
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Immunol., 06 February 2025
Sec. Inflammation
Volume 16 - 2025 | https://doi.org/10.3389/fimmu.2025.1546474
Sepsis, a heterogeneous illness produced by a dysregulated host response to infection, remains a severe mortality risk. Recent discoveries in sepsis research have stressed phenotyping as a feasible strategy for tackling heterogeneity and enhancing therapy precision. Sepsis phenotyping has moved from traditional stratifications based on severity and prognosis to dynamic, phenotype-driven therapeutic options. This review covers recent progress in connecting sepsis subgroups to personalized treatments, with a focus on phenotype-based therapeutic predictions and decision-support systems. Despite ongoing challenges, such as standardizing phenotyping frameworks and incorporating findings into clinical practice, this topic has enormous promise. By investigating phenotypic variation in therapy responses, we hope to uncover new biomarkers and phenotype-driven therapeutic solutions, laying the groundwork for more effective therapies and, ultimately improving patient outcomes.
Sepsis is a primary cause of mortality among critically sick patients worldwide, accounting for approximately 49 million cases and 11 million deaths each year, representing nearly 20% of all deaths worldwide (1–3). It results from an anomalous immunological response to infection that precipitates organ failure (1, 4). Sepsis exhibits high clinical heterogeneity, making the discovery of effective treatments exceedingly challenging (5). Despite advances in understanding the cellular and molecular causes of sepsis, its complicated pathophysiology remains a problem (6). This heterogeneity arises from diverse pathogens, infection sites, and host immune responses that interact intricately to shape the clinical presentation (7–11). The phenotypical variability in sepsis is reflected across various clinical manifestation. Hemodynamic alterations, including blood pressure, cardiac output, and systemic vascular resistance, are prevalent indicators of sepsis-induced circulatory dysfunction (12, 13). Coagulation diseases, including disseminated intravascular coagulation (DIC), platelet failure, and microthrombosis, intensify organ damage (14, 15). Neurological manifestations encompass sepsis-associated encephalopathy and delirium, reflecting both direct and indirect impacts on the central nervous system (16, 17). Metabolic instability, including hyperlactatemia and mitochondrial dysfunction, underscores the systemic nature of sepsis (18, 19). Respiratory issues, especially acute respiratory distress syndrome (ARDS) and hypoxemia, often predominate the clinical trajectory, however other systems, including renal and hepatic, exhibit varied responses based on the patient’s phenotype (13, 20–22).
Given this multifaceted heterogeneity, understanding how these clinical factors interplay to shape sepsis phenotypes is essential. Recent advancements in big data methodologies, artificial intelligence, and high-throughput multi-omics data, along with disease phenotyping, offer novel pathways for enhancing the comprehension of sepsis and formulating more personalized treatment strategies (23). Phenotypic approaches employ computational tools to analyze clinical, biomarker, and genetic data, aiming to discover more homogeneous subpopulations within sepsis patients (23, 24). This technology facilitates precise therapy delivery, guaranteeing that the appropriate patient receives the exact treatment at the designated time. In other fields, such as oncology, phenotypic methodologies have enhanced therapeutic accuracy (25, 26). Leveraging these advancements, sepsis treatment could be revolutionized, perhaps leading to the identification of new biomarkers and therapeutic targets that correspond to phenotypic differences in treatment response, thus enhancing patient outcomes.
This review examines current advancements in sepsis phenotyping (Figure 1), highlighting the potential of these methods to tackle sepsis heterogeneity and optimize treatment protocols. We aim to establish a foundation for the identification of novel biomarkers and targeted treatments based on differential responses among sepsis subtypes, ultimately enhancing patient outcomes.
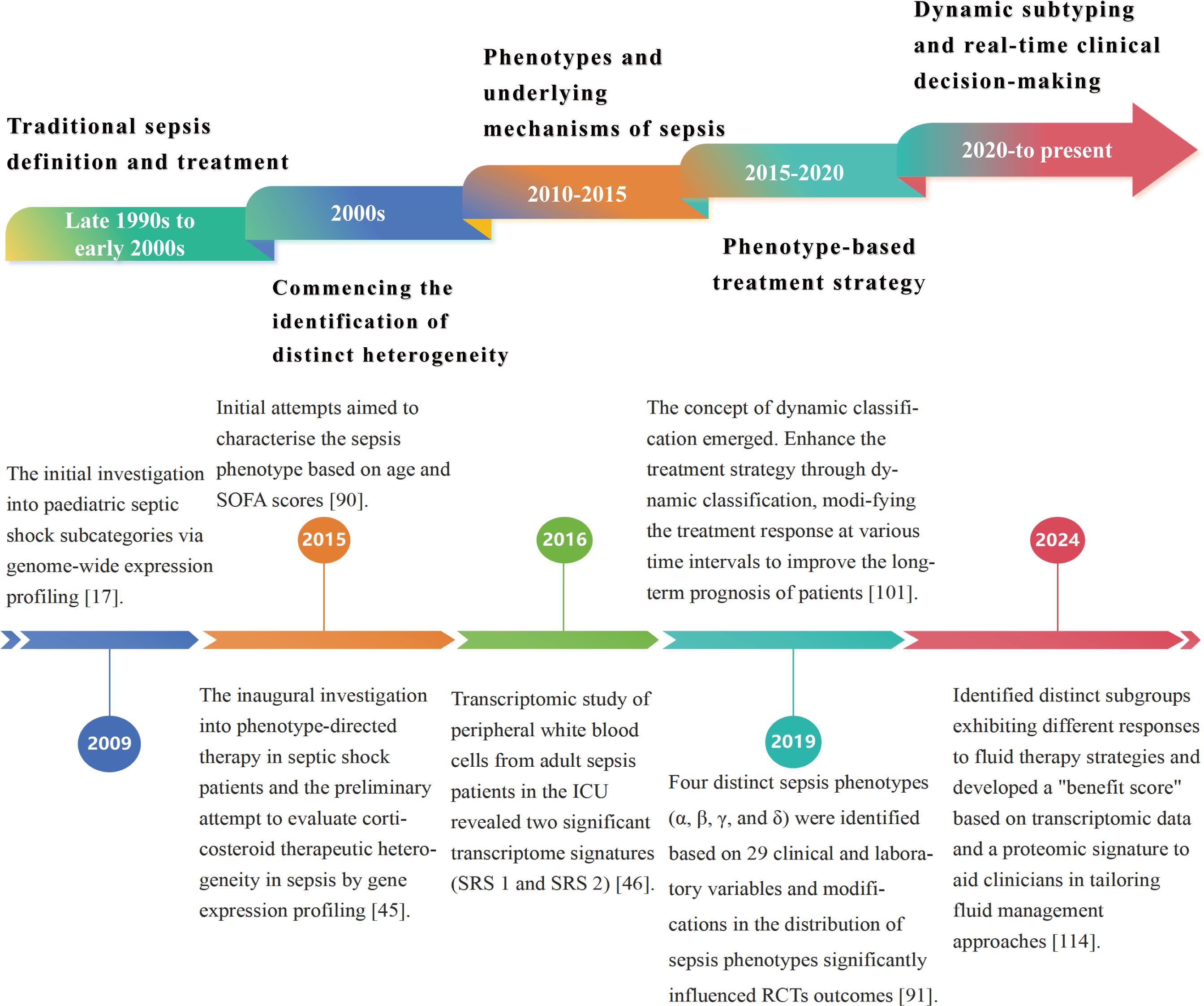
Figure 1. Chronology of the developmental history of sepsis classification. The evolution of sepsis phenotypic research from static categorization to dynamic classification has led to increasingly customized and precise treatment approaches for sepsis. Precise phenotypic classification and phenotype-based therapy strategies enhance understanding of sepsis heterogeneity, identify novel therapeutic targets, and optimize clinical treatment protocols, ultimately improving patient outcomes. Future research will focus on improving treatment outcomes through real-time monitoring and dynamic modifications to treatment protocols, aimed at addressing existing challenges in precision therapy.
Sepsis is a complex clinical disease defined by a variety of immunological responses, infection locations, and patient characteristics (7, 27). It is now regarded as a coexisting, dynamic proinflammatory and immunosuppressive route driven by pathogen virulence, host immunological regulation, and pharmaceutical interventions (27–30). Sepsis-induced immunological dysregulation, which includes genetic and metabolic changes to immune cells, causes organ failure and increases the chance of re-infection after pathogen clearance (31–36). Patient demographics, comorbidities, infection characteristics, and treatment timing all contribute to sepsis heterogeneity, which has an impact on clinical symptoms and treatment outcomes (Figure 2). Despite advances in sepsis research, a lack of specialized therapy options limits further success (36, 37).
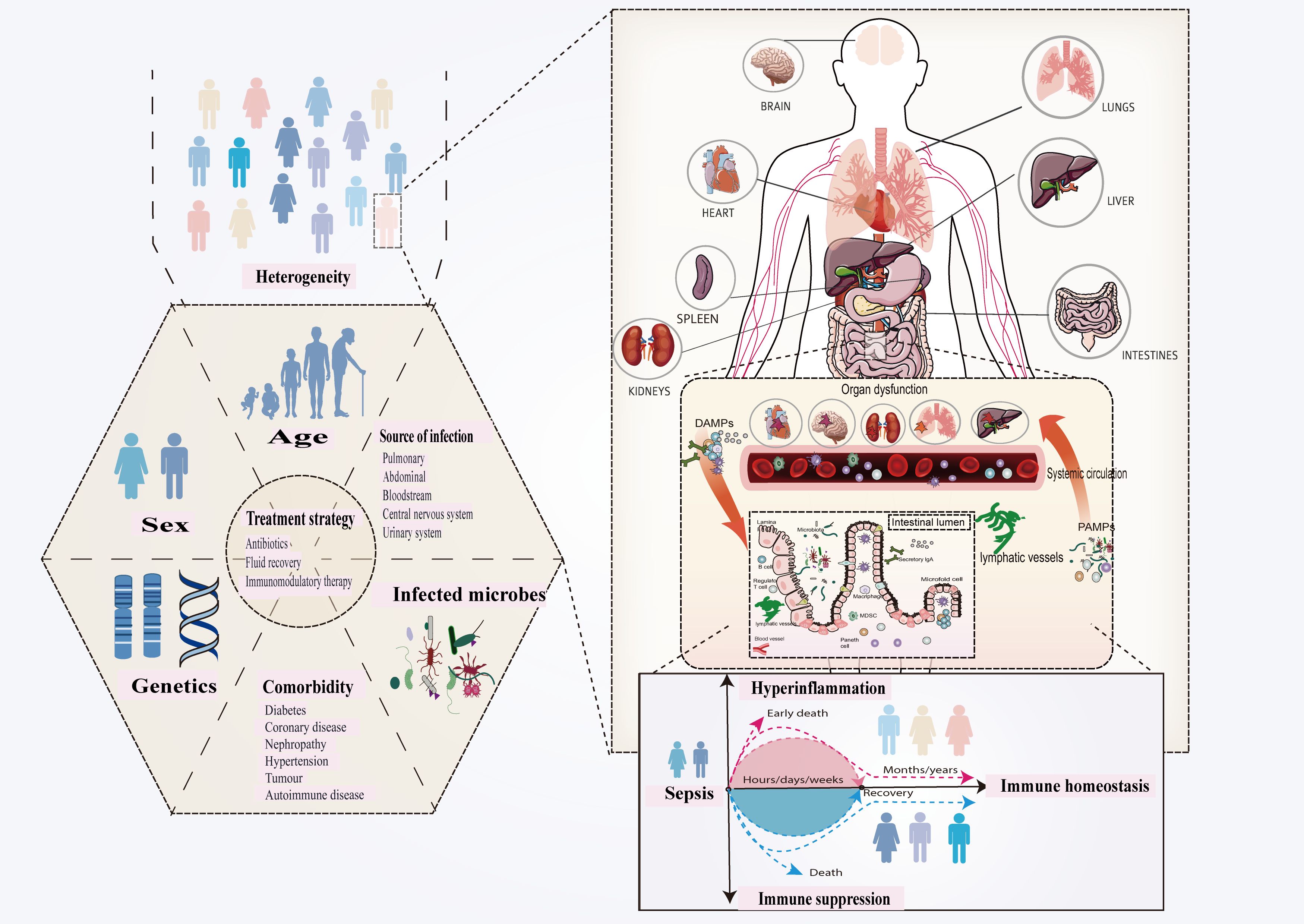
Figure 2. Sepsis is defined as a dysregulated host response to infection that leads to organ dysfunction. Innate immunity is inherently structured to swiftly respond to conserved molecules known as PAMPs and DAMPs, which are produced by infections and hosts, respectively. The complexities of pathogen-host interactions increase the variability of sepsis manifestations. The observed heterogeneity primarily results from pre-existing individual health conditions, variability in pathogenic bacteria, differences in infection sites, and variations in host immune responses. This variety underpins differences in clinical presentation, prognosis, and treatment outcomes, highlighting the importance of understanding sepsis diversity. Improving phenotype-based therapy techniques, identifying novel markers and targets through diverse therapeutic responses, and stratifying patients can all improve results and steer newer drugs. Pathogen-associated molecular patterns (PAMPs) and damage-associated molecular patterns (DAMPs).
In recent decades, extensive clinical trials designed to enhance sepsis treatment have predominantly failed to demonstrate significant clinical efficacy (38–46). The trials examined several medications, including hormone therapy, immunomodulators, antibiotics, and fluid resuscitation methods. Despite these efforts, various medications, such as recombinant activated protein C, have not reliably decreased mortality or enhanced organ function, with some associated with adverse side effects (38, 40, 46). Likewise, the reliance on therapies targeting individual pathways, such as anti-inflammatory cytokines like IL-1 and IL-6, fails to address the complex interplay of pro-inflammatory and anti-inflammatory responses in sepsis (41, 42). The primary reason for these failures lies in the assumption that sepsis is a homogenous condition, treatable with a “one-size-fits-all” approach. Research on antibiotics and fluid resuscitation has underscored the intricacies of sepsis management, revealing a lack of consensus regarding optimal treatment protocols (43–45, 47–49). Many RCTs lack robust stratification of patients by phenotype, leading to diluted efficacy signals and conflicting results. An example of this heterogeneity can be seen in patients with macrophage activation syndrome (MAS), a hyperinflammatory condition associated with sepsis. Studies have shown that these patients may benefit from IL-1 receptor antagonists, highlighting the importance of targeted therapies for specific subgroups (41). These insights underscore the need to move beyond conventional RCT designs and develop phenotype-driven approaches that tailor interventions to patient-specific profiles.
The failures of these trials highlight the importance of understanding sepsis heterogeneity at the molecular and clinical levels. Stratifying patients by genetic, molecular, and clinical phenotypes could enhance the precision of trials and allow the identification of subgroup-specific responses. And adaptive designs that allow modifications based on interim data can better accommodate the dynamic nature of sepsis and its heterogeneous population. Meanwhile, addressing the interplay of multiple pathways using combination therapies or network-driven strategies may prove more effective than single-target interventions. By addressing these issues, future trials can better align with the complexity of sepsis and pave the way for more effective treatments.
Progress in genomic and transcriptomic technologies has revealed that sepsis has several subphenotypes, each associated with distinct clinical consequences. These findings highlight the promise of phenotype-driven therapies to surpass the limitations of traditional sepsis treatments. Initial research utilizing genetic analysis identified numerous sepsis subphenotypes exhibiting diverse clinical outcomes. An investigation of whole genome expression in whole blood RNA from 98 adolescent septic shock patients in 2009 uncovered numerous novel findings (24). This is the inaugural phenotypic investigation of septic shock patients, revealing three subphenotypes characterized by distinct immune response patterns and varying degrees of illness severity (24). Subsequent research corroborated these findings, indicating that gene expression profiling can delineate sepsis subgroups with varying treatment responses (50–52). In a 2015 study, researchers employed NanoString nCounter to quantify messenger RNA (mRNA) for 100 categorized genes, demonstrating gene expression mosaicism. Notwithstanding the enhancement, the model only identified subphenotypes A and B in the two cohorts (n=168 and n=132) (52). Subtype A activates glucocorticoid receptors to a lesser extent than subtype B (52). This prompted researchers to investigate adjuvant corticosteroid therapy and its prognostic implications. Adjuvant corticosteroids were administered to 52 (43%) of 120 patients with subphenotype A and 104 (58%) of 180 patients with subtype B. Corticosteroid therapy markedly decreased mortality in subtype A, but not in subtype B (52). This research is the first investigation of phenotype-directed therapy in septic shock patients by gene expression profiling to ascertain therapeutic heterogeneity in sepsis. Nonetheless, the administration of corticosteroids is non-randomized.
Furthermore, recent studies have shown demonstrated diversity in sepsis recovery (53–55). Research indicates that long-term outcomes differ, with numerous survivors facing persistent physical and psychological challenges (56–62). A 2021 study identified three distinct paths of depressive symptoms in septic shock survivors, highlighting the importance of personalized post- intensive care unit (ICU) care (59). Another study of clinical and biomarker data from 467 septic shock survivors identified two subgroups post-ICU discharge: Type A, characterized by low multi-organ dysfunction, and Type B, with a higher one-year mortality rate (34% vs. 16%) (55). Enhancing patient outcomes necessitates comprehending sepsis heterogeneity and developing phenotype-based therapeutics. The integration of epigenomics, transcriptomics, proteomics, metabolomics and cytomics with AI technologies is essential for advancing precision therapies in sepsis. This integration aids in identifying biomarkers and molecular signatures associated with sepsis subphenotypes, enhancing patient classification and treatment development. By integrating genetic and clinical data, researchers might enhance their comprehension of sepsis heterogeneity and develop stratified trial designs that consider subphenotype responses. This methodology establishes a robust framework for the identification of novel biomarkers and therapeutic targets, facilitating personalized treatments that enhance clinical outcomes and optimize therapeutic interventions.
Sepsis exhibits significant variability and presents management challenges; thus, defining its subphenotypes is essential for improving treatment. Recently emerging multi-omics technologies, including epigenomics, transcriptomics, proteomics, metabolomics, and cytomics, have enabled the molecular classification of sepsis (Figure 3). When integrated with vital signs, biochemical markers, and evaluations of organ function, these technologies elucidate the pathophysiological underpinnings of sepsis. Epigenomic changes, such as DNA methylation and histone irregularities, influence immunological responses and disease outcomes. Transcriptomic and proteomic analyses reveal gene expression patterns and protein biomarkers associated with clinical variability in sepsis patients, facilitating the identification of biomarkers and therapeutic targets. Moreover, artificial intelligence (AI) and machine learning (ML) are extensively employed to analyze these high-dimensional datasets to identify novel disease patterns and predict patient outcomes (Table 1). K-means clustering and latent profile analysis have been employed to identify clinically significant subgroups, categorizing patients according to clinical and genomic data to facilitate more personalized therapeutic decisions (63–65). Supervised learning methods such as random forests and logistic regression can identify the onset of sepsis and enhance fluid resuscitation and antibiotic treatment (64, 66–70). AI/ML methodologies integrated with clinical data can design individualized therapy regimens that enhance clinical outcomes (71–77). Artificial Intelligence and Machine Learning exhibit potential (78, 79); yet, their incorporation into clinical practice necessitates thorough validation and external verification to ensure their dependability and relevance across diverse healthcare environments.
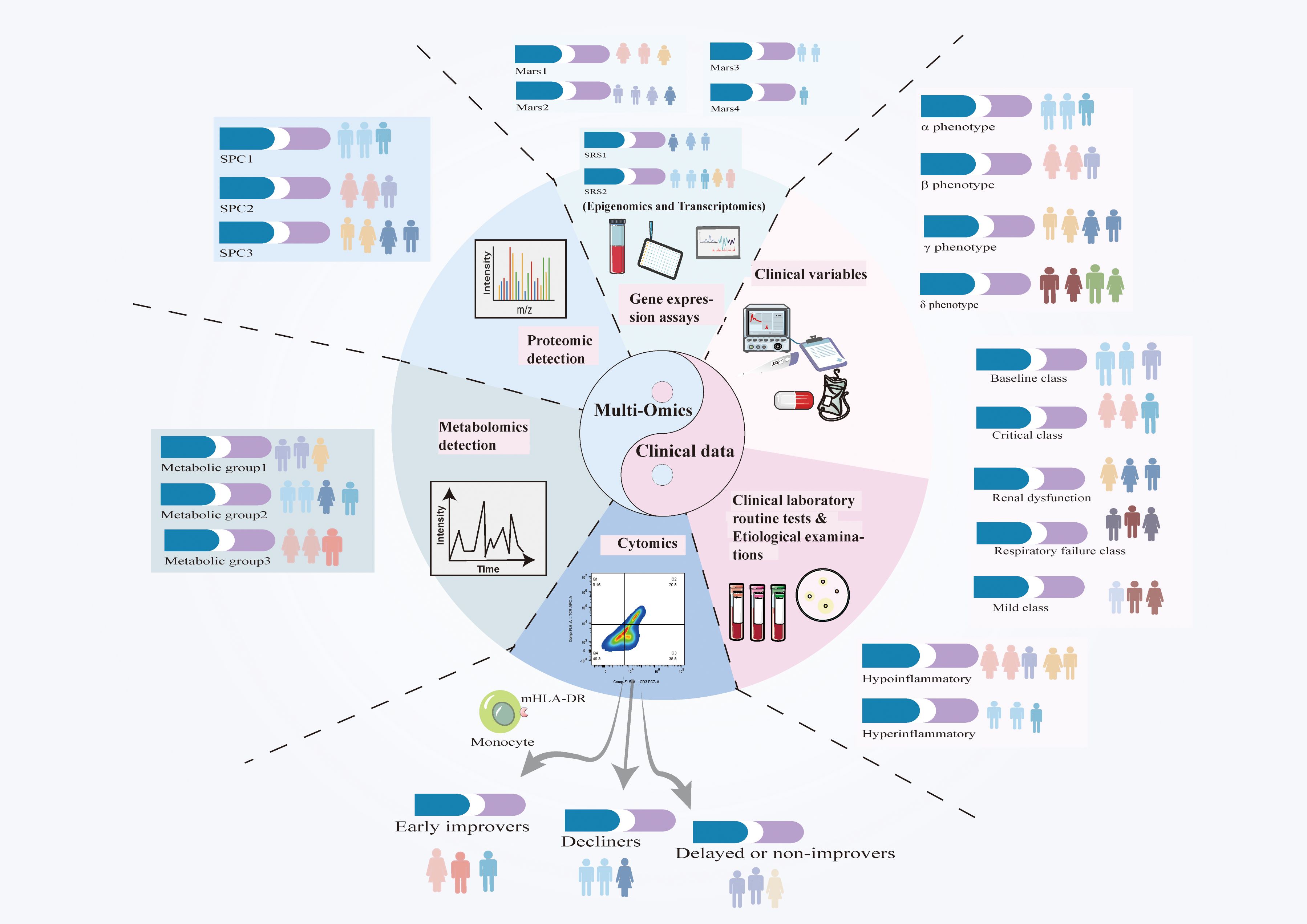
Figure 3. Advancements in big data and artificial intelligence (AI) have enhanced the integration of clinical, biomarker, and molecular data, facilitating a deeper understanding of sepsis heterogeneity. Although classifications of sepsis subtypes differ due to variations in study designs and methodologies, these technologies facilitate the identification of novel biomarkers and therapeutic targets. The integration of multi-omics data with artificial intelligence is essential for advancing precision medicine, refining patient stratification, and directing phenotype-based therapy. By integrating genetic and clinical data, researchers can enhance the comprehension of sepsis heterogeneity and formulate stratified trials, facilitating the development of personalized medications that optimize patient outcomes.
Sepsis is complex, thus knowing its molecular basis is crucial. Integrating Epigenomics, transcriptomics, proteomics, metabolomics, and cytomics data offers a comprehensive view of sepsis phenotyping (Figure 3). These methodologies delineate molecular subphenotypes, categorize patients, and formulate personalized treatment strategies. The following discussion addresses each omics technology and their potential synergy in the understanding and management of sepsis.
Epigenomics examines heritable modifications in gene expression that do not include alterations to the DNA sequence, including DNA methylation, histone modifications, and non-coding RNA. These epigenetic modifications significantly influence the immune response to sepsis. One study identified unique DNA methylation patterns in sepsis patients, correlating hypermethylation of immune regulatory genes with impaired immunological function (80). Another investigation identified histone modifications associated with sepsis-induced immune tolerance, offering insights into potentially reversible mechanisms (81). These findings underscore the epigenome’s pivotal role in defining molecular subtypes of sepsis and its potential therapeutic implications. Nonetheless, despite its promise, epigenomics possesses constraints. Epigenetic modifications can be affected by numerous environmental and temporal factors, complicating the establishment of definitive causality in sepsis. Moreover, the transient nature of many epigenetic modifications, coupled with the intricate relationships within the epigenome, complicates the ability to reach definitive findings. Epigenomics provides essential insights into the regulatory mechanisms of sepsis, sometimes complemented by transcriptomics, which captures real-time gene expression alterations and reveals more dynamic molecular endophenotypes of the condition.
Transcriptomics focuses on gene expression profiling to classify molecular endophenotypes of sepsis, which reveal significant variations in treatment responses and outcomes. Cohort studies employing unsupervised clustering to assess blood leukocyte gene expression data typically discern molecular subphenotypes of sepsis (82, 83). Comprehending the diverse forms of sepsis and tailoring treatment to specific phenotypes can enhance patient outcomes. Multi-omics approaches, particularly transcriptomics, have provided significant insights into the immunological dysregulation and genetic markers that characterize sepsis subphenotypes (82–84).
Transcriptomics-based studies have identified several molecular subphenotypes associated with distinct immune responses in sepsis patients. For example, a transcriptomic analysis of peripheral white blood cells from 265 adult sepsis patients in the ICU with community-acquired pneumonia was published in 2016. This investigation identified two significant transcriptome signatures—SRS 1 and SRS 2—that are associated with a bad and favorable prognosis, respectively (82). These signatures were confirmed across numerous cohorts and contributed to the development of gene classifiers capable of accurately predicting sepsis subphenotypes (82, 85, 86). Importantly, SRS 1 was associated with immunosuppression, whereas SRS 2 was associated with a more active immune response (82). Moreover, dynamic changes in SRS subgroup membership were observed during ICU stays (85).
A significant issue in transcriptomics research has been the absence of standardized protocols, leading to variability among studies. Recent initiatives to enhance research methodologies and replicate results across cohorts have yielded more dependable and reproducible outputs. For example, a gene expression analysis utilizing whole blood mRNA microarray data from diverse infection sources, including pneumonia and peritonitis, identified similarities and differences in immune responses, contributing to the characterization of sepsis subphenotypes (Mars1-4) (83). The Mars1 endophenotype, characterized by diminished gene expression in innate and adaptive immune functions such as toll-like receptor and T-cell receptor signaling pathways, was linked to unfavorable prognosis (83). Endophenotype indicators, specifically BPGM and TAP 2, were identified to enhance clinical application, predicting Mars1 (83). TAP 2 and TAP 1 constitute the TAP complex, responsible for transporting exogenous protein fragments (peptides) to the endoplasmic reticulum. The peptide is presented on the cell surface by major histocompatibility complex (MHC) class I proteins and identified by CD8+ T lymphocytes. If the peptide is detrimental, the immune system combats the intruders (87). These findings underscore the significance of personalized treatment strategies informed by molecular markers, rather than broad-spectrum medicines.
Research indicates that sepsis subphenotypes influence the effectiveness of immunomodulatory therapies (Table 2). A retrospective examination of gene expression in 176 septic shock patients from the VANISH clinical study, utilizing mRNA data, identified two SRS subphenotypes (SRS 1 and SRS 2) and assessed their impact on reactions to vasoactive drugs and steroids (86). The VANISH clinical trials randomly allocated patients to receive either norepinephrine or vasopressin within six hours of shock onset, thereafter administering hydrocortisone or a placebo (43). Patients displaying the immunosuppressive SRS 1 signature had a neutral response to steroid therapy, while those with the immune-active SRS 2 signature encountered worsened results (86). The restricted sample size may have negatively impacted the SRS 2 subgroup, resulting in an increased mortality rate. Numerous studies have investigated transcriptome-derived phenotypes, particularly with corticosteroid therapy. In 2020, researchers analyzed and incorporated 12 GEO and ArrayExpress datasets containing whole blood gene expression data from 1,613 adult sepsis patients (88). This research identified two sepsis subtypes (Class 1 and Class 2) by the application of deep learning methodologies and autoencoders for feature extraction. Class 1, characterized by immunosuppression, exhibited a significantly higher death rate compared to Class 2 (88). The research additionally examined the influence of sepsis subtypes on hydrocortisone response by a secondary analysis of an independent VANISH trial dataset (43, 88). The use of hydrocortisone elevated mortality rates in Class 2 patients, but not in Class 1 patients (88). This discovery underscores the necessity for comprehensive investigations into sepsis subphenotypes to enhance clinical treatment options and deepen our understanding of the complexity and treatment responsiveness of sepsis.
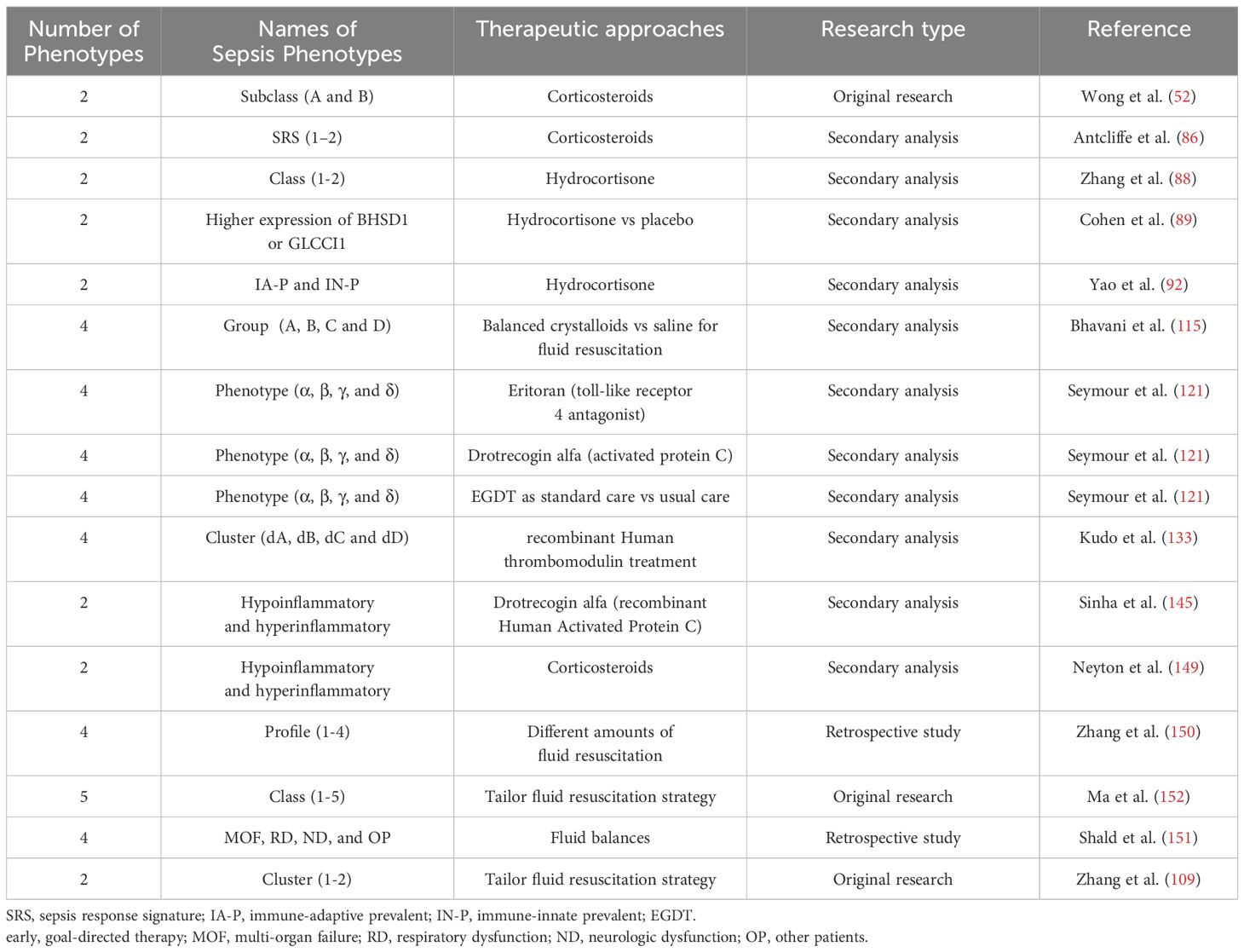
Table 2. Principal papers referenced in the text concerning the potential for phenotype-guided therapy in sepsis.
A further work in 2021 analyzed the correlation between corticosteroid responsiveness in septic shock patients and adrenocortical gene expression (89). A thorough analysis of whole blood RNA sequencing data from 697 patients across 28 medical-surgical ICUs involved in the ADRENAL experiment revealed no correlation between adrenal cortical gene expression levels and mortality (46, 89). A logistic regression analysis demonstrated an atypical occurrence: those treated with hydrocortisone with elevated Glucocorticoid-induced transcript 1 (GLCCI1) gene expression levels recovered from shock more rapidly than those with increased beta-hydroxysteroid dehydrogenase 1 (BHSD1) gene expression (89). And a 2022 study using a classifier based on 33 gene expression patterns to classify 13 sepsis datasets into three categories (90–92). The researchers subsequently identified a minimal collection of predictive genes from differentially expressed genes (DEGs) within each subclass to accurately classify subclasses and reclassify gene expression patterns into immune-adaptive, immune-innate, and immune-coagulant subphenotypes (92). Given that the coagulant malfunction phenotype may not directly influence the response to glucocorticoid therapy, the researchers re-evaluated 117 patients from the VANISH clinical trial with septic shock, categorizing them as immune-adaptive or immune-innate, and investigated the impact of these subtypes on steroid therapy (92). The research indicated that glucocorticoids elevated death rates in the high immune adaptability cohort, supporting previous results from smaller studies and demonstrating that transcriptomically generated phenotypes could influence treatment choices (92). Patients in septic shock may exhibit varying corticosteroid responses attributable to gene expression differences. There is increased research on transcriptome-derived phenotypes and immune cell characteristics in individuals with sepsis (93). These findings assist in prognostic predictions and the assessment of treatments such as corticosteroids and vasoactive agents, which influence patients variably based on their immunological condition.
Thus, transcriptomics research has enhanced our comprehension of sepsis heterogeneity and demonstrated the potential for phenotype-based therapeutics. Precision medicine in sepsis enables the customization of therapy based on the patient’s molecular profile, enhancing diagnosis and outcomes. The discovery and clinical application of molecular classifiers are essential for enhancing therapeutic regimens and patient survival rates. Transcriptomics offers valuable insights into gene expression related to sepsis, although it does not directly reflect functional outcomes. Proteomics examines proteins, the functional outcomes of gene expression, to enhance comprehension of sepsis heterogeneity.
The heterogeneity of patient reactions in sepsis complicates clinical care and impedes the development of effective, personalized therapeutics (6, 94, 95). Proteomics provides insights into the functional protein landscape in sepsis, unveiling key biomarkers associated with disease severity and outcomes (96, 97). By identifying important plasma proteins and their connections with disease severity, organ dysfunction, and patient outcomes, proteomics aids in the understanding of the mechanisms behind sepsis heterogeneity and the development of precision medicine solutions.
Recent studies have shown the effectiveness of proteomic markers in stratifying patients with sepsis. In 2022, researchers employed proteomics to differentiate between children with sepsis who developed ARDS and those who did not (97). The study identified significant markers, including S100A8, S100A12, and superoxide dismutase (SOD), in plasma that were higher in individuals with ARDS (97). S100A8/A9, a calcium-binding heterodimer predominantly located in neutrophils and monocytes, possesses pro-inflammatory and immunosuppressive characteristics and is integral to the etiology of sepsis. It is considered a potential biomarker for sepsis and its related organ damage (98). Therapeutic approaches targeting S100A8/A9 may help reduce tissue damage resulting from inflammation and enhance prognosis. The findings facilitated the creation of a proteomic signature that effectively distinguished between sepsis with and without ARDS, offering essential insights into the relevant immune systems and pinpointing potential therapeutic targets. Another large-scale study employed high-throughput mass spectrometry to examine plasma proteomes from over 1,600 sepsis patients, merging data with leukocyte transcriptomes to identify protein indicators and co-expression modules linked to sepsis severity (96). This study identified three distinct proteomic subgroups (SPC 1, SPC 2, and SPC 3) that were associated with clinical outcomes such as organ failure and death (96). For example, SPC 1 patients, who had the most severe sickness symptoms, had significantly higher mortality rates than the other subgroups (96). This classification stresses the importance of proteome profiling in identifying high-risk individuals and guiding treatment decisions based on the specifics of their sepsis phenotype.
Proteomics elucidates sepsis heterogeneity and facilitates the development of biomarkers that more accurately forecast disease progression and treatment outcomes. This establishes the foundation for individualized sepsis therapy that focuses on particular metabolic pathways. Proteomics may facilitate the identification of novel pharmacological targets, enhance patient classification, and optimize sepsis management, hence improving patient outcomes. Proteomics emphasizes functional molecules and proteins; nonetheless, it inadequately encompasses the biochemical dynamics of sepsis, which are crucial for understanding the systemic changes associated with the disease. Metabolomics bridges this gap by demonstrating the metabolic alterations associated with sepsis in a more dynamic manner.
Metabolomics examines metabolites, the end products of cellular processes, offering a snapshot of the biochemical changes during sepsis. Investigating metabolic alterations associated with sepsis phenotypes may reveal novel biomarkers for early identification, facilitate the prediction of organ failure, and enhance therapeutic strategies for specific patient subgroups, ultimately improving prognosis (99, 100). A 2015 study demonstrated the efficacy of metabolomics in the early identification of sepsis by differentiating between children requiring intensive care and those suitable for emergency room treatment (99). A combination of 14 metabolites and three protein mediators accurately indicated patients for ICU admission, highlighting the significance of metabolomics in shaping triage decisions and enhancing early intervention (99). While these findings are intriguing, they require validation in larger cohorts.
Current study has utilized metabolomics to identify organ dysfunction early in sepsis, in addition to detecting metabolic abnormalities. Lipid mediator profiling indicated that non-survivors exhibited elevated levels of inflammatory and pro-resolving mediators, correlating with severe respiratory failure and ARDS (3, 100–107). A 2021 study identified three distinct metabolic groups in patients with early sepsis and sepsis-related ARDS, each associated with different mortality rates linked to plasma lipid levels (107). Patients with high lipid levels exhibited the lowest mortality rates and a diminished risk of organ dysfunction (107). Notwithstanding these promising outcomes, the therapeutic efficacy of the notion remains uncertain, necessitating further validation. A recent study distinguished sepsis from septic shock by analyzing plasma metabolites within 48 hours of ICU admission, uncovering distinct metabolic patterns that may facilitate early diagnosis and treatment timing (108). Nonetheless, additional comprehensive validation studies and improvement of metabolite combinations remain necessary for early risk prediction.
Metabolomics has the potential to transform sepsis treatment by enhancing early detection, forecasting organ failure, and distinguishing phenotypic subgroups with varying reactions to medication. Similar to transcriptomics and proteomics, metabolomic data alone cannot adequately capture the complexity of sepsis phenotypes. Metabolomics reflects the biochemical changes occurring during sepsis, it does not account for the cellular processes and immune system dysfunction that drive disease progression. Cytomics, which focuses on the phenotypic and functional diversity of immune cells, is essential for completing the biological picture of sepsis phenotyping, providing deeper insights into immune dysregulation and aiding in the development of more personalized therapeutic strategies.
Cytomics investigates the phenotypic and functional heterogeneity of cells, especially immune cells, offering real-time insights on immunological dysregulation in sepsis. Advancements in single-cell RNA sequencing (scRNA-seq) and immune cell profiling have enhanced the understanding of the pathophysiology and heterogeneity of sepsis. Specific immune cell subsets and statuses, such as MS1 cells, identified as one of the four mononuclear states, have been linked to increased mortality and organ dysfunction in sepsis patients in research, serving as potential biomarkers for sepsis prediction and therapeutic intervention (83–85). Flow cytometry and other immune profiling methodologies enhance scRNA-seq by enabling more efficient and cost-effective assessments of immune dysfunction, which may be monitored in real-time to predict patient outcomes. For example, monocyte human leukocyte antigen-DR (mHLA-DR), part of MHC-II expressed by antigen-presenting cells, is commonly measured by flow cytometry (86–88). Studies have shown that changes in the mHLA-DR expression pattern are strongly associated with a poor outcome in individuals with septic shock (89–91). These findings underscore the significance of mHLA-DR as a predictive biomarker and highlight the necessity of incorporating data from several immunological markers to obtain a holistic understanding of immune responses in sepsis. Ultimately, although each omics method provides valuable insights, the integration of these diverse datasets is essential for comprehending the complete complexity of sepsis and formulating effective, phenotype-driven therapeutics.
Sepsis heterogeneity is observable by the integration of epigenomic, transcriptomic, proteomic, metabolomic, and cytomic data. Each omics layer elucidates the molecular causes of sepsis, although their integration is the most potent. Researchers can identify stable biomarkers, molecular layer interactions, and dynamic changes by integrating this information (109). Transcriptomics quantifies gene expression, proteomics analyzes post-translational changes, and metabolomics examines downstream metabolic processes. These data augment our comprehension of sepsis etiology at the systemic level. Multi-omics integration revolutionizes sepsis research. High-dimensional datasets necessitate the utilization of machine learning and network-based technologies for their integration and interpretation. The integration of multi-omics enhances the classification of sepsis subphenotypes, predictions of therapy responses, and the development of the illness. Multi-omics investigations have elucidated patterns of immune dysregulation and metabolic abnormalities in sepsis, facilitating tailored therapeutic approaches (109, 110). As these technologies advance, they have the potential to revolutionize sepsis management through early detection, targeted therapy, and improved long-term outcomes.
Understanding the variability in sepsis patient responses is essential for improving therapeutic outcomes, and AI and ML techniques are increasingly utilized to identify distinct sepsis subphenotypes based on clinical indicators (111, 112). These approaches offer novel mechanistic pathways, therapeutic targets, and biomarkers essential for developing sepsis-specific phenotype-driven therapies. For instance, unsupervised machine learning approaches such as cluster analysis allow for the classification of sepsis patients into subphenotypes without prior assumptions, utilizing clinical data such as vital signs, white blood cell counts, and biochemical markers (111, 113, 114). A 2022 study identified and validated several novel sepsis subphenotypes based on changes in vital signs over time (115). This multicenter retrospective study assessed vital signs, including body temperature, heart rate, respiratory rate, and blood pressure, in 20,729 hospitalized patients with suspected infections within the initial eight hours post-admission (115). Four sepsis subphenotypes (Groups A, B, C, and D) exhibited variations in clinical manifestations, laboratory findings, organ impairment, and prognosis (115). The secondary analysis of the SMART trial compared balanced crystalloids to normal saline in critically ill patients and demonstrated that subphenotypes influenced mortality outcomes (115, 116). Balanced crystalloids decreased mortality in Group D relative to normal saline (115). The observations indicate that sepsis subphenotypes characterized by easily obtainable vital signs may exhibit varied responses to pharmacological interventions, potentially impacting future clinical trials. Phenotype-driven therapeutic approaches customize interventions and enhance patient outcomes. Notwithstanding these advancements, the application of AI in sepsis diagnosis and treatment remains impeded by challenges such as subgroup selection and biases in training datasets (117–119). AI and ML can enhance sepsis classification and identify individualized patient response patterns, facilitating precision therapy. These approaches can revolutionize sepsis therapy by facilitating earlier, more precise actions that improve patient outcomes when integrated with genetic data.
Researchers have progressively employed various omics data (Figure 3) in conjunction with standard clinical and laboratory test data—such as white blood cell counts and biochemical markers—utilizing sophisticated big data algorithms and artificial intelligence methods to categorize sepsis phenotypes (120–123). These projects have produced substantial insights into the diversity of sepsis and its clinical implications. A 2015 study utilizing self-organizing maps (SOMs) and K-means clustering found four clinical phenotypes among 2,533 individuals with severe sepsis or septic shock, each with unique organ failure patterns and fatality rates (120). A 2020 study utilizing SOM and cluster analysis on the Medicine Information Database III (MIME-III) identified four distinct sepsis subtypes, each defined by specific organ failures, underscoring the necessity for personalized treatment strategies (124). A notable study published in 2019 using consensus K-means clustering to assess sepsis patients (n=20,189) from 12 medical institutions and three randomized clinical trials (n=4,737) (121). Within six hours of admission, 29 clinical and laboratory data points from electronic health records were employed to identify four distinct phenotypes (α, β, γ, and δ), each defined by specific clinical characteristics and associated mortality risks (121). The research examined 43,086 cases, involving 31,160 unique people, to evaluate phenotypic repeatability. Simulations of three randomized controlled trials demonstrated that altering the distribution of α and δ traits greatly affected trial outcomes (44, 121, 125, 126). This study highlights the significance of accounting for phenotypic dispersion in clinical trial design, as it can affect the efficacy of treatment interventions (121, 127, 128). This study underscores the need of identifying appropriate phenotypic classifications and administering prompt therapeutic interventions, as drugs may prove ineffective or harmful if not congruent with specific patient morphologies.
Nonetheless, machine learning methodologies exhibit a deficiency in transparency, and phenotypic consistency varies across studies. Subphenotypes are typically identified using cross-sectional clinical and biochemical data, which constrains our understanding of their temporal dynamics. Recent studies have examined dynamic sepsis subtyping according to the temporal progression of the disease (129–131). A 2022 study analyzed 642 patients across 20 hospitals, used 23 baseline clinical markers, and identified clustering through heat maps at 0, 6, 24, and 72 hours (129). Five organ failure symptoms (Types L1, L2, M, H1, and H2) exhibiting varied patterns and durations were validated in an independent sample of 381 individuals from 11 institutions (129). The two high-risk phenotypes, H1 and H2, had comparable mortality rates; however, there were significant discrepancies in baseline disease severity scores, clinical characteristics (including age and organ failure patterns), and plasma marker levels (129). Another study employed 72-hour sequential organ failure assessment (SOFA) score trajectories for sepsis patients, utilizing dynamic temporal warping (DTW) and hierarchical agglomerative clustering (HAC) to delineate subgroups based on trajectory data (130). The researchers constructed a random forest model to forecast subtype affiliation at 6 and 24 hours post-ICU admission (130). The study comprised 4,678 sepsis patients in the development cohort and 3,665, 12,282, and 4,804 patients in the validation cohorts. Four groups were identified: rapidly deteriorating, gradually declining, rapidly enhancing, and gradually improving. Distinct baselines, organ dysfunction, and clinical outcomes were present for each category. Although the rapidly deteriorating subtype exhibited a lower SOFA score at ICU admission (mean: 4.5) compared to the rapidly improving subtype (mortality rate: 5.5%, mean SOFA: 5.5), it nonetheless had the highest in-hospital mortality rate at 28.3% (130). This research identified four unique sepsis subphenotypes, each exhibiting distinct natural histories, illustrating host-pathogen interactions throughout standard treatment. Comprehending these dynamic trajectories is essential for formulating and forecasting clinical trial interventions, as it uncovers discrepancies in natural history and treatment effectiveness. Other studies employed population-based trajectory modeling to classify sepsis according to variations in body temperature, uncovering groupings with markedly distinct inflammatory profiles and fatality rates (131, 132). These strategies enhance our comprehension of sepsis progression and facilitate the development of more tailored therapeutic choices.
Alongside standard clinical indicators, phenotypic assessments encompass coagulation profiles and hemodynamic evaluations. K-means clustering was employed in machine learning including 3,694 patients from three Japanese multicenter studies conducted in 2021 (133). The research examined the reactions of different phenotypes to recombinant human thrombomodulin (rhTM) injections and the subsequent clinical outcomes (133). Four distinct coagulation phenotypes were identified: dA, dB, dC, and dD. In the dA phenotypic group, rhTM therapy decreased 28-day and in-hospital mortality, although the absence of standardization or randomization (133). These findings highlight the therapeutic potential of phenotype-driven therapies and the importance of establishing successful therapy windows. Current research faces issues in repeatability and transparency in machine learning approaches. However, collected evidence suggests that phenotype-based interventions may have a considerable impact on sepsis therapy (134–137). Integrating multi-omics data with dynamic clinical signs, such as alterations in inflammatory markers or hemodynamic parameters, is expected to enhance phenotypic classification. Longitudinal tracking of clinical changes enables researchers to associate genetic findings with disease progression and get a deeper understanding of sepsis features. These advancements facilitate the identification of novel biomarkers and therapeutic targets for sepsis subphenotypes, enhancing patient outcomes and customized treatment.
Sepsis, akin to other intricate diseases, exhibits heterogeneity, complicating treatment and diminishing the effectiveness of various research trials (94, 138). Distinguishing distinct biological phenotypes that may exhibit varied responses to therapies is essential for enhancing therapeutic outcomes (129, 139–144). Two principal subphenotypes in sepsis—hyperinflammatory and hypoinflammatory—have shown significant in forecasting disease progression, treatment responses, and patient outcomes (109, 145–147).
These two inflammatory types were accurately identified in numerous sepsis cohorts by latent class analysis (LCA) of clinical and biomarker data (145). The hyperinflammatory phenotype is characterized by elevated proinflammatory cytokines, utilization of vasoactive medications, heightened risk of bacteremia, and increased death rate (145). The hypoinflammatory phenotype is characterized by diminished inflammatory markers, a lower risk of bacteremia, and an improved prognosis (145). Clinical and treatment responses differ by phenotype, suggesting that phenotype-specific strategies may enhance patient outcomes. The secondary analysis of the PROWESS SHOCK trial (N=1680) assessing recombinant human activated protein C (drotrecogin alfa) in septic shock revealed that hyperinflammatory patients exhibited superior survival rates compared to hypoinflammatory patients, who demonstrated diminished survival rates (40, 145). No treatment interactions were identified in the secondary analysis of the VASST trial, which compared norepinephrine to early antidiuretic hormone therapy in cases of septic shock (145, 148). Nevertheless, distinctions consistent with other cohort studies emphasize the significance of phenotype in therapeutic classification.
Further investigation has examined the impact of these variables on mortality associated with sepsis-related ARDS (146). Hypoinflammatory patients predominantly succumbed to respiratory failure, while hyperinflammatory patients primarily perished from circulatory failure (146). The data indicate that addressing individual inflammatory profiles may enhance treatment customization and effectiveness. Genomic and microbiomic data suggest that hyperinflammatory individuals have elevated bacterial numbers and immunological activity, impacting metabolic and T cell response genes (149). Researchers employed this methodology on VANISH trial participants with transcriptome data (N=117), classifying them as hypoinflammatory or hyperinflammatory and evaluating phenotype-specific treatment benefits by logistic regression models (43, 149). Steroid medication influenced various phenotypes differently, with hypoinflammatory patients perhaps encountering adverse outcomes (149). The diverse sepsis phenotypes underscore the necessity of tailored treatment according to clinical severity and genetic factors. Phenotype-based predictive enrichment in clinical trials is advantageous as hyperinflammatory and hypoinflammatory phenotypes respond differentially to treatment.
Classifying patients based on their inflammatory profiles may aid in identifying individuals who benefit most from various medications, thereby improving the efficacy and safety of sepsis treatment. Defining and characterizing sepsis hyperinflammatory and hypoinflammatory subphenotypes clarifies clinical heterogeneity and facilitates phenotype-guided therapy. Examining biomarkers and therapeutic targets related to these traits could improve patient outcomes, inform clinical decision-making, and increase the effectiveness of sepsis treatment.
Effective fluid management is essential in sepsis and septic shock to enhance hemodynamic stability and organ perfusion (28). A phenotype-driven strategy for fluid therapy is essential, since both excessive and inadequate fluid resuscitation can adversely affect patient outcomes. Research indicates that fluid resuscitation affects mortality risk and recovery across many sepsis subtypes (150, 151). A 2018 Latent Profile Analysis of 14,993 sepsis patients showed four categories regarding fluid balance and mortality risk (150). The results indicated that responses to fluid resuscitation varied markedly among different morphologies. Subtype 2 got a lesser volume of fluid compared to subtype 3, which necessitated a greater volume within 24 hours. Notably, augmenting fluid administration during the initial 48 hours elevated mortality in subtype 4, while concurrently reducing mortality in subtype 3, which exhibited hallmarks of circulatory shock, including diminished mean arterial pressure and heightened vasopressor requirements (150). These findings underscore the necessity of tailoring fluid resuscitation to the individual characteristics of each patient.
A dynamic treatment regime (DTR) model enhanced fluid resuscitation for septic shock in 2021. More than 1,400 patients were assessed to delineate five phenotypes, ranging from the most severely ill (Phenotype 2) to the healthiest (Phenotype 5) (152). Early, vigorous fluid resuscitation succeeded by a de-resuscitation period to mitigate fluid excess was determined to be the optimal fluid management strategy. Phenotype-specific fluid quantities and norepinephrine dosages enhanced mortality outcomes, with certain patients gaining advantages from early administration, while others, particularly the critically ill, benefitted from delayed treatment (152). This study indicates that treatment for sepsis should be tailored to its varied phenotypes. A 2022 study investigated fluid balance in patients with clinical sepsis to validate these findings (151). Patients were classified into categories of multiple organ failure (MOF), respiratory dysfunction (RD), nervous system dysfunction (ND), and others (151). The data indicated considerable phenotypic differences in fluid balance and mortality. MOF exhibited the highest mortality rate at 48.4%, whilst OP demonstrated the lowest at 13.7% (151). At different time intervals post-therapy, MOF and ND patients exhibited the highest fluid volumes balances, indicating that fluid management must be customized for each phenotype.
A recent study proposed utilizing transcriptome data to guide fluid management in septic shock patients through a ‘benefit score’ (109). A comprehensive review of multi-omics data from 494 septic shock patients revealed distinct subgroups with varying responses to fluid therapy strategies and established a “benefit score” derived from transcriptomic data and a proteomic signature to assist clinicians in customizing fluid management approaches (109). This study employs a systems biology approach utilizing multi-omics data to enhance our comprehension of sepsis heterogeneity and to offer novel strategies for personalized treatment. These findings are valuable; nevertheless, prospective validation, rapid detection methods, and cost-effectiveness analysis are necessary prior to their implementation in clinical practice.
These studies underscore the importance of accurate fluid management in sepsis treatment, along with comprehending the molecular and clinical diversity of sepsis phenotypes. Identifying and targeting phenotype-specific fluid resuscitation methods enables the optimization of treatment, diminishes complications, and ultimately enhances patient outcomes in sepsis and septic shock.
Investigations into sepsis subphenotypes have yielded essential understanding of the disease’s heterogeneity and shown the potential for phenotype-directed therapeutic strategies. The results underscore the need for additional validation and enhancement in converting subphenotypic insights into effective treatment strategies that enhance patient outcomes. A deeper comprehension of sepsis heterogeneity is essential for developing phenotype-specific treatments. Current subphenotypic research are significant, but they are limited by a lack of standardized validation across diverse demographics and clinical circumstances. Numerous studies depend on retrospective or cohort data (Table 3); nevertheless, prospective research is crucial for creating effective, real-time phenotyping instruments that can immediately impact treatment decision-making (153–155). Clinical trials have shown that subphenotypes are associated with different treatment responses, emphasizing the necessity of screening these phenotypes ahead of time to optimize therapy and enhance prognosis. Illustrations from various domains exemplify the pragmatic efficacy of phenotype-guided therapy. In a chronic pain management study, 60 patients were randomly divided into two groups: one receiving opioid medicine guided by CYP2D6, μ-opioid receptor (OPRM1), and catechol-O-methyltransferase (COMT) genotyping, and the other receiving standard care (156). The genotype-guided group experienced significantly better pain relief and quality of life compared to the control group. This case underscores the significance of integrating genetic and phenotypic data to enhance therapeutic outcomes. Similar strategies could be adapted for sepsis management, emphasizing the need for prospective studies to evaluate their applicability.
Sepsis phenotyping has several challenges, including data heterogeneity, validation difficulties, and the integration of emerging techniques such as ML and AI. The variability in patient demographics, clinical environments, and sampling methods restricts the generalizability and reliability of phenotyping studies (157, 158). Establishing uniform terminology such as “phenotype,” “subphenotype,” and “subgroup” is essential for enhancing clarity and collaboration. Standardized methods, precise nomenclature, and cohesive research methodologies are essential to resolve overlaps among subphenotypes and enhance classification systems for precision medicine.
A notable challenge is the limited accessibility of contemporary multi-omics technologies, attributed to elevated costs and technical intricacies, especially in resource-limited settings (1). Phenotypic models should be tested across diverse populations to guarantee their global applicability and fairness in sepsis research. Interdisciplinary collaboration integrating clinical, biological, and computational expertise is crucial for synthesizing diverse datasets and improving phenotype-guided methodologies. International efforts to standardize procedures and disseminate data can accelerate discoveries and facilitate access to advanced methodologies.
Emerging tools, including machine learning and artificial intelligence, can identify underlying diversity in treatment responses, even within large studies that yield neutral results (130, 159–161). Despite the potential of machine learning and artificial intelligence to find variability in treatment responses and uncover novel biomarkers, obstacles such as insufficient transparency, scalability issues, and challenges in integrating into clinical workflows hinder their widespread adoption (162, 163). Developing economical, interpretable AI models and scalable platforms is essential for practical implementation. Moreover, robust ethical frameworks for data protection and governance are essential for the proper utilization of patient data. Surmounting these obstacles will advance sepsis phenotyping, facilitating more tailored therapies and improved patient outcomes.
A comprehensive understanding of sepsis requires extending beyond blood-based evaluations to incorporate cross-organ system analyses that elucidate the complex interactions influencing disease progression (164). Although omics technologies provide significant molecular insights, the gap between these findings and clinical results highlights the necessity for more thorough phenotypic evaluations. Sepsis often impacts many organ systems, resulting in differing levels of dysfunction; therefore, it is essential to investigate gene expression, protein function, and metabolite regulation across tissues (13). Broadening phenotypic studies to encompass organ-specific abnormalities, particularly in the lungs, liver, and kidneys, will enhance our comprehension of sepsis etiology and facilitate the creation of therapies aimed at organ-specific harm (20, 165, 166). Future study ought to integrate multi-omics methodologies with cross-organ studies to develop a whole comprehension of sepsis and propel advancements in detection and treatment.
The evolving characteristics of sepsis phenotypes present both challenges and opportunities for precision treatment (167). Determining the optimal intervention window and tracking phenotypic changes is critical for tailoring therapy to individual patients and improving long-term outcomes (Figure 4). Nevertheless, the majority of contemporary research employs single time-point cross-sectional data, which constrains their ability to differentiate between ephemeral chemical states and persistent phenotypic characteristics. For example, phenotypes identified in early hyperinflammatory stages may shift to immunosuppressive states during later stages of sepsis progression (147). Without longitudinal sampling, it is challenging to distinguish transient changes from stable molecular characteristics, potentially leading to misclassification. This limitation raises the concern that current subtyping studies may reflect transient stages of sepsis progression rather than stable, distinct patient subtypes. longitudinal sampling, coupled with multi-omics integration, can surmount these limitations by monitoring molecular and clinical markers over time. This method facilitates the discovery of stable phenotypes, improves categorization systems, and directs the development of stage-specific therapies. Emphasizing dynamic studies will allow researchers to customize treatment strategies to the evolving characteristics of sepsis, enhancing patient outcomes and advancing the field of precision medicine.
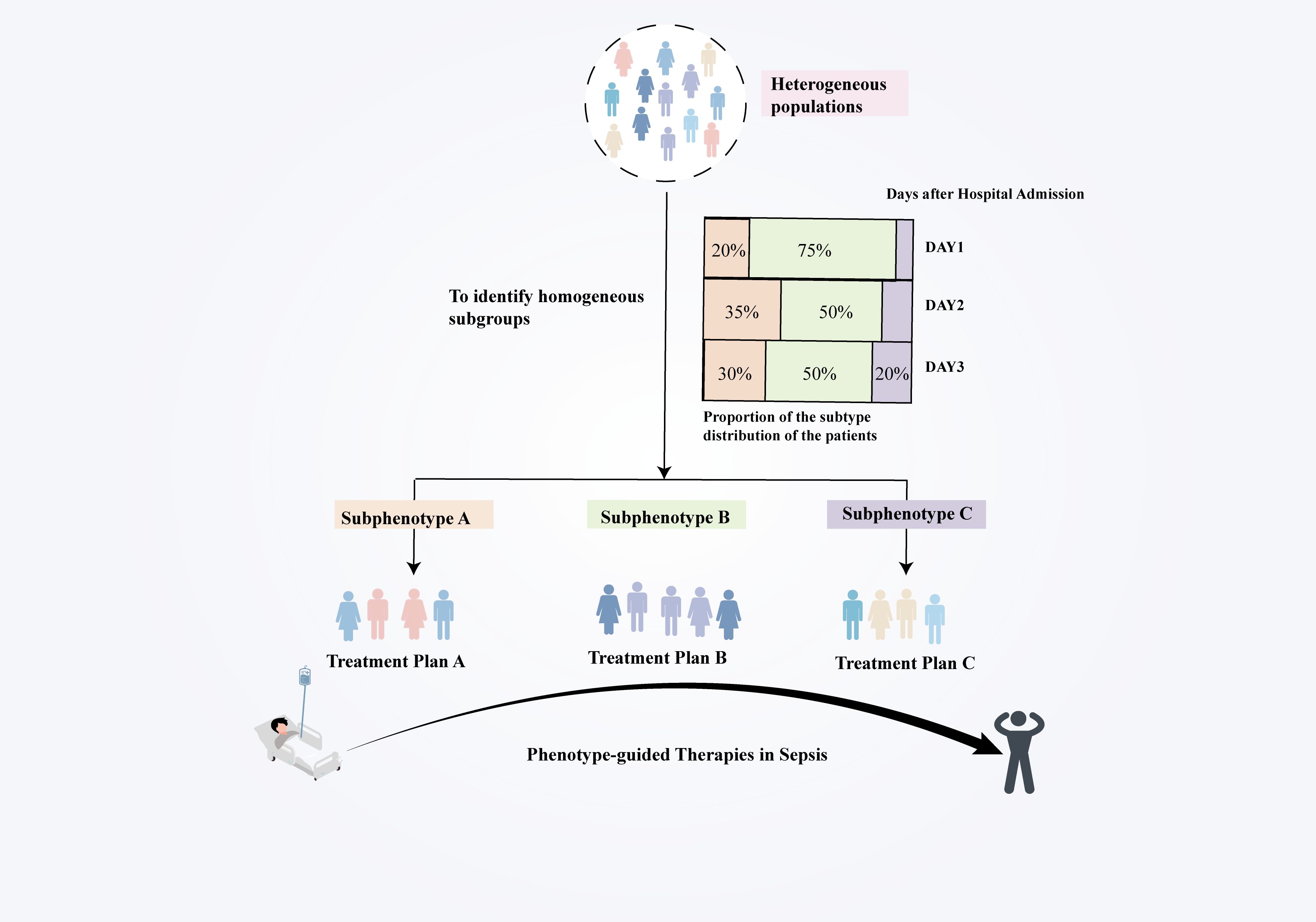
Figure 4. Dynamic subtyping and real-time decision-making address sepsis heterogeneity by combining clinical, biomarker, and molecular data to identify more homogeneous subpopulations. This methodology promotes phenotype-driven medicines, facilitates the discovery of new markers and targets through various treatment responses, and improves patient outcomes by providing accurate pharmaceuticals to the appropriate individuals at the right time.
Understanding the heterogeneity of sepsis is pivotal for advancing phenotype-based therapeutic strategies and improving patient outcomes. This review synthesizes advances in clustering algorithms, multi-omics technologies, and artificial intelligence, providing a comprehensive framework for identifying distinct sepsis subtypes with varied treatment responses. These insights lay the groundwork for discovering novel biomarkers and therapeutic targets, underscoring the transformative potential of precision medicine to tailor treatments based on phenotypic diversity. However, challenges such as phenotyping standardization, dynamic data integration, and clinical translation must be addressed through interdisciplinary collaboration and innovative research. By bridging these gaps, future efforts can fully implement phenotype-driven therapies, transforming sepsis management, enhancing survival rates, and establishing a new paradigm for precision medicine in complex diseases.
XZ: Data curation, Investigation, Validation, Visualization, Writing – original draft, Writing – review & editing. WZ: Conceptualization, Formal analysis, Funding acquisition, Project administration, Resources, Supervision, Writing – review & editing. HZ: Writing – review & editing, Data curation, Investigation. XL: Conceptualization, Formal analysis, Funding acquisition, Project administration, Resources, Software, Supervision, Writing – review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by the National key research and development plan of China (grant number 2022YFC2009804).
All authors gave thanks to the West China Hospital, Sichuan University and West China Tianfu Hospital for data supply that made the study possible.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declare that no Generative AI was used in the creation of this manuscript.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
AI, artificial intelligence; BHSD1, beta-hydroxysteroid dehydrogenase 1; ARDS, acute respiratory distress syndrome; COMT, catechol-O-methyltransferase; DEGs, differentially expressed genes; DTW, dynamic time warping; DTR, dynamic treatment regime; DAMPs, damage-associated molecular patterns; DIC, disseminated intravascular coagulation; GLCCI1, Glucocorticoid-induced transcript 1; HAC, hierarchical agglomerative clustering; ICUs, intensive care unit; LCA, latent class analysis; mHLA-DR, monocyte human leukocyte antigen-DR; MIME-III, Medicine Information Database III; MHC, major histocompatibility complex; mRNA, messengerRNA; MAS, macrophage activation syndrome; ML, Machine learning; OPRM1, μ-opioid receptor; PAMPs, Pathogen-associated molecular patterns; qRT-PCR, quantitative real-time PCR; rhTM, recombinant human thrombomodulin; RCTs, randomized clinical trials; SOFA, sequential organ failure assessment; SRS, sepsis response signature; scRNA-seq, single-cell RNA sequencing; SOD, superoxide dismutase; SOMs, self-organizing maps.
1. Rudd KE, Johnson SC, Agesa KM, Shackelford KA, Tsoi D, Kievlan DR, et al. Global, regional, and national sepsis incidence and mortality, 1990-2017: Analysis for the global burden of disease study. Lancet (2020) 395(10219):200–11. doi: 10.1016/S0140-6736(19)32989-7
2. Sakr Y, Jaschinski U, Wittebole X, Szakmany T, Lipman J, Namendys-Silva SA, et al. Sepsis in intensive care unit patients: Worldwide data from the intensive care over nations audit. Open Forum Infect Dis (2018) 5(12):ofy313. doi: 10.1093/ofid/ofy313
3. Singer M, Deutschman CS, Seymour CW, Shankar-Hari M, Annane D, Bauer M, et al. The third international consensus definitions for sepsis and septic shock (Sepsis-3). Jama (2016) 315(8):801–10. doi: 10.1001/jama.2016.0287
4. Levy MM, Artigas A, Phillips GS, Rhodes A, Beale R, Osborn T, et al. Outcomes of the surviving sepsis campaign in intensive care units in the USA and europe: A prospective cohort study. Lancet Infect Dis (2012) 12(12):919–24. doi: 10.1016/S1473-3099(12)70239-6
5. Leligdowicz A, Matthay MA. Heterogeneity in sepsis: New biological evidence with clinical applications. Crit Care (London England) (2019) 23(1):80. doi: 10.1186/s13054-019-2372-2
6. van der Poll T, Shankar-Hari M, Wiersinga WJ. The immunology of sepsis. Immunity (2021) 54(11):2450–64. doi: 10.1016/j.immuni.2021.10.012
7. Gotts JE, Matthay MA. Sepsis: Pathophysiology and clinical management. BMJ (Clinical Res ed) (2016) 353:i1585. doi: 10.1136/bmj.i1585
8. Gyawali B, Ramakrishna K, Dhamoon AS. Sepsis: The evolution in definition, pathophysiology, and management. SAGE Open Med (2019) 7:2050312119835043. doi: 10.1177/2050312119835043
9. Pickkers P, Kox M. Towards precision medicine for sepsis patients. Crit Care (London England) (2017) 21(1):11. doi: 10.1186/s13054-016-1583-z
10. Prescott HC, Angus DC. Enhancing recovery from sepsis: A review. Jama (2018) 319(1):62–75. doi: 10.1001/jama.2017.17687
11. Drawnel FM, Zhang JD, Kung E, Aoyama N, Benmansour F, Araujo Del Rosario A, et al. Molecular phenotyping combines molecular information, biological relevance, and patient data to improve productivity of early drug discovery. Cell Chem Biol (2017) 24(5):624–34 e3. doi: 10.1016/j.chembiol.2017.03.016
12. Morelli A, Passariello M. Hemodynamic coherence in sepsis. Best Pract Res Clin Anaesthesiol (2016) 30(4):453–63. doi: 10.1016/j.bpa.2016.10.009
13. Lelubre C, Vincent JL. Mechanisms and treatment of organ failure in sepsis. Nat Rev Nephrol (2018) 14(7):417–27. doi: 10.1038/s41581-018-0005-7
14. Giustozzi M, Ehrlinder H, Bongiovanni D, Borovac JA, Guerreiro RA, Gasecka A, et al. Coagulopathy and sepsis: Pathophysiology, clinical manifestations and treatment. Blood Rev (2021) 50:100864. doi: 10.1016/j.blre.2021.100864
15. Assinger A, Schrottmaier WC, Salzmann M, Rayes J. Platelets in sepsis: An update on experimental models and clinical data. Front Immunol (2019) 10:1687. doi: 10.3389/fimmu.2019.01687
16. Fan TH, Premraj L, Roberts J, Lydston M, Robba C, Hager D, et al. In-hospital neurologic complications, neuromonitoring, and long-term neurologic outcomes in patients with sepsis: A systematic review and meta-analysis. Crit Care Med (2024) 52(3):452–63. doi: 10.1097/CCM.0000000000006096
17. Sekino N, Selim M, Shehadah A. Sepsis-associated brain injury: Underlying mechanisms and potential therapeutic strategies for acute and long-term cognitive impairments. J Neuroinflamm (2022) 19(1):101. doi: 10.1186/s12974-022-02464-4
18. Gattinoni L, Vasques F, Camporota L, Meessen J, Romitti F, Pasticci I, et al. Understanding lactatemia in human sepsis. potential impact for early management. Am J Respir Crit Care Med (2019) 200(5):582–9. doi: 10.1164/rccm.201812-2342OC
19. Willmann K, Moita LF. Physiologic disruption and metabolic reprogramming in infection and sepsis. Cell Metab (2024) 36(5):927–46. doi: 10.1016/j.cmet.2024.02.013
20. Qiao X, Yin J, Zheng Z, Li L, Feng X. Endothelial cell dynamics in sepsis-induced acute lung injury and acute respiratory distress syndrome: Pathogenesis and therapeutic implications. Cell Commun Signal (2024) 22(1):241. doi: 10.1186/s12964-024-01620-y
21. Peerapornratana S, Manrique-Caballero CL, Gomez H, Kellum JA. Acute kidney injury from sepsis: Current concepts, epidemiology, pathophysiology, prevention and treatment. Kidney Int (2019) 96(5):1083–99. doi: 10.1016/j.kint.2019.05.026
22. Strnad P, Tacke F, Koch A, Trautwein C. Liver - guardian, modifier and target of sepsis. Nat Rev Gastroenterol Hepatol (2017) 14(1):55–66. doi: 10.1038/nrgastro.2016.168
23. Shivade C, Raghavan P, Fosler-Lussier E, Embi PJ, Elhadad N, Johnson SB, et al. A review of approaches to identifying patient phenotype cohorts using electronic health records. J Am Med Inform Assoc (2014) 21(2):221–30. doi: 10.1136/amiajnl-2013-001935
24. Wong HR, Cvijanovich N, Lin R, Allen GL, Thomas NJ, Willson DF, et al. Identification of pediatric septic shock subclasses based on genome-wide expression profiling. BMC Med (2009) 7(1):34. doi: 10.1186/1741-7015-7-34
25. Aerts HJ, Velazquez ER, Leijenaar RT, Parmar C, Grossmann P, Carvalho S, et al. Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach. Nat Commun (2014) 5:4006. doi: 10.1038/ncomms5006
26. Sadanandam A, Lyssiotis CA, Homicsko K, Collisson EA, Gibb WJ, Wullschleger S, et al. A colorectal cancer classification system that associates cellular phenotype and responses to therapy. Nat Med (2013) 19(5):619–25. doi: 10.1038/nm.3175
27. Cicchinelli S, Pignataro G, Gemma S, Piccioni A, Picozzi D, Ojetti V, et al. Pamps and damps in sepsis: A review of their molecular features and potential clinical implications. Int J Mol Sci (2024) 25(2). doi: 10.3390/ijms25020962
28. Cecconi M, Evans L, Levy M, Rhodes A. Sepsis and septic shock. Lancet (2018) 392(10141):75–87. doi: 10.1016/S0140-6736(18)30696-2
29. Angus DC, van der Poll T. Severe sepsis and septic shock. N Engl J Med (2013) 369(9):840–51. doi: 10.1056/NEJMra1208623
30. Hotchkiss RS, Monneret G, Payen D. Sepsis-induced immunosuppression: From cellular dysfunctions to immunotherapy. Nat Rev Immunol (2013) 13(12):862–74. doi: 10.1038/nri3552
31. Zaborin A, Smith D, Garfield K, Quensen J, Shakhsheer B, Kade M, et al. Membership and behavior of ultra-Low-Diversity pathogen communities present in the gut of humans during prolonged critical illness. mBio (2014) 5(5):e01361–14. doi: 10.1128/mBio.01361-14
32. Andersson U, Tracey KJ. Reflex principles of immunological homeostasis. Annu Rev Immunol (2012) 30(1):313–35. doi: 10.1146/annurev-immunol-020711-075015
33. Yang L, Xie M, Yang M, Yu Y, Zhu S, Hou W, et al. Pkm2 regulates the warburg effect and promotes Hmgb1 release in sepsis. Nat Commun (2014) 5(1):4436. doi: 10.1038/ncomms5436
34. Carson WF, Cavassani KA, Dou Y, Kunkel SL. Epigenetic regulation of immune cell functions during post-septic immunosuppression. Epigenetics (2011) 6(3):273–83. doi: 10.4161/epi.6.3.14017
35. Krumholz HM. Post-hospital syndrome–an acquired, transient condition of generalized risk. N Engl J Med (2013) 368(2):100–2. doi: 10.1056/NEJMp1212324
36. Scales DC, Fischer HD, Li P, Bierman AS, Fernandes O, Mamdani M, et al. Unintentional continuation of medications intended for acute illness after hospital discharge: A population-based cohort study. J Gen Intern Med (2016) 31(2):196–202. doi: 10.1007/s11606-015-3501-5
37. Wang W, Liu CF. Sepsis heterogeneity. World J Pediatr (2023) 19(10):919–27. doi: 10.1007/s12519-023-00689-8
38. Sprung CL, Annane D, Keh D, Moreno R, Singer M, Freivogel K, et al. Hydrocortisone therapy for patients with septic shock. N Engl J Med (2008) 358(2):111–24. doi: 10.1056/NEJMoa071366
39. Fujii T, Salanti G, Belletti A, Bellomo R, Carr A, Furukawa TA, et al. Effect of adjunctive vitamin C, glucocorticoids, and vitamin B1 on longer-term mortality in adults with sepsis or septic shock: A systematic review and a component network meta-analysis. Intensive Care Med (2022) 48(1):16–24. doi: 10.1007/s00134-021-06558-0
40. Ranieri VM, Thompson BT, Barie PS, Dhainaut JF, Douglas IS, Finfer S, et al. Drotrecogin alfa (Activated) in adults with septic shock. N Engl J Med (2012) 366(22):2055–64. doi: 10.1056/NEJMoa1202290
41. Shakoory B, Carcillo JA, Chatham WW, Amdur RL, Zhao H, Dinarello CA, et al. Interleukin-1 receptor blockade is associated with reduced mortality in sepsis patients with features of macrophage activation syndrome: Reanalysis of a prior phase iii trial. Crit Care Med (2016) 44(2):275–81. doi: 10.1097/CCM.0000000000001402
42. Investigators R-C, Gordon AC, Mouncey PR, Al-Beidh F, Rowan KM, Nichol AD, et al. Interleukin-6 receptor antagonists in critically ill patients with covid-19. N Engl J Med (2021) 384(16):1491–502. doi: 10.1056/NEJMoa2100433
43. Gordon AC, Mason AJ, Thirunavukkarasu N, Perkins GD, Cecconi M, Cepkova M, et al. Effect of early vasopressin vs norepinephrine on kidney failure in patients with septic shock: The vanish randomized clinical trial. Jama (2016) 316(5):509–18. doi: 10.1001/jama.2016.10485
44. Pro CI, Yealy DM, Kellum JA, Huang DT, Barnato AE, Weissfeld LA, et al. A randomized trial of protocol-based care for early septic shock. N Engl J Med (2014) 370(18):1683–93. doi: 10.1056/NEJMoa1401602
45. Young P, Bailey M, Beasley R, Henderson S, Mackle D, McArthur C, et al. Effect of a buffered crystalloid solution vs saline on acute kidney injury among patients in the intensive care unit: The split randomized clinical trial. Jama (2015) 314(16):1701–10. doi: 10.1001/jama.2015.12334
46. Venkatesh B, Finfer S, Cohen J, Rajbhandari D, Arabi Y, Bellomo R, et al. Adjunctive glucocorticoid therapy in patients with septic shock. N Engl J Med (2018) 378(9):797–808. doi: 10.1056/NEJMoa1705835
47. Sawyer RG, Claridge JA, Nathens AB, Rotstein OD, Duane TM, Evans HL, et al. Trial of short-course antimicrobial therapy for intraabdominal infection. N Engl J Med (2015) 372(21):1996–2005. doi: 10.1056/NEJMoa1411162
48. Harris PNA, Tambyah PA, Lye DC, Mo Y, Lee TH, Yilmaz M, et al. Effect of piperacillin-tazobactam vs meropenem on 30-day mortality for patients with e coli or klebsiella pneumoniae bloodstream infection and ceftriaxone resistance: A randomized clinical trial. Jama (2018) 320(10):984–94. doi: 10.1001/jama.2018.12163
49. Li HK, Rombach I, Zambellas R, Walker AS, McNally MA, Atkins BL, et al. Oral versus intravenous antibiotics for bone and joint infection. N Engl J Med (2019) 380(5):425–36. doi: 10.1056/NEJMoa1710926
50. Wong HR, Wheeler DS, Tegtmeyer K, Poynter SE, Kaplan JM, Chima RS, et al. Toward a clinically feasible gene expression-based subclassification strategy for septic shock: Proof of concept. Crit Care Med (2010) 38(10):1955–61. doi: 10.1097/CCM.0b013e3181eb924f
51. Wong HR, Cvijanovich NZ, Allen GL, Thomas NJ, Freishtat RJ, Anas N, et al. Validation of a gene expression-based subclassification strategy for pediatric septic shock. Crit Care Med (2011) 39(11):2511–7. doi: 10.1097/CCM.0b013e3182257675
52. Wong HR, Cvijanovich NZ, Anas N, Allen GL, Thomas NJ, Bigham MT, et al. Developing a clinically feasible personalized medicine approach to pediatric septic shock. Am J Respir Crit Care Med (2015) 191(3):309–15. doi: 10.1164/rccm.201410-1864OC
53. Taylor SP, Bray BC, Chou SH, Burns R, Kowalkowski MA. Clinical subtypes of sepsis survivors predict readmission and mortality after hospital discharge. Ann Am Thorac Soc (2022) 19(8):1355–63. doi: 10.1513/AnnalsATS.202109-1088OC
54. Puthucheary ZA, Gensichen JS, Cakiroglu AS, Cashmore R, Edbrooke L, Heintze C, et al. Implications for post critical illness trial design: Sub-phenotyping trajectories of functional recovery among sepsis survivors. Crit Care (London England) (2020) 24(1):577. doi: 10.1186/s13054-020-03275-w
55. Soussi S, Sharma D, Juni P, Lebovic G, Brochard L, Marshall JC, et al. Identifying clinical subtypes in sepsis-survivors with different one-year outcomes: A secondary latent class analysis of the frog-icu cohort. Crit Care (London England) (2022) 26(1):114. doi: 10.1186/s13054-022-03972-8
56. Yang R, Han D, Zhang L, Huang T, Xu F, Zheng S, et al. Analysis of the correlation between the longitudinal trajectory of sofa scores and prognosis in patients with sepsis at 72 hour after admission based on group trajectory modeling. J Intensive Med (2022) 2(1):39–49. doi: 10.1016/j.jointm.2021.11.001
57. Iwashyna TJ, Ely EW, Smith DM, Langa KM. Long-term cognitive impairment and functional disability among survivors of severe sepsis. Jama (2010) 304(16):1787–94. doi: 10.1001/jama.2010.1553
58. Herridge MS, Tansey CM, Matte A, Tomlinson G, Diaz-Granados N, Cooper A, et al. Functional disability 5 years after acute respiratory distress syndrome. N Engl J Med (2011) 364(14):1293–304. doi: 10.1056/NEJMoa1011802
59. Boede M, Gensichen JS, Jackson JC, Eissler F, Lehmann T, Schulz S, et al. Trajectories of depression in sepsis survivors: An observational cohort study. Crit Care (London England) (2021) 25(1):161. doi: 10.1186/s13054-021-03577-7
60. Schmidt K, Worrack S, Von Korff M, Davydow D, Brunkhorst F, Ehlert U, et al. Effect of a primary care management intervention on mental health-related quality of life among survivors of sepsis: A randomized clinical trial. Jama (2016) 315(24):2703–11. doi: 10.1001/jama.2016.7207
61. Schmidt KF, Schwarzkopf D, Baldwin LM, Brunkhorst FM, Freytag A, Heintze C, et al. Long-term courses of sepsis survivors: Effects of a primary care management intervention. Am J Med (2020) 133(3):381–5 e5. doi: 10.1016/j.amjmed.2019.08.033
62. Schmidt K, Thiel P, Mueller F, Schmuecker K, Worrack S, Mehlhorn J, et al. Sepsis survivors monitoring and coordination in outpatient health care (Smooth): Study protocol for a randomized controlled trial. Trials (2014) 15:283. doi: 10.1186/1745-6215-15-283
63. Cai D, Greco M, Wu Q, Cheng Y. Sepsis-induced coagulopathy subphenotype identification by latent class analysis. Balkan Med J (2023) 40(4):244–51. doi: 10.4274/balkanmedj.galenos.2023.2023-4-6
64. Huang JZ, Ng MK, Rong H, Li Z. Automated variable weighting in k-means type clustering. IEEE Trans Pattern Anal Mach Intell (2005) 27(5):657–68. doi: 10.1109/TPAMI.2005.95
65. Liu G, Wu R, He J, Xu Y, Han L, Yu Y, et al. Clinical phenotyping of septic shock with latent profile analysis: A retrospective multicenter study. J Crit Care (2025) 85:154932. doi: 10.1016/j.jcrc.2024.154932
66. Wang K, Liu Q, Mo S, Zheng K, Li X, Li J, et al. A decision tree model to help treatment decision-making for severe spontaneous intracerebral hemorrhage. Int J Surg (2024) 110(2):788–98. doi: 10.1097/JS9.0000000000000852
67. Sun B, Lei M, Wang L, Wang X, Li X, Mao Z, et al. Prediction of sepsis among patients with major trauma using artificial intelligence: A multicenter validated cohort study. Int J Surg (2024) 111(1):467–80. doi: 10.1097/JS9.0000000000001866
68. Michelson AP, Oh I, Gupta A, Puri V, Kreisel D, Gelman AE, et al. Developing machine learning models to predict primary graft dysfunction after lung transplantation. Am J Transplant (2024) 24(3):458–67. doi: 10.1016/j.ajt.2023.07.008
69. Dieckhaus H, Brocidiacono M, Randolph NZ, Kuhlman B. Transfer learning to leverage larger datasets for improved prediction of protein stability changes. Proc Natl Acad Sci USA (2024) 121(6):e2314853121. doi: 10.1073/pnas.2314853121
70. van den Berg MAM, Medina O, Loohuis IIP, van der Flier MM, Dudink JJ, Benders M, et al. Development and clinical impact assessment of a machine-learning model for early prediction of late-onset sepsis. Comput Biol Med (2023) 163:107156. doi: 10.1016/j.compbiomed.2023.107156
71. Bhavani SV, Xiong L, Pius A, Semler M, Qian ET, Verhoef PA, et al. Comparison of time series clustering methods for identifying novel subphenotypes of patients with infection. J Am Med Inform Assoc (2023) 30(6):1158–66. doi: 10.1093/jamia/ocad063
72. Wang Y, Li X, Wong KC, Chang Y, Yang S. Evolutionary multiobjective clustering algorithms with ensemble for patient stratification. IEEE Trans Cybern (2022) 52(10):11027–40. doi: 10.1109/TCYB.2021.3069434
73. Kijpaisalratana N, Sanglertsinlapachai D, Techaratsami S, Musikatavorn K, Saoraya J. Machine learning algorithms for early sepsis detection in the emergency department: A retrospective study. Int J Med Inform (2022) 160:104689. doi: 10.1016/j.ijmedinf.2022.104689
74. De Zuani M, Hortova-Kohoutkova M, Andrejcinova I, Tomaskova V, Sramek V, Helan M, et al. Human myeloid-derived suppressor cell expansion during sepsis is revealed by unsupervised clustering of flow cytometric data. Eur J Immunol (2021) 51(7):1785–91. doi: 10.1002/eji.202049141
75. Lauritsen SM, Kalor ME, Kongsgaard EL, Lauritsen KM, Jorgensen MJ, Lange J, et al. Early detection of sepsis utilizing deep learning on electronic health record event sequences. Artif Intell Med (2020) 104:101820. doi: 10.1016/j.artmed.2020.101820
76. Turki T, Wei Z. Boosting support vector machines for cancer discrimination tasks. Comput Biol Med (2018) 101:236–49. doi: 10.1016/j.compbiomed.2018.08.006
77. Loftus TJ, Brakenridge SC, Croft CA, Smith RS, Efron PA, Moore FA, et al. Neural network prediction of severe lower intestinal bleeding and the need for surgical intervention. J Surg Res (2017) 212:42–7. doi: 10.1016/j.jss.2016.12.032
78. Yang J, Hao S, Huang J, Chen T, Liu R, Zhang P, et al. The application of artificial intelligence in the management of sepsis. Med Rev (2021) (2023) 3(5):369–80. doi: 10.1515/mr-2023-0039
79. Li C, Zhou R, Chen G, Hao X, Zhu T. Knowledge mapping and research hotspots of artificial intelligence on icu and anesthesia: From a global bibliometric perspective. Anesthesiology Perioperative Sci (2023) 1(4). doi: 10.1007/s44254-023-00031-5
80. Cao L, Zhu T, Lang X, Jia S, Yang Y, Zhu C, et al. Inhibiting DNA methylation improves survival in severe sepsis by regulating nf-kappab pathway. Front Immunol (2020) 11:1360. doi: 10.3389/fimmu.2020.01360
81. Falcao-Holanda RB, Brunialti MKC, Jasiulionis MG, Salomao R. Epigenetic regulation in sepsis, role in pathophysiology and therapeutic perspective. Front Med (2021) 8:685333. doi: 10.3389/fmed.2021.685333
82. Davenport EE, Burnham KL, Radhakrishnan J, Humburg P, Hutton P, Mills TC, et al. Genomic landscape of the individual host response and outcomes in sepsis: A prospective cohort study. Lancet Respir Med (2016) 4(4):259–71. doi: 10.1016/S2213-2600(16)00046-1
83. Scicluna BP, van Vught LA, Zwinderman AH, Wiewel MA, Davenport EE, Burnham KL, et al. Classification of patients with sepsis according to blood genomic endotype: A prospective cohort study. Lancet Respir Med (2017) 5(10):816–26. doi: 10.1016/S2213-2600(17)30294-1
84. Cajander S, Kox M, Scicluna BP, Weigand MA, Mora RA, Flohe SB, et al. Profiling the dysregulated immune response in sepsis: Overcoming challenges to achieve the goal of precision medicine. Lancet Respir Med (2024) 12(4):305–22. doi: 10.1016/S2213-2600(23)00330-2
85. Burnham KL, Davenport EE, Radhakrishnan J, Humburg P, Gordon AC, Hutton P, et al. Shared and distinct aspects of the sepsis transcriptomic response to fecal peritonitis and pneumonia. Am J Respir Crit Care Med (2017) 196(3):328–39. doi: 10.1164/rccm.201608-1685OC
86. David B, Antcliffe KLB, Al-Beidh F, Santhakumaran S, Brett SJ, Hinds CJ, et al. Erratum: Transcriptomic signatures in sepsis and a differential response to steroids. from the vanish randomized trial. Am J Respir Crit Care Med (2022) 206(12):1572–3. doi: 10.1164/rccm.v206erratum10
87. Mantel I, Sadiq BA, Blander JM. Spotlight on tap and its vital role in antigen presentation and cross-presentation. Mol Immunol (2022) 142:105–19. doi: 10.1016/j.molimm.2021.12.013
88. Zhang Z, Pan Q, Ge H, Xing L, Hong Y, Chen P. Deep learning-based clustering robustly identified two classes of sepsis with both prognostic and predictive values. EBioMedicine (2020) 62:103081. doi: 10.1016/j.ebiom.2020.103081
89. Cohen J, Blumenthal A, Cuellar-Partida G, Evans DM, Finfer S, Li Q, et al. The relationship between adrenocortical candidate gene expression and clinical response to hydrocortisone in patients with septic shock. Intensive Care Med (2021) 47(9):974–83. doi: 10.1007/s00134-021-06464-5
90. Sweeney TE, Azad TD, Donato M, Haynes WA, Perumal TM, Henao R, et al. Unsupervised analysis of transcriptomics in bacterial sepsis across multiple datasets reveals three robust clusters. Crit Care Med (2018) 46(6):915–25. doi: 10.1097/CCM.0000000000003084
91. Sweeney TE, Liesenfeld O, Wacker J, He YD, Rawling D, Remmel M, et al. Validation of inflammopathic, adaptive, and coagulopathic sepsis endotypes in coronavirus disease 2019. Crit Care Med (2021) 49(2):e170–e8. doi: 10.1097/CCM.0000000000004786
92. Yao L, Rey DA, Bulgarelli L, Kast R, Osborn J, Van Ark E, et al. Gene expression scoring of immune activity levels for precision use of hydrocortisone in vasodilatory shock. Shock (2022) 57(3):384–91. doi: 10.1097/SHK.0000000000001910
93. Zhang S, Wu Z, Chang W, Liu F, Xie J, Yang Y, et al. Classification of patients with sepsis according to immune cell characteristics: A bioinformatic analysis of two cohort studies. Front Med (2020) 7:598652. doi: 10.3389/fmed.2020.598652
94. Marshall JC. Why have clinical trials in sepsis failed? Trends Mol Med (2014) 20(4):195–203. doi: 10.1016/j.molmed.2014.01.007
95. Stanski NL, Wong HR. Prognostic and predictive enrichment in sepsis. Nat Rev Nephrol (2020) 16(1):20–31. doi: 10.1038/s41581-019-0199-3
96. Mi Y, Burnham KL, Charles PD, Heilig R, Vendrell I, Whalley J, et al. High-throughput mass spectrometry maps the sepsis plasma proteome and differences in patient response. Sci Transl Med (2024) 16(750):eadh0185. doi: 10.1126/scitranslmed.adh0185
97. Yehya N, Fazelinia H, Taylor DM, Lawrence GG, Spruce LA, Thompson JM, et al. Differentiating children with sepsis with and without acute respiratory distress syndrome using proteomics. Am J Physiol Lung Cell Mol Physiol (2022) 322(3):L365–L72. doi: 10.1152/ajplung.00164.2021
98. Wang Q, Long G, Luo H, Zhu X, Han Y, Shang Y, et al. S100a8/A9: An emerging player in sepsis and sepsis-induced organ injury. Biomedicine Pharmacotherapy = Biomedecine Pharmacotherapie (2023) 168:115674. doi: 10.1016/j.biopha.2023.115674
99. Mickiewicz B, Thompson GC, Blackwood J, Jenne CN, Winston BW, Vogel HJ, et al. Development of metabolic and inflammatory mediator biomarker phenotyping for early diagnosis and triage of pediatric sepsis. Crit Care (London England) (2015) 19(1):320. doi: 10.1186/s13054-015-1026-2
100. Liu Z, Yin P, Amathieu R, Savarin P, Xu G. Application of lc-Ms-Based metabolomics method in differentiating septic survivors from non-survivors. Anal Bioanal Chem (2016) 408(27):7641–9. doi: 10.1007/s00216-016-9845-9
101. Reisinger AC, Posch F, Hackl G, Marsche G, Sourij H, Bourgeois B, et al. Branched-chain amino acids can predict mortality in icu sepsis patients. Nutrients (2021) 13(9). doi: 10.3390/nu13093106
102. Liu Z, Triba MN, Amathieu R, Lin X, Bouchemal N, Hantz E, et al. Nuclear magnetic resonance-based serum metabolomic analysis reveals different disease evolution profiles between septic shock survivors and non-survivors. Crit Care (London England) (2019) 23(1):169. doi: 10.1186/s13054-019-2456-z
103. Wang J, Sun Y, Teng S, Li K. Prediction of sepsis mortality using metabolite biomarkers in the blood: A meta-analysis of death-related pathways and prospective validation. BMC Med (2020) 18(1):83. doi: 10.1186/s12916-020-01546-5
104. Lee SH, Park MS, Park BH, Jung WJ, Lee IS, Kim SY, et al. Prognostic implications of serum lipid metabolism over time during sepsis. BioMed Res Int (2015) 2015:789298. doi: 10.1155/2015/789298
105. Khaliq W, Grossmann P, Neugebauer S, Kleyman A, Domizi R, Calcinaro S, et al. Lipid metabolic signatures deviate in sepsis survivors compared to non-survivors. Comput Struct Biotechnol J (2020) 18:3678–91. doi: 10.1016/j.csbj.2020.11.009
106. Cirstea M, Walley KR, Russell JA, Brunham LR, Genga KR, Boyd JH. Decreased high-density lipoprotein cholesterol level is an early prognostic marker for organ dysfunction and death in patients with suspected sepsis. J Crit Care (2017) 38:289–94. doi: 10.1016/j.jcrc.2016.11.041
107. Dalli J, Colas RA, Quintana C, Barragan-Bradford D, Hurwitz S, Levy BD, et al. Human sepsis eicosanoid and proresolving lipid mediator temporal profiles: Correlations with survival and clinical outcomes. Crit Care Med (2017) 45(1):58–68. doi: 10.1097/CCM.0000000000002014
108. Lodge S, Litton E, Gray N, Ryan M, Millet O, Fear M, et al. Stratification of sepsis patients on admission into the intensive care unit according to differential plasma metabolic phenotypes. J Proteome Res (2024) 23(4):1328–40. doi: 10.1021/acs.jproteome.3c00803
109. Zhang Z, Chen L, Sun B, Ruan Z, Pan P, Zhang W, et al. Identifying septic shock subgroups to tailor fluid strategies through multi-omics integration. Nat Commun (2024) 15(1):9028. doi: 10.1038/s41467-024-53239-9
110. Chen Q, Liang X, Wu T, Jiang J, Jiang Y, Zhang S, et al. Integrative analysis of metabolomics and proteomics reveals amino acid metabolism disorder in sepsis. J Trans Med (2022) 20(1):123. doi: 10.1186/s12967-022-03320-y
111. Ke X, Zhang F, Huang G, Wang A. Interpretable machine learning to optimize early in-hospital mortality prediction for elderly patients with sepsis: A discovery study. Comput Math Methods Med (2022) 2022:4820464. doi: 10.1155/2022/4820464
112. Giamarellos-Bourboulis EJ, Aschenbrenner AC, Bauer M, Bock C, Calandra T, Gat-Viks I, et al. The pathophysiology of sepsis and precision-Medicine-Based immunotherapy. Nat Immunol (2024) 25(1):19–28. doi: 10.1038/s41590-023-01660-5
113. Vranas KC, Jopling JK, Sweeney TE, Ramsey MC, Milstein AS, Slatore CG, et al. Identifying distinct subgroups of icu patients: A machine learning approach. Crit Care Med (2017) 45(10):1607–15. doi: 10.1097/CCM.0000000000002548
114. Castela Forte J, Perner A, van der Horst ICC. The use of clustering algorithms in critical care research to unravel patient heterogeneity. Intensive Care Med (2019) 45(7):1025–8. doi: 10.1007/s00134-019-05631-z
115. Bhavani SV, Semler M, Qian ET, Verhoef PA, Robichaux C, Churpek MM, et al. Development and validation of novel sepsis subphenotypes using trajectories of vital signs. Intensive Care Med (2022) 48(11):1582–92. doi: 10.1007/s00134-022-06890-z
116. Semler MW, Self WH, Wanderer JP, Ehrenfeld JM, Wang L, Byrne DW, et al. Balanced crystalloids versus saline in critically ill adults. N Engl J Med (2018) 378(9):829–39. doi: 10.1056/NEJMoa1711584
117. Komorowski M, Green A, Tatham KC, Seymour C, Antcliffe D. Sepsis biomarkers and diagnostic tools with a focus on machine learning. EBioMedicine (2022) 86:104394. doi: 10.1016/j.ebiom.2022.104394
118. Sharma M, Taweesedt PT, Surani S. Utilizing artificial intelligence in critical care: Adding a handy tool to our armamentarium. Cureus (2021) 13(6):e15531. doi: 10.7759/cureus.15531
119. Qin Y, Caldino Bohn RI, Sriram A, Kernan KF, Carcillo JA, Kim S, et al. Refining empiric subgroups of pediatric sepsis using machine-learning techniques on observational data. Front Pediatr (2023) 11:1035576. doi: 10.3389/fped.2023.1035576
120. Knox DB, Lanspa MJ, Kuttler KG, Brewer SC, Brown SM. Phenotypic clusters within sepsis-associated multiple organ dysfunction syndrome. Intensive Care Med (2015) 41(5):814–22. doi: 10.1007/s00134-015-3764-7
121. Seymour CW, Kennedy JN, Wang S, Chang CH, Elliott CF, Xu Z, et al. Derivation, validation, and potential treatment implications of novel clinical phenotypes for sepsis. Jama (2019) 321(20):2003–17. doi: 10.1001/jama.2019.5791
122. Ding M, Luo Y. Unsupervised phenotyping of sepsis using nonnegative matrix factorization of temporal trends from a multivariate panel of physiological measurements. BMC Med Inform Decis Mak (2021) 21(Suppl 5):95. doi: 10.1186/s12911-021-01460-7
123. Papin G, Bailly S, Dupuis C, Ruckly S, Gainnier M, Argaud L, et al. Clinical and biological clusters of sepsis patients using hierarchical clustering. PloS One (2021) 16(8):e0252793. doi: 10.1371/journal.pone.0252793
124. Ibrahim ZM, Wu H, Hamoud A, Stappen L, Dobson RJB, Agarossi A. On classifying sepsis heterogeneity in the icu: Insight using machine learning. J Am Med Inform Assoc (2020) 27(3):437–43. doi: 10.1093/jamia/ocz211
125. Bernard GR, Vincent JL, Laterre PF, LaRosa SP, Dhainaut JF, Lopez-Rodriguez A, et al. Efficacy and safety of recombinant human activated protein c for severe sepsis. N Engl J Med (2001) 344(10):699–709. doi: 10.1056/NEJM200103083441001
126. Opal SM, Laterre PF, Francois B, LaRosa SP, Angus DC, Mira JP, et al. Effect of eritoran, an antagonist of Md2-Tlr4, on mortality in patients with severe sepsis: The access randomized trial. Jama (2013) 309(11):1154–62. doi: 10.1001/jama.2013.2194
127. Knaus WA, Marks RD. New phenotypes for sepsis: The promise and problem of applying machine learning and artificial intelligence in clinical research. Jama (2019) 321(20):1981–2. doi: 10.1001/jama.2019.5794
128. Hasegawa D, Nishida O. Patient selection in sepsis: Precision medicine using phenotypes and its implications for future clinical trial design. J Thorac Dis (2019) 11(9):3672–5. doi: 10.21037/jtd.2019.09.31
129. Dahmer MK, Yang G, Zhang M, Quasney MW, Sapru A, Weeks HM, et al. Identification of phenotypes in paediatric patients with acute respiratory distress syndrome: A latent class analysis. Lancet Respir Med (2022) 10(3):289–97. doi: 10.1016/S2213-2600(21)00382-9
130. Xu Z, Mao C, Su C, Zhang H, Siempos I, Torres LK, et al. Sepsis subphenotyping based on organ dysfunction trajectory. Crit Care (London England) (2022) 26(1):197. doi: 10.1186/s13054-022-04071-4
131. Bhavani SV, Carey KA, Gilbert ER, Afshar M, Verhoef PA, Churpek MM. Identifying novel sepsis subphenotypes using temperature trajectories. Am J Respir Crit Care Med (2019) 200(3):327–35. doi: 10.1164/rccm.201806-1197OC
132. Bhavani SV, Spicer A, Sinha P, Malik A, Lopez-Espina C, Schmalz L, et al. Distinct immune profiles and clinical outcomes in sepsis subphenotypes based on temperature trajectories. Intensive Care Med (2024) 50(12):2094–104. doi: 10.1007/s00134-024-07669-0
133. Kudo D, Goto T, Uchimido R, Hayakawa M, Yamakawa K, Abe T, et al. Coagulation phenotypes in sepsis and effects of recombinant human thrombomodulin: An analysis of three multicentre observational studies. Crit Care (London England) (2021) 25(1):114. doi: 10.1186/s13054-021-03541-5
134. Papathanakos G, Andrianopoulos I, Xenikakis M, Papathanasiou A, Koulenti D, Blot S, et al. Clinical sepsis phenotypes in critically ill patients. Microorganisms (2023) 11(9). doi: 10.3390/microorganisms11092165
135. Madushani R, Patel V, Loftus T, Ren Y, Li HJ, Velez L, et al. Early biomarker signatures in surgical sepsis. J Surg Res (2022) 277:372–83. doi: 10.1016/j.jss.2022.04.052
136. Baek MS, Kim JH, Kwon YS. Cluster analysis integrating age and body temperature for mortality in patients with sepsis: A multicenter retrospective study. Sci Rep (2022) 12(1):1090. doi: 10.1038/s41598-022-05088-z
137. Thomas-Ruddel DO, Hoffmann P, Schwarzkopf D, Scheer C, Bach F, Komann M, et al. Fever and hypothermia represent two populations of sepsis patients and are associated with outside temperature. Crit Care (London England) (2021) 25(1):368. doi: 10.1186/s13054-021-03776-2
138. Matthay MA, McAuley DF, Ware LB. Clinical trials in acute respiratory distress syndrome: Challenges and opportunities. Lancet Respir Med (2017) 5(6):524–34. doi: 10.1016/S2213-2600(17)30188-1
139. Reddy K, Sinha P, O'Kane CM, Gordon AC, Calfee CS, McAuley DF. Subphenotypes in critical care: Translation into clinical practice. Lancet Respir Med (2020) 8(6):631–43. doi: 10.1016/S2213-2600(20)30124-7
140. Sinha P, Calfee CS. Phenotypes in acute respiratory distress syndrome: Moving towards precision medicine. Curr Opin Crit Care (2019) 25(1):12–20. doi: 10.1097/MCC.0000000000000571
141. Famous KR, Delucchi K, Ware LB, Kangelaris KN, Liu KD, Thompson BT, et al. Acute respiratory distress syndrome subphenotypes respond differently to randomized fluid management strategy. Am J Respir Crit Care Med (2017) 195(3):331–8. doi: 10.1164/rccm.201603-0645OC
142. Calfee CS, Delucchi KL, Sinha P, Matthay MA, Hackett J, Shankar-Hari M, et al. Acute respiratory distress syndrome subphenotypes and differential response to simvastatin: Secondary analysis of a randomised controlled trial. Lancet Respir Med (2018) 6(9):691–8. doi: 10.1016/S2213-2600(18)30177-2
143. Sinha P, Furfaro D, Cummings MJ, Abrams D, Delucchi K, Maddali MV, et al. Latent class analysis reveals covid-19-Related acute respiratory distress syndrome subgroups with differential responses to corticosteroids. Am J Respir Crit Care Med (2021) 204(11):1274–85. doi: 10.1164/rccm.202105-1302OC
144. Sinha P, Delucchi KL, Thompson BT, McAuley DF, Matthay MA, Calfee CS, et al. Latent class analysis of ards subphenotypes: A secondary analysis of the statins for acutely injured lungs from sepsis (Sails) study. Intensive Care Med (2018) 44(11):1859–69. doi: 10.1007/s00134-018-5378-3
145. Sinha P, Kerchberger VE, Willmore A, Chambers J, Zhuo H, Abbott J, et al. Identifying molecular phenotypes in sepsis: An analysis of two prospective observational cohorts and secondary analysis of two randomised controlled trials. Lancet Respir Med (2023) 11(11):965–74. doi: 10.1016/S2213-2600(23)00237-0
146. Evrard B, Sinha P, Delucchi K, Hendrickson CM, Kangelaris KN, Liu KD, et al. Causes and attributable fraction of death from ards in inflammatory phenotypes of sepsis. Crit Care (London England) (2024) 28(1):164. doi: 10.1186/s13054-024-04943-x
147. van Amstel RBE, Bartek B, Vlaar APJ, Gay E, van Vught LA, Cremer OL, et al. Temporal transitions of the hyperinflammatory and hypoinflammatory phenotypes in critical illness. Am J Respir Crit Care Med (2024). doi: 10.1164/rccm.202406-1241OC
148. Russell JA, Walley KR, Singer J, Gordon AC, Hebert PC, Cooper DJ, et al. Vasopressin versus norepinephrine infusion in patients with septic shock. N Engl J Med (2008) 358(9):877–87. doi: 10.1056/NEJMoa067373
149. Neyton LPA, Sinha P, Sarma A, Mick E, Kalantar K, Chen S, et al. Host and microbe blood metagenomics reveals key pathways characterizing critical illness phenotypes. Am J Respir Crit Care Med (2024) 209(7):805–15. doi: 10.1164/rccm.202308-1328OC
150. Zhang Z, Zhang G, Goyal H, Mo L, Hong Y. Identification of subclasses of sepsis that showed different clinical outcomes and responses to amount of fluid resuscitation: A latent profile analysis. Crit Care (London England) (2018) 22(1):347. doi: 10.1186/s13054-018-2279-3
151. Shald EA, Erdman MJ, Ferreira JA. Impact of clinical sepsis phenotypes on mortality and fluid status in critically ill patients. Shock (2022) 57(1):57–62. doi: 10.1097/SHK.0000000000001864
152. Ma P, Liu J, Shen F, Liao X, Xiu M, Zhao H, et al. Individualized resuscitation strategy for septic shock formalized by finite mixture modeling and dynamic treatment regimen. Crit Care (London England) (2021) 25(1):243. doi: 10.1186/s13054-021-03682-7
153. Sinha P, Calfee CS, Cherian S, Brealey D, Cutler S, King C, et al. Prevalence of phenotypes of acute respiratory distress syndrome in critically ill patients with covid-19: A prospective observational study. Lancet Respir Med (2020) 8(12):1209–18. doi: 10.1016/S2213-2600(20)30366-0
154. Kotsaki A, Pickkers P, Bauer M, Calandra T, Lupse M, Wiersinga WJ, et al. Immunosep (Personalised immunotherapy in sepsis) international double-blind, double-dummy, placebo-controlled randomised clinical trial: Study protocol. BMJ Open (2022) 12(12):e067251. doi: 10.1136/bmjopen-2022-067251
155. Ates HC, Alshanawani A, Hagel S, Cotta MO, Roberts JA, Dincer C, et al. Unraveling the impact of therapeutic drug monitoring Via machine learning for patients with sepsis. Cell Rep Med (2024) 5(8):101681. doi: 10.1016/j.xcrm.2024.101681
156. Agullo L, Aguado I, Muriel J, Margarit C, Gomez A, Escorial M, et al. Pharmacogenetic guided opioid therapy improves chronic pain outcomes and comorbid mental health: A randomized, double-blind, controlled study. Int J Mol Sci (2023) 24(13). doi: 10.3390/ijms241310754
157. Cummings MJ, Bakamutumaho B, Tomoiaga AS, Owor N, Jain K, Price A, et al. A transcriptomic classifier model identifies high-risk endotypes in a prospective study of sepsis in uganda. Crit Care Med (2024) 52(3):475–82. doi: 10.1097/CCM.0000000000006023
158. DeMerle KM, Angus DC, Baillie JK, Brant E, Calfee CS, Carcillo J, et al. Sepsis subclasses: A framework for development and interpretation. Crit Care Med (2021) 49(5):748–59. doi: 10.1097/CCM.0000000000004842
159. Goligher EC, Lawler PR, Jensen TP, Talisa V, Berry LR, Lorenzi E, et al. Heterogeneous treatment effects of therapeutic-dose heparin in patients hospitalized for covid-19. Jama (2023) 329(13):1066–77. doi: 10.1001/jama.2023.3651
160. Kent DM, Paulus JK, van Klaveren D, D'Agostino R, Goodman S, Hayward R, et al. The predictive approaches to treatment effect heterogeneity (Path) statement. Ann Intern Med (2020) 172(1):35–45. doi: 10.7326/M18-3667
161. Aldrighetti CM, Niemierko A, Van Allen E, Willers H, Kamran SC. Racial and ethnic disparities among participants in precision oncology clinical studies. JAMA Netw Open (2021) 4(11):e2133205. doi: 10.1001/jamanetworkopen.2021.33205
162. Cadario R, Longoni C, Morewedge CK. Understanding, explaining, and utilizing medical artificial intelligence. Nat Hum Behav (2021) 5(12):1636–42. doi: 10.1038/s41562-021-01146-0
163. He J, Baxter SL, Xu J, Xu J, Zhou X, Zhang K. The practical implementation of artificial intelligence technologies in medicine. Nat Med (2019) 25(1):30–6. doi: 10.1038/s41591-018-0307-0
164. Flores C, Villar J. The road to precision medicine in sepsis: Blood transcriptome endotypes. Lancet Respir Med (2017) 5(10):767–8. doi: 10.1016/S2213-2600(17)30297-7
165. Srdic T, Durasevic S, Lakic I, Ruzicic A, Vujovic P, Jevdovic T, et al. From molecular mechanisms to clinical therapy: Understanding sepsis-induced multiple organ dysfunction. Int J Mol Sci (2024) 25(14). doi: 10.3390/ijms25147770
166. Tang F, Zhao XL, Xu LY, Zhang JN, Ao H, Peng C. Endothelial dysfunction: Pathophysiology and therapeutic targets for sepsis-induced multiple organ dysfunction syndrome. Biomedicine Pharmacotherapy = Biomedecine Pharmacotherapie (2024) 178:117180. doi: 10.1016/j.biopha.2024.117180
Keywords: sepsis heterogeneity, phenotyping, personalized therapy, phenotype-driven treatment, biomarkers, therapeutic target identification
Citation: Zhang X, Zhang W, Zhang H and Liao X (2025) Sepsis subphenotypes: bridging the gaps in sepsis treatment strategies. Front. Immunol. 16:1546474. doi: 10.3389/fimmu.2025.1546474
Received: 17 December 2024; Accepted: 20 January 2025;
Published: 06 February 2025.
Edited by:
Daolin Tang, University of Texas Southwestern Medical Center, United StatesReviewed by:
Xiaopei Shen, Fujian Medical University, ChinaCopyright © 2025 Zhang, Zhang, Zhang and Liao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xuelian Liao, bGlhb3h1ZWxpYW5Ac2N1LmVkdS5jbg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.