
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 09 June 2023
Sec. Microbial Physiology and Metabolism
Volume 14 - 2023 | https://doi.org/10.3389/fmicb.2023.1165701
 Zhilei Jia
Zhilei Jia Mengdi Yan
Mengdi Yan Xiaobei Li
Xiaobei Li Qiuyan Sun
Qiuyan Sun Gen Xu
Gen Xu Shuai Li
Shuai Li Wenchao Chen
Wenchao Chen Zhimin Shi
Zhimin Shi Zhonghai Li*
Zhonghai Li* Mei Chen*
Mei Chen* Xiaoming Bao
Xiaoming BaoIn this study, a phosducin-like protein, PoPlp1, was identified and functionally studied in the cellulase-producing strain Penicillium oxalicum 114-2. PoPlp1 was proven to participate in several biological processes, including mycelium development, conidiation, and expression of cellulases and amylases. With deletion of Poplp1, morphology and development varied significantly in ΔPoplp1. Colony growth, glucose utilization, and the hydrolysis capability of starch and cellulose were limited, whereas conidiation was enhanced. Based on detection of the levels of expression of transcription factors involved in asexual development, we conjectured that PoPlp1 is involved in conidiation via the major factor BrlA. We explored the effect of PoPlp1 on cellulase and amylase expression and observed that cellulase and amylase activity and major gene transcription levels were all dramatically reduced in ΔPoplp1. Deletion of PoPlp1 caused a decrease in intracellular cAMP levels, and the cellulase gene expression level of ΔPoplp1 was restored to a certain extent through external addition of cAMP. These findings demonstrate that PoPlp1 may affect cellulase and amylase expression by regulating cAMP concentration. To comprehensively explore the mechanism of PoPlp1 in regulating multiple biological processes, we performed a comparative transcriptomic analysis between strains P. oxalicum 114-2 and ΔPoplp1. The major cellulase and amylase genes were all downregulated, congrent with the results of real-time quantitative polymerase chain reaction analysis. The genes involved in the G protein–cAMP signaling pathway, including several G-protein-coupled receptors, one regulator of G protein signaling, and two cAMP phosphodiesterases, were disrupted by deletion of PoPlp1. These results confirm the positive function of PoPlp1 in the G protein–cAMP signaling pathway. This functional analysis of PoPlp1 will be very beneficial for further study of the regulatory mechanisms of cellulase expression and other biological processes in P. oxalicum 114-2 via the G protein–cAMP signaling pathway.
Penicillium oxalicum is a filamentous fungus expressing relatively complete and effective lignocellulose-degrading enzymes, which are widely used in various industries, in agriculture, and so on. P. oxalicum is also used as a model strain for study of the regulatory mechanisms of cellulase synthesis and asexual development; this research benefits from its mature genetic engineering system. The efficient synthesis of cellulase and several developmental processes (e.g., mycelial growth and conidiation) are co-regulated synergistically by multiple transcription factors in P. oxalicum (Li et al., 2015, 2020; Guo et al., 2021; Xu et al., 2022). The regulation of these processes in filamentous fungi is usually triggered by environmental signals (i.e., soluble carbon sources and light), which act on target genes through an intracellular signaling pathway, i.e., the G protein–cAMP signaling pathway (Hu et al., 2013; Cabrera et al., 2022; Schmoll and Hinterdobler, 2022).
The G protein–cAMP signaling pathway is involved in multiple biological processes, such as development, morphogenesis, induced enzyme synthesis, and secondary metabolism (Lafon et al., 2006; Hinterdobler et al., 2020; Cabrera et al., 2022). Heterotrimeric G proteins (consisting of α, β, and γ subunits), G-protein-coupled receptors (GPCRs), regulators of G protein signaling (RGS), the secondary messenger cAMP, and cAMP-dependent protein kinase (PKA) all play pivotal roles in the G protein–cAMP signaling pathway. Phosducin and phosducin-like proteins (PhLPs) are positive regulators of Gβγ heterodimer, and they act as co-chaperones with the cytosolic chaperonin complex for Gβγ folding and assembly in mammals, plants, and fungi (Lukov et al., 2005; Willardson and Howlett, 2007; Castellano and Sablowski, 2008; Gao et al., 2013). Three PhLPs (PhnA, PhnB, and PhnC) are present in Aspergillus nidulans (Yu, 2006). PhnA is necessary for Gβγ function. Deletion of phnA results in reduced biomass, asexual sporulation in liquid-submerged culture, and defective fruiting body formation; these effects are almost identical to those of the deletion of Gβγ subunits (Seo and Yu, 2006). In Trichoderma reesei, PhLP1 (GNB1 and GNG1) also acts as an important node in heterotrimeric G protein signaling, which influences light responsiveness, sexual development, and glycoside hydrolase gene transcription (Tisch et al., 2011). In the plant pathogens Fusarium graminearum and Cryphonectria parasitica, the putative phosducin-like homolog BDM1 acts on sporulation, germ tube development, and mycelial morphology through the G protein signaling pathway. The virulence of plants has also been found to be significantly reduced after the deletion of BDM1 (Salamon et al., 2010; Horevaj and Bluhm, 2012).
In P. oxalicum 114-2, damage to the G protein–cAMP pathway inhibits the growth, development, and expression of amylase and cellulase (Hu et al., 2013); this similarly occurs in other cellulase-producing filamentous fungi, such as T. reesei and A. nidulans (Amore et al., 2013). In an earlier study, a Gα subunit, PGA3, was found to be essential in the regulatory process for control of the cAMP level in P. oxalicum 114-2. However, further regulatory mechanisms and roles of other factors in the pathway have not been confirmed. In order to establish a clear understanding of the effect of the G protein–cAMP pathway on the expression of cellulase and amylase in P. oxalicum, we identified a regulatory protein PoPlp1 (PDE_07458) in the G protein–cAMP pathway and investigated its roles in regulation of development and cellulase expression.
The wild-type (WT) strain of P. oxalicum 114-2 (CGMCC 5302) was stored in our laboratory. Spores of P. oxalicum strains were cultured on potato dextrose agar (PDA) medium at 30°C for 4 days and harvested using sterile water. Counting of the spores indicated that the final concentration was over 1010/mL. For mycelium cultivation, 108 spores/mL spores were inoculated in liquid minimal mineral salt (MMS) medium containing 2% glucose and incubated at 30°C and 200 rpm for 24 h. For cellulase production and real-time quantitative polymerase chain reaction (RT-qPCR), exactly 0.5 g pre-cultured mycelia were transferred to 50 mL enzyme production medium (liquid MMS medium containing 1% wheat bran and 1% microcrystalline cellulose). Cellulase production was performed in a 300 mL flask at 30°C and 200 rpm.
Deletion of the Poplp1 gene was performed by the homologous recombination method. The up- and downstream homologous flanking DNA sequences were amplified from the genomic DNA of P. oxalicum 114-2 with primers 7458-F1/7458hph-R and 7458hph-F/7458-R1. The hygromycin resistance gene hph was amplified from plasmid pSilent-1 using primers hph-F/hph-R. The three fragments were fused by double-joint PCR (Yu et al., 2004), and the full-length deletion cassette was amplified by primers 7458-F2/7458-R2. The deletion cassette was transformed into P. oxalicum 114-2 by the polyethylene glycol-mediated method (Li et al., 2010) to obtain the Poplp1 gene knock-out strain ΔPoplp1. Poplp1 deletion was confirmed using three pairs of primers: primers inside the target gene Poplp1 7458-F/R, and primer pairs 7458-F1/hphyz-R and hph-yzF/7458-R1.
For the complement of the Poplp1 gene, the integrated expression cassette was amplified from the genomic DNA of P. oxalicum 114-2 with primers 7458-F1/7458-ptraR. The selected marker gene ptrA was amplified from plasmid pME2892 with primers ptrA-F/ptra-7458-R, and downstream homologous flanking DNA sequences were amplified from the genomic DNA of P. oxalicum 114-2 with primers 7458-ptraF/7458-R0. The three fragments were fused, and the complement cassette was amplified using primers 7458-F3/7458-R3. The cassette was transformed into the strain ΔPoplp1 using the abovementioned method, and the complement strain CPoplp1 was thereby constructed. Complementation of Poplp1 was confirmed by primers 7458-F/7458-R.
Supplementary Table S1 lists all primers used in the construction of strains ΔPoplp1 and CPoplp1.
Phenotype analysis was performed on three kinds of sole-carbon-source medium plates (MMS medium containing 2% glucose, 1% cellulose, or 1% starch) and two kinds of compound medium plates (PDA and 10% wheat bran medium) (Li et al., 2015). The plates were inoculated with exactly 1 μL of fresh spores (a total of 105 spores) at a concentration of 108/mL and incubated for 3 days at 30°C.
To measure the biomass of the strains and glucose consumption in the growth medium of the strain, we transferred 0.5 g pre-cultured mycelia to 20 mL of MMS medium containing 2% glucose. All cultures were sampled every 12 h. The mycelia and culture medium were separated by centrifugation and collected separately. Mycelia were dried for 4 h to constant weight at 70°C, and the glucose content of the medium was determined by the 3,5-dinitrosalicylic acid method. Three biological triplicates were performed, and the mean values and corresponding standard deviations were calculated.
The spore production capacity of P. oxalicum 114-2 and its mutants was measured on a PDA medium. Exactly 250 μL of fresh spores at a concentration of 108 spores/mL were spread across the plates. The plates were incubated at 30°C, and the number of spores within a fixed area was counted at 24, 30, 36, 48, 60, and 72 h, separately for each strain. Three biological triplicates were performed, and the mean values and corresponding standard deviations were calculated.
To observe the mycelial morphology, we spread 120 μL of fresh spores at a concentration of 108/mL evenly onto PDA plates. Several cover glasses were inserted into the medium slant and the plates were incubated at 30°C. The cover glasses were then removed at 24 and 48 h and observed under an optical microscope at 400× magnification.
For the enzyme activity assay, all strains were incubated in the enzyme production medium. Fresh spores were inoculated at 108/mL into liquid MMS medium containing 2% glucose and incubated at 30°C and 200 rpm for 24 h. Next, the mycelia were collected by vacuum pump filtration. Exactly 0.5 g mycelium was transferred to a 50 mL measure of enzyme-producing medium and incubated for 6 days in a 300 mL bottle at 30°C and 200 rpm. Fermentation broths were sampled and measured every 24 h from 72 to 144 h. For the filter paper activity (FPA), endoglucanase, cellobiohydrolase, β-glucosidase, and amylase activity assays, Whatman™ 1 filter paper, sodium carboxymethyl cellulose (CMC-Na, Sigma), p-nitrophenyl-β-D-cellobioside (pNPC, Sigma), p-nitrophenyl-β-D-glucopyranoside (pNPG, Sigma), and starch (Sigma) were used as substrates, respectively (Chen et al., 2013). One unit of enzyme activity was defined as the amount of enzyme required to produce 1 μmol glucose or p-nitrophenyl (Sigma) per minute under the assayed conditions (Wood and Bhat, 1988). Three biological triplicates were performed, and the mean values and corresponding standard deviations were calculated.
For RT-qPCR, 0.5 g pre-incubated mycelia of the three strains was induced in 50 mL of cellulase production medium for 4 h. Subsequently, the mycelia were collected for RNA extraction using TRIzol reagent (Invitrogen), in accordance with the operation manual. cDNA and RT-qPCR reaction systems were synthesized using the PrimeScript™ RT reagent kit with gDNA Eraser (Perfect Real Time) and TB Green® Premix Ex Taq™ II (Tli RNaseH Plus) (TAKARA), respectively, following the manufacturers' protocols. The RT-qPCR reaction procedure was performed under the following conditions on a LightCycler®480 System (Roche): 95°C for 2 min; 40 cycles at 95°C for 10 s and 61°C for 30 s. The melting curves were measured with a temperature gradient of 0.1°C per second from 65 to 95°C. The expression levels of all genes were calculated using relative quantification, with actin as the reference gene. Three biological triplicates were performed, and the mean values and corresponding standard deviations were calculated. Supplementary Table S2 lists the primers used in RT-qPCR.
For each of the three strains, 0.5 g pre-incubated mycelia was cultured in carbon-free MMS medium for 2 h and then transferred to 50 mL of cellulose-inducing medium for 4 and 24 h, respectively. The mycelia were collected by vacuum filtration and ground into powder. The cell contents of 0.1 g of ground mycelia were extracted with 1 mL of HCl (0.1 M). Intracellular protein concentration was determined by the Bradford method (Bradford, 1976); intracellular cAMP concentration was measured by following the instructions of the microbial cAMP enzyme-linked immunosorbent assay kit (Jiangsu Enzyme-Biao Biotechnology Co., Ltd.). The intracellular cAMP concentration for each of the analyzed samples was standardized to the concentration of intracellular protein, with the cAMP concentration of the wild-type strain taken to be 100%. Three biological triplicates were performed, and the mean values and corresponding standard deviations were calculated.
The weighed cAMP powder was added directly to the cellulose induction medium, and 0.5 g hyphae were transferred to the cellulose induction medium with cAMP for 4 h. Samples were collected and their gene expression levels were measured. The expression levels of all genes were calculated using relative quantification, with actin as the reference gene. Three biological triplicates were performed, and the mean values and corresponding standard deviations were calculated.
For the transcriptome analysis, liquid MMS medium containing 2% glucose was inoculated with spores at 108/mL and incubated at 30°C with agitation at 200 rpm for 24 h. Subsequently, the mycelia were collected by vacuum pump filtration and transferred to a liquid MMS medium without a carbon source. After 2 h of carbon starvation, the mycelia were collected, and 0.5 g mycelia were transferred to 50 mL of liquid MMS medium, including 1% microcrystalline cellulose, and incubated at 30°C with agitation at 200 rpm for 4 h. The RNA of P. oxalicum 114-2 and ΔPoplp1 was extracted using the TRIzol reagent (Invitrogen, USA) in accordance with the manufacturer's protocol. Digital gene expression profiling experiments were performed by Novogene (China) on an Illumina NovaSeq 6000 platform (Illumina, USA). Gene expression levels were indicated by the expected number of fragments per kilobase of transcript sequence per million base pairs sequenced (reads per kb per million reads). The raw RNA-Seq data were deposited at the National Center for Biotechnology Information (NCBI) Sequence Read Archive with the series reference number PRJNA910646. Genes with an adjusted P-value ≤0.05 as identified by DESeq2 and fold change ≥ 2 were considered to be differentially expressed. Gene Ontology and Kyoto Encyclopedia of Genes and Genomes enrichment analyses of differentially expressed genes were implemented using the R package clusterProfiler (Yu et al., 2012).
Equal-variance one-tailed t-tests were conducted to test comparisons for statistical significance. The mean, corresponding standard deviation, and P-value for the corresponding statistical test were calculated in all quantitative analyses. P-values≤ 0.05 were considered to represent statistical significance.
PhLPs have been identified as downregulators of G protein signaling and they also regulate multiple physiological processes. Here, the PhLP PoPlp1 was screened and analyzed in P. oxalicum 114-2; it contains a phosducin domain with a length of 159 amino acids (120–159). The complete amino acid sequences of Phlp homolog proteins from Penicillium species, Aspergillus species, Trichoderma species, Neurospora crassa, and Saccharomyces cerevisiae were downloaded from the NCBI database. The phylogenetic tree of Phlp homolog proteins was analyzed using MEGA-X software via the neighbor-joining method (Figure 1) (Kumar et al., 2018). Sequence alignment of the amino acids of Phlp homologs was completed via the ClustalW multiple alignment function using a biological editing tool. The sequence alignment results showed that the N-terminus and C-terminus of Phlp in Penicillium species, Aspergillus species, and Trichoderma species were highly conserved (Supplementary Figure S1). The amino acid sequence of PoPlp1 exhibited the closest relationship with other Penicillium species (OKP10650.1, KAF3384507.1), with sequence similarities above 80%. Meanwhile, it shared 56% identity with the homologs of A. nidulans FGSC (PhnA, XP_657686.1), which is essential in Gβγ-mediated signaling for the vegetative growth and development of A. nidulans (Seo and Yu, 2006). In the case of Phlp1 from T. reesei QM6a (EGR50146.1), which influences glycoside hydrolase gene transcription and sexual development, PoPlp1 also exhibited 39% identity (Tisch et al., 2011).
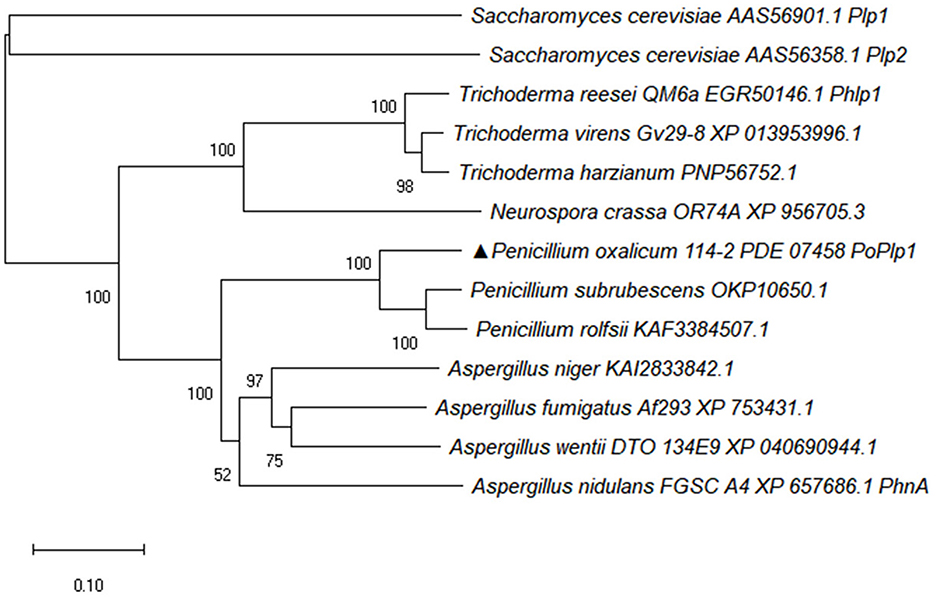
Figure 1. Phylogenetic analysis of PoPlp1. The complete protein sequences of Phlp homolog proteins from Penicillium species, Aspergillus species, Trichoderma species, N. crassa, and S. cerevisiae were downloaded from the NCBI database. The phylogenetic tree of Phlp homolog proteins was analyzed using the MEGA-X software via the neighbor-joining method.
In view of the results of sequence alignment, we determined whether PoPlp1 played differential roles in regulating growth and development in P. oxalicum 114-2.
To confirm the functions of PoPlp1 in G protein signaling regulation, we deleted and complemented it in the P. oxalicum 114-2 strain through homologous recombination (Figure 2A). Deletion of PoPlp1 was verified by PCR, Southern blot, and RT-qPCR (Figures 2B–D).
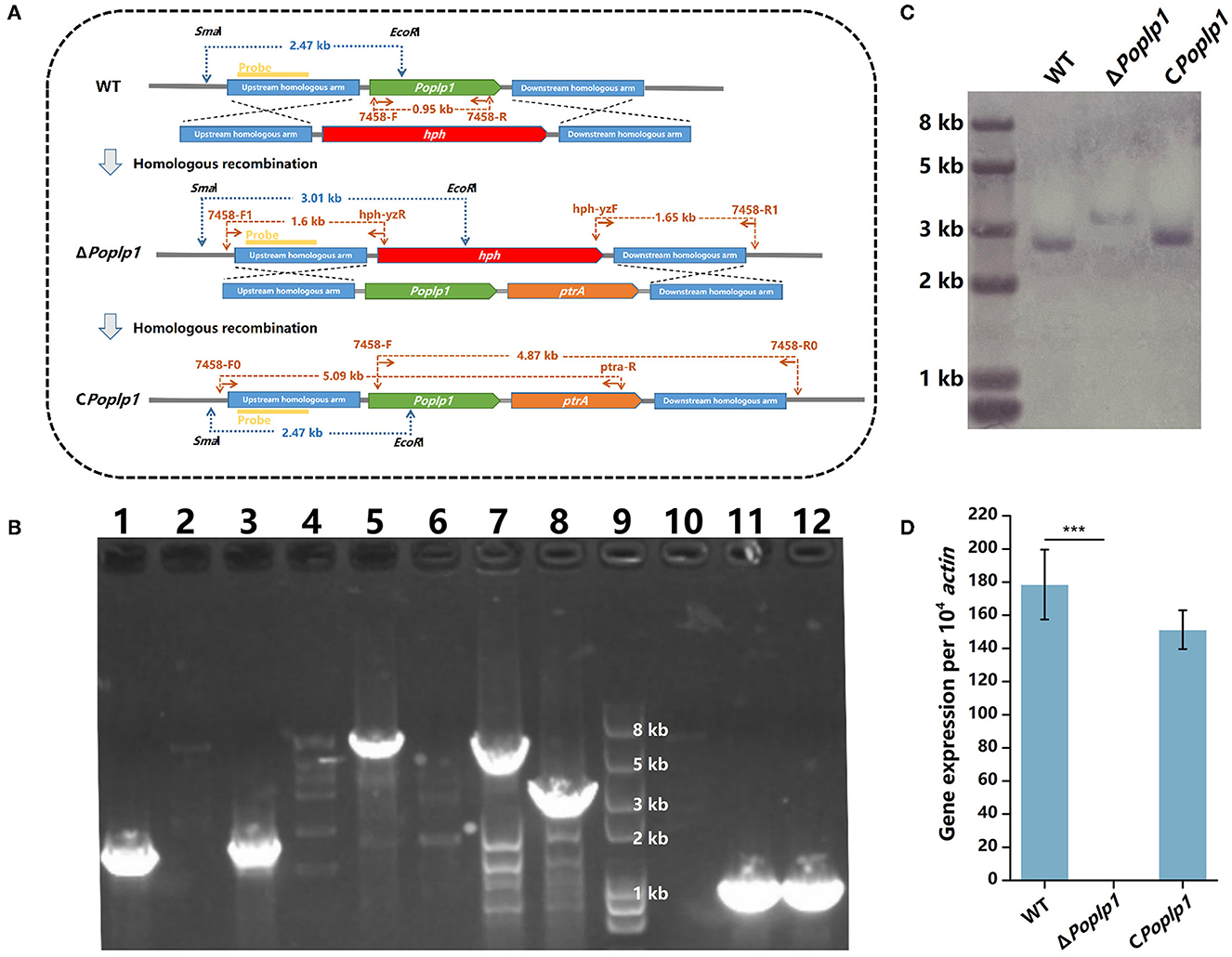
Figure 2. Deletion and verification of PoPlp1. (A) Strategy for the construction and verification of ΔPoplp1. The positions of the primers, probes for Southern blot analysis, and DNA fragment sizes are marked at the corresponding positions. (B) PCR analysis of deletion and complementation of Poplp1. PCR confirmation of Poplp1 deletion was performed using three pairs of primers (lanes 1–4); Poplp1 complementation was confirmed using three pairs of primers (lanes 5–8). (1) Primer pairs 7458-F1/hph-yzR (lanes 1–2) and hph-yzF/7458-R1 (lanes 3–4) (Supplementary Table S1). The theoretical length of the DNA fragment of the deletion strain in each case was 1.60 and 1.65 kb. (2) Primers inside the target gene Poplp1 7458-F/R (lanes 10–12). No DNA fragments could be detected in ΔPoplp1. PCR complementation of Poplp1 was verified using primers inside the target gene Poplp1 7458-F/R (lane 11). A 0.95 kb-length DNA fragment should be detected. (3) Primer pairs 7458-F0/ptra-R (lanes 5–6) and 7458-F/7458-R0 (lanes 7–8). The theoretical length of the DNA fragment of the complement strain in each case was 5.09 and 4.87 kb. Lanes 2, 4, 6, and 8 are the results for P. oxalicum 114-2. Lanes 1, 3, and 10 are the results for Poplp1-deletion strain ΔPoplp1. Lanes 5, 7, and 10 are the results for the Poplp1-complement strain CPoplp1. Lane 9 is the Trans2K Plus II DNA marker (TransGen Biotech, China). (C) Southern blot analysis of ΔPoplp1. The genomic DNA of P. oxalicum 114-2, ΔPoplp1, and CPoplp1 was digested by SmaI and EcoRI. A 0.8 kb-length fragment upstream of the target gene was amplified as the hybridization probe. P. oxalicum 114-2 and CPoplp1 genomic DNA were digested by SmaI and EcoRI and a 2.47 kb-length fragment containing the complete probe sequence was generated, while a 3.01 kb-length fragment containing the probe sequence was generated from the ΔPoplp1 genomic DNA. (D) qRT-PCR analysis of the gene expression levels of Poplp1 in the mutant strains. Poplp1 expression in P. oxalicum 114-2, ΔPoplp1, and CPoplp1 was tested using primers RT-7458-F/R inside Poplp1 (Supplementary Table S2). T-tests were conducted to analyze differences in gene expression levels for significance (*P < 0.05).
The influence of PoPlp1 on the development of P. oxalicum 114-2, including colony phenotype, growth rate, conidiation, and mycelial morphological characteristics, was examined. The colony growth of mutant ΔPoplp1 on the solid medium containing glucose as the sole carbon source was significantly restricted (Figure 3A), and the colony diameter of ΔPoplp1 was considerably smaller than that of P. oxalicum 114-2. Consistent with this finding, the growth rate of mycelia was also affected by use of a liquid medium with glucose as the only carbon source at 24–48 h (Figure 3B). This outcome may be attributable to deletion of the Poplp1 gene, which affected the rate of glucose uptake, with a significantly slower rate of glucose consumption observed in the culture medium of ΔPoplp1 than in that of the original strain at 12–48 h (Figure 3C). In the case of the solid medium with starch or cellulose as the sole carbon source, the diameter of the transparent zone was significantly smaller than in the case of P. oxalicum 114-2, which indicates that the hydrolysis abilities of ΔPoplp1 for starch and cellulose were affected (Figure 3A). Possibly, the lack of Poplp1 affected the synthesis of starch and cellulose hydrolytic enzymes.
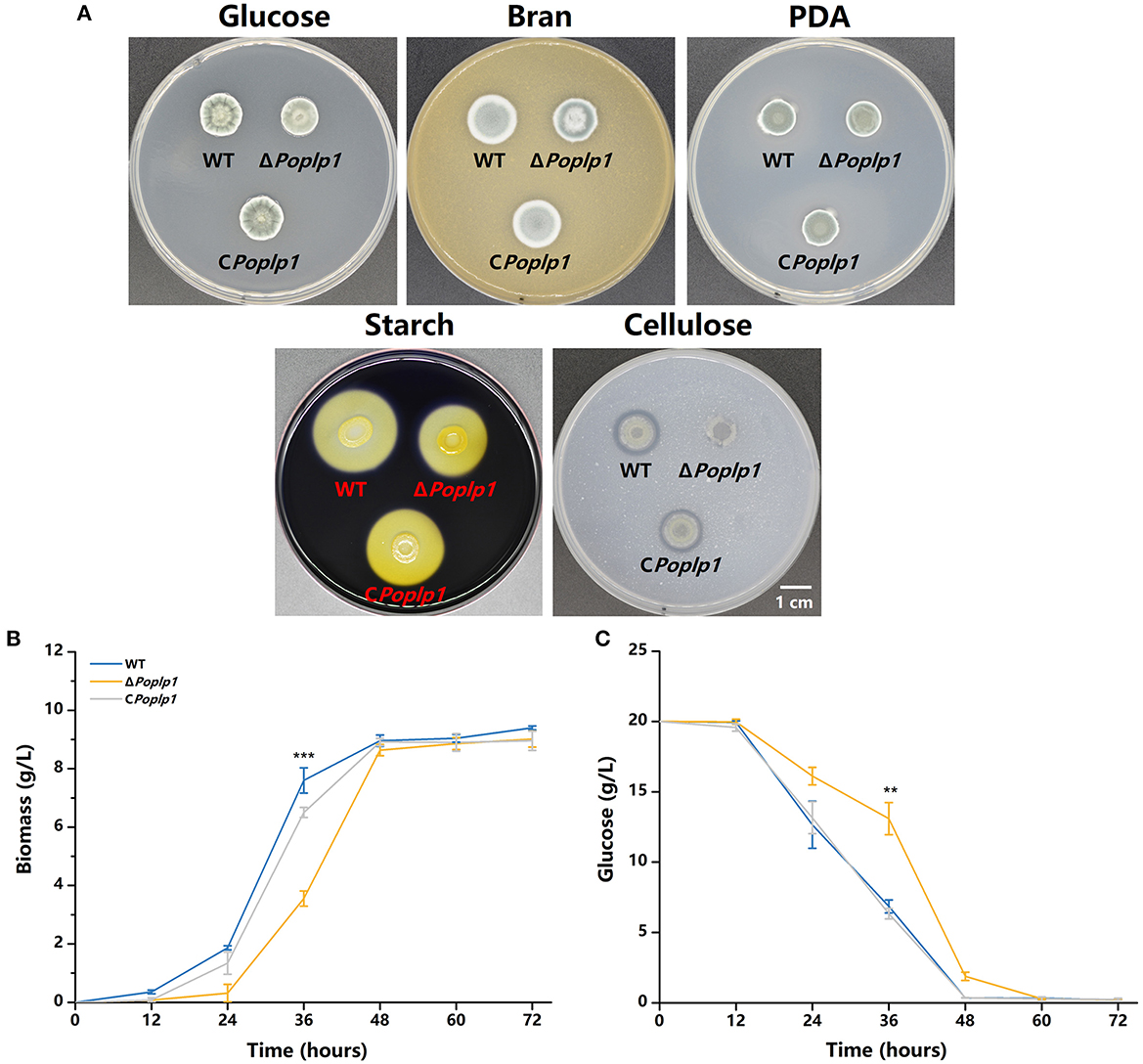
Figure 3. Effect of PoPlp1 on the growth and phenotype of P. oxalicum 114-2. (A) Colony phenotypes of P. oxalicum 114-2, ΔPoplp1, and CPoplp1. Phenotype analysis was performed on MMS medium plates containing 2% glucose, 1% cellulose, or 1% starch, PDA plates, and 10% wheat bran medium plates. Plates were inoculated with 1 μL of suspension liquid at 108 spores/mL and incubated for 3 days at 30°C. (B) Effects of the Poplp1 gene on biomass in glucose culture. The dry weight of mycelia of all strains in the glucose medium was measured every 12 h. (C) The rate of glucose consumption by P. oxalicum 114-2, ΔPoplp1, and CPoplp1 strains. Residual glucose in the culture medium was detected every 12 h. Three biological triplicates were performed, and the mean values and corresponding standard deviations were calculated; t-tests were conducted to analyze the differences in growth and glucose consumption level between P. oxalicum 114-2 and ΔPoplp1 for significance (*P < 0.05, **P < 0.01, ***P < 0.005).
To examine the influence of PoPlp1 on conidiation, we measured the conidiation rate of all strains on the PDA plate. Conidiation occurred 6 h earlier for ΔPoplp1 than for P. oxalicum 114-2. At 72 h, the number of conidia of ΔPoplp1 was twice that of the number for the original strain (Figure 4A). Mycelium morphology at 24 and 48 h indicated that strain ΔPoplp1 produced conidiophores at 24 h, whereas the parental strain did not (Figure 4B). At 48 h, strain ΔPoplp1 produced more conidium chains, which resulted in an increase in conidia (Figure 4B).
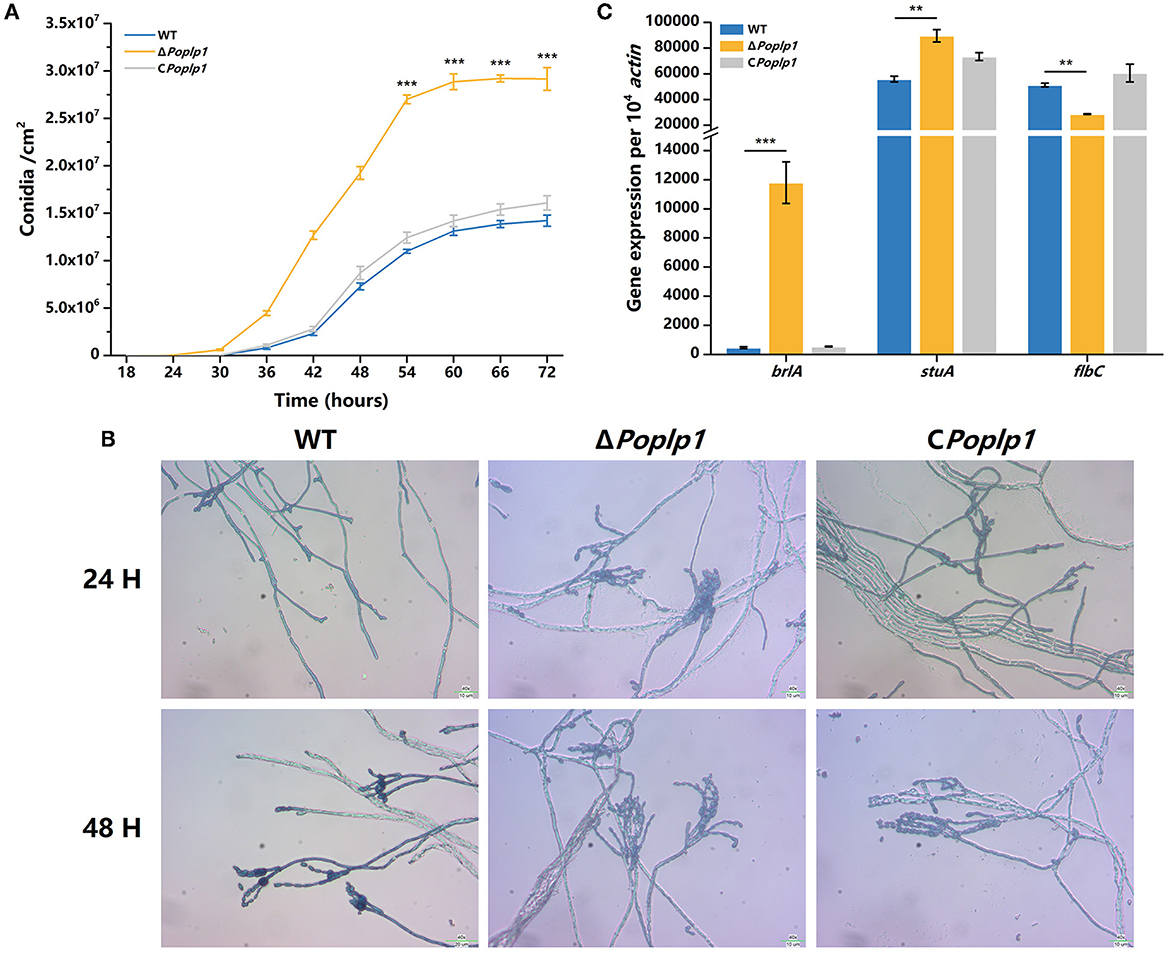
Figure 4. Effect of PoPlp1 on conidiation in P. oxalicum 114-2. (A) Determination of sporulation rate in P. oxalicum 114-2, ΔPoplp1, and C Poplp1. The number of spores within a fixed area of the PDA medium were counted every 6 h. (B) Microscopic observations of spore and mycelia morphology on PDA medium. The microscopic morphology of spores and mycelia was observed by optical microscope at 400× magnification after culturing for 24 and 48 h, respectively. (C) Analysis of gene expression levels of conidiation-related transcription regulators. The gene expression levels of BrlA, StuA, and FlbC in strains P. oxalicum 114-2, ΔPoplp1, and CPoplp1 were detected by RT-qPCR. Three biological triplicates were performed, and the mean values and corresponding standard deviations were calculated. The levels of expression of all genes were calculated using relative quantification, with actin as the reference gene; t-tests were conducted to analyze the differences in sporulation rate and gene expression level between P. oxalicum 114-2 and ΔPoplp1 for significance (*P < 0.05, **P < 0.01, ***P < 0.005).
In P. oxalicum 114-2, several key transcriptional regulators of asexual development have been identified; these include BrlA, StuA, and FlbC, which can regulate levels of expression of pigmentation-related and spore wall protein-related genes (Qin et al., 2013; Yao et al., 2016; Li et al., 2019). To explore the mechanism by which PoPlp1 affects conidiation, we analyzed the transcription levels of three genes in strains 114-2, ΔPoplp1, and CPoplp1 by RT-PCR. After deletion of Poplp1, the levels of expression of brlA and stuA were significantly enhanced in ΔPoplp1 and were nearly 25.0- and 1.6-fold higher, respectively, than those occurring in P. oxalicum 114-2 (Figure 4C). Meanwhile, expression of flbC decreased significantly, undergoing a 44.2% decrease compared with that of P. oxalicum 114-2. Given that BrlA is a crucial regulator of conidiation (Qin et al., 2013), we conclude that PoPlp1 is involved in conidiation through regulation of transcription levels of the key regulator BrlA.
According to the results of phenotypic analysis of the cellulose medium, PoPlp1 affected the synthesis of cellulose hydrolytic enzymes in P. oxalicum. Given that PoPlp1 acts as a G protein regulator, we focused on the mechanism by which PoPlp1 regulates cellulase synthesis in P. oxalicum. Its effects on the expression of cellulases were determined by detection of activity toward filter paper (indicating overall cellulase activity), CMC-Na (endoglucanase), pNPC (cellobiohydrolase activity), and pNPG (β-glucosidase). In ΔPoplp1, all detected enzyme activities were reduced. This strain exhibited the highest FPA after fermentation on the fifth day, and its activity was reduced to 15% of that occurring in P. oxalicum 114-2 (Figure 5A). Similarly, the endoglucanase activity of ΔPoplp1 reached its peak on the fifth day and decreased to 34% of that of P. oxalicum 114-2 (Figure 5B). The cellobiohydrolase and β-glucosidase activity of ΔPoplp1 continually increased, with values reaching 34 and 36%, respectively, of those of P. oxalicum 114-2 on the sixth day (Figures 5C, D). Activity measurement showed that PoPlp1 plays an important role in the production of cellulase, and the absence of PoPlp1 dramatically reduced the production of cellulases.
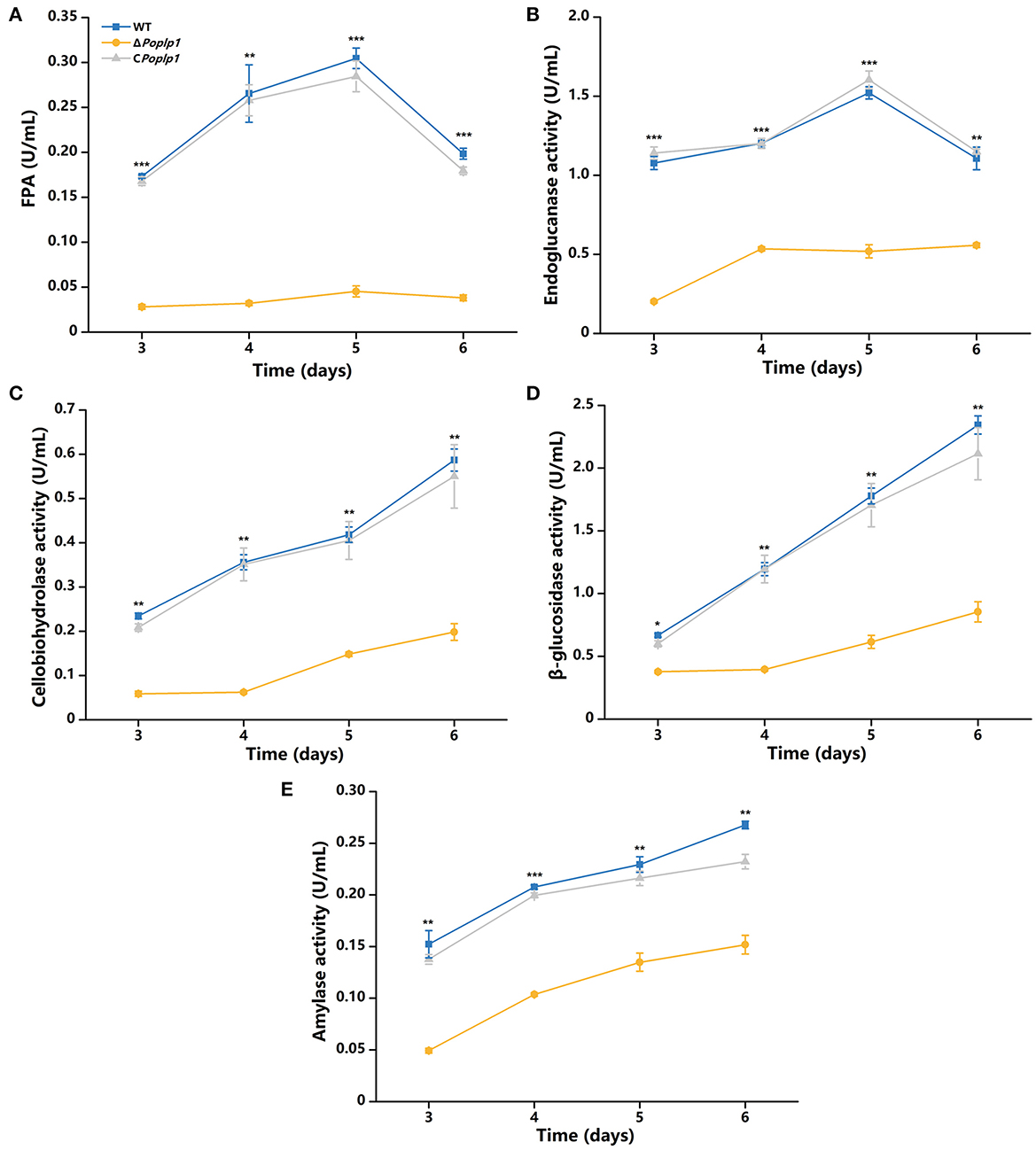
Figure 5. Analysis of the cellulase activity of P. oxalicum 114-2, ΔPoplp1, and CPoplp1. (A) FPA; (B) endoglucanase activity; (C) cellobiohydrolase activity; (D) β-glucosidase activity; and (E) amylase activity. All strains were sampled and measured every 24 h from day 3 to 6. Three biological triplicates were performed, and the mean values and corresponding standard deviations were calculated; t-tests were conducted to analyze differences in the cellulase activity expression levels between P. oxalicum 114-2 and ΔPoplp1 for significance (*P < 0.05, **P < 0.01, ***P < 0.005).
To further investigate the mechanism underlying the involvement of PoPlp1 in regulating cellulase expression, we analyzed the transcriptional levels of the main cellulase genes and regulators under cellulose induction by RT-qPCR. In ΔPohpl1, expression of the major cellobiohydrolase and endoglucanase genes cbh1 and eg1 was substantially reduced, reaching 0.6% and 1.4%, respectively, of the levels of expression observed in P. oxalicum 114-2 after inducement by cellulose for 4 h (Figure 6B). The major extracellular β-glucosidase gene bgl1 in ΔPoplp1 was also expressed at a lower level than in P. oxalicum 114-2; this was reduced by 76.4% in the absence of PoPlp1. These results were consistent with the reduction of FPA, cellobiohydrolase, and endoglucanase activity in ΔPoplp1, indicating that PoPlp1 affects cellulase production at the gene expression level.
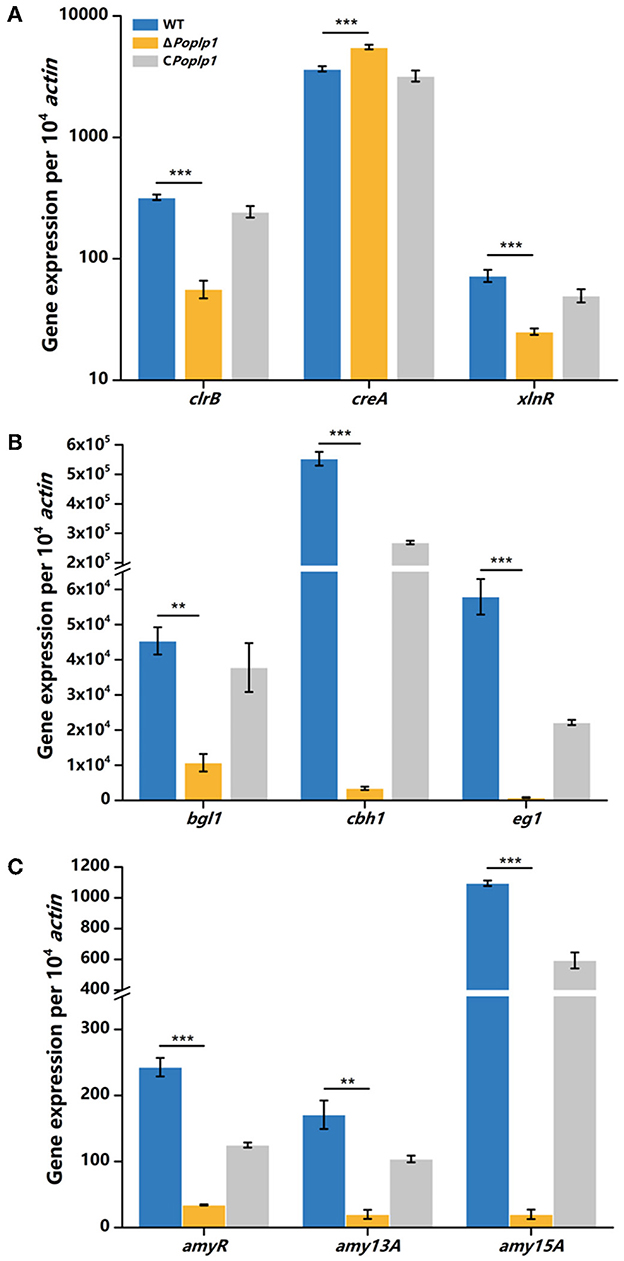
Figure 6. qRT-PCR analysis of the major cellulases and transcriptional regulators of P. oxalicum 114-2, ΔPoplp1, and CPoplp1. Expression levels of (A) the four cellulase genes, (B) the major transcriptional regulators, and (C) amylase genes were measured after induction by cellulose for 4 h. Three biological triplicates were performed, and the mean values and corresponding standard deviations were calculated. The levels of expression of all genes were calculated using relative quantification, with actin as the reference gene; t-tests were conducted to analyze the differences in gene expression levels between P. oxalicum 114-2 and ΔPoplp1 for significance (*P < 0.05, **P < 0.01, ***P < 0.005).
Cellulase gene expression was regulated by multiple transcriptional factors in P. oxalicum 114-2, including CreA, ClrB, AmyR, and XlnR (Li et al., 2015). CreA and ClrB are the most important transcription repressor and activator of cellulases in P. oxalicum 114-2, respectively. AmyR and XlnR are involved in co-regulation of cellulase expression as the major activators of amylase and xylanase, respectively. The RT-qPCR results showed that the gene expression level of CreA increased to 1.5-fold that of P. oxalicum 114-2 when induced by cellulose for 4 h, whereas activators ClrB and XlnR decreased to 18 and 34%, respectively, of the levels observed for P. oxalicum 114-2 (Figure 6A). Nevertheless, expression of the cellulase gene repressor AmyR decreased noticeably, and was reduced to 14% in P. oxalicum 114-2, which was inconsistent with the downregulation of amylase genes amy13A and amy15A (Figure 6C). We assume that PoPlp1 may regulate expression of cellulase and amylase genes in coordination with multiple cellulolytic transcription factors.
Given the role of PoPlp1 as a potential G protein regulator and the influence of intracellular cAMP concentration on cellulase and amylase expression in P. oxalicum 114-2 (Hu et al., 2013), we hypothesized that PoPlp1 regulates cellulase and amylase expression by affecting the levels of expression of transcriptional regulators through the G protein–cAMP signaling pathway. In this study, to investigate the mechanism underlying PoPlp1 regulation of cellulase expression, we measured intracellular cAMP levels. When the strains were cultured in the cellulase enzyme production medium, the intracellular cAMP concentration of ΔPoplp1 was significantly decreased compared with that of P. oxalicum 114-2, which was consistent with the decrease in cellulase enzyme activity observed in ΔPoplp1. The cAMP concentration in ΔPoplp1 decreased by 17.2 and 36.2% at 120 and 144 h, respectively (Figure 7A). At the same time, cellulase enzyme activity exhibited the highest levels during this period. Under the cellulose-inducing condition, the intracellular cAMP concentration of ΔPoplp1 was decreased by 13.3 and by 8.6% after 4 and 24 h, respectively (Figure 7B). This finding was also consistent with the downregulated expression of cellulase genes observed in ΔPoplp1. These results showed that PoPlp1 regulates the G protein–cAMP signaling pathway by affecting intracellular cAMP levels, which leads to a signal transduction cascade of cellulase and amylase expression.
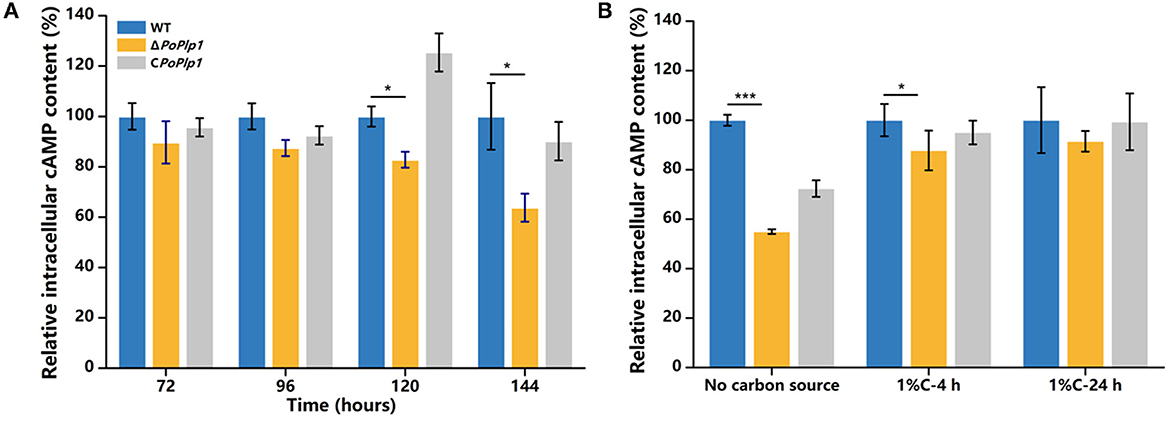
Figure 7. Variation in intracellular cAMP level induced by deletion of PoPlp1. (A) Intracellular cAMP levels under enzyme production conditions. (B) Intracellular cAMP levels under cellulose-inducing conditions. The intracellular cAMP concentrations of all the detected samples were standardized with reference to the same level of intracellular protein. The cAMP concentration of the wild-type strain was considered to be 100%. Three biological triplicates were performed, and the mean values and corresponding standard deviations were calculated; t-tests were used to analyze the differences in intracellular cAMP levels between P. oxalicum 114-2 and ΔPoplp1 for significance (*P < 0.05, **P < 0.01, ***P < 0.005).
To further investigate the effect of cAMP on the ΔPoplp1 cellulase gene and its transcription regulatory factors, we cultured all strains in cellulose medium supplemented with 4 mM cAMP for 4 h and measured the levels of expression of cellulase genes and related transcription regulatory factors. Under this condition, with addition of cAMP, gene expression of the main cellulase transcription activator clrB in ΔPoplp1 was decreased by 42.7% compared with that observed in P. oxalicum 114-2; in contrast, without addition of cAMP, gene expression of clrB was decreased by 82.4% (Figure 8A). This finding indicates that exogenous addition of cAMP effectively restores expression of the clrB gene. The level of expression of the main cellulase transcription inhibitor CreA showed an overall downward trend after the addition of cAMP. However, compared with WT strains, the exogenous addition of cAMP did not effectively restore the gene expression level of CreA (Figure 8B). The gene expression of the other two main transcription factors (XlnR and AmyR) showed two opposing trends (Figures 8C, D). In addition, for ΔPoplp1, three major cellulase genes exhibited varying degrees of upregulation after the exogenous addition of cAMP. Moreover, compared with WT strains, their expression levels exhibited varying degrees of recovery (Figures 8E–G). These results indicate that the effect of cAMP on the expression of cellulase genes and their transcription factors is a complex process. However, the addition of cAMP after deletion of Poplp1 can restore the expression of cellulase genes to a certain extent. This outcome further indicates that downregulation of the cellulase gene in ΔPoplp1 is caused by a decrease in intracellular cAMP content, which strongly supports the above conclusion.
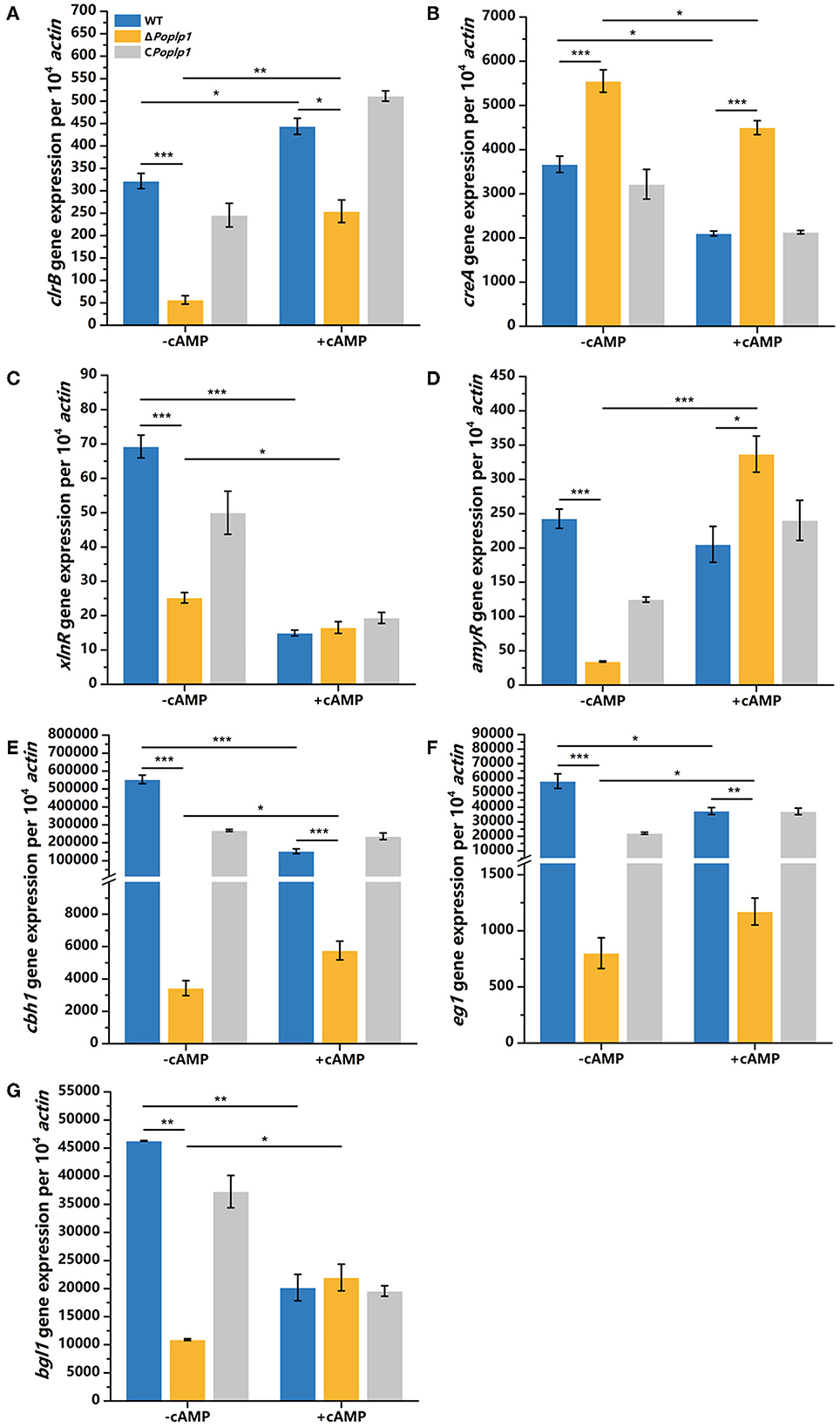
Figure 8. qRT-PCR analysis of the major cellulases and transcriptional regulators of P. oxalicum 114-2, ΔPoplp1, and CPoplp1 with the addition of cAMP. Levels of expression of (A) clrB, (B) creA, (C) xlnR, (D) amyR, (E) cbh1, (F) eg1, and (G) bgl1 were observed after induction with 4 mM cAMP cellulose for 4 h. Three biological triplicates were performed, and the mean values and corresponding standard deviations were calculated. The expression levels of all genes were calculated using relative quantification, with actin as the reference gene; t-tests were conducted to analyze differences in gene expression levels for significance (*P < 0.05, **P < 0.01, ***P < 0.005).
To comprehensively explore the functions of PoPlp1, we performed a comparative transcriptomic analysis between strains P. oxalicum 114-2 and ΔPoplp1. After deletion of Poplp1, 158 were genes upregulated and 237 were downregulated. Moreover, the differentially expressed genes were mainly involved in pathways involving the biosynthesis of secondary metabolites (82 genes), starch and sucrose metabolism (23 genes), tyrosine metabolism (17 genes), and phenylalanine metabolism (16 genes) (Supplementary Figure S2).
Considering the regulatory capacity of PoPlp1 toward cellulases, the expression levels of 80 annotated cellulolytic genes were analyzed. The results showed that the expression of most cellulase and hemicellulase genes was substantially downregulated in ΔPoplp1 (Figure 9). Among these genes, the major cellobiohydrolase gene cbh1, endo-β-1,4-glucanase gene eg1, and extracellular β-glucosidase gene bgl1 were all dramatically downregulated in ΔPoplp1, which was consistent with the results of RT-qPCR.
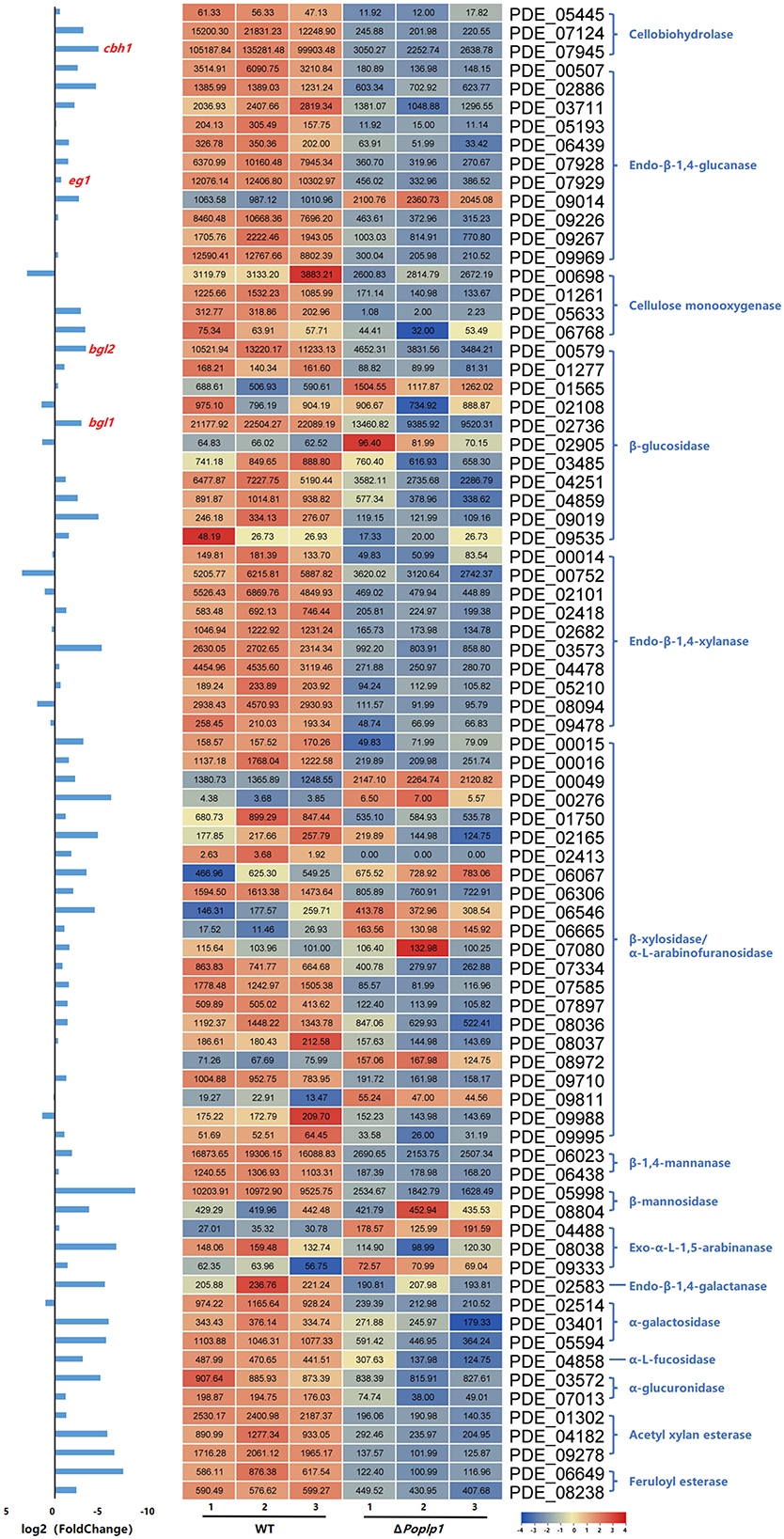
Figure 9. Expression of 80 annotated cellulose hydrolase genes in P. oxalicum 114-2 and ΔPoplp1. Levels of expression (RPKM) of the 80 cellulose hydrolase genes are indicated by a green–yellow–red color scale. The minimum value is 0, the median is 100, and the maximum value is 8,054.
The genes involved in the G protein–cAMP signaling pathway were analyzed. Although the absence of PoPlp1 probably inhibited the activity of G proteins, the gene expression levels of heterotrimeric G proteins (consisting of three Gα, one Gβ, and one Gγ subunits) hardly changed (Table 1). However, the RT-qPCR results showed that deletion of Poplp1 resulted in significant downregulation of Gβ gene expression levels, which indicates that deletion of Poplp may have caused a lack of G beta-folding and a decrease in G protein signaling (Supplementary Figure S3). Meanwhile, the gene expression levels of multiple GPCRs (including one Class III GPCR, two Class IX GPCRs, and several PTH11-like GPCRs) were significantly upregulated in ΔPoplp1. This result may be attributable to the interruption of G protein signaling in ΔPoplp1, leading to an invalid response by the target genes to external signals. In addition, the expression of GPCRs was upregulated to enhance reception of these signals. Moreover, a Gα regulator, FlbA (PDE_03092), which is also considered to be involved in conidium production (Xie et al., 2022), was dramatically downregulated in ΔPoplp1. For the cAMP signaling proteins involved, the transcription levels of adenylate cyclase (PDE_08988) and PKA (catalytic subunit PDE_03213 and regulatory subunit PDE_04688) were barely disrupted by PoPlp1. Nevertheless, the expression of a low-affinity cAMP phosphodiesterase (PDE_00536) doubled compared with that observed in the wild-type strain 114-2, while a high-affinity cAMP phosphodiesterase (PDE_02983) was reduced by half in ΔPoplp1.
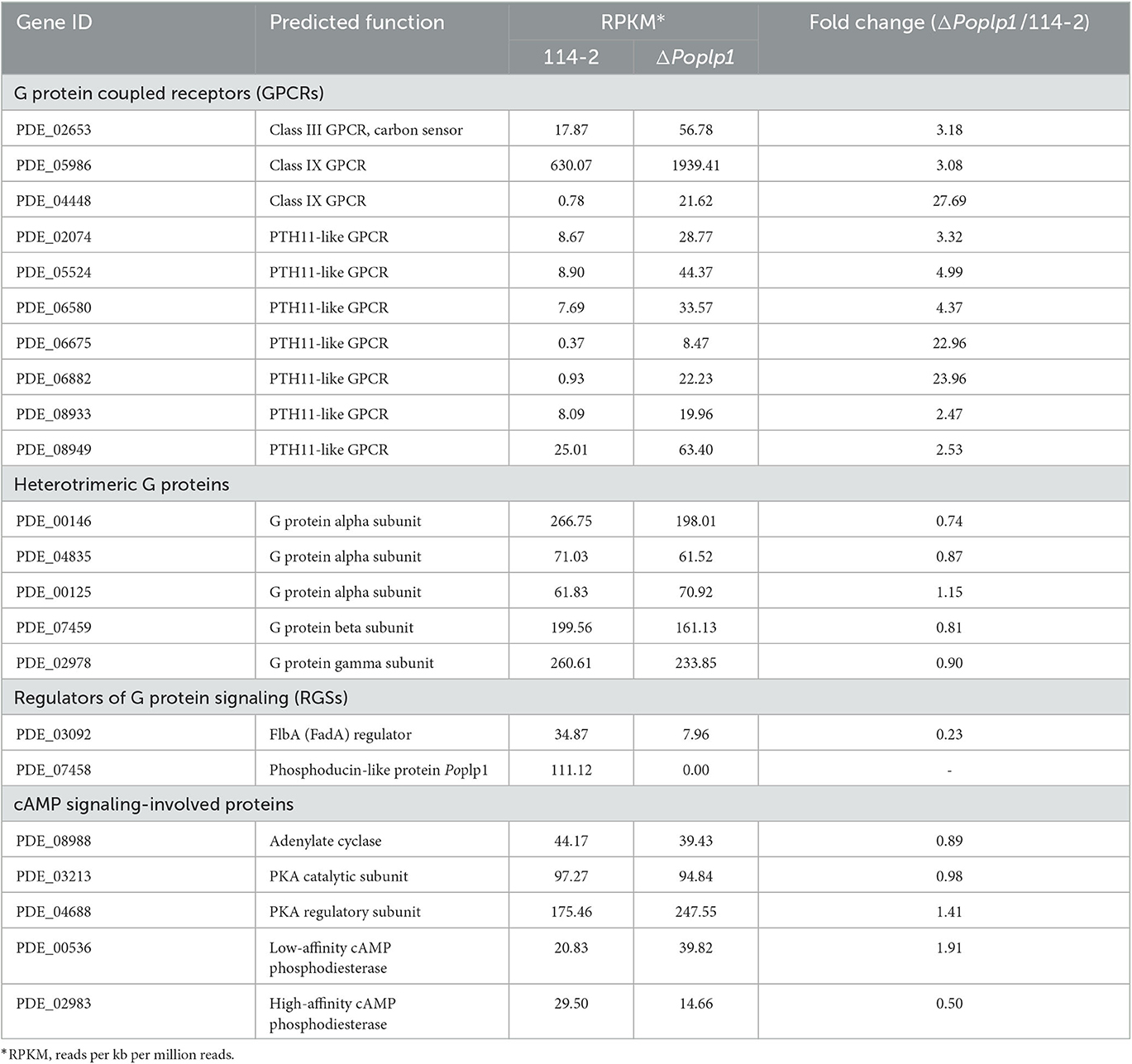
Table 1. Transcription levels of genes involving in the G protein–cAMP signaling pathway in P. oxalicum 114-2 and ΔPoplp1.
The results of this comparative transcriptome analysis provide a comprehensive understanding of the regulatory network involved in PoPlp1, which is highly beneficial for study of the regulatory mechanisms of cellulase expression and other biological processes in P. oxalicum 114-2.
In numerous species, the G protein–cAMP signaling pathway is involved in multiple biological processes, such as development, morphogenesis, glycoside hydrolase synthesis, and secondary metabolism. A previous study has also confirmed its roles in regulating growth, development, and expression of amylase and cellulase in P. oxalicum 114-2 (Hu et al., 2013). However, a comprehensive and deep understanding of the regulatory mechanisms has been lacking. In the present study, a G protein regulator, PoPlp1, was identified and functionally studied in P. oxalicum 114-2. PoPlp1 is conserved with PhLPs in A. nidulans and T. reesei and is involved in growth, conidiation, sexual development, and glycoside hydrolase gene transcription (Seo and Yu, 2006; Tisch et al., 2011). After deletion of Poplp1, phenotypes with respect to these functions were significantly altered in ΔPoplp1, including limitation of colony growth and glucose utilization, the hydrolysis capability of starch and cellulose, advancement of conidiation time, and enhancement of conidiation ability and conidium chain (Figures 3, 4). Through detection of the levels of gene expression of several sporulation-related transcription regulators, we observed that expression of the key regulator BrlA, whose absence can completely block conidiation (Qin et al., 2013), was significantly enhanced in ΔPoplp1. Therefore, we hypothesize that PoPlp1 is involved in conidiation through regulation of the transcription levels of BrlA. Deletion of Poplp also led to a decrease in intracellular cAMP content. When cAMP was added to the culture medium, the expression of brlA relative to P. oxalicum 114-2 did not significantly recover. However, the increase in cAMP content resulted in a joint decrease in the expression of the brlA gene in WT and knockout strains (Supplementary Figure S4). This result indicates that the regulation of BrlA by PoPlp1 is not caused by changes in intracellular cAMP content, but the level of intracellular cAMP can affect the expression of brlA. Another unidentified conidiation regulator, FlbA, was downregulated in ΔPoplp1 according to the transcriptome analysis (Table 1). FlbA is a negative regulator of G protein; it interacts with Gα subunits and positively modulates conidiophore formation in Aspergillus (Yu et al., 1996; Krijgsheld et al., 2013; Xie et al., 2022). However, conidiation was significantly enhanced along with the reduced expression of FlbA in P. oxalicum ΔPoplp1, which was contrary to the results observed in Aspergillus. The function of FlbA in P. oxalicum is worth studying to further explain the regulatory mechanism of the G protein–cAMP signaling pathway in development.
Based on the phenotype of reduced hydrolysis capability for starch and cellulose in ΔPoplp1, the activities of FPA, cellobiohydrolase, endoglucanase, β-glucosidase, and amylase were all dramatically decreased (Figure 5). Most of the annotated glycoside hydrolase genes, including cbh1, eg1, and bgl1, were also downregulated in ΔPoplp1, according to the transcriptome analysis (Figures 6, 9). After deletion of Poplp1, expression of the main cellulase transcription activator clrB was significantly downregulated, whereas that of the main cellulase transcription inhibitor creA was significantly upregulated (Figure 6). This finding indicates that changes in the levels of expression of transcription factors may be the direct cause of the reduced gene expression of cellulase and amylase. The regulation of the G protein–cAMP signaling pathway to cellulase and amylase genes is carried out via control of cAMP levels (Hu et al., 2013). The results of the measurement of intracellular cAMP showed that deletion of PoPlp1 caused a decrease in intracellular cAMP levels of ΔPoplp1 (Figure 7). When cAMP was added to the culture medium, the level of expression of the main cellulase transcription activating factor ClrB was significantly restored, and those of the main cellulase genes also exhibited varying degrees of recovery (Figure 8). This finding indicates that the decrease in the levels of cellulase gene expression caused by Poplp1 deficiency can be partially restored by the exogenous addition of cAMP. This result confirmed the positive function of PoPlp1 in the G protein–cAMP signaling pathway, and indicated that PoPlp1 may affect cellulase and amylase expressions by regulating the cAMP levels.
To comprehensively explore the mechanisms underlying the involvement of PoPlp1 in the regulation of multiple biological processes, we performed a comparative transcriptomic analysis between strains P. oxalicum 114-2 and ΔPoplp1. The genes involved in the G protein–cAMP signaling pathway were analyzed. The results indicated that PoPlp1 may function in the G protein–cAMP signaling pathway by influencing the activity of G proteins, rather than their expression levels. The gene expression levels of heterotrimeric G proteins (consisting of three Gα, one Gβ, and one Gγ subunits) hardly changed (Table 1). However, the RT-qPCR results showed that the deletion of Poplp1 resulted in significant downregulation of Gβ gene expression (Supplementary Figure S3), which implies that deletion of Poplp may have caused a deficiency in G beta-folding and a decrease in G protein signaling. GPCRs are important components for signal reception and the main upstream components of the G protein signaling pathway (Syrovatkina et al., 2016). In the transcriptome data, several GPCRs (including one Class III GPCR, two Class IX GPCRs, and several PTH11-like GPCRs) exhibited significantly enhanced expression in ΔPoplp1, which enhances signal reception in cases of G protein signaling obstruction. The negative RGS FlbA binding to Gα was dramatically downregulated in Δpoplp1, which increased the G protein activity by reducing the level of expression of Gα inhibitory factors. Given that the downstream G protein signaling pathway directly acts on adenylate cyclase (Sun et al., 2022), the upstream signal of G protein was blocked, which generally leads to a chain reaction downstream. However, the transcription level of adenylate cyclase did not change significantly, whereas that of intracellular cAMP decreased significantly. Therefore, the deletion of PoPlp1 does not affect the transcription level of adenylate cyclase, but does affect the synthesis of intracellular cAMP by adenylate cyclase. Reduction of the cAMP level will affect the regulation of multiple downstream biological pathways through a change in the target protein phosphorylation level (Sun et al., 2022). Nevertheless, the expression of a low-affinity cAMP phosphodiesterase (PDE_00536) was doubled compared with that observed in WT strain 114-2, whereas a high-affinity cAMP phosphodiesterase (PDE_02983) was reduced by half in ΔPoplp1. This may be attributable to mechanisms operating to maintain the levels of secondary messengers (i.e., cAMP and cGMP) to ensure the necessary physiological regulation (Azevedo et al., 2014). The comparative transcriptome results provide a comprehensive picture of the regulatory network with which PoPlp1 is involved, which will helpful in further investigation of the functions of PoPlp1 and the G protein–cAMP signaling pathway.
In this study, a G protein-positive regulator, PoPlp1, was identified and functionally studied in P. oxalicum 114-2. PoPlp1 was found to participate in several biological processes, including mycelium development, conidiation, and the expression of cellulases and amylases. The findings of this study are very beneficial for further study of the regulatory mechanisms underlying cellulase expression and other biological processes in P. oxalicum 114-2 via the G protein–cAMP signaling pathway.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.
ZL conceived and designed the experiments. ZJ, MY, XL, QS, GX, SL, and WC performed the experiments. ZJ, MC, and ZS analyzed the data. MC and ZL drafted the manuscript. XB revised the manuscript. All authors read and approved the final manuscript.
This study was funded by the Natural Science Foundation of Shandong Province (ZR2020MC013), the Project of Qilu University of Technology (2022PYI005), and the Shandong and Chongqing Science and Technology Cooperation Project.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2023.1165701/full#supplementary-material
Amore, A., Giacobbe, S., and Faraco, V. (2013). Regulation of cellulase and hemicellulase gene expression in fungi. Curr. Genomics 14, 230–249. doi: 10.2174/1389202911314040002
Azevedo, M. F., Faucz, F. R., Bimpaki, E., Horvath, A., Levy, I., de Alexandre, R. B., et al. (2014). Clinical and molecular genetics of the phosphodiesterases (PDEs). Endocr. Rev. 35, 195–233. doi: 10.1210/er.2013-1053
Bradford, M. M. (1976). A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72, 248–254. doi: 10.1016/0003-2697(76)90527-3
Cabrera, I. E., Oza, Y., Carrillo, A. J., Collier, L. A., Wright, S. J., Li, L., et al. (2022). Regulator of G protein signaling proteins control growth, development and cellulase production in Neurospora crassa. J. Fungi 8, 1076. doi: 10.3390/jof8101076
Castellano, M. M., and Sablowski, R. (2008). Phosducin-like protein 3 is required for microtubule-dependent steps of cell division but not for meristem growth in Arabidopsis. Plant Cell 20, 969–981. doi: 10.1105/tpc.107.057737
Chen, M., Qin, Y., Cao, Q., Liu, G., Li, J., Li, Z., et al. (2013). Promotion of extracellular lignocellulolytic enzymes production by restraining the intracellular beta-glucosidase in Penicillium decumbens. Bioresour. Technol. 137, 33–40. doi: 10.1016/j.biortech.2013.03.099
Gao, X., Sinha, S., Belcastro, M., Woodard, C., Ramamurthy, V., Stoilov, P., et al. (2013). Splice isoforms of phosducin-like protein control the expression of heterotrimeric G proteins. J. Biol. Chem. 288, 25760–25768. doi: 10.1074/jbc.M113.486258
Guo, H., Xu, G., Wu, R., Li, Z., Yan, M., Jia, Z., et al. (2021). A homeodomain-containing transcriptional factor PoHtf1 regulated the development and cellulase expression in Penicillium oxalicum. Front. Microbiol. 12, 671089. doi: 10.3389/fmicb.2021.671089
Hinterdobler, W., Beier, S., Monroy, A. A., Berger, H., Dattenbock, C., and Schmoll, M. (2020). The G-protein coupled receptor GPR8 regulates secondary metabolism in Trichoderma reesei. Front. Bioeng. Biotechnol. 8, 558996. doi: 10.3389/fbioe.2020.558996
Horevaj, P., and Bluhm, B. H. (2012). BDM1, a phosducin-like gene of Fusarium graminearum, is involved in virulence during infection of wheat and maize. Mol. Plant. Pathol. 13, 431–444. doi: 10.1111/j.1364-3703.2011.00758.x
Hu, Y., Liu, G., Li, Z., Qin, Y., Qu, Y., and Song, X. (2013). G protein-cAMP signaling pathway mediated by PGA3 plays different roles in regulating the expressions of amylases and cellulases in Penicillium decumbens. Fungal. Genet. Biol. 58–59, 62–70. doi: 10.1016/j.fgb.2013.08.002
Krijgsheld, P., Nitsche, B. M., Post, H., Levin, A. M., Muller, W. H., Heck, A. J., et al. (2013). Deletion of flbA results in increased secretome complexity and reduced secretion heterogeneity in colonies of Aspergillus niger. J. Proteome Res. 12, 1808–1819. doi: 10.1021/pr301154w
Kumar, S., Stecher, G., Li, M., Knyaz, C., and Tamura, K. (2018). MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35, 1547–1549. doi: 10.1093/molbev/msy096
Lafon, A., Han, K. H., Seo, J. A., Yu, J. H., and d'Enfert, C. (2006). G-protein and cAMP-mediated signaling in Aspergilli: a genomic perspective. Fungal. Genet. Biol. 43, 490–502. doi: 10.1016/j.fgb.2006.02.001
Li, C. X., Zhao, S., Luo, X. M., and Feng, J. X. (2020). Weighted gene co-expression network analysis identifies critical genes for the production of cellulase and xylanase in Penicillium oxalicum. Front. Microbiol. 11, 520. doi: 10.3389/fmicb.2020.00520
Li, Y., Hu, Y., Zhao, K., Pan, Y., Qu, Y., Zhao, J., et al. (2019). The indispensable role of histone methyltransferase PoDot1 in extracellular glycoside hydrolase biosynthesis of Penicillium oxalicum. Front. Microbiol. 10, 2566. doi: 10.3389/fmicb.2019.02566
Li, Z., Yao, G., Wu, R., Gao, L., Kan, Q., Liu, M., et al. (2015). Synergistic and dose-controlled regulation of cellulase gene expression in Penicillium oxalicum. PLoS Genet. 11, e1005509. doi: 10.1371/journal.pgen.1005509
Li, Z. H., Du, C. M., Zhong, Y. H., and Wang, T. H. (2010). Development of a highly efficient gene targeting system allowing rapid genetic manipulations in Penicillium decumbens. Appl. Microbiol. Biotechnol. 87, 1065–1076. doi: 10.1007/s00253-010-2566-7
Lukov, G. L., Hu, T., McLaughlin, J. N., Hamm, H. E., and Willardson, B. M. (2005). Phosducin-like protein acts as a molecular chaperone for G protein betagamma dimer assembly. EMBO J. 24, 1965–1975. doi: 10.1038/sj.emboj.7600673
Qin, Y., Bao, L., Gao, M., Chen, M., Lei, Y., Liu, G., et al. (2013). Penicillium decumbens BrlA extensively regulates secondary metabolism and functionally associates with the expression of cellulase genes. Appl. Microbiol. Biotechnol. 97, 10453–10467. doi: 10.1007/s00253-013-5273-3
Salamon, J. A., Acuna, R., and Dawe, A. L. (2010). Phosphorylation of phosducin-like protein BDM-1 by protein kinase 2 (CK2) is required for virulence and G beta subunit stability in the fungal plant pathogen Cryphonectria parasitica. Mol. Microbiol. 76, 848–860. doi: 10.1111/j.1365-2958.2010.07053.x
Schmoll, M., and Hinterdobler, W. (2022). Tools for adapting to a complex habitat: G-protein coupled receptors in Trichoderma. Prog. Mol. Biol. Transl. Sci. 193, 65–97. doi: 10.1016/bs.pmbts.2022.06.003
Seo, J. A., and Yu, J. H. (2006). The phosducin-like protein PhnA is required for Gbetagamma-mediated signaling for vegetative growth, developmental control, and toxin biosynthesis in Aspergillus nidulans. Eukaryot. Cell. 5, 400–410. doi: 10.1128/EC.5.2.400-410.2006
Sun, Z. B., Yu, S. F., Wang, C. L., and Wang, L. (2022). cAMP Signalling Pathway in Biocontrol Fungi. Curr. Issues Mol. Biol. 44, 2622–2634. doi: 10.3390/cimb44060179
Syrovatkina, V., Alegre, K. O., Dey, R., and Huang, X. Y. (2016). Regulation, signaling, and physiological functions of G-proteins. J. Mol. Biol. 428, 3850–3868. doi: 10.1016/j.jmb.2016.08.002
Tisch, D., Kubicek, C. P., and Schmoll, M. (2011). The phosducin-like protein PhLP1 impacts regulation of glycoside hydrolases and light response in Trichoderma reesei. BMC Genomics 12, 613. doi: 10.1186/1471-2164-12-613
Willardson, B. M., and Howlett, A. C. (2007). Function of phosducin-like proteins in G protein signaling and chaperone-assisted protein folding. Cell. Signal. 19, 2417–2427. doi: 10.1016/j.cellsig.2007.06.013
Wood, T. M., and Bhat, K. M. (1988). Methods for measuring cellulase activities. Methods Enzymol. 160, 87–112. doi: 10.1016/0076-6879(88)60109-1
Xie, R., Yang, K., Tumukunde, E., Guo, Z., Zhang, B., Liu, Y., et al. (2022). Regulator of G protein signaling contributes to the development and aflatoxin biosynthesis in Aspergillus flavus through the regulation of Gα activity. Appl. Environ. Microbiol. 88, e0024422. doi: 10.1128/aem.00244-22
Xu, G., Guo, H., Yan, M., Jia, Z., Li, Z., Chen, M., et al. (2022). An actin-like protein PoARP9 involves in the regulation of development and cellulase and amylase expression in Penicillium oxalicum. J. Appl. Microbiol. 132, 2894–2905. doi: 10.1111/jam.15466
Yao, G., Li, Z., Wu, R., Qin, Y., Liu, G., and Qu, Y. (2016). Penicillium oxalicum PoFlbC regulates fungal asexual development and is important for cellulase gene expression. Fungal. Genet. Biol. 86, 91–102. doi: 10.1016/j.fgb.2015.12.012
Yu, G., Wang, L. G., Han, Y., and He, Q. Y. (2012). ClusterProfiler: an R package for comparing biological themes among gene clusters. OMICS.16, 284–287. doi: 10.1089/omi.2011.0118
Yu, J. H. (2006). Heterotrimeric G protein signaling and RGSs in Aspergillus nidulans. J. Microbiol. 44, 145–154.
Yu, J. H., Hamari, Z., Han, K. H., Seo, J. A., Reyes-Dominguez, Y., and Scazzocchio, C. (2004). Double-joint PCR: a PCR-based molecular tool for gene manipulations in filamentous fungi. Fungal. Genet. Biol. 41, 973–981. doi: 10.1016/j.fgb.2004.08.001
Keywords: Penicillium oxalicum, cellulase, development, G protein-cAMP signaling pathway, phosducin-like protein
Citation: Jia Z, Yan M, Li X, Sun Q, Xu G, Li S, Chen W, Shi Z, Li Z, Chen M and Bao X (2023) Phosducin-like protein PoPlp1 impacts cellulase and amylase expression and development in Penicillium oxalicum via the G protein–cAMP signaling pathway. Front. Microbiol. 14:1165701. doi: 10.3389/fmicb.2023.1165701
Received: 14 February 2023; Accepted: 15 May 2023;
Published: 09 June 2023.
Edited by:
Hector Riveros-Rosas, National Autonomous University of Mexico, MexicoReviewed by:
Barry Willardson, Brigham Young University, United StatesCopyright © 2023 Jia, Yan, Li, Sun, Xu, Li, Chen, Shi, Li, Chen and Bao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhonghai Li, bHpobHpoQHZpcC4xMjYuY29t; Mei Chen, Y2hlbm1laV8xNkAxNjMuY29t
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.