- 1College of Food and Biological Engineering, Anhui Key Laboratory of Intensive Processing of Agriculture Products, Hefei University of Technology, Hefei, China
- 2Department of Biological, Food and Environment Engineering, Hefei University, Hefei, China
Aflatoxin B1 (AFB1) contaminates rice during harvest or storage and causes a considerable risk to human and animal health. In this study, Trametes versicolor AFB1–degrading enzyme (TV–AFB1D) gene recombinantly expressed in engineered E. coli BL21 (DE3) and Saccharomyces cerevisiae. The TV–AFB1D enzymatic characteristics and AFB1 degradation efficiency in contaminated rice were investigated. Results showed that the size of recombinant TV-AFB1D expressing in E. coli BL21 (DE3) and S. cerevisiae was appropriately 77 KDa. The kinetic equation of TV-AFB1D was y = 0.01671x + 1.80756 (R2 = 0.994, Km = 9.24 mM, and Vmax = 553.23 mM/min). The Kcat and Kcat/Km values of TV-AFB1D were 0.07392 (s−1) and 8 M−1 s−1, respectively. The AFB1 concentration of contaminated rice decreased from 100 μg/ml to 32.6 μg/ml after treatment at 32°C for 5 h under the catabolism of TV-AFB1D. S. cerevisiae engineered strains carrying aldehyde oxidase 1 (AOX1) and Cauliflower mosaic virus 35 S (CaMV 35 S) promoters caused the residual AFB1 contents, respectively, decreased to 3.4 and 2.9 μg/g from the initial AFB1 content of 7.4 μg/g after 24 h of fermentation using AFB1-contaminated rice as substrate. The AFB1 degradation rates of S. cerevisiae engineered strains carrying AOX1 and CaMV promoters were 54 and 61%, respectively. Engineered S. cerevisiae strains integrated with TV-AFB1D expression cassettes were developed to simultaneously degrade AFB1 and produce ethanol using AFB1-contaminated rice as substrate. Thus, TV-AFB1D has significant application potential in the AFB1 decomposition from contaminated agricultural products.
Introduction
Aflatoxins (AFs) are a group of secondary metabolites produced by Aspergillus flavus and Aspergillus parasiticus that have highly teratogenic, carcinogenic, and mutagenic properties (Lee et al., 2022). AF contamination has posed a serious risk (Yang et al., 2020) and huge economic losses to food safety (Garcia-Cela et al., 2019), as well as human and animal health (Javed et al., 2021). AFs are classified as AFB1, AFB2, AFG1, AFG2, AFM1, AFM2, AFP1, and AFQ1. AFB1 is the most toxic with acute toxicity, teratogenicity, mutagenicity, and carcinogenicity (Cao et al., 2022). AFs are the most frequent contaminants of peanuts, corn, wheat, rice, nuts, milk, and their by-products (Shi et al., 2018; Hajian et al., 2020). More than 10% of the global exposure to AFs was from maize, peanuts, rice, sorghum, and wheat (JECFA, 2017), which caused remarkable economic losses (Pitt and Miller, 2017). The climate change due to latitude differences and the carbon utilization pattern of A. flavus were the main reasons of aflatoxin contamination (Mohale et al., 2013; Van der Fels-Klerx et al., 2016). In addition, the adverse storage conditions of high humidity and high temperature caused excessive aflatoxin levels (Villers, 2014).
The biological degradation of Afs is an emerging biotechnological strategy that is considered an inexpensive and safe practice. The decontaminant activity of microorganisms is associated with fermentation processing and the binding capacity of the cell wall to the contaminant (Wochner et al., 2018). The biological degradation strategy depends on the adsorption of Lactobacillus (Luo et al., 2020) and probiotics (Zoghi et al., 2014) as the microbiological adsorbents and the degradation of AFs using AF oxidases, including laccases (Song et al., 2021), peroxidases (Loi et al., 2020), and lactonases (Pereyra et al., 2019). The enzymatic degradation effectively removes AF toxicity through the destruction of the furan ring and coumarin structure with the formation of nontoxic degradation products (Wang et al., 2019b). The overexpression of AF-degrading enzymes is regarded as an effective way to obtain enzymes that could be used for AF detoxification (Yang et al., 2021). However, the enzymatic separation, purification, and catalytic processes expend great cost during production (Schmidt et al., 2021). Current measures based on direct detoxification approaches, such as physical adsorption, chemical decomposition, and enzymatic degradation (Peng et al., 2018), still have great difficulties in realizing AF detoxification in the fields of food and feed (Kumar et al., 2021). Therefore, the degradation of AFB1 and the expression of AFB1-degrading enzyme during the fermentation of microorganisms would be conducive to decreasing the operating costs.
In the present study, the AFB1-degrading enzyme from Trametes versicolor (TV-AFB1D) was expressed in the recombinant E. coli BL21(DE3). The enzymatic characteristics and catalytic effect were investigated. In addition, TV-AFB1D was used to construct engineered S. cerevisiae strains by Clustered Regularly Interspaced Short Palindromic Repeats–Cas 9 (CRISPR-Cas9) technology. Two TV-AFB1D cassettes were integrated into the hexokinase 2 (HXK2) locus of S. cerevisiae. The degradation of AFB1 in AFB1–contaminated rice was investigated during S. cerevisiae fermentation. An alternative strategy for simultaneous AFB1 degradation and ethanol production by TV–AFB1D–engineered S. cerevisiae strains were explored using the AFB1-contaminated rice as the substrate.
Materials and methods
Plasmids, primers, and reagents
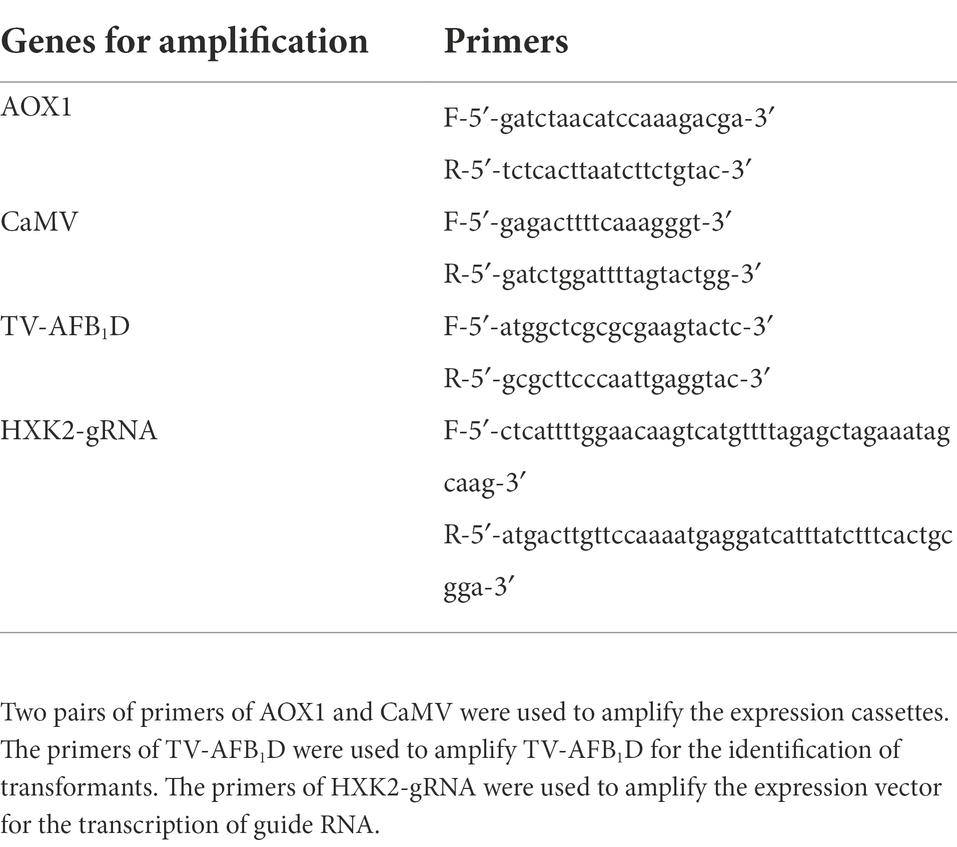
pET-28a (+) expression kit, pEASY-T1 cloning plasmid, RT-PCR kit, E. coli BL21(DE3), ProteinIso Ni-IDA Resin, and E. coli DH 5α were from Transgen Biotech. The gel imaging system, SDS-PAGE, PCR amplification, and electrophoresis devices were manufactured by Bio-Rad Company (United States). Chirascan qCD was manufactured by Applied Photophysics Ltd. (United Kingdom). Plasmid Cas9-NAT carrying nourseothricin and ampicillin resistance genes was obtained from Addgene. The S. cerevisiae HXK2 guide RNA (gRNA) expression vector (HXK2-gRNA) for the expression of the 20 bp gRNA was obtained through the amplification of the gRNA-Trp-Hyb plasmid (Addgene) using the designed primers in Table 1. A. flavus AFB1 was from FERMENTEK (Israel). Primer synthesis and gene sequencing were performed by Sangon Company (China). The analytically pure reagents were from Aladdin Reagent Company (China).
Engineered Escherichia coli BL21(DE3) construction
In this study, TV-AFB1D sequences from NCBI reference sequence NW_007360323.1 was synthesized by Sangon Company (China). TV-AFB1D was inserted into a pET-28a (+) expression vector backbone by double-enzyme XbaI and BamHI digestion and integration approach. T7 promoter primer and T7 terminator primer were used to amplify the insertion sequence by sequencing. The recombinant expression vector embracing the target gene possessed the correct insertion direction and location. The downstream sequence was close to the T7 terminator primer. The confirmed expression vector was transformed into E. coli BL21(DE3) for recombinant expression.
Recombinant expression of TV-AFB1D in E. coli BL21(DE3)
Isopropyl β-D-Thiogalactoside (IPTG) was used to induce the expression of the target gene in engineered E. coli BL21(DE3). The engineered E. coli BL21(DE3) was inoculated into a 100 ml Erlenmeyer flask loaded with 10 ml of LB liquid medium containing 90 μg/ml of kanamycin at 37°C with a shaking speed of 200 rpm. Different concentrations of IPTG were added to induce gene expression when cell concentration reached 0.5 OD600. After centrifugation of fermented broth at 9,000 rpm for 10 min, the supernatant and cells were obtained from the stratified solution to further detect the activity of the recombinant. The composition of fermentation broth mainly included LB liquid medium (10 mg/ml of tryptone, 5 mg/ml of yeast extract, 10 mg/ml of NaCl), 0.19 mg/ml of IPTG (0.8 mmol/l), 90 μg/ml of kanamycin, 0.5 OD600 of TV-AFB1D engineering E. coli BL21(DE3) cells, recombinant TV-AFB1D, and water.
Enzymatic characteristics of recombinant TV-AFB1D
The conditions of 32°C and pH 7 were maintained to investigate the effect of pH value and temperature on TV-AFB1D activity, respectively. The optimum temperature and pH value, and kinetic parameters were determined according to the enzyme activities under the different conditions. Vmax (mM/min) and Km (mM) were calculated based on the Lineweaver–Burk method. The kinetic equation was drawn with 1/V0 and 1/[S] as the ordinate and abscissa, respectively. The Arrhenius plot approach was used to investigate the effect of temperature on TV-AFB1D stabilization by analyzing the relationship between the initial reaction speed and temperature (Fan et al., 2000).
Secondary structure determination
The effect of temperature on the secondary structure of TV-AFB1D was investigated using low temperature (4°C), optimum temperature (32°C), and high temperature (70°C). Circular dichroism spectroscopy (CD) was used to investigate the effect of temperature on the secondary structure. Chirascan qCD was applied to record the CD spectrum values with 0.5 mg/ml of TV-AFB1D in a path length cuvette. CDPro software analyzed the data from 190 nm to 260 nm with a time-per-point of 1 s and an interval of 1 nm (Yi et al., 2019).
TV-AFB1D expression cassette assembly for transformation in Saccharomyces cerevisiae
The TV-AFB1D expression cassettes assembled using the promoters of inducible aldehyde oxidase 1 (AOX1, NCBI Sequence ID: LT727205.1) and constitutive Cauliflower mosaic virus (CaMV) 35S (NCBI Sequence ID: X04879.1) were synthesized according to the detailed sequences. The inducible expression cassette was assembled with the AOX1 promoter, TV-AFB1D, and AOX1 terminator, whereas the constitutive expression cassette was assembled using the CaMV 35S promoter, TV-AFB1D, and CaMV poly-A signal sequences. The primers for AOX1 and CaMV amplification in Table 1 were used to identify the insertion gene. The primers for HXK2-gRNA amplification were used to obtain an expression vector for the transcription of a 20 bp gRNA.
Integration of TV-AFB1D by CRISPR-Cas9 approach in S. cerevisiae
The TV-AFB1D expression cassettes were integrated into the S. cerevisiae genome by HXK2 knockout using the CRISPR-Cas9 technology (Figure 1). The nuclease expressed by the Cas9 vector was cut off by double-stranded DNA guided by gRNA. Then, the TV-AFB1D expression cassettes were inserted into the cut site as the donor DNA to repair the break. The TV-AFB1D expression cassettes were integrated into the genomic DNA of S. cerevisiae by the CRISPR-Cas9 approach through the following steps (Yang et al., 2018). Cas9 plasmid containing nourseothricin resistance gene was transformed into S. cerevisiae by the LiAc/ssDNA/PEG method (Zhang et al., 2014). The 50 μl transformation solution was sucked out and coated on the solid yeast extract peptone dextrose (YPD) medium with the components of 1% yeast extract, 2% yeast extract, and 2% glucose (w/v) containing 100 μg/ml nourseothricin. The S. cerevisiae transformants were used for gene integration. The HXK2-gRNA plasmid and TV-AFB1D expression cassettes were transformed into S. cerevisiae integrated with the Cas9 plasmid. The transformation solution was incubated at 30°C on a solid YPD medium containing 100 μg/ml nourseothricin and 300 μg/ml hygromycin B for 48 h, and the putative transformants were screened out for further identification.
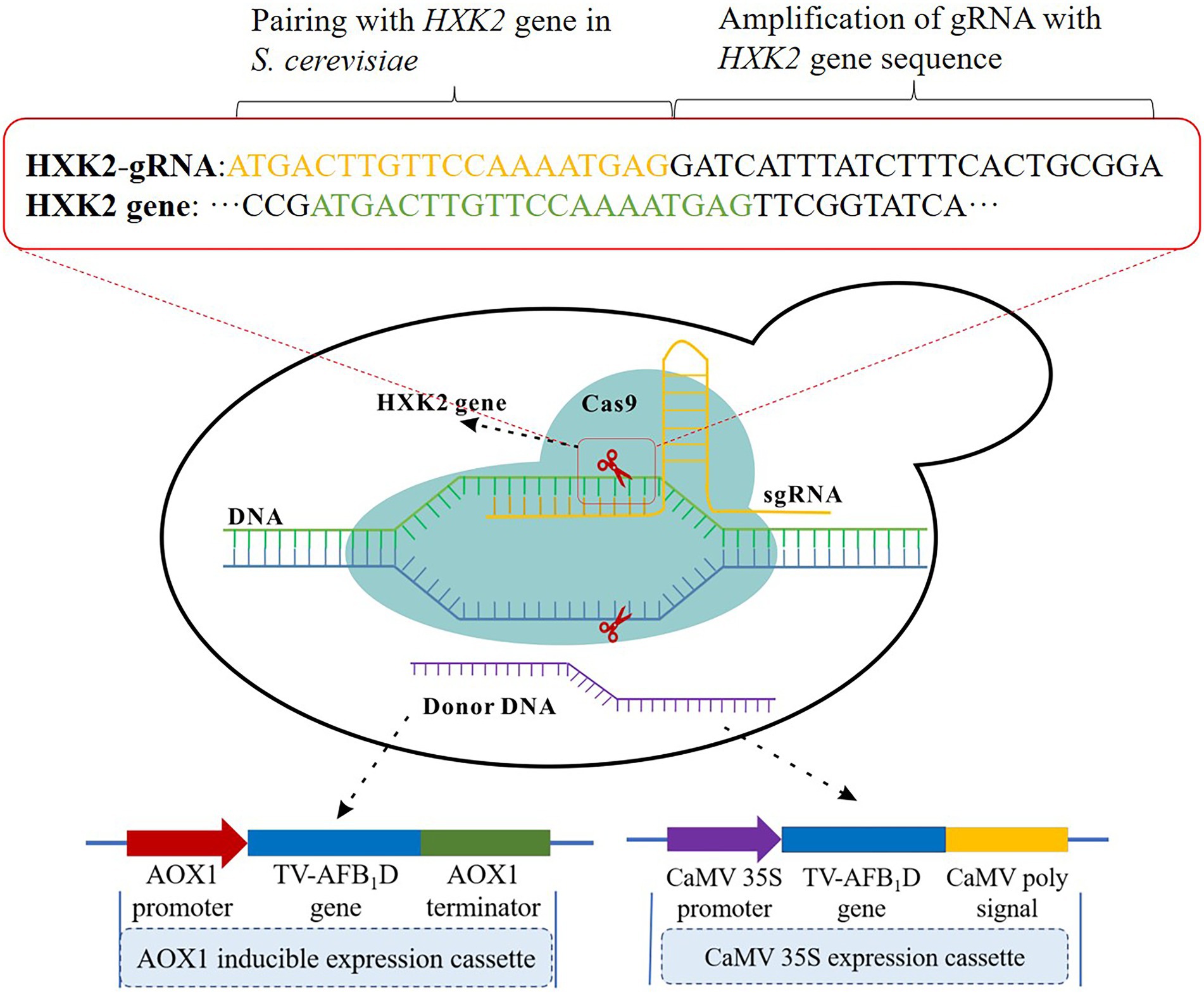
Figure 1. Construction strategy of TV-AFB1D engineered Saccharomyces cerevisiae. Two kinds of expression cassettes containing AXO1 and CaMV promoters were assembled by the promoter, TV-AFB1D, and terminator. The integration locus of expression cassettes as donor DNA was in the HXK2 of S. cerevisiae recognized by 20-bp guide RNA using the CRISPR-Cas9 technology.
Identification and recombinant expression of TV-AFB1D in S. cerevisiae
The putative S. cerevisiae transformants were incubated in a YPD medium at 30°C with a shaking speed of 200 rpm. The primers for TV-AFB1D were used to amplify the genomic DNA of the S. cerevisiae transformants. After confirmation, a profile analysis of the recombinant TV-AFB1D was performed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS–PAGE). The recombinant TV-AFB1D of the engineered S. cerevisiae in the supernatant was concentrated through an ultrafiltration membrane (Singh et al., 2020). The S. cerevisiae transformants after confirmation were inoculated into 500 ml triangular flasks with 200 ml of YPD medium with a shaking speed of 200 rpm at 30°C. The recombinant TV-AFB1D of the AOX1-engineered S. cerevisiae was induced after the addition of methanol to a final concentration of 0.8% (v/v). The supernatant was collected after centrifugation at 7,500 rpm for 20 min.
Cell proliferation of engineered S. cerevisiae
The cell proliferation of S. cerevisiae was investigated in a YPD medium at 30°C with a shaking speed of 200 rpm. S. cerevisiae concentration was determined using the absorbance value under the wavelength of 600 nm. When the OD600 value reached 1, 1 ml of broth was sucked out and poured into a 250 ml triangular bottle containing 100 ml of YPD medium. The effect of gene knockout on the growth of engineered S. cerevisiae was investigated according to the OD600 values (Oh and Jin, 2020).
Determination of glucose and ethanol concentrations
The supernatant of the fermentation broth was obtained after centrifugation at 10,000 rpm for 10 min. Trichloroacetic acid (10% v/v) was added to the supernatant in equal proportion for protein removal after treatment at 4°C for 12 h. The supernatant was further purified by filtration through a 0.45 μm filter membrane. Glucose and ethanol concentrations were determined by high-performance liquid chromatography (HPLC) using a Waters series HPLC system equipped with a refractive detector and SUGAR SH1011 column, a mobile phase of 0.01 mol/l H2SO4, a flow rate of 0.8 ml/min, and a column temperature of 50°C.
Saccharification, fermentation, and in situ degradation of AFB1
The saccharification of rice power was carried out by the double-enzyme method. The starch of rice was hydrolyzed by the mixture of 20 g rice power, 100 ml of water, and 0.02 g α-amylase (40 U/mg) at 70°C for 30 min. After the pH value was adjusted to 4.5, 0.08 g glucoamylase (50 U/mg) was added for the release of glucose at 60°C for 4 h. The saccharified liquid was used to prepare the fermentation broth containing 1% yeast extract (w/v) and 2% peptone (w/v). The two kinds of engineered S. cerevisiae strains were inoculated into the fermentation broth with a shaking speed of 200 rpm at 30°C. In the fermentation processing of the inducible AOX1-engineered S. cerevisiae, 0.8% methanol (v/v) was added for the induction of TV-AFB1D.
AFB1 extraction and HPLC determination
AFB1 extraction was performed by batch micro-solid phase extraction (Chmangui et al., 2021). The rice powder (5 g) was soaked in a 50 ml tube containing an 84% acetonitrile solution (v/v). The supernatant was collected by centrifugation at 6,000 rpm for 10 min. Fat, protein, pigment, and carbohydrate in the extraction solution were removed by a solid-phase column for AF purification (Zhang et al., 2020). AFB1 detection was performed by the HPLC method using the following parameters: a mobile phase of 4:6 methanol–water, C18 reversed-phase column with a UV detector, a detection wavelength of 365 nm, and a flow rate of 0.6 ml/min. AFB1 content was determined by the external standard method (Munoz-Solano and Gonzalez-Penas, 2020).
Data analysis
All the data were presented in the form of mean ± standard deviation with three replicates. Statistical analyses were performed by Origin 9 Software.
Results
Expression of recombinant TV-AFB1D in E. coli BL21(DE3)
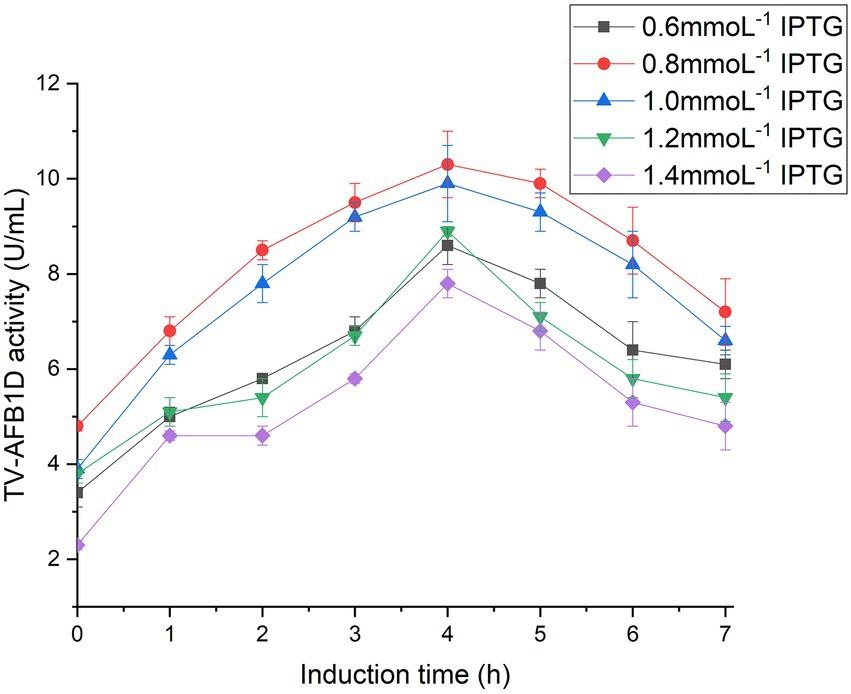
TV-AFB1D was integrated into a pET-28a (+) expression plasmid. The expression vector was confirmed by sequencing. The correct pET-28a (+) vector embracing TV-AFB1D was transformed into E. coli BL21(DE3). The size of TV-AFB1D expressed in engineered E. coli BL21(DE3) was appropriately 77 kDa in the presence of different concentrations of IPTG (Figure 2A). The expression of engineered E. coli BL21(DE3) was induced in the presence of IPTG. The concentration of 0.8 mmol/l of IPTG could effectively induce the expression of TV-AFB1D in engineered E. coli BL21(DE3). Thus, the expression of the recombinant TV-AFB1D in the engineered E. coli BL21(DE3) at 37°C was investigated after the addition of 0.8 mmol/l of IPTG. The highest activity of TV-AFB1D reached 9.3 U/ml for 4 h of induction (Figure 2B). As the control, the wild-type E. coli BL21(DE3) could not produce any TV-AFB1D activity. IPTG concentration and induction time were used to investigate TV-AFB1D activity under the same induction conditions (Figure 3). The combined parameters of 0.8 mmol/l of IPTG and 4 h induction led to the highest TV-AFB1D activity among the set combinations of 0.6–1.4 mmol/l of IPTG and 0–7 h induction time.
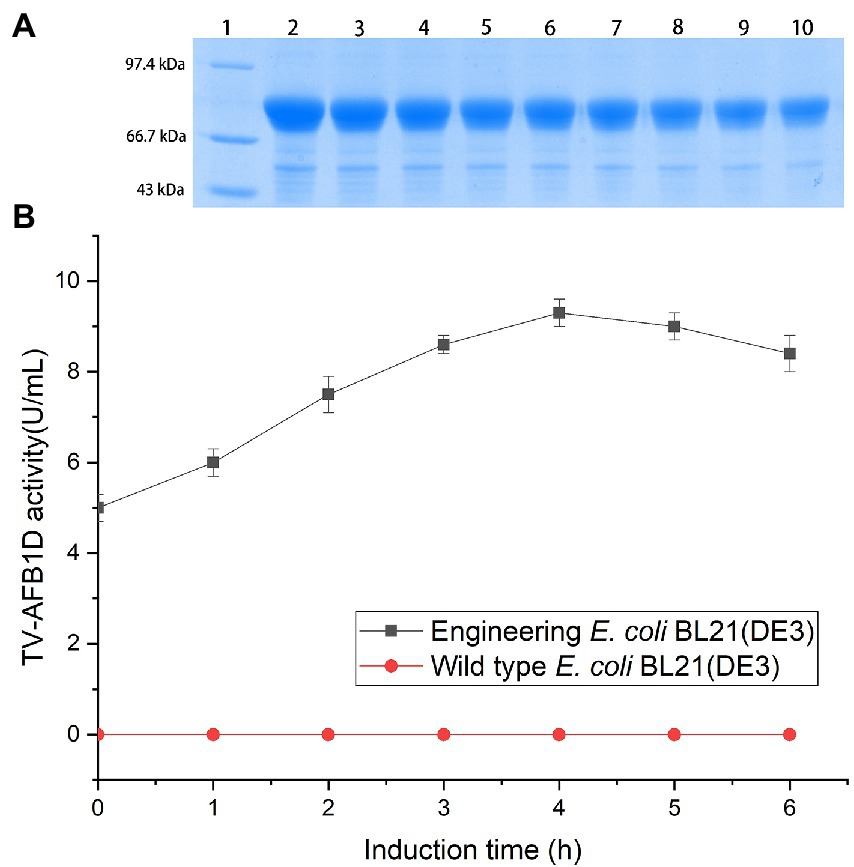
Figure 2. Expression of TV-AFB1D in E. coli BL21(DE3) by IPTG induction (A) Profile of TV-AFB1D expressed in engineered E. coli BL21(DE3) induced by IPTG via SDS-PAGE approach. Note: Lane 1 indicated protein marker; Lane 2–10 indicated the recombinant TV-AFB1D in engineered E. coli BL21(DE3) induced by 1.4, 1.2, 1, 0.8, 0.6, 0.4, 0.3, 0.2, 0.1 mmol/l, respectively. (B) Expression of TV-AFB1D in the transformant and wild-type E. coli BL21(DE3).
Effect of temperature and pH values on TV-AFB1D activity
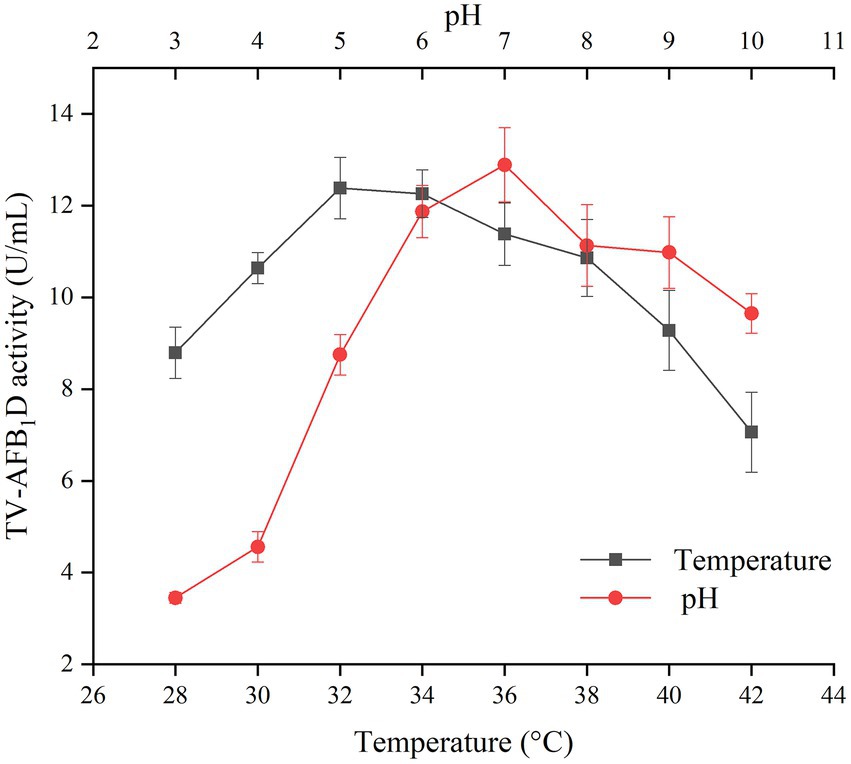
The effects of temperature and pH values on the activity of recombinant TV-AFB1D were investigated to optimize the catalytic conditions (Figure 4). Both temperature and pH values resulted in the changes of TV-AFB1D activity. The activity of recombinant TV-AFB1D reached 12.38 U/ml at 32°C, which was the highest among those during the temperatures of 28–42°C. In addition, the activity of TV-AFB1D was 12.89 U/ml when pH value was adjusted to 7, which was the highest among those at pH values of 3–10. Too high and low temperature and pH values would decrease the activity of recombinant TV-AFB1D. Therefore, TV-AFB1D had characteristics of the optimal temperature of 32°C and pH value of 7.
Determination of kinetic parameters
The multiple kinetic parameters of TV-AFB1D were determined according to their specific calculation formulas in Figure 5. The Lineweaver–Burk equation was used to draw the kinetic equations of TV-AFB1D with a formula of y = 0.01671x + 1.80756 (R2 = 0.994; Figure 5A). In addition, other parameters of Km = 9.24 mM, Vmax = 553.23 mM/min, Kcat = 0.07392 (s−1), and Kcat/Km = 8 M−1 s−1 were calculated according to the relation between substrate concentrations and reaction rate (Figure 5B). Further, Arrhenius plot method was used to determine the relationship between the initial catalytic speed and reaction temperature of TV-AFB1D with a formula of ln v = 16.51–3338.73/T (R2 = 0.999;. Figure 5C).
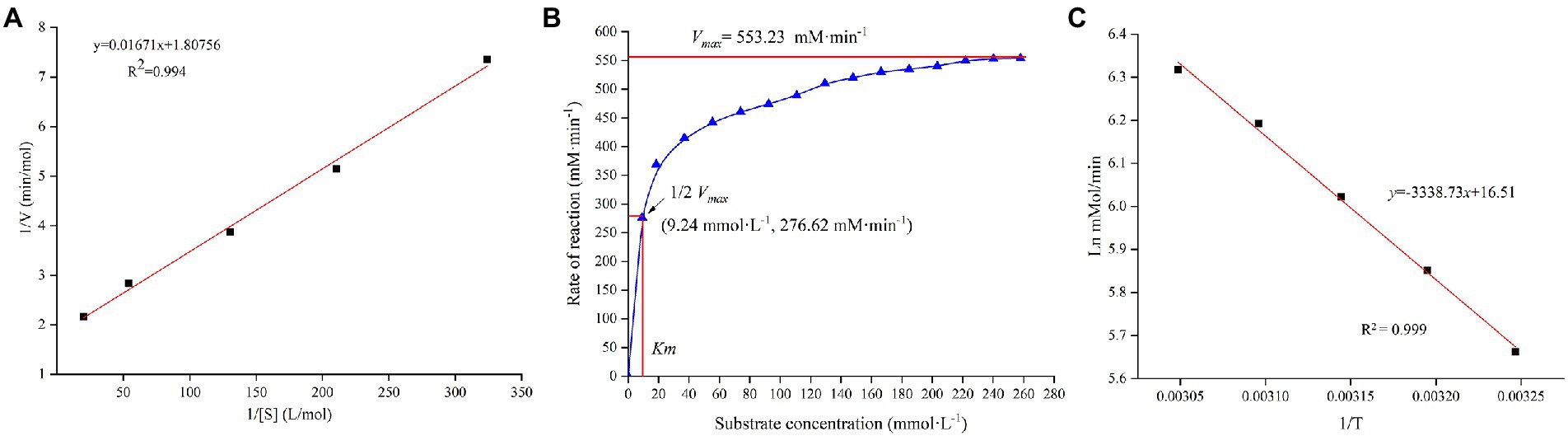
Figure 5. Enzymatic characteristics of recombinant TV-AFB1D. (A) The kinetic equation of TV-AFB1D drawn by Lineweaver–Burk equation based on the double reciprocal of substrate concentration and catalytic rate; (B) Correlation between substrate concentration and reaction rate of TV-AFB1D; (C) Arrhenius plot equation of TV-AFB1D based on the reciprocal of temperature.
Effect of temperature on the secondary structure
The CD spectrum method was used to analyze the secondary structure of TV-AFB1D at 4°C, 32°C, and 70°C (Figure 6A). At 32°C, the highest peak value [θ] was 41.01 deg.·cm2·dmol−1·10−3, which was 1.34-fold at 4°C (30.52 deg.·cm2·dmol−1·10−3) and 2.82-fold at 70°C (14.56 deg.·cm2·dmol−1·10−3). The types of the secondary structure are calculated in Figure 6B. The percentages of random coil and helix at 32°C were highest among the temperatures. The percentage of antiparallel structure at 32°C was lower than those at 4°C and 70°C. Three different treatment temperatures led to similar percentages of parallel and beta-turn structures. Therefore, the percentages of the random coil, helix, and antiparallel structures could be the main factors affecting the activity and stability of TV-AFB1D.
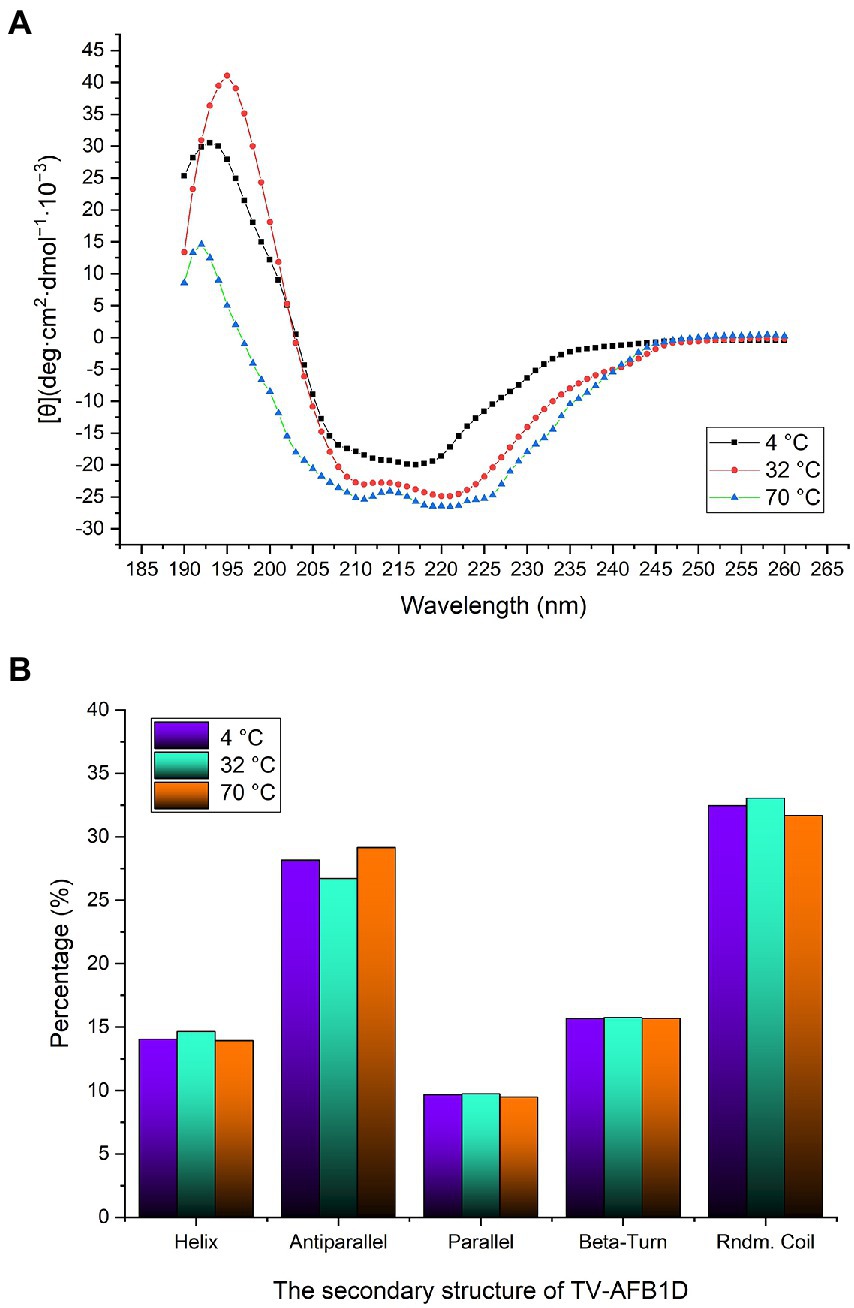
Figure 6. Determination of TV-AFB1D secondary structure. (A) CD spectrum method determined TV-AFB1D secondary structure; (B) Types and percentages of the secondary structure of TV-AFB1D at 4, 32 and 70°C.
Effect of treatment time on the concentration of AFB1
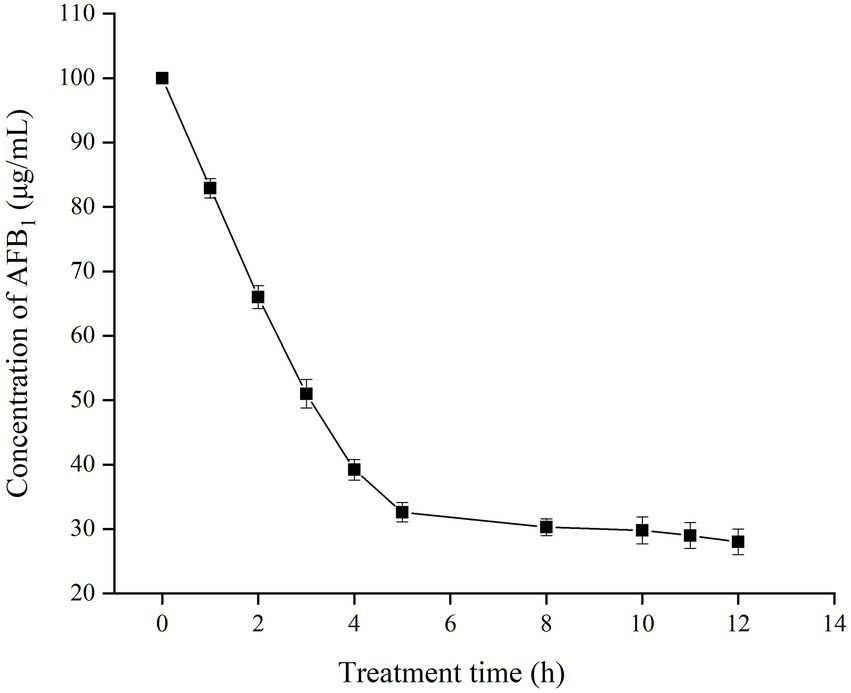
The effect of treatment time on the residual concentration of AFB1 was investigated in the presence of TV-AFB1D (Figure 7). The content of AFB1 gradually decreased from 100 μg/ml to 32.6 μg/ml after catabolism for 5 h. Then, the decrease rate of AFB1 content substantially slowed down with the next treatment of 5–12 h. Therefore, 67.4% of AFB1 was eliminated by TV-AFB1D after 5 h of catabolism.
Transformation of TV-AFB1D expression cassettes into S. cerevisiae
AOX1 inducible and CaMV constitutive expression cassettes have sizes of 3,663 and 3,048 bp, respectively (Figure 8A). After sequencing confirmation, the two expression cassettes were transformed into wild-type S. cerevisiae on the screening medium containing nourseothricin and hygromycin B. The primers designed for TV-AFB1D sequences were used to amplify the genomic DNA of the putative transformants for transformant identification. The sequences of the isolated DNA were further confirmed by the sequencing method. The engineered strains were constructed by integrating the two TV-AFB1D expression cassettes into the S. cerevisiae genome.
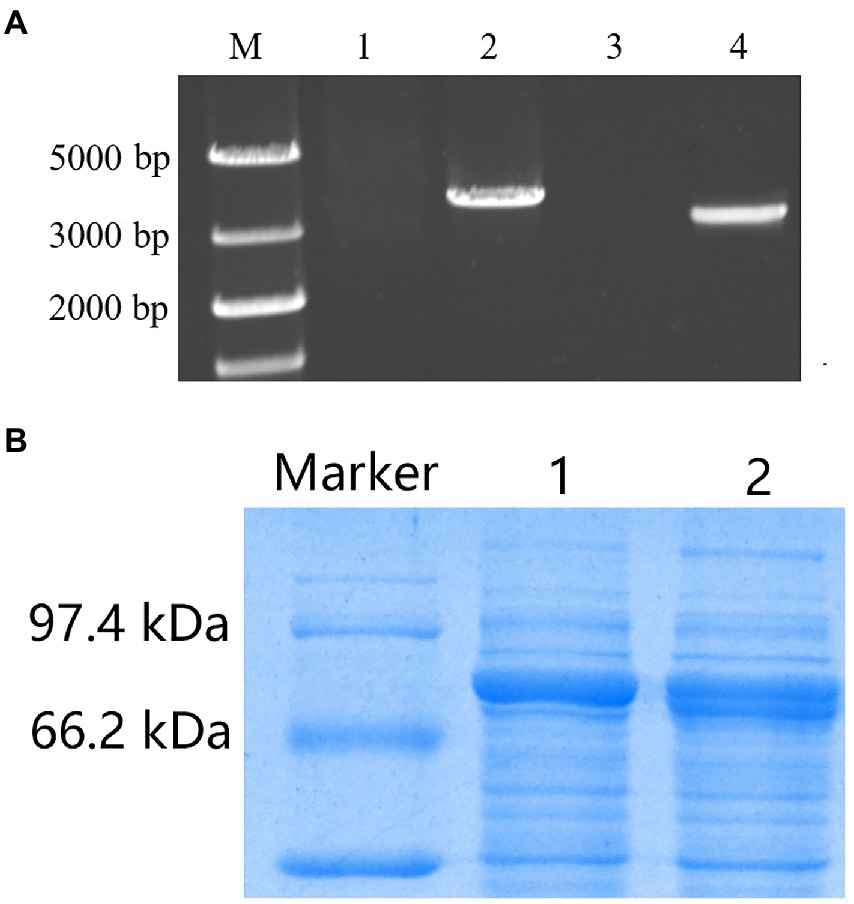
Figure 8. Recombinant expression of TV-AFB1D in Saccharomyces cerevisiae. (A) Amplification of two kinds of TV-AFB1D expression cassettes. Lanes 1 and 2 represented the control and AOX1 expression cassette with a size of 3,663 bp, respectively; lanes 3 and 4 represented the control and CaMV expression cassette with a size of 3,048 bp, respectively; (B) Recombinant TV-AFB1D by the SDS-PAGE approach. Lanes 1 and 2 represented AXO1- and CaMV-engineered strains of S. cerevisiae, respectively.
Profile analysis of recombinant TV-AFB1D by SDS–PAGE
SDS–PAGE was used to analyze the profiles of recombinant TV-AFB1D (Figure 8B). The recombinant TV-AFB1D was expressed by the two kinds of engineered S. cerevisiae with a size of approximately 77 kDa. In addition, the wild-type S. cerevisiae could not express recombinant TV-AFB1D. Thus, engineered S. cerevisiae could express the recombinant TV-AFB1D under the control of AOX1 and CaMV promoters.
Detection of AFB1 catabolism by recombinant TV-AFB1D
The HPLC method was used to detect AFB1 and its catabolites by recombinant TV-AFB1D (Figure 9). AFB1 possessed a chromatographic peak after the run time of 45 min. The catabolites of AFB1 by recombinant TV-AFB1D were also detected by HPLC with a run time of 22 min. The result indicated that a new product was formed under the catabolism of recombinant TV-AFB1D.
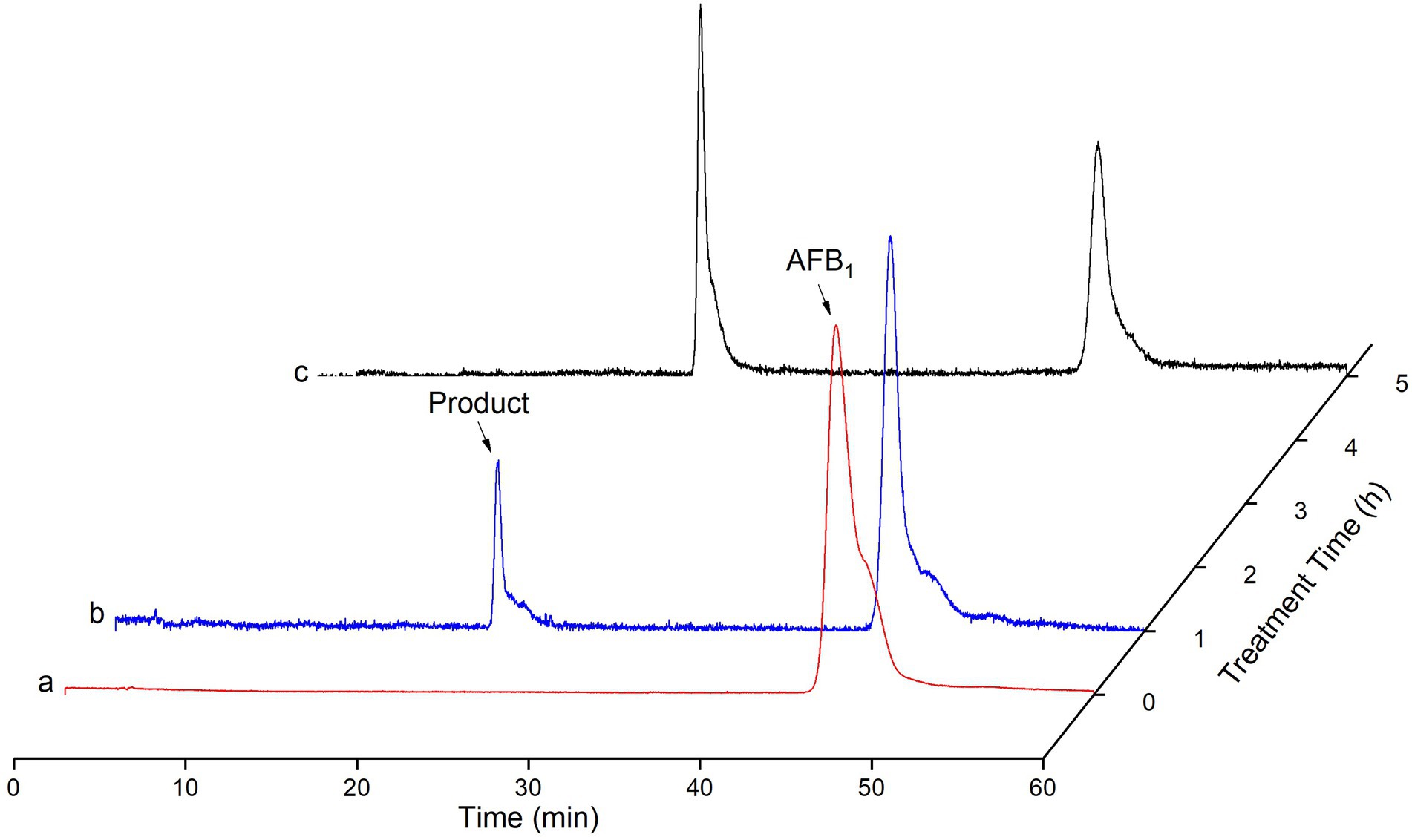
Figure 9. Determination of AFB1 degraded by TV-AFB1D using HPLC approach. a, b, and c, respectively, represented HPLC standard AFB1, products after catabolism for 2 h and 4 h.
HXK2 knockout affecting the growth, ethanol production, and glucose consumption of S. cerevisiae
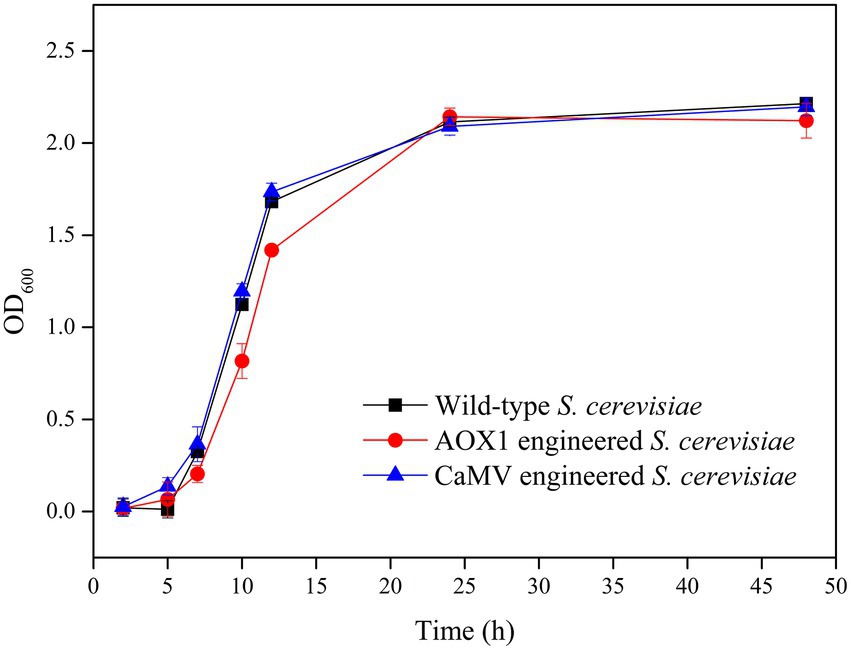
The absorbance of the fermentation broth at the wavelength of 600 nm was determined to investigate the effect of the gene integration of TV-AFB1D expression cassettes on the growth of S. cerevisiae mutants with HXK2 knockout (Figure 10). The results showed that S. cerevisiae mutants integrated with AOX1 and CaMV expression cassettes had similar growth curves as the wild-type S. cerevisiae during fermentation for 48 h. Therefore, the HXK2 mutation of S. cerevisiae by gene knockout did not affect the cell proliferation of the engineered strains.
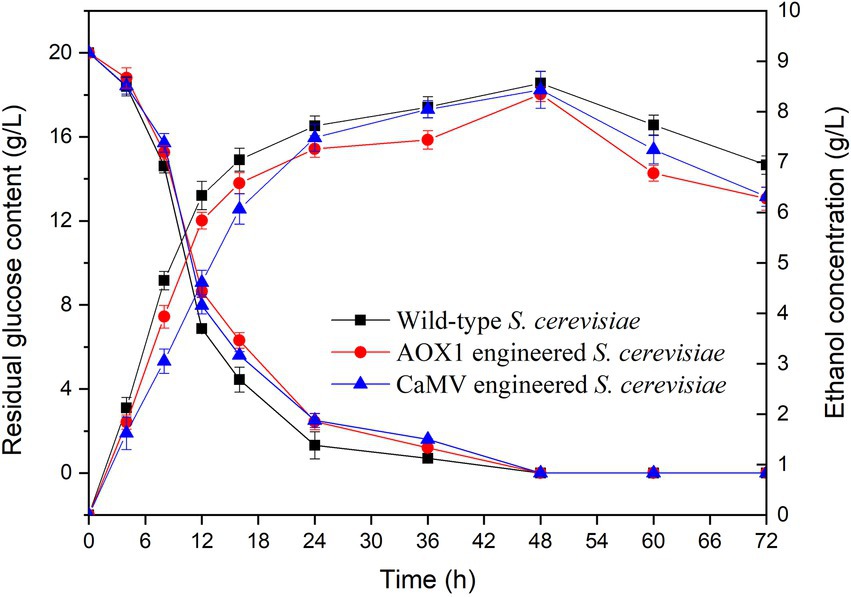
The residual glucose contents and ethanol concentrations of the engineered and wild-type S. cerevisiae strains were determined during fermentation for 72 h (Figure 11). During the initial fermentation (0–24 h), all the strains had a rapid increase in ethanol concentration from 0 g/l to 7.5 g/l. All the strains reached the highest ethanol concentrations after fermentation for 48 h. The ethanol concentrations from engineered S. cerevisiae strains carrying AOX1 (8.35 g/l) and CaMV (8.43 g/l) promoters were 97.4 and 98.4% of that in the wild-type strain (8.57 g/l), respectively. The integration of TV-AFB1D expression cassettes did not affect ethanol production during fermentation.
Residual glucose contents were investigated during the fermentation of the wild-type and engineered S. cerevisiae strains for 72 h (Figure 11). The results indicated that the two kinds of engineered S. cerevisiae strains had similar trends in glucose content as that of the wild-type S. cerevisiae. The residual glucose contents markedly decreased during fermentation time at 0–24 h. In addition, the residual glucose contents slightly decreased during the subsequent fermentation at 24–48 h. Almost all the glucose was consumed by the tested strains during fermentation at 48–72 h. The integration of TV-AFB1D expression cassettes by HXK2 knockout did not affect the glucose consumption of the engineered S. cerevisiae strains.
In situ catabolism of AFB1 and fermentation of AFB1-contaminated rice
The starch of AFB1-contaminated rice was converted into glucose under the catabolism of α-amylase and glucoamylase. The saccharification solution was used to prepare the liquid broth for ethanol production by S. cerevisiae fermentation. The residual AFB1 concentrations of S. cerevisiae in the fermentation broth were investigated during the fermentation for 48 h (Figure 12). The results indicated that the two kinds of engineered strains possessed higher AFB1 degradation efficiencies than the wild-type S. cerevisiae. The concentrations of residual AFB1 markedly decreased during the initial fermentation time of 0–24 h and slightly decrease at 24–48 h of fermentation. The concentrations of residual AFB1 from the wild-type strain (6.1 μg/g), AOX1-engineered strain (3.4 μg/g), and CaMV-engineered strain (2.9 μg/g) after 24 h of fermentation were lower than the initial AFB1 concentration of 7.4 μg/g. The AOX1- and CaMV-engineered S. cerevisiae strain resulted in the degradation of 54 and 61% of AFB1, respectively, which were higher than the wild-type S. cerevisiae (18% AFB1). Thus, both engineered strains could degrade AFB1 during the fermentation of the saccharification solution from AFB1-contaminated rice for ethanol production.
Discussion
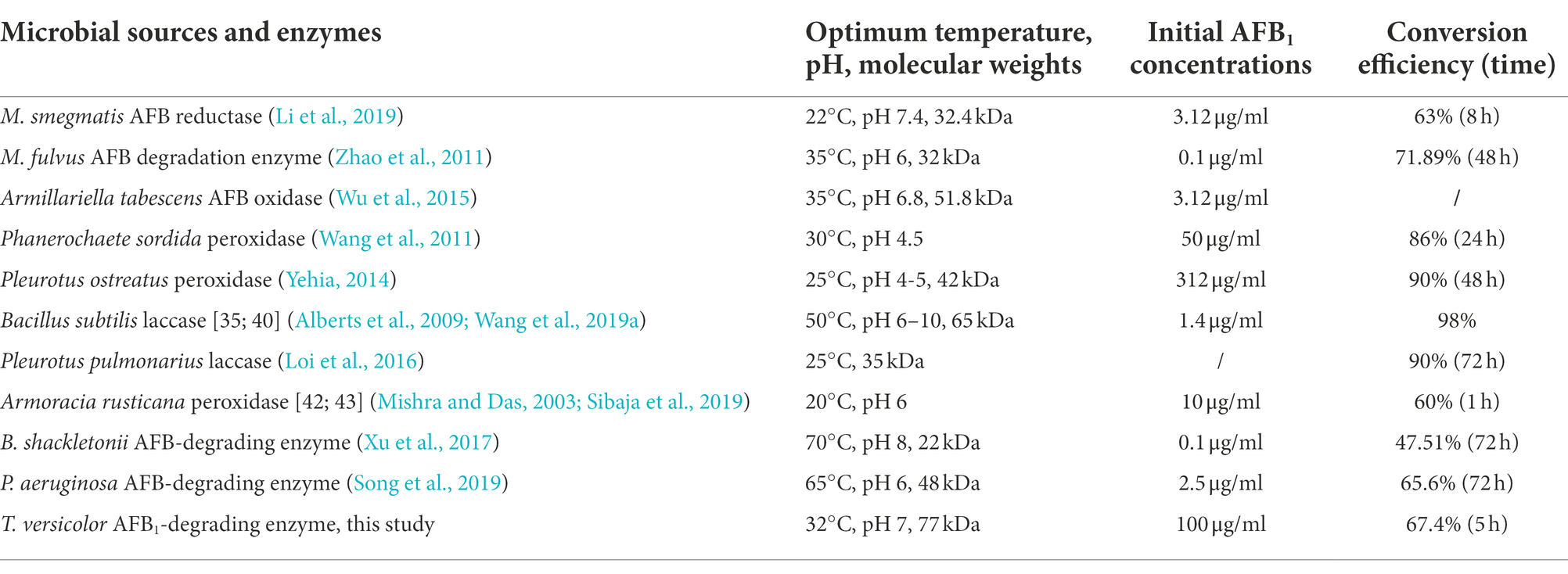
AFB1 widely exists in nature and even breaks out in certain environments as a chemically stable mycotoxin. It threatens food and animal feed security and causes an economic challenge and health hazard to consumers. Enzymatic hydrolysis is an effective detoxification approach of AFB1 owing to its nutrition maintenance without toxicity residue. The specific enzymes can directly degrade aflatoxins without the shortcoming of the application of a whole microorganism. The F420H2-dependent reductases reduced the α,β-unsaturated ester and destabilized the lactone ring (Taylor et al., 2010). Various enzymes such as peroxidases (Yehia, 2014), laccases (Alberts et al., 2009), oxidases (Wu et al., 2015), and reductases (Taylor et al., 2010) have been identified as responsible for AFB1 degradation (Table 2). The optimum temperature of 32°C and pH value of 7 could maintain better enzymic activity and stability of TV-AFB1D compared with other enzymes. The previously reported enzymes from different host bacteria demonstrate the optimum temperature of 20–70°C and pH values of 4.5–10 (Table 2). The optimum temperature and pH of AFB1 degradation enzyme are identical to the fermentation conditions of S. cerevisiae (32°C and pH 7). This coincidence can facilitate yeast fermentation and AFB1 degradation by TV-AFB1D. In addition, TV-AFB1D can convert 67.4% of AFB1 into other compounds in 5 h, whereas most of the other enzymes reached a high conversion rate after processing for 48–72 h. Therefore, the recombinant TV-AFB1D has good application value for its high catalytic efficiency.
The cell wall of yeast as mycotoxin adsorbents could reduce the exposure of animals to mycotoxins to some extent (Yiannikouris et al., 2021). However, the adsorption capacity is affected by the composition, thickness, mannan-oligosaccharide, and β-glucan content of the cell walls of yeast (Pereyra et al., 2018). In the present study, we demonstrated the expression of TV-AFB1D and in situ AFB1 degradation during the ethanol fermentation of engineered S. cerevisiae strains. The TV-AFB1D-engineered S. cerevisiae strains are capable of AFB1 degradation during the fermentation of AFB1-contaminated rice for ethanol production without the addition of any other detoxification agents. Based on the design concept and study result, we proposed a new strategy for the AFB1 degradation of AFB1-contaminated rice by TV-AFB1D-engineered S. cerevisiae strains during the fermentation for ethanol production (Figure 13). In this strategy, the engineered S. cerevisiae strains integrated with TV-AFB1D expression cassettes were constructed by CRISPR-Cas9 knockout. The gene integration in the HXK2 locus did not markedly affect the growth and ethanol production of the engineered strains. The application of this strategy in AFB1-contaminated grain could effectively decrease the AFB1 concentration during fermentation without the addition of extra enzymes, microorganisms, and adsorbents. Therefore, the TV-AFB1D-engineered S. cerevisiae strains could degrade about 60% of AFB1 from AFB1-contaminated rice by in situ AFB1 degradation. TV-AFB1D has application potential in the AFB1 degradation from contaminated agricultural products.
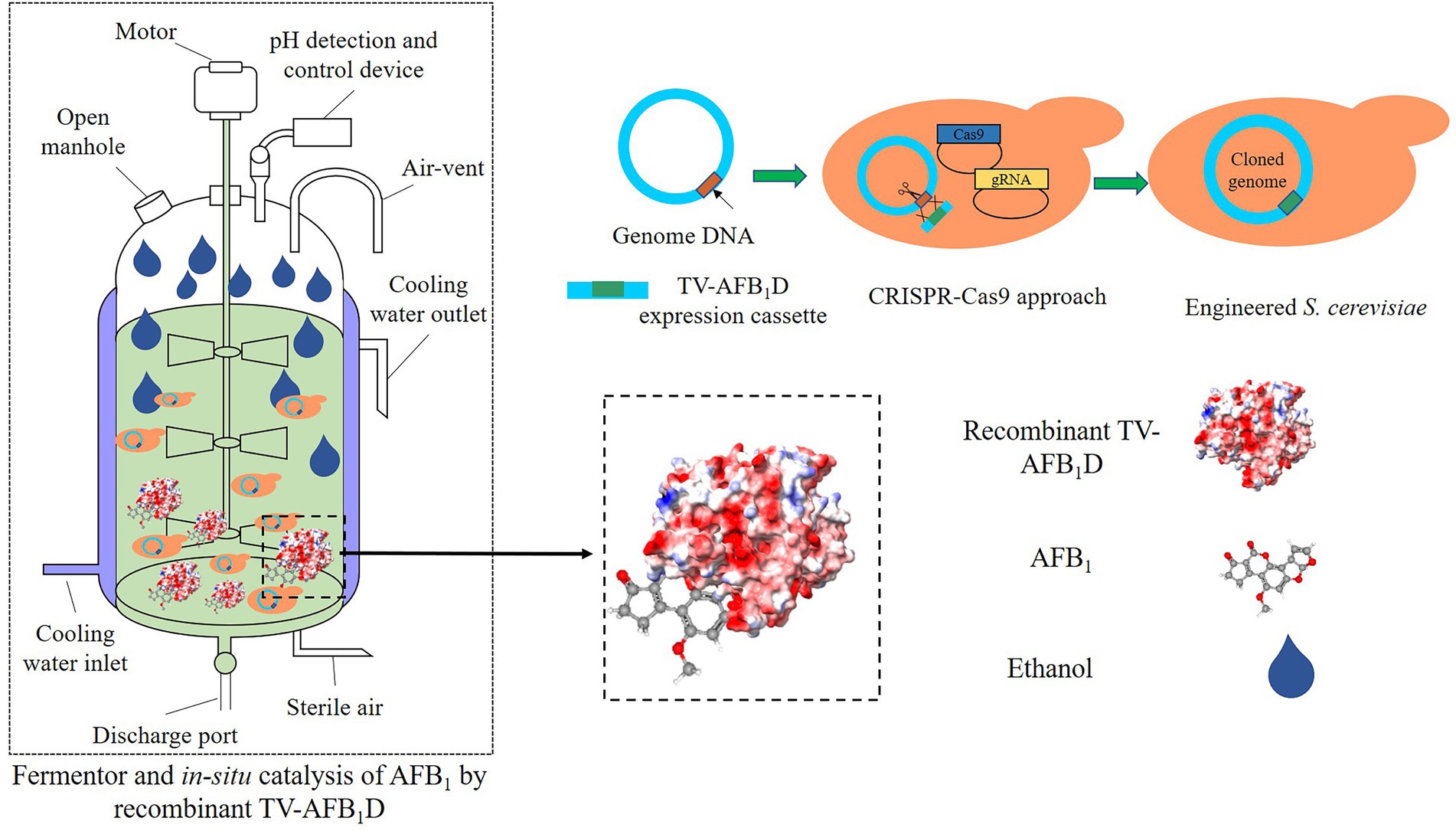
Figure 13. Strategy for AFB1 degradation and ethanol production by TV-AFB1D-engineered S. cerevisiae using AFB1-contaminated rice as material.
Conclusion
The recombinant TV-AFB1D with a size of appropriately 77 KDa expressed in engineered E. coli BL21(DE3) and S. cerevisiae. The kinetic equation of TV-AFB1D was y = 0.01671x + 1.80756. The concentration of AFB1 from contaminated rice decreased from the initial 100 μg/ml to the final 32.6 μg/ml after catabolism for 5 h in the presence of TV-AFB1D. After fermentation for 24 h, the AFB1 contents of the wild-type, AOX1-engineered, and CaMV-engineered S. cerevisiae strains were 6.1, 3.4, and 2.9 μg/g from the initial concentration of 7.4 μg/g, respectively. In addition, an alternative strategy was proposed to degrade the AFB1 from AFB1-contaminated grain using TV-AFB1D-engineered S. cerevisiae strains during the fermentation processing. This measure has various advantages of reducing the steps of detoxification treatment, reducing production costs, and ensuring the safety of downstream products. Therefore, TV-AFB1D-engineered S. cerevisiae has an important development value to produce ethanol by simultaneous detoxification and fermentation with mycotoxin-contaminated crops as substrates.
Data availability statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Author contributions
PY provided conceptualization and performed writing. SL performed the experiment. ShuJ and SuJ provided software. ZZ and ShtJ provided resources. WX, JC, and WW performed investigation. All authors contributed to the article and approved the submitted version.
Funding
This research was funded by the Natural Science Foundation of Anhui Province (1908085MC80) and Major Science and Technology Projects of Anhui Province (202003c08020001 and 202103b06020014).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.960882/full#supplementary-material
References
Alberts, J., Gelderblom, W., Botha, A., and Zyl, W. (2009). Degradation of aflatoxin B1 by fungal laccase enzymes. Int. J. Food Microbiol. 135, 47–52. doi: 10.1016/j.ijfoodmicro.2009.07.022
Cao, W. Y., Yu, P., Yang, K. P., and Cao, D. L. (2022). Aflatoxin B1: metabolism, toxicology, and its involvement in oxidative stress and cancer development. Toxicol. Mech. Methods 1, 1–25. doi: 10.1080/15376516.2021.2021339
Chmangui, A., Jayasinghe, G., Driss, M. R., Touil, S., Bermejo-Barrera, P., Bouabdallah, S., et al. (2021). Assessment of trace levels of aflatoxins AFB1 and AFB2 in non-dairy beverages by molecularly imprinted polymer based micro solid-phase extraction and liquid chromatography-tandem mass spectrometry. Anal. Methods 13, 3433–3443. doi: 10.1039/D1AY00793A
Fan, Y., McPhie, P., and Miles, E. (2000). Regulation of tryptophan synthase by temperature, monovalent cations, and an allosteric ligand. Evidence from Arrhenius plots, absorption spectra, and primary kinetic isotope effects. Biochemistry 39, 4692–4703. doi: 10.1021/bi9921586
Garcia-Cela, E., Kiaitsi, E., Sulyok, M., Krska, R., Medina, A., Damico, I. P., et al. (2019). Influence of storage environment on maize grain: CO2 production, dry matter losses and aflatoxins contamination. Food Addit. Contam. Part A-Chem. 36, 175–185. doi: 10.1080/19440049.2018.1556403
Hajian, N., Salami, M., Mohammadian, M., Moghadam, M., Emam, Z., and Djomeh, (2020). Production of low-fat camel milk functional ice creams fortified with camel milk casein and its antioxidant hydrolysates. Appl. Food Biotechnol. 7, 95–102. doi: 10.22037/afb.v7i2.27779
Javed, A., Naeem, I., Benkerroum, N., Riaz, M., Akhtar, S., Ismail, A., et al. (2021). Occurrence and health risk assessment of aflatoxins through intake of eastern herbal medicines collected from four districts of southern Punjab-Pakistan. Int. J. Environ. Res. Public Health 18, 1–20. doi: 10.3390/ijerph18189531
JECFA (2017). Evaluation of certain contaminants in food. Eighty-third report of the Joint FAO/WHO Expert Committee on Food Additives (JECFA). Food and Agriculture Organization of the United Nations, World Health Organization, WHO Technical Report Series 1002, World Health Organization, Geneva.
Kumar, V., Bahuguna, A., Ramalingam, S., Dhakal, G., Shim, J. J., and Kim, M. (2021). Recent technological advances in mechanism, toxicity, and food perspectives of enzyme-mediated aflatoxin degradation. Crit. Rev. Food Sci. Nutr. 1, 1–18. doi: 10.1080/10408398.2021.2010647
Lee, S. Y., Woo, S. Y., Tian, F., Jeong, A. Y., Park, S. B., and Chun, H. S. (2022). Contamination characteristics and risk assessment of aflatoxins in homemade soybean paste, a traditional fermented soybean food, in South Korea. J. Hazard. Mater. 424, 1–11. doi: 10.1016/j.jhazmat.2021.127576
Li, C., Li, W., Hsu, I., Liao, Y., Yang, C., Taylor, M., et al. (2019). Recombinant aflatoxin-degrading F420H2-dependent reductase from mycobacterium smegmatis protects mammalian cells from aflatoxin toxicity. Toxins 11, 1–16. doi: 10.3390/toxins11050259
Loi, M., Fanelli, F., Zucca, P., Liuzzi, V., Quintieri, L., Cimmarusti, M., et al. (2016). Aflatoxin B1 and M1 degradation by Lac2 from Pleurotus pulmonarius and redox mediators. Toxins 8, 1–16. doi: 10.3390/toxins8090245
Loi, M., Renaud, J. B., Rosini, E., Pollegioni, L., Vignali, E., Haidukowski, M., et al. (2020). Enzymatic transformation of aflatoxin B-1 by Rh_DypB peroxidase and characterization of the reaction products. Chemosphere 250, 1–7. doi: 10.1016/j.chemosphere.2020.126296
Luo, Y., Liu, X. J., Yuan, L., and Li, J. K. (2020). Complicated interactions between bio-adsorbents and mycotoxins during mycotoxin adsorption: current research and future prospects. Trends Food Sci. Technol. 96, 127–134. doi: 10.1016/j.tifs.2019.12.012
Mishra, H. N., and Das, C. (2003). A review on biological control and metabolism of aflatoxin. Crit. Rev. Food Sci. Nutr. 43, 245–264. doi: 10.1080/10408690390826518
Mohale, S., Magan, N., and Medina, A. (2013). Comparison of growth, nutritional utilisation patterns, and niche overlap indices of toxigenic and atoxigenic Aspergillus flavus strains. Fungal Biol. 117, 650–659. doi: 10.1016/j.funbio.2013.07.002
Munoz-Solano, B., and Gonzalez-Penas, E. (2020). Mycotoxin determination in animal feed: An LC-FLD method for simultaneous quantification of aflatoxins, ochratoxins and zearelanone in this matrix. Toxins 12, 1–16. doi: 10.3390/toxins12060374
Oh, E. J., and Jin, Y. S. (2020). Engineering of Saccharomyces cerevisiae for efficient fermentation of cellulose. FEMS Yeast Res. 20, 1–11. doi: 10.1093/femsyr/foz089
Peng, Z., Chen, L. K., Zhu, Y. L., Huang, Y., Hu, X. L., Wu, Q. H., et al. (2018). Current major degradation methods for aflatoxins: a review. Trends Food Sci. Technol. 80, 155–166. doi: 10.1016/j.tifs.2018.08.009
Pereyra, C. M., Gil, S., Cristofolini, A., Bonci, M., Makita, M., Monge, M. P., et al. (2018). The production of yeast cell wall using an agroindustrial waste influences the wall thickness and is implicated on the aflatoxin B1 adsorption process. Food Res. Int. 111, 306–313. doi: 10.1016/j.foodres.2018.05.026
Pereyra, M. L. G., Martinez, M. P., and Cavaglieri, L. R. (2019). Presence of aiiA homologue genes encoding for N-acyl homoserine lactone-degrading enzyme in aflatoxin B1-decontaminating bacillus strains with potential use as feed additives. Food Chem. Toxicol. 124, 316–323. doi: 10.1016/j.fct.2018.12.016
Pitt, J. I., and Miller, J. D. (2017). A concise history of mycotoxin research. J. Agric. Food Chem. 65, 7021–7033. doi: 10.1021/acs.jafc.6b04494
Schmidt, M. A., Mao, Y., Opoku, J., and Mehl, H. L. (2021). Enzymatic degradation is an effective means to reduce aflatoxin contamination in maize. BMC Biotechnol. 21, 1–10. doi: 10.1186/s12896-021-00730-6
Shi, H. T., Li, S. L., Bai, Y. Y., Prates, L. L., Lei, Y. G., and Yu, P. Q. (2018). Mycotoxin contamination of food and feed in China: occurrence, detection techniques, toxicological effects and advances in mitigation technologies. Food Control 91, 202–215. doi: 10.1016/j.foodcont.2018.03.036
Sibaja, K., Garcia, S., Feltrin, A., Remedi, R., Cerqueira, M., Badiale-Furlong, E., et al. (2019). Aflatoxin biotransformation by commercial peroxidase and its application in contaminated food. J. Chem. Technol. Biotechnol. 94, 1187–1194. doi: 10.1002/jctb.5865
Singh, R. S., Singh, T., and Kennedy, J. F. (2020). Purification, thermodynamics and kinetic characterization of fungal endoinulinase for the production of fructooligosaccharides from inulin. Int. J. Biol. Macromol. 164, 3535–3545. doi: 10.1016/j.ijbiomac.2020.09.001
Song, Y. Y., Wang, Y. A., Guo, Y. P., Qiao, Y. Y., Ma, Q. G., Ji, C., et al. (2021). Degradation of zearalenone and aflatoxin B1 by Lac2 from Pleurotus pulmonarius in the presence of mediators. Toxicon 201, 1–8. doi: 10.1016/j.toxicon.2021.08.003
Song, J., Zhang, S., Xie, Y., and Li, Q. (2019). Purification and characteristics of an aflatoxin B1 degradation enzyme isolated from Pseudomonas aeruginosa. FEMS Microbiol. Lett. 366, 1–7. doi: 10.1093/femsle/fnz034
Taylor, M., Jackson, C., Tattersall, D., French, N., Peat, T., Newmann, J., et al. (2010). Identification and characterization of two families of F420H2-dependent reductases from Mycobacteria that catalyse aflatoxin degradation. Mol. Microbiol. 78, 561–575. doi: 10.1111/j.1365-2958.2010.07356.x
Van der Fels-Klerx, H. J., Liu, C., and Battilani, P. (2016). Modelling climate change impacts on mycotoxin contamination. World Mycotoxin J. 9, 717–726. doi: 10.3920/WMJ2016.2066
Villers, P. (2014). Aflatoxins and safe storage. Front. Microbiol. 5, 1–6. doi: 10.3389/fmicb.2014.00158
Wang, X., Bai, Y., Huang, H., Tu, T., Wang, Y., Wang, Y., et al. (2019a). Degradation of aflatoxin B1 and zearalenone by bacterial and fungal laccases in presence of structurally defined chemicals and complex natural mediators. Toxins 11, 1–17. doi: 10.3390/toxins11100609
Wang, J., Ogata, M., Hirai, H., and Kawagishi, H. (2011). Detoxification of aflatoxin B1 by manganese peroxidase from the white-rot fungus Phanerochaete sordida YK-624. FEMS Microbiol. Lett. 314, 164–169. doi: 10.1111/j.1574-6968.2010.02158.x
Wang, X. L., Qin, X., Hao, Z. Z., Luo, H. Y., Yao, B., and Su, X. Y. (2019b). Degradation of four major mycotoxins by eight manganese peroxidases in presence of a dicarboxylic acid. Toxins 11, 1–16. doi: 10.3390/toxins11100566
Wochner, K. F., Becker-Algeri, T. A., Colla, E., Badiale-Furlong, E., and Drunkler, D. A. (2018). The action of probiotic microorganisms on chemical contaminants in milk. Crit. Rev. Microbiol. 44, 112–123. doi: 10.1080/1040841X.2017.1329275
Wu, Y., Lu, F., Jian, H., Tan, C., Yao, D., Xie, C., et al. (2015). The furofuran-ring selectivity, hydrogen peroxide production and low km value are the three elements for highly effective detoxification of aflatoxin oxidase. Food Chem. Toxicol. 76, 125–131. doi: 10.1016/j.fct.2014.12.004
Xu, L., Ahmed, M., Sangare, L., Zhao, Y., Selvaraj, J., Xing, F., et al. (2017). Novel aflatoxin-degrading enzyme from bacillus shackletonii L7. Toxins 9, 1–15. doi: 10.3390/toxins9010036
Yang, P. Z., Lu, S. H., Xiao, W., Zheng, Z., Jiang, S. W., Jiang, S. T., et al. (2021). Activity enhancement of Trametes versicolor aflatoxin B-1-degrading enzyme (TV-AFB(1)D) by molecular docking and site-directed mutagenesis. Food Bioprod. Process. 129, 168–175. doi: 10.1016/j.fbp.2021.08.007
Yang, P. Z., Wu, Y., Zheng, Z., Cao, L. L., Zhu, X. X., Mu, D. D., et al. (2018). CRISPR-Cas9 approach constructing cellulase sestc-engineered Saccharomyces cerevisiae for the production of orange peel ethanol. Front. Microbiol. 9, 1–7. doi: 10.3389/fmicb.2018.02436
Yang, B. L., Zhang, C. X., Zhang, X. J., Wang, G., Li, L., Geng, H. R., et al. (2020). Survey of aflatoxin B1 and heavy metal contamination in peanut and peanut soil in China during 2017-2018. Food Control 118, 1–8. doi: 10.1016/j.foodcont.2020.107372
Yehia, R. (2014). Aflatoxin detoxification by manganese peroxidase purified from Pleutorus ostreatus. Braz. J. Microbiol. 45, 127–134. doi: 10.1590/S1517-83822014005000026
Yi, J., Bai, R., An, Y., Liu, T. T., Liang, J. H., Tian, X. G., et al. (2019). A natural inhibitor from Alisma orientale against human carboxylesterase 2: kinetics, crcular dichroism spectroscopic analysis, and docking simulation. Int. J. Biol. Macromol. 133, 184–189. doi: 10.1016/j.ijbiomac.2019.04.099
Yiannikouris, A., Apajalahti, J., Siikanen, O., Dillon, G. P., and Moran, C. A. (2021). Saccharomyces cerevisiae cell wall-based adsorbent reduces aflatoxin B1 absorption in rats. Toxins 13, 1–20. doi: 10.3390/toxins13030209
Zhang, G. C., Kong, H., Kim, J. J., Liu, J. H. D. C., and Jin, Y. S. (2014). Construction of a quadruple auxotrophic mutant of an industrial poolyploid Saccharomyces cerevisiae strain by using RNA-guided Cas9 nuclease. Appl. Environ. Microbiol. 80, 7694–7701. doi: 10.1128/AEM.02310-14
Zhang, B., Yu, L. T., Liu, Z. J., Lu, H. Y., Fu, X. L., and Du, D. L. (2020). Rapid determination of aflatoxin B1 by an automated immunomagnetic bead purification sample pretreatment method combined with high-performance liquid chromatography. J. Sep. Sci. 43, 3509–3519. doi: 10.1002/jssc.202000293
Zhao, L., Guan, S., Gao, X., Ma, Q., Lei, Y., Bai, X., et al. (2011). Preparation, purification and characteristics of an aflatoxin degradation enzyme from Myxococcus fulvus ANSM068. J. Appl. Microbiol. 110, 147–155. doi: 10.1111/j.1365-2672.2010.04867.x
Keywords: Saccharomyces cerevisiae, Escherichia coli BL21 (DE3), aflatoxin B1, aflatoxin degrading enzyme, enzymatic characteristics, CRISPR-Cas9
Citation: Yang P, Xiao W, Lu S, Jiang S, Jiang S, Chen J, Wu W, Zheng Z and Jiang S (2022) Characterization of a Trametes versicolor aflatoxin B1-degrading enzyme (TV-AFB1D) and its application in the AFB1 degradation of contaminated rice in situ. Front. Microbiol. 13:960882. doi: 10.3389/fmicb.2022.960882
Edited by:
Nídia S. Caetano, Instituto Superior de Engenharia do Porto (ISEP), PortugalCopyright © 2022 Yang, Xiao, Lu, Jiang, Jiang, Chen, Wu, Zheng and Jiang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Peizhou Yang, yangpeizhou@hfut.edu.cn; yangpeizhou@163.com
 Peizhou Yang
Peizhou Yang