- 1Fisheries College of Guangdong Ocean University, Zhanjiang, Guangdong, China
- 2Guangdong Provincial Key Laboratory of Aquatic Animal Disease Control and Healthy Culture, Zhanjiang, Guangdong, China
Tumor necrosis factor receptor-associated factors (TRAFs) have been studied in a few mollusks and participate in various biological processes, like apoptosis, immune response, stress, and inflammatory response. However, TRAFs’ function and mechanism of pearl oysters (Pinctada fucata martensii) are still unclear. In this study, the novel PmTRAF2 and PmTRAF4 from P. f. martensii were cloned by rapid amplification of complementary DNA ends and their mRNA expression were analyzed by quantitative real-time PCR (qPCR). The interacting protein of PmTRAF2 was verified by the yeast two-hybrid assay. The result shows that full-length of PmTRAF2 and PmTRAF4 cDNA were 2055 bp and 2365 bp, respectively. The deduced PmTRAF2 and PmTRAF4 proteins contain TRAF-type zinc finger domain and MATH domain, while PmTRAF4 lacks a RING finger domain. Multiple sequence alignment revealed that PmTRAF2 and PmTRAF4 had high homology with the ortholog of other species. Phylogenic analysis indicated that PmTRAF4 clustered with the homolog protein of Mytilus edulis and Mytilus galloprovincialis, and PmTRAF2 has the closest genetic relationship to Crassostrea gigas TRAF2. The qPCR analysis revealed that PmTRAF2 and PmTRAF4 were expressed in all six tissues, and both of them were significantly expressed in hepatopancreas and gill (p < 0.01). Under lipopolysaccharide (LPS) stimulation, polyinosinic acid (PolyI:C) stimulation, and nucleus insertion surgery, the transcripts of PmTRAF2, PmTRAF3, PmTRAF4 and PmTRAF6 in hepatopancreas were markedly changed at corresponding time points. These results have indicated that these genes may play a role in P. f. martensii innate immunity. Yeast two-hybrid assays show that PmTRAF2 interacts with PmTRAF6 but not PmTRAF3, potentially affecting downstream immune signaling pathways. Our findings provide new perspectives for further investigation of TRAFs’ immune mechanisms in bivalves.
1 Introduction
Tumor necrosis factor receptor-associated factors (TRAFs) are a family of cytoplasmic adapters that play important roles in innate and acquired immunity, promoting intracellular signal transduction via receptors, such as tumor necrosis factor receptor (TNFR), toll-like receptors-interleukin-1binding receptors (TLR/IL-1R), the Epstein–Barr virus protein LMP1, and the RIG-I–like receptor family, potentiating the recruitment of signal transduction proteins (Arch et al., 1998; Evans et al., 2018; Guo et al., 2020). Seven members of the TRAF family, TRAF1-7, have been identified and participated in a host of signaling pathways relative to immunity. TRAF2, a prototypical TRAF, combines with TNFR1, TNFR2, CD40, and LMP-1 to induce downstream signaling factors, participating in cell survival and apoptosis, immunity, and infection process (Han et al., 2003). TRAF1, TRAF2, TRAF3, TRAF5, and TRAF6 can be recruited by CD40, whose cytoplasmic tail has two TRAF-interacting motifs; TRAF2 and TRAF3 bind to PxQxT motif to positively regulate the downstream factor nuclear factor kappa-light-chain-enhancer of activated B cells (NF-κB), while TRAF6 binds to another motif to activate NF-κB (Bishop, 2004; Hauer et al., 2005). In addition, TRAF4 modulated mitogen-activated protein kinase, c-Jun N-terminal kinase, Wnt/β-catenin, and NF-κB signaling pathways via interacting with other molecules in mammals subsequently exerting functions in immunity (Xu et al., 2002; Wang et al., 2014; Lalani et al., 2018).
Interacting with receptors to modulate pathways, TRAFs can function as a regulatory factor in physiological processes. TRAF1 can regulate pathways that can be mediated by TRAF2, and TRAF3 can increase TRAF6-mediated NF-κB transcriptional activity while inhibiting the TRAF2/5-mediated NF-κB signal pathway (Hauer et al., 2005; Xie et al., 2006; Foight and Keating, 2016). These findings have revealed that different members of the TRAFs’ family interactions with others can result in different effects. TRAF members can interact to form homo- and hetero-dimers with distinct regulatory functions involved in signal transduction (Arron et al., 2002). In the CD40 signal pathway, TRAF1, TRAF2, TRAF3, and TRAF6 can form homo-dimers and bind CD40 receptors. TRAF3 and TRAF5 can form hetero-oligomers (Arron et al., 2002; Xie et al., 2006). These findings indicate that TRAFs may regulate immune-related pathways through their interactions. However, at present, the specific mechanism of the mutual regulation and binding of TRAFs is still unclear.
In recent years, most TRAFs from bivalves in marine mollusks have been identified and studied. Four Chlamys nobilis TRAF genes were identified, and their messenger RNA (mRNA) expression levels in hemolymph were significantly increased by exposure to Vibrio parahaemolyticus compared to the control group, which indicates that TRAFs are involved in host immunity (Zhang et al., 2021). HcTRAF6 expression level was significantly up-regulated in gills and hemocytes in response to immune stimulation (Aeromonas hydrophila and lipopolysaccharide (LPS) stimulation) in Hyriopsis cumingii (Huang et al., 2018). In the oyster, TRAF2 and TRAF6 from Crassostrea gigas and TRAF6 from Mizuhopecten yessoensis were induced to express upon bacterial infection (He et al., 2013; Huang et al., 2016; Mao et al., 2017). Although TRAFs can have an impact in response to immune-related stimulation, their functions and molecular mechanisms in different bivalves still need to be explored.
The oyster Pinctada fucata martensii, a type of seawater pearl oyster, relies primarily on innate immunity to defend against invading pathogens (Zhang et al., 2022). In previous studies, the TRAF3, TRAF6, and TRAF7 genes from the pearl oyster were identified (Huang et al., 2012; Jiao et al., 2014; Lei et al., 2016); Huang et al. and Jiao et al. have observed that TRAF3 and TRAF6 are involved in the innate immune response to V. alginolyticus and LPS stimulation, respectively. However, less is well understood about two P. f. martensii TRAFs, TRAF2 (PmTRAF2) and TRAF4 (PmTRAF4), and the role of these genes in the immune response remains obscure. In this study, PmTRAF2 and PmTRAF4 were identified in P. f. martensii hemocyte transcriptome, and their expression levels were analyzed in different tissues. The expression levels of PmTRAF2, PmTRAF3, PmTRAF4, and PmTRAF6 were analyzed in the hepatopancreas in response to LPS and polyinosinic acid (PolyI:C) stimulation, and nucleus insertion surgery. In addition, we used yeast two-hybrid to verify the interacting proteins. Our study offers new insights to further study the function of TRAF and the mechanism of shellfish immunity.
2 Materials and methods
2.1 Experimental sample
P. f. martensii (about 6-8 cm in shell length) were procured from Xuwen Breeding Base, Zhanjiang, Guangdong Province, China, and were temporarily cultured in seawater at 20-25°C for about 3 d to adapt to the indoor environment and conduct follow-up experiments.
2.2 Stimulation experiments and tissue sample collection
A total of 250 oysters P. f. martensii were randomly divided into four groups: 10 oysters in the tissue quantification group, 60 oysters in the LPS stimulation group, 60 oysters in the PolyI:C stimulation group, 60 oysters in the phosphate-buffered saline (PBS) group (control group), and 60 oysters in the nuclear insertion surgery.
In the tissue quantification group, mantle, gills, adductor muscle, hepatopancreas, hemolymph, and gonad were collected from each pearl oyster. The hemolymph was centrifuged at 3500 rpm for 5 min at 4°C, and the pellet (hemocytes) was collected and resuspended with 1 mL of Trizol reagent (Thermo-Fisher Scientific, Waltham, MA, USA).
The adductor muscle injection method was used in LPS and PolyI:C stimulation experiments. Each pearl oyster in the LPS stimulation group was injected with 100 μL LPS (10 μg/mL, diluted with sterile PBS), while the control group was injected with 100 μL PBS and the PolyI:C stimulation group was injected with 100 μL PolyI:C (10 μg/mL, diluted with sterile PBS) (Liang et al., 2022). At 0, 12, 24, 48, 72, and 96 h exposed to LPS and PBS, hepatopancreas were collected with 10 pearl oysters at each time point. All samples were stored at -80°C.
Sixty oysters were used to perform nucleus insertion surgery by an experienced technician at Xuwen Breeding Base, China. In nucleus insertion surgery, a piece of mantle graft from a donor oyster with a spherical shell bead or nucleus is transplanted into the gonad of the host oyster (Wu et al., 2020). At 0, 12, 24, 48, and 72 h after insertion, hepatopancreas samples were collected with 10 pearl oysters.
2.3 Cloning and sequence analysis
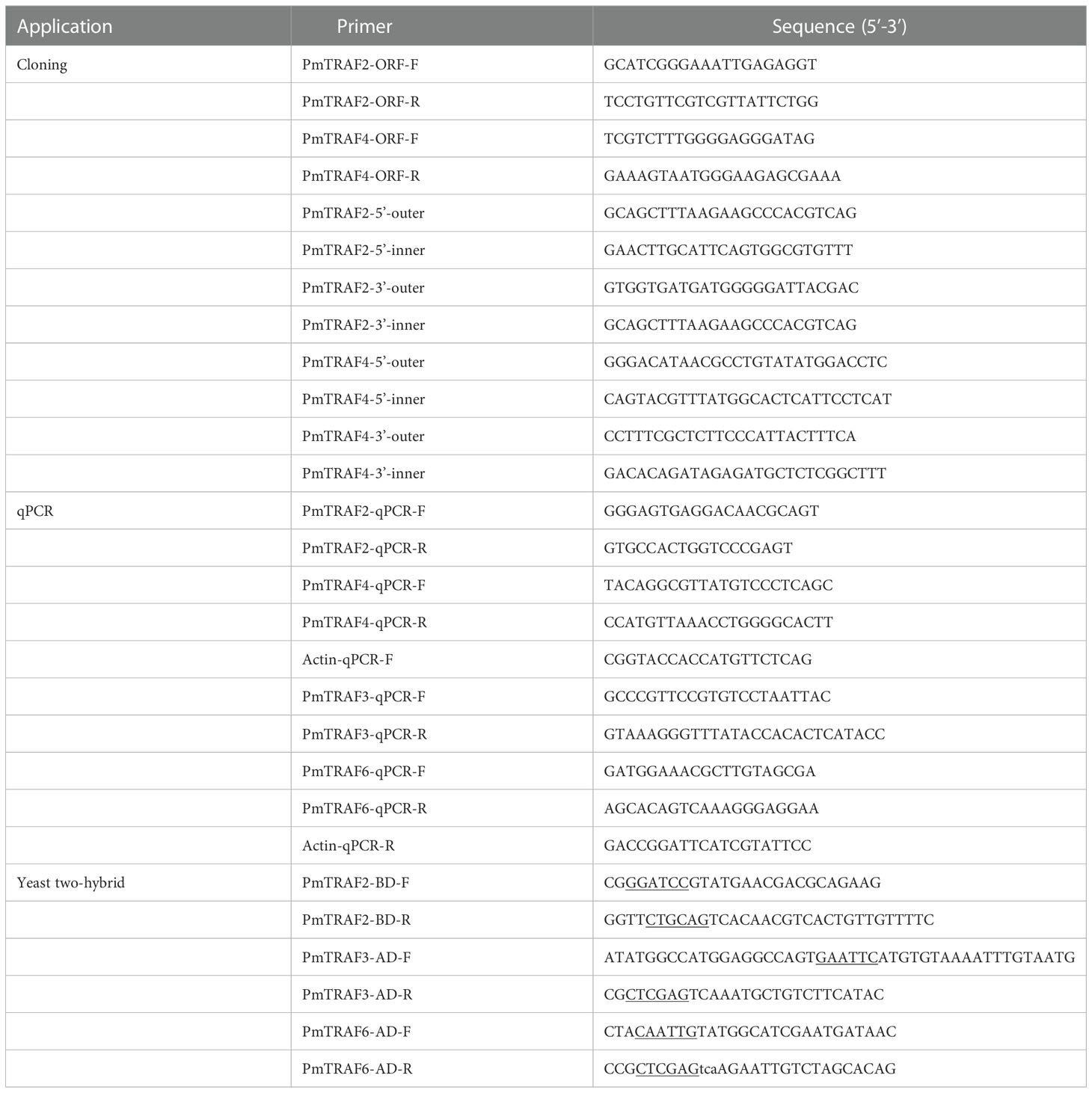
The partial sequences of PmTRAF2 and PmTRAF4 were from the hemocyte transcriptome of P. f. martensii in a previous study (Wang et al., 2017). To obtain the full-length sequences, we used Rapid amplification of complementary DNA (cDNA) ends to amplify the 5′ and 3′ ends of PmTRAF2 and PmTRAF4 (Liang et al., 2022). The primers are presented in Table 1. DNAMAN8.0 software was used to align and splice full-length nucleotide sequences, and open reading frames (ORF) finder was employed to predict ORF. The theoretical isoelectric point and molecular weight of amino acid sequences were predicted by ExPASy-ProtParam (https://web.expasy.org/protparam/). Protein domains were carried out on the SMART online sites. After performing multiple sequence alignments with ClustalW, the phylogenetic tree was constructed by MEGAX software using the neighbor-joining method.
2.4 Quantitative real-time PCR (qPCR) assay
Total RNA was extracted from the isolated P.f. martensii tissues (mantle, gills, adductor muscle, hepatopancreas, hemolymph, and gonad) and purified using Trizol reagent (Thermo-Fisher Scientific, Waltham, MA, USA) according to the manufacturer’s instructions. The purified RNA was treated with RNase-free Dnase (Promega, Promega, Madison, WI, USA) to eliminate DNA contamination. RNA integrity was assessed via electrophoresis on a 1% agarose gel stained with ethidium bromide. The RNA concentration was determined based on the OD260/OD280 ratio using a NanoDrop 2000 spectrophotometer (Thermo-Fisher Scientific, Waltham, MA, USA). RNA aliquots (2 μg) were used to synthesize cDNA using M-MLV reverse transcriptase (Promega, Promega, Madison, WI, USA).
To explore the potential role of PmTRAF2 and PmTRAF4, we performed qPCR to detect their tissue-specific expression in all tissues and their temporal expression in the hepatopancreas after LPS stimulation. Reagents were mixed according to the instructions of the Dy NAmo Flash SYBR Green qPCR kit (ThermoFisher Scientific, Waltham, MA, USA), and fluorescence detection was performed on an Applied Biosystems 7500/7500 Fast Real-Time system. The PCR reaction steps were as follows: 95°C for 5 min; 95°C for 10 s, 60°C for 15 s, 72°C for 15 s, 40 cycles; 95°C for 10 s, 60°C for 60 s, 95°C for 1 s; 37°C for 30 s. The target gene’s relative expression was calculated via the 2-ΔΔCT method. The qPCR primers are shown in Table 1. Actin was an internal reference primer. After completing all runs, data from three replicates of each sample were analyzed using 7500 System SDS software 2.0.1 (ABI, Applied Biosystems, Inc., Waltham, MA, USA). The baseline was automatically set by the software.
2.5 The yeast two-hybrid assay
To verify whether PmTRAF2 interacts with PmTRAF3 or PmTRAF6, we performed a yeast two-hybrid experiment using the GAL4 system. The coding sequence of the PmTRAF2 gene was inserted into the pGBKT7 vector, while the coding sequences of the PmTRAF3 and PmTRAF6 genes were respectively inserted into the pGADT7 and sequenced to screen for the correct plasmid. Two plasmids (0.3 µg each) and denatured salmon sperm DNA (0.1 mg) were added to yeasts. Then, 600 µL PEG/LiAc solution was added to yeasts and put in the water bath at 30°C for 45 min. Then, 70 μL DMSO was added and put in the water bath at 42°C for 20 min and placed in ice for 2 min. Transformed yeasts were obtained after centrifuging at 700 g for 5 min at room temperature. The pGBKT7-PmTRAF2/pGADT7-PmTRAF3 and pGBKT7-PmTRAF2/pGADT7-PmTRAF6 plasmids were co-transformed into yeast AH109 as experimental groups, respectively, while PGBKT7-53/PGADT7-T was used as a positive control group, pGBKT7-lam/pGADT7-T as a negative control group, and pGBKT7-PmTRAF2/pGADT7, pGADT7-PmTRAF3/pGBKT7, and pGADT7-PmTRAF6/pGBKT7 as self-activation assay groups. The co-transformed yeast cells were dispersed on selective SD/-Leu/-Trp and SD/-Ade/-His/-Leu/-Trp plates and cultured at 30°C for 5 d. Yeasts grown on SD/-Ade/-His/-Leu/-Trp plates were transferred to new SD/-Ade/-His/-Leu/-Trp plates and cultured for 2-4 d. If the yeasts turn blue, it indicates successful protein interactions.
2.6 Data analysis
All experimental results on qPCR were expressed as a mean ± standard deviation (S.D.) and were analyzed using SPSS 19.0 (IBM, Chicago, IL, USA). Differences in gene expression levels in the tissue quantification group and nucleus insertion surgery group were determined by a one-way analysis of variance with Duncan’s Multiple Range test. Differences in gene expression levels between different groups at the same time point were analyzed by T-test. Statistical significance was defined at p < 0.05.
3 Results
3.1 Sequence analysis and characterization of PmTRAF2 and PmTRAF4
The full-length sequences of PmTRAF2 and PmTRAF4 are obtained using RACE (Figure 1). The nucleotide sequence of PmTRAF2 was 2055 bp, containing a 5’-untranslated region (UTR; 183 bp), a 3’ UTR (288 bp) with a polyA tail (27 bp), and an ORF (1584 bp), encoding 527 amino acids (Figure 1A). The predicted molecular mass and theoretical isoelectric point (pI) of PmTRAF2 were 60.46 kD and 8.06, respectively. A RING finger domain, two TRAF-type zinc finger domains, a TRAF_BIRC3_bd domain, and a MATH domain were identified in the deduced PmTRAF2 protein.

Figure 1 The full-length sequences and deduced amino acids of PmTRAF2 (A) and PmTRAF4 (B). Amino acids with double-underscore stand for the RING finger domain, light shadows for the TRAF-type zinc finger domain, light shadows with boxes for the TRAF_BIRC3_bd domain, and deep shades of gray for the MATH domain. * indicates the translation stop codon.
The nucleotide sequence of PmTRAF4 was 2365 bp, containing a 5’ UTR (127 bp), a 3’ UTR (825 bp) with a polyA tail (20 bp), and ORF (1413 bp), encoding 470 amino acids with a predicted molecular mass of 54.70 kD and theoretical isoelectric point (pI) of 7.15 (Figure 1B). The deduced amino acid sequence was composed of three TRAF-type zinc finger and a MATH domain.
3.2 Multiple sequence alignment and phylogenic relationships
PmTRAF2 was compared with TRAF2 sequences from C. gigas, Homo sapiens, Mus musculus, Oncorhynchus mykiss, and Columba livia, showing that PmTRAF2 shared the highest homology with TRAF2 from C. gigas (47.1%; Figure 2A). The comparison of PmTRAF4 with TRAF4 amino acid sequences from H. sapiens, Branchiostoma belcheri, Mytilus coruscus, Aplysia californica, and Cryptotermes secundus indicated that PmTRAF4 was highly similar to M. coruscus TRAF4 (70.4%) and A. californica TRAF4 (65.1%), and TRAF4 sequences were highly conserved (Figure 2B).
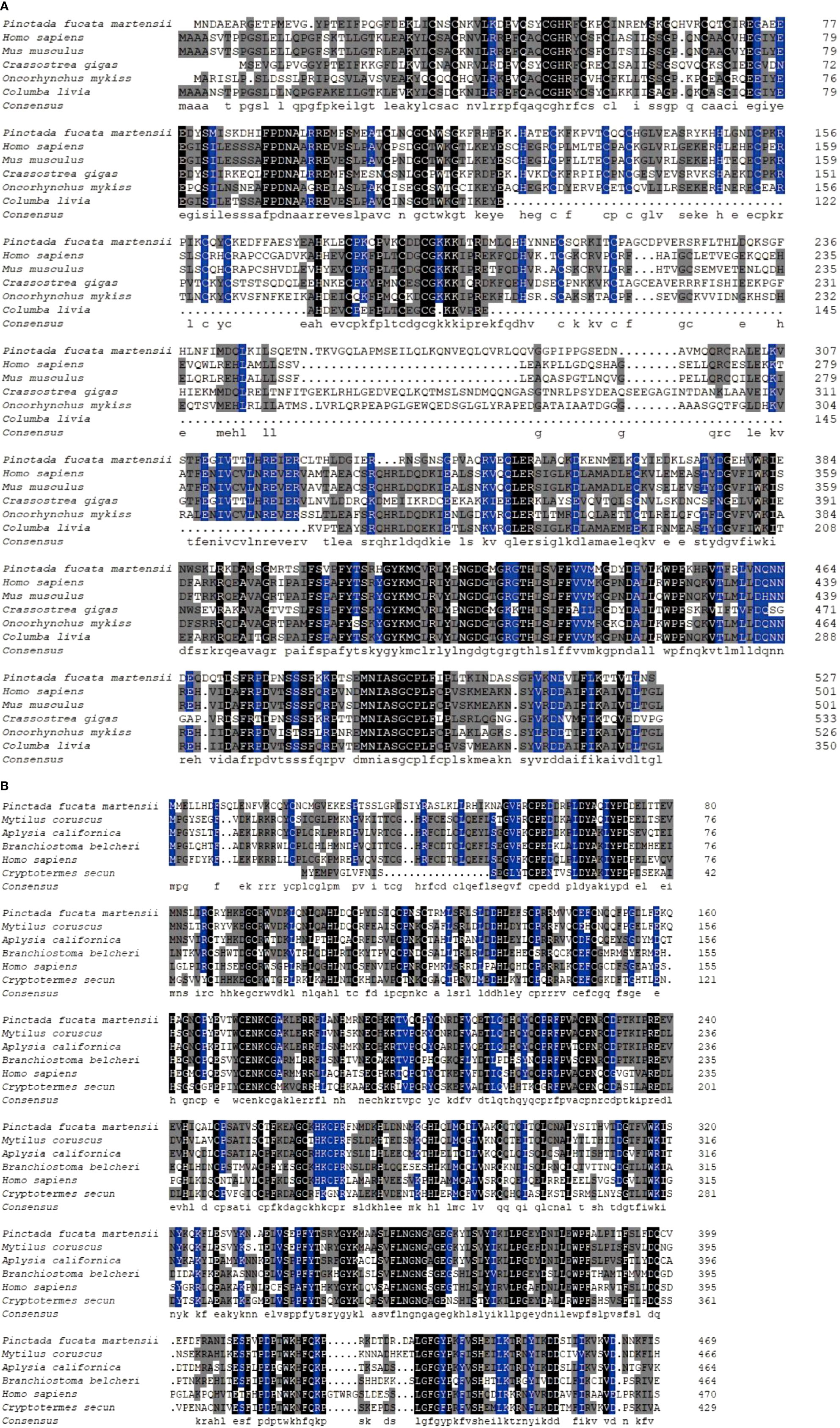
Figure 2 Alignment of PmTRAF2 (A) and PmTRAF4 (B) with their corresponding homologous amino acid sequences from other species, respectively. Identical residues are shown in black, while conserved residues in all sequences with more than 75% and 50% similarity are highlighted in blue and gray. The GenBank accession numbers of TRAF4 ortholog sequences are as follows: Mytilus coruscus (Accession no.: CAC5367166.1), Aplysia californica (Accession no.: XP_005107744.2), Branchiostoma belcheri (Accession no.: ABN04153.1), Homo sapiens (Accession no.: NP_004286.2), and Cryptotermes secundus (Accession no.: PNF16644.1). The GenBank accession numbers of TRAF2 ortholog sequences are as follows: Homo sapiens (Accession no.: NP_066961.2), Mus musculus (Accession no.: AAH03801.1), Crassostrea gigas (Accession no.: XP_034302657.1), Oncorhynchus mykiss (Accession no.: NP_001117865.1), and Columba livia (Accession no.: PKK17224.1).
Our phylogenetic analysis revealed that proteins TRAF2 and TRAF4 from vertebrates and invertebrates clustered into two major branches. All TRAF4 and TRAF2 had a highly conserved structural domain composition (Figure 3B). PmTRAF4 clustered with Mytilus edulis TRAF4 and Mytilus galloprovincialis TRAF4 (Figure 3A). Furthermore, PmTRAF2 clustered with C. gigas TRAF2, and a distinct division between vertebrate and invertebrate TRAF2 sequences was observed in the TRAF2 branch (Figure 3A).
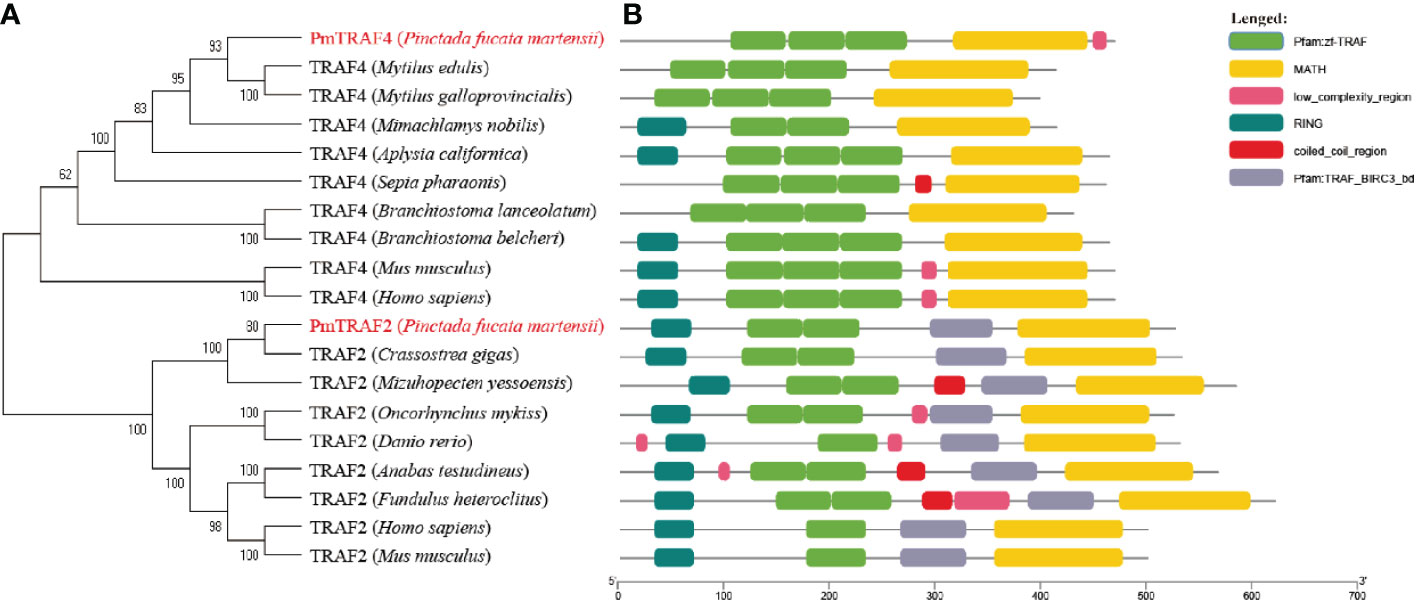
Figure 3 Phylogenic analysis (A) and protein domain information (B) of PmTRAF2 and PmTRAF4. The GenBank accession numbers of TRAF4 ortholog sequences are as follows: Mytilus edulis (Accession no.: CAG2230844.1), Mytilus galloprovincialis (Accession no.: VDI05192.1), Mimachlamys nobilis (Accession no.: QOS44542.1), Aplysia californica (Accession no.: XP_005107744.2), Sepia pharaonis (Accession no.: CAE1235462.1), Branchiostoma lanceolatum (Accession no.: CAH1232131.1), Branchiostoma belcheri (Accession no.: ABN04153.1), Homo sapiens (Accession no.: NP_004286.2), and Mus musculus (Accession no.: NP_033449.2). The GenBank accession numbers of TRAF2 ortholog sequences are as follows: Crassostrea gigas (Accession no.: XP_034302657.1), Mizuhopecten yessoensis (Accession no.: OWF39470.1), Oncorhynchus mykiss (Accession no.: NP_001117865.1), Danio rerio (Accession no.: XP_005165557.1), Anabas testudineus (Accession no.: XP_026198148.1), Fundulus heteroclitus (Accession no.: JAR83986.1), Homo sapiens (Accession no.: NP_066961.2), and Mus musculus (Accession no.: AAH03801.1).
3.3 Expression levels of PmTRAF2 and PmTRAF4
The qPCR analysis showed that PmTRAF2 and PmTRAF4 were expressed in adductor muscle, hemocytes, gill, mantle, hepatopancreas, and gonads, and both genes were significantly expressed in hepatopancreas and gill tissues (p < 0.01, Figure 4).
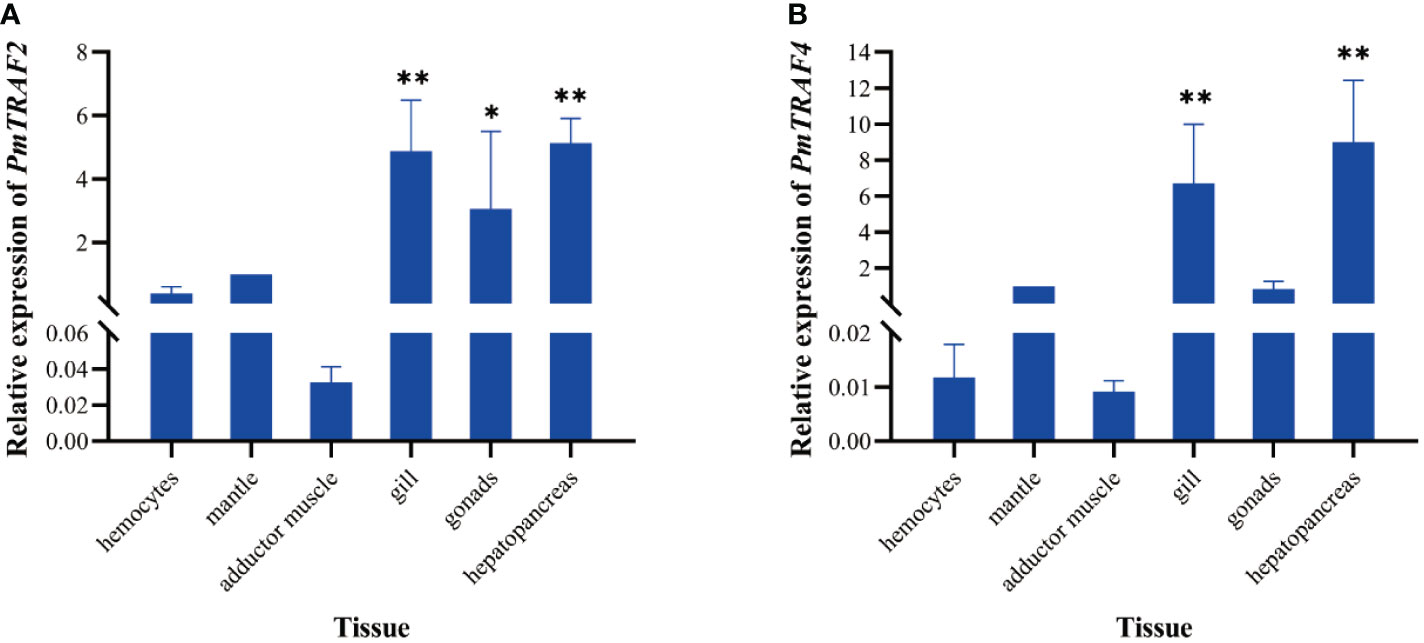
Figure 4 Expression distribution of PmTRAF2 (A) and PmTRAF4 (B) in different tissues. Error bars are expressed as the mean ± S.D. (n = 5). Asterisks denote significant differences: **p < 0.01; *p < 0.05.
3.4 Expression of PmTRAFs response to LPS stimulation, PolyI:C stimulation, and nuclear insertion surgery
After LPS stimulation, the expression of PmTRAF2 in the hepatopancreas of the LPS group was significantly up-regulated compared with the PBS group at 24 h, 48 h, and 96 h (p < 0.01); PmTRAF4 expression was significantly up-regulated at 96 h (p < 0.05); PmTRAF3 and PmTRAF6 were significantly expressed at 12, 24 and 96 h compared with the control group. Additionally, PmTRAF3 was significantly expressed at 48 h after the LPS challenge (Figure 5A).
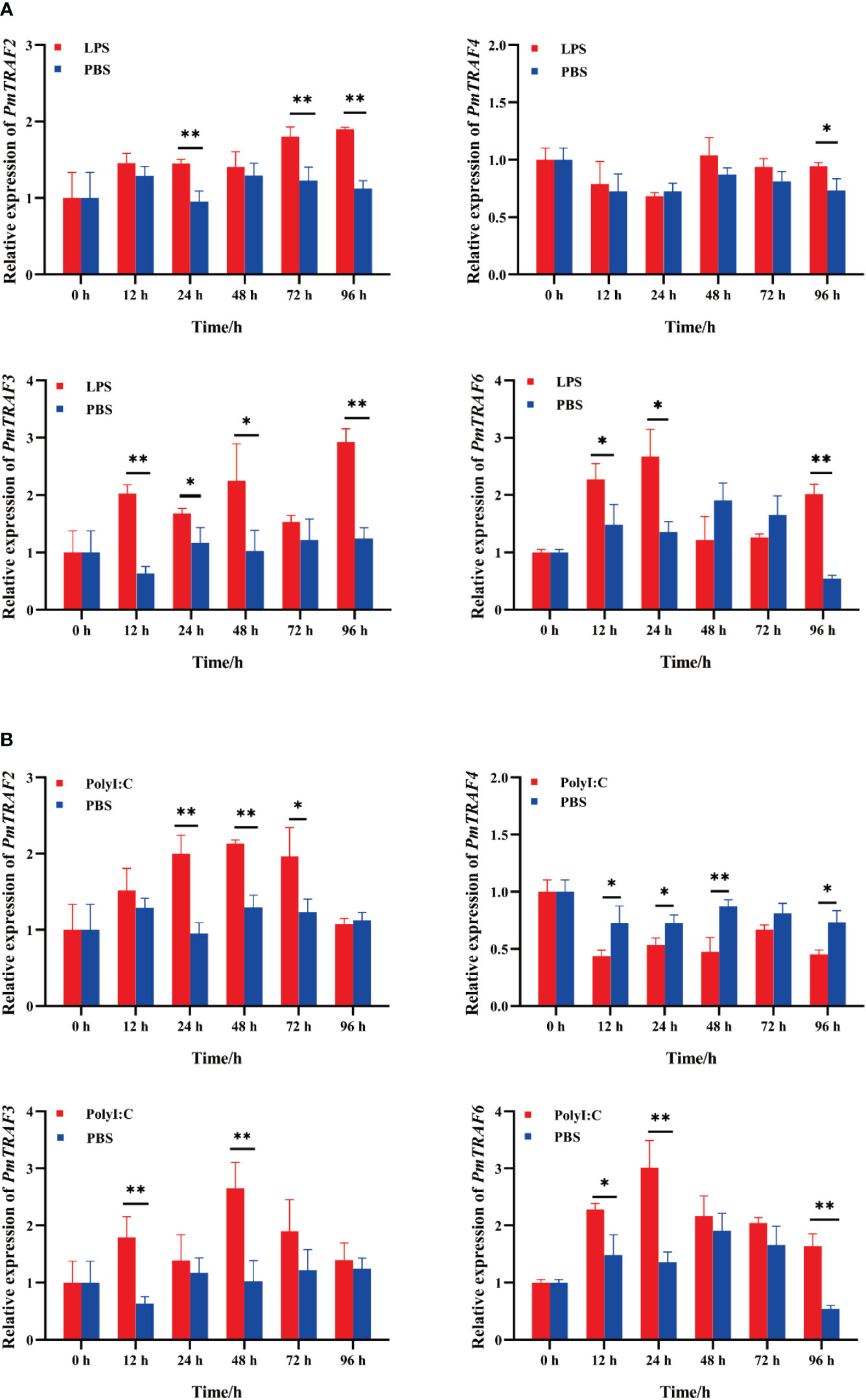
Figure 5 The mRNA expression of PmTRAFs in hepatopancreas after LPS challenge (A) and PolyI:C challenge (B). Relative expression was expressed as the mean ± S.D. (n = 5). Significant differences in relative expression levels between two groups at the same time point are denoted by asterisks: **p < 0.01; *p < 0.05.
Compared with the control group, the mRNA expression levels of PmTRAF3 and PmTRAF6 in the hepatopancreas began to increase at 6 h after PolyI:C stimulation and reached maximum expression level at 48 h and 24 h, respectively (Figure 5B). At 24, 48, and 72 h exposure to PolyI:C stimulation, PmTRAF2 mRNA expression was rapidly increased. However, PmTRAF4 mRNA was significantly decreased (Figure 5B).
In nuclear insertion surgery, the mRNA expression trends of PmTRAF2 and PmTRAF6 genes in the hepatopancreas are the same, showing decreased, then increased, and then decreased (Figure 6). PmTRAF4 mRNA reached maximum expression level at 6 h. PmTRAF3 mRNA rapidly increased at 6 h, decreased at 24 h and 48 h, and rapidly increased at 72 h and 96 h (Figure 6).
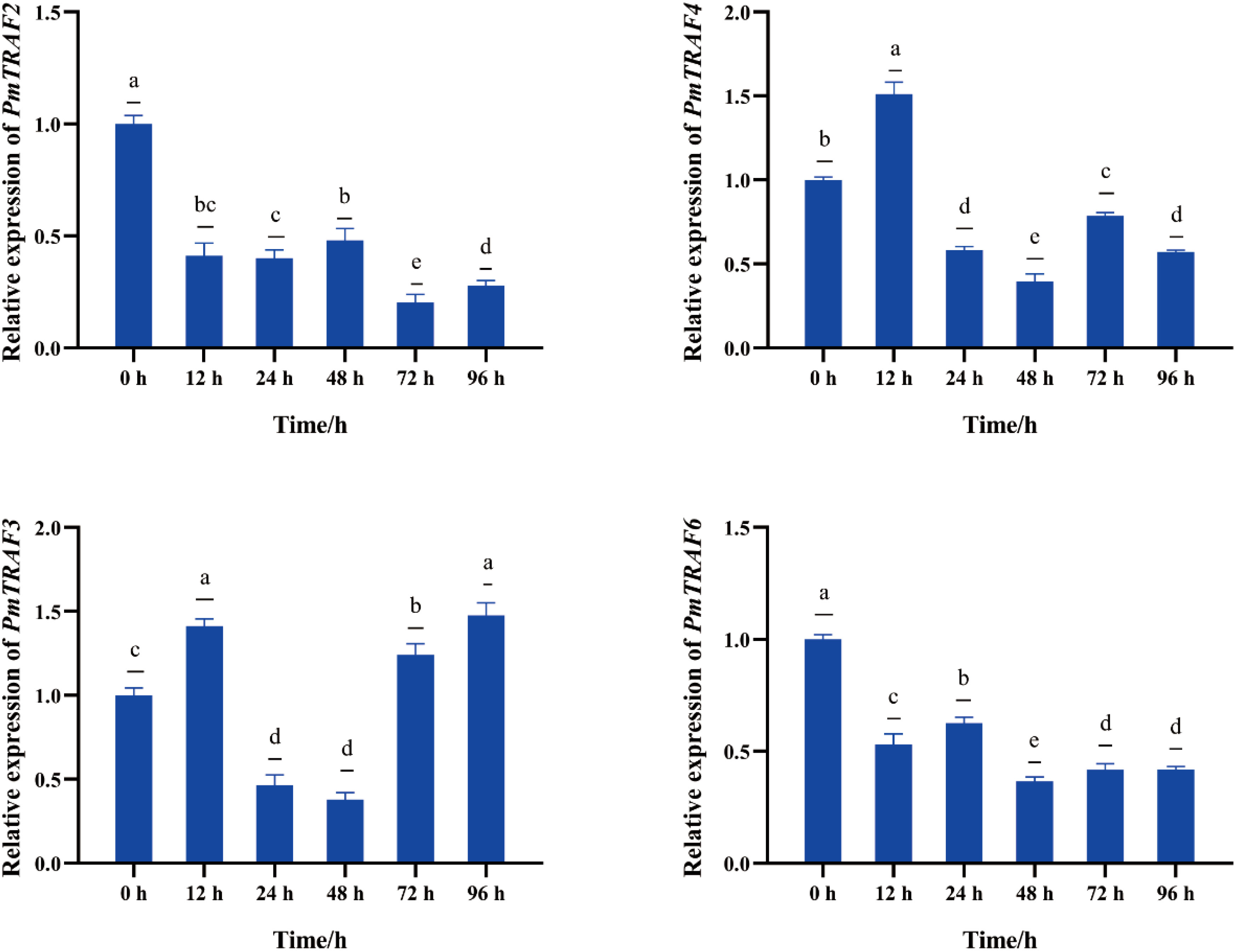
Figure 6 The mRNA expression of PmTRAFs in nuclear insertion surgery. Relative expression was expressed as the mean ± S.D. (n = 5). Different superscript letters indicate a significant difference in the groups (p < 0.05).
3.5 PmTRAF2 interacts with PmTRAF6
In this experiment, a yeast two-hybrid assay was performed to study the interaction between PmTRAF2 and PmTRAF3 or PmTRAF6. AH109 of pGBKT7-PmTRAF2/pGADT7 co-transformed growth, pGADT7-PmTRAF3/pGBKT7 co-transformed growth, and pGADT7-PmTRAF6/pGBKT7 co-transformed growth grew on the SD/-Leu/-Trp medium but not on the SD/-Leu/-Trp/-His/-Ade/x-a-gal medium, indicating that PmTRAF2, PmTRAF3, and PmTRAF6 did not activate themselves (Figure 7). The yeasts of the experimental, positive control, and negative control groups grew on SD/-Leu/-Trp medium, except that yeasts of the positive control group and experimental group pGBKT7-PmTRAF2/pGADT7-PmTRAF6 grew on the SD/-Leu/-Trp/-His/-Ade/x-a-gal medium and turned blue, while the negative control group and experimental group pGBKT7-PmTRAF2/pGADT7-PmTRAF3 did not grow, indicating an interaction between PmTRAF2 and PmTRAF6 (Figure 7).
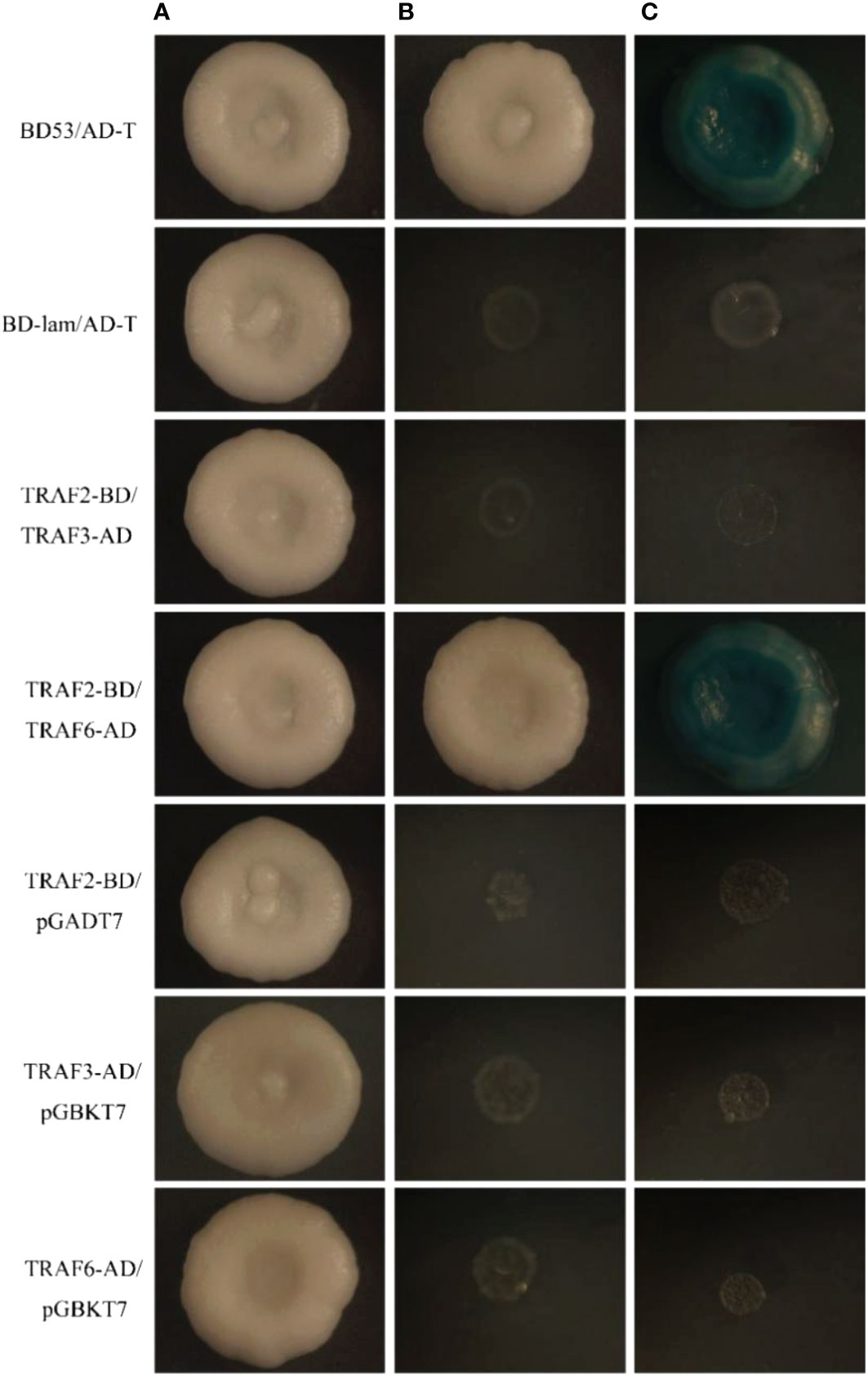
Figure 7 The yeast two-hybrid assay of PmTRAF2, PmTRAF3, and PmTRAF6. Co-transformed yeasts were cultured on SD/-Leu/-Trp medium (A) and SD/-Leu/-Trp/-His/-Ade medium (B). Protein interactions were verified with SD/-Leu/-Trp/-His/-Ade medium containing x-a-gal (C). AD represents pGADT7, and BD denotes pGBKT7. PGBKT7-53/PGADT7-T is the positive control group. pGBKT7-lam/pGADT7-T is used as the negative control group; pGBKT7-PmTRAF2/pGADT7- PmTRAF3 and pGBKT7-PmTRAF2/pGADT7-PmTRAF6 are expressed as experimental groups.
4 Discussion
Worsening coastal water quality has brought various diseases to P. f. martensii, which lacks a specific immune system. Thus, studying the role of immune-related genes in pathogen response may improve the survival of P. f. martensii (He et al., 2019b). TRAFs are major signal transducers of the TNF receptor superfamily and other receptors, which can regulate various cellular activities and innate immune responses, including cell survival, proliferation, and death (Jiao et al., 2014). In the present study, we cloned two novel TRAFs, PmTRAF2, and PmTRAF4, in P. f. martensii. The protein domain of PmTRAF2 and orthologs of other species contain the typical RING finger domain, TRAF-type zinc finger domain, TRAF_BIRC3_bd domain, and MATH domain (TRAF-C) (Figure 3B). The MATH domain is crucial for homo- and hetero-oligomerization of TRAFs and the interaction of upstream and downstream factors, while the RING finger domain is involved in the activation of downstream signaling pathways (Zapata et al., 2007; Ha et al., 2009), which helps to investigate the immune-related signaling pathways involved in PmTRAF2. The PmTRAF4 TRAF-type zinc finger domains and MATH domain and their numbers are similar to those of other mollusk and vertebrate TRAF4. However, PmTRAF4 lacks the RING finger domain, and its deletion may affect the transmission of downstream signaling proteins. The results of the phylogenetic analysis revealed that TRAF2 and TRAF4 were highly conserved across all species, and PmTRAF2 and PmTRAF4 were closer to bivalves’ homologs, which suggests that PmTRAF2 and PmTRAF4 may have similar functions to other homologs.
TRAFs genes were detected and expressed at high levels in various immune-related tissues (gill, hemocytes, and hepatopancreas) in mollusks, suggesting their involvement in the host immune response. For instance, the mRNA of Patinopecten yessoensis’s intact TRAFs (TRAF2, TRAF3, TRAF4, TRAF6, and TRAF7) were mainly abundant in gills and hemocytes (Wang et al., 2015). In C. nobilis, high expression levels of CnTRAF2, CnTRAF3, and CnTRAF4 were observed in hemocytes, whereas CnTRAF6 was highly expressed in gills (Zhang et al., 2021). C. gigas TRAF3-S and TRAF3-L transcripts were up-regulated in hemolymph and muscle relative to the gill of the internal reference sample (Huang et al., 2014). Consistent with the above results, the mRNA transcripts of PmTRAF2 and PmTRAF4 were highly expressed in hepatopancreas and gills, indicating that they operated in immune defense. TRAF2 is one of the major recruitment targets for TRADD and TNFR5 proteins, and all three genes from P. f. martensii share high expression in gills and hepatopancreas (Hsu et al., 1996; He et al., 2019a; Wu et al., 2020). It can be speculated that PmTRADD and PmTNFR5 can recruit PmTRAF2 and play a role in apoptosis and other immune responses.
To investigate the function of TRAF family (PmTRAF2, PmTRAF3, PmTRAF4 and PmTRAF6) in P.f. martensii’s immune response, their expression levels were examined in the hepatopancreas after three immune stimulation, including nuclear insertion surgery, LPS challenge, and PolyI:C challenge. Several reports have indicated that the TNF–TNFR system (TNFR, TRAF, and NF-κB family members) responded to the nuclear insertion surgery. These genes may be involved in allograft immunity in pearl oysters (Wu et al., 2020). Here, in hepatopancreas tissues, PmTRAF3 and PmTRAF4 transcripts with significant up-regulation at different times after nuclear insertion surgery, and PmTRAF2 and PmTRAF6 with down-regulation at all times, indicating that four TRAFs might participate in the immune defense at different time. The expression pattern of PmTRAF2 was similar to PmTRAF6, which suggests that they may have the same regulation in response to nuclear insertion surgery. PmTRAF3 and PmTRAF4 transcripts were rapidly increased at 6 h, which illustrates that PmTRAF3 and PmTRAF4 may quickly involve in immune response after nuclear insertion surgery. In addition, four TRAFs mRNA in hepatopancreas were decreased at a short time (24 h and 48 h). However, in a longer period of time (5 d to 30 d), PmTRAF2 transcripts in hemocytes were significantly upregulated on 5 d, 15 d, and 30 d after nuclear insertion surgery, while PmTRAF3 was significantly upregulated on 15 d (Wu et al., 2020). The difference between the two results indicates that the downregulation of PmTRAFs mRNA in the hepatopancreas may be caused by time or that different tissues have different immune responses, which requires more experiments to verify.
Pathogen-associated molecular patterns (PAMPs) are highly conserved molecular structures that are common in pathogenic microorganisms and serve as ligand receptors for pattern recognition receptor (PRR) binding, and LPS and PolyI:C were typical PRR moleculars (Shen et al., 2022). PmTRAF4 transcript was downregulated within 96 h exposure to PolyI:C. Contrary to this result, the transcript level of Larimichthys crocea TRAF4 in gill was markedly upregulated at 6 h post PolyI:C (Chen et al., 2022). In Patinopecten yessoensis, TRAF4 was upregulated in the hemocytes post-bacterial infection (Micrococcus luteus and Vibrio anguillarum) (Wang et al., 2015). The above different phenomena suggest that PmTRAF4 may have different expression patterns in innate defenses in response to pathogens. Furthermore, TRAF4 can inhibit innate immune signaling pathways, such as TRAF4 inhibits nucleotide-binding oligomerization domain 2-induced NF-κB activation (Marinis et al., 2011); TRAF4 inhibits the toll-like receptors signaling pathway by associating with p47phox and interacting with TRAF6 and toll-IL-1 receptor domain-containing adaptor-inducing IFN-β (Takeshita et al., 2005). Whether TRAF4 can inhibit the immune response to pathogens is worth exploring. PmTRAF2, PmTRAF3, and PmTRAF6 were significantly induced to express at multiple time points within 96 h under LPS and PolyI:C, which suggests that their involvement in the innate immune system may be highly sensitive to the invasion of pathogenic microorganisms. Post LPS and PolyI:C infection, the expression levels of PmTRAF3 and PmTRAF6 increased sharply at 12 h, whereas PmTRAF2 at 24 h, which may indicate that PmTRAF3 and PmTRAF6 were more sensitive to LPS and PolyI:C model pathogen invasion than PmTRAF2. However, PmTRAF2 was responsive to LPS challenge in the gill tissue of P. f. martensii, with expression significantly up-regulated at 6 h and 12 h relative to the control group, which may be caused by the ability of gills to recognize and defend against LPS model pathogens rapidly (Wu et al., 2020; Zhang et al., 2022). Our results speculate that PmTRAFs (PmTRAF2, PmTRAF3, PmTRAF4 and PmTRAF6) may be the important molecule in the host’s immune defense.
Based on our studies, PmTRAF2, PmTRAF3, and PmTRAF6 may exert a similar action at the transcript level. Therefore, these may have a coordinative effect on signaling pathways. TRAF2, TRAF3, or TRAF6 have been found to mediate CD40-induced activation of NF-κB (Lu et al., 2003). Thus studying the relationship between them is important. Furthermore, in a previous report, TRAFs members can participate in signal transduction by interacting to form homo- and hetero-dimers with distinct regulatory functions (Arron et al., 2002). The TRAF-C structural domain of TRAF3 interacts with the TRAF-N structural domain and zinc fingers 4 and 5 of TRAF2 to form multimers, which inhibits the capability of TRAF2 to induce the NF-κB activation (He et al., 2004). Using RNA interference and immunoblotting, the researchers found for the first time in nonhemopoietic cells that TRAF6 can regulate CD40 signaling by associating with TRAF2 (Davies et al., 2005). Therefore, yeast two-hybrid assays were performed to understand the relationship between TRAF2 and TRAF3, and TRAF6 in the present study, which is crucial for subsequent exploration of the downstream signaling. The yeast two-hybrid assays have revealed that PmTRAF2 interacts with PmTRAF6 while not interacting with PmTRAF3. The TRAF domain is crucial for homo- and hetero-oligomerization of TRAFs. However, TRAF6 forms a heterodimer by binding to the RING domain of TRAF2 via its RING domain (Das et al., 2021). How PmTRAF2 and PmTRAF6 form a heterodimer and their molecular mechanism for downstream signaling still need to be explored.
5 Conclusion
The novel PmTRAF2 and PmTRAF4 were identified from P. f. martensii and had highly conserved regions of amino acid sequences. PmTRAF2 and PmTRAF4 with abundant expression levels in hepatopancreas and gill. Transcripts of PmTRAF2, PmTRAF3, PmTRAF6, and PmTRAF4 were markedly changed post- LPS injection, PolyI:C injection, and nuclear insertion surgery, indicating that these genes may play a role in P. f. martensii innate immunity. PmTRAF2 interacts with PmTRAF6 instead of PmTRAF3, potentially affecting downstream immune signaling pathways, such as NF-κB signaling pathways. The above results provide new perspectives for further investigation of TRAF’s immune mechanisms in bivalves.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/supplementary material.
Author contributions
MZ: Investigation and designed the study, Analyzed all data, Writing - Original draft preparation; CS: Analyzed all sequencing data; HL: Conceptualization, Methodology, Writing- Reviewing and Editing; YW: Investigation, Analyzed all sequencing data; BL: Prepared the samples, Conclusion. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the grants from the National Natural Science Fundation of China (No. 31472306), Guangdong Natural Science Fundation of China (No. 2021A1515010962), Science and technology Special Fund of Guangdong Province (2021A05250), Special Fund for Harbor Construction and Fishery Industry Development of Guangdong Province (No. A201608B15) and Innovation Team Project from the department of Education of Guangdong Province (Grant No. 2021KCXTD026).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Arch R. H., Gedrich R. W., Thompson C. B. (1998). Tumor necrosis factor receptor-associated factors (TRAFs)–a family of adapter proteins that regulates life and death. Genes Dev. 12 (18), 2821–2830. doi: 10.1101/gad.12.18.2821
Arron J. R., Walsh M. C., Choi Y. (2002). TRAF-mediated TNFR-family signaling. Curr. Protoc. Immunol. 51 (1), 11.19 D. 11–11.19 D. 14. doi: 10.1002/0471142735.im1109ds51
Bishop G. A. (2004). The multifaceted roles of TRAFs in the regulation of b-cell function. Nat. Rev. Immunol. 4 (10), 775–786. doi: 10.1038/nri1462
Chen Y., Li Y., Li P. T., Luo Z. H., Zhang Z. P., Wang Y. L., et al. (2022). Novel findings in teleost TRAF4, a protein acts as an enhancer in TRIF and TRAF6 mediated antiviral and inflammatory signaling. Front. Immunol. 13. doi: 10.3389/fimmu.2022.944528
Das A., Middleton A. J., Padala P., Ledgerwood E. C., Mace P. D., Day C. L. (2021). The structure and ubiquitin binding properties of TRAF RING heterodimers. J. Mol. Biol. 433 (8), 166844. doi: 10.1016/j.jmb.2021.166844
Davies C. C., Mak T. W., Young L. S., Eliopoulos A. G. (2005). TRAF6 is required for TRAF2-dependent CD40 signal transduction in nonhemopoietic cells. Mol. Cell. Biol. 25 (22), 9806–9819. doi: 10.1128/MCB.25.22.9806-9819.2005
Evans S., Tzeng H.-P., Veis D. J., Matkovich S., Weinheimer C., Kovacs A., et al. (2018). TNF receptor–activated factor 2 mediates cardiac protection through noncanonical NF-κB signaling. JCI Insight 3 (3), e98278. doi: 10.1172/jci.insight.98278
Foight G. W., Keating A. E. (2016). Comparison of the peptide binding preferences of three closely related TRAF paralogs: TRAF2, TRAF3, and TRAF5. Protein Sci. 25 (7), 1273–1289. doi: 10.1002/pro.2881
Guo Y., Xu Y., Xiong D., Zhou Y., Kang X., Meng C., et al. (2020). Molecular characterisation, expression and functional feature of TRAF6 in the king pigeon (Columba livia). Innate Immun. 26 (6), 490–504. doi: 10.1177/1753425920920930
Ha H., Han D., Choi Y. (2009). TRAF-mediated TNFR-family signaling. Curr. Protoc. Immunol. 87 (1), 11.19 D. 11–11.19 D. 19. doi: 10.1002/0471142735.im1109ds87
Han S., Yoon K., Lee K., Kim K., Jang H., Lee N. K., et al. (2003). TNF-related weak inducer of apoptosis receptor, a TNF receptor superfamily member, activates NF-κB through TNF receptor-associated factors. Biochem. Biophys. Res. Commun. 305 (4), 789–796. doi: 10.1016/S0006-291X(03)00852-0
Hauer J., Püschner S., Ramakrishnan P., Simon U., Bongers M., Federle C., et al. (2005). TNF receptor (TNFR)-associated factor (TRAF) 3 serves as an inhibitor of TRAF2/5-mediated activation of the noncanonical NF-κB pathway by TRAF-binding TNFRs. Proc. Natl. Acad. Sci. 102 (8), 2874–2879. doi: 10.1073/pnas.0500187102
He L., Grammer A. C., Wu X., Lipsky P. E. (2004). TRAF3 forms heterotrimers with TRAF2 and modulates its ability to mediate NF-κB activation. J. Biol. Chem. 279 (53), 55855–55865. doi: 10.1074/jbc.M407284200
He J., Liang H., Chen S., Fang X., Huang X. (2019a). Gene cloning and tissue expression analysis of TRADD from Pinctada martensii. J. Guangdong Ocean Univ. 39 (4), 13–19. doi: 10.3969/j.issn.1673-9159.2019.04.003
He J., Liang H., Zhu J., Fang X. (2019b). Separation, identification and gene expression analysis of PmAMP-1 from Pinctada fucata martensii. Fish Shellfish Immunol 92, 728–735. doi: 10.1016/j.fsi.2019.07.002
He C., Wang Y., Liu W., Gao X., Chen P., Li Y., et al. (2013). Cloning, promoter analysis and expression of the tumor necrosis factor receptor-associated factor 6 (TRAF6) in Japanese scallop (Mizuhopecten yessoensis). Mol. Biol. Rep. 40 (8), 4769–4779. doi: 10.1007/s11033-013-2573-8
Hsu H., Shu H. B., Pan M. G., Goeddel D. V. (1996). TRADD–TRAF2 and TRADD–FADD interactions define two distinct TNF receptor 1 signal transduction pathways. Cell 84 (2), 299–308. doi: 10.1016/S0092-8674(00)80984-8
Huang D., Bai Z., Shen J., Zhao L., Li J. (2018). Identification of tumor necrosis factor receptor-associated factor 6 in the pearl mussel Hyriopsis cumingii and its involvement in innate immunity and pearl sac formation. Fish Shellfish Immunol. 80, 335–347. doi: 10.1016/j.fsi.2018.06.035
Huang X. D., Liu W. G., Guan Y. Y., Shi Y., Wang Q., Zhao M., et al. (2012). Molecular cloning, characterization and expression analysis of tumor necrosis factor receptor-associated factor 3 (TRAF3) from pearl oyster Pinctada fucata. Fish Shellfish Immunol. 33 (3), 652–658. doi: 10.1016/j.fsi.2012.06.026
Huang B., Zhang L., Du Y., Li L., Qu T., Meng J., et al. (2014). Alternative splicing and immune response of Crassostrea gigas tumor necrosis factor receptor-associated factor 3. Mol. Biol. Rep. 41 (10), 6481–6491. doi: 10.1007/s11033-014-3531-9
Huang B., Zhang L., Du Y., Li L., Tang X., Zhang G. (2016). Molecular characterization and functional analysis of tumor necrosis factor receptor-associated factor 2 in the pacific oyster. Fish Shellfish Immunol. 48, 12–19. doi: 10.1016/j.fsi.2015.11.027
Jiao Y., Tian Q., Du X., Wang Q., Huang R., Deng Y., et al. (2014). Molecular characterization of tumor necrosis factor receptor-associated factor 6 (TRAF6) in pearl oyster Pinctada martensii. Genet. Mol. Res. 13 (4), 10545–10555. doi: 10.4238/2014.December.12.17
Lalani A. I., Zhu S., Gokhale S., Jin J., Xie P. (2018). TRAF molecules in inflammation and inflammatory diseases. Curr. Pharmacol. Rep. 4 (1), 64–90. doi: 10.1007/s40495-017-0117-y
Lei Q., Cui S., Liang H., Deng Y. (2016). Clone and expression analysis of TRAF7 (tumor necrosis factor receptor-associated factor 7) gene cDNA from Pinctada martensii. Genomics Appl. Biol. 35 (1), 79–86. doi: 10.13417/j.gab.035.000079
Liang H., Zhang M., Shen C., He J., Lu J., Guo Z. (2022). Cloning and functional analysis of a trypsin-like serine protease from Pinctada fucata martensii. Fish Shellfish Immunol. 126, 327–335. doi: 10.1016/j.fsi.2022.05.058
Lu L. F., Cook W. J., Lin L. L., Noelle R. J. (2003). CD40 signaling through a newly identified tumor necrosis factor receptor-associated factor 2 (TRAF2) binding site. J. Biol. Chem. 278 (46), 45414–45418. doi: 10.1074/jbc.M309601200
Mao F., Li J., Zhang Y., Xiang Z., Zhang Y., Yu Z. (2017). Molecular cloning and functional analysis of tumor necrosis factor receptor-associated factor 6 (TRAF6) in Crossastrea gigas. Fish Shellfish Immunol. 68, 37–45. doi: 10.1016/j.fsi.2017.06.049
Marinis J. M., Homer C. R., McDonald C., Abbott D. W. (2011). A novel motif in the crohn's disease susceptibility protein, NOD2, allows TRAF4 to down-regulate innate immune responses. J. Biol. Chem. 286 (3), 1938–1950. doi: 10.1074/jbc.M110.189308
Shen C., Zhang M., Liang H., He J., Zhang B., Liang B. (2022). Gene cloning and functional study of PmKSPI from Pinctada fucata martensii. Fish Shellfish Immunol 131, 1157–1165. doi: 10.1016/j.fsi.2022.11.021
Takeshita F., Ishii K. J., Kobiyama K., Kojima Y., Coban C., Sasaki S., et al. (2005). TRAF4 acts as a silencer in TLR-mediated signaling through the association with TRAF6 and TRIF. Eur. J. Immunol. 35 (8), 2477–2485. doi: 10.1002/eji.200526151
Wang A., Wang J., Ren H., Yang F., Sun L., Diao K., et al. (2014). TRAF4 participates in wnt/β-catenin signaling in breast cancer by upregulating β-catenin and mediating its translocation to the nucleus. Mol. Cell. Biochem. 395 (1), 211–219. doi: 10.1007/s11010-014-2127-y
Wang J., Wang R., Wang S., Zhang M., Ma X., Liu P., et al. (2015). Genome-wide identification and characterization of TRAF genes in the yesso scallop (Patinopecten yessoensis) and their distinct expression patterns in response to bacterial challenge. Fish Shellfish Immunol. 47 (1), 545–555. doi: 10.1016/j.fsi.2015.09.050
Wang W., Wu Y., Lei Q., Liang H., Deng Y. (2017). Deep transcriptome profiling sheds light on key players in nucleus implantation induced immune response in the pearl oyster Pinctada martensii. Fish Shellfish Immunol. 69, 67–77. doi: 10.1016/j.fsi.2017.08.011
Wu Y., He J., Yao G., Liang H., Huang X. (2020). Molecular cloning, characterization, and expression of two TNFRs from the pearl oyster Pinctada fucata martensii. Fish Shellfish Immunol 98,147–159. doi: 10.1016/j.fsi.2020.01.010
Xie P., Hostager B. S., Munroe M. E., Moore C. R., Bishop G. A. (2006). Cooperation between TNF receptor-associated factors 1 and 2 in CD40 signaling. J. Immunol. 176 (9), 5388–5400. doi: 10.4049/jimmunol.176.9.5388
Xu Y. C., Wu R. F., Gu Y., Yang Y.-S., Yang M.-C., Nwariaku F. E., et al. (2002). Involvement of TRAF4 in oxidative activation of c-jun n-terminal kinase. J. Biol. Chem. 277 (31), 28051–28057. doi: 10.1074/jbc.M202665200
Zapata J. M., Martínez-García V., Lefebvre S. (2007). Phylogeny of the TRAF/MATH domain. Adv. Exp. Med. Biol. 597, 1–24. doi: 10.1007/978-0-387-70630-6_1
Zhang M., Lu J., Liang H., Zhang B., Liang B., Zou H. (2022). The succinylome of Pinctada fucata martensii implicates lysine succinylation in the allograft-induced stress response. Fish Shellfish Immunol. 127, 585–593. doi: 10.1016/j.fsi.2022.07.009
Keywords: Pinctada fucata martensii, PmTRAF2, PmTRAF4, innate immunity, yeast two-hybrid assay
Citation: Zhang M, Shen C, Liang H, Wu Y and Liang B (2023) Molecular cloning and function of two tumor necrosis factor receptor-associated factors genes (TRAF2 and TRAF4) from Pinctada fucata martensii. Front. Mar. Sci. 9:1082975. doi: 10.3389/fmars.2022.1082975
Received: 01 November 2022; Accepted: 27 December 2022;
Published: 13 January 2023.
Edited by:
Yang Jin, Carnegie Institution for Science, United StatesReviewed by:
Thomas Harvey, Norwegian University of Life Sciences, NorwayChunsheng Liu, Hainan University, China
Copyright © 2023 Zhang, Shen, Liang, Wu and Liang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Haiying Liang, empsaWFuZ2h5QDEyNi5jb20=
 Meizhen Zhang1
Meizhen Zhang1 Chenghao Shen
Chenghao Shen Haiying Liang
Haiying Liang Bidan Liang
Bidan Liang