- 1Key Laboratory of Medicinal Chemistry and Molecular Diagnosis of Ministry of Education, College of Life Science, Hebei University, Baoding, China
- 2Hainan Key Laboratory of Research and Development of Natural Product from Li Folk Medicine, Hainan Institute for Tropical Agricultural Resources, Institute of Tropical Bioscience and Biotechnology, CATAS, Haikou, China
- 3Affiliated Hospital of Medical College, Hebei University of Engineering, Handan, China
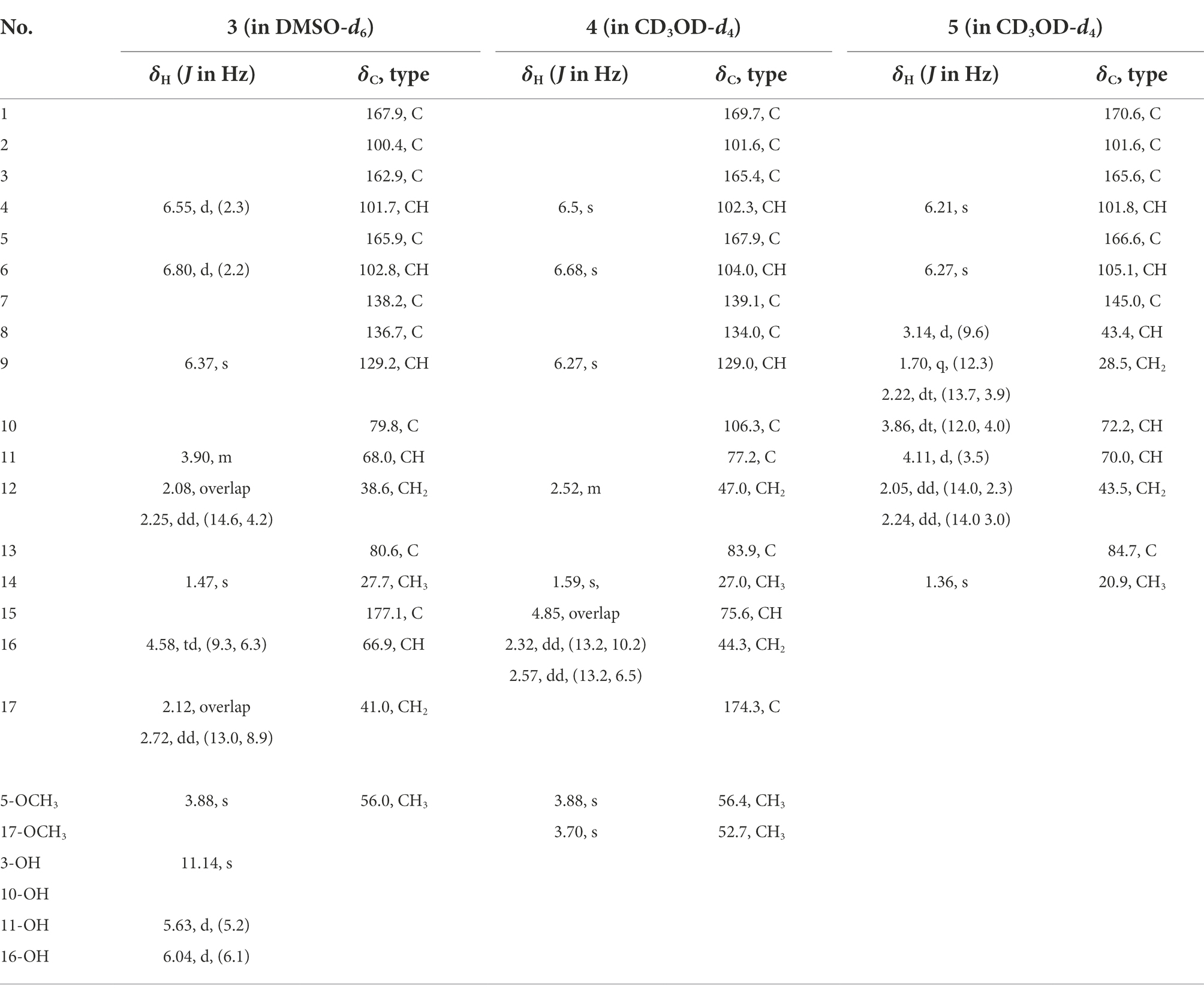
Five new polyketides named alternafurones A (1) and B (2), alternapyrones M-O (3–5), together with fourteen known ones (6–19), were isolated from the desert-plant-derived fungus Alternaria sp. HM 134. The structures of the new compounds were elucidated from spectroscopic data and ECD spectroscopic analyses. Alternafurones A and B represent polyketides with an unprecedented 6/5/6 skeleton core. Compounds 1, 2 and 4 showed definite inhibitory activities against isocitrate dehydrogenase 1 gene (IDH1 R132h) with IC50 values of 29.38, 19.41 and 14.14 μg/ml, respectively. Seven compounds (6, 7, 9–12, 14) showed potent protein tyrosine phosphatase 1B (PTP1B) inhibitory activity with IC50 values ranging from 0.97 μg/ml to 89.80 μg/ml.
Introduction
Desert regions are extreme ecological environments with large temperature, fluctuations and low precipitation (Eida et al., 2019). The desert has unique extremophiles that aren’t present in other environments and as a result microbes that coevolve in this unique niche may have developed unique or novel secondary metabolites to interact with each other (Yang C. L. et al., 2019; Ameen et al., 2021). Song et al. (2019) obtained the new skeleton dimers from the desert plant endophytic fungus Trematosphaeria terricola, which showed antitumor activity. The antibacterial new ansamycin-type polyketides from Streptomyces sp. (Rateb et al., 2011) and cytotoxic resorcylic acid lactones (Zhang et al., 2021) from Chaetosphaeronema hispidulur were obtained from endophytic fungi of desert environment.
Although some progress had been made in the study of desert fungi, it is still an underdeveloped domain. Thus, we attempted to mine the novel natural products focus on the endophytic fungi in the desert area. An endophytic fungus, Alternaria sp. HM 134 was isolated from the endophytic fungi of Aster pekinensis Kitag. in Ordos. The Alternaria sp. was a recognized plant pathogen, which led to serious crop corruption and a wide range of plant diseases (Zhao et al., 2020). The mycotoxins in Alternaria sp. had been widely studied, leading to the definition of a series of secondary metabolites, including polyketones (Lu et al., 2021), dibenzopyrones (Chen et al., 2022), anthraquinones (Chen et al., 2014) and cephalochromin (Song et al., 2021). These metabolites have cytotoxic, antiviral and enzyme inhibitory activities (Xu et al., 2019; Tan et al., 2020; Saxena, 2021; Zhang et al., 2022).
In this study, chemical investigation on the fermentation broth of Alternaria sp. HM 134 led to the identification of five new polyketides (1–5) and fourteen known ones (6–19). All the isolated compounds were tested for their inhibitory activities against isocitrate dehydrogenase (IDH) and potent protein tyrosine phosphatase (PTPs). Compound 9 has significant inhibitory activity against PTP1B. Herein, the details of the isolation, structure identification, and bioactivity of these compounds were described.
Materials and methods
General experimental procedure
Ultraviolet (UV) spectra were measured on a UV-3600 spectrometer. ECD spectra were measured on Bio-Logic MOS-450 spectropolarimeter. IR spectra were recorded on a Thermo Nicolet iS 10 spectrometer. The NMR spectra were recorded on a Bruker AM-600 spectrometer with TMS as an internal standard. HRESIMS spectra were obtained on a Thermo U3000 spectrometer fitted with an ESI source. Semipreparative HPLC was performed on Thermo UltiMate 3,000 machine equipped with a 5C18-MS column (5 μM, 250 × 10 mm,COSMOSIL, JPN). ECD spectroscopic analyses was supported by the High-Performance Computing Center of Hebei University.
Fungal material
The HM 134 was isolated from the desert plant Aster pekinensis Kitag. in Ordos. The strain was identified as Alternaria sp. based on microscopic examination and by internal transcribed spacer (ITS1-4) sequencing. The ITS sequence has been deposited in GenBank1 with accession number No. MK478900. The purified strain was cultivated in a PDA medium plate (containing 200 g potatoes, boil 20 min, take filtrate; 20 g glucose; 20 g agar in 1 l water) at 28°C for 7 days. Then, it was cut into small pieces and cultured in PDB medium plate (containing 200 g potatoes, boil 20 min, take filtrate; 20 g glucose;in 1 l water) for 5 days.
Fermentation, extraction, and isolation
The fermentation was carried out in 100 flasks (500 ml), each containing 80 g of rice and 100 ml H2O, autoclaving at 15 psi for 30 min. After cooling to room temperature, each flask was inoculated with 20 ml of the spore inoculum and incubated at room temperature for 30 days. The fermented material was extracted successively with EtOAc for three times, then the EtOAc solutions were combined and evaporated under reduced pressure to get 170 g of crude extract. The extract was fractionated by a silica gel VLC column using different solvents of increasing polarity, from EtOAc-petroleum to yield eight fractions (Fr.1–8). Fraction Fr.3 was chromatographed over C18 reversed-phase (RP-18) silica gel using MeOH/H2O (30:70, 40:60, 50:50, 60:40, 70:30, 90:10) to collect six subfractions (SF.3a − 3f). SF.3d was separated on a semipreparative reversed-phase (RP) HPLC column using MeOH/H2O = 60:40 (2.5 ml/min) to give 4 (tR = 16.3 min, 3 mg). SF.3c was separated on a semipreparative reversed-phase (RP) HPLC column using MeOH/H2O = 40:60 (2.5 ml/min) to give 5 (tR = 18.6 min, 2 mg). The precipitated compound 3 was obtained in SF.3e. Fraction Fr.4 was chromatographed over C18 reversed-phase (RP-18) silica gel using MeCN/H2O (10,90, 20:80, 30:70, 40:60, 50:50, 60:40, 70:30) to collect seven subfractions (SF.4a − 4 g). SF.4b was separated on a semipreparative reversed-phase (RP) HPLC column using MeCN/H2O = 18:82 (2.5 ml/min) to give 1 (tR = 21.2 min, 3 mg). SF.4b was separated on a semipreparative reversed-phase (RP) HPLC column using MeCN/H2O = 18:82 (2.5 ml/min) to give 2 (tR = 22 min, 2 mg).
Spectroscopic data
Alternafurone A (1): Yellow oil; [α]25D + 57.3 (c 0.1, MeOH); UV (MeOH) λmax (log ε): 218 (5.18), 257 (2.01), 293 (0.78) nm; IR (KBr) vmax cm−1: 3422, 2,945, 1,698, 1,672, 1,512, 1,458, 1,016. 1H and 13C NMR data see Table 1; HRESIMS m/z 291.0876 [M - H]− (calcd for C15H15O6−, 291.0874).
Alternafurone B (2): Yellow oil; [α]25D + 49.4 (c 0.1, MeOH); UV (MeOH) λmax (log ε): 246 (4.20), 278 (1.62), 320 (0.99) nm; IR (KBr) vmax cm−1: 3455, 2,963, 1714, 1,675, 1,516, 1,445, 1,033. 1H and 13C NMR data see Table 1; HRESIMS m/z 291.0879 [M - H]− (calcd for C15H15O6−, 291.0874).
Alternapyrone M (3): White solid; [α]25D + 39.1 (c 0.1, MeOH); UV (DMSO) λmax (log ε): 243 (2.86), 278 (0.88), 316 (0.49) nm; IR (KBr) vmax cm−1: 3526, 1710, 1,663, 1,618, 1,463, 1,012. 1H and 13C NMR data see Table 2; HRESIMS m/z 361.0932[M - H]− (calcd for C18H17O8−, 361.0929).
Alternapyrone N (4): Brown oil; [α]25D -13.2 (c 0.1, MeOH); UV (MeOH) λmax (log ε): 208 (2.76), 271 (2.05), 295 (1.05) nm; IR (KBr) vmax cm−1: 3428, 2,948, 1717, 1,578, 1,438, 1,162. 1H and 13C NMR data see Table 2; HRESIMS m/z 391.1039[M - H]− (calcd for C19H19O9−, 391.1035).
Alternapyrones O (5): Pink oil; [α]25D -9.1 (c 0.1, MeOH); UV (MeOH) λmax (log ε): 213 (2.80), 269 (1.13), 302 (0.62) nm; IR (KBr) vmax cm−1: 3679, 2,942, 1718, 1,586, 1,435, 1,014. 1H and 13C NMR data see Table 2; HRESIMS m/z 279.0873[M - H]− (calcd for C14H15O6−, 279.0876).
Computational section
The initial configuration was retrieved by Molecular Operating Environment (MOE) using MMFF94 molecular mechanics force field. Gauss 16 software was used for density functional theory calculation (Frisch et al., 2019). These conformations were optimized with B3LYP/6-31G (d) in gas phase, and the conformations with Boltzmann-population of over 1% were retained for the next operation. The remaining configurations were further optimized with B3LYP/6-31G (d) in gas phase, and all configurations were guaranteed to be frequency analyzed at the same level to avoid repeated configurations. The ECD spectra were calculated by the Time-dependent Density functional theory (TDDFT) methodology at the B3LYP/6–31 + g (d, p) level in methanol. ECD spectra were simulated using SpecDis 1.71 (Bruhn et al., 2013) with σ = 0.30 eV.
Inhibition of protein tyrosine phosphatase (PTPs) assay
Protein tyrosine phosphatase (PTPs) plays an important role in many human threatening diseases, such as PTP1B, an effective target for the treatment of type II diabetes; CD45, an effective target for the treatment of leukemia; and TCPTP, a reliable target for the treatment of cancer involved in multiple signaling pathways (Ruddraraju and Zhang, 2017). Nitrophenyl phosphate (pNPP) was used as substrate to determine the enzyme inhibition test. Fifty microliter reaction buffer (pH 6.5) including 50 mM HEPES, 100 mM NaCl, 1 mM EDTA, and 1 mM dithiothreitol (DTT) and compounds was added to 96 well plates, and incubate at room temperature for 15 min. Further, 50 μl reaction buffer with 50 mM pNPP was added and incubated at 37°C for 60 min. Na3VO4 was used as positive control and DMSO as the negative control. The phosphatase activity was determined by absorbance measured at 405 nm. The test was repeated three times for each compound. The IC50 value was derived from three independent experiments.
Isocitrate dehydrogenase inhibition assay
IDH1 R132h model was used to screen the inhibitory activity of the compounds. DMSO was used to configure the compounds into different concentrations as test samples. 2 mm NADPH (3 μl) and 1 m α-Kg (0.1 μl) was added to IDH1 buffer (pH 6.5; 96 μl) solution as reaction substrate in the reaction system. Then, the test samples (2.0 μl), IDH1 R132h (0.4 μl), PMS (0.1 μl) and 15 mM WST-8 (1.0 μl) were successively added to the above mixture and reacted at room temperature for 60 min. AGI-5198 was used as positive control and DMSO as the negative control. The absorption peak of 450 nm was detected by microplate reader. The test was repeated three times for each test samples, and the IC50 was calculated by GraphPad Prism.
Results and discussion
Isolation and structure elucidation
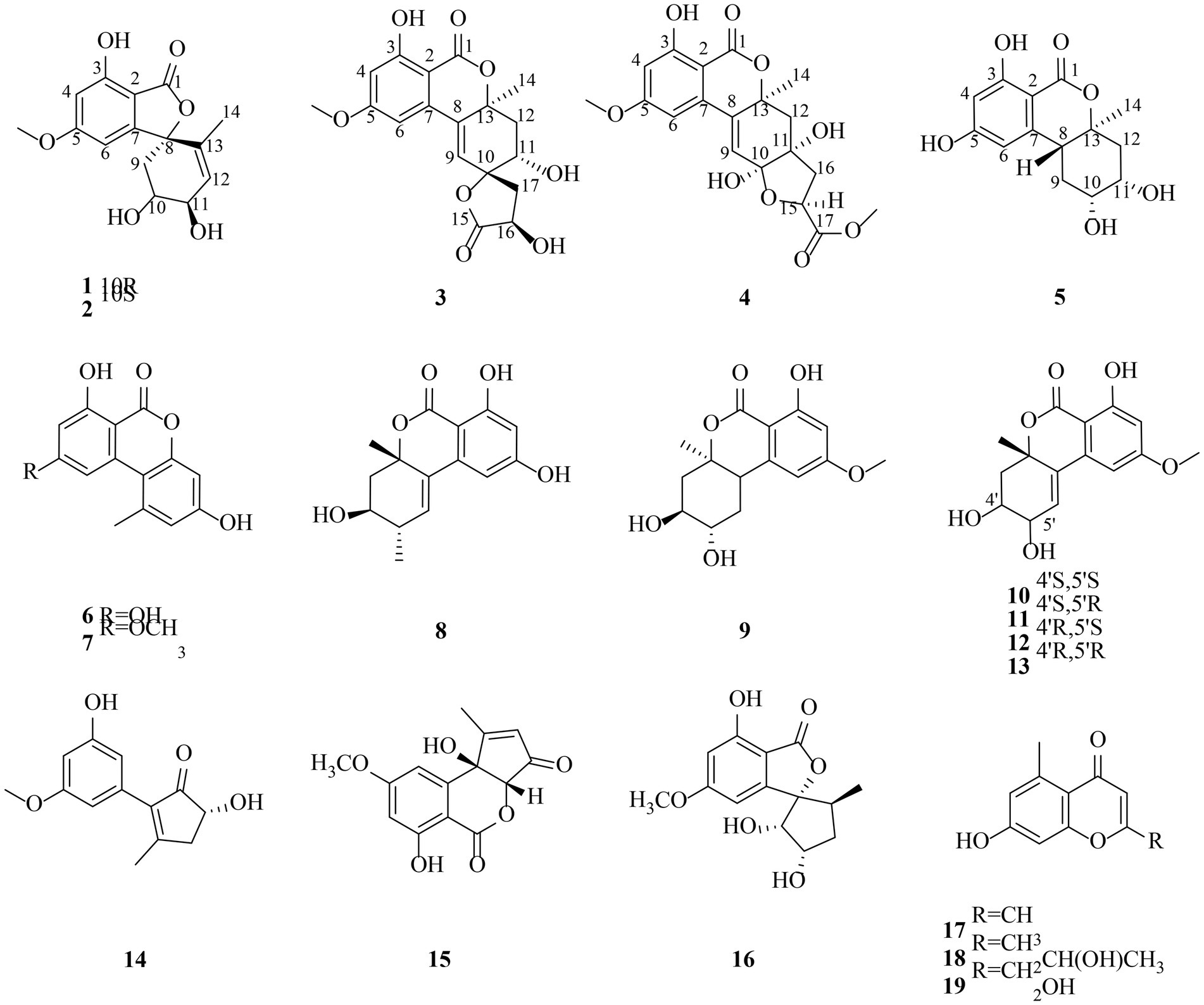
The fungus Alternaria sp.HM 134 was isolated from the desert plant Aster pekinensis Kitag. in Ordos and identified by internal transcribed spacer (ITS1-4) sequencing. The strain was grown in solid medium containing 80 g of rice and 100 ml H2O and incubated at room temperature for 30 days. The fermented material was extracted with EtOAc and the crude extract was separated and purified by a variety of separation and analysis methods. Finally, 19 natural products were isolated and identified from this fungus (Figure 1), including five new polyketides named alternafurones A (1) and B (2), alternapyrones M-O (3–5), and were tested for their inhibitory activities against PTPs and IDH for the first time.
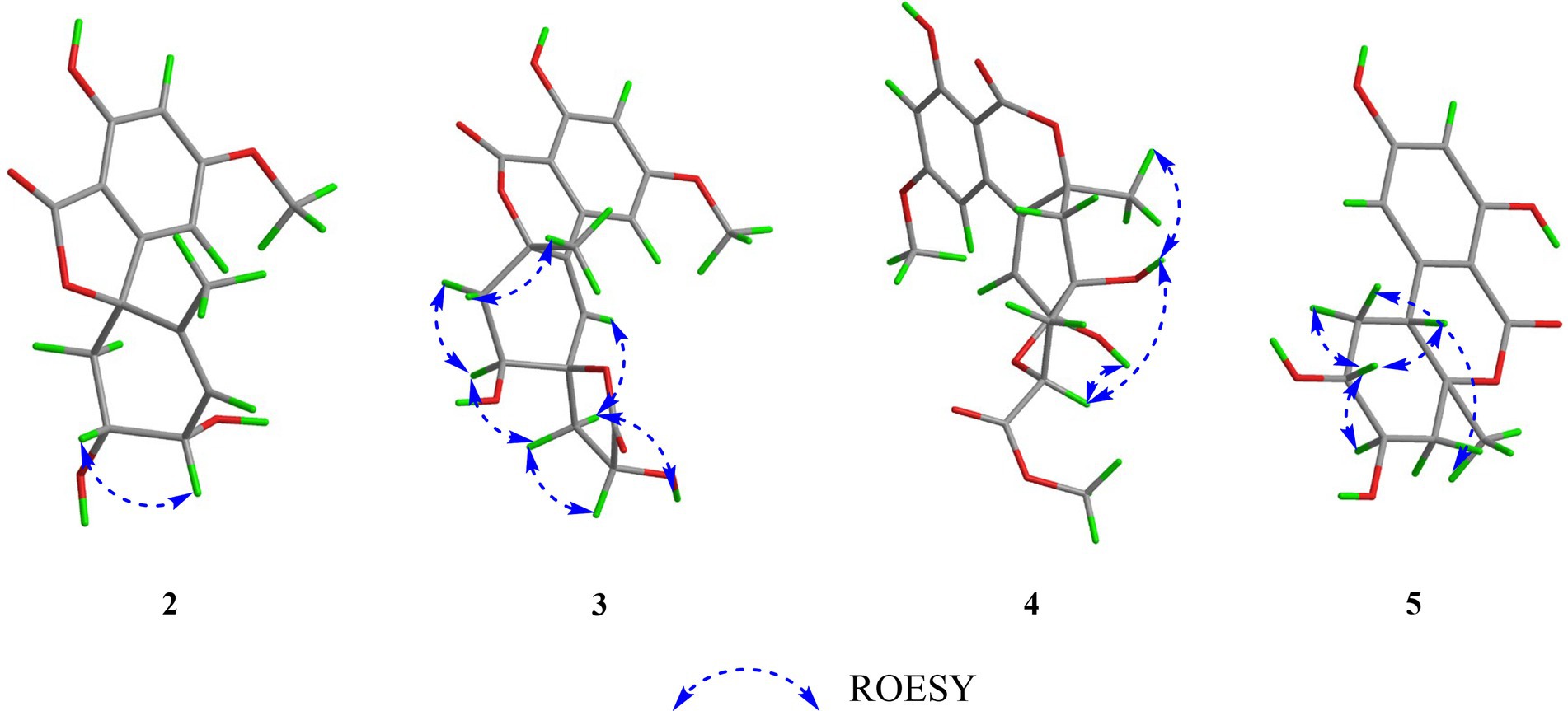
Compound 1 was obtained as yellow oil, and its molecular formula was determined to be C15H16O6 by HRESIMS and NMR data (Table 1), indicating an index of hydrogen defificiency of 8. The 1H NMR spectrum indicated two aromatic protons [δH 6.44 (H-4) and 6.39 (H-6)], an olefinic proton (δH 6.44,H-12), one methoxy (δH 3.84, H-15), and one methyl (δH 1.40, H-14). The 13C NMR spectrum revealed 15 carbon resonances that were classified by HSQC spectrum as, one carbonyl, one methyl, one methoxy, one methylene, five methines (two oxygenated and three orefinic) and six nonprotonated carbons. A comparison of the NMR data of 1 with those of alternatains A (16; Pang et al., 2018) indicated that they shared similar isobenzofuranone skeleton, except that the five-membered ring via the spiro carbon C-8 in alternatains A was replaced by a six-membered ring. This deduction was confirmed by HMBC correlations (Figure 2) from H-9 (δH 1.98, 2.47) to C-7 (δC 157.8) and C-8 (δC 87.3), from H-12 (δH 5.86) and H-14 (δH1.40) to C-8, as well as sequential COSY correlations of H-9/H-10/H-11/H-12. The whole connectivity of 6/5/6 skeleton core in 1 was further demonstrated by other HMBC correlations (Figure 2) and analysis of its molecular formula. The linkage of methoxy (δH/C 3.84/56.6) at C-5 was based on HMBC correlations from methoxy protons to C-5 and from H-6 (δH 6.39) to C-8 (δC 87.3) and C-5. Thus, the gross structure of 1 was resolved as shown. The relative configuration was established by analysis of ROESY data. According to the ROESY spectrum, the absence of ROESY correlation between H-10 (4.13, 1H) and H-11 (4.25, 1H) suggested that 10-OH is opposite to 11-OH. In order to ascertain the absolute configuration of 1, its electronic circular dichroism (ECD) spectrum was determined in MeOH and simulated at the CAM-B3LYP/6–311++G(2d, p) level after conformational optimization at the same level via Gaussian 05 software. The Boltzmann-weighted ECD curve agreed well with the experimental one (Figure 3A), and the absolute configurations of stereocenters C-8, C-10, and C-11 were assigned to be 8S,10R,11R.
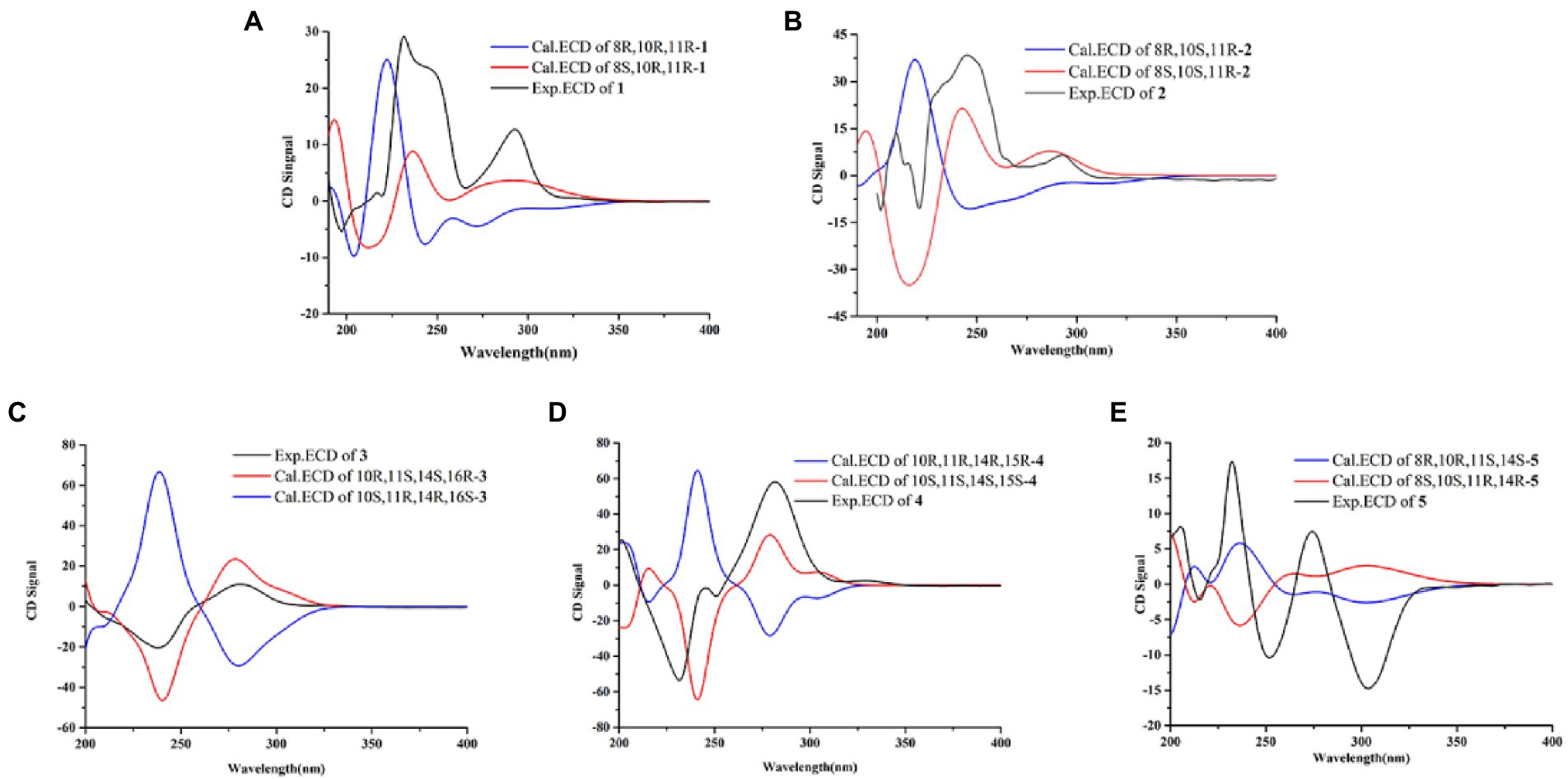
Figure 3. Experimental and calculated ECD curves for compound 1 (A), compound 2 (B), compound 3 (C), compound 4 (D), compound 5 (E).
Compound 2 had the same molecular formula C15H16O6 as that of 1 by HRESIMS and NMR data (Table 1) Further comparing its closely similar NMR data with those of compound 1 suggested that they possessed the same planar structure with 6/5/6 skeleton core. The main difference of their NMR data was reflected in the chemical shifts of C-9-C-13 in six-membered ring, which may be resulted from the different configuration of C-10 and C-11. The whole connectivity of compound 2 was also further demonstrated by other HMBC correlations (Figure 2). The relative configuration of six-membered ring was established by analysis of ROESY data. The obvious ROESY correlation of H-10 (3.87, 1H) and H-11 (4.07, 1H) indicated the same face of these two protons (Figure 4). The calculated ECD curve for (8S,10S,11R)-2 matched well with the experimental spectrum (Figure 3B), assigning the 8S,10S,11R absolute configuration of 2.
The molecular formula of compound 3 was determined to be C18H18O8 by HRESIMS and NMR data (Table 2), indicating 10 degrees of unsaturation. The 1H and 13C NMR data of 3, with the aid of a HSQC spectrum, showed a total of 18 carbon signals comprising two ester carbonyls, eight olefinic or aromatic carbons with three protonated, two sp3 methylenes, two sp3 oxygenated methines, two sp3 non-protonated oxygenated carbons, one methoxy and one methyl. Detail analyses of the 1D and 2D NMR data revealed that the structure of 3 had a 6/6/6 skeleton system, which was similar to compound alternatain D (Yang H. Y. et al., 2019). The obvious structural difference between them is that the carboxyl group in alternatain D was replaced an ester carbonyl via OH-10 to firm the five-membered spiro ring in 3. This deduction was supported by the COSY cross-peaks (Figure 2) of OH-11/H-11/H-12 and H-16/H-17, combined the analysis of its molecular formula. The whole connectivity of compound 3 with 6/6/6/5 skeleton system was also further demonstrated by other HMBC correlations from H-16 (δH 4.58) to C-15 (δC 177.1) and C-17 (δC 41.0), from H-14 to C-8, C-13 and C-12. The relative configuration was established by analysis of ROESY data, revealing the key correlations of H-17β (2.12) with H-9 (6.37) and 16-OH, H-17α (2.72) with H-16 (4.58) and H-11, H-12β (2.25) with H-11 and H-12α (2.08) with H-14 (Figure 4). These ROESY correlations showed the relative configuration of 3 as shown in Figure 4, suggesting that its absolute configuration was determined as 10R,11S,14S,16R or 10S,11R,14R,16S (Figure 4). Comparison of its experimental ECD spectrum with the calculated data of 3 (Figure 3), the absolute configuration of compound 3 was finally determined as 10R,11S,14S,16R.
Compound 4 had the molecular formula C19H20O9 as established from its HRESIMS and NMR data (Table 2), indicative of an index of hydrogen deficiency of 10. Its 1H NMR spectrum showed three olefinic or aromatic protons [δH 6.50 (H-4), 6.68 (H-6), 6.27 (H-9)], one methyl (δH 1.59, H-14), two methoxys (δH 3.88 and 3.70). The 13C NMR data of 4 showed a total of 19 carbon signals comprising two ester carbonyls, eight olefinic or aromatic carbons with three protonated, two sp3 methylenes, two sp3 oxygenated methines, two sp3 non-protonated oxygenated carbons, two methoxys and one methyl. Detail comparison of its 1D and 2D NMR data with that of alternatain C (Yang H. Y. et al., 2019) showed that they had the same 6/6/6/5 skeleton system. The structural difference between them is that the hemiketal hydroxy at C-15 in alternatain C was linked at C-10 and one additional methyl connected the carboxyl group to firm ester carbonyl in 4. This deduction was comfirmed by the key HMBC correlations from H-9 and H-12 to C-10, from the methoxy protons to C-17, and the COSY cross-peaks (Figure 2) of H-15/H-16. The whole connectivity of compound 4 was also further demonstrated by other HMBC correlations (Figure 2). The ROESY spectrum of compound 4 was obtained by dissolution of the sample with DMSO (see Supplementary Material, Figure S27), and the key correlations of 10-OH (5.78, 1H)/H-15 (4.72, 1H), H-15/11-OH (6.17, 1H), and 11-OH/H-14 (1.51, 3H) assigned its relative configuration (Figure 4). The absolute configuration of 4 was determined based on comparing its experimental ECD spectrum with those of calculated data (Figure 3D), indicating the stereocenters of 10S,11S,14S,15S.
The molecular formula of compound 5 was determined to be C14H16O6 by HRESIMS and NMR data (Table 2), establishing an index of hydrogen deficiency of 7. 1H NMR showed two aromatic protons [δH 6.27 (1H, s, H-6), 6.21 (1H, d, H-4)] and one methyl [δH 1.36 (3H, s, 14-CH3)]. The 13C NMR spectrum revealed 14 carbon resonances that were classified by HSQC spectrum as one carbonyl, one methyl, two sp3 methylenes, five methines (two olefinic and two oxygenated) and five nonprotonated carbons. Analyses of the NMR data revealed that the structure of 5 was closely similar to dihydroaltenuene A (Jiao et al., 2006). The only difference between them was that the methoxy at C-8 in dihydroaltenuene A was replaced by a hydroxy in 5, which was comfirmed by the molecular weight difference 14 and the HMBC correlation of the methoxy with C-8. The relative configuration was established by the key ROESY correlations of H-11 (4.11)/H-10 (3.86), H-10/H-8 (3.14), H-9α/H-8 and H-9β/H-14 (1.36). The absolute configuration was assigned to be 8R,10R,11S,13S by comparison of its experimental ECD spectrum with the calculated ECD curves of 5 (Figure 3E).
In addition to the five compounds described above, we also isolated 14 known ones (Figure 1), alternariol (6; Tanahashi et al., 1997), alternariol 9-methyl ether (7; Meng et al., 2012) altenuene (8; Xiao et al., 2014), 3-epidihydroaltenuene A (9; Tian et al., 2016), 5-hydroxyepialtenuene (10; Jin et al., 2013), 5′-epialtenuene (11; Bradburn et al., 1994), 4′-epialtenuene (12; Aly et al., 2008) isoaltenuene (13; He et al., 2012), (+)-nigrosporaol A (14; He et al., 2016), (3aR,9bR)-6,9b-dihydroxy-8-methoxy-1-methyl-cyclopentene[c]isochromen-3,5-dion (15; Jiao et al., 2013), alternatains A (16; Yang H. Y. et al., 2019), 2,5-dimethyl-7-hydroxychromone (17; Kashiwada et al., 2008), 2-(2’S-hydroxypropyl)-5-methyl-7-hydroxychromone (18; Khamthong et al., 2012), 7-hydroxy-2-hydroxymethyl-5-methyl-4H-chromen-4-one (19; Kimura et al., 2008). All of these compounds were identified by comparing their 1H and 13C NMR data with those reported in the literatures.
Protein tyrosine phosphatase inhibition assay
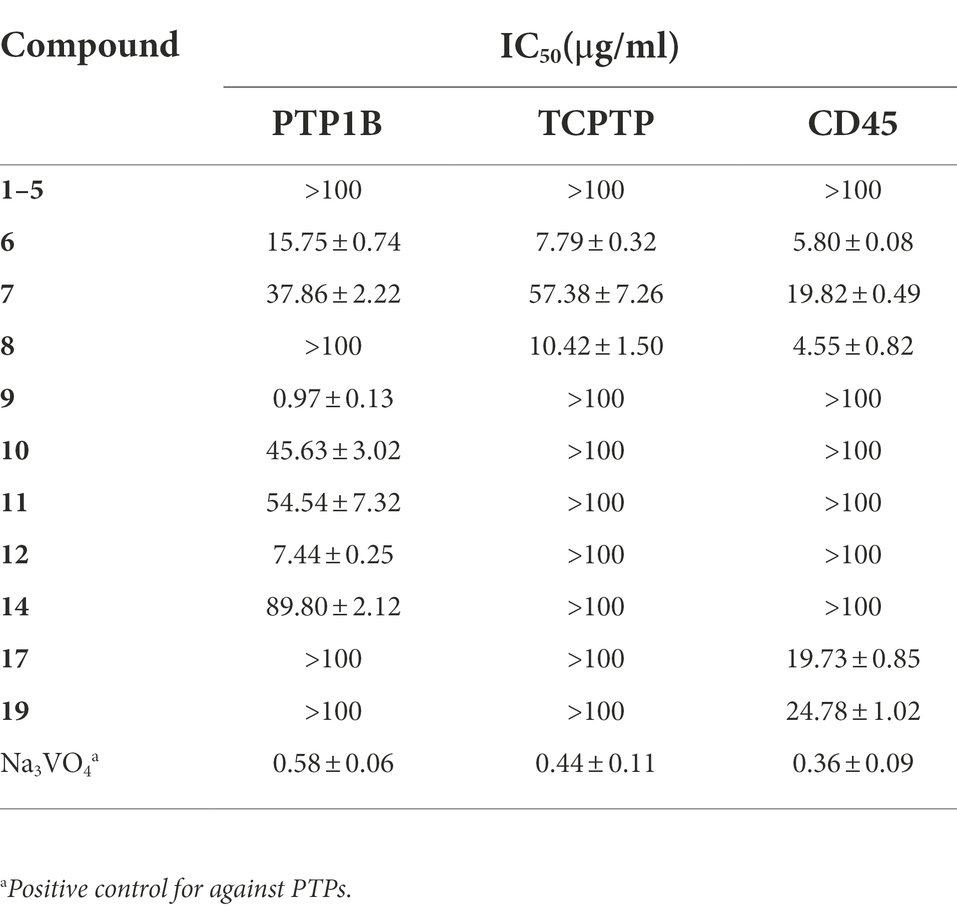
Protein tyrosine phosphatase (PTPs) plays an important role in many human threatening diseases, such as PTP1B, an effective target for the treatment of type II diabetes; CD45, an effective target for the treatment of leukemia; and TCPTP, a reliable target for the treatment of cancer involved in multiple signaling pathways (Zhang, 2003; Vintonyak et al., 2009). All the compounds were tested for their in vitro inhibitory activities against PTP1B, CD45 and TCPTP. The results showed that the five new compounds had no inhibitory activity against the three enzymes of the PTPs family compared with Na3VO4, as shown in Table 3. Compounds 9 and 12 had inhibitory activity against PTP1B compared with other compounds. It was noteworthy that the IC50 value (0.97 μg/ml) of compound 9 was similar to the positive control with IC50 of 0.58 μg/ml. Compound 6 and 8 had definite inhibitory activity against TCPTP and CD45. For the three enzymes, compound 6 had a wide range of inhibitory activities.
Biological activity against isocitrate dehydrogenase
We have learned that isocitrate dehydrogenase (IDH) gene mutations are closely related to the occurrence and development of tumors (Fujii et al., 2016). IDH mutations mainly occur in malignant tumors such as glioma, acute myeloid leukemia, chondrosarcoma, and intrahepatic cholangiocarcinoma. IDH mutation ultimately promote the occurrence and development of tumors (Gondim et al., 2019). Therefore, inhibition of IDH1 R132h activity was screened using the above compounds. AGI-5198 was used as a positive control in this experiment with the IC50 value of 0.04 μg/ml. The results showed that compounds 1, 2 and 4 had a low level of IDH1 R132h inhibitory activities, with IC50 of 29.38, 19.41 and 14.14 μg/ml, as shown in Table 4.
Conclusion
In this work, nineteen polyketides, including 5 new ones, were isolated from the endophytic fungus Alternaria sp. HM134 from the desert plant Aster pekinensis Kitag. in Ordos. Although most isolated polyketides have been found in the genus Alternaria in non-desert environments, the skeleton of alternafurones A (1) and B (2) comprised an unprecedented 6/5/6 skeleton core, which enriched the diversity of polyketone chemical structure. Meanwhile, three compounds (1, 2 and 4) showed inhibitory activities against IDH1 R132h. Compounds 6–8 had inhibitory activity against TCPTP and five compounds (6–8, 17, 19) showed inhibitory activity against CD45. Seven compounds (6, 7, 9–12, 14) showed inhibitory activity against PTP1B, among which compound 9 had a specific and significant inhibition. Furthermore, polyketides are reported with significant PTP1B inhibitory activity for the first time, which may provide new options for the development of therapeutic agents for diabetes and cancer.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary material, further inquiries can be directed to the corresponding authors.
Author contributions
DL, YZ, and ZW contributed to the conception and design of the study. LY and FK determined the plane structure and absolute configuration. ZL and LM wrote the first draft of the manuscript and performed all of the experimental work. QM contributed to isolation of compounds. ZL contributed to bioactivity assay. DL, YZ, and ZL improved the manuscript. All authors contributed to manuscript revision as well as read and approved the submitted version.
Funding
This research was supported by Youth program of National Natural Science Foundation (32100569), Financial Fund of the Ministry of Agriculture and Rural Affairs, P. R. of China (NFZX2021) and Post-graduate’s Innovation Fund Project of Hebei University (HBU2021bs005).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Footnotes
References
Aly, A. H., Edradaebel, R. A., Indriani, I. D., Wray, V., Müller, W. E. G., Ebel, R., et al. (2008). Cytotoxic metabolites from the fungal endophyte Alternaria sp. and their subsequent detection in its host plant Polygonum senegalense. J. Nat. Prod. 71, 972–980. doi: 10.1021/np070447m
Ameen, F., Stephenson, S. L., AlNadhari, S., and Yassin, M. A. (2021). Isolation, identification and bioactivity analysis of an endophytic fungus isolated from Aloe vera collected from Asir desert. Saudi Arabia. Bioprocess Biosyst. Eng. 44, 1063–1070. doi: 10.1007/s00449-020-02507-1
Bradburn, N., Coker, R. D., Blunden, G., Turner, C. H., and Crabb, T. A. (1994). 5′-epialtenuene and neoaltenuene, dibenzo-α-pyrones from Alternaria alternata cultured on rice. Phytochemistry 35, 665–669. doi: 10.1016/S0031-9422(00)90583-1
Bruhn, T., Schaumloffel, A., Hemberger, Y., and Bringmann, G. (2013). SpecDis: quantifying the comparison of calculated and experimental electronic circular dichroism spectra. Chirality 25, 243–249. doi: 10.1002/chir.22138
Chen, Y., Liu, C., Kumaravel, K., Nan, L., and Tian, Y. (2022). Two new sulfate-modified dibenzopyrones with anti-foodborne bacteria activity from sponge-derived fungus Alternaria sp. SCSIOS02F49. Front. Microbiol. 13:879674. doi: 10.3389/fmicb.2022.879674
Chen, B., Shen, Q., Zhu, X., and Lin, Y. (2014). The anthraquinone derivatives from the fungus Alternaria sp. XZSBG-1 from the saline lake in Bange, Tibet, China. Molecules 19, 16529–16542. doi: 10.3390/molecules191016529
Eida, A. A., Ziegler, M., Lafi, F. F., Michell, C. T., Voolstra, C. R., Saad, M. M., et al. (2019). Aesert plant bacteria reveal host influence and beneficial plant growth properties. PLoS One 13:12. doi: 10.1371/journal.pone.0208223
Frisch, M. J., Trucks, G. W., Schlegel, H. B., Scuseria, G. E., Robb, M. A., Fox, D. J., et al. (2019). Gaussian 16, Revision C.01. Wallingford CT: Gaussian, Inc.
Fujii, T., Khawaja, M. R., Dinardo, C. D., Atkins, J. T., and Janku, F. (2016). Targeting isocitrate dehydrogenase (IDH) in cancer. Discov. Med. 21, 373–380.
Gondim, D. D., Gener, M. A., Curless, K. L., Cohen-Gadol, A. A., Hattab, E. M., and Cheng, L. (2019). Determining IDH-mutational status in gliomas using IDH1-R132H antibody and polymerase chain reaction. Appl. Immunohistochem. Mol. Morphol. 27, 722–725. doi: 10.1097/PAI.0000000000000702
He, J. W., Chen, G. D., Gao, H., Yang, F., Li, X. X., Yao, X. S., et al. (2012). Heptaketides with antiviral activity from three endolichenic fungal strains Nigrospora sp. Alternaria sp. and Phialophora sp. Fitoterapia 83, 1087–1091. doi: 10.1016/j.fitote.2012.05.002
He, J. W., Wang, C. X., Yang, L., Chen, G. D., Hu, D., Gao, H., et al. (2016). A pair of new polyketide enantiomers from three endolichenic fungal strains Nigrospora sphaerica, Alternaria alternata, and Phialophora sp. Nat. Prod. Commun. 11, 829–831. doi: 10.1177/1934578x1601100633
Jiao, P., Gloer, J. B., Campbell, J., and Shearer, C. A. (2006). Altenuene derivatives from an unidentified freshwater fungus in the family Tubeufiaceae. J. Nat. Prod. 69, 612–615. doi: 10.1021/np0504661
Jiao, Y., Zhang, X., Wang, L., Li, G., Zhou, J. C., and Lou, H. X. (2013). Metabolites from Penicillium sp. an endophytic fungus from the liverwort Riccardia multifida (L.) S. Gray. Phytochem. Lett. 6, 14–17. doi: 10.1016/j.phytol.2012.10.005
Jin, P. F., Zuo, W. J., Guo, Z. K., Mei, W. L., and Dai, H. F. (2013). Metabolites from the endophytic fungus Penicillium sp. FJ-1 of ceriops tagal. Yao Xue Xue Bao 48, 1688–1691.
Kashiwada, Y., Nonaka, G., and Nishioka, I. (2008). Studies on rhubarb (Rhei rhizoma). V. isolation and characterization of chromone and chromanone derivatives. Chem. Pharm. Bull. 32, 3493–3500. doi: 10.1248/cpb.32.3493
Khamthong, N., Rukachaisirikul, V., Tadpetch, K., Kaewpet, M., Phongpaichit, S., Sakayaroj, J., et al. (2012). Tetrahydroanthraquinone and xanthone derivatives from the marine-derived fungus Trichoderma aureoviride PSU-F95. Arch. Pharm. Res. 35, 461–468. doi: 10.1007/s12272-012-0309-2
Kimura, Y., Shiojima, K., Nakajima, H., and Hamasaki, T. (2008). Structure and biological activity of plant growth regulators produced by Penicillium sp. no.31f. Biosci. Biotechnol. Biochem. 56, 1138–1139. doi: 10.1271/bbb.56.1138
Lu, X., Tang, X. Y., Wang, H. X., Huang, W. J., Feng, W. X., and Feng, B. M. (2021). Polyketone metabolites isolated from Rhodiola tibetica endohytic fungus Alternaria sp. HJT-Y7 and their SARS-CoV-2 virus inhibitory activitives. Bioorg. Chem. 116, 105309–101058. doi: 10.1016/j.bioorg.2021.105309
Meng, X. J., Mao, Z. L., Lou, J. F., Xu, L., Zhong, L. Y., Wang, M. G., et al. (2012). Benzopyranones from the endophytic fungus Hyalodendriella sp. ponipodef12 and their bioactivities. Molecules 17, 11303–11314. doi: 10.3390/molecules171011303
Pang, X. Y., Lin, X. P., Wang, P., Zhou, X. F., Yang, B., Liu, Y. H., et al. (2018). Perylenequione derivatives with anticancer activities isolated from the marine sponge-derived fungus, Alternaria sp. SCSIO41014. Mar. Drugs 16:280. doi: 10.3390/md16080280
Rateb, M. E., Houssen, W. E., Arnold, M., Abdelrahman, M. H., Deng, H., and Harrison, W. T. (2011). Chaxamycins A-D, bioactive ansamycins from a hyper-arid desert Streptomyces sp. J. Nat. Prod. 74, 1491–1499. doi: 10.1021/np200320u
Ruddraraju, K. V., and Zhang, Z. Y. (2017). Covalent inhibition of protein tyrosine phosphatases. Mol. BioSyst. 13, 1257–1279. doi: 10.1039/c7mb00151g
Saxena, S. (2021). “Biologically active secondary metabolites from endophytic Alternaria species”, Endophytes. eds. R. H. Patil and V. L. Maheshwari (Springer Singapore), 1–20.
Song, B., Li, L. Y., Shang, H., Liu, Y., Yu, M., Ding, G., et al. (2019). Trematosphones A and B, two unique dimeric structures from the desert plant endophytic fungus Trematosphaeria terricola. Org. Lett. 21, 2139–2142. doi: 10.1021/acs.orglett.9b00454
Song, X. M., Zhou, X. M., Li, Y. L., Huang, L. B., Chen, C. C., Gao, Y., et al. (2021). Two new cephalochromin derivative from the Alternaria sp. ZG22. Nat. Prod. Res. 35, 3370–3375. doi: 10.1080/14786419.2019.1700248
Tan, Y. Z., Guo, Z. K., Zhu, M. Y., Shi, J., Li, W., Ge, H. M., et al. (2020). Anti-inflammatory spirobisnaphthalene natural products from a plant-derived endophytic fungus Edenia gomezpompae. Chin. Chem. Lett. 31, 1406–1409. doi: 10.1016/j.cclet.2020.03.059
Tanahashi, T., Kuroishi, M., Kuwahara, A., Nagakura, N., and Hamada, N. (1997). Four phenolics from the cultured lichen mycobiont of Graphis scripta var. pulverulenta. Chem. Pharm. Bull. 45, 1183–1185. doi: 10.1002/chin.199751259
Tian, J., Fu, L. Y., Zhang, Z. H., Dong, X. J., Xu, D., Zhou, L. G., et al. (2016). Dibenzo-α-pyrones from the endophytic fungus Alternaria sp. Samif01: isolation, structure elucidation, and their antibacterial and antioxidant activities. Nat. Prod. Res. 31, 387–396. doi: 10.1080/14786419.2016.1205052
Vintonyak, V. V., Antonchick, A. P., Rauh, D., and Waldmann, H. (2009). The therapeutic potential of phosphatase inhibitors. Curr. Opin. Chem. Biol. 13, 272–283. doi: 10.1016/j.cbpa.2009.03.021
Xiao, J., Zhang, Q., Gao, Y. Q., Tang, J. J., Zhang, A. L., and Gao, J. M. (2014). Secondary metabolites from the endophytic Botryosphaeria dothidea of Melia azedarach and their antifungal, antioxidant, and cytotoxic activities. J. Agric. Food Chem. 62, 3584–3590. doi: 10.1021/jf500054f
Xu, J., Hu, Y. W., Qu, W., Chen, M. H., Zhou, L. S., Zhang, J., et al. (2019). Cytotoxic and neuroprotective activities of constituents from Alternaria alternate, a fungal endophyte of Psidium littorale. Bioorg. Chem. 90:103046. doi: 10.1016/j.bioorg.2019.103046
Yang, H. Y., Qi, B. W., Ding, N., Jiang, F. F., Jia, F. F., Shi, S. P., et al. (2019). Polyketides from Alternaria alternata MT-47, an endophytic fungus isolated from Huperzia serrata. Fitoterapia 137:104282. doi: 10.1016/j.fitote.2019.104282
Yang, C. L., Wu, H. M., Liu, C. L., Zhang, X., Guo, Z. K., Ge, H. M., et al. (2019). Bialternacins A-F, aromatic polyketide dimers from an endophytic Alternaria sp. J. Nat. Prod. 82, 792–797. doi: 10.1021/acs.jnatprod.8b00705
Zhang, Z. Y. (2003). Mechanistic studies on protein tyrosine phosphatases. Prog. Nucleic Acid Res. Mol. Biol. 73, 171–220. doi: 10.1016/S0079-6603(03)01006-7
Zhang, X., Liu, X. X., Xing, Y. N., Zhang, M., Zhao, Y., Jiao, R. H., et al. (2022). Alternatones A and B, two polyketides possessing novel skeletons from entophyte Alternaria alternate L-10. J. Asian Nat. Prod. Res. 24, 353–360. doi: 10.1080/10286020.2021.1935893
Zhang, X. Y., Tan, X. M., Yu, M., Yang, J., Sun, B. D., Qin, J. C., et al. (2021). Bioactive metabolites from the desert plant-associated endophytic fungus Chaetomium globosum (Chaetomiaceae). Phytochemistry 185, 112701–112707. doi: 10.1016/j.phytochem.2021.112701
Keywords: desert-plant-derived fungus, polyketides, protein tyrosine phosphatase inhibitory activities, isocitrate dehydrogenase inhibitory activities, Alternaria sp.
Citation: Li Z, Meng L, Ma QY, Wang Z, Zhao YX and Luo DQ (2022) Polyketides with IDH1 R132h and PTP1B inhibitory activities from the desert-plant-derived fungus Alternaria sp. HM 134. Front. Microbiol. 13:975579. doi: 10.3389/fmicb.2022.975579
Edited by:
Dipesh Dhakal, University of Florida, United StatesReviewed by:
Xu-Wen Li, Shanghai Institute of Materia Medica (CAS), ChinaJay D. Keasling, University of California, Berkeley, United States
Copyright © 2022 Li, Meng, Ma, Wang, Zhao and Luo. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhen Wang, d2FuZ3poZW5AaGViZXUuZWR1LmNu; Youxing Zhao, emhhb3lvdXhpbmdAaXRiYi5vcmcuY24=; Duqiang Luo, ZHVxaWFuZ2x1b0AxNjMuY29t
†These authors have contributed equally to this work
 Zhuang Li1†
Zhuang Li1† Zhen Wang
Zhen Wang Youxing Zhao
Youxing Zhao Duqiang Luo
Duqiang Luo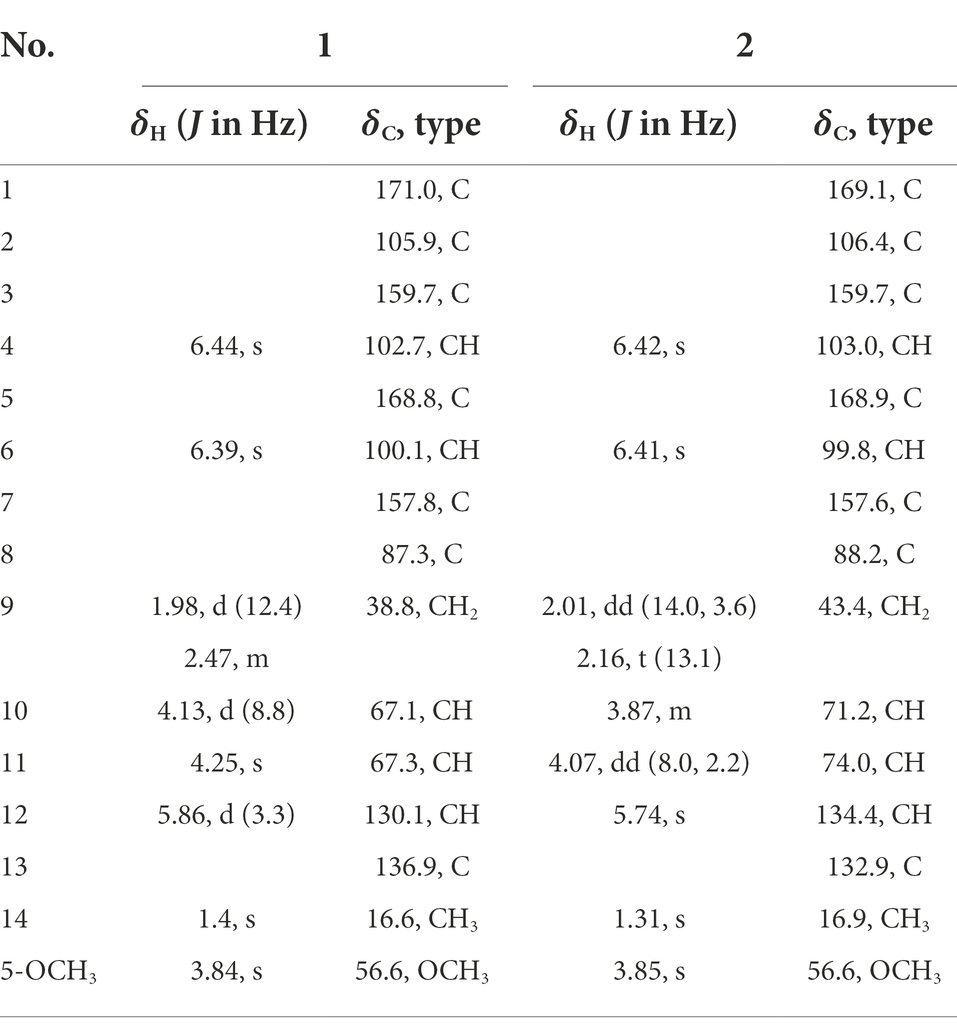
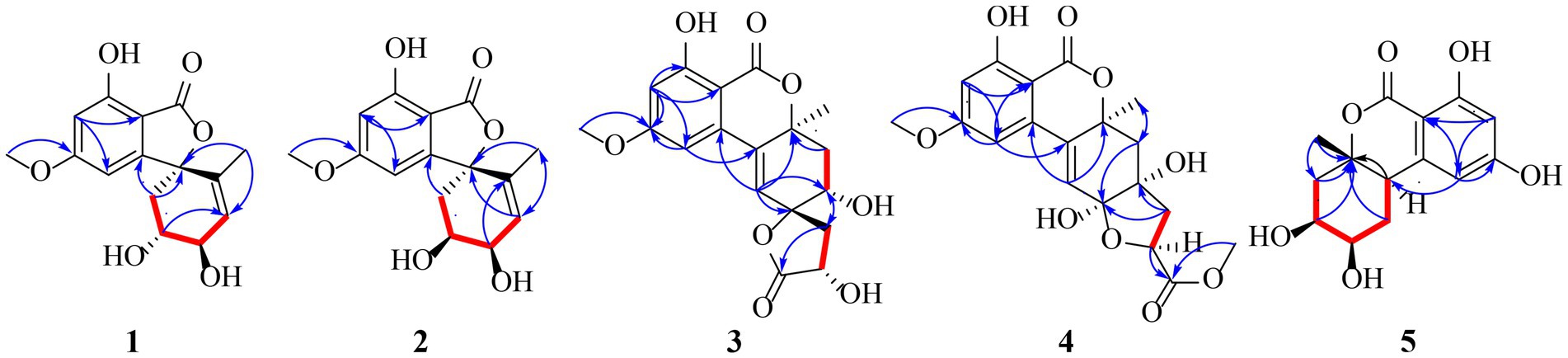
 ) and HMBC (
) and HMBC ( ) correlations of 1-5.
) correlations of 1-5.