
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 30 June 2016
Sec. Food Microbiology
Volume 7 - 2016 | https://doi.org/10.3389/fmicb.2016.01018
This article is part of the Research Topic Microbiota of grapes: positive and negative role on wine quality View all 20 articles
 Angela Capece1
Angela Capece1 Lisa Granchi2*
Lisa Granchi2* Simona Guerrini2
Simona Guerrini2 Silvia Mangani2
Silvia Mangani2 Rossana Romaniello1
Rossana Romaniello1 Massimo Vincenzini2
Massimo Vincenzini2 Patrizia Romano1
Patrizia Romano1Numerous studies, based on different molecular techniques analyzing DNA polymorphism, have provided evidence that indigenous Saccharomyces cerevisiae populations display biogeographic patterns. Since the differentiated populations of S. cerevisiae seem to be responsible for the regional identity of wine, the aim of this work was to assess a possible relationship between the diversity and the geographical origin of indigenous S. cerevisiae isolates from two different Italian wine-producing regions (Tuscany and Basilicata). For this purpose, sixty-three isolates from Aglianico del Vulture grape must (main cultivar in the Basilicata region) and from Sangiovese grape must (main cultivar in the Tuscany region) were characterized genotypically, by mitochondrial DNA restriction analysis and MSP-PCR by using (GTG)5 primers, and phenotypically, by determining technological properties and metabolic compounds of oenological interest after alcoholic fermentation. All the S. cerevisiae isolates from each region were inoculated both in must obtained from Aglianico grape and in must obtained from Sangiovese grape to carry out fermentations at laboratory-scale. Numerical analysis of DNA patterns resulting from both molecular methods and principal component analysis of phenotypic data demonstrated a high diversity among the S. cerevisiae strains. Moreover, a correlation between genotypic and phenotypic groups and geographical origin of the strains was found, supporting the concept that there can be a microbial aspect to terroir. Therefore, exploring the diversity of indigenous S. cerevisiae strains can allow developing tailored strategies to select wine yeast strains better adapted to each viticultural area.
Traditionally, Saccharomyces cerevisiae is the predominant yeast species in spontaneous wine fermentations and thus it is the main responsible for the chemical and sensory properties of wines (Pretorius, 2000; Fleet, 2003; Romano et al., 2003; Cocolin et al., 2004; Camarasa et al., 2011). During the last decades, a large number of surveys, based on different molecular techniques analyzing DNA polymorphism, have demonstrated that this species is characterized by a high genetic diversity (Frezier and Dubourdieu, 1992; Querol et al., 1994; Guillamón et al., 1996; Sabate et al., 1998; Pramateftaki et al., 2000; Torija et al., 2001; Schuller et al., 2005; Agnolucci et al., 2007; Romano et al., 2008; Sun et al., 2009; Csoma et al., 2010; Mercado et al., 2011; Capece et al., 2013). In spite of the occurrence of a high number of different S. cerevisiae strains at the beginning of the fermentation, it was pointed out that, usually, only few strains (from one to three) dominate the process in the latter stages. Some S. cerevisiae strains were isolated over several years in the same cellar as predominant microbiota in wine fermentations (Sabate et al., 1998; Gutièrrez et al., 1999; Augruso et al., 2006) so that the existence of a “winery effect” was suggested (Vezinhet et al., 1992). Alternatively, specific S. cerevisiae strains were widespread in different cellars of the same wine-producing region (Versavaud et al., 1995; Blanco et al., 2006) and they were considered representative of an oenological area (Guillamón et al., 1996; Torija et al., 2001). More recently, biogeographical characterization of S. cerevisiae wine yeasts carried out at scales above 100 km, has revealed the presence of regional population with specific genotype but no differentiation within the region (Knight and Goddard, 2015). These findings suggest that specific native strains could be associated with a terroir, a term that classically includes only grape variety, climate and soil as fundamental factors determining the typical nature of wines (Van Leeuwen and Seguin, 2006) and that might be revised including also a “microbial aspect” (Bokulich et al., 2014; Taylor et al., 2014). Since it is well established that chemical and sensory properties of some wines reflect their geographic origin (Villanova and Sieiro, 2006; Callejon et al., 2010) it was interesting to determine whether regionally defined S. cerevisiae genotypes actually exhibit specific metabolic profiles (or phenotypes) able to modulate the wine quality, thus contributing to terroir-associated wine characteristics. Indeed, in a recent study Knight et al. (2015) demonstrated significant correlation between the region of isolation of S. cerevisiae and aroma profile in wines. The evidence that certain regions have “signature” S. cerevisiae populations that can produce significantly different chemical and sensory profiles of wine is of relevance to the wine industry because it may link territory, environment, and final products for wine valorisation (Torija et al., 2001; Romano et al., 2003; Aa et al., 2006; Camarasa et al., 2011; Pretorius et al., 2012; Tofalo et al., 2013). For this reason, the demand of indigenous S. cerevisiae, which could be representative of a specific oenological area, is increasing (Orlić et al., 2010). In fact, each strain of S. cerevisiae is able to produce different types and quantities of secondary compounds, which are determinant on the desirable organoleptic characteristics of a wine (Pretorius, 2000; Romano et al., 2003; Barrajón et al., 2011; Scacco et al., 2012). Since to perform a better control of the alcoholic fermentation in the modern winemaking the use of yeast starter cultures is diffused, selecting the proper yeast strain can be critical for the development of the desired wine style. Moreover, by using these selected yeast starter cultures, that are better adapted to the environmental conditions, the must fermentation can occur in the correct way (Callejon et al., 2010). In this perspective, the goal of this study was to investigate a possible relationship between the diversity and the geographical origin of indigenous S. cerevisiae isolated from two different Italian wine-producing regions (Tuscany and Basilicata) considering two regional grape varieties usually used to produce Controlled Designation of Origin (DOC) wines. Such studies are of great interest in order to establish the existence of typical S. cerevisiae strains that would then be useful as inocula in the vinifications carried out in the specific oenological areas (Gutièrrez et al., 1999). The use of autochthonous yeast strains, besides assuring the maintenance of the typical sensory properties of the wines of any given region, can contribute to promote or retain the natural S. cerevisiae biodiversity.
Sixty-three Saccharomyces cerevisiae isolates were used. The yeasts were previously isolated from spontaneously fermented grape musts of two varieties: “Aglianico del Vulture,” Basilicata region (coded with R1-R33) and “Sangiovese,” Tuscany region (coded with R34-R63). The isolates were maintained on YPD medium [1% (w/v) yeast extract, 2% (w/v) peptone, 2% (w/v) glucose, 2% (w/v) agar].
Differentiation between the 63 indigenous S. cerevisiae isolates was performed by two molecular methods: microsatellite-primed PCR (MSP-PCR) by using the synthetic oligonucleotide (GTG)5 (Orlić et al., 2010) and mitochondrial DNA restriction analysis (mtDNA-RFLP) by using the restriction endonucleases RsaI according to Granchi et al. (2003). Genomic DNA was extracted using a synthetic resin (Instagene Bio-Rad Matrix), following the protocol described in Capece et al. (2011). Amplification reactions were performed in a final volume of 50 μL containing 10 μL 5X Buffer (Promega), 4.0 μL of 25 mM MgCl2 (Promega), 1 μL of 10mM dNTP (Promega), 5 μL of 5 μM primer, 0.25 μL (5 U/μL) of Taq DNA polymerase (Promega) and 5 μL of the extracted DNA, by adding sterile water until final volume. The thermal cycler was programmed as follows: initial denaturation at 95°C for 5 min, 35 cycles at 94°C for 1 min for denaturing, 1 min at 52°C, 2 min at 72°C for extension and a final step at 72°C for 5 min. PCR products were analyzed by electrophoresis in 1.2% (w/v) agarose gel. The obtained profiles were submitted to cluster analysis using “Complete Linkage” method with Pearson distance by FPQuest software v.4.5 (Bio-Rad).
DNA digestions were performed with the enzyme RsaI and restriction DNA fragments were separated on 0.8% (w/v) agarose gels containing ethidium bromide (1 μg mL-1) by electrophoresis in 1X⋅TBE buffer (90 mM Tris-borate, 2 mM, EDTA pH 8.0) at 4 V cm-1 for 6 h. The obtained patterns were submitted to pairwise comparison with the Dice coefficient (SD) (Sneat and Sokal, 1973) and cluster analysis with unweighted pair group method (UPGMA) by GelCompar 4.0 software (Applied Math, Kortrijk, Belgium).
The 63 isolates were submitted to screening for some phenotypic properties, such as sulfur dioxide, ethanol and copper resistance and fermentative performance. The SO2 and ethanol resistance was evaluated on agarized grape must (pH 3.6), added with increasing doses of K2S2O5 (100–300 mg L-1) and ethanol (10–18% vol/vol), respectively. Copper resistance was evaluated on agarized synthetic medium, containing 6.7 g L-1 YNB (Yeast Nitrogen Base without amino acids and sulfate), 20 g L-1 glucose, added with increasing doses of CuSO4 (50, 100, 200, 300, 400, and 500 μmolL-1). The strain resistance to the three compounds was evaluated on the basis of positive growth after incubation at 26°C for 24 h, in comparison to the control (the medium without the compound). The degree of resistance of each strain was reported as minimal dose of compounds allowing the growth. All the tests were carried out in duplicate.
The fermentative performance of the 63 S. cerevisiae isolates was tested in inoculated fermentations in two different grape musts, “Aglianico del Vulture” and “Sangiovese” possessing, respectively, the following physico-chemicals characteristics: pH: 3.7 and 3.2; sugars (g L-1): 227 and 214; yeast assimilable nitrogen (mgL-1): (130 ± 1.4) and (120 ± 2.5). The fermentations were performed according to Capece et al. (2012): 130-mL Erlenmeyer flasks were filled with 100 mL of the two grape musts and added with 50 mg L-1 of SO2. Each strain was inoculated in grape must at a concentration of 106 cells mL-1, from a pre-culture grown for 48 h in the same must. The fermentation was performed at 26°C and the fermentative course was monitored by measuring weight loss, determined by carbon dioxide evolution during the process. At the end of the process, indicated by constant weight of the samples, the wine samples were refrigerated at 4°C to clarify the wine, racked and stored at -20°C until required for analysis. All the experiments were performed in duplicate. Fermentation vigor was measured as weight loss after 2 days of incubation at 26°C, whereas the fermentative power was defined as total weight loss, detected at the end of the process.
In grape musts, α-amino acid and ammonium concentrations were determined by the NOPA procedure (Dukes and Butzke, 1998) and enzymatic assay according to the manufacturer’s instructions (STEROGLASS s.r.l., Perugia), respectively. Glucose, fructose, ethanol, glycerol, 2,3-butanediol and acetic acid concentrations in experimental wines were determined by HPLC, according to Schneider et al. (1987) and Granchi et al. (1998), utilizing a MetaCarb H Plus Column (8 μm particle, 300 × 7.8 mm; Varian Inc.) and a Pro-star 210 chromatograph equipped with a Refractive Index Detector, in series (Varian Inc.). Higher alcohols (1-propanol, isobutanol, n-butanol, 2-methyl-1-butanol, 3-methyl-1-butanol), acetoin, diacetyl, acetaldehyde and ethyl acetate were analyzed by gas chromatography equipped with glass column (6.6% CW 20M BA 80/120 225, 2 m × 6 × 2 mm) as described by Romano et al. (1999).
The raw data obtained by HPLC and GC analysis were subjected to Principal Component Analysis (PCA) and t-test by Statistica software (version 7, StatSoft, Tulsa, OK, USA).
Genetic polymorphism in S. cerevisiae isolates from Aglianico del Vulture and Sangiovese grape musts was evaluated by MSP-PCR analysis with the (GTG)5 primer and mt-DNA-RFLP. MSP-PCR profiles contained a variable number of bands, some of which were common to numerous isolates. The dendrogram resulting from MSP-PCR analysis (Figure 1), by considering a similarity coefficient of about 65%, revealed the presence of two clusters (A and B). For the majority of isolates, the distribution was related to yeast origin; in fact, the cluster A grouped isolates from Sangiovese grape must (except R28 and R29 strains), whereas the cluster B grouped all the isolates from Aglianico del Vulture grape must, except the Bs sub-cluster, which contains 9 S. cerevisiae isolates from Sangiovese grape must.
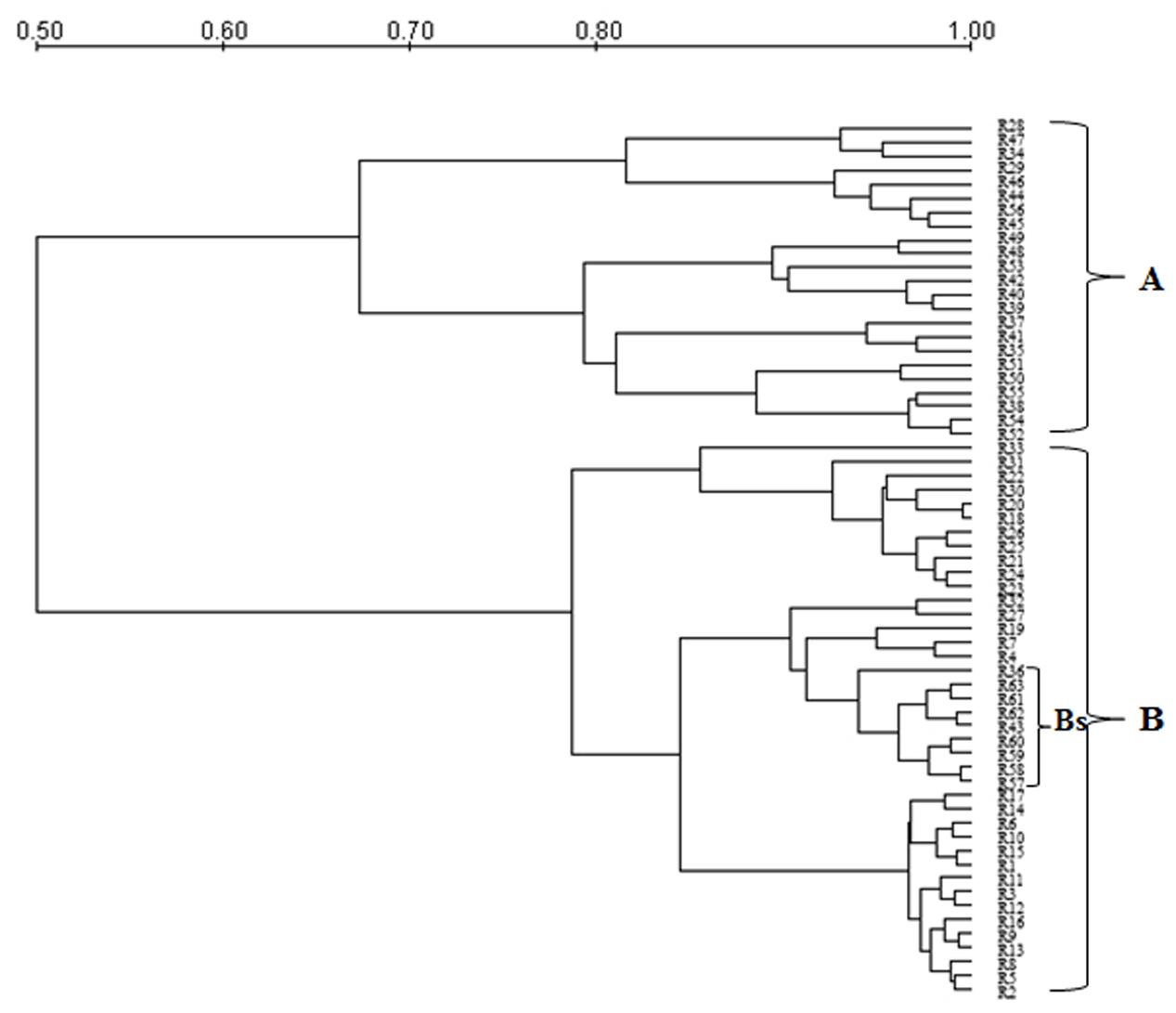
FIGURE 1. Dendrogram from UPGMA clustering analysis, based on Pearson coefficient, of the profiles obtained by MSP-PCR of the S. cerevisiae isolates from the Aglianico del Vulture (cluster B) and Sangiovese (cluster A) grape must. Bs indicates a sub-cluster including 9 S. cerevisiae isolates from Sangiovese.
The mitochondrial DNA-RFLP analysis revealed the presence of 24 different patterns, i.e., 24 strains, among the 63 isolates analyzed, confirming the high polymorphism found in S. cerevisiae populations. The resulting dendrogram from UPGMA analysis of the patterns obtained with RsaI, reported in Figure 2, indicated that S. cerevisiae isolates at 40% similarity grouped into four clusters, I, II, III, and IV. All the isolates from Aglianico del Vulture grape must, except for R6, were included in the clusters I, II, and III while all the isolates from Sangiovese grape must, except for R58, were comprised in the cluster IV. Therefore, the analysis pointed out a possible grouping of the assayed S. cerevisiae isolates according to their geographic origin.
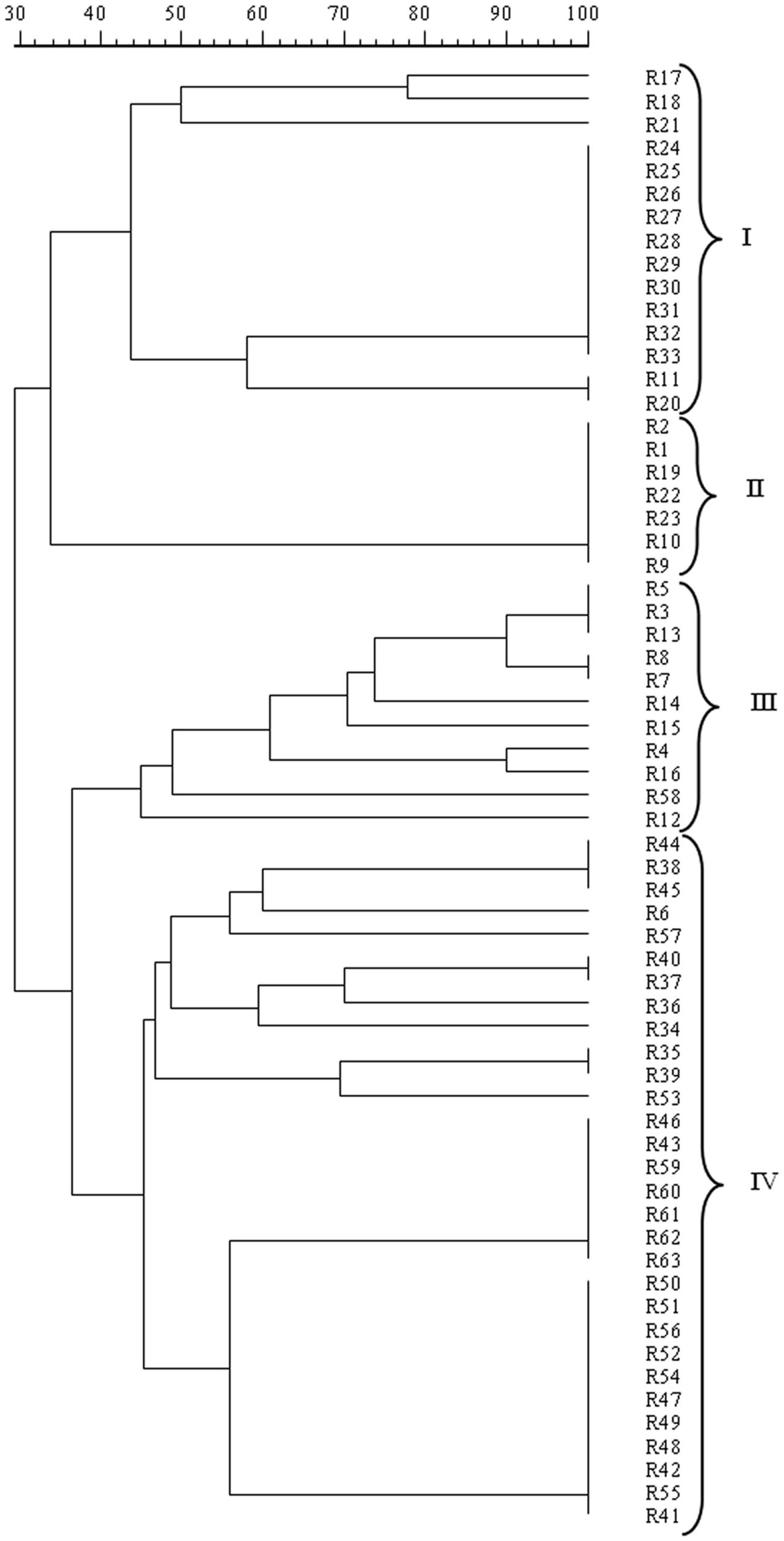
FIGURE 2. Dendrogram from UPGMA clustering analysis, based on Dice coefficient of mtDNA RsaI restriction patterns of the S. cerevisiae isolates from Aglianico del Vulture (clusters I, II, and III) and Sangiovese (cluster IV) grape must.
As regards the evaluation of technological characters, the isolates exhibited a high tolerance to ethanol; almost all isolates tolerated 16% v/v of this compound (only few strains developed until 18% v/v of ethanol), whereas a certain variability was found for sulfur dioxide (Figure 3A) and copper (Figure 3B) resistance. Although the most of isolates exhibited a low tolerance to sulfur dioxide (100 mg L-1of SO2), all the yeasts developing to the highest tested doses of SO2 were isolated from “Sangiovese” (except one). As regards copper tolerance, the isolates were distributed in different classes of resistance. A high number of Sangiovese isolates tolerated 200 μM CuSO4, whereas the majority of “Aglianico” isolates developed between 200 and 400 μM CuSO4. However, also for copper tolerance, the yeasts growing at the highest copper doses (500 μM CuSO4) were mainly Sangiovese isolates.
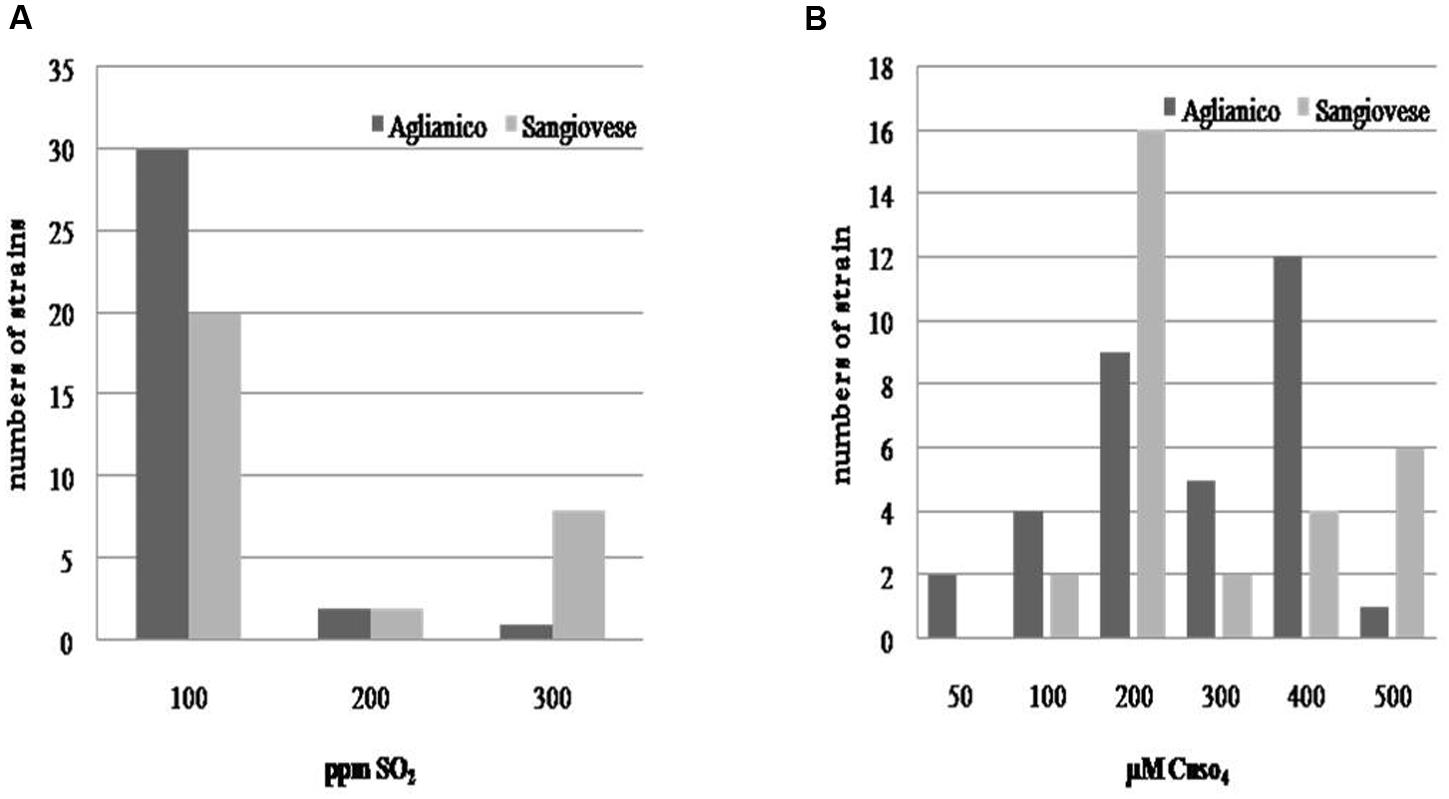
FIGURE 3. Distribution of S. cerevisiae according to their resistance to sulfur dioxide (A) and copper (B).
The wild strains were tested in inoculated fermentations at laboratory scale. Two different red musts, “Aglianico” (the isolation grape must of the R1-R33 isolates) and “Sangiovese” (the isolation grape must of the R34-R63 isolates), were used to evaluate strain fermentative performance. The data related to fermentative vigor of Aglianico isolates (Figure 4A) indicate that these isolates have shown a different vigor in the two musts; in particular, the isolates exhibited a lower vigor in Sangiovese must (mean value 2.76 g CO2/100 mL) than in Aglianico (mean value 3.74 g CO2/100 mL). Furthermore, these isolates showed higher variability in Aglianico than in Sangiovese must and the highest fermentative vigor was found in Aglianico isolates fermenting the same isolation grape must (8.24 g CO2/100 mL, Figure 4A). Also Sangiovese isolates (Figure 4B) showed different fermentative vigor in the two grape musts. The results obtained for Sangiovese isolates were similar to data found in Aglianico yeasts, with highest vigor and variability in Aglianico fermentation, although the maximum fermentative vigor of Sangiovese isolates (5.48 g CO2/100 mL, Figure 4B) was lower than maximum value showed by Aglianico isolates.
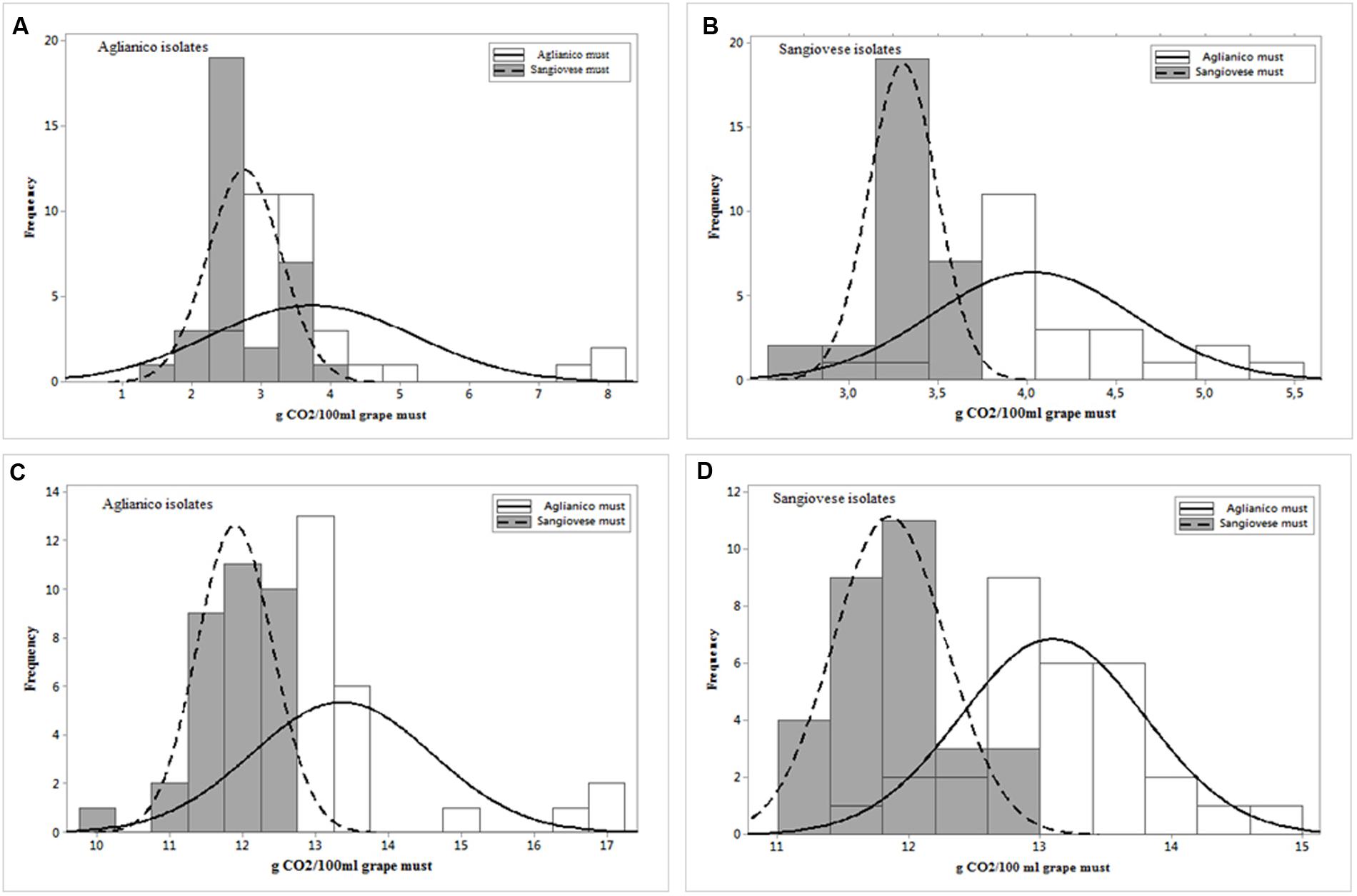
FIGURE 4. Distribution of S. cerevisiae isolated from Aglianico del Vulture and Sangiovese according to fermentative vigor (g CO2/100 mL after 2 days of fermentation) (A,B, respectively) and according to fermentative power (g CO2/100 mL at the end of fermentation) (C,D, respectively) in Aglianico del Vulture and Sangiovese grape must.
The data related to fermentative power, showed in Figures 4C,D, confirmed the results obtained for fermentative vigor, with different fermentative behavior in the two musts. As already found for fermentative vigor, Aglianico isolates (Figure 4C) showed higher fermentative power than Sangiovese isolates (Figure 4D), and both isolates groups showed the best performance in Aglianico grape must, with the highest value of 17.23 g CO2/100mL for Aglianico isolates (Figure 4C) and 14.8 g CO2/100 mL for Sangiovese isolates (Figure 4D). Furthermore, the isolates exhibited higher variability for this parameter in Aglianico than in Sangiovese must (values ranging between 11.76–17.23 and 9.96–12.7 g CO2/100 mL, respectively). Finally, the best value was exhibited in Aglianico must by strains isolated from this variety.
The content of 14 yeast metabolic compounds (ethanol, glycerol, acetic acid, acetaldehyde, 1-propanol, 3-methyl-isobutanol, n-butanol, 2-methyl-1-butanol, 3-methyl-1-butanol, diacetyl, acetoin, meso and racemic 2,3-butanediol, ethyl acetate) was determined in the experimental wines obtained at the end of the alcoholic fermentations of Aglianico and Sangiovese grape musts.
Statistical analysis of metabolites produced (Table 1) demonstrated that, independently of the grape must variety fermented (Aglianico or Sangiovese), some compounds showed significant differences which may be related to the different origin of the yeast strains carrying out the fermentative process. In particular, S. cerevisiae isolates from Aglianico produced significantly higher amounts of acetic acid, acetaldehyde, acetoin, 1-butanol and 2,3-butanediols, while S. cerevisiae isolates from Sangiovese yielded higher concentrations of 2-methyl-1-butanol and 3-methyl-1-butanol (Table 1). The metabolic profiles obtained confirm the wide phenotypic variability within S. cerevisiae species, but, in any case, wine composition, independently of the grape variety, was markedly characterized by metabolites of the fermenting yeast strain.
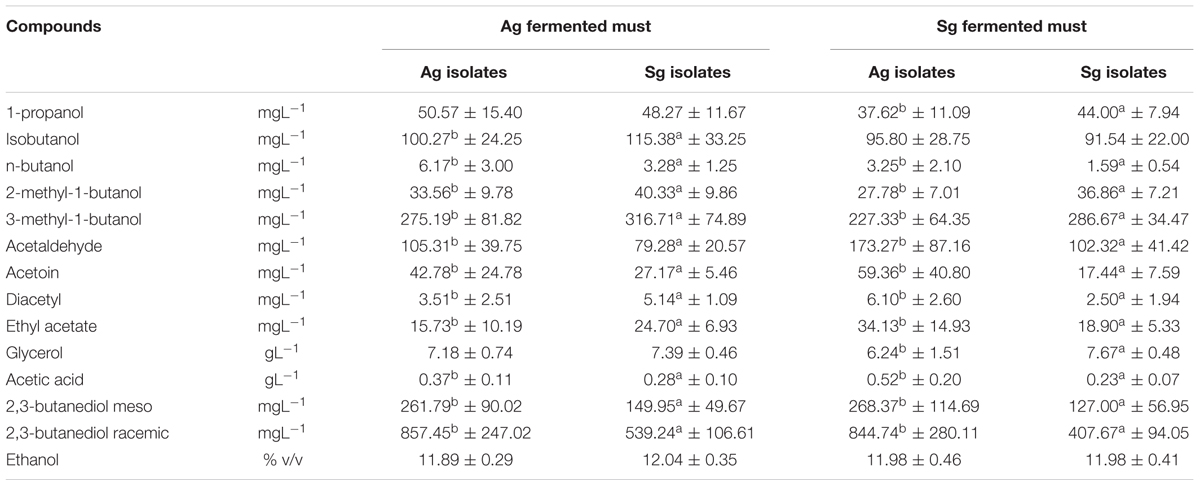
TABLE 1. Statistical analysis (t-test p < 0.05) of metabolites from fermentations of Aglianico del Vulture grape must (Ag fermented must) and Sangiovese grape must (Sg fermented must) carried out by S. cerevisiae isolates from Aglianico del Vulture (Ag) and Sangiovese (Sg), (Values as means ± SD) (different letters indicate significant differences among metabolites produced in the same must).
Principal component analysis (PCA) was applied to the matrix of multivariate data comprising concentrations of the metabolic compounds. Residual sugars resulted lower than 2 gL-1 in all experimental fermented wines, with the exception for five Sangiovese wines obtained with the strains R2, R4, R16, R22, and R23 originating from Aglianico del Vulture and two Aglianico wines produced by the strains R2 and R53 (originating from Aglianico and from Sangiovese must, respectively). Therefore, these seven wine samples were not included in the PCA analysis.
Figures 5A,B show PCA scores and loadings biplots, respectively, for all the experimental wines deriving from both grape musts fermentation by the 56 S. cerevisiae strains. Examination of the data by PCA showed that PC1 and PC2 accounted for 54% of variation in the dataset. Along the first component, independently of the fermented must, most of the wine samples grouped into two clusters according to the geographic origin of the fermenting yeast strains. In particular, 95% of wines produced by S. cerevisiae isolates from Sangiovese grouped in a cluster on the right of the plot, whereas 70% of wines obtained by S. cerevisiae isolates from Aglianico del Vulture were grouped into a more scattered cluster on the left of the plot. The first principal component correlated positively with 2-methyl-1-butanol, 3-methyl-1-butanol and glycerol and negatively with 2,3-butanediols.
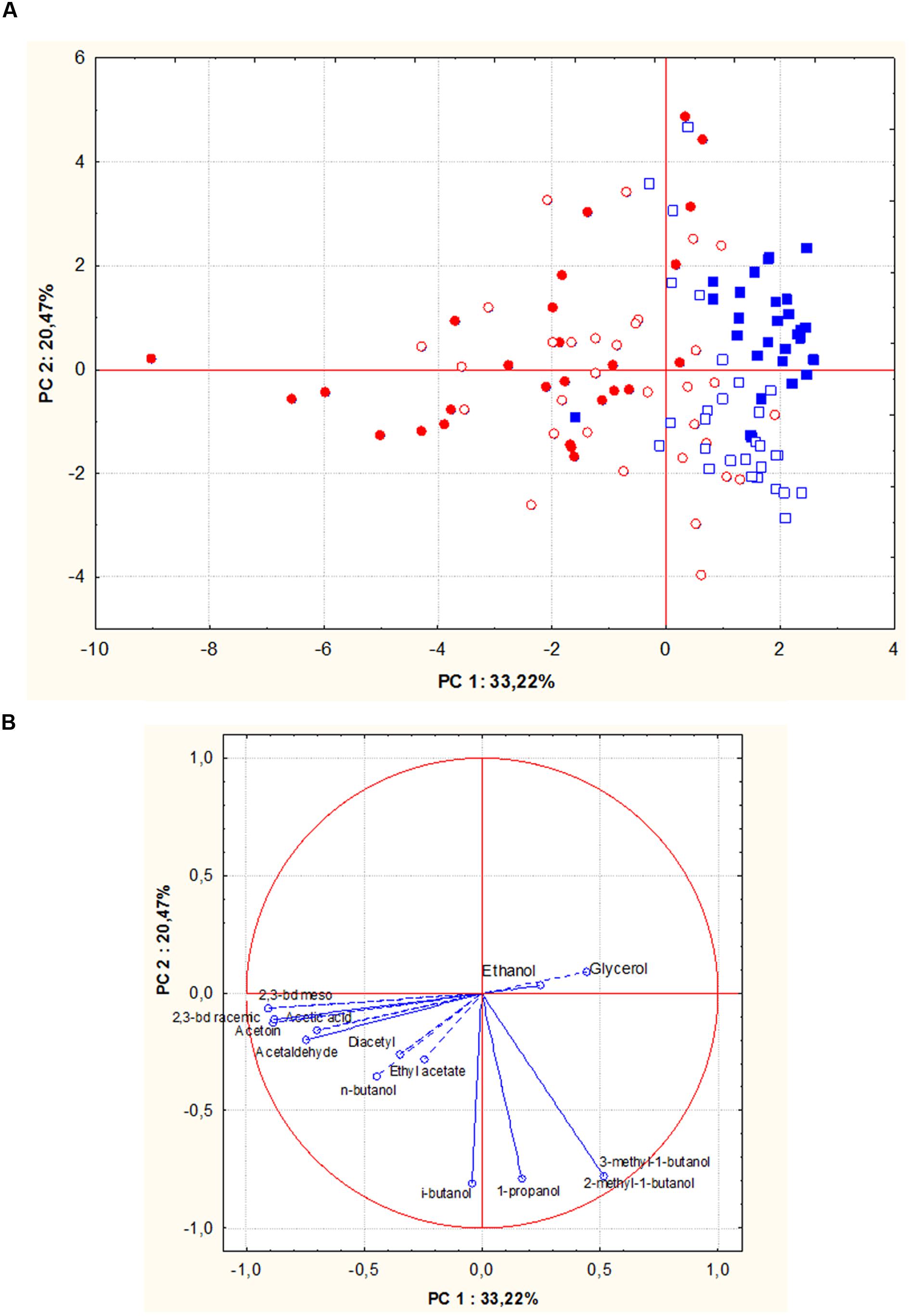
FIGURE 5. Principal component analysis (PCA) plot based on by-products of alcoholic fermentations in Aglianico del Vulture and Sangiovese wine samples produced by S. cerevisiae isolates from Aglianico del Vulture and Sangiovese grape must. (Blue square = wines obtained by isolates from Sangiovese: open symbols indicate Aglianico wines and close symbols indicate Sangiovese wines; red circle = wines obtained by isolates from Aglianico: open symbols indicate Aglianico wines and close symbols indicate Sangiovese wines); the scores (A) and variable loadings (B) for the two first principal components.
In addition, in order to evaluate whether S. cerevisiae isolates could group according to their geographic origin, all assayed oenological properties, including technological characters and by-products of alcoholic fermentations of both grape musts, were combined and analyzed by PCA. The biplot of the parameters considered, pointed out that along the first component all the Sangiovese isolates, except one, were positioned in the left quadrants while 88% of Aglianico isolates grouped on the right quadrants (Figures 6A,B). Therefore, a good relationship between S. cerevisiae isolates and their geographical origin was confirmed.
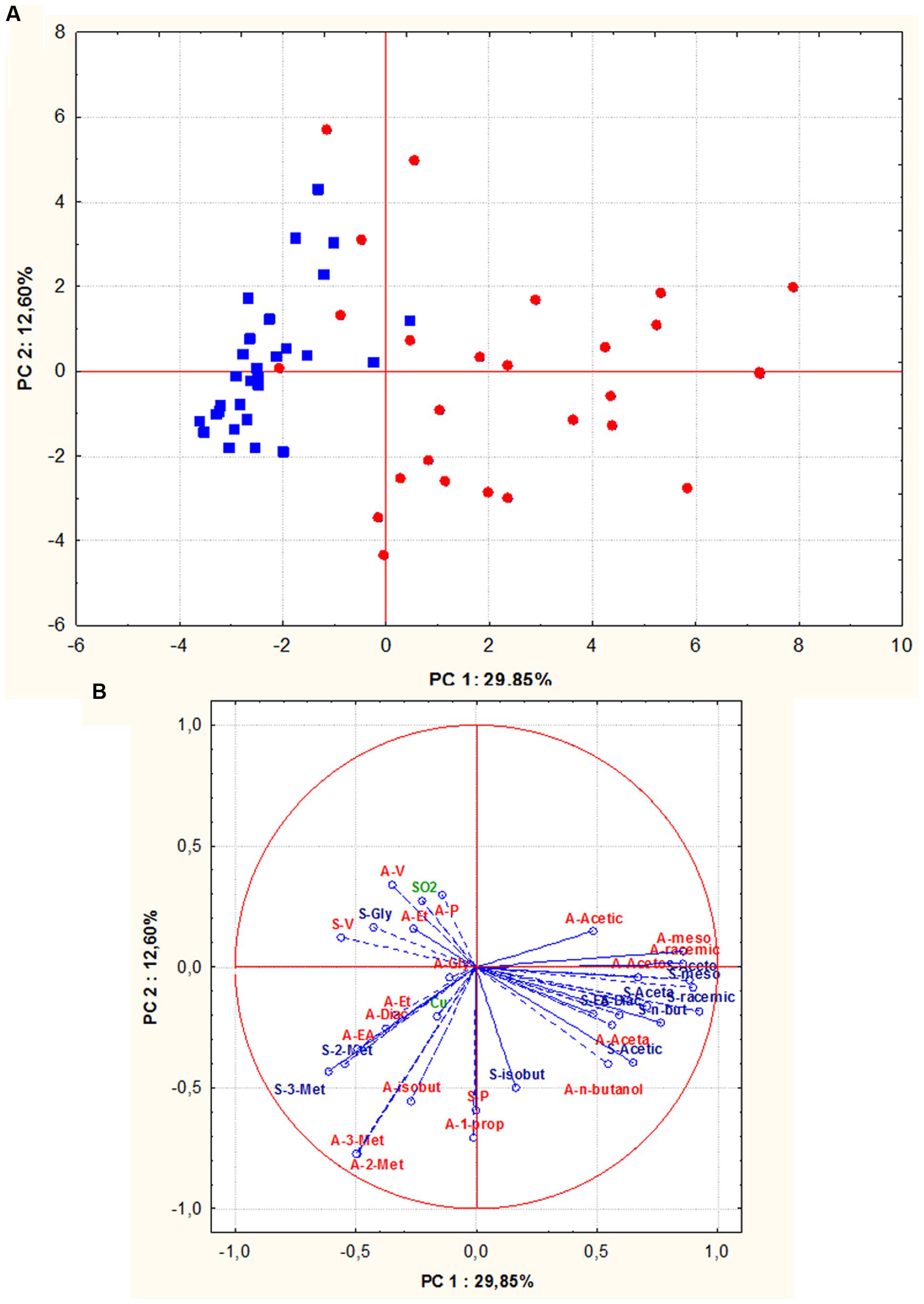
FIGURE 6. Principal component analysis (PCA) of oenological properties (resistance to SO2 and copper, fermentative vigor and power, by-products of alcoholic fermentation) of S. cerevisiae isolates (red circle = isolates from Aglianico del Vulture, blue square = isolates from Sangiovese), the scores (A) and variable loadings (B) for the two first principal components. Abbreviations for chemical measures in loadings are: red “A” refers to Aglianico del Vulture, blu “S” refers to Sangiovese, V, fermentative vigor; P, fermentative power; SO2, SO2 resistance; Cu, copper resistance; Et, ethanol; Acetic, acetic acid; meso, meso 2,3 butanediol; racemic, racemic 2,3 butanediol; Gly, glycerol; Aceta, acetaldehyde; Diac, diacetyl; Aceto, acetoin; EA, ethyl acetate; 3-met, 3- methyl 2 butanol; 2-met, 2- methyl 2 butanol; n-butanol, normal butanol; isobut, isobutanol; 1-prop, 1-propanol.
The concept of terroir for wine is classically considered as the result from the interaction between specific Vitis vinifera varieties and the local soils, geography, climate and agricultural practices (Van Leeuwen and Seguin, 2006). Recently, there is limited but increasing evidence showing that the microorganisms that influence vine growth, fermentation and wine style (as S. cerevisiae does) also exhibit regional differentiation (Lopandic et al., 2007; Gayevskiy and Goddard, 2012; Bokulich et al., 2014; Taylor et al., 2014; Knight and Goddard, 2015), supporting the concept that there could be a microbial aspect to terroir. In the present study, 63 S. cerevisiae isolates from two different grape musts (Aglianico del Vulture and Sangiovese) were characterized in order to assess the influence of geographic origin of these yeasts on their genetic and phenotypic patrimony. The results obtained by molecular fingerprinting using MSP-PCR with (GTG)5 and RFLP-mtDNA methods confirmed applicability and sensitivity of these methodologies for identification of different S. cerevisiae strains (Schuller et al., 2007; Orlić et al., 2010). Furthermore, these techniques were able to detect genetic differences between “Aglianico” and “Sangiovese” strains, resulting suitable methods to differentiate S. cerevisiae isolates based on their provenience as most of the isolates grouped according to their origin of isolation. The high genetic polymorphism found in the yeasts analyzed using the MSP-PCR and RFLP-mtDNA could be a result of a constant adaptation to the ecological conditions they are exposed to. Studies based on genetic and microbiological analyses suggest that in S. cerevisiae a significant part of the mechanisms affecting this genetic polymorphism occur during the vegetative phase of its growth cycle, where meiosis is a rare event (Puig et al., 2000; Perez-Ortin et al., 2002; Aa et al., 2006). That is to say, if yeasts reproduce clonally and they are constantly adapting to their particular environment, there must be a link between the genetic similarity of the strains and their geographic origin. It is well known that geographic or ecological isolation is one of the mechanisms involved in the process of speciation (Dobzhansky, 1951) as it creates a barrier for the genetic flux, so that strains coming from the same microenvironment will be more similar to each other than those from other geographic origin (Martínez et al., 2007). Our results, although based on the analysis of a small number of isolates, confirm that the wine production areas represent a reservoir of natural yeasts with peculiar genotypic profile, selected by the natural environment and by the interactions between yeasts and its environment (Guillamón et al., 1996; Martínez et al., 2007).
Further step of yeast characterization was addressed to identify potential relationships between strain origin and fermentation phenotype. Indeed, the yeast strain diversity might significantly affect the fermentation performance (Schuller et al., 2012; Tofalo et al., 2014). The data related to fermentative performance of the two groups of S. cerevisiae isolates, tested in the two isolation grape musts, seem to suggest that this parameter is influenced by both fermentation medium and source of yeast isolation. In fact, the isolates showed different fermentative performance in the two musts, with best results in Aglianico grape must, and Aglianico isolates exhibited higher fermentative performances than Sangiovese isolates (Figures 4A–D). As regards the fermentation substrate, the different yeast behavior could be correlated to the different composition of grape must. It has been underlined that wine fermentation conditions represent a combination of various stresses (osmotic, ethanol, acidic, nutrient limitation) that accentuate the metabolic differences between strains. Our results showed a certain influence of isolation origin (the grapes variety, in our case) on strain performance during the fermentation and might support the hypothesis that the autochthonous yeast strains are better adapted to the ecological and technological features of their own winegrowing area. These relationships between the origin and the phenotypes of some yeast strains could be due to physiological and metabolic adaptations in response to specific environmental conditions (Camarasa et al., 2011). Probably, “in the isolation grape must the strains are able to express their own better characteristics because they are better adapted to metabolize the precursors present in this grape must” (Capece et al., 2012). Indeed, in Sangiovese must five S. cerevisiae isolates from Aglianico were unable to complete the alcoholic fermentation. Furthermore, our data indicate significant correlations between the geographic relatedness of S. cerevisiae isolates and their effect on content of some compounds in the resulting wines in agreement with the results obtained by Knight et al. (2015). The PCA of experimental wines obtained by inoculating the S. cerevisiae isolates in Aglianico and Sangiovese grape musts (Figure 5) revealed that the samples were mainly grouped according to the geographic origin of the yeast strains. To our knowledge, few researches reporting the correlation between strain origin and fermentation phenotype are available until now. A study, aimed to analyze the variability of 36 S. cerevisiae strains, isolated from different grape varieties and from two very distant Italian zones (Mauriello et al., 2009), demonstrated that production of volatile aromatic compounds (VOC) allowed to differentiate the yeasts in function of isolation area. Indeed, S. cerevisiae isolated from Southern Italy grapes were able to produce more volatile compounds than those from Northern Italy. In a study performed on regionally genetically differentiated population of S. cerevisiae in New Zealand, Knight et al. (2015) demonstrated that these populations differentially affected wine phenotype. By evaluating the correlation between S. cerevisiae genetic distance and volatile chemical profile of wines, obtained by inoculating these strains, the authors found that these factors are correlated, confirming “the significant relationship existing between the genetic relatedness of natural S. cerevisiae sub-populations and their effect on resulting wine phenotypes” (Knight et al., 2015). However, these authors found that the chemicals responsible for the differences between regions are not consistently from any particular class. On the contrary, our results show that the production level of some compounds is correlated with yeast origin, independently from fermentation substrate. In particular, in both grape musts tested, S. cerevisiae from Aglianico produced significantly higher amounts of acetic acid, acetoin, acetaldehyde, n-butanol and 2,3-butanediols, while experimental wines produced by inoculating S. cerevisiae isolates from Sangiovese contained higher concentrations of 2-methyl-1-butanol and 3-methyl-1-butanol (Table 1). These results might be correlated to genotypic characteristics of yeasts. In fact, it is reported that the types and concentrations of metabolites produced by S. cerevisiae are significantly influenced by yeast genotype (Camarasa et al., 2011; Pretorius et al., 2012; Richter et al., 2013), while not exclusively genetically determined.
These findings support the need to unveil the indigenous S. cerevisiae population of specific areas and explore its natural biodiversity in order to produce valuable wines styles. The natural biodiversity of grape berries, grape juice, and winery environment, well correlated to each specific terroir, show a unique composition and represent great resources to winemaking. In fact, this work demonstrated that indigenous microorganisms are better adapted to the “chemical environment” of the grape must coming from a specific wine-producing area. The use of these strains as specific starter cultures can give distinct regional characteristics to wines. This suggests that safeguarding and exploiting natural biodiversity can allow the development of modern winemaking practices and the diversification of wine production, with tangible economic imperatives.
AC contributed to the design of the work, to the molecular characterization of yeasts, to the interpretation of data for the work, to draft the work and revising it; LG contributed to the design of the work, to the interpretation of data for the work, to draft the work and revising it; SG contributed to the molecular characterization of yeasts, to statistical analysis of data and to the interpretation of data for the work; SM contributed to chemical analysis of must and experimental wines, to statistical elaboration of data, RR contributed to the molecular characterization of yeasts, to the management of experimental fermentation, to the statistical elaboration of data; MV contributed to the design of the work, to the draft of the work and revising it, PR contributed to the design of the work, to the draft of the work and revising it, and ensured that that questions related to the accuracy or integrity of any part of the work were appropriately investigated and resolved.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Aa, E., Townsend, J. P., Adams, R. I., Nielsen, K. M., and Taylor, J. W. (2006). Population structure and gene evolution in Saccharomyces cerevisiae. FEMS Yeast Res. 6, 702–715. doi: 10.1111/j.1567-1364.2006.00059.x
Agnolucci, M., Scarano, S., Santoro, S., Sassano, C., Toffanin, A., and Nuti, M. (2007). Genetic and phenotypic diversity of autochthonous Saccharomyces spp. strains associated to natural fermentation of ‘Malvasia delle Lipari’. Lett. Appl. Microbiol. 45, 657–662. doi: 10.1111/j.1472-765X.2007.02244.x
Augruso, S., Granchi, L., and Vincenzini, M. (2006). Biodiversità intraspecifica di Saccharomyces cerevisiae e ambiente vitivinicolo. VQ 7, 38–45.
Barrajón, N., Capece, A., Arévalo-Villena, M., Briones, A., and Romano, P. (2011). Co-inoculation of different Saccharomyces cerevisiae strains and influence on volatile composition of wines. Food Microbiol. 28, 1080–1086. doi: 10.1016/j.fm.2011.02.016
Blanco, P., Ramilo, A., Cerdeira, M., and Orriols, I. (2006). Genetic diversity of wine Saccharomyces cerevisiae strains in an experimental winery from Galicia (NW Spain). Antonie Van Leeuwenhoek 89, 351–357. doi: 10.1007/s10482-005-9038-6
Bokulich, N. A., Thorngate, J. H., Richardson, P. M., and Mills, D. A. (2014). Microbial biogeography of wine grapes is conditioned by cultivar, vintage, and climate. Proc. Natl. Acad. Sci. U.S.A. 111, E139–E148. doi: 10.1073/pnas.1317377110
Callejon, R. M., Clavijo, A., Ortigueira, P., Troncoso, A. M., Paneque, P., and Morales, M. L. (2010). Volatile and sensory profile of organic red wines produced by different selected autochthonous and commercial Saccharomyces cerevisiae strains. Anal. Chem. Acta 660, 68–75. doi: 10.1016/j.aca.2009.09.040
Camarasa, C., Sanchez, I., Brial, P., Bigey, F., and Dequin, S. (2011). Phenotypic landscape of Saccharomyces cerevisiae during wine fermentation: evidence for origin-dependent metabolic traits. PLoS ONE 6:e25147. doi: 10.1371/journal.pone.0025147
Capece, A., Pietrafesa, R., and Romano, P. (2011). Experimental approach for target selection of wild wine yeasts from spontaneous fermentation of “Inzolia” grapes. World J. Microbiol. Biotechnol. 27, 2775–2783. doi: 10.1007/s11274-011-0753-z
Capece, A., Romaniello, R., Siesto, G., and Romano, P. (2012). Diversity of Saccharomyces cerevisiae yeasts associated to spontaneously fermenting grapes from an Italian “heroic vine-growing area.” Food Microbiol. 31, 159–166. doi: 10.1016/j.fm.2012.03.010
Capece, A., Siesto, G., Romaniello, R., Lagreca, V. M., Pietrafesa, R., Calabretti, A., et al. (2013). Assessment of competition in wine fermentation among wild Saccharomyces cerevisiae strains isolated from Sangiovese grapes in Tuscany region. LWT - Food Sci. Technol. 54, 485–492. doi: 10.1016/j.lwt.2013.07.001
Cocolin, L., Pepe, V., Comitini, F., Comi, G., and Ciani, M. (2004). Enological and genetic traits of Saccharomyces cerevisiae isolated from former and modern wineries. FEMS Yeast Res. 5, 237–245. doi: 10.1016/j.femsyr.2004.08.005
Csoma, H., Zakany, N., Capece, A., Romano, P., and Sipiczki, M. (2010). Biological diversity of Saccharomyces yeasts of spontaneously fermenting wines in four wine regions: comparative genotypic and phenotypic analysis. Int. J. Food Microbiol. 140, 239–248. doi: 10.1016/j.ijfoodmicro.2010.03.024
Dukes, B. C., and Butzke, C. E. (1998). Rapid determination of primary amino acids in grape juice using an o-phthaldialdehyde /N-acetyl-Lcysteine spectrophotometric assay. Am. J. Enol. Vitic. 49, 125–134.
Fleet, G. H. (2003). Yeast interactions and wine flavor. Int. J. Food Microbiol. 86, 11–22. doi: 10.1016/S0168-1605(03)00245-9
Frezier, V., and Dubourdieu, D. (1992). Ecology of yeast strain Saccharomyces cerevisiae during spontaneous fermentation in a Bordeaux winery. Am. J. Enol. Vitic. 53, 375–380.
Gayevskiy, V., and Goddard, M. R. (2012). Geographic delineations of yeast communities and populations associated with vines and wines in New Zealand. ISME J. 6, 1281–1290. doi: 10.1038/ismej.2011.195
Granchi, L., Ganucci, D., Messini, A., Rosellini, D., and Vincenzini, M. (1998). Dynamics of yeast populations during the early stages of natural fermentation for the production of Brunello di Montalcino wines. Food Technol. Biotechnol. 36, 313–318.
Granchi, L., Ganucci, D., Viti, C., Giovannetti, L., and Vincenzini, M. (2003). Saccharomyces cerevisiae biodiversity in spontaneous commercial fermentations of grape musts with adequate and inadequate assimilable-nitrogen content. Lett. Appl. Microbiol. 36, 54–58. doi: 10.1046/j.1472-765X.2003.01263.x
Guillamón, J. M., Barrio, E., and Querol, A. (1996). Characterization of Wine Yeast Strains of the Saccharomyces genus on the basis of molecular markers: relationships between genetic distance and geographic or ecological origin. Syst. Appl. Microbiol. 19, 122–132. doi: 10.1016/S0723-2020(96)80019-1
Gutièrrez, A. R., Santamarìa, R., Epifanio, S., Garijo, P., and Lopez, R. (1999). Ecology of spontaneous fermentation in one winery during 5 consecutive years. Lett. Appl. Microbiol. 29, 411–415. doi: 10.1046/j.1472-765X.1999.00657.x
Knight, S., and Goddard, M. R. (2015). Quantifying separation and similarity in a Saccharomyces cerevisiae metapopulation. ISME J. 9, 361–370. doi: 10.1038/ismej.2014.132
Knight, S., Klaere, S., Fedrizzi, B., and Goddard, M. R. (2015). Regional microbial signatures positively correlate with differential wine phenotypes: evidence for a microbial aspect to terroir. Sci. Rep. 5:14233. doi: 10.1038/srep14233
Lopandic, K., Gangl, H., Wallner, E., Tscheik, G., Leitner, G., Querol, A., et al. (2007). Genetically different wine yeasts isolated from Austrian vine-growing regions influence wine aroma differently and contain putative hybrids between Saccharomyces cerevisiae and Saccharomyces kudriavzevii. FEMS Yeast Res. 7, 953–965. doi: 10.1111/j.1567-1364.2007.00240.x
Martínez, C., Cosgaya, P., Vásquez, C., Gac, S., and Ganga, A. (2007). High degree of correlation between molecular polymorphism and geographic origin of wine yeast strains. J. Appl. Microbiol. 103, 2185–2195. doi: 10.1111/j.1365-2672.2007.03493.x
Mauriello, G., Capece, A., D’Auria, M., Garde-Cerdán, T., and Romano, P. (2009). SPME-GC method as a tool to differentiate VOC profiles in Saccharomyces cerevisiae wine yeasts. Food Microbiol. 26, 246–252. doi: 10.1016/j.fm.2009.01.003
Mercado, L., Sturm, M. E., Rojo, M. C., Ciklic, I., Martínez, C., and Combina, M. (2011). Biodiversity of Saccharomyces cerevisiae populations in Malbec vineyards from the “Zona Alta del Río Mendoza” region in Argentina. Int. J. Food Microbiol. 151, 319–326. doi: 10.1016/j.ijfoodmicro.2011.09.026
Orlić, S., Vojvoda, T., Babić, K. H., Arroyo-López, F. N., Jeromel, A., Kozina, B., et al. (2010). Diversity and oenological characterization of indigenous Saccharomyces cerevisiae associated with Zilavka grapes. World J. Microbiol. Biotechnol. 26, 1483–1489. doi: 10.1007/s11274-010-0323-9
Perez-Ortin, J. E., Querol, A., Puig, S., and Barrio, E. (2002). Molecular characterization of a chromosomal rearrangement involved in the adaptive evolution of yeast strains. Genome Res. 12, 1533–1539. doi: 10.1101/gr.436602
Pramateftaki, P. V., Lanaridis, P., and Typas, M. A. (2000). Molecular identification of wine yeasts at species or strain level: a case study with strains from two vine-growing areas of Greece. J. Appl. Microbiol. 89, 236–248. doi: 10.1046/j.1365-2672.2000.01102.x
Pretorius, I. S. (2000). Tailoring wine yeast for the new millennium: novel approaches to the ancient art of winemaking. Yeast 16, 675–729. doi: 10.1002/1097-0061(20000615)16:8<675::AID-YEA585>3.0.CO;2-B
Pretorius, I. S., Curtin, C. D., and Chambers, P. J. (2012). The winemaker’s bug. Winemakers Bug 3, 149–158. doi: 10.4161/bbug.19687
Puig, S., Querol, A., Barrio, E., and Pérez-Ortin, J. (2000). Mitotic recombination and genetic change in Saccharomyces cerevisiae during wine fermentation. Appl. Environ. Microbiol. 66, 2057–2061. doi: 10.1128/AEM.66.5.2057-2061.2000
Querol, A., Barrio, E., and Ramon, D. (1994). Population dynamics of wine yeast strains in natural fermentations. Int. J. Food Microbiol. 21, 315–323. doi: 10.1016/0168-1605(94)90061-2
Richter, C. L., Dunn, B., Sherlock, G., and Pugh, T. (2013). Comparative metabolic footprinting of a large number of commercial wine yeast strains in Chardonnay fermentations. FEMS Yeast Res. 13, 394–410. doi: 10.1111/1567-1364.12046
Romano, P., Capece, A., Serafino, V., Romaniello, R., and Poeta, C. (2008). Biodiversity of wild strains of Saccharomyces cerevisiae as tool to complement and optimize wine quality. World J. Microbiol. Biotechnol. 24, 1797–1802. doi: 10.1007/s11274-008-9672-z
Romano, P., Fiore, C., Paraggio, M., Caruso, M., and Capece, A. (2003). Function of yeast species and strains in wine flavour. Int. J. Food Microbiol. 86, 169–180. doi: 10.1016/S0168-1605(03)00290-3
Romano, P., Marchese, R., Laurita, C., Salzano, G., and Turbanti, L. (1999). Biotechnological suitability of Saccharomycodes ludwigii for fermented beverages. World J. Microbiol. Biotechnol. 15, 451–545. doi: 10.1023/A:1008948623024
Sabate, J., Cano, J., Querol, A., and Guillamon, J. M. (1998). Diversity of Saccharomyces strains in wine fermentations: analysis for two consecutive years. Lett. Appl. Microbiol. 26, 452–455. doi: 10.1046/j.1472-765X.1998.00369.x
Scacco, A., Oliva, D., Di Maio, S., Polizzotto, G., Genna, G., Tripodi, G., et al. (2012). Indigenous Saccharomyces cerevisiae strains and their influence on the quality of Cataratto, Inzolia and Grillo white wines. Food Res. Int. 46, 1–9. doi: 10.1016/j.foodres.2011.10.038
Schneider, A., Gerbi, V., and Redoglia, M. (1987). A rapid HPLC method for separation and determination of major organic acids in grape musts and wines. Am. J. Enol. Vitic. 38, 151–155.
Schuller, D., Alves, H., Dequin, S., and Casal, M. (2005). Ecological survey of Saccharomyces cerevisiae strains from vineyards in the Vinho Verde Region of Portugal. FEMS Microbiol. Ecol. 51, 167–177. doi: 10.1016/j.femsec.2004.08.003
Schuller, D., Cardoso, F., Sousa, S., Gomes, P., Gomes, A. C., Santos, M. A. S., et al. (2012). Genetic diversity and population structure of Saccharomyces cerevisiae strains isolated from different grape varieties and winemaking regions. PLoS ONE 7:e32507. doi: 10.1371/journal.pone.0032507
Schuller, D., Pereira, L., Alves, H., Cambon, B., Dequin, S., and Casal, M. (2007). Genetic characterization of commercial Saccharomyces cerevisiae isolates recovered from vineyard environments. Yeast 24, 625–636. doi: 10.1002/yea.1496
Sneat, P. H. A., and Sokal, R. R. (ed.). (1973). Numerical Taxonomy. The Principles and Practice of Numerical Classification. San Francisco, CA: W. H. Freeman and Company.
Sun, H., Ma, H., Hao, M., Pretorius, I. S., and Chen, S. (2009). Identification of yeast population dynamics of spontaneous fermentation in Beijing wine region, China. Ann. Microbiol. 59, 69–76. doi: 10.1007/BF03175601
Taylor, M. W., Tsai, P., Anfang, N., Ross, H. A., and Goddard, M. R. (2014). Pyrosequencing reveals regional differences in fruit-associated fungal communities. Environ. Microbiol. 16, 2848–2858. doi: 10.1111/1462-2920.12456
Tofalo, R., Perpetuini, G., Fasoli, G., Schirone, M., Corsetti, A., and Suzzi, G. (2014). Biodiversity study of wine yeasts belonging to the “terroir” of Montepulciano d’Abruzzo “Colline Teramane” revealed Saccharomyces cerevisiae strains exhibiting atypical and “unique 5.8S-ITS restriction patterns”. Food Microbiol. 39, 7–12. doi: 10.1016/j.fm.2013.10.001
Tofalo, R., Perpetuini, G., Schirone, M., Fasoli, G., Aguzzi, I., Corsetti, A., et al. (2013). Biogeographical characterization of Saccharomyces cerevisiae wine yeast by molecular methods. Front. Microbiol. 4:166. doi: 10.3389/fmicb.2013.00166
Torija, M., Rozès, N., Poblet, M., Guillamón, J. M., and Mas, A. (2001). Yeast population dynamics in spontaneous fermentations: comparison between two different wine-producing areas over a period of three years. Antonie van Leeuwenhoek 79, 345–352. doi: 10.1023/A:1012027718701
Van Leeuwen, C., and Seguin, G. (2006). The concept of terroir in viticulture. J. Wine Res. 17, 1–10. doi: 10.1080/09571260600633135
Versavaud, A., Courcoux, P., Roulland, C., Dulau, L., and Hallet, J. N. (1995). Genetic diversity and geographical distribution of wild Saccharomyces cerevisiae strains from the wine-producing area of Charentes, France. Appl. Environ. Microbiol. 61, 3521–3529.
Vezinhet, F., Hallet, J. N., Valade, M., and Poulard, A. (1992). Ecological survey of wine strains by molecular methods of identification. Am. J. Enol. Vitic. 43, 83–86.
Keywords: Saccharomyces cerevisiae, wine, terroir, Aglianico del Vulture, Sangiovese, yeast diversity, genotyping, fermentation products
Citation: Capece A, Granchi L, Guerrini S, Mangani S, Romaniello R, Vincenzini M and Romano P (2016) Diversity of Saccharomyces cerevisiae Strains Isolated from Two Italian Wine-Producing Regions. Front. Microbiol. 7:1018. doi: 10.3389/fmicb.2016.01018
Received: 29 February 2016; Accepted: 15 June 2016;
Published: 30 June 2016.
Edited by:
Sandra Torriani, Università degli Studi di Verona, ItalyReviewed by:
Jyoti Prakash Tamang, Sikkim University, IndiaCopyright © 2016 Capece, Granchi, Guerrini, Mangani, Romaniello, Vincenzini and Romano. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lisa Granchi, bGlzYS5ncmFuY2hpQHVuaWZpLml0
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.