
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 14 March 2022
Sec. Terrestrial Microbiology
Volume 13 - 2022 | https://doi.org/10.3389/fmicb.2022.848750
This article is part of the Research Topic Rhizosphere Microbiology: Toward a Clean and Healthy Soil Environment View all 25 articles
 Tingting Wang1
Tingting Wang1 Silu Sun1
Silu Sun1 Yanxing Xu1
Yanxing Xu1 Michael Gatheru Waigi1
Michael Gatheru Waigi1 Emmanuel Stephen Odinga1
Emmanuel Stephen Odinga1 Galina K. Vasilyeva2
Galina K. Vasilyeva2 Yanzheng Gao1
Yanzheng Gao1 Xiaojie Hu1*
Xiaojie Hu1*The increasing antibiotic resistance genes (ARGs) in fertilizer-amended soils can potentially enter food chains through their transfer in a soil–vegetable system, thus, posing threats to human health. As nitrogen is an essential nutrient in agricultural production, the effect of nitrogen (in the forms NH4+-N and NO3−-N) on the distribution of ARGs (blaTEM-1, sul1, cmlA, str, and tetO) and a mobile genetic element (MGE; tnpA-4) in a soil–Chinese cabbage system was investigated. Not all the tested genes could transfer from soil to vegetable. For transferable ones (blaTEM-1, sul1, and tnpA-4), nitrogen application influenced their abundances in soil and vegetable but did not impact their distribution patterns (i.e., preference to either leaf or root tissues). For ARGs in soil, effects of nitrogen on their abundances varied over time, and the positive effect of NH4+-N was more significant than that of NO3−-N. The ARG accumulation to vegetables was affected by nitrogen application, and the nitrogen form was no longer a key influencing factor. In most cases, ARGs were found to prefer being enriched in roots, and nitrogen application may slightly affect their migration from root to leaf. The calculated estimated human intake values indicated that both children and adults could intake 106–107 copies of ARGs per day from Chinese cabbage consumption, and nitrogen application affected ARG intake to varying degrees. These results provided a new understanding of ARG distribution in vegetables under the agronomic measures such as nitrogen application, which may offer knowledge for healthy vegetable cultivation in future.
Antibiotics play a vital role in protecting human health and promoting the development of animal husbandry. However, the overuse and abuse of antibiotics have caused increases in resistant bacteria and antibiotic resistance genes (ARGs) in the environments (Zhu et al., 2017a; Guo et al., 2018; Zeng et al., 2019; Sun et al., 2020a; Yue et al., 2021). ARGs have been widely regarded as an emerging contaminant (Pruden et al., 2006), which would remain, transfer, and spread in different environmental media. Soil is known as an ARG reservoir, and ARGs in soil have been found to be transferred to vegetable endophytes (Yang et al., 2018), which might then enter into the human body during vegetable consumption, thus, threatening human health (Zhu et al., 2017b).
It is worth noting that some agricultural management measures could regulate ARG distribution in a soil–vegetable system. Under the application of organic fertilizers (such as manure), ARGs in the soil and the fertilizers would migrate into the plant, thus facilitating the ARG contamination (He et al., 2016). For example, Zhou et al. (2019) reported that the total ARG abundances in phyllosphere of rice was enhanced by ~20% and ~40% after a short term (within 1 month) of fertilization of pig manure collected from farms using reduced and standard antibiotic practices, respectively. Chen et al. (2018) discovered that a long term (over 10 years) of chicken manure application to soil caused a 2,638-fold ARG enrichment in the phyllosphere of maize. For inorganic fertilizers, even though they do not contain exogenous ARGs as the organic fertilizers, they could still alter ARG abundance in soil through changing the bacterial community composition (Forsberg et al., 2014). It was reported that addition of 100–200 mg kg−1 nitrogen fertilizer (ammonium and nitrate nitrogen) significantly increased the relative abundance of ARGs in soil (Sun et al., 2020b). As one of the typical inorganic fertilizers, nitrogen is an essential nutrient for plant growth, and applied widely and heavily in agricultural production (74 kg year−1 hectare−1 in the world; Cui et al., 2018). However, until now, studies have merely focused on their regulation on ARGs in the soil, and instead ignored their influence on the distribution and abundances of ARGs in soil–vegetable system.
The transfer of ARGs in soil–vegetable systems is a complex process. Bacteria have been reported could drive the transmission of ARGs from soil to plants via internal pathways (Cerqueira et al., 2019a; Zhang et al., 2019). Numerous evidences manifest that the rhizosphere is a hotspot for the spatial transmission of antibiotic resistant bacteria (ARB) from soil to vegetable and also for the horizontal transfer of ARGs from soil microbiome to vegetable endophytes (Wang et al., 2015; Yang et al., 2018). For the spatial transmission of ARB, the bacterium in the soil could secrete cell wall degrading enzymes to break through root barrier and enter the plant. They may also enter plant through natural wounds on the root surface or following the root infection (Chi et al., 2005; Liu et al., 2017; Wang et al., 2021). For gene exchanges between soil and endophytic bacteria, ARGs on mobile genetic elements (MGEs) may transfer inside of plant when soil bacteria and vegetable endophytes closely contact with each other in the rhizosphere (Barlow, 2009).
To this end, this study examined the impact of nitrogen (NH4+-N and NO3−-N) on the distribution of ARGs in the soil–Chinese cabbage (Brassica chinensis L.) system. The ARG abundances in soil, vegetable roots and vegetable leaves were determined by real-time quantitative polymerase chain reaction (RT-qPCR). Bioaccumulation factor (BAF) and transfer factor (TF) was determined to examine the enrichment and transferable ability of ARGs in Chinese cabbage. We further estimated the daily intake of ARGs (EIARGs) to determine the exposure of ARGs to human after consumption of Chinese cabbage. This study may provide new insight to the distribution of ARGs in the soil–vegetable system under nitrogen application and improve our understanding on antibiotic resistance risks in agricultural production.
Soil used in the pot experiment was collected from a dairy farm (31°53′6′′N, 119°15′30′′E) in Jurong, Jiangsu Province, China. The soil is a typical yellow-brown earth with the pH value of 6.6 and the moisture content of 27.5%, which contains 21.8 g kg−1 of organic matter. Other physicochemical properties of the soil were described in Supplementary Table S1. The soil samples were used for pot experiments after passing through a 2-mm sieve. Seven treatments were set up according to the nitrogen application level in the pots: no nitrogen (CK), 25 mg kg−1 ammonium nitrogen (LA), 100 mg kg−1 ammonium nitrogen (MA), 200 mg kg−1 ammonium nitrogen (HA), 25 mg kg−1 nitrate nitrogen (LN), 100 mg kg−1 nitrate nitrogen (MN), and 200 mg kg−1 nitrate nitrogen (HN) were added to the potting soil, respectively. The NH4Cl and NaNO3 were applied representing the ammonium nitrogen and nitrate nitrogen, respectively. Chinese cabbage seeds were soaked overnight and planted in the soil mixed with amendments. Deionization water was sprayed into the soil to keep the soil water content at 70% of the field water holding capacity. The potted vegetables were managed in the greenhouse and the soil water content was controlled by daily watering. The pots were arranged in the greenhouse with a temperature of 25 ± 2°C in daylight (12 h) and 15 ± 2°C at night (12 h) and humidity of 60 ± 5%. All treatments were conducted in triplicate. Photos of vegetable growth at 15 days (seedling period), 35 days (growing period), and 60 days (maturing period) under different treatments were shown in Supplementary Figure S1.
Soil was sampled at 15, 35, and 60 days. A medicine spoon sterilized with 70% ethanol was used to collect the soil from each pot. The soil samples were stored at −80°C before the procedures of DNA extraction and chemical analysis. The vegetable samples were harvested at 15, 35, and 60 days, washed with running sterile water to remove their surface attaching soil and debris, and then divided into roots and leaves. All vegetable samples were stored at −80°C.
For soil samples, the DNA was extracted by a DNeasy Power Soil kit (Qiagen, Germany). For vegetable samples, DNA was extracted according to the method in a previous study (Chen et al., 2018), with some modifications. Around 0.5 g of the surface-disinfected vegetable sample was put into a sterile mortar. After that, the liquid nitrogen was added to the mortar, followed by an immediate trituration to vegetable samples. Then, the sample was suspended with 10 ml of sterile phosphate buffered saline (PBS). The DNA was extracted from the sample suspensions by applying the FastDNA Spin Kit for Soil (MP Biomedicals, LLC, Solon, OH, United States). The concentrations of DNA extracted from soil and vegetables were measured by Nanodrop 2000 spectrophotometer (Thermo, Waltham, MA, United States).
Five ARGs and a MGE were selected as the target genes: β-lactam resistance gene (blaTEM-1), chloramphenicol resistance gene (cmlA), aminoglycoside resistance gene (str), sulfonamide resistance gene (sul1), tetracycline resistance gene (tetO), and IS6 transposase (tnpA-4). The six genes had relatively higher abundances in the collected soil samples (according to the pre-test study) and were in frequent detection in the farmland soil as reported in previous studies (Knapp et al., 2010; Luo et al., 2010; Chen et al., 2019; Sun et al., 2019). Their background abundances in soil samples were shown in Supplementary Table S2. The abundance of 16S rRNA gene was also determined to quantitatively estimate the abundance of bacteria. The ARG abundances in soil and vegetable samples were determined through QuantStudio 3 Real-Time PCR System with AceQ qPCR SYBR Green Master Mix (Applied Biosystems, Foster City, CA, United States). The reaction mixture (20 μl) consisted of 10 μl of SYBR Green Master Mix (Vazyme, Nanjing, China), 0.8 μl of each primer at 10 mM (Supplementary Table S3), 1 μl of DNA template, and 7.4 μl of sterile double-distilled water (Supplementary Table S4). The reaction conditions are described in Supplementary Table S5.
Bioaccumulation factor (BAF) was used to quantify the ARG enrichment capacity of roots and leaves of Chinese cabbage. Transfer factor (TF) was used to examine the transfer ability of ARGs from Chinese cabbage roots to leaves.
where RAcabbage is the relative abundance of ARGs in Chinese cabbage roots or leaves, RAleaf is the relative abundance of ARGs in Chinese cabbage leaves, RAroot is the relative abundance of ARGs in Chinese cabbage roots, RAsoil is the relative abundance of ARGs in soil.
The estimated daily intake of ARGs (EIARGs) was used to determine the copy number of ARGs consumed by eating Chinese cabbage. The daily consumption of vegetables is about 276 and 228 g (of which 15% were Chinese cabbage) for adult and child, respectively (Wang et al., 2018; Mei et al., 2021).
where GCN/g is gene copy number of ARGs per gram of Chinese cabbage, Wintake is the weight of vegetable daily intake, Rcabbage is the rate of Chinese cabbage in raw vegetables.
The quantitative data of soil and vegetable ARGs were exported by QuantStudio Design & Analysis Software. Statistical analyses were performed using Microsoft Excel 2019 and IBM SPSS statistics 25. Differences in the gene abundances were assessed using Duncan multiple comparisons (p < 0.05). Data visualization were performed using Origin 2019.
All the tested genes including the five ARGs (blaTEM-1, cmlA, str, sul1, and tetO) and the MGE (tnpA-4) could be detected in the soil. Among them, only two types of ARGs (blaTEM-1 and sul1) and the tnpA-4 gene could be detected in Chinese cabbage (Figure 1), whose abundances in soil were relatively higher. This suggests that some ARGs in soil could not be transferred into vegetables. A previous study also found that only 60% of ARGs in soil were plant-transferable, and the ARG abundances decreased gradually from soil to plant (Cerqueira et al., 2019b; Xiao et al., 2021). ARGs in soil could transfer to vegetable through two major ways and the rhizosphere is a hotspot for their transfer (Van Elsas et al., 2003; Wang et al., 2015; Yang et al., 2018). Through rhizosphere some soil bacteria that carrying ARGs (i.e., ARB) could get into the vegetable and thus bring the ARGs from soil to the inside of vegetables. After entering vegetables, some ARB could grow well and the abundance of their carrying ARGs would thus increase. However, some other ARB cannot compete with the native plant endophytes and their growth would be largely suppressed, which might thus inhibit the propagation of their harboring ARGs (Xu et al., 2021). This might be a reason for the unsuccessful transfer of some ARGs (e.g., cmlA, str, and tetO in this study). In addition, rhizosphere is also regarded as a hotspot for gene exchange between the soil and plant microbiome (Chen et al., 2017). That is, soil bacteria harbored ARGs could transfer to vegetable endophytes by horizontal gene transfer (HGT) in the vegetable rhizosphere. In this study, the tnpA-4 gene (a MGE) had high abundances in both soil and vegetable samples, indicating that HGT also contributed to the transmission of ARGs from soil to vegetables (Heuer et al., 2011).
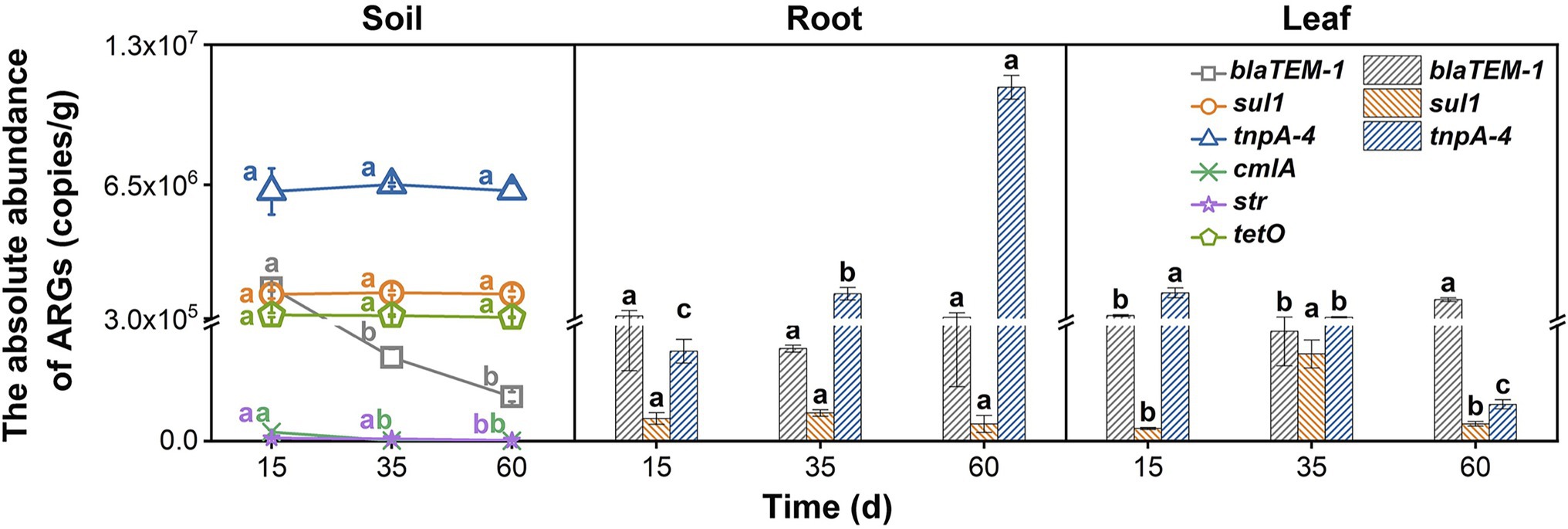
Figure 1. The absolute abundance of ARGs in the soil, roots, and leaves at three growth periods (15, 35, and 60 days) with no nitrogen application (CK). Error bars represent standard deviations of triplicate samples. Different lowercase letters above the bars indicated significant differences among growth periods at p < 0.05.
Nitrogen application greatly influenced the abundances of ARGs and the MGE in soil. But the influencing effect varied over time. For sul1, at 35 days, nitrogen obviously inhibited its abundance, which was only 53%–84% of that under the control treatment. However, at 60 days, the abundances of sul1 with nitrogen application (particularly NH4+-N) was increased 16%–77% than the control treatment (Figure 2). On the contrary, for tnpA-4, its abundance reached the highest under MA, HA, LN, and MN treatments at 35 days, which increased by 202%–360% as compared with the control treatment. However, with cultivation time going (at 60 days), tnpA-4 abundances in these treatments significantly decreased to the level similar to the control treatment (Figure 2). Previous study reported that microbial community in soil would give a quick response to the change of their growing environment (such as the change of nitrogen concentration; Geisseler and Scow, 2014), thus resulting in a better or worse growth of some population. But this alteration of microbial community was not stable at this stage (e.g., <40 days; Stark et al., 2007; Fierer et al., 2012). A period of 60 days was sufficient to stabilize the soil microbial community structure (Stark et al., 2007). This may offer an explanation for the changing influencing effect over time, and also illustrated that the results at 60 days might obtain more reference values in the practical agricultural activities. Furthermore, we particularly compared the differences in the effects on ARG abundances in soil between the NH4+-N and NO3−-N at 60 days. Obviously, NH4+-N had a more significant positive effect on soil ARGs than NO3−-N (Figure 2). It is known that NH4+-N is typically used as a primary nitrogen source for soil bacteria (Rahman et al., 2018). Therefore, soil bacteria including those with abundances positively correlated to the ARG abundances (e.g., Nitrosospira and Nitrosomonas), usually grow better under the NH4+-N exposure than under that of NO3−-N (Sun et al., 2020b, 2021). The better propagation of these soil bacteria would correspondingly increase the abundances of associated ARGs. Therefore, the promoting effect of NH4+-N on ARG abundance was stronger than NO3−-N.
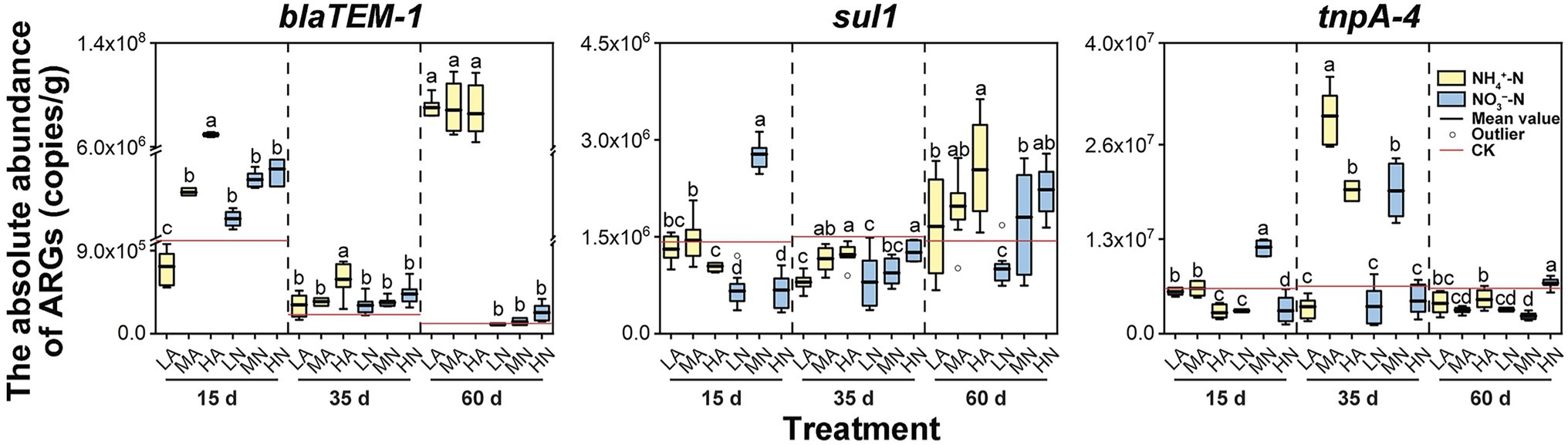
Figure 2. The absolute abundance of ARGs (blaTEM-1, sull, and tnpA-4) in the soil at three periods (15, 35, and 60 days) with nitrogen treatments: 25 mg kg−1 NH4+-N (LA), 100 mg kg−1 NH4+-N (MA), 200 mg kg−1 NH4+-N (HA), 25 mg kg−1 NO3−-N (LN), 100 mg kg−1 NO3−-N (MN), and 200 mg kg−1 NO3−-N (HN). Errors bars represent standard deviations of triplicate samples. Different lowercase letters above indicated significant among treatments at p < 0.05.
Nitrogen application also impact the ARG and MGE accumulation to vegetables. In the early growing stage of Chinese cabbage (15 days), application of NH4+-N and NO3−-N generally decreased blaTEM-1 abundance but increased sul1 and tnpA-4 abundances in roots as compared to the control treatment. However, with Chinese cabbage growing (60 day), in general, the nitrogen treatment enhanced the accumulation of blaTEM-1 and sul1 to roots but inhibited that of tnpA-4. As shown in Figure 3, blaTEM-1 abundances increased from 15 to 60 days under NH4+-N and NO3−-N treatments. But under control treatment, the blaTEM-1 abundances decreased by 16% from 15 to 60 days. On the contrary, for tnpA-4, although at 15 days their abundances in nitrogen treatments were higher than that in control group, with vegetable growing to 60 days, the abundance in control group turned to be higher than those under the nitrogen treatments, suggesting tnpA-4 accumulation in roots were inhibited by nitrogen. This trend showed a similar pattern to that of the abundances of blaTEM-1, sul1 and tnpA-4 in soil under the nitrogen application (i.e., blaTEM-1 and sul1 abundances were increased and tnpA abundance was decreased at 60 days; Figure 2). Cerqueira et al. (2019a) also discovered that the abundance of ARGs in the roots of legume plants was highly associated with that in the soil. For ARGs in leaves, as compared the ARG abundances at 15 days with those at 60 days, generally, nitrogen application promoted blaTEM-1 accumulation, and had no obvious effect on sul1 and tnpA-4 accumulation. For example, under the control treatment, the abundance of blaTEM-1 at 15 days in the leaves was very closed to that at 60 days, which means the accumulation of this gene to the vegetable leave is limited. However, under the treatment of nitrogen, the abundance of blaTEM-1 increased up to 80.44-fold higher from 15 to 60 days, indicating a stronger accumulation of this gene into Chinese cabbage leaves under nitrogen application. Similar to that on soil ARGs, we also compared the differences in the influencing degrees of NH4+-N and NO3−-N on ARG abundances in vegetables at 60 days. It was found that the differences in the regulating effects between NH4+-N and NO3−-N appeared lack of regularity, suggesting the nitrogen forms were no longer a crucial influencing factor on ARG abundances in vegetables. It is known that most vegetables prefer to utilize NO3−-N than NH4+-N (Zhang et al., 2016) and thus would adsorb more NO3−-N from soil compared to NH4+-N. The higher level of NO3−-N in vegetable might offset the stronger regulation of NH4+-N on the soil-derived ARB inside the Chinese cabbage, which might make the nitrogen forms no longer a crucial influencing factor.
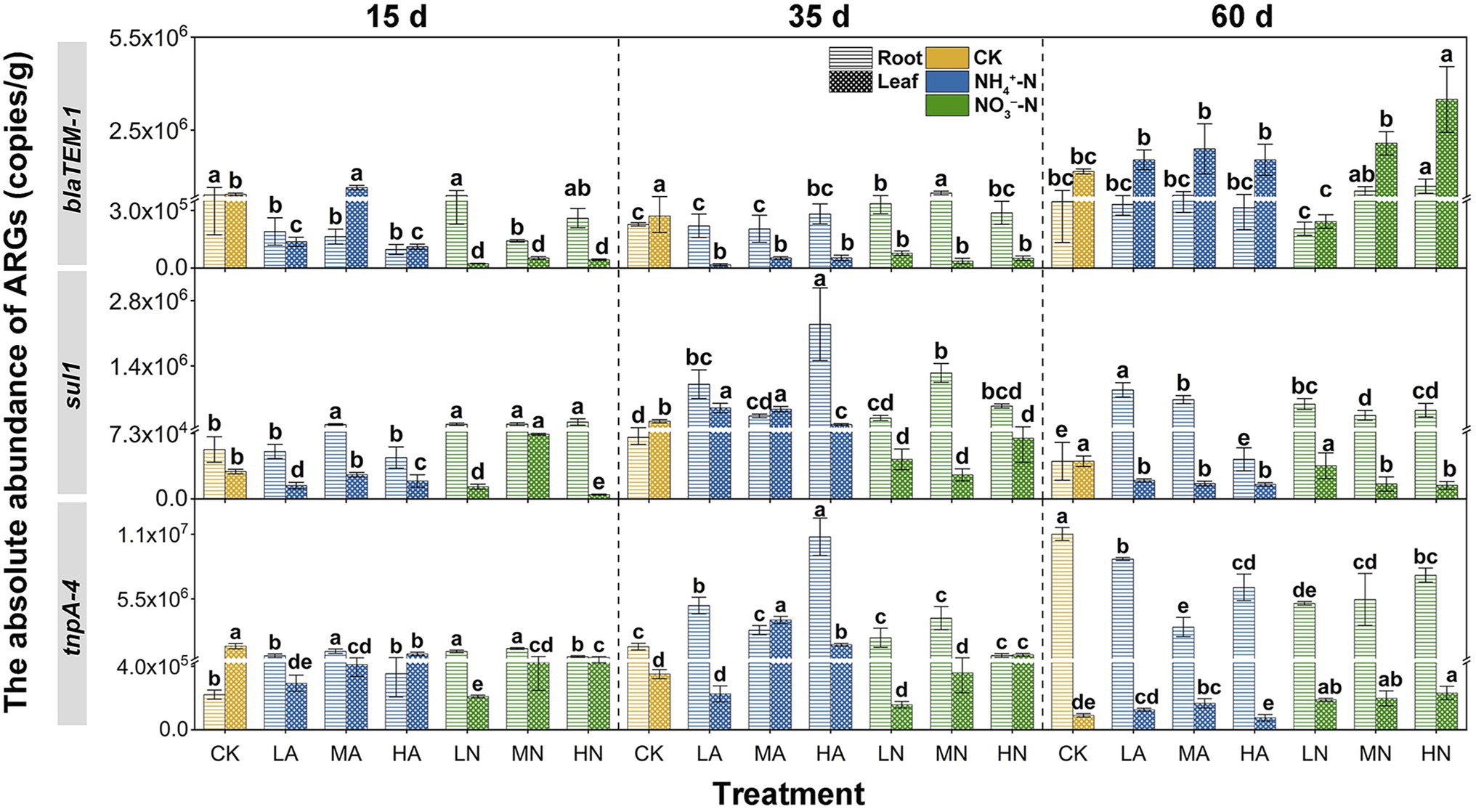
Figure 3. The absolute abundance of ARGs (blaTEM-1, sull, and tnpA-4) in Chinese cabbage (roots and leaves) at three periods (15, 35, and 60 days) under different treatments: no nitrogen (CK), 25 mg kg−1 NH4+-N (LA), 100 mg kg−1 NH4+-N (MA), 200 mg kg−1 NH4+-N (HA), 25 mg kg−1 NO3−-N (LN), 100 mg kg−1 NO3−-N (MN), and 200 mg kg−1 (HN). Errors bars represent standard deviations of triplicate samples. Different lowercase letters above the bars indicated significant differences among treatments at p < 0.05.
Overall, the bacterial community composition and HGT majorly control the ARG distribution in various environmental niches (Barlow, 2009; Forsberg et al., 2014). According to a previous study, nitrogen application could alter the bacterial community structure (Fierer et al., 2012), which might be a mechanism accounting for the regulation of nitrogen on ARGs in soil and vegetable in this study. Besides that, the MGE tnpA-4 abundances were also influenced by nitrogen application to certain extents, implying the HGT process affected by nitrogen application should be another mechanism.
Although nitrogen application influenced the ARG and MGE abundances in vegetables to various degrees, it could not influence their distribution patterns (i.e., preference to either leaf or root tissues) in Chinese cabbage. As shown in Figure 1, different ARGs had different distribution patterns in vegetables under control treatment. For example, as the vegetables grew by 60 days, a large amount of blaTEM-1 accumulated in the leaves, and its abundance was 3.38 times higher than that in the roots. But for tnpA-4 and sul1, they were enriched in the roots. Particularly for tnpA-4, its abundance in roots was even 121.87 times higher than that in leaves. It is reported that the reason for the accumulation of tnpA-4 in roots might be that this gene normally acts as a MGE (Cerqueira et al., 2019b), and the rhizosphere is the hotspot of HGT (Zhang et al., 2019). Previous studies have shown that the selective enrichment of foreign ARB in plant tissues may be the cause of different preferences of ARGs. For example, Bacteroidetes in rhizosphere could migrate to plant roots but rarely could further migrate to the leaves (Guo et al., 2021). Interestingly, after nitrogen application, be it in NH4+-N or NO3−-N forms, or even at low or high levels, could not alter the distribution patterns of these ARGs. As shown in Figure 3, for the LA, MA, HA, LN, MN, and HN treatments, blaTEM-1 always preferred being in the leaves than in the roots. Previous studies also indicated a similar preference for blaTEM-1 in plant leaves (Cerqueira et al., 2019a). TnpA-4 and sul1 were still enriched in vegetable roots (Figure 3). For each ARG, their endophyte host commonly has a relatively fixed ecological niche (Karmakar et al., 2019), which might not be easily changed by external factors such as nitrogen application.
In our study, BAF was analyzed to determine the ARG enrichment capacity in different vegetable parts. The TF was also calculated to examine the transfer capacity of ARGs from roots to leaves. In Figure 4A, the blue colors in the root column were darker than those in the leaf column, illustrating the ARG enrichment capacities in roots were higher than those in leaves for both the control and nitrogen treatments (except for blaTEM-1 gene at 60 days). This was in accordance with previous studies reported that rhizosphere is the main point of ARG migration from soil to plant (Van Elsas et al., 2003). Microbes played as an important carrier for ARGs to migrate into plants (Cordovez et al., 2019). They first move along the rhizosphere to the root surface and then colonize inside the root (Edwards et al., 2015). Therefore, root plays an indispensable role in the transfer of ARGs in the soil–vegetable system. However, for the blaTEM-1 gene at 60 days, which was a special case, their enrichment capacities in leaves were higher than roots. As shown in Figure 4B, without or with nitrogen application (except for HA), the TFs of blaTEM-1 gene at 60 days were significantly higher than those of sul1 and tnpA-4. For example, for the control treatment, the TF of the blaTEM-1 gene was close to 6.70, much higher than those for sul1 and tnpA-4. This indicated that blaTEM-1 had a higher transfer capacity (i.e., TFs) in Chinese cabbage, which made them easier to accumulate in leaves. Moreover, we found nitrogen application influenced the ARG transfer capacity in vegetable, but the influencing effect lacked regularity. For instance, the transportability of blaTEM-1 was enhanced in LA and HN treatments, but those of sul1 were enhanced in HA treatment, while tnpA-4 was in MA and HN treatments. The influencing effect might not be intense, as the distribution patterns of ARGs in vegetables could not be altered by nitrogen application as described above.
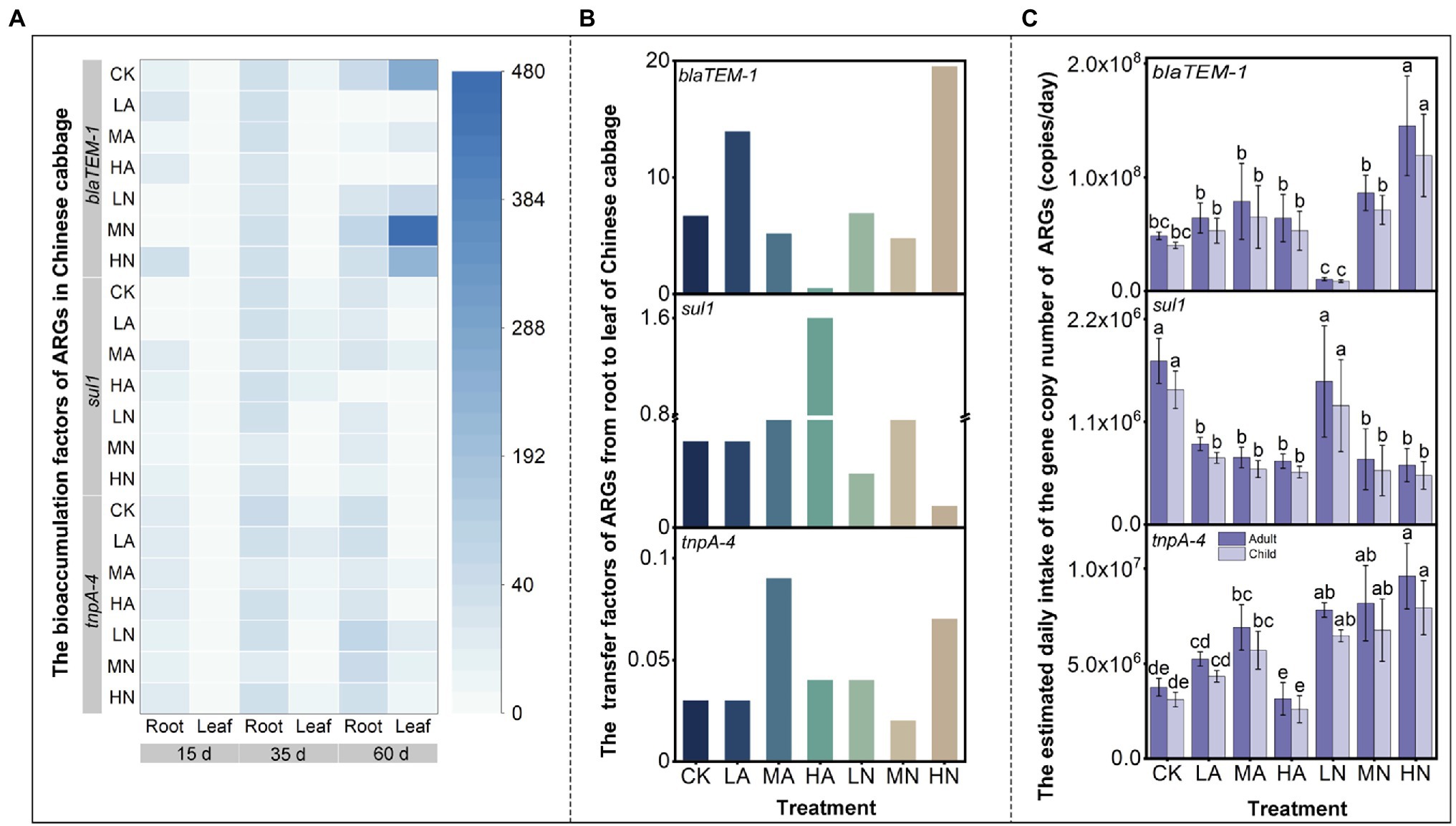
Figure 4. (A) Heatmap shows the bioaccumulation factors (BAF) of ARGs in the roots and leaves of Chinese cabbage. The color of white to blue represents the BAF values of the corresponding ARGs in different treatments and times. (B) The transfer factors (TF) of ARGs from roots to leaves at 60 days. (C) The estimated daily intake (EI) of ARGs by eating matured Chinese cabbage leaves (at 60 days). Different lowercase letters above the bars indicated significant differences among treatments at p < 0.05.
We calculated the estimated daily intakes (EIs) of ARGs from the consumption of Chinese cabbage based on the study of Mei et al. (2021). The study investigated the ARG intake from eating the raw carrots, and reported that children and adults would intake 2.7 × 107 and 3.2 × 107 copies of ARGs per day, respectively. In this study, it was calculated that children and adults may intake up to 3.99 × 107 and 4.83 × 107 copies of blaTEM-1, 3.09 × 106 and 3.74 × 106 copies of tnpA-4, and 1.44 × 106 and 1.75 × 106 copies of sul1 per day by consuming Chinese cabbage leaves under control treatment, respectively. The highest EI values of blaTEM-1 may be attributed to the highest transfer capacity of this gene from root to leaf in Chinese cabbage (Figure 4B). Until now, there was no solid evidence demonstrating that consuming foods harboring ARGs would lead to an increase of ARGs in human body. However, HGT is recognized as a common event in human oral bacteria (Mira, 2008), and plasmids (a common vector of ARGs) was proven could get from mouth to gut of mammalian and persisted in gut for hours (Wilcks et al., 2004). These potentially indicated that vegetable-carrying ARGs particularly for those on MGEs had a probability to enter human body, which might transfer to the pathogens and thus threatened human health. Martínez et al. (2015) also pointed out that ARGs would pose a substantial risk for the dissemination of resistance if they reside on MGEs. In this study, we found NO3−-N application increased EI tnpA-4 (i.e., the MGE) by 110%–160% (Figure 4C), which might increase the potential of ARG transfer from vegetables to human pathogens, enlarging health risks. Besides tnpA-4, application of NH4+-N increased EIblaTEM-1 by 30%–60% (Figure 4C) but reduced EIsul1 by 50%–60% (Figure 4C). This manifests that nitrogen application may differentially affect the intake of ARGs in human. Therefore, in future agricultural practice, it is suggested that nitrogen application should carefully refer to the ARG types in soil.
The results in this study provided a new understanding on ARG distribution in soil-vegetable system under the agronomic measures of nitrogen application. For those genes that could transfer from soil to vegetables (blaTEM-1, sul1, and tnpA-4), nitrogen application could not change their distribution patterns in vegetables but could regulate their abundance. Under most circumstances, ARGs preferred being enriched in the roots of Chinese cabbage. When human consumed the Chinese cabbage leaves, the effect of nitrogen application on ARG intake varies with ARG types. These results are helpful in understanding the distribution and transfer of ARGs in the soil-vegetable system and provide data support for the health risk assessment of ARGs in future.
The original contributions presented in the study are included in the article/Supplementary Material, and further inquiries can be directed to the corresponding author.
TW, SS, YG, and XH designed the study. TW and SS conducted the experiments and drafted the manuscript. YX, MW, EO, GV, and XH proofread the manuscript. All authors contributed to the article and approved the submitted version.
This work was funded by the National Natural Science Foundation of China (42107024 and 41925029), the Natural Science Foundation of Jiangsu Province of China (BK20210413), and the National Science Foundation for Postdoctoral Scientists of China (2021M691609).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.848750/full#supplementary-material
Barlow, M. (2009). What antimicrobial resistance has taught us about horizontal gene transfer. Methods Mol. Biol. 532, 397–411. doi: 10.1007/978-1-60327-853-9_23
Cerqueira, F., Matamoros, V., Bayona, J. M., Berendonk, T. U., Elsinga, G., Hornstra, L. M., et al. (2019b). Antibiotic resistance gene distribution in agricultural fields and crops: a soil-to-food analysis. Environ. Res. 177:108608. doi: 10.1016/j.envres.2019.108608
Cerqueira, F., Matamoros, V., Bayona, J., Elsinga, G., Hornstra, L. M., and Piña, B. (2019a). Distribution of antibiotic resistance genes in soils and crops. A field study in legume plants (Vicia faba L.) grown under different watering regimes. Environ. Res. 170, 16–25. doi: 10.1016/j.envres.2018.12.007
Chen, Q., An, X., Zheng, B., Ma, Y., and Su, J. (2018). Long-term organic fertilization increased antibiotic resistome in phyllosphere of maize. Sci. Total Environ. 645, 1230–1237. doi: 10.1016/j.scitotenv.2018.07.260
Chen, Q. L., An, X. L., Zhu, Y. G., Su, J. Q., Gillings, M. R., Ye, Z. L., et al. (2017). Application of struvite alters the antibiotic resistome in soil, rhizosphere, and phyllosphere. Environ. Sci. Technol. 51, 8149–8157. doi: 10.1021/acs.est.7b01420
Chen, Q. L., Cui, H. L., Su, J. Q., Penuelas, J., and Zhu, Y. G. (2019). Antibiotic resistomes in plant microbiomes. Trends Plant Sci. 24, 530–541. doi: 10.1016/j.tplants.2019.02.010
Chi, F., Shen, S.H., Cheng, H.P., Jing, Y.X., Yanni, Y.G., and Dazzo, F.B. (2005). Ascending migration of endophytic rhizobia, from roots to leaves, inside rice plants and assessment of benefits to rice growth physiology. Appl. Environ. Microbiol. 71, 7271–7278.2005. doi: 10.1128/AEM.71.11.7271-7278.2005
Cordovez, V., Dini-Andreote, F., Carrión, V. J., and Raaijmakers, J. M. (2019). Ecology and evolution of plant microbiomes. Annu. Rev. Microbiol. 73, 69–88. doi: 10.1146/annurev-micro-090817-062524
Cui, E. P., Gao, F., Liu, Y., Fan, X. Y., Li, Z. Y., Du, Z. J., et al. (2018). Amendment soil with biochar to control antibiotic resistance genes under unconventional water resources irrigation: proceed with caution. Environ. Pollut. 240, 475–484. doi: 10.1016/j.envpol.2018.04.143
Edwards, J., Johnson, C., Santos-Medellín, C., Lurie, E., Podishetty, N. K., Bhatnagar, S., et al. (2015). Structure, variation, and assemblyof the root-associated microbiomes of rice. Proc. Natl. Acad. Sci. U. S. A. 112, E911–E920. doi: 10.1073/pnas.1414592112
Fierer, N., Lauber, C. L., Ramirez, K. S., Zaneveld, J., Bradford, M. A., and Knight, R. (2012). Comparative metagenomic, phylogenetic and physiological analyses of soil microbial communities across nitrogen gradients. ISME J. 6, 1007–1017. doi: 10.1038/ismej.2011.159
Forsberg, K. J., Patel, S., Gibson, M. K., Lauber, C. L., Knight, R., Fierer, N., et al. (2014). Bacterial phylogeny structures soil resistomes across habitats. Nature 509, 612–616. doi: 10.1038/nature13377
Geisseler, D., and Scow, K. M. (2014). Long-term effects of mineral fertilizers on soil microorganisms – a review. Soil Biol. Biochem. 75, 54–63. doi: 10.1016/j.soilbio.2014.03.023
Guo, Y., Qiu, T., Gao, M., Sun, Y., Cheng, S., Gao, H., et al. (2021). Diversity and abundance of antibiotic resistance genes in rhizosphere soil and endophytes of leafy vegetables: focusing on the effect of the vegetable species. J. Hazard. Mater. 415:125595. doi: 10.1016/j.jhazmat.2021.125595
Guo, X., Stedtfeld, R. D., Hedman, H., Eisenberg, J. N. S., Trueba, G., Yin, D., et al. (2018). Antibiotic resistome associated with small-scale poultry production in rural Ecuador. Environ. Sci. Technol. 52, 8165–8172. doi: 10.1021/acs.est.8b01667
He, L., Ying, G., Liu, Y., Su, H., Chen, J., Liu, S., et al. (2016). Discharge of swine wastes risks water quality and food safety: antibiotics and antibiotic resistance genes from swine sources to the receiving environments. Environ. Int. 92-93, 210–219. doi: 10.1016/j.envint.2016.03.023
Heuer, H., Schmitt, H., and Smalla, K. (2011). Antibiotic resistance gene spread due to manure application on agricultural fields. Curr. Opin. Microbiol. 14, 236–243. doi: 10.1016/j.mib.2011.04.009
Karmakar, R., Bindiya, S., and Hariprasad, P. (2019). Convergent evolution in bacteria from multiple origins under antibiotic and heavy metal stress, and endophytic conditions of host plant. Sci. Total Environ. 650, 858–867. doi: 10.1016/j.scitotenv.2018.09.078
Knapp, C. W., Dolfing, J., Ehlert, P. A. I., and Graham, D. W. (2010). Evidence of increasing antibiotic resistance gene abundances in archived soils since 1940. Environ. Sci. Technol. 44, 580–587. doi: 10.1021/es901221x
Liu, H., Carvalhais, L. C., Crawford, M., Singh, E., Dennis, P. G., Pieterse, C. M. J., et al. (2017). Inner plant values: diversity, colonization and benefits from endophytic bacteria. Front. Microbiol. 8:2552. doi: 10.3389/fmicb.2017.02552
Luo, Y., Mao, D., Rysz, M., Zhou, Q., Zhang, H., Xu, L., et al. (2010). Trends in antibiotic resistance genes occurrence in the Haihe River, China. Environ. Sci. Technol. 44, 7220–7225. doi: 10.1021/es100233w
Martínez, J. L., Coque, T. M., and Baquero, F. (2015). What is a resistance gene? Ranking risk in resistomes. Nat. Rev. Microbiol. 13, 116–123. doi: 10.1038/nrmicro3399
Mei, Z., Xiang, L., Wang, F., Xu, M., Fu, Y., Wang, Z., et al. (2021). Bioaccumulation of manure-borne antibiotic resistance genes in carrot and its exposure assessment. Environ. Int. 157:106830. doi: 10.1016/j.envint.2021.106830
Mira, A. (2008). “Horizontal gene transfer in oral bacteria,” in Molecular Oral Microbiology. ed. A. H. Rogers (Norfolk: Caister Academic Press), 62–64.
Pruden, A., Pei, R., Storteboom, H., and Carlson, K. H. (2006). Antibiotic resistance genes as emerging contaminants: studies in northern Colorado. Environ. Sci. Technol. 40, 7445–7450. doi: 10.1021/es060413l
Rahman, M. M., Shan, J., Yang, P., Shang, X., Xia, Y., and Yan, X. (2018). Effects of long-term pig manure application on antibiotics, abundance of antibiotic resistance genes (ARGs), anammox and denitrification rates in paddy soils. Environ. Pollut. 240, 368–377. doi: 10.1016/j.envpol.2018.04.135
Stark, C., Condron, L. M., Stewart, A., Di, H. J., and O’Callaghan, M. (2007). Influence of organic and mineral amendments on microbial soil properties and processes. Appl. Soil Ecol. 35, 79–93. doi: 10.1016/j.apsoil.2006.05.001
Sun, Y., Clarke, B., Clarke, J., and Li, X. (2021). Predicting antibiotic resistance gene abundance in activated sludge using shotgun metagenomics and machine learning. Water Res. 202:117384. doi: 10.1016/j.watres.2021.117384
Sun, J., Jin, L., He, T., Wei, Z., Liu, X., Zhu, L., et al. (2020a). Antibiotic resistance genes (ARGs) in agricultural soils from the Yangtze River Delta, China. Sci. Total Environ. 740:140001. doi: 10.1016/j.scitotenv.2020.140001
Sun, S., Lu, C., Liu, J., Williams, M. A., Yang, Z., Gao, Y., et al. (2020b). Antibiotic resistance gene abundance and bacterial community structure in soils altered by ammonium and nitrate concentrations. Soil Biol. Biochem. 149:107965. doi: 10.1016/j.soilbio.2020.107965
Sun, Y., Qiu, T., Gao, M., Shi, M., Zhang, H., and Wang, X. (2019). Inorganic and organic fertilizers application enhanced antibiotic resistome in greenhouse soils growing vegetables. Ecotoxicol. Environ. Saf. 179, 24–30. doi: 10.1016/j.ecoenv.2019.04.039
Van Elsas, J. D., Turner, S., and Bailey, M. J. (2003). Horizontal gene transfer in the phytosphere. New Phytol. 157, 525–537. doi: 10.1046/j.1469-8137.2003.00697.x
Wang, C., Hu, R., Strong, P. J., Zhuang, W., Huang, W., Luo, Z., et al. (2021). Prevalence of antibiotic resistance genes and bacterial pathogens along the soil–mangrove root continuum. J. Hazard. Mater. 408:124985. doi: 10.1016/j.jhazmat.2020.124985
Wang, F. H., Qiao, M., Chen, Z., Su, J. Q., and Zhu, Y. G. (2015). Antibiotic resistance genes in manure-amended soil and vegetables at harvest. J. Hazard. Mater. 299, 215–221. doi: 10.1016/j.jhazmat.2015.05.028
Wang, J., Xia, K., Waigi, M. G., Gao, Y., Odinga, E. S., Ling, W., et al. (2018). Application of biochar to soils may result in plant contamination and human cancer risk due to exposure of polycyclic aromatic hydrocarbons. Environ. Int. 121, 169–177. doi: 10.1016/j.envint.2018.09.010
Wilcks, A., Van Hoek, A. H. A. M., Joosten, R. G., Jacobsen, B. B. L., and Aarts, H. J. M. (2004). Persistence of DNA studied in different ex vivo and in vivo rat models simulating the human gut situation. Food Chem. Toxicol. 42, 493–502. doi: 10.1016/j.fct.2003.10.013
Xiao, Z., Zhang, Y., Li, G., Carter, L. J., Wang, H., Ding, J., et al. (2021). Application of pharmaceutical waste sludge compost alters the antibiotic resistome in soil under the Chinese cabbage system. J. Clean. Prod. 291:125229. doi: 10.1016/j.jclepro.2020.125229
Xu, Y., Li, H., Shao, Z., Li, X., Zheng, X., and Xu, J. (2021). Fate of antibiotic resistance genes in farmland soil applied with three different fertilizers during the growth cycle of pakchoi and after harvesting. J. Environ. Manag. 289:112576. doi: 10.1016/j.jenvman.2021.112576
Yang, L., Liu, W., Zhu, D., Hou, J., Ma, T., Wu, L., et al. (2018). Application of biosolids drives the diversity of antibiotic resistance genes in soil and lettuce at harvest. Soil Biol. Biochem. 122, 131–140. doi: 10.1016/j.soilbio.2018.04.017
Yue, Z., Zhang, J., Zhou, Z., Ding, C., Wan, L., Liu, J., et al. (2021). Pollution characteristics of livestock faeces and the key driver of the spread of antibiotic resistance genes. J. Hazard. Mater. 409:124957. doi: 10.1016/j.jhazmat.2020.124957
Zeng, Q., Sun, J., and Zhu, L. (2019). Occurrence and distribution of antibiotics and resistance genes in greenhouse and open-field agricultural soils in China. Chemosphere 224, 900–909. doi: 10.1016/j.chemosphere.2019.02.167
Zhang, Y. J., Hu, H. W., Chen, Q. L., Singh, B. K., Yan, H., Chen, D., et al. (2019). Transfer of antibiotic resistance from manure-amended soils to vegetable microbiomes. Environ. Int. 130:104912. doi: 10.1016/j.envint.2019.104912
Zhang, J., Wang, J., Müller, C., and Cai, Z. (2016). Ecological and practical significances of crop species preferential N uptake matching with soil N dynamics. Soil Biol. Biochem. 103, 63–70. doi: 10.1016/j.soilbio.2016.08.009
Zhou, S. Y. D., Zhu, D., Giles, M., Yang, X. R., Daniell, T., Neilson, R., et al. (2019). Phyllosphere of staple crops under pig manure fertilization, a reservoir of antibiotic resistance genes. Environ. Pollut. 252, 227–235. doi: 10.1016/j.envpol.2019.05.098
Zhu, B., Chen, Q., Chen, S., and Zhu, Y. (2017b). Does organically produced lettuce harbor higher abundance of antibiotic resistance genes than conventionally produced? Environ. Int. 98, 152–159. doi: 10.1016/j.envint.2016.11.001
Keywords: antibiotic resistance genes, ammonium nitrogen, nitrate nitrogen, soil-vegetable system, transmission
Citation: Wang T, Sun S, Xu Y, Waigi MG, Odinga ES, Vasilyeva GK, Gao Y and Hu X (2022) Nitrogen Regulates the Distribution of Antibiotic Resistance Genes in the Soil–Vegetable System. Front. Microbiol. 13:848750. doi: 10.3389/fmicb.2022.848750
Received: 05 January 2022; Accepted: 31 January 2022;
Published: 14 March 2022.
Edited by:
Bin Ma, Zhejiang University, ChinaReviewed by:
Xiao Liu, Helmholtz Association of German Research Centres (HZ), GermanyCopyright © 2022 Wang, Sun, Xu, Waigi, Odinga, Vasilyeva, Gao and Hu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiaojie Hu, aHV4aWFvamllQG5qYXUuZWR1LmNu
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.