
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 27 May 2015
Sec. Antimicrobials, Resistance and Chemotherapy
Volume 6 - 2015 | https://doi.org/10.3389/fmicb.2015.00483
 Alicia G. Beukers1,2
Alicia G. Beukers1,2 Rahat Zaheer2
Rahat Zaheer2 Shaun R. Cook2
Shaun R. Cook2 Kim Stanford3
Kim Stanford3 Alexandre V. Chaves1
Alexandre V. Chaves1 Michael P. Ward1
Michael P. Ward1 Tim A. McAllister2*
Tim A. McAllister2*Tylosin phosphate is a macrolide commonly administered to cattle in North America for the control of liver abscesses. This study investigated the effect of in-feed administration of tylosin phosphate to cattle at subtherapeutic levels and its subsequent withdrawal on macrolide resistance using enterococci as an indicator bacterium. Fecal samples were collected from steers that received no antibiotics and steers administered tylosin phosphate (11 ppm) in-feed for 197 days and withdrawn 28 days before slaughter. Enterococcus species isolated from fecal samples were identified through sequencing the groES-EL intergenic spacer region and subject to antimicrobial susceptibility testing, identification of resistance determinants and pulsed-field gel electrophoresis profiling. Tylosin increased (P < 0.05) the proportion of eryR and tylR enterococci within the population. Just prior to its removal, the proportion of eryR and tylR resistant enterococci began decreasing and continued to decrease after tylosin was withdrawn from the diet until there was no difference (P > 0.05) between treatments on d 225. This suggests that antibiotic withdrawal prior to slaughter contributes to a reduction in the proportion of macrolide resistant enterococci entering the food chain. Among the 504 enterococci isolates characterized, Enterococcus hirae was found to predominate (n = 431), followed by Enterococcus villorum (n = 32), Enterococcus faecium (n = 21), Enterococcus durans (n = 7), Enterococcus casseliflavus (n = 4), Enterococcus mundtii (n = 4), Enterococcus gallinarum (n = 3), Enterococcus faecalis (n = 1), and Enterococcus thailandicus (n = 1). The diversity of enterococci was greater in steers at arrival than at exit from the feedlot. Erythromycin resistant isolates harbored the erm(B) and/or msrC gene. Similar PFGE profiles of eryR E. hirae pre- and post-antibiotic treatment suggest that increased abundance of eryR enterococci after administration of tylosin phosphate reflects selection for strains that were already present within the gastrointestinal tract of cattle at arrival.
Subtherapeutic administration of antibiotics in livestock feed has come under increasing scrutiny due to concerns that such a practice increases the emergence of antibiotic resistant bacteria (Aarestrup, 1999). This concern is particularly relevant for bacteria that reside in livestock and are associated with clinical infections in humans.
Enterococci are commensal bacteria of the human and bovine gastrointestinal tract, but are also associated with nosocomial and community-acquired infections in humans (Poh et al., 2006; Franz et al., 2011). Enterococcus faecalis and Enterococcus faecium are the two species most frequently associated with enterococcal infections in humans, being responsible for as much as a third of the nosocomial infections worldwide (Werner et al., 2008). Whereas in cattle, Enterococcus hirae, a species not commonly associated with human infections is predominately isolated from bovine feces (Anderson et al., 2008; Jackson et al., 2010; Zaheer et al., 2013).
In North America, tylosin phosphate is commonly included in cattle feed for the control of liver abscesses (Page and Gautier, 2012). Previous research has shown therapeutic and subtherapeutic administrations of macrolides to cattle increases the proportion of erythromycin resistant enterococci in bovine feces (Jacob et al., 2008; Zaheer et al., 2013). In 2005, the WHO identified macrolides as critically important antimicrobials for which management strategies are urgently required to reduce the prevalence of bacterial resistance (Collignon et al., 2009). Macrolides are part of the MLSB (macrolide-lincosamide-stretogramin B) superfamily with each antibiotic having slight structural differences, but resistance to one member of the family can cross-select for resistance to other drugs in the family. Consequently, if the inclusion of tylosin in feed leads to tylosin resistant enterococci in cattle it may also select for enterococci that are resistant to other macrolides such as erythromycin, an antibiotic important for the treatment of bacterial infections in humans (Roberts, 2008; Desmolaize et al., 2011).
Enterococci resistant to macrolides commonly carry the resistance determinant erm(B), an rRNA methlyase that confers cross-resistance to MLSB antibiotics, or msrC, a macrolide efflux pump (Portillo et al., 2000). Very little is known about the nature and resistance characteristics of enterococci isolated from feedlot cattle. If E. hirae is consistently found as the predominant species in cattle feces, administering macrolides to cattle may not pose as a significant risk because this species is not commonly associated with human infections. Furthermore, antibiotics are often withdrawn prior to slaughter to reduce the risk of residues contaminating meat. In this study, we hypothesized that withdrawal of tylosin prior to slaughter would be an effective method of reducing the risk of resistant enterococci entering the food chain.
The objectives of this study were to determine the prevalence of macrolide resistant enterococci recovered from cattle continuously fed tylosin phosphate, and following its withdrawal. The recovered enterococci were characterized through species identification, antimicrobial susceptibility testing, identification of resistance determinants and pulsed-field gel electrophoresis (PFGE) profiling.
The enterococci isolates investigated in this study were a subset of those archived during a larger study. Full methodological details have been described previously (Alexander et al., 2008; Sharma et al., 2008) and are summarized briefly below.
British crossbred steers (150 ± 20 kg) were randomly assigned to 10 pens (10 steers per pen) at the Lethbridge Research Centre feedlot (Lethbridge, Alberta, Canada). Steers were obtained from a single ranch (Deseret Ranches, Raymond, Alberta, Canada) and received no antibiotics prior to the beginning of the experiment.
Five pens of cattle each were randomly assigned to one of two treatments: (i) control, no antibiotics (denoted CON); (ii) tylosin phosphate (Tylan®, Elanco Animal Health; treatment denoted T11) at 11 ppm in the diet. Tylosin was administered continuously for 197 days, starting on arrival at the feedlot and was withdrawn from the diet 28 days prior to slaughter (Figure 1). To avoid cross contamination between diets, tylosin was mixed with 5 kg of supplement and manually spread over the surface of the feed during the morning feeding. Steers were fed once daily to ensure that all feed allotted to each pen was consumed. Steers in CON and T11 treatments were housed in opposite sides of the feed alley to ensure that steers in different treatments did not have direct contact with one another. The animals involved in this study were cared for according to the guidelines set out by the Canadian Council on Animal Care (Canadian Council on Animal Care, 2003).
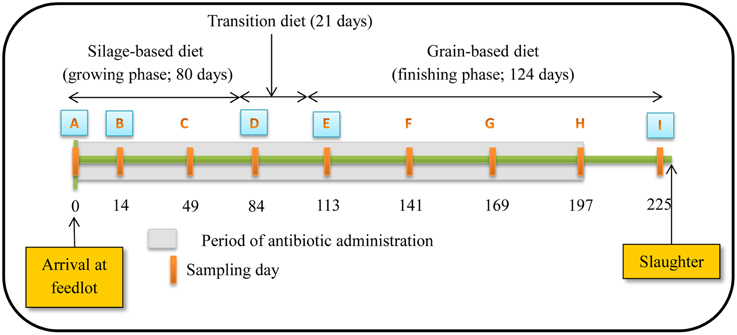
Figure 1. Schematic representation of experiment timeline (Figure reproduced from Sharma et al., 2008). Numbers indicate day of feeding period. Periodic orange rectangles indicate points where fecal samples were collected from steers. A, B, D, E and I represent points where isolates were selected for assessing antibiotic susceptibility, PFGE profiles and identifying resistance determinants. Grey shaded area represents the period that tylosin was administered in the diet.
Steers were fed diets typical of the western Canadian feedlot industry during a growing and finishing period. For the growing period, a silage-based diet consisting of 70% barley silage, 25% barley grain, and 5% supplement on a dry-matter (DM) basis was fed for the first 80 days (Figure 1). Cattle were transitioned from the silage-based growing diet to a grain-based finishing diet (85% barley grain, 10% barley silage, and 5% supplement on a DM basis) over 21 days and maintained on this diet for a further 124 days until slaughtered. A common watering bowl was shared between adjacent pens on the same treatment.
The study occurred from November 2004 to July 2005. Rectal fecal samples were collected from each steer upon arrival at the feedlot and monthly thereafter until slaughter (Figure 1). Proportion of steers positive for macrolide resistant enterococci, CFU counts and the proportion of macrolide resistant enterococci in steers were estimated at all 9 sampling dates with enterococci isolates from 5 of these dates used for assessing antimicrobial susceptibility, identifying resistance determinants and PFGE profiles. The five sampling dates were selected to include isolates prior to administration of tylosin, during the growing and finishing feeding periods and post-withdrawal of tylosin from the diet.
On each sampling date, fecal grab samples were collected and immediately transported to the lab within 1 h after collection. At the lab, fecal slurries were created by mixing feces (10 g) with 90 ml of 1 × phosphate-buffered saline in a stomacher bag (Fisher Scientific, Ottawa, Ontario, Canada) and using a Stomacher (2 min, 230 rpm, room temperature; Seward Ltd., Worthing, West Sussex, United Kingdom). Slurries were serially diluted 10-fold and 100 μL of the appropriate dilution plated in duplicate onto Bile-Esculin-Azide (BEA; BD, Franklin Lakes, New Jersey, USA) agar containing no antibiotics or onto BEA amended with erythromycin (8 μg/mL; BEAE), or tylosin (32 μg/mL; BEAT) to select for enterococci resistant to erythromycin or tylosin. The breakpoint for erythromycin was based on the Clinical and Laboratory Standards Institute (CLSI) guidelines whilst an arbitrary value, based on avoiding plate over growth and the levels used by Davies and Roberts (1999), was selected for tylosin. Plates were incubated for 48 h at 37°C and colonies from BEA, BEAE, and BEAT were enumerated. Two isolates from control plates and four isolates from antibiotic selective plates were streaked onto Trypticase soy agar (TSA; BD), incubated for 24 h, transferred to 20% glycerol in brain heart infusion broth (BD) and stored at −80°C until processed.
A total of 1029 presumptive enterococci isolates representing one isolate from each steer fecal sample were revived on the same media from which they were initially isolated (BEA, BEAE or BEAT; BD). Cultures were grown over 36 h at 37°C and two colonies were selected and suspended in 75 μL of TE (10 mM Tris, 1 mM EDTA, pH 8.0). Samples were heat lysed for 5 min using a thermomixer set at 98°C with shaking at 1000 RPM, followed by centrifugation at 10,000 × g for 5 min. The supernatant containing the genomic DNA was used as a source of template for all PCR reactions. Simultaneously, a subset of presumptive enterococci consisting of ~50% isolates of each category including treatment type, media type and sampling day were randomly selected for species identification. In this manner, 519 presumptive enterococci isolates were selected (Table 1). All of the 1029 isolates were screened by PCR with Enterococcus specific groES-EL primers Ent-ES-211-233-F and Ent-EL-74-95-R (Zaheer et al., 2012) for confirmation as Enterococcus spp. whereas the 519 selected isolates for species identification were further processed for sequencing of the groES-EL PCR product. Occasionally, the sequence results of the groES-EL PCR product varied from publically available databases. In order to characterize those Enterococcus spp. isolates correctly, multilocus sequencing including 16S rRNA, atpA, pheS, and rpoA genes was used to identify species. Detailed methodology can be found in the supplementary information (Supplementary Figure 1 and Supplementary Table 1). In cases where an isolate did not generate the groES-EL PCR product, i.e., was not an Enterococcus spp., PCR amplification and sequencing of the 16S rRNA gene using primers 27F (5′-AGAGTTTGATCMTGGCTCAG-3′) and 1492R (5′-TACGGYTACCTTGTTACGACTT-3′) was conducted for taxonomic identification.
A subset of 171 isolates representing major species (~25% coverage) and all minor species were subject to antimicrobial susceptibility testing. These selected isolates were subject to PCR-based identification of resistance determinants and PFGE profiling.
Disk susceptibility tests were conducted on 171 characterized enterococci isolates according to the CLSI documents M02-A11 and M100-S24 (Clinical and Laboratory Standards Institute, 2014a,b). The antimicrobials tested, suppliers and resistance breakpoints applied are listed in Table 2. Reference strains Staphylococcus aureus ATCC 25923® and E. faecalis ATCC 29212® were used as quality controls. Resulting zones of inhibition were read using the BioMic V3 imaging system (Giles Scientific, Inc., Santa Barbara, CA, USA) and classified as sensitive or resistant based on CLSI interpretive criteria (Clinical and Laboratory Standards Institute, 2014b), except for tigecycline which used EUCAST interpretive criteria (The European Committee on Antimicrobial Susceptibility Testing, (EUCAST), 2014). Neither EUCAST nor CLSI defined breakpoints exist for enterococci with tylosin, however the quality control range of tylosin disks (30 μg) has recently been acknowledged for S. aureus ATCC 25923® (Buß et al., 2014). Tylosin minimum inhibitory concentration (MIC) were established for a sub-set of isolates containing erm(B) or msrC, both genes or neither gene according to CLSI documents M100-S24 and M07-A9, with results reported in the supplementary information (Supplementary Figure 2). Isolates exhibiting a high MIC (≥128 μg/mL) to tylosin also contained the resistance determinant erm(B). Therefore, isolates harboring the resistance determinant erm(B) were given the designation of resistant to tylosin.
Of selected isolates, 125 isolates displaying intermediate or complete resistance to erythromycin were screened for the presence of macrolide resistance determinants. Isolates were first screened by PCR for the commonly found macrolide resistance determinants in enterococci, erm(B) and msrC (Portillo et al., 2000). For erm(B), PCR primers and reaction conditions were used as described by Chen et al. (2007). For msrC PCR, the forward and reverse primers, msrC_F1 (5′-TCGTTTTGTCATGAGACAAACAG-3′) and msrC_R1 (5′-AAATTAGTCGGTTCATCTAACAG-3′), respectively were used. A 20 μL PCR reaction using 2 μL of template DNA was prepared with the following reaction conditions: initial denaturation for 5 min at 95°C, followed by 35 cycles of denaturation for 30 s at 94°C, annealing for 30 s at 53°C, extension for 30 s at 72°C with a final extension for 10 min at 72°C. The PCR reaction product (5 μL) was resolved on a 2% agarose gel, and visualized for the presence of a 191 bp PCR product. An environmental sample, showing positive amplification for msrC and verified by DNA sequencing, was used as a positive control.
A subset of 40 isolates containing erm(B) or msrC or both genes and consisting of all identified species with a variety of PFGE profiles were further screened for the presence of other macrolide resistance determinants. These included erm(A), erm(C), erm(F), and erm(T) with primers and reaction conditions as described by Chen et al. (2007).
Isolates displaying intermediate or complete resistance to doxycycline were further screened for the tetracycline resistance determinants tet(B), tet(C), tet(L), and tet(M). A 20 μL PCR reaction using 2 μL of template DNA was prepared with products resolved on a 2% agarose gel. For tet(B), primers as described by Peak et al. (2007) were used with the following reaction conditions; initial denaturation for 5 min at 95°C, followed by 35 cycles of denaturation for 30 s at 94°C, annealing for 30 s at 60°C, extension for 30 s at 72°C, and a final extension for 10 min at 72°C. Primers and reaction conditions for tet(C), tet(L), and tet(M) were as described by Ng et al. (2001). The expected product size for tet(B), tet(C), tet(L), and tet(M) were 205, 418, 267, and 406 bp, respectively.
For all PCR reactions, the commercially available HotStarTaq Plus Master Mix Kit (Qiagen Canada, Inc., Mississauga, ON, Canada) was used according to manufacturer's instructions. Plasmids containing the corresponding gene fragments were used as positive controls (Alexander et al., 2009; Zaheer et al., 2013).
One-hundred and seventy-one isolates were subjected to PFGE profiling with SmaI restriction enzyme using a modified procedure of PulseNet USA (Center for Disease Control and Prevention, 2012). Briefly, bacteria grown overnight on brain-heart infusion-agar (BHI-agar; BD) were harvested using sterile swabs and suspended in TE buffer to an OD of 1.85 at 610 nm. An aliquot (400 μL) of cell suspension was transferred to a 1.5 mL microfuge tube containing 20 μL of lysozyme (50 mg/mL; Sigma-Aldrich, Co., St Louis, Mo, USA), gently mixed and incubated at 55°C for 45 min. An equal volume of 1.2% molten SeaKem Gold agarose (Lornza, Rockland, Maine, USA) in TE buffer was added and the mixture dispensed in duplicate into re-useable plug molds (Bio-Rad Laboratories, Hercules, CA, USA) and allowed to solidify at room temperature. Duplicate plugs were added to 2 mL microfuge tubes containing 1.8 mL cell lysis buffer [50 mM Tris; 50 mM EDTA; 1% sodium sarcosyl] and 9 μL of Proteinase K (20 mg/mL; Sigma-Aldrich, Co., St Louis, Mo, USA) and incubated for 2 h at 55°C with agitation (300 rpm). Plugs were washed twice in sterile, deionized H2O (1.8 mL) and three times in TE (1.8 mL) for 10 min each using a thermomixer set at 50°C and 300 rpm. Restriction digestion and electrophoresis conditions were as described by Zaheer et al. (2013). Gels were photographed using an AlphaImager gel documentation system (Alpha Innotech Corp., St. Leandro, CA, USA) and banding patterns analyzed with BioNumerics V6.6 software (Applied Maths Inc., Austin, TX, USA), using Dice coefficient and the unweighted pair group method (UPGMA). Optimization and band tolerance were both set at 1%. Salmonella serotype Braenderup digested with XbaI was included in each gel as a control reference and for normalization of band fragments.
Enumeration data were used to determine the proportion of steers positive for macrolide resistant enterococci and the proportion of macrolide resistant Enterococcus in the total population. For the purposes of enumeration, esculin hydrolyzing colonies observed on BEA, BEAE, and BEAT plates were assumed to be enterococci.
Data were analyzed using commercially available statistical analysis software (SAS Systems for Windows, version 9.3, SAS Institute Inc, Cary, NC, USA). Prior to analysis, enumeration data were normalized through a log transformation. When enumeration data for the antibiotic selective media exceeded that of the non-selective media for each sampling point, it was assumed that 100% of the population was resistant to the respective antibiotic. The MIXED procedure of SAS was used to assess CFU counts over time and the proportion of macrolide resistant enterococci in the total population. The CFU counts over time were analyzed with media type, day and media type × day in the model as fixed effects while for the proportion of macrolide resistant enterococci in the total population, day, treatment and day × treatment interaction were included in the model as fixed effects. For both analyses, day was included as a repeated measure. Results were considered significant when P < 0.05. For most sampling days, 50 samples were collected, but due to conflicts with other experiments in the feedlot facility, only 30 samples were collected on day 49, 141, 169, and 197.
Upon arrival at the feedlot, 28 and 24% (CON and T11, respectively) of the steers were positive for eryR enterococci, whilst 44 and 38% (CON and T11, respectively) were positive for tylR enterococci, even though steers did not previously receive antibiotics (Figure 2).
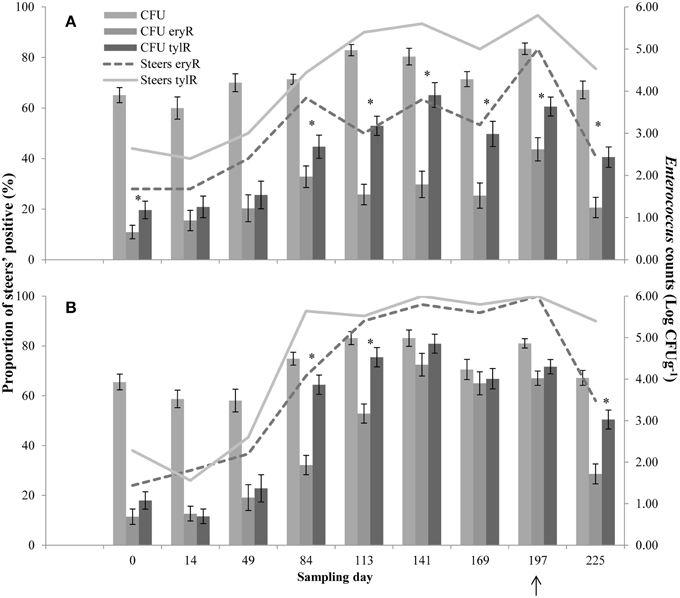
Figure 2. Proportion of steers positive for eryR enterococci (Steers eryR) or tylR enterococci (Steers tylR) and Enterococcus counts (log CFUg−1) of, total population (CFU), eryR enterococci (CFU eryR) or tylR enterococci (CFU tylR) for CON (A) or T11 (B) treatments. Arrow indicates when antibiotics were withdrawn from the diet. An “*” indicates days for which there was a significant difference between eryR and tylR Enterococcus counts (P < 0.05). For each treatment (day 0, 14, 84, 113, and 225 n = 50; day 49, 141, 169, and 197 n = 30).
For the control group, the counts of tylR enterococci were higher (P < 0.05) than the counts of eryR enterococci on d 0, 84, 113, 141, 169, 197, and 225 (Figure 2A). Whilst for the tylosin treatment, the counts of tylR enterococci were higher (P < 0.05) than the counts of eryR enterococci for d 84, 113, and 225 (Figure 2B). In general, the counts of eryR enterococci in the tylosin treatment group and counts of tylR enterococci in both treatment groups increased over the sampling period as the cattle were transitioned from a silage-based growing diet to a grain-based finishing diet (Figure 2). The increased counts of macrolide resistant enterococci over the experiment were due to an increase in the proportion of macrolide resistant enterococci within the total population.
No difference (P > 0.05) was observed between control and tylosin-fed steers on d 0, 14, 49, and 84 for the proportion of eryR enterococci or d 0, 14, and 49 for the proportion of tylR enterococci (Figures 3A,B, respectively). On d 113, 141, 169, and 197, the proportion of eryR enterococci was higher (P < 0.001) for steers fed tylosin compared to controls. The proportion of tylR enterococci, resistance was higher (P < 0.001) for steers fed tylosin compared to controls on d 84, 113, 141, 169, and 197. After withdrawal of tylosin on d 197, the proportion of eryR or tylR enterococci decreased until there was no difference (P > 0.05) between tylosin-fed and control steers on d 225 (Figures 3A,B, respectively).
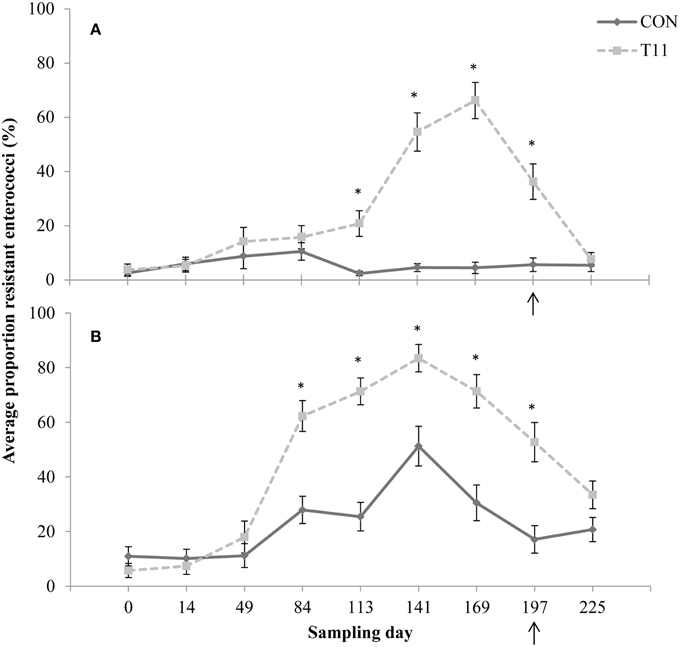
Figure 3. Proportion of erythromycin-resistant (A) or tylosin-resistant (B) fecal enterococci isolates for both treatments across all sampling days. Arrow indicates when antibiotics were withdrawn from the diet. Line styles distinguish the treatment. An “*” indicates days for which there was a significant difference between treatments (P < 0.05). For each treatment (day 0, 14, 84, 113, and 225 n = 50; day 49, 141, 169, and 197 n = 30).
Of the 1029 isolates analyzed, 95.2% were confirmed as enterococci by PCR. Of the 519 isolates speciated, 504 were identified as E. hirae (n = 431), Enterococcus villorum (n = 32), E. faecium (n = 21), Enterococcus durans (n = 7), Enterococcus casseliflavus (n = 4), Enterococcus mundtii (n = 4), Enterococcus gallinarum (n = 3), E. faecalis (n = 1), and Enterococcus thailandicus (n = 1). The remaining 15 non-enterococci were identified as Lactobacillus spp. (n = 3), Aerococcus spp. (n = 9), Streptococcus spp. (n = 2), and Staphylococcus epidermids (n = 1) as determined by 16S rRNA sequencing. All the species identified were represented by the 231 isolates originally recovered from BEA, whereas only six species (E. hirae, E. villorum, E. faecium, E. durans, E. casseliflavus, and E. gallinarum) were isolated from BEAE and BEAT (Figure 4). Variants of the groES-EL sequence for two isolates of E. faecium and single isolates of E. thailandicus and E. villorum have been submitted to the NCBI database (Accession numbers KP993544, KP993545, KP993546, and KP993547, respectively). The diversity of enterococci tended to be greater in steers upon arrival than at exit from the feedlot. A greater diversity of enterococci species were isolated from non-selective BEA compared with either BEAE or BEAT, with similar proportions of most species occurring in control and tylosin-fed steers. E. hirae was the predominant species isolated from both control and tylosin-fed steers across all sampling dates (Figure 4).
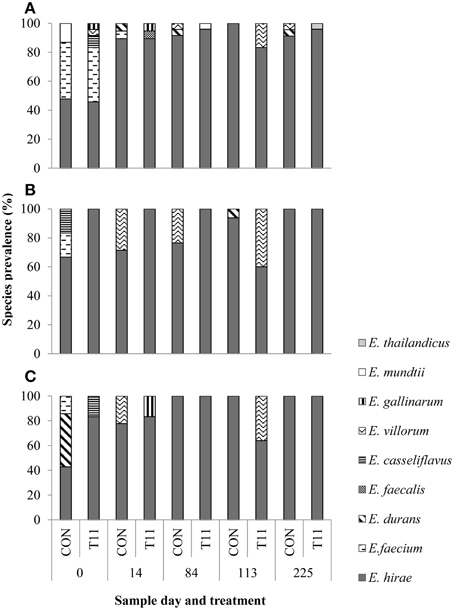
Figure 4. Species distribution of characterized isolates from (A) BEA (bile esculin azide agar), (B) BEAE (bile esculin azide agar amended with erythromycin [8 μg/mL]) and (C) BEAT (bile esculin azide agar amended with tylosin [32 μg/mL]). Prevalence was calculated by dividing the number of isolates for each species by the total number of isolates from each sample day and treatment.
A subset (n = 171) of enterococci representing all of the isolated Enterococcus species were tested for antibiotic susceptibility (Table 3). Resistance to ampicillin, gentamicin, linezolid, streptomycin or tigecycline was not detected in any of the isolates. Vancomycin resistance was also absent in all isolates except for one which displayed intermediate resistance. One isolate of E. casseliflavus exhibited ERY-TYL-Q-D-van resistance and one isolate of E. durans exhibited ERY-TYL-q-d (lower case denotes intermediate resistance and upper case complete resistance). One isolate of E. faecium was ERY-DOX-TYL-q-d resistant, with other single isolates exhibiting intermediate ery-nit, ery-lvx or dox-nit-lvx-q-d resistance. Two isolates of E. gallinarum showed ery-TYL resistance and a number of E. hirae isolates were resistant to ERY-TYL (n = 27), ery-TYL (n = 27), ERY-dox-TYL (n = 8), or ERY-TYL-q-d (n = 7). With one exception, all E. villorum isolates exhibited ERY-TYL (n = 31) resistance.
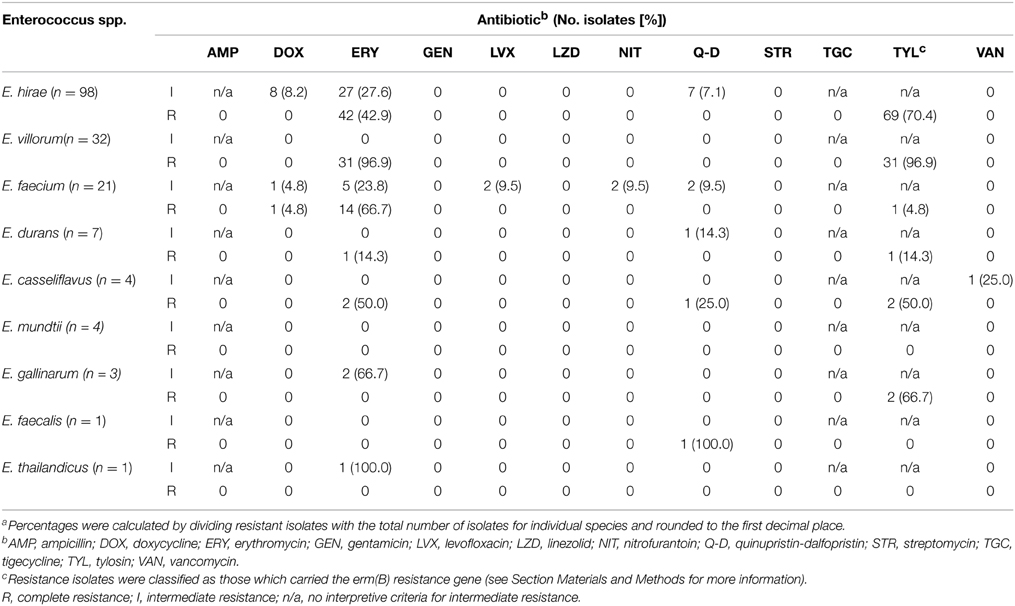
Table 3. Number of enterococci isolates (percentage of total speciesa) showing intermediate or complete resistance to antibiotics pooled across treatments, isolation media and sample date.
In general, isolates grown on BEAT also exhibited erythromycin resistance. An exception to this was three isolates of E. durans isolated on BEAT, which remained susceptible to erythromycin.
Of the 125 enterococci isolates displaying intermediate or complete resistance to erythromycin, the erm(B) gene was detected in 106 isolates representing E. hirae, E. durans, E. faecium, E. villorum, E. gallinarum, and E. casseliflavus. Of the 19 erythromycin-resistant E. faecium isolates obtained all except one lacked erm(B), but all were positive for msrC. The isolate identified as E. thailandicus displayed intermediate resistance to erythromycin, but was negative for all of the macrolide resistance determinants tested. None of the isolates tested positive for the other macrolide resistance determinants.
A total of 10 isolates displayed intermediate or complete resistance to doxycycline. None of the isolates were positive for tet(B) or tet(C). All 10 isolates were positive for tet(M) and 9 were positive for tet(L).
The PFGE profiles of E. faecium, E. villorum and erythromycin resistant E. hirae are displayed in Figures 5–7, respectively. E. faecium had at least 16 isolates from different steers with the same PFGE profile, suggesting the presence of a clonal population. Isolates from this clonal population were isolated only on day 0 (Figure 5). The similarity (>95%) of PFGE profiles of E. villorum also suggested clonality (Figure 6). Unlike E. faecium, these profiles appeared on day 14 of the trial and persisted until the end of the experiment. PFGE profiles of erythromycin resistant E. hirae produced 8 clusters with >85% similarity (Figure 7).
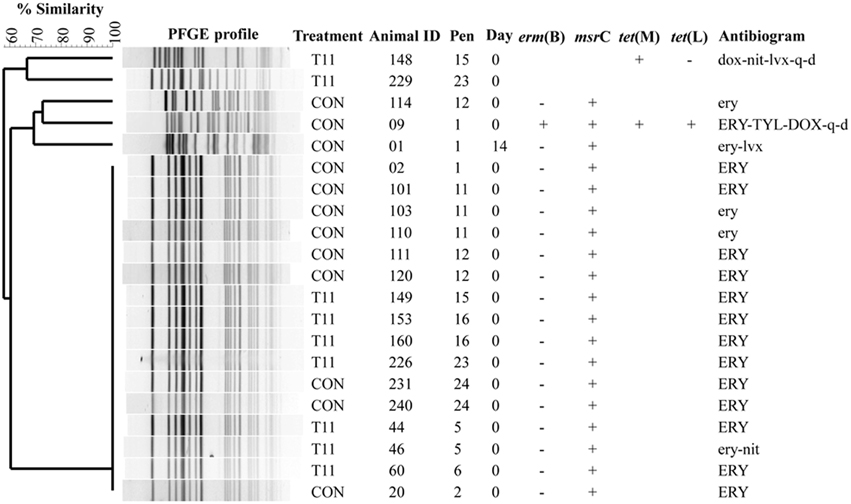
Figure 5. Dendrogram of PFGE SmaI profiles from isolates identified as Enterococcus faecium. A “+” indicates PCR positive and “−” indicates PCR negative to the respective genes. A “blank” space indicates the gene was not screened for in the respective isolate. For the antibiogram, upper case denotes complete resistance and lower case denotes incomplete resistance.
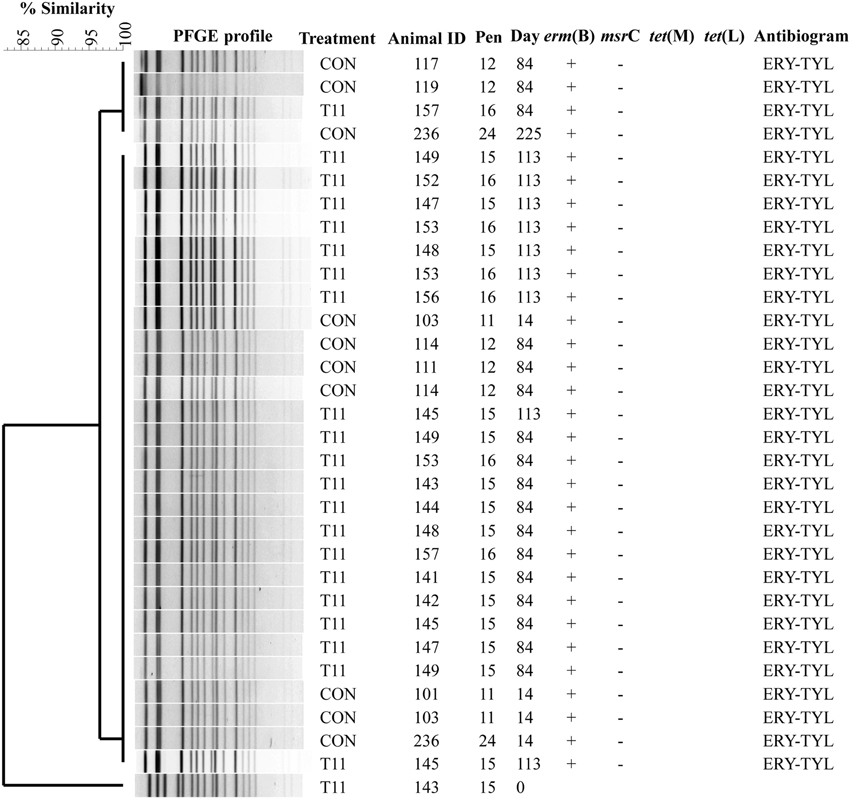
Figure 6. Dendrogram of PFGE SmaI profiles from isolates identified as Enterococcus villorum. A “+” indicates PCR positive and “−” indicates PCR negative to the respective genes. A “blank” space indicates the gene was not screened for in the respective isolate. For the antibiogram, upper case denotes complete resistance and lower case denotes incomplete resistance.
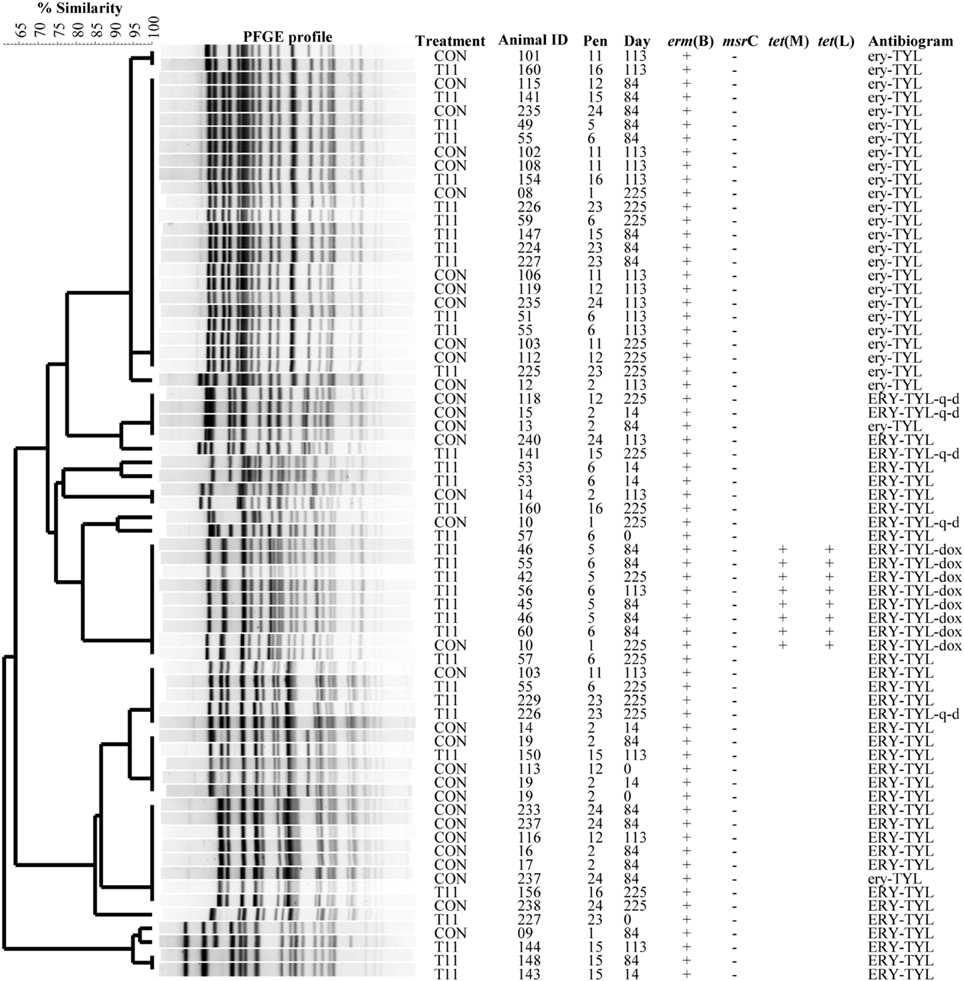
Figure 7. Dendrogram of PFGE SmaI profiles from isolates identified as erythromycin resistant Enterococcus hirae. A “+” indicates PCR positive and “−”indicates PCR negative to the respective genes. A “blank” space indicates the gene was not screened for in the respective isolate. For the antibiogram, upper case denotes complete resistance and lower case denotes incomplete resistance.
Enterococci are ubiquitous in nature and are frequently isolated from the gastrointestinal tract of mammals, including humans (Franz et al., 2011). Of the enterococci recovered from this study E. hirae was revealed to be the predominant species isolated, an observation consistent with previous studies (Anderson et al., 2008; Jackson et al., 2010; Zaheer et al., 2013).
Enterococci have been described as a “drug resistance gene trafficker” due to the ease with which they can acquire and transfer resistance genes (Werner et al., 2013). They have emerged as a serious threat to human health, particularly due to the acquisition of vancomycin resistance, increasing the difficulty of successful treatment (Center for Disease Control and Prevention, 2013). Of the 171 isolates examined for antibiotic resistance, only one isolate displayed intermediate resistance to vancomycin. This isolate was identified as E. cassiflavus, an outcome that likely reflects the intrinsic resistance of E. casseliflavus and E. gallinarum to low levels of vancomycin (Hollenbeck and Rice, 2012). This observation is encouraging, as the enterococci isolated from beef cattle do not appear to represent a significant source of vancomycin resistance.
E. faecium and E. faecalis are the two species most commonly associated with nosocomial human infections (Ruoff et al., 1990; Werner et al., 2008; Sievert et al., 2013). These species have been isolated from cattle (Kuhn et al., 2003; Anderson et al., 2008; Jackson et al., 2010), but they do not predominate, with our study suggesting that their prevalence declines after cattle enter the feedlot. Although E. hirae, as well as other enterococcal species (i.e., Enterococcus avium, E. durans, E. casseliflavus, E. gallinarum, and Enterococcus raffinosus) can cause clinical infections in humans, they are rare and thought to be more opportunistic in nature than those caused by E. faecium and E. faecalis (Ruoff et al., 1990; Alfouzan et al., 2014). Presence of E. hirae predominantly in the bovine gastrointestinal tract suggests that cattle do not present a significant source of Enterococcus that could colonize and infect humans.
In the absence of selection, the predominant resistance phenotype observed in the enterococci recovered from cattle was to erythromycin or tylosin, including isolates recovered pre- and post- antibiotic treatment. Despite no prior treatment with antimicrobials, steers harbored eryR (28 and 24%, CON and T11 respectively) and tylR (44 and 38%, CON and T11 respectively) enterococci upon arrival at the feedlot (Figure 2). This suggests that naturally occurring resistance determinants coding for macrolide resistance are already present and circulating in bovine gut enterococci populations.
For some days, the counts of tylR enterococci were higher (P < 0.05) than eryR enterococci for both treatment groups (Figure 2). It would be expected that similar counts would be obtained for both eryR and tylR enterococci as the same resistance mechanism confers resistance to both antibiotics (Roberts, 2008; Desmolaize et al., 2011). Enterococci with both intermediate and complete resistance to erythromycin were isolated from tylosin plates; erythromycin plates only selected for enterococci with complete resistance to erythromycin, explaining some of the discrepancy seen between enumeration data for the two media. Isolates from tylosin media with intermediate resistance to erythromycin also carried the erm(B) gene. It appears that the MIC breakpoint for erythromycin may be too high, therefore missing enterococci with intermediate resistance which also carry a resistance determinant. Conversely, the MIC breakpoint for tylosin may be too low thereby selecting for isolates that contain resistance determinants that may be compromised, resulting in an intermediate resistance phenotype. The fact that three isolates of E. durans from the tylosin media remained susceptible to erythromycin supports this theory. It is possible however, that these isolates carry a resistance determinant not screened for. It would be worthwhile to further explore the likely genetic differences between the resistance determinant(s) from complete and intermediate tylosin resistant isolates to identify the linkage between antimicrobial resistance (AMR) genotype and phenotype.
As the trial progressed, the number of steers positive for macrolide resistant enterococci increased in both treatment groups. This increase, even in the control group may be a reflection of increased transmission between steers due to close proximity in the feedlot environment. Likewise, the changing population dynamics of enterococci in the gastrointestinal tract of cattle may also contribute to increased transmission. Increased shedding of macrolide resistant enterococci would increase the likelihood of cattle being exposed to macrolide resistant enterococci and thus also increase the detection of positive cattle. Similarly, an increase in the proportion of the population that are macrolide resistant would increase the chances of isolating macrolide resistant enterococci. For a steer to be considered positive in this study, isolation of a single macrolide resistant enterococci colony was required. In order to make an assessment of resistance development it is important to look at resistance as a proportion of the total enterococci population.
The CFU counts of the overall enterococci population remained relatively constant over the experiment for both treatments (Figure 2). This trend was also true for CFU counts of eryR enterococci in the control group (Figure 2A), whilst the CFU counts of eryR enterococci in the tylosin treatment group tended to increase during the period of tylosin administration before dropping off on d 197, presumably due to its withdrawal from the diet (Figure 2B). This trend was also observed for the CFU counts of tylR enterococci for both treatments, with possible differences between eryR and tylR CFU being attributed to the selection of intermediate resistant enterococci on the tylosin media (Figure 2). A delay between the increase of CFU counts and tylosin administration can be seen, with increases coinciding with the transition from a silage-based diet to a grain-based diet.
High-grain diets tend to increase the amount of starch available in the lower intestinal tract, changing the nutrient availability for bacterial growth (Callaway et al., 2009). Previous researchers have reported a 1 (Scott et al., 2000) to 3 log (Diez-Gonzalez et al., 1998) increase in Escherichia coli when cattle were transitioned from a forage- to a grain-based diet. Changes that occur in the gastrointestinal environment of cattle as a result of increased starch in the diet alter the composition of the microbiome (Shanks et al., 2011). It is possible that the transition to a grain-based diet created conditions ideal for proliferation of macrolide resistant enterococci. Although not seen with the CFU of eryR enterococci, the increase of tylR enterococci in both the control and tylosin treatment group, suggest factors other than administration of tylosin may have been selecting for macrolide resistant enterococci.
Increases in eryR enterococci in cattle as a result of the administration of tylosin has been previously documented (Jacob et al., 2008; Zaheer et al., 2013), but these authors did not study the effect of withdrawal of tylosin from the diet. As in previous studies, there was an increase in the proportion of eryR and tylR resistant enterococci isolated from cattle administered tylosin. The proportion of eryR and tylR resistant enterococci for the tylosin treatment began decreasing just prior to removal of tylosin from the diet and continued to decrease after its withdrawal, until no difference (P > 0.05) was observed between treatments on d 225 (Figure 3). It appears that withdrawal of tylosin phosphate prior to slaughter contributes to a reduction in the proportion of macrolide resistant enterococci entering the food chain. However, the possibility that other unknown factors such as stress, age and diet may also be influencing this decline cannot be eliminated. It would be interesting to investigate this phenomenon further to determine why this reduction is occurring prior to the withdrawal of tylosin from the diet.
A decrease in Enterococcus species diversity was observed as the experiment progressed, with E. hirae being the predominant species identified. Transitioning of the diet from a forage- to a grain-based diet alters the fecal microbiome of cattle (Shanks et al., 2011). Diet may be a contributing factor in the shift in species diversity seen in this study, but it is also possible that other factors, such as age, may be influencing the fecal microbial community (Devriese et al., 1992).
In this study, E. thaliandicus and E. villorum were identified using multilocus sequencing of 16S rRNA, atpA, pheS, and rpoA genes after the discovery of groES-EL PCR products that varied from publically available databases (Supplementary Figure 1). To our knowledge, these species have not been previously isolated from cattle. E. thailandicus was first isolated in 2008 from fermented sausage in Thailand (Tanasupawat et al., 2008) and has been found in swine feces (Liu et al., 2013). E. villorum was first isolated in 2001 from piglets (Vancanneyt et al., 2001). Traditional methods of identifying Enterococcus species rely on biochemical tests which are unreliable for atypical species or species that have not been previously isolated (Deasy et al., 2000; Jackson et al., 2004). Molecular techniques have the advantage of being able to differentiate between closely related enterococci species.
Erythromycin resistant enterococci possessed either erm(B) or msrC or both resistance genes. Isolates designated as tylosin resistant possessed erm(B). Other macrolide resistance determinants were absent in the subset of isolates screened and it is possible that isolates not screened may have contained macrolide resistance determinants other than erm(B) or msrC. Presence of at least one resistance determinant in these isolates however confirmed the association between resistance phenotype and genotype.
Eight isolates of E. hirae and one isolate of E. faecium displayed complete resistance to erythromycin and either complete or intermediate resistance to doxycycline. These isolates were all positive for erm(B), tet(L), and tet(M). The resistance genes erm(B) and tet(M) are often associated with the transposon Tn1545 (Clewell et al., 1995; Rice, 1998). The transposon integrase gene (int gene) of Tn916/Tn1545 family of transposons has been previously detected in enterococci (De Leener et al., 2004). The identification of erm(B) and tet(M) in the same isolate in this study could possibly suggest the presence of mobile genetic elements. It would be worthwhile to investigate this further as many erm genes are often linked with other antibiotic resistance genes, tetracycline in particular (Roberts et al., 1999). Linkage of macrolide and other resistance genes is potentially problematic as administrating tylosin to cattle may not only select for macrolide resistance, but also for resistance to antibiotics such as tetracycline. Co-selection of tetracycline resistance upon the administration of tylosin has been suggested to occur within the fecal microbial communities of beef cattle (Chen et al., 2008). Linkage of these genes on mobile genetic elements increases the potential for the transfer of genes conferring resistance to multiple antibiotics (Hegstad et al., 2010; Tremblay et al., 2012).
Pulsed-field gel electrophoresis revealed a predominate cluster of E. faecium containing msrC and displaying a similar AMR profile of intermediate or complete resistance to erythromycin. Sequencing of msrC revealed that all isolates within this cluster had identical sequences. However, there were sequence differences in the msrC gene among these isolates and isolates with unique PFGE profiles (Figure 5). The four newly identified sequences have been submitted to the NCBI sequence database (Accession numbers KP775623, KP775624, KP775625, and KP775626).
Similar PFGE profiles were seen pre- and post-antibiotic treatment for erythromycin resistant E. hirae, highlighting that administration of tylosin selected for erythromycin resistant enterococci already present in the bovine gastrointestinal tract. These same profiles were still present after d 225; 28 days after tylosin had been removed from the diet. This suggests that although administration of tylosin increased the proportion of macrolide resistant enterococci in beef cattle it does not appear to be promoting the transfer of resistance between isolates. Once the selection pressure is removed (withdrawal of tylosin), the proportion of macrolide resistant enterococci returned to levels seen before antibiotic treatment.
Few studies have investigated the role that administration of tylosin in the feed of beef cattle has on the development of macrolide resistance in enterococci. This study demonstrated that administering tylosin to cattle increases the proportion of macrolide resistant enterococci. Withdrawal of tylosin from the diet appears to contribute to the decline in macrolide resistant enterococci but may not be the only factor influencing this decline. Furthermore, transitioning cattle to a grain based diet appears to alter the species population of enterococci to one in favor of E. hirae, a species not commonly associated with infection in humans. PFGE profiling of erythromycin resistant E. hirae suggest that antibiotic administration selects resistant strains already present in the intestinal microbial population.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
This study was conducted with funding from Beef Cattle Research Council (BCRC) Beef Cluster and Agriculture and Agri-Food Canada (AAFC). A. Beukers was supported by an Australian Postgraduate Award from the Australian Government. We thank Ruth Barbieri, Jay Yanke, Wendi Smart, and animal handling staff for their technical assistance.
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2015.00483/abstract
Aarestrup, F. M. (1999). Association between the consumption of antimicrobial agents in animal husbandry and the occurrence of resistant bacteria among food animals. Int. J. Antimicrob. Agents 12, 279–285. doi: 10.1016/S0924-8579(99)90059-6
Alexander, T. W., Reuter, T., Sharma, R., Yanke, L. J., Topp, E., and McAllister, T. A. (2009). Longitudinal characterization of resistant Escherichia coli in fecal deposits from cattle fed subtherapeutic levels of antimicrobials. Appl. Environ. Microbiol. 75, 7125–7134. doi: 10.1128/AEM.00944-09
Alexander, T. W., Yanke, L. J., Topp, E., Olson, M. E., Read, R. R., Morck, D. W., et al. (2008). Effect of subtherapeutic administration of antibioitics on the prevalence of antibiotic-resistant Escherichia coli bacteria in feedlot cattle. Appl. Environ. Microbiol. 74, 4405–4416. doi: 10.1128/AEM.00489-08
Alfouzan, W., Al-Sheriday, S., Al-Jabban, A., Dhar, R., Al-Mutairi, A. R., and Udo, E. (2014). A case of multiple splenic abscesses due to Enterococcus hirae. JMM Case Rep. 1, 3. doi: 10.1099/jmmcr.0.001214
Anderson, J. F., Parrish, T. D., Akhtar, M., Zurek, L., and Hirt, H. (2008). Antibiotic resistance of enterococci in American bison (Bison bison) from a nature preserve compared to that of enterococci in pastured cattle. Appl. Environ. Microbiol. 74, 1726–1730. doi: 10.1128/AEM.02164-07
Buß, M., Feßler, A. T., Turnidge, T. P., and Schwarz, S. (2014). Quality control ranges for tylosin 30 μg and 15 μg discs applicable to Staphylococcus aureus ATCC® 25923. J. Antimicrob. Chemother. 69, 277–280. doi: 10.1093/jac/dkt309
Callaway, T. R., Carr, M. A., Edrington, T. S., Andersen, S. R., and Nisbet, D. J. (2009). Diet, Escherichia coli O157:H7, and cattle: a review after 10 years. Curr. Issues Mol. Biol. 11, 67–80.
Canadian Council on Animal Care. (2003). Guide to the Care and Use of Experimental Animals, Vol. 1, 2nd Edn. Ottawa: Canadian Council on Animal Care.
Center for Disease Control and Prevention. (2012). Unified Pulsed-Field Gel Electrophoresis (PFGE) Protocol for Gram Positive Bacteria. Available online at: http://www.cdc.gov/hai/pdfs/labSettings/Unified_PFGE_Protocol.pdf (Accesed August 20, 2014).
Center for Disease Control and Prevention. (2013). Antibiotic resistance threats in the United States, 2013. Available online at: http://www.cdc.gov/drugresistance/threat-report-2013/pdf/ar-threats-2013-508.pdf (Accessed December 2, 2014).
Chen, J., Fluharty, F. L., St-Pierre, N., Morrison, M., and Yu, Z. (2008). Technical note: occurrence in fecal microbiota of genes conferring resistance to both macrolide-lincosamide-streptogramin B and tetracyclines concomitant with feeding of beef cattle with tylosin. J. Anim. Sci. 86, 2385–2391. doi: 10.2527/jas.2007-0705
Chen, J., Yu, Z. T., Michel, F. C., Wittum, T., and Morrison, M. (2007). Development and application of real-time PCR assays for quantification of erm genes conferring resistance to macrolides-lincosamides-streptogramin B in livestock manure and manure management systems. Appl. Environ. Microbiol. 73, 4407–4416. doi: 10.1128/AEM.02799-06
Clewell, D. B., Flannagan, S. E., and Jaworski, D. D. (1995). Unconstrained bacterial promiscuity: the Tn916-Tn1545 family of conjugative transposons. Trends Microbiol. 3, 229–236. doi: 10.1016/S0966-842X(00)88930-1
Clinical and Laboratory Standards Institute. (2014a). Performance Standards for Antimicrobial Disk Susceptibility Tests;Approved Standard-Eleventh Edition M02-A11. Wayne, PA: Clinical and Laboratory Standards Institute.
Clinical and Laboratory Standards Institute. (2014b). Performance Standards for Antimicrobial Susceptibility Testing; Twenty-Fourth Informational Supplement M100-S24. Wayne, PA: Clinical and Laboratory Standards Institute.
Collignon, P., Powers, J. H., Chiller, T. M., Aidara-Kane, A., and Aarestrup, F. M. (2009). World Health Organization ranking of antimicrobials according to their importance in human medicine: a critical step for developing risk management strategies for the use of antimicrobials in food production animals. Clin. Infect. Dis. 49, 132–141. doi: 10.1086/599374
Davies, R., and Roberts, T. A. (1999). Antimicrobial susceptibility of enterococci recovered from commercial swine carcasses: effect of feed additives. Lett. Appl. Microbiol. 29, 327–333. doi: 10.1046/j.1472-765X.1999.00634.x
Deasy, B. M., Rea, M. C., Fitzgerald, G. F., Cogan, T. M., and Beresford, T. P. (2000). A rapid PCR based method to distinguish between Lactococcus and Enterococcus. System. Appl. Microbiol. 23, 510–522. doi: 10.1016/S0723-2020(00)80025-9
De Leener, E., Martel, A., Decostere, A., and Haesebrouck, F. (2004). Distribution of the erm(B) gene, tetracycline resistance genes, and Tn1545-like transposons in macrolide- and lincosamide-resistant enterococci from pigs and humans. Microb. Drug Resist. 10, 341–345. doi: 10.1089/mdr.2004.10.341
Desmolaize, B., Rose, S., Warrass, R., and Douthwaite, S. (2011). A novel Erm monomethyltransferase in antibiotic-resistant isolates of Mannheimia haemolytica and Pasteurella multocida. Mol. Microbiol. 80, 184–194. doi: 10.1111/j.1365-2958.2011.07567.x
Devriese, L. A., Laurier, L., Deherdt, P., and Haesebrouck, F. (1992). Enterococcal and streptococcal species isolated from feces of calves, young cattle and dairy-cows. J. Appl. Bacteriol. 72, 29–31. doi: 10.1111/j.1365-2672.1992.tb04877.x
Diez-Gonzalez, F., Callaway, T. R., Kizoulis, M. G., and Russell, J. B (1998). Grain feeding and the dissemination of acid-resistant Escherchia coli from cattle. Science 281, 1666–1668. doi: 10.1126/science.281.5383.1666
Franz, C., Huch, M., Abriouel, H., Holzapfel, W., and Galvez, A. (2011). Enterococci as probiotics and their implications in food safety. Int. J. Food Microbiol. 151, 125–140. doi: 10.1016/j.ijfoodmicro.2011.08.014
Hegstad, K., Mikalsen, T., Coque, T. M., Werner, G., and Sundsfjord, A. (2010). Mobile genetic elements and their contribution to the emergence of antimicrobial resistant enterococcus faecalis and Enterococcus faecium. Clin. Microbiol. Infect. 16, 541–554. doi: 10.1111/j.1469-0691.2010.03226.x
Hollenbeck, B. L., and Rice, L. B. (2012). Intrinsic and acquired resistance mechanisms in enterococcus. Virulence 3, 421–433. doi: 10.4161/viru.21282
Jackson, C. R., Fedorka-Cray, J., and Barrett, J. B. (2004). Use of a genus- and spcies-specific multiplex PCR for identification of enterococci. J. Clin. Microbiol. 42, 3558–3565. doi: 10.1128/JCM.42.8.3558-3565.2004
Jackson, C. R., Lombard, J. E., Dargatz, D. A., and Fedorka-Cray, P. J. (2010). Prevalence, species distribution and antimicrobial resistance of enterococci isolated from US dairy cattle. Lett. Appl. Microbiol. 52, 41–48. doi: 10.1111/j.1472-765X.2010.02964.x
Jacob, M. E., Fox, J. T., Narayanan, S. K., Drouillard, J. S., Renter, D. G., and Nagaraja, T. G. (2008). Effects of feeding wet corn distillers grains with solubles with or without monensin and tylosin on the prevalence and antimicrobial susceptibilities of fecal foodborne pathogenic and commensal bacteria in feedlot cattle. J. Anim. Sci. 86, 1182–1190. doi: 10.2527/jas.2007-0091
Kuhn, I., Iversen, A., Burman, L. G., Olsson-Liljequist, B., Franklin, A., Finn, M., et al. (2003). Comparison of enterococcal populations in animals, humans, and the environment - a European study. Int. J. Food Microbiol. 88, 133–145. doi: 10.1016/S0168-1605(03)00176-4
Liu, Y., Wang, Y., Schwarz, S., Li, Y., Shen, Z., Zhang, Q., et al. (2013). Transferable multiresistance plasmids carrying cfr in Enterococcus spp. from swine and fram environment. Antimicrob. Agents Chemother. 57, 42–48. doi: 10.1128/AAC.01605-12
Ng, L. K., Martin, I., Alfa, M., and Mulvey, M. (2001). Multiplex PCR for the detection of tetracycline resistant genes. Mol. Cell. Probes 15, 209–215. doi: 10.1006/mcpr.2001.0363
Page, S. W., and Gautier, P. (2012). Use of antimicrobial agents in Livestock. Rev. Sci. Tech. 31, 145–188.
Peak, N., Knapp, C. W., Yang, R. K., Hanfelt, M. M., Smith, M. S., Aga, D. S., et al. (2007). Abundance of six tetracycline resistance genes in wastewater lagoons at cattle feedlots with different antibiotic use strategies. Enivoron. Microbiol. 9, 143–151. doi: 10.1111/j.1462-2920.2006.01123.x
Poh, C. H., Oh, H. M. L., and Tan, A. L. (2006). Epidemiology and clinical outcome of enterococcal bacteraemia in an acute care hospital. J. Infect. 52, 383–386. doi: 10.1016/j.jinf.2005.07.011
Portillo, A., Ruiz-Larrea, F., Zarazaga, M., Alonso, A., Martinez, J. L., and Torres, C. (2000). Macrolide resistance genes in Enterococcus spp. Antimicrob. Agents Chemother. 44, 967–971. doi: 10.1128/AAC.44.4.967-971.2000
Rice, L. B. (1998). Tn916 family conjugative transposons and dissemination of antimicrobial resistance determinants. Antimicrob. Agents Chemother. 42, 1871–1877.
Roberts, M. C. (2008). Update on macrolide-lincosamide-streptogramin, ketolide, and oxazolidinone resistance genes. FEMS Microbiol. Lett. 282, 147–159. doi: 10.1111/j.1574-6968.2008.01145.x
Roberts, M. C., Sutcliffe, J., Courvalin, P., Jensen, L. B., Rood, J., and Seppala, H. (1999). Nomenclature for macrolide and macrolide-lincosamide-streptogramin B resistance determinants. Antimicrob. Agents Chemother. 43, 2823–2830.
Ruoff, K. L., de la Maza, L., Murtagh, M. J., Spargo, J. D., and Ferraro, M. J. (1990). Species identities of enterococci isolated from clinical specimens. J. Clin. Microbiol. 28, 435–437.
Scott, T., Wilson, C., Bailey, D., Klopfenstein, T., Milton, T., Moxley, R., et al. (2000). Influence of diet on total and acid-resistant Escherichia coli and colonic pH. Nebraska Beef Rep. 39–41. Available online at: http://digitalcommons.unl.edu/animalscinbcr
Shanks, O. C., Kelty, C. A., Archibeque, S., Jenkins, M., Newton, R. J., McLellan, S. L., et al. (2011). Community structures of fecal bacteria in cattle from different animal feeding operations. Appl. Environ. Microbiol. 77, 2992–3001. doi: 10.1128/AEM.02988-10
Sharma, R., Munns, K., Alexander, T., Entz, T., Mirzaagha, P., Yanke, L. J., et al. (2008). Diversity and distribution of commensal fecal Escherichia coli bacteria in beef cattle administered selected subtherapeutic antimicrobials in a feedlot setting. Appl. Environ. Microbiol. 74, 6178–6186. doi: 10.1128/AEM.00704-08
Sievert, D. M., Ricks, P., Edwards, J. R., Schneider, A., Patel, J., Srinivasan, A., et al. (2013). Antimicrobial-resistant pathogens associated with healthcare-associated infections: summary of data reported to the National Healthcare Safety Network at the Centers for Disease Control and Prevention, 2009-2010. Infect. Control Hosp. Epidemiol. 34, 1–14. doi: 10.1086/668770
Tanasupawat, S., Sukontasing, S., and Lee, J. (2008). Enterococcus thailandicus sp. nov., isolated from fermented sausage (‘mum’) in Thailand. Int. J. Syst. Evol. Microbiol. 58, 1630–1634. doi: 10.1099/ijs.0.65535-0
The European Committee on Antimicrobial Susceptibility Testing, (EUCAST). (2014). Breakpoint Tables for Interpretation of MICs and Zone Diameters. Version 4.0. Available online at: www.eucast.org (Accessed November 5, 2014).
Tremblay, C., Letellier, A., Quessy, S., Daignault, D., and Archambault, M. (2012). Antibiotic-resistant Enterococcus facalis in abattoir pigs and plasmid colocalization and cotransfer of tet(M) and erm(B) genes. J. Food Prot. 75, 1595–1602. doi: 10.4315/0362-028X.JFP-12-047
Vancanneyt, M., Snauwaert, C., Cleenwerck, I., Baele, M., Descheemaeker, P., Goossens, H., et al. (2001). Enterococcus villorum sp. nov., an enteroadherent bacterium associated with diarrhoea in piglets. Int. J. Syst. Evol. Microbio. 51, 393–400.
Werner, G., Coque, T. M., Franz, C. M. A. P., Grohmann, E., Hegstad, K., Jensen, L., et al. (2013). Antibiotic resistant enterococci – tales of a drug resistance gene trafficker. Int. J. Med. Microbiol. 303, 306–379. doi: 10.1016/j.ijmm.2013.03.001
Werner, G., Coque, T. M., Hammerum, A. M., Hope, R., Hryniewicz, W., Johnson, A., et al. (2008). Emergence and spread of vancomycin resistance among enterococci in Europe. Euro Surveill. 13, 19046.
Zaheer, R., Cook, S. R., Klima, C. L., Stanford, K., Alexander, T., Topp, E., et al. (2013). Effect of subtherapeutic vs. therapeutic administration of macrolides on antimicrobial resistance in Mannheimia haemolytica and enterococci isolated from beef cattle. Front. Microbiol. 4:133. doi: 10.3389/fmicb.2013.00133
Keywords: enterococci, antimicrobial resistance, subtherapeutic macrolides, beef cattle, tylosin, erythromycin
Citation: Beukers AG, Zaheer R, Cook SR, Stanford K, Chaves AV, Ward MP and McAllister TA (2015) Effect of in-feed administration and withdrawal of tylosin phosphate on antibiotic resistance in enterococci isolated from feedlot steers. Front. Microbiol. 6:483. doi: 10.3389/fmicb.2015.00483
Received: 27 February 2015; Accepted: 02 May 2015;
Published: 27 May 2015.
Edited by:
Satoru Suzuki, Ehime University, JapanReviewed by:
Anuradha Ghosh, Kansas State University, USACopyright © 2015 Beukers, Zaheer, Cook, Stanford, Chaves, Ward and McAllister. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Tim A. McAllister, Lethbridge Research Centre, Agriculture and Agri-Food Canada, 5403-1 st Ave. South, Lethbridge, AB T1J 4B1, Canada,dGltLm1jYWxsaXN0ZXJAYWdyLmdjLmNh
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.