
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 13 August 2021
Sec. Evolutionary and Genomic Microbiology
Volume 12 - 2021 | https://doi.org/10.3389/fmicb.2021.677842
 Anusha Rai1†
Anusha Rai1† Uppada Jagadeeshwari2†
Uppada Jagadeeshwari2† Gupta Deepshikha1
Gupta Deepshikha1 Nandardhane Smita1
Nandardhane Smita1 Chintalapati Sasikala2*
Chintalapati Sasikala2* Chintalapati Venkata Ramana1*
Chintalapati Venkata Ramana1*The genus Roseomonas is a significant group of bacteria which is invariably of great clinical and ecological importance. Previous studies have shown that the genus Roseomonas is polyphyletic in nature. Our present study focused on generating a lucid understanding of the phylogenetic framework for the re-evaluation and reclassification of the genus Roseomonas. Phylogenetic studies based on the 16S rRNA gene and 92 concatenated genes suggested that the genus is heterogeneous, forming seven major groups. Existing Roseomonas species were subjected to an array of genomic, phenotypic, and chemotaxonomic analyses in order to resolve the heterogeneity. Genomic similarity indices (dDDH and ANI) indicated that the members were well-defined at the species level. The Percentage of Conserved Proteins (POCP) and the average Amino Acid Identity (AAI) values between the groups of the genus Roseomonas and other interspersing members of the family Acetobacteraceae were below 65 and 70%, respectively. The pan-genome evaluation depicted that the pan-genome was an open type and the members shared 958 core genes. This claim of reclassification was equally supported by the phenotypic and chemotaxonomic differences between the groups. Thus, in this study, we propose to re-evaluate and reclassify the genus Roseomonas and propose six novel genera as Pararoseomonas gen. nov., Falsiroseomonas gen. nov., Paeniroseomonas gen. nov., Plastoroseomonas gen. nov., Neoroseomonas gen. nov., and Pseudoroseomonas gen. nov.
The genus Roseomonas comes under the family Acetobacteraceae, the class Alphaproteobacteria, and the phylum Proteobacteria (Rihs et al., 1993). Altogether, there are 46 validly published genera names under the family Acetobacteraceae to date.1 Members of the genus Roseomonas are metabolically diverse and prevalent (Rihs et al., 1993; Han et al., 2003). They may have fundamental functions in supporting various ecological and biogeographical processes. They are highly ubiquitous and have been widely isolated from clinical specimens like blood, wounds, and genitourinary samples (Bibashi et al., 2000; Han et al., 2003) and environmental samples like contaminated oil sediments (Subhash and Lee, 2018), water (Gallego et al., 2006), soil (Kim and Ka, 2014), and air (Kim et al., 2013). Members of this genus are aerobic, Gram-negative, pink pigmented, and non-fermentative (Rihs et al., 1993). At the time of writing, the genus Roseomonas comprised 45 validly described species names and two sub-species names.2 The genus Roseomonas was first described by Rihs et al. (1993). The type species of this genus is Roseomonas gilardii (Rihs et al., 1993) and the species name was validated in the year 1998. “Roseomonas aceris” (Tonouchi and Tazawa, 2014), “Roseomonas baikalica” (Andreeva et al., 2007), “Roseomonas chloroacetimidivorans” (Chu et al., 2016), and “Candidatus Roseomonas massiliae” (Greub et al., 2004) are not valid names. “Roseomonas hellenica” was described recently (Rat et al., 2021). Furthermore, R. fauriae, described by Rihs et al. (1998), is a later heterotypic synonym of Azospirillum brasilense (Tarrand et al., 1978; Helsel et al., 2006).
Phylotaxogenomics of the class Alphaproteobacteria was extensively studied recently by Hördt et al. (2020), which showed that several taxa under it, including the members of the family Acetobacteraceae, were disordered. One of the major discussions under this communication is the re-evaluation of the phylogenetic relationships between the members of the genus Roseomonas. This is based on the fact that the genus Roseomonas was distinctly polyphyletic with members like Roseomonas stagni and Roseomonas lacus interspersed in between other sister groups of Humitalea and Rubritepida in the family Acetobacteraceae (Hördt et al., 2020). Furthermore, the same study revealed that Rhodovarius lipocyclicus nested within the genus Roseomonas based on the constrained comprehensive tree (CCT) (Hördt et al., 2020). Likewise, a study conducted by Romano-Bertrand et al. (2016) depicted that the members of the genus Roseomonas were organized into seven different clades based on the 16S rRNA gene phylogeny and ecology.
Ambiguity has emerged due to the poor discriminatory power of the16S rRNA gene marker for absolute resolution at the genus/species level (Fox et al., 1992; Janda and Abbott, 2007; Olm et al., 2020). Over the decade, meaningful deductions of unresolved and longstanding phylogenetic relationships between members of the genera like Mycobacterium (Gupta et al., 2018), Corallococcus (Livingstone et al., 2018), Roseobacter (Wirth and Whitman, 2018), Rhodobacter (Suresh et al., 2019), and Lactobacillus (Wittouck et al., 2019) have been worked out well. This resolution has been successfully elucidated owing to the robust calculated parameters and integrated comparative genomics that included average amino acid identity (AAI), average nucleotide identity (ANI), genomic signatures, and pan-genome analysis (McInerney et al., 2017). In this study, we revisited the current status of the genus Roseomonas based on definite and congruent genomic evidence. Here, on the basis of phylogenomic studies like phylogenetic, pan-genomic, and taxogenomic analysis, we have proposed to re-classify the genus Roseomonas into Roseomonas genus sensu-stricto along with six novel genera (gen. nov.).
The genome sequences affiliated to the family Acetobacteraceae, which were available publicly at the NCBI and JGI databases, were retrieved (Supplementary Table 1). A total of 34 type strains and 28 other related strains affiliated to the genus Roseomonas were considered in this study (Table 1). As the genome sequence of the type species of the genus Roseomonas was not available at the time of writing, R. gilardii subsp. rosea was taken as a representative for the type species in this analysis.
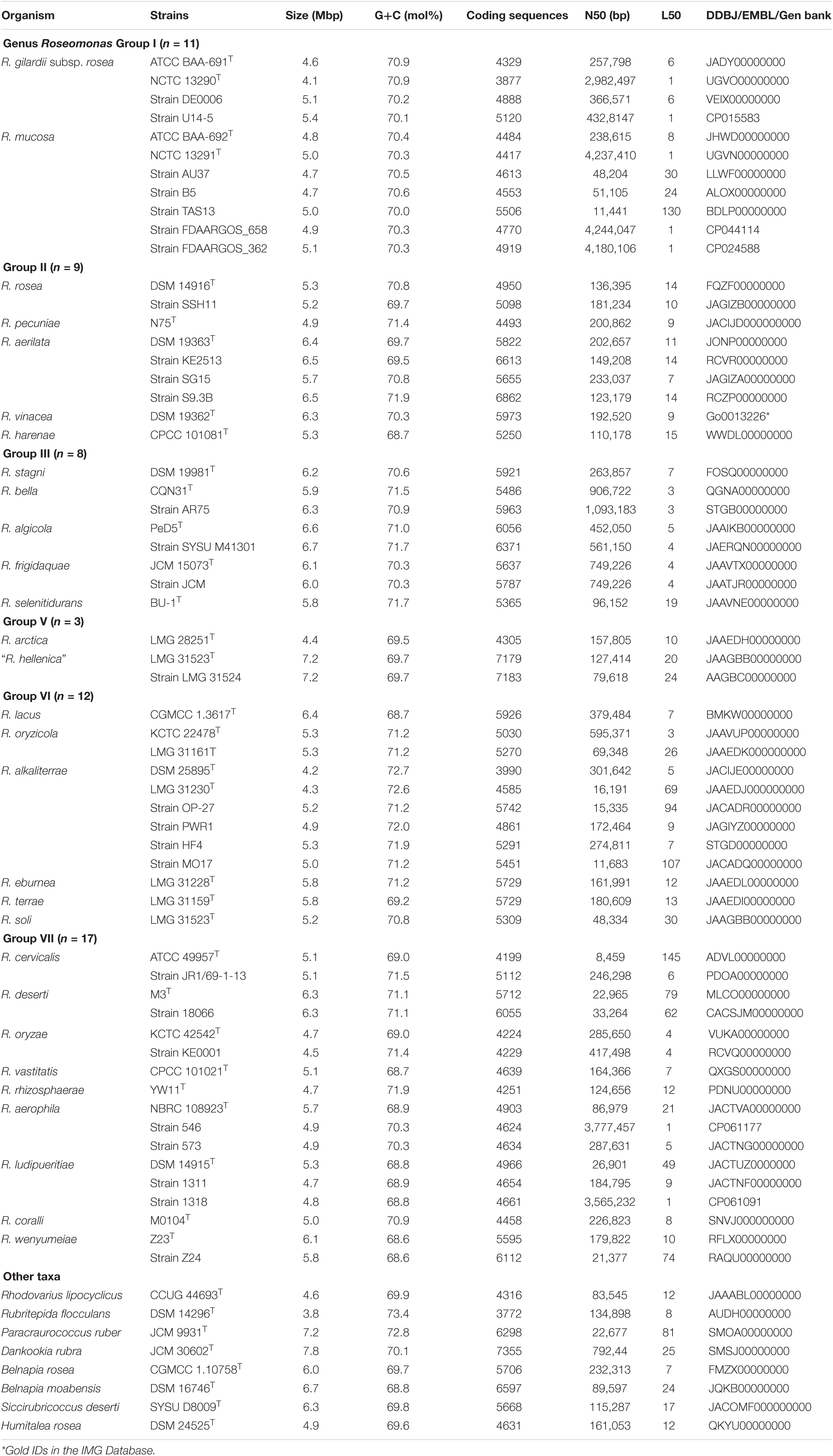
Table 1. Genome characteristics of the members of the genus Roseomonas and closely related strains in the family Acetobacteraceae.
For the phylogenetic analysis, 16S rRNA gene sequences of type strains of all the validly published species names, effectively but not validly published names like “R. aceris” R-1T, “R. chloroacetimidivorans” BUT-13T, and “candidatus Roseomonas massiliae” of the genus Roseomonas, were taken from the NCBI database. For Roseomonas aquatica and Roseomomas fluminis, cloned sequences were also taken to understand the phylogenetic position and confirm their status within the genus Roseomonas. In addition, the 16S rRNA gene sequences of representative members and cloned sequences of 11 other genera affiliated to the family of Acetobacteraceae were also accounted for the phylogenetic analysis. The 16S rRNA sequence of Elioraea tepidiphila DSM 17972T was used as an outgroup. MUSCLE algorithm (Edgar, 2004) of MEGA7 was used for sequence alignments and the phylogenetic analysis of the sequences was performed using MEGA7 (Kumar et al., 2016). Distances were calculated using Kimura two-parameter in a deletion manner (Kimura, 1980). Neighbor-joining (NJ), maximum likelihood (ML), and maximum parsimony (MP) methods in the MEGA7 software were used to reconstruct phylogenetic trees. Percentage support values were obtained using a bootstrap procedure (Felsenstein, 1985).
As for the phylogenomic tree reconstruction, 62 genomes of members of the genus Roseomonas (34 type strains, 28 other related strains) and 135 genomes representative of 43 other genera of the family Acetobacteraceae (as per the NCBI database) were considered for the phylogenomic study (Supplementary Table 1). A total of 92 core genes were identified in 197 genomes and were retrieved for phylogenomic analysis using the Up-to-date Bacterial Core Gene (UBCG) tool (Na et al., 2018). The genes considered for the phylogenomic tree analysis are given in Supplementary Table 2, and Stella humosa DSM 5900T was used as an outgroup. The RAXML-based phylogenomic tree was constructed using the concatenated sequences of the 92 core genes of 197 genomes.
Bacterial Pan-genome Analysis (BPGA) pipeline (Chaudhari et al., 2016) was applied for the analysis of the genomic diversity of Roseomonas. The default parameters were set to check the conserved, accessory, and strain-specific genomic traits between the members of the genus Roseomonas. The same process was also carried out for each of the proposed genera (n ≥ 3) in this study.
Robust parameters like average nucleotide identity (ANI) and digital DNA-DNA hybridization (dDDH) were computed for the precise delineation of the members of the genus Roseomonas. Whereas, AAI and Percentage of Conserved Proteins (POCP) values were calculated within the members of the genus Roseomonas as well as with the members of the other genera in the family Acetobactereaceae. OrthoANI tool (Yoon et al., 2017) and Genome-to-Genome Distance calculator 2.13 (Auch et al., 2010) were used for calculating the ANI and dDDH values, respectively. AAI was calculated using the AAI calculator developed by Kostas lab4 (Rodriguez and Konstantinidis, 2014). POCP was calculated as described by Qin et al. (2014). The obtained AAI and POCP values were used to construct a heatmap using an online tool, Morpheus,5 and were clustered hierarchically based on the Euclidean distance.
The identification and annotation of biosynthetic gene clusters (BCGs) for the members of the genus Roseomonas were carried out with the antiSMASH 5.0 web server (Blin et al., 2019) which applies the Hidden Markov Models for the identification of BCGs. For the detection, strictness level was set to “relaxed.” Genomes were submitted to the KBase online software (Arkin et al., 2018)6 using the default parameters to generate genomic data which were later manually analyzed to gather the group-specific gene clusters.
To understand the pathogenicity of the members of the genus Roseomonas, genomes were submitted individually to the IslandViewer 4 server (Bertelli et al., 2017) in the .gbk format where Salmonella enterica Serovar Typhimurium LT2 was given as the reference genome. Virulence factors were also checked by submitting the genome sequences (.fas format) to the VirulenceFinder 2.07 hosted by the Center for Genomic Epidemiology (CGE) against four genera of Listeria, Staphylococcus, Escherichia, and Enterococcus. The threshold for percent identity (%ID) between input and matching gene in the database was 90% with a minimum length of 60%.
Phenotypic and chemotaxonomic characterization of each described species for taxon delineation was obtained from the original species descriptions, Bergey’s Manual of Systematic Bacteriology (Weyant and Whitney, 2005), and from other references as specified.
The 16S rRNA gene-based phylogenetic tree showed that the genus Roseomonas was polyphyletic and was segregated into seven major clades (Figure 1). These seven major clades were defined as Groups I–VII on the basis of the clade formation. The validly described members of each of the clade were as follows: Group I (n = 4), Group II (n = 8), Group III (n = 10), Group IV (n = 2), Group V (n = 2), Group VI (n = 7), and Group VII (n = 16) (Figure 1). Group I shall be considered as Roseomonas genus sensu stricto. The pairwise 16S rRNA gene identity was calculated using the pairwise nucleotide sequence alignment for taxonomy.8 The members of a group delineated by the phylogenetic tree shared at least 93.5–99.2% of the 16S rRNA gene identity (Supplementary Table 3).

Figure 1. NJ phylogenetic tree based on the 16S rRNA gene sequences showing the phylogenetic relationship between the members of the genus Roseomonas and other closely related members of the family Acetobacteraceae. Elioraea tepidiphila DSM 17972T was used as an outgroup. Numbers at nodes represent bootstrap values (given as percentages of 1,000 replications) of >50% shown at branch points (NJ/ML/ME). The GenBank accession numbers for the 16S rRNA gene sequences are shown in parentheses. Bar, 0.01 accumulated changes per nucleotide substitutions (red color indicates the availability of their genomes).
The phylogenomic tree was reconstructed using 92 core genes (Supplementary Table 2) of 197 genomes by the UBCG tool (Na et al., 2018). The study by Na et al. (2018) showed that these 92 core genes (Supplementary Table 2) were found as a single copy in more than 95% of the genome sequences. Furthermore, the UBCG-based phylogenomic tree (92 genes) could separate E. coli/Shigella spp. better than the 16S rRNA gene alone (Na et al., 2018). UBCG-based phylogenomic tree (Figure 2) further confirmed the heterogeneity of the genus Roseomonas. Genome sequences were not available for the members of Group IV including R. aquatica and R. fluminis. Furthermore, as per this study, Roseomonas aeriglobus and a genomospecies of Roseomonas cladded outside of the family Acetobacteraceae. Therefore, the genomes of these members were not considered for further study.
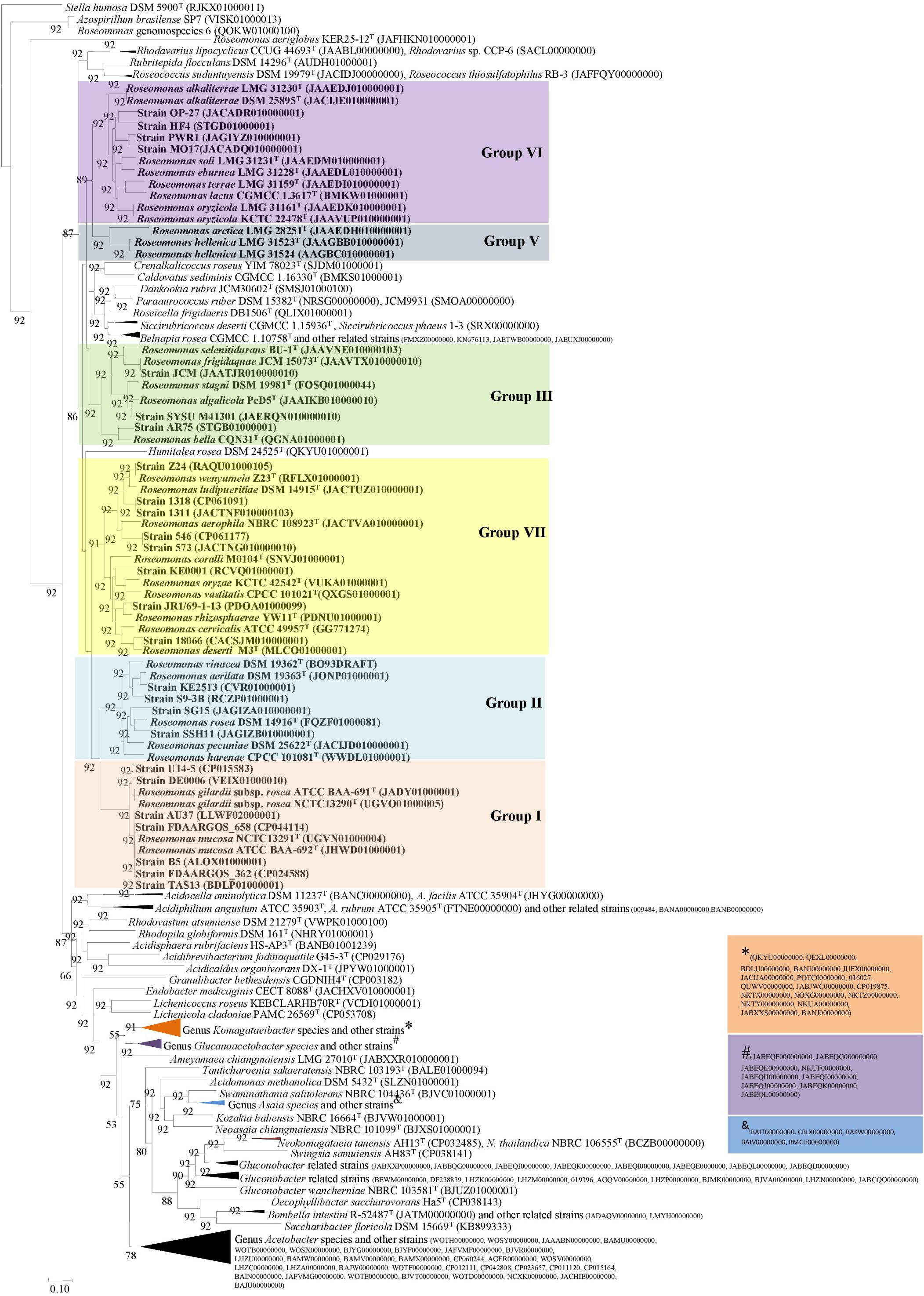
Figure 2. Phylogenomic tree constructed using 92 core genes tool based on the Up-to-date Bacterial Core Gene (UBCG) (Na et al., 2018). The tree was generated using the MEGA7 software (NJ) with Stella humosa DSM 5900T (RJKX00000000) as an outgroup.
The genome information of the type strains used in this study are given in Table 1. Groups I (n = 11), II (n = 9), III (n = 8), V (n = 3), VI (n = 12), and VII (n = 17) have genome sizes of 4.1–5.4 Mbp, 4.9–6.5 Mbp, 5.8–6.7 Mbp, 4.4–7.2 Mbp, 4.2–6.4 Mbp, and 4.5–6.3 Mbp, respectively (Table 1). The genomic G+C content (mol%) of the taxa belonging to Groups I (n = 11), II (n = 9), III (n = 8), V (n = 3), VI (n = 12), and VII (n = 17) were 70.1–70.9%, 68.7–71.9%, 70.3–71.7%, 69.5–69.7%, 68.7–72.7%, and 68.6–71.9%, respectively (Table 1). On the average taxa between the groups, Group I had the lowest and Group V had the highest genome size of 4.8 and 6.3 Mbp, respectively. Similarly, the lowest (69.6 mol%) average genomic G+C content was observed with the taxa of Group V, whereas the highest (71.2 mol%) was observed with the taxa of Group VI. When the genome size of the members of the genus Roseomonas was compared with the taxa of the other genera in the family Acetobacteraceae, Dankookia (n = 1) had the highest genome size of 7.8 Mbp. On the contrary, the genus Rubritepida (n = 1) had the lowest genome size of 3.8 Mbp with the highest (73.4 mol%) genomic G+C (Table 1).
To examine the distribution of genes and genomic diversity across the genus Roseomonas, genome sequences were given as input in the BPGA tool. The analyzed data of the genus Roseomonas are given in Supplementary Table 4. Genus Roseomonas members have 958 core genes (19.1%), 219,753 accessory genes (72.8%), and 24,320 strain-specific genes (8.1%) (Figure 3A). For the diagrammatic representation of the pan-genome for the genus Roseomonas, only the type strains were considered. The core-pan plot (Figure 3B) showed an open pan-genome for the genus Roseomonas as it did not level off into a plateau and extended with the increase in the number of genomes. The core genome was conserved at the genus level as the plot leveled off. The KEGG distribution (%) of genes based on function is illustrated in Figure 3C for the genus Roseomonas. Members of Groups I, II, III, V, VI, and VII had core genes between the range of 1,573–3,152 (Supplementary Table 4). The diagrammatic representation of the core and pan genome plot for each of the group are given in Supplementary Figures 1A–F. For each of the group, the KEGG distribution (%) of genes based on function is illustrated in Supplementary Figures 2A–F.
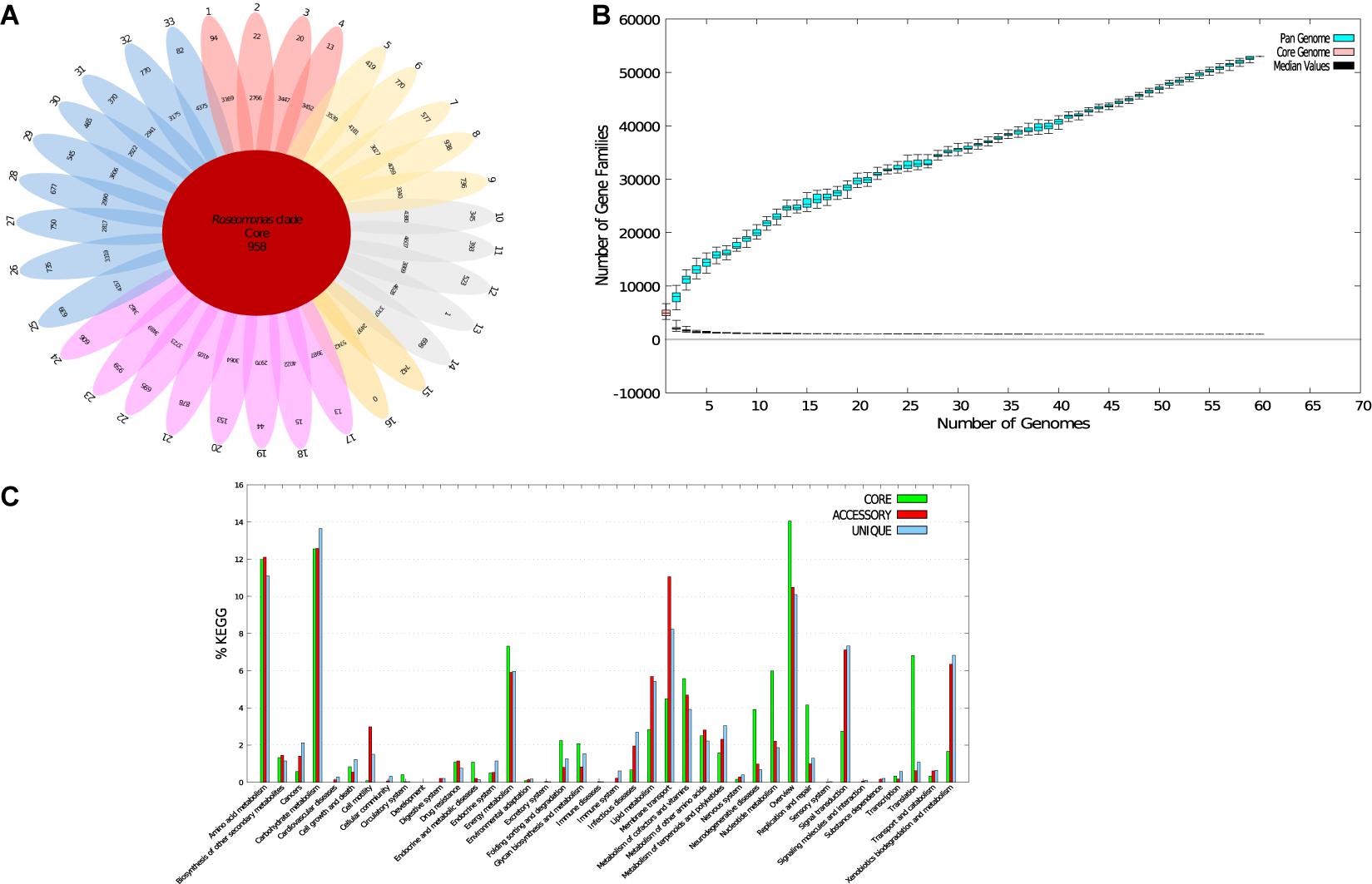
Figure 3. Core-pan-genome analysis of Roseomonas type strains. (A) Flower-pot diagram representing core, accessory, and unique genomes of the genomes of all strains. (B) Core-pan-plot for all the genomes. (C) KEGG distribution of core, accessory, and unique genes (1. R. gilardi subsp. rosea ATCC BAA-691T; 2. R. gilardi subsp. rosea NCTC 13290T; 3. R. mucosa ATCC BAA-692T; 4. R. mucosa NCTC 13291T; 5. R. rosea DSM 14916 T; 6. R. aerilata DSM 19363T; 7. R. pecuniae N75T; 8. R. sp. vinacea DSM 19362T; 9. R. harenae CPCC 101081T; 10. R. stagni DSM 19981T; 11. R. algicola PeD5 T; 12. R. bella CQN31T; 13. R. frigidaquae JCM 15073T; 14. R. selenitidurans BU-1T; 15. R. arctica LMG 28251T; 16. “R. hellenica” LMG 31523T; 17. R. oryzicola KCTC 22478T; 18. R. oryzicola LMG 31230T; 19. R. alkaliterrae DSM 25895 T; 20. R. alkaliterrae LMG 31230T; 21. R. lacus CGMCC 1.3617T; 22. R. eburnea LMG 31228T; 23. R. terrae LMG 31159T; 24. R. soli LMG 31523T; 25. R. deserti M3T; 26. R. aerophila NBRC 108923T; 27. R. cervicalis ATCC 49957 T; 28. R. coralli M0104T; 29. R. ludipueritiae DSM 14915T; 30. R. oryzae KCTC 42542 T; 31. R. rhizosphaerae YW11T; 32. R. vastitatis CPCC 101021T; 33. R. wenyumeiae Z23T).
At an intra-group level, members of a group (consisting both type and non-type strains) defined by phylotaxogenomics shared dDDH and ANI values of at least 21–100% and 78.0–99.9%, respectively (Supplementary Table 5). At an inter-group level, members of different groups did not show dDDH and ANI values above 20.8 and 76.8%, respectively. POCP and AAI values were calculated between the members of the groups (intra-group) as well as at the inter-group level (Supplementary Tables 6, 7) as they are considered pivotal and accurate for genus delineation (Qin et al., 2014). POCP values for Group I (n = 11), Group II (n = 9), Group III (n = 8), Group V (n = 3), Group VI (n = 12), and Group VII (n = 17) taxa were 80–98.5%, 65.2–71.7%, 66.9–98.7%, 65.8–99.3%, 66.4–99.2%, and 65.2–99.4%, respectively. While the AAI values were 91.9–99.0%, 76.7–84.8%, 71.8–99.9%, 73.8–99.9%, 77.0–99.9%, and 70.2–99.9% for the taxa of the respective groups. The POCP and AAI heatmaps (Supplementary Figures 3, 4) support the division of the six groups. The AAI values between Roseomonas fauriae (later heterotypic synonym of Azospirillum brasilense) and the members of Group I are in between 51.2 and 51.5%, thus making them distinct species under different genera.
The BCGs prediction based on the antiSMASH 5.0 web server indicated an average Roseomonas genome consisted of seven BCGs (420 BCGs were found in 60 genomes). Group I (n = 11) members had 65 BCGs (clustered into 11 types of BCG families); Group II (n = 9), 72 (19 BCGs families); Group III (n = 8), 59 (13 BCG families); Group V (n = 3), 35 (9 BCG families); Group VI (n = 12), 68 (14 BCG families); and Group VII (n = 17), 124 (24 BCG families) (Supplementary Table 8). Terpenes were the most prevalent ones (two per genome on an average) but also an average of one type1 polyketide synthases (TPKS) per genome. The members of Group I exclusively had non-ribosomal peptide synthetases (NRPS), beta-lactone (fengymycin), and thioamitides (asukamycin). The members of Groups I and II both had phosphonate/terpene (phosphinothricintripeptide) BCG. All the members of Group II possessed a redox-cofactor BCG, whereas the members of both Groups II and III shared type 3 polyketide synthases (T3PKS). Group V members had NRPS/T1PKS and arylpolene BCGs. As for Group VI, eight genomes (out of 17) had NRPS and five genomes had arylpolene (xanthomonadin) BCGs. As for the Group VII, four genomes had unique ribosomally synthesized and post-translationally modified peptides (RiPPs) BCG exclusively.
The genome annotation showed that 226, 58, and 82 gene clusters were unique to Groups I, II, and III, respectively (Supplementary Table 9). Members of Group V comprised of 67 unique gene clusters, Group VI consisted of 74 unique gene clusters, and Group VII comprised16 unique gene clusters (Supplementary Table 9). As predicted by the IslandViewer 4 server and VirulenceFinder 2.0, the draft genomes of the members of the genus Roseomonas did not possess any genes related to its pathogenicity and virulence.
Correlation between the genomic studies and phenotypic characters are desirable for supporting taxa delineation. Members of Groups II and VI can be distinguished from those of Group V by being non-motile. Members of Groups II and III can be differentiated from those of Groups I, IV, V, VI, and VII in showing variable catalase activity. Members of Groups I and II show variable oxidase activity, whereas members of the other groups are either oxidase positive (Groups III, VI, VII) or negative (Groups IV, V). Members of Groups VI and VII have a high NaCl tolerance than members of the other groups. NaCl tolerance was used as a differentiating taxonomic character for delineating the genus Swaminathania biochemically from the other genera of the family Acetobacteraceae like Acetobacter, Gluconobacter, and Kozakia (Loganathan and Nair, 2004). Members of Groups II and III were variable for starch hydrolysis, while those of Groups IV, V, VI, and VII were negative. Members of Group VI were variably positive for casein hydrolysis, whereas members of the rest of the groups were negative. Members of Groups I, IV, and VI were urease positive, whereas members of Groups II, III, and VII were variable in reaction. Members of Groups II, III, V, and VII showed a negative reaction for nitrate reduction, whereas those of the other groups were variable. Similarly, Groups II and III can be differentiated from the other groups in showing variable activity of gelatin hydrolysis. Group VII can be distinguished from Groups II, III, and IV in the positive utilization of D-glucose. Members of Groups V, VI, and VII could utilize L-arabinose for growth, whereas members of Groups II, III, and IV could not utilize the same. Members of Groups III, VI, and VII could utilize sucrose, whereas those of Groups IV and V were negative. Members of Group VI contained Q-9 as the unique respiratory quinone which differs from those of the other groups which contained Q-10. Groups III and VII members can be distinguished from those of Groups I, II, IV, V, and VI in having a glycolipid. Similarly, Groups V and VII can be distinguished from each other in having an unidentified lipid as one of the polar lipids. Analysis of polar lipids is a significant chemotaxonomic aid and has often been used as a differential character for reclassification. The phenotypic differentiating characteristics between the genus Roseomonas and other closely related genera of the family Acetobacteraceae are given in Table 2.
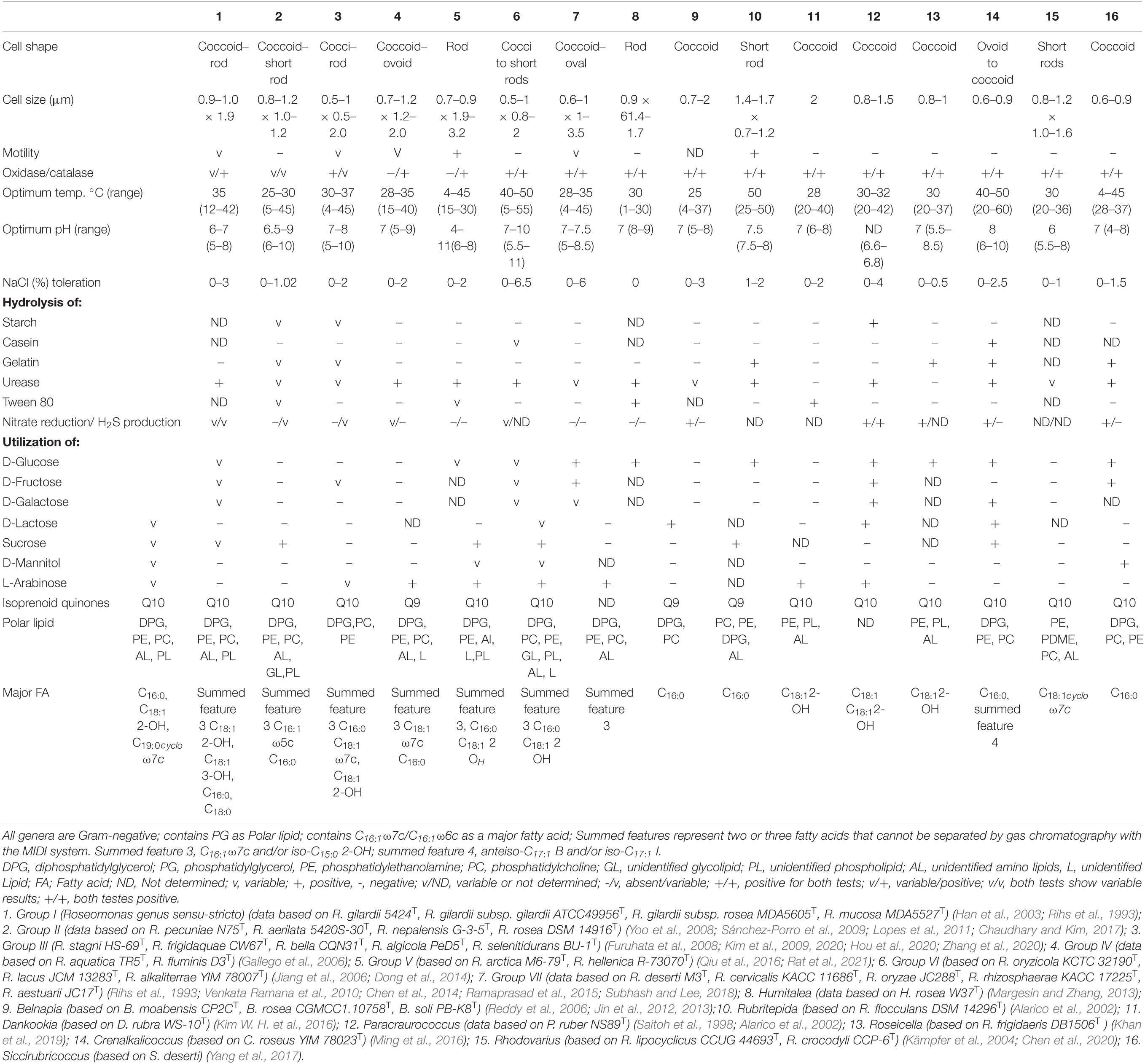
Table 2. Characteristics differentiating the Roseomonas groups from other closely related genera in the family Acetobacteraceae.
Genus Roseomonas has been widely studied as the members have clinical and environmental significances (Rihs et al., 1993; Han et al., 2003; Romano-Bertrand et al., 2016; Subhash et al., 2016). Six novel species names were validly published in the years 2020–2021 (see text footnote 2). The supremacy of the genomic era has identified whole-genome sequence data being more definitive and conclusive for taxa delineation than the 16S rRNA gene marker. Conventionally, a polyphasic approach was adopted for the demarcation of the Roseomonas species. As per the work conducted by Romano-Bertrand et al. (2016), they showed that the genus Roseomonas was segregated into seven different clades based mainly on the 16S rRNA gene phylogeny and ecology. Likewise, the study carried out by Hördt et al. (2020) showed that the genus Roseomonas was polyphyletic and consequently called in for a major reclassification. Taxogenomics is considered more precise and superior for the taxa delineation as it strengthens data reproducibility and reliability (Chun and Rainey, 2014; Lalucat et al., 2020). To determine the absolute phylogenetic standing between the current members of genus Roseomonas, a comprehensive taxogenomic analysis was carried out.
The 16S rRNA gene-based phylogenetic tree showed that the genus Roseomonas was polyphyletic and was segregated into seven major clades: Groups I to VII (Figure 1). The cladding patterns of Groups I, II, III, VI, and VII members in this study are comparable to the cladding pattern observed by Romano-Bertrand et al. (2016) in terms of the species composition. Group IV member, R. aquatica (Group IV), cladded outside the designated clade observed by Romano-Bertrand et al. (2016) whereas, R. fluminis which also belongs to this group was described later in 2018 by Ko et al. (2018). Furthermore, R. arctica (Group V) was shown to be affiliated to the R. stagni (Group III) clade, whereas “R. hellenica” is not included by them as it was described later (Rat et al., 2021). It was suggested that the whole genome phylogenies could provide a better resolution rather than a single gene like the 16S rRNA gene-based phylogenies for taxonomic delineation (Logan et al., 2009). However, in the case of Roseomonas, the phylogenomic tree based on 92 concatenated genes also showed the same cladding pattern (Figure 2; except for Group IV for which the genome sequences are not available).
Nouioui et al. (2018) and Hördt et al. (2020) have shown that genome size variation can be applied as a reliable taxonomic marker. They showed that the genome size appeared to be genus-specific for the members of the phylum Actinobacteria and the class Alphaproteobacteria. Both studies implied that the genome size and G+C content (mol%) were phylogenetically conserved. Hence, the genome sizes have been formally added to the descriptions. In this context, Group I had the smallest genome size as compared to the other groups. Analysis of pan and core genomes using the BPGA pipeline illustrated that the pan-genome of the genus Roseomonas is open and consists of 958 core genes, summing up to 19.1% of the total pan-genome (Supplementary Table 3). As for the BCGs, only groups of the genus Roseomonas could produce specific secondary metabolites, i.e., NRPS, fengymycin, and asukamycic (Group I); redox-cofactor (Group II); and NRPS/T1PKS and arylpolene (Group V) (Supplementary Table 8). The genome annotation also revealed the composition of the gene clusters unique to each group with the highest number of clusters unique to Group I and lowest in Group VII (Supplementary Table 9).
Chun et al. (2018) have proposed minimum standards based on an overall genome-related index (OGRI) like dDDH and ANI for species delineation. Wayne et al. (1984) prescribed a cut-off of 70% dDDH value for species delineation. In the present scenario, ≥95% of ANI between the two strains concludes that both belong to the same species whereas for values <95%, two strains are considered as different species (<75% for different genera) (Richter and Rosselló-Móra, 2009; Rodriguez and Konstantinidis, 2014; Rosselló-Móra and Amann, 2015). Both indices (dDDH and ANI) are consistent at the intra-group level with the recommended standards confirming that all members of Roseomonas are well-described at the species level (Supplementary Table 3). For the POCP values, <50% were considered a cut-off for genera delineation (Qin et al., 2014). POCP values between the groups of the genus Roseomonas were <65% (40–65%) (Figure 4 and Supplementary Table 6) and with the other genera of the members of the family Acetobacteraceae were also <70% (20–60%) (Supplementary Table 6). Thus, for the species of the genus Roseomonas, the calculated values of POCP for genus delineation were not in agreement with the ones observed by Qin et al. (2014). Although a proposed genus boundary of the POCP value for prokaryotic lineages was assigned as 50%, many studies later showed exceptions at the inter-genera comparison. The universal cut-off of 50% is considered conservative, as being only an index of relatedness (Liu et al., 2020) as reflected in its ineffectiveness in delineating different genera of the family Methylococcaceae (Orata et al., 2018), Bacillaceae (Aliyu et al., 2016), Burkholderiaceae (Lopes-Santos et al., 2017), and Rhodobacteraceae (Wirth and Whitman, 2018). Luo et al. (2014) have shown that AAI values of related but different genera ranged from 60 to 80%. In our study, AAI values were below 70% between the different groups of the genus Roseomonas (Figure 4 and Supplementary Table 7) and other genera members of the family Acetobacteraceae (Supplementary Table 7), hence in congruence with the work of Luo et al. (2014).
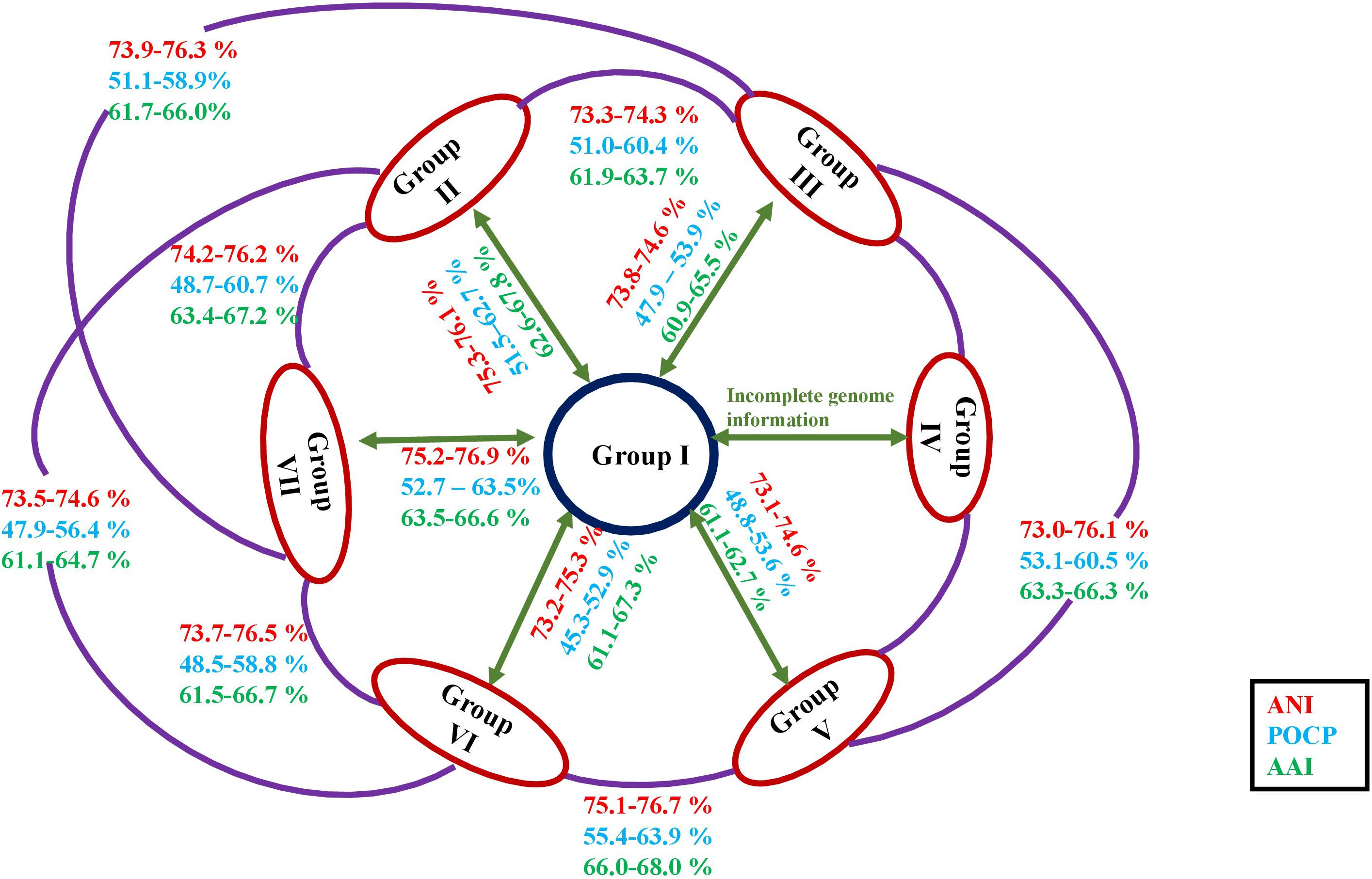
Figure 4. Diagrammatic representation of the ANI, AAI, and POCP values in between the members of each group of the genus Roseomonas.
The study conducted by Hördt et al. (2020) showed that certain members of the genus Roseomonas like R. stagni were interspersed with H. rosea and R. lacus with Rubritepida flocculans. The distinction between the mentioned species pairs was clarified by the dDDH, ANI, AAI, and POCP values. dDDH, ANI, AAI, and POCP values between R. stagni and H. rosea and R. lacus and R. flocculans were 19.4, 74.1, 64.2, and 64.2 and 19.8, 74.8, 66.6, and 57.1%, respectively. Similarly, the study also showed that Rhodovarius lipocyclicus was nested within the genus Roseomonas (Hördt et al., 2020). However, R. lipocyclicus is a distinct member of the genus Rhodovarius from Roseomonas as the AAI and POCP values are 50.8–66.8% and 53.2–64.7%, respectively, below the recommended cut-off for genus delineation. Thus, the above indices clearly differentiate the Roseomonas species from the genera of Humitalea, Rubritepida, and Rhodovarius. Therefore, the above detailed discussion entails the formation of seven different groups within the genus Roseomonas in congruence with the findings given by Romano-Bertrand et al. (2016) and Hördt et al. (2020).
Members of the genus Roseomonas have a ubiquitous distribution in the environment. Group I members were the only ones to have been isolated from a human blood sample (R. gilardii, R. mucosa, R. gilardii subsp. rosea) (Rihs et al., 1993; Han et al., 2003) except for R. elaeocarpi which was isolated from olive phyllosphere (Damtab et al., 2016). An examination of the literature demonstrated that the strains of Roseomonas have been widely reported in human infections and isolated from clinical samples like sputum, wounds, and genitourinary sites (Wallace et al., 1990; Rihs et al., 1993). It was also found to be associated with immunocompromised patients (Marin et al., 2001). Other reports of bacteremia caused by the Roseomonas species were in vertebral osteomyelitis (Nahass et al., 1995), infected tooth (Diesendorf et al., 2017), endocarditis (Shao et al., 2019), and peritonitis (Malini et al., 2016). However, many reports in case studies suggests that the majority of the patients had underlying diseases, malignancy being the most common factor (Wang et al., 2012; Michon et al., 2014). Pathogenic genes and virulence factors are potent segments of the genomic islands. They become part of the genome as a consequence of horizontal gene transfer, and these genes are observed to confer their pathogenicity and virulence to the bacteria (da Silva Filho et al., 2018). The above-mentioned reports can be substantiated with the predicted facts that the genomes of the members do not suggest any kind of genes responsible toward its pathogenicity and virulence. Thus, it may be concluded that Roseomonas strains are incidental and not causative of pathogenicity. In addition to being associated with human clinical samples, Roseomonas spp. were also isolated from various sources like soil (Kim and Ka, 2014), sediment (He et al., 2014), sludges (Wang et al., 2016), contaminated soil (Subhash and Lee, 2018), freshwater (Baik et al., 2012), and also human sources like blood (Rihs et al., 1993; Figure 1).
In an ecological context, the presence of accessory genes in the members shows their divergence due to environmental adaptations. The presence of accessory genes shows the acquisition of genes in response to the selective pressure (Brito et al., 2015) or for the colonization of the new ecological habitats (McInerney et al., 2017). The cosmopolitan distribution of the members of the groups may be attributed to the gain of accessory genes for their survival which in turn may be attributed to their variable genomic sizes, simple organization, or horizontal gene transfer rates (Kuo and Ochman, 2009; Aherfi et al., 2018). Also, the formation of ecotypes based on the varied isolation sources may not be feasible for groups of the genus Roseomonas. This is also evident from the distinctive genomic features observed during the pan-genome evaluation for each of the groups, which may have resulted in response to the environmental changes.
The extensive taxogenomics study performed substantiates the separation of the respective groups of the genus Roseomonas into separate genera as suggested by Romano-Bertrand et al. (2016) and Hördt et al. (2020). Considering the fact that Roseomonas is a very divergent genus, henceforth members of this genus cannot be clustered under one phylogenetic genus, necessitating the creation of six new genera. For each of the novel genera, emended description and reclassification are given. Phylogenetic trees based on the 16S rRNA gene (Figure 1) and whole genome sequences (Figure 2) agree with the formation of seven different clades as separate genera. Although the 16S rRNA gene studies have certain drawbacks, this approach is still quite accountable for the delineation of the members of the genus Roseomonas. This study would be a first attempt in re-evaluating the genus Roseomonas and reclassifying it by creating six new genera: Group II as Pararoseomonas gen. nov., Group III as Falsiroseomonas gen. nov., Group IV as Paeniroseomonas gen. nov., Group V as Plastoroseomonas gen. nov., Group VI as Neoroseomonas gen. nov., and Group VII as Pseudoroseomonas gen. nov. The genus delineation for Group IV is based on the 16S rRNA gene analysis (in the absence of genome sequences) and phenotypic characteristics. The genus delineation constructed from the 16S rRNA gene sequences and phenotypic characters provide sufficient resolution to distinguish the Group IV members from other groups of the genus Roseomonas. However, the availability of the genome sequences may further substantiate the reclassification in the future. The species within each of the newly defined genera have been defined accurately as per the recommended standards for ANI and dDDH (Supplementary Table 5). As for the genus delineation, the inter-group AAI and POCP values agree with the standards given for genus determination (Supplementary Tables 6, 7). The justification for genus delineation based on the 16S rRNA gene identity, AAI and POCP, and other phenotypic characters are given in Figure 4 and Table 2.
The description is the same as the one given by Rihs et al. (1993) and Sánchez-Porro et al. (2009) for the genus Roseomonas except for a few modifications. H2S formation is variable and the genome-based G+C content is ∼70 mol%.
The type species is Roseomonas gilardii.
Pararoseomonas (Pa.ra.ro.se.o.mo’nas. Gr. pref. para-, next to, resembling; N.L. fem. n. Roseomonas, a bacterial genus; N.L. fem. n. Pararoseomonas, next to Roseomonas).
Members are aerobic, Gram-negative, coccoid or short rods, and non-motile. Variable in both oxidase and catalase. Polar lipids consist of diphosphatidylglycerol, phosphatidylglycerol, phosphatidylethanolamine, phosphatidylcholine, unidentified phospholipid, and unidentified aminolipids. Summed feature 3, C18:1 2-OH, C18:1 3-OH, C16:0, C18:0 are the major fatty acids. Members have a genomic size of 4.9–6.4 Mbp and a genomic G+C content of 69.7–71.4 mol%. The genus delineation is based on the 16S rRNA gene, 92 core genes (phylogenomics), AAI indices, POCP values, and phenotypic and genomic features.
The type species is Pararoseomonas rosea.
Pararoseomonas rosea (ro’se.a. L. fem. adj. rosea rose-colored, pink).
The description is the same as that given for Roseomonas rosea by Sánchez-Porro et al. (2009). AJ488505 is the accession number for the 16S rRNA gene sequence and FQZF00000000 for the genome sequence. The type strain is 173/96T (=CIP 107419T = DSM 14916T).
Pararoseomonas vinacea (vi.na’ce.a. L. fem. adj. vinacea, of or belonging to wine or to the grape, referring to the colony color).
The description is same as that given for Roseomonas vinacea by Zhang et al. (2008). The accession numbers for the 16S rRNA gene and genome sequences are MT760143 and BO93DRAFT, respectively. The type strain is CPCC 100056T (=KCTC 22045T = CCM 7468T).
Pararoseomonas nepalensis (ne.pal.en’sis. N.L. fem. adj. nepalensis, pertaining to Nepal, the country where source soil samples were collected).
The description is the same as the one given for Roseomonas nepalensis by Chaudhary and Kim (2017). The accession number for the 16S rRNA gene sequence is KX129819. The type strain is G-3-5T (=JCM 31470T = KACC 18908T).
Pararoseomonas aeriglobus (a.e.ri.glo’bus Gr. masc. n. aêr, air; L. masc. n. globus, a sphere; N.L. masc. n. aeriglobus, a sphere living in air).
The description of Pararoseomonas aeriglobus is the same as that of Roseomonas aeriglobus as given by Lee and Jeon (2018). The accession number for the 16S rRNA gene sequence is KY864922. The type strain is KER25-12T (=KACC 19282T = JCM 32049T).
Pararoseomonas aerilata (ae.ri.la’ta. Lat. masc. n. aêr, air; L. fem. perf. part. lata, carried; N.L. fem. part. adj. aerilata, airborne).
The description is the same as that of Roseomonas aerilata as given by Yoo et al. (2008). Accession numbers of the 16S rRNA gene and genome sequences are EF661571 and JONP00000000, respectively. The type strain is 5420S-30T (=KACC 12521T = DSM 19363T).
Pararoseomonas radiodurans (ra.di.o.du’rans. L. masc. n. radius, a beam or ray; N.L. pref. radio-, pertaining to radiation; L. pres. part. durans, enduring; N.L. part. adj. radiodurans, resisting radiation).
The description is the same as that given by Kim et al. (2018) for Roseomonas radiodurans. The accession number for the 16S rRNA gene sequence is KY887689.
The type strain is 17Sr1-1T (=KCTC 52899T = NBRC 112872T).
Pararoseomonas pecuniae (pe.cu’ni.ae. L. gen. n. pecuniae, of/from money or a coin, referring to the source of isolation of the type strain).
The description given is the same as that of Roseomonas pecuniae as given by Lopes et al. (2011). GU168019 is the accession number for the 16S rRNA gene sequence. The accession number for the genome sequence is JACIJD00000000. The type strain is N75T (=LMG 25481T = CIP 110074T).
Pararoseomonas harenae (ha.re’nae. L. gen. fem. n. harenae, of sand, referring to the isolation of the type strain from desert sand).
The description is the same as that of Roseomonas harenae as given by Deng et al. (2020). MN883396 and WWDL00000000 are the accession numbers for the 16S rRNA gene and genome sequences, respectively. The type strain is CPCC 101081T (=KCTC 62852T = NBRC 113512T).
Falsiroseomonas (Fal’si.ro.se.o.mo’nas. L. adj. falsus, false; N.L. fem. n. Roseomonas, a bacterial genus name; N.L. fem. n. Falsiroseomonas, false Roseomonas).
Members of this genus are aerobic, Gram-negative, and coccoid-rod in shape. Motility varies within the genus. Oxidase positive and catalase variable. Polar lipids consist of diphosphatidylglycerol, phosphatidylglycerol, phosphatidylethanolamine, phosphatidylcholine, unidentified phospholipid, unidentified glycolipid, and unidentified aminolipid. Summed feature 3, C16:1 ω5c and C16:0 are the major fatty acids. Members have a genomic size of 5.8–6.6 Mb and a genomic G+C content of 70–72 mol%. The genus delineation is based on the 16S rRNA gene, 92 core genes (phylogenomics), AAI, POCP values, and phenotypic and genomic features.
The type species is Falsiroseomonas stagni.
Falsiroseomonas stagni (stag’ni. L. gen. neut. n. stagni, of a pond, indicating the site of isolation of this organism).
The species description is the same as that of Roseomonas stagni as given by Furuhata et al. (2008). AB369258 is the accession number for the 16S rRNA gene sequence and FOSQ00000000 for the genome sequence. The type strain is HS-69T (=DSM 19981T = JCM 15034T = KCTC 22213T).
Falsiroseomonas bella (bel’la. L. fem. adj. bella, pretty).
The description is the same as that given by Zhang et al. (2020) for Roseomonas bella. MG601506 is the accession number for the 16S rRNA gene sequence and QGNA00000000 for the genome sequence. The type strain is CQN31T (=KCTC 62447T = MCCC 1H00309T).
Falsiroseomonas wooponensis (woo.po.nen’sis. N.L. masc./fem. adj. wooponensis, of or belonging to Woopo wetland, South Korea, the geographical origin of the type strain of the species).
The description is the same as that of Roseomonas wooponensis as given by Lee et al. (2015). KF619243 is the accession number for the 16S rRNA gene sequence. The type strain is WW53T (=KCTC 32534T = JCM 19527T).
Falsiroseomonas terricola (ter.ri’co.la. L. fem. n. terra, earth, soil; L. masc./fem. suff. -cola, inhabitant, dweller; from L. masc./fem. n. incola, dweller; N.L. masc./fem. n. terricola, a dweller upon earth, soil-dweller, referring to the isolation of the type strain from soil).
The description is the same as that given for Roseomonas terricola by Kim et al. (2017). FJ654263 is the accession number for the 16S rRNA gene sequence. The type strain is EM302T (=KACC 13942T = KCTC 42906T = NBRC 111477T).
Falsiroseomonas selenitidurans (se.le.ni.ti.du’rans. N.L. neut. n. selenitum, selenite; L. v. duro, withstand; N.L. part. adj. selenitidurans, withstanding selenite).
The description is the same as that given for Roseomonas selenitidurans by Hou et al. (2020). MT107132 is the accession number for the 16S rRNA gene sequence and JAAVNE00000000 for the genome sequence. The type strain is BU-1T (=GDMCC 1.1776T = KACC 21750 T).
Falsiroseomonas frigidaquae (fri.gi.da’quae. L. masc. adj. frigidus, cold; L. fem. n. aqua, water; N.L. gen. fem. n. frigidaquae, from/of cold water, referring to the isolation of the type strain from a water-cooling system).
The description is the same as that given by Kim et al. (2009) for Roseomonas frigidaquae. EU210160 is the accession number for the 16S rRNA gene sequence and JAAVTX00000000 for the genome sequence. The type strain is CW67T (=JCM 15073T = KCTC 22211T).
Falsiroseomonas tokyonensis (to.ky.o.nen’sis. N.L. masc./fem. adj. tokyonensis, of Tokyo, from where the strain was isolated).
The description is the same as the one given for Roseomonas tokyonensis by Furuhata et al. (2013). AB297501 is the gene accession number for the 16S rRNA gene sequence. The type strain is K-20T (=JCM 14634T = KCTC 32152T).
Falsiroseomonas riguiloci (ri.gu.i.lo’ci. L. masc. adj. riguus, well-watered; L. masc. n. locus, a site; N.L. gen. n. riguiloci, from a well-watered place where the type strain was isolated).
The description is the same as that given by Baik et al. (2012) for Roseomonas riguiloci. HQ436503 is the accession number for the 16S rRNA gene sequence. The type strain is 03SU10-PT (=KCTC 23339T = JCM 17520T = DSM 29515T).
Falsiroseomonas algicola (al.gi’co.la. L. fem. n. alga (gen. algae), an alga; L. masc./fem. suff. -cola, dweller; from L. masc./fem. n. incola, an inhabitant; N.L. masc./fem. n. algicola, an inhabitant of algae).
The description is the same as the one given by Kim et al. (2020) for Roseomonas algicola. MK342491 is the accession number for the 16S rRNA gene sequence and JAAIKB00000000 for the genome sequence. The type strain is PeD5T (=JCM 33309T = KACC 19925T).
Falsiroseomonas arcticisoli (arc’ti.cus arc.ti.ci.so’li L. masc. adj. arcticus, northern; L. neut. n. solum, soil; N.L. gen. n. arcticisoli, of soil from the Arctic).
The description is the same as the one given by Kim M. C. et al. (2016) for Roseomonas arcticisoli. KP274055 is the accession number for the 16S rRNA gene sequence. The type strain is MC 3624T (=CCTCC AB 2014278T = LMG 28637T).
Paeniroseomonas (Pa.e.ni.ro.se.o.mo’nas. L. adv. paene, almost; - i-, connecting vowel; N.L. fem. n. Roseomonas, a bacterial genus; N.L. fem. n. Paeniroseomonas, almost a Roseomonas).
Members of this genus are strictly aerobic, Gram-negative, and coccoid-rod shaped. Cells variable motile. Oxidase negative and catalase positive. Diphosphatidylglycerol, phosphatidylglycerol, phosphatidylethanolamine, and phosphatidylcholine are the major polar lipids. Summed feature 3, C18:1 ω7c, C18:1 2-OH, and C16:0 are the major fatty acids. G+C% content of 68.6–73.1 (mol%). The genus delineation is based on the 16S rRNA gene-based phylogeny and phenotypic features.
The type species is Paeniroseomonas aquatica.
Paeniroseomonas aquatica (a.qua’ti.ca L. fem. adj. aquatica, found in water, aquatic).
The description is the same as that given by Gallego et al. (2006) for Roseomonas aquatica. AM231587 is the accession number for the 16S rRNA gene sequence. The type strain is TR53T (=CECT 7131T = JCM 13556T = DSM 19438T).
Paeniroseomonas fluminis (flu’mi.nis L. gen. neut. n. fluminis, of a river).
The description is the same as that of Roseomonas fluminis as given by Ko et al. (2018). KY649439 is the accession number for the 16S rRNA gene sequence. The type strain is D3T (=JCM 31968T = KACC 19269T).
Plastoroseomonas (Plas.to.ros.e.o.mo’nas. Gr. adj. plastos, false; N.L. fem. n. Roseomonas, a bacterial genus; N.L. fem. n. Plastoroseomonas, a false Roseomonas).
Members of the genus are aerobic, Gram-negative, and rod shaped. Cells are motile. Oxidase negative and catalase positive. Diphosphatidylglycerol, phosphatidylglycerol, phosphatidylethanolamine, unidentified aminolipid, and unidentified lipid are the major polar lipids. Summed feature 3, C18:1 2-OH and C16:0 are the major fatty acids. Genomic size of members ranges from 4.4 to 7.2 Mb and G+C content is ∼70 mol%. The genus delineation is based on the 16S rRNA gene, 92 core genes (phylogenomic), AAI indices, POCP values, and genomic and phenotypic features.
The type species is Plastoroseomonas arctica.
Plastoroseomonas arctica (arc’ti.ca. L. fem. adj. arctica, northern, from the Arctic, referring to the site where the type strain was isolated).
The description is the same as that of Roseomonas arctica as given by Qiu et al. (2016). KJ647399 and JAAEDH000000000 are the accession numbers for the 16S rRNA gene and genome sequences, respectively. The type strain is M6-79T (=CCTCC AB 2013101T = LMG 28251T).
“Plastoroseomonas hellenica” (hel-lé-ni-ka. Gr. adj. ellenikos, Greek, L. fem. adj. hellenica, Greek, pertaining to Greece, the country from where the bacterium was first isolated).
The description is the same as that of Roseomonas hellenica as given by Rat et al. (2021). MN647549 and JAAGBB000000000 are the accession numbers for the 16S rRNA gene and genome sequences, respectively. The type strain is R-73080T (=LMG 31523T = CECT 30032T).
Pseudoroseomonas (Pseu.do.ro.se.o.mo’nas. Gr. adj. pseudês, false; N.L. fem. n. Roseomonas, a bacterial genus; N.L. fem. n. Pseudoroseomonas, false Roseomonas).
Members of the genus are aerobic, Gram-negative, and coccoid to short rods in shape. Cells are non-motile. Oxidase positive and catalase positive. Diphosphatidylglycerol, phosphatidylglycerol, phosphatidylethanolamine, unidentified aminolipid, unidentified lipid, and unidentified phospholipid are the major polar lipids. Summed feature 3, C18:1 2-OH and C16:0 are the major fatty acids. Genomic size of members ranges from 4.2 to 6.4 Mb and G+C% content is 68.7–72.7%. The genus delineation is based on the 16S rRNA gene, 92 core genes (phylogenomic), AAI indices, POCP values, and phenotypic and genomic features.
The type species is Pseudoroseomonas cervicalis.
Pseudoroseomonas cervicalis [cer.vi.ca’lis. L. fem. n. cervix (gen. cervicis), neck; L. masc./fem. Adj. suff. -alis, suffix denoting pertaining to; N.L. fem. adj. cervicalis, pertaining to cervix, from the cervix].
The description is the same as that of Roseomonas cervicalis as given by Rihs et al. (1993). AF533353 is the accession number for the 16S rRNA gene sequence and ADVL00000000 for the genome sequence. The type strain is E7107T (=ATCC 49957T = CIP 104027T).
Pseudoroseomonas suffusca (suf.fus’ca. L. fem. adj. suffusca, light brown, referring to the color of colonies).
The description is the same as that of Roseomonas suffusca as given by Subhash and Lee (2017). LT009497 is the accession number for the 16S rRNA gene sequence. The type strain is S1T (=KEMB 563-465T = JCM 31176T).
Pseudoroseomonas rubra (ru’bra. L. fem. adj. rubra, red).
The description is the same as that of Roseomonas rubra as given by Subhash et al. (2016). LT009499 is the accession number for the 16S rRNA gene sequence. The type strain is S5T (=JCM 31177T = KEMB 563-468T).
Pseudoroseomonas hibiscisoli (hi.bis.ci.so’li. N.L. masc. n. Hibiscus, Mugunghwa/Hibiscus syriacus; L. neut. n. solum, soil; N.L. gen. neut. n. hibiscisoli, of soil of a Hibiscus, the source of the type strain).
The description is the same as that of Roseomonas hibiscisoli as given by Yan et al. (2017). KX456186 is the accession number for the 16S rRNA gene sequence. The type strain is THG-N2.22T (=KACC 18935T = CCTCC AB 2016176T).
Pseudoroseomonas rhizosphaerae (rhi.zo.sphae’rae. Gr. fem. n. rhiza, a root; Gr. fem. n. sphaîra, a ball, a sphere; N.L. fem. n. rhizosphaera, the rhizosphere; N.L. gen. fem. N. rhizosphaerae, of the rhizosphere).
The description is the same as that of Roseomonas rhizosphaerae as given by Chen et al. (2014). KC904962 is the accession number for the 16S rRNA gene sequence and PDNU00000000 for the genome sequence. The type strain is YW11T (= KACC 17225T = CCTCC AB2013041T).
Pseudoroseomonas aestuarii (aes.tu.a’ri.i. L. gen. n. aestuarii, of an estuary, the habitat from which the type strain was isolated).
The description is the same as that of Roseomonas aestuarii as given by Venkata Ramana et al. (2010). AB682256 is the accession number for the 16S rRNA gene sequence. The type strain is JC17T (=CCUG 57456T = KCTC 22692T = NBRC 105654T).
Pseudoroseomonas aerofrigidensis (a.e.ro.fri.gi.den’sis. Gr. masc. n. aêr, air; L. masc. adj. frigidus, cold, cool, chilled; L. masc./fem. adj. suff. -ensis, suffixes used in the sense of “belonging to” or “coming from”; N.L. fem. adj. aerofrigidensis, pertaining to cooling air, as the strain was isolated from an air conditioner).
The description is the same as that of Roseomonas aerofrigidensis as given by Hyeon and Jeon (2017). KY126356 is the accession number for the 16S rRNA gene sequence. The type strain is HC1T (=JCM 31878T = KACC 19097T).
Pseudoroseomonas oryzae (o.ry’zae. L. gen. fem. n. oryzae, of rice, pertaining to the isolation of the type strain from rice paddy soil).
The description is to the same as that of Roseomonas oryzae, as described by Ramaprasad et al. (2015). LN810637 is the accession number for the 16S rRNA gene sequence and VUKA00000000 for the genome sequence. The type strain is JC288T (=KCTC 42542T = LMG 28711T).
Pseudoroseomonas vastitatis (vas.ti.ta’tis. L. gen. fem. n. vastitatis, of a desert, referring to the isolation source of the type strain).
The description is the same as that of Roseomonas vastitatis as given by Zhao et al. (2020). MK421542 is the accession number for the 16S rRNA gene sequence and QXGS00000000 for the genome sequence. The type strain is CPCC 101021T (=J1A743T = KCTC 62043T).
Pseudoroseomonas globiformis (glo.bi.for’mis. L. masc. n. globus, sphere; L. fem. n. forma, shape; N.L. masc./fem. adj. globiformis, of spherical shape).
The description is to the same as that of Roseomonas globiformis as given by Fang et al. (2018). MG589944 is the accession number for the 16S rRNA gene sequence. The type strain is CPCC 100847T (=KCTC 52094T).
Pseudoroseomonas wenyumeiae (wen.yu.mei’ae. N.L. gen. fem. n. wenyumeiae, of Yumei Wen, a famous microbiologist, for her contribution to the Hepatitis B vaccine and anti-HBs complex research and her fundamental role in immunology in China).
The description is the same as that of Roseomonas wenyumeiae as given by Tian et al. (2019). MH974806 is the accession number for the 16S rRNA gene sequence and RFLX00000000 for the genome sequence. The type strain is Z23T (=CGMCC 1.16540T = DSM 106207T).
Pseudoroseomonas ludipueritiae [lu.di.pu.e.ri’ti.ae. L. masc. n. ludus, a place of exercise or practice, a school for elementary instruction; L. masc. n. puer (gen. pueri), a child; N.L. gen. n. ludipueritiae, of a playing place of childhood, intended to mean a kindergarten].
The description is the same as that of Roseomonas ludipueritia as given by Kämpfer et al. (2003) and Sánchez-Porro et al. (2009). AJ488504 is the accession number for the 16S rRNA gene sequence and JACTUZ0000000 for the genome sequence. The type strain is 170-96T (=CIP 107418T = DSM 14915T).
Pseudoroseomonas aerophila (a.e.ro’phi.la. Gr. masc. n. aêr, air; Gr. masc. adj. philos, loving; N.L. fem. adj. aerophila, air-loving).
The description is the same as that of Roseomonas aerophile as given by Kim et al. (2013). JX275860 is the accession number for the 16S rRNA gene sequence and JACTVA00000000 for the genome sequence. The type strain is 7515T-07T (=KACC 16529T = NBRC 108923T).
Pseudoroseomonas musae (mu’sae. L. gen. fem. n. musae, of Musa, isolated from leaf of banana Musa sp).
The description is the same as that of Roseomonas musae as given by Nutaratat et al. (2013). AB594201 is the accession number for the 16S rRNA gene sequence. The type strain is PN1T (=BCC 44863T = NBRC 107870T).
Pseudoroseomonas coralli (co.ral’li. L. gen. n. coralli, of coral, from which the organism was isolated).
The description is the same as that of Roseomonas coralli as given by Li et al. (2021). MN336179 is the accession number for the 16S rRNA gene sequence and SNVJ000000000 for the genome sequence. The type strain is M0104T (=KCTC 62359T = MCCC 1K03632T).
Pseudoroseomonas deserti (de.ser’ti. L. gen. neut. n. deserti, of a desert).
The description is the same as that of Roseomonas deserti as given by Subhash and Lee (2018). LT837512 is the accession number for the 16S rRNA gene sequence and MLCO00000000 for the genome sequence. The type strain is M3T (=KEMB 2255-459T = JCM 31275T).
Neoroseomonas (Ne.o.ro.se.o.mo’nas. Gr. adj. neos, new; N.L. fem. n. Roseomonas, a bacterial genus; N.L. fem. n. Neoroseomonas, to refer to the fact that it is a new group of Roseomonas).
Members are aerobic, Gram-negative, and coccoid to short rod in shape. Motility is variable within the members. Oxidase and catalase positive. Diphosphatidylglycerol, phosphatidylglycerol, phosphatidylcholine, phosphatidylethanolamine, unidentified glycolipid, unidentified aminolipid, unidentified lipid, and unidentified phospholipid are the major polar lipids. Summed feature 3, C18:1 2-OH and C16:0 are the major fatty acids. Genome size of members ranges from 4.7 to 6.3 Mb and G+C% content is 68.8–71.5%. The genus delineation is based on the 16S rRNA gene, 92 core genes (phylogenomic), AAI indices, POCP values, and phenotypic and genomic features.
The type species is Neoroseomonas lacus.
Neoroseomonas lacus (la’cus. L. gen. masc. n. lacus, of a lake, indicating the site of isolation of this organism).
The description is the same as that of Roseomonas lacu as given by Jiang et al. (2006). AJ786000 is the accession number for the 16S rRNA gene sequence and BMKW00000000 for the genome sequence. The type strain is TH-G33T (=CGMCC 1.3617T = JCM 13283T).
Neoroseomonas terrae (ter’rae. L. gen. fem. n. terrae, of the soil).
The description is the same as that of Roseomonas terrae as given by Yoon et al. (2007). EF363716 and JAAEDI000000000 are the accession numbers for the 16S rRNA gene and genome sequences, respectively. The type strain is DS-48T (=KCTC 12874T = JCM 14592T).
Neoroseomonas eburnea (e.bur’ne.a. L. fem. adj. eburnea, white as ivory).
The description is the same as that of Roseomonas eburnean as given by Wang et al. (2016). KF254767 and JAAEDL000000000 are the accession numbers for the 16S rRNA gene and genome sequences, respectively. The type strain is BUT-5T (=CCTCC AB2013276T = KACC 17166T).
Neoroseomonas alkaliterrae (al.ka.li.ter’rae. Arabic masc. n. al-qaliy, the ashes of saltwort; N.L. neut. N. alkali, alkali; L. gen. fem. n. terrae, of the soil or earth; N.L. gen. fem. n. alkaliterrae, of alkaline soil).
The description is the same as that of Roseomonas alkaliterrae as given by Dong et al. (2014). KF771274 is the accession number for the 16S rRNA gene sequence and JACIJE00000000 for the genome sequence. The type strain is YIM 78007T (=BCRC 80644T = JCM 19656T = DSM 25895T).
Neoroseomonas oryzicola (o.ry.zi’co.la. L. fem. n. oryza, rice; L. masc./fem. suff. -cola, an inhabitant; from L. masc./fem. n. incola, dweller; N.L. masc./fem. n. oryzicola, an inhabitant of rice).
The description is the same as that of Roseomonas oryzicola as given by Chung et al. (2015). EU707562 is the accession number for the 16S rRNA gene sequence and JAAVUP00000000 for the genome sequence. The type strain is YC6724T (=KCTC 22478T = NBRC 109439T).
Neoroseomonas soli (so’li. L. gen. neut. n. soli, of soil, the source of the type strain).
The description is the same as that of Roseomonas soli as given by Kim and Ka (2014). JN575264 and JAAEDM00000000 are the accession numbers for the 16S rRNA gene and genome sequences, respectively. The type strain is 5N26T (=KACC 16376T = NBRC 109097T).
Neoroseomonas sediminicola (se.di.mi.ni.co’la. L. neut. n. sedimen -inis, sediment; L. masc./fem. suff. -cola, inhabitant, dweller; N.L. masc./fem. n. sediminicola, sediment-dweller, referring to the source of the type strain).
The description is the same as that of Roseomonas sediminicola as given by He et al. (2014). JQ349047 is the accession number for the 16S rRNA gene sequence. The type strain is FW-3T (=KACC 16616T = JCM 18210T).
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
AR, UJ, and NS designed the studies under the supervision of CS and CR. UJ, AR, NS, and GD performed the genomic and phylogenetic analysis. AR and UJ wrote the manuscript. CR and CS supervised the study, contributed to the text preparation, and revised the manuscript. All authors read and approved the final version of the manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We thank Bernhard Schink of University of Konstanz, Germany for correcting the protologues. AR thanks DST, Government of India for the award of the INSPIRE Junior/Senior Research Fellowship. UJ thank TEQIP III, JNTUH, for the award of the research assistantship. CR thank the Department of Biotechnology, Government of India for the award of the Tata Innovation Fellowship. The financial support received from CSIR is acknowledged. CS thank UGC for the award of the mid-career grant and AICTE and for the research project under the Research Promotion Scheme. The infrastructural facilities funded by DST-FIST (UoH and JNTUH), UGC-SAP (DRS; UoH), TEQIP, and AICTE (from JNTUH) are acknowledged.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.677842/full#supplementary-material
NCBI, National Center for Biotechnology Information; ANI, Average Nucleotide Identity; AAI, Average Amino acid Identity; POCP, Percentage of Conserved Proteins; dDDH, digital DNA-DNA Hybridization; BLAST, Basic Local Alignment Search Tool; MUSCLE, MUltiple Sequence Comparison by Log-Expectation; KCTC, Korean Collection for Type Cultures; CGMCC, China General Microbiological Culture Collection Centre; ATCC, American Type Culture Collection; CIP, Institute Pasteur Collection; DSM, Deutsche Sammlung von Mikroorganismen und Zellkulturen GmbH; JCM, Japan Collection of Microorganisms-RIKEN BioResource Center; KACC, Korean Agricultural Culture Collection; NBRC, NITE Biological Resource Center; LMG, Laboratorium voor Microbiologie; MCCC, Marine Culture Collection of China; GDMCC, Guangdong Microbial Culture Collection Center; CECT Colección Española de Cultivos Tipo; KEMB, Korea Environmental Microorganisms Bank; CCUG, Culture Collection University of Göteborg; BCC, BIOTEC Culture Collection; UBCG, Up-to-date Bacterial Core Gene set.
Aherfi, S., Andreani, J., Baptiste, E., Oumessoum, A., Dornas, F. P., Andrade, A. C., et al. (2018). A large open pangenome and a small core genome for giant pandoraviruses. Front. Microbiol. 9:1486. doi: 10.3389/fmicb.2018.01486
Alarico, S., Rainey, F. A., Empadinhas, N., Schumann, P., Nobre, M. F., and da Costa, M. S. (2002). Rubritepida Flocculans gen. nov., sp. nov., a new slightly thermophilic member of the alpha-1 subclass of the Proteobacteria. Syst. Appl. Microbiol. 25, 198–206. doi: 10.1078/0723-2020-00116
Aliyu, H., Lebre, P., Blom, J., Cowan, D., and De Maayer, P. (2016). Phylogenomic re-assessment of the thermophilic genus Geobacillus. Syst. Appl. Microbiol. 39, 527–533. doi: 10.1016/j.syapm.2016.09.004
Andreeva, I. S., Pechurkina, N. I., Morozova, O. V., Riabchikova, E. I., Belikov, S. I., Puchkova, L. I., et al. (2007). The new eubacterium Roseomonas baikalica sp. nov. isolated from core samples collected by deep-hole drilling of the bottom of Lake Baikal. Mikrobiologiia 76, 552–559.
Arkin, A. P., Cottingham, R. W., Henry, C. S., Harris, N. L., Stevens, R. L., Maslov, S., et al. (2018). KBase: the united states department of energy systems biology knowledgebase. Nat. Biotechnol. 36:566. doi: 10.1038/nbt.4163
Auch, A.-F., Klenk, H.-P., and Göker, M. (2010). Standard operating procedure for calculating genome-to-genome distances based on high-scoring segment pairs. Stand. Genomic Sci. 2, 142–148. doi: 10.4056/sigs.541628
Baik, K. S., Park, S. C., Choe, H. N., Kim, S. N., Moon, J. H., and Seong, C. N. (2012). Roseomonas riguiloci sp. nov., isolated from wetland freshwater. Int. J. Syst. Evol. Microbiol. 62, 3024–3029. doi: 10.1099/ijs.0.036186-0
Bertelli, C., Laird, M. R., Williams, K. P., Lau, B. Y., Hoad, G., Winsor, G. L., et al. (2017). IslandViewer 4: expanded prediction of genomic islands for larger-scale datasets. Nucleic Acids Res. 45, W30–W35. doi: 10.1093/nar/gkx343
Bibashi, E., Sofianou, D., Kontopoulou, K., Mitsopoulos, E., and Kokolina, E. (2000). Peritonitis due to Roseomonas fauriae in a patient under- going continuous ambulatory peritoneal dialysis. J. Clin. Microbiol. 38, 456–457.
Blin, K., Shaw, S., Steinke, K., Villebro, R., Ziemert, N., Lee, S. Y., et al. (2019). antiSMASH 5.0: updates to the secondary metabolite genome mining pipeline. Nucleic Acids Res. 47, 81–87. doi: 10.1093/nar/gkz310
Brito, A. F., Braconi, C. T., Weidmann, M., Dilcher, M., Alves, J. M., Gruber, A., et al. (2015). The pangenome of the Anticarsia gemmatalis multiple nucleopolyhedrovirus (AgMNPV). Genome Biol. Evol. 27, 94–108. doi: 10.1093/gbe/evv231
Chaudhari, N. M., Gupta, V. K., and Dutta, C. (2016). BPGA-an ultra-fast pan-genome analysis pipeline. Sci. Rep. 6:24373. doi: 10.1038/srep24373
Chaudhary, D. K., and Kim, J. (2017). Roseomonas nepalensis sp. nov., isolated from oil-contaminated soil. Int. J. Syst. Evol. Microbiol. 67, 981–987. doi: 10.1099/ijsem.0.001727
Chen, Q., Sun, L. N., Zhang, X. X., He, J., Kwon, S. W., Zhang, J., et al. (2014). Roseomonas rhizosphaerae sp. nov., a triazophos-degrading bacterium isolated from soil. Int. J. Syst. Evol. Microbiol. 64, 1127–1133. doi: 10.1099/ijs.0.057000-0
Chen, W. M., Xie, Y. R., Sheu, D. S., Tsai, J. M., and Sheu, S. Y. (2020). Rhodovarius crocodyli sp. nov., isolated from a crocodile pond. Int. J. Syst. Evol. Microbiol. 70, 5141–5148. doi: 10.1099/ijsem.0.004396
Chu, C. W., Chen, Q., Wang, C. H., Wang, H. M., Sun, Z. G., He, Q., et al. (2016). Roseomonas chloroacetimidivorans sp. nov., a chloroacetamide herbicide-degrading bacterium isolated from activated sludge. Antonie Van Leeuwenhoek. 109, 611–618. doi: 10.1007/s10482-016-0664-y
Chun, J., Oren, A., Ventosa, A., Christensen, H., Arahal, D. R., da Costa, M. S., et al. (2018). Proposed minimal standards for the use of genome data for the taxonomy of prokaryotes. Int. J. Syst. Evol. Microbiol. 68, 461–466. doi: 10.1099/ijsem.0.002516
Chun, J., and Rainey, F. A. (2014). Integrating genomics into the taxonomy and systematics of the Bacteria and Archaea. Int. J. Syst. Evol. Microbiol. 64, 316–324. doi: 10.1099/ijs.0.054171-0
Chung, E. J., Yoon, H. S., Kim, K. H., Jeon, C. O., and Chung, Y. R. (2015). Roseomonas oryzicola sp. nov., isolated from the rhizosphere of rice (Oryza sativa L.). Int. J. Syst. Evol. Microbiol. 65, 4839–4844. doi: 10.1099/ijsem.0.000656
da Silva Filho, A. C., Raittz, R. T., Guizelini, D., De Pierri, C. R., Augusto, D. W., et al. (2018). Comparative analysis of genomic Island prediction tools. Front. Genet. 12:619. doi: 10.3389/fgene.2018.00619
Damtab, J., Nutaratat, P., Boontham, W., Srisuk, N., Duangmal, K., Yurimoto, H., et al. (2016). Roseomonas elaeocarpi sp. nov., isolated from olive (Elaeocarpus hygrophilus Kurz.) phyllosphere. Int. J. Syst. Evol. Microbiol. 66, 474–480. doi: 10.1099/ijsem.0.000748
Deng, Y., Sun, Y., Wang, H., Yu, L. Y., and Zhang, Y. Q. (2020). Roseomonas harenae sp. nov., from desert gravel soil. Int. J. Syst. Evol. Microbiol. 70, 5711–5716. doi: 10.1099/ijsem.0.004467
Diesendorf, N., Köhler, S., Geißdörfer, W., Grobecker-Karl, T., Karl, M., and Burkovski, A. (2017). Characterisation of Roseomonas mucosa isolated from the root canal of an infected tooth. BMC Res. Notes 10:212. doi: 10.1186/s13104-017-2538-4
Dong, L., Ming, H., Yin, Y. R., Duan, Y., Zhou, E.-M., Nie, G.-X., et al. (2014). Roseomonas alkaliterrae sp. nov., isolated from an alkali geothermal soil sample in Tengchong. Yunnan, south-west China. Antonie van Leeuwenhoek. 105, 899–905. doi: 10.1007/s10482-014-0144-1
Edgar, R. C. (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797. doi: 10.1093/nar/gkh340
Fang, X. M., Bai, J. L., Zhang, D. W., Su, J., Zhao, L. L., Zhang, Y. Q., et al. (2018). Roseomonas globiformis sp. nov., an airborne bacteria isolated from an urban area of Beijing. Int. J. Syst. Evol. Microbiol. 68, 3301–3306. doi: 10.1099/ijsem.0.002985
Felsenstein, J. (1985). Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39, 783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x
Fox, G. E., Wisotzkey, J. D., and Jurtshuk, P. Jr. (1992). How close is close: 16S rRNA sequence identity may not be sufficient to guarantee species identity. Int. J. Syst. Bacteriol. 42, 166–170. doi: 10.1099/00207713-42-1-166
Furuhata, K., Ishizaki, N., Edagawa, A., and Fukuyama, M. (2013). Roseomonas tokyonensis sp. nov. isolated from a biofilm sample obtained from a cooling tower in Tokyo. Japan. Biocontrol Sci. 18, 205–209. doi: 10.4265/bio.18.205
Furuhata, K., Miyamamoto, H., Goto, K., Kato, Y., Hara, M., and Fukuyama, M. (2008). Roseomonas stagni sp. nov., isolated from pond water in Japan. J. Gen. Appl. Microbiol. 54, 167–171. doi: 10.2323/jgam.54.167
Gallego, V., Sanchez-Porro, C., Garcia, M. T., and Ventosa, A. (2006). Roseomonas aquatica sp. nov., isolated from drinking water. Int. J. Syst. Evol. Microbiol. 56, 2291–2295. doi: 10.1099/ijs.0.64379-0
Greub, G., La Scola, B., and Raoult, D. (2004). Amoebae-resisting bacteria isolated from human nasal swabs by amoebal coculture. Emerg. Infect. Dis. 10, 470–477. doi: 10.3201/eid1003.020792
Gupta, R. S., Lo, B., and Son, J. (2018). Phylogenomics and comparative genomic studies robustly support division of the genus Mycobacterium into an emended genus Mycobacterium and four novel genera. Front. Microbiol. 9:67. doi: 10.3389/fmicb.2018.00067
Han, X. Y., Pham, A. S., Tarrand, J. J., Rolston, K. V., Helsel, L. O., and Levett, P. N. (2003). Bacteriologic characterization of 36 strains of Roseomonas species and proposal of Roseomonas mucosa sp. nov. and Roseomonas gilardii subsp rosea subsp. nov. Am. J. Clin. Pathol. 120, 256–264. doi: 10.1309/731V-VGVC-KK35-1Y4J
He, D., Kim, J. K., Jiang, X. Y., Park, H. Y., Sun, C., Yu, H. S., et al. (2014). Roseomonas sediminicola sp. nov., isolated from fresh water. Antonie Van Leeuwenhoek. 105, 191–197. doi: 10.1007/s10482-013-0065-4
Helsel, L. O., Hollis, D. G., Steigerwalt, A. G., and Levett, P. N. (2006). Reclassification of Roseomonas fauriae Rihs et al. 1998 as a later heterotypic synonym of Azospirillum brasilense Tarrand et al. 1979. Int. J. Syst. Evol. Microbiol. 56, 2753–2755. doi: 10.1099/ijs.0.64549-0
Hördt, A., López, M. G., Meier-Kolthoff, J. P., Schleuning, M., Weinhold, L. M., Tindall, B. J., et al. (2020). Analysis of 1,000+ Type-Strain genomes substantially improves taxonomic classification of Alphaproteobacteria. Front. Microbial. 11:468. doi: 10.3389/fmicb.2020.00468
Hou, X., Liu, H., Wei, S., Ding, Z., Sang, F., Zhao, Y., et al. (2020). Roseomonas selenitidurans sp. nov., isolated from urban soil, and emended description of Roseomonas frigidaquae. Int. J. Syst. Evol. Microbiol. 70, 5937–5942. doi: 10.1099/ijsem.0.004496
Hyeon, J. W., and Jeon, C. O. (2017). Roseomonas aerofrigidensis sp. nov., isolated from an air conditioner. Int. J. Syst. Evol. Microbiol. 67, 4039–4044. doi: 10.1099/ijsem.0.002246
Janda, J. M., and Abbott, S. L. (2007). 16S rRNA gene sequencing for bacterial identification in the diagnostic laboratory: pluses, perils, and pitfalls. J. Clin. Microbiol. 45, 2761–2764. doi: 10.1128/JCM.01228-07
Jiang, C.-Y., Dai, X., Wang, B.-J., Zhou, Y.-G., and Liu, S.-J. (2006). Roseomonas lacus sp. nov., isolated from freshwater lake sediment. Int. J. Syst. Evol. Microbiol. 56, 25–28. doi: 10.1099/ijs.0.63938-0
Jin, L., Lee, H. G., No, K. J., Ko, S. R., Kim, H. S., Ahn, C. Y., et al. (2013). Belnapia soli sp. nov., a proteobacterium isolated from grass soil. Int. J. Syst. Evol. Microbiol. 63, 1955–1959. doi: 10.1099/ijs.0.045302-0
Jin, R., Su, J., Liu, H. Y., Wei, Y. Z., Li, Q. P., Zhang, Y.-Q., et al. (2012). Description of Belnapia rosea sp. nov. and emended description of the genus Belnapia Reddy et al., 2006. Int. J. Syst. Evol. Microbiol. 62, 705–709. doi: 10.1099/ijs.0.031021-0
Kämpfer, P., Andersson, M. A., Jäckel, U., and Salkinoja-Salonen, M. (2003). Teichococcus ludipueritiae gen. nov. sp. nov., and Muricocccus roseus gen. nov. sp. nov. representing two new genera of the alpha-1 subclass of the Proteobacteria. Syst. Appl. Microbiol. 26, 23–29. doi: 10.1078/072320203322337272
Kämpfer, P., Busse, H. J., Rosséllo-Mora, R., Kjellin, E., and Falsen, E. (2004). Rhodovarius lipocyclicus gen. nov. sp. nov., a new genus of the alpha-1 subclass of the Proteobacteria. Syst. Appl. Microbiol. 27, 511–516. doi: 10.1078/0723202041748235
Khan, S. A., Jeong, S. E., Jung, H. S., Quan, Z. X., and Jeon, C. O. (2019). Roseicella frigidaeris gen. nov., sp. nov., isolated from an air-conditioning system. Int. J. Syst. Evol. Microbiol. 69, 1384–1389. doi: 10.1099/ijsem.0.003322
Kim, D. U., and Ka, J. O. (2014). Roseomonas soli sp. nov., isolated from an agricultural soil cultivated with Chinese cabbage (Brassica campestris). Int. J. Syst. Evol. Microbiol. 64, 1024–1029. doi: 10.1099/ijs.0.053827-0
Kim, D. U., Lee, H., Kim, S. G., and Ka, J. O. (2017). Roseomonas terricola sp. nov., isolated from agricultural soil. Int. J. Syst. Evol. Microbiol. 67, 4836–4841. doi: 10.1099/ijsem.0.002389
Kim, H. M., Khan, S. A., Han, D. M., Chun, B. H., and Jeon, C. O. (2020). Roseomonas algicola sp. nov., isolated from a green alga. Pediastrum duplex. Int. J. Syst. Evol. Microbiol. 11, 5634–5639. doi: 10.1099/ijsem.0.004454
Kim, J. Y., Kim, D. U., Kang, M. S., Jang, J. H., Kim, S. J., Kim, M. J., et al. (2018). Roseomonas radiodurans sp. nov., a gamma-radiation-resistant bacterium isolated from gamma ray-irradiated soil. Int. J. Syst. Evol. Microbiol. 68, 2443–2447. doi: 10.1099/ijsem.0.002852
Kim, M. C., Rim, S., Pak, S., Ren, L., Zhang, Y., Chang, X., et al. (2016). Roseomonas arcticisoli sp. nov., isolated from Arctic tundra soil. Int. J. Syst. Evol. Microbiol. 66, 4057–4064. doi: 10.1099/ijsem.0.001310
Kim, W. H., Kim, D. H., Kang, K., and Ahn, T. Y. (2016). Dankookia rubra gen. nov., sp. nov., an alphaproteobacterium isolated from sediment of a shallow stream. J. Microbiol. 54, 420–425. doi: 10.1007/s12275-016-6054-3
Kim, M. S., Baik, K. S., Park, S. C., Rhee, M. S., Oh, H.-M., and Seong, C. N. (2009). Roseomonas frigidaquae sp. nov., isolated from a water-cooling system. Int. J. Syst. Evol. Microbiol. 59, 1630–1634. doi: 10.1099/ijs.0.004812-0
Kim, S. J., Weon, H. Y., Ahn, J. H., Hong, S. B., Seok, S. J., Whang, K. S., et al. (2013). Roseomonas aerophila sp. nov., isolated from air. Int. J. Syst. Evol. Microbiol. 63, 2334–2337. doi: 10.1099/ijs.0.046482-0
Kimura, M. (1980). A simple method for estimating evolutionary rate of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 16, 111–120. doi: 10.1007/BF01731581
Ko, Y., Yim, J., Hwang, W. M., Kang, K., and Ahn, T. Y. (2018). Roseomonas fluminis sp. nov. isolated from sediment of a shallow stream. Int. J. Syst. Evol. Microbiol. 68, 782–787. doi: 10.1099/ijsem.0.002578
Kumar, S., Glen, S., and Koichiro, T. (2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Kuo, C. H., and Ochman, H. (2009). The fate of new bacterial genes. FEMS Microbiol. Rev. 33, 38–43. doi: 10.1111/j.1574-6976.2008.00140.x
Lalucat, J., Mulet, M., Gomila, M., and García-Valdés, E. (2020). Genomics in bacterial taxonomy: impact on the genus Pseudomonas. Genes 29, 11–139. doi: 10.3390/genes11020139
Lee, J. H., Kim, M. S., Baik, K. S., Kim, H. M., Lee, K. H., and Seong, C. N. (2015). Roseomonas wooponensis sp. nov., isolated from wetland freshwater. Int. J. Syst. Evol. Microbiol. 65, 4049–4054. doi: 10.1099/ijsem.0.000536
Lee, Y., and Jeon, C. O. (2018). Roseomonas aeriglobus sp. nov., isolated from an air-conditioning system. Antonie Van Leeuwenhoek 111, 343–351. doi: 10.1007/s10482-017-0956-x
Li, F., Huang, Y., Hu, W., Li, Z., Wang, Q., Huang, S., et al. (2021). Roseomonas coralli sp. nov., a heavy metal resistant bacterium isolated from coral. Int. J. Syst. Evol. Microbiol. 71:004624. doi: 10.1099/ijsem.0.004624
Liu, A., Zhang, Y. J., Cheng, P., Peng, Y. J., Blom, J., and Xue, K. J. (2020). Whole genome analysis calls for a taxonomic rearrangement of the genus Colwellia. Antonie Van Leeuwenhoek 113, 919–931. doi: 10.1007/s10482-020-01405-6
Livingstone, P. G., Morphew, R. M., and Whitworth, D. E. (2018). Genome Sequencing and pan-genome analysis of 23 Corallococcus spp. strains reveal unexpected diversity, with particular plasticity of predatory gene sets. Front. Microbiol. 9:3187. doi: 10.3389/fmicb.2018.03187
Logan, N. A., Berge, O., Bishop, A. H., Busse, H. J., De Vos, P., Fritze, D., et al. (2009). Proposed minimal standards for describing new taxa of aerobic, endospore-forming bacteria. Int. J. Syst. Evol. Microbiol. 59, 2114–2121. doi: 10.1099/ijs.0.013649-0
Loganathan, P., and Nair, S. (2004). Swaminathania salitolerans gen. nov., sp. nov., a salt-tolerant, nitrogen-fixing and phosphate-solubilizing bacterium from wild rice (Porteresia coarctata Tateoka). Int. J. Syst. Evol. Microbiol. 54, 1185–1190. doi: 10.1099/ijs.0.02817-0
Lopes, A., Espirito, S. C., Grass, G., Chung, A. P., and Morais, P. V. (2011). Roseomonas pecuniae sp. nov., isolated from the surface of a copper-alloy coin. Int. J. Syst. Evol. Microbiol. 61, 610–615. doi: 10.1099/ijs.0.020966-0
Lopes-Santos, L., Castro, D. B. A., Ferreira-Tonin, M., Corrêa, D. B. A., Weir, B. S., Park, D., et al. (2017). Reassessment of the taxonomic position of Burkholderia andropogonis and description of Robbsia andropogonis gen. nov., comb. nov. Antonie Van Leeuwenhoek 110, 727–736. doi: 10.1007/s10482-017-0842-6
Luo, C., Rodriguez-R, L. M., and Konstantinidis, K. T. (2014). MyTaxa: an advanced taxonomic classifier for genomic and metagenomic sequences. Nucleic Acids Res. 42:e73. doi: 10.1093/nar/gku169
Malini, S., Goh, B. L., and Lim, T. S. (2016). A rare case of Roseomonas gilardii peritonitis in a patient on continuous ambulatory peritoneal dialysis. Perit. Dial. Int. 36:578. doi: 10.3747/pdi.2016.00103
Margesin, R., and Zhang, D. C. (2013). Humitalea rosea gen. nov., sp. nov., an aerobic bacteriochlorophyll-containing bacterium of the family Acetobacteraceae isolated from soil. Int. J. Syst. Evol. Microbiol. 63, 1411–1416. doi: 10.1099/ijs.0.043018
Marin, M. E., Marco, D. J., Diabar, E., Fernandez, C. L., Greco, G., Flores, Y., et al. (2001). Catheter-related bacteremia caused by Roseomonas gilardii in an immunocompromised patient. Int. J. Infect. Dis. 5, 170–171. doi: 10.1016/s1201-9712(01)90095-5
McInerney, J. O., McNally, A., and O’Connell, M. J. (2017). Why prokaryotes have pangenomes. Nat. Microbiol. 28, 2–17040. doi: 10.1038/nmicrobiol.2017.40
Michon, A. L., Saumet, L., Bourdier, A., Haouy, S., Sirvent, N., and Marchandin, H. (2014). Bacteremia due to imipenem-resistant Roseomonas mucosa in a child with acute lymphoblastic leukemia. J. Pediatr. Hematol. Oncol. 36, e165–e168.
Ming, H., Duan, Y. Y., Yin, Y. R., Meng, X.-L., Li, S., Zhou, E.-M., et al. (2016). Crenalkalicoccus roseus gen. nov., sp. nov., a thermophilic bacterium isolated from alkaline hot springs. Int. J. Syst. Evol. Microbiol. 66, 2319–2326. doi: 10.1099/ijsem.0.001029
Na, S. I., Kim, Y. O., Yoon, S. H., Ha, S. M., Baek, I., and Chun, J. (2018). UBCG: up-to-date bacterial core gene set and pipeline for phylogenomic tree reconstruction. J. Microbiol. 56, 281–285. doi: 10.1007/s12275-018-8014-6
Nahass, R. G., Wisneski, R., Herman, D. J., Hirsh, E., and Goldblatt, K. (1995). Vertebral osteomyelitis due to Roseomonas species: case report and review of the evaluation of vertebral osteomyelitis. Clin. Infect. Dis. 21, 1474–1476. doi: 10.1093/clinids/21.6.1474
Nouioui, I., Carro, L., Garcia-Lopez, M., Meier-Kolthoff, J. P., Woyke, T., Kyrpides, N. C., et al. (2018). Genome-based taxonomic classification of the phylum Actinobacteria. Front. Microbiol. 9:2007. doi: 10.3389/fmicb.2018.02007
Nutaratat, P., Srisuk, N., Duangmal, K., Yurimoto, H., Sakai, Y., Muramatsu, Y., et al. (2013). Roseomonas musae sp. nov., a new bacterium isolated from a banana phyllosphere. Antonie Van Leeuwenhoek. 103, 617–624. doi: 10.1007/s10482-012-9845-5
Olm, M. R., Crits-Christoph, A., Diamond, S., Lavy, A., Matheus Carneyali, P. B., and Banfield, J. F. (2020). Consistent metagenome-derived metrics verify and delineate bacterial species boundaries. mSystems 5:e00731-19. doi: 10.1128/mSystems.00731-719
Orata, F. D., Meier-Kolthoff, J. P., Sauvageau, D., and Stein, L. Y. (2018). Phylogenomic analysis of the gammaproteobacterial methanotrophs (Order Methylococcales) calls for the reclassification of members at the genus and species levels. Front. Microbiol. 9:3162. doi: 10.3389/fmicb.2018.03162
Qin, Q., Xie, B., Zhang, X., Chen, X., Zhou, B., Zhou, J., et al. (2014). A proposed genus boundary for the prokaryotes based on genomic insights. J. Bacteriol. 196, 2210–2215. doi: 10.1128/jb.01688-14
Qiu, X., Qu, Z., Jiang, F., Lin, Y., Zhang, Y., Chang, X., et al. (2016). Roseomonas arctica sp. nov., isolated from arctic glacial foreland soil. Int. J. Syst. Evol. Microbiol. 66, 1218–1223. doi: 10.1099/ijsem.0.000857
Ramaprasad, E. V. V., Sasikala, C., and Ramana, C. V. (2015). Roseomonas oryzae sp. nov., isolated from paddy rhizosphere soil. Int. J. Syst. Evol. Microbiol. 65, 3535–3540. doi: 10.1099/ijsem.0.000449
Rat, A., Naranjo, H. D., Lebbe, L., Cnockaert, M., Krigas, N., Grigoriadou, K., et al. (2021). Roseomonas hellenica sp. nov., isolated from roots of wild-growing Alkanna tinctoria. Syst. Appl. Microbiol. 44:126206. doi: 10.1016/j.syapm.2021.126206
Reddy, G. S., Nagy, M., and Garcia-Pichel, F. (2006). Belnapia moabensis gen. nov., sp. nov., an alphaproteobacterium from biological soil crusts in the Colorado Plateau, USA. Int. J. Syst. Evol. Microbiol. 56, 51–58. doi: 10.1099/ijs.0.63764-0
Richter, M., and Rosselló-Móra, R. (2009). Shifting the genomic gold standard for the prokaryotic species definition. Proc. Natl. Acad. Sci. U S A. 106, 19126–19131. doi: 10.1073/pnas.0906412106
Rihs, J. D., Brenner, D. J., Weaver, R. E., Steigerwalt, A. G., Hollis, D. G., and Yu, V. L. (1993). Roseomonas, a new genus associated with bacteremia and other human infections. J. Clin. Microbiol. 31, 3275–3283. doi: 10.1128/JCM.31.12.3275-3283.1993
Rodriguez, L. M., and Konstantinidis, K. T. (2014). Bypassing cultivation to identify bacterial species. Microbe 9, 111–117.
Romano-Bertrand, S., Bourdier, A., Aujoulat, F., Michon, A. L., Masnou, A., Parer, S., et al. (2016). Skin microbiota is the main reservoir of Roseomonas mucosa, an emerging opportunistic pathogen so far assumed to be environmental. Clin. Microbiol. Infect. 22:737.e1-7. doi: 10.1016/j.cmi.2016.05.024
Rosselló-Móra, R., and Amann, R. (2015). Past and future species definitions for bacteria and Archaea. Syst. Appl. Microbiol. 38, 209–202. doi: 10.1016/j.syapm.2015.02.001
Saitoh, S., Suzuki, T., and Nishimura, Y. (1998). Proposal of Craurococcus roseus gen. nov., sp. nov. and Paracraurococcus ruber gen. nov., sp. nov., novel aerobic bacteriochlorophyll a-containing bacteria from soil. Int. J. Syst. Evol. Microbiol. 48, 1043–1047. doi: 10.1099/00207713-48-3-1043
Sánchez-Porro, C., Gallego, V., Busse, H. J., Kämpfer, P., and Ventosa, A. (2009). Transfer of Teichococcus ludipueritiae and Muricoccus roseus to the genus Roseomonas, as Roseomonas ludipueritiae comb. nov. and Roseomonas rosea comb. nov., respectively, and emended description of the genus Roseomonas. Int. J. Syst. Evol. Microbiol. 59, 1193–1198. doi: 10.1099/ijs.0.004820-0
Shao, S., Guo, X., Guo, P., Cui, Y., and Chen, Y. (2019). Roseomonas mucosa infective endocarditis in patient with systemic lupus erythematosus: case report and review of literature. BMC Infect. Dis. 19: 140. doi: 10.1186/s12879-019-3774-0
Subhash, Y., Bang, J. J., You, T. H., and Lee, S. S. (2016). Roseomonas rubra sp. nov., isolated from lagoon sediments. Int. J. Syst. Evol. Microbiol. 66, 3821–3827. doi: 10.1099/ijsem.0.001271
Subhash, Y., and Lee, S. S. (2017). Roseomonas suffusca sp. nov., isolated from lagoon sediments. Int. J. Syst. Evol. Microbiol. 67, 2390–2396. doi: 10.1099/ijsem.0.001966
Subhash, Y., and Lee, S. S. (2018). Roseomonas deserti sp. nov., isolated from crude oil contaminated desert sand. Int. J. Syst. Evol. Microbiol. 68, 675–680. doi: 10.1099/ijsem.0.002565
Suresh, G., Lodha, T. D., Indu, B., Sasikala, Ch, and Ramana, Ch. V (2019). Taxogenomics resolves conflict in the genus Rhodobacter: a two and half decades pending thought to reclassify the genus Rhodobacter. Front. Microbiol. 10:2480. doi: 10.3389/fmicb.2019.02480
Tarrand, J. J., Krieg, N. R., and Döbereiner, J. (1978). A taxonomic study of the Spirillum lipoferum group, with descriptions of a new genus, Azospirillum gen. nov. and two species, Azospirillum lipoferum (Beijerinck) comb. nov. and Azospirillum brasilense sp. nov. Can. J. Microbiol. 24, 967–980. doi: 10.1139/m78-160
Tian, Z., Lu, S., Jin, D., Yang, J., Pu, J., Lai, X. H., et al. (2019). Roseomonas wenyumeiae sp. nov., isolated from faeces of Tibetan antelopes (Pantholops hodgsonii) on the Qinghai-Tibet Plateau. Int. J. Syst. Evol. Microbiol. 69, 2979–2986. doi: 10.1099/ijsem.0.003479
Tonouchi, A., and Tazawa, D. (2014). Roseomonas aceris sp. nov. isolated from a mono maple tree in the Shirakami Mountains in Japan. J. Gen. Appl. Microbiol. 60, 38–43. doi: 10.2323/jgam.60.38
Venkata Ramana, V., Sasikala, C. H., Takaichi, S., and Ramana, Ch. V. (2010). Roseomonas aestuarii sp. nov., a bacteriochlorophyll-a containing alphaproteobacterium isolated from an estuarine habitat of India. Syst. Appl. Microbiol. 33, 198–203. doi: 10.1016/j.syapm.2009.09.004
Wallace, P. L., Hollis, D. G., Weaver, R. E., and Moss, C. W. (1990). Biochemical and characterization of pink-pigmented oxidative bacteria. J. Clin. Microbiol. 28, 689–693.
Wang, C., Deng, S., Liu, X., Yao, L., Shi, C., Jiang, J., et al. (2016). Roseomonas eburnea sp. nov., isolated from activated sludge. Int. J. Syst. Evol. Microbiol. 66, 385–390. doi: 10.1099/ijsem.0.000728
Wang, C. M., Lai, C. C., Tan, C. K., Huang, Y. C., Chung, K. P., Lee, M. R., et al. (2012). Clinical characteristics of infections caused by Roseomonas species and antimicrobial susceptibilities of the isolates. Diagn. Microbiol. Infect. Dis. 72, 199–203. doi: 10.1016/j.diagmicrobio.2011.11.013
Wayne, L. G., Brenner, D. J., Colwell, R. R., Grimont, P. A. D., Kandler, O., Krichevsk, M. I., et al. (1984). International committee on systematic bacteriology. report of the ad hoc committee on reconciliation of approaches to bacterial systematics. Int J. Syst. Bacteriol. 37, 463–464.
Weyant, R. S., and Whitney, A. M. (2005). “Roseomonas,” in Bergey’s Manual of Systematic Bacteriology, part C, eds D. J. Brenner, N. R. Krieg, J. T. Staley, and G. M. Garrity (New York, NY: Springer), 88–92. doi: 10.1007/978-0-387-29298-4_1
Wirth, J. S., and Whitman, W. B. (2018). Phylogenomic analyses of a clade within the roseobacter group suggest taxonomic reassignments of species of the genera Aestuariivita, Citreicella, Loktanella, Nautella, Pelagibaca, Ruegeria, Thalassobius, Thiobacimonas and Tropicibacter, and the proposal of six novel genera. Int. J. Syst. Evol. Microbiol. 68, 2393–2411. doi: 10.1099/ijsem.0.002833
Wittouck, S., Wuyts, S., and Lebeer, S. (2019). Towards a genome-based reclassification of the genus Lactobacillus. Appl. Environ. Microbiol. 85:e02155-18. doi: 10.1128/AEM.02155-18
Yan, Z. F., Lin, P., Li, C. T., Kook, M., Wang, Q. J., and Yi, T. H. (2017). Roseomonas hibiscisoli sp. nov., isolated from the rhizosphere of Mugunghwa (Hibiscus syriacus). Int. J. Syst. Evol. Microbiol. 67, 2873–2878. doi: 10.1099/ijsem.0.002036
Yang, Z. W., Salam, N., Hua, Z. S., Liu, B. B., Han, M. X., Fang, B. Z., et al. (2017). Siccirubricoccus deserti gen. nov., sp. nov., a proteobacterium isolated from a desert sample. Int. J. Syst. Evol. Microbiol. 67, 4862–4867. doi: 10.1099/ijsem.0.002397
Yoo, S. H., Weon, H. Y., Noh, H. J., Hong, S. B., Lee, C. M., Kim, B. Y., et al. (2008). Roseomonas aerilata sp. nov., isolated from an air sample. Int. J. Syst. Evol. Microbiol. 58, 482–485. doi: 10.1099/ijs.0.65385-0
Yoon, J. H., Kang, S. J., Oh, H. W., and Oh, T. K. (2007). Roseomonas terrae sp. nov. Int. J. Syst. Evol. Microbiol. 57, 2485–2488. doi: 10.1099/ijs.0.65113-0
Yoon, S. H., Ha, S. M., Lim, J. M., Kwon, S. J., and Chun, J. (2017). A large-scale evaluation of algorithms to calculate average nucleotide identity. Antonie Van Leeuwenhoek 110, 1281–1286.
Zhang, J. Y., Jiang, X. B., Zhu, D., Wang, X. M., Du, Z. J., and Mu, D. S. (2020). Roseomonas bella sp. nov., isolated from lake sediment. Int. J. Syst. Evol. Microbiol. 70, 5473–5478. doi: 10.1099/ijsem.0.004436
Zhang, Y. Q., Yu, L. Y., Wang, D., Liu, H. Y., Sun, C. H., Jiang, W., et al. (2008). Roseomonas vinacea sp. nov., a Gram-negative coccobacillus isolated from a soil sample. Int. J. Syst. Evol. Microbiol. 58, 2070–2074. doi: 10.1099/ijs.0.65789-0
Keywords: phylotaxogenomics, average amino acid Identity (AAI), percentage of conserved proteins (POCP), reclassification, Roseomonas
Citation: Rai A, Jagadeeshwari U, Deepshikha G, Smita N, Sasikala C and Ramana CV (2021) Phylotaxogenomics for the Reappraisal of the Genus Roseomonas With the Creation of Six New Genera. Front. Microbiol. 12:677842. doi: 10.3389/fmicb.2021.677842
Received: 08 March 2021; Accepted: 08 June 2021;
Published: 13 August 2021.
Edited by:
Iain Sutcliffe, Northumbria University, United KingdomReviewed by:
Wen-Jun Li, Sun Yat-sen University, ChinaCopyright © 2021 Rai, Jagadeeshwari, Deepshikha, Smita, Sasikala and Ramana. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chintalapati Sasikala, c2FzaTQ0OUB5YWhvby5pZQ==; c2FzaWthbGEuY2hAZ21haWwuY29t; Chintalapati Venkata Ramana, Y3ZyNDQ5QGdtYWlsLmNvbQ==
†These authors have contributed equally to this work and share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.