
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 13 June 2022
Sec. Infectious Agents and Disease
Volume 13 - 2022 | https://doi.org/10.3389/fmicb.2022.928025
This article is part of the Research Topic Innovative Tools to Support the Elimination of Neglected Tropical Diseases (NTDs) View all 14 articles
Chlamydia psittaci is the causative agent of psittacosis, a worldwide zoonotic disease. A rapid, specific, and sensitive diagnostic assay would be benefit for C. psittaci infection control. In this study, an assay combining recombinase-aided amplification and a lateral flow strip (RAA-LF) for the detection of active C. psittaci infection was developed. The RAA-LF assay targeted the CPSIT_RS02830 gene of C. psittaci and could be accomplished in 15 min at a single temperature (39°C). The analytical sensitivity of the assay was as low as 1 × 100 copies/μl and no cross-reaction with some other intracellular pathogens was observed. Moreover, all feces samples from mice infected with C. psittaci at day-1 post-infection were positive in the RAA-LF assay. In conclusion, the RAA-LF assay provides a convenient, rapid, specific and sensitive method for detection of active C. psittaci infection and it is also suitable for C. psittaci detection in field.
Zoonoses are major and global challenges for public health (Xiaoping and Lynn, 2021). Chlamydia psittaci, an obligate intracellular Gram-negative bacterium with a unique developmental cycle, is the causative agent of psittacosis, a worldwide zoonotic disease. More than 467 bird species and many nonavian domestic animals, including swine, dogs, horses, cattle, sheep, goats, and cats, as well as rodents are susceptible to C. psittaci (Pantchev et al., 2010; Lenzko et al., 2011; Knittler and Sachse, 2015; Gu et al., 2020). C. psittaci infection in humans could occasionally occur after inhalation of C. psittaci-contaminated aerosols from infected birds/nonavian animals (Wolff et al., 2018). Due to low awareness of the disease and atypical clinical presentation in majority of the cases, psittacosis in humans is underestimated (Gu et al., 2020).
The burden of C. psittaci infection is also likely underestimated due to diagnostic challenges: the bacterium requires intracellular growth and is a zoonotic human pathogen responsible for atypical pneumonias that may be caused by various pathogens such as Mycoplasma pneumonia, Legionella pneumophila and Coxiella burnetii (Cunha, 2006; Jiao et al., 2014). To date, several assays have been developed for C. psittaci detection. Conventional methods including pathogen isolation, indirect immunofluorescence, enzyme linked immunosorbent assay (ELISA), complement fixation, outer membrane protein A (ompA) sequencing and restriction fragment length polymorphism analysis have been used (Souriau and Rodolakis, 1986; Sanderson et al., 1994; Mitchell et al., 2009a,b; Opota et al., 2015), but these methods are laborious, low sensitive, time-consuming and also require a trained personnel. The PCR-based diagnostic methods such as conventional PCR, multi-PCR, and real-time PCR have been developed to detect C. psittaci (Hewinson et al., 1997; Messmer et al., 1997; Wolff et al., 2018). Although these PCR-based methods are highly sensitive, the requirement of expensive instruments and skilled operators make the use of these methods difficult in resource-limited settings.
Recombinase-aided amplification (RAA) assay is a new isothermal amplification technology for pathogens detection with the advantages of rapidity, simplicity, and low cost (Chen et al., 2018; Duan et al., 2018; Yan et al., 2018; Wang et al., 2020; Ruichen Lv et al., 2022). This amplification process only requires approximately 15–30 min at 39°C and has been successfully applied for detection of African Swine Fever Virus (Fan et al., 2020), Klebsiella pneumoniae (Hou et al., 2021), human norovirus GII.4 (Qin et al., 2021), duck circovirus (Li et al., 2021), Trypanosoma evansi (Li et al., 2020), etc. Additionally, the result of RAA assay can be observed on the lateral flow strip (LF) by naked eyes, providing a convenient and rapid diagnostic assay for the detection of the target pathogens, and thus it is suitable for clinical application and/or testing in field.
For Chlamydia spp. detection, urethral swabs, choanal cleft swabs, pharyngeal swab, and cloacal swabs from animals or human are often used for DNA extraction and testing (Holland et al., 1990; Dumke et al., 2015; Mina et al., 2019; Feodorova et al., 2022). One route of transmission of C. psittaci is a inhalation of aerosolized bird feces from the environment (Szeredi et al., 2005), and fresh feces samples from animals are also used for detection of C. psittaci (Takashima et al., 1996; Sachse et al., 2012; McGovern et al., 2021). Another advantage for the choice of fresh feces as a key specimen for detection of C. psittaci maybe that a feces collection is more convenient than a blood collection: rapid, harmless, and suitable for detection in filed. In the present study, a convenient and rapid recombinase-aided amplification and a lateral flow strip (RAA-LF) assay was developed for the detection of active C. psittaci infection, and its sensitivity and specificity were investigated. In addition, the effectiveness of the RAA-LF assay was evaluated with feces samples from mice experimentally infected with C. psittaci.
Chlamydia psittaci strain 6BC was cultured in buffalo green monkey kidney (BGMK) cells, and then the whole DNA were extracted from the cells infected with the strain using a QIAamp Blood and Tissue Mini DNA kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions. The DNA sample was eluted in 100 μl of nuclease-free water and determined by quantitative PCR (qPCR) as described previously (Menard et al., 2006).
DNA samples of Anaplasma phagocytophilum, Rickettsia heilongjiangensis, Rickettsia rickettsii, Rickettsia sibirica, Rickettsia canada, Rickettsia australis, Rickettsia typhi, Rickettsia prowazekii, Ehrlichia chaffeensis, C. burnetii (strain Henzerling and QIYI), L. pneumophila, Listeria monocytogenes, Salmonella typhimurium, Staphylococcus aureus, Vibrio cholera, Bartonella henselae, Bartonella quintana, Streptococcus suis, Shigella sonnei and Chlamydia trachomatis were preserved in our laboratory.
Deoxyribonucleic acid samples extracted from fresh feces of C. psittaci-infected mice were also provided. Briefly, specific-pathogen free (SPF) BALB/c mice (female, 6–8 weeks old) were divided into two groups with four mice per group and each mouse was infected with 1 × 105 C. psittaci organisms suspended in 50 μl phosphate buffer saline (PBS) via intranasal administration. All individuals in Group 2 were intraperitoneally administered with tetracycline (40 mg per kg) at day 7 post-infection. Fresh feces samples from mice in both groups were collected at days 1, 3, 7, 10, 14, 17 and/or 20 post-infection, and DNA was extracted from each feces sample using a QIAamp Blood and Tissue Mini DNA kit (Qiagen, Hilden, Germany). The purified DNA was eluted in 100 μl of nuclease-free water.
The nucleotide sequences of the CPSIT_RS02830 gene of several C. psittaci strains were aligned and their conserved regions were selected to design the primers and probes. The designed primers and probe were blasted against nucleotide database (GenBank) in the National Center for Biotechnology Information1 to confirm their specificity. The primers and probe were synthesized by GenScript Biotech (Nanjing, China).
The RAA assays were performed in 50-μl reaction volumes using a commercial RAA kit (ZC Biosicence, Hangzhou, China). The reaction mixture contained 4 μl of extracted DNA template, 2.5 μl of distilled water, 37.9 μl of reaction buffer A, 2 μl of primer F (2.5 μm), 2 μl of primer R (2.5 μm), and 0.6 μl of probe (2.5 μm). The reaction mixture was added to a tube containing the RAA enzyme mix, and then 1 μl of reaction buffer B was added to initiate the reaction. The tubes were transferred to a metal bath (Coyote, Beijing, China) at 39°C for 15 min. A negative control (blank) was included in each run. After the reaction, 10 μl of the amplified product was diluted in 90 μl PBS, and then 50 μl of the diluted sample was transferred to the sample pad of a Milenia GenLine HybriDetect strip (Milenia Biotec, Gieben, Germany) and incubated 3 min at room temperature. Results were judged visually by naked eyes, and a positive result was determined when both the low line of test (T) and the upper line of control (C) developed. Only C line occurrence indicated a negative result, and if the C line did not develop, the strip should be replaced. The assays were performed in triplicate.
The optimal RAA-LF reaction time and temperature were determined by examining different time settings ranging from 5 to 30 min and various temperature settings ranging from 37 to 42°C. For evaluation of the specificity, 1 × 105 copies/μl of C. psittaci DNA and at least 1 × 104 copies/μl of DNA templates from other bacteria were utilized in a 50 μl RAA reaction mixture. For evaluation of the sensitivity, the RAA-LF reaction was evaluated using 10-fold serial dilutions of C. psittaci DNA, ranging from 1 × 100 copy to 1 × 105 copies/μl.
To evaluate the performance of the RAA-LF assays for the detection of active C. psittaci infection, specific-pathogen free (SPF) BALB/c mice (n = 8, female, 6–8 weeks old) were divided into two groups with four mice per group, and each mouse was infected with 1 × 105 C. psittaci strain 6BC suspended in 50 μl phosphate buffer saline (PBS) via intranasal administration. Fresh feces samples from mice in both the groups were collected at days 1, 3, 7, 10, 14, 17, and/or 20 post-infection and DNA extracted from fresh feces obtained from experimentally infected mice was subjected to RAA-LF or qPCR. Fresh feces collected from naïve mice (n = 4, female, 6–8 weeks old) were used as negative control. RAA-LF amplification was performed as mentioned above, and qPCR was performed as previously described (Menard et al., 2006).
Screening of specific primer pairs and probes is the critical step for RAA assay. In the present study, the conservation of CPSIT_RS02830 gene sequence among 20 C. psittaci strains was evaluated by alignment (Figure 1A). As a result, the sequence between primers F and R was aligned with the corresponding sequence in all 20 C. psittaci strains but without that of another Chlamydia species such as C. trachomatis or C. abortus, and only one base in the probe mismatched with that of several strains, indicating the target sequence was conserved and suitable for the detection for C. psittaci (Figure 1B and Table 1).
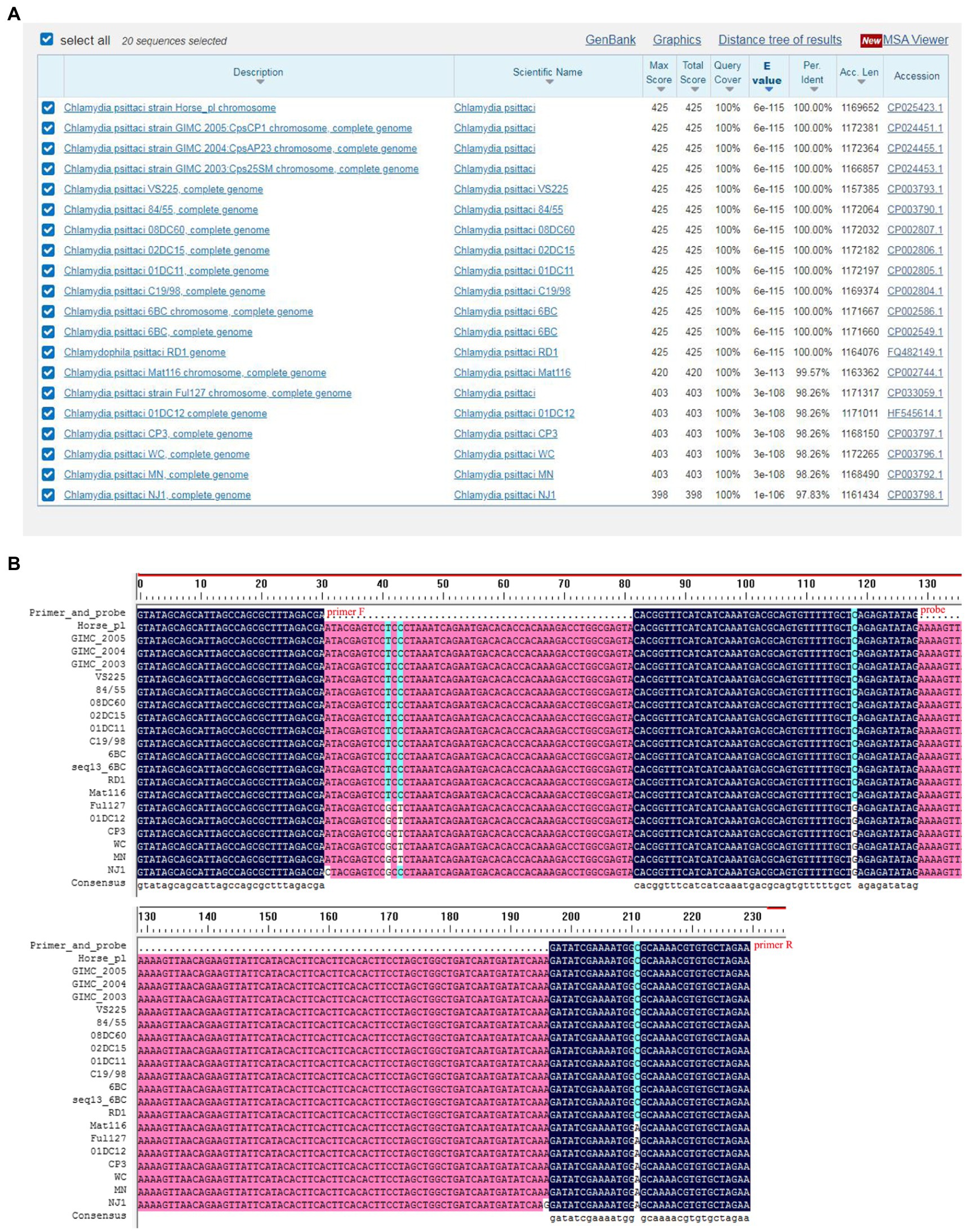
Figure 1. Alignment of the conservation of the CPSIT_RS02830 gene. The conservative sequence of the CPSIT_RS02830 gene in Chlamydia psittaci was blasted against the National Center for Biotechnology Information nucleotide database (A). The target sequences of the CPSIT_RS02830 gene was aligned between the forward and reverse primers among various C. psittaci strains (B).
The RAA-LF reaction temperature was tested at a wide range of temperatures from 37 to 42°C. The results showed that the reaction performs well at 38–42°C after 3 min incubation on the lateral flow strip (Figure 2A). Then the RAA-LF amplification was achieved at times ranging from 5 to 30 min at 39°C. The results showed that a bright T line could be detected when the amplification time was 10 min, and it became brighter as the amplified time increased to 30 min (Figure 2B).
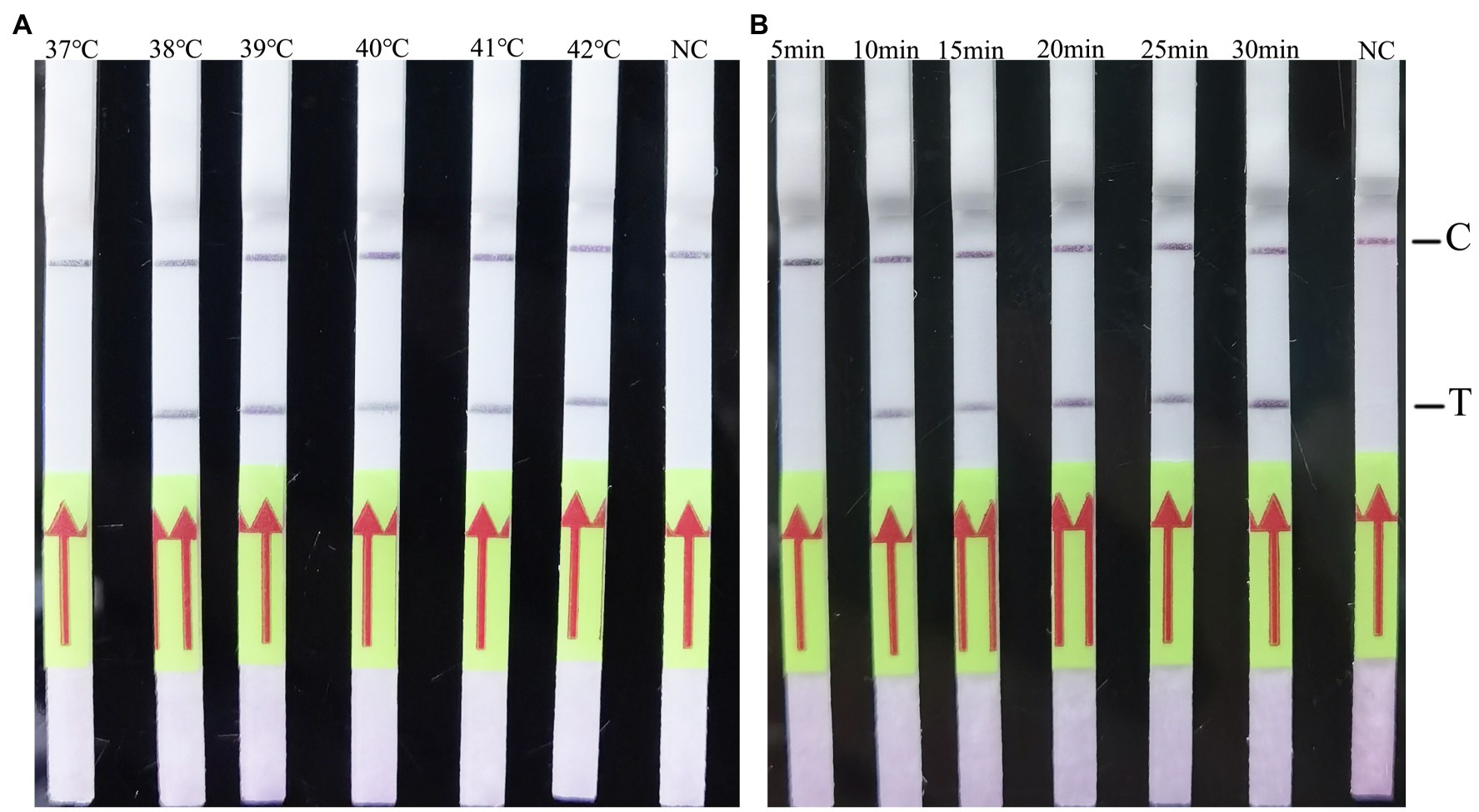
Figure 2. Evaluation of the amplification temperatures and amplification times on the RAA-LF assay. 1 × 105 copies/μl of C. psittaci DNA was utilized in a 50 μl RAA reaction mixture and deionized water was provide as negative control. A positive result was determined when both the low line of test (T) and the upper line of control (C) developed. Only C line occurrence indicated a negative result. The RAA-LF worked well in a wide range of amplification temperature from 37 to 42°C (A). After 5–30 min of isothermal amplification reaction, the positive reaction was visible on the test strip (B).
To evaluate the specificity of the developed RAA-LF assay, DNA samples belong to Rickettsia, Legionella, and several other pathogenic bacteria were assessed. Our results showed that a clearly T line could be observed by the naked eyes only for DNA sample of C. psittaci, and there was no cross-reactivity with DNA samples of A. phagocytophilum, R. heilongjiangensis, R. rickettsia, R. sibirica, R. Canada, R. australis, R. typhi, R. prowazekii, E. chaffeensis, C. burnetii (Henzerling and QIYI), L. pneumophila, L. monocytogenes, S. typhimurium, S. aureus, V. cholera, B. henselae, B. quintana, S. suis, S. sonnei and C. trachomatis (Figure 3).
Figure 3. Analytical specificity of the RAA-LF assay. Line 1: Anaplasma phagocytophilum; Line 2: Rickettsia heilongjiangensis; Line 3: R. rickettsia; Line 4: R. sibirica; Line 5: R. Canada; Line 6: R. australis; Line 7: R. typhi; Line 8: R. prowazekii; Line 9: Ehrlichia chaffeensis; Line 10: Coxiella burnetii (Henzerling); Line 11: C. burnetii (QIYI); Line 12: Legionella pneumophila; Line 13: Listeria monocytogenes; Line 14: Salmonella typhimurium; Line 15: Staphylococcus aureus; Line 16: Vibrio cholera; Line 17: Bartonella henselae; Line 18: Bartonella quintana; Line 19: Streptococcus suis; Line 20: Shigella sonnei; Line 21: Chlamydia trachomatis; NC: negative control; PC: C. psittaci. A positive result was determined when both the low line of test (T) and the upper line of control (C) developed. Only C line occurrence indicated a negative result.
To evaluate the sensitivity of the developed RAA-LF assay, 10-fold serial dilutions of C. psittaci DNA, ranging from 1 × 100 copy to 1 × 105 copies/μl were used. Results showed that the developed RAA-LF assay was highly sensitive with a detection limit as low as 1 × 100 copies/μl of C. psittaci (Figure 4).
Figure 4. Analytical sensitivity of the RAA-LF assay. Ten-fold serial dilutions of C. psittaci genomic DNA were evaluated by the RAA-LF assay, and deionized water was provided as negative control. A positive result was determined when both the low line of test (T) and the upper line of control (C) developed. Only C line occurrence indicated a negative result. Lines 0–5: 1 × 100 copy to 1 × 105 copies/μl. NC: negative control.
In the experiment, BALB/c mice infected with C. psittaci were divided into two groups. Mice in group 1 were left untreated, while mice in group 2 were treated with tetracycline at day-7 post-infection. Fresh feces samples from mice of both the groups were collected at days 1, 3, 7, 10, 14, 17, and/or 20 post-infection, and DNA extracted from feces samples were detected by the developed RAA-LF assay.
As shown in Figure 5, the two techniques did not yield identical results for all of the feces samples. All of the feces samples were detected positive in the RAA-LF assay. However, none of the feces samples from the infected mice at day 1 post-infection was detected positive in C. psittaci-specific qPCR (Figure 5; Supplementary Table S1 and Supplementary Figure S1), suggesting that the RAA-LF assay is more sensitive for detection of an early C. psittaci infection compared with qPCR. All of the feces samples in group 1, which contained around 103–104 copies/μl of genomic DNA as determined in qPCR at days 3, 7, 10, and 14 post-infection (Supplementary Table S1), were also detected positive in the RAA-LF assay. Unfortunately, all mice in group 1 died at day 15 post-infection.
Figure 5. Results of the evaluation of qPCR and the RAA-LF assay for experimentally infected samples. BALB/c mice infected with Chlamydia psittaci were divided into two groups of four individuals. Group 1 was left untreated, while Group 2 was treated with tetracycline at 7 days post-infection. All mice in group 1 died at day 15 post-infection and all mice in group 2 were survival (A). Fresh feces samples from group 1 (B) and group 2 (C) were collected at days 1, 3, 7, 10, 14, 17 and/or 20 post-infection, and the extracted DNA from fresh feces samples were evaluated by qPCR and the RAA-LF assay. The results were displayed as the percentages of mice that scored positive (orange) or negative (green).
In group 2, all mice were survival and their feces samples were detected with decreasing levels of C. psittaci from day 7 post-infection (Supplementary Table S1). All of the feces samples from day 3 post-infection were detected positive in C. psittaci-specific qPCR and RAA-LF assay, indicating that the RAA-LF is a suitable assay for visual detection of active C. psittaci infection under the experimental condition. The feces samples from naïve mice were also detected and there was no positive result in the RAA-LF assay (Supplementary Figure S1).
Chlamydia psittaci is a zoonotic agent of systemic wasting disease in birds and atypical pneumonia in mammalians including humans, constituting a public health risk (Knittler and Sachse, 2015). Human infections caused by C. psittaci are underestimated, which is mainly due to the difficulty in diagnosing this disease. A convenient, rapid, specific and sensitive method for the detection of C. psittaci infection is needed in order to achieve an effective psittacosis control. In the present study, we have developed a convenient and rapid RAA-LF assay for detection of active C. psittaci infection.
Evaluation of specificity of a detection assay using unrelated bacteria DNA is of equal importance as using related bacteria DNA (Sails et al., 2003; Gumaa et al., 2019). The RAA-LF assay was evaluated using the whole DNA samples of C. psittaci or another intracellular pathogenic bacteria which are typically neglected in causing fever of unknown origin (Rickettsia spp. or Legionella spp.), or another unrelated pathogenic bacterium (S. typhimurium, S. aureus etc.). Specially, DNA of V. cholera, a pathogenic bacterium which is commonly transmitted by drinking water contaminated by the patients’ feces, was also used as an unrelated sample in the present study. As a result, only DNA samples of C. psittaci were detected positive in the RAA-LF assay, suggesting that this RAA-LF assay performs a good specificity. Further studies should focus on verifying potential cross-reactivity with DNA from other species belong to chlamydia spp., like C. abortus.
The developed RAA-LF assay is also highly sensitive and the detected limitation is as low as 1 × 100 copies/μl of C. psittaci genomic DNA, which is almost 20-fold more sensitive compared with that of the recombinant polymerase amplification (RPA/RAA) assay developed by Pang et al. (2021), making it useful for assessing the samples from the patients in the early acute phase of C. psittaci infection. Actually, three forward primers, three reverse primers, and two probes targeting the conservation of CPSIT_RS02830 gene sequence have been designed and combined to generate 18 different combinations, and then these combinations have been screened for the RAA-LF assay (data not shown). The best combination is the one that descried in Table 1. This may be one explanation of the high sensitivity of the developed RAA-LF assay. In addition, compared with conventional PCR and real-time PCR requiring more than 1 h to complete in a well-equipped laboratory (Herring, 1991; Rasmussen et al., 1992; Sykes et al., 2001; Angen et al., 2021), the time required of the RAA-LF assay is also shorter and could be completed in less than 20 min. Moreover, the RAA-LF assay works well in a wide range of temperature (38–42°C) which is lower than the ones required in loop-mediated isothermal amplification (LAMP) assay (Njiru, 2012). Therefore, this RAA-LF assay does not require any expensive instrument and can be performed in minimally equipped laboratories and even in field.
The RAA-LF assay was evaluated for its potential to detect active C. psittaci infection at various time points in an experimental mouse model. The results showed that the RAA-LF assay exhibits a potential suitable for direct detection of early C. psittaci infection. Feces, as the primary test sample, has potential for PCR inhibition and ultimately reduced analytical and diagnostic sensitivity (Acharya et al., 2017), what’s important is that the complex compositions of feces seems to have not much effects on the test results in the present study. The C. psittaci could be detected from fresh feces samples of infected mice at day 1 post-infection, also indicating the available for assessing patient samples in the acute phase. Moreover, an important feature of the method is the results can also be visualized with naked eyes. Therefore, the RAA-LF assay is more suitable for usage under field conditions.
However, there are several limitations in evaluation of the developed RAA-LF assay. Firstly, for lacking of genomic DNA, no various strains of C. psittaci or other Chlamydia spp. strains except C. trachomatis were tested in the present study. Secondly, no clinical samples of patients with psittacosis were used to verify the developed RAA-LF assay. And the addressing of these limitations will contribute to the further preciseness of this study.
A method for the detection of active C. psittaci infection based on a RAA-LF was developed in the present study. The RAA-LF assay can be performed within 20 min using a portable device and positively detect as low as 1 × 100 copies/μl of DNA. In addition, the assay results can be visualized with naked eyes. Therefore, it is a convenient, rapid, sensitive and specific assay for detection of active C. psittaci infection, and it is suitable for use in minimally equipped laboratories even field settings.
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
The animal study was reviewed and approved by the Institutional Animal Care and Use Committee (IACUC) of the Academy of Military Medical Science.
This study was conceived and designed by JJ and XX. Samples collection and laboratory work were performed by PH, XO, and YY. Experimental data analysis was performed by YQ and WW. The manuscript was drafted by JJ and XX, and edited by BW. All authors contributed to the article and approved the submitted version.
This work was supported by the National Natural Science Foundation of China [grant numbers 32000139 and 31970178] and the National Key Research and Development Program of China [grant numbers 2019YFC1200500 and 2019YFC1200602].
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.928025/full#supplementary-material
Supplementary Figure S1 | Evaluation of the RAA-LF assay for detection of active Chlamydia psittaci infection in mice. Fresh feces samples from group 1 (A), group 2 (B) and naïve mice (C) were tested by the developed RAA-LF assay. 1, 3 7, 10, 14, 17, 20: days at post-infection. N: negative control. N1–N4: naïve mice. A positive result was determined when both the low line of test (T) and the upper line of control (C) developed. Only C line occurrence indicated a negative result.
Acharya, K. R., Dhand, N. K., Whittington, R. J., and Plain, K. M. (2017). PCR inhibition of a quantitative PCR for detection of Mycobacterium avium subspecies Paratuberculosis DNA in feces: diagnostic implications and potential solutions. Front. Microbiol. 8:115. doi: 10.3389/fmicb.2017.00115
Angen, O., Johannesen, T. B., Petersen, R. F., Uldum, S. A., and Schnee, C. (2021). Development of a species-specific real-time PCR test for Chlamydia psittaci and its employment in the investigation of zoonotic transmission from racing pigeons in Denmark. Diagn. Microbiol. Infect. Dis. 100:115341. doi: 10.1016/j.diagmicrobio.2021.115341
Chen, C., Li, X. N., Li, G. X., Zhao, L., Duan, S. X., Yan, T. F., et al. (2018). Use of a rapid reverse-transcription recombinase aided amplification assay for respiratory syncytial virus detection. Diagn. Microbiol. Infect. Dis. 90, 90–95. doi: 10.1016/j.diagmicrobio.2017.10.005
Cunha, B. A. (2006). The atypical pneumonias: clinical diagnosis and importance. Clin. Microbiol. Infect. 12, 12–24. doi: 10.1111/j.1469-0691.2006.01393.x
Duan, S., Li, G., Li, X., Chen, C., Yan, T., Qiu, F., et al. (2018). A probe directed recombinase amplification assay for detection of MTHFR A1298C polymorphism associated with congenital heart disease. BioTechniques 64, 211–217. doi: 10.2144/btn-2018-2010
Dumke, R., Schnee, C., Pletz, M. W., Rupp, J., Jacobs, E., Sachse, K., et al. (2015). Mycoplasma pneumoniae and Chlamydia spp. infection in community-acquired pneumonia, Germany, 2011-2012. Emerg. Infect. Dis. 21, 426–434. doi: 10.3201/eid2103.140927
Fan, X., Li, L., Zhao, Y., Liu, Y., Liu, C., Wang, Q., et al. (2020). Clinical validation of two Recombinase-based isothermal amplification assays (RPA/RAA) for the rapid detection of African swine fever virus. Front. Microbiol. 11:1696. doi: 10.3389/fmicb.2020.01696
Feodorova, V. A., Saltykov, Y. V., Kolosova, A. A., Rubanik, L. V., Poleshchuk, N. N., and Motin, V. L. (2022). Emergence of novel Chlamydia trachomatis sequence types among Chlamydia patients in the Republic of Belarus. Microorganisms 10:478. doi: 10.3390/microorganisms10020478
Gu, L., Liu, W., Ru, M., Lin, J., Yu, G., Ye, J., et al. (2020). The application of metagenomic next-generation sequencing in diagnosing Chlamydia psittaci pneumonia: a report of five cases. BMC Pulm. Med. 20:65. doi: 10.1186/s12890-020-1098-x
Gumaa, M. M., Cao, X., Li, Z., Lou, Z., Zhang, N., Zhang, Z., et al. (2019). Establishment of a recombinase polymerase amplification (RPA) assay for the detection of Brucella spp. Infection. Mol Cell Probes. 47:101434. doi: 10.1016/j.mcp.2019.101434
Herring, A. J. (1991). PCR test for Chlamydia psittaci. Vet. Rec. 128:555. doi: 10.1136/vr.128.23.555-b
Hewinson, R. G., Griffiths, P. C., Bevan, B. J., Kirwan, S. E., Field, M. E., Woodward, M. J., et al. (1997). Detection of Chlamydia psittaci DNA in avian clinical samples by polymerase chain reaction. Vet. Microbiol. 54, 155–166. doi: 10.1016/S0378-1135(96)01268-0
Holland, S. M., Gaydos, C. A., and Quinn, T. C. (1990). Detection and differentiation of Chlamydia trachomatis, Chlamydia psittaci, and Chlamydia pneumoniae by DNA amplification. J. Infect. Dis. 162, 984–987. doi: 10.1093/infdis/162.4.984
Hou, L., Li, D., Zhang, N., Zhao, J., Zhao, Y., and Sun, X. (2021). Development of an isothermal recombinase aided amplification assay for the rapid and visualized detection of Klebsiella pneumoniae. J. Sci. Food Agric. doi: 10.1002/jsfa.11737 [Epub ahead of print]
Jiao, J., Xiong, X., Qi, Y., Gong, W., Duan, C., Yang, X., et al. (2014). Serological characterization of surface-exposed proteins of Coxiella burnetii. Microbiology (Reading) 160, 2718–2731. doi: 10.1099/mic.0.082131-0
Knittler, M. R., and Sachse, K. (2015). Chlamydia psittaci: update on an underestimated zoonotic agent. Pathog. Dis. 73, 1–15. doi: 10.1093/femspd/ftu007
Lenzko, H., Moog, U., Henning, K., Lederbach, R., Diller, R., Menge, C., et al. (2011). High frequency of chlamydial co-infections in clinically healthy sheep flocks. BMC Vet. Res. 7:29. doi: 10.1186/1746-6148-7-29
Li, Z., Pinto Torres, J. E., Goossens, J., Stijlemans, B., Sterckx, Y. G., and Magez, S. (2020). Development of a recombinase polymerase amplification lateral flow assay for the detection of active Trypanosoma evansi infections. PLoS Negl. Trop. Dis. 14:e0008044. doi: 10.1371/journal.pntd.0008044
Li, X., Wang, C., Wang, W., Zhang, Z., Zhang, Z., Wang, C., et al. (2021). Research note: development of rapid isothermal amplification assay for detection of duck circovirus. Poult. Sci. 100:101339. doi: 10.1016/j.psj.2021.101339
McGovern, O. L., Kobayashi, M., Shaw, K. A., Szablewski, C., Gabel, J., Holsinger, C., et al. (2021). Use of real-time PCR for Chlamydia psittaci detection in human specimens During an outbreak of psittacosis–Georgia and Virginia, 2018. MMWR Morb. Mortal. Wkly Rep. 70, 505–509. doi: 10.15585/mmwr.mm7014a1
Menard, A., Clerc, M., Subtil, A., Megraud, F., Bebear, C., and De Barbeyrac, B. (2006). Development of a real-time PCR for the detection of Chlamydia psittaci. J. Med. Microbiol. 55, 471–473. doi: 10.1099/jmm.0.46335-0
Messmer, T. O., Skelton, S. K., Moroney, J. F., Daugharty, H., and Fields, B. S. (1997). Application of a nested, multiplex PCR to psittacosis outbreaks. J. Clin. Microbiol. 35, 2043–2046. doi: 10.1128/jcm.35.8.2043-2046.1997
Mina, A., Fatemeh, A., and Jamshid, R. (2019). Detection of Chlamydia psittaci genotypes among birds in Northeast Iran. J. Avian Med. Surg. 33, 22–28. doi: 10.1647/2017-334
Mitchell, S. L., Budhiraja, S., Thurman, K. A., Lanier Thacker, W., and Winchell, J. M. (2009a). Evaluation of two real-time PCR chemistries for the detection of Chlamydophila pneumoniae in clinical specimens. Mol. Cell. Probes 23, 309–311. doi: 10.1016/j.mcp.2009.07.005
Mitchell, S. L., Wolff, B. J., Thacker, W. L., Ciembor, P. G., Gregory, C. R., Everett, K. D., et al. (2009b). Genotyping of Chlamydophila psittaci by real-time PCR and high-resolution melt analysis. J. Clin. Microbiol. 47, 175–181. doi: 10.1128/JCM.01851-08
Njiru, Z. K. (2012). Loop-mediated isothermal amplification technology: towards point of care diagnostics. PLoS Negl. Trop. Dis. 6:e1572. doi: 10.1371/journal.pntd.0001572
Opota, O., Jaton, K., Branley, J., Vanrompay, D., Erard, V., Borel, N., et al. (2015). Improving the molecular diagnosis of Chlamydia psittaci and Chlamydia abortus infection with a species-specific duplex real-time PCR. J. Med. Microbiol. 64, 1174–1185. doi: 10.1099/jmm.0.000139
Pang, Y., Cong, F., Zhang, X., Li, H., Chang, Y. F., Xie, Q., et al. (2021). A recombinase polymerase amplification-based assay for rapid detection of Chlamydia psittaci. Poult. Sci. 100, 585–591. doi: 10.1016/j.psj.2020.11.031
Pantchev, A., Sting, R., Bauerfeind, R., Tyczka, J., and Sachse, K. (2010). Detection of all Chlamydophila and Chlamydia spp. of veterinary interest using species-specific real-time PCR assays. Comp. Immunol. Microbiol. Infect. Dis. 33, 473–484. doi: 10.1016/j.cimid.2009.08.002
Qin, Z., Xue, L., Cai, W., Gao, J., Jiang, Y., Yang, J., et al. (2021). Development of a recombinase-aided amplification assay for rapid detection of human norovirus GII.4. BMC Infect. Dis. 21:248. doi: 10.1186/s12879-021-05942-x
Rasmussen, S. J., Douglas, F. P., and Timms, P. (1992). PCR detection and differentiation of Chlamydia pneumoniae, Chlamydia psittaci and Chlamydia trachomatis. Mol. Cell. Probes 6, 389–394. doi: 10.1016/0890-8508(92)90032-S
Ruichen Lv, N. L., Wang, J., Li, Y., and Qi, Y. (2022). Recombinase polymerase amplification for rapid detection of zoonotic pathogens: an overview. Zoonoses 2, 1–12. doi: 10.15212/ZOONOSES-2022-0002
Sachse, K., Kuehlewind, S., Ruettger, A., Schubert, E., and Rohde, G. (2012). More than classical Chlamydia psittaci in urban pigeons. Vet. Microbiol. 157, 476–480. doi: 10.1016/j.vetmic.2012.01.002
Sails, A. D., Fox, A. J., Bolton, F. J., Wareing, D. R., and Greenway, D. L. (2003). A real-time PCR assay for the detection of Campylobacter jejuni in foods after enrichment culture. Appl. Environ. Microbiol. 69, 1383–1390. doi: 10.1128/AEM.69.3.1383-1390.2003
Sanderson, T. P., Andersen, A. A., Miller, L. D., Andrews, J. J., Janke, B. H., Larson, D. L., et al. (1994). An indirect microimmunofluorescence test for detection of chlamydial antibodies in ovine fetal fluids. J. Vet. Diagn. Investig. 6, 315–320. doi: 10.1177/104063879400600306
Souriau, A., and Rodolakis, A. (1986). Rapid detection of Chlamydia psittaci in vaginal swabs of aborted ewes and goats by enzyme linked immunosorbent assay (ELISA). Vet. Microbiol. 11, 251–259. doi: 10.1016/0378-1135(86)90027-1
Sykes, J. E., Allen, J. L., Studdert, V. P., and Browning, G. F. (2001). Detection of feline calicivirus, feline herpesvirus 1 and Chlamydia psittaci mucosal swabs by multiplex RT-PCR/PCR. Vet. Microbiol. 81, 95–108. doi: 10.1016/S0378-1135(01)00340-6
Szeredi, L., Hotzel, H., and Sachse, K. (2005). High prevalence of chlamydial (Chlamydophila psittaci) infection in fetal membranes of aborted equine fetuses. Vet. Res. Commun. 29, 37–49. doi: 10.1007/s11259-005-0835-1
Takashima, I., Imai, Y., Itoh, N., Kariwa, H., and Hashimoto, N. (1996). Polymerase chain reaction for the detection of Chlamydia psittaci in the feces of budgerigars. Microbiol. Immunol. 40, 21–26. doi: 10.1111/j.1348-0421.1996.tb03312.x
Wang, Y., Cui, Y., Yu, Z., Li, Y., Bai, C., Sun, P., et al. (2020). Development of a recombinase-aided amplification assay for detection of orf virus. J. Virol. Methods 280:113861. doi: 10.1016/j.jviromet.2020.113861
Wolff, B. J., Morrison, S. S., and Winchell, J. M. (2018). Development of a multiplex TaqMan real-time PCR assay for the detection of Chlamydia psittaci and Chlamydia pneumoniae in human clinical specimens. Diagn. Microbiol. Infect. Dis. 90, 167–170. doi: 10.1016/j.diagmicrobio.2017.11.014
Xiaoping, D., and Lynn, S. (2021). Emerging and re-emerging zoonoses are major and global challenges for public health. Zoonoses 1, 1–2. doi: 10.15212/ZOONOSES-2021-0001
Keywords: Chlamydia psittaci, detection, recombinase-aided amplification, lateral flow strip, infection
Citation: Jiao J, Qi Y, He P, Wan W, OuYang X, Yu Y, Wen B and Xiong X (2022) Development of a Lateral Flow Strip-Based Recombinase-Aided Amplification for Active Chlamydia psittaci Infection. Front. Microbiol. 13:928025. doi: 10.3389/fmicb.2022.928025
Received: 25 April 2022; Accepted: 27 May 2022;
Published: 13 June 2022.
Edited by:
Wei Wang, Jiangsu Institute of Parasitic Diseases (JIPD), ChinaReviewed by:
Valentina A. Feodorova, Federal Research Center for Virology and Microbiology, RussiaCopyright © 2022 Jiao, Qi, He, Wan, OuYang, Yu, Wen and Xiong. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiaolu Xiong, eGlvbmd4aWFvbHU2MjRAc29odS5jb20=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.