
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 14 February 2024
Sec. Antimicrobials, Resistance and Chemotherapy
Volume 15 - 2024 | https://doi.org/10.3389/fmicb.2024.1361550
Aspergillus fungi are renowned for producing a diverse range of natural products with promising biological activities. These include lovastatin, itaconic acid, terrin, and geodin, known for their cholesterol-regulating, anti-inflammatory, antitumor, and antibiotic properties. In our current study, we isolated three dimeric nitrophenyl trans-epoxyamides (1–3), along with fifteen known compounds (4–18), from the culture of Aspergillus terreus MCCC M28183, a deep-sea-derived fungus. The structures of compounds 1–3 were elucidated using a combination of NMR, MS, NMR calculation, and ECD calculation. Compound 1 exhibited moderate inhibitory activity against human gastric cancer cells MKN28, while compound 7 showed similar activity against MGC803 cells, with both showing IC50 values below 10 μM. Furthermore, compound 16 exhibited moderate potency against Vibrio parahaemolyticus ATCC 17802, with a minimum inhibitory concentration (MIC) value of 7.8 μg/mL. This promising research suggests potential avenues for developing new pharmaceuticals, particularly in targeting specific cancer cell lines and combating bacterial infections, leveraging the unique properties of these Aspergillus-derived compounds.
Aspergillus terreus, a prolific filamentous fungus, synthesizes an array of secondary metabolites with distinct biological functions. Lovastatin, a prominent metabolite, exhibits a unique hexahydronaphthalene ring and a 2-methylbutyric acid moiety, efficaciously inhibiting HMG-CoA reductase for cholesterol regulation (Huang et al., 2021). Itaconic acid, another metabolite, serves as an industrial precursor and exhibits potential anti-inflammatory properties (Diankristanti and Ng, 2023). Terrein, with its 4H-furan-3-one framework, demonstrates multifaceted biological activities, including antitumor and anti-inflammatory effects, and inhibits melanin synthesis, suggesting therapeutic applications in hyper-pigmentation (Zhao et al., 2016). The dihydroxyphenylalanine (DOPA) pigments, geodin and epi-geodin, are noted for their antibiotic activity and metal ion chelation capabilities (Boruta et al., 2021). This spectrum of metabolites, each with unique structural and pharmacological properties, underscores the importance of Aspergillus terreus in pharmaceutical and industrial applications. Ongoing research in this domain is crucial for advancing natural product chemistry and drug discovery, underscoring the significant role of fungi in producing bioactive compounds.
In exploring marine-derived bioactive compounds, a cytotoxic dihydrobenzofuran was extracted from the deep-sea fungus Aspergillus terreus CC-S06-18 (Wang et al., 2020). Following this, extensive large-scale fermentation enabled the identification and characterization of eighteen secondary metabolites. This included three novel dimeric nitrophenyl trans-epoxyamides (1–3) and an additional fifteen compounds (4–18) (Figure 1). Subsequently, each of these compounds underwent evaluation to determine their cytotoxic and antibacterial activities.
The Aspergillus terreus strain, initially isolated from a seawater sample taken at a depth of 5,250 meters in the Pacific Ocean and originally labeled as CC-S06-18, has had its ITS sequence submitted to GenBank, under the sequence number MN463005. Additionally, this strain is currently preserved in the Marine Culture Collection of China (MCCC), with the preservation number M28183.
The fermentation process was conducted statically using two hundred 1-liter Erlenmeyer flasks, each filled with 30 grams of millet, 70 grams of rice, and 100 milliliters of seawater. Following the inoculation, the flasks were left to incubate at room temperature for 28 days. Subsequently, the product of fermentation was subjected to three rounds of extraction using ethyl acetate. The final step involved filtering and concentrating the extract, resulting in a total yield of 85 grams.
The purification process of the crude extract was systematically executed using silica gel column chromatography with a petroleum ether and ethyl acetate mixture as the eluent, progressing through gradients of 9.5:0.5 to 8.0:2.0 (v/v). This approach resulted in six fractions (Frs1-6). Subsequent purification of Fr3 via Sephadex LH-20 chromatography in MeOH yielded five subfractions (Fr3.1-Fr3.5). Specifically, Fr3.1 was further refined using the same chromatographic technique to isolate 8 (136.8 mg). Meanwhile, Fr3.4 and Fr3.5 underwent RP-HPLC with a 20–80% MeCN in H2O gradient over 20.0 min at a 15 mL/min flow rate. This procedure led to the isolation of 15 (1.0 mg, tR = 13.3 min) and 14 (2.2 mg, tR = 18.5 min) from Fr3.4, and 17 (2.3 mg, tR = 16.0 min) from Fr3.5. Fr4 was similarly fractionated using Sephadex LH-20 in MeOH, resulting in four subfractions (Fr4.1-Fr4.4). The RP-HPLC purification of Fr4.2, with a nuanced protocol involving 20–80% MeCN, followed by 80–100% MeCN, and finally 100% MeCN, yielded 9 (1.3 mg, tR = 24.6 min). The subfractions Fr4.3 and Fr4.4 were also purified by RP-HPLC, leading to the isolation of 18 (1.1 mg, tR = 16.4 min) and 6 (1.4 mg, tR = 5.5 min), respectively. Further, Fr5 was divided into three subfractions (Fr5.1-Fr5.3) via Sephadex LH-20 in MeOH. The RP-HPLC purification of Fr5.2, with a modified gradient of 30–80% MeCN in H2O, followed by 80–100% MeCN, and finally 100% MeCN, resulted in the isolation of 13 (4.3 mg, tR = 18.4 min), 1 (2.6 mg, tR = 16.1 min), 4 (1.1 mg, tR = 24.5 min), and 2 (1.0 mg, tR = 27.1 min). Fr5.3 was processed similarly to yield 10 (1.3 mg, tR = 9.0 min) and 16 (1.2 mg, tR = 15.7 min). Finally, Fr6 was subdivided into eight subfractions (Fr6.1-Fr6.8) using Sephadex LH-20 in MeOH, with Fr6.1, Fr6.2, and Fr6.3 undergoing RP-HPLC purification (30–60% MeCN in H2O, then 60–100% MeCN) to yield 11 (1.6 mg, tR = 13.5 min), 7 (2.1 mg, tR = 9.9 min), and 5 (0.8 mg, tR = 14.0 min) respectively. Fr6.8 was further purified by RP-HPLC with a similar gradient, culminating in 100% MeCN, to isolate 3 (1.2 mg, tR = 25.0 min) and 12 (1.6 mg, tR = 31.2 min).
A white amorphous powder; [α]25 D − 14.2 (c 0.1, MeOH); UV (MeOH) λmax (log ε): 270 (4.00) nm; ECD (c 0.1 mg/mL, MeOH) λmax (∆ε): 218 (−7.93), 263 (1.71) nm; HRESIMS m/z 607.2017 [M + Na]+ (calcd for C28H32N4O10Na, 607.2016); 1H and 13C NMR data, Table 1.
A white amorphous powder; [α]25 D + 11.3 (c 0.1, MeOH); UV (MeOH) λmax (log ε): 270 (3.89) nm; ECD (c 0.1 mg/mL, MeOH) λmax (∆ε): 217 (−7.06), 246 (2.10) nm; HRESIMS m/z 555.1711 [M − H]− (calcd for C26H27N4O10, 555.1727); 1H and 13C NMR data, Table 1.
A white amorphous powder; [α]25 D − 62.0 (c 0.1, MeOH); UV (MeOH) λmax (log ε): 269 (4.06) nm; ECD (c 0.1 mg/mL, MeOH) λmax (∆ε): 209 (−10.28), 255 (1.68) nm; HRESIMS m/z 593.1859 [M + Na]+ (calcd for C27H30N4O10Na, 593.1860); 1H and 13C NMR data, Table 1.
Initially, MGC803 and MKN28 cell lines were dissociated and counted. Following this, 3 × 103 cells from each line were seeded into each well of 96-well plates and incubated overnight to promote cell attachment. During the incubation period, the edge wells of the plate were filled with PBS. Post-attachment, the cells were exposed to increasing concentrations of the test compounds for a growth period of 72 h. Subsequently, 10 μL of CCK8 reagent was added to each well, followed by an incubation at 37°C of 1 h. The absorbance was measured at 450 nm using a microplate reader (Tecan, Morrisville, NC, United States) according to the manufacturer’s instructions. The CCK8 experimental setup was conducted in triplicate. Data were analyzed with GraphPad Prism software to determine IC50 of each compound (Wang et al., 2018, 2019; Hong et al., 2022; Lv et al., 2022).
Antimicrobial activity testing against Salmonella enteritidis (CICC 21482), Vibrio vulnificus (MCCC 1A08743), Staphylococcus aureus (CICC 10384), Vibrio parahaemolyticus (ATCC 17802), Escherichia coli (CICC 10302), and Vibrio parahaemolyticus (vp-HL) (Yang et al., 2022) was conducted using a microdilution method in 96-well plates with resazurin as a growth marker. Resazurin sodium salt was dissolved in sterile water to give a 2.0 mg/mL solution. A tested bacterial strain, during its mid-logarithmic phase and with an initial inoculum of 1 × 105 CFU/mL, was introduced into plates containing serial dilutions of the test compound and a 10% resazurin solution. These plates were covered with foil and incubated at 37°C with shaking for 24 h. Following incubation, a visual inspection was conducted to detect any color change from blue to pink, indicating bacterial growth. The Minimum Inhibitory Concentration (MIC) was determined as the lowest concentration at which no color change occurred. All experiments were conducted in triplicate, and the average MIC values were calculated for the test compound (Wang et al., 2018, 2019; Hong et al., 2022; Lv et al., 2022).
Conformers were initially generated using CREST software (Grimme, 2019; Pracht et al., 2020) and subsequently underwent refinement via DFT optimization at the B3LYP/6-31G(d) level in Gaussian 16 (Frisch et al., 2016). This process targeted conformers within a 10 kcal/mol energy window. Frequency analysis was employed to verify the local minimum status of each conformer. Electronic energies were then recalculated at the more advanced M062X/6–311 + G(2d,p) level. The conformer populations were assessed using Boltzmann distribution, focusing on those that accounted for more than 2% of the total for in-depth analysis.
The GIAO method was used for the calculation of NMR shielding constants at mPW1PW91-SCRF/6–31 + G(d,p) level with tetramethylsilane (TMS) as a reference (Willoughby et al., 2014). For each candidate, linear regression parameters a and b (δcal = aδexp + b) were calculated, along with the correlation coefficient (R2), Mean Absolute Error (MAE, Σn |δcal − δexp|/n), and Corrected Mean Absolute Error (CMAE, Σn |δcorr − δexp|/n), where δcorr = (δcal − b)/a.
ECD calculations were done via TDDFT at the Cam-B3LYP/6-311G(d) level, calculating 36 excited states per conformer (Pescitelli and Bruhn, 2016). Multiwfn 3.6 software was used to generate ECD curves (Lu and Chen, 2012).
Compound 1, a white powder, was analyzed using HRESIMS and demonstrated a sodium adduct ion peak with m/z of 607.2017 [M + Na]+. This peak suggested the molecular formula of C28H32N4O10, indicative of 15 degrees of unsaturation. Examination of the 13C NMR and DEPT spectra revealed 14 carbon atoms: comprising two methyl groups (δC 19.2, C-13 and δC 15.3, C-14), a single methoxyl group (δC 54.9, C-15), three methines linked to oxygen or nitrogen (δC 86.4, C-1; δC 47.6, C-2; δC 62.7, C-6), an oxygen-bearing quaternary carbon (δC 65.0, C-5), four aromatic methines (δC 124.3, C-9/11 and δC 128.8, C-8/12), two quaternary aromatic carbons (δC 142.7, C-7 and δC 148.9, C-10), and a carbonyl carbon atom (δC 172.3, C-4) (Table 1). The 1H NMR data supplemented these observations, showing signals for three methyl groups (δH 3.43, OCH3-1; δH 1.31, H-13; δH 1.39, H-14), three methines related to oxygen or nitrogen (δH 4.46, H-6; δH 5.63, H-1; δH 4.66, H-2), along with four aromatic methines (δH 7.68, d, H-8/12; δH 8.33, d, J = 8.6 Hz, H-9/11) (Table 1). Considering the NMR data and its molecular formula, compound 1 was theorized to be a symmetrically structured dimer.
The planar configuration of compound 1 was established through a comprehensive analysis using both 1D and 2D NMR spectroscopy. The existence of a p-nitrophenyl group was initially inferred from HMBC spectral correlations, including signals from H-9/H-11 to C-7 (δC 142.7) and C-10 (δC 148.9), and from H-8/H-12 to C-10. These findings were further corroborated by COSY spectral correlations, specifically between H-9 and H-8, and H-11 and H-12 (Figure 2). Further, the structure was identified to contain a 2-methyl-2,3-epoxyamide group, evidenced by HMBC correlations linking H-6 to C-4 and C-5, and H3-14 to C-4, C-5, and C-6, with the chemical shifts of C-5 (δC 65.0) and C-6 (δC 62.7) confirming the presence of an epoxy group (Figure 2). The isopropyl segment in the structure was pinpointed through COSY correlations from the methyl group H3-13 to H-1 and H-2, and its attachment to N-3 was established by HMBC correlations from H-2 to C-4 (Figure 2). As compound 1 was presumed to be a dimer, the molecular formula of its monomer was deduced to be C14H16N2O5. This monomer comprised a 2-methyl-2,3-epoxyamide group, an isopropyl group, and a 1,4-disubstituted phenyl, along with a methoxy group attached to the C-1 position. Considering the remaining atomic composition of one nitrogen and two oxygens in the monomer, it was concluded that a nitro group was attached at the C-10 position. All these data led to the confirmation of subunit A in the structure, as shown in Figure 2. Additionally, the HMBC correlations between H-1 and C-4′, coupled with the chemical shifts of C-1/1′ (δC 86.4) (Figure 2), suggested that two subunit A units are linked via two C-N bonds, forming a dimer with a central symmetrical framework, as represented in Figure 2.
Key NOESY correlations, specifically from H-1,1′ to H3-13/13′, as well as between H-2,2′ and H3-15,15′, demonstrated that H-1/1′ and H3-13/13′ resided on the same piperazine ring, suggesting an α-orientation. Further NOESY correlations between H-8,8′ and H3-14,14′ suggested that H-6,6′ and H3-14,14′ occupy different faces of the oxirane ring (Figure 2). This led to the deduction of two possible relative configurations of (1S*,1’S*,2S*,2’S*,5S*,5’S*,6R*,6’R*)-1 (1a) and (1S*,1’S*,2S*,2’S*,5R*,5’R*,6S*,6’S*)-1 (1b). A conformational search was conducted for these configurations, followed by geometry optimization. The lowest energy conformers identified were subjected to NMR calculations at the mPW1PW91-SCRF/6–31 + G(d,p) level using GIAO method. 1a had a DP4+ possibility of 100%, with its calculated 13C NMR data closely aligned with the experimental results, exhibiting R2, MAE, and CMAE values of 0.9990, 1.8, and 1.2, respectively (Figure 3A). The absolute configuration was definitely identified as 1S,1’S,2S,2’S,5S,5’S,6R,6’R through comparative analysis of the experimental ECD spectrum and the CAM-B3LYP/6-311G(d) calculated spectra for both stereoisomers in MeOH. This identification was affirmed as the calculated ECD for (1S,1’S,2S,2’S,5S,5’S,6R,6’R) closely matched the experimental spectrum (Figure 3B).
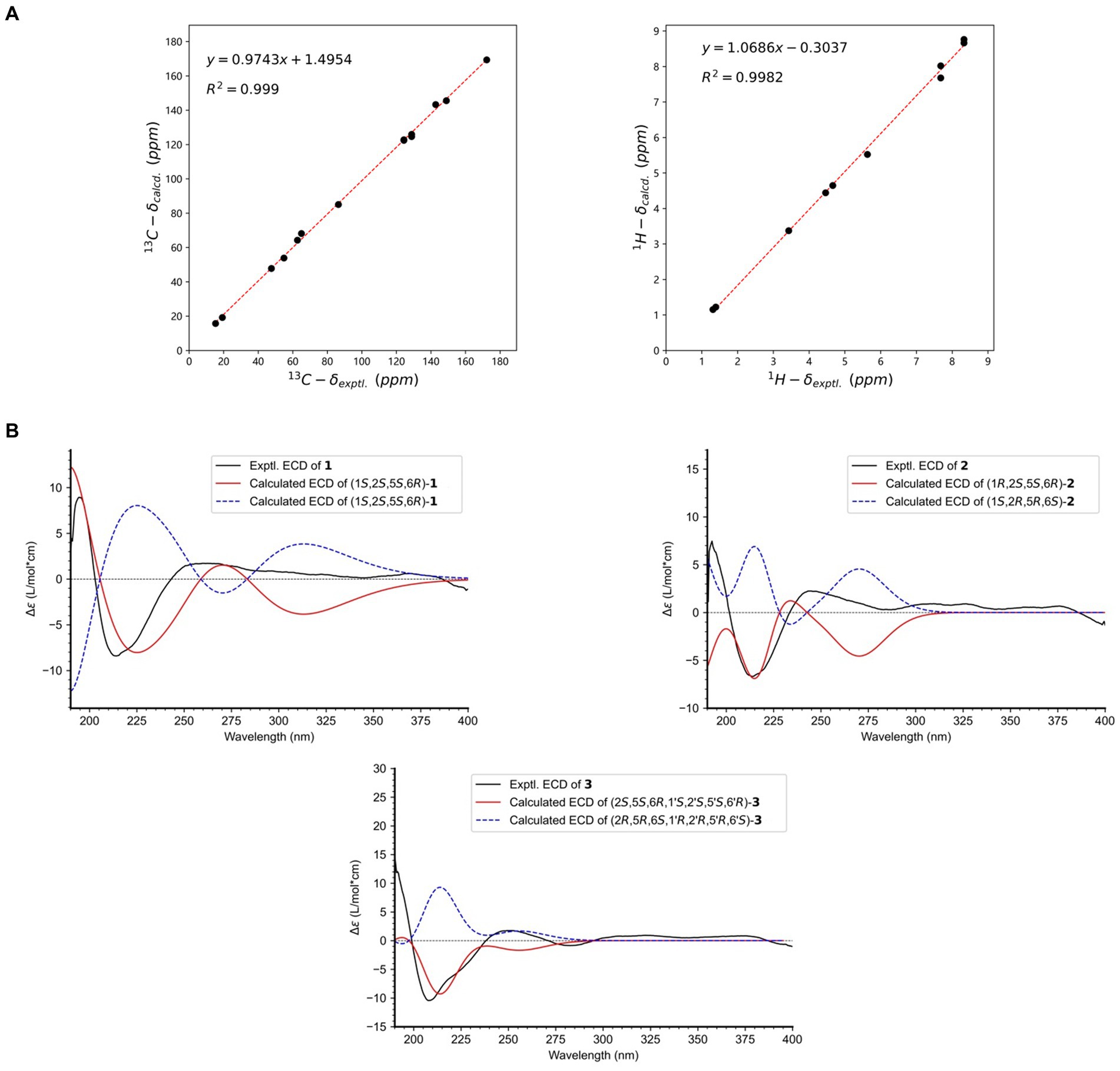
Figure 3. (A) Linear regression analysis between the experimental and calculated NMR data of 1; (B) Experimental and calculated ECD spectra of 1–3.
Compound 2 has been established to possess the molecular formula C26H28N4O10, which was derived from HRESIMS data exhibiting a deprotonated ion peak at a m/z of 555.1711 [M − H]−. This indicated a reduction of 28 atomic mass units (amu) compared to compound 1, along with the presence of 15 degrees of unsaturation. A comparative analysis of NMR data between compounds 1 and 2 revealed that the primary variations are concentrated in the piperazine ring. Notably, the chemical shifts at C-1 and C-2 altered from δC 86.4 to δC 91.3, and from δC 47.6 to δC 58.9, respectively. The observation of 13 unique carbon resonances in the 13C NMR spectrum, in conjunction with its molecular weight, indicates that compound 2 shared the same symmetrical framework as compound 1, as detailed in Table 1. In the NOESY spectrum, correlations between H-1,1′ and H-2,2′ in compound 2 suggested that H3-13/13′ and H-1/1′ were positioned on opposite sides of the piperazine ring. Furthermore, NOESY correlations between H-8,8′ and H3-14,14′ implied that H-6,6′ and H3-14,14′ were situated on different faces of the oxirane ring, paralleling the pattern observed in compound 1, as depicted in Figure 2. Considering the biosynthetic pathway, compound 2 is likely the 1,1′-epimer derivative of compound 1, characterized by a hydroxyl group attached to both C-1′ and C-1. Finally, the absolute configuration of compound 2 is designated as 1R,1’R,2S,2’S,5S,5’S,6R,6’R by ECD calculation at CAM-B3LYP/6-311G(d) (Figure 3B).
Compound 3, isolated as a white powder, had a molecular formula of C27H30N4O10. This was established from the HRESIMS data, which showed a sodium adduct ion peak at m/z 593.1859 [M + Na]+, indicating a reduction of 14 amu from 1 and 15 degrees of unsaturation. A comparison of the 1H NMR and 13C NMR data revealed similarities between 1 and 3. The most significant difference was the substitution of the nitrogenated methine group (δC 86.4, C-1) in 1 with a formyl group (δC 198.3) in 3, as detailed in Table 1. This alteration was corroborated by COSY correlations of H3-13/H-2/H-1 and HMBC correlations of H-1/C-13, H-2/C-1′, and H-2/C-4, as illustrated in Figure 2. The coupling constant 3JH1’ − H2 of 9.2 Hz indicated that H-1′ and H-2′ were positioned on opposite faces of the octatomic ring, which was formed by a hydrogen bond between the formyl and the amide group. In the NOESY spectrum, correlations between H3-13/H-1′, H3-13’/H-1′, and H-2/H-2′ indicated that H3-13, H-1′, and H3-13′ are located on the same side of the octatomic ring. Similarly, the positioning of H-6/6′ and H3-14/14′ on opposite faces of the oxirane ring was evidenced by NOESY correlations between H-8/8′ and H3-14/14′. Considering the biosynthetic pathway, the absolute configurations of C-2 and C-2′ were supposed to be S. Compound 3 was assigned the absolute configuration as 2S,5S,6R,1’S,2’S,5’S,6’R, which was corroborated by ECD calculations at the CAM-B3LYP/6-311G(d) level, as illustrated in Figure 3B.
The biosynthetic pathway, as proposed in Figure 4, began with L-Phenylalanine undergoing a sequence of transformations: deamination, oxidation, methylation, and aryl nitration, leading to the formation of intermediate a. Acylation of intermediate a with L-Alanine resulted in intermediate b. Two units of b underwent a condensation reaction by removing two molecules of water to give intermediate c, which was reduced to yield 2. Intermediate c underwent reduction and methylation to yield 1. Similarly, two units of b condensed by eliminating one molecule of water, followed by reduction and methylation steps, resulting in 3 (Figure 4).
A comparison of NMR data with previously reported data in the literature led to the identification of the remaining fifteen known compounds, numbered 4–18. They were questin (4) (Liu et al., 2015), ω-hydroxyemodin (5) (Khamthong et al., 2012), penicillixanthone (6) (Rukachaisirikul et al., 2014), isorhodoptilometrin (7) (Kim et al., 2022), emodin (8) (Khaliq et al., 2023), citrinin H1 (9) (Ngan et al., 2017), 3,5-dichloroasterric acid (10) (Lee et al., 2002), dihydrogeodin (11) (Hamed et al., 2019), dimethoxyphtalide (12) (Wei et al., 2020), anodendroic acid (13) (Prompanya et al., 2016), varioloid B (14) (Zhang et al., 2015), italicum (15) (Tomás-Barberán et al., 1990), 46-dichloro-5-methylbenzene-13-diol (16) (Slavov et al., 2010), 4-hydroxybenzaldehyde (17) (Hsu et al., 2009), and 2-phenylethanol (18) (Pehlivan et al., 2011).
We performed a CCK8 assay to investigate the effects of all compounds on the viability of two gastric cancer cell lines, MKN28 and MGC803. 1 exhibited a moderate inhibitory effect on the growth of MKN28 cells, with an IC50 value of 7.4 μM. Similarly, 7 displayed a moderate inhibitory effect on MGC803 cells, with an IC50 value of 2.5 μM. The remaining compounds did not achieve a 50% inhibition rate within the tested concentration range, indicating that their IC50 values exceeded 10 μM. Paclitaxel served as the positive control, with IC50 values of 0.32 and 0.7 nM for MKN28 and MGC803, respectively.
The in vitro antibacterial activities of compounds 1–18 were tested against six bacterial strains. Among them, Compound 16 showed potency against Vibrio parahaemolyticus ATCC 17802, exhibiting a MIC of 7.8 μg/mL. This potency was compared to the positive control, chloramphenicol, which had an MIC of 1.1 μg/mL.
This study details the characterization of three new compounds (1–3) and fifteen known compounds (4–18) from the deep-sea fungus Aspergillus terreus. Compound 1 was identified as a symmetrical dimer with a complex structure, including a p-nitrophenyl moiety, a 2-methyl-2,3-epoxyamide group, and an isopropyl fragment. Its absolute configuration was determined using DFT methods and ECD spectrum analysis. 2, structurally similar to 1 but differing in the piperazine ring, is considered an epimer of 1. Compound 3 shares a similar structure with 1, but with a formyl group replacing a nitrogenated methine group. The biogenetic pathways of these compounds are proposed, involving precursors like L-phenylalanine and L-alanine. The study also reports the biological activities of these compounds. Specifically, compounds 1 and 7 exhibit moderate growth inhibitory effects on gastric cancer cell lines, while compound 16 shows moderate antibacterial properties. The study emphasizes the need for extensive research on the biological activities and potential therapeutic applications of these compounds, particularly those showing promise in preliminary tests. Developing synthetic methods for these compounds could facilitate further studies and potential pharmaceutical applications. Investigating the mechanisms behind their observed biological activities could lead to the discovery of new drug targets or therapeutic strategies.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.
Ethical approval was not required for the studies on humans in accordance with the local legislation and institutional requirements because only commercially available established cell lines were used. Ethical approval was not required for the studies on animals in accordance with the local legislation and institutional requirements because only commercially available established cell lines were used.
XH: Conceptualization, Formal analysis, Funding acquisition, Writing – original draft, Writing – review & editing, Supervision. YW: Formal analysis, Writing – review & editing, Methodology. GL: Writing – review & editing, Resources. ZS: Writing – review & editing, Project administration, Investigation. JX: Data curation, Methodology, Supervision, Validation, Writing – review & editing. J-JQ: Formal analysis, Writing – review & editing. WW: Conceptualization, Formal analysis, Funding acquisition, Project administration, Writing – original draft, Writing – review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This research was funded by the National Key Research and Development Program of China (2022YFC2804100), Xiamen Southern Oceanographic Center Project (22GYY007HJ07), Marine Organism Application Technology Collaborative Innovation Center Project (XTZX-HYSW-1801), Natural Science Foundation of Fujian Province (2021J01509), Research Projects for High-level Talents of Xiamen Ocean Vocational College (140008), and Open Funding Project of Key Laboratory of Marine Biogenetic Resources, Third Institute of Oceanography, Ministry of Natural Resources (HY202303, HY202307).
The authors also thank the assistant from the High-Field Nuclear Magnetic Resonance Research Center of Xiamen University for providing kind help during the experiments.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2024.1361550/full#supplementary-material
Boruta, T., Gornicka, A., Grzybowska, I., Stefaniak, I., and Bizukojc, M. (2021). Exploring the extremes: applying high concentration of yeast extract leads to drastic morphological changes and elimination of (+)-geodin and asterric acid production in aspergillus terreus submerged cultures. Biotechnol. Lett. 43, 61–71. doi: 10.1007/s10529-020-03018-5
Diankristanti, P. A., and Ng, I. S. (2023). Microbial itaconic acid bioproduction towards sustainable development: insights, challenges, and prospects. Bioresour. Technol. 384:129280. doi: 10.1016/j.biortech.2023.129280
Frisch, M. J., Trucks, G. W., Schlegel, H. B., Scuseria, G. E., Robb, M. A., Cheeseman, J. R., et al. (2016). "Gaussian 16 rev. C.01 ". (Wallingford, CT).
Grimme, S. (2019). Exploration of chemical compound, conformer, and reaction space with meta-dynamics simulations based on tight-binding quantum chemical calculations. J. Chem. Theory Comput. 15, 2847–2862. doi: 10.1021/acs.jctc.9b00143
Hamed, A., Ismail, M., El-Metwally, M. M., Frese, M., Stammler, H. G., Sewald, N., et al. (2019). X-ray, structural assignment and molecular docking study of dihydrogeodin from aspergillus terreus TM8. Nat. Prod. Res. 33, 117–121. doi: 10.1080/14786419.2018.1431642
Hong, X., Guan, X., Lai, Q., Yu, D., Chen, Z., Fu, X., et al. (2022). Characterization of a bioactive meroterpenoid isolated from the marine-derived fungus Talaromyces sp. Appl. Microbiol. Biotechnol. 106, 2927–2935. doi: 10.1007/s00253-022-11914-1
Hsu, P. J., Miller, J. S., and Berger, J. M. (2009). Bakuchiol, an antibacterial component of Psoralidium tenuiflorum. Nat. Prod. Res. 23, 781–788. doi: 10.1080/14786410902840158
Huang, X., Men, P., Tang, S., and Lu, X. (2021). Aspergillus terreus as an industrial filamentous fungus for pharmaceutical biotechnology. Curr. Opin. Biotechnol. 69, 273–280. doi: 10.1016/j.copbio.2021.02.004
Khaliq, T., Waseem, M. A., Mir, S. A., Sultan, P., Malik, F. A., and Hassan, Q. P. (2023). Isolation and characterisation of pharmaceutically versatile molecules from Rumex dentatus and evaluation of their cytotoxic activity against human cancer cell lines. Nat. Prod. Res. 37, 857–862. doi: 10.1080/14786419.2022.2092864
Khamthong, N., Rukachaisirikul, V., Tadpetch, K., Kaewpet, M., Phongpaichit, S., Preedanon, S., et al. (2012). Tetrahydroanthraquinone and xanthone derivatives from the marine-derived fungus Trichoderma aureoviride PSU-F95. Arch. Pharm. Res. 35, 461–468. doi: 10.1007/s12272-012-0309-2
Kim, D. C., Quang, T. H., Tien, N. T., Kim, K. W., Kim, Y. C., Ngan, N. T. T., et al. (2022). Anti-neuroinflammatory effect of oxaline, isorhodoptilometrin, and 5-hydroxy-7-(2′-hydroxypropyl)-2-methyl-chromone obtained from the marine fungal strain penicillium oxalicum CLC-MF05. Arch. Pharm. Res. 45, 90–104. doi: 10.1007/s12272-022-01370-w
Lee, H. J., Lee, J. H., Hwang, B. Y., Kim, H. S., and Lee, J. J. (2002). Fungal metabolites, asterric acid derivatives inhibit vascular endothelial growth factor (VEGF)-induced tube formation of HUVECs. J. Antibiot. (Tokyo) 55, 552–556. doi: 10.7164/antibiotics.55.552
Liu, D., Yan, L., Ma, L., Huang, Y., Pan, X., Liu, W., et al. (2015). Diphenyl derivatives from coastal saline soil fungus aspergillus iizukae. Arch. Pharm. Res. 38, 1038–1043. doi: 10.1007/s12272-014-0371-z
Lu, T., and Chen, F. (2012). Multiwfn: a multifunctional wavefunction analyzer. J. Comput. Chem. 33, 580–592. doi: 10.1002/jcc.22885
Lv, D., Xia, J., Guan, X., Lai, Q., Zhang, B., Lin, J., et al. (2022). Indole diketopiperazine alkaloids isolated from the marine-derived fungus aspergillus chevalieri MCCC M23426. Front. Microbiol. 13:950857. doi: 10.3389/fmicb.2022.950857
Ngan, N. T., Quang, T. H., Kim, K. W., Kim, H. J., Sohn, J. H., Kang, D. G., et al. (2017). Anti-inflammatory effects of secondary metabolites isolated from the marine-derived fungal strain penicillium sp. SF-5629. Arch. Pharm. Res. 40, 328–337. doi: 10.1007/s12272-017-0890-5
Pehlivan, L., Métay, E., Laval, S., Dayoub, W., Delbrayelle, D., Mignani, G., et al. (2011). Reduction of aromatic and aliphatic esters using the reducing systems MoO2(acac)2 or V(O)(OiPr)3 in combination with 1,1,3,3-tetramethyldisiloxane. Eur. J. Org. Chem. 2011, 7400–7406. doi: 10.1002/ejoc.201101016
Pescitelli, G., and Bruhn, T. (2016). Good computational practice in the assignment of absolute configurations by TDDFT calculations of ECD spectra. Chirality 28, 466–474. doi: 10.1002/chir.22600
Pracht, P., Bohle, F., and Grimme, S. (2020). Automated exploration of the low-energy chemical space with fast quantum chemical methods. Phys. Chem. Chem. Phys. 22, 7169–7192. doi: 10.1039/c9cp06869d
Prompanya, C., Dethoup, T., Gales, L., Lee, M., Pereira, J. A., Silva, A. M., et al. (2016). New polyketides and new benzoic acid derivatives from the marine sponge-associated fungus Neosartorya quadricincta KUFA 0081. Mar. Drugs 14:134. doi: 10.3390/md14070134
Rukachaisirikul, V., Satpradit, S., Klaiklay, S., Phongpaichit, S., Borwornwiriyapan, K., and Sakayaroj, J. (2014). Polyketide anthraquinone, diphenyl ether, and xanthone derivatives from the soil fungus penicillium sp. PSU-RSPG99. Tetrahedron 70, 5148–5152. doi: 10.1016/j.tet.2014.05.105
Slavov, N., Cvengros, J., Neudorfl, J. M., and Schmalz, H. G. (2010). Total synthesis of the marine antibiotic pestalone and its surprisingly facile conversion into pestalalactone and pestalachloride a. Angew. Chem. Int. Ed. 49, 7588–7591. doi: 10.1002/anie.201003755
Tomás-Barberán, F., Iniesta-Sanmartín, E., Tomás-Lorente, F., and Rumbero, A. (1990). Antimicrobial phenolic compounds from three Spanish Helichrysum species. Phytochemistry 29, 1093–1095. doi: 10.1016/0031-9422(90)85410-H
Wang, W., Liao, Y., Chen, R., Hou, Y., Ke, W., Zhang, B., et al. (2018). Chlorinated azaphilone pigments with antimicrobial and cytotoxic activities isolated from the deep sea derived fungus Chaetomium sp. NA-S01-R1. Mar. Drugs 16:61. doi: 10.3390/md16020061
Wang, W., Liao, Y., Zhang, B., Gao, M., Ke, W., Li, F., et al. (2019). Citrinin monomer and dimer derivatives with antibacterial and cytotoxic activities isolated from the deep sea-derived fungus penicillium citrinum NLG-S01-P1. Mar. Drugs 17:46. doi: 10.3390/md17010046
Wang, W., Yang, J., Liao, Y. Y., Cheng, G., Chen, J., Mo, S., et al. (2020). Aspeterreurone a, a cytotoxic dihydrobenzofuran-phenyl acrylate hybrid from the deep-sea-derived fungus aspergillus terreus CC-S06-18. J. Nat. Prod. 83, 1998–2003. doi: 10.1021/acs.jnatprod.0c00189
Wei, C., Deng, Q., Sun, M., and Xu, J. (2020). Cytospyrone and cytospomarin: two new polyketides isolated from mangrove endophytic fungus, Cytospora sp. Molecules 25:4224. doi: 10.3390/molecules25184224
Willoughby, P. H., Jansma, M. J., and Hoye, T. R. (2014). A guide to small-molecule structure assignment through computation of ((1)H and (1)(3)C) NMR chemical shifts. Nat. Protoc. 9, 643–660. doi: 10.1038/nprot.2014.042
Yang, F., Xu, L., Huang, W., and Li, F. (2022). Highly lethal Vibrio parahaemolyticus strains cause acute mortality in Penaeus vannamei post-larvae. Aquaculture 548:737605. doi: 10.1016/j.aquaculture.2021.737605
Zhang, P., Li, X.-M., Wang, J.-N., and Wang, B.-G. (2015). Oxepine-containing diketopiperazine alkaloids from the algal-derived endophytic fungus Paecilomyces variotii EN-291. Helv. Chim. Acta 98, 800–804. doi: 10.1002/hlca.201400328
Keywords: Aspergillus , secondary metabolites, cytotoxic, antibacterial, deep-sea
Citation: Huang X, Wang Y, Li G, Shao Z, Xia J, Qin J-J and Wang W (2024) Secondary metabolites from the deep-sea derived fungus Aspergillus terreus MCCC M28183. Front. Microbiol. 15:1361550. doi: 10.3389/fmicb.2024.1361550
Received: 26 December 2023; Accepted: 02 February 2024;
Published: 14 February 2024.
Edited by:
Dewu Zhang, Chinese Academy of Medical Sciences, ChinaReviewed by:
Mallique Qader, University of Illinois at Chicago, United StatesCopyright © 2024 Huang, Wang, Li, Shao, Xia, Qin and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jinmei Xia, eGlhamlubWVpQHRpby5vcmcuY24=; Jiang-Jiang Qin, anFpbkB1Y2FzLmFjLmNu; Weiyi Wang, d3l3YW5nQHRpby5vcmcuY24=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.