- 1Large Pelagics Research Center, Gloucester, MA, United States
- 2Pacific Islands Fisheries Group, Kailua, HI, United States
Striped marlin, Kajikia audax, have been in overfished condition in the Western and Central North Pacific, and overfishing is still occurring, prompting an urgent need to devise conservation and management measures based on the best, current information on biology and ecology of this species. Despite conventional tagging efforts off Hawaii, striped marlin movements are not known across the broader Pacific, and little research has been conducted since 2005. To address this gap, 31 popup satellite archival tags (PSAT) were deployed on striped marlin (138-192 cm eye fork length) between 2016 and 2019 via the Hawaii-based longline fleet. To complement tagging efforts, 148 fin clips were also collected for genetic analyses during June-August 2017. Tag attachments ranged from 1-365 days (median = 74), where mechanical failures and non-reporting tags lowered expected data returns. Striped marlin tracks revealed extensive spatial use of the Central Pacific, spanning 15°S to 43°N and 122 to 170°W, showing diverse seasonal dispersal patterns and individual movements, and some coincided in time and space with known spawning grounds. Genetic profiles of 55 Hawaii-landed striped marlin were assigned to two genetic groups: Australia, New Zealand and Hawaii (n = 19), and Hawaii alone (n=36), suggesting the Hawaii-based longline fleet interacted with individuals from multiple populations.
A year-long track confirmed migration between the Central North Pacific and Australia (>9,400 km), and combined with genetic results, is the first to document regional connectivity. By combining tools from conventional and electronic tags, biological sampling, and genomic techniques, a more holistic understanding emerges, suggesting that striped marlin should be collectively managed. Under global warming scenarios and changing pelagic ecosystems, integrative knowledge is critical for designing effective management strategies for rebuilding sustainable populations across the Pacific Ocean.
Introduction
Striped marlin, Kajikia audax, have been in overfished condition in the Western and Central North Pacific since the mid-1990s (ISC, 2019; Sculley, 2021). The majority of catches of striped marlin are taken as bycatch by international fleets, namely Japan and Chinese Taipei. However, a continued decline in the spawning stock biomass and excess fishing mortality relative to the maximum sustainable yield call for additional catch reductions or other management measures that impact international and Hawaii-based fleets (Brodziak, 2021). In Hawaii, striped marlin are important to the islands’ economy and cultures, small boat fisheries and recreation charter vessels in particular, and stricter management measures could bring hardship to local communities. In addition, the prohibition of sales of Hawaii-landed billfish to the US mainland market by the 2018 amendments to the Billfish Conservation Act of 2012, coupled with detrimental impacts from the COVID-19 pandemic, have further reduced economic returns. Even though a pan-North Pacific genetic stock exists (Purcell and Edmands, 2011; Mamoozadeh et al., 2020), tagging to date has shown that trans-Pacific movement is rare. Only ten out of 21,000+ conventional tags released on striped marlin traveled from Southern California or Mexico to Hawaii (Anonymous, 2016). No eastward movement from Japan to Hawaii or to the North American west coast (e.g., California and Mexico) has been documented in previous tag records.
A current challenge is the identification of the source and composition of striped marlin in the Central North Pacific (CNP), as optimal management requires an understanding of the spatial aspects of stock structure. In Hawaii, juveniles and larger individuals appear differentially in the catches of the fishing fleets, and in different seasons (Sculley, 2019), so an outstanding question is whether these size classes have different origins but mix in the CNP. Additionally, the coexistence of separate genetic units of immature and mature fish in Hawaii (Purcell and Edmands, 2011) further complicates interpretation of the level of mixing and mechanisms of isolation for fish of various sizes and origins. Tagging effort for striped marlin around Hawaii is almost nonexistent north of 22°N and south of 15°N (Bromhead et al., 2004). Published work is limited to a single acoustic tagging study (Brill et al., 1993) and one satellite tagging study (Domeier, 2006), providing data for a combined total of 12 striped marlin. Consequently, details regarding longer-term movements (i.e., >9 months) are lacking. Although a striped marlin spawning area is found off Kona, Hawaiʻi (Hyde et al., 2006) and active female spawners were mostly sampled in a T-shaped area between 25-30°N and 150-165°W, and 20-30°N and 155-160°W during May to July (Humphreys and Brodziak, 2019), the stock composition of the assemblage is unknown. Details of migration pathways and connectivity of the spawners to those elsewhere remains undetermined. To address these gaps, we deployed popup satellite archival tags (PSAT) on striped marlin intercepted by the Hawaii-based longline fishery in order to delineate the extent of their movements in relation to local fisheries, and to establish their potential connectivity to other parts of the Pacific with genetic profiling. Combined with results from analysis of vertical movements in relation to oceanographic features, known spawning areas, and interactions with predators in a companion publication (Lam et al., submitted), we hope to update the understanding of striped marlin in the CNP in times of changing climate and ecosystems, information essential to effective international fisheries management.
Materials and Methods
Electronic Tagging
Thirty-one PSATs (4 X-Tags; Microwave Telemetry, Inc.; 27 MiniPATs; Wildlife Computers Inc.) were deployed on striped marlin between 2016 and 2019 by Hawaii-based longline vessels operating in the Central North Pacific (Table 1). Tag tethers and anchors were constructed according to materials and methods developed for bluefin tuna tagging (Lutcavage et al., 1999; Galuardi and Lutcavage, 2012), and performed well on sailfish (Lam et al., 2016). At the start of the project, we trained participating longline captains in safe fish handling and optimal placement of PSAT anchors, and they or trained crew members conducted all releases. Upon retrieval of a longline set, an individual that was undamaged and assessed to be in good condition was tagged on deck (n = 2) or alongside the vessel via a custom-built tagging pole (n = 29). The tagger implanted the black nylon “umbrella” dart through the musculature, into the pterygiophores at the anterior dorsal region. Prior to release, hooks were either removed, or fishing line cut as close to the jaw as possible. Whole weight (W) was estimated by the vessel’s captain to the nearest five pounds (1 lb ~ 0.4536 kg) when tagged along the vessel, and eye fork length (EFL) was measured to the nearest centimeter when tagged on deck. W (in kg) was then converted to EFL (in cm) or lower jaw fork length (LJFL, in cm) following Hsu et al. (2019):
and
PSATs were programmed to record relative light level, external temperature, pressure (depth) for 12-month missions. Archived depth and temperature time series were sub-sampled to 5-minute (MiniPAT) and 15-minute (X-Tag) resolution by manufacturer routines for transmission through the Argos satellites. Transmitted light data represented either truncated light curves around sunset and sunrise (MiniPAT) or detected time stamps of sunrise and sunset (X-Tag). All tags had a failsafe release if the tag experienced any constant depth for 3 (X-Tag) or 4 (MiniPAT) days, a hallmark of post-release mortality or tag shedding. MiniPAT firmware also included detection and transmission of a hardware failure message (“pin broke”) indicating that the tag’s nosecone pin had broken, separating the tag from its tether, causing premature release. Once a PSAT popped off and began transmitting, positions with Argos location classes LC 1, 2, and 3 were noted, and the first of these positions was selected as the tag’s reporting position. Returned data were imported into and managed through Tagbase (Lam and Tsontos, 2011).
Conventional Tagging
Ten single-anchor dart tags (Hallprint Fish Tags) were deployed on striped marlin between 15 and 50 lbs. estimated whole weight, but none had been recovered to date. In order to support comparison of previous tagging efforts with PSAT tracks, recovered striped marlin releases from the NOAA Southwest Fisheries Science Center Cooperative Billfish Tagging Program or CBTP (Heberer et al., 2021) were accessed for records to October 2021 (fisheries.noaa.gov/inport/item/2327). Quality control steps to remove questionable recovery records followed that of Fitchett (2019). These release-recapture trajectories were analyzed in conjunction with our electronic tagging results.
Geolocation
State-space Kalman filter models, TrackIt (Lam et al., 2010) and Ukfsst (Lam et al., 2008) were used to reconstruct positions based on transmitted light data and sea surface temperature (SST) matching with NOAA Optimum Interpolation SST V2 (esrl.noaa.gov/psd/data/gridded/data.noaa.oisst.v2.html) for MiniPAsT and X-tags, respectively. Tracks were only reconstructed for fish at liberty for over 14 days. All estimated positions, except for one fish (PG01), were placed in the ocean, and thus required no further correction. For PG01, bathymetric correction (Galuardi et al., 2010) was applied to generate the position estimates during the last two months of its deployment when it approached Australia. The last position of a given reconstructed track represented one of the three scenarios: (1) PSAT completed its mission and reported on schedule to an orbiting Argos satellite, (2) pin broke – where tag released and reported immediately to a satellite, and (3) tag was either shed or ingested by a predator – where an estimated track was trimmed to the day before shedding or predation occurred. Predation was inferred if sensor data indicated ingestion (i.e., sustained low light levels, increased temperature indicative of warm-bodied predator, depth pattern changes) before the tag reported.
Horizontal Movement
From the reconstructed tracks and their associated error, tracks were regularized to a daily resolution using the R package, crawl (Johnson et al., 2008). A utilization distribution (UD) was then generated (Galuardi and Lutcavage, 2012), with UD values between 0 and 50% representing high use areas. Linear displacement was calculated between the release position and the last track position using the ‘distGeo’ function in R package, geosphere (cran.r-project.org/web/packages/geosphere/index.html). Similarly, release and recapture positions were used to calculate linear displacement for recovered CBTP conventional tags.
To aid the interpretation of fish dispersal, the following geographical information system resources were accessed: bathymetric values from Smith & Sandwell Topography (0.0167° resolution, version 11.1), and sea surface height (variable ‘zos’) and temperature (variable ‘thetao’) monthly mean values [product ID GLOBAL_REANALYSIS_PHY_001_030] from the Copernicus Marine Service Global Monitoring and Forecasting Centre (CMEMS; marine.copernicus.eu). Additionally, daily sea surface temperature [product ID METOFFICE-GLO-SST-L4-NRT-OBS-SST-MON-V2], and chlorophyll-a [product ID GLOBAL_REANALYSIS_BIO_001_029] were obtained from CMEMS. Surface temperature isotherms at 24, 18.5 and 17.5°C were also obtained from World Ocean Atlas 2013 monthly dataset (Locarnini et al., 2013). These isotherms were selected for their association with striped marlin observed in fisheries catch distribution (Ueyanagi and Wares, 1975; Howell et al., 2008; Lien et al., 2013; Su et al., 2015) and in track trajectories of electronic tagging (Sippel et al., 2007; Sippel et al., 2011).
In order to identify movements in relation to fisheries management jurisdictions, management boundaries of the International Scientific Committee for Tuna and Tuna-like Species in the North Pacific Ocean (ISC), Inter American Tropical Tuna Commission (IATTC) and Western and Central Pacific Fisheries Commission (WCPFC) were accessed through RAM Legacy Stock Boundary Database (marine.rutgers.edu/~cfree/ram-legacy-stock-boundary-database).
To investigate connectivity of tracked striped marlin with conspecifics tagged in other Pacific locations, additional data were requested for fish carrying tags 13P0434, 15P0239, 16P1157, 16P1159 and L330-3034 in the International Game Fish Association (IFGA) Great Marlin Race (igfa.org/the-great-marlin-race), tag 990317 in Domeier et al. (2019), tag 63292 in Sippel et al. (2011) via Animal Tracking Network’s public repository (coastwatch.pfeg.noaa.gov/erddap/info/gtoppAT/index.html), and lastly, tag 03P0523 in Lam et al. (2015). Selected metadata from these individuals were appended to Table 1 for completeness and to aid the comparison with striped marlin tagged in the present study.
Genetic Sampling and Analyses
Collection of biological samples from tagged and released fish was not feasible due to operational constraints of tagging from longline vessels, so genetic sampling (n = 148) was conducted between June 30 and August 3, 2017 at the United Fish Agency’s Honolulu Fish Auction. A small clip (~1 cm2) was taken from the dorsal or caudal fin and stored in 95% ethanol. Fish weight with head and gut removed was noted to the nearest pound, and multiplied by 1.37 to estimate whole weight (Ito, 2019) for conversion to EFL (Eqn. 1). Fin clip samples were extracted for DNA by the Commonwealth Scientific and Industrial Research Organization (CSIRO) and extracts genotyped by Diversity Arrays Technology Pty. Ltd. These samples were evaluated along with samples collected off east Australia (n = 34 in 1994 & 41 in 2017) and New Zealand (n = 57 in 2018 & 15 in 2019) and analyzed in a separate study (Evans et al., 2021).
Centroids of fishing activities were requested for the vessels that landed the genetically sampled fish from NOAA Pacific Islands Fisheries Center (PIFSC) Fisheries Research and Monitoring Division. Only non-confidential locations, resolved at a 5°x5° grid that is routinely used in assessment, were obtained to protect the intellectual property and identity of vessel operators.
Results
Release dates of tagged fish reflected the two longline fishing seasons (April-July and December-February) for striped marlin around Hawaii (Table 1). Whole weight of PSAT-tagged striped marlin was 35 ± 4 kg (mean ± sd) and eye fork length was 149 ± 6 cm. Returned light data from reporting PSAT tags allowed reconstruction of seventeen tracks with their associated error (Appendix 1). Among the four X-tags deployed, one failed to report, one shed prematurely after 102 days, and two completed one-year missions, for a mean of 277 days at liberty (Table 1). Of the 27 MiniPATs deployed, seven (26%) did not report, five had nosecone pin breakage (19%), six indicated a mortality event (22%), and ten (37%) shed prematurely (Table 1). MiniPAT-tagged fish were tracked for 2 to 294 days = 76 ± 81); nosecone pin breakage occurred between 53 and 99 days. Three mortality events inferred from sensor data (two predated, one sunk) occurred between two and six days post-release. Although no electronic or conventional tag has been recovered to date, PG23 was recaptured by a local fisherman at 21.36°N, 158.39°W, three months after the tag had popped off.
Horizontal Movements
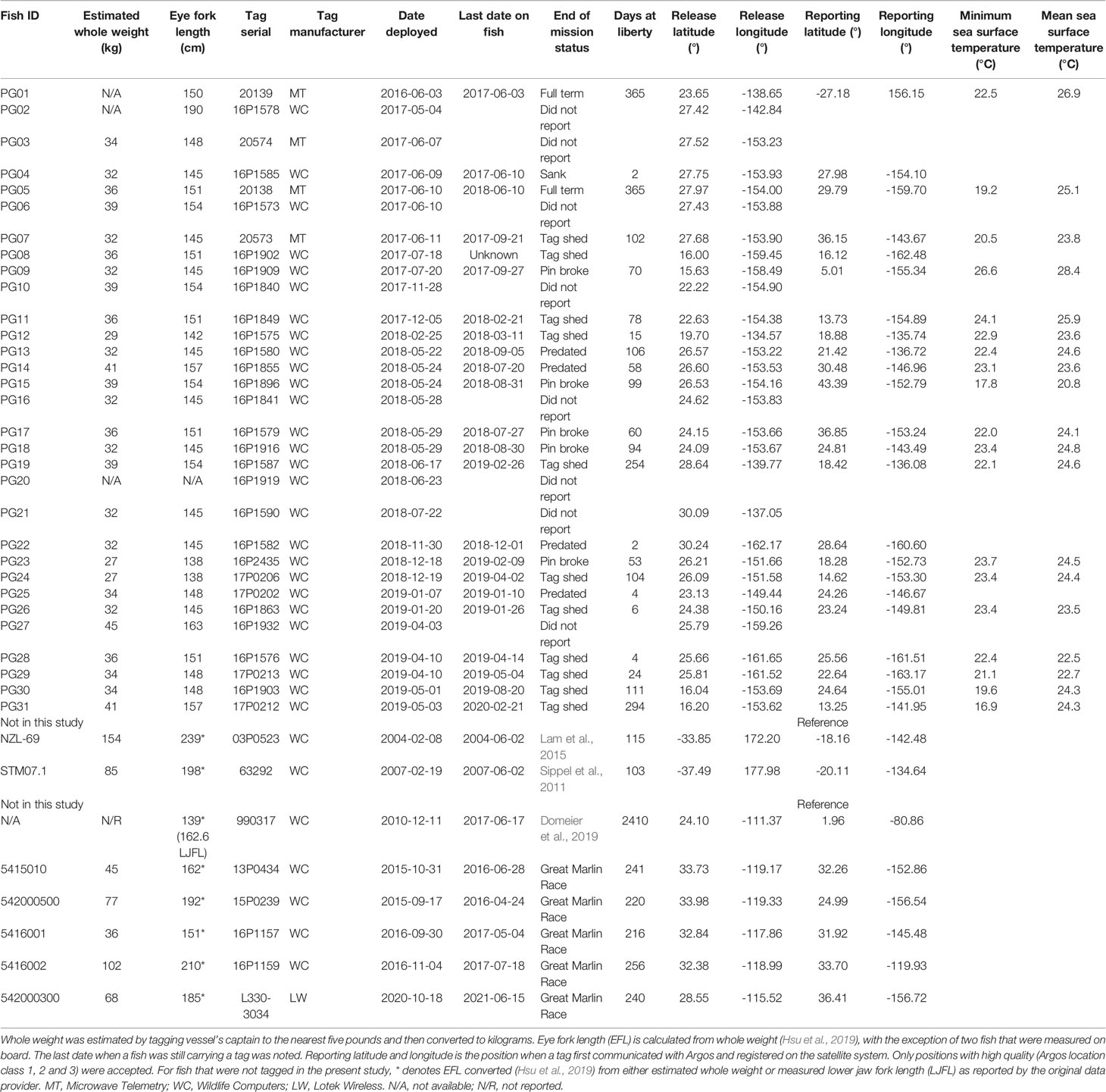
Striped marlin were distributed over a broad area in the CNP between 3-45°N and 121-169°W (Figure 1). Linear displacement was 1,987 ± 2,080 km (mean ± sd), and ranged between 152 (PG12) and 9,410 km (PG01). Daily displacement rate averaged 37 ± 13 km day-1, with a maximum of 229 km day-1 (Table 2). Most tagged marlin dispersed in the north-south direction, and while their individual trajectories varied, most consistently followed SST frontal dynamics (Appendices 2, 3). Tracked marlin crossed multiple fracture zones in the Northeast Pacific, from Surveyor Fracture Zone as warming waters expanded north in the summer, to Clipperton Fracture Zone in the south (Figure 1). Upon nearing a fracture zone, some fish either remained in the vicinity, or traveled along its longitudinal boundary for up to 4 weeks, while others (e.g., PG09, 29, 30 & 31) were associated with seamounts. Generally, striped marlin remained offshore except in springtime (March to May) when they approached the Main Hawaiian Islands (MHI), first appearing off Hawaiʻi and then MHI windward and leeward coasts (Figure S1). For example, over one year, PG05 spent ten months ranging widely from 8-38°N and 147-163°W before reaching the MHI the following April (Figure 2A).
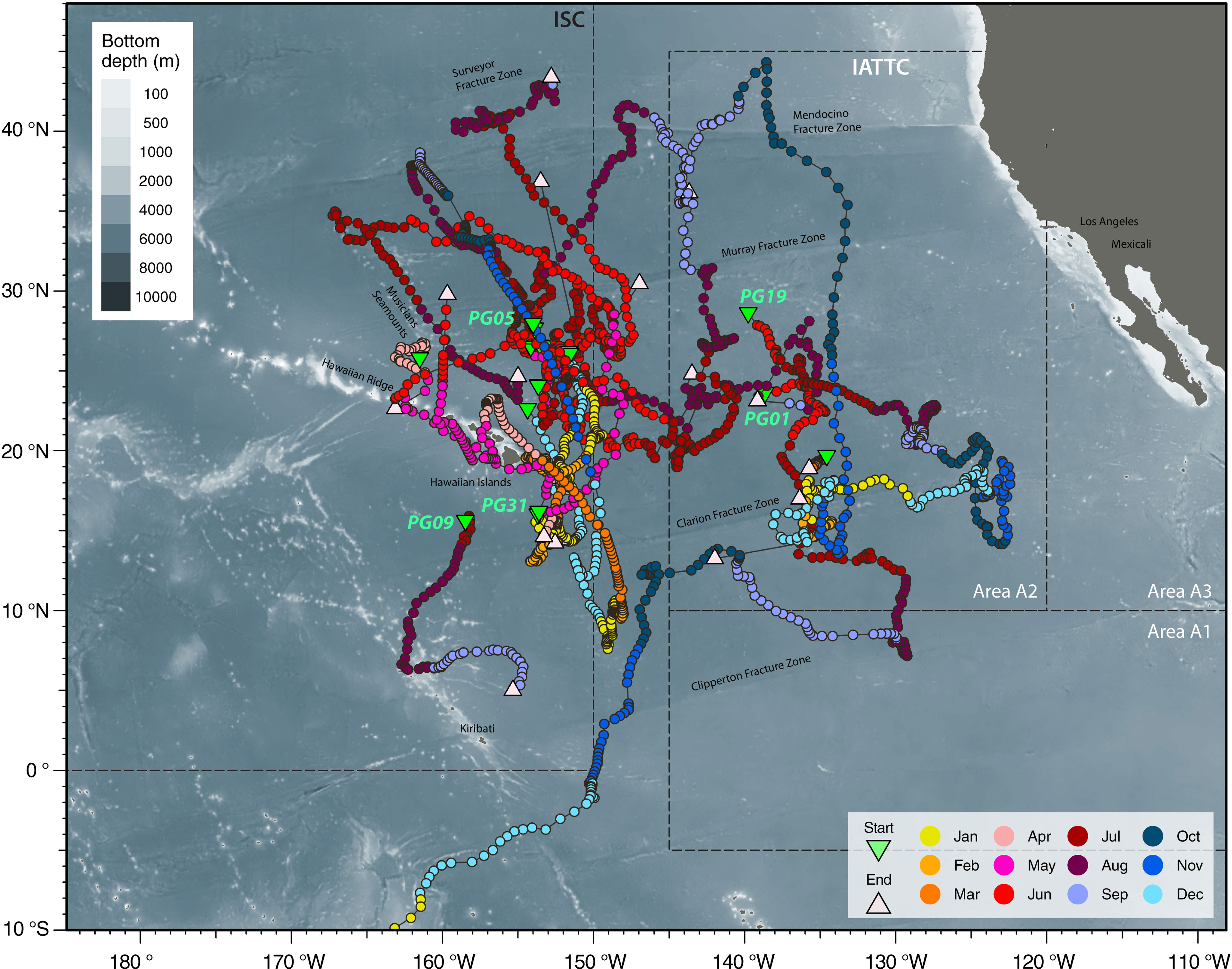
Figure 1 Horizontal movements of striped marlin tagged in the Central North Pacific. Positions are color-coded by months, and selected starting positions are labeled with their Fish ID (refer to Table 1 for details) as they are also featured in other figures or the reporting of results. Note the track of PG01 is clipped to the map extent to maintain a zoom level that is easier to read. Tagging locations, green triangle; last known fish or reporting locations, light pink triangle; management boundaries of International Scientific Committee for Tuna and Tuna-like Species in the North Pacific Ocean (ISC) and Inter American Tropical Tuna Commission (IATTC), dashed lines. Selected cities, islands, bathymetric features and management units are labeled for reference.
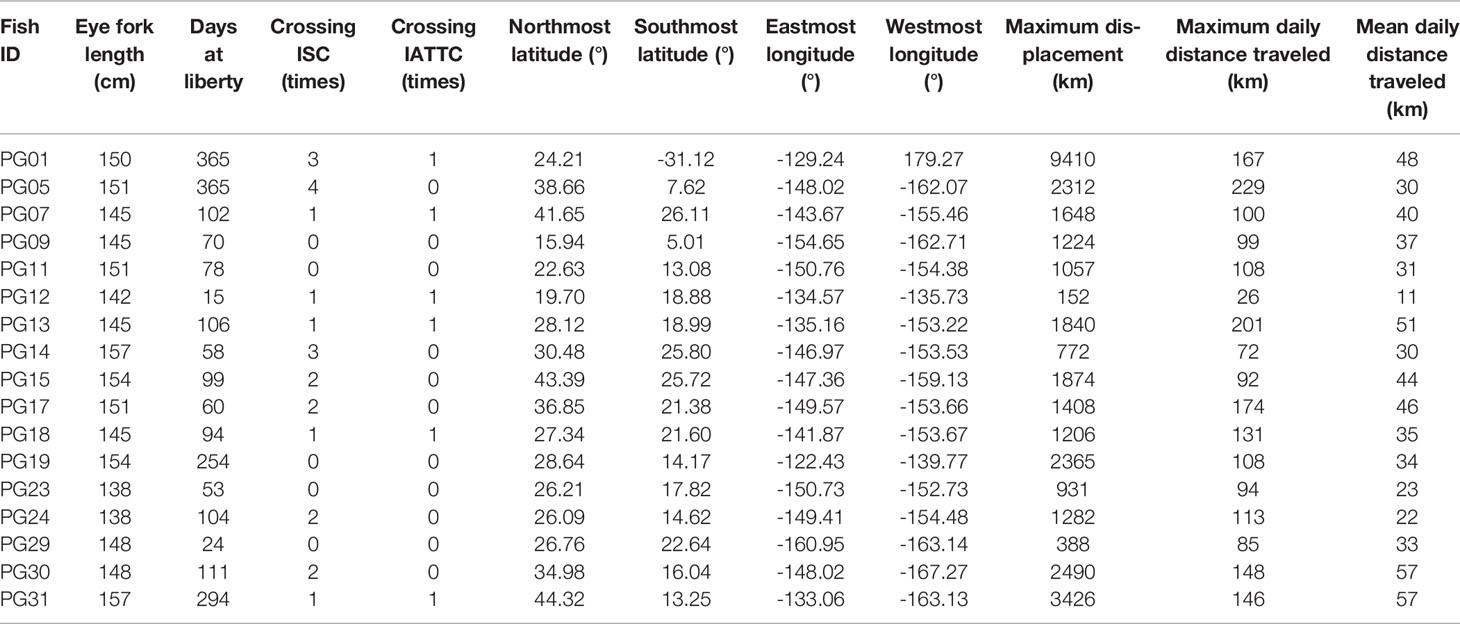
Table 2 Movement characteristics of tagged striped marlin relative to the management boundary of International Scientific Committee for Tuna and Tuna-like Species in the North Pacific Ocean (ISC) at 150°W and of Inter American Tropical Tuna Commission (IATTC) at 145°W.
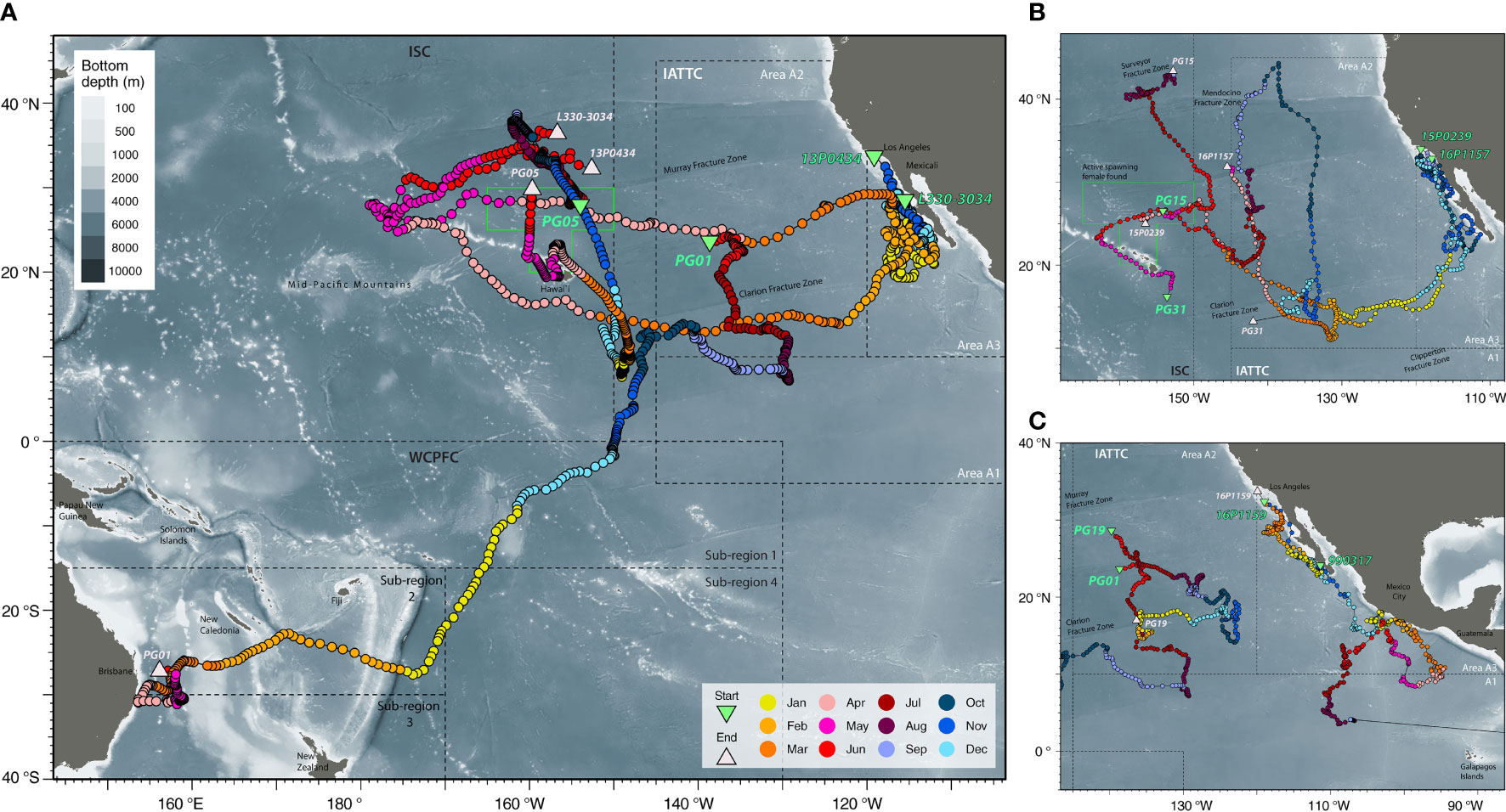
Figure 2 Spatiotemporal patterns of connectivity. (A) Long-distance dispersals of striped marlin tagged in the Central North Pacific [PG01 & 05] and the Eastern Pacific [13P0434 & L330-3034] with reference to a spawning ground north of the Main Hawaiian Islands (green box; Humphreys and Brodziak, 2019); (B) Trajectories of striped marlin tagged in the Central North Pacific [PG15, PG31] and off Southern California [15P0239 & 16P1157]; (C) Striped marlin movements in the Central North Pacific [PG01 & 19] and off the Southern California [16P1159] in relation to a conspecific [990317] tagged off Baja California that was later recaptured in Ecuador. Positions are color-coded by months, and starting and reporting positions are labeled with their Fish ID or tag serial (refer to Table 1 for details). Tagging locations, green triangle; last known fish or reporting locations, light pink triangle; management boundaries of International Scientific Committee for Tuna and Tuna-like Species in the North Pacific Ocean (ISC), Inter American Tropical Tuna Commission (IATTC) and Western and Central Pacific Fisheries Commission (WCPFC), dashed lines. Selected cities, islands, bathymetric features and management units are labeled for reference.
Another year-long track (PG01) revealed trans-Pacific movement not previously observed for striped marlin. Tagged inside the IATTC management area, PG01 spent four months there, and over the next five months, continued southwest across the CNP to the South Pacific (Figure 2A). In January, it sped through the West Pacific Warm Pool (>30°C SST) between 0 and 15°S, perhaps aided by the counterclockwise flow of the South Equatorial Current. Arriving after the summer spawning season in the Coral Sea (Kopf et al., 2012), PG01 associated with SSTs of 24-28°C during February-April and 23°C in May off the central east coast of Australia (Figure 3A), before its tag released on schedule.
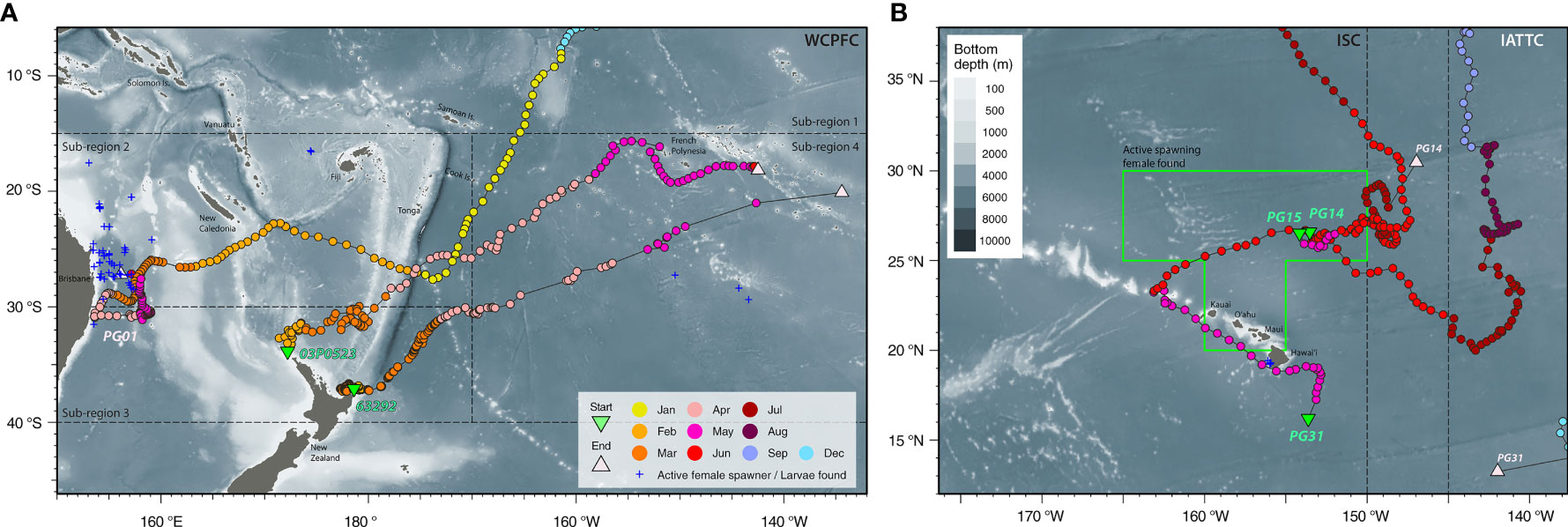
Figure 3 Movements in relation to known locations with reproductive activities. Striped marlin tagged in the Central North Pacific: (A) PG01 moved through the South Pacific and reached the east coast of Australia after the summer spawning season. Blue crosses represent ripening and spawning female locations in Kopf et al. (2012); (B) PG14, 15 & 31 were present in a known area with active female spawners (green box; Humphreys and Brodziak, 2019) during the spawning months of May and June. Blue crosses represent larval locations in Hyde et al. (2006). Note portions of their tracks cannot be shown in the map at the current zoom level. Positions are color-coded by months, and starting and reporting positions are labeled with their Fish ID or tag serial (refer to Table 1 for details). Tagging locations, green triangle; last known fish or reporting locations, light pink triangle; management boundaries of International Scientific Committee for Tuna and Tuna-like Species in the North Pacific Ocean (ISC), Inter American Tropical Tuna Commission (IATTC) and Western and Central Pacific Fisheries Commission (WCPFC), dashed lines. Selected cities, islands, bathymetric features and management units are labeled for reference.
Fish caught and released at similar times and locations (PG13, 14, 15, 17 & 18) mostly remained near a SST front (~25.5°N, 153°W) for about three weeks before dispersing (Appendix 2). Notably, after two months or so, PG13 and PG15 were over 2,700 km apart. Similarly, PG30 and PG31, also released close together, undertook different trajectories that are visualized as loops extending to 30° in longitude and latitude (Appendix 3 and Figure 2B), with movements aligned with SST fronts that generally sloped southeast across the CNP. Apart from this duo, loop-like trajectories were also observed for other marlin (Figures 2, S2).
Seasonal Distribution and SST Associations
Spatial distribution of our tagged striped marlin varied throughout the year, and coincided with the 24°C SST isotherm at multiple locations (Figure 4). Fish were relatively aggregated in waters southeast of the MHI between 10-20°N in January and February (Figure 4A), and remained mostly southeast and northwest off the Hawaiian coast in March and April (Figure 4B). Striped marlin favored the 24° C isotherm when near the MHI and similarly, when they dispersed northeast to 21-28°N and 148-155°W (Figure 4C). Other fish occupied waters further east to ~138°W, roughly halfway between the MHI and California, and to the south of Clarion Fracture Zone. In July and August, striped marlin were distributed over the broader CNP (Figures 4D). At that time, some tagged fish remained near known spawning grounds, while others traveled to 40°N, with cooler conditions (~18°C SST). Striped marlin were most distant from the MHI during September and October (Figure 4E), and by November, returned to lower latitudes, associating again with the 24°C isotherm (Figure 4F). Presence in the lower latitudes between 0 and 10°N occurred mostly between July and December.
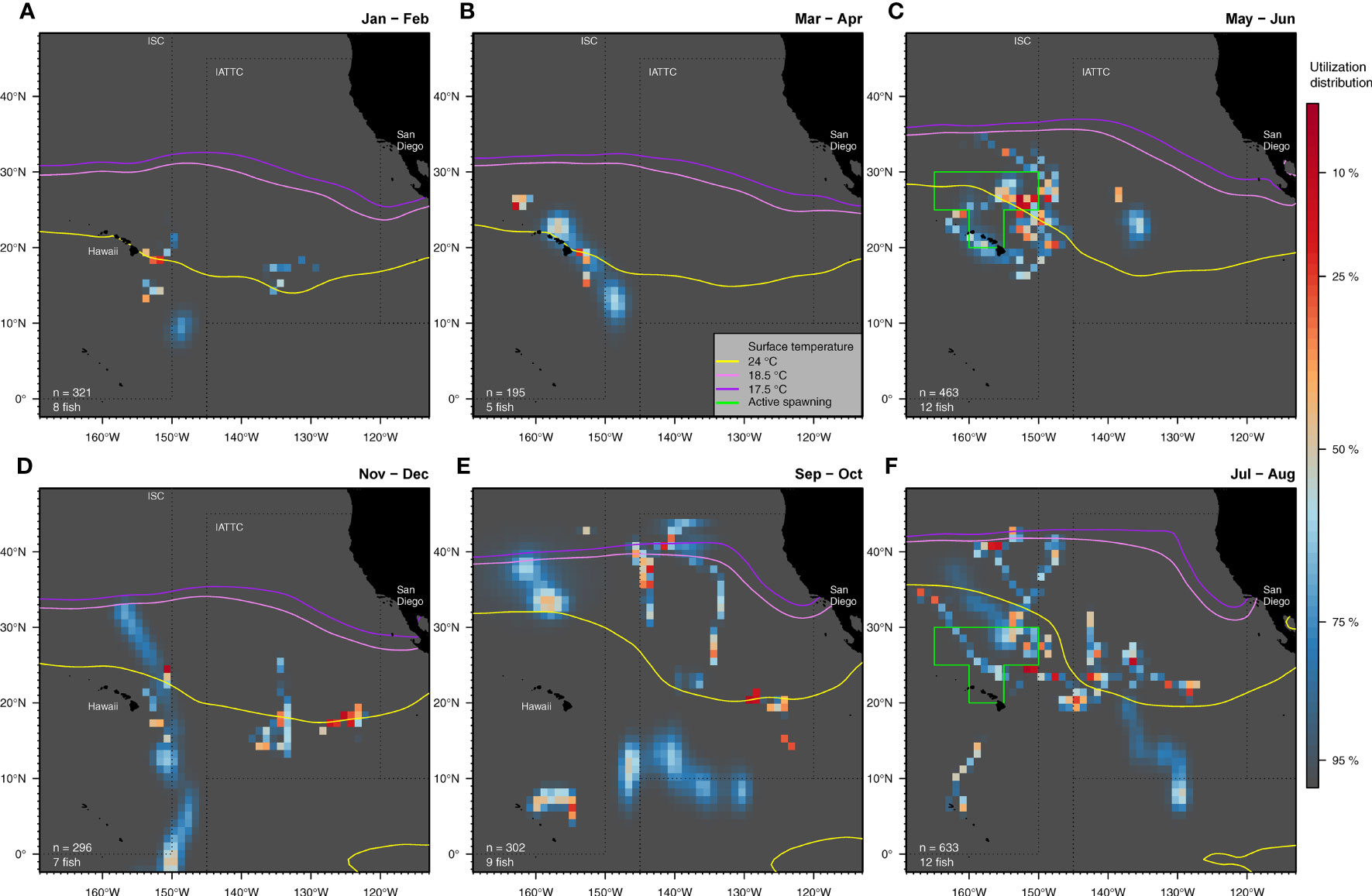
Figure 4 Spatial use of the Central North Pacific by tagged striped marlin summarized bimonthly: January-February (A), March-April (B), May-June (C), July-August (F), September-October (E) and November-December (D). Utilization distribution (UD) is represented in false color. UD values of less than 50% signify high use areas. In each panel, monthly climatological surface temperature isotherms at 17.5 (violet), 18.5 (lavender) and 24°C (yellow) are contoured. For clarity, only isotherms from the second month of each period are shown. The extent of an area with active female spawners (Humphreys and Brodziak, 2019) during the spawning season (May-August) is outlined in green. The number of individuals and positions available are indicated for each bimonthly period.
Movements and Fisheries Management Boundaries
Tracked striped marlin crossed multiple fisheries management boundaries within the maximum observation period of one year (Figures 1, 2; Table 2). Among the fourteen striped marlin tagged within the ISC management area west of 150°W, four fish (29%) remained west of the boundary, five (36%) ventured east beyond the boundary but later returned inside ISC area, one (7%) tag reported between 145 and 150°W, an area not under management, and four (29%) traveled east and were located in areas under IATTC’s jurisdiction (Table 2). Of three fish tagged inside IATTC areas, two remained inside, and one marlin entered the South Pacific, under WCPFC jurisdiction (Figure 2A).
Movement Patterns From Conventional and Electronic Tagging
Conventional tags, as well as biologically sampled fish, can provide additional information on the timing when an individual from a nearby or faraway site ventured to a particular location (Figure 5). Striped marlin tagged around the MHI (<1,000 km linear displacement) showed similar seasonal destinations to those observed by PSATs (Figure 4). An exception was the forays to waters just south of the MHI between January and March, most of which took under 90 days to complete, but were not observed in PSAT-tagged fish. Their presence coincided with the general high availability to fishing efforts around the MHI during this time of the year. The recapture of striped marlin from California (conventional tag A013173, H041215 & H046722; 214-320 days at liberty) in waters north of the MHI during the spawning months of May and June, was also noticeable (Figure 5). Their recapture locations were similar to the summer recaptures originating from the MHI (500-1,000 km linear displacement), and in the vicinity of PSATs 13P0434 and 15P0239 (Figure 2). Other long-ranged (>2,000 km) dispersals from California either had destinations near/along the tracks of 13P0434 and 16P1157 in the IATTC management areas (Figure 2), or west of the MHI near the Mid-Pacific Mountains (12-16°N and 175-180°W).
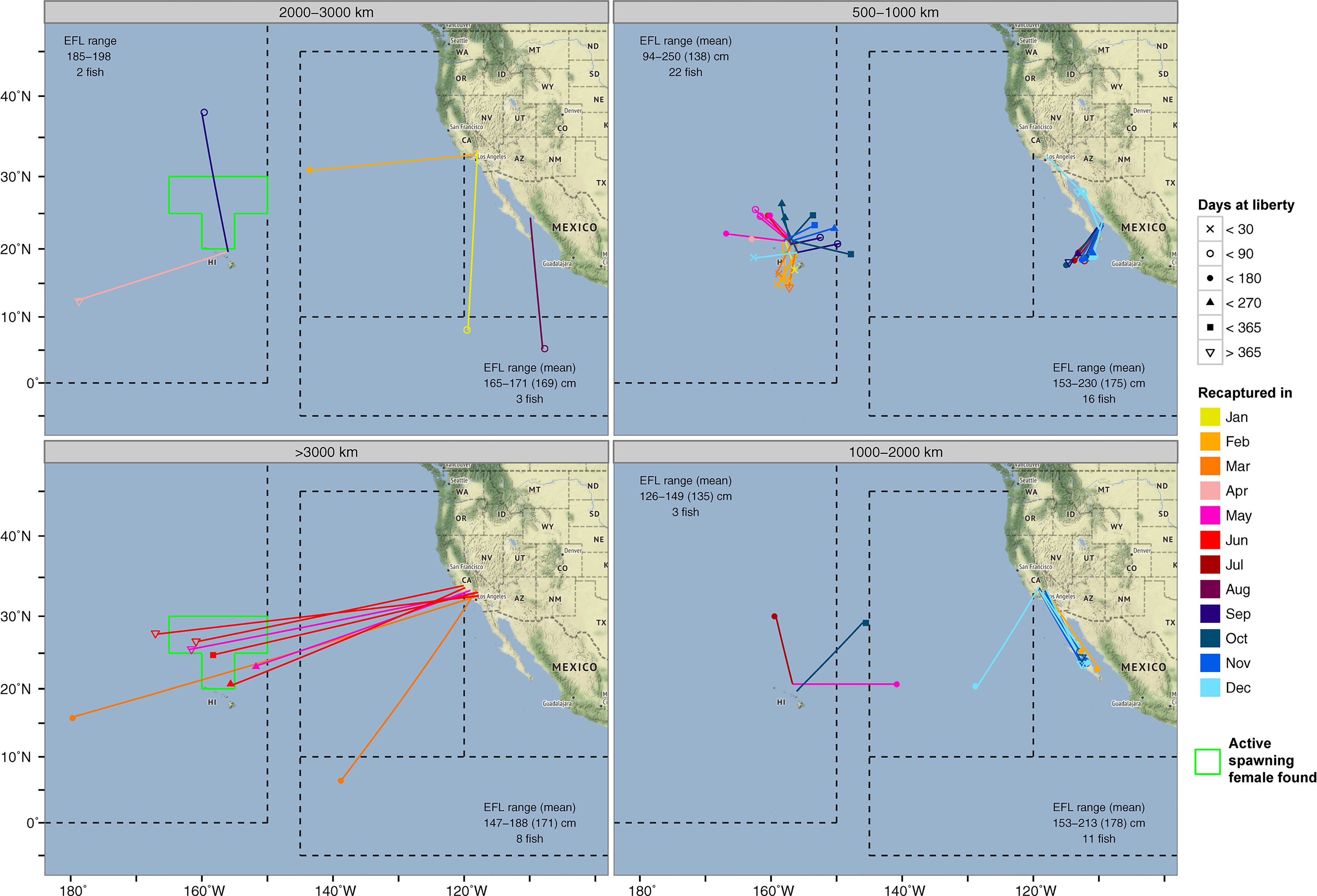
Figure 5 Long-distance (>500 km) conventional tag recaptures of striped marlin from NOAA Southwest Fisheries Science Center Cooperative Billfish Tagging Program. The month in which a tag was recaptured is color-coded. Icons at the recapture location indicate the number of days post-release. The extent of an area with active female spawners (Humphreys and Brodziak, 2019) during the spawning season is outlined in green, and displayed for recaptures >2,000 km. The number of tag recaptures and eye fork length statistics are indicated in each panel.
Genetic Profiles
After performing all the necessary quality control on sequenced DNAs, 55 Hawaii-landed striped marlin were analyzed in conjunction with 67 and 57 individuals caught off east Australia and New Zealand, respectively (Evans et al., 2021). Based on the best mixture model, landed fish were assigned to two genetic groups: Australia, New Zealand and Hawaii (AU-NZ-HI) comprising the first group (n = 19), and a second (n = 36) as Hawaii (HI) alone. Notably, 17 (89%) individuals assigned to AU-NZ-HI were sampled prior to July 11, while 26 (72%) HI fish were sampled afterward (Table 3). In addition, individuals assigned to AU-NZ-HI (172 ± 12 cm EFL) were significantly (Student’s t-test, p < 0.0001) larger than those of HI (145 ± 9 cm EFL). Review of non-confidential logbook records showed striped marlin from both genetic groups mixed in an area centered at 27.5°N, 150°W, between June and mid-July, where longline vessels also concentrated. The centroid of fishing activities then shifted south to 17.5°N, 157°W after July 11.
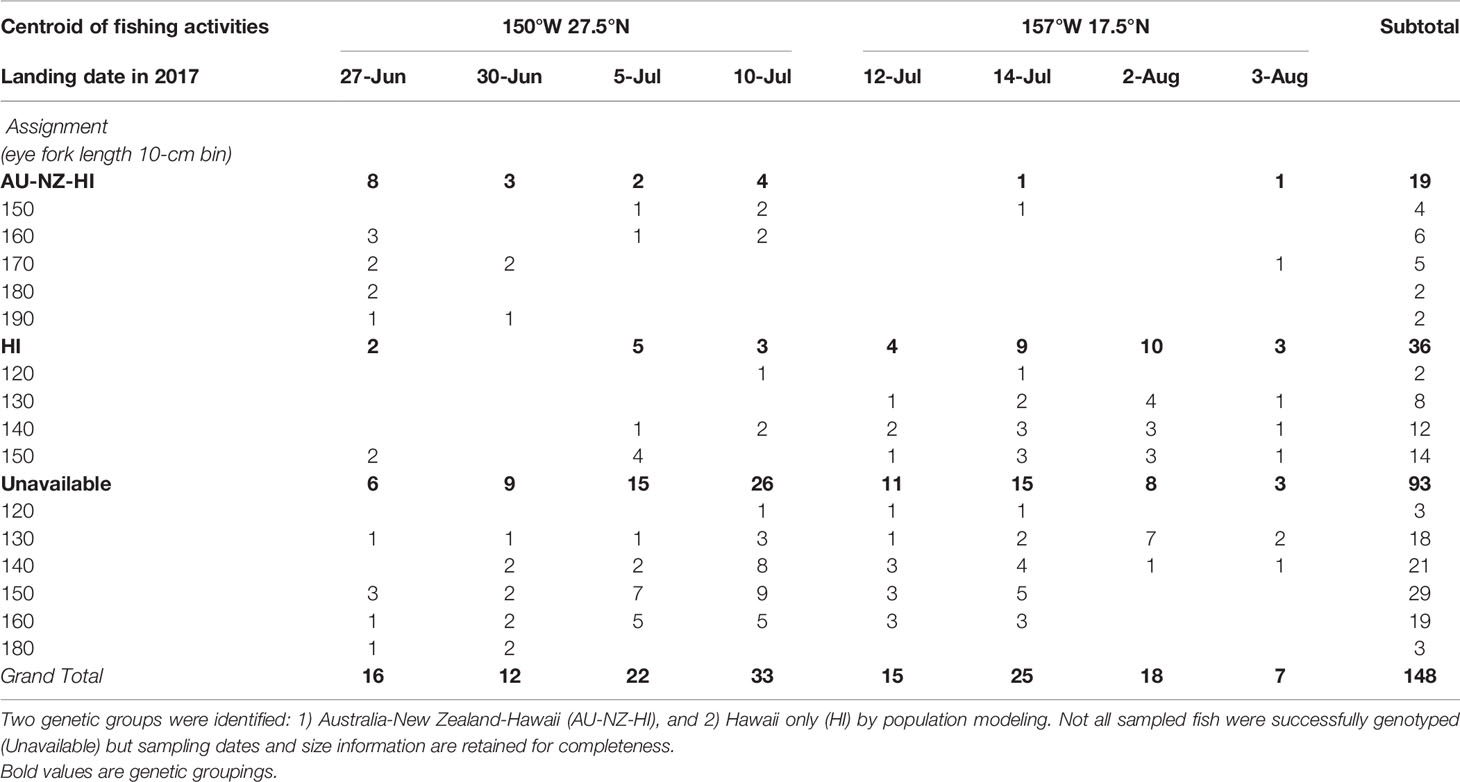
Table 3 Assignment results of genetically sampled striped marlin in relation to the approximate location of fishing activities (5°-grid cell), sampling date on the auction floor and size classes in 10-cm eye fork length (converted from dressed weight) intervals.
Discussion
The work reported here represents the most extensive effort to date to characterize striped marlin movements in the Central North Pacific. Previous studies have relied heavily on the recreational fleets for access to striped marlin, biasing sampling to coastal waters. A strong working partnership with fishermen collaborators, important to solving complex fisheries challenges (Hare, 2020), has enabled us to successfully tag fish over a broad area of striped marlin pelagic habitat. While we did not follow a cohort of individuals released from the same coastal aggregation through time, two full-year tracks delivered long-term movement datasets for billfish rivaled only by archival tagging of striped marlin (Domeier et al., 2019) and PSAT tagging of sailfish (Lam et al., 2016). Striped marlin tagged in the CNP exhibited a diversity of dispersal patterns that had not previously been documented, yet they fell into the characteristic horseshoe-shaped (⊃) distribution long established by fisheries catch statistics (Bromhead et al., 2004).
Reproductive Activities
While current PSATs lack physiological sensors with the capability to detect spawning, nine of our tracked marlin (PG05, 07, 13, 14, 15, 17, 18, 30 & 31) visited a spawning ground in May-June (Figure 3B and Appendices 1, 3), where active spawning females concentrated at a median latitude of 27.1°N (Humphreys and Brodziak, 2019). Notably, eye fork length at 50% maturity was determined at 152.2-153.6 cm (Humphreys and Brodziak, 2021), and the smallest mature female was 146 cm EFL (Humphreys and Brodziak, 2019). During its tracked period, PG05 spent time on a spawning ground during June in two consecutive years (Appendix 1). Prior to reaching this spawning ground, PG05 & 31 also trekked past Kona, Hawaiʻi where larvae had previously been collected (Hyde et al., 2006). Taking these lines of evidence together, it is plausible that some of our tracks depict striped marlin entering a spawning ground for reproduction.
Patterns of Connectivity
Long fundamental to fisheries management, spatial jurisdictions and static boundaries ideally would capture either fishing patterns or genetic populations as separate, self-contained management units, or stocks (Cadrin, 2020). Striped marlin in the Pacific Ocean have at least three genetic populations (Mamoozadeh et al., 2020): pan-North Pacific (NPO), Eastern Pacific (ECPO) and Western South Pacific (WSPO). Unlike the Pacific-wide representation of sampling locations in Purcell and Edmands (2011) and Mamoozadeh et al. (2020), genetic analyses of our Hawaii-landed fish were conducted only with other samples collected off east Australia and New Zealand (Evans et al., 2021). Despite this limitation, population modeling identified just two genetic groups: AU-NZ-HI and HI only, and rejected any additional groupings. The AU-NZ-HI group may be part of the WSPO population, while the HI only group may belong to the NPO population. Further investigation will be necessary to clarify the relevance of our genetic sampling results for determining population structure for the broader Pacific.
Albeit a developing story, we identify the Central North Pacific as a dynamic hub for mixing: striped marlin of different genetic identities may be present throughout the year, likely foraging in or transiting throughout the broader region. Some of these fish, perhaps joined by pulses of new arrivals (e.g., from California; Figure 2), might then converge on spawning grounds for reproduction in the summer. After the spawning season, between August and October, fish would disperse to forage elsewhere at higher latitudes or in the Eastern Pacific (Figure 4), chasing squid, lancetfish, small tuna and other epipelagic fish (Moteki et al., 2001; Kitchell et al., 2004; Shimose et al., 2010; Lin and Zhu, 2020), before returning to productive waters off the MHI in winter and spring (Figure 4). Pooling DNA collections for re-analysis with common methodology and protocols, as well as sampling at a high temporal resolution over multiple years, and with explicit location information, will be sensible next steps to clear up ambiguities in population structure among genetic studies, and reconcile connectivity patterns between electronic tagging and genetic profiling.
Mixing, Gene Flow, and Management Considerations
Our genetic results have provided evidence for a size-based partitioning of striped marlin habitat, where fish >160 cm EFL from different genetic groups mixed north of MHI in June, whereas smaller fish (<160 cm EFL) were found mostly south of the MHI in July (Table 3). Yet even in mixing areas, gene flow must be limited, a condition necessary for the existence of at least three genetically distinct Pacific populations. Resolving how the Central North Pacific is utilized by one or more striped marlin populations, and whether distinct spawning areas exist for different populations in various parts of the Pacific will provide more information regarding the assemblage composition off Hawaii, and possible isolation mechanisms that prevent gene flow.
Our tracks have shown that striped marlin may cross management boundaries and jurisdictions multiple times within one year. Presumably, the Hawaii-based longline fishery interacts with striped marlin from different populations, at least in fishing grounds north of MHI. Explicit genetic connectivity among populations might not be especially relevant to the implementation of fisheries management measures: barring major technological breakthroughs, fleets have no means of verifying the genetic identity of a captured fish. Consequently, they can’t be selective about retaining or releasing individuals from a specific population or stock. Given an emerging picture of complex mixing patterns and frequent border crossings, effective management of striped marlin may require dissolving static management boundaries (Koubrak and Vanderzwaag, 2020), and move towards a coordinated, cross-jurisdictional management in the Pacific, with the full acknowledgement of mixing.
Tag failure, as reported in this study and others (e.g. Lam et al., 2020), has increasingly become a costly impediment to our ability to track and observe billfish and tuna (Musyl et al., 2011; Lutcavage et al., 2015). To ensure the least amount of bias introduced into stock assessment and fisheries management, stakeholders must encourage tag manufacturers to provide reliable products with consistent performance, and push for innovation in tracking technologies so that research dollars can be spent most effectively.
Pacific-Wide Movement Scenarios
Prior to this study, tagging had failed to replicate patterns of connectivity among genetic populations. Earlier conventional and electronic tagging results (Ortiz et al., 2003; Domeier, 2006) concluded striped marlin were localized to their regional aggregations, and large-scale movements were rare (Bromhead et al., 2004). Our first documented trans-Pacific movement along with other examples of longer-term tracks from the Eastern Pacific (Figures 2, S2) provided new evidence in support of a broader connectivity, complementary to that indicated by genetic analyses. When a growing number of longer-term tracks are collectively evaluated, as we have attempted here, they show when and where mixing could occur, offering a spatiotemporal perspective unattainable by genetic or chemical techniques. In fact, PG01 revealed one route for half of a “roundtrip” migration between the CNP and Oceania (Figure 2A). The putative return leg had been carried out by 36% of fish in Evans et al. (2021) and 19% (4 out of 21) in Mamoozadeh et al. (2020). With a proper, targeted tagging design, this type of cyclic, trans-Pacific migration could finally be elucidated.
Given the financial and technical constraints commonly faced by fisheries scientists, we had a limited window to observe long-term, large-scale movements: plagued by hardware issues, only 10 PSATs lasted over 90 days, and genetic samples were collected over a span of 37 days between June and August. PSATs deployed on our largest individuals (168 & 190 cm EFL) failed to report, squandering the unique opportunity to follow a size class less frequently captured by the Hawaii-based longline fishery (Ito, 2019). Consequently, exchanges between the CNP and Oceania may be higher than currently realized, or more frequent during specific times or seasons. We further postulate that habitats northeast of Hawai`i and between Clarion and Clipperton Fracture Zones (130-145°W) is a hotspot for mixing, with individuals arriving from the Eastern Pacific (Figure 2). Combined tracking and genetic results reported here suggest a Pacific-wide movement corridor linking Oceania, Hawaii and the Central North Pacific with the Eastern Pacific. Intriguingly, three fish (out of 37) sampled off Ecuador clustered with striped marlin in the Western South Pacific (WSPO) population (Mamoozadeh et al., 2020), indicating a possible route for striped marlin traveling east across the South Pacific into the Eastern Pacific via French Polynesia (Figure 3A). Mapping out additional movement pathways, such as one connecting Japan or Taiwan to Hawaii or the South Pacific, should be a research priority. Complexities of ontogenetic changes in dispersal patterns, spawning site selection, and biological or environmental drivers impacting movements are yet to be investigated, further restricting our ability to fully understand striped marlin migration.
Conclusions
Electronic tagging results have documented diverse seasonality in striped marlin movements, distributed over a vast area spanning from 3 to 45°N and 122 to 170°W in the Central North Pacific. A year-long track revealed trans-Pacific movement not previously observed for striped marlin, challenging previous notions that striped marlin are highly localized in their regional, coastal aggregations. Our work suggests their potential connectivity to other regions may have been previously overlooked due to the lack of sufficiently long-term deployments of PSATs. Continued striped marlin tagging efforts integrated with genetic profiling are highly encouraged, and recommended with our first and exciting discovery of cyclic migration between the North Pacific and Oceania.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Ethics Statement
Ethical review and approval was not required for the animal study because in the US, it is accepted practice that placing a dart type external fish tag aboard a commercial fishing vessel does not require review by IAUC committees. Tag and release, with a small bio-compatible dart, conducted in the manner stated in our paper, is routine in US recreational and commercial fisheries, and does not require permitting by fisheries regulators.
Author Contributions
CL, ML, and CT designed and executed the research. CL performed the analyses. All authors prepared, revised and approved the manuscript.
Funding
We are grateful to the funding provided by NOAA Saltonstall-Kennedy Grant (award # NA15NMF4270324).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We are indebted to our accomplished tagging partners, Captains Steve Sexton, Tim Jones, Steve Gates, Joel Ralston and their crews, and the United Fishing Agency for support of bio-sampling. We thank Dr. Michael Domeier and Dr. R. Keller Kopf for sharing their original tagging and maturity data, respectively, and Dr. Nadya Mamoozadeh for sharing metadata and helpful insights in our far-reaching discussions. The International Game Fishing Association and Stanford University, especially Jason Schratweister and Leah Baumwell, graciously shared the Great Marlin Race data, with Dr. Jonathan Dale patiently entertained multiple data requests. Genetic sample processing and analyses performed by CSIRO were supported by FRDC Project No 2016/018 (frdc.com.au/project/2016-018). The Western Pacific Regional Fishery Management Council, and namely, Dr. Mark Fitchett, greatly facilitated the genetic work. We thank NOAA scientists Dr. Keith Bigelow, Russell Ito and Dr. Christopher Tokita for providing public access to non-confidential logbook records, and Liana Heberer in compiling conventional tag recoveries.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2022.879463/full#supplementary-material
Supplementary Figure 1 | Spatial use of the Central North Pacific by striped marlin between March and May. Utilization distribution (UD) is represented in false color. UD values of less than 50% signify high use areas. The number of individuals and positions available are indicated for each month.
Supplementary Figure 2 | Horizontal movements of striped marlin tagged in the Eastern North Pacific. Positions are color-coded by months, and starting and reporting positions are labeled with their tag serial (refer to for details). The extent of an area with active female spawners (Humphreys and Brodziak, 2019) during the spawning season (May-August) is outlined in green. Tagging locations, green triangle; reporting locations, light pink triangle; management boundaries of International Scientific Committee for Tuna and Tuna-like Species in the North Pacific Ocean (ISC) and Inter American Tropical Tuna Commission (IATTC), and Western and Central Pacific Fisheries Commission (WCPFC), dashed lines. Tracks were reconstructed following the same procedures outlined in Materials and methods. Tag data courtesy of Great Marlin Race of the International Game Fishing Association.
Appendix 2 | Animation of five striped marlin (PG13, 14, 15, 17 & 18) released in May 2018. Sea surface temperature imagery is OSTIA global foundation Sea Surface Temperature (product ID SST_GLO_SST_L4_NRT_OBSERVATIONS_010_001) obtained from European Union Copernicus Marine Environment Monitoring Service (marine.copernicus.eu/services-portfolio/access-to-products).
Appendix 3 | Animation of three striped marlin (PG30 & 31) released in June 2018. Sea surface temperature imagery is OSTIA global foundation Sea Surface Temperature (product ID SST_GLO_SST_L4_NRT_OBSERVATIONS_010_001) obtained from European Union Copernicus Marine Environment Monitoring Service (marine.copernicus.eu/services-portfolio/access-to-products).
References
Anonymous. (2016). The Southwest Fisheries Science Center's 2016 Billfish Newsletter. Available at: www.fisheries.noaa.gov/resource/document/2016-billfish-newsletter.
Brill R. W., Holts D. B., Chang R. K. C., Sullivan S., Dewar H., Carey F. G. (1993). Vertical and Horizontal Movements of Striped Marlin (Tetrapturus Audax) Near the Hawaiian-Islands, Determined by Ultrasonic Telemetry, With Simultaneous Measurement of Oceanic Currents. Marine Biol. 117, 567–574. doi: 10.1007/BF00349767
Brodziak J. (2021). “Some Rebuilding Analyses for the Western and Central North Pacific Ocean Striped Marlin Stock,” in ISC/21/BILLWG-01/04 (ISC Billfish Working Group Workshop Webinar).
Bromhead D., Pepperell J., Wise B., Findlay J. (2004). “Striped Marlin: Biology and Fisheries. Final Report to the Australian Fisheries Management Authority and the Fisheries Research Fund,” in Bureau of Rural Sciences(Canberra, Australia: Bureau of Rural Sciences).
Cadrin S. X. (2020). Defining Spatial Structure for Fishery Stock Assessment. Fish Res. 221, 105397. doi: 10.1016/j.fishres.2019.105397
Domeier M. L. (2006). An Analysis of Pacific Striped Marlin (Tetrapturus Audax) Horizontal Movement Patterns Using Pop-Up Satellite Archival Tags. Bull. Marine Sci. 79, 811–825(815).
Domeier M. L., Ortega-Garcia S., Nasby-Lucas N., Offield P. (2019). First Marlin Archival Tagging Study Suggests New Direction for Research. Marine Freshwater Res. 70, 603–608. doi: 10.1071/MF18160
Evans K., Grewe P., Foster S., Gunasekara R., Lansdell M. (2021). Determination of the Spatial Dynamics and Movement Rates of the Principal Target Species Within the Eastern Tuna and Billfish Fishery and Connectivity With the Broader Western and Central Pacific Ocean - Beyond Tagging (Hobart, Australia: Fisheries Research and Development Corporation).
Fitchett M. D. (2019). “Estimating Age and Growth of Central North Pacific Striped Marlin Using Tagging Data and Direct Observations of Age,” in ISC/19/BILLWG-1/12 (Honolulu, USA: ISC Billfish Working Group).
Galuardi B., Lutcavage M. (2012). Dispersal Routes and Habitat Utilization of Juvenile Atlantic Bluefin Tuna, Thunnus Thynnus, Tracked With Mini PSAT and Archival Tags. PloS One 7, e37829. doi: 10.1371/journal.pone.0037829
Galuardi B., Royer F., Golet W., Logan J., Neilson J., Lutcavage M. (2010). Complex Migration Routes of Atlantic Bluefin Tuna (Thunnus Thynnus) Question Current Population Structure Paradigm. Can. J. Fish Aquat. Sci. 67, 966–976. doi: 10.1139/F10-033
Hare J. A. (2020). Ten Lessons From the Frontlines of Science in Support of Fisheries Management. ICES J. Marine Sci. 77, 870–877. doi: 10.1093/icesjms/fsaa025
Heberer L. N., Wraith J. A., Kohin S., Gu Y., Nasby-Lucas N., Dewar H. (2021). "The Southwest Fisheries Science Center Cooperative Billfish Tagging Program Operations and Database", in NOAA Technical Memorandum NMFS-SWFSC-640 (U.S. Department of Commerce), 46 pp..
Howell E. A., Kobayashi D. R., Parker D. M., Balazs G. H., Polovina A. (2008). TurtleWatch: A Tool to Aid in the Bycatch Reduction of Loggerhead Turtles Caretta Caretta in the Hawaii-Based Pelagic Longline Fishery. Endanger Species Res. 5, 267–278. doi: 10.3354/esr00096
Hsu J., Chang Y. J., Yeh S. Y., Sun C. L., Yeh S. Z. (2019). “Catch and Size Data of Striped Marlin (Kajikia Audax) by the Taiwanese Fisheries in the Western and Central North Pacific Ocean During 1958-2017,” in ISC/19/BILLWG-1/03 (Honolulu, USA: ISC Billfish Working Group).
Humphreys R., Brodziak J. (2019). “Reproductive Maturity of Striped Marlin, Kajikia Audax, in the Central North Pacific Off Hawaii,” in ISC/19/BILLWG-2/02 (Honolulu, USA: ISC Billfish Working Group).
Humphreys R., Brodziak J. (2021). “Revised Analyses of the Reproductive Maturity of Female Striped Marlin, Kajikia Audax, in the Central North Pacific Off Hawaii,” in ISC/21/BILLWG-3/07. (Webinar; ISC Billfish Working Group)
Hyde J. R., Humphreys R., Musyl M., Lynn E., Vetter R. (2006). A Central North Pacific Spawning Ground for Striped Marlin, Tetrapturus Audax. Bull. Marine Sci. 79, 683–690.
ISC. (2019). “Stock Assessment Report for Striped Marlin (Kajikia Audax) in the Western and Central North Pacific Ocean Through 2017,” in 19th Meeting of the International Scientific Committee for Tuna and Tuna-Like Species in the North Pacific Ocean Taipei, Taiwan July 11-15, 2019. Annex 11 (Taipei, Taiwan).
Ito R. Y. (2019). “U.S. Commercial Fisheries for Marlins in the North Pacific Ocean,” in ISC/19/BILLWG-1/01 (Honolulu, USA: ISC Billfish Working Group).
Johnson D. S., London J. M., Lea M.-A., Durban J. W. (2008). Continuous-Time Random Walk Model for Animal Telemetry Data. Ecology 89, 1208–1215. doi: 10.1890/07-1032.1
Kitchell J. F., Kaplan I. C., Cox S. P., Martell S. J. D., Essington T. E., Boggs C. H., et al. (2004). Ecological and Economic Components of Alternative Fishing Methods to Reduce by-Catch of Marlin in a Tropical Pelagic Ecosystem. Bull. Marine Sci. 74, 607–619.
Kopf R. K., Davie P. S., Bromhead D. B., Young J. W. (2012). Reproductive Biology and Spatiotemporal Patterns of Spawning in Striped Marlin Kajikia Audax. J. Fish Biol. 81, 1834–1858. doi: 10.1111/j.1095-8649.2012.03394.x
Koubrak O., Vanderzwaag D. L. (2020). Are Transboundary Fisheries Management Arrangements in the Northwest Atlantic and North Pacific Seaworthy in a Changing Ocean? Ecol. Soc. 25, 42. doi: 10.5751/ES-11835-250442
Lam C. H., Galuardi B., Mendillo A., Chandler E., Lutcavage M. E. (2016). Sailfish Migrations Connect Productive Coastal Areas in the West Atlantic Ocean. Sci. Rep. 6, 38163. doi: 10.1038/srep38163
Lam C. H., Kiefer D. A., Domeier M. L. (2015). Habitat Characterization for Striped Marlin in the Pacific Ocean. Fish Res. 166, 80–91. doi: 10.1016/j.fishres.2015.01.010
Lam C. H., Nielsen A., Sibert J. R. (2008). Improving Light and Temperature Based Geolocation by Unscented Kalman Filtering. Fish Res. 91, 15–25. doi: 10.1016/j.fishres.2007.11.002
Lam C. H., Nielsen A., Sibert J. R. (2010). Incorporating Sea-Surface Temperature to the Light-Based Geolocation Model TrackIt. Marine Ecol. Prog. Ser. 419, 71–84. doi: 10.3354/meps08862
Lam C. H., Tam C., Kobayashi D. R., Lutcavage M. E. (2020). Complex Dispersal of Adult Yellowfin Tuna From the Main Hawaiian Islands. Front. Marine Sci. 7, 138. doi: 10.3389/fmars.2020.00138
Lam C. H., Tsontos V. M. (2011). Integrated Management and Visualization of Electronic Tag Data With Tagbase. PloS One 6, e21810. doi: 10.1371/journal.pone.0021810
Lien Y.-H., Su N.-J., Sun C.-L., Punt A., Yeh S.-Z., Dinardo G. (2013). Spatial and Environmental Determinants of the Distribution of Striped Marlin (Kajikia Audax) in the Western and Central North Pacific Ocean. Environ. Biol. Fishes 1-10.
Lin Q., Zhu J.-F. (2020). Topology-Based Analysis of Pelagic Food Web Structure in the Central and Eastern Tropical Pacific Ocean Based on Longline Observer Data. Acta Oceanologica Sin. 39, 1–9. doi: 10.1007/s13131-020-1592-2
Locarnini R. A., Mishonov A. V., Antonov J. I., Boyer T. P., Garcia H. E., Baranova O. K., et al. (2013). World Ocean Atlas 2013, Volume 1: Temperature (Washington, D.C: U.S. Government Printing Office).
Lutcavage M. E., Brill R. W., Skomal G. B., Chase B. C., Howey P. W. (1999). Results of Pop-Up Satellite Tagging of Spawning Size Class Fish in the Gulf of Maine: Do North Atlantic Bluefin Tuna Spawn in the Mid-Atlantic? Can. J. Fish Aquat. Sci. 56, 173–177. doi: 10.1139/f99-016
Lutcavage M., Lam C. H., Galuardi B. (2015). Seventeen Years and $3 Million Dollars Later: Performance of PSAT Tags Deployed on Atlantic Bluefin and Bigeye Tuna. Collect. Vol. Sci. Pap. ICCAT. 71 (4), 1757–1765
Mamoozadeh N. R., Graves J. E., Mcdowell J. R. (2020). Genome-Wide SNPs Resolve Spatiotemporal Patterns of Connectivity Within Striped Marlin (Kajikia Audax), a Broadly Distributed and Highly Migratory Pelagic Species. Evol Appl. 13, 677–698. doi: 10.1111/eva.12892
Moteki M., Arai M., Tsuchiya K., Okamoto H. (2001). Composition of Piscine Prey in the Diet of Large Pelagic Fish in the Eastern Tropical Pacific Ocean. Fish Sci. 67, 1063–1074. doi: 10.1046/j.1444-2906.2001.00362.x
Musyl M. K., Domeier M. L., Nasby-Lucas N., Brill R. W., Mcnaughton L. M., Swimmer J. Y., et al. (2011). Performance of Pop-Up Satellite Archival Tags. Marine Ecol. Prog. Ser. 433, 1–U58. doi: 10.3354/meps09202
Ortiz M., Prince E. D., Serafy J. E., Holts D. B., Davy K. B., Pepperell J. G., et al. (2003). Global Overview of the Major Constituent-Based Billfish Tagging Programs and Their Results Since 1954. Marine Freshwater Res. 54, 489–507. doi: 10.1071/MF02028
Purcell C. M., Edmands S. (2011). Resolving the Genetic Structure of Striped Marlin, Kajikia Audax, in the Pacific Ocean Through Spatial and Temporal Sampling of Adult and Immature Fish. Can. J. Fish Aquat. Sci. 68, 1861–1875. doi: 10.1139/f2011-104
Sculley M. (2019). “Striped Marlin (Kajikia Audax) Length Data Available From 1995-2017 in the Hawaii-Based Longline Fishery,” in ISC/19/BILLWG-1/04(Honolulu, USA: ISC Billfish Working Group).
Sculley M. (2021). “Update to the 2019 Western and Central North Pacific Ocean Striped Marlin Stock Assessment,” in ISC/21/BILLWG-01/02 (ISC Billfish Working Group Workshop Webinar).
Shimose T., Yokawa K., Saito H. (2010). Habitat and Food Partitioning of Billfishes (Xiphioidei). J. Fish Biol. 76, 2418–2433. doi: 10.1111/j.1095-8649.2010.02628.x
Sippel T. J., Davie P. S., Holdsworth J. C., Block B. A. (2007). Striped Marlin (Tetrapturus Audax) Movements and Habitat Utilization During a Summer and Autumn in the Southwest Pacific Ocean. Fish Oceanogr 16, 459–472. doi: 10.1111/j.1365-2419.2007.00446.x
Sippel T., Holdsworth J., Dennis T., Montgomery J. (2011). Investigating Behaviour and Population Dynamics of Striped Marlin (Kajikia Audax) From the Southwest Pacific Ocean With Satellite Tags. PloS One 6, e21087. doi: 10.1371/journal.pone.0021087
Su N.-J., Sun C.-L., Punt A. E., Yeh S.-Z., Dinardo G. (2015). Environmental Influences on Seasonal Movement Patterns and Regional Fidelity of Striped Marlin Kajikia Audax in the Pacific Ocean. Fish Res. 166, 59–66. doi: 10.1016/j.fishres.2014.07.017
Ueyanagi S., Wares P. G. (1975). “Synopsis of Biological Data on Striped Marlin, Tetrapturus Audax (Philippi 1887),” in Proceedings of the International Billfish Symposium, Kailua–Kona, Hawaii, 9–12 August 1972, Part 3. Species synopses. Eds. Shomura R. S., Williams F. (Seattle, WA: National Marine Fisheries Service), 132–159.
Keywords: PSAT, tagging, dispersal, stock mixing, Main Hawaiian Islands, Central North Pacific
Citation: Lam CH, Tam C and Lutcavage ME (2022) Connectivity of Striped Marlin From the Central North Pacific Ocean. Front. Mar. Sci. 9:879463. doi: 10.3389/fmars.2022.879463
Received: 19 February 2022; Accepted: 30 March 2022;
Published: 28 April 2022.
Edited by:
David M. P. Jacoby, University of Lancaster, United KingdomReviewed by:
Jorge Paramo, University of Magdalena, ColombiaAaron Carlisle, University of Delaware, United States
Copyright © 2022 Lam, Tam and Lutcavage. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chi Hin Lam, dGFndHVuYUBnbWFpbC5jb20=
 Chi Hin Lam
Chi Hin Lam Clayward Tam
Clayward Tam Molly E. Lutcavage1,2
Molly E. Lutcavage1,2