
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Microbiol. , 17 March 2023
Sec. Infectious Agents and Disease
Volume 14 - 2023 | https://doi.org/10.3389/fmicb.2023.1107496
This article is part of the Research Topic Reviews in Neglected Tropical Infectious diseases View all 5 articles
 Carlos Gaspar-Castillo1
Carlos Gaspar-Castillo1
 Mario H. Rodríguez1
Mario H. Rodríguez1
 Vianney Ortiz-Navarrete2
Vianney Ortiz-Navarrete2
 Celia M. Alpuche-Aranda1
*
Celia M. Alpuche-Aranda1
*
 Jesus Martinez-Barnetche1
*
Jesus Martinez-Barnetche1
*
Dengue and Zika are arthropod-borne viral diseases present in more than 100 countries around the world. In the past decade, Zika emerged causing widespread outbreaks in new regions, where dengue has been endemic-epidemic for a long period. The wide and extensive dissemination of the mosquito vectors, Aedes aegypti, and Ae. albopictus, favor the co-existence of both infections in the same regions. Together with an important proportion of asymptomatic infections, similar clinical manifestations, and a short time window for acute infection confirmatory tests, it is difficult to differentially estimate both dengue and Zika incidence and prevalence. DENV and ZIKV flavivirus share high structural similarity, inducing a cross-reactive immune response that leads to false positives in serological tests particularly in secondary infections. This results in overestimation of recent Zika outbreaks seroprevalence in dengue endemic regions. In this review, we address the biological basis underlying DENV and ZIKV structural homology; the structural and cellular basis of immunological cross reactivity; and the resulting difficulties in measuring dengue and Zika seroprevalence. Finally, we offer a perspective about the need for more research to improve serological tests performance.
Dengue is the most common arthropod-borne viral disease transmitted by Aedes aegypti and A. albopictus mosquito vectors. Dengue serocomplex belongs to the genus Flavivirus (Flaviviridae family) and is composed of four serotypes DENV 1–4 (DENVs). The endemic circulation of different serotypes, alternating with epidemic cycles, lead to a high disease burden in tropical and subtropical countries. An estimated 390 million annual dengue infections occur globally, of which more than 70% are reported in South-East Asia, 16% in Africa and 14% in Americas (Bhatt et al., 2013). Although 75% of cases are asymptomatic (Bhatt et al., 2013), approximately 2 million progress to severe dengue (Gubler, 2012) that causes life threatening plasma leakage and hemorrhagic complications (Guzman et al., 2016).
Zika virus (ZIKV) is closely related to DENVs (Faye et al., 2014). ZIKV caused extensive outbreaks during the past decade. ZIKV shares the same mosquito vector species as DENVs, so that dengue endemic areas were also susceptible to Zika outbreaks (Duffy et al., 2009; Sanchez, 2016; Aubry et al., 2017). A large outbreak occurred in Yap Islands, Micronesia in 2007, reaching an attack rate of 66% (Aubry et al., 2017). Then, ZIKV spread across the Pacific in 2013–2017, affecting French Polynesia and most of Latin America (Sanchez, 2016; Aubry et al., 2017). Like dengue, up to 80% of Zika cases are asymptomatic, however, its association with rare but important clinical outcomes, including congenital microcephaly, miscarriages and Guillain-Barré Syndrome in adults (Brasil et al., 2016; Cao-Lormeau et al., 2016; Aubry et al., 2017), increases the concern about its transition to an endemic stage.
Most developing countries monitor infectious disease incidence by symptomatic-based surveillance to identify the population at risk, with the intention to focus efforts on control (Murray and Cohen, 2017). In general, this strategy allows the identification of populations more affected by the disease and could guide control efforts (De Vries et al., 2021). However, flaviviral infection incidence is considerably underreported due to the high proportion of asymptomatic and mild disease cases, and similar clinical manifestations confound the etiologic diagnosis (Ioos et al., 2014). Pathogen detection by molecular testing (qPCR) or antigen detection (i.e., NS1) have a good performance only during a short-time window (Hunsperger et al., 2016; Judice et al., 2018) and the increasing number of private low-cost healthcare facilities with a questionable level of participation in the disease surveillance system (Berntsen and Syverud, 2009; Ahmadi et al., 2013). Thus, underreporting represents a missed opportunity to stablish a reliable denominator to monitor epidemic dynamics.
Serological surveys provide better estimations of the exposure to pathogens. For many infectious diseases, pathogen-specific IgG population-based seroprevalence estimation is a good epidemiological tool that provides a denominator to calculate pathogen exposure as indirect measure of incidence, independently of clinical manifestations and access to the health system (Cutts and Hanson, 2016). The capability of IgG to persist for a long time in bloodstream (Guzmán and Kourí, 2004), makes it a reliable biomarker that provides useful indicators for retrospective monitoring of the epidemic magnitude. Moreover, seroprevalence estimation, particularity in the context of dengue, could contribute to guide prioritization of vaccination based on previous exposure (WHO, Immunization, Vaccines, and Biologicals, 2017; Aguiar and Stollenwerk, 2018; WHO, 2019).
However, DENVs and ZIKV share a high degree of protein structural similarity, including the NS1 and E proteins, which are commonly used as antigens in indirect serological tests aimed to measure virus-specific IgG. Moreover, the induced B cell repertoire contains highly cross-reactive antibodies (crAbs), which does not correlate with their neutralizing capacity and breadth, affecting the diagnostic performance of serological tests and the consequent inaccurate seroprevalence estimation. Most of the commercial tests designed after the Zika outbreaks in the Americas were validated in volunteers or travelers who lived in non-endemic areas. However, they have poor performance for dengue and Zika detection in dengue endemic areas, although in dengue non-endemic regions the specificity is above 90% (Table 1).
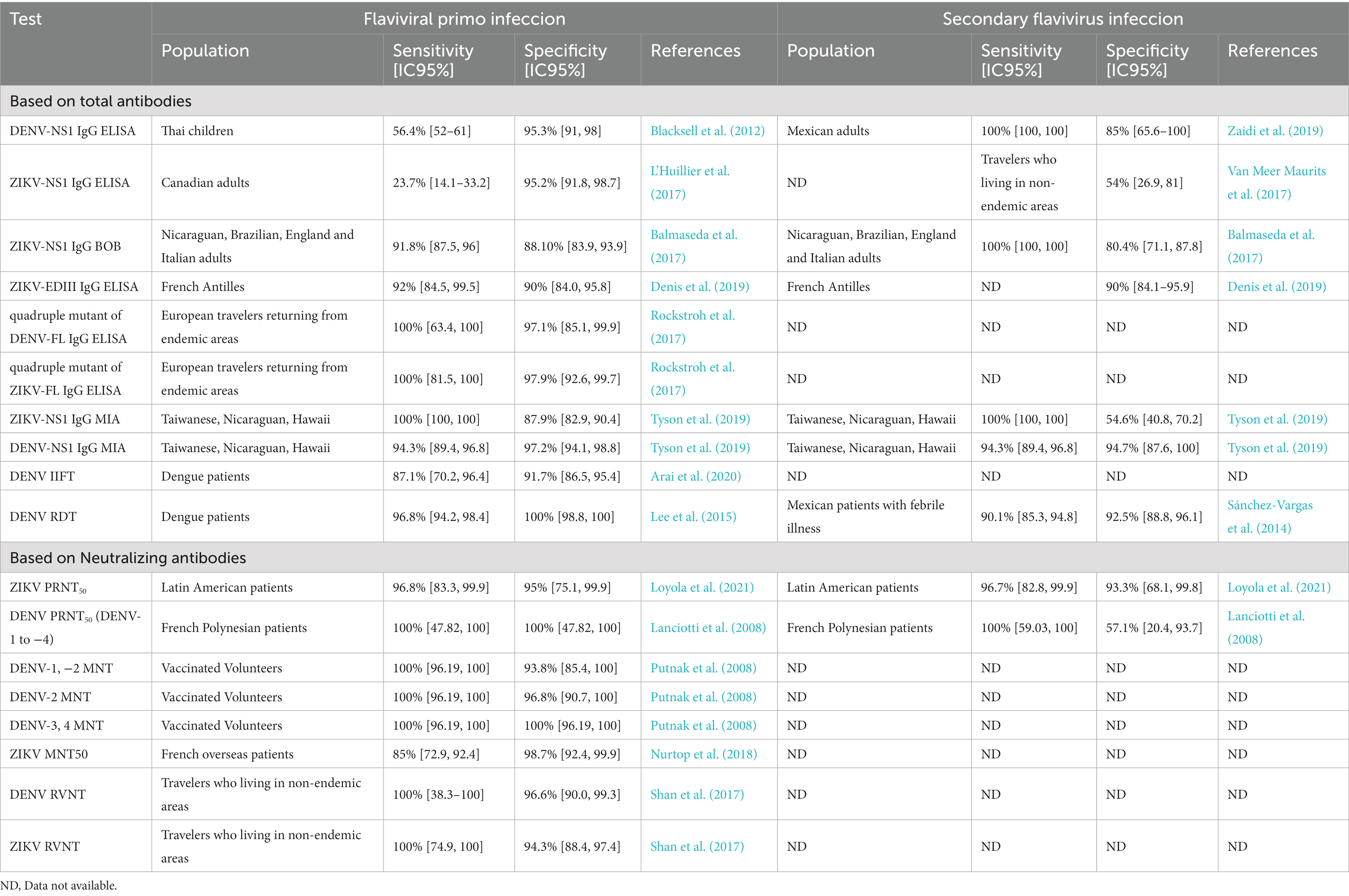
Table 1. Diagnostic sensibility and specificity of dengue and Zika serological tests obtained in endemic and non-endemic regions.
This integrative review aims to provide a broad overview of the structural and immunological basis of the cross-reactive antibody response to dengue and Zika infections, and their implications in epidemiological serosurveillance, as well as insights into the design of novel, more specific serological tests.
Evidence points to DENVs originating from sylvatic strains infecting non-human primates and many forest-dwelling Aedes vectors different from Ae. aegypti and Ae. albopictus (Rudnick, 1965; Robin et al., 1980). Phylogenetic analyses of E protein sequences of historical sylvatic and endemic isolates of DENV suggests that the ancestor of all DENV could have originated in West Africa (Rudnick, 1965; Robin et al., 1980), approximately 1,000 years ago (Rambaut, 2000; Twiddy et al., 2003). Endemic DENV-2 genotypes diverged from West African sylvatic strains about 400–600 years ago, and then sylvatic and epidemic DENV-1 and -4 strains diverged around 100–200 years ago (Twiddy et al., 2003; Weaver and Vasilakis, 2009). Slave trade from Africa to other continents and more recently, human mobility, urbanization and trade routes have contributed to global DENV spread and diversification (Smith, 1956).
ZIKV was first isolated in Uganda in 1947 from a rhesus monkey and its origin is still unclear, mainly due to limited sampling. Like DENV, ZIKV could have originated in a widely enzootic cycle among multiple animal species in Africa (Gutiérrez-Bugallo et al., 2019). Phylogenetic analyses of E and NS5 sequences showed that the common ancestor of ZIKV diverged from Spondweni virus before 1800s (Pettersson et al., 2016), and the common ancestor of all ZIKV lineages could have originated at the beginning of 1900s, forming the Ugandan lineage. This lineage probably spread in West African countries around 1935 (West African lineage), and in South East Asia around 1945. ZIKV reached Micronesia around 1960 (Asian lineage) (Faye et al., 2014). The pathway of ZIKV dissemination from Africa to other continents is unclear. The recent emergence of Zika in Brazil was associated with the massive movement and gathering of people motivated by the FIFA Soccer World Cup or Canoe Sprint World Championships in 2014 (Faria et al., 2016), supporting human movement and trade routes as major determinants for the rapid spread of Zika.
There is little evidence to estimate a precise divergence time between DENVs and ZIKV, but it is plausible that divergence occurred before 1,000 years ago, when the common ancestor of DENVs originated (Figure 1). It is also proposed that ZIKV could have diverged from DENVs, based on the close relationship between ZIKV and DENV-4 (Ratfi and Nizam, 2016), implicating that ZIKV origin is more recent. Nevertheless, both theories point to a relatively short divergence time, which explains the high structural similarity between both viruses (Barba-Spaeth et al., 2016).
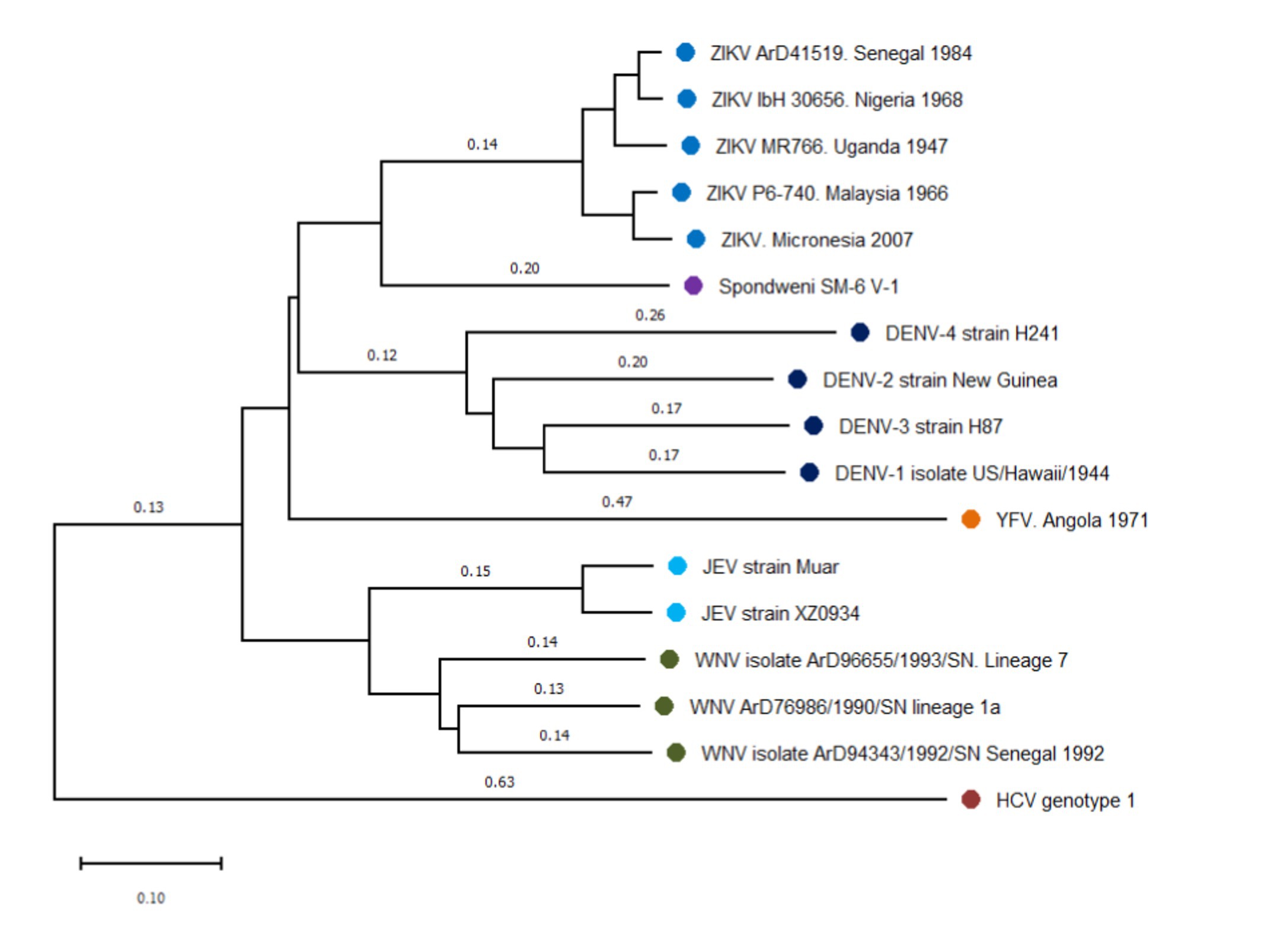
Figure 1. Phylogenetic N-J tree of flaviviruses related to dengue and Zika virus. The phylogeny was represented using reference sequences of different flaviviruses, showing the close relationship among dengue (DENV) and Zika virus (ZIKV). Four dengue serotypes form the dengue serocomplex, that is presented in this tree by DENV-1 (EU848545), DENV-2 (AF038403), DENV-3 (M93130), and DENV-4 (AY947539). ZIKV and Spondwenii virus constitute a Spondwenii serocomplex, that is represented by the first ZIKV isolated from rhesus monkey (NC_012532), two strains of ZIKV West African lineage: ArD41519 (HQ234501) and ibH30656 (HQ234500) and two of Asian lineage P6-740 (HQ234499) and isolated strains from Micronesia 2007 (U545988). Spondwenii virus was represented for SM-6 V-1 strain (DQ859064). Japanese Encephalitis serocomplex was represented for Japanese Encephalitis virus (JEV) Muar (HM596272) and XZ0934 strains (JF915894), and West Nile Virus (WNV) lineage 7 (KY703855), lineage 1a (KY703854), and strains from Senegal, 1992 (KY703856). Yellow Fever virus (YFV) forms a distinct group, represented by AY968064. Hepatitis C virus (HCV) strain (NC_004102) was used to outroot the tree. Distance represents base substitutions per site.
Flaviviruses are enveloped single-stranded positive-sense RNA viruses that share the same basic genomic organization and molecular structure (Lindenbach et al., 2011). RNA genome of flaviviruses encode a single polyprotein, which is cleaved into seven non-structural (NS) proteins (NS1, NS2A, NS2B, NS3, NS4A, NS4B, and NS5) implicated in controlling critical steps of the viral life cycle; and three structural proteins (the capsid [C], precursor membrane [prM] and envelope protein [E]) constituents of the viral particle (Figures 2A,B; Lindenbach et al., 2011). This review is focused on NS1 and E, both highly immunodominant antigens used as targets for antibody detection in the majority of serological tests.
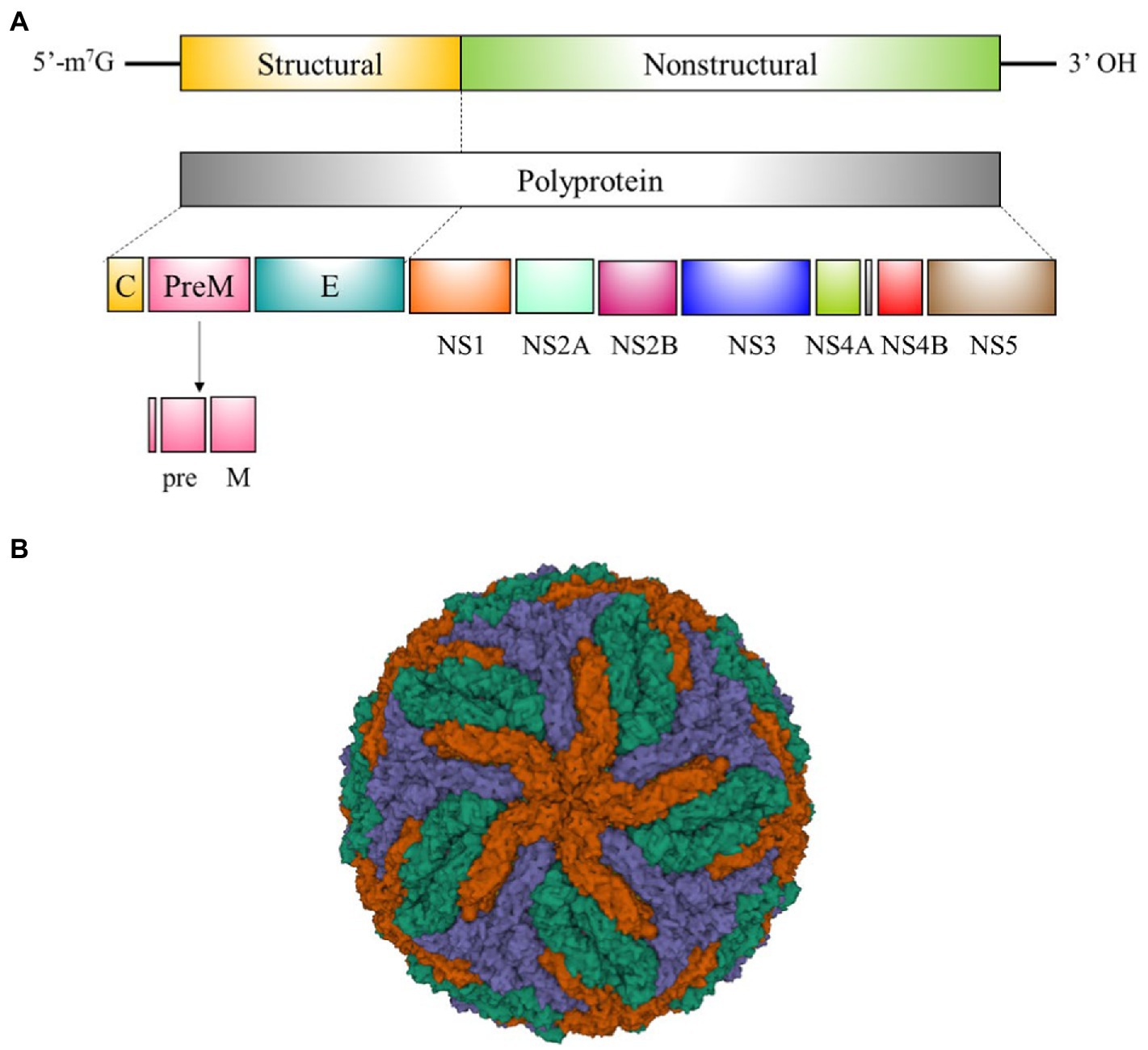
Figure 2. Flavivirus genome organization and structure. (A) Schematic representation of flavivirus genome organization and processing of the polypeptide into mature viral proteins. (B) Cryogenic electron microscopy structure of the dengue virus (DENV). E:M heterodimers of the same color are equivalent by icosahedral symmetry. Heterodimers of different colors are quasi-equivalent, with red E:M dimers falling on the icosahedral 5-fold axes, blue on the 3-fold, and green on the 2-fold (PDB 3 J27) (Zhang et al., 2013).
NS1 is highly conserved among flaviviruses. It is constituted by around 352 amino acids and has a molecular weight of between 46 and 55 kDa. The NS1 of most flaviviruses has N-linked glycosylation sites at asparagine 130 and 207 and cysteine residues which form disulfide bonds at the carboxy terminal region. The NS1 contains three distinct domains: A N-terminal β-roll, a wing domain, and a C-terminal β-ladder, that have approximately a amino acid sequence of 30, 150, and 170 residues, respectively (Figure 3A; Akey et al., 2014).
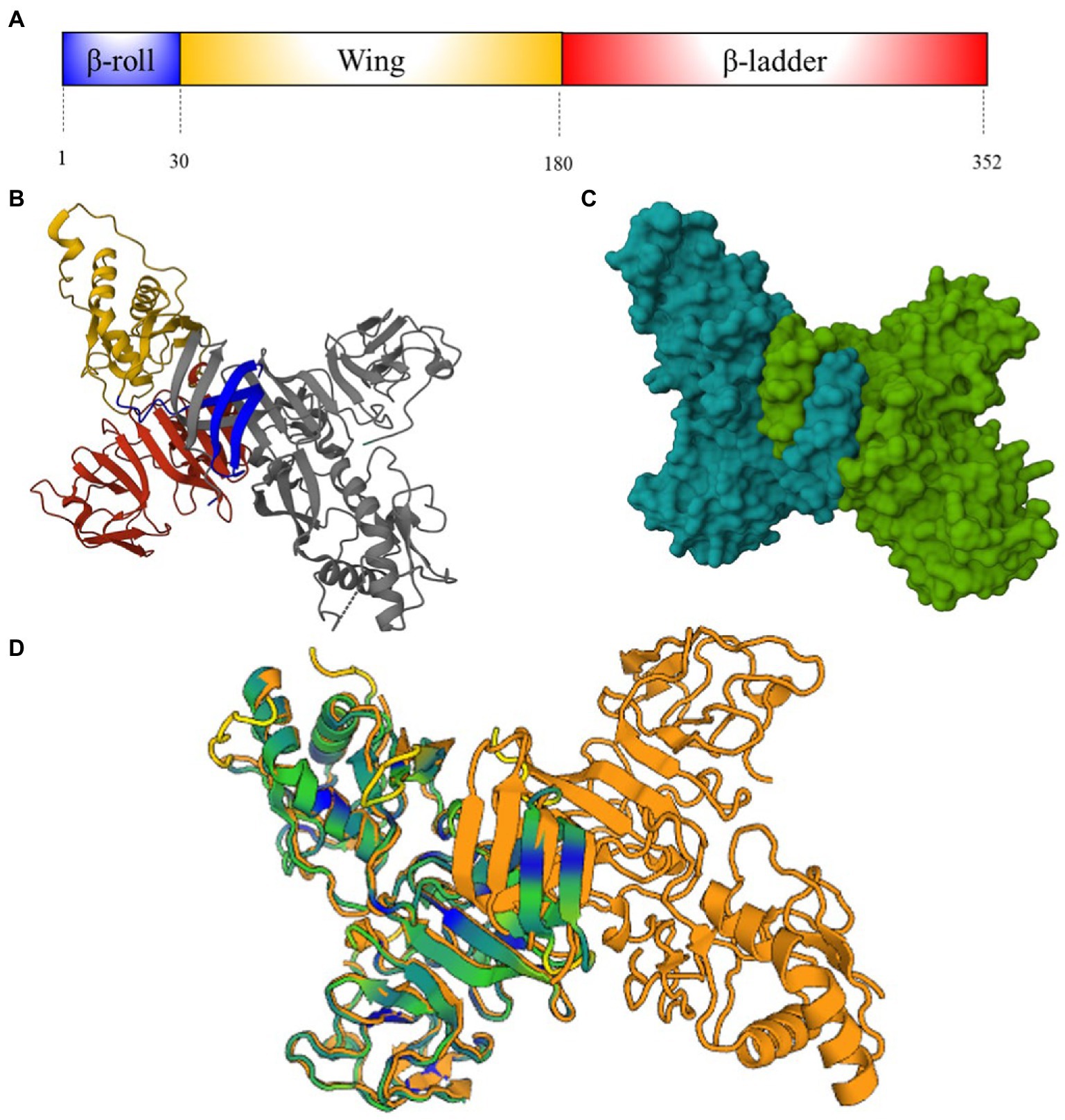
Figure 3. NS1 protein structure. (A) Schematic representation of ZIKV NS1-protein showing β-roll (blue), wing (yellow) and β-ladder domain (red). Numbers below represent the residue positions (B) Ribbon molecular structure of ZIKV NS1 cross-shaped dimer (PDB: 5K6K) (Brown et al., 2016). Domains on one monomer are represented as (A), another domain is showed in gray. (C) Molecular structure of ZIKV NS1 cross-shaped dimer (PDB 5K6K). (D) DENV (4O6B) and ZIKV (5K6K) NS1 superposition [Dali server: (Holm, 2022)]. ZIKV-NS1 sequence A (green) is super posited on DENV-2-NS1 sequence A and B (orange) (Akey and Smith, 2014). Cyan and blue regions indicate conserved motifs and conserved amino acid sequences.
NS1 is synthesized in the endoplasmic reticulum (ER) as a monomer is anchored in the plasma membrane as a dimer. The C-terminal disulfide bonds of this protein are crucial for dimerization. NS1 dimers are cross-shaped attached by the intertwined β-roll domains, the β-ladders interact with each other, and the wing domains are end to end (Figures 3B,C). The quaternary structure of NS1 on the cellular membrane has two faces: The hydrophilic “outer” face that contains glycosylation sites and the hydrophobic “inner” face that belongs to the β-roll domain and an adjacent “greasy finger” loop. The NS1 is secreted as hexamer, in a barrel-like structure, attached by the hydrophobic inner face (Youn et al., 2012).
In the particular case of DENVs and ZIKV, alignment of their NS1 protein sequences reveals 51–53% similarity (Wen and Shresta, 2019). Crystal structure of ZIKV-NS1 showed differences on the wing domain flexible loop (residues 108–129) that contain three highly conserved tryptophan residues (residues 123–124) on the inner hydrophobic face. Together with other tryptophan residues on the wing domain and greasy fingers loop form a hydrophobic protrusion, which is an additional site for cellular membrane interaction (Brown et al., 2016). Negative charged regions displayed on the wing and central β-ladder domains of the outer hydrophilic face of ZIKV-NS1 are absent in DENV-2 and West Nile Virus (WNV) NS1. These differences do not modify the protein folding and the hexameric configuration of NS1 among DENVs and ZIKV (Figure 3D), although they could represent differential sites for antibody recognition and vaccines designing (Akey et al., 2014).
The E protein is highly conserved among flaviviruses. The monomer consists of approximately 500 residues and has a molecular weight of 50 kDa (Dai et al., 2016). DENV and ZIKV E protein contains one or two glycosylated asparagine residues (Asn 67 and/or Asn153-154) and α-helices at C- terminus (Dai et al., 2016). For both viruses, E is formed by three different domains (EDI-EDIII). The EDI is a central ß-barrel domain with 130 residues in three segments (residues 1–51, 132–192, and 280–295) and contains a glycan loop (GL, residues 147–161). The EDII has a finger-like conformation formed by two segments (residues 52–131 and 193–279), that include a highly hydrophobic fusion loop (FL, residues 98–109); and EDIII has a immunoglobulin-like fold (residues 296–403; Figure 4A; Dai et al., 2016).
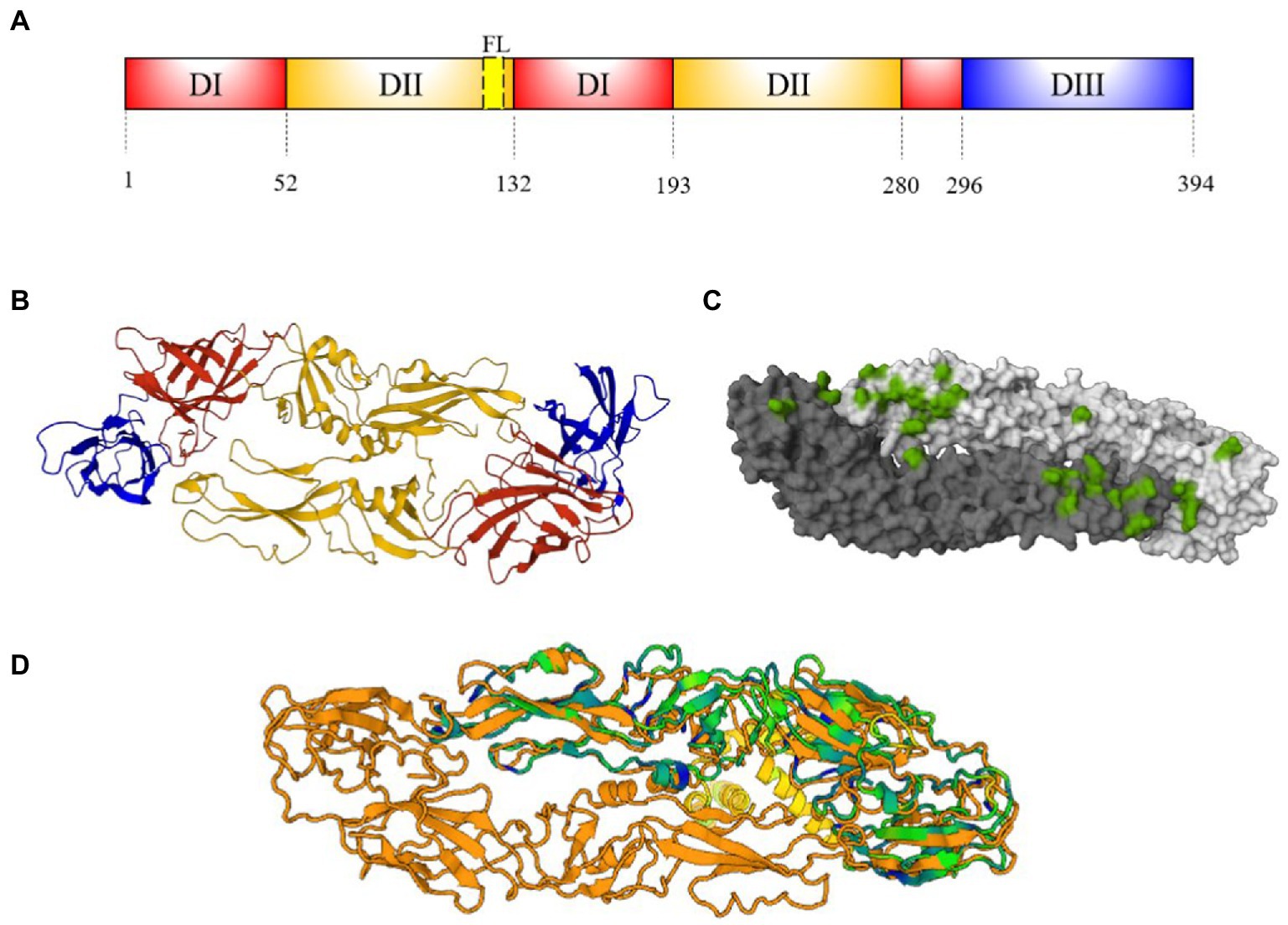
Figure 4. E protein structure. (A) Schematic representation of E-protein ectodomain showing EDI (red), EDII (orange), FL (yellow) and EDIII (blue). Numbers below represent the residue positions. (B) Ribbon structure of DENV E antiparallel dimer (PDB: 1UZG) (Modis et al., 2005). Domains on dimer are represented as (A,C) Footprint of EDE2 Fab C8 mAb on a DENV-2 antiparallel dimer (4UTC) representation shown in green (Barba-Spaeth et al., 2016). (D) DENV (1UZG) and ZIKV (5IRE) E dimer superposition [Dali server: (Holm, 2022)]. ZIKV-NS1 sequence A (green) (Sirohi et al., 2016) is super posited on DENV-3-E sequence A and B (orange). Cyan and blue regions indicate conserved motifs and conserved amino acid sequences, yellow motifs indicate DENV-E intracellular regions.
The E proteins are assembled in 60 trimeric protrusions anchored to preM in ER, forming immature virions with spiky surfaces (Lindenbach et al., 2011). During the virus maturation, low pH in trans-Golgi networks induces a reorganization of E-preM heterodimers, producing 90 antiparallel E homodimers (Figure 2B), connected through EDII and EIII (Figures 4B,C). This organization produces the exposure and cleavage of pre-M and herringbone pattern formed by E homodimers in the mature virion (Lindenbach et al., 2011).
The EDI, EDII, and EDIII of ZIKV and DENV share 35, 51, and 29% amino acid similarity, respectively (Dai et al., 2016). Structural studies on flavivirus E protein reported a N67 glycosylation motif on EDI present only in DENV. The N164 glycosylation motif is only present at ZIKV-GL. The latter is larger than DENV, due to the presence of four additional amino acids (Sirohi et al., 2016). The highly similarity into EDII is brought by FL, which is highly conserved among flaviviruses (Stettler et al., 2016). On the other hand, EDIII of ZIKV and DENVs differs in 52 residues, including three additional amino acids. It also includes surface-exposed, internally facing and cryptic residues not displayed in DENVs (Conlan et al., 2019). The E protein quaternary structure is identical in both mature virion species (Figure 4D; Sirohi et al., 2016). Thus, the reduced similarity between EDIII of DENV and ZIKV make it an attractive antigen candidate to improve specificity of serological assays aimed at discriminating seroprevalence between the two infections.
Antibodies are soluble forms of the membrane anchored B cell antigen receptor (BCR). Each naïve B cell bears a unique BCR and ligand (antigen) - binding capacity, which can be clonally selected by the cognate antigen, resulting in antibody secretion by alternative splicing of the BCR transcript. Antigen specificity and cross-reactivity depend on antigen binding site (paratope) structure. The paratope is composed by three hypervariable loops called Complementarity Determining Regions (CDR-H1, -H2, and -H3) of the Heavy and 3 CDR’s of the Light chain, at the Variable region of both chains. CDR1 and 2 have limited variability because they are encoded in germline V segments, while the CDR3 structure is highly variable because it is determined by V(D)J recombination (Davis and Bjorkman, 1988; Schroeder and Cavacini, 2010).
All known CDR1, 2 and CDR-L3 adopt a limited number of standard canonical structures (Chothia and Lesk, 1987; Lara-Ochoa et al., 1996). Only the CDR-H3 loop, is highly diverse and does not adopt a canonical structure. Current evidence suggests that all CDR’s, except CDR-H3, are rather cross-reactive structures that contribute to stabilize the antigen-paratope interaction, and the major determinant for the affinity is the CDR-H3 (Davis, 2004). Under this model, an epitope will be cross-reactive if critical residues that bind to the CDR-H3 are conserved in a variant epitope, despite little or non-conservation of contact residues of the remaining CDR’s. Conversely, variant-specific epitope recognition will occur if the variant epitope has mutated in critical residues that bind to the CDR-H3, despite conservation of the remaining CDR contacts.
A common denominator in some viral infections such as HIV, influenza and flaviviruses is the rapid antigenic variation of the causative agent and the consequent escape from neutralizing antibodies. Yet, in such cases, a small fraction of the responding B cells have strong biases in the use of particular VH segments (Gorny et al., 2009; Wrammert et al., 2011; Wu et al., 2011; Lingwood et al., 2012; Cortina-Ceballos et al., 2015; Dunand and Wilson, 2015; Godoy-Lozano et al., 2016; Nielsen et al., 2020; Robbiani et al., 2020; Voss et al., 2021). These “stereotyped” responses are produced in genetically unrelated individuals and are directed toward highly conserved viral epitopes (structural convergence), hence these are cross-reactive and in some cases, broadly protective. This type of epitope recognition differs with the model proposed by Davis, because the contribution of the germline encoded CDR1 and 2 to overall affinity is higher, explaining the biased V-gene segment use. Thus, germline conservation of certain CDR structures may be an additional source of broad, “innate” cross-reactivity against several viral capsid epitopes.
Somatic hypermutation (SHM) is an antigen and T-cell dependent (TD) process that promotes genetic diversification of the V-region followed by affinity-dependent selection of B cells. SHM tends to prevent cross-reactivity by fine-tuning antigen-paratope interaction (Davis, 2004). Affinity matured antibody responses toward TD antigens have important implications for population immunity because they are associated with germinal center (GC) response as the major site of antibody affinity maturation, class switch and the generation of long lived plasma cells (LLPC) and memory B (mB) cells (Goodnow et al., 2010).
However, repeated antigen stimulation and the consequent extensive SHM, in the GC pathway may also contribute to cross-reactivity and polyreactivity as has been demonstrated in HIV (Gorny et al., 2009; Wu et al., 2011) and possibly is implicated in dengue progressively cross-reactive and cross-protective response after repeated infections with different serotypes (Zompi et al., 2012; Priyamvada et al., 2016; Patel et al., 2017; Durham et al., 2019).
Geometrically arrayed protein multimers included in viral capsids (Figure 2B), are capable of strong BCR crosslinking in a T-independent (TI) fashion (Zinkernagel, 1996). Thus, in these cases, in the absence of SHM and GC formation, responses against TI antigens are more cross-reactive than TD responses because increased avidity of the interaction.
In recent years, it has become clear that the extrafollicular response (GC-independent) is an important source of non-classical mB cells and plasmablasts secreting cross-reactive unmutated IgG. Thus, EF contributes to cross-reactivity mainly through stereotyped use of certain V-gene segments. Although viral infections induce EF and GC responses, for some acute viral infections that progress to severe systemic inflammation such as severe dengue and severe COVID-19, suppression of GC reaction occurs (Aye et al., 2014; Kaneko et al., 2020), thereby promoting cross-reactivity.
Antibody responses to flavivirus are guided to both structural and non-structural proteins, including NS1, NS3, NS5, pre-M, C, and E proteins (Falconi-Agapito et al., 2021; Khare and Kuhn, 2022). The NS1 and E proteins are targets of neutralizing, non-neutralizing protective and enhancing antibodies, so most research related to the antibody response, including cross reactivity, is focused on them. Only NS1 and E are used as antigenic targets in the available serological tests (Balmaseda et al., 2017; L’Huillier et al., 2017; Safronetz et al., 2017; Lee et al., 2018; Denis et al., 2019; Tyson et al., 2019).
High-throughput B cell repertoire sequencing in acute and post-convalescent dengue infections show a marked stereotyped B cell response with important dominance of VH1 and 3 family segments. The precise antigen causing such stereotyped responses is still unknown (Parameswaran et al., 2013; Godoy-Lozano et al., 2016). More recently, using scRNA-seq coupled to BCR sequencing, a disproportionate use of IGHV1-69 against DENV non-FL epitopes in EF derived plasmablasts was described in primary and secondary dengue infection (Rouers et al., 2021). Similarly, mAbs against ZIKV using unmutated V-gene segments have been described (Magnani et al., 2017; Gao et al., 2019), indicating that the immune response to ZIKV can be stereotyped and may contribute to cross-reactivity.
In flaviviruses, exposed surfaces are commonly more variable and immunodominant than conserved cryptic domains, thus strong neutralizing Abs (nAbs) are usually type-specific (i.e., non-cross reactive), whereas weak nAbs recognize cryptic epitopes that are highly cross-reactive (Campos et al., 2018). A low proportion of the potent and highly specific nAbs are elicited to EDIII of DENV and ZIKV (Wahala et al., 2009; Gallichotte et al., 2015). Stereotyped responses of anti-ZIKV EDIII antibodies using IGHV3-23/IGKV1-5 are cross-neutralizing for ZIKV and DENV-1, whereas those using different IGHV segments only neutralize ZIKV (Robbiani et al., 2017).
In contrast, a high proportion of poorly neutralizing Abs are directed to the highly conserved EDI/EDII interphase and the FL (EDII), and are highly cross reactive (Priyamvada et al., 2017; Montoya et al., 2018; Premkumar et al., 2018). In fact, cross reactivity and poor neutralizing capacity of anti-FL antibodies are associated with antibody-dependent enhancement (ADE) (Rey et al., 2018), which is associated with heterologous serotype infections (Montoya et al., 2018).
The dimerization on the E monomers creates conformational epitopes along the dimer interphase called EDE-1 and EDE-2 (Figure 4C; Dejnirattisai et al., 2014; Rouvinski et al., 2015). These epitopes are highly conserved within the EDI/DIII–EDII interphase. Antibodies toward conformational epitopes are cross-neutralizing to both, DENV and ZIKV (Dejnirattisai et al., 2014; Rouvinski et al., 2015). Anti-EDE-1 antibody binding is not affected by protein E N153 glycosylation, whereas binding to EDE-2 is dependent on glycosylation. Both types of antibodies potently neutralize DENV, and anti-EDE-1 also strongly neutralizes ZIKV (Dejnirattisai et al., 2016).
Studies in influenza A virus neutralization indicate that the human B cell repertoire generates antibodies capable of broad neutralization targeting cryptic, highly conserved, conformational epitopes, suggesting that such lymphocytes have a selective disadvantage compared to type-specific B cell clones targeting type-specific epitopes (Wrammert et al., 2011; Guthmiller et al., 2020). Studies in hyperendemic dengue regions indicate that the generation of broadly neutralizing responses depend on sequential reinfection by different serotypes (Duffy et al., 2009; Guerbois et al., 2016; Aubry et al., 2017). After a primary infection, most neutralizing B cells are type-specific, and cross-neutralizing mB cells are rare. Subsequent heterologous reinfections induce the recruitment of such mB cells to GC’s, reshaping specificity by SHM. After two or more heterologous reinfections initial subdominant cross neutralizing mB cells start predominating. Thus, as in HIV infection under highly active antiretroviral therapy (HAART) (Gorny et al., 2009; Wu et al., 2011) in recurrent dengue infections GC’s are central for a slowly evolving clonal selection process that results in a broadly neutralizing response (Zompi et al., 2012; Priyamvada et al., 2016; Patel et al., 2017; Durham et al., 2019).
Recent studies have shown that there is a highly cross-reactive antibody and mB cell responses against E in convalescent ZIKV infection in people with previous DENV infection, but that cross-reactivity wanes rather quickly, after which the response becomes ZIKV-specific (Collins et al., 2017; Andrade et al., 2019). These findings suggest that despite the structural similarity, ZIKV should be considered apart from the DENV serocomplex (Collins et al., 2017).
Anti-NS1 antibodies can confer Fcγ-mediated protection and inhibit severe flavivirus infections through Fcγ-receptor mediated cytotoxicity (Schlesinger et al., 1993; Krishna et al., 2009), complement activation, and other undetermined Fc-independent mechanisms (Chung et al., 2006, 2007). However, anti-NS1 antibodies also participate in severe dengue pathogenesis by promoting bleeding diathesis through inhibition of thrombin activity and enhancement of fibrinolysis (Chuang et al., 2014, 2016).
Most antibodies directed to the wing domain of anti-NS1 protein are cross-reactive, as seen with mouse monoclonal antibodies (mAb), which cross-react with DENVs, JEV and Kunjin virus (Falconar and Young, 1991). Other crAbs directed against the NS1 β-ladder, cross-react with ZIKV and DENV-2 (Falconar and Young, 1991).
Current evidence about pan-anti-flavivirus antibodies has contributed to our knowledge about the antigen-binding sites of antibodies elicited by NS1. The 2B7 human mAb binds to overlapped epitopes at the β-ladder region on the “inner” face and at the wing domain of the protein, blocking NS1 binding to endothelial cells (Biering et al., 2021). CDR-H3 recognition of highly conserved residues, particularly R299 and T301 on NS1 represents a highly cross-reactive response among all dengue serotypes, ZIKV and WNV, despite the rest of CDRs-binding sites are non-conserved among flavivirus (Biering et al., 2021). In contrast, 22NS1, a mouse mAb against the WNV NS1-β-ladder, is non-protective against other flavivirus. The basis of such specificity is the lack of conservation of critical contact sites of the CDR-H3, despite some non-CDR-H3 contact sites being conserved among other flaviviruses (Edeling et al., 2014).
Accuracy of different diagnostic tests depend on the infection stage. During the viremia, i.e., the first 5–7 days posterior to the infection onset. Specific RNA can be detected by qPCR in blood, serum and plasma, with specificities above to 95%. NS1 can also be detected, although its high similarity makes it difficult to discern among flaviviral infections (Brown et al., 2016; Wen and Shresta, 2019). Anti-flavivirus IgM titres in acute sera are measured to serologically confirm infection, however its short half-life (<120 days posterior onset infection) limits their applicability to a biomarker of active infections and extended cross-reactivity is also present in these serological tests. In contrast, IgGs achieve measurable levels at around seven days posterior the infection onset and persist for more than 3 years in bloodstream (Nascimento et al., 2018). Thus, for clinical diagnosis, IgGs detection is useless the during acute phase, but their high concentration and long half-life make them the most commonly used antibodies in seroprevalence studies (Siegrist, 2018).
In addition, the determination of IgG as a marker of DENV pre-exposure has been considered relevant in studies of the dengue vaccine Dengvaxia® since the evidence of an excess risk of severe dengue in seronegative vaccine recipients compared to seropositive non-vaccinated individuals (Aguiar and Stollenwerk, 2018; WHO, 2019).
The implementation of tests in epidemiological surveillance requires a rigorous validation to determine intrinsic capabilities, i.e., the diagnostic sensibility (DSe) and diagnostic specificity (DSp) (Trevethan, 2017). High DSp increases the likelihood of a true-negative result (Trevethan, 2017), which is ideal when the goal is to discriminate between past infections, for example to determinate seroprevalences in scenarios of dengue endemic regions with recent Zika outbreaks.
A growing number of Enzyme-Linked Immunosorbent Assays (ELISA) based on anti-NS1 IgG detection that are available in the market (L’Huillier et al., 2017; Safronetz et al., 2017), are summarized in Table 1. However, NS1 and E preparations used in commercial ELISA, may contain highly conserved epitopes that may detect crAbs binding, affecting the analytical specificity.
A blockade-of-binding (BOB)-ELISA, which is a competitive ligand-binding assay, produces better results than conventional ELISAs. Using a mAb to ZIKV-NS1 (ZKA3), showed an overall DSp of 80.4% in a panel of secondary dengue infections and 95.9% in dengue patients and healthy volunteers (Balmaseda et al., 2017). Although the ZKA3 epitope is not fully revealed, defined as S2 (Stettler et al., 2016), it is likely that it binds to a non-conserved NS1 region among flavivirus, highlighting the applicability of type specific Abs to improve prevalence estimation. However, there is still much room for specificity improvement, which will require testing novel, more specific epitopes.
Other techniques are being tested to improve performance. The magnetic immunoassay (MIA) utilizes microspheres coated with NS1 (Beck et al., 2015). In primary infections, anti-NS1 IgG MIA showed a DSp of 86–87.9% (Wong et al., 2017) and 97.2% (Tyson et al., 2019) for ZIKV and DENVs, respectively. However, in secondary dengue infections the DSp of ZIKV-NS1 MIA dropped to 54.6%, while it performed better (>90%) for DENVs (Tyson et al., 2019). It is possible that NS1 antigen coupling facilitates the exposure of antigenic hydrophilic epitopes. The NS1 array around beads can also favor better antibody binding than that obtained in solid-phase of conventional ELISA. The analytical specificity is improved for paired antigen of DENVs and ZIKV running out in the same well, and the limit of detection is improved with more resolution using fluorescence intensity than optical densities of ELISA. High-throughput and less clinical volume samples makes MIA a better assay. Unfortunately, antigens used in MIA are not commercially available and the reading systems (Luminex) are not cost-efficient in studies of large numbers of samples required for seroprevalence estimation.
Serological tests based on E protein are less available. Although poor or null DSp is obtained using a whole DENV-E protein in MIA (Wong et al., 2017), its performance significantly improves using EDIII, suggesting that this epitope is less cross-reactive. Similarity, the DSp of anti-ZIKV EDIII IgG MIA was 90% in sera from patients with previous flavivirus infections (Rockstroh et al., 2017). Although there is no data on serological tests for DENVs, Modis et al. (2005) reported that EDIII 622–637 peptide is a DENV-specific sequence, indicating that it is a potential candidate for serological tests in formats with high optical resolution.
Other serological methods are the indirect Immunoflourescent Test (IIFT) and rapid diagnostic tests (RDT). IIFT is a microscopic method that can visualize viral proteins expressed in infected cells monolayers via antigen–antibody binding. IIFT is commercially available with specificity up to 91% (Arai et al., 2020) reported in dengue patients. However, there are not validation studies conducted in populations with other flavivirus co-circulation including ZIKV. In addition, the use of IIFT for population – scale surveys implies increasing throughput, which seems unfeasible in low and middle-income countries. RDT is a screening method that could provide results at the point-of-care. In a systematic review of 10 studies conducted in dengue endemic areas, the specificity of the dengue RDT IgG component ranges from 65 to 100% (Luo et al., 2019). However, most tests were compared to immunoassays, which present a significantly high false-positive rate, preventing a correct validation (Luo et al., 2019). More robust validation studies involving well-characterized samples from vaccinated volunteers and patients with a well-documented history of flavivirus infections are needed.
The plaque reduction neutralization test (PRNT) is currently the gold standard test to determine previous flavivirus infections. PRNT and its derivatives measure nAbs that block infections of susceptible cell monolayers (Russell and Nisalak, 1967; Russell et al., 1967). However, the presence of potent cross-neutralizing antibodies in recurrent infections (Zompi et al., 2012; Priyamvada et al., 2016; Patel et al., 2017; Durham et al., 2019) affects the specificity of these tests. PRNT shows a DSp of 100% in both, dengue and Zika naïve serum samples, but a total cross-neutralization among secondary dengue infection. In the case of ZIKV PRNT50, the DSp is 25% with previous dengue infections (Duffy et al., 2009; Swanstrom et al., 2016; Lee et al., 2018). Although higher cut-off points have been used (e.g., PRNT60 or PRNT90) (Rainwater-Lovett et al., 2012), to reduce cross-reactivity (Roehrig et al., 2008), it has not been evaluated in endemic areas with more than one flavivirus co-circulation. In addition, these tests usually have a laborious and time consuming methodology, expensive facilities and laboratory supplies, and specialized training, limiting its use in national surveillance of dengue and Zika in endemic countries (Thomas et al., 2009; Lee et al., 2018).
Peptide microarrays represent a powerful tool for mapping type-specific Abs against antigens other than NS1 and E proteins. Using a panel of sera from patients with a well-documented history of different flavivirus infections, immobilized overlapping linear peptides can identify potentially relevant epitopes for surveillance (Viedma et al., 2020). A single 20-residue peptide from ZIKV NS2B was identified in a high-density microarray, that in an ELISA test showed a 95.9% specificity and 96% sensitivity in dengue immune patients (Mishra et al., 2018). More studies are needed to determinate its feasibility and performance for diagnostic and surveillance purposes.
The short divergence time between DENVs and ZIKV is a major determinant of their structural similarity, explaining a high proportion of cross-reactive over type-specific antibodies, leading to the consideration by some that ZIKV is a “fifth DENV serotype” (Harrison, 2016). Despite the enormous B cell repertoire diversity, some structural constraints imposed by germline IGHV segments may contribute to the induction of cross-reactive and polyreactive antibodies, independently of SHM. In addition, the co-circulation of multiple serotypes and viral species in endemic regions cause sequential heterologous infections, which result in the generation of highly cross reactive responses by clonal selection and extensive SHM in GC’s.
There is a considerable knowledge of structural and immunological E and NS1 protein epitopes, because they represent targets for protective (neutralizing) or disease enhancing antibody responses. However, further research is needed to define novel non-conserved linear epitopes in E (i.e., E DIII) and NS1 proteins, as well as other flaviviral antigens that may be irrelevant in terms of protection or pathology, but sufficiently divergent to increase the specificity of serological tests, regardless of the technological platform. Another challenge is the availability of population cohorts in endemic areas with adequate documentation of previous flaviviral exposure, which is required for extensive validation of test performance and eventual use for seroprevalence estimation. Although cross-reactivity impedes accurate dengue and Zika prevalence estimation, it does not modify decision making in terms of public health intervention, because this is focused on vector control, which in this case is the same for both diseases. However, accurate seroprevalence estimation will be required if widespread vaccination against dengue and Zika becomes a goal.
CG-C performed bibliographic research, drafted the manuscript, made figures, and table. JM-B and CA-A performed bibliographic research and drafted the manuscript. VO-N and MHR drafted the manuscript and provided significant insight. All authors contributed to the article and approved the submitted version.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Aguiar, M., and Stollenwerk, N. (2018). Dengvaxia: age as surrogate for Serostatus. Lancet Infect. Dis. 18:245. doi: 10.1016/S1473-3099(17)30752-1
Ahmadi, A., Nedjat, S., Gholami, J., and Majdzadeh, R. (2013). Disease surveillance and private sector in the metropolitans: a troublesome collaboration. Int. J. Prev. Med. 4, 1036–1044.
Akey, D. L., Clay Brown, W., Dutta, S., Konwerski, J., Jose, J., Jurkiw, T. J., et al. (2014). Flavivirus NS1 structures reveal surfaces for associations with membranes and the immune system. Science 343, 881–885. doi: 10.1126/science.1247749
Akey, D. L., and Smith, J. L. (2014). Dengue Type2 virus non-structural protein 1 (NS1) form 1 crystal. Science 343, 881–885. doi: 10.2210/pdb4o6b/pdb
Andrade, P., Gimblet-Ochieng, C., Modirian, F., Collins, M., Cárdenas, M., Katzelnick, L. C., et al. (2019). Impact of pre-existing dengue immunity on human antibody and memory B cell responses to Zika. Nat. Commun. 10:938. doi: 10.1038/s41467-019-08845-3
Arai, K. E., Bo, C. R. D., Aguirra, A. P. M., da Silva, S., Rodrigues, S., and Mangueira, C. L. P. (2020). Performance evaluation of an indirect immunofluorescence kit for the serological diagnosis of dengue. Einstein 18:eAO5078. doi: 10.31744/einstein_journal/2020AO5078
Aubry, M., Teissier, A., Huart, M., Merceron, S., Vanhomwegen, J., Roche, C., et al. (2017). Zika Virus Seroprevalence, French Polynesia, 2014–2015. Emerg. Infect. Dis. 23, 669–672. doi: 10.3201/eid2304.161549
Aye, K. S., Charngkaew, K., Win, N., Wai, K. Z., Moe, K., Punyadee, N., et al. (2014). Pathologic highlights of dengue hemorrhagic fever in 13 autopsy cases from Myanmar. Hum. Pathol. 45, 1221–1233. doi: 10.1016/j.humpath.2014.01.022
Balmaseda, A., Stettler, K., Medialdea-Carrera, R., Collado, D., Jin, X., Zambrana, J. V., et al. (2017). Antibody-based assay discriminates Zika virus infection from other Flaviviruses. Proc. Natl. Acad. Sci. 114, 8384–8389. doi: 10.1073/pnas.1704984114
Barba-Spaeth, G., Dejnirattisai, W., Rouvinski, A., Vaney, M. C., Medits, I., Sharma, A., et al. (2016). Structural basis of potent Zika-dengue virus antibody cross-neutralization. Nature 536, 48–53. doi: 10.1038/nature18938
Beck, C., Desprès, P., Paulous, S., Vanhomwegen, J., Lowenski, S., Nowotny, N., et al. (2015). A high-performance multiplex immunoassay for Serodiagnosis of Flavivirus-associated neurological diseases in horses. Biomed. Res. Int. 2015, 1–13. doi: 10.1155/2015/678084
Berntsen, C. F., and Syverud, K. H. (2009). The Dengue Prevention, Surveillance and Control Programme in the State of Morelos, Mexico, University of Oslo: Oslo.
Bhatt, S., Gething, P. W., Brady, O. J., Messina, J. P., Farlow, A. W., Moyes, C. L., et al. (2013). The global distribution and burden of dengue. Nature 496, 504–507. doi: 10.1038/nature12060
Biering, S. B., Akey, D. L., Wong, M. P., Brown, W. C., Lo, N. T. N., Puerta-Guardo, H., et al. (2021). Structural basis for antibody inhibition of Flavivirus NS1–triggered endothelial dysfunction. Science 371, 194–200. doi: 10.1126/science.abc0476
Blacksell, S. D., Jarman, R. G., Gibbons, R. V., Tanganuchitcharnchai, A., Mammen, M. P., Nisalak, A., et al. (2012). Comparison of seven commercial antigen and antibody enzyme-linked Immunosorbent assays for detection of acute dengue infection. Clin. Vaccine Immunol. 19, 804–810. doi: 10.1128/CVI.05717-11
Brasil, P., Pereira, J. P., Moreira, M. E., Ribeiro Nogueira, R. M., Damasceno, L., Wakimoto, M., et al. (2016). Zika virus infection in pregnant women in Rio de Janeiro. N. Engl. J. Med. 375, 2321–2334. doi: 10.1056/NEJMoa1602412
Brown, W. C., Akey, D. L., Konwerski, J. R., Tarrasch, J. T., Skiniotis, G., Kuhn, R. J., et al. (2016). Extended surface for membrane Association in Zika Virus NS1 structure. Nat. Struct. Mol. Biol. 23, 865–867. doi: 10.1038/nsmb.3268
Campos, S., Luis, J., Mongkolsapaya, J., and Screaton, G. R. (2018). The immune response against flaviviruses. Nat. Immunol. 19, 1189–1198. doi: 10.1038/s41590-018-0210-3
Cao-Lormeau, V.-M., Blake, A., Mons, S., Lastère, S., Roche, C., Vanhomwegen, J., et al. (2016). Guillain-Barré syndrome outbreak associated with Zika virus infection in French Polynesia: a case-control study. Lancet 387, 1531–1539. doi: 10.1016/S0140-6736(16)00562-6
Chothia, C., and Lesk, A. M. (1987). Canonical structures for the Hypervariable regions of immunoglobulins. J. Mol. Biol. 196, 901–917. doi: 10.1016/0022-2836(87)90412-8
Chuang, Y.-C., Lin, J., Lin, Y.-S., Wang, S., and Yeh, T.-M. (2016). Dengue virus nonstructural protein 1–induced antibodies cross-react with human plasminogen and enhance its activation. J. Immunol. 196, 1218–1226. doi: 10.4049/jimmunol.1500057
Chuang, Y.-C., Lin, Y.-S., Liu, H.-S., and Yeh, T.-M. (2014). Molecular mimicry between dengue virus and coagulation factors induces antibodies to inhibit thrombin activity and enhance fibrinolysis. J. Virol. 88, 13759–13768. doi: 10.1128/JVI.02166-14
Chung, K. M., Nybakken, G. E., Thompson, B. S., Engle, M. J., Marri, A., Fremont, D. H., et al. (2006). Antibodies against West Nile virus nonstructural protein NS1 prevent lethal infection through fc γ receptor-dependent and -independent mechanisms. J. Virol. 80, 1340–1351. doi: 10.1128/JVI.80.3.1340-1351.2006
Chung, K. M., Thompson, B. S., Fremont, D. H., and Diamond, M. S. (2007). Antibody recognition of cell surface-associated NS1 triggers fc-gamma receptor-mediated phagocytosis and clearance of West Nile virus-infected cells. J. Virol. 81, 9551–9555. doi: 10.1128/JVI.00879-07
Collins, M. H., McGowan, E., Jadi, R., Young, E., Lopez, C. A., Baric, R. S., et al. (2017). Lack of durable cross-neutralizing antibodies against Zika virus from dengue virus infection. Emerg. Infect. Dis. 23, 773–781. doi: 10.3201/eid2305.161630
Conlan, S., Lau, A. F., Deming, C., Spalding, C. D., Lee-Lin, S., Thomas, P. J., et al. (2019). Role of Zika virus envelope protein domain III as a target of human neutralizing antibodies. MBio 10:19. doi: 10.1128/MBIO.01485-19
Cortina-Ceballos, B., Godoy-Lozano, E. E., Sámano-Sánchez, H., Aguilar-Salgado, A., Del Castillo, M., Velasco-Herrera, C. V.-C., et al. (2015). Reconstructing and mining the B cell repertoire with ImmunediveRsity. MAbs 7, 516–524. doi: 10.1080/19420862.2015.1026502
Cutts, F. T., and Hanson, M. (2016). Seroepidemiology: an underused tool for designing and monitoring vaccination Programmes in low- and middle-income countries. Trop. Med. Int. Health 21, 1086–1098. doi: 10.1111/tmi.12737
Dai, L., Song, J., Xishan, L., Deng, Y. Q., Musyoki, A. M., Cheng, H., et al. (2016). Structures of the Zika virus envelope protein and its complex with a Flavivirus broadly protective antibody. Cell Host Microbe 19, 696–704. doi: 10.1016/j.chom.2016.04.013
Davis, M. M. (2004). The evolutionary and structural ‘logic’ of antigen receptor diversity. Semin. Immunol. 16, 239–243. doi: 10.1016/j.smim.2004.08.003
Davis, M. M., and Bjorkman, P. J. (1988). T-cell antigen receptor genes and T-cell recognition. Nature 334, 395–402.
Vries, L.De, Koopmans, M., Morton, A., and van Baal, P. (2021). The economics of improving global infectious disease surveillance. BMJ Glob. Health, 6::e006597. doi: 10.1136/bmjgh-2021-006597
Dejnirattisai, W., Supasa, P., Wongwiwat, W., Rouvinski, A., Barba-Spaeth, G., Duangchinda, T., et al. (2016). Dengue virus Sero-cross-reactivity drives antibody-dependent enhancement of infection with Zika virus. Nat. Immunol. 17, 1102–1108. doi: 10.1038/ni.3515
Dejnirattisai, W., Wongwiwat, W., Supasa, S., Zhang, X., Dai, X., Rouvinsky, A., et al. (2014). A new class of highly potent, broadly neutralizing antibodies isolated from Viremic patients infected with dengue virus. Nat. Immunol. 16, 170–177. doi: 10.1038/ni.3058
Denis, J., Attoumani, S., Gravier, P., Tenebray, B., Garnier, A., Briolant, S., et al. (2019). High specificity and sensitivity of Zika EDIIIbased ELISA diagnosis highlighted by a large human reference panel. PLoS Negl. Trop. Dis. 13:e0007747. doi: 10.1371/journal.pntd.0007747
Duffy, M. R., Tai-Ho Chen, W., Hancock, T., Powers, A. M., Kool, J. L., Lanciotti, R. S., et al. (2009). Zika virus outbreak on Yap Island, Federated States of Micronesia. N. Engl. J. Med. 360, 2536–2543. doi: 10.1056/NEJMoa0805715
Dunand, C. J. H., and Wilson, P. C. (2015). Restricted, canonical, stereotyped and convergent immunoglobulin responses. Philos. Trans. R. Soc. Lond. Ser. B Biol. Sci. 370:20140238. doi: 10.1098/rstb.2014.0238
Durham, N. D., Agrawal, A., Waltari, E., Croote, D., Zanini, F., Fouch, M., et al. (2019). Broadly neutralizing human antibodies against dengue virus identified by single B cell Transcriptomics. elife 8:52384. doi: 10.7554/eLife.52384
Edeling, M. A., Diamond, M. S., and Fremont, D. H. (2014). Structural basis of Flavivirus NS1 assembly and antibody recognition. Proc. Natl. Acad. Sci. U. S. A. 111, 4285–4290. doi: 10.1073/pnas.1322036111
Falconar, A. K. I., and Young, P. R. (1991). Production of dimer-specific and dengue virus group cross-reactive mouse monoclonal antibodies to the dengue 2 virus non-structural glycoprotein NS1. J. Gen. Virol. 72, 961–965. doi: 10.1099/0022-1317-72-4-961
Falconi-Agapito, F., Kerkhof, K., Merino, X., Michiels, J., Van Esbroeck, M., Bartholomeeusen, K., et al. (2021). Dynamics of the magnitude, breadth and depth of the antibody response at epitope level following dengue infection. Front. Immunol. 12:686691. doi: 10.3389/fimmu.2021.686691
Faria, N. R., Azevedo, R. D. S. D. S., Kraemer, M. U. G., Souza, R., Cunha, M. S., Hill, S. C., et al. (2016). Zika virus in the Americas: early epidemiological and genetic findings. Science 352, 345–349. doi: 10.1126/science.aaf5036
Faye, O., Freire, C. C. M., Iamarino, A., Faye, O., Juliana, V. C., de Oliveira, M., et al. (2014). Molecular evolution of Zika virus during its emergence in the 20(Th) century. PLoS Negl. Trop. Dis. 8, –e2636. doi: 10.1371/journal.pntd.0002636
Gallichotte, E. N., Widman, D. G., Yount, B. L., Wahala, W. M., Durbin, A., Whitehead, S., et al. (2015). A new quaternary structure epitope on dengue virus serotype 2 is the target of durable type-specific neutralizing antibodies. MBio 6:15. doi: 10.1128/MBIO.01461-15/ASSET/4E4CE46E-9EA1-44DA-A9FD-87FCFD8BA52B/ASSETS/GRAPHIC/MBO0051524830004.JPEG
Gao, F., Lin, X., He, L., Wang, R., Wang, H., Shi, X., et al. (2019). Development of a potent and protective Germline-like antibody lineage against Zika virus in a convalescent human. Front. Immunol. 10:2424. doi: 10.3389/fimmu.2019.02424
Godoy-Lozano, E. E., Téllez-Sosa, J., Sánchez-González, G., Sámano-Sánchez, H., Aguilar-Salgado, A., Salinas-Rodríguez, A., et al. (2016). Lower IgG somatic Hypermutation rates during acute dengue virus infection is compatible with a germinal center-independent B cell response. Genome Med. 8:23. doi: 10.1186/s13073-016-0276-1
Goodnow, C. C., Vinuesa, C. G., Randall, K. L., Mackay, F., and Brink, R. (2010). Control systems and decision making for antibody production. Nat. Immunol. 11, 681–688. doi: 10.1038/ni.1900
Gorny, M. K., Wang, X.-H., Williams, C., Volsky, B., Revesz, K., Witover, B., et al. (2009). Preferential use of the VH5-51 gene segment by the human immune response to code for antibodies against the V3 domain of HIV-1. Mol. Immunol. 46, 917–926. doi: 10.1016/j.molimm.2008.09.005
Gubler, D. J. (2012). The economic burden of dengue. Am. J. Trop. Med. Hyg. 86, 743–744. doi: 10.4269/ajtmh.2012.12-0157
Guerbois, M., Fernandez-Salas, I., Azar, S. R., Danis-Lozano, R., Alpuche-Aranda, C. M., Leal, G., et al. (2016). Outbreak of Zika virus infection, Chiapas state, Mexico, 2015, and first confirmed transmission by Aedes Aegypti mosquitoes in the Americas. J. Infect. Dis. 214, 1349–1356. doi: 10.1093/infdis/jiw302
Guthmiller, J. J., Lan, L. Y.-L., Fernández-Quintero, M. L., Han, J., Utset, H. A., Bitar, D. J., et al. (2020). Polyreactive broadly neutralizing B cells are selected to provide defense against pandemic threat influenza viruses. Immunity 53, 1230–1244.e5. doi: 10.1016/j.immuni.2020.10.005
Gutiérrez-Bugallo, G., Piedra, L. A., Rodriguez, M., Bisset, J. A., Lourenço-de-Oliveira, R., Weaver, S. C., et al. (2019). Vector-borne transmission and evolution of Zika virus. Nat. Ecol. Evol. 3, 561–569. doi: 10.1038/s41559-019-0836-z
Guzman, M. G., Gubler, D. J., Izquierdo, A., Martinez, E., and Halstead, S. B. (2016). Dengue infection. Nat. Rev. Dis. Primers. 2, –16055. doi: 10.1038/nrdp.2016.55
Guzmán, M. G., and Kourí, G. (2004). Dengue diagnosis, advances and challenges. Int. J. Infect. Dis. 8, 69–80. doi: 10.1016/j.ijid.2003.03.003
Harrison, S. C. (2016). Immunogenic cross-talk between dengue and Zika viruses. Nat. Immunol. 17, 1010–1012. doi: 10.1038/ni.3539
Holm, L. (2022). Dali server: structural unification of protein families. Nucleic Acids Res. 50, W210–W215. doi: 10.1093/nar/gkac387
Hunsperger, E. A., Muñoz-Jordán, J., Beltran, M., Colón, C., Carrión, J., Vazquez, J., et al. (2016). Performance of dengue diagnostic tests in a single-specimen diagnostic algorithm. J. Infect. Dis. 214, 836–844. doi: 10.1093/infdis/jiw103
Ioos, S., Mallet, H.-P., Leparc Goffart, I., Gauthier, V., Cardoso, T., and Herida, M. (2014). Current Zika virus epidemiology and recent epidemics. Med. Mal. Infect. 44, 302–307. doi: 10.1016/j.medmal.2014.04.008
Judice, C. C., Tan, J. J. L., Parise, P. L., Kam, Y.-W., Milanez, G. P., Leite, J. A., et al. (2018). Efficient detection of Zika virus RNA in patients’ blood from the 2016 outbreak in Campinas, Brazil. Sci. Rep. 8:4012. doi: 10.1038/s41598-018-22159-2
Kaneko, N., Kuo, H.-H., Boucau, J., Farmer, J. R., Allard-Chamard, H., Mahajan, V. S., et al. (2020). Loss of Bcl-6-expressing T follicular helper cells and germinal centers in COVID-19. Cells 183, 143–157.e13. doi: 10.1016/j.cell.2020.08.025
Khare, B., and Kuhn, R. J. (2022). The Japanese encephalitis antigenic complex viruses: from structure to immunity. Viruses 14:2213. doi: 10.3390/v14102213
Krishna, V. D., Rangappa, M., and Satchidanandam, V. (2009). Virus-specific Cytolytic antibodies to nonstructural protein 1 of Japanese encephalitis virus effect reduction of virus output from infected cells. J. Virol. 83, 4766–4777. doi: 10.1128/JVI.01850-08
L’Huillier, A. G., Hamid-Allie, A., Kristjanson, E., Papageorgiou, L., Hung, S., Wong, C. F., et al. (2017). Evaluation of Euroimmun anti-Zika virus IgM and IgG enzyme-linked Immunosorbent assays for Zika virus serologic testing. J. Clin. Microbiol. 55, 2462–2471. doi: 10.1128/JCM.00442-17
Lanciotti, R. S., Kosoy, O. L., Laven, J. J., Velez, J. O., Lambert, A. J., Johnson, A. J., et al. (2008). Genetic and serologic properties of Zika virus associated with an epidemic, yap state, Micronesia, 2007. Emerg. Infect. Dis. 14, 1232–1239. doi: 10.3201/eid1408.080287
Lara-Ochoa, F., Almagro, J. C., Vargas-Madrazo, E., and Conrad, M. (1996). Antibody-antigen recognition: a canonical structure paradigm. J. Mol. Evol. 43, 678–684. doi: 10.1007/BF02202116
Lee, J., Kim, Y.-E., Kim, H.-Y., Sinniah, M., Chong, C.-K., and Song, H.-O. (2015). Enhanced performance of an innovative dengue IgG/IgM rapid diagnostic test using an anti-dengue EDI monoclonal antibody and dengue virus antigen. Sci. Rep. 5:18077. doi: 10.1038/srep18077
Lee, W. T., Wong, S. J., Kulas, K. E., Dupuis, A. P., Payne, A. F., Kramer, L. D., et al. (2018). Development of Zika virus serological testing strategies in New York state. J. Clin. Microbiol. 56:17. doi: 10.1128/JCM.01591-17
Lindenbach, B. D., Thiel, H.-J., and Rice, C. M. (2011). “Flaviviridae: The Viruses and Their Replication,” in Fields Virology. eds. D. M. Knipe and O. M. Howley. 5th Edn., (Philadelphia: Lippincot William & Wilkins), 1101–1151.
Lingwood, D., McTamney, P. M., Yassine, H. M., Whittle, J. R. R., Guo, X., Boyington, J. C., et al. (2012). Structural and genetic basis for development of broadly neutralizing influenza antibodies. Nature 489, 566–570. doi: 10.1038/nature11371
Loyola, S., Huaman, A., Popuche, D., Castillo, E., Ampuero, J. S., Silva, M., et al. (2021). Evaluation of two serological assays for diagnosing Zika virus infection. Diagnostics 11:1696. doi: 10.3390/DIAGNOSTICS11091696
Luo, R., Fongwen, N., Kelly-Cirino, C., Harris, E., Wilder-Smith, A., and Peeling, R. W. (2019). Rapid diagnostic tests for determining dengue Serostatus: a systematic review and key informant interviews. Clin. Microbiol. Infect. 25, 659–666. doi: 10.1016/j.cmi.2019.01.002
Magnani, D. M., Silveira, C. G. T., Rosen, B. C., Ricciardi, M. J., Pedreño-Lopez, N., Gutman, M. J., et al. (2017). A human inferred Germline antibody binds to an Immunodominant epitope and neutralizes Zika virus. PLoS Negl. Trop. Dis. 11:e0005655. doi: 10.1371/journal.pntd.0005655
Mishra, N., Caciula, A., Price, A., Thakkar, R., Ng, J., Chauhan, L. V., et al. (2018). Diagnosis of Zika virus infection by peptide Array and enzyme-linked immunosorbent assay. MBio 9:18. doi: 10.1128/mbio.00095-18
Modis, Y., Ogata, S., Clements, D., and Harrison, S. C. (2005). Variable surface epitopes in the crystal structure of dengue virus type 3 envelope glycoprotein. J. Virol. 79, 1223–1231. doi: 10.1128/JVI.79.2.1223-1231.2005
Montoya, M., Collins, M., Dejnirattisai, W., Katzelnick, L. C., Puerta-Guardo, H., Jadi, R., et al. (2018). Longitudinal analysis of antibody cross-neutralization following Zika virus and dengue virus infection in Asia and the Americas. J. Infect. Dis. 218, 536–545. doi: 10.1093/infdis/jiy164
Murray, J., and Cohen, A. L. (2017). Infectious disease surveillance. Int. Encycl. Public Health 2017, 222–229. doi: 10.1016/B978-0-12-803678-5.00517-8
Nascimento, E. J. M., Huleatt, J. W., Cordeiro, M. T., Castanha, P. M. S., George, J. K., Grebe, E., et al. (2018). Development of antibody biomarkers of long term and recent dengue virus infections. J. Virol. Methods 257, 62–68. doi: 10.1016/j.jviromet.2018.04.009
Nielsen, S. C. A., Yang, F., Jackson, K. J. L., Hoh, R. A., Röltgen, K., Jean, G. H., et al. (2020). Human B cell clonal expansion and convergent antibody responses to SARS-CoV-2. Cell Host Microbe 28, 516–525.e5. doi: 10.1016/j.chom.2020.09.002
Nurtop, E., Villarroel, P. M. S., Pastorino, B., Ninove, L., Drexler, J. F., Roca, Y., et al. (2018). Combination of ELISA screening and Seroneutralisation tests to expedite Zika virus Seroprevalence studies. Virol. J. 15:192. doi: 10.1186/s12985-018-1105-5
Parameswaran, P., Liu, Y., Roskin, K. M., Jackson, K. K. L., Dixit, V. P., et al. (2013). Convergent antibody signatures in human dengue. Cell Host Microbe 13, 691–700. doi: 10.1016/j.chom.2013.05.008
Patel, B., Longo, P., Miley, M. J., Montoya, M., Harris, E., and de Silva, A. M. (2017). Dissecting the human serum antibody response to secondary dengue virus infections. PLoS Negl. Trop. Dis. 11:e0005554. doi: 10.1371/JOURNAL.PNTD.0005554
Pettersson, J. H. O., Eldholm, V., Seligman, S. J., Lundkvist, Å., Falconar, A. K., Gaunt, M. W., et al. (2016). How did Zika virus emerge in the Pacific Islands and Latin America? MBio 7:16. doi: 10.1128/MBIO.01239-16
Premkumar, L., Collins, M., Graham, S., Liou, G. J. A., Lopez, C. A., Jadi, R., et al. (2018). Development of envelope protein antigens to serologically differentiate Zika virus infection from dengue virus infection. J. Clin. Microbiol. 56:17. doi: 10.1128/JCM.01504-17
Priyamvada, L., Cho, A., Onlamoon, N., Zheng, N.-Y., Huang, M., Kovalenkov, Y., et al. (2016). B cell responses during secondary dengue virus infection are dominated by highly cross-reactive, memory-derived Plasmablasts. J. Virol. 90, 5574–5585. doi: 10.1128/JVI.03203-15
Priyamvada, L., Hudson, W., Ahmed, R., and Wrammert, J. (2017). Humoral cross-reactivity between Zika and dengue viruses: implications for protection and pathology. Emerg. Microbes Infect. 6, 1–6. doi: 10.1038/emi.2017.42
Putnak, J. R., De La Barrera, R., Burgess, T., Pardo, J., Dessy, F., Gheysen, D., et al. (2008). Comparative evaluation of three assays for measurement of dengue virus neutralizing antibodies. Am. J. Trop. Med. Hyg. 79, 115–122. doi: 10.4269/ajtmh.2008.79.115
Rainwater-Lovett, K., Rodriguez-Barraquer, I., Cummings, D. A. T., and Lessler, J. (2012). Variation in dengue virus plaque reduction neutralization testing: systematic review and pooled analysis. BMC Infect. Dis. 12:233. doi: 10.1186/1471-2334-12-233
Rambaut, A. (2000). Estimating the rate of molecular evolution: incorporating non-contemporaneous sequences into maximum likelihood phylogenies. Bioinformatics 16, 395–399. doi: 10.1093/bioinformatics/16.4.395
Ratfi, N. S., and Nizam, S. (2016). Phylogenetic analysis of Zika and dengue virus. Malaysian Appl. Biol. 45, 159–161.
Rey, F. A., Stiasny, K., Vaney, M.-c., Dellarole, M., and Heinz, F. X. (2018). The bright and the dark side of human antibody responses to Flaviviruses: lessons for vaccine design. EMBO Rep. 19, 206–224. doi: 10.15252/embr.201745302
Robbiani, D. F., Bozzacco, L., Keeffe, J. R., Khouri, R., Olsen, P. C., Gazumyan, A., et al. (2017). Recurrent potent human neutralizing antibodies to Zika virus in Brazil and Mexico. Cells 169, 597–609.e11. doi: 10.1016/j.cell.2017.04.024
Robbiani, D. F., Gaebler, C., Muecksch, F., Lorenzi, J. C. C., Wang, Z., Cho, A., et al. (2020). Convergent antibody responses to SARS-CoV-2 in convalescent individuals. Nature 584, 437–442. doi: 10.1038/s41586-020-2456-9
Robin, Y., Cornet, M., Heme, G., and Le Gonidec, G. (1980). Isolement Du Virus de La Dengue Au Sénégal. Annales de l’Institut Pasteur. Virologie 131, 149–154. doi: 10.1016/0769-2617(80)90029-5
Rockstroh, A., Moges, B., Barzon, L., Sinigaglia, A., Palù, G., Kumbukgolla, W., et al. (2017). Specific detection of dengue and Zika virus antibodies using envelope proteins with mutations in the conserved fusion loop. Emerg. Microbes Infect. 6:e99. doi: 10.1038/emi.2017.87
Roehrig, J. T., Hombach, J., and Barrett, A. D. T. (2008). Guidelines for plaque-reduction neutralization testing of human antibodies to dengue viruses. Viral Immunol. 21, 123–132. doi: 10.1089/vim.2008.0007
Rouers, A., Appanna, R., Chevrier, M., Lum, J., Lau, M. C., Tan, L., et al. (2021). CD27hiCD38hi Plasmablasts are activated B cells of mixed origin with distinct function. IScience 24:102482. doi: 10.1016/j.isci.2021.102482
Rouvinski, A., Guardado-Calvo, P., Barba-Spaeth, G., Duquerroy, S., Vaney, M. C., Kikuti, C. M., et al. (2015). Recognition determinants of broadly neutralizing human antibodies against dengue viruses. Nature 520, 109–113. doi: 10.1038/nature14130
Rudnick, A. (1965). Studies of the ecology of dengue in Malaysia: a preliminary report. J. Med. Entomol. 2, 203–208. doi: 10.1093/jmedent/2.2.203
Russell, P. K., and Nisalak, A. (1967). Dengue virus identification by the plaque reduction neutralization test. J. Immunol. 99, 291–296.
Russell, P. K., Nisalak, A., Sukhavachana, P., and Vivona, S. (1967). A plaque reduction test for dengue virus neutralizing antibodies. J. Immunol. 99, 285–290. doi: 10.4049/jimmunol.99.2.285
Safronetz, D., Sloan, A., Stein, D. R., Mendoza, E., Barairo, N., Ranadheera, C., et al. (2017). Evaluation of 5 commercially available Zika virus immunoassays. Emerg. Infect. Dis. 23, 1577–1580. doi: 10.3201/eid2309.162043
Sanchez, J. D. (2016). “OPS/OMS.” Pan American Health Organization/World Health Organization. Available at: http://www.paho.org/hq/index.php?option=com_content&view=article&id=11599&Itemid=41691&lan (Accessed January 20, 2016)
Sánchez-Vargas, L. A., Sánchez-Marce, E. E., and Vivanco-Cid, H. (2014). Evaluation of the SD BIOLINE dengue duo rapid test in the course of acute and convalescent dengue infections in a Mexican endemic region. Diagn. Microbiol. Infect. Dis. 78, 368–372. doi: 10.1016/j.diagmicrobio.2013.12.019
Schlesinger, J. J., Foltzer, M., and Chapman, S. (1993). The fc portion of antibody to yellow fever virus NS1 is a determinant of protection against YF encephalitis in mice. Virology 192, 132–141. doi: 10.1006/viro.1993.1015
Schroeder, H. W. Jr., and Cavacini, L. (2010). Structure and function of immunoglobulins. J. Allergy Clin. Immunol. 125, S41–S52. doi: 10.1016/j.jaci.2009.09.046
Shan, C., Ortiz, D. A., Yang, Y., Wong, S. J., Kramer, L. D., Shi, P. Y., et al. (2017). Evaluation of a novel reporter virus neutralization test for serological diagnosis of Zika and dengue virus infection. J. Clin. Microbiol. 55, 3028–3036. doi: 10.1128/JCM.00975-17
Siegrist, C.-A. (2018). “Vaccine Immunology” in Plotkin’s Vaccines. eds. S. A. Plotkin, W. A. Orestein, P. A. Offit, and K. M. Edwards (Amsterdam: Elsevier), 16–34.e7.
Sirohi, D., Chen, Z., Sun, L., Klose, T., Pierson, T. C., Rossmann, M. G., et al. (2016). The 3.8 Å resolution Cryo-EM structure of Zika virus. Science 352, 467–470. doi: 10.1126/science.aaf5316
Smith, C. E. G. (1956). The history of dengue in tropical Asia and its probable relationship to the mosquito Aedes Aegypti - PubMed. J. Trop. Med. Hyg. 59, 243–251.
Stettler, K., Beltramello, M., Espinosa, D. A., Graham, V., Cassotta, A., Bianchi, S., et al. (2016). Specificity, cross-reactivity, and function of antibodies elicited by Zika virus infection. Science 353, 823–826. doi: 10.1126/science.aaf8505
Swanstrom, J. A., Plante, J. A., Plante, K. S., Young, E. F., McGowan, E., Gallichotte, E. N., et al. (2016). Dengue virus envelope dimer epitope monoclonal antibodies isolated from dengue patients are protective against Zika virus. MBio 7, e01123–e01116. doi: 10.1128/mBio.01123-16
Thomas, S. J., Nisalak, A., Anderson, K. B., Libraty, D. H., Kalayanarooj, S., Vaughn, D. W., et al. (2009). Dengue plaque reduction neutralization test (PRNT) in primary and secondary dengue virus infections: how alterations in assay conditions impact performance. Am. J. Trop. Med. Hyg. 81, 825–833. doi: 10.4269/ajtmh.2009.08-0625
Trevethan, R. (2017). Sensitivity, specificity, and predictive values: foundations, Pliabilities, and pitfalls in research and practice. Front. Public Health 5:307. doi: 10.3389/fpubh.2017.00307
Twiddy, S. S., Holmes, E. C., and Rambaut, A. (2003). Inferring the rate and time-scale of dengue virus evolution. Mol. Biol. Evol. 20, 122–129. doi: 10.1093/molbev/msg010
Tyson, J., Tsai, W. Y., Tsai, J. J., Mässgård, L., Stramer, S. L., Lehrer, A. T., et al. (2019). A high-throughput and multiplex microsphere immunoassay based on non-structural protein 1 can discriminate three Flavivirus infections. PLoS Negl. Trop. Dis. 13:e0007649. doi: 10.1371/journal.pntd.0007649
Van Meer Maurits, P. A., Mögling, R., Klaasse, J., Chandler, F. D., Pas, S. D., Van Der Eijk, A. A., et al. (2017). Re-evaluation of routine dengue virus serology in travelers in the era of Zika virus emergence. J. Clin. Virol. 92, 25–31. doi: 10.1016/j.jcv.2017.05.001
Viedma, M., del Pilar, M., Kose, N., Parham, L., Balmaseda, A., Kuan, G., et al. (2020). Peptide arrays incubated with three collections of human sera from patients infected with mosquito-borne viruses. F1000 Res. 8:1875. doi: 10.12688/f1000research.20981.3
Voss, W. N., Hou, Y. J., Johnson, N. V., Delidakis, G., Kim, J. E., Javanmardi, K., et al. (2021). Prevalent, protective, and convergent IgG recognition of SARS-CoV-2 non-RBD spike epitopes. Science 372, 1108–1112. doi: 10.1126/science.abg5268
Wahala, W. M. P. B., Kraus, A. A., Haymore, L. B., Accavitti-Loper, M. A., and de Silva, A. M. (2009). Dengue virus neutralization by human immune sera: role of envelope protein domain III-reactive antibody. Virology 392, 103–113. doi: 10.1016/j.virol.2009.06.037
Weaver, S. C., and Vasilakis, N. (2009). Molecular evolution of dengue viruses: contributions of Phylogenetics to understanding the history and epidemiology of the preeminent Arboviral disease. Infect. Genet. Evol. 9, 523–540. doi: 10.1016/j.meegid.2009.02.003
Wen, J., and Shresta, S. (2019). Antigenic cross-reactivity between Zika and dengue viruses: is it time to develop a universal vaccine? Curr. Opin. Immunol. 59, 1–8. doi: 10.1016/j.coi.2019.02.001
WHO, Immunization, Vaccines, and Biologicals . (2017). A Guide to the Design and Conduct of Dengue Serosurveys. Geneva: World Health Organization.
WHO . (2019). Dengue vaccine: WHO position paper, September 2018–recommendations. Vaccine 37, 4848–4849. doi: 10.1016/j.vaccine.2018.09.063
Wong, S. J., Furuya, A., Zou, J., Xie, X., Dupuis, A. P., Kramer, L. D., et al. (2017). A multiplex microsphere immunoassay for Zika virus diagnosis. EBioMedicine 16, 136–140. doi: 10.1016/j.ebiom.2017.01.008
Wrammert, J., Koutsonanos, D., Li, G.-M., Edupuganti, S., Sui, J., Morrissey, M., et al. (2011). Broadly cross-reactive antibodies dominate the human B cell response against 2009 pandemic H1N1 influenza virus infection. J. Exp. Med. 208, 181–193. doi: 10.1084/jem.20101352
Wu, X., Zhou, T., Zhu, J., Zhang, B., Georgiev, I., Wang, C., et al. (2011). Focused evolution of HIV-1 neutralizing antibodies revealed by structures and deep sequencing. Science 333, 1593–1602. doi: 10.1126/science.1207532
Youn, S., Li, T., McCune, B. T., Edeling, M. A., Fremont, D. H., Cristea, I. M., et al. (2012). Evidence for a genetic and physical interaction between nonstructural proteins NS1 and NS4B that modulates replication of West Nile virus. J. Virol. 86, 7360–7371. doi: 10.1128/JVI.00157-12
Zaidi, M. B., Cedillo-Barron, L., González, M. E., Almeida, Y., Garcia-Cordero, J., Campos, F. D., et al. (2019). Serological tests reveal significant cross-reactive human antibody responses to Zika and dengue viruses in the Mexican population. Acta Trop. 201, –105201. doi: 10.1016/j.actatropica.2019.105201
Zhang, X., Ge, P., Xuekui, Y., Brannan, J. M., Bi, G., Zhang, Q., et al. (2013). Cryo-EM structure of the mature dengue virus at 3.5-Å resolution. Nat. Struct. Mol. Biol. 20, 105–110. doi: 10.1038/nsmb.2463
Zinkernagel, R. M. (1996). Immunology taught by viruses. Science 271, 173–178. doi: 10.1126/science.271.5246.173
Keywords: dengue, Zika, cross-reactivity, antibody, serosurveillance
Citation: Gaspar-Castillo C, Rodríguez MH, Ortiz-Navarrete V, Alpuche-Aranda CM and Martinez-Barnetche J (2023) Structural and immunological basis of cross-reactivity between dengue and Zika infections: Implications in serosurveillance in endemic regions. Front. Microbiol. 14:1107496. doi: 10.3389/fmicb.2023.1107496
Received: 25 November 2022; Accepted: 24 February 2023;
Published: 17 March 2023.
Edited by:
Suvankar Ghorai, Raiganj University, IndiaReviewed by:
Karen Kerkhof, National Institute for Public Health and the Environment, NetherlandsCopyright © 2023 Gaspar-Castillo, Rodríguez, Ortiz-Navarrete, Alpuche-Aranda and Martinez-Barnetche. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jesus Martinez-Barnetche, am1iYXJuZXRAaW5zcC5teA==; Celia M. Alpuche-Aranda, Y2VsaWEuYWxwdWNoZUBpbnNwLm14
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.