
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Immunol. , 18 July 2024
Sec. Molecular Innate Immunity
Volume 15 - 2024 | https://doi.org/10.3389/fimmu.2024.1452609
This article is part of the Research Topic Molecular Innate Immunity in Aquatic Animals and Their Response to Epidemic Diseases View all 6 articles
 Jin-Min Pan1†
Jin-Min Pan1† Yu Liang1†
Yu Liang1† Ke-Cheng Zhu1,2,3
Ke-Cheng Zhu1,2,3 Hua-Yang Guo1,2,3
Hua-Yang Guo1,2,3 Bao-Suo Liu1,2,4
Bao-Suo Liu1,2,4 Nan Zhang1,2,3
Nan Zhang1,2,3 Lin Xian1,2,3
Lin Xian1,2,3 Teng-Fei Zhu1,2,3
Teng-Fei Zhu1,2,3 Dian-Chang Zhang1,2,3*
Dian-Chang Zhang1,2,3*Galectins (Gals) are a type of S-type lectin that are widespread and evolutionarily conserved among metazoans, and can act as pattern recognition receptors (PRRs) to recognize pathogen-associated molecular patterns (PAMPs). In this study, 10 Gals (ToGals) were identified in the Golden pompano (Trachinotus ovatus), and their conserved domains, motifs, and collinearity relationships were analyzed. The expression of ToGals was regulated following infection to Cryptocaryon irritans and Streptococcus agalactiae, indicating that ToGals participate in immune responses against microbial pathogens. Further analysis was conducted on one important member, Galectin-3, subcellular localization showing that ToGal-3like protein is expressed both in the nucleus and cytoplasm. Recombinant protein obtained through prokaryotic expression showed that rToGal-3like can agglutinate red blood cells of rabbit, carp and golden pompano and also agglutinate and kill Staphylococcus aureus, Bacillus subtilis, Vibrio vulnificus, S. agalactiae, Pseudomonas aeruginosa, and Aeromonas hydrophila. This study lays the foundation for further research on the immune roles of Gals in teleosts.
Teleost, as aquatic vertebrates, possess a relatively lower complexity compared to mammals and primarily defend against the myriad of pathogens in their environment through innate immunity. Renowned for its rapid response, this form of immunity includes physical barriers such as skin, mucus, and gills, which constitute the first line of defense in the teleost immune system (1). Research on teleosts has revealed that their mucus contains a combination of agglutinins, lysozymes, antimicrobial peptides, and complement proteins, all of which work together to directly neutralize pathogens (2). When pathogens encounter these physical barriers, Pattern Recognition Receptors (PRRs) recognize Pathogen-Associated Molecular Patterns (PAMPs) on the pathogens, thereby activating the innate immune response (3).
lectins not only play a crucial role within physical barriers but are also key components of PRRs. Initially, animal agglutinins were classified into calcium-dependent C-type lectins and S-type lectins Galectins are a part of the S-type lectins (4). Based on their molecular structure, galectins can be divided into three types: proto-type, which contains only one Carbohydrate Recognition Domain (CRD); tandem repeat type, which features two homologous CRDs at both the N and C termini; and chimeric type like Galectin-3 (5). Galectin-related protein (GRP) has similar structural domains and is often analyzed together with Gals (6).
Trachinotus ovatus, commonly known as golden pompano, is an economically important fish species in the South China Sea. In 2023, it ranked second in marine fish production in China, renowned for its firm flesh and rapid growth (7). In previous studies on Golden pompano, extensive research was conducted on PRRs, such as the classic Toll-like Receptor (TLRs) and NOD-like Receptor (NLRs) (8–10). as well as on proteins with antimicrobial functions like antimicrobial peptides (11, 12). However, these PRRs-Galectins, which also have the antimicrobial functions, have not yet been identified in Golden pompano.
Under high-density aquaculture conditions, outbreaks of bacterial and parasitic infections are common. Among these pathogens, Streptococcus agalactiae and Cryptocaryon irritans pose significant threats to economically important fish species. Teleosts, whether marine or freshwater species, typically exhibit enlarged spleens and livers and accumulate large amounts of fluid in the abdominal cavity following infection with S. agalactiae (13). For Golden pompano, S. agalactiae is one of the most significant bacterial diseases (14). C. irritans is a ciliated obligate parasite predominantly infesting the outer body surfaces of fish, such as gills, skin, and fins, causing illness in marine teleosts. Commonly referred to as white spot disease due to its visually detectable white cysts, C. irritans can inflict substantial damage on host organs. This damage includes respiratory difficulties, abnormal behavior, mechanical injuries, and susceptibility to secondary bacterial infections (15). More seriously, the mortality rate for golden pompano infected with C. irritans can reach 100% (16).
This study has identified the T. ovatus galectins (ToGals) in whole genome wide. The conserved domain, collinearity and evolutionary relationship were analyzed. Moreover, we conducted an infection test for S. agalactiae and C. irritans, analyzing the differences of expression patterns following infection. Additionally, we conducted subcellular localization of a member of ToGals - ToGal3like, in the Golden pompano muscle cell line (GPM). At last, the recombination ToGal3like protein was also expressed using a prokaryotic expression system, and its hemagglutination and bacterial agglutination activities were analyzed. The objective of this study is to provide guidance for the prevention and control of pathogenic microbes in T. ovatus, and to further lay a foundation for research into the functions of Gals in T. ovatus and other teleost species.
To identify the ToGals, amino acid sequences of Galectins from closely related species, the Yellowtail kingfish and the Great amberjack, were initially download from the Ensembl database. Based on these sequences, the genome of T. ovatus was searched using TblastN (https://blast.ncbi.nlm.nih.gov/) (e-value of <1e-10), revealing several candidate genes. Additionally, Hidden Markov Models (HMM) of the Gal-lectin domain were retrieved from the Pfam database (http://pfam.sanger.ac.uk/). Subsequently, genes containing the Gal-lect domain were screened using the HMM searcher (P < 0.05) (17). Ultimately, by integrating both methods, the candidate ToGals genes have been identified.
To enhance the understanding of ToGals, the full mRNA and coding sequences (CDS) for these genes were retrieved using the GTF annotations of the T. ovatus genome (18). Protein sequences were analyzed using ProtParam on the ExPASy (https://expassy.org/protparam/) to determine the molecular weights (MW) and isoelectric points (PI) of the ToGals. Domains within the proteins were defined using a comprehensive search on the NCBI-CDD (https://www.ncbi.nlm.nih.gov/Structure/cdd/cdd.shtml). Using Meme for Linux (https://meme-suite.org/meme/) (19), we identified five key motifs across ToGals mRNA. Comparative analysis was conducted to examine the variances in the total Gals copy numbers between T. ovatus and other species. Finally, we analyzed the collinearity of ToGals using McscanX (20) and visualized it with Circos (21).
For the phylogenetic analysis of ToGals, amino acid sequences of galectins from sapiens, Mus musculus, Danio rerio, Scophthalmus maximus, Takifugu rubripes, and Pseudoplatystoma corruscans, along with the sequences of ToGals, were downloaded from NCBI to create a galectin amino acid library. The sequences were aligned using MAFFT (https://mafft.cbrc.jp/alignment/software/). A maximum likelihood (ML) tree was then constructed using FastTree. The final visualization of the phylogenetic tree was done using Chiplot (https://www.chiplot.online/).
The T. ovatus used in this experiment were all sourced from marine cages in DaPeng Bay, Shenzhen, and were transported to an experimental concrete pond for acclimatization for half a month prior to the experiment, with a salinity of 15‰, pH 8.0 ± 0.2, and water temperature at 28.5 ± 1.5°C. The Committee of the South China Sea Fisheries Research Institute, Chinese Academy of Fisheries Sciences (no. SCSFRI96-254), approved the animal protocols, and all experiments were performed under the applicable standards.
S. iniae was identified from infected T. ovatus in an aquaculture pond in Dapeng, Shenzhen. After purification and identification, 100 mL of the bacterial suspension were spread on BHI agar plates and incubated overnight at 28°C. A bacterial colony was then selected and cultured in BHI liquid medium on a shaker to expand. Following 16S rDNA sequencing performed by Guangzhou Ruibo Biotechnology, the bacteria were confirmed to be S. iniae via BLAST. Prior to infection, S. iniae was cultured for 24 hours in BHI liquid medium under specific conditions (28°C and 140 × g). The sediment was separated by centrifugation at 6200 × g for 8 m, then diluted and spread on plates at five different concentrations, ranging from 1.0 × 106 to 1.0 × 1010 CFU/ml. The concentrations were quantified by measuring colony-forming units per milliliter. The 120-hour lethal dose 50 (LD50) was found to be 2.0 × 107 CFU per T. ovatus, obtained by dilution in PBS (22). In the actual experiment, six tanks each containing 140 L of fresh seawater were used. Three hundred healthy T. ovatus were evenly and randomly divided into control and experimental groups, with three replicates each. The fish in the experimental group were injected with a bacterial mixture of 2.0 × 107 CFU/fish (volume of 200 mL), while the control group received an injection of 200 mL of sterile PBS. Samples were taken immediately after infection, and then 48 and 96 hours later. At each time point, samples were collected from the liver, spleen, and kidneys of nine randomly selected fish from each group. After anesthetizing the T. ovatus with 40 mg/L MS-222, samples from three T. ovatus were pooled into one sample. The samples were then rapidly frozen in liquid nitrogen and stored at -80°C.
C. irritans were sourced from T. ovatus infected at the Dapeng bay. The organism was identified as C. irritans via microscopic examination and was further propagated using T. ovatus as host organisms. Prior to the infection experiments, skin samples from nine unexposed, healthy T. ovatus were collected to serve as a control group (BFS).
The infection method for C. irrita was carried out according to Dan et al. (23). Preliminary studies proceeded to establish the LD50 of C. irritans infection of T. ovatus (24). This involved placing the fish in 0.5 m3 plastic cylinders containing water at a density of 8000 tomite/L. Following a brief 10-second infection to this infectious dose, the fish were relocated to a concrete holding tank. Skin samples, including those from areas where C. irritans trophonts attached areas (TAS) and adjacent areas (NRS), were collected 48 h after infection. The samples were rapidly frozen in liquid nitrogen and stored at -80°C, with three replicates for each group. These specimens were then sent to Novogene (TJ, China) for RNA sequencing analysis.
Using RNA-seq data, we computed FPKM values for each experimental group and focused our analysis on the identified ToGals to examine their expression variations among different groups. To pinpoint ToGals that exhibited statistically significant changes in expression, we set a criterion of |log2(FoldChange)| > 1, with a Padj no greater than 0.05. We also applied the Benjamini–Hochberg correction to the P-values to reduce the risk of false positives. For effective presentation, we visualized the expression data using Chiplot (https://www.chiplot.online/).
Primers F 5’ CGGCAGCCATATGGATCTCTCAGA 3’ and R 5’ TGGTGGTGCTCGAGTTAGGGCAAG 3’, with the underlined positions indicating the restriction sites, were used to amplify the cDNA fragment ToGal-3like ORF for subcellular localization. Subsequently, the amplified ORF was cloned into the pEGFP-N3 vector, which had been digested with the restriction enzymes Xho I and BamH I, to construct pEGFP-N3-ToGal3like. For subcellular localization, the Golden pompano Muscle cell line (GPM) from our research group has been used (19). The specific steps are as follows: the GPM were revived and passaged three times to ensure stability before being cultured in a six-well plate. pEGFP-N3 and pEGFP-N3-ToGal3like were each transfected into three wells using Lipo8000 (Beyotime, Shanghai, China). Twenty-four hours later, observations were made using the inverted fluorescence microscope (Leica, DMi8, Germany).
To express the ToGal-3like protein in prokaryotic cells, the plasmid (pet28a) was ligated with the ORF of ToGal-3like transformed into Rosetta (DE3) Super Competent Cells. Once the bacterial culture reached an optical density (OD) of 0.8, IPTG was added to a final concentration of 0.1 mM, followed by incubation at 37°C with shaking for 4 hours. After centrifugation, the pellet was resuspended in 40 μL of 1×loading buffer. A 10μL aliquot was then subjected to SDS-PAGE for analysis. Once the appropriate expression was confirmed, large-scale expression was initiated. In brief, the IPTG-induced bacteria were resuspended in NTA-0 buffer, treated with lysozyme, and incubated in an ice bath for 30 minutes. The lysed bacteria were then subjected to ultrasonication, followed by centrifugation to separate the supernatant and pellet, which were further analyzed by SDS-PAGE. The remaining sample was stored at 4°C for subsequent use. For the final step of protein purification, the supernatant was passed through a Ni-NTA column (Invitrogen™), and the purified rToGal-3like was analyzed again using SDS-PAGE.
Blood was collected from rabbits, carp (freshwater fish) and golden pompano (male: female = 1:1). After centrifugation, the blood samples were washed three times using PBS and then resuspended to produce a 2% RBCs (red blood cells) solution. To this, either 100 μl of 2% RBCs solution was mixed with 250 μl/ml rToGal-8 solution or 100 μl of a solution with a final concentration of 10 mM CaCl2 and 250 μl/ml rToGal-3like. For the control group, 6 x HIS tag protein was used under the same conditions. After incubation for one hour, the samples were observed under an optical microscope. Each group was replicated three times.
In a nutshell, strains of Staphylococcus aureus, Bacillus subtilis, Vibrio vulnificus, S. agalactiae, Pseudomonas aeruginosa, and Aeromonas hydrophila were isolated and cultured on agar plates. Following the selection of individual colonies, sequencing was carried out for identification. Post-identification, they were further cultivated in LB liquid medium until they reached the logarithmic growth phase. Bacterial cells were harvested by centrifugation at 6000 × g for 5 minutes and subsequently resuspended in physiological saline. Treatment began with a starting concentration of 1mg/ml of rToGal-3like, which was serially diluted in a twofold manner. The rToGal-3like solution was then combined with the bacterial suspension and incubated at room temperature for 2 h. Following this, the Live/Dead® BacLight™ Bacterial Viability Kit (Invitrogen) was introduced to the mixture. After blending the dye with an equal volume of the bacterial and protein solution, it was kept in dark conditions for an incubation period of 15 minutes. The same concentration BSA solution served as the control group. Observations and image captures were carried out using a Leica inverted fluorescence microscope (DMI8). Each group was replicated three times.
Using homologous genes from other species and Hidden Markov Models of the CRD domains, this study identified the longest transcripts and amino acid sequences of 10 ToGals members (Table 1) through screening with BLAST and HMM searcher software. Their mRNA lengths range from 917 bp (Galectin-2b) to 5957 bp (GRPb), with the longest being GRPb and the shortest being Galectin-2b. The range of isoelectric points is from 5.00 (Galectin-3like) to 9.90 (GRPa).
Subsequently, this study tallied the copy numbers of Galectin members in Golden pompano, humans, mice, chickens, zebrafish, pufferfish, and turbot (Figure 1). Overall, from fish to mammals and birds, the number of Galectins is similar, ranging from 8 (in chickens) to 16 (in zebrafish), with humans also possessing 15 Galectins. Galectin-7 and Galectin-6 are exclusive to mammals and can be found in teleosts where GRP has three copies.
ToGals motifs and domains have been visualized (Figure 2), revealing that they all possess Gal-bind lectin domains (Figure 2B). Among these, three GRPs and two Galectins - Galectin-2 and Galectin-3 - each have one Gal-bind lectin domain. Galectin-8 and Galectin-9, however, each have two such domains. The N-terminal Gal-bind lectins correspond to Motifs 1, 2, 3, and 5, while the C-terminal Gal-bind lectins correspond to Motifs 1, 2, 3, and 4 (Figure 2A).
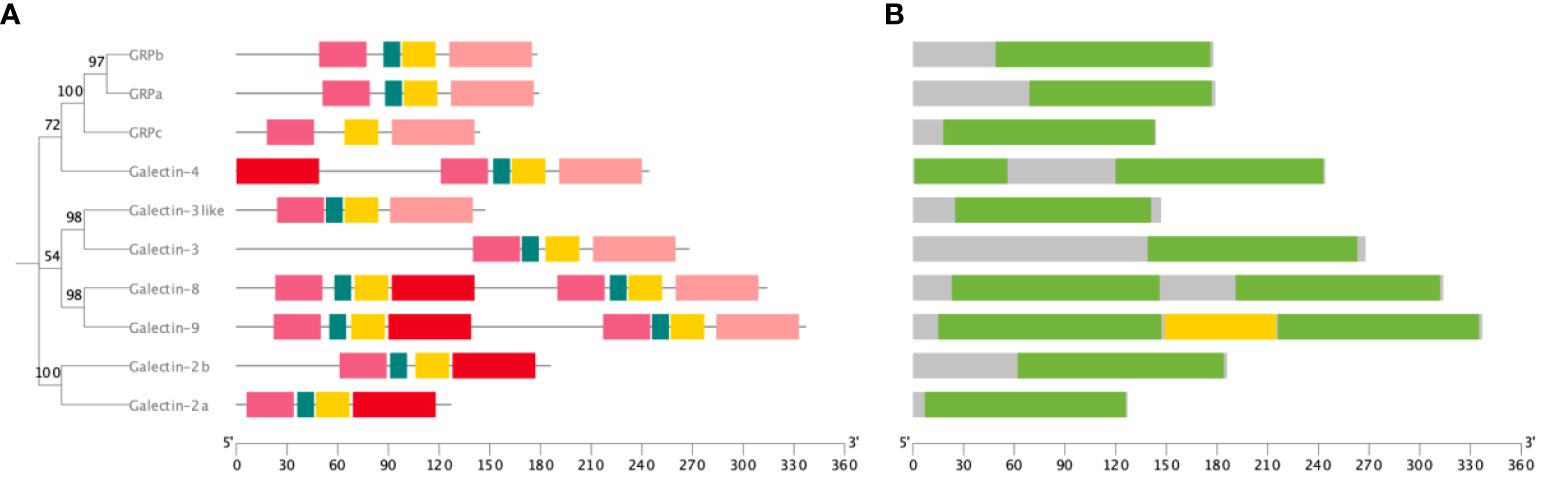
Figure 2 Motif and domain of ToGals. (A) Motif of ToGals. 5 conserved motifs are established by MEME database and represented by 5 different colors.; (B) Main domains in ToGals.
The 10 ToGals were localized on 7 chromosomes. Galectin-4 is located on LG5, and GRPb on LG6. LG9 contains both Galectin-2b and GRPb; Galectin-8 is located on LG13, and Galectin-2a on LG16. GRPc and Galectin-3like are positioned on LG18, while Galectin-8 and Galectin-3 are on LG19 (Figure 3). Subsequently, the analysis of collinearity within ToGals was conducted. Five pairs of collinear relationships were identified. It was observed that within ToGals, Galectin-2a and Galectin-2b, as well as GRPa and GRPb, exhibit collinearity. Moreover, Galectin-4, GRPb, and Galectin-2a all displayed collinearity with a region on LG22 (Figure 4).
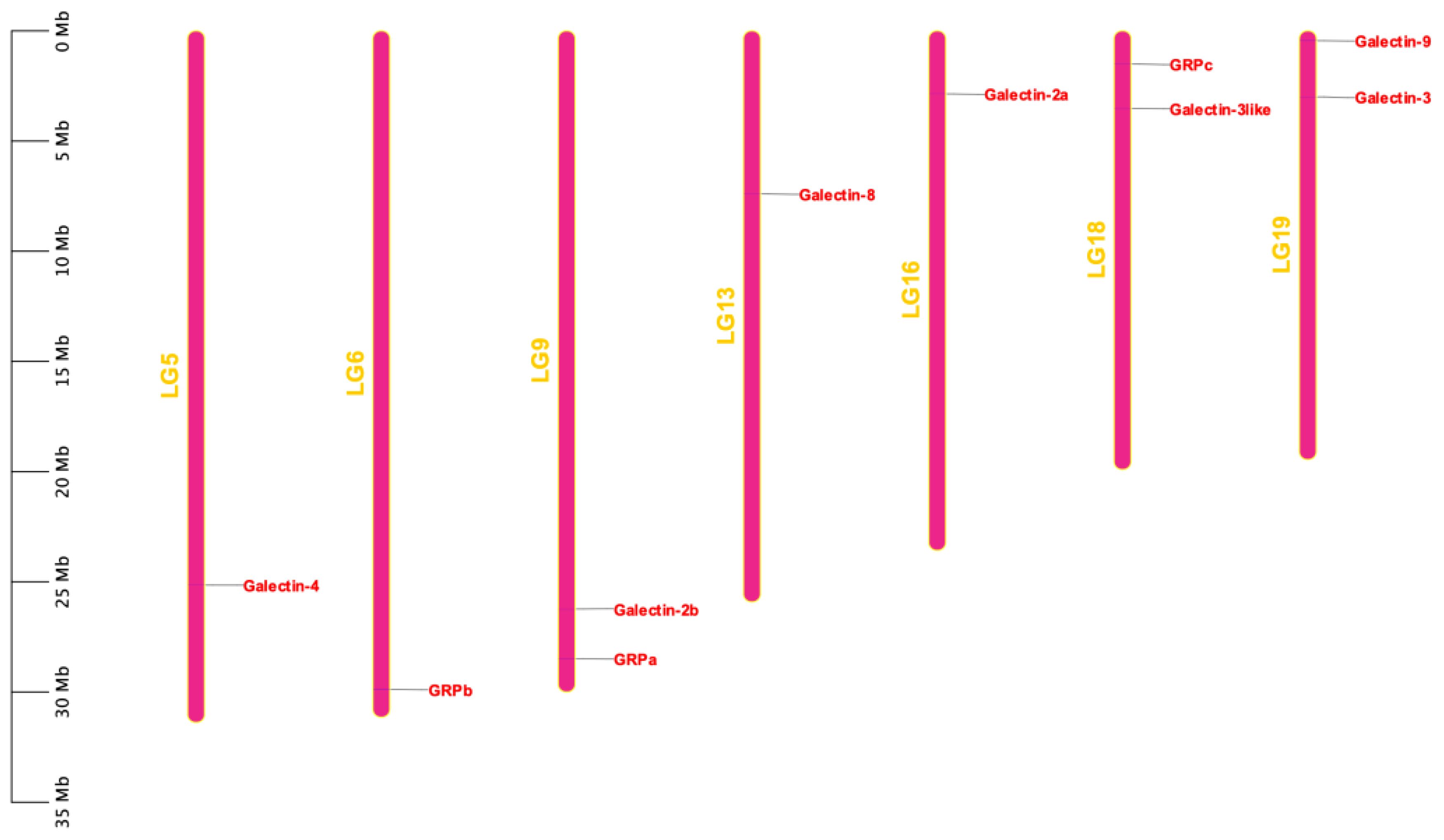
Figure 3 Chromosomal locations of the ToGals. The location of ToGals in the genome is indicated by a red line and a red label, The size of a chromosome is represented by its relative length.
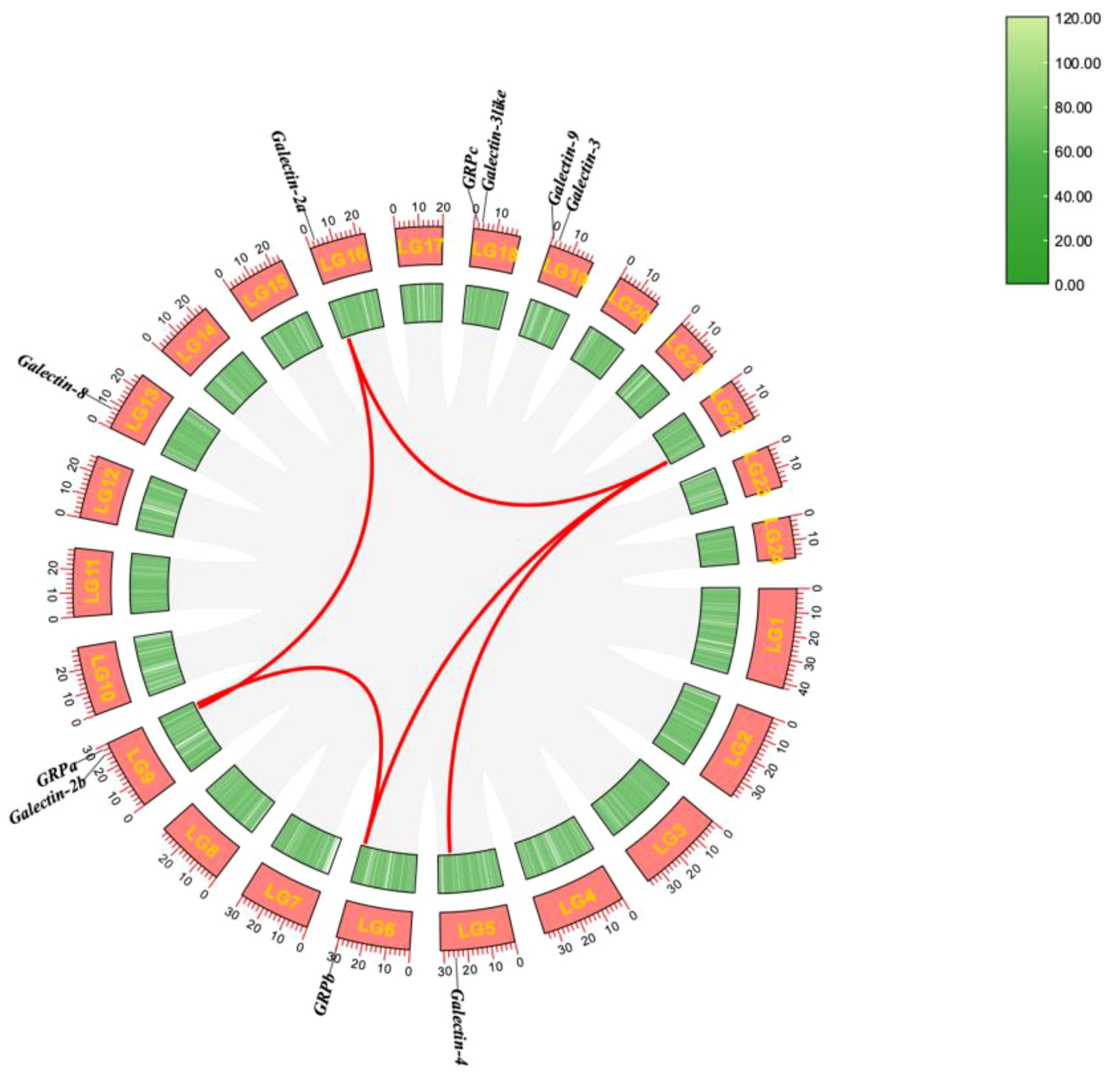
Figure 4 Synteny analysis of the ToGals genes in Golden pompano. Gray lines represent all synteny blocks in the Golden pompano genome. Tandem and segmental duplicates are exhibited with red lines.
Phylogenetic analysis was conducted using FastTree (Figure 5). Initially, ToGals grouped together with the direct orthologues from other teleost fish, while human and mouse Galectins formed another distinct clade. Subsequently, within the subfamilies of Galectins, each grouped into a separate branch. The Gal-8, Gal-9, and Gal-4 subfamilies, which are tandem repeat-type galectins, clustered together. Gal-2a and Gal-2b, which are prototypical galectins, formed another branch. Outgroups comprised GRP, GRPa, GRPb, and GRPc, each forming their own branch. This tree aligns with the traditional evolutionary relationships and the evolutionary relations among subfamilies.
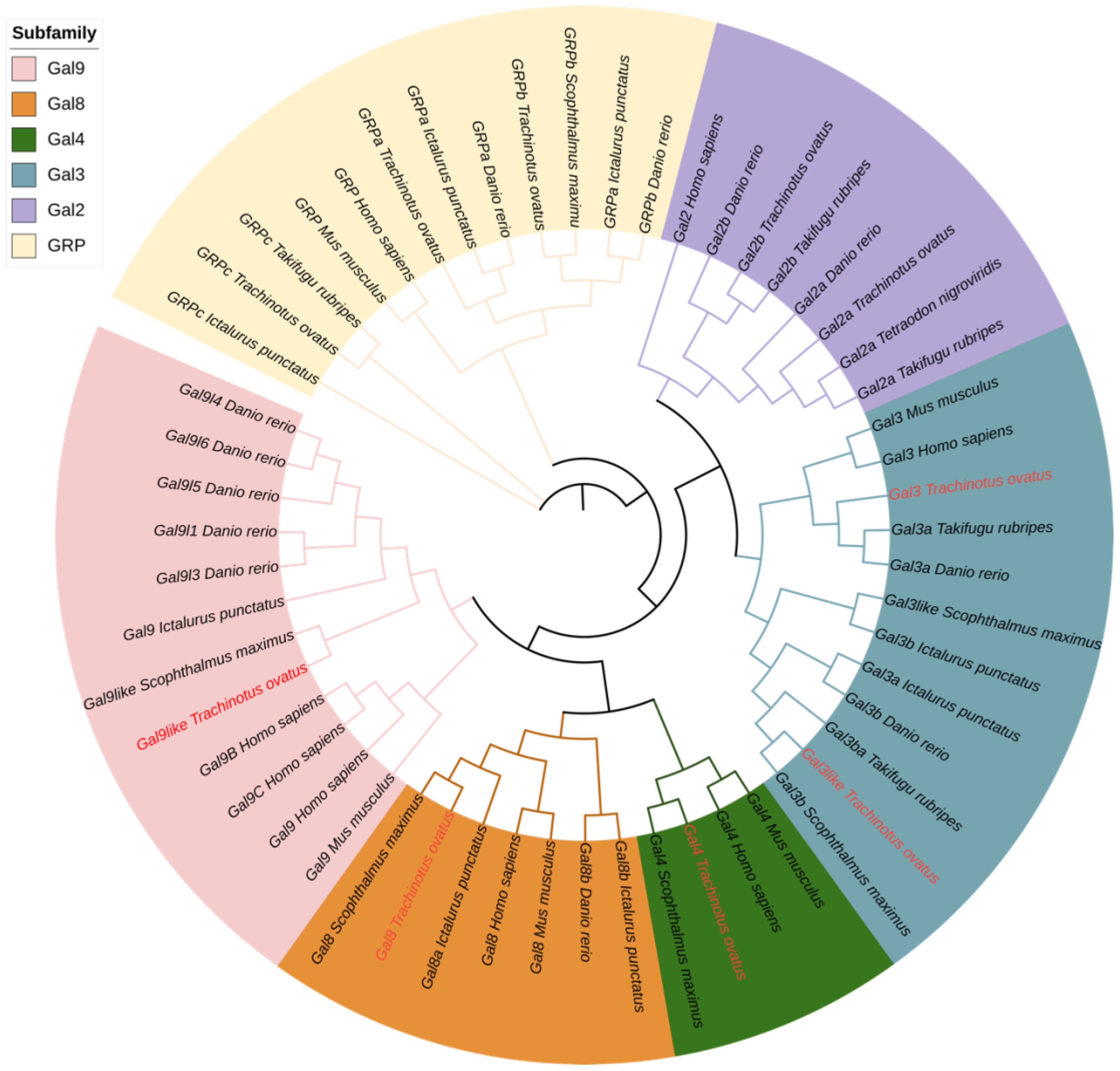
Figure 5 Phylogenetic tree analysis of Gals from different species. The phylogenetic tree was divided into six groups, each with a different color. ToGals labs were marked in red.
We analyzed the expression levels of ToGals in the spleen, liver, and kidneys following infection with S. iniae (Figure 6A). The results showed that overall, at all three time points, the expression levels of Galectin-9, Galectin-2a, and Galectin-3like were relatively high across all three tissues. Conversely, GRPc and Galectin-3 exhibited lower expression levels. Interestingly, Galectin-2b had lower expression levels in the liver and spleen, but high levels in the kidneys, with a trend of initial decrease followed by an increase from 0h to 96h. Overall, the expression levels of ToGals in the liver were lower than in the spleen and kidneys.
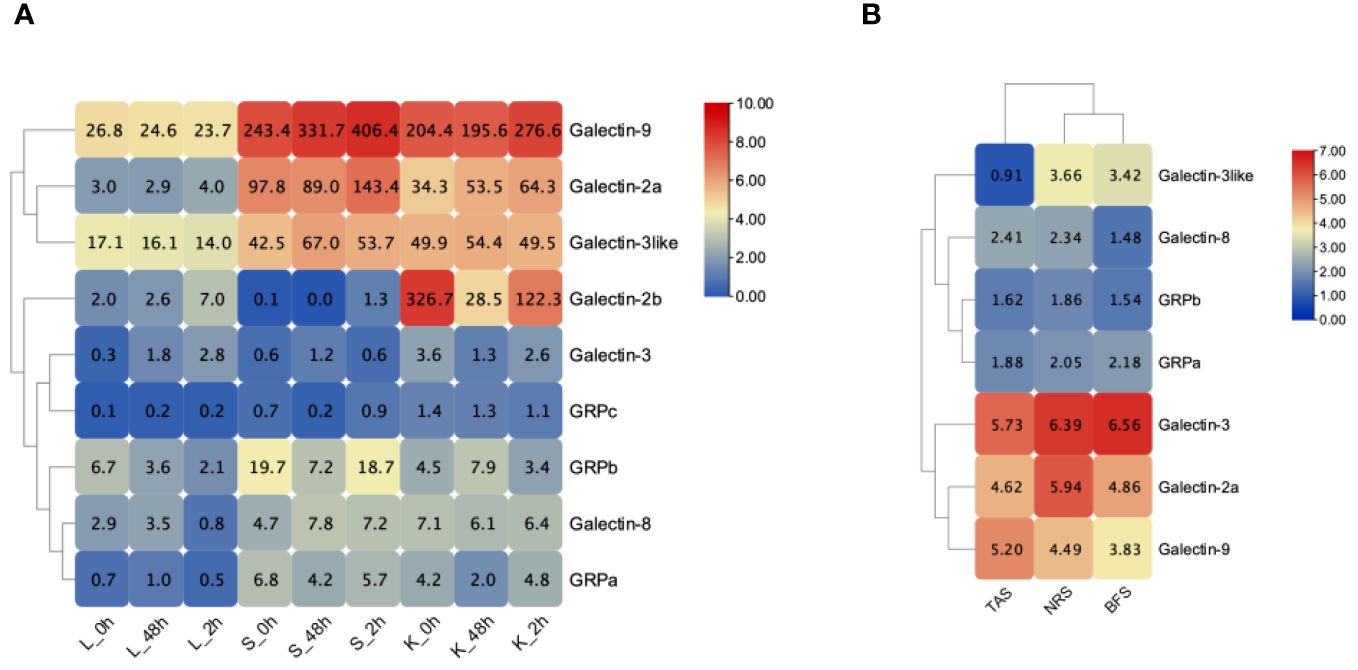
Figure 6 Expression pattern of ToGals after infection after pathogens. (A), S. agalactiae infection after 0, 48, 96 h in Liver (Li), spleen (Sp) and kidney (Ki); (B), Expression pattern of the skin before infection of C. irritans (BFS), the areas containing the trophont attached skin (TAS) and nearby region skin (NRS). The numerical representation in heat map means FPKM.
Following infection with C. irritans, ToGals displayed different expression patterns (Figure 6B). In various tissues, Galectin-3, Galectin-2a and Galectin-9 generally had higher expression levels, while GRPa was lower. They also showed varying trends; for Galectin-9 and Galectin-8, the highest expression levels were observed in TAS, whereas other members had higher expression in NRS or BFS.
To study the subcellular localization of the ToGal-3like protein, the recombinant plasmid pEGFP-N3-ToGal3like was constructed and transfected into GPM (Figure 7), after transfection and DAPI staining, all plasmids expressed green fluorescence overall, while the nuclei exhibited blue fluorescence. ToGal3like was distributed both in the cytoplasm and the nucleus.
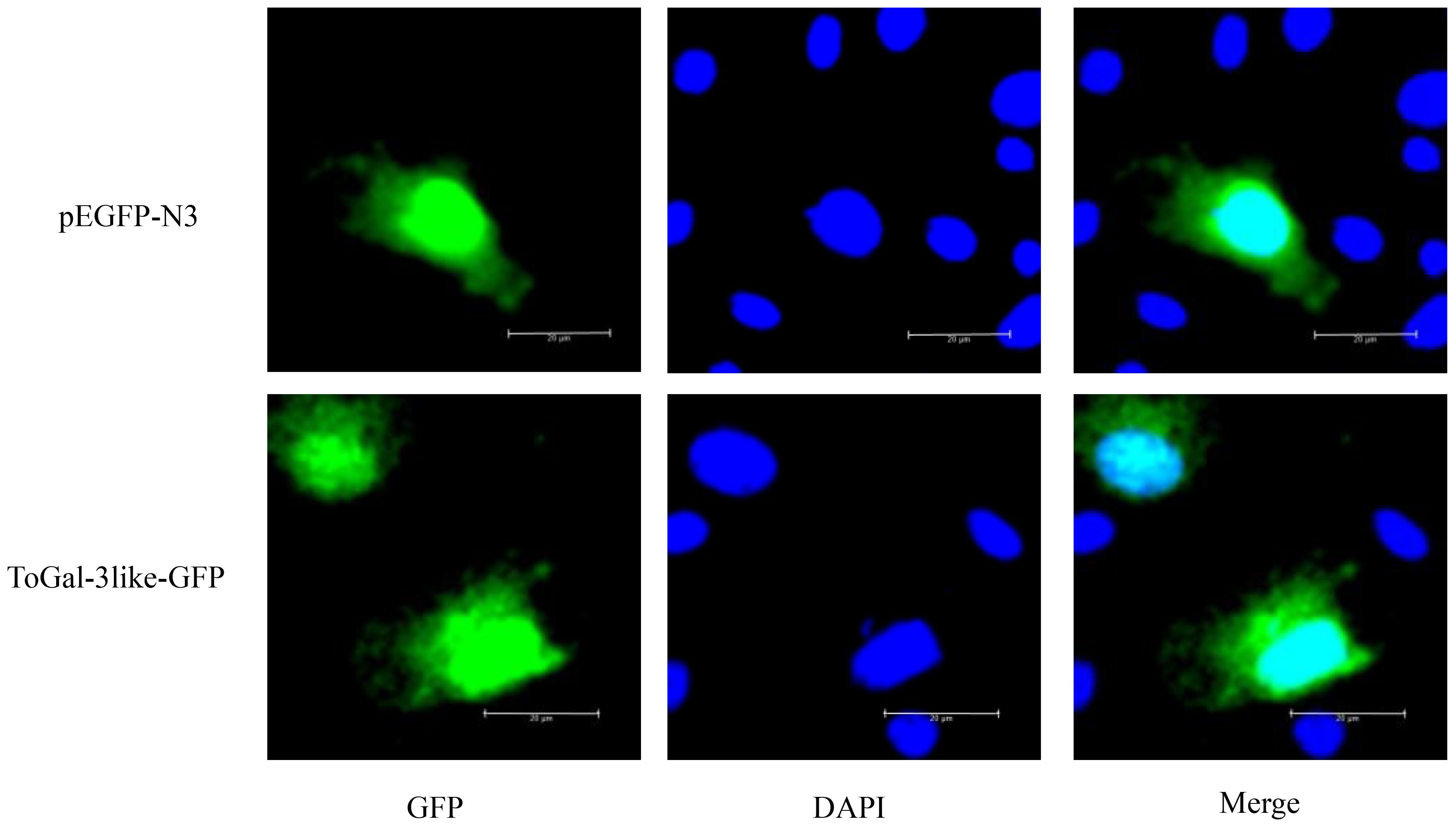
Figure 7 The subcellular localization of ToGal-3like in GPM cells. GPM cells were transfected with pEGFP-N3-ToGal3like, pEGFP-N3. After 24 h, the cells were fixed and the nuclei stained with 4, 6-diamidino-2-phenylindole (DAPI). The left green fluorescence protein panels are pEGFP-N3 and ToGal-3like fusion protein and GFP expression profile under fluorescence, the middle DAPI panels are the cell nucleus stained with DAPI, while the right panels are the combined images of pEGFP-N3, ToGal-3like fusion proteins and GFP with cell nucleus. Bar = 20 μm.
Successfully identified recombinant positive clones using antibiotic selection, where clones resistant to kanamycin were matched with ToGal-3like, confirmed by plasmid sequencing. After small-scale expression and IPTG induction, SDS-PAGE analysis showed effective expression of rToGal-3like in the E. coli system (Supplementary Figure 1A). For large-scale expression, the selected positive clones were cultured, lysed by sonication, and the resultant rToGal-3like (including His tag) displayed the expected MW of approximately 33.48 kDa on SDS-PAGE (Supplementary Figure 1B). At last, we purified a total of 500ug/ml of rToGal-3like with 90% purity using Ni-NTA affinity chromatography from the concentrated supernatant, which MW was then verified on SDS-PAGE (Supplementary Figure 1C).
The 2% RBC of rabbit, carp, and golden pompano were prepared using the centrifugation and resuspension method. The rToGal-3like was added to observe hemagglutination. The hemagglutination assay demonstrated that rToGal-3like could hemagglutination the RBC of rabbit, carp, and golden pompano, and this activity was independent of Ca2+ presence (Figure 8). In the control group, under both the presence and absence of Ca2+, there was no agglutination of RBC from any of the three species. Thus, ToGal-3like possesses agglutinating activity of RBC.
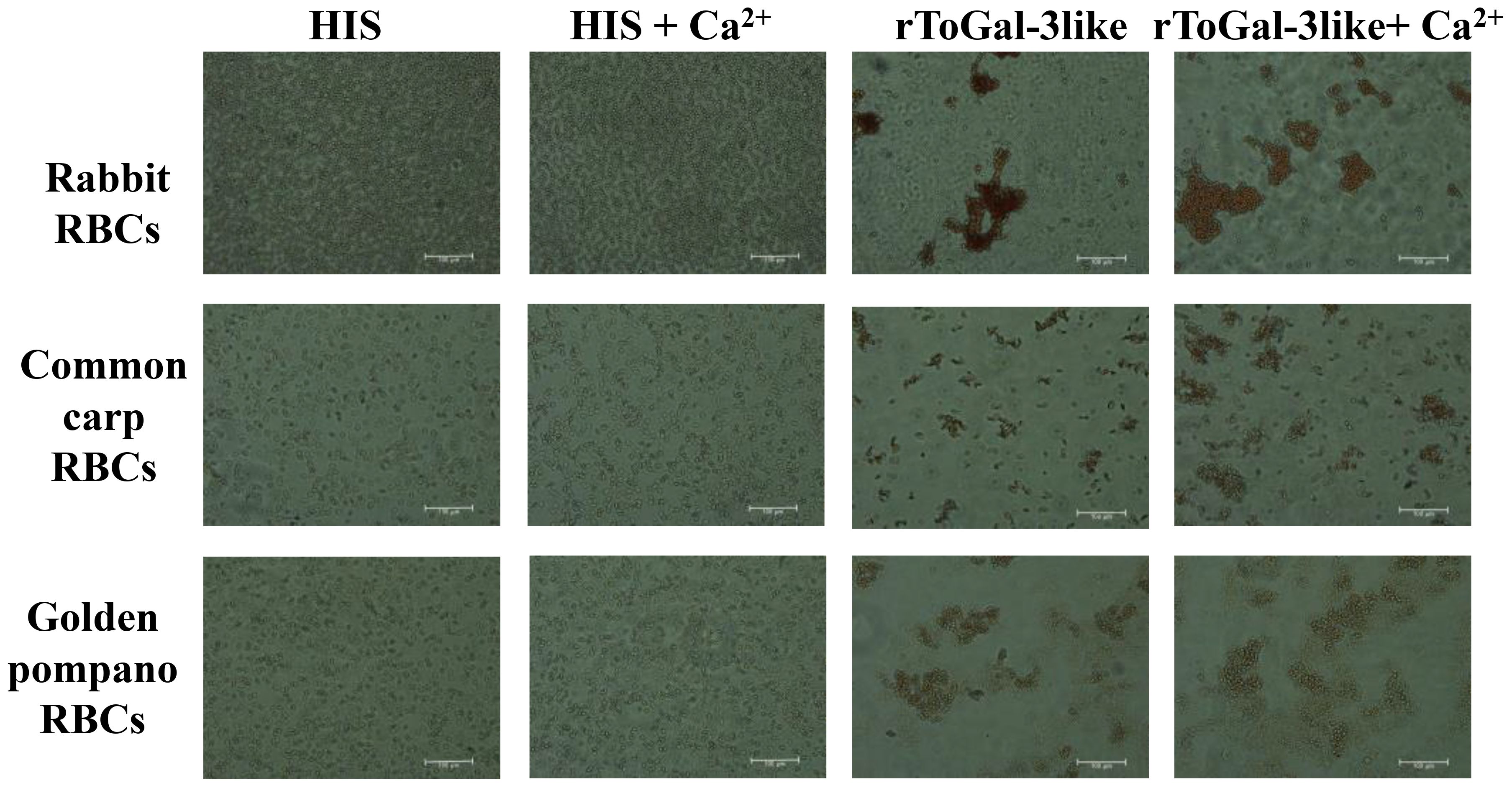
Figure 8 Hemagglutination of rToGal-3like. RBCs from rabbit, Common Carp, and Golden pompano have been agglutination by rToGal-3like. The His protein groups were as negative controls.
In this study, bacterial agglutination assays with rToGal-3like were conducted using four Gram-negative bacteria: V. vulnificus, S. agalactiae, P. aeruginosa and A. hydrophila, as well as two Gram-positive bacteria: S. aureus and B. subtilis (Figure 9). It was observed that in the control group under SYTO9 imaging, bacteria remained agglomerated, appearing as scattered green fluorescence. In contrast, in the test group treated with rToGal-3like, clear bacterial agglutination was observable under SYTO9 imaging, and most bacteria emitted red fluorescence when stained with Pi. rToGal-3like demonstrated the ability to agglutinate and kill both Gram-negative and Gram-positive bacteria.
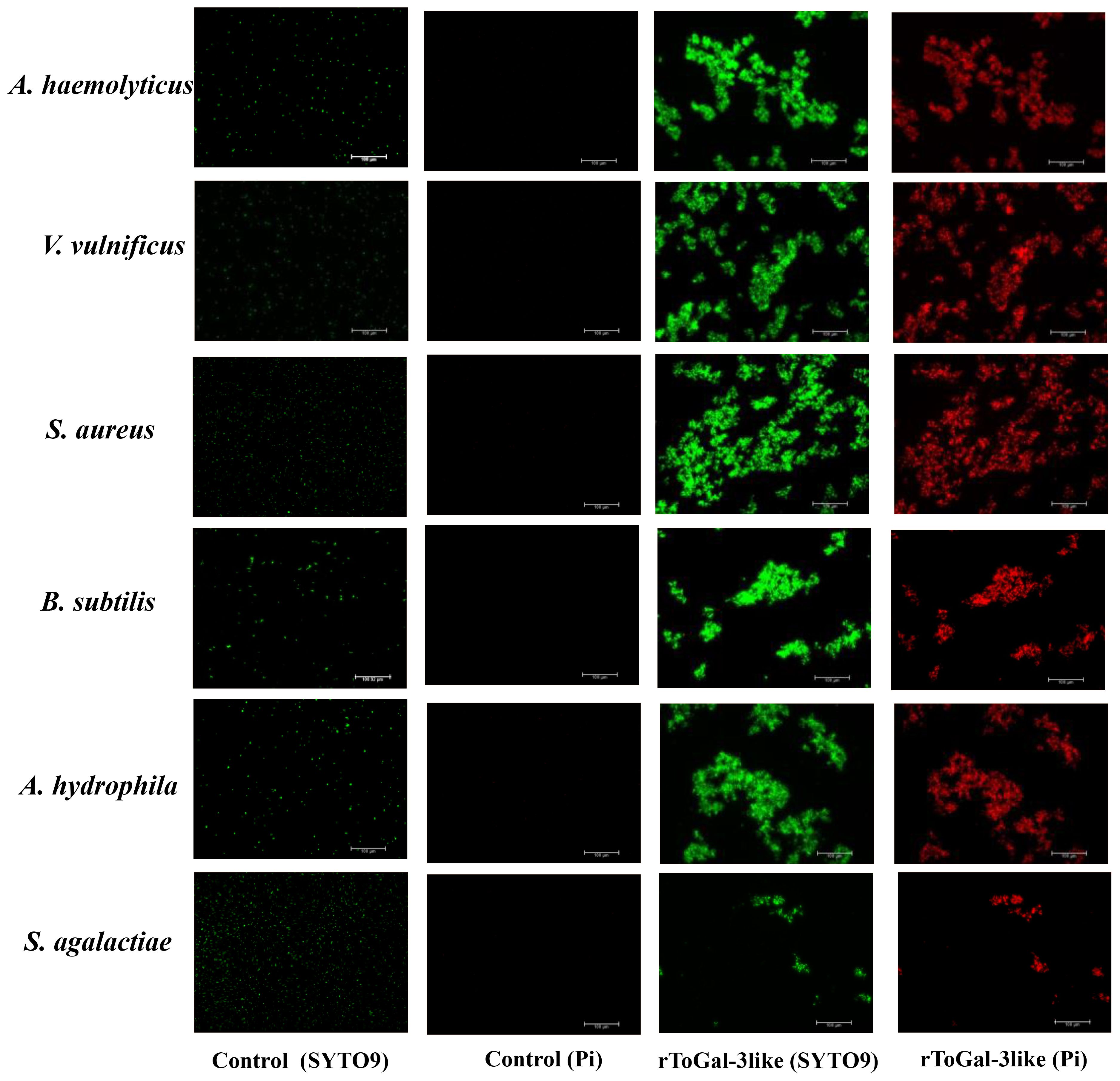
Figure 9 Bacterial agglutinations by rToGal-3like. All the bacteria stained by SYTO 9 were in green and dead bacteria stained by Pi were in red. Agglutination occurred in S. aureus, B subtilis, V. vulnificus, S. agalactiae, A haemolyticus and A hydrophila.
This study analyzed the number of Gals members in the genomes of three teleost fish—golden pompano, turbot, and zebrafish—and two mammals—humans and mice—as well as chickens. The number of ToGals was similar across species, but Gal-6 and Gal-7 were absent in teleost fish. Gal-6 has also been identified in insects such as the Egyptian mosquito, where it has been found to block the binding of Cry11Aa in the widely used agricultural insecticide BT to ALP1 (Alkaline Phosphatase 1) and APN2 (Aminopeptidase N2), thereby neutralizing the toxicity of the insecticide (25). Gal-7 was first discovered about 30 years ago in epithelial tissues involved in apoptotic responses (26). Their functions are similar to those of other Galectins, which may be the reason for their evolutionary loss (27).
The prediction of conserved domains in ToGals revealed that the tandem repeat ToGals, such as ToGal-8, ToGal-9 and ToGal-4, have two CRDs, but the conserved motifs corresponding to these homologous CRDs are not identical. They are connected by a hinge region, and the coordination between the two CRDs and the linker region is crucial for the protein’s function. According to studies by Hsu et al (28) and Levy et al. (29). the C-terminal CRD tends to bind with hydrophilic glycoproteins, while the N-terminal CRD more often associates with hydrophobic glycoproteins or glycolipids, a characteristic that appears to be common among all isotypes. This finding is also supported by the research of Ideo et al (30). Furthermore, Yoshida et al. discovered that the C-terminal and N-terminal CRDs respectively show higher binding affinity to oligosaccharides on N-glycan branches and to sugars that are 3′-O-sulfated or 3′-O-sialylated (31). These functional differences are likely a significant reason for the variations in the conserved motifs between the two CRDs.
This study also analyzed the expression patterns of ToGals following an S. agalactiae infection. Initially, ToGal-3 expression in the spleen and liver was upregulated and subsequently downregulated, mirroring the response seen in grass carp injected with GCRV and various PAMPs, where Gal-3 expression notably increased before decreasing (Zhu et al., 2020). Moreover, Gal-3’s role in antimicrobial immune responses includes pathogen recognition and regulation of monocytes/macrophages’ activity (32). Similarly, post-infection, ToGal-2a expression rose in the spleen and kidneys, a pattern also observed in Sinonovacula constricta and S.maximus after Vibrio anguillarum infection. Additionally, following the injection of rOnGal-2 in Nile tilapia, both antioxidant and antibacterial activities were enhanced (33). These observations underscore the critical role of Gals in antimicrobial processes.
Following infection with C. irritans, the various members of ToGals exhibited distinct expression patterns. Notably, although ToGal-3like showed overall lower expression levels compared to ToGal-3 at all sites, it consistently displayed lower expression in TAS compared to NRS and BFS. Gal-3 has been identified to play a role as a PRR in humans infected with Leishmania amazonensis, controlling parasite invasion, proliferation, and the formation of phagosomes (34). The absence of Gal-3 is known to enhance intracellular parasite replication in vitro, increase systemic parasitemia in vivo, and decrease the recruitment of leukocytes, with observations showing reduced secretion of pro-inflammatory cytokines in the spleens and hearts of infected Gal-3 knockout mice (35). ToGal-9 was found to have the highest expression in TAS. While there is limited research on the role of Galectin-9 in teleost fish following parasitic infections, studies have found that in mice, the expression of Tim-3/Galectin-9 is upregulated following malaria-induced acute lung injury, where Gal-9 plays a crucial immunoregulatory role by inducing apoptosis or inhibiting effector functions through binding to its receptor Tim-3 (36).
The subcellular localization results indicate that ToGal-3like is distributed in both the cytoplasm and nucleus of GPM. Numerous observations have been reported on the distribution of Gal-3 in the nucleus and cytoplasm (37, 38). The COOH-terminal end of Gal-3 (the last 28 aa) is crucial for nuclear localization, Gal-3 shuttles between the cytoplasm and nucleus based on targeting signals recognized by importins for nuclear localization and exportin-1 (CRM1) for nuclear export (39). In fact, many ligands of Gal-3 in both the cytoplasm and nucleus have been reported. For example, in the cytoplasm, Gal-3 interacts with the apoptosis inhibitor (Bcl-2), and this interaction may involve the anti-apoptotic activity of Gal-3 (40). In the nucleus, Gal-3 is an essential precursor mRNA splicing factor; the protein integrates into the spliceosome by binding to the U1 small nuclear ribonucleoprotein (snRNP) complex (41). Thus, ToGal-3 plays important roles in both the nucleus and cytoplasm.
Furthermore, Galectins also function in the extracellular matrix, where bacterial agglutination is another vital defensive action against invading pathogens. Galectins can trap pathogens in the extracellular matrix before they enter host cells, which may then facilitate the phagocytic action of macrophages (42). In this study, four Gram-negative and two Gram-positive bacteria underwent agglutination under the influence of rToGal-3like. Previous research has shown that the recombinant protein of Gal-3 in Nile tilapia can agglutinate S. agalactiae and A. hydrophila (32). Lipopolysaccharide (LPS) is the primary PAMP in Gram-negative bacteria and a major ligand for galectins. LPS consists of three main components: lipid A, core polysaccharide, and O antigen (43), with galectins primarily interacting with the β-galactoside polysaccharides in the O-antigen region. Although Gram-positive bacteria lack β-galactoside polysaccharides, Gals can bind to hydrophobic ligands in the peptidoglycan (PGN) of the cell wall (44, 45). This might explain why rToGal-3like showed agglutination/binding activity with both Gram-positive and Gram-negative bacteria in this study. Most importantly, this foreshadowed the potential function of ToGal-3like as a PRR in innate immunity.
This study systematically identified and characterized 10 ToGals genes in golden pompano. Their motifs, protein domains, and collinearity relationships were analyzed. The regulation of ToGals following infection with S. agalactiae and C. irritans indicates their involvement in immune responses against parasites and bacteria. A key member, ToGal-3like, was selected for further analysis. Subcellular localization results showed that ToGal-3like protein is expressed in both the cytoplasm and nucleus. Finally, the recombinant rToGal-3like was obtained through prokaryotic expression. rToGal-3like has the ability to agglutinate various RBCs without the need for Ca2+ and can also agglutinate various Gram-negative and Gram-positive bacteria. This study lays a scientific foundation for further research on the function of Gals in teleost, particularly highlighting the potential role of Gals as PRRs in innate immunity. However, more experimental data are needed to reveal or validate the findings of this study.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/genbank/, GCA_900607315.1.
The animal study was approved by The Committee of the South China Sea Fisheries Research Institute, Chinese Academy of Fisheries Sciences. The study was conducted in accordance with the local legislation and institutional requirements.
J-MP: Data curation, Formal analysis, Methodology, Visualization, Writing – original draft, Writing – review & editing. YL: Writing – original draft, Writing – review & editing. K-CZ: Methodology, Writing – review & editing. H-YG: Conceptualization, Writing – review & editing. B-SL: Methodology, Writing – review & editing. NZ: Conceptualization, Writing – review & editing. LX: Software, Writing – review & editing. T-FZ: Software, Writing – review & editing. D-CZ: Conceptualization, Writing – original draft.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This study was supported by the National Key Research and Development Program of China (2022YFD2400505), the National Natural Science Foundation of China (U20A2064), Seed Industry Revitalization Project of Special Fund for Rural Revitalization Strategy in Guangdong Province (2022SPY02002).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2024.1452609/full#supplementary-material
1. Smith NC, Rise ML, Christian SL. A comparison of the innate and adaptive immune systems in cartilaginous fish, ray-finned fish, and lobe-finned fish. Front Immunol. (2019) 10:2292. doi: 10.3389/fimmu.2019.02292
2. Salinas I. The mucosal immune system of teleost fish. Biol (Basel). (2015) 4:525–39. doi: 10.3390/biology4030525
3. Liao Z, Su J. Progresses on three pattern recognition receptor families (TLRs, RLRs and NLRs) in teleost. Dev Comp Immunol. (2021) 122:104131. doi: 10.1016/j.dci.2021.104131
4. Drickamer K. Two distinct classes of carbohydrate-recognition domains in animal lectins. J Biol Chem. (1988) 263:9557–60. doi: 10.1016/S0021-9258(19)81549-1
5. Zhang D, Jiang S, Hu Y, Cui S, Guo H, Wu K, et al. A multidomain galectin involved in innate immune response of pearl oyster Pinctada fucata. Dev Comp Immunol. (2011) 35:1–6. doi: 10.1016/j.dci.2010.08.007
6. Tian M, Xu D, Fu Q, Zhang L, Yang N, Xue T, et al. Galectins in turbot (Scophthalmus maximus L.): Characterization and expression profiling in mucosal tissues. Fish Shellfish Immunol. (2021) 109:71–81. doi: 10.1016/j.fsi.2020.12.004
7. Pan J-M, Liu M-J, Guo H-Y, Zhu K-C, Liu B-S, Zhang N, et al. Early development and allometric growth patterns of Trachinotus ovatus (Linnaeus, 1758). Aquaculture. (2023) 575:739804. doi: 10.1016/j.aquaculture.2023.739804
8. Wu M, Zhu K-C, Guo H-Y, Guo L, Liu B, Jiang S-G, et al. Characterization, expression and function analysis of the TLR3 gene in golden pompano (Trachinotus ovatus). Dev Comp Immunol. (2021) 117:103977. doi: 10.1016/j.dci.2020.103977
9. Wu M, Guo L, Zhu K-C, Guo H-Y, Liu B-S, Zhang N, et al. Molecular characterization of toll-like receptor 14 from golden pompano Trachinotus ovatus (Linnaeus, 1758) and its expression response to three types of pathogen-associated molecular patterns. Comp Biochem Physiol B-Biochemistry Mol Biol. (2019) 232:1–10. doi: 10.1016/j.cbpb.2019.02.010
10. Pan J-M, Liang Y, Zhu K-C, Guo H-Y, Liu B-S, Zhang N, et al. Identification of the NOD-like receptor family of golden pompano and expression in response to bacterial and parasitic exposure reveal its key role in innate immunity. Dev Comp Immunol. (2024) 152:105123. doi: 10.1016/j.dci.2023.105123
11. Liang Y, Pan J-M, Zhu K-C, Xian L, Guo H-Y, Liu B-S, et al. Genome-Wide Identification of Trachinotus ovatus Antimicrobial Peptides and Their Immune Response against Two Pathogen Challenges. Mar Drugs. (2023) 21:505. doi: 10.3390/md21100505
12. Liu B, Liu G-D, Guo H-Y, Zhu K-C, Guo L, Zhang N, et al. Characterization and functional analysis of liver-expressed antimicrobial peptide-2 (LEAP-2) from golden pompano Trachinotus ovatus (Linnaeus 1758). Fish Shellfish Immunol. (2020) 104:419–30. doi: 10.1016/j.fsi.2020.06.029
13. Zhu Z-X, Yao Y-Y, Ai C-H, Yang G, Liang X-Y, Liu T-D, et al. Dietary supplementation of Patchouli oil increases the resistance against Streptococcus agalactiae infection in GIFT tilapia as revealed by transcriptome and 16s amplicon profiling. Aquaculture Rep. (2023) 33:101754. doi: 10.1016/j.aqrep.2023.101754
14. Gao J, Liu M, Guo H, Zhu K, Liu B, Liu B, et al. ROS Induced by Streptococcus agalactiae Activate Inflammatory Responses via the TNF-α/NF-κB Signaling Pathway in Golden Pompano Trachinotus ovatus (Linnaeus, 1758). Antioxidants. (2022) 11:1809. doi: 10.3390/antiox11091809
15. Cheng J, Liu P, Yang Y, Liu Y, Xia Y. Functional role of TrIL-1β in Takifugu rubripes defense against Cryptocaryon irritans infection. Int J Biol Macromolecules. (2024) 269:132167. doi: 10.1016/j.ijbiomac.2024.132167
16. Zhu K-C, Liu J, Liu B-S, Guo H-Y, Zhang N, Guo L, et al. Functional characterization of four ToRac genes and their association with anti-parasite traits in Trachinotus ovatus (Linnaeus, 1758). Aquaculture. (2022) 560:738514. doi: 10.1016/j.aquaculture.2022.738514
17. Chen C, Chen H, Zhang Y, Thomas HR, Frank MH, He Y, et al. TBtools: an integrative toolkit developed for interactive analyses of big biological data. Mol Plant. (2020) 13:1194–202. doi: 10.1016/j.molp.2020.06.009
18. Zhang D-C, Guo L, Guo H-Y, Zhu K-C, Li S-Q, Zhang Y, et al. Chromosome-level genome assembly of golden pompano (Trachinotus ovatus) in the family Carangidae. Sci Data. (2019) 6:216. doi: 10.1038/s41597-019-0238-8
19. Bailey TL, Elkan C. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc Int Conf Intell Syst Mol Biol. (1994) 2:28–36.
20. Wang Y, Tang H, DeBarry JD, Tan X, Li J, Wang X, et al. MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. (2012) 40:e49. doi: 10.1093/nar/gkr1293
21. Krzywinski M, Schein J, Birol İ, Connors J, Gascoyne R, Horsman D, et al. Circos: An information aesthetic for comparative genomics. Genome Res. (2009) 19:1639–45. doi: 10.1101/gr.092759.109
22. Gao J, Guo H-Y, Liu M-J, Zhu K-C, Liu B, Liu B-S, et al. Transcriptome Analysis of the Immune Process of Golden Pompano (Trachinotus ovatus) Infected with Streptococcus agalactiae. Fishes. (2023) 8:52. doi: 10.3390/fishes8010052
23. Dan XM, Li AX, Lin XT, Teng N, Zhu XQ. A standardized method to propagate Cryptocaryon irritans on a susceptible host pompano Trachinotus ovatus. Aquaculture. (2006) 258:127–33. doi: 10.1016/j.aquaculture.2006.04.026
24. Liu B, San L, Guo H, Zhu K, Zhang N, Yang J, et al. Transcriptomic Analysis Reveals Functional Interaction of mRNA-lncRNA-miRNA in Trachinotus ovatus Infected by Cryptocaryon irritans. Int J Mol Sci. (2023) 24:15886. doi: 10.3390/ijms242115886
25. Hu X, Chen H, Xu J, Zhao G, Huang X, Liu J, et al. Function of Aedes aEgypti galectin-6 in modulation of Cry11Aa toxicity. Pesticide Biochem Physiol. (2020) 162:96–104. doi: 10.1016/j.pestbp.2019.09.010
26. Madsen P, Rasmussen HH, Flint T, Gromov P, Kruse TA, Honoré B, et al. Cloning, expression, and chromosome mapping of human galectin-7. J Biol Chem. (1995) 270:5823–9. doi: 10.1074/jbc.270.11.5823
27. Albalat R, Cañestro C. Evolution by gene loss. Nat Rev Genet. (2016) 17:379–91. doi: 10.1038/nrg.2016.39
28. Hsu DK, Yang R, Liu F. Galectins in Apoptosis. In: Methods in Enzymology. Boston, MA, USA: Academic Press (2006). p. 256–73. doi: 10.1016/S0076-6879(06)17018-4
29. Levy Y, Auslender S, Eisenstein M, Vidavski RR, Ronen D, Bershadsky AD, et al. It depends on the hinge: a structure-functional analysis of galectin-8, a tandem-repeat type lectin. Glycobiology. (2006) 16:463–76. doi: 10.1093/glycob/cwj097
30. Ideo H, Seko A, Ishizuka I, Yamashita K. The N-terminal carbohydrate recognition domain of galectin-8 recognizes specific glycosphingolipids with high affinity. Glycobiology. (2003) 13:713–23. doi: 10.1093/glycob/cwg094
31. Yoshida H, Yamashita S, Teraoka M, Itoh A, Nakakita S, Nishi N, et al. X-ray structure of a protease-resistant mutant form of human galectin-8 with two carbohydrate recognition domains. FEBS J. (2012) 279:3937–51. doi: 10.1111/j.1742-4658.2012.08753.x
32. Niu J, Huang Y, Liu X, Luo G, Tang J, Wang B, et al. Functional characterization of galectin-3 from Nile tilapia (Oreochromis niloticus) and its regulatory role on monocytes/macrophages. Fish Shellfish Immunol. (2019) 95:268–76. doi: 10.1016/j.fsi.2019.10.043
33. Niu J, Liu X, Zhang Z, Huang Y, Tang J, Wang B, et al. The in vivo roles of galectin-2 from Nile tilapia (Oreochromis niloticus) in immune response against bacterial infection. Fish Shellfish Immunol. (2020) 106:473–9. doi: 10.1016/j.fsi.2020.08.011
34. Oliveira RM, Teixeira TL, Rodrigues CC, da Silva AA, Borges BC, Brígido RTS, et al. Galectin-3 plays a protective role in Leishmania (Leishmania) amazonensis infection. Glycobiology. (2021) 31:1378–89. doi: 10.1093/glycob/cwab062
35. da Silva AA, Teixeira TL, Teixeira SC, MaChado FC, Dos Santos MA, Tomiosso TC, et al. Galectin-3: A Friend but Not a Foe during Trypanosoma cruzi Experimental Infection. Front Cell Infect Microbiol. (2017) 7:463. doi: 10.3389/fcimb.2017.00463
36. Liu J, Xiao S, Huang S, Pei F, Lu F. Upregulated Tim-3/galectin-9 expressions in acute lung injury in a murine malarial model. Parasitol Res. (2016) 115:587–95. doi: 10.1007/s00436-015-4775-6
37. Zhu D, Huang R, Chu P, Chen L, Li Y, He L, et al. Characterization and expression of galectin-3 in grass carp (Ctenopharyngodon idella). Dev Comp Immunol. (2020) 104:103567. doi: 10.1016/j.dci.2019.103567
38. Joo H-G, Goedegebuure PS, Sadanaga N, Nagoshi M, von Bernstorff W, Eberlein TJ. Expression and function of galectin-3, a β-galactoside-binding protein in activated T lymphocytes. J Leukocyte Biol. (2001) 69:555–64. doi: 10.1189/jlb.69.4.555
39. Haudek KC, Spronk KJ, Voss PG, Patterson RJ, Wang JL, Arnoys EJ. Dynamics of galectin-3 in the nucleus and cytoplasm. Biochim Biophys Acta (BBA) - Gen Subj. (2010) 1800:181–9. doi: 10.1016/j.bbagen.2009.07.005
40. Yang RY, Hsu DK, Liu FT. Expression of galectin-3 modulates T-cell growth and apoptosis. Proc Natl Acad Sci. (1996) 93:6737–42. doi: 10.1073/pnas.93.13.6737
41. Haudek KC, Voss PG, Locascio LE, Wang JL, Patterson RJ. A Mechanism for Incorporation of Galectin-3 into the Spliceosome through Its Association with U1 snRNP. Biochemistry. (2009) 48:7705–12. doi: 10.1021/bi900071b
42. Hou F, Liu Y, He S, Wang X, Mao A, Liu Z, et al. A galectin from shrimp Litopenaeus vannamei is involved in immune recognition and bacteria phagocytosis. Fish Shellfish Immunol. (2015) 44:584–91. doi: 10.1016/j.fsi.2015.03.017
43. Reyes RE, González CR, Jiménez RC, Herrera MO, Andrade AA. Mechanisms of O-Antigen Structural Variation of Bacterial Lipopolysaccharide (LPS). In: The Complex World of Polysaccharides. London, UK: IntechOpen (2012). doi: 10.5772/48147
44. Mey A, Leffler H, Hmama Z, Normier G, Revillard JP. The animal lectin galectin-3 interacts with bacterial lipopolysaccharides via two independent sites. J Immunol. (1996) 156:1572–7. doi: 10.4049/jimmunol.156.4.1572
Keywords: Galectins, Golden pompano, subcellular localization, protein function, bacterial agglutination
Citation: Pan J-M, Liang Y, Zhu K-C, Guo H-Y, Liu B-S, Zhang N, Xian L, Zhu T-F and Zhang D-C (2024) Genome-wide characterization, phylogenetic and expression analysis of Galectin gene family in Golden pompano Trachinotus ovatus. Front. Immunol. 15:1452609. doi: 10.3389/fimmu.2024.1452609
Received: 21 June 2024; Accepted: 08 July 2024;
Published: 18 July 2024.
Edited by:
Linwei Yang, Fujian Agriculture and Forestry University, ChinaReviewed by:
Min Yang, South China Agricultural University, ChinaCopyright © 2024 Pan, Liang, Zhu, Guo, Liu, Zhang, Xian, Zhu and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Dian-Chang Zhang, emhhbmdkY2hAc2NzZnJpLmFjLmNu
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.