- 1Department of Dermatology, The Affiliated Hospital of the Non-Commissioned Officer (NCO) School, The Army Medical University, Shijiazhuang, Hebei, China
- 2Institute of Immunology, Department of Basic Medicine, The Army Military Medical University, Chongqing, China
- 3Chronic Disease Research Center, Medical College, Dalian University, Dalian, Liaoning, China
- 4Department of Otolaryngology, The Second Affiliated Hospital of the Army Military Medical University, Chongqing, China
MS4A (membrane-spanning 4-domain, subfamily A) molecules are categorized into tetraspanins, which possess four-transmembrane structures. To date, eighteen MS4A members have been identified in humans, whereas twenty-three different molecules have been identified in mice. MS4A proteins are selectively expressed on the surfaces of various immune cells, such as B cells (MS4A1), mast cells (MS4A2), macrophages (MS4A4A), Foxp3+CD4+ regulatory T cells (MS4A4B), and type 3 innate lymphoid cells (TMEM176A and TMEM176B). Early research confirmed that most MS4A molecules function as ion channels that regulate the transport of calcium ions. Recent studies have revealed that some MS4A proteins also function as chaperones that interact with various immune molecules, such as pattern recognition receptors and/or immunoglobulin receptors, to form immune complexes and transmit downstream signals, leading to cell activation, growth, and development. Evidence from preclinical animal models and human genetic studies suggests that the MS4A superfamily plays critical roles in the pathogenesis of various diseases, including cancer, infection, allergies, neurodegenerative diseases and autoimmune diseases. We review recent progress in this field and focus on elucidating the molecular mechanisms by which different MS4A molecules regulate the progression of tumors, Alzheimer’s disease, and autoimmune diseases. Therefore, in-depth research into MS4A superfamily members may clarify their ability to act as candidate biomarkers and therapeutic targets for these diseases. Eighteen distinct members of the MS4A (membrane-spanning four-domain subfamily A) superfamily of four-transmembrane proteins have been identified in humans, whereas the MS4A genes are translated into twenty-three different molecules in mice. These proteins are selectively expressed on the surface of various immune cells, such as B cells (MS4A1), macrophages (MS4A4A), mast cells (MS4A2), Foxp3+CD4+ regulatory T cells (MS4A4B), type 3 innate lymphoid cells (TMEM176A and TMEM176B) and colonic epithelial cells (MS4A12). Functionally, most MS4A molecules function as ion channels that regulate the flow of calcium ions [Ca2+] across cell membranes. Recent studies have revealed that some MS4A proteins also act as molecular chaperones and interact with various types of immune receptors, including pattern recognition receptors (PRRs) and immunoglobulin receptors (IgRs), to form signaling complexes, thereby modulating intracellular signaling and cellular activity. Evidence from preclinical animal models and human genetic studies suggests that MS4A proteins play critical roles in various diseases (2). Therefore, we reviewed the recent progress in understanding the role of the MS4A superfamily in diseases, particularly in elucidating its function as a candidate biomarker and therapeutic target for cancer.
1 MS4A genes and structural characteristics of MS4A proteins
The human MS4A superfamily consists of eighteen different members (MS4A115 and MS4A18), which are encoded by genomic loci located on chromosome 11q12, in contrast, the MS4A genes in mice are located on chromosome 19 and encode twenty-three different molecules (1). TMEM176A (HCA112) and TMEM176B (LR8/TORID), whose genes are specifically located at 7q36.1 on chromosome 7, are also categorized as members of the MS4A superfamily because they have similar four-transmembrane protein structures, but their proteins only share ~16% amino acid sequence similarity with MS4A molecules (3). Genome-wide association studies (GWAS) have identified susceptibility genes for atopy and allergic diseases within this genomic region, indicating that mutations in MS4A genes might predispose individuals to related diseases (4). The MS4A genes first appeared in cartilaginous fish and are expressed in tissues beyond the immune system. These genes have since evolved and are now present in various vertebrates, including mammals, birds, reptiles, and amphibians. The TMEM176 gene, however, is expressed primarily in mammals and bony fish (3).
Most MS4A proteins consist of 200~300 amino acids, with a molecular weight of approximately 22~35 kDa. According to transmembrane prediction databases, nearly all identified MS4A proteins, except for MS4A4E and MS4A6E, possess a four-transmembrane structure, therefore, they are classified as tetraspanins (2). Tetraspanins can interact with other proteins to form tetraspanin-enriched microdomains (TEMs) on the cellular membrane, which play critical roles in regulating cellular physiological processes (5). MS4A molecules typically contain two extracellular loops and one intracellular loop, and both the N- and C-termini are located intracellularly. Among the different MS4A proteins, the first three transmembrane regions exhibit high homology: the first transmembrane region of the extracellular loop has approximately thirteen amino acids and consists of conserved amino acid sequences such as VLGAIQIL, LGAXQI, and LSLG. The second extracellular loop varies in length from 1046 amino acids, with its transmembrane region containing conserved sequences, such as GYPFWG and FIISGSLS, and this region exhibits significant heterogeneity between different molecules. Interestingly, the conserved sequences SLX2NX2 and SX3AX2G are commonly found in the third transmembrane region (6, 7). In addition to MS4A8B and MS4A12, most MS4A molecules possess two cysteine residues in the second extracellular loop that can form disulfide bonds. Additionally, the intracellular segments of MS4A proteins generally contain SH2 and SH3 domain binding sites (7). Notably, the intracellular segment of MS4A2 includes an immunoreceptor tyrosine-based activation motif (ITAM), whereas MS4A8B and TMEM176B contain an immunoreceptor tyrosine-based inhibitory motif (ITIM), these structures facilitate the formation of signal transduction complexes. Therefore, MS4A proteins can regulate cytoskeletal remodeling, transcriptional responses, signal transduction cascades, and cellular differentiation (8).
2 Characteristic expression of MS4A molecules in cell subsets and tissues
Most MS4A molecules are expressed on the surface of immune cells. For example, MS4A1 (CD20) is expressed primarily on the membrane of precursors and mature B cells, but its expression is lost on plasma cells. The high-affinity IgE receptor (FcϵRI) is a tetramer that consists of one α subunit, one β subunit, and two γ subunits. MS4A2 (FcϵRβ) serves as the β subunit of both the high-affinity FcϵRI and the low-affinity IgG receptor (FcγRIII), which are predominantly expressed on the surface of mast cells and basophils (6). MS4A3 (HTm4) is selectively expressed on various myeloid and lymphoid progenitor cells within the hematopoietic system (1). MS4A5 (TETM4.1) is expressed on the surface of precursor monocytes (2). Although MS4A8B (L985P) has been detected in B-cell lines, such as BJAB, DAUDI, and SB, it is expressed primarily on ciliated cells of the bronchial mucosal epithelium (6, 7). MS4A12 is exclusively expressed in the mucosal epithelial cells of colonic tissue and in colon cancer cell lines (9). Notably, TMEM176A (HCA112) and TMEM176B (LR8 or TORID) are highly expressed on immature or resting dendritic cells (DCs), as well as on the surface of helper T cells and type 3 innate lymphoid cells (ILC-3) (10). Moreover, MS4A4B appears to be restricted to Foxp3+ regulatory T cells (Tregs) based on a yeast split-ubiquitin Treg library screen (11). Notably, some MS4A proteins may exist as homo- and/or heterodimers, for example, TMEM176A and TMEM176B have been shown to interact with each other in DCs. Additionally, MS4A4A associates with itself and/or with two other members of the MS4A family, including MS4A6A and MS4A7, to form characteristic molecular clusters on the surface of macrophages (12). Here, we list the distributions of several major MS4A molecules in cell types and tissues (Table 1).
3 The predominant biofunctions of MS4A molecules
3.1 MS4A proteins act as ion channels
Early studies confirmed that MS4A molecules function primarily as ion channels that regulate the exchange of calcium ions [Ca2+] between the intracellular and extracellular environments. For example, MS4A1 is present in both homodimers and heterodimers on human B lymphoblastoid cells, where it serves as an ion channel to regulate [Ca2+] flow across the cell membrane, as evidenced by the increase in [Ca2+] conductance after transfection with MS4A1 (64, 65). MS4A1 crosslinks with BCR and positively regulates BCR-induced cytoplasmic [Ca2+] mobilization (66). MS4A2 contains an ITAM motif ((D/E)-XXYXXL-(X)7–9YXX-L/I) in its C-terminus, and crosslinking of the IgE-bound FcϵRI receptor by an antigen can phosphorylate tyrosine residues in the ITAMs of MS4A2, triggering downstream signal cascades that result in the mobilization of intracellular calcium stores (67); conversely, the downregulation of MS4A2 reduces [Ca2+] influx upon IgE cross-linking (67). Human mast cells express MS4A4A, and silencing MS4A4A via RNAi reduces [Ca2+] influx upon IgE crosslinking, suggesting that MS4A4A can also regulate store-operated [Ca2+] entry (68). MS4A12 is a novel component of store-operated [Ca2+] entry in intestinal cells, and loss of MS4A12 in LoVo colon cancer cells attenuates epidermal growth factor receptor-mediated signaling, thereby promoting colonic carcinoma migration (9). Recent two-photon imaging of olfactory epithelial cells has also demonstrated that different MS4A proteins can recognize specific chemical ligands, thereby increasing the activity of the calcium indicator GCaMP6 (69). Additionally, Xenopus oocytes express TMEM176B, which initiates an inward current activated by acidification of the extracellular solution to pH 5, suggesting that TMEM176B might function as an acid-sensitive, nonselective, monovalent cation channel (61). Because TMEM176A also induces an inward current activated by acidification of the extracellular solution in Xenopus oocytes, TMEM176A may act as a cation channel (70).
3.2 MS4A proteins function as molecular chaperones
Recent studies have suggested that some MS4A molecules function as chaperones that interact with pattern recognition receptors (PRRs) and/or immunoglobulin receptors (IgRs). For example, MS4A1 interacts with membrane proteins, such as major histocompatibility complex (MHC) class I proteins, MHC class II proteins, tetraspanins (CD53, CD81, and CD82), and the BCR, thereby promoting B-cell activation and antibody class switching (Figure 1A) (71, 72). MS4A2 associates with FcϵRIα and FcϵRIγ to form a tetrameric receptor complex (αβγ2) on mast cells at high density, therefore, MS4A2 acts as a signal amplifier through its ability to increase Lyn-dependent phosphorylation of FcϵRIγ (Figure 1B) (73). Human MS4A3 (HTm4) regulates the cell cycle of hematopoietic cells and controls their differentiation into various cell types through binding to the cyclin-dependent kinase-associated (CDK-associated) phosphatase-CDK2 (KAP-CDK2) complex (74), and the overexpression of MS4A3 causes cell cycle arrest at the G(0)/G(1) phase (Figure 1C). MS4A4A4A interacts with and colocalizes with the β-glucan receptor Dectin-1 in lipid rafts, and the MS4A4A/Dectin-1 complex can trigger downstream spleen tyrosine kinase (SYK) signaling cascades to induce macrophage and NK cell activation in response to dectin-1 ligands (12, 75). Additionally, MS4A4A on mast cells facilitates trafficking of the receptor tyrosine kinase KIT to caveolin-1-rich microdomains through endocytic recycling rather than degradation pathways, thus promoting KIT signaling in endosomes (Figure 1D) (76). MS4A4B interacts with the glucocorticoid-induced tumor necrosis factor receptor (GITR) on Foxp3+CD4+ Tregs, forming a membrane signaling complex (MS4A4B/GITR) that enhances IL-2 secretion in response to triggering with GITR ligands or anti-GITR Abs (Figure 1E) (11). In mice, MS4A4B is highly expressed on T cells and influences T-cell apoptosis by controlling the activity of caspases 3, 8 and 9. Conversely, knocking down Ms4a4b with siRNA or shRNA promotes apoptosis in naïve T cells or the T32 cell line, whereas the overexpression of Ms4a4b reduces EL-4 cell apoptosis (77, 78).
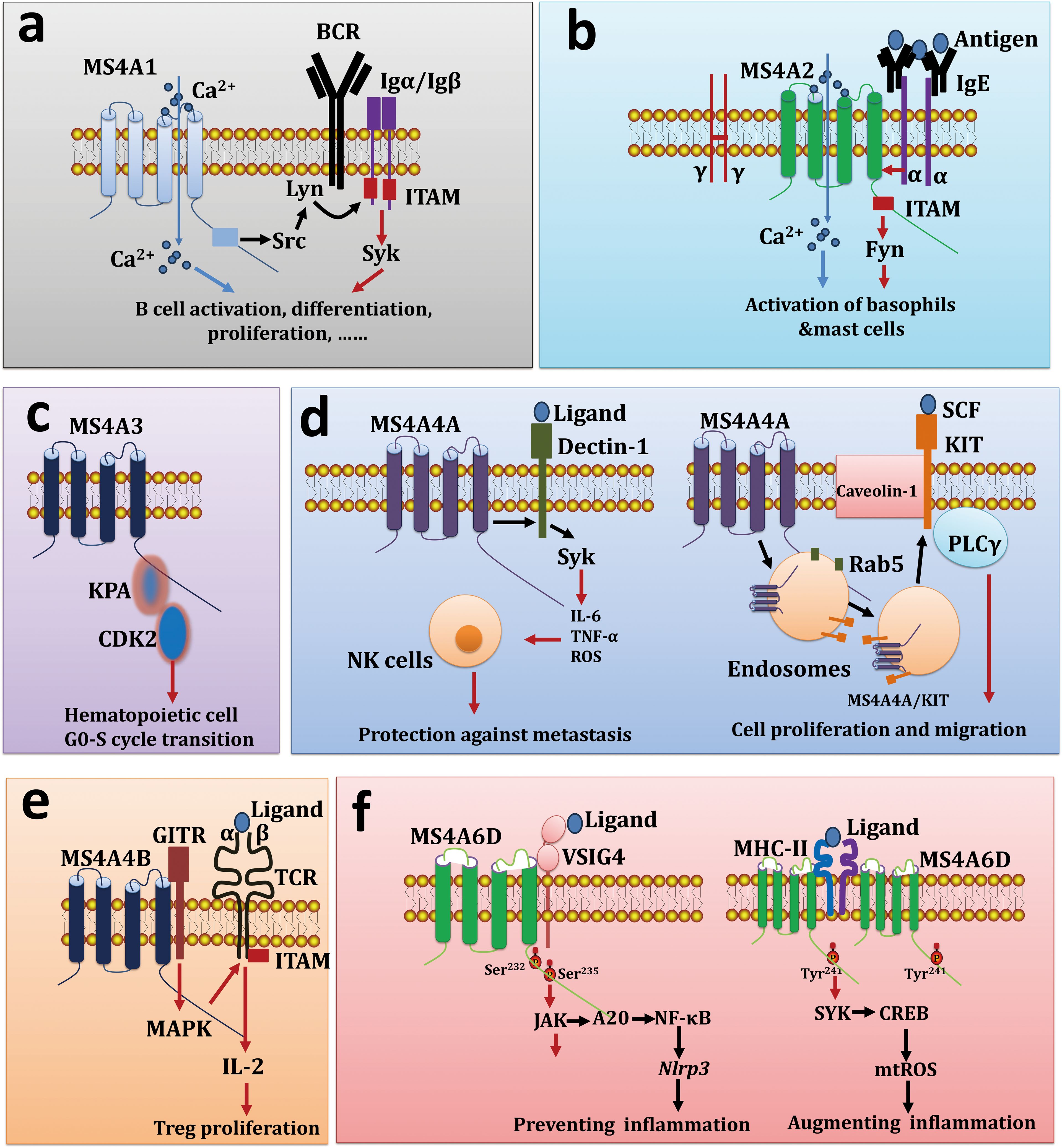
Figure 1. MS4A proteins function as molecular chaperones. (A) MS4A1 interacts with BCR to induce B cell activation, differentiation, and proliferation. (B) MS4A2 as a receptor for IgE and triggers the activation of basophils as well as mast cells. (C) MS4A3 promotes hematopoietic cell G0-S cycle transition via interaction with KPA and CDK2. (D) MS4A4A/Dectin-1 complex induce NK cell activation and protection against metastasis (left), on the other hand, MS4A4A/KIT complex also can controls cell proliferation and migration (right). (E) MS4A4B crosslink with GITR and control Treg proliferation. (F) VSIG4 forms a complex with MS4A6D, regulating the Jak-STAT1-A20-NF-κB pathway, which in turn influences the transcription of Nlrp3 and its substrate Il1b genes (left), on the other hand, MHC-II forms a complex with MS4A6D, regulating the SYK-CREB-SDHB pathway, which affects mitochondrial mtROS secretion, leading to macrophage inflammatory responses.
Our recent research revealed that MS4A6D, a novel member of the MS4A superfamily, is restrictively expressed in tissue-resident macrophages and DCs in mice. MS4A6D interacts with VSIG4 to form a cell surface signaling complex, activating the STAT3-A20-NF-κB signaling pathway and thereby inhibiting the transcription of the Nlrp3 and Il1b genes in peritoneal macrophages (Figure 1F) (79). Recent studies have shown that MS4A6D on the surface of monocyte−macrophages cross-links with MHC-II molecules under inflammatory conditions, activating downstream SYK signaling and leading to the release of the inflammatory cytokines IL-1, IL-6, TNFα, and mitochondrial reactive oxygen species (mtROS) (80). Notably, we found that MS4A6D primarily exists as a homodimer, with the cysteine at position 237 playing a decisive role in dimer formation (Figure 1F). Therefore, MS4A6D plays a crucial role in macrophage activation and the progression of inflammatory diseases.
4 Associations between MS4A molecules and diseases
Evidence from preclinical animal models and human genetic studies indicates that most members of the MS4A superfamily play critical roles in various pathological disorders, including cancer, infectious diseases, and neurodegenerative diseases. As a result, some MS4A members might serve as candidate biomarkers and therapeutic targets for specific diseases.
4.1 MS4A superfamily molecules and Alzheimer’s disease
Alzheimer’s disease (AD) is the most common cause of dementia worldwide, with the prevalence continuing to grow in part because of the aging population. The progression of this neurodegenerative disease is characterized by two hallmark pathologies: β-amyloid (Aβ) plaque deposition and neurofibrillary tangles of hyperphosphorylated Tau (81). AD predominantly affects elderly individuals, especially those who are older than 85 years. Nearly 43% of elderly individuals are speculated to suffer from AD in 2050, creating a substantial burden on patients and society (82). Removing aggregated Aβ from the brains of symptomatic patients can slow the progression of AD, but the clinical benefit achieved in these trials has been modest, highlighting the need for a deeper understanding of the pathogenesis of disease mechanisms.
GWAS have revealed a correlation between mutations in the MS4A gene cluster, particularly the rs610932 and rs4938933 loci of the MS4A4A gene, which are more susceptible to AD (83, 84). Additionally, polymorphisms at the rs1562990 locus, which is located between the MS4A4E and MS4A4A genes, have also been suggested to be linked with AD susceptibility (85). Astrocytes and microglia affect AD progression by clearing pathological proteins, and single-nucleus RNA sequencing (snRNA-Seq) studies have shown that the MS4A6A gene in microglia affects Aβ plaque deposition and Tau protein phosphorylation (86). The triggering receptor expressed on myeloid cells 2 (TREM2), which can regulate microglial activation and Aβ phagocytic function, is found on the surface of microglia. Furthermore, single-nucleotide polymorphisms (SNPs) within the MS4A gene cluster, such as rs1582763, are associated with increased levels of soluble TREM2 (sTREM2) in cerebrospinal fluid; remarkably, sTREM2 can reduce AD risk and delay AD onset; conversely, the rs6591561 locus is linked to decreased sTREM2 levels, leading to increased AD risk (30). Additionally, the rs667897 locus within the MS4A gene cluster promotes the expression of the MS4A6A gene, thereby increasing AD risk (87). Therefore, a deeper understanding of the role of MS4A molecules in the pathogenesis of AD would help develop new preventive and therapeutic strategies.
4.2 MS4A superfamily molecules and autoimmune diseases
Autoimmune diseases (AuDs) are characterized by a loss of immune tolerance to self-antigens, leading to strong immune responses and thus resulting in tissue damage. AuDs can be classified into organ-specific and systemic autoimmune diseases. Organ-specific AuDs include Hashimoto’s thyroiditis, Graves’ disease, and myasthenia gravis, whereas systemic ADs include rheumatoid arthritis (RA), systemic lupus erythematosus (SLE), and Sjögren’s syndrome (SS) (88). Approximately 5%~8% of the global population is affected by AuDs.
Several members of the MS4A family are implicated in the progression of AuDs. For example, MS4A1 is expressed primarily on the surface of B cells, which exacerbates the progression of AuDs. Rituximab, a monoclonal antibody (mAb) that targets MS4A1, has been approved by the U.S. Food and Drug Administration (FDA) and the European Medicine Agency (EMA) for the treatment of RA in the clinic (89). Additionally, MS4A4A, a macrophage marker within the MS4A family, has also been detected in the synovial tissue of early RA patients (90). The MS4A2 molecule is the β subunit of FcϵRI, and mice with Ms4a2 gene knockout are protected from both skin and systemic allergic reactions (91). Human MS4A2 is not only associated with allergies but also linked to an increased risk of allergic rhinitis (91). Recent studies have shown that MS4A4A expression is associated with cutaneous systemic sclerosis, polyangiitis, and Kawasaki disease (92, 93). Our research has also identified MS4A6D as a potential therapeutic target for various inflammatory diseases, including colitis and psoriasis (94, 95). Therefore, certain MS4A family members may serve as novel therapeutic targets or immune interventions for AuDs.
4.3 MS4A superfamily molecules and tumors
In addition to being involved in controlling the pathogenesis of AD and autoimmune disorders, MS4A molecules are also associated with the occurrence and progression of hematological malignancies. We reviewed the different MS4A molecules in certain solid tumors in much greater detail (Table 1).
4.3.1 MS4A1
MS4A1 is a characteristic marker of B cells and is closely related to the treatment and prognosis of lymphomas and lymphocytic leukemias. Rituximab, a mAb against MS4A1, was approved for the treatment of relapsed B-cell lymphomas and relapsed non-Hodgkin’s lymphoma (96). Recently, MS4A1 has also been detected in other cancer tissues, including glioblastoma, mucinous colorectal adenocarcinoma, lymphomas, esophageal cancer, ovarian cancer, glioma, and lung adenocarcinoma, indicating that the therapeutic application of Rituximab may extend beyond B-cell lymphomas (97). Mechanistically, the antitumor activity of Rituximab is attributed primarily to the induction of apoptosis, complement-dependent cytotoxicity (CDC), and antibody-dependent cell-mediated cytotoxicity (ADCC). Recent studies have suggested that antibody-dependent cellular phagocytosis (ADCP) may be the predominant mechanism by which Rituximab clears cancer cells (98). However, because Rituximab is a chimeric human-mouse anti-MS4A1 mAb, it contains approximately 30% mouse-derived sequences, which can elicit a human anti-mouse immune response (HAMA) when it is introduced into the human body, this side effect significantly restricts its clinical application. To reduce the immunogenicity of the mouse-derived sequences in Rituximab, antibody humanization techniques, such as “complementarity-determining region (CDR) grafting” have been employed (99). This technique involves grafting the CDRs of mouse antibodies onto the framework regions of human antibodies, thereby reducing the immunogenicity associated with the mouse antibody framework. In addition to Rituximab, other therapeutic strategies that target MS4A1, such as CAR-T cell therapy and antibody−drug conjugates (ADCs), are under active development (100, 101). CAR-T cell therapies have shown promise in overcoming resistance in relapsed hematologic malignancies, with several companies and institutions currently conducting clinical trials.
4.3.2 MS4A3
MS4A3 (HTm4) is expressed on bone marrow-derived macrophage precursors, DCs, and monocytes in peripheral blood, making it a key regulator of the cell cycle in hematopoietic cells. Abnormal expression of MS4A3 leads to increased kinase-associated phosphatase activity, causing cells to arrest in the G0/G1 phase. Recent studies have revealed a close association between MS4A3 and tumorigenesis, with significant differences in MS4A3 expression observed in prostate cancer, ovarian cancer, and breast cancer tissues compared with normal tissues (26–29). The transcription factor EVI-1 (ecotropic virus integration site 1) is highly expressed in myeloid leukemia, and research by Heller G et al. revealed that overexpression of EVI-1 in myeloid leukemia cells suppresses MS4A3 expression, thereby promoting tumor growth (102), suggesting a potential link between MS4A3 and tumor development.
4.3.3 MS4A6A
MS4A6A (also known as CDA01, MS4A6, 4SPAN3, or CD20L3) is a prominent member of the MS4A gene family (103). MS4A6A is expressed on the surface of classical CD14+CD16-monocytes and M2 macrophages, with minimal expression on CD14-CD16+ M1 macrophages and CD14+CD16-transitional macrophages, suggesting that MS4A6A might play a critical role in tissue repair (36). MS4A6A is significantly expressed in lung-infiltrated macrophages and can serve as a prognostic marker for non-small cell lung cancer (NSCLC) (37). In lung adenocarcinoma, researchers have reported a positive correlation between MS4A6A expression and the infiltration of immune cells, such as macrophages and DCs, within the tumor microenvironment (37, 104). We also detected the expression of MS4A6A in tissues from breast ductal carcinoma in situ (DCIS), and multicolor fluorescence staining revealed that MS4A6A is expressed mainly in infiltrated CD68+ macrophages. Moreover, survival is better among DCIS patients with high levels of MS4A6A-positive cells than among patients with low numbers of these cells. Our study revealed that MS4A6A can function as a prognostic marker in various malignant tumors, including DCIS, because of its role in tumorigenesis and tumor immunity. However, the precise role of MS4A6A in cancer progression remains unclear, and the relationship between MS4A6A expression in tumor tissues and immune cell infiltration needs further investigation.
4.3.4 MS4A7
MS4A7 (CFFM4) is expressed primarily in monocyte/macrophage-containing tissues, such as the spleen, liver, and lungs. Moreover, research indicates that MS4A7 is expressed in various cancer tissues, including glioblastoma, lung adenocarcinoma, esophageal cancer, and glioma. In triple-negative breast cancer (TNBC), MS4A7 has been identified as a prognostic factor, and a predictive model that incorporates the MS4A7, SPARC, and CD300C genes has demonstrated strong prognostic accuracy (105). In gastric cancer, low mRNA transcription levels of MS4A7 are associated with better overall survival, whereas high mRNA transcription levels of MS4A6A suggest a better prognosis (51). Functionally, the regulatory role of MS4A7 in the differentiation of monocytic leukemia cells may be related to the activation of the p38 MAPK pathway.
4.3.5 MS4A8B
MS4A8B is a recently identified member of the MS4A family that has been implicated in cell differentiation and tumorigenesis. Its murine homolog, MS4A8A, is expressed on tumor-associated macrophages in breast cancer and melanoma. Overexpression of MS4A8A in the RAW264.7 cell line has been shown to increase breast tumor growth in a mouse model of breast cancer. Additionally, MS4A8B is expressed on the surface of intestinal epithelial cells, and its expression is elevated in colorectal cancer. Immunohistochemical studies demonstrated that MS4A8B was upregulated in small cell lung cancer, and its levels have also been linked to the progression of prostate cancer (106). Silencing Ms4a8b in prostate cancer cell lines leads to cell cycle arrest, suggesting that MS4A8B promotes G1/S cell cycle transition (71), conversely, the overexpression of MS4A8A has been shown to significantly reduce the proliferation and migration rates of mouse colorectal cancer cells (33). Clinical studies have also revealed significant differences in MS4A8B protein expression across benign prostate tissue, adjacent prostate cancer tissue, prostatic intraepithelial neoplasia, prostate carcinoma in situ, and metastatic prostate cancer lymph nodes. Research has revealed that MS4A8B protein expression is associated with tumor recurrence, Gleason scores, and proliferation indices (45), indicating that MS4A8B expression is related to postsurgical recurrence and metastasis in prostate cancer patients.
4.3.6 MS4A12
MS4A12 is specifically expressed in colonic tissue, and early immunohistochemical studies revealed that MS4A12 is expressed exclusively in colorectal cancer cells, with no expression in adjacent stromal or nontumor epithelial cells. The rate of MS4A12 positivity in colorectal cancer is as high as 63%, and MS4A12 expression is regulated by the transcription factor caudal type homeobox 2 (CDX2), which can influence the proliferation and cell cycle of colorectal cancer cells (9). Drew J et al. analyzed six colorectal cancer and six colorectal adenoma tissue samples and reported significant differences in MS4A12 expression between normal colon tissue, inflammatory polyps, and colorectal cancer tissues, suggesting that MS4A12 expression may be related to the degree of colonocyte differentiation (107). Additionally, Dalerba P et al. reported that patients with negative MS4A12 expression had significantly reduced survival rates (108), indicating that the unique expression pattern of MS4A12 in the colon may make it a novel target for colorectal cancer immunotherapy.
4.3.7 TMEM176A and TMEM176B
TMEM176A and TMEM176B are unique members of the MS4A family with distinct expression characteristics, and recent studies have improved our understanding of their structures, distribution patterns, biological functions, and associations with various clinical diseases, including cancer. TMEM176A and TMEM176B interact with each other and function as ion channels that play critical roles in regulating antigen cross-presentation in DCs (10). Abnormal DNA methylation of CpG islands in human TMEM176A and TMEM176B is associated with breast cancer development (109). In hepatocellular carcinoma (HCC) tissues, the transcription of the 5’ and 3’ introns of the gene that encodes human TMEM176B is significantly reduced (110). Additionally, the expression levels of TMEM176A and TMEM176B differ significantly between cancerous and normal tissues in breast cancer, lymphoma, skin cancer, and liver cancer, suggesting their potential as diagnostic markers for tumors (111). Knockdown of the TMEM176A gene has been shown to inhibit the proliferation, migration, and invasion of colorectal cancer cells (112), indicating a possible role for TMEM176A in the invasion and metastasis of colorectal cancer. In breast cancer cells, the expression of TMEM176B is crucial for AKT/mTOR signaling, angiogenesis, KRAS signaling, epithelial−mesenchymal transition (EMT), and the regulation of estrogen and interferon response genes (62), therefore, therapeutic antibodies that target TMEM176B may inhibit tumor cell proliferation. These findings suggest that TMEM176A and TMEM176B could serve as novel targets for immunotherapy in certain cancers.
5 Perspective
MS4A family proteins play crucial regulatory roles in cell growth, survival, and activation. These proteins function physiologically as ion channels or signal modulators of immune receptors, often existing as homomeric and heteromeric complexes within lipid raft microdomains. Several MS4A members have significant physiological functions in various diseases, including Alzheimer’s disease, autoimmune disorders, and cancer. Future research should focus on elucidating the molecular mechanisms of signal transduction mediated by MS4A, with particular emphasis on the biological functions of various splicing variants of MS4A proteins. Understanding the mechanisms of action of MS4A family molecules will have profound implications for a wide range of diseases, including malignant tumors.
Author contributions
YC: Investigation, Writing – original draft, Writing – review & editing. XJL: Writing – original draft. BL: Conceptualization, Writing – original draft. YY: Methodology, Writing – original draft. LF: Investigation, Writing – review & editing. QZ: Methodology, Writing – review & editing. XYL: Funding acquisition, Writing – review & editing. XZ: Writing – review & editing, Data curation, Formal analysis.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by Hebei Province Medical Science Research Project (No. 20251847); The Key Project of the Youth Doctoral Program at the Second Affiliated Hospital of the Army Military Medical University (No. 2023YQB031).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Adra CN, Lelias JM, Kobayashi H, Kaghad M, Morrison P, Rowley JD, Lim B. Cloning of the cDNA for a hematopoietic cell-specific protein related to CD20 and the beta subunit of the high-affinity IgE receptor: evidence for a family of proteins with four membrane-spanning regions. Proc Natl Acad Sci U.S.A. (1994) 91:5. doi: 10.1073/pnas.91.21.10178
2. Mattiola I, Mantovani A, Locati M. The tetraspan MS4A family in homeostasis, immunity, and disease. Trends Immunol. (2021) 42:764–81. doi: 10.1016/j.it.2021.07.002
3. Zuccolo J, Bau J, Childs SJ, Goss GG, Sensen CW, Deans JP. Phylogenetic analysis of the MS4A and TMEM176 gene families. PLoS One. (2010) 5:e9369. doi: 10.1371/journal.pone.0009369
4. Deichmann KA, Starke B, Schlenther S, Heinzmann A, Sparholt SH, Forster J, Kuehr. Linkage J. and association studies of atopy and the chromosome 11q13 region. J Med Genet. (1999) 36:5.
5. Murru L, Moretto E, Martano G, Passafaro M. Tetraspanins shape the synapse. Mol Cell Neurosci. (2018) 91:76–81. doi: 10.1016/j.mcn.2018.04.001
6. Liang Y, Tedder TF. Identification of a CD20-, FcepsilonRIbeta-, and HTm4-related gene family: sixteen new MS4A family members expressed in human and mouse. Genomics. (2001) 72:8. doi: 10.1006/geno.2000.6472
7. Kuek LE, Leffler M, Mackay GA, Hulett MD. The MS4A family: counting past 1, 2 and 3. Immunol Cell Biol. (2016) 94:11–23. doi: 10.1038/icb.2015.48
8. Leffler M, Hulett MD. Identification and characterisation of new members of the MS4A family, a novel family of tetraspan genes expressed in leukocytes. Tissue Antigens. (2005) 66:475–.
9. Koslowski M, Türeci Ö, Huber C, Sahin U. Selective activation of tumor growth-promoting Ca channel MS4A12 in colon cancer by caudal type homeobox transcription factor CDX2. Mol Cancer. (2009) 8:77. doi: 10.1186/1476-4598-8-77
10. Condamine T, Le Texier L, Howie D, Lavault A, Hill M, Halary F, et al. Tmem176B and Tmem176A are associated with the immature state of dendritic cells. J Leukoc Biol. (2010) 88:507–15. doi: 10.1189/jlb.1109738
11. Howie D, Nolan KF, Daley S, Butterfield E, Adams E, Garcia-Rueda H, et al. MS4A4B is a GITR-associated membrane adapter, expressed by regulatory T cells, which modulates T cell activation. J Immunol. (2009) 183:4197–204. doi: 10.4049/jimmunol.0901070
12. Mattiola I, Tomay F, De Pizzol M, Silva-Gomes R, Savino B, Gulic T, et al. The macrophage tetraspan MS4A4A enhances dectin-1-dependent NK cell-mediated resistance to metastasis. Nat Immunol. (2019) 20:1012–22. doi: 10.1038/s41590-019-0417-y
13. Mudd TW, Lu CW, Klement JD, Liu KB. MS4A1 expression and function in T cells in the colorectal cancer tumor microenvironment. Cell Immunol. (2021) 360:104260. doi: 10.1016/j.cellimm.2020.104260
14. Li SL, Fang Y. MS4A1 as a potential independent prognostic factor of breast cancer related to lipid metabolism and immune microenvironment based on TCGA database analysis. Med Sci Monitor. (2022) 28:e934597. doi: 10.12659/MSM.934597
15. Jiang HC, Park SJ, Wang IH, Bear DM, Nowlan A, Greer PL. CD20/MS4A1 is a mammalian olfactory receptor expressed in a subset of olfactory sensory neurons that mediates innate avoidance of predators. Nat Commun. (2024) 15:3360. doi: 10.1038/s41467-024-47698-3
16. Rushton CK, Arthur SE, Alcaide M, Cheung M, Jiang A, Coyle KM, et al. Genetic and evolutionary patterns of treatment resistance in relapsed B-cell lymphoma. Blood Adv. (2020) 4:2886–98. doi: 10.1182/bloodadvances.2020001696
17. Zhang WW, Ling YW, Li ZD, Peng XC, Ren YZ. Peripheral and tumor-infiltrating immune cells are correlated with patient outcomes in ovarian cancer. Cancer Med-Us. (2023) 12:10045–61. doi: 10.1002/cam4.v12.8
18. Bisgin A, Sonmezler O, Boga I, Yilmaz M. The impact of rare and low-frequency genetic variants in common variable immunodeficiency (CVID). Sci Rep-Uk. (2021) 11:8308. doi: 10.1038/s41598-021-87898-1
19. Luque A, Serrano I, Ripoll E, Malta C, Gom M, Blom AM, et al. Noncanonical immunomodulatory activity of complement regulator C4BP(β-) limits the development of lupus nephritis. Kidney Int. (2020) 97:551–66. doi: 10.1016/j.kint.2019.10.016
20. Zou ZN, Ha YP, Liu SG, Huang BW. Identification of tumor-infiltrating immune cells and microenvironment -relevant genes in nasopharyngeal carcinoma based on gene expression profiling. Life Sci. (2020) 263:118505. doi: 10.1016/j.lfs.2020.118620
21. Koyuncu D, Tavolara T, Gatti DM, Gower AC, Ginese ML, Kramnik I, et al. B cells in perivascular and peribronchiolar granuloma-associated lymphoid tissue and B-cell signatures identify asymptomatic lung infection in Diversity Outbred mice. Infect Immun. (2024) 92:e0026323. doi: 10.1128/iai.00263-23
22. Dekkers KF, Slieker RC, Ioan-Facsinay A, van Iterson M, BIOS consortium, Ikram MA, et al. Lipid-induced transcriptomic changes in blood link to lipid metabolism and allergic response. Nat Commun. (2023) 14:544. doi: 10.1038/s41467-022-35663-x
23. Chen X, Liu Q, Chen N, Ma J, Wu X, Zhang H, et al. Diagnostic biomarker for type 2 diabetic peripheral neuropathy via comprehensive bioinformatics analysis. J Diabetes. (2024) 16:e13506. doi: 10.1111/1753-0407.13506
24. Silva-Gomes R, Mapelli SN, Boutet M-A, Mattiola I, Sironi M, Grizzi F, et al. Differential expression and regulation of MS4A family members in myeloid cells in physiological and pathological conditions. J Leukocyte Biol. (2022) 111:817–36. doi: 10.1002/JLB.2A0421-200R
25. Li S, Zhao Z, Yang H, Wang D, Sun W, Li S, et al. Construction and validation of a nomogram for the preoperative prediction of lymph node metastasis in gastric cancer. Cancer Control. (2021) 28:10732748211027160. doi: 10.1177/10732748211027160
26. Zhao H, Pomicter AD, Eiring AM, Franzini A, Ahmann J, Hwang J-Y, et al. MS4A3 promotes differentiation in chronic myeloid leukemia by enhancing common β-chain cytokine receptor endocytosis. Blood. (2022) 139:761–78. doi: 10.1182/blood.2021011802
27. Verma AK, Zheng J, Mack M, Ginhoux F, Perlman S. Differential effects of prostaglandin D2 signaling on macrophages and microglia in murine coronavirus encephalomyelitis. mBio. (2021) 12:e0196921. doi: 10.1128/mBio.01969-21
28. Yang S, Cao C, Xie Z, Zhou Z. Analysis of potential hub genes involved in the pathogenesis of Chinese type 1 diabetic patients. Ann Transl Med. (2020) 8:295. doi: 10.21037/atm.2020.02.171
29. Rodrigues PF, Trsan T, Cvijetic G, Khantakova D, Panda SK, Liu Z, Ginhoux F, et al. Progenitors of distinct lineages shape the diversity of mature type 2 conventional dendritic cells. Immunity. (2024) 57:21. doi: 10.1016/j.immuni.2024.05.007
30. Deming Y, Filipello F, Cignarella F, Cantoni C, Hsu S, Mikesell R, et al. The MS4A gene cluster is a key modulator of soluble TREM2 and Alzheimer's disease risk. Sci Transl Med. (2019) 11:eaau2291. doi: 10.1126/scitranslmed.aau2291
31. Li Y, Shen Z, Chai Z, Zhan Y, Zhang Y, Liu Z, et al. Targeting MS4A4A on tumour-associated macrophages restores CD8+T-cell-mediated antitumour immunity. Gut. (2023) 72:2307–20. doi: 10.1136/gutjnl-2022-329147
32. Li X, Liu J, Zeng M, Yang K, Zhang S, Liu Y, et al. GBP2 promotes M1 macrophage polarization by activating the notch1 signaling pathway in diabetic nephropathy. Front Immunol. (2023) 14. doi: 10.3389/fimmu.2023.1127612
33. Zhou H, Shen Y, Zheng G, Zhang B, Wang A, Zhang J, et al. Integrating single-cell and spatial analysis reveals MUC1-mediated cellular crosstalk in mucinous colorectal adenocarcinoma. Clin Transl Med. (2024) 14:e1701. doi: 10.1002/ctm2.v14.5
34. Zeng Y, Tan P, Ren C, Gao L, Chen Y, Hu S, Tang N, et al. Comprehensive analysis of expression and prognostic value of MS4As in glioma. Front Genet. (2022) 13. doi: 10.3389/fgene.2022.795844
35. Hou M, Xu G, Ran M, Luo W, Wang H. APOE-ϵ4 carrier status and gut microbiota dysbiosis in patients with alzheimer disease. Front Neurosci. (2021) 15. doi: 10.3389/fnins.2021.619051
36. Zhang C, Liu H, Tan Y, Xu Y, Li Y, Tong S, et al. MS4A6A is a new prognostic biomarker produced by macrophages in glioma patients. Front Immunol. (2022) 13. doi: 10.3389/fimmu.2022.865020
37. Li Z, Zhou B, Zhu X, Yang F, Jin K, Dai J, et al. Differentiation-related genes in tumor-associated macrophages as potential prognostic biomarkers in non-small cell lung cancer. Front Immunol. (2023) 14. doi: 10.3389/fimmu.2023.1123840
38. Pei Y, Risal S, Jiang H, Lu H, Lindgren E, Stener-Victorin E, et al. Transcriptomic survey of key reproductive and metabolic tissues in mouse models of polycystic ovary syndrome. Commun Biol. (2023) 6:69. doi: 10.1038/s42003-022-04362-0
39. Harwood JC, Leonenko G, Sims R, Escott-Price V, Williams J, Holmans P. Defining functional variants associated with Alzheimer's disease in the induced immune response. Brain Commun. (2021) 3:fcab083. doi: 10.1093/braincomms/fcab083
40. Jun G, Ibrahim-Verbaas CA, Vronskaya M, Lambert J-C, Chung J, Naj AC, et al. A novel Alzheimer disease locus located near the gene encoding tau protein. Mol Psychiatr. (2016) 21:108–17. doi: 10.1038/mp.2015.23
41. Zhou L, Qiu X, Meng Z, Liu T, Chen Z, Zhang P, et al. Hepatic danger signaling triggers TREM2 macrophage induction and drives steatohepatitis via MS4A7-dependent inflammasome activation. Sci Transl Med. (2024) 16:eadk1866. doi: 10.1126/scitranslmed.adk1866
42. Ni B, Huang G, Yang R, Wang Z, Song H, Li K, et al. The short isoform of MS4A7 is a novel player in glioblastoma microenvironment, M2 macrophage polarization, and tumor progression. J Neuroinflamm. (2023) 20:80. doi: 10.1186/s12974-023-02766-1
43. Pang JY, Yu Q, Chen YZ, Yuan HJ, Sheng MM, Tang WR. Integrating Single-cell RNA-seq to construct a Neutrophil prognostic model for predicting immune responses in non-small cell lung cancer. J Transl Med. (2022) 20:531. doi: 10.1186/s12967-022-03723-x
44. Liu ZT, Shen JT, Lei YJ, Huang YC, Zhao GQ, Zheng CH, et al. Molecular subtyping based on immune cell marker genes predicts prognosis and therapeutic response in patients with lung adenocarcinoma. BMC Cancer. (2023) 23:1141. doi: 10.1186/s12885-023-11579-7
45. Leyten GH, Hessels D, Smit FP, Jannink SA, de Jong H, Melchers WJ, et al. Identification of a candidate gene panel for the early diagnosis of prostate cancer. Clin Cancer Res. (2015) 21:3061–70. doi: 10.1158/1078-0432.CCR-14-3334
46. Wang S, Zhang MJ, Wu ZZ, Zhu SW, Wan SC, Zhang BX, et al. GSDME is related to prognosis and response to chemotherapy in oral cancer. J Dent Res. (2022) 101:848–58. doi: 10.1177/00220345211073072
47. Suenaga M, Schirripa M, Cao S, Zhang W, Cremolini C, Lonardi S, et al. Clinical significance of enterocyte-specific gene polymorphisms as candidate marker of oxaliplatin-based treatment for metastatic colorectal cancer. J Clin Oncol. (2018) 36:285–95. doi: 10.1200/JCO.2018.36.15_suppl.12066
48. Han J, Zhang X, Liu Y, Jing L, Liu YB, Feng L. CLCA4 and MS4A12 as the significant gene biomarkers of primary colorectal cancer. Biosci Rep. (2020) 40:BSR20200963. doi: 10.1042/BSR20200963
49. Kumamoto K, Nakachi Y, Mizuno Y, Yokoyama M, Ishibashi K, Kosugi C, et al. Expressions of 10 genes as candidate predictors of recurrence in stage III colon cancer patients receiving adjuvant oxaliplatin-based chemotherapy. Oncol Lett. (2019) 18:1388–94. doi: 10.3892/ol.2019.10437
50. Kaneko T, Toshimori K, Iida H. Subcellular localization of MS4A13 isoform 2 in mouse spermatozoa. Reproduction. (2017) 154:843–57. doi: 10.1530/REP-17-0477
51. Sun L, Zhang YL, Zhang C. Distinct expression and prognostic value of MS4A in gastric cancer. Open Med-Warsaw. (2018) 13:178–88. doi: 10.1515/med-2018-0028
52. Zheng ZJ, Li HP, Yang RJ, Guo H. Role of the membrane-spanning 4A gene family in lung adenocarcinoma. Front Genet. (2023) 14. doi: 10.3389/fgene.2023.1162787
53. Lee HA, Ahn EH, Kim JH, Kim JO, Ryu CS, Lee JY, et al. Association study of frameshift and splice variant polymorphisms with risk of idiopathic recurrent pregnancy loss. Mol Med Rep. (2018) 18:2417–26. doi: 10.3892/mmr.2018.9202
54. Xin S, Mueller C, Pfeiffer S, Kraft VAN, Merl-Pham J, Bao X, et al. MS4A15 drives ferroptosis resistance through calcium-restricted lipid remodeling. Cell Death Differ. (2022) 29:670–86. doi: 10.1038/s41418-021-00883-z
55. Fang Y, Yu HY, Zhou HE. MS4A15 acts as an oncogene in ovarian cancer through reprogramming energy metabolism. Biochem Bioph Res Co. (2022) 598:47–54. doi: 10.1016/j.bbrc.2022.01.128
56. Guo Y, Cao F, Hu S, Liu S, Sun H, Li A, et al. TMEM176A acts as a tumor suppressor gene in pancreatic cancer by inhibiting ERK signaling. Discovery Med. (2020) 30:145–53.
57. Li H, Yang W, Zhang M, He T, Zhou F, Herman JG, et al. Methylation of TMEM176A, a key ERK signaling regulator, is a novel synthetic lethality marker of ATM inhibitors in human lung cancer. Epigenomics-Uk. (2021) 13:1403–19. doi: 10.2217/epi-2021-0217
58. Picotto G, Morse LR, Nguyen N, Saltzman J, Battaglino R. TMEM176A and TMEM176B are candidate regulators of inhibition of dendritic cell maturation and function after chronic spinal cord injury. J Neurotraum. (2020) 37:528–33. doi: 10.1089/neu.2019.6498
59. Lancien M, Bienvenu G, Salle S, Gueno L, Feyeux M, Merieau E, et al. Dendritic cells require TMEM176A/B ion channels for optimal MHC class II antigen presentation to naive CD4 T cells. J Immunol. (2021) 207:421–35. doi: 10.4049/jimmunol.2000498
60. Grunin M, Hagbi-Levi S, Rinsky B, Smith Y, Chowers I. Transcriptome analysis on monocytes from patients with neovascular age-related macular degeneration. Sci Rep-Uk. (2016) 6:29046. doi: 10.1038/srep29046
61. Segovia M, Russo S, Jeldres M, Mahmoud YD, Perez V, Duhalde M, et al. Targeting TMEM176B enhances antitumor immunity and augments the efficacy of immune checkpoint blockers by unleashing inflammasome activation. Cancer Cell. (2019) 35:767–81. doi: 10.1016/j.ccell.2019.04.003
62. Kang C, Rostoker R, Ben-Shumel S, Rashed R, Duty JA, Demircioglu D, Antoniou IM, et al. TMEM176B regulates AKT/mTOR signaling and tumor growth in triple-negative breast cancer. Cells. (2021) 10:3430. doi: 10.3390/cells10123430
63. Jing L, An Y, Cai T, Xiang J, Li B, Guo J, Ma X, et al. A subpopulation of CD146+ macrophages enhances antitumor immunity by activating the NLRP3 inflammasome. Cell Mol Immunol. (2023) 20:15. doi: 10.1038/s41423-023-01047-4
64. Bubien JK, Zhou LJ, Bell PD, Frizzell RA, Tedder TF. Transfection of the CD20 cell surface molecule into ectopic cell types generates a Ca2+ conductance found constitutively in B lymphocytes. J Cell Biol. (1993) 121:1121–32. doi: 10.1083/jcb.121.5.1121
65. Li HD, Ayer LM, Lytton J, Deans JP. Store-operated cation entry mediated by CD20 in membrane rafts. J Biol Chem. (2003) 278:42427–34. doi: 10.1074/jbc.M308802200
66. Polyak MJ, Li HD, Shariat N, Deans JP. CD20 homo-oligomers physically associate with the B cell antigen receptor - Dissociation upon receptor engagement and recruitment of phosphoproteins and calmodulin-binding proteins. J Biol Chem. (2008) 283:18545–52. doi: 10.1074/jbc.M800784200
67. Hiraoka S, Furumoto Y, Koseki H, Takagaki Y, Taniguchi M, Okumura K, et al. Fc receptor beta subunit is required for full activation of mast cells through Fc receptor engagement. Int Immunol. (1999) 11:199–207. doi: 10.1093/intimm/11.2.199
68. Arthur GK, Ehrhardt-Humbert LC, Snider DB, Jania C, Tilley SL, Metcalfe DD, Cruse G. The FcϵRIβ homologue, MS4A4A, promotes FcϵRI signal transduction and store-operated Ca2+ entry in human mast cells. Cell Signal. (2020) 71:109617. doi: 10.1016/j.cellsig.2020.109617
69. Greer PL, Bear DM, Lassance J-M, Bloom LM, Tsukahara T, Pashkovski SL, et al. A family of non-GPCR chemosensors defines an alternative logic for mammalian olfaction. Cell. (2016) 165:1734–48. doi: 10.1016/j.cell.2016.05.001
70. Drujont L, Lemoine A, Moreau A, Bienvenu G, Lancien M, Cens T, et al. RORγt cells selectively express redundant cation channels linked to the Golgi apparatus. Sci Rep-Uk. (2016) 6:23682. doi: 10.1038/srep23682
71. Deans J, Deng LL, Zuccolo J. MS4A8B oligomerizes with CD20 (MS4A1) and modulates B cell receptor-stimulated calcium entry. FASEB J. (2008) 22:675–87. doi: 10.1096/fasebj.22.1_supplement.1066.10
72. Wright CM, Francis SMS, Tan ME, Martins MU, Winterford C, Davidson MR, et al. MS4A1 dysregulation in asbestos-related lung squamous cell carcinoma is due to CD20 stromal lymphocyte expression. PLoS One. (2012) 7:e34943. doi: 10.1371/journal.pone.0034943
73. Donnadieu E, Jouvin MH, Kinet JP. A second amplifier function for the allergy-associated Fc(epsilon)RI-beta subunit. Immunity. (2000) 12:9. doi: 10.1016/S1074-7613(00)80203-4
74. Donato JL, Ko J, Kutok JL, Cheng T, Shirakawa T, Mao XQ, et al. Human HTm4 is a hematopoietic cell cycle regulator. J Clin Invest. (2002) 109:8. doi: 10.1172/JCI0214025
75. Mattiola I, Tomay F, De Pizzol M, Silva-Gomes R, Savino B, Gulic T, et al. The macrophage tetraspan MS4A4A enhances Dectin-1-dependent NK cell-mediated resistance to metastasis. Eur J Immunol. (2019) 49:1517–8. doi: 10.1038/s41590-019-0417-y
76. Cruse G, Beaven MA, Music SC, Bradding P, Gilfillan AM, Metcalfe DD. The CD20 homologue MS4A4 directs trafficking of KIT toward clathrin-independent endocytosis pathways and thus regulates receptor signaling and recycling. Mol Biol Cell. (2015) 26:1711–27. doi: 10.1091/mbc.E14-07-1221
77. Xu H, Yan Y, Li Z, Zhang G-X, Williams M, Carey G, et al. Anti-MS4a4B treatment abrogates MS4a4B-mediated protection in T cells and ameliorates experimental autoimmune encephalomyelitis. J Immunol. (2012) 188:1106–19. doi: 10.4049/jimmunol.188.Supp.51.13
78. Yan Y, Li Z, Zhang G-X, Williams MS, Carey GB, Zhang J, et al. Anti-MS4a4B treatment abrogates MS4a4B-mediated protection in T cells and ameliorates experimental autoimmune encephalomyelitis. Apoptosis. (2013) 18:1106–19. doi: 10.1007/s10495-013-0870-2
79. Li J, Diao B, Guo S, Huang X, Yang C, Feng Z, et al. VSIG4 inhibits proinflammatory macrophage activation by reprogramming mitochondrial pyruvate metabolism. Nat Commun. (2017) 8:1322. doi: 10.1038/s41467-017-01327-4
80. Feng Z, Huang X, Wang C, Yang C, Wu Y, Chen Y. MS4A6D promotes NLRP3 inflammasome activation through inducing calcium mobilization in macrophages. Eur J Immunol. (2019) 49:41–.
81. Scheltens P, De Strooper B, Kivipelto M, Holstege H, Chételat G, Teunissen CE, Cummings J, et al. Alzheimer's disease. Lancet. (2021) 397:1577–90. doi: 10.1016/S0140-6736(20)32205-4
82. Khan S, Barve KH, Kumar MS. Recent advancements in pathogenesis, diagnostics and treatment of alzheimer's disease. Curr Neuropharmacol. (2020) 18:20. doi: 10.2174/1570159X18666200528142429
83. Hollingworth P, Harold D, Sims R, Gerrish A, Lambert JC, Carrasquillo MM, et al. Common variants at ABCA7, MS4A6A/MS4A4E, EPHA1, CD33 and CD2AP are associated with Alzheimer's disease. Nat Genet. (2011) 43:429–35. doi: 10.1038/ng.803
84. Naj AC, Jun G, Beecham GW, Wang LS, Vardarajan BN, Buros J, et al. Common variants at MS4A4/MS4A6E, CD2AP, CD33 and EPHA1 are associated with late-onset Alzheimer's disease. Nat Genet. (2011) 43:5. doi: 10.1038/ng.801
85. Antúnez C, Boada M, González-Pérez A, Gayán J, Ramírez-Lorca R. The membrane-spanning 4-domains, subfamily A (MS4A) gene cluster contains a common variant associated with Alzheimer's disease. Genome Med. (2011) 3:33. doi: 10.1186/gm249
86. Smith AM, Davey K, Tsartsalis S, Khozoie C, Fancy N, Tang SS, et al. Diverse human astrocyte and microglial transcriptional responses to Alzheimer's pathology. Acta Neuropathol. (2022) 143:75–91. doi: 10.1007/s00401-021-02372-6
87. Lacher SE, Alazizi A, Wang X, Bell DA, Pique-Regi R, Luca F, et al. A hypermorphic antioxidant response element is associated with increased expression and Alzheimer's disease. Redox Biol. (2018) 14:686–93. doi: 10.1016/j.redox.2017.10.018
88. Li JY, Zhao MJ, Luo WJ, Huang JQ, Zhao B, Zhou ZG. B cell metabolism in autoimmune diseases: signaling pathways and interventions. Front Immunol. (2023) 14. doi: 10.3389/fimmu.2023.1232820
89. Boppana TK, Mittal S, Madan K, Mohan A, Hadda V, Guleria R. Rituximab for rheumatoid arthritis-related interstitial lung disease: A systematic review and meta-analysis. Arch Rheumatol. (2024) 39:317–29. doi: 10.46497/ArchRheumatol
90. Wierczeiko A, Butto T, Mündnich S, Marchand V, Motorin Y, Helm M, et al. A call for gene expression analysis in whole blood of patients with rheumatoid arthritis (RA) as a biomarker for RA-associated interstitial lung disease. J Rheumatol. (2024) 51:4. doi: 10.3899/jrheum.2023-0588
91. Dombrowicz D, Flamand V, Brigman KK, Koller BH, Kinet J P. Abolition of anaphylaxis by targeted disruption of the high affinity immunoglobulin E receptor alpha chain gene. Cell Death Differ. (1993) 75:969–76. doi: 10.1016/0092-8674(93)90540-7
92. Amo G, García-Menaya J, Campo P, Cordobés C, Plaza Serón MC, Ayuso P, et al. A nonsynonymous FCER1B SNP is associated with risk of developing allergic rhinitis and with igE levels. Sci Rep. (2016) 6:19724. doi: 10.1038/srep19724
93. Rice LM, Ziemek J, Stratton EA, McLaughlin SR, Padilla CM, Mathes AL, Christmann RB, et al. A longitudinal biomarker for the extent of skin disease in patients with diffuse cutaneous systemic sclerosis. Arthritis Rheumatol. (2015) 67:3004–15. doi: 10.1002/art.v67.11
94. Chen Y, Li S, Huang X, Wang C, Pan Y, Xiang Q, et al. Tetraspan MS4A6D is a coreceptor of MHC class II antigen (MHC-II) that promotes macrophages-derived inflammation. Mol Immunol. (2023) 160:121–32. doi: 10.1016/j.molimm.2023.07.003
95. Yang J, Zhang N, Luo T, Yang M, Shen W, Tan Z, Xia Y, et al. TCellSI: A novel method for T cell state assessment andits applications in immune environment prediction. iMeta. (2024) 3(5):e231. doi: 10.1002/imt2.v3.5
96. Zhou K, Wang T, Pan L, Xu W, Jin J, Zhang W, et al. Improved efficacy and safety of zanubrutinib versus ibrutinib in patients with relapsed/refractory chronic lymphocytic leukemia (R/R CLL) in China: a subgroup of ALPINE. Ann Hematol. (2024) 103(10):4183–91. doi: 10.1007/s00277-024-05823-8
97. Atallah-Yunes SA, Salman O, Robertson MJ. Post-transplant lymphoproliferative disorder: Update on treatment and novel therapies. Brit J Haematol. (2023) 201:383–95. doi: 10.1111/bjh.18763
98. Van Wagoner CM, Rivera-Escalera F, Jaimes-Delgadillo NC, Chu CC, Zent CS, Elliott MR. Antibody-mediated phagocytosis in cancer immunotherapy. Immunol Rev. (2023) 319:128–41. doi: 10.1111/imr.13265
99. Riechmann L, Clark M, Waldmann H, Winter G. Reshaping human antibodies for therapy. Nat Commun. (1988) 332:323–7. doi: 10.1038/332323a0
100. Tong C, Zhang Y, Liu Y, Wang Y, Wu ZQ, Han WD. Optimized tandem CD19/CD20 CAR-engineered T cells in refractory/relapsed B-cell lymphoma (vol 136, pg 1632, 2020). Blood. (2023) 141:1896–644. doi: 10.1182/blood.2020005278
101. Zhang YJ. Safety and efficacy of optimized tandem CD19/CD20 CAR-engineered T cells in patients with relapsed/refractory non-Hodgkin lymphoma. J Clin Oncol. (2020) 38:15. doi: 10.1200/JCO.2020.38.15_suppl.3034
102. Heller G, Rommer A, Steinleitner K, Etzler J, Hackl H, Heffeter P, et al. EVI1 promotes tumor growth via transcriptional repression of MS4A3. J Hematol Oncol. (2015) 8:28. doi: 10.1186/s13045-015-0124-6
103. Bitting K, Hedgespeth B, Ehrhardt-Humbert LC, Arthur GK, Schubert AG, Bradding P, et al. Identification of redundancy between human FcϵRIβ and MS4A6A proteins points toward additional mechanisms for FcERI trafficking and signaling. J Immunol. (2022) 208:1204–17. doi: 10.1111/all.15595
104. Zhou X, Xu R, Lu T, Xu R, Wang C, Peng B, et al. Identification of immunotherapy biomarkers for improving the clinical outcome of homologous recombination deficiency patients with lung adenocarcinoma. Aging-Us. (2023) 15:8090–112. doi: 10.18632/aging.204957
105. Wu H, Feng J, Zhong W, Zouxu X, Xiong Z, Huang W, et al. Model for predicting immunotherapy based on M2 macrophage infiltration in TNBC. Front Immunol. (2023) 14. doi: 10.3389/fimmu.2023.1151800
106. Ye L, Yao X-D, Wan F-N, Qu Y-Y, Liu Z-Y, Shen X-X, et al. MS4A8B promotes cell proliferation in prostate cancer. Prostate. (2014) 74:911–22. doi: 10.1002/pros.22802
107. Drew JE, Farquharson AJ, Mayer CD, Vase HF, Coates PJ, Steele RJ, et al. Predictive gene signatures: molecular markers distinguishing colon adenomatous polyp and carcinoma. PLoS One. (2014) 9:e113071. doi: 10.1371/journal.pone.0113071
108. Dalerba P, Kalisky T, Sahoo D, Rajendran PS, Rothenberg ME, Leyrat AA, et al. Single-cell dissection of transcriptional heterogeneity in human colon tumors. Nat Biotechnol. (2011) 29:1120–U11. doi: 10.1038/nbt.2038
109. Wang Y, Zhang Y, Herman JG, Linghu EQ, Guo MZ. Epigenetic silencing of TMEM176A promotes esophageal squamous cell cancer development. Oncotarget. (2017) 8:70035–48. doi: 10.18632/oncotarget.19550
110. Hodo Y, Hashimoto S, Honda M, Yamashita T, Suzuki Y, Sugano S, et al. Comprehensive gene expression analysis of 5'-end of mRNA identified novel intronic transcripts associated with hepatocellular carcinoma. Genomics. (2010) 95:217–23. doi: 10.1016/j.ygeno.2010.01.004
111. Cuajungco MP, Podevin W, Valluri VK, Bui Q, Nguyen VH, Taylor K. Abnormal accumulation of human transmembrane (TMEM)-176A and 176B proteins is associated with cancer pathology. Acta Histochem. (2012) 114:705–12. doi: 10.1016/j.acthis.2011.12.006
Keywords: MS4A, Alzheimer’s disease, autoimmune diseases, tumors, immunotherapy
Citation: Luo X, Luo B, Fei L, Zhang Q, Liang X, Chen Y and Zhou X (2024) MS4A superfamily molecules in tumors, Alzheimer’s and autoimmune diseases. Front. Immunol. 15:1481494. doi: 10.3389/fimmu.2024.1481494
Received: 16 August 2024; Accepted: 12 November 2024;
Published: 09 December 2024.
Edited by:
Xiangpeng Dai, Jilin University, ChinaReviewed by:
Jin Hou, Second Military Medical University, ChinaXun Sun, China Medical University, China
Copyright © 2024 Luo, Luo, Fei, Zhang, Liang, Chen and Zhou. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xueqin Zhou, MjkwODE5ODdAcXEuY29t; Yongwen Chen, Y2hlbnlvbmd3ZW5AdG1tdS5lZHUuY24=; Xinyu Liang, Njc1ODUxMDY0QHFxLmNvbQ==
†These authors have contributed equally to this work
 Xuejiao Luo1†
Xuejiao Luo1† Yongwen Chen
Yongwen Chen