
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Immunol. , 17 February 2025
Sec. Cancer Immunity and Immunotherapy
Volume 16 - 2025 | https://doi.org/10.3389/fimmu.2025.1538658
This article is part of the Research Topic m6A Methylation and Cancer Immunity View all articles
Hepatocellular carcinoma (HCC) is the most prevalent primary liver malignancy and a leading cause of cancer-related deaths globally. The asymptomatic progression of early-stage HCC often results in diagnosis at advanced stages, significantly limiting therapeutic options and worsening prognosis. Immunotherapy, with immune checkpoint inhibitors (ICIs) at the forefront, has revolutionized HCC treatment. Nevertheless, tumor heterogeneity, immune evasion, and the presence of immunosuppressive components within the tumor immune microenvironment (TIME) continue to compromise its efficacy. Furthermore, resistance or non-responsiveness to ICIs in some patients underscores the urgent need to unravel the complexities of the TIME and to design innovative strategies that enhance immunotherapeutic outcomes. Emerging evidence has revealed the pivotal role of N6-methyladenosine (m6A), a prominent RNA methylation modification, in shaping the TIME in HCC. By regulating RNA stability and translation, m6A influences immune-related factors, including cytokines and immune checkpoint molecules. This modification governs PD-L1 expression, facilitating immune escape and contributing to resistance against ICIs. Advances in this field have also identified m6A-related regulators as promising biomarkers for predicting immunotherapy response and as potential therapeutic targets for optimizing treatment efficacy. This review examines the regulatory mechanisms of m6A modification within the TIME of HCC, with a focus on its impact on immune cells and cytokine dynamics. It also explores the therapeutic potential of targeting m6A pathways to improve immunotherapy efficacy and outlines emerging directions for future research. These insights aim to provide a foundation for developing novel strategies to overcome immune resistance and advance HCC treatment.
Hepatocellular carcinoma (HCC) is the most common type of primary liver cancer globally and a leading cause of cancer-related deaths. Its high incidence is often linked to chronic hepatitis virus infections (HBV or HCV), liver cirrhosis, excessive alcohol consumption, and non-alcoholic fatty liver disease (1). Due to the lack of early symptoms, many HCC patients are diagnosed at advanced stages, leading to limited treatment options and poor prognosis. Recently, immunotherapy, particularly immune checkpoint inhibitors, has brought new hope for HCC treatment (2). However, challenges like liver cancer heterogeneity, immune evasion, and immunosuppressive cells in the tumor microenvironment (TME) undermine immune responses (3). Additionally, some patients show resistance or no response, limiting its effectiveness (4).Thus, a deeper understanding of the HCC immune microenvironment and immune evasion mechanisms is crucial for improving immunotherapy outcomes.
Post-transcriptional modifications (PTMs) are critical regulatory processes that occur after RNA is transcribed from DNA. These modifications are widely found in various types of RNA, including messenger RNA (mRNA), transfer RNA (tRNA), ribosomal RNA (rRNA), and non-coding RNA. PTMs play a critical role in regulating gene expression by modulating RNA stability, splicing, translation, transport, and degradation (5). Key types of RNA modifications include N6-methyladenosine (m6A), 5-methylcytosine (m5C), pseudouridine (Ψ), N1-methyladenosine (m1A), and adenosine-to-inosine (A-to-I) editing (6). Among these, m6A is the most well-known and extensively studied, predominantly found in mRNA. m6A is dynamically regulated by specific enzymes: “writers” (e.g., METTL3), “erasers” (e.g., FTO and ALKBH5), and “readers” (e.g., YTHDF1) (7). This dynamic modification allows RNA to quickly respond to internal and external signals, influencing its fate and function.
In recent years, m6A modification has emerged as a key regulator in the immunotherapy of HCC, attracting growing attention. By modulating the expression of immune checkpoint molecules, such as PD-1 and PD-L1, m6A facilitates tumor immune evasion, thereby compromising the anti-tumor activity of T cells (8). Moreover, m6A influences the polarization of tumor-associated macrophages (TAMs), reshaping immune responses within the TME (9). Given the dynamic and reversible nature of m6A, targeting this modification presents a promising therapeutic strategy for HCC immunotherapy (10, 11). Such interventions could enhance treatment sensitivity and foster the development of novel therapeutics. This review aims to elucidate the pivotal role of m6A modification in HCC immunotherapy, dissect its underlying mechanisms in regulating tumor immune evasion and remodeling of the TME, and assess its potential as a therapeutic target for improved clinical outcomes.
The regulation of m6A modification is dynamically controlled by three distinct classes of proteins: “writers,” “erasers,” and “readers.” Acting in concert, these proteins mediate the deposition, removal, and interpretation of m6A marks, ensuring the reversible and precise nature of this epitranscriptomic modification. Collectively, they establish a complex regulatory network that governs RNA metabolism and gene expression, underpinning a wide array of critical cellular processes (Figure 1).

Figure 1. The dynamicregulation of m6A modification. The regulation of m6A modification is mediated by three distinct classes of proteins that collaboratively maintain its dynamic and reversible nature. “Writers,” including METTL3 and METTL14, catalyze the addition of m6A marks, while “erasers,” represented by FTO and ALKBH5, remove these modifications. “Readers,” encompassing the YTH and IGF2BP protein families, decode m6A marks to influence RNA processes, including stability, splicing, and translation. Collectively, these regulators establish a sophisticated epitranscriptomic network that fine-tunes RNA metabolism and gene expression.
The installation of m6A marks is driven by the m6A methyltransferase complex, with METTL3 and METTL14 functioning as the core catalytic components responsible for methylating the N6 position of adenosine at specific RNA sites (12, 13). METTL3 accounts for catalyzing approximately 95% of m6A modifications on mRNA (14). Supporting proteins such as WTAP, KIAA1429, and RBM15 contribute to site specificity and enhance methylation efficiency (15, 16). This writer complex typically modifies regions near the 5’ and 3’ untranslated regions (UTRs), regulating key processes like RNA stability, splicing, and translation efficiency (17).
m6A modification is dynamic and reversible, with its removal facilitated by demethylases, or “erasers.” The primary enzymes responsible for this demethylation are FTO (fat mass and obesity-associated protein) and ALKBH5 (18). FTO was the first demethylase identified (19), with a crucial role in reversing m6A modifications, while ALKBH5 contributes to post-transcriptional regulation of RNA, influencing RNA stability, splicing, and translation (20, 21). By removing m6A, these erasers restore RNA to its unmodified state, affecting its stability and translational capacity.
The biological effects of m6A modification are mediated by “reader” proteins, which recognize and bind to m6A-modified sites, thereby dictating the fate of the RNA. Major reader proteins include the YTHDF1/2/3 and YTHDC1/2 families, which regulate RNA degradation, transport, splicing, and translation (22, 23). For example, YTHDF1 enhances protein synthesis by promoting mRNA translation, whereas YTHDF2 directs the degradation of m6A-modified RNA. In addition, the IGF2BP family stabilizes m6A-marked mRNAs, further refining gene expression at the post-transcriptional level (24, 25).
As a dynamic and reversible RNA methylation process, m6A exerts a profound influence on HCC progression initiation and progression by modulating RNA stability, splicing, translation, and degradation. This biochemical modification governs the expression of both oncogenes and tumor suppressor genes, thereby affecting HCC cell proliferation and apoptosis. The writer enzyme METTL3 facilitates tumor growth by stabilizing oncogene mRNA, whereas erasers such as FTO and ALKBH5 suppress tumor progression by removing m6A marks (26–29). Additionally, m6A enhances HCC cell migration and invasion by regulating metastasis-related genes, including VEGFA and ZEB1, and promoting epithelial-mesenchymal transition (EMT), thus elevating metastatic potential (30, 31). m6A modification also sustains HCC cell stem-like properties by modulating stemness-associated genes, such as SOX2 and KLF4, which contribute to drug resistance and recurrence (32, 33). Furthermore, m6A facilitates immune evasion by modulating immune checkpoint molecules like PD-L1, helping HCC cells escape immune surveillance (34, 35). Given the dynamic and reversible nature of m6A, targeting m6A regulatory enzymes (e.g., METTL3 and FTO) presents a promising therapeutic approach. Modulating m6A modification offers an opportunity to influence HCC proliferation, metastasis, and immune evasion, creating new avenues for HCC treatment.
The TME of HCC possesses unique immunosuppressive characteristics that support tumor growth and enable immune evasion. This TME includes regulatory T cells (Tregs), myeloid-derived suppressor cells (MDSCs), and M2-polarized tumor-associated macrophages (TAMs), all of which release immunosuppressive factors like IL-10 and TGF-β to inhibit anti-tumor responses (3, 36–38). Tumor cells further promote immune escape by expressing PD-L1, which interacts with PD-1 on T cells, reducing T-cell-mediated anti-tumor activity. Hypoxia within the TME intensifies immunosuppression and drives tumor resistance, while the liver’s immune-tolerant nature provides an ideal environment for tumor cell survival and spread (39, 40). Thus, the TME, though challenging, presents a promising target for immunotherapy in HCC.
In HCC, high expression levels of m6A writers, particularly METTL3, are associated with poor prognosis, as they enhance PD-L1 levels and subsequently inhibit T cell-mediated anti-tumor responses (41, 42). METTL3-mediated m6A modification also upregulates the non-coding RNA TUG1, which increases the expression of PD-L1 and CD47 via miR-141 and miR-340 sponging and interaction with YBX1, a transcriptional regulator. This regulation suppresses CD8+ T cell activation, aiding immune evasion and contributing to tumor progression (42). Another recent study found that METTL3 stabilizes SMPDL3A via m6A modification, promoting HCC growth and immune evasion by interacting with IGF2BP1. Knockdown of METTL3 activated CD8+ T cells, increasing TNFα/IFN-γ production and reducing HCC cell survival. SMPDL3A overexpression reverses these effects (43). Additionally, METTL3 facilitates immune escape in non-alcoholic fatty liver disease-associated HCC (NAFLD-HCC) by promoting cholesterol biosynthesis, which further impairs CD8+ T cell function. Targeting METTL3 in conjunction with PD-1 blockade has demonstrated synergistic effects in restoring CD8+ T cell cytotoxicity (44). WTAP, another m6A writer, stabilizes PD-L1 mRNA, which promotes immune evasion and enhances aerobic glycolysis in HCC cells. This effect suppresses the tumor-killing function of CD8+ T cells, but can be reversed through WTAP knockdown, thus improving T cell anti-tumor activity (45). Similarly, the m6A “eraser” FTO stabilizes GPNMB mRNA, which inhibits CD8+ T cell activation through SDC4 receptor binding in small extracellular vesicles (sEVs). Knocking down FTO enhances CD8+ T cell recruitment and bolsters anti-tumor responses (28). BMI1, independent of its classical role, influences CD127+KLRG1+ memory CD8+ T cell differentiation in HCC by regulating TCF1 expression. BMI1 interacts with YTHDF2 to prevent m6A-driven degradation of TCF1 mRNA. This regulation shifts T cells towards memory and effector states, enhancing their tumor-killing abilities. Meanwhile, tumor cell-intrinsic BMI1 expression downregulates BMI1 in T cells. Liver-specific BMI1 knockdown effectively restores CD8+ T cell functionality and supports immunotherapy efficacy in HCC (46). In NASH-HCC, YTHDF1, an m6A reader, promotes MDSC accumulation and suppresses CD8+ T cell functionality via IL-6 secretion. By binding to m6A-modified EZH2 mRNA, YTHDF1 enhances IL-6 production. siRNA targeting of YTHDF1 has shown potential in enhancing anti-PD-1 therapy, making YTHDF1 a promising target in immune-based HCC therapies (47). Exosomal circCCAR1, stabilized by WTAP-mediated m6A modification, promotes HCC growth by forming a feedback loop with miR-127-5p and WTAP. When absorbed by CD8+ T cells, circCCAR1 induces dysfunction by stabilizing PD-1 and promoting resistance to anti-PD1 therapy. This effect is further enhanced through PD-L1 transcription activation by EP300-induced CCAR1 and β-catenin interaction, which reinforces immune evasion (48).
Macrophages in the HCC TME are primarily polarized to the M2 phenotype, which secretes immunosuppressive cytokines like IL-10 and TGF-β, promoting tumor growth and suppressing anti-tumor T cell activity. METTL3- and METTL16-mediated m6A modification stabilizes ZNNT1 in HCC, which enhances macrophage recruitment and polarization to M2 via the osteopontin (OPN)/S100A9 feedback loop, ultimately supporting immune evasion and tumor progression (49). Similarly, the lncRNA miR4458HG, through the m6A reader IGF2BP2, stabilizes key glycolytic mRNAs like HK2 and GLUT1, promoting glucose metabolism and tumor-associated macrophage polarization in HCC, thereby contributing to immune suppression (50). In contrast, circFUT8 downregulation via M1 macrophage-derived exosomal miR-628-5p limits METTL14-mediated m6A modification, reducing circFUT8’s influence on the circFUT8/miR-552-3p/CHMP4B pathway, and suppressing HCC progression by inhibiting tumor growth and immune suppression (51). Additionally, ALKBH5 overexpression in HCC enhances tumor progression by upregulating MAP3K8 through m6A modification, leading to JNK and ERK pathway activation. This triggers IL-8 expression, attracting macrophages and amplifying immune evasion (52). In HBV-related HCC, the METTL3-mediated upregulation of lncRNA MAAS in M2 macrophages drives tumor progression. HBV-associated antigen HBeAg elevates MAAS, which is then transferred to HCC cells through exosomes. Inside HCC cells, MAAS stabilizes c-Myc, promoting cell cycle progression and proliferation, thus highlighting an m6A-regulated feedback loop contributing to HCC malignancy and immune escape (53). Together, these findings underscore m6A modification’s regulatory role in macrophage polarization in HCC, providing multiple potential targets for therapeutic intervention.
m6A modification significantly impacts the antigen-presenting function of dendritic cells (DCs), with downstream effects on T cell activation and antitumor immunity. The m6A reader YTHDF1 specifically modulates DC function by influencing neoantigen presentation. In YTHDF1-deficient mice, increased cross-presentation of tumor antigens by DCs strengthens CD8+ T cell activation and antitumor responses. Mechanistically, YTHDF1 binds m6A-modified transcripts of lysosomal proteases, enhancing cathepsin translation and thus limiting cross-presentation. Inhibiting cathepsins in wild-type DCs similarly boosts antigen presentation, demonstrating YTHDF1’s role as a potential immunotherapy target, especially in conjunction with PD-L1 checkpoint inhibitors (54). Under radiotherapy, YTHDF1 upregulation in DCs reduces IFN-I production via STING degradation, impairing DC cross-priming and diminishing T cell activation. Loss of YTHDF1 not only amplifies radiotherapy efficacy in melanoma models but also induces long-lasting immunity, marked by robust CD4+ and CD8+ T cell responses and IFN-γ secretion, as seen in gastric cancer studies (55, 56). Collectively, these findings position YTHDF1 and m6A modulation in DCs as promising targets to enhance T cell-mediated antitumor immunity and improve cancer immunotherapies.
MDSCs, known for their immunosuppressive function, contribute to T-cell inhibition and facilitate tumor immune evasion. Elevated m6A levels enhance the immunosuppressive nature of MDSCs, promoting their accumulation and activation within the TME. In colorectal cancer (CRC), high m6A levels and reduced ALKBH5 expression in MDSCs correlate with increased immunosuppressive capacity. ALKBH5 overexpression reduces m6A levels on Arg-1 mRNA, destabilizing its expression and diminishing MDSC-mediated suppression (57). METTL3 also plays a key role in MDSC regulation. In CRC models, METTL3 promotes MDSC migration by activating the m6A-BHLHE41-CXCL1 pathway. Silencing METTL3 reduces MDSC accumulation and promotes CD4+ and CD8+ T-cell proliferation, suppressing CRC growth. Mechanistic studies reveal that METTL3 induces BHLHE41 expression in an m6A-dependent manner, which in turn activates CXCL1, guiding MDSC migration through the CXCR2 axis (58). In ovarian cancer, Mettl3 deficiency within myeloid cells shifts immune balance from M1 to M2 macrophage polarization, enhancing pro-tumorigenic inflammation via cytokines including IL-1β, CCL2, and CXCL2 (59). This modulation underscores Mettl3’s role in maintaining immune response homeostasis within the TME.
Figure 2 illustrates the m6A-mediated regulation of immune cells within the HCC microenvironment. Although m6A modification is a pivotal regulator of immune evasion and tumor progression in HCC, significant gaps hinder its therapeutic translation. Current research overly focuses on PD-1/PD-L1 blockade in T cells, neglecting alternative immune checkpoints and downstream pathways. The role of m6A-modified non-coding RNAs in immune dysfunction also demands further investigation to identify novel targets. In macrophages, the mechanisms driving M2 polarization and their interaction with immune networks remain poorly understood, and the feedback roles of METTL3 and ALKBH5 within the TME lack clarity. Advancing m6A targeted immunotherapy necessitates a broader investigation into immune interactions, alternative pathways, and HCC specific mechanisms.
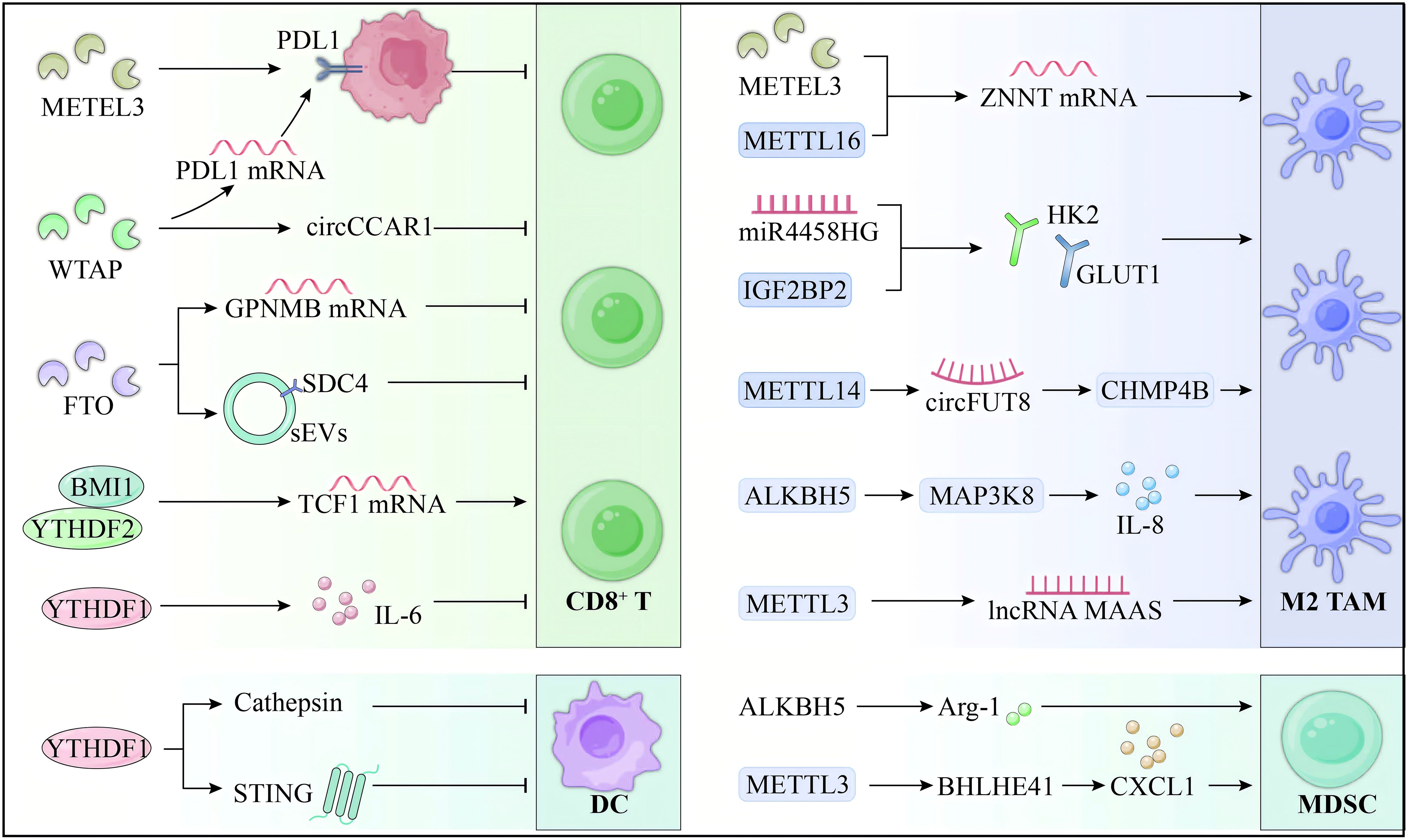
Figure 2. m6A regulation of immune cells in HCC microenvironment. m6A modification plays a critical role in modulating the TIME of HCC, driving immune evasion and affecting therapeutic responses. In T cells, METTL3 promotes PD-L1 expression and suppresses CD8+ T cell activity, while FTO stabilizes GPNMB, further impairing their cytotoxicity. In TAMs, m6A drives M2 polarization through METTL3 and ALKBH5, enhancing immunosuppressive cytokine production. In DCs, YTHDF1 limits antigen presentation, reducing CD8+ T cell activation. Additionally, m6A enhances MDSC immunosuppressive functions. The role of m6A in regulating immune cells within the HCC TIME underscores its potential as a promising therapeutic target to improve immunotherapy outcomes.
m6A modification plays a crucial role in modulating interleukin (IL) signaling, significantly influencing HCC progression and inflammation. One key interleukin, IL-6, is highly regulated by m6A modification. Enhanced m6A methylation stabilizes IL-6 mRNA, increasing its expression in the HCC microenvironment, which promotes tumor cell proliferation and survival. A notable feedback loop involving exosomal SLC16A1-AS1 stabilizes SLC16A1 mRNA via m6A, enhancing lactate influx and activating the c-Raf/ERK pathway. This pathway induces M2 macrophage polarization, which in turn secretes IL-6, activating STAT3 signaling. STAT3 activation leads to METTL3 upregulation, further stabilizing SLC16A1-AS1 through m6A modification, promoting HCC growth, invasion, and glycolysis (60). YTHDF1 overexpression in NASH-HCC promotes IL-6 secretion through m6A-modified EZH2 (47), resulting in MDSC recruitment and CD8+ T-cell suppression. Similarified GNAS upregulation by LPS stimulation enhances IL-6 expression by elevating m6A methylation of mRNA, while GNAS knockdown diminishes IL-6, thereby inhibiting tumor growth (61).
Beyond IL-6, reduced YTHDF2 in HCC increases m6A-modified IL-11 mRNA, exacerbating inflammation and metastasis by impairing the degradation of IL11 and SERPINE2 mRNAs (62). Additionally, upregulated m6A-modified RNA AC026356.1 enhances cancer progression by binding IGF2BP1, stabilizing IL11 mRNA, and activating IL11/STAT3 signaling (63). In chronic liver inflammation, MeRIP-seq analysis revealed reduced m6A methylation of IL-17RA mRNA, linked to inflammation-driven HCC. The demethylase FTO, rather than METTL3, primarily mediates this reduced methylation (64). Furthermore, in alcoholic steatohepatitis (ASH), chronic alcohol intake induces Kupffer cell pyroptosis and increases IL-1β release. Silencing METTL3 alleviates this inflammation by regulating pri-miR-34A splicing (65), highlighting METTL3 as a key modulator of interleukin-driven inflammation and a potential therapeutic target.
m6A modification has emerged as a pivotal regulator of TGF-β signaling, profoundly influencing cancer progression and immune modulation. In HCC, TGF-β-induced METTL3-mediated m6A modification destabilizes ITIH1 mRNA, disrupting fibronectin and focal adhesion kinase signaling, thus driving tumor growth and invasion (66). Additionally, TGF-β induces m6A modification that destabilizes PCDHGA9 mRNA, facilitating HCC progression and metastasis (67). In gastric carcinoma, aberrant overexpression of the m6A regulator WTAP stabilizes TGF-β mRNA, promoting cell migration, EMT, and resistance to chemoradiotherapy (68). Similarly, in breast cancer, YTHDC1 enhances the stability of m6A-modified SMAD3 mRNA, potentiating TGF-β signaling and enabling lung metastasis. Loss of YTHDC1 disrupts EMT and cell migration, highlighting its critical role in TGF-β-mediated tumor progression (69).
In addition to malignancies, m6A modification interconnects metabolic and inflammatory pathways in liver diseases. In NASH, LPS-induced NF-κB activation upregulates METTL3/METTL14, driving hypermethylation of TGF-β1 mRNA at its 5’ UTR, enhancing translation and linking inflammation to fibrosis (70). Correspondingly, the liver progenitor-specific gene RALYL mitigates m6A modification on TGF-β2 mRNA, stabilizing its expression and activating the PI3K/AKT and STAT3 pathways to promote tumorigenicity and metastasis in HCC (71).
As key anti-tumor cytokines, the expression of interferons is also regulated by m6A modification. In radiation-induced liver diseases (RILD), irradiation triggers ALKBH5 to demethylate m6A residues in the 3’ UTR of HMGB1, activating the STING-IRF3 pathway and promoting the production of type I interferon (IFN), which contributes to hepatocyte apoptosis. Loss of ALKBH5 or silencing of HMGB1 reduces IFN levels and inflammation. Also, YTHDF2 facilitates the degradation of m6A-modified HMGB1, further linking m6A to the regulation of liver inflammation and apoptosis (72). In liver cancer, MATR3 inhibits IFN signaling by binding to DHX58 mRNA, recruiting YTHDF2, and promoting mRNA degradation. MATR3 knockout restores IFN signaling, enhancing CD8+ T cell-mediated tumor elimination (73). In HBV-related HCC, elevated serum pgRNA levels are associated with poor prognosis. IFN-α-2a enhances m6A modification of pgRNA, leading to its destabilization and reduced tumorigenicity, suggesting that targeting m6A-modified pgRNA may improve IFN signaling and offer a potential therapeutic strategy for HCC (74).
Multiple studies have established that m6A RNA modification regulates chemokine expression by modulating mRNA stability and translation. In acute liver failure (ALF), tristetraprolin (TTP) upregulation enhances m6A modification of CCL2 and CCL5, leading to mRNA destabilization and reduced expression. This m6A-mediated degradation, driven by enzymes such as METTL14, alleviates liver injury (75). In intrahepatic cholangiocarcinoma (ICC), hepatocyte-secreted CCL3 promotes metastasis via VIRMA-mediated m6A modification, which upregulates SIRT1 and drives tumor progression (76). Similarly, hepatitis B virus (HBV) surface antigens (SHBs) increase CCR9 expression through KIAA1429-mediated m6A modification, stabilizing CCR9 mRNA and facilitating HCC progression and regorafenib resistance (77). This stabilization positions CCR9 as a critical therapeutic target and prognostic biomarker in HBV-related HCC. Additionally, the m6A reader protein YTHDF2 stabilizes Cx3cl1 mRNA in peritumoral hepatocytes, enhancing CD8+ T cell recruitment and activation via the cGAS-STING pathway (78). The m6A-dependent process bolsters immune responses, improves immunotherapy efficacy, and suppresses liver tumor growth. In metabolic-associated fatty liver disease (MAFLD), reduced METTL14 expression diminishes GLS2 translation via the m6A/YTHDF1 axis, exacerbating oxidative stress and recruiting pro-fibrotic Cx3cr1+ macrophages. These macrophages activate hepatic stellate cells via the CX3CR1/MyD88/NF-κB signaling axis, contributing to liver fibrosis (79). Restoring METTL14 expression or inhibiting MyD88 signaling alleviates fibrosis, offering promising avenues for intervention in MAFLD.
m6A modification critically regulates inflammatory cytokines and chemokines, shaping HCC progression and immune evasion (Figure 3). Stabilization of IL-6 mRNA by m6A promotes tumor proliferation, while aberrant m6A modifications of IL-11 and IL-17RA exacerbate inflammation and metastasis. TGF-β signaling and interferon pathways, modulated by m6A, further drive invasion, immune suppression, and resistance. Chemokines such as CCL2 and CCL3 are similarly affected, altering the TME and therapy responses. However, the current understanding is limited by the lack of precise, in vivo models to dissect m6A’s temporal and spatial effects. Future research should integrate multi-omics, single-cell technologies, and m6A targeted therapeutics to refine mechanistic insights and clinical applications.
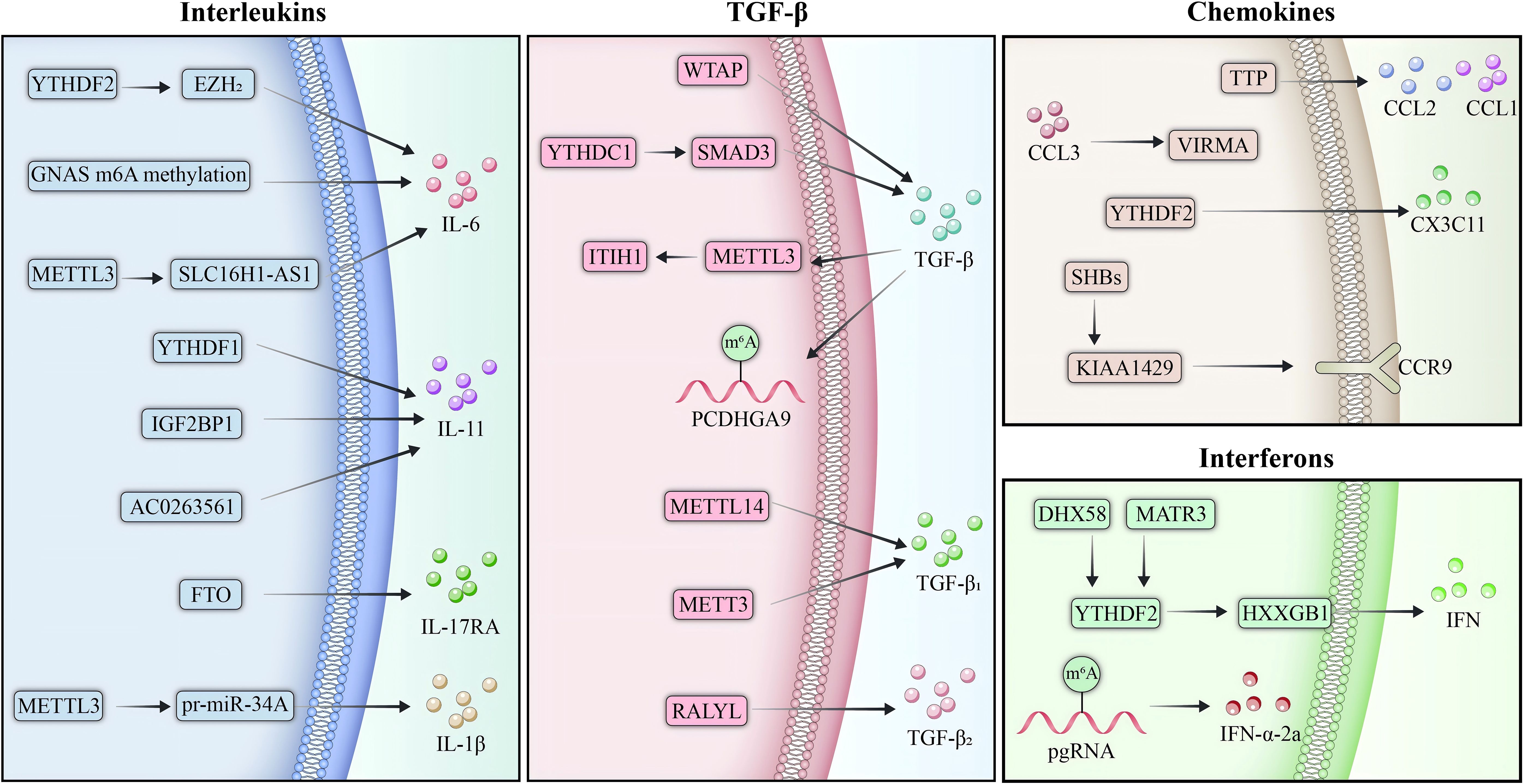
Figure 3. m6A regulation of inflammatory and chemokine signaling in HCC. m6A modification serves as a pivotal regulator of inflammatory cytokines and chemokines in the TME of HCC, shaping immune responses and driving tumor progression. Stabilization of IL-6 mRNA by m6A enhances STAT3 activation, promoting immune suppression through M2 macrophage polarization and recruitment of MDSCs. Similarly, reduced YTHDF2 expression leads to the stabilization of m6A-modified IL-11 mRNA, further amplifying inflammation and metastasis. TGF-β signaling is tightly modulated by m6A, where METTL3-mediated destabilization of tumor-suppressive transcripts facilitates tumor growth and links inflammation to fibrosis. In addition to cytokines, m6A regulates chemokine system, with KIAA1429 stabilizing CCR9 mRNA in HBV-related HCC, driving metastasis and resistance to therapy. Conversely, YTHDF2-stabilized Cx3cl1 promotes CD8+ T cell recruitment and activation, underscoring m6A’s dual role in HCC.
ICIs, including PD-1/PD-L1 inhibitors (e.g., nivolumab and pembrolizumab) and CTLA-4 inhibitors (e.g., ipilimumab), have emerged as a central immunotherapy strategy for liver cancer, acting by restoring T-cell activity through the blockade of immune checkpoint molecules. Despite their promise, patient response rates to ICIs remain suboptimal, with resistance being a frequent challenge. Recent studies highlight the pivotal role of m6A modification in modulating PD-L1 expression and immune cell function within the TME. Targeting m6A may attenuate tumor immune evasion mechanisms, thereby enhancing the anti-tumor efficacy of ICIs in liver cancer.
The m6A methyltransferases METTL3 and METTL14 intricately orchestrate immune responses by selectively modulating specific mRNA targets. METTL3 amplifies the translation of SCAP mRNA in NAFLD-HCC, promoting cholesterol biosynthesis while simultaneously impairing the cytotoxic capacity of CD8+ T cells. Inhibition of METTL3 synergizes with anti-PD-1 therapy, thereby restoring immune functionality (79). Similarly, METTL14 facilitates the degradation of Siah2 mRNA through m6A modification and YTHDF2-mediated decay in cholangiocarcinoma (CCA). Siah2 promotes PD-L1 ubiquitination, enhancing immune evasion. Suppressing Siah2 significantly improves the therapeutic efficacy of immune checkpoint inhibitors (ICIs) (80). In intrahepatic cholangiocarcinoma (ICC), the m6A demethylase ALKBH5 has been shown to diminish m6A methylation on PD-L1 mRNA, shielding it from YTHDF2-mediated degradation. This stabilization sustains PD-L1 expression, suppressing T-cell activation and reshaping the tumor immune microenvironment (TIME) to favor immune escape. Notably, tumors with elevated nuclear ALKBH5 expression exhibit heightened sensitivity to anti-PD-1 therapy (81).
The m6A readers YTHDF1 and YTHDF2 play distinct but complementary roles in modulating immune responses. YTHDF1 enhances the translation of EZH2 mRNA in NASH-HCC, which promotes IL-6 secretion and the recruitment of MDSCs, collectively impairing CD8+ T-cell function (47). Strikingly, YTHDF1 silencing, when combined with anti-PD-1 therapy, significantly suppresses tumor progression. Moreover, circRHBDD1 functions as a cofactor for YTHDF1, directing it to PIK3R1 mRNA, thereby driving aerobic glycolysis and contributing to resistance against anti-PD-1 therapy (82). Conversely, YTHDF2 is essential for maintaining mitochondrial fitness and chromatin remodeling in CD8+ T cells via m6A-dependent RNA decay, processes critical for sustaining T-cell polyfunctionality (83). Loss of YTHDF2 diminishes the efficacy of ICIs, whereas inhibition of IKZF1/3 restores T-cell functionality, underscoring its therapeutic relevance.
Circular RNAs (circRNAs) stabilized by m6A modifications have also been identified as critical mediators of immune evasion and therapeutic resistance. In HCC, circCCAR1, stabilized through WTAP-mediated m6A modification, forms a feedback loop with miR-127-5p, upregulating WTAP expression. Secreted via exosomes, circCCAR1 stabilizes PD-1 expression in CD8+ T cells, inducing dysfunction and resistance to ICIs (48). Similarly, circRHBDD1 facilitates immune escape by recruiting YTHDF1 to PIK3R1 mRNA (82), thereby enhancing glycolysis—a vulnerability that can be therapeutically targeted to improve the efficacy of PD-1 blockade. One recent study revealed that TAM transfer m6A-modified circPETH via extracellular vesicles to HCC cells, where it encodes circPETH-147aa. This protein promotes glycolysis and metastasis by facilitating PKM2-catalyzed ALDOA-S36 phosphorylation and impairs CD8+ T cell function by stabilizing SLC43A2 mRNA, driving resistance to immune checkpoint blocker (ICB). Norathyriol, a small molecule targeting circPETH-147aa, reverses these effects, enhancing anti-PD1 therapy and restoring CD8+ T-cell activity, highlighting its potential in overcoming ICB resistance in HCC (84). Another study demonstrated lncRNA TUG1, upregulated by METTL3-mediated m6A modification, drives HCC immune evasion. The combination of TUG1-siRNA therapy with anti-PD-L1 antibodies exhibits synergistic tumor suppression effects (42).
Additionally, m6A modifies IDO1 expression through the IFN-γ/JAK1/STAT1 pathway, promoting immune escape and PD-L1 upregulation. IDO1 inhibition with Abrine reduces PD-L1 expression, enhances macrophage phagocytosis, and, when combined with anti-PD-1 therapy, improves T cell responses, suppressing tumor growth (85).
The intricate interplay between m6A regulatory pathways and immune responses unveils numerous therapeutic opportunities (Table 1). Targeting m6A writers (METTL3/14), erasers (ALKBH5), readers (YTHDF1/2), and circRNA-driven pathways (circCCAR1, circRHBDD1) represents a promising strategy to overcome resistance to ICIs.
m6A modification levels show promise as potential biomarkers for predicting HCC patient responses to immunotherapy. Integrating m6A-related gene expression with immunotherapy regimens could provide a basis for personalized treatment, improving patients’ responses to immunotherapy. Machine learning (ML) is a powerful tool for predicting immunotherapy efficacy in HCC, combining multi-omics data such as genomics, immune checkpoint expression, and TIME features. The inclusion of m6A-related gene signatures improves the accuracy of these ML models, facilitating better patient stratification based on tumor mutation burden (TMB), immune infiltration, and m6A modification patterns. The integration of m6A data with clinical and radiomic features offers a more comprehensive approach to personalized immunotherapy, providing insights into the likelihood of response to treatments like anti-PD-1 and CTLA-4 inhibitors. Further research is needed to validate these models for clinical use. Table 2 summarizes recent m6A-related risk models associated with predicting immunotherapy efficacy.
m6A modification, a prevalent form of RNA methylation, is emerging as a critical regulator of HCC progression and immune response. Acting as an epigenetic mechanism, m6A influences various cellular processes, including immune cell function, inflammatory cytokine production, and immune checkpoint regulation, ultimately shaping the TIME and impacting the efficacy of immunotherapy. Understanding how m6A alters the immune landscape in HCC reveals its potential as a therapeutic target, particularly for enhancing immunotherapy outcomes.
The TIME within HCC is composed of various immune cells, including T cells, macrophages, and dendritic cells, all of which interact with tumor cells to drive both tumor progression and immune evasion. m6A modification plays a central role in modulating the mRNA stability and translation of molecules that mediate these interactions. For instance, METTL3, a key m6A methyltransferase, promotes the expression of IL-6 (99), a cytokine known to recruit myeloid-derived suppressor cells (MDSCs) and inhibit CD8+ T cell cytotoxicity. This contributes to the suppression of antitumor immunity and supports tumor survival and growth. The m6A reader protein YTHDF1 further exacerbates this effect by enhancing the translation of EZH2 mRNA, which in turn amplifies IL-6 secretion and impairs T cell function (47). These findings suggest that m6A regulators, such as METTL3 and YTHDF1, represent potential therapeutic targets that could help restore immune surveillance and enhance the immune system’s ability to fight tumors.
In addition to cytokine regulation, m6A also influences immune checkpoint expression, which is central to the success of immunotherapy. PD-1/PD-L1 interactions dampen T cell activity and promote immune evasion in many cancers, including HCC. m6A modification is involved in regulating PD-L1 expression, thus influencing its interaction with immune cells. In intrahepatic cholangiocarcinoma, the demethylase ALKBH5 stabilizes PD-L1 mRNA through demethylation, maintaining its expression and suppressing T cell activation (81). Moreover, circRNAs stabilized by m6A enhance PD-L1 expression in HCC, leading to T cell dysfunction and contributing to resistance to ICIs (48, 82). These findings highlight the therapeutic potential of targeting m6A pathways to sensitize tumors to ICIs and improve immunotherapy responses.
Given the pivotal role of m6A in modulating the TIME and regulating immune checkpoints, it holds significant promise as a therapeutic target for enhancing immunotherapy. Preclinical studies have demonstrated that inhibiting METTL3 or YTHDF1 can synergize with anti-PD-1 therapy, significantly improving antitumor response and overall treatment efficacy (47, 100, 101). Furthermore, modulating m6A levels in inflammatory pathways, particularly those involving IL-6/STAT3 signaling, could reduce chronic inflammation and inhibit tumorigenesis, addressing fundamental drivers of HCC progression. This suggests that targeting m6A may not only enhance immune responses but also improve the TME, providing a more comprehensive approach for cancer treatment.
In conclusion, the intricate involvement of m6A in shaping the immune landscape and regulating immune checkpoints underscores its potential as a powerful therapeutic target in HCC. By strategically targeting m6A pathways, it may be possible to reprogram the TIME, enhance the effectiveness of ICIs, and provide novel strategies for treating HCC. Future research should focus on refining the specificity of m6A-targeting drugs, exploring their integration into combination therapies, and identifying biomarkers that can predict patient responses. These efforts could pave the way for more personalized and effective treatment options, offering new hope for patients with HCC.
FL: Conceptualization, Data curation, Formal analysis, Investigation, Visualization, Writing – original draft, Writing – review & editing. QL: Data curation, Validation, Visualization, Writing – original draft, Writing – review & editing. XL: Investigation, Validation, Writing – original draft, Writing – review & editing. YW: Investigation, Supervision, Validation, Writing – original draft, Writing – review & editing. RC: Investigation, Validation, Writing – original draft, Writing – review & editing. sz: Investigation, Validation, Writing – original draft, Writing – review & editing. SJ: Conceptualization, Formal analysis, Funding acquisition, Supervision, Writing – original draft, Writing – review & editing. jw: Conceptualization, Formal analysis, Funding acquisition, Methodology, Resources, Writing – original draft, Writing – review & editing.
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This study was supported by National Natural Science Foundation of China (grant no. 82074360), High-Level Traditional Chinese Medicine Fundamental Theory Key Discipline Construction Project of National Administration of Traditional Chinese Medicine (ZYYZDXK-2023118), Project of National Administration of Traditional Chinese Medicine (No. GZY-KJS-SD-2023-026), Natural Science Foundation of Shandong Province (grant no. ZR2022MH319 and ZR2022LZY027), and Key Research and Development Program of Jining City (grant no. 2023YXNS223 and 2022YXNS170).
Thanks to the editors and reviewers for their hard work and important comments.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declare that no Generative AI was used in the creation of this manuscript.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
1. Devarbhavi H, Asrani SK, Arab JP, Nartey YA, Pose E, Kamath PS. Global burden of liver disease: 2023 update. J Hepatol. (2023) 79:516–37. doi: 10.1016/j.jhep.2023.03.017
2. Rizzo A, Brunetti O, Brandi G. Hepatocellular carcinoma immunotherapy: predictors of response, issues, and challenges. Int J Mol Sci. (2024) 25:11091. doi: 10.3390/ijms252011091
3. Yin Y, Feng W, Chen J, Chen X, Wang G, Wang S, et al. Immunosuppressive tumor microenvironment in the progression, metastasis, and therapy of hepatocellular carcinoma: from bench to bedside. Exp Hematol Oncol. (2024) 13:72. doi: 10.1186/s40164-024-00539-x
4. Childs A, Aidoo-Micah G, Maini MK, Meyer T. Immunotherapy for hepatocellular carcinoma. JHEP Rep. (2024) 6:101130. doi: 10.1016/j.jhepr.2024.101130
5. Feng Q, Wang D, Xue T, Lin C, Gao Y, Sun L, et al. The role of RNA modification in hepatocellular carcinoma. Front Pharmacol. (2022) 13:984453. doi: 10.3389/fphar.2022.984453
6. Liu WW, Zheng SQ, Li T, Fei YF, Wang C, Zhang S, et al. RNA modifications in cellular metabolism: implications for metabolism-targeted therapy and immunotherapy. Signal Transduct Target Ther. (2024) 9:70. doi: 10.1038/s41392-024-01777-5
7. Chen D, Gu X, Nurzat Y, Xu L, Li X, Wu L, et al. Writers, readers, and erasers RNA modifications and drug resistance in cancer. Mol Cancer. (2024) 23:178. doi: 10.1186/s12943-024-02089-6
8. Liu L, Liang L, Li H, Shao W, Yang C, Lin F, et al. The role of m6A-mediated PD-1/PD-L1 in antitumor immunity. Biochem Pharmacol. (2023) 210:115460. doi: 10.1016/j.bcp.2023.115460
9. Chen J, Zhou Y, Wu M, Yuan Y, Wu W. m6A modification mediates exosomal LINC00657 to trigger breast cancer progression via inducing macrophage M2 polarization. Clin Breast Cancer. (2023) 23:546–60. doi: 10.1016/j.clbc.2023.04.007
10. Liu Q, Qi J, Li W, Tian X, Zhang J, Liu F, et al. Therapeutic effect and transcriptome-methylome characteristics of METTL3 inhibition in liver hepatocellular carcinoma. Cancer Cell Int. (2023) 23:298. doi: 10.1186/s12935-023-03096-1
11. Zha X, Gao Z, Li M, Xia X, Mao Z, Wang S. Insight into the regulatory mechanism of m(6)A modification: From MAFLD to hepatocellular carcinoma. BioMed Pharmacother. (2024) 177:116966. doi: 10.1016/j.biopha.2024.116966
12. Bokar JA, Shambaugh ME, Polayes D, Matera AG, Rottman FM. Purification and cDNA cloning of the AdoMet-binding subunit of the human mRNA (N6-adenosine)-methyltransferase. RNA. (1997) 3:1233–47.
13. Zaccara S, Ries RJ, Jaffrey SR. Reading, writing and erasing mRNA methylation. Nat Rev Mol Cell Biol. (2019) 20:608–24. doi: 10.1038/s41580-019-0168-5
14. Poh HX, Mirza AH, Pickering BF, Jaffrey SR. Alternative splicing of METTL3 explains apparently METTL3-independent m6A modifications in mRNA. PloS Biol. (2022) 20:e3001683. doi: 10.1371/journal.pbio.3001683
15. Ilieva M, Uchida S. Epitranscriptomics in fibroblasts and fibrosis. Am J Physiol Cell Physiol. (2022) 322:C1110–6. doi: 10.1152/ajpcell.00121.2022
16. Jiang X, Liu B, Nie Z, Duan L, Xiong Q, Jin Z, et al. The role of m6A modification in the biological functions and diseases. Signal Transduct Target Ther. (2021) 6:74. doi: 10.1038/s41392-020-00450-x
17. Lan Q, Liu PY, Haase J, Bell JL, Huttelmaier S, Liu T. The critical role of RNA m(6)A methylation in cancer. Cancer Res. (2019) 79:1285–92. doi: 10.1158/0008-5472.CAN-18-2965
18. Guo J, Zhao L, Duan M, Yang Z, Zhao H, Liu B, et al. Demethylases in tumors and the tumor microenvironment: Key modifiers of N(6)-methyladenosine methylation. BioMed Pharmacother. (2024) 174:116479. doi: 10.1016/j.biopha.2024.116479
19. Zhang D, Wornow S, Peehl DM, Rankin EB, Brooks JD. The controversial role and therapeutic development of the m6A demethylase FTO in renal cell carcinoma. Transl Oncol. (2022) 25:101518. doi: 10.1016/j.tranon.2022.101518
20. Shu W, Huang Q, Chen R, Lan H, Yu L, Cui K, et al. Complicated role of ALKBH5 in gastrointestinal cancer: an updated review. Cancer Cell Int. (2024) 24:298. doi: 10.1186/s12935-024-03480-5
21. Qu J, Yan H, Hou Y, Cao W, Liu Y, Zhang E, et al. RNA demethylase ALKBH5 in cancer: from mechanisms to therapeutic potential. J Hematol Oncol. (2022) 15:8. doi: 10.1186/s13045-022-01224-4
22. Zou Z, He C. The YTHDF proteins display distinct cellular functions on m(6)A-modified RNA. Trends Biochem Sci. (2024) 49:611–21. doi: 10.1016/j.tibs.2024.04.001
23. Yen YP, Chen JA. The m(6)A epitranscriptome on neural development and degeneration. J BioMed Sci. (2021) 28:40. doi: 10.1186/s12929-021-00734-6
24. Duan M, Liu H, Xu S, Yang Z, Zhang F, Wang G, et al. IGF2BPs as novel m(6)A readers: Diverse roles in regulating cancer cell biological functions, hypoxia adaptation, metabolism, and immunosuppressive tumor microenvironment. Genes Dis. (2024) 11:890–920. doi: 10.1016/j.gendis.2023.06.017
25. Ramesh-Kumar D, Guil S. The IGF2BP family of RNA binding proteins links epitranscriptomics to cancer. Semin Cancer Biol. (2022) 86:18–31. doi: 10.1016/j.semcancer.2022.05.009
26. Hu J, Chen K, Hong F, Gao G, Dai X, Yin H. METTL3 facilitates stemness properties and tumorigenicity of cancer stem cells in hepatocellular carcinoma through the SOCS3/JAK2/STAT3 signaling pathway. Cancer Gene Ther. (2024) 31:228–36. doi: 10.1038/s41417-023-00697-w
27. Yu F, Fang P, Fang Y, Chen D. Circ_0027791 contributes to the growth and immune evasion of hepatocellular carcinoma via the miR-496/programmed cell death ligand 1 axis in an m6A-dependent manner. Environ Toxicol. (2024) 39:3721–33. doi: 10.1002/tox.24188
28. Chen A, Zhang VX, Zhang Q, Sze KM, Tian L, Huang H, et al. Lyu X et al. Targeting the oncogenic m6A demethylase FTO suppresses tumourigenesis and potentiates immune response in hepatocellular carcinoma. Gut. (2024) 74(1):90–102. doi: 10.1136/gutjnl-2024-331903
29. Chen Y, Zhao Y, Chen J, Peng C, Zhang Y, Tong R, et al. ALKBH5 suppresses Malignancy of hepatocellular carcinoma via m(6)A-guided epigenetic inhibition of LYPD1. Mol Cancer. (2020) 19:123. doi: 10.1186/s12943-020-01239-w
30. Xu X, Wu S, Zhang Y, Fan W, Lin X, Chen K, et al. m6A modification of VEGFA mRNA by RBM15/YTHDF2/IGF2BP3 contributes to angiogenesis of hepatocellular carcinoma. Mol Carcinog. (2024) 63:2174–89. doi: 10.1002/mc.v63.11
31. Wang M, Yang Y, Yang J, Yang J, Han S. circ_KIAA1429 accelerates hepatocellular carcinoma advancement through the mechanism of m(6)A-YTHDF3-Zeb1. Life Sci. (2020) 257:118082. doi: 10.1016/j.lfs.2020.118082
32. Qin S, Jin H, Li Y, Chen X, He J, Xiao J, et al. Comprehensive analysis of IGF2BP3 with expression features, prognosis, immune modulation and stemness in hepatocellular carcinoma and pan-cancer. J Cancer. (2024) 15:2845–65. doi: 10.7150/jca.92768
33. Zang Y, Tian Z, Wang D, Li Y, Zhang W, Ma C, et al. METTL3-mediated N(6)-methyladenosine modification of STAT5A promotes gastric cancer progression by regulating KLF4. Oncogene. (2024) 43:2338–54. doi: 10.1038/s41388-024-03085-2
34. Shi Y, Wang Y, Zhang W, Niu K, Mao X, Feng K, et al. N6-methyladenosine with immune infiltration and PD-L1 in hepatocellular carcinoma: novel perspective to personalized diagnosis and treatment. Front Endocrinol (Lausanne). (2023) 14:1153802. doi: 10.3389/fendo.2023.1153802
35. Wen J, Xue L, Wei Y, Liang J, Jia W, Yong T, et al. YTHDF2 is a therapeutic target for HCC by suppressing immune evasion and angiogenesis through ETV5/PD-L1/VEGFA axis. Adv Sci (Weinh). (2024) 11:e2307242. doi: 10.1002/advs.202307242
36. Tian X, Yan T, Liu F, Liu Q, Zhao J, Xiong H, et al. Link of sorafenib resistance with the tumor microenvironment in hepatocellular carcinoma: Mechanistic insights. Front Pharmacol. (2022) 13:991052. doi: 10.3389/fphar.2022.991052
37. Kong Y, Guo L. Sunitinib suppresses M2 polarization of macrophages in tumor microenvironment to regulate hepatocellular carcinoma progression. J Biochem Mol Toxicol. (2023) 37:e23333. doi: 10.1002/jbt.23333
38. Ruishi X, Linyi X, Yunfan B, Wenbo Y, Xiaoying Z, Xiaoxue F, et al. New perspectives on chemokines in hepatocellular carcinoma therapy: a critical pathway for natural products regulation of the tumor microenvironment. Front Immunol. (2024) 15:1456405. doi: 10.3389/fimmu.2024.1456405
39. Bigos KJ, Quiles CG, Lunj S, Smith DJ, Krause M, Troost EG, et al. Tumour response to hypoxia: understanding the hypoxic tumour microenvironment to improve treatment outcome in solid tumours. Front Oncol. (2024) 14:1331355. doi: 10.3389/fonc.2024.1331355
40. Sin SQ, Mohan CD, Goh RMW, You M, Nayak SC, Chen L, et al. Hypoxia signaling in hepatocellular carcinoma: Challenges and therapeutic opportunities. Cancer Metastasis Rev. (2023) 42:741–64. doi: 10.1007/s10555-022-10071-1
41. Kong F, Wang K, Wang L. Systematic analysis of the expression profile and prognostic significance of m6A regulators and PD-L1 in hepatocellular carcinoma. Discovery Oncol. (2022) 13:131. doi: 10.1007/s12672-022-00595-x
42. Xi Q, Yang G, He X, Zhuang H, Li L, Lin B, et al. M(6)A-mediated upregulation of lncRNA TUG1 in liver cancer cells regulates the antitumor response of CD8(+) T cells and phagocytosis of macrophages. Adv Sci (Weinh). (2024) 11:e2400695. doi: 10.1002/advs.202400695
43. Xu W, Tao M, Liu Y, Yan J, Hu J, Wang L. METTL3-mediated SMPDL3A promotes cell growth, metastasis and immune process of hepatocellular carcinoma by regulating LRPPRC. Cell Signal. (2024) 127:111543. doi: 10.1016/j.cellsig.2024.111543
44. Pan Y, Chen H, Zhang X, Liu W, Ding Y, Huang D, et al. METTL3 drives NAFLD-related hepatocellular carcinoma and is a therapeutic target for boosting immunotherapy. Cell Rep Med. (2023) 4:101144. doi: 10.1016/j.xcrm.2023.101144
45. Yu F, Feng Y, Wang Q, Sun J. N(6)-methyladenosine (m(6)A) writer WTAP potentiates hepatocellular carcinoma immune evasion and aerobic glycolysis. Cell Biochem Biophys. (2024) 82:2321–31. doi: 10.1007/s12013-024-01342-5
46. Wang S, Xu N, Wang J, Chen Y, Li W, Chen H, et al. BMI1-induced CD127+KLRG1+ memory T cells enhance the efficacy of liver cancer immunotherapy. Cancer Lett. (2023) 571:216336. doi: 10.1016/j.canlet.2023.216336
47. Wang L, Zhu L, Liang C, Huang X, Liu Z, Huo J, et al. Targeting N6-methyladenosine reader YTHDF1 with siRNA boosts antitumor immunity in NASH-HCC by inhibiting EZH2-IL-6 axis. J Hepatol. (2023) 79:1185–200. doi: 10.1016/j.jhep.2023.06.021
48. Hu Z, Chen G, Zhao Y, Gao H, Li L, Yin Y, et al. Exosome-derived circCCAR1 promotes CD8 + T-cell dysfunction and anti-PD1 resistance in hepatocellular carcinoma. Mol Cancer. (2023) 22:55. doi: 10.1186/s12943-023-01759-1
49. Wei H, Li W, Yang M, Fang Q, Nian J, Huang Y, et al. METTL3/16-mediated m(6)A modification of ZNNT1 promotes hepatocellular carcinoma progression by activating ZNNT1/osteopontin/S100A9 positive feedback loop-mediated crosstalk between macrophages and tumour cells. Clin Immunol. (2024) 261:109924. doi: 10.1016/j.clim.2024.109924
50. Ye Y, Wang M, Wang G, Mai Z, Zhou B, Han Y, et al. lncRNA miR4458HG modulates hepatocellular carcinoma progression by activating m6A-dependent glycolysis and promoting the polarization of tumor-associated macrophages. Cell Mol Life Sci. (2023) 80:99. doi: 10.1007/s00018-023-04741-8
51. Wang L, Yi X, Xiao X, Zheng Q, Ma L, Li B. Exosomal miR-628-5p from M1 polarized macrophages hinders m6A modification of circFUT8 to suppress hepatocellular carcinoma progression. Cell Mol Biol Lett. (2022) 27:106. doi: 10.1186/s11658-022-00406-9
52. You Y, Wen D, Zeng L, Lu J, Xiao X, Chen Y, et al. ALKBH5/MAP3K8 axis regulates PD-L1+ macrophage infiltration and promotes hepatocellular carcinoma progression. Int J Biol Sci. (2022) 18:5001–18. doi: 10.7150/ijbs.70149
53. Tao L, Li D, Mu S, Tian G, Yan G. LncRNA MAPKAPK5_AS1 facilitates cell proliferation in hepatitis B virus -related hepatocellular carcinoma. Lab Invest. (2022) 102:494–504. doi: 10.1038/s41374-022-00731-9
54. Han D, Liu J, Chen C, Dong L, Liu Y, Chang R, et al. Anti-tumour immunity controlled through mRNA m(6)A methylation and YTHDF1 in dendritic cells. Nature. (2019) 566:270–4. doi: 10.1038/s41586-019-0916-x
55. Bai X, Wong CC, Pan Y, Chen H, Liu W, Zhai J, et al. Loss of YTHDF1 in gastric tumors restores sensitivity to antitumor immunity by recruiting mature dendritic cells. J Immunother Cancer. (2022) 10:e003663. doi: 10.1136/jitc-2021-003663
56. Wen C, Wang L, Piffkó A, Chen D, Yu X, Zawieracz K, et al. YTHDF1 loss in dendritic cells potentiates radiation-induced antitumor immunity via STING-dependent type I IFN production. J Clin Invest. (2024) 134(23):e181612. doi: 10.1172/JCI181612
57. Feng L, Li M, Ma J, Wang W, Wang S, Mao Z, et al. ALKBH5 regulates arginase 1 expression in MDSCs and their immunosuppressive activity in tumor-bearing host. Noncoding RNA Res. (2024) 9:913–20. doi: 10.1016/j.ncrna.2024.03.003
58. Chen H, Pan Y, Zhou Q, Liang C, Wong CC, Zhou Y, et al. METTL3 inhibits antitumor immunity by targeting m(6)A-BHLHE41-CXCL1/CXCR2 axis to promote colorectal cancer. Gastroenterology. (2022) 163:891–907. doi: 10.1053/j.gastro.2022.06.024
59. Wang J, Ling D, Shi L, Li H, Peng M, Wen H, et al. METTL3-mediated m6A methylation regulates ovarian cancer progression by recruiting myeloid-derived suppressor cells. Cell Biosci. (2023) 13:202. doi: 10.1186/s13578-023-01149-6
60. Hu Y, Li Y, Xiong H, Zhang Y, Wang F, Zhuo W, et al. Exosomal SLC16A1-AS1-induced M2 macrophages polarization facilitates hepatocellular carcinoma progression. Int J Biol Sci. (2024) 20:4341–63. doi: 10.7150/ijbs.94440
61. Ding H, Zhang X, Su Y, Jia C, Dai C. GNAS promotes inflammation-related hepatocellular carcinoma progression by promoting STAT3 activation. Cell Mol Biol Lett. (2020) 25:8. doi: 10.1186/s11658-020-00204-1
62. Hou J, Zhang H, Liu J, Zhao Z, Wang J, Lu Z, et al. YTHDF2 reduction fuels inflammation and vascular abnormalization in hepatocellular carcinoma. Mol Cancer. (2019) 18:163. doi: 10.1186/s12943-019-1082-3
63. Wei H, Yang J, Lu R, Huang Y, Huang Z, Huang L, et al. m(6)A modification of AC026356.1 facilitates hepatocellular carcinoma progression by regulating the IGF2BP1-IL11 axis. Sci Rep. (2023) 13:19124. doi: 10.1038/s41598-023-45449-w
64. Gan X, Dai Z, Ge C, Yin H, Wang Y, Tan J, et al. FTO promotes liver inflammation by suppressing m6A mRNA methylation of IL-17RA. Front Oncol. (2022) 12:989353. doi: 10.3389/fonc.2022.989353
65. Pan XS, Li BW, Wang LL, Li N, Lin HM, Zhang J, et al. Kupffer cell pyroptosis mediated by METTL3 contributes to the progression of alcoholic steatohepatitis. FASEB J. (2023) 37:e22965. doi: 10.1096/fj.202300059RR
66. Liao Z, Zhang H, Liu F, Wang W, Liu Y, Su C, et al. m(6)A-dependent ITIH1 regulated by TGF-beta acts as a target for hepatocellular carcinoma progression. Adv Sci (Weinh). (2024) 11:e2401013. doi: 10.1002/advs.202401013
67. Wang H, Guan W, Zhang X, Wu Y, Ou Y, Zhang Y, et al. TGF-beta promotes hepatocellular carcinoma metastasis through through m6A modification of PCDHGA9. Cancer Biother Radiopharm. (2024) 39:644–53. doi: 10.1089/cbr.2023.0144
68. Liu Y, Da M. Wilms tumor 1 associated protein promotes epithelial mesenchymal transition of gastric cancer cells by accelerating TGF-beta and enhances chemoradiotherapy resistance. J Cancer Res Clin Oncol. (2023) 149:3977–88. doi: 10.1007/s00432-022-04320-7
69. Tan B, Zhou K, Liu W, Prince E, Qing Y, Li Y, et al. RNA N(6) -methyladenosine reader YTHDC1 is essential for TGF-beta-mediated metastasis of triple negative breast cancer. Theranostics. (2022) 12:5727–43. doi: 10.7150/thno.71872
70. Feng Y, Dong H, Sun B, Hu Y, Yang Y, Jia Y, et al. METTL3/METTL14 transactivation and m(6)A-dependent TGF-beta1 translation in activated kupffer cells. Cell Mol Gastroenterol Hepatol. (2021) 12:839–56. doi: 10.1016/j.jcmgh.2021.05.007
71. Wang X, Wang J, Tsui YM, Shi C, Wang Y, Zhang X, et al. RALYL increases hepatocellular carcinoma stemness by sustaining the mRNA stability of TGF-beta2. Nat Commun. (2021) 12:1518. doi: 10.1038/s41467-021-21828-7
72. Chen G, Zhao Q, Yuan B, Wang B, Zhang Y, Li Z, et al. ALKBH5-modified HMGB1-STING activation contributes to radiation induced liver disease via innate immune response. Int J Radiat Oncol Biol Phys. (2021) 111:491–501. doi: 10.1016/j.ijrobp.2021.05.115
73. Xiao Z, Chen H, Xu N, Chen Y, Wang S, Xu X. MATR3 promotes liver cancer progression by suppressing DHX58-mediated type I interferon response. Cancer Lett. (2024) 604:217231. doi: 10.1016/j.canlet.2024.217231
74. Ding WB, Wang MC, Yu J, Huang G, Sun DP, Liu L, et al. HBV/pregenomic RNA increases the stemness and promotes the development of HBV-related HCC through reciprocal regulation with insulin-like growth factor 2 mRNA-binding protein 3. Hepatology. (2021) 74:1480–95. doi: 10.1002/hep.31850
75. Xiao P, Li M, Zhou M, Zhao X, Wang C, Qiu J, et al. TTP protects against acute liver failure by regulating CCL2 and CCL5 through m6A RNA methylation. JCI Insight. (2021) 6:e149276. doi: 10.1172/jci.insight.149276
76. Zhou S, Yang K, Chen S, Lian G, Huang Y, Yao H, et al. CCL3 secreted by hepatocytes promotes the metastasis of intrahepatic cholangiocarcinoma by VIRMA-mediated N6-methyladenosine (m(6)A) modification. J Transl Med. (2023) 21:43. doi: 10.1186/s12967-023-03897-y
77. Lv Z, Liu L, You J, Zhou P, Su Y, Zhao K, et al. Small HBV surface antigen drives regorafenib resistance in HCC via KIAA1429-dependent m6A modification of CCR9. J Med Virol. (2024) 96:e29894. doi: 10.1002/jmv.29894
78. Yang Z, Wang X, Fu Y, Wu W, Hu Z, Lin Q, et al. YTHDF2 in peritumoral hepatocytes mediates chemotherapy-induced antitumor immune responses through CX3CL1-mediated CD8(+) T cell recruitment. Mol Cancer. (2024) 23:186. doi: 10.1186/s12943-024-02097-6
79. Wang YF, Zhang WL, Li ZX, Liu Y, Tan J, Yin HZ, et al. METTL14 downregulation drives S100A4(+) monocyte-derived macrophages via MyD88/NF-kappaB pathway to promote MAFLD progression. Signal Transduct Target Ther. (2024) 9:91. doi: 10.1038/s41392-024-01797-1
80. Zheng H, Zheng WJ, Wang ZG, Tao YP, Huang ZP, Yang L, et al. Decreased expression of programmed death ligand-L1 by seven in absentia homolog 2 in cholangiocarcinoma enhances T-cell-mediated antitumor activity. Front Immunol. (2022) 13:845193. doi: 10.3389/fimmu.2022.845193
81. Qiu X, Yang S, Wang S, Wu J, Zheng B, Wang K, et al. M(6)A demethylase ALKBH5 regulates PD-L1 expression and tumor immunoenvironment in intrahepatic cholangiocarcinoma. Cancer Res. (2021) 81:4778–93. doi: 10.1158/0008-5472.CAN-21-0468
82. Cai J, Chen Z, Zhang Y, Wang J, Zhang Z, Wu J, et al. CircRHBDD1 augments metabolic rewiring and restricts immunotherapy efficacy via m(6)A modification in hepatocellular carcinoma. Mol Ther Oncolytics. (2022) 24:755–71. doi: 10.1016/j.omto.2022.02.021
83. Zhang H, Luo X, Yang W, Wu Z, Zhao Z, Pei X, et al. YTHDF2 upregulation and subcellular localization dictate CD8 T cell polyfunctionality in anti-tumor immunity. Nat Commun. (2024) 15:9559. doi: 10.1038/s41467-024-53997-6
84. Lan T, Gao F, Cai Y, Lv Y, Zhu J, Liu H, et al. The protein circPETH-147aa regulates metabolic reprogramming in hepatocellular carcinoma cells to remodel immunosuppressive microenvironment. Nat Commun. (2025) 16:333. doi: 10.1038/s41467-024-55577-0
85. Liang X, Gao H, Xiao J, Han S, He J, Yuan R, et al. Abrine, an IDO1 inhibitor, suppresses the immune escape and enhances the immunotherapy of anti-PD-1 antibody in hepatocellular carcinoma. Front Immunol. (2023) 14:1185985. doi: 10.3389/fimmu.2023.1185985
86. Ma E, Li J, Shen C, Gu Y, Zhang X, Li L, et al. The m(6)A-related gene signature stratifies poor prognosis patients and characterizes immunosuppressive microenvironment in hepatocellular carcinoma. Front Immunol. (2023) 14:1227593. doi: 10.3389/fimmu.2023.1227593
87. Yan X, Qi Y, Yao X, Yin L, Wang H, Fu J, et al. N6-methyladenosine regulators in hepatocellular carcinoma: investigating the precise definition and clinical applications of biomarkers. Biol Direct. (2024) 19:103. doi: 10.1186/s13062-024-00554-2
88. Zou R, Liu Y, Qiu S, Lu Y, Chen Y, Yu H, et al. The identification of N6-methyladenosine-related miRNAs predictive of hepatocellular carcinoma prognosis and immunotherapy efficacy. Cancer biomark. (2023) 38:551–66. doi: 10.3233/CBM-230263
89. Wu X, Wang Y, Tao L, Zhou Y, Zhang X, Wang L, et al. Comprehensive molecular analyses of a 7-m6A-related lncRNAs signature for prognosis, tumor immunity and therapeutic effect in patients with hepatocellular carcinoma. Cell Mol Biol (Noisy-le-grand). (2024) 70:186–93. doi: 10.14715/cmb/2024.70.1.25
90. Zhang C, Zhang W, Shui Y, Li P, Tian Z, Duan S, et al. Implications of m6A-associated snRNAs in the prognosis and immunotherapeutic responses of hepatocellular carcinoma. Front Immunol. (2022) 13:1001506. doi: 10.3389/fimmu.2022.1001506
91. Liu D, Zhou X, Zhao J. Prognostic signature and immune efficacy of m(1)A-, m(5)C-, m(6)A-, m(7)G-, and DNA methylation-related regulators in hepatocellular carcinoma. J Cancer. (2024) 15:4287–300. doi: 10.7150/jca.95730
92. Ren M, Fan B, Cao G, Zong R, Feng L, Sun H. Exploration and validation of a combined Hypoxia and m6A/m5C/m1A regulated gene signature for prognosis prediction of liver cancer. BMC Genomics. (2023) 24:776. doi: 10.1186/s12864-023-09876-3
93. Xiao Y, Li J, Wu J. Development and validation of a novel prognostic signature based on m6A/m5C/m1A-related genes in hepatocellular carcinoma. BMC Med Genomics. (2023) 16:177. doi: 10.1186/s12920-023-01611-x
94. Liu T, Sun L, Li ZZ, Yang K, Chen JM, Han XY, et al. The m6A/m5C/m1A regulator genes signature reveals the prognosis and is related with immune microenvironment for hepatocellular carcinoma. BMC Gastroenterol. (2023) 23:147. doi: 10.1186/s12876-023-02776-6
95. Zhou D, Wang Y, Wei W, Zhou W, Gu J, Kong Y, et al. m6A regulator-mediated methylation modification highlights immune infiltration patterns for predicting risk in hepatocellular carcinoma. J Cancer Res Clin Oncol. (2023) 149:3661–80. doi: 10.1007/s00432-022-04255-z
96. Jiang H, Ning G, Wang Y, Lv W. Identification of an m6A-related signature as biomarker for hepatocellular carcinoma prognosis and correlates with sorafenib and anti-PD-1 immunotherapy treatment response. Dis Markers. (2021) 2021:5576683. doi: 10.1155/2021/5576683
97. Li L, Xie R, Lu G. Identification of m6A methyltransferase-related lncRNA signature for predicting immunotherapy and prognosis in patients with hepatocellular carcinoma. Biosci Rep. (2021) 41:BSR20210760. doi: 10.1042/BSR20210760
98. Cai J, Zhou M, Xu J. N6-methyladenosine (m6A) RNA methylation regulator SNRPC is a prognostic biomarker and is correlated with immunotherapy in hepatocellular carcinoma. World J Surg Oncol. (2021) 19:241. doi: 10.1186/s12957-021-02354-8
99. Huang J, Wang Y, Zhou Y. METTL3 and METTL14 regulate IL-6 expression via RNA m6A modification of zinc transporter SLC39A9 and DNA methylation of IL-6 in periodontal ligament cells. Biochim Biophys Acta Mol Cell Res. (2024) 1871:119605. doi: 10.1016/j.bbamcr.2023.119605
100. Bao Y, Zhai J, Chen H, Wong CC, Liang C, Ding Y, et al. Targeting m(6)A reader YTHDF1 augments antitumour immunity and boosts anti-PD-1 efficacy in colorectal cancer. Gut. (2023) 72:1497–509. doi: 10.1136/gutjnl-2022-328845
Keywords: hepatocellular carcinoma, tumor immune microenvironment, m6A modification, immune checkpoint inhibitors, immunotherapy resistance
Citation: Liu F, Liu Q, Li X, Wang Y, Cao R, Zhang S, Jiang S and Wu J (2025) m6A epitranscriptomic modification in hepatocellular carcinoma: implications for the tumor microenvironment and immunotherapy. Front. Immunol. 16:1538658. doi: 10.3389/fimmu.2025.1538658
Received: 03 December 2024; Accepted: 27 January 2025;
Published: 17 February 2025.
Edited by:
Qun Chen, University of Oklahoma, United StatesReviewed by:
Yu’e Liu, Harvard Medical School, United StatesCopyright © 2025 Liu, Liu, Li, Wang, Cao, Zhang, Jiang and Wu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jianlin Wu, d3VqaWFubGluMjAxOEAxNjMuY29t; Shulong Jiang, am5zbGppYW5nQG1haWwuam5tYy5lZHUuY24=
†These authors have contributed equally to this work and share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.