
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 01 December 2022
Sec. Antimicrobials, Resistance and Chemotherapy
Volume 13 - 2022 | https://doi.org/10.3389/fmicb.2022.1059997
 Shuang Liu1,2†
Shuang Liu1,2† Lei Zhang1,2†
Lei Zhang1,2† Chunlin Feng2
Chunlin Feng2 Jin Zhu3
Jin Zhu3 Anqi Li2
Anqi Li2 Jingxuan Zhao2
Jingxuan Zhao2 Yuan Zhang2
Yuan Zhang2 Mengdi Gao1,2
Mengdi Gao1,2 Weina Shi2
Weina Shi2 Qiaoling Li1,2
Qiaoling Li1,2 Xueya Zhang1,2
Xueya Zhang1,2 Hailin Zhang1
Hailin Zhang1 Teng Xu4
Teng Xu4 Junwan Lu5
Junwan Lu5 Qiyu Bao1,2,5*
Qiyu Bao1,2,5*In this study, we identified and characterized a novel chromosomally-encoded class B metallo-β-lactamase (MBL) gene designated blaWUS-1 in a carbapenem-resistant isolate Myroides albus P34 isolated from sewage discharged from an animal farm. Comparative analysis of the deduced amino acid sequence revealed that WUS-1 shares the highest amino acid similarities with the function-characterized MBLs MUS-1 (AAN63647.1; 70.73%) and TUS-1 (AAN63648.1; 70.32%). The recombinant carrying blaWUS-1 exhibited increased MICs levels against a number of β-lactam antimicrobials such as carbenicillin, ampicillin and imipenem, and β-lactamase inhibitors (clavulanic acid and tazobactam). The metallo-β-lactamase WUS-1 could also hydrolyze these antimicrobials and the hydrolytic activities could be inhibited by EDTA. Genetic context analysis of blaWUS-1 revealed that no mobile genetic element was found in its surrounding region. The plasmid pMA84474 of Myroides albus P34 harbored 6 resistance genes (blaOXA-347, aadS, blaMYO-1, ereD, sul2 and ermF) within an approximately 17 kb multidrug resistance (MDR) region. These genes, however, were all related to mobile genetic elements.
Bacteria of the genus Myroides are peculiar opportunistic and extensively antibiotic-resistant pathogens, which originally belonged to the genus Flavobacterium (Bergey et al., 1923). They are well known for their resistance to antibiotics that are generally used in the treatment of infections caused by gram-negative bacteria (Holmes et al., 1979). Myroides strains have been gradually implicated as important nosocomial pathogens, as infections caused by Myroides spp. have been increasingly reported. The earliest recorded isolation of the species, previously named Flavobacterium breve, was from the human intestine by Stutzer (Bergey et al., 1923). In 1984, Holmes et al. had suggested that F. odoratum should be classified in a separate genus on the basis of its unique phenotypic features (Holmes et al., 1984). Differing from most Flavobacterium strains, being nonsaccharolytic and failing to produce indole, researchers suggested that F. odoratum should be excluded from the emended genus Flavobacterium based on its unique genotypic, chemotaxonomic, and phenotypic data (Bernardet et al., 1996). Vancanneyt et al. conducted a general polyphasic taxonomy analysis and placed F. odoratum into a separate new genus, Myroides (Vancanneyt et al., 1996). The first two species to be delineated were Myroides odoratus comb. nov. and Myroides odoratimimus sp. nov.
Members of the genus Myroides are nonmotile, nonfermentative, obligately aerobic, gram-negative rods that produce a characteristic fruity odor and yellow pigment. Although the strains are widely distributed in environmental ecosystems, such as oil, and fresh and marine waters (Maneerat et al., 2006; Yoon et al., 2006), they have also been previously reported in insect guts (Spiteller et al., 2000). The species has also been reported to be frequently isolated from human urine, wound discharge, sputum, and blood (Ahamed et al., 2018; Cruz-Choappa et al., 2021; O'Neal et al., 2022), but it is not normally a part of the human microflora (Shewan and McMeekin, 1983). Definite cases of human infection caused by this organism are rather rare and generally occur in immunocompromised individuals. However, while infections are sometimes life-threatening, they rarely affect immunocompetent hosts (Maraki et al., 2012). Myroides organisms have been considered to be responsible for cases of pneumonia, endocarditis, urinary tract infections, ventriculitis, bacteremia, and soft-tissue infections (Macfarlane et al., 1985; Ferrer et al., 1995; Green et al., 2001; Crum-Cianflone et al., 2014; Deepa et al., 2014; Faraz et al., 2022). In addition, nosocomial outbreaks such as urinary tract infections and other central venous catheter-associated bloodstream infections caused by Myroides spp. due to contaminated water have been reported (Douce et al., 2008; Ktari et al., 2012; Kutlu et al., 2020). Infections caused by Myroides spp. are not common, but antibiotic therapy is troublesome due to their resistance to virtually all β-lactam antimicrobials, including carbapenems. Reference reports indicated that Myroides strains exhibit variable susceptibility to β-lactam antibiotics (Holmes et al., 1979), with a decreased susceptibility to cephalosporins and carbapenems (Mammeri et al., 2002), and MBLs are found to play a significant role. MBLs differ structurally from the other β-lactamases based on their requirement for active-site zinc ions for activity, and their catalysis proceeds by directly attacking hydroxide ions (Wang et al., 1999; Crowder et al., 2006). MBLs show little or no susceptibility to mechanism-based inhibitors of serine enzymes, such as clavulanic acid, sulbactam or tazobactam, but can be inactivated by metal chelators such as EDTA (Drawz and Bonomo, 2010). MBLs were discovered approximately 50 years ago and were initially identified as clinically irrelevant enzymes that were chromosomally encoded in occasional bacilli such as Bacillus and Aeromonas (Lim et al., 1988; Walsh et al., 1994). They were not taken seriously, resulting in far less study compared to other β-lactamases, until the metalloenzyme IMP-1 was discovered in Japan in the 1990s and the encoding gene was found to be located on an integron. Horizontal transfer is mediated by the movement of integrons and the insertion and removal of gene boxes (Watanabe et al., 1991). At present, a variety of metalloenzymes that can be horizontally transferred have been found (De Oliveira et al., 2020; Li et al., 2022). With the widespread application of carbapenems, the potential spread of metalloenzymes among pathogenic bacteria is a frightening possibility, which emphasizes the importance of understanding their properties.
In this study, the first complete genome of a Myroides albus strain is presented. We characterized WUS-1 by researching its catalytic abilities and impacts on antimicrobial susceptibility. In addition, multiple sequence alignments and phylogenetic analysis were performed to investigate the role of WUS-1 in the intrinsic resistance mechanisms of this unusual opportunistic pathogen to β-lactam antibiotics. We are looking forward to providing valuable information for the study of Myroides drug resistance characteristics.
Myroides albus P34 was isolated from sewage discharged from an animal farm in Wenzhou, Zhejiang Province, China. The sewage samples were inoculated onto the blood plate, and incubated for 24 h at 35°C. Individual colonies of different color and size were randomly selected and preliminarily confirmed by a Vitek-60 microorganism autoanalysis system (bioMe’rieux Corporate, Craponne, France). Further identification was performed by sequencing the 16S rRNA gene and performing ANI analyses. E. coli DH5α and E. coli BL21 were used as the hosts for the resistance gene cloning and protein overexpression, respectively. The blaWUS-1 gene was first cloned into the pUCP24 plasmid, and then the pCold I vector was used to induce cold shock expression of histidine-tagged WUS-1. All the strains and plasmids used in this work are listed in Table 1.
Antimicrobial susceptibility testing was conducted using the agar dilution method following the guidelines of the Clinical and Laboratory Standards Institute (CLSI). Antimicrobials used in this study included carbenicillin, ampicillin, ticarcillin, amoxicillin-clavulanate, ampicillin-sulbactam, piperacillin-tazobactam, cefazolin, cefoxitin, ceftazidime, cefepime, aztreonam, imipenem, meropenem, florfenicol, chloramphenicol, nalidixic acid, tetracycline, streptomycin, kanamycin, netilmicin, gentamicin, and amikacin. When neither the CLSI M100 (32nd Edition, 2022) nor European Enterobacteriaceae Antimicrobial Susceptibility Test Committee (European Committee on Antimicrobial Susceptibility Testing. [EUCAST], 2021) provided exact breakpoints for Myroides spp., the minimum inhibitory concentration (MIC) values were interpreted following the criteria for non-Enterobacteriaceae bacteria (such as A. faecalis, Elizabethkingia or Chryseobacterium; CLSI, 32nd Edition, 2022). E. coli ATCC 25922 was used as a quality control in each MIC test.
Genomic DNA extraction of Myroides albus P34 was performed using the AxyPrep Bacterial Genomic DNA Miniprep kit (Axygen Scientific, Union City, CA, United States) and was sequenced on both the Illumina HiSeq 2500 and PacBio RS II platforms by Shanghai Personal Biotechnology Co., Ltd. (Shanghai, China). The long PacBio reads were initially assembled by Canu (Koren et al., 2017), and then the sequence generated by the Illumina HiSeq 2500 platform was aligned to the primary assembly to correct for possible assembly errors using BWA v0.7.12 (Li and Durbin, 2009) and Pilon (Walker et al., 2014). Potential open reading frames (ORFs) were predicted using Prokka v1.14.6 (Seemann, 2014). Further functional annotation of predicted proteins was performed using DIAMOND (Buchfink et al., 2015) and searching against the NCBI nonredundant protein database with an e-value threshold of 1e-5. Resistance genes were annotated using the Resistance Gene Identifier (RGI) of the Comprehensive Antibiotic Resistance Database (CARD; McArthur et al., 2013). The ANI was calculated using FastANI v1.31 (Jain et al., 2018). Mobile genetic elements (MGEs) were detected using ISFinder (Siguier et al., 2006) and INTEGRALL (Moura et al., 2009). CGView Server (Petkau et al., 2010) was used to depict the circular map of the plasmid. Multiple amino acid sequence alignments and neighbor-joining phylogenetic tree constructions for WUS-1 and other class B1 MBLs sharing the closest amino acid sequence similarities to WUS-1 were performed using MAFFT (Katoh and Standley, 2013) and MEGAX (Kumar et al., 2018), respectively. Linear maps of genomic structure and structural comparisons of multiple genomes were portrayed using genoPlotR (Guy et al., 2010). The promoter region of blaWUS-1 was predicted by BPROM.1 The molecular weight and pI value of WUS-1 were predicted using the Expasy ProtParam Tool,2 and the putative signal peptide cleavage site was predicted by SignalP 5.0 (Almagro Armenteros et al., 2019).
The strain Myroides albus P34 was screened for carbapenemase production by the modified Hodge test, in the meantime, the mCIM and eCIM tests following the CLSI M100 (32nd Edition, 2022) specifications were performed for carbapenemase phenotypic detection.
Amplification of the chromosomal blaWUS-1 gene with its promoter region was performed using primers (Table 2) that contained BamHI and HindIII restriction endonuclease adapters at the 5′ ends. The PCR product was eluted from the agarose gel, digested with the restriction endonucleases BamHI and HindIII (Takara Bio, Inc., Dalian, China), and ligated into the pUCP24 vector using the T4 DNA ligase cloning kit (Takara Bio, Inc., Dalian, China). Recombinant plasmids were introduced into E. coli DH5α by the calcium chloride method. A single transformant was selected on a Luria-Bertani (LB) agar plate containing 40 mg/L gentamicin, and the cloned DNA fragment of the recombinant plasmid was confirmed by both PCR and PCR product Sanger sequencing (Shanghai Sunny Biotechnology Co., Ltd., Shanghai, China). Furthermore, antimicrobial susceptibility testing for the transformants was carried out as described above.
The PCR amplification product without a signal peptide but containing a thrombin cleavage site and a pair of flanking restriction endonuclease adapters (a BamHI site and a thrombin cleavage site were included in the orf-blaWUS-1-F forward primer, and a HindIII site was included in the orf-blaWUS-1-R reverse primer; shown in Table 2) was digested with BamHI and HindIII and then cloned into plasmid pCold I (digested with the same restriction enzymes). The recombinant plasmids were subsequently transformed into E. coli BL21 by the calcium chloride method, and the transformants were selected on LB agar plates containing 100 mg/L ampicillin. The transformant was further screened by PCR and sequencing and cultured in LB liquid medium with 100 mg/L ampicillin at 37°C overnight. Then, the overnight culture was diluted in LB liquid medium at a ratio of 1:100 and further cultured until it reached an OD600 between 0.6 and 0.8. Protein expression was induced by incubating for 12 h at 16°C with isopropyl-β-d-thiogalactoside (IPTG; Sigma Chemicals Co., St. Louis, MO, United States) at a final concentration of 1 mM. Cells were harvested and resuspended and then disrupted sonically. The supernatant containing the β-lactamase was purified by a BeyoGold His-tag Purification Resin kit (Beyotime, Shanghai, China) according to the manufacturer’s instructions. Then, the His-tag was removed by incubating with thrombin (GenScript, Nanjing, China) for 12 h at 16°C. Finally, protein purification kits were applied to remove the free His-tag from the incubated protein. The purity and relative molecular mass of enzymes were checked using SDS–PAGE analysis, and the protein concentration was determined spectrophotometrically with a BCA Protein Assay Kit (Beyotime, Shanghai, China).
The enzymatic activities of purified WUS-1 were measured by a continuous spectrophotometric assay with a SpectraMax multifunctional microplate reader (M5, Molecular Devices, America). The rates of hydrolysis were calculated by monitoring the absorbance drop in a 200 μL reaction mixture containing 10 mM phosphate buffer (pH 7.4) and 50 μM ZnCl2 at 30°C (Mammeri et al., 2002). Consequent data were analyzed by GraphPad Prism 9 software (GraphPad Software, San Jose, CA, United States) using nonlinear regression of initial reaction rates with the Michaelis–Menten equation to calculate the steady-state kinetic parameters (kcat and Km). The enzyme kinetic parameters were calculated following the formula.
Where v is velocity, Vmax is maximum velocity when all the enzymes are complexed to the substrate, [S] is substrate concentration and Km is Michaelis–Menten constant. The wavelengths are shown in Supplementary Table S1. To investigate the effects of inhibitors, the enzyme was preincubated with various concentrations of EDTA for 5 min at 30°C before the addition of substrate (100 μM imipenem; Mammeri et al., 2002). Fifty percent inhibitory concentrations (IC50) were required to reduce the hydrolysis of 100 μM imipenem by 50%, and the results were determined by nonlinear regression in Prism software (Chen et al., 2019).
The colony of Myroides albus P34 is medium sized with fruity odor and yellow pigment. It is positive for oxidase, urease and phosphatase, but negtative for catabolizing glucose, lactose, maltose, mannitol, sucrose, aesculin, or indole. It was preliminary identified as Myroides spp. The 16S rRNA gene of isolate P34 had the closest relationship with that of Myroides albus BIT-d1 (MK734183.1) with 100% coverage and 100% identity. ANI analysis showed that it shared the highest identity (99.38%) with the Myroides albus strain BIT-d1 Scaffold1 (NZ_WMJX01000001). Therefore, this isolate was designated Myroides albus P34. The complete genome of Myroides albus P34 consisted of a 3,701,500 bp chromosome and one circular plasmid of approximately 84,474 bp in length. The chromosome and the plasmid encoded 3,218 and 95 coding sequences (CDSs) with an average GC content of 34.18 and 31.07%, respectively (Supplementary Table S2). Sequence homology analysis showed that the plasmid shared the highest similarity to the plasmid p63039 of Myroides odoratimimus PR63039 (CP013691.1) at 91% coverage and 99.71% identity. Similar to most strains of the genus Myroides, in vitro susceptibility tests showed that Myroides albus P34 exhibited resistance to all antimicrobials tested except for chloramphenicol, with especially high MIC levels for carbapenems (imipenem >512 mg/L and meropenem >512 mg/L, Table 3). The modified Hodge test and the mCIM and eCIM tests (zone diameter of 6 mm for the mCIM test and 20 mm for the eCIM test with a zone diameter difference of 14 mm) were all positive, suggesting metallo-β-lactamase production. To investigate whether any novel MBL gene was encoded in the Myroides albus P34 genome, we checked the whole genome annotation result and found that in addition to one plasmid-encoded, function-characterized MBL gene (blaMYO-1), the P34 genome also harbored six chromosomally encoded hypothetical MBL genes. We analyzed the sequence structures of the predicted MBL genes and found that one (finally designated blaWUS-1) contained conserved motifs of an Ambler class B1 β-lactamase. We then cloned the gene and determined its function.
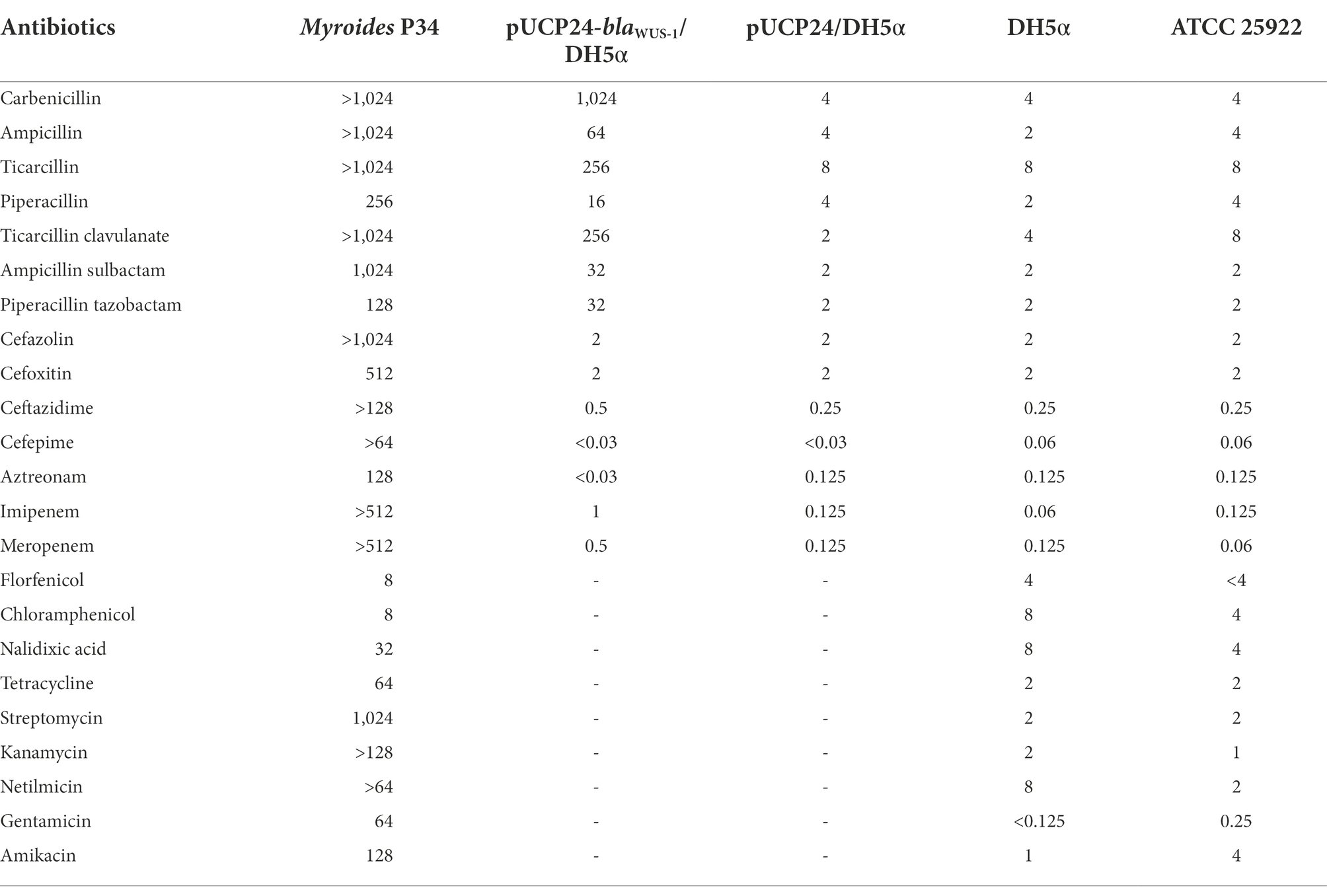
Table 3. MICs of 22 antimicrobials for P34, the recombinant carrying blaWUS-1 and the control strains (mg/L).
The blaWUS-1 gene is 741 bp in length and encodes a predicted protein of 246 amino acids with a calculated molecular mass of ca. 28.64 kDa. The predicted cleavage site of the signal peptide is found between positions 19 and 20 (alanine and glutamine residues). After removal of the signal peptide, the enzyme is a ca. 26.33 kDa protein with a predicted pI value of 7.09. Nine functionally characterized protein sequences with the highest similarity (ranging from 38.36–70.73%) to WUS-1 were retrieved from the NCBI nonredundant (NR) protein database, and they were all class B1 β-lactamases, of which the β-lactamase MUS-1 (AAN63647.1) ranked the first. The phylogenetic tree for these proteins was constructed and showed that WUS-1 is most closely related to MUS-1 from M. odoratimimus CIP 103073 (100% coverage and 70.73% identity), TUS-1 from M. odoratus CIP 103105 (100% coverage and 70.32% identity) and MUS-2 from M. odoratimimus 35a (100% coverage and 69.92% identity; Figure 1). Alignment result of the deduced amino acid sequence of WUS-1 with the other 4 closest relatives (identities ranging from 42.33–70.73%) is shown in Figure 2. It showed that they shared two strictly conserved motifs of the subclass B1 metallo-β-lactamases that interact with the Zn2+ cofactor or with the water molecule located in the active site. The Zn1 binding site, also known as the 3H site, contains three histidine residues, His94, His96 and His157, while the ligands for the Zn2 or DCH site include Asp98, Cys176 and His218 (Garau et al., 2004).
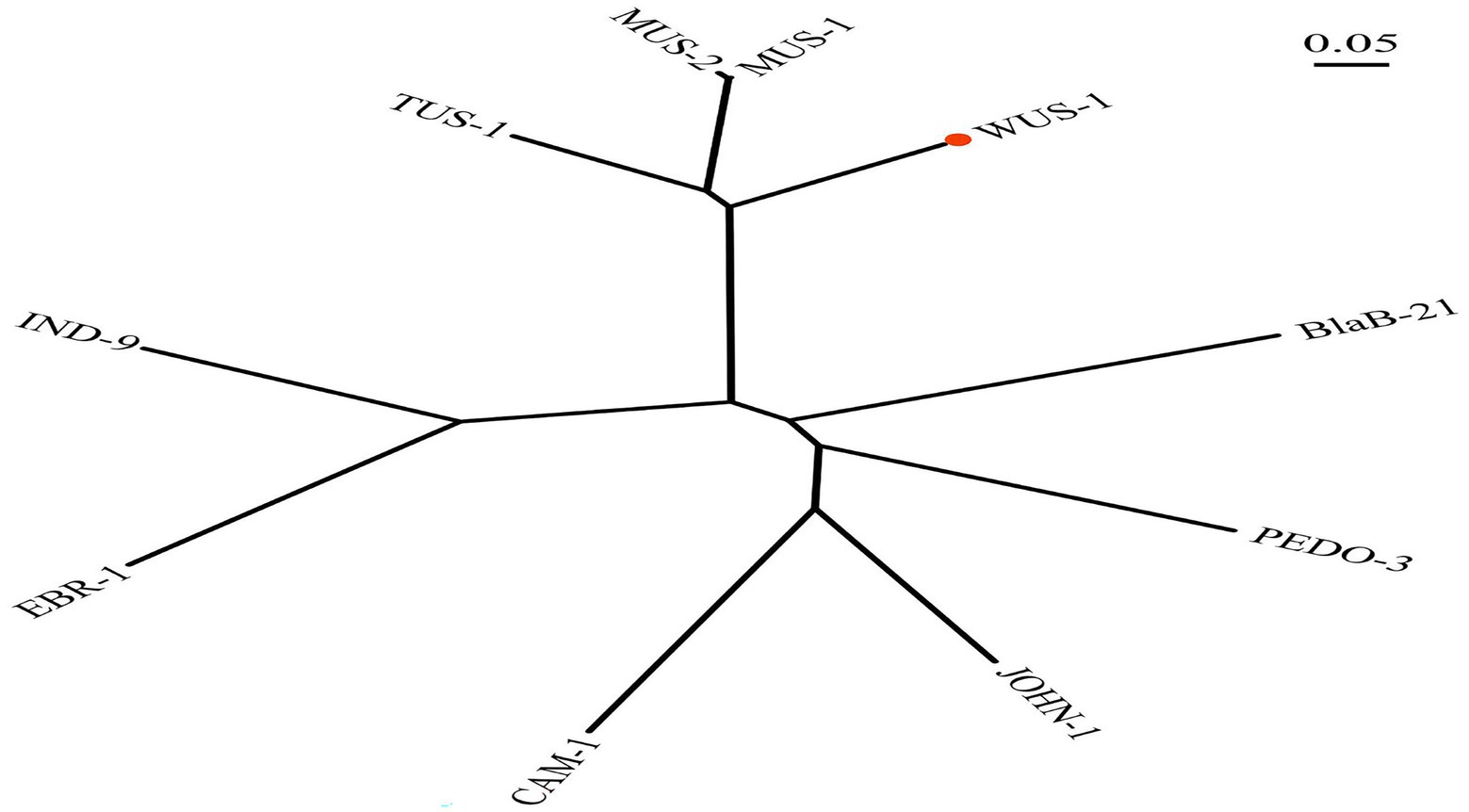
Figure 1. Phylogenetic tree showing the relationship of WUS-1 to other class B β-lactamases. WUS-1 from this study is marked with a red dot. The sequences and their accession numbers are as follows: MUS-1 (AAN63647.1), MUS-2 (AKN19901.1), TUS-1 (AAN63648.1), IND9 (ACZ65153.1), EBR-1 (AAN32638.1), CAM-1 (AVX51087.1), JOHN-1 (AAK38324.1), PEDO-3 (AJP77076.1), and BlaB-21 (AIL46641.1).

Figure 2. Amino acid alignment of WUS-1 with other function-characterized class B β-lactamases. Dots indicate amino acid residues identical to those of WUS-1. Amino acid residues that may be involved in Zn2+ binding are shaded in black. The predicted signal sequence cleavage site for WUS-1 is indicated by a black arrow. The sequences and their accession numbers are as follows: MUS-1 (AAN63647.1), TUS-1 (AAN63648.1), MUS-2 (AKN19901.1), and JOHN-1 (AAK38324.1).
When searching for WUS-1 homologous proteins (>70% amino acid similarity) in the NCBI NR database, 23 functionally uncharacterized sequences (percent identities between 70.2 and 98.37%) were retrieved (Figure 3), all of which were from the Myroides species. The closest relative of WUS-1 was a hypothetical class B enzyme from Myroides sp. Loew2-1 (WP_160339331.1), and they shared an identity of 98.37% with a coverage of 100%.
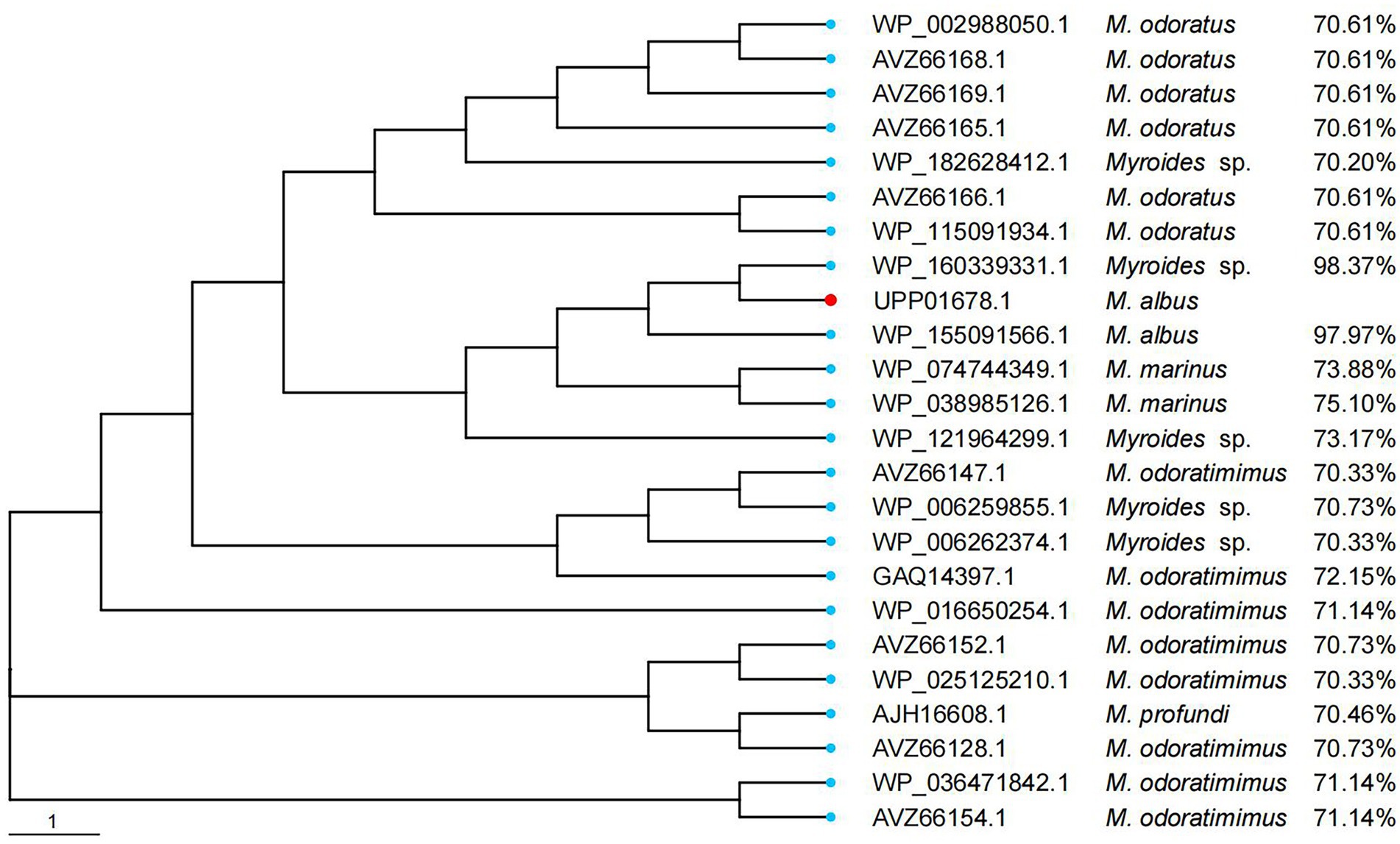
Figure 3. Phylogenetic analysis of WUS-1 with other putative class B β-lactamases (≥70% amino acid similarity). WUS-1 from this study is represented with a red dot.
The recombinant strain pUCP24-blaWUS-1/DH5α showed increased MIC levels for ampicillin (16-fold), carbenicillin (256-fold), ticarcillin (32-fold), imipenem (8-fold) and meropenem (4-fold) in comparison with the control strains (DH5α carrying the vector pUCP24), whereas no MIC level change was observed for expanded spectrum cephalosporins or aztreonam (Table 3). Clavicularulanic acid and sulbactam did not restore the activity of β-lactams. WUS-1 displayed extremely low catalytic efficiencies (kcat/Km) for cefazolin and cefoxitin (Table 4). In accordance with other subclass B1 β-lactamases, the catalytic efficiencies of WUS-1 for cephalosporins were lower than those of penicillins or carbapenems (Mammeri et al., 2002; Vessillier et al., 2002; Naas et al., 2003). The highest catalytic efficiency was observed with ampicillin (kcat/Km rate of 10.06 μM−1·s−1). The catalytic efficiency for carbenicillin (1.93 μM−1·s−1) was lower than that of carbapenems. Carbapenems behaved as good substrates, and the high kcat/Km ratios observed with imipenem (3.99 μM−1·s−1) and meropenem (3.24 μM−1·s−1) always resulted from very high turnover rates. Cefoxitin (0.32 μM−1·s−1) and cefazolin (0.57 μM−1·s−1) were poorly hydrolyzed by WUS-1, and aztreonam was not hydrolyzed at all. The result of the β-lactamase activity inhibition analysis, as measured by the IC50 (50% inhibitory concentration), demonstrated that the activity of WUS-1 was inhibited by EDTA (IC50 of 71.28 μM for WUS-1).
The blaWUS-1 gene was located on the chromosome of Myroides albus P34. To compare the genetic environment of blaWUS-1 with those of homologous genes, a comparative genomic analysis was carried out on a nucleotide sequence of approximately 21 kb in length containing the upstream and downstream flanking regions of the blaWUS-1 gene. Five chromosome segments with blaWUS-1 equivalent genes that shared the highest sequence similarities in the center were found from the NCBI database (similarities ranging between 78.89 and 81.01%), and they were all from Myroides spp. strains (Figure 4). The genes upstream and downstream of the blaWUS-1-related region were quite different from those sequences carrying counterpart genes. However the orfD-blaWUS-1-orfP locus was relatively conserved and flanked by a pair of 9 bp imperfect inverted repeats (IRs), suggesting that sequence variation might result from rearrangement mediated by the inverted repeats.
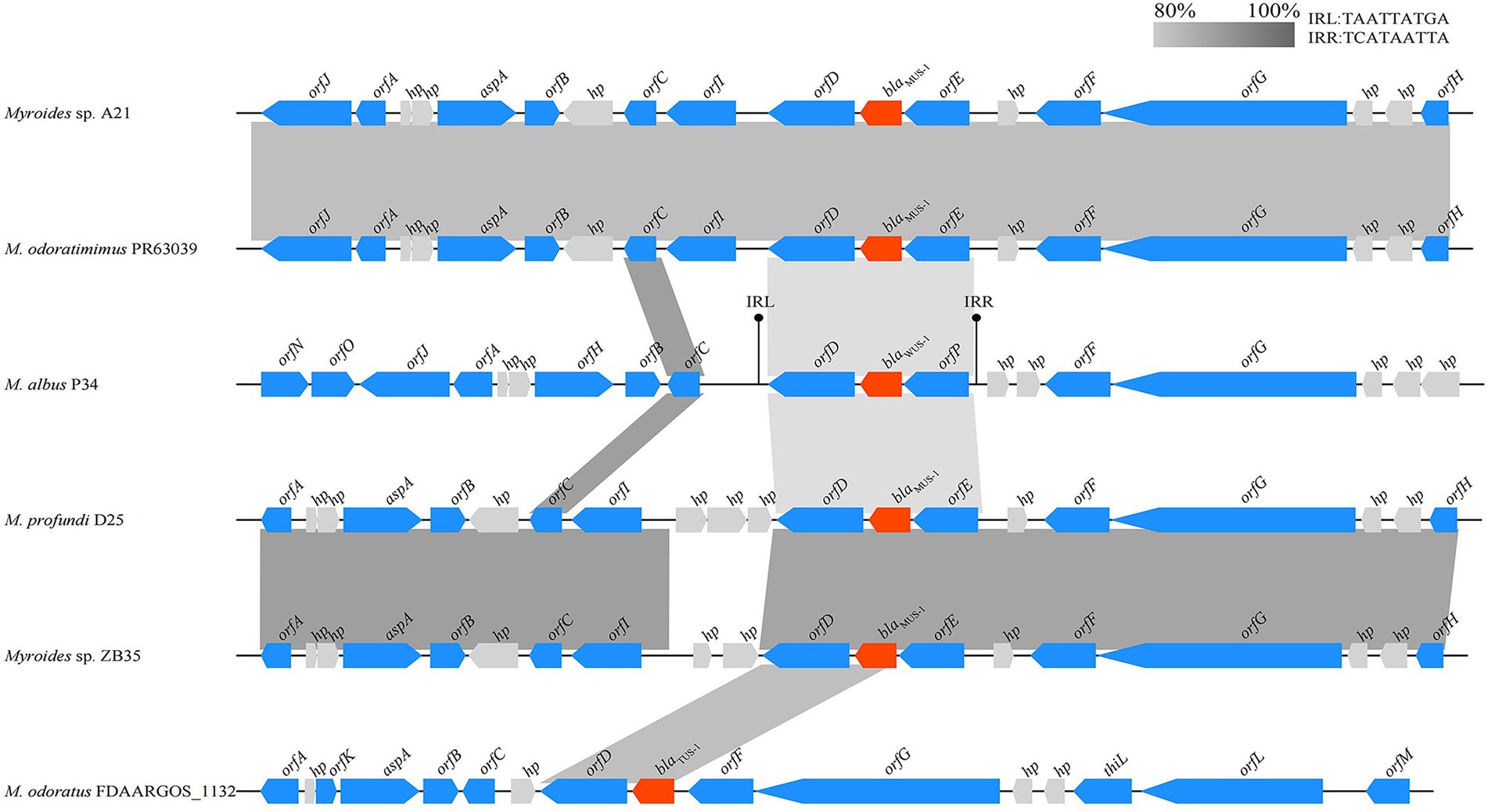
Figure 4. Comparison of the genetic environment of the blaWUS-1 gene with those sequences carrying its homologous genes. Genes are shown as arrows and colored based on gene function classification. Genes without functional annotation are colored in gray. Shading denotes homologous regions between them. The accession numbers are as follows: Myroides sp. A21 chromosome (CP010327.1), Myroides odoratimimus PR63039 chromosome (NZ_CP013690.1), M. profundi D25 chromosome (NZ_CP010817.1), Myroides sp. ZB35 chromosome (NZ_CP017769.1) and M. odoratus FDAARGOS_1132 chromosome (CP068107.1).
Analyzing the complete genome sequence of Myroides albus P34, we found that the isolate contained a circular plasmid designated pMA84474, which shared the highest identity with the plasmid p63039 of Myroides odoratimimus PR63039 (CP013691.1, 91% coverage and 99.71% identity) retrieved from the NCBI nucleotide database. The annotation result showed that there were six drug-resistance genes and a series of mobile genetic elements densely distributed in the multidrug resistance (MDR) region (20.2–37.5 kb) of pMA84474 (Figure 5), and the six resistance genes included two β-lactam resistance genes (blaOXA-347 and blaMYO-1), one aminoglycoside drug resistance gene (aadS), two macrolide resistance genes (ereD and ermF), and one sulfonamide resistance gene (sul2). Comparative genomic analysis showed that the MDR region of pMA84474 shared the highest nucleotide sequence similarity with sequences in the plasmid p63039 (CP013691.1, 59.0% coverage and 93.69% identity) and the chromosomes of Myroides odoratimimus PR63039 (CP013690.1, 77.0% coverage and 94.83% identity) and Myroides odoratimimus G13 (CP037427.1, 66.0% coverage and 99.56% identity). The five resistance genes were clustered in a mobile genetic element-related unit flanked by a pair of IS91 family transposase ISWz1 sequences (ISWz1-aadS-blaMYO-1-IS4-ereD-ermF-blaOXA-347-intI-ISWz1). IS91 is the prototype element of a family of bacterial insertion sequences and is able to perform one-ended transposition with high efficiency, which raises the possibility of IS91-mediated gene spreading without the need to assemble a compound transposon (Garcillan-Barcia and de la Cruz, 2002).
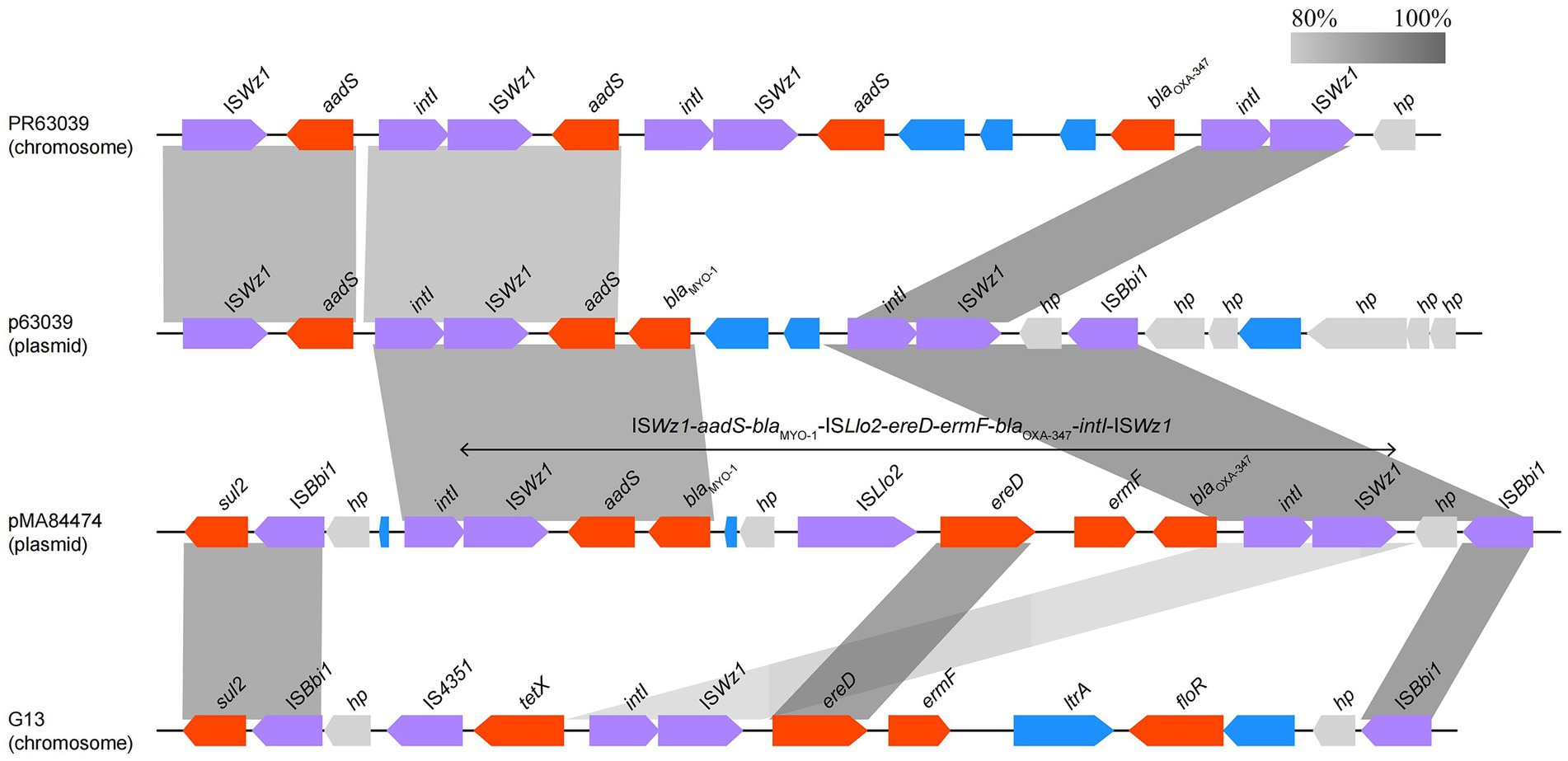
Figure 5. Comparison of the MDR region of pMA84474 with the related sequences. Genes are denoted by arrows and colored according to gene function classification. Shading indicates the homologous regions. The sequences and accession numbers are Myroides odoratimimus PR63039 chromosome (NZ_CP013690.1), Myroides odoratimimus p63039 plasmid (NZ_CP013691.1), and Myroides odoratimimus G13 chromosome (NZ_CP037427.1).
In this study, we identified a novel chromosomal class B metallo-β-lactamase gene, blaWUS-1, in the environmental Myroides albus isolate P34. Phylogenetic analysis showed that WUS-1 establishes a new branch in the class B β-lactamase family.
MBLs are Zn2+-dependent β-lactamases. They are classified as class B β-lactamases based on the Ambler molecular structure classification (Ambler, 1980) or as group 3 according to the Bush-Jacoby-Medeiros functional classification (Bush et al., 1995). MBLs can be further divided based on either structure (subclasses B1, B2, and B3; Garau et al., 2004, Frere et al., 2005, Galleni et al., 2001) or function (subgroups 3a, 3b, and 3c; Rasmussen and Bush, 1997). Sequence analysis revealed that WUS-1 contains two zinc ion binding sites and belongs to subclass B1 (Frere et al., 2005). Furthermore, WUS-1 demonstrated high catalytic efficiencies with penicillin and had kcat/Km values comparable to those of imipenem. Taking into consideration the biochemical criteria established by Rasmussen and Bush (Rasmussen and Bush, 1997), WUS-1 belongs to functional subgroup 3a. Most MBLs display an extremely broad substrate profile and are able to inactivate most clinically useful β-lactam antibiotics (Crowder et al., 2006). There are two Zn2+ in the active site of MBLs in subgroups B1 and B3, which can hydrolyze penicillins, cephalosporin and carbapenem antibiotics. The active site of MBLs in the B2 subgroup contains one Zn2+, which is dedicated to hydrolyzing carbapenem antibiotics (Rasmussen and Bush, 1997; Garau et al., 2004). Despite only sharing 70.73% amino acid sequence identity with the function-characterized B1 metallo-β-lactamases, WUS-1 had key conserved residues of the subclass B1 enzymes (Palzkill, 2013). The blaWUS-1 gene exhibits similar drug resistance phenotypes with the other two subclass B1 β-lactamase genes blaMUS-1 and blaTUS-1, which all showed resistance to carbapenems and semisynthetic penicillins and could not be inhibited by mechanism-based inhibitors (Mammeri et al., 2002). However, the MIC levels of the cloned blaWUS-1 gene against the carbapenems (imipenem and meropenem) were higher (increased by 4- and 2-fold, respectively) than those of the blaMUS-1 or blaTUS-1 genes (Mammeri et al., 2002), but different from these two enzymes, WUS-1 did not affect MIC levels against cefazolin and cefoxitin, which may be because the activity in vivo was simply too low to promote activity in vitro. Kinetic analysis showed that WUS-1 presented catalytic behavior similar to that of the closely related class B1 metallo-β-lactamases, which was in accordance with their detected MICs against the β-lactam antimicrobials.
In addition to the novel metallo-β-lactamase gene blaWUS-1 encoded by the chromosome, this isolate also has two β-lactamase genes, blaMYO-1 and blaOXA-347, carried on a plasmid. No mobile genetic element (MGE) was found in the surrounding areas of the blaWUS-1 gene, except for a pair of IRs. In contrast to the blaWUS-1 gene, the two β-lactamase genes together with a number of other resistance genes on the plasmid are all related to mobile genetic elements. All these findings suggest the possible transmission of resistance genes among bacteria of different species or genera.
In this study, we reported a complete genome sequence of a Myroides albus isolate P34 and characterized a novel chromosomally-encoded class B β-lactamase gene, blaWUS-1, which confers resistance to some β-lactam antimicrobials, including carbapenems. Carbapenems are used extensively in clinical treatment, and growing resistance to these drugs is a unignorable problem. The production of metalloenzymes plays an important role in the resistance to carbapenems. It remains to be further studied whether these bacteria are reservoirs of such a variety of metalloenzyme genes. Discovering novel resistance genes will be helpful for explaining at least part of their intrinsic resistance to antimicrobials and for finding ways to treat infectious diseases more effectively.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found at: https://www.ncbi.nlm.nih.gov/genbank/, CP102754, CP102755, and UPP01678.1.
HZ, JL, TX, and QB conceived and designed the experiments. SL, LZ, AL, JZ, YZ, MG, WS, QL, and JXZ performed the experiments. SL CF, JL, and QB contributed to data analysis and interpretation. SL, LZ, TX, and QB contributed to drafting of the manuscript. All authors contributed to the article and approved the submitted version.
This study was supported by the Science and Technology Project of Wenzhou City, China (N20210001), Zhejiang Provincial Natural Science Foundation of China (LY19C060002 and LQ17H190001), National Natural Science Foundation of China (81960381 and 81973382), and the Science and Technology Project of Jinhua City, China (2022-2-013).
The authors would like to acknowledge all study participants and individuals who contributed to this study.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.1059997/full#supplementary-material
Ahamed, I., Annapandian, V. M., and Muralidhara, K. D. (2018). Myroides odoratimimus urinary tract infection. Saudi J. Kidney Dis. Transpl. 29, 1220–1222. doi: 10.4103/1319-2442.243957
Almagro Armenteros, J. J., Tsirigos, K. D., Sonderby, C. K., Petersen, T. N., Winther, O., Brunak, S., et al. (2019). SignalP 5.0 improves signal peptide predictions using deep neural networks. Nat. Biotechnol. 37, 420–423. doi: 10.1038/s41587-019-0036-z
Ambler, R. P. (1980). The structure of beta-lactamases. Philos. Trans. R. Soc. Lond. Ser. B Biol. Sci. 289, 321–331. doi: 10.1098/rstb.1980.0049
Bergey, D. H., Harrison, F. C., Breed, R. S., Hammer, B. W., and Huntoon, F. M. (1923). Bergey’s manual of determinative bacteriology. 1st Edn. Baltimore: The Williams & Wilkins.
Bernardet, J. F., Segers, P., Vancanneyt, M., Berthe, F., Kersters, K., and Vandamme, P. (1996). Cutting a Gordian knot: emended classification and description of the genus Flavobacterium, emended description of the family Flavobacteriaceae, and proposal of Flavobacterium hydatis nom. nov. (Basonym, Cytophaga aquatilis Strohl and Tait 1978). Int. J. Syst. Bacteriol. 46, 128–148. doi: 10.1099/00207713-46-1-128
Buchfink, B., Xie, C., and Huson, D. H. (2015). Fast and sensitive protein alignment using DIAMOND. Nat. Methods 12, 59–60. doi: 10.1038/nmeth.3176
Bush, K., Jacoby, G. A., and Medeiros, A. A. (1995). A functional classification scheme for beta-lactamases and its correlation with molecular structure. Antimicrob. Agents Chemother. 39, 1211–1233. doi: 10.1128/AAC.39.6.1211
Chen, Q., Zhou, W., Qian, C., Shen, K., Zhu, X., Zhou, D., et al. (2019). OXA-830, a novel chromosomally encoded extended-Spectrum class D beta-lactamase in Aeromonas simiae. Front. Microbiol. 10:2732. doi: 10.3389/fmicb.2019.02732
Crowder, M. W., Spencer, J., and Vila, A. J. (2006). Metallo-beta-lactamases: novel weaponry for antibiotic resistance in bacteria. Acc. Chem. Res. 39, 721–728. doi: 10.1021/ar0400241
Crum-Cianflone, N. F., Matson, R. W., and Ballon-Landa, G. (2014). Fatal case of necrotizing fasciitis due to Myroides odoratus. Infection 42, 931–935. doi: 10.1007/s15010-014-0626-0
Cruz-Choappa, R., Lopez, E., Ocara, M., and Leiva, Y. (2021). Myroides odoratimimus, a rare cause of soft tissue infection and osteomyelitis. Case report. Rev. Chil. Infectol. 38, 297–299. doi: 10.4067/S0716-10182021000200297
De Oliveira, D. M. P., Forde, B. M., Kidd, T. J., Harris, P. N. A., Schembri, M. A., Beatson, S. A., et al. (2020). Antimicrobial resistance in ESKAPE pathogens. Clin. Microbiol. Rev. 33, e00181-19. doi: 10.1128/CMR.00181-19
Deepa, R., Venkatesh, K. G., Parveen, J. D., Banu, S. T., and Jayalakshmi, G. (2014). Myroides odoratus and Chryseobacterium indologenes: two rare isolates in the immunocompromised. Indian J. Med. Microbiol. 32, 327–330. doi: 10.4103/0255-0857.136592
Douce, R. W., Zurita, J., Sanchez, O., and Cardenas Aldaz, P. (2008). Investigation of an outbreak of central venous catheter-associated bloodstream infection due to contaminated water. Infect. Control Hosp. Epidemiol. 29, 364–366. doi: 10.1086/533543
Drawz, S. M., and Bonomo, R. A. (2010). Three decades of beta-lactamase inhibitors. Clin. Microbiol. Rev. 23, 160–201. doi: 10.1128/CMR.00037-09
European Committee on Antimicrobial Susceptibility Testing. [EUCAST] (2021). Breakpoint Tables for interpretation of MICs and Zone diameters. Available at: http://www.eucast.org.
Faraz, A., Fathima, K., Kazmi, S. Y., Al Motery, A. S., Ghaffar, U. B., and Farhan, M. A. (2022). Recurrent urinary tract infection in a renal transplant patient by pan-resistant Myroides Spp. J. Coll. Physicians Surg. Pak. 32, S34–S36. doi: 10.29271/jcpsp.2022.Supp1.S34
Ferrer, C., Jakob, E., Pastorino, G., and Juncos, L. I. (1995). Right-sided bacterial endocarditis due to Flavobacterium odoratum in a patient on chronic hemodialysis. Am. J. Nephrol. 15, 82–84. doi: 10.1159/000168806
Frere, J. M., Galleni, M., Bush, K., and Dideberg, O. (2005). Is it necessary to change the classification of beta-lactamases? J. Antimicrob. Chemother. 55, 1051–1053. doi: 10.1093/jac/dki155
Galleni, M., Lamotte-Brasseur, J., Rossolini, G. M., Spencer, J., Dideberg, O., Frere, J. M., et al. (2001). Standard numbering scheme for class B beta-lactamases. Antimicrob. Agents Chemother. 45, 660–663. doi: 10.1128/AAC.45.3.660-663.2001
Garau, G., Garcia-Saez, I., Bebrone, C., Anne, C., Mercuri, P., Galleni, M., et al. (2004). Update of the standard numbering scheme for class B beta-lactamases. Antimicrob. Agents Chemother. 48, 2347–2349. doi: 10.1128/AAC.48.7.2347-2349.2004
Garcillan-Barcia, M. P., and de la Cruz, F. (2002). Distribution of IS91 family insertion sequences in bacterial genomes: evolutionary implications. FEMS Microbiol. Ecol. 42, 303–313. doi: 10.1111/j.1574-6941.2002.tb01020.x
Green, B. T., Green, K., and Nolan, P. E. (2001). Myroides odoratus cellulitis and bacteremia: case report and review. Scand. J. Infect. Dis. 33, 932–934. doi: 10.1080/00365540110077065
Guy, L., Kultima, J. R., and Andersson, S. G. (2010). genoPlotR: comparative gene and genome visualization in R. Bioinformatics 26, 2334–2335. doi: 10.1093/bioinformatics/btq413
Holmes, B., Owen, R. J., and McMeekin, T. A. (1984). “Genus Flavobacterium (Bergey, Harrison, Breed, Hammer and Huntoon 1923),” in In the Bergey's manual of systematic bacteriology. eds. N. R. Kring and J. G. Holt (Baltimore: Williams and Wilkins), 353–361.
Holmes, B., Snell, J. J., and Lapage, S. P. (1979). Flavobacterium odoratum: a species resistant to a wide range of antimicrobial agents. J. Clin. Pathol. 32, 73–77. doi: 10.1136/jcp.32.1.73
Jain, C., Rodriguez, R. L., Phillippy, A. M., Konstantinidis, K. T., and Aluru, S. (2018). High throughput ANI analysis of 90K prokaryotic genomes reveals clear species boundaries. Nat. Commun. 9:5114. doi: 10.1038/s41467-018-07641-9
Katoh, K., and Standley, D. M. (2013). MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30, 772–780. doi: 10.1093/molbev/mst010
Koren, S., Walenz, B. P., Berlin, K., Miller, J. R., Bergman, N. H., and Phillippy, A. M. (2017). Canu: scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res. 27, 722–736. doi: 10.1101/gr.215087.116
Ktari, S., Mnif, B., Koubaa, M., Mahjoubi, F., Ben Jemaa, M., Mhiri, M. N., et al. (2012). Nosocomial outbreak of Myroides odoratimimus urinary tract infection in a Tunisian hospital. J. Hosp. Infect. 80, 77–81. doi: 10.1016/j.jhin.2011.09.010
Kumar, S., Stecher, G., Li, M., Knyaz, C., and Tamura, K. (2018). MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35, 1547–1549. doi: 10.1093/molbev/msy096
Kutlu, H. H., Avci, M., Dal, T., Ari, O., and Durmaz, R. (2020). A healthcare-associated outbreak of urinary tract infections due to Myroides odoratimimus. Jpn. J. Infect. Dis. 73, 421–426. doi: 10.7883/yoken.JJID.2019.536
Li, H., and Durbin, R. (2009). Fast and accurate short read alignment with burrows-wheeler transform. Bioinformatics 25, 1754–1760. doi: 10.1093/bioinformatics/btp324
Li, Y., Zhu, Y., Zhou, W., Chen, Z., Moran, R. A., Ke, H., et al. (2022). Alcaligenes faecalis metallo-beta-lactamase in extensively drug-resistant Pseudomonas aeruginosa isolates. Clin. Microbiol. Infect. 28, 880.e1–880.e8. doi: 10.1016/j.cmi.2021.11.012
Lim, H. M., Pene, J. J., and Shaw, R. W. (1988). Cloning, nucleotide sequence, and expression of the Bacillus cereus 5/B/6 beta-lactamase II structural gene. J. Bacteriol. 170, 2873–2878. doi: 10.1128/jb.170.6.2873-2878.1988
Macfarlane, D., Baumthureen, P., and Crandon, I. (1985). Flavobacterium odoratum ventriculitis treated with intraventricular cefotaxime. J. Inf. Secur. 11, 233–238. doi: 10.1016/s0163-4453(85)93228-1
Mammeri, H., Bellais, S., and Nordmann, P. (2002). Chromosome-encoded beta-lactamases TUS-1 and MUS-1 from Myroides odoratus and Myroides odoratimimus (formerly Flavobacterium odoratum), new members of the lineage of molecular subclass B1 metalloenzymes. Antimicrob. Agents Chemother. 46, 3561–3567. doi: 10.1128/AAC.46.11.3561-3567.2002
Maneerat, S., Bamba, T., Harada, K., Kobayashi, A., Yamada, H., and Kawai, F. (2006). A novel crude oil emulsifier excreted in the culture supernatant of a marine bacterium, Myroides sp. strain SM1. Appl. Microbiol. Biotechnol. 70, 254–259. doi: 10.1007/s00253-005-0050-6
Maraki, S., Sarchianaki, E., and Barbagadakis, S. (2012). Myroides odoratimimus soft tissue infection in an immunocompetent child following a pig bite: case report and literature review. Braz. J. Infect. Dis. 16, 390–392. doi: 10.1016/j.bjid.2012.06.004
McArthur, A. G., Waglechner, N., Nizam, F., Yan, A., Azad, M. A., Baylay, A. J., et al. (2013). The comprehensive antibiotic resistance database. Antimicrob. Agents Chemother. 57, 3348–3357. doi: 10.1128/AAC.00419-13
Moura, A., Soares, M., Pereira, C., Leitao, N., Henriques, I., and Correia, A. (2009). INTEGRALL: a database and search engine for integrons, integrases and gene cassettes. Bioinformatics 25, 1096–1098. doi: 10.1093/bioinformatics/btp105
Naas, T., Bellais, S., and Nordmann, P. (2003). Molecular and biochemical characterization of a carbapenem-hydrolysing beta-lactamase from Flavobacterium johnsoniae. J. Antimicrob. Chemother. 51, 267–273. doi: 10.1093/jac/dkg069
O'Neal, M., Labay, C. E., Harris, J. E., Musick, W. L., Cernoch, P. L., Grimes, K. A., et al. (2022). Extensively drug-resistant Myroides odoratus in critically ill patients: A case series and literature review. Case Rep. Infect. Dis. 2022, 6422861–6422867. doi: 10.1155/2022/6422861
Palzkill, T. (2013). Metallo-beta-lactamase structure and function. Ann. N. Y. Acad. Sci. 1277, 91–104. doi: 10.1111/j.1749-6632.2012.06796.x
Petkau, A., Stuart-Edwards, M., Stothard, P., and Van Domselaar, G. (2010). Interactive microbial genome visualization with GView. Bioinformatics 26, 3125–3126. doi: 10.1093/bioinformatics/btq588
Rasmussen, B. A., and Bush, K. (1997). Carbapenem-hydrolyzing beta-lactamases. Antimicrob. Agents Chemother. 41, 223–232. doi: 10.1128/AAC.41.2.223
Seemann, T. (2014). Prokka: rapid prokaryotic genome annotation. Bioinformatics 30, 2068–2069. doi: 10.1093/bioinformatics/btu153
Shewan, J. M., and McMeekin, T. A. (1983). Taxonomy (and ecology) of Flavobacterium and related genera. Annu. Rev. Microbiol. 37, 233–252. doi: 10.1146/annurev.mi.37.100183.001313
Siguier, P., Perochon, J., Lestrade, L., Mahillon, J., and Chandler, M. (2006). ISfinder: the reference Centre for bacterial insertion sequences. Nucleic Acids Res. 34, D32–D36. doi: 10.1093/nar/gkj014
Spiteller, D., Dettner, K., and Bolan, W. (2000). Gut bacteria may be involved in interactions between plants, herbivores and their predators: microbial biosynthesis of N-acylglutamine surfactants as elicitors of plant volatiles. Biol. Chem. 381, 755–762. doi: 10.1515/BC.2000.096
Vancanneyt, M., Segers, P., Torck, U., Hoste, B., Bernardet, J. F., Vandamme, P., et al. (1996). Reclassification of Flavobacterium odoratum (Stutzer 1929) strains to a new genus, Myroides, as Myroides odoratus comb. nov. and Myroides odoratimimus sp. nov. Int. J. Syst. Bacteriol. 46, 926–932. doi: 10.1099/00207713-46-4-926
Vessillier, S., Docquier, J. D., Rival, S., Frere, J. M., Galleni, M., Amicosante, G., et al. (2002). Overproduction and biochemical characterization of the Chryseobacterium meningosepticum BlaB metallo-beta-lactamase. Antimicrob. Agents Chemother. 46, 1921–1927. doi: 10.1128/AAC.46.6.1921-1927.2002
Walker, B. J., Abeel, T., Shea, T., Priest, M., Abouelliel, A., Sakthikumar, S., et al. (2014). Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS One 9:e112963. doi: 10.1371/journal.pone.0112963
Walsh, T. R., Hall, L., Assinder, S. J., Nichols, W. W., Cartwright, S. J., MacGowan, A. P., et al. (1994). Sequence analysis of the L1 metallo-β-lactamase from Xanthomonas maltophilia. Biochim. Biophys. Acta - Gene Sturct. Exp. 1218, 199–201. doi: 10.1016/0167-4781(94)90011-6
Wang, Z., Fast, W., Valentine, A. M., and Benkovic, S. J. (1999). Metallo-β-lactamase: structure and mechanism. Curr. Opin. Chem. Biol. 3, 614–622. doi: 10.1016/s1367-5931(99)00017-4
Watanabe, M., Iyobe, S., Inoue, M., and Mitsuhashi, S. (1991). Transferable imipenem resistance in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 35, 147–151. doi: 10.1128/AAC.35.1.147
Keywords: metallo-β-lactamase, Myroides albus, blaWUS-1, kinetic analysis, antimicrobial resistance
Citation: Liu S, Zhang L, Feng C, Zhu J, Li A, Zhao J, Zhang Y, Gao M, Shi W, Li Q, Zhang X, Zhang H, Xu T, Lu J and Bao Q (2022) Characterization and Identification of a novel chromosome-encoded metallo-β-lactamase WUS-1 in Myroides albus P34. Front. Microbiol. 13:1059997. doi: 10.3389/fmicb.2022.1059997
Received: 02 October 2022; Accepted: 14 November 2022;
Published: 01 December 2022.
Edited by:
Shangshang Qin, Zhengzhou University, ChinaReviewed by:
Hong-Ning Wang, Sichuan University, ChinaCopyright © 2022 Liu, Zhang, Feng, Zhu, Li, Zhao, Zhang, Gao, Shi, Li, Zhang, Zhang, Xu, Lu and Bao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Qiyu Bao, YmFvcXlAZ2Vub21pY3MuY24=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.