- 1Université de Tours, INRAE, Infectiologie et Santé Publique, BRMF, Tours, France
- 2Service de Bactériologie-Virologie, Centre Hospitalier Régional Universitaire de Tours, Tours, France
Streptococci form a wide group of bacteria and are involved in both human and animal pathologies. Among pathogenic isolates, differences have been highlighted especially concerning their adaptation and virulence profiles. CRISPR-Cas systems have been identified in bacteria and many streptococci harbor one or more systems, particularly subtypes I-C, II-A, and III-A. Since the demonstration that CRISPR-Cas act as an adaptive immune system in Streptococcus thermophilus, a lactic bacteria, the diversity and role of CRISPR-Cas were extended to many germs and functions were enlarged. Among those, the genome editing tool based on the properties of Cas endonucleases is used worldwide, and the recent attribution of the Nobel Prize illustrates the importance of this tool in the scientific world. Another application is CRISPR loci analysis, which allows to easily characterize isolates in order to understand the interactions of bacteria with their environment and visualize species evolution. In this review, we focused on the distribution, diversity and roles of CRISPR-Cas systems in the main pathogenic streptococci.
Introduction
Streptococci are cocci Gram positive bacteria arranged typically in pairs or chains. They form a large group of bacteria, composed of 103 species (Patel and Gupta, 2018). They are widely distributed and are particularly found at the surface of mucous membranes, like upper respiratory, gastro-intestinal, or genito-urinary tracts of humans and many animals. Streptococci are also present in the environment, soil, plants, food or dairy products (Hardie and Whiley, 1997; Kok and Hutkins, 2018). Some Streptococcus species belong to the microbial flora and are considered opportunistic pathogens that can lead to significant health problems in humans or animals (Krzyściak et al., 2013). Streptococcus pyogenes (Cunningham, 2000; González-Abad and Alonso Sanz, 2020) and Streptococcus pneumoniae (Musher, 1992; Weiser et al., 2018) are more particularly involved in human pathologies and can cause invasive infections. Other streptococci belong specifically to animal microbiota, like Streptococcus suis, which is an important cause of various clinical diseases in swine and domestic animals (Staats et al., 1997; Haas and Grenier, 2018). Also, several species are involved in both human and animal pathology, like Streptococcus agalactiae, which represents a major human pathogen causing invasive neonatal infections (Raabe and Shane, 2019), after an initial description in bovine mastitis (Nocard and Mollereau, 1887). More recently, it is also a major pathogen in farm fishes with important economic consequences (Mian et al., 2009; Eto et al., 2020). Finally some streptococci are essential as lactic ferments in agro-food industries (Mercenier et al., 1994; Nagaoka, 2019).
Streptococci were initially classified according to their hemolysis on blood agar which can be complete, incomplete or absent (Schottmuller, 1903). Then Rebecca Lancefield classified beta-hemolytic streptococci based on antigenic properties of the polysaccharide C, a carbohydrate present in the bacterial cell wall (Lancefield, 1933). Many improvements were made in the following years and now, with molecular techniques, like 16S rRNA gene sequence analysis or whole genome sequencing, many more precise classifications have emerged. Nevertheless, despite the number of techniques currently available, none provide a perfect distinction between isolates, especially for clinical isolates belonging to the viridans group. However, in routine laboratory practice, bacterial culture identification is easier and faster since the use of mass spectrometry (Carbonnelle et al., 2011). Matrix Assisted Laser Desorption Ionization Time-Of-Flight (MALDI-TOF) is used daily for bacterial identification and gives reliable results thanks to a relatively rich spectral database which compares the bacterial spectrum obtained for each sample. Despite this powerful technology, some gaps exist. This is the case for the Streptococcus pneumoniae species which is difficult to differentiate from the Streptococcus mitis and Streptococcus oralis group because their spectra are very similar (Marín et al., 2017). Additional tests such as the use of optochin discs are therefore required to complete identification.
Over the past 10 years, CRISPR-Cas systems (Clustered Regularly Interspaced Short Palindromic Repeats and CRISPR—associated proteins) have been described in approximately half of bacterial species, including many streptococcal species, which have been model organisms for their study (Barrangou et al., 2007). Indeed, after several years of research, the immune role of these new systems has been elucidated in Streptococcus thermophilus (Barrangou et al., 2007; Horvath and Barrangou, 2010). The initial discoveries for these systems date from 1987 with the analysis of the iap gene of Escherichia coli (Ishino et al., 1987). They described a region composed of short repeated DNA sequences interspaced with variable sequences. The acronym “CRISPR” was finally approved in 2002 (Jansen et al., 2002). CRISPR loci are composed of short DNA repeats called direct repeats (DR) separated by unique variable DNA sequences called spacers. DR are partially palindromic which give them the ability to form a stable secondary structure in order to interact with other proteins (Kunin et al., 2007; Al-Attar et al., 2011). On the other hand, spacers were suspected to have an extra-chromosomic origin as they share sequence homologies with foreign mobile genetic elements (MGE), such as bacteriophages or plasmids (Bolotin et al., 2005; Mojica et al., 2005; Pourcel et al., 2005). At the 5′ end of the CRISPR locus, the leader is an Adenine and Thymine rich region, which contains the transcription promoter of the CRISPR locus. Generally close to the CRISPR locus, cas operon encodes proteins involved in the system’s functionality. The cas gene composition is different between systems and gives rise to complex classification. Currently, CRISPR-Cas systems are separated into two classes, six types and 33 subtypes (Makarova et al., 2020). Class 1 systems possess effector modules composed of multiple Cas proteins, while class 2 systems carry a single, multidomain crRNA-binding protein (CRISPR-RNA binding protein) like Cas9, the signature protein of type II systems. Indeed, all systems contain two conserved proteins, Cas1 and Cas2, and a signature protein which is specific of a type.
CRISPR-Cas systems operate in three stages, involving different actors depending on the type of system. During the first stage, called adaptation, a small piece of DNA from a foreign MGE is integrated as a new spacer at the leader end of the CRISPR locus. The second stage, called expression, corresponds to the transcription of the whole CRISPR array into a pre-crRNA, which will be processed subsequently, by many actors, in mature crRNA. A crRNA is composed of a spacer and a part of the adjacent direct repeat (Brouns et al., 2008). During the last stage, called interference, crRNA guides Cas nucleases effector complex toward complementary foreign nucleic acids. After specific recognition, which frequently involves the protospacer adjacent motif (PAM; Mojica et al., 2009), Cas nucleases lead to invader DNA degradation (Garneau et al., 2010; Gasiunas et al., 2012; Sternberg et al., 2014). The sequence homology between spacer and MGE allows the system to fight against MGEs already encountered (Garneau et al., 2010), which is a very particular and innovative mechanism. The recent discoveries enabling us to understand these systems’ mechanisms, and particularly their simplicity of action, have inspired researchers to develop new genetic tools. Today, CRISPR-Cas9 (derived from S. pyogenes type II-A system) is internationally used in genetic engineering for genome editing (Jiang et al., 2013). This tool allows to edit genome easily by introducing mutations or deletions in DNA sequences at very specific positions, and thus appears very promising for gene therapy (Hsu et al., 2013; Gori et al., 2015; Broeders et al., 2020) as shown by the recent attribution of the Nobel Prize to Emmanuelle Charpentier and Jennifer Doudna in 2020.
CRISPR-Cas systems were first considered as the adaptive immune system of bacteria (Garneau et al., 2010; Magadán et al., 2012) and in streptococci, S. thermophilus systems were among the best described and studied for many years. This species belongs to the large Salivarius group (Facklam, 2002). It is a thermophilic lactic acid bacteria used in the food industry in combination with Lactobacillus spp. in order to produce dairy products, like cheese or yogurts (Hols et al., 2005). The choice of strains used for dairy product manufacturing is essential as susceptibility to virulent phages can impair milk fermentation and lead to significant economic losses. Dairy industries had to adapt production techniques and select for bacteriophages-insensitive mutants (BIMs). Therefore, various typing methods have been developed to characterize them. Pulse-field gel electrophoresis (PFGE) was the first method used (Boutrou et al., 1995) and then, Random Amplification Polymorphic DNA (RAPD) was set up (Moschetti et al., 1998). More recently, four CRISPR-Cas systems were described in S. thermophilus and their analyses were proposed as a new typing method. CRISPR1 locus (type II-A) is ubiquitous, CRISPR2 (type III-A) and CRISPR3 (type II-A) loci are present in about 40% of strain (Horvath et al., 2008). The last locus, CRISPR4, is a type I-E system and is present in few strains (Horvath and Barrangou, 2010). Only CRISPR1 and CRISPR3 loci seem to be active, as they can acquire new spacers (Barrangou et al., 2013). The main advantage of CRISPR-typing in S. thermophilus is the presence of four different CRISPR loci, which allows better discrimination between isolates (Barrangou and Dudley, 2016). With this technique, producers can have an idea of meetings between phages and bacteria, and adapt strains used for milk fermentation. Other streptococcal species were investigated in order to search for CRISPR loci close to those of S. thermophilus. Results have shown that a homolog of CRISPR3 seems to be present in most streptococci species like S. agalactiae, S. mutans or S. pyogenes, instead of CRISPR1, whose homologs are found in only a few species, like Streptococcus vestibularis and S. suis. Homologs of the CRISPR2 system are more rare (Horvath et al., 2008). Tools have now been developed to find CRISPR loci in isolates and with the whole genome sequencing of bacteria, databases have been constituted to reference known systems (Grissa et al., 2007b; Couvin et al., 2018).
Global analysis of CRISPR-Cas systems, and particularly the spacer content, can thus provide information on species classification but also on their evolution. Previous studies have shown that old spacers, at the trailer end, are conserved between strains whereas new ones, at the leader end, are more variable (Barrangou and van der Oost, 2012). For these reasons, CRISPR loci analysis has been proposed as a promising new typing tool for many bacteria and particularly streptococci (Karimi et al., 2018). It is already used in many species, particularly in Mycobacterium tuberculosis (Brudey et al., 2006; Zhang et al., 2010) or in Yersinia pestis (Pourcel et al., 2005; Cui et al., 2008) and it could be useful for streptococci.
As the Streptococcus genus comprises many species, that could contain one or more CRISPR-Cas systems, the aim of this review was to summarize knowledge about these systems and particularly their roles and applications in the well characterized pathogenic Streptococcus species. The presence and/or the types of CRISPR-Cas systems have been determined using the CRISPRCasfinder database, available at https://crisprcas.i2bc.paris-saclay.fr/ (Grissa et al., 2007a). The database contains more than 26,000 bacterial genomes, including 820 of streptococci, corresponding to 50 different species. Some differences could exist between results obtained with the CRISPRCasfinder database and data in the literature, in the subtypes of CRISPR-Cas systems and their distribution in the population. Indeed, this tool is based on a bioinformatical genome analysis and not on CRISPR-Cas systems functionality. Information concerning the distribution of CRISPR-Cas systems, along with the habitat and the pathogenicity of each Streptococcus species is resumed in Table 1. For more details in this review, we have chosen to focus on human pathogenic streptococci for which CRISPR-Cas systems have been already studied and described in literature (Table 2). This review was organized following the 16S rRNA classification described by Thompson et al. (2013) (Figure 1).
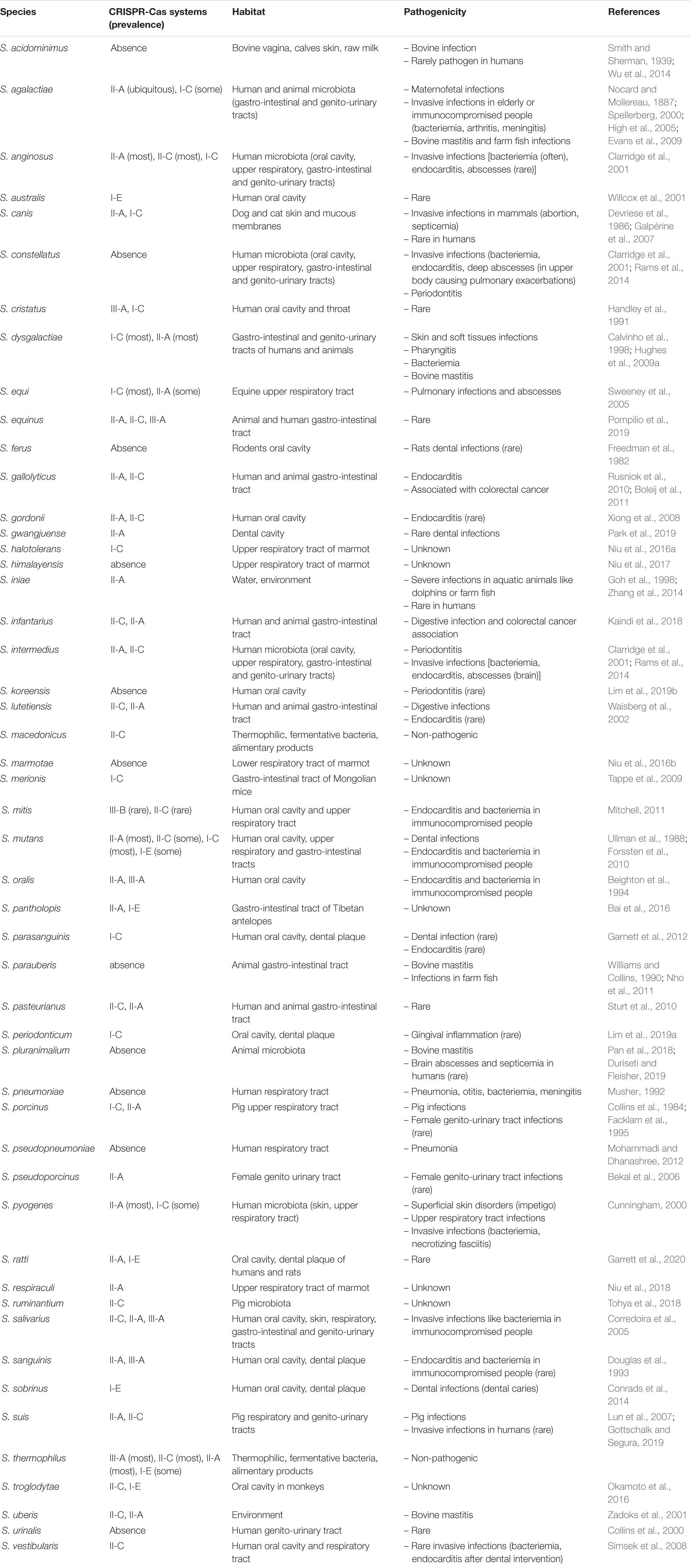
Table 1. Distribution of CRISPR-Cas systems in streptococci, with habitat and pathogenicity of all these species, based on CRISPRCasfinder analysis, available at https://crisprcas.i2bc.paris-saclay.fr/MainDb/StrainList (Grissa et al., 2007a).
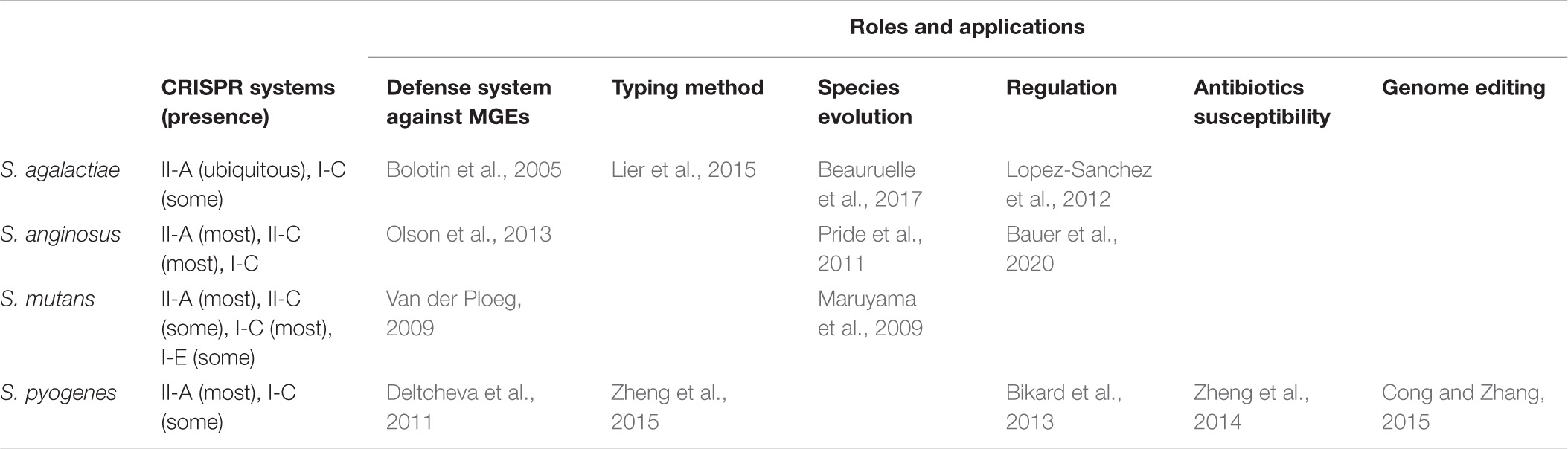
Table 2. Roles and applications derived from CRISPR-Cas systems of pathogenic streptococci, described in literature.
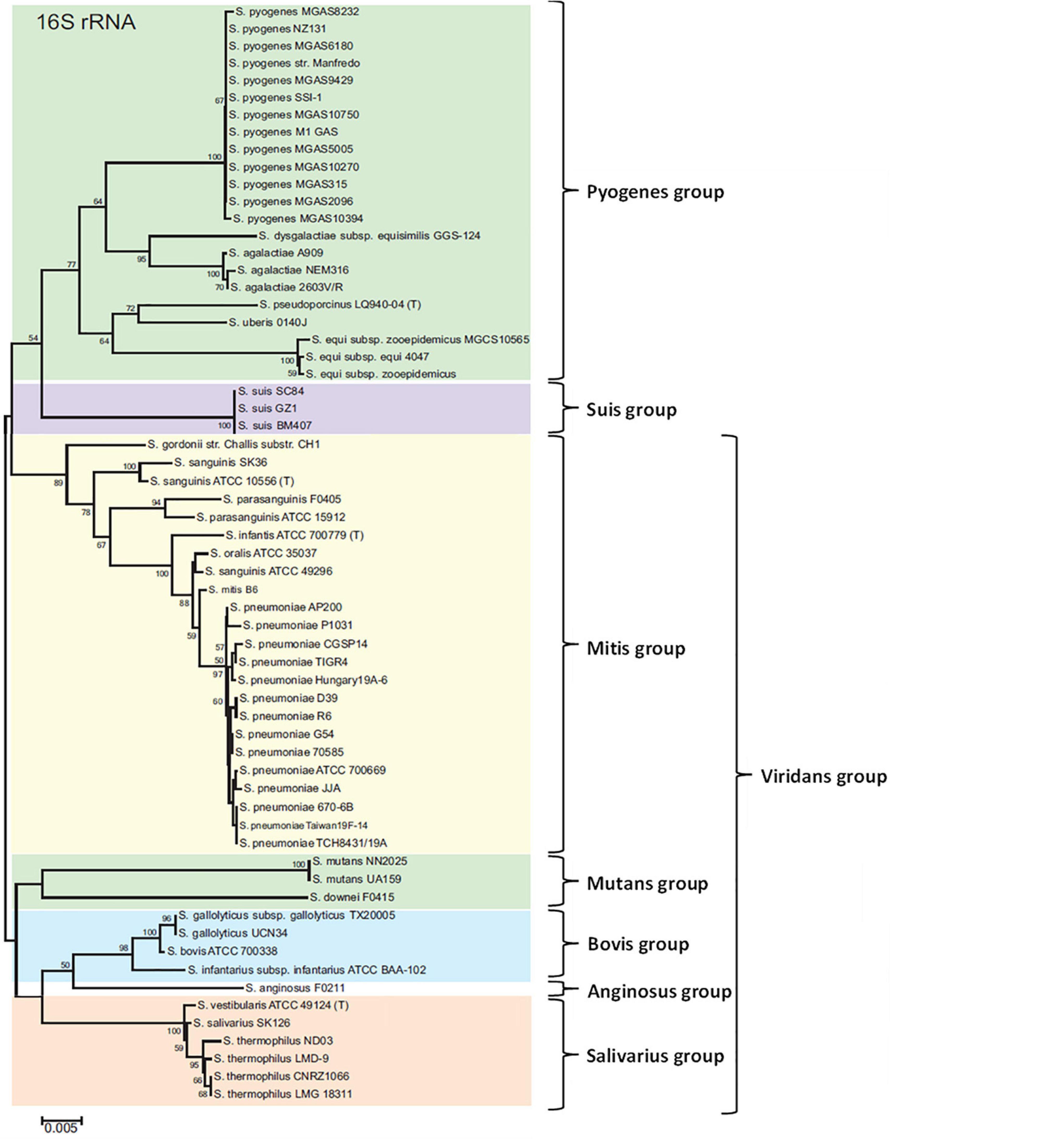
Figure 1. Seven groups formed from a neighbor-joining tree based on 16S rRNA gene sequences of Streptococcus. The numbers at the nodes indicate the values of bootstrap statistics after 2,000 replications, and values below 50% are not shown. Bars at 0.005% estimated sequence divergence. Adapted from Thompson et al. (2013).
Pyogenes Group
Streptococcus agalactiae
Streptococcus agalactiae, or group B Streptococcus, was originally described in bovine mastitis (Nocard and Mollereau, 1887; Wanger and Dunny, 1987). Since the 1960’s, it has appeared in human medicine as a major pathogen in maternofetal infections (Bergqvist and Hurvell, 1970; McCracken, 1973; Raabe and Shane, 2019). This species belongs to the commensal microbiota and colonizes the gastro-intestinal and genito-urinary tracts in about 30% of people (Van der Mee-Marquet et al., 2008). Today, it is still the leading cause of neonatal infections in developed countries (Verani and Schrag, 2010; Furfaro et al., 2018). Moreover, it is also increasingly involved in adult infections, especially in elderly and immunocompromised people, causing bacteremia, meningitis, arthritis, etc. (Farley and Strasbaugh, 2001; High et al., 2005).
The typing of S. agalactiae isolates is important as pathogenesis and virulence are different according to strains, especially in infants (Poyart et al., 2008). Firstly, ten serotypes have been described by analyzing capsular polysaccharides, and serotype III has been identified as the most associated with neonatal infections. As the discrimination with this method was not sufficient, multilocus sequence typing (MLST), which consists in sequencing seven housekeeping genes, has been developed (Jones et al., 2003). MLST is the current reference method used to distinguish S. agalactiae isolates, and more than 1,600 different sequence types (ST) are now described. This method has a good discriminating power but, with the discovery and research on CRISPR-Cas systems, recent studies propose CRISPR-typing as a promising alternative method (Beauruelle et al., 2017, 2021).
Two CRISPR loci have been identified in S. agalactiae, the CRISPR1 locus, which is a type II-A system, and the CRISPR2 locus, which belongs to type I-C systems, based on the current classification (Table 1; Lopez-Sanchez et al., 2012). The first one is ubiquitous and dynamic, whereas the second is present in a minority of strains. Firstly, the sequencing of the CRISPR1 locus has shown that direct repeats were usually highly conserved between strains, whereas the terminal repeat has many variants, which defined different groups. These groups correlate well with the phylogenetic lineages determined using the MLST method (Lier et al., 2015). Moreover, the analysis of the spacers content has shown a high degree of polymorphism which could probably lead to very precise classification. In order to confirm this hypothesis, the CRISPR-typing method has been compared to existing techniques. It has been shown that CRISPR-typing correlates very well with the MLST method and could even be more precise thanks to the analysis of the most recent spacers and according to that, identified subgroups within a same ST (Lier et al., 2015). Other studies have shown the high discriminating power of CRISPR-typing. An initial study, which followed the vaginal carriage of S. agalactiae in women for 11 years, demonstrated that CRISPR-typing correlates well with the results obtained by the MLST method and gives more information about colonizing strains, such as MGEs encountered in the vaginal environment (Beauruelle et al., 2017). Indeed, CRISPR-typing makes it possible to classify strains into ST like MLST, but it is more precise because it highlights differences in strains within the same ST according to the spacers present in the locus. Another study, exploring the diversity of strains in vaginal swabs of pregnant women by CRISPR-typing, demonstrated that some women carried isolates belonging to the same phylogenetic group (sharing the same CRISPR1 locus), others carried homogeneous population of isolates with few differences at the CRISPR locus, and some women are colonized at the vaginal level by isolates belonging to different phylogenetic groups (Beauruelle et al., 2018).
Thus, the S. agalactiae CRISPR1-Cas system appears to be a useful tool for typing isolates but also for providing information about the evolution of this species (Table 2). It has also been shown that it could be involved in the differences of virulence observed between strains by integrating or not integrating new spacers from MGEs. Indeed, virulence genes are frequently acquired by horizontal gene transfer (Koonin et al., 2001) and CRISPR-Cas systems can be a barrier to this transfer. The S. agalactiae genome is divided in a “core genome,” which is the conserved part of the genome shared by all the strains, and a “dispensable genome,” which is a variable part and can correspond to horizontal gene acquisition (Tettelin et al., 2005). According to the CRISPR-Cas system’s efficiency and its spacers contents, strains are more or less susceptible to invaders and consequently to the acquisition of new genes. However, these genes can improve the pathogenicity and the survival of bacteria, as they can correspond to resistance or virulence genes. Differences have been highlighted according to ST. For example, strains belonging to ST17, considered as hypervirulent because more involved in neonatal infections, have less spacers than other STs (Lier et al., 2015). More recently, specificity in the locus organization has been highlighted for these strains and the system’s functionality has been explored (Pastuszka et al., 2021). Thanks to its CRISPR1-Cas system, S. agalactiae can not only defend itself against invaders but also regulate its own genome. It was shown that spacer diversity is important and some of them target MGEs integrated in the bacterial genome, in order to regulate what is called the “mobilome” of S. agalactiae (Lopez-Sanchez et al., 2012). Other spacers target genes from the core genome, and in order to differentiate between self- and non-self-targeting, the PAM plays an essential role and allows fine regulation (Mojica et al., 2009).
In S. agalactiae, like in most streptococci, CRISPR-Cas systems have many important roles and their analyses can provide a lot of information about species and the differences observed between strains.
Streptococcus pyogenes
Streptococcus pyogenes, according to the Lancefield classification, is a group A Streptococcus involved in human pathology. It is a strictly human bacteria, which belongs to the commensal microbiota of the skin, or may be found in the upper respiratory tract, according to age (Hartley et al., 1945; DeMuri and Wald, 2014). Initially, it was implicated in mild illnesses like impetigo, a superficial skin disorder, or infections of the upper respiratory tract, such as pharyngitis (Stevens and Bryant, 2016). Since the 1980’s, an increased incidence of invasive infections, like bacteremia or necrotizing fasciitis, has been highlighted (Carapetis et al., 2005; Lamagni et al., 2008).
Two CRISPR loci have been described in S. pyogenes, CRISPR1 and CRISPR2 (Nozawa et al., 2011), belonging, respectively, to type II-A and type I-C systems (Table 1; Marraffini, 2016). CRISPR analysis is an element which provides a better understanding of the evolution of species because it reflects meetings of bacteria over time and their adaptation to them (Table 2; Le Rhun et al., 2019). Among streptococci, S. pyogenes possesses less spacers than other species, and some strains have completely lost their CRISPR loci. These findings have demonstrated that the strains, which have no or few spacers, contain more virulence factors acquired from MGEs (Nozawa et al., 2011). The loss of a CRISPR-Cas system could favorize an adaptation leading to a better bacterial permeability, allowing to acquire new virulence factors in order to improve their pathogenicity. But CRISPR-Cas systems are still active because it was shown that some strains are resistant to phages infection, thanks to a recent spacer acquisition. The presence of a spacer with homology with a phage sequence conferred resistance against prophage insertion to these strains. CRISPR-Cas system protects bacteria from phages lysis but this can also lead to a potential decrease in virulence. Indeed, it has been shown that up to 14% of the S. pyogenes genome is encoded by prophage insertions, often providing them virulence factors, like exotoxins, for example (Fischetti, 2007), leading to the scarlet fever (Yamada et al., 2019). The balance between defense mechanisms protecting bacteria integrity, and invasion permission in order to be more virulent, is very important and changing over time, leading to species evolution.
Streptococcus pyogenes is a major pathogen involved in severe invasive infections. Methods used to characterize species have been developed in order to better understand virulence differences observed between isolates. Firstly, sera agglutination was used to serotype isolates, based on antigenic properties. M protein, encoded by emm gene, a surface protein and a virulence factor carried by the strain was used initially. A few years later, it was decided to sequence a part of this gene in order to establish a more precise classification (Beall et al., 2000; McMillan et al., 2013b). Emm-typing is currently the reference method, and more than 200 emm-types have been described (McMillan et al., 2013a; Sanderson-Smith et al., 2014). Emm1 was the most prevalent type (Hoe et al., 1999), but since the 2000s other types, like emm28 and emm89, are emerging worldwide (O’Brien et al., 2002; Ikebe et al., 2007; Luca-Harari et al., 2009). All emm1, emm12, and emm28 strains possess two CRISPR loci and emm3 strains have no CRISPR2 locus. Analyzing the composition of CRISPR loci, has enabled to show relations between spacer content and emm-types (Zheng et al., 2015). Indeed, they have highlighted that most spacers are emm-type specific and so, they propose CRISPR sequencing as a new typing method. The analysis of all the CRISPR loci present in a strain, or of each locus alone, have a higher Wallace coefficient than the emm-typing, proving the good congruence of both methods (Severiano et al., 2011). These results place the CRISPR-typing method as a good alternative to characterize S. pyogenes strains, and a better technique to differentiate isolates classified in the same emm-type. As previously mentioned, sequencing only one locus may not be sufficient to differentiate subtypes, and a combination of both loci is necessary. For example, the CRISPR1 locus analysis only is not able to differentiate emm3 and emm4 types, and the sequencing of the CRISPR2 locus is essential. However, emm-typing will remain useful, particularly for strains which do not contain CRISPR loci. It has been established that CRISPR loci of S. pyogenes acquire few spacers compared to other streptococci, which places this typing method as a good tool for this species. Indeed, there is not much spacer deletion or spacer insertion in loci, so less variability between regions in the world, giving stability and reliability to CRISPR-typing (Nozawa et al., 2011).
Studies have also shown that analyzing S. pyogenes CRISPR arrays can provide additional information concerning isolates, particularly reflecting encounters and evolution of species. For example, it may predict macrolide susceptibility (Zheng et al., 2014). This observation is in relation with the adaptive immune defense role of the system. Strains with a lot of spacers are generally more susceptible to erythromycin. Indeed, the resistance gene to this antibiotic is carried by a MGE, which will be cleaved in the event of sequence homology between this invader DNA and spacers present in the locus. Fighting against invasion by MGEs also prevents bacteria from acquiring new genes, which may support resistance or virulence genes.
However and rapidly, the CRISPR-Cas9 system of S. pyogenes has been used in genetic engineering because of its ease of use (Cong and Zhang, 2015) and the Nobel Prize was awarded for this application in 2020. Many discoveries have enabled the development of this application and particularly the demonstration of the role and importance of tracrRNA (Deltcheva et al., 2011). This extensive use has made CRISPR-Cas systems popular and interesting for many people. Nevertheless, more than its principal use in genome editing, CRISPR-Cas systems are very important in S. pyogenes and their analyses are promising in strain classification and for understanding the evolution of the species.
Other Members
The pyogenes group comprises other species that can be involved in human pathologies, among which Streptococcus dysgalactiae, Streptococcus pseudoporcinus, Streptococcus porcinus, Streptococcus equi, etc. These species can carry a type I-C and/or a type II-A CRISPR-Cas system, which are the most frequent in Streptococci, but no data is currently available about their role (Table 1).
Suis Group
Streptococcus suis
Streptococcus suis can lead to invasive infections in humans, especially in people working with pigs (Hughes et al., 2009b). According to the CRISPR finding tool, strains can carry type II-A and/or type II-C CRISPR-Cas systems (Table 1). As demonstrated in a study highlighting defense systems against invading DNA in this species, three CRISPR-Cas loci were identified (Okura et al., 2017).
Viridans Group
The viridans group is composed of five subgroups: anginosus group, bovis group, mitis group, mutans group and salivarius group.
Anginosus (or Milleri) Group
Streptococcus anginosus
S. anginosus is part of the human microbiota and is found in the oral cavity, the gastro-intestinal and the genito-urinary tracts (Whiley et al., 1992). Since many years, it has been increasingly involved in deep infections, like bacteremia and abscesses (Reißmann et al., 2010; Siegman-Igra et al., 2012), and is therefore considered as an important pathogen.
CRISPR loci have been described in S. anginosus previously. Most strains carry a type II-A and/or a type II-C system (Table 1). Few strains possess a type I-C system. Similarities have been demonstrated with systems found in other oral streptococci, like S. mutans or S. sanguinis, which suggest horizontal gene transfer between species (Olson et al., 2013). Sequence homologies between spacers and foreign genetic elements have been highlighted for all these CRISPR-Cas systems (Table 2).
Within this species, strains can be alpha-hemolytic, beta-hemolytic or non-hemolytic (Jensen et al., 2013). Beta-hemolysis of some strains is due to the presence of a virulence factor encoded by sag genes (Asam et al., 2013). This hemolysin seems to be acquired from a S. pyogenes genetic locus, encoding a bacteriocin-like peptide, the streptolysin S (Nizet et al., 2000). An inverse correlation has been shown between the presence of a CRISPR-Cas system and the presence of sag genes in S. anginosus isolates (Bauer et al., 2020). Indeed, the presence of a CRISPR locus, and especially its spacer content, gives the strain the ability to fight against integration in its genome of foreign DNA pieces if homology exists. This avoids the transfer and acquisition of genetic elements like virulence factors from MGEs, such as hemolysin for example. Thus, it is possible to classify strains and predict their virulence by analyzing their CRISPR arrays. CRISPR-typing has also been used in a study to follow the presence of streptococci in the salivary microbiota, in order to visualize differences, over time, in a single subject and between subjects. The aim of this study was to better understand the evolution of bacteria and their encounters with viruses or other MGEs (Pride et al., 2011). CRISPR arrays analysis seems to be a promising typing tool in this species, but must be continued in order to enhance knowledge.
Other Members
The large anginosus group, which is also called the milleri group, includes two other important species in human pathology. Streptococcus constellatus and Streptococcus intermedius belong to the human microbiota and can be involved in severe invasive infections like bacteremia, endocarditis, or deep abscesses (Tran et al., 2008; Chrastek et al., 2020). Concerning the presence of CRISPR-Cas systems, two types can be found in S. intermedius, type II-A and type II-C, like in S. anginosus but they are less well studied (Ben Zakour et al., 2012). However, no CRISPR-Cas system has been identified in S. constellatus (Table 1).
Bovis Group
Species of this group are particularly associated with digestive infections and colorectal cancers (Boleij et al., 2011; Kaindi et al., 2018) and can be responsible for endocarditis (Rusniok et al., 2010). Moreover, thanks to the CRISPR finder tool, two CRISPR-Cas systems can generally be found, a type II-A and a type II-C (Table 1). Loci organization of CRISPR-Cas systems has been analyzed in S. gallolyticus but it is no longer studied (Lin et al., 2011).
Mitis Group
The mitis group comprises many species including the well-known human pathogen Streptococcus pneumoniae and many other commensal species belonging to the human oral cavity and that can lead to invasive infections, especially in immunocompromised people (Mitchell, 2011; Weiser et al., 2018; Basaranoglu et al., 2019). A wide range of different CRISPR-Cas systems can be identified depending on the species (Table 1).
Streptococcus mitis
According to the most recent version of the CRISPR finder tool, it has been shown that Streptococcus mitis strains can carry type II-C or type III-B CRISPR-Cas systems, but it seems extremely rare. It has been described that some strains carry CRISPR-like sequences, but they are not well characterized (Maeda et al., 2011). Indeed, these sequences share similarities with CRISPR loci, suggesting that strains which carry them could share a common ancestor.
Streptococcus pneumoniae
Streptococcus pneumoniae belongs to the human respiratory tract and is an important pathogen in human medicine. Indeed, it can be responsible for many common infections like otitis or pneumonia, but also for more invasive infections, like bacteremia and meningitis (Musher, 1992; Weiser et al., 2018). S. pneumoniae strains generally do not possess CRISPR locus (Table 1 and Figure 2), but can sometimes carry a remnant system. Indeed, homologous sequences to those coding CRISPR-Cas systems have been found, but no functional CRISPR-Cas system has been actually described in this species. It is supposed that S. pneumoniae possessed a CRISPR-Cas system a few years ago, but for some reason it lost it. Several explanations are plausible. The most likely hypothesis is that S. pneumoniae is a naturally competent bacteria making numerous genetic exchanges via horizontal gene transfers in order to adapt to its environment. CRISPR interference can be an obstacle to these transfers, so the loss of a CRISPR-Cas system can be beneficial for species evolution. Other hypotheses can be that S. pneumoniae is not often in contact with phages, so it is not useful, or because the system is too inordinate for the growth and survival of bacteria (Bikard et al., 2012).
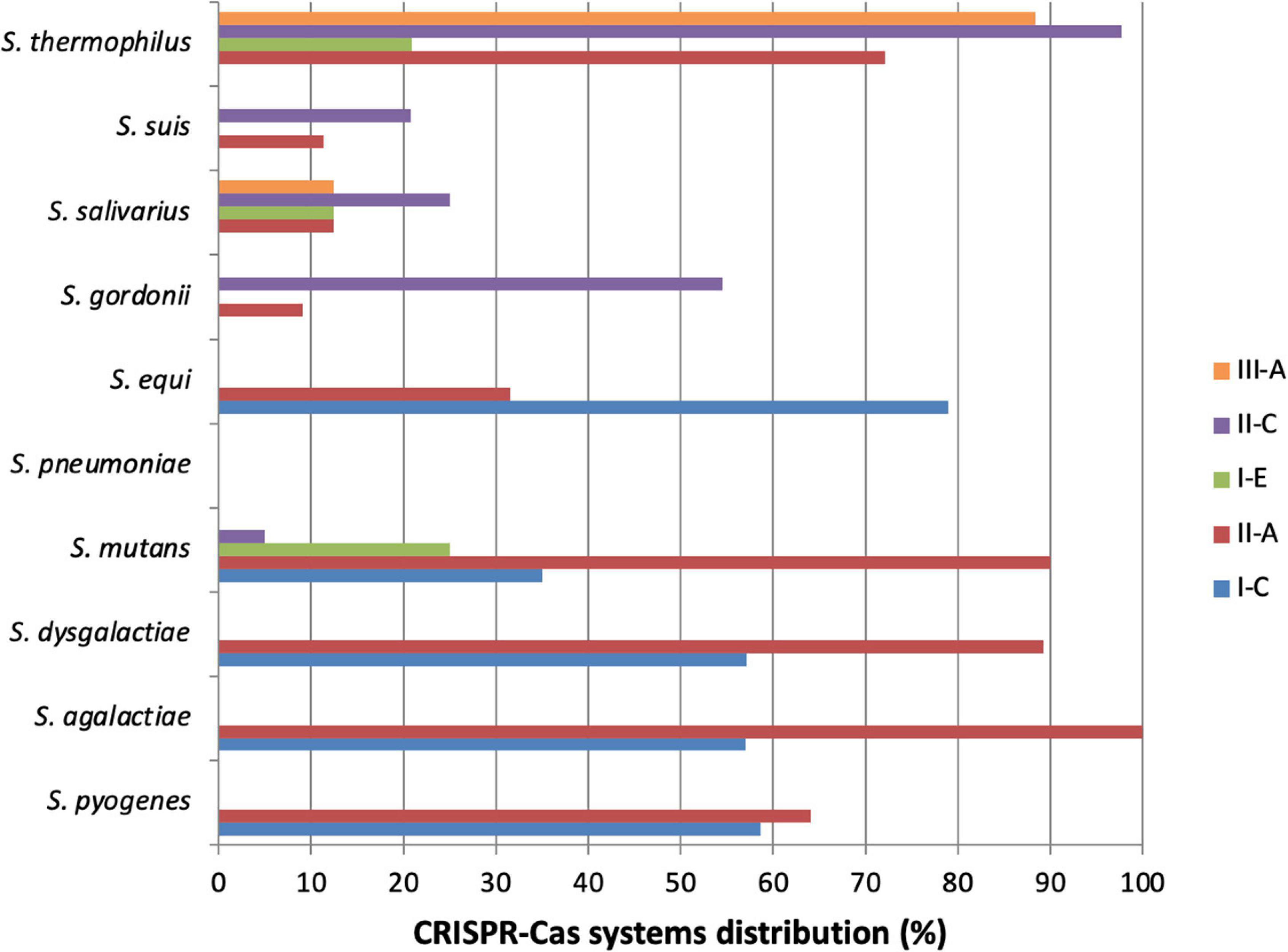
Figure 2. Distribution of CRISPR-Cas systems in Streptococcus species. This graph represents the distribution of CRISPR-Cas systems in some Streptococcus species. It is based on the data available on “CRISPR finder” online, considering the tool limits. Particularly for S. thermophilus, it is important to notice that type II-C CRISPR-Cas systems found are actually type II-A. Only species of which at least 10 strains have been sequenced and analyzed are represented. S. thermophilus (n = 43), S. suis (n = 53), S. salivarius (n = 16), S. gordonii (n = 11), S. equi (n = 19), S. pneumoniae (n = 141), S. mutans (n = 20), S. dysgalactiae (n = 28), S. agalactiae (n = 121), S. pyogenes (n = 220).
Mutans Group
The mutans group is composed of several streptococci but CRISPR-Cas systems have been already described and studied in S. mutans only.
Streptococcus mutans
Streptococcus mutans belongs to the human buccal cavity. Like other buccal streptococci, it is involved in dental infections, and particularly in dental caries (Hamada and Slade, 1980). Because of the increasing antibiotic resistance in bacteria, research focused on phage therapy to eliminate S. mutans from the dental plaque of people prone to dental caries. Lytic phages have been isolated (Delisle and Rostkowski, 1993) but S. mutans has already developed resistance to some of them. Studying the origin of these resistances led to the discovery of CRISPR loci in this species (Shibata et al., 2009; Serbanescu et al., 2015).
Two CRISPR loci have been described in S. mutans (Haft et al., 2005). The CRISPR1 locus is a type II-A system and the CRISPR2 locus is a type I-C system (Table 1). Spacers of these two loci have been shown to share similarities with phage sequences (Van der Ploeg, 2009). Moreover, strains resistant to some phages have specific spacers that sensitive strains do not have. The CRISPR1 locus (Mosterd and Moineau, 2020) is more present than the CRISPR2 in strains but none is ubiquitous. A great variability in their spacer content has been highlighted and shown that they are a good tool for S. mutans typing, especially the CRISPR2 locus, which is more variable and best reflects the evolution of this species (Table 2; Maruyama et al., 2009). Since the description of these two systems, two other CRISPR-Cas systems have been integrated in CRISPR databases, which are a type II-C system and a type I-E system. The typing of S. mutans strains is interesting in human medicine. A very high number of STs have been described, showing that this species is very diverse (Nakano et al., 2007). This diversity between isolates was confirmed using the CRISPR arrays analysis. Its information provides insight into the bacteria encounters and resistance genes that they carried. In another study, it was also shown that S. mutans CRISPR-Cas systems were involved in strain virulence. Indeed, it was shown that cas genes were more transcribed when strains were submitted to stress and also during biofilm formation, which indicates their implication for bacteria persistence in its environment (Louwen et al., 2014). A more recent study analyzed the clinical strains of S. mutans, isolated from early childhood dental caries. They found that the spacer content of the CRISPR1 and CRISPR2 loci was very diverse (Chen et al., 2017). They also showed that strains carrying a CRISPR-Cas system were more virulent than strains without a CRISPR-Cas system, because they had a better ability to produce extracellular polysaccharides and form biofilm on teeth surfaces. This was supported by another study, which showed that strains deleted for cas3 (type I CRISPR-Cas system) were less virulent and produced a finer biofilm than wild type strains. They also showed that cas3 deleted mutants were more susceptible to fluoride, which can be a treatment used to reduce biofilm formation (Tang et al., 2019).
Further studies are required for this species in order to better understand S. mutans strains evolution and perhaps adapt treatment strategies.
Salivarius Group
Species from this group like Streptococcus vestibularis or Streptococcus salivarius belong to the human microbiota and are usually found in the oral cavity. They can be involved in bacteremia in immunocompromised people, but it is still rare (Corredoira et al., 2005). According to the CRISPR finder tool, strains can carry type II-A, type II-C or type III-A CRISPR-Cas systems (Table 1), and sequence similarities between spacers and extrachromosomal elements have been shown (Bolotin et al., 2005).
Streptococcus thermophilus is also a member of this group. It is not a pathogen for humans or animals, so is not detailed in this review, but cannot be ignored when referring to CRISPR-Cas systems, because it was the first studied Streptococcus.
Discussion
Streptococci are a wide group of bacteria and an important genus in human and animal medicine (Lamagni et al., 2008; Haas and Grenier, 2018; Raabe and Shane, 2019). For each species, differences in evolution and virulence have been highlighted, depending on the isolated strain. Since their discovery, research on CRISPR-Cas systems has shown that sequencing CRISPR loci can provide numerous isolate information, about their virulence, foreign MGEs, adaptation and evolution (Bolotin et al., 2005; Horvath et al., 2008; Mojica et al., 2009). A great majority of streptococci contain one or more CRISPR loci, or at least sequences which have similarities with CRISPR loci but cannot be qualified as such (Moore et al., 2008). Only two types of CRISPR-Cas systems can be found for most bacterial species, but all three types can be found specifically for streptococci (Figure 2), and particularly the subtypes I-C, II-A and III-A (Louwen et al., 2014; Bauer et al., 2020). Subtype II-A is particularly present in streptococci, like in many commensal and pathogenic bacteria in humans (Chylinski et al., 2014). Thanks to existing databases presence of CRISPR-Cas systems is relatively well known in streptococci. To improve knowledges, the exact distribution and the study of the genes conservation inside species, and between species that contain the same type of CRISPR-Cas system should be of interest. If strong identities are found, this could expand our knowledge of the function of the system in one species to other species for which little data are currently available. Large-scale bioinformatic analyses will be necessary to complete this work.
One of the most developed applications of the CRISPR-Cas systems is their use in genome editing. Because of the simple multi-domain single-protein effector action, class 2 CRISPR-Cas systems are the most extensively used for genome editing. First applications were derived from Cas9 endonucleases, but Cas12 endonucleases were rapidly used (Anzalone et al., 2020). Cas13 is also used, but it preferentially targets RNA. Some differences exist between these nucleases, such as the nucleic acid cleavage site, and the choice of whether to use one or the other depends on the desired result. With these nucleases, double stranded or single stranded DNA can be cut or modified, depending on the Cas variant used. This plasticity and this wide range of possibilities means that CRISPR-Cas derivatives are very promising tools for treating some diseases by gene therapy. Techniques have now been improved and multiplex gene editing is almost available. Cas proteins have already been modified in order to improve the affinity with a targeted DNA and recognize more variable PAM sequences. Techniques were also improved to limit off-target and be more precise in order to insert modifications in DNA sequences. Thanks to these applications, progress has been made in better understanding some diseases, by introducing mutations in genes and observing the consequences, in order to highlight the function of these genes and their implications (Wang et al., 2016). Research is ongoing for use in gene therapy, such as treating cystic fibrosis (Schwank et al., 2013) or beta-thalassemia (Xie et al., 2014). Work must be continued in order to improve use and answer all given queries before being able to cure diseases.
More than this important and great use, numerous studies have shown that analyzing CRISPR arrays can provide a lot of information about streptococci isolates. Its main role of immune defense system confers an ability to fight against MGEs via integrated DNA sequences as spacers into its locus and the analysis of the spacer contents shows an encounter of streptococci with these MGEs. CRISPR-Cas systems can regulate their ability to acquire, or not, new genes from other bacteria, phages or various MGEs, and so, to acquire, or not, virulence or resistance genes. This could partly explain the differences in virulence observed between strains. This virulence involvement has already been demonstrated for other bacterial species (Louwen et al., 2014; Ahmed et al., 2018). Francisella novicida uses its CRISPR-Cas system to repress the expression of a lipoprotein that will not be recognized by the host immune system, and thus progress in infections (Sampson and Weiss, 2014). Pseudomonas aeruginosa can also be cited as an example. It was established that the type I-F CRISPR-Cas system is able to repress some genes in order to escape the host defense immune system and reduce the amount of cytokines produced by the host (Li et al., 2016; Wiedenheft and Bondy-Denomy, 2017). With this review, we can see that this link between CRISPR-Cas systems and bacterial virulence exists also in streptococci, as proved with S. pyogenes (Nozawa et al., 2011), S. mutans (Chen et al., 2017) or S. anginosus (Bauer et al., 2020). A link has also been established between CRISPR-Cas systems and antibiotics resistance, as demonstrated for S. pyogenes and the macrolides susceptibility. Furthermore, with the increasing dissemination of antibiotics resistance genes, two new uses of CRISPR-Cas9 genome editing have been developed. The first one consists in programming Cas9 endonuclease to target and cleave essential genes of bacteria and, therefore, kill a targeted bacterial population (Bikard et al., 2014; Gomaa et al., 2014). The second consists in using a “dead” Cas9 (dCas9), which is able to bind to a targeted element but not cleave it. This binding could repress the gene transcription and could be interesting in targeting antibiotics resistance genes without killing bacteria (Bikard et al., 2013).
In addition, this review shows that, especially in streptococci but also in other bacteria, sequencing CRISPR loci is also a relatively easy way to characterize and classify isolates precisely. Indeed, many studies have demonstrated that using CRISPR as a typing method is a promising tool and must be developed (Pourcel et al., 2005; Brudey et al., 2006; Beauruelle et al., 2021). More than precisely classifying strains, it can give a lot of information about species evolution.
Finally, the adaptive immune role of CRISPR-Cas systems, providing bacteria with a strong protection against phages and other MGEs was first elucidated in a Streptococcus species, S. thermophilus. However, phages, stuck in a perpetual evolutionary arms race with bacteria, have developed different mechanisms to counter these bacterial defenses. The simplest way was to acquire mutations in the targeted sequences, until the discovery of small proteins, capable of antagonizing the CRISPR-Cas systems, the anti-CRISPR (Acr) proteins. The first Acrs were described in Pseudomonas aeruginosa prophages with efficacy against type I-E and I-F CRISPR-Cas systems (Bondy-Denomy et al., 2013). Indeed, Acrs have been frequently identified in temperate phages, genomic islands or prophages, suggesting that their presence would be necessary for these elements to be maintained in a bacterial genome carrying a CRISPR-Cas system. Streptococci may contain many prophages and, not surprisingly, bioinformatic tools can predict Acrs in their genomes, like in S. pyogenes and other streptococcal species carrying a type II-A CRISPR-Cas system (Eitzinger et al., 2020). Depending on the Acrs types, different stages of CRISPR-Cas systems can be blocked. In consequence, Acrs can have a more or less broad activity spectrum depending on the Cas proteins targeted, and the inhibitory mechanism involved. Since their discovery, Acr genes have been identified in many bacteria, with a wide range of activities, inhibiting diverse class 1 or class 2 systems. Some of them are even able to block the activity of SpCas9, the widely used genome-editing tool, especially Acrs described in S. thermophilus virulent phages (Hynes et al., 2017; Rauch et al., 2017). All these elements make Acrs a promising tool to control Cas9 activities in genome editing.
Many tools have been developed since the discovery of the CRISPR-Cas systems, but a lot of studies remain to be performed on streptococcal species that have not been explored yet, in order to improve our knowledge of these systems, and of their more recently discovered anti-systems, and definitely develop new applications.
Author Contributions
CL, AP, and PL designed the study and wrote the manuscript. BL and LM provided critical feedback of the manuscript. All authors contributed to the article and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We are very grateful to Philippe Horvath for his critical reading of the manuscript.
References
Ahmed, W., Hafeez, M. A., Ahmad, R., and Mahmood, S. (2018). CRISPR-Cas system in regulation of immunity and virulence of bacterial pathogens. Gene Rep. 13, 151–157. doi: 10.1016/j.genrep.2018.10.004
Al-Attar, S., Westra, E. R., van der Oost, J., and Brouns, S. J. J. (2011). Clustered regularly interspaced short palindromic repeats (CRISPRs): the hallmark of an ingenious antiviral defense mechanism in prokaryotes. Biol. Chem. 392:42. doi: 10.1515/bc.2011.042
Anzalone, A. V., Koblan, L. W., and Liu, D. R. (2020). Genome editing with CRISPR–Cas nucleases, base editors, transposases and prime editors. Nat. Biotechnol. 38, 824–844. doi: 10.1038/s41587-020-0561-9
Asam, D., Mauerer, S., Walheim, E., and Spellerberg, B. (2013). Identification of β-haemolysin-encoding genes in Streptococcus anginosus. Mol. Oral Microbiol. 28, 302–315. doi: 10.1111/omi.12026
Bai, X., Xiong, Y., Lu, S., Jin, D., Lai, X., Yang, J., et al. (2016). Streptococcus pantholopis sp. nov., isolated from faeces of the Tibetan antelope (Pantholops hodgsonii). Int. J. Syst. Evol. Microbiol. 66, 3281–3286. doi: 10.1099/ijsem.0.001189
Barrangou, R., Coûté-Monvoisin, A.-C., Stahl, B., Chavichvily, I., Damange, F., Romero, D. A., et al. (2013). Genomic impact of CRISPR immunization against bacteriophages. Biochem. Soc. Trans. 41, 1383–1391. doi: 10.1042/BST20130160
Barrangou, R., and Dudley, E. G. (2016). CRISPR-Based Typing and Next-Generation Tracking Technologies. Annu. Rev. Food Sci. Technol. 7:395. doi: 10.1146/annurev-food-022814-015729
Barrangou, R., Fremaux, C., Deveau, H., Richards, M., Boyaval, P., Moineau, S., et al. (2007). CRISPR provides acquired resistance against viruses in prokaryotes. Science 315, 1709–1712. doi: 10.1126/science.1138140
Barrangou, R., and van der Oost, J. (2012). CRISPR-Cas Systems: RNA-mediated Adaptive Immunity in Bacteria and Archaea. New York, NY: Springer Science & Business Media.
Basaranoglu, S. T., Ozsurekci, Y., Aykac, K., Aycan, A. E., Bıcakcigil, A., Altun, B., et al. (2019). Streptococcus mitis/oralis Causing Blood Stream Infections in Pediatric Patients. Jpn. J. Infect. Dis. 72, 1–6. doi: 10.7883/yoken.JJID.2018.074
Bauer, R., Neffgen, N., Grempels, A., Furitsch, M., Mauerer, S., Barbaqadze, S., et al. (2020). Heterogeneity of Streptococcus anginosus ß-hemolysis in relation to CRISPR/Cas. Mol. Oral Microbiol. 35, 56–65. doi: 10.1111/omi.12278
Beall, B., Gherardi, G., Lovgren, M., Facklam, R. R., Forwick, B. A., and Tyrrell, G. J. (2000). emm and sof gene sequence variation in relation to serological typing of opacity-factor-positive group A streptococci. Microbiology 146, 1195–1209. doi: 10.1099/00221287-146-5-1195
Beauruelle, C., Pastuszka, A., Horvath, P., Perrotin, F., Mereghetti, L., and Lanotte, P. (2017). CRISPR: A Useful Genetic Feature to Follow Vaginal Carriage of Group B Streptococcus. Front. Microbiol. 8:1981. doi: 10.3389/fmicb.2017.01981
Beauruelle, C., Pastuszka, A., Mereghetti, L., and Lanotte, P. (2018). Group B Streptococcus Vaginal Carriage in Pregnant Women as Deciphered by Clustered Regularly Interspaced Short Palindromic Repeat Analysis. J. Clin. Microbiol. 56:17. doi: 10.1128/JCM.01949-17
Beauruelle, C., Treluyer, L., Pastuszka, A., Cochard, T., Lier, C., Mereghetti, L., et al. (2021). CRISPR Typing Increases the Discriminatory Power of Streptococcus agalactiae Typing Methods. Front. Microbiol. 12:675597. doi: 10.3389/fmicb.2021.675597
Beighton, D., Carr, A. D., and Oppenheim, B. A. (1994). Identification of viridans streptococci associated with bacteraemia in neutropenic cancer patients. J. Med. Microbiol. 40, 202–204. doi: 10.1099/00222615-40-3-202
Bekal, S., Gaudreau, C., Laurence, R. A., Simoneau, E., and Raynal, L. (2006). Streptococcus pseudoporcinus sp. nov., a Novel Species Isolated from the Genitourinary Tract of Women. J. Clin. Microbiol. 44, 2584–2586. doi: 10.1128/JCM.02707-05
Ben Zakour, N., Beatson, S., van den Broek, A., Thoday, K., and Fitzgerald, R. (2012). Comparative Genomics of the Staphylococcus intermedius Group of Animal Pathogens. Front. Cell. Infect. Microbiol. 2:44. doi: 10.3389/fcimb.2012.00044
Bergqvist, G., and Hurvell, B. (1970). Neonatal infections caused by Streptococcus agalactiae (Lancefield group B). Acta Pathol. Microbiol. Scand. [B] Microbiol. Immunol. 78:270.
Bikard, D., Euler, C. W., Jiang, W., Nussenzweig, P. M., Goldberg, G. W., Duportet, X., et al. (2014). Exploiting CRISPR-Cas nucleases to produce sequence-specific antimicrobials. Nat. Biotechnol. 32, 1146–1150. doi: 10.1038/nbt.3043
Bikard, D., Hatoum-Aslan, A., Mucida, D., and Marraffini, L. A. (2012). CRISPR Interference Can Prevent Natural Transformation and Virulence Acquisition during In Vivo Bacterial Infection. Cell Host Microbe 12, 177–186. doi: 10.1016/j.chom.2012.06.003
Bikard, D., Jiang, W., Samai, P., Hochschild, A., Zhang, F., and Marraffini, L. A. (2013). Programmable repression and activation of bacterial gene expression using an engineered CRISPR-Cas system. Nucleic Acids Res. 41, 7429–7437. doi: 10.1093/nar/gkt520
Boleij, A., van Gelder, M. M. H. J., Swinkels, D. W., and Tjalsma, H. (2011). Clinical Importance of Streptococcus gallolyticus Infection Among Colorectal Cancer Patients: Systematic Review and Meta-analysis. Clin. Infect. Dis. 53, 870–878. doi: 10.1093/cid/cir609
Bolotin, A., Quinquis, B., Sorokin, A., and Ehrlich, S. D. (2005). Clustered regularly interspaced short palindrome repeats (CRISPRs) have spacers of extrachromosomal origin. Microbiol. Read. Engl. 151, 2551–2561. doi: 10.1099/mic.0.28048-0
Bondy-Denomy, J., Pawluk, A., Maxwell, K. L., and Davidson, A. R. (2013). Bacteriophage genes that inactivate the CRISPR/Cas bacterial immune system. Nature 493, 429–432. doi: 10.1038/nature11723
Boutrou, R., Thuault, D., and Bourgeois, C. M. (1995). Identification and characterization of Streptococcus thermophilus strains by pulsed-field gel electrophoresis. J. Appl. Bacteriol. 79, 454–458. doi: 10.1111/j.1365-2672.1995.tb03161.x
Broeders, M., Herrero-Hernandez, P., Ernst, M. P. T., van der Ploeg, A. T., and Pijnappel, W. W. M. P. (2020). Sharpening the Molecular Scissors: Advances in Gene-Editing Technology. iScience 23:100789. doi: 10.1016/j.isci.2019.100789
Brouns, S. J. J., Jore, M. M., Lundgren, M., Westra, E. R., Slijkhuis, R. J. H., Snijders, A. P. L., et al. (2008). Small CRISPR RNAs Guide Antiviral Defense in Prokaryotes. Science 321, 960–964. doi: 10.1126/science.1159689
Brudey, K., Driscoll, J. R., Rigouts, L., Prodinger, W. M., Gori, A., Al-Hajoj, S. A., et al. (2006). Mycobacterium tuberculosis complex genetic diversity: mining the fourth international spoligotyping database (SpolDB4) for classification, population genetics and epidemiology. BMC Microbiol. 6:23. doi: 10.1186/1471-2180-6-23
Calvinho, L. F., Almeida, R. A., and Oliver, S. P. (1998). Potential virulence factors of Streptococcus dysgalactiae associated with bovine mastitis. Vet. Microbiol. 61, 93–110. doi: 10.1016/S0378-1135(98)00172-2
Carapetis, J. R., Steer, A. C., Mulholland, E. K., and Weber, M. (2005). The global burden of group A streptococcal diseases. Lancet Infect. Dis. 5, 685–694. doi: 10.1016/S1473-3099(05)70267-X
Carbonnelle, E., Mesquita, C., Bille, E., Day, N., Dauphin, B., Beretti, J.-L., et al. (2011). MALDI-TOF mass spectrometry tools for bacterial identification in clinical microbiology laboratory. Clin. Biochem. 44, 104–109. doi: 10.1016/j.clinbiochem.2010.06.017
Chen, J., Li, T., Zhou, X., Cheng, L., Huo, Y., Zou, J., et al. (2017). Characterization of the clustered regularly interspaced short palindromic repeats sites in Streptococcus mutans isolated from early childhood caries patients. Arch. Oral Biol. 83, 174–180. doi: 10.1016/j.archoralbio.2017.07.023
Chrastek, D., Hickman, S., Sitaranjan, D., Vokshi, I., Kakisi, O., Kadlec, J., et al. (2020). Streptococcus constellatus Causing Empyema and Sepsis, Necessitating Early Surgical Decortication. Case Rep. Infect. Dis. 2020:e4630809. doi: 10.1155/2020/4630809
Chylinski, K., Makarova, K. S., Charpentier, E., and Koonin, E. V. (2014). Classification and evolution of type II CRISPR-Cas systems. Nucleic Acids Res. 42, 6091–6105. doi: 10.1093/nar/gku241
Clarridge, J. E., Attorri, S., Musher, D. M., Hebert, J., and Dunbar, S. (2001). Streptococcus intermedius, Streptococcus constellatus, and Streptococcus anginosus (“Streptococcus milleri Group”) Are of Different Clinical Importance and Are Not Equally Associated with Abscess. Clin. Infect. Dis. 32, 1511–1515. doi: 10.1086/320163
Collins, M. D., Farrow, J. A. E., Katic, V., and Kandler, O. (1984). Taxonomic studies on streptococci of serological groups E, P, U and V: Description of Streptococcus porcinus sp. nov. Syst. Appl. Microbiol. 5, 402–413. doi: 10.1016/S0723-2020(84)80041-7
Collins, M. D., Hutson, R. A., Falsen, E., Nikolaitchouk, N., LaClaire, L., and Facklam, R. R. (2000). An unusual Streptococcus from human urine, Streptococcus urinalis sp. nov. Int. J. Syst. Evol. Microbiol. 50(Pt 3), 1173–1178. doi: 10.1099/00207713-50-3-1173
Cong, L., and Zhang, F. (2015). ““Genome Engineering Using CRISPR-Cas9 System,”,” in Chromosomal Mutagenesis Methods in Molecular Biology, ed. S. M. Pruett-Miller (New York, NY: Springer), 197–217. doi: 10.1007/978-1-4939-1862-1_10
Conrads, G., de Soet, J. J., Song, L., Henne, K., Sztajer, H., Wagner-Döbler, I., et al. (2014). Comparing the cariogenic species Streptococcus sobrinus and S. mutans on whole genome level. J. Oral Microbiol. 6:26189. doi: 10.3402/jom.v6.26189
Corredoira, J. C., Alonso, M. P., García, J. F., Casariego, E., Coira, A., Rodriguez, A., et al. (2005). Clinical characteristics and significance of Streptococcus salivarius bacteremia and Streptococcus bovis bacteremia: a prospective 16-year study. Eur. J. Clin. Microbiol. Infect. Dis. 24, 250–255. doi: 10.1007/s10096-005-1314-x
Couvin, D., Bernheim, A., Toffano-Nioche, C., Touchon, M., Michalik, J., Néron, B., et al. (2018). CRISPRCasFinder, an update of CRISRFinder, includes a portable version, enhanced performance and integrates search for Cas proteins. Nucleic Acids Res. 46, W246–W251. doi: 10.1093/nar/gky425
Cui, Y., Li, Y., Gorgé, O., Platonov, M. E., Yan, Y., Guo, Z., et al. (2008). Insight into Microevolution of Yersinia pestis by Clustered Regularly Interspaced Short Palindromic Repeats. PLoS One 3:e2652. doi: 10.1371/journal.pone.0002652
Cunningham, M. W. (2000). Pathogenesis of Group A Streptococcal Infections. Clin. Microbiol. Rev. 13:42.
Delisle, A. L., and Rostkowski, C. A. (1993). Lytic bacteriophages ofStreptococcus mutans. Curr. Microbiol. 27, 163–167. doi: 10.1007/BF01576015
Deltcheva, E., Chylinski, K., Sharma, C. M., Gonzales, K., Chao, Y., Pirzada, Z. A., et al. (2011). CRISPR RNA maturation by trans -encoded small RNA and host factor RNase III. Nature 471, 602–607. doi: 10.1038/nature09886
DeMuri, G. P., and Wald, E. R. (2014). The Group A Streptococcal Carrier State Reviewed: Still an Enigma. J. Pediatr. Infect. Dis. Soc. 3, 336–342. doi: 10.1093/jpids/piu030
Devriese, L. A., Hommez, J., Kilpper_Balz, R., and Schleifer, K.-H. (1986). Streptococcus canis sp. nov.: A Species of Group G Streptococci from Animals. Int. J. Syst. Evol. Microbiol. 36, 422–425. doi: 10.1099/00207713-36-3-422
Douglas, C. W. I., Heath, J., Hampton, K. K., and Preston, F. E. (1993). Identity of viridans streptococci isolated from cases of infective endocarditis. J. Med. Microbiol. 39, 179–182. doi: 10.1099/00222615-39-3-179
Duriseti, P., and Fleisher, J. (2019). Streptococcus pluranimalium infective endocarditis and brain abscess. IDCases 18:e00587. doi: 10.1016/j.idcr.2019.e00587
Eitzinger, S., Asif, A., Watters, K. E., Iavarone, A. T., Knott, G. J., Doudna, J. A., et al. (2020). Machine learning predicts new anti-CRISPR proteins. Nucleic Acids Res. 48, 4698–4708. doi: 10.1093/nar/gkaa219
Eto, S. F., Fernandes, D. C., Moraes, A. C., de, Alecrim, J. V., da, C., et al. (2020). Meningitis Caused by Streptococcus agalactiae in Nile Tilapia (Oreochromis niloticus): Infection and Inflammatory Response. Anim. Open Access J. MDPI 10:E2166. doi: 10.3390/ani10112166
Evans, J. J., Klesius, P. H., Pasnik, D. J., and Bohnsack, J. F. (2009). Human Streptococcus agalactiae Isolate in Nile Tilapia (Oreochromis niloticus). Emerg. Infect. Dis. 15, 774–776. doi: 10.3201/eid1505.080222
Facklam, R. (2002). What Happened to the Streptococci: Overview of Taxonomic and Nomenclature Changes. Clin. Microbiol. Rev. 15, 613–630. doi: 10.1128/CMR.15.4.613-630.2002
Facklam, R., Elliott, J., Pigott, N., and Franklin, A. R. (1995). Identification of Streptococcus porcinus from human sources. J. Clin. Microbiol. 33, 385–388.
Farley, M. M., and Strasbaugh, L. J. (2001). Group B Streptococcal Disease in Nonpregnant Adults. Clin. Infect. Dis. 33, 556–561. doi: 10.1086/322696
Fischetti, V. A. (2007). In vivo acquisition of prophage in Streptococcus pyogenes. Trends Microbiol. 15, 297–300. doi: 10.1016/j.tim.2007.05.003
Forssten, S. D., Björklund, M., and Ouwehand, A. C. (2010). Streptococcus mutans, Caries and Simulation Models. Nutrients 2, 290–298. doi: 10.3390/nu2030290
Freedman, M. L., Coykendall, A. L., and O’Neill, E. M. (1982). Physiology of “mutans-like” Streptococcus ferus from wild rats. Infect. Immun. 35, 476–482. doi: 10.1128/iai.35.2.476-482.1982
Furfaro, L. L., Chang, B. J., and Payne, M. S. (2018). Perinatal Streptococcus agalactiae Epidemiology and Surveillance Targets. Clin. Microbiol. Rev. 31:18. doi: 10.1128/CMR.00049-18
Galpérine, T., Cazorla, C., Blanchard, E., Boineau, F., Ragnaud, J.-M., and Neau, D. (2007). Streptococcus canis infections in humans: Retrospective study of 54 patients. J. Infect. 55, 23–26. doi: 10.1016/j.jinf.2006.12.013
Garneau, J. E., Dupuis, M.-E., Villion, M., Romero, D. A., Barrangou, R., Boyaval, P., et al. (2010). The CRISPR/Cas bacterial immune system cleaves bacteriophage and plasmid DNA. Nature 468, 67–71. doi: 10.1038/nature09523
Garnett, J. A., Simpson, P. J., Taylor, J., Benjamin, S. V., Tagliaferri, C., Cota, E., et al. (2012). Structural insight into the role of Streptococcus parasanguinis Fap1 within oral biofilm formation. Biochem. Biophys. Res. Commun. 417, 421–426. doi: 10.1016/j.bbrc.2011.11.131
Garrett, S. C., Olson, S., and Graveley, B. R. (2020). Complete Genome Sequence of Streptococcus ratti JH145. Microbiol. Resour. Announc. 9:20. doi: 10.1128/MRA.00144-20
Gasiunas, G., Barrangou, R., Horvath, P., and Siksnys, V. (2012). Cas9–crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria. Proc. Natl. Acad. Sci. 109, E2579–E2586. doi: 10.1073/pnas.1208507109
Goh, S. H., Driedger, D., Gillett, S., Low, D. E., Hemmingsen, S. M., Amos, M., et al. (1998). Streptococcus iniae, a Human and Animal Pathogen: Specific Identification by the Chaperonin 60 Gene Identification Method. J. Clin. Microbiol. 36, 2164–2166. doi: 10.1128/JCM.36.7.2164-2166.1998
Gomaa, A. A., Klumpe, H. E., Luo, M. L., Selle, K., Barrangou, R., and Beisel, C. L. (2014). Programmable removal of bacterial strains by use of genome-targeting CRISPR-Cas systems. mBio 5, e928–e913. doi: 10.1128/mBio.00928-13
González-Abad, M. J., and Alonso Sanz, M. (2020). [Invasive Streptococcus pyogenes infections (2011–2018): EMM-type and clinical presentation]. An. Pediatr. 92, 351–358. doi: 10.1016/j.anpedi.2019.10.014
Gori, J. L., Hsu, P. D., Maeder, M. L., Shen, S., Welstead, G. G., and Bumcrot, D. (2015). Delivery and Specificity of CRISPR/Cas9 Genome Editing Technologies for Human Gene Therapy. Hum. Gene Ther. 26, 443–451. doi: 10.1089/hum.2015.074
Gottschalk, M., and Segura, M. (2019). Streptococcosis in Diseases of Swine. Hoboken, NJ: John Wiley & Sons, Ltd, 934–950. doi: 10.1002/9781119350927.ch61
Grissa, I., Vergnaud, G., and Pourcel, C. (2007b). The CRISPRdb database and tools to display CRISPRs and to generate dictionaries of spacers and repeats. BMC Bioinformatics 8:172. doi: 10.1186/1471-2105-8-172
Grissa, I., Vergnaud, G., and Pourcel, C. (2007a). CRISPRFinder: a web tool to identify clustered regularly interspaced short palindromic repeats. Nucleic Acids Res. 35, W52–W57. doi: 10.1093/nar/gkm360
Haas, B., and Grenier, D. (2018). Understanding the virulence of Streptococcus suis: A veterinary, medical, and economic challenge. Med. Mal. Infect. 48, 159–166. doi: 10.1016/j.medmal.2017.10.001
Haft, D. H., Selengut, J., Mongodin, E. F., and Nelson, K. E. (2005). A Guild of 45 CRISPR-Associated (Cas) Protein Families and Multiple CRISPR/Cas Subtypes Exist in Prokaryotic Genomes. PLoS Comput. Biol. 1:10060. doi: 10.1371/journal.pcbi.0010060
Hamada, S., and Slade, H. D. (1980). Biology, immunology, and cariogenicity of Streptococcus mutans. Microbiol. Rev. 44, 331–384. doi: 10.1128/mr.44.2.331-384.1980
Handley, P., Coykendall, A., Beighton, D., Hardie, J., and Whiley, R. (1991). Streptococcus crista sp. nov., a viridans streptococcus with tufted fibrils, isolated from the human oral cavity and throat. Int. J. Syst. Bacteriol. 41, 543–547. doi: 10.1099/00207713-41-4-543
Hardie, J. M., and Whiley, R. A. (1997). Classification and overview of the genera Streptococcus and Enterococcus. J. Appl. Microbiol. 83, 1S–11S. doi: 10.1046/j.1365-2672.83.s1.1.x
Hartley, G., Enders, J. F., Mueller, J. H., and Schoenbach, E. B. (1945). Absence of clinical disease in spite of a high incidence of carriers of group A hemolytic streptococci of a single type; failure of thyrithricin ti influence the carrier rate. J. Clin. Invest. 24, 92–96. doi: 10.1172/JCI101584
High, K. P., Edwards, M. S., and Baker, C. J. (2005). Group B Streptococcal Infections in Elderly Adults. Clin. Infect. Dis. 41, 839–847. doi: 10.1086/432804
Hoe, N., Nakashima, K., Grigsby, D., Pan, X., Dou, S. J., Naidich, S., et al. (1999). Rapid molecular genetic subtyping of serotype M1 group A Streptococcus strains. Emerg. Infect. Dis. 5, 254–263. doi: 10.3201/eid0502.990210
Hols, P., Hancy, F., Fontaine, L., Grossiord, B., Prozzi, D., Leblond-Bourget, N., et al. (2005). New insights in the molecular biology and physiology of Streptococcus thermophilus revealed by comparative genomics. FEMS Microbiol. Rev. 29, 435–463. doi: 10.1016/j.femsre.2005.04.008
Horvath, P., and Barrangou, R. (2010). CRISPR/Cas, the immune system of bacteria and archaea. Science 327, 167–170. doi: 10.1126/science.1179555
Horvath, P., Romero, D. A., Coûté-Monvoisin, A.-C., Richards, M., Deveau, H., Moineau, S., et al. (2008). Diversity, Activity, and Evolution of CRISPR Loci in Streptococcus thermophilus. J. Bacteriol. 190, 1401–1412. doi: 10.1128/JB.01415-07
Hsu, P. D., Scott, D. A., Weinstein, J. A., Ran, F. A., Konermann, S., Agarwala, V., et al. (2013). DNA targeting specificity of RNA-guided Cas9 nucleases. Nat. Biotechnol. 31, 827–832. doi: 10.1038/nbt.2647
Hughes, J. M., Wilson, M. E., Wertheim, H. F. L., Nghia, H. D. T., Taylor, W., and Schultsz, C. (2009b). Streptococcus suis: An Emerging Human Pathogen. Clin. Infect. Dis. 48, 617–625. doi: 10.1086/596763
Hughes, J. M., Wilson, M. E., Brandt, C. M., and Spellerberg, B. (2009a). Human Infections Due to Streptococcus dysgalactiae Subspecies equisimilis. Clin. Infect. Dis. 49, 766–772. doi: 10.1086/605085
Hynes, A. P., Rousseau, G. M., Lemay, M.-L., Horvath, P., Romero, D. A., Fremaux, C., et al. (2017). An anti-CRISPR from a virulent streptococcal phage inhibits Streptococcus pyogenes Cas9. Nat. Microbiol. 2, 1374–1380. doi: 10.1038/s41564-017-0004-7
Ikebe, T., Hirasawa, K., Suzuki, R., Ohya, H., Isobe, J., Tanaka, D., et al. (2007). Distribution of emm genotypes among group A streptococcus isolates from patients with severe invasive streptococcal infections in Japan, 2001–2005. Epidemiol. Infect. 135, 1227–1229. doi: 10.1017/S0950268807007984
Ishino, Y., Shinagawa, H., Makino, K., Amemura, M., and Nakata, A. (1987). Nucleotide sequence of the iap gene, responsible for alkaline phosphatase isozyme conversion in Escherichia coli, and identification of the gene product. J. Bacteriol. 169, 5429–5433. doi: 10.1128/jb.169.12.5429-5433.1987
Jansen, R., Embden, J. D. A., van, Gaastra, W., and Schouls, L. M. (2002). Identification of genes that are associated with DNA repeats in prokaryotes. Mol. Microbiol. 43, 1565–1575. doi: 10.1046/j.1365-2958.2002.02839.x
Jensen, A., Hoshino, T., and Kilian, M. (2013). Taxonomy of the Anginosus group of the genus Streptococcus and description of Streptococcus anginosus subsp. whileyi subsp. nov. and Streptococcus constellatus subsp. viborgensis subsp. nov. Int. J. Syst. Evol. Microbiol. 63, 2506–2519. doi: 10.1099/ijs.0.043232-0
Jiang, W., Bikard, D., Cox, D., Zhang, F., and Marraffini, L. A. (2013). RNA-guided editing of bacterial genomes using CRISPR-Cas systems. Nat. Biotechnol. 31, 233–239. doi: 10.1038/nbt.2508
Jones, N., Bohnsack, J. F., Takahashi, S., Oliver, K. A., Chan, M.-S., Kunst, F., et al. (2003). Multilocus sequence typing system for group B streptococcus. J. Clin. Microbiol. 41, 2530–2536. doi: 10.1128/jcm.41.6.2530-2536.2003
Kaindi, D. W. M., Kogi-Makau, W., Lule, G. N., Kreikemeyer, B., Renault, P., Bonfoh, B., et al. (2018). Colorectal cancer-associated Streptococcus infantarius subsp. infantarius differ from a major dairy lineage providing evidence for pathogenic, pathobiont and food-grade lineages. Sci. Rep. 8:9181. doi: 10.1038/s41598-018-27383-4
Karimi, Z., Ahmadi, A., Najafi, A., and Ranjbar, R. (2018). Bacterial CRISPR Regions: General Features and their Potential for Epidemiological Molecular Typing Studies. Open Microbiol. J. 12, 59–70. doi: 10.2174/1874285801812010059
Kok, C. R., and Hutkins, R. (2018). Yogurt and other fermented foods as sources of health-promoting bacteria. Nutr. Rev. 76, 4–15. doi: 10.1093/nutrit/nuy056
Koonin, E. V., Makarova, K. S., and Aravind, L. (2001). Horizontal Gene Transfer in Prokaryotes: Quantification and Classification. Annu. Rev. Microbiol. 55, 709–742. doi: 10.1146/annurev.micro.55.1.709
Krzyściak, W., Pluskwa, K. K., Jurczak, A., and Kościelniak, D. (2013). The pathogenicity of the Streptococcus genus. Eur. J. Clin. Microbiol. Infect. Dis. 32, 1361–1376. doi: 10.1007/s10096-013-1914-9
Kunin, V., Sorek, R., and Hugenholtz, P. (2007). Evolutionary conservation of sequence and secondary structures in CRISPR repeats. Genome Biol. 8:R61. doi: 10.1186/gb-2007-8-4-r61
Lamagni, T. L., Darenberg, J., Luca-Harari, B., Siljander, T., Efstratiou, A., Henriques-Normark, B., et al. (2008). Epidemiology of Severe Streptococcus pyogenes Disease in Europe. J. Clin. Microbiol. 46, 2359–2367. doi: 10.1128/JCM.00422-08
Lancefield, R. C. (1933). A serological differenciation of human and other groups of hemolytic streptococci. J. Exp. Med. 57, 571–595. doi: 10.1084/jem.57.4.571
Le Rhun, A., Escalera-Maurer, A., Bratovič, M., and Charpentier, E. (2019). CRISPR-Cas in Streptococcus pyogenes. RNA Biol. 16, 380–389. doi: 10.1080/15476286.2019.1582974
Li, R., Fang, L., Tan, S., Yu, M., Li, X., He, S., et al. (2016). Type I CRISPR-Cas targets endogenous genes and regulates virulence to evade mammalian host immunity. Cell Res. 26, 1273–1287. doi: 10.1038/cr.2016.135
Lier, C., Baticle, E., Horvath, P., Haguenoer, E., Valentin, A.-S., Glaser, P., et al. (2015). Analysis of the type II-A CRISPR-Cas system of Streptococcus agalactiae reveals distinctive features according to genetic lineages. Front. Genet. 6:214. doi: 10.3389/fgene.2015.00214
Lim, Y. K., Park, S.-N., Shin, J. H., Ji, S., Jo, E., Chang, Y.-H., et al. (2019b). Streptococcus koreensis sp. nov., Isolated from Human Subgingival Dental Plaque of Periodontitis Lesion. Curr. Microbiol. 76, 1531–1536. doi: 10.1007/s00284-019-01778-6
Lim, Y. K., Park, S.-N., Shin, J. H., Chang, Y.-H., Shin, Y., Paek, J., et al. (2019a). Streptococcus periodonticum sp. nov., Isolated from Human Subgingival Dental Plaque of Periodontitis Lesion. Curr. Microbiol. 76, 835–841. doi: 10.1007/s00284-019-01695-8
Lin, I.-H., Liu, T.-T., Teng, Y.-T., Wu, H.-L., Liu, Y.-M., Wu, K.-M., et al. (2011). Sequencing and Comparative Genome Analysis of Two Pathogenic Streptococcus gallolyticus Subspecies: Genome Plasticity, Adaptation and Virulence. PLoS One 6:20519. doi: 10.1371/journal.pone.0020519
Lopez-Sanchez, M.-J., Sauvage, E., Da Cunha, V., Clermont, D., Ratsima Hariniaina, E., Gonzalez-Zorn, B., et al. (2012). The highly dynamic CRISPR1 system of Streptococcus agalactiae controls the diversity of its mobilome: Streptococcus agalactiae CRISPR dynamics. Mol. Microbiol. 85, 1057–1071. doi: 10.1111/j.1365-2958.2012.08172.x
Louwen, R., Staals, R. H. J., Endtz, H. P., van Baarlen, P., and van der Oost, J. (2014). The Role of CRISPR-Cas Systems in Virulence of Pathogenic Bacteria. Microbiol. Mol. Biol. Rev. 78, 74–88. doi: 10.1128/MMBR.00039-13
Luca-Harari, B., Darenberg, J., Neal, S., Siljander, T., Strakova, L., Tanna, A., et al. (2009). Clinical and microbiological characteristics of severe Streptococcus pyogenes disease in Europe. J. Clin. Microbiol. 47, 1155–1165. doi: 10.1128/JCM.02155-08
Lun, Z.-R., Wang, Q.-P., Chen, X.-G., Li, A.-X., and Zhu, X.-Q. (2007). Streptococcus suis: an emerging zoonotic pathogen. Lancet Infect. Dis. 7, 201–209. doi: 10.1016/S1473-3099(07)70001-4
Maeda, Y., Goldsmith, C. E., Coulter, W. A., Mason, C., Dooley, J. S. G., Lowery, C. J., et al. (2011). Prevalence of clustered regulatory interspaced short palindromic repeat (CRISPR)-like sequences in mitis-group streptococci. Br. J. Biomed. Sci. 68, 65–68. doi: 10.1080/09674845.2011.11730325
Magadán, A. H., Dupuis, M.-E., Villion, M., and Moineau, S. (2012). Cleavage of Phage DNA by the Streptococcus thermophilus CRISPR3-Cas System. PLoS One 7:40913. doi: 10.1371/journal.pone.0040913
Makarova, K. S., Wolf, Y. I., Iranzo, J., Shmakov, S. A., Alkhnbashi, O. S., Brouns, S. J. J., et al. (2020). Evolutionary classification of CRISPR-Cas systems: a burst of class 2 and derived variants. Nat. Rev. Microbiol. 18, 67–83. doi: 10.1038/s41579-019-0299-x
Marín, M., Cercenado, E., Sánchez-Carrillo, C., Ruiz, A., Gómez González, A., Rodríguez-Sánchez, B., et al. (2017). Accurate Differentiation of Streptococcus pneumoniae from other Species within the Streptococcus mitis Group by Peak Analysis Using MALDI-TOF MS. Front. Microbiol. 8:698. doi: 10.3389/fmicb.2017.00698
Marraffini, L. A. (2016). “The CRISPR-Cas system of Streptococcus pyogenes: function and applications,” in Streptococcus pyogenes: Basic Biology to Clinical Manifestations, eds J. J. Ferretti, D. L. Stevens, and V. A. Fischetti (Oklahoma City (OK): University of Oklahoma Health Sciences Center).
Maruyama, F., Kobata, M., Kurokawa, K., Nishida, K., Sakurai, A., Nakano, K., et al. (2009). Comparative genomic analyses of Streptococcus mutans provide insights into chromosomal shuffling and species-specific content. BMC Genomics 10:358. doi: 10.1186/1471-2164-10-358
McCracken, G. H. (1973). Group B streptococci: The new challenge in neonatal infections. J. Pediatr. 82, 703–706. doi: 10.1016/S0022-3476(73)80603-1
McMillan, D. J., Sanderson-Smith, M. L., Smeesters, P. R., and Sriprakash, K. S. (2013b). Molecular markers for the study of streptococcal epidemiology. Curr. Top. Microbiol. Immunol. 368, 29–48. doi: 10.1007/82_2012_278
McMillan, D. J., Drèze, P.-A., Vu, T., Bessen, D. E., Guglielmini, J., Steer, A. C., et al. (2013a). Updated model of group A Streptococcus M proteins based on a comprehensive worldwide study. Clin. Microbiol. Infect. Off. Publ. Eur. Soc. Clin. Microbiol. Infect. Dis. 19, E222–E229. doi: 10.1111/1469-0691.12134
Mercenier, A., Pouwels, P. H., and Chassy, B. M. (1994). ““Genetic engineering of lactobacilli, leuconostocs and Streptococcus thermophilus,”,” in Genetics and Biotechnology of Lactic Acid Bacteria, eds M. J. Gasson and W. M. De Vos (Dordrecht: Springer Netherlands), 252–293. doi: 10.1007/978-94-011-1340-3_6
Mian, G. F., Godoy, D. T., Leal, C. A. G., Yuhara, T. Y., Costa, G. M., and Figueiredo, H. C. P. (2009). Aspects of the natural history and virulence of S. agalactiae infection in Nile tilapia. Vet. Microbiol. 136, 180–183. doi: 10.1016/j.vetmic.2008.10.016
Mitchell, J. (2011). Streptococcus mitis: walking the line between commensalism and pathogenesis. Mol. Oral Microbiol. 26, 89–98. doi: 10.1111/j.2041-1014.2010.00601.x
Mohammadi, J. S., and Dhanashree, B. (2012). Streptococcus pseudopneumoniae: an emerging respiratory tract pathogen. Indian J. Med. Res. 136, 877–880.
Mojica, F. J. M., Díez-Villaseñor, C., García-Martínez, J., and Almendros, C. (2009). Short motif sequences determine the targets of the prokaryotic CRISPR defence system. Microbiol. Read. Engl. 155, 733–740. doi: 10.1099/mic.0.023960-0
Mojica, F. J. M., Díez-Villaseñor, C., García-Martínez, J., and Soria, E. (2005). Intervening sequences of regularly spaced prokaryotic repeats derive from foreign genetic elements. J. Mol. Evol. 60, 174–182. doi: 10.1007/s00239-004-0046-3
Moore, J. E., Mason, C. K., Coulter, W. A., McCarron, P., Leggett, P., Montgomery, J., et al. (2008). Comparison of clustered, regularly interspaced short palindrome repeats (CRISPRs) in viridans streptococci (Streptococcus gordonii, S. mutans, S. sanguinis, S. thermophilus) and in S. pneumoniae. Br. J. Biomed. Sci. 65, 104–108. doi: 10.1080/09674845.2008.11978109
Moschetti, G., Blaiotta, G., Aponte, M., Catzeddu, P., Villani, F., Deiana, P., et al. (1998). Random amplified polymorphic DNA and amplified ribosomal DNA spacer polymorphism: powerful methods to differentiate Streptococcus thermophilus strains. J. Appl. Microbiol. 85, 25–36. doi: 10.1046/j.1365-2672.1998.00461.x
Mosterd, C., and Moineau, S. (2020). Characterization of a Type II-A CRISPR-Cas System in Streptococcus mutans. mSphere 5:20. doi: 10.1128/mSphere.00235-20
Musher, D. M. (1992). Infections Caused by Streptococcus pneumoniae: Clinical Spectrum, Pathogenesis, Immunity, and Treatment. Clin. Infect. Dis. 14, 801–809. doi: 10.1093/clinids/14.4.801
Nagaoka, S. (2019). Yogurt Production. Methods Mol. Biol. Clifton NJ 1887, 45–54. doi: 10.1007/978-1-4939-8907-2_5
Nakano, K., Lapirattanakul, J., Nomura, R., Nemoto, H., Alaluusua, S., Grönroos, L., et al. (2007). Streptococcus mutans clonal variation revealed by multilocus sequence typing. J. Clin. Microbiol. 45, 2616–2625. doi: 10.1128/JCM.02343-06
Nho, S. W., Hikima, J., Cha, I. S., Park, S. B., Jang, H. B., Castillo, C. S., et al. (2011). Complete Genome Sequence and Immunoproteomic Analyses of the Bacterial Fish Pathogen Streptococcus parauberis. J. Bacteriol. 193, 3356–3366. doi: 10.1128/JB.00182-11
Niu, L., Hu, S., Lu, S., Lai, X.-H., Yang, J., Jin, D., et al. (2018). Isolation and characterization of Streptococcus respiraculi sp. nov. from Marmota himalayana (Himalayan marmot) respiratory tract. Int. J. Syst. Evol. Microbiol. 68, 2082–2087. doi: 10.1099/ijsem.0.002806
Niu, L., Lu, S., Hu, S., Jin, D., Lai, X., Yang, J., et al. (2016a). Streptococcus halotolerans sp. nov. isolated from the respiratory tract of Marmota himalayana in Qinghai-Tibet Plateau of China. Int. J. Syst. Evol. Microbiol. 66, 4211–4217. doi: 10.1099/ijsem.0.001337
Niu, L., Lu, S., Hu, S., Jin, D., Lai, X., Yang, J., et al. (2016b). Streptococcusmarmotae sp. nov., isolated from the respiratory tract of Marmota himalayana. Int. J. Syst. Evol. Microbiol. 66, 4315–4322. doi: 10.1099/ijsem.0.001350
Niu, L., Lu, S., Lai, X.-H., Hu, S., Chen, C., Zhang, G., et al. (2017). Streptococcus himalayensis sp. nov., isolated from the respiratory tract of Marmota himalayana. Int. J. Syst. Evol. Microbiol. 67, 256–261. doi: 10.1099/ijsem.0.001609
Nizet, V., Beall, B., Bast, D. J., Datta, V., Kilburn, L., Low, D. E., et al. (2000). Genetic Locus for Streptolysin S Production by Group A Streptococcus. Infect. Immun. 68, 4245–4254. doi: 10.1128/IAI.68.7.4245-4254.2000
Nocard, E., and Mollereau, R. (1887). Sur une mammite contagieuse des vaches laitières. Ann. Inst. Pasteur 1:109.
Nozawa, T., Furukawa, N., Aikawa, C., Watanabe, T., Haobam, B., Kurokawa, K., et al. (2011). CRISPR Inhibition of Prophage Acquisition in Streptococcus pyogenes. PLoS One 6:e19543. doi: 10.1371/journal.pone.0019543
O’Brien, K. L., Beall, B., Barrett, N. L., Cieslak, P. R., Reingold, A., Farley, M. M., et al. (2002). Epidemiology of Invasive Group A Streptococcus Disease in the United States, 1995–1999. Clin. Infect. Dis. 35, 268–276. doi: 10.1086/341409
Okamoto, M., Naito, M., Miyanohara, M., Imai, S., Nomura, Y., Saito, W., et al. (2016). Complete genome sequence of Streptococcus troglodytae TKU31 isolated from the oral cavity of a chimpanzee (Pan troglodytes). Microbiol. Immunol. 60, 811–816. doi: 10.1111/1348-0421.12453
Okura, M., Nozawa, T., Watanabe, T., Murase, K., Nakagawa, I., Takamatsu, D., et al. (2017). A Locus Encoding Variable Defense Systems against Invading DNA Identified in Streptococcus suis. Genome Biol. Evol. 9, 1000–1012. doi: 10.1093/gbe/evx062
Olson, A. B., Kent, H., Sibley, C. D., Grinwis, M. E., Mabon, P., Ouellette, C., et al. (2013). Phylogenetic relationship and virulence inference of Streptococcus Anginosus Group: curated annotation and whole-genome comparative analysis support distinct species designation. BMC Genomics 14:895. doi: 10.1186/1471-2164-14-895
Pan, Y., An, H., Fu, T., Zhao, S., Zhang, C., Xiao, G., et al. (2018). Characterization of Streptococcus pluranimalium from a cattle with mastitis by whole genome sequencing and functional validation. BMC Microbiol. 18:1327. doi: 10.1186/s12866-018-1327-0
Park, S.-N., Lim, Y. K., Shin, J. H., Chang, Y.-H., Shin, Y., Paek, J., et al. (2019). Streptococcus gwangjuense sp. nov., Isolated from Human Pericoronitis. Curr. Microbiol. 76, 799–803. doi: 10.1007/s00284-019-01687-8
Pastuszka, A., Beauruelle, C., Camiade, E., Rousseau, G. M., Moineau, S., Mereghetti, L., et al. (2021). Functional Study of the Type II-A CRISPR-Cas System of Streptococcus agalactiae Hypervirulent Strains. CRISPR J. 4, 233–242. doi: 10.1089/crispr.2020.0145
Patel, S., and Gupta, R. S. (2018). Robust demarcation of fourteen different species groups within the genus Streptococcus based on genome-based phylogenies and molecular signatures. Infect. Genet. Evol. 66, 130–151. doi: 10.1016/j.meegid.2018.09.020
Pompilio, A., Di Bonaventura, G., and Gherardi, G. (2019). An Overview on Streptococcus bovis/Streptococcus equinus Complex Isolates: Identification to the Species/Subspecies Level and Antibiotic Resistance. Int. J. Mol. Sci. 20:20030480. doi: 10.3390/ijms20030480
Pourcel, C., Salvignol, G., and Vergnaud, G. (2005). CRISPR elements in Yersinia pestis acquire new repeats by preferential uptake of bacteriophage DNA, and provide additional tools for evolutionary studies. Microbiology 151, 653–663. doi: 10.1099/mic.0.27437-0
Poyart, C., Réglier-Poupet, H., Tazi, A., Billoët, A., Dmytruk, N., Bidet, P., et al. (2008). Invasive Group B Streptococcal Infections in Infants, France. Emerg. Infect. Dis. 14, 1647–1649. doi: 10.3201/eid1410.080185
Pride, D. T., Sun, C. L., Salzman, J., Rao, N., Loomer, P., Armitage, G. C., et al. (2011). Analysis of streptococcal CRISPRs from human saliva reveals substantial sequence diversity within and between subjects over time. Genome Res. 21, 126–136. doi: 10.1101/gr.111732.110
Raabe, V. N., and Shane, A. L. (2019). Group B Streptococcus (Streptococcus agalactiae). Microbiol. Spectr. 7:2018. doi: 10.1128/microbiolspec.GPP3-0007-2018
Rams, T. E., Feik, D., Mortensen, J. E., Degener, J. E., and Winkelhoff, A. J. (2014). Antibiotic Susceptibility of Periodontal Streptococcus constellatus and Streptococcus intermedius Clinical Isolates. J. Periodontol. 85, 1792–1798. doi: 10.1902/jop.2014.130291
Rauch, B. J., Silvis, M. R., Hultquist, J. F., Waters, C. S., McGregor, M. J., Krogan, N. J., et al. (2017). Inhibition of CRISPR-Cas9 with Bacteriophage Proteins. Cell 168, 150.e–158.e. doi: 10.1016/j.cell.2016.12.009
Reißmann, S., Friedrichs, C., Rajkumari, R., Itzek, A., Fulde, M., Rodloff, A. C., et al. (2010). Contribution of Streptococcus anginosus to Infections Caused by Groups C and G Streptococci, Southern India. Emerg. Infect. Dis. 16, 656–663. doi: 10.3201/eid1604.090448
Rusniok, C., Couvé, E., Cunha, V. D., Gana, R. E., Zidane, N., Bouchier, C., et al. (2010). Genome Sequence of Streptococcus gallolyticus: Insights into Its Adaptation to the Bovine Rumen and Its Ability To Cause Endocarditis. J. Bacteriol. 192, 2266–2276. doi: 10.1128/JB.01659-09
Sampson, T. R., and Weiss, D. S. (2014). CRISPR-Cas systems: new players in gene regulation and bacterial physiology. Front. Cell. Infect. Microbiol. 4:37. doi: 10.3389/fcimb.2014.00037
Sanderson-Smith, M., De Oliveira, D. M. P., Guglielmini, J., McMillan, D. J., Vu, T., Holien, J. K., et al. (2014). A Systematic and Functional Classification of Streptococcus pyogenes That Serves as a New Tool for Molecular Typing and Vaccine Development. J. Infect. Dis. 210, 1325–1338. doi: 10.1093/infdis/jiu260
Schottmuller, H. (1903). Die Artumterscheidung der fur den Menschen pathogenen Streptokokken durch Blutagar. Munch Med Wochenschr 50:849.
Schwank, G., Koo, B.-K., Sasselli, V., Dekkers, J. F., Heo, I., Demircan, T., et al. (2013). Functional repair of CFTR by CRISPR/Cas9 in intestinal stem cell organoids of cystic fibrosis patients. Cell Stem Cell 13, 653–658. doi: 10.1016/j.stem.2013.11.002
Serbanescu, M. A., Cordova, M., Krastel, K., Flick, R., Beloglazova, N., Latos, A., et al. (2015). Role of the Streptococcus mutans CRISPR-Cas Systems in Immunity and Cell Physiology. J. Bacteriol. 197, 749–761. doi: 10.1128/JB.02333-14
Severiano, A., Pinto, F. R., Ramirez, M., and Carriço, J. A. (2011). Adjusted Wallace Coefficient as a Measure of Congruence between Typing Methods. J. Clin. Microbiol. 49, 3997–4000. doi: 10.1128/JCM.00624-11
Shibata, Y., Yamashita, Y., and van der Ploeg, J. R. (2009). The serotype-specific glucose side chain of rhamnose-glucose polysaccharides is essential for adsorption of bacteriophage M102 to Streptococcus mutans. FEMS Microbiol. Lett. 294, 68–73. doi: 10.1111/j.1574-6968.2009.01546.x
Siegman-Igra, Y., Azmon, Y., and Schwartz, D. (2012). Milleri group streptococcus—a stepchild in the viridans family. Eur. J. Clin. Microbiol. Infect. Dis. 31, 2453–2459. doi: 10.1007/s10096-012-1589-7
Simsek, A. D., Sezer, S., Ozdemir, N. F., and Mehmet, H. (2008). Streptococcus vestibularis bacteremia following dental extraction in a patient on long-term hemodialysis: a case report. NDT Plus 1, 276–277. doi: 10.1093/ndtplus/sfn071
Spellerberg, B. (2000). Pathogenesis of neonatal Streptococcus agalactiae infections. Microbes Infect. 2, 1733–1742. doi: 10.1016/s1286-4579(00)01328-9
Staats, J. J., Feder, I., Okwumabua, O., and Chengappa, M. M. (1997). Streptococcus Suis: Past and Present. Vet. Res. Commun. 21, 381–407. doi: 10.1023/A:1005870317757
Sternberg, S. H., Redding, S., Jinek, M., Greene, E. C., and Doudna, J. A. (2014). DNA interrogation by the CRISPR RNA-guided endonuclease Cas9. Nature 507, 62–67. doi: 10.1038/nature13011
Stevens, D. L., and Bryant, A. E. (2016). Impetigo, Erysipelas and Cellulitis. University of Oklahoma Health Sciences Center. Available online at: https://www.ncbi.nlm.nih.gov/sites/books/NBK333408/ (accessed August 10, 2020).
Sturt, A. S., Yang, L., Sandhu, K., Pei, Z., Cassai, N., and Blaser, M. J. (2010). Streptococcus gallolyticus Subspecies pasteurianus (Biotype II/2), a Newly Reported Cause of Adult Meningitis. J. Clin. Microbiol. 48, 2247–2249. doi: 10.1128/JCM.00081-10
Sweeney, C. R., Timoney, J. F., Newton, J. R., and Hines, M. T. (2005). Streptococcus equi Infections in Horses: Guidelines for Treatment, Control, and Prevention of Strangles. J. Vet. Intern. Med. 19, 123–134. doi: 10.1892/0891-6640(2005)19<123:seiihg>2.0.co;2
Tang, B., Gong, T., Zhou, X., Lu, M., Zeng, J., Peng, X., et al. (2019). Deletion of cas3 gene in Streptococcus mutans affects biofilm formation and increases fluoride sensitivity. Arch. Oral Biol. 99, 190–197. doi: 10.1016/j.archoralbio.2019.01.016
Tappe, D., Pukall, R., Schumann, P., Gronow, S., Spiliotis, M., Claus, H., et al. (2009). Streptococcus merionis sp. nov., isolated from Mongolian jirds (Meriones unguiculatus). Int. J. Syst. Evol. Microbiol. 59, 766–770. doi: 10.1099/ijs.0.65823-0
Tettelin, H., Masignani, V., Cieslewicz, M. J., Donati, C., Medini, D., Ward, N. L., et al. (2005). Genome analysis of multiple pathogenic isolates of Streptococcus agalactiae: Implications for the microbial “pan-genome.”. Proc. Natl. Acad. Sci. 102, 13950–13955. doi: 10.1073/pnas.0506758102
Thompson, C. C., Emmel, V. E., Fonseca, E. L., Marin, M. A., and Vicente, A. C. P. (2013). Streptococcal taxonomy based on genome sequence analyses. F1000Research 2:67. doi: 10.12688/f1000research.2-67.v1
Tohya, M., Sekizaki, T., and Miyoshi-Akiyama, T. (2018). Complete Genome Sequence of Streptococcus ruminantium sp. nov. GUT-187T (=DSM 104980T =JCM 31869T), the Type Strain of S. ruminantium, and Comparison with Genome Sequences of Streptococcus suis Strains. Genome Biol. Evol. 10, 1180–1184. doi: 10.1093/gbe/evy078
Tran, M. P., Caldwell-McMillan, M., Khalife, W., and Young, V. B. (2008). Streptococcus intermedius causing infective endocarditis and abscesses: a report of three cases and review of the literature. BMC Infect. Dis. 8:154. doi: 10.1186/1471-2334-8-154
Ullman, R. F., Miller, S. J., Strampfer, M. J., and Cunha, B. A. (1988). Streptococcus mutans endocarditis: report of three cases and review of the literature. Heart Lung J. Crit. Care 17, 209–212.
Van der Mee-Marquet, N., Fourny, L., Arnault, L., Domelier, A.-S., Salloum, M., Lartigue, M.-F., et al. (2008). Molecular Characterization of Human-Colonizing Streptococcus agalactiae Strains Isolated from Throat, Skin, Anal Margin, and Genital Body Sites. J. Clin. Microbiol. 46, 2906–2911. doi: 10.1128/JCM.00421-08
Van der Ploeg, J. R. (2009). Analysis of CRISPR in Streptococcus mutans suggests frequent occurrence of acquired immunity against infection by M102-like bacteriophages. Microbiology 155, 1966–1976. doi: 10.1099/mic.0.027508-0
Verani, J. R., and Schrag, S. J. (2010). Group B streptococcal disease in infants: progress in prevention and continued challenges. Clin. Perinatol. 37, 375–392. doi: 10.1016/j.clp.2010.02.002
Waisberg, J., Matheus, C., de, O., and Pimenta, J. (2002). Infectious endocarditis from Streptococcus bovis associated with colonic carcinoma: case report and literature review. Arq. Gastroenterol. 39, 177–180. doi: 10.1590/s0004-28032002000300008
Wang, H., La Russa, M., and Qi, L. S. (2016). CRISPR/Cas9 in Genome Editing and Beyond. Annu. Rev. Biochem. 85, 227–264. doi: 10.1146/annurev-biochem-060815-014607
Wanger, A. R., and Dunny, G. M. (1987). Identification of a Streptococcus agalactiae protein antigen associated with bovine mastitis isolates. Infect. Immun. 55, 1170–1175. doi: 10.1128/iai.55.5.1170-1175.1987
Weiser, J. N., Ferreira, D. M., and Paton, J. C. (2018). Streptococcus pneumoniae: transmission, colonization and invasion. Nat. Rev. Microbiol. 16, 355–367. doi: 10.1038/s41579-018-0001-8
Whiley, R. A., Beighton, D., Winstanley, T. G., Fraser, H. Y., and Hardie, J. M. (1992). Streptococcus intermedius, Streptococcus constellatus, and Streptococcus anginosus (the Streptococcus milleri group): association with different body sites and clinical infections. J. Clin. Microbiol. 30, 243–244. doi: 10.1128/jcm.30.1.243-244.1992
Wiedenheft, B., and Bondy-Denomy, J. (2017). CRISPR control of virulence in Pseudomonas aeruginosa. Cell Res. 27, 163–164. doi: 10.1038/cr.2017.6
Willcox, M. D., Zhu, H., and Knox, K. W. (2001). Streptococcus australis sp. nov., a novel oral streptococcus. Int. J. Syst. Evol. Microbiol. 51, 1277–1281. doi: 10.1099/00207713-51-4-1277
Williams, A. M., and Collins, M. D. (1990). Molecular taxonomic studies on Streptococcus uberis types I and II. Description of Streptococcus parauberis sp. nov. J. Appl. Bacteriol. 68, 485–490. doi: 10.1111/j.1365-2672.1990.tb02900.x
Wu, C., Liang, B., Gong, Y., Zhang, L., Zou, Y., and Ge, J. (2014). Streptococcus acidominimus causing invasive disease in humans: a case series. J. Med. Case Reports 8:57. doi: 10.1186/1752-1947-8-57
Xie, F., Ye, L., Chang, J. C., Beyer, A. I., Wang, J., Muench, M. O., et al. (2014). Seamless gene correction of β-thalassemia mutations in patient-specific iPSCs using CRISPR/Cas9 and piggyBac. Genome Res. 24, 1526–1533. doi: 10.1101/gr.173427.114
Xiong, Y. Q., Bensing, B. A., Bayer, A. S., Chambers, H. F., and Sullam, P. M. (2008). Role of the serine-rich surface glycoprotein GspB of Streptococcus gordonii in the pathogenesis of infective endocarditis. Microb. Pathog. 45, 297–301. doi: 10.1016/j.micpath.2008.06.004
Yamada, S., Shibasaki, M., Murase, K., Watanabe, T., Aikawa, C., Nozawa, T., et al. (2019). Phylogenetic relationship of prophages is affected by CRISPR selection in Group A Streptococcus. BMC Microbiol. 19:1393. doi: 10.1186/s12866-019-1393-y
Zadoks, R. N., Allore, H. G., Barkema, H. W., Sampimon, O. C., Gröhn, Y. T., and Schukken, Y. H. (2001). Analysis of an Outbreak of Streptococcus uberis Mastitis. J. Dairy Sci. 84, 590–599. doi: 10.3168/jds.S0022-0302(01)74512-2
Zhang, B., Zhang, J., and Sun, L. (2014). Streptococcus iniae SF1: complete genome sequence, proteomic profile, and immunoprotective antigens. PLoS One 9:e91324. doi: 10.1371/journal.pone.0091324
Zhang, J., Abadia, E., Refregier, G., Tafaj, S., Boschiroli, M. L., Guillard, B., et al. (2010). Mycobacterium tuberculosis complex CRISPR genotyping: improving efficiency, throughput and discriminative power of “spoligotyping” with new spacers and a microbead-based hybridization assay. J. Med. Microbiol. 59, 285–294. doi: 10.1099/jmm.0.016949-0
Zheng, P.-X., Chan, Y.-C., Chiou, C.-S., Chiang-Ni, C., Wang, S.-Y., Tsai, P.-J., et al. (2015). Clustered Regularly Interspaced Short Palindromic Repeats Are emm Type-Specific in Highly Prevalent Group A Streptococci. PLS One 10:e0145223. doi: 10.1371/journal.pone.0145223
Zheng, P.-X., Chiang-Ni, C., Wang, S.-Y., Tsai, P.-J., Kuo, C.-F., Chuang, W.-J., et al. (2014). Arrangement and number of clustered regularly interspaced short palindromic repeat spacers are associated with erythromycin susceptibility in emm12, emm75 and emm 92 of group A streptococcus. Clin. Microbiol. Infect. 20, 516–523. doi: 10.1111/1469-0691.12379
Keywords: CRISPR, CRISPR role, CRISPR application, streptococci, pathogenic streptococci
Citation: Lemaire C, Le Gallou B, Lanotte P, Mereghetti L and Pastuszka A (2022) Distribution, Diversity and Roles of CRISPR-Cas Systems in Human and Animal Pathogenic Streptococci. Front. Microbiol. 13:828031. doi: 10.3389/fmicb.2022.828031
Received: 02 December 2021; Accepted: 10 January 2022;
Published: 31 January 2022.
Edited by:
Fumito Maruyama, Hiroshima University, JapanReviewed by:
Meichen Pan, North Carolina State University, United StatesTatiana Castro Abreu Pinto, Federal University of Rio de Janeiro, Brazil
Copyright © 2022 Lemaire, Le Gallou, Lanotte, Mereghetti and Pastuszka. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Philippe Lanotte, cGhpbGlwcGUubGFub3R0ZUB1bml2LXRvdXJzLmZy
 Coralie Lemaire
Coralie Lemaire Brice Le Gallou
Brice Le Gallou Philippe Lanotte
Philippe Lanotte Laurent Mereghetti
Laurent Mereghetti Adeline Pastuszka
Adeline Pastuszka