
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 18 April 2022
Sec. Food Microbiology
Volume 13 - 2022 | https://doi.org/10.3389/fmicb.2022.856802
This article is part of the Research Topic Control of Specific Spoilage Organism in Seafood View all 5 articles
 Dangfeng Wang1,2,3
Dangfeng Wang1,2,3 Fangchao Cui2,3
Fangchao Cui2,3 Likun Ren4
Likun Ren4 Xiqian Tan2,3
Xiqian Tan2,3 Xinran Lv2,3
Xinran Lv2,3 Qiuying Li2,3
Qiuying Li2,3 Jianrong Li1,2,3*
Jianrong Li1,2,3* Tingting Li5*
Tingting Li5*Pseudomonas fluorescens is a common specific spoilage organism (SSO) of aquatic products. The spoilage ability of SSO can be regulated by the quorum sensing (QS) system. However, the QS system in P. fluorescens and their relationship with the spoilage potential have not been systematically analyzed. In the present study, the complete genome of P. fluorescens PF08 isolated from spoilage turbot was sequenced. The identification of key genes that involved in the QS, enzyme synthesis, sulfur, and amino acid metabolism explained the spoilage potential of P. fluorescens PF08. Results of quantitative real-time PCR revealed the key role of the P. fluorescens PF08 QS system in regulating the transcription of spoilage-related genes and its sensitivity to environmental stress. These findings provide insight into the spoilage features of P. fluorescens PF08 from a genomic perspective. The knowledge may be valuable in the development of new strategies for the targeted inhibition of aquatic product spoilage based on QS interference.
Food spoilage is a complex process that is caused mainly by the metabolic activities of specific spoilage organisms (SSOs). SSOs are regulated by a common cell-to-cell communication mechanism named quorum sensing (QS) (Bai and Rai, 2011). In the food industry, microbial spoilage has attracted increasing attention because of the subsequent economic losses and safety problems. When the self-secreted QS signaling molecules reach a certain threshold level with the increased density of the contaminating bacteria during food storage, bacteria will regulate the secretion of a series of spoilage-required factors such as extracellular proteases and lipases, resulting in protein degradation and off-flavor in food (Papenfort and Bassler, 2016). Detection of the key regulators of the QS system in spoilage bacteria and blocking or interfering with the operation of the system is a feasible strategy to control food spoilage while avoiding drug resistance caused by the survival pressure (Ding et al., 2017).
Pseudomonas fluorescens is a common Gram-negative bacterium and a major spoilage organism frequently associated with the spoilage of aquatic products (Speranza et al., 2013). Moreover, P. fluorescens is a psychrotroph and can grow well at temperatures as low as 4°C. Thus, spoilage caused by P. fluorescens cannot be effectively controlled by the low temperature environment in cold chain transportation (Packiavathy et al., 2014). Similar to most Gram-negative bacteria, the secretion of spoilage factors in P. fluorescens can be regulated by QS based on N-acyl-homoserine lactones (AHLs) (Brachmann et al., 2013). The genetic background and genomic information of the QS system have been intensively studied in P. aeruginosa (Chan et al., 2014), but little related information is available for P. fluorescens. The availability of the genome sequence of P. fluorescens would be helpful to clarify the key factors of QS-related system in P. fluorescens. This information could also be expected to improve the understanding of the QS-regulated spoilage in P. fluorescens and to inform new strategies for quality control of aquatic products.
The genomes of different P. fluorescens strains have been sequenced and uploaded to the Genome database of GeneBank1. Most of the genomic information came from P. fluorescens isolated from soil, sewage, or the plant rhizosphere. There is scant information concerning the relationship between the genome information and food spoilage. Therefore, in the present study, the entire genome of P. fluorescens strain PF08, which we previously reported as the SSO of refrigerated turbot (Scophthalmus maximus) (Li T. et al., 2019), was sequenced and analyzed. Several QS-related genes were found in the P. fluorescens PF08 genome. Analysis of the genome identified several genes and gene clusters involved in the protease production, sulfur, and amino acid metabolism, stress response, biofilm formation, motility, and drug resistance. The findings revealed the spoilage potential of PF08 and its excellent adaptability to external survival pressure exerted by food preservation. Moreover, the QS system in PF08 was inhibited or activated by cinnamaldehyde or C4-HSL in this study and the regulatory effect of the QS system on spoilage-related genes in PF08 was evaluated by analyzing the changes of transcription level of these key genes. The effects of environmental conditions on the transcription of QS regulatory genes of rhlI/R were also determined. The data provide insight into the spoilage features of P. fluorescens PF08 from a genomic perspective.
The N-butanoyl-L-homoserine lactone (C4-HSL) standard was purchased from Sigma-Aldrich (St. Louis, MO, United States). It was dissolved in methanol at a concentration of 2 mg/mL for use.
Pseudomonas fluorescens PF08 (GenBank accession number: CP032618) was isolated from refrigerated spoiled turbots in Jinzhou, China. Aeromonas salmonicida CS12 and Staphylococcus aureus were kept in our laboratory. All the bacteria were cultured in Luria-Bertani (LB) broth (1% peptone, 1% NaCl, 0.5% yeast extract, pH 7.0 ± 0.2) at 28°C with agitation (160 rpm) until an optical density at 595 nm (OD595) of 1 was reached.
Ten mL of overnight culture of P. fluorescens PF08 (OD595 = 1) was centrifugated at 8000 × g for 10 min. Then, the cells were collected and transported to the Beijing Novogene Bioinformatics Technology Co., Ltd., in dry ice for genome sequencing and assembly. The experimental details are described in the Supporting Information.
The interspersed repetitive sequences of the genome were predicted using RepeatMasker (Saha et al., 2008)2. Tandem repeats were analyzed using the tandem repeats finder. Transfer RNA (tRNA) genes were predicted using the tRNAscan-SE (Lowe and Eddy, 1997). Ribosomal RNA (rRNA) genes were analyzed using the rRNAmmer (Lagesen et al., 2007). The IslandPath-DIOMB program (Hsiao et al., 2003) was used to predict genomics islands. Transposon PSI was used to predict transposons based on the homologous Blast method. CRISPRFinder was used for the CRISPR identification (Grissa et al., 2007).
The gene functions of the genome were annotated through Blast searching the whole genome against seven databases (E-value < 1e-5, minimal 30 alignment length percentage > 40%), including the Gene Ontology (GO), Kyoto Encyclopedia of Genes and Genomes (KEGG), Clusters of Orthologous Groups (COG), Non-Redundant (NR) protein databases, Transporter Classification Database (TCDB), Swiss-Prot, and TrEMBL. The Antibiotic Resistance Genes Database (ARDB) was used for drug resistance analysis.
A multiple alignment of whole genome sequences from PF08 and other Pseudomonas strains compared in this analysis was generated using CVTree 3. The tree was constructed using the alignment-free method based on K-tuple counting and background subtraction with a bootstrap value of 1,000.
An overnight culture of P. fluorescens PF08 (OD595 = 1) was diluted 1:100 in fresh LB broth (pH 7.0 ± 0.2). C4-HSL, and cinnamaldehyde were used as the agonist and inhibitor of the QS system and added to the broth at final concentration of 20 μg/mL or 0.1 μL/mL, respectively (Li et al., 2018). The broth containing both of 20 μg/mL of C4-HSL and 0.1 μL/mL of cinnamaldehyde was used as control group. The samples were incubated at 28°C until OD595 was 1. After centrifugation at 8000 × g for 10 min, the pelleted cells were examined to determine the transcription levels of key genes (rhlI, rhlR, pilG, pilJ, fliL, flhA, algA, algX, aprX, aprA, serP, pvdA, pvdE, ahpC, sspA, cusS) using quantitative real-time (qPCR). Samples incubated in LB broth containing equivalent concentration of methanol at 28°C were the blank group.
AB medium (3 g/L K2HPO4, 0.3 g/L MgSO4⋅7H2O, 1 g/L NH4Cl, 0.15 g/L KCl, 1 g/L NaH2PO4, 0.01 g/L CaCl2, 0.0025 g/L FeSO4⋅7H2O, 5 g/L casein hydrolyzed amino acids) was prepared as previously described (Zimmer et al., 2014). The overnight culture of P. fluorescens PF08 (OD595 = 1) was diluted 1:100 in prepared AB culture medium, followed by the addition of 5 g/L of different carbon sources (glucose, xylose, sucrose, fructose, lactose, or maltose). All samples were incubated at 28°C until an OD595 of 1 was reached. After centrifugation at 8000 × g for 10 min, the cells were collected for the analysis of rhlI/R transcription level using qPCR. Samples incubated in LB broth at 28°C were the control.
An overnight culture of P. fluorescens PF08 (OD595 = 1) was diluted 1:100 in fresh LB broth (pH 7.0 ± 0.2) containing different mass concentrations of NaCl (0.5, 1, 3, or 5%). All samples were incubated at 28°C until an OD595 of 1 was reached. After centrifugation at 8000 × g for 10 min, the precipitates were collected for further analysis of rhlI/R transcription level using qPCR.
An overnight culture of P. fluorescens PF08 (OD595 = 1) was diluted 1:100 in fresh LB broth. The pH was adjusted to 5, 6, 7, 8, or 9 by addition of HCl or NaOH. All samples were incubated at 28°C until OD595 was 1. After centrifugation at 8,000 × g for 10 min, the cells were collected for further analysis of rhlI/R transcription level using qPCR.
An overnight culture of P. fluorescens PF08 (OD595 = 1) was diluted 1:100 in fresh LB broth (pH 7.0 ± 0.2). The samples were incubated at 4, 15, 25, or 37°C until OD595 was 1. After centrifugation at 8000 × g for 10 min, the cells were collected for further analysis of rhlI/R transcription level using qPCR. Samples incubated in LB broth at 25°C were the control.
The qPCR analysis was performed as we previously described (Li X. et al., 2019), and the experimental details are described in the Supporting Information.
The growth inhibition of S. aureus and A. salmonicida CS12 was tested using the Oxford cup assay as previously described (Diao et al., 2014). One milliliter of overnight culture of P. fluorescens PF08 was inoculated to 100 mL of fresh LB broth and incubated at 28°C for 24 h. After centrifugation at 8000 × g for 10 min, the supernatant was collected. Metabolites were extracted using 100 mL of ethyl acetate. The organic phase was evaporated in a rotary evaporator and redissolved in 1 mL of 50% (v/v) methanol for further assay.
To prepare the test plates, the overnight cultures of S. aureus or A. salmonicida CS12 were diluted 1:100 in 20 mL of LB nutrient agar. Agar was poured into two plates containing autoclaved Oxford cups. Once the medium had solidified, 200 μL of ethyl acetate extract of P. fluorescens PF08 was added to the wells, and the plates were incubated for 24 h at 28°C. The antibacterial activity of the samples was evaluated by observing the growth inhibition zone. A control test was performed using 200 μL of 50% (v/v) methanol.
Data with normal distribution were analyzed by one-way analysis of variance (ANOVA) with t-test using the IBM SPSS Statistics program (version 20.0). The qPCR tests were performed for three times and comparisons of distributions between mean values of different groups were made by Duncan’s test at a statistical significance level of p < 0.05. Graphs were prepared in Graphpad Prism (version 8.02).
The genome of P. fluorescens PF08 is composed of a 6,030,573 bp chromosome (Figure 1A) with a G + C content of 60.7%. The genome analysis predicted a total of 5,600 genes consisting of 5,514 coding sequences. Genome phylogenetic tree based on the whole genome sequences of PF08 was performed and the result was shown in Figure 2. Further predicted were 66 tRNA genes, 16 rRNA genes, 4 ncRNA genes, and 164 pseudo genes. The size of the coding genes generally ranged from 200 to 1500 bp (Figure 3A). These features were similar to those of other P. fluorescens strains (Table 1), indicating that PF08 conformed to the basic features of P. fluorescens. Furthermore, PF08 was found to contain 69 tandem repeat sequences and 8 gene islands with an average length of 20,743 bp. Among the predicted genes, approximately 11 genes were related to mobile elements predicted as transposons and phage. Three CRISPR locations were also identified. The alignments of the genome sequences of P. fluorescens PF08 to those of P. fluorescens UK4 was performed using the Mauve program because of their close relationship based on the 16S rRNA genes. The alignments of some type strains of other Pseudomonads, including P. fluorescens UK4, P. aeruginosa PA01, and P. putida NBRC 14164, were also analyzed as control. As shown in Supplementary Figure 1, the genome sequences of P. fluorescens PF08 and P. fluorescens UK4 showed overall similarity. But the translocations between some fragments suggested differences in their metabolic patterns.
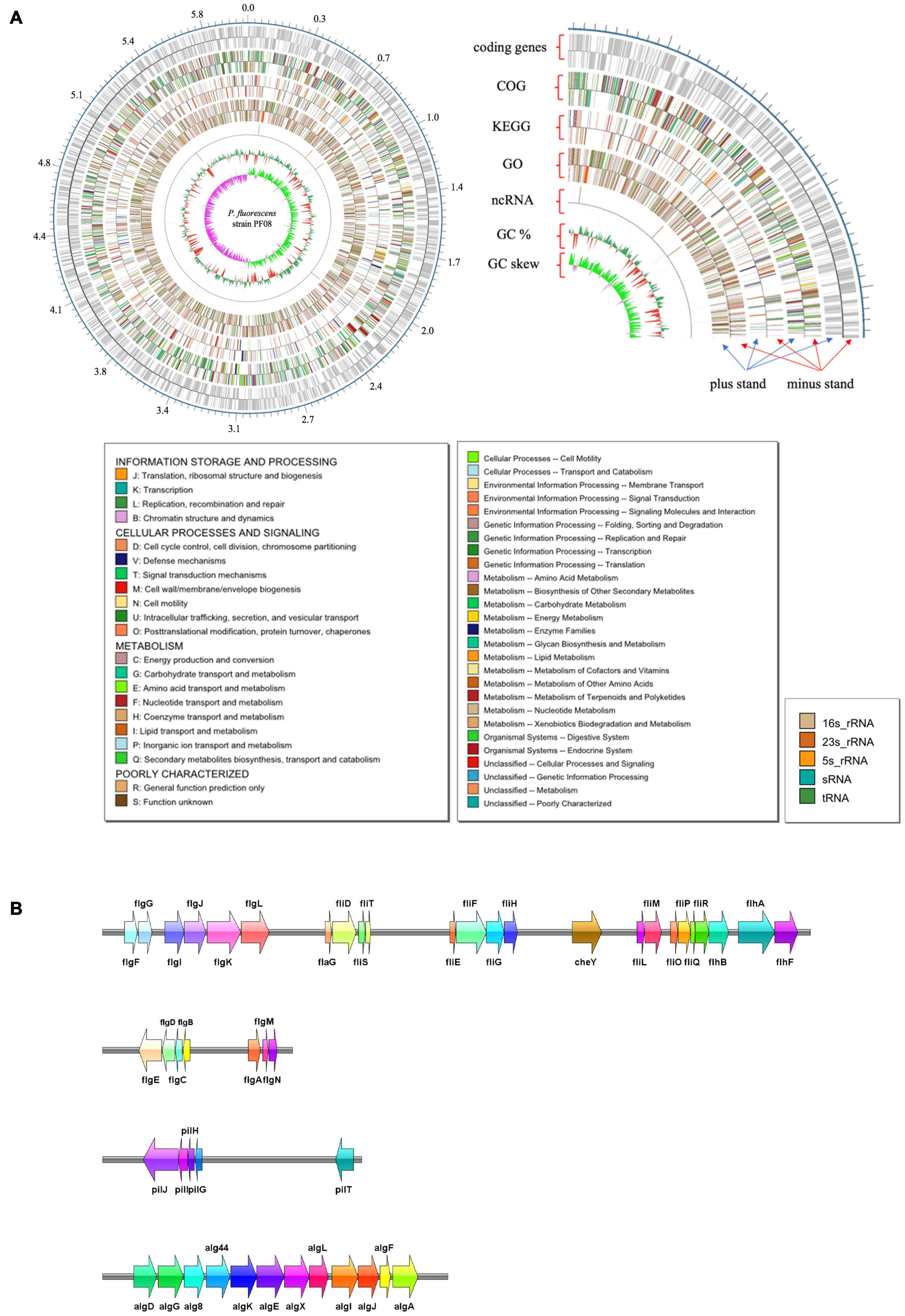
Figure 1. Graphical map of the completed chromosome of Pseudomonas fluorescens strain PF08. Numbers represent megabases (Mb). The type shown on the legend is the type that appears in the annotation result. The color of the box on the left corresponds to the color on the map (A). The spoilage-related gene clusters in the genome of P. fluorescens strain PF08 (B).
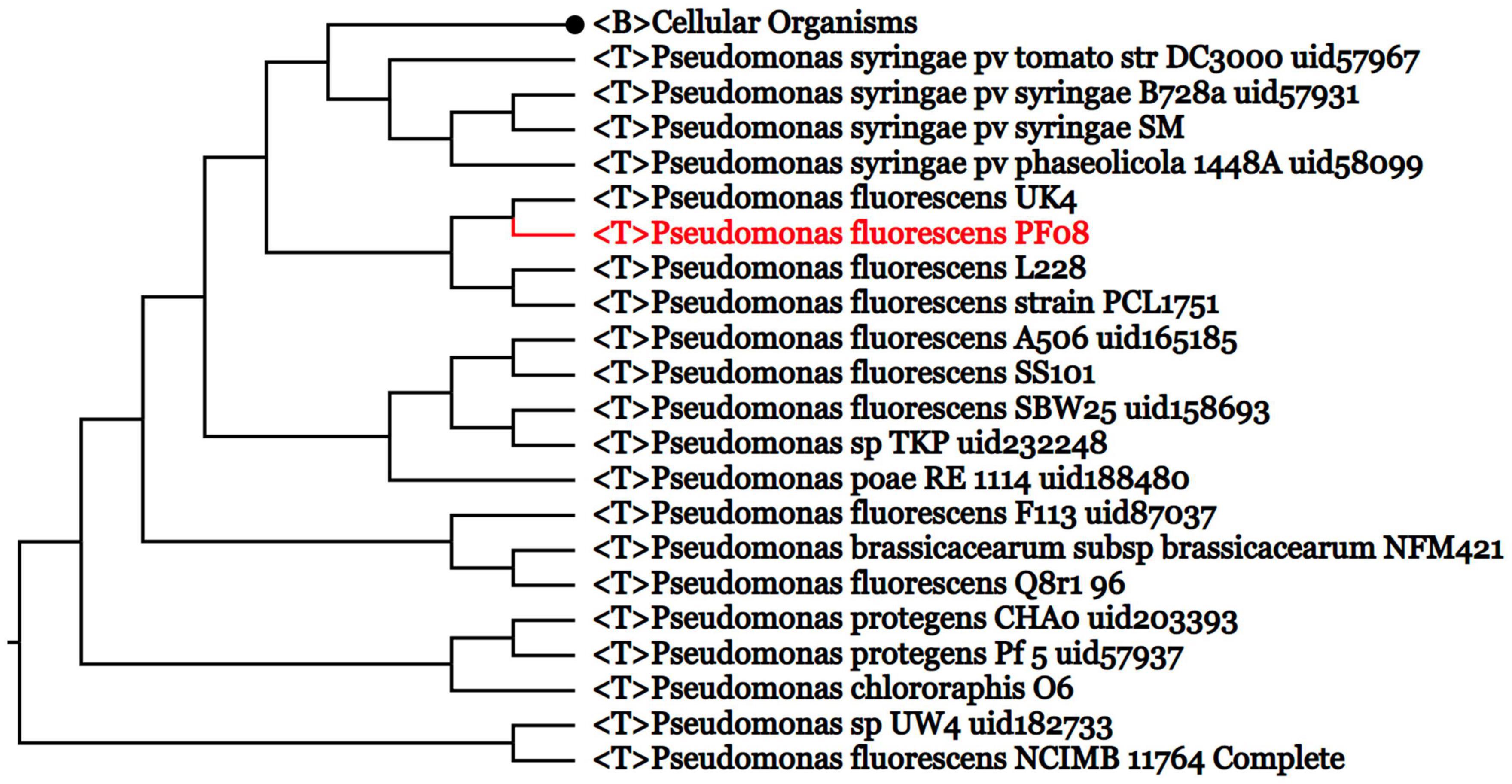
Figure 2. Genome phylogenetic tree based on the whole genome sequences of PF08 and some Pseudomonas strains compared in the study.
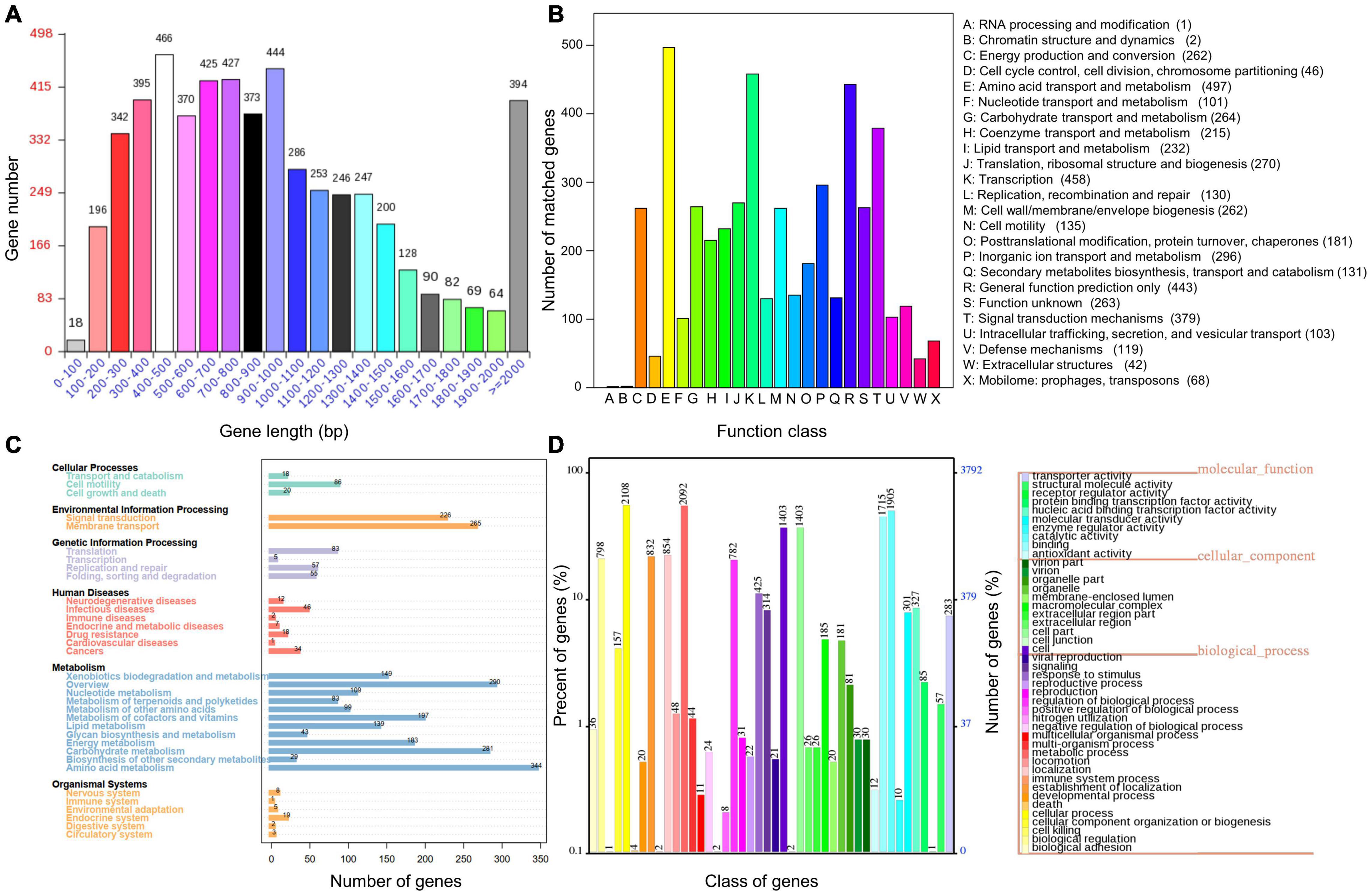
Figure 3. Distribution of gene sizes in Pseudomonas fluorescens PF08 (A) and the functional categories annotated using COG (B), KEGG (C), and GO (D).
To reveal more comprehensive genetic information of PF08, the genome was functionally annotated using the COG, GO, and KEGG databases. The results are shown in Figure 3. COG clustering showed that P. fluorescens PF-08 had more coding sequences involved in amino acid transport and metabolism, transcription, general function prediction, signal transduction mechanism, and lipid transport, with 497, 458, 443, 379, and 232 sequences, respectively (Figure 3B). The genes of P. fluorescens PF-08 mainly comprised functions of amino acid metabolism (344), carbohydrate metabolism (281), signal transduction (226), and transmembrane transport (265) (Figure 3C). The knowledge that the bacterial signal transduction process can be directly involved in the QS system indicates that PF-08 has a potential for QS signaling molecular transduction.
The annotation information obtained by comparing PF08 genome with the GO database using IPRscan revealed that the functions of the genes in P. fluorescens PF-08 were mainly distributed in the categories of cellular process, metabolic process, catalytic activity, and binding. The numbers of annotations were 2108, 2092, 1905, and 1715, accounting for 12.55, 12.46, 11.35, and 10.21% of the total number of annotations, respectively. The findings indicate that strain PF-08 has pronounced activities of biometabolism, enzyme catalysis, and functional factor binding (Figure 3D).
The LuxI/R type system, which is mainly composed of AHLs LuxI and specific receptor protein LuxR, is one of the most typical QS regulatory systems. The system has been found in many Gram-negative bacteria, including Burkholderia, Escherichia coli, Hafnia alvei, and Pseudoalteromonas (Weiss et al., 2008; Choudhary et al., 2013; Yu et al., 2019; Zhu et al., 2019). In this system, sufficient LuxI-secreted AHLs can interact with LuxR to form a complex that combines with the promoters of the downstream target genes to regulate their expression. This mediates the production of spoilage-required factors, including a variety of proteases (Galloway et al., 2011). Presently, the LuxI/R type system RhlI/R was detected in an organized operon in the genome of P. fluorescens PF08. In this system, rhlI encodes the AHLs synthetase, and rhlR encodes the response regulator. The RhlI/R system is reported to work as part of a cascade QS regulatory system to regulate the production of pyocyanin and chitinase in conjunction with the LasI/R system, in which the expression of rhlI/R is also positively regulated by the LasI/R system (Defoirdt, 2018). Interestingly, in this study, the genes encoding LasI/R were not found in the genome of P. fluorescens PF08, suggesting that the QS system RhlI/R may act as an independent system to accomplish the regulation of related factors, which is uncommon. This is consistent with previous observations (Steindler et al., 2009), who found that the two QS systems are not hierarchically organized and are both important for the colonization of P. aeruginosa on the rice rhizosphere.
During evolution, some Gram-negative bacteria have lost one or more LuxI-type AHL synthetases and have retained only unpaired LuxR-type regulator proteins. These proteins, termed LuxR solos or Orphan proteins, are still able to sense AHLs and regulate bacterial behavior accordingly. Thus, they have important roles in bacterial eavesdropping and inter- and intraspecific signaling (Hudaiberdiev et al., 2015). Therefore, exploring the luxR solo genes in the genome is valuable to understand the QS properties of P. fluorescens PF08. Eleven genes encoding LuxR family regulatory proteins were identified in the genome of PF08 in this study (Table 2). The response regulators PprA and PprB were identified as a two-component regulatory system of P. aeruginosa with the function to regulate virulence factor production and cell motility. Furthermore, some QS regulatory genes in P. aeruginosa, such as the las system and rhl system, are also mediated by this system, and most PprB-activated genes are activated by C4-HSL (Dong et al., 2005). In the P. fluorescens PF08 genome, only a separate gene, pprB, encoding the regulator PprB was identified. It was annotated as a luxR solo type regulator in the NR database. The presence of pprB explains the greater sensitivity of PF08 to C4-HSL than to other AHLs, which we previously demonstrated (Li T. et al., 2019). The hybrid histidine kinase RcsC and the response regulator RcsB are homologous to environmentally responsive two-component regulators capable of promoting biofilm formation and the appearance of fimbriae (Stout and Gottesman, 1990; Nicastro et al., 2009). In this study, the rcsB gene was identified as a luxR solo type regulator in the genome of PF08, which is helpful to further understand the regulation process of biofilm formation in PF08. The sdiA gene predicted to encode LuxR homologs to regulate the ftsQAZ locus and virulence factors EspD in E. coli and Salmonella enterica has also been identified in PF08 (Kanamaru et al., 2000). In addition, some genes encoding putative LuxR solo type regulator, such as the environmental sensing gene agmR and the virulence regulator gene bvgA, were also annotated in the PF08 genome. These findings could genetically explain the ability of PF08 to enhance its virulence and spoilage factors through AHLs eavesdropping (Li T. et al., 2019).
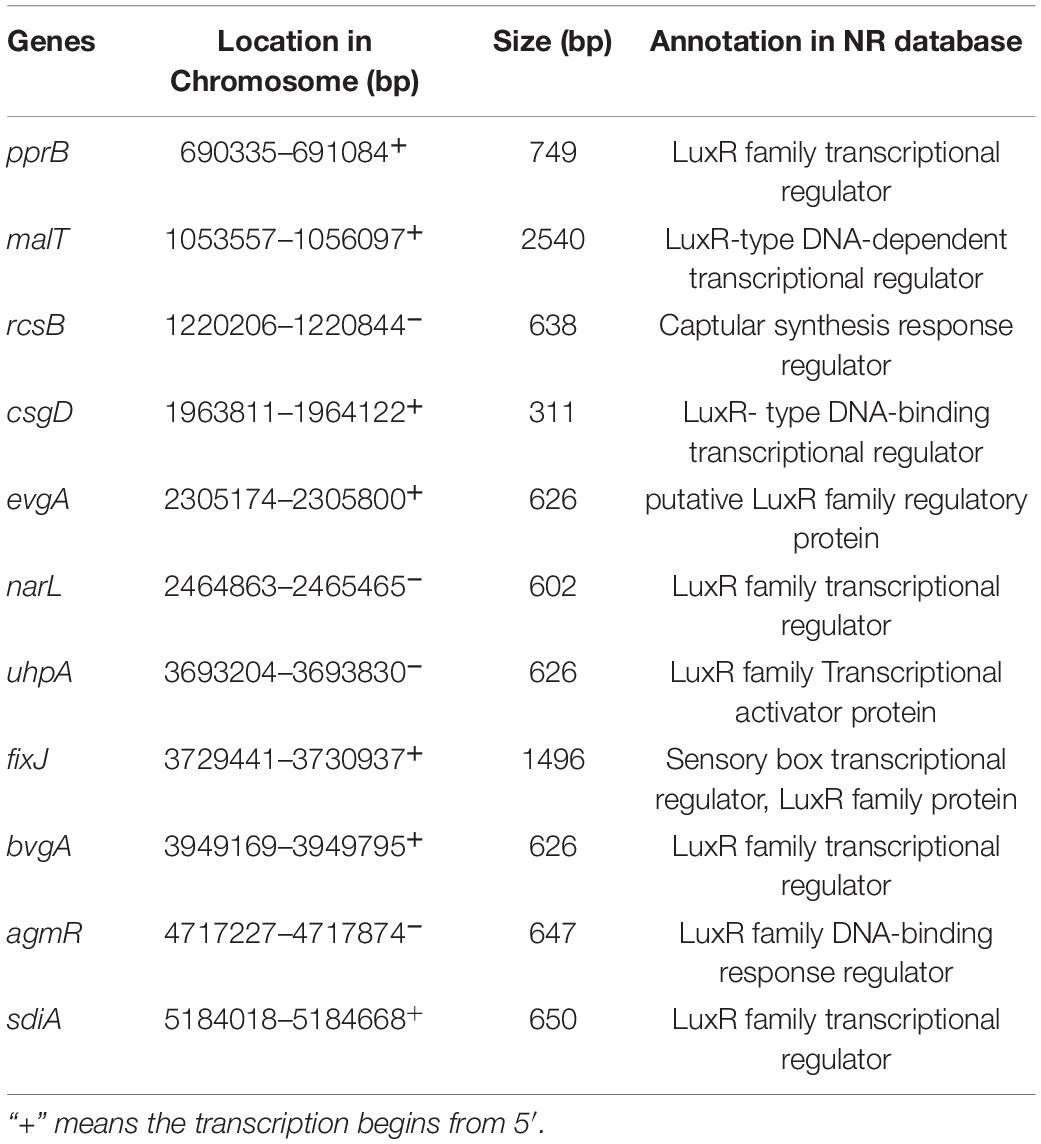
Table 2. Genes encoding predicted LuxR solo type proteins in the genome of Pseudomonas fluorescens PF08.
Flagellar driven motility is an effective way for bacteria to escape immune system components and fungicides. The motility is mainly activated by the QS system under environmental stress (Díaz et al., 2011). Pili can mediate the adhesion of bacteria to different surfaces, which is essential to initiate biofilm formation. Thus, pili pose risks in medical facilities and the food industry. Our preliminary study observed the good motility and biofilm formation ability of P. fluorescens (Li et al., 2018). Presently, we explored the genes associated with biofilm formation and motility of P. fluorescens PF08. A total of 34 genes related to flagella were identified. Of these, 13 belonged to the flg gene cluster located in 3 flagellar operons. These genes regulate the synthesis of flagella by sensing the integrity of the flagellar structure (Chilcott and Hughes, 2000). Gene clusters including fliD/S/T/E/F/G/H and fliL/M/O/P/Q/R encode the flagellar assembly proteins. These clusters were detected in P. fluorescens PF08 (Figure 1B). The FliF protein forms the M-ring embedded in the cytoplasmic membrane of the basal part of the flagellar. The integral membrane proteins located in the MS-C loop (FlhA, FlhB, FliP, FliQ, FliR, and FliO) and soluble proteins (FliL, FliH, and FliJ) form the output device of flagella. FlhA and FlhB proteins comprise the platform for the attachment of FliL and FliH. The corresponding response of bacteria to external environmental changes is called chemotaxis. Chemotaxis is regulated by a two-component system involving the methyl-accepting chemotaxis protein (MCP). MCP can sense the changes of chemical signals in the external environment and adjusts the motility of bacterial flagella by regulating the two-component system (Kulasekara et al., 2013). The histidine kinase CheA and response regulator CheY that perform signal transduction are important components of the two-component regulatory system. The genes encoding CheA and CheY were detected in P. fluorescens PF08 in this study, indicating that the bacteria can adapt to environmental changes.
Concerning biofilm establishment, gene clusters involved in the biosynthesis of type IV pili were detected in the P. fluorescens PF08 genome in this study. The genes can promote bacterial adhesion and biofilm formation. Some of these genes are highly conserved in Pseudomonas sp. Two examples are pilG and pilJ, which are required for pili extension and twitching motility (Darzins, 1993; DeLange et al., 2007). Cyclic bis (3′–5′) diguanylic acid (c-di-GMP) is a second messenger molecule commonly found in bacteria. c-di-GMP improves the synthesis of cellulose, a major biofilm component, and therefore plays an important role in regulating biofilm formation (Tischler and Camilli, 2004). Genes encoding the key enzymes in the synthesis and metabolism of c-di-GMP were detected in this study. These enzymes included diguanylate cyclase (GGDEF domain), c-di-GMP synthetase, and phosphodiesterase (EAL domain). Their presence suggests the existence of the c-di-GMP regulatory pathway in P. fluorescens PF08. Alginate and certain extracellular proteins form the reticular skeleton that maintains the mechanical stability of biofilms. These components are crucial for the extracellular polymer (Laue et al., 2006). In the initial stage of extracellular polymer formation, a high content of alginate is important in the attachment of bacteria to the surface. The presence of a series of alginate synthesis genes, including alg8, alg44, algA, algK, algE, algQ, and algX, and the aforementioned genes explains the strong biofilm formation and motility ability of P. fluorescens at the genetic level, which was confirmed in our previous study (Li et al., 2018).
Extracellular proteases secreted by spoilage bacteria can decompose proteins in food matrix into small nitrogen-containing substances to maintain their own growth, accompanied by rapid deterioration of food quality including taste and flavor. Protease is the most important spoilage factor. Therefore, knowledge of protease-related genes is necessary to better understand and inhibit food spoilage caused by P. fluorescens PF08. Presently, a total of 13 protease-encoding genes, including the metalloprotease aprX, alkaline protease aprA, ATP-dependent Clp protease clpP, Zn-dependent metalloprotease sprT, aspartyl protease asP1, and serine protease serP, were identified in the PF08 genome. Among these, alkaline proteases are the most important type of proteases that cause food spoilage. Scatamburlo et al. (2015) demonstrated that the alkaline proteases from Pseudomonas sp. cause the spoilage of goat milk. Similarly, bacterial metalloproteases have been implicated in the spoilage of diverse foods. Reducing the activity of metalloprotease AprX can improve the quality of ultra-high temperature-treated milk during storage (Zhang et al., 2019). Furthermore, the serine protease SerP in H. alvei H4 may be crucial in the spoilage of instant sea cucumber (Zhu et al., 2020). In addition to these typical proteases, other genes encode some spoilage-related lipases in PF08, such as lipase D (lipD), phospholipase (rssA), lysophospholipase (pldB), and triacylglycerol esterase (estA). These lipases can degrade fats and lipoproteins of aquatic products to produce free fatty acids, glycerol, and choline, thus accelerating spoilage (Glantz et al., 2020). Choline can be further converted into harmful end products like dimethylamine and trimethylamine, which detrimentally affect the flavor of aquatic products. The foregoing genes found in the P. fluorescens PF08 genome confirmed the spoilage potential of the bacterium from a genetic perspective.
Some metabolic pathways in bacteria are responsible for further metabolizing proteins, carbohydrates, and lipids. Various substances produced by their degradation form volatile components, such as trimethylamine, putrescine, and hydrogen sulfide. These volatiles are associated with putrid off-odors in aquatic products and are markers of spoilage. Pathways of amino acid metabolism, sulfur metabolism, and amine metabolism are therefore considered to be involved in food spoilage. The genetic information related to these relevant metabolic pathways was presently analyzed to explain the spoilage potential of P. fluorescens PF08. As shown in Figure 3C, amino acid metabolism was the primary function of P. fluorescens PF08 in KEGG annotations, suggesting good potential for protein degradation. The Cyc gene cluster is an important gene cluster involved in sulfur metabolism. Genes in the cluster encode key enzymes, including adenylylsulfate kinase CysC and phosphoadenosine phosphosulfate reductase CysH. These enzymes catalyze the multi-step reduction of adenosine-5’-phosphosulfate to hydrogen sulfide in PF08. Other genes associated with sulfur metabolism that were detected in the PF08 genome included glpE (encoding thiosulfate sulfurtransferase), ssuD (encoding alkanesulfonate monooxygenase), and sseA (encoding 3-mercaptopyruvate sulfurtransferase). Their presence is indicative of sulfur metabolic activity of PF08.
Amine metabolism of SSOs is another important metabolic pathway involved in the spoilage of aquatic products. This pathway was also analyzed in this study. Putrescine is one of the main causes of the unpleasant smell of rotten aquatic products. The genes encoding a series of proteins responsible for the production of putrescine, such as putrescine transport system permease protein, putrescine-binding periplasmic protein E, putrescine aminotransferase, and gamma-glutamylputrescine oxidase, were identified in the ammonia metabolism pathway of P. fluorescens PF08. Genes encoding trimethylamine dehydrogenase and spermidine synthase were also detected. These enzymes are involved in trimethylamine metabolism and spermidine production, respectively. The results demonstrate that PF08 has the potential for amine metabolism. Interestingly, although Gloria et al. (2011) detected cadaverine and histamine in spoilage milk inoculated with P. fluorescens; however, no genes related to cadaverine and histamine production were found in P. fluorescens PF08 in this study.
The continuous consumption of proteins and ions caused by microbial metabolism leads to the irreversible deterioration of the food microenvironment. To adapt to this change of the environment, microorganisms adjust their metabolic activities to maintain their normal growth by secreting some proteins involved in homeostasis. We have demonstrated that P. fluorescens PF08 is the SSO of turbot during low temperature storage, suggesting that this bacterium has a particular advantage in interbacterial competition during this process (Li T. et al., 2019). Investigation of genes related to the stress response in the P. fluorescens PF08 genome is helpful to reveal the reasons behind this advantage. RpoS and RelA were detected in the genome. They are considered typical environmental response regulators involved in stress survival in bacteria. Liu et al. (2018) found the RpoS-deficient mutant strains of P. fluorescens were less tolerant to environmental factors of high temperature, low pH, and starvation. The QS system of the mutant strains was also suppressed. In P. fluorescens PF08, the rpoS gene may play a similar role. Pyoverdine is a low molecular weight organic compound that can bind Fe2+ efficiently under low iron stress conditions to maintain the necessary Fe2+ uptake in cells. The synthesis of pyoverdine in bacteria is also regulated by the QS system (Xu et al., 2019). The pvdA and pvdE genes regulate the biosynthesis of pyoverdine. The presence of both genes in PF08 indicate that PF08 may have a certain growth advantage in low iron stress. Other genes identified in PF08 included some general stress response-related genes, including the antioxidant protein-coding gene ahpC, glutathione peroxidase-coding gene bsaA, starvation stringent protein-coding genes sspA and sspB, and heavy metal detoxification-related genes cusS and merR. The existence of these genes may be a potential reason for the environmental adaptability and competitive advantage of PF08.
The accumulation of antibiotics in the environment and animal food farming processes imposes antibiotic pressure on bacteria, which had driven the evolution of a series of resistance pathways. Bacterial drug resistance can be transmitted by conjugation transfer, which poses a great challenge to food safety. More than 40 drug resistance-related genes were found in the genome of PF08. Hydrolysis of antibiotics into harmless metabolites by secretion of some key enzymes is the main pathway of bacterial drug resistance. β-lactamase and penicillin amidase encoded in PF08 can accelerate the metabolism of intracellular penicillin, thereby enhancing the tolerance of PF08 to penicillin. The gene encoding D-amino-acid oxidase (DAO), that can degrade cephalosporin, was also detected in PF08. In addition, the rapid efflux through the transport system of antibiotics that are not degraded is another effective means of bacterial survival. A total of 12 ABC-type multidrug transport system-related genes and 24 multidrug efflux pump genes were identified in the PF08 genome. The system composed of these coding genes can efficiently recognize specific lipophilic antibiotics and transport them out of the cells. The discovery of these genes has contributed to a better understanding of the drug resistance of PF08.
The antibacterial properties of the metabolites of PF08 were also investigated. This antibacterial property was implied by the observed advantage of PF08 in competing with other bacteria for survival during turbot spoilage. As shown in Supplementary Figure 2, the ethyl acetate extract of PF08 produced significant zones of growth inhibition in plates inoculated with S. aureus and A. salmonicida. The findings indicated the bacteriostasis of metabolites by PF08. Indole alkaloids are one of the marine alkaloids. They are derived mainly from marine organisms, such as sponges and mosses, which have significant antibacterial, antitumor, and other biological activities. The genes involving in the biosynthesis of indole alkaloids were detected in PF08, indicating the potential of PF08 to synthesize alkaloids. Polyketides are natural compounds produced by bacteria and fungi. The various polyketides include macrolides, tetracyclines, and polyethers. These compounds have antibacterial and anti-infection activities. In bacteria, the biosynthesis of polyketides is regulated by polyketide synthase. Construction of the carbocyclic scaffold catalyzed by polyketide cyclase is a key step in the biosynthesis. The genes encoding polyketide synthase and polyketide cyclase were detected in PF08, as were antibiotic biosynthesis monooxygenase (ABM) superfamily genes. Qin et al. (2019) identified the non-catalytic role of ABM superfamily proteins in polyketide synthesis in maintaining the fidelity of the biosynthetic pathway. The results suggest that PF08 may have the ability to secrete this kind of antibacterial substance. These antimicrobial substances produced by PF08 are mostly lipophilic and can be transported extracellularly using their own drug efflux pump system to reduce intracellular accumulation and thus inhibit the growth of competing microorganisms without damaging themselves.
The QS system has been widely reported as a key factor in the regulation of food spoilage (Liu et al., 2018). However, the role of QS system in the regulation of spoilage-related genes in P. fluorescens has not been systematically reported. Therefore, the effect of C4-HSL, the typical AHL synthesized by the RhlI protein, on the transcription of rhlI/R and other typical spoilage-related genes, including pilG, pilJ, fliL, flhA, algA, algX, aprX, aprA, serP, pvdA, pvdE, ahpC, sspA, and cusS, in PF08 was further analyzed in this study to explore the regulatory role of QS system on the spoilage potential of PF08. The results are presented in Figure 4A. As an agonist of the QS system, C4-HSL significantly increased the transcription of rhlI/R genes in PF08, suggesting that the QS system was successfully activated because of the feedback regulation mechanism of the Lux-type QS system. Transcription of some genes related to motility and biofilm formation, such as pilG, pilJ, flhA, algA, and algX, was increased with the addition of C4-HSL. More importantly, the transcription of genes, including aprX, aprA, and serP, which encode proteases that are active in spoilage was all promoted after incubating with C4-HSL. The findings suggest, at the genetic level, the regulatory role of QS on the spoilage potential of PF08. The addition of C4-HSLs also promoted the transcription of pvdA, pvdE, ahpC, and cusS related to stress response. This might make PF08 more competitive than other bacteria during turbot spoilage. Interestingly, the transcription of sspA that encodes starvation stringent protein was inhibited after adding C4-HSL. These results suggested that the addition of C4-HSL put the bacteria in a more “excited” state.
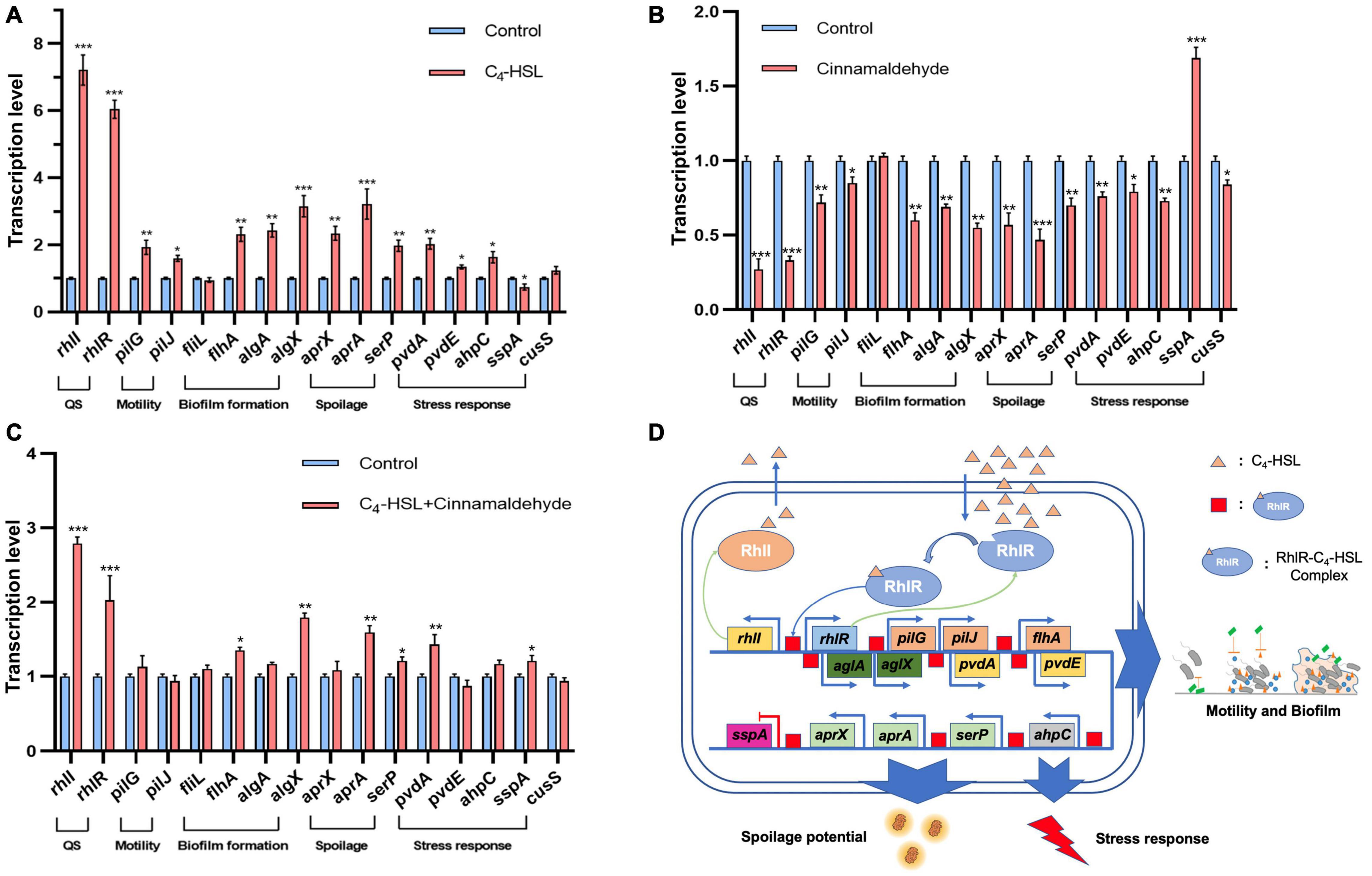
Figure 4. Transcription levels of typical genes related to spoilage and stress response in Pseudomonas fluorescens PF08 after incubation with 20 μg/mL C4-HSL (A), or 0.1 μL/mL cinnamaldehyde (B), or 20 μg/mL C4-HSL and 0.1 μL/mL cinnamaldehyde (C) until an optical density at 595 nm (OD 595) of 1 was reached. Schematic diagram of the regulation of QS system in PF08 on the spoilage potential (D). Data are presented as means ± SD (n = 3, *P < 0.05, **P < 0.01, ***P < 0.001).
Although we found that the transcription of some key spoilage related genes changed synchronously with the promotion of QS system. However, this is not enough to demonstrate their correlation with QS system. The QS inhibitory effect of cinnamaldehyde on Pseudomonas has been wildly reported (Topa et al., 2020). Our previous study also showed that cinnamaldehyde can inhibit the QS system of P. fluorescens by targeting LuxR-type QS receptor without influencing AHLs (Li et al., 2018). Therefore, cinnamaldehyde was used as a QS inhibitor here and the transcription of these key genes in cinnamaldehyde treated PF08 was further analyzed. As shown in Figure 4B, the downregulation of the transcription of rhlI and rhlR gene proved the inhibition of the QS system in PF08 caused by cinnamaldehyde. Moreover, the transcription of pilG, pilJ, flhA, algA, algX, aprX, aprA, pvdA, pvdE, ahpC, and cusS were all down-regulated by cinnamaldehyde. Correspondingly, the transcription level of sspA increased after the treatment of cinnamaldehyde. More importantly, when cinnamaldehyde and C4-HSL were added simultaneously, the transcription promotion of these key genes caused by C4-HSL was significantly weakened (Figure 4C). The results demonstrate the correlation between the transcription of spoilage related genes and the QS system (rhlI/R) in PF08. Although knockout of the QS system may be a more direct method, since C4-HSL and cinnamaldehyde have been proved to act directly on the QS system, we believe that the spoilage potential of PF08 is likely to be regulated by QS system by regulating the transcription of relevant genes (Figure 4D).
Specific spoilage organisms mainly regulate the production of spoilage-required factors, such as protease, by the QS system, which can sense environmental changes during food spoilage and provides feedback by initiating the transcription of relevant genes. Therefore, the growth environment may change the characteristics of bacteria and may further affect their spoilage potential by stimulating the QS system. The effects of different environmental conditions on the transcription of rhlI and rhlR genes in PF08 were evaluated using qPCR. As shown in Figure 5, the environmental factors could affect the transcription of rhlI and rhlR in PF08. Carbon is an essential and basic nutrient for microbial growth. The carbon source can be a potential factor affecting the transcription of QS systems owing to the different metabolic pathways of bacteria for the different sources. As shown in Figure 5A, the relative transcription of rhlI was the highest during culture with lactose as the carbon source. Growth was 5.6-fold higher than that of the control group. The relative transcription of rhlI was the lowest when xylose was the carbon source. The transcription of rhlI decreased sequentially when the carbon sources were sucrose, maltose, glucose, and fructose. Similar to the rhlI gene, the relative transcription of rhlR was the highest and lowest when lactose and xylose were the respective carbon sources. However, the effect of carbon source on rhlR transcription was not significant compared with that on rhlI transcription and was mostly inhibitory.
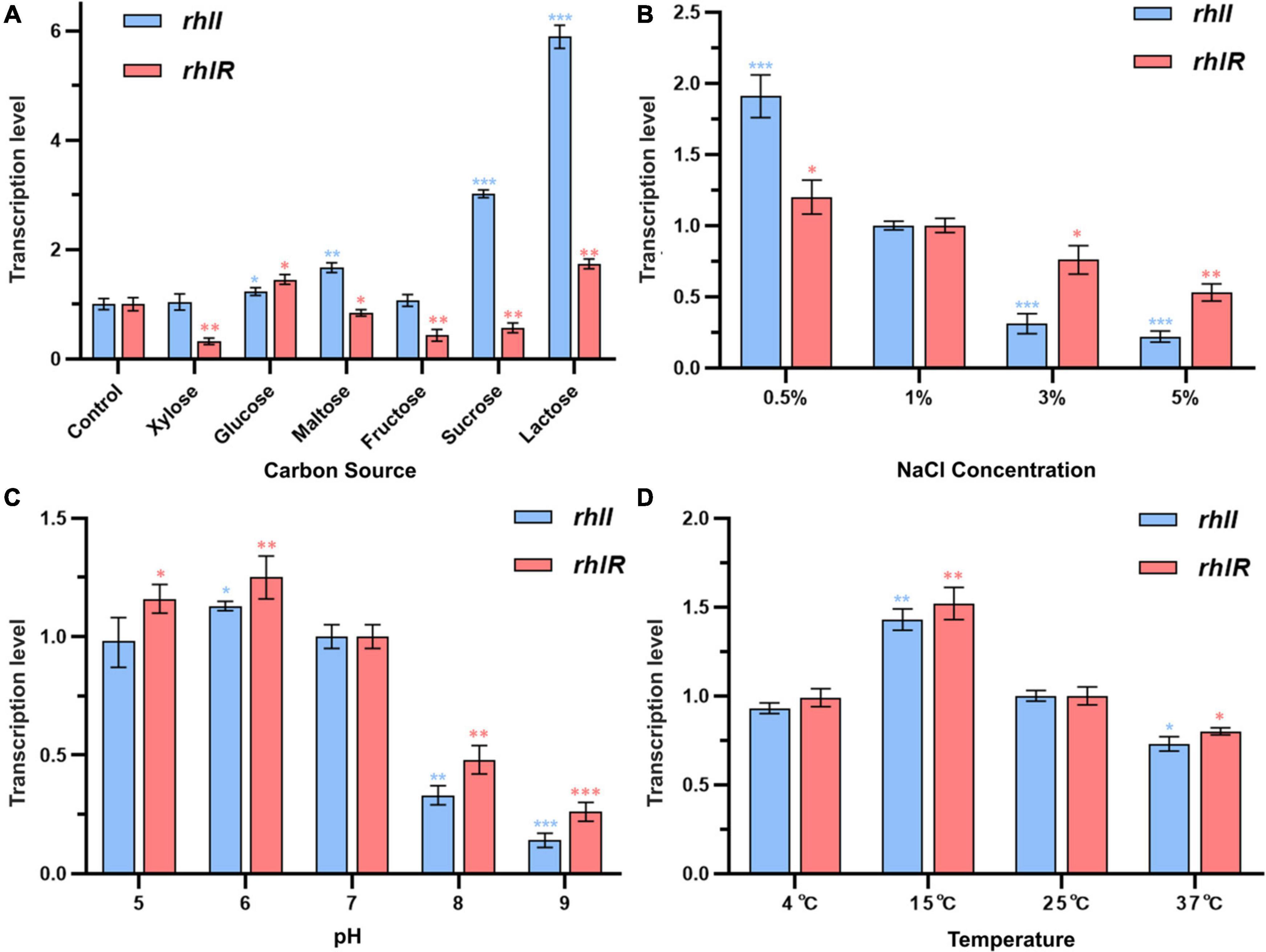
Figure 5. Transcription levels of rhlI/R genes in Pseudomonas fluorescens PF08 after incubation in different conditions until an optical density at 595 nm (OD 595) of 1 was reached. (A) Different carbon sources in culture medium; (B) different NaCl concentrations of the culture medium; (C) different pH of the culture medium; (D) different temperatures during incubation. Data are presented as means ± SD (n = 3, *P < 0.05, **P < 0.01, ***P < 0.001).
Figure 5B depicts the transcription of rhlI and rhlR genes in PF08 cultured in different NaCl concentrations. The transcription of the rhlI and rhlR genes decreased significantly with increasing NaCl concentration and was the lowest when the NaCl concentration was 5% (w/v). The transcription of rhlI and rhlR was only 22 and 53% of that apparent at 1% NaCl, respectively. In contrast, the transcription of rhlI and rhlR increased when the NaCl concentration < 1%. The respective transcription was 1.9 and 1.2-times higher, respectively, than the expression at 1% NaCl. The results showed that the concentration of NaCl in the culture environment could affect the transcription of rhlI and rhlR genes in P. fluorescens, and that a low NaCl concentration was more beneficial to the transcription of the two genes. Similarly, Yarwood and Schlievert (2003) showed that the secretion of microbial AHLs is susceptible to stress from low or high NaCl concentrations in the environment.
The transcription of rhlI and rhlR genes in PF08 cultured in different pH conditions is shown in Figure 5C. An acidic environment had little effect on the transcription of rhlI and rhlR genes. However, when the culture environment was alkaline, rhlI and rhlR transcription was significantly inhibited. When the initial pH of the medium was 9, the transcription of rhlI and rhlR genes in PF08 had the lowest values, which were 14 and 16% of those at pH 7. Our previous study demonstrated that the secretion of AHLs from P. fluorescens was significantly reduced in alkaline environments. We suggested that this reflects the destruction of the lactone ring of AHLs in alkaline environments, resulting in a reduction in their chemical stability (Yang et al., 2017). The results obtained here demonstrate that this may be due to the synergistic inhibition of the transcription of AHLs synthetase gene rhlI under alkaline conditions.
Temperature is another important environmental factor affecting the growth and metabolism of bacteria. Previous studies have shown that the bacterial QS system is affected by temperature. Therefore, the effects of culture temperature on the transcription of rhlI and rhlR genes in PF08 were explored. The results are shown in Figure 5D. When the culture temperature was 15°C, the relative transcription levels of rhlI and rhlR genes were the highest (1.43 and 1.52-times higher, respectively, than those at 25°C). In contrast, the relative transcription levels of rhlI and rhlR genes in PF08 cultured at 37°C were only 73 and 80% of the values at 25°C. This is consistent with the observations of Zhu et al. (2020), who found that the culture temperature could affect the expression of luxI/R genes in H. alvei H4, with expression suppressed at high temperatures. Interestingly, the relative transcription of rhlI and rhlR genes remained essentially unchanged when incubated at 4°C, reaching 93 and 99% of the values at 25°C. This may reflect that P. fluorescens is a psychrotrophic bacterium. The results suggests that PF08 may still effectively regulate the production of spoilage-required factors through the QS system at the storage temperature of aquatic products (4°C).
The findings expand the knowledge of genes associated with QS system and spoilage potential, including protease production, biofilm formation, and sulfur and amine metabolism, in the genome of P. fluorescens PF08. Furthermore, the regulatory effect of the QS system on the transcription of typical genes related to the spoilage potential of PF08 was demonstrated. The influence of environmental factors on the transcription level of the rhlI/R system in PF08 was evaluated. The results further clarify the relationship between QS system and the spoilage potential of PF08 and will inform the development of QS-targeted spoilage control strategies.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
DW conceived and designed the experiments, analyzed the data, contributed reagents, materials, and analysis tools, authored or reviewed drafts of the manuscript, and approved the final draft. FC conceived and designed the experiments. LR analyzed the data, contributed reagents, materials, and analysis tools, prepared the figures and/or tables. XT, XL, and QL conceived and designed the experiments. JL conceived and designed the experiments, approved the final draft, had overall responsibility for this project. TL conceived and designed the experiments, performed the experiments, analyzed the data, prepared the figures, and/or tables, authored or reviewed drafts of the manuscript. All the authors contributed to the article and approved the submitted version.
This study was supported by the National Key Research and Development Program of China (2019YFD0901702).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We thank the Novogene Bioinformatics Technology Co., Ltd. for the work in genome sequencing and analysis. We also thank the executive editor at Editage Inc. for English editing.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.856802/full#supplementary-material
Supplementary Figure 1 | Alignment of the genomes of Pseudomonas fluorescens PF08 and different type strains of other Pseudomonas. P. fluorescens UK4 (A), P. aeruginosa PA01 (B), P. putida NBRC 14164 (C), P. syringe BIM B-268 (D). The same color represents the colinear regions of similarity.
Supplementary Figure 2 | The inhibitory effect of ethyl acetate extract of Pseudomonas fluorescens PF08 on the growth of Staphylococcus aureus and Aeromonas salmonicida CS12. (a) Metabolites of PF08; (b) Equal amount of 50% (v/v) methanol used as control.
Bai, A. J., and Rai, V. R. (2011). Bacterial quorum sensing and food industry. Rev. Food Sci. 10, 183–193. doi: 10.1111/j.1541-4337.2011.00150.x
Brachmann, A. O., Brameyer, S., Kresovic, D., Hitkova, I., Kopp, Y., Manske, C., et al. (2013). Pyrones as bacterial signaling molecules. Nat. Chem. Biol. 9, 573–578. doi: 10.1038/nchembio.1295
Chan, K. G., Yin, W. F., and Lim, Y. L. (2014). Complete genome sequence of Pseudomonas aeruginosa strain YL84, a quorum-sensing strain isolated from compost. Genome Announc. 2, e246–e214. doi: 10.1128/genomeA.00246-14
Chilcott, G. S., and Hughes, K. T. (2000). Coupling of flagellar gene expression to flagellar assembly in Salmonella enterica serovar typhimurium and Escherichia coli. Microbiol. Mol. Biol. Rev. 64, 694–708. doi: 10.1128/mmbr.64.4.694-708.2000
Choudhary, S., Hudaiberdiev, Z., Gelencsér, B., Gonçalves Coutinho, V., Venturi, S., and Pongor, S. (2013). The organization of the quorum sensing luxI/R family genes in Burkholderia. Int. J. Mol. Sci. 14, 13727–13747. doi: 10.3390/ijms140713727
Darzins, A. (1993). The pilG gene product, required for Pseudomonas aeruginosa pilus production and twitching motility, is homologous to the enteric, single-domain response regulator chey. J. Bacteriol. 175, 5934–5944. doi: 10.1128/jb.175.18.5934-5944.1993
Defoirdt, T. (2018). Quorum-sensing systems as targets for antivirulence therapy. Trends Microbiol. 26, 313–328. doi: 10.1016/j.tim.2017.10.005
DeLange, P. A., Collins, T. L., Pierce, T. L., and Robinson, J. B. (2007). PilJ localizes to cell poles and is required for type iv pilus extension in Pseudomonas aeruginosa. Curr. Microbiol. 55, 389–395. doi: 10.1007/s00284-007-9008-5
Diao, W. R., Zhang, L. L., Feng, S. S., and Xu, J. G. (2014). Chemical composition, antibacterial activity, and mechanism of action of the essential oil from Amomum kravanh. J. Food Prot. 77, 1740–1746. doi: 10.4315/0362-028X.JFP-14-014
Díaz, C., Schilardi, P. L., Salvarezza, R. C., and Fernández Lorenzo de Mele, M. (2011). Have flagella a preferred orientation during early stages of biofilm formation: AFM study using patterned substrates. Colloids Surf. B: Biointerfaces 82, 536–542. doi: 10.1016/j.colsurfb.2010.10.013
Ding, T., Li, T., Wang, Z., and Li, J. (2017). Curcumin liposomes interfere with quorum sensing system of Aeromonas sobria and in silico analysis. Sci. Rep. 7:18612. doi: 10.1038/s41598-017-08986-9
Dong, Y. H., Zhang, X. F., Soo, H.-M. L., Greenberg, E. P., and Zhang, L. H. (2005). The two-component response regulator PprB modulates quorum-sensing signal production and global gene expression in Pseudomonas aeruginosa. Mol. Microbiol. 56, 1287–1301. doi: 10.1111/j.1365-2958.2005.04612.x
Galloway, W. R. J. D., Hodgkinson, J. T., Bowden, S. D., Welch, M., and Spring, D. R. (2011). Quorum sensing in Gram-negative bacteria: small-molecule modulation of AHL and AI-2 quorum sensing pathways. Chem. Rev. 111, 28–67. doi: 10.1021/cr100109t
Glantz, M., Rosenlöw, M., Lindmark-Månsson, H., Buhelt Johansen, L., Hartmann, J., Höjer, A., et al. (2020). Impact of protease and lipase activities on quality of Swedish raw milk. Int. Dairy J. 107:104724. doi: 10.1016/j.idairyj.2020.104724
Gloria, M. B. A., Saraiva, P. R., Rigueira, J. C. S., and Brandão, S. C. C. (2011). Bioactive amines changes in raw and sterilised milk inoculated with Pseudomonas fluorescens stored at different temperatures. Int. J. Dairy Technol. 64, 45–51. doi: 10.1111/j.1471-0307.2010.00637.x
Grissa, I., Vergnaud, G., and Pourcel, C. (2007). CRISPRFinder: a web tool to identify clustered regularly interspaced short palindromic repeats. Nucl. Acids Res. 35(Suppl. 2), W52–W57. doi: 10.1093/nar/gkm360
Hsiao, W., Wan, I., and Jones, S. J. (2003). IslandPath: aiding detection of genomic islands in prokaryotes. Bioinformatics 19, 418–420. doi: 10.1093/bioinformatics/btg004
Hudaiberdiev, S., Choudhary, K. S., Vera Alvarez, R. V., Gelencsér, Z., Ligeti, B., and Lamba, B. (2015). Census of solo luxR genes in prokaryotic genomes. Front. Cell. Infect. Microbiol. 5:20. doi: 10.3389/fcimb.2015.00020
Kanamaru, K., Kanamaru, K., Tatsuno, I., Tobe, T., and Sasakawa, C. (2000). SdiA, an Escherichia coli homologue of quorum-sensing regulators, controls the expression of virulence factors in enterohaemorrhagic Escherichia coli O157:H7. Mol. Microbiol. 38, 805–816. doi: 10.1046/j.1365-2958.2000.02171.x
Kulasekara, B. R., Kamischke, C., Kulasekara, H. D., Christen, M., Wiggins, P. A., and Miller, S. I. (2013). C-di-gmp heterogeneity is generated by the chemotaxis machinery to regulate flagellar motility. eLife 2:e01402. doi: 10.7554/eLife.01402
Lagesen, K., Hallin, P., and Rødland, E. A. (2007). RNAmmer: consistent and rapid annotation of ribosomal RNA genes. Nucl. Acids Res. 35, 3100–3108. doi: 10.1093/nar/gkm160
Laue, H., Schenk, A., Li, H., Lambertsen, L., Neu, T. R., Molin, S., et al. (2006). Contribution of alginate and levan production to biofilm formation by Pseudomonas syringae. Microbiology 152, 2909–2918. doi: 10.1099/mic.0.28875-0
Li, T., Wang, D., Liu, N., Ma, Y., Ding, T., Mei, Y., et al. (2018). Inhibition of quorum sensing-controlled virulence factors and biofilm formation in Pseudomonas fluorescens by cinnamaldehyde. Int. J. Food Microbiol. 269, 98–106. doi: 10.1016/j.ijfoodmicro.2018.01.023
Li, T., Wang, D., Ren, L., Mei, Y., Ding, T., Li, Q., et al. (2019). Involvement of exogenous N-acyl-homoserine lactones in spoilage potential of Pseudomonas fluorescens isolated from refrigerated turbot. Front. Microbiol. 10:2716. doi: 10.3389/fmicb.2019.02716
Li, X., Zhang, G., Zhu, Y., Bi, J., Hao, H., and Hou, H. (2019). Effect of the luxI/R gene on ahl-signaling molecules and QS regulatory mechanism in Hafnia alvei H4. AMB Express 9:197. doi: 10.1186/s13568-019-0917-z
Liu, L., Yan, Y., Feng, L., and Zhu, J. (2018). Quorum sensing AsaI mutants affect spoilage phenotypes, motility, and biofilm formation in a marine fish isolate of Aeromonas salmonicida. Food Microbiol. 76, 40–51. doi: 10.1016/j.fm.2018.04.009
Lowe, T. M., and Eddy, S. R. (1997). TRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25, 955–964. doi: 10.1093/nar/25.5.955
Nicastro, G. G., Boechat, A. L., Abe, C. M., Kaihami, G. H., and Baldini, R. L. (2009). Pseudomonas aeruginosa PA14 cupD transcription is activated by the RcsB response regulator, but repressed by its putative cognate sensor RcsC. FEMS Microbiol. Lett. 301, 115–123. doi: 10.1111/j.1574-6968.2009.01803.x
Packiavathy, I. A. S. V., Priya, S., Pandian, S. K., and Ravi, A. V. (2014). Inhibition of biofilm development of uropathogens by curcumin - an anti-quorum sensing agent from Curcuma longa. Food Chem. 148, 453–460. doi: 10.1016/j.foodchem.2012.08.002
Papenfort, K., and Bassler, B. L. (2016). Quorum sensing signal-response systems in Gram-negative bacteria. Nat. Rev. Microbiol. 14, 576–588. doi: 10.1038/nrmicro.2016.89
Qin, Z., Devine, R., Hutchings, M. I., and Wilkinson, B. (2019). A role for antibiotic biosynthesis monooxygenase domain proteins in fidelity control during aromatic polyketide biosynthesis. Nat. Commun. 10:3611. doi: 10.1038/s41467-019-11538-6
Saha, S., Bridges, S., and Magbanua, Z. V. (2008). Empirical comparison of ab initio repeat finding programs. Nucl. Acids Res. 36, 2284–2294. doi: 10.1093/nar/gkn064
Scatamburlo, T. M., Yamazi, A. K., Cavicchioli, V. Q., Pieri, F. A., and Nero, L. A. (2015). Spoilage potential of Pseudomonas species isolated from goat milk. J. Dairy Sci. 98, 759–764. doi: 10.3168/jds.2014-8747
Speranza, B., Bevilacqua, A., Conte, A., Del Nobile, M. A., Sinigaglia, M., and Corbo, M. R. (2013). Use of desirability approach to predict the inhibition of Pseudomonas fluorescens, Shewanella putrefaciens and Photobacterium phosphoreum in fish fillets through natural antimicrobials and modified atmosphere packaging. Food Bioprocess Technol. 6, 2319–2330. doi: 10.1007/s11947-012-0889-3
Steindler, L., Bertani, I., De Sordi, L., Schwager, S., Eberl, L., Las, V., et al. (2009). LasI/R and RhlI/R quorum sensing in a strain of Pseudomonas aeruginosa beneficial to plants. Appl. Environ. Microbiol. 75, 5131–5140. doi: 10.1128/AEM.02914-08
Stout, V., and Gottesman, S. (1990). Rcsb and RcsC: a two-component regulator of capsule synthesis in Escherichia coli. J. Bacteriol. 172, 659–669. doi: 10.1128/jb.172.2.659-669.1990
Tischler, A. D., and Camilli, A. (2004). Cyclic diguanylate (c-di-gmp) regulates Vibrio cholerae biofilm formation. Mol. Microbiol. 53, 857–869. doi: 10.1111/j.1365-2958.2004.04155.x
Topa, S. H., Palombo, E. A., Kingshott, P., and Blackall, L. L. (2020). Activity of cinnamaldehyde on quorum sensing and biofilm susceptibility to antibiotics in Pseudomonas aeruginosa. Microorganisms 8:455. doi: 10.3390/microorganisms8030455
Weiss, L. E., Badalamenti, J. P., Weaver, L. J., Tascone, A. R., Weiss, P. S., and Richard, T. L. (2008). Engineering motility as a phenotypic response to LuxI/R-dependent quorum sensing in Escherichia coli. Biotechnol. Bioeng. 100, 1251–1255. doi: 10.1002/bit.21862
Xu, Z., Zhang, H., Yu, H., Dai, Q., Xiong, J., Sheng, H., et al. (2019). Allicin inhibits Pseudomonas aeruginosa virulence by suppressing the rhl and pqs quorum-sensing systems. Can. J. Microbiol. 65, 563–574. doi: 10.1139/cjm-2019-0055
Yang, B., Li, T. T., and Li, J. R. (2017). Isolation, identification and quorum sensing of dominant spoilage bacteria from Scophthalmus maximus. J. Chin. Inst. Food Sci. Technol. 17, 139–145.
Yarwood, J. M., and Schlievert, P. M. (2003). Quorum sensing in Staphylococcus infections. J. Clin. Invest. 112, 1620–1625. doi: 10.1172/JCI20442
Yu, Z., Yu, D., Mao, Y., Zhang, M., Ding, M., Zhang, J., et al. (2019). Identification and characterization of a LuxI/R-type quorum sensing system in Pseudoalteromonas. Res. Microbiol. 170, 243–255. doi: 10.1016/j.resmic.2019.07.001
Zhang, C., Bijl, E., Svensson, B., and Hettinga, K. (2019). The extracellular protease AprX from Pseudomonas and its spoilage potential for UHT milk: a review. Compr. Rev. Food Sci. Food Saf. 18, 834–852. doi: 10.1111/1541-4337.12452
Zhu, Y. L., Hou, H. M., Zhang, G. L., Wang, Y. F., and Hao, H. S. (2019). AHLs regulate biofilm formation and swimming motility of Hafnia alvei H4. Front. Microbiol. 10:1330. doi: 10.3389/fmicb.2019.01330
Zhu, Y., Sang, X., Li, X., Zhang, Y., Hao, H., Bi, J., et al. (2020). Effect of quorum sensing and quorum sensing inhibitors on the expression of serine protease gene in Hafnia alvei H4. Appl. Microbiol. Biotechnol. 104, 7457–7465. doi: 10.1007/s00253-020-10730-9
Keywords: Pseudomonas fluorescens PF08, complete genome, spoilage ability, quorum sensing, specific spoilage organism
Citation: Wang D, Cui F, Ren L, Tan X, Lv X, Li Q, Li J and Li T (2022) Complete Genome Analysis Reveals the Quorum Sensing-Related Spoilage Potential of Pseudomonas fluorescens PF08, a Specific Spoilage Organism of Turbot (Scophthalmus maximus). Front. Microbiol. 13:856802. doi: 10.3389/fmicb.2022.856802
Received: 17 January 2022; Accepted: 18 February 2022;
Published: 18 April 2022.
Edited by:
Spiros Paramithiotis, Agricultural University of Athens, GreeceReviewed by:
Junjie Zhang, Zhengzhou University of Light Industry, ChinaCopyright © 2022 Wang, Cui, Ren, Tan, Lv, Li, Li and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jianrong Li, bGlqcjY0OTFAMTYzLmNvbQ==; Tingting Li, NjQ4NTgyNjc3QHFxLmNvbQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.