- 1Department of Neurosurgery, The First Affiliated Hospital of Soochow University, Suzhou, China
- 2Department of Neurosurgery, The Affiliated Xuzhou Municipal Hospital of Xuzhou Medical University, Xuzhou, China
- 3Department of Neurosurgery, The Second Affiliated Hospital of Guilin Medical University, Guangxi, China
- 4Molecular and Experimental Surgery, Clinic for General-, Visceral -, Vascular- and Transplantation Surgery, Medical Faculty and University Hospital Magdeburg, Otto-von-Guericke University, Magdeburg, Germany
- 5Nantong University, Nantong, China
- 6Chifeng Municipal Hospital, Chifeng, China
Background: The tumor microenvironment plays a crucial role in the progression of both glioma and glioma-induced autoimmune encephalitis. However, there remains a significant lack of effective therapeutic targets for these diseases.
Method: We collected 54 CT images of glioma patients and 54 glioma-induced autoimmune encephalitis patients, respectively. Radiomics features were extracted from tumors and encephalitis regions using Python, followed by dimensionality reduction via random forest and lasso regression, and construction of radiomics-based risk scores. Genomic data matched with clinical information were analyzed to identify key prognostic genes significantly associated with risk scores. Gene expression was validated by immunohistochemistry using our clinical samples. Immune infiltration was evaluated using five algorithms (MCP-counter, EPIC, TIMER, QUANT and IPS). The association between hub genes and immune checkpoint markers as well as immunoregulation-related genes was also analyzed using Spearman correlation.
Results: We identified 980 radiomics features both in glioma and encephalitis patient images and selected four key features through lasso regression to build a radiomics-based risk score. COL22A1 was strongly correlated with the risk score and identified as the hub prognostic gene. COL22A1 expression was higher in glioblastoma tissues and cell lines, and correlated with clinical factors such as higher age, WHO grade, and IDH mutation status. Immune infiltration analysis indicated associations with diverse immune and stromal cell populations, including CD8+T cells, macrophages, and CAFs. COL22A1 was also positively correlated with immune checkpoints and immune-regulated genes.
Conclusion: Our study highlights the critical role of COL22A1 in gliomas and glioma-Induced Autoimmune Encephalitis, demonstrating its strong association with poor prognosis and its significant involvement in tumor immune regulation.
Introduction
Glioblastoma (GBM) represents the most common and severe type of malignant primary brain tumor, making up roughly 30% of all primary brain tumors (1). Glioma-Induced Autoimmune Encephalitis is a Common Paraneoplastic Syndrome of GBM. This disease predominantly affects the elderly and shows a higher prevalence in men compared to women, with annual incidence rates ranging between 0.59 and 5 cases per 100,000 individuals (2, 3). However, There remains a significant lack of effective therapeutic targets for these diseases.
Gliomas are classified into four grades based on histopathology, from low-grade (I and II) to high-grade (III and IV, with IV being GBM), reflecting their levels of malignancy (4). Recent advancements have introduced genetic criteria such as TERT promoter mutations, EGFR amplification, and chromosomal changes for a more precise diagnosis and tailored treatment of glioblastoma IDH-wildtype (5). Despite advances in understanding its molecular biology and pathogenesis, GBM remains extremely difficult to manage, with nearly inevitable tumor recurrence. Patients typically have a median survival of approximately 15 months and a five-year survival rate of 6.8%, underscoring the urgent need for new treatment approaches for GBM patients (1, 6). The primary treatment modalities for GBM include surgical resection, chemotherapy, and radiation therapy. Advances in surgical resection, such as frameless stereotaxy and brain mapping, have enhanced the precision of tumor removal, significantly influencing survival rates (7, 8). Standard post-surgical treatments, including temozolomide and radiation therapy, target residual cancer cells, extending progression-free survival but not fully eradicating the disease (6, 9). Improved radiation techniques now more accurately focus on cancer cells while protecting healthy tissue, addressing the challenges of hypoxic tumor environments to enhance treatment efficacy (10). Recent progress in comprehending the molecular and genetic underpinnings of GBM has spurred the creation of novel treatments targeting specific genetic alterations and signaling cascades. For instance, immunotherapy has shown potential as a treatment option, though overall efficacy remains moderate in GBM clinical trials, with certain patients achieving extended survival following therapy (11). The ReACT Phase II trial demonstrates that rindopepimut, combined with bevacizumab, significantly enhances survival and induces a strong immune response against EGFRvIII in patients with relapsed GBM, showing promising results in both progression-free and OS rates (12). In addition, another study reveals that the recombinant poliovirus/rhinovirus chimera PVSRIPO acts as a powerful intratumoral immune adjuvant in GBM, enhancing antitumor immunity by promoting dendritic cell and T cell infiltration and inducing specific cytotoxic T lymphocyte responses against tumor antigens (13). However, GBM employs a variety of mechanisms to evade immune detection and suppression, including an intact blood-brain barrier (BBB) that restricts immune cell entry, a tumor microenvironment (TME) that suppresses immune responses, and the manipulation of immune checkpoints and other key pathways (14–16). As a result, phase III clinical trials involving immune checkpoint inhibitors (ICIs) and vaccine therapies for GBM have yielded disappointing outcomes (17).
Autoimmune encephalitis (AE), characterized by brain inflammation due to an aberrant immune response targeting self-antigens in the central nervous system, represents a diverse group of disorders (18). While infections are the most common triggers, recent scholarly reports suggest that gliomas may also induce autoimmune encephalitis through mechanisms such as altered immune system functions and disrupted immune surveillance (19). The presence of gliomas might lead to an increased production of autoantibodies against brain antigens, potentially triggering this autoimmune condition. This underscores the intricate relationship between tumor-related mechanisms and immune system dysfunction, highlighting the critical need for meticulous clinical evaluation and monitoring in glioma patients. Therefore, elucidating the immune complexities of GBM and identifying its immunosuppressive factors are key to enhancing immunotherapy. Similarly, recognizing what triggers gliomas to cause autoimmune encephalitis is vital for improving diagnostic precision and refining treatment approaches, ultimately minimizing misdiagnosis and delays in therapy.
In this study, we identified COL22A1 as a novel shared biomarker in glioblastoma (GBM) and its induced autoimmune encephalitis (AE) through radiogenomics. Building on this foundation, we further explored the immunological characteristics of this gene, providing valuable insights for the precise diagnosis and treatment of both diseases.
Materials and methods
Data obtain and prepared
We collected 54 CT images of glioma patients and 54 Glioma-Induced Autoimmune Encephalitis patients from our hospital. All images were normalized and low-quality images were removed. The genomic information of the patients was obtained by matching clinical information from the corresponding public databases. In addition, clinical tissue samples and baseline data were collected for subsequent validation of core gene expression.
Radiomics feature extra and related riskscore
We used 3D slice to first label the DICOM-formatted glioma images and Glioma-Induced Autoimmune Encephalitis images. In the next step, we used Python to perform feature extraction for both groups and used Random Forest to perform dimensionality reduction analysis on the common features. Further, we performed further feature selection on the results extracted by Random Forest, where we used lasso regression. Finally, we built Radiomics related Riskscore based on the regression coefficients and feature expressions from lasso regression.
Identify hub prognosistic gene associated with riskscore
We performed batch survival analysis on the matched genomic data and defined genes as significant for influencing the prognosis of gliomas if HR was greater than 3 and p less than 0.05. Subsequently, we correlated these screened genes with riskscore by Spearman method and identified novel and strongly correlated candidate genes.
Expression validate and survival analysis of hub gene
We first verified the expression of the gene in tissue samples from public database. Then we download cell line expression data of this gene from The Cancer Cell Line Encyclopedia(CCLE) database, ggplot2 was used for visualization. Subsequently, we collected 20 gliomas as well as paired paraneoplastic tissue samples from the hospital’s brain center and performed immunohistochemical analyses, and the experimental steps were performed in strict accordance with standardized procedures. Further, we performed a correlation analysis with the clinical data based on the differences in the expression of the gene at the protein level in the tumor samples. Finally, we evaluated the effect of the gene on the survival events of the patients, and the outcome indicators were Overall Survival(OS), Disease-Free Survival(DFS), and Progression-Free Survival(PFS), respectively. In addition, we likewise analyzed the correlation of core genes with patient clinical information in public databases for clinical variables including age, gender, race, WHO classification, IDH status, and 1q/19q codeletion.
Enrichment analysis of hub gene
To explore the potential biological mechanism of this gene, we performed single-gene GSEA analysis. We first categorized the samples into high and low groups by the median expression value, followed by single-gene GSEA analysis using the R package cluster, where an absolute NES value greater than 1 was considered a meaningful enrichment result. Subsequently, we used the ggplot2 package to visualize the enrichment results of interest.
Immune infiltration analysis of hub gene
To explore the relationship between the hub gene and immune cell infiltration within the tumor microenvironment, we conducted a comprehensive immune infiltration analysis using five distinct algorithms, they are Microenvironment Cell Populations counter(MCP-counter), Estimating the Proportions of Immune and Cancer cells(EPIC), Tumor Immune Estimation Resource(TIMER), Quantify the immune cell types(QUANT), and Immunophenoscore(IPS). These algorithms were selected due to their complementary strengths in quantifying immune cell proportions and activity across diverse datasets. Gene expression data for the hub gene were extracted from TCGA datasets, and normalized as appropriate for each algorithm. Statistical analyses were conducted to assess the correlation between hub gene expression and immune infiltration levels, using Spearman correlation. These analyses provided a multi-faceted view of immune infiltration and its association with the hub gene, offering insights into its potential role in immune regulation within the tumor microenvironment.
Immune check points and immune regulate factor with hub genes
To investigate the association between the hub gene and immune checkpoint molecules, we analyzed the correlation between hub gene expression and key immune checkpoint markers, including BTLA, CD274, CD96, PDCD1LG2, ICOS, IDO1, CD86, LAG3, and PDCD1. Gene expression data were obtained from TCGA public database, and normalized prior to analysis. Spearman correlation analyses were conducted to determine the strength and direction of the association between the hub gene and immune checkpoint molecules. Additionally, we visualized these relationships using scatter plots, generated in ggplot2, to better understand the interaction between the hub gene and immune checkpoints.
Results
Radiomics selection and risksocre build
The identification process of core genes is shown in Figure 1A. From all patients image, we collect 980 radiomics features and after conduct random forest a total of 131 radiomics features were selected. We show the top 5 importance radiomics features in Figure 1B. Further, our study found that lasso regression analysis of the above results suggested that a total of 4 Radiomics features were selected (Figures 1C, D). We then performed risk score calculation based on the regression coefficients and feature expression.
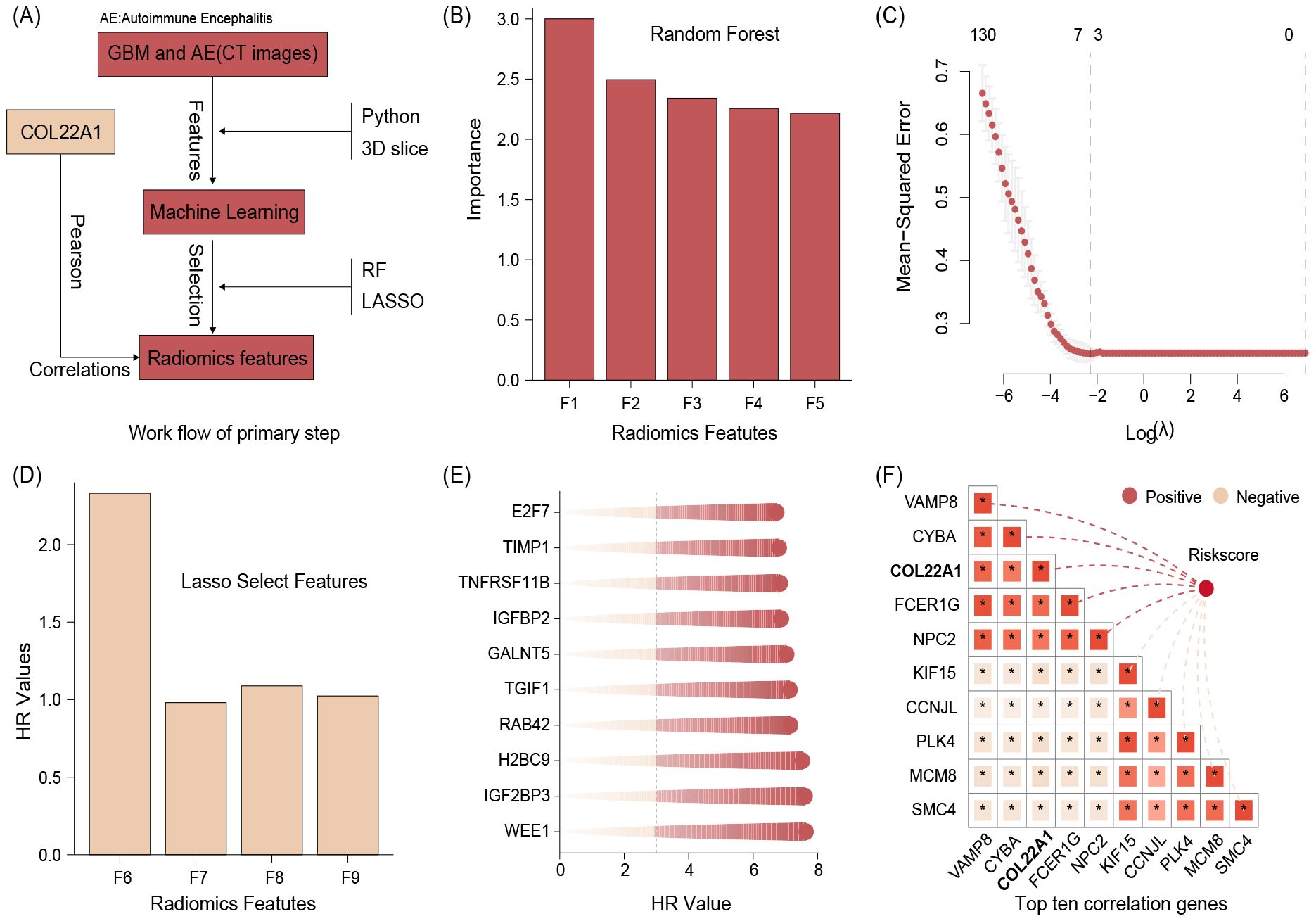
Figure 1. The identification process of core genes (A),The top 5 importance radiomics features (B). Lasso regression results suggested that a total of 4 Radiomics features were selected (C, D). We then performed risk score calculation based on the regression coefficients and feature expression. The top 10 glioma prognostic genes (E). The correlation between prognostic genes and riskscore, and the results suggested that the top 10 genes were VAMP8, CYBA, COL22A1, FCER1G, NPC2, KIF15, CCNJL, PLK4, MCM8, and SMC4 (F).
COL22A1 was positive with radiomics related risksocre
On the other hand, we screened a total of 2113 glioma prognostic genes with HR greater than 3 and p less than 0.05 by batch survival analysis and showed the top 10 gene names in Figure 1E. Finally, we performed correlation analysis between the above genes and riskscore, and the results suggested that the top 10 genes were VAMP8, CYBA, COL22A1, FCER1G, NPC2, KIF15, CCNJL, PLK4, MCM8, and SMC4 (Figure 1F). Synthesizing the results of the literature review and correlation analysis, we finally identified COL22A1 as a follow-up study.
COL22A1 was high expression in GBM and negative with survival rate
The transcriptome data analysis results validation that COL22A1 was high expression in GBM tissues while compared with normal tissues (Figure 2A). And the Cell lines results show that this gene expression high in SNU626 cell lines and low expression in SW1783 cell line (Figure 2B). Our center IHC results also demonstrate that this gene high expression in GBM tissues(Figure 2C). We further analyzed samples with high and low tumors suggested by immunohistochemical results. The results suggested that patients with high expression had greater age and were more male, more obese patients, as well as high WHO grade (Figure 2D). In addition, the survival analysis show that high expression of COL22A1 was negative with survival rate, no matter OS,DFS or PFI (Figures 2E-G).
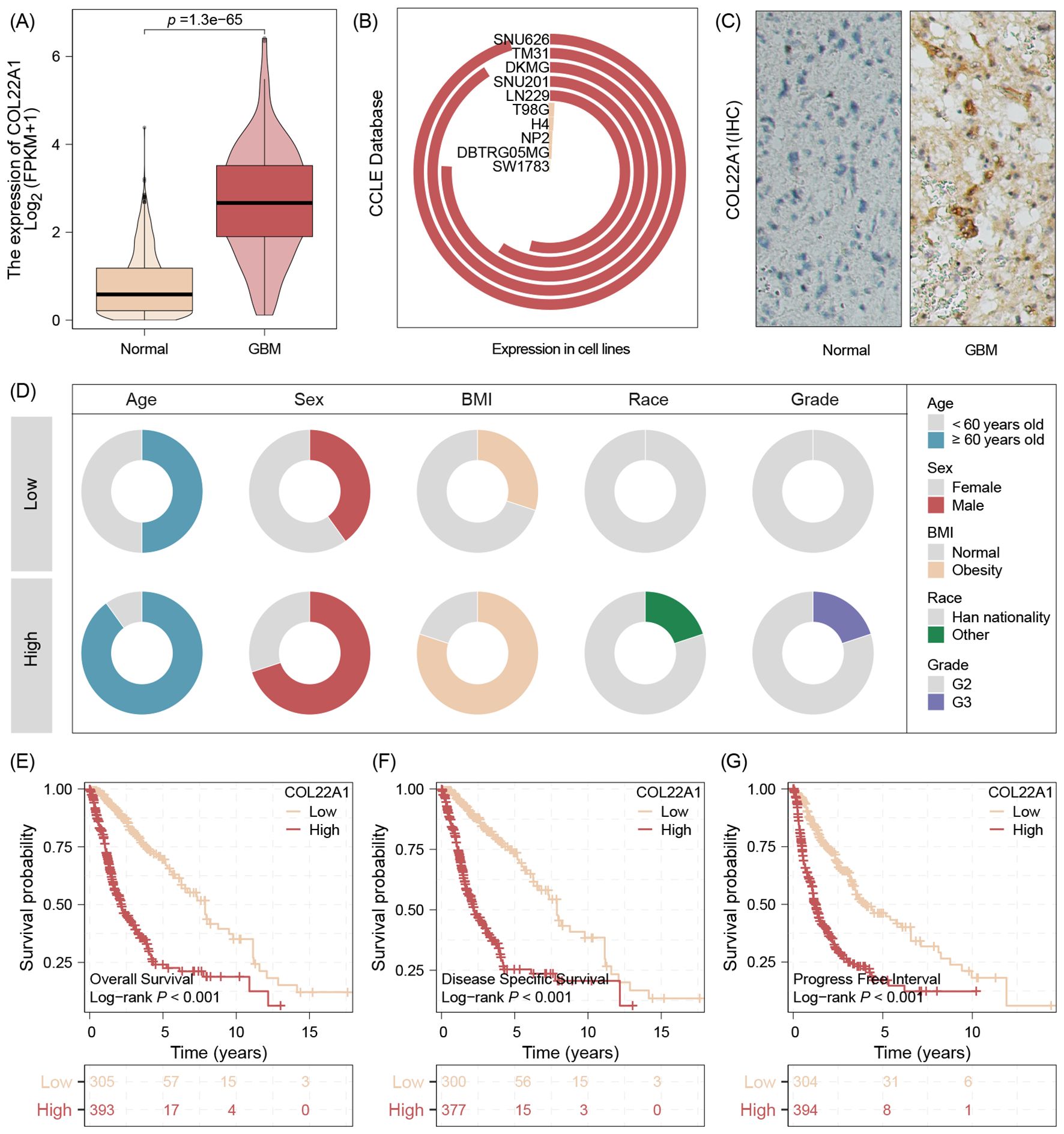
Figure 2. The transcriptome data analysis results validation that COL22A1 was high expression in GBM tissues while compared with normal tissues (A). And the Cell lines results show that this gene expression high in SNU626 cell lines and low expression in SW1783 cell line (B). The IHC results also demonstrate that this gene high expression in GBM tissues (C). Patients with high expression had greater age and were more male, more obese patients, as well as high WHO grade (D). The survival analysis show that high expression of COL22A1 was negative with survival rate, no matter OS,DFS or PFI (E-G).
COL22A1 was associated with clinical factors
The results of the correlation analysis suggest that patients with high expression of the gene usually imply a higher age (Figure 3A). However, there was no difference in the expression of the gene by sex or race (Figures 3B, C). Interestingly, we observed a similar gradual increase in the expression of this gene as the WHO Grade increased (Figure 3D). In addition, we found that the expression of this gene was elevated in the WT and 1p/19q codeletion groups of IDH and was statistically different between the groups (Figures 3E, F).
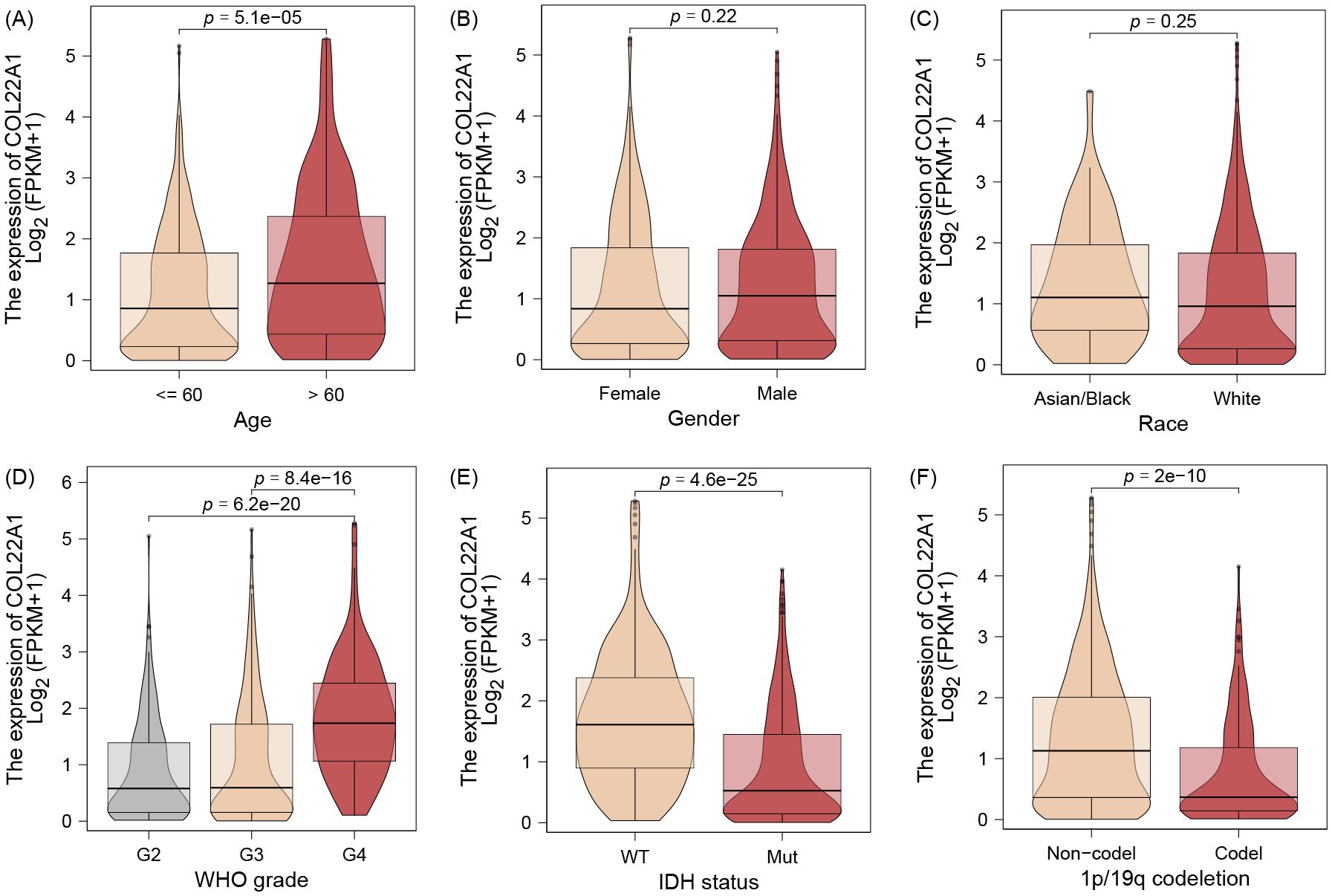
Figure 3. The results of the correlation analysis suggest that patients with high expression of the gene usually imply a higher age (A). However, there was no difference in the expression of the gene by sex or race (B, C). Interestingly, we observed a similar gradual increase in the expression of this gene as the WHO Grade increased (D). In addition, we found that the expression of this gene was elevated in the WT and 1p/19q codeletion groups of IDH and was statistically different between the groups (E, F).
COL22A1 was involved in multi-immune pathway and classical pathway
The GSEA analysis results show that, this gene could be enrichment in multi-immune pathway, such as T cell receptor signaling pathway (Figure 4A), intestinal immune network for lga production (Figure 4B), and Natural killer cell mediated cytotoxicity (Figure 4C). In addition, the enrichment results also demonstrate that COL22A1 was involved in cell cycle (Figure 4D), DNA replication (Figure 4E), P53 signaling pathway (Figure 4F), Adipocytokine signaling pathway (Figure 4G), Tgf Beta signaling pathway (Figure 4H), and hedgehog signaling pathway (Figure 4I).
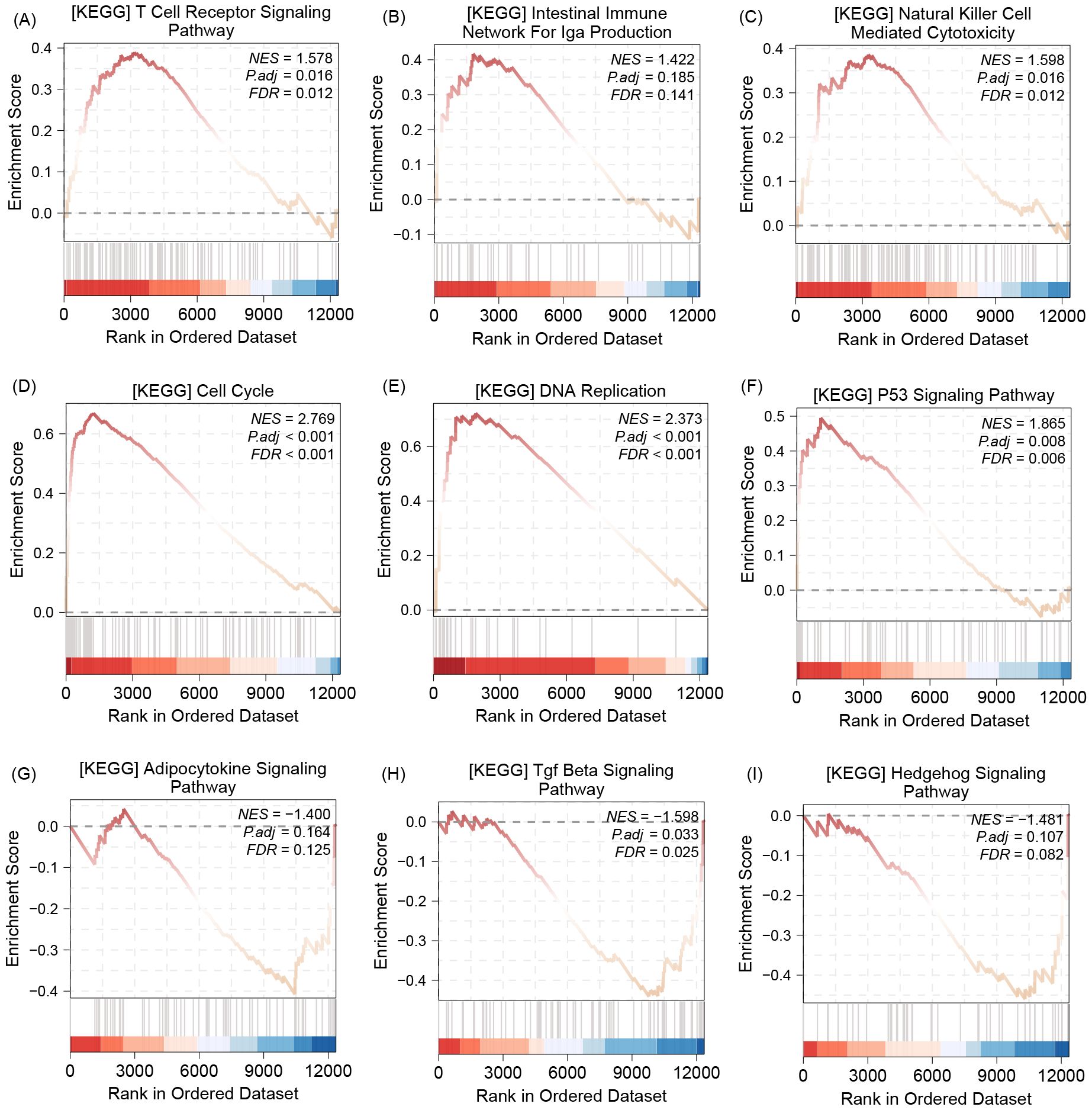
Figure 4. The GSEA analysis results show that, this gene could be enrichment in multi-immune pathway, such as T cell receptor signaling pathway (A), intestinal immune network for lga production (B), and Natural killer cell mediated cytotoxicity (C). In addition, the enrichment results also demonstrate that COL22A1 was involved in cell cycle (D), DNA replication (E),P53 signaling pathway (F), Adipocytokine signaling pathway (G), Tgf Beta signaling pathway (H), and hedgehog signaling pathway (I).
COL22A1 was related to immune infiltration
The expression of COL22A1 was found to be associated with immune infiltration across multiple analytical platforms. In the MCP-counter analysis, COL22A1 expression showed a positive correlation with CD8+ T cells, endothelial cells, and fibroblasts (Figure 5A). Similarly, the EPIC analysis revealed a negative correlation with B cells and CD4+ T cells, while showing a positive correlation with cancer-associated fibroblasts (CAFs) and macrophages (Figure 5B). TIMER analysis further confirmed a positive correlation between COL22A1 expression and macrophages as well as dendritic cells (DCs) (Figure 5C). In the QUANT analysis, COL22A1 expression was negatively correlated with B cells and positively correlated with M2 macrophages (Figure 5D). Finally, IPS analysis demonstrated a positive correlation with MHC molecules and endothelial cells (ECs), and a negative correlation with suppressive cells (SCs) and tumor-associated angiogenic factors (CPs) (Figure 5E). These findings collectively suggest that COL22A1 plays a significant role in modulating the tumor immune microenvironment by interacting with diverse immune and stromal cell populations.
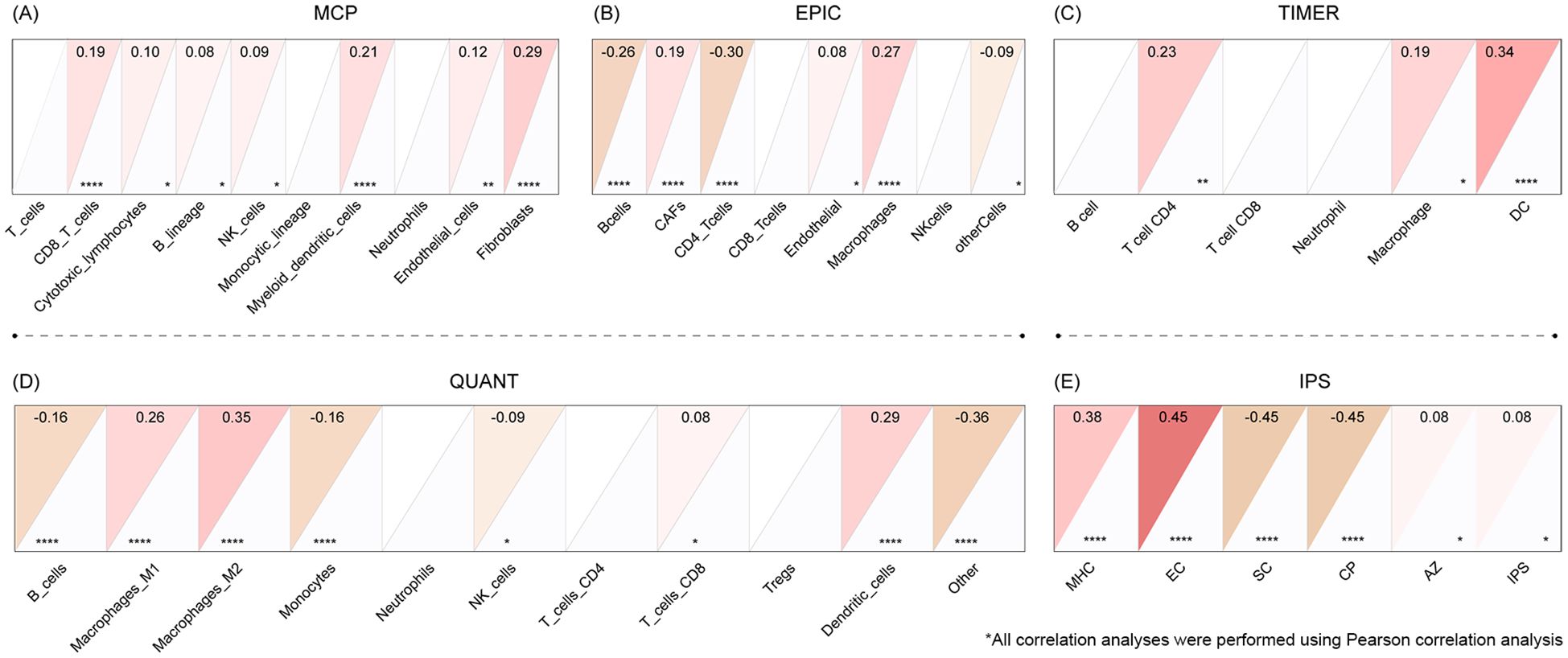
Figure 5. The expression of COL22A1 was found to be associated with immune infiltration across multiple analytical platforms. In the MCP-counter analysis, COL22A1 expression showed a positive correlation with CD8+ T cells, endothelial cells, and fibroblasts (A). Similarly, the EPIC analysis revealed a negative correlation with B cells and CD4+ T cells, while showing a positive correlation with cancer-associated fibroblasts (CAFs) and macrophages (B). TIMER analysis further confirmed a positive correlation between COL22A1 expression and macrophages as well as dendritic cells (DCs) (C). In the QUANT analysis, COL22A1 expression was negatively correlated with B cells and positively correlated with M2 macrophages (D). Finally, IPS analysis demonstrated a positive correlation with MHC molecules and endothelial cells (ECs), and a negative correlation with suppressive cells (SCs) and tumor-associated angiogenic factors (CPs) (E).
COL22A1 was associated with immune checkpoints and immune regulated genes
The expression of COL22A1 was positively correlated with several immune checkpoints and immune-regulated genes, suggesting its involvement in immune modulation. Specifically, COL22A1 showed significant positive correlations with immune checkpoint molecules, including BTLA, CD274, CD96, PDCD1LG2, ICOS, IDO1, CD86, LAG3, and PDCD1 (Figures 6A-I). Additionally, COL22A1 exhibited correlations with immune-regulated genes, further supporting its role in modulating the immune microenvironment. These findings highlight the potential importance of COL22A1 in immune escape mechanisms and its relevance as a candidate for therapeutic targeting in immuno-oncology (Figure 6J).
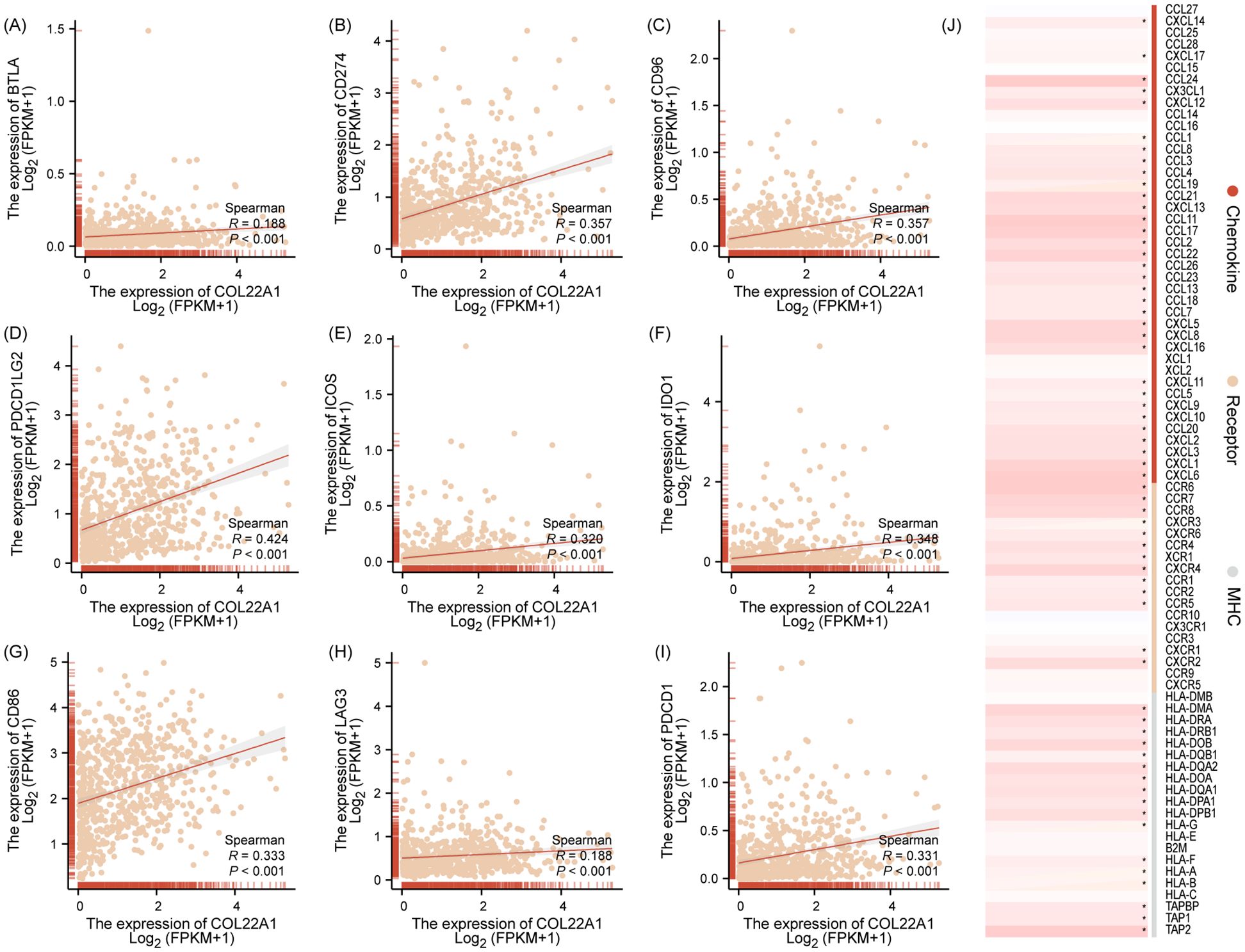
Figure 6. COL22A1 showed significant positive correlations with immune checkpoint molecules, including BTLA, CD274, CD96, PDCD1LG2, ICOS, IDO1, CD86, LAG3, and PDCD1 (A–I). Additionally, COL22A1 exhibited correlations with immune-regulated genes, further supporting its role in modulating the immune microenvironment (J).
Discussion
In recent years, the application of multi-omics has revolutionized the diagnosis and prognostic prediction of cancers, offering unprecedented insights into complex biological systems and their interactions in various oncological conditions. Correspondingly, an increasing number of studies are applying multi-omic approaches to improve the diagnosis and prognostic stratification of GBM, facilitating more precise and targeted therapies (20). Yan et al (21). developed and validated a deep-learning imaging signature that significantly predicts survival outcomes in GBM patients, correlates with key biological pathways and genetic alterations, and outperforms traditional clinical molecular approaches in risk stratification, demonstrating an improved C index and net reclassification improvement. In addition, a previous study, by integrating clinical, radiomic, and genetic data, significantly enhanced the accuracy of predicting OS in IDH-wildtype GBM patients, with the combined omics approach improving the concordance index and achieving AUCs of 0.78 in the discovery cohort and 0.75 in the replication cohort (22). As in our study, we have integrated radiomics and genomics to identify five genes (VAMP8, CYBA, COL22A1, FCER1G, and NPC2) positively correlated with a risk score designed to predict the aggressiveness and potential outcomes of GBM, as well as its potential to trigger AE. This approach enhances our understanding of GBM’s complex behavior and its implications for AE, providing a more robust predictive model for clinical outcomes. Among these genes, COL22A1 was identified as the hub gene, indicating its critical influence on the regulatory networks affecting GBM behavior and patient prognosis, as well as its potential role in driving AE. This highlights COL22A1’s importance not only in GBM progression but also in its potential to activate immune mechanisms involved in AE.
Collagen type XXII alpha 1 chain (COL22A1), a member of the collagen family, is known for its role in the structural matrix of connective tissues (23). Like many in its family, COL22A1 and related collagen products are implicated in promoting tumor development, indicating its potential significance in cancer biology and tumor progression (24). In a recent study, COL22A1 was identified as part of a group of extracellular matrix molecules that show elevated expression in the TME of GBM, suggesting its involvement in regulating the complex angiogenic processes within GBM and potentially impacting patient survival outcomes (25). In addition, COL22A1 was found to be overexpressed and to play a pro-oncogenic role in GBM, as demonstrated by impaired proliferation, migration, and invasion of glioma cells following COL22A1 silencing, underscoring its significance in tumor progression and as a biomarker for predicting patient survival (26). In our study, COL22A1 was also shown to be highly expressed in multiple GBM cell lines and verified at the protein level. Furthermore, the expression of COL22A1 is associated with common clinical risk factors such as older age, male gender, and higher tumor grade. Additionally, high expression of COL22A1 correlates with poorer patient prognosis. These findings confirm the potential role of COL22A1 in the progression of GBM and highlight its significance as a marker for prognosis and a target for therapeutic intervention. However, current research does not establish a direct link between the gene COL22A1 and AE nor its impact on the immune profile of GBM, highlighting an area for future studies to explore potential connections that could provide deeper insights into the genetic and immunological underpinnings of these conditions.
Immunotherapy has dramatically altered the landscape of cancer treatment, with significant successes using checkpoint inhibitors and CAR T cells for various cancers (27), while GBM has consistently demonstrated robust resistance due to its unique intrinsic and adaptive immune evasion mechanisms (14, 28). For example, the CheckMate 143 trial, the first large randomized study targeting programmed death-1 (PD-1) signaling in GBM, demonstrated that nivolumab did not extend OS in patients with recurrent GBM compared to bevacizumab (29). The TME of GBM may partly explain the suboptimal outcomes observed, as a study has shown that the clinical response to anti-PD-1 immunotherapy in GBM is linked to specific genetic alterations, immune-related expression profiles, and the degree of immune cell infiltration, which collectively reflect the tumor’s clonal evolution and adaptive immune landscape during treatment (22, 30). Notably, we found that the expression of COL22A1 positively correlates with common immune checkpoints such as PD-1 (PDCD1), PD-L1 (CD274), and LAG3 in GBM, suggesting that COL22A1 may play a significant role in regulating immune evasion mechanisms in GBM. Additionally, DEGs between groups with high and low COL22A1 expression were enriched in immune-related pathways such as the T Cell Receptor Signaling and Natural Killer Cell-Mediated Cytotoxicity pathways, suggesting a significant role for COL22A1 in modulating immune responses in the TME. Moreover, a previous study has revealed that changes in the extracellular matrix (ECM) components, such as the loss of Hyaluronan and Proteoglycan Link Protein 1 (HAPLN1), significantly impacted immune responses and cell migration within the TME in aging skin (24). Given that COL22A1 is known for its role in ECM organization and has been associated with tumor progression in other studies (31, 32), It is plausible that COL22A1 could similarly influence the structural and functional dynamics of the ECM, potentially affecting interactions between immune cells and cancer cells. However, establishing a definitive link between COL22A1 and the immune response requires further exploration.
Overall, we utilized machine learning to analyze radiomics data, constructing a risk model integrated with genomic data that identifies COL22A1 as a hub gene. This model is designed not only to diagnose and predict the prognosis of GBM but also to assess the potential for GBM to drive AE. Building on this foundation, we further explored the relationship between COL22A1 and the TME, discovering correlations with multiple immune cell checkpoints, chemokines, and MHC loci that influence immune responses, enriched in immune-related pathways. These findings highlight COL22A1’s potential as a therapeutic target and lay the groundwork for developing new immunotherapeutic strategies that could address both GBM prognosis and the likelihood of associated AE. However, our study has several limitations. Firstly, the impact of COL22A1 on the survival of GBM patients and their response to immunotherapy has not been validated with real-world data. Secondly, the interaction between the tumor and its microenvironment is a complex process, and we did not explore the effects of COL22A1 on the GBM and AE TME at the single-cell level. Additionally, although our research shows that COL22A1 expression correlates with common immune checkpoints and MHC loci, suggesting its potential as a target for immune evasion, it remains unclear whether this effect is mediated through immune checkpoints or influences the interactions between immune and tumor cells via the extracellular matrix. Furthermore, the specific immune mechanisms through which GBM drives AE via COL22A1 are still unclear. Further studies are needed to clarify these mechanisms.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Ethics statement
The studies involving humans were approved by The Medical Ethics Committee of Xuzhou Municipal Hospital approved this study. The studies were conducted in accordance with the local legislation and institutional requirements. The participants provided their written informed consent to participate in this study.
Author contributions
BY: Data curation, Investigation, Project administration, Resources, Writing – review & editing, Conceptualization, Formal analysis, Methodology, Software, Validation, Visualization, Writing – original draft. QC: Investigation, Methodology, Project administration, Resources, Software, Writing – original draft, Writing – review & editing. DW: Formal analysis, Methodology, Software, Visualization, Writing – review & editing. LD: Formal analysis, Investigation, Methodology, Resources, Software, Writing – review & editing. JQ: Methodology, Software, Validation, Visualization, Writing – review & editing. RD: Formal analysis, Investigation, Methodology, Visualization, Writing – review & editing. WS: Data curation, Investigation, Project administration, Resources, Writing – review & editing. UK: Data curation, Funding acquisition, Investigation, Project administration, Resources, Supervision, Writing – review & editing. ZY: Conceptualization, Funding acquisition, Project administration, Resources, Supervision, Visualization, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. Guangxi Medicine and Health Science Research Program Project (No.Z-C20221005).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2025.1562070/full#supplementary-material
Supplementary Table 1 | Random Forest results of Top 5 features.
Supplementary Table 2 | Lasso regression results.
References
1. Ostrom QT, Price M, Neff C, Cioffi G, Waite KA, Kruchko C, et al. CBTRUS statistical report: primary brain and other central nervous system tumors diagnosed in the United States in 2015–2019. Neuro-oncology. (2022) 24:v1–v95. doi: 10.1093/neuonc/noac202
2. Pellerino A, Caccese M, Padovan M, Cerretti G, Lombardi G. Epidemiology, risk factors, and prognostic factors of gliomas. Clin Trans Imaging. (2022) 10:467–75. doi: 10.1007/s40336-022-00489-6
3. Verdugo E, Puerto I, Medina M. An update on the molecular biology of glioblastoma, with clinical implications and progress in its treatment. Cancer Commun. (2022) 42:1083–111. doi: 10.1002/cac2.v42.11
4. Rong L, Li N, Zhang Z. Emerging therapies for glioblastoma: current state and future directions. J Exp Clin Cancer Res. (2022) 41:142. doi: 10.1186/s13046-022-02349-7
5. Louis DN, Perry A, Wesseling P, Brat DJ, Cree IA, Figarella-Branger D, et al. The 2021 WHO classification of tumors of the central nervous system: a summary. Neuro-oncology. (2021) 23:1231–51. doi: 10.1093/neuonc/noab106
6. Stupp R, Mason WP, van den Bent MJ, Weller M, Fisher B, Taphoorn MJ, et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med. (2005) 352:987–96. doi: 10.1056/NEJMoa043330
7. Watts C, Sanai N. Surgical approaches for the gliomas. Handb Clin neurology. (2016) 134:51–69. doi: 10.1016/B978-0-12-802997-8.00004-9
8. Kircher MF, de la Zerda A, Jokerst JV, Zavaleta CL, Kempen PJ, Mittra E, et al. A brain tumor molecular imaging strategy using a new triple-modality MRI-photoacoustic-Raman nanoparticle. Nat Med. (2012) 18:829–34. doi: 10.1038/nm.2721
9. Aasland D, Götzinger L, Hauck L, Berte N, Meyer J, Effenberger M, et al. Temozolomide induces senescence and repression of DNA repair pathways in glioblastoma cells via activation of ATR–CHK1, p21, and NF-κB. Cancer Res. (2019) 79:99–113. doi: 10.1158/0008-5472.CAN-18-1733
10. Rockwell S, Dobrucki IT, Kim EY, Marrison ST, Vu VT. Hypoxia and radiation therapy: past history, ongoing research, and future promise. Curr Mol Med. (2009) 9:442–58. doi: 10.2174/156652409788167087
11. Wang Z, Zhao Y, Zhang L. Emerging trends and hot topics in the application of multi-omics in drug discovery: A bibliometric and visualized study. Curr Pharm Anal. (2024) 21:20–32. doi: 10.1016/j.cpan.2024.12.001
12. Reardon DA, Schuster J, Tran DD, Fink KL, Nabors LB, Li G, et al. ReACT: Overall survival from a randomized phase II study of rindopepimut (CDX-110) plus bevacizumab in relapsed glioblastoma. Am Soc Clin Oncol. (2015) 62:198–9. doi: 10.1200/jco.2015.33.15_suppl.2009
13. Brown MC, Holl EK, Boczkowski D, Dobrikova E, Mosaheb M, Chandramohan V, et al. Cancer immunotherapy with recombinant poliovirus induces IFN-dominant activation of dendritic cells and tumor antigen–specific CTLs. Sci Trans Med. (2017) 9:eaan4220. doi: 10.1126/scitranslmed.aan4220
14. Desbaillets N, Hottinger AF. Immunotherapy in glioblastoma: a clinical perspective. Cancers. (2021) 13:3721. doi: 10.3390/cancers13153721
15. Glaviano A, Lau HS, Carter LM, Lee EHC, Lam HY, Okina E, et al. Harnessing the tumor microenvironment: targeted cancer therapies through modulation of epithelial-mesenchymal transition. J Hematol Oncol. (2025) 18:6. doi: 10.1186/s13045-024-01634-6
16. Gao Z, Bai Y, Lin A, Jiang A, Zhou C, Cheng Q, et al. Gamma delta T-cell-based immune checkpoint therapy: attractive candidate for antitumor treatment. Mol Cancer. (2023) 22:31. doi: 10.1186/s12943-023-01722-0
17. Liau LM, Ashkan K, Tran DD, Campian JL, Trusheim JE, Cobbs CS, et al. First results on survival from a large Phase 3 clinical trial of an autologous dendritic cell vaccine in newly diagnosed glioblastoma. J Trans Med. (2018) 16:1–9. doi: 10.1186/s12967-018-1507-6
18. Uy CE, Binks S, Irani SR. Autoimmune encephalitis: clinical spectrum and management. Pract neurology. (2021) 21:412–23. doi: 10.1136/practneurol-2020-002567
19. Consoli S, Dono F, Evangelista G, Corniello C, Onofrj M, Thomas A, et al. Case Report: Brain tumor’s pitfalls: two cases of high-grade brain tumors mimicking autoimmune encephalitis with positive onconeuronal antibodies. Front Oncol. (2023) 13:1254674. doi: 10.3389/fonc.2023.1254674
20. Karati D, Kumar D. Molecular insight into the apoptotic mechanism of cancer cells: an explicative review. Curr Mol Pharmacol. (2024) 17:e18761429273223. doi: 10.2174/0118761429273223231124072223
21. Yan J, Sun Q, Tan X, Liang C, Bai H, Duan W, et al. Image-based deep learning identifies glioblastoma risk groups with genomic and transcriptomic heterogeneity: a multi-center study. Eur Radiology. (2023) 33:904–14. doi: 10.1007/s00330-022-09066-x
22. Zhao J, Chen AX, Gartrell RD, Silverman AM, Aparicio L, Chu T, et al. Immune and genomic correlates of response to anti-PD-1 immunotherapy in glioblastoma. Nat Med. (2019) 25:462–9. doi: 10.1038/s41591-019-0349-y
23. Kehlet S, Karsdal M. Type XXII collagen. In: Biochemistry of Collagens, Laminins and Elastin. Elsevier (2016). p. 135–7.
24. Kaur A, Ecker BL, Douglass SM, Kugel CH III, Webster MR, Almeida FV, et al. Remodeling of the collagen matrix in aging skin promotes melanoma metastasis and affects immune cell motility. Cancer discovery. (2019) 9:64–81. doi: 10.1158/2159-8290.CD-18-0193
25. Barbosa LC, MaChado GC, Heringer M, Ferrer VP. Identification of established and novel extracellular matrix components in glioblastoma as targets for angiogenesis and prognosis. Neurogenetics. (2024) 25:1–14. doi: 10.1007/s10048-024-00763-x
26. Liu H, Zeng Z, Sun P. Prognosis and immunoinfiltration analysis of angiogene-related genes in grade 4 diffuse gliomas. Aging (Albany NY). (2023) 15:9842. doi: 10.18632/aging.205054
27. Peng S, Lin A, Jiang A, Zhang C, Zhang J, Cheng Q, et al. CTLs heterogeneity and plasticity: implications for cancer immunotherapy. Mol Cancer. (2024) 23:58. doi: 10.1186/s12943-024-01972-6
28. Liu L, Xie Y, Yang H, Lin A, Dong M, Wang H, et al. HPVTIMER: A shiny web application for tumor immune estimation in human papillomavirus-associated cancers. Imeta. (2023) 2:e130. doi: 10.1002/imt2.v2.3
29. Filley AC, Henriquez M, Dey M. Recurrent glioma clinical trial, CheckMate-143: the game is not over yet. Oncotarget. (2017) 8:91779. doi: 10.18632/oncotarget.21586
30. Manohar SM. At the crossroads of TNF α Signaling and cancer. Curr Mol Pharmacol. (2024) 17:e060923220758. doi: 10.2174/1874467217666230908111754
31. Li X, Jin Y, Xue J. Unveiling collagen’s role in breast cancer: insights into expression patterns, functions and clinical implications. Int J Gen Med. (2024) 17:1773–87. doi: 10.2147/IJGM.S463649
32. Leal A, Taranto P, Blasi PB, Garrote CT, Moura F. Abstract PS18-41: Somatic mutational landscapes of invasive ductal and lobular carcinomas in the GENIE consortium cohort: Real-world gene actionability assessment of 8,756 breast cancer patients. Cancer Res. (2021) 81:PS18–41-PS18-41. doi: 10.1158/1538-7445.SABCS20-PS18-41
Keywords: artificial intelligence, tumor microenvironment, COL22A1, autoimmune encephalitis, radiomics
Citation: Yan B, Chen Q, Wang D, Ding L, Qu J, Du R, Shi W, Kahlert UD and Yu Z (2025) Artificial intelligence-based radiogenomics reveals the potential immunoregulatory role of COL22A1 in glioma and its induced autoimmune encephalitis. Front. Immunol. 16:1562070. doi: 10.3389/fimmu.2025.1562070
Received: 16 January 2025; Accepted: 17 February 2025;
Published: 06 March 2025.
Edited by:
Minghua Ren, First Affiliated Hospital of Harbin Medical University, ChinaCopyright © 2025 Yan, Chen, Wang, Ding, Qu, Du, Shi, Kahlert and Yu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wenjie Shi, d2VuamllLnNoaUBtZWQub3ZndS5kZQ==; Ulf D. Kahlert, dWxmLmthaGxlcnRAbWVkLm92Z3UuZGU=; Zhengquan Yu, emhlbmdxdWFuLnl1QG5ldXJvc2NpLmNvbS5jbg==
†These authors have contributed equally to this work
 Bingchao Yan1,2†
Bingchao Yan1,2† Renfei Du
Renfei Du Wenjie Shi
Wenjie Shi Ulf D. Kahlert
Ulf D. Kahlert