
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Microbiol. , 11 January 2024
Sec. Food Microbiology
Volume 14 - 2023 | https://doi.org/10.3389/fmicb.2023.1241259
This article is part of the Research Topic New Insight into the Interplay Between Probiotics, Prebiotics, Postbiotics, and Gut Microbiota in Glycolipid Metabolism, Inflammation, and Associated Metabolic Disorders View all 8 articles
 Olga Ignatyeva*
Olga Ignatyeva* Darya Tolyneva
Darya Tolyneva Aleksandr Kovalyov
Aleksandr Kovalyov Lorena Matkava
Lorena Matkava Mikhail Terekhov
Mikhail Terekhov Daria Kashtanova
Daria Kashtanova Anzhelika Zagainova
Anzhelika Zagainova Mikhail Ivanov
Mikhail Ivanov Vladimir Yudin
Vladimir Yudin Valentin Makarov
Valentin Makarov Anton Keskinov
Anton Keskinov Sergey Kraevoy
Sergey Kraevoy Sergey Yudin
Sergey YudinBackground: As the field of probiotic research continues to expand, new beneficial strains are being discovered. The Christensenellaceae family and its newly described member, Christensenella minuta, have been shown to offer great health benefits. We aimed to extensively review the existing literature on these microorganisms to highlight the advantages of their use as probiotics and address some of the most challenging aspects of their commercial production and potential solutions.
Methods: We applied a simple search algorithm using the key words “Christensenellaceae” and “Christensenella minuta” to find all articles reporting the biotherapeutic effects of these microorganisms. Only articles reporting evidence-based results were reviewed.
Results: The review showed that Christensenella minuta has demonstrated numerous beneficial properties and a wider range of uses than previously thought. Moreover, it has been shown to be oxygen-tolerant, which is an immense advantage in the manufacturing and production of Christensenella minuta-based biotherapeutics. The results suggest that Christensenellaceae and Christensenella munita specifically can play a crucial role in maintaining a healthy gut microbiome. Furthermore, Christensenellaceae have been associated with weight management. Preliminary studies suggest that this probiotic strain could have a positive impact on metabolic disorders like diabetes and obesity, as well as inflammatory bowel disease.
Conclusion: Christensenellaceae and Christensenella munita specifically offer immense health benefits and could be used in the management and therapy of a wide range of health conditions. In addition to the impressive biotherapeutic effect, Christensenella munita is oxygen-tolerant, which facilitates commercial production and storage.
We are home to trillions of microorganisms. They live virtually on and in every part of our bodies, affecting our metabolism, immune system, and many vital physiological functions. One could even argue that this coexistence and, hence, coevolution have shaped our species. These highly complex, self-regulating microbial communities form the microbiome, which is often referred to as one of the body’s organs. It has long been considered a promising therapeutic target that can be regulated with probiotics, or “good microorganisms”.
Probiotic foods such as fermented dairy products became a staple long before people were aware of the benefits they offered. The first deliberate efforts to “harness the power” of the microbiome came soon after the successful isolation of several bacterial species from human feces. This newly acquired ability to isolate, culture, and produce beneficial bacterial strains ushered in the era of probiotic drugs. Since then, they have proven effective in treating many infectious and non-communicable diseases.
Most common probiotic drugs contain various strains of lyophilized lactobacilli and/or bifidobacteria; less frequently, Streptococcus, Bacillus, and others. Since the first successful cultivation of fecal microorganisms, these bacteria have long been considered to be the most important representatives of the human gut microbiome. More recently, the advent of culture-independent molecular techniques, such as polymerase chain reaction (PCR) and especially DNA sequencing, has allowed for a much closer examination of the commensal bacteria. These scientific advances enabled the detection of hundreds of new microorganisms that could not be detected using conventional techniques. Many of these microorganisms were eventually successfully cultured and studied in more detail. More importantly, some of these new representatives of the human microbiome, such as Akkermansia, Faecalibacterium, Christensenella, Anaerobutyricum, Bacteroides, Roseburia, etc., have proven crucial to human health and well-being. This could represent a breakthrough in the history of probiotic medicine and pave the way for next-generation probiotics (NGPs).
In this review, we present an overview of the Christensenellaceae family and focus on Christensenella minuta (C. minuta) as an NGP candidate that could offer tremendous health benefits.
C. minuta (DSM 22607) was first isolated from the feces of a healthy Japanese donor by Morotomi et al. (2012). It was designated as the first member of the new Christensenellaceae family in the Clostridiales order in the Firmicutes phylum. The genus name honors Professor Henrik Christensen, an outstanding Danish microbiologist. The species name reflects the small size of the bacterial cells and their colonies (Morotomi et al., 2012).
C. minuta is a small (ranging in size from 0.5 mm to 1.9 mm), gram-negative, non-sporulating, and non-motile bacterium (Figure 1A) that forms circular, almost colorless colonies (Figure 1B). It was originally described as strictly anaerobic; however, later studies demonstrated that it can tolerate oxygen for several hours (Kropp et al., 2021). Contrary to popular belief, exposure to atmospheric air does not instantly kill the bacterium but rather reduces its viability. C. minuta grows most rapidly at 37–40° and pH 7.5 (Morotomi et al., 2012). It is not particularly fastidious and can grow on various media, such as Gifu Anaerobic Medium (GAM Broth), Wilkins–Chalgren Anaerobe Agar, and Schaedler Anaerobe Agar (Morotomi et al., 2012).
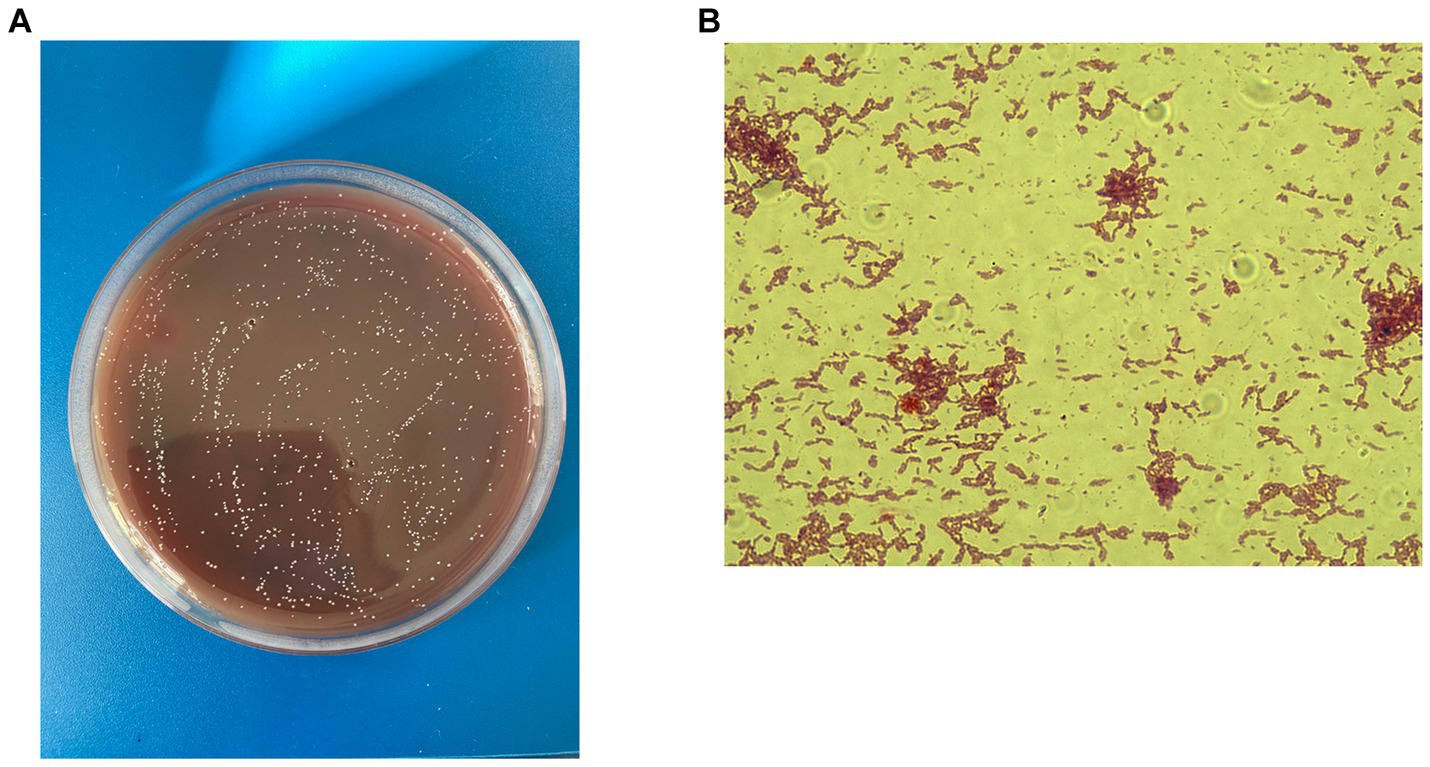
Figure 1. C. minuta VKM B-3687D isolated at the Centre for Strategic Planning and Management of Biomedical Health Risks: (A) Gram staining; x 100 and (B) colonies grown on the Schaedler agar.
C. minuta can ferment many sugars, including glucose, salicin, xylose, arabinose, rhamnose, and mannose (Morotomi et al., 2012). The main fermentation products include acetic and butyric acids. Additionally, it has been demonstrated to convert organic substrates via fermentation to create large volumes of H2 (Ruaud et al., 2020). C. minuta does not produce catalase, oxidase, urease, or indole, and does not reduce nitrates (Morotomi et al., 2012). Recently, DSM 22607, one of C. minuta strains, has been shown to produce a novel bile salt hydrolase (BSH), which could expand the biotherapeutic range of this microorganism (Dejean et al., 2021).
Metagenomic studies have shown that members of the family Christensenellaceae are found on all continents (Escobar et al., 2014; Morton et al., 2015; Oki et al., 2016; Barrett et al., 2018; Brooks et al., 2018). They live in the microbiome of a wide variety of animals, from cockroaches (Richards et al., 2017) and lizards (Baldo et al., 2018) to birds (Crisol-Martinez et al., 2017) and mammals (Kamke et al., 2016; McKenzie et al., 2017), including humans. These bacteria are primarily found in the gastrointestinal tract but have also been found in the respiratory tract (Huang et al., 2010; Gangneux et al., 2020) and genitourinary tract in primates (Adapen et al., 2022).
Another potential therapeutic advantage of Christensenellaceae is their high heritability, as demonstrated in a large-scale UK study on the gut microbiome of twins (Goodrich et al., 2014). This family has also been described as the hub of a co-occurrence network, i.e., the one with the largest number of connections with other highly heritable taxa. This finding has several implications. First, members of the family Christensenellaceae are “hardwired” into the host’s metabolism. Second, they are likely to affect the abundance of other taxa and promote the growth of beneficial commensals such as Methanobacteriaceae. Therefore, this low-abundance family, accounting for just about 0.01% of the human microbiome (Goodrich et al., 2014), may hypothetically be a crucial microbiome “builder” and health promoter.
Numerous studies have shown that Christensenellaceae are abundant in the healthy microbiome, suggesting that their abundance could be a reliable health marker and a promising biotherapeutic target.
The Christensenellaceae family was isolated and included in the reference databases in 2012. Since then, the benefits offered by its members have been recognized worldwide. This family has been repeatedly described as the “microbial satellite” of health and leanness (Goodrich et al., 2014). It has also been linked to healthy aging (Biagi et al., 2016). Within just 10 years, ample evidence has been accumulated, indicating that Christensenellaceae are significantly depleted in many diseases (Figure 2).
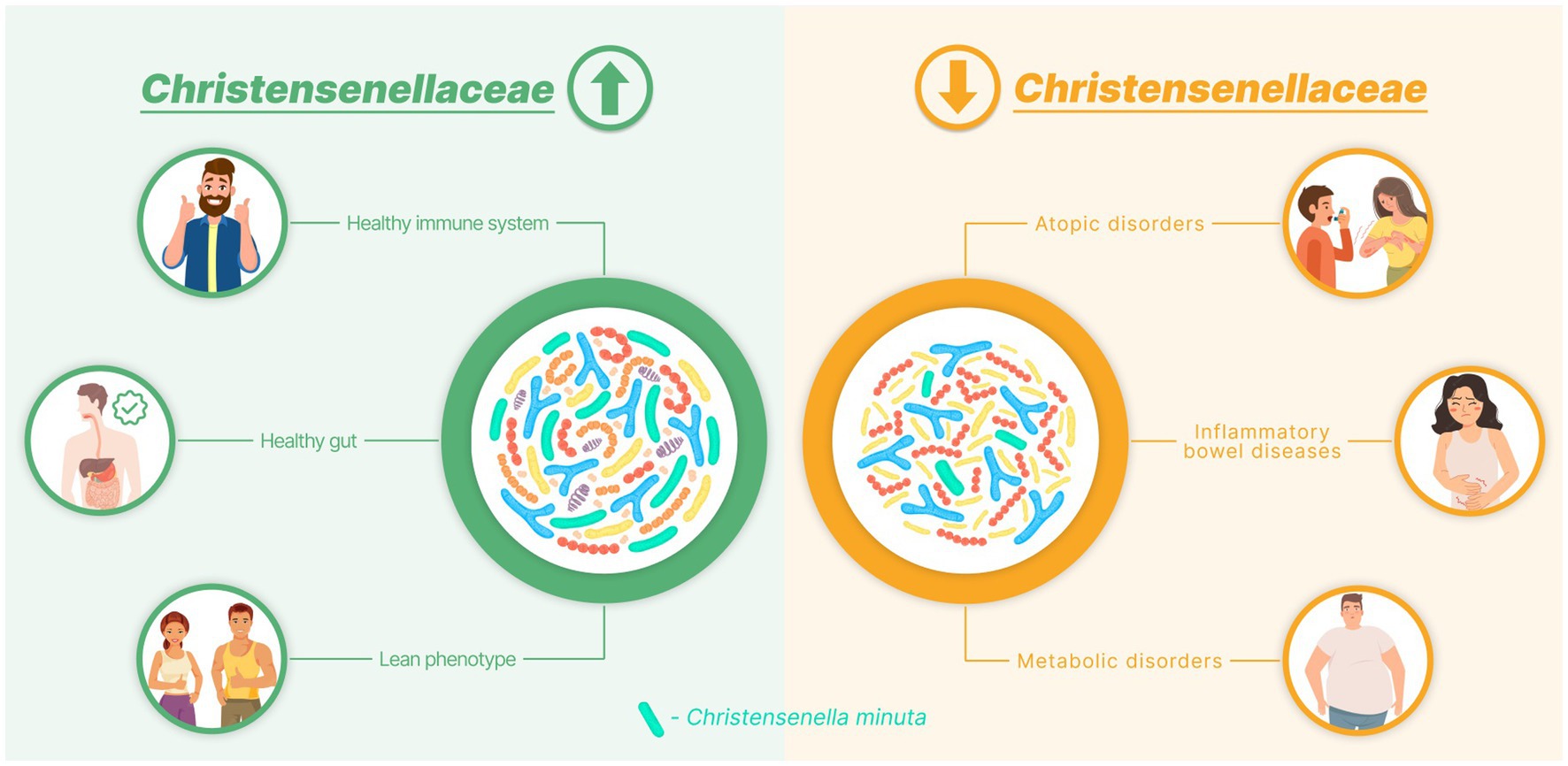
Figure 2. Diagram demonstrating the association between the Christensenellaceae abundance and human health.
The Christensenellaceae family has been highlighted in several studies of human metabolism and its markers. Metabolic syndrome and obesity have been commonly reported as disorders associated with Christensenellaceae deficiency in both adults (Lim et al., 2017; He et al., 2018; Peters et al., 2018) and children (Alcazar et al., 2022). Multiple pieces of evidence indicate that the abundance of this family correlates with numerous metabolic markers. Sowah et al. detected a negative correlation between the abundance of Christensenellaceae in the gut microbiome and several metabolic indicators, such as body mass index (BMI), visceral adipose tissue (VAT), liver fat, insulin resistance (HOMA-IR), and serum levels of triglycerides (Sowah et al., 2022). Other studies have shown that Christensenellaceae negatively correlate with total cholesterol, low-density lipoproteins, and apolipoprotein B (He et al., 2018; Lopez-Contreras et al., 2018). The family was underrepresented in patients with atherogenic dyslipidemia (Lopez-Montoya et al., 2022). Not surprisingly, depletion of Christensenellaceae has been observed in pathologies normally associated with metabolic syndrome, such as hypertension (Gomez-Arango et al., 2016; Calderon-Perez et al., 2020), prediabetes (Org et al., 2017; Jayanama et al., 2022), and diabetic retinopathy (Liu et al., 2022).
The second most common group of disorders associated with levels of Christensenellaceae are gastrointestinal pathologies, primarily inflammatory bowel diseases (IBDs). Numerous studies have documented the depletion of Christensenellaceae in patients with Crohn’s disease (CD) and ulcerative colitis (UC) (Pascal et al., 2017; Imhann et al., 2018; Kennedy et al., 2018; Zakrzewski et al., 2019; Teofani et al., 2022). Interestingly, a significant decrease in the abundance of Christensenellaceae was observed in patients with quiescent CD immediately prior to flares (Braun et al., 2019), possibly indicating their role in disease progression. A lower abundance of Christensenellaceae was also observed in patients with diarrhea (Brunkwall et al., 2021).
Christensenellaceae have also been linked to bronchial asthma and atopic diseases. Hu et al. found that children whose fecal microbiome was rich in Christensenellaceae were less likely to develop eczema and sensitization to inhalant allergens (Hu et al., 2021). Remarkably, the abundance of Christensenellaceae in the household environment may play an important role in bronchial asthma. Gangneux et al. analyzed the metagenomic profiles of dust collected from the houses of healthy children and children and adults with asthma. They found that Christensenellaceae was significantly overrepresented in the dust from “healthy” houses, whereas it was depleted in the dust from “asthmatic” houses (Gangneux et al., 2020). This is hardly direct evidence of the impact of Christensenellaceae in dust on the likelihood of developing asthma. However, this peculiar finding may reflect differences in the microbial profiles of healthy people and those with bronchial asthma.
Kidney stones (Stanford et al., 2020), affective disorders (Vinberg et al., 2019; Coello et al., 2021), thyroid cancer (Lu et al., 2022), mucous membrane pemphigoid (Low et al., 2022), polycystic ovary syndrome (Dong et al., 2021), and recurrent aphthous stomatitis (Wang et al., 2022) have also been linked to a lower abundance of Christensenellaceae.
However, a higher abundance of Christensenellaceae is not generally associated with a healthy phenotype. There is evidence, though scarce, of a possible link between this family and various pathologies. In a study by Xu et al., a higher abundance of Christensenellaceae increased the risk of death in critically ill patients with neurological disorders admitted to an intensive care unit (Xu et al., 2019). This taxon was significantly enriched in patients with Parkinson’s disease (Shen et al., 2021) and particularly in those with poor clinical profiles (Barichella et al., 2019). An increased abundance of Christensenellaceae has also been observed in people with Alzheimer’s disease (Verhaar et al., 2021; Kaiyrlykyzy et al., 2022), multiple sclerosis (Galluzzo et al., 2021), and anorexia nervosa (Prochazkova et al., 2021).
Despite the numerous documented beneficial properties of this microorganism, there is evidence to suggest that C. minuta and neurocognitive health could be negatively correlated. As an active producer of short-chain fatty acids, C. minuta would be expected to have a positive effect on the gut-brain axis and, by extension, neurocognitive status. However, this may not always be the case. Waters and Ley (2019) also noted this surprising correlation and hypothesized it could be linked to delayed gastrointestinal transit time and constipation commonly seen in patients with Parkinson’s disease and multiple sclerosis (Wiesel et al., 2001; Pedrosa Carrasco et al., 2018). Notably, increased abundance of Christensenellaceae was positively correlated with prolonged transit time (Oki et al., 2016; Roager et al., 2016), providing insight into the relationship between this microorganism and neurocogntive status. Therefore, the observed correlation between elevated Christensenellaceae levels and neurodegenerative conditions may be indirect, with an alternative pathway directly linking the two.
Information on the prevalence of C. minuta in health and disease is summarized in Table 1.
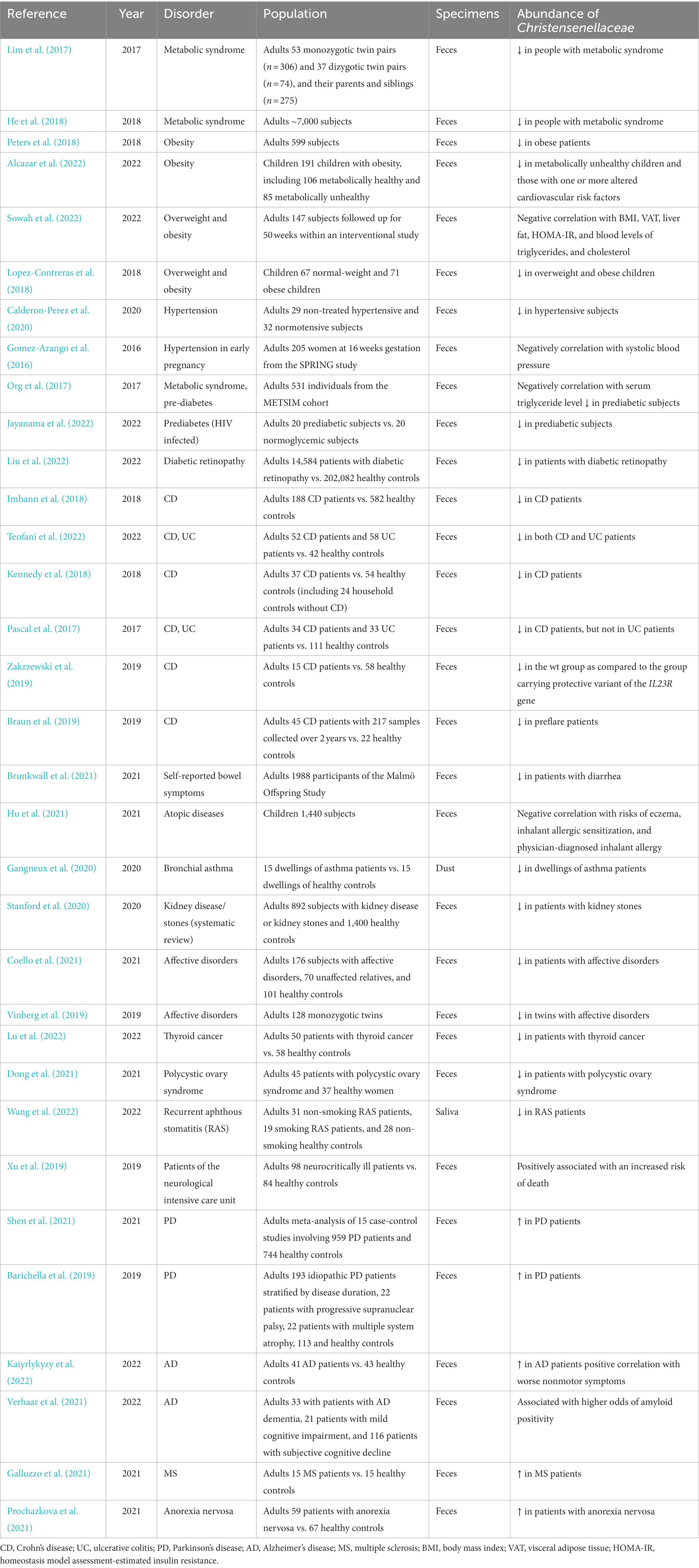
Table 1. Summary of the studies reporting the association between the Christensenellaceae family and a healthy phenotype or disorder.
The above metagenomic studies demonstrated that Christensenellaceae could be a source of a highly potent probiotic drug that would benefit many patient groups, particularly those with metabolic disorders and inflammatory gastrointestinal pathologies. This inspired new research into the properties and therapeutic capacity of C. minuta.
All mechanisms underlying the probiotic activity of C. minuta have yet to be fully understood; however, there have been many positive developments. Kropp et al. tested C. minuta DSM 22607 in a series of in vitro and in vivo experiments, and their results were extremely promising. First, both the bacterium and its supernatant demonstrated a strong anti-inflammatory potential provided by their ability to limit IL-8 production in HT-29 cells. The supernatant also inhibited the NF-kB signaling pathway, whereas the bacterium did not. Second, C. minuta also showed the ability to protect the intestinal barrier in TNF-α-compromised Caco-2 cells, as demonstrated by transepithelial electrical resistance (TEER) (Kropp et al., 2021).
These results were confirmed in murine models of dinitrobenzene sulfonic acid (DNBS)- and trinitrobenzene sulfonic acid (TNBS)-induced colitis. In both experiments, C. minuta displayed distinctive anti-inflammatory properties and protected the colonic tissues as effectively as 5-aminosalicylic acid (5-ASA). The bacterium reduced both macroscopic and microscopic chemical damage, decreased immune cell infiltration (ICI) in the colon, limited oxidative stress, and lowered the secretion of pro-inflammatory cytokines and the expression of lipocalin-2 (Kropp et al., 2021).
The experiments also shed some light on the metabolic phenotype of C. minuta, particularly its ability to produce large amounts of acetate and moderate amounts of butyrate, which supports the findings of Morotomi et al. (Kropp et al., 2021). Importantly, C. minuta has been reported to produce both short-chain fatty acids (SCFAs), while most microorganisms can only produce butyrate or acetate.
Relizani et al. isolated and tested thirty-two new C. minuta strains from nine donors to identify the best candidate probiotic drug. They analyzed the anti-inflammatory and protective properties of these strains in a series of experiments and selected five main candidates. All five candidates prevented NF-kB pathway activation after TNF-α stimulation and induced IL-10 production in in vitro cell models. In animal models, two of the five strains significantly improved the inflammatory lesions caused by TNBC and had pronounced local anti-inflammatory effects. In addition, the authors demonstrated the ability of the C. minuta strains to stimulate IL-10 production by human-derived PBMC in an in vitro model (Relizani et al., 2022).
Mazier et al. conducted a thorough investigation of the anti-obesity potential of C. minuta DSM22607 (Mazier et al., 2021). They found that daily administration of 2 × 109 colony-forming units (CFU) of C. minuta prevented weight gain and hyperglycemia in mice fed a high-fat diet (HFD) but did not affect their food intake. Surprisingly, there was no statistically significant difference in weight gain between the animals consuming the probiotic strain and the animals feeding on normal chow; however, the HFD-fed mice receiving the vehicle demonstrated a significant and rapid weight gain. This strongly suggests that C. minuta limited fat accumulation by altering metabolism rather than by affecting feeding behavior. These findings correlated with the observations made on serum metabolic markers, namely a decrease in leptin and resistin levels in HFD mice. C. minuta may have disrupted hepatic lipogenesis, as demonstrated by a reduced expression of the Gck gene encoding glucokinase. In addition, the probiotic strain had a strong protective effect on gut permeability by upregulating the Ocln and Zo1 genes, which encode major tight-junction proteins. This may also have contributed to C. minuta’s anti-obesity effect by limiting systemic inflammation caused by leaky gut.
In the same study (Mazier et al., 2021), the authors also attempted to assess the effects of a C. minuta biotherapeutic on gut microbiome composition in a murine model and in a SHIME® model inoculated with human feces from obese individuals. In mice daily gavaged with C. minuta DSM22607, its abundance in feces reached 20% on average, which resulted in a lower alpha-diversity compared with the controls. To avoid bias, the authors excluded the Christensenellaceae family from the analysis and found that the microbial profile in the HFD-DSM22607 mice fell somewhere between those of the mice fed with normal chow and the mice fed with an HFD vehicle, with no significant difference in beta-diversity. Meanwhile, the microbial profiles of these two animal groups differed significantly. The authors concluded that C. minuta tended to limit HFD-related shifts in the gut microbiome. Some valuable results were observed in the SHIME® model. C. minuta failed to engraft in the microbial ecosystem of this in vitro gut model after the treatment; it, however, improved the microbial diversity in the “distal colon” compartment and significantly enriched both compartments with SCFAs, which was sustained even during the washout period. Finally, C. minuta increased the levels of litocholic acid and unconjugated bile salt acid, indicating the production of a bile salt hydrolase.
Pan et al. have further corroborated the evidence for the beneficial effects of C. minuta and its critical role in metabolic processes. The authors used two strains of the genus Christensenella (C. minuta DSM 22607 and C. timonesis DSM 102800) to treat streptozotocin (STZ)-induced type 2 diabetes mellitus in mice. Numerous metabolic indicators were improved by both strains. The gavaged probiotic bacteria lowered blood glucose levels, limited oxidative stress, promoted the restoration of damaged pancreatic islet and liver cells, and suppressed the expression of several pro-inflammatory cytokines and TLR4 in the liver and colon. Importantly, the treatment with C. minuta and C. timonesis also upregulated Zonula occludens-1 and Claudin-1 in the colon, thereby strengthening the intestinal barrier. This finding was supported by the decreased serum LPS levels in the test subjects. Both strains also had a profound effect on metabolism by stimulating the expression of proglucagon, increasing serum levels of glucagon-like peptide-1 (GLP-1), and limiting hepatic gluconeogenesis. Finally, both Christensenella strains altered the composition of the gut microbiome by increasing the abundance of many beneficial microorganisms, such as Bifidobacterium and Phascolarctobacterium. Overall, C. minuta and C. timonesis improved metabolic processes in T2DM mice and mitigated inflammatory responses (Pan et al., 2022).
We can only hypothesize about the exact mechanisms underlying the beneficial effects of the Christensenella species. However, all of the above studies have laid a solid foundation for further research.
Based on the above results, we hypothesized about the mechanisms that could underlie the effects of the Christensenella family and its metabolites, including the ability to control inflammation, improve metabolism, and modulate the entire microbiome. Here, we will attempt to summarize what is known about Christensenella and outline some possible research trajectories and their implications.
Short-chain fatty acids (SCFAs) are primarily derived from the fermentation of indigestible carbohydrates by gut microbes, including C. minuta. This species produces acetate and butyrate (Kropp et al., 2021), valuable metabolites acting as postbiotics, which likely contribute to the therapeutic effects of C. minuta. Numerous studies have demonstrated the beneficial effects of SCFAs both locally in the intestine and at the systemic level.
Acetate and butyrate can serve as fuel for adenosine triphosphate (ATP) in intestinal epithelial cells and support homeostasis. Moreover, both SCFAs synthesized by C. minuta are presumably involved in maintaining the intestinal epithelial barrier. For example, acetate and butyrate can activate the nucleotide-binding oligomerization domain 3 (NLRP3) inflammasome by binding to G-protein-coupled receptors GPR43 and GPR109A. This increases IL-18 release, facilitating the repair of epithelial cells (Macia et al., 2015). Butyrate stabilizes hypoxia-inducible factor (HIF) (Kelly and Colgan, 2016), a key molecule for barrier protection and tissue regeneration, upregulates tight junction proteins (Peng et al., 2009; Wang et al., 2012; Miao et al., 2016), and increases mucin production by goblet cells (Gaudier et al., 2004), resulting in a stronger intestinal barrier. Interestingly, butyrate can also have the opposite effect. Kaiko et al. found that butyrate can inhibit the proliferation of intestinal stem cells through the forkhead box O3 (Foxo3) (Kaiko et al., 2016). On the one hand, this impedes epithelial regeneration and wound healing. On the other hand, it can be a control mechanism for excessive cell division and tumor growth prevention. Therefore, as a key producer of such an important metabolite, C. minuta could offer additional benefits.
The beneficial effects of SCFAs are not limited to the intestinal barrier. Many authors have reported their immunomodulating potential. For instance, both acetate and butyrate showed their ability to inhibit LPS-induced release of tumor necrosis factor-α (TNF-α) and interferon-γ (IFN-γ) in human leucocytes (Cox et al., 2009). In the study by Park et al., butyrate inhibited IFN-γ-induced production of nitric oxide synthase (NOS), TNF-α, and IL-6 in murine macrophages and increased IL-10 release, which indicates a high anti-inflammatory potency of this SCFA (Park et al., 2016). Butyrate could also suppress the LPS-induced production of chemokines, namely CCL3, CCL4, CCL5, CXCL9, CXCL10, and CXCL11, thus affecting leukocyte trafficking and limiting the production of IL-6 and IL-12p40 (Nastasi et al., 2015). It has also been demonstrated to promote regulatory T-cell production and differentiation, which is critical for immune regulation and homeostasis (Arpaia et al., 2013).
The profiles of acetate and butyrate would not be complete without mentioning their contribution to the gut-brain axis and their metabolic effects. These SCFAs, produced by the gut microbiome, interact with enteroendocrine cells in the colonic mucosa and induce the release of glucagon-like peptide-1 (GLP-1) and peptide YY (PYY) (Freeland and Wolever, 2010; Christiansen et al., 2018), known as anorexic hormones. These hormones enter the systemic circulation and exert their effects on many organs and tissues, most importantly – in the stomach and pancreas. Together, GLP-1 and PYY prevent rapid gastric emptying, inhibit acid secretion and motility, and slow gastrointestinal transit, which collectively results in poor appetite and reduced food intake. GLP-1 also stimulates insulin secretion and prevents β-cell exhaustion in the pancreas (Rondas et al., 2013).
Bile salt hydrolases (BSHs) are a group of bacterial enzymes involved in the biotransformation of bile acids (BAs). BAs are produced in the liver by cholesterol oxidation, then stored in the gallbladder, and finally excreted into the duodenum after food intake. Before reaching the intestine, BAs are conjugated to either taurine or glycine, making them less hydrophobic and less toxic. After fulfilling their function, most BAs are reabsorbed and migrate to the liver, where they can be reused. However, about 5% of BAs are deconjugated in the gut by enzymes produced by BSH-competent commensal microorganisms, such as C. minuta, and eventually excreted in the feces. Dejean et al. showed that two strains of C. minuta (DSM33407 and DSM) carried an active BSH that can hydrolyze both taurine- and glycine-conjugated BAs, with DSM being more active (Dejean et al., 2021).
The relationship between the gut microbiome and BAs could be essential to both. After their deconjugation, BAs become significantly more toxic and can affect or even shape bacterial communities in the gut. These communities, in turn, can cause changes in BA metabolism and extend the effects of BAs well beyond the intestine. Gadaleta et al. hypothesized that BSH-mediated deconjugation of BAs can, to some extent, disrupt BA recycling. As a result, BAs are reabsorbed and transported to the liver at a much slower rate, while their de novo synthesis increases, consuming cholesterol and lowering its levels in blood serum (Gadaleta et al., 2022). The exact mechanisms underlying the effect of microbiome-derived BSH have yet to be fully understood; however, there is increasing evidence that BAs and the gut microbiome interact very closely and are both affected.
The mystery of C. minuta’s anti-inflammatory properties is likely to be explained, at least in part, by its unusual lipopolysaccharides (LPSs). Yang et al. conducted a detailed assessment of LPSs in C. minuta and obtained interesting results. They compared the genetics of LPS biosynthesis in E. coli and C. minuta; C. minuta lacked some genes encoding key virulence-related LPSs, suggesting that its LPSs might be less toxic. Furthermore, C. minuta presumably has a shorter O-antigen repeat chain, which is important for LPS recognition by the immune system. Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) of LPSs in E. coli and C. minuta confirmed the differences in their structures (Yang et al., 2018). The authors also examined the effect of LPSs in E. coli and C. minuta on murine macrophages. LPSs from both bacteria demonstrated the ability to stimulate proliferation and phagocytosis. However, LPSs of E. coli were effective even at much lower concentrations. Then, the authors measured the expression of key proteins involved in the NF-κB pathway after the macrophages were exposed to LPSs and found that C. minuta LPSs were a relatively weak trigger of this important inflammation pathway. Furthermore, LPS-induced production of cytokines, nitric oxide, and reactive oxygen species was substantially lower in the macrophages stimulated by C. minuta LPS than in those stimulated by E. coli LPS (Yang et al., 2018).
Therefore, C. minuta LPSs, being distinctively different from those of more common gram-negative bacteria, have lower ability to elicit innate immune responses by interacting with toll-like receptors (TLRs). These findings may explain some of the effects observed in the experiments by Kropp et al. and Relizani et al., but they do not clarify the mechanism underlying the production of anti-inflammatory cytokines. Overall, the study by Yang et al. is an important step towards solving the mystery of C. minuta.
The beneficial effects of C. minuta and the Christensenellaceae family are likely due to their specific ability to interact with numerous other bacterial communities in the gut. Besides its direct effects, C. minuta can also affect the host indirectly by promoting or restricting the growth of certain taxa. The co-occurrence network constructed by Li et al. revealed that the Christensenellaceae family was positively correlated with many taxa, including Oscillospira, Ruminococcus, Coprococcus, Prevotella, Akkermansia, Roseburia, etc. In contrast, several genera such as Klebsiella, Streptococcus, Fusobacterium, Magamonas, and Blautia were inversely correlated with Christensenellaceae. Furthermore, there was a significant association between a higher abundance of Christensenellaceae and increased microbial richness and diversity (Li et al., 2020). Notably, several of the co-occurring taxa have been proposed as new-generation probiotics (Oscillospira, Roseburia) (Yang et al., 2021; Zhang et al., 2022) or are currently being used in this capacity (Akkermansia).1 Interestingly, the bacterial genera usually depleted in the presence of Christensenellaceae include several noxious taxa, for instance, opportunistic pathogens, such as Klebsiella and Streptococcus, which are known to cause infections in humans and animals, and Fusobacterium – a microorganism that can contribute to cancer (Rubinstein et al., 2019). Therefore, we hypothesize that C. minuta can modulate the gut microbiome by supporting the growth of beneficial species and inhibiting potentially harmful ones.
It is likely that different bacterial species interact through the transfer of metabolites. Ruaud et al. extensively studied the co-occurrence of the Christensenellaceae and the Methanobacteriaceae in the domain Archaea, the abundance of which was also inversely correlated with body mass index (Goodrich et al., 2014). They conducted a meta-analysis of 1,821 samples from 10 independent studies and confirmed a strong positive correlation at both the family level (between Christensenellaceae and Methanobacteriaceae) and the species level (between C. minuta and Methanobrevibacter smithii). Additionally, the authors co-cultured these two microorganisms to study their physical and metabolic interactions in vitro. They found that the strong co-occurrence patterns observed in the metagenomic studies were largely confirmed in the in vitro experiments. As an active H2 producer, C. minuta effectively supported the growth of M. smithii, which depends on H2 supply. In the co-culture, the amount of H2 released by C. minuta, was presumably sufficient to ensure the viability of M. smithii, equivalent to that in a monoculture with H2 excess. Moreover, M. smithii tended to form small inclusions within C. minuta flocs on solid media, as demonstrated by confocal microscopy, suggesting their mutually beneficial interaction. M. smithii, in turn, modulated the metabolism of C. minuta, causing a shift in SCFA production from butyrate to acetate. Based on the observed increase in acetate production, the authors conjectured that C. minuta could be an effective supplier of this substrate to other butyrate producers and directly facilitate their growth. They also hypothesized that methane production by Methanobacteriaceae results in carbon loss and less energy available to the host, which may in part explain the association between this bacterial family and the lean phenotype (Ruaud et al., 2020).
Members of the Christensenellaceae family are important components of an extremely complex, self-regulating community of intestinal microorganisms. Presumably, they interact in different ways. Some of them have been discovered, but most have not.
Probiotics are living organisms with specific requirements and often a high sensitivity to oxygen. Therefore, manufacturing next-generation probiotics on an industrial scale is an extremely demanding and time-consuming process, which differs drastically from a regular laboratory workflow. Cultured bacteria are not as resilient. For a probiotic drug to be effective, the bacteria must be kept alive by fine-tuning the environment (pH levels, temperature, atmospheric oxygen) and ensuring their viability and integrity throughout the process (drying, freezing, etc.). This applies in particular to anaerobic microorganisms.
Another challenge is ensuring that the drug is properly digested in the body. To reach their destination in the small and large intestine, beneficial bacteria must be resistant to low pH levels in the stomach. This potential challenge is usually addressed by overage, which compensates for losses during storage and drug passage through the gastrointestinal tract. Moreover, it is crucial to ensure freedom from allergens and/or potentially harmful substances. Therefore, any additional components (e.g., cryoprotectants) should be selected and handled with great care. Possible impurities from the culture media must be removed as far as possible. Quality control should be implemented at every stage of the production chain. Long-term stability is one of the most desirable properties of commercial drug products. An ideal probiotic drug is one that is moisture-free, has a good shelf life at ambient temperature for at least 12 months, and will produce significant numbers of viable microorganisms by the end of its shelf life. This can be achieved either through the use of protective techniques or by ensuring that the probiotic is highly resilient to stressful environmental conditions. Various techniques have been developed to ensure better survival of probiotic microorganisms, including spray drying, freeze drying, microencapsulation with alginates or other polysaccharides, etc. Microencapsulation has proven to be effective for a number of probiotic strains, such as Akkermansia muciniphila (77, 78), Lactobacillus plantarum (79), Lactobacillus rhamnosus (80, 81), Bifidobacterium bifidum (82) and others. Advances in delivery systems have enabled the incorporation of microorganisms into functional foods. For example, Marcial-Coba managed to incorporate microencapsulated A. muciniphila into dark chocolate, improving bacterial survival (83). Afzaal et al. incorporated microencapsulated L. acidophilus into ice cream and recovered viable bacteria after 120 days of storage (84). Several authors have also successfully used cheese to deliver various probiotic strains (85–87).
To the best of our knowledge, no attempt has been made to cultivate C. minuta on an industrial scale, and there have been no reports of the processing or stabilization of C. minuta as a probiotic drug or the development of an optimal drug formulation. C. minuta is a very promising probiotic agent, not only because of its therapeutic potential but also because it is relatively low-maintenance. It grows well on some fairly basic liquid and solid media and, most importantly, can tolerate oxygen for up to 24 h (2). This gives C. minuta a significant advantage over many other beneficial but strictly anaerobic microorganisms. Lower sensitivity to atmospheric oxygen provides more time for effective processing of raw C. minuta biomass, i.e., freeze-drying microencapsulation or incorporation into functional foods.
Another potentially promising form of C. minuta-based biotherapeutics is pasteurized C. minuta and its supernatant, which contains bacterial metabolites. To date, either live bacteria or their supernatants have been tested in all in vivo and in vitro experiments with C. minuta (2, 54, 55). Surprisingly, pasteurized C. minuta has remained unnoticed. Pasteurization is not a brand-new technique. It helps eliminate some challenges related to drug storage and delivery to the intestine. Pasteurized forms of another anaerobic strain, Akkermansia muciniphila, have been extensively tested and have demonstrated some exciting probiotic properties in both animal models (88) and humans (89). Therefore, pasteurized C. minuta and its supernatant are particularly attractive as candidates for biotherapeutics. Scaling up their production would likely be easier and cheaper than that of viable strains. Given the above benefits, further studies must include all three forms of C. minuta to identify an optimal form and improve our understanding of the mechanisms underlying its impressive effects.
A new candidate NGP, C. minuta, has drawn the attention of the international scientific community since its isolation about 12 years ago. Many reports have suggested that there is a strong association between the species and the healthy and lean phenotype. However, this suggestion begs the question – which of the two is the cause and which is the effect? Do changes in the microbiome in general and in the abundance of Christensenellaceae in particular cause a disease or at least contribute to its development? Or does the disease, caused by reasons other than the microbiome, trigger microbial shifts? The causative role of Christensenellaceae in human diseases and its extent have yet to be established. One might think that the association between the depletion of this bacterial taxon and a disease is solely due to lifestyle and diet; hence, the microbial profile is an effect rather than the cause. There are a number of findings supporting this hypothesis. For instance, the abundance of Christensenellaceae increased in overweight patients after dietary changes (Aleman et al., 2018; Deledda et al., 2022; Jian et al., 2022) and increased consumption of animal products (David et al., 2014), but decreased in patients on a low-protein diet (Lai et al., 2019). Hence, the question of causality remains.
Nevertheless, some significant effects of C. minuta have been reported in more recent studies (Kropp et al., 2021; Mazier et al., 2021; Pan et al., 2022; Relizani et al., 2022), including the prevention of HFD-induced weight gain, restriction of gut inflammation, and preservation of gut permeability. Clearly, causality is a complex problem. Reducing it to a straightforward equation is of little value. Evidently, the microbiome and disease have a reciprocal relationship in which each affects the other. Hence, the emergence of any disorder is an intricate process involving numerous variables, and the microbiome is just one of them. Our commensal microbial communities are undoubtedly altered by pathological changes, and this can affect how a disease develops and progresses.
The initial in vivo and in vitro experiments have demonstrated the exceptional value of C. minuta as a candidate NGP. Because of its relative tolerance to atmospheric oxygen and, consequently, much easier handling, C. minuta has an advantage over other anaerobic strains.
The formulation and manufacturing of a commercial NGP derived from C. minuta present a number of scientific and legal challenges. First and foremost, further research should corroborate the current results and should explore new hypotheses. We believe many diseases could also be treated with C. minuta-derived NGP; however, metabolic disorders and inflammatory bowel disease should probably still be its primary targets. C. minuta may prove to be of great benefit to patients with atopic disorders because it can improve gut permeability and mitigate systemic inflammation. Patients with asthma (Benard et al., 1996), eczema (Jackson et al., 1981; Pike et al., 1986), and food allergies (Schrander et al., 1990) are more likely to have a “leaky gut,” which is thought to contribute to disease pathogenesis and trigger flares. Given its ability to limit intestinal stem cell proliferation (Kaiko et al., 2016), a C. minuta-derived probiotic may also be beneficial for patients with malignancies, especially colon cancer. Hence, we believe C. minuta should be used to treat a variety of conditions, which calls for more research using animal models.
Second, C. minuta in forms other than live strains also deserves more consideration. Compared to viable C. minuta, the C. minuta metabolites (supernatant) have demonstrated impressive effects both in in vitro and in vivo experiments (Kropp et al., 2021). Surprisingly, no reports on experiments using pasteurized C. minuta exist, suggesting a gap in this field of study. In our opinion, both pasteurized C. minuta and its postbiotic (supernatant) merit further consideration, along with viable C. minuta. Indeed, testing several therapeutic forms of C. minuta rather than one live form would greatly facilitate unraveling its mechanisms of action. For example, a more detailed analysis of the C. minuta metabolites using high-performance liquid chromatography or other techniques could shed some light on how this microorganism has become so important for human health and would expedite the development of a formulation. Given the challenges associated with the large-scale production of NGPs, especially live biotherapeutics, selecting the most optimal and cost-effective form is crucial.
Third, it is critical to determine whether any form of C. minuta can produce long-term effects on the gut microbiome and on the entire body. So far, very few studies have attempted to analyze the effects of C. minuta-based treatment. Mazier et al. examined the effects of C. minuta in a SHIME® model and found no C. minuta engraftment into the microbial community of the simulated intestine, although some effects lasted during a one-week wash-out period (Mazier et al., 2021). We believe that more studies on animals with a longer wash-out period are needed to gain a better understanding of the long-term effects of a C. minuta-based pro/postbiotic.
Finally, we would like to emphasize the potential utility of mixed biotherapeutics containing multiple microbial species. The Christensenellaceae family has demonstrated strong correlations with other potentially beneficial strains; therefore, we hypothesize that combining it with other NGPs, such as Akkermansia muciniphila and Roseburia faecis, or even with conventional probiotic strains, such as bifidobacteria and lactobacilli, may enhance the treatment effects. In fact, people with disorders often experience depletion of many bacterial taxa, which could be remedied by a combination of probiotics. These could be mixtures of at least one viable probiotic strain and at least one postbiotic from a different strain (or strains). To our knowledge, only mixtures of conventional probiotic strains have been tested (Chapman et al., 2006) or formulated so far, whereas combinations of NGPs remain largely uninvestigated. We surmise that this approach will advance biotherapeutics and pave the way for new multicomponent and multi-purpose pro/postbiotics, which are highly likely to include C. minuta as one of the most important representatives of a healthy gut microbiome.
Once the best or most cost-effective form/combination has been identified, other, more practical challenges should be addressed, such as scaling up the production, safety, and clinical trials. Clearly, only randomized placebo-controlled trials will suffice to demonstrate the efficacy of any C. minuta-containing biotherapeutic. Since biotherapeutic drugs are based on living microorganisms, there will be additional regulatory challenges, and the development and registration processes will be complicated. To date, only one pharmaceutical company, YSOPIA Bioscience (France), has attempted to reach these final stages with their single-strain live Xla1 biotherapeutic containing C. minuta DSM 33407. They reported entering the first in-human clinical trials in 2021 and shared the legal challenges they had met in advancing to the clinical trials stage (Paquet et al., 2021).
Thus, significant progress has been made in developing effective and reliable NGPs, and a C. minuta-derived biotherapeutic seems to be within reach. However, the promising findings of numerous studies must be confirmed in clinical trials.
OI, DT, AKo, and LM: literature search, manuscript preparation, writing, and editing. MT: figures and manuscript editing. DK, AZ, MI, VY, VM, AKe, SK, and SY: manuscript discussion and editing. VY, VM, AKe, SK, and SY: overall supervision. All authors contributed to the article and approved the submitted version.
The author(s) declare that no financial support was received for the research, authorship, and/or publication of this article.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Adapen, C., Reot, L., Nunez, N., Cannou, C., and Marlin, R. (2022). Local innate markers and vaginal microbiota composition are influenced by hormonal cycle phases. Front. Immunol. 13:841723. doi: 10.3389/fimmu.2022.841723
Alcazar, M., Escribano, J., Ferre, N., Closa-Monasterolo, R., and Selma-Royo, M. (2022). Gut microbiota is associated with metabolic health in children with obesity. Clin. Nutr. 41, 1680–1688. doi: 10.1016/j.clnu.2022.06.007
Aleman, J. O., Bokulich, N. A., Swann, J. R., Walker, J. M., and De Rosa, J. C. (2018). Fecal microbiota and bile acid interactions with systemic and adipose tissue metabolism in diet-induced weight loss of obese postmenopausal women. J. Transl. Med. 16:244. doi: 10.1186/s12967-018-1619-z
Arpaia, N., Campbell, C., Fan, X., Dikiy, S., van der Veeken, J., and deRoos, P. (2013). Metabolites produced by commensal bacteria promote peripheral regulatory T-cell generation. Nature 504, 451–455. doi: 10.1038/nature12726
Baldo, L., Riera, J. L., Mitsi, K., and Pretus, J. L. (2018). Processes shaping gut microbiota diversity in allopatric populations of the endemic lizard Podarcis lilfordi from Menorcan islets (Balearic Islands). FEMS Microbiol. Ecol. 94:e186. doi: 10.1093/femsec/fix186
Barichella, M., Severgnini, M., Cilia, R., Cassani, E., Bolliri, C., and Caronni, S. (2019). Unraveling gut microbiota in Parkinson's disease and atypical parkinsonism. Mov. Disord. 34, 396–405. doi: 10.1002/mds.27581
Barrett, H. L., Gomez-Arango, L. F., Wilkinson, S. A., McIntyre, H. D., and Callaway, L. K. (2018). A vegetarian diet is a major determinant of gut microbiota composition in early pregnancy. Nutrients 10:890. doi: 10.3390/nu10070890
Benard, A., Desreumeaux, P., Huglo, D., Hoorelbeke, A., and Tonnel, A. B. (1996). Increased intestinal permeability in bronchial asthma. J. Allergy Clin. Immunol. 97, 1173–1178. doi: 10.1016/s0091-6749(96)70181-1
Biagi, E., Franceschi, C., Rampelli, S., Severgnini, M., Ostan, R., and Turroni, S. (2016). Gut microbiota and extreme longevity. Curr. Biol. 26, 1480–1485. doi: 10.1016/j.cub.2016.04.016
Braun, T., Di Segni, A., BenShoshan, M., Neuman, S., and Levhar, N. (2019). Individualized dynamics in the gut microbiota precede Crohn's disease flares. Am. J. Gastroenterol. 114, 1142–1151. doi: 10.14309/ajg.0000000000000136
Brooks, A. W., Priya, S., Blekhman, R., and Bordenstein, S. R. (2018). Gut microbiota diversity across ethnicities in the United States. PLoS Biol. 16:e2006842. doi: 10.1371/journal.pbio.2006842
Brunkwall, L., Ericson, U., Nilsson, P. M., Orho-Melander, M., and Ohlsson, B. (2021). Self-reported bowel symptoms are associated with differences in overall gut microbiota composition and enrichment of Blautia in a population-based cohort. J. Gastroenterol. Hepatol. 36, 174–180. doi: 10.1111/jgh.15104
Calderon-Perez, L., Gosalbes, M. J., Yuste, S., Valls, R. M., and Pedret, A. (2020). Gut metagenomic and short chain fatty acids signature in hypertension: a cross-sectional study. Sci. Rep. 10:6436. doi: 10.1038/s41598-020-63475-w
Chapman, T. M., Plosker, G. L., and Figgitt, D. P. (2006). VSL#3 probiotic mixture: a review of its use in chronic inflammatory bowel diseases. Drugs 66, 1371–1387. doi: 10.2165/00003495-200666100-00006
Christiansen, C. B., Gabe, M. B. N., Svendsen, B., Dragsted, L. O., and Rosenkilde, M. M. (2018). The impact of short-chain fatty acids on GLP-1 and PYY secretion from the isolated perfused rat colon. Am. J. Physiol. Gastrointest. Liver Physiol. 315, G53–G65. doi: 10.1152/ajpgi.00346.2017
Coello, K., Hansen, T. H., Sorensen, N., Ottesen, N. M., and Miskowiak, K. W. (2021). Affective disorders impact prevalence of Flavonifractor and abundance of Christensenellaceae in gut microbiota. Prog. Neuro-Psychopharmacol. Biol. Psychiatry 110:110300. doi: 10.1016/j.pnpbp.2021.110300
Cox, M. A., Jackson, J., Stanton, M., Rojas-Triana, A., Bober, L., and Laverty, M. (2009). Short-chain fatty acids act as antiinflammatory mediators by regulating prostaglandin E(2) and cytokines. World J. Gastroenterol. 15, 5549–5557. doi: 10.3748/wjg.15.5549
Crisol-Martinez, E., Stanley, D., Geier, M. S., Hughes, R. J., and Moore, R. J. (2017). Sorghum and wheat differentially affect caecal microbiota and associated performance characteristics of meat chickens. PeerJ 5:e3071. doi: 10.7717/peerj.3071
David, L. A., Maurice, C. F., Carmody, R. N., Gootenberg, D. B., Button, J. E., and Wolfe, B. E. (2014). Diet rapidly and reproducibly alters the human gut microbiome. Nature 505, 559–563. doi: 10.1038/nature12820
Dejean, G., Tudela, H., Bruno, L., Kissi, D., Rawadi, G., and Claus, S. P. (2021). Identifying a novel bile salt hydrolase from the keystone gut bacterium Christensenella minuta. Microorganisms 9:1252. doi: 10.3390/microorganisms9061252
Deledda, A., Palmas, V., Heidrich, V., Fosci, M., Lombardo, M., and Cambarau, G. (2022). Dynamics of gut microbiota and clinical variables after ketogenic and Mediterranean diets in drug-naive patients with type 2 diabetes mellitus and obesity. Meta 12:e92. doi: 10.3390/metabo12111092
Dong, S., Jiao, J., Jia, S., Li, G., and Zhang, W. (2021). 16S rDNA full-length assembly sequencing technology analysis of intestinal microbiome in polycystic ovary syndrome. Front. Cell. Infect. Microbiol. 11:634981. doi: 10.3389/fcimb.2021.634981
Escobar, J. S., Klotz, B., Valdes, B. E., and Agudelo, G. M. (2014). The gut microbiota of Colombians differs from that of Americans, Europeans and Asians. BMC Microbiol. 14:311. doi: 10.1186/s12866-014-0311-6
Freeland, K. R., and Wolever, T. M. (2010). Acute effects of intravenous and rectal acetate on glucagon-like peptide-1, peptide YY, ghrelin, adiponectin and tumour necrosis factor-alpha. Br. J. Nutr. 103, 460–466. doi: 10.1017/S0007114509991863
Gadaleta, R. M., Cariello, M., Crudele, L., and Moschetta, A. (2022). Bile salt hydrolase-competent probiotics in the management of IBD: unlocking the bile acid code. Nutrients 14:53212. doi: 10.3390/nu14153212
Galluzzo, P., Capri, F. C., Vecchioni, L., Realmuto, S., and Scalisi, L. (2021). Comparison of the intestinal microbiome of Italian patients with multiple sclerosis and their household relatives. Life 11:620. doi: 10.3390/life11070620
Gangneux, J. P., Sassi, M., Lemire, P., and Le Cann, P. (2020). Metagenomic characterization of indoor dust bacterial and fungal microbiota in homes of asthma and non-asthma patients using next generation sequencing. Front. Microbiol. 11:1671. doi: 10.3389/fmicb.2020.01671
Gaudier, E., Jarry, A., Blottiere, H. M., de Coppet, P., and Buisine, M. P. (2004). Butyrate specifically modulates MUC gene expression in intestinal epithelial goblet cells deprived of glucose. Am. J. Physiol. Gastrointest. Liver Physiol. 287, G1168–G1174. doi: 10.1152/ajpgi.00219.2004
Gomez-Arango, L. F., Barrett, H. L., McIntyre, H. D., Callaway, L. K., and Morrison, M. (2016). Increased systolic and diastolic blood pressure is associated with altered gut microbiota composition and butyrate production in early pregnancy. Hypertension 68, 974–981. doi: 10.1161/HYPERTENSIONAHA.116.07910
Goodrich, J. K., Waters, J. L., Poole, A. C., Sutter, J. L., Koren, O., and Blekhman, R. (2014). Human genetics shape the gut microbiome. Cells 159, 789–799. doi: 10.1016/j.cell.2014.09.053
He, Y., Wu, W., Wu, S., Zheng, H. M., Li, P., and Sheng, H. F. (2018). Linking gut microbiota, metabolic syndrome and economic status based on a population-level analysis. Microbiome 6:172. doi: 10.1186/s40168-018-0557-6
Hu, C., van Meel, E. R., Medina-Gomez, C., Kraaij, R., Barroso, M., and Kiefte-de Jong, J. (2021). A population-based study on associations of stool microbiota with atopic diseases in school-age children. J. Allergy Clin. Immunol. 148, 612–620. doi: 10.1016/j.jaci.2021.04.001
Huang, Y. J., Kim, E., Cox, M. J., Brodie, E. L., Brown, R., and Wiener-Kronish, J. P. (2010). A persistent and diverse airway microbiota present during chronic obstructive pulmonary disease exacerbations. OMICS 14, 9–59. doi: 10.1089/omi.2009.0100
Imhann, F., Vich Vila, A., Bonder, M. J., Fu, J., Gevers, D., and Visschedijk, M. C. (2018). Interplay of host genetics and gut microbiota underlying the onset and clinical presentation of inflammatory bowel disease. Gut 67, 108–119. doi: 10.1136/gutjnl-2016-312135
Jackson, P. G., Lessof, M. H., Baker, R. W., Ferrett, J., and MacDonald, D. M. (1981). Intestinal permeability in patients with eczema and food allergy. Lancet 1, 1285–1286. doi: 10.1016/s0140-6736(81)92459-4
Jayanama, K., Phuphuakrat, A., Pongchaikul, P., Prombutara, P., and Nimitphong, H. (2022). Association between gut microbiota and prediabetes in people living with HIV. Curr Res Microb Sci 3:100143. doi: 10.1016/j.crmicr.2022.100143
Jian, C., Silvestre, M. P., Middleton, D., Korpela, K., Jalo, E., and Broderick, D. (2022). Gut microbiota predicts body fat change following a low-energy diet: a PREVIEW intervention study. Genome Med. 14:54. doi: 10.1186/s13073-022-01053-7
Kaiko, G. E., Ryu, S. H., Koues, O. I., Collins, P. L., Solnica-Krezel, L., and Pearce, E. J. (2016). The colonic crypt protects stem cells from microbiota-derived metabolites. Cells 165, 1708–1720. doi: 10.1016/j.cell.2016.05.018
Kaiyrlykyzy, A., Kozhakhmetov, S., Babenko, D., Zholdasbekova, G., and Alzhanova, D. (2022). Study of gut microbiota alterations in Alzheimer’s dementia patients from Kazakhstan. Sci. Rep. 12:15115. doi: 10.1038/s41598-022-19393-0
Kamke, J., Kittelmann, S., Soni, P., Li, Y., Tavendale, M., and Ganesh, S. (2016). Rumen metagenome and metatranscriptome analyses of low methane yield sheep reveals a Sharpea-enriched microbiome characterised by lactic acid formation and utilisation. Microbiome 4:56. doi: 10.1186/s40168-016-0201-2
Kelly, C. J., and Colgan, S. P. (2016). Breathless in the gut: implications of luminal O2 for microbial pathogenicity. Cell Host Microbe 19, 427–428. doi: 10.1016/j.chom.2016.03.014
Kennedy, N. A., Lamb, C. A., Berry, S. H., Walker, A. W., Mansfield, J., and Parkes, M. (2018). The impact of NOD2 variants on fecal microbiota in Crohn’s disease and controls without gastrointestinal disease. Inflamm. Bowel Dis. 24, 583–592. doi: 10.1093/ibd/izx061
Kropp, C., Le Corf, K., Relizani, K., Tambosco, K., and Martinez, C. (2021). The keystone commensal bacterium Christensenella minuta DSM 22607 displays anti-inflammatory properties both in vitro and in vivo. Sci. Rep. 11:11494. doi: 10.1038/s41598-021-90885-1
Lai, S., Molfino, A., Testorio, M., Perrotta, A. M., Currado, A., and Pintus, G. (2019). Effect of Low-protein diet and inulin on microbiota and clinical parameters in patients with chronic kidney disease. Nutrients 11:e6. doi: 10.3390/nu11123006
Li, X., Li, Z., He, Y., Li, P., Zhou, H., and Zeng, N. (2020). Regional distribution of Christensenellaceae and its associations with metabolic syndrome based on a population-level analysis. PeerJ 8:e9591. doi: 10.7717/peerj.9591
Lim, M. Y., You, H. J., Yoon, H. S., Kwon, B., Lee, J. Y., and Lee, S. (2017). The effect of heritability and host genetics on the gut microbiota and metabolic syndrome. Gut 66, 1031–1038. doi: 10.1136/gutjnl-2015-311326
Liu, K., Zou, J., Fan, H., Hu, H., and You, Z. (2022). Causal effects of gut microbiota on diabetic retinopathy: a Mendelian randomization study. Front. Immunol. 13:930318. doi: 10.3389/fimmu.2022.930318
Lopez-Contreras, B. E., Moran-Ramos, S., Villarruel-Vazquez, R., Macias-Kauffer, L., and Villamil-Ramirez, H. (2018). Composition of gut microbiota in obese and normal-weight Mexican school-age children and its association with metabolic traits. Pediatr. Obes. 13, 381–388. doi: 10.1111/ijpo.12262
Lopez-Montoya, P., Cerqueda-Garcia, D., Rodriguez-Flores, M., Lopez-Contreras, B., and Villamil-Ramirez, H. (2022). Association of gut microbiota with atherogenic dyslipidemia, and its impact on serum lipid levels after bariatric surgery. Nutrients 14:545. doi: 10.3390/nu14173545
Low, L., Suleiman, K., Shamdas, M., Bassilious, K., Poonit, N., and Rossiter, A. E. (2022). Gut Dysbiosis in ocular mucous membrane pemphigoid. Front. Cell. Infect. Microbiol. 12:780354. doi: 10.3389/fcimb.2022.780354
Lu, G., Yu, X., Jiang, W., Luo, Q., Tong, J., and Fan, S. (2022). Alterations of gut microbiome and metabolite profiles associated with anabatic lipid Dysmetabolism in thyroid Cancer. Front Endocrinol 13:893164. doi: 10.3389/fendo.2022.893164
Macia, L., Tan, J., Vieira, A. T., Leach, K., Stanley, D., and Luong, S. (2015). Metabolite-sensing receptors GPR43 and GPR109A facilitate dietary fibre-induced gut homeostasis through regulation of the inflammasome. Nat. Commun. 6:6734. doi: 10.1038/ncomms7734
Mazier, W., Le Corf, K., Martinez, C., Tudela, H., and Kissi, D. (2021). A new strain of Christensenella minuta as a potential biotherapy for obesity and associated metabolic diseases. Cells 10:823. doi: 10.3390/cells10040823
McKenzie, V. J., Song, S. J., Delsuc, F., Prest, T. L., Oliverio, A. M., and Korpita, T. M. (2017). The effects of captivity on the mammalian gut microbiome. Integr. Comp. Biol. 57, 690–704. doi: 10.1093/icb/icx090
Miao, W., Wu, X., Wang, K., Wang, W., Wang, Y., and Li, Z. (2016). Sodium butyrate promotes reassembly of tight junctions in Caco-2 monolayers involving inhibition of MLCK/MLC2 pathway and phosphorylation of PKCbeta2. Int. J. Mol. Sci. 17:696. doi: 10.3390/ijms17101696
Morotomi, M., Nagai, F., and Watanabe, Y. (2012). Description of Christensenella minuta gen. Nov., sp. nov., isolated from human faeces, which forms a distinct branch in the order Clostridiales, and proposal of Christensenellaceae fam. Nov. Int. J. Syst. Evol. Microbiol. 62, 144–149. doi: 10.1099/ijs.0.026989-0
Morton, E. R., Lynch, J., Froment, A., Lafosse, S., Heyer, E., and Przeworski, M. (2015). Variation in rural African gut microbiota is strongly correlated with colonization by Entamoeba and subsistence. PLoS Genet. 11:e1005658. doi: 10.1371/journal.pgen.1005658
Nastasi, C., Candela, M., Bonefeld, C. M., Geisler, C., Hansen, M., and Krejsgaard, T. (2015). The effect of short-chain fatty acids on human monocyte-derived dendritic cells. Sci. Rep. 5:16148. doi: 10.1038/srep16148
Oki, K., Toyama, M., Banno, T., Chonan, O., Benno, Y., and Watanabe, K. (2016). Comprehensive analysis of the fecal microbiota of healthy Japanese adults reveals a new bacterial lineage associated with a phenotype characterized by a high frequency of bowel movements and a lean body type. BMC Microbiol. 16:284. doi: 10.1186/s12866-016-0898-x
Org, E., Blum, Y., Kasela, S., Mehrabian, M., Kuusisto, J., and Kangas, A. J. (2017). Relationships between gut microbiota, plasma metabolites, and metabolic syndrome traits in the METSIM cohort. Genome Biol. 18:70. doi: 10.1186/s13059-017-1194-2
Pan, T., Zheng, S., Zheng, W., Shi, C., Ning, K., and Zhang, Q. (2022). Christensenella regulated by Huang-qi-Ling-Hua-san is a key factor by which to improve type 2 diabetes. Front. Microbiol. 13:1022403. doi: 10.3389/fmicb.2022.1022403
Paquet, J. C., Claus, S. P., Cordaillat-Simmons, M., Mazier, W., Rawadi, G., and Rinaldi, L. (2021). Entering first-in-human clinical study with a single-strain live biotherapeutic product: input and feedback gained from the EMA and the FDA. Front Med 8:716266. doi: 10.3389/fmed.2021.716266
Park, J. H., Kotani, T., Konno, T., Setiawan, J., Kitamura, Y., and Imada, S. (2016). Promotion of intestinal epithelial cell turnover by commensal bacteria: role of short-chain fatty acids. PLoS One 11:e0156334. doi: 10.1371/journal.pone.0156334
Pascal, V., Pozuelo, M., Borruel, N., Casellas, F., Campos, D., and Santiago, A. (2017). A microbial signature for Crohn’s disease. Gut 66, 813–822. doi: 10.1136/gutjnl-2016-313235
Pedrosa Carrasco, A. J., Timmermann, L., and Pedrosa, D. J. (2018). Management of constipation in patients with Parkinson’s disease. NPJ Parkinsons Dis 4:6. doi: 10.1038/s41531-018-0042-8
Peng, L., Li, Z. R., Green, R. S., Holzman, I. R., and Lin, J. (2009). Butyrate enhances the intestinal barrier by facilitating tight junction assembly via activation of AMP-activated protein kinase in Caco-2 cell monolayers. J. Nutr. 139, 1619–1625. doi: 10.3945/jn.109.104638
Peters, B. A., Shapiro, J. A., Church, T. R., Miller, G., Trinh-Shevrin, C., and Yuen, E. (2018). A taxonomic signature of obesity in a large study of American adults. Sci. Rep. 8:9749. doi: 10.1038/s41598-018-28126-1
Pike, M. G., Heddle, R. J., Boulton, P., Turner, M. W., and Atherton, D. J. (1986). Increased intestinal permeability in atopic eczema. J. Invest. Dermatol. 86, 101–104. doi: 10.1111/1523-1747.ep12284035
Prochazkova, P., Roubalova, R., Dvorak, J., Kreisinger, J., Hill, M., and Tlaskalova-Hogenova, H. (2021). The intestinal microbiota and metabolites in patients with anorexia nervosa. Gut Microbes 13, 1–25. doi: 10.1080/19490976.2021.1902771
Relizani, K., Le Corf, K., Kropp, C., Martin-Rosique, R., and Kissi, D. (2022). Selection of a novel strain of Christensenella minuta as a future biotherapy for Crohn’s disease. Sci. Rep. 12:6017. doi: 10.1038/s41598-022-10015-3
Richards, C., Otani, S., Mikaelyan, A., and Poulsen, M. (2017). Pycnoscelus surinamensis cockroach gut microbiota respond consistently to a fungal diet without mirroring those of fungus-farming termites. PLoS One 12:e0185745. doi: 10.1371/journal.pone.0185745
Roager, H. M., Hansen, L. B., Bahl, M. I., Frandsen, H. L., and Carvalho, V. (2016). Colonic transit time is related to bacterial metabolism and mucosal turnover in the gut. Nat. Microbiol. 1:16093. doi: 10.1038/nmicrobiol.2016.93
Rondas, D., Bugliani, M., D’Hertog, W., Lage, K., and Masini, M. (2013). Glucagon-like peptide-1 protects human islets against cytokine-mediated beta-cell dysfunction and death: a proteomic study of the pathways involved. J. Proteome Res. 12, 4193–4206. doi: 10.1021/pr400527q
Ruaud, A., Esquivel-Elizondo, S., de la Cuesta-Zuluaga, J., Waters, J. L., and Angenent, L. T. (2020). Syntrophy via interspecies H(2) transfer between Christensenella and Methanobrevibacter underlies their global Cooccurrence in the human gut. MBio 11:19. doi: 10.1128/mBio.03235-19
Rubinstein, M. R., Baik, J. E., Lagana, S. M., Han, R. P., Raab, W. J., and Sahoo, D. (2019). Fusobacterium nucleatum promotes colorectal cancer by inducing Wnt/beta-catenin modulator Annexin A1. EMBO Rep. 20:7638. doi: 10.15252/embr.201847638
Schrander, J. J., Unsalan-Hooyen, R. W., Forget, P. P., and Jansen, J. (1990). [51Cr]EDTA intestinal permeability in children with cow’s milk intolerance. J. Pediatr. Gastroenterol. Nutr. 10, 189–192. doi: 10.1097/00005176-199002000-00008
Shen, T., Yue, Y., He, T., Huang, C., and Qu, B. (2021). The association between the gut microbiota and Parkinson’s disease, a meta-analysis. Front. Aging Neurosci. 13:636545. doi: 10.3389/fnagi.2021.636545
Sowah, S. A., Milanese, A., Schubel, R., Wirbel, J., and Kartal, E. (2022). Calorie restriction improves metabolic state independently of gut microbiome composition: a randomized dietary intervention trial. Genome Med. 14:30. doi: 10.1186/s13073-022-01030-0
Stanford, J., Charlton, K., Stefoska-Needham, A., Ibrahim, R., and Lambert, K. (2020). The gut microbiota profile of adults with kidney disease and kidney stones: a systematic review of the literature. BMC Nephrol. 21:215. doi: 10.1186/s12882-020-01805-w
Teofani, A., Marafini, I., Laudisi, F., Pietrucci, D., Salvatori, S., and Unida, V. (2022). Intestinal taxa abundance and diversity in inflammatory bowel disease patients: an analysis including covariates and confounders. Nutrients 14:260. doi: 10.3390/nu14020260
Verhaar, B. J. H., Hendriksen, H. M. A., de Leeuw, F. A., Doorduijn, A. S., and van Leeuwenstijn, M. (2021). Gut microbiota composition is related to AD pathology. Front. Immunol. 12:794519. doi: 10.3389/fimmu.2021.794519
Vinberg, M., Ottesen, N. M., Meluken, I., Sorensen, N., and Pedersen, O. (2019). Remitted affective disorders and high familial risk of affective disorders associate with aberrant intestinal microbiota. Acta Psychiatr. Scand. 139, 174–184. doi: 10.1111/acps.12976
Wang, X., Luo, N., Mi, Q., Kong, W., and Zhang, W. (2022). Influence of cigarette smoking on oral microbiota in patients with recurrent aphthous stomatitis. J. Investig. Med. 70, 805–813. doi: 10.1136/jim-2021-002119
Wang, H. B., Wang, P. Y., Wang, X., Wan, Y. L., and Liu, Y. C. (2012). Butyrate enhances intestinal epithelial barrier function via up-regulation of tight junction protein Claudin-1 transcription. Dig. Dis. Sci. 57, 3126–3135. doi: 10.1007/s10620-012-2259-4
Waters, J. L., and Ley, R. E. (2019). The human gut bacteria Christensenellaceae are widespread, heritable, and associated with health. BMC Biol. 17:83. doi: 10.1186/s12915-019-0699-4
Wiesel, P. H., Norton, C., Glickman, S., and Kamm, M. A. (2001). Pathophysiology and management of bowel dysfunction in multiple sclerosis. Eur. J. Gastroenterol. Hepatol. 13, 441–448. doi: 10.1097/00042737-200104000-00025
Xu, R., Tan, C., Zhu, J., Zeng, X., Gao, X., Wu, Q., et al. (2019). Dysbiosis of the intestinal microbiota in neurocritically ill patients and the risk for death. Crit. Care 23:195. doi: 10.1186/s13054-019-2488-4
Yang, Y., Gu, H., Sun, Q., and Wang, J. (2018). Effects of Christensenella minuta lipopolysaccharide on RAW 264.7 macrophages activation. Microb. Pathog. 125, 411–417. doi: 10.1016/j.micpath.2018.10.005
Yang, J., Li, Y., Wen, Z., Liu, W., and Meng, L. (2021). Oscillospira – a candidate for the next-generation probiotics. Gut Microbes 13:1987783. doi: 10.1080/19490976.2021.1987783
Zakrzewski, M., Simms, L. A., Brown, A., Appleyard, M., Irwin, J., and Waddell, N. (2019). IL23R-protective coding variant promotes beneficial Bacteria and diversity in the Ileal microbiome in healthy individuals without inflammatory bowel disease. J. Crohns Colitis 13, 451–461. doi: 10.1093/ecco-jcc/jjy188
Keywords: Christensenellaceae, Christensenella, probiotics, metabolic disorders, inflammatory bowel diseases
Citation: Ignatyeva O, Tolyneva D, Kovalyov A, Matkava L, Terekhov M, Kashtanova D, Zagainova A, Ivanov M, Yudin V, Makarov V, Keskinov A, Kraevoy S and Yudin S (2024) Christensenella minuta, a new candidate next-generation probiotic: current evidence and future trajectories. Front. Microbiol. 14:1241259. doi: 10.3389/fmicb.2023.1241259
Received: 17 July 2023; Accepted: 26 December 2023;
Published: 11 January 2024.
Edited by:
Haiying Cai, Zhejiang University of Science and Technology, ChinaReviewed by:
Guangqiang Wang, University of Shanghai for Science and Technology, ChinaCopyright © 2024 Ignatyeva, Tolyneva, Kovalyov, Matkava, Terekhov, Kashtanova, Zagainova, Ivanov, Yudin, Makarov, Keskinov, Kraevoy and Yudin. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Olga Ignatyeva, b2lnbmF0aWV2YUBjc3BmbWJhLnJ1
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.