a particular impact on mRNA function (Motorin and Helm, 2022; Boulias and Greer, 2023). Additionally, m6A modifications have been identified in non-coding RNAs (lncRNAs, circRNAs, miRNAs, etc.) (Meyer et al., 2012; Lv et al., 2020). m6A modification is a dynamic and reversible epigenetic process (Fu et al., 2014; Meyer and Jaffrey, 2014; Roundtree et al., 2017a) mediated by three functional proteins, namely, methyltransferases (referred to as “writers”), demethylases (referred to as “erasers”), and m6A-binding proteins (referred to as “readers”), which are responsible for methylation, demethylation, and RNA binding, respectively (Wu et al., 2017; Flamand et al., 2023; Wang et al., 2023). Accumulating evidence suggests that m6A modification plays a critical role in regulating cellular mRNA function and is widespread among the genomic RNAs of DNA and RNA viruses, affecting viral infection (Zhao et al., 2020; Yu et al., 2021). The initial identification of m6A residues in all mRNAs was performed in the IAV, with eight m6A modification sites in the HA mRNA (Krug et al., 1976). Moreover, the distribution of m6A modification sites varies among different influenza virus mRNAs (Narayan et al., 1987). Recent studies have highlighted the significant role of m6A modification in regulating the gene expression and replication of IAV, although the precise regulatory mechanism involved remains unclear (Imam et al., 2020). This review focuses on recent studies that elucidate the molecular mechanisms of m6A modification and its underlying regulatory role in viral genomes and cellular antiviral responses during IAV infection. These findings provide a deeper understanding of the detailed interaction patterns between host proteins and viral proteins and offer new insights for future investigations into therapeutic interventions for IAV infection.
Over the past decade, an increasing body of evidence has confirmed the involvement of m6A modification in regulating various physiological and pathological processes in mammalian cells. These processes include stem cell differentiation, cell reprogramming, stress response, circadian rhythm, DNA damage repair, metabolism, tumorigenesis, and inflammatory response (Shi et al., 2019; Wang et al., 2020; Huang W. et al., 2021). m6A modifications have been detected in different tissues and organs, with the highest levels observed in the brain, heart, and kidneys (Meyer et al., 2012). Three key proteins play vital roles in the m6A modification process. Methyltransferases mediate RNA methylation through stable multicomponent complexes in the nucleus (Murakami and Jaffrey, 2022; Sendinc and Shi, 2023). Specifically, methyltransferase-like 3 (METTL3) acts as the only subunit with catalytic activity, whereas its homolog, METTL14, serves as an allosteric activator that supports the catalytic activity of METTL3. These two proteins interact closely in the crystal structure, forming the stable heterodimer METTL3-METTL14 methyltransferase core complex, which plays a crucial role in substrate RNA recognition (Liu et al., 2014; Wang X. et al., 2016). In addition to the METTL3–METTL14 complex, the two CCCH-type zinc finger domains in the METTL3 structure also contribute to the complete enzymatic activity of the methyltransferase during the process of substrate RNA recognition and catalysis (Wang P. et al., 2016). In contrast, another homolog of METTL3, called METTL4, does not contain a catalytic domain and lacks catalytic activity. However, it can increase the catalytic efficiency of METTL3 (Luo et al., 2022). The m6A modification process is also regulated by the splicing factor Wilms tumor 1-associated protein (WTAP) (Ping et al., 2014). The m6A methyltransferase complex (MTC) consists of METTL3, METTL14, and WTAP. METTL3 contributes to the catalytic activity of MTC, whereas METTL14 recognizes and binds to its substrate RNA. WTAP is responsible for recruiting METTL3 and METTL14 to nuclear speckles for localization within subcellular structures (Sun et al., 2020). Additionally, several other proteins, including METTL16, Vir-like m6A methyltransferase-associated protein (VIRMA), zinc finger CCCH-type containing 13 (ZC3H13), and RNA-binding motif 15 (RBM15)/15B, are also involved in the RNA methylation process as part of the MTC (Zhang et al., 2019; Wang et al., 2021; Zhu W. et al., 2021; Lin et al., 2022; Satterwhite and Mansfield, 2022). Dominissini proposed a novel technique known as methylated RNA immunoprecipitation sequencing (MeRIP-seq). This innovative method combines immunoprecipitation using an anti-m6A affinity antibody with high-throughput RNA sequencing of the precipitated RNA fragments. The application of MeRIP-seq enabled Dominissini et al. to demonstrate, for the first time, that m6A modification sites exhibit greater sequence specificity than other regions (Dominissini et al., 2012). These m6A modification sites are not randomly distributed within transcripts; they are often found in the conserved RRACH motif of RNA (R = G/A, H = U/A/C) and are primarily enriched in stop codons and 3′-untranslated regions (3′-UTRs). Upon recognition and binding of the m6A modification sites on the targeted RNA, the MTC transfers methyl groups from S-adenosylmethionine (SAM) to the RNA, thereby influencing the functionality of both cellular- and virus-encoded mRNAs. Consequently, SAM is converted to S-adenosylhomocysteine (SAH) via demethylation. Conversely, demethylases are capable of removing methyl groups from the targeted RNA, effectively reversing the m6A modification process and dynamically regulating RNA methylation. Fat mass and obesity-associated protein (FTO) was the first demethylase identified to mediate RNA demethylation. Nuclear RNA serves as the primary substrate for FTO (Jia et al., 2011; Bayoumi and Munir, 2020). Furthermore, alkylation repair homolog protein 5 (ALKBH5) has been identified as another demethylase in mammals that exhibits demethylation activity comparable to that of FTO (Zheng et al., 2013). Both FTO and ALKBH5 regulate nuclear export, metabolism, and gene expression by removing methylation residues from mRNAs. During the process of RNA methylation, m6A-modified transcripts are specifically recognized and bound by various reader proteins, including the YTH domain family (YTHDC1–2 and YTHDF1–3) (Gokhale et al., 2016), IGF2BP1–3 (Müller et al., 2019), eIF3 (Choe et al., 2018), and hnRNPC/G/A2B1 proteins (Huang X.-T. et al., 2021). At the nuclear stage, m6A residues can bind to specific nuclear reader proteins, which can affect mRNA splicing, nuclear export, and other nuclear processes. Upon export to the cytoplasm, m6A residues bind to specific cytoplasmic reader proteins, thereby influencing the structural stability, degradation, translation, and localization of mRNA (Figure 1) (Zaccara et al., 2019). Using the linear amplification of complementary DNA ends and sequencing (LACE-seq) method (Su et al., 2021), Qiao demonstrated that the nucleus-localized RNA-binding protein (RBP) YTHDC1 specifically recognizes the m6A site through its C-terminal YTH domain. This recognition enables YTHDC1 to regulate the selective splicing and nuclear export of mRNA in skeletal muscle stem cells (Qiao et al., 2023). YTHDC1, in conjunction with hnRNPG, synergistically promotes mRNA splicing, regulates isomer diversity, and enhances mRNA output by binding to the THO nuclear export complex. These proteins also interact with the nuclear export adapter protein SRSF3, facilitating RNA binding to SRSF3 and nuclear RNA export factor 1 (NXF1), thereby mediating methylated mRNA export from the nucleus to the cytoplasm in HeLa cells. Deletion of YTHDC1 results in prolonged residence time for nuclear mRNA containing m6A, leading to the accumulation of transcripts in the nucleus and depletion in the cytoplasm (Roundtree et al., 2017b; Qiao et al., 2023).
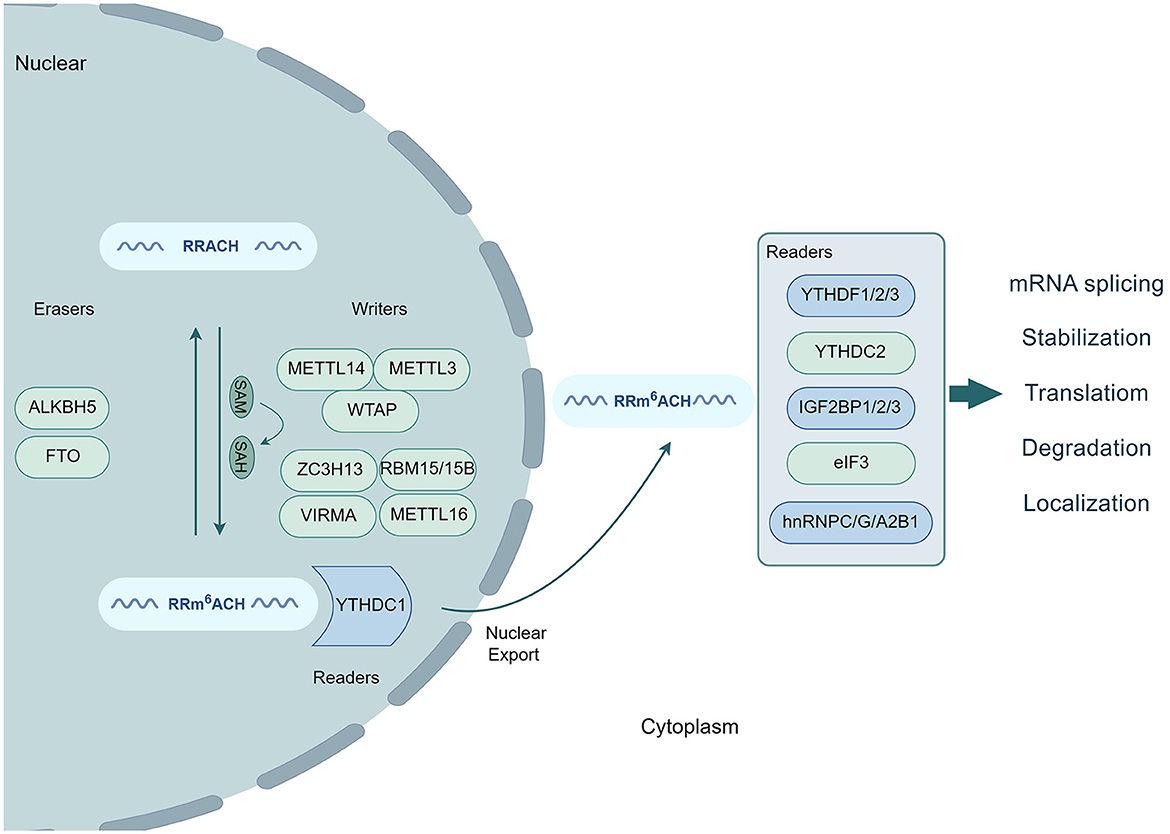
Figure 1. Process of m6A modification. The conserved RRACH motif of RNA undergoes methylation by writers (METTL3, METTL14, WTAP, ZC3H13, VIRMA, RBM15/15B, and METTL16) along with SAM conversion to SAH. This modification is reversibly controlled by erasers (FTO and ALKBH5) that demethylate RNA. Readers, such as YTHDC1/2, YTHDF1/2/3, IGF2BP1/2/3, eIF3, and hnRNPC/G/A2B1, recognize and bind to m6A-modified transcripts, influencing mRNA splicing, stabilization, translation, degradation, and localization.
In addition to YTHDC1, YTHDF1 also recruits argonaute family protein 2 (AGO2) via its YTH domain to promote the solvation and degradation of targeted mRNAs. However, the absence of YTHDF1 results in the formation of AGO2/RNA patches, delayed mRNA degradation, and cytoplasmic deposition (Li et al., 2022). The protein translation initiation factor EIF3C serves as a direct target for YTHDF1. m6A-modified EIF3C mRNA binds to YTHDF1, which then regulates EIF3C mRNA translation in an m6A-dependent manner. This promotes the synthesis of the oncogenic protein EIF3C, thereby facilitating tumor occurrence and metastasis in ovarian cancer (Liu et al., 2020). SUMOylation of YTHDF2 significantly enhances its mRNA binding ability. YTHDF2 selectively binds to m6A-methylated mRNAs to regulate mRNA degradation, such as increasing suppressor of cytokine signaling 2 (SOCS2) mRNA degradation, to promote METTL3-induced carcinogenesis (Chen et al., 2018; Hou et al., 2021). With respect to protein translation regulation, IGF2BP3 interacts with chromosome condensation 2 (RCC2) mRNA in an m6A-dependent manner to regulate its stability. This interaction contributes to the protein expression of RCC2 (Zhang N. et al., 2022) (Figure 1).
IAV mRNA transcripts containing multiple internal m6A modification sites were initially detected, establishing a foundation for further investigation of viral epitranscriptomic regulation (Krug et al., 1976). The impact of m6A modification on virus gene expression and viral replication has been reported, as it regulates the splicing, stability, translation, and secondary structure of viral RNA (Figure 2). Previous research has indicated that the addition of m6A residues to IAV mRNA transcripts is crucial for promoting viral gene expression and replication. The absence of methyltransferase METTL3 significantly reduces the transcription of the NP, NS1, and M2 genes, as well as protein expression in IAV, leading to a decrease in the production of viral particles in a human lung epithelial cell line (A549 cells). Conversely, overexpression of the reading protein YTHDF2 has a noticeable positive effect on IAV gene expression and replication. However, the overexpression of YTHDF1 and YTHDF3 does not significantly affect IAV replication or the production of viral particles (Courtney et al., 2017). The role of YTHDF2 during IAV infection contradicts the promotion of intracellular mRNA degradation. The facilitation of intracellular mRNA degradation by YTHDF2 may diminish the transcription of host antiviral genes. As a result, the overexpression of YTHDF2 increases the expression level of viral mRNA and protein and the release of infectious virus particles, thereby promoting viral replication. By utilizing photoactivatable enhanced ribonucleoside crosslinking and immunoprecipitation (PAR-CLIP) and photo-crosslinking-assisted m6A sequencing (PA-m6A seq) technology (Hafner et al., 2010; Chen et al., 2015), the binding sites of the YTHDF protein were found to congregate into a sense mRNA open reading frame (ORF) and minus strand vRNA fragments encoding the viral structural proteins HA, NP, NA, and M, whereas their presence was limited to mRNA and vRNA fragments encoding the three subunits of RNA-dependent polymerase (PB1, PB2, and PA). These findings indicate the high selectivity of m6A modification in IAV RNA (Courtney et al., 2017; Bayoumi and Munir, 2021). Coupled with the enrichment of multiple highly conserved RRACH motifs in HA mRNA, the expression of HA mRNA and protein significantly decreases in m6A-deficient IAV mutants that target HA-modified sites without substantial effects on other viral mRNAs and proteins. Additionally, the pathogenicity and viral load of IAV mutants decrease. The m6A reader protein YTHDC1 has been reported to regulate IAV mRNA splicing by interacting with the NS1 protein in the nucleus. Increased expression of YTHDC1 was observed in A549 cells during the early and late stages of IAV infection. YTHDC1 directly targets and binds to the 3′ splicing site of NS mRNA, inhibiting the splicing of the NS segment dependent on the NS1 protein. Consequently, the expression of nuclear export protein (NEP) decreases, ultimately promoting the replication and pathogenicity of IAV, both in vitro and in vivo (Zhu Y. et al., 2023). In addition to the m6A modification, other modifications, such as 5-methylcytidine (m5C), N4-acetylcytidine (ac4C), and 2′ O-methylated nucleoside (Nm), can influence various steps in the viral replication cycle. These modifications promote viral replication by increasing the stability and translation of viral mRNA and preventing host RNA-specific innate immune factor melanoma differentiation-associated protein 5 (MDA5) from recognizing and binding to viral RNA (Netzband and Pager, 2020; Tsai and Cullen, 2020) (Figure 2).
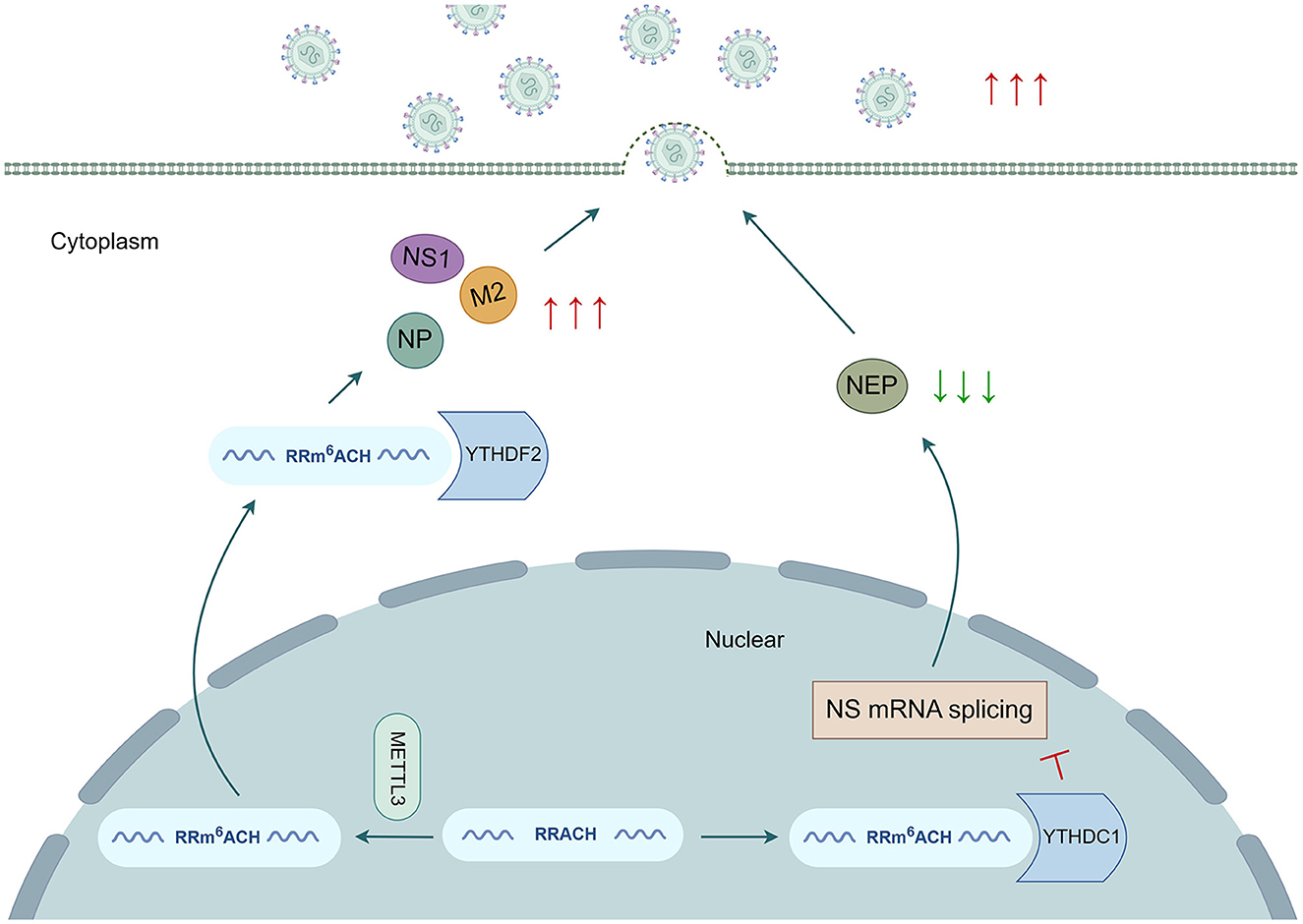
Figure 2. m6A modifications have an impact on the replication cycles of the influenza A virus. m6A modifications are involved in regulating various stages of IAV gene expression and replication. METTL3 and YTHDF2 enhance the expression of the NP, NS1, and M2 genes and the production of viral particles. Moreover, YTHDC1 reduces NEP gene expression by hindering NS mRNA splicing, thereby facilitating IAV replication.
Innate immune signaling functions as the primary defense mechanism of host cells against viral infections. PRRs, such as RIG-I-like receptors (RLRs) and Toll-like receptors (TLRs), play crucial roles in identifying viral PAMPs. Upon recognition, these receptors trigger signaling cascade responses, leading to the production of interferon, inflammatory cytokines, and chemokines. These initial antiviral responses serve to combat viral infection (Iwasaki and Pillai, 2014; Onomoto et al., 2021; Zhou et al., 2021). Recent studies have highlighted the significant regulatory role of m6A modification in cellular mRNA function in response to IAV infection (Liu et al., 2019; Zhao et al., 2020). It has been confirmed that TANK-binding kinase 1 (TBK1) in the type I interferon signaling pathway can enhance the catalytic activity of METTL3 and promote the generation of interferon regulatory factor 3 (IRF3), thereby improving the antiviral immunity of host cells. In Mettl3-deficient mice infected with IAV, the production of IFN-β significantly decreased, inflammatory cells extensively infiltrated the lung tissue, and viral replication increased. These results indicate that METTL3 plays a crucial role in the host defense against viruses (Chen et al., 2022). Interestingly, another study revealed that m6A serves as a negative regulator of type I interferons. The methylation of IFN-β mRNA was mediated by METTL3 and YTHDF2 in THP-1 cells infected with various viruses, such as IAV, leading to decreased stability and increased degradation of IFN-β mRNA and resulting in reduced IFN-α and IFN-β production and downregulation of the host innate immune response. This process benefits viral replication and is conserved across various viral infections. However, following the depletion of METTL3 or YTHDF2 in IAV-infected cells, the expression of IFN-β and interferon-stimulated gene 15 (ISG15) mRNA significantly increased, while the expression of the IAV gene was inhibited (Winkler et al., 2019). METTL14 depletion increased both IFN-β mRNA production and stability in response to viral infection, leading to decreased virus protein expression and reproduction. In contrast, ALKBH5 depletion decreased the production of IFN-β mRNA without causing detectable changes in IFN-β mRNA decay, stimulating viral replication (Rubio et al., 2018). Current research shows that m6A can serve as either an inhibitor or a promoter for the expression of IFN-β mRNA. This conflict may be explained by the fact that the expression of IFN-β mRNA is significantly induced in the early stage of viral infection to activate the innate immune response, and starts to be degraded in the late stage to prevent inflammatory damage. Upon viral infection, METTL3 enhances the nuclear export and translation of antiviral signaling molecule mRNAs, such as IRF3, and promotes the induction of IFN-β and inflammatory cytokines against viral infection. Subsequently, m6A modifications (METTL3 and YTHDF2) promote the instability of IFN-β and ISG15 mRNA and control inflammation to maintain homeostasis. m6A modification has been confirmed to participate in the degradation of IFN-β mRNA in the termination of the innate response (Zhu J. et al., 2023). Furthermore, host cells actively reprogram cellular metabolism to resist viral infection in an interferon-independent manner through m6A modification. During viral infection, the enzymatic activity of ALKBH5 is reduced in host cells, which leads to the downregulation of the α-ketoglutarate dehydrogenase (OGDH)-itaconate pathway associated with viral infection mediated by YTHDF2. This subsequently inhibits viral replication (Liu et al., 2019). Overall, m6A modification plays an emerging role in the cellular defense against invading pathogens via virus–host interactions.
To date, numerous studies have demonstrated the effectiveness of small-molecule inhibitors targeting methylases and demethylases for antiviral therapy. One such inhibitor is 3-denitroadenosine (3-DAA), which inhibits SAH hydrolase. This inhibitor significantly reduces the m6A modification of viral RNA without affecting mRNA capping by depleting intracellular SAM and thus inhibiting the replication of various DNA and RNA viruses (Fustin et al., 2013; Kennedy et al., 2016). Consequently, 3-DAA shows promise as a broad-spectrum antiviral drug (Bray et al., 2000; Yanagi et al., 2022). However, it is important to note that 3-DAA also inhibits the methylation of histones and DNA, highlighting the need for more specific methylase inhibitors to eliminate off-target effects.
Another highly selective demethylase inhibitor is meclofenamic acid (MA), which primarily targets FTO rather than ALKBH5. MA competitively binds to FTO with m6A-methylated RNA in a dose-dependent manner, thus inhibiting the demethylation function of FTO (Huang et al., 2015). Furthermore, the metal protein nanoparticle GSTP1-MT3 (Fe2+) has demonstrated effective antiviral effects by increasing the expression of methyltransferases such as METTL14 and WTAP in virus-infected macrophages. GSTP1-MT3 (Fe2+) regulates m6A modification levels and inhibits the replication of the H1N1 (WSN) virus in various cells, providing potential avenues for the development of antiviral drugs that specifically target m6A modifications (Zhu X.-J. et al., 2021).
Previous studies have identified METTL3 as an efficient therapeutic target for the treatment of various tumors (Xu and Ge, 2022). For example, STM2457, the first bioavailable small-molecule inhibitor of METTL3, has been shown to significantly impede the development and progression of acute myeloid leukemia (AML) by directly binding to the enzymatic activity of METTL3 and inhibiting the catalytic activity of the METTL3/METTL14 MTC (Yankova et al., 2021). Importantly, compared to other RNA, DNA, and protein methyltransferases, STM2457 exhibits high selectivity for METTL3, thus reducing its off-target effects. However, it remains unknown whether small-molecule inhibitors targeting METTL3, such as STM2457, contribute to multiple viral infections and host immune responses.
In the future, the development of more selective and potent small inhibitors or drugs for m6A-related proteins may offer more effective therapeutic strategies to treat IAV infections and combat IAV evasion. Consequently, targeting m6A modifications may be a promising direction for future investigations of anti-IAV strategies.
Dominissini proposed a novel approach called MeRIP-seq, which combines immunoprecipitation using an anti-m6A affinity antibody and high-throughput RNA sequencing of precipitated RNA fragments. Their research showed that m6A modification sites exhibit greater sequence specificity than other regions.
The latest sequencing technology, LACE-seq, utilizes reverse transcription termination of immunopurified protein RNA complexes mediated by RBPs to amplify cDNA ends. This step is followed by high-throughput sequencing and analysis of the products. LACE-seq allows sensitive identification of binding sites and regulatory mechanisms between functional proteins and RNA.
PAR-CLIP is a photoactivatable ribonucleoside-dependent technology used to identify m6A modification sites. This process involves the insertion of a photoactivatable ribonucleoside into RNA transcripts, leading to thymidine-to-cytidine transitions at the UV crosslinking sites of the anti-m6A affinity antibody and m6A-modified RNA. High-throughput sequencing of these transitions enables single-base resolution recognition of m6A modification sites.
PA-m6A-seq follows a procedure similar to that of PAR-CLIP but has the added ability to recognize single methylation sequences in a 23-base region and accurately identify transcriptome-wide m6A methylation sites.
During m6A modification, the core MTC METTL3-METTL14 is responsible for initially recognizing the RNA substrate and transferring methyl groups to the RNA for methylation with the help of SAM. m6A-modified transcripts are then recognized and bound by reading proteins such as YTHDF1 and YTHDC1, which subsequently affect various aspects of the mRNA, including selective splicing processing, nuclear output, stability, translation, and localization. Concurrently, demethylases, such as FTO and ALKBH5, can remove methyl residues from RNA, reversing m6A methylation and dynamically regulating the balance of mRNA methylation. In the context of IAV infection, viral RNA undergoes specific modifications by methyltransferases and demethylases within infected host cells. m6A modification is involved in regulating multiple steps of the viral life cycle, particularly viral RNA transcription, and assembly, thereby playing a significant role in both viral infection and cellular antiviral responses. Notably, the replication of viral RNA has been shown to depend on m6A modification during IAV infection. The presence of METTL3 promotes the m6A modification of viral RNA, increasing the levels of viral mRNA and protein in addition to the release of virions, thereby enhancing viral replication and pathogenicity. Furthermore, m6A modification also regulates the mRNA transcription of various antiviral molecules associated with innate immune signaling pathways in host cells, thereby influencing splicing, stability, and translation in response to viral infection. By predominantly targeting the type I interferon signaling pathway, m6A modification has been identified as a regulator of the host immune response. The potential mechanism underlying this regulation is that m6A modification can promote mRNA nuclear export, stability, and translation of antiviral immune molecules (cGAS, IFI16, and STING), activate the downstream factor IRF3, induce the expression of IFN-β, and thus enhance the host antiviral immune response during the early stage of viral infection. However, at later stages, the m6A modification of IFN-β mRNA acts as a negative regulatory factor, promoting the degradation of IFN-β mRNA and alleviating IFN-β expression and inflammatory damage. As a result, the host's antiviral innate immune response is weakened. Additionally, the m6A modification of IAV RNA plays a crucial role in inhibiting the recognition and binding of the innate immune signal RIG-I to viral RNA, thus participating in viral immune evasion.
Current research indicates that m6A modification plays a crucial role in regulating the replication of IAV and the innate immune response to antiviral activity. However, there are numerous unresolved questions and controversies surrounding the precise regulation and facilitation of m6A modification proteins in cells infected by IAV. Therefore, it is crucial to further investigate the role of m6A modification in anti-IAV innate immunity and to explore the implications of various mapping techniques. Multiple studies have demonstrated that inhibitors targeting methyltransferases and demethylases can effectively disrupt IAV RNA replication by targeting m6A modifications. This discovery may provide innovative insights for the development of therapeutic interventions for IAV based on m6A modification.
m6A modification is the predominant form of epigenetic transcription modification in eukaryotes, and substantial evidence supports its significant role in regulating IAV infection. This review explores the mechanisms of three key proteins, methyltransferases, demethylases, and reading proteins, and the regulatory impact of m6A modification on IAV infection. These findings have the potential to identify valuable targets for anti-IAV strategies and serve as a foundation for implementing preventive or therapeutic measures.
XL: Conceptualization, Investigation, Writing – original draft. WC: Funding acquisition, Supervision, Writing – review & editing. KL: Supervision, Writing – review & editing. JS: Supervision, Writing – review & editing, Conceptualization, Funding acquisition.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This study was supported by the Natural Science Foundation of Guangdong Province (2022A1515012144 and 2023A1515012055) and the Shantou University Medical College Scientific Research Initiation Grant (510858047).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Ann, J., Papenburg, J., Bouhy, X., Rhéaume, C., Hamelin, M.-È., Boivin, G., et al. (2012). Molecular and antigenic evolution of human influenza A/H3N2 viruses in Quebec, Canada, 2009-2011. J. Clini. Virol. 53, 88–92. doi: 10.1016/j.jcv.2011.09.016
Bayoumi, M., and Munir, M. (2020). Structural insights into m6A-erasers: a step toward understanding molecule specificity and potential antiviral targeting. Front. Cell Dev. Biol. 8:587108. doi: 10.3389/fcell.2020.587108
Bayoumi, M., and Munir, M. (2021). Evolutionary conservation of the DRACH signatures of potential N6-methyladenosine (m6A) sites among influenza A viruses. Sci. Rep. 11:4548. doi: 10.1038/s41598-021-84007-0
Boulias, K., and Greer, E. L. (2023). Biological roles of adenine methylation in RNA. Nat. Rev. Genet. 24, 143–160. doi: 10.1038/s41576-022-00534-0
Bray, M., Driscoll, J., and Huggins, J. W. (2000). Treatment of lethal Ebola virus infection in mice with a single dose of an S-adenosyl-L-homocysteine hydrolase inhibitor. Antiviral Res. 45, 135–147. doi: 10.1016/S0166-3542(00)00066-8
Brennan, K., and Bowie, A. G. (2010). Activation of host pattern recognition receptors by viruses. Curr. Opin. Microbiol. 13, 503–507. doi: 10.1016/j.mib.2010.05.007
Carrique, L., Fan, H., Walker, A. P., Keown, J. R., Sharps, J., Staller, E., et al. (2020). Host ANP32A mediates the assembly of the influenza virus replicase. Nature 587, 638–643. doi: 10.1038/s41586-020-2927-z
Chen, J., Wei, X., Wang, X., Liu, T., Zhao, Y., Chen, L., et al. (2022). TBK1-METTL3 axis facilitates antiviral immunity. Cell Rep. 38:110373. doi: 10.1016/j.celrep.2022.110373
Chen, K., Lu, Z., Wang, X., Fu, Y., Luo, G.-Z., Liu, N., et al. (2015). High-resolution N(6)-methyladenosine (m(6) A) map using photo-crosslinking-assisted m(6) A sequencing. Angewandte Chemie 54, 1587–1590. doi: 10.1002/anie.201410647
Chen, M., Wei, L., Law, C.-T., Tsang, F. H.-C., Shen, J., Cheng, C. L.-H., et al. (2018). RNA N6-methyladenosine methyltransferase-like 3 promotes liver cancer progression through YTHDF2-dependent posttranscriptional silencing of SOCS2. Hepatology 67, 2254–2270. doi: 10.1002/hep.29683
Cho, J.-H., Zhao, B., Shi, J., Savage, N., Shen, Q., Byrnes, J., et al. (2020). Molecular recognition of a host protein by NS1 of pandemic and seasonal influenza A viruses. Proc. Natl. Acad. Sci. USA. 117, 6550–6558. doi: 10.1073/pnas.1920582117
Choe, J., Lin, S., Zhang, W., Liu, Q., Wang, L., Ramirez-Moya, J., et al. (2018). mRNA circularization by METTL3-eIF3h enhances translation and promotes oncogenesis. Nature 561, 556–560. doi: 10.1038/s41586-018-0538-8
Chou, Y.-Y., Vafabakhsh, R., Doganay, S., Gao, Q., Ha, T., Palese, P., et al. (2012). One influenza virus particle packages eight unique viral RNAs as shown by FISH analysis. Proc. Natl. Acad. Sci. USA. 109, 9101–9106. doi: 10.1073/pnas.1206069109
Courtney, D. G., Kennedy, E. M., Dumm, R. E., Bogerd, H. P., Tsai, K., Heaton, N. S., et al. (2017). Epitranscriptomic enhancement of influenza A virus gene expression and replication. Cell Host Microbe 22, 377–386.e5. doi: 10.1016/j.chom.2017.08.004
Dominissini, D., Moshitch-Moshkovitz, S., Schwartz, S., Salmon-Divon, M., Ungar, L., Osenberg, S., et al. (2012). Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature 485, 201–206. doi: 10.1038/nature11112
Downey, J., Pernet, E., Coulombe, F., Allard, B., Meunier, I., Jaworska, J., et al. (2017). RIPK3 interacts with MAVS to regulate type I IFN-mediated immunity to influenza A virus infection. PLoS Pathog. 13:e1006326. doi: 10.1371/journal.ppat.1006326
Fan, H., Walker, A. P., Carrique, L., Keown, J. R., Serna Martin, I., Karia, D., et al. (2019). Structures of influenza A virus RNA polymerase offer insight into viral genome replication. Nature 573, 287–290. doi: 10.1038/s41586-019-1530-7
Flamand, M. N., Tegowski, M., and Meyer, K. D. (2023). The proteins of mRNA modification: writers, readers, and erasers. Annu. Rev. Biochem. 92, 145–173. doi: 10.1146/annurev-biochem-052521-035330
Fu, Y., Dominissini, D., Rechavi, G., and He, C. (2014). Gene expression regulation mediated through reversible m6A RNA methylation. Nat. Rev. Genetics 15, 293–306. doi: 10.1038/nrg3724
Fustin, J.-M., Doi, M., Yamaguchi, Y., Hida, H., Nishimura, S., Yoshida, M., et al. (2013). RNA-methylation-dependent RNA processing controls the speed of the circadian clock. Cell 155, 793–806. doi: 10.1016/j.cell.2013.10.026
Gokhale, N. S., Mcintyre, A. B. R., Mcfadden, M. J., Roder, A. E., Kennedy, E. M., Gandara, J. A, et al. (2016) N6-methyladenosine in flaviviridae viral RNA genomes regulates infection. Cell Host Microbe 20, 654–665. doi: 10.1016/j.chom.2016.09.015.
Gubareva, L. V., Kaiser, L., and Hayden, F. G. (2000). Influenza virus neuraminidase inhibitors. Lancet 355, 827–835. doi: 10.1016/S0140-6736(99)11433-8
Hafner, M., Landthaler, M., Burger, L., Khorshid, M., Hausser, J., Berninger, P., et al. (2010). Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell 141, 129–141. doi: 10.1016/j.cell.2010.03.009
Herold, S., Becker, C., Ridge, K. M., and Budinger, G. R. S. (2015). Influenza virus-induced lung injury: pathogenesis and implications for treatment. Eur. Respir. J. 45, 1463–1478. doi: 10.1183/09031936.00186214
Hou, G., Zhao, X., Li, L., Yang, Q., Liu, X., Huang, C., et al. (2021). SUMOylation of YTHDF2 promotes mRNA degradation and cancer progression by increasing its binding affinity with m6A-modified mRNAs. Nucleic Acids Res. 49, 2859–2877. doi: 10.1093/nar/gkab065
Huang, W., Chen, T.-Q., Fang, K., Zeng, Z.-C., Ye, H., Chen, Y.-Q., et al. (2021). N6-methyladenosine methyltransferases: functions, regulation, and clinical potential. J. Hematol. Oncol. 14:117. doi: 10.1186/s13045-021-01129-8
Huang, X.-T., Li, J.-H., Zhu, X.-X., Huang, C.-S., Gao, Z.-X., Xu, Q.-C., et al. (2021). HNRNPC impedes m6A-dependent anti-metastatic alternative splicing events in pancreatic ductal adenocarcinoma. Cancer Lett. 518, 196–206. doi: 10.1016/j.canlet.2021.07.016
Huang, Y., Yan, J., Li, Q., Li, J., Gong, S., Zhou, H., et al. (2015). Meclofenamic acid selectively inhibits FTO demethylation of m6A over ALKBH5. Nucleic Acids Res. 43, 373–384. doi: 10.1093/nar/gku1276
Imam, H., Kim, G.-W., and Siddiqui, A. (2020). Epitranscriptomic(N6-methyladenosine) modification of viral RNA and virus-host interactions. Front. Cell. Infect. Microbiol. 10:584283. doi: 10.3389/fcimb.2020.584283
Iwasaki, A., and Pillai, P. S. (2014). Innate immunity to influenza virus infection. Nat. Rev. Immunol. 14, 315–328. doi: 10.1038/nri3665
Jia, G., Fu, Y., and He, C. (2013). Reversible RNA adenosine methylation in biological regulation. Trends Genet. 29, 108–115. doi: 10.1016/j.tig.2012.11.003
Jia, G., Fu, Y., Zhao, X., Dai, Q., Zheng, G., Yang, Y., et al. (2011). N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat. Chem. Biol. 7, 885–887. doi: 10.1038/nchembio.687
Kennedy, E. M., Bogerd, H. P., Kornepati, A. V. R., Kang, D., Ghoshal, D., Marshall, J. B., et al. (2016). Posttranscriptional m(6)A editing of HIV-1 mRNAs enhances viral gene expression. Cell Host Microbe 19, 675–685. doi: 10.1016/j.chom.2016.04.002
Krug, R. M., Morgan, M. A., and Shatkin, A. J. (1976). Influenza viral mRNA contains internal N6-methyladenosine and 5'-terminal 7-methylguanosine in cap structures. J. Virol. 20, 45–53. doi: 10.1128/jvi.20.1.45-53.1976
Li, J., Chen, K., Dong, X., Xu, Y., Sun, Q., Wang, H., et al. (2022). YTHDF1 promotes mRNA degradation via YTHDF1-AGO2 interaction and phase separation. Cell Prolif. 55:e13157. doi: 10.1111/cpr.13157
Lin, X., Wang, F., Chen, J., Liu, J., Lin, Y.-B., Li, L., et al. (2022). N6-methyladenosine modification of CENPK mRNA by ZC3H13 promotes cervical cancer stemness and chemoresistance. Military Med. Res. 9:19. doi: 10.1186/s40779-022-00378-z
Liu, J., Yue, Y., Han, D., Wang, X., Fu, Y., Zhang, L., et al. (2014). A METTL3-METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nat. Chem. Biol. 10, 93–95. doi: 10.1038/nchembio.1432
Liu, Q., Zhou, Y.-H., and Yang, Z.-Q. (2016). The cytokine storm of severe influenza and development of immunomodulatory therapy. Cell. Mol. Immunol. 13, 3–10. doi: 10.1038/cmi.2015.74
Liu, T., Wei, Q., Jin, J., Luo, Q., Liu, Y., Yang, Y., et al. (2020). The m6A reader YTHDF1 promotes ovarian cancer progression via augmenting EIF3C translation. Nucleic Acids Res. 48, 3816–3831. doi: 10.1093/nar/gkaa048
Liu, Y., You, Y., Lu, Z., Yang, J., Li, P., Liu, L., et al. (2019). N 6-methyladenosine RNA modification-mediated cellular metabolism rewiring inhibits viral replication. Science 365, 1171–1176. doi: 10.1126/science.aax4468
Luo, Q., Mo, J., Chen, H., Hu, Z., Wang, B., Wu, J., et al. (2022). Structural insights into molecular mechanism for N6-adenosine methylation by MT-A70 family methyltransferase METTL4. Nat. Commun. 13:5636. doi: 10.1038/s41467-022-33277-x
Lv, Z., Sun, L., Xu, Q., Xing, C., and Yuan, Y. (2020). Joint analysis of lncRNA mA methylome and lncRNA/mRNA expression profiles in gastric cancer. Cancer Cell Int. 20:464. doi: 10.1186/s12935-020-01554-8
McGeoch, D., Fellner, P., and Newton, C. (1976). Influenza virus genome consists of eight distinct RNA species. Proc. Natl. Acad. Sci. USA. 73, 3045–3049. doi: 10.1073/pnas.73.9.3045
Meyer, K. D., and Jaffrey, S. R. (2014). The dynamic epitranscriptome: N6-methyladenosine and gene expression control. Nat. Rev. Mol. Cell Biol. 15, 313–326. doi: 10.1038/nrm3785
Meyer, K. D., Saletore, Y., Zumbo, P., Elemento, O., Mason, C. E., Jaffrey, S. R., et al. (2012). Comprehensive analysis of mRNA methylation reveals enrichment in 3' UTRs and near stop codons. Cell 149, 1635–1646. doi: 10.1016/j.cell.2012.05.003
Mora, R., Rodriguez-Boulan, E., Palese, P., and García-Sastre, A. (2002). Apical budding of a recombinant influenza A virus expressing a hemagglutinin protein with a basolateral localization signal. J. Virol. 76, 3544–3553. doi: 10.1128/JVI.76.7.3544-3553.2002
Motorin, Y., and Helm, M. (2022). RNA nucleotide methylation: 2021 update. Wiley Interdiscip. Rev. RNA 13:e1691. doi: 10.1002/wrna.1691
Müller, S., Glaß, M., Singh, A. K., Haase, J., Bley, N., Fuchs, T, et al. (2019) IGF2BP1 promotes SRF-dependent transcription in cancer in a m6A- miRNA-dependent manner. Nucleic Acids Res. 47, 375–390. doi: 10.1093/nar/gky1012.
Murakami, S., and Jaffrey, S. R. (2022). Hidden codes in mRNA: control of gene expression by m6A. Mol. Cell 82, 2236–2251. doi: 10.1016/j.molcel.2022.05.029
Narayan, P., Ayers, D. F., Rottman, F. M., Maroney, P. A., and Nilsen, T. W. (1987). Unequal distribution of N6-methyladenosine in influenza virus mRNAs. Mol. Cell. Biol. 7, 1572–1575. doi: 10.1128/MCB.7.4.1572
Netzband, R., and Pager, C. T. (2020). Epitranscriptomic marks: emerging modulators of RNA virus gene expression. Wiley Interdiscip. Rev. RNA 11:e1576. doi: 10.1002/wrna.1576
Onomoto, K., Onoguchi, K., and Yoneyama, M. (2021). Regulation of RIG-I-like receptor-mediated signaling: interaction between host and viral factors. Cell. Mol. Immunol. 18, 539–555. doi: 10.1038/s41423-020-00602-7
Peacock, T. P., Sheppard, C. M., Staller, E., and Barclay, W. S. (2019). Host determinants of influenza RNA synthesis. Annu. Rev. Virol. 6, 215–233. doi: 10.1146/annurev-virology-092917-043339
Ping, X.-L., Sun, B.-F., Wang, L., Xiao, W., Yang, X., and Wang, W.-J. (2014) Mammalian WTAP is a regulatory subunit of the RNA N6-methyladenosine methyltransferase. Cell Res. 24, 177–189. doi: 10.1038/cr.2014.3.
Qiao, Y., Sun, Q., Chen, X., He, L., Wang, D., Su, R., et al. (2023). Nuclear m6A reader YTHDC1 promotes muscle stem cell activation/proliferation by regulating mRNA splicing and nuclear export. Elife 12:e82703. doi: 10.7554/eLife.82703.sa2
Roundtree, I. A., Evans, M. E., Pan, T., and He, C. (2017a). Dynamic RNA modifications in gene expression regulation. Cell 169, 1187–1200. doi: 10.1016/j.cell.2017.05.045
Roundtree, I. A., Luo, G.-Z., Zhang, Z., Wang, X., Zhou, T., Cui, Y., et al. (2017b). YTHDC1 mediates nuclear export of N6-methyladenosine methylated mRNAs. Elife 6:e31311. doi: 10.7554/eLife.31311
Rubio, R. M., Depledge, D. P., Bianco, C., Thompson, L., and Mohr, I. (2018). RNA m6 A modification enzymes shape innate responses to DNA by regulating interferon β. Genes Dev. 32, 1472–1484. doi: 10.1101/gad.319475.118
Saletore, Y., Meyer, K., Korlach, J., Vilfan, I. D., Jaffrey, S., Mason, C. E., et al. (2012). The birth of the epitranscriptome: deciphering the function of RNA modifications. Genome Biol. 13:175. doi: 10.1186/gb-2012-13-10-175
Satterwhite, E. R., and Mansfield, K. D. (2022). RNA methyltransferase METTL16: targets and function. Wiley Interdiscip. Rev. RNA 13:e1681. doi: 10.1002/wrna.1681
Sendinc, E., and Shi, Y. (2023). RNA m6A methylation across the transcriptome. Mol. Cell 83, 428–441. doi: 10.1016/j.molcel.2023.01.006
Shi, H., Wei, J., and He, C. (2019). Where, when, and how: context-dependent functions of RNA methylation writers, readers, and erasers. Mol. Cell 74, 640–650. doi: 10.1016/j.molcel.2019.04.025
Skehel, J. J., and Wiley, D. C. (2000). Receptor binding and membrane fusion in virus entry: the influenza hemagglutinin. Annu. Rev. Biochem. 69, 531–569. doi: 10.1146/annurev.biochem.69.1.531
Su, R., Fan, L.-H., Cao, C., Wang, L., Du, Z., Cai, Z., et al. (2021). Global profiling of RNA-binding protein target sites by LACE-seq. Nat. Cell Biol. 23, 664–675. doi: 10.1038/s41556-021-00696-9
Sun, H.-L., Zhu, A. C., Gao, Y., Terajima, H., Fei, Q., Liu, S., et al. (2020). Stabilization of ERK-phosphorylated METTL3 by USP5 increases m6A methylation. Mol. Cell 80, 633–647. doi: 10.1016/j.molcel.2020.10.026
Taubenberger, J. K., and Kash, J. C. (2010). Influenza virus evolution, host adaptation, and pandemic formation. Cell Host Microbe 7, 440–451. doi: 10.1016/j.chom.2010.05.009
Te Velthuis, A. J. W., and Fodor, E. (2016). Influenza virus RNA polymerase: insights into the mechanisms of viral RNA synthesis. Nat. Rev. Microbiol. 14, 479–493. doi: 10.1038/nrmicro.2016.87
Tsai, K., and Cullen, B. R. (2020). Epigenetic and epitranscriptomic regulation of viral replication. Nat. Rev. Microbiol. 18, 559–570. doi: 10.1038/s41579-020-0382-3
Wang, P., Doxtader, K. A., and Nam, Y. (2016). Structural basis for cooperative function of Mettl3 and Mettl14 methyltransferases. Mol. Cell 63, 306–317. doi: 10.1016/j.molcel.2016.05.041
Wang, T., Kong, S., Tao, M., and Ju, S. (2020). The potential role of RNA N6-methyladenosine in cancer progression. Mol. Cancer 19:88. doi: 10.1186/s12943-020-01204-7
Wang, X., Feng, J., Xue, Y., Guan, Z., Zhang, D., Liu, Z., et al. (2016). Structural basis of N(6)-adenosine methylation by the METTL3-METTL14 complex. Nature 534, 575–578. doi: 10.1038/nature18298
Wang, X., Tian, L., Li, Y., Wang, J., Yan, B., Yang, L., et al. (2021). RBM15 facilitates laryngeal squamous cell carcinoma progression by regulating TMBIM6 stability through IGF2BP3 dependent. J. Exp. Clin. Cancer Res. 40:80. doi: 10.1186/s13046-021-01871-4
Wang, Y., Wang, Y., Patel, H., Chen, J., Wang, J., Chen, Z.-S., et al. (2023). Epigenetic modification of m6A regulator proteins in cancer. Mol. Cancer 22:102. doi: 10.1186/s12943-023-01810-1
Winkler, R., Gillis, E., Lasman, L., Safra, M., Geula, S., Soyris, C., et al. (2019). mA modification controls the innate immune response to infection by targeting type I interferons. Nat. Immunol. 20, 173–182. doi: 10.1038/s41590-018-0275-z
World Health Organization (2023). Influenza (Seasonal). Available online at: https://www.who.int/news-room/fact-sheets/detail/influenza-(seasonal)# (accessed October 3, 2023).
Worobey, M., Han, G.-Z., and Rambaut, A. (2014). Genesis and pathogenesis of the 1918 pandemic H1N1 influenza A virus. Proc. Natl. Acad. Sci. USA. 111, 8107–8112. doi: 10.1073/pnas.1324197111
Wu, B., Li, L., Huang, Y., Ma, J., and Min, J. (2017). Readers, writers and erasers of N6-methylated adenosine modification. Curr. Opin. Struct. Biol. 47, 67–76. doi: 10.1016/j.sbi.2017.05.011
Xu, P., and Ge, R. (2022). Roles and drug development of METTL3 (methyltransferase-like 3) in anti-tumor therapy. Eur. J. Med. Chem. 230:114118. doi: 10.1016/j.ejmech.2022.114118
Yanagi, Y., Watanabe, T., Hara, Y., Sato, Y., Kimura, H., Murata, T., et al. (2022). EBV exploits RNA m6A modification to promote cell survival and progeny virus production during lytic cycle. Front. Microbiol. 13:870816. doi: 10.3389/fmicb.2022.870816
Yang, J., Liu, S., Du, L., and Jiang, S. (2016). A new role of neuraminidase (NA) in the influenza virus life cycle: implication for developing NA inhibitors with novel mechanism of action. Rev. Med. Virol. 26, 242–250. doi: 10.1002/rmv.1879
Yankova, E., Blackaby, W., Albertella, M., Rak, J., De Braekeleer, E., and Tsagkogeorga, G. (2021) Small-molecule inhibition of METTL3 as a strategy against myeloid leukaemia. Nature 593, 597–601. doi: 10.1038/s41586-021-03536-w.
Yoo, J.-K., Kim, T. S., Hufford, M. M., and Braciale, T. J. (2013). Viral infection of the lung: host response and sequelae. J. Allergy Clin. Immunol. 132, 1263–1276. doi: 10.1016/j.jaci.2013.06.006
Yu, P.-L., Cao, S.-J., Wu, R., Zhao, Q., and Yan, Q.-G. (2021). Regulatory effect of m A modification on different viruses. J. Med. Virol. 93:6100–6115. doi: 10.1002/jmv.27246
Zaccara, S., Ries, R. J., and Jaffrey, S. R. (2019). Reading, writing and erasing mRNA methylation. Nat. Rev. Mol. Cell Biol. 20, 608–624. doi: 10.1038/s41580-019-0168-5
Zhang, C., Fu, J., and Zhou, Y. (2019). A review in research progress concerning m6A methylation and immunoregulation. Front. Immunol. 10:922. doi: 10.3389/fimmu.2019.00922
Zhang, H., Alford, T., Liu, S., Zhou, D., and Wang, J. (2022). Influenza virus causes lung immunopathology through down-regulating PPARγ activity in macrophages. Front. Immunol. 13:958801. doi: 10.3389/fimmu.2022.958801
Zhang, N., Shen, Y., Li, H., Chen, Y., Zhang, P., Lou, S., et al. (2022). The m6A reader IGF2BP3 promotes acute myeloid leukemia progression by enhancing RCC2 stability. Exp. Mol. Med. 54, 194–205. doi: 10.1038/s12276-022-00735-x
Zhao, Y., Shi, Y., Shen, H., and Xie, W. (2020). m6A-binding proteins: the emerging crucial performers in epigenetics. J. Hematol. Oncol. 13:35. doi: 10.1186/s13045-020-00872-8
Zheng, G., Dahl, J. A., Niu, Y., Fedorcsak, P., Huang, C.-M., and Li, C. J. (2013) ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism mouse fertility. Mol. Cell 49, 18–29. doi: 10.1016/j.molcel.2012.10.015.
Zhou, R., Liu, L., and Wang, Y. (2021). Viral proteins recognized by different TLRs. J. Med. Virol. 93, 6116–6123. doi: 10.1002/jmv.27265
Zhu, J., Liu, S., Fang, J., Cui, Z., Wang, B., Wang, Y., et al. (2023). Enzymolysis-based RNA pull-down identifies YTHDC2 as an inhibitor of antiviral innate response. Cell Rep. 42:113192. doi: 10.1016/j.celrep.2023.113192
Zhu, W., Wang, J.-Z., Wei, J.-F., and Lu, C. (2021). Role of m6A methyltransferase component VIRMA in multiple human cancers (Review). Cancer Cell Int. 21:172. doi: 10.1186/s12935-021-01868-1
Zhu, X.-J., Feng, J.-Q., Zheng, M.-Z., Yang, Z.-R., Zhao, L., Zhang, W., et al. (2021). Metal-protein nanoparticles facilitate anti-VSV and H1N1 viruses through the coordinative actions on innate immune responses and METTL14. Macromol. Biosci. 21:e2000382. doi: 10.1002/mabi.202000382
Keywords: influenza A virus, RNA modification, m6A methylation, immune escape, anti-IAV strategy
Citation: Liu X, Chen W, Li K and Sheng J (2024) RNA N6-methyladenosine methylation in influenza A virus infection. Front. Microbiol. 15:1401997. doi: 10.3389/fmicb.2024.1401997
Received: 16 March 2024; Accepted: 30 May 2024;
Published: 18 June 2024.
Edited by:
Rahul K. Suryawanshi, Gladstone Institutes, United StatesReviewed by:
Mohsan Ullah Goraya, Huaqiao University, ChinaCopyright © 2024 Liu, Chen, Li and Sheng. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jiangtao Sheng, anRzaGVuZ0BzdHUuZWR1LmNu; Kangsheng Li, a3NsaTIwMTNAeWVhaC5uZXQ=
†ORCID: Kangsheng Li orcid.org/0000-0002-6981-9263
Jiangtao Sheng orcid.org/0000-0003-2438-7345
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.