
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol., 29 November 2023
Sec. Microorganisms in Vertebrate Digestive Systems
Volume 14 - 2023 | https://doi.org/10.3389/fmicb.2023.1270974
This article is part of the Research TopicWomen in Microorganisms in Vertebrate Digestive Systems: 2023View all 17 articles
Introduction: The aim of this work was to characterize a new strain of Ligilactobacillus salivarius (CNCM I-4866) (CNCM I-4866) to address its potential as probiotic with a special focus on intestinal inflammation. Potential anti-inflammatory abilities of this strain were evaluated through in vivo and in vitro experiments.
Methods: Firstly, the strain was tested in a murine acute inflammation colitis model induced by DNBS. In vitro characterization was then performed with diverse tests: modulation capability of intestinal permeability; study of the impact on immunity profile through cytokines dosage; capacity to inhibit pathogens and adhere to intestinal cells lines. Production of metabolites, antibiotic resistance and survival to gastro-intestinal tract conditions were also tested.
Results: In vitro assay has shown a reduction of colonic damage and markers of inflammation after treatment with CNCM I-4866. Transcriptomic analysis performed on colons showed the capacity of the strain to down-regulate pro-inflammatory cytokines. L. salivarius CNCM I-4866 exerted anti-inflammatory profile by reducing IL-8 production by TNF-α stimulated cell and modulated cytokines profile on peripheral blood mononuclear cells (PBMC). It protected intestinal integrity by increasing trans-epithelial electrical resistance (TEER) on Caco-2 TNF-α inflamed cells. Additionally, L. salivarius CNCM I-4866 displayed inhibition capacity on several intestinal pathogens and adhered to eukaryotic cells. Regarding safety and technical concerns, CNCM I-4866 was highly resistant to 0.3% of bile salts and produced mainly L-lactate. Finally, strain genomic characterization allowed us to confirm safety aspect of our strain, with no antibiotic gene resistance found.
Discussion: Taken together, these results indicate that L. salivarius CNCM I-4866 could be a good probiotic candidate for intestinal inflammation, especially with its steady anti-inflammatory profile.
Inflammatory bowel diseases (IBDs), including ulcerative colitis and Crohn’s disease, are common chronic gastrointestinal diseases, mainly in Western countries (Aldars-Garcia et al., 2021). IBDs are characterized by inflammation bursting of the gastrointestinal tract driven by over-stimulation of the immune system. This deleterious inflammatory response includes the overproduction of reactive oxygen species, damage to the intestinal epithelial barrier, and an imbalance in the immune response with the secretion of pro-inflammatory cytokines (Jakubczyk et al., 2020).
Even if IBD development is multifactorial, it has been shown that IBDs are linked with deregulation in the intestinal microbiota (Glassner et al., 2020). The intestinal microbiota is composed of more than trillions of different microorganisms, such as bacteria, fungi, or viruses. The majority of intestinal bacteria belong to four phyla: Bacillota, Bacteroidota, Pseudomonadota, and Actinomycetes (Thursby and Juge, 2017). The gut microbiota has many functions, such as host nutrient metabolism, immunomodulation, and protection against pathogens (Jandhyala et al., 2015). In the case of IBD, both the prevalence of pathobionts and/or the lack of key bacteria can promote IBD development and maintenance (Frank et al., 2007).
In the Bacillota phylum, the family Lactobacillaceae is a widely diverse group of Gram-positive bacteria, harboring 25 genera and almost 200 species. They exert an anaerobic metabolism, producing lactic acid through the fermentation of sugars, but are able to tolerate the presence of oxygen. Lactobacilli are widely studied for their potential beneficial properties. For example, some strains are shown to have a beneficial impact on restoring intestinal permeability (Chamignon et al., 2020), which is increased during inflammation. Another key role of some Lactobacillus strains is their ability to modulate the immune response in an inflammatory state by decreasing pro-inflammatory cytokine production or enhancing the levels of anti-inflammatory cytokines, such as IL-10 (Alard et al., 2018). Some Lactobacillus members are also known to be high producers of exopolysaccharides (EPSs), which have been recognized to directly infer the health-promoting properties of Lactobacilli (Juraskova et al., 2022; Martin et al., 2023).
Ligilactobacillus salivarius, formerly named Lactobacillus salivarius, is a homo-fermentative species. Many studies have been conducted on L. salivarius because of its useful properties for human health (Chaves et al., 2017). Indeed, beyond their capacity to alleviate inflammation-induced colitis in the murine model (Peran et al., 2005; Iyer et al., 2022), some L. salivarius strains are well known to exert antimicrobial activity (Tinrat et al., 2011; Messaoudi et al., 2013). Thereby, several strains of the Lactobacillaceae family were characterized these last years as potential probiotics for IBD, serving as complements to treatments in order to reduce disease-associated symptoms. Probiotics are defined as “live microorganisms, that, when administered in adequate amounts, confer a health benefit on the host” (Hill et al., 2014). Due to the wide range of applications of probiotics and their various mechanisms of action, the EFSA has proposed “Guidance on the characterization of microorganisms used as feed additives or as production organisms” (Rychen et al., 2018), suggesting safety and effectiveness criteria for their evaluation. These criteria include mostly physiological characterization and safety assessment by genomic analysis, particularly research for antibiotic resistance. Other key features to ensure the strain’s ability to exert its beneficial activity are resistance to the gastrointestinal tract or adhesion to the intestinal mucosa.
In this study, we have assessed the probiotic capacities of L. salivarius CNCM I-4866 to target gut inflammation as the first step to characterize its potential beneficial effects on IBD management. Murine acute colitis assays and in vitro experiments were conducted to target some potentially beneficial effects observed in vivo (restoration of intestinal permeability and immunomodulatory response). Technological and safety parameters and genomic characterization were also explored to validate the safe status of this strain and its industrial interest.
Ligilactobacillus salivarius CNCM I-4866 was isolated by SORBIAL company from the rumen of grazing lamb. Ligilactobacillus salivarius CNCM I-4866 and Lacticaseibacillus rhamnosus GG were grown in Man Rogosa and Sharpe (MRS) (Biokar, Solabia, France) at 37°C in aerobic conditions.
Bacterial cultures were centrifuged at 8000xg, washed twice in DPBS (Gibco, Thermo Fisher, USA), and resuspended in DPBS at an established concentration. These aliquots were used for in vitro and in vivo assays described below.
The human colon adenocarcinoma cell lines HT-29 and Caco-2 were obtained from the American Type Culture Collection (ATCC, United Kingdom). HT-29 cells were grown in Dulbecco’s modified Eagle’s medium supplemented with GlutaMAX (DMEM) (Gibco, Thermo Fisher) and 10% (v/v) of heat-inactivated fetal bovine serum (FBS, Eurobio, France). The Caco-2 cell line was cultured on DMEM GlutaMAX medium, 10% FBS, 1% non-essential amino acids (Gibco, Thermo Fisher), and 0.1% penicillin/streptomycin (Gibco, Thermo Fisher). The cultures were maintained at 37°C under 10% CO2, and the medium was changed every 2 days.
For DNBS assays, 54 6-week-old male C57BL/6JrJ mice were obtained from Janvier Laboratory (Janvier, France) and maintained under specific pathogen-free (SPF) conditions in the animal facilities of the IERP Experimental Unit, INRAE. They were housed in four or five cages. Experiments were performed in accordance with European Union legislation on animal welfare and were approved by COMETHEA, a local committee on animal experimentation (no. 16744–201807061805486), and in compliance with the ARRIVE relevant guidelines. After a 7-day acclimation period, 54 mice were divided into 3 groups (n = 8 or n = 10 mice/group): the vehicle control group [no inflammation; Ethanol (EtOH)-Vehicle], the inflamed control group (inflammation-induced; DNBS-Vehicle), and the treated group (DNBS-CNCM I-4866). For 10 days, intra-gastric administration of DPBS (Vehicle) (Gibco, Thermo Fisher) (200 μL), 16% (v/v) glycerol, or the bacteria resuspended in DPBS (109 CFU/mL in 200 μL) was performed. Gavages were performed daily until the end of the experiment. After 7 days, the mice were anesthetized with an intraperitoneal injection of 0.1% ketamine and 0.06% xylazine. Subsequently, an intra-rectal injection of 2,4-dinitrobenzenesulfonic acid hydrate (Sigma, Switzerland) at a 2.75 mg/mice concentration dissolved in 30% ethanol in DPBS was injected. The vehicle control group received an intra-rectal injection of 30% ethanol in DPBS alone. After 3 days of the injection, blood was collected from the sub-mandibular vein, and mice were euthanized by cervical dislocation. The experiment procedure is presented in Figure 1.
Macroscopic scores in terms of Wallace scores (Wallace et al., 1989), microscopic scores in terms of Ameho scores (Ameho et al., 1997), and myeloperoxidase (MPO) activity levels were determined on the colon samples as described before (Barone et al., 2018). The levels of lipocalin-2 (Mouse Lipocalin-2, R&D Systems, USA) and sCD14 (Mouse sCD14, R&D Systems, USA) were determined using ELISA, according to the manufacturer’s instructions.
Transcriptomic analysis was performed on the colon samples of mice. In brief, samples were conserved in RNA at −80°C, and RNA was extracted afterward with the RNeasy kit (RNeasy kit, Qiagen, the Netherlands), following the manufacturer’s instructions. Transcriptomic analysis was performed by the GENOM’IC platform at Cochin Institute using the 3’tag method. FASTQ files were then aligned using the STAR algorithm (version 2.7.6a) on the Ensembl release 101 reference. Reads were then counted using RSEM (v1.3.1), and the statistical analyses on the read counts were performed with R (version 3.6.3) and the DESeq2 package (DESeq2_1.26.0) to determine the proportion of differentially expressed genes between two conditions. The standard DESeq2 normalization method (DESeq2’s median of ratios with the DESeq function) was used, with pre-filter of reads and genes (reads uniquely mapped on the genome, or up to 10 different loci with a count adjustment, and genes with at least 10 reads in at least 3 different samples). Following the package recommendations, the Wald test with the contrast function and the Benjamin–Hochberg FDR control procedure were used to identify the differentially expressed genes. Selected gene lists (|log2FoldChange| > 1.5 and value of p < 0.05) were loaded into ingenuity pathway analysis (IPA) to analyze pathways and generate data.
HT-29 cells were seeded into 24-well plates (1 × 105 cells per well). After 6 days, when confluence was reached, the medium was replaced by a DMEM GlutaMAX medium with 5% FBS. After 24 h, on day 7, co-incubation with the bacterial cells was performed at a multiplicity of infection (MOI) of 40 in DMEM GlutaMAX, 0.1% penicillin/streptomycin, and 5% FBS and supplemented or not with TNF-α at a final concentration of 5 ng/mL (PeproTech, USA). DPBS was used as a negative control and butyrate at 10 mM as a positive control. After 6 h of co-incubation, supernatants were recovered and stored at −80°C. Interleukin (IL)-8 concentrations were quantified using the Human IL-8 ELISA MAX Standard Set (BioLegend, USA), according to the manufacturer’s instructions. The absorbance was measured at 450 nm using the Infinite M200 Pro (TECAN, Switzerland).
Caco-2 cells were grown on Transwell inserts and kept at 37°C under 10% CO2 until 80% confluence was reached. The medium was changed every 2 days. When optimal transepithelial resistance (TEER) values were reached (REMS AutoSampler, World Precision Instruments, USA), the fresh medium was added. Then, the strain L. salivarius CNCM I-4866 and Lacticaseibacillus rhamnosus GG (used as a positive control; Chamignon et al., 2020) at MOI 40 or the control (DPBS) were added to the apical compartment of the cells. After 3 h, 100 ng/mL of TNF-α was added to the basal compartment of Transwell plates. TEER was measured just before and 24 h after the treatments. The results were normalized to basal TEER as follows:
Human peripheral blood mononuclear cells (PBMCs) isolated from the blood of healthy donors were obtained from StemCells (StemCells, Canada) and stored in liquid nitrogen. Five donors were selected according to the next selective criteria as follows: male individuals, BMI between 20 and 30, non-smokers, and with no allergies or diseases, such as asthma. After thawing, PBMCs were washed twice with Roswell Park Memorial Institute GlutaMAX medium (RPMI) (Gibco, Thermo Fisher) containing 10% FBS, and DNase I was added to avoid aggregate formation. Then, cells were centrifuged at 200 g for 15 min at room temperature, and the supernatants were discarded. The washing step was performed twice, and then PBMCs were counted using the trypan blue method. Next, PBMCs were seeded at a rate of 1 million per 24-well plate. In total, 50 μL of fresh bacteria cultured in MRS (Difco, Thermo Fisher) was added at an MOI of 10, and co-cultures were maintained at 37°C in 5% CO2 for 24 h. Escherichia coli TG1 was used as a control (Sokol et al., 2008). Finally, supernatants were collected, and interleukin-10 (IL-10), interleukin-12 (IL-12), and tumor necrosis factor (TNF-α) were quantified by ELISA using specific kits (Mabtech, Sweden), according to the manufacturer’s guidelines.
Pathogen inhibition capacity against eight pathogens was determined. Salmonella typhimurium, Salmonella enteritidis, Listeria monocytogenes EDGE, Escherichia coli ATCC 700928, Staphylococcus aureus CNRZ 875, and Clostridium perfringens ATCC 13124 were obtained from the INRAE internal collection. Helicobacter pylori 26,695 was kindly provided by the Pasteur Institute (Dr. Hilde de Reuse team). Campylobacter jejuni BF was provided by the INRAE/ONIRIS Nantes collection. The first six strains were cultivated on Mueller Hinton (Thermo Fisher) at 37°C under aerobic conditions. H. pylori and C. jejuni were cultivated on Mueller Hinton agar supplemented with 5% sheep blood (Thermo Fisher) at 37°C in a micro-aerophilic atmosphere (bioMérieux, France). To perform inhibition tests, a lawn of each pathogen from a fresh suspension was made on Mueller Hinton. Holes were made in the agar with P100 sterile tips, to which 50 μL of filtered supernatants from a stationary-phase culture or control medium alone (MRS) was added. The results were read after 48 h of incubation as the diameter of inhibition (mm). To assess whether inhibition was due to acid production by the L. salivarius CNCM I-4866 strain, we performed the assay with supernatants neutralized at pH 7 with sodium hydroxide.
To assess bacterial adhesion capacity, Caco-2, HT-29, and its derivative, HT-29 MTX, were used. Cells were seeded into a 24-well tissue culture plate at a concentration of 1 × 105 cells/well, and adhesion was performed for 7 days for Caco-2 and HT-29 ATCC. For HT-29 MTX, after confluence (6 days), plates were incubated for an additional 14 days to allow cell differentiation (the medium was changed every day). In all cases, after 7 days or 21 days, wells were washed twice with DPBS, and fresh media without antibiotics were added. Each bacterial suspension was added at MOI 40 from a stationary-phase culture. After 3 h of incubation, monolayers were washed three times with DPBS to remove any bacteria that were not attached to the cells. Afterward, bacteria were disassociated by covering the monolayer with 150 μL of a 1% (v/v) Triton (Triton X-100, Sigma) solution in DPBS. Subsequently, 300 μL of DMEM was added in order to stop the reaction, and the number of viable adherent bacteria was determined by plating serial dilutions on MRS agar plates. Adhesion was expressed as the percentage of adhered bacteria with respect to the number of input bacteria, or DPBS as a negative control.
The adhesion test was also performed on mucin (Porcine gastric mucin, type III, Sigma). Mucin was prepared at 10 mg/mL in sterile DPBS and put on a 96-well plate overnight at 4°C. Adhesion assay was performed as described above, with an additional incubation at room temperature for 90 min after adding Triton solution.
D-lactate and L-lactate were measured in the supernatant of the bacterial culture at the stationary phase. The supernatant was precipitated with trichloroacetic acid (10%) and centrifuged at 20,000 g for 5 min at 4°C. Acid supernatants were neutralized with TEA 0.1 M at pH 9.15. Lactate was then measured with an enzymatic kit according to the manufacturer’s instructions (Biosentec, France).
Phenotypic resistance to antibiotics was assessed according to the EFSA recommendations (European Food Safety Authority, 2018). Lactic acid bacteria susceptibility test medium (LSM agar) was prepared with 90% IST (Iso-Sensitest broth, Oxoid, United Kingdom), 10% MRS broth, and 1.5% granulated agar. Bacterial suspensions were streaked on plates to obtain a lawn, and antibiotic strips (bioMérieux) were used. The inhibition area with the corresponding concentration (minimum inhibitory concentration, MIC) was then determined and compared with the EFSA guidelines.
Genomic DNA was extracted from 5 mL of culture with the first step of enzymatic lysis with the following cocktail: mutanolysin at 233.3 U/mL; lysostaphin at 13.3 U/mL, and lysozyme at 50 mg/mL, followed by incubation with RNAse A (Qiagen) at 10 mg/mL and proteinase K (Euromedex, France) at 50 mg/mL. Purification was performed with a DNA extraction kit (Genomic DNA Buffer Set and Genomic Tips, Qiagen), according to the manufacturer’s instructions. DNA was resuspended in TE buffer, and the concentration was measured with NanoDrop (NanoDrop 1,000, Thermo Fisher). The genome was sequenced by Eurofins Genomics (France) using whole-genome sequencing with de novo assembly, with the PacBio method on single-molecule real-time (SMRT) cells, a 240-mn collection time, a mean length superior to 6,000 bp, and a genome coverage of 100 X. The analysis of the obtained reads began with a quality check and de novo assembly of contigs. Contigs were then circularized and mapped when possible.
The genome is available at NCBI (BioSample accession ID: SAMN37542358). The presence of antibiotic gene resistance was searched online on two databases, namely, CARD1 and ResFinder.2 According to the “EFSA statement on the requirements for whole genome sequence analysis of microorganisms intentionally used in the food chain” (European Food Safety Authority, 2021), only hits with 80% identity and 70% length were reported. The presence of prophages in the genomes was determined in silico using Phaster.3 Only intact prophages were considered. Potential bacteriocin activity was determined in silico using BAGEL4.4
An induction assay was performed to establish if the intact prophage found with Phaster was active. In brief, induction with mitomycin C (Sigma) at 1 μg/mL was performed on culture at the beginning of the exponential phase. When the culture reached the stationary phase, 1 mL of culture was centrifuged (8,000 g, 10 mn, 4°C), and the supernatant was then filtered (0.22 μM) before being frozen at −20°C. Listeria ivanovii WSCL 3009 (Institute for Food and Health from the Technical University of Munich, Germany) was used as a receptor strain for B025 prophage. Two different protocols were performed with MRS agar (1.5%) and semi-solid MRS agar (0.75%), supplemented with 2 mM of CaCl2 to increase phage absorption. On spot assay, 100 μL of receptor bacteria at the beginning of the exponential phase culture was poured with semi-solid agar on top of solid agar. In total, 10 μL of inducted supernatant or control was spotted on the surface of the Petri dish. On double lawn assay, 100 μL of receptor bacteria at the beginning of exponential phase culture was mixed with 100 μL of inducted supernatant or control. After 15 min of incubation at room temperature, 3 mL of semi-solid agar was added and then poured on solid agar. After 48 h of incubation, the presence of potential inhibition halos, indicating the presence of active phages, was observed.
Potential bacteriocin activity was determined using a sensitive strain to enterolysin (which is the bacteriocin predicted for our strain): Lactococcus lactis IL403. The bacterial suspension was collected from the fresh colony of sensitive bacteria in sterile peptone water (McFarland 1) and streaked on LSM medium (medium prepared as described before). In total, 10 μL of supernatant was added to the agar. Three conditions were tested for each supernatant: filtered only; filtered and pH adjusted to 6; and filtered with pH 6 and 1 mg/mL of catalase, which inhibits potential H2O2 action. A bacterial growth medium was used as a negative control. Potential inhibition spots were observed after 48 h of incubation.
The hemolytic activity of our strain was determined by using blood agar, which was streaked with our strain and incubated at 37°C for 48 h. After incubation, the hemolytic activity was evaluated and classified based on the lysis of red blood cells in the medium around the colonies.
Tolerance to bile salts was studied to mimic the passage of the strains in the gastrointestinal tract. From stationary-phase culture, bacterial cells were exposed to 0% or 0.3% bile salts (Oxgall Powder, Sigma) for 1 h before viable cells were counted. Second, viable counts were performed after 24 h of growth of the L. salivarius CNCM I-4866 strain in media containing 0% or 0.3% bile salts.
Tolerance to acidic conditions was tested in the same conditions for bile salts by following the growth in media with a modified pH or after 1 h of exposition to a low pH and counting viable cells. We have performed assays at pH 2 and pH 4.
The production of potential exopolysaccharides (EPSs) by L. salivarius CNCM I-4866 was quantified with the Ropy phenotype test. In brief, a loop was used to observe EPS filament from a fresh colony on agar.
To determine bacterial structures, transmission electron microscopy (TEM) was performed by the microscopy and imaging platform (MIMA2, INRAE). The bacterial pellet obtained from an exponential phase was washed twice with phosphate buffer and recovered with 2% glutaraldehyde (EMS) in sodium cacodylate 0.1 M buffer. Suspensions were incubated for 1 h at room temperature. The pellet was then washed with sodium cacodylate in 0.1 M buffer (Fluka) and supplemented with 0.2 M sucrose. The sample was conserved at 4°C before being processed. The Hitachi HT7700 (Hitachi High-Tech, Japan) was used for microscopic observations.
DNBS-induced colitis was performed to observe a potential protective effect of the strain on acute inflammation. A significant decrease was observed in the macroscopic scores of the treated group compared with the DNBS-Vehicle group, indicating lower inflammation in the treated group (Figure 2A). Microscopic scores and myeloperoxidase (MPO) activity determinations (Figures 2B,C) showed a tendency to recovery (with p-values of 0.0798 and 0.1038), reflecting an improvement in colonic epithelial structure and reduced immune cell infiltration.
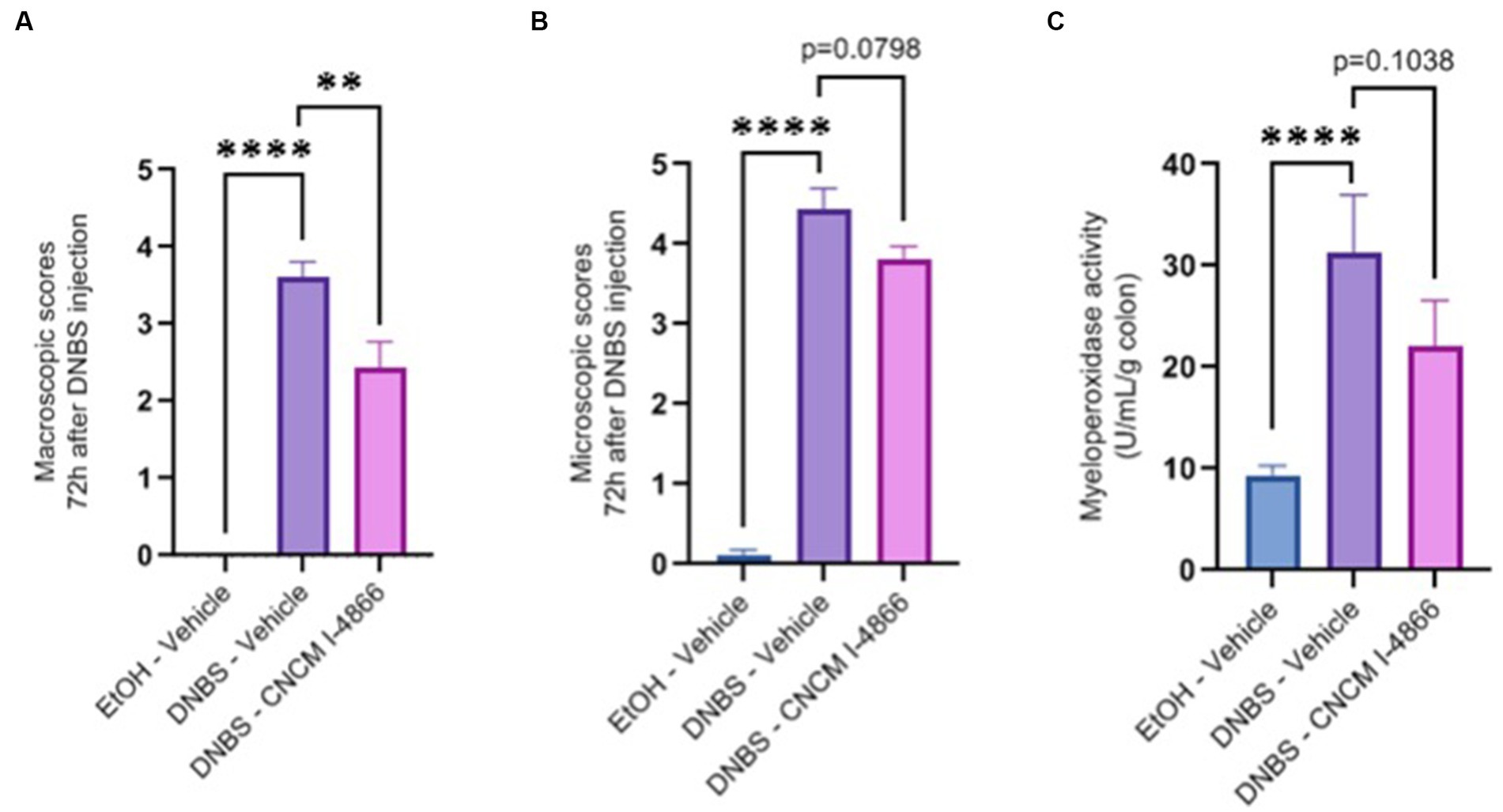
Figure 2. Effects of L. salivarius CNCM I-4866 on DNBS-induced colitis. (A) Colon macroscopic scores (Wallace scores); (B) colon microscopic scores (Ameho scores); and (C) levels of MPO activity in the colon. Results of Mann–Whitney U-tests compared with the DNBS-Vehicle group with the EtOH-Vehicle and treated groups: *p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001.
Intestinal permeability is known to increase in cases of inflammation (Salvo-Romero et al., 2015). Ligilactobacillus salivarius I-4866 showed a tendency to decrease sCD14, an indicator of permeability, compared with the DNBS-Vehicle group (Figure 3A). Moreover, L. salivarius CNCM I-4866 treatment tended to decrease LCN-2 concentration in serum (Figure 3B), a biological marker of inflammation.
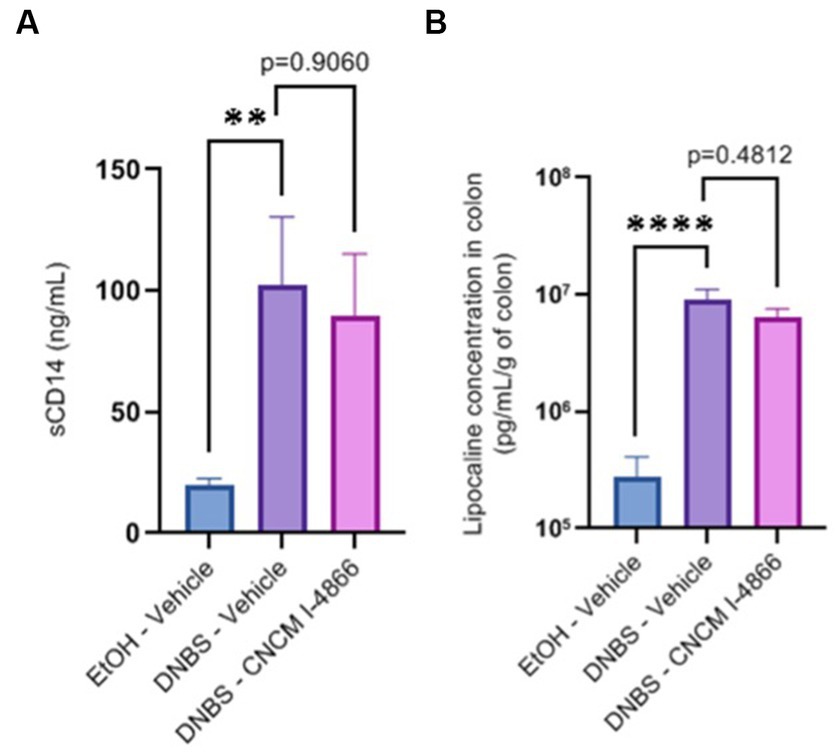
Figure 3. Effects of the L. salivarius 4,866 strain on inflammation markers in DNBS-induced colitis. (A) Levels of sCD14 in serum and (B) levels of lipocalin in the colon. Results of Mann–Whitney U-tests compared with the DNBS-Vehicle group with the EtOH-Vehicle and treated groups: *p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001.
Comparative colonic transcriptomic analysis revealed that 22 genes were modulated between the DNBS-CNCM I-4866 and DNBS-Vehicle groups (Figure 4A). Among them, inflammatory cytokines were found. IL-1β was one of the genes linked to inflammation that was less expressed in the CNCM I-4866-treated group. Analysis of the specific signaling pathways modulated between these two groups (Figure 4B) also revealed that the tumor environment pathway is the top activated pathway in DNBS-Vehicle (Figure 4B). In addition, several pro-inflammatory cytokines (IL-1α, TNF, IL-1β, and IFNγ, among others) were also found as upstream regulators with significant z-scores (Figure 4C) when comparing the DNBS-Vehicle and DNBS-CNCM I-4866 groups. In other words, these results indicated that CNCM I-4866 treatment moderated the production of pro-inflammatory cytokines, as mentioned above. On the other hand, IL-10, an anti-inflammatory cytokine, had a negative z-score, indicating that it was downregulated in the DNBS-Vehicle group compared with the treated group. These results suggest that IL-10 could be more expressed in the colon of mice that have received CNCM I-4866.
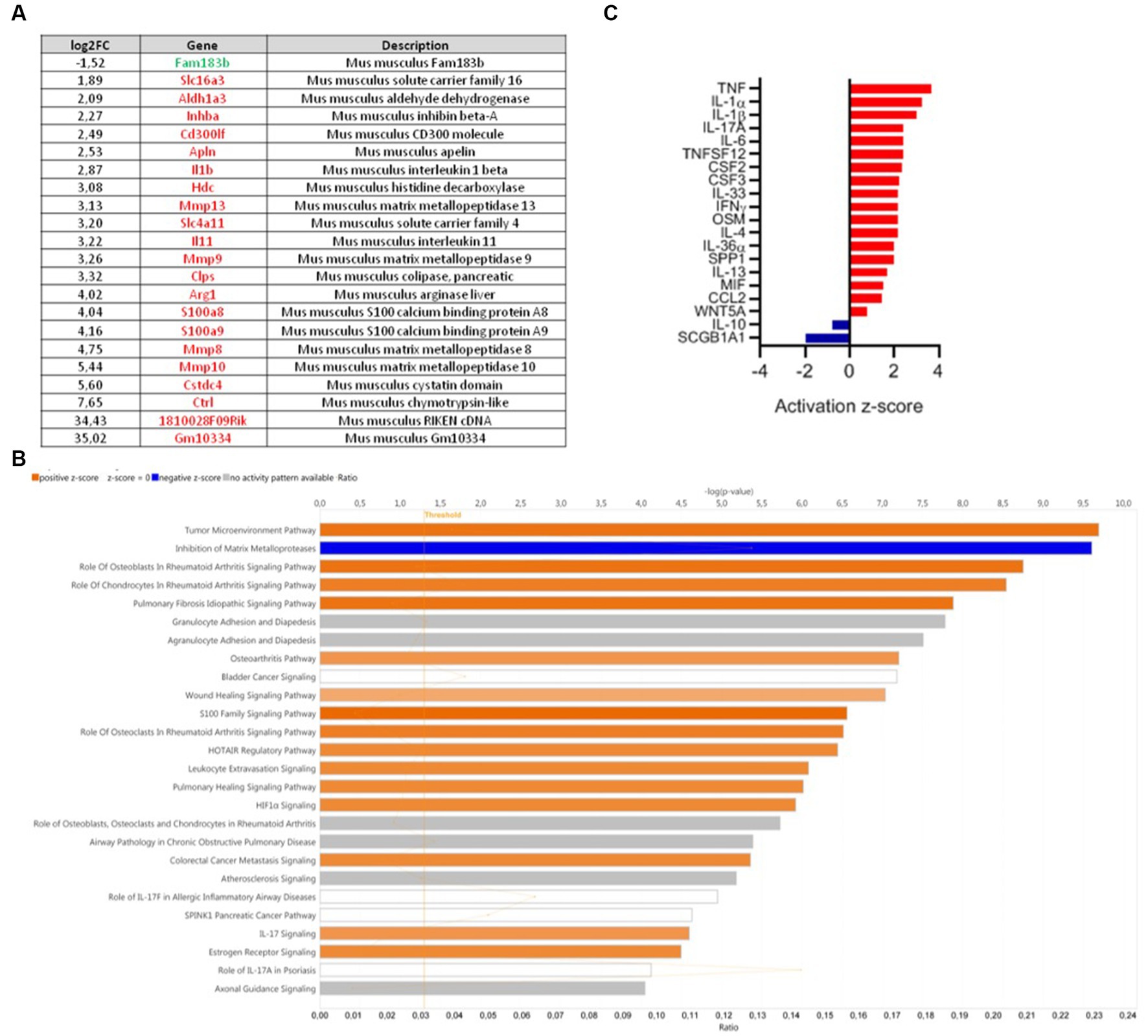
Figure 4. Transcriptomic analysis of mouse colons from the DNBS-Vehicle or DNBS-CNCM I-4866-treated group. (A) Modulation of genes between the DNBS-Vehicle group and DNBS-CNCM I-4866-treated group (adjusted value of p < 0.05 and |log2FoldChange| > 1.5). Upregulated genes are in red and downregulated genes in green; (B) IPA canonical pathway display of the genes modulated in a comparison of DNBS-Vehicle versus DNBS-CNCM I-4866: the y-axis displays the -log of the value of p, which is calculated by a right-tailed Fisher’s exact test. The orange- and blue-colored bars indicate predicted pathway activation or predicted inhibition, respectively. The orange points interconnected by a thin line represent the ratio; (C) Top 20 of affected upstream regulators (only cytokines are represented here) based on IPA. Red indicates activation, while blue indicates suppression.
The capacity of L. salivarius CNCM I-4866 to modulate TNF-α-induced secretion of IL-8, a major pro-inflammatory cytokine, and limit inflammation was conducted on HT-29 cells. In a similar way to the positive control butyrate (Lenoir et al., 2020), our strain has shown great ability to reduce IL-8 production on inflamed HT-29 cells (Figure 5A).

Figure 5. In vitro beneficial capacities of the L. salivarius CNCM I-4866 strain. (A) IL-8 production by HT-29 cells exposed to TNF-α in the presence of L. salivarius CNCM I-4866 and butyrate (positive control); (B) TEER ratio of Caco-2 cells monolayer exposed to TNF-α in the presence of L. salivarius CNCM I-4866 and L. rhamnosus GG. The results of Mann–Whitney U-tests comparing the DPBS control group with the other groups: *p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001.
TEER results have shown that L. salivarius CNCM I-4866 treatment was able to maintain barrier integrity in Caco-2 cells challenged with TNF-α with a slight lower effect that the well-known probiotic candidate L. rhamnosus GG (Figure 5B).
Immunomodulatory effect analysis was also conducted on PBMC from five human donors with the same criteria. Pro-inflammatory cytokines (IL-12) and anti-inflammatory cytokines (IL-10) were dosed after co-incubation of PBMC cells with L. salivarius CNCM I-4866, L. rhamnosus GG, and the control E. coli TG1 (Figures 6A,B). E. coli TG1 is known to produce IL-10 and not IL-12, thus having an elevated IL-10/IL-12 ratio (Sokol et al., 2008). Ligilactobacillus salivarius CNCM I-4866 showed an anti-inflammatory profile based on its high production of IL-10 and high ratio of IL-10/IL-12 (Figure 6D). Moreover, TNF-α production was measured (Figure 6C) in order to determine the TNF-α /IL-10 ratio, which is also an indicator of the inflammatory profile. It appeared to be relatively low for L. salivarius CNCM I-4866 compared with L. rhamnosus GG and equivalent to the E. coli TG1 ratio (Figure 6E).
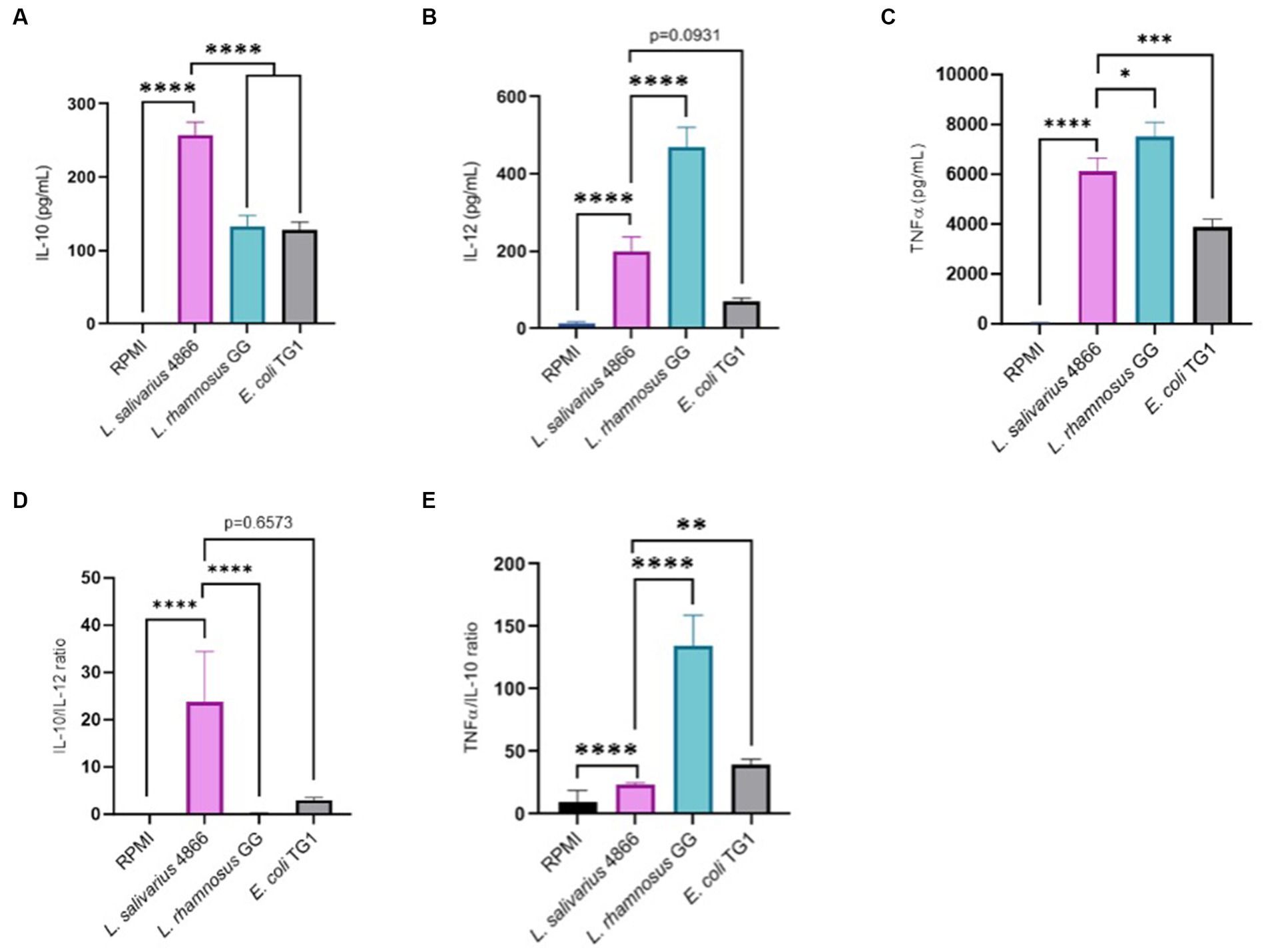
Figure 6. Anti-inflammatory profile based on co-incubation with human PBMCs. (A) IL-10 production by L. salivarius CNCM I-4866 strain after co-incubation with human PBMCs for five donors. L. rhamnosus GG and E. coli TG1 are used as controls with known effects; (B) IL-12 production by L. salivarius CNCM I-4866 strain after co-incubation with human PBMCs, for 5 donors. L. rhamnosus GG and E. coli TG1 are used as controls with known effects; (C) TNF-α production by L. salivarius CNCM I-4866 strain after co-incubation with human PBMCs, for 5 donors. L. rhamnosus GG and E. coli TG1 are used as controls with known effects; (D) IL-10/IL-12 ratio for L. salivarius CNCM I-4866 after co-incubation with human PBMCs. A low ratio is a marker of the pro-inflammatory profile, whereas a high ratio is a marker of the anti-inflammatory profile; (E) TNFα/IL-10 ratio for L. salivarius CNCM I-4866 after co-incubation with human PBMCs. A low ratio is a marker of an anti-inflammatory profile, whereas a high ratio is a marker of a pro-inflammatory profile. Results of Mann–Whitney U-tests comparing L. salivarius 4,866 with other groups: *p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001.
We have studied L. salivarius CNCM I-4866 ability to adhere to several cell lines: HT-29, HT-29 MTX, and Caco-2, as well as porcine mucin (Figure 7A). Comparatively to the reference strain L. rhamnosus GG, which is known to have a good adhesion capacity (Tuomola and Salminen, 1998; Ayeni et al., 2011), L. salivarius CNCM I-4866 has shown a good capacity to adhere to HT-29 and Caco-2 cells. Adhesion to HT-29 MTX cells and porcine mucin was more moderate but similar to L. rhamnosus GG control.
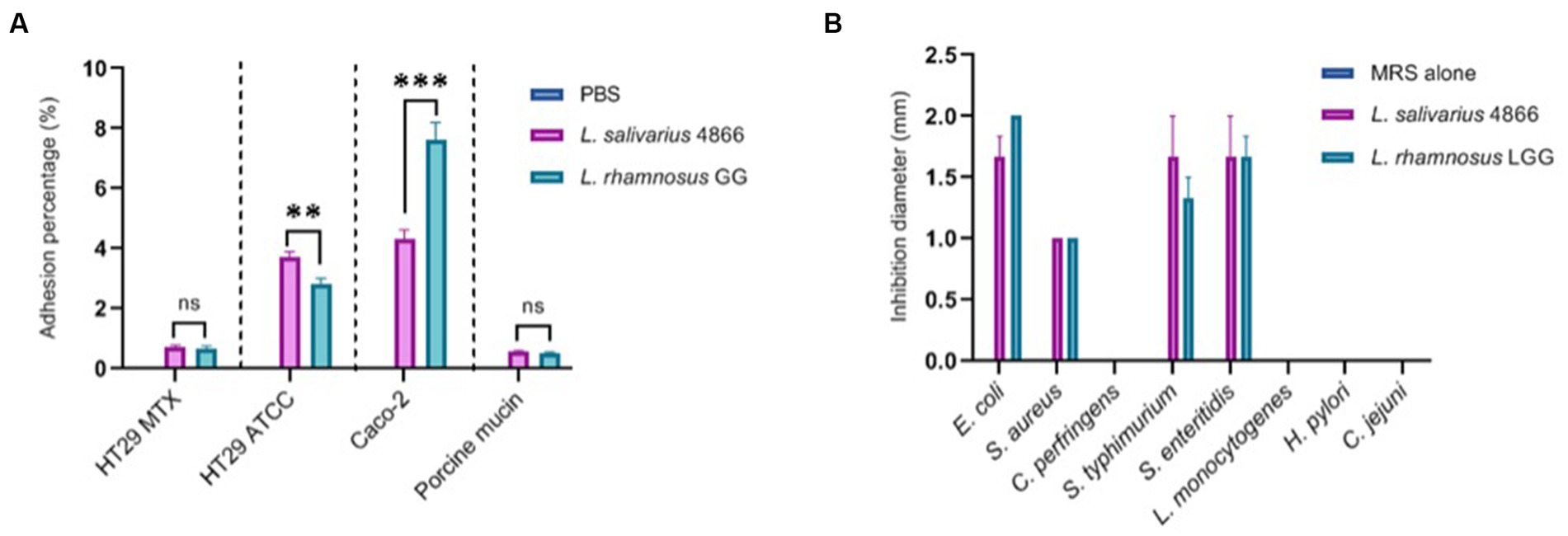
Figure 7. Ligilactobacillus salivarius CNCM I-4866 adhesion and pathogen inhibition in vitro (A) Adhesion percentage to HT-29 MTX, HT-29, Caco-2 cells, and to porcine mucin. Percentage is the ratio of viable count (CFU/ml) after co-incubation compared with the initial inoculum. L. rhamnosus GG was used as a positive control; (B) pathogen inhibition ability of eight pathogens in the L. salivarius CNCM I-4866 supernatant. L. rhamnosus GG was used as a positive control. Results of Mann–Whitney U-tests comparing L. salivarius 4866 and L. rhamnosus GG: **p < 0.01, ***p < 0.001.
The ability of the strain supernatant to inhibit eight pathogens was also tested. Ligilactobacillus salivarius CNCM I-4866 supernatant inhibited four strains: Escherichia coli, Staphylococcus aureus, Salmonella typhimurium, and S. enteritidis (Figure 7B). This inhibition effect was lost when the pH of the supernatant was increased by adding sodium hydroxide, which indicates that the inhibition effect was probably due to lactic acid production.
No significant difference was found after growth in media with 0.3% bile salts (Figure 8A) or after 1 h of exposition to 0.3% bile salts. Concerning tolerance to acidic conditions, L. salivarius CNCM I-4866 growth was found to be slightly impacted by pH 4 and inhibited at pH 2 (Figure 8B). Moreover, no hemolytic activity was found after the growth of L. salivarius CNCM I-4866 on blood agar.
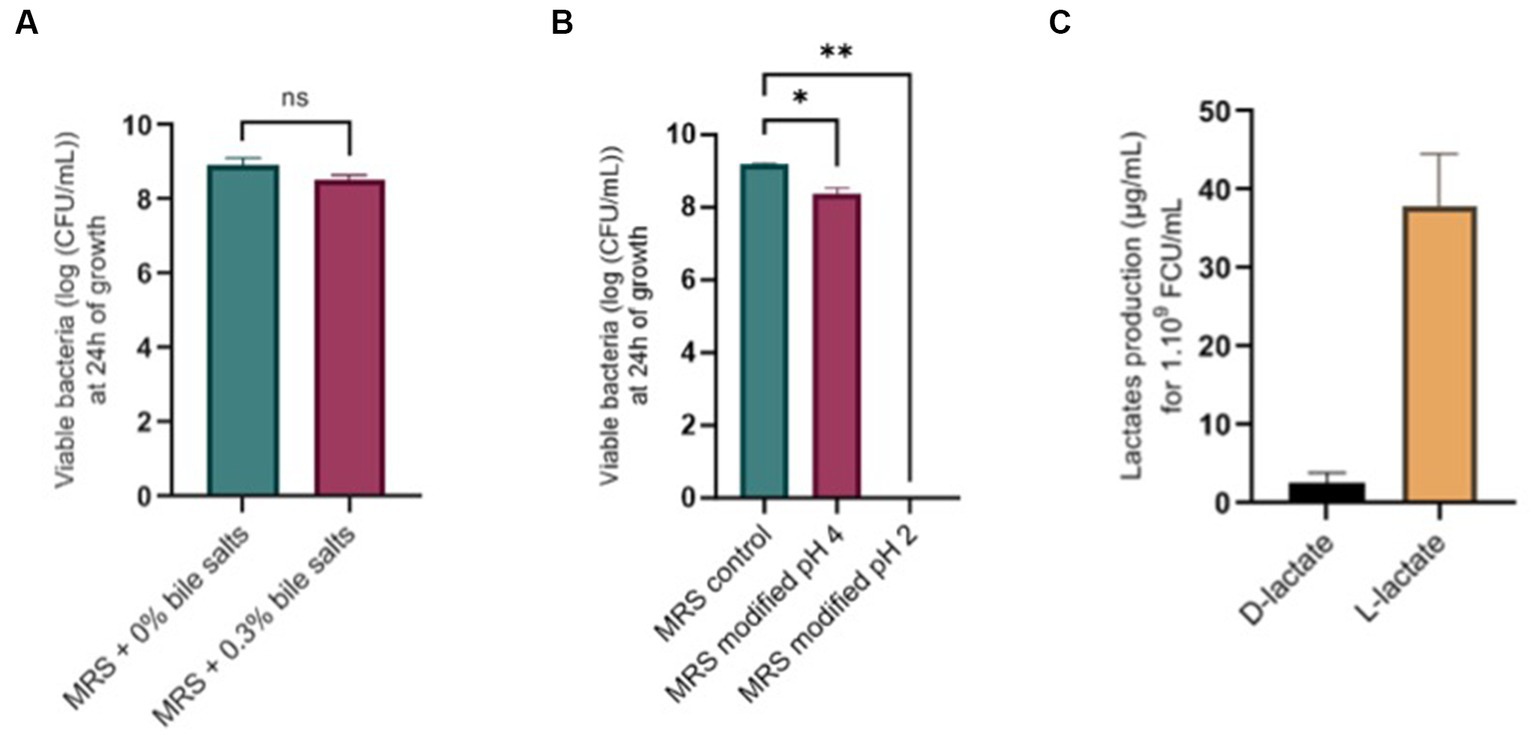
Figure 8. Physiological potentialities of L. salivarius CNCM I-4866. (A) Survival of L. salivarius CNCM I-4866 after 24-h growth in media with 0.3% or 0% bile salts, expressed in log of CFU/ml; (B) Survival of L. salivarius CNCM I-4866 after 24-h growth in media at pH 4 or pH 2, expressed in log of CFU/ml; (C) D-lactate and L-lactate production of L. salivarius CNCM I-4866. Results of Mann–Whitney U-tests: *p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001.
We have measured D-lactate and L-lactate production from the strain L. salivarius CNCM I-4866 which produced mostly L-lactate (Figure 8C).
Finally, potential exopolysaccharide production was first assessed by the Ropy test. The Ropy test was positive (data not shown). This test was completed with a global analysis of the structure of L. salivarius CNCM I-4866 by transmission electron microscopy (Figures 9A,B). This result has shown a structure that is compatible with an EPS layer, suggesting EPS production by the strain.
To ensure the health safety of L. salivarius CNCM I-4866, phenotypic resistance to several antibiotics was determined according to the EFSA recommendations (Table 1). The results showed that the strain is sensitive to ampicillin, gentamicin, streptomycin, erythromycin, clindamycin, tetracycline, and chloramphenicol and resistant to kanamycin.
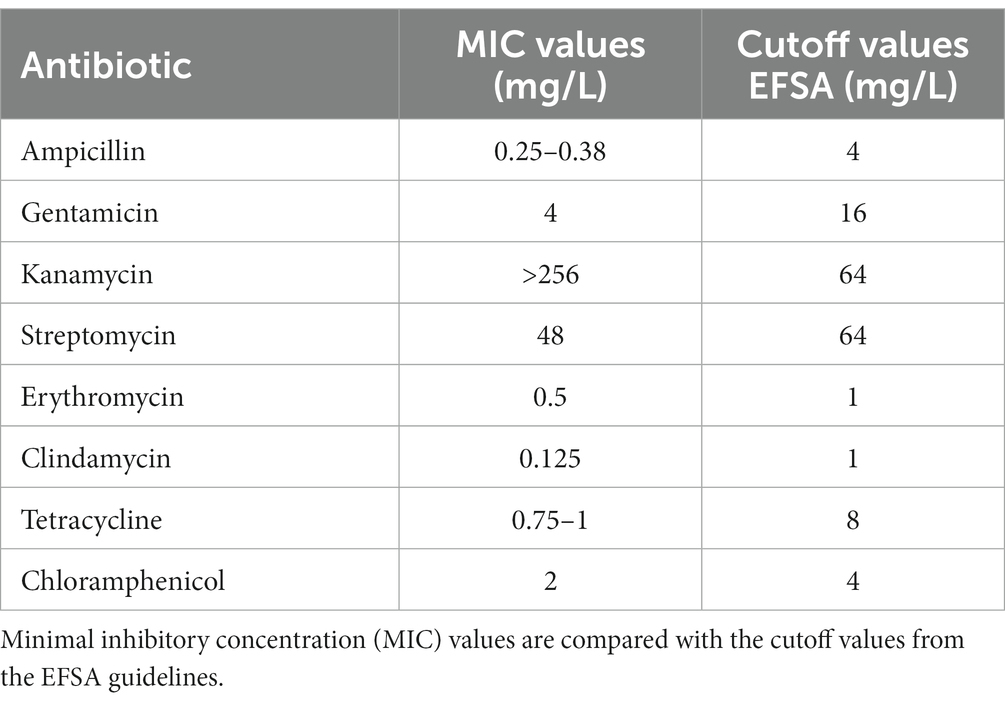
Table 1. Phenotypic antibiotic resistance according to the EFSA recommendations for L. salivarius CNCM I-4866.
Furthermore, the L. salivarius CNCM I-4866 genome was sequenced by whole-genome sequencing. Potential genes for antibiotic resistance were searched in online databases. No gene responsible for antibiotic resistance was found according to our thresholds.
Potential phage or bacteriocin production has been determined in silico and in vitro. In silico Phaster analysis highlighted the presence of an intact prophage, B025, a Listeria prophage (score of 120, length of 43.7 Kb, and region from 121,049 to 164,766 with a percentage of GC of 33.63%). Induction assay and research on lytic plaques with a sensitive strain for this phage have shown that this prophage was not active in our conditions (data not shown). Moreover, in silico, the bacteriocin enterolysin was found to be potentially active. After testing on a sensitive strain, no halo was observed, indicating that the bacteriocin was not active.
In Western countries, IBDs are widespread chronic diseases, with no curative treatment available for the moment (Aldars-Garcia et al., 2021). Therapies based on supplementation with beneficial microorganisms have been pointed out as a potential co-treatment in the management of the symptoms. This approach could be performed with traditional probiotics such as lactic acid bacteria (Doron et al., 2005; Cheng et al., 2020). In this study, we characterized a Lactobacillaceae strain, L. salivarius CNCM I-4866, as a potential probiotic strain to manage and moderate intestinal inflammation.
First, the anti-inflammatory properties of our strain were assessed in a DNBS-induced colitis model. This acute inflammation displays many related features of Crohn’s disease, making it an approaching model for this pathology (Wallace et al., 1995). After treatment with CNCM I-4866, colon macroscopic scores improved significantly compared with DNBS control. These results were in accordance with microscopic scores, including damage to colonic epithelial structure and immune cell infiltration, which L. salivarius CNCM I-4866 tended to improve. To assess the immunomodulatory effects of the strain in our DNBS model, colonic MPO, an enzyme found in the intracellular granules of neutrophils, and serum LCN-2, a non-invasive marker of inflammation, were quantified. Administration of L. salivarius CNCM I-4866 tended to decrease both markers when compared with the DNBS control. Colonic inflammation has been proven to be linked with a dysfunction of intestinal barrier function and, therefore, an increase in permeability as it is observed in IBD patients (Michielan and D’Inca, 2015; Salvo-Romero et al., 2015). By determining sCD14 levels in serum, we have observed that L. salivarius CNCM I-4866 tends to maintain intestinal barrier integrity in cases of inflammation. This model is a robust model for probiotic identification as it has employed other bacteria with well-known probiotic capacities, such as Faecalibacterium prausnitzii (Martin et al., 2014) or several Lactobacillus strains (Benitez-Cabello et al., 2020).
Additionally, colonic transcriptome analysis by comparing the DNBS-Vehicle group with the DNBS-L. salivarius CNCM I-4866-treated group has revealed that genes implied in inflammation and, more precisely, pro-inflammatory cytokine production were less expressed when mice were treated with CNCM I-4866. Conversely, based on the z-score, the expression of the anti-inflammatory cytokine IL-10 was higher in the treated group. The major pathway upregulated in DNBS-Vehicle is the tumor environment pathway, which includes not only cancer cells but also many immune cells that occur during inflammation development. Thereby, IL-1β is a major inflammatory cytokine that mediates other pro-inflammatory cytokines, such as TNF-α or IL-12 (Wang et al., 2017). As these cytokines were upregulated in the control DNBS group (DNBS-Vehicle) compared with the DNBS-CNCM I-4866 group, we can assume that our probiotic candidate has the ability to pacify gene expression linked to immune response. A previous study has shown an equivalent pattern of immunity modulation in a DSS-induced colitis model with a Lactobacillus plantarum strain (Wu et al., 2022). To summarize, L. salivarius CNCM I-4866 seems to alleviate inflammatory bursting caused by DNBS colitis.
Taken together, these in vivo findings indicate that the L. salivarius CNCM I-4866 strain could be a good probiotic candidate to manage and reduce intestinal inflammation found in IBD patients. To go further on the understanding of the beneficial properties of CNCM I-4866 and, more specifically, immunomodulatory effects, two cellular models were used: TNF-α-activated HT-29 cells and PBMCs. Consequently, the anti-inflammatory effect was evaluated by measuring pro-inflammatory cytokine (IL-8) concentration in a co-incubation model of HT-29 cells with L. salivarius CNCM I-4866 bacteria after the TNF-α challenge. IL-8 secretion was shown to be increased in IBD patients and correlated with mucosal inflammation (Daig et al., 1996). De Oliveira et al. (2013) described that this interleukin is responsible for neutrophil activation in the case of inflammation. Previous studies have already evaluated probiotic aptitude with this parameter (Martin et al., 2019; Maillard et al., 2023). Our strain has shown a good ability to reduce IL-8 production as compared with inflamed cells alone, confirming the anti-inflammatory properties observed in vivo.
To have a better picture of the impact of L. salivarius CNCM I-4866 on immunity at the peripheral level, we have determined its capacity to regulate the production of IL-10, IL-12, and TNF-α on human PBMC cells. High secretion of IL-10 was measured with CNCM I-4866 by comparing it with controls. Rogler and Andus (1998) have described the importance of this cytokine in immune homeostasis in IBD patients as an anti-inflammatory response. On the other hand, the pro-inflammatory cytokine IL-12 leads to a Th1 immune-mediated response with the differentiation of T-helper cells. The IL-10/IL-12 ratio is described as a reliable indicator for establishing the inflammatory profile of a strain, with a high ratio associated with an anti-inflammatory profile (Foligne et al., 2007). By comparing it with the well-known probiotic L. rhamnosus GG, CNCM I-4866 produced significantly more IL-10 and less IL-12, revealing a high IL-10/IL-12 ratio. These results are in accordance with those of Foligne et al. (2007), who have shown that L. salivarius Ls33 has a pronounced anti-inflammatory profile based on the IL-10/IL-12 ratio. The low TNF-α/IL-10 ratio, compared with L. rhamnosus GG, allowed us to confirm this anti-inflammatory pattern. These outcomes, taken with in vivo and IL-8 results, point out a strong anti-inflammatory profile of L. salivarius CNCM I-4866.
As mentioned above, intestinal permeability is impacted in the case of inflammation and, therefore, in IBD pathologies. In the DNBS model, CNCM I-4866 tends to maintain barrier permeability by using the sCD14 marker. Furthermore, we have evaluated this capacity in an in vitro model. Caco-2 cells are exposed to TNF-α, which disrupts tight junctions and increases epithelial barrier permeability. L. salivarius CNCM I-4866 treatment was able to restore barrier integrity in Caco-2 cells challenged with TNF-α. A study has shown that L. rhamnosus GG could attenuate permeability dysfunction induced by TNF-α and IFNγ by inhibiting the NF-κB pathway (Donato et al., 2010). Lactobacillus plantarum MB452 was also found to enhance intestinal barrier function by modulating tight junction proteins (Anderson et al., 2010). For CNCM I-4866, the in vivo effect on permeability was not pronounced, but in vitro assay with TEER highlighted the beneficial property of maintaining the permeability of our strain. Additional experiments should be carried out for a better understanding of the partial transferability of these in vitro results to the in vivo preclinical context and to further analyze the underlying mechanisms.
Rossi et al. (2015) and Hidalgo-Cantabrana et al. (2016) have shown that EPS production by probiotics candidates was a key parameter for them to exert their anti-inflammatory properties. To continue with a deeper characterization of L. salivarius CNCM I-4866, we have thus determined EPS production. EPS secretion is known to be a criterion for an adequate probiotic candidate as it has health benefits (Juraskova et al., 2022). With electronic transmission microscopy, we have observed that our strain possesses a potential EPS. This observation is supported by the Ropy test, indicating that CNCM I-4866 produces Ropy-linked EPS. Several mechanisms are known to be implied in beneficial effects exerted by EPS, for example modulation of intestinal microbiota (Salazar et al., 2008).
Even if the underlying mechanisms are not well known, it is known that IBDs are linked to a microbiota imbalance between commensal and pathogenic bacteria. Indeed, some pathogen populations are increased in the case of IBD, such as Salmonella, Escherichia coli, or Listeria monocytogenes (Axelrad et al., 2021). As L. salivarius species are well known to exert antimicrobial activity (Tinrat et al., 2011; Messaoudi et al., 2013), we have evaluated this capacity for CNCM I-4866 against eight intestinal pathogens. L. salivarius CNCM I-4866 was able to inhibit two Salmonella strains: one E. coli strain and one S. aureus strain. This property, potentially due to lactic acid production, is an interesting feature that can be considered for further applications. In a previous study, Kang et al. (2017) highlighted the anti-microbial mechanisms of L. salivarius strains against S. aureus, such as the secretion of anti-staphylococcal proteins.
Beyond its health-beneficial properties, we wanted to ensure that L. salivarius CNCM I-4866 was a good probiotic candidate (European Food Safety Authority, 2018). As a potential human probiotic, tolerance to bile salts is essential, as it will allow the bacteria to reach the lower intestinal tract. Thereby, we have shown that L. salivarius CNCM I-4866 could resist 0.3% bile salts, corresponding to the physiological concentration in the human gastrointestinal tract (Chateau et al., 1994; Prete et al., 2020). This capacity constitutes an advantage for the in vivo survival of the strain. In previous studies, probiotic candidate strains were screened on this parameter, and Lactobacillus strains have also shown good survival at 0.3% bile salts (Khiralla et al., 2015). The ability to tolerate bile salts is commonly known due to bile salt hydrolase activities (Noriega et al., 2006). However, Pan et al. (2021) have described the fact that, for some L. salivarius strains, other mechanisms could be responsible for this property. The growth of our strain was impacted by the low pH (pH 4 and pH 2) that mimic the passage in the gastrointestinal tract and, more specifically, gastric conditions. However, probiotic strains are often administered orally in a protective vehicle that ensures the viability of strain during its passage through the gastrointestinal tract (Tee et al., 2014).
Adhesion to the intestinal mucosa constitutes a key parameter in selecting a probiotic, as it allows the strain to persist and exert its health-beneficial effects for a longer period. L. salivarius CNCM I-4866 possessed good adhesion capacities on several cell lines and mucus at a similar or even better degree than the well-known probiotic, L. rhamnosus GG. Adhesion mechanisms are well described and can be either specific to adhesion proteins (fibronectin, collagen, mucin, and laminin) or unspecific to binding to hydrophobic surfaces (de Wouters et al., 2015). Additionally, CbpA protein was identified in L. salivarius REN as essential for its adhesion to the HT-29 line (Wang et al., 2017). For L. rhamnosus GG, functional analysis has revealed that SpaCBA pili act as an essential factor in adhesion and immunomodulation (Lebeer et al., 2012), as is the case of SpaFED pili for L. rhamnosus CNCM-I3690 (Martin et al., 2023).
Regarding safety concerns, antibiotic resistance constitutes a major issue nowadays, and the risk of resistance gene dissemination should be limited to its maximum (Li et al., 2020). Following the EFSA recommendations (European Food Safety Authority, 2021), no antibiotic-resistance gene was found in our strain. Nevertheless, phenotypic antibiotic resistance has highlighted the resistance to kanamycin. It is well described that Lactobacillus are frequently resistant to kanamycin due to intrinsic resistance (Anisimova and Yarullina, 2019; Campedelli et al., 2019). As no gene is detected, L. salivarius CNCM I-4866 is validated on safety aspects. Additionally, no hemolytic activity was found for our strain. Lactic acid bacteria, as their name suggests, are high producers of lactate. Iraporda et al. (2016) have established that L-lactate treatment could alleviate intestinal inflammation in a mouse TNBS model. However, it has been shown that D-lactate accumulation can lead to acidosis in people with short bowel syndrome (Mack, 2004). As L. salivarius CNCM I-4866 produces mostly L-lactate and very little D-lactate, this strain is suitable for these patients.
In conclusion, our study has shown that a new strain, L. salivarius CNCM I-4866, displays strong anti-inflammatory capacities in vitro and in vivo. Even if further research could be useful to better understand the mechanisms involved or to test this strain on moderate inflammation, CNCM I-4866 is confirmed to be a promising probiotic candidate to alleviate inflammation at the preclinical level on a DNBS model, mimicking IBD and, more specifically, Crohn’s disease. Nevertheless, a human clinical trial should be performed to confirm its potential.
The data presented in the study are deposited in the biosamplehelp@ncbi.nlm.nih.gov repository, accession number SAMN37542358.
Ethical approval was not required for the studies on humans in accordance with the local legislation and institutional requirements because only commercially available established cell lines were used. The animal study was approved by Experiments were performed in accordance with European Union legislation on animal welfare and were approved by COMETHEA, our local committee on animal experimentation (n° 16744–201807061805486) and in compliance with the ARRIVE relevant guidelines. The study was conducted in accordance with the local legislation and institutional requirements.
CC: Conceptualization, Formal analysis, Investigation, Methodology, Validation, Writing – original draft. SC: Investigation, Validation, Methodology, Writing – review & editing. CK: Investigation, Validation, Methodology, Writing – review & editing. LM: Investigation, Validation, Methodology, Writing – review & editing. FC: Investigation, Validation, Methodology, Writing – review & editing. PL: Conceptualization, Funding acquisition, Validation, Writing – review & editing. RM: Conceptualization, Funding acquisition, Investigation, Methodology, Project administration, Supervision, Validation, Writing – review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This project was funded by the SORBIAL company. The SORBIAL company played no role in data collection and interpretation.
This study has benefited from the facilities and expertise of @BRIDGe (Université Paris-Saclay, INRAE, AgroParisTech, GABI, 78350 Jouy-en-Josas, France) and MIMA2-MET (Université Paris-Saclay, INRAE, AgroParisTech, GABI, 78350 Jouy-en-Josas, France). The authors wish to thank the staff of the INRAE Infectiology of Fishes and Rodents Facility (IERP-UE907, Jouy-en-Josas Research Center, France), where animal experiments have been performed. The IERP Facility belongs to the National Distributed Research Infrastructure for the Control of Animal and Zoonotic Emerging Infectious Diseases through in vivo investigation. The authors also want to thank Hilde de Reuse from the Pasteur Institute for kindly providing us with the Helicobacter pylori strain and ONIRIS Nantes for providing the Campylobacter jejuni strain.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Alard, J., Peucelle, V., Boutillier, D., Breton, J., Kuylle, S., Pot, B., et al. (2018). New probiotic strains for inflammatory bowel disease management identified by combining in vitro and in vivo approaches. Benef Microbes 9, 317–331. doi: 10.3920/BM2017.0097
Aldars-Garcia, L., Chaparro, M., and Gisbert, J. P. (2021). Systematic review: the gut microbiome and its potential clinical application in inflammatory bowel disease. Microorganisms 9:977. doi: 10.3390/microorganisms9050977
Ameho, C. K., Adjei, A., Harrison, E. K., Takeshita, K., Morioka, T., Arakaki, Y., et al. (1997). Prophylactic effect of dietary glutamine supplementation on interleukin 8 and tumour necrosis factor α production in trinitrobenzene sulphonic acid induced colitis. Gut 41, 487–493. doi: 10.1136/gut.41.4.487
Anderson, R. C., Cookson, A. L., McNabb, W. C., Park, Z., McCann, M. J., Kelly, W. J., et al. (2010). Lactobacillus plantarum MB452 enhances the function of the intestinal barrier by increasing the expression levels of genes involved in tight junction formation. BMC Microbiol. 10:316. doi: 10.1186/1471-2180-10-316
Anisimova, E. A., and Yarullina, D. R. (2019). Antibiotic resistance of LACTOBACILLUS strains. Curr. Microbiol. 76, 1407–1416. doi: 10.1007/s00284-019-01769-7
Axelrad, J. E., Cadwell, K. H., Colombel, J. F., and Shah, S. C. (2021). The role of gastrointestinal pathogens in inflammatory bowel disease: a systematic review. Ther. Adv. Gastroenterol. 14:17562848211004493. doi: 10.1177/17562848211004493
Ayeni, F. A., Sanchez, B., Adeniyi, B. A., de Los Reyes-Gavilan, C. G., Margolles, A., and Ruas-Madiedo, P. (2011). Evaluation of the functional potential of Weissella and Lactobacillus isolates obtained from Nigerian traditional fermented foods and cow's intestine. Int. J. Food Microbiol. 147, 97–104. doi: 10.1016/j.ijfoodmicro.2011.03.014
Barone, M., Chain, F., Sokol, H., Brigidi, P., Bermudez-Humaran, L. G., and Langella, P. (2018). A versatile new model of chemically induced chronic colitis using an outbred murine strain. Front. Microbiol. 9:565. doi: 10.3389/fmicb.2018.00565
Benitez-Cabello, A., Torres-Maravilla, E., Bermudez-Humaran, L., Langella, P., Martin, R., Jimenez-Diaz, R., et al. (2020). Probiotic properties of Lactobacillus strains isolated from Table olive biofilms. Probiotics Antimicrob Proteins 12, 1071–1082. doi: 10.1007/s12602-019-09604-y
Campedelli, I., Mathur, H., Salvetti, E., Clarke, S., Rea, M. C., and Torriani, S. (2019). Genus-wide assessment of antibiotic resistance in Lactobacillus spp. Appl. Environ. Microbiol. 85:e01738-18. doi: 10.1128/AEM.01738-18
Chamignon, C., Gueneau, V., Medina, S., Deschamps, J., Gil-Izquierdo, A., Briandet, R., et al. (2020). Evaluation of the probiotic properties and the capacity to form biofilms of various Lactobacillus strains. Microorganisms 8:1053. doi: 10.3390/microorganisms8071053
Chateau, N., Deschamps, A. M., and Sassi, H. (1994). Heterogeneity of bile salts resistance in the Lactobacillus isolates of a probiotic consortium. Lett. Appl. Microbiol. 18, 42–44. doi: 10.1111/j.1472-765X.1994.tb00796.x
Chaves, B. D., Brashears, M. M., and Nightingale, K. K. (2017). Applications and safety considerations of Lactobacillus salivarius as a probiotic in animal and human health. J. Appl. Microbiol. 123, 18–28. doi: 10.1111/jam.13438
Cheng, F. S., Pan, D., Chang, B., Jiang, M., and Sang, L. X. (2020). Probiotic mixture VSL#3: an overview of basic and clinical studies in chronic diseases. World J. Clin. Cases 8, 1361–1384. doi: 10.12998/wjcc.v8.i8.1361
Daig, R., Andus, T., Aschenbrenner, E., Falk, W., Schölmerich, J., and Gross, V. (1996). Increased interleukin 8 expression in the colon mucosa of patients with inflammatory bowel disease. Gut 38, 216–222. doi: 10.1136/gut.38.2.216
de Oliveira, S., Reyes-Aldasoro, C. C., Candel, S., Renshaw, S. A., Mulero, V., and Calado, A. (2013). Cxcl8 (IL-8) mediates neutrophil recruitment and behavior in the zebrafish inflammatory response. J. Immunol. 190, 4349–4359. doi: 10.4049/jimmunol.1203266
de Wouters, T., Jans, C., Niederberger, T., Fischer, P., and Ruhs, P. A. (2015). Adhesion potential of intestinal microbes predicted by Physico-chemical characterization methods. PLoS One 10:e0136437. doi: 10.1371/journal.pone.0136437
Donato, K. A., Gareau, M. G., Wang, Y. J. J., and Sherman, P. M. (2010). Lactobacillus rhamnosus GG attenuates interferon-gamma and tumour necrosis factor-alpha-induced barrier dysfunction and pro-inflammatory signalling. Microbiology (Reading) 156, 3288–3297. doi: 10.1099/mic.0.040139-0
Doron, S., Snydman, D. R., and Gorbach, S. L. (2005). Lactobacillus GG: bacteriology and clinical applications. Gastroenterol. Clin. N. Am. 34, 483–498. doi: 10.1016/j.gtc.2005.05.011
European Food Safety Authority (2018). Guidance on the characterisation of microorganisms used as feed additives or as production organisms. EFSA J. 16:e05206.
European Food Safety Authority (2021). EFSA statement on the requirements for whole genome sequence analysis of microorganisms intentionally used in the food chain. EFSA J. 19:e06506. doi: 10.2903/j.efsa.2021.6506
Foligne, B., Nutten, S., Grangette, C., Dennin, V., Goudercourt, D., Poiret, S., et al. (2007). Correlation between in vitro and in vivo immunomodulatory properties of lactic acid bacteria. World J. Gastroenterol. 13, 236–243. doi: 10.3748/wjg.v13.i2.236
Frank, D. N., St Amand, A. L., Feldman, R. A., Boedeker, E. C., Harpaz, N., and Pace, N. R. (2007). Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc. Natl. Acad. Sci. U. S. A. 104, 13780–13785. doi: 10.1073/pnas.0706625104
Glassner, K. L., Abraham, B. P., and Quigley, E. M. M. (2020). The microbiome and inflammatory bowel disease. J. Allergy Clin. Immunol. 145, 16–27. doi: 10.1016/j.jaci.2019.11.003
Hidalgo-Cantabrana, C., Algieri, F., Rodriguez-Nogales, A., Vezza, T., Martinez-Camblor, P., Margolles, A., et al. (2016). Effect of a ropy exopolysaccharide-producing Bifidobacterium animalis subsp. lactis strain orally administered on DSS-induced colitis mice model. Front. Microbiol. 7:868. doi: 10.3389/fmicb.2016.00868
Hill, C., Guarner, F., Reid, G., Gibson, G. R., Merenstein, D. J., Pot, B., et al. (2014). Expert consensus document. The international scientific Association for Probiotics and Prebiotics consensus statement on the scope and appropriate use of the term probiotic. Nat. Rev. Gastroenterol. Hepatol. 11, 506–514. doi: 10.1038/nrgastro.2014.66
Iraporda, C., Romanin, D. E., Bengoa, A. A., Errea, A. J., Cayet, D., Foligne, B., et al. (2016). Local treatment with lactate prevents intestinal inflammation in the TNBS-induced colitis model. Front. Immunol. 7:651. doi: 10.3389/fimmu.2016.00651
Iyer, N., Williams, M. A., O'Callaghan, A. A., Dempsey, E., Cabrera-Rubio, R., Raverdeau, M., et al. (2022). Lactobacillus salivarius UCC118 dampens inflammation and promotes microbiota recovery to provide therapeutic benefit in a DSS-induced colitis model. Microorganisms 10:1383. doi: 10.3390/microorganisms10071383
Jakubczyk, D., Leszczynska, K., and Gorska, S. (2020). The effectiveness of probiotics in the treatment of inflammatory bowel disease (IBD)-a critical review. Nutrients 12:1973. doi: 10.3390/nu12071973
Jandhyala, S. M., Talukdar, R., Subramanyam, C., Vuyyuru, H., Sasikala, M., and Nageshwar Reddy, D. (2015). Role of the normal gut microbiota. World J. Gastroenterol. 21, 8787–8803. doi: 10.3748/wjg.v21.i29.8787
Juraskova, D., Ribeiro, S. C., and Silva, C. C. G. (2022). Exopolysaccharides produced by lactic acid Bacteria: from biosynthesis to health-promoting properties. Foods 11:156. doi: 10.3390/foods11020156
Kang, M. S., Lim, H. S., Oh, J. S., Lim, Y. J., Wuertz-Kozak, K., Harro, J. M., et al. (2017). Antimicrobial activity of Lactobacillus salivarius and Lactobacillus fermentum against Staphylococcus aureus. Pathog. Dis. 75. doi: 10.1093/femspd/ftx0009
Khiralla, G. M., Mohamed, E. A. H., Farag, A. G., and Elhariry, H. (2015). Antibiofilm effect of Lactobacillus pentosus and Lactobacillus plantarum cell-free supernatants against some bacterial pathogens. J. Biotech. Res. 6, 86–95.
Lebeer, S., Claes, I., Tytgat, H. L., Verhoeven, T. L., Marien, E., von Ossowski, I., et al. (2012). Functional analysis of Lactobacillus rhamnosus GG pili in relation to adhesion and immunomodulatory interactions with intestinal epithelial cells. Appl. Environ. Microbiol. 78, 185–193. doi: 10.1128/AEM.06192-11
Lenoir, M., Martin, R., Torres-Maravilla, E., Chadi, S., Gonzalez-Davila, P., Sokol, H., et al. (2020). Butyrate mediates anti-inflammatory effects of Faecalibacterium prausnitzii in intestinal epithelial cells through Dact3. Gut Microbes 12, 1–16. doi: 10.1080/19490976.2020.1826748
Li, T., Teng, D., Mao, R., Hao, Y., Wang, X., and Wang, J. (2020). A critical review of antibiotic resistance in probiotic bacteria. Food Res. Int. 136:109571. doi: 10.1016/j.foodres.2020.109571
Mack, D.R. (2004). D(−)-lactic acid-producing probiotics, D(−)-lactic acidosis and infants. Can. J. Gastroenterol.. 2004;18, 671–675. doi: 10.1155/2004/342583
Maillard, F., Meynier, M., Mondot, S., Pepke, F., Galbert, C., Torres Maravilla, E., et al. (2023). From in vitro to in vivo: a rational flowchart for the selection and characterization of candidate probiotic strains in intestinal disorders. Microorganisms 11:906. doi: 10.3390/microorganisms11040906
Martin, R., Benitez-Cabello, A., Kulakauskas, S., Viana, M. V. C., Chamignon, C., Courtin, P., et al. (2023). Over-production of exopolysaccharide by Lacticaseibacillus rhamnosus CNCM I-3690 strain cutbacks its beneficial effect on the host. Sci. Rep. 13:6114. doi: 10.1038/s41598-023-32116-3
Martin, R., Chain, F., Miquel, S., Lu, J., Gratadoux, J. J., Sokol, H., et al. (2014). The commensal bacterium Faecalibacterium prausnitzii is protective in DNBS-induced chronic moderate and severe colitis models. Inflamm. Bowel Dis. 20, 417–430. doi: 10.1097/01.MIB.0000440815.76627.64
Martin, R., Chamignon, C., Mhedbi-Hajri, N., Chain, F., Derrien, M., Escribano-Vazquez, U., et al. (2019). The potential probiotic Lactobacillus rhamnosus CNCM I-3690 strain protects the intestinal barrier by stimulating both mucus production and cytoprotective response. Sci. Rep. 9:5398. doi: 10.1038/s41598-019-41738-5
Messaoudi, S., Manai, M., Kergourlay, G., Prevost, H., Connil, N., Chobert, J. M., et al. (2013). Lactobacillus salivarius: bacteriocin and probiotic activity. Food Microbiol. 36, 296–304. doi: 10.1016/j.fm.2013.05.010
Michielan, A., and D’Inca, R. (2015). Intestinal permeability in inflammatory bowel disease: pathogenesis, clinical evaluation, and therapy of leaky gut. Mediat. Inflamm. 2015:628157. doi: 10.1155/2015/628157
Noriega, L., Cuevas, I., Margolles, A., and De los Reyes-Gavilán, C. G. (2006). Deconjugation and bile salts hydrolase activity by Bifidobacterium strains with acquired resistance to bile. Int. Dairy J. 16, 850–855. doi: 10.1016/j.idairyj.2005.09.008
Pan, Q., Shen, X., Yu, L., Tian, F., Zhao, J., Zhang, H., et al. (2021). Comparative genomic analysis determines the functional genes related to bile salt resistance in Lactobacillus salivarius. Microorganisms 9:2038. doi: 10.3390/microorganisms9102038
Peran, L., Camuesco, D., Comalada, M., Nieto, A., Concha, A., Diaz-Ropero, M. P., et al. (2005). Preventative effects of a probiotic, Lactobacillus salivarius ssp. salivarius, in the TNBS model of rat colitis. World J. Gastroenterol. 11, 5185–5192. doi: 10.3748/wjg.v11.i33.5185
Prete, R., Long, S. L., Gallardo, A. L., Gahan, C. G., Corsetti, A., and Joyce, S. A. (2020). Beneficial bile acid metabolism from Lactobacillus plantarum of food origin. Sci. Rep. 10:1165. doi: 10.1038/s41598-020-58069-5
Rogler, G., and Andus, T. (1998). Cytokines in inflammatory bowel disease. World J. Surg. 22, 382–389. doi: 10.1007/s002689900401
Rossi, O., Khan, M. T., Schwarzer, M., Hudcovic, T., Srutkova, D., Duncan, S. H., et al. (2015). Faecalibacterium prausnitzii strain HTF-F and its extracellular polymeric matrix attenuate clinical parameters in DSS-induced colitis. PLoS One 10:e0123013. doi: 10.1371/journal.pone.0123013
Rychen, G., Aquilina, G., Azimonti, G., Bampidis, V., Bastos, M., Bories, G., et al. (2018). Guidance on the characterisation of microorganisms used as feed additives or as production organisms. EFSA J. 16:e05206. doi: 10.2903/j.efsa.2018.5206
Salazar, N., Gueimonde, M., Hernandez-Barranco, A. M., Ruas-Madiedo, P., and de los Reyes-Gavilan, C. (2008). Exopolysaccharides produced by intestinal Bifidobacterium strains act as fermentable substrates for human intestinal Bacteria. Appl. Environ. Microbiol. 74, 4737–4745. doi: 10.1128/AEM.00325-08
Salvo-Romero, E., Alonso-Cotoner, C., Pardo-Camacho, C., Casado-Bedmar, M., and Vicario, M. (2015). The intestinal barrier function and its involvement in digestive disease. Rev. Esp. Enferm. Dig. 107, 686–696. doi: 10.17235/reed.2015.3846/2015
Sokol, H., Pigneur, B., Watterlot, L., Lakhdari, O., Bermudez-Humaran, L. G., Gratadoux, J. J., et al. (2008). Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proc. Natl. Acad. Sci. U. S. A. 105, 16731–16736. doi: 10.1073/pnas.0804812105
Tee, W. F., Nazzarudin, R., Tan, Y. N., and Ayob, M. K. (2014). Effects of encapsulation on the viability of potential probiotic Lactobacillus plantarum exposed to high acidity condition and presence of bile salts. Food Sci. Technol. Int. 20, 399–404. doi: 10.1177/1082013213488775
Thursby, E., and Juge, N. (2017). Introduction to the human gut microbiota. Biochem. J. 474, 1823–1836. doi: 10.1042/BCJ20160510
Tinrat, S., Sumarn, S., and Mullika, T. C. (2011). Isolation and characterization of Lactobacillus salivarius MTC 1026 as a potential probiotics. J. Gen. Appl. Microbiol. 57, 365–378. doi: 10.2323/jgam.57.365
Tuomola, E. M., and Salminen, S. J. (1998). Adhesion of some probiotic and dairy Lactobacillus strains to Caco-2 cell cultures. Int. J. Food Microbiol. 41, 45–51. doi: 10.1016/S0168-1605(98)00033-6
Wallace, J. L., Le, T., Carter, L., Appleyard, C. B., and Beck, P. L. (1995). (1995). Hapten-induced chronic colitis in the rat: alternatives to Trinitrobenzene sulfonic acid. J. Pharmacol. Toxicol. Methods 33, 237–239. doi: 10.1016/1056-8719(95)00001-X
Wallace, JL.M.W ., Morris, GP., and Beck, PL (1989). Inhibition of leukotriene synthesis markedly accelerates healing in a rat model of inflammatory bowel disease. Gastroenterology 96, 29–36. doi: 10.1016/0016-5085(89)90760-9
Wang, R., Jiang, L., Zhang, M., Zhao, L., Hao, Y., Guo, H., et al. (2017). The adhesion of Lactobacillus salivarius REN to a human intestinal epithelial cell line requires S-layer proteins. Sci. Rep. 7:44029. doi: 10.1038/srep44029
Wang, M., Zhao, J., Zhang, L., Wei, F., Lian, Y., Wu, Y., et al. (2017). Role of tumor microenvironment in tumorigenesis. J. Cancer 8, 761–773. doi: 10.7150/jca.17648
Wu, Y., Jha, R., Li, A., Liu, H., Zhang, Z., Zhang, C., et al. (2022). Probiotics (Lactobacillus plantarum HNU082) supplementation relieves ulcerative colitis by affecting intestinal barrier functions, immunity-related gene expression, gut microbiota, and metabolic pathways in mice. Microbiol Spectr. 10:e0165122. doi: 10.1128/spectrum.01651-22
Keywords: Ligilactobacillus salivarius , probiotic, DNBS inflammation, in vitro , anti-inflammatory
Citation: Carbonne C, Chadi S, Kropp C, Molimard L, Chain F, Langella P and Martin R (2023) Ligilactobacillus salivarius CNCM I-4866, a potential probiotic candidate, shows anti-inflammatory properties in vitro and in vivo. Front. Microbiol. 14:1270974. doi: 10.3389/fmicb.2023.1270974
Received: 01 August 2023; Accepted: 23 October 2023;
Published: 29 November 2023.
Edited by:
Edoardo Pasolli, University of Naples Federico II, ItalyReviewed by:
Biswaranjan Pradhan, Indian Institute of Technology Bhubaneswar, IndiaCopyright © 2023 Carbonne, Chadi, Kropp, Molimard, Chain, Langella and Martin. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Rebeca Martin, cmViZWNhLm1hcnRpbi1yb3NpcXVlQGlucmFlLmZy
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.