
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 28 January 2022
Sec. Food Microbiology
Volume 12 - 2021 | https://doi.org/10.3389/fmicb.2021.772546
Plantaricin is a broad-spectrum bacteriocin produced by Lactiplantibacillus plantarum with significant food industry application potential. It was found that the plantaricin production of L. plantarum RX-8 was enhanced when co-culturing with Bacillus subtilis BS-15. This study, therefore, set out to explore how B. subtilis BS-15 induces biosynthesis of plantaricin. The effect of co-culturing with B. subtilis BS-15 on cell growth, plantaricin production, quorum-sensing (QS) signal molecule PlnA/autoinducer-2 (AI-2) secretion, as well as plantaricin biosynthesis gene cluster and AI-2 synthesis-associated gene expression, was investigated in bacteriocin-producer L. plantarum RX-8. When L. plantarum RX-8 and B. subtilis BS-15 were co-inoculated in Man–Rogosa–Sharp (MRS) for 20 h at an inoculum ratio of 1:1 (106:106 CFU/ml), the greatest plantaricin output (2,048 AU/ml) was obtained, rising by 32-fold compared with the monoculture of L. plantarum RX-8. Additionally, co-culture increased PlnA-inducing activity and AI-2 activity by 8- and 1.14-fold, respectively, over monoculture. RT-qPCR findings generated every 4 h (4–32 h) demonstrated that B. subtilis BS-15 remarkably improved the transcription of plnABCD and plnEF, and increased pfs and luxS transcription, even when using 200 mM D-ribose, a kind of AI-2 inhibitor. Based on the above findings, co-culturing with B. subtilis BS-15 as an environmental stimulus could activate the plantaricin induction via the PlnA-mediated intraspecies QS system and the AI-2-mediated interspecies QS system. Moreover, the inducing effect of PlnA and AI-2 in co-culture was independent. Differential proteomics analysis of B. subtilis BS-15 in co-culture indicated that bacteriocin-inducing regulatory mechanism may be related to flagellar assembly, peptidoglycan biosynthesis, anaerobic respiration, glycine cleavage system, or thiamin pyrophosphate biosynthesis.
Lactiplantibacillus plantarum is a common and versatile lactic acid bacteria (LAB) that is found in a wide variety of fermented foods and human gastrointestinal tract (Corsetti and Valmorri, 2001). In the mixed-culture food fermentations, L. plantarum not only contribute significantly to food preservation and fortification, as well as flavors and texture, but also secretes a wide range of beneficial compounds, including bacteriocins (da Silva Sabo et al., 2014), short-chain fatty acids (Osei Sekyere et al., 2020), and polysaccharides (Zhang et al., 2020). Plantaricin is the name given to the bacteriocins produced by L. plantarum. Their presence in food is generally considered safe for consumers due to their inactivation by pancreatic or gastric enzymes (Simons et al., 2020). Plantaricin has been found to be efficient against pathogenic and spoilage microorganisms when being used as a food biopreservative ingredient (Zhu et al., 2014; Todorov et al., 2018; Le et al., 2019; Lei et al., 2020). However, industrial production and use of plantaricin are challenging to achieve due to the low production and high cost associated with laboratory culture (Gutiérrez-Cortés et al., 2018). Moreover, the capability of L. plantarum to biosynthesize plantaricin may be gradually lost in a noncompetitive environment (Vriezen et al., 2009).
Several strategies were used to enhance plantaricin biosynthesis, including the screening for high-yield strains, optimizing their production conditions, physical and chemical mutagenesis, and heterologous expression. Nevertheless, they have some drawbacks, including inadequate specificity, restricted growth potential, genetic instability, and low safety (Man and Xiang, 2021). Recent research indicates that microbial co-culture fermentations are favorable for bacteriocin production and are extensively used among L. plantarum strains that produce bacteriocin (Maldonado-Barragán et al., 2013). For example, in a test of screening bacteriocin-inducing strains under co-culture conditions, 10 out of 45 strains were able to cause L. plantarum J23 to produce plantaricin (Rojo-Bezares et al., 2007), 41 out of 82 strains were able to stimulate L. plantarum NC8 to produce plantaricin (Maldonado et al., 2004), and 4 out of 76 strains could induce L. plantarum KLDS1.0391 to produce plantaricin (Man et al., 2012). Co-culturing with certain bacterial strains as an environmental stimulus may be a feasible technique for improving plantaricin production in the food sector due to the possibility of synergistic exploitation of the metabolic pathways of all engaged strains.
Understanding the mechanism of induction regulation behind bacteriocin production via co-culture is crucial for ensuring its effective application in the food industry (Chanos and Mygind, 2016). Until present, only the mechanism of plantaricin biosynthesis in monoculture has been thoroughly researched. As previously documented, plantaricin biosynthesis in monoculture is density-dependently regulated via an interspecies quorum-sensing (QS) system mediated by autoinducing peptide (AIP) (Sturme et al., 2007; Diep et al., 2009). The plnABCD operon encodes the QS system, which consists of an AIP gene plnA, a membrane-located histidine protein kinase (HPK) gene plnB and two response regulator (RR) genes plnC and plnD (Sturme et al., 2007). The AIP, also known as PlnA in L. plantarum, is responsible for determining the cell density of its producer (Diep et al., 2009). When PlnA reaches a level detectable by the extracellular part of the histidine kinase (PlnB), PlnB will catalyze the autophosphorylation of the cytoplasmic domain (Meng et al., 2021). Following this, the two RRs phosphorylate and attach to plnEF, which encodes the bacteriocin structural genes. Subsequently, the bacteriocin is secreted from the cell via the ABC-transport system (Zhao et al., 2021).
However, the bacteriocin-inducing regulation mechanism in co-culture with specific bacteria has been challenging, as both the AIP-mediated interspecies QS system and autoinducer-2 (AI-2)-mediated intraspecies QS system exist simultaneously (Sieuwerts et al., 2008). Some research has demonstrated a positive correlation between the AI-2 activity of L. plantarum and bacteriocin production in co-culture (Di Cagno et al., 2009; Man et al., 2012, 2014). Moreover, the HPK and RR interspecies QS system continue to play core roles in the induction in co-culture conditions. In one instance, the PLNC8 was used to stimulate the production of bacteriocins by L. plantarum NC8 after co-culture with certain Gram-positive bacteria (Maldonado et al., 2004). When the plNC8-plNC8HK-plnD and plnBCD genes were removed from L. plantarum NC8 and WCFS1, respectively, the potential of these strains to generate bacteriocin through the co-culture with inducing bacterial strains was abolished (Maldonado-Barragán et al., 2013). However, the regulation intensity of AIP-mediated interspecies QS system/AI-2-mediated intraspecies QS system on plantaricin induction and their relationship in co-culture remain unknown.
Bacillus subtilis BS-15 was extracted from Chinese traditional cereal vinegar brewing. In previous studies, it was discovered that the co-culture with B. subtilis BS-15 had enhanced the plantaricin production of L. plantarum RX-8. This breakthrough in plantaricin production makes industrial production and food preservation applications possible (Gutiérrez-Cortés et al., 2018). This study, therefore, set out to explore how B. subtilis BS-15 induces biosynthesis of plantaricin. To this end, this paper analyzed the influence of co-culturing with B. subtilis BS-15 on the growth of bacteriocin-producer L. plantarum RX-8, plantaricin production, QS signal molecule PlnA/AI-2 secretion, as well as plantaricin biosynthesis gene cluster and AI-2 synthesis-related gene expression. Furthermore, the addition of the QS inhibition D-ribose helped assess the effect of AI-2-mediated interspecies QS system on plantaricin production of L. plantarum RX-8 in co-culture with B. subtilis BS-15. Finally, differential proteomics was used to investigate the changes in the protein level of bacteriocin-inducer B. subtilis BS-15 in co-culture.
The L. plantarum RX-8 strain was isolated from the traditional Chinese Sichuan Pao Cai and has been preserved in the China General Microbiological Culture Collection Center (CGMCC 20852). Listeria monocytogenes 35152 (ATCC 35152) and Vibrio harveyi BB170 (ATCC BAA-1117) were obtained from the American Type Culture Collection (ATCC). The stable bacteriocin-producer L. plantarum RX-8 was grown in de Man–Rogosa–Sharp (MRS) broth at 37°C. B. subtilis BS-15, isolated from Chinese traditional cereal vinegar brewing, is grown in tryptic soy broth (TSB) at 37°C and preserved in CGMCC (CGMCC 20851). L. monocytogenes 35152, grown in TSB at 37°C, was used as the indicator strain to detect the inhibitory and induction activities in bacteriocin production. V. harveyi BB170 was a reporter strain to detect the AI-2 activity of co-culture supernatants and grown in marine broth (MB) media at 30°C, 180 rpm. All strains were stored at −80°C in 40% (v/v) glycerol and resuscitated twice in their corresponding broth medium before being used.
A two-compartment separated co-culture system in Transwell plates was used to evaluate the effect of the secretion of B. subtilis BS-15 on the plantaricin production of L. plantarum RX-8. To be specific, L. plantarum RX-8 cells were suspended in co-culture medium and placed in the bottom wells of Transwell plates (Corning, United States), while B. subtilis BS-15 cells were placed in the upper wells, separated from the bottom wells by a membrane with a 0.4-μm pore size. To establish the optimal inducible co-culture, two strains were co-inoculated in MRS at different inoculum ratios (Table 1). D-ribose is a kind of competitive inhibitor, which could inhibit AI-2-induced intracellular communication. In order to study the role of AI-2-induced QS regulation system in co-cultures, D-ribose was added, and then the viable cell count, plantaricin production, QS signal molecule activity, as well as bacteriocin biosynthesis gene cluster and AI-2 synthesis-related gene expression of harvested co-culture at pre-fixed incubation times were compared. The monoculture of L. plantarum RX-8 was used as control.

Table 1. Scheme of the inoculation of L. plantarum RX-8 and B. subtilis BS-15 in co-culture systems with different inoculum ratios.
The twofold serial dilution assayed the production level of plantaricin in co-culture/monoculture through agar well diffusion (Zhu et al., 2014). L. monocytogenes 35152 (107 CFU/ml) was used as the indicator strain. After a 24-h stationary incubation at 37°C, the cells were removed by centrifugation at 10,000 rpm for 10 min at 4°C to obtain cell-free supernatants (CFSs). Then the 51.6-g ammonium sulfate was added to 100 ml of CFS with continuous stirring by a magnetic stirrer for 1 h. After storing at 4°C overnight, the protein precipitate was pelleted by centrifugation at 12,000 rpm for 10 min at 4°C and solubilized in 20 mM sodium phosphate buffer pH 6.0 (10 ml per 100-ml culture). Then 100 μl of the 10-fold concentrated plantaricin was placed into each well. The plates were held at 4°C for 8 h to allow the bacteriocin to completely diffuse, and these plates were incubated overnight at 37°C. The bacteriocin activity was expressed in arbitrary units (AU/ml), which is represented as reciprocal of the highest dilution showing a distinct zone of inhibition and calculated according to the equation:
where a is 2 (dilution factor), n is the reciprocal of the highest dilution that resulted in inhibition of the indicator strain, b is 100 μl (sample volume in each well), and c is 10 (sample concentration fold). Furthermore, the relative bacteriocin activity was defined as the ratio of bacteriocin activity in co-culture to that in monoculture. It was used to evaluate the influence of B. subtilis BS-15 on plantaricin production in co-culture.
The nonbacteriocin-producing (Bac–) culture is used to analyze the inducing activity of PlnA. Due to the effect of intraspecies QS on bacteriocin production in monoculture, L. plantarum loses the ability to produce bacteriocins when inoculated on liquid media with an inoculum size less than the specified threshold (Maldonado-Barragán et al., 2009). To prepare the nonbacteriocin-producing (Bac–) culture, the inoculum size of L. plantarum RX-8 in monoculture was diluted. Afterward, the 100-ml 24-h-old culture was centrifuged for 2 min at 8,000 rpm and resuspended it in 1 ml of fresh MRS medium after removing the supernatants to get the initial concentration of 1.4 × 1011 CFU/ml of L. plantarum RX-8 cell suspension. The cell suspension was then serially diluted 10-fold for six times, and 1% (v/v) of each dilution was inoculated in sterile MRS broth to generate the final inoculum size of approximately 109, 108, 107, 106, 105, and 104 CFU/ml, respectively. After incubation at 37°C for 24 h, the samples were centrifuged for 10 min at 10,000 rpm to remove the cells, and the CFS was subsequently measured for plantaricin production using the agar well diffusion method as described above.
PlnA was semipurified according to the method of Maldonado-Barragán et al. (2009) with some modifications. As described above, after a 24-h-old co-culture/monoculture of L. plantarum RX-8, cells were removed by centrifugation (10,000 rpm, 10 min), and the pH was adjusted to 7.0. The PlnA present in the supernatant fraction was concentrated by ammonium sulfate precipitation (80% saturation). After the mixture had been stirred overnight at 4°C, the precipitate was pelleted by centrifugation (12,000 rpm, 10 min). The collected precipitate was then dissolved in 20 mM sodium phosphate buffer pH 7.0 and desalted using PD10 gel filtration column (GE Healthcare, WI, United States) equilibrated with the same buffer. The mixture was then applied into a SP-Sepharose Fast Flow cation exchange column (GE Healthcare, WI, United States) equilibrated with 20 mM sodium phosphate buffer (pH 7.0). The fraction with charged cationic property was eluted with a linear salt gradient (0 to 1 M of NaCl) at a flow rate of 1 ml/min and then desalted by a Sephadex G15 column (Sigma-Aldrich, Shanghai, China) preequilibrated with 50 mM sodium acetate buffer pH 5.2. The mixture was further purified by Sephadex G25 column (Sigma-Aldrich, Shanghai, China), and the active fraction was eluted employing the same acetate buffer at a flow rate of 0.5 ml/min. Different fractions were collected and concentrated in a Vacuum Rotary Evaporation Concentrator (Jiaimu, Beijing, China).
The inducing activity tests selected the active fraction containing PlnA. Briefly, each 50-μl concentrated fraction was added to Bac– culture and cultured for 24 h at 37°C. The recovered bacteriocin-producing activity was defined as the PlnA-inducing activity. The relative PlnA-inducing activity was calculated by the ratio of semipurified PlnA-inducing activity in co-culture CFS to that in monoculture CFS.
The cells in the co-culture/monoculture sample were removed by centrifuging at 10,000 rpm, 4°C, for 10 min. The CFS was filtrated with a 0.22-μm sterile filter and adjusted to pH 7.0. Then the neutralizing CFS was concentrated 10-fold by a Vacuum Rotary Evaporation Concentrator (Jiaimu, China). The reporter strain V. harveyi BB170 was diluted 1:5,000 with fresh Autoinducer Bioassay (AB) medium, and the 10-fold concentrated CFS sample was added to the diluted BB170 culture at 1:10 (v/v). The mixture was incubated at 30°C for 4 h with agitation (180 rpm), and 200-μl aliquots were added to white 96-well plates (Thermo, United States) to detect AI-2 activity. The suspension of strain BB170 in AB medium (1:5,000) was used as a blank control. Relative luminescence units (RLU) were measured using a Multi-Detection Plate Reader (SpectraMax i3, Molecular Devices, United States) in luminescence mode. The relative AI-2 activity was defined as the ratio of RLU (the value of CFS)/(the value of blank control), and it was used to evaluate the effect on AI-2 activity of strain RX-8 under different culture conditions.
D-ribose inhibition of co-culture AI-2 activity was also determined by using the BB170 reporter strain. D-ribose (0, 10, 50, 100, and 200 mM) was added to the co-culture system and incubated at 37°C for 24 h to find out the appropriate concentration of D-ribose with optimal inhibitory effect. Then as described above, the AI-2 activity of the co-culture sample with D-ribose was detected according to RLU. The co-culture supernatant without D-ribose was used as a control.
The PCR has tested and determined the existences of plnA gene encoding AIP, plnB gene encoding HPK, plnC and plnD gene encoding RRs, AI-2 synthase genes (pfs and luxS), and bacteriocin structural genes (plnEF) in L. plantarum RX-8. This study designed the primers based on known sequences of corresponding genes in L. plantarum from GenBank (Supplementary Table 1). The following conditions were applied for PCRs: denaturation at 95°C for 5 min, followed by 30 cycles of 94°C for 30 s, annealing at 53°C for 30 s, polymerization at 72°C for 10 s, and final elongation at 72°C for 10 min. The study identified the amplicons obtained by agarose gel electrophoresis.
Quantitative real-time reverse transcription polymerase chain reaction (RT-qPCR) determined the expression level of plantaricin biosynthesis gene clusters (plnA, plnB, plnC, plnD, plnE, and plnF) and AI-2 synthesis-related genes (pfs and luxS). First, the cells were harvested from monoculture/co-culture and stored in liquid nitrogen for RNA extraction. According to the recommendations of the manufacturer, the total RNA of L. plantarum RX-8 were extracted with the RNAprep Pure Cell/Bacteria Kit (Tiangen Biotech, China). The FastQuant RT Kit (with gDNase; Tiangen Biotech, China) synthesized the first-strand cDNA with the FQ-RT Primer Mix. The RT-qPCR monitoring of gene expression was realized by SYBR Green PCR master mix (Tiangen Biotech, China) with the software of CFX96 Real-Time PCR Detection System 3.0 version (Bio-Rad, Hercules, CA, United States). Primer Premier V6.1 (Supplementary Table 2) designed primers for plnA, plnB, plnC, plnD, plnE, plnF, luxS, pfs, and 16S rRNA genes (NCBI accession No. AL935263). The transcription of total RNA was reversed into cDNA, which was used for qPCR with unigene-specific primers. The amplification program was performed by the SuperReal PreMix Plus (SYBR Green) for the fluorophore SYBR green with fluorescein in a process including 95°C for 5 min, followed by 30 cycles of 94°C for 30 s, 53°C for 30 s, and 72°C for 10 s. The comparative threshold cycle (CT) method calculated the relative abundance of transcripts. The 16S rRNA genes were used as the housekeeping reference gene. RT-qPCR was triplicated for each sample.
Both the positive and negative bacteriocin-inducing culture were fermented at 37°C for 20 h, and the fermentation broth was centrifuged at 10,000 rpm for 10 min at 4°C Then the cells of B. subtilis BS-15 were sent to Beijing YM Biological Institute (YMBio, Beijing, China) for proteomics analysis, which applied t-test and set the p-value at 0.05 as the threshold to determine significance in the two groups of regulated proteins. The GengGO/MetaCore software1 and KEGG website2 analyzed the pathway enrichment with the false discovery rate (FDR) cutoff at 0.001. The Benjamini–Hochberg method adjusted the p-values for multiple test corrections. In the GO category and KEGG pathway analysis, the cut-off is less than 0.01.
All experiments were performed in triplicates. Representative data and images are presented in this paper. The data analysis involved the one-way ANOVA engaged in the Duncan test that used SPSS Statistical package Version 16.0 (SPSS Inc., Chicago, IL, United States). Statistical significance was considered at p < 0.05, and the results were expressed as mean ± SD.
Six inoculum volumes have different effects on the production of plantaricin. As shown in Figure 1, 105–109 CFU/ml had positive influences on plantaricin biosynthesis, while when the volume is 104 CFU/ml, the influences on plantaricin biosynthesis turned to be negative. In addition, L. plantarum RX-8 could not produce bacteriocins when being inoculated in liquid media with less than 105 CFU/ml of inoculum, but the production ability would be restored when being added with CFS from a previous bacteriocin-producing (Bac+) culture (data not shown). The results in this section showed that the threshold inoculum size was 105 CFU/ml, and the inoculum size of 104 CFU/ml could be used for preparing non-bacteriocin-producing (Bac–) culture.
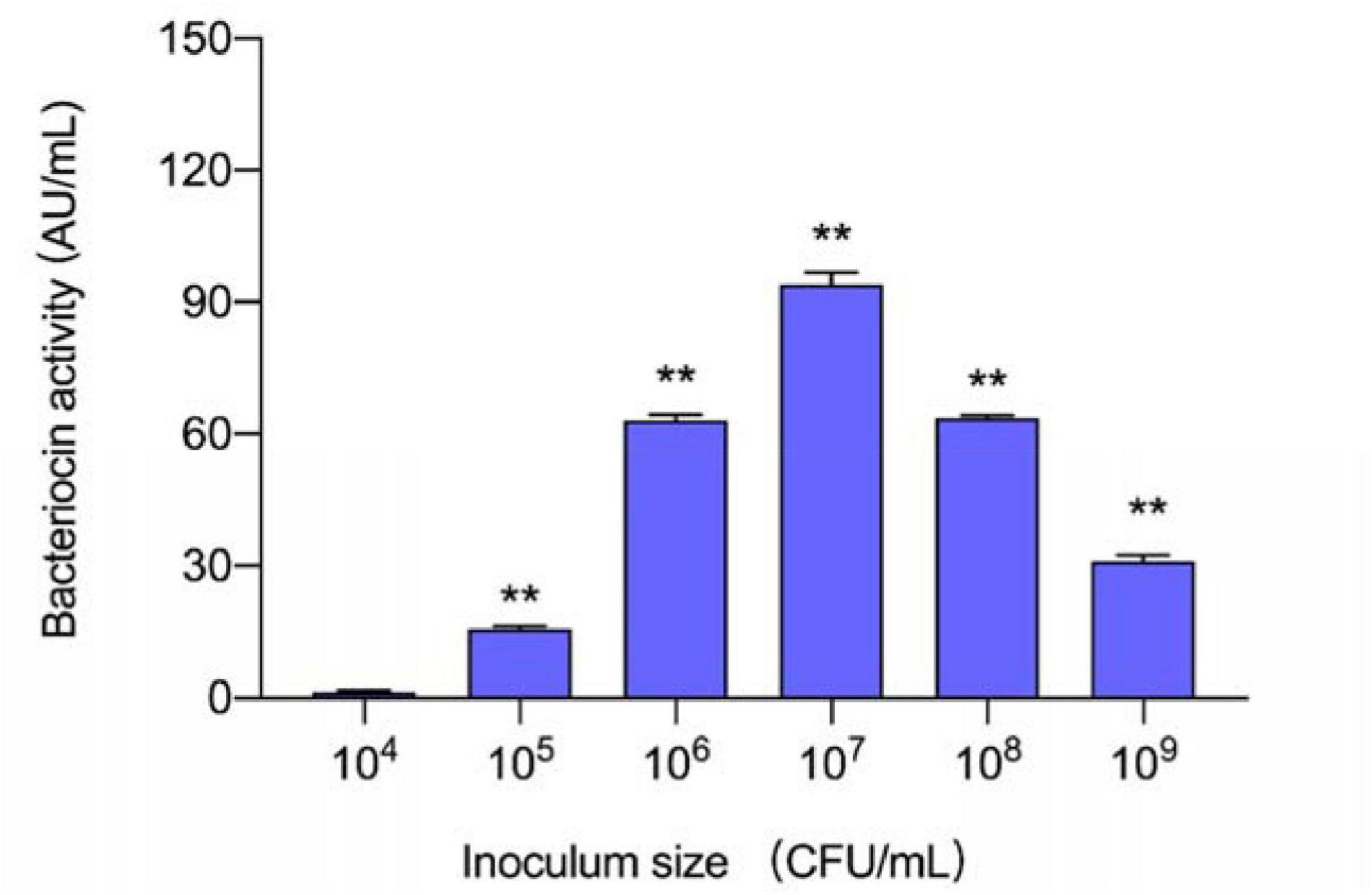
Figure 1. Effect of different inoculum sizes of L. plantarum RX-8 on the production of plantaricin. **p < 0.01.
To discover the appropriate inducible co-culture, several treatments (A–I) with varying inoculum ratios of L. plantarum RX-8 and B. subtilis BS-15 were designed to evaluate bacteriocin-inducing activity (Figure 2). The maximum plantaricin output of 1,024 AU/ml was obtained in the co-culture system, which increased by eightfold compared with monoculture (Control 2), due to the 1:1 ratio (106:106 CFU/ml) of L. plantarum RX-8 to B. subtilis BS-15. For the same inoculum size (106 CFU/ml) of L. plantarum RX-8, incubating 106 CFU/ml of B. subtilis BS-15 (treatment D) could exert the most positive bacteriocin-inducing activity, which was negative for 104 CFU/ml of B. subtilis BS-15 (treatment F). These findings indicate that the appropriate inoculation of B. subtilis BS-15 is critical for a robust inducing effect in the co-culture. Due to the apparent disparities between treatments D and F, two treatments were selected for subsequent experiments to assay the relationship between PlnA/AI-2 secretion and plantaricin production. Treatment D, a 1:1 inoculum ratio (106:106 CFU/ml) of L. plantarum RX-8 and B. subtilis BS-15 in fresh MRS broth at 37°C, was selected as a positive bacteriocin-inducing co-culture (PC). As for treatment F, the co-culture of L. plantarum RX-8 and B. subtilis BS-15 at a 100:1 inoculum ratio (106:104 CFU/ml) in fresh MRS broth at 37°C was designated as a negative bacteriocin-inducing co-culture (NC).
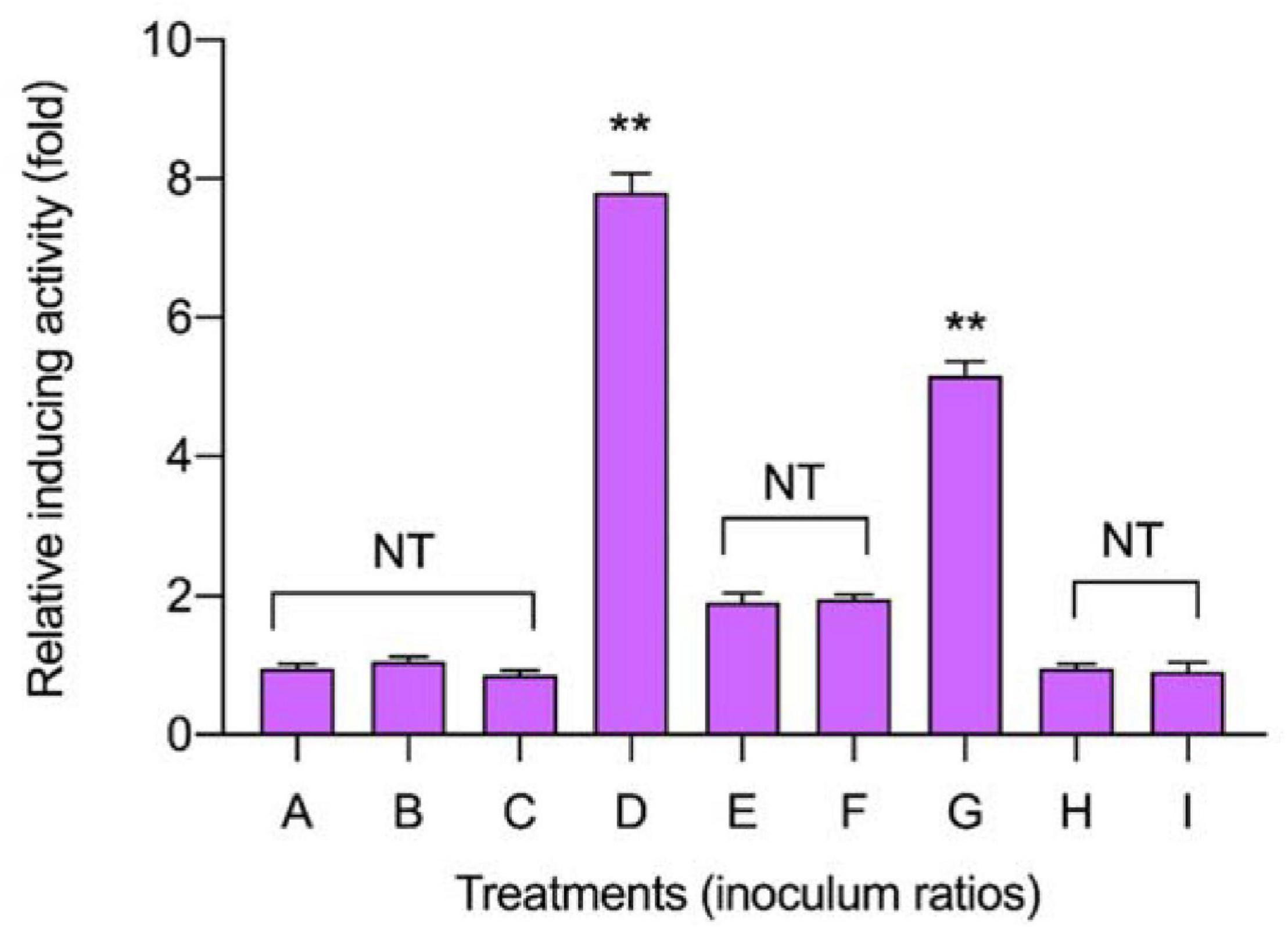
Figure 2. Effects of different inoculum ratios on the production of plantaricin in co-culture. The relative bacteriocin activity was defined as the ratio of bacteriocin activity in co-culture to that in monoculture. The inoculum ratios of A–I are shown in Table 1. NT indicates that there are no significant differences in the results in the control; *p < 0.05 and the **p < 0.01.
The secretion of QS signal molecules PlnA and AI-2 in mono-culture and co-culture (PC and NC) are shown in Figure 3. In co-culture CFS, the activity of the two QS signal molecules composed of one part of L. plantarum RX-8 culture CFS (PC-1 and NC-1) and one part of B. subtilis BS-15 culture CFS (PC-2 and NC-2) were not significantly different. It was found that both QS signal molecules increased in PC over 24 h, but the secretion remained unchanged in NC. Meanwhile, the production of plantaricin showed a similar rule in PC, NC, and control. According to Figure 3, the activity of QS signal molecules and plantaricin production in PC were higher than those in NC. Besides, there were no significant difference in the activity of two QS signal molecules and plantaricin production. It can be deduced that the QS system engaged in the induction of the production of plantaricin.
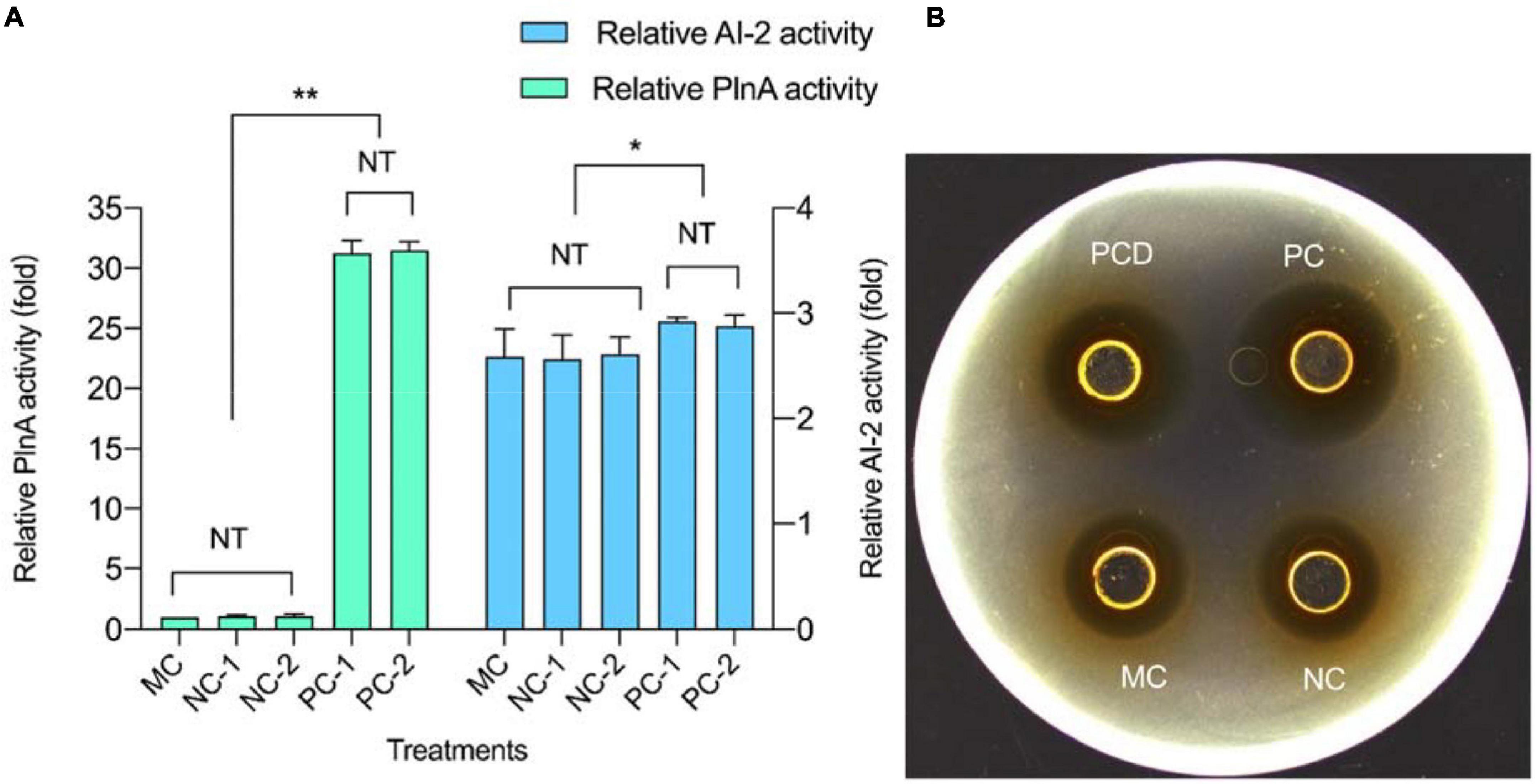
Figure 3. Quorum-sensing (QS) signal molecule secretion and plantaricin production at 24 h in co-culture. (A) QS signal molecules. (B) Plantaricin production. PC means positive bacteriocin-inducing co-culture of L. plantarum RX-8 and B. subtilis BS-15 (106:106 CFU/ml) at 37°C for 24 h, PCD means PC with 200 mM D-ribose, and NC means negative bacteriocin-inducing co-culture of L. plantarum RX-8 and B. subtilis BS-15 (106:104 CFU/ml) at 37°C for 24 h. MC means monoculture of L. plantarum RX-8 (106 CFU/ml) at 37°C for 24 h. “1” represents CFS from the upper well, and “2” represents CFS from the bottom well. NT indicates that there were no significant differences in the results between the control; *p < 0.05 and **p < 0.01. The relative AI-2 activity (fold) is defined as (the RLU of CFS)/(the RLU of blank control). The relative PlnA activity (fold) is defined as (the bacteriocin activity of co-culture)/(the bacteriocin activity of monoculture).
D-ribose bound competitively to the AI-2 transporter (LuxP) or AI-2 receptors (Ryu et al., 2016) of L. plantarum RX-8, thus, inhibiting AI-2-induced intracellular communication. As shown in Figure 4, all D-ribose inhibited AI-2 activity regardless of the concentration, whereas 200 mM D-ribose reached more than 90% inhibition (92.76%). Therefore, 200 mM D-ribose was selected for further analysis, and PC with 200 mM D-ribose was defined as PCD.
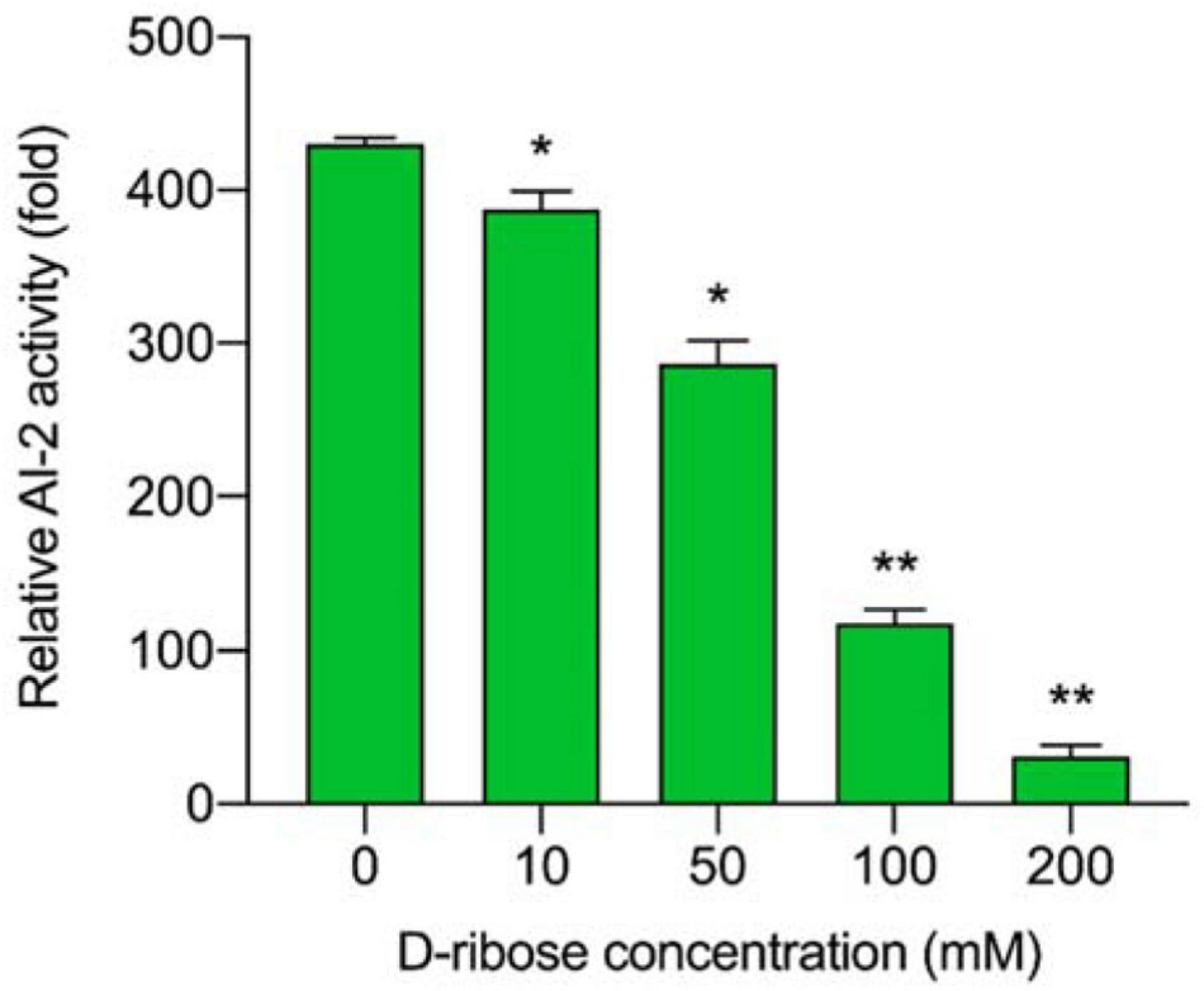
Figure 4. The inhibitory effect of different D-ribose concentrations on AI-2 activity. The relative AI-2 activity is defined as (the RLU of CFS)/(the RLU of blank control). *p < 0.05 and **p < 0.01.
The viable cell amount of L. plantarum RX-8/B. subtilis BS-15 and the relative bacteriocin production in monoculture (MC) and co-culture (PC and PCD) from 4 to 32 h are shown in Figure 5. As shown in Figure 5A, the viable cell counts of L. plantarum RX-8 in PC and PCD were increased slightly than in MC, while the viable cell counts of the B. subtilis BS-15 in PC and PCD were significantly lower than in MC (p < 0.05), which is easy to understand as the plantaricin and other antibacterial substances (e.g., lactic acid) in medium could depress the survival of B. subtilis BS-15. The results in Figure 5B illustrated that L. plantarum RX-8 began to biosynthesize plantaricin at 8 h in PC and PCD, while the initial production time of plantaricin was 12 h in MC. At the 20th hour, the plantaricin production in all treatments reached the highest point and stabilized in the 20- to 28-h period before decreasing at the 32th hour. Compared with the slight increase in viable cell counts of L. plantarum RX-8, the bacteriocin activity of L. plantarum RX-8 in PC and PCD was promoted effectively. Specifically, plantaricin production in PC finally reached 2,048 AU/ml at 20 h, 32 times greater than that in MC, and for PCD, the statistics were 1,024 AU/ml and 16 times greater. A certain competitive relationship existed between L. plantarum RX-8 and B. subtilis BS-15, which may induce the plantaricin production as an environmental stimulus. Meanwhile, after adding AI-2 inhibitor, the reduction of bacteriocin-inducing activity is 25% (8 h), 62.5% (12 h), 50% (16–24 h), 68.75% (28 h), and 60% (32 h), respectively. These results showed that the proportion of regulation intensity of AI-2 on the bacteriocin-inducing activity in the 32-h culture period was variable. It was presumed that the AI-2-mediated intraspecies interspecies QS system played a more important role in late-exponential and stationary phases than that in early-exponential phase.
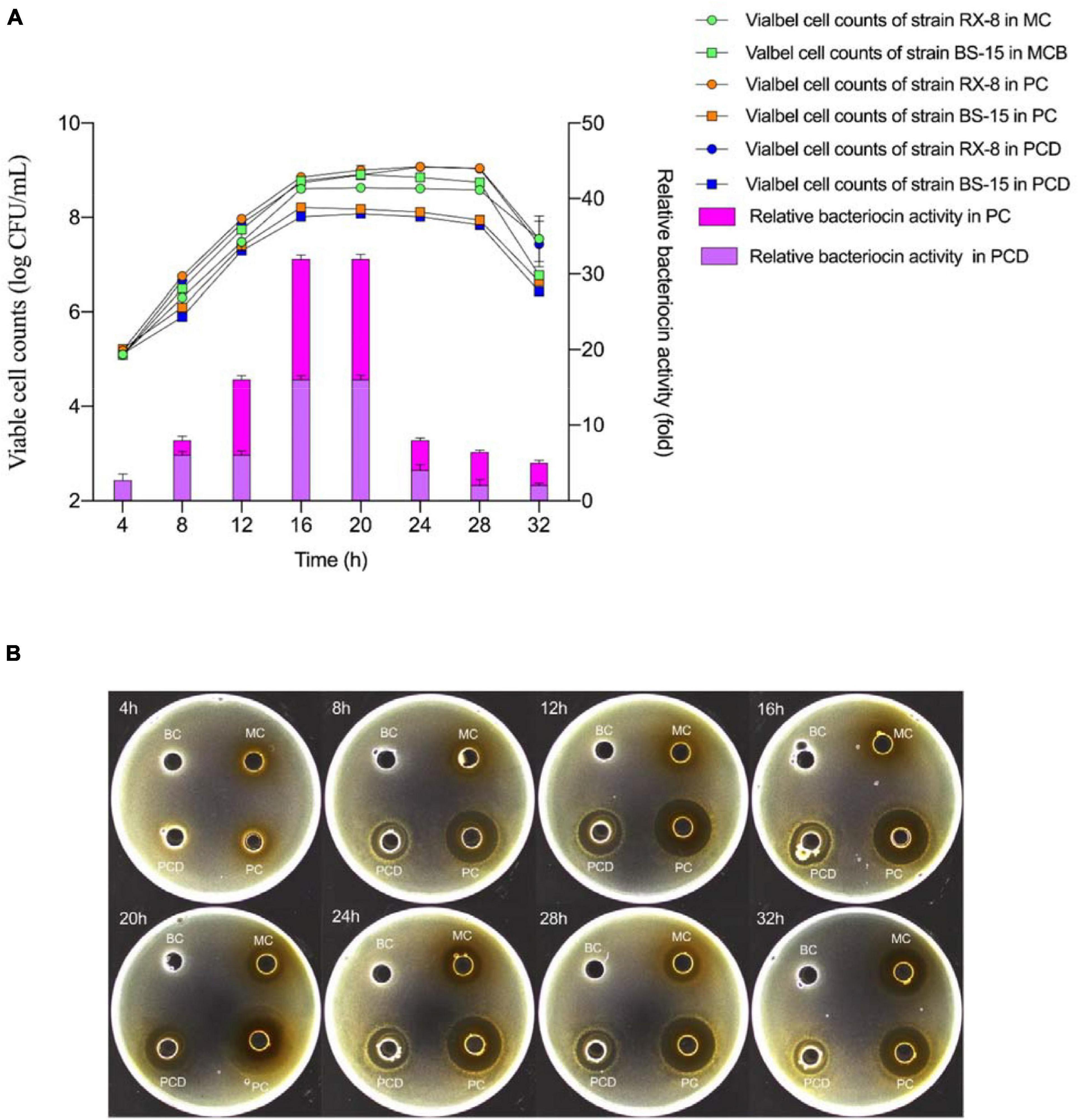
Figure 5. Viable cell amount and plantaricin production in co-culture. (A) Viable counts (left) and relative bacteriocin activity (right). (B) The plantaricin production at every 4 h from 4 to 32 h. PC means positively bacteriocin-inducing co-culture of L. plantarum RX-8 and B. subtilis BS-15 (106:106 CFU/ml) at 37°C in 4–32 h. PCD means PC with 200 mM D-ribose. MC means monoculture of L. plantarum RX-8 (106 CFU/ml) at 37°C in 4–32 h, MCB means monoculture of B. subtilis BS-15 (106 CFU/ml) at 37°C in 4–32 h. BC means blank control (20 mM sodium phosphate buffer, pH 6). The relative bacteriocin activity was defined as the ratio of bacteriocin activity in co-culture to that in monoculture.
PlnA-inducing activity and AI-2 activity in monoculture (MC) and co-culture (PC and PCD) were analyzed every 4 h. As shown in Figure 6A, the PlnA-inducing activity in all treatments were increased from 8 to 20 h, peaked at 20 h, followed by a sudden decrease at 32 h. Combined with the results in Figure 5, the changing tendency of PlnA activity in MC, PC, and PCD was consistent with the variety of cell counts of L. plantarum RX-8 from 4 to 32 h. As a signal molecule of intraspecies QS system, the concentration of PlnA was enhanced with the increase in viable cell counts. Differently, the PlnA-inducing activity was dramatically increased in PC and PCD, whereas the viable cell counts of L. plantarum RX-8 were slightly increased compared with that in MC. Hence, it can be deduced that B. subtilis BS-15 probably produces some certain compounds, which could promote PlnA secretion of L. plantarum RX-8. PlnA-inducing activity was no difference in PC and PCD, while it was greater in PC and PCD than in MC from 8 to 32 h. These results suggested that more PlnA in co-culture was secreted to enhance plantaricin production, and inhibiting AI-2 activity would not influence PlnA-induced intraspecies communication. As shown in Figure 6B, AI-2 activity in MC and PC steadily increased from 4 to 32 h, while the AI-2 activity maintained a low level stably owing to the presence of D-ribose.
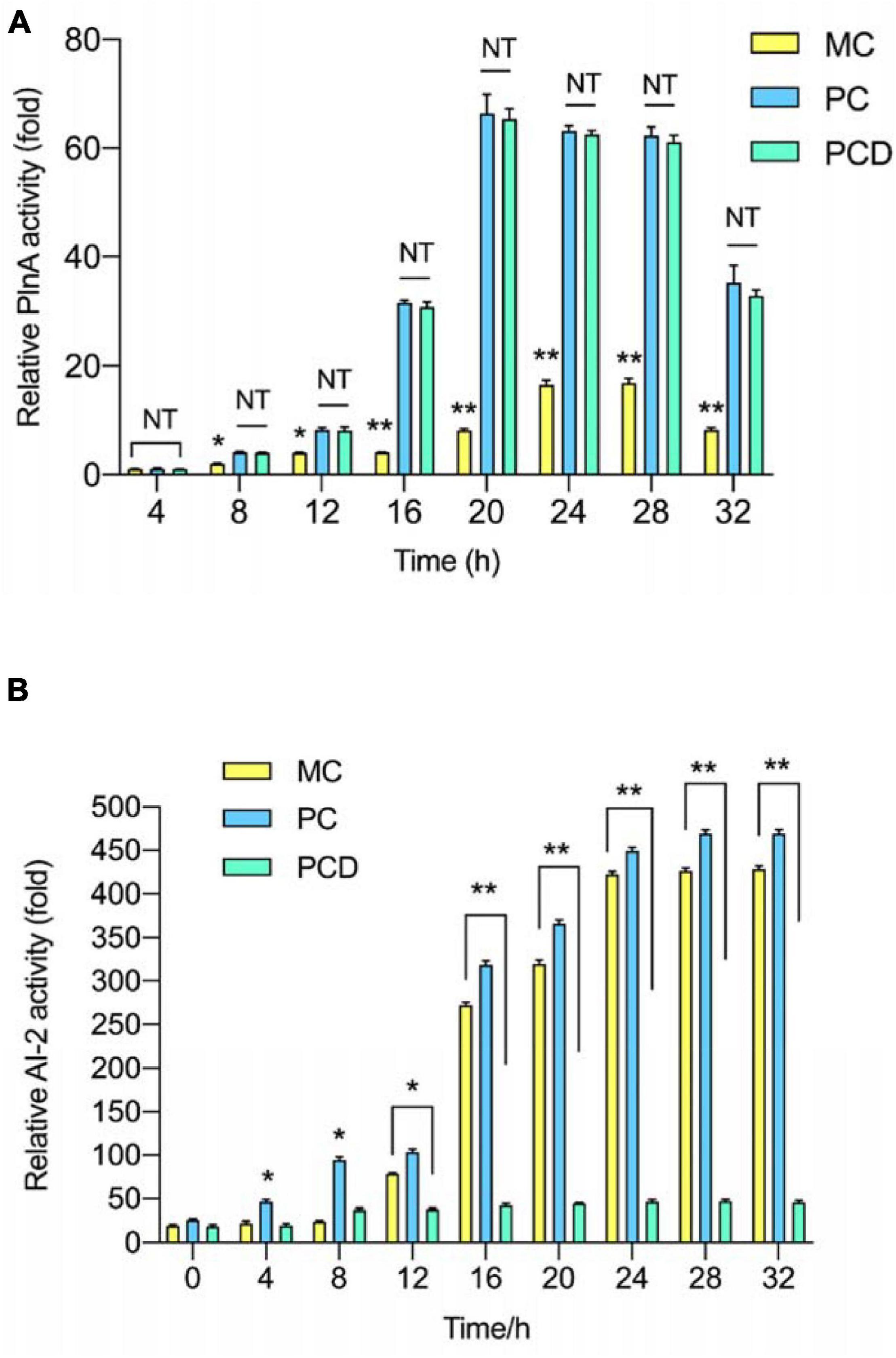
Figure 6. QS signal molecules activity in co-culture. (A) Relative PlnA activity. (B) Relative AI-2 activity. PC means positive bacteriocin-inducing co-culture of L. plantarum RX-8 and B. subtilis BS-15 (106:106 CFU/ml) at 37°C in 4–32 h. PCD means PC with 200 mM D-ribose. MC means monoculture of L. plantarum RX-8 (106 CFU/ml) at 37°C in 4–32 h. NT indicates that there were no significant differences in the results of control; *p < 0.05 and **p < 0.01.
To explore why B. subtilis BS-15 could enhance the biosynthesis of plantaricin (Figure 7), the genes of plnABCDEF, pfs, and luxS in L. plantarum RX-8 were determined by PCR (Supplementary Figure 1). Meanwhile, these genes were detected at the transcriptional level in PC, PCD, and MC. According to Figure 7, the transcriptional levels of plnE and plnF in co-culture significantly increased in 8–32 h compared with monoculture (p < 0.05), and the highest values of 8.32-fold (plnE) and 8.40-fold (plnF) in PC were detected at 20 h. A similar tendency was observed in co-culture plantaricin production. Moreover, the expression of plnABCD, pfs, and luxS was positively correlated with plantaricin production in co-culture, indicating that B. subtilis BS-15 probably induced plantaricin effect by upregulating the expression of these genes.
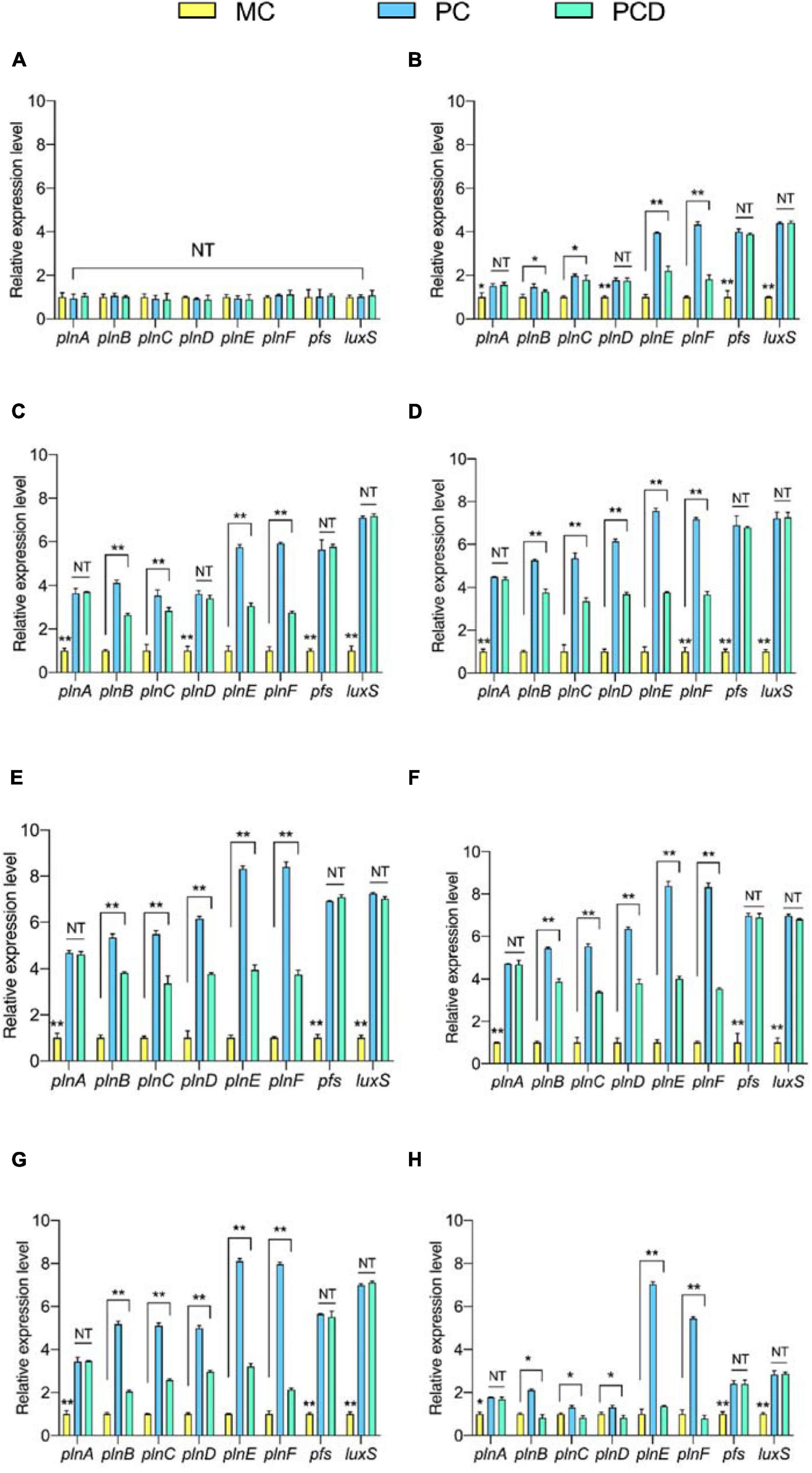
Figure 7. The relative transcriptional levels of genes plnABCDEF, pfs, and luxS at 4 h (A), 8 h (B), 12 h (C), 16 h (D), 20 h (E), 24 h (F), 28 h (G), and 32 h (H), respectively. The yellow bar (left) means MC of L. plantarum RX-8 (106 CFU/ml) without B. subtilis BS-15 at 37°C in 4∼32 h, the blue bar (middle) means PC, of positive bacteriocin-inducing co-culture of L. plantarum RX-8 and B. subtilis BS-15 (106:106 CFU/ml) at 37°C in 4–32 h, and the green bar (right) means PCD, PC with 200 mM D-ribose. NT indicates that there were no significant differences in the results of the control; *p < 0.05 and **p < 0.01.
In contrast to the decreasing trend found for interspecies QS signal AI-2 activity after the addition of D-ribose, the expression levels of pfs and luxS in PCD were considerably upregulated (p < 0.05). This fully verified D-ribose inhibits AI-2 by competitive binding to specific receptors rather than influencing the expression of key AI-2 biosynthesis enzymes (LuxS and Pfs). While 92.76% of AI-2 activity was lost following the addition of 200 mM D-ribose, the relative expression of plnE and plnF declined by only 48.02 and 42.53%, and the plantaricin production was decreased by only about 50% in PCD at 20 h. Additionally, similar changes in plnBCD transcriptional levels were observed in PCD. These findings suggested that the AI-2-mediated interspecies QS system had a role in the regulation of plantaricin induction when co-cultured with B. subtilis BS-15, but it was not exclusive to induction.
Given that the transcriptional levels of plnA and plnEF in co-culture were elevated even after the addition of D-ribose, it was hypothesized that B. subtilis BS-15 probably activated the plnA transcription by secreting a specific metabolite and that the PlnA-mediated intraspecies QS system played an important role in the plantaricin induction in co-culture. There was no significant difference in plnA transcriptional level between PC and PCD at all tested times (p > 0.05), indicating that the inducing activities of PlnA and AI-2 were independent.
The label-free proteomics analysis detected 211 proteins from B. subtilis BS-15, which were significantly (p < 0.05) different in PC and NC at 20 h. Meanwhile, the expression of 165 proteins was upregulated, whereas 46 proteins were downregulated (Figure 8A). The 211 differentially expressed proteins were functionally identified using the GO category system and KEGG pathway analysis, with the findings displayed in Figure 8B and Supplementary Figure 2.
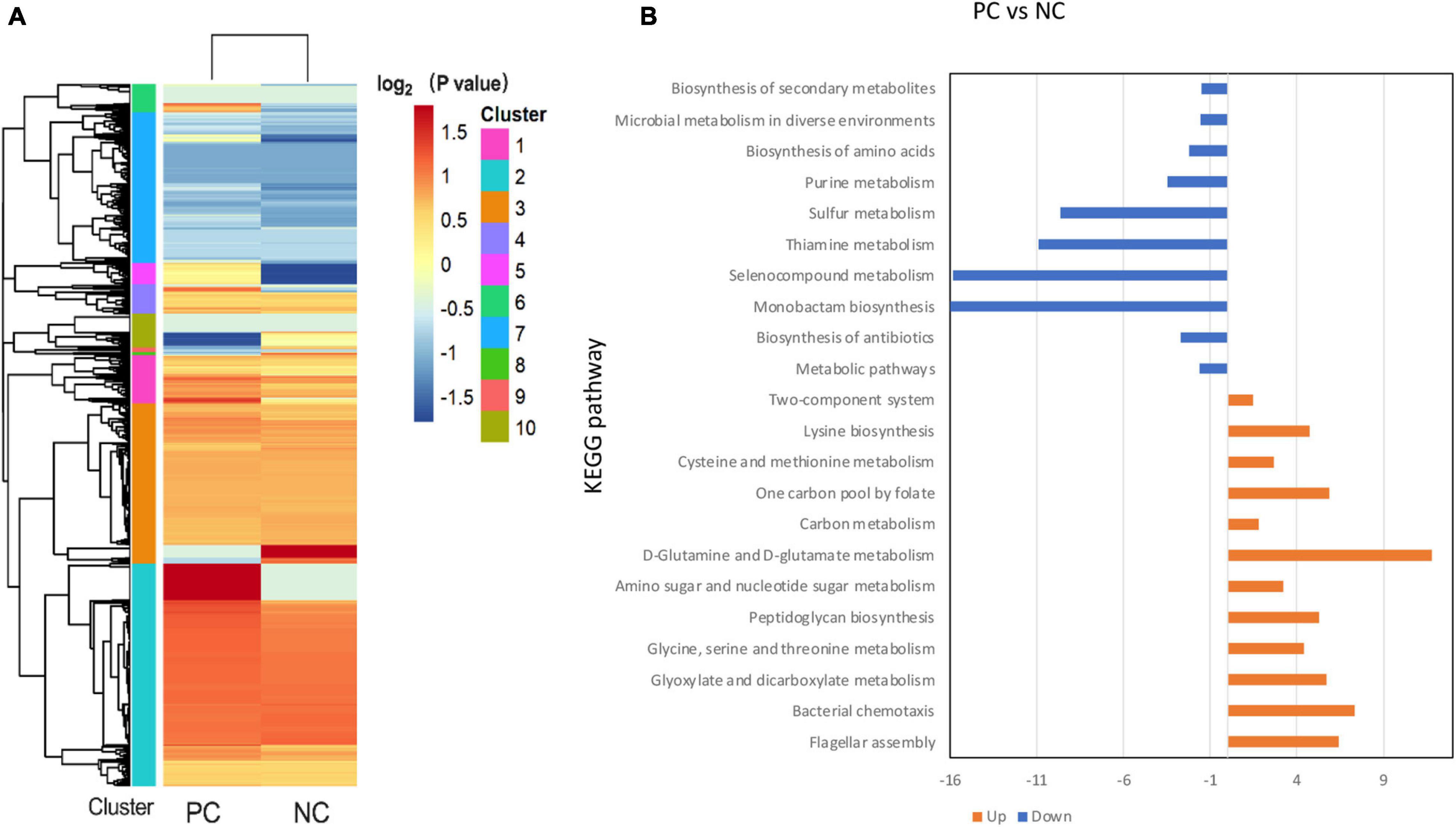
Figure 8. Categorical proteomics summary of B. subtilis BS-15 in co-culture. (A) Heatmap of different identified proteins. (B) Identified proteins were mapped to different KEGG pathways. PC means positive bacteriocin-inducing co-culture of L. plantarum RX-8 and B. subtilis BS-15 (106:106 CFU/ml) at 37°C for 20 h, and NC means negative bacteriocin-inducing co-culture of L. plantarum RX-8 and B. subtilis BS-15 (106:104 CFU/ml) at 37°C for 20 h.
To be more precise, these up-regulated proteins are mainly involved in flagellar assembly (MotB, FliM, FliG, FliY, and CheA), anaerobic respiration (CydA, CydB, and NarH), peptidoglycan (PG) biosynthesis (MurB, MurC, and MurD), and glycine cleavage system (GCS) (GcvPA, GcvPB, GcvH, GcvT, and PurU). In terms of the flagellar assembly pathway, CheA is a core protein of chemotaxis in B. subtilis that senses the chemical environment (Arapov et al., 2020), while MotB, FliM, FliG, and FliY control the direction of flagellar rotation based on chemotaxis levels (Phillips et al., 2015). It is likely that B. subtilis BS-15 dodged attack under PC by changing the directions of flagellar rotation, thus, moving to more favorable conditions. In the case of anaerobic respiration, CydA and CydB would be active only in insufficient oxygen (Larsson et al., 2005). NarH, a subunit of respiratory nitrate reductase, is required for the use of nitrate (NO3–) or nitrite (NO2–) as electron acceptors in low oxygen availability (Sonenshein et al., 2002). As a result, it is likely that B. subtilis BS-15 in PC survived by adapting its respiration pathway in response to environment changes. The upregulation of MurB, MurC, and MurD at protein level suggested that there were more PG in PC than in NC, as these proteins are involved in the synthesis of PG precursors (Real and Henriques, 2006; Vasudevan et al., 2007). The GCS catalyzed the oxidative decarboxylation of glycine and was dependent on tetrahydrofolate (THF) biosynthesis (Yang et al., 2020). It is helpful to obtain energy and 5,10-methylene-tetrahydrofolate, a C1 unit that is necessary for the production of serine, thymidine, and purines (Kikuchi et al., 2008). Almost every protein in the GCV system showed high upregulation, and PurU, a key enzyme of THF in PC, also increased. It can be deduced that the PC induces the GCS system. The functions of downregulated proteins mainly gathered in thiamin pyrophosphate biosynthesis (ThiC and ThiG), chorismate mutase (AroH), and ATP-sulfurylase (Sat). These pathways are associated with energy metabolism (Simonics et al., 2002; Pratap et al., 2017), indicating that B. subtilis BS-15 in NC may have been subjected to energy limitation.
Plantaricin is a broad-spectrum bacteriocin produced by L. plantarum RX-8 with significant food industry application potential to control foodborne pathogens. Low plantaricin production has been considered a major limitation restricting its commercialization. Considering induction of bacteriocin production by co-culture is widespread among plantaricin-producing L. plantarum strains, co-culturing with specific bacterial strains as an environmental stimulus may be a promising strategy to improve plantaricin production in food industry (Chanos and Mygind, 2016). In our study, co-cultured plantaricin production was 32 times higher than the plantaricin generated by the monoculture of L. plantarum RX-8. To facilitate the screening of effective bacteriocin-inducer strains and promote large-scale production and application of plantaricin in the food industry, preliminary study on the induction regulation mechanism of bacteriocin production by co-culture was carried out.
In the present study, B. subtilis BS-15 enhanced the plantaricin production of L. plantarum RX-8 without cell-to-cell physical contact between the two strains. A similar finding was reported in L. plantarum DC400, where bacteriocin increased due to the co-culturing with Fructilactobacillus sanfranciscensis DPPMA174 or Furfurilactobacillus rossiae A7 in a double-culture vessel apparatus separated by a 0.4-μm membrane filter (Di Cagno et al., 2009). It is indicated that cell-to-cell direct physical contact in co-culture is not necessary for plantaricin-inducing effect.
Considering the complexity of the interactions between L. plantarum RX-8 and B. subtilis BS-15, the inoculum ratio of the two bacteria in co-culture was a critical parameter. As observed, different inoculum ratios may have a distinct effect on the induction of plantaricin production in co-culture, implying that the inoculum ratio in co-culture was a key parameter in bacteriocin-inducing activity. According to Ariana and Hamedi (2017), the nisin synthesis may be induced at three different inoculum ratios (1:1, 1:2 and 2:1) of Yarrowia lipolytica ATCC 18942 and bacteriocin-producing Lactococcus lactis subsp. lactis UTMC 106. The largest amount of nisin was produced at a 1:1 inoculum ratio (Ariana and Hamedi, 2017). The co-culture bacteriocin-producing Lactobacillus delbrueckii subsp. bulgaricus BB18 and Streptococcus thermophilus 11A had similar behaviors. Bacteriocin production was dependent on the initial ratio between the two microorganisms, and bacteriocin activity in the initial ratio of 1:1 increased by 36% compared with that of 3:1 (Simova et al., 2008).
In order to ascertain the regulation intensity of AIP-mediated interspecies QS system/AI-2-mediated intraspecies QS system on plantaricin induction and their relationship in co-culture, the study analyzed the effect of co-culturing with B. subtilis BS-15 on bacteriocin-producer L. plantarum RX-8 cell growth, plantaricin production, QS signal molecule PlnA/AI-2 secretion, as well as plantaricin biosynthesis gene cluster and AI-2 synthesis-related gene expression. Results indicated that while the viable cell counts of L. plantarum RX-8 were slightly higher in co-culture (PC) than in monoculture (MC), the bacteriocin activity, PlnA inducing, and AI-2 activity were stronger in PC than in MC. It was concluded that there is a strong and positive correlation between QS signal molecule activity and plantaricin production in co-culture, but the correlation between viable cell counts and plantaricin production was not apparent. According to Man et al. (2014), when L. plantarum KLDS1.0391 was co-cultured with Lactobacillus helveticus KLDS1.9207, cell numbers were slightly higher than in monoculture, whereas the AI-2 activity was enhanced effectively (Man et al., 2014). A similar relationship between PLNC8 activity and cell number of L. plantarum NC8 was observed in co-culture with inducer strains (Maldonado et al., 2004). Furthermore, when AI-2 inhibitor was added, bacteriocin activity decreased in comparison with that in PC. As a result, it was found that AI-2 was involved in the bacteriocin-inducing activity of B. subtilis BS-15 for L. plantarum RX-8. Similar findings were observed in the co-culture of L. plantarum KLDS1.0391 and L. helveticus KLDS1.9207 (Man et al., 2014), as well as in L. plantarum DC400 that was co-cultured with F. sanfranciscensis DPPMA174 or F. rossiae A7 (Di Cagno et al., 2009). In accordance with the bacteriocin activity, the PlnA-inducing activity was dramatically increased in PC and PCD compared with MC. Like AI-2, PlnA is a crucial parameter for co-culture-induced bacteriocin synthesis. When co-culturing with another type of AIP, PlNC8 had the same effect (Maldonado et al., 2004). Moreover, PlnA-inducing activity in PC is not greatly different from that in PCD (p > 0.05). A similar pattern was also observed at the transcriptional level of the plnA gene. That indicated that AI-2 inhibitor had no effect on PlnA production in co-culture. Therefore, the inducing impact of PlnA and AI-2 was independent in co-culture. Interestingly, the PCD massively boosted the PlnA-inducing activities, while the viable cell counts of L. plantarum RX-8 only increased marginally. Apart from the effect of viable cell counts, it can be deduced that B. subtilis BS-15 likely produces some compounds that promote the secretion of PlnA of L. plantarum RX-8. In the case of Paracin 1.7, the CFS of a B. subtilis strain could improve the output of Lacticaseibacillus paracasei HD17 of Paracin 1.7 in co-culture, and the active compounds in the B. subtilis culture supernatant were several peptides between 15 and 45 kDa (Ge et al., 2014). Due to the fact that the inducing compounds from B. subtilis BS-15 have not been discovered or defined, further research will be required. The findings related to plantaricin biosynthesis gene cluster and AI-2 synthesis-related gene expression have further proved the above findings.
It is hypothesized that the mechanism by which L. plantarum RX-8 response to intraspecific/interspecific QS signal molecules (PlnA/AI-2) could be two paths: One is that AI-2 activates the transcription of genes plnBCD, followed by an increase in the transcription of genes plnEF, resulting in a significant increase in plantaricin production. Another possibility is that a specific metabolite from B. subtilis BS-15 activated the transcription of plnA, and then more PlnA was released to enhance the transcription of the three-component regulatory system (coded by plnBCD) and finally led to a higher plantaricin production. Man et al. (2012, 2014) discovered that AI-2 acted as an indirect trigger of upregulation of genes plNC8HK–plnD, and enhance bacteriocin production in L. plantarum KLDS1.0391 (Man et al., 2012, 2014). The capability of exogenous bacteria to induce bacteriocin production is apparently very strain specific (Rojo-Bezares et al., 2007). Possibly, the active substance released by B. subtilis BS-15 might lose its inducing effect when it is co-cultured with other bacteriocin-producing strains.
The effect of varying inoculum ratios of L. plantarum RX-8 and B. subtilis BS-15 on plantaricin production showed that the proper inoculation of B. subtilis BS-15 was vital for a successful inducing effect in co-culture. This study carried out differential proteomics on B. subtilis BS-15 between PC and NC to explore how bacteriocin-inducing B. subtilis BS-15 activated PlnA/AI-2. Given the change in B. subtilis BS-15 at the protein level, it is likely that B. subtilis BS-15 in PC detected changes in the chemical environment before moving to more favorable conditions by changing the direction of flagellar motor rotation to avoid plantaricin attack (Cannistraro et al., 2011), thus, producing more PG to guarantee its survival (Larsson et al., 2005). Moreover, the terminal oxidases and other respiratory enzymes of B. subtilis BS-15 in PC were altered to conserve energy, and the GCV system generated ATP and NADH through utilizing glycine as a nitrogen source. Additionally, the GCV system is capable of generating C1 units, which are used to produce purines, histidine, thymine, and methionine (Kikuchi et al., 2008). It was hypothesized that activating the GCV system would disrupt a crucial function of B. subtilis BS-15, resulting in modification or synthesis of signal molecules that may regulate bacteriocin production of L. plantarum RX-8. It is required to be determined in future research through gene disruption mutants or metabolomics analysis. Due to the fact that the strongly downregulated proteins (thiamin pyrophosphate biosynthesis, chorismate mutase, and ATP-sulfurylase) were related to energy and metabolic pathways, it could be concluded that B. subtilis BS-15 in NC lacked conserving energy and suffered from thiamin pyrophosphate and energy shortage.
The co-culturing of B. subtilis BS-15 has been proven to be an alternative for the induction of plantaricin production. When L. plantarum RX-8 and B. subtilis BS-15 were co-inoculated in MRS for 20 h at 1:1 inoculum ratio (106:106 CFS/ml), plantaricin achieved the largest output (2048 AU/ml) in co-culture. In comparison with the monoculture of L. plantarum RX-8, plantaricin production in co-culture with B. subtilis BS-15 improved by 32 times. Both the PlnA-mediated intraspecies QS system and the AI-2-mediated interspecies QS system play important roles in plantaricin induction when co-culturing with B. subtilis BS-15, and the inducing effect of PlnA and AI-2 was independent in co-culture. It is possible that AI-2 activates the transcription of plantaricin biosynthesis genes, resulting in a significant increase in plantaricin production. Meanwhile, the inducing strain BS-15 appears to activate the secretion of PlnA, triggering the QS system, which further increases plantaricin production. Differential proteomics analysis of B. subtilis BS-15 in co-culture indicated that the bacteriocin-inducing regulatory mechanism might be related to flagellar assembly, peptidoglycan biosynthesis, anaerobic respiration, glycine cleavage system, and thiamin pyrophosphate biosynthesis. We are currently performing a detailed study on the bacteriocin-inducing regulatory mechanism in co-culture, using integrated transcriptomic and proteomic methods.
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
GL conceptualized the idea, made corrections, and approved the research article for publication. XL, YS, and JZ performed the experiments. RN, XH, and JD analyzed the results and wrote the first draft of the manuscript. YL and RN revised the article. All authors contributed to the article and approved the submitted version.
This project was funded by Natural Science Foundation of China (Grant No. 31871772), Beijing Natural Science Foundation (Grant No. 6192003), School Level Cultivation Fund of Beijing Technology and Business University for Distinguished and Excellent Young Scholars (Grant No. BTBUYP2022), 2021 Postgraduate Scientific Research Ability Improvement Project of Beijing Technology and Business University, Graduate Innovation Project of School of Food and Health in 2021.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.772546/full#supplementary-material
Arapov, T. D., Saldaña, R. C., Sebastian, A. L., Ray, W. K., Helm, R. F., and Scharf, B. E. (2020). Cellular stoichiometry of chemotaxis proteins in Sinorhizobium meliloti. J. Bacteriol. 202:e00141-20. doi: 10.1128/JB.00141-20
Ariana, M., and Hamedi, J. (2017). Enhanced production of nisin by co-culture of Lactococcus lactis sub sp. lactis and Yarrowia lipolytica in molasses based medium. Biotechnol. J. 256, 21–26. doi: 10.1016/j.jbiotec.2017.07.009
Cannistraro, V. J., Glekas, G. D., Rao, C. V., and Ordal, G. W. (2011). Cellular stoichiometry of the chemotaxis proteins in Bacillus subtilis. J. Bacteriol. 193, 3220–3227. doi: 10.1128/JB.01255-10
Chanos, P., and Mygind, T. (2016). Co-culture-inducible bacteriocin production in lactic acid bacteria. Appl. Microbiol. Biotechnol. 100, 4297–4308. doi: 10.1007/s00253-016-7486-8
Corsetti, A., and Valmorri, S. (2001). “Lactic acid bacteria | Lactobacillus spp.: Lactobacillus plantarum,” in Encyclopedia of Dairy Sciences, 2nd Edn. ed. John W. Fuquay (Cambridge, MA: Academic Press), 111–118.
da Silva Sabo, S., Vitolo, M., González, J. M. D., and Oliveira, R. P. S. (2014). Overview of Lactobacillus plantarum as a promising bacteriocin producer among lactic acid bacteria. Food Res. Int. 64, 527–536. doi: 10.1016/j.foodres.2014.07.041
Di Cagno, R., De Angelis, M., Coda, R., Minervini, F., and Gobbetti, M. (2009). Molecular adaptation of sourdough Lactobacillus plantarum DC400 under co-cultivation with other lactobacilli. Res. Microbiol. 160, 358–366. doi: 10.1016/j.resmic.2009.04.006
Diep, D. B., Straume, D., Kjos, M., Torres, C., and Nes, I. F. (2009). An overview of the mosaic bacteriocin pln loci from Lactobacillus plantarum. Peptides 30, 1562–1574. doi: 10.1016/j.peptides.2009.05.014
Ge, J., Fang, B., Wang, Y., Song, G., and Ping, W. (2014). Bacillus subtilis enhances production of Paracin1.7, a bacteriocin produced by Lactobacillus paracasei HD1-7, isolated from Chinese fermented cabbage. Ann. Microbiol. 64, 1735–1743. doi: 10.1007/s13213-014-0817-z
Gutiérrez-Cortés, C., Suarez, H., Buitrago, G., Nero, L. A., and Todorov, S. D. (2018). Enhanced bacteriocin production by Pediococcus pentosaceus 147 in co-culture with Lactobacillus plantarum LE27 on cheese whey broth. Front. Microbiol. 9:2952. doi: 10.3389/fmicb.2018.02952
Kikuchi, G., Motokawa, Y., Yoshida, T., and Hiraga, K. (2008). Glycine cleavage system: reaction mechanism, physiological significance, and hyperglycinemia. Proc. Jpn. Acad. Ser. B 84, 246–263. doi: 10.2183/pjab.84.246
Larsson, J. T., Rogstam, A., and von Wachenfeldt, C. (2005). Coordinated patterns of cytochrome bd and lactate dehydrogenase expression in Bacillus subtilis. Microbiology 151, 3323–3335. doi: 10.1099/mic.0.28124-0
Le, N., Bach, L., Nguyen, D., Le, T., Pham, K., Nguyen, D., et al. (2019). Evaluation of factors affecting antimicrobial activity of bacteriocin from Lactobacillus plantarum microencapsulated in alginate-gelatin capsules and its application on pork meat as a bio-preservative. Int. J. Environ. Res. Public Health 16:1017. doi: 10.3390/ijerph16061017
Lei, S., Zhao, R., Sun, J., Ran, J., Ruan, X., and Zhu, Y. (2020). Partial purification and characterization of a broad−spectrum bacteriocin produced by a Lactobacillus plantarum zrx03 isolated from infant’s feces. Food. Sci. Nutr. 8, 2214–2222. doi: 10.1002/fsn3.1428
Maldonado, A., Ruiz-Barba, J. L., and Jiménez-Díaz, R. (2004). Production of plantaricin NC8 by Lactobacillus plantarum NC8 is induced in the presence of different types of gram-positive bacteria. Arch. Microbiol. 181, 8–16. doi: 10.1007/s00203-003-0606-8
Maldonado-Barragán, A., Caballero-Guerrero, B., Lucena-Padrós, H., and Ruiz-Barba, J. L. (2013). Induction of bacteriocin production by coculture is widespread among plantaricin-producing Lactobacillus plantarum strains with different regulatory operons. Food Microbiol. 33, 40–47. doi: 10.1016/j.fm.2012.08.009
Maldonado-Barragán, A., Ruiz-Barba, J. L., and Jiménez-Díaz, R. (2009). Knockout of three-component regulatory systems reveals that the apparently constitutive plantaricin-production phenotype shown by Lactobacillus plantarum on solid medium is regulated via quorum sensing. Int. J. Food Microbiol. 130, 35–42. doi: 10.1016/j.ijfoodmicro.2008.12.033
Man, L.-L., Meng, X.-C., and Zhao, R.-H. (2012). Induction of plantaricin MG under co-culture with certain lactic acid bacterial strains and identification of LuxS mediated quorum sensing system in Lactobacillus plantarum KLDS1.0391. Food Control 23, 462–469. doi: 10.1016/j.foodcont.2011.08.015
Man, L.-L., Meng, X.-C., Zhao, R.-H., and Xiang, D.-J. (2014). The role of plNC8HK-plnD genes in bacteriocin production in Lactobacillus plantarum KLDS1.0391. Int. Dairy J. 34, 267–274. doi: 10.1016/j.idairyj.2013.08.009
Man, L. L., and Xiang, D. J. (2021). LuxS-mediated quorum sensing system in Lactobacillus plantarum NMD-17 from koumiss: induction of plantaricin MX in co-cultivation with certain lactic acid bacteria. Folia Microbiol. 66, 855–871. doi: 10.1007/s12223-021-00890-0
Meng, F., Lu, F., Du, H., Nie, T., Zhu, X., Connerton, I. F., et al. (2021). Acetate and auto−inducing peptide are independent triggers of quorum sensing in Lactobacillus plantarum. Mol. Microbiol. 116, 298–310. doi: 10.1111/mmi.14709
Osei Sekyere, J., Maningi, N. E., and Fourie, P. B. (2020). Mycobacterium tuberculosis, antimicrobials, immunity, and lung–gut microbiota crosstalk: current updates and emerging advances. Ann. N. Y. Acad. Sci. 1467, 21–47. doi: 10.1111/nyas.14300
Phillips, A. M., Calvo, R. A., and Kearns, D. B. (2015). Functional activation of the flagellar type III secretion export apparatus. PLoS Genet. 11:e1005443. doi: 10.1371/journal.pgen.1005443
Pratap, S., Dev, A., Kumar, V., Yadav, R., Narwal, M., Tomar, S., et al. (2017). Structure of chorismate mutase-like domain of DAHPS from Bacillus subtilis complexed with novel inhibitor reveals conformational plasticity of active site. Sci. Rep. 7:6364. doi: 10.1038/s41598-017-06578-1
Real, G., and Henriques, A. O. (2006). Localization of the Bacillus subtilis murB gene within the dcw cluster is important for growth and sporulation. J. Bacteriol. 188, 1721–1732. doi: 10.1128/JB.188.5.1721-1732.2006
Rojo-Bezares, B., Saenz, Y., Navarro, L., Zarazaga, M., Ruizlarrea, F., and Torres, C. (2007). Coculture-inducible bacteriocin activity of Lactobacillus plantarum strain J23 isolated from grape must. Food Microbiol. 24, 482–491. doi: 10.1016/j.fm.2006.09.003
Ryu, E.-J., Sim, J., Sim, J., Lee, J., and Choi, B.-K. (2016). D-galactose as an autoinducer 2 inhibitor to control the biofilm formation of periodontopathogens. J. Microbiol. 54, 632–637. doi: 10.1007/s12275-016-6345-8
Sieuwerts, S., de Bok, F. A., Hugenholtz, J., and van Hylckama Vlieg, J. E. (2008). Unraveling microbial interactions in food fermentations: from classical to genomics approaches. Appl. Environ. Microbiol. 74, 4997–5007. doi: 10.1128/AEM.00113-08
Simonics, T., Bánszky, L., and Maráz, A. (2002). Genetics of sulphate assimilation in Schizosaccharomyces pombe. Acta Microbiol. Immunol. Hung. 49, 279–283. doi: 10.1556/amicr.49.2002.2-3.15
Simons, A., Alhanout, K., and Duval, R. E. (2020). Bacteriocins, antimicrobial peptides from bacterial origin: overview of their biology and their impact against multidrug-resistant bacteria. Microorganisms 8:639. doi: 10.3390/microorganisms8050639
Simova, E. D., Beshkova, D. M., Angelov, M. P., and Dimitrov, Z. P. (2008). Bacteriocin production by strain Lactobacillus delbrueckii ssp. bulgaricus BB18 during continuous prefermentation of yogurt starter culture and subsequent batch coagulation of milk. J. Ind. Microbiol. Biotechnol. 35, 559–567. doi: 10.1007/s10295-008-0317-x
Sonenshein, A. L., Hoch, J. A., and Losick, R. (eds) (2002). Bacillus Subtilis and its Closest Relatives. Washington, DC: ASM Press.
Sturme, M. H. J., Francke, C., Siezen, R. J., de Vos, W. M., and Kleerebezem, M. (2007). Making sense of quorum sensing in lactobacilli: a special focus on Lactobacillus plantarum WCFS1. Microbiology 153(Pt 12), 3939–3947. doi: 10.1099/mic.0.2007/012831-0
Todorov, S. D., de Paula, O. A. L., Camargo, A. C., Lopes, D. A., and Nero, L. A. (2018). Combined effect of bacteriocin produced by Lactobacillus plantarum ST8SH and vancomycin, propolis or EDTA for controlling biofilm development by Listeria monocytogenes. Rev. Argent. Microbiol. 50, 48–55. doi: 10.1016/j.ram.2017.04.011
Vasudevan, P., Weaver, A., Reichert, E. D., Linnstaedt, S. D., and Popham, D. L. (2007). Spore cortex formation in Bacillus subtilis is regulated by accumulation of peptidoglycan precursors under the control of sigma K. Mol. Microbiol. 65, 1582–1594. doi: 10.1111/j.1365-2958.2007.05896.x
Vriezen, J. A., Valliere, M., and Riley, M. A. (2009). The evolution of reduced microbial killing. Mol. Biol. Evol. 1, 400–408. doi: 10.1093/gbe/evp042
Yang, H., Liu, Y., Li, J., Liu, L., Du, G., and Chen, J. (2020). Systems metabolic engineering of Bacillus subtilis for efficient biosynthesis of 5−methyltetrahydrofolate. Biotechnol. Bioeng. 117, 2116–2130. doi: 10.1002/bit.27332
Zhang, Z., Fan, S., Huang, D., Xiong, T., Nie, S., and Xie, M. (2020). Polysaccharides from fermented Asparagus officinalis with Lactobacillus plantarum NCU116 alleviated liver injury via modulation of glutathione homeostasis, bile acid metabolism, and SCFA production. Food Funct. 11, 7681–7695. doi: 10.1039/D0FO01435D
Zhao, D., Meng, F., Zhou, L., Lu, F., Bie, X., Sun, J., et al. (2021). Maltose effective improving production and regulatory biosynthesis of plantaricin EF in Lactobacillus plantarum 163. Appl. Microbiol. Biotechnol. 105, 2713–2723. doi: 10.1007/s00253-021-11218-w
Keywords: bacteriocin, co-culture, quorum sensing, biopreservation, proteome
Citation: Liu G, Nie R, Liu Y, Li X, Duan J, Hao X, Shan Y and Zhang J (2022) Bacillus subtilis BS-15 Effectively Improves Plantaricin Production and the Regulatory Biosynthesis in Lactiplantibacillus plantarum RX-8. Front. Microbiol. 12:772546. doi: 10.3389/fmicb.2021.772546
Received: 08 September 2021; Accepted: 11 November 2021;
Published: 28 January 2022.
Edited by:
Paloma López, Margarita Salas Center for Biological Research, Spanish National Research Council (CSIC), SpainReviewed by:
Djamel Drider, Lille University of Science and Technology, FranceCopyright © 2022 Liu, Nie, Liu, Li, Duan, Hao, Shan and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Guorong Liu, bGl1Z3Vvcm9uZzE5ODNAMTI2LmNvbQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.