- 1Key Laboratory for Sustainable Utilization of Marine Fisheries Resources of Ministry of Agriculture, Yellow Sea Fisheries Research Institute, Chinese Academy of Fishery Sciences, Qingdao, China
- 2Function Laboratory for Marine Fisheries Science and Food Production Processes, Qingdao National Laboratory for Marine Science and Technology, Qingdao, China
The ridgetail white prawn, Exopalaemon carinicauda is an important cultured seawater species in China. As female E.carinicauda grows faster than males, it is significant to search for the differentially expressed genes (DEGs) between males and females. However, there is no public available E.carinicauda genome data, and genes related to E. carinicauda sex differences are unclear. In this study, the transcriptome sequencing for ovary and testis tissues of E. carinicauda were conducted, and 20,891 DEGs were identified including 11,709 up-regulated DEGs and 9,182 downregulated DEGs. The functional categories related to meiosis and reproduction were enriched as well as the steroidogenesis KEGG pathway was clustered. Furthermore, the genes related to male reproduction and cell cycle were dug out which were verified by real-time PCR. In addition, two-color fluorescent in situ hybridization result showed that foxj1b might play roles during early stage of the ovary development. Therefore, our result provides clues for the study of genes related to reproduction and sex difference in E.carinicauda.
Introduction
The ridgetail white prawn, Exopalaemon carinicauda which belongs to long arm shrimp family and white shrimp genus, mainly distributed in the southeast coastal areas of China (Xu et al., 2010). As a kind of small and important economic shrimp, E. carinicauda processes delicious meat quality, with characteristics of high protein content and low-fat level, which is popular with majority of consumers (Wang et al., 2020). The high-resolution genetic linkage map of E. carinicauda was constructed by our laboratory and 52 sex-related QTL markers were identified, which is an important work for explicating the sex determination mechanism (Lv et al., 2020). However, genes related to E. carinicauda sex differences including spermatogenesis and oogenesis are still unknown. Therefore, it is necessary to excavate the DEGs between male and female in E. carinicauda.
Previous studies have shown that there are sexual dimorphisms in economic traits in many aquatic animals (Wan et al., 2021). As for growth characteristics, males grow faster than females in tilapia and yellow catfish (Huang et al., 2022), while females of half-smooth tongue sole showed a growth advantage, with females growing about 30% faster than males (Liao et al., 2014). Among crustaceans, male Eriocheir sinensis has a growth advantage over females, but female E. sinensis are favourite among consumers for their greater taste (Du et al., 2019); Female Fenneropenaeus chinensis are more popular than males with breeders since they are larger than males at the same period (Wang Q. et al., 2019). While Procambarus clarkii males are larger than female individuals in the same sexual morphotype (Hamasaki et al., 2020; Shen et al., 2022). Similar to most aquatic animals, the growth trait of E. carinicauda also shows dimorphism, female individuals are larger than concurrent males (Lv et al., 2020). Therefore, searching the differentially expressed genes related to valuable characters between males and females is significant for molecular breeding and superior seed cultivation.
Transcriptome sequencing is not only an important means of studying gene structure and expression patterns (Wang Q. et al., 2019), but a bridge to infer gene functions based on phenotype. Due to the particularity of species, it is challenging to conduct gene function research in economic aquatic animals compared to model organisms. Thus, transcriptome sequencing is essential in searching for genes related to some purpose for non-vertebrate animals. For example, Wang et al. obtained the differential expression factors of muscle and gonads of E. sinensis between females and males by transcriptome sequencing, which is important for the study of growth traits between the sexes (Wang B. et al., 2019). And Zhang et al. found the sex-determining gene in Portunus trituberculatus by transcriptome sequencing, which provided an important basis for the systematic study of sex determination mechanisms in P. trituberculatus (Zhang et al.,2022).
There was no high-quality genome data which had been published since E. carinicauda processed a large complex genome (Yuan et al., 2017; Li et al., 2019). In this study, we conducted the transcriptome sequencing of ovary and testis to identify the differentially expressed genes and significantly enriched pathways between male and female. Furthermore, we found some genes related to male reproduction, cell cycle and ovary development, which was verified by real-time PCR and two-color fluorescent in situ hybridization. Our study would be helpful for further investigating genes related to E. carinicauda reproduction and sex difference.
Materials and methods
Bioethics statement
The animal study was reviewed and approved by Ethics Committee of Yellow Sea Fisheries Research Institute. Written informed consent was obtained from the owners for the participation of their animals in this study.
Experiment animals and sample collection
The E. carinicauda used in this study was produced by RiZhao Haichen Aquaculture Co., Ltd and the samples including ovary and testis were taken from the live and healthy shrimp. The total RNA was extracted from the samples by TRIzol reagent according to the manufacturer’s instructions. The genome DNA was removed by DNase I. The purity and concentration of RNA was determined and quantified by 2100 Bioanalyzer (Agilent Technologies, Inc., Santa Clara CA, USA) and ND-2000 (NanoDrop Thermo Scientific, Wilmington, DE, USA). Quality-qualified RNA sample (OD260/280 = 1.8~2.2, OD260/230≥2.0, RIN≥8.0, 28S:18S≥1.0, Total>1μg) was used for subsequent constructing sequencing libraries.
Library preparation and Illumina Hiseq NovaSeq 6000 sequencing
RNA purification, reverse transcription, library construction and sequencing were performed at Shanghai Majorbio Bio-pharm Biotechnology Co., Ltd. (Shanghai, China). The construction of the sequencing library used the Illumina TruSeq™ RNA sample preparation Kit (Illumina, San Diego, CA). The purification of mRNA was by oligo-dT-attached magnetic beads and then fragmented by fragmentation buffer. Taking these short fragments as templates, double-stranded cDNA was synthesized using a SuperScript double-stranded cDNA synthesis kit (Invitrogen, CA) with random hexamer primers (Illumina). Then the synthesized cDNA was subjected to end-repair, phosphorylation and ‘A’ base addition according to Illumina’s library construction protocol. Libraries were size selected for cDNA target fragments of 200–300 bp on 2% Low Range Ultra Agarose followed by PCR amplified using Phusion DNA polymerase (New England Biolabs, Boston, MA) for 15 PCR cycles. After quantified by TBS380, two RNAseq libraries were sequenced in single lane on an Illumina Hiseq xten/NovaSeq 6000 sequencer (Illumina, San Diego, CA) for 2×150bp paired-end reads.
De novo assembly and annotation
The raw paired end reads were trimmed by SeqPrep (https://github.com/jstjohn/SeqPrep) and quality was controlled by Sickle (https://github.com/najoshi/sickle) with default parameters. Then clean data from the gonads including ovary and testis were assembled denovo with Trinity (http://trinityrnaseq.sourceforge.net/). All the assembled transcripts would be aligned by six databases including the NCBI protein nonredundant (NR), COG, Swiss-Prot, Pfam, COG, GO and KEGG according to the sequence similarity. The function of the transcript was annotated by BLAST2GO (http://www.blast2go.com/b2ghome) including biological processes, molecular functions and cellular components. And the pathway analysis was performed using the Kyoto Encyclopedia of Genes and Genomes (KEGG, http://www.genome.jp/kegg/).
Differential expression analysis and functional enrichment
The expression level of each transcript was calculated according to the transcripts per million reads (TPM) method in order to identify DEGs between ovary and testis. The software RSEM (http://deweylab.biostat.wisc.edu/rsem/) was used to quantify gene abundances for further analysis of the gene expression level. Essentially, differential expression analysis was performed using the DESeq2. Genes satisfied with Q value ≤ 0.05, DEGs with |log2FC|>1 and Q value<=0.05 DESeq2/Qvalue<=0.001(DEGseq) were considered to be significantly different expressed genes. The software Goatools (https://github.com/tanghaibao/Goatools) were used to identify DEGs were significantly enriched in GO terms including biological processes, molecular functions and cellular components. And the software KOBAS (http://kobas.cbi.pku.edu.cn/home.do) was used to do KEGG pathway enrichment analysis with metabolic pathways at Bonferroni-corrected P-value ≤ 0.05 compared with the whole-transcriptome background.
Checking the expression level by real-time PCR
In order to check the accuracy of the transcriptome data, the DEGs were verified by real-time fluorescent quantitative PCR (real-time PCR). Total RNA from ovary and testis of E.carinicauda was extracted with TRIzol (Invitrogen, 15596026) according to the manufacturer’s instructions. The quality and concentration of the obtained RNA were analyzed by agarose gel electrophoresis and spectrophotometer (Thermo, USA) respectively. Then, the genome DNA was removed by DNase I and cDNA were obtained using the ReverTra Ace reverse transcriptase kit (Toyobo, Japan) and random primers. The fluorescence quantitative PCR assay was performed using the SYBR green mix (Toyobo, Japan) according to the manufacturer’s instructions: 2×SYBR Green real-time master mix, 10 µL; cDNA, 2µL; F, 0.4µL; R, 0.4µL; H2O, 7.2µL. The PCR amplification conditions were 95°C for 3min; 45cycles × (95°C for 15s; 55°C for 20s; 72°C for 30s); 65°C for 0.06s; 95°C, 0.5s. The primers used in this study was designed by Primer5 and the sequences were listed in Figure S1.
Two-color fluorescent in situ hybridization
We examined the expression of foxj1b in the gonads by two-color fluorescent in situ hybridization. Gonads including testis and ovary with different stages were fixed in 4%PFA overnight and dehydrated by 30% sucrose. After being embedded with O.C.T. compound (SAKURA, 4583), the gonad was sectioned by a freezing microtome (Leica, CM1950). Probes were designed and synthesized according to the cDNA sequence of the foxj1b and vasa, and labeled with digoxin (Roche Diagnostics, 11277073910) and fluorescein labeling (Roche Diagnostics, 11685619910) respectively. And they were purified by the clean-up purification kit (Sigma, S5059-70EA). Two-color fluorescent in situ hybridization experiments were then carried out as described (Lauter et al., 2011), and the sections was dealt with prehybridization, permeabilization, hybridization, antibody incubation, staining reaction and stop reaction. The nucleus was stained with DAPI. The section was photographed by laser confocal microscope (Leica, Sp8). Primer sequences used for synthetic probes were listed in Figure S1.
Results
De novo assembly of E. carinicauda gonad transcriptome
We obtained 48.22Gb raw data from 6 samples transcriptome sequencing including ovary and testis and the raw data were upload into the NCBI SRA database with the accession number PRJNA856985. After RNA sequencing based on the Illumina platform and sequence assembly, a total of 47.22 Gb clean data were generated. And the average of each sample was over 6.71Gb with the percentage of Q30 over 94.4% (Supplementary Table 1). The clean data was done de novo assembly with Trinity and the optimized evaluation of the assembly results showed that there were 105,440 unigenes and 191,647 transcripts with the average length of N50 was 1,719bp. The clean reads were aligned to the reference sequence obtained from the trinity assembly, and the alignment rate for this analysis ranged from 75.25% to 76.62%.
Identification of DEGs in ovary and testis as well as GO and KEGG analysis
Through the expression quantity matrix analysis, the ovary samples were significantly clustered together, so were the testis samples, which indicates a good reproducibility between the parallel samples (Figure 1A). The PCA plot showed clearly that the principal components were similar between different replicates in the same sample (Figure 1B). By analyzing the expression of the samples, we can see that there were 33,495 transcripts expressed in the EcF groups and 48,719 transcripts expressed in the EcM groups as well as 17,532 transcripts were shared in both kinds of groups (Figure 1C).
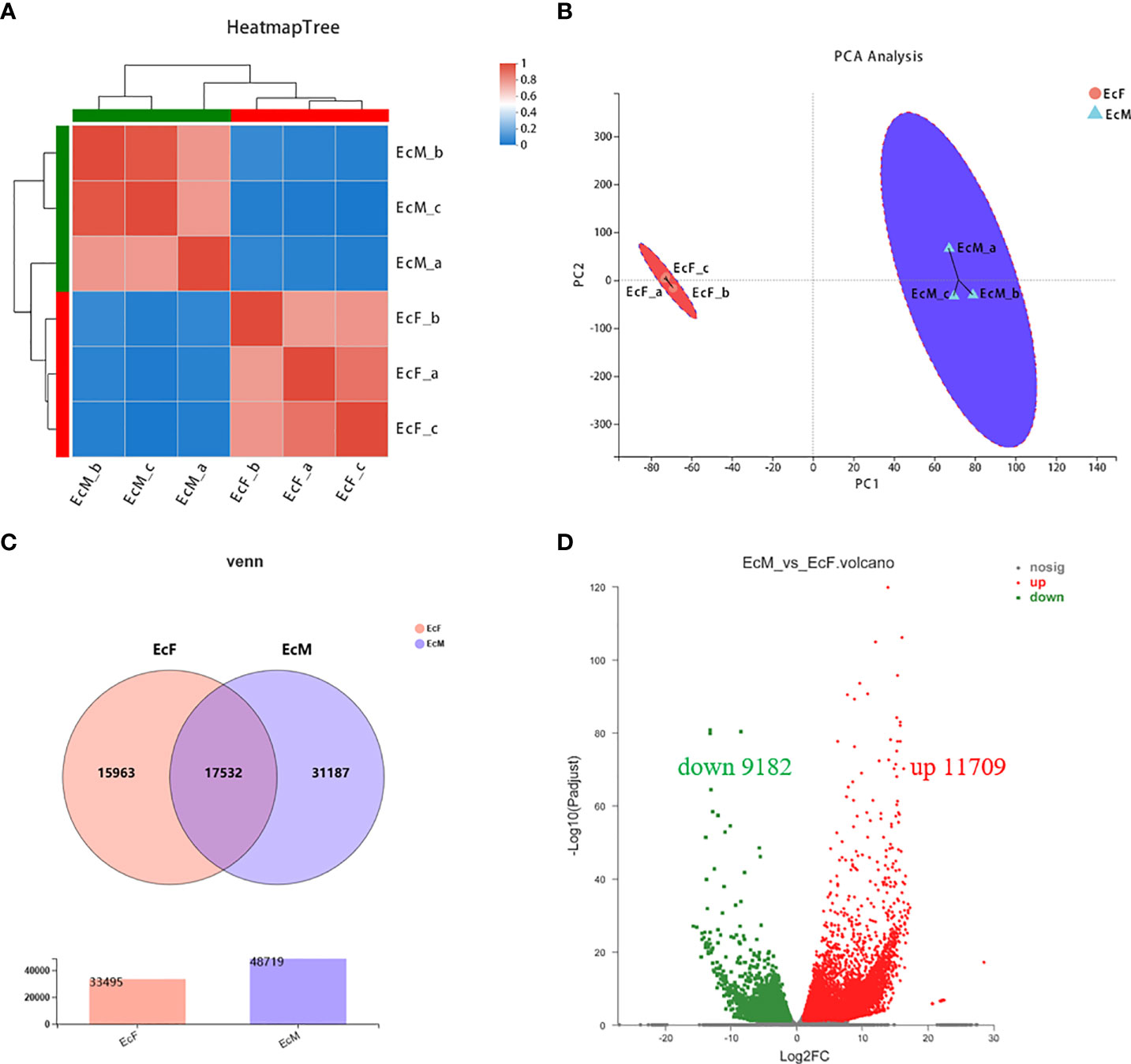
Figure 1 The profile of transcriptome sequence between ovary and testis in E carinicauda. (A) Heatmap Tree of the 3 ovary and 3 testis samples. The right and lower sides are sample names while the left and upper sides are sample clusters. (B) Principal component analysis (PCA) of the 3 replicated ovary samples and 3 replicated testis samples. The distance of each sample point represents the similarity of the sample, the closer distance indicates the higher similarity between the samples. (C) Venn diagram of DEGs in comparison between EcF and EcM group. (D) Volcano plot of DEGs identified from ovary and testis. Red dots indicate the significantly upregulated transcript. Green dots mean the significantly downregulated transcript. Black dots are the non-significant difference of transcript.
Analysis of DEGs in E. carinicauda ovary and testis by transcriptome sequencing helped us to have a better understanding of the gonad development. We screened the DEGs by the principle of FDR<=0.05 and |log2FC|>1. There were 20,891 differentially expressed genes including 11,709 upregulated genes and 9,182 down-regulated genes (EcM-vs-EcF) (Figure 1D). The heatmap of the 25 genes with the top expression level in testis was shown in Figure 2A, while the other 25 genes with the highest expression level in ovary were listed in Figure 2B.
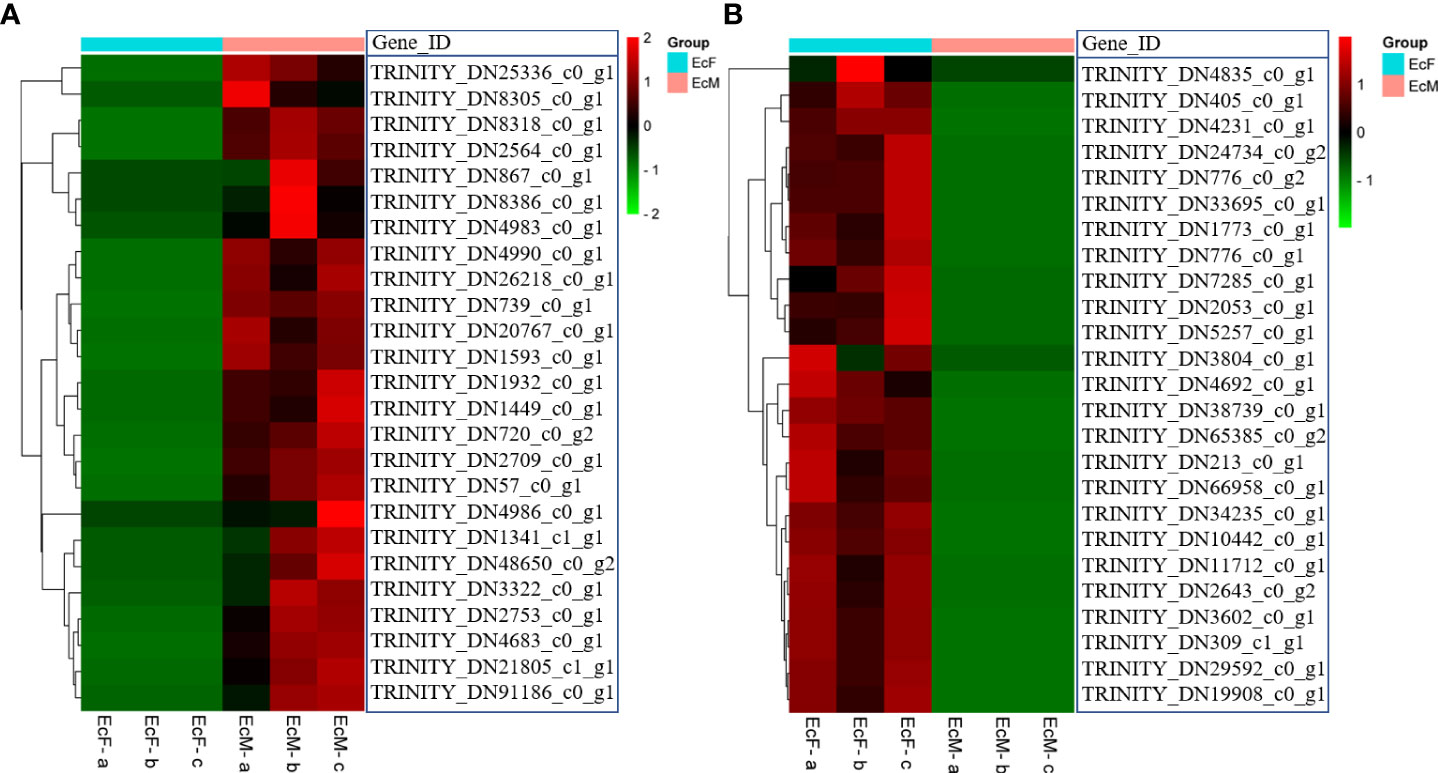
Figure 2 Heatmap of the top 25 highly expressed genes in ovary and testis. (A) The top 25 highly expressed genes in the testis; (B) The top 25 highly expressed genes in the ovary; Each column represents one sample and each row represents one transcript. The color in the figure represents the expression level of the transcript in this sample. The red color represents the high expression level of the transcript in the sample; The green color represents the lower expression level.
The top 20 GO terms enrichment analysis of the DEGs were listed in Figure 3 which included metabolic process, cellular metabolic process and organic cyclic compound metabolic process. And the functional categories were mainly related to meiosis and reproduction including DNA recombination and cellular aromatic compound metabolic process. The KEGG pathway analysis of the top 20 pathways was shown in Figure 4 which was mainly involved in steroidogenesis including ubiquinone and other terpenoid-quinone biosynthesis, fatty acid elongation and fatty acid degradation.
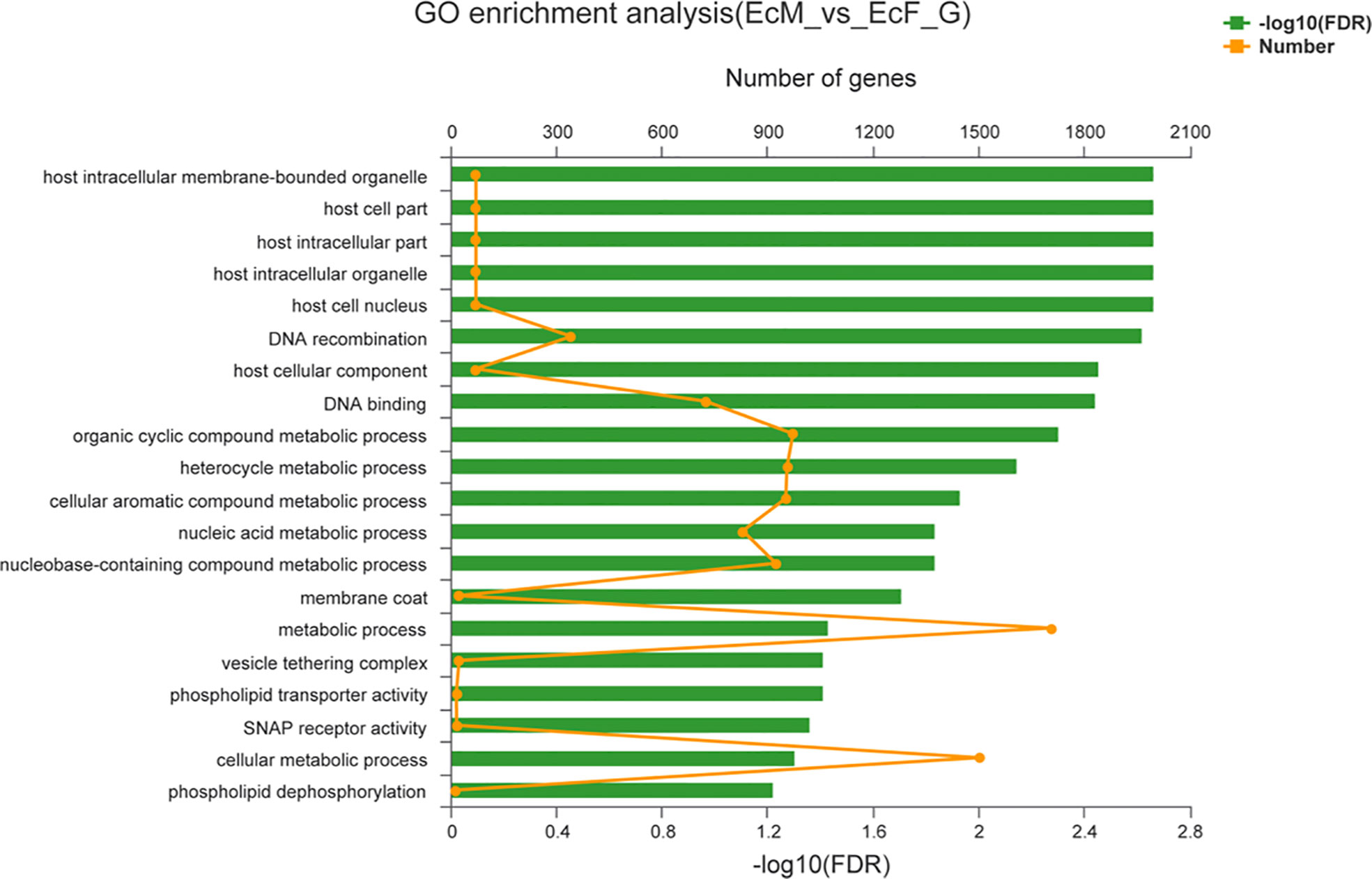
Figure 3 Column diagrams of top 20 enriched biological processes of GO classifications. The ordinate represents the GO term and the upper abscissa indicates the number of transcripts of the GO term on the alignment, corresponding to different points on the broken line; the lower abscissa indicates the significance level of enrichment, the lower abscissa indicates the height of the column, where the smaller the FDR, the greater the-log10 (FDR) value, the more significantly enriched the GO term is.
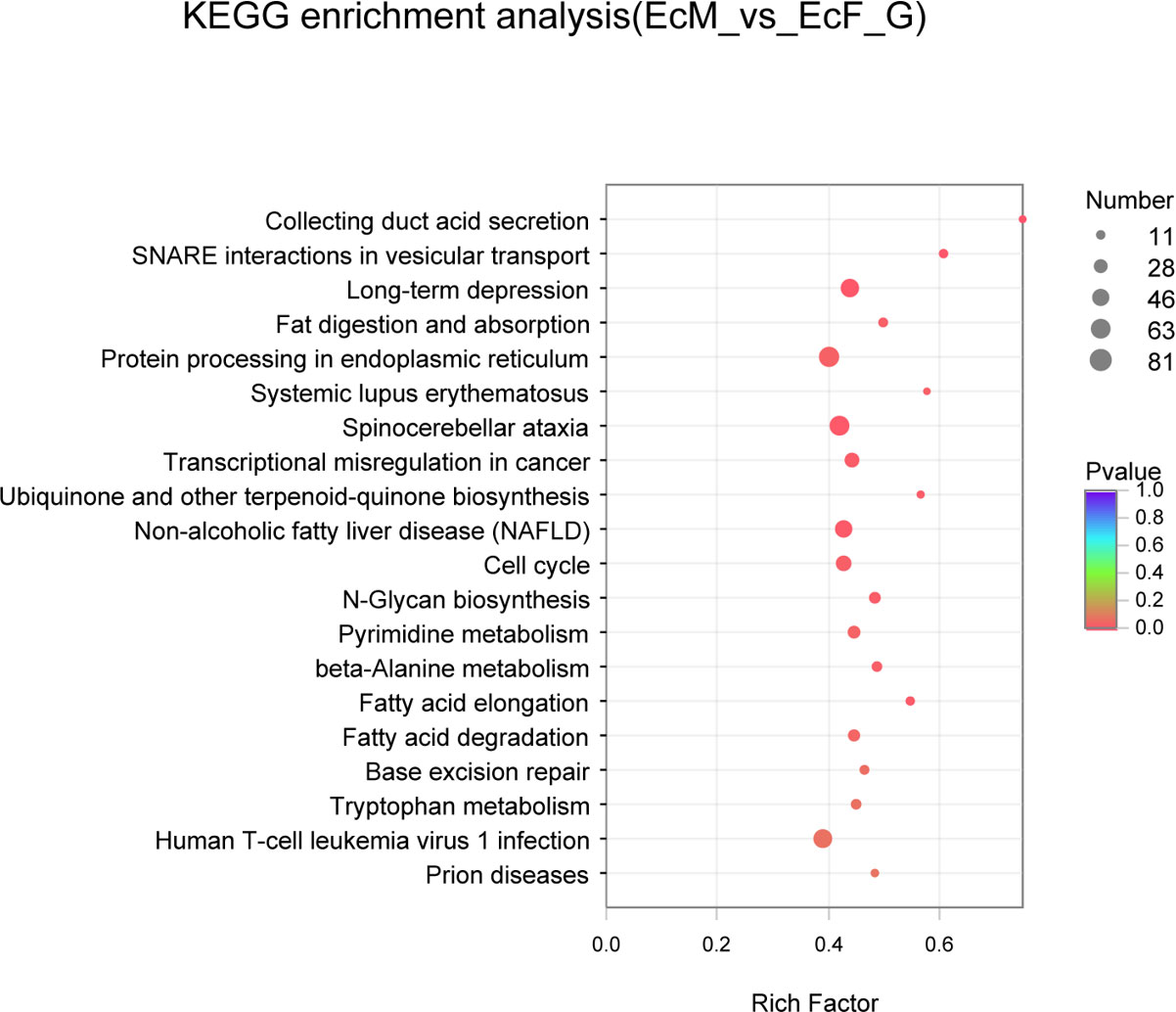
Figure 4 Scatter plots of KEGG pathway enrichment analysis about the top 20 significant pathways of DEGs in EcF-vs-EcM. The ordinate represents the pathway and the abscissa indicates the rich factor, where the bigger the rich factor, the more significantly enriched the KEGG pathway is.
Differentially expressed genes related to male reproduction
Combined with the annotation of the NR database, we found a number of genes specifically expressed in testis (Figure 5A). Then, we checked the results by real-time PCR and the expression level of these genes was in accord with the transcriptome data (Figure 5B) which indicated the accuracy of the transcriptome sequencing. These genes showed a significantly higher expression level in males than in females (*P<0.05, **P<0.01, ***P<0.001, ****P<0.0001). The annotations of these genes are mainly related to spermatogenesis and male reproduction including some proteins which involved in this process.
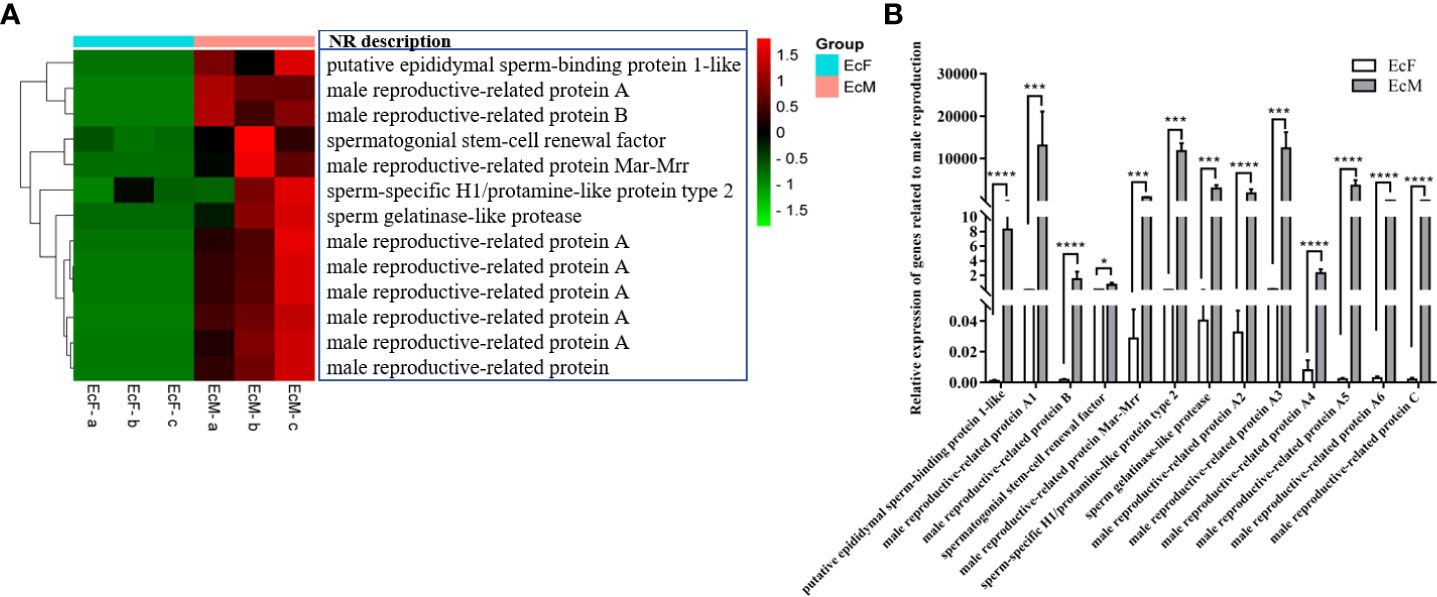
Figure 5 Differentially expressed genes (DEGs) related to male reproduction. (A) Heatmap of DEGs related to male reproduction. Each column in the figure represents one sample and each row represents one transcript, the color in the figure indicates the high or low expression of transcript in that sample, the red color represents the higher expression of the transcript in the sample, the green color indicates the lower expression. (B) The real-time PCR validation of gene expression related to male reproduction (*P<0.05, ***P<0.001, ****P<0.0001). The data analysis was conducted by 2-∆CT and 18S was chosen as reference gene.
Differentially expressed genes related to cell cycle
As is known to us, spermatogenesis and oogenesis show different characteristics and they contain various events. We identified some DEGs related to cell cycle and the functions related to anaphase promoting factors, spindle assembly checkpoints and cell division. Some of them are highly expressed in testis while others show a high level in the ovaries and they present their own characteristics (Figure 6A). And this result was further examined by real-time PCR (Figure 6B). Just as meiosis in other species, the complete of ovary meiosis need more time since oogenesis need to accumulate much vitellogenin while the spermatogenesis process doesn’t need (Tsukimura, 2001; Wei et al., 2021). Consequently, the cell proliferation and differentiation in testis and ovary of E. carinicauda showed a sexual dimorphism.
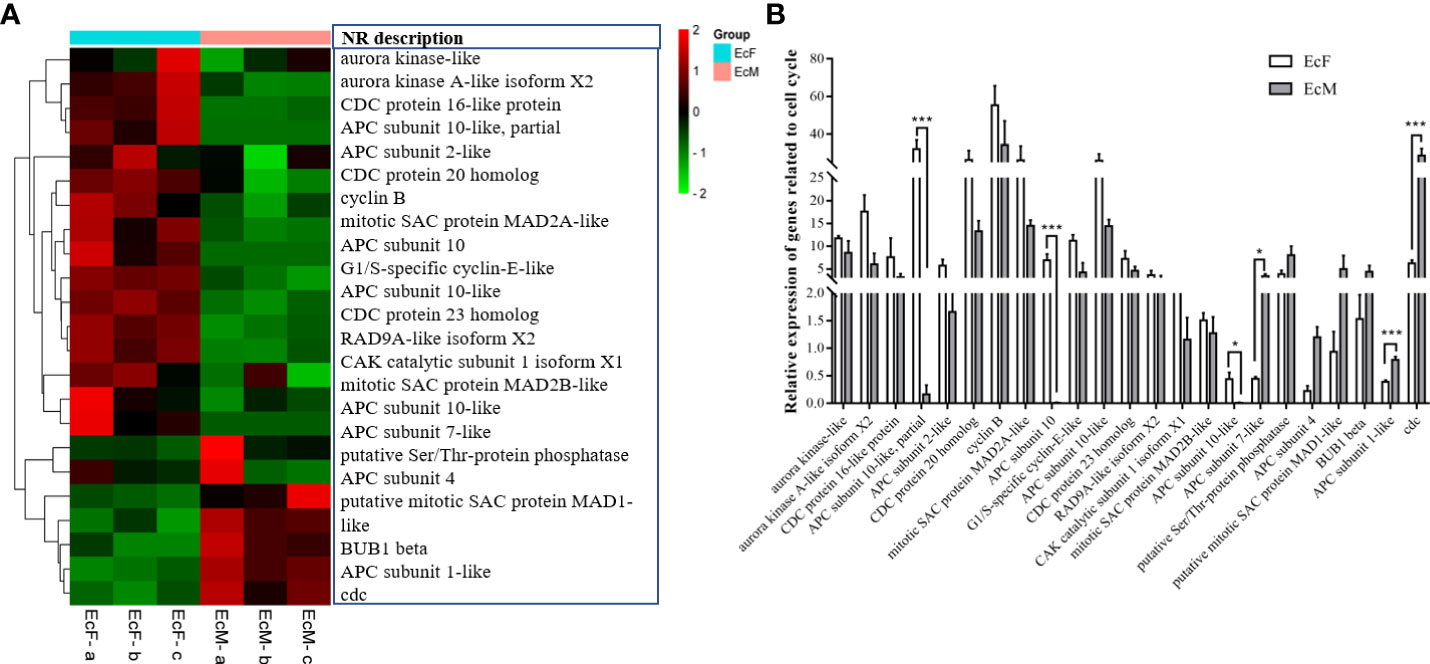
Figure 6 Differentially expressed genes (DEGs) related to cell cycle. (A) Heatmap of DEGs related to cell cycle. Each column in the figure represents one sample and each row represents one transcript, the color in the figure indicates the high or low expression of transcript in that sample, the red color represents the higher expression of the transcript in the sample, the green color indicates the lower expression level transcripts. (B) Validation of gene expression related to cell cycle by real-time PCR (*P<0.05, ***P<0.001). The data analysis was conducted by 2-∆CT and 18S was chosen as reference gene.
Excavation and identification of the transcription factor foxj1b associated with ovarian differentiation
Transcription factors are proteins that can bind to specific DNA sequences and generally process one or more functional domains (Lambert et al., 2018). Usually, the transcription factors play functions by two ways. Besides binding to promoter upstream regions of the gene, the transcription factor could also form complexes with other transcription factors to affect gene transcription (Pope & Medzhitov, 2018). There are 23 families including 567 transcription factor members (Figure 7A). And we found foxj1b which comes from the fork head family might be associated with ovary differentiation. We did a preliminary validation by two-color fluorescent in situ hybridization using ovary section samples in various stage (Figure 7B). The results showed that foxj1b is mainly expressed in early development of the ovary (Ovary I) and late ovulation stage (Ovary III). There were little signals in testis and mature ovary (Ovary II).
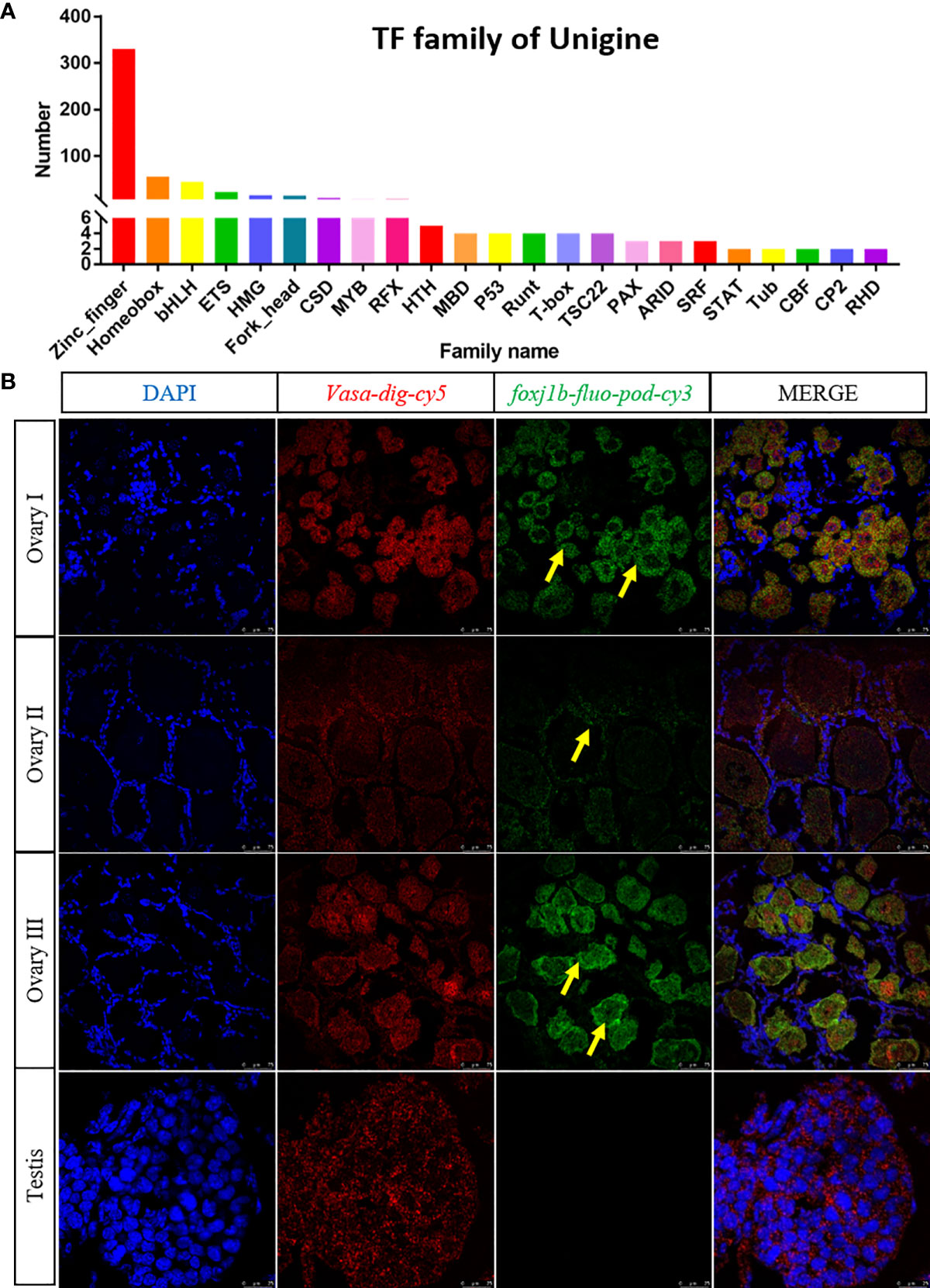
Figure 7 checking the expression profile of the transcription factor foxj1b by two-color fluorescent in situ hybridization. (A) Statistics of transcription factors in E.carinicauda. The abscissa stands for different families of transcription factors; the ordinate means the number of transcripts falling to the family. (B) Mapping the expression of foxj1b and vasa by two-color fluorescent in situ hybridization in gonads. The blue signal: DAPI; the red signal: vasa; the green signal: foxj1b.
Discussion
There are several characteristics in E. carinicauda including small body size which is fit to feed under laboratory conditions and whole transparent body as well as high fecundity which are suitable to genetic operation (Yuan et al., 2017). Therefore, E. carinicauda is a popular species in genetic study. So far, E. carinicauda is the only reported decapoda crustacean species which is suitable for gene editing (Gui et al., 2016). According to the result of the genome survey, the structure of the E. carinicauda genome was big and complicated (Yuan et al., 2017; Li et al., 2019). And there were no published E. carinicauda genome data at present. Therefore, it is essential to enrich and re-annotate the E. carinicauda genome by diverse omics data, especially for important tissues with significant differences between males and females. As for the study of gene function, the accurate annotation of the genome is essential and significant. However, there is still a long way to go on the study of E. carinicauda functional genomics research. In this study, we obtained DEGs between ovary and testis by transcriptome sequencing, including 11,709 up-regulated genes and 9,182 down-regulated genes, which could provide clue for the study of E. carinicauda reproduction development.
Spermiogenesis is an important process in male reproduction (Staub & Johnson, 2018). The first stage of spermiogenesis is the spermatogonia division and proliferation, which could produce spermatocytes. And the second stage is the spermatocyte meiosis that could form a haploid sperm cell. The third stage is that the sperm cell metamorphoses into sperm. We identified several genes related to spermiogenesis (Figure S2) although their functions need to be further confirmed. Compared with spermiogenesis, the distinctive events in oogenesis are the synthesis of VTG and the process of oocyte maturation (Subramoniam, 2010). There are several transcripts which might be related to the synthesis of VTG in our transcriptome result (Figure S3). These findings offered clues for the study of E. carinicauda spermiogenesis and oogenesis.
Transcription factors regulate the target gene expression by their specific functional domains. Currently, they play increasingly important roles in sex determination and differentiation, embryo development, and organ formation. Sry is the first transcription factor identified in humans for sex determination, which could regulate the expression of Sox9 (Sinclair et al., 1990; Vining et al., 2021). Then transcription factors such as amh, dmy and foxl2 were successively found to play roles in sex differentiation and ovary development respectively (Matsuda et al., 2002; Georges et al., 2014; Song et al., 2021). For instance, amh is a sex-determining gene in a chicken and dmy could determine the sex medaka (Matsuda et al., 2002; Smith & Sinclair, 2004). While foxl2 belongs to the fork head family which could play roles in ovary differentiation in mammals and zebrafish (Yang et al., 2017; Tucker, 2021). However, there were no genes which were reported related to ovary differentiation in E. carinicauda. We found 567 transcription factors by transcriptome sequencing (Figure 7A). After a comparison of the expression level between males and females, we focused on the transcription factor foxj1b. The two-color fluorescent in situ hybridization results showed that the foxj1b was mainly expressed in early development and late ovulation stage of the ovary but the expression level was very low in mature ovary and no signal could be detected in testis (Figure 7B). Our results suggested that foxj1b might play roles in ovary early development in E. carinicauda by two-color fluorescent in situ hybridization for the first time. However, the specific function of foxj1b in E. carinicauda still needs to be further studied by gene editing or overexpression.
In conclusion, we discovered 105,440 differentially expressed genes in testis and ovary by transcriptome sequencing, including genes related to reproduction and cell cycle as well as GO terms enrichment and KEGG pathway analysis. Furthermore, the results were verified by real-time PCR and two-color fluorescent in situ hybridization. Our results would lay the foundation for subsequent studies of gene functions related to male reproduction and ovary development in E. carinicauda.
Data availability statement
The datasets presented in this study can be found in online repositories. The data presented in the study are deposited in the NCBI SRA repository, accession number PRJNA856985.
Ethics statement
The animal study was reviewed and approved by Ethics Committee of Yellow Sea Fisheries Research Institute.
Author contributions
JL designed the study. SJ and LJ performed the experiments and analyzed the data. SJ wrote the manuscript. JJL, JW and JTL helped in manuscript revising. JL and PL checked the experiment design and reviewed the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This research was financially supported by National Key R&D Program of China (2019YFD0900403), China Agriculture Research System of MOF and MARA (CARS-48), Postdoctoral Applied Research Project of Qingdao (2021), National Natural Science Foundation of China (32072974) and Central Public-interest Scientific Institution Basal Research Fund of CAFS (2020TD46).
Acknowledgments
We are grateful to Ms. Jiawei Wang (Majorbio Co., Ltd) for her kind help in professionally operating Majorbio Cloud Platform.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2022.995790/full#supplementary-material
Abbreviations
DEGs, the differentially expressed genes; NR, Non-Redundant Protein Sequence Database; COG, Database of Clusters of Orthologous Genes; Pfam, The Pfam protein families database; GO, Gene Ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes; PCA, Principal component analysis; FDR, False Discovery Rate; EcF, the ovary group of female Exopalaemon carinicauda; EcM, the testis group of male Exopalaemon carinicauda.
References
Du J., Liu Y., Song C., Cui Z. (2019). Discovery of sex-related genes from embryonic development stage based on transcriptome analysis in Eriocheir sinensis. Gene 710, 1–8. doi: 10.1016/j.gene.2019.05.021
Georges A., Auguste A., Bessiere L., Vanet A., Todeschini A. L., Veitia R. A. (2014). FOXL2: a central transcription factor of the ovary. J. Mol. Endocrinol. 52 (1), R17–R33. doi: 10.1530/JME-13-0159
Gui T., Zhang J., Song F., Sun Y., Xie S., Yu K., et al. (2016). CRISPR/Cas9-mediated genome editing and mutagenesis of EcChi4 in Exopalaemon carinicauda. G3 (Bethesda) 6 (11), 3757–3764. doi: 10.1534/g3.116.034082
Hamasaki K., Osabe N., Nishimoto S., Dan S., Kitada S. (2020). Sexual dimorphism and reproductive status of the red swamp crayfish Procambarus clarkii. Zool Stud. 59, e7. doi: 10.6620/ZS.2020.59-07
Huang P., Guo W., Wang Y., Xiong Y., Ge S., Gong G., et al. (2022). Genome-wide association study reveals the genetic basis of growth trait in yellow catfish with sexual size dimorphism. Genomics 114 (3), 110380. doi: 10.1016/j.ygeno.2022.110380
Lambert S. A., Jolma A., Campitelli L. F., Das P. K., Yin Y., Albu M., et al. (2018). The human transcription factors. Cell 172 (4), 650–665. doi: 10.1016/j.cell.2018.01.029
Lauter G., Söll I., Hauptmann G. (2011). Multicolor fluorescent in situ hybridization to define abutting and overlapping gene expression in the embryonic zebrafish brain. Neural Dev. 6, 10. doi: 10.1186/1749-8104-6-10
Liao X., Xu G., Chen S. L. (2014). Molecular method for sex identification of half-smooth tongue sole (Cynoglossus semilaevis) using a novel sex-linked microsatellite marker. Int. J. Mol. Sci. 15 (7), 12952–12958. doi: 10.3390/ijms150712952
Li J., Lv J., Liu P., Chen P., Wang J., Li J. (2019). Genome survey and high-resolution backcross genetic linkage map construction of the ridgetail white prawn Exopalaemon carinicauda applications to QTL mapping of growth traits. BMC Genomics 20, 598. doi: 10.1186/s12864-019-5981-x
Lv J., Lu X., Ti X., Liu P., Li J., Li J. (2020). QTL mapping and marker identification for sex determination in the ridgetail white prawn, Exopalaemon carinicauda. Genomics 112 (6), 5240–5247. doi: 10.1016/j.ygeno.2020.09.037
Matsuda M., Nagahama Y., Shinomiya A., Sato T., Matsuda C., Kobayashi T., et al. (2002). DMY is a y-specific DM-domain gene required for male development in the medaka fish. Nature 417 (6888), 559–563. doi: 10.1038/nature751
Pope S. D., Medzhitov R. (2018). Emerging principles of gene expression programs and their regulation. Mol. Cell 71 (3), 389–397. doi: 10.1016/j.molcel.2018.07.017
Shen Y., Wang Q., Wang W., Li Y. (2022). Exploration of an XX/XY sex determination system and development of PCR-based sex-specific markers in Procambarus clarkii based on next-generation sequencing data. Front. Genet. 13, 850983. doi: 10.3389/fgene.2022.850983
Sinclair A. H., Berta P., Palmer M. S., Hawkins J. R., Griffiths B. L., Smith M. J., et al. (1990). A gene from the human sex-determining region encodes a protein with homology to a conserved DNA-binding motif. Nature 346 (6281), 240–244. doi: 10.1038/346240a0
Smith C. A., Sinclair A. H. (2004). Sex determination: insights from the chicken. Bioessays 26 (2), 120–132. doi: 10.1002/bies.10400
Song W., Xie Y., Sun M., Li X., Fitzpatrick C. K., Vaux F., et al. (2021). A duplicated amh is the master sex-determining gene for sebastes rockfish in the Northwest pacific. Open Biol. 11 (7), 210063. doi: 10.1098/rsob.210063
Staub C., Johnson L. (2018). Review: Spermatogenesis in the bull. Animal 12 (s1), s27–s35. doi: 10.1017/S1751731118000435
Subramoniam T. (2010). Mechanisms and control of vitellogenesis in crustaceans. Fisheries Sci. 77 (1), 1–21. doi: 10.1007/s12562-010-0301-z
Tsukimura B. J. A. Z. (2001). Crustacean vitellogenesis: Its role in oocyte development. Amer. Zool. 41, 465–476. doi: 10.1093/icb/41.3.465
Vining B., Ming Z., Bagheri-Fam S., Harley V. (2021). Diverse regulation but conserved function: SOX9 in vertebrate sex determination. Genes (Basel) 12 (4), 486. doi: 10.3390/genes12040486
Wang Q., He Y., Li J. (2019). Conjoint analysis of SMRT- and illumina-based RNA-sequencing data of Fenneropenaeus chinensis provides insight into sex-biased expression genes involved in sexual dimorphism. Front. Genet 10, 1175. doi: 10.3389/fgene.2019.01175
Wang B., Kumar V., Olson A., Ware D. (2019). Reviving the transcriptome studies: An insight into the emergence of single-molecule transcriptome sequencing. Front. Genet. 10, 384. doi: 10.3389/fgene.2019.00384
Wang J., Li J., Ge Q., Chen Z., Li J. (2020). Effects of inbreeding on genetic characteristic, growth, survival rates, and immune responses of a new inbred line of exopalaemon carinicauda. Int. J. Genomics 2020, 1–11. doi: 10.1155/2020/5735968
Wan Z. Y., Lin V. C. L., Hua Y. G. (2021). Pomc plays an important role in sexual size dimorphism in tilapia. Mar. Biotechnol. (NY) 23 (2), 201–214. doi: 10.1007/s10126-020-10015-2
Wei L. L., Chen T. T., Luo B. Y., Qiu G. F. (2021). Evidences for red pigment concentrating hormone (RPCH) and beta-pigment dispersing hormone (beta-PDH) inducing oocyte meiotic maturation in the Chinese mitten crab, Eriocheir sinensis. Front. Endocrinol. (Lausanne) 12, 802768. doi: 10.3389/fendo.2021.802768
Xu W., Xie J., Shi H., Li C. (2010). Hematodinium infections in cultured ridgetail white prawns, Exopalaemon carinicauda, in eastern China. Aquaculture 300 (1-4), 25–31. doi: 10.1016/j.aquaculture.2009.12.024
Yang Y. J., Wang Y., Li Z., Zhou L., Gui J. F. (2017). Sequential, divergent, and cooperative requirements of Foxl2a and Foxl2b in ovary development and maintenance of zebrafish. Genetics 205 (4), 1551–1572. doi: 10.1534/genetics.116.199133
Yuan J., Gao Y., Zhang X., Wei J., Liu C., Li F., et al. (2017). Genome sequences of marine shrimp exopalaemon carinicauda holthuis provide insights into genome size evolution of caridea. Mar. Drugs 15 (7), 213. doi: 10.3390/md15070213
Keywords: Exopalaemon carinicauda, reproduction, gonad, transcriptome, foxj1b
Citation: Jia S, Jin L, Lv J, Wang J, Li J, Liu P and Li J (2022) Comparative transcriptomic analysis and validation of the ovary and testis in the ridgetail white prawn (Exopalaemon carinicauda). Front. Mar. Sci. 9:995790. doi: 10.3389/fmars.2022.995790
Received: 16 July 2022; Accepted: 21 September 2022;
Published: 10 October 2022.
Edited by:
Yunyan Deng, Institute of Oceanology (CAS), ChinaReviewed by:
Yafei Duan, South China Sea Fisheries Research Institute, ChinaXiaofang Lai, Jiangsu Ocean Universiity, China
Muhammad Adil Farooq, Khwaja Fareed University of Engineering and Information Technology (KFUEIT), Pakistan
Copyright © 2022 Jia, Jin, Lv, Wang, Li, Liu and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jian Li, bGlqaWFuQHlzZnJpLmFjLmNu
†These authors have contributed equally to this work and share first authorship
 Shaoting Jia
Shaoting Jia Ling Jin1,2†
Ling Jin1,2† Jiajia Wang
Jiajia Wang Jitao Li
Jitao Li Ping Liu
Ping Liu