- 1Department of Microbiology, Faculty of Biological and Veterinary Sciences, Nicolaus Copernicus University, Toruń, Poland
- 2Center for Modern Interdisciplinary Technologies, Nicolaus Copernicus University, Toruń, Poland
- 3Department of Cellular and Molecular Biology, Faculty of Biological and Veterinary Sciences, Nicolaus Copernicus University, Toruń, Poland
Contamination of vegetables with human pathogenic microorganisms (HPMOs) is considered one of the most important problems in the food industry, as current nutritional guidelines include increased consumption of raw or minimally processed organic vegetables due to healthy lifestyle promotion. Vegetables are known to be potential vehicles for HPMOs and sources of disease outbreaks. In this study, we tested the susceptibility of radish (Raphanus sativus) to colonization by different HPMOs, including Escherichia coli PCM 2561, Salmonella enterica subsp. enterica PCM 2565, Listeria monocytogenes PCM 2191 and Bacillus cereus PCM 1948. We hypothesized that host plant roots containing bactericidal compounds are less prone to HPMO colonization than shoots and leaves. We also determined the effect of selected pathogens on radish growth to check host plant–microbe interactions. We found that one-week-old radish is susceptible to colonization by selected HPMOs, as the presence of the tested HPMOs was demonstrated in all organs of R. sativus. The differences were noticed 2 weeks after inoculation because B. cereus was most abundant in roots (log10 CFU – 2.54), S. enterica was observed exclusively in stems (log10 CFU – 3.15), and L. monocytogenes and E. coli were most abundant in leaves (log10 CFU – 4.80 and 3.23, respectively). The results suggest that E. coli and L. monocytogenes show a higher ability to colonize and move across the plant than B. cereus and S. enterica. Based on fluorescence in situ hybridization (FISH) and confocal laser scanning microscopy (CLSM) approach HPMOs were detected in extracellular matrix and in some individual cells of all analyzed organs. The presence of pathogens adversely affected the growth parameters of one-week-old R. sativus, especially leaf and stem fresh weight (decreased by 47–66 and 17–57%, respectively). In two-week-old plants, no reduction in plant biomass development was noted. This observation may result from plant adaptation to biotic stress caused by the presence of HPMOs, but confirmation of this assumption is needed. Among the investigated HPMOs, L. monocytogenes turned out to be the pathogen that most intensively colonized the aboveground part of R. sativus and at the same time negatively affected the largest number of radish growth parameters.
Introduction
Vegetables are vital sources of carbohydrates, vitamins (e.g., B, C, K), minerals (e.g., calcium, potassium, and magnesium) and fiber. Consumption of raw plants (400 g per day) ensures health, prevents diseases (e.g., heart disease, cancer, diabetes and obesity) and is the basis of a balanced diet (Alemu et al., 2018; Balali et al., 2020). Current changes in nutritional guidelines resulting in increased demand for fresh or minimally processed vegetables have been linked to a higher incidence of foodborne illnesses in the past three decades (Mogren et al., 2018). Contamination of vegetables by human pathogenic microorganisms (HPMOs) can occur at the preharvest stage (e.g., soil amendment with improperly composted manure, soil irrigation with sewage or contaminated surface water, insects, dust or feces of wild animals) or during postharvest handling (e.g., washing, slicing, soaking, packing and food preparation) (Luna-Guevara et al., 2019; Jacob and Melotto, 2020). Plants can be colonized by HPMOs through natural openings, e.g., stomata, lenticels, hydathodes, sites of lateral root formation but also at sites of physical and chemical damage (Chahar et al., 2021). Raw food-borne HPMOs include Escherichia coli O157-H7 (Shiga toxin-producing bacteria), Salmonella spp., Shigella spp., Listeria monocytogenes, Clostridium botulinum, Campylobacter spp., B. cereus, Aeromonas hydrophila, Vibrio cholerae, and Yersinia enterocolitica.
The level of plant colonization by HPMOs can depend on many different aspects, e.g., differences in the metabolic traits of various plant species and genotypes, the stage of plant development and fruit maturity, the resistance of bacteria to antagonistic chemical compounds present in plant tissues and environmental conditions (Luna-Guevara et al., 2019; Jacob and Melotto, 2020). Notably, E. coli, S. enterica and L. monocytogenes are known to spread through contaminated water and soil directly to vegetables (Andino and Hanning, 2015; Carstens et al., 2019). HPMOs expelled from the host gut to the environment have different abilities to survive and multiply. E. coli, L. monocytogenes and S. enterica can persist in soil for different time periods, ranging from 3 days to 7–25 weeks or even a year, respectively (Winfield and Groisman, 2003; Miceli and Settanni, 2019). The colonization of plants by HPMOs is a complex process in which an important role is played by the ability of bacteria to attach to the plant surface, aggregate and/or form biofilms (Gorski et al., 2003; Jacob and Melotto, 2020). E. coli and Salmonella sp. can produce molecules important for the process of attachment and biofilm matrix formation in response to contact with plant leaves (Yaron and Römling, 2014). L. monocytogenes and B. cereus, due to their motility associated with flagellar presence and high potential to form biofilms, can efficiently colonize plant surfaces and tissues (Gorski et al., 2009; Majed et al., 2016; Lin et al., 2022). The mechanisms activated during plant invasion by specific HPMOs are still unclear; however, it was revealed that motile bacteria can more often colonize extracellular (apoplastic) spaces than the interior of plant cells (Holden et al., 2009). HPMOs show different specificities in the colonization of plant organs. Several works have confirmed the spread of pathogens in whole plants, while others have shown that pathogens behave like organ-specific microorganisms, e.g., colonizing only plant roots, irrespective of inoculation techniques and plant growth conditions (Cooley et al., 2003; Jablasone et al., 2005; Burris et al., 2020).
Based on genomic analysis six different subgroups of E. coli exist. They are able to colonize various ecological niches and can exhibit commensalistic or pathogenic lifestyle (van Elsas et al., 2011). Pathogenic E. coli can be responsible for numerous human infections associated with the consumption of contaminated vegetables. This microbe causes diarrhoeal disease of different severities, but cases of death are also noted (Mogren et al., 2018; Carstens et al., 2019). E. coli O157:H7 (infectious dose 50–100 cfu/g or mL) is one of the most common foodborne pathogens and contributes to several outbreaks (Sun et al., 2019; Puligundla and Lim, 2022), e.g., after consumption of romaine lettuce (58 persons/9 states) (Gupta and Madramootoo, 2017), baby spinach (26 states in the USA) (FDA, 2007), fresh washed spinach (2006: 199 cases/102 hospitalizations/3 deaths), ready-to-eat salads (2013: 33 cases/7 hospitalizations) and alfalfa sprouts (2016, 11cases/2 hospitalizations) and lettuce (Laven, 2006; Luna-Guevara et al., 2019).
According to the World Health Organization (WHO) report, the nontyphoidal S. enterica serotype is a common cause of foodborne diseases (ca. 79,000,000 cases in 2010) (Havelaar et al., 2015). Outbreaks of S. enterica have been related to different vegetables, such as romaine lettuce (2017, U.S. – 151 cases), onion (U.S., 2022–1,040 cases), raw almonds (2004, USA – 29 cases), black pepper (2010, USA – 272 cases), pistachios (2010, USA – 11 cases), iceberg lettuce (2008, Finland – 103/2 cases/deaths), baby spinach (2007, Sweden – 102 cases), tomatoes (2011, Denmark – 43 cases, Germany, Italy, Austria and Belgium – 28 cases), cucumber (2014, USA – 275/1 cases/death) and cantaloupe (2012, U.S. – 261/3 cases/deaths) (Denny et al., 2007; Lienemann et al., 2011; CDC, 2012; Müller et al., 2016; Clark, 2017; Dyda et al., 2020; Li et al., 2020; CDC, 2022).
Listeria monocytogenes has been found in various plant species, e.g., in sprouts of alfa-alfa, broccoli, cabbage, cantaloupe and radish. Microbe occurrence was noted both in high-income country, e.g., USA and Norway, and in developing countries, including India and Malaysia (Gorski et al., 2004, 2008, 2009; Ponniah et al., 2010; Walsh et al., 2014; Kljujev et al., 2018). The risk of getting ill from the consumption of contaminated vegetables is high because of the small infectious dose (ranging from 107 to 109 CFU in healthy hosts and 105 to 107 CFU in case of individuals at high risk of infection) (Farber et al., 1996). Several outbreaks were associated with the consumption of L. monocytogenes-contaminated: cantaloupe (2011, U.S. – 147/33 cases/deaths), sprouts (2014, U.S. – 5/2 cases/deaths), salad mix (2015–2016, U.S. – 19/1 cases/deaths) (CDC, 2012; Laksanalamai et al., 2012; CDC, 2016; Self et al., 2016; Gu et al., 2021).
Bacillus cereus includes highly versatile bacterial strains, inhabiting different environments including soil, plants, gut of healthy individuals and others (Ceuppens et al., 2013; Kulkova et al., 2022). They can posses various metabolic properties that predispose them to a pathogenic or commensal lifestyle. It has been noticed that specific strains of B. cereus can act as plant growth promoting bacteria having a beneficial influence on their hosts (Ceuppens et al., 2013; Kulkova et al., 2022). B. cereus comprise well-known food-borne pathogen; however, due to the sparse symptoms associated with infection, only a few reports exist (Dierick et al., 2005). The occurrence of B. cereus in fresh products reaches up to 37.5%, making this bacterium the most frequent foodborne pathogen (Kim et al., 2016). Analysis of different vegetables, including lettuce (Qu et al., 2021), onion, parsley, basil, coriander (Gdoura-Ben Amor et al., 2018), cucumbers, tomatoes, lettuce (Rosenquist et al., 2005; Yu et al., 2019), garlic chives, perilla leaf, romaine lettuce (Park et al., 2018), fresh peppers, carrots, zucchini, garlic (Valero et al., 2002), and broccoli (Flores-Urban et al., 2014), collected from China (Yu et al., 2019), Korea (Park et al., 2018), Tunisia (Gdoura-Ben Amor et al., 2018), and Mexico (Flores-Urban et al., 2014), confirmed the presence of B. cereus. B. cereus-related food poisoning outbreaks are estimated to be approximately 3.9–5.9% (reports for 2011–2015), some of which have ben related to the consumption of vegetables such as tomatoes (2007, France, 4 cases), potatoes (2008, France, 28 cases), salad (2010, France, 44 cases) and carrots (2011, France, 3 cases) (Glasset et al., 2016; Jessberger et al., 2018).
Existing studies have shown that many vegetables are efficiently colonized by HPMOs. However, there are reports that some plants, e.g., cauliflower, broccoli, and okara byproducts, have antimicrobial potential against different food-borne pathogens, including S. enterica serovar Typhimurium, E. coli O157: H7 and B. cereus (Sanz-Puig et al., 2015). In the case of L. monocytogenes, similar observations were made for tomatoes and carrots (Gorski et al., 2003). Radish (annual herb, member of Brassicaceae, commonly consumed by humans) may belong to the group of vegetables less susceptible to colonization by HPMOs. In 1947, it was found that raphanin contained in radish seeds has bacteriostatic activity against different bacteria (Staphylococcus and E. coli) (former Bact. coli) (Ivánovics and Horváth, 1947). Over the next years, knowledge about the antibacterial compounds contained in radish seeds was extended. The following pathogens, i.e., Streptococcus pyogenes, E. coli, Salmonella enteritidis 110, Cronobacter sakazakii KCTC 2949, B. cereus ATCC 10876 and Staphylococcus aureus ATCC 6538 are sensitive to radish metabolites (Jadoun et al., 2016; Lim et al., 2019). The effect of red-peel radish root extract on 52 food-borne bacteria confirmed the presence of compounds with antimicrobial activity (Jadoun et al., 2016; Kaymak et al., 2018). The authors emphasized the potential role of these compounds in preventing microbial contamination of food (Kaymak et al., 2018).
The main aim of our research was to determine the interactions between radish and selected species of HPMOs: E. coli PCM 2561, S. enterica subsp. enterica PCM 2565, L. monocytogenes PCM 2191 and B. cereus PCM 1948. We hypothesized that (i) radish roots, rich in bactericidal substances, would be the plant organ least susceptible to colonization by the tested food-borne pathogens compared to leaves and shoots and (ii) HPMOs would have a negative effect on plant growth. In this study, we used one-week- and two-week-old radish organs (roots, stems and leaves) to examine their colonization level by the tested species with culture-dependent plating and culture-independent qPCR methods. Using the FISH technique, we also determined in situ the colonization of bacteria in the organs of all variants of plant cultivation. Additionally, the impact of pathogens on plant growth parameters (root, aboveground plant and plant length, fresh and dry weight of leaves, stem, root and whole plant) was investigated.
Materials and methods
Experimental design
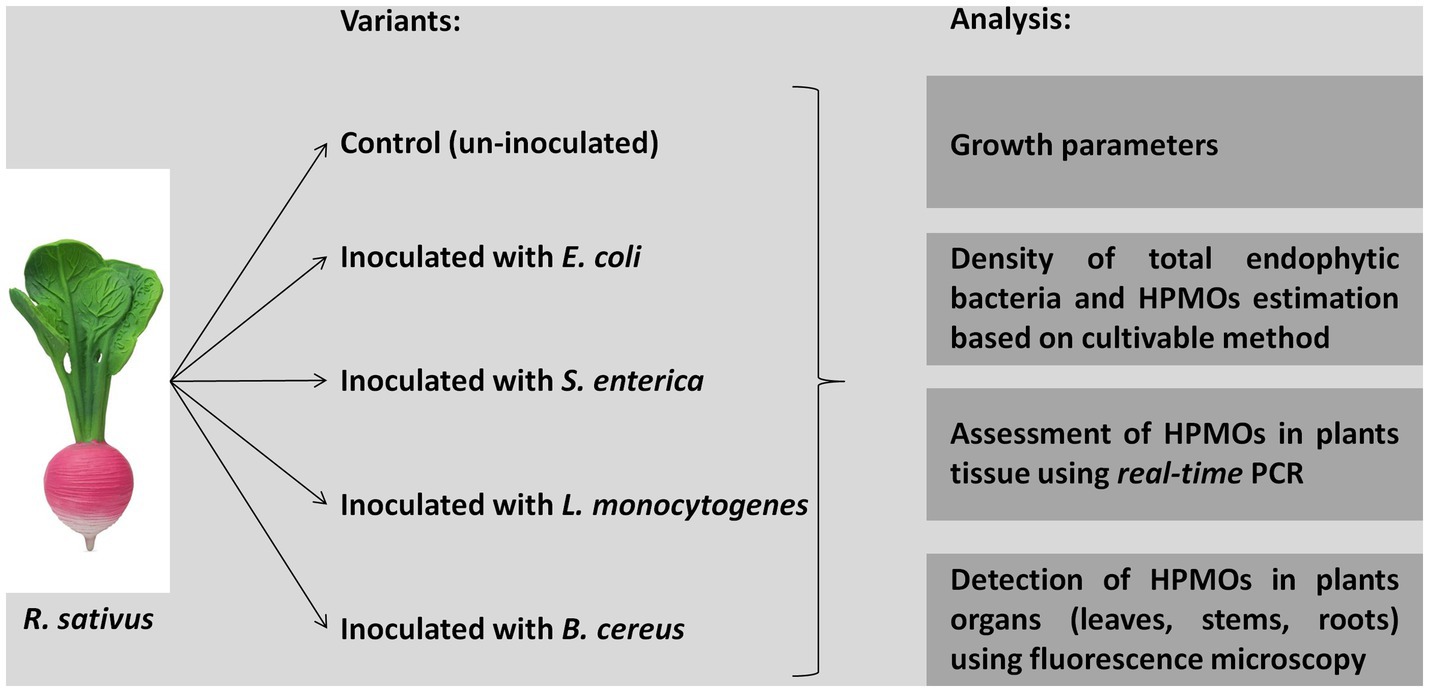
Raphanus sativus seeds were obtained from Plantico Company. Healthy and uniform-sized seeds were surface sterilized with 70% ethanol for 2 min, rinsed three times with sterile distilled water, treated with 30% sodium hypochlorite for 2 min and washed three times with sterile distilled water. The liquid obtained from the last washing was used to assess the efficiency of sterilization by plating on R2A (Difco) and Martin (BTL) agar media. Bacterial strains, i.e., E. coli Group A PCM 2561, S. enterica subsp. enterica serovar Choleraesuis PCM 2565, L. monocytogenes serovar 01/2 PCM 2191 and B. cereus PCM 1948, were cultivated in liquid trypticase soy broth (TSB, BD) with shaking at 37°C for 24 h. Then, suspensions containing 1.5 × 108 cells/ml (equivalent to McFarland OD = 0.5) were prepared. Surface-sterilized seeds were incubated in suspensions of the respective HPMO strains for 45 min with shaking (120 rpm) (Figure 1). As a control, sterilized distilled water was used instead of bacterial suspension. Treated seeds were cultivated in MS medium (Duchefa) in plastic rectangular shaped containers (107 × 94 × 96 mm, Duchefa, 10 plants/container, 10 containers/one variant) under controlled environmental conditions (temperature: 22°C ± 2°C and light/dark: 16 h/8 h). After 1 and 2 weeks, plants were carefully removed from the containers, surface sterilized with 70% ethanol for 2 min, rinsed with sterile distilled water and dried on sterile tissue paper. Collected plants were analyzed as shown in Figure 1.
Plant growth parameters analysis
Different growth parameters were analyzed, including total plant length, shoot and root length, fresh and dry weight (72 h drying at 85°C) of leaves, stems, roots and plants.
Estimation of total endophytic bacteria and selected HPMOs density in different organs of Raphanus sativus
Separated plant organs (leaves, stems or roots; five biological replicates per variant) were homogenized using a mortar and pestle with sterile distilled water in a 1:9 ratio (sample weight:volume of sterile distilled water). Serial dilutions (10−1 −10−3) of all homogenates were plated in triplicate on trypticase soy agar (TSA, BD) and appropriate selective media for S. enterica (Chromogenic, Salmonella LAB-AGAR TM and Salmonella Chromogenic Supplement, Biomaxima), E. coli (E. coli Chromogenic Medium, Biomaxima), B. cereus (B. cereus Selective LAB – AGAR™ Base and B. cereus Supplement, Biomaxima), and L. monocytogenes (Chromogenic Listeria acc. to Ottaviani and Agostii LAB -AGAR™ Base and Chromogenic Listeria Supplement acc. to ISO 11290, Biomaxima). Prepared plates were incubated for two (selective media) and three (TSA medium) days at 37°C. Colonies were counted each day. After 24 h, all colonies of E. coli, B. cereus and L. monocytogenes were fully grown, while the highest density of S. enterica was observed after two or three (in the case of TSA medium) days of cultivation. Colony forming units (CFU) were counted and calculated per g of fresh plant weight using plates with 30–300 colonies.
Assessment of HPMOs in plant tissue using real-time PCR
Plant material collection and genomic DNA isolation
Surface sterilized roots, stems and leaves were packed separately in 1.5 mL Eppendorf tubes and stored at −80°C. Three biological replicates were prepared per plant organ: roots, stems and leaves (nine samples per variant were obtained in total). Genomic DNA isolation was performed using a Plant and Fungi DNA Purification Kit (EURx, Poland) following the manufacturer’s procedure with a modified homogenization step (FastPrep-24 bead-beater, three cycles of 20 s at 4.5 m/s). DNA was quantified using a Qubit™ dsDNA HS Assay Kit (Invitrogen™). DNA was stored at −80°C.
Real-time quantitative PCR assay
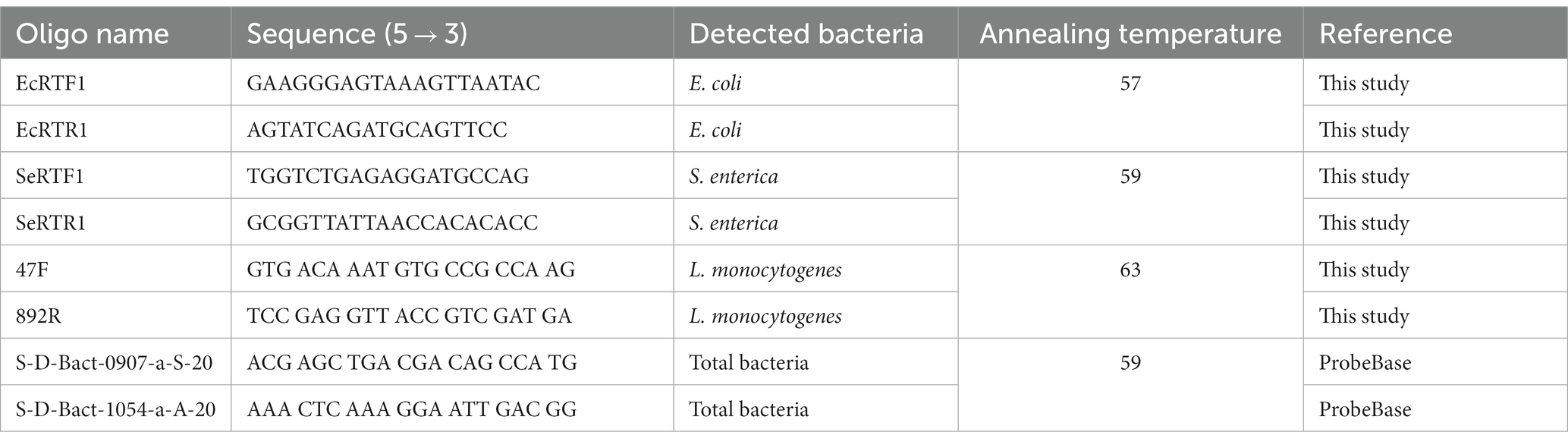
Absolute quantifications of each investigated HPMO (due to lack of specific primers for B. cereus this microorganism was not tested with qPCR) and total 16S rRNA gene copy (expressed as pEF copy number) determination in roots, stems and leaves of R. sativus were carried out using LightCycler 480 and LightCycler 480 SYBR Green I Master Kit (Roche). qPCR was performed in a total volume of 10 μL, which contained 5 μL of 2x SYBR Green I Master Mix (Roche), 3.5 μL of H2O, 0.25 μL of each specific primer (10 pmol/μl, Table 1) and 1 μL of DNA template (20 ng/μl for stems and leaves and 6 ng/μl for roots). The cycling conditions included initial denaturation for 5 min at 95°C, then 40 cycles of 10 s of denaturation at 95°C, 20 s of annealing at the optimal temperature for the primer pair (Table 1), 20 s of elongation at 72°C, and finally a melting curve with a continuous temperature increase from 65°C to 95°C. Positive (DNA of specific strains) and negative (molecular grade water) controls were analyzed in parallel with experimental samples. All qPCR analyses were performed in three biological and technical replicates (nine results for each variant were obtained).
Standard curves for quantifications of 16S rDNA copy number for each strain were prepared based on amplicons generated with 27F (5′AGA GTT TGA TCM TGG CTC AG 3′) and 1492R (5′ TAC GGY TAC CTT GTT ACG AC 3′) universal primers (Lane, 1991). PCRs were prepared using 2x Plus Taq Mix (Qiagen). Annealing conditions for the primer pair used in PCR included incubation for 30 s at 50°C. Amplicons were purified with AMPure XP (Beckman Coulter) and checked for their concentration with a Qubit™ dsDNA HS Assay Kit (Invitrogen™, USA) and Qubit fluorometer (ThermoFisher Scientific, USA). Standard curves were prepared using 10-fold dilutions of amplicons containing 10 million, 1 million, 100,000, 10,000, 1,000, and 100 copies per reaction. Based on the standard curve, the reaction efficiency was determined for each primer set. The results of absolute quantification were expressed as copies of specific bacterial 16S rRNA genes in 1 ng of total plant DNA. To determine the fold change in HPMO quantity in plant organs, the values were then calculated against the control samples. Furthermore, the percentage of HPMOs was calculated in relation to the total copy number of bacterial 16S rRNA genes.
FISH detection of HPMOs in plant organs
The roots, stems and leaves of R. sativus seedlings (three per variant) were fixed in 4% paraformaldehyde (Polyscience, USA) and 0.25% glutaraldehyde (Sigma-Aldrich, USA) in phosphate-buffered saline (PBS) buffer pH 7.2 overnight at 4°C and after dehydration were embedded in BMM resin (butyl methacrylate, methyl methacrylate, 0.5% benzoyl ethyl ether) (Sigma-Aldrich, USA) with 10 mM DDT (Thermo Fisher Scientific, USA) according to Niedojadło et al. (2015). The material was cut on a Leica UCT ultramicrotome into serial cross semithin sections and collected on Thermo Scientific™ Polysine (Thermo Fisher Scientific, USA) adhesion microscope slides. Before FISH reaction, the resin was removed with two changes of acetone and washed in distilled water and 4xSSC (Saline-Sodium Citrate buffer, Sigma-Aldrich, USA) and incubated with lysozyme 1 mg/mL (Thermo Fisher Scientific, USA). A E. coli, L. monocytogenes and S. enterica specific sequences targeting the 16S rRNA were used as specific FISH probes (Genomed, Poland) and were resuspended in hybridization buffer (Sigma-Aldrich, USA) with 30% v/v formamide at a concentration of 50 pmoL/mL. The following antisense DNA oligonucleotides labeled with Cy3 were used for the reactions for the detection of: E. coli 5′-Cy3-ACATCCGATGGCAAGAGGCCCGAA GGT-3′, L. monocytogenes 5′-Cy3-CGATAGCCGAAACCATCTTTCA AAAGCGTGG-3′, S. enterica 5′-GCTGCGGTTATTAACCACAAC ACCTTCCTC-3′. Hybridization was performed overnight at 42°C in a humidified chamber. After reaction the material was stained for DNA detection with Hoechst 33342 (1,1,000) (Invitrogen, USA) and mounted in ProLong Gold Antifade reagent (Invitrogen, USA). The positive control reactions were performed using only embedded bacteria. Two types of the negative control reactions were carried out: the samples were incubated only with hybridization buffer or reaction was performed on the samples from an uninoculated R. sativus seedling.
The images were captured with Olympus FV3000 confocal laser scanning microscope (CLSM). The optimized pinhole, long exposure (400 Hz), and 63X (numerical aperture 1.4) Plan Apochromat DIC H oil immersion lens were used. The images were collected simultaneously in the blue (Hoechst 33342) and red (Cy3) channels. To minimize bleed-through between the channels, we employed low laser power (1.5% of maximum power) and sequential collection. For all probes and plants organs, the obtained data were corrected for background autofluorescence, as determined by negative-control signal intensities. All image acquisition was performed using constant parameters (laser power, detector gain, emission band, and resolution).
Statistical analyses
One-way ANOVA and Newman–Keuls post hoc tests were used to compare growth parameters (including fresh and dry weight of leaves, stems, roots, plant, as well as length of roots and shoots) and colonization level (total density of endophytes and HPMOs in leaves, stems and roots) between treatments (i.e., uninoculated R. sativus – Ctr and plants inoculated with HPMOs: L. monocytogenes PCM 2191/L.m., B. cereus PCM 1948/B.c., E. coli PCM 2561/E.c. and S. enterica subsp. enterica PCM 2565/S.e.) depending on the cultivation time (1 or 2 weeks). Calculations were performed with the Statistica 10.0 software package (StatSoft, 2006).
The effect of treatment (Ctr, L.m., B.c., E.c, S.e.) on the growth parameters of R. sativus and the rate of its colonization by endophytes and HPMOs was checked with multivariate statistical analysis including principal component analysis (PCA) (R software). The analyses were performed for samples collected after the first and the second week of the experiment. For each PCA, the statistical significance was checked using the ‘arleyc/PCAtest’ R package. Statistically significant results were used to prepare the PCA plot.
Results
HPMOs affect the growth of Raphanus sativus
Inoculation of R. sativus with HPMOs (L.m., B.c., E.c, S.e.) negatively affected the investigated plant growth parameters; however, the level of impact depended on the plant age (1 or 2 weeks) and the species of pathogen (Figures 2–4). Changes in radish growth parameters in response to the presence of specific bacterial strain were more evident in one-week-old seedlings than in two-week-old seedlings (Figures 2–4). One-week-old seedlings upon inoculation were characterized by both lower total fresh biomass and decreased fresh weight of leaves compared to the control (Ctr, noninoculated plants) (Figure 2). Among the tested HPMOs, L. monocytogenes influenced the highest number of growth parameters in one-week-old radish, including root length (decreased by 39% compared to the control variant), stem fresh weight (reduced by 57%) and dry weight of leaves, stem and whole plant (decreased by 66, 57 and 43%, respectively) (Figures 2–4). The remaining strains negatively affected only some growth parameters, e.g., B. cereus reduced plant length (by 45%), while E. coli decreased stem fresh (by 51%) and dry (by 31%) weight (Figures 2–4). In the case of two-week-old plants, S. enterica and B. cereus decreased the length of whole plants (23 and 17%) and roots (33 and 22%), and E. coli reduced stem fresh (49%) and dry (48%) weight (Figures 2–4).
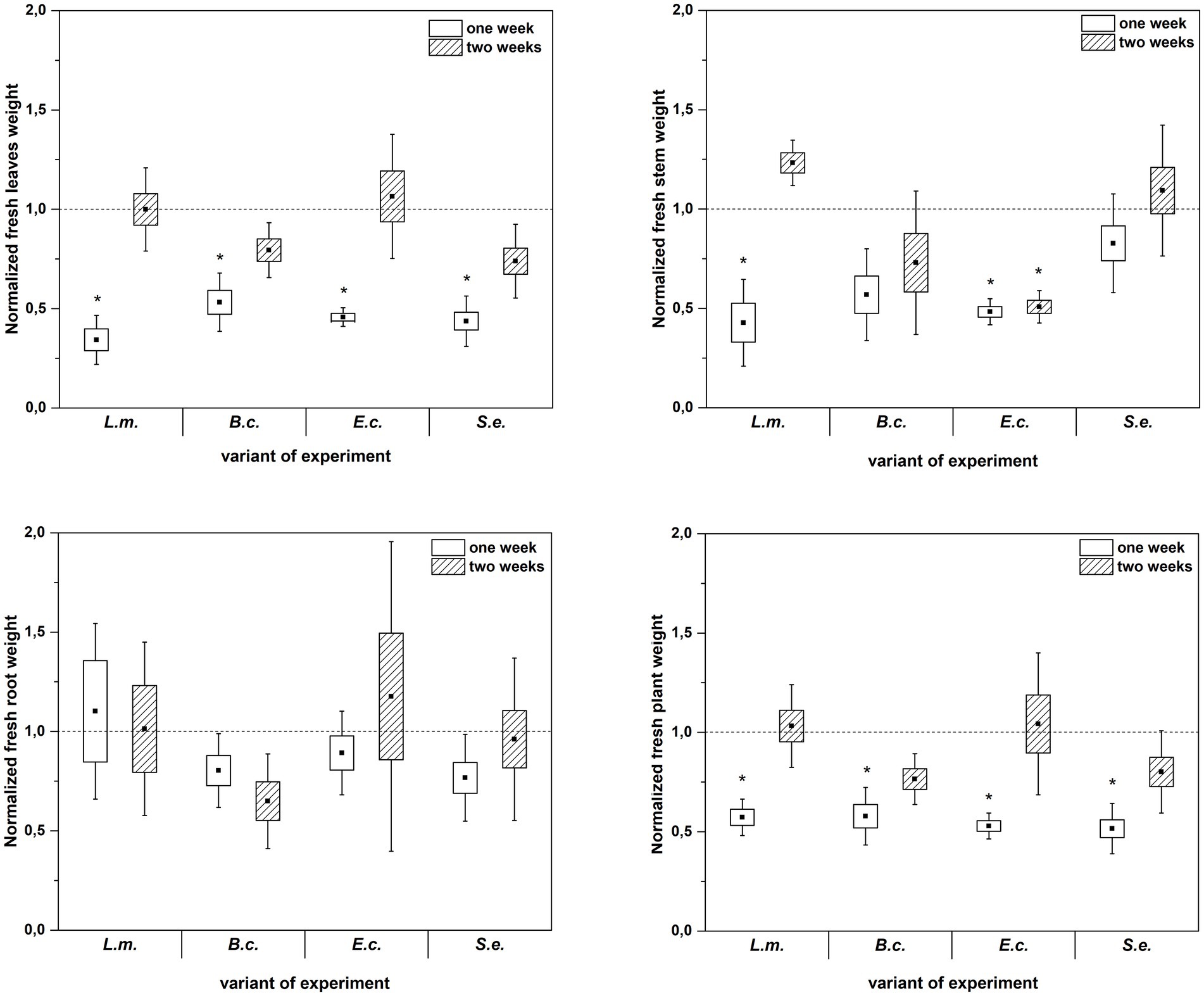
Figure 2. Normalized fresh leaves, stems, roots and plant weight of R. sativus inoculated with L. monocytogenes PCM 2191 – L.m., B. cereus PCM 1948 – B.c., E. coli PCM 2561 – E.c. and S. enterica subsp. enterica PCM 2565 – S.e. after 1 and 2 weeks of plant cultivation. Significant differences (p < 0.05, one-way ANOVA with Newman–Keuls post hoc comparisons) between the control (noninoculated) and inoculated variants at each time of R. sativus cultivation are denoted by different marks (*).
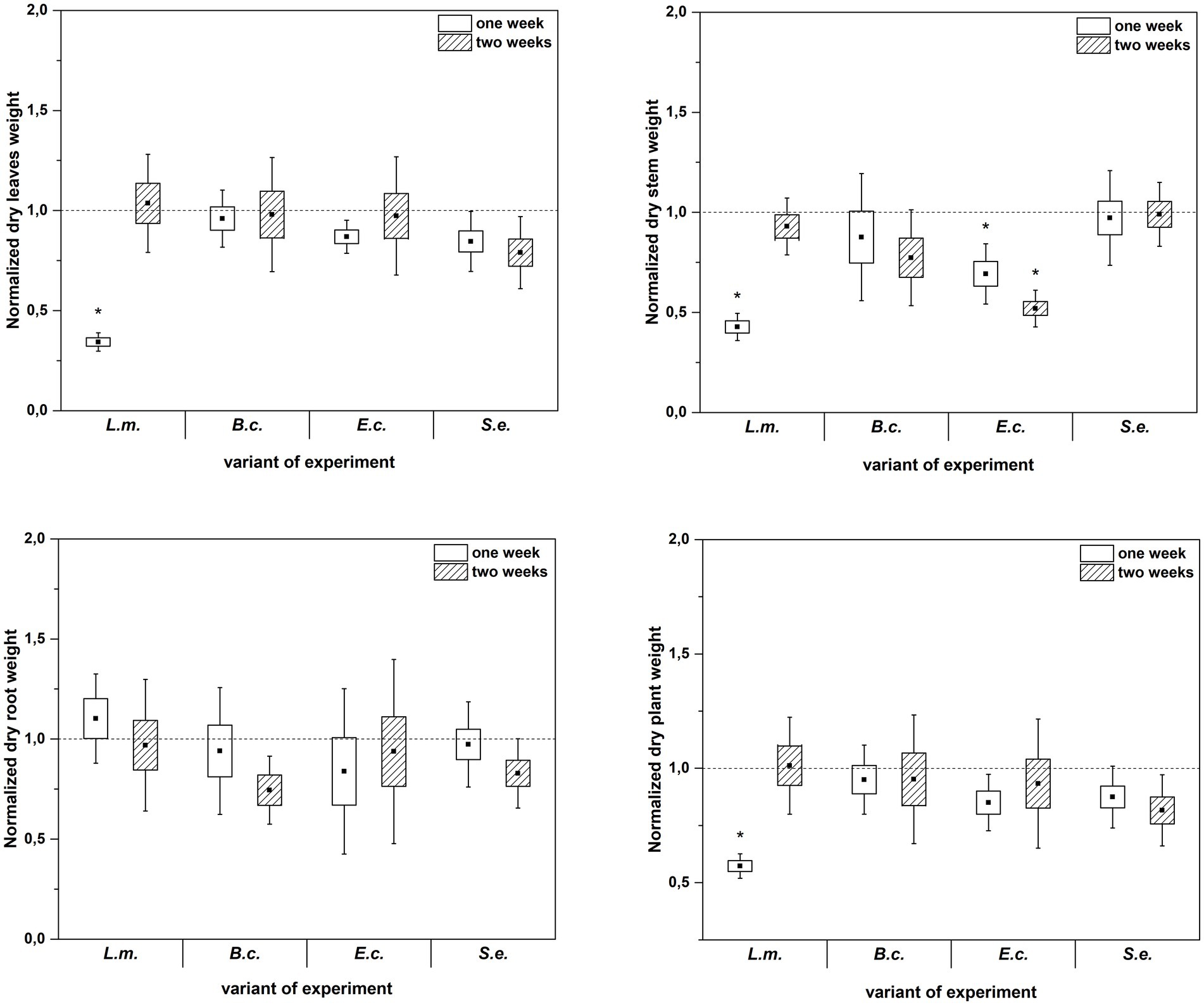
Figure 3. Normalized dry leaves, stem, root and plant weight of R. sativus inoculated with L. monocytogenes PCM 2191 – L.m., B. cereus PCM 1948 – B.c., E. coli PCM 2561 – E.c. and S. enterica subsp. enterica PCM 2565 – S.e. after 1 and 2 weeks of plant cultivation. Significant differences (p < 0.05, one-way ANOVA with Newman–Keuls post hoc comparisons) between the control (noninoculated) and inoculated variants at each time of R. sativus cultivation are denoted by different marks (*).
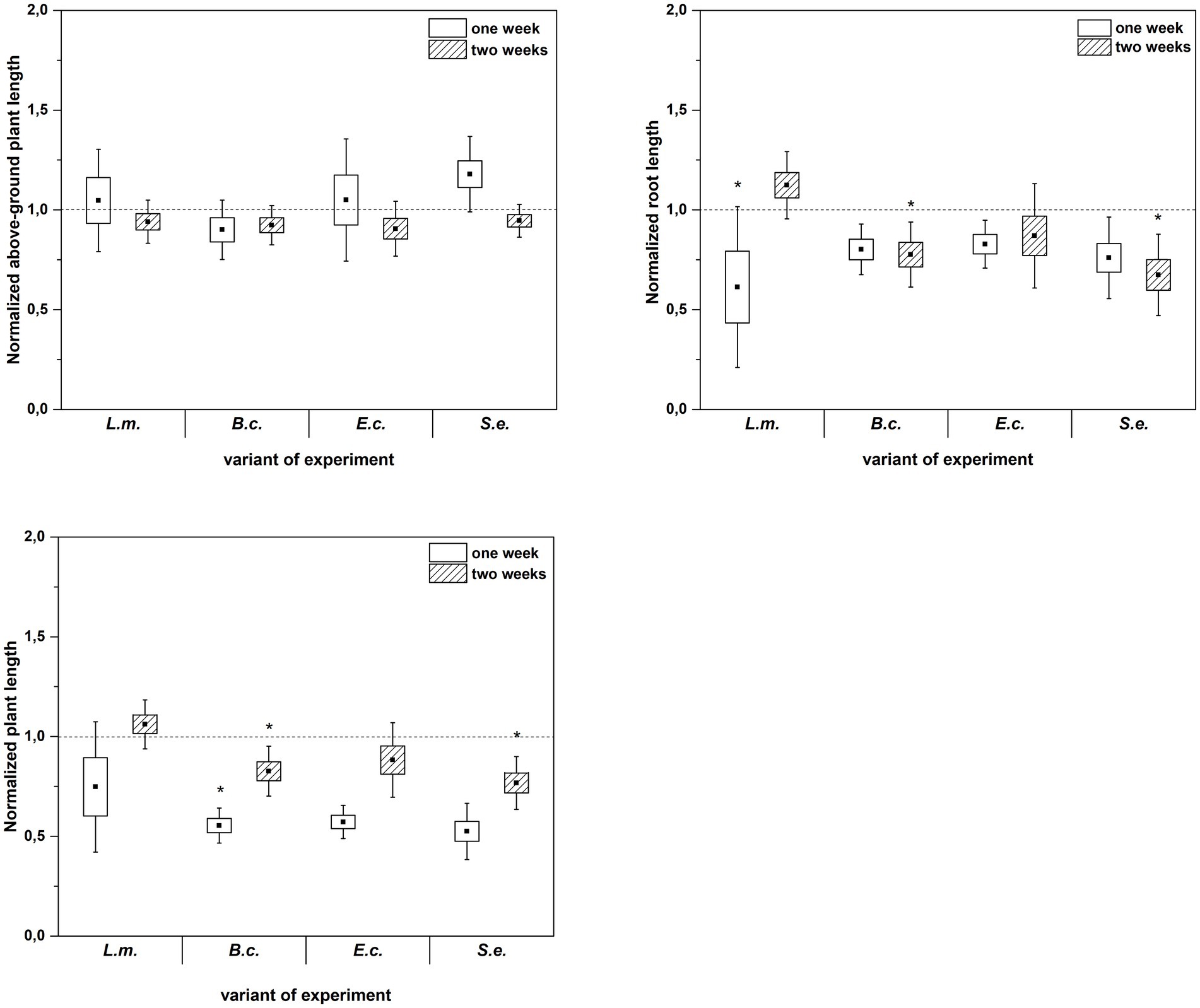
Figure 4. Normalized aboveground plant length, root length and plant length of R. sativus inoculated with L. monocytogenes PCM 2191 – L.m., B. cereus PCM 1948 – B.c., E. coli PCM 2561 – E.c. and S. enterica subsp. enterica PCM 2565 – S.e. after 1 and 2 weeks of plant cultivation. Significant differences (p < 0.05, one-way ANOVA with Newman–Keuls post hoc comparisons) between the control (noninoculated) and inoculated variants at each time of R. sativus cultivation are denoted by different marks (*).
Abundance of endophytes and HPMOs in plant tissues of Raphanus sativus: cultivation-dependent and cultivation-independent methods
Cultivation-dependent methods revealed that the total numbers of endophytes and HPMOs in the studied R. sativus organs (roots – R, stems – S and leaves – L) were associated with the plant age and the bacterial strain chosen for inoculation (Supplementary Figure S1 and Tables 2, 3).
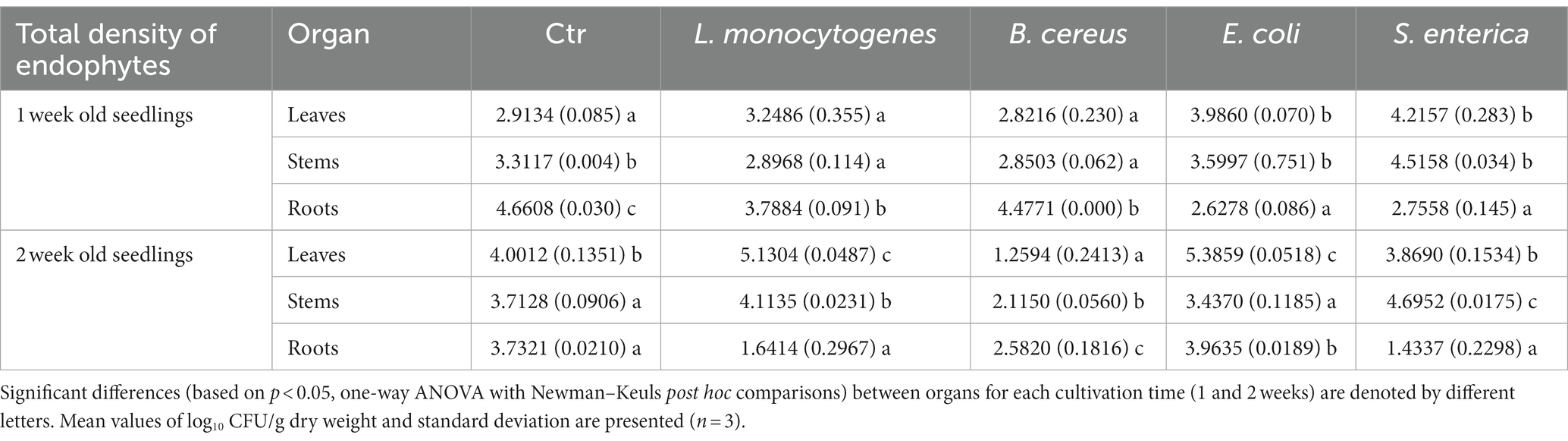
Table 2. Total density of endophytic bacteria in leaves, stems and roots of R. sativus for control (Ctr, uninoculated plants) and variants inoculated with L. monocytogenes, B. cereus, E. coli and S. enterica after 1 and 2 weeks of cultivation.
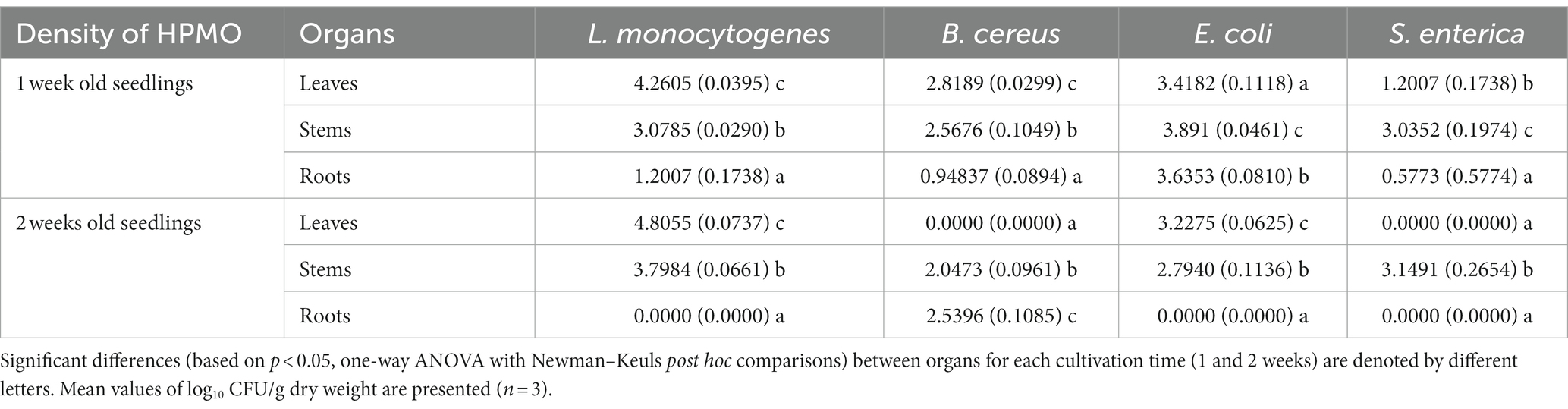
Table 3. Density of HPMO in leaves, stems and roots of R. sativus for variants inoculated with L. monocytogenes, B. cereus, E. coli and S. enterica after 1 and 2 weeks of cultivation.
The time of plant cultivation significantly affected the total abundance of endophytes. Higher numbers of endophytes were noted in the roots of one-week-old R. sativus in almost all variants (with the exception of plants inoculated with E. coli) compared to stems and leaves. The aboveground parts of R. sativus were more intensively colonized after 2 weeks of cultivation, with the exception of leaves of plants treated with S. enterica and stems of plants inoculated with E. coli, where a similar total density of endophytic bacteria was observed (Supplementary Figures S1A–C and Table 2).
The inoculation of plants with HPMOs resulted in a decrease in the total density of endophytes in the roots of one-week- and two-week-old R. sativus compared to the noninoculated control variant (for L.m.: 18.7% vs. 56.0%, B.c.: 3.9% vs. 30.8%, S.e: 40.9% vs. 61.6%). Interestingly, in the case of E. coli, the amount of bacteria was similar regardless of the cultivation time (Supplementary Figure S1C and Table 2). In stems and leaves of one-week-old plants, a similar density of endophytes was noted for all tested variants with the exception of S. enterica-treated plants (increases of 36.4 and 44.7% were observed in stems and leaves, respectively). The presence of HPMOs significantly changed the total density of endophytes in the aboveground parts of two-week-old plants. The number of endophytes in stems and leaves increased in L.m. variant (by 10.8 and 28.2%, respectively). Opposite observations were made for the B.c. variant, where a decrease in the abundance of bacteria was noted (by 43.1% in stems and 68.5% in leaves) (Supplementary Figures S1A,B and Table 2).
HPMOs were found in all organs of one-week-old plants, while in the case of two-week-old R. sativus, no pathogens were detected in specific organs. The absence of L.m., E.c. and S.e. was observed in roots. Surprisingly, the leaves of plants inoculated with B.c. and S.e. were pathogen-free (Supplementary Figures S1D–F and Table 3). In general, HPMOs (excluding B.c.) more efficiently colonized the aboveground parts of plants, regardless of their age. B.c. preferentially inhabited the roots of two-week-old plants (Supplementary Figures S1D–F and Table 3). L. monocytogenes showed the highest ability to colonize R. sativus leaves at both cultivation times and stems of two-week-old plants compared to other tested HPMOs (Supplementary Figures S1D,E and Table 3).
Principal component analysis (PCA) showed a dispersion of the investigated variants in the ordination space of PC1-PC2. The PC1 and PC2 axes explained nearly 75% (49.52 and 25.10%, respectively) of the total variance. PC1 was significant (p value 0.01) in explaining the variance between the samples. The variables having significant loadings on PC1 included root and whole plant length, leaf and stem fresh weight and HPMO content in leaves (all parameters are marked in the PCA plot with asterisks). The inoculation with L. monocytogenes and E. coli exerted a similar effect on one-week-old plants, as both were associated (positively correlated) with root fresh weight and elevated HPMO density in leaves and to a lesser extent in roots (Figure 5). The impact of S. enterica was different because a negative correlation of this pathogen with all tested parameters was observed (Figure 5). Ordination analysis for samples collected after the second week was not statistically significant.
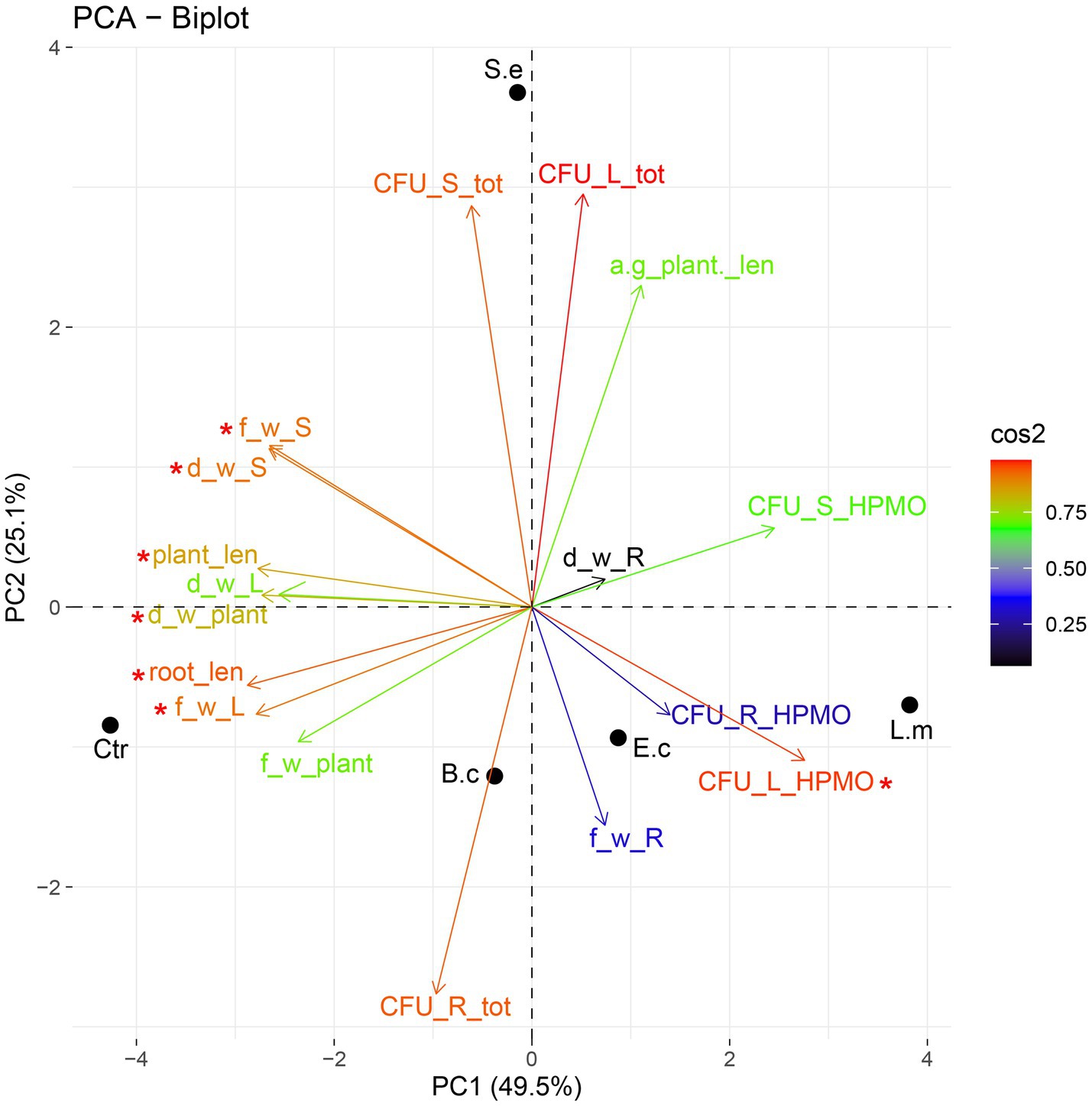
Figure 5. PCA ordination of growth parameters and density of endophytes after one (A) and two (B) weeks of R. sativus cultivation. a-g_plant _len, aboveground plant length; root_len, root length; plant_len, plant length; f_w_L, fresh leaf weight; f_w_R, fresh root weight; f_w_S, fresh stem weight; f_w_plant, fresh plant weight; d_w_L, dry leaf weight; d_w_R, dry root weight; d_w_S, dry stem weight; d_w_plant, dry plant weight; CFU_L_tot, total density of endophytes in leaves; CFU_S_tot, total density of endophytes in stems; CFU_R_tot, total density of endophytes in roots; CFU_L_HPMO, density of HPMOs in leaves; CFU_S_HPMO, density of HPMOs in stems; CFU_R_HPMO density of HPMOs in roots.
The culture-independent method (qPCR analysis) partially confirmed the results obtained with the use of microbiological media. In this assay, inoculation with L. monocytogenes also showed no effect on the total 16S rRNA gene copy number in either the first or the second week of plant cultivation (with the exception of two-week-old plants leaves). Similarly, inoculation with E. coli caused an increase in the total 16S rRNA gene copy number in leaves of two-week-old R. sativus compared to the uninoculated variant (Figure 6). Only the presence of S. enterica was associated with a significantly increased 16S rRNA gene copy number in stems of one-week-old plants (Figure 6). Contrary to the results based on cultivation, qPCR showed no effect of inoculation with tested strains on 16S rRNA gene copy number in stems of two-week-old R. sativus. Additionally, inoculation with S. enterica did not influence the 16S rRNA gene copy number in leaves of one-week-old plants compared to control variants, but an increase in that number was noticed after 2 weeks (Figure 6).
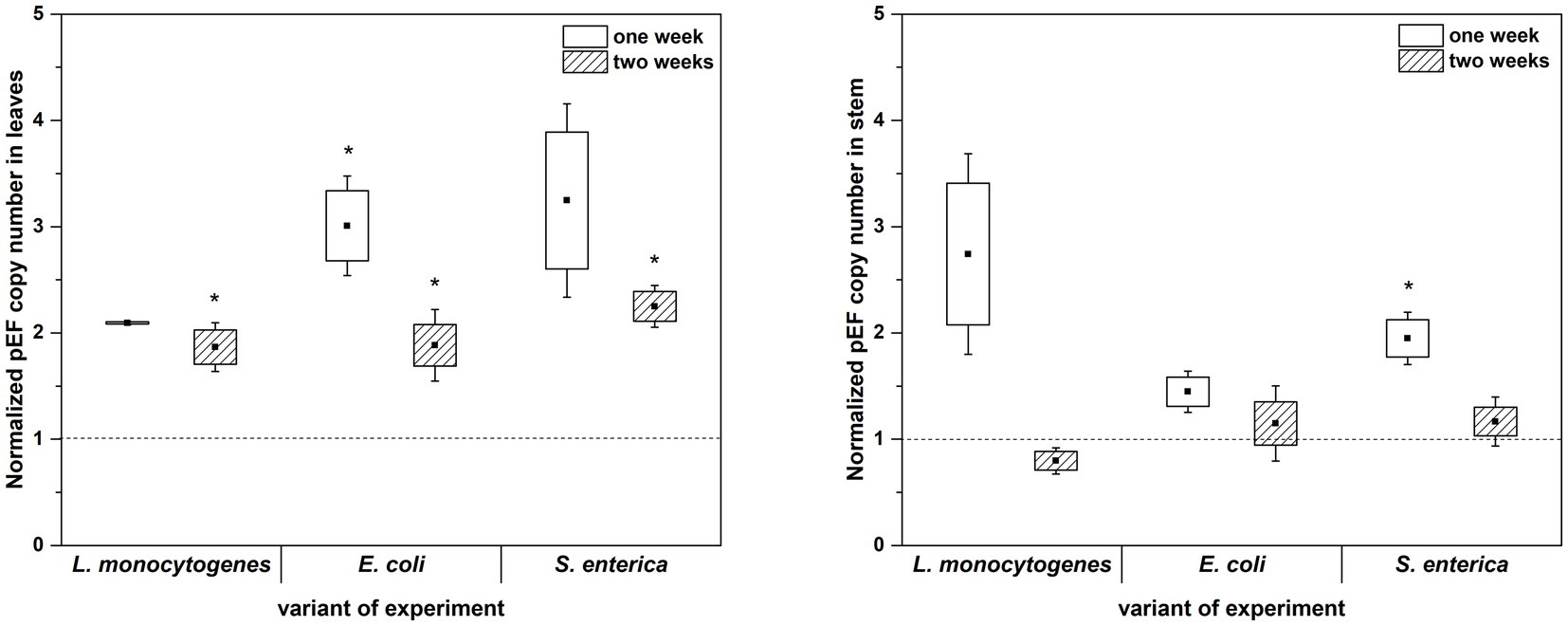
Figure 6. Normalized pEF copy number in leaves and stems of R. sativus after 1 and 2 weeks of plant cultivation. Significant differences (p < 0.05, one-way ANOVA with Newman–Keuls post hoc comparisons) between control and inoculated (inoculated with L. monocytogenes PCM 2191, E. coli PCM 2561 and S. enterica subsp. enterica PCM 2565) treatments are denoted by different marks (*).
qPCR analysis showed a higher proportion of L. monocytogenes and E. coli in relation to the total number of bacteria in plant leaves (1 week: L.m. – 0.41% and E.c. – 0.77%; 2 weeks: L.m. – 0.75% and E.c. – 20.43%) compared to stems (1 week: L.m. – 0.02% and E.c. – 0.24%; 2 weeks: L.m. – 0.01% and E.c. – 3.57%) (Figure 7). The distribution of S. enterica in leaves (1 week: 4.46% and 2 weeks: 3.80%) and stems (1 week: 3.36% and 2 weeks: 4.31%) was similar in both tested cultivation variants (1 and 2 weeks) (Figure 7). The relative level of L. monocytogenes in leaves and stems was the lowest in comparison to the other investigated pathogens. Based on the qPCR results, E. coli had the highest ability to colonize leaves of two-week-old R. sativus, while S. enterica preferentially colonized the stems of both one-week-old and two-week-old plants (Figure 7).
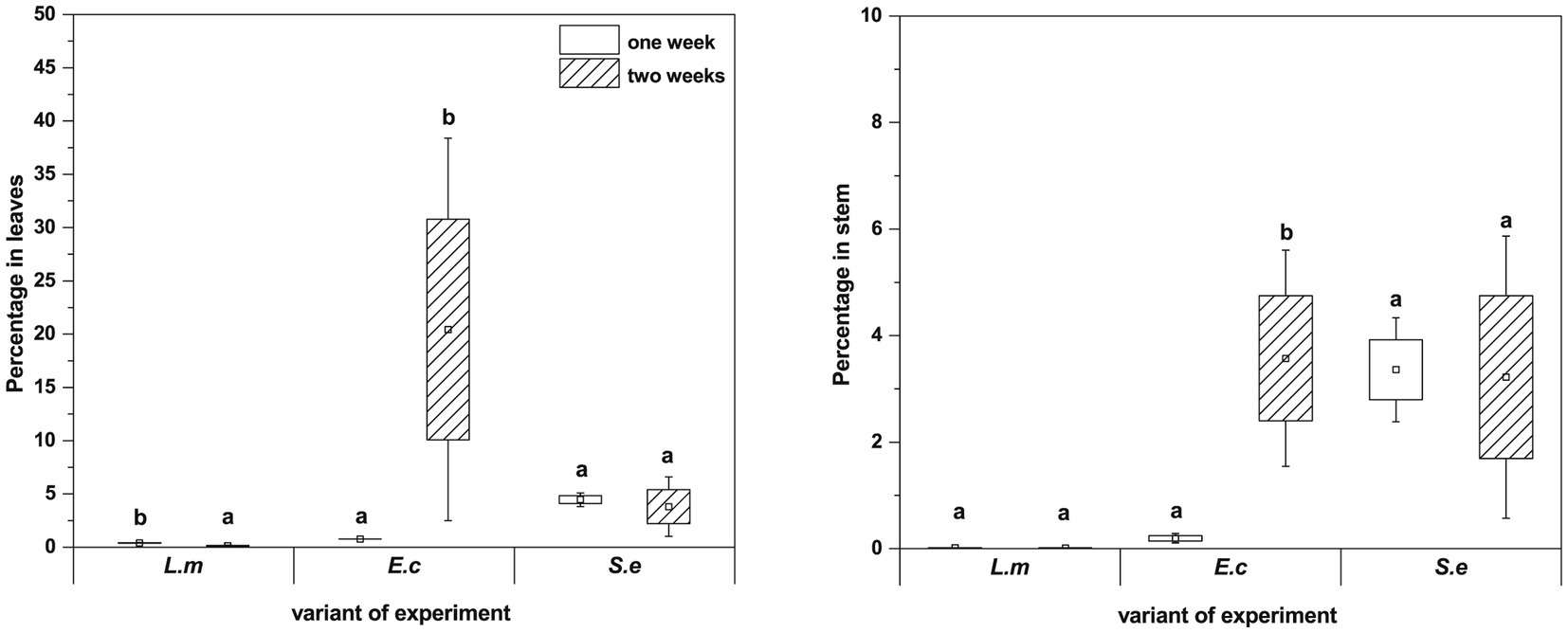
Figure 7. Significant differences (p < 0.05, one-way ANOVA with Newman–Keuls post hoc comparisons) between percentage share of HPMO (L. monocytogenes, E. coli, S. enterica) in stem and leaves of R. sativus obtained for two tested times of plant cultivation (1 and 2 weeks). The presented values were calculated in relation to total density of bacteria (based on pEF gene copy number) amounting 100%. Standard error (box) and standard deviation (whiskars) are shown.
Visualization of HPMOs colonization in plant organs of Raphanus sativus
To verify and visualize the bacterial colonization pattern of R. sativus roots, stems and leaves we used a E. coli, L. monocytogenes and S. enterica specific sequences targeting the 16S rRNA as specific probes to FISH technique. At first, we optimized the FISH protocol for embedded bacteria detection as positive control of reactions (Figures 8A,E,I) and next we localized the bacteria in semithin sections of inoculated one-week and two-week-old plants. FISH-CLSM analysis confirmed in situ presence of all bacteria populations in all organs (roots Figures 8B,F,J and stems Figures 8C,D,G,H,K,L). The fluorescence spots indicating the presence of bacteria were mainly visible in the extracellular matrix of all cell layers of roots, stems and leaves. We also detected bacteria in some the individual cells (for example, Figures 8C,D,G). Similarly, the strong autofluorescence of chloroplasts in leaves completely made impossible the precise localization of bacteria in the apoplast or inside the cells (not shown). Therefore, in this variant of organs further studies of intracellular presence of bacteria at the ultrastructure level in electron microscopy are necessary. In all the negative control reactions no FISH signals were observed. In an uninoculated R. sativus 1-week seedling root and shoot only the autofluorescence of cell walls and the chloroplasts are visible (Supplementary Figure S2).
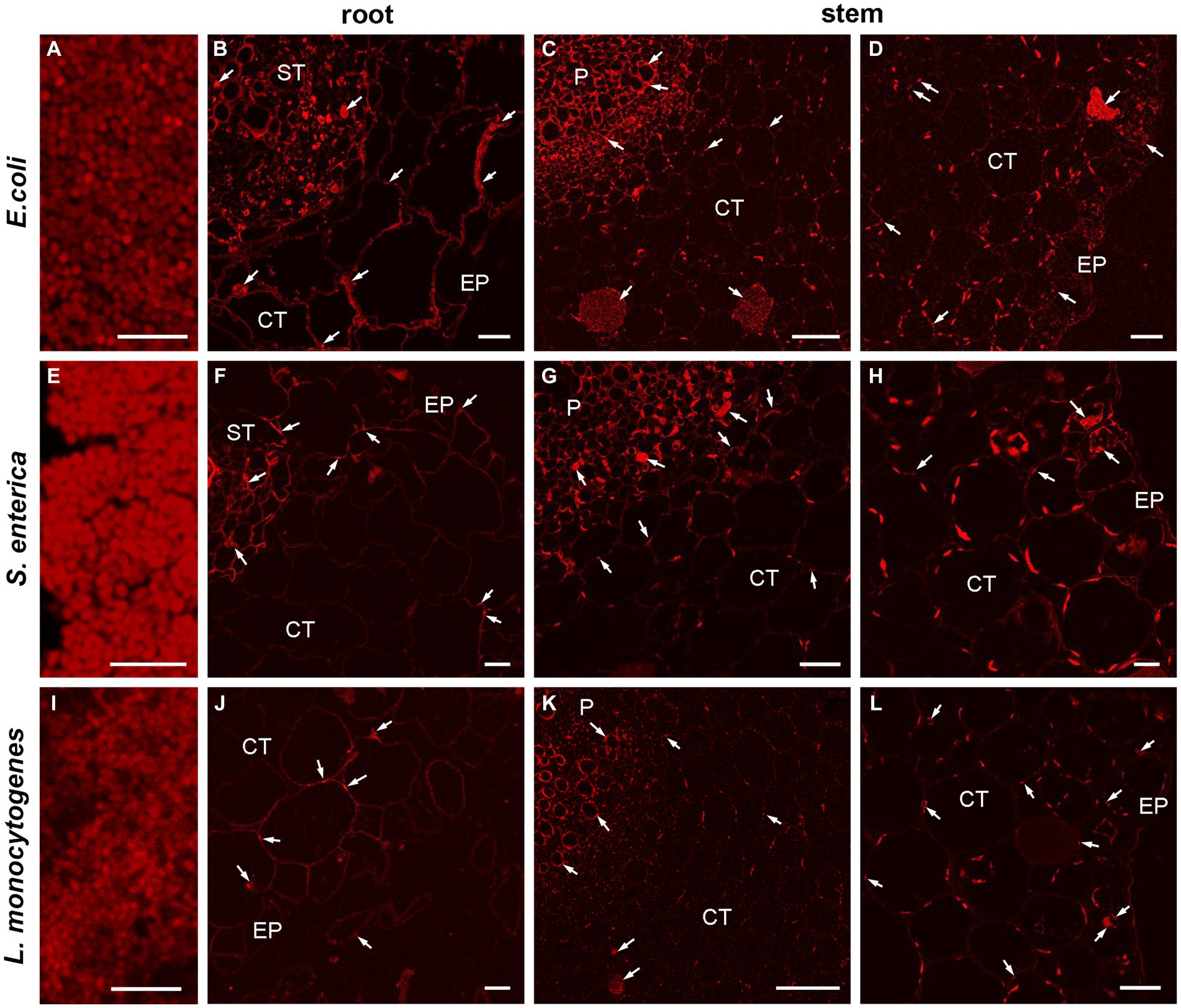
Figure 8. CLSM (confocal laser scanning microscopy) analysis of bacterial (E. coli, L. monocytogenes, S. enterica) colonization pattern of R. sativus 1-week seedling root and shoot detected by FISH. The representative images of the positive control of FISH reaction, bars 5 μm (A,E,I), the localization of bacteria in root and in shoot, bars 20 μm (B,D,G,L), bars 10 μm (F,H,J) and bars 50 μm (C,K). The arrows indicate the presence of bacteria in analyzed organs: in the extracellular matrix or inside the cells (arrows). The autofluorescence of the chloroplasts are visible (C,D,G,H,K,L), EP, epidermis; CT, cortex tissue; ST, stele tissue; P, pith.
Discussion
Plants are an attractive niche for many microorganisms, including bacteria, fungi, archaea, viruses, and algae (Truong et al., 2021). Many studies have examined the colonization of various vegetables, e.g., tomatoes, lettuce, cucumbers, and parsley, by individual HPMOs; however, in the case of red radish, there is a lack of comparative studies regarding the interaction of radish with different microbes, including Gram-negative and Gram-positive bacteria (Szymczak et al., 2014; Truong et al., 2021).
In this study, we checked the colonization of R. sativus var. radicula organs by selected pathogens. The first critical choice we made regarded the method of bacterial inoculation. We decided to inoculate the plant seeds directly by short-term incubation in OD-adjusted HPMO suspensions. Seed inoculation is one of the most effective methods of bacterial delivery to plants (O’Callaghan, 2016; Luna-Guevara et al., 2019). Numerous studies have shown that vegetable contamination by HPMOs can originate from pathogen inoculation into the soil, substrate or water used for plant watering (Islam et al., 2004a,b; Jechalke et al., 2019). Our results, based on culture-dependent and culture-independent methods, revealed the ability of all tested HPMOs, including E. coli, S. enterica, L. monocytogenes and B. cereus, to colonize one- and two-week-old radish. Torres et al. (2005) stressed that adhesion is the most important step in the colonization of plants by inoculated pathogens. All studied pathogens are characterized by high adhesion and biofilm formation capacity (Gorski et al., 2003; Yaron and Römling, 2014; Antequera-Gómez et al., 2019; Elpers et al., 2020; Lin et al., 2022). In the adhesion process, an important role is assigned to motility and chemotaxis due to flagellar rotation ability and the presence of the lipopolysaccharide (LPS) layer (Elpers et al., 2020). Liu et al. (2005) confirmed the ability of S. enterica and enterohemorrhagic E. coli (EHEC) to attach to vegetable seeds. Gorski et al. (2003) revealed high adherence of L. monocytogenes to radish slices, and the highest ability was observed in the range of temperatures (20 and 30°C) corresponding to the conditions in which we incubated the seeds in the bacterial suspension (26°C).
The next step of plant colonization after the adhesion of pathogens to the seed surface is their translocation to internal tissues (Melotto et al., 2014). Many scientists have reported a more intensive colonization of internal plant tissues after seed inoculation compared to seedling inoculation (Schoeller et al., 2002; Kim et al., 2018). It is associated with the occurrence of cracks during root development. The cracks are potential places for pathogens to enter the plant. Using these natural openings, bacterial invaders may slow the immune system reaction or even evade plant immune responses (Truong et al., 2021). Our studies showed a high level of one-week R. sativus tissue colonization by the tested HPMOs (up to approximately 103 and 104 CFU/g dry weight of shoots and leaves, respectively), which corresponds to the abundance of endophytes observed in plants growing under natural conditions (Compant et al., 2021). A rapid increase in the level of plant colonization by Salmonella and E. coli O157:H7, reaching from 0.1 log CFU/g to as high as 6 log units, was observed under sprouting conditions (Howard and Hutcheson, 2003). Schoeller et al. (2002) determined the growth and survival capacity of alfalfa sprouts treated with L. monocytogenes. In this investigation, plants showed a drastic increase in the density of pathogenic bacteria 1 day postinoculation of seeds. In the same study, the inoculation of sprouted seeds with L. monocytogenes performed later (on Day 5) did not result in such an increase (2002). Similarly, Kim et al. (2018) compared the growth dynamics of the S. enterica population after inoculation of alfalfa seeds or fully germinated sprouts. The authors found a higher number of pathogens in the case of seed inoculation. Undoubtedly, the choice of inoculation method (in our case, direct seed inoculation) has a key impact on the level of internal plant tissue colonization by bacteria.
In nature, a higher number of bacteria is observed in the roots (from 105 to 107 cultivable bacteria per gram) than in the aboveground parts (103–104) (Compant et al., 2021). Our results for the uninoculated variant, where only endophytes were present inside the plants, showed a high density of bacteria in all radish organs following the trend of roots (ap. 104.5) > shoots > leaves (ap. 103) in the first week after sowing the seeds. Seeds of most plant species are not axenic and are characterized by the presence of bacteria and/or fungi (Truyens et al., 2015; Pitzschke, 2018). We also observed morphologically different bacterial colonies during germination of surface-sterilized radish seeds. Schoeller et al. (2002) observed a drastic increase in the total number of bacteria after 24 h of sprout development from uninoculated seeds (from ca. 3.5 log CFU/g to ca. 8.0 log CFU/g). The longer cultivation time was associated with a more equal distribution of bacteria in shoots (ap. 103.7) and roots (ap. 103.7) of two-week-old R. sativus, while leaves were characterized by the highest density of bacteria (104). It is possible that the presence of compounds with antimicrobial effects in radish roots may cause the movement of bacteria toward the aboveground parts of plants (Beevi et al., 2009). Moreover, in variants inoculated with HPMOs, the interaction between the tested strains and autochthonous endophytes can significantly shape the total number of endophytic microbes, which in turn may affect the process of plant colonization by HPMO (Truong et al., 2021). The presence of HPMOs significantly decreased the total density of bacteria in the roots of one-week R. sativus, while in the shoots and leaves, no effect was noted. The exception was a variant treated with S. enterica, where an increase in the total density of bacteria in stems and leaves of the week-old radish was observed.
The results of our research confirmed the ability of the tested pathogens not only to colonize plants but also to move from the roots toward the aerial parts. In situ studies using FISH-CLSM revealed the presence of HPMOs primarily in apoplast and some cells of roots, stems and leaves in the one-week-old inoculated seedlings. Penetration ability via apoplast has been observed for many HPMOs including E. coli serotype O157:H7 (in case of lettuce and spinach through roots and leaves), L. monocytogenes (in romanian lettuce) and S. enterica (tomatoes) (Shenoy et al., 2017; Wright et al., 2017; Zarkani and Schikora, 2021). The translocation of multiple Salmonella serovars (including S. Javiana, S. Newport, S. Poona and S. Montevideo) to the lower stems of cucumber (3–5 cm) 1 week post inoculation into the root zone was observed by Burris et al. (2020). In the case of tomatoes, 7 days after inoculation, distant migration of S. enterica (reaching up to 10 cm) from the soil to the shoots was shown (Zheng et al., 2013). There are also reports confirming the ability of L. monocytogenes and E. coli to translocate from the root zone to the aboveground part (Quilliam et al., 2012; Hofmann et al., 2014; Standing et al., 2014). It was previously discussed that this phenomenon depends on various factors, including the type of substrate and cultivar, time of inoculation and serotype applied (Hirneisen et al., 2012; Burris et al., 2020; Esmael et al., 2023). In this study, E. coli showed the highest capacity to colonize roots and stems of one-week-old plants. Most likely, the short time of E. coli multiplication (approximately 20 min) influenced the obtained result (Dewachter et al., 2018). Our observation is in line with results described by Standing et al. (2014). The authors, who compared the average density of internalized E. coli O157:H7, L. monocytogenes, S. enterica subsp. enterica serovar Typhimurium and S. aureus in one-week-old lettuce roots and leaves, found the highest abundance of E. coli in the plant roots, while leaves were characterized by the highest number of L. monocytogenes (2013). Similarly, we also observed that L. monocytogenes exhibited the highest ability to translocate along the plant and colonize leaves in both one- and two-week-old plants. Distant translocation of bacteria can be associated with flagellum presence and may occur via plant vasculature (Melotto et al., 2014). Shenoy et al. (2017) demonstrated the high mobility of L. monocytogenes in the vascular system after inoculation of romaine lettuce seeds.
The raphanin contained in radish seeds and leaves has an antimicrobial effect against several bacteria, including E. coli, Pseudomonas pyocyaneus, Salmonella typhi, Bacillus subtilis, S. aureus, streptococci and pneumococci, as well as Listeria, Micrococcus, Enterococcus, Lactobacillus and Pedicoccus species (Shukla et al., 2011). Our research showed a significant effect of plant age (one- or two-week-old plants were investigated) on colonization by the tested HPMOs. Selected HPMOs had the ability to colonize all organs of one-week R. sativus; however, in the case of two-week-old plants, we noted significant changes. Most likely, the concentration of antimicrobial compounds in leaves and roots of R. sativus increases with the age of the plant. Among the tested HPMOs, S. enterica was the most sensitive strain, and its presence was detected only in stems of two-week-old R. sativus. A previous study considering the impact of R. sativus extracts on bacterial growth confirmed the higher sensitivity of S. enterica compared to other pathogens (Lim et al., 2019). Lim et al. (2019) indicated that R. raphanistrum subsp. sativus (radish) extract inhibited the growth of S. enteritidis 110, Cronobacter sakazakii KCTC 2949, B. cereus ATCC 10876, and Staphylococcus aureus ATCC 6538, while it showed no effect against L. monocytogenes ATCC 51776 and E. coli 23,716. The roots of two-week-old plants seemed to be a difficult niche for most bacterial strains compared to stems and leaves, as only spore-forming B. cereus was able to survive in that organ (Moteshareie et al., 2022). Among extracts prepared from roots, shoots and leaves of R. sativus, the first one showed the greatest antibacterial activity against foodborne pathogens, including B. subtilis, S. aureus, Staphylococcus epidermidis, Enterococcus faecalis, Salmonella typhimurium, Enterobacter aerogenes, Enterobacter cloacae, and E. coli (Beevi et al., 2009). Moreover, Shukla et al. (2011) checked the effect of radish root juice on Klebsiella pneumoniae, Staphylococcus aureus, Pseudomonas aeruginosa, Enterococcus faecalis and E. coli growth. The authors observed a greater antibacterial effect on gram-negative than gram-positive bacteria, which is in line with our results showing that gram-negative strains avoided the colonization of radish roots (Shukla et al., 2011).
A plant is a good alternative host for HPMOs, and its colonization can be crucial for the survival of pathogens, first, because the plant provides a refuge for bacteria when it enters the soil, and second, the presence in the plant gives the pathogen a chance for returning to herbivorous and omnivorous hosts (Brandl et al., 2013). Rhizosphere and endophytic microorganisms significantly influence the growth of the host plant (Szymańska et al., 2016). Moreover, HPMOs are also considered important regulators of plant productivity (Nautiyal et al., 2010; Afzal et al., 2019). The plant-HPMO interaction is not fully understood, but it has been indicated that plants can recognize human enteric pathogens and activate basic defense signaling pathways (Brandl et al., 2013). The results of our research suggested that in the first week, the presence of HPMOs may cause a pronounced plant stress reaction, which was manifested, among the others, by the decrease in plant fresh weight. After 2 weeks, the plant response was less noticeable, which may indicate that the plants are adapting to the stress caused by the presence of pathogens. Jechalke et al. (2019) showed that lettuce responded to the presence of S. typhimurium 14,028 s through upregulation of genes associated with the stress response and genes related to the plant immune response. Among the pathogens studied by us, L. monocytogenes exerted the most unfavorable effect on the radish growth parameters. Similar to our results, Klerks et al. (2007) observed the inhibition of lettuce seedling growth and their biomass reduction after inoculation with Salmonella Dublin. Furthermore, the negative impact of HPMOs on many different plants, e.g., tomato, romaine lettuce, and Medicago truncatula, was previously found (Jayaraman et al., 2014; Deering et al., 2015; Simko et al., 2015). Furthermore, the symptoms noticed in plants after HPMO inoculation included chlorosis, wilting, tissue necrosis or root growth inhibition, which was also observed by researchers (Klerks et al., 2007; Schikora et al., 2008; Berger et al., 2011).
In summary, we conclude that all tested HPMOs (E. coli, S. enterica, L. monocytogenes and B. cereus) have the ability to colonize radish no later than in the first week of its growth. E. coli and L. monocytogenes were characterized by the highest ability to migrate along the plant (from roots, through shoot to leaves) and to colonize the above-ground plant organs. Plant age significantly influenced the distribution of HPMOs in R. sativus organs. The tested HPMOs did not colonize the two-week-old radish roots (with the exception of B. cereus, which most likely survived due to the formation of spores). Only L. monocytogenes and E. coli preferred to colonize the leaves of two-week-old radish. Limited colonization of roots and leaves of two-week-old R. sativus by HPMOs could be related to the presence of compounds with antibacterial properties in these radish organs. On the other hand, the presence of pathogens in plant organs inhibits the growth of radish, which is manifested by a decrease in growth parameters after the first week. Nevertheless, R. sativus can adapt to the presence of pathogens, causing no further decrease in the growth rate of two-week-old plants.
Data availability statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Author contributions
SS: Conceptualization, Data curation, Formal analysis, Funding acquisition, Investigation, Methodology, Project administration, Resources, Visualization, Writing – original draft. ED-S: Investigation, Methodology, Supervision, Visualization, Writing – review & editing. MS: Investigation, Methodology, Writing – review & editing. KN: Investigation, Methodology, Visualization, Writing – review & editing. JM: Investigation, Writing – review & editing. KH: Supervision, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This study was financially supported by a grant from the National Science Centre (Poland) – Miniatura3 (DEC-2019/03/X/NZ9/00751). Costs of publication proof reading were financially supported by the Emerging Field “Plant productivity and food safety: Soil science, Microbiology, Agricultural Genetics and Food quality” (Excellence Initiative – Research University, IDUB). The publication fee was funded by the Excellence Initiative – Research University publication grant for SS.
Acknowledgments
The authors are very grateful to Michał Świdziński from Department of Cellular and Molecular Biology, Faculty of Biological and Veterinary Sciences, Nicolaus Copernicus University (Lwowska 1, 87-100 Toruń, Poland) for technical assistance to KN.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2024.1296372/full#supplementary-material
Supplementary Figure S1 | Density of cultivable endophytes (total density of endophytes; (A-C) and human pathogenic microorganism (HPMO, including L. monocytogenes PCM 2191 – L.m., B. cereus PCM 1948 – B.c., E. coli PCM 2561 – E.c. and S. enterica subsp. enterica PCM 2565 – S.e.; (D-F) in the roots, stems and leaves of R. sativus after one and two weeks of plant cultivation. Significant differences (p < 0.05, one-way ANOVA with Newman-Keuls post hoc comparisons) between treatments (Ctr, control, noninoculated; L.m., inoculated with L. monocytogenes PCM 2191; B.c., inoculated with B. cereus PCM 1948; E.c., inoculated with E. coli PCM 2561 and S.e., inoculated with S. enterica subsp. enterica PCM 2565) at each R. sativus organ are denoted by different letters (including capital and small letters in the case of one- and two-week-old plants, respectively); differences between values obtained for each variant of experiment (Ctr, L.m., B.c., E.c. and S.e.) in the case of tree tested organs are marked with different marks (*). The mean ± standard deviation are presented (n = 3).
Supplementary Figure S2 | The representative CLSM (confocal laser scanning microscopy) images of the negative control of FISH reaction of bacterial (E. coli, S. enterica, L. monocytogenes) colonization of an uninoculated R. sativus 1-week seedling root and shoot, bars 50 µm (A, B, D, E, G, H) and bars 30 µm (C, F, I). Only the autofluorescence of cell walls and the chloroplasts are visible, EP – epidermis, CT – cortex tissue, ST – stele tissue, P – pith.
References
Afzal, I., Shinwari, Z. K., Sikandar, S., and Shahzad, S. (2019). Plant beneficial endophytic bacteria: mechanisms, diversity, host range and genetic determinants. Microbiol. Res. 221, 36–49. doi: 10.1016/j.micres.2019.02.001
Alemu, G., Mama, M., and Siraj, M. (2018). Bacterial contamination of vegetables sold in Arba Minch Town, Southern Ethiopia. BMC Res. Notes 11:775. doi: 10.1186/s13104-018-3889-1
Andino, A., and Hanning, I. (2015). Salmonella enterica: survival, colonization, and virulence differences among serovars. ScientificWorldJournal 2015:520179. doi: 10.1155/2015/520179
Antequera-Gómez, M. L., Díaz-Martínez, L., Guadix, J. A., Sánchez-Tévar, A. M., Sopeña-Torres, S., Hierrezuelo, J., et al. (2019). Sporulation is dispensable for the vegetable-associated life cycle of the human pathogen Bacillus cereus. Microb. Biotechnol. 14, 1550–1565. doi: 10.1111/1751-7915.13816
Balali, G. I., Yar, D. D., Dela, V. G. A., and Adjei-Kusi, P. (2020). Microbial contamination, an increasing threat to the consumption of fresh fruits and vegetables in Today's world. Int. J. Microbiol 2020:3029295. doi: 10.1155/2020/3029295
Beevi, S. S., Mangamoori, L. N., Dhand, V., and Ramakrishna, D. S. (2009). Isothiocyanate profile and selective antibacterial activity of root, stem, and leaf extracts derived from Raphanus sativus L. Foodborne Pathog. Dis. 6, 129–136. doi: 10.1089/fpd.2008.0166
Berger, C. N., Brown, D. J., Shaw, R. K., Minuzzi, F., Feys, B., and Frankel, G. (2011). Salmonella enterica strains belonging to O serogroup 1,3,19 induce chlorosis and wilting of Arabidopsis thaliana leaves. Environ. Microbiol. 13, 1299–1308. doi: 10.1111/j.1462-2920.2011.02429
Brandl, M. T., Cox, C. E., and Teplitski, M. (2013). Salmonella interactions with plants and their associated microbiota. Phytopathology 103, 316–325. doi: 10.1094/PHYTO-11-12-0295-RVW
Burris, K. P., Simmons, O. D. III, Webb, H. M., Deese, L. M., Moore, R. G., Jaykus, L. A., et al. (2020). Colonization and internalization of Salmonella enterica and its prevalence in cucumber plants. Front. Microbiol. 11:1135. doi: 10.3389/fmicb.2020.01135
Carstens, C. K., Salazar, J. K., and Darkoh, C. (2019). Multistate outbreaks of foodborne illness in the United States associated with fresh produce from 2010 to 2017. Front. Microbiol. 10:2667. doi: 10.3389/fmicb.2019.02667
CDC (2012). Multistate outbreak of Salmonella Typhimurium and Salmonella Newport infections linked to cantaloupe (final update). Available at: https://www.cdc.gov/salmonella/typhimurium-cantaloupe-08-12/index.html
CDC (2016). Multistate outbreak of listeriosis linked to packaged salads produced at Springfield, Ohio dole processing facility (Final Update). Available at: https://www.cdc.gov/listeria/outbreaks/baggedsalads-01-16/index.html (Accessed October 31, 2017).
CDC (2022). Salmonella outbreak linked to onions. Available at: https://www.cdc.gov/salmonella/oranienburg-09-21/index.html
Ceuppens, S., Boon, N., and Uyttendaele, M. (2013). Diversity of Bacillus cereus group strains is reflected in their broad range of pathogenicity and diverse ecological lifestyles. FEMS Microbiol. Ecol. 84, 433–450. doi: 10.1111/1574-6941.12110
Chahar, M., Kroupitski, Y., Gollop, R., Belausov, E., Melotto, M., and Sela-Saldinger, S. (2021). Determination of Salmonella enterica leaf internalization varies substantially according to the method and conditions used to assess bacterial localization. Front. Microbiol. 12:622068. doi: 10.3389/fmicb.2021.622068
Clark, M. (2017). Multistate outbreak of Salmonella enteritidis linked to romaine lettuce. Available at: http://www.outbreakdatabase.com/details/2017-multistate-outbreak-of-salmonella-enteritidis-linked-to-romaine-lettuce/?state=60&country=US
Compant, S., Cambon, M. C., Vacher, C., Mitter, B., Samad, A., and Sessitsch, A. (2021). The plant endosphere world–bacterial life within plants. Environ. Microbiol. 23, 1812–1829. doi: 10.1111/1462-2920.15240
Cooley, M. B., Miller, W. G., and Mandrell, R. E. (2003). Colonization of Arabidopsis thaliana with Salmonella enterica and enterohemorrhagic Escherichia coli O157:H7 and competition by Enterobacter asburiae. Appl. Environ. Microbiol. 69, 4915–4926. doi: 10.1128/AEM.69.8.4915-4926.2003
Deering, A. J., Jack, D. R., Pruitt, D. R., and Mauer, L. J. (2015). Movement of Salmonella serovar typhimurium and E. coli O157:H7 to ripe tomato fruit following various routes of contamination. Microorganisms 3, 809–825. doi: 10.3390/microorganisms3040809
Denny, J., Threlfall, J., Takkinen, J., Lofdahl, S., Westrell, T., Varela, C., et al. (2007). Multinational Salmonella paratyphi B variant Java (Salmonella Java) outbreak, August-December 2007. Euro Surveill. 12:E071220.2. doi: 10.2807/esw.12.51.03332-en
Dewachter, L., Verstraeten, N., Fauvart, M., and Michiels, J. (2018). An integrative view of cell cycle control in Escherichia coli. FEMS Microbiol. Rev. 42, 116–136. doi: 10.1093/femsre/fuy005
Dierick, K., Coillie, E. V., Swiecicka, I., Meyfroidt, G., Devlieger, H., Meulemans, A., et al. (2005). Fatal family outbreak of Bacillus cereus-associated food poisoning. J. Clin. Microbiol. 43, 4277–4279. doi: 10.1128/JCM.43.8.4277-4279.2005
Dyda, A., Nguyen, P. Y., Chughtai, A. A., and MacIntyre, C. R. (2020). Changing epidemiology of Salmonella outbreaks associated with cucumbers and other fruits and vegetables. Glob. Biosec. 1. doi: 10.31646/gbio.49
Elpers, L., Kretzschmar, J., Nuccio, S. P., Bäumler, A. J., and Hensel, M. (2020). Factors required for adhesion of Salmonella enterica serowar typhimurium to corn salad (Valerianella locusta). Appl. Environ. Microbiol. 86:e02757-19. doi: 10.1128/AEM.02757-19
Esmael, A., Al-Hindi, R. R., Albiheyri, R. S., Alharbi, M. G., Filimban, A. A. R., Alseghayer, M. S., et al. (2023). Fresh produce as a potential vector and reservoir for human bacterial pathogens: revealing the ambiguity of interaction and transmission. Microorganisms 11:753. doi: 10.3390/microorganisms11030753
Farber, J. M., Ross, W. H., and Harwig, J. (1996). Health risk assessment of Listeria monocytogenes in Canada. Int. J. Food Microbiol. 30, 145–156. doi: 10.1016/0168-1605(96)01107-5
FDA (2007). FDA finalizes report on 2006 spinach outbreak. Available at: http://www.fda.gov/NewsEvents/Newsroom/PressAnnouncements/2007/ucm108873.htm (Accessed March 2014).
Flores-Urban, K. A., Natividad-Bonifacio, I., Vazquez-Quinones, C. R., Vazquez-Salinas, C., and Quinones-Ramirez, E. I. (2014). Detection of toxigenic Bacillus cereus strains isolated from vegetables in Mexico City. J. Food Prot. 77, 2144–2147. doi: 10.4315/0362-028x.jfp-13-479
Gdoura-Ben Amor, M., Siala, M., Zayani, M., Grosset, N., Smaoui, S., Messadi-Akrout, F., et al. (2018). Isolation, identification, prevalence, and genetic diversity of Bacillus cereus group bacteria from different foodstuffs in Tunisia. Front. Microbiol. 9:447. doi: 10.3389/fmicb.2018.00447
Glasset, B., Herbin, S., Guillier, L., Cadel-Six, S., Vignaud, M., Grout, J., et al. (2016). Bacillus cereus - induced food-borne outbreaks in France, 2007 to 2014: epidemiology and genetic characterisation. Euro Surveill. 21:30413. doi: 10.2807/1560-7917
Gorski, L., Duhé, J. M., and Flaherty, D. (2009). The use of flagella and motility for plant colonization and fitness by different strains of the foodborne pathogen Listeria monocytogenes. PLoS One 4:e5142. doi: 10.1371/journal.pone.0005142
Gorski, L., Flaherty, D., and Duhé, J. M. (2008). Comparison of the stress response of Listeria monocytogenes strains with sprout colonization. J. Food Prot. 71, 1556–1562. doi: 10.4315/0362-028X-71.8.1556
Gorski, L., Palumbo, J. D., and Mandrell, R. E. (2003). Attachment of Listeria monocytogenes to radish tissue is dependent upon temperature and flagellar motility. Appl. Environ. Microbiol. 69, 258–266. doi: 10.1128/AEM.69.1.258-266.2003
Gorski, L., Palumbo, J. D., and Nguyen, K. D. (2004). Strain-specific differences in the attachment of Listeria monocytogenes to alfalfa sprouts. J. Food Prot. 67, 2488–2495. doi: 10.4315/0362-028X-67.11.2488
Gu, G., Strawn, L. K., Ottesen, A. R., Ramachandran, P., Reed, E. A., Zheng, J., et al. (2021). Correlation of Salmonella enterica and Listeria monocytogenes in irrigation water to environmental factors, fecal indicators, and bacterial communities. Front. Microbiol. 11:557289. doi: 10.3389/fmicb.2020.557289
Gupta, D., and Madramootoo, C. A. (2017). Escherichia coli contamination on ready-to-eat (RTE), lettuce. Expo Health 9, 249–259. doi: 10.1007/s12403-016-0236-4
Havelaar, A. H., Kirk, M. D., Torgerson, P. R., Gibb, H. J., Hald, T., Lake, R. J., et al. (2015). World Health Organization global estimates and regional comparisons of the burden of foodborne disease in 2010. PLoS Med. 12:e1001923. doi: 10.1371/journal.pmed.1001923
Hirneisen, K. A., Sharma, M., and Kniel, K. E. (2012). Human enteric pathogen internalization by root uptake into food crops. Foodborne Pathog. Dis. 9, 396–405. doi: 10.1089/fpd.2011.1044
Hofmann, A., Fischer, D., Hartmannand, A., and Schmid, M. (2014). Colonization of plants by human pathogenic bacteria in the course of organic vegetable production. Front. Mcrobiol. 5:191. doi: 10.3389/fmicb.2014.00191
Holden, N., Pritchard, L., and Toth, I. (2009). Colonization outwith the colon: plants as an alternative environmental reservoir for human pathogenic enterobacteria. FEMS Microbiol. Rev. 33, 689–703. doi: 10.1111/j.1574-6976.2008.00153.x
Howard, M. B., and Hutcheson, S. W. (2003). Growth dynamics of Salmonella enterica strains on alfalfa sprouts and in waste seed irrigation water. Appl. Environ. Microbiol. 69, 548–553. doi: 10.1128/AEM.69.1.548-553.2003
Islam, M., Morgan, J., Doyle, M. P., Phatak, S. C., Millner, P., and Jiang, X. (2004a). Fate of Salmonella enterica serovar typhimurium on carrots and radishes grown in fields treated with contaminated manure composts or irrigation water. Appl. Environ. Microbiol. 70, 2497–2502. doi: 10.1128/AEM.70.4.2497-2502.2004
Islam, M., Morgan, J., Doyle, M. P., Phatak, S. C., Millner, P., and Jiang, X. (2004b). Persistence of Salmonella enterica serovar typhimurium on lettuce and parsley and in soils on which they were grown in fields treated with contaminated manure composts or irrigation water. Foodborne Pathog. Dis. 1, 27–35. doi: 10.1089/153531404772914437
Ivánovics, G., and Horváth, S. (1947). Raphanin, an antibacterial principle of the radish (Raphanus sativus). Nature 160, 297–298. doi: 10.1038/160297a0
Jablasone, J., Warriner, K., and Griffiths, M. (2005). Interactions of Escherichia coli O157:H7, Salmonella typhimurium and Listeria monocytogenes plants cultivated in a gnotobiotic system. Int. J. Food Microbiol. 99, 7–18. doi: 10.1016/j.ijfoodmicro.2004.06.011
Jacob, C., and Melotto, M. (2020). Human pathogen colonization of lettuce dependent upon plant genotype and defense response activation. Front. Plant Sci. 10:1769. doi: 10.3389/fpls.2019.01769
Jadoun, J., Yazbak, A., Rushrush, S., Rudy, A., and Azaizeh, H. (2016). Identification of a new antibacterial sulfur compound from Raphanus sativus seeds. Evid. Based Complement. Alternat. Med. 2016:9271285. doi: 10.1155/2016/9271285
Jayaraman, D., Valdés-Lòpez, O., Kaspar, C. W., and Ané, J. M. (2014). Response of Medicago truntula seedlings to colonization by Salmonella enterica and Escherichia coli O157:H7. PLoS One 9:e87970. doi: 10.1371/journal.pone.0087970
Jechalke, S., Schierstaedt, J., Becker, M., Flemer, B., Grosch, R., Smalla, K., et al. (2019). Salmonella establishment in agricultural soil and colonization of crop plants depend on soil type and plant species. Front. Microbiol. 10:967. doi: 10.3389/fmicb.2019.00967
Jessberger, N., Dietrich, R., Granum, P. E., and Märtlbauer, E. (2018). The Bacillus cereus food infection as multifactorial process. Toxins 12:701. doi: 10.3390/toxins12110701
Kaymak, H. C., Yilmaz, S. O., Ercisli, S., and Guvenc, I. (2018). Antibacterial activities of red colored radish types (Raphanus sativus L.). Rom. Biotechnol. Lett. 23, 13744–13749. doi: 10.26327/RBL2018.144
Kim, H. J., Koo, M., Hwang, D., Choi, J. H., Kim, S. M., and Oh, S. W. (2016). Contamination patterns and molecular typing of Bacillus cereus in fresh-cut vegetable salad processing. Appl. Biol. Chem. 59, 573–577. doi: 10.1007/s13765-016-0198-z
Kim, W. I., Ryu, S. D., Kim, S. R., Kim, H. J., Lee, S., and Kim, J. (2018). Population changes and growth modeling of Salmonella enterica during alfalfa seed germination and early sprout development. Food Sci. Biotechnol. 27, 1865–1869. doi: 10.1007/s10068-018-0412-3
Klerks, M. K., Franz, E., van Gent-Pelzer, M., Zijlstra, C., and van Bruggen, A. H. C. (2007). Differential interaction of Salmonella enterica serovars with lettuce cultivars and plant-microbe factors influencing the colonization efficiency. ISME 1, 620–631. doi: 10.1038/ismej.2007.82
Kljujev, I., Raicevic, V., Jovicic-Petrovic, J., Vujovic, B., Mirkovic, M., and Rothballer, M. (2018). Listeria monocytogenes – danger for health safety vegetable production. Microb. Pathog. 120, 23–31. doi: 10.1016/j.micpath.2018.04.034
Kulkova, I., Dobrzyński, J., Kowalczyk, P., Bełżecki, G., and Kramkowski, K. (2022). Plant growth promotion using Bacillus cereus. Int. J. Mol. Sci. 24:9759. doi: 10.3390/ijms24119759
Laksanalamai, P., Joseph, L. A., Silk, B. J., Burall, L. S., Tarr, C. L., Gerner-Smidt, P., et al. (2012). Genomic characterization of Listeria monocytogenes strains involved in a multistate listeriosis outbreak associated with cantaloupe in US. PLoS One 7:e42448. doi: 10.1371/journal.pone.0042448
Lane, D. J. (1991). “16S/23S rRNA sequencing” in Nucleic acid techniques in bacterial systematic. eds. E. Stackebrandt and M. Goodfellow (New York: John Wiley and Sons), 115–175.
Laven, R. (2006). Diagnosis of calf diarrhoea: a different perspective? UK Vet Livestock 11, 36–38. doi: 10.1111/j.2044-3870.2006.tb00004.x
Li, Y., Salazar, J. K., He, Y., Desai, P., Porwollik, S., Chu, W., et al. (2020). Mechanisms of Salmonella attachment and survival on in-shell black peppercorns, almonds, and hazelnuts. Front. Microbiol. 11:582202. doi: 10.3389/fmicb.2020.582202
Lienemann, T., Niskanen, T., Guedes, S., Siitonen, A., Kuusi, M., and Rimhanen-Finne, R. (2011). Iceberg lettuce as suggested source of a nationwide outbreak caused by two Salmonella serotypes, Newport and reading, in Finland in 2008. J. Food Prot. 74, 1035–1040. doi: 10.4315/0362-028X.JFP-10-455
Lim, H. W., Song, K. Y., Chon, J. W., Jeong, D., and Seo, K. H. (2019). Antimicrobial action of Raphanus raphanistrum subsp. sativus (radish) extracts against foodborne bacteria present in various milk products: a preliminary study. J. Milk Sci. Biotechnol. 37, 187–195. doi: 10.22424/jmsb.2019.37.3.187
Lin, Y., Briandet, R., and Kovács, A. T. (2022). Bacillus cereus sensu lato biofilm formation and its ecological importance. Biofilms 4:100070. doi: 10.1016/j.bioflm.2022.100070
Liu, D., Lawrence, M. L., Ainsworth, A. J., and Austin, F. W. (2005). Comparative assessment of acid, alkali and salt tolerance in Listeria monocytogenes virulent and avirulent strains. FEMS Microbiol. Lett. 243, 373–378. doi: 10.1016/j.femsle.2004.12.025
Luna-Guevara, J. J., Arenas-Hernandez, M. M. P., de la Peńa, C. M., Silva, J. L., and Luna-Guevara, M. L. (2019). The role of pathogenic E. coli in fresh vegetables: behavior, contamination factors, and preventive measures. Int. J. Microbiol. 2019:2894328. doi: 10.1155/2019/2894328
Majed, R., Faille, C., Kallassy, M., and Gohar, M. (2016). Bacillus cereus biofilms—same, only different. Front. Microbiol. 7:1054. doi: 10.3389/fmicb.2016.01054
Melotto, M., Panchal, S., and Roy, D. (2014). Plant inna te immunity against human bacterial pathogens. Front. Microbiol. 5:411. doi: 10.3389/fmicb.2014.00411
Miceli, A., and Settanni, L. (2019). Influence of agronomic practices and pre-harvest conditions on the attachment and development of Listeria monocytogenes in vegetables. Ann. Microbiol. 69, 185–199. doi: 10.1007/s13213-019-1435-6
Mogren, L., Windstam, S., Boqvist, S., Vågsholm, I., Söderqvist, K., Rosberg, A. K., et al. (2018). The hurdle approach–a holistic concept for controlling food safety risks associated with pathogenic bacterial contamination of leafy green vegetables – review. Front. Microbiol. 9:1965. doi: 10.3389/fmicb.2018.01965
Moteshareie, H., Hassen, W. M., Dirieh, Y., Groulx, E., Dubowski, J. J., and Tayabali, A. F. (2022). Rapid, sensitive, and selective quantification of Bacillus cereus spores using xMAP technology. Microorganisms 10:1408. doi: 10.3390/microorganisms10071408
Müller, L., Kjelsø, C., Frank, C., Jensen, T., Torpdahl, M., Søborg, B., et al. (2016). Outbreak of Salmonella Strathcona caused by datterino tomatoes, Denmark, 2011. Epidemiol. Infect. 144, 2802–2811. doi: 10.1017/S0950268816000121
Nautiyal, C. S., Rehman, A., and Chauhan, P. S. (2010). Environmental Escherichia coli occur as natural plant growth-promoting soil bacterium. Arch. Microbiol. 192, 185–193. doi: 10.1007/s00203-010-0544-1
Niedojadło, K., Hyjek, M., and Bednarska-Kozakiewicz, E. (2015). Spatial and temporal localization of homogalacturonans in Hyacinthus orientalis L. ovule cells before and after fertilization. Plant Cell Rep. 34, 97–109. doi: 10.1007/s00299-014-1690-8
O’Callaghan, M. (2016). Microbial inoculation of seed for improved crop performance: issues and opportunities. Appl. Microbiol. Biotechnol. 100, 5729–5746. doi: 10.1007/s00253-016-7590-9
Park, K. M., Jeong, M., Park, K. J., and Koo, M. (2018). Prevalence, enterotoxin genes, and antibiotic resistance of Bacillus cereus isolated from raw vegetables in Korea. J. Food Prot. 81, 1590–1597. doi: 10.4315/0362-028x.Jfp-18-205
Pitzschke, A. (2018). Molecular dynamics in germinating, endophyte-colonized quinoa seeds. Plant Soil 422, 135–154. doi: 10.1007/s11104-017-3184-2
Ponniah, J., Robin, T., Paie, M. S., Radu, S., Ghazali, F. M., Kqueen, C. Y., et al. (2010). Listeria monocytogenes in raw salad vegetables sold at retail level in Malaysia. Food Control 21, 774–778. doi: 10.1016/j.foodcont.2009.09.008
Puligundla, S., and Lim, L. (2022). Biocontrol approaches against Escherichia coli O157:H7 in foods. Foods 11:756. doi: 10.3390/foods11050756
Qu, Y., Wei, C., Dai, X., Bai, Y., Zhao, X., Lan, Q., et al. (2021). The possible transmission and potential enterotoxicity of Bacillus cereus on lettuce farms in five Chinese provinces. Front. Microbiol. 12:746632. doi: 10.3389/fmicb.2021.746632
Quilliam, R. S., Williams, A. P., and Jones, D. L. (2012). Lettuce cultivar mediates both phyllosphere and rhizosphere activity of Escherichia coli O157:H7. PLoS One 7:e33842. doi: 10.1371/journal.pone.0033842
Rosenquist, H., Smidt, L., Andersen, S. R., Jensen, G. B., and Wilcks, A. (2005). Occurrence and significance of Bacillus cereus and Bacillus thuringiensis in ready-to-eat food. FEMS Microbiol. 250, 129–136. doi: 10.1016/j.femsle.2005.06.054
Sanz-Puig, M., Pina-Perez, M. C., Criado, M. N., Rodrigo, D., and Martinez-Lopez, A. (2015). Antimicrobial potential of cauliflower, broccoli, and okara byproducts against foodborne bacteria. Foodborne Pathog. Dis. 12, 39–46. doi: 10.1089/fpd.2014.1801
Schikora, A., Carreri, A., Charpentier, E., and Hirt, H. (2008). The dark side of the salad: Salmonella ryphimurium overcomes the innate immune response of Arabidopsis thaliana and shows an endopathogenic lifestyle. PLoS One 3:e2279. doi: 10.1371/journal.pone.0002279
Schoeller, N. P., Ingham, S. C., and Ingham, B. H. (2002). Assessment of the potential for Listeria monocytogenes survival and growth during alfalfa sprout production and use of ionizing radiation as a potential intervention treatment. J. Food Prot. 65, 1259–1266. doi: 10.4315/0362-028X-65.8.1259
Self, J. L., Conrad, A., Stroika, S., Jackson, A., Burnworth, L., Beal, J., et al. (2016). Notes from the field: outbreak of listeriosis associated with consumption of packaged salad - United States and Canada, 2015-2016. MMWR Morb. Mortal. Wkly Rep. 65, 879–881. doi: 10.15585/mmwr.mm6533a6
Shenoy, A. G., Oliver, H. F., and Deering, A. J. (2017). Listeria monocytogenes internalizes in romaine lettuce grown in greenhouse conditions. J. Food Prot. 80, 573–581. doi: 10.4315/0362-028X.JFP-16-095
Shukla, S., Chatterji, S., Yadav, D. K., and Watal, G. (2011). Antimicrobial efficacy of Raphanus sativus root juice. Int J Pharm Pharm Sci 3, 89–92.
Simko, I., Zhou, Y., and Brandl, M. T. (2015). Downy mildew disease promotes the colonization of romaine lettuce by Escherichia coli O157:H7 and Salmonella enterica. Microbiology 15:19. doi: 10.1186/s12866-015-0360-5
Standing, T. A., du Plessis, E., Duvenage, S., and Korsten, L. (2014). Internalisation potential of Escherichia coli O157:H7, Listeria monocytogenes, Salmonella enterica subsp. enterica serovar typhimurium and Staphylococcus aureus in lettuce seedlings and mature plants. J. Water Health 11, 210–223. doi: 10.2166/wh.2013.164
Sun, Y., Wang, D., Ma, Y., Guan, H., Liang, H., and Zhao, X. (2019). Elucidating Escherichia coli O157:H7 colonization and internalization in cucumbers using an inverted fluorescence microscope and hyperspectral microscopy. Microorganisms 7:499. doi: 10.3390/microorganisms7110499
Szymańska, S., Płociniczak, T., Piotrowska-Seget, Z., and Hrynkiewicz, K. (2016). Endphytic and rhizosphere bacteria associated with the roots of the halophyte Salicornia europaea L.:community structure and metabolic potential. Microbiol. Res. 192, 37–51. doi: 10.1016/j.micres.2016.05.012
Szymczak, B., Szymczak, M., Sawicki, W., and Dąbrowski, W. (2014). Anthropogenic impact on the presence of L. monocytogenes in soil, fruits, and vegetables. Folia Microbiol. 59, 23–29. doi: 10.1007/s12223-013-0260-8
Torres, A. G., Zhou, X., and Kaper, J. B. (2005). Adherence of diarrheagenic Escherichia coli strains to epithelial cells. Infect. Immun. 73, 8–29. doi: 10.1128/IAI.73.1.18-29.2005
Truong, H. N., Garmyn, D., Gal, L., Fournier, C., Sevellec, Y., Jeandroz, S., et al. (2021). Plants as a realized niche for Listeria monocytogenes. Microbiology 10:e1255. doi: 10.1002/mbo3.1255
Truyens, S., Weyens, N., Cuypers, A., and Vangronsveld, J. (2015). Bacterial seed endophytes: genera, vertical transmission and interaction with plants. Environ. Microbiol. Rep. 7, 40–50. doi: 10.1111/1758-2229.12181
Valero, M., Hernandez-Herrero, L. A., Fernandez, P. S., and Salmeron, M. C. (2002). Characterization of Bacillus cereus isolates from fresh vegetables and refrigerated minimally processed foods by biochemical and physiological tests. Food Microbiol. 19, 491–499. doi: 10.1006/yfmic.507
van Elsas, J. D., Semenov, A. V., Costa, R., and Trevors, J. T. (2011). Survival of Escherichia coli in the environment: fundamental and public health aspects. ISME J. 5, 173–183. doi: 10.1038/ismej.2010.80
Walsh, K. A., Bennett, S. D., Mahovic, M., and Gould, L. H. (2014). Outbreaks associated with cantaloupe, watermelon, and honeydew in the United States, 1973–2011. Foodborne Pathog. Dis. 11, 945–952. doi: 10.1089/fpd.2014.1812
Winfield, M. D., and Groisman, E. A. (2003). Role of nonhost environments in the lifestyles of Salmonella and Escherichia coli. Appl. Environ. Microbiol. 69, 3687–3694. doi: 10.1128/AEM.69.7.3687-3694.2003
Wright, K. M., Crozier, L., Marshall, J., Mermet, B., Holmes, A., and Holden, N. J. (2017). Differences in internalization and growth of Escherichia coli O157:H7 within the apoplast of edible plants, spinach and lettuce, compared with the model species Nicotiana benthamiana. Microb. Biotechnol. 10, 555–569. doi: 10.1111/1751-7915.12596
Yaron, S., and Römling, U. (2014). Biofilm formation by enteric pathogens and its role in plant colonization and persistence. Microb. Biotechnol. 7, 496–516. doi: 10.1111/1751-7915.12186
Yu, P., Yu, S., Wang, J., Guo, H., Zhang, Y., Liao, X., et al. (2019). Bacillus cereus isolated from vegetables in China: incidence, genetic diversity, virulence genes, and antimicrobial resistance. Front. Microbiol. 10:948. doi: 10.3389/fmicb.2019.00948
Zarkani, A. A., and Schikora, A. (2021). Mechanisms adopted by Salmonella to colonize plant hosts. Food Microbiol. 99:103833. doi: 10.1016/j.fm.2021.103833
Keywords: human pathogenic microorganisms (HPMOs), Raphanus sativus, Escherichia coli, Salmonella enterica, Listeria monocytogenes, Bacillus cereus
Citation: Szymańska S, Deja-Sikora E, Sikora M, Niedojadło K, Mazur J and Hrynkiewicz K (2024) Colonization of Raphanus sativus by human pathogenic microorganisms. Front. Microbiol. 15:1296372. doi: 10.3389/fmicb.2024.1296372
Edited by:
Md. Motaher Hossain, Bangabandhu Sheikh Mujibur Rahman Agricultural University, BangladeshReviewed by:
Cedric Nicolas Berger, Cardiff University, United KingdomShaikhul Islam, Bangladesh Wheat and Maize Research Institute (BWMRI), Bangladesh
Copyright © 2024 Szymańska, Deja-Sikora, Sikora, Niedojadło, Mazur and Hrynkiewicz. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sonia Szymańska, c29uaWFzekB1bWsucGw=
 Sonia Szymańska
Sonia Szymańska Edyta Deja-Sikora
Edyta Deja-Sikora Marcin Sikora2
Marcin Sikora2 Katarzyna Niedojadło
Katarzyna Niedojadło Katarzyna Hrynkiewicz
Katarzyna Hrynkiewicz