
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 08 January 2024
Sec. Food Microbiology
Volume 14 - 2023 | https://doi.org/10.3389/fmicb.2023.1301841
This article is part of the Research Topic Zoonotic Antimicrobial Resistance and Virulence: One Health Integrated Approaches to Monitor and Reduce Food Chain Hazards View all 8 articles
Raw meat diets (RMD) for dogs are an increasingly popular alternative pet food choice, however studies worldwide have demonstrated them to be contaminated with zoonotic and antimicrobial resistant (AMR) bacteria, including bacteria resistant to critically important antibiotics. Despite this, few data exist surrounding the presence of these bacteria in RMD in the United Kingdom. The present study aimed to identify the most commonly selected RMD and non-raw diets (NRMD) by United Kingdom dog owners. Additionally, it investigated the presence of AMR-Enterobacteriaceae in samples of pre-prepared RMD and cooked commercial kibble dog foods. An online survey investigating diet preferences of United Kingdom dog owners was open for 6 weeks between February–March 2020. From this, the top 10 brands of pre-prepared raw and cooked kibble diets were ascertained and 134 samples purchased (110 RMD, 24 kibble) and subjected to microbiological testing. Bacterial enumeration of E. coli and other Enterobacteriaceae was undertaken, and the presence of Salmonella spp. and AMR-E. coli within samples determined. Whole genome sequencing was undertaken on Salmonella spp. and third-generation cephalosporin-resistant 3GCR-E. coli isolates. Pre-prepared RMD was most commonly selected by dog owners who fed RMD, and cooked commercial complete dry food was most frequently fed by owners who fed NRMD. Damaged and leaking packaging was observed in samples of RMD, alongside variability in information provided surrounding product traceability. Counts of E. coli and other Enterobacteriaceae exceeding >5,000 CFU/g were identified in samples of RMD. AMR-, extended-spectrum beta-lactamase (ESBL)-producing and 3GCR-E. coli was isolated from 39, 14 and 16% of RMD samples, respectively. Multiple antimicrobial resistance genes were identified in 3GCR-E. coli isolates. Of the ESBL encoding genes, blaCTX-M-15 was most commonly identified. S. enterica was isolated from 5% of RMD samples. No Enterobacteriaceae were isolated from any of the cooked kibble samples. The present study suggests that pre-prepared RMD available for dogs in the United Kingdom can be contaminated with zoonotic and AMR-Enterobacteriaceae. RMDs, therefore, are potentially an important One Health concern. Veterinary and medical professionals, pet food retailers and pet owners should be aware of these risks; and stringent hygiene measures should be practiced if owners choose to feed RMD.
There is a diverse range of diets currently available for dog owners to select from to feed their pets, and while conventional cooked proprietary diets (including dry and semi-moist kibbles, tins and trays of wet food) make up the majority of pet dog diets, there is increasing availability of alternative options such as home-made diets, vegetarian/vegan foods, insect-based and raw meat-based diets (RMD). Indeed, many dogs are fed diets where non-conventional options comprise some or all of the ration (Dodd et al., 2020).
RMD are an increasingly popular diet choice. Broadly, RMD are composed of uncooked animal-derived material including muscle, internal organs, bones, skin and tendons (Freeman et al., 2013). The 2022 PDSA Animal Welfare (PAW) report that surveyed a sample of dog owners which was demographically representative of the United Kingdom population, stated that 7% of dogs in the United Kingdom were fed RMD, with 5% fed pre-prepared RMD and 2% fed a home-made or home-prepared diet, although the true prevalence within the United Kingdom dog-owning population may be higher (PDSA, 2022). Additional data, such as market share, are limited regarding the actual proportion of RMD fed to dogs in the United Kingdom. However investigation of the Animal and Plant Health Agency (APHA) surveillance data from 2008 to 2018 indicated that the number of production plants registered to produce RMD increased greatly in that time period suggesting a response to increased demand (Withenshaw et al., 2020).
Raw materials which are classed as category 3 animal by-products by the Department for Environment and Rural Affairs (Defra) are allowed to be utilized in RMD for pets (Defra/APHA, 2018) This may include meat and carcasses passed fit for human consumption at the slaughterhouse, and animal material originally intended for human consumption but rejected for commercial reasons. It can also include material from animals which passed an antemortem inspection but was subsequently deemed unfit for human consumption (Defra/APHA, 2019). Regular product sampling must be undertaken for both Enterobacteriaceae and Salmonella spp. (Defra/APHA, 2020).
A variety of bacterial pathogens have been isolated from raw pet food worldwide including Escherichia coli, Salmonella spp., Campylobacter spp., Listeria spp. and Clostridium spp. (Weese et al., 2005; Strohmeyer et al., 2006; Mehlenbacher et al., 2012; Nemser et al., 2014; Bojanić et al., 2017; Van Bree et al., 2018; Hellgren et al., 2019; Bottari et al., 2020; Kananub et al., 2020; Treier et al., 2021). In the United Kingdom there have been limited studies specifically investigating the presence of bacterial pathogens in RMD, however there have been a number of recalls involving RMD reported by the Food Standards Agency due to the presence of Salmonella spp. in particular (Food Standards Agency, 2021a,b, 2022, all accessed March 2023). The number of Salmonella spp. isolations associated with raw pet food increased up to the year 2018, and this has been linked to a concurrent increase in the number of plants registered to produce RMD (Withenshaw et al., 2020). Between 2014 and 2018, the number of Salmonella spp. isolations from RMD sampled by the APHA ranged from 26 to 244 isolations per year, compared to 4–27 isolations per year for cooked commercial kibble-based food, with the highest number of isolations occurring in both RMD and cooked kibble in 2018 (Withenshaw et al., 2020). In addition, in 2017 a cluster of human cases of Shiga-toxin producing E. coli O157:H7 in the United Kingdom was epidemiologically linked to the provision of contaminated RMD containing tripe (Kaindama et al., 2020).
Escherichia coli makes up part of the normal mammalian commensal intestinal flora (Johnson and Russo, 2002), and as such, is utilized as an indicator of fecal contamination of food products (Strohmeyer et al., 2006). The EU absolute threshold for numbers of E.coli in raw pet food is 5 × 103 CFU/g [Commission Regulation (EU) No 142/2011] at the point of production, however numbers in raw pet food samples in Europe commonly exceed this (Davies et al., 2019). RMD products are often described as comprising ‘human grade’ meat which may lead to the perception of a better microbiological quality. However, an Italian study which sampled raw meat pet diet products (N = 112) which were of ‘human grade. but no longer intended for human consumption due to defects, manufacturing problems or commercial reasons, identified the presence of E. coli in 100% (N = 52) of poultry samples, 100% (N = 30) of pork samples and 93% (N = 28) of beef samples tested (Bacci et al., 2019), as well as Salmonella spp. in 12% (N = 6) of poultry and 13% (N = 4) of pork samples.
Alongside the zoonotic disease concerns, there is increasing interest surrounding the potential for raw pet foods to be a source of antimicrobial-resistant (AMR) bacteria. Of particular interest is the presence of transmissible extended-spectrum beta lactamase (ESBL)-producing, and third generation cephalosporin resistant (3GCR), Enterobacteriaceae. Such resistances are of concern; not only do they confer resistance to beta-lactam antibiotics, but also hydrolyse third generation cephalosporins including cefotaxime, ceftiofur, cefpodoxime and ceftazidime, which are highest priority critically important antibiotics (WHO, 2019), and are increasingly associated with multidrug resistance (Livermore and Hawkey, 2005; Livermore, 2008; Wedley et al., 2017). A high prevalence of ESBL-producing and 3GCR-Enterobacteriaceae has been reported in pre-prepared RMD in European studies (Nilsson, 2015; Van Bree et al., 2018; Nüesch-Inderbinen et al., 2019), as well as from meat products previously intended for the human food chain, but destined for pet food production (Bacci et al., 2019). Furthermore, RMD-feeding has been demonstrated to be a risk factor for canine fecal shedding of AMR-bacteria (Van den Bunt et al., 2020), and studies from the United Kingdom and Sweden observed that dogs fed RMD were significantly more likely to shed AMR-E. coli than those fed conventional cooked diets (Runesvärd et al., 2020; Groat et al., 2022).
Despite the interest in alternative diet choices and the growing canine and public health concerns regarding RMD, there are few studies investigating this aspect of dog ownership in the United Kingdom. Importantly, despite studies globally identifying microbiological and AMR risks associated with RMD products, currently there are no data surrounding pre-prepared RMD available for dogs in the United Kingdom. The aims of the present study were firstly to identify the most common RMD and non-raw meat-based diets (NRMD) selected by United Kingdom dog owners, their preferred treat types and from where owners obtained their dog’s food. Secondly the study aimed to investigate the prevalence of E. coli, other Enterobacteriaceae and Salmonella spp. within both commonly-purchased pre-prepared RMD and NRMD, as well as the prevalence of AMR- and ESBL-producing E. coli within the most commonly fed diets.
An online survey titled ‘A Dog’s Dinner: A survey investigating dog food selection by United Kingdom dog owners’ was created using JISC online software. The survey was available to dog owners from the whole of the United Kingdom, regardless of dog food preference and some findings including participant demographics, as well as methods of dissemination and participant recruitment, have been published (Morgan et al., 2022). The survey was advertised via social media, at Crufts Dog Show 2020 and via letters to veterinary news publications, and was available online for approximately 6 weeks from the 19th of February to the 31st of March 2020.
A sub-section of the questionnaire involved questions specifically regarding the diet fed, including the types of food chosen, the sources from where foods were obtained, the preferred meat types (RMD only), preferred treat types and the preferred brands chosen. Dog owners were directed to either a raw-feeding or non-raw feeding specific set of questions, depending on their answer to the question “Do you feed any raw animal material to your dog(s).” Owners were requested to complete this section once on behalf of all dogs in the household if they were fed the same diet, or individually for each dog that was fed differently, up to a possible total of 10 dogs per owner.
Ethical approval was granted by the University of Liverpool Veterinary Research Ethics Committee (approval number VREC913).
The sample size of participants required to achieve statistical power for the online survey was calculated to be 1,066, using an estimated prevalence of raw feeding of 50%, with 3% precision.
Analysis was undertaken using SPSS 27 [IBM Corp. (released 2020). IBM SPSS Statistics for Windows, Version 27.0. Armonk, NY: (IBM Corp.). The frequency and percentage] of responses from participants feeding either RMD or NRMD were computed. RMD were classed as those including raw animal material more than once weekly, and NRMD were classed as all diets comprising cooked material (e.g., kibble, cans, trays and sachets of cooked commercial wet food, home cooked diets, vegetarian diets, etc).
Descriptive analysis (frequency, percentage) was undertaken for both raw and non-raw food choices. Type of food preferred, source of food and types of treats were compared, and included options provided in the survey and those identified in the free text answers provided by owners. In addition, sources of non-pre-prepared raw meat were determined. Finally, the top 20 most frequently utilized brands of pre-prepared RMD and of NRMD were identified from the free text answers provided by dog owners.
The 10 most frequently utilized brands each of RMD and NRMD identified from the survey were sampled. Where a RMD brand was not readily available for purchase (due to reasons such as lack of availability, inability to courier small order sizes, or availability only as a subscription service), another brand from the top 20 was selected instead. Samples (RMD 9–15 samples per brand, total 110 samples, NRMD 1–3 samples per brand, total 24 samples) were purchased approximately monthly, one brand at a time, directly from their brand websites (RMD) or from pet shops online and in person (NRMD) between August 2020–October 2021. Sample flavors were selected based on availability at the time of purchase, and to reflect a range of meat types available per brand. All RMD samples were pre-prepared items and were supplied frozen. All NRMD samples were cooked kibble.
Each food sample was assigned a unique number, and the brand, sample type (RMD, NRMD), batch number/lot code (where present), country of origin of ingredients and whether the product was produced in the United Kingdom was recorded. Sample packets were inspected for packaging material type and any evidence of damage or leakage. To ensure no cross-contamination of samples, RMD samples were stored frozen as per manufacturer instructions and defrosted fully in a refrigerated unit prior to testing within separate containers. NRMD samples were stored at room temperature and bags were opened only at the time of sampling. All samples tested were used within the ‘use-by’ date where this was provided. Three brands did not have a ‘use-by’ date provided, however all samples were tested within 1 week of their delivery to the laboratory.
The amount of food (25 g) to be tested was collected aseptically using sterile instruments from multiple sites within the food sample, and homogenized via stomaching in a sterile plastic stomaching bag for 1 min with 225 mL of buffered peptone water (BPW), at room temperature. Approximately 20 mL of homogenate was poured into a sterile universal tube and incubated aerobically at 37°C for 18 ± 2 h. The remainder of the RMD sample was placed into a sterile sealable bag and repeat frozen at -20°C for further testing at a later date if required. NRMD bags were re-sealed and stored at room temperature.
Following incubation, a 5 μL loopful of BPW homogenate was used to inoculate one each of a chromogenic Harlequin E. coli/Coliform Agar (HECA) (Neogen, United Kingdom) and a HECA plate infused with 1 μg/mL cefotaxime (HECA+Cx), and incubated overnight at 37°C. Further homogenate (100 μL) was added to 10 mL of Rappaport Vasiliadis Broth (RVB) and incubated overnight at 42°C for Salmonella species culture.
Following incubation, the HECA plates were analyzed for the presence of typical E. coli colonies (dark blue-violet, 0.1 mm-2 mm diameter), and four such colonies were picked and plated onto nutrient agar (NA) (Neogen, United Kingdom). The HECA+Cx plates were analyzed for both typical blue E. coli and rose-pink colonies indicative of other Enterobacteriaceae (e.g., Enterobacter spp.) and 2 colonies of each (if present) were picked and plated onto NA. One 5 μL loopful of the incubated RVB was plated to Harlequin Chromogenic Agar for Salmonella Esterase (CASE) (Neogen, United Kingdom). The NA and CASE plates were incubated for 18 ± 2 h at 37°C.
Isolates which were phenotypically identified as E. coli underwent PCR for the uspA gene to confirm them as E. coli prior to undergoing whole genome sequencing. Methods were as per Anastasi et al. (2010). Primers used were CCGATACGCTGCCAATCAGT (forward) and ACGCAGACCGTAGGCCAGAT (reverse), with an amplicon size of 884 base pairs.
Following incubation, the CASE plates were analyzed for the presence of suspected Salmonella spp. (turquoise blue/green colonies) and if present, two colonies were picked and plated onto NA before overnight incubation at 37°C. Confirmation of Salmonella spp. was then undertaken via matrix-assisted laser desorption/ionization-time of flight mass spectrometry (MALDI-TOF).
The E. coli isolates from plain HECA plates, and Salmonella spp. isolates, underwent antimicrobial susceptibility testing (AST) via the disk diffusion method using seven antibiotic disks chosen based on European Committee on Antimicrobial Susceptibility Testing (EUCAST) recommendations (EUCAST, 2022). Isolates were inoculated into sterile saline using a 5 μL loop to 0.5 McFarland units then a sterile swab was used to spread the inoculated saline onto Muller-Hinton agar (Neogen, United Kingdom) and antibiotic disks were applied. Plates were incubated aerobically at 37°C for 18 ± 2 h. Antimicrobials tested were ampicillin 10 μg, amoxycillin-clavulanic acid 20 μg/10 μg, ciprofloxacin 5 μg, tigecycline 15 μg, trimethoprim-sulphamethoxazole 1.25 μg/23.75 μg, amikacin 30 μg and meropenem 10 μg (MAST Group Ltd., Liverpool United Kingdom). A susceptible control strain of E. coli (ATCC 25922) was also tested to ensure disk efficacy.
Following incubation, zones of inhibition (ZOI) for each antibiotic disk were measured to the nearest millimeter. EUCAST clinical breakpoints (EUCAST, 2022) were used for interpretation for all antibiotics other than amoxycillin-clavulanic acid, where the breakpoint used for interpretation was as recommended by the Clinical and Laboratory Standards Institute (2020).
Multidrug resistance (MDR) was defined as demonstrated phenotypic resistance to three or more classes of antibiotics tested (Magiorakos et al., 2012).
The E. coli isolates from HECA+Cx plates initially underwent extended-spectrum beta-lactamase (ESBL) double-disk test using paired disks of cefotaxime 5 μg and cefotaxime 5 μg + clavulanic acid 10 μg, and ceftazidime 10 μg and ceftazidime 10 μg + clavulanic acid 10 μg (EUCAST ESBL detection set, MAST Group Ltd., Liverpool United Kingdom). Plates were incubated at 37°C for 18 ± 2 h. Isolates which were deemed ESBL-positive, whereby the ZOI surrounding the cephalosporin +clavulanic acid disk was a minimum of 5 mm diameter larger than the ZOI for the corresponding cephalosporin disk alone for ≥1 antibiotic pair, were continued to the full AST as described. Non-ESBL producing 3GCR isolates which did not demonstrate a typical positive result for ESBL production on the double disk test, but which demonstrated a pattern suggestive of AmpC production whereby there was no, or minimal, ZOI present surrounding the clavulanic acid disk(s), were also continued to full AST.
Bacterial enumeration was undertaken for food samples using the Miles and Misra method. An initial suspension was made up to a 1/10 dilution (25 g food in 225 mL BPW) and 1 mL was then added to 9 mL BPW to make a 1/100 dilution. Three 20 μL drops of the 1/100 dilution broth were placed onto a section of a HECA plate, followed by three 20 μL drops of the 1/10 dilution broth onto a separate section. Plates were incubated overnight at 37°C.
Individual blue E. coli colonies and rose-pink colonies indicative of other Enterobacteriaceae were counted and an average of the counts of the three drops per dilution was calculated, followed by calculating the colony forming units (CFU)/g for each sample.
Bacterial counts were compared to the acceptable levels for laboratory testing of E. coli and Enterobacteriaceae presence in animal by-products (ABP) as published by Defra (Defra/APHA, 2020) accessed July 2021. For the purpose of the present study, the acceptable reference levels were those presented for one sub-sample tested per sample, where samples would fail if one sub-sample tested had greater than 5,000 CFU/g E. coli or Enterobacteriaceae.
DNA extraction was performed on phenotypically ESBL-producing E. coli isolates using the QIAamp® DNA mini kit (Qiagen, Crawley, United Kingdom).
Genomic DNA samples were submitted to the Center for Genomic Research, University of Liverpool for Illumina NEBNext Ultra II FS DNA Library Prep, which was completed following the manufacturer’s protocol. Each library was quantified using Qubit and the size distribution assessed using the Fragment Analyzer. These final libraries were pooled in equimolar amounts using the Qubit and Fragment analyzer data.
The quantity and quality of the pool was assessed by Bioanalyzer and subsequently by qPCR using the Illumina Library Quantification Kit from Kapa (KK4854) on a Roche Light Cycler LC480II according to manufacturer’s instructions.
Following calculation of the molarity using qPCR data, template DNA was diluted to 300pM and denatured for 8 min at room temperature using freshly diluted 0.2 N sodium hydroxide (NaOH) and the reaction was subsequently terminated by the addition of 400 mM TrisCl pH = 8. To improve sequencing quality control 1% PhiX was spiked-in. The libraries were sequenced on the Illumina® NovaSeq 6,000 platform (Illumina®, San Diego, United States) following the standard workflow over 1 lane of an S4 flow cell, generating 2 × 150 bp paired-end reads.
Following sequencing, reads were assembled into contigs using SPAdes and contigs smaller than <200 bps were removed. Quality control (QC) was undertaken on assemblies, and those which passed QC were subject to multi-locus sequence typing (MLST) by submitting locus allele sequences to pubmlst.org. eBURST analysis was performed to group similar isolates based on the sharing of alleles, giving each isolate a e-BURST group assignment.
Gene prediction was carried out using Prokka. Detection of AMR genes was undertaken using Resistance Gene identifier (RGI),1 and plasmids were identified using PlasmidFinder and the Enterobacteriaceae plasmid marker database.
DNA extraction and WGS was performed on Salmonella spp. isolates by the United Kingdom Health Security Agency, Gastrointestinal Bacteria Reference Unit.
Following DNA extraction, isolates were prepared for sequencing with Nextera XT DNA preparation kits, and sequenced on the Illumina HiSeq 2,500 platform in rapid run mode to produce 100 bp paired-end reads. Trimmomatic v0.40 (Bolger et al., 2014) was used to quality trim fastq reads with bases removed from the trailing end that fell below a PHRED score of 30. The Metric Orientated Sequence Type (most) v1 (Tewolde et al., 2016) was used for sequence type (ST) assignment and serotype was assigned using a combination of the Salmonella MLST database and SeqSero2 (Achtman et al., 2012; Ashton et al., 2016; Zhang et al., 2019). FASTQ sequences were deposited in the National Center for Biotechnology Information (NCBI) Sequence Read Archive under the BioProject accession number PRJNA248792.2 AMR determinants were sought using Genefinder v1–5, as previously described (Neuert et al., 2018) and using ResFinder 4.1.3 Known acquired resistance genes and resistance-conferring mutations relevant to β-lactams (including carbapenems), fluoroquinolones, aminoglycosides, chloramphenicol, macrolides, sulphonamides, tetracyclines, trimethoprim, rifamycins and fosfomycin, and acquired genes associated with colistin resistance, were included in the analysis (Neuert et al., 2018). β-Lactamase variants were determined with 100% identity using the reference sequences downloaded from the Lahey4 or National Center for Biotechnology Information (NCBI)5 β-lactamase data resources. Reference sequences for acquired resistance genes were curated from those described in the Comprehensive Antimicrobial Resistance Database6 and the ResFinder datasets.7 Chromosomal mutations were based on previously published variations in the quinolone-resistance-determining regions (QRDRs) of gyrA, gyrB, parC and parE, which are associated with resistance to quinolones.
In total, 1831 dog owners completed the online survey (n = 915 RMD-feeding, 916 NRMD-feeding), providing dietary information for 3,212 dogs (n = 1754 RMD-fed, 1,458 NRMD-fed).
The most popular types of food for dogs fed RMD (n = 1754) were pre-prepared raw meat and/or bone diets (78.1%, n = 1,369), raw eggs (62.8%, n = 1,102) and DIY/home-prepared raw meat and/or bone diets (58.8%, n = 1,032), whereas the most popular type of food for dogs fed NRMD (n = 1,458) was overwhelmingly cooked commercial complete dry food (91.1%, n = 1,326; Table 1).
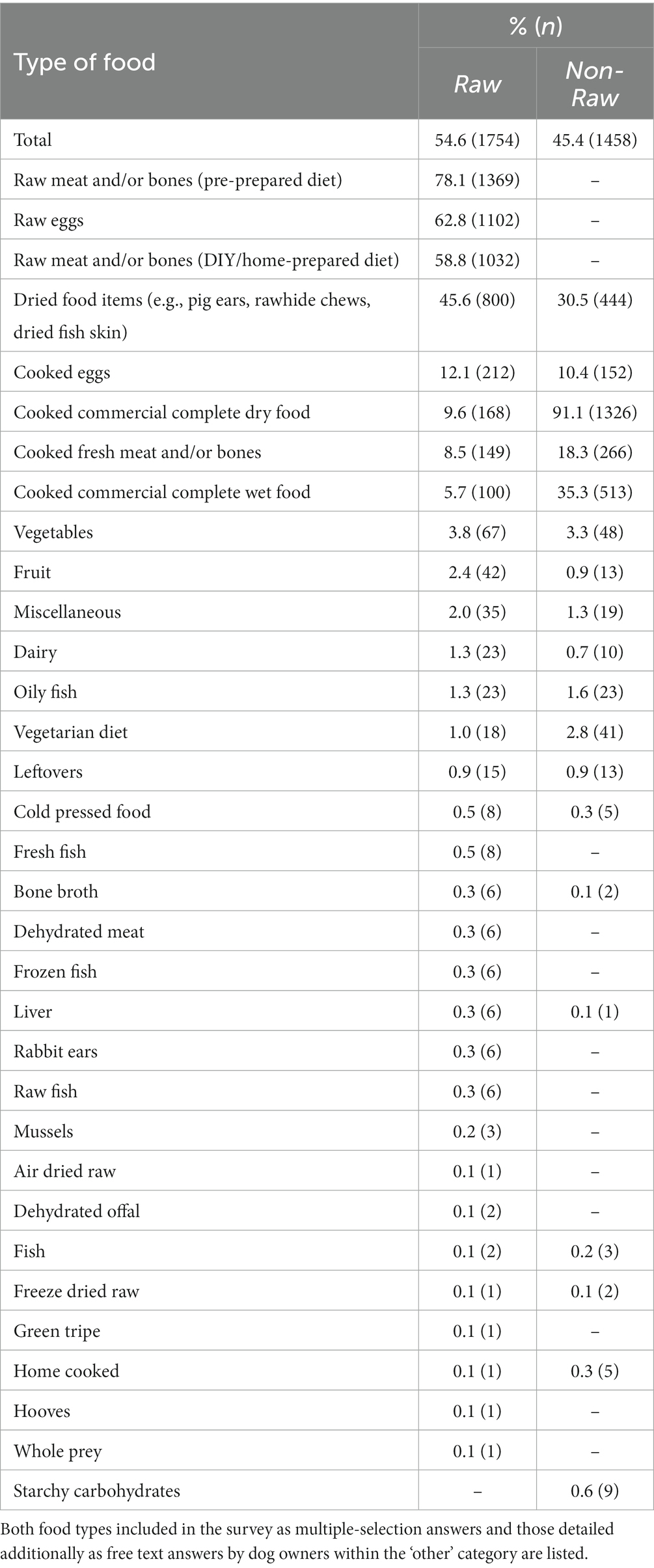
Table 1. Frequency (n) and percentage (%) of types of food provided to dogs fed RMD (n = 1754) and those fed NRMD (n = 1,458).
The main sources of food provided to dogs fed RMD were shop bought, pre-prepared frozen raw food (55.1%, n = 966), raw food from an online supplier (48.2%, n = 846) and fresh raw meat from the butcher or supermarket (41.2%, n = 723). The main source of food for dogs fed NRMD was shop bought or purchased online cooked dry kibble (84.6%, n = 1,233; Table 2). The predominant sources of non-pre-prepared raw meat for those who fed RMD were the supermarket (38.4%, n = 673) and butcher (37.8%, n = 663) (Appendix Table A1).
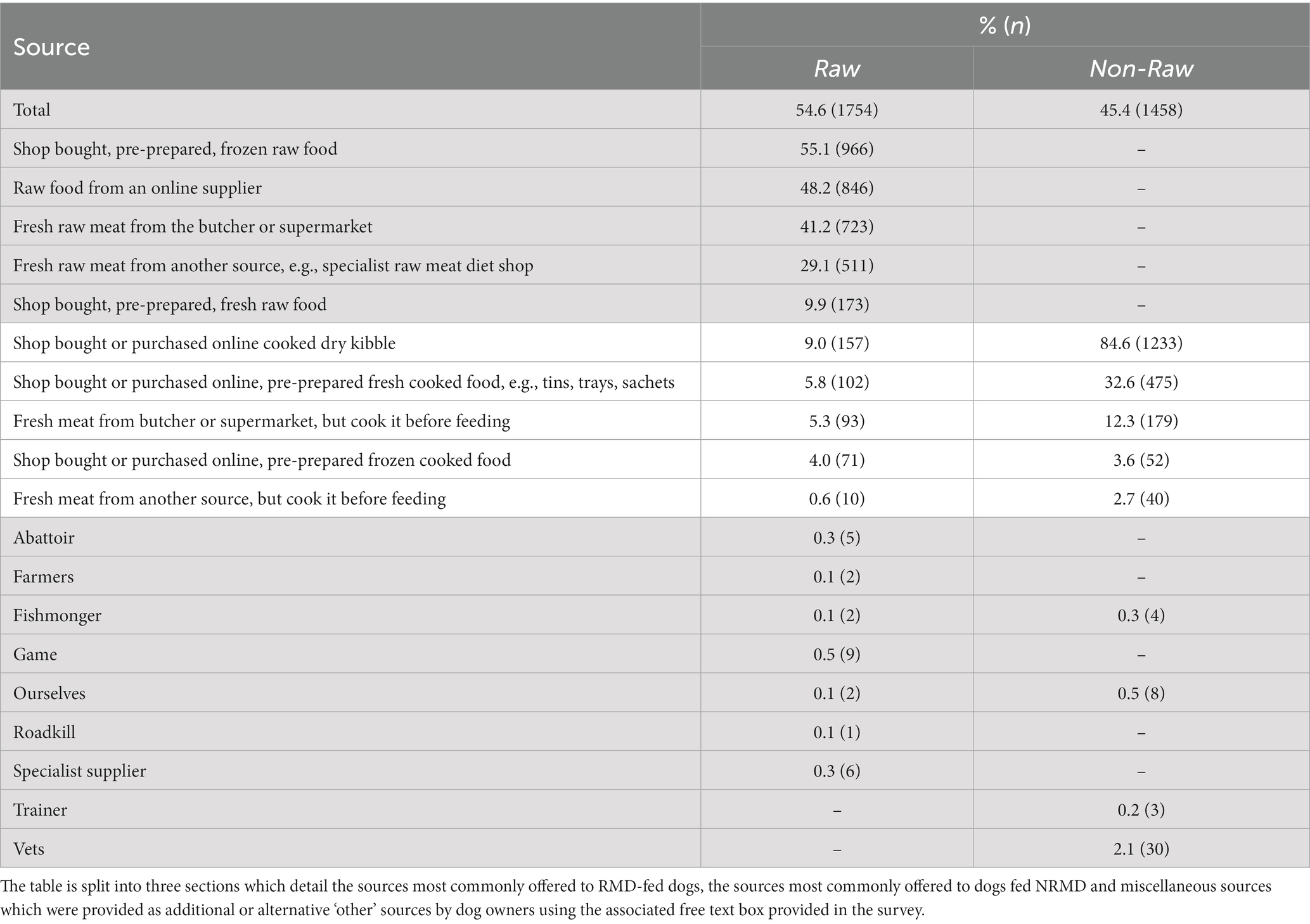
Table 2. Frequency (n) and percentage (%) of sources of the food provided to dogs fed RMD (n = 1754) diet and those fed NRMD (n = 1,458) diet.
The most commonly fed types of raw meat provided to RMD-fed dogs (n = 1754) as either a pre-prepared commercial raw diet or part of a non-pre-prepared DIY/home-prepared meal were offal (83.0%, n = 1,456), beef (82.6%, n = 1,448), lamb (79.0%, n = 1,386), chicken (78.2%, n = 1,372), turkey (75.0%, n = 1,315) and duck (72.8%, n = 1,277; Table 3). Types of raw meat that were represented at less than 2% were excluded.
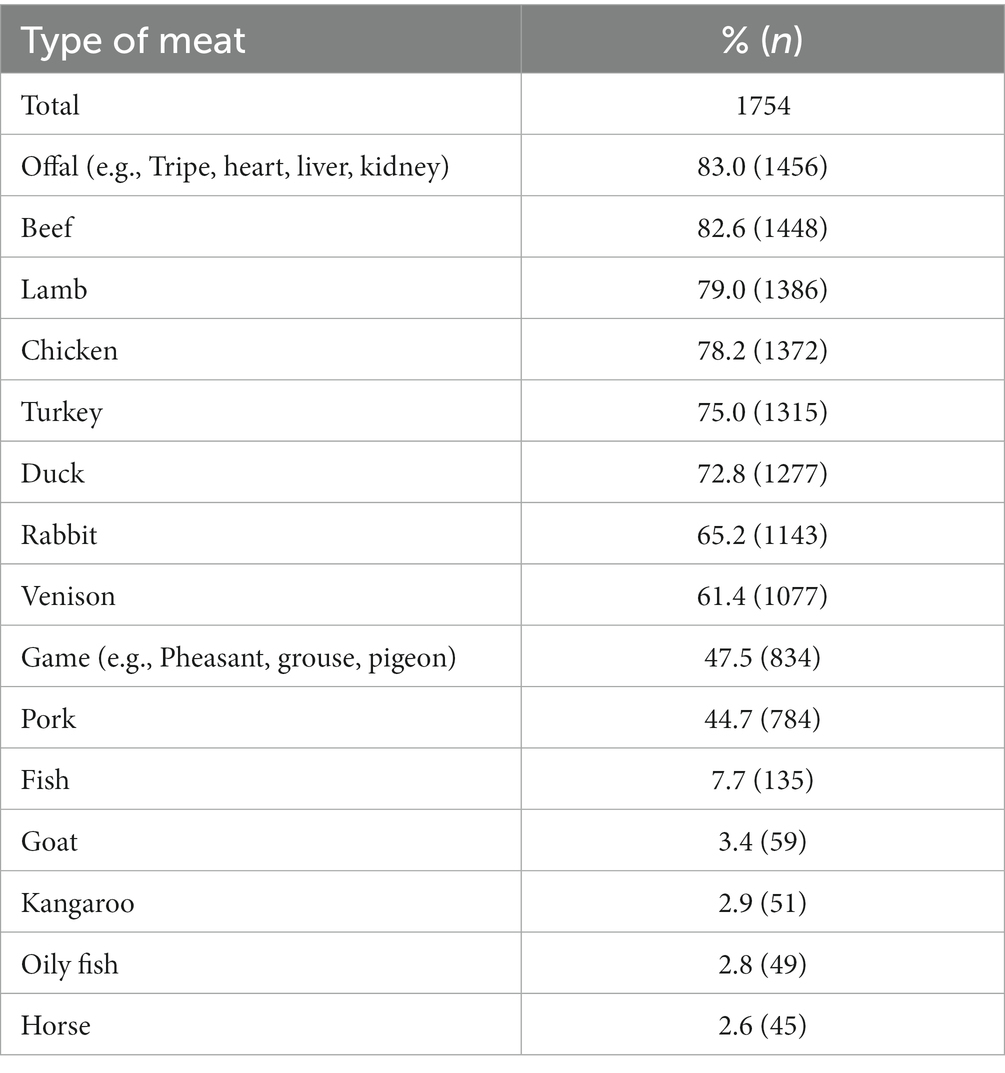
Table 3. Frequency (n) and percentage (%) of types of meat provided to dogs fed RMD (n = 1754), either as part of pre-prepared commercial raw diet or non-pre-prepared meat (meat types represented at <2% were excluded).
The preferred types of treats given to dogs fed RMD and to those fed NRMD are detailed in Appendix Table A2. The most popular types of treat for dogs fed RMD were freeze-dried meat/fish treats (56.8%, n = 997), raw bones (56.2%, n = 986) and dried treats such as chicken feet, pig ears and rawhide (55.5%, n = 973). By far, the most popular type of treat for dogs fed NRMD was shop bought cooked treats/biscuits (78.7%, n = 1,148).
Of the 10 RMD brands studied, 6 had batch numbers present on the sample packs, although it was not always clearly stated as some were present on sticky labels which became loose, or had printed numbers on the packets which were presumed to be batch numbers, although were not explicitly labeled as such (Appendix Table A3). Five brands clearly stated that the meat ingredients were sourced from the United Kingdom, and five had an unknown meat source but terminology such as ‘organic’ and ‘ethically sourced’ were used instead (Appendix Table A3). Whether the products were made in the United Kingdom was not clear for all brands, and only two brands stated specifically that they were made in the United Kingdom, however others stated they used British ingredients or used terminology such as ‘packed in the United Kingdom. The sample packs themselves were not swabbed for evidence of contamination, however samples from four brands were damaged on arrival and as such did not have sealed contents, and samples from eight brands did not have leakproof packaging. Samples from two brands were presented in cardboard packaging which subsequently became compromised on defrosting. Figures 1–3 demonstrate some of the damaged and contaminated packaging observed in this study. Figure 4 demonstrates fluid leakage in the bottom of a defrosting box following defrosting of a sample from one brand tested.

Figure 1. Shattered rigid plastic packaging and open film seal from samples from two different brands of RMD.
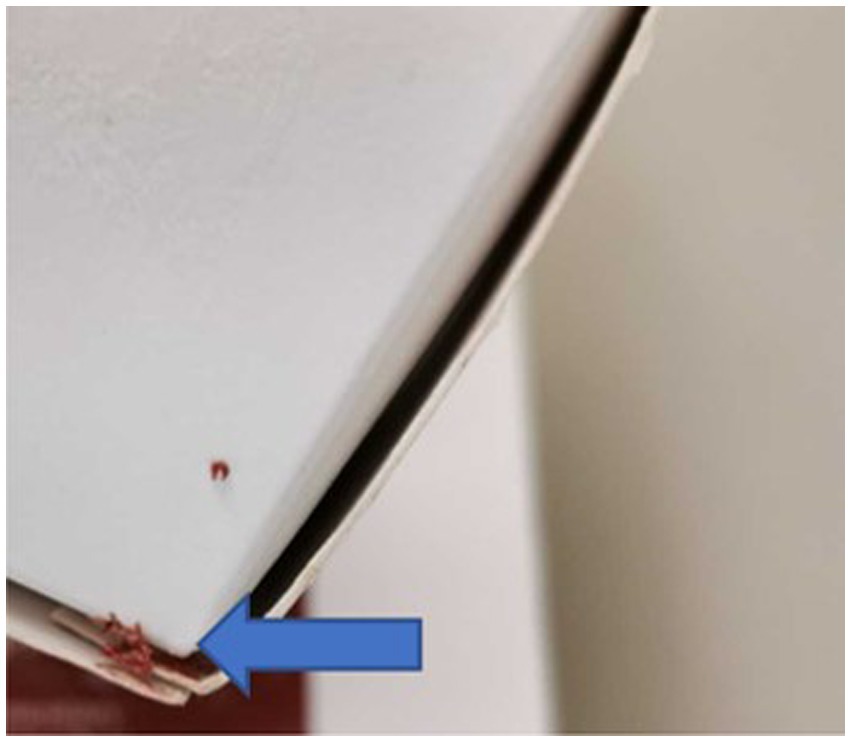
Figure 2. Frozen raw material evident on outside of cardboard packaging of RMD sample prior to defrosting.
Enumeration of E. coli and other Enterobacteriaceae was undertaken on 110 RMD samples and 24 NRMD samples from 10 brands each (Table 4, full enumeration results presented in Appendix Table A4). No bacteria were isolated from any of the NRMD samples.
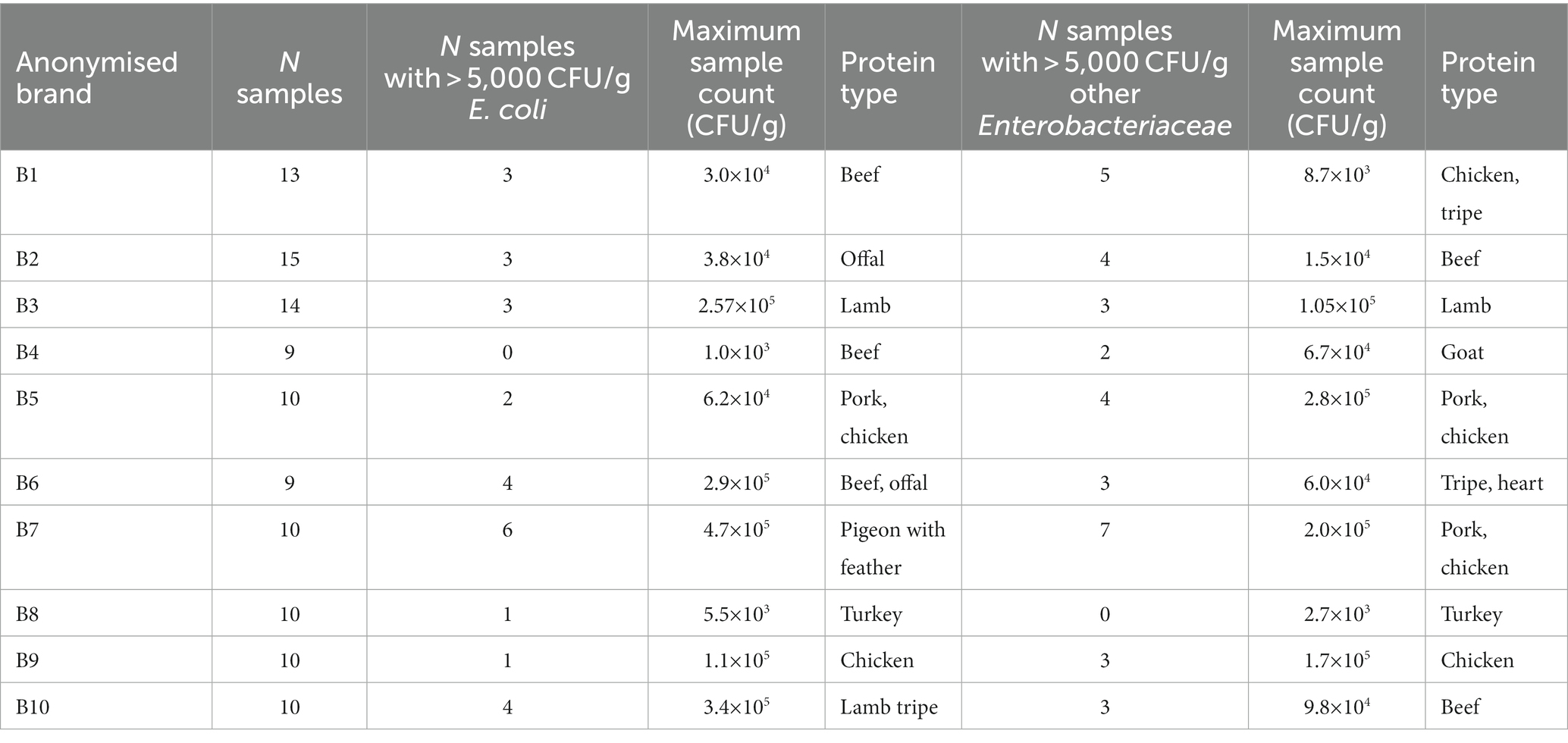
Table 4. Bacterial enumeration from RMD samples, illustrating the number of samples per brand tested, the number of samples with >5,000 CFU/g of E. coli and other Enterobacteriaceae, the maximum CFU/g of E. coli and other Enterobacteriaceae isolated within a sample from each brand, and the RMD ingredients within the sample associated with this count.
Among RMD samples, 24.5% (27/110) had counts for E. coli, and 30.9% (34/110) had counts for other Enterobacteriaceae, greater than 5,000 CFU/g, and therefore would fail Defra testing. Additionally, 20.0% (22/110) of samples had counts of both E. coli and other Enterobacteriaceae present within the same sample which each exceeded 5,000 CFU/g. Of the brands tested, 80% (8/10) had at least one sample tested which had counts of both E. coli and other Enterobacteriaceae greater than 5,000 CFU/g, and for one brand, 60% (6/10) of samples tested had E. coli counts greater than 5,000 CFU/g, and 70% (7/10) had counts of other Enterobacteriaceae greater than 5,000 CFU/g. The highest CFU/g for E. coli was associated with minced feathered pigeon, and the highest CFU/g for other Enterobacteriaceae was associated with a pork and chicken mix.
The majority of tested RMD samples grew E. coli (99.1%, 109/110) and isolates with phenotypic resistance to at least one class of antibiotic were isolated from 39.1% (43/110) of samples (Table 5). Fluoroquinolone (ciprofloxacin)-resistant E. coli was present in 8.2% of samples, and multidrug-resistant (MDR) E. coli was isolated from 7.3% (8/110) of samples. No resistance to tigecycline or meropenem was observed.

Table 5. Percentage (%) and number (N) of RMD samples with resistance to antibiotics, an antimicrobial-resistant (AMR) phenotype, and a multidrug-resistant (MDR) phenotype detected, and their associated component protein ingredients.
AMR E. coli was isolated from a number of different meat proteins, while MDR E. coli was isolated from samples containing goat, turkey and chicken only.
3GCR-E. coli (including ESBL-producing and non-ESBL producing E. coli) was identified from 16.4% (18/110) of samples of RMD, and phenotypic ESBL-producing E. coli (as determined by a positive double-disk test result) was isolated from 13.6% (15/110) of samples (Table 6).
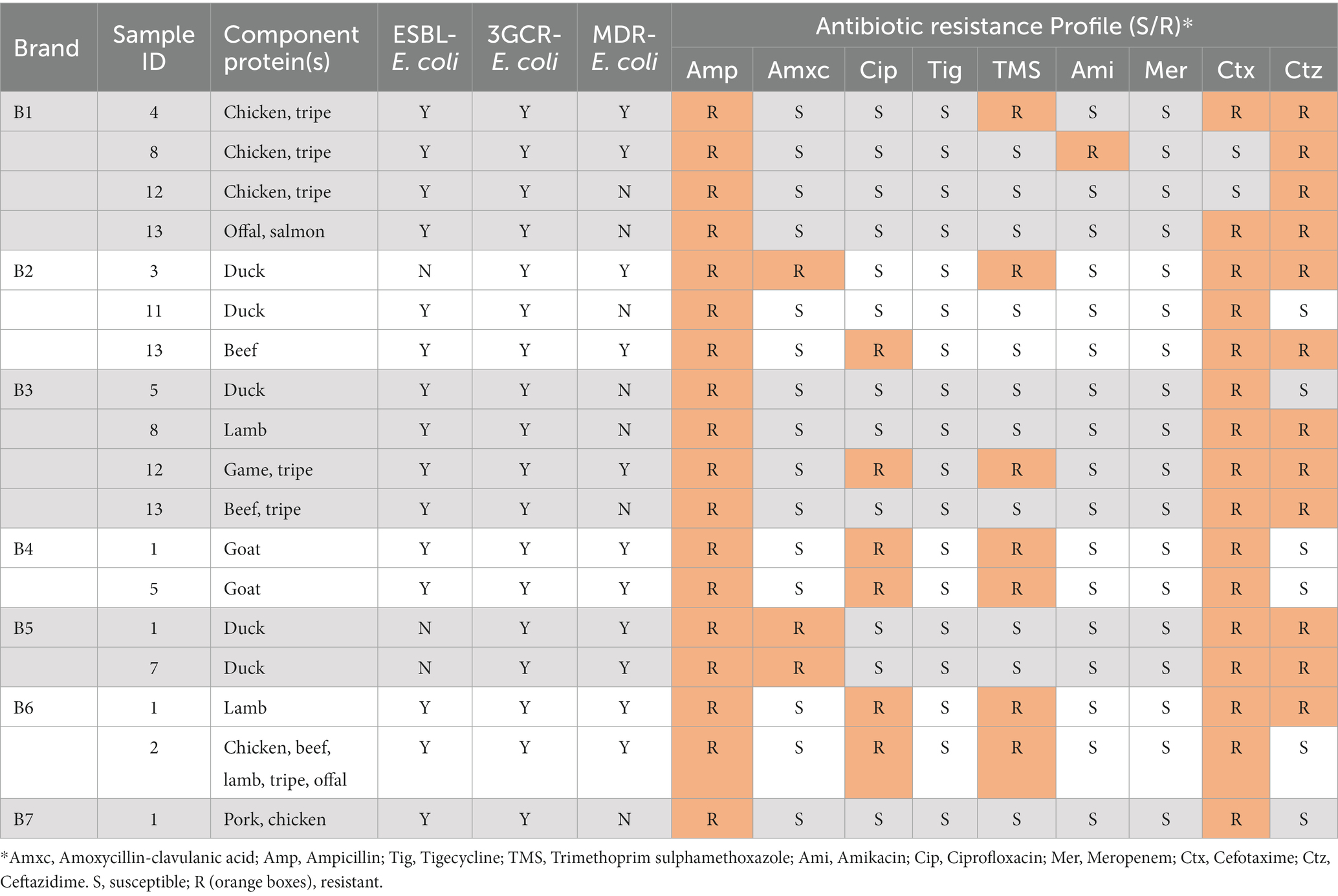
Table 6. Antimicrobial resistances observed among 3GCR-E. coli iisolates from RMD samples-aggregated sample level data.
From the 18 samples yielding 3GCR-E. coli, antimicrobial resistances were identified in the following proportions: Ampicillin 100% (n = 18), amoxycillin-clavulanate 17% (n = 3), ciprofloxacin 33% (n = 6), TMS 39% (n = 7), cefotaxime 89% (n = 16), ceftazidime 67% (n = 12). No resistance to tigecycline or meropenem was observed.
MDR-ESBL-producing E. coli was isolated from 10.0% (11/110) of samples, and fluoroquinolone resistant ESBL-producing E. coli was isolated from 5.5% (6/110). Resistances to ciprofloxacin and to TMS were observed in 40% (6/15) of samples with ESBL-producing E. coli present.
Of the samples where ESBL-producing E. coli was present, 46.7% (7/15) included tripe and/or offal as a component ingredient, and 33.3% (5/15) were composed, at least in part, of chicken. 3GCR-E. coli was most frequently isolated from samples containing offal/tripe (38.9%, 7/18) and duck (27.8%, 5/18) (Appendix Table A6).
3GCR-E. coli isolates which demonstrated ESBL-production on the double disk test, or were non-ESBL producing and suspected of having AmpC, and which demonstrated a unique resistance phenotype within a sample underwent WGS (n = 17) Of these, 13 were phenotypic ESBL-producing E. coli, as determined by the double-disk test, and four were suspected to have their ESBL phenotype ‘masked’ due to the presence of pAmpC genes. Representative isolates were sent from all food samples except brand B4 as isolates were not available.
Eleven distinct sequence types (STs) were identified. The most frequently observed ST was ST10 (n = 4). Food samples with ST10 contained duck, lamb, beef, tripe, pork and chicken. Other STs represented by more than one isolate were ST58, ST69 and ST1629 (n = 2 for each). There was no distinct relationship between the food protein types and the STs observed, other than for ST1629 where both E. coli isolates were from a combined chicken and tripe product (Table 7).
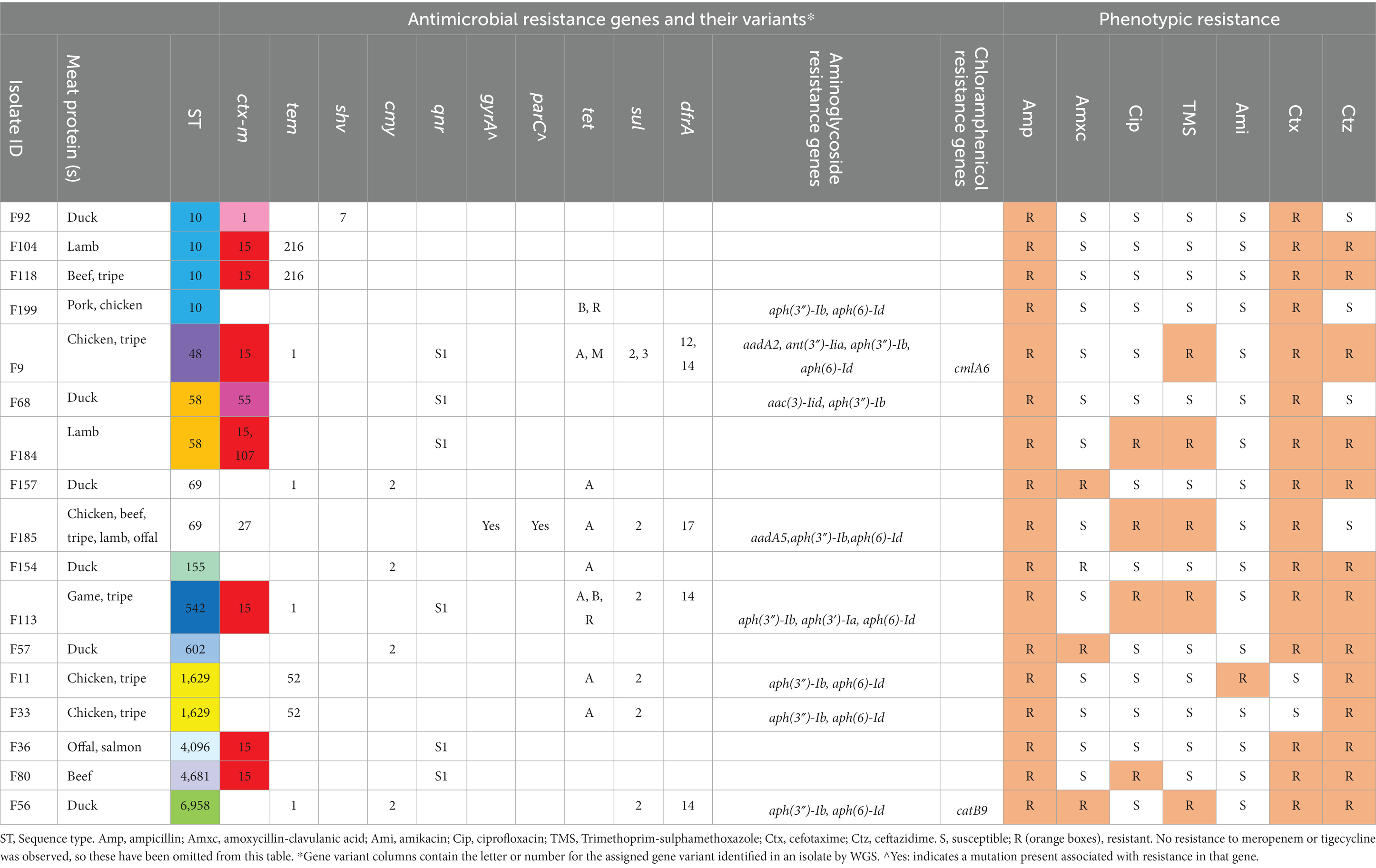
Table 7. Food protein, sequence type, phenotypic antimicrobial resistance as determined by disk diffusion and resistance genes present for ESBL-producing/3GCR- E. coli isolates from raw food samples.
Multiple AMR genes were identified (Table 7). In terms of ESBL-encoding genes, blaCTX-M genes were present in 10 isolates (59%). The most frequently identified was blaCTX-M-15, present in seven isolates (41%), which were associated with a range of STs. The blaCTX-M-1 gene was identified in one isolate, which was ST10. The blaCTX-M-27 and blaCTX-M-55 genes were present in one isolate each (ST69 and ST58, respectively). One isolate, which was ST58, carried both blaCTX-M-15 and blaCTX-M-107 genes. blaTEM genes were identified in 47% (8/17) of isolates, however the only ESBL-encoding variant isolated was blaTEM-52, which was identified in two isolates (both ST 1629). The ESBL-encoding blaSHV-7 gene was identified in one isolate (ST10) only. The blaOXA gene was not observed in any of the isolates. Five isolates did not have blaCTX-M, blaTEM or blaSHV ESBL genes present, however four of these did have the AmpC gene blaCMY-2 present (Table 7). These isolates were 3GCR, and demonstrated phenotypic amoxycillin-clavulanic acid resistance on AST.
Of the 10 MDR isolates, four were associated with the presence of blaCTX-M-15, and were ST48, ST58, ST542 and ST4681. The isolates were associated with raw food samples containing chicken (n = 1), tripe (n = 2), lamb (n = 1), game (n = 1) and beef (n = 1). The qnrS1 gene, associated with quinolone resistance, was present in 35% (6/17) of isolates. Of these, five isolates were associated with concurrent presence of blaCTX-M-15 and one isolate was associated with concurrent blaCTX-M-1. However, only three of the isolates which carried the qnrS1 gene demonstrated phenotypic fluoroquinolone resistance. Additionally, one ST69 isolate which demonstrated phenotypic fluoroquinolone resistance carried both gyrA and parC gene variants, alongside concurrent blaCTX-M-27. In terms of trimethoprim-sulphamethoxazole (TMS) resistance, the dfr gene (trimethoprim resistance) was found in 24% (4/17) of isolates, and the sul gene (sulphomethoxazole resistance) in 35% (6/17) of isolates. All isolates which carried the dfr gene also carried the sul gene, and coincided with phenotypic TMS resistance. Interestingly, two of the isolates (ST48 and ST542) which carried both dfr and sul genes, and demonstrated phenotypic TMS resistance, also carried qnrS1 and blaCTX-M-15.
Multiple genes encoding aminoglycoside modifying enzymes were present, however only one isolate (ST1629) demonstrated phenotypic resistance to amikacin, the test aminoglycoside, where resistance genes aph(3″)-Ib and aph(6)-Id were present. Additional genes of interest present which were not specifically tested for phenotypic resistance included those encoding chloramphenicol and fosfomycin resistance.
Incompatibility (Inc) group plasmids associated with ESBL genes of interest in the ESBL-producing E. coli isolates in this study are presented in Appendix Table A7. IncF was the most frequently identified plasmid group (n = 7 IncF types). Within this, plasmid type IncFIB was identified most commonly. Multiple IncF plasmids were associated with blaCTX-M-15 presence, as well as IncH and IncI group plasmids. One isolate carried blaCTX-M-55 (ST58), and as well as being associated with IncF group plasmids, it was the only isolate associated with plasmid IncX1. Four isolates carried blaCMY-2, three of which were associated with IncFIB and IncFIC, with the fourth isolate (ST602) being linked to IncI2(Delta) only. All but two of the MDR food isolates were associated with the presence of IncF plasmids. For the two that were not associated with IncF plasmids, one (ST602) was associated with IncI2(Delta), the other did not have an identified Inc. group plasmid present.
Of the RMD samples, 17.3% (19/110) had turquoise colonies present on CASE agar, indicating presumptive Salmonella spp. No presumptive Salmonella spp. colonies were isolated from NRMD samples.
Following this, five (4.5%, 5/110) RMD samples from two different brands (brands 2 and 10) were confirmed to have Salmonella enterica present (Table 8). There was a diverse range of S. enterica serotypes; S. Kottbus, S. typhimurium (monophasic), S. Indiana and S. Enteriditis. In addition, a separate subspecies, S. diarizonae, was identified.
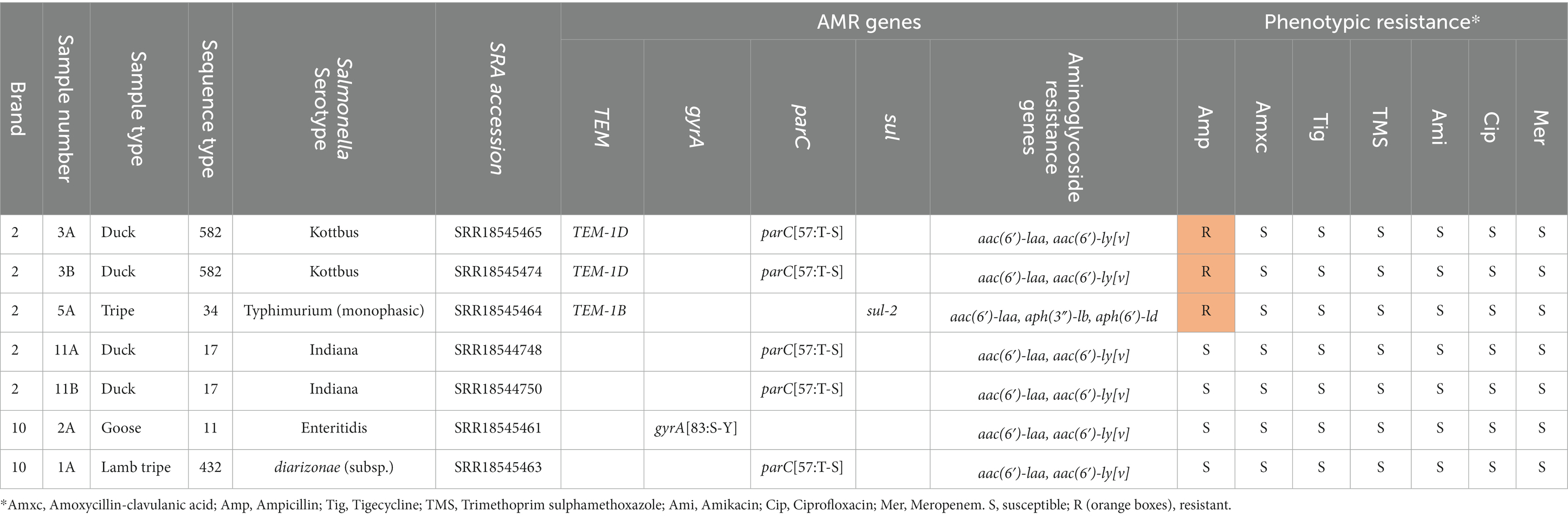
Table 8. Sample number, raw meat protein type, brand, sequence type and S. enterica serotype, alongside antimicrobial resistance (AMR) genes identified and susceptibility results of isolates confirmed as Salmonella isolated from raw food samples in this study.
Within each brand, each specific S. enterica serotype was associated with a specific food protein type. Two samples which contained duck from brand B2 were separately contaminated with two different serotypes (S. Kottbus and S. Indiana). S. Kottbus and S. typhimurium isolates demonstrated resistance to ampicillin on AST, and on WGS were found to harbor blaTEM-1 genes. Although all isolates harbored the aminoglycoside resistance gene aac(6′)-laa, no phenotypic resistance to amikacin was observed. No further phenotypic antimicrobial resistance was observed in any of the isolates.
The present study has provided important information regarding diet choices for pet dogs in the United Kingdom, and contributes to the growing body of evidence regarding the microbiological concerns surrounding RMD. RMD samples were contaminated with potentially pathogenic bacteria with zoonotic potential, and were associated with the presence of E. coli which demonstrated resistance to highest priority critically important antibiotics (HPCIAs), which are important in both human and veterinary medicine.
While conventional commercial cooked diets remain the staple diet for the majority of dogs worldwide other choices are increasing in popularity. A survey of pet owners in the United States and Australia identified that home prepared diets, raw food and table scraps comprised approximately a quarter of the diet for 17.3% of dogs, with provision of bones and raw food at least weekly for nearly a quarter of dogs (Laflamme et al., 2008). A more recent survey of dog owners from the United States, Canada, Australia, New Zealand and the United Kingdom also observed that while conventional commercial feeds were provided to the majority of pet dogs, only 13% of dogs were fed conventional commercial feeds exclusively, with many being provided additions of homemade food and/or RMD (Dodd et al., 2020). Additionally, 40.3% of the respondents of a recent internet-based survey of pet food preferences of dog owners in Brazil indicated that they fed RMD, with the majority adopting the diet within the previous year, further suggesting increasing popularity of this diet choice (Viegas et al., 2020). In the present study, approximately 50% of United Kingdom respondents indicated that they fed RMD items at least once per week, which is a higher proportion than reported previously.
While there was a broader range of food types provided to dogs fed RMD than NRMD, the most common type of RMD provided to dogs in the present study was pre-prepared raw meat and/or bones. The more frequent use of pre-prepared diets may reflect the concerns of owners regarding correct diet formulation and the desire to ensure adequate nutrition, but may also reflect convenience, brand familiarity and the increasing use of internet resources and social media for dietary information with the ready use of targeted advertising via these communication streams. Cooked commercial complete dry food was by far the most commonly provided food type for dogs fed NRMD, with >90% of NRMD-fed dogs being provided this as at least a component of the diet. This is a similar finding to previous research (Laflamme et al., 2008; Dodd et al., 2020).
Although over half of the RMD in this study was reportedly purchased frozen from a shop, nearly 50% was also purchased from an online supplier, indicating the importance of internet-based resources. There is a greater availability of products and choice online, and an added convenience of delivery straight to the consumer, and this result echoes the increasing desire for online shopping among people in general (Brand et al., 2020). However, purchasing food through this method could potentially pose further risks as delivery relies on the cold chain remaining uninterrupted, and if disruption or delay occurs at any point the RMD could be exposed to warmer temperatures, thus allowing proliferation of potentially harmful and AMR bacteria. Additionally, packaging may be damaged in transit, resulting in content leakage and contamination of external packaging. This in itself may pose a risk of transmission of potentially harmful bacteria to facilities where the products are stored on defrosting or prepared, and directly to those handling the products. The external packaging of products was not swabbed in the present study; however, this could be a consideration for future research.
The most frequently fed RMD protein sources were offal such as tripe, liver and kidney (83.0%), as well as beef, lamb, chicken, turkey and duck. These results are broadly similar to the study by Morelli et al. (2019) who observed that beef, chicken and turkey were preferred, with type of offal analyzed separately, and Groat et al. (2022), who observed that most dogs fed RMD were fed a mix of meats, with chicken, red meat and tripe being the most frequently chosen. In the present study, approximately half of dogs fed RMD were fed freeze-dried meat/fish, dried foodstuffs such as pig ears, chicken feet and hide and raw bones as treats, which may again echo the desire indicated previously by owners feeding RMD to provide non-processed products which are perceived as ‘more natural’ in general (Bulochova and Evans, 2021b).
Escherichia coli was isolated from all but one of the RMD samples tested, however no E. coli was cultured from any of the NRMD kibble samples. This is in agreement with recent research from the USA where similarly, no E. coli was isolated from samples tested of commercially available conventional diets with no uncooked components (Gibson et al., 2022). Freezing raw meat is often believed to reduce or negate the risks associated with any microbiological contaminants present. However, as demonstrated in this study, this is not the case for E. coli and other Enterobacteriaceae, or Salmonella spp. Importantly, the freezing process did not kill these bacteria.
In the present study, E. coli and other Enterobacteriaceae were present at >5,000 CFU/g, therefore exceeding Defra/APHA sub-sample acceptable thresholds, in a quarter and a third of RMD samples tested, respectively. Nine out of 10 brands tested had at least one sample tested which had counts of E. coli or other Enterobacteriaceae which were greater than those deemed acceptable by Defra/APHA. This highlights that pre-prepared RMD samples tested were frequently contaminated with bacteria which can be pathogenic and cause zoonotic disease, often to a concerningly high degree. This finding is in agreement with those of previous studies (Weese et al., 2005). In one Swedish study, bacteria belonging to the family Enterobacteriaceae, including E. coli, was present at a level which exceeded EU regulations for raw meat intended for pet food production in 52% of RMD samples (Hellgren et al., 2019), and in a study from Switzerland, 73% of samples tested exceeded these limits (Nüesch-Inderbinen et al., 2019). This was also the case for frozen commercially available RMD in Thailand (Kananub et al., 2020) and in Italy, where samples were found to be contaminated with Salmonella spp., E. coli O157:H7, Listeria monocytogenes and Campylobacter spp., despite freezing (Bottari et al., 2020). In another study from Italy, RMD products purchased online and received frozen were found to be highly contaminated with E. coli immediately following defrosting, as well as having Listeria spp., Clostridia spp. and Yersinia spp. present, and were suggested to be of poor microbiological quality initially, but demonstrated distinct worsening of quality if products were improperly refrigerated, or not utilized immediately following defrosting (Morelli et al., 2020).
All brands of food tested in the present study were received frozen, stored at -20°C, then defrosted overnight in the fridge prior to testing. While some bacterial multiplication could have occurred during the defrosting process, this is unlikely due to the refrigeration throughout, and rapid processing of samples once defrosted. If there were any breaks in the cold chain during the packing and delivery process, this may have allowed bacterial multiplication. However, this mirrors the process by which owners would receive the foods, therefore it is representative of the microbiological quality of the products received by consumers. All packs were received with insulating packing of different types, and some were more successful at keeping foodstuffs frozen than others, with some leakage of package contents identified in some cases. Nevertheless, it is most likely that a high degree of bacterial contamination was already present in the samples, and this highlights the importance of safe storage (refrigeration at 0-4°C) and defrosting processes for these diets. Additionally, it highlights that RMD products may have poor microbiological quality prior to freezing, thus more needs to be done in manufacturing to minimize contamination, both at source by reviewing the raw materials utilized or during the production process. Previous research has demonstrated that dog owners utilize a number of different methods for defrosting and preparing RMD (Bulochova and Evans, 2021a; Morgan et al., 2022), with poor practices regularly employed, potentially indicating some confusion as to appropriate measures for defrosting RMD. Defrosting processes have been demonstrated previously to be important for food safety, and time–temperature abuse has been shown to be an important factor in the increase in bacteria in contaminated raw meat products, thus increasing the risk of foodborne disease (Roccato et al., 2015). Not all brands tested included detailed instructions for safe defrosting of the product on their product packaging. This highlights an area where improvement is needed regarding safe handling of RMD.
It is concerning that RMD were frequently contaminated with AMR E. coli. AMR is a global One Health threat; it has been estimated that in 2015, bacterial AMR infections accounted for 33,000 human deaths in the European Union (EU) and European Economic Area (EEA), with the burden highest in those <1 year and > 65 years old (Cassini et al., 2019). A more recent study estimated that in 2019, 4.95 million deaths globally were associated with bacterial AMR, and 1.27 million deaths were directly attributable (Murray et al., 2022). Furthermore, the United Kingdom 2016 Review on Antimicrobial Resistance estimated that without intervention, as many as 10 million human deaths worldwide could be attributable to AMR by 2050 (O’Neill, 2016).
Approximately 16% of RMD samples tested had 3GCR-E. coli present, and 14% had ESBL-producing E. coli present. Additionally, 10% of samples tested had MDR ESBL-producing E. coli present, with phenotypic resistance to TMS and/or ciprofloxacin observed alongside ESBL-production within many of these isolates. AMR E. coli was not isolated from any NRMD samples, a finding similar to that of Baede et al. (2017). It is concerning that E. coli which demonstrated concurrent phenotypic resistance to both fluoroquinolones and third generation cephalosporins (cefotaxime, ceftazidime) was isolated from approximately 6% of RMD samples. Both of these antibiotic classes are HPCIAs as determined by the World Health Organization (Veterinary Medicines Directorate, 2015; Collignon et al., 2016). The presence of ESBL-producing and 3GCR-E. coli was associated most frequently with samples containing offal/tripe and poultry meat (chicken and duck respectively), however there was no distinct link between meat type and the presence of phenotypic fluoroquinolone resistance, with resistance demonstrated in E. coli isolated from RMD samples containing a range of proteins. These meat types were often mixed in combinations in the food samples, however there were single-protein samples of goat, lamb and beef.
The prevalence of 3GCR and ESBL-producing E. coli in pre-prepared RMD samples in the present study is lower than that previously reported by smaller studies in Europe. A study of 51 samples of RMD available in Switzerland observed that approximately 61% of samples tested had ESBL-producing E. coli present, with the majority of affected samples involving products of cattle or poultry origin (Nüesch-Inderbinen et al., 2019). An additional smaller study of 35 samples from eight brands available in The Netherlands reported that 80% of RMD samples had ESBL-producing E. coli isolated (Van Bree et al., 2018). Finally, a study from Sweden identified that 23% of 39 samples tested had 3GCR- E. coli present (Nilsson, 2015), and all of the 3GCR E. coli harbored the blaCMY-2 gene, which was also the most frequently observed blaCMY gene in the present study.
There remains limited evidence currently regarding the AMR risks specifically from pre-prepared RMD available in the United Kingdom and elsewhere for comparison. However, there are studies of AMR- E. coli contamination in meats destined for the human food chain and for pet food. One national study of meat samples purchased from retailers for human consumption in the United Kingdom identified that 65% of chicken samples had ESBL-producing E. coli present (Day et al., 2019), another more localized study identified ESBL-producing E. coli in 18% of meat products from United Kingdom supermarkets, with the majority of products being chicken (Ludden et al., 2019). While the majority of products were of United Kingdom origin, products were also imported from a range of other countries, highlighting the multinational origin of meat products entering both the human and pet food chains. Finally, a study from Italy identified ESBL-producing and MDR E. coli in meat products that were originally packaged at a mass retailer for human consumption but became pet foods once they were deemed no longer suitable for human consumption (Bacci et al., 2019). It is important to note that meat sold for human consumption would be intended to be cooked, which would mitigate the risk of AMR-bacteria (James et al., 2021). A concern regarding DIY/home-prepared raw diets is that the meats used are still likely to harbor zoonotic and AMR bacteria, whereas pre-prepared RMDs should undergo testing to ensure bacteria do not exceed acceptable levels; it is not possible to measure the risk posed by meats from unknown sources prepared within the home. However, it could also be argued that there is potential for more opportunity for cross-contamination within pre-prepared diets in the manufacturing process, particularly in minced products where more than one protein type is included.
In the present study, while a United Kingdom origin for meat ingredients was stated on the sample packets for 50% of the RMD brands tested, the remainder did not specifically state the country of origin of the meats used. Additionally, 60% of the brands tested had a batch number clearly present on the sample packets, but whether the food was produced in the United Kingdom was not clear for a number of brands. This is a concern because it would seem that there is a lack of traceability and provenance of product present, which would prove an issue if there was an outbreak of disease potentially associated with the product. The importance of traceability of RMD ingredients was highlighted when raw hare meat intended for use in RMD was found to be contaminated with Brucella suis, and had been imported into the United Kingdom from The Netherlands after originating in Argentina (Frost, 2017). Therefore, this is potentially an area of RMD production which requires attention.
On whole genome sequencing, a number of sequence types (STs) were identified in ESBL-producing E. coli isolates, with the most frequently encountered being ST10. ST10 E. coli is frequently associated with ESBL-genes, in particular, blaCTX-M genes (Oteo et al., 2009; Cormier et al., 2019). Other STs of interest which were identified in ESBL-producing E. coli isolated from RMD samples in this study were ST58, ST69 and ST155. E. coli ST58 and ST69 are globally disseminated uropathogens and have previously been associated with blaESBL and AMR gene carriage (Novais et al., 2013; De Souza da-Silva et al., 2020; Reid et al., 2022). E. coli ST58 has been isolated from livestock and food sources previously (Reid et al., 2022), including raw pet food (Nüesch-Inderbinen et al., 2019). E. coli ST155 is an important ExPEC (extraintestinal pathogenic E. coli) strain with zoonotic potential, previously identified in beef cattle faces, chicken meat and human blood in the United Kingdom, as well as in RMD samples (Ludden et al., 2019; Nüesch-Inderbinen et al., 2019).
A variety of AMR genes were identified in the ESBL-producing E. coli isolates from RMD samples. The predominant blaESBL gene identified was blaCTX-M-15, present in 41% of isolates. Presence of the blaCTX-M-15 gene was frequently observed alongside co-carriage of additional plasmid-mediated resistance genes such as qnrS1, which mediates fluoroquinolone resistance, and genes encoding resistance to other antibiotic classes such as tetracyclines, trimethoprim-sulphamethoxazole and aminoglycosides. The predominance of blaCTX-M-15 is of concern as it is carried on mobile transferrable genetic elements which frequently harbor resistance genes to other antimicrobials, including fluoroquinolones, thus increasing the risk of conferring MDR (Baba Ahmed-Kazi Tani et al., 2013). Only one isolate demonstrated the presence of blaCTX-M-1. Again, there is little data available from pre-prepared RMDs for comparison, however this finding contrasts with the findings of Nüesch-Inderbinen et al. (2019), who observed that blaCTX-M-1 was the most frequently detected blaESBL gene in ESBL-producing E. coli isolated from RMD samples commercially available in Switzerland, although blaCTX-M-15 was the second-most frequently detected blaESBL gene.
Isolates harboring the blaCTX-M-15 gene in the present study were not associated with any specific meat protein type. However, the blaCTX-M-15 gene has also been isolated from ESBL-producing E. coli from livestock in the United Kingdom, including from pig meat (Veterinary Medicines Directorate, 2022), and from faces of chicken, beef and pigs (Ludden et al., 2019).
Although blaCTX-M genes were the predominant blaESBL genes in this study, blaTEM-52 was also isolated from two RMD samples containing a combination of chicken and tripe. This ESBL-gene has previously been observed in E. coli isolated from United Kingdom produced broiler chickens and turkeys (Randall et al., 2011). Finally, the plasmid-mediated AmpC (pAmpC) resistance gene blaCMY-2 was identified in four samples, all of which were raw duck, were single-protein, and obtained from two different suppliers. In all cases, isolates were resistant to ampicillin, amoxycillin-clavulanic acid and a third-generation cephalosporin. This gene has been identified in broilers and chicken meat within Europe previously (Voets et al., 2013; Solà-Ginés et al., 2015), and from a ducks in China (Ma et al., 2012; Zheng et al., 2022), however to the authors’ knowledge this is the first report of blaCMY-2 being present in products containing duck meat in the United Kingdom, although the country of origin of the meat was unknown, again highlighting the importance and need for improved traceability of RMD ingredients.
It is concerning that such a range of AMR genes was present, frequently in combination, within RMDs in the present study. These genes are potentially transmissible through mechanisms such as mobile plasmids and as such these isolates could act as a reservoir for MDR. There are a multitude of RMD brands in the United Kingdom, which utilize meat products sourced from both within the United Kingdom and abroad, therefore larger scale studies are required to investigate the problem with regards to AMR E. coli presence in United Kingdom-fed RMDs further, nevertheless the findings of the current study indicate that contamination with AMR E. coli is also a problem with RMD fed to dogs in the United Kingdom. This is concerning from an animal health and welfare point of view, but also presents a potential public health risk. Dogs fed RMD have been shown to shed AMR E. coli in their faces, and the provision of a raw diet has been demonstrated to be a risk factor for the carriage of ESBL-producing E. coli by healthy dogs (Schmidt et al., 2015; Wedley et al., 2017; Runesvärd et al., 2020; Van den Bunt et al., 2020; Groat et al., 2022).
Five (4.5%) RMD samples from two brands were contaminated with S. enterica, with five different serotypes/subspecies identified, each associated with a unique meat protein type, however most isolates were susceptible to antimicrobials. The reported prevalence of Salmonella spp. contamination in RMD in studies in Europe, Canada and the USA is wide ranging, from 4 to 25% (Weese et al., 2005; Strohmeyer et al., 2006; Mehlenbacher et al., 2012; Nemser et al., 2014; Hellgren et al., 2019; Nüesch-Inderbinen et al., 2019), therefore the prevalence identified in the present study is at the lower end of that range. However, no meat containing Salmonella spp. should be present within pet food at retail, and samples which test positive for Salmonella spp. at production should be removed from entering sale, therefore the presence of any Salmonella spp. contamination is concerning. S. Enteriditis and S. typhimurium (monophasic) are among the top five serotypes resulting in human infection reported to the United Kingdom Health Security Agency (UKHSA) (Chattaway et al., 2019), however all serotypes present in the RMD samples in the present study have the potential to cause disease in humans, again highlighting the public health concerns associated with RMD. Concerningly, a study of experimentally-inoculated raw meat identified that Salmonella spp. persisted in pet food bowls despite standard cleaning methods, including bleach, scrubbing with soap and washing in the dishwasher (Weese and Rousseau, 2006). Furthermore, while much of Salmonella spp. contamination occurred in samples containing poultry, S. enterica subspecies diarizonae was isolated from a sample containing lamb tripe. This is unsurprising as S. enterica subsp. diarizonae is commonly associated with reptiles and sheep, and although uncommon, invasive human infections with S. enterica subsp. diarizonae have been reported (Giner-Lamia et al., 2019), demonstrating zoonotic potential. It is noteworthy that dried raw treats were frequently chosen by respondents in present study, however Salmonella spp. has also been isolated from dried raw pet treats in the United Kingdom (Morgan et al., 2023).
The presence of Salmonella spp. in RMD samples is not only a risk to public health, but it also poses a risk to animal health and welfare. The provision of Salmonella-contaminated RMD to pets has been implicated as a cause of mesenteric lymphadenitis in two dogs (Binagia and Levy, 2020), diarrhea and death in Greyhound puppies (Morley et al., 2006), enterocolitis and death in a series of puppies and kittens (Jones et al., 2019) as well as being highly suspected as the cause of two cases of salmonellosis in cats (Giacometti et al., 2017).
There were some limitations to the present study. Approximately 50% of respondents indicated that they fed RMD at least once per week. However, this is unlikely to be representative of the diet choices of the dog owning population in the United Kingdom, with RMD likely being over-represented due in part to the participant recruitment methods and self-selection bias due to a high uptake of the survey within the RMD feeding community, therefore this finding must be interpreted with caution. Nevertheless, this result may be indicative of the interest surrounding RMD and, as in other countries, the popularity of RMD within the United Kingdom is likely to be increasing. Additional research is required to validate this further.
The food brands chosen to test were selected based on the preferences of the dog owners who responded to the survey, not on market share, therefore may not be fully representative of the possible levels of contamination present in dog foods (RMD and NRMD) available in the United Kingdom. There are a multitude of brands available, and different brands which were not sampled may have different levels of contamination. Due to time and financial constraints and product availability at the time of purchase, only a limited number of samples were tested per brand, particularly of NRMD. This may have underestimated the contamination present within a brand, or indeed may have overestimated if a particularly contaminated batch was tested. Additionally, for RMD, bacterial proliferation could have occurred due to a break in the cold chain in transit during the order packing and delivery process, or during defrosting, although this was deemed unlikely as discussed earlier. Furthermore, the present study only tested pre-packaged samples of RMD, and did not include home-prepared/DIY diets, which may have differing levels of contamination as discussed earlier. It also focused on the presence of Enterobacteriaceae, and did not screen for other bacteria with zoonotic potential such as Campylobacter spp. or Listeria spp. There may be an underestimation of the presence of Salmonella spp. in the food samples as the method of isolating Salmonella spp. using the CASE agar is likely to have selected only for S. enterica, which may mean that a small number of other Salmonella subspecies could have been missed.
There was a slight deviation from EUCAST AST methods in that the incubator temperature utilized in the present study was 37°C, instead of 35 ± 1°C, however all isolates which were phenotypically ESBL-producing on the double disk test were subjected to WGS for confirmation of the presence of ESBL resistance genes. WGS was only undertaken on 3GCR and ESBL-producing E. coli isolates, and further WGS on non-ESBL E. coli was beyond the scope of the present study. There may be further resistance genes of interest in the AMR- E. coli isolates which were not 3GCR/ESBL-producing, and this warrants further investigation. Furthermore, analysis of E. coli and Salmonella spp. virulence factors present would provide further depth surrounding the potential health risks associated with RMD. Finally, we were only able to identify the plasmids present which were associated with ESBL gene presence, however further in-depth investigation is required to determine which plasmids genes were carried on specifically, and how transmissible these may be.
In conclusion, this study has demonstrated a number of concerns surrounding RMDs for dogs available in the United Kingdom. RMD samples were frequently contaminated with fecal bacteria with potential to cause disease in both humans and animals. In addition, there were concerning levels of AMR-E. coli present, with resistance to more than one highest priority critically important antibiotic class demonstrated. Furthermore, concerns surrounding packaging damage, leaks and limited traceability in some cases were evident. Therefore, RMD for pets could pose an important human and animal health risk. Pre-prepared RMDs are often sold as ‘human-grade. which may suggest a perceived greater level of quality and safety, however all meats which are utilized within RMDs are graded as at least Defra category 3 ABPs, and as demonstrated, this does not negate the microbiological risks. It is crucial that veterinary professionals, medical staff, pet food retailers and dog owners are aware of these risks, and if dog owners do choose to feed a RMD, it is vital that strict hygiene measures are practiced throughout the food storage, defrosting and preparation processes, including using separate food storage and preparation facilities, practicing thorough hand washing, and disinfection of food bowls and food preparation areas after feeding.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.
GM: Data curation, Formal analysis, Investigation, Supervision, Writing – original draft. GP: Conceptualization, Formal analysis, Funding acquisition, Supervision, Visualization, Writing – review & editing. ET: Data curation, Writing – original draft. MC: Data curation, Formal analysis, Investigation, Methodology, Software, Visualization, Writing – review & editing. VS: Funding acquisition, Supervision, Writing – review & editing. NW: Conceptualization, Funding acquisition, Methodology, Project administration, Supervision, Visualization, Writing – review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This study was joint funded by the Veterinary Medicines Directorate and the University of Liverpool as part of a wider PhD project.
MC is affiliated to the National Institute for Health Research Health Protection Research Unit (NIHR HPRU) in Genomics and Enabling Data at University of Warwick in partnership with the United Kingdom Health Security Agency (UKHSA) MC is based at UKHSA. With thanks to the Veterinary Microbiology Diagnostic Laboratory at the University of Liverpool for undertaking MALDI-TOF analysis, to the Leahurst laboratory technicians at the University of Liverpool for preparing the media required for this study, and to the Veterinary Medicines Directorate (VMD) for their review and feedback on this paper. Finally, the authors extend our great thanks to all of the dog owners who participated in the survey component of this study.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The views expressed are those of the author(s) and not necessarily those of the NIHR, the Department of Health and Social Care or the United Kingdom Health Security Agency.
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2023.1301841/full#supplementary-material
1. ^https://card.mcmaster.ca/analyze/rgi
2. ^www.ncbi.nlm.nih.gov/bioproject/?term=248792
3. ^https://cge.food.dtu.dk/services/ResFinder/
5. ^www.ncbi.nlm.nih.gov/pathogens/betalactamase-data-resources
Achtman, M., Wain, J., Weill, F. X., Nair, S., Zhou, Z., Sangal, V., et al. (2012). Multilocus sequence typing as a replacement for serotyping in Salmonella enterica. PLoS Pathog. 8:e1002776. doi: 10.1371/journal.ppat.1002776
Anastasi, E. M., Matthews, B., Gundogdu, A., Vollmerhausen, T. L., Ramos, N. L., Stratton, H., et al. (2010). Prevalence and persistence of Escherichia coli strains with uropathogenic virulence characteristics in sewage treatment plants. Appl. Environ. Microbiol. 76, 5882–5886. doi: 10.1128/AEM.00141-10
Ashton, P. M., Nair, S., Peters, T. M., Bale, J. A., Powell, D. G., Painset, A., et al. (2016). (2016) ‘identification of Salmonella for public health surveillance using whole genome sequencing. PeerJ 4, e1752–e1718. doi: 10.7717/peerj.1752
Baba Ahmed-Kazi Tani, Z., Decre, D., Genel, N., Boucherit-Otmani, Z., Arlet, G., Drissi, M., et al. (2013). Molecular and epidemiological characterization of enterobacterial multidrug-resistant strains in Tlemcen Hospital (Algeria) (2008-2010). Microb. Drug. Resist. 19, 185–190. doi: 10.1089/mdr.2012.0161
Bacci, C., Vismarra, A., Dander, S., Barilli, E., and Superchi, P. (2019). Occurrence and antimicrobial profile of bacterial pathogens in former foodstuff meat products used for pet diets. J. Food Prot. 82, 316–324. doi: 10.4315/0362-028X.JFP-18-352
Baede, V. O., Broens, E. M., Spaninks, M. P., Timmerman, A. J., Graveland, H., Wagenaar, J. A., et al. (2017). Raw pet food as a risk factor for shedding of extended-spectrum beta-lactamase-producing Enterobacteriaceae in household cats. PLoS One 12:e0187239. doi: 10.1371/journal.pone.0187239
Binagia, E. M., and Levy, N. A. (2020). Salmonella mesenteric lymphadenitis causing septic peritonitis in two dogs. Vet. Med. 11, 25–30. doi: 10.2147/vmrr.s238305
Bojanić, K., Midwinter, A. C., Marshall, J. C., Rogers, L. E., Biggs, P. J., and Acke, E. (2017). Isolation of Campylobacter spp. from client-owned dogs and cats, and retail raw meat pet food in the Manawatu, New Zealand. Zoonoses Public Health 64, 438–449. doi: 10.1111/zph.12323
Bolger, A. M., Lohse, M., and Usadel, B. (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. doi: 10.1093/bioinformatics/btu170
Bottari, B., Bancalari, E., Barera, A., Ghidini, S., and Gatti, M. (2020). Evaluating the presence of human pathogens in commercially frozen, biologically appropriate raw pet food sold in Italy. Vet. Rec. 187:e50. doi: 10.1136/vr.105893
Brand, C., Schwanen, T., and Anable, J. (2020). “Online omnivores” or “willing but struggling”? Identifying online grocery shopping behavior segments using attitude theory. J. Retail. Consum. Serv. 57:102195. doi: 10.1016/j.jretconser.2020.102195
Bulochova, V., and Evans, E. W. (2021a). Exploring food safety perceptions and self-reported practices of pet owners providing raw meat⇓based diets to pets. J. Food Prot. 84, 912–919. doi: 10.4315/JFP-20-338
Bulochova, V., and Evans, E. W. (2021b). Raw meat–based pet feeding and food safety: Netnography study of pet owner comments and review of manufacturers’ information provision. J. Food Prot. 84, 2099–2108. doi: 10.4315/jfp-21-158
Cassini, A., Högberg, L. D., Plachouras, D., Quattrocchi, A., Hoxha, A., Simonsen, G. S., et al. (2019). Attributable deaths and disability-adjusted life-years caused by infections with antibiotic-resistant bacteria in the EU and the European economic area in 2015: a population-level modelling analysis. Lancet Infect. Dis. 19, 56–66. doi: 10.1016/S1473-3099(18)30605-4
Chattaway, M. A., Dallman, T. J., Larkin, L., Nair, S., McCormick, J., Mikhail, A., et al. (2019). The transformation of reference microbiology methods and surveillance for Salmonella with the use of whole genome sequencing in England and Wales. Front. Public Health 7, 1–12. doi: 10.3389/fpubh.2019.00317
Clinical and Laboratory Standards Institute. (2020) CLSI M100-ED29: 2021 performance standards for antimicrobial susceptibility testing. 30th Edn. Wayne, PA: CLSI.
Collignon, P. C., Conly, J. M., Andremont, A., McEwen, S., Aidara-Kane, A., Agerso, Y., et al. (2016). World health organization ranking of antimicrobials according to their importance in human medicine: a critical step for developing risk management strategies to control antimicrobial resistance from food animal production. Clin. Infect. Dis. 63, 1087–1093. doi: 10.1093/cid/ciw475
Cormier, A., Zhang, P. L. C., Chalmers, G., Weese, J. S., Deckert, A., Mulvey, M., et al. (2019). Diversity of CTX-M-positive Escherichia coli recovered from animals in Canada. Vet. Microbiol. 231, 71–75. doi: 10.1016/j.vetmic.2019.02.031
Davies, R. H., Lawes, J. R., and Wales, A. D. (2019). Raw diets for dogs and cats: a review, with particular reference to microbiological hazards. J. Small Anim. Pract. 60, 329–339. doi: 10.1111/jsap.13000
Day, M. J., Hopkins, K. L., Wareham, D. W., Toleman, M. A., Elviss, N., Randall, L., et al. (2019). Extended-spectrum β-lactamase-producing Escherichia coli in human-derived and foodchain-derived samples from England, Wales, and Scotland: an epidemiological surveillance and typing study. Lancet Infect. Dis. 19, 1325–1335. doi: 10.1016/S1473-3099(19)30273-7
de Souza da-Silva, A. P., de Sousa, V. S., de Araújo Longo, L. G., Caldera, S., Baltazar, I. C. L., Bonelli, R. R., et al. (2020). Prevalence of fluoroquinolone-resistant and broad-spectrum cephalosporin-resistant community-acquired urinary tract infections in Rio de Janeiro: impact of Escherichia coli genotypes ST69 and ST131. Infect. Genet. Evol. 85:104452. doi: 10.1016/j.meegid.2020.104452
Defra/APHA. (2018). Animal by-product categories, site approval, hygiene and disposal. Available at: https://www.gov.uk/guidance/animal-by-product-categories-site-approval-hygiene-and-disposal.
Defra/APHA. (2019). Using animal by-products to make pet food. Available at: https://www.gov.uk/guidance/using-animal-by-products-to-make-pet-food.
Defra/APHA. (2020). Laboratory testing requirements for animal by-products (ABPs). Available at: https://www.gov.uk/guidance/laboratory-testing-requirements-for-animal-by-products-abps.
Dodd, S., Cave, N., Abood, S., Shoveller, A. K., Adolphe, J., and Verbrugghe, A. (2020). An observational study of pet feeding practices and how these have changed between 2008 and 2018. Vet. Rec. 186, 643–649. doi: 10.1136/vr.105828
EUCAST. (2022) European committee on antimicrobial susceptibility testing, the European committee on antimicrobial susceptibility testing. Breakpoint tables for interpretation of MICs and zone diameters. Version 12.0, 2022. Available at: http://www.eucast.org.
Food Standards Agency. (2021a). Happy Hounds Wales Ltd recalls frozen raw dog food products due to the presence of Salmonella. Available at: https://www.food.gov.uk/news-alerts/alert/fsa-prin-37-2021).
Food Standards Agency. (2021b). Natural Instinct recalls several products containing duck because salmonella has been found in the products. Available at: https://www.food.gov.uk/news-alerts/alert/fsa-prin-39-2021.
Food Standards Agency. (2022). Dogs Choice UK recalls frozen raw dog foods because of the presence of Salmonella. Available at: https://www.food.gov.uk/news-alerts/alert/fsa-prin-09-2022.
Freeman, L. M., Chandler, M. L., Hamper, B. A., and Weeth, L. P. (2013). Current knowledge about the risks and benefits of raw meat-based diets for dogs and cats. J. Am. Vet. Med. Assoc. 243, 1549–1558. doi: 10.2460/javma.243.11.1549
Frost, A. (2017). Feeding of raw Brucella suis -infected meat to dogs in the UK. Vet. Rec. 181:484. doi: 10.1136/vr.j4972
Giacometti, F., Magarotto, J., Serraino, A., and Piva, S. (2017). Highly suspected cases of salmonellosis in two cats fed with a commercial raw meat-based diet: health risks to animals and zoonotic implications. BMC Vet. Res. 13, 1–6. doi: 10.1186/s12917-017-1143-z
Gibson, J. F., Parker, V. J., Howard, J. P., Snell, C. M., Cross, E. W., Pagliughi, L. B., et al. (2022). Escherichia coli pathotype contamination in raw canine diets. Am. J. Vet. Res. 83, 3–8. doi: 10.2460/ajvr.21.10.0166
Groat, E. F., Williams, N. J., Pinchbeck, G., Warner, B., Simpson, A., and Schmidt, V. M. (2022). UK dogs eating raw meat diets have higher risk of Salmonella and antimicrobial-resistant Escherichia coli faecal carriage. J. Small Anim. Pract. 63, 435–441. doi: 10.1111/jsap.13488
Hellgren, J., Hästö, L. S., Wikström, C., Fernström, L. L., and Hansson, I. (2019). Occurrence of Salmonella, Campylobacter, Clostridium and Enterobacteriaceae in raw meat-based diets for dogs. Vet. Rec. 184:442. doi: 10.1136/vr.105199
James, C., Dixon, R., Talbot, L., James, S. J., Williams, N., and Onarinde, B. A. (2021). Assessing the impact of heat treatment of food on antimicrobial resistance genes and their potential uptake by other bacteria–a critical review. Antibiotics 10. doi: 10.3390/antibiotics10121440
Johnson, J. R., and Russo, T. A. (2002). Extraintestinal pathogenic Escherichia coli: “the other bad E. coli”. J. Lab. Clin. Med. 139, 155–162. doi: 10.1067/mlc.2002.121550
Jones, J. L., Wang, L., Ceric, O., Nemser, S. M., Rotstein, D. S., Jurkovic, D. A., et al. (2019). Whole genome sequencing confirms source of pathogens associated with bacterial foodborne illness in pets fed raw pet food. J. Vet. Diagn. Investig. 31, 235–240. doi: 10.1177/1040638718823046
Kaindama, L., Jenkins, C., Aird, H., Jorgensen, F., Stoker, K., and Byrne, L. (2020). A cluster of Shiga toxin producing Escherichia coli O157:H7 highlights raw pet food as an emerging potential source of infection in humans. Epidemiol. Infect. 149:e124. doi: 10.1017/S0950268821001072
Kananub, S., Pinniam, N., Phothitheerabut, S., and Krajanglikit, P. (2020). Contamination factors associated with surviving bacteria in Thai commercial raw pet foods. Vet. World 13, 1988–1991. doi: 10.14202/vetworld.2020.1988-1991
Laflamme, D. P., Abood, S. K., Fascetti, A. J., Fleeman, L. M., Freeman, L. M., Michel, K. E., et al. (2008). Timely topics in nutrition: pet feeding practices of dog and cat owners in the United States and Australia. J. Am. Vet. Med. Assoc. 232, 687–694. doi: 10.2460/javma.232.5.687
Livermore, D. M. (2008). Defining an extended-spectrum β-lactamase. Clin. Microbiol. Infect. 14, 3–10. doi: 10.1111/j.1469-0691.2007.01857.x
Livermore, D. M., and Hawkey, P. M. (2005). CTX-M: changing the face of ESBLs in the UK. J. Antimicrob. Chemother. 56, 451–454. doi: 10.1093/jac/dki239
Ludden, C., Raven, K. E., Jamrozy, D., Gouliouris, T., Blane, B., Coll, F., et al. (2019). One health genomic surveillance of Escherichia coli demonstrates distinct lineages and mobile genetic elements in isolates from humans versus livestock. MBio 10, 1–12. doi: 10.1128/mBio.02693-18
Ma, J., Liu, J. H., Lv, L., Zong, Z., Sun, Y., Zheng, H., et al. (2012). Characterization of extended-spectrum β-lactamase genes found among Escherichia coli isolates from duck and environmental samples obtained on a duck farm. Appl. Environ. Microbiol. 78, 3668–3673. doi: 10.1128/AEM.07507-11
Magiorakos, A. P., Srinivasan, A., Carey, R. B., Carmeli, Y., Falagas, M. E., Giske, C. G., et al. (2012). Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 18, 268–281. doi: 10.1111/j.1469-0691.2011.03570.x
Mehlenbacher, S., Churchill, J., Olsen, K. E., and Bender, J. B. (2012). Availability, brands, labelling and Salmonella contamination of raw pet food in the Minneapolis/St. Paul area. Zoonoses Public Health 59, 513–520. doi: 10.1111/j.1863-2378.2012.01491.x
Morelli, G., Bastianello, S., Catellani, P., and Ricci, R. (2019). Raw meat-based diets for dogs: survey of owners’ motivations, attitudes and practices. BMC Vet. Res. 15, 1–10. doi: 10.1186/s12917-019-1824-x
Morelli, G., Catellani, P., Miotti Scapin, R., Bastianello, S., Conficoni, D., Contiero, B., et al. (2020). Evaluation of microbial contamination and effects of storage in raw meat-based dog foods purchased online. J. Anim. Physiol. Anim. Nutr. 104, 690–697. doi: 10.1111/jpn.13263
Morgan, G., Saal, M., Corr, A., Jenkins, C., Chattaway, M. A., Pinchbeck, G., et al. (2023). Isolation of Salmonella species of public health concern from commonly fed dried meat dog treats. Vet. Rec. 192. doi: 10.1002/vetr.2642
Morgan, G., Williams, N., Schmidt, V., Cookson, D., Symington, C., and Pinchbeck, G. (2022). A Dog’s dinner: factors affecting food choice and feeding practices for UK dog owners feeding raw meat- based or conventional cooked diets. Prev. Vet. Med. 208:105741. doi: 10.1016/J.PREVETMED.2022.105741
Morley, P. S., Strohmeyer, R. A., Tankson, J. D., Hyatt, D. R., Dargatz, D. A., and Fedorka-Cray, P. J. (2006). Evaluation of the association between feeding raw meat and Salmonella enterica infections at a greyhound breeding facility. J. Am. Vet. Med. Assoc. 228, 1524–1532. doi: 10.2460/javma.228.10.1524
Murray, C., Ikuta, K. S., Sharara, F., Swetschinski, L., Robles Aguilar, G., Gray, A., et al. (2022). Global burden of bacterial antimicrobial resistance in 2019: a systematic analysis. Lancet 399, 629–655. doi: 10.1016/S0140-6736(21)02724-0
Nemser, S. M., Doran, T., Grabenstein, M., McConnell, T., McGrath, T., Pamboukian, R., et al. (2014). Investigation of Listeria, Salmonella, and toxigenic Escherichia coli in various pet foods. Foodborne Pathog. Dis. 11, 706–709. doi: 10.1089/fpd.2014.1748
Neuert, S., Nair, S., Day, M. R., Doumith, M., Ashton, P. M., Mellor, K. C., et al. (2018). Prediction of phenotypic antimicrobial resistance profiles from whole genome sequences of non-typhoidal Salmonella enterica. Front. Microbiol. 9, 1–11. doi: 10.3389/fmicb.2018.00592
Nilsson, O. (2015). Hygiene quality and presence of ESBL-producing Escherichia coli in raw food diets for dogs. Infect. Ecol. Epidemiol. 5:28758. doi: 10.3402/iee.v5.28758
Novais, Â., Vuotto, C., Pires, J., Montenegro, C., Donelli, G., Coque, T. M., et al. (2013). Diversity and biofilm-production ability among isolates of Escherichia coli phylogroup D belonging to ST69, ST393 and ST405 clonal groups. BMC Microbiol. 13:144. doi: 10.1186/1471-2180-13-144
Nüesch-Inderbinen, M., Treier, A., Zurfluh, K., and Stephan, R. (2019). Raw meat-based diets for companion animals: a potential source of transmission of pathogenic and antimicrobial-resistant Enterobacteriaceae. R. Soc. Open Sci. 6. doi: 10.1098/rsos.191170
O’Neill, J. (2016). Tackling drug-resistant infections globally: final report and recommendations. The review on antimicrobial resistance. Available at: https://amrreview.org/sites/default/files/160518_Finalpaperwithcover.pdf.
Oteo, J., Diestra, K., Juan, C., Bautista, V., Novais, A., Pérez-Vázquez, M., et al. (2009). Extended-spectrum beta-lactamase-producing Escherichia coli in Spain belong to a large variety of multilocus sequence typing types, including ST10 complex/a, ST23 complex/a and ST131/B2. Int. J. Antimicrob. Agents 34, 173–176. doi: 10.1016/j.ijantimicag.2009.03.006
PDSA. (2022) PDSA animal wellbeing PAW report 2022: The essential insight into the wellbeing of UK pets, PDSA Animal Wellbeing Report. Available at: https://www.pdsa.org.uk/media/4371/paw-2018-full-web-ready.pdf.
Randall, L. P., Clouting, C., Horton, R. A., Coldham, N. G., Wu, G., Clifton-Hadley, F. A., et al. (2011). Prevalence of Escherichia coli carrying extended-spectrum β-lactamases (CTX-M and TEM-52) from broiler chickens and turkeys in Great Britain between 2006 and 2009. J. Antimicrob. Chemother. 66, 86–95. doi: 10.1093/jac/dkq396
Reid, C. J., Cummins, M. L., Börjesson, S., Brouwer, M. S. M., Hasman, H., Hammerum, A. M., et al. (2022). A role for ColV plasmids in the evolution of pathogenic Escherichia coli ST58. Nat. Commun. 13, 683–615. doi: 10.1038/s41467-022-28342-4
Roccato, A., Uyttendaele, M., Cibin, V., Barrucci, F., Cappa, V., Zavagnin, P., et al. (2015). Effects of domestic storage and thawing practices on Salmonella in poultry-based meat preparations. J. Food Prot. 78, 2117–2125. doi: 10.4315/0362-028X.JFP-15-048
Runesvärd, E., Wikström, C., Fernström, L. L., and Hansson, I. (2020). Presence of pathogenic bacteria in faeces from dogs fed raw meat-based diets or dry kibble. Vet. Rec. 187, 1–6. doi: 10.1136/vr.105644
Schmidt, V. M., Pinchbeck, G. L., Nuttall, T., McEwan, N., Dawson, S., Williams, N. J., et al. (2015). Antimicrobial resistance risk factors and characterisation of faecal E. coli isolated from healthy Labrador retrievers in the United Kingdom. Prev. Vet. Med. 119, 31–40. doi: 10.1016/j.prevetmed.2015.01.013
Solà-Ginés, M., Cameron-Veas, K., Badiola, I., Dolz, R., Majó, N., Dahbi, G., et al. (2015). Diversity of multi-drug resistant avian pathogenic Escherichia coli (APEC) causing outbreaks of colibacillosis in broilers during 2012 in Spain. PLoS One 10, 1–14. doi: 10.1371/journal.pone.0143191
Strohmeyer, R. A., Morley, P. S., Hyatt, D. R., Dargatz, D. A., Scorza, A. V., and Lappin, M. R. (2006). Evaluation of bacterial and protozoal contamination of commercially available raw meat diets for dogs. J. Am. Vet. Med. Assoc. 228, 537–542. doi: 10.2460/javma.228.4.537
Tewolde, R., Dallman, T., Schaefer, U., Sheppard, C. L., Ashton, P., Pichon, B., et al. (2016). MOST: a modified MLST typing tool based on short read sequencing. PeerJ 4, e2308–e2310. doi: 10.7717/peerj.2308
Giner-Lamia, J., Vinuesa, P., Betancor, L., Silva, C., Bisio, J., Soleto, L., et al. (2019). Genome analysis of Salmonella enterica subsp. diarizonae isolates from invasive human infections reveals enrichment of virulence-related functions in lineage ST1256. BMC Genomics 20, 99–14. doi: 10.1186/s12864-018-5352-z
Treier, A., Stephan, R., Stevens, M. J. A., Cernela, N., and Nüesch-Inderbinen, M. (2021). High occurrence of Shiga toxin-producing Escherichia coli in raw meat-based diets for companion animals–a public health issue. Microorganisms 9. doi: 10.3390/microorganisms9081556
Van Bree, F. P. J., Bokken, G. C. A. M., Mineur, R., Franssen, F., Opsteegh, M., van der Giessen, J. W. B., et al. (2018). Zoonotic bacteria and parasites found in raw meat-based diets for cats and dogs. Vet. Rec. 182:50. doi: 10.1136/vr.104535
Van den Bunt, G., Fluit, A. C., Spaninks, M. P., Timmerman, A. J., Geurts, Y., Kant, A., et al. (2020). Faecal carriage, risk factors, acquisition and persistence of ESBL-producing Enterobacteriaceae in dogs and cats and co-carriage with humans belonging to the same household. J. Antimicrob. Chemother. 75, 342–350. doi: 10.1093/jac/dkz462
Veterinary Medicines Directorate. (2015). UK-VARSS 2015 highlights report. Veterinary antibiotic resistance and sales surveillance report (UK-VARSS 2015).
Veterinary Medicines Directorate. (2022) UK - veterinary antibiotic resistance and sales surveillance report: 2021. Supplementary material 3 – resistance data. Veterinary Antibiotic Resistance and Sales Surveillance Report (UK-VARSS 2021). Available at: https://www.gov.uk/government/publications/veterinary-antimicrobial-resistance-and-sales-surveillance-2021.
Viegas, F. M., Ramos, C. P., Xavier, R. G. C., Lopes, E. O., Júnior, C. A. O., Bagno, R. M., et al. (2020). Fecal shedding of Salmonella spp., Clostridium perfringens, and Clostridioides difficile in dogs fed raw meat-based diets in Brazil and their owners’ motivation. PLoS One 15, e0231275–e0231213. doi: 10.1371/journal.pone.0231275
Voets, G. M., Fluit, A. C., Scharringa, J., Schapendonk, C., van den Munckhof, T., Leverstein-van Hall, M. A., et al. (2013). Identical plasmid AmpC beta-lactamase genes and plasmid types in E. coli isolates from patients and poultry meat in the Netherlands. Int. J. Food Microbiol. 167, 359–362. doi: 10.1016/j.ijfoodmicro.2013.10.001
Wedley, A. L., Dawson, S., Maddox, T. W., Coyne, K. P., Pinchbeck, G. L., Clegg, P., et al. (2017). Carriage of antimicrobial resistant Escherichia coli in dogs: prevalence, associated risk factors and molecular characteristics. Vet. Microbiol. 199, 23–30. doi: 10.1016/j.vetmic.2016.11.017
Weese, J. S., and Rousseau, J. (2006). Survival of Salmonella Copenhagen in food bowls following contanimation with experimentally inoculated raw meat: effects of time, cleaning, and disinfection. Can. Vet. J. 47, 887–889.
Weese, J. S., Rousseau, J., and Arroyo, L. (2005). Bacteriological evaluation of commercial canine and feline raw diets. Can. Vet. J. 46, 513–516.
WHO. (2019). Critically important antimicrobials for human medicine, 6th revision. Geneva: World Health Organisation. Available at: https://apps.who.int/iris/bitstream/handle/10665/312266/9789241515528-eng.pdf.
Withenshaw, S. M., Lawes, J. R., Teale, C., and Davies, R. H. (2020). Industry expansion and Salmonella surveillance trends for raw meat pet foods in Great Britain over the last 10 years. In: Society for Veterinary Epidemiology and Preventative Medicine, pp. 33–45. Available at: https://www.researchgate.net/publication/340647945_INDUSTRY_EXPANSION_AND_SALMONELLA_SURVEILLANCE_TRENDS_FOR_RAW_MEAT_PET_FOOD_IN_GREAT_BRITAIN_OVER_THE_LAST_TEN_YEARS#fullTextFileContent.
Zhang, S., den Bakker, H., Li, S., Chen, J., Dinsmore, B. A., Lane, C., et al. (2019). SeqSero2: rapid and improved salmonella serotype determination using whole-genome sequencing data. Appl. Environ. Microbiol. 85, e01746–e01719. doi: 10.1128/AEM.01746-19
Keywords: raw, dog, food, Enterobacteriaceae, antimicrobial resistance, pet, E. coli
Citation: Morgan G, Pinchbeck G, Taymaz E, Chattaway MA, Schmidt V and Williams N (2024) An investigation of the presence and antimicrobial susceptibility of Enterobacteriaceae in raw and cooked kibble diets for dogs in the United Kingdom. Front. Microbiol. 14:1301841. doi: 10.3389/fmicb.2023.1301841
Received: 11 October 2023; Accepted: 18 December 2023;
Published: 08 January 2024.
Edited by:
Mariana Carmen Chifiriuc, University of Bucharest, RomaniaReviewed by:
Gunaraj Dhungana, Nepal Health Research Council, NepalCopyright © 2024 Morgan, Pinchbeck, Taymaz, Chattaway, Schmidt and Williams. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Genever Morgan, R2VuZXZlci5Nb3JnYW4yQGxpdmVycG9vbC5hYy51aw==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.