
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
METHODS article
Front. Microbiol. , 02 July 2021
Sec. Food Microbiology
Volume 12 - 2021 | https://doi.org/10.3389/fmicb.2021.687691
This article is part of the Research Topic Emerging Frontiers in the Formation of Viable but Non-Culturable Microorganisms and Biofilms During Food Processing View all 25 articles
Pediococcus acidilactici may significantly reduce the pH-value, and thus has different influence, including serving as a probiotic in human microbiota but a spoilage in human food as it could change the flavor. Pediococcus acidilactici is also capable of entering into the viable but non-culturable (VBNC) state causing false negative results of standard culture-based detection method. Thus, development of detection method for VBNC state P. acidilactici is of great significance. In this study, propidium monoazide (PMA) combined with cross priming amplification (CPA) was developed to detect the VBNC cells of P. acidilactici and applied on the detection in different systems. With detection limit of 104 cells/ml, high sensitivity, and 100% specificity, PMA-CPA can successfully detect VBNC cells of P. acidilactici and be applied in with high robustness.
Pediococcus acidilactici may significantly reduce the pH-value, and thus has different influence, including serving as a probiotic in human microbiota but a spoilage in human food. Most commonly, P. acidilactici has been considered to be a probiotic bacteria for human beings, and also a typical type of lactic acid bacteria (LAB), which have been well-documented to be widely existing in human microbiota. Such probiotic bacteria have been well-studied to significantly aid in the microbiota balance, and once altered, various types of human diseases could occur. One important example is the correlation between human microbiota and urinary tract infection, in which microbiota in a few different sites, including intestine and vagina, has been found to play a role in the occurrence of urinary tract infection (Foxman, 2010; Whiteside et al., 2015; Stapleton, 2017; Paalanne et al., 2018; Magruder et al., 2020). Therefore, probiotic LAB is an important type of human probiotic and significantly aids in the prevention of urinary tract infection. In addition, accurate detection of probiotic commensal has significantly raised the public interest as this serves as an important indicator. However, a major concern still remains, as a large proportion of such probiotic commensal are hard to culture, which has further raised the aim of this study. As a commonly existing human bacteria, P. acidilactici is also one of LAB that are frequently used due to their capacity to produce bacteriocin to inhibit the growth of other spoilage bacteria (Salminen et al., 2004; Zhong et al., 2013). However, with improper processing, P. acidilactici produces acids including lactic acid, malic acid, citric acid, propionic acid, acetic acid, and short-chain fatty acids through fermentation, resulting in the significant decrease of pH-value, and the change in morphology and flavor (Olaoye et al., 2008; Xu et al., 2009). In consequence, the shelf life of the food would be shorter and the deterioration of the food occurs. Furthermore, it has been proved that P. acidilactici is able to enter into the viable but non-culturable (VBNC) state, which causes the false negative detection by the standard culture-based detection methods (Fakruddin et al., 2013; Ramamurthy et al., 2014; Xie et al., 2017a,b; Xu et al., 2017a,b; Li et al., 2020). Thus, accurate detection of P. acidilactici cells in the VBNC state is in need (Ding et al., 2017; Truchado et al., 2020).
Nowadays, the detection of the VBNC state is currently based on molecular detection techniques and the fluorescence microscopy with dying kit (Xu et al., 2011a,b,c; Zhao et al., 2018a,b; Dong et al., 2020). In the past few years, PCR based methodologies have been developed for rapid detection of viable bacterial cells, in combination with use of different substances including propidium monoazide (PMA) (Zhong et al., 2016; Bao et al., 2017a,b,c; Jia et al., 2018; Liu et al., 2018b,c; Zhong and Zhao, 2018). However, PCR based methodologies require strict temperature change process and determination step, such as gel electrophoresis or hybridization, which significantly reduces rapidity and simplicity in operation of such methods (Miao et al., 2016, 2017a,b,c; Zhao et al., 2020). In addition, the PMA-PCR has a lower sensitivity compared to the novel isothermal amplification methods with complex procedure and higher cost (Xu et al., 2008a,b; Liu et al., 2019).
Cross priming amplification (CPA) was developed to detect the target DNA with exponential amplification (Xu et al., 2007, 2010; Bai et al., 2015). Cross priming amplification can be completed under constant temperature with the advantages of simplicity, rapidity, high sensitivity, and cost-efficiency (Liu et al., 2015, 2017; Zheng et al., 2020). The expensive heating machine can be replaced by the simple water bath or other heating block. The whole amplification procedure can be completed within 1 h by one pair of primer, which spans three distinct sequences of a target gene (Lin et al., 2016; Zhang et al., 2019). The products of CPA can be measured based on the turbidity, electrophoresis of amplicons, and DNA-specific fluorescent reaction in tubes, such as SYBR Green-I (Xu et al., 2012b,c, 2016a,b,c; Lin et al., 2017).
In a previous study (Li et al., 2020), we had studied the key conditions of VBNC formation of P. acidilactici, and then based on the key conditions, we had discovered the reduction of VBNC formation. However, a major concern still remains, as rapid detection of P. acidilactici is highly required, especially direct detection and real-time surveillance from food products. Consequently, in this study, first we developed a rapid, sensitive, and specific detection assay on P. acidilactici based on CPA methodology. Then, we had further developed a PMA-CPA assay to directly identify the VBNC cells of P. acidilactici. Last, we had applied the established methodology on three different types of food products.
A total of 20 bacterial strains, which are common foodborne pathogens and spoilage bacteria, were tested in this study, including a P. acidilactici strain, and 19 non-target bacteria (Table 2), including Escherichia coli, Salmonella, Staphylococcus aureus, Listeria monocytogenes, Vibrio parahaemolyticus, Lactobacillus casei, Lactobacillus acetotolerans, Lactobacillus plantarum, and Pseudomonas aeruginosa. In brief, after inoculation of single colony on the plate for culturing at 37°C overnight, DNA extraction, VBNC induction, as well as serial dilution were further performed based on the bacterial suspension.
The culture of P. acidilactici in the log phase was used to induce into the VBNC state under the freezing condition (−20°C). The changing on the culturable cell number was used as an index to investigate the culturability until the culturable number was below 1 cell/ml; the cells can be regarded as those that entered into the VBNC state (Wang et al., 2011; Miao et al., 2018). The VBNC cells were finally determined by Live/Dead BacLight bacterial viability kit (Thermo Fisher Scientific, USA) (Liu et al., 2018a) in combination with fluorescence microscopy. The growth curves, including total cell numbers, culturable cell numbers, and viable cell numbers, were determined and further analyzed as described previously.
The CPA primer was designed to distinguish P. acidilactici based on pheS gene, which is a housekeeping gene. Primers used in this study were designed using Primer Premier 5 (Table 1) (Singh et al., 2018; Xu et al., 2018). The crude DNA from P. acidilactici and other bacterial strains used as templates for CPA amplification was prepared from overnight culture in MRS broth. DNA was extracted using whole genomic DNA extraction kit (Dongsheng Biotech, Guangzhou) according to the manufacturer's instructions. The DNA concentration and quality were measured using a Nano Drop 2000 (Thermo Fisher Scientific Inc., Waltham, MA, USA). The qualified DNA samples were stored at −20°C until further use (Wen et al., 2020).
Cross priming amplification reaction was carried out in a total 26 μl reaction mixture containing 20 mM Tris-HCl, 10 mM (NH4)2SO4, 10 mM KCl, 8 mM MgSO4, 0.1% Tween 20, 0.7 M betaine (Sigma, USA), 1.4 mM of dNTP mix, 8 U of Bst DNA polymerase large fragment (NEB, USA), 1.0 μM primer of 2a/1s, 0.5 μM (each) primer of 2a and 3a, 0.6 μM (each) primer of 4s and 5a, 1 μl DNA template, and 1 μl mixed chromogenic agent, and the total reaction mixture was made up of 26 μl with nuclease free water (Xu et al., 2012a). The mixed chromogenic agent was composed of 0.13 mM calcein and 15.6 mM MnCl2·4H2O. Mixture without DNA template was used as negative control (Xu et al., 2012a). The reaction mixtures are maintained at 63°C for 1 h followed by 80°C for 2 min. The amplified products were detected by electrophoresis on 1.5% agarose gel with ethidium bromide staining.
The specificity of CPA assay was evaluated by amplifying genomic DNA extracted from P. acidilactici and 19 non-target bacteria (Table 2). Nuclease free water was added instead of DNA template as blank control. The CPA reaction was conducted under the corresponding conditions mentioned above.
The evaluation of the CPA assays was conducted by using serially 10-fold diluted genomic DNA and applied in food samples with 10–108 cells/ml. The CPA products were detected by electrophoresis on 1.5% agarose gel with ethidium bromide staining (Xu et al., 2019). All the tests were performed in triplicate. The developed CPA assay was used to detect the VBNC cells of P. acidilactici combined with the PMA dying. Furthermore, the PMA-CPA was applied to detect the VBNC cells in the food samples using pheS gene as target. The products were analyzed by 1.5% agarose gel electrophoresis and fluorescent dye (MgCl2 and calcein). The ladder-like bands were observed under UV light. The color of fluorescent dye that changes reaction system from orange to green indicates positive result (Figure 1).
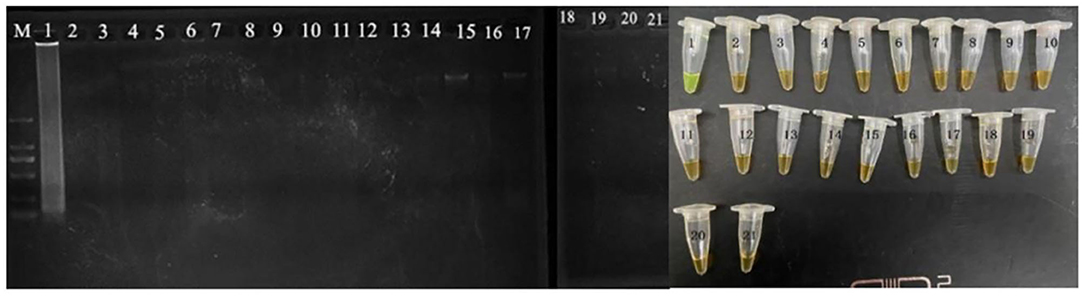
Figure 1. Specificity of CPA detection for different strains with pheS genes by 1.5% agarose gel electrophoresis and mixed chromogenic agent; M-DNA marker; lane/tube 1, P. acidilactici BM-PA17927; lane/tube 2–20, non-P. acidilactici BM-PA17927 strains of E. coli, Salmonella enteric, Vibrio parahaemolyticus, Pseudomonas aeruginosa, Listeria monocytogenes, Staphylococcus aureus, and Lactobacillus casei; lane 21, negative control.
In this study, three major types of rice/flour products in China were selected, which were mantou, rice noodle, and Cantonese pastry. Mantou, rice noodle, and Cantonese pastry are among the top list of common food in China, with a large consumption market. The sample processing had been performed as follows. Each sample of mantou, rice noodle, and Cantonese pastry weighs 25 g, and a total of 100 μl with bacterial suspension was added to each sample. All mantou, rice noodle, and Cantonese pastry samples were further stored and subjected to CFU counting at different time points. For the first few days, CFU was performed for each day, and after 3 days, CFU was performed every 3 days.
In order to further confirm if the established PMA-CPA methodology is capable of detection in the P. acidilactici VBNC state, the food samples (mantou, rice noodle, and Cantonese pastry) contaminated with P. acidilactici in the VBNC state was used as template. The PMA-treated bacteria suspension was centrifuged at 10,000 r/min for 5 min, and supernatant was removed. DNA was then extracted using a bacterial whole genomic DNA extraction kit (Dongsheng Biotech, Guangzhou). All samples including mantou, rice noodle, and Cantonese pastry were processed according to the standard procedure, and the PMA-CPA was performed accordingly.
A novel and simple nucleic acid isothermal amplification CPA has been developed to detect the species specific gene pheS. According to the results, the CPA based assay is capable of detecting P. acidilactici cells. According to the evaluation of the CPA assay conducted by series of DNA, the detection limit of CPA was 52 pg/μl for pheS gene (Figure 2).

Figure 2. Sensitivity of the CPA assay in genomic DNA of P. acidilactici with pheS genes by 1.5% agarose gel electrophoresis and mixed chromogenic agent; M-DNA marker; 1–6 refer to 52, 5.2 ng/μl, 520, 52, 5.2 pg/μl, and 520 fg/μl; 7 refers to negative control.
The developed CPA assay was utilized in detection of food samples. The bacteria were inoculated into food samples with the concentration of 10–108 cells/ml. The results showed that the detection limit of application was 104 cells/ml (Figure 3). Compared with previously reported PCR based methodologies, the CPA assays had shown significant advantages in terms of sensitivity, specificity, rapidity, and simplicity in operation.
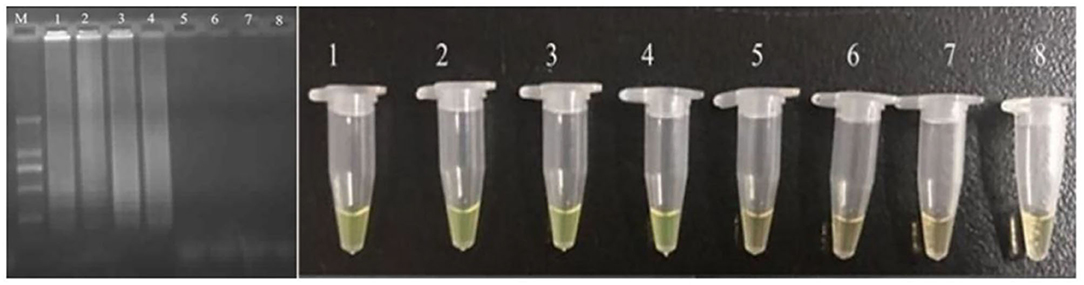
Figure 3. Sensitivity for pheS in artificially contaminated food samples. M-DNA marker; 1–7 refer to 107, 106, 105, 104, 103, 102, and 10 cells/ml; 8 refers to negative control.
The cells considered may enter into the VBNC state when the culturable cell number in the induction solution is under 1 cell/ml. In order to determine whether P. acidilactici cells in the induction solution enter the VBNC state, the LIVE/DEAD® BacLight™ bacterial viability kit (Thermo Fisher Scientific, USA) was used (Berney et al., 2007; You et al., 2012). For these assays, 500 μl of P. acidilactici culture sample was obtained and centrifuged at 5,000 r/min for 15 min. Subsequently, the P. acidilactici cells were washed twice with PBS. The supernatant was removed, and the pellet was resuspended in 500 μl of PBS. The cell suspensions were incubated with 1.5 μl of dye mixture containing SYTO 9 and PI for 30 min at room temperature in the dark. The mixture (5 μl) was observed under a fluorescent microscope. The viable cells including normal and VBNC state showed green, and the dead cells are in red. For PMA-CPA assay, the VBNC cells were added to a 1.5 ml centrifuge tube and were thoroughly mixed and left at room temperature for 10 min. Subsequently, the centrifuge tube was placed on a crushed ice box, and light treatment was performed for 15 min at 15 cm from a 650 W halogen lamp for PMA and DNA binding. The PMA molecules remaining after the treatment are passivated. The extracted DNA was detected by CPA. The results showed that the VBNC cells can be detected successfully by PMA-CPA (Figure 4A).
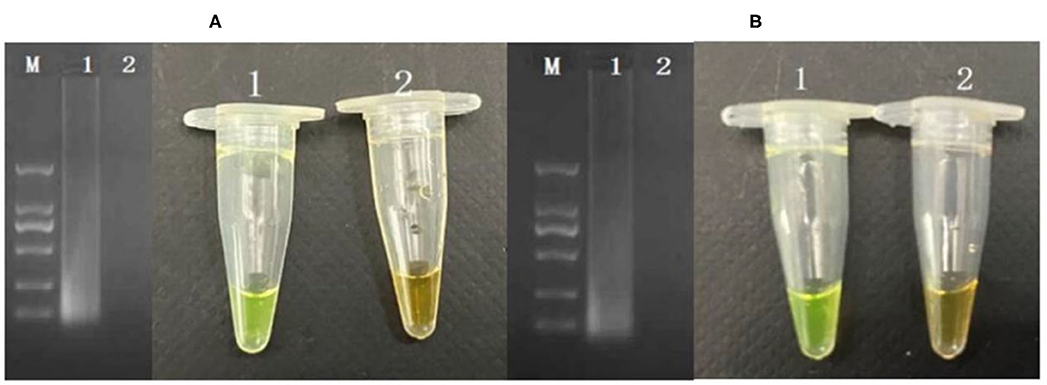
Figure 4. Application of detection of VBNC cells in pure cultures (A) and food samples (B). M-DNA marker; 1: VBNC cells in pure culture; 2: dead cells in pure culture.
The developed PMA-CPA assay as above had been further applied to detect VBNC cells of P. acidilactici from food samples including mantou, rice noodle, and Cantonese pastry. Firstly, formation of VBNC cells had been performed in artificially contaminated food samples including mantou, rice noodle, and Cantonese pastry. Following a rapid processing method, the samples had been further subjected to PMA-CPA detection. According to the results (Figure 4B), food samples of mantou, rice noodle, and Cantonese pastry containing P. acidilactici had yielded positive results. However, samples containing either none of strains (negative control) or non-target microorganisms had yielded negative results. The results demonstrated the fact that the developed method can be used in direct detection of VBNC cells of food samples (Figure 4B).
As concluded, from a widely existed human probiotic bacteria and the interest for its accurate detection as an important indicator of the health status of human microbiota, including intestine and vagina, which will be significantly correlated with urinary tract infection, a major concern had been raised as how to accurately detect the probiotic commensal LAB as they are mostly hard to culture. Consequently, in this study, we had firstly developed a rapid, sensitive, and specific detection assay on P. acidilactici based on CPA methodology, which had obtained high sensitivity and specificity. Then, we had connected the CPA assay with PMA processing to achieve a PMA-CPA assay to directly identify the VBNC cells of P. acidilactici. Thirdly, the developed PMA-CPA method had been further applied for direct detection of VBNC P. acidilactici cells to show its robustness.
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
YG conceived the study and participated in its design and coordination. KW and YY performed the experimental work and collected the data. LC organized the database. TH wrote the manuscripts. YZ revised the manuscript. All authors contributed to manuscript revision and read and approved the submitted manuscript.
This work was supported by the National Key Research and Development Program of China (2016YFD04012021) and National Key Research and Development Program of China (No. 2017YFC1601202).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Bai, Z., Xie, H., You, Q., Pickerill, S., Zhang, Y., Li, T., et al. (2015). Isothermal cross-priming amplification implementation study. Lett. Appl. Microbiol. 60, 205–209. doi: 10.1111/lam.12342
Bao, X., Jia, X., Chen, L., Peters, B. M., Lin, C., Chen, D., et al. (2017a). Effect of polymyxin resistance (pmr) on biofilm formation of Cronobacter sakazakii. Microb. Pathog. 106, 16–19. doi: 10.1016/j.micpath.2016.12.012
Bao, X., Yang, L., Chen, L., Li, B., Li, L., Li, Y., et al. (2017b). Virulent and pathogenic features on the Cronobacter sakazakii polymyxin resistant pmr mutant strain s-3. Microb. Pathog. 110, 359–364. doi: 10.1016/j.micpath.2017.07.022
Bao, X., Yang, L., Chen, L., Li, B., Li, L., Li, Y., et al. (2017c). Analysis on pathogenic and virulent characteristics of the Cronobacter sakazakii strain BAA-894 by whole genome sequencing and its demonstration in basic biology science. Microb. Pathog.109, 280–286. doi: 10.1016/j.micpath.2017.05.030
Berney, M., Hammes, F., Bosshard, F., Weilenmann, H. U., and Egli, T. (2007). Assessment and interpretation of bacterial viability by using the LIVE/DEAD BacLight kit in combination with flow cytometry. Am. Soc. Microbiol. 73, 3283–3290. doi: 10.1128/AEM.02750-06
Ding, T., Suo, Y., Xiang, Q., Zhao, X., Chen, S., Ye, X., et al. (2017). Significance of viable but nonculturable Escherichia coli: induction, detection, and control. J. Microbiol. Biotechnol. 27, 417–428. doi: 10.4014/jmb.1609.09063
Dong, K., Pan, H., Yang, D., Rao, L., Zhao, L., Wang, et al. (2020). Induction, detection, formation, and resuscitation of viable but non-culturable state microorganisms. Comprehen. Rev. Food Sci. Food Saf. 19, 149–183. doi: 10.1111/1541-4337.12513
Fakruddin, M. D., Mannan, K. S. B., and Andrews, S. (2013). Viable but nonculturable bacteria: food safety and public health perspective. ISRN Microbiol. 2013:703813. doi: 10.1155/2013/703813
Foxman, B. (2010). The epidemiology of urinary tract infection. Nat. Rev. Urol. 7, 653–660. doi: 10.1038/nrurol.2010.190
Jia, X., Hua, J., Liu, L., Xu, Z., and Li, Y. (2018). Phenotypic characterization of pathogenic Cronobacter spp. strains. Microb. Pathogen. 121, 232–237. doi: 10.1016/j.micpath.2018.05.033
Li, Y., Huang, T.-Y., Mao, Y., Chen, Y., Shi, F., Peng, R., et al. (2020). Effect of environmental conditions on the formation of the viable but nonculturable state of Pediococcus acidilactici BM-PA17927 and its control and detection in food system. Front. Microbiol. 11:586777. doi: 10.3389/fmicb.2020.586777
Lin, S., Li, L., Li, B., Zhao, X., Lin, C., Deng, Y., et al. (2016). Development and evaluation of quantitative detection of N-epsilon-carboxymethyl-lysine in Staphylococcus aureus biofilm by LC-MS method. Basic Clin. Pharmacol. Toxicol. 118: 33.
Lin, S., Yang, L., Chen, G., Li, B., Chen, D., Li, L., et al. (2017). Pathogenic features and characteristics of food borne pathogens biofilm: biomass, viability and matrix. Microb. Pathog. 111, 285–291. doi: 10.1016/j.micpath.2017.08.005
Liu, J., Deng, Y., Soteyome, T., Li, Y., Su, J., Li, L., et al. (2018a). Induction and recovery of the viable but nonculturable state of hop-resistance Lactobacillus brevis. Front. Microbiol. 9:2076. doi: 10.3389/fmicb.2018.02076
Liu, J., Zhou, R., Li, L., Peters, B. M., Li, B., Lin, C. W., et al. (2017). Viable but non-culturable state and toxin gene expression of enterohemorrhagic Escherichia coli O157 under cryopreservation. Res. Microbiol. 168, 188–193. doi: 10.1016/j.resmic.2016.11.002
Liu, L., Lu, Z., Li, L., Li, B., Zhang, X., Zhang, X., et al. (2018b). Physical relation and mechanism of ultrasonic bactericidal activity on pathogenic E. coli with WPI. Microb. Pathog. 117, 73–79. doi: 10.1016/j.micpath.2018.02.007
Liu, L., Xu, R., Li, L., Li, B., Zhang, X., Zhang, X., et al. (2018c). Correlation and in vitro mechanism of bactericidal activity on E. coli with whey protein isolate during ultrasonic treatment. Microb. Pathog. 115, 154–158. doi: 10.1016/j.micpath.2017.12.062
Liu, L., Ye, C., Soteyome, T., Zhao, X., Xia, J., Xu, W., et al. (2019). Inhibitory effects of two types of food additives on biofilm formation by foodborne pathogens. Microbiol. Open. 8:e853. doi: 10.1002/mbo3.853
Liu, W., Dong, D., Yang, Z., Zou, D., Chen, Z., Yuan, J., et al. (2015). Polymerase Spiral Reaction (PSR): a novel isothermal nucleic acid amplification method. Sci. Rep. 5:12723. doi: 10.1038/srep12723
Magruder, M., Edusei, E., Zhang, L., Albakry, S., Satlin, M. J., et al. (2020). Gut commensal microbiota and decreased risk for Enterobacteriaceae bacteriuria and urinary tract infection. Gut Microbes. 12:1805281. doi: 10.1080/19490976.2020.1805281
Miao, J., Chen, L., Wang, J., Wang, W., Chen, D., Li, L., et al. (2017a). Current methodologies on genotyping for nosocomial pathogen methicillin-resistant Staphylococcus aureus (MRSA). Microb. Pathog. 107, 17–28. doi: 10.1016/j.micpath.2017.03.010
Miao, J., Chen, L., Wang, J., Wang, W., Chen, D., Li, L., et al. (2017b). Evaluation and application of molecular genotyping on nosocomial pathogen-methicillin-resistant Staphylococcus aureus isolates in Guangzhou representative of Southern China. Microb. Pathog. 107, 397–403. doi: 10.1016/j.micpath.2017.04.016
Miao, J., Liang, Y., Chen, L., Wang, W., Wang, J., Li, B., et al. (2017c). Formation and development of Staphylococcus biofilm: with focus on food safety. J. Food Saf. 7:e12358. doi: 10.1111/jfs.12358
Miao, J., Peters, B. M., Li, L., Li, B., Zhao, X., Xu, Z., et al. (2016). Evaluation of ERIC-PCR for fingerprinting methicillin-resistant Staphylococcus aureus strains. Basic Clin. Pharmacol. Toxicol. 118:33.
Miao, J., Wang, W., Xu, W., Su, J., Li, L., Li, B., et al. (2018). The fingerprint mapping and genotyping systems application on methicillin-resistant Staphylococcus aureus. Microb. Pathog.125, 246–251. doi: 10.1016/j.micpath.2018.09.031
Olaoye, O. A., Onilude, A. A., and Dodd, C. E. R. (2008). Identification of Pediococcus spp. from beef and evaluation of their lactic acid production in varying concentrations of different carbon sources. Adv. Nat. Appl. Sci. 2, 197–207.
Paalanne, N., Husso, A., Salo, J., Pieviläinen, O., Tejesviet, M. V., et al. (2018). Intestinal microbiome as a risk factor for urinary tract infections in children. Eur. J. Clin. Microbiol. Infect. Dis. 37, 1881–1891 doi: 10.1007/s10096-018-3322-7
Ramamurthy, T., Ghosh, A., Pazhani, G. P., and Shinoda, S. (2014). Current perspectives on Viable but Non-Culturable (VBNC) pathogenic bacteria. Front. Publ. Health. 2:103. doi: 10.3389/fpubh.2014.00103
Salminen, S., Wright, A. V., and Ouweland, A. (2004). Lactic Acid Bacteria Microbiological and Functional Aspects, 3rd Edn., Revised and Expanded. New York, NY: Marcel Dekker. doi: 10.1201/9780824752033
Singh, V. K., Mangalam, A. K., Dwivedi, S., and Naik, S. (2018). Primer Premier: program for design of degenerate primers from a protein sequence. Biotechnoques 24, 318–319. doi: 10.2144/98242pf02
Stapleton, A. E. (2017). “The vaginal microbiota and urinary tract infection,” in Urinary Tract Infections, eds M. A. Mulvey, D. J. Klumpp, and A. E. Stapleton (Washington, DC: American Society for Microbiology). doi: 10.1128/9781555817404.ch5
Truchado, P., Gil, M. I., Larrosa, M., and Allende, A. (2020). Detection and quantification methods for Viable but Non-culturable (VBNC) cells in process wash water of fresh-cut produce: industrial validation. Front. Microbiol. 11:673. doi: 10.3389/fmicb.2020.00673
Wang, L., Zhao, X., Chu, J., Li, Y., Li, Y., Li, C., et al. (2011). Application of an improved loop-mediated isothermal amplification detection of Vibrio parahaemolyticus from various seafood samples. Afr. J. Microbiol. Res. 5, 5765–5771 doi: 10.5897/AJMR11.1237
Wen, S., Feng, D., Chen, D., Yang, L., and Xu, Z. (2020). Molecular epidemiology and evolution of Haemophilus influenzae. Infect. Genet. Evolut. 80:104205. doi: 10.1016/j.meegid.2020.104205
Whiteside, S. A., Razvi, H., Dave, S., Reid, G., and Burton, J. P. (2015). The microbiome of the urinary tract—a role beyond infection. Nat. Rev. Urol. 12, 81–90. doi: 10.1038/nrurol.2014.361
Xie, J., Peters, B. M., Li, B., Li, L., Yu, G., Xu, Z., et al. (2017a). Clinical features and antimicrobial resistance profiles of important Enterobacteriaceae pathogens in Guangzhou representative of Southern China, 2001-2015. Microb. Pathog. 107, 206–211. doi: 10.1016/j.micpath.2017.03.038
Xie, J., Yang, L., Peters, B. M., Chen, L., Chen, D., Li, B., et al. (2017b). A 16-year retrospective surveillance report on the pathogenic features and antimicrobial susceptibility of Pseudomonas aeruginosa isolated from Guangzhou representative of Southern China. Microb. Pathog. 110:37–41. doi: 10.1016/j.micpath.2017.06.018
Xu, G., Hu, L., Zhong, H., Wang, H., Yusa, S., Weiss, T. C., et al. (2012a). Cross priming amplification: mechanism and optimization for isothermal DNA amplification. Sci. Rep. 2:246. doi: 10.1038/srep00246
Xu, W., Gao, J., Zheng, H., Yuan, C., Hou, J., Zhang, L., et al. (2019). Establishment and application of polymerase spiral reaction amplification for salmonella detection in food. J. Microbiol. Biotechnol. 29, 1543–1552. doi: 10.4014/jmb.1906.06027
Xu, Z., Gui, Z., Li, L., Li, B., Su, J., Zhao, X., et al. (2012b). Expression and purification of gp41-gp36 fusion protein and application in serological screening assay of HIV-1 and HIV-2. Afr. J. Microbiol. Res. 6, 6295–6299. doi: 10.5897/AJMR12.1075
Xu, Z., Hou, Y., Peters, B. M., Chen, D., Li, B., and Li, L. (2016b). Chromogenic media for MRSA diagnostics. Mol. Biol. Rep. 43, 1205–1212. doi: 10.1007/s11033-016-4062-3
Xu, Z., Hou, Y., Qin, D., Liu, X., Li, B., Li, L., et al. (2016a). Evaluation of current methodologies for rapid identification of methicillin-resistant staphylococcus aureus strains. Basic Clin. Pharmacol. Toxicol. 118:33.
Xu, Z., Li, L., Alam, M. J., Li, L., and Yamasaki, S. (2008a). Integron-bearing methicillin-resistant coagulase-negative staphylococci in South China, 2001-2004. FEMS Microbiol. Lett. 278, 223–230. doi: 10.1111/j.1574-6968.2007.00994.x
Xu, Z., Li, L., Alam, M. J., Zhang, L., Yamasaki, S., and Shi, L. (2008b). First confirmation of integron-bearing methicillin-resistant Staphylococcus aureus. Curr. Microbiol. 57, 264–268. doi: 10.1007/s00284-008-9187-8
Xu, Z., Li, L., Chu, J., Peters, B. M., Harris, M. L., Li, B., et al. (2012c). Development and application of loop-mediated isothermal amplification assays on rapid detection of various types of Staphylococci strains. Food Res. Int. 47, 166–173. doi: 10.1016/j.foodres.2011.04.042
Xu, Z., Li, L., Shi, L., and Shirliff, M. E. (2011a). Class 1 integron in staphylococci. Mol. Biol. Rep. 38, 5261–5279. doi: 10.1007/s11033-011-0676-7
Xu, Z., Li, L., Shirliff, M. E., Alam, M. J., Yamasaki, S., and Shi, L. (2009). Occurrence and characteristics of class 1 and 2 integrons in Pseudomonas aeruginosa isolates from patients in Southern China. J. Clin. Microbiol. 47, 230–234. doi: 10.1128/JCM.02027-08
Xu, Z., Li, L., Shirliff, M. E., Peters, B. M., Li, B., and Peng, Y. (2011b). Resistance class 1 integron in clinical methicillin-resistant Staphylococcus aureus strains in southern China, 2001-2006. Clin. Microbiol. Infect. 17, 714–718. doi: 10.1111/j.1469-0691.2010.03379.x
Xu, Z., Li, L., Shirliff, M. E., Peters, B. M., Peng, Y., et al. (2010). First report of class 2 integron in clinical Enterococcus faecalis and class 1 integron in Enterococcus faecium in South China. Diagn. Microbiol. Infect. Dis. 68, 315–317. doi: 10.1016/j.diagmicrobio.2010.05.014
Xu, Z., Li, L., Zhao, X., Chu, J., Li, B., Shi, L., et al. (2011c). Development and application of a novel multiplex polymerase chain reaction (PCR) assay for rapid detection of various types of Staphylococci strains. Afr. J. Microbiol. Res. 5, 1869–1873. doi: 10.5897/AJMR11.437
Xu, Z., Liang, Y., Lin, S., Chen, D., Li, B., Li, L., et al. (2016c). Crystal violet and XTT assays on Staphylococcus aureus biofilm quantification. Curr. Microbiol. 73, 474–482. doi: 10.1007/s00284-016-1081-1
Xu, Z., Shi, L., Zhang, C., Zhang, L., Li, X., Cao, Y., et al. (2007). Nosocomial infection caused by class 1 integron-carrying Staphylococcus aureus in a hospital in South China. Clin. Microbiol. Infect. 13, 980–984. doi: 10.1111/j.1469-0691.2007.01782.x
Xu, Z., Xie, J., Peters, B. M., Li, B., Li, L., Yu, G., et al. (2017a). Longitudinal surveillance on antibiogram of important gram-positive pathogens in Southern China, 2001 to 2015. Microb. Pathog. 103, 80–86. doi: 10.1016/j.micpath.2016.11.013
Xu, Z., Xie, J., Yang, L., Chen, D., Peters, B. M., and Shirtliff, M. E. (2018). Complete sequence of pCY-CTX, a plasmid carrying a phage-like region and ISEcp1-mediated Tn2 element from Enterobacter cloacae. Microb. Drug Resist. 24, 307–313. doi: 10.1089/mdr.2017.0146
Xu, Z., Xu, X., Yang, L., Li, B., Li, L., Li, X., et al. (2017b). Effect of aminoglycosides on the pathogenic characteristics of microbiology. Microb. Pathog. 113, 357–364. doi: 10.1016/j.micpath.2017.08.053
You, R., Gui, Z., Xu, Z., Shirtliff, M. E., Yu, G., Zhao, X., et al. (2012). Methicillin-resistance Staphylococcus aureus detection by an improved rapid PCR assay. Afr. J. Microbiol. Res. 6, 7131–7133. doi: 10.5897/AJMR12.708
Zhang, J., Biao, D. I., Shan, H., Liu, J., Zhou, Y., Chen, H., et al. (2019). Rapid detection of Bacillus cereus using cross-priming amplification. J. Food Prot. 82, 1744–1750. doi: 10.4315/0362-028X.JFP-19-156
Zhao, L., Lv, X., Cao, X., Zhang, J., Gu, X., Zeng, X., et al. (2020). Improved quantitative detection of VBNC Vibrio parahaemolyticus using immunomagnetic separation and PMAxx-qPCR. Food Control. 110:106962. doi: 10.1016/j.foodcont.2019.106962
Zhao, X., Li, M., and Xu, Z. (2018a). Detection of foodborne pathogens by surface enhanced raman spectroscopy. Front. Microbiol. 9:1236. doi: 10.3389/fmicb.2018.01236
Zhao, X., Yu, Z., and Xu, Z. (2018b). Study the features of 57 confirmed CRISPR loci in 38 strains of Staphylococcus aureus. Front. Microbiol. 9:1591. doi: 10.3389/fmicb.2018.01591
Zheng, F., Li, S., Wang, S., Feng, T., Jiang, Z., and Pan, J. (2020). Cross-priming isothermal amplification combined with nucleic acid test strips for detection of meat species. Anal. Biochem. 597:113572. doi: 10.1016/j.ab.2020.113672
Zhong, J., and Zhao, X. (2018). Detection of viable but non-culturable Escherichia coli O157:H7 by PCR in combination with propidium monoazide. 3 Biotech. 8:28. doi: 10.1007/s13205-017-1052-7
Zhong, N., Gui, Z., Liu, X., Jiangrong, H. U. K., Gao, X., et al. (2013). Solvent-free enzymatic synthesis of 1, 3-diacylglycerols by direct esterification of glycerol with saturated fatty acids. Lipids Health Dis. 12:65. doi: 10.1186/1476-511X-12-65
Keywords: Pediococcus acidilactici, viable but non-culturable, artificially contaminated food, PMA-CPA, rapid detection, safety control
Citation: Guan Y, Wang K, Zeng Y, Ye Y, Chen L and Huang T (2021) Development of a Direct and Rapid Detection Method for Viable but Non-culturable State of Pediococcus acidilactici. Front. Microbiol. 12:687691. doi: 10.3389/fmicb.2021.687691
Received: 29 March 2021; Accepted: 31 May 2021;
Published: 02 July 2021.
Edited by:
Yang Deng, Qingdao Agricultural University, ChinaReviewed by:
Yuting Tian, Fujian Agriculture and Forestry University, ChinaCopyright © 2021 Guan, Wang, Zeng, Ye, Chen and Huang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yu Guan, Z3Vhbnl1MTk4Ny40LjE5QGdtYWlsLmNvbQ==; Tengyi Huang, aC1odHlAaG90bWFpbC5jb20=
†These authors share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.