- Department of Biochemistry and Microbiology, Faculty of Food and Biochemical Technology, University of Chemistry and Technology, Prague, Czechia
In terms of the number and diversity of living units, the prokaryotic empire is the most represented form of life on Earth, and yet it is still to a significant degree shrouded in darkness. This microbial “dark matter” hides a great deal of potential in terms of phylogenetically or metabolically diverse microorganisms, and thus it is important to acquire them in pure culture. However, do we know what microorganisms really need for their growth, and what the obstacles are to the cultivation of previously unidentified taxa? Here we review common and sometimes unexpected requirements of environmental microorganisms, especially soil-harbored bacteria, needed for their replication and cultivation. These requirements include resuscitation stimuli, physical and chemical factors aiding cultivation, growth factors, and co-cultivation in a laboratory and natural microbial neighborhood.
Introduction
The planet we know today is largely the result of the microbial activity in the biosphere. Earth’s smallest and simplest organisms created the conditions for the development of the vast number of life forms we all know. The microscopic world is even vaster, and its diversity is stunning, but it is very difficult to reach. Even though its existence has been acknowledged for several centuries, it has been very challenging to study its roles. A crucial advance in the study of “the unreachables” arose in the days of Robert Koch at the end of the 19th century. He established a causative relationship between a microbe and its impact on a host (disease). Koch’s postulates demanded the presence of a microorganism in pure culture, isolated from the host, to confirm the link between the pathogen and the disease. From this point on, microbes were no longer considered scientific curiosities, but rather modelers of our bodies and Earth’s ecosystems (Turnbaugh et al., 2007; Graham et al., 2016; Gilbert et al., 2018). Much more efforts have been taken over the following decades to study microorganisms: these progressed from the description of and fight against the most critical human and plant pathogens, which dramatically improved our quality of life, to the later investigations on the community composition of different environments, the most advanced of which used marker gene or metagenome sequencing (Lane et al., 1985; Lynch et al., 2012). In recent years, sequencing technologies have addressed many environmental and human health-associated issues, such as the analysis of microbial responses to contamination (Hemme et al., 2010), the discovery of novel taxa to be used for bioremediation, the discovery of novel producers of antibiotics (Ling et al., 2015), or revealing the co-occurrence of antibiotic resistance genes in different environments (Li et al., 2015), to name a few.
Microorganisms live in virtually any environment, including those considered extreme due to their high temperature, pH, salinity, or concentration of pollutants (Mirzaie et al., 2015; Mehetre et al., 2018; Panda et al., 2018; Power et al., 2018; Maza et al., 2019). The physiological and biochemical potential of microbes living within these extreme environments is enormous. Thriving at the limits of life, extremophilic and extremotolerant microorganisms can provide enzymes such as the widely used Taq polymerase isolated from Thermus aquaticus (Brock, 1967; Brock and Freeze, 1969); or uncommon metabolites, such as previously unknown lipids (Schneider et al., 2019), unusual polyunsaturated fatty acids (Řezanka et al., 2019), antioxidants, pigments (Asker et al., 2012), bioactive natural compounds and other secondary metabolites with a wide range of applications (Lewis et al., 2010; Manivasagan et al., 2014). Microbes can also offer improved bioremediation possibilities (Pascoal et al., 2020), can assimilate unusual substrates (Xu R. et al., 2018) including toxic compounds, or resist and detoxify several antibiotics (Rettedal et al., 2014; McLain et al., 2016).
In order to fully describe these microorganisms and reveal their vast potential, it is necessary to obtain them in pure culture. Moreover, cultivation provides context to the metagenomic data (Nichols, 2007) and helps us verify metagenome-based conclusions on microbial interactions (microbe-microbe, microbe-plant, microbe-environment). However, bringing environmental microbes to pure culture under standard laboratory conditions has proven to be a very challenging task. Cultivation can be labor-intensive, tiring, time-consuming, and may not ensure success; but it can be rewarding if all the factors required for microbial growth are included (Figure 1). Here we discuss some generalities that elucidate the phenomenon of unculturability, with special attention paid to soil, being a habitat that harbors the greatest diversity of microorganisms, to build a foundation upon which to review some of the recent strategies to better reach “the unreachables.”
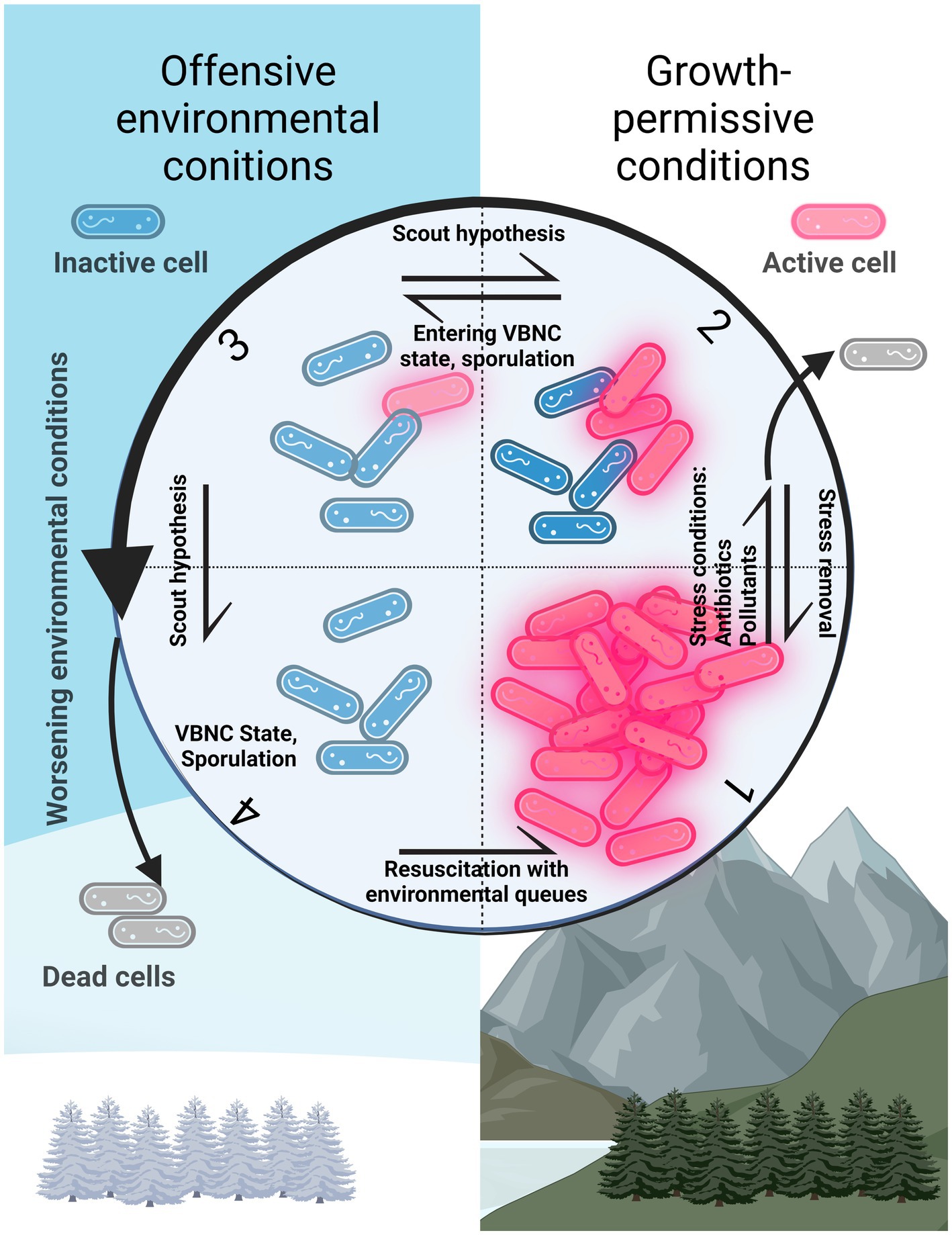
Figure 1. Different activity states of cells in the environment. 1: Active community or population. 2: In the presence of substances such as pollutants and antibiotics, a portion of the population dies and some cells can persist. These latter cells can then divide again when the substance is removed. 3: Cells in the viable but non-culturable state (VBNC). If a cell stochastically awakens in growth-permissive conditions (section number 2), the population starts replicating. If not, the cells die off (section number 4). 4: Cells in the VBNC state. Cells can resuscitate if environmental conditions become growth-permissive again (section number 1). This represents the resuscitation mediated by present environmental queues. Created with BioRender.com.
Why do you not grow?
If it is alive, no microorganism is unreachable: we just do not know how to recreate their natural environment in order to obtain a pure culture (Watve et al., 2000; Stewart, 2012). With this in mind, the key step toward successful cultivation would be to replicate essential aspects of the microorganism’s natural existence as thoroughly as possible (Figure 1). Some of the environmental variables are easily discovered and can be readily incorporated into cultivation methodologies, but many other factors that influence growth are much more obscure, and including them in cultivation strategies is not as straightforward.
The environment in which microorganisms exist is usually different from the one we create for them in the laboratory. Microorganisms live under what Koch (1971) called a “feast and famine existence.” As a consequence, the growth dynamic observed under nutrient-rich laboratory conditions does not necessarily exist in nature, where environmental changes are common and poor nutritional conditions need to be withstood for longer periods of time (Koch, 2001; Pinto et al., 2015). Microorganisms can be categorized by their resource intake characteristics either as oligotrophs or copiotrophs (Meyer, 1994; Fierer et al., 2010). The main distinguishing parameters between these categories, as Ho et al. (2017) states, are their growth kinetics, substrate affinity, and efficiency at substrate utilization. Copiotrophs have higher Michaelis–Menten kinetics and maximal growth rate. Conversely, oligotrophs are slow-growing but have higher substrate utilization efficiency, and thus higher biomass yields per substrate molecule utilized. Oligotrophs thrive in environments with low nutrient flows, but not in substrate-rich/diverse environments. Copiotrophs, on the other hand, can utilize highly concentrated substrates rapidly and react promptly to substrate changes; they nevertheless lack the necessary regulatory mechanism of starvation, and are thus generally unable to grow in nutrient-poor sites (Ho et al., 2017).
The proportion of copiotrophs to oligotrophs in the environment, as well as under laboratory conditions, is governed by a dynamic process called succession (Fierer et al., 2010). Microbial communities change over time after they colonize a certain environment. For heterotrophic bacteria, organic carbon can be constantly supplied, i.e., exogenous succession, or present all at once at the initial colonization point, i.e., endogenous succession (Fierer et al., 2010). In the initial stage of endogenous succession, when nutrients are plentiful, copiotrophs are more abundant in the community; oligotrophs become dominant when highly concentrated substrates are depleted (Song et al., 2016). Both the changing environmental conditions in nature and an inappropriate choice of growth conditions in the laboratory hinder the ability of microorganisms to replicate and could thus render them dormant and seemingly unculturable.
Do not wake up until it is beautiful outside
The low number of microbes cultivated in the laboratory compared with the total number of microorganisms observed under the microscope hinted at the existence of other states in which microorganisms may exist in nature, apart from being alive (replicating) or “dead” (non-replicating). This discrepancy, known as the “great plate count anomaly,” is a large difference, by several orders of magnitude, between the viable plate counts and the total direct microscopic counts (Staley and Konopka, 1985). This phenomenon reveals our failure to isolate all cells from a particular environment in pure cultures. Just as cells wait in a quiescent state for environmental conditions to be favorable again and start replicating (Kaprelyants et al., 1994), they can be waiting for these optimal conditions when deposited in the laboratory environment. Grandly said, microbes can be unreachable because they are “sleeping” (Xu et al., 1982).
The term “sleeping cells” encompasses several dormancy or quiescence phenomena that can cause unculturability under laboratory conditions. Dormancy is “any rest period or reversible interruption of the phenotypic development of an organism” (Sussman and Halvorson, 1966), or simply a state of metabolic inactivity as defined by Kell et al. (1998): cells exhibit negligible metabolic activity but can later transit to a growing state. This inactivity can be caused by the advent of unfavorable conditions, for example, the famine period in the dual feast-famine existence. Several dormancy phenomena have been identified, which suggests the existence of a “dormancy continuum,” where some states of dormancy can be deeper than others (Ayrapetyan et al., 2015). The most well-known state of dormancy is sporulation (Morrison and Rettger, 1930; Keep et al., 2006), in which some bacterial and fungal cells form spores as a survival strategy and outlast deleterious conditions. Spores then germinate when environmental conditions become favorable again.
Another dormancy-related phenomenon is that of “persistent cells,” first coined by Bigger (1944). This phenotype was already described in a study by Hobby et al. (1942), who observed that after exposing an infection-causing community to penicillin, 1% of the cells persisted. Persistent cells are non-growing phenotypic variants, completely dormant cells or cells inactivating genes selectively, frequently occurring in bacterial and fungal biofilms as small subpopulations (Harriott, 2019). They usually appear during the stationary phase or rarely in the exponential phase, and exhibit high tolerance to antibiotics (Wood et al., 2013). They avoid the antibiotic’s effects without undergoing genetic changes, so they play a significant role in population survival and biofilm re-creation (Lewis, 2010). In environmental biofilms, they create a subpopulation that supports biofilm survival against stress conditions such as starvation or other factors causing dormancy (Balaban, 2011; Carvalho et al., 2018).
Another common dormancy phenomenon is the viable but non-culturable (VBNC) state, believed to be widespread throughout gram-negative bacteria (Giagnoni et al., 2018). VBNC is a survival strategy that is similar to sporulation but present in non-sporulating cells (Mukamolova et al., 2003). It can be triggered by deleterious environmental changes, such as oxygen or substrate concentration changes or pH changes (Du et al., 2007). The inoculation of cells from their environment into artificial media can potentially trigger such a state. For example, when cultivating oligotrophs, the usage of a nutrient-rich medium can lead to cellular death; this may be a result of a depletion of energy for balanced growth or by osmotic shock caused by the sudden intake of non-metabolic complex substrates (Ho et al., 2017). In the VBNC state, cells do not replicate but remain viable after being exposed to stressful conditions (Xu et al., 1982). VBNC cells are also different from metabolically active and dividing cells, since they perform respiration and gene expression at low rates (del Mar et al., 2000; Shleeva et al., 2004; Li et al., 2014). They are also able to change their adhesion properties and virulence potential (Rahman et al., 1994; Du et al., 2007). Furthermore, their lower metabolic rate, strengthened cell wall and higher peptidoglycan cross-linking confer them better physical and chemical resistance, as opposed to normally dividing cells (Signoretto et al., 2000). When activity rates are reduced, VBNC cells also reduce their size (Biosca et al., 1996), increase their surface-to-volume ratio (Li et al., 2014), and, as a consequence, their nutrient intake increases (Baker et al., 1983). This size reduction was observed in Burkholderia pseudomallei and Vibrio cholerae cells when changing from rods during exponential growth to cocci in the VBNC state (Inglis and Sagripanti, 2006; Senoh et al., 2010).
Dormancy can be one of the many reasons for unculturability, which biases the picture of the community observed via culturing methods. Fortunately, dormant cells are not totally unculturable but can be more challenging to culture because not only must their growth conditions be elucidated but also their resuscitation mechanisms. Two different mechanisms are thought to resuscitate microorganisms from dormant stages: either they depend on some environmental queue to do so or they do not, the latter situation being called the scout hypothesis (Epstein, 2009; Buerger et al., 2012). This stochastic reactivation of growth is the consequence of phenotypic variation within the dormant population (Sturm and Dworkin, 2015), which resembles the idea of the “dormancy continuum” previously mentioned. In both cases, knowing which factors are present in the environments where microorganisms dwell can teach us what is necessary for their effective culturing in the laboratory (Figure 2). If they wake up stochastically, they still need environment-resembling conditions where they can thrive after awakening. If they need environmental stimuli, then these would need to be included in in vitro cultivations for microorganisms to resuscitate and grow (Figure 2).
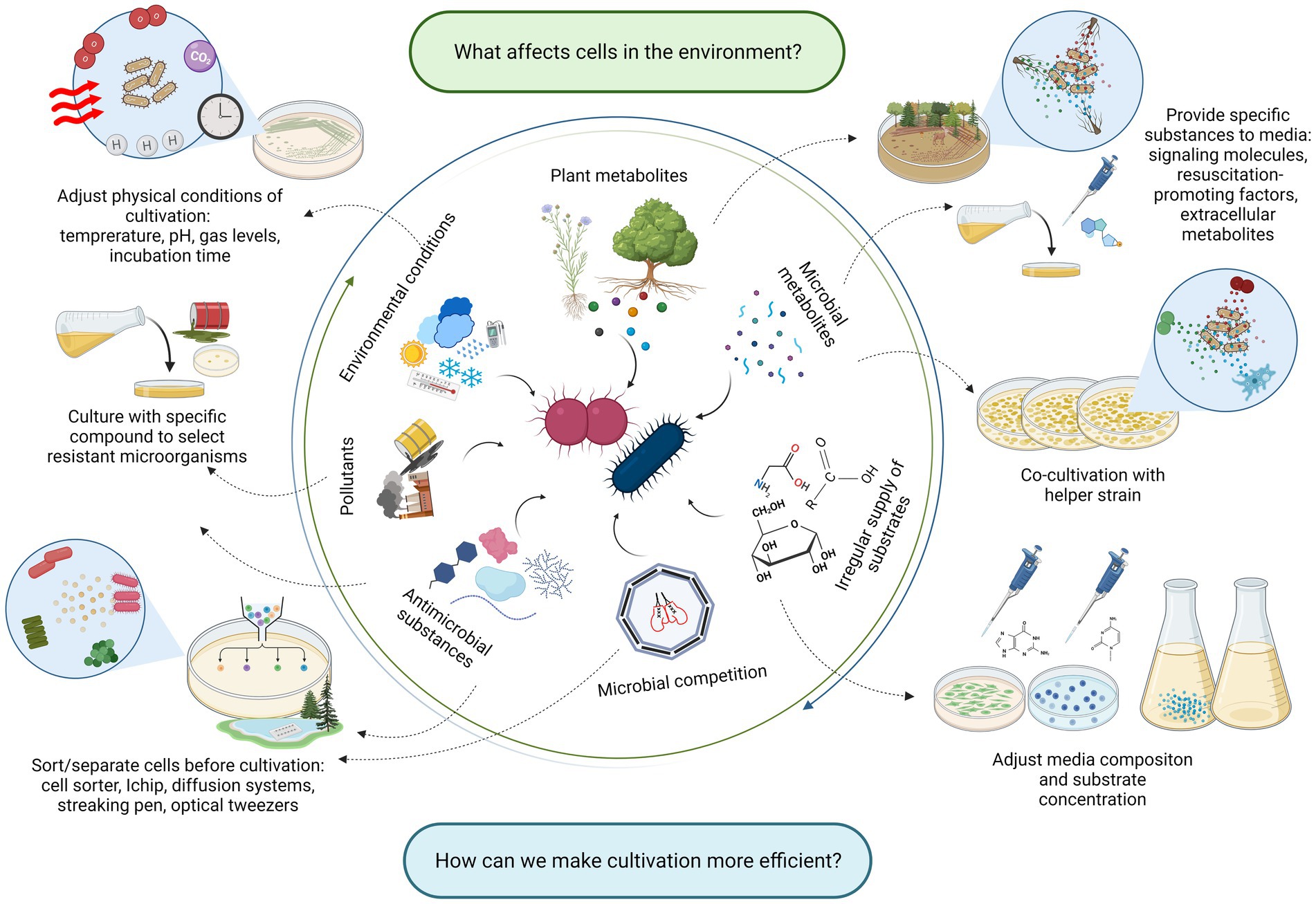
Figure 2. The list of factors affecting microorganisms in their environment (inner circle), and strategic approaches reflecting these factors in the cultivation (outer circle). Created with BioRender.com.
The stimuli needed to resuscitate microorganisms from dormancy include physical and chemical stimuli, which can be provided by the environment or by organisms to which yet-unculturable microbes are associated (Zhang et al., 2021). In this sense, the conditions needed to support growth in the laboratory medium can overlap with those to resuscitate microbes from dormancy but both phenomena correspond to different physiological processes, namely the exit from a reduced metabolic existence, after which comes the ability to replicate. Zhang et al. (2021) reviewed the factors that play a role in the resuscitation of VBNC organisms such as the addition of metabolites to minimize oxidative stress, quorum sensing autoinducers or temperature changes. In the present manuscript, the focus will be on those factors aiding the growth of microorganisms in the laboratory environment and what cultivation implies for modern microbiology.
A helping hand from the environment – Physical and chemical factors
Temperature, pH, osmotic pressure, and oxygen and nutrient concentrations are ever-changing factors in the environment (Puspita et al., 2012). These changing conditions are stress factors that shape the composition of microbial communities as well as the environments in which they live. Soil pH has a major impact since it influences soil chemistry, including the availability of organic matter, redox conditions, and oxygen availability (Anderson et al., 2018). Energy-yielding metabolisms such as microbial respiration (Jin and Kirk, 2018) and the hydroxylated lipid membrane composition (Wang et al., 2016) also respond strongly to pH changes. The impact of pH on soil chemistry even shapes the assembly of microbial communities on a global scale (Feng et al., 2014; Tripathi et al., 2018). In this regard, according to a cross-continental phylogenetic survey of over eighty soils representing a wide range of ecosystems, soil pH was significantly correlated with the overall bacterial community composition (Lauber et al., 2009). The pH has been shown to significantly influence the community structure of other environments such as lakes (Ren et al., 2015), permafrost (Ren et al., 2018), and animal microbiomes (Sylvain et al., 2016). Even small changes in this variable can thwart growth on an artificial medium since some microorganisms have a very narrow zone of pH tolerance (Rousk et al., 2010). Adamberg et al. (2003) used a pH-auxostat to study the growth rate decrease of different lactic acid bacterial strains. A pH decrease from 6 to 4.3 was enough to slow down the bacterial growth rate, and ATP production was also lowered. However, microbial growth is not only affected by drastic changes in pH disabling microbial growth, but also by suboptimal pH, at which cell growth is detectable but the growth rate is significantly decreased, as was shown in the cultivation study of Bacillus termoamylovorans when pH changes by ~1.5 from the optimal pH for its growth caused a significant reduction in the growth rate and thus caused a reduction in energy yield per glucose molecule consumed (Combet-Blanc et al., 1995). However, there are cases when the microbes themselves, intentionally and unintentionally, are able to adjust the pH of their near environment, even by excreting basic metabolites or enzymes, and thus shape the microbial community and subsequently determine the interactions between individual species of the consortium (Ratzke and Gore, 2018).
Oxygen concentration also shapes the composition of entire microbial niches: whether it is oxygen-requiring algae, microaerophilic or facultatively anaerobic purple non-sulfur photoheterotrophs, anaerobic green-sulfur bacteria, or any chemotrophs, the development of individual subpopulations is impacted based on their relationship to oxygen. Not just the simple dichotomy of aerobic and anaerobic conditions is important, but also small, specific changes in oxygen concentration matter. For instance, Coxiella burnetii, the intracellular pathogenic agent of Q-fever, infects mammalian cells at a microaerobic concentration of O2 ~ 3%. Omsland et al. (2009) successfully cultivated an axenic culture of Coxiella burnetii on an improved acidified citrate cysteine medium under an oxygen tension of 2.5%–5%. Because of its ability to grow at lower oxygen levels, the hitherto uncultured Coxiella burnetii was able to utilize up to 17 different substrates and form visible colonies in the absence of host cells. Recently, C. burnetii was cultured in a modular hypoxic chamber that maintains the required O2 concentration (2.5%) without constant airflow, which greatly reduces the evaporation of the medium (Miller et al., 2020).
Oxygen concentration also induces oxidative stress caused by reactive oxygen species. Generally, the ideal oxygen conditions depend on oxidative stress sensitivity and the need for a reduced form of a nutrient (Vallejo Esquerra et al., 2017). Since reactive oxygen species often have a lethal effect on cells, it is desirable to reduce their concentration to a minimum. Oxidative stress during cultivation can be reduced by procedures such as autoclaving the agar and the phosphates separately (Tanaka et al., 2014; Kato et al., 2018, 2020) or by adding catalase or pyruvate to media (Bogosian et al., 2000; Tanaka et al., 2014).
Another decisive factor that enhances cultivation success is the choice of substrates and notably their concentration. Differing carbon concentrations create niches that are occupied by different bacteria (Eichorst et al., 2011; Wu et al., 2020). In environments prone to drastic environmental changes such as soil or water, selective pressure favors cells with a low metabolic cost existence (Mukamolova et al., 2003). Diluted, low-carbon media favor slow-growers and increase the overall diversity, thus increasing the chances of culturing unknown taxa. Low-carbon media have successfully increased the culturing of microorganisms coming from a wide range of environments, such as sea sponges (Karimi et al., 2019; Gutleben et al., 2020), aquatic environments (Imazaki and Kobori, 2010; Sun et al., 2019), or soils (Janssen et al., 2002; Molina-Menor et al., 2021). Aquatic environments offer the advantage of using the water directly from the source as part of the cultivation media. Applying this strategy, Kapinusova et al. (2022) isolated over 100 bacterial species, including several novel species of Alphaproteobacteria, Betaproteobacteria, Flavobacteriia, and even a member of a novel genus of Thermoleophilia (Kapinusova et al., 2022). A similar strategy combined with a prolonged incubation time was used for the culturomics of the world-renowed thermal springs of Karlovy Vary (Smrhova et al., 2022) and led to the acquisition of several thermotolerant strains of the Bacillota phylum and isolation of novel microorganisms of Bacilli, Gammaproteobacteria, and Actinomycetia classes. The dilution-to-extinction technique, based on the cultivation of soil oligotrophic microorganisms on media containing 100-fold diluted nutrients, resulted in the isolation of a wide spectrum of the most abundant soil representatives, and also of members of two previously undescribed actinobacterial lineages (Bartelme et al., 2020). The combination of the above-mentioned factors into one modified cultivation procedure, namely an adjusted N2/CO2 atmosphere (80:20), low substrate concentrations, the temperature corresponding to the original environment, etc., led to the successful isolation of members belonging to the OP5 phylum (Mori et al., 2008), first described by the 16S rRNA gene analysis in a hot spring in Yellowstone National Park (Hugenholtz et al., 1998).
Since many unreachables are slow-growers, prolonged incubation times can lead to their successful cultivation. Prolonged cultivations, usually coupled with culturing diluted cell suspensions, have proved to be useful in many studies (Eilers et al., 2001; Connon and Giovannoni, 2002; Rappé et al., 2002; Kakumanu and Williams, 2012; Adam et al., 2018; Bender et al., 2020). In a study by Davis et al. (2005), autochthonous soil cells, as well as non-native cells from constructed consortia, were counted on six different media at 7-day intervals. Cell counts increased even after 12 weeks of incubation. Another successful example of prolonged cultivation, and an important microbiological milestone, was the isolation of the previously uncultured archaeon Candidatus Prometheoarchaeum syntrophicum MK-D1 (Imachi et al., 2020). This extremely slow-growing Asgard archaeon, related to the Lokiarchaeota, was isolated from a 2,533 m deep-water sediment in the Nankai trough, Japan. Aiming to achieve deep-sea microbial cultivation, Imachi et al. (2020) set up a methane-fed-continuous bioreactor in which the enrichment cultivation ran for 2,000 days, resulting in the isolation of this archaeon from a symbiotic culture. The growth of some organisms from cold and oligotrophic environments, such as those isolated from Antarctica, can only be seen in culture after prolonged incubation times (Pulschen et al., 2017; Tahon and Willems, 2017). These organisms form very small colonies which often have to be observed under a microscope (Pulschen et al., 2017).
Longer incubations in a Petri dish or batch liquid medium can be problematic because the composition of the medium tends to change over time, either because of the action of the organism’s metabolism or other processes, such as water evaporation. Even though microbial species with apparently long cultivation times can have these incubations shortened upon subculturing (Buerger et al., 2012), their initial isolation from the natural environment could fail if they are cultured together with a faster-growing species. Slow-growing microorganisms can be disadvantaged mainly when microorganisms from complex consortia are attempted to be cultured together. Physically separating or sorting the microorganisms before their culturing is a helping strategy to overcome this problem and it is discussed further in the text.
Periodically varying conditions exist in nature, from the feast and famine cycles (Koch, 1971) to alternating oxic and anoxic periods (Dorofeev et al., 2019) and seasonality (Steiner et al., 2020), all of which can affect microbial communities. Besides culturing in continuous cultures (open systems) or batch cultures (closed systems), cyclic cultivation can be useful for microorganisms with a cyclic type of metabolism. This metabolism is divided into two phases: first, energy and carbon sources are accumulated, which are then used in the second phase to biosynthesize biomass (Dorofeev et al., 2014). Any of the above-mentioned culture parameters (e.g., temperature, oxygen, or substrate concentration) can be the cycling factor in the cultivation strategy (Dorofeev et al., 2014).
Some growth-influencing factors can be more enigmatic. One such factor is acoustic vibration, which is useful as a cultivation enhancement in several biotechnological studies (Bochu et al., 2003; Avhad and Rathod, 2015; Huang et al., 2017). By causing (i) cavitation and repairable damage in microbial cells, (ii) loosening of microbial aggregates in liquid cultures, and (iii) an increase in cell membrane permeability, ultrasonic low-intensity waves (∼20 kHz) can increase the substrate intake in microbial cells and subsequently enhance microbial proliferation (Huang et al., 2017, 2021), and thus can help in the cultivation of the unreachables.
Similarly, all the aforementioned culturing parameters can be combined in a high-throughput fashion to describe as much of the community composition as possible using cultivation, with each condition used being “a different aspect of the community’s picture.” This approach is referred to as culturomics (Greub, 2012). Bacteria obtained in culture are massively characterized using MALDI TOF-MS, or 16S rRNA gene sequencing (Strejcek et al., 2018; Nowrotek et al., 2019).
A helping hand from the surroundings – Carrier particles
Many prokaryotes prefer to live attached to surfaces rather than in a dispersed, single-celled planktonic state (Mills, 2003; Flemming and Wingender, 2010; Hemkemeyer et al., 2018). In soils, different particle size fractions (PSFs) have a different impact on the concentration, chemical composition, and availability of organic matter (Christensen, 1992; Hemkemeyer et al., 2018). Organic matter is associated with fine-sized particles such as silt and clay; nevertheless, the sand fraction contains most of the free particulate organic matter (POM; Christensen, 2001), and therefore represents the fraction with the highest availability of substrates. The reported reduction in diversity among larger-sized fractions can be caused by low nutrient availability, protozoan grazing, and competition with fungi (Sessitsch et al., 2001). Hence, Hemkemeyer et al. (2018) observed the suitability of different PSFs and their associated POM to harbor microbial communities differing in their structure, functional potential, and sensitivity to environmental conditions. Genetic fingerprinting showed very strong preferences of the observed bacterial communities (up to 56% OTUs) for specific PSFs, while the archaeal populations did not exhibit significant preferences. Members of Bacteroidota and Alphaproteobacteria preferred the sand-sized fraction with POM, while Actinomycetota and Betaproteobacteria preferred fine silt, Planctomycetales clay, and Gemmatimonadales coarse silt (Hemkemeyer et al., 2018).
If cells prefer living in close contact with surfaces, it can result in it being difficult for them to grow in liquid media. Surfaces composed of different materials such as glass, steel, or synthetic polymeric substances such as polyurethane foams can enhance the cultivation of biofilm-forming bacteria from different natural environments (Yasumoto-Hirose et al., 2006; Gich et al., 2012; Dellagnezze et al., 2016). Liquid media provide many advantages compared to solid media: they guarantee a homogenous distribution of nutrients and oxygen, while also facilitating the manipulation of cultures. Aiming to combine the benefits of liquid media while meeting the requirements of microorganisms that live attached to surfaces, liquid media can be improved by adding a small amount of gelling agents such as gellan gum, xanthan gum, or carrageenan (Das et al., 2015), glass beads (Nguyen et al., 2005; Droce et al., 2013), or sand (Suman et al., 2019). Adding these supplementary solid agents can help the microorganisms to attach to the surface but still live and divide in the liquid or semiliquid medium.
A helping hand from your neighbors – Growth factors
Trace elements from the environment, apart from the carbon source, are necessary to guarantee growth in vitro. To give a simple example, genera of the slow-growing Acidobacteriota living in manganese-enriched environments benefit from the addition of this element into their growth medium (Costa et al., 2020). Complex matrices, such as soil, harbor many phylogenetically diverse microorganisms (Bahram et al., 2018) that not only participate in important biogeochemical cycles (Louca et al., 2019), but also create conditions that enable the growth of other microorganisms by sharing metabolites and essential growth substances (Schink, 2002). These molecules include those that play a role in quorum sensing, biofilm community cooperation, or in the mutualism between plants and plant-growth promoting organisms (Jacoby et al., 2017), such as rhizobacteria and endophytes (Papik et al., 2020). If a metabolite is available in the environment, microorganisms can lose the metabolic capability of producing it and thus become metabolically dependent on their neighborhood (Pande and Kost, 2017). The absence of neighbors in pure culture, and consequently the absence of the necessary metabolites, is then one of the reasons behind unculturability (Pande and Kost, 2017). Bacteria living in certain environments, such as endophytes, benefit from the use of highly specialized growth medium containing the environment’s original metabolites (Gerna et al., 2022).
With the above said, some bacteria can only grow in a pure medium when in co-culture with another community member, also called a helper strain, which can be a phylogenetically different bacterium or even a different organism such as an amoeba (Boilattabi et al., 2021). Co-culturing can be achieved either by direct culturing of the helper strain together with the bacterium of interest or by using spent supernatants as a proxy for the helper strain (Stewart, 2012). Spent supernatants are the media where the helper strain grew, so the supernatants contain the metabolites that are potentially essential for other members of the community. Microbes can also be cultured together with the host from their natural environments (Knobloch et al., 2019; Lopez Marin et al., 2021). High-throughput co-culture is also now possible with devices such as microscale microbial incubators (Ge et al., 2016), micro-petri dishes (Ingham et al., 2007), microfluidic devices (Frimat et al., 2011; Burmeister et al., 2019), or agarose-based microwell chips (Zhang et al., 2019), where hundreds of single cells can grow in parallel in individual compartments, sharing metabolites and necessary substances for growth. The latter approach has proved very helpful in culturing bacteria directly related to human health, such as antibiotic-resistant pathogens from the human gut (Versluis et al., 2019).
Metabolites from associated bacteria can provide nutrients or trigger other stimuli necessary for growth. As was previously mentioned, when water and nutrients are on the wane and the surrounding conditions are unfavorable, some cells can enter dormancy. Dormant cells can be resuscitated by different resuscitation stimuli (Pinto et al., 2015). There can be many sources of such stimuli, but they often include substances such as amino acids and peptides (Nichols et al., 2008; Pinto et al., 2011), metabolites such as N-acyl homoserine lactones (Batchelor et al., 1997), or resuscitation promoting factors (Mukamolova et al., 2006; Pinto et al., 2013; Lopez Marin et al., 2021). For example, in a study by Bruns et al. (2002), the signaling molecules cAMP and N-(butyryl)-DL-homoserine lactone (BHL) increased total bacterial counts in highly diluted inocula from aquatic environments by several orders of magnitude. Thanks to this effort, the previously uncultured bacterial clone G100, Citreicella manganoxidans, belonging to the Rhodobacteraceae family, was cultured (Bruns et al., 2002; Wirth and Whitman, 2018). Less ambitious but still hopeful results were provided by the follow-up studies of Bruns, where the addition of cAMP led to a 10% increase in MPN values (Bruns et al., 2003). Yet, in several studies where signaling compounds were used for increasing cultivation yields, the influence of cAMP on culturability has been disproven (Pernthaler et al., 2003; Sangwan et al., 2005).
The resuscitation promoting factor (Rpf) produced by Micrococcus luteus promotes bacterial resuscitation and growth in the same producing organism (Mukamolova et al., 2006), but can influence taxa distributed along several other phyla, such as Pseudomonadota and Bacteroidota (Su et al., 2018; Lopez Marin et al., 2021; Su et al., 2021). This small protein (16–17 kDa) with a lysozyme-like structure (Cohen-Gonsaud et al., 2005) promotes bacterial cell growth even at picomolar concentrations (Mukamolova et al., 1998; Sexton et al., 2015). Rpf-like encoding genes are distributed among other prokaryotic genomes, especially in G + C rich gram-positive Actinomycetota (Nikitushkin et al., 2016), but Rpf-like proteins extend to other bacterial phyla, such as Bacillota (Shah and Dworkin, 2010) and Pseudomonadota (Li et al., 2020). The addition of Rpf during cultivation has resulted in the isolation of novel bacteria, such as organisms of the genera Rhodococcus and Arthrobacter, or of the family Alcaligenaceae (Su et al., 2013, 2015, 2018, 2021). Lopez Marin (Lopez Marin et al., 2021) isolated 51 novel bacterial species belonging mainly to the phyla Actinomycetota, Pseudomonadota, and Bacteroidota on reasoner’s 2A (R2A) agar and an agar made from the soil’s water-soluble fraction after supplementing Micrococcus luteus Rpf-containing supernatant to soils. Some of these species were members of novel genera, such as Pedomonas mirosovicensis of the family Sphingosinicellaceae, or Solicola gregarius of the family Nocardioidaceae (Lopez Marin et al., 2022, 2023). Spent supernatants containing growth factors have also aided the cultivation of Chloroflexota strains (Xian et al., 2020) or Leucobacter, the growth of which was supported through the action of zincmethylphyrins and coproporphyrins produced by Sphingopyxis sp. (Bhuiyan et al., 2016).
Do you want to stay in your neighborhood?
The identification of specific substances promoting cell growth is not an easy task. To bypass the search for crucial growth factors, microorganisms can be co-cultured with growth-promoting microorganisms or can be cultivated in situ in the environments they come from Bollmann et al. (2007) and Remenár et al. (2015). In situ cultivation allows for the isolation of microorganisms that are more adapted to the original environment than those originating from the same habitat but obtained on standard agar media (Jung et al., 2016). Several innovative devices have been envisioned to deal with in situ cultivation. In an early attempt, Kaeberlein et al. (2002) developed a diffusion chamber that allowed the nutrients from the natural environment to migrate to the site where bacteria were inoculated. Seawater solidified with agar was sandwiched between two polycarbonate membranes, which allowed the flow of nutrients from the natural environment to the agar while at the same time isolating the inoculum from the natural environment (Kaeberlein et al., 2002). Diffusion chambers have since increased the diversity of culturable bacteria (Bollmann et al., 2007), including those that are difficult to culture, such as members of the phylum Verrucomicrobiota (Pascual et al., 2017) or bacteria highly resistant to heavy metals (Remenár et al., 2015).
A similar device to the diffusion chamber is the soil substrate membrane system (SSMS), which allows the growth of colonies over a membrane (made of materials such as polycarbonate), through which the nutrients and growth factors of the natural environment permeate and reach these colonies (Ferrari et al., 2005). Using the SSMS, Ferrari et al. (2005) isolated previously uncultured members of the genera Aminomonas, Nocardia, Pseudomonas, and Enterobacter. This membrane system has also been used to recover hydrocarbon-degrading bacteria from diesel-spiked polar soils (van Dorst et al., 2016) and was proven to recover rarer bacterial taxa from ice-free polar desert compared to conventional cultivation approaches (Pudasaini et al., 2017).
Later modifications of the diffusion chamber have been designed to culture microorganisms in the natural environment but using liquid media instead. One such early device was the hollow-fiber membrane chamber developed by Aoi et al. (2009). It is composed of hollow polyvinylidene tubes where microbes are inoculated and grown. The tubes are porous, so they allow the transport of molecules from the natural environment to the inside of the tube. In comparison with standard petri dish methods, the hollow-fiber membrane chamber technique yielded a higher ratio of novel phylotypes, mostly of Pseudomonadota, Actinomycetota, Bacteroidota, and Spirochaetota, and also resulted in an overall higher diversity of the recovered isolates (Aoi et al., 2009). Another liquid medium-based diffusion chamber is a bioreactor separated from the surrounding environment by a polycarbonate membrane (Chaudhary et al., 2019; Chaudhary and Kim, 2019). With this device, 35 previously uncultured bacteria belonging to the phyla Pseudomonadota, Bacillota, Bacteroidota, and Actinomycetota were isolated; the largest number of novel isolates was obtained when soil extract was used for the preparation of the medium (Chaudhary et al., 2019). Diffusion chambers have been manufactured in 3D printers, which increases their customization possibilities for their use in different applications (Wilson et al., 2019).
Diffusion chamber devices have been subject to further modifications. One such example is the so-called microbial trap, which consists of two semipermeable membranes with agar or gellan gum “sandwiched” between them (Gavrish et al., 2008). Filamentous Actinomycetota can access the medium from the outside through the semipermeable membranes. A similar trap was designed by Jung et al. (2013), with the difference that the trap’s access size can be modified. This latter trap has been used to culture various microorganisms from extreme environments, such as saline lakes (Jung et al., 2013) and hot springs (Jung et al., 2018). Yet another modification to the microbial trap uses sub-micrometer constrictions, where microorganisms compete to reach a chamber with nutrients going through a thin opening that allows only one bacterium to access and form a pure culture (Tandogan et al., 2014). Both groups of devices, diffusion chambers and microbial traps, have been shown to help reduce cultivation bias by culturing bacterial representatives which metagenomics approaches identified as the main representatives in a specific community (Pathak et al., 2020).
A successful high-throughput modification of the diffusion chamber technique is a system of multiple diffusion chambers called the isolation chip (iChip), first coined by Nichols et al. (2010). It consists of an assembly of three flat plates, a central one, and two symmetrical external plates. The external polyoxymethylene plates are provided with a set of 384 holes, since every chamber in the central plate is designed to capture, ideally, just one cell. The inoculated central plate is covered, as with Bollman’s device (Bollmann et al., 2007), with a standard polycarbonate membrane, which permits the flow of nutrients from the environment and at the same time keeps the cells inside the chambers. The external plates prevent the cells from migrating in and out, and also keep them literally trapped inside their chambers. This chip can then be placed in the natural environment to serve as a cultivation chamber in situ (Berdy et al., 2017). Among others, the Antarctic bacterium Aequorivita sp., possessing antimicrobial and anthelmintic activity, was isolated using the iChip system (Esposito et al., 2018; Liu et al., 2021). The iChip has also aided in the cultivation of antibiotic-producing bacteria, such as the bacterium Eleftheria terrae, which produces the antibiotic teixobactin (Ling et al., 2015).
Devices similar to the iChip have been used recently to culture fastidious bacteria. The diffusion sandwich system, a device based on the iChip, led to a successful culturing of Pseudomonas soli which can produce xantholysin congeners (Pascual et al., 2014) or the gellan gum-degrading bacterium Luteolibacter gellanilyticus (Pascual et al., 2017). Acuna (Acuna et al., 2020) used microwell chambers, devices similar to the iChip in design, to culture rhizobacterial populations. Rhizosphere microorganisms were also cultured in situ using the Rhizochip, an acrylic device with holes, in which microorganisms are randomly and not evenly inoculated, and placed into a plant rhizosphere (Gurusinghe et al., 2019). All these examples show that when the unreachables stay in their environments, we are more likely to reach them in cultures.
Want to be sorted or isolated before cultivation?
Because of the enormous number of microorganisms awaiting cultivation, it is natural to assume that automation and high-throughput culturability will be more and more common. Organisms in a community can be individually sorted and cultured under a broad range of conditions. Among these sorting approaches are the preselection of cells by their size, shape, or by any other characteristic. This results in the division of the total microbial community into several subpopulations consisting of similar microorganisms. Such a separation requires equipment such as optical tweezers, flow cytometry coupled with sorting cell assays, or the integration of both methods (Tewari Kumar et al., 2020).
In 2002, Zengler and his team presented a method involving microdroplets of solidified agarose for encapsulating single bacterial cells. The encapsulated cells were then grown in a column with low nutrient media, and thus were able to grow “together but apart” (Zengler et al., 2002). This high-throughput cultivation method resulted in the growth and successful isolation of newly identified Planctomycetales and Alphaproteobacteria (Zengler et al., 2002). An advantage of this microdroplet cultivation is the broad range of environments to which the technology can be applied. Later, in 2005, Zengler presented an improved version of the method, Diversa’s high-throughput cultivation using microcapsules, by which it is possible to obtain more than 10,000 bacterial and fungal isolates from a matrix (Zengler et al., 2005). More recently, alginate microbeads have been successfully used for the high-throughput culturing of bacteria that usually resist cultivation such as Verrucomicrobiota and Epsilonproteobacteria (Ji et al., 2012), and also to cultivate anaerobes (Börner et al., 2013). Analogously to the co-culture strategy, bacteria grow in the presence of other members of the community, just separated from each other in individual capsules or drops. Encapsulated microorganisms can then be sorted, for example by using fluorescence-activated cell sorting, according to their phenotype of interest (Eun et al., 2011) or other distinguishing properties such as the presence or absence of growth in each droplet (Zang et al., 2013; Ota et al., 2019), their growth rate (Akselband et al., 2006; Ota et al., 2019), chemotactic motility (Dong et al., 2016), or their metabolic activity (Espina, 2020).
Focusing on slow-growing microorganisms after sorting can result in the cultivation of rare taxa (Watterson et al., 2020). Jian and coworkers developed a microbial microdroplet culture system, where cells are cultured in water-in-oil droplets placed in Teflon tubes. This system uses up to 200 droplets with a volume of 2 μL, in which microbes are cultured in a high-throughput fashion (Jian et al., 2020). The droplets can be manipulated to meet the needs of different experimental designs. Microbe-harboring beads or droplets (or, in general, sorted cells) can also be cultured in their natural environments, which can be achieved by encapsulating the beads inside an extra polysulfonate membrane to isolate the encapsulated cells from the environment (Ben-Dov et al., 2009) or, more recently, using devices such as the Microbe Domestication Pod (Alkayyali et al., 2021). The pod, which holds agarose microbeads containing encapsulated cells, is placed in the environment, allowing the encapsulated microorganisms to be cultured individually but guaranteeing cell-to-cell communication and the presence of important environmental necessities.
Sorted droplets can also be placed in microwell slides in order to facilitate downstream cultivation and analysis (Bai et al., 2014). The sorting of cells in compartments can also be exploited to research cell-to-cell interactions among encapsulated bacteria (Ohan et al., 2019) and biofilm formation or growth (Chang et al., 2015; Jin et al., 2018). The elucidated interactions can cast light upon each cell’s needs for growth, and thus on its effective cultivation. Devices such as the SlipChip, composed of two conjoined plates, allow the duplication of a microbial colony so that half of it can be further preserved or cultured, while the other half can be used for destructive analyses (Ma et al., 2014).
Another way to sort the unreachables is to separate them while growing on a petri dish. Cultures can be sprayed onto medium plates instead of being spread with a hockey stick. This procedure effectively compartmentalizes microorganisms in droplets, hence the aggregation of cells and interspecies competition, once they land on the medium, is significantly reduced (Huang et al., 2021). Gao and co-workers developed a microbe observation and cultivation array (MOCA) that allows the recovery of microbes on a small scale and does not require any complex equipment (Gao et al., 2013). MOCA involves a petri dish with arrays of oil-covered droplets of cells. The oil covering provides a separation between cells and thus enables the cultivation of multiple separated droplets of cells (Gao et al., 2013). Several marine microorganisms were isolated using this technique, including Pseudoalteromonas spp. and previously uncultured members of the genera Shewanella and Colwellia (Gao et al., 2013). Compared to conventional approaches, MOCA offers an easy system for compact, parallel cultivation and multiple variations of different media on a relatively small scale.
“Streaking pen” developed by Jiang and his group is a robust, high-throughput method based on a simple streaking and picking strategy to achieve single-cell cultivation on microfluidic streak plates. Using this technique, a previously unknown fluoranthene-degrading Blastococcus species was isolated (Jiang et al., 2016), and so were novel species of bacteria from a marine sediment (Xu B. et al., 2018; Hu et al., 2020). This method has also been used to culture termite-associated bacteria of the genera Burkholderia, Micrococcus, and Dysgonomonas (Zhou et al., 2019). In general, cell sorting enables the design of complex experiments using just a few plates, and thus represents a great experimental simplification that allows for a better examination of individual subpopulations and, as a result, increases the chances of culturing novel taxa.
Let us seek information about the cultivation of the unreachables in the (meta)genome
Successful cultivation of just a few novel taxa while adding “vital” molecules to the media, trying different media and cultivation conditions, or the combination of all the above, is a lengthy and material-consuming way to find the requirements for microbial growth of specific taxa, given the vast diversity of the unreachables. Nowadays, the metagenome has become a promising source of information on cultivation needs, since it reveals “who is there and what their roles are” (Remenár et al., 2015; Nowrotek et al., 2019). In other words, why try dozens of media or condition combinations, when each cell’s growth requirements can be found in its genome?
As was mentioned earlier, a common phenomenon in a community is the loss of the ability to metabolize certain compounds if these are provided by other organisms (Pande and Kost, 2017). Such a gene loss can be ultimately seen in the genome (Carini et al., 2013). Reconstruction of the metabolic pathways through genomic information reveals the bacterium’s deficiencies or needs, which can be provided in the medium (Liu et al., 2022). For example, the nutritional requirements of Pelagibacter ubique, most likely the most abundant bacterium on Earth, were determined in part by its absence of genes for assimilatory sulfate reduction and its need for reduced sulfur compounds for growth (Carini et al., 2013). Karnachuk et al. (2020) isolated a thermophilic spirochete thanks to information from a metagenome-assembled genome which suggested the presence of 12 alpha-amylase hydrolases. This bacterium was then cultured using a medium composed mainly of starch (Karnachuk et al., 2020).
Metagenomic data can also be used to create co-occurrence network approaches based on network inference techniques in order to model the abundance or roles of specific community members in an environment (Faust and Raes, 2012). These relationships can be exploited in co-culture approaches, which can represent these relationships, e.g., by using spent media from other culturable bacteria in the community (Xian et al., 2020). Information contained in an RNA sequence (metatranscriptome) can be even more useful because it reflects the necessary genes being expressed in a given environment and time. For instance, the metatranscriptome of the leech Hirudo verbana was characterized, revealing the expression of genes coding for sulfated-mucin desulfatases and sialidases (Bomar et al., 2011). A medium with added mucin then allowed the cultivation of a Rickenella-like leech symbiont in vitro (Bomar et al., 2011).
A metagenome is a very complex collection of information, so discerning specific, individual genomes out of this mixture is often a difficult task, and techniques that provide a link between identity and function can help to discern which specific organisms carry which metabolic activity. One such technique is stable isotope probing (SIP), a method that links certain metabolic capabilities to individual community members. Upon probing with stable isotopes, the metagenome of these community members can be separated and sequenced to reveal their identity (Uhlik et al., 2013). SIP in tandem with metagenomics helped culture different bacteria with, for example, biodegradative functions. The bacterium Polaromonas naphthalenivorans was isolated in a pure culture after its role in the degradation of naphthalene was determined by SIP (Jeon et al., 2003, 2004). A similar approach was followed for isolating novel phenanthrene- and biphenyl-degrading Ralstonia populations (Li et al., 2019), novel isoprenedegrading bacteria belonging to different genera (Larke-Mejía et al., 2019), or hydrocarbon-degrading bacteria from the sea basin (Mishamandani et al., 2014; Gutierrez et al., 2015) or oil spills (Gutierrez et al., 2013). In these examples, the stable isotope-labeled, or “heavy” molecule used for the biodegradation analysis was also included in the cultivation efforts, but “heavy” genomes could also highlight other requirements that the degrading bacteria may need.
Finally, metabolic needs can be elucidated by single-cell genomics (Wurch et al., 2016), which can be boosted with the cell-sorting approaches described earlier. Single-cell genomic information has enabled metabolic reconstruction and aided the isolation of difficult-to-culture organisms such as symbiotic archaea (Wurch et al., 2016). Additionally, Cross et al. (2019), using single-cell genomic data, developed a method to capture specific microorganisms using antibody engineering. These antibodies are designed based on membrane-associated proteins, whose sequences can be found in the genome. The antibodies are labeled with a fluorescent dye, and thus the cells to which the antibody binds can be sorted by flow cytometry and cultivated in different media (Cross et al., 2019).
Conclusion and future perspectives
In recent years, a large number of microbes have been cultured employing the procedures discussed here in. Over the last two decades, in particular, a great deal of effort has been spent to improve culturing work, and many new taxa have been described; in fact, more bacteria have been cultured and described in the first 20 years of the 21st century than in all previous years of microbiological research combined (Figure 3; Parte et al., 2020). The high-throughput sequencing revolution that enabled the analysis of the metagenome has great potential to aid the cultivation progress. There is an unavoidable synergy between culture-independent and culture-dependent knowledge: as our knowledge of metagenomes increases, so does our knowledge of what microbes need to grow. The majority of Earth’s environments still harbor mainly hitherto uncultured microorganisms (Lloyd et al., 2018). Just as an ebb primarily uncovers areas close to the shore, and maybe never reveals the perpetually hidden abyss, so the phylogenetically distant cells, or “phylogenetically divergent non-cultured cells” as described by Lloyd et al. (2018), may remain undiscovered. These unreachables are the real “dark matter” of the microbial world and keep on shaping our planet right under everyone’s noses. But in theory, nothing is impossible to culture, and what we do not successfully culture today can be brought to culture tomorrow. Just like the Yellowstone National Park’s Obsidian Pool gave us a hint of the then so-called OP5 or OP10 phylum (Hugenholtz et al., 1998), whose members were isolated more than a decade later (Mori et al., 2008; Lee et al., 2011; Tamaki et al., 2011), other environments will reveal their secret inhabitants via culture-independent, omics-based approaches, after which culturing will be applied in search of their objectification. But not just simple, low-scale culturing; automatized, high-throughput culturomics will be needed. Sorting technologies such as those based on microfluidic systems could already be coupled with machine learning systems (Srikanth et al., 2021) so that growth needs can be elucidated and a high number of microorganisms can be cultured in the shortest time possible. This is the same as with many other big questions that still afflict us: it seems that machines and algorithms are coming to the rescue. So many microbes will be unreachable no more, and the time for this is already being reached.
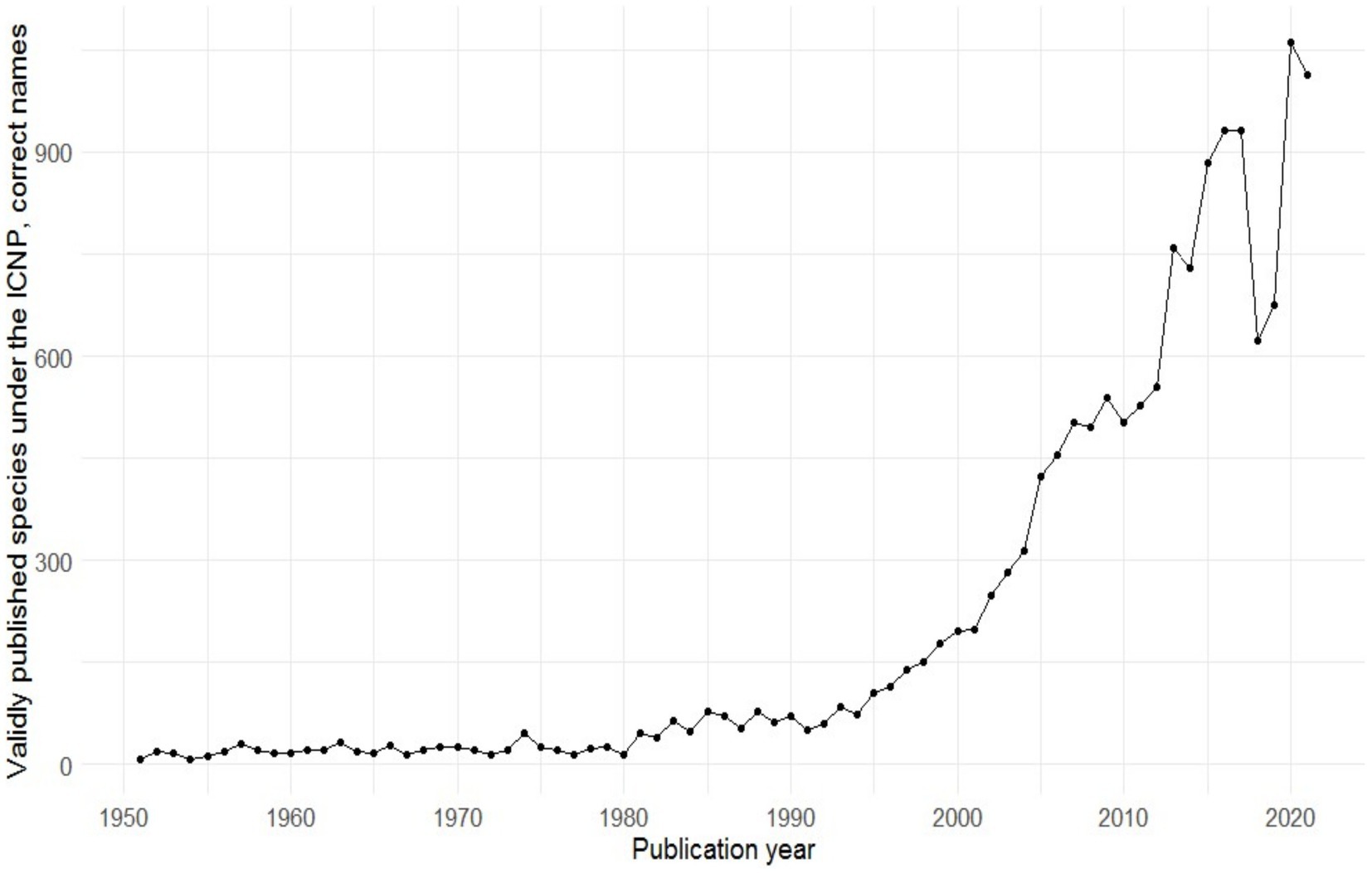
Figure 3. The number of validly published species within the last 70 years (Parte et al., 2020).
As mentioned in the previous section, there may be an interest in culturing a specific organism from the environment and techniques have been proposed to tackle this challenge (Cross et al., 2019). Throughout the course of microbiological research, several taxa have been categorized as “most wanted” because of the important roles they play, such as in the human microbiome (Almeida et al., 2016) or other environments in the biosphere (Steele et al., 2011). At the same time, some microorganisms exist as obligate symbionts: their genomes have been reduced because of the loss of functional genes, and these lost functions can be guaranteed by the host (Moran and Bennett, 2014). Entire bacterial phyla such as the Candidate phyla radiation are thought to be composed mostly of symbionts (Castelle and Banfield, 2018). Should we force them to try to exist by themselves in a pure culture, despite their loss of basic structural features such as cell wall components and extremely small genomes (<200 Kbp) with maybe no possibility of growing away from their host, or should we better make more flexible regulations of what is required to propose new prokaryotic species? Cultivation is made difficult not only because of the intricacies needed for the growth of microorganisms in the laboratory but by placing unreal requirements for their study through culturing.
There are calls to reform the one species-one publication formula (Rosselló-Móra and Amann, 2015) and, due to the diversity of bacteria in the environment, it is not difficult to imagine that it may be impossible to describe all bacterial species using the polyphasic approach employed today for circumscribing new species, even if all microbes were culturable. Recent estimates suggest that the number of different bacterial taxa in the biosphere (established with a 16S rRNA gene similarity cutoff of 97%) is 2.2–4.3 million (Louca et al., 2019). New bacterial descriptions are also constrained by journal capabilities (Tamames and Rosselló-Móra, 2012). In order to give an identity to the mass of uncultured microorganisms, the availability of a pure culture is maybe not necessary anymore. High-quality genome sequences are being proposed as nomenclatural types (instead of viable anexic cultures in culture collections), and a new classification system, the SeqCode, is being developed to exist (at least temporarily) parallel to the International Code of Nomenclature of Prokaryotes (Hedlund et al., 2022; Whitman et al., 2022). The requirements of a pure axenic culture of the ICNP as the only type material possible for naming new microbial species has been criticized as self-limiting, hindering microbiological research and raising the costs associated with naming new taxa (Palmer et al., 2022). If the “dream of a phylogenetic system” was materialized upon the bases of genomics (Woese, 1992), the development of a reliable system based on genomics must be pursued and supported.
These recent developments in prokaryotic systematics will not negatively affect the importance of cultivation because microbiology is a science whose reach extends far beyond taxonomy and the basic knowledge of microbes. It is expected that, by 2024, the economic value of the global microbes and the microbial market will exceed USD 675.2 billion (Estevinho et al., 2020). These figures are reached by allocating organisms in high-value biotechnological industries which produce the goods previously mentioned in the introduction. The “dark matter of life” conceals not only the answer to “who is there,” but also “what are they doing.” This second question is still what may be most relevant contributing to the advancement of technology. The future of cultivation is one that begins with its strengths: the ability to select and culture microorganisms relevant to their functions and technological potential. But we must be open-minded enough to not limit our horizons with just apparent and obvious applications: a world of possibilities can be opened with each microorganism isolated and studied.
Author contributions
GK and MLM contributed equally to the literature research and writing of the review, all under OU’s guidance and supervision. All authors approved the final version.
Funding
Financial support is acknowledged of the Czech Science Foundation under grant no. 22-00132S and INTER-EXCELLENCE program of the Ministry of Education, Youth and Sports of the Czech Republic under grant no. LTAUSA19028.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Acuna, J. J., Marileo, L. G., Araya, M. A., Rilling, J. I., Larama, G. A., Mora, M. L., et al. (2020). In situ cultivation approach to increase the culturable bacterial diversity in the rhizobiome of plants. J. Soil Sci. Plant Nutr. 20, 1411–1426. doi: 10.1007/s42729-020-00222-0
Adam, D., Maciejewska, M., Naômé, A., Martinet, L., Coppieters, W., Karim, L., et al. (2018). Isolation, characterization, and antibacterial activity of hard-to-culture Actinobacteria from cave moonmilk deposits. Antibiotics 7:28. doi: 10.3390/antibiotics7020028
Adamberg, K., Kask, S., Laht, T.-M., and Paalme, T. (2003). The effect of temperature and pH on the growth of lactic acid bacteria: a pH-auxostat study. Int. J. Food Microbiol. 85, 171–183. doi: 10.1016/S0168-1605(02)00537-8
Akselband, Y., Cabral, C., Castor, T. P., Chikarmane, H. M., and McGrath, P. (2006). Enrichment of slow-growing marine microorganisms from mixed cultures using gel microdrop (GMD) growth assay and fluorescence-activated cell sorting. J. Exp. Mar. Biol. 329, 196–205. doi: 10.1016/j.jembe.2005.08.018
Alkayyali, T., Pope, E., Wheatley, S. K., Cartmell, C., Haltli, B., Kerr, R. G., et al. (2021). Development of a microbe domestication pod (MD pod) for in situ cultivation of micro-encapsulated marine bacteria. Biotechnol. Bioeng. 118, 1166–1176. doi: 10.1002/bit.27633
Almeida, M., Pop, M., Le Chatelier, E., Prifti, E., Pons, N., Ghozlane, A., et al. (2016). Capturing the most wanted taxa through cross-sample correlations. ISME J. 10, 2459–2467. doi: 10.1038/ismej.2016.35
Anderson, C. R., Peterson, M. E., Frampton, R. A., Bulman, S. R., Keenan, S., and Curtin, D. (2018). Rapid increases in soil pH solubilise organic matter, dramatically increase denitrification potential and strongly stimulate microorganisms from the Firmicutes phylum. PeerJ 6:e6090. doi: 10.7717/peerj.6090
Aoi, Y., Kinoshita, T., Hata, T., Ohta, H., Obokata, H., and Tsuneda, S. (2009). Hollow-fiber membrane chamber as a device for in situ environmental cultivation. Appl. Environ. Microbiol. 75, 3826–3833. doi: 10.1128/AEM.02542-08
Asker, D., Awad, T. S., Beppu, T., and Ueda, K. (2012). “Isolation, characterization, and diversity of novel radiotolerant carotenoid-producing bacteria” in Microbial carotenoids from bacteria and microalgae: methods and protocols, methods in molecular biology. eds. Barredo and José-Luis (Springer), 21–60.
Avhad, D. N., and Rathod, V. K. (2015). Ultrasound assisted production of a fibrinolytic enzyme in a bioreactor. Ultrason. Sonochem. 22, 257–264. doi: 10.1016/j.ultsonch.2014.04.020
Ayrapetyan, M., Williams, T. C., and Oliver, J. D. (2015). Bridging the gap between viable but non-culturable and antibiotic persistent bacteria. Trends Microbiol. 23, 7–13. doi: 10.1016/j.tim.2014.09.004
Bahram, M., Hildebrand, F., Forslund, S. K., Anderson, J. L., Soudzilovskaia, N. A., Bodegom, P. M., et al. (2018). Structure and function of the global topsoil microbiome. Nature 560, 233–237. doi: 10.1038/s41586-018-0386-6
Bai, Y., Weibull, E., Joensson, H. N., and Andersson-Svahn, H. (2014). Interfacing picoliter droplet microfluidics with addressable microliter compartments using fluorescence activated cell sorting. Sens Actuators B Chem. 194, 249–254. doi: 10.1016/j.snb.2013.12.089
Baker, R. M., Singleton, F. L., and Hood, M. A. (1983). Effects of nutrient deprivation on Vibrio cholerae. Appl. Environ. Microbiol. 46, 930–940. doi: 10.1128/aem.46.4.930-940.1983
Balaban, N. (2011). Persistence: mechanisms for triggering and enhancing phenotypic variability. Curr. Opin. Genet. Dev. 21, 768–775. doi: 10.1016/j.gde.2011.10.001
Bartelme, R. P., Custer, J. M., Dupont, C. L., Espinoza, J. L., Torralba, M., Khalili, B., et al. (2020). Influence of substrate concentration on the culturability of heterotrophic soil microbes isolated by high-throughput dilution-to-extinction cultivation. mSphere 5, e00024–e00020. doi: 10.1128/mSphere.00024-20
Batchelor, S. E., Cooper, M., Chhabra, S. R., Glover, L. A., Stewart, G. S., Williams, P., et al. (1997). Cell density-regulated recovery of starved biofilm populations of ammonia-oxidizing bacteria. Appl. Environ. Microbiol. 63, 2281–2286. doi: 10.1128/aem.63.6.2281-2286.1997
Behera, H. T., Mojumdar, A., and Ray, L. (2022). “Chapter 9 – biology, genetic aspects and oxidative stress response of actinobacteria and strategies for bioremediation of toxic metals” in Microbial biodegradation and bioremediation. eds. S. Das and H. R. Dash 2nd ed (Amsterdam: Elsevier), 181–192.
Bender, K. E., Glover, K., Archey, A., and Barton, H. A. (2020). The impact of sample processing and media chemistry on the culturable diversity of bacteria isolated from a cave. Int. J. Speleol. 49, 209–220. doi: 10.5038/1827-806X.49.3.2337
Ben-Dov, E., Kramarsky-Winter, E., and Kushmaro, A. (2009). An in situ method for cultivating microorganisms using a double encapsulation technique. FEMS Microbiol. 68, 363–371. doi: 10.1111/j.1574-6941.2009.00682.x
Berdy, B., Spoering, A. L., Ling, L. L., and Epstein, S. S. (2017). In situ cultivation of previously uncultivable microorganisms using the ichip. Nat. Protoc. 12, 2232–2242. doi: 10.1038/nprot.2017.074
Bhuiyan, M. N. I., Takai, R., Mitsuhashi, S., Shigetomi, K., Tanaka, Y., Kamagata, Y., et al. (2016). Zincmethylphyrins and coproporphyrins, novel growth factors released by Sphingopyxis sp., enable laboratory cultivation of previously uncultured Leucobacter sp. through interspecies mutualism. J. Antibiot. 69, 97–103. doi: 10.1038/ja.2015.87
Bigger, J. (1944). Treatment of staphyloeoeeal infections with penicillin by intermittent sterilisation. Lancet. 244, 497–500. doi: 10.1016/S0140-6736(00)74210-3
Biosca, E. G., Amaro, C., Marco-Noales, E., and Oliver, J. D. (1996). Effect of low temperature on starvation-survival of the eel pathogen Vibrio vulnificus biotype 2. Appl. Environ. Microbiol. 62, 450–455. doi: 10.1128/aem.62.2.450-455.1996
Bochu, W., Lanchun, S., Jing, Z., Yuanyuan, Y., and Yanhong, Y. (2003). The influence of Ca2+ on the proliferation of S. cerevisiae and low ultrasonic on the concentration of Ca2+ in the S. cerevisiae cells. Colloids Surf. B. Biointerfaces 32, 35–42. doi: 10.1016/S0927-7765(03)00129-2
Bogosian, G., Aardema, N. D., Bourneuf, E. V., Morris, P. J., and O'Neil, J. P. (2000). Recovery of hydrogen peroxide-sensitive culturable cells of Vibrio vulnificus gives the appearance of resuscitation from a viable but nonculturable state. J. Bacteriol. 182, 5070–5075. doi: 10.1128/JB.182.18.5070-5075.2000
Boilattabi, N., Barrassi, L., Bouanane-Darenfed, A., and La Scola, B. (2021). Isolation and identification of legionella spp. from hot spring water in Algeria by culture and molecular methods. J. Appl. Microbiol. 130, 1394–1400. doi: 10.1111/jam.14871
Bollmann, A., Lewis, K., and Epstein, S. S. (2007). Incubation of environmental samples in a diffusion chamber increases the diversity of recovered isolates. Appl. Environ. Microbiol. 73, 6386–6390. doi: 10.1128/AEM.01309-07
Bomar, L., Maltz, M., Colston, S., and Graf, J. (2011). Directed culturing of microorganisms using metatranscriptomics. MBio 2, e00012–e00011. doi: 10.1128/mBio.00012-11
Börner, R. A., Aliaga, M. T. A., and Mattiasson, B. (2013). Microcultivation of anaerobic bacteria single cells entrapped in alginate microbeads. Biotechnol. Lett. 35, 397–405. doi: 10.1007/s10529-012-1094-1
Brock, T. D. (1967). Life at high temperatures: evolutionary, ecological, and biochemical significance of organisms living in hot springs is discussed. Science 158, 1012–1019. doi: 10.1126/science.158.3804.1012
Brock, T. D., and Freeze, H. (1969). Thermus aquaticus gen. n. and sp. n., a nonsporulating extreme thermophile. J. Bacteriol. 98, 289–297. doi: 10.1128/jb.98.1.289-297.1969
Bruns, A., Cypionka, H., and Overmann, J. (2002). Cyclic AMP and acyl homoserine lactones increase the cultivation efficiency of heterotrophic bacteria from the Central Baltic Sea. Appl. Environ. Microbiol. 68, 3978–3987. doi: 10.1128/AEM.68.8.3978-3987.2002
Bruns, A., Nübel, U., Cypionka, H., and Overmann, J. (2003). Effect of signal compounds and incubation conditions on the culturability of freshwater bacterioplankton. Appl. Environ. Microbiol. 69, 1980–1989. doi: 10.1128/AEM.69.4.1980-1989.2003
Buerger, S., Spoering, A., Gavrish, E., Leslin, C., Ling, L., and Epstein, S. S. (2012). Microbial scout hypothesis, stochastic exit from dormancy, and the nature of slow growers. Appl. Environ. Microbiol. 78, 3221–3228. doi: 10.1128/AEM.07307-11
Burmeister, A., Hilgers, F., Langner, A., Westerwalbesloh, C., Kerkhoff, Y., Tenhaef, N., et al. (2019). A microfluidic co-cultivation platform to investigate microbial interactions at defined microenvironments. Lab Chip 19, 98–110. doi: 10.1039/C8LC00977E
Carini, P., Steindler, L., Beszteri, S., and Giovannoni, S. J. (2013). Nutrient requirements for growth of the extreme oligotroph ‘Candidatus Pelagibacter ubique’ HTCC1062 on a defined medium. ISME J. 7, 592–602. doi: 10.1038/ismej.2012.122
Carvalho, G., Balestrino, D., Forestier, C., and Mathias, J.-D. (2018). How do environment-dependent switching rates between susceptible and persister cells affect the dynamics of biofilms faced with antibiotics? NPJ Biofilms Microbiomes 4:6. doi: 10.1038/s41522-018-0049-2
Castelle, C. J., and Banfield, J. F. (2018). Major new microbial groups expand diversity and alter our understanding of the tree of life. Cells 172, 1181–1197. doi: 10.1016/j.cell.2018.02.016
Chang, C. B., Wilking, J. N., Kim, S. H., Shum, H. C., and Weitz, D. A. (2015). Monodisperse emulsion drop microenvironments for bacterial biofilm growth. Small 11, 3954–3961. doi: 10.1002/smll.201403125
Chaudhary, D. K., Khulan, A., and Kim, J. (2019). Development of a novel cultivation technique for uncultured soil bacteria. Sci. Rep. 9:6666. doi: 10.1038/s41598-019-43182-x
Chaudhary, D. K., and Kim, J. (2019). Experimental setup for a diffusion bioreactor to isolate unculturable soil bacteria. Bio Protoc. 9:e3388. doi: 10.21769/BioProtoc.3388
Cho, J.-C., and Giovannoni, S. J. (2004). Cultivation and growth characteristics of a diverse group of oligotrophic marine Gammaproteobacteria. Appl. Environ. Microbiol. 70, 432–440. doi: 10.1128/AEM.70.1.432-440.2004
Christensen, B. T. (1992). “Physical fractionation of soil and organic matter in primary particle size and density separates” in Advances in soil science. ed. B. A. Stewart (New York, NY: Springer), 1–90.
Christensen, B. T. (2001). Physical fractionation of soil and structural and functional complexity in organic matter turnover. Eur. J. Soil Sci. 52, 345–353. doi: 10.1046/j.1365-2389.2001.00417.x
Cohen-Gonsaud, M., Barthe, P., Bagnéris, C., Henderson, B., Ward, J., Roumestand, C., et al. (2005). The structure of a resuscitation-promoting factor domain from mycobacterium tuberculosis shows homology to lysozymes. Nat. Struct. 12, 270–273. doi: 10.1038/nsmb905
Combet-Blanc, Y., Kalamba, K. K., and Kergoat, P. Y. (1995). Effect of pH on bacillus thermoamylovorans growth and glucose fermentation. Appl. Environ. Microbiol. 61, 656–659. doi: 10.1128/aem.61.2.656-659.1995
Connon, S. A., and Giovannoni, S. J. (2002). High-throughput methods for culturing microorganisms in very-low-nutrient media yield diverse new marine isolates. Appl. Environ. Microbiol. 68, 3878–3885. doi: 10.1128/AEM.68.8.3878-3885.2002
Costa, O. Y. A., Oguejiofor, C., Zühlke, D., Barreto, C. C., Wünsche, C., Riedel, K., et al. (2020). Impact of different trace elements on the growth and proteome of two strains of Granulicella, class "Acidobacteriia". Front. Microbiol. 11:1227. doi: 10.3389/fmicb.2020.01227
Crane, K. W., and Grover, J. P. (2010). Coexistence of mixotrophs, autotrophs, and heterotrophs in planktonic microbial communities. J. Theor. Biol. 262, 517–527. doi: 10.1016/j.jtbi.2009.10.027
Cross, K. L., Campbell, J. H., Balachandran, M., Campbell, A. G., Cooper, C. J., Griffen, A., et al. (2019). Targeted isolation and cultivation of uncultivated bacteria by reverse genomics. Nat. Biotechnol. 37, 1314–1321. doi: 10.1038/s41587-019-0260-6
Das, N., Triparthi, N., Basu, S., Bose, C., Maitra, S., and Khurana, S. (2015). Progress in the development of gelling agents for improved culturability of microorganisms. Front. Microbiol. 6:698. doi: 10.3389/fmicb.2015.00698
Davis, K. E., Joseph, S. J., and Janssen, P. H. (2005). Effects of growth medium, inoculum size, and incubation time on culturability and isolation of soil bacteria. Appl. Environ. Microbiol. 71, 826–834. doi: 10.1128/AEM.71.2.826-834.2005
del Mar, L. M., Pierobon, S., Tafi, M. C., Signoretto, C., and Canepari, P. (2000). mRNA detection by reverse transcription-PCR for monitoring viability over time in an enterococcus faecalis viable but nonculturable population maintained in a laboratory microcosm. Appl. Environ. Microbiol. 66, 4564–4567. doi: 10.1128/AEM.66.10.4564-4567.2000
Dellagnezze, B. M., Vasconcellos, S. P., Melo, I. S., Santos Neto, E. V., and Oliveira, V. M. (2016). Evaluation of bacterial diversity recovered from petroleum samples using different physical matrices. Braz. J. Microbiol. 47, 712–723. doi: 10.1016/j.bjm.2016.04.004
Dong, L., Chen, D.-W., Liu, S.-J., and Du, W. (2016). Automated chemotactic sorting and single-cell cultivation of microbes using droplet microfluidics. Sci. Rep. 6:24192. doi: 10.1038/srep24192
Dorofeev, A. G., Grigor’eva, N. V., Kozlov, M. N., Kevbrina, M. V., Aseeva, V. G., and Nikolaev, Y. A. (2014). Approaches to cultivation of “nonculturable” bacteria: cyclic cultures. Microbiology 83, 450–461. doi: 10.1134/S0026261714050087
Dorofeev, A. G., Nikolaev, Y. A., Mardanov, A. V., and Pimenov, N. V. (2019). Cyclic metabolism as a mode of microbial existence. Microbiology 88, 402–415. doi: 10.1134/S0026261719040052
Droce, A., Sørensen, J. L., Giese, H., and Sondergaard, T. E. (2013). Glass bead cultivation of fungi: combining the best of liquid and agar media. J. Microbiol. Methods 94, 343–346. doi: 10.1016/j.mimet.2013.07.005
Du, M., Chen, J., Zhang, X., Li, A., Li, Y., and Wang, Y. (2007). Retention of virulence in a viable but nonculturable Edwardsiella tarda isolate. Appl. Environ. Microbiol. 73, 1349–1354. doi: 10.1128/AEM.02243-06
Dworkin, J., and Shah, I. M. (2010). Exit from dormancy in microbial organisms. Nat. Rev. Microbiol. 8, 890–896. doi: 10.1038/nrmicro2453
Eichorst, S. A., Kuske, C. R., and Schmidt, T. M. (2011). Influence of plant polymers on the distribution and cultivation of bacteria in the phylum Acidobacteria. Appl. Environ. Microbiol. 77, 586–596. doi: 10.1128/AEM.01080-10
Eilers, H., Pernthaler, J., Peplies, J., Glöckner, F. O., Gerdts, G., and Amann, R. (2001). Isolation of novel pelagic bacteria from the German bight and their seasonal contributions to surface picoplankton. Appl. Environ. Microbiol. 67, 5134–5142. doi: 10.1128/AEM.67.11.5134-5142.2001
Epstein, S. S. (2013). The phenomenon of microbial uncultivability. Curr. Opin. Microbiol. 16, 636–642. doi: 10.1016/j.mib.2013.08.003
Espina, L. (2020). An approach to increase the success rate of cultivation of soil bacteria based on fluorescence-activated cell sorting. PLoS One 15:e0237748. doi: 10.1371/journal.pone.0237748
Esposito, F. P., Ingham, C. J., Hurtado-Ortiz, R., Bizet, C., Tasdemir, D., and de Pascale, D. (2018). Isolation by miniaturized culture chip of an Antarctic bacterium Aequorivita sp. with antimicrobial and anthelmintic activity. Biotechnol. Rep. 20:e00281. doi: 10.1016/j.btre.2018.e00281
Estevinho, L. M., Combarros-Fuertes, P., and Paula, V. B. (2020). Recent advances in applied microbiology: editorial. Microorganisms 8:1364. doi: 10.3390/microorganisms8091364
Eun, Y. J., Utada, A. S., Copeland, M. F., Takeuchi, S., and Weibel, D. B. (2011). Encapsulating bacteria in agarose microparticles using microfluidics for high-throughput cell analysis and isolation. ACS Chem. Biol. 6, 260–266. doi: 10.1021/cb100336p
Faust, K., and Raes, J. (2012). Microbial interactions: from networks to models. Nat. Rev. Microbiol. 10, 538–550. doi: 10.1038/nrmicro2832
Feng, Y., Grogan, P., Caporaso, J. G., Zhang, H., Lin, X., Knight, R., et al. (2014). pH is a good predictor of the distribution of anoxygenic purple phototrophic bacteria in Arctic soils. Soil Biol. Biochem. 74, 193–200. doi: 10.1016/j.soilbio.2014.03.014
Ferrari, B. C., Binnerup, S. J., and Gillings, M. (2005). Microcolony cultivation on a soil substrate membrane system selects for previously uncultured soil bacteria. Appl. Environ. Microbiol. 71, 8714–8720. doi: 10.1128/AEM.71.12.8714-8720.2005
Fierer, N., Nemergut, D., Knight, R., and Craine, J. M. (2010). Changes through time: integrating microorganisms into the study of succession. Res. Microbiol. 161, 635–642. doi: 10.1016/j.resmic.2010.06.002
Filiatrault, M. J. (2011). Progress in prokaryotic transcriptomics. Curr. Opin. Microbiol. 14, 579–586. doi: 10.1016/j.mib.2011.07.023
Flemming, H.-C., and Wingender, J. (2010). The biofilm matrix. Nat. Rev. Microbiol. 8, 623–633. doi: 10.1038/nrmicro2415
Frimat, J.-P., Becker, M., Chiang, Y.-Y., Marggraf, U., Janasek, D., Hengstler, J. G., et al. (2011). A microfluidic array with cellular valving for single cell co-culture. Lab Chip 11, 231–237. doi: 10.1039/C0LC00172D
Gao, W., Navarroli, D., Naimark, J., Zhang, W., Chao, S.-h., and Meldrum, D. R. (2013). Microbe observation and cultivation array (MOCA) for cultivating and analyzing environmental microbiota. Microbiome 1:4. doi: 10.1186/2049-2618-1-4
Gavrish, E., Bollmann, A., Epstein, S., and Lewis, K. (2008). A trap for in situ cultivation of filamentous actinobacteria. J. Microbiol. Methods 72, 257–262. doi: 10.1016/j.mimet.2007.12.009
Ge, Z., Girguis, P. R., and Buie, C. R. (2016). Nanoporous microscale microbial incubators. Lab Chip 16, 480–488. doi: 10.1039/C5LC00978B
Gerna, D., Clara, D., Allwardt, D., Mitter, B., and Roach, T. (2022). Tailored media are key to unlocking the diversity of endophytic bacteria in distinct compartments of germinating seeds. Microbiol. Spectr. 10, e00172–e00122. doi: 10.1128/spectrum.00172-22
Giagnoni, L., Arenella, M., Galardi, E., Nannipieri, P., and Renella, G. (2018). Bacterial culturability and the viable but non-culturable (VBNC) state studied by a proteomic approach using an artificial soil. Soil Biol. Biochem. 118, 51–58. doi: 10.1016/j.soilbio.2017.12.004
Gich, F., Janys, M. A., König, M., and Overmann, J. (2012). Enrichment of previously uncultured bacteria from natural complex communities by adhesion to solid surfaces. Environ. Microbiol. 14, 2984–2997. doi: 10.1111/j.1462-2920.2012.02868.x
Gilbert, J. A., Blaser, M. J., Caporaso, J. G., Jansson, J. K., Lynch, S. V., and Knight, R. (2018). Current understanding of the human microbiome. Nat. Med. 24, 392–400. doi: 10.1038/nm.4517
Goers, L., Freemont, P., and Polizzi, K. M. (2014). Co-culture systems and technologies: taking synthetic biology to the next level. J. R. Soc. Interface 11:20140065. doi: 10.1098/rsif.2014.0065
Graham, E. B., Knelman, J. E., Schindlbacher, A., Siciliano, S., Breulmann, M., Yannarell, A., et al. (2016). Microbes as engines of ecosystem function: when does community structure enhance predictions of ecosystem processes? Front. Microbiol. 7:214. doi: 10.3389/fmicb.2016.00214
Greub, G. (2012). Culturomics: a new approach to study the human microbiome. Clin. Microbiol. Infect. 18, 1157–1159. doi: 10.1111/1469-0691.12032
Grover, S. C., Skirtach, A. G., Gauthier, R. C., and Grover, C. P. (2001). Automated single-cell sorting system based on optical trapping. J. Biomed. Opt. 6, 14–22. doi: 10.1117/1.1333676
Gurusinghe, S., Brooks, T. L., Barrow, R. A., Zhu, X., Thotagamuwa, A., Dennis, P. G., et al. (2019). Technologies for the selection, culture and metabolic profiling of unique rhizosphere microorganisms for natural product discovery. Molecules 24:1955. doi: 10.3390/molecules24101955
Gutierrez, T., Biddle, J. F., Teske, A., and Aitken, M. D. (2015). Cultivation-dependent and cultivation-independent characterization of hydrocarbon-degrading bacteria in Guaymas Basin sediments. Front. Microbiol. 6:695. doi: 10.3389/fmicb.2015.00695
Gutierrez, T., Singleton, D. R., Berry, D., Yang, T., Aitken, M. D., and Teske, A. (2013). Hydrocarbon-degrading bacteria enriched by the Deepwater horizon oil spill identified by cultivation and DNA-SIP. ISME J. 7, 2091–2104. doi: 10.1038/ismej.2013.98
Gutleben, J., Loureiro, C., Ramírez Romero, L. A., Shetty, S., Wijffels, R. H., Smidt, H., et al. (2020). Cultivation of bacteria from Aplysina aerophoba: effects of oxygen and nutrient gradients. Front. Microbiol. 11:175. doi: 10.3389/fmicb.2020.00175
Handelsman, J., Rondon, M. R., Brady, S. F., Clardy, J., and Goodman, R. M. (1998). Molecular biological access to the chemistry of unknown soil microbes: a new frontier for natural products. Chem. Biol. 5, R245–R249. doi: 10.1016/S1074-5521(98)90108-9
Harriott, M. M. (2019). “Biofilms and antibiotics” in Reference module in biomedical sciences (Elsevier), 1–11.
Hedlund, B. P., Chuvochina, M., Hugenholtz, P., Konstantinidis, K. T., Murray, A. E., Palmer, M., et al. (2022). SeqCode: a nomenclatural code for prokaryotes described from sequence data. Nat. Microbiol. 7, 1702–1708. doi: 10.1038/s41564-022-01214-9
Hemkemeyer, M., Dohrmann, A. B., Christensen, B. T., and Tebbe, C. C. (2018). Bacterial preferences for specific soil particle size fractions revealed by community analyses. Front. Microbiol. 9:149. doi: 10.3389/fmicb.2018.00149
Hemme, C. L., Deng, Y., Gentry, T. J., Fields, M. W., Wu, L., Barua, S., et al. (2010). Metagenomic insights into evolution of a heavy metal-contaminated groundwater microbial community. ISME J. 4, 660–672. doi: 10.1038/ismej.2009.154
Hett, E. C., Chao, M. C., Deng, L. L., and Rubin, E. J. (2008). A mycobacterial enzyme essential for cell division synergizes with resuscitation-promoting factor. PLoS Pathog. 4:e1000001. doi: 10.1371/journal.ppat.1000001
Ho, A., Di Lonardo, D. P., and Bodelier, P. L. (2017). Revisiting life strategy concepts in environmental microbial ecology. FEMS Microbiol. 93:fix0006. doi: 10.1093/femsec/fix0006
Hobby, G. L., Meyer, K., and Chaffee, E. (1942). Observations on the mechanism of action of penicillin. Proc. Soc. Exp. Biol. Med. 50, 281–285. doi: 10.3181/00379727-50-13773
Hu, B., Xu, B., Yun, J., Wang, J., Xie, B., Li, C., et al. (2020). High-throughput single-cell cultivation reveals the underexplored rare biosphere in deep-sea sediments along the southwest Indian ridge. Lab Chip 20, 363–372. doi: 10.1039/C9LC00761J
Huang, G., Chen, S., Dai, C., Sun, L., Sun, W., Tang, Y., et al. (2017). Effects of ultrasound on microbial growth and enzyme activity. Ultrason. Sonochem. 37, 144–149. doi: 10.1016/j.ultsonch.2016.12.018
Huang, X., Li, P., Zhou, M., Li, Y., Ou, X., Chen, P., et al. (2021). A high-throughput ultrasonic spraying inoculation method promotes colony cultivation of rare microbial species. Environ. Microbiol. 23, 1275–1285. doi: 10.1111/1462-2920.15386
Hugenholtz, P., Pitulle, C., Hershberger, K. L., and Pace, N. R. (1998). Novel division level bacterial diversity in a Yellowstone hot spring. J. Bacteriol. 180, 366–376. doi: 10.1128/JB.180.2.366-376.1998
Imachi, H., Nobu, M. K., Nakahara, N., Morono, Y., Ogawara, M., Takaki, Y., et al. (2020). Isolation of an archaeon at the prokaryote–eukaryote interface. Nature 577, 519–525. doi: 10.1038/s41586-019-1916-6
Imazaki, I., and Kobori, Y. (2010). Improving the culturability of freshwater bacteria using FW70, a low-nutrient solid medium amended with sodium pyruvate. Can. J. Microbiol. 56, 333–341. doi: 10.1139/W10-019
Ingham, C. J., Sprenkels, A., Bomer, J., Molenaar, D., van den Berg, A., van Hylckama Vlieg, J. E., et al. (2007). The micro-petri dish, a million-well growth chip for the culture and high-throughput screening of microorganisms. Proc. Natl. Acad. Sci. U. S. A. 104, 18217–18222. doi: 10.1073/pnas.0701693104
Inglis, T. J. J., and Sagripanti, J.-L. (2006). Environmental factors that affect the survival and persistence of Burkholderia pseudomallei. Appl. Environ. Microbiol. 72, 6865–6875. doi: 10.1128/AEM.01036-06
Jacoby, R., Peukert, M., Succurro, A., Koprivova, A., and Kopriva, S. (2017). The role of soil microorganisms in plant mineral nutrition-current knowledge and future directions. Front. Plant Sci. 8:1617. doi: 10.3389/fpls.2017.01617
Janssen, P. H., Yates, P. S., Grinton, B. E., Taylor, P. M., and Sait, M. (2002). Improved culturability of soil bacteria and isolation in pure culture of novel members of the divisions Acidobacteria, Actinobacteria, Proteobacteria, and Verrucomicrobia. Appl. Environ. Microbiol. 68, 2391–2396. doi: 10.1128/AEM.68.5.2391-2396.2002
Jeon, C. O., Park, W., Ghiorse, W. C., and Madsen, E. L. (2004). Polaromonas naphthalenivorans sp. nov., a naphthalene-degrading bacterium from naphthalene-contaminated sediment. Int. J. Syst. Evol. Microbiol. 54, 93–97. doi: 10.1099/ijs.0.02636-0
Jeon, C. O., Park, W., Padmanabhan, P., DeRito, C., Snape, J. R., and Madsen, E. L. (2003). Discovery of a bacterium, with distinctive dioxygenase, that is responsible for in situ biodegradation in contaminated sediment. Proc. Natl. Acad. Sci. U. S. A. 100, 13591–13596. doi: 10.1073/pnas.1735529100
Ji, S., Zhao, R., Yin, Q., Zhao, Y., Liu, C., Xiao, T., et al. (2012). Gel microbead cultivation with a subenrichment procedure can yield better bacterial cultivability from a seawater sample than standard plating method. J. Ocean Univ. China 11, 45–51. doi: 10.1007/s11802-012-1869-y
Jian, X., Guo, X., Wang, J., Tan, Z. L., Xing, X., Wang, L., et al. (2020). Microbial microdroplet culture system (MMC): an integrated platform for automated, high-throughput microbial cultivation and adaptive evolution. Biotechnol. Bioeng. 117, 1724–1737. doi: 10.1002/bit.27327
Jiang, C.-Y., Dong, L., Zhao, J.-K., Hu, X., Shen, C., Qiao, Y., et al. (2016). High throughput single-cell cultivation on microfluidic streak plates. Appl. Environ. Microbiol. 85, 2210–2218. doi: 10.1128/AEM.03588-15
Jin, Q., and Kirk, M. F. (2018). pH as a primary control in environmental microbiology: 1 Thermodynamic perspective. Front. Environ. Sci. 6:21. doi: 10.3389/fenvs.2018.00021
Jin, Z., Nie, M., Hu, R., Zhao, T., Xu, J., Chen, D., et al. (2018). Dynamic sessile-droplet habitats for controllable cultivation of bacterial biofilm. Small 14:e1800658. doi: 10.1002/smll.201800658
Jung, D., Aoi, Y., and Epstein, S. S. (2016). In situ cultivation allows for recovery of bacterial types competitive in their natural environment. Microbes Environ. 31, 456–459. doi: 10.1264/jsme2.ME16079
Jung, D., Seo, E.-Y., Epstein, S. S., Joung, Y., Yim, J. H., Lee, H. K., et al. (2013). A new method for microbial cultivation and its application to bacterial community analysis in Buus Nuur, Mongolia. Fundam. Appl. Limnol. 182, 171–181. doi: 10.1127/1863-9135/2013/0391
Jung, D., Seo, E. Y., Owen, J. S., Aoi, Y., Yong, S., Lavrentyeva, E. V., et al. (2018). Application of the filter plate microbial trap (FPMT), for cultivating thermophilic bacteria from thermal springs in Barguzin area, eastern Baikal, Russia. Biosci. Biotechnol. Biochem. 82, 1624–1632. doi: 10.1080/09168451.2018.1482194
Kaeberlein, T., Lewis, K., and Epstein, S. S. (2002). Isolating "uncultivable" microorganisms in pure culture in a simulated natural environment. Science 296, 1127–1129. doi: 10.1126/science.1070633
Kakumanu, M. L., and Williams, M. A. (2012). Soil diffusion system enriches the growth of diverse and previously uncultivated bacterial taxa. Soil Sci. Soc. Am. J. 76, 463–474. doi: 10.2136/sssaj2011.0227
Kapinusova, G., Jani, K., Smrhova, T., Pajer, P., Jarosova, I., Suman, J., et al. (2022). Culturomics of bacteria from radon-saturated water of the world’s oldest radium mine. Microbiol. Spectr. 10, e01995–e01922. doi: 10.1128/spectrum.01995-22
Kaprelyants, A. S., Mukamolova, G. V., and Kell, D. B. (1994). Estimation of dormant Micrococcus luteus cells by penicillin lysis and by resuscitation in cell-free spent culture medium at high dilution. FEMS Microbiol. Lett. 115, 347–352. doi: 10.1111/j.1574-6968.1994.tb06662.x
Karimi, E., Keller-Costa, T., Slaby, B. M., Cox, C. J., da Rocha, U. N., Hentschel, U., et al. (2019). Genomic blueprints of sponge-prokaryote symbiosis are shared by low abundant and cultivatable Alphaproteobacteria. Sci. Rep. 9:1999. doi: 10.1038/s41598-019-38737-x
Karnachuk, O. V., Lukina, A. P., Kadnikov, V. V., Sherbakova, V. A., Beletsky, A. V., Mardanov, A. V., et al. (2020). Targeted isolation based on metagenome-assembled genomes reveals a phylogenetically distinct group of thermophilic spirochetes from deep biosphere. Environ. Microbiol. 23, 3585–3598. doi: 10.1111/1462-2920.15218
Kato, S., Terashima, M., Yama, A., Sato, M., Kitagawa, W., Kawasaki, K., et al. (2020). Improved isolation of uncultured anaerobic bacteria using medium prepared with separate sterilization of agar and phosphate. Microbes Environ. 35:n/a. doi: 10.1264/jsme2.ME19060
Kato, S., Yamagishi, A., Daimon, S., Kawasaki, K., Tamaki, H., Kitagawa, W., et al. (2018). Isolation of previously uncultured slow-growing bacteria by using a simple modification in the preparation of agar media. Appl. Environ. Microbiol. 84, e00807–e00818. doi: 10.1128/AEM.00807-18
Keep, N. H., Ward, J. M., Cohen-Gonsaud, M., and Henderson, B. (2006). Wake up! Peptidoglycan lysis and bacterial non-growth states. Trends Microbiol. 14, 271–276. doi: 10.1016/j.tim.2006.04.003
Kell, D. B., Kaprelyants, A. S., Weichart, D. H., Harwood, C. R., and Barer, M. R. (1998). Viability and activity in readily culturable bacteria: a review and discussion of the practical issues. Antonie Van Leeuwenhoek 73, 169–187. doi: 10.1023/A:1000664013047
Knobloch, S., Jóhannsson, R., and Marteinsson, V. (2019). Co-cultivation of the marine sponge Halichondria panicea and its associated microorganisms. Sci. Rep. 9:10403. doi: 10.1038/s41598-019-46904-3
Koch, A. L. (1971). “The adaptive responses of Escherichia coli to a feast and famine existence” in Advances in microbial physiology. eds. A. H. Rose and J. F. Wilkinson, vol. 6 (Academic Press), 147–217.
Konopka, A. (2009). What is microbial community ecology? ISME J. 3, 1223–1230. doi: 10.1038/ismej.2009.88
Lagier, J. C., Dubourg, G., Million, M., Cadoret, F., Bilen, M., Fenollar, F., et al. (2018). Culturing the human microbiota and culturomics. Nat. Rev. Microbiol. 16, 540–550. doi: 10.1038/s41579-018-0041-0
Lane, D. J., Pace, B., Olsen, G. J., Stahl, D. A., Sogin, M. L., and Pace, N. R. (1985). Rapid determination of 16S ribosomal RNA sequences for phylogenetic analyses. Proc. Natl. Acad. Sci. U. S. A. 82, 6955–6959. doi: 10.1073/pnas.82.20.6955
Larke-Mejía, N. L., Crombie, A. T., Pratscher, J., McGenity, T. J., and Murrell, J. C. (2019). Novel isoprene-degrading Proteobacteria from soil and leaves identified by cultivation and metagenomics analysis of stable isotope probing experiments. Front. Microbiol. 10:2700. doi: 10.3389/fmicb.2019.02700
Lauber, C. L., Hamady, M., Knight, R., and Fierer, N. (2009). Pyrosequencing-based assessment of soil pH as a predictor of soil bacterial community structure at the continental scale. Appl. Environ. Microbiol. 75, 5111–5120. doi: 10.1128/AEM.00335-09
Lee, K. C., Dunfield, P. F., Morgan, X. C., Crowe, M. A., Houghton, K. M., Vyssotski, M., et al. (2011). Chthonomonas calidirosea gen. Nov., sp. nov., an aerobic, pigmented, thermophilic micro-organism of a novel bacterial class, Chthonomonadetes classis nov., of the newly described phylum Armatimonadetes originally designated candidate division OP10. Int. J. Syst. Evol. Microbiol. 61, 2482–2490. doi: 10.1099/ijs.0.027235-0
Lewis, K. (2010). Persister cells. Annu. Rev. Microbiol. 64, 357–372. doi: 10.1146/annurev.micro.112408.134306
Lewis, K., Epstein, S., D'Onofrio, A., and Ling, L. L. (2010). Uncultured microorganisms as a source of secondary metabolites. J. Antibiot. 63, 468–476. doi: 10.1038/ja.2010.87
Li, Y., Chen, J., Wang, Y., Ma, D., and Rui, W. (2020). The effects of the recombinant YeaZ of Vibrio harveyi on the resuscitation and growth of soil bacteria in extreme soil environment. PeerJ 8:e10342. doi: 10.7717/peerj.10342
Li, J., Luo, C., Zhang, D., Cai, X., Jiang, L., and Zhang, G. (2019). Stable-isotope probing-enabled cultivation of the indigenous bacterium Ralstonia sp. strain M1, capable of degrading phenanthrene and biphenyl in industrial wastewater. Appl. Environ. Microbiol. 85, e00511–e00519. doi: 10.1128/aem.00511-19
Li, L., Mendis, N., Trigui, H., Oliver, J. D., and Faucher, S. P. (2014). The importance of the viable but non-culturable state in human bacterial pathogens. Front. Microbiol. 5:258. doi: 10.3389/fmicb.2014.00258
Li, B., Yang, Y., Ma, L., Ju, F., Guo, F., Tiedje, J. M., et al. (2015). Metagenomic and network analysis reveal wide distribution and co-occurrence of environmental antibiotic resistance genes. ISME J. 9, 2490–2502. doi: 10.1038/ismej.2015.59
Ling, L. L., Schneider, T., Peoples, A. J., Spoering, A. L., Engels, I., Conlon, B. P., et al. (2015). A new antibiotic kills pathogens without detectable resistance. Nature 517, 455–459. doi: 10.1038/nature14098
Liu, S., Moon, C. D., Zheng, N., Huws, S., Zhao, S., and Wang, J. (2022). Opportunities and challenges of using metagenomic data to bring uncultured microbes into cultivation. Microbiome 10:76. doi: 10.1186/s40168-022-01272-5
Liu, X., Wang, M., Nie, Y., and Wu, X. L. (2021). Isolation chip increases culturable bacterial diversity and reduces cultivation bias. Curr. Microbiol. 78, 2025–2032. doi: 10.1007/s00284-021-02474-0
Lloyd, K. G., Steen, A. D., Ladau, J., Yin, J., Crosby, L., and Neufeld, J. D. (2018). Phylogenetically novel uncultured microbial cells dominate earth microbiomes. mSystems 3, e00055–e00018. doi: 10.1128/mSystems.00055-18
Lopez Marin, M. A., Strejcek, M., Junkova, P., Suman, J., Santrucek, J., and Uhlik, O. (2021). Exploring the potential of Micrococcus luteus culture supernatant with resuscitation-promoting factor for enhancing the culturability of soil bacteria. Front. Microbiol. 12:1715. doi: 10.3389/fmicb.2021.685263
Lopez Marin, M. A., Suman, J., Jani, K., Ulbrich, P., Cajthaml, T., Filipova, A., et al. (2023). Solicola gregarius gen. nov., sp. nov., a soil actinobacterium isolated after enhanced cultivation with Micrococcus luteus culture supernatant. Int J Syst Evol Microbiol. 73. doi: 10.1099/ijsem.0.005678
Lopez Marin, M. A., Suman, J., Jani, K., Ulbrich, P., Cajthaml, T., Pajer, P., et al. (2022). Pedomonas mirosovicensis gen. Nov., sp. nov., a bacterium isolated from soil with the aid of Micrococcus luteus culture supernatant containing resuscitation-promoting factor. Int. J. Syst. Evol. Microbiol. 72. doi: 10.1099/ijsem.0.005467
Louca, S., Mazel, F., Doebeli, M., and Parfrey, L. W. (2019). A census-based estimate of Earth's bacterial and archaeal diversity. PLoS Biol. 17:e3000106. doi: 10.1371/journal.pbio.3000106
Lynch, M. D., Bartram, A. K., and Neufeld, J. D. (2012). Targeted recovery of novel phylogenetic diversity from next-generation sequence data. ISME J. 6:2067. doi: 10.1038/ismej.2012.50
Ma, L., Datta, S. S., Karymov, M. A., Pan, Q., Begolo, S., and Ismagilov, R. F. (2014). Individually addressable arrays of replica microbial cultures enabled by splitting SlipChips. Integr. Biol. (Camb) 6, 796–805. doi: 10.1039/C4IB00109E
Manivasagan, P., Kang, K. H., Sivakumar, K., Li-Chan, E. C., Oh, H. M., and Kim, S. K. (2014). Marine actinobacteria: an important source of bioactive natural products. Environ. Toxicol. Pharmacol. 38, 172–188. doi: 10.1016/j.etap.2014.05.014
Marchesi, J. R., and Ravel, J. (2015). The vocabulary of microbiome research: a proposal. Microbiome 3:31. doi: 10.1186/s40168-015-0094-5
Maza, F., Maldonado, J., Vásquez-Dean, J., Mandakovic, D., Gaete, A., Cambiazo, V., et al. (2019). Soil bacterial communities from the Chilean Andean highlands: taxonomic composition and culturability. Front. Bioeng. Biotechnol. 7:10. doi: 10.3389/fbioe.2019.00010
McLain, J. E., Cytryn, E., Durso, L. M., and Young, S. (2016). Culture-based methods for detection of antibiotic resistance in agroecosystems: advantages, challenges, and gaps in knowledge. J. Environ. Qual. 45, 432–440. doi: 10.2134/jeq2015.06.0317
Mehetre, G., Shah, M., Dastager, S. G., and Dharne, M. S. (2018). Untapped bacterial diversity and metabolic potential within Unkeshwar hot springs, India. Arch. Microbiol. 200, 753–770. doi: 10.1007/s00203-018-1484-4
Meyer, O. (1994). “Functional groups of microorganisms” in Biodiversity and ecosystem function. eds. Schulze, Ernst-Detlef, Mooney and A. Harold (Berlin, Heidelberg: Springer), 67–96.
Miller, C. N., Khan, M., Ahmed, S. A., Kota, K., Panchal, R. G., and Hale, M. L. (2020). Development of a Coxiella burnetii culture method for high-throughput assay to identify host-directed therapeutics. J. Microbiol. Methods 169:105813. doi: 10.1016/j.mimet.2019.105813
Mills, A. L. (2003). Keeping in touch: microbial life on soil particle surfaces. Adv. Agron. 78, 2–45. doi: 10.1016/S0065-2113(02)78001-2
Mirzaie, A., Mehrabadi, J. F., Amirmozafari, N., and Nejadsattari, T. (2015). Isolation and characterization of a new gamma and UV radiation resistant bacterium from soil samples of an Iranian radioactive site and analysis of its pigment. Microbiology 84, 449–452. doi: 10.1134/S0026261715030133
Mishamandani, S., Gutierrez, T., and Aitken, M. D. (2014). DNA-based stable isotope probing coupled with cultivation methods implicates Methylophaga in hydrocarbon degradation. Front. Microbiol. 5:76. doi: 10.3389/fmicb.2014.00076
Molina-Menor, E., Gimeno-Valero, H., Pascual, J., Peretó, J., and Porcar, M. (2021). High culturable bacterial diversity from a european desert: the Tabernas desert. Front. Microbiol. 11:583120. doi: 10.3389/fmicb.2020.583120
Moran, N. A., and Bennett, G. M. (2014). The tiniest tiny genomes. Annu. Rev. Microbiol. 68, 195–215. doi: 10.1146/annurev-micro-091213-112901
Mori, K., Sunamura, M., Yanagawa, K., Ishibashi, J.-i., Miyoshi, Y., Iino, T., et al. (2008). First cultivation and ecological investigation of a bacterium affiliated with the candidate phylum OP5 from hot springs. Appl. Environ. Microbiol. 74, 6223–6229. doi: 10.1128/AEM.01351-08
Morrison, E. W., and Rettger, L. F. (1930). Bacterial spores I. a study in heat resistance and dormancy. J. Bacteriol. 20, 299–311. doi: 10.1128/jb.20.5.299-311.1930
Mukamolova, G. V., Kaprelyants, A. S., Kell, D. B., and Young, M. (2003). Adoption of the transiently non-culturable state-a bacterial survival strategy? Adv. Microb. Physiol. 47, 66–131. doi: 10.1016/S0065-2911(03)47002-1
Mukamolova, G. V., Kaprelyants, A. S., Young, D. I., Young, M., and Kell, D. B. (1998). A bacterial cytokine. Proc. Natl. Acad. Sci. U. S. A. 95, 8916–8921. doi: 10.1073/pnas.95.15.8916
Mukamolova, G. V., Murzin, A. G., Salina, E. G., Demina, G. R., Kell, D. B., Kaprelyants, A. S., et al. (2006). Muralytic activity of Micrococcus luteus Rpf and its relationship to physiological activity in promoting bacterial growth and resuscitation. Mol. Microbiol. 59, 84–98. doi: 10.1111/j.1365-2958.2005.04930.x
Mukamolova, G. V., Turapov, O. A., Young, D. I., Kaprelyants, A. S., Kell, D. B., and Young, M. (2002). A family of autocrine growth factors in mycobacterium tuberculosis. Mol. Microbiol. 46, 623–635. doi: 10.1046/j.1365-2958.2002.03184.x
Nguyen, L. D., Kalachová, L., Novotná, J., Holub, M., Kofroňová, O., Benada, O., et al. (2005). Cultivation system using glass beads immersed in liquid medium facilitates studies of Streptomyces differentiation. Appl. Environ. Microbiol. 71, 2848–2852. doi: 10.1128/AEM.71.6.2848-2852.2005
Nichols, D. (2007). Cultivation gives context to the microbial ecologist. FEMS Microbiol 60, 351–357. doi: 10.1111/j.1574-6941.2007.00332.x
Nichols, D., Cahoon, N., Trakhtenberg, E. M., Pham, L., Mehta, A., Belanger, A., et al. (2010). Use of ichip for high-throughput in situ cultivation of “uncultivable” microbial species. Appl. Environ. Microbiol. 76, 2445–2450. doi: 10.1128/AEM.01754-09
Nichols, D., Lewis, K., Orjala, J., Mo, S., Ortenberg, R., O'Connor, P., et al. (2008). Short peptide induces an “uncultivable” microorganism to grow in vitro. Appl. Environ. Microbiol. 74, 4889–4897. doi: 10.1128/AEM.00393-08
Nikitushkin, V. D., Demina, G. R., and Kaprelyants, A. S. (2016). Rpf proteins are the factors of reactivation of the dormant forms of Actinobacteria. Biochem. Mosc. 81, 1719–1734. doi: 10.1134/S0006297916130095
Nowrotek, M., Jałowiecki, Ł., Harnisz, M., and Płaza, G. A. (2019). Culturomics and metagenomics: in understanding of environmental resistome. Front. Environ. Sci. Eng. 13:40. doi: 10.1007/s11783-019-1121-8
Ohan, J., Pelle, B., Nath, P., Huang, J.-H., Hovde, B., Vuyisich, M., et al. (2019). High-throughput phenotyping of cell-to-cell interactions in gel microdroplet pico-cultures. BioTechniques 66, 218–224. doi: 10.2144/btn-2018-0124
Omsland, A., Cockrell, D. C., Howe, D., Fischer, E. R., Virtaneva, K., Sturdevant, D. E., et al. (2009). Host cell-free growth of the Q fever bacterium Coxiella burnetii. Proc. Natl. Acad. Sci. U. S. A. 106, 4430–4434. doi: 10.1073/pnas.0812074106
Ota, Y., Saito, K., Takagi, T., Matsukura, S., Morita, M., Tsuneda, S., et al. (2019). Fluorescent nucleic acid probe in droplets for bacterial sorting (FNAP-sort) as a high-throughput screening method for environmental bacteria with various growth rates. PLoS One 14:e0214533. doi: 10.1371/journal.pone.0214533
Palmer, M., Sutcliffe, I., Venter, S. N., and Hedlund, B. P. (2022). It is time for a new type of type to facilitate naming the microbial world. New Microbes New Infect. 47:100991. doi: 10.1016/j.nmni.2022.100991
Panda, A. K., Bisht, S. S., Rana, M., De Mandal, S., and Kumar, N. S. (2018). “Biotechnological potential of thermophilic Actinobacteria associated with hot springs” in New and future developments in microbial biotechnology and bioengineering. eds. Bhim Pratap Singh, Vijai Kumar Gupta, and Ajit Kumar Passari (Elsevier), 155–164.
Pande, S., and Kost, C. (2017). Bacterial unculturability and the formation of intercellular metabolic networks. Trends Microbiol. 25, 349–361. doi: 10.1016/j.tim.2017.02.015
Papik, J., Folkmanova, M., Polivkova, M., Suman, J., and Uhlik, O. (2020). The invisible life inside plants: deciphering the riddles of endophytic bacterial diversity. Biotechnol. Adv. 44:107614. doi: 10.1016/j.biotechadv.2020.107614
Parte, A. C., Carbasse, J. S., Meier-Kolthoff, J. P., Reimer, L. C., and Göker, M. (2020). List of prokaryotic names with standing in nomenclature (LPSN) moves to the DSMZ. Int. J. Syst. Evol. Microbiol. 70, 5607–5612. doi: 10.1099/ijsem.0.004332
Pascoal, F., Magalhães, C., and Costa, R. (2020). The link between the ecology of the prokaryotic rare biosphere and its biotechnological potential. Front. Microbiol. 11:231. doi: 10.3389/fmicb.2020.00231
Pascual, J., García-López, M., Carmona, C., Sousa Tda, S., de Pedro, N., Cautain, B., et al. (2014). Pseudomonas soli sp. nov., a novel producer of xantholysin congeners. Syst. Appl. Microbiol. 37, 412–416. doi: 10.1016/j.syapm.2014.07.003
Pascual, J., García-López, M., González, I., and Genilloud, O. (2017). Luteolibacter gellanilyticus sp. nov., a gellan-gum-degrading bacterium of the phylum Verrucomicrobia isolated from miniaturized diffusion chambers. Int. J. Syst. Evol. Microbiol. 67, 3951–3959. doi: 10.1099/ijsem.0.002227
Pathak, A., Jaswal, R., Xu, X., White, J. R., Edwards, B. 3rd, Hunt, J., et al. (2020). Characterization of bacterial and fungal assemblages from historically contaminated metalliferous soils using metagenomics coupled with diffusion chambers and microbial traps. Front. Microbiol. 11:1024. doi: 10.3389/fmicb.2020.01024
Pernthaler, J., Pernthaler, A., and Amann, R. (2003). Automated enumeration of groups of marine picoplankton after fluorescence in situ hybridization. Appl. Environ. Microbiol. 69, 2631–2637. doi: 10.1128/AEM.69.5.2631-2637.2003
Pinto, D., Almeida, V., Almeida Santos, M., and Chambel, L. (2011). Resuscitation of Escherichia coli VBNC cells depends on a variety of environmental or chemical stimuli. J. Appl. Microbiol. 110, 1601–1611. doi: 10.1111/j.1365-2672.2011.05016.x
Pinto, D., Santos, M. A., and Chambel, L. (2015). Thirty years of viable but nonculturable state research: unsolved molecular mechanisms. Crit. Rev. Microbiol. 41, 61–76. doi: 10.3109/1040841X.2013.794127
Pinto, D., São-José, C., Santos, M. A., and Chambel, L. (2013). Characterization of two resuscitation promoting factors of listeria monocytogenes. Microbiology 159, 1390–1401. doi: 10.1099/mic.0.067850-0
Power, J. F., Carere, C. R., Lee, C. K., Wakerley, G. L. J., Evans, D. W., Button, M., et al. (2018). Microbial biogeography of 925 geothermal springs in New Zealand. Nat. Commun. 9:2876. doi: 10.1038/s41467-018-05020-y
Pudasaini, S., Wilson, J., Ji, M., van Dorst, J., Snape, I., Palmer, A. S., et al. (2017). Microbial diversity of Browning peninsula, eastern Antarctica revealed using molecular and cultivation methods. Front. Microbiol. 8:591. doi: 10.3389/fmicb.2017.00591
Pulschen, A. A., Bendia, A. G., Fricker, A. D., Pellizari, V. H., Galante, D., and Rodrigues, F. (2017). Isolation of uncultured bacteria from Antarctica using long incubation periods and low nutritional media. Front. Microbiol. 8:1346. doi: 10.3389/fmicb.2017.01346
Puspita, I. D., Kamagata, Y., Tanaka, M., Asano, K., and Nakatsu, C. H. (2012). Are uncultivated bacteria really uncultivable? Microbes Environ. 27, 356–366. doi: 10.1264/jsme2.ME12092
Rahman, I., Shahamat, M., Kirchman, P., Russek-Cohen, E., and Colwell, R. (1994). Methionine uptake and cytopathogenicity of viable but nonculturable Shigella dysenteriae type 1. Appl. Environ. Microbiol. 60, 3573–3578. doi: 10.1128/aem.60.10.3573-3578.1994
Rappé, M. S., Connon, S. A., Vergin, K. L., and Giovannoni, S. J. (2002). Cultivation of the ubiquitous SAR11 marine bacterioplankton clade. Nature 418:630. doi: 10.1038/nature00917
Ratzke, C., and Gore, J. (2018). Modifying and reacting to the environmental pH can drive bacterial interactions. PLoS Biol. 16:e2004248. doi: 10.1371/journal.pbio.2004248
Remenár, M., Karelová, E., Harichová, J., Zámocký, M., Kamlárová, A., and Ferianc, P. (2015). Isolation of previously uncultivable bacteria from a nickel contaminated soil using a diffusion-chamber-based approach. Appl. Soil Ecol. 95, 115–127. doi: 10.1016/j.apsoil.2015.06.013
Ren, B., Hu, Y., Chen, B., Zhang, Y., Thiele, J., Shi, R., et al. (2018). Soil pH and plant diversity shape soil bacterial community structure in the active layer across the latitudinal gradients in continuous permafrost region of northeastern China. Sci. Rep. 8:5619. doi: 10.1038/s41598-018-24040-8
Ren, L., Jeppesen, E., He, D., Wang, J., Liboriussen, L., Xing, P., et al. (2015). pH influences the importance of niche-related and neutral processes in lacustrine bacterioplankton assembly. Appl. Environ. Microbiol. 81, 3104–3114. doi: 10.1128/AEM.04042-14
Rettedal, E. A., Gumpert, H., and Sommer, M. O. A. (2014). Cultivation-based multiplex phenotyping of human gut microbiota allows targeted recovery of previously uncultured bacteria. Nat. Commun. 5:4714. doi: 10.1038/ncomms5714
Řezanka, T., Gharwalová, L., Nováková, G., Kolouchová, I., Uhlík, O., and Sigler, K. (2019). Kocuria bacterial isolates from radioactive springs of Jáchymov spa (Joachimsthal) as sources of polyunsaturated fatty acids. Lipids 54, 177–187. doi: 10.1002/lipd.12136
Rice, S. A., McDougald, D., and Kjelleberg, S. (2000). Vibrio vulnificus: a physiological and genetic approach to the viable but nonculturable response. J. Infect. Chemother. 6, 115–120. doi: 10.1007/PL00012150
Rosero-Chasoy, G., Rodríguez-Jasso, R. M., Aguilar, C. N., Buitrón, G., Chairez, I., and Ruiz, H. A. (2021). Microbial co-culturing strategies for the production high value compounds, a reliable framework towards sustainable biorefinery implementation – an overview. Bioresour. Technol. 321:124458. doi: 10.1016/j.biortech.2020.124458
Rosselló-Móra, R., and Amann, R. (2015). Past and future species definitions for bacteria and Archaea. Syst. Appl. Microbiol. 38, 209–216. doi: 10.1016/j.syapm.2015.02.001
Rousk, J., Bååth, E., Brookes, P. C., Lauber, C. L., Lozupone, C., Caporaso, J. G., et al. (2010). Soil bacterial and fungal communities across a pH gradient in an arable soil. ISME J. 4, 1340–1351. doi: 10.1038/ismej.2010.58
Sangwan, P., Kovac, S., Davis, K. E. R., Sait, M., and Janssen, P. H. (2005). Detection and cultivation of soil Verrucomicrobia. Appl. Environ. Microbiol. 71, 8402–8410. doi: 10.1128/AEM.71.12.8402-8410.2005
Schink, B. (2002). Synergistic interactions in the microbial world. Antonie Van Leeuwenhoek 81, 257–261. doi: 10.1023/A:1020579004534
Schneider, Y. K.-H., Ø Hansen, K., Isaksson, J., Ullsten, S., H Hansen, E., and Hammer Andersen, J. (2019). Anti-bacterial effect and cytotoxicity assessment of lipid 430 isolated from Algibacter sp. Molecules 24:3991. doi: 10.3390/molecules24213991
Seeger, S., Monajembashi, S., Hutter, K. J., Futterman, G., Wolfrum, J., and Greulich, K. (1991). Application of laser optical tweezers in immunology and molecular genetics. Cytom. J. Int. Soc. Anal. Cytol 12, 497–504. doi: 10.1002/cyto.990120606
Senoh, M., Ghosh-Banerjee, J., Ramamurthy, T., Hamabata, T., Kurakawa, T., Takeda, M., et al. (2010). Conversion of viable but nonculturable Vibrio cholerae to the culturable state by co-culture with eukaryotic cells. Microbiol. Immunol. 54, 502–507. doi: 10.1111/j.1348-0421.2010.00245.x
Sessitsch, A., Weilharter, A., Gerzabek, M. H., Kirchmann, H., and Kandeler, E. (2001). Microbial population structures in soil particle size fractions of a long-term fertilizer field experiment. Appl. Environ. Microbiol. 67, 4215–4224. doi: 10.1128/AEM.67.9.4215-4224.2001
Sexton, D. L., St-Onge, R. J., Haiser, H. J., Yousef, M. R., Brady, L., Gao, C., et al. (2015). Resuscitation-promoting factors are cell wall-lytic enzymes with important roles in the germination and growth of Streptomyces coelicolor. J. Bacteriol. 197, 848–860. doi: 10.1128/JB.02464-14
Shah, I. M., and Dworkin, J. (2010). Induction and regulation of a secreted peptidoglycan hydrolase by a membrane Ser/Thr kinase that detects muropeptides. Mol. Microbiol. 75, 1232–1243. doi: 10.1111/j.1365-2958.2010.07046.x
Shleeva, M., Mukamolova, G. V., Young, M., Williams, H. D., and Kaprelyants, A. S. (2004). Formation of ‘non-culturable’cells of mycobacterium smegmatis in stationary phase in response to growth under suboptimal conditions and their Rpf-mediated resuscitation. Microbiology 150, 1687–1697. doi: 10.1099/mic.0.26893-0
Signoretto, C., del Mar, L. M., Tafi, M. C., and Canepari, P. (2000). Cell wall chemical composition of Enterococcus faecalis in the viable but nonculturable state. Appl. Environ. Microbiol. 66, 1953–1959. doi: 10.1128/AEM.66.5.1953-1959.2000
Smrhova, T., Jani, K., Pajer, P., Kapinusova, G., Vylita, T., Suman, J., et al. (2022). Prokaryotes of renowned Karlovy Vary (Carlsbad) thermal springs: phylogenetic and cultivation analysis. Environ. Microbiol. 17:48. doi: 10.1186/s40793-022-00440-2
Song, W., Kim, M., Tripathi, B. M., Kim, H., and Adams, J. M. (2016). Predictable communities of soil bacteria in relation to nutrient concentration and successional stage in a laboratory culture experiment. Environ. Microbiol. 18, 1740–1753. doi: 10.1111/1462-2920.12879
Srikanth, S., Dubey, S. K., Javed, A., and Goel, S. (2021). Droplet based microfluidics integrated with machine learning. Sens Actuator A Phys. 332:113096. doi: 10.1016/j.sna.2021.113096
Staley, J. T., and Konopka, A. (1985). Measurement of in situ activities of nonphotosynthetic microorganisms in aquatic and terrestrial habitats. Annu. Rev. Microbiol. 39, 321–346. doi: 10.1146/annurev.mi.39.100185.001541
Steele, J. A., Countway, P. D., Xia, L., Vigil, P. D., Beman, J. M., Kim, D. Y., et al. (2011). Marine bacterial, archaeal and protistan association networks reveal ecological linkages. ISME J. 5, 1414–1425. doi: 10.1038/ismej.2011.24
Steiner, P. A., Geijo, J., Fadeev, E., Obiol, A., Sintes, E., Rattei, T., et al. (2020). Functional seasonality of free-living and particle-associated prokaryotic communities in the coastal Adriatic Sea. Front. Microbiol. 11:584222. doi: 10.3389/fmicb.2020.584222
Stewart, E. J. (2012). Growing unculturable bacteria. J. Bacteriol. 194, 4151–4160. doi: 10.1128/JB.00345-12
Strejcek, M., Smrhova, T., Junkova, P., and Uhlik, O. (2018). Whole-cell MALDI-TOF MS versus 16S rRNA gene analysis for identification and dereplication of recurrent bacterial isolates. Front. Microbiol. 9:1294. doi: 10.3389/fmicb.2018.01294
Sturm, A., and Dworkin, J. (2015). Phenotypic diversity as a mechanism to exit cellular dormancy. Curr. Biol. 25, 2272–2277. doi: 10.1016/j.cub.2015.07.018
Su, X., Li, S., Xie, M., Tao, L., Zhou, Y., Xiao, Y., et al. (2021). Enhancement of polychlorinated biphenyl biodegradation by resuscitation promoting factor (Rpf) and Rpf-responsive bacterial community. Chemosphere 263:128283. doi: 10.1016/j.chemosphere.2020.128283
Su, X., Shen, H., Yao, X., Ding, L., Yu, C., and Shen, C. (2013). A novel approach to stimulate the biphenyl-degrading potential of bacterial community from PCBs-contaminated soil of e-waste recycling sites. Bioresour. Technol. 146, 27–34. doi: 10.1016/j.biortech.2013.07.028
Su, X., Wang, Y., Xue, B., Zhang, Y., Mei, R., Zhang, Y., et al. (2018). Resuscitation of functional bacterial community for enhancing biodegradation of phenol under high salinity conditions based on Rpf. Bioresour. Technol. 261, 394–402. doi: 10.1016/j.biortech.2018.04.048
Su, X., Zhang, Q., Hu, J., Hashmi, M. Z., Ding, L., and Shen, C. (2015). Enhanced degradation of biphenyl from PCB-contaminated sediments: the impact of extracellular organic matter from Micrococcus luteus. Appl. Microbiol. Biotechnol. 99, 1989–2000. doi: 10.1007/s00253-014-6108-6
Suman, J., Zubrova, A., Rojikova, K., Pechar, R., Svec, P., Cajthaml, T., et al. (2019). Pseudogemmobacter bohemicus gen. Nov., sp. nov., a novel taxon from the Rhodobacteraceae family isolated from heavy-metal-contaminated sludge. Int. J. Syst. Evol. Microbiol. 69, 2401–2407. doi: 10.1099/ijsem.0.003493
Sun, J., Guo, J., Yang, Q., and Huang, J. (2019). Diluted conventional media improve the microbial cultivability from aquarium seawater. J. Microbiol. 57, 759–768. doi: 10.1007/s12275-019-9175-7
Sussman, A. S., and Halvorson, H. O. Spores: their dormancy and germination, New York and London Harper & Row.; (1966) 356.
Sylvain, F.-É., Cheaib, B., Llewellyn, M., Gabriel Correia, T., Barros Fagundes, D., Luis Val, A., et al. (2016). pH drop impacts differentially skin and gut microbiota of the Amazonian fish tambaqui (Colossoma macropomum). Sci. Rep. 6:32032. doi: 10.1038/srep32032
Tahon, G., and Willems, A. (2017). Isolation and characterization of aerobic anoxygenic phototrophs from exposed soils from the Sør Rondane Mountains, East Antarctica. Syst. Appl. Microbiol. 40, 357–369. doi: 10.1016/j.syapm.2017.05.007
Tamaki, H., Tanaka, Y., Matsuzawa, H., Muramatsu, M., Meng, X. Y., Hanada, S., et al. (2011). Armatimonas rosea gen. Nov., sp. nov., of a novel bacterial phylum, Armatimonadetes phyl. Nov., formally called the candidate phylum OP10. Int. J. Syst. Evol. Microbiol. 61, 1442–1447. doi: 10.1099/ijs.0.025643-0
Tamames, J., and Rosselló-Móra, R. (2012). On the fitness of microbial taxonomy. Trends Microbiol. 20, 514–516. doi: 10.1016/j.tim.2012.08.012
Tanaka, T., Kawasaki, K., Daimon, S., Kitagawa, W., Yamamoto, K., Tamaki, H., et al. (2014). A hidden pitfall in the preparation of agar media undermines microorganism cultivability. Appl. Environ. Microbiol. 80, 7659–7666. doi: 10.1128/AEM.02741-14
Tandogan, N., Abadian, P. N., Epstein, S., Aoi, Y., and Goluch, E. D. (2014). Isolation of microorganisms using sub-micrometer constrictions. PLoS One 9:e101429. doi: 10.1371/journal.pone.0101429
Tewari Kumar, P., Decrop, D., Safdar, S., Passaris, I., Kokalj, T., Puers, R., et al. (2020). Digital microfluidics for single bacteria capture and selective retrieval using optical tweezers. Micromachines 11:308. doi: 10.3390/mi11030308
Thompson, J. N. (2020). “Population ecology” in Encyclopedia Britannica. Available at: https://www.britannica.com/science/population-ecology (Accessed November 18, 2020).
Tripathi, B. M., Stegen, J. C., Kim, M., Dong, K., Adams, J. M., and Lee, Y. K. (2018). Soil pH mediates the balance between stochastic and deterministic assembly of bacteria. ISME J. 12, 1072–1083. doi: 10.1038/s41396-018-0082-4
Turnbaugh, P. J., Ley, R. E., Hamady, M., Fraser-Liggett, C. M., Knight, R., and Gordon, J. I. (2007). The human microbiome project. Nature 449, 804–810. doi: 10.1038/nature06244
Uhlik, O., Leewis, M.-C., Strejcek, M., Musilova, L., Mackova, M., Leigh, M. B., et al. (2013). Stable isotope probing in the metagenomics era: a bridge towards improved bioremediation. Biotechnol. Adv. 31, 154–165. doi: 10.1016/j.biotechadv.2012.09.003
Vallejo Esquerra, E., Yang, H., Sanchez, S. E., and Omsland, A. (2017). Physicochemical and nutritional requirements for axenic replication suggest physiological basis for Coxiella burnetii niche restriction. Front. Cell. Infect. Microbiol. 7:190. doi: 10.3389/fcimb.2017.00190
van Dorst, J. M., Hince, G., Snape, I., and Ferrari, B. C. (2016). Novel culturing techniques select for heterotrophs and hydrocarbon degraders in a subantarctic soil. Sci. Rep. 6:36724. doi: 10.1038/srep36724
Versluis, D., de J Bello González, T., Zoetendal, E. G., Passel, M. W. J. V., and Smidt, H. (2019). High throughput cultivation-based screening on porous aluminum oxide chips allows targeted isolation of antibiotic resistant human gut bacteria. PLoS One 14:e0210970. doi: 10.1371/journal.pone.0210970
Wang, C., Bendle, J., Yang, Y., Yang, H., Sun, H., Huang, J., et al. (2016). Impacts of pH and temperature on soil bacterial 3-hydroxy fatty acids: development of novel terrestrial proxies. Org. Geochem. 94, 21–31. doi: 10.1016/j.orggeochem.2016.01.010
Watterson, W. J., Tanyeri, M., Watson, A. R., Cham, C. M., Shan, Y., Chang, E. B., et al. (2020). Droplet-based high-throughput cultivation for accurate screening of antibiotic resistant gut microbes. eLife 9:e56998. doi: 10.7554/eLife.56998
Watve, M., Shejval, V., Sonawane, C., Rahalkar, M., Matapurkar, A., Shouche, Y., et al. (2000). The 'K' selected oligophilic bacteria: a key to uncultured diversity? Curr. Sci. 78, 1535–1542.
Whitman, W. B., Chuvochina, M., Hedlund, B. P., Hugenholtz, P., Konstantinidis, K. T., Murray, A. E., et al. (2022). Development of the SeqCode: a proposed nomenclatural code for uncultivated prokaryotes with DNA sequences as type. Syst. Appl. Microbiol. 45:126305. doi: 10.1016/j.syapm.2022.126305
Wilson, L., Iqbal, K. M., Simmons-Ehrhardt, T., Bertino, M. F., Shah, M. R., Yadavalli, V. K., et al. (2019). Customizable 3D printed diffusion chambers for studies of bacterial pathogen phenotypes in complex environments. J. Microbiol. Methods 162, 8–15. doi: 10.1016/j.mimet.2019.05.002
Wirth, J. S., and Whitman, W. B. (2018). Phylogenomic analyses of a clade within the roseobacter group suggest taxonomic reassignments of species of the genera Aestuariivita, Citreicella, Loktanella, Nautella, Pelagibaca, Ruegeria, Thalassobius, Thiobacimonas and Tropicibacter, and the proposal of six novel genera. Int. J. Syst. Evol. Microbiol. 68, 2393–2411. doi: 10.1099/ijsem.0.002833
Woese, C. R. (1992). “Prokaryote systematics: the evolution of a science” in The Prokayotes. eds. A. Balows, H. G. Trüper, M. Dworkin, W. Harder and K-H. Schleifer (New York, NY: Springer), 3–18.
Wood, T. K., Knabel, S. J., and Kwan, B. W. (2013). Bacterial persister cell formation and dormancy. Appl. Environ. Microbiol. 79, 7116–7121. doi: 10.1128/AEM.02636-13
Wu, X., Spencer, S., Gushgari-Doyle, S., Yee, M. O., Voriskova, J., Li, Y., et al. (2020). Culturing of “unculturable” subsurface microbes: natural organic carbon source fuels the growth of diverse and distinct bacteria from groundwater. Front. Microbiol. 11:610001. doi: 10.3389/fmicb.2020.610001
Wurch, L., Giannone, R. J., Belisle, B. S., Swift, C., Utturkar, S., Hettich, R. L., et al. (2016). Genomics-informed isolation and characterization of a symbiotic Nanoarchaeota system from a terrestrial geothermal environment. Nat. Commun. 7, 1–10. doi: 10.1038/ncomms12115
Xian, W.-D., Salam, N., Li, M.-M., Zhou, E.-M., Yin, Y.-R., Liu, Z.-T., et al. (2020). Network-directed efficient isolation of previously uncultivated Chloroflexi and related bacteria in hot spring microbial mats. NPJ Biofilms Microbiomes 6, 1–10. doi: 10.1038/s41522-020-0131-4
Xu, B., Hu, B., Wang, J., Lan, Y., Zhu, Y., Dai, X., et al. (2018). Virgibacillus indicus sp. nov. and Virgibacillus profundi sp. nov, two moderately halophilic bacteria isolated from marine sediment by using microfluidic streak plates. Int. J. Syst. Evol. Microbiol. 68, 2015–2023. doi: 10.1099/ijsem.0.002782
Xu, H.-S., Roberts, N., Singleton, F. L., Attwell, R. W., Grimes, D. J., and Colwell, R. R. (1982). Survival and viability of nonculturable Escherichia coli and vibrio cholerae in the estuarine and marine environment. Microb. Ecol. 8, 313–323. doi: 10.1007/BF02010671
Xu, R., Zhang, K., Liu, P., Han, H., Zhao, S., Kakade, A., et al. (2018). Lignin depolymerization and utilization by bacteria. Bioresour. Technol. 269, 557–566. doi: 10.1016/j.biortech.2018.08.118
Yasumoto-Hirose, M., Nishijima, M., Ngirchechol, M. K., Kanoh, K., Shizuri, Y., and Miki, W. (2006). Isolation of marine bacteria by in situ culture on media-supplemented polyurethane foam. Mar. Biotechnol. 8, 227–237. doi: 10.1007/s10126-005-5015-3
Zang, E., Brandes, S., Tovar, M., Martin, K., Mech, F., Horbert, P., et al. (2013). Real-time image processing for label-free enrichment of Actinobacteria cultivated in picolitre droplets. Lab Chip 13, 3707–3713. doi: 10.1039/c3lc50572c
Zengler, K., Toledo, G., Rappé, M., Elkins, J., Mathur, E. J., Short, J. M., et al. (2002). Cultivating the uncultured. Proc. Natl. Acad. Sci. U. S. A. 99, 15681–15686. doi: 10.1073/pnas.252630999
Zengler, K., Walcher, M., Clark, G., Haller, I., Toledo, G., Holland, T., et al. (2005). High-throughput cultivation of microorganisms using microcapsules. Methods Enzymol. 397, 124–130. doi: 10.1016/S0076-6879(05)97007-9
Zhang, Y. (2014). Persisters, persistent infections and the Yin–Yang model. Emerg Microbes Infect 3, 1–10. doi: 10.1038/emi.2014.3
Zhang, X.-H., Ahmad, W., Zhu, X.-Y., Chen, J., and Austin, B. (2021). Viable but nonculturable bacteria and their resuscitation: implications for cultivating uncultured marine microorganisms. Mar. Life Sci. Technol. 3, 189–203. doi: 10.1007/s42995-020-00041-3
Zhang, L., Chen, P., Zhou, Z., Hu, Y., Sha, Q., Zhang, H., et al. (2019). Agarose-based microwell array chip for high-throughput screening of functional microorganisms. Talanta 191, 342–349. doi: 10.1016/j.talanta.2018.08.090
Zhang, A., Sun, H., Wang, P., Han, Y., and Wang, X. (2012). Modern analytical techniques in metabolomics analysis. Analyst 137, 293–300. doi: 10.1039/C1AN15605E
Zhou, N., Sun, Y. T., Chen, D. W., Du, W., Yang, H., and Liu, S. J. (2019). Harnessing microfluidic streak plate technique to investigate the gut microbiome of Reticulitermes chinensis. MicrobiologyOpen 8:e00654. doi: 10.1002/mbo3.654
Glossary
Keywords: environmental microbiome, microbial ecology, dormancy, VBNC, growth factors, cultivation techniques, improved cultivation, difficult-to-culture microorganisms
Citation: Kapinusova G, Lopez Marin MA and Uhlik O (2023) Reaching unreachables: Obstacles and successes of microbial cultivation and their reasons. Front. Microbiol. 14:1089630. doi: 10.3389/fmicb.2023.1089630
Edited by:
Romy Chakraborty, Berkeley Lab (DOE), United StatesReviewed by:
Slava Epstein, Northeastern University, United StatesBrendan Paul Burns, University of New South Wales, Australia
Copyright © 2023 Kapinusova, Lopez Marin and Uhlik. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ondrej Uhlik, b25kcmVqLnVobGlrQHZzY2h0LmN6
†These authors have contributed equally to this work
 Gabriela Kapinusova
Gabriela Kapinusova Marco A. Lopez Marin
Marco A. Lopez Marin Ondrej Uhlik
Ondrej Uhlik