- 1Department of Cellular and Molecular Medicine, Herbert Wertheim College of Medicine, Florida International University, Miami, FL, United States
- 2The Ronald O. Perelman Department of Dermatology, NYU Grossman School of Medicine, New York, NY, United States
- 3Department of Medical Biochemistry, Faculty of Medicine, South Valley University, Qena, Egypt
Galectin-9 (Gal-9) is a tandem-repeat galectin with diverse roles in immune homeostasis, inflammation, malignancy, and autoimmune diseases. In cancer, Gal-9 displays variable expression patterns across different tumor types. Its interactions with multiple binding partners, both intracellularly and extracellularly, influence key cellular processes, including immune cell modulation and tumor microenvironment dynamics. Notably, Gal-9 binding to cell-specific glycoconjugate ligands has been implicated in both promoting and suppressing tumor progression. Here, we provide insights into Gal-9 and its involvement in immune homeostasis and cancer biology with an emphasis on multiple myeloma (MM) pathophysiology, highlighting its complex and context-dependent dual functions as a pro- and anti-tumorigenic molecule and its potential implications for therapy in MM patients.
1 Introduction
Galectins are a family of 15 β-galactoside-binding lectins widely expressed by a wide range of mammalian cells, including immune cells (1). Galectins have been categorized into three main groups based on their molecular structures (Figure 1) (2). These groups include proto-type galectins (galectin (Gal)-1, -2, -5, -7, -10, -11, -13, -14, and -15), characterized by a single carbohydrate-recognition domain (CRD) that can form homodimers; chimera-type galectins (Gal-3), featuring a single CRD and an amino-terminal polypeptide tail rich in proline, glycine, and tyrosine residues for oligomer formation; and tandem-repeat galectins (Gal-4, -6, -8, -9, and -12), consisting of two CRDs connected by a peptide linker of variable length, ranging from 5 to over 50 amino acids (3, 4).
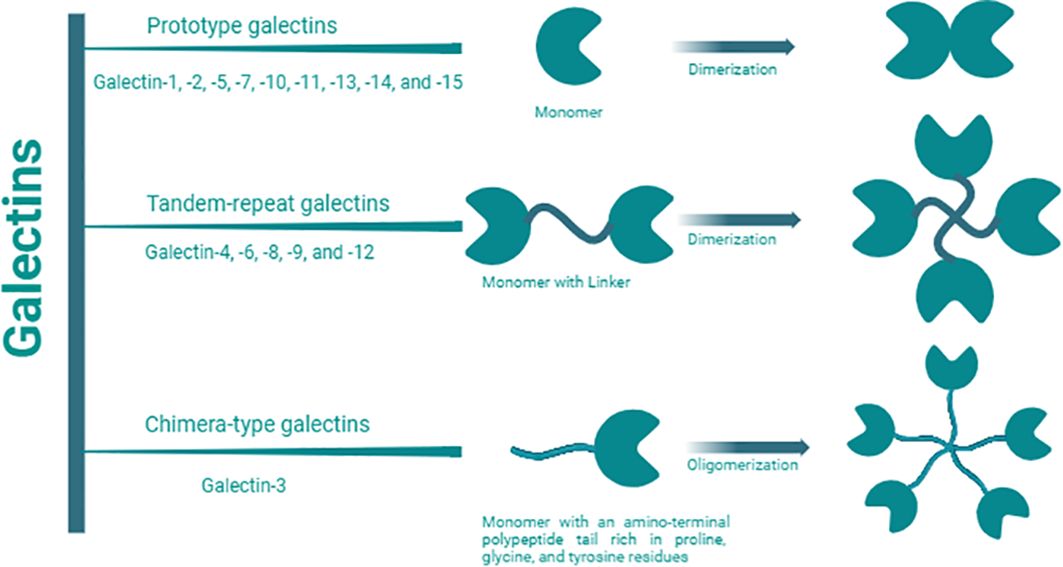
Figure 1. Galectin classification. Galectin members are categorized into three main groups: proto-type, chimera-type, and tandem repeat based on the number, structures, and orientation of carbohydrate recognition domains (CRDs). They specifically recognize and bind to β-galactoside on glycoconjugates (proteins, lipids, or other molecules) through these CRDs. Proto-type Gal-1, -2, -5, -7, -10, -11, -13, -14, and -15, are characterized by a single CRD which can form homodimers; chimera-type Gal-3, is composed of one CRD and an amino-terminal polypeptide tail rich in proline, glycine, and tyrosine residues for oligomer formation; and tandem-repeat Gal-4, -6, -8, -9, and -12, consist of two distinct CRDs connected by a peptide linker of variable length, ranging from 5 to over 50 amino acids.
Extensive research has established galectins as important regulators of immune homeostasis (5), inflammation (6), malignancy (7–9), and autoimmune diseases (10). Considerable advancements have been achieved in understanding how galectins influence both arms of the immune response (11). In the realm of innate immunity, galectins regulate granulocyte chemotaxis, dendritic cell maturation, mast cell activation, and many other activities (6). In adaptive immunity, galectins are widely recognized for their effects on T cell function, where they differentially modulate T cell development, activation, differentiation, and effector function (6, 12, 13). However, their roles in B cell development and activation have been receiving more attention as galectins appear to have profound effects on B cell responses (11). To date, Gal-1, -3, -8, and -9 have been identified as regulators of B cell signaling (11, 14–17).
Gal-9 has been observed to exhibit both tumor-promoting and tumor-suppressing roles across various cancer types, including multiple myeloma (MM) (18, 19). This dual functionality underscores the necessity for a thorough examination of current research on Gal-9 involvement in cancer progression. In this review, we present the latest insights on this topic with particular emphasis on the role of Gal-9 in driving MM progression and its potential therapeutic implications. We urge for a more comprehensive understanding of the precise mechanisms through which Gal-9 influences MM to guide the development of future therapeutic strategies.
2 Galectin-9 (Gal-9)
2.1 Gal-9 structure, expression, and secretion
Gal-9, a tandem-repeat galectin with a molecular weight of 34 to 39 kDa, has emerged as a multifaceted molecule with substantial implications in various physiological and pathological processes (20, 21). In humans, Gal-9 consists of two distinct CRDs connected by a linker peptide (Figure 2) (20, 22). Depending on the length of the peptide, this lectin is categorized into three types: long form (58 amino acids), medium form (26 amino acids), and short form (14 amino acids) (20). Gal-9 shows widespread distribution across multiple organs, including the liver, small intestine, thymus, kidney, spleen, lung, cardiac, and skeletal muscles, with minimal detectability in reticulocytes and brain tissues (23). Gal-9 is primarily expressed by various immune cells including T cells, B cells, macrophages, and mast cells (24, 25). Moreover, under both normal and pathophysiological conditions, Gal-9 can also be detected in endothelial cells, fibroblasts, and astrocytes (14, 17, 26–28).
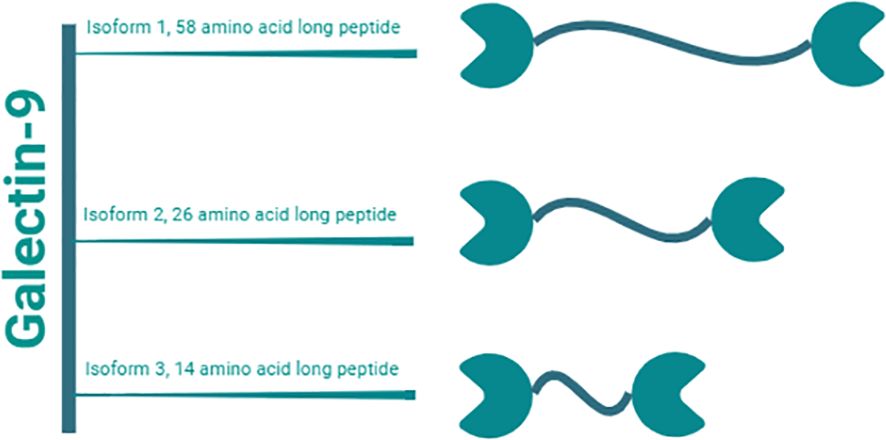
Figure 2. Gal-9 isoforms. Gal-9 is a tandem-repeat galectin with a molecular weight of 34 to 39 kDa. It consists of two distinct CRDs linked by a peptide of variable lengths. Humans have three natural isoforms of this lectin, which differ in the length of this interdomain peptide. These isoforms are termed i) long form Gal-9 (58 amino acids), ii) medium form Gal-9 (26 amino acids), and short form Gal-9 (14 amino acids).
Gal-9 can be found either extracellularly or intracellularly within the nucleus (29–31) as well as in the cytoplasm (21, 29, 32). While cell surface Gal-9 plays various roles in different cellular processes such as cell aggregation and apoptosis, its cytoplasmic functions are still not fully understood (31, 32). Gal-9 secretion into the extracellular space does not follow the classical secretory pathway due to the absence of the signal secretion peptide essential for its transport into the endoplasmic reticulum (33, 34). The exact mechanism by which Gal-9 is secreted is not fully understood; however, it is proposed to be released from the cell via non-classical secretory pathways involving matrix-metalloproteinases and protein kinase C (PKC) (20). These pathways may include translocation directly across the plasma membrane, release via exosome packaging, or export via lysosomes, endosomes, and microvesicles (35–37).
2.2 Gal-9 and its glycoconjugate ligands
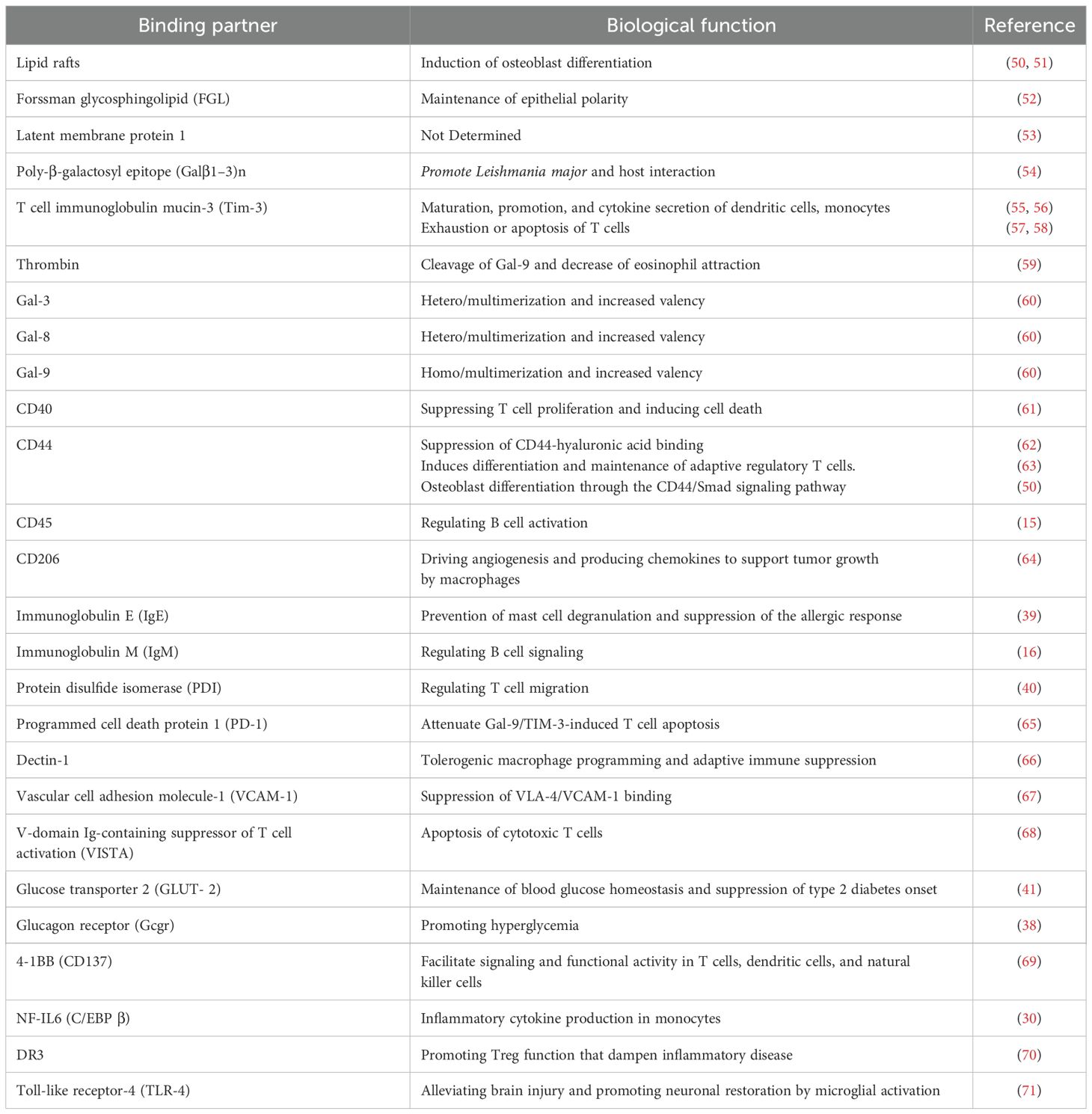
Gal-9 binds to β-galactoside-bearing glycoconjugates on the cell surface through its CRDs, influencing diverse cellular functions and signaling pathways (38–43). The N- and C-terminal CRDs of Gal-9 exhibit only 39% amino acid similarity and interact with distinct sets of carbohydrate ligands (28, 44). The varying specificity and affinity of both the N- and C-terminal CRDs for specific glycoconjugates directly influence Gal-9 affinity toward its binding partners (45, 46). For instance, the N-terminal CRD is suggested to play a predominant role in activating dendritic cell function, while the C-terminal CRD is mainly involved in triggering T-cell death (47). Moreover, variations in the length of the linker domain between the two CRDs affect their rotational flexibility, facilitating the formation of higher-order multimers and increasing Gal-9 valency (48, 49). Earl et al. experimentally demonstrated that incorporating the linker region from Gal-9 into constructs of Gal-1 dimer alters their potency in killing T cells, while the glycan-binding specificity of the CRD remains unchanged compared to the wild-type galectin (49). The broad range of functions carried out by Gal-9 stems from its ability to interact with multiple partners (Table 1). These binding partners range from intracellular cytosolic/nuclear proteins to canonical membrane glycoproteins uniquely expressed in a cell- or tissue-specific manner (30, 43, 62). Moreover, glycosylation of these Gal-9 glycoprotein ligands is often contextualized in relation to the coordinated expression of glycosyltransferases associated with cell differentiation or a malignant or metastatic cell transition (15, 72).
2.3 Gal-9 role in immune homeostasis
Galectins are now well-recognized for playing critical roles in both innate and adaptive immune responses (73). Gal-9 initiates innate immunity by promoting the maturation of dendritic cells, as evidenced by the increased expression of Th1 cytokines and co-stimulatory molecules, including HLA-DR, CD83, CD80, CD54, and CD40. Subsequently, these matured dendritic cells migrate to lymph nodes, where they trigger the activation of T cells (55). In addition, Gal-9 acts as a chemoattractant for eosinophils, modulates signal-dependent chemotaxis of neutrophils, and enhances phagocytosis (27, 74). In monocytes, intracellular Gal-9 induces the transcription of the proinflammatory cytokines IL-1α, IL-1β, and IFN-γ (30). Conversely, Gal-9 also promotes the expansion of immunosuppressive macrophages (75).
In adaptive T cell-mediated immune responses, Gal-9 plays a crucial role in regulating T cell development and homeostasis (76). It has diverse immunomodulatory effects depending on the concentration, receptors, and skewing signals available for the interactions (77). At high concentrations, Gal-9 induces apoptosis of activated T cells (CD8+ and CD4+), while it increases cytokine production by activated T cells at low concentrations (77, 78). Additionally, Gal-9 facilitates the differentiation of naïve T cells into regulatory T cells (Tregs) by amplifying Foxp3 expression, nevertheless inhibiting the production of Th17 cells, thus participating in Th17/Treg immune regulatory functions (79).
Similarly, galectins are also crucial in signal transduction and the regulation of B cell development, differentiation, activation, and antibody production (14). In particular, Gal-9 can compromise B cell activation transmitted via the B cell receptor (15, 16). In humans, the interaction between Gal-9 and appropriated glycosylated CD45 on human naïve/memory B cells inhibits calcium signaling through a Lyn-CD22-SHP-1 pathway, ultimately reducing B cell activation (15). Additionally, both recombinant and mesenchymal stem cell-derived Gal-9 can attenuate B cell proliferation and the formation of antibody-secreting cells in a dose-dependent fashion (80). These findings are further solidified by studies from Hu et al. that show human cord blood-derived stem cells directly modulate activated B cells through a Gal-9-mediated mechanism, resulting in significant suppression of B cell proliferation and noticeable phenotypic changes (81). In addition, Gal-9 can bridge human circulating and naive B cells to vascular endothelial cells, coordinating their pace of transendothelial migration (17). Furthermore, these interactions induce a global transcriptional response in gene families associated with the regulation of naive B cell signaling and membrane/cytoskeletal dynamics (17). Among the key immunoregulators elevated by Gal-9 binding is the signaling lymphocytic activation molecule family member 7 (SLAMF7), while its cytosolic adapter EAT-2 necessary for cell activation is unaffected (17). Moreover, Gal-9 can encourage the survival traits of human naïve and circulating B cells by inducing the phosphorylation of the pro-survival factor, pERK (17).
3 Gal-9 involvement in malignancy
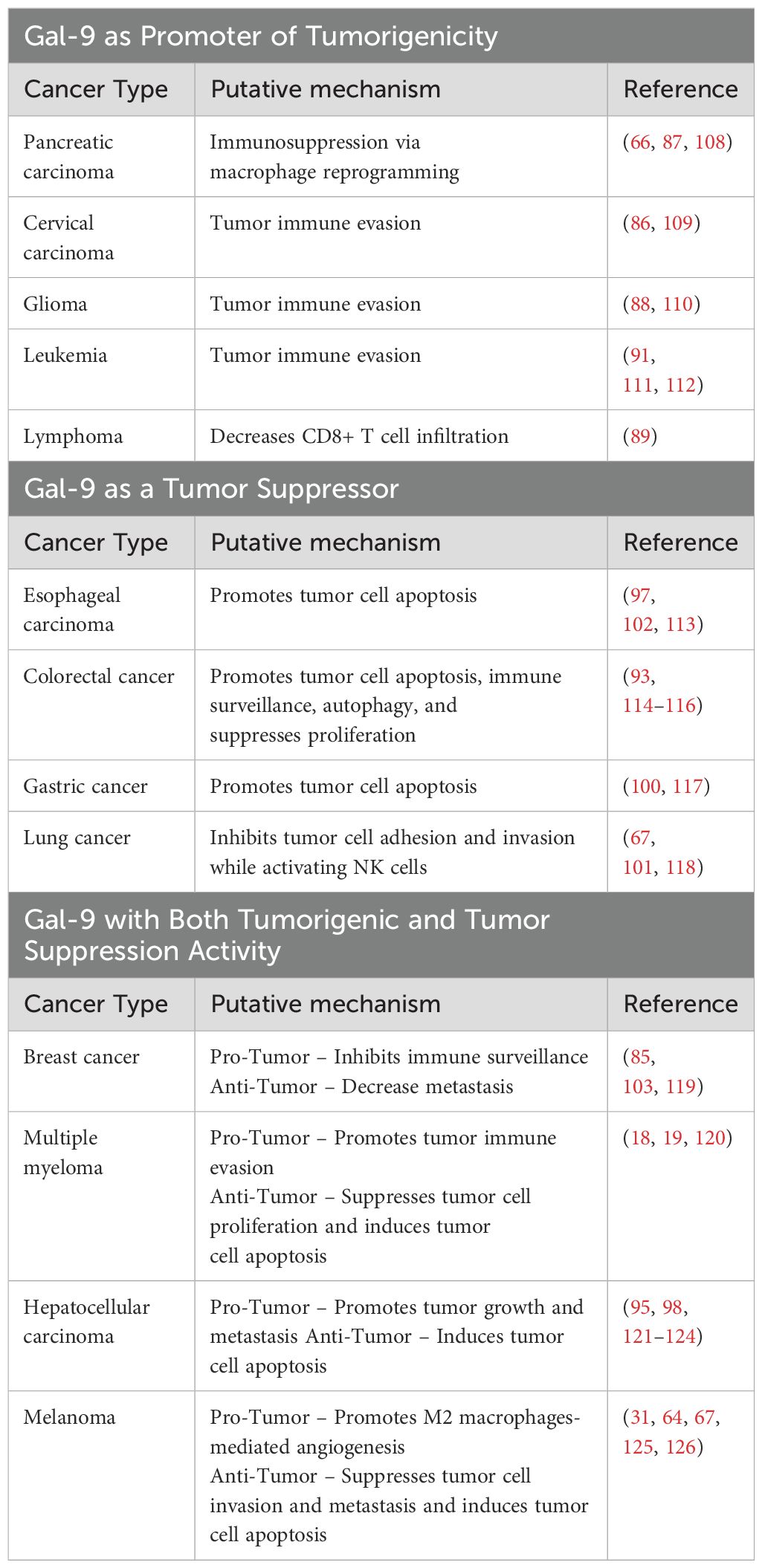
Central to the development of effective therapies for eliminating cancer is the exploration of malignancy-associated molecules and their role in driving tumor development, metastasis, and relapse (82). The patterns of Gal-9 expression and its roles in various types of cancer have emerged as a compelling area of research, aiming to deepen our comprehension of mechanisms underlying cancer initiation and progression and identify novel therapeutic targets (83, 84). Gal-9 exhibits substantial variability in its expression levels across different types of cancer (83). Compared to healthy tissues, Gal-9 is upregulated in pancreatic carcinoma, breast cancer, glioblastoma multiforme, cervical carcinoma, chronic lymphocytic leukemia, acute myeloid leukemia (AML), and cutaneous T cell lymphoma (CTCL) (85–91). In B-cell acute lymphoblastic leukemia (B-ALL) patients, while serum Gal-9 levels remain unchanged compared to healthy controls, Lee et al. demonstrated that adipocyte-secreted factors in obese patients upregulate Gal-9 surface expression on B-ALL cells (91, 92). Conversely, Gal-9 expression is notably downregulated in esophageal carcinoma, renal cell carcinoma, hepatocellular carcinoma, colon cancer, gastric cancer, prostate cancer, lung cancer, melanoma, and adrenal carcinoma compared to healthy counterparts (31, 93–97).
Additionally, there has been a growing interest in exploring the prognostic value of Gal-9 expression levels in patients with cancer (83). Many studies suggest an inverse relationship between Gal-9 expression levels and cancer progression for many solid tumors, including esophageal carcinoma, gastric cancer, hepatocellular carcinoma, colorectal cancer, lung cancer, breast cancer, and melanoma (93, 98–105). In these instances, administering exogenous Gal-9 can effectively impede the growth of these tumors, representing a promising anti-cancer treatment (83). However, contradictory findings exist where a few studies associate high tissue and plasma Gal-9 with poor survival and clinical outcomes (90, 106). Indeed, Gal-9 and its binding partner, T cell immunoglobulin mucin-3 (Tim-3), have been implicated in negatively regulating the cellular immune response by inducing T cell apoptosis and exhaustion, fostering an immunosuppressive tumor microenvironment (TME) (18, 43). Interestingly, in non-small cell lung carcinoma, high Gal-9 expression on cancer cells is associated with longer overall survival, whereas elevated Gal-9 levels on tumor-infiltrating lymphocytes predict shorter recurrence-free survival (101). Hence, these opposing effects are likely reflected by context-dependent roles of Gal-9 in cancer, influenced by its localization within the TME, by cancer-specific responses, by tumor heterogeneity, by variability in the expression profiles of Gal-9 glycoconjugates ligands, and by diversity of experimental results driven by differing methods of analysis and inadequate clinical sample sizes (107). These variabilities of Gal-9 expression not only influence tumor development and progression, but also patient prognosis (Table 2).
3.1 Gal-9 and multiple myeloma (MM)
3.1.1 Characteristics and diagnosis of MM
MM is a clonal B cell malignancy, accounting for approximately 1% of all cancers and 10% of hematologic malignancies (127). It is characterized by abnormal accumulation of ≥10% malignant clonal plasma cells in the bone marrow (128–132). MM is commonly diagnosed in elderly individuals, with a median age at diagnosis around 66 years (133). There is a clear sex disparity in MM incidence across all ages and racial/ethnic groups, with males showing higher incidence and mortality rates compared to females. However, the underlying mechanisms and the impact of sex on patient outcomes remain poorly understood and underexplored (134, 135). Data from a recent study suggest that individuals with MM have a median survival rate of about 7.5 years (136). Most individuals with MM exhibit the production of a monoclonal immunoglobulin protein, also termed M-protein, generated by aberrant clonal plasma cells. Nonetheless, in 15–20% of cases, MM cells exclusively release monoclonal free light chains, and in fewer than 3% of patients, these cells do not secrete any monoclonal protein (133, 137).
The malignant clonal plasma cells in MM cause typical end-organ damage, such as hypercalcemia, renal failure, anemia, and lytic bone lesions referred to as “CRAB” symptoms (138–140). The presence of one or more of the CRAB symptoms sets MM apart from other plasma cell disorders, such as smoldering multiple myeloma (SMM) and monoclonal gammopathy of undetermined significance (MGUS) (138). SMM is identified by 10-60% bone marrow plasma cell proliferation and/or elevated M-protein ≥3 g/dL, without CRAB symptoms (141), while MGUS is characterized by a clonal bone marrow plasma cells <10% and a serum M-protein level <3 g/dL in asymptomatic patients (142). Both SMM and MGUS are recognized as precursors to MM, with an annual progression risk of 10% and 1%, respectively (143).
The surface antigens CD138, CD38, CD45, CD19, CD56, CD117, CD20, CD28, CD27, and CD81 are the most widely used markers for characterizing normal and malignant plasma cells (144). BM-derived myeloma cells are typically defined by their high expression of CD38, CD138, and CD56, low expression of CD45, and absence of CD19 (145). In addition to its crucial role in establishing MM diagnosis, numerous studies have reported the prognostic and therapeutic significance of plasma cell immunophenotypic features in MM patients (146). For instance, the upregulation of CD19 and its regulatory partner CD81 are linked to poorly differentiated plasma cell clones associated with poor outcomes in MM patients (147). Similarly, the expression of CD45 on clonal plasma cells is associated with an aggressive phenotype of MM (148). B cell maturation antigen (BCMA/CD269) is another surface marker that is primarily expressed by plasma cells and mature B lymphocytes (149, 150), and is upregulated on malignant plasma cells in MM (151, 152). BCMA is a target antigen for the FDA approved chimeric antigen receptor-T (CAR-T) cell therapies for MM (153).
3.1.2 Mechanistic roles of Gal-9 across diverse malignancies
In breast cancer, Gal-9 overexpression enhances cancer cell adhesion and invasion through Focal Adhesion Kinase activation, which upregulates the pro-invasive protein S100A4, and facilitates immune evasion via the Tim-3-Gal-9 pathway by transferring Gal-9 to the cell surface (85, 154–156). In contrast, recombinant protease-resistant Gal-9 (rGal-9) exerts anti-proliferative effects in gastric cancer by inducing apoptosis, reducing the phosphorylation of VEGFR-3 and IGF-1R, and altering miRNA expression (100). In colorectal cancer, Gal-9 promotes extensive intra-tumoral NK cell infiltration through Rho/ROCK1 signaling, correlating with improved prognosis (93, 157, 158). Similarly, in mice with lung cancer, Gal-9 induces macrophage differentiation into plasmacytoid dendritic cell-like macrophages, potentially enhancing NK cell activation, and extending longevity (118). Gal-9 also inhibits tumor growth in hepatocellular carcinoma by inducing apoptosis via MicroRNAs mediated miR-1246-DYRK1A-caspase-9 axis (121), and suppresses esophageal squamous cell carcinoma proliferation both in vitro and in vivo through Jun NH2-terminal kinase (JNK) and p38 MAPK pathways (113). However, Gal-9 may support tumor immunosuppression in glioblastoma by promoting M2 tumor-associated macrophage activity (88). In contrast, in pancreatic ductal adenocarcinoma, Gal-9’s binding to Dectin-1-expressing macrophages fosters immunosuppression and tumor progression, though exogenous rGal-9 can induce apoptosis by promoting cytochrome release and altering miRNAs expression in this context (66, 159). In cervical cancer, the Tim-3-Gal-9 pathway facilitates immune escape by promoting regulatory T cell activity, with epigenetic regulation through the H3K9me3-specific histone methyltransferase (SUV39H1) - DNA methyltransferase 3 alpha (DNMT3A) axis (86, 109). While serum Gal-9 levels are elevated in patients with advanced cutaneous T-cell lymphoma, the exogenous administration of rGal-9 induces apoptosis by Tim-3 independent activation of caspase-3 and caspase-9, and suppresses in vivo tumor growth (89). Finally, in AML, the Tim-3-Gal-9 autocrine loop promotes leukemic stem cell self-renewal and disease progression through the NF-κB, β-catenin, and PKC/mTOR pathways (91, 111). These findings underscore the complex and context-dependent roles of Gal-9 in cancer biology, highlighting its potential therapeutic importance in MM.
3.1.3 The BM microenvironment in MM
The BM consists of a heterogeneous population of cells, including hematopoietic stem cells and their myeloid and lymphoid progeny, along with endothelial cells and cells derived from mesenchymal stromal cells, such as adipocytes, chondrocytes and osteoblasts, all of which are embedded in the extracellular matrix (ECM) (160). In MM, the crosstalk between MM cells and the surrounding BM microenvironment activates intracellular signaling pathways that impact tumor behavior, including tumor growth, progression, angiogenesis, immune evasion, and drug resistance (161, 162). This crosstalk is facilitated by direct cell-cell contact through surface adhesion molecules, secreted soluble cytokines and growth factors, and exosome-mediated intercellular communication (163).
Different molecular complexes on MM cells interact with ECM proteins, such as syndecan-1 (CD138) with collagen type 1 (164) and very late antigen-4 (VLA-4) with fibronectin (165), or with their binding partners on various non-tumor cells, such as VLA-4 with vascular cell adhesion molecule-1 (VCAM-1) and lymphocyte function-associated antigen-1 (LFA-1) with intercellular adhesion molecule-1 (ICAM-1) (166). These interactions activate various downstream signaling pathways associated with tumor progression and poor outcomes in MM patients such as NF-κB, ERK, and phosphoinositide 3-kinases (PI3K)/Akt pathways (163). Cytokines and growth factors, synthesized and released by bone marrow stromal cells (BMSCs) or MM cells, are also implicated in triggering the major signaling pathways that promote the survival and dissemination of MM cells and mediate drug resistance (166). Key MM-associated cytokines include interleukin-6 (IL-6), IL-10, IL-17a, insulin-like growth factor-1 (IGF1), vascular endothelial growth factor (VEGF), fibroblast growth factor (FGF), transforming growth factor-β (TGF-β), and B cell-activating factor (BAFF) (167–169). Cytokines are also involved in disrupting normal bone remodeling in MM, leading to increased bone resorption and formation of osteolytic lesions, which are present in approximately 80% of MM patients (170). Both BMSCs and MM cells contribute to the pathogenesis of myeloma osteolytic lesions through secretion of osteoclast activating factors, such as receptor activator of nuclear factor-κB (RANKL), tumor necrosis factor-α (TNF-α), macrophage inflammatory protein-1α (MIP-1α), macrophage colony-stimulating factor (MCSF), IL-3, and IL-6 (171) as well as osteoblast inhibitory factors such as the Wnt antagonists Dickkopf-1 (DKK1), secreted frizzled-related protein-2 (sFRP-2), Runt-related transcription factor 2 (Runx2), and TGF-β (172).
Characteristically, MM cells foster an immunosuppressive bone marrow environment not only by inhibiting antitumor effector cells and disrupting antigen presentation, but also by encouraging the expansion of regulatory immune cells (173), such as myeloid-derived suppressor cells (174, 175), Tregs (176, 177), and regulatory B cells (178, 179). The myeloma microenvironment also shows a significant increase in Th17 T cells, probably due to the myeloma cell-derived IL-6 and TGF-β production (180). Increased IL-17 production by Th17 T cells has been demonstrated to suppress cytotoxic T cell activity, promote myeloma cell growth in vitro and in vivo through IL-17 receptors (IL17R), and play a crucial role in MM-associated bone disease (180, 181). Moreover, dendritic cells (DCs) exhibit functional impairment in MM patients, characterized by the inability to upregulate CD80 expression in response to stimulation by human CD40LT and IL-2, possibly due to their inhibition by the elevated levels of TGFβ1 and IL-10 in the myeloma microenvironment (182, 183). Importantly, studies have demonstrated an upregulated programmed death ligand-1 (PD-L1) on malignant plasma cells is reported to increase as MGUS progresses to MM (184), combined with elevated expression of its receptor programmed death 1 (PD-1) on several MM-associated immune cell subsets, including T cells, B cells, NK cells, and DCs (185). The interaction between PD-L1 and PD-1 represents a major contributor to the immunosuppressive characteristics observed in the MM microenvironment and is shown to be counteracted by PD-L1 or PD-1 blockade (186, 187).
3.1.4 Dual role of Gal-9 in MM
Data from MM studies on the role of Gal-9 are sparse and controversial, with evidence documenting both pro-tumorigenic and anti-tumorigenic activities (Figure 3) (18, 19). Investigations conducted by Lee et al. on BM aspirate samples obtained from 109 newly diagnosed MM patients indicate that Gal-9 expression is an independent predictor of poor survival in patients who exhibit elevated PD-L1 levels (107). In a recent study performed on blood samples from 60 newly diagnosed MM patients and 40 healthy controls, Zhang et al. have revealed upregulated expression levels of Tim-3 on CD4+ T cell surfaces, elevated Gal-9 mRNA in peripheral blood mononuclear cells, and increased serum levels of Gal-9 in MM patients compared to healthy donors (18). The researchers have demonstrated that Tim-3, upon binding with Gal-9, disrupts the balance between CD4+ T cell subsets (Th1, Th2, Th17, and Treg cells) and impacts secretion of their cytokines, leading to inhibition of the cytotoxic function of Th1 cells and promotion of Th2 and Th17 cells involvement in the immune escape of MM (18).
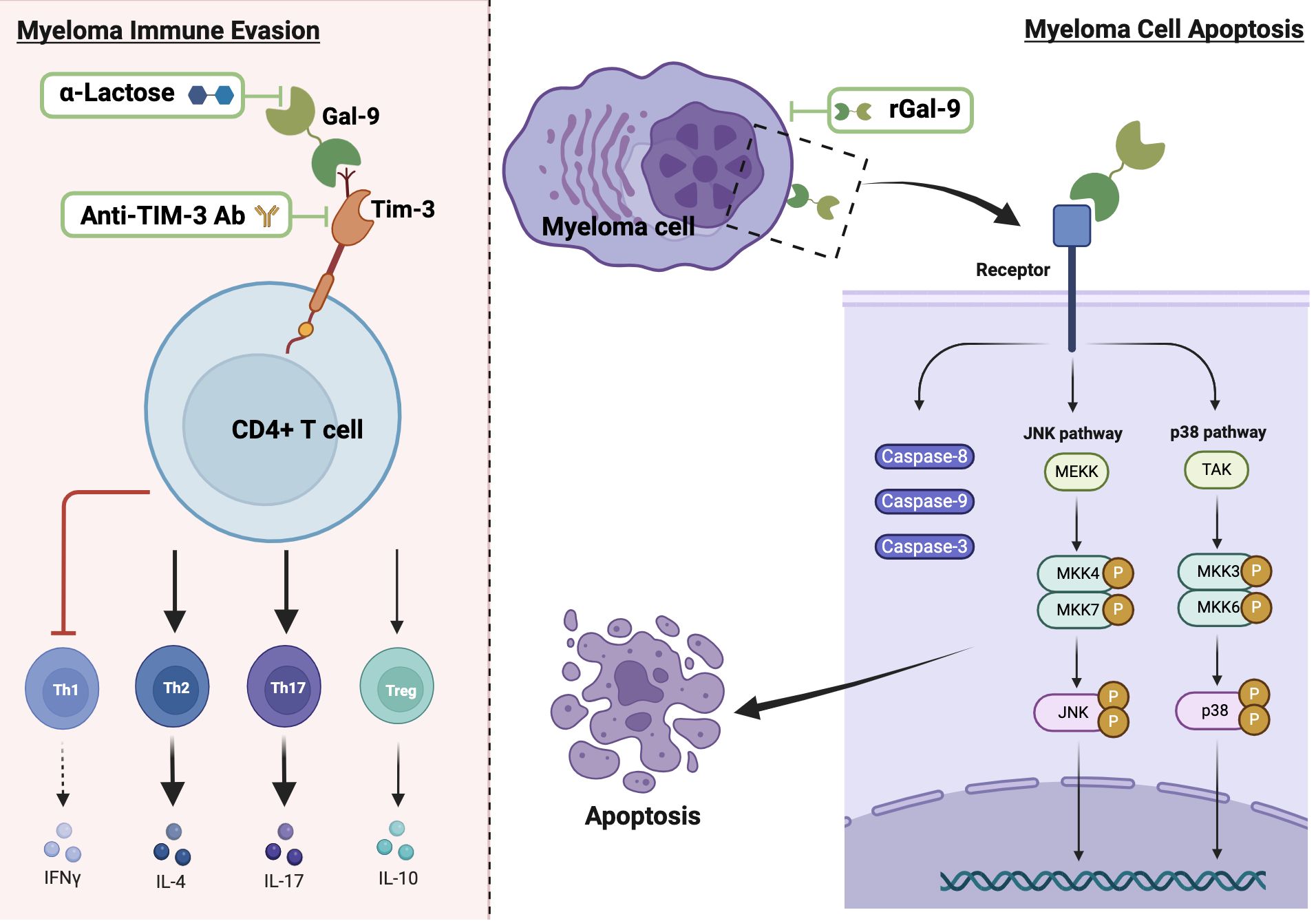
Figure 3. The dual role of Gal-9 in MM and potential therapeutic strategies. The left panel depicts the pro-tumorigenic effect of Gal-9, showing its binding to Tim-3 on CD4+ T cells, which leads to disrupted T cell subset balance by inhibiting Th1 and enhancing the immune response of Th2 and Th17 cells. The right panel illustrates the anti-tumorigenic effect of Gal-9, demonstrating its binding to myeloma cells and subsequent activation of apoptotic pathways through caspases and JNK/p38 MAPK signaling. Thick black arrows indicate activation and bar-ended red lines represent inhibition. Potential therapeutic strategies are shown within green frames.
On the other hand, earlier studies led by Kobayashi et al. report that rGal-9 effectively suppresses the proliferation of various human B-lymphoblast myeloma cell lines (HMCL), namely IM9, AMO-1, KMS-12-BM, NCI-H929, and RPMI8226, and its effectiveness is directly associated with the binding affinity of Gal-9 to each HMCL (19). Their data also demonstrate that Gal-9 exerts a dose-dependent pro-apoptotic effect on primary myeloma cells derived from 10 MM patients, including cells from treatment-resistant patients with poor prognosis and chromosomal abnormalities (19). Mechanistically, the authors suggest that Gal-9 induces apoptosis through the activation of caspase-8, -9, and -3, associated with the activation of JNK and p38 MAPK signaling pathways, while inhibiting JNK or p38 MAPK pathways result in attenuated the anti-proliferative effect of Gal-9, highlighting the crucial role of these pathways in mediating the anti-myeloma cell effect of Gal-9 (19). In contrast, data from another study reveal that Gal-9 induces T cell apoptosis with no effect on MM cells (120). The dual role of Gal-9 extends beyond MM; for instance, in hepatocellular carcinoma, its downregulation in hepatocytes promotes tumor growth and metastasis, while its overexpression in Kupffer cells and endothelial cells suppresses the anti-tumor immune response by inducing T cell apoptosis or senescence (98, 121–123). This duality may be due to the loss of Gal-9 expression during tumor progression after its initial upregulation helps establish a tumorigenic environment (122). In malignant melanoma, high Gal-9 expression suppresses the invasion and metastasis by blocking adhesion to endothelium and ECM (67). However, in metastatic malignant melanoma patients, Gal-9 binding to CD206 expressing M2 macrophages promotes tumor growth by increased angiogenesis and chemokine production (64). Further research is required to explore how Gal-9 contributes to each of the cancer hallmarks in MM patients, considering its cellular and subcellular localizations and its interacting partners within the TME. Additionally, the potential significance of individual Gal-9 splice variants in regulating MM progression needs to be elucidated.
3.1.5 Clinical implications of Gal-9 in MM
The standard treatment for MM involves a variety of drug combinations aiming to enhance patient survival and quality of life (188, 189). These include alkylating agents (melphalan, cyclophosphamide), corticosteroids (dexamethasone, prednisone), anthracyclines (doxorubicin and liposomal doxorubicin), proteasome inhibitors (PIs; bortezomib, carfilzomib, ixazomib), immunomodulatory drugs (IMiDs; thalidomide, lenalidomide, pomalidomide), monoclonal antibodies (mAbs; Daratumumab, isatuximab, elotuzumab), CAR-T cell therapies, nuclear export inhibitors, histone deacetylase inhibitors (iHDACs), and autologous stem cell transplantation (ASCT) (190–192). The selection of therapy throughout the disease course is influenced by factors such as age, performance status, comorbidities, eligibility for stem cell transplantation, and the risk stratification of MM patients (189). The strategic use of these treatment modalities has significantly enhanced the overall survival rates in MM patients (193–195). However, challenges, such as drug intolerability, drug resistance, and disease-relapse remain prevalent, necessitating the ongoing development of novel therapeutic strategies (196).
Prior studies have underscored the critical role of Gal-9 and its binding partner Tim-3 on CD4+ T cell surfaces in the development of treatment resistance in both hematologic and solid malignancies (197). In AML, patients who failed to respond to chemotherapy exhibited elevated expression levels of Gal-9 and Tim-3, suggesting that targeting the Gal-9/Tim-3 axis could potentially enhance the effectiveness of induction chemotherapy and improve remission rates in AML patients (198). Furthermore, a growing body of evidence from both preclinical studies and clinical trials indicates that combination therapy targeting Gal-9/Tim-3 alongside anti-PD-1/PD-L1 treatment produces superior outcomes in cancer patients compared to either approach alone (83).
These promising results in various cancer types spark interest in exploring the potential of Gal-9/Tim-3 targeting in MM. While comprehensive studies specifically examining the combination of Gal-9 inhibitors with standard anti-MM therapies are currently lacking, the substantial evidence of Gal-9 contribution to an immunosuppressive TME in MM strongly suggests the potential for synergistic therapeutic effects (18). For instance, blocking Gal-9/TIM-3 interactions could potentially enhance the efficacy of FDA-approved BCMA-targeted CAR-T cell therapies by prolonging their survival and activity within the TME, resulting in a more effective elimination of MM cells (199). This approach could be particularly beneficial in MM, where maintaining CAR-T cell function in the hostile TME remains a significant challenge (200). Likewise, combining Gal-9 inhibition with the lenalidomide, a standard anti-MM therapy that enhances the cytotoxic activities of T-cells and NK-cells against tumor cells, could potentially augment the immune response against MM (201). However, this combination strategy warrants further investigation.
In this regard, blocking Tim-3 using mAbs or small molecules has shown promise in preclinical studies (202–204). Similarly, α-Lactose and its derivatives have been used for targeting Gal-9 (205, 206). However, concerns about their non-selective inhibition of galectins and widespread presence in food and pharmaceutical preparations emphasize the need for alternative Gal-9 inhibitors (206). Recently, researchers have developed novel Gal-9-neutralizing antibodies that protect T cells from Gal-9-induced cell death and enhance T cell-mediated tumor cell killing (207).
On the other hand, considering the pro-apoptotic effect of rGal-9 on MM cells, there is a possibility of targeting MM through the development of stable rGal-9 formulations for delivery to myeloma cells (19). Indeed, Kobayashi et al. have demonstrated significant efficacy and relative safety of Gal-9 as a tumor growth inhibitor in MM xenograft models, suggesting its potential as a therapeutic approach for MM (19). Notably, exogenous administration of Gal-9 has been shown to effectively induce tumor cell apoptosis in other types of cancer, such as esophageal cancer (113), hepatocellular carcinoma (121), gastric cancer (100), chronic myeloid leukemia (208), and CTCL (89). Furthermore, rGal-9 administration has been shown to inhibit tumor progression by impeding the in vivo metastasis of highly metastatic melanoma and colon cancer cells (67). Prior studies have also demonstrated that exogenous Gal-9 treatment significantly increases the expression of the B cell immunoregulatory factor SLAMF7, which represents a promising immunotherapeutic target for MM (17). Accordingly, combining rGal-9 with anti-SLAMF7 therapy holds the potential for synergistically enhancing the targeting of MM cells (209, 210). However, alterations in the cell glycome during the transition from normal plasma cells to myeloma cells likely impact Gal-9 binding affinity and modify intracellular signaling activation, highlighting the necessity for thorough investigation of this hypothesis (211, 212). Additionally, combining rGal-9 with proteasome inhibitors, which disrupts protein degradation in myeloma cells, could yield synergistic effects in inducing apoptosis through the simultaneous targeting of complementary cell death pathways (213). Further research is needed to fully elucidate the potential of Gal-9-based therapy, identify patients who would benefit most, and develop novel combination strategies for MM patients to overcome current treatment challenges and improve patient outcomes.
4 Conclusions
Gal-9 is a ubiquitous molecule predominately expressed by immune and stromal cells of tissues in the immune system. It interacts with a diverse array of intracellular, cell membrane and extracellular ligands, influencing numerous cellular processes that govern development/regulation of immune responses and development/progression of a malignancy. A growing body of evidence, in fact, indicates that the Gal-9 - ligand axis considerably contributes to the pathophysiology of many cancer types. In MM, the Gal-9 ligand-binding activity causes both tumor-promoting and tumor-suppressive properties depending on the appropriately glycosylated ligands on discrete cell types in the TME. Further research is warranted to elucidate the precise mechanisms underlying the dual role of Gal-9 in MM, paving the way for targeting Gal-9 in combination with currently available anti-MM therapies to enhance the treatment efficacy. Translating these findings into clinical applications holds promise for improving patient care and outcomes of this challenging hematologic malignancy.
Author contributions
RS: Writing – original draft, Writing – review & editing. NM: Writing – original draft, Writing – review & editing. CD: Conceptualization, Data curation, Formal analysis, Funding acquisition, Investigation, Methodology, Project administration, Resources, Software, Supervision, Validation, Visualization, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This review was supported by the NIH/National Cancer Institute (NCI) Alliance of Glycobiologists for Cancer Research: Biological Tumor Glycomics Laboratory U01 grant CA225644 (CD) and NIH/NCI R01 grant CA282520 (CD).
Acknowledgments
The figures were created using Biorender illustrating tool (Biorender.com).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Author disclaimer
The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
References
1. Rabinovich GA, Toscano MA. Turning 'sweet' on immunity: galectin-glycan interactions in immune tolerance and inflammation. Nat Rev Immunol. (2009) 9:338–52. doi: 10.1038/nri2536
2. Nabi IR, Shankar J, Dennis JW. The galectin lattice at a glance. J Cell Sci. (2015) 128:2213–9. doi: 10.1242/jcs.151159
3. Varki A, Cummings RD, Esko JD, Stanley P, Hart GW, Aebi M, et al. Essentials of glycobiology. 4th ed. Cold Spring Harbor: Cold Spring Harbor Laboratory Press (2022).
4. Heusschen R, Griffioen AW, Thijssen VL. Galectin-9 in tumor biology: A jack of multiple trades. Biochim Biophys Acta (BBA) - Rev Cancer. (2013) 1836:177–85. doi: 10.1016/j.bbcan.2013.04.006
5. Rabinovich GA, Croci DO. Regulatory circuits mediated by lectin-glycan interactions in autoimmunity and cancer. Immunity. (2012) 36:322–35. doi: 10.1016/j.immuni.2012.03.004
6. Liu FT, Rabinovich GA. Galectins: regulators of acute and chronic inflammation. Ann N Y Acad Sci. (2010) 1183:158–82. doi: 10.1111/j.1749-6632.2009.05131.x
7. Rabinovich GA, Conejo-Garcia JR. Shaping the immune landscape in cancer by galectin-driven regulatory pathways. J Mol Biol. (2016) 428:3266–81. doi: 10.1016/j.jmb.2016.03.021
8. Dimitroff CJ. Galectin-binding O-glycosylations as regulators of Malignancy. Cancer Res. (2015) 75:3195–202. doi: 10.1158/0008-5472.CAN-15-0834
9. Cedeno-Laurent F, Dimitroff CJ. Galectins and their ligands: negative regulators of anti-tumor immunity. Glycoconj J. (2012) 29:619–25. doi: 10.1007/s10719-012-9379-0
10. Toscano MA, Martinez Allo VC, Cutine AM, Rabinovich GA, Marino KV. Untangling galectin-driven regulatory circuits in autoimmune inflammation. Trends Mol Med. (2018) 24:348–63. doi: 10.1016/j.molmed.2018.02.008
11. Liu F-T, Stowell SR. The role of galectins in immunity and infection. Nat Rev Immunol. (2023) 23:479–94. doi: 10.1038/s41577-022-00829-7
12. Cedeno-Laurent F, Dimitroff CJ. Galectin-1 research in T cell immunity: past, present and future. Clin Immunol. (2012) 142:107–16. doi: 10.1016/j.clim.2011.09.011
13. Perillo NL, Pace KE, Seilhamer JJ, Baum LG. Apoptosis of T cells mediated by galectin-1. Nature. (1995) 378:736–9. doi: 10.1038/378736a0
14. Giovannone N, Smith LK, Treanor B, Dimitroff CJ. Galectin-glycan interactions as regulators of B cell immunity. Front Immunol. (2018) 9:2839. doi: 10.3389/fimmu.2018.02839
15. Giovannone N, Liang J, Antonopoulos A, Geddes Sweeney J, King SL, Pochebit SM, et al. Galectin-9 suppresses B cell receptor signaling and is regulated by I-branching of N-glycans. Nat Commun. (2018) 9:3287. doi: 10.1038/s41467-018-05770-9
16. Cao A, Alluqmani N, Buhari FHM, Wasim L, Smith LK, Quaile AT, et al. Galectin-9 binds IgM-BCR to regulate B cell signaling. Nat Commun. (2018) 9:3288. doi: 10.1038/s41467-018-05771-8
17. Chakraborty A, Staudinger C, King SL, Erickson FC, Lau LS, Bernasconi A, et al. Galectin-9 bridges human B cells to vascular endothelium while programming regulatory pathways. J Autoimmun. (2021) 117:102575. doi: 10.1016/j.jaut.2020.102575
18. Zhang R, Chen S, Luo T, Guo S, Qu J. Activated Tim-3/Galectin-9 participated in the development of multiple myeloma by negatively regulating CD4 T cells. Hematology. (2024) 29:2288481. doi: 10.1080/16078454.2023.2288481
19. Kobayashi T, Kuroda J, Ashihara E, Oomizu S, Terui Y, Taniyama A, et al. Galectin-9 exhibits anti-myeloma activity through JNK and p38 MAP kinase pathways. Leukemia. (2010) 24:843–50. doi: 10.1038/leu.2010.25
20. Chabot S, Kashio Y, Seki M, Shirato Y, Nakamura K, Nishi N, et al. Regulation of galectin-9 expression and release in Jurkat T cell line cells. Glycobiology. (2002) 12:111–8. doi: 10.1093/glycob/12.2.111
21. Hirashima M, Kashio Y, Nishi N, Yamauchi A, Imaizumi TA, Kageshita T, et al. Galectin-9 in physiological and pathological conditions. Glycoconj J. (2002) 19:593–600. doi: 10.1023/B:GLYC.0000014090.63206.2f
22. Lau LS, Mohammed NB, Dimitroff CJ. Decoding strategies to evade Immunoregulators Galectin-1,-3, and-9 and their ligands as novel Therapeutics in cancer Immunotherapy. Int J Mol Sci. (2022) 23:15554. doi: 10.3390/ijms232415554
23. Wada J, Ota K, Kumar A, Wallner EI, Kanwar YS. Developmental regulation, expression, and apoptotic potential of galectin-9, a beta-galactoside binding lectin. J Clin Invest. (1997) 99:2452–61. doi: 10.1172/JCI119429
24. Seki M, Sakata KM, Oomizu S, Arikawa T, Sakata A, Ueno M, et al. Beneficial effect of galectin 9 on rheumatoid arthritis by induction of apoptosis of synovial fibroblasts. Arthritis Rheumatism: Off J Am Coll Rheumatol. (2007) 56:3968–76. doi: 10.1002/art.v56:12
25. Wiener Z, Kohalmi B, Pocza P, Jeager J, Tolgyesi G, Toth S, et al. TIM-3 is expressed in melanoma cells and is upregulated in TGF-beta stimulated mast cells. J Invest Dermatol. (2007) 127:906–14. doi: 10.1038/sj.jid.5700616
26. Yoshida H, Imaizumi T, Kumagai M, Kimura K, Satoh C, Hanada N, et al. Interleukin-1beta stimulates galectin-9 expression in human astrocytes. Neuroreport. (2001) 12:3755–8. doi: 10.1097/00001756-200112040-00030
27. Matsumoto R, Hirashima M, Kita H, Gleich GJ. Biological activities of ecalectin: a novel eosinophil-activating factor. J Immunol. (2002) 168:1961–7. doi: 10.4049/jimmunol.168.4.1961
28. Sato M, Nishi N, Shoji H, Seki M, Hashidate T, Hirabayashi J, et al. Functional analysis of the carbohydrate recognition domains and a linker peptide of galectin-9 as to eosinophil chemoattractant activity. Glycobiology. (2002) 12:191–7. doi: 10.1093/glycob/12.3.191
29. Thijssen VL, Hulsmans S, Griffioen AW. The galectin profile of the endothelium: altered expression and localization in activated and tumor endothelial cells. Am J Pathol. (2008) 172:545–53. doi: 10.2353/ajpath.2008.070938
30. Matsuura A, Tsukada J, Mizobe T, Higashi T, Mouri F, Tanikawa R, et al. Intracellular galectin-9 activates inflammatory cytokines in monocytes. Genes to Cells. (2009) 14:511–21. doi: 10.1111/j.1365-2443.2009.01287.x
31. Kageshita T, Kashio Y, Yamauchi A, Seki M, Abedin MJ, Nishi N, et al. Possible role of galectin-9 in cell aggregation and apoptosis of human melanoma cell lines and its clinical significance. Int J Cancer. (2002) 99:809–16. doi: 10.1002/ijc.10436
32. Fujita K, Iwama H, Oura K, Tadokoro T, Samukawa E, Sakamoto T, et al. Cancer therapy due to apoptosis: galectin-9. Int J Mol Sci. (2017) 18:74. doi: 10.3390/ijms18010074
33. Wada J, Kanwar YS. Identification and characterization of galectin-9, a novel β-galactoside-binding mammalian lectin. J Biol Chem. (1997) 272:6078–86. doi: 10.1074/jbc.272.9.6078
34. Cooper D, Barondes SH. Evidence for export of a muscle lectin from cytosol to extracellular matrix and for a novel secretory mechanism. J Cell Biol. (1990) 110:1681–91. doi: 10.1083/jcb.110.5.1681
35. Bänfer S, Jacob R. Galectins in intra-and extracellular vesicles. Biomolecules. (2020) 10:1232. doi: 10.3390/biom10091232
36. Popa SJ, Stewart SE, Moreau K. Unconventional secretion of annexins and galectins. In: Seminars in cell & developmental biology. Amsterdam, Netherlands: Elsevier (2018).
37. Hughes RC. Secretion of the galectin family of mammalian carbohydrate-binding proteins. Biochim Biophys Acta (BBA)-General Subj. (1999) 1473:172–85. doi: 10.1016/S0304-4165(99)00177-4
38. Johswich A, Longuet C, Pawling J, Rahman AA, Ryczko M, Drucker DJ, et al. N-glycan remodeling on glucagon receptor is an effector of nutrient sensing by the hexosamine biosynthesis pathway. J Biol Chem. (2014) 289:15927–41. doi: 10.1074/jbc.M114.563734
39. Niki T, Tsutsui S, Hirose S, Aradono S, Sugimoto Y, Takeshita K, et al. Galectin-9 is a high affinity IgE-binding lectin with anti-allergic effect by blocking IgE-antigen complex formation. J Biol Chem. (2009) 284:32344–52. doi: 10.1074/jbc.M109.035196
40. Bi S, Hong PW, Lee B, Baum LG. Galectin-9 binding to cell surface protein disulfide isomerase regulates the redox environment to enhance T-cell migration and HIV entry. Proc Natl Acad Sci United States America. (2011) 108:10650–5. doi: 10.1073/pnas.1017954108
41. Ohtsubo K, Takamatsu S, Minowa MT, Yoshida A, Takeuchi M, Marth JD. Dietary and genetic control of glucose transporter 2 glycosylation promotes insulin secretion in suppressing diabetes. Cell. (2005) 123:1307–21. doi: 10.1016/j.cell.2005.09.041
42. John S, Mishra R. Galectin-9: From cell biology to complex disease dynamics. J Biosci. (2016) 41:507–34. doi: 10.1007/s12038-016-9616-y
43. Zhu C, Anderson AC, Schubart A, Xiong H, Imitola J, Khoury SJ, et al. The Tim-3 ligand galectin-9 negatively regulates T helper type 1 immunity. Nat Immunol. (2005) 6:1245–52. doi: 10.1038/ni1271
44. Tureci O, Schmitt H, Fadle N, Pfreundschuh M, Sahin U. Molecular definition of a novel human galectin which is immunogenic in patients with Hodgkin's disease. J Biol Chem. (1997) 272:6416–22. doi: 10.1074/jbc.272.10.6416
45. Yoshida H, Teraoka M, Nishi N, Nakakita S-i, Nakamura T, Hirashima M, et al. X-ray structures of human galectin-9 C-terminal domain in complexes with a biantennary oligosaccharide and sialyllactose. J Biol Chem. (2010) 285:36969–76. doi: 10.1074/jbc.M110.163402
46. Nagae M, Nishi N, Murata T, Usui T, Nakamura T, Wakatsuki S, et al. Structural analysis of the recognition mechanism of poly-N-acetyllactosamine by the human galectin-9 N-terminal carbohydrate recognition domain. Glycobiology. (2008) 19:112–7. doi: 10.1093/glycob/cwn121
47. Li Y, Feng J, Geng S, Geng S, Wei H, Chen G, et al. The N-and C-terminal carbohydrate recognition domains of galectin-9 contribute differently to its multiple functions in innate immunity and adaptive immunity. Mol Immunol. (2011) 48:670–7. doi: 10.1016/j.molimm.2010.11.011
48. Bi S, Earl LA, Jacobs L, Baum LG. Structural features of galectin-9 and galectin-1 that determine distinct T cell death pathways. J Biol Chem. (2008) 283:12248–58. doi: 10.1074/jbc.M800523200
49. Earl LA, Bi S, Baum LG. Galectin multimerization and lattice formation are regulated by linker region structure. Glycobiology. (2011) 21:6–12. doi: 10.1093/glycob/cwq144
50. Tanikawa R, Tanikawa T, Hirashima M, Yamauchi A, Tanaka Y. Galectin-9 induces osteoblast differentiation through the CD44/Smad signaling pathway. Biochem Biophys Res Commun. (2010) 394:317–22. doi: 10.1016/j.bbrc.2010.02.175
51. Tanikawa R, Tanikawa T, Okada Y, Nakano K, Hirashima M, Yamauchi A, et al. Interaction of galectin-9 with lipid rafts induces osteoblast proliferation through the c-Src/ERK signaling pathway. J Bone Mineral Res. (2008) 23:278–86. doi: 10.1359/jbmr.071008
52. Mishra R, Grzybek M, Niki T, Hirashima M, Simons K. Galectin-9 trafficking regulates apical-basal polarity in Madin-Darby canine kidney epithelial cells. Proc Natl Acad Sci United States America. (2010) 107:17633–8. doi: 10.1073/pnas.1012424107
53. Pioche-Durieu C, Keryer C, Souquère S, Bosq J, Faigle W, Loew D, et al. In nasopharyngeal carcinoma cells, Epstein-Barr virus LMP1 interacts with galectin 9 in membrane raft elements resistant to simvastatin. J Virol. (2005) 79:13326–37. doi: 10.1128/JVI.79.21.13326-13337.2005
54. Pelletier I, Hashidate T, Urashima T, Nishill N, Nakamura T, Futai M, et al. Specific recognition of Leishmania major poly-β-galactosyl epitopes by galectin-9: Possible implication of galectin-9 in interaction between L. major and host cells. J Biol Chem. (2003) 278:22223–30. doi: 10.1074/jbc.M302693200
55. Dai SY, Nakagawa R, Itoh A, Murakami H, Kashio Y, Abe H, et al. Galectin-9 induces maturation of human monocyte-derived dendritic cells. J Immunol. (2005) 175:2974–81. doi: 10.4049/jimmunol.175.5.2974
56. Anderson AC, Anderson DE, Bregoli L, Hastings WD, Kassam N, Lei C, et al. Promotion of tissue inflammation by the immune receptor Tim-3 expressed on innate immune cells. Science. (2007) 318:1141–3. doi: 10.1126/science.1148536
57. Oomizu S, Arikawa T, Niki T, Kadowaki T, Ueno M, Nishi N, et al. Cell surface galectin-9 expressing Th cells regulate Th17 and Foxp3+ Treg development by galectin-9 secretion. PloS One. (2012) 7:e48574. doi: 10.1371/journal.pone.0048574
58. Koguchi K, Anderson DE, Yang L, O'Connor KC, Kuchroo VK, Hafler DA. Dysregulated T cell expression of TIM3 in multiple sclerosis. J Exp Med. (2006) 203:1413–8. doi: 10.1084/jem.20060210
59. Nishi N, Itoh A, Shoji H, Miyanaka H, Nakamura T. Galectin-8 and galectin-9 are novel substrates for thrombin. Glycobiology. (2006) 16:15C–20C. doi: 10.1093/glycob/cwl028
60. Miyanishi N, Nishi N, Abe H, Kashio Y, Shinonaga R, Nakakita SI, et al. Carbohydrate-recognition domains of galectin-9 are involved in intermolecular interaction with galectin-9 itself and other members of the galectin family. Glycobiology. (2007) 17:423–32. doi: 10.1093/glycob/cwm001
61. Vaitaitis GM, Wagner DH Jr. Galectin-9 controls CD40 signaling through a Tim-3 independent mechanism and redirects the cytokine profile of pathogenic T cells in autoimmunity. PloS One. (2012) 7:e38708. doi: 10.1371/journal.pone.0038708
62. Katoh S, Ishii N, Nobumoto A, Takeshita K, Dai SY, Shinonaga R, et al. Galectin-9 inhibits CD44-hyaluronan interaction and suppresses a murine model of allergic asthma. Am J Respir Crit Care Med. (2007) 176:27–35. doi: 10.1164/rccm.200608-1243OC
63. Wu C, Thalhamer T, Franca RF, Xiao S, Wang C, Hotta C, et al. Galectin-9-CD44 interaction enhances stability and function of adaptive regulatory T cells. Immunity. (2014) 41:270–82. doi: 10.1016/j.immuni.2014.06.011
64. Enninga EAL, Chatzopoulos K, Butterfield JT, Sutor SL, Leontovich AA, Nevala WK, et al. CD206-positive myeloid cells bind galectin-9 and promote a tumor-supportive microenvironment. J Pathol. (2018) 245:468–77. doi: 10.1002/path.2018.245.issue-4
65. Yang R, Sun L, Li C-F, Wang Y-H, Yao J, Li H, et al. Galectin-9 interacts with PD-1 and TIM-3 to regulate T cell death and is a target for cancer immunotherapy. Nat Commun. (2021) 12:832. doi: 10.1038/s41467-021-21099-2
66. Daley D, Mani VR, Mohan N, Akkad N, Ochi A, Heindel DW, et al. Dectin 1 activation on macrophages by galectin 9 promotes pancreatic carcinoma and peritumoral immune tolerance. Nat Med. (2017) 23:556–67. doi: 10.1038/nm.4314
67. Nobumoto A, Nagahara K, Oomizu S, Katoh S, Nishi N, Takeshita K, et al. Galectin-9 suppresses tumor metastasis by blocking adhesion to endothelium and extracellular matrices. Glycobiology. (2008) 18:735–44. doi: 10.1093/glycob/cwn062
68. Yasinska IM, Meyer NH, Schlichtner S, Hussain R, Siligardi G, Casely-Hayford M, et al. Ligand-receptor interactions of galectin-9 and VISTA suppress human T lymphocyte cytotoxic activity. Front Immunol. (2020) 11:580557. doi: 10.3389/fimmu.2020.580557
69. Madireddi S, Eun SY, Lee SW, Nemčovičová I, Mehta AK, Zajonc DM, et al. Galectin-9 controls the therapeutic activity of 4-1BB-targeting antibodies. J Exp Med. (2014) 211:1433–48. doi: 10.1084/jem.20132687
70. Madireddi S, Eun S-Y, Mehta AK, Birta A, Zajonc DM, Niki T, et al. Regulatory T cell–mediated suppression of inflammation induced by DR3 signaling is dependent on galectin-9. J Immunol. (2017) 199:2721–8. doi: 10.4049/jimmunol.1700575
71. Liang T, Ma C, Wang T, Deng R, Ding J, Wang W, et al. Galectin-9 promotes neuronal restoration via binding TLR-4 in a rat intracerebral hemorrhage model. NeuroMolecular Med. (2021) 23:267–84. doi: 10.1007/s12017-020-08611-5
72. Costa AF, Campos D, Reis CA, Gomes C. Targeting glycosylation: a new road for cancer drug discovery. Trends Cancer. (2020) 6:757–66. doi: 10.1016/j.trecan.2020.04.002
73. Vasta GR, Ahmed H, Nita-Lazar M, Banerjee A, Pasek M, Shridhar S, et al. Galectins as self/non-self recognition receptors in innate and adaptive immunity: an unresolved paradox. Front Immunol. (2012) 3:199. doi: 10.3389/fimmu.2012.00199
74. Robinson BS, Arthur CM, Evavold B, Roback E, Kamili NA, Stowell CS, et al. The sweet-side of leukocytes: galectins as master regulators of neutrophil function. Front Immunol. (2019) 10:1762. doi: 10.3389/fimmu.2019.01762
75. Arikawa T, Saita N, Oomizu S, Ueno M, Matsukawa A, Katoh S, et al. Galectin-9 expands immunosuppressive macrophages to ameliorate T-cell-mediated lung inflammation. Eur J Immunol. (2010) 40:548–58. doi: 10.1002/eji.200939886
76. Wiersma VR, de Bruyn M, Helfrich W, Bremer E. Therapeutic potential of Galectin-9 in human disease. Medicinal Res Rev. (2013) 33:E102–26. doi: 10.1002/med.2013.33.issue-S1
77. Gooden MJ, Wiersma VR, Samplonius DF, Gerssen J, van Ginkel RJ, Nijman HW, et al. Galectin-9 activates and expands human T-helper 1 cells. PloS One. (2013) 8:e65616. doi: 10.1371/journal.pone.0065616
78. Kashio Y, Nakamura K, Abedin MJ, Seki M, Nishi N, Yoshida N, et al. Galectin-9 induces apoptosis through the calcium-calpain-caspase-1 pathway. J Immunol. (2003) 170:3631–6. doi: 10.4049/jimmunol.170.7.3631
79. Seki M, Oomizu S, Sakata K-m, Sakata A, Arikawa T, Watanabe K, et al. Galectin-9 suppresses the generation of Th17, promotes the induction of regulatory T cells, and regulates experimental autoimmune arthritis. Clin Immunol. (2008) 127:78–88. doi: 10.1016/j.clim.2008.01.006
80. Ungerer C, Quade-Lyssy P, Radeke HH, Henschler R, Königs C, Köhl U, et al. Galectin-9 is a suppressor of T and B cells and predicts the immune modulatory potential of mesenchymal stromal cell preparations. Stem Cells Dev. (2014) 23:755–66. doi: 10.1089/scd.2013.0335
81. Hu W, Song X, Yu H, Fan S, Shi A, Sun J, et al. Suppression of B-cell activation by human cord blood-derived stem cells (CB-SCs) through the galectin-9-dependent mechanism. Int J Mol Sci. (2024) 25:1830. doi: 10.3390/ijms25031830
82. Tobin NP, Foukakis T, De Petris L, Bergh J. The importance of molecular markers for diagnosis and selection of targeted treatments in patients with cancer. J Internal Med. (2015) 278:545–70. doi: 10.1111/joim.2015.278.issue-6
83. Lv Y, Ma X, Ma Y, Du Y, Feng J. A new emerging target in cancer immunotherapy: Galectin-9 (LGALS9). Genes Dis. (2023) 10:2366–82. doi: 10.1016/j.gendis.2022.05.020
84. Rodrigues CF, Santos FA, Amorim LAA, da Silva ALC, Marques LGA, Rocha BAM. Galectin-9 is a target for the treatment of cancer: A patent review. Int J Biol Macromolecules. (2024) 254:127768. doi: 10.1016/j.ijbiomac.2023.127768
85. Yasinska IM, Sakhnevych SS, Pavlova L, Teo Hansen Selnø A, Teuscher Abeleira AM, Benlaouer O, et al. The Tim-3-Galectin-9 pathway and its regulatory mechanisms in human breast cancer. Front Immunol. (2019) 10:1594. doi: 10.3389/fimmu.2019.01594
86. Chen Z, Dong D, Zhu Y, Pang N, Ding J. The role of Tim-3/Galectin-9 pathway in T-cell function and prognosis of patients with human papilloma virus-associated cervical carcinoma. FASEB J. (2021) 35:e21401. doi: 10.1096/fj.202000528RR
87. Seifert AM, Reiche C, Heiduk M, Tannert A, Meinecke A-C, Baier S, et al. Detection of pancreatic ductal adenocarcinoma with galectin-9 serum levels. Oncogene. (2020) 39:3102–13. doi: 10.1038/s41388-020-1186-7
88. Yuan F, Ming H, Wang Y, Yang Y, Yi L, Li T, et al. Molecular and clinical characterization of Galectin-9 in glioma through 1,027 samples. J Cell Physiol. (2020) 235:4326–34. doi: 10.1002/jcp.v235.5
89. Nakajima R, Miyagaki T, Kamijo H, Oka T, Shishido-Takahashi N, Suga H, et al. Possible therapeutic applicability of galectin-9 in cutaneous T-cell lymphoma. J Dermatol Sci. (2019) 96:134–42. doi: 10.1016/j.jdermsci.2019.09.004
90. Wdowiak K, Gallego‑Colon E, Francuz T, Czajka‑Francuz P, Ruiz-Agamez N, Kubeczko M, et al. Increased serum levels of Galectin−9 in patients with chronic lymphocytic leukemia. Oncol Lett. (2019) 17:1019–29. doi: 10.3892/ol.2018.9656
91. Kikushige Y, Miyamoto T, Yuda J, Jabbarzadeh-Tabrizi S, Shima T, Takayanagi S-i, et al. A TIM-3/Gal-9 autocrine stimulatory loop drives self-renewal of human myeloid leukemia stem cells and leukemic progression. Cell Stem Cell. (2015) 17:341–52. doi: 10.1016/j.stem.2015.07.011
92. Lee M, Hamilton JA, Talekar GR, Ross AJ, Michael L, Rupji M, et al. Obesity-induced galectin-9 is a therapeutic target in B-cell acute lymphoblastic leukemia. Nat Commun. (2022) 13:1157. doi: 10.1038/s41467-022-28839-y
93. Wang Y, Sun J, Ma C, Gao W, Song B, Xue H, et al. Reduced expression of galectin-9 contributes to a poor outcome in colon cancer by inhibiting NK cell chemotaxis partially through the Rho/ROCK1 signaling pathway. PloS One. (2016) 11:e0152599. doi: 10.1371/journal.pone.0152599
94. Lahm H, André S, Hoeflich A, Fischer JR, Sordat B, Kaltner H, et al. Comprehensive galectin fingerprinting in a panel of 61 human tumor cell lines by RT-PCR and its implications for diagnostic and therapeutic procedures. J Cancer Res Clin Oncol. (2001) 127:375–86. doi: 10.1007/s004320000207
95. Chen S, Pu J, Bai J, Yin Y, Wu K, Wang J, et al. EZH2 promotes hepatocellular carcinoma progression through modulating miR-22/galectin-9 axis. J Exp Clin Cancer Res. (2018) 37:1–12. doi: 10.1186/s13046-017-0670-6
96. Yang J, Zhu L, Cai Y, Suo J, Jin J. Role of downregulation of galectin-9 in the tumorigenesis of gastric cancer. Int J Oncol. (2014) 45:1313–20. doi: 10.3892/ijo.2014.2494
97. Hou N, Ma J, Li W, Zhao L, Gao Q, Mai L. T−cell immunoglobulin and mucin domain−containing protein−3 and galectin−9 protein expression: Potential prognostic significance in esophageal squamous cell carcinoma for Chinese patients. Oncol Lett. (2017) 14:8007–13. doi: 10.3892/ol.2017.7188
98. Zhang Z-Y, Dong J-H, Chen Y-W, Wang X-Q, Li C-H, Wang J, et al. Galectin-9 acts as a prognostic factor with antimetastatic potential in hepatocellular carcinoma. Asian Pacific J Cancer Prev. (2012) 13:2503–9. doi: 10.7314/APJCP.2012.13.6.2503
99. Jiang J, Jin M-S, Kong F, Cao D, Ma H-X, Jia Z, et al. Decreased galectin-9 and increased Tim-3 expression are related to poor prognosis in gastric cancer. PloS One. (2013) 8:e81799. doi: 10.1371/journal.pone.0081799
100. Takano J, Morishita A, Fujihara S, Iwama H, Kokado F, Fujikawa K, et al. Galectin-9 suppresses the proliferation of gastric cancer cells in vitro. Oncol Rep. (2016) 35:851–60. doi: 10.3892/or.2015.4452
101. He Y, Jia K, Dziadziuszko R, Zhao S, Zhang X, Deng J, et al. Galectin-9 in non-small cell lung cancer. Lung Cancer. (2019) 136:80–5. doi: 10.1016/j.lungcan.2019.08.014
102. Akashi E, Fujihara S, Morishita A, Tadokoro T, Chiyo T, Fujikawa K, et al. Effects of galectin-9 on apoptosis, cell cycle and autophagy in human esophageal adenocarcinoma cells. Oncol Rep. (2017) 38:506–14. doi: 10.3892/or.2017.5689
103. Irie A, Yamauchi A, Kontani K, Kihara M, Liu D, Shirato Y, et al. Galectin-9 as a prognostic factor with antimetastatic potential in breast cancer. Clin Cancer Res. (2005) 11:2962–8. doi: 10.1158/1078-0432.CCR-04-0861
104. Wang K, Chen Z, Wu R, Yin J, Fan M, Xu X. Prognostic role of high gal-9 expression in solid tumours: a meta-analysis. Karger S, Basel AG, editors. Basel, Switzerland: Karger (2018) p. 993–1002.
105. Zhou X, Sun L, Jing D, Xu G, Zhang J, Lin L, et al. Galectin-9 expression predicts favorable clinical outcome in solid tumors: a systematic review and meta-analysis. Front Physiol. (2018) 9:452. doi: 10.3389/fphys.2018.00452
106. Labrie M, De Araujo LOF, Communal L, Mes-Masson A-M, St-Pierre Y. Tissue and plasma levels of galectins in patients with high grade serous ovarian carcinoma as new predictive biomarkers. Sci Rep. (2017) 7:13244. doi: 10.1038/s41598-017-13802-5
107. Lee B-H, Park Y, Kim J-H, Kang K-W, Lee S-J, Kim SJ, et al. Prognostic value of galectin-9 relates to programmed death-ligand 1 in patients with multiple myeloma. Front Oncol. (2021) 11:669817. doi: 10.3389/fonc.2021.669817
108. Mondragón L, Kroemer G, Galluzzi L. Immunosuppressive γδ T cells foster pancreatic carcinogenesis. In: OncoImmunology. United Kingdom: Taylor & Francis (2016).
109. Zhang L, Tian S, Zhao M, Yang T, Quan S, Yang Q, et al. SUV39H1-DNMT3A-mediated epigenetic regulation of Tim-3 and galectin-9 in the cervical cancer. Cancer Cell Int. (2020) 20:1–15. doi: 10.1186/s12935-020-01380-y
110. Liu Z, Han H, He X, Li S, Wu C, Yu C, et al. Expression of the galectin-9−Tim-3 pathway in glioma tissues is associated with the clinical manifestations of glioma. Oncol Lett. (2016) 11:1829–34. doi: 10.3892/ol.2016.4142
111. Silva IG, Yasinska IM, Sakhnevych SS, Fiedler W, Wellbrock J, Bardelli M, et al. The Tim-3-galectin-9 secretory pathway is involved in the immune escape of human acute myeloid leukemia cells. EBioMedicine. (2017) 22:44–57. doi: 10.1016/j.ebiom.2017.07.018
112. Pang N, Alimu X, Chen R, Muhashi M, Ma J, Chen G, et al. Activated Galectin-9/Tim3 promotes Treg and suppresses Th1 effector function in chronic lymphocytic leukemia. FASEB J. (2021) 35:e21556. doi: 10.1096/fj.202100013R
113. Chiyo T, Fujita K, Iwama H, Fujihara S, Tadokoro T, Ohura K, et al. Galectin-9 induces mitochondria-mediated apoptosis of esophageal cancer in vitro and in vivo in a xenograft mouse model. Int J Mol Sci. (2019) 20:2634. doi: 10.3390/ijms20112634
114. Yang Q, Hou C, Huang D, Zhuang C, Jiang W, Geng Z, et al. miR−455−5p functions as a potential oncogene by targeting galectin−9 in colon cancer. Oncol Lett. (2017) 13:1958–64. doi: 10.3892/ol.2017.5608
115. Sakhnevych SS, Yasinska IM, Fasler-Kan E, Sumbayev VV. Mitochondrial defunctionalization supresses Tim-3-Galectin-9 secretory pathway in human colorectal Cancer cells and thus can possibly affect tumor immune escape. Front Pharmacol. (2019) 10:342. doi: 10.3389/fphar.2019.00342
116. Wiersma VR, de Bruyn M, Wei Y, van Ginkel RJ, Hirashima M, Niki T, et al. The epithelial polarity regulator LGALS9/galectin-9 induces fatal frustrated autophagy in KRAS mutant colon carcinoma that depends on elevated basal autophagic flux. Autophagy. (2015) 11:1373–88. doi: 10.1080/15548627.2015.1063767
117. Long B, Yu Z, Zhou H, Ma Z, Ren Y, Zhan H, et al. Clinical characteristics and prognostic significance of galectins for patients with gastric cancer: a meta-analysis. Int J Surg. (2018) 56:242–9. doi: 10.1016/j.ijsu.2018.06.033
118. Kadowaki T, Arikawa T, Shinonaga R, Oomizu S, Inagawa H, Soma G, et al. Galectin-9 signaling prolongs survival in murine lung-cancer by inducing macrophages to differentiate into plasmacytoid dendritic cell-like macrophages. Clin Immunol. (2012) 142:296–307. doi: 10.1016/j.clim.2011.11.006
119. Yamauchi A, Kontani K, Kihara M, Nishi N, Yokomise H, Hirashima M. Galectin-9, a novel prognostic factor with antimetastatic potential in breast cancer. Breast J. (2006) 12:S196–200. doi: 10.1111/j.1075-122X.2006.00334.x
120. An G, Acharya C, Feng X, Wen K, Zhong M, Zhang L, et al. Osteoclasts promote immune suppressive microenvironment in multiple myeloma: therapeutic implication. Blood. (2016) 128:1590–603. doi: 10.1182/blood-2016-03-707547
121. Fujita K, Iwama H, Sakamoto T, Okura R, Kobayashi K, Takano J, et al. Galectin-9 suppresses the growth of hepatocellular carcinoma via apoptosis in vitro and in vivo. Int J Oncol. (2015) 46:2419–30. doi: 10.3892/ijo.2015.2941
122. Golden-Mason L, Rosen HR. Galectin-9: Diverse roles in hepatic immune homeostasis and inflammation. Hepatology. (2017) 66:271–9. doi: 10.1002/hep.29106
123. Yang Q, Jiang W, Zhuang C, Geng Z, Hou C, Huang D, et al. microRNA-22 downregulation of galectin-9 influences lymphocyte apoptosis and tumor cell proliferation in liver cancer. Oncol Rep. (2015) 34:1771–8. doi: 10.3892/or.2015.4167
124. Sideras K, Biermann K, Verheij J, Takkenberg BR, Mancham S, Hansen BE, et al. PD-L1, Galectin-9 and CD8+ tumor-infiltrating lymphocytes are associated with survival in hepatocellular carcinoma. Oncoimmunology. (2017) 6:e1273309. doi: 10.1080/2162402X.2016.1273309
125. Holderried TA, de Vos L, Bawden EG, Vogt TJ, Dietrich J, Zarbl R, et al. Molecular and immune correlates of TIM-3 (HAVCR2) and galectin 9 (LGALS9) mRNA expression and DNA methylation in melanoma. Clin Epigenet. (2019) 11:1–15. doi: 10.1186/s13148-019-0752-8
126. Wiersma VR, De Bruyn M, Van Ginkel RJ, Sigar E, Hirashima M, Niki T, et al. The glycan-binding protein galectin-9 has direct apoptotic activity toward melanoma cells. J Invest Dermatol. (2012) 132:2302. doi: 10.1038/jid.2012.133
127. Rajkumar SV. Multiple myeloma: 2022 update on diagnosis, risk stratification, and management. Am J Hematol. (2022) 97:1086–107. doi: 10.1002/ajh.26590
128. Waxman AJ, Mink PJ, Devesa SS, Anderson WF, Weiss BM, Kristinsson SY, et al. Racial disparities in incidence and outcome in multiple myeloma: a population-based study. Blood. (2010) 116:5501–6. doi: 10.1182/blood-2010-07-298760
129. Buadi F, Hsing AW, Katzmann JA, Pfeiffer RM, Waxman A, Yeboah ED, et al. High prevalence of polyclonal hypergamma-globulinemia in adult males in Ghana, Africa. Am J Hematol. (2011) 86:554–8. doi: 10.1002/ajh.22040
130. Kazandjian D, Hill E, Hultcrantz M, Rustad EH, Yellapantula V, Akhlaghi T, et al. Molecular underpinnings of clinical disparity patterns in African American vs. Caucasian American multiple myeloma patients. Blood Cancer J. (2019) 9:15. doi: 10.1038/s41408-019-0177-9
131. Landgren O, Graubard BI, Katzmann JA, Kyle RA, Ahmadizadeh I, Clark R, et al. Racial disparities in the prevalence of monoclonal gammopathies: a population-based study of 12,482 persons from the National Health and Nutritional Examination Survey. Leukemia. (2014) 28:1537–42. doi: 10.1038/leu.2014.34
132. Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer statistics, 2021. CA Cancer J Clin. (2021) 71:7–33. doi: 10.3322/caac.21654
133. Kyle RA, Gertz MA, Witzig TE, Lust JA, Lacy MQ, Dispenzieri A, et al. Review of 1027 patients with newly diagnosed multiple myeloma. In: Mayo Clinic Proceedings. Amsterdam, Netherlands: Elsevier (2003).
134. Castañeda-Avila MA, Suárez-Ramos T, Torres-Cintrón CR, Epstein MM, Gierbolini-Bermúdez A, Tortolero-Luna G, et al. Multiple myeloma incidence, mortality, and survival differences at the intersection of sex, age, and race/ethnicity: A comparison between Puerto Rico and the United States SEER population. Cancer Epidemiol. (2024) 89:102537. doi: 10.1016/j.canep.2024.102537
135. Bird S, Cairns D, Menzies T, Boyd K, Davies F, Cook G, et al. Sex Differences in Multiple Myeloma Biology but not Clinical Outcomes: Results from 3894 Patients in the Myeloma XI Trial. Clin Lymphoma Myeloma Leuk. (2021) 21:667–75. doi: 10.1016/j.clml.2021.04.013
136. Abdallah NH, Smith AN, Geyer S, Binder M, Greipp PT, Kapoor P, et al. Conditional survival in multiple myeloma and impact of prognostic factors over time. Blood Cancer J. (2023) 13:78. doi: 10.1038/s41408-023-00852-4
137. Drayson M, Tang LX, Drew R, Mead GP, Carr-Smith H, Bradwell AR. Serum free light-chain measurements for identifying and monitoring patients with nonsecretory multiple myeloma. Blood J Am Soc Hematol. (2001) 97:2900–2. doi: 10.1182/blood.V97.9.2900
138. Rajkumar SV. Multiple myeloma: 2020 update on diagnosis, risk-stratification and management. Am J Hematol. (2020) 95:548–67. doi: 10.1002/ajh.25791
139. Rajkumar SV, Dimopoulos MA, Palumbo A, Blade J, Merlini G, Mateos MV, et al. International Myeloma Working Group updated criteria for the diagnosis of multiple myeloma. Lancet Oncol. (2014) 15:e538–48. doi: 10.1016/S1470-2045(14)70442-5
140. Michels TC, Petersen KE. Multiple myeloma: diagnosis and treatment. Am Family physician. (2017) 95:373–383A.
141. Rajkumar SV, Landgren O, Mateos M-V. Smoldering multiple myeloma. Blood J Am Soc Hematol. (2015) 125:3069–75. doi: 10.1182/blood-2014-09-568899
142. Kyle RA, Therneau TM, Rajkumar SV, Offord JR, Larson DR, Plevak MF, et al. A long-term study of prognosis in monoclonal gammopathy of undetermined significance. N Engl J Med. (2002) 346:564–9. doi: 10.1056/NEJMoa01133202
143. Mateos M-V, Landgren O. MGUS and smoldering multiple myeloma: diagnosis and epidemiology. Plasma Cell Dyscrasias. (2016) p:3–12. doi: 10.1007/978-3-319-40320-5_1
144. Bonsignore M, Trusso S, De Pasquale C, Ferlazzo G, Allegra A, Innao V, et al. A multivariate analysis of Multiple Myeloma subtype plasma cells. Spectrochimica Acta Part A: Mol Biomolecular Spectrosc. (2021) 258:119813. doi: 10.1016/j.saa.2021.119813
145. Draube A, Pfister R, Vockerodt M, Schuster S, Kube D, Diehl V, et al. Immunomagnetic enrichment of CD138 positive cells from weakly infiltrated myeloma patients samples enables the determination of the tumor clone specific IgH rearrangement. Ann Hematol. (2001) 80:83–9. doi: 10.1007/s002770000245
146. Wang HW, Lin P. Flow cytometric immunophenotypic analysis in the diagnosis and prognostication of plasma cell neoplasms. Cytometry Part B: Clin Cytometry. (2019) 96:338–50. doi: 10.1002/cyto.b.21844
147. Paiva B, Puig N, Cedena M-T, de Jong BG, Ruiz Y, Rapado I, et al. Differentiation stage of myeloma plasma cells: biological and clinical significance. Leukemia. (2017) 31:382–92. doi: 10.1038/leu.2016.211
148. Gonsalves WI, Timm MM, Rajkumar SV, Morice WG, Dispenzieri A, Buadi FK, et al. The prognostic significance of CD45 expression by clonal bone marrow plasma cells in patients with newly diagnosed multiple myeloma. Leukemia Res. (2016) 44:32–9. doi: 10.1016/j.leukres.2016.03.003
149. Carpenter RO, Evbuomwan MO, Pittaluga S, Rose JJ, Raffeld M, Yang S, et al. B-cell maturation antigen is a promising target for adoptive T-cell therapy of multiple myeloma. Clin Cancer Res. (2013) 19:2048–60. doi: 10.1158/1078-0432.CCR-12-2422
150. Dagar G, Gupta A, Masoodi T, Nisar S, Merhi M, Hashem S, et al. Harnessing the potential of CAR-T cell therapy: progress, challenges, and future directions in hematological and solid tumor treatments. J Trans Med. (2023) 21:449. doi: 10.1186/s12967-023-04292-3
151. Xu Y, Mao X, Que Y, Xu M, Li C, Almeida VDF, et al. The exploration of B cell maturation antigen expression in plasma cell dyscrasias beyond multiple myeloma. BMC Cancer. (2023) 23:123. doi: 10.1186/s12885-023-10591-1
152. Shah N, Chari A, Scott E, Mezzi K, Usmani SZ. B-cell maturation antigen (BCMA) in multiple myeloma: rationale for targeting and current therapeutic approaches. Leukemia. (2020) 34:985–1005. doi: 10.1038/s41375-020-0734-z
153. Yu B, Jiang T, Liu D. BCMA-targeted immunotherapy for multiple myeloma. J Hematol Oncol. (2020) 13:1–24. doi: 10.1186/s13045-020-00962-7
154. Pally D, Banerjee M, Hussain S, Kumar RV, Petersson A, Rosendal E, et al. Galectin-9 signaling drives breast cancer invasion through extracellular matrix. ACS Chem Biol. (2022) 17:1376–86. doi: 10.1021/acschembio.1c00902
155. Pylayeva Y, Gillen KM, Gerald W, Beggs HE, Reichardt LF, Giancotti FG. Ras-and PI3K-dependent breast tumorigenesis in mice and humans requires focal adhesion kinase signaling. J Clin Invest. (2009) 119:252–66. doi: 10.1172/JCI37160
156. Sieg DJ, Hauck CR, Schlaepfer DD. Required role of focal adhesion kinase (FAK) for integrin-stimulated cell migration. J Cell Sci. (1999) 112:2677–91. doi: 10.1242/jcs.112.16.2677
157. Coca S, Perez‐Piqueras J, Martinez D, Colmenarejo A, Saez MA, Vallejo C, et al. The prognostic significance of intratumoral natural killer cells in patients with colorectal carcinoma. Cancer: Interdiscip Int J Am Cancer Soc. (1997) 79:2320–8. doi: 10.1002/(SICI)1097-0142(19970615)79:12<2320::AID-CNCR5>3.0.CO;2-P
158. Tartter PI, Steinberg B, Barron DM, Martinelli G. The prognostic significance of natural killer cytotoxicity in patients with colorectal cancer. Arch Surg. (1987) 122:1264–8. doi: 10.1001/archsurg.1987.01400230050009
159. Okura R, Fujihara S, Iwama H, Morishita A, Chiyo T, Watanabe M, et al. MicroRNA profiles during galectin−9−induced apoptosis of pancreatic cancer cells. Oncol Lett. (2018) 15:407–14. doi: 10.3892/ol.2017.7316
160. Batsali AK, Georgopoulou A, Mavroudi I, Matheakakis A, Pontikoglou CG, Papadaki HA. The role of bone marrow mesenchymal stem cell derived extracellular vesicles (MSC-EVs) in normal and abnormal hematopoiesis and their therapeutic potential. J Clin Med. (2020) 9:856. doi: 10.3390/jcm9030856
161. Podar K, Chauhan D, Anderson KC. Bone marrow microenvironment and the identification of new targets for myeloma therapy. Leukemia. (2009) 23:10–24. doi: 10.1038/leu.2008.259
162. Podar K, Richardson PG, Hideshima T, Chauhan D, Anderson KC. The Malignant clone and the bone-marrow environment. Best Pract Res Clin Haematol. (2007) 20:597–612. doi: 10.1016/j.beha.2007.08.002
163. Hideshima T, Anderson KC. Signaling pathway mediating myeloma cell growth and survival. Cancers. (2021) 13:216. doi: 10.3390/cancers13020216
164. Dhodapkar MV, Sanderson RD. Syndecan-1 (CD 138) in myeloma and lymphoid Malignancies: a multifunctional regulator of cell behavior within the tumor microenvironment. Leukemia lymphoma. (1999) 34:35–43. doi: 10.3109/10428199909083378
165. Bou Zerdan M, Nasr L, Kassab J, Saba L, Ghossein M, Yaghi M, et al. Adhesion molecules in multiple myeloma oncogenesis and targeted therapy. Int J Hematologic Oncol. (2022) 11:IJH39. doi: 10.2217/ijh-2021-0017
166. Hervás-Salcedo R, Martín-Antonio B. A journey through the inter-cellular interactions in the bone marrow in multiple myeloma: Implications for the next generation of treatments. Cancers. (2022) 14:3796. doi: 10.3390/cancers14153796
167. Jasrotia S, Gupta R, Sharma A, Halder A, Kumar L. Cytokine profile in multiple myeloma. Cytokine. (2020) 136:155271. doi: 10.1016/j.cyto.2020.155271
168. Gu J, Huang X, Zhang Y, Bao C, Zhou Z, Jin J. Cytokine profiles in patients with newly diagnosed multiple myeloma: Survival is associated with IL-6 and IL-17A levels. Cytokine. (2021) 138:155358. doi: 10.1016/j.cyto.2020.155358
169. Aksoy O, Lind J, Sunder-Plaßmann V, Vallet S, Podar K. Bone marrow microenvironment-induced regulation of Bcl-2 family members in multiple myeloma (MM): therapeutic implications. Cytokine. (2023) 161:156062. doi: 10.1016/j.cyto.2022.156062
170. Mukkamalla SKR, Malipeddi D. Myeloma bone disease: a comprehensive review. Int J Mol Sci. (2021) 22:6208. doi: 10.3390/ijms22126208
171. Galson DL, Silbermann R, Roodman GD. Mechanisms of multiple myeloma bone disease. BoneKEy Rep. (2012) 1:135. doi: 10.1038/bonekey.2012.135
172. Xi H, An R, Li L, Wang G, Tao Y, Gao L. Myeloma bone disease: Progress in pathogenesis. Prog Biophysics Mol Biol. (2016) 122:149–55. doi: 10.1016/j.pbiomolbio.2016.08.003
173. Ho M, Goh CY, Patel A, Staunton S, O’Connor R, Godeau M, et al. Role of the bone marrow milieu in multiple myeloma progression and therapeutic resistance. Clin Lymphoma Myeloma Leukemia. (2020) 20:e752–68. doi: 10.1016/j.clml.2020.05.026
174. Görgün GT, Whitehill G, Anderson JL, Hideshima T, Maguire C, Laubach J, et al. Tumor-promoting immune-suppressive myeloid-derived suppressor cells in the multiple myeloma microenvironment in humans. Blood. (2013) 121:2975–87. doi: 10.1182/blood-2012-08-448548
175. Ramachandran IR, Martner A, Pisklakova A, Condamine T, Chase T, Vogl T, et al. Myeloid-derived suppressor cells regulate growth of multiple myeloma by inhibiting T cells in bone marrow. J Immunol. (2013) 190:3815–23. doi: 10.4049/jimmunol.1203373
176. Brimnes MK, Vangsted AJ, Knudsen LM, Gimsing P, Gang A, Johnsen HE, et al. Increased level of both CD4+ FOXP3+ regulatory T cells and CD14+ HLA-DR–/low myeloid-derived suppressor cells and decreased level of dendritic cells in patients with multiple myeloma. Scandinavian J Immunol. (2010) 72:540–7. doi: 10.1111/j.1365-3083.2010.02463.x
177. Lad D, Huang Q, Hoeppli R, Garcia R, Xu L, Levings M, et al. Evaluating the role of Tregs in the progression of multiple myeloma. Leukemia Lymphoma. (2019) 60(9):2134–42. doi: 10.1080/10428194.2019.1579324
178. Zhang L, Tai Y, Ho M, Xing L, Chauhan D, Gang A, et al. Regulatory B cell-myeloma cell interaction confers immunosuppression and promotes their survival in the bone marrow milieu. Blood Cancer J. (2017) 7:e547–7. doi: 10.1038/bcj.2017.24
179. Zou Z, Guo T, Cui J, Zhang L, Pan L. Onset of regulatory B cells occurs at initial stage of B cell dysfunction in multiple myeloma. Blood. (2019) 134:1780. doi: 10.1182/blood-2019-128703
180. Noonan K, Marchionni L, Anderson J, Pardoll D, Roodman GD, Borrello I. A novel role of IL-17-producing lymphocytes in mediating lytic bone disease in multiple myeloma. Blood. (2010) 116:3554–63. doi: 10.1182/blood-2010-05-283895
181. Prabhala RH, Pelluru D, Fulciniti M, Prabhala HK, Nanjappa P, Song W, et al. Elevated IL-17 produced by TH17 cells promotes myeloma cell growth and inhibits immune function in multiple myeloma. Blood. (2010) 115:5385–92. doi: 10.1182/blood-2009-10-246660
182. Brown RD, Pope B, Murray A, Esdale W, Sze DM, Gibson J, et al. Dendritic cells from patients with myeloma are numerically normal but functionally defective as they fail to up-regulate CD80 (B7-1) expression after huCD40LT stimulation because of inhibition by transforming growth factor-beta1 and interleukin-10. Blood. (2001) 98:2992–8. doi: 10.1182/blood.V98.10.2992
183. Ratta M, Fagnoni F, Curti A, Vescovini R, Sansoni P, Oliviero B, et al. Dendritic cells are functionally defective in multiple myeloma: the role of interleukin-6. Blood. (2002) 100:230–7. doi: 10.1182/blood.V100.1.230
184. Liu J, Hamrouni A, Wolowiec D, Coiteux V, Kuliczkowski K, Hetuin D, et al. Plasma cells from multiple myeloma patients express B7-H1 (PD-L1) and increase expression after stimulation with IFN-γ and TLR ligands via a MyD88-, TRAF6-, and MEK-dependent pathway. Blood J Am Soc Hematol. (2007) 110:296–304. doi: 10.1182/blood-2006-10-051482
185. Benson DM Jr, Bakan CE, Mishra A, Hofmeister CC, Efebera Y, Becknell B, et al. The PD-1/PD-L1 axis modulates the natural killer cell versus multiple myeloma effect: a therapeutic target for CT-011, a novel monoclonal anti–PD-1 antibody. Blood J Am Soc Hematol. (2010) 116:2286–94. doi: 10.1182/blood-2010-02-271874
186. Hallett WH, Jing W, Drobyski WR, Johnson BD. Immunosuppressive effects of multiple myeloma are overcome by PD-L1 blockade. Biol Blood marrow Transplant. (2011) 17:1133–45. doi: 10.1016/j.bbmt.2011.03.011
187. Rosenblatt J, Glotzbecker B, Mills H, Keefe W, Wellenstein K, Vasir B, et al. CT-011, anti-PD-1 antibody, enhances ex-vivo T cell responses to autologous dendritic/myeloma fusion vaccine developed for the treatment of multiple myeloma. Blood. (2009) 114:781. doi: 10.1182/blood.V114.22.781.781
188. Cowan AJ, Green DJ, Kwok M, Lee S, Coffey DG, Holmberg LA, et al. Diagnosis and management of multiple myeloma: a review. Jama. (2022) 327:464–77. doi: 10.1001/jama.2022.0003
189. Rajkumar SV, Kumar S. Multiple myeloma current treatment algorithms. Blood Cancer J. (2020) 10:94. doi: 10.1038/s41408-020-00359-2
190. Cejalvo MJ, de la Rubia J. Which therapies will move to the front line for multiple myeloma? Expert Rev Hematol. (2017) 10:383–92. doi: 10.1080/17474086.2017.1317589
191. Rajkumar SV. Multiple myeloma: Every year a new standard? Hematological Oncol. (2019) 37:62–5. doi: 10.1002/hon.2586
192. Sheykhhasan M, Ahmadieh-Yazdi A, Vicidomini R, Poondla N, Tanzadehpanah H, Dirbaziyan A, et al. CAR T therapies in multiple myeloma: Unleashing the future. Cancer Gene Ther. (2024) 31:667–86. doi: 10.1038/s41417-024-00750-2
193. Kumar SK, Dispenzieri A, Lacy MQ, Gertz MA, Buadi FK, Pandey S, et al. Continued improvement in survival in multiple myeloma: changes in early mortality and outcomes in older patients. Leukemia. (2014) 28:1122–8. doi: 10.1038/leu.2013.313
194. Kumar SK, Rajkumar SV, Dispenzieri A, Lacy MQ, Hayman SR, Buadi FK, et al. Improved survival in multiple myeloma and the impact of novel therapies. Blood J Am Soc Hematol. (2008) 111:2516–20. doi: 10.1182/blood-2007-10-116129
195. Rajkumar SV. Treatment of multiple myeloma. Nat Rev Clin Oncol. (2011) 8:479–91. doi: 10.1038/nrclinonc.2011.63
196. Das S, Juliana N, Yazit NAA, Azmani S, Abu IF. Multiple myeloma: challenges encountered and future options for better treatment. Int J Mol Sci. (2022) 23:1649. doi: 10.3390/ijms23031649
197. Kandel S, Adhikary P, Li G, Cheng K. The TIM3/Gal9 signaling pathway: An emerging target for cancer immunotherapy. Cancer Lett. (2021) 510:67–78. doi: 10.1016/j.canlet.2021.04.011
198. Dama P, Tang M, Fulton N, Kline J, Liu H. Gal9/Tim-3 expression level is higher in AML patients who fail chemotherapy. J immunotherapy Cancer. (2019) 7:1–7. doi: 10.1186/s40425-019-0611-3
199. Cao X, Jin X, Zhang X, Utsav P, Zhang Y, Guo R, et al. Small-molecule compounds boost CAR-T cell therapy in hematological Malignancies. Curr Treat Options Oncol. (2023) 24:184–211. doi: 10.1007/s11864-023-01049-4
200. Lindo L, Wilkinson LH, Hay KA. Befriending the hostile tumor microenvironment in CAR T-cell therapy. Front Immunol. (2021) 11:618387. doi: 10.3389/fimmu.2020.618387
201. Kim SY, Park S-S, Lim J-Y, Lee JY, Yoon J-H, Lee S-E, et al. Prognostic role of the ratio of natural killer cells to regulatory T cells in patients with multiple myeloma treated with lenalidomide and dexamethasone. Exp Hematol. (2022) 110:60–8. doi: 10.1016/j.exphem.2022.03.012
202. Acharya N, Sabatos-Peyton C, Anderson AC. Tim-3 finds its place in the cancer immunotherapy landscape. J immunotherapy Cancer. (2020) 8:000911. doi: 10.1136/jitc-2020-000911
203. Zeidan AM, Komrokji RS, Brunner AM. TIM-3 pathway dysregulation and targeting in cancer. Expert Rev Anticancer Ther. (2021) 21:523–34. doi: 10.1080/14737140.2021.1865814
204. Zhang D, Jiang F, Zaynagetdinov R, Huang H, Sood VD, Wang H, et al. Identification and characterization of M6903, an antagonistic anti–TIM-3 monoclonal antibody. Oncoimmunology. (2020) 9:1744921. doi: 10.1080/2162402X.2020.1744921
205. Hong Y, Si X, Liu W, Mai X, Zhang Y. Lactose and its derivatives are a new class of immune checkpoint inhibitors for cancer therapy. bioRxiv. (2023) 2023.12:19.572484. doi: 10.1101/2023.12.19.572484
206. Bailly C, Thuru X, Quesnel B. Modulation of the Gal-9/TIM-3 immune checkpoint with α-lactose. Does anomery of lactose matter? Cancers. (2021) 13:6365. doi: 10.3390/cancers13246365
207. Yang R, Sun L, Li C-F, Wang Y-H, Xia W, Liu B, et al. Development and characterization of anti-galectin-9 antibodies that protect T cells from galectin-9-induced cell death. J Biol Chem. (2022) 298:101821. doi: 10.1016/j.jbc.2022.101821
208. Kuroda J, Yamamoto M, Nagoshi H, Kobayashi T, Sasaki N, Shimura Y, et al. Targeting activating transcription factor 3 by Galectin-9 induces apoptosis and overcomes various types of treatment resistance in chronic myelogenous leukemia. Mol Cancer Res. (2010) 8:994–1001. doi: 10.1158/1541-7786.MCR-10-0040
209. Malaer JD, Mathew PA. CS1 (SLAMF7, CD319) is an effective immunotherapeutic target for multiple myeloma. Am J Cancer Res. (2017) 7:1637–41.
210. Kikuchi J, Hori M, Iha H, Toyama-Sorimachi N, Hagiwara S, Kuroda Y, et al. Soluble SLAMF7 promotes the growth of myeloma cells via homophilic interaction with surface SLAMF7. Leukemia. (2020) 34:180–95. doi: 10.1038/s41375-019-0525-6
211. Moehler TM, Seckinger A, Hose D, Andrulis M, Moreaux J, Hielscher T, et al. The glycome of normal and Malignant plasma cells. PloS One. (2013) 8:e83719. doi: 10.1371/journal.pone.0083719
212. Andrulis M, Ellert E, Mandel U, Clausen H, Lehners N, Raab MS, et al. Expression of Mucin-1 in multiple myeloma and its precursors: correlation with glycosylation and subcellular localization. Histopathology. (2014) 64:799–806. doi: 10.1111/his.2014.64.issue-6
Keywords: galectins, multiple myeloma, galectin-9, tumor microenvironment, therapy
Citation: Shil RK, Mohammed NBB and Dimitroff CJ (2024) Galectin-9 – ligand axis: an emerging therapeutic target for multiple myeloma. Front. Immunol. 15:1469794. doi: 10.3389/fimmu.2024.1469794
Received: 24 July 2024; Accepted: 09 September 2024;
Published: 25 September 2024.
Edited by:
Laia Querol Cano, Radboud University Medical Centre, NetherlandsReviewed by:
Priyanka S. Rana, Case Western Reserve University, United StatesReshmi Parameswaran, Case Western Reserve University, United States
Copyright © 2024 Shil, Mohammed and Dimitroff. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Charles J. Dimitroff, Y2RpbWl0cm9mZkBmaXUuZWR1
 Rajib K. Shil
Rajib K. Shil Norhan B. B. Mohammed2,3
Norhan B. B. Mohammed2,3 Charles J. Dimitroff
Charles J. Dimitroff