- 1Biotechnology Division, CSIR-Institute of Himalayan Bioresource Technology, Palampur, Himachal Pradesh, India
- 2Academy of Scientific and Innovative Research (AcSIR), Ghaziabad, Uttar Pradesh, India
Present study reports a novel and robust GH9 processive endoglucanase β-1,4-glucanase from Bacillus sp. PCH94 (EGaseBL) with thermo-alkali stable properties. The EGaseBL gene was cloned in pET-28b(+) and expressed in Escherichia coli BL21(DE3) cells. The recombinant protein was purified 94-fold with a yield of 67.8%. The biochemical characterization revealed an active enzyme at a wide pH (4.0–10.0) and temperature (4–100°C). It showed a Km and Vmax of 1.10 mg/ml and 208.24 IU/mg, respectively, using β-glucan as a substrate. The EGaseBL showed dual activities for endoglucanase (134.17 IU/mg) and exoglucanase (28.76 IU/mg), assayed using substrates β-glucan and Avicel, respectively. The enzyme is highly stable in neutral and alkaline pH and showed a half-life of 11.29 h, and 8.31 h in pH 7.0 and 9.0, respectively. The enzyme is also compatible with commercial detergents (Tide, Surf, Ghadi, Raj, and Healing tree) of the Indian market and retained > 85% enzyme activity. Concisely, robustness, extreme functionality, and detergent compatibility endorse EGaseBL as a potential bioresource for the detergent industry, in addition to its implications for the bioethanol industry.
Highlights
– Cloning, expression, and purification of putative novel GH9 family β-1,4-glucanase.
– Processive endoglucanase with CBM3 domain and bi-functional (endo/exo) activity.
– Broad pH-temperature active and stable enzyme.
– Compatible with commercial detergent powders.
Introduction
The global detergent market is growing at a CAGR of 6.3% after an estimated value of 154.34 billion in 2021.1 The detergent industry has experienced a revolution with the use of enzyme additives such as cellulase, amylase, and protease in detergents (Cavaco-Paulo et al., 1997; Niyonzima, 2019). These hydrolytic enzymes led to detergent formulations with efficient stain removal properties. Among them, alkaline cellulases have been the key additives in detergents for fabric softening and color brightness (Cavaco-Paulo, 1998; Niyonzima, 2019). In addition to the detergent industry, microbial enzymes are the hotspot to deal with a global challenge in alternate energy sources (Zhu et al., 2020; Antar et al., 2021). Among cellulases, the endoglucanase (EG; EC 3.2.1.4) is the key enzyme that carries out the breakdown of β-1,4-glycosidic bonds in cellulose chains. They belong to 12 glycosyl hydrolase (GH) superfamilies: 5, 6, 7, 8, 9, 12, 44, 45, 48, 51, 74, and 124 (Kim et al., 2016; Kumar and Naraian, 2018).
Endoglucanases (EGs) are required in various industries like biorefinery, paper, and pulp, and detergent for applications such as biomass conversion, biopulping, deinking, and biopolishing, respectively (Bhat, 2000; Belghith et al., 2001; Chandel et al., 2007, 2012; Kuhad et al., 2011; Kumar et al., 2018; Golgeri et al., 2022). The cellulolytic enzymes depolymerize the cellulosic biomass into simpler sugars, which are further fermented to produce bioethanol (Zhu et al., 2020). Also, in the detergent industry, cellulase assists in the softness, color brightness, and defibring of clothes (Miettinen-Oinonen and Suominen, 2002; Sadhu et al., 2013; Kumar and Naraian, 2018; Niyonzima, 2019). Nevertheless, the different industrial bioprocesses need endoglucanases to work under different conditions. For instance, the biorefinery needs cellulolytic enzymes to work at elevated temperatures and the pulp industry requires acidic cellulases. In contrast, the detergent industry requires an enzyme that works in alkaline environment (Kuhad et al., 2011; Ladeira et al., 2015; Bagewadi et al., 2016).
The endoglucanases act on their substrates through two action mechanisms, processive and non-processive. Most GH9 family EGs are processive, containing a catalytic domain (CD) and a carbohydrate-binding module (CBM; Konar et al., 2022). The CBMs are responsible for the processive nature of EGs (Chiriac et al., 2010). The processive endoglucanases (PEGs) show both endo- exo- bi-functional activities (Lv et al., 2021; Wu et al., 2022). It generates a higher diversity of hydrolysis products than exoglucanases (Wu and Wu, 2020). The first report of PEGs from Thermomonospora fusca was reported in 1993 (Irwin et al., 1993), followed by several others. The majority of PEGs studied so far belong to Clostridium sp., while Paenibacillus, Bacillus, Cellulomonas, Ruminococcus, and Thermobifida are among the other genera (Reverbel-Leroy et al., 1997; Wilson, 2004; Mejia-Castillo et al., 2008; Chiriac et al., 2010; Wu and Wu, 2020; Gavande et al., 2022). The bi-functional PEGs, combined with other cellulases, are more suitable for cellulose-based bioprocess development for bioethanol generation (Akram et al., 2018). The PEGs also have potential applications in deinking and biostoning (Wu et al., 2007; Liu et al., 2017).
PEGs are emerging as game-changing players in the cellulose-based industries. The lower stability and incompetence to work in extreme conditions is the biggest bottleneck of commercial cellulases (Han et al., 2020). Therefore, discovering robust cellulases compatible with multi-industrial applications is of prime necessity. Previously, we have isolated Bacillus sp. PCH94 from compost in the Western Himalayas and studied its biomass hydrolysis potential on damaged rice grain waste (Thakur et al., 2018, 2021). The current study investigated the biochemical characteristics of a recombinant processive EGaseBL. Here, we subjected the purified EGaseBL enzyme to varied pH and temperature conditions to assess its compatibility with industrial bioprocess standards. The multi-dimensional properties of the enzyme are key stipulations for biorefinery, detergent, and textile industries.
Materials and methods
Bioinformatic analysis of β-1,4-glucanase sequence
The Bacillus licheniformis strain PCH94 was isolated in our lab and studied for its cellulolytic potential (Thakur et al., 2018, 2021). The gene EGaseBL was obtained from a whole-genome sequence of Bacillus sp. PCH94 (Thakur et al., 2021) and submitted to the NCBI GenBank database with an accession number OM867537. The NCBI ORF finder program was used to identify the open reading frame in the gene sequence.2 The gene sequence was translated using the ExPASy translate tool.3 The protein sequence identity was confirmed by the NCBI protein BLAST (blastp) tool with the protein databank (PDB) database as a reference. The prediction of signal peptide in protein sequence was performed by SignalP server 5.0. The NCBI conserved domain database (CDD) was used to predict the conserved domains.4 The physicochemical properties like isoelectric point (pI) and molecular weight (MW) of the protein sequence of EGaseBL were identified using ProtParam.5 The phylogenetic tree was constructed to study the evolutionary relationship of EGaseBL protein sequence through Mega 7.0. The protein structure analysis was carried out using the RaptorX Property web server (Wang et al., 2016). The homology model of EGaseBL protein was generated using the SWISS-MODEL server (Waterhouse et al., 2018).
Cloning and heterologous expression of EGaseBL in pET-28b(+)/Escherichia coli BL21(DE3)
The EGaseBL gene was amplified from genomic DNA using gene-specific primers, inserted with restriction enzymes sites (indicated with underline) for NcoI (Forward primer 5′-CATG CCATGG GC ATG AAA GCG CTT TGT TTG GC-3′) and XhoI (Reverse primer 5′-CCG CTCGAG GTA ACC GGG CTC ATG TCC GAA-3′). The PCR to amplify the gene was carried out under the following conditions: initial denaturation at 95°C for 2 min; 35 cycles of 95°C for 30s; 58°C for 30s; 72°C for 2 min; and a final extension at 72°C for 7 min. The amplified gene product was purified using the FavorPrep PCR purification kit (FAVORGEN Biotech Corp, Taiwan) as per the manufacturer’s instructions. The final EGaseBL gene product and pET-28b(+) vector were further subjected to double restriction digestion with Fast digest NcoI and XhoI restriction enzymes. The digested products were purified and ligated using T4 DNA ligase. The ligated product was transformed into competent BL21 (DE3) cells using the heat-shock method. The colonies were isolated on LB plates with 50 μg/ml kanamycin, and positive transformants were screened through the colony PCR technique. The desired cloned gene in positive transformants was also confirmed through T7 promoter-based sequencing in ABI PRISM™ 3,130 × l Genetic Analyzer (Applied Biosystems, United States).
The expression of EGaseBL gene was carried out under optimized conditions, i.e., 1.0 ml of overnight grown seed culture was inoculated in a 50 ml LB broth containing Kanamycin (50 μg/ml) and incubated at 37°C/200 rpm. The culture was grown until OD600 reached 0.4–0.6, supplemented with 0.1 mM Isopropyl β-D-1-Thiogalactopyranoside (IPTG), and transferred to 28°C/200 rpm for 24 h. The cells were harvested, centrifuged, and the pellet was re-suspended in lysis buffer containing Tris–HCl buffer (25 mM/pH 8.0), 300 mM of NaCl, and 5 mM imidazole. The cells were homogenized using an ultrasonic sonicator (Ultrasonic Processor SKL-150D) with 10s on/off, 5 cycles. After sonication, the lysed cells solution was centrifuged at 12000 g for 10 min at 4°C, and cell-free supernatant (CFS) was used as a crude enzyme for further studies. The expression of the desired protein was visualized on a 10% SDS-PAGE.
Enzyme activity assay
The enzyme activity of the EGaseBL enzyme was determined using the DNSA method for reducing sugars (Miller, 1959). The reaction conditions were as follows: incubation temperature: 50°C, time: 60 min, pH: 8.0/25 mM, and substrate: 1.0% (w/v) barley β-glucan (prepared in 25 mM pH 8.0 Tris–HCl buffer), enzyme: substrate ratio-1:2 (v/v). The enzyme-specific activity of β-1,4-glucanase was defined as μ moles of reducing sugar released from β-glucan per minute per mg of the enzyme under the assay conditions. Each reaction with its controls was carried out in triplicates in all the experiments. The observed values were subjected to standard deviation (SD), and ± 2 SD was considered significant.
Purification of recombinant EGaseBL and its proteomic analysis
The EGaseBL protein was expressed and produced in 1.0 liter LB broth media. The HisPur Cobalt superflow resin was used for the purification of recombinant EGaseBL. The chromatography column was equilibrated using a basic buffer containing 25 mM Tris–HCl pH 8.0, 300 mM NaCl, and 5 mM imidazole. The CFS was filtered through a 0.45 μM syringe filter and loaded on the pre-equilibrated column. The CFS was further subjected to IMAC (Immobilized metal affinity chromatography) resin-based purification using HisPur™ cobalt Superflow Agarose. The CFS was allowed to pass through the gravity column twice for the maximum binding. The column was washed twice with the basic buffer using 10 column volumes to remove non-specific proteins. The desired EGaseBL protein bound to the resin was eluted by using elution buffer, i.e., Tris–HCl pH 8.0, 300 mM NaCl, and 150 mM imidazole. The total amount of protein was estimated by using the Bradford assay (Bradford, 1976). The purity of the eluted fraction was visualized on a 10% SDS-PAGE gel.
Biochemical characterization of purified EGaseBL
The one factor at a time (OFAT) scheme was used to study the biochemical characteristics of purified EGaseBL and obtain its highest activity. The reaction pH range was calculated using varied buffer systems consisting of sodium citrate (pH 3.0–5.0), potassium phosphate (pH 6.0–7.0), Tris–HCl (pH 8.0–10.0), and sodium carbonate–bicarbonate (pH 9.0–10.0). The reaction conditions were as follows: temperature: 50°C, time: 60 min, and substrate: 1.0% (w/v) barley β-glucan. The buffer system with the highest specific activity was considered the optimum pH for the reaction. Furthermore, the optimum reaction temperature was assessed under optimized pH in 60 min, and 1.0% (w/v) barley β-glucan substrate. The reactions were incubated at a temperature range of 4–100°C. The enzyme concentration in the reaction was optimized under optimum pH and temperature conditions. The enzyme concentration was varied from 0.1 to 5.0 μg per reaction and incubated for 30 min. Finally, the incubation time was optimized from a varying range from 5 to 30 min under previously optimized conditions.
Assessment of kinetic parameters of purified EGaseBL
The kinetic parameters like Michaelis constant (Km) and maximum reaction velocity (Vmax) were estimated on EGaseBL activity on β-glucan under optimized reaction conditions. The reactions were carried out at 60°C/ pH 7.0 buffer (25 mM Potassium Phosphate). The substrate concentration varied from 0.05 to 0.5% to a constant enzyme concentration. The Michaelis–Menten equation and double-reciprocal Lineweaver–Burk plots were used for calculating Km and Vmax. The multi-substrate-specificity of EGaseBL was also studied on β-glucan, Avicel, filter paper, cellobiose, beechwood xylan, and starch.
pH and thermal stability of purified EGaseBL
The purified enzyme was incubated in various buffer systems for 5 h to investigate the effect of pH on enzyme activity. The three different buffers (25 mM), i.e., Citrate buffer pH 5.0, Potassium Phosphate buffer pH 7.0, and sodium carbonate–bicarbonate pH 9.0, were used. The enzyme reaction was performed every hour, and the half-life (t1/2) of the enzyme at different pH levels was calculated. Similarly, to study the half-life of EGaseBL at different temperatures, the enzyme was incubated at 50, 60, 70, and 80°C.
Detergent stability of purified EGaseBL in commercial detergents
The stability of purified EGaseBL was studied in various domestic Indian detergent powders such as Tide, Surf, Ghadi, Raj, and Healing tree. The detergent solution concentration was kept at 1.0% to stimulate washing conditions. Before enzyme addition, detergents were pre-incubated in boiling water for 10 min to deactivate existing enzymes (Niyonzima, 2019). The enzyme was incubated in a 1.0% detergent solutions for 30 min at room temperature. The enzyme reaction was performed following the incubation, and residual enzyme activity was calculated.
Results and discussion
Genome mining and bioinformatic analysis of EGaseBL gene
The gene EGaseBL consisted of 1,941 bp and was mined from the whole-genome sequence of Bacillus sp. PCH94 (Thakur et al., 2021). The gene’s nucleotide sequence was submitted to NCBI GenBank with the accession number OM867537. The ExPASy translate server revealed that EGaseBL protein comprises 647 amino acids. The NCBI protein blast showed 53.08% similarity in the PDB database. The phylogenetic analysis of the EGaseBL protein sequence with the closest matches in the PDB server was carried out. The EGaseBL made a separate clade to its closest matches with protein sequences of Bacillus pumilus and Bacillus sp. (Figure 1). The percentage (%) similarity and phylogenetic analysis of the protein sequence of EGaseBL endorse it to be a putative novel protein. It has a molecular weight of 72.6 kDa and a theoretical pI of 5.34. The sequence contains 83 negatively and 61 positively charged residues. The instability index (II) is 28.88, classifying this protein as stable. The protein is extracellular in nature, as predicted by SignalP server. Further protein sequence analysis revealed that the protein belongs to the Glycosyl hydrolase-9 (GH9) family as predicted from the SWISS-MODEL server (Figure 2). Also, it contains a carbohydrate-binding module, i.e., CBM3 (Figure 3). In fact, the presence of CBM along with catalytic domains is a key feature of PEGs. The PEGs act as traditional EGs without CBM3 (Kumar et al., 2019). Further, the protein sequence analysis performed by the RaptorX Property web server predicted 34% alpha-helix, 13% beta-sheet, and 52% coil regions (Supplementary Figure S1).
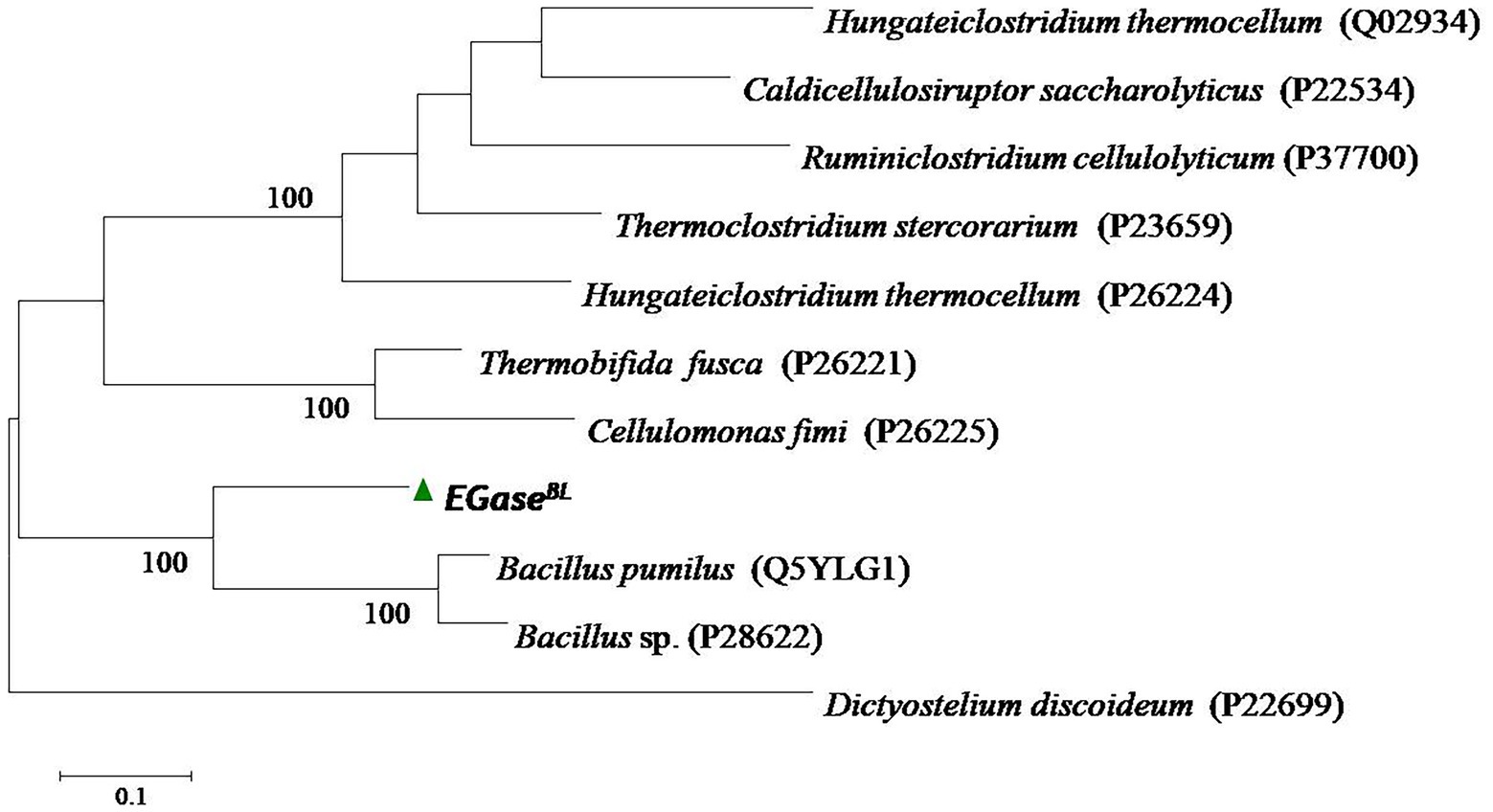
Figure 1. The phylogenetic analysis of the protein sequence of β-1,4-glucanase (EGaseBL) of Bacillus sp. PCH94 with the closest matches in the Protein Data Bank (PDB) database.
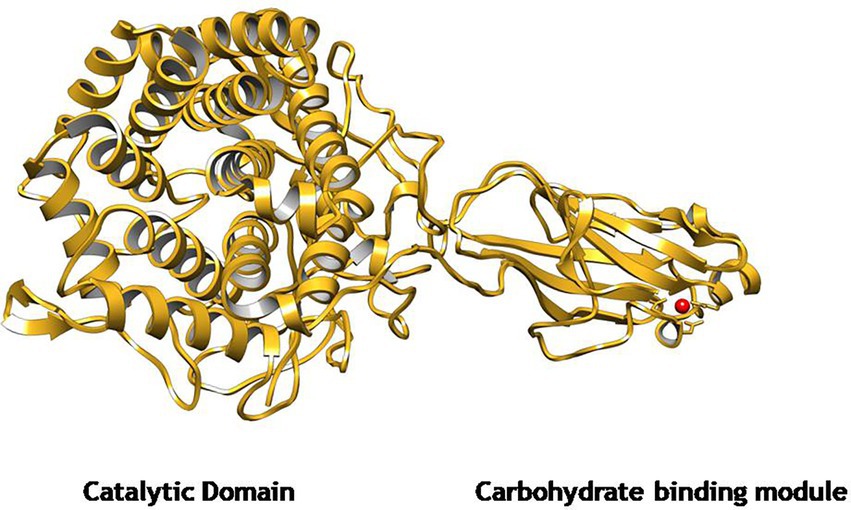
Figure 2. The predicted homology model of GH9 family β-1,4-glucanase (EGaseBL) using Endoglucanase 9G from Clostridium cellulolyticum as a template in SWISS-MODEL server. The structure depicts a catalytic domain (CD) and a carbohydrate-binding module (CBM3).

Figure 3. The conserved domain prediction in the protein sequence of GH9 β-1,4-glucanase using the NCBI database.
Cloning, heterologous expression, and purification of gene encoding β-1,4-glucanase
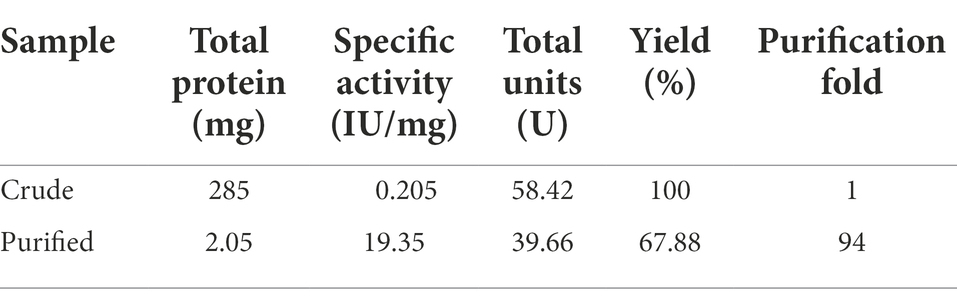
The β-1,4-glucanase (EGaseBL) gene was amplified from the genomic DNA of Bacillus sp. PCH94 using gene-specific primers. The pET-28b(+)-EGaseBL construct was transformed in the expression host E. coli BL21(DE3) (Supplementary Figure S2). The positive transformants with the target gene were induced with 0.1 mM IPTG in LB media at 37°C and expressed under optimized conditions, i.e., 200 rpm/ 28°C. The maximum expression of EGaseBL was observed after 36 h at 28°C. The cell-free supernatant’s (CFS’s) quantitative estimation of β-1,4-glucanase activity showed the specific activity of 0.205 IU/mg on the β-glucan substrate. The use of affinity protein purification resulted in a single-step 94-fold purification of protein with a yield of 67.8%. Under unoptimized enzyme reaction conditions, the purified protein showed 19.35 IU/mg specific activity (Table 1). The purified protein was visualized on 10% SDS-PAGE, which revealed a distinct ~ 72 kDa band of purified EGaseBL (Figure 4).
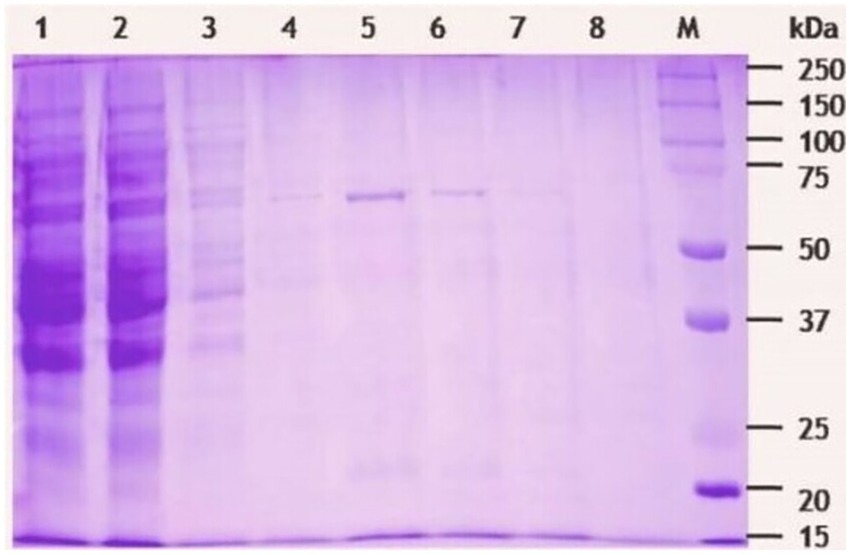
Figure 4. SDS-PAGE (10%) analysis of β-1,4-glucanase (EGaseBL) purified using HisPur Cobalt superflow resin. The lane 1 to 9 represents total lysate, flow through, wash, elute 1(10 mM imidazole), elute 2 (50 mM imidazole), elute 3 (100 mM imidazole), elute 4 (150 mM imidazole), blank, and protein marker, respectively.
Previously, the EGs belonging to different GH families have been cloned from various sources. Earlier, 63.5 and 64 kDa GH5 recombinant endoglucanases from Paenibacillus sp. were purified using the Ni-NTA column, resulting in 22.33 and 44.9% yields with activities of 7.55 and 3.63 IU/mg, respectively (Dhar et al., 2015a,b). A 41.56 kDa GH8 EGs was purified using High-Q and CHT-II column chromatography revealed 2.2 IU/mg activity with 15.9% recovery (Na et al., 2015). Similarly, a 59 kDa GH5 family recombinant EG from Bacillus subtilis was purified using the Ni-NTA column, giving the highest activity of 7.65 IU/mg (Guan et al., 2017). The purification scheme used in this study for EGaseBL has resulted in a higher enzyme recovery, purification fold, and specific activity than in previous studies. The highly purified enzyme can lead to efficient substrate conversion with high specificity.
Biochemical characterization of purified EGaseBL
The buffer pH is a key parameter for optimum activity of enzyme. So, the enzyme activity was examined from pH 4.0 to 10.0. The highest specific activity of 24.06 IU/mg was obtained at pH 7.0 in 25 mM Tris–HCl buffer (Figure 5A). The relative activity of 70 and 64% was observed at pH 5.0 and 9.0, respectively. The activity decreased to ~0.8 and 19% at pH 4.0 and 10.0, respectively. Furthermore, the effect of temperature on EGaseBL activity was studied in a varied temperature range of 4–100°C, with the highest activity of 25.72 IU/mg observed at 60°C (Figure 5B). Relative activity of more than 50% was observed at a temperature range of 30–80°C. Also, 3.9 and 17.8% of relative activity was observed at 4°C and 100°C, respectively. The enzyme load plays a significant role in converting substrate into products. Hence, the effect of varied enzyme concentrations in the reaction was studied. The enzyme concentrations varied from 0.1 to 5.0 μg, and 0.5 μg per reaction was observed as the optimum enzyme concentration to achieve the highest specific activity of 115.7 IU/mg (Figure 5C). Further, the initial reaction rate was studied by optimizing the incubation time of the reaction. The incubation time of 10 to 30 min with 10 min intervals was studied. After 10 min of incubation, the specific activity of 130.6 IU/mg was achieved (Figure 5D). Concisely, biochemical characterization led to optimized reaction conditions, which are as follows: pH 7.0 Tris–HCl buffer, temperature 60°C, enzyme concentration 0.5 μg, and reaction time 10 min. Similarly, there are reports for the recombinant EG with a pH range of 4.0 to 9.0 and a temperature range of 20–70°C (Furtado et al., 2011). The rumen metagenome-derived EG showed a pH range of 4.0–10.0 and a temperature range of 20–70°C (Song et al., 2017). The EG from Thermotoga maritima showed a pH range of 4.0–8.0 (optimum pH 5.0) and a temperature range of 30–100°C (optimum temperature 95°C; Wang et al., 2020). The EGaseBL in the present study has shown a broader window of pH and temperature than the earlier reported studies. Such wider pH and temperature range has definitive advantages in the varied industrial bioprocesses.
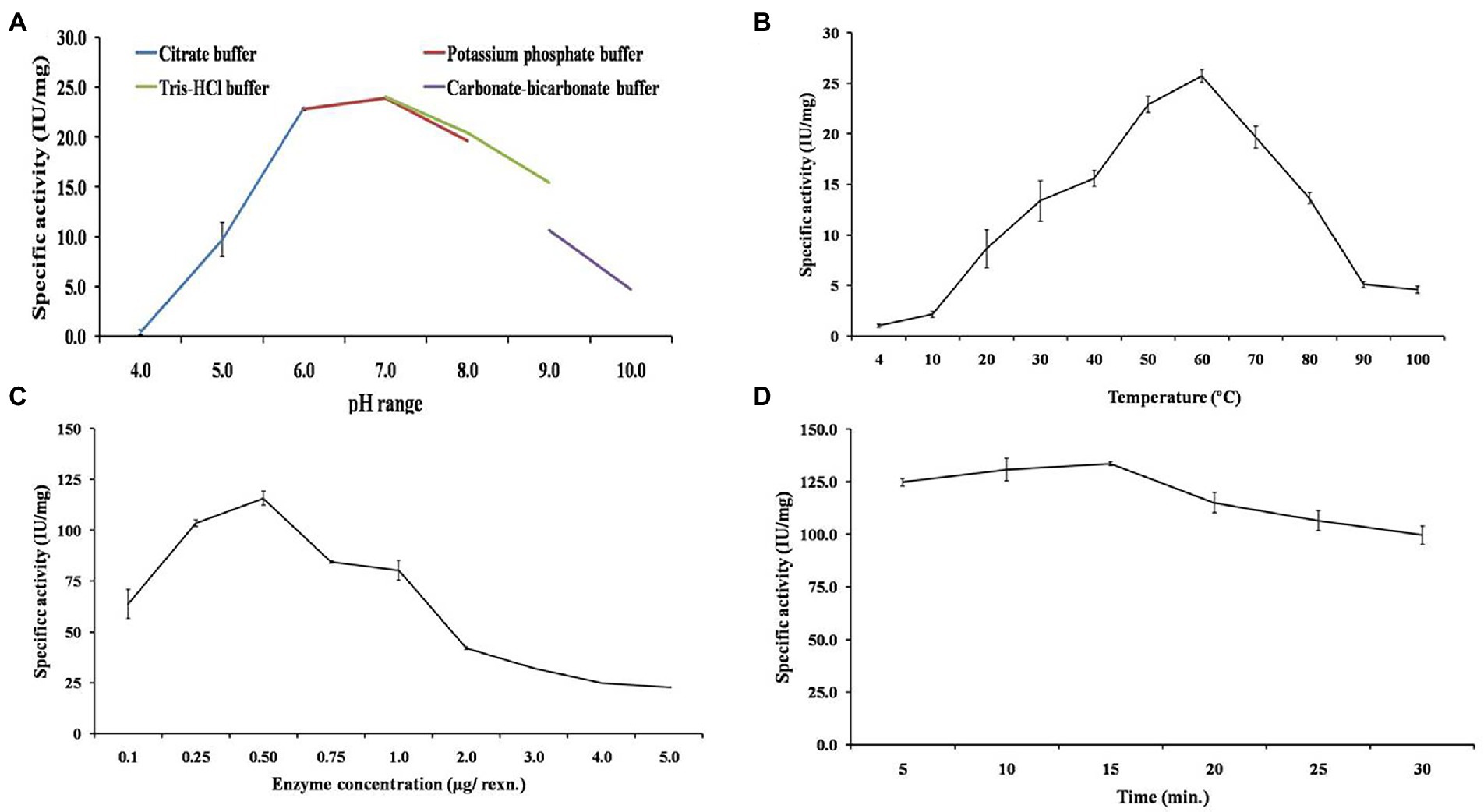
Figure 5. Biochemical characterization of purified β-1,4-glucanase (EGaseBL) to obtain the optimum enzymatic conditions. The various parameters such as (A) optimum pH, (B) optimum temperature, (C) enzyme concentration, and (D) incubation time, were shown.
Investigation of kinetic parameters and substrate-specificity
The Lineweaver–Burk double-reciprocal plot revealed that the EGaseBL follows Michaelis–Menten kinetics for β-glucan (Figure 6A). The Km and Vmax values of purified EGaseBL were 1.10 mg/ml and 208.24 IU/mg, respectively. The EG of Raoultella ornithinolytica showed Km of 8.6 mg/ml and Vmax of 74.9 IU/mg for β-glucan (Scapin et al., 2017). The recombinant EG of Enterobacter sp. had Km and Vmax of 13.5 mg/ml and 109.6 IU/mg, respectively, on β-glucan (Ontañon et al., 2019). A GH8 EG from Bacillus subtilis was estimated for Km (1.78 mg/ml) and Vmax (50.09 IU/mg) on CMC as a substrate (Huang et al., 2021). The EGaseBL has shown low Km and high Vmax values compared to these recent studies. The lower Km suggests a high affinity for β-glucan, representing that it would take less substrate to reach Vmax.
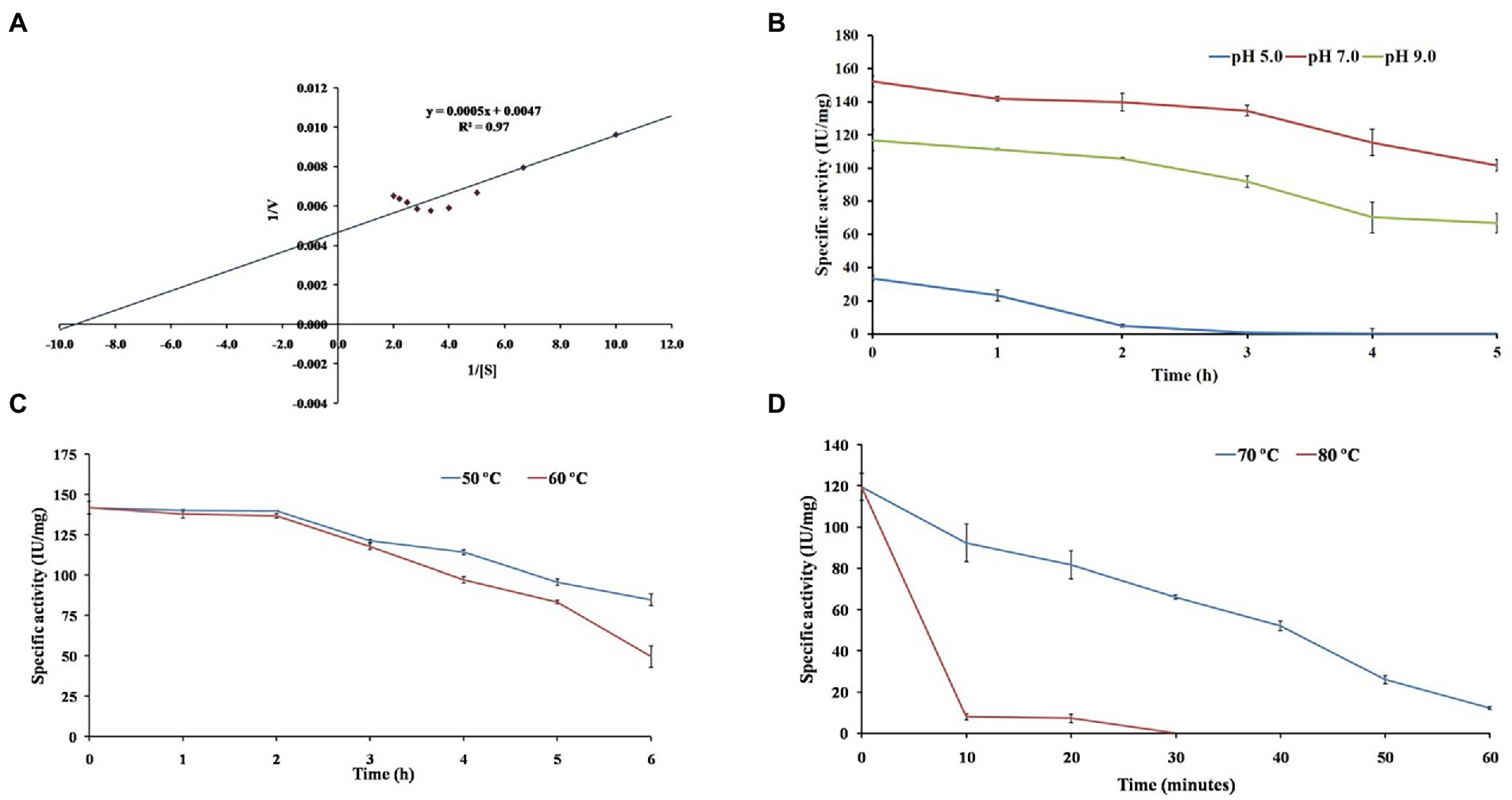
Figure 6. The figure depicts investigating β-1,4-glucanase (EGaseBL) for (A) enzyme kinetics on β-glucan substrate, (B) pH stability at pH 5.0, 7.0 and 9.0, (C) thermal stability at 50 and 60°C, and (D) thermal stability at 70 and 80°C.
The substrate-specificity of purified EGaseBL was studied on polysaccharides like β-glucan, Avicel, beechwood xylan, starch, cellobiose, and filter paper. The enzyme showed a specific activity of 134.17 IU/mg and 28.76 IU/mg on β-glucan and Avicel, respectively. While, with the rest of the other substrates, no enzymatic activity could be detected. The results imply that the enzyme has dual endoglucanase and avicelase/exoglucanase activity. The bi-functional activity profile and presence of CBM3 confirm the processive nature of EGaseBL.
pH and thermostability profile of purified EGaseBL
The EGaseBL has shown wide pH and temperature working range. Therefore, the prolonged effect of pH and temperature on the enzyme was studied. The half-life (t1/2) of 0.80, 11.29, and 8.31 h was observed at pH of 5.0, 7.0, and 9.0 (Figure 6B). The enzyme could retain 90% residual activity after 1.0 h of incubation at pH 7.0 and 9.0. Interestingly, more than 75, and 60% activity at pH 7.0 and 9.0, respectively, were retained even after 5.0 h of incubation. The current results suggested that enzyme has the highest stability at neutral pH and is more stable at alkaline than acidic pH. The stability of EGaseBL in an alkaline environment is suitable for application as an additive in the detergent industry.
In addition to pH, thermal stability is a key factor for EGs, especially for their application in biomass saccharification. Therefore, thermal stability was evaluated by incubating the purified EGaseBL at 50, 60, 70, and 80°C. The half-life (t1/2) of 14.41 h, 8.63 h, 27 min, and > 5 min was observed at 50, 60, 70, and 80°C, respectively (Figures 6C,D). The enzyme showed optimum activity at 60°C and retained > 80% residual activity after 4.0 h of incubation. Previously, a β-1,4-glucanase from Paenibacillus sp. revealed a half-life of 60 min at 60°C (Na et al., 2015). The half-life of 3.0 h at 50°C was reported from the EG of Raoultella ornithinolytica (Scapin et al., 2017). The glucanase from Bacillus sp. lost 50% residual activity after incubation at 60°C for 10 min (Jang et al., 2021). Comparatively, the EGaseBL has shown much higher stability than the β-1,4-glucanases discussed above. Thermostability enables the enzyme to hydrolyze at a higher temperature more efficiently. Hence, it lowers the risk of contamination compared to mesophilic enzymes in a process (Akram et al., 2018).
Detergent compatibility of purified EGaseBL
The EGs are key additives to the enzyme cocktails used in the detergent powders. Most detergent brands instruct to keep clothes pre-soaked in detergent water for 20–30 min before washing. Therefore, we studied the compatibility/stability of purified EGaseBL by incubating it for 30 min in detergent powders available in the local market. The enzyme retained 91.1, 81.5, 85.9, 87.7, and 94.6% activity in Tide, Surf, Ghadi, Raj, and Healing tree, respectively (Figure 7). Previously, the residual activity of 72, 65, and 57% were reported for Ariel, Surf Excel, and Tide; respectively (Sadhu et al., 2013). Thermo-tolerant endoglucanase showed 50% residual activity in Surf Excel, Tide, Ariel, Wheel, and Patanjali detergent powders after 1.0 h (Joshi et al., 2021). The higher stability of EGaseBL in commercial detergents suggested it as a potential additive in the detergent industry.
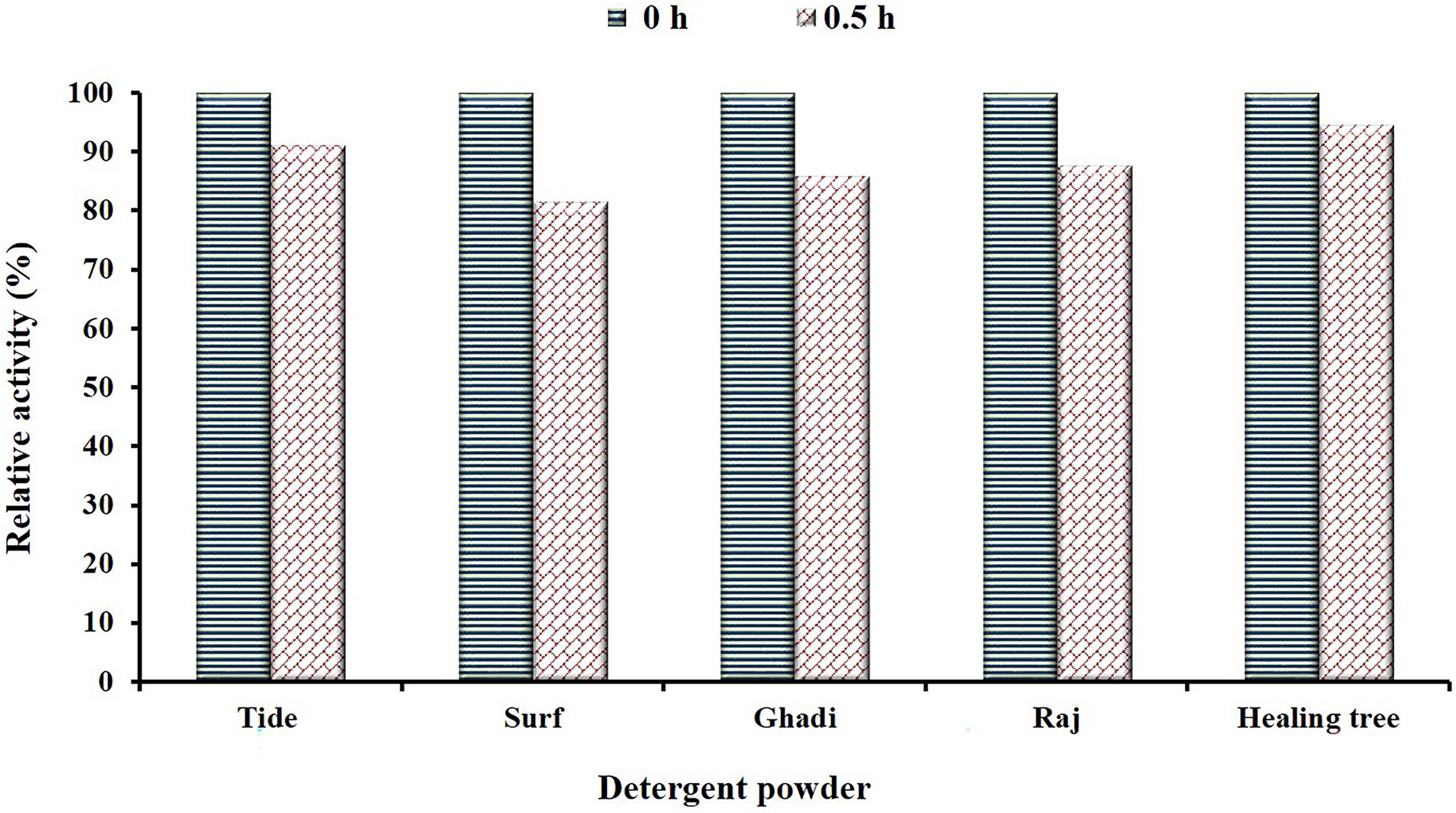
Figure 7. Depicts the stability/compatibility of β-1,4-glucanase (EGaseBL) in commercial detergents (Tide, Surf, Ghadi, Raj, and Healing tree) available in the local market. The enzyme was incubated for 30 min in detergents at a room temperature condition.
Conclusion
A novel thermo-alkali stable and detergent compatible β-1,4-glucanase (EGaseBL) was cloned, expressed, and purified. The sequence analysis, presence of CBM module, and bi-functional activity confirm the processive nature of the GH9 family EGaseBL. The recombinant protein was highly purified to 94 fold with a recovery of 67.7%. Also, the physicochemical characterization revealed the robust properties of EGaseBL activity and stability in broad pH and temperature ranges. The enzyme revealed remarkable stability (> 85%) in commercial detergents for 30 min. The compatibility and stability of EGaseBL in commercial detergents are promising for the detergent industry. In addition, the kinetics analysis showed a low Km (1.10 mg/ml) and high Vmax (208.6 IU/mg) on the β-glucan substrate. The ability of EGaseBL for β-glucan and Avicel hydrolysis and broad pH/temperature stability makes it too a potential player in the biorefinery industry. Therefore, the recombinant EGaseBL has shown promise for varied industrial applications, and its future explorations could lead to astonishing bioprocess developments.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.
Ethics statement
The authors declare that this research did not involve human participants and/or animals. The authors declare that this work involved no unusual hazard, animal, or human subjects.
Author contributions
VT: designed, performed experiments, data analysis, and manuscript writing. DS: conceived the study, research methodology and design, data interpretation, manuscript writing, and research supervision. All authors contributed to the article and approved the submitted version.
Funding
VT duly received support from the Senior Research Fellowship (award no. 31/54(0144)/2019-EMR-1) from the Council of Scientific and Industrial Research (CSIR), New Delhi, India. DS received financial support from CSIR (grant MLP0143), New Delhi, under Niche Creating Project and Department of Biotechnology, New Delhi, in the form of GAP0130 under “Himalayan Bioresources Mission.”
Acknowledgments
The Director, CSIR-IHBT, Palampur (HP), India, is duly acknowledged for providing the necessary facilities to carry out the work.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.1058249/full#supplementary-material
Footnotes
1. ^https://www.polarismarketresearch.com/industry-analysis/laundry-detergent-market
2. ^https://www.ncbi.nlm.nih.gov/orffinder/
3. ^https://web.expasy.org/translate/
4. ^https://www.ncbi.nlm.nih.gov/Structure/cdd/docs/cdd_search.html
References
Akram, F., Haq, I. U., Imran, W., and Mukhtar, H. (2018). Insight perspectives of thermostable endoglucanases for bioethanol production: a review. Renew. Energy 122, 225–238. doi: 10.1016/j.renene.2018.01.095
Antar, M., Lyu, D., Nazari, M., Shah, A., Zhou, X., and Smith, D. L. (2021). Biomass for a sustainable bioeconomy: an overview of world biomass production and utilization. Renew. Sust. Energ. Rev. 139:110691. doi: 10.1016/j.rser.2020.110691
Bagewadi, Z. K., Mulla, S. I., and Ninnekar, H. Z. (2016). Purification and characterization of endo β-1, 4-d-glucanase from Trichoderma harzianum strain HZN11 and its application in production of bioethanol from sweet sorghum bagasse. 3. Biotech 6, 101–110. doi: 10.1007/s13205-016-0421-y
Belghith, H., Ellouz-Chaabouni, S., and Gargouri, A. (2001). Biostoning of denims by Penicillium occitanis (Pol6) cellulases. J. Biotechnol. 89, 257–262. doi: 10.1016/S0168-1656(01)00309-1
Bhat, M. K. (2000). Cellulases and related enzymes in biotechnology. Biotechnol. Adv. 18, 355–383. doi: 10.1016/S0734-9750(00)00041-0
Bradford, M. M. (1976). A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72, 248–254. doi: 10.1016/0003-2697(76)90527-3
Cavaco-Paulo, A. (1998). Mechanism of cellulase action in textile processes. Carbohydr. Polym. 37, 273–277. doi: 10.1016/S0144-8617(98)00070-8
Cavaco-Paulo, A., Cortez, J., and Almeida, L. (1997). The effect of cellulase treatment in textile washing processes. J. Soc. Dye. Colour. 113, 218–222. doi: 10.1111/j.1478-4408.1997.tb01902.x
Chandel, A. K., Chandrasekhar, G., Silva, M. B., and Silvério Da Silva, S. (2012). The realm of cellulases in biorefinery development. Crit. Rev. Biotechnol. 32, 187–202. doi: 10.3109/07388551.2011.595385
Chandel, A. K., Rudravaram, R., Rao, L. V., Ravindra, P., and Narasu, M. L. (2007). Industrial enzymes in bioindustrial sector development: an Indian perspective. J. Commer. Biotechnol. 13, 283–291. doi: 10.1057/palgrave.jcb.3050065
Chiriac, A. I., Cadena, E. M., Vidal, T., Torres, A. L., Diaz, P., and Pastor, F. I. J. (2010). Engineering a family 9 processive endoglucanase from Paenibacillus barcinonensis displaying a novel architecture. Appl. Microbiol. Biotechnol. 86, 1125–1134. doi: 10.1007/s00253-009-2350-8
Dhar, H., Kasana, R. C., Dutt, S., and Gulati, A. (2015b). Cloning and expression of low temperature active endoglucanase EG5C from Paenibacillus sp. IHB B 3084. Int. J. Biol. Macromol. 81, 259–266. doi: 10.1016/j.ijbiomac.2015.07.060
Dhar, H., Kasana, R. C., and Gulati, A. (2015a). Heterologous expression and characterization of detergent stable endoglucanase EG5B from Paenibacillus sp. IHB B 3084. J. Mol. Catal. B Enzym. 120, 9–15. doi: 10.1016/j.molcatb.2015.06.009
Furtado, G. P., Ribeiro, L. F., Santos, C. R., Tonoli, C. C., De Souza, A. R., Oliveira, R. R., et al. (2011). Biochemical and structural characterization of a β-1,3-1,4-glucanase from Bacillus subtilis 168. Process Biochem. 46, 1202–1206. doi: 10.1016/j.procbio.2011.01.037
Gavande, P. V., Nath, P., Kumar, K., Ahmed, N., Fontes, C. M., and Goyal, A. (2022). Highly efficient, processive and multifunctional recombinant endoglucanase RfGH5_4 from Ruminococcus flavefaciens FD-1 v3 for recycling lignocellulosic plant biomasses. Int. J. Biol. Macromol. 209, 801–813. doi: 10.1016/j.ijbiomac.2022.04.059
Golgeri, M. D. B., Mulla, S. I., Bagewadi, Z. K., Tyagi, S., Hu, A., Sharma, S., et al. (2022). A systematic review on potential microbial carbohydrases: current and future perspectives. Crit. Rev. Food Sci. Nutr. 1-18, 1–18. doi: 10.1080/10408398.2022.2106545
Guan, X., Chen, P., Xu, Q., Qian, L., Huang, J., and Lin, B. (2017). Expression, purification and molecular characterization of a novel endoglucanase protein from Bacillus subtilis SB13. Protein Expr. Purif. 134, 125–131. doi: 10.1016/j.pep.2017.04.009
Han, C., Liu, Y., Liu, M., Wang, S., and Wang, Q. (2020). Improving the thermostability of a thermostable endoglucanase from Chaetomium thermophilum by engineering the conserved noncatalytic residue and N-glycosylation site. Int. J. Biol. Macromol. 164, 3361–3368. doi: 10.1016/j.ijbiomac.2020.08.225
Huang, Z., Ni, G., Zhao, X., Wang, F., and Qu, M. (2021). Characterization of a GH8 β-1,4-Glucanase from Bacillus subtilis B111 and its saccharification potential for agricultural straws. J. Microbiol. Biotechnol. 31, 1446–1454. doi: 10.4014/jmb.2105.05026
Irwin, D. C., Spezio, M., Walker, L. P., and Wilson, D. B. (1993). Activity studies of eight purified cellulases: specificity, synergism, and binding domain effects. Biotechnol. Bioeng. 42, 1002–1013. doi: 10.1002/bit.260420811
Jang, W. J., Lee, G. H., Lee, J. M., Kim, T. Y., Jeon, M. H., Kim, Y. H., et al. (2021). Improving enzyme activity, thermostability and storage stability of β-1,3-1,4-glucanase with poly-γ-glutamic acid produced by Bacillus sp. SJ-10. Enzym. Microb. Technol. 143:109703. doi: 10.1016/j.enzmictec.2020.109703
Joshi, N., Kaushal, G., and Singh, S. P. (2021). Biochemical characterization of a novel thermo-halo-tolerant GH5 endoglucanase from a thermal spring metagenome. Biotechnol. Bioeng. 118, 1531–1544. doi: 10.1002/bit.27668
Kim, D. Y., Lee, M. J., Cho, H. Y., Lee, J. S., Lee, M. H., Chung, C. W., et al. (2016). Genetic and functional characterization of an extracellular modular GH6 endo-β-1, 4-glucanase from an earthworm symbiont, Cellulosimicrobium funkei HY-13. Antonie Van Leeuwenhoek, 109, 1–12. doi: 10.1007/s10482-015-0604-2
Konar, A., Aich, S., Katakojwala, R., Datta, S., and Mohan, S. V. (2022). A processive GH9 family endoglucanase of Bacillus licheniformis and the role of its carbohydrate-binding domain. Appl. Microbiol. Biotechnol. 106, 6059–6075. doi: 10.1007/s00253-022-12117-4
Kuhad, R. C., Gupta, R., and Singh, A. (2011). Microbial cellulases and their industrial applications. Enzyme Res. 2011, 1–10. doi: 10.4061/2011/280696
Kumar, A., and Naraian, R. (2018). Differential expression of the microbial β-1,4-xylanase, and β-1,4-endoglucanase genes. In: New and Future Developments in Microbial Biotechnology and Bioengineerin. (Microbial Genes Biochemistry and Applications. Elsevier), 95–111.
Kumar, K., Singal, S., and Goyal, A. (2019). Role of carbohydrate binding module (CBM3c) of GH9 β-1,4 endoglucanase (Cel9W) from Hungateiclostridium thermocellum ATCC 27405 in catalysis. Carbohydr. Res. 484:107782. doi: 10.1016/j.carres.2019.107782
Kumar, N. V., Rani, M. E., Gunaseeli, R., and Kannan, N. D. (2018). Paper pulp modification and deinking efficiency of cellulase-xylanase complex from Escherichia coli SD5. Int. J. Biol. Macromol. 111, 289–295. doi: 10.1016/j.ijbiomac.2017.12.126
Ladeira, S. A., Cruz, E., Delatorre, A. B., Barbosa, J. B., and Martins, M. L. L. (2015). Cellulase production by thermophilic bacillus sp. SMIA-2 and its detergent compatibility. Electron. J. Biotechnol. 18, 110–115. doi: 10.1016/j.ejbt.2014.12.008
Liu, M., Yang, S., Long, L., Wu, S., and Ding, S. (2017). The enzymatic deinking of waste papers by engineered bifunctional chimeric neutral lipase–endoglucanase. BioResources 12, 6812–6831.
Lv, K., Shao, W., Pedroso, M. M., Peng, J., Wu, B., Li, J., et al. (2021). Enhancing the catalytic activity of a GH5 processive endoglucanase from Bacillus subtilis BS-5 by site-directed mutagenesis. Int. J. Biol. Macromol. 168, 442–452. doi: 10.1016/j.ijbiomac.2020.12.060
Mejia-Castillo, T., Hidalgo-Lara, M. E., Brieba, L. G., and Ortega-Lopez, J. (2008). Purification, characterization and modular organization of a cellulose-binding protein, CBP105, a processive β-1,4-endoglucanase from Cellulomonas flavigena. Biotechnol. Lett. 30, 681–687. doi: 10.1007/s10529-007-9589-x
Miettinen-Oinonen, A., and Suominen, P. (2002). Enhanced production of Trichoderma reesei endoglucanases and use of the new cellulase preparations in producing the stonewashed effect on denim fabric. Appl. Environ. Microbiol. 68, 3956–3964. doi: 10.1128/AEM.68.8.3956-3964.2002
Miller, G. L. (1959). Use of dinitrosalicylic acid reagent for determination of reducing sugar. Anal. Chem. 31, 426–428. doi: 10.1021/ac60147a030
Na, H. B., Jung, W. K., Jeong, Y. S., Kim, H. J., Kim, S. K., Kim, J., et al. (2015). Characterization of a GH family 8 β-1,3-1,4-glucanase with distinctive broad substrate specificity from Paenibacillus sp. X4. Biotechnol. Lett. 37, 643–655. doi: 10.1007/s10529-014-1724-x
Niyonzima, F. N. (2019). Detergent-compatible bacterial cellulases. J. Basic Microbiol. 59, 134–147. doi: 10.1002/jobm.201800436
Ontañon, O. M., Ghio, S., Díaz, M., de Villegas, R., Garrido, M. M., Talia, P. M., et al. (2019). A thermostable GH8 endoglucanase of Enterobacter sp. R1 is suitable for β-glucan deconstruction. Food Chem. 298:124999. doi: 10.1016/j.foodchem.2019.124999
Reverbel-Leroy, C., Pages, S., Belaich, A., Belaich, J. P., and Tardif, C. (1997). The processive endocellulase CelF, a major component of the clostridium cellulolyticum cellulosome: purification and characterization of the recombinant form. J. Bacteriol. 179, 46–52. doi: 10.1128/jb.179.1.46-52.1997
Sadhu, S., Saha, P., Sen, S. K., Mayilraj, S., and Maiti, T. K. (2013). Production, purification and characterization of a novel thermotolerant endoglucanase (CMCase) from Bacillus strain isolated from cow dung. Springerplus 2, 1–10. doi: 10.1186/2193-1801-2-10
Scapin, S. M. N., Souza, F. H. M., Zanphorlin, L. M., De Almeida, T. S., Sade, Y. B., Cardoso, A. M. H., et al. (2017). Structure and function of a novel GH8 endoglucanase from the bacterial cellulose synthase complex of Raoultella ornithinolytica. PLoS One 12, e0176550–e0176518. doi: 10.1371/journal.pone.0176550
Song, Y. H., Lee, K. T., Baek, J. Y., Kim, M. J., Kwon, M. R., Kim, Y. J., et al. (2017). Isolation and characterization of a novel endo-β-1,4-glucanase from a metagenomic library of the black-goat rumen. Braz. J. Microbiol. 48, 801–808. doi: 10.1016/j.bjm.2017.03.006
Thakur, V., Kumar, V., Kumar, S., and Singh, D. (2018). Diverse culturable bacterial communities with cellulolytic potential revealed from pristine habitat in Indian trans-Himalaya. Can. J. Microbiol. 64, 798–808. doi: 10.1139/cjm-2017-0754
Thakur, V., Kumar, V., Kumar, V., and Singh, D. (2021). Genomic insights driven statistical optimization for production of efficient cellulase by Himalayan thermophilic Bacillus sp. PCH94 using agricultural waste. Waste Biomass Valorization 12, 6917–6929. doi: 10.1007/s12649-021-01491-1
Wang, S., Li, W., Liu, S., and Xu, J. (2016). RaptorX-property: a web server for protein structure property prediction. Nucleic Acids Res. 44, W430–W435. doi: 10.1093/nar/gkw306
Wang, S., Lin, R., Ren, Y., Zhang, T., Lu, H., Wang, L., et al. (2020). Non-chromatographic purification of thermostable endoglucanase from Thermotoga maritima by fusion with a hydrophobic elastin-like polypeptide. Protein Expr. Purif. 173:105634. doi: 10.1016/j.pep.2020.105634
Waterhouse, A., Bertoni, M., Bienert, S., Studer, G., Tauriello, G., Gumienny, R., et al. (2018). SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res. 46, W296–W303. doi: 10.1093/nar/gky427
Wilson, D. B. (2004). Studies of Thermobifida fusca plant cell wall degrading enzymes. Chem. Rec. 4, 72–82. doi: 10.1002/tcr.20002
Wu, S., Ding, S., Zhou, R., and Li, Z. (2007). Comparative characterization of a recombinant Volvariella volvacea endoglucanase I (EG1) with its truncated catalytic core (EG1-CM), and their impact on the bio-treatment of cellulose-based fabrics. J. Biotechnol. 130, 364–369. doi: 10.1016/j.jbiotec.2007.05.016
Wu, M., Lv, K., Li, J., Wu, B., and He, B. (2022). Coevolutionary analysis reveals a distal amino acid residue pair affecting the catalytic activity of GH5 processive endoglucanase from Bacillus subtilis BS-5. Biotechnol. Bioeng. 119, 2105–2114. doi: 10.1002/bit.28113
Wu, S., and Wu, S. (2020). Processivity and the mechanisms of Processive endoglucanases. Appl. Biochem. Biotechnol. 190, 448–463. doi: 10.1007/s12010-019-03096-w
Keywords: glucanase, hydrolysis, thermostability, industrial applications, biorefinery
Citation: Thakur V and Singh D (2022) A thermo-alkali stable and detergent compatible processive β-1,4-glucanase from Himalayan Bacillus sp. PCH94. Front. Microbiol. 13:1058249. doi: 10.3389/fmicb.2022.1058249
Edited by:
Madan L. Verma, Indian Institute of Information Technology, Una, IndiaReviewed by:
Sikandar I. Mulla, REVA University, IndiaShashi Kant Bhatia, Konkuk University, South Korea
Copyright © 2022 Thakur and Singh. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Dharam Singh, ZGhhcmFtc2luZ2hAaWhidC5yZXMuaW4=;ZGhhcmFtczE0QGdtYWlsLmNvbQ==
 Vikas Thakur
Vikas Thakur Dharam Singh
Dharam Singh