- 1Global Research Team, Basecamp Research, London, United Kingdom
- 2Jack Gilbert Lab, Scripps Institution of Oceanography, University of California, San Diego, San Diego, CA, United States
- 3Underwater Cultural Heritage Unit, Heritage Malta, Kalkara, Malta
- 4Department of Classics & Archaeology, University of Malta, Msida, Malta
The anthropogenic influences of historical shipwrecks on the marine environment remain largely uncharacterized, particularly for wrecks located at depths of ~50 meters and lost over 80 years ago. Below 50 meters, the marine environment transitions from the euphotic to the mesophotic zone, where reduced light, significant changes in oxygen levels, and increased pressure may alter microbial communities and metabolic pathways. Understanding these shifts can provide insights into the adaptation and evolution of microorganisms. Sediment samples were collected by a team of divers from nine wreck sites off the coast of Malta, varying in depth from 5 to 100 meters. All sampling was conducted with the consent and authorization of the Government of Malta for the collection of marine genetic resources. Chemical analyses and metabarcoding techniques were employed to assess how shipwrecks influence microbial diversity and community composition in the surrounding sediments. Alpha (α) diversity of microbial communities was higher in sediments adjacent to the shipwrecks compared to those collected over 100 meters away from any wreck site. Across all nine wreck sites, sediment size emerged as the most significant driver of microbial diversity, followed by frame type and the presence of oil leaks. This study demonstrates that shipwrecks, varying in depth, frame type, and country of origin, significantly influence the microbial ecosystems and sediment chemistry of their surrounding environment. The findings suggest that shipwrecks act as focal points for ecological and chemical changes, contributing to the dynamic interactions between historical artifacts and marine ecosystems.
1 Introduction
It is estimated that over 3 million ship and plane wrecks lie on the seabed of the Earth’s oceans and the majority of these wrecks remain at the site they were lost, due both to the cost and logistical challenges of removing sunken vessels and also the important underwater cultural heritage that historical wrecks can embody (Gambin et al., 2021a). Wrecks are key influencers of marine ecosystems as they form ecological hotspots by acting as artificial reefs that create habitats that foster biodiversity across many trophic levels (Paxton et al., 2023). However, a wreck’s influence on biodiversity is not exclusively positive: wreck cargo can include potentially hazardous materials such as unexploded ordnance (UXO) or crude oil that can pose threats to marine ecosystem health (Wolska et al., 2012; Rogowska and Namiesnik, 2013). The influences of wreck sites on the marine environment depend heavily on variation across all elements of the marine environment, from depth to temperature, and from salinity to current. As well as the characteristics of the wreck itself, including its age, frame type, and cargo. The geo-location of the wrecks also vary; some of the wrecks in this study lay within the harbour context, whilst others lie offshore. Fully understanding the influences that wrecks have on marine environments requires a broader study of wrecks and their immediate environment with the goal of building insights that can inform the course of action for wreck management to support marine biodiversity and preserve important underwater heritage.
Heritage Malta, the national agency charged with the protection, management and promotion of cultural heritage in Malta, curates over 90 sites and museums. This includes 19 historic wreck sites, located around Malta and Gozo at depths varying from 55 metres to 115 metres. The types and dates of site also vary, from an ancient 3rd century BC site to WWI battleships and minesweepers, WWII destroyers and various types of aircraft datable to WWII and the Cold War. This variety has placed Malta in the unique position of curating a global heritage (Gambin et al., 2021b). National legislation defines “remains found on the seabed that are situated in the territorial waters of Malta and are at least fifty years old” (Act No. XIX of 2019) as part of the island’s underwater cultural heritage. This contrasts with the UNESCO Convention on the Protection of the Underwater Cultural Heritage, which only considers remains over 100-years-old to be underwater cultural heritage (UNESO 2001) (Gambin et al., 2021a, c).
Heritage Malta has implemented a management system that looks beyond the cultural value of these sites to include numerous other elements. Relevant to this paper is the work being done to understand the microbial diversity surrounding this high density of (relatively) shallow wreck sites. An integral part of the work being done by Heritage Malta is the 3D documentation of these wrecks, important from a scientific perspective as from an outreach perspective. From a scientific perspective, these 3D datasets act as baseline records on the condition of the site and its state of preservation. When sites are located at 50m or deeper, time is of the essence, whereas the 3D data set can be explored indefinitely and datasets can be compared and changes to the site over time can be highlighted (Gambin et al., 2018). What can be noted from these surveys is the high concentration of marine life on these wrecks, despite the presence of pollutants such as munitions and unexploded ordnance (UXO).
Previous studies incorporating marine life and shipwrecks have primarily focused on the larger-scale ecological effects of shipwrecks, such as their role as biodiversity hotspots and their influence on macroscopic populations (Paxton et al., 2024; Lengkeek et al., 2013; Meyer-Kaiser et al., 2022; Asner et al., 2022). However, the role of wrecks’ on the microbial communities in the surrounding sediments has been less studied, especially in wrecks below 50m. Given the crucial roles that microbes play in ecosystem functionality, from nutrient cycling to degradation of pollutants, a systematic understanding is important. Shipwrecks are often noted to have a positive impact on the surrounding ecology, as they act as artificial reefs, providing a solid surface in an aquatic ecosystem which is of particular importance in microbial communities (Connell, 2000; Church et al., 2009). Conversely, the microbial communities may have a positive or a negative impact on the shipwrecks themselves, as they may contribute to the wreck’s preservation or deterioration (Price et al., 2020). Previous studies have indicated that shipwrecks create their own specialised niche that is associated to that shipwreck and differ significantly from surrounding sediment and the aquatic environment (Cullimore and Johnston, 2008; Hamdan et al., 2018; Mugge et al., 2019). When looking at the impact of wrecks that contain contaminants such as oil or UXO, this area of research has hugely increased in the last two decades, especially on wrecks from the two world wars, as there are over 8,600 wrecks on the seabed, containing an estimated 2.5 - 20.4 million tonnes of crude oil (Thomas et al., 2021). As the crude oil contains heavy metals and polycyclic aromatic hydrocarbons (PAH’s), it is noted that certain microbes will thrive in these environments, including hydrocarbon-degrading bacteria (Head et al., 2006). Oil pollutants often alter the microbial community composition surrounding a shipwreck significantly, whilst also reducing the species richness & diversity as selecting for oil-degrading bacteria can occur (Thomas et al., 2021), however other factors can also have a significant impact on the microbial community in the sediments around shipwrecks [eg. depth (Hamdan et al., 2021), grain size (Cammen, 1982) and frame type (Moseley et al., 2022)].
This study aimed to fill this gap by examining the microbial diversity in sediments surrounding archaeological wrecks around Malta and Gozo, and comparing those to a wreck’s construction materials and its contents. We also analysed how factors such as a wreck’s country of origin, frame type, the presence of oil leaks, and their depth influence microbial communities and surrounding ecological processes. This research not only contributes to the field of marine ecology by evaluating long-term ecological implications, but also provides insights that could inform the management and preservation of underwater cultural heritage sites by examining the interactions between wrecks and their surrounding environment.
2 Results
2.1 Overview of wreck sites assessed
Ten ship and aeroplane wreck sites were sampled within Malta’s territorial waters. Distances of these wrecks from the shoreline range from less than 0.1 nautical mile to approximately 5.4 nautical miles offshore. At each site, a minimum of five samples were collected, supplemented by two samples taken 100 metres away from any wreck site (None Wreck Site). The wrecks were situated at depths ranging from 5 metres to 100 metres on the seabed and were located to the north, northeast, and southeast of Malta’s coastline (Figure 1).
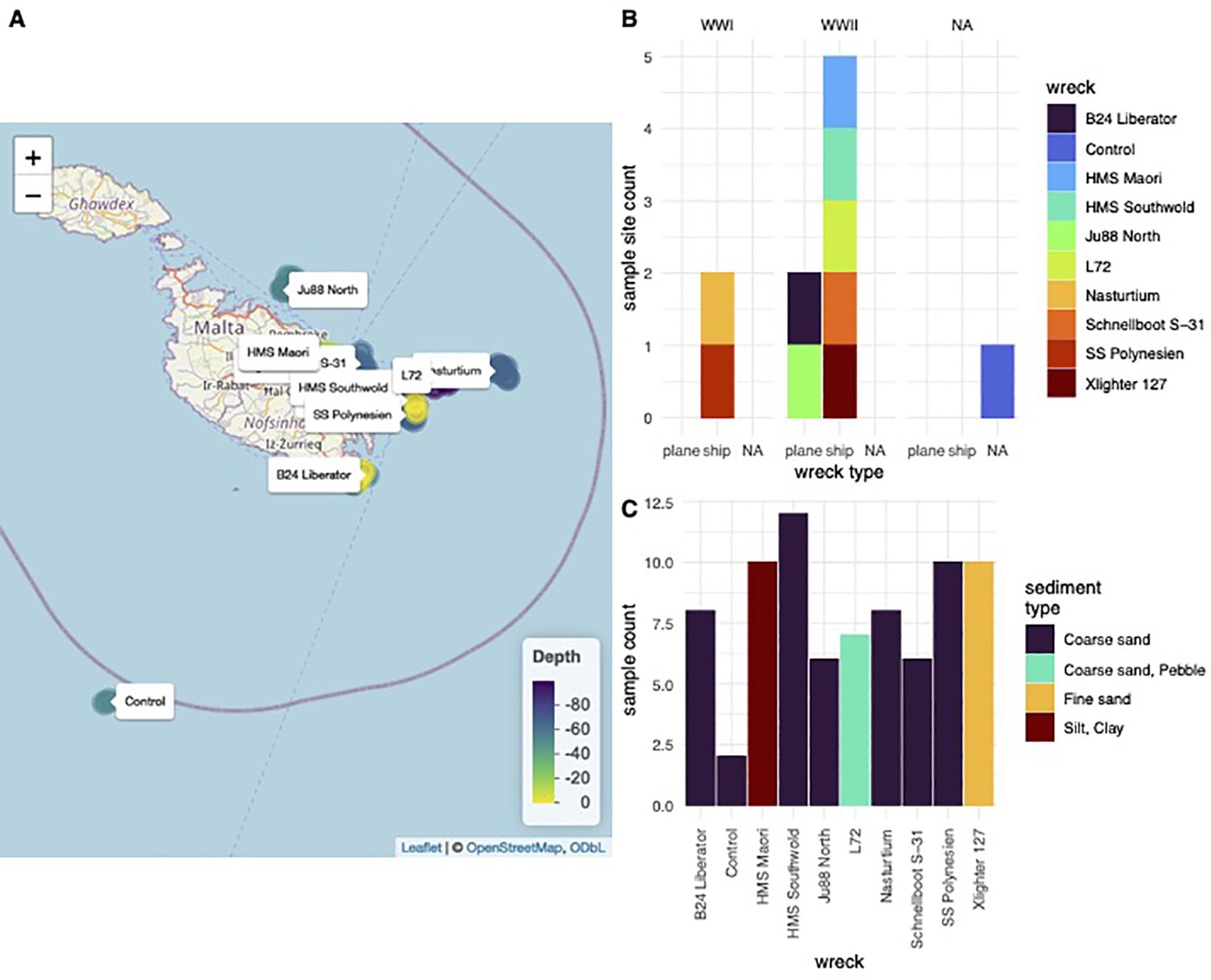
Figure 1. Sampled sites overview. (A) map of wreck locations by depth (B) type (plane/ship) and age (WWI/WWII) of wrecks (C) samples taken per wreck site and sediment type of the wreck site.
The ages of the wrecks varied, with two dating back to World War I (HMS Nasturtium, 1916, and SS Polynesien, 1918), and the remaining eight from World War II (all lost in 1942). The construction materials and countries of origin of these wrecks were diverse, including steel, wood, and aluminium, and originated from Britain, France, Poland, and Germany (Figure 1). The seabed compositions also varied; the majority of the wrecks (eight out of ten) rested on a coarse sand seabed, one on fine sand (Xlighter 127), and one on a silt/clay seabed (HMS Maori) (Figure 1).
2.2 Microbial diversity of the surrounding sediment
The alpha diversity scores showed a significant difference across sediment type, frame type and oil leak presence/absence (Figure 2). In this study, frame type is characterised by its primary wreck material. Although these wrecks are made of multiple materials, the complete characterisation of the wreck component parts is a significant piece of study outside of the scope of this work and therefore a consequence, the primary wreck material has been reported on going forward. Oil leak presence in this study is characterised by bunker oil or heavy fuel oil. Due to decades underwater, the oil drums containing these oils are actively leaching into the surrounding environment. These oils include High-Molecular-Weight Hydrocarbons and Polycyclic Aromatic Hydrocarbons (PAHs) and also contain high amounts of heavy metals, either as a result of impurities in the refining process or as additives. Common heavy metals in these wrecks include lead, cadmium, nickel, arsenic, and vanadium.
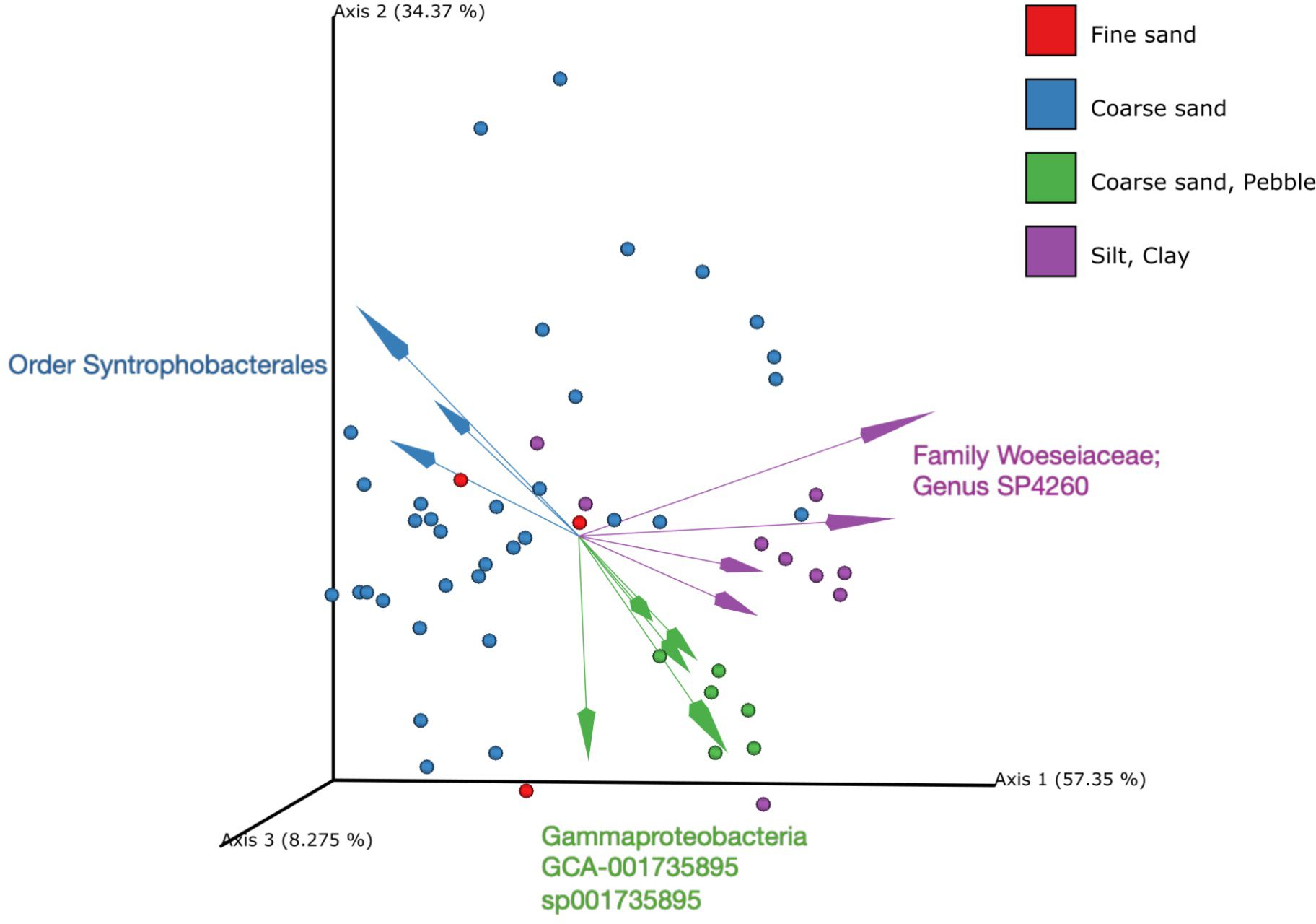
Figure 2. Taxonomic overview of the samples. Robust aitchison’s distance PCA with coloured dots representing sediment type and coloured arrows representing taxa. Sediment type is a significant factor in explaining composition (adonis, p = 0.001).
By adonis test, the primary diversity of the samples can be explained by sediment type (30.3% of variation) with frame (aluminium, steel, steel/wood, wood/alloy, None Wreck Site: 17.6%), oil leaks (Y/N: 8.9%), and wreck country of origin (American, British, France, Germany, Poland, None Wreck Site: 6.8%) following (Figure 2). Sediment type was classified according to size using the Wentworth scale (Appendix I); the smallest size sediment sampled was clay and the largest in this study was pebbles (Table 1).
The most abundant phyla in the sediment samples across all wreck sites was Pseudomonadota (Figure 2). There are some additional taxa to note that are in higher abundance at certain wreck sites. These include the Schnellboot S-31, which shows a higher abundance of Burkledeliareos & Staphylococcaceae (order). Additionally, the Ju88 North displays a higher abundance of Lokiarchaeota across five of the six sediment samples taken at this wreck (Appendix II).
Sediment types are differentiated primarily by the ratio of Woeseiaceae genus SP4260 to Gammaproteobacteria genus GCA-001735895 and the ratio between Woeseiaceae genus SP4260 to Syntrophobacterales. Woeseiaceae is a key species driving variation across the sediment types and is compared against Gammaproteobacteria genus GCA-001735895 and Syntrophobacterales as references to normalise its abundance across samples (Figure 3).
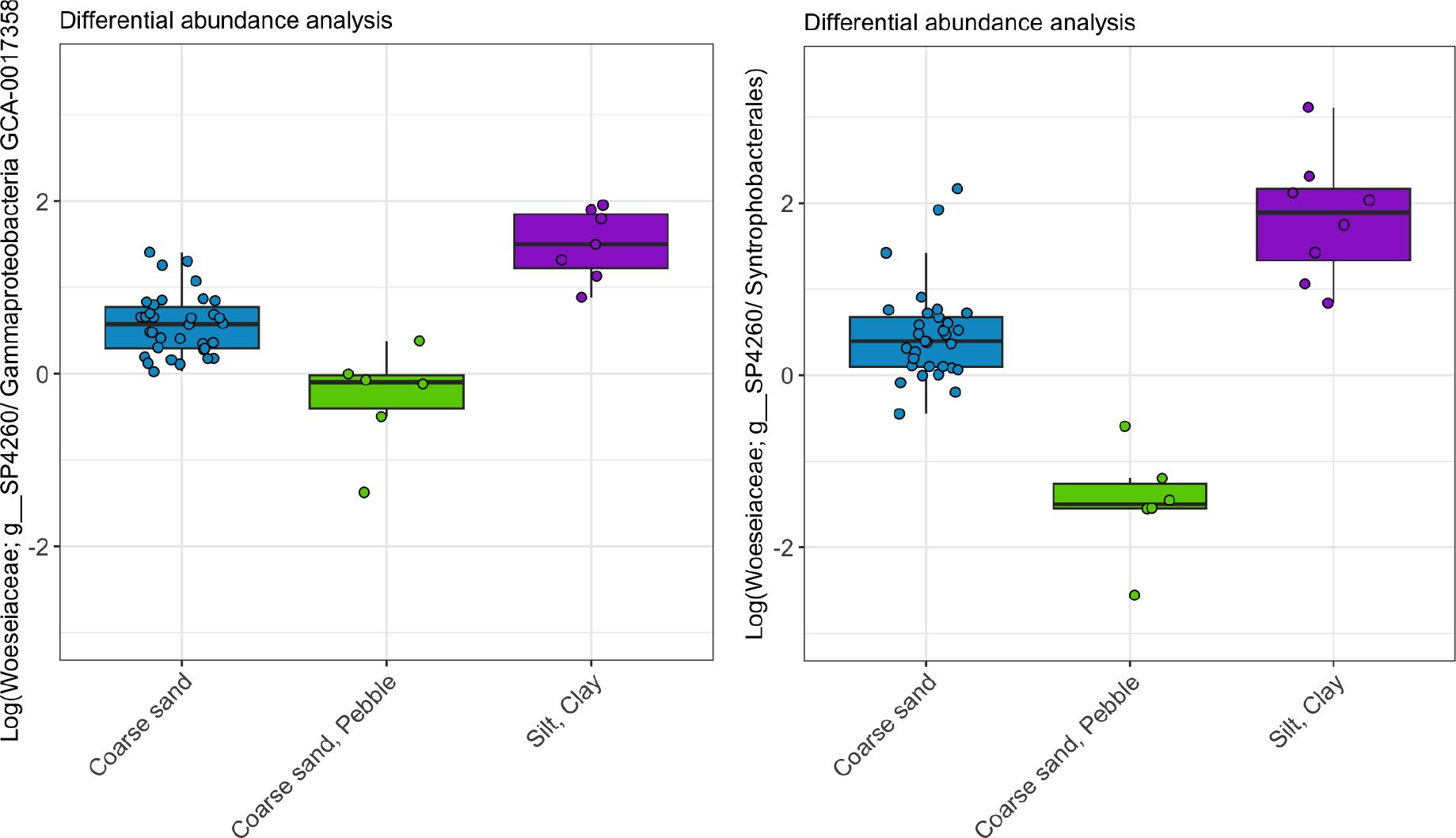
Figure 3. Differential abundance analysis of key species driving variation across the different sediment types. Left plot displays the log-ratio of Woeseiaceae genus SP4260 to Gammaproteobacteria genus GCA-001735895 for each sample, separated by sediment type. All sediment types are significantly different from one another (TukeyHSD, p < 0.001). The right plot in Figure 3 displays the log-ratio of Woeseiaceae genus SP4260 to Syntrophobacterales for each sample, separated by sediment type. All sediment types are significantly different from one another (p < 0.001).
Overall, Silt/Clay has a higher ratio of Woeseiaceae while Coarse sand/pebble has a lower ratio of Woeseiaceae (Figure 3). Coarse sand falls in between these two sediment types and fine sand was not included due to low sample numbers. ANOVA tests show that each sediment type is significantly different from one another with respect to these ratios (Woeseiaceae genus SP4260 to Gammaproteobacteria genus GCA-001735895, p values < 0.001; Woeseiaceae genus SP4260 to Syntrophobacterales, p values < 0.001).
2.3 Microbial diversity in ‘Coarse Sand Sediment’
By Robust aitchison’s distance PCA within the sediment type of ‘Coarse Sand’, the primary diversity of the samples can be explained by frame (Steel, Wood/Steel, Aluminium & Wood/Aluminium: 38.2% of variation) with country of origin (American, British, France, Germany, Poland: 15.9%) and oil leak (Y/N: 5.4%) following (Figure 4).
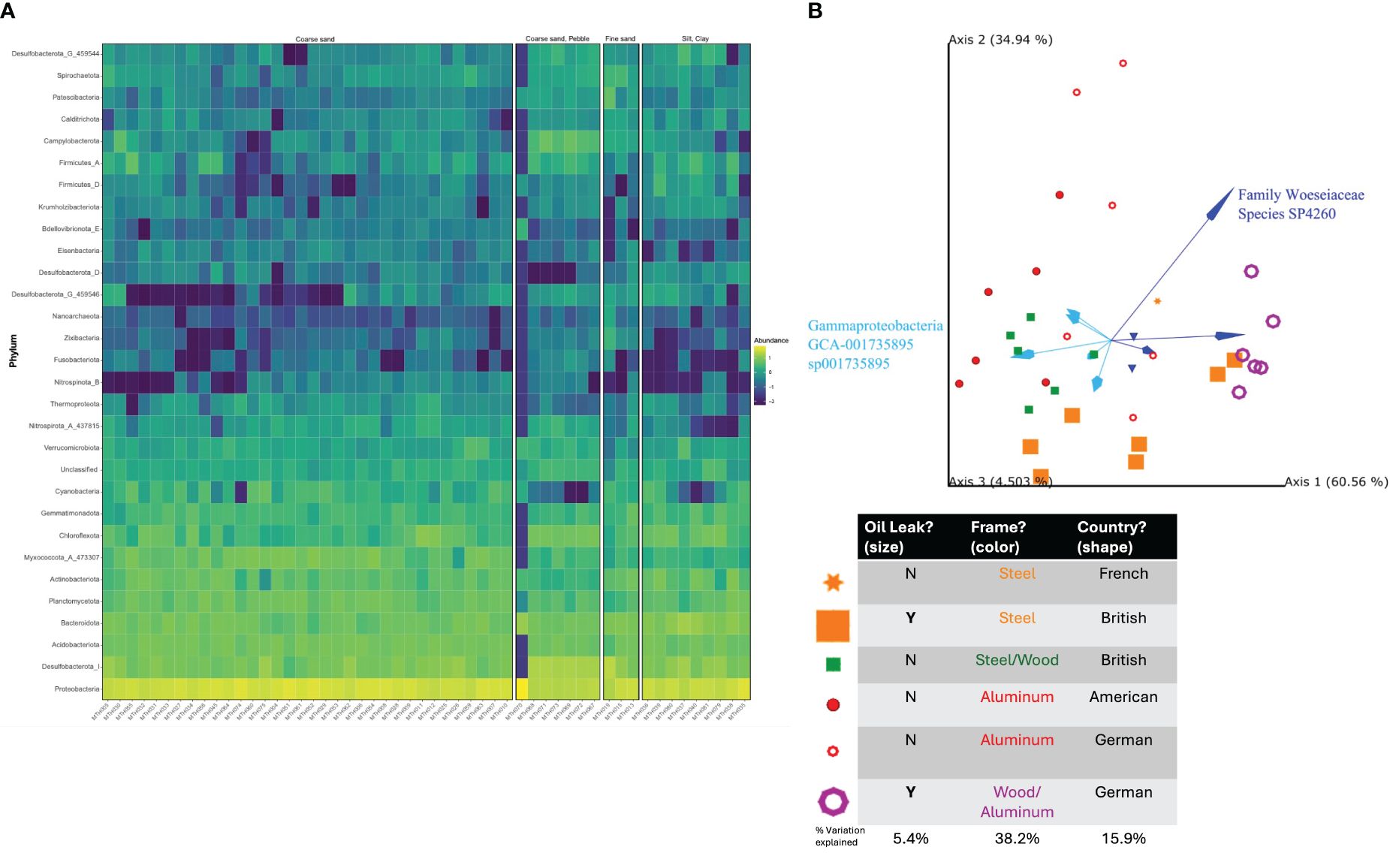
Figure 4. (A) Heatmap of most abundant phyla by sediment type. (B) Robust aitchison’s distance PCA of taxonomic diversity across factors of wreck frame type, country origin and oil leaks presence, within the coarse sand sample types.
2.4 Chemical content of the sediment
Differences in the chemical composition of the sample could be largely explained by the differences in sediment type (Figure 5A). The sediment samples were sent for XRF analysis and it was hypothesised that the presence of a wreck would influence sediment chemical signature from this analysis. However, no significant associations were found between the wreck frame type, the age of the wreck (WWI or WWII), and the presence of UXO or coal on a given sediment sample’s chemical composition (Figure 5C).
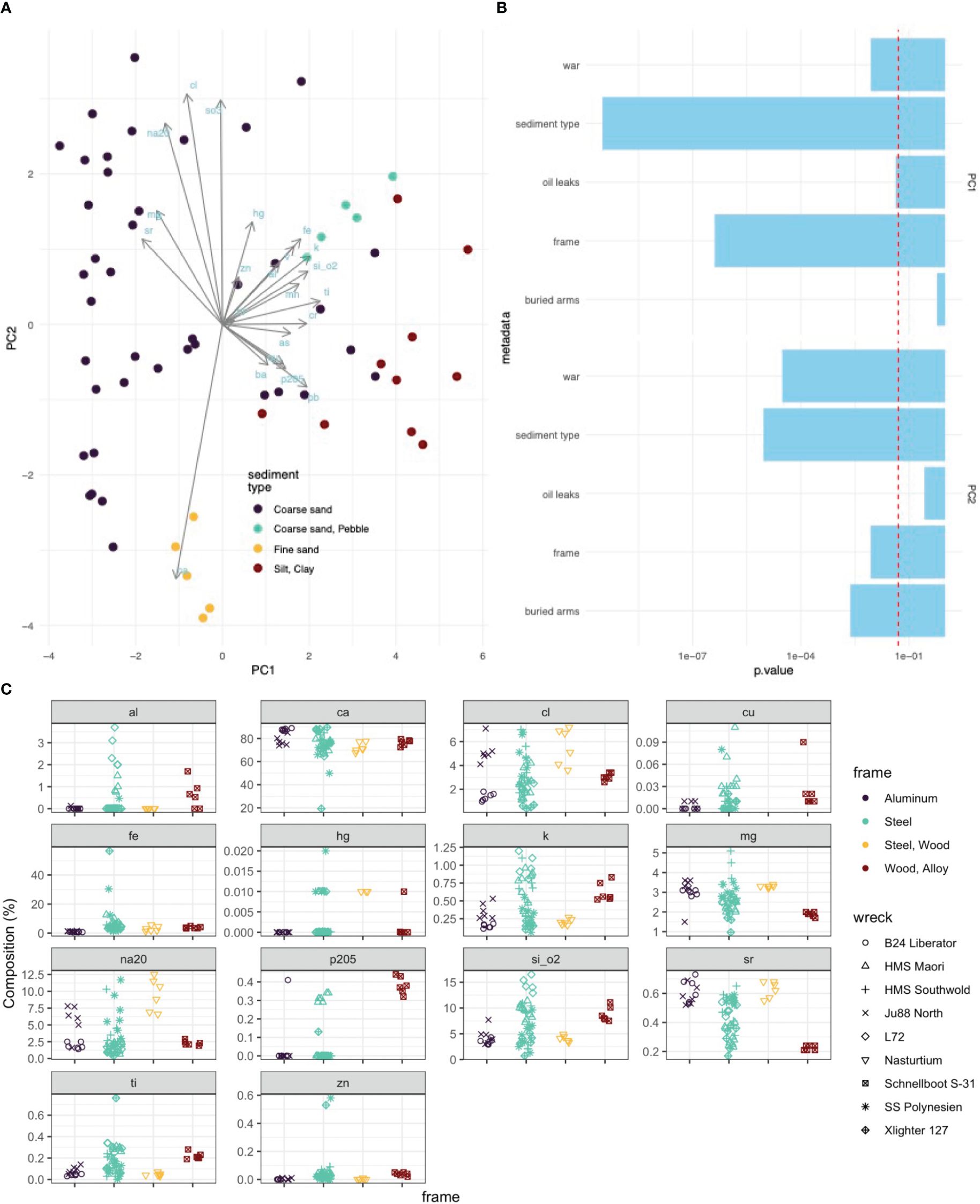
Figure 5. Chemical composition of the samples. (A) PCA (B) ANOVA of the categorical variables versus the two primary principle components. (C) Chemical composition data from XRF analysis of key elements associated with wreck material.
Although the frame type did not show a systematic and significant effect on the chemical analysis of the surrounding sediment, there is some evidence that leaching occurred. Specifically, samples from two of the steel-framed wrecks, the SS Polynesien and the Xlighter 127, exhibited iron signature spikes of up to 60% in some of the sediment samples (Figure 5C). Interestingly, this was not observed across all the samples taken from these wreck sites, suggesting that a wreck’s high iron content is not in general a confounding characteristic affecting the sediment in an entire wreck site. In contrast, the predominantly wood and aluminium-framed wrecks consistently showed much lower iron percentages, around 2-5% (Figure 5B).
It was anticipated that this same heightened signature might be observed for aluminium at the sites of aluminium-framed wrecks, however this was not observed (Figure 5C).
3 Discussion
3.1 The sediment microbiome shows indication of wreck influence
Although across all wreck types, sediment grain size is the biggest driver of microbial diversity, distinct and quantifiable influences from a wreck’s features on the surrounding environment were observed. Over a third of the variation in microbial diversity recorded in the ‘Coarse Sand’ samples is attributed to wreck frame type.
Wooden wrecks often introduce more organic material into the marine environment, which serves as a good substrate for microbial colonisation and increases the diversity of microbial communities (Hampel et al., 2022; Moseley et al., 2022). The type of wood can also influence the diversity of microbes, with oak often supporting the highest. Diversity scores associated with mahogany wrecks are less well known. In this study, the wrecks with wooden planking on their frames (Schnellboot S-31), or with wooden elements incorporated into their structure (HMS Nasturtium) presented differing taxa to the other wrecks whose frames wholly consisted of steel or aluminium.
Steel and aluminium wrecks are often seen to influence microbial diversity in an adverse nature compared to wood as these metal framed wrecks do not provide organic substrates that would lead to productive biofilm growth. Instead, microbial communities on these frame types are often influenced by corrosion processes and the release of metal ions, which can sometimes select for metal-tolerant and metal utilising microbes (Price et al., 2020). Across the majority of steel and aluminium framed wrecks we observed high abundances of Pseudomonadota and Cyanobacteria, both including taxa that can tolerate and interact with these metals, playing crucial roles in the biogeochemical cycling of metals in these environments.
When focusing on Schnellboot S-31, a higher abundance of a strain of bacteria degrading polyaromatic hydrocarbons (PAHs), Burkholderia (Kim et al., 2003), was recorded in the sediment samples. This correlates with the presence of oil leaks and UXO on this wreck. Staphylococcaceae was also recorded in higher abundance at this wreck compared to other wrecks sampled. This species is known for its heavy metal tolerance and resistance (Dissanayake et al., 2023), which could be due to the corroded metal frame type of the Schnellboot S-31.
The archaea group Lokiarchaeota at Ju88 - North was recorded across 4 of samples at this wreck. Lokiarchaeota, a group within the Asgard archaea, have been identified in various extreme environments, including deep-sea sediments near hydrothermal vents (Spang et al., 2015) and other marine environments (Liu et al., 2012). Their presence at wreck sites may be due to the unique conditions that influence microbial communities differently than natural seabed environments, emphasising a high ecological plasticity (Yin et al., 2021). The Ju88 is one of the most northern wreck sites and sits below 55m with an aluminium frame: these factors indicate conditions that Lokiarchaeota thrive in: anoxic, higher pressure and a slightly more nutrient-poor but chemically rich environment compared to some of the shallower wrecks sampled.
Research has shown that shipwrecks which consist of frames made out of mixed materials (of wood and metal) significantly influence the dispersal of microbial biofilms, including archaea. The study of wooden wrecks in the Gulf of Mexico highlighted how these structures can increase microbial richness and alter community composition, suggesting similar impacts might be observed with Lokiarchaeota at metal wreck sites (Moseley et al., 2022).
3.2 The influence of grain size on microbial diversity
A clear difference found in this study was the difference in microbial diversity across sediment types: thirty percent of the primary bacterial diversity could be explained by sediment type across all wreck depths and frame type. This is in keeping with previous literature which has highlighted that the influence of sediment grain size on microbial diversity in marine sediments is often significantly influenced by grain size due to the physical and chemical differences in the environment it creates (Lin et al., 2023). This can be due to the porosity, permeability and surface area of the sediments, which all can contribute to oxygen penetration, nutrient availability and habitat structure (Snelgrove et al., 2000).
When looking at the influence of grain size on microbial diversity from depths between 5 to 100 metres near wreck sites around Maltas’ coastlines, there is a notable gap that this study begins to address.
Wrecks in this study that lay on the fine-grained sediments (silt and clay samples) tend to retain more organic matter, creating more anoxic conditions (Franco et al., 2021) and we therefore record a higher abundance of anaerobic microorganisms (eg Desulfobacter and Pseudomonas).
In contrast, the wrecks laying on coarser grain size (such as the coarse sand samples), allow for better oxygen diffusion and therefore support a higher abundance of aerobic microbial communities (eg. cyanobacteria) (Lin et al., 2023).
3.3 Chemical characterization of sediments
Previous studies have shown that wrecks that have been present on the seabed for over 60 years leach micropollutants into the sediment, from both coal and oil stores as aromatic compound content and from the wreck itself in the form of heavy metals (Van Landuyt et al., 2022). Although this study does not conclusively rule out the influence of these wrecks on seabed chemistry, it is important to consider the following: (1) XRF is a broad analytical technique and specific chemical signatures may require more targeted follow-up analyses; (2) more comparisons with matched samples are necessary for a comprehensive understanding.
To further determine the impact of wrecks on chemical signatures in the sediment, additional samples collected in a radial pattern at increasing distances from the wrecks would be necessary.
4 Conclusions
In conclusion, this study demonstrated that the local microbial communities of marine sediments around World War I and World War II ship and aeroplane wrecks are influenced by a number of varying factors associated with each wreck. Although the sediment type that the wreck rests on is the main driver of microbial diversity, the data show that bacteria are influenced by the frame composition of the wreck, the wreck’s country of origin & the presence of oil leaks.
To investigate further the full spatial impact that wrecks are having on the local marine microbial ecology, radial sampling (distance away from wreck) and more in depth chemical analysis (to analyse the presence of PAHs and/or heavy metals in the sediment) would be required.
From this initial study of nine wreck sites around Malta & Gozo, it can be shown that across a multitude of wreck characteristics, the frame type and country of origin, along with a high influence from sediment type, played the most significant role in surrounding wreck microbial diversity that wrecks are influencing diversity at the lowest trophic level. Despite the fact that all sites in this study are within a limited geographical area, this study shows the potential for diversity even within a small area and continues to highlight the cultural and biological importance of archaeological ship and aeroplane wrecks as an integral part of Malta’s underwater cultural heritage.
5 Materials and methods
5.1 Wreck selection and sampling
Based on the documentation of wreck sites performed by Heritage Malta (Figure 6), ten wreck sites were chosen to be sampled (Figure 7) over a 7 month period (13th March 2023 - 18th October 2023). For this study, 77 samples were collected ranging from depths of 5 - 100m deep, with 5 of these wreck sites containing UXO. The wrecks also varied by year lost, hull type, surrounding sediment composition, coal storage and oil leaks. These characteristics and features were noted in an effort to understand what drivers are influencing the microbial diversity and chemical data of the sediments surrounding the wrecks.

Figure 6. Photographs of rebreather divers collecting sediment samples from around two of the wreck sites included in this study (left: Schnellboot S-31 at 65m, right: B24 Liberator at 54m).

Figure 7. An example of sediment samples taken from HMS Nasturtium at the bow, stern, port and starboard locations, 2m away from the wreck hull. The location of the coal storage and buried UXO on the wreck was identified by the Underwater Cultural Heritage Unit prior to sampling for this study.
Six to eight sampling points were decided at each wreck, taken 2m away from the wreck hull at the port, starboard, stern and bow by a team of rebreather divers. At each sampling point, two falcon tubes of surface sediment (2cm²) were collected at a sampling depth of approximately 3cm. Once the samples were returned to the surface and onboard, the samples were frozen at -20°C prior to processing for microbial and chemical analysis.
5.1.1 Wrecks included in the study
A total of ten wreck sites were sampled during this study (Table 2), which included a variety of factors considered potentially relevant for identifying drivers of microbial diversity (eg. depth of each wreck, frame type and origin country of wreck). Visual inspections by divers recorded the presence of UXO on 5 of these wrecks and that 3 of these wrecks contained leaks from oil barrels sunk with the wreck. The majority of these wrecks were lost in WWII (in 1942), however two of the wrecks included in this study were lost in WWI (the HMS Nasturtium & the SS Polynesien). A brief description of each wreck will be presented in the Appendix IIII.
5.2 DNA extraction and 16S amplicon sequencing
Subsamples of approximately 20g were taken from -20°C frozen sediment samples. DNA extraction was performed using Qiagen PowerMax kits, as per the producer’s protocol. The DNA was cleaned and size selection applied with SPRI beads.
The 16S rRNA V3-V4 region was amplified with primers 341F (Herlemann ISME J. 2011) and 785R (Klindworth Nucleic Acids Res. 2013) using Phusion HF polymerase. The DNA was sequenced on an Illumina MiSeq on PE300 cycles to achieve a minimum of 30,000 read pairs per sample. Fastqs were quality controlled with phiX removal and primer trimming using cutadapt version 1.12.
5.3 Chemical, pH, conductivity, TDS & salinity analysis
5.3.1 XRF
X-ray fluorescence (XRF) analysis was used to identify the elemental composition of the samples. Samples were dried in a controlled environment to prevent contamination and moisture interference and then homogenised using a mechanical grinder to obtain a fine powder, ensuring uniform particle size. Approximately 5 grams of each powdered sample were then pressed into a pellet using a hydraulic press with a force of 10 tons for 30 seconds. This process ensured the creation of stable and uniform pellets, suitable for XRF analysis.
Samples were analysed using an PANalytical X-Ray Fluorescence Epsilon 3XL Energy Dispersive Spectrometer. Elemental relative abundance was determined by analysing the characteristic X-ray emissions from the sample and the obtained spectra were processed to identify and quantify the presence of various elements within the samples.
5.3.2 pH, conductivity, TDS & salinity analysis
The Palintest Sensor 7000 multiparameter probe was used to measure pH, TDS, conductivity, and salinity. The probe was calibrated before each use with standard solutions. pH calibration was performed using standard buffer solutions at pH 4.0, 7.0, and 10.0. For TDS, conductivity, and salinity, calibration standards with known concentrations were used to ensure the instrument’s accuracy across the expected range of measurements.
Each sediment sample was shaken with di-ionised water (20ml for pH, 50ml for TDS, conductivity, and salinity) to ensure homogeneity before measurement. The probe was rinsed with distilled water and then submerged into the sample. The following parameters were recorded: pH - the probe measured the hydrogen ion concentration, providing an immediate readout of the sample’s pH level; Total Dissolved Solids (TDS) - the TDS was measured by the probe’s ability to detect the total concentration of dissolved ions in the water, reported in mg/L; Electrical Conductivity - conductivity measurements, recorded in µS/cm; Salinity - based on the conductivity measurements, with results expressed in parts per thousand (ppt).
5.4 Data analysis
5.4.1 Bioinformatics analysis
Bioinformatics analysis of raw 16S amplicon sequence reads was conducted using the Quantitative Insights into Microbial Ecology 2 (QIIME2) platform (Bolyen et al., 2019). Sequences were trimmed to 250 base pairs and quality controlled using DADA2 (Callahan et al., 2016) for paired-end sequences at default settings. The resulting feature table (output from DADA2) contains amplicon sequence variants. Taxonomy was classified using the greengenes2 database [McDonald et al., 2024 (https://www.nature.com/articles/s41587-023-01845-1)].
Raw 16S sequences have been deposited to EBI and are available under ENA number PRJEB78985 (https://www.ebi.ac.uk/ena).
5.4.2 Statistical data analysis
Analysis and visualization of the sample metadata (including location, chemical composition, and so on) were carried out in R (R Core Team, 2024) using packages including leaflet (Graul and Graul, 2016), tidyverse (Wickham et al., 2019). PCA and ANOVA of the chemical composition were carried out in R using prcomp. Data were first cleaned so that only selected chemicals which were detected in samples (Al, As, Ba, Ca, Cd, Cl, Co, Cr, Cu, Fe, Hg, K, Mg, Mn, Na20, Ni, P205, Pb, SO3, SiO2, Sr, Ti, V, Zn, and Zr) were analysed, and NAs were set to zero. Principal Component Analysis (PCA) was performed on cleaned and scaled data, and anova was carried out with base R functions. Abundance heatmaps were generated with the qiime2R package in R (Bolyen et al., 2019) using ggplot. The OTU tables were summarised on the phylum level before plotting.
Beta diversity analysis was conducted with the QIIME2 DEICODE plugin, which performs a robust calculation of Aitchison distance (RPCA) [Martino et al., 2019 (doi:10.1128/mSystems.00016-19)]. Using a robust Aitchison’s distance PCA allowed us to accurately analyse and interpret the compositional data, addressing the specific statistical challenges posed by such data types which are inherently dominated by zero values. This methodology not only improved the robustness of our findings but also ensured that the principal components were reflective of the true underlying patterns in the dataset. The distances between samples were plotted in a Principal Component’s Analysis biplot using the Emperor visualisation tool (https://doi.org/10.1186/2047-217X-2-16, https://doi.org/10.1016/j.chom.2016.12.009), with arrows (loadings) corresponding to important features driving the differences between samples. Adonis with 999 permutations was run in QIIME2 to determine the significance of sediment type, oil leaks, frame type and country source in differentiating community composition.
Differential abundance analysis was performed on the taxa highlighted in the RPCA biplot. Qurro (https://doi.org/10.1093/nargab/lqaa023) was used to visualise log ratios of Woeseiaceae genus SP4260: Gammaproteobacteria genus GCA-001735895 and Woeseiaceae genus SP4260: Syntrophobacterales. These values were re-plotted as boxplots in R using ggplot (https://ggplot2.tidyverse.org) and ANOVA (Chambers et al., 2017) tests were run to determine statistical differences between sediment types.
A cut-off of p-value < 0.05 was used to test for significant differences in all statistical tests, including adonis with Benjamini-Hochberg adjustment for multiple comparisons and ANOVA with Tukey’s HSD (Miller and Miller, 1981; Yandell, 1997).
Further analysis and visualization was carried out on R (R Core Team, 2024) using packages including the tidyverse (Wickham et al., 2019), phyloseq (McMurdie and Holmes, 2013), and leaflet (Graul and Graul, 2016).
We also applied the robust Aitchison’s distance PCA to solely the ‘Coarse Sand’ dataset and visualised the biplot in Emperor. Adonis with 999 permutations was run in QIIME2 to determine the significance of oil leaks, frame type and country source in differentiating community composition amongst just the coarse sand samples.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ebi.ac.uk/ena, PRJEB78985.
Author contributions
EB: Conceptualization, Data curation, Methodology, Project administration, Supervision, Writing – original draft, Writing – review & editing. NG: Conceptualization, Funding acquisition, Supervision, Writing – original draft. EK: Data curation, Formal analysis, Methodology, Visualization, Writing – original draft. JM: Data curation, Writing – review & editing. NP: Data curation, Formal analysis, Methodology, Visualization, Writing – original draft. MS: Conceptualization, Project administration, Supervision, Writing – review & editing. PO: Data curation, Formal analysis, Funding acquisition, Supervision, Visualization, Writing – original draft. TG: Conceptualization, Methodology, Project administration, Supervision, Writing – review & editing.
Funding
The author(s) declare that no financial support was received for the research, authorship, and/or publication of this article.
Acknowledgments
We would like to express our gratitude to the Heritage Malta dive team for their invaluable assistance throughout this project, whose expertise in collecting samples from these depths have made this study possible. We would also like to express our thanks to the broader Basecamp Research team for their continuous support and collaboration. We would also like to thank the Superintendence of Cultural Heritage for its continuous work in ensuring the protection and accessibility of Malta’s cultural heritage sites, as defined by the Cultural Heritage Act. Lastly, we would like to extend our appreciation to Tom Dietz for his thorough review and insightful feedback - we are thankful for your time and dedication. This work was carried out under IRCC permit ABSCH-IRCC-MT-263440-2 from the Maltese Plant Protection Directorate - we would like to express our gratitude to Louis Fresta of the PPD for support in this process.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2024.1480265/full#supplementary-material
References
Asner G. P., Giardina S. F., Balzotti C., Drury C., Hopson S., Martin R. E. (2022). Are sunken warships biodiversity havens for corals? Diversity 14, 139. doi: 10.3390/d14020139
Bolyen E., Rideout J. R., Dillon M. R., Bokulich N. A., Abnet C. C., Al-Ghalith G. A., et al. (2019). Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 37 (8), 852–857.
Callahan B. J., McMurdie P. J., Rosen M. J., Han A. W., Johnson A. J. A., Holmes S. P. (2016). DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 13 (7), 581–583.
Cammen L. M. (1982). Effect of particle size on organic content and microbial abundance within four marine sediments. Mar. Ecol. Prog. Ser. 9, 273–290. doi: 10.3354/meps009273
Chambers J. M., Freeny A. E., Heiberger R. M. (2017). “Analysis of variance; designed experiments,” in Statistical Models in S. Eds. Chambers J. M., Hastie T. J. (Routledge: Wadsworth & Brooks/Cole).
Church R. A., Warren D. J., Irion J. B. (2009). Analysis of deepwater shipwrecks in the Gulf of Mexico: artificial reef effect of six world war II shipwrecks. Oceanography 22, 50–63. doi: 10.5670/oceanog.2009.38
Connell S. D. (2000). Floating pontoons create novel habitats for subtidal epibiota. J. Exp. Mar. Biol. Ecol. 247, 183–194. doi: 10.1016/S0022-0981(00)00147-7
Cullimore D., Johnston L. (2008). Microbiology of concretions, sediments and mechanisms influencing the preservation of submerged archaeological artifacts. Int. J. Hist. Archaeol. 12, 120–132. doi: 10.1007/s10761-008-0045-y
Dissanayake D. M. D. C. D., Kumari W. M. N. H., Chandrasekharan N. V., Wijayarathna C. D. (2023). Isolation of heavy metal-resistant Staphylococcus epidermidis strain TWSL_22 and evaluation of heavy metal bioremediation potential of recombinant E. coli cloned with isolated cadD. FEMS Microbiol. Lett. 370, fnad092. doi: 10.1093/femsle/fnad092
Franco N. R., Giraldo M.Á., López-Alvarez D., Gallo-Franco J. J., Dueñas L. F., Puentes V., et al. (2021). Bacterial composition and diversity in deep-sea sediments from the Southern Colombian Caribbean Sea. Diversity 13. doi: 10.3390/d13010010
Gambin T., Bravo-Morata Rodríguez A., Sausmekat M. (2021a). From discovery to public consumption: the process of mapping and evaluating underwater cultural heritage in Malta. Heritage 4, 2732–2745. doi: 10.3390/heritage4040154
Gambin T., Chemisky B., Drap P. (2018). Exploring the Phoenician shipwreck off Xlendi bay, Gozo. A report on methodologies used for the study of a deep-water site. Underwater Technol. Int. J. Soc. Underwater. 35, 71–86. doi: 10.3723/ut.35.071
Gambin T., Hyttinen K., Sausmekat M., Wood J. (2021b). Making the invisible visible: underwater Malta—A virtual museum for submerged cultural heritage. Remote Sens 13, 1558. doi: 10.3390/rs13081558
Gambin T., Sausmekat M., Kovacevic D. (2021c). The innovative and state of the art public access management of Malta’s underwater cultural heritage. Heritage 4, 3365–3381. doi: 10.3390/heritage4040187
Graul C., Graul M. C. (2016). Package ‘leafletR’. Interactive Web-Maps Based on the Leaflet JavaScript Library. Available online at: https://github.com/chgrl/leafletR (accessed April 1, 2016).
Hamdan L. J., Hampel J. J., Moseley R. D., et al. (2021). Deep-sea shipwrecks represent island-like ecosystems for marine microbiomes. ISME J. 15, 2883–2891. doi: 10.1038/s41396-021-00978-y
Hamdan L. J., Salerno J. L., Reed A., Joye S. B., Damour M. (2018). The impact of the Deepwater horizon blowout on historic shipwreck-associated sediment microbiomes in the Northern Gulf of Mexico. Sci. Rep. 8, 9057. doi: 10.1038/s41598-018-27350-z
Hampel J. J., Moseley R. D., Mugge R. L., Ray A., Damour M., Jones D., et al. (2022). Deep-sea wooden shipwrecks influence sediment microbiome diversity. Limnol. Oceanogr. 67, 482–497. doi: 10.1002/lno.12008
Head I., Jones D., Röling W. (2006). Marine microorganisms make a meal of oil. Nat. Rev. Microbiol. 4, 173–182. doi: 10.1038/nrmicro1348
Kim T. J., Lee E. Y., Kim Y. J., Cho K. S., Ryu H. W. (2003). Degradation of polyaromatic hydrocarbons by Burkholderia cepacia 2A-12. World J. Microbiol. Biotechnol. 19, 411–417. doi: 10.1023/A:1023998719787
Lengkeek W., Coolen J. W. P., Gittenberger A., Schrieken N. (2013). Ecological relevance of shipwrecks in the North Sea. Nederlandse Faunistische Mededelingen 41, 49–57.
Lin J., Zhou X., Lu X., Xu Y., Wei Z., Ruan A. (2023). Grain size distribution drives microbial communities vertically assemble in nascent lake sediments. Environ. Res. 227, 115828.
Liu L. Y., Wang J. Z., Wei G. L., Guan Y. F., Zeng E. Y. (2012). Polycyclic aromatic hydrocarbons (PAHs) in continental shelf sediment of China: Implications for anthropogenic influences on coastal marine environment. Environ. pollut. 167, 155–162.
Martino C., Morton J. T., Marotz C. A., Thompson L. R., Tripathi A., Knight R., et al. (2019). A novel sparse compositional technique reveals microbial perturbations. MSystems 4 (1), 10–1128.
McDonald D., Jiang Y., Balaban M., Cantrell K., Zhu Q., Gonzalez A., et al. (2024). Greengenes2 unifies microbial data in a single reference tree. Nat. Biotechnol. 42 (5), 715–718.
McMurdie P. J., Holmes S. (2013). phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PloS One 8, e61217. doi: 10.1371/journal.pone.0061217
Meyer-Kaiser K. S., Mires C. H., Haskell B. (2022). Invertebrate communities on shipwrecks in Stellwagen Bank National Marine Sanctuary. Mar. Ecol. Prog. Ser. 685, 19–29. doi: 10.3354/meps13987
Miller R. G., Miller R. G. (1981). Normal univariate techniques. Simultaneous statistical inference, 37–108.
Moseley R. D., Hampel J. J., Mugge R. L., Hamdan L. J. (2022). Historic wooden shipwrecks influence dispersal of deep-sea biofilms. Front. Mar. Sci. 9. doi: 10.3389/fmars.2022.873445
Mugge R. L., Brock M. L., Salerno J. L., Damour M., Church R. A., Lee J. S., et al. (2019). Deep-sea biofilms, historic shipwreck preservation and the deepwater horizon spill. Front. Mar. Sci. 6. doi: 10.3389/fmars.2019.00048
Paxton A. B., McGonigle C., Damour M., Holly G., Caporaso A., Campbell P., et al. (2024). Shipwreck ecology: Understanding the function and processes from microbes to megafauna. BioScience 741 (1), 12–24. doi: 10.1093/biosci/biad084
Paxton A. B., Riley T. N., Steenrod C. L., et al. (2023). What evidence exists on the performance of nature-based solutions interventions for coastal protection in biogenic, shallow ecosystems? A systematic map protocol. Environ. Evid. 12, 11. doi: 10.1186/s13750-023-00303-4
Price K. A., Garrison C. E., Richards N., Field E. K. (2020). A shallow water ferrous-hulled shipwreck reveals a distinct microbial community. Front. Microbiol. 11. doi: 10.3389/fmicb.2020.01897
R Core Team (2024). R: A language and environment for statistical computing (Vienna, Austria: R Foundation for Statistical Computing). Available at: https://www.R-project.org/.
Rogowska J., Namieśnik J. (2013). “Environmental Risk Assessment of WWII Shipwreck Pollution,” in Wastewater Reuse and Management.. Eds. Sharma S., Sanghi R. (Dordrecht: Springer Netherlands) doi: 10.1007/978-94-007-4942-9_16
Snelgrove P. V. R., Austen M. C., Boucher G., Heip C., Hutchings P. A., King G. M., et al. (2000). Linking Biodiversity Above and Below the Marine Sediment–Water Interface: The organisms living on the ocean floor are linked to those living in the ocean above, but whether or how the biodiversity in these two realms is linked remains largely unknown. BioScience 50, 1076–1088. doi: 10.1641/0006-3568(2000)050[1076:LBAABT]2.0.CO;2
Spang A., Saw J. H., Jørgensen S. L., Zaremba-Niedzwiedzka K., Martijn J., Lind A. E., et al. (2015). Complex archaea that bridge the gap between prokaryotes and eukaryotes. Nature 521, 173–179. doi: 10.1038/nature14447
Thomas G., Bolam S., Brant J., Brash R., Goodsir F., Ellis C., et al. (2021). Evaluation of polycyclic aromatic hydrocarbon pollution from the HMS royal oak shipwreck and effects on sediment microbial community structure. Front. Mar. Sci. 8. doi: 10.3389/fmars.2021.650139
Van Landuyt J., Kundu K., Van Haelst S., Neyts M., Parmentier K., De Rijcke M., et al. (2022). 80 years later: Marine sediments still influenced by an old war ship. Front. Mar. Sci. 9, 1017136.
Wickham H., Averick M., Bryan J., Chang W., McGowan L. D., François R., et al. (2019). Welcome to the tidyverse. J. Open Source Software 4, 1686. doi: 10.21105/joss.01686
Wolska L., Mechlińska A., Rogowska J., Namieśnik J. (2012). Sources and fate of PAHs and PCBs in the marine environment. Crit. Rev. Environ. Sci. Technol. 42 (11), 1172–1189. doi: 10.1080/10643389.2011.556546
Keywords: wreck, microbial, monitoring, diversity, ecosystem, Malta
Citation: Bolton E, Greenhalgh N, Kunselman E, Mifsud J, Patel N, Sausmekat MP, Oldach P and Gambin T (2024) Deepening the understanding of wreck ecology: a comparative study of marine sediment microbiomes across 10 Maltese wreck sites. Front. Mar. Sci. 11:1480265. doi: 10.3389/fmars.2024.1480265
Received: 26 August 2024; Accepted: 19 November 2024;
Published: 06 December 2024.
Edited by:
Jin Zhou, Tsinghua University, ChinaReviewed by:
Dr. Sujata Dabolkar, Government College of Arts, Science and Commerce, IndiaErin Field, East Carolina University, United States
Copyright © 2024 Bolton, Greenhalgh, Kunselman, Mifsud, Patel, Sausmekat, Oldach and Gambin. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Emma Bolton, ZW1tYUBiYXNlY2FtcC1yZXNlYXJjaC5jb20=
 Emma Bolton
Emma Bolton Nadine Greenhalgh
Nadine Greenhalgh Emily Kunselman2
Emily Kunselman2 Jasmine Mifsud
Jasmine Mifsud Neem Patel
Neem Patel Maja Pace Sausmekat
Maja Pace Sausmekat Phoebe Oldach
Phoebe Oldach Timmy Gambin
Timmy Gambin