- 1Department of Clinical Microbiology and Infection Control, The University of Hong Kong - Shenzhen Hospital, Shenzhen, China
- 2Department of Microbiology, Li Ka Shing Faculty of Medicine, The University of Hong Kong, Pokfulam, Hong Kong SAR, China
- 3Department of Pathology, The University of Hong Kong - Shenzhen Hospital, Shenzhen, China
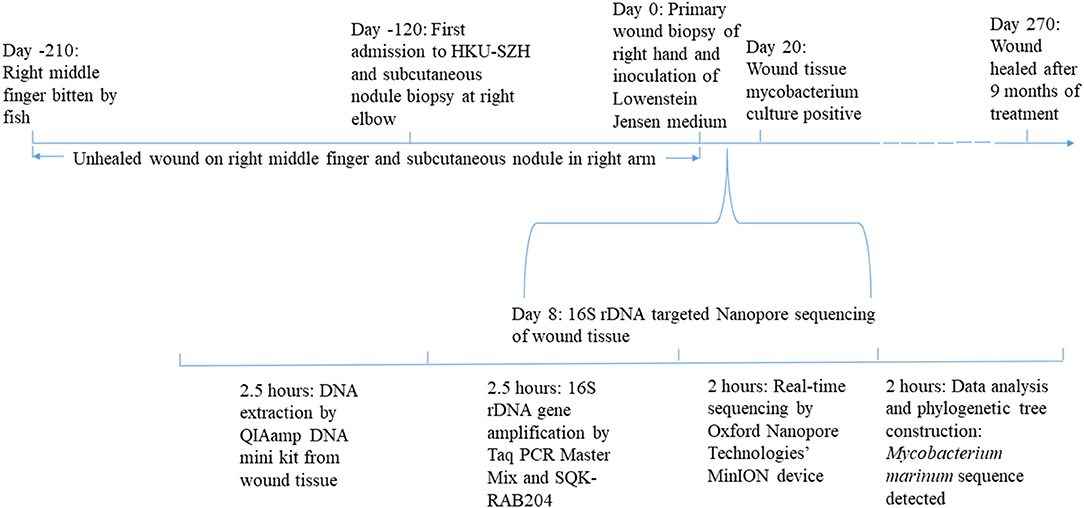
We present the first report of histology- and culture-proven Mycobacterium marinum infection diagnosed by next-generation sequencing (NGS). It took <2 days to make a microbiological diagnosis using the Oxford Nanopore Technologies' MinION device, compared to 20 days for the mycobacterium to be isolated from the tissue biopsy. NGS is particularly useful for culture-negative and slow-growing microorganism infections, such as mycobacterial, fungal and partially treated pyogenic bacterial infections. Due to its low equipment cost, short turn-around-time and portable size, the Oxford Nanopore Technologies' MinION device is a useful platform for NGS in routine clinical microbiology laboratories.
Introduction
Mycobacterium marinum is a pigmented slow-growing non-tuberculous mycobacterium associated with skin and soft tissue infections. The bacterium is commonly found in both fresh or saltwater in many parts of the world (1). Most infections develop 2–3 weeks after direct or indirect contact with contaminated water or fish as papules or ulcerations on the hands and arms, which may progress to ascending lesions. As the bacterium grows optimally at 28–30°C, it is not associated with deep-seated infections.
Traditionally, M. marinum infections were diagnosed in the laboratory by culturing and identifying the bacterium as well as histology. As the bacterium is slow-growing, it often takes 2–3 weeks, and in our experience, sometimes up to 6 weeks, to isolate it from the clinical specimen, and a few more weeks for identifying it to the species level. Therefore, methods for rapid identification of the bacterium are crucial for helping the clinicians to start the specific anti-mycobacterial regimen on a timely basis. In this article, we report the first case of M. marinum diagnosed by the Oxford Nanopore Technologies' MinION device, a handy platform of next-generation sequencing (NGS) that can be used in routine clinical microbiology laboratories.
Case Description
A 38-year-old Chinese man was admitted because of unhealed wound at the right hand in August 2020. The patient was the owner of a seafood market in Shenzhen, China. Four months before admission, his right middle finger was bitten by a grouper. Topical disinfection was performed. In the next 4 months, he started to recognize small masses developing adjacent to the wound at the back of the right hand as well as in the right elbow and right arm. There was no fever or other systematic symptoms. When the patient was admitted, examination showed the poorly healed primary wound at the back of the right middle finger and a secondary lesion at the back of his right hand (Figure 1A), as well as a 5 × 6 cm subcutaneous nodule at the right elbow and 2 supratrochlear lymph nodes of 1 cm in diameter. The total white cell count was 4.89 × 109/L (normal range, 3.89–9.93 × 109/L) with normal differential count. Liver and renal function tests were normal. C-reactive protein was 1.23 mg/L (normal range, 0–5 mg/L). The subcutaneous nodule and overlying skin at the right elbow were excised for histology and bacterial, fungal and mycobacterial culture. Histological examination revealed chronic granulomatous inflammation but all the culture results were negative. No antibiotic was prescribed.
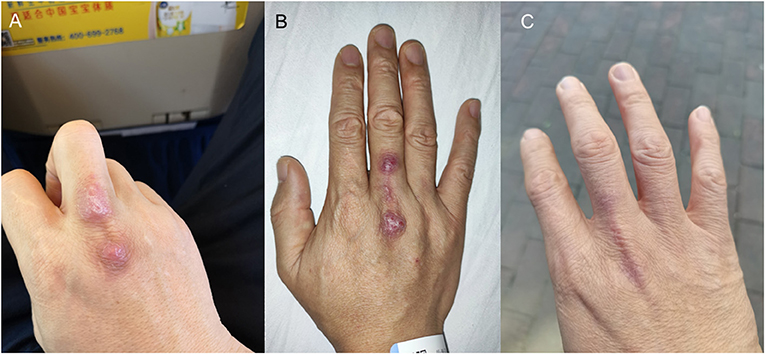
Figure 1. Photos of the patient's right hand. (A) Poorly healed primary wound and secondary lesion at the back of the right hand. (B) Ulcerated lesions at the back of the right hand. (C) Healed wounds after 9 months of treatment.
From August to November, the wound on the right middle finger and hand did not heal well and another new painful nodule gradually developed at the right elbow. Examination revealed 2 ulcerated lesions at the back of the right hand (Figure 1B), multiple subcutaneous nodules at the right elbow and palpable supratrochlear, right axillary and cervical lymph nodes. The total white cell count was 3.61 × 109/L (normal range, 3.89–9.93 × 109/L). Liver and renal function tests were normal. C-reactive protein was 17.44 mg/L (normal range, 0–5 mg/L). A biopsy of the primary wound on the right hand was performed. Histological examination showed acanthosis cell layer hyperplasia, granular layer thickening with mild hyperkeratosis and parakeratosis, lymphocytic infiltration in epithelium, proliferation of small vessels in the corium layer with lymphocytic and plasmocytic infiltration, epithelioid cells and Langerhans multinucleated giant cells around appendages in middle and deep dermis but no necrosis and vasculitis (Figure 2A). Ziehl-Neelsen stain was negative. 16S rDNA gene targeted NGS of the tissue samples using the Oxford Nanopore Technologies' MinION device revealed sequences of M. marinum (Figure 2B). Mycobacterial culture using Lowenstein Jensen medium was positive with yellow colonies (Strain_HKU-SZH_1120) after 20 days of incubation (Figure 3). Matrix-assisted laser desorption ionization time-of-flight mass spectrometry (MALDI-TOF MS) (top match score 2.283) and 16S rDNA gene sequencing confirmed the identification as M. marinum. MALDI-TOF MS was performed by the direct transfer method using the MALDI-TOF MS spectrometer (Bruker Daltonik) and the spectrum was analyzed with IVD MALDI Biotyper 2.3 and reference Mycobacteria library 6.0 (Bruker Daltonik) (2). 16S rDNA gene sequencing was performed according to our previous publications (3, 4). Oral rifampin, minocycline and clarithromycin were prescribed for a total of 9 months. All the wounds were healed and the infection has remained in remission at the time of writing, 2 months from stopping the antibiotics (Figure 1C).
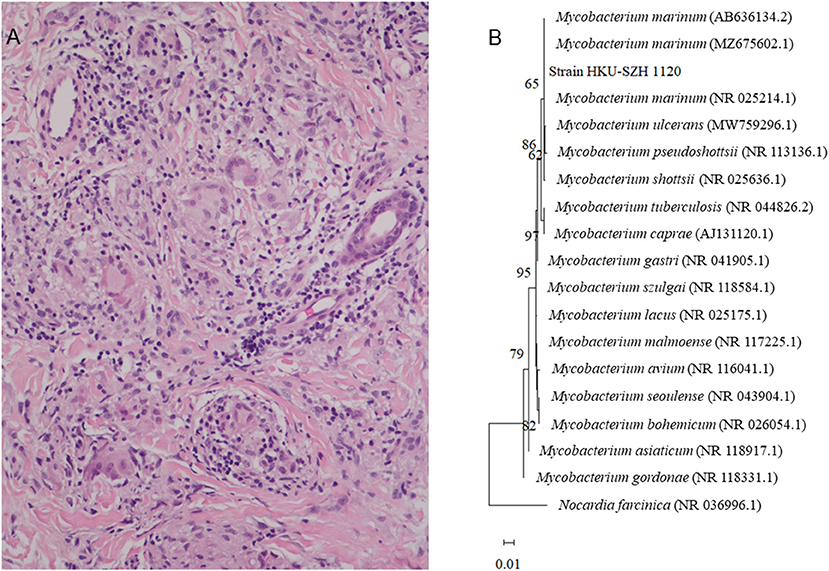
Figure 2. (A) Histological section of the dermis, showing granuloma with Langerhans multinucleated giant cells aggregation with surrounding infiltration of lymphocytes (H&E ×200). (B) Phylogenetic tree showing the relationships among Strain_HKU-SZH_1120 and other closely related Mycobacterium species. A total of 1,231 nucleotide positions in each 16S rRNA gene were included in the analysis. The tree was constructed using the Maximum Likelihood method and Tamura 3-parameter model and rooted using Norcardia facinica (NR 036996.1). The bootstrap values calculated from 1,000 trees were shown when they were >60%. The scale bar indicates the estimated number of substitutions per 100 bases. The names and accession numbers (in parentheses) were presented as cited in the GenBank database.
Discussion
When NGS technologies first appeared in the market, they were mainly used for genome sequencing (5). With the advancement of sequencing chemistries and computational capacity, NGS technologies have matured into clinical applications in the recent years (6). In the clinical setting for infectious diseases, NGS is used most often for patients who have fever without localizing features or culture-negative infections (7, 8). In addition, it is also useful for rapid diagnosis of infections caused by slow-growing microorganisms. For example, we have recently reported its usefulness in rapid diagnosis of fungal infections (9). As for mycobacteria, NGS has been used for the diagnosis of tuberculosis and a number of non-tuberculous mycobacterial infections (10–15). Since many mycobacteria, such as M. chimaera, M. yongonense, M. avium complex, M. kubicae, M. leprae, and in the present case, M. marinum, are slow-growing or non-cultivable in artificial medium and the anti-mycobacterial therapeutic regimen for different mycobacteria are radically different, it is of crucial importance to identify the mycobacteria involved to the species level in a timely basis in order to prescribe the specific and effective medications for the treatment of the infections. Furthermore, early diagnosis can also avoid unnecessary investigations and side effects resulting from inappropriate medications and hence reduce the related costs. In our experience, it usually takes around 6–12 weeks to isolate and identify a slow-growing mycobacterium from clinical specimens using conventional methods. On the other hand, it will only take up to a few days, even if the samples are sent out to private laboratories, if NGS technology is used. Therefore, this has undeniably important impact in the management of mycobacterial infections.
The Oxford Nanopore Technologies' MinION device is becoming a useful platform for NGS in routine clinical microbiology laboratories. When NGS was first used for laboratory diagnosis of infectious disease, usually more than 10 samples were processed in one sequencing run in order to reduce the sequencing cost per sample. However, the recent invention of Oxford Nanopore Technologies' MinION device has expedited the use of NGS in laboratory diagnosis, due to its low equipment cost, short turn-around-time and portable size (16, 17). When this technology platform was first made commercially available in 2015, its sequencing error rate was still very high (18). After several rounds of improvement, the sequencing error rate has reached the acceptable range (19, 20). This device is small enough to be held by one hand and can process one specimen at a time, making it particularly useful for routine use in clinical microbiology laboratories. The cost of the Oxford Nanopore Technologies' MinION device is inexpensive compared to the traditional NGS machines; and in our laboratory, the cost of sequencing one sample is ~90 USD. This is in fact more economical than sending the specimen out to private laboratories in our hospital setting. The turn-around-time for using the Oxford Nanopore Technologies' MinION device is <2 days, compared to 2–4 days if the sample is sent out. Furthermore, if the demand for the service is increased, the technology is also easily scalable. Such flexibility enables it to fit in clinical microbiology laboratories that handle different specimen volumes. All these advantages have made the Oxford Nanopore Technologies' MinION device an emerging technology platform in clinical microbiology laboratories.
Data Availability Statement
The 16S rDNA gene sequence of Strain_HKU-SZH_1120 has been deposited in GenBank (Accession Number: OM095442).
Ethics Statement
Written informed consent was obtained from the individual(s) for the publication of any potentially identifiable images or data included in this article.
Author Contributions
FX and PW wrote the manuscript. FX reviewed the clinical data. SL supervised the microbiological investigations. YM, JI, and W-MC processed and analyzed the 16S rDNA gene sequence data. MZ and MG reviewed the pathological data. All authors have read and approved the final version of the manuscript.
Funding
This study was partly supported by Sanming Project of Medicine in Shenzhen, China (SZSM201911014).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Tebruegge M, Curtis N. Mycobacterium marinum infection. Adv Exp Med Biol. (2011) 719:201–10. doi: 10.1007/978-1-4614-0204-6_17
2. Lau SK, Tang BS, Teng JL, Chan TM, Curreem SO, Fan RY, et al. Matrix-assisted laser desorption ionisation time-of-flight mass spectrometry for identification of clinically significant bacteria that are difficult to identify in clinical laboratories. J Clin Pathol. (2014) 67:361–6. doi: 10.1136/jclinpath-2013-201818
3. Woo PC, Tsoi HW, Leung KW, Lum PN, Leung AS, Ma CH, et al. Identification of Mycobacterium neoaurum isolated from a neutropenic patient with catheter-related bacteremia by 16S rRNA sequencing. J Clin Microbiol. (2000) 38:3515–7. doi: 10.1128/JCM.38.9.3515-3517.2000
4. Woo PC, Leung KW, Wong SS, Chong KT, Cheung EY, Yuen KY. Relatively alcohol-resistant mycobacteria are emerging pathogens in patients receiving acupuncture treatment. J Clin Microbiol. (2002) 40:1219–24. doi: 10.1128/JCM.40.4.1219-1224.2002
5. Jarvie T. Next generation sequencing technologies. Drug Discov Today Technol. (2005) 2:255–60. doi: 10.1016/j.ddtec.2005.08.003
6. Voelkerding KV, Dames SA, Durtschi JD. Next-generation sequencing: from basic research to diagnostics. Clin Chem. (2009) 55:641–58. doi: 10.1373/clinchem.2008.112789
7. Simner PJ, Miller S, Carroll KC. Understanding the promises and hurdles of metagenomic next-generation sequencing as a diagnostic tool for infectious diseases. Clin Infect Dis. (2018) 66:778–88. doi: 10.1093/cid/cix881
8. Wilson MR, Naccache SN, Samayoa E, Biagtan M, Bashir H, Yu G, et al. Actionable diagnosis of neuroleptospirosis by next-generation sequencing. N Engl J Med. (2014) 370:2408–17. doi: 10.1056/NEJMoa1401268
9. Tsang CC, Teng JLL, Lau SKP, Woo PCY. Rapid genomic diagnosis of fungal infections in the age of next-generation sequencing. J Fungi (Basel). (2021) 7:636. doi: 10.3390/jof7080636
10. Satta G, Lipman M, Smith GP, Arnold C, Kon OM, McHugh TD. Mycobacterium tuberculosis and whole-genome sequencing: how close are we to unleashing its full potential? Clin Microbiol Infect. (2018) 24:604–9. doi: 10.1016/j.cmi.2017.10.030
11. van Ingen J, Kohl TA, Kranzer K, Hasse B, Keller PM, Katarzyna Szafrańska A, et al. Global outbreak of severe Mycobacterium chimaera disease after cardiac surgery: a molecular epidemiological study. Lancet Infect Dis. (2017) 17:1033–41. doi: 10.1016/S1473-3099(17)30324-9
12. Wang L, Liu D, Yung L, Rodriguez GD, Prasad N, Segal-Maurer S, et al. Co-infection with 4 species of mycobacteria identified by using next-generation sequencing. Emerg Infect Dis. (2021) 27:2948–50. doi: 10.3201/eid2711.203458
13. Zhu H, Zhu M, Lei JH, Xiao YL, Zhao LM. Metagenomic next-generation sequencing can clinch diagnosis of non-tuberculous mycobacterial infections: a case report. Front Med (Lausanne). (2021) 8:679755. doi: 10.3389/fmed.2021.679755
14. Hendrix J, Epperson LE, Durbin D, Honda JR, Strong M. Intraspecies plasmid and genomic variation of Mycobacterium kubicae revealed by the complete genome sequences of two clinical isolates. Microb Genom. (2021) 7:mgen000497. doi: 10.1099/mgen.0.000497
15. Stefani MMA, Avanzi C, Bührer-Sékula S, Benjak A, Loiseau C, Singh P, et al. Whole genome sequencing distinguishes between relapse and reinfection in recurrent leprosy cases. PLoS Negl Trop Dis. (2017) 11:e0005598. doi: 10.1371/journal.pntd.0005598
16. Taylor TL, Volkening JD, DeJesus E, Simmons M, Dimitrov KM, Tillman GE, et al. Rapid, multiplexed, whole genome and plasmid sequencing of foodborne pathogens using long-read nanopore technology. Sci Rep. (2019) 9:16350. doi: 10.1038/s41598-019-52424-x
17. Walter MC, Zwirglmaier K, Vette P, Holowachuk SA, Stoecker K, Genzel GH, et al. MinION as part of a biomedical rapidly deployable laboratory. J Biotechnol. (2017) 250:16–22. doi: 10.1016/j.jbiotec.2016.12.006
18. Laver T, Harrison J, O'Neill PA, Moore K, Farbos A, Paszkiewicz K, et al. Assessing the performance of the oxford nanopore technologies MinION. Biomol Detect Quantif. (2015) 3:1–8. doi: 10.1016/j.bdq.2015.02.001
19. Alvarez JR, Skachkov D, Massey SE, Kalitsov A, Velev JP. DNA/RNA transverse current sequencing: intrinsic structural noise from neighboring bases. Front Genet. (2015) 6:213. doi: 10.3389/fgene.2015.00213
Keywords: Mycobacterium marinum, non-tuberculosis mycobacteria, next-generation sequencing, Oxford Nanopore, MinION
Citation: Xing F, Lo SKF, Ma Y, Ip JD, Chan W-M, Zhou M, Gong M, Lau SKP and Woo PCY (2022) Rapid Diagnosis of Mycobacterium marinum Infection by Next-Generation Sequencing: A Case Report. Front. Med. 9:824122. doi: 10.3389/fmed.2022.824122
Received: 28 November 2021; Accepted: 12 January 2022;
Published: 04 February 2022.
Edited by:
Jessica L. Jones, United States Food and Drug Administration, United StatesReviewed by:
Josephine Bryant, University of Cambridge, United KingdomAdwoa Asante-Poku, University of Ghana, Ghana
Copyright © 2022 Xing, Lo, Ma, Ip, Chan, Zhou, Gong, Lau and Woo. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Patrick C. Y. Woo, cGN5d29vJiN4MDAwNDA7aGt1Lmhr
 Fanfan Xing
Fanfan Xing Simon K. F. Lo1
Simon K. F. Lo1 Miaozi Gong
Miaozi Gong Susanna K. P. Lau
Susanna K. P. Lau