
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Mar. Sci. , 20 June 2024
Sec. Marine Biology
Volume 11 - 2024 | https://doi.org/10.3389/fmars.2024.1404183
Large yellow croaker (Larimichthys crocea), the most developing fish of China, suffering from both biotic and abiotic stressors. A genome-wide study was performed for the first time to investigate the roles of Toll-like receptor genes in large yellow croaker (LcTLRs) working in hypoxia response and Aeromonas hydrophila infection. 12 TLR genes were identified and annotated, and phylogenetic tree, analysis of structure and motifs demonstrated that TLRs were highly conserved. To further investigate the LcTLRs under environmental stress, expression pattern analysis of hypoxic response and Aeromonas hydrophila infection revealed that there were nine and six members of LcTLRs showing significant differential expression, indicating that they were concerned in hypoxia stress and disease responses. Meanwhile, their expression levels were validated utilizing qPCR. Taken together, a greater appreciation and understanding of TLRs function in the reaction to biotic and abiotic stress would ultimately lead to more efficiently environmental adaptation in large yellow croaker.
Large yellow croaker (Larimichthys crocea), the native species of East Asia, has the highest aquaculture yield among marine fish in China (257,683 tons in 2022) (MOA, 2023). However, the intensive net-cage inshore cultivation model, the degrading farming environment, outbreaks of various abiotic and biotic stressors like hypoxia and Aeromonas hydrophila infection, and other factors, had caused significant economic losses to the aquaculture industry (Ding et al., 2022; Zhang et al., 2022). Many previous studies had confirmed that Toll-like receptor genes played a vital role in innate immunity (Ni et al., 2022; Yao et al., 2023; Gao et al., 2024), however, research on their role in environmental adaptability was still in its early stages. Therefore, the research of biotic and abiotic environmental factors was on the front-page of L.crocea’s survival. This experiment would lay a foundation for the specific role of Toll-like receptor genes in the host environmental adaptation to resistance to hypoxic response and Aeromonas hydrophila infection.
Dissolved oxygen (DO), or molecular oxygen dissolved in water, is the primary source of oxygen for a variety of aquatic organisms. As L. crocea cultures grow and their densities rise, the culture environment degrades and the water exchange capacity eventually drops, leaving large areas with low DO levels. Marine ecosystems are more vulnerable to hypoxic stress than terrestrial environments are (Lee et al., 2024). Low dissolved oxygen levels have a negative impact on fish physiological and biochemical processes, including apoptosis (Liu et al., 2022b), energy metabolism (Jaworski et al., 2019), and other processes, which can result in a variety of illnesses and injuries (Shun et al., 2024). Reactive oxygen species (ROS) are produced by the organism from part of the oxygen taken up by fish respiration. A tiny percentage of ROS control gene expression and cell activity through their involvement in cell signaling pathways (Shi et al., 2024). According to Abdel-Tawwab et al. (2019), fish that experience oxidative stress due to excessive ROS production during hypoxia eventually undergo apoptosis and sustain tissue damage. Aquatic species’ growth, survival, behavior, immunity, and reproduction are all significantly impacted by such hypoxic environments. According to Ding et al. (2020), hypoxia in L.crocea causes a rise in mortality and is now the primary barrier to the growth of its sustainable mariculture. On the other hand, the breeding of L.crocea was threatened by bacterial diseases (Mu et al., 2018). Previous studies had shown that the diseased L. crocea had suffered a variety of severe illness symptoms after Gram-negative bacteria A. hydrophila infection (Chen et al., 2010; Mu et al., 2010; Zhang et al., 2022), as well as the verification of TLRs differential expression following an A. hydrophila challenge in common carp (Gong et al., 2017). Therefore, in order to allow large yellow croak culture population, exploit a better ecological opportunity under the stress of environmental factors such as hypoxia and A. hydrophila infection, researchers were committed to exploring the molecular mechanism of resistance regulation of TLR genes.
To date, studies have shown that TLRs bear the responsibility of playing important roles in innate immunity in organisms (Verma et al., 2024). Innate immune cells, such as dendritic cells (DCs), are activated when the immune system detects microorganisms through a variety of receptors known as pattern recognition receptors (PRRs), which are encoded by germ-line genes and recognize a wide range of microbial structures (Wang et al., 2024). Up to now, many PRRs have been found, including C-type lectin receptors, scavenger receptors (SR), intracellular receptors, lipopolysaccharides (LPS) and β-1, toll-like receptors (TLRs), and Down syndrome cell adhesion molecules (Dscam) (Tran et al., 2019). Among these PRRs, the role of TLRs in innate immune responses during evolution is thought to be highly conserved, usually acting as transmembrane receptors that recognized different microbial components and directly activated immune cells. Exposure of immune cells to the ligands of receptors can activate intracellular signaling cascades, and then rapidly induce the expression of a variety of overlapping and unique genes involved in inflammation and immune responses. After pathogen infection, pathogen-associated molecular patterns (PAMPs) are recognized, myeloid differentiation factor 88 (MyD88) and adaptors containing TIR domains are recruited by TLRs to induce interferon-beta (IFN-β) level (Dias et al., 2022; Ryan and O’Neill, 2023), thereby leading to the activation of protein 1 (AP-1) transcription factor and nuclear factor kappa-B (NF-κB) (Wang et al., 2017). This induces the production of pro-inflammatory cytokines, such as TNF-α and IL1b, to activate innate and adaptive immune responses (Vidya et al., 2018). Structurally, TLR consists of several domains, including N-terminal signaling domains, multiple extracellular leucine-rich repeat (LRRs) domains, transmembrane domains, and intracellular Toll/interleukin-1 receptor (TIR). The TIR domain functions as a protein scaffold, attracting downstream molecules and initiating signaling cascades that activate the expression of target genes, such as antimicrobial peptides, while the extracellular LRR domain recognizes the PAMP ligands of pathogenic microorganisms.
TLRs are a family of germline-encoded PRRs conserved in function. They are essential for recognizing PAMPs like flagellin, lipoprotein, nucleic acid, LPS, and lipoteichoic acid (Tran et al., 2019). For instance, bacterial flagellin stimulates TLR5, while double-stranded RNA (dsRNA) can activate TLR3 (Adams et al., 2024). Similar in location, structure, and function, TLR7 and TLR8 are recognized to detect single-stranded RNA (ssRNA) from viruses (Wallach et al., 2023). Furthermore, it has been discovered that TLR9 and TLR21 can both identify CpG DNA in fish and that they react preferentially to CpG DNA with distinct CpG (Yeh et al., 2013). Following ligand identification, TLR uses the Toll/Interleukin-1 receptor (TIR) domain and TIR-containing cohesive molecules, such Myd88 and other TIR-domain-containing convergent proteins, to initiate downstream signaling cascades and an immunological response (Sahoo et al., 2012). Activation of the TLR signaling pathway triggers the production of co-stimulatory molecules and inflammatory cytokines, which effectively trigger the host’s antiviral or antimicrobial immune responses (Li et al., 2022).
In order to carry out a thorough identification of the TLR family members in L. crocea, all TLR sequences of nine teleosts, including channel catfish (Ictalurus punctatus), fugu (Takifugu rubripes), grass carp (Ctenopharyngodon idella), Japanese flounder (Paralichthys olivaceus), medaka (Oryzias latipes), Nile tilapia (Oreochromis niloticus), rainbow trout (Oncorhynchus mykiss), spotted gar (Lepisosteus oculatus), spotted sea bass (Lateolabrax maculatus) and zebrafish (Danio rerio) were downloaded from NCBI (http://www.ncbi.nlm.nih.gov) and Ensembl (http://www.ensembl.org) databases (Yan et al., 2021). The non-redundant TLRs full-length coding sequences were utilized as queries to search against L. crocea genomic resources by TBLASTN and BLASTP, with an e-value threshold of e-value < e−10 to acquire the candidates, after the redundant and incomplete sequences were manually removed (El-Gebali et al., 2019). The conserved TLR domain was then confirmed and verified by submitting the putative TLR sequences to the Pfam and SMART databases (Letunic and Bork, 2018). The ExPASy ProtParam tool (http://web.expasy.org/protparam/) was utilized to assess the molecular weight, theoretical isoelectric point, and amount of amino acids (Duvaud et al., 2021). Ultimately, every potential TLR gene extracted from the L. crocea genome was renamed based on their homology to TLRs found in zebrafish, and then added to the GenBank database.
A phylogenetic tree was created using all of the amino acid sequences of TLR from L. crocea and other teleosts. Multiple sequence alignment was performed by ClustalW with default settings (Edgar, 2004). Using the JTT (Jones-Taylor-Thornton) matrix-based model, phylogenetic analysis was carried out through the MEGA 7 program and the Maximum Likelihood approach (Kumar et al., 2016). The Evolview website (https://evolgenius.info//evolview) was then used to show the phylogenetic tree (Zhang et al., 2012).
Using the Gene Structure Display Server (GSDS, http://gsds.cbi.pku.edu.cn) online analysis tool, the exon/intron structures were visualized by aligning the coding sequences with the corresponding genomic DNA sequences. To display the gene structure of L. crocea, including the length and position of the introns and exons, the gff file was uploaded to the GSDS program (GenBank No. GCA_000972845.2). The conserved protein motif was analyzed using the MEME website (http://meme-suite.org/tools/meme), with the motif number set to 10 and the other parameters left at default (Bailey et al., 2009). The TBtools program was then used to create the phylogenetic tree, conserved motifs, and gene structure (Chen et al., 2020).
A total of 300 healthy L. crocea were obtained and maintained in indoor tanks at the same conditions as the culture cages (25°C, salinity: 23–25, DO concentration: 7.8 ± 0.5 mg/L). The fish were fed compound feed twice a day, at 6:00 and 17:00, up until twelve hours before the trials began. Following a temporary raising period of 14 days, nitrogen gas was blasted into the tanks to conduct hypoxia-exposure studies. Using a DO meter (YSI, Canada), the appropriate concentration of dissolved oxygen was measured. In the control group, the DO concentration was 7.8 ± 0.5 mg/L. Over the course of ten minutes, the oxygen concentration in the tank was reduced in the experimental group from 7.8 ± 0.5 mg/L to 1.6 ± 0.2 mg/L. For the purpose of extracting RNA from the gills, heart, spleen, and head kidney at 0, 6, 24, and 48 hours following the hypoxic test, six subjects were chosen at random. To create templates for additional transcriptome analysis, equivalent molar ratios of RNA from two subjects were combined into one replicate at each time point, for a total of three repetitions. Raw sequencing read data had been deposited in the Sequence Read Archive (SRA) of NCBI under the BioProject accession numbers PRJNA574876 and PRJNA576086.
A total of 300 healthy L. crocea were acclimated for 2 weeks, and feeding was stopped one day before the experiment. In a preliminary experiment (DO 7.5–8.5 mg/L, 23 ± 0.5°C), the experimental group was injected intraperitoneally into 60 fish, with three experimental replicates per group, at the 24-hour half-lethal dose (LD50) of A. hydrophila, which was found to be 8 × 106 CFU·mL−1 (0.5 mL). The control group was injected with 0.5 ml PBS. At 0, 3, 12, and 24 hours, the survival rates of L. crocea were 100.0%, 100.0%, 77.5% and 56.0%, respectively. Three individuals from each group were chosen randomly, and the head kidney tissues were taken for RNA sequencing. Fish were put down with 0.05% MS-222 (3-aminobenzoic acid ethyl ester methanesulfonate, Sigma, USA) before to sampling. The NCBI Sequence Read Archive received the raw data and assigned it the accession number PRJNA764439.
In order to corroborate the TLR gene expression patterns after both hypoxic and A. hydrophila infection stressors above, using primers created by Primer Premier 5.0 software for qRT-PCR and reported in Supplementary Table 1, we identified genes that were differentially expressed. Utilizing β-actin as the internal control, we computed the relative expression level using the 2-△△CT technique (n = 3). Tukey’s multiple range test and one-way ANOVA were used for the statistical analysis (SPSS, version 22.0).
Following the TLR gene screening and validation in channel catfish, fugu, grass carp, Japanese flounder, medaka, Nile tilapia, rainbow trout, spotted gar, spotted sea bass and zebrafish, 12 TLR genes in L. crocea (LcTLRs) were identified. Table 1 displayed the TLR gene copy counts for eleven different teleost species. The probable molecular weights of the proteins ranged from 50,904 kDa (LcTLR2b) to 121,457 kDa (LcTLR9), and their theoretical isoelectric points (pI) varied from 5.98 (LcTLR2b) to 8.94 (LcTLR5). The proteins’ lengths varied from 439 (LcTLR2b) to 1056 (LcTLR9) amino acids. Table 2 provided specific information on 12 LcTLRs.
139 full protein sequences from the teleosts mentioned above were used to create a phylogenetic tree and examine the evolutionary relationships between the eleven teleost TLR genes. Figure 1 illustrated the similar genes from these species grouped together, demonstrating the relative conservation of TLR genes. LcTLR3, LcTLR5, LcTLR21, LcTLR22, and LcTLR23 were clustered into a branch; LcTLR1, LcTLR2, and LcTLR14 were grouped together; LcTLR7, LcTLR8, and LcTLR9 were clustered into a branch.
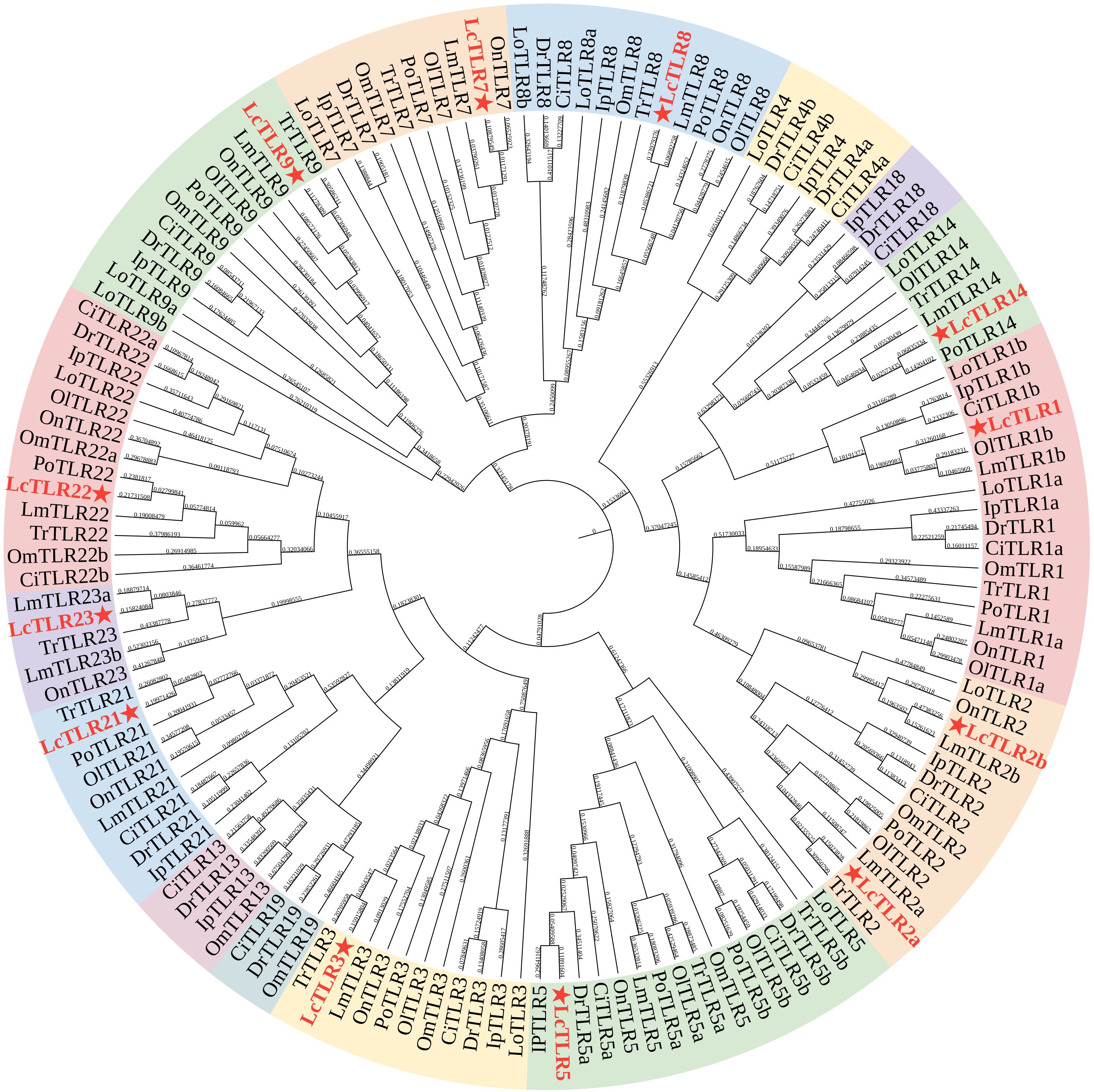
Figure 1 Phylogenetic tree of eleven teleost species’ TLR gene families. A red asterisk was used to identify the LcTLRs. The Ip, Ictalurus punetaus, Tr, Takifugu rubripes; Ci, Ctenopharyngodon idella; Po, Paralichthys olivaceus; On, Oreochromis niloticus; Om, Oncorhynchus mykiss; Ol, Oryzias latipes; Lo, Lepisosteus oculatus; Lm, Lateolabrax maculatus; Dr, Danio rerio and Lc, Larimichthys crocea.
We examined the CDS structure and conserved motifs of the TLR family to gain a deeper understanding of the structural conservation of the TLR gene family. Ten conserved motifs were predicted from the TLRs of L. crocea, as Figure 2 illustrated. The numbers and placements of these motifs were more similar in proteins that were closer in relationship. There were more motifs in LcTLR7, LcTLR8, and LcTLR9 than in LcTLR5, which has the fewest motifs. Motifs 4, 8, and 9 were present in all 12 LcTLRs, and motif 5 was present in most LcTLRs. Of all the LcTLRs, LcTLR2a had the most introns, and LcTLR14 had the longest length of introns.
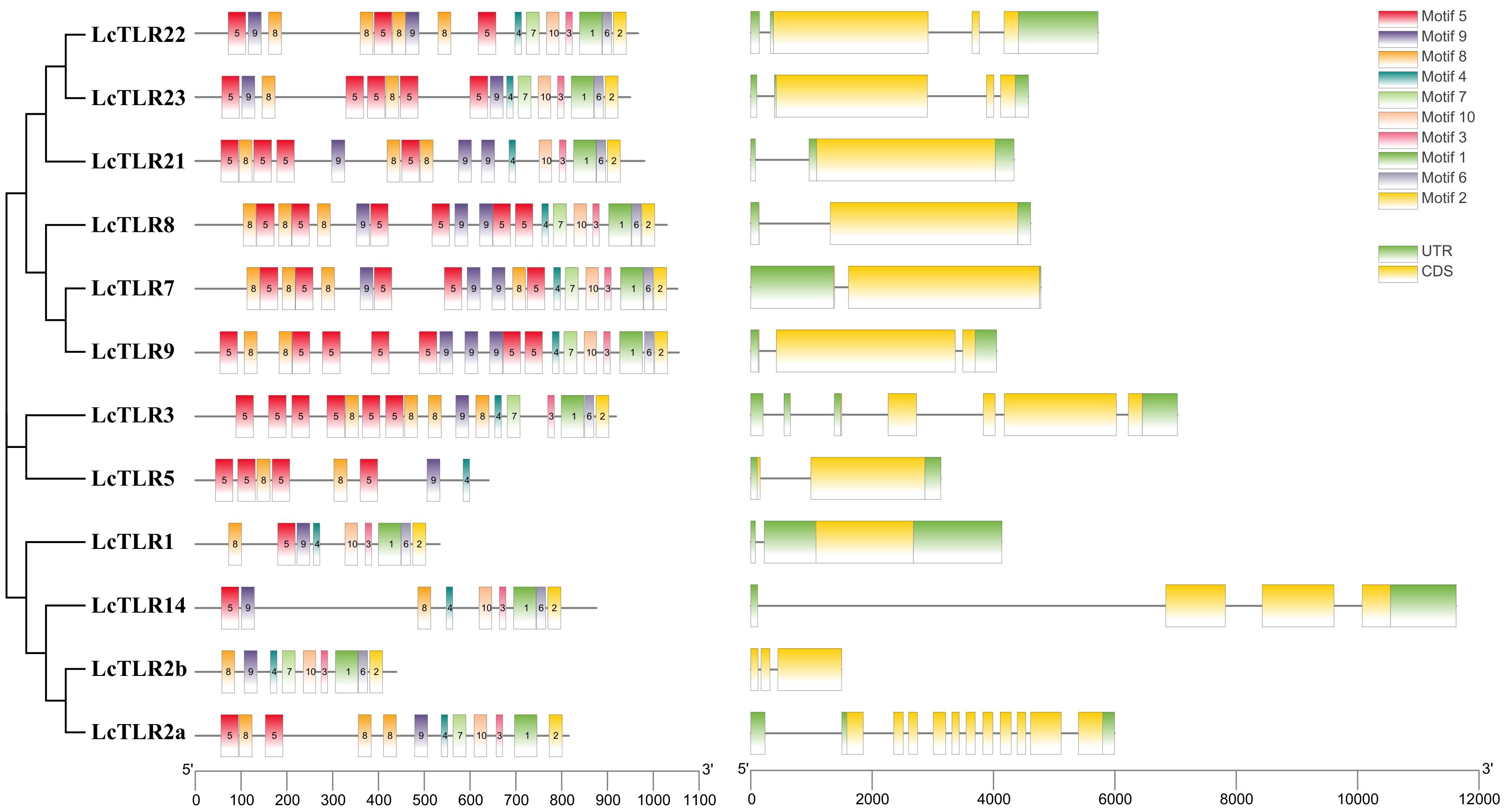
Figure 2 The arrangement of 12 LcTLRs’ conserved motif, gene structure, and evolutionary relationship. The phylogenetic relationships are displayed in the figure on the left. The distribution of conserved motifs is depicted by the middle picture, and each colored rectangular box represents a motif. Rectangular boxes with yellow backgrounds stand for CDSs, whereas green backgrounds stand for UTRs. The scale can be used to infer the length of the exons.
Based on RNA-seq data, the expression profile of LcTLRs in four hypoxia stress-affected tissues (the gill, heart, kidney and spleen) was examined to learn more about the function of the LcTLRs gene in the hypoxic response. Eight LcTLR genes were involved in gill (P<0.05, |log2 fold change|>1), as Figure 3 illustrates. There was a considerable up-regulation of LcTLR1 at 48 h and a noticeable up-regulation of LcTLR5 at 24 h. The expression level of LcTLR2a, LcTLR3, LcTLR14 and LcTLR22 showed a decreasing trend at different time points, while LcTLR9 and LcTLR21 were increased.
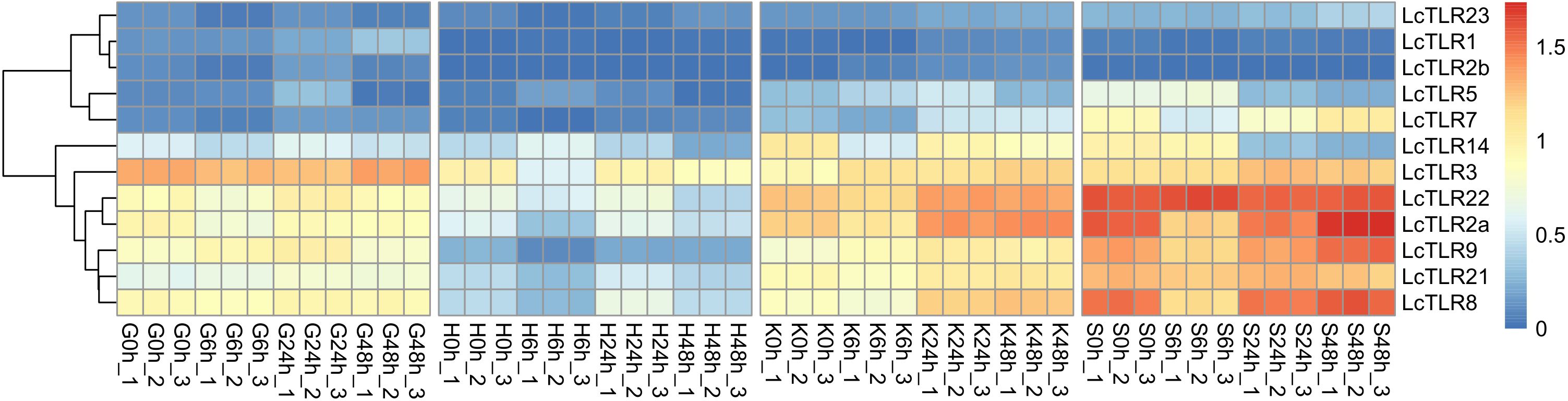
Figure 3 Heatmap of LcTLRs expression under hypoxia stress. Every cell in the heatmap represents an expression level that has been log10 (FPKM+1) normalized. G stands for gill, H for heart, K for kidney and S for spleen. 0 h as control groups, 6 h, 24 h and 48 h represent after infection treatment 6, 24 and 48 hours. There are three biological replications: X-1, X-2, and X-3.
Eight LcTLR genes exerted actions in the heart under hypoxia challenge. The results of the examination showed that whereas LcTLR5 was dramatically up-regulated at 6 h, LcTLR2a, LcTLR3, and LcTLR9 were remarkably down-regulated. It is noteworthy that there was a considerable up-regulation of LcTLR8 expression at 24 hours, but a decrease at 6 hours.
During hypoxic stress, nine LcTLRs took involved in the kidney’s immunological response. Following a 6-hour drop, LcTLR2a increased till 48 hours later. The expression of LcTLR3, LcTLR5, LcTLR7, LcTLR8 and LcTLR9 increased persistently, and there was a discernible rise in LcTLR7 and LcTLR8 at 48 hours. After 6 hours, LcTLR14 was down-regulated and continued to decline.
The spleen regulated nine out of the LcTLR genes. At 6 h, there was a noticeable drop in LcTLR2a, LcTLR7, and LcTLR8 and then showed an increasing trend step by step. LcTLR5 and LcTLR14 were signally down-regulated at both 24 h and 48 h. Supplementary Table 2 revealed the P-value and log2 fold change.
Using A. hydrophila infection transcriptome sequencing data in L. crocea, the involvement of TLR genes in the kidney was visualized in Figure 4. Supplementary Table 3 displayed P-value and log2 fold change values. The majority of TLR genes were markedly down-regulated (P < 0.05, |log2 fold change|>1) in bacterial infections. At 3 h after bacterial infection, LcTLR3, LcTLR7, LcTLR8, LcTLR9 and LcTLR14 were remarkedly down-regulated; At 12 h after bacterial infection, LcTLR3, LcTLR7, LcTLR9 and LcTLR14 were significantly down-regulated. At 24 h after bacterial infection, the expressions of LcTLR3, LcTLR7, LcTLR9 and LcTLR14 showed similar downward trends like above two-time points. Interestingly, only LcTLR5 was visibly elevated at 3 h and then started a decreasing trend until it returned to normal at 24 h. Notably, LcTLR2b did not participate in immune inflammatory response from beginning to end.
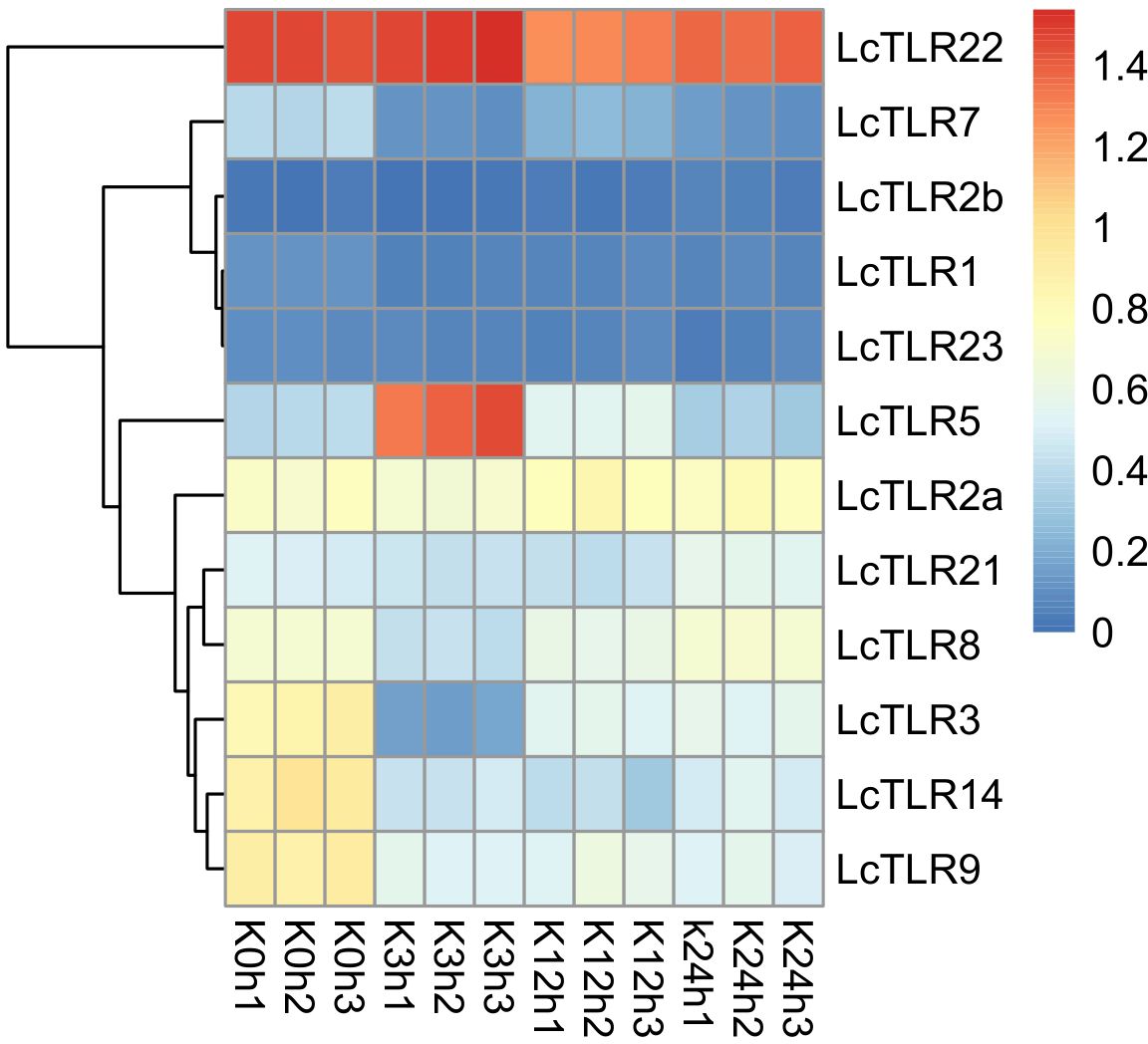
Figure 4 Heatmap illustrating the expression of LcTLRs following infection with Aeromonas hydrophila. Every cell in the heatmap represents an expression level that has been log10 (FPKM+1) normalized. K is kidney. K0h as control groups, K3, K12 and K24 represent after infection treatment 3, 12 and 24 hours, X1, X2 and X3 are the three biological replications.
After A. hydrophila infection and hypoxic stress, expression profiles of LcTLR2a, LcTLR3, LcTLR5, LcTLR8, LcTLR9, and LcTLR22 were created using qRT-PCR to confirm the differentially expressed TLR genes. The expression trends of the qRT-PCR data were essentially in line with the RNA-Seq findings, as illustrated in Figure 5. In general, the qPCR findings proved that the results of RNA-seq analysis were accurate and reliable.
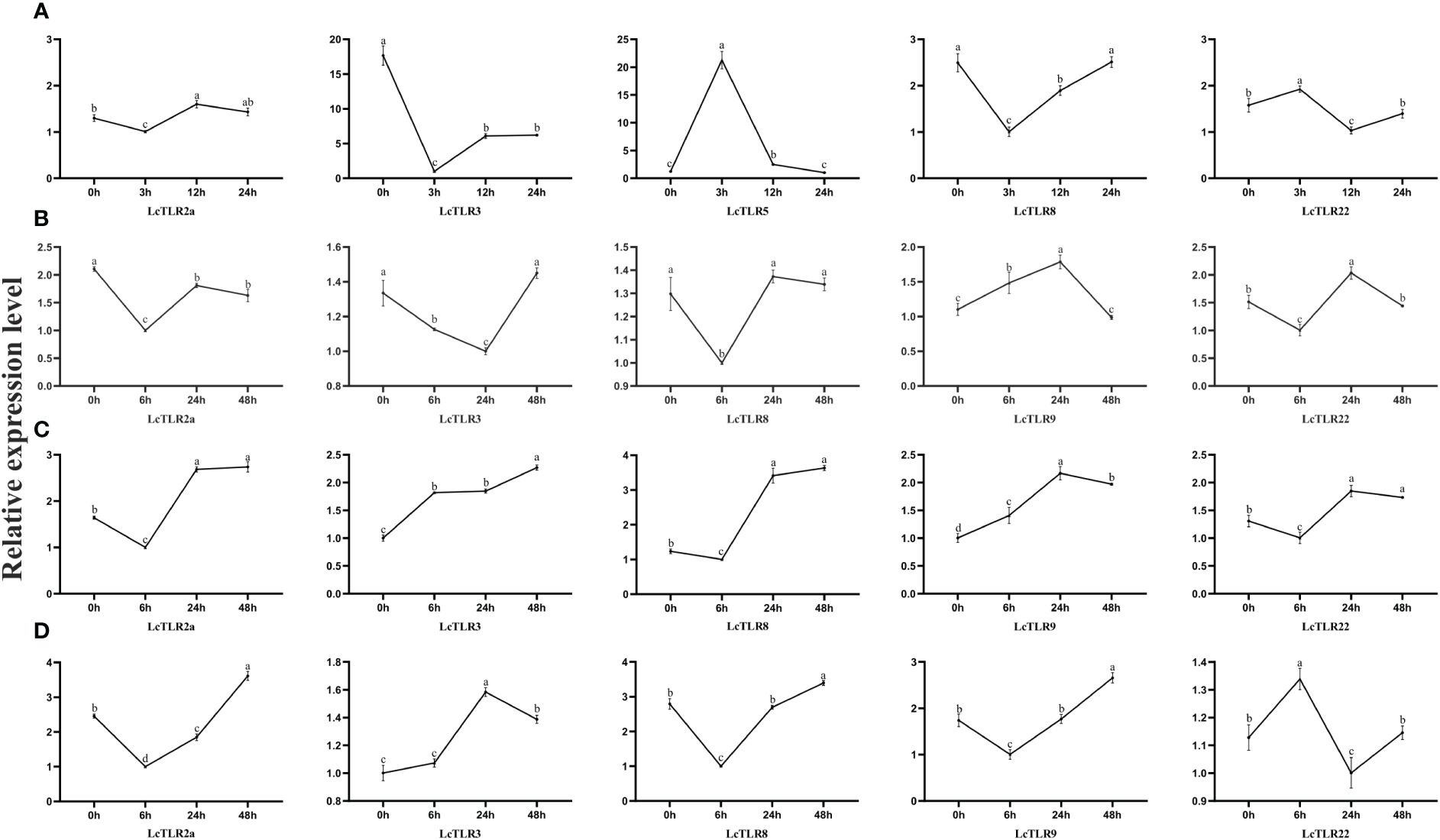
Figure 5 Validation of the expression levels of LcTLRs after A. hydrophila infection and hypoxic stress. (A–D) stands for four groups. Group A represents A. hydrophila infection, the remaining three groups represent gill, kidney and spleen after hypoxia stress, respectively.
It is now evident that TLRs promote and regulate the quality of the adaptive immune response in addition to playing a critical role in the innate identification of microorganisms (Manicassamy and Pulendran, 2009). The discovery of 21 distinct TLR types in a range of fish species in recent years has spurred interest in investigating TLRs’ potential as targets for increasing fish immunity and disease resistance (Mahapatra et al., 2023). Twelve TLRs in all were found in the L. crocea examined in this investigation. The number was the same as the closely related species T. rubripes and O. latipes (Xie et al., 2023). The majority of genes were carried by L. crocea, which reached a high level of evolutionary conservativeness. Besides, four “fish-specific” family members (Rebl et al., 2010) of TLRs were identified in L. crocea, including TLR21, 22, 23. These findings corroborated those of earlier research, which revealed that fish TLRs had a great deal of diversity and distinctive traits, most likely due to their varied evolutionary histories and the varied settings they live in (Palti, 2011).
Members of the TLR family in teleosts were divided into six clades based on sequence homology comparison (Palti, 2011; Fan et al., 2019). Additionally, TLRs 18–20 and 22–28 are thought to be special to fish (Palti, 2011). While the majority of fish TLRs have not been documented to recognize comparable PAMPs, the fish TLRs have unique characteristics resulting from their varied evolutionary backgrounds and intricate aquatic environments. Notably, even though L. crocea contained most TLR genes, we found that the large yellow croaker’s genome lacked the TLR4 and TLR13 genes, and we also noticed the similar situation in the genome of D. rerio, P. olivaceus and O. niloticus etc., indicating that gene gain and loss events might occur frequently in fish.
The phylogeny of gene families may be further explained by structural analysis. Here, we found that LcTLR14 had a longer intron structure while others had shorter introns. Besides, it was discovered that the majority of the TLR genes in Cyprinus carpio and Patinopecten yessoensis have intron-less architectures (Gong et al., 2017; Xing et al., 2017). The splicing process of post-transcriptional modification will take longer for gene architectures with longer or more introns (Jeffares et al., 2008), which is not good for the efficient regulation of genes. It was discovered that under stress, genes with short or no introns might express themselves quickly (Guo et al., 2015; Liu et al., 2022a). These findings implied that the TLR gene family might have evolved in a significant way toward gene structural simplification as a result of environmental adaption.
Studies have shown that environmental stressors just like hypoxia stress and bacterial infection could affect TLR expression. In the kidneys, spleen, brain, and fish gills, hypoxia typically results in cell necrosis, apoptosis, and inflammation while also encouraging the growth of new blood vessels (Harper and Wolf, 2009). However, little is known about how TLRs function in fish in response to hypoxia. Prior research has demonstrated that in both human and mouse dendritic cells (DCs), hypoxia preferentially upregulated the expressions of TLR2 and TLR6 (Kuhlicke et al., 2007). According to Stridh et al. (2011), hypoxia in neonatal mice led to an increase in TLR1 and TLR2 expression in the brain. Additionally, research on Tibetan schizothoracine fish (Gymnocypris eckloni) revealed that while TLR2 and TLR3 expressions increased in the head kidney, TLR8a, TLR12 and TLR19 in the spleen, and TLR1 expressions increased in the gill, the majority of TLRs’ mRNA expressions either significantly decreased or remained unchanged under acute hypoxia (Qi et al., 2017). In this study, the expression of LcTLR1, LcTLR9, and LcTLR21 in gill, LcTLR5 in heart, LcTLR3, LcTLR5, LcTLR7, LcTLR8 and LcTLR9 in kidney, and LcTLR2a, LcTLR7, LcTLR8 in spleen were significantly up-regulated, whereas the expressions of other TLRs in the gill, heart, kidney and spleen declined dramatically or remained unchanged in a severe hypoxic environment. This result was consistent with Tibetan schizothoracine fish TLRs expression in response to acute hypoxia. The cessation of the energy-saving mechanism of TLR protein synthesis leads to the reduction of TLR mRNA (Roesner et al., 2006), and TLRs are normally involved in innate immune responses regulated by the expression of damage-associated molecular patterns (DAMP), which are secreted by necrotic or damaged cells (Mkaddem et al., 2010). Thus, the unique function of TLRs in mediating the innate immune response to host defense against hypoxia-induced tissue damage might be connected to tissue-specific TLR expression under hypoxic settings. TLRs offer a great model for examining the natural selection imposed by pathogenic microorganisms on the organism genome because of their direct placement at the host–environment interface and potential for coevolutionary dynamics with their pathogenic counterparts (Barreiro et al., 2009).
There had suggested an idea called TLR/TLR cross-talk, which means that, in comparison to DCs stimulated with individual TLR4 ligand and TLR7 ligand, TLR/TLR synergy, such as simultaneous stimulation of human monocyte-derived DCs with LPS (TLR4 agonist) and R484 (TLR7 agonist), leads in a synergistic rise in anti-inflammatory cytokine IL-10 (Napolitani et al., 2005; Manicassamy and Pulendran, 2009). In this study, A. hydrophila infection was performed to evaluate L. crocea TLR gene alterations, and our results were similar to a research of L. crocea in response to Cryptocaryon irritans infection, demonstrating that TLR5 was distinctly up-regulated following infection (Zhang et al., 2020). This might due to the function of TLR5 responding to PAMP, such as lipids and bacterial proteins (Yang et al., 1999). TLR genes that were up-regulated during the bacterial infection were generally thought to be crucial for identifying bacterial ligands (Zhang et al., 2017). Besides, LcTLR3, LcTLR7, LcTLR9 and LcTLR14 were remarkedly down-regulated after 12 h in our study. The role of TLR3 in immune responses was complex and differed between species. TLR3 in the spleen did not significantly alter following injection of Vibrio parahaemolyticus in the prior research of Pseudosciaena crocea, suggesting that bacterial infection of the spleen might not be the initial cause of the PcTLR3 response (Huang et al., 2011). After bath challenge with a Gram-negative bacterium, rainbow trout’s spleen showed a similar outcome (Rodriguez et al., 2005). However, TLR3 was shown to be significantly down-regulated in the kidney of channel catfish following an infection with Edwardsiella tarda. This finding, along with our results, suggested that this protein might be the most susceptible to bacterial infection, as the subpopulation of phagocytes expressing these genes might quickly migrate from the kidney to the infection sites (Zhang et al., 2013). This variation in expression changes of TLR3 might be due to differences in TLR3-mediated inflammatory responses among different immune organs, further hinting that apart from their recognition of dsRNA, TLR3 was also engaged in fish immune responses to bacterial infections. SmTLR7 was markedly down-regulated in the gill and intestine of turbot (Scophthalmus maximus) following infection with both E. tarda and Vibrio anguillarum (Wang et al., 2022). After A. hydrophila infection, the expression level of MaTLR14 was up-regulated in the liver, spleen, foregut, and hindgut of Asian swamp eels (Monopterus albus) (Liu et al., 2022b). All of these findings revealed that different Gram-negative bacterial components regulated TLRs, suggesting that the immune response to bacterial infection might involve numerous TLR-mediated signaling cascades at the same time. Notably, LcTLR2 had no significant change throughout the process, which was inconsistent with the previous study showing that the LcTLR2 transcripts increased significantly after Vibrio parahaemolyticus, Polyinosinic-polycytidylic acid (Poly(I:C)) and LPS immune challenges (p < 0.05) in the spleen, liver and head-kidney (Fan et al., 2015). This might be due to differences in immune response induced by A. hydrophila and V. parahaemolyticus.
Together with A. hydrophila infection and hypoxic stress, this study revealed that there were six and nine members of LcTLRs showing significant differential expression in immune responses, respectively. These results provided insights into TLRs regulation to biological and abiotic stresses, implying that TLR expressions specific to particular tissues mediated innate immune responses for host defense against tissue damage or physiological changes brought on by bacterial infection and hypoxic stress.
The larger yellow croaker (Larimichthys crocea) was found to harbor a total of 12 TLR genes in this study. Analysis of the TLR genes’ phylogenetic relationships, gene structures, and motif compositions revealed that these genes were highly conserved among different species. Expression pattern analysis and qPCR validation of A. hydrophila infection and hypoxic stress revealed that there were six and nine members of LcTLRs showing significant differential expression in immune responses, respectively. Notably, LcTLR5 was significantly expressed under both biological and abiotic stresses, suggesting that it might play a significant role in the effort to combat environmental stressors. In summary, the identification of TLRs and their roles would advance our knowledge of fish disease resistance mechanisms and open up new avenues for therapeutic intervention to control immune responses.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
The animal study was approved by the Institutional Animal Care and Use Committee at the Zhejiang Laboratory Animal Research Center and Ningbo University. The study was conducted in accordance with the local legislation and institutional requirements.
T-YY: Writing – original draft, Methodology, Formal analysis. Q-TM: Writing – original draft, Investigation. X-YS: Writing – original draft, Conceptualization. CR: Writing – original draft, Data curation. Q-PX: Writing – original draft, Formal analysis. X-BW: Writing – review & editing, Supervision, Project administration, Funding acquisition.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. The work was supported by the Natural Science Foundation of Shandong Province (No. ZR2022MD064) and the Scientific Research Foundation for Introduced Talents of Ningbo University (ZX2022000602).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2024.1404183/full#supplementary-material
Abdel-Tawwab M., Monier M., Hoseinifar S., Faggio C. (2019). Fish response to hypoxia stress: growth, physiological, and immunological biomarkers. Fish. Physiol. Biochem. 45, 997–1013. doi: 10.1007/s10695-019-00614-9
Adams C. S., Kim H., Burtner A. E., Lee D. S., Dobbins C., Criswell C., et al. (2024). De novo design of protein minibinder agonists of TLR3. bioRxiv. [Preprint]. 2024.04.17.589973. doi: 10.1101/2024.04.17.589973
Bailey T., Boden M., Buske F., Frith M., Grant C., Clementi L., et al. (2009). MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 37, W202–W208. doi: 10.1093/nar/gkp335
Barreiro L., Ben-Ali M., Quach H., Laval G., Patin E., Pickrell J., et al. (2009). Evolutionary dynamics of human Toll-like receptors and their different contributions to host defense. PLoS Genet 5, e1000562. doi: 10.1016/j.molp.2020.06.009
Chen C., Chen H., Zhang Y., Thomas H., Frank M., He Y., et al. (2020). TBtools: an integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 13, 1194–1202. doi: 10.1016/j.molp.2020.06.009
Chen X., Wu Z., Yu S., Wang S., Peng X. (2010). Beta2-microglobulin is involved in the immune response of large yellow croaker to Aeromonas hydrophila: a proteomic based study. Fish. Shellfish. Immunol. 28, 151–158. doi: 10.1016/j.fsi.2009.10.015
Dias B. T., Goundry A., Vivarini A. C., Costa T. F. R., Mottram J. C., Lopes U. G., et al. (2022). Toll-like receptor- and protein kinase R-induced type I interferon sustains infection of leishmania donovani in macrophages. Front. Immunol. 13, 801182. doi: 10.3389/fimmu.2022.801182
Ding J., Liu C., Luo S., Zhang Y., Gao X., Wu X., et al. (2020). Transcriptome and physiology analysis identify key metabolic changes in the liver of the large yellow croaker (Larimichthys crocea) in response to acute hypoxia. Ecotoxicol. Environ. Saf. 189, 109957. doi: 10.1016/j.ecoenv.2019.109957
Ding J., Zhang Y., Wang J., Liu C., Gao X., Wu Y., et al. (2022). Genome-wide association study identified candidate SNPs and genes associated with hypoxia tolerance in large yellow croaker (Larimichthys crocea). Aquaculture 560, 738472. doi: 10.1016/j.aquaculture.2022.738472
Duvaud S., Gabella C., Lisacek F., Stockinger H., Ioannidis V., Durinx C. (2021). Expasy the Swiss bioinformatics resource portal, as designed by its users. Nucleic Acids Res. 49, W216–W227. doi: 10.1093/nar/gkab225
Edgar R. (2004). MUSCLE: multiple sequence alignment with high accuracy and high Throughput. Nucleic Acids Res. 32, 1792–1797. doi: 10.1093/nar/gkh340
El-Gebali S., Mistry J., Bateman A., Eddy S., Luciani A., Potter S., et al. (2019). The Pfam protein families database in 2019. Nucleic Acids Res. 47, D427–D432. doi: 10.1093/nar/gky995
Fan H., Wang L., Wen H., Wang K., Qi X., Li J., et al. (2019). Genome-wide identification and characterization of toll-like receptor genes in spotted sea bass (Lateolabrax maculatus) and their involvement in the host immune response to Vibrio harveyi infection. Fish. Shellfish. Immunol. 92, 782–791. doi: 10.1016/j.fsi.2019.07.010
Fan Z., Jia Q., Yao C. (2015). Characterization and expression analysis of Toll-like receptor 2 gene in large yellow croaker, Larimichthys crocea. Fish. Shellfish. Immunol. 44, 129–137. doi: 10.1016/j.fsi.2015.01.037
Gao S., Huang W., Peng S., Zhou J., Zhan H., Lu T., et al. (2024). Molecular characterization and expression analysis of nine toll like receptor (TLR) genes in Scortum barcoo under Streptococcus agalactiae infection. Int. J. Biol. Macromol. 254, 127667. doi: 10.1016/j.ijbiomac.2023.127667
Gong Y., Feng S., Li S., Zhang Y., Zhao Z., Hu M., et al. (2017). Genome-wide characterization of Toll-like receptor gene family in common carp (Cyprinus carpio) and their involvement in host immune response to Aeromonas hydrophila infection. Comp. Biochem. Physiol. Part D. Genomics Proteomics 24, 89–98. doi: 10.1016/j.cbd.2017.08.003
Guo M., Liu H., Lu P., Zhai F., Wang H., Gong H., et al. (2015). Genome-wide analysis of the CaHsp20 gene family in pepper: comprehensive sequence and expression profile analysis under heat stress. Front. Plant Sci. 6, 806. doi: 10.3389/fpls.2015.00806
Harper C., Wolf J. (2009). Morphologic effects of the stress response in fish. ILAR. J. 50, 387–396. doi: 10.1093/ilar.50.4.387
Huang X., Wang Z., Yao C. (2011). Characterization of Toll-like receptor 3 gene in large yellow croaker, Pseudosciaena crocea. Fish. Shellfish. Immunol. 31, 98–106. doi: 10.1016/j.fsi.2011.04.009
Jaworski L., Kleinhans K., Jackson A. (2019). Effects of oxygen concentration and culture time on porcine nucleus pulposus cell metabolism: an in vitro study. Front. Bioeng. Biotechnol. 7. doi: 10.3389/fbioe.2019.00064
Jeffares D., Penkett J., Bahler J. (2008). Rapidly regulated genes are intron poor. Trends Genet. 24, 375–378. doi: 10.1016/j.tig.2008.05.006
Kuhlicke J., Frick J., Morote-Garcia J., Rosenberger P., Eltzschig H. (2007). Hypoxia inducible factor (HIF)-1 coordinates induction of Toll-like receptors TLR2 and TLR6 during hypoxia. PloS One 2, 1364. doi: 10.1371/journal.pone.0001364
Kumar S., Stecher G., Tamura K. (2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets, Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Lee Y., Kim D. H., Lee J. S., Kim H. S., Maszczyk P., Wang M., et al. (2024). Combined exposure to hypoxia and nanoplastics leads to negative synergistic oxidative stress-mediated effects in the water flea Daphnia magna. Mar. pollut. Bull. 202, 116306. doi: 10.1016/j.marpolbul.2024.116306
Letunic I., Bork P. (2018). 20 years of the SMART protein domain annotation resource. Nucleic Acids Res. 46, D493–D496. doi: 10.1093/nar/gkx922
Li S., Yang Z., Tian H., Ren S., Zhang W., Wang A. (2022). Effects of dietary carbohydrate/lipid ratios on non-specific immune responses, antioxidant capacity, hepatopancreas and intestines histology, and expression of TLR-MAPK/NF-κB signaling pathway-related genes of Procambarus clarkia. Fish. Shellfish. Immunol. 124, 219–229. doi: 10.1016/j.fsi.2022.04.005
Liu R., Qi Y., Feng H., Niu Y., Zhang F., Yang G., et al. (2022b). Fish-specific Toll-like receptor 14 (TLR14) from Asian swamp eel (Monopterus albus) is involved in immune response to bacterial infection. Fish. Shellfish. Immunol. 124, 313–323. doi: 10.1016/j.fsi.2022.04.010
Liu D., Qin Z., Wei M., Kong D., Zheng Q., Bai S., et al. (2022a). Genome-wide analyses of heat shock protein superfamily provide new insights on adaptation to sulfide-rich environments in Urechis unicinctus (Annelida, Echiura). Int. J. Mol. Sci. 23, 2715. doi: 10.3390/ijms23052715
Mahapatra S., Ganguly B., Pani S., Saha A., Samanta M. (2023). A comprehensive review on the dynamic role of toll-like receptors (TLRs) in frontier aquaculture research and as a promising avenue for fish disease management. Int. J. Biol. Macromol. 253, 126541. doi: 10.1016/j.ijbiomac.2023.126541
Manicassamy S., Pulendran B. (2009). Modulation of adaptive immunity with Toll-like receptors. Semin. Immunol. 21, 185–193. doi: 10.1016/j.smim.2009.05.005
Mkaddem S., Bens M., Vandewalle A. (2010). Differential activation of Toll-like receptor-mediated apoptosis induced by hypoxia. Oncotarget 1, 741–750. doi: 10.18632/oncotarget.v1i8
Mu Y., Ding F., Cui P., Ao J., Hu S., Chen X. (2010). Transcriptome and expression profiling analysis revealed changes of multiple signaling pathways involved in immunity in the large yellow croaker during Aeromonas hydrophila infection. BMC Genomics 11, 506. doi: 10.1186/1471-2164-11-506
Mu Y., Huo J., Guan Y., Fan D., Xiao X., Wei J., et al. (2018). An improved genome assembly for Larimichthys crocea reveals hepcidin gene expansion with diversified regulation and function. Commun. Biol. 1, 195. doi: 10.1038/s42003-018-0207-3
Napolitani G., Rinaldi A., Bertoni F., Sallusto F., Lanzavecchia A. (2005). Selected Toll-like receptor agonist combinations synergistically trigger a T helper type 1-polarizing program in dendritic cells. Nat. Immunol. 6, 769–776. doi: 10.1038/ni1223
Ni L., Xu Y., Zhao Q., Shao F., Peng Z. (2022). Genome-wide characterization of the Toll-like receptor gene family in Triplophysa rosa and expression profiles in response to Aeromonas hydrophila infection. Aquaculture 555, 738208. doi: 10.1016/j.aquaculture.2022.738208
Palti Y. (2011). Toll-like receptors in bony fish: from genomics to function. Dev. Comp. Immunol. 35, 1263–1272. doi: 10.1016/j.dci.2011.03.006
Qi D., Xia M., Chao Y., Zhao Y., Wu R. (2017). Identification, molecular evolution of toll-like receptors in a Tibetan schizothoracine fish (Gymnocypris eckloni) and their expression profiles in response to acute hypoxia. Fish. Shellfish. Immunol. 68, 102–113. doi: 10.1016/j.fsi.2017.07.014
Rebl A., Goldammer T., Seyfert H. (2010). Toll-like receptor signaling in bony fish. Vet. Immunol. Immunopathol. 134, 139–150. doi: 10.1016/j.vetimm.2009.09.021
Rodriguez M., Wiens G., Purcell M., Palti Y. (2005). Characterization of Toll-like receptor 3 gene in rainbow trout (Oncorhynchus mykiss). Immunogenetics 57, 510–519. doi: 10.1007/s00251-005-0013-1
Roesner A., Hankeln T., Burmester T. (2006). Hypoxia induces a complex response of globin expression in zebrafish (Danio rerio). J. Exp. Biol. 209, 2129–2137. doi: 10.1242/jeb.02243
Ryan T. A. J., O’Neill L. A. J. (2023). An emerging role for type I interferons as critical regulators of blood coagulation. Cells 12, 778. doi: 10.3390/cells12050778
Sahoo B., Basu M., Swain B., Maharana J., Dikhit M., Jayasankar P., et al. (2012). Structural insights of rohu TLR3, its binding site analysis with fish reovirus dsRNA, poly I:C and zebrafish TRIF. Int. J. Biol. Macromol. 51, 531–543. doi: 10.1016/j.ijbiomac.2012.06.005
Shi B., Zhang Z., Xing J., Liu Q., Cai J., Zhang Z. (2024). Perfluorooctane sulfonate causes pyroptosis and lipid metabolism disorders through ROS-mediated NLRP3 inflammasome activation in grass carp hepatocyte. Aquat. Toxicol. 267, 106839. doi: 10.1016/j.aquatox.2024.106839
Shun C., Wen-Ping J., Shi-Li L., Jian-Bo Z., Mei-Li C., Xiao-Ying H., et al. (2024). Effect of dissolved oxygen and ammonia nitrogen on Culter alburnus: Physiology, biochemistry, and molecular analyses. Heliyon 10, e27921. doi: 10.1016/j.heliyon.2024.e27921
Stridh L., Smith P., Naylor A., Wang X., Mallard C. (2011). Regulation of toll-like receptor 1 and -2 in neonatal mice brains after hypoxia-ischemia. J. Neuroinflamm. 8, 45. doi: 10.1186/1742-2094-8-45
Tran N., Kong T., Zhang M., Li S. (2019). Pattern recognition receptors and their roles on the innate immune system of mud crab (Scylla paramamosain). Dev. Comp. Immunol. 102, 103469. doi: 10.1016/j.dci.2019.103469
Verma S., Menon R., Sowdhamini R. (2024). Structural insights into the role of deleterious mutations at the dimeric interface of Toll-like receptor interferon-β related adaptor protein. Proteins. doi: 10.1002/prot.26707
Vidya M., Kumar V., Sejian V., Bagath M., Krishnan G., Bhatta R. (2018). Toll-like receptors: significance, ligands, signaling pathways, and functions in mammals. Int. Rev. Immunol. 37, 20–36. doi: 10.1080/08830185.2017.1380200
Wallach T., Raden M., Hinkelmann L., Brehm M., Rabsch D., Weidling H., et al. (2023). Distinct SARS-CoV-2 RNA fragments activate Toll-like receptors 7 and 8 and induce cytokine release from human macrophages and microglia. Front. Immunol. 13, 1066456. doi: 10.3389/fimmu.2022.1066456
Wang K., Huang H., Zhan Q., Ding H., Li Y. (2024). Toll-like receptors in health and disease. MedComm. (2020). . 5, e549. doi: 10.1002/mco2.549
Wang X., Liu S., Yang Y., Fu Q., Abebe A., Liu Z. (2017). Identification of NF-kappa B related genes in channel catfish and their expression profiles in mucosal tissues after columnaris bacterial infection. Dev. Comp. Immunol. 70, 27–38. doi: 10.1016/j.dci.2017.01.003
Wang B., Liu X., Zhao J., Cao M., Yu Z., Fu Q., et al. (2022). Characterization, evolution and expression analysis of Toll-like receptor 7 (TLR7) in turbot (Scophthalmus maximus L.). Fish. Shellfish. Immunol. 125, 9–16. doi: 10.1016/j.fsi.2022.04.035
Xie Q., Zhan W., Shi J., Liu F., Niu B., He X., et al. (2023). Whole-genome assembly and annotation for the little yellow croaker (Larimichthys polyactis) provide insights into the evolution of hermaphroditism and gonochorism. Mol. Ecol. Resour. 23, 632–658. doi: 10.1111/1755-0998.13731
Xing Q., Liao H., Xun X., Wang J., Zhang Z., Yang Z., et al. (2017). Genome-wide identification, characterization and expression analyses of TLRs in Yesso scallop (Patinopecten yessoensis) provide insight into the disparity of responses to acidifying exposure in bivalves. Fish. Shellfish. Immunol. 68, 280–288. doi: 10.1016/j.fsi.2017.07.020
Yan W., Qiao Y., Qu J., Liu X., Zhang Q., Wang X. (2021). The hsp40 Gene Family in Japanese Flounder: identification, Phylogenetic Relationships, Molecular Evolution Analysis, and Expression Patterns. Front. Mar. Sci. 7. doi: 10.3389/fmars.2020.596534
Yang R., Mark M., Gurney A., Godowski P. (1999). Signaling events induced by lipopolysaccharide-activated toll-like receptor 2. J. Immunol. 163, 639–643. doi: 10.4049/jimmunol.163.2.639
Yao T., Wang R., Han P., Liu X., Wang X. (2023). Identification of olive flounder (Paralichthys olivaceus) toll-like receptor genes: involvement in immune response to temperature stress and Edwardsiella tarda infection. Fish. Shellfish. Immunol. 138, 108841. doi: 10.1016/j.fsi.2023.108841
Yeh D., Liu Y., Lo Y., Yuh C., Yu G., Lo J., et al. (2013). Toll-like receptor 9 and 21 have different ligand recognition profiles and cooperatively mediate activity of CpG-oligodeoxynucleotides in zebrafish. Proc. Natl. Acad. 110, 20711–20716. doi: 10.1073/pnas.1305273110
Zhang H., Gao S., Lercher M., Hu S., Chen W. (2012). EvolView, an online tool for visualizing, annotating and managing phylogenetic trees. Nucleic Acids Res. 40, W569–W572. doi: 10.1093/nar/gks576
Zhang J., Liu S., Rajendran K., Sun L., Zhang Y., Sun F., et al. (2013). Pathogen recognition receptors in channel catfish: III phylogeny and expression analysis of Toll-like receptors. Dev. Comp. Immunol. 40, 185–194. doi: 10.1016/j.dci.2013.01.009
Zhang Y., Shen W., Ding J., Gao X., Wu X., Zhu J. (2022). Comparative transcriptome analysis of head kidney of aeromonas hydrophila-infected hypoxia-tolerant and normal large yellow croaker. Mar. Biotechnol. (NY). 24, 1039–1054. doi: 10.1007/s10126-022-10158-4
Zhang X., Zhang G., Shi Z., Yuan Y., Zheng H., Lin L., et al. (2017). Expression analysis of nine Toll-like receptors in yellow catfish (Pelteobagrus fulvidraco) responding to Aeromonas hydrophila challenge. Fish. Shellfish. Immunol. 63, 384–393. doi: 10.1016/j.fsi.2017.02.021
Keywords: large yellow croaker, toll-like receptors genes, stress response, environmental adaptation, innate immune
Citation: Yao T-Y, Miao Q-T, Sun X-Y, Rui C, Xie Q-P and Wang X-B (2024) The identification of toll-like receptor genes in large yellow croaker (Larimichthys crocea): provides insights into its environmental adaptation to biological and abiotic stresses. Front. Mar. Sci. 11:1404183. doi: 10.3389/fmars.2024.1404183
Received: 20 March 2024; Accepted: 06 June 2024;
Published: 20 June 2024.
Edited by:
Yinnan Mu, Fujian Agriculture and Forestry University, ChinaReviewed by:
Liang Guo, Hunan Normal University, ChinaCopyright © 2024 Yao, Miao, Sun, Rui, Xie and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xu-Bo Wang, d2FuZ3h1Ym9AbmJ1LmVkdS5jbg==; Qing-Ping Xie, eGllcXBAemFhcy5hYy5jbg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.