- 1College of Life Sciences, Guangxi Normal University, Key Laboratory of Ecology of Rare and Endangered Species and Environmental Protection (Guangxi Normal University), Ministry of Education, Guilin, China
- 2Guangxi Key Laboratory of Landscape Resources Conservation and Sustainable Utilization in Lijiang River Basin (Guangxi Normal University), Guilin, China
- 3School of Life Sciences, Sun Yat-sen University, Guangzhou, China
Carotenoids are widely distributed and structurally diverse, which have significant roles in the photosynthesis of plants. As a precursor of vitamin A, carotenoids are also antioxidants that reduce various chronic diseases, which are beneficial for human health. Currently, the existing studies concerned the biological roles of APETALA2 (AP2)/ethylene-responsive factor (ERF) genes originated from higher plants. The AP2 superfamily of the transcriptional regulator was identified in higher plants, which was related to growth, development, carotenoid metabolism, and responses to various stresses. However, the regulatory mechanisms of the AP2-modulating carotenoid metabolism have not been reported in microalgae, which remain to be elucidated. Dunaliella parva AP2 (i.e., DpAP2), an important transcription factor, promotes carotenoid accumulation by binding to the promoter of target gene. Here, we identified an important AP2/ERF transcription factor, DpAP2, which could promote carotenoid accumulation by binding to the promoter of target gene. To demonstrate the function of DpAP2, the interacting proteins were identified by the yeast two-hybrid system. The results showed that DpAP2 could interact with three proteins with different activities (DNA-binding transcription factor activity, protein kinase activity, and alpha-D-phosphohexomutase activity); these proteins may be associated with multiple biological processes. This paper laid a good foundation for a deep understanding of the regulatory mechanisms of DpAP2 and genetic engineering breeding in D. parva.
Introduction
Microalgae have been utilized commercially in the Far East for healthy foodstuffs such as Arthrospira platensis and in the United States for wastewater treatment (Oswald, 2003). At present, in response to the increasing concern around global warming, the microalgal culture was again considered for a carbon-neutral process to obtain third-generation biofuels such as biodiesel (Gilmour, 2019). Biomass productivity, oil content, and energy-intensive harvesting were the major constraints for biodiesel production from microalgae on a large scale (Shahid et al., 2020). Nitrogen limitation has been widely used to increase oil content in microalgae (Kumari et al., 2021; Zhang et al., 2021). When Scenedesmus acuminatus grew under nitrogen-limited conditions, oil productivity was enhanced (Zhang et al., 2021). Nitrogen limitation increased oil content in Chlorella vulgaris, which might be related to the upregulation of mRNA levels of several oil biosynthesis genes (Kumari et al., 2021). Our previous study also showed that nitrogen limitation could induce an increase in oil content from 25% to 40% in D. parva (Shang et al., 2016). However, nitrogen limitation also hampered the growth, photosynthetic antennae size, and maximum photosynthetic efficiency in algae (Jiang et al., 2021; Kamalanathan et al., 2021), which was also demonstrated by our previous study (Shang et al., 2016). The biochemical engineering approach depended on physiological stresses such as nitrogen limitation to channel metabolic fluxes to lipid accumulation. However, an inherited shortcoming of the biochemical engineering approach was the reduced cell division, which limited the application of this approach (Ratledge, 2002). Compared with the biochemical engineering approach, the transcription factor engineering approach was an emerging technology to improve the yield of the specific metabolites by overexpressing transcription factors regulating the metabolic pathways related to the accumulation of target metabolites (Courchesne et al., 2009).
A comprehensive utilization of microalgae could significantly reduce the cost of microalgae biofuels, which included microalgae cultivation using wastewaters in outdoor raceway ponds (Arora et al., 2016; Gao et al., 2018; Keerthana et al., 2020) and the production of bioactive substances such as polysaccharides (Li et al., 2019), antiviral substances (Fritzsche et al., 2021), bioactive peptides (Donadio et al., 2021), and carotenoids (du Preez et al., 2021; Todorović et al., 2021).
Carotenoid biosynthesis involves a complex pathway (Narang et al., 2021). Our previous study also cloned and characterized several carotenoid synthesis genes such as GGPS encoding geranylgeranyl diphosphate synthase (Shang et al., 2016b), Psy encoding phytoene synthase, and Pds encoding phytoene desaturase (Shang et al., 2018). In D. parva, the PSY gene played important roles in carotenoid metabolism (Ismaiel et al., 2018). These genes played a key role in the carotenoid biosynthesis pathway because they catalyzed the rate-limiting step. The current studies focused on the regulators (especially transcription factors) of the metabolic pathway rather than the key enzyme (Li et al., 2021). A lot of transcription factors (TFs) regulating the carotenoid biosynthesis pathway were identified in higher plants, which included MADS-box (Kang et al., 2021), NAC (Gong et al., 2021), AP2/ERF (Dang et al., 2021), and MYB families (Shi et al., 2021). These TFs can bind to the promoter of the target gene. The putative carotenoid biosynthetic pathway in microalgae is shown in Figure 1. The metabolic enzymes were encoded by the structural genes in the carotenoid biosynthesis pathway, which included zeta-carotene desaturase (ZDS), carotenoid isomerase (CRTISO), ζ-carotene isomerase (ZISO), lycopene β-cyclase (LCY-β), beta-carotene hydroxylase (CHY-β), lycopene ϵ-cyclase (LCY-ϵ), and zeaxanthin epoxidase (ZEP) (Ampomah-Dwamena C et al., 2019).
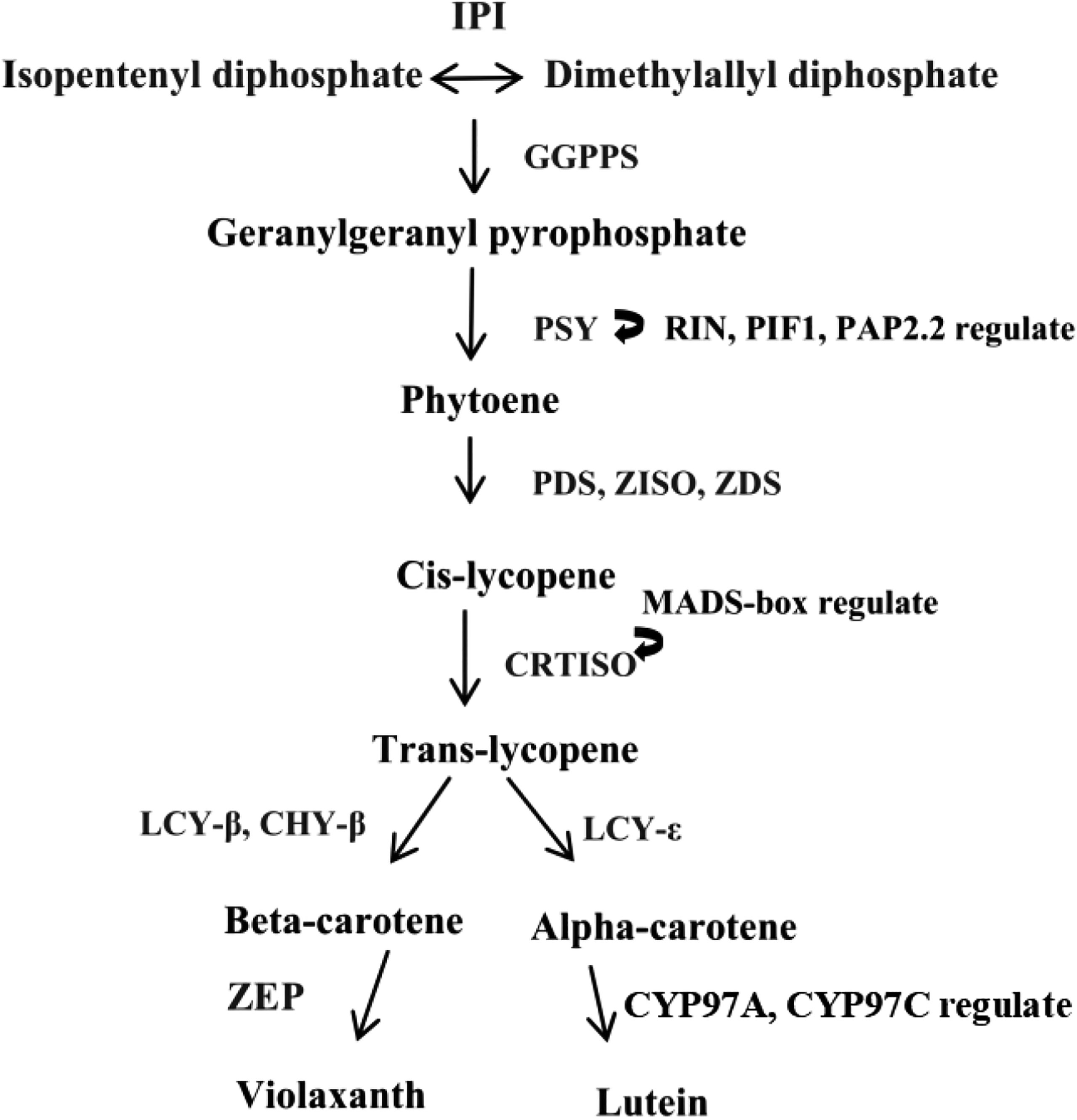
Figure 1 A graphic representation depicting the putative carotenoid biosynthetic pathway in microalgae. IPI, isopentenyl diphosphate isomerase; GGPPS, geranylgeranyl diphosphate synthase; PSY, phytoene synthase; PDS, phytoene desaturase; ZDS, zeta-carotene desaturase; CRTISO, carotenoid isomerase; ZISO, ζ-carotene isomerase; LCY-β, lycopene β-cyclase; CHY-β, beta-carotene hydroxylase; LCY-ϵ, lycopene ϵ-cyclase; ZEP, zeaxanthin epoxidase.
The APETALA2/ethylene-responsive factor (AP2/ERF) superfamily in plants contains four subfamilies including AP2, CBF/DREB, ERF, and RAV according to the number of AP2/ERF domains and the sequence (Xing et al., 2021). TFs played important regulatory roles in the development, growth, and responses to various stresses in plants. The AP2/ERF, a complex TF family, is one of the largest families, which efficiently regulates carotenoid accumulation in plants. In Auxenochlorella protothecoides, AP2, ERF, and R2R3-MYB promoted triacylglycerol accumulation by triggering the lipid biosynthesis pathway (Xing et al., 2021). Previous studies have reported that AP2/ERF TF could modulate carotenoid accumulation. In apple, MdAP2-34 was able to enhance carotenoid accumulation by binding to the MdPSY2-1 promoter (Dang et al., 2021). SIERF6 could exactly regulate carotenoid accumulation in tomato (Lee et al., 2012). The TFs (AP2, AP2-like) related to phytohormones potentially affected carotenoid metabolism in apricot (Zhang et al., 2019). RAP2.2 could efficiently regulate carotenoid accumulation by binding to the selected site of the promoter region in tomato (Koul et al., 2019). In Arabidopsis, RAP2.2 could also accelerate carotenoid accumulation by binding to the PSY promoter (Welsch et al., 2007). In addition, we have overexpressed DpAP2 genes in D. parva in our subsequent experiments. The total carotenoid content increased to 0.85 μg/g of dry cell weight. However, the regulatory mechanism of AP2/ERF TF for carotenoid metabolism is currently unknown in D. parva.
D. parva is an oleaginous halophilic green alga without a cell wall, which can also accumulate large amounts of carotenoids (mainly β-carotene) (Shang et al., 2016). The simultaneous production of biodiesel and carotenoids using D. parva is more attractive compared with the model microalgae. As a new type of bioreactor, D. parva has been widely used because of its characteristics including rapid growth, rapid reproduction, easy culture, the ease of controlling pollution, high efficiency, and low price (Shang et al., 2016). D. parva is rich in natural carotenoids, folic acid, calcium, and other trace elements required for human health. Carotenoids can be utilized as a colorant, provitamin, precursor of abscisic acid, chemopreventive substance against cancer, and antioxidant (Mohsin et al., 2021). AP2/ERF TF plays significant roles in carotenoid accumulation in plants, but these roles remain unknown in D. parva.
In a previous study, we identified a gene fragment DpAP2 encoding AP2/ERF TF. The role of DpAP2 is still unclear in carotenoid metabolism in D. parva. In view of a correlation between DpAP2 and carotenoid production, it is of interest to study the function of DpAP2 at the molecular level. Here, we reported the cloning of the full-length complementary DNA (cDNA) of DpAP2. Then, the DpAP2-interacting proteins were identified by yeast two-hybrid systems and subjected to bioinformatics analysis and further discussion.
Materials and Methods
Microalgal Material and Growth Conditions
D. parva FACHB-815 was purchased from Freshwater Algae Culture Collection at the Institute of Hydrobiology (Wuhan, China). The cells of D. parva grew in the Dunaliella medium under the light intensity of 34 μmol of photons m−2 s−1 illumination at 26°C with 14 h light/10 h dark cycle. The algae bottles were gently swirled one or three times each day by hand.
First-Strand cDNA Synthesis
Total RNA was extracted from D. parva cells using a Total RNA Extractor (Trizol) (Sangon Biotech, Shanghai, China). To ensure that the enough and qualified samples were obtained, the RNA quantity and quality were determined with a Nanophotometer NP80 Spectrophotometer (Geneflow, Lichfield, United Kingdom). First-stand cDNA was synthesized using PrimeScript II Reverse Transcriptase (Takara Bio, Dalian, China) and Oligo (dT) primer. A total amount of 1.5 μg of total RNA was used for reverse transcription reaction.
Full-Length cDNA Cloning
An attempt was made to gain full-length cDNA by the rapid amplification of cDNA ends (RACE) method. Based on the previous 582 bp cDNA fragment of DpAP2, four specific primers (Table 1), AP2(3w)/AP2(3n) for 3’ RACE and AP2(5w)/AP2(5n) for 5’ RACE, were designed. The 5’ and 3’ RACE were firstly performed using the 5’/3’-Full RACE Kit (Takara) according to the manufacturer’s protocol. The outer and inner PCR were conducted with LA Taq (Takara) under the following conditions, respectively: 94°C 3 min, 25 cycles (94°C 30 s, 55°C 30 s, 72°C 2 min), 72°C 10 min for outer PCR, and 94°C 3 min, 35 cycles (94°C 30 s, 55°C 30 s,72°C 2 min), 72°C 10 min for inner PCR. The resulting products were detected with 1% agarose gel electrophoresis and ligated into the pMD19-T vector. However, the resulting 5’ nucleotide sequence was shorter than the expected 5’ end sequence. In order to obtain the entire 5′ sequence, the second 5’ RACE was carried out using the specific primer AP2(GSP) and Long Primer from the SMARTer RACE 5’/3’ Kit (Table 1) (Zhu et al., 2001). The primer AP2(GSP) was designed according to the obtained sequence in the first 5’ RACE. According to the assembled full-length sequence, specific primers AP2(N) and AP2(C) (Table 1) were synthesized to obtain the accurate full-length cDNA of DpAP2. PCR products were purified, cloned, and sequenced as described above (Abid et al., 2012).
Construction of cDNA Library of D. parva
Total RNA was isolated from D. parva using the MiniBEST Plant RNA Extraction Kit (Takara). This kit is more efficient, fast, and convenient. The construction of the cDNA library of D. parva requires a high concentration and purity of RNA. Up to 10 µg of total RNA with high purity could be extracted from 50~100 mg plant tissue by this kit. The degradation and contamination of RNA were detected by 1% agarose gel electrophoresis. The RNA concentration was determined with the Nanophotometer NP80 Spectrophotometer. The cDNA library was constructed according to the instructions from the SMART cDNA Library Construction Kit (Takara) (Mendelsohn and Brent, 1994). First-strand cDNA was synthesized using CDS (complementary determining region) III Oligo (dT) primer and MMLV reverse transcriptase. Double-strand cDNA was synthesized by LD PCR with specific primers (5’ PCR Primer and 3’ PCR Primer) (Table 1). The cDNA size was fractionated by CHROMA SPIN+TE-400 Column to select cDNA molecules (>200 bp). Finally, cDNA fragments were cloned into the pGADT7-Rec plasmid through homologous recombination in vivo, transformed into the competent yeast strain Y187, and selected on the selective medium by the yeast two-hybrid assay.
Subtraction Efficiency of the Library
The purified double-strand cDNA and pGADT7-Rec were co-transformed into Y187 yeast competent cells. Then, all liquids were combined in a single sterile flask. The library broth was diluted 10-2 and 10-4 times, and 100 μl was spread onto the SD/–Leu plate for 3–5 days. The number of colonies on plates were counted. PCR amplification was used to evaluate recombination efficiency (Zheng et al., 2005). PCR was performed with universal sequencing primers designed for PGADT7-REC using 24 colonies as templates, which were randomly selected on SD/-Leu plates. PCR was performed using Green Taq Mix 12.5 μl, T7 Primer (10 μM) 1 μl, 3’AD Primer (10 μM) 1 μl, yeast culture 1 μl, ddH2O 9.5 μl. The procedures were as follows: 94°C for 3 min, 35 cycles (98°C for 10 s, 42°C for 30 s, 72°C for 2 min), 72°C for 10 min. PCR products were analyzed by 1% agarose gel electrophoresis. The size and recombination rate of inserted fragments were measured and analyzed.
Generation of Yeast Bait Strain
All plasmids and strains used in the two-hybrid experiment were included in the Matchmaker Gold Yeast Two-Hybrid System Kit (Takara). To confirm the binding region required for the interaction of DpAP2, cDNA fragments corresponding to N-terminus (67–252 and 349–537 bp) and C-terminus (537–2,331 bp) of DpAP2 were amplified by PCR, respectively. A PCR-amplified bait was obtained using primers AP2-JHN2 and AP2-JHC2 that contained 24 bp homology to the bait sequence and 15 bp homology to the linear end of pGBKT7. The 5’ terminus of DpAP2 was cloned in-frame into the pGBKT7 plasmid at BamH I and EcoR I sites. In order to confirm that the fusion construct was in frame, the construct was sequenced using the T7 Primer. It is imperative to confirm that the bait does not autonomously activate reporter genes and is not toxic in Y2HGold in the absence of a prey protein. The pGBKT7 and pGBKT7-AP2N3 plasmids were used in a series of transformation tests to exclude false activation and the toxicity of reporter gene of the system (Chung et al., 2021). The yeast strains Y2HGold and Y187 were transformed using a slightly modified method. In brief, one colony was inoculated into 3 ml of the YPDA medium at 30°C with shaking at 250 rpm for 8–12 h. Then, 5 µl of the culture was transferred to 50 ml of the YPDA medium, and it was continued to be incubated until OD600 reaches 0.15–0.3. Cells were centrifuged at 700 g for 5 min at room temperature. The supernatant was discarded and each pellet was resuspended in 30 ml of sterile ddH2O and centrifuged again using the same condition. Cells were resuspended in freshly prepared LiAc/TE. Plasmid DNA, carrier DNA, and DMSO were added; then, the mixture was centrifuged with 14,000 g at room temperature after incubation. The yeast sediment was resuspended in YPD plus the medium in a shaker at 30°C for 90 min. Yeast cells were resuspended in 0.9% NaCl, and 100 µl of 1/10 and 1/100 diluted cells were spread on the SD selection medium plate at 30°C for 3–6 days.
Library Screening
Library screening was done via the yeast mating of one aliquot of Y187 cDNA library with 5 ml (an OD600 : Optical Density (OD) of at least 2.0) of the Y2HGold culture transformed with pGBKT7-AP2N3 (Zhang et al., 2020). Two cultures were combined in a sterile 2 L flask, and 45 ml of the 2×YPDA liquid medium (with 50 µg/ml kanamycin) was added. The culture was incubated at 30°C with slow shaking (30–50 rpm) for 20–24 h to prevent cells from settling at the base of the flask. After 20 h, we checked for the presence of zygotes under the phase contrast microscope (×40). The mating culture was centrifuged at 1,000 g for 10 min, washed with 100 ml of 0.5×YPDA containing 50 μg/ml of kanamycin, and resuspended in 10 ml of 0.5×YPDA with 50 μg/ml of kanamycin. For the mated culture, 100 μl of undiluted, 1:100 and 1:10,000 diluted cultures were spread on each of the SD/-Trp, SD/-Leu, and SD/-Leu/-Trp (DDO) agar plates to calculate the mating efficiency. The remaining culture was spread on SD/-Leu/-Trp (with X-α-Gal/AbA) and SD/-Ade/-His/-Leu/-Trp (with X-α-Gal/AbA) and incubated at 30°C for 3–5 days. Colonies were numbered and restreaked on the SD/-Ade/-His/-Leu/-Trp (with X-α-Gal/AbA) (QDO/X/A) master plate and then grown for another 4–6 days. All QDO/X/A-positive interactions must be further analyzed to identify duplicates and verify the genuineness of the interaction.
Yeast Colony PCR
Positive colonies were selected and incubated on the QDO/X/A liquid medium. PCR amplification was performed with primers T7 and 3’AD under the following conditions: 94°C for 3 min; 35 cycles (98°C for 10 s, 42°C for 30 s, and 72°C for 2 min). Plasmid was isolated from yeast using the MiniBEST Plasmid Purification Kit (Takara) and transformed into Escherichia coli. Positive colonies were subjected to sequencing and similarity searches in the National Center for Biotechnology Information (NCBI) database. All identified colonies were checked for the right frame and orientation.
Results and Discussion
Full-Length cDNA Cloning
A 582 bp cDNA fragment of differentially expressed DpAP2 gene associated with the regulation of carotenoid biosynthesis in D. parva was obtained by a comparison between SCH-5.0mMA (nitrogen-sufficient control sample) and SCH-0.5mMA (nitrogen limitation-treated sample) using transcriptome technology. To obtain the full-length cDNA, we performed 5’ RACE and 3’ RACE with gene-specific primers designed from the obtained 582 bp fragment. Then, using primers AP2(GSP) and the Long Primer, a 405 bp cDNA fragment was obtained. The full-length cDNA was 3,129 bp with an open reading frame (ORF) of 2,331 bp encoding 776 amino acids, a 3’ untranslated region (UTR) of 582 bp, and a 5’ UTR of 216 bp (Figure 2). The agarose gel electrophoresis analysis of the product amplified by primers AP2(N) and AP2 (C) detected a band of approximately 2,331 bp (Figure 3). The Blastx search suggested that the PCR product was homologous to a number of known AP2 genes. The detailed sequence of DpAP2 is shown in Supplementary Figure 1.

Figure 2 Full-length cDNA of the DpAP2 gene. The graphic was constructed base on a complete sequence of the DpAP2 gene using the DOG 2.0 program. The full-length cDNA was 3,129 bp with an ORF of 2,331 bp encoding 776 amino acids, a 3’ UTR of 582 bp, and a 5’ UTR of 216 bp.
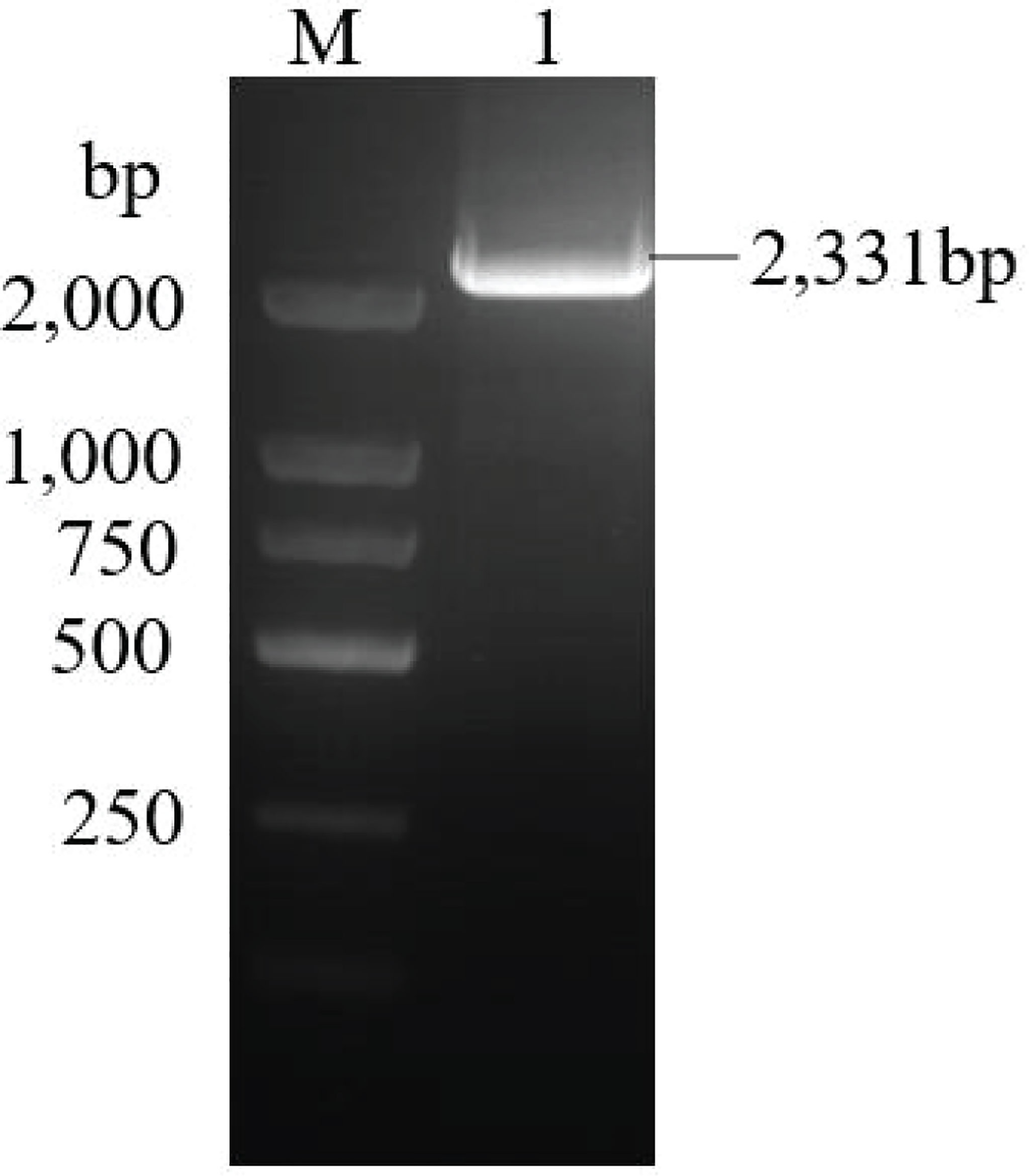
Figure 3 ORF amplification of the DpAP2 gene. M: DL 2000 Marker, 1: ORF of the DpAP2 gene. The ORF of DpAP2 gene was amplified by primers AP2(N) and AP2 (C).
Phylogenetic Analysis of AP2
The conserved amino acid residues in the AP2 domain from D. parva and Arabidopsis thaliana AP2/ERF superfamily proteins were identified by MEME online software (Figure 4). The AP2 subfamily in D. parva contains two AP2 domains such as AP2-R1 and AP2-R2. The results displayed that both D. parva and A. thaliana have RAYD, WLG/YLG, AA, and YRG elements. The analysis of the AP2 domain between D. parva and A. thaliana suggested that the domain was highly conserved. The AP2-R1 domain includes G22, E29, F24, L21, and T26, and the AP2-R2 domain includes E15, S16, F7, and H17. We constructed an evolutionary tree to analyze the evolutionary relationship of AP2/ERF TFs (Figure 5). The results indicated that Scenedesmus sp. And D. parva shared the closest relationship, which was consistent with the traditional classification.
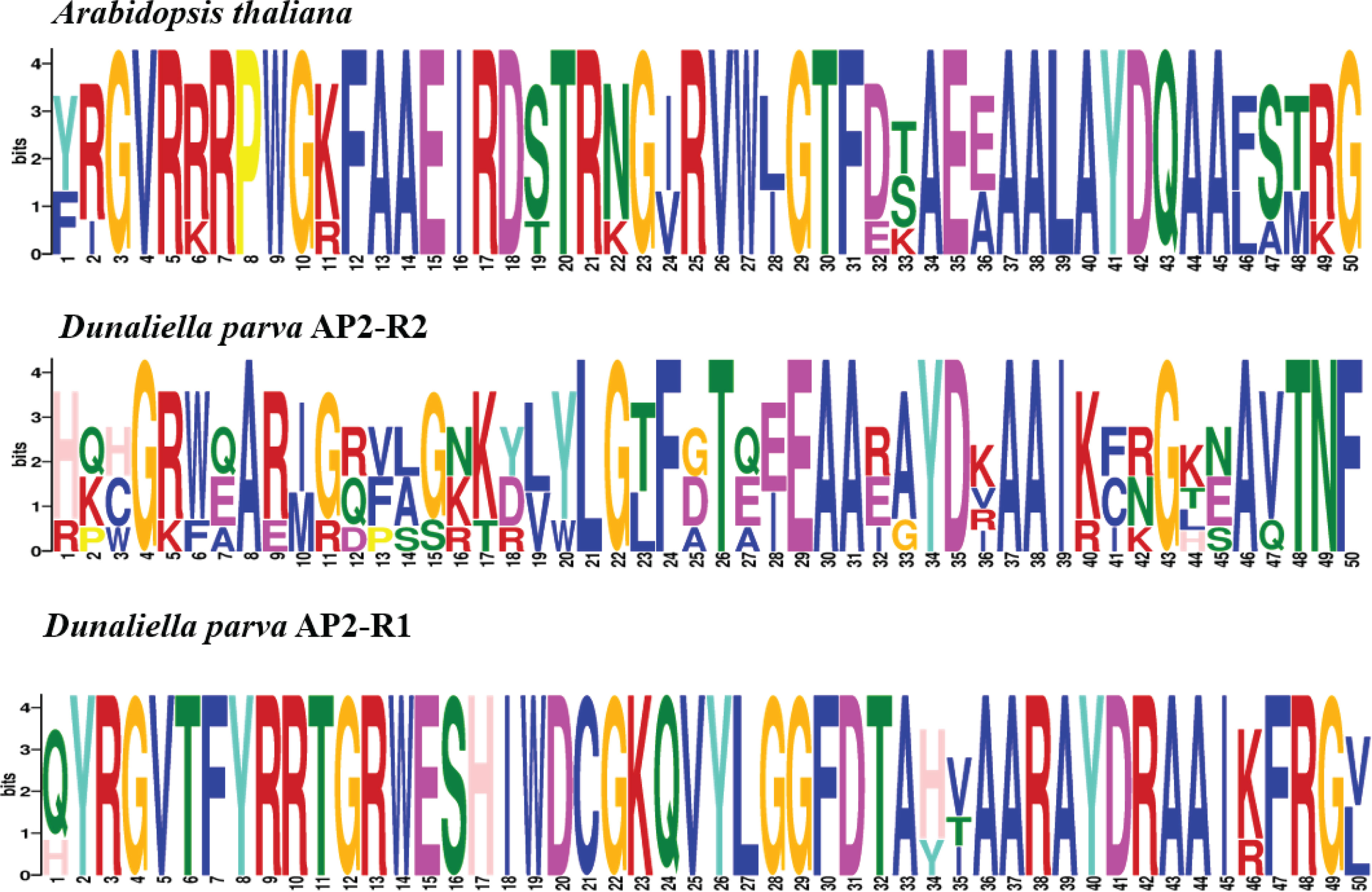
Figure 4 The conserved amino acid residues in the AP2 domain from the D. parva and A. thaliana AP2 subfamily. The domain was analyzed by MEME software. The horizontal number indicates the amino acid site; each site represents a stack. The height of character represents the frequency of the amino acid.
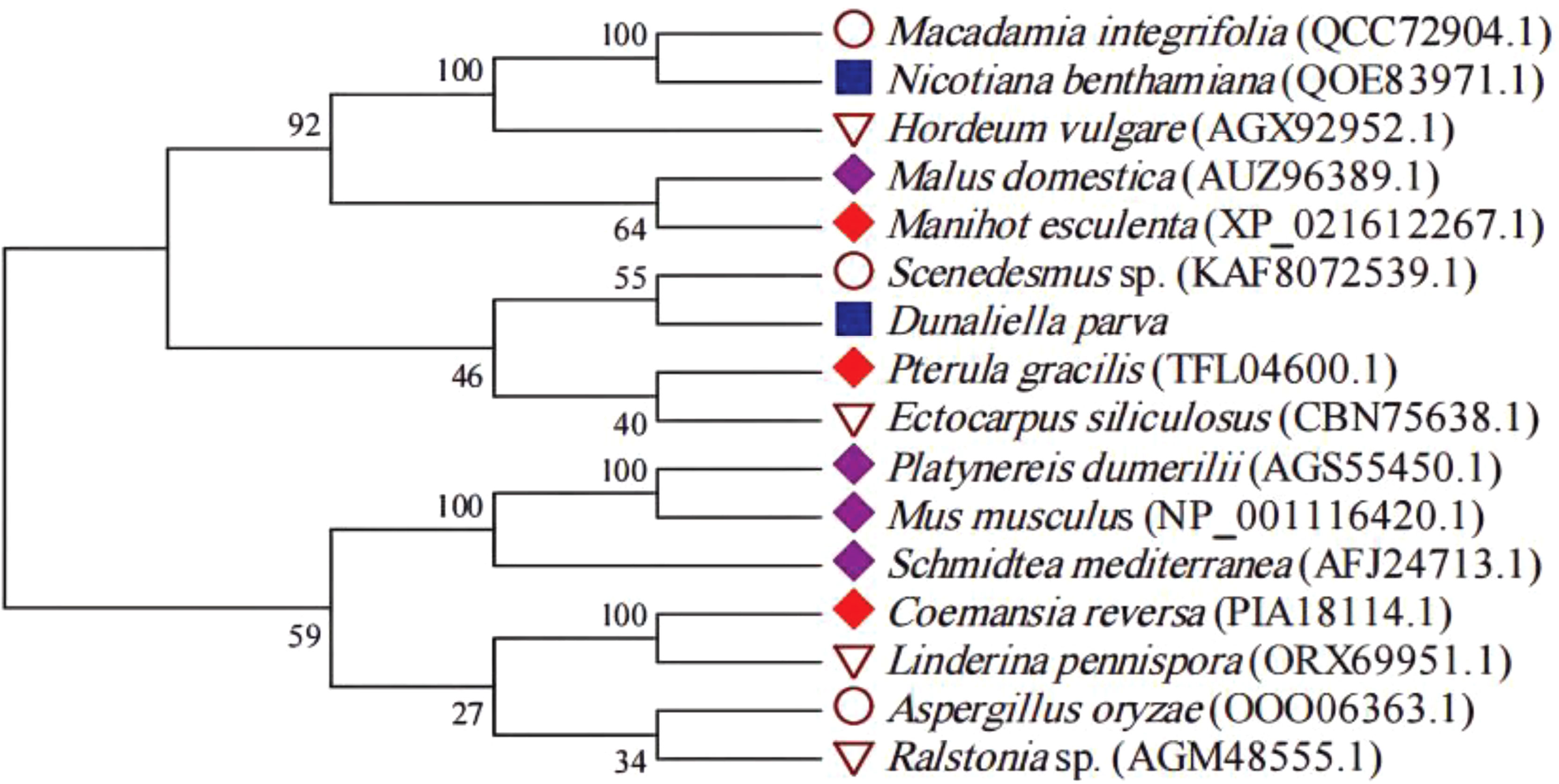
Figure 5 Phylogenetic tree of AP2 proteins. The tree was constructed based on the complete protein sequences of AP2 proteins by the neighbor-joining method of the MEGA program.
Construction of Two-Hybrid cDNA Library
In order to hunt for the interaction partners of DpAP2, we generated an appropriate cDNA library well suited for the yeast two-hybrid system. Total RNA was extracted from D. parva cells (Figure 6). Approximately 2.0 µg of total RNA was transcribed to form first-strand cDNA using SMART technology, and second-strand cDNA was amplified using long-distance PCR with the universal primers (Figure 7). The CHROMA SPIN TE-400 Column was used to select DNA molecules more than 200 bp that were likely to encode the translated region of mRNAs. The cDNA was cloned into yeast strain Y187 along with AD plasmid pGADT7-Rec. Plasmid pGADT7-Rec and cDNA fragments were combined in vivo through homologous recombination. Transformation efficiency was identified by calculating the number of colonies on a plate with a 1:1,000 dilution of transformation (Figure 8). PCR was performed to evaluate recombination efficiency (Figure 9). Transformation efficiency could be calculated with the data shown in Table 2. Cell density was more than 2 × 107 per milliliter of the cDNA library.
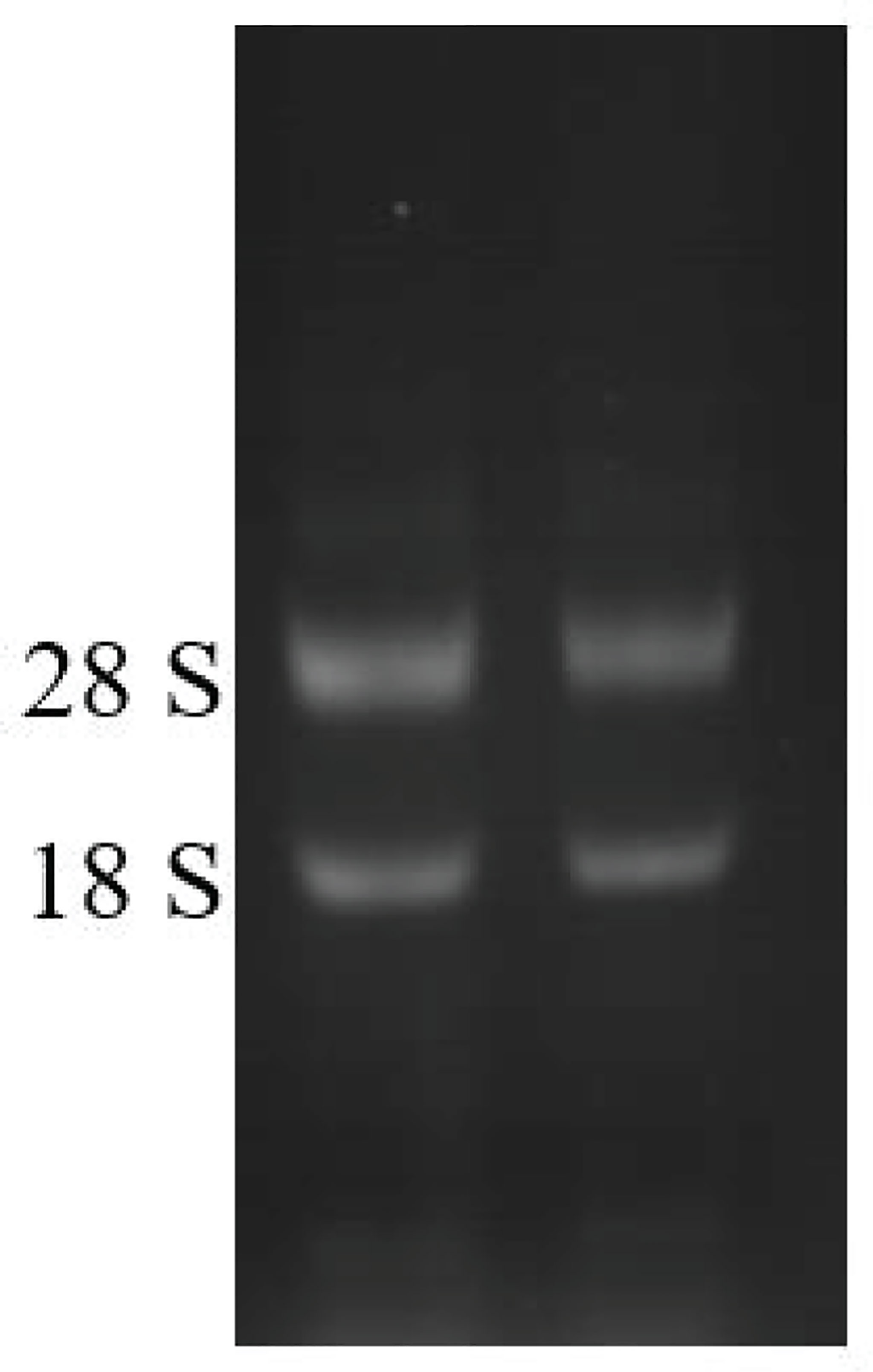
Figure 6 Total RNA from D. parva cells. Lanes 1 and 2 represented the total RNA of D. parva cells. The integrity of RNA was detected by 1% agarose gel electrophoresis.
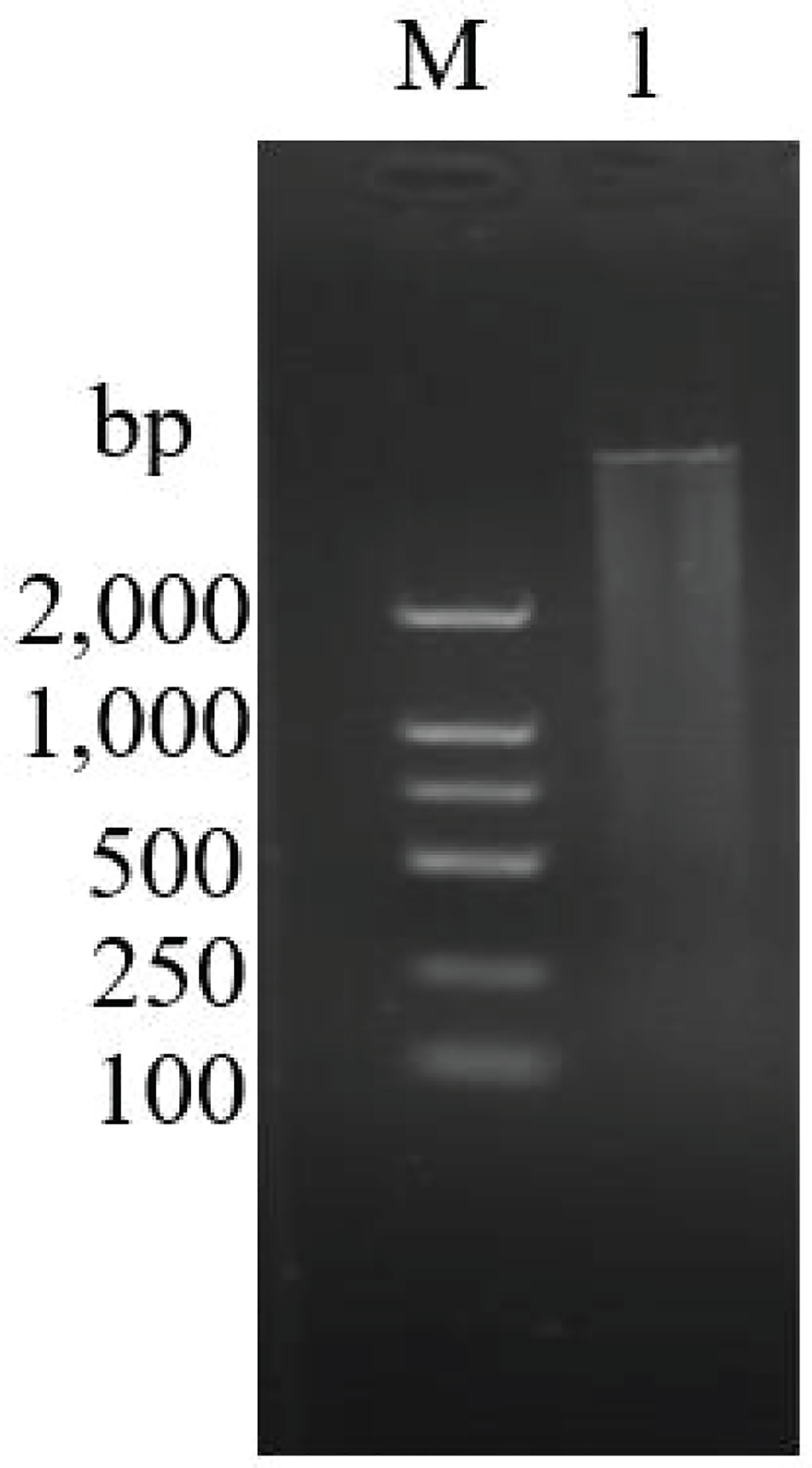
Figure 7 Long-distance PCR for second-strand cDNA synthesis. M: DL 2000 Marker. 1: Second-strand cDNA. The length of cDNA molecules was more than 200 bp.
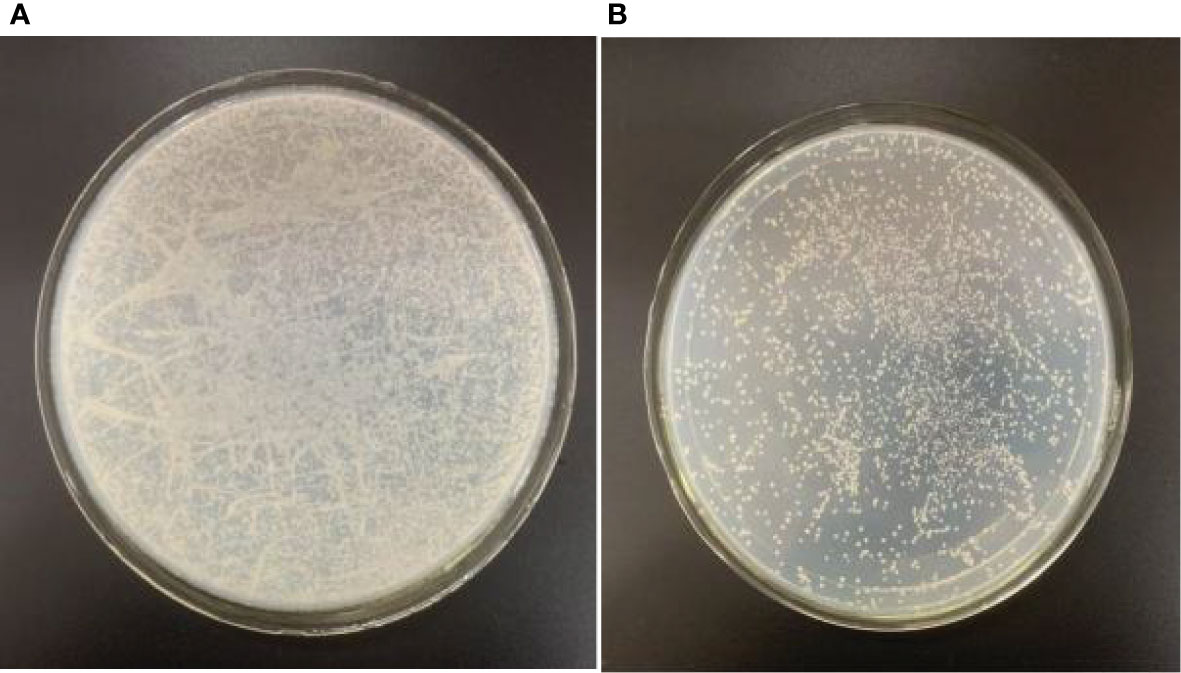
Figure 8 Colony number was counted for the cDNA library. (A) 1/1,000 dilution. (B) 1/10,000 dilution. The colony number on plate was approximately 600 under the condition of 1:10,000 dilution.
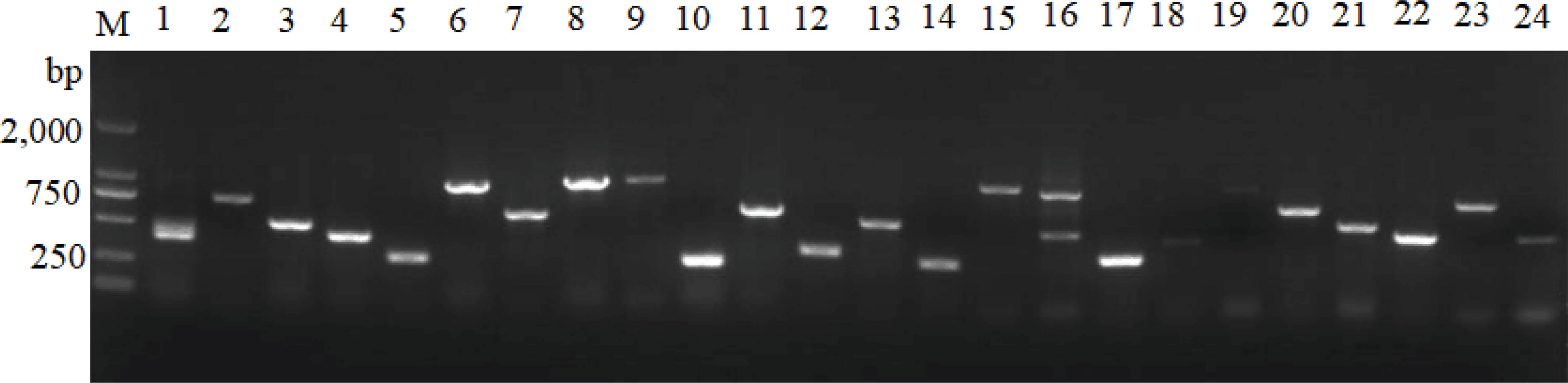
Figure 9 Identification of the integrity of the recombinant sequence of the cDNA library. M: DL 2000 Marker. 1–24: PCR verification of positive colonies. Positive colonies were selected to evaluate recombination efficiency by PCR amplification with primers T7 and 3’AD.
Generation of Bait Strain Y2HGold
To confirm the binding region required for the interaction of DpAP2, the cDNA fragments of the N-terminus (67–252 and 349–537 bp) and C-terminus (537–2,331 bp) was amplified using gene-specific primers, respectively (Figure 10). Plasmid pGBKT7-AP2N3 (537–2,331 bp) was transformed into the yeast strain Y2HGold. pGBKT7 and pGBKT7-AP2N3 were used in a series of transformation tests to exclude the false activation and toxicity of the reporter gene of the system. Then, the self-activation of the recombinant vector was determined by testing the autoactivation of Aureobasidin A and beta-galactosidase reporter. The recombinant bait plasmid had no toxic effect on yeast Y2HGold cells without the self-activation of reporter genes. The yeast strains of Y2HGold-pGBKT7-AP2N3 and Y2HGold-pGBKT7 could grow on the SD/-Trp medium without a significant difference in the colony size and number, which suggested that plasmid pGBKT7-AP2N3 had no effect on yeast growth and no toxicity to yeast (Figure 11). Furthermore, the blue Y2HGold-pGBKT7-AP2N3 colonies were detected on the SD/-Trp/X-α-Gal agar medium; however, they were absent on the SD/-Ade/-Trp/X-α-gal plate. Therefore, the DpAP2 protein had no autonomous activation effect. Yeast strains Y2HGold and Y187 were transformed by a slightly modified method.
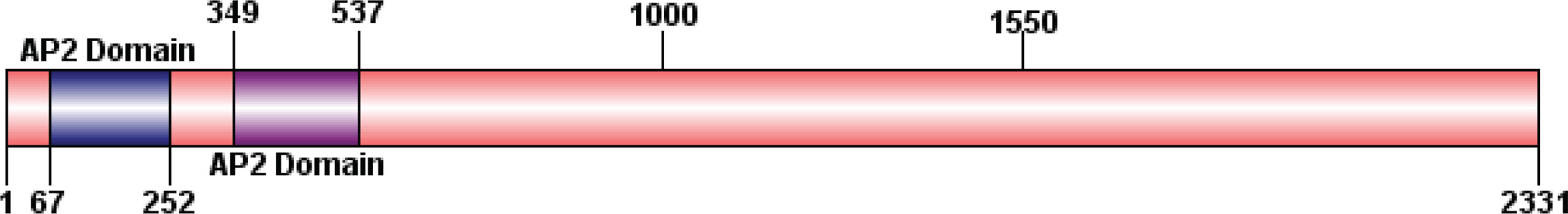
Figure 10 Prediction of conserved domains in DpAP2. The diagram was constructed based on a complete protein sequence of DpAP2 using the DOG 2.0 program. The conserved domain was analyzed by the NCBI CD-Search tool.
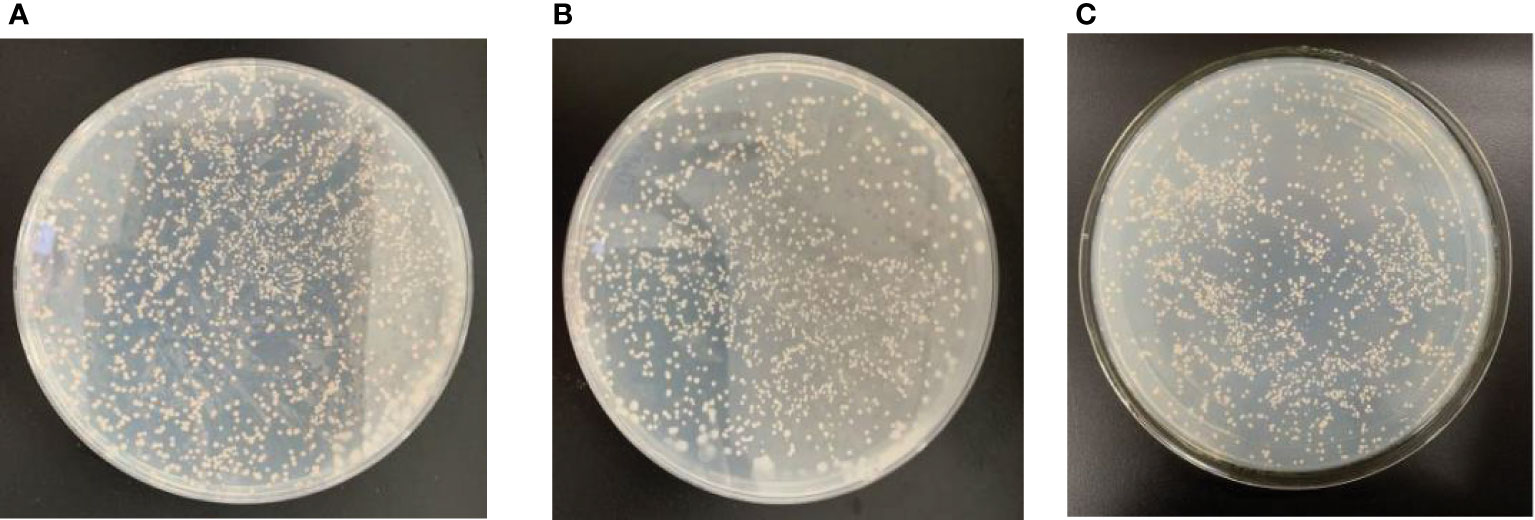
Figure 11 The toxicity assays of DpAP2 protein in yeast cells. (A) PGBKT7-AP2N3. (B) pGBKT7-AP2N2. (C) PGBKT7. Yeast strains containing plasmids pGBKT7-AP2N3 and pGBKT7 could grow without significant differences in the colony size and number, which suggested that plasmid pGBKT7-AP2N3 had no effect on yeast growth and no toxicity to yeast.
Screening of Positive Colonies
One aliquot of cDNA library containing more than 2 × 107 transformed Y187 cells were mated with more than 1 × 108 Y2HGold cells transformed with pGBKT7-AP2N3. The zygotes were detected under a phase contrast microscope (40×) after 24 h of co-incubation, which showed that yeast mating was successful. The mating culture was centrifuged and resuspended on the SD/-Leu/-Trp (with X-α-Gal/AbA) plate. The mating efficiency was counted by analyzing the SD/-Leu, SD/-Trp and SD/-Leu/-Trp plates on which different dilutions of the mating mixture were spread. The mating efficiency of the two-hybrid screen was approximately 2%. Many positive colonies grew on the SD/-Leu/-Trp/X-α-Gal/AbA agar medium and were restreaked for three times on SD/-Ade/-His/-Leu/-Trp/X-α-Gal/AbA plates (Figure 12). Finally, 34 putative positive yeast colonies were obtained.
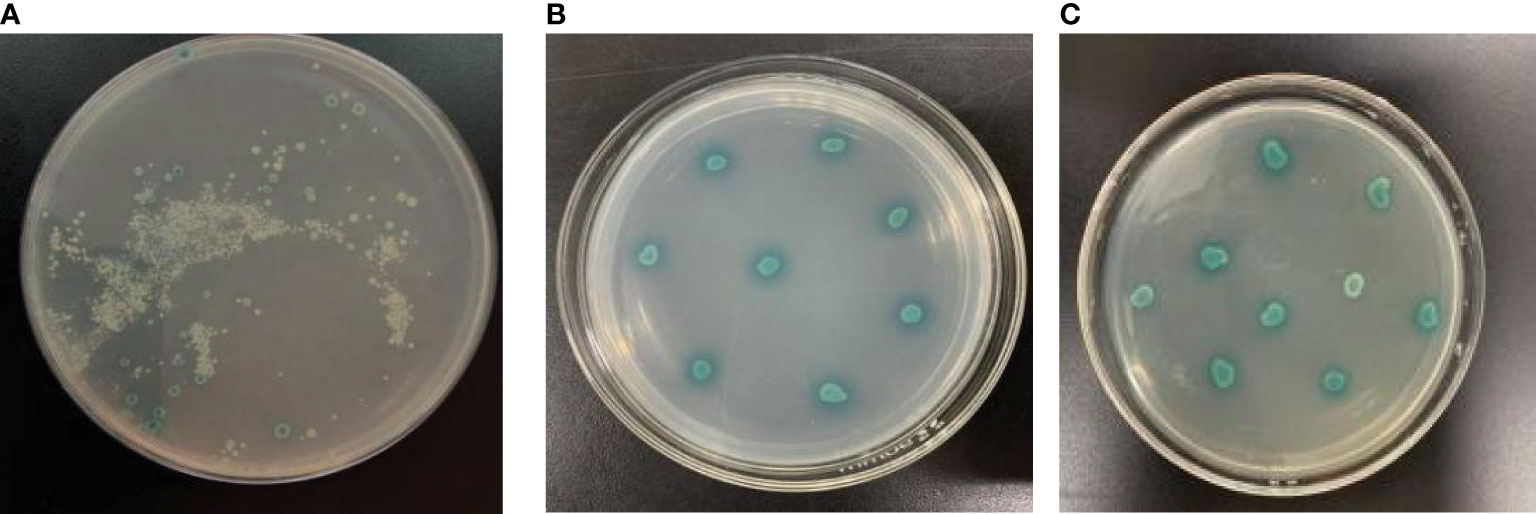
Figure 12 Screening of positive colonies. The positive colonies were restreaked for three times on SD/-Ade/-His/-Leu/-Trp/X-α-Gal/AbA plates. (A) The initial screening. (B) The second screening. (C) The third screening.
Bioinformatical Analysis of Sequencing Information
To select for stable expression of interaction partners, the colonies of mating screen were restreaked on the 4×Dropout medium, and 34 positive yeast colonies were obtained. A total of 34 cDNA inserts were verified by a colony PCR using universal primers T7 and 3’AD, and 24 cDNA fragments were determined. Plasmid was isolated from 24 yeast strains and purified, then transformed into E. coli DH5α (Figure 13). The transformants containing only pGADT7-DpAP2 plasmids from the cDNA library were obtained on the Luria-Bertani (LB) agar medium with 50 μg/ml of ampicillin. The sequencing results were subjected to the Blastx search. Three proteins interacting with DpAP2 were identified, including protein 1 (alpha-D-phosphohexomutase), protein 2 (protein serine/threonine kinase activity), and protein 3 (DNA-binding transcription factor activity) in D. parva (Table 3). The detailed sequences of three proteins interacting with DpAP2 are shown in Supplementary Figure 2. The functions of three interacting proteins of DpAP2 were analyzed by NetGO 2.0 software, which are shown in Supplementary Figure 3.
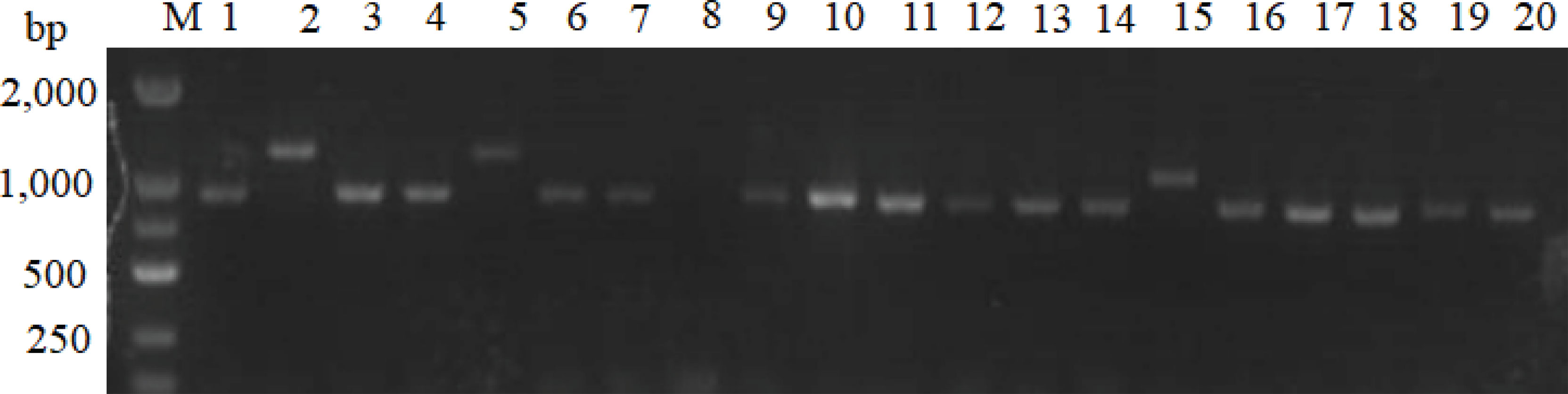
Figure 13 Verification of positive colonies by PCR. M: DL2000 Marker. 1-20: PCR verification of positive yeast colonies.
The yeast two-hybrid system was widely used to identify the interacting protein of specific proteins. Chen et al. found that the AP2/ERF transcription factor SlERF.F5 functioned in leaf senescence in tomato, and SlERF.F5 and SlMYC2 (a transcription factor downstream of the jasmonic acid receptor) could physically interact by the yeast two-hybrid experiment (Chen et al., 2022). Transcription factor SHE1 was identified as an interacting protein of cucumber mosaic virus 1a protein in the yeast two-hybrid system (Yoon and Palukaitis, 2021). AP2/ERF family transcription factors ORA59 and RAP2.3 interacted in the nucleus identified by yeast two-hybrid technology and functioned together in ethylene responses (Kim et al., 2018). Zander et al. reported that at least 17 plant-specific glutaredoxins interacted with TGA2 by the yeast two-hybrid system in Arabidopsis (Zander et al., 2012).
The ubiquitous and ancient alpha-D-phosphohexomutases are a large enzyme superfamily that exists in three domains of organisms (Backe et al., 2020). Enzymes in alpha-D-phosphohexomutases superfamily catalyze the reversible conversion of phosphosugars, such as glucose 1-phosphate and glucose 6-phosphate (Stiers et al., 2017). The phosphoglucomutase 5 (PGM5) of the alpha-D-phosphohexomutase family is a structural muscle protein in humans (Gong et al., 2020). In the Atlantic herring, PGM5 is a gene closely related to ecological adaptation to the brackish Baltic Sea (Gustafsson et al., 2020). In our study, D. parva also lives in the brackish environment. Perhaps protein 1 (alpha-D-phosphohexomutase) is related to the halophilic characteristic in D. parva. However, a detailed understanding of its function is lacking in D. parva.
The interacting protein 2 of DpAP2 with protein serine/threonine kinase activity was also identified. The serine/threonine kinase Akt is a key factor regulating glucose and lipid energy metabolism, which is activated in response to various stimuli such as cell stress and various hormones and drugs (Miao et al., 2022). The eukaryotic-like serine/threonine protein kinases play important roles in cell growth and signal transduction in Mycobacterium tuberculosis (Burastero et al., 2022). Protein kinase G regulates the carbon and nitrogen metabolism by the phosphorylation of the glycogen accumulation regulator (GarA) at Thr21 (Burastero et al., 2022). Protein kinase B is related to the formation of the cell shape, cell wall synthesis, and phosphorylation of GarA at Thr22 (Burastero et al., 2022). In a word, protein serine/threonine kinase plays an important role in cell metabolism. However, the function of protein 2 remains unclear in D. parva.
The analysis of interacting protein 3 by NetGO 2.0 software indicated that protein 3 had a DNA-binding transcription factor activity. Transcription factors are proteins that help turn specific target genes “on” or “off” by binding to the nearby DNA. The previous studies have suggested that the AP2/ERF transcription factor could interact with other transcription factors, which was consistent with our study (Kim et al., 2018; Chen et al., 2022). However, the function and interacting target genes of protein 3 remain unclear in D. parva. ChIP and ChIP-Seq will be used to identify the target genes of protein 3 in the future.
Conclusion
DpAP2 is a key regulator of carotenoid biosynthesis. The DpAP2 encoding TF DpAP2 has been identified in our former study. However, the target proteins of DpAP2 remain unknown. The interacting proteins of DpAP2 were identified by the yeast two-hybrid system in order to further demonstrate the function of DpAP2 in this study. The results showed that three target proteins were identified. This study laid a good foundation for the further understanding of the regulatory mechanism of carotenoid biosynthesis related to DpAP2.
Data Availability Statement
The data presented in the study are deposited in the National Center for Biotechnology Information repository, accession number ON548534, ON548535, ON548536, ON548537.
Author Contributions
CS is the corresponding author. CS contributed to the conception, design, data acquisition, and drafting of the article. BP is the co-first author. BP contributed to the data acquisition and drafting of the article. JZ, LY, SG, YL, and HW contributed to partial data acquisition and drafting of the article. All authors read and approved the final manuscript.
Funding
This study was financially supported by National Natural Science Foundation of China (31860010), Innovation Project of Guangxi Graduate Education (YCSW2022178 and XJCY2022011), National Training Program of Innovation and Entrepreneurship for Undergraduates (202210602035), Guangxi Key Research and Development Program (AB21220057, 2021AB27009), Research Funds of the Guangxi Key Laboratory of Landscape Resources Conservation and Sustainable Utilization in Lijiang River Basin, Guangxi Normal University (LRCSU21Z0207), Research Funds of Key Laboratory of Ecology of Rare and Endangered Species and Environmental Protection (Guangxi Normal University), Ministry of Education, China (ERESEP2022Z11).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2022.907065/full#supplementary-material
Supplementary Figure 1 | Full-length cDNA sequence of DpAP2 gene. The initiation and termination codons were ATG and TAG, respectively. The ORF of DpAP2 was 2,331 bp
Supplementary Figure 2 | Full-length cDNA encoding three interacting proteins of DpAP2. The initiation and termination codons were underlined. (A) Interacting protein of DpAP2 (alpha-D-phosphohexomutase activity). (B) Interacting protein of DpAP2 (protein kinase activity). (C) Interacting protein of DpAP2 (DNA-binding transcription factor activity).
Supplementary Figure 3 | Function prediction of three interacting proteins of DpAP2 by NetGO 2.0 software. (A) Interacting protein of DpAP2 (alpha-D-phosphohexomutase activity). (B) Interacting protein of DpAP2 (protein kinase activity). (C) Interacting protein of DpAP2 (DNA-binding transcription factor activity).
References
Abid G., Muhovski Y., Jacquemin J. M., Mingeot D., Sassi K., Toussaint A., et al. (2012). Characterization and Expression Profile Analysis of a Sucrose Synthase Gene From Common Bean (Phaseolus Vulgaris L.) During Seed Development. Mol. Biol. Rep. 39, 1133–1143. doi: 10.1007/s11033-011-0842-y
Ampomah-Dwamena C., Thrimawithana A. H., Dejnoprat S., Lewis D., Espley R. V., Allan A. C. (2019). A Kiwifruit (Actinidia Deliciosa) R2R3-MYB Transcription Factor Modulates Chlorophyll and Carotenoid Accumulation. New Phytol. 221, 309–325. doi: 10.1111/nph.15362
Arora N., Patel A., Sartaj K., Pruthi P. A., Pruthi V. (2016). Bioremediation of Domestic and Industrial Wastewaters Integrated With Enhanced Biodiesel Production Using Novel Oleaginous Microalgae. Environ. Sci. Pollut. Res. Int. 23, 20997–21007. doi: 10.1007/s11356-016-7320-y
Backe P. H., Laerdahl J. K., Kittelsen L. S., Dalhus B., Mørkrid L., Bjørås M. (2020). Structural Basis for Substrate and Product Recognition in Human Phosphoglucomutase-1 (PGM1) Isoform 2, a Member of the α-D-Phosphohexomutase Superfamily. Sci. Rep. 10, 5656. doi: 10.1038/s41598-020-62548-0
Burastero O., Cabrera M., Lopez E. D., Defelipe L. A., Arcon J. P., Durán R., et al. (2022). Specificity and Reactivity of Mycobacterium Tuberculosis Serine/Threonine Kinases PknG and PknB. J. Chem. Inf. Model. 62, 1723–1733. doi: 10.1021/acs.jcim.1c01358
Chen Y., Feng P., Tang B., Hu Z., Xie Q., Zhou S., et al. (2022). The AP2/ERF Transcription Factor SlERF.F5 Functions in Leaf Senescence in Tomato. Plant Cell Rep. 41, 1181–1195. doi: 10.1007/s00299-022-02846-1
Chung I. Y. W., Li L., Tyurin O., Gagarinova A., Wibawa R., Li P., et al. (2021). Structural and Functional Study of Legionella Pneumophila Effector RavA. Protein Sci. 30, 940–955. doi: 10.1002/pro.4057
Courchesne N. M., Parisien A., Wang B., Lan C.Q. (2009). Enhancement of Lipid Production Using Biochemical, Genetic and Transcription Factor Engineering Approaches. J. Biotechnol. 141, 31–41. doi: 10.1016/j.jbiotec.2009.02.018
Dang Q., Sha H., Nie J., Wang Y., Yuan Y., Jia D. (2021). An Apple (Malus Domestica) AP2/ERF Transcription Factor Modulates Carotenoid Accumulation. Hortic. Res. 8, 223. doi: 10.1038/s41438-021-00694-w
Donadio G., Santoro V., Dal Piaz F., De Tommasi N. (2021). Food Matrices Affect the Peptides Produced During the Digestion of Arthrospira Platensis-Based Functional Aliments. Nutrients 13, 3919. doi: 10.3390/nu13113919
du Preez R., Majzoub M. E., Thomas T., Panchal S. K., Brown L. (2021). Nannochloropsis Oceanica as a Microalgal Food Intervention in Diet-Induced Metabolic Syndrome in Rats. Nutrients 13, 3991. doi: 10.3390/nu13113991
Fritzsche S., Blenk P., Christian J., Castiglione K., Becker A. M. (2021). Inhibitory Properties of Crude Microalgal Extracts on the In Vitro Replication of Cyprinid Herpesvirus 3. Sci. Rep. 11, 23134. doi: 10.1038/s41598-021-02542-2
Gao F., Peng Y. Y., Li C., Yang G. J., Deng Y. B., Xue B., et al. (2018). Simultaneous Nutrient Removal and Biomass/Lipid Production by Chlorella Sp. In Seafood Processing Wastewater. Sci. Total Environ. 640-641, 943–953. doi: 10.1016/j.scitotenv.2018.05.380
Gilmour D. J. (2019). Microalgae for Biofuel Production. Adv. Appl. Microbiol. 109, 1–30. doi: 10.1016/bs.aambs.2019.10.001
Gong J., Zeng Y., Meng Q., Guan Y., Li C., Yang H., et al. (2021). Red Light-Induced Kumquat Fruit Coloration Is Attributable to Increased Carotenoid Metabolism Regulated by FcrNAC22. J. Exp. Bot. 72, 6274–6290. doi: 10.1093/jxb/erab283
Gong J., Zeng Y., Meng Q., Guan Y., Li C., Yang H., et al. (2020). Structure and Characterization of Phosphoglucomutase 5 From Atlantic and Baltic Herring-An Inactive Enzyme With Intact Substrate Binding. Biomolecules 10, 1631. doi: 10.3390/biom10121631
Gustafsson R., Eckhard U., Ye W., Enbody E. D., Pettersson M., Jemth P., et al. (2020). Structure and Characterization of Phosphoglucomutase 5 from Atlantic and Baltic Herring-An Inactive Enzyme with Intact Substrate Binding. Biomolecules 10, 1631. doi: 10.3390/biom10121631
Ismaiel M. M. S., El-Ayouty Y. M., Said A. A., Fathey H. A. (2018). Transformation of Dunaliella Parva With PSY Gene: Carotenoids Show Enhanced Antioxidant Activity Under Polyethylene Glycol and Calcium Treatments. Biocatal. Agric. Biotechnol. 16, 378–384. doi: 10.1016/j.bcab.2018.09.011
Jiang X., Zhang Y., Hutchins D. A., Gao K. (2021). Nitrogen-Limitation Exacerbates the Impact of Ultraviolet Radiation on the Coccolithophore Gephyrocapsa Oceanica. J. Photochem. Photobiol. B. 226, 112368. doi: 10.1016/j.jphotobiol.2021.112368
Kamalanathan M., Hillhouse J., Claflin N., Rodkey T., Mondragon A., Prouse A., et al. (2021). Influence of Nutrient Status on the Response of the Diatom Phaeodactylum Tricornutum to Oil and Dispersant. PLoS One 16, e0259506. doi: 10.1371/journal.pone.0259506
Kang J., Gong J., Zhang L., Gao Z., Xie Q., Hu Z., et al. (2021). A Novel E6-Like Gene, E6-2, Affects Fruit Ripening in Tomato. Plant Sci. 313, 111066. doi: 10.1016/j.plantsci.2021.111066
Keerthana S., Sekar S., Kumar S. D., Santhanam P., Divya M., Krishnaveni N., et al. (2020). Scenedesmus Pecsensis Cultivation in Rice Mill Effluent Using Commercial Scale Nutrient Sources. Bioresource Tech. Rep. 9, 100379. doi: 10.1016/j.biteb.2019.100379
Kim N. Y., Jang Y. J., Park O. K. (2018). AP2/ERF Family Transcription Factors ORA59 and RAP2.3 Interact in the Nucleus and Function Together in Ethylene Responses. Front. Plant Sci. 9. doi: 10.3389/fpls.2018.01675
Kumari K., Samantaray S., Sahoo D., Tripathy B. C. (2021). Nitrogen, Phosphorus and High CO2 Modulate Photosynthesis, Biomass and Lipid Production in the Green Alga Chlorella Vulgaris. Photosynth. Res. 148, 17–32. doi: 10.1007/s11120-021-00828-0
Koul A., Sharma D., Kaul S., Dhar M. K.. (2019). Identification and In Silico Characterization of Cis-Acting Elements of Genes Involved in Carotenoid Biosynthesis in Tomato. 3 Biotech. 9, 287. doi: 10.1007/s13205-019-1798-1
Lee J. M., Joung J. G., McQuinn R., Chung M. Y., Fei Z., Tieman D., et al. (2012). Combined Transcriptome, Genetic Diversity and Metabolite Profiling in Tomato Fruit Reveals That the Ethylene Response Factor SlERF6 Plays an Important Role in Ripening and Carotenoid Accumulation. Plant J. 70, 191–204. doi: 10.1111/j.1365-313X.2011.04863.x
Li S., Ji L., Shi Q., Wu H., Fan J. (2019). Advances in the Production of Bioactive Substances From Marine Unicellular Microalgae Porphyridium Spp. Bioresour. Technol. 292, 122048. doi: 10.1016/j.biortech.2019.122048
Li R., Tan Y., Zhang H. (2021). Regulators of Starch Biosynthesis in Cereal Crops. Molecules 26, 7092. doi: 10.3390/molecules26237092
Mendelsohn A. R., Brent R. (1994). Biotechnology Applications of Interaction Traps/Two-Hybrid Systems. Curr. Opin. Biotechnol. 5, 482–486. doi: 10.1016/0958-1669(94)90061-2
Miao R., Fang X., Wei J., Wu H., Wang X., Tian J. (2022). Akt: A Potential Drug Target for Metabolic Syndrome. Front. Physiol. 13. doi: 10.3389/fphys.2022.822333
Mohsin A., Hussain M. H., Zaman W. Q., Mohsin M. Z., Zhang J., Liu Z., et al. (2021). Advances in Sustainable Approaches Utilizing Orange Peel Waste to Produce Highly Value-Added Bioproducts. Crit. Rev. Biotechnol. 2, 1–20. doi: 10.1080/07388551.2021.2002805
Narang P. K., Dey J., Mahapatra S. R., Roy R., Kushwaha G. S., Misra N., et al. (2021). Genome-Based Identification and Comparative Analysis of Enzymes for Carotenoid Biosynthesis in Microalgae. World J. Microbiol. Biotechnol. 38, 8. doi: 10.1007/s11274-021-03188-y
Oswald W. J. (2003). My Sixty Years in Applied Algology. J. Appl. Phycol. 15, 99–106. doi: 10.1023/A:1023871903434
Ratledge C. (2002). Regulation of Lipid Accumulation in Oleaginous Micro-Organisms. Biochem. Soc Trans. 30, 1047–1050. doi: 10.1042/bst0301047
Shahid A., Rehman A. U., Usman M., Ashraf M. U. F., Javed M. R., Khan A. Z., et al. (2020). Engineering the Metabolic Pathways of Lipid Biosynthesis to Develop Robust Microalgal Strains for Biodiesel Production. Biotechnol. Appl. Biochem. 67, 41–51. doi: 10.1002/bab.1812
Shang C., Bi G., Yuan Z., Wang Z., Alam M. A., Xie J. (2016). Discovery of Genes for Production of Biofuels Through Transcriptome Sequencing of Dunaliella Parva. Algal Res. 13, 318–326. doi: 10.1016/j.algal.2015.12.012
Shang C., Wang W., Zhu S., Wang Z., Qin L., Alam M. A., et al. (2018). The Responses of Two Genes Encoding Phytoene Synthase (Psy) and Phytoene Desaturase (Pds) to Nitrogen Limitation and Salinity Up-Shock With Special Emphasis on Carotenogenesis in Dunaliella Parva. Algal Res. 32, 1–10. doi: 10.1016/j.algal.2018.03.002
Shang C., Xu X., Yuan Z., Wang Z., Hu L., Alam M. A., et al. (2016). Cloning and Differential Expression Analysis of Geranylgeranyl Diphosphate Synthase Gene From Dunaliella Parva. J. Appl. Phycol. 28, 2397–2405. doi: 10.1007/s10811-015-0767-2
Shi Y., Pang X., Liu W., Wang R., Su D., Gao Y., et al. (2021). SlZHD17 is Involved in the Control of Chlorophyll and Carotenoid Metabolism in Tomato Fruit. Hortic. Res. 8, 259. doi: 10.1038/s41438-021-00696-8
Stiers K. M., Muenks A. G., Beamer L. J. (2017). Biology, Mechanism, and Structure of Enzymes in the α-D-Phosphohexomutase Superfamily. Adv. Protein Chem. Struct. Biol. 109, 265–304. doi: 10.1016/bs.apcsb.2017.04.005
Todorović B., Grujić V. J., Krajnc A. U., Kranvogl R., Ambrožič-Dolinšek J. (2021). Identification and Content of Astaxanthin and its Esters From Microalgae Haematococcus Pluvialis by HPLC-DAD and LC-QTOF-MS After Extraction With Various Solvents. Plants (Basel). 10, 2413. doi: 10.3390/plants10112413
Welsch R., Maass D., Voegel T., Dellapenna D., Beyer P. (2007). Transcription Factor RAP2.2 and its Interacting Partner SINAT2: Stable Elements in the Carotenogenesis of Arabidopsis Leaves. Plant Physiol. 145, 1073–1085. doi: 10.1104/pp.107.104828
Xing G., Li J., Li W., Lam S. M., Yuan H., Shui G., et al. (2021). AP2/ERF and R2R3-MYB Family Transcription Factors: Potential Associations Between Temperature Stress and Lipid Metabolism in Auxenochlorella Protothecoides. Biotechnol. Biofuels 14, 22. doi: 10.1186/s13068-021-01881-6
Yoon J. Y., Palukaitis P. (2021). Cucumber Mosaic Virus 1a Protein Interacts With the Tobacco SHE1 Transcription Factor and Partitions Between the Nucleus and the Tonoplast Membrane. Plant Pathol. J. 37, 182–193. doi: 10.5423/PPJ.FT.03.2021.0045
Zander M., Chen S., Imkampe J., Thurow C., Gatz C. (2012). Repression of the Arabidopsis Thaliana Jasmonic Acid/Ethylene-Induced Defense Pathway by TGA-Interacting Glutaredoxins Depends on Their C-Terminal ALWL Motif. Mol. Plant 5, 831–840. doi: 10.1093/mp/ssr113
Zhang L., Zhang Q., Li W., Zhang S., Xi W. (2019). Identification of Key Genes and Regulators Associated With Carotenoid Metabolism in Apricot (Prunus Armeniaca) Fruit Using Weighted Gene Coexpression Network Analysis. BMC Genomics 20, 876. doi: 10.1186/s12864-019-6261-5
Zhang H., Zhao L., Chen Y., Zhu M., Xu Q., Wu M., et al. (2021). Trophic Transition Enhanced Biomass and Lipid Production of the Unicellular Green Alga Scenedesmus Acuminatus. Front. Bioeng. Biotechnol. 9. doi: 10.3389/fbioe.2021.638726
Zhang Z. M., Zhuang M., Wang B. T., Jin L., Jin F. J. (2020). Identification and Characterization of a DevR-Interacting Protein in Aspergillus Oryzae. Fungal Biol. 124, 155–163. doi: 10.1016/j.funbio.2020.01.001
Zheng M., Wu Y. J., Cai W. M., Weng H. L., Liu R. H. (2005). Construction of a Hepatic Stellate Cells Subtracted cDNA Library of Differentially Expressed Genes in Normal Mice and Mice With Schistosomiasis Japonica. J. Zhejiang Univ. Sci. B. 6, 280–287. doi: 10.1631/jzus.2005.B0280
Keywords: Dunaliella parva, AP2, yeast two-hybrid system, interacting proteins, carotenoid biosynthesis
Citation: Shang C, Pang B, Zhang J, Yu L, Gan S, Li Y and Wu H (2022) Identification of Interacting Proteins of Transcription Factor DpAP2 Related to Carotenoid Biosynthesis From Marine Microalga Dunaliella parva. Front. Mar. Sci. 9:907065. doi: 10.3389/fmars.2022.907065
Received: 29 March 2022; Accepted: 19 May 2022;
Published: 23 June 2022.
Edited by:
Shuhao Huo, Jiangsu University, ChinaReviewed by:
Xupeng Cao, Dalian Institute of Chemical Physics (CAS), ChinaJichang Han, Ningbo University, China
Ang Ren, Nanjing Agricultural University, China
Qiuning Liu, Yancheng Teachers University, China
Copyright © 2022 Shang, Pang, Zhang, Yu, Gan, Li and Wu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Changhua Shang, c2hhbmdjaGFuZ2h1YUBtYWlsYm94Lmd4bnUuZWR1LmNu
†These authors have contributed equally to this work
 Changhua Shang
Changhua Shang Bingbing Pang1,2†
Bingbing Pang1,2† Lihong Yu
Lihong Yu

