
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Immunol., 25 November 2024
Sec. Vaccines and Molecular Therapeutics
Volume 15 - 2024 | https://doi.org/10.3389/fimmu.2024.1448489
Cancer continues to pose an alarming threat to global health, necessitating the need for the development of efficient therapeutic solutions despite massive advances in the treatment. mRNA cancer vaccines have emerged as a hopeful avenue, propelled by the victory of mRNA technology in COVID-19 vaccines. The article delves into the intricate mechanisms and formulations of cancer vaccines, highlighting the ongoing efforts to strengthen mRNA stability and ensure successful translation inside target cells. Moreover, it discusses the design and mechanism of action of mRNA, showcasing its potential as a useful benchmark for developing efficacious cancer vaccines. The significance of mRNA therapy and selecting appropriate tumor antigens for the personalized development of mRNA vaccines are emphasized, providing insights into the immune mechanism. Additionally, the review explores the integration of mRNA vaccines with other immunotherapies and the utilization of progressive delivery platforms, such as lipid nanoparticles, to improve immune responses and address challenges related to immune evasion and tumor heterogeneity. While underscoring the advantages of mRNA vaccines, the review also addresses the challenges associated with the susceptibility of RNA to degradation and the difficulty in identifying optimum tumor-specific antigens, along with the potential solutions. Furthermore, it provides a comprehensive overview of the ongoing research efforts aimed at addressing these hurdles and enhancing the effectiveness of mRNA-based cancer vaccines. Overall, this review is a focused and inclusive impression of the present state of mRNA cancer vaccines, outlining their possibilities, challenges, and future predictions in the fight against cancer, ultimately aiding in the development of more targeted therapies against cancer.
Cancer remains a significant global health challenge, with estimates projecting 34 million new cases by 2070 (1). Despite advancements in treatment, current therapies exhibit limitations, emphasizing the need for innovative solutions for cancer treatment. (2). The concept of mRNA cancer vaccines has emerged as a promising avenue for immunotherapy, following the success of mRNA technology in producing effective COVID-19 vaccines during the pandemic. (3–6). Immunotherapy, such as immune checkpoint inhibitors and CAR-T therapies, have revolutionized cancer treatment by harnessing the power of the immune system. (7). Recently, these approaches, combined with mRNA vaccines, offer a personalized and effective strategy against cancer, providing a more targeted approach (8, 9).
The mRNA vaccines offer a novel prophylactic strategy, delivering genetic instructions directly to the cells for inducing precise protein production and triggering robust immune responses to actively combat cancer (10). A typical mRNA vaccine harbors synthetic mRNA molecules designed to encode cancer-specific antigens (11). In this regard, various optimization strategies are employed to ensure efficient translation of mRNA vaccines within human cells (12). The advent of lipid nanoparticles, frequently employed as delivery vehicles to encapsulate mRNA to prevent it from degradation and facilitate cellular uptake, has significantly revolutionized the field of mRNA vaccines (13). Once inside the cells, the mRNA gets translated into the antigen protein by utilizing the cellular translation machinery (14). Consequently, the antigen protein gets processed by the antigen-presenting cells and presented to the immune cells for eliciting a robust innate and adaptive immune response (15). The successful development of the first mRNA cancer vaccine in 1995, which encoded the carcinoembryonic antigen in mice, marked a significant milestone, which prompted scientists to critically explore the potential of immunotherapy against cancer (16). The mRNA vaccines hold a distinctive advantage over conventional virus-based vaccines, owing to their enhanced safety, cost-effectiveness, purity, dismal vaccine resistance, and integration concerns (17). Moreover, the mRNA vaccines encoding full-length tumor antigens can generate broad-spectrum T-cell-mediated immune responses regardless of the Human Leucocyte Antigen (HLA) types (18, 19), hence elevating their therapeutic potential (20). Furthermore, multiple strategies, including mRNA encapsulation in the immune cell-specific nanoparticles and self-amplifying mRNA (saRNA) designs, aim to improve vaccine efficacy and minimize the potential side effects (21, 22). Additionally, novel approaches involving the incorporation of mRNAs encoding multiple antigens in the vaccine are substantially explored to broaden the protection against various diseases (23). The remarkable advancements in mRNA vaccine technology hold great promise for addressing health concerns through the practical utility of these vaccines in nearby future.
The current review aims to emphasize the significance of mRNA vaccines as an innovative immunotherapeutic approach against cancer. It will address the intricate mechanisms and formulations of these groundbreaking vaccines, highlighting their effectiveness, mechanism of immune system activation, and future safety considerations. As the field of mRNA vaccines continues to expand, the upcoming research strives to optimize these vaccines by improving the efficacy, minimizing the side effects, addressing the inherent susceptibility of mRNA to enzymatic degradation, and ensuring successful mRNA translation in the target cells. Consequently, these attempts will remarkably elevate the applicability of mRNA vaccines against diverse cancers.
Recently, mRNA, a molecule central to cellular protein translation, has emerged as a notable platform in remarkably revolutionizing cancer immunotherapy. Compared to DNA-based vaccines, mRNA vaccines offer marked benefits making them a promising therapeutic choice (24). In this regard, mRNA vaccines harbor a potentially enhanced therapeutic effectiveness due to their equivalent tendency of translation in the dividing and non-dividing cells. Moreover, unlike DNA-based vaccines, mRNA vaccines evade the requirement of integration into the host genome, and showcase their effects via cytosolic translation of Tumor-Associated Antigens (TAA), hence minimizing the subsequent cellular damage. Consequently, mRNA appears to be an exceptionally competent candidate for vaccine development. Despite abundant advantages, the challenges regarding the mode of delivery, stability, and specificity of mRNA vaccines persist. Therefore, comprehensive research is crucial to address these challenges and fully harness the potential of this impressive technology for targeting cancer. Table 1 illustrates an extensive review of the historical aspects encompassing the advent and advancement of mRNA vaccine technology for cancer.
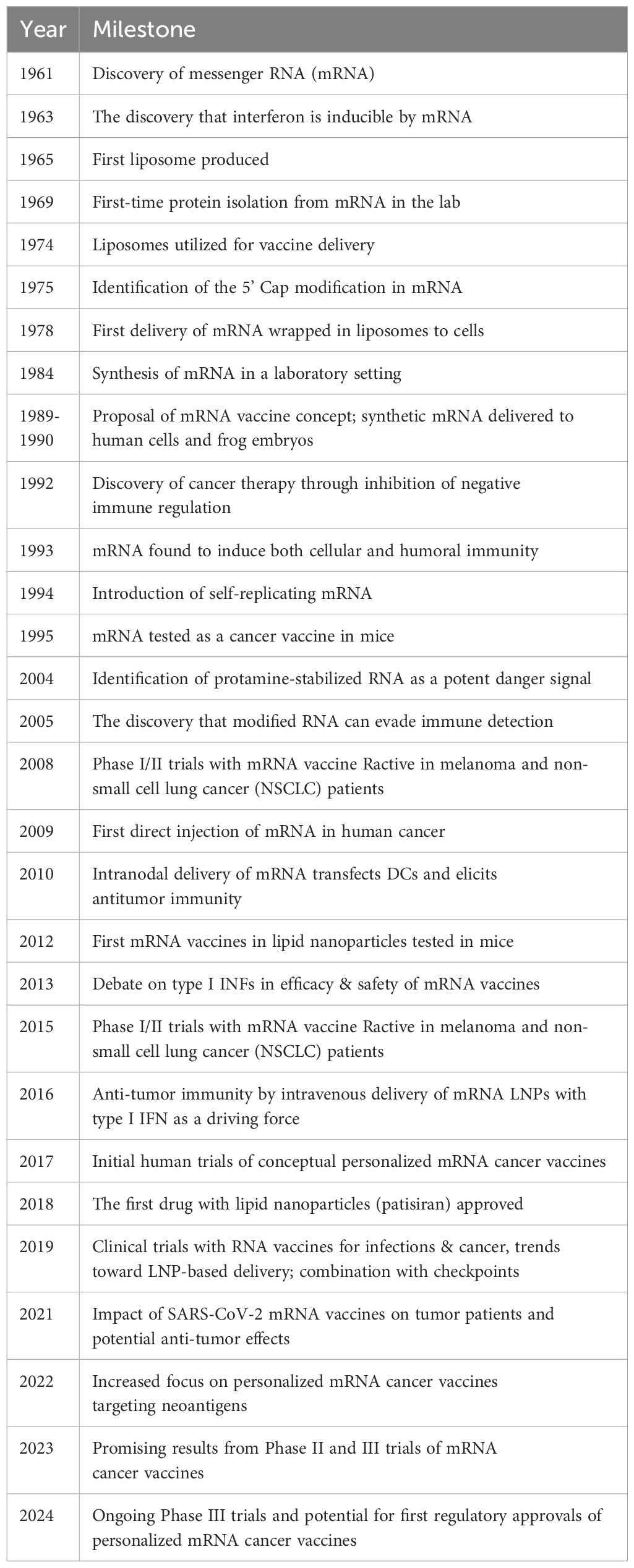
Table 1. Timeline of the advent and advancements of the mRNA vaccine technology adapted by Li et al. (25).
For designing an mRNA vaccine, the foremost step is the synthetic design of the mRNA. In this regard, a typical mRNA design includes an open reading frame (ORF) encoding the antigen sequence, flanked by 5’ and 3’ untranslated regions (UTRs) accompanying certain artificial modifications to promote efficacy and cellular uptake (26). Additionally, a sophisticated innovation in this technology employs the self-amplifying mRNA (SAM), enabling sustained mRNA augmentation within the host cells. This consequently ensures enhanced production of desired protein aided via the cellular ribosomal machinery. Eventually, the residual mRNA templates are subjected to degradation, hence reducing the potential risk of metabolite toxicity (21). Nonetheless, it is imperative to critically evaluate the potential safety concerns associated with the modifications executed in the vaccine design. Hence, extended research is crucial to ensure the safety profile of the advanced mRNA vaccine platforms.
Careful identification and optimization of specific mRNA sequences of target antigens is essential for developing potent mRNA vaccines for cancer. Possible targeted antigens for the development of mRNA vaccine are shown in Table 2. In this context, precise sequence designing, followed by efficient synthesis and strategic choice to targeted delivery, is paramount. To address this, multiple strategies are devised for the selection of the target antigen. This mostly involves the utilization of full-length cancer-specific mutant proteins or neoantigens as target TAAs for designing mRNA sequences (71). However, another approach involves a multi-epitope strategy, where immunogenic peptides from multiple TAAs are encoded within the mRNA molecule to stimulate a wide range of immune responses (72). Furthermore, the recent progression in personalized medicine has unlocked new avenues for crafting personalized mRNA vaccines aimed at engineering personalized mRNA sequences to integrate patient-specific mutations in the neoantigens (73). Following the meticulous design of the mRNA sequence, specific nucleotide modifications, such as uridine substitution and the addition of pseudouridine, are introduced to achieve maximal stability, translation efficiency, and precise immunogenicity (26). Additionally, codon optimization is performed using algorithms to prioritize codons readily translated by human cells, thereby maximizing protein expression (26).
The in vitro transcription (IVT) is a widely applied procedure for synthesizing the mRNA. It utilizes specialized enzymes and optimized reaction conditions for producing large quantities of designed mRNA sequences (74). Additionally, enzymatic amplification, such as rolling circle amplification, is frequently administered for potentially higher mRNA yields (75). The optimization of the carrier vehicles is equally crucial for ensuring the targeted delivery of the vaccine in the human system. In this regard, lipid nanoparticles (LNPs) are usually employed for encapsulation of mRNA to ensure protection, stability, and targeted delivery (76). Moreover, LNPs are carefully designed based on the tumor-specific ligands for efficient delivery and reduced systemic harmful effects. Consequently, a reliable and optimized mRNA can be crafted to induce a robust immune response against cancer. Although, multi-epitope strategy and personalized vaccines offer promising therapeutic avenues against cancer, a careful investigation is crucial to avoid potential immunodominance or off-target effects.
Chemical modifications involve the introduction of chemically modified bases in the mRNA sequence. In this regard, most frequent chemical modifications involve the addition of pseudouridine (Ψ), N1-methyl pseudouridine (m1Ψ), 5-methylcytidine (m5C), N6-methyladenosine (m6A), 2-thiouridine (s2U), and 5-methyluridine (m5U). These modifications render potentially elevated efficiency, stability, and enhanced translational capacity of the resulting mRNA vaccine (77). An overview of modified mRNA with modifications for mRNA vaccine production is shown in Figure 1.
Pseudouridine (Ψ), an isomer of uridine characterized by the replacement of a characteristic nitrogen-carbon glycosidic bond in the nucleotide with a carbon-carbon covalent bond, is most commonly employed for chemical modifications in the mRNA vaccine (78). This modification is introduced into the mRNA sequence via pseudouridine synthase (PUS) enzyme (79). Thermodynamic analysis revealed the impact of this modification on the overall stability of mRNA. In this regard, the replacement of uridine with Ψ makes the mRNA molecule relatively stable, whereas the level of stabilization varies depending upon the precise location of the modified base within the molecule, the specific base pair subjected to modification, and the orientation of neighboring Watson-Crick base pairs (80). Moreover, the incorporation of Ψ is shown to boost the translational capacity of the mRNA molecule (81). This enhanced translational capacity is achieved by three mechanisms that protect mRNA from decay in the cytosol. Firstly, this modification directly confers resistance to the mRNA molecule against RNase L-mediated degradation (82). Secondly, Ψ modification masks protein kinase R (PKR) activation, ultimately corresponding to a reduction in the PKR-mediated phosphorylation of the eukaryotic translation initiation factor 2 α subunit (eIF2α) (83, 84). Finally, this modification evades the activation of intracellular receptor 2’-5’-oligoadenylate synthase (OAS), which consequently prevents OAS-mediated RNase L activation.
The methylated derivative of Ψ, N1-methyl pseudouridine (m1Ψ), is also utilized for replacing the typical mRNA bases. The incorporation of these bases not only renders the resulting mRNA molecule markedly stable but also contributes to their safety by making them to elicit a negligible immune response in the cell, hence reducing their cytotoxic potential (85, 86). Besides, the mRNA molecules harboring modified bases, Ψ and m1Ψ, are found to be inherently inert or minimally reactogenic, hence avoiding excessive immune stimulation (81, 83, 87, 88). Moreover, individual or combinatorial introduction of chemically modified bases has been shown to generate a substantially reduced Toll-Like receptor (TLR) – mediated immunogenicity (89). In this regard, accumulated evidence indicates that m1Ψ outperforms Ψ in achieving elevated protein production and evading TLR3 activation (90). Also, the mRNA modified with m1Ψ escapes TLR7 recognition, hence leading to dismal expression of inflammation-producing genes (91, 92). Furthermore, m1Ψ modification contributes to reduced immune response generation mediated by intracellular Retinoic acid-inducible Gene-I-Like (RIG-I-Like) receptors owing to its ability to alter the mRNA secondary structure (92, 93). Moreover, a direct correlation between m1Ψ-based mRNA modification and enhanced size and proportion of ribosomes is observed (85). This provides a direct link between m1Ψ modification and boosted translational efficiency by developing productive poly-ribosomal interactions as a consequence of rapid initiation and relatively slow progression of translation.
5-methylcytidine (m5C), a methylated derivative of cytidine, is frequently observed in multiple RNA species as a natural epigenetic modification (94). This modification is exploited in mRNA vaccine technology as well and confers impressive stability, immunogenicity, efficiency, and sustenance in the cellular environment to the mRNA vaccines (95, 96). In the cellular translation initiation sites, RNA methyltransferases of NSUN and DNMT2 families are found to be substantially enriched for potentially catalyzing the methylation of cytidine bases (97). Similar to the Ψ modification, m5C modification also adds to the mRNA stability and protects it from enzymatic degradation by inducing alterations in the secondary and tertiary structural forms of mRNA (26). A study conducted by Kormann et al. revealed that incorporation of 25% m5C and 25% thiouridine generates 5 times higher protein levels in comparison to the unmodified mRNA (98). Hence, by modulating the level of methylation in the cytidine bases of mRNA sequence via methyltransferases and demethylases, the mRNA vaccines can be efficiently optimized (99).
N6-methyladenosine (m6A), a methylated derivative of adenosine, is another frequent modification observed in naturally existing mRNA transcripts (100). In the cellular environment, this modification regulates mRNA metabolism at various levels, including splicing, nucleus-to-cytosolic export, and stable translation by recruiting reader proteins, including YTHDF1 and YTHDF3 (101). Moreover, with the induction of m6A modification at the specific regions, such as the 5’ UTR or on the bases neighboring the stop codon, the translation efficiency can improve the translation efficiency manifold by active recruitment of translation initiation factors (102). Taken together, this modification significantly boosts mRNA efficacy and safety, and, therefore, serves as a promising tool for improving mRNA-based vaccines.
2-thiouridine (s2U) is among the novel modifications recently explored for their potential in promoting the activity of mRNA vaccines, and is found to be highly promising (103). Based on the findings of various studies, s2U modification in the IVT mRNA markedly evades recognition from TLR3 and TLR7, hence actively reducing the induction of subsequent inflammatory pathways (104, 105). Furthermore, this modification contributes to mRNA stability by providing resistance against enzymatic degradation. Moreover, the s2U modification promotes mRNA translation efficiency by preventing the activation of PKR, which ultimately results in the inhibition of eIF2α phosphorylation and facilitates protein synthesis (83).
5-methyluridine (m5U), is another chemically modified base often assessed in the context of mRNA vaccine technology (89). In line with the immunosuppressive mechanisms adopted by the modifications discussed above, m5U also ensures precise immune response generation by evading recognition from TLR3 (90). This promotes mitigation of the innate immune response, hence facilitating efficient delivery of mRNA vaccine into the target cells. Additionally, the incorporation of m5U bases in the mRNA vaccine design renders elevated stability and translation efficiency to the mRNA molecule, consequently improving the vaccine potency (105). After the successful in vitro modification of raw mRNA sequence, it is packaged within the delivery vehicles to facilitate the vaccine delivery, protection from degradation, and active cellular uptake (106).
5’ Cap and Poly(A) tail mark the key characteristics of eukaryotic mRNA. The 5’ cap, also called “cap 0”, includes an addition of a guanosine molecule methylated at the N7 position to the 5’ end of the first mRNA nucleotide through a 5’,5’-triphosphate bridge (107–109). The addition of the 5’ cap to the mRNA is crucial for appropriate translation initiation followed by ribosome recognition. It protects mRNA from exonuclease-mediated degradation and also facilitates subsequent transcription, polyadenylation, splicing, and mRNA transport from the nucleus to the cytoplasm (110, 111). Several studies have reported that most eukaryotic viral and parasitic mRNAs also harbor a 5’ cap feature (112–114). The addition of a 5’ cap is a well-regulated enzymatic process, and careful monitoring is performed at the cellular level to ensure appropriate capping, as faulty capping can result in premature mRNA decay and compromised translational efficiency. In this regard, the capping machinery involves three specific enzymes, namely RNA triphosphatase, RNA guanylyl transferase, and RNA (guanine-7)-methyltransferase (115). The capping process typically occurs co-transcriptionally, when the pre-mRNA transcript reaches a length of approximately 20-25 nucleotides, which begin to protrude from the RNA exit channel of RNA polymerase II, leading to the addition of cap 0 (116, 117). Following this, further methylation steps are executed involving the addition of methyl groups to the first and/or second transcribed nucleotides, resulting in cap 1 or cap 2, respectively (112). The O-methylation in cap 2 is crucial for demarcating self mRNA from the foreign molecules (118). The 5’ capping marks the natural mechanism of mRNA to cope with the cellular environment and ensure an enhanced level of translation. Hence, it should be considered as a modification mechanism during IVT mRNA synthesis for designing a stable and efficient vaccine.
Additionally, poly(A) tail is another characteristic feature of eukaryotic mRNA, which critically regulates its lifespan (119, 120). It involves the addition of multiple adenosine residues at the 3’ end of the mRNA molecule by a specialized enzyme, Poly(A) Polymerase. This post-transcriptional modification is incorporated into the IVT mRNA vaccine synthesis as well, where the tail length corresponds to overall mRNA stability and efficiency. To achieve this goal, IVT employs various strategies. In this regard, one primarily utilized procedure involves poly(A) tailing of mRNA mediated by recombinant poly(A) polymerase. However, the enzymatic tailing adds poly(A) tails of varying lengths to the mRNA transcripts, thereby resulting in heterogeneous mRNAs concerning the tail lengths. Alternatively, poly(A) tail can be encoded in the template vector designed for in vitro transcribing the mRNA molecule to produce synthetic mRNA with uniform tail length. Nevertheless, it is critically challenging to determine the precise duration (121). Regardless of this, IVT attempts to optimize this modification and administers the recombinant poly(A) polymerase to insert modified nucleotides into the tail, preventing poly(A) specific nucleases from deadenylating the poly(A) tail Additionally, poly (A) tailing in IVT mRNA synthesis renders additional benefits to the synthetic mRNA molecule. It has been observed that, for synthetic mRNA, extending the poly (A) tail to 120 bases progressively boosted the level of protein expression, whereas further extension in the tail was not implicated in further uplifting the protein expression levels (98, 122). Therefore, it is imperative to carefully optimize the addition of poly(A) tail in the mRNA molecule for acquiring substantial benefits.
5’ and 3’ UTRs comprise essential regulatory elements of mRNA contributing significantly to the mRNA stability, modulation of complex structures of mRNA, ribosomal recognition, and association of mRNA with the translation machinery (123). Moreover, UTRs can alter the rate of mRNA decay by modulating interactions with various RNA-binding proteins. Studies have reported that skipping the start codon by altering the non-canonical start codons in the 5’ UTR can disrupt translation by various mechanisms, including prevention of stable secondary structure formation, inability to recruit ribosomal machinery and masked codon recognition (124). On the other hand, precise 5’ UTR modification can enhance mRNA stability and translation accuracy. Likewise, 3’ UTR sequences play an equally crucial role in determining mRNA stability and efficiency. In this regard, by imparting considerable stability to the mRNA molecule, 3’ UTR sequences may enable an extended duration of gene expression. In particular, the 3’ UTR of α-globin mRNA is found to harbor discontinuous stretches of pyrimidine-rich regions, which render it extra stablility. This results in the synthesis of messenger ribonucleoprotein α-complex upon recognition of this stretch by cytosolic proteins (125–127). To aid this process, an integral component of α-complex, α-globin poly(C) binding protein (αCP), maintains the attachment of poly(A) binding protein on the poly(A) tail (128). This ultimately renders stability to mRNA by preventing deadenylation of the poly(A) tail. Collectively, UTRs substantially impact mRNA molecules, thereby demanding careful monitoring and optimization of UTR sequences during IVT following comprehensive research and analysis.
During IVT mRNA synthesis, ensuring high-quality production is imperative as it has a direct impact on the efficacy of the vaccine and the subsequent manufacturing steps. Hence, quality control is a critical parameter of IVT mRNA synthesis and should be monitored during various synthesis steps. In this regard, the synthesis process generally includes multiple steps, including a selection of the target antigen sequence, DNA template creation, IVT mRNA synthesis, mRNA purification, and LNP formulation (129). IVT employs several RNA polymerases, including T3, T7, and SP6, for artificially synthesizing high yields of relatively longer mRNA transcripts with length ranging in kilobases (130, 131). However, upon the introduction of modified bases in the IVT mRNA for the first time, the necessity of rigorous purification of the underlying mRNA transcripts for effective immune response generation was emphasized. To address this, various purification techniques were explored. In this regard, size-based mRNA chromatographic purification via high-performance liquid chromatography (HPLC) can eliminate bigger and smaller by-products, including double-stranded RNA, mRNA trances from non-linearized DNA templates, and abortive transcripts (104, 132). This comprehensive purification significantly boosted the output by potentially removing the impurities that may be implicated in negatively stimulating the immune system. Nonetheless, the advancements in the IVT mRNA synthesis and purification techniques have significantly improved the quality and efficacy of mRNA vaccines, the substantial challenges in optimization persist and need to be addressed by thorough research activity.
Among the core challenges associated with mRNA vaccines, the compromised delivery of mRNA inside the target cell and the subsequent risk of immune rejection are paramount. Additionally, failure to induce the required immune response is another significant limitation. To address these challenges, self-amplifying mRNA technology holds substantial promise. This technology employs an advanced approach involving the incorporation of additional sequences, such as viral RNA replication genes, in the mRNA sequence alongside the target protein sequence (3, 133). Upon expression in the target cell, these additional sequences facilitate replication of the target mRNA, thereby boosting the target protein production (21). The enzymatic process, In Vitro Transcription (IVT), remains the gold standard method for synthesizing mRNA from a DNA template within a controlled laboratory setting. In this regard, by utilizing DNA-dependent RNA polymerases, often derived from bacteriophages, IVT eliminates the requirement of an intermediate step involving plasmid DNA, streamlining the overall production process (134). Consequently, the generated mRNA is inherently fragile and necessitates the protection for safe and directed delivery to the target antigen-presenting cells within the body. To overcome this hurdle, specialized LNPs are employed as carriers to encapsulate the designed mRNA molecules (135). The LNPs ensure the safe, efficient, and targeted delivery facilitating the intracellular release of the mRNA (13). Upon the intracellular uptake in the target cells, the mRNA gets translated into the desired protein through the cellular ribosomal machinery (13, 136). The generated proteins are subsequently processed via the proteasomal machinery into shorter antigenic peptides, which are then loaded onto Major Histocompatibility Complex (MHC) molecules in the endoplasmic reticulum (137). MHC-I and MHC-II molecules presenting the antigenic peptides on the cellular surface of APCs engage with the T-cell receptors (TCRs), hence activating the Cytotoxic T-cells (CTLs) and Helper T-cells (HTLs), respectively. Additionally, B-cells recognize and internalize intact antigens through their B-cell receptors (BCRs). After internalization, B-cells process these antigens and present the derived peptides on their MHC class II molecules to helper T-cells. The activation of helper T-cells by this interaction, along with co-stimulatory signals, triggers the differentiation of B cells into antibody-producing plasma cells and memory B cells. Plasma cells then produce neutralizing antibodies that can bind to and neutralize circulating tumor antigens (138). Hence, the mRNA vaccine prompts an immune response within the host and offers a defense against the disruptive influence of the tumor cells (139). Figure 2 illustrates the entire process of immune activation mediated by mRNA vaccine administration.
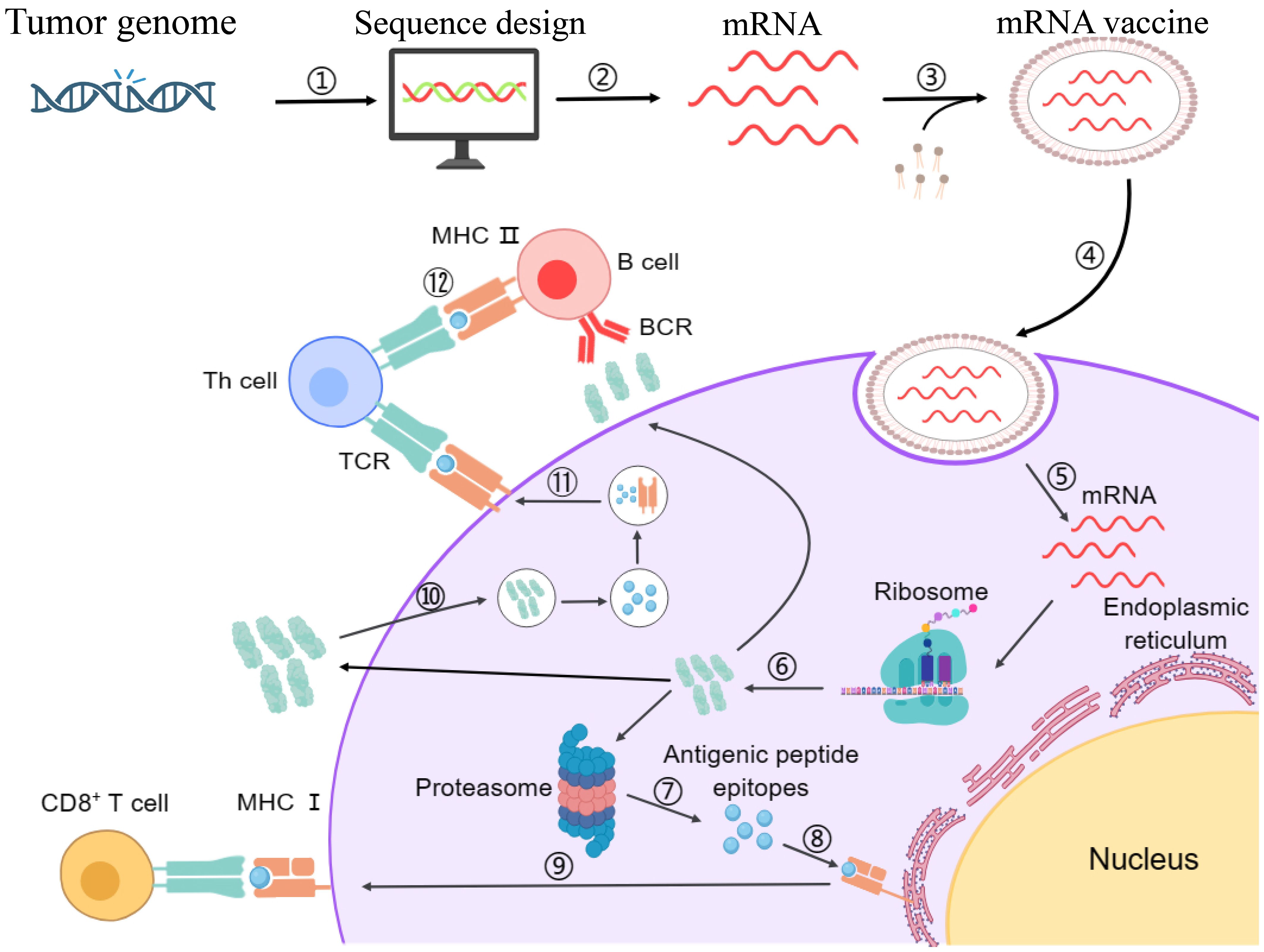
Figure 2. Schematic representation of designing mechanism of mRNA vaccine and its mode of action inside the cells. Step 1: The marked antigen sequence is designed and then introduced into the plasmid DNA vector when the tumor genome has been accomplished. Step 2: Artificial mRNA designed by in vitro transcription using the linearized plasmid DNA template is purified. Step 3: The purified mRNA is combined with delivery intermediaries to produce the mRNA vaccine. Step 4: Endocytosis takes the mRNA vaccine up inside the cells. Step 5: Release of the marked mRNA into the cytoplasm. Step 6: The ribosome translates the mRNA into protein. Step 7: The proteasome complex breaks down the protein product into antigenic peptide epitopes. Step 8: In the endoplasmic reticulum, the antigenic epitopes are loaded onto MHC class I molecules. Step 9: MHC class I molecules deliver CD8+ T cells antigenic peptides. Interchangeably, the protein product is designed, captivated by the cell, and then uncovered to an endosomal degradation process in step 10. Step 11: MHC class II molecules present the antigenic fragments to T-helper cells on the cell surface. Step 12: T-helper cells prompt B cells to make antibodies that neutralize target-specific cancer antigens. MHC, main histocompatibility complex, BCR and TCR, T cell and B cell surface receptor. The figure illustrates the overview of modified mRNA for mRNA vaccine production. The structure of mRNA is presented, starting from the 5’ cap, followed by the 5’ untranslated region (UTR), the coding region with chemically modified bases, and the 3’ untranslated region (UTR) with a poly(A) tail. The modifications shown include methylation and isomerization of bases: N6-methyladenosine (m6A), 5-methyluridine (m5U), 2-thiouridine (s2U), pseudouridine (Ψ), N1-methylpseudouridine (m1Ψ), and 5-methylcytidine (m5C), among others. These modifications occur at adenosine, uridine, and cytidine bases, providing specific structural and functional benefits.
Nucleic acid vaccines are vulnerable to degradation by nucleases in the body, hence requiring specific methods to improve their distribution (140). Moreover, delivering naked mRNA is inefficient due to rapid degradation and poor cellular uptake (141). In this regard, vectors can serve as specialized transport systems that guard nucleic acids from degradation and promote their cellular uptake (142). Gene delivery vectors are broadly classified into two categories: viral and non-viral vectors (143). Viral vectors refer to the carrier systems obtained from the viruses, whereas the non-viral vectors generally integrate artificial particles (144, 145). In terms of eliciting the immune response, the viral vectors have been shown to surpass the non-viral vectors and offer relatively higher efficacy (146). However, non-viral vectors appear comparatively less effective, and potentially less likely to induce immune reactions (147). Moreover, cellular uptake of mRNA remains another significant hurdle owing to the inherent negative charges on both, mRNA and cell membrane, coupled with the relative size of mRNA molecules, and their susceptibility to degradation by ribonucleases in the skin and bloodstream (148). Hence, to address these limitations, strategies for delivering mRNA into the cells have been extensively explored and pinpointed robust delivery carriers, including polymers, peptides, and lipid-based materials (149). In this context, liposomes have emerged as a promising drug delivery system due to their capacity to encapsulate and transport poorly water-soluble drugs (135). Doxil, a liposomal formulation of the antitumor agent doxorubicin, was the initial liposomal drug to receive clinical approval, leading to a shift in the trends in vaccine delivery, with lipid-based nanoparticles being widely utilized in the formulations (150). Moreover, numerous cationic lipid amphiphiles have been developed in recent years and evaluated as carriers of nucleic acids. The primary difference between these cationic lipids and their natural equivalents involves the presence of an additional ionizable (cationic) head group rather than a zwitterionic or a usually anionic head group. Natural and cationic lipids exhibit comparable molecular features, featuring a hydrophobic region consisting of two alkyl chains or cholesterol, and a linker joins their positively charged polar head groups to the hydrophobic moiety. Regarding their vaccine delivery potential, ionizable lipids are a superior choice compared to nonionizable cationic lipids because they are relatively less hazardous, remaining neutral in the circulation and exhibiting a positive charge only upon entering the cell due to pH changes (151). These delivery vehicles can be further optimized to increase the efficacy and safety of mRNA vaccine and thorough research is required to develop more relevant and innovative delivery carriers for mRNA vaccine.
Additionally, for the delivery of mRNA vaccines, many polymer-based vectors have been developed, including poly(l-lysine) (PLL), poly(amido-amine) (PAA), poly (beta amino-esters) (PBAEs), and poly(ethylenimine) (152, 153). However, only PEI has been extensively used in clinical studies. (154). Recently, new lipid-polymer complexes known as charge-altering releasable transporters (CARTs) have been established for the active distribution of mRNA molecules. (155). Moreover, mRNA and cytosine-phosphate-guanine (CpG), a synthetic toll-like receptor-9 agonist, are combined in CARTs to prepare a nanoparticle formulation that effectively transports antigen-coding mRNA to APCs (156). After efficient delivery of the mRNA into the target cells, the mRNA translation, followed by antigen processing and presentation triggers a robust immune response for effectively eradicating existing tumors (157). Specifically, the delivery of antigen-coding mRNA for melanoma immunotherapy, exemplified by the technology behind the Pfizer-BioNTech COVID-19 vaccine, has demonstrated marked effectiveness in clinical trials (158). Furthermore, polymer-based carriers offer a hopeful strategy for mRNA delivery. Nanoparticles made from polymers like poly lactic-co-glycolic acid (PLGA) can provide sustained release of the mRNA and can be potentially combined with adjuvants to further improve the immune response (159). Hence, mRNA encapsulated in a fatty shell offers a promising strategy for facilitating protected mRNA transport and cellular uptake (160). In this regard, mRNA-1273 (Moderna) successfully employed this delivery strategy (161).
LNP formulation typically involves a combination of four major constituents, including ionizable cationic lipids, phospholipids, cholesterol, and PEGylated lipids (162–164). Ionizable lipids play a crucial role in LNP formulations for mRNA delivery (165). Compared to traditional cationic lipid nanoparticles, the ionizable lipid-based LNPs (iLNPs) maintain electrical neutrality under physiological conditions, which helps reduce rapid elimination from the bloodstream and minimize immune system stimulation after intravenous administration (166, 167). Representative ionizable lipids include DLin-DMA, DLin-KC2-DMA (168), and DLin-MC3-DMA (169), synthesized through rational design; C12-200 and cKK-E12 (170), identified via high throughput screening of combinatorial libraries; and next-generation ionizable lipids, such as DLin-MC3-DMA derivative L319 (Alnylam and AlCana Technologies) (171), most of which are biodegradable. The key advantage of ionizable lipids is their ability to undergo protonation at acidic pH, allowing them to interact with the negatively charged mRNA through electrostatic forces (167). Following the cellular uptake and exposure to the acidic endosomal environment, the ionizable lipids become positively charged, hence disrupting the endosomal membrane structure and facilitating the release of mRNA into the cytosol (172). This remarkable alteration in the physiochemical properties of ionizable lipids in a pH-responsive manner offers an impressive potential for mRNA delivery (173), (174). Following this, phospholipids make up the second integral component of LNP formulation, where they significantly contribute to the stability and fluidic nature of LNPs. Precisely, phospholipids, including DOPE and DSPC, generally contribute to as much as 10-30% of the entire lipid composition within the LNP (175). Furthermore, the polarity and degree of unsaturation of the head and tail groups of the phospholipids, respectively, significantly affect the overall design of mRNA-LNPs, and should, therefore, be carefully optimized (167). In this regard, phospholipids harboring head regions equipped with quaternary amino groups coupled with unsaturated fatty acid tails can improve the efficiency of delivery and enhance the potential of mRNA endosomal escape (176). Thirdly, cholesterol serves to be a core component of LNP formulation by contributing significantly to the LNP lipid composition (177). Cholesterol promotes LNP formulation efficiency by various means. In this regard, it offers fluidity for maintaining the structural integrity of LNP during the integration of bulk cargo (178). Moreover, in addition to facilitating mRNA release followed by successful cellular uptake, cholesterol also stabilizes LNP by minimizing the association of proteins on the LNP surface (178). Finally, PEGylated lipids, such as ALC-0159 and DMG-PEG2000, mark the fourth crucial LNP component. Unlike the aforementioned three components, less proportion of PEGylated lipids, generally 0.5-3%, is integrated into the LNPs. PEGylated lipids render ‘stealth’ properties to the LNPs, impressively elevating the in vivo circulation duration (179). Moreover, the PEG component provides steric hindrance, preventing aggregation, reducing non-specific uptake, and evading clearance by the mononuclear phagocyte system, thereby enhancing the delivery efficacy of the LNPs (180). Nonetheless, all these elements are essential and contribute to the stability, transfection efficiency, and safety of LPNs (179). Key components of LNP for mRNA vaccine formulation are shown in Figure 3.
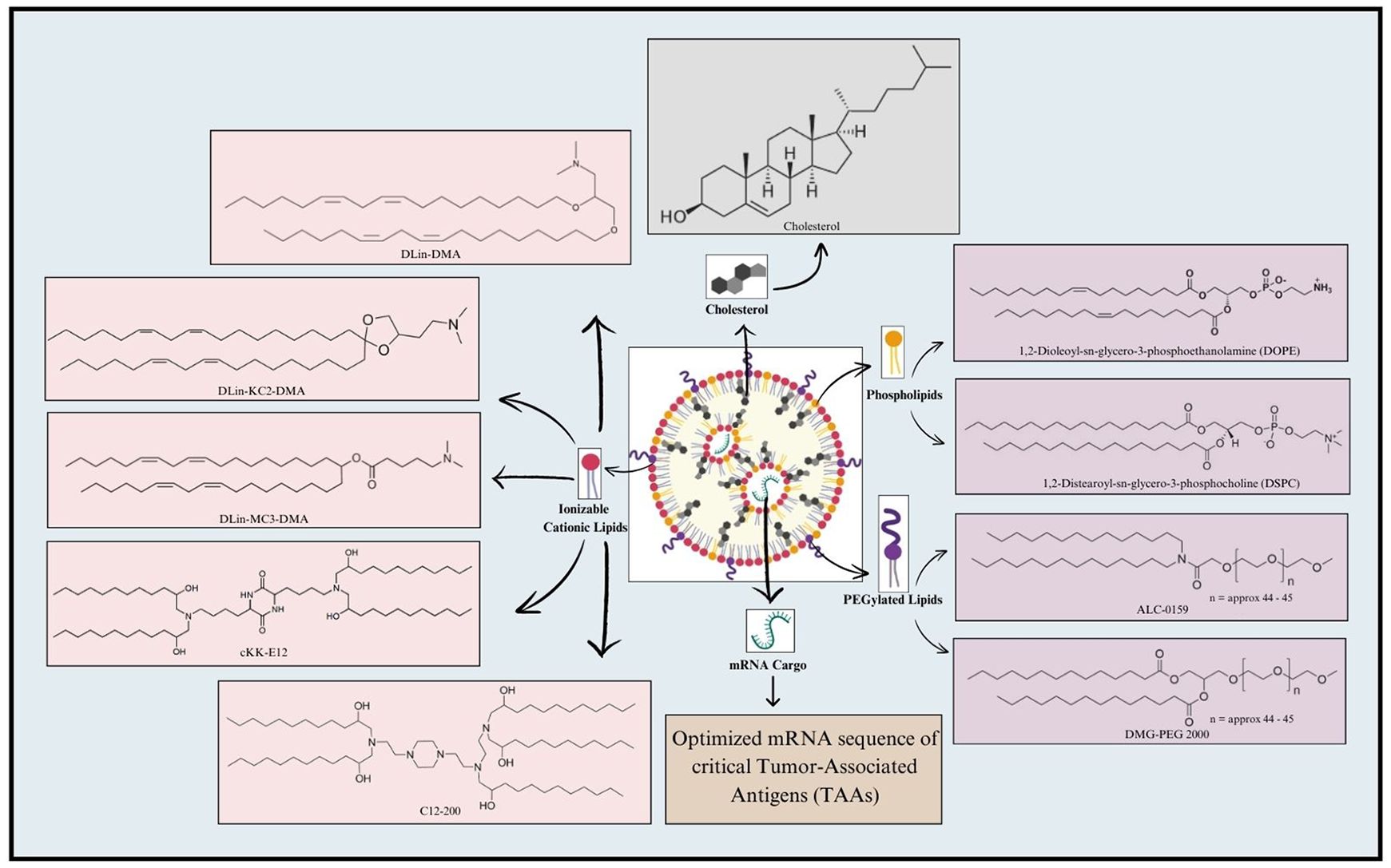
Figure 3. Key components involved in mRNA-LNP formulation. A typical LNP includes four key components, namely Ionizable cationic lipids, Phospholipids, PEGylated lipids, and Cholesterol. Ionizable cationic lipids generally employed for LNP formulation include DLin-DMA; DLin-KC2-DMA; DLin-MC3-DMA; cKK-E12; and C12-200. Phospholipids, such as 1,2-Dioleoyl-sn-glycero-3-phosphoethanolamine (DOPE) and 1,2-Distearoyl-sn-glycero-3-phosphocholine (DSPC), are frequently utilized in this regard. Additionally, PEGylated lipids used for LNP formulation include ALC-0159 and DMG-PEG 2000. Cholesterol is also incorporated in the LNP to maintain its structural integrity. Finally, the mRNA cargo loaded within LNP includes the mRNA sequence of critical genes encoding Tumor-Associated Antigens (TAAs).
For mRNA-LNP synthesis, different lipids and mRNA are usually dissolved in the ethanol and acidic aqueous phases (such as pH 4.0 citrate buffer) with a microfluidic device at a volume ratio of 1:3, and the execution of the self-assembly process is enabled. Then, during the formation phase, negatively charged mRNA is allowed to establish electrostatic interactions with the ionizable cationic lipid after the protonation lipid becomes positively charged. Moreover, to stabilize the produced mRNA-LNP, other helper lipids such as cholesterol, phospholipids, and PEGylated lipids self-assemble on them (181). Consequently, the ionizable lipids become uncharged and less hazardous at physiological pH. Following this, the mRNA-LNP solution is buffer exchanged to a neutral pH (182). The latest examples of mRNA vaccines utilizing these LNP components include Moderna COVID-19 vaccine (mRNA-1273) and Pfizer-BioNTech’s COVID-19 vaccine (BNT162b2. Moderna COVID-19 vaccine uses the ionizable lipid SM-102, DSPC, cholesterol, and DMG-PEG2000, whereas Pfizer-BioNTech COVID-19 vaccine employs the ionizable lipid ALC-0315, DSPC, cholesterol, and ALC-0159 (a PEG-linked lipid) (183). Hence, these advanced LNP formulations have undoubtedly played a crucial role in the success and rapid development of mRNA-based COVID-19 vaccines.
Furthermore, when evaluating the efficiency of mRNA-LNP complexes, a critical assessment of the zeta potential is essential, as it significantly affects the delivery efficiency and biodistribution of LNPs (184). In this regard, LNPs undergo a transition from a net positive charge at low pH to a negative charge at high pH, as detected by the broad ZP titration curve (185). This charge transition is important for the endosomal escape of the mRNA payload. At low endosomal pH, the positively charged LNP can form ion pairs with anionic endosomal phospholipids, destabilizing the endosomal membrane and releasing the mRNA. Additionally, the breadth of the ZP titration curve (over ~4 pH units) indicates that the LNPs maintain a positive charge across the entire endosomal pH range, facilitating the membrane disruption and cytosolic delivery of mRNA (185, 186). The net charge of the LNPs, as measured by ZP, also determines the in vivo biodistribution and targeting of LNPs (187). In this regard, cationic LNPs tend to accumulate in the lungs after intravenous (IV) administration, whereas, reducing the cationic lipid content for crafting more negatively charged LNPs leads to preferential targeting of the spleen after IV injection (188). Additionally, for intramuscular (IM) administration, more negatively charged LNPs exhibit higher off-target expression in the liver compared to less negatively charged formulations (185). The positive charge of LNPs facilitates their cellular uptake by interacting with negatively charged heparin sulfate proteoglycans on the cell membrane. These positively charged LNPs also demonstrated correlated potency for intramuscular (IM) administration in mice (189). Moreover, LNPs with a positive charge throughout the endosomal pH range are more efficient for in vitro mRNA transfection in HEK293 cells (185). In addition, the positive charge is beneficial for IM delivery, likely due to interactions with negatively charged proteoglycans in the extracellular matrix of muscle tissue. Conversely, for intravascular (IV) administration, negatively charged LNPs are favored for hepatocyte targeting (190) The negative charge facilitates passive ApoE-mediated targeting and binding in the bloodstream (191). Nonetheless, positively charged LNPs have potential applications in mRNA delivery for in vivo muscle gene expression and IV administration, as they can target the spleen (192).
Numerous newly synthesized materials are being utilized to create PEG-free nano-carriers to boost biocompatibility. A recently developed material, tB-UC18 (comprising a benzene-ring scaffold and three unsaturated lipid tails), has been employed to self-assemble with the aid of 1,2-dioleoyl-sn-glycerol-3-phosphoethanolamine, forming lipid-like nanoassemblies (LLNs) (193). These LLNs have demonstrated thermostability and resistance to nuclease degradation. Moreover, when mRNA was encapsulated in LLNs, the delivery system maintained thermal stability for at least two weeks without PEG lipids. In this regard, a newly developed PEG-free mRNA vaccine, PFTCmvac, exhibited a marked immunogenic potential. The vaccine was capable of generating broad-spectrum adaptive immunity with negligible side effects in mice and evasion of complement system activation in the human serum. This strategy can be further exploited in the mRNA vaccine technology for developing PEG-free nano-carriers (194).
Recently, a novel drug delivery strategy, namely Lipid-polymer hybrid nanoparticles (LPHNPs), has emerged that integrates the beneficial aspects of liposome carriers coupled with biodegradation polymeric nanoparticles (195). This carrier features an innovative structure encompassing three components, including a polymeric core, a surrounding lipid monolayer, and an outermost PEGylated layer (196). The LPHNP formulation involves a sophisticated combination of biodegradable polymer, zwitterionic phospholipids, ionizable lipids, PEG-lipids, and cholesterol (196). The LPHNP-based hybrid nano-carrier system offers striking benefits, including improved transfection properties, targeted mRNA delivery, enhanced stability, and upgraded mRNA release kinetics (174, 197). Moreover, the biocompatible nature of LPHNPs renders them significant resistance against degradation and boosts their ability to facilitate mRNA encapsulation, stability, and cellular uptake (174, 196, 198). Additionally, their optimized hydrodynamic parameters including sizes within the 200-250 nm range further add to their benefits (199). Hence, these nanoparticles significantly widen the therapeutic window of mRNA vaccines in combating diverse cancers and genetic disorders (197, 198).
Taken together, the incorporation of innovative delivery methods can substantially revolutionize the mRNA vaccine technology by offering better optimization parameters. In addition, vaccine administration parameters should also be carefully optimized. In this regard, the administration of SARS-CoV-2 mRNA-LNP via a needle-free injection method ensured a minimally invasive procedure, while achieving enhanced immunogenicity compared to conventional needle-based injection methods (200). In the future, further optimizations and modifications can be executed to improve delivery, stability, targeting, immune stimulation, cost-effectiveness, and yield for wider applicability of mRNA vaccines in clinical set-up (186).
The efficient intracellular delivery of mRNA payloads is a crucial requirement for the success of mRNA-based therapies (H. Liu et al.). After cellular internalization, mRNA-loaded lipid nanoparticles (LNPs) must escape the endosomal compartment and release the mRNA into the cytoplasm, where it can then be translated into the desired protein (201, 202). In this regard, two primary mechanisms have been proposed, including “membrane destabilization” and “proton sponge effect”.
Firstly, membrane destabilization involves the protonation of ionizable lipids in the acidic pH of endosomes, followed by interactions with anionic lipids on the endosomal membrane. This induces a non-bilayer, hexagonal structure that can disrupt the membrane and release the mRNA payload. (168). Various studies have demonstrated that a pKa range of 6.2 to 6.5 is optimal for effective in vivo silencing in hepatocytes, with the highest potency observed at a pKa of 6.44 (169). Moreover, the optimal pKa for protein expression from mRNA-LNPs can be altered by route of administration. In this regard, a study showed that vaccine administration via the intravenous route exhibited a pKa lower than that required for protein expression compared to the administration via the intramuscular route (185, 189). Secondly, the ‘proton sponge effect’ functions by the buffering capacity of LNPs to activate the proton pumps, increasing the membrane potential, This leads to the influx of chloride ions, increasing the osmotic pressure and causing the endosome to swell and burst, thereby releasing the mRNA (203).
After the successful release of mRNA from the endosome and into the cytoplasm, it interacts with the cellular translation machinery to be expressed as a functional protein (204). This process involves ribosome recognition in which the mRNA must be in an unstructured, single-stranded form that can be accessed by the ribosome, the cellular organelle responsible for protein synthesis. Certain mRNA design elements, such as the 5’ cap structure and 3’ poly(A) tail, can enhance mRNA stability and promote efficient translation initiation by the ribosome. Moreover, the mRNA sequence and codon optimization can also impact translation efficiency. Therefore, by integrating the endosomal escape mechanisms and the subsequent mRNA translation process, mRNA-based vaccines can effectively deliver the genetic payload and express the target proteins within the cell.
With the recent innovations in the delivery methods, a lot of new approaches are employed to increase the efficacy of mRNA-based therapeutics for cancer treatment. Recently, Miao et al. created a range of ionizable lipid-like materials and validated the best candidate formulations as mRNA delivery vehicles for vaccination against cancer (205). These LNP formulations were used to deliver ovalbumin (OVA) mRNA to the OVA-expressing B16F10 mouse melanoma model. The egg white protein OVA is commonly used as a model antigen owing to its potential of enhanced cytotoxic lymphocyte-based recognition of neoantigens (206). After the initial two doses, the LNP-based mRNA vaccines significantly suppressed tumor growth and produced a robust antigen-specific cytotoxic T-cell response. Additionally, rather than activating TLRs, these LNP formulations stimulated adaptive immune cells via the stimulator of interferon genes (STING) pathway, which resulted in strong antigen expression, and local production of pro-inflammatory cytokines (207). Furthermore, recently a formulation was designed based on a common tumor suppressor gene, phosphatase, and tensin homolog deleted on chromosome ten (PTEN), which is mutated or deleted in a variety of human cancers. In this regard, the lipid materials G0-C14, polylactic-co-glycolic acid (PLGA), and lipid-PEG were used to synthesize mRNA-LNP, and PTEN was reintroduced into cancer cells to restore its ability to serve as a tumor suppressor (208). Another research, focused to craft and refine a platform of mRNA nanoparticles, targeted at C-X-C-Motif Receptor 4 (CXCR4) to effectively increase p53 expression in models of hepatic cell carcinoma (HCC). They combined anti-PD-1 immunotherapy with CXCR4-targeting p53 mRNA nanoparticles, which revealed the massive potential of this combination strategy in enhancing the liver cancer anti-tumor immune response and suppressing tumor growth (209). The researchers demonstrate immense confidence in the potential of this therapy to effectively treat immunosuppressive cancers, alongside liver cancer, and are eager to apply the potential findings from animal models to human clinical trials.
Recently, lipid calcium phosphate nanoparticles (LCPs) were used in the development of a vaccine against melanoma to deliver two essential components, including siRNA directed against the immunological checkpoint PD-L1 and mRNA encoding the tumor antigen TRP-2. Following their loading onto the dendritic cells (DCs), these LCPs were injected into mice. The outcomes demonstrated the effective transport of siRNA and mRNA to DCs in the lymph nodes, which prompted CD8+ T cells to mount a targeted immune response against TRP-2, which significantly slowed down tumor development in mice models (157). Overall, the promising results indicate that LNP-based mRNA vaccines have the potential to become an effective strategy for cancer immunotherapy. To completely comprehend the mechanics and long-term impacts of these formulations, additional meticulous research and critical evaluation are required.
Therapeutic mRNA cancer vaccines have emerged as a promising novel approach to cancer immunotherapy (25), offering high specificity, better efficacy, and fewer side effects compared to traditional therapeutic strategies (210). In this regard, multiple therapeutic mRNA cancer vaccines are subjected to stringent evaluation in preclinical and clinical trials, with promising early-phase results (211). Recently, preclinical research has been conducted on mRNA melanoma vaccines using orthotopic mice models, where B16F10 melanoma cells were employed as an experimental cell line. The findings revealed strong stimulation of T cells immune response in the mice model upon vaccine administration, suggesting the promising effectiveness of the vaccine in clinical settings (212). Moreover, immune checkpoint inhibitors, including monoclonal antibodies to counter CTLA-4, PD-1, and PD-L1, have been permitted for therapeutic use. These drugs are explicitly intended to address melanoma, suggesting a synergistic or combined mRNA vaccine approach against melanoma, which can further enhance both efficacy and strength of immune response (213). Furthermore, an mRNA vaccine designed by integrating several MHC classes I and II-restricted neoepitopes produced from B16F10 melanoma cells was shown to be efficacious by Kreiter et al. in 2015 (214). In a mouse model, this vaccination effectively generated strong tumor-specific CD4+ and CD8+ T cell responses, which resulted in considerable tumor rejection (60–80% survival). Expanding on this achievement, Chen et al. created a new formulation of lipid nanoparticles (113-O12B) that demonstrated improved lymph node targeting compared to conventional LNP formulations (215). Notably, an mRNA vaccine expressing a Trp2 180-188-specific epitope was more effective owing to the enhanced delivery method, resulting in a 40% complete response rate in mice challenged with melanoma (216). The injectable vaccination Melanoma FixVac BNT111, administered by liposomal RNA (RNA-LPX), was the subject of a recent clinical trial (Lipo-MERIT, NCT02410733) (217). This first-ever human Phase I trial assessed the vaccine safety and effectiveness in patients with metastatic melanoma. Four frequently occurring, non-mutated tumor-associated antigens in melanoma were targeted by melanoma FixVac BNT111. The trial’s initial dose-escalation phase established the safety and tolerability of the vaccination. Also, encouraging findings from an intermediate analysis were found (218).
Recently, a novel adjuvant therapy strategy for melanoma combining anti-PD-1 medication with a personalized mRNA-based cancer vaccine has been constructed. The neoepitopes have been shown to efficiently induce robust anticancer immunity in vivo (219). In the KEYNOTE-942 stage II test, patients with resected high-risk stage IB–IV melanoma were randomized to receive either pembrolizumab alone or the mRNA-4157 vaccine, leading to enhanced immune response generation (220). Another study examined the immunogenic potential of GM-GSF as an adjuvant along with the mRNA vaccination encoding six melanoma-specific TAA genes in a phase I/II clinical trial involving 21 melanoma patients in 2009. The data showed that immunization significantly reduced the number of immunosuppressive cells (Foxp3+/CD4+T cells) without creating any adverse side effects (221). Furthermore, an mRNA vaccine, namely Lipo-MERIT, combining PD-L1 with four TAAs, was introduced to melanoma. Consequently, it effectively triggered IFN-γ production and encouraged the recruitment of T cells specific to the antigen (NCT02410733) (222). Afterward, recently developed lymph node-targeted LNP-based mRNA vaccination enhanced antitumor immunity and produced robust CD8+ T cell responses in B16F10 melanoma-bearing mice (215). Additionally, Christian et al. demonstrated high efficacy in mouse tumor models by encoding a variety of cytokines, including GM-CSF, IL-15, interleukin-12 (IL-12) single chain, and interferon-α (IFN-α), in an mRNA combination through multiple intratumoral injections against melanoma. Furthermore, when the tumor growth was effectively controlled, the majority of them completely disappeared. This was further linked to several factors, including the formation of immunological memory, amplified granzyme B+, T-cell infiltration, intratumoral IFN-γ induction, systemic antigen-specific T-cell growth, and anticancer activity of mRNAs encoding four different cytokines. This defensive response was also evident in metastatic areas external to the initial tissue, where mRNA-encoded cytokines are generated in vivo (215). This offers a promising hope for combining cytokines with mRNA vaccine as an adjuvant therapy for melanoma.
Recently, a phase II clinical trial of the GP2 peptide-based vaccine, under the vaccine ID NCT00524277, showed groundbreaking outcomes of a 100% 5-year survival rate in patients with HER2+ breast cancer (223). Although no breast cancer vaccine has yet been permitted for either treatment or inhibition, this evokes a general interest that breast cancer mRNA vaccine would have great implications in significantly improving the survival rate. Another ongoing clinical trial (NCT03289962) targeting TNBC with RO7198457 (individualized mRNA vaccination) + atezolizumab (anti-PD-L1) has shown that tumors overexpressing HER2 show less response to immune checkpoint blockade (224). However, by revitalizing T-cells, HER2-specific mRNA vaccination may enhance the sensitivity of cancer cells to immune checkpoint inhibitors (225). Moreover, research has revealed that the mRNA vaccination and the anti-CTLA-4 monoclonal antibody can work in conjunction to produce a strong CTL response against TNBC (226). Moreover, another study has revealed the effectiveness of mannose-modified lipid-core nanoparticles (LCP) in delivering MUC1 TAA encoding mRNA to the dendritic cells, triggering a robust immune response against TNBC cells (227). In addition, new research investigated the efficacy of VRP-HER2, a viral-based HER2 RNA vaccine, in a mouse model. Based on the findings, a considerable induction of HER2-specific T-cells mediated by VRP-HER2 was revealed, substantially restricting tumor growth. The HER2-specific T-cells induced by this vaccination were CD8+ T-cells capable of expressing perforin, which has been previously reported to be implicated in promoting disease-free survival in breast cancer patients (228). Additionally, clinical trials of TriMix vaccine (NCT03788083) are in progress to evaluate the safety and efficacy parameters of intratumoral mRNA vaccine against breast cancer (229). Besides this, individualized mRNA neoantigen vaccine (iNeST) is also under assessment in amalgamation with added lipoplex-formulated mRNA-producing TAAs (BNT114), and RNA encrypting p53 in patients with TNBC (NCT02316457). Currently, CARvac, an mRNA lipoplex vaccine encoding claudin-6 protein (CLDN6), is under inspection in a phase 1/2 clinical trial (NCT04503278) for patients with advanced solid CLDN6-positive tumors. Moreover, this vaccine can also be combined with mRNA technology to further boost its efficacy. To improve CAR T-cell treatment, BNT211, an autologous CLDN6, and CARVac are simultaneously injected intravenously. Early findings showed that four (57%) of the seven evaluable patients treated with a combination of CARVac and CLDN6 CAR T-cell therapy exhibited a partial response, while one patient (14%) remained stable at the 6-week evaluation (230, 231). BioNTech is currently developing BNT114, a pre-made set of common cancer antigens. Conversely, BNT-122 is a personalized vaccine targeting a patient-specific tumor mutation (211). Following this, the phase I clinical trial of AVX901, an amplifying mRNA vaccine based on a self-amplifying VEEV vector articulating HER2, has been completed. Next, phase II clinical investigation aims at assessing the efficacy of AVX901 in combination with pembrolizumab (232). Moreover, NCT01837602, a phase 0 clinical trial, is now investigating the safety and feasibility of anti-cMet chimeric antigen receptor T cells (CAR-T cells) to treat patients with metastatic breast cancer (233). TNBC patients have undergone evaluations of the safety, immunogenicity, and effectiveness of a customized mRNA vaccine made with neoantigen liposomes (NCT02316457), where the preliminary results reflect a strong generation of poly-epitopic T-cell response (234). Recently, a stage 1 clinical trial of a custom-made mRNA vaccine, autogenous cevumeran, displayed that it can initiate improved immune responses in 50% of PDAC patients after medical resection, which increases the hope of treating this deadly disease. In this regard, the vaccine design is equipped with 20 neoantigens and is transported using LNPs via circulatory injection in amalgamation with chemotherapy and immune checkpoint therapy (235). The Phase I clinical trial results confirmed the clinical benefits for patients with surgically resected PDAC (NCT04161755) and the ability of autogene cevumeran to expand neoantigen-specific and functional CD8+ T cells (236). Moreover, MUC1 is distinguishably an overexpressed TAA in pancreatic cancer, making it a superlative target of therapeutic design (237). Furthermore, the high frequency of KRAS point mutations in pancreatic cancer suggests it to be a potential target for therapeutic strategy against pancreatic cancer (238). In this regard, a personalized mRNA vaccine can be constructed by targeting patient-specific KRAS mutations. Moreover, an ongoing medical trial has recruited patients for appraisal of the safety and acceptability of mRNA5671/V941, either unaccompanied or in grouping with pembrolizumab, for the treatment of progressive or metastatic pancreatic cancer (NCT03948763) (107). Another mRNA-based vaccine trial is undergoing in clinical settings to assess the protection and effectiveness of progressive adjuvant atezolizumab (Genentech) and autogene cevumeran in patients with PDAC (235).
Following this, another clinical trial to appraise the protection and worth of a customized mRNA neoantigen vaccine (iNeST), in grouping with mFOLFIRINOX, a first line of therapeutic combination for PDAC, in patients with surgically resectable PDAC is in progress. The main aim for designing this vaccine is to intensify neoantigen-specific T-cells repressed by PD-1 signaling and major naive T-cells in response to vaccine neoantigens. The iNeST has been shown to be considerably safe with marked feasibility and dismal side effects in patients. It has also been shown to produce significant neoantigen-specific T-cells in 50% of unselected patients with resectable PDAC. Furthermore, vaccine-induced T-cells are shown to be resilient and can last up to two years after receiving mFOLFIRINOX treatment (235). Additional ongoing clinical trials for solid tumors, including PDAC, are mounting on Phase 2 with the reference IDs: NCT03289962 and NCT04161755 (236). Afterward, mRNA-4650, an mRNA vaccine targeting KRAS mutations, developed by Moderna and Merck, was used in amalgamation with or without pembrolizumab for the treatment of pancreatic carcinoma to create adjuvant or combinational therapy. The result of the therapy evaluated that the LNP distribution system for mRNA-4650 was well endured and provoked an anti-tumor immune response (19). Likewise, hTERT vaccination shows promise in inducing an immune response against cancer, with a case study demonstrating complete remission in a pancreatic cancer patient following DC vaccination with hTERT mRNA. This successful treatment also identified novel epitopes that can be incorporated into future hTERT vaccines for broader application (239). The latest mRNA-based and peptide-based combinational vaccines have been constructed to incorporate specific antigens and mRNA. In this regard, one such vaccine, mRNA-5671/V941, targets four KRAS mutations, including G12D-, G12V-, G13D-, and G12C-, is tested in combination with pembrolizumab in stage I experimental trial for patients with KRAS-mutant advanced colorectal cancer under the clinical reference ID of NCT03948763 (240–242). During the trials, the vaccine was administered intramuscularly in LNPs for nine cycles every three weeks. Consequently, significant antitumor response was observed with marked tolerance of the formulation in vivo. Upon translation and processing of the antigen protein, peptide epitopes were presented via MHCs and enabled the generation of T-cell mediated immune responses (6). Additionally, two NSCLC-specific mRNA vaccines, CV9201 (NCT00923312) and CV9202 (NCT01915524) were evaluated in clinical trials to assess their therapeutic potential against lung cancer in combination with checkpoint inhibitors. CV9201 targets five TAAs specifically linked to NSCLC, whereas CV9202 is a self-adjuvating mRNA vaccine that targets six antigens commonly expressed in NSCLC (NY-ESO-1, MAGEC1, MAGEC2, 5 T4, survivin, and MUC1) (243). By facilitating antigen expression and stimulating TLR7/8 and intracellular RNA sensors, they aim to activate immune cells against NSCLC cells (244). Based on the initial results, the vaccines were shown to be safe and efficient in producing antigen-specific T-cell immune responses in 63% (for CV9201) and 84% (for CV9202) of individuals. Nevertheless, neither progression-free survival nor overall survival was observed to be enhanced by CV9201 (245, 246). Moreover, a vaccination trial, NCT00004604, is carried out to evaluate the safety and dose-limiting toxicity of a vaccine including DCs injected with CEA-encoding mRNA. Phase I data indicated that, while all patients felt malaise and subcutaneous nodules at the injection site, no major toxicities were noted. As a result, it was established that giving CEA mRNA-transfected DCs was safe (247, 248). Recently, multi-epitope neoantigen was synthesized as a KRAS mRNA vaccine, mRNA-1521, that wholly covered all predominant KRAS mutations to attain a broad-spectrum immune response. Following this, immune response and anti-tumor effectiveness were measured in the colon cancer Balb/c mice model. It was noted that prophylactic immunization with mRNA-1521 pointedly repressed tumor growth in the mouse model of colon cancer. The repressive effects of the vaccine were additionally improved upon the combination with anti-PD-1 antibodies (249). mRNA-1521 also provoked specific T and B cell responses vital for anti-cancer immunotherapy (249). A single intratumorally administered injection of (murine) IL-12 mRNA-LNP, under the vaccine ID (NCT03946800), produced a tumor clearance rate of approximately 86% in MC38 mouse models with colon adenocarcinoma, and almost all of the mice were resistant to tumor recurrence (250). A vaccination based on OMV-LL-mRNA has been shown to dramatically slow the growth of melanoma and induce 37.5% full remission in a colon cancer model. After 60 days, OMV-LL-mRNA markedly protected the mice from tumor challenges by inducing a long-term immunological memory (251). Furthermore, the mRNA cancer vaccination expressing the MC38 neoantigen successfully suppressed MC38 colon cancer mice and prevented tumor recurrence when given in concert with immune checkpoint blockade therapy (152, 252). Finally, phase I/II clinical trials are currently ongoing to evaluate the therapeutic potential of a KRAS-targeting mRNA vaccine, ELI-002 against operable colorectal cancer (242). The successful findings revealed by these clinical trials boost the confidence of researchers in the clinical applicability of mRNA vaccines against cancers. Nonetheless, extended evaluation is imperative to unfold the long-term effects of these vaccines before their utilization in cancer therapeutics. mRNA vaccine for different cancer types is shown in Table 3.
Cancer is declared the deadliest clinicopathological condition encompassing the whole globe owing to its prevalence and impact. However, recent advancements in molecular medicine, particularly the development of mRNA vaccines and cancer immunotherapy, offer great promise for effectively treating cancer. Numerous clinical trials are underway for the formulation of novel vaccines against cancer, either as standalone or administered in combinational therapies. Although, initial vaccine trials have reported promising results, a substantial follow-up alongside critical improvements is still needed before a finalized product can be made accessible to the general public. Hence, enhancing the vaccine technology to boost efficacy and safety remains a pivotal challenge to be addressed shortly.
UL: Writing – original draft, Writing – review & editing. WA: Software, Visualization, Writing – review & editing. Z-XX: Conceptualization, Methodology, Supervision, Writing – review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by the National Key Research and Development Program of China (2023YFE0109800) and the National Natural Science Foundation of China (82020108024).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
1. Soerjomataram I, Bray F. Planning for tomorrow: global cancer incidence and the role of prevention 2020–2070. Nat Rev Clin Oncol. (2021) 18:663–72. doi: 10.1038/s41571-021-00514-z
2. Gupta S, Shukla S. Limitations of immunotherapy in cancer. Cureus. (2022) 14(10):e30856. doi: 10.7759/cureus.30856
3. Wang Y, Zhang Z, Luo J, Han X, Wei Y, Wei X. mRNA vaccine: a potential therapeutic strategy. Mol cancer. (2021) 20:33. doi: 10.1186/s12943-021-01311-z
4. Heine A, Juranek S, Brossart P. Clinical and immunological effects of mRNA vaccines in Malignant diseases. Mol cancer. (2021) 20:1–20. doi: 10.1186/s12943-021-01339-1
5. Qu Y, Xu J, Zhang T, Chen Q, Sun T, Jiang C. Advanced nano-based strategies for mRNA tumor vaccine. Acta Pharm Sin B. (2024) 14:170–89. doi: 10.1016/j.apsb.2023.07.025
6. Wei J, Hui AM. The paradigm shift in treatment from Covid-19 to oncology with mRNA vaccines. Cancer Treat Rev. (2022) 107:102405. doi: 10.1016/j.ctrv.2022.102405
7. Al-Haideri M, Tondok SB, Safa SH, Maleki AH, Rostami S, Jalil AT, et al. CAR-T cell combination therapy: the next revolution in cancer treatment. Cancer Cell Int. (2022) 22:365. doi: 10.1186/s12935-022-02778-6
8. Wu J, Wu W, Zhou B, Li B. Chimeric antigen receptor therapy meets mRNA technology. Trends Biotechnol. (2024) 42:228–40. doi: 10.1016/j.tibtech.2023.08.005
9. Wu Q, Bazzini AA. Translation and mRNA Stability Control. Annu Rev Biochem. (2023) 92:227–45. doi: 10.1146/annurev-biochem-052621-091808
10. Pardi N, Hogan MJ, Weissman D. Recent advances in mRNA vaccine technology. Curr Opin Immunol. (2020) 65:14–20. doi: 10.1016/j.coi.2020.01.008
11. Gao M, Zhang Q, Feng XH, Liu J. Synthetic modified messenger RNA for therapeutic applications. Acta biomaterialia. (2021) 131:1–5. doi: 10.1016/j.actbio.2021.06.020
12. Wadhwa A, Aljabbari A, Lokras A, Foged C, Thakur A. Opportunities and challenges in the delivery of mRNA-based vaccines. Pharmaceutics. (2020) 12(2):102.
13. Zong Y, Lin Y, Wei T, Cheng Q. Lipid nanoparticle (LNP) enables mRNA delivery for cancer therapy. Adv Mater. (2023) 35:2303261. doi: 10.1002/adma.202303261
14. Van Hoecke L, Roose K. How mRNA therapeutics are entering the monoclonal antibody field. J Trans Med. (2019) 17:54. doi: 10.1186/s12967-019-1804-8
15. Linares-Fernández S, Lacroix C, Exposito JY, Verrier B. Tailoring mRNA vaccine to balance innate/adaptive immune response. Trends Mol Med. (2020) 26:311–23. doi: 10.1016/j.molmed.2019.10.002
16. Conry RM, LoBuglio AF, Wright M, Sumerel L, Pike MJ, Johanning F, et al. Characterization of a messenger RNA polynucleotide vaccine vector. Cancer Res. (1995) 55:1397–400.
17. Bukhari MH, Syed M, Zain S. The differences between traditional vaccines and RNA vaccines: safety, efficacy, reliability and future of COVID-19 vaccines. Lahore, Pakistan: Annals of King Edward Medical University (2021) 27(2). doi: 10.21649/akemu.v27i2.4531
18. Thess A, Grund S, Mui BL, Hope MJ, Baumhof P, Fotin-Mleczek M, et al. Sequence-engineered mRNA without chemical nucleoside modifications enables an effective protein therapy in large animals. Mol Ther. (2015) 23:1456–64. doi: 10.1038/mt.2015.103
19. Cafri G, Gartner JJ, Zaks T, Hopson K, Levin N, Paria BC, et al. mRNA vaccine–induced neoantigen-specific T cell immunity in patients with gastrointestinal cancer. J Clin Invest. (2020) 130:5976–88. doi: 10.1172/JCI134915
20. Esposito M, Minnai F, Copetti M, Miscio G, Perna R, Piepoli A, et al. Human leukocyte antigen variants associate with BNT162b2 mRNA vaccine response. Commun Med. (2024) 4:63. doi: 10.1038/s43856-024-00490-2
21. Maruggi G, Ulmer JB, Rappuoli R, Yu D. Self-amplifying mRNA-based vaccine technology and its mode of action. Cham: Springer International Publishing (2021) p. 31–70.
22. Brader ML, Williams SJ, Banks JM, Hui WH, Zhou ZH, Jin L. Encapsulation state of messenger RNA inside lipid nanoparticles. Biophys J. (2021) 120:2766–70. doi: 10.1016/j.bpj.2021.03.012
23. Firdessa-Fite R, Creusot RJ. Nanoparticles versus dendritic cells as vehicles to deliver mRNA encoding multiple epitopes for immunotherapy. Mol Therapy-Methods Clin Dev. (2020) 16:50–62. doi: 10.1016/j.omtm.2019.10.015
24. Kuhn NA, Beissert T, Simon P, Vallazza B, Buck J, Davies BP, et al. mRNA as a versatile tool for exogenous protein expression. Curr Gene Ther. (2012) 12:347–61. doi: 10.2174/156652312802762536
25. Li Y, Wang M, Peng X, Yang Y, Chen Q, Liu J, et al. mRNA vaccine in cancer therapy: Current advance and future outlook. Clin Trans Med. (2023) 13:e1384. doi: 10.1002/ctm2.v13.8
26. Kim SC, Sekhon SS, Shin WR, Ahn G, Cho BK, Ahn JY, et al. Modifications of mRNA vaccine structural elements for improving mRNA stability and translation efficiency. Mol Cell toxicol. (2022) 1:1–8. doi: 10.1007/s13273-021-00171-4
27. Nazli O, Bozdag AD, Tansug T, Kir R, Kaymak E. The diagnostic importance of CEA and CA 19-9 for the early diagnosis of pancreatic carcinoma. Hepato-gastroenterology. (2000) 47:1750–2.
28. Satake K, Chung YS, Yokomatsu H, Nakata B, Tanaka H, Sawada T, et al. A clinical evaluation of various tumor markers for the diagnosis of pancreatic cancer. Int J pancreatol. (1990) 7:25–36. doi: 10.1007/BF02924217
29. Imaoka H, Mizuno N, Hara K, Hijioka S, Tajika M, Tanaka T, et al. Prognostic impact of carcinoembryonic antigen (CEA) on patients with metastatic pancreatic cancer: A retrospective cohort study. Pancreatology. (2016) 16(5):859–64.
30. Omar N, Yan B, Salto-Tellez M. HER2: An emerging biomarker in non-breast and non-gastric cancers. Pathogenesis. (2015) 2:1–9. doi: 10.1016/j.pathog.2015.05.002
31. Bryant KL, Mancias JD, Kimmelman AC, Der CJ. KRAS: feeding pancreatic cancer proliferation. Trends Biochem Sci. (2014) 39(2):91–100. doi: 10.1016/j.tibs.2013.12.004
32. Eser S, Schnieke A, Schneider G, Saur D. Oncogenic KRAS signalling in pancreatic cancer. Br J Cancer. (2014) 111(5):817–22. doi: 10.1038/bjc.2014.215
33. Ordóñez NG. Application of mesothelin immunostaining in tumor diagnosis. Am J Surg pathol. (2003) 27:1418–28. doi: 10.1097/00000478-200311000-00003
34. Argani P, Iacobuzio-Donahue C, Ryu B, Rosty C, Goggins M, Wilentz RE, et al. Mesothelin is overexpressed in the vast majority of ductal adenocarcinomas of the pancreas: identification of a new pancreatic cancer marker by serial analysis of gene expression (SAGE). Clin Cancer Res. (2001) 7:3862–8.
35. Qu CF, Li Y, Song YJ, Rizvi SM, Raja C, Zhang D, et al. MUC1 expression in primary and metastatic pancreatic cancer cells for in vitro treatment by 213Bi-C595 radioimmunoconjugate. Br J cancer. (2004) 91:2086–93. doi: 10.1038/sj.bjc.6602232
36. Li D, Xie K, Wolff R, Abbruzzese JL. Pancreatic cancer. Lancet. (2004) 363:1049–57. doi: 10.1016/S0140-6736(04)15841-8
37. Mello SS, Flowers BM, Mazur PK, Lee JJ, Müller F, Denny SK, et al. Multifaceted role for p53 in pancreatic cancer suppression. Proc Natl Acad Sci. (2023) 120(10):e2211937120.
38. Konduri S, Colon J, Baker CH, Safe S, Abbruzzese JL, Abudayyeh A, et al. Tolfenamic acid enhances pancreatic cancer cell and tumor response to radiation therapy by inhibiting survivin protein expression. Mol Cancer Ther. (2009) 8:533–42. doi: 10.1158/1535-7163.MCT-08-0405
39. Seki K, Suda T, Aoyagi Y, Sugawara S, Natsui M, Motoyama H, et al. Diagnosis of pancreatic adenocarcinoma by detection of human telomerase reverse transcriptase messenger RNA in pancreatic juice with sample qualification. Clin Cancer Res. (2001) 7:1976–81.
40. Doi Y, Yashiro M, Yamada N, Amano R, Noda S, Hirakawa K. VEGF-A/VEGFR-2 signaling plays an important role for the motility of pancreas cancer cells. Ann Surg Oncol. (2012) 19:2733–43. doi: 10.1245/s10434-011-2181-6
41. Costache MI, Ioana M, Iordache S, Ene D, Costache CA, Săftoiu A. VEGF expression in pancreatic cancer and other malignancies: a review of the literature. Romanian J Internal Med. (2015) 53(3):199–208.
42. Davis L, Tarduno A, Lu YC. Neoantigen-reactive T cells: the driving force behind successful melanoma immunotherapy. Cancers. (2021) 13:6061. doi: 10.3390/cancers13236061
43. Berset M, Cerottini JP, Guggisberg D, Romero P, Burri F, Rimoldi D, et al. Expression of melan-a/MART-1 antigen as a prognostic factor in primary cutaneous melanoma. Int J cancer. (2001) 95:73–7. doi: 10.1002/1097-0215(20010120)95:1<73::AID-IJC1013>3.0.CO;2-S
44. Fernandes BF, Odashiro AN, Saraiva VS, Logan P, Antecka E, Burnier MN Jr. Immunohistochemical expression of melan-A and tyrosinase in uveal melanoma. J carcinogen. (2007) 6:6. doi: 10.1186/1477-3163-6-6
45. Ohsie SJ, Sarantopoulos GP, Cochran AJ, Binder SW. Immunohistochemical characteristics of melanoma. J cutaneous pathol. (2008) 35:433–44. doi: 10.1111/j.1600-0560.2007.00891.x
46. Mann JE, Hasson N, Su DG, Adeniran AJ, Smalley KS, Djureinovic D, et al. GP100 expression is variable in intensity in melanoma. Cancer Immunology Immunother. (2024) 73(10):191.
47. Barrow C, Browning J, MacGregor D, Davis ID, Sturrock S, Jungbluth AA, et al. Tumor antigen expression in melanoma varies according to antigen and stage. Clin Cancer Res. (2006) 12:764–71. doi: 10.1158/1078-0432.CCR-05-1544
48. Giavina-Bianchi MH, Giavina-Bianchi PF, Festa C. Melanoma: tumor microenvironment and new treatments. Anais brasileiros dermatol. (2017) 92:156–66. doi: 10.1590/abd1806-4841.20176183
49. Ruault M, van der Bruggen P, Brun ME, Boyle S, Roizès G, Sario AD. New BAGE (B melanoma antigen) genes mapping to the juxtacentromeric regions of human chromosomes 13 and 21 have a cancer/testis expression profile. Eur J Hum Genet. (2002) 10:833–40. doi: 10.1038/sj.ejhg.5200891
50. Hugdahl E, Kalvenes MB, Mannelqvist M, Ladstein RG, Akslen LA. Prognostic impact and concordance of TERT promoter mutation and protein expression in matched primary and metastatic cutaneous melanoma. Br J cancer. (2018) 118:98–105. doi: 10.1038/bjc.2017.384
51. Shah S, Chen B. Testing for HER2 in breast cancer: a continuing evolution. Pathol Res Int. (2011) 2011:903202. doi: 10.4061/2011/903202
52. Bilous M, Ades C, Armes J, Bishop J, Brown R, Cooke B, et al. Predicting the HER2 status of breast cancer from basic histopathology data: an analysis of 1500 breast cancers as part of the HER2000 International Study. breast. (2003) 12(2):92–8.
53. Siroy A, Abdul-Karim FW, Miedler J, Fong N, Fu P, Gilmore H, et al. MUC1 is expressed at high frequency in early-stage basal-like triple-negative breast cancer. Hum pathol. (2013) 44:2159–66. doi: 10.1016/j.humpath.2013.04.010
54. Jansen K, Kornfeld L, Lennartz M, Blessin NC, Rico S, Kind S, et al. 2355P Carcinoembryonic antigen (CEA) expression in human tumors: A tissue microarray study on 15,413 tumors. Ann Oncol. (2023) 34:S1197. doi: 10.1016/j.annonc.2023.09.1383
55. Sugita Y, Wada H, Fujita S, Nakata T, Sato S, Noguchi Y, et al. NY-ESO-1 expression and immunogenicity in Malignant and benign breast tumors. Cancer Res. (2004) 64:2199–204. doi: 10.1158/0008-5472.CAN-03-3070
56. Raghavendra A, Kalita-de Croft P, Vargas AC, Smart CE, Simpson PT, Saunus JM, et al. Expression of MAGE-A and NY-ESO-1 cancer/testis antigens is enriched in triple-negative invasive breast cancers. Histopathology. (2018) 73:68–80. doi: 10.1111/his.2018.73.issue-1
57. Situ D, Wang J, Ma Y, Zhu Z, Hu Y, Long H, et al. Expression and prognostic relevance of MUC1 in stage IB non-small cell lung cancer. Med Oncol. (2011) 28:596–604. doi: 10.1007/s12032-010-9752-4
58. Bag AK, Schultz AR, Goala P, Karreth FA, Adeegbe DO. Investigating the role of KLRG1 in regulatory T-cells and implications for anti-tumor immunity in Non-small Cell Lung Cancer. J Immunol. (2021) 206:57–02. doi: 10.4049/jimmunol.206.Supp.57.02
59. Chen Y, Shen L, Chen B, Han X, Yu Y, Yuan X, et al. The predictive prognostic values of CBFA2T3, STX3, DENR, EGLN1, FUT4, and PCDH7 in lung cancer. Ann Trans Med. (2021) 9(10):843. doi: 10.21037/atm-21-1392
60. Lynch TJ, Bell DW, Sordella R, Gurubhagavatula S, Okimoto RA, Brannigan BW, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non–small-cell lung cancer to gefitinib. New Engl J Med. (2004) 350:2129–39. doi: 10.1056/NEJMoa040938
61. Al Olayan A, Al Hussaini H, Jazieh AR. The roles of epidermal growth factor receptor (EGFR) inhibitors in the management of lung cancer. J infect Public Health. (2012) 5:S50–60. doi: 10.1016/j.jiph.2012.09.004
62. Reita D, Pabst L, Pencreach E, Guérin E, Dano L, Rimelen V, et al. Direct targeting KRAS mutation in non-small cell lung cancer: focus on resistance. Cancers. (2022) 14:1321. doi: 10.3390/cancers14051321
63. Baqar AR, Wilkins S, Staples M, Lee CH, Oliva K, McMurrick P. The role of preoperative CEA in the management of colorectal cancer: A cohort study from two cancer centres. Int J Surge. (2019) 64:10–5. doi: 10.1016/j.ijsu.2019.02.014
64. Betge J, Schneider NI, Harbaum L, Pollheimer MJ, Lindtner RA, Kornprat P, et al. MUC1, MUC2, MUC5AC, and MUC6 in colorectal cancer: expression profiles and clinical significance. Virchows Archiv. (2016) 469:255–65. doi: 10.1007/s00428-016-1970-5
65. Galizia G, Lieto E, Ferraraccio F, De Vita F, Castellano P, Orditura M, et al. Prognostic significance of epidermal growth factor receptor expression in colon cancer patients undergoing curative surgery. Ann Surg Oncol. (2006) 13:823–35. doi: 10.1245/ASO.2006.05.052
66. Dinu D, Dobre M, Panaitescu E, Bîrlă R, Iosif C, Hoara P, et al. Prognostic significance of KRAS gene mutations in colorectal cancer-preliminary study. J Med Life. (2014) 7:581.
67. Awad H, Saleh T, Alhesa A, Al Shboul S, Yousef R, Obeid A, et al. Beta Catenin Expression in Colorectal Carcinoma and its relation to survival and prognostic factors. Ann Cancer Res Ther. (2024) 32:10–6. doi: 10.4993/acrt.32.10
68. Al-Maghrabi H, Al-Mansouri Z, Al-Maghrabi J. Survivin expression is associated with lymph node metastasis and short survival in patients with colorectal adenocarcinoma. Int J Clin Exp Pathol. (2024) 17:39. doi: 10.62347/ZCUD7995
69. Shia J, Klimstra DS, Nitzkorski JR, Low PS, Gonen M, Landmann R, et al. Immunohistochemical expression of folate receptor α in colorectal carcinoma: patterns and biological significance. Hum pathol. (2008) 39:498–505. doi: 10.1016/j.humpath.2007.09.013
70. Snook AE, Magee MS, Waldman SA. GUCY2C-targeted cancer immunotherapy: past, present and future. Immunol Res. (2011) 51:161–9. doi: 10.1007/s12026-011-8253-7
71. Yao R, Xie C, Xia X. Recent progress in mRNA cancer vaccines. Hum Vaccines Immunotherapeutics. (2024) 20(1):2307187.
72. Zhang L. Multi-epitope vaccines: a promising strategy against tumors and viral infections. Cell Mol Immunol. (2018) 15:182–4. doi: 10.1038/cmi.2017.92
73. Esprit A, de Mey W, Bahadur Shahi R, Thielemans K, Franceschini L, Breckpot K. Neo-antigen mRNA vaccines. Vaccines. (2020) 8:776. doi: 10.3390/vaccines8040776
74. Loomis KH, Lindsay KE, Zurla C, Bhosle SM, Vanover DA, Blanchard EL, et al. In vitro transcribed mRNA vaccines with programmable stimulation of innate immunity. Bioconjugate Chem. (2018) 29:3072–83. doi: 10.1021/acs.bioconjchem.8b00443
75. Abe N, Matsumoto K, Nishihara M, Nakano Y, Shibata A, Maruyama H, et al. Rolling circle translation of circular RNA in living human cells. Sci Rep. (2015) 5:16435. doi: 10.1038/srep16435
76. Kon E, Elia U, Peer D. Principles for designing an optimal mRNA lipid nanoparticle vaccine. Curr Opin Biotechnol. (2022) 73:329–36. doi: 10.1016/j.copbio.2021.09.016
77. Li B, Luo X, Dong Y. Effects of chemically modified messenger RNA on protein expression. Bioconjugate Chem. (2016) 27:849–53. doi: 10.1021/acs.bioconjchem.6b00090
78. Schwartz S, Bernstein DA, Mumbach MR, Jovanovic M, Herbst RH, León-Ricardo BX, et al. Transcriptome-wide mapping reveals widespread dynamic-regulated pseudouridylation of ncRNA and mRNA. Cell. (2014) 159:148–62. doi: 10.1016/j.cell.2014.08.028
79. Uddin MB, Wang Z, Yang C. Dysregulations of functional RNA modifications in cancer, cancer stemness and cancer therapeutics. Theranostics. (2020) 10:3164. doi: 10.7150/thno.41687
80. Kierzek E, Malgowska M, Lisowiec J, Turner DH, Gdaniec Z, Kierzek R. The contribution of pseudouridine to stabilities and structure of RNAs. Nucleic Acids Res. (2014) 42:3492–501. doi: 10.1093/nar/gkt1330
81. Karikó K, Muramatsu H, Welsh FA, Ludwig J, Kato H, Akira S, et al. Incorporation of pseudouridine into mRNA yields superior nonimmunogenic vector with increased translational capacity and biological stability. Mol Ther. (2008) 16:1833–40. doi: 10.1038/mt.2008.200
82. Anderson BR, Muramatsu H, Jha BK, Silverman RH, Weissman D, Karikó K. Nucleoside modifications in RNA limit activation of 2′-5′-oligoadenylate synthetase and increase resistance to cleavage by RNase L. Nucleic Acids Res. (2011) 39:9329–38. doi: 10.1093/nar/gkr586
83. Anderson BR, Muramatsu H, Nallagatla SR, Bevilacqua PC, Sansing LH, Weissman D, et al. Incorporation of pseudouridine into mRNA enhances translation by diminishing PKR activation. Nucleic Acids Res. (2010) 38:5884–92. doi: 10.1093/nar/gkq347
84. Nallagatla SR, Bevilacqua PC. Nucleoside modifications modulate activation of the protein kinase PKR in an RNA structure-specific manner. Rna. (2008) 14:1201–13. doi: 10.1261/rna.1007408
85. Svitkin YV, Cheng YM, Chakraborty T, Presnyak V, John M, Sonenberg N. N1-methyl-pseudouridine in mRNA enhances translation through eIF2α-dependent and independent mechanisms by increasing ribosome density. Nucleic Acids Res. (2017) 45:6023–36. doi: 10.1093/nar/gkx135
86. Parr CJ, Wada S, Kotake K, Kameda S, Matsuura S, Sakashita S, et al. N 1-Methylpseudouridine substitution enhances the performance of synthetic mRNA switches in cells. Nucleic Acids Res. (2020) 48:e35. doi: 10.1093/nar/gkaa070
87. Karikó K, Muramatsu H, Keller JM, Weissman D. Increased erythropoiesis in mice injected with submicrogram quantities of pseudouridine-containing mRNA encoding erythropoietin. Mol Ther. (2012) 20:948–53. doi: 10.1038/mt.2012.7
88. Boros G, Miko E, Muramatsu H, Weissman D, Emri E, Rózsa D, et al. Transfection of pseudouridine-modified mRNA encoding CPD-photolyase leads to repair of DNA damage in human keratinocytes: A new approach with future therapeutic potential. J Photochem Photobiol B: Biol. (2013) 129:93–9. doi: 10.1016/j.jphotobiol.2013.09.010
89. Karikó K, Buckstein M, Ni H, Weissman D. Suppression of RNA recognition by Toll-like receptors: the impact of nucleoside modification and the evolutionary origin of RNA. Immunity. (2005) 23:165–75. doi: 10.1016/j.immuni.2005.06.008
90. Andries O, Mc Cafferty S, De Smedt SC, Weiss R, Sanders NN, Kitada T. N1-methylpseudouridine-incorporated mRNA outperforms pseudouridine-incorporated mRNA by providing enhanced protein expression and reduced immunogenicity in mammalian cell lines and mice. J Controlled Release. (2015) 217:337–44. doi: 10.1016/j.jconrel.2015.08.051
91. Nance KD, Meier JL. Modifications in an emergency: the role of N1-methylpseudouridine in COVID-19 vaccines. ACS Cent sci. (2021) 7:748–56. doi: 10.1021/acscentsci.1c00197
92. Freund I, Eigenbrod T, Helm M, Dalpke AH. RNA modifications modulate activation of innate toll-like receptors. Genes. (2019) 10:92. doi: 10.3390/genes10020092
93. Mauger DM, Cabral BJ, Presnyak V, Su SV, Reid DW, Goodman B, et al. mRNA structure regulates protein expression through changes in functional half-life. Proc Natl Acad Sci. (2019) 116:24075–83. doi: 10.1073/pnas.1908052116
94. Gao Y, Fang J. RNA 5-methylcytosine modification and its emerging role as an epitranscriptomic mark. RNA Biol. (2021) 18:117–27. doi: 10.1080/15476286.2021.1950993
95. Song H, Zhang J, Liu B, Xu J, Cai B, Yang H, et al. Biological roles of RNA m5C modification and its implications in Cancer immunotherapy. biomark Res. (2022) 10:15. doi: 10.1186/s40364-022-00362-8
96. Weng Y, Li C, Yang T, Hu B, Zhang M, Guo S, et al. The challenge and prospect of mRNA therapeutics landscape. Biotechnol adv. (2020) 40:107534. doi: 10.1016/j.biotechadv.2020.107534
97. Xue C, Zhao Y, Li L. Advances in RNA cytosine-5 methylation: detection, regulatory mechanisms, biological functions and links to cancer. biomark Res. (2020) 8:1–3. doi: 10.1186/s40364-020-00225-0
98. Kormann MS, Hasenpusch G, Aneja MK, Nica G, Flemmer AW, Herber-Jonat S, et al. Expression of therapeutic proteins after delivery of chemically modified mRNA in mice. Nat Biotechnol. (2011) 29:154–7. doi: 10.1038/nbt.1733
99. Zhang T, Zhao F, Li J, Sun X, Zhang X, Wang H, et al. Programmable RNA 5-methylcytosine (m5C) modification of cellular RNAs by dCasRx conjugated methyltransferase and demethylase. Nucleic Acids Res. (2024) 52:2776–91. doi: 10.1093/nar/gkae110
100. Desrosiers R, Friderici K, Rottman F. Identification of methylated nucleosides in messenger RNA from Novikoff hepatoma cells. Proc Natl Acad Sci. (1974) 71:3971–5. doi: 10.1073/pnas.71.10.3971
101. Wang S, Lv W, Li T, Zhang S, Wang H, Li X, et al. Dynamic regulation and functions of mRNA m6A modification. Cancer Cell Int. (2022) 22:48. doi: 10.1186/s12935-022-02452-x
102. Lin S, Choe J, Du P, Triboulet R, Gregory RI. The m6A methyltransferase METTL3 promotes translation in human cancer cells. Mol Cell. (2016) 62:335–45. doi: 10.1016/j.molcel.2016.03.021
103. Roy B. Effects of mRNA modifications on translation: an overview. RNA Modif: Methods Protoc. (2021) 2298:327–56. doi: 10.1007/978-1-0716-1374-0_20
104. Karikó K, Muramatsu H, Ludwig J, Weissman D. Generating the optimal mRNA for therapy: HPLC purification eliminates immune activation and improves translation of nucleoside-modified, protein-encoding mRNA. Nucleic Acids Res. (2011) 39:e142. doi: 10.1093/nar/gkr695
105. Liu A, Wang X. The pivotal role of chemical modifications in mRNA therapeutics. Front Cell Dev Biol. (2022) 10:901510. doi: 10.3389/fcell.2022.901510
106. Melamed JR, Hajj KA, Chaudhary N, Strelkova D, Arral ML, Pardi N, et al. Lipid nanoparticle chemistry determines how nucleoside base modifications alter mRNA delivery. J Controlled Release. (2022) 341:206–14. doi: 10.1016/j.jconrel.2021.11.022
107. Phan T, Fan D, Melstrom LG. Developing Vaccines in Pancreatic Adenocarcinoma: Trials and Tribulations. Current Oncology. (2024) 12:5746–63.
108. Xu S, Yang K, Li R, Zhang L. mRNA vaccine era—mechanisms, drug platform and clinical prospection. Int J Mol Sci. (2020) 21:6582. doi: 10.3390/ijms21186582
109. Shuman S. The mRNA capping apparatus as drug target and guide to eukaryotic phylogeny. In: Cold Spring Harbor symposia on quantitative biology, vol. 66. Cold Spring Harbor, New York, USA: Cold Spring Harbor Laboratory Press (2001). 66:301–12.
110. Gu M, Lima CD. Processing the message: structural insights into capping and decapping mRNA. Curr Opin Struct Biol. (2005) 15:99–106. doi: 10.1016/j.sbi.2005.01.009
111. Lewis JD, Izaurflde E. The role of the cap structure in RNA processing and nuclear export. Eur J Biochem. (1997) 247:461–9. doi: 10.1111/j.1432-1033.1997.00461.x
112. Furuichi Y, Shatkin AJ. Viral and cellular mRNA capping: past and prospects. Advances in virus research. (2000) 55:135–84. doi: 10.1016/S0065-3527(00)55003-9
113. Sonenberg N, Hinnebusch AG. Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell. (2009) 136:731–45. doi: 10.1016/j.cell.2009.01.042
114. Shuman S. What messenger RNA capping tells us about eukaryotic evolution. Nat Rev Mol Cell Biol. (2002) 3:619–25. doi: 10.1038/nrm880
115. Wang SP, Deng L, Ho CK, Shuman S. Phylogeny of mRNA capping enzymes. Proc Natl Acad Sci. (1997) 94:9573–8. doi: 10.1073/pnas.94.18.9573
116. Shatkin AJ. Capping of eucaryotic mRNAs. Cell. (1976) 9:645–53. doi: 10.1016/0092-8674(76)90128-8
117. Shatkin AJ, Manley JL. The ends of the affair: capping and polyadenylation. Nat Struct Biol. (2000) 7:838–42. doi: 10.1038/79583
118. Daffis S, Szretter KJ, Schriewer J, Li J, Youn S, Errett J, et al. 2′-O methylation of the viral mRNA cap evades host restriction by IFIT family members. Nature. (2010) 468:452–6. doi: 10.1038/nature09489
119. Park JE, Yi H, Kim Y, Chang H, Kim VN. Regulation of poly (A) tail and translation during the somatic cell cycle. Mol Cell. (2016) 62:462–71. doi: 10.1016/j.molcel.2016.04.007
120. Jalkanen AL, Coleman SJ, Wilusz J. Determinants and implications of mRNA poly (A) tail size–does this protein make my tail look big? In: Seminars in cell & developmental biology, vol. 34. Cambridge, Massachusetts, USA: Academic Press (2014). 36:24–32. doi: 10.1016/j.semcdb.2014.05.018
121. Kieleczawa J. Fundamentals of sequencing of difficult templates—an overview. J biomol tech: JBT. (2006) 17:207.
122. Diebold SS, Kaisho T, Hemmi H, Akira S, Reis e Sousa C. Innate antiviral responses by means of TLR7-mediated recognition of single-stranded RNA. Science. (2004) 303:1529–31. doi: 10.1126/science.1093616
123. Tanguay RL, Gallie DR. Translational efficiency is regulated by the length of the 3′ untranslated region. Mol Cell Biol. (1996) 16:146–56. doi: 10.1128/MCB.16.1.146
124. Leppek K, Das R, Barna M. Functional 5′ UTR mRNA structures in eukaryotic translation regulation and how to find them. Nat Rev Mol Cell Biol. (2018) 19:158–74. doi: 10.1038/nrm.2017.103
125. Vinhas V, Freire M, Bacellar O, Cunha S, Rocha H, Carvalho EM. Characterization of T cell responses to purified leishmania antigens in subjects infected with Leishmania chagasi. Braz J Med Biol Research= Rev Bras Pesquisas Medicas e Biol. (1994) 27:1199–205.
126. Troye-Blomberg M, Olerup O, Perlmann H, Larsson A, Elghazali G, Fogdell A, et al. Characterization of regulatory T-cell responses in humans induced by the P. Falciparum blood stage antigen Pf155/RESA. Behring Inst Mitt. (1994) 95:97–105.
127. Yu J, Russell JE. Structural and functional analysis of an mRNP complex that mediates the high stability of human beta-globin mRNA. Mol Cell Biol. (2001) 21(17):5879–5888. doi: 10.1128/MCB.21.17.5879-5888.2001
128. Wang Z, Kiledjian M. The poly (A)-binding protein and an mRNA stability protein jointly regulate an endoribonuclease activity. Mol Cell Biol. (2000) 20(17):6334–41. doi: 10.1128/.20.17.6334-6341.2000
129. Zhang J, Liu Y, Li C, Xiao Q, Zhang D, Chen Y, et al. Recent advances and innovations in the preparation and purification of in Vitro-Transcribed-mRNA-Based molecules. Pharmaceutics. (2023) 15:2182. doi: 10.3390/pharmaceutics15092182
130. Beckert B, Masquida B. Synthesis of RNA by in vitro transcription. RNA: methods and protocols. Methods Mol Biol (Clifton, NJ). (2011) 703:29–41. doi: 10.1007/978-1-59745-248-9_3
131. Thiel V, Herold J, Schelle B, Siddell SG. Infectious RNA transcribed in vitro from a cDNA copy of the human coronavirus genome cloned in vaccinia virus. J Gen Virol. (2001) 82:1273–81. doi: 10.1099/0022-1317-82-6-1273
132. Pascolo S. Messenger RNA-based vaccines. Expert Opin Biol Ther. (2004) 4:1285–94. doi: 10.1517/14712598.4.8.1285
133. Iavarone C, O’hagan DT, Yu D, Delahaye NF, Ulmer JB. Mechanism of action of mRNA-based vaccines. Expert Rev Vaccines. (2017) 16:871–81. doi: 10.1080/14760584.2017.1355245
134. Pardi N, Hogan MJ, Porter FW, Weissman D. mRNA vaccines—a new era in vaccinology. Nat Rev Drug discover. (2018) 17:261–79. doi: 10.1038/nrd.2017.243
135. Tenchov R, Bird R, Curtze AE, Zhou Q. Lipid nanoparticles─from liposomes to mRNA vaccine delivery, a landscape of research diversity and advancement. ACS nano. (2021) 15:16982–7015. doi: 10.1021/acsnano.1c04996
136. Wei J, Hui A. Review of ribosome interactions with SARS-CoV-2 and COVID-19 mRNA vaccine. Life. (2022) 12:57. doi: 10.3390/life12010057
137. Lung P, Yang J, Li Q. Nanoparticle formulated vaccines: opportunities and challenges. Nanoscale. (2020) 12:5746–63.
138. Mei Y, Wang X. RNA modification in mRNA cancer vaccines. Clin Exp Med. (2023) 23:1917–31. doi: 10.1007/s10238-023-01020-5
139. Oberli MA, Reichmuth AM, Dorkin JR, Mitchell MJ, Fenton OS, Jaklenec A, et al. Lipid nanoparticle assisted mRNA delivery for potent cancer immunotherapy. Nano letters. (2017) 17:1326–35. doi: 10.1021/acs.nanolett.6b03329
140. Lee Y, Choe J, Park OH, Kim YK. Molecular mechanisms driving mRNA degradation by m6A modification. Trends Genet. (2020) 36:177–88. doi: 10.1016/j.tig.2019.12.007
141. Glover DJ, Lipps HJ, Jans DA. Towards safe, non-viral therapeutic gene expression in humans. Nat Rev Genet. (2005) 6:299–310. doi: 10.1038/nrg1577
142. Qin M, Du G, Sun X. Recent advances in the noninvasive delivery of mRNA. Accounts Chem Res. (2021) 54:4262–71. doi: 10.1021/acs.accounts.1c00493
143. Butt MH, Zaman M, Ahmad A, Khan R, Mallhi TH, Hasan MM, et al. Appraisal for the potential of viral and nonviral vectors in gene therapy: A review. Genes. (2022) 13:1370. doi: 10.3390/genes13081370
144. Sharma D, Arora S, Singh J, Layek B. A review of the tortuous path of nonviral gene delivery and recent progress. Int J Biol macromol. (2021) 183:2055–73. doi: 10.1016/j.ijbiomac.2021.05.192
145. Li X, Le Y, Zhang Z, Nian X, Liu B, Yang X. Viral vector-based gene therapy. Int J Mol Sci. (2023) 24:7736. doi: 10.3390/ijms24097736
146. Travieso T, Li J, Mahesh S, Mello JD, Blasi M. The use of viral vectors in vaccine development. NPJ Vaccines. (2022) 7:75. doi: 10.1038/s41541-022-00503-y
147. Wang Y, Bruggeman KF, Franks S, Gautam V, Hodgetts SI, Harvey AR, et al. Is viral vector gene delivery more effective using biomaterials? Adv Healthc Mater. (2021) 10:2001238. doi: 10.1002/adhm.202001238
148. Kowalski PS, Rudra A, Miao L, Anderson DG. Delivering the messenger: advances in technologies for therapeutic mRNA delivery. Mol Ther. (2019) 27:710–28. doi: 10.1016/j.ymthe.2019.02.012
149. Yang W, Mixich L, Boonstra E, Cabral H. Polymer-based mRNA delivery strategies for advanced therapies. Adv Healthc Mater. (2023) 12:2202688. doi: 10.1002/adhm.202202688
150. Barenholz YC. Doxil®—The first FDA-approved nano-drug: Lessons learned. J Controlled release. (2012) 160:117–34. doi: 10.1016/j.jconrel.2012.03.020
151. Sun D, Lu ZR. Structure and function of cationic and ionizable lipids for nucleic acid delivery. Pharm Res. (2023) 40:27–46. doi: 10.1007/s11095-022-03460-2
152. Liu X, Huang P, Yang R, Deng H. mRNA Cancer vaccines: construction and boosting strategies. ACS nano. (2023) 17:19550–80. doi: 10.1021/acsnano.3c05635
153. Karlsson J, Rhodes KR, Green JJ, Tzeng SY. Poly (beta-amino ester) s as gene delivery vehicles: challenges and opportunities. Expert Opin Drug delivery. (2020) 17:1395–410. doi: 10.1080/17425247.2020.1796628
154. Li M, Li Y, Peng K, Wang Y, Gong T, Zhang Z, et al. Engineering intranasal mRNA vaccines to enhance lymph node trafficking and immune responses. Acta biomaterialia. (2017) 64:237–48. doi: 10.1016/j.actbio.2017.10.019
155. McKinlay CJ, Vargas JR, Blake TR, Hardy JW, Kanada M, Contag CH, et al. Charge-altering releasable transporters (CARTs) for the delivery and release of mRNA in living animals. Proc Natl Acad Sci. (2017) 114:E448–56. doi: 10.1073/pnas.1614193114
156. Haabeth OA, Lohmeyer JJ, Sallets A, Blake TR, Sagiv-Barfi I, Czerwinski DK, et al. An mRNA SARS-CoV-2 vaccine employing charge-altering releasable transporters with a TLR-9 agonist induces neutralizing antibodies and T cell memory. ACS Cent Sci. (2021) 7:1191–204. doi: 10.1021/acscentsci.1c00361
157. Wang Y, Zhang L, Xu Z, Miao L, Huang L. mRNA vaccine with antigen-specific checkpoint blockade induces an enhanced immune response against established melanoma. Mol Ther. (2018) 26:420–34. doi: 10.1016/j.ymthe.2017.11.009
158. Xu Z, Fisher DE. mRNA melanoma vaccine revolution spurred by the COVID-19 pandemic. Front Immunol. (2023) 14:1155728. doi: 10.3389/fimmu.2023.1155728
159. Sharifnia Z, Bandehpour M, Hamishehkar H, Mosaffa N, Kazemi B, Zarghami N. In-vitro transcribed mRNA delivery using PLGA/PEI nanoparticles into human monocyte-derived dendritic cells. Iranian J Pharm Res: IJPR. (2019) 18:1659. doi: 10.22037/ijpr.2019.1100872
160. Patel S, Ashwanikumar N, Robinson E, DuRoss A, Sun C, Murphy-Benenato KE, et al. Boosting intracellular delivery of lipid nanoparticle-encapsulated mRNA. Nano letters. (2017) 17:5711–8. doi: 10.1021/acs.nanolett.7b02664
161. Xiao Y, Shi J. Lipids and the emerging RNA medicines. Chem Rev. (2021) 121:12109–11. doi: 10.1021/acs.chemrev.1c00778
162. Schoenmaker L, Witzigmann D, Kulkarni JA, Verbeke R, Kersten G, Jiskoot W, et al. mRNA-lipid nanoparticle COVID-19 vaccines: Structure and stability. Int J pharm. (2021) 601:120586. doi: 10.1016/j.ijpharm.2021.120586
163. Kulkarni JA, Cullis PR, van der Meel R. Lipid nanoparticles enabling gene therapies: from concepts to clinical utility. Nucleic Acid Ther. (2018) 28:146–57. doi: 10.1089/nat.2018.0721
164. Kulkarni JA, Witzigmann D, Chen S, Cullis PR, van der Meel R. Lipid nanoparticle technology for clinical translation of siRNA therapeutics. Accounts Chem Res. (2019) 52:2435–44. doi: 10.1021/acs.accounts.9b00368
165. Tilstra G, Couture-Senécal J, Lau YM, Manning AM, Wong DS, Janaeska WW, et al. Iterative design of ionizable lipids for intramuscular mRNA delivery. J Am Chem Society. (2023) 145:2294–304. doi: 10.1021/jacs.2c10670
166. Kim M, Jeong M, Hur S, Cho Y, Park J, Jung H, et al. Engineered ionizable lipid nanoparticles for targeted delivery of RNA therapeutics into different types of cells in the liver. Sci Adv. (2021) 7:eabf4398. doi: 10.1126/sciadv.abf4398
167. Tang X, Zhang Y, Han X. Ionizable lipid nanoparticles for mRNA delivery. Adv NanoBiomed Res. (2023) 3:2300006. doi: 10.1002/anbr.202300006
168. Semple SC, Akinc A, Chen J, Sandhu AP, Mui BL, Cho CK, et al. Rational design of cationic lipids for siRNA delivery. Nat Biotechnol. (2010) 28:172–6. doi: 10.1038/nbt.1602
169. Jayaraman M, Ansell SM, Mui BL, Tam YK, Chen J, Du X, et al. Maximizing the potency of siRNA lipid nanoparticles for hepatic gene silencing in vivo. Angewandte Chemie. (2012) 124:8657–61. doi: 10.1002/ange.201203263
170. Dong Y, Love KT, Dorkin JR, Sirirungruang S, Zhang Y, Chen D, et al. Lipopeptide nanoparticles for potent and selective siRNA delivery in rodents and nonhuman primates. Proc Natl Acad Sci. (2014) 111:3955–60. doi: 10.1073/pnas.1322937111
171. Maier MA, Jayaraman M, Matsuda S, Liu J, Barros S, Querbes W, et al. Biodegradable lipids enabling rapidly eliminated lipid nanoparticles for systemic delivery of RNAi therapeutics. Mol Ther. (2013) 21:1570–8. doi: 10.1038/mt.2013.124
172. Hafez IM, Maurer N, Cullis PR. On the mechanism whereby cationic lipids promote intracellular delivery of polynucleic acids. Gene Ther. (2001) 8:1188–96. doi: 10.1038/sj.gt.3301506
173. Sabnis S, Kumarasinghe ES, Salerno T, Mihai C, Ketova T, Senn JJ, et al. A novel amino lipid series for mRNA delivery: improved endosomal escape and sustained pharmacology and safety in non-human primates. Mol Ther. (2018) 26:1509–19. doi: 10.1016/j.ymthe.2018.03.010
174. Hou X, Zaks T, Langer R, Dong Y. Lipid nanoparticles for mRNA delivery. Nat Rev Mater. (2021) 6:1078–94. doi: 10.1038/s41578-021-00358-0
175. Guevara ML, Persano F, Persano S. Advances in lipid nanoparticles for mRNA-based cancer immunotherapy. Front Chem. (2020) 8:589959. doi: 10.3389/fchem.2020.589959
176. Kauffman KJ, Dorkin JR, Yang JH, Heartlein MW, DeRosa F, Mir FF, et al. Optimization of lipid nanoparticle formulations for mRNA delivery in vivo with fractional factorial and definitive screening designs. Nano letters. (2015) 15:7300–6. doi: 10.1021/acs.nanolett.5b02497
177. Fenton OS, Kauffman KJ, McClellan RL, Kaczmarek JC, Zeng MD, Andresen JL, et al. Customizable lipid nanoparticle materials for the delivery of siRNAs and mRNAs. Angewandte Chemie Int Edition. (2018) 57:13582–6. doi: 10.1002/anie.201809056
178. Eygeris Y, Patel S, Jozic A, Sahay G. Deconvoluting lipid nanoparticle structure for messenger RNA delivery. Nano letters. (2020) 20:4543–9. doi: 10.1021/acs.nanolett.0c01386
179. Álvarez-Benedicto E, Farbiak L, Ramírez MM, Wang X, Johnson LT, Mian O, et al. Optimization of phospholipid chemistry for improved lipid nanoparticle (LNP) delivery of messenger RNA (mRNA). Biomater sci. (2022) 10:549–59. doi: 10.1039/D1BM01454D
180. Zhang C, Ma Y, Zhang J, Kuo JC, Zhang Z, Xie H, et al. Modification of lipid-based nanoparticles: An efficient delivery system for nucleic acid-based immunotherapy. Molecules. (2022) 27:1943. doi: 10.3390/molecules27061943
181. Larson NR, Hu G, Wei Y, Tuesca AD, Forrest ML, Middaugh CR. pH-dependent phase behavior and stability of cationic lipid–mRNA nanoparticles. J Pharm Sci. (2022) 111:690–8. doi: 10.1016/j.xphs.2021.11.004
182. Zhang L, More KR, Ojha A, Jackson CB, Quinlan BD, Li H, et al. Effect of mRNA-LNP components of two globally-marketed COVID-19 vaccines on efficacy and stability. NPJ Vaccines. (2023) 8:156. doi: 10.1038/s41541-023-00751-6
183. Jung HN, Lee SY, Lee S, Youn H, Im HJ. Lipid nanoparticles for delivery of RNA therapeutics: Current status and the role of in vivo imaging. Theranostics. (2022) 12:7509. doi: 10.7150/thno.77259
184. Mehta M, Bui TA, Yang X, Aksoy Y, Goldys EM, Deng W. Lipid-based nanoparticles for drug/gene delivery: An overview of the production techniques and difficulties encountered in their industrial development. ACS Mater Au. (2023) 3:600–19. doi: 10.1021/acsmaterialsau.3c00032
185. Carrasco MJ, Alishetty S, Alameh MG, Said H, Wright L, Paige M, et al. Ionization and structural properties of mRNA lipid nanoparticles influence expression in intramuscular and intravascular administration. Commun Biol. (2021) 4:956. doi: 10.1038/s42003-021-02441-2
186. Chatterjee S, Kon E, Sharma P, Peer D. Endosomal escape: A bottleneck for LNP-mediated therapeutics. Proc Natl Acad Sci. (2024) 121:e2307800120. doi: 10.1073/pnas.2307800120
187. Albertsen CH, Kulkarni JA, Witzigmann D, Lind M, Petersson K, Simonsen JB. The role of lipid components in lipid nanoparticles for vaccines and gene therapy. Adv Drug delivery Rev. (2022) 188:114416. doi: 10.1016/j.addr.2022.114416
188. Khalil IA, Younis MA, Kimura S, Harashima H. Lipid nanoparticles for cell-specific in vivo targeted delivery of nucleic acids. Biol Pharm Bullet. (2020) 43:584–95. doi: 10.1248/bpb.b19-00743
189. Hassett KJ, Benenato KE, Jacquinet E, Lee A, Woods A, Yuzhakov O, et al. Optimization of lipid nanoparticles for intramuscular administration of mRNA vaccines. Mol Therapy-Nucleic Acids. (2019) 15:1–1. doi: 10.1016/j.omtn.2019.01.013
190. Gyanani V, Goswami R. Key design features of lipid nanoparticles and electrostatic charge-based lipid nanoparticle targeting. Pharmaceutics. (2023) 15:1184. doi: 10.3390/pharmaceutics15041184
191. Dilliard SA, Siegwart DJ. Passive, active and endogenous organ-targeted lipid and polymer nanoparticles for delivery of genetic drugs. Nat Rev Mater. (2023) 8:282–300. doi: 10.1038/s41578-022-00529-7
192. Meyer RA, Neshat SY, Green JJ, Santos JL, Tuesca AD. Targeting strategies for mRNA delivery. Mater Today Adv. (2022) 14:100240. doi: 10.1016/j.mtadv.2022.100240
193. Li M, Huang Y, Wu J, Li S, Mei M, Chen H, et al. A PEG-lipid-free COVID-19 mRNA vaccine triggers robust immune responses in mice. Mater Horizons. (2023) 10:466–72. doi: 10.1039/D2MH01260J
194. Qu Y, Xu J, Zhang T, Chen Q, Sun T, Jiang C. Advanced nano-based strategies for mRNA tumor vaccine. Acta Pharmaceutica Sinica B. (2024) 14(1):170–89. doi: 10.1016/j.apsb.2023.07.025
195. Karati D, Mukherjee S, Prajapati B, Bose A, Paul S, Elossaily GM, et al. A review on lipid-polymer hybrid nanocarriers in cancer. J Drug Delivery Sci Technol. (2024) 29:105827. doi: 10.1016/j.jddst.2024.105827
196. Mukherjee A, Waters AK, Kalyan P, Achrol AS, Kesari S, Yenugonda VM. Lipid–polymer hybrid nanoparticles as a next-generation drug delivery platform: state of the art, emerging technologies, and perspectives. Int J nanomed. (2019) 19:1937–52. doi: 10.2147/IJN.S198353
197. Awadeen RH, Boughdady MF, Zaghloul RA, Elsaed WM, Abu Hashim II, Meshali MM. Formulation of lipid polymer hybrid nanoparticles of the phytochemical Fisetin and its in vivo assessment against severe acute pancreatitis. Sci Rep. (2023) 13:19110. doi: 10.1038/s41598-023-46215-8
198. Gajbhiye KR, Salve R, Narwade M, Sheikh A, Kesharwani P, Gajbhiye V. Lipid polymer hybrid nanoparticles: a custom-tailored next-generation approach for cancer therapeutics. Mol Cancer. (2023) 22:160. doi: 10.1186/s12943-023-01849-0
199. Kliesch L, Delandre S, Gabelmann A, Koch M, Schulze K, Guzmán CA, et al. Lipid–polymer hybrid nanoparticles for mRNA delivery to dendritic cells: impact of lipid composition on performance in different media. Pharmaceutics. (2022) 14:2675. doi: 10.3390/pharmaceutics14122675
200. Mao S, Li S, Zhang Y, Long L, Peng J, Cao Y, et al. A highly efficient needle-free-injection delivery system for mRNA-LNP vaccination against SARS-CoV-2. Nano Today. (2023) 48:101730. doi: 10.1016/j.nantod.2022.101730
201. Liu H, Chen MZ, Payne T, Porter CJ, Pouton CW, Johnston AP. Beyond the endosomal bottleneck: understanding the efficiency of mRNA/LNP delivery. Adv Funct Mater. (2024) 34(39):2404510. doi: 10.1002/adfm.202404510
202. Kauffman KJ, Webber MJ, Anderson DG. Materials for non-viral intracellular delivery of messenger RNA therapeutics. J Control Release. (2016) 240:227–34. doi: 10.1016/j.jconrel.2015.12.032
203. Behr JP. The proton sponge: a trick to enter cells the viruses did not exploit. Chimia. (1997) 51:34. doi: 10.2533/chimia.1997.34
204. Wu Q, Bazzini AA. Translation and mRNA stability control. Annu Rev Biochem. (2023) 92:227–45. doi: 10.1146/annurev-biochem-052621-091808
205. Miao L, Li L, Huang Y, Delcassian D, Chahal J, Han J, et al. Delivery of mRNA vaccines with heterocyclic lipids increases anti-tumor efficacy by STING-mediated immune cell activation. Nat Biotechnol. (2019) 37:1174–85. doi: 10.1038/s41587-019-0247-3
206. He M, Huang L, Hou X, Zhong C, Bachir ZA, Lan M, et al. Efficient ovalbumin delivery using a novel multifunctional micellar platform for targeted melanoma immunotherapy. Int J Pharma. (2019) 560:1–0. doi: 10.1016/j.ijpharm.2019.01.027
207. Neill B, Romero AR, Fenton OS. Advances in nonviral mRNA delivery materials and their application as vaccines for melanoma therapy. ACS Appl Bio Mater. (2023) 7(8):4894–913. doi: 10.1021/acsabm.3c00721
208. Islam MA, Xu Y, Tao W, Ubellacker JM, Lim M, Aum D, et al. Author Correction: Restoration of tumour-growth suppression in vivo via systemic nanoparticle-mediated delivery of PTEN mRNA. Nat Biomed Eng. (2018) 2(12):968. doi: 10.1038/s41551-018-0331-x
209. Xiao Y, Chen J, Zhou H, Zeng X, Ruan Z, Pu Z, et al. Combining p53 mRNA nanotherapy with immune checkpoint blockade reprograms the immune microenvironment for effective cancer therapy. Nat Commun. (2022) 13:758. doi: 10.1038/s41467-022-28279-8
210. Echaide M, Chocarro de Erauso L, Bocanegra A, Blanco E, Kochan G, Escors D. mRNA vaccines against SARS-CoV-2: Advantages and caveats. Int J Mol Sci. (2023) 24:5944. doi: 10.3390/ijms24065944
211. Lorentzen CL, Haanen JB, Met Ö, Svane IM. Clinical advances and ongoing trials of mRNA vaccines for cancer treatment. Lancet Oncol. (2022) 23:e450–8. doi: 10.1016/S1470-2045(22)00372-2
212. Boehm DT, Landreth KM, Sen-Kilic E, Lee KS, Misra B, Bobbala S, et al. Intratumoral administration of mRNA-1273 vaccine delays melanoma growth in mice. bioRxiv. (2024). doi: 10.1101/2024.05.06.592840
213. Curran MA, Montalvo W, Yagita H, Allison JP. PD-1 and CTLA-4 combination blockade expands infiltrating T cells and reduces regulatory T and myeloid cells within B16 melanoma tumors. Proc Natl Acad Sci. (2010) 107:4275–80. doi: 10.1073/pnas.0915174107
214. Kreiter S, Vormehr M, Van de Roemer N, Diken M, Löwer M, Diekmann J, et al. Mutant MHC class II epitopes drive therapeutic immune responses to cancer. Nature. (2015) 520:692–6. doi: 10.1038/nature14426
215. Chen J, Ye Z, Huang C, Qiu M, Song D, Li Y, et al. Lipid nanoparticle-mediated lymph node–targeting delivery of mRNA cancer vaccine elicits robust CD8+ T cell response. Proc Natl Acad Sci. (2022) 119:e2207841119. doi: 10.1073/pnas.2207841119
216. Sharbi-Yunger A, Grees M, Cafri G, Bassan D, Eichmüller SB, Tzehoval E, et al. A universal anti-cancer vaccine: Chimeric invariant chain potentiates the inhibition of melanoma progression and the improvement of survival. Int J cancer. (2019) 144:909–21. doi: 10.1002/ijc.31795
217. Sahin U, Oehm P, Derhovanessian E, Jabulowsky RA, Vormehr M, Gold M, et al. An RNA vaccine drives immunity in checkpoint-inhibitor-treated melanoma. Nature. (2020) 585:107–12. doi: 10.1038/s41586-020-2537-9
218. Kieser RE, Khan S, Bejar N, Kiss DL. The dawning of a new enterprise: RNA therapeutics for the skin. J Dermatol skin sci. (2023) 5:4. doi: 10.29245/2767-5092/2023/1.1168
219. Ugurel S, Röhmel J, Ascierto PA, Flaherty KT, Grob JJ, Hauschild A, et al. Survival of patients with advanced metastatic melanoma: the impact of novel therapies. Eur J cancer. (2016) 53:125–34. doi: 10.1016/j.ejca.2015.09.013
220. Khattak A, Weber JS, Meniawy T, Taylor MH, Ansstas G, Kim KB, et al. Distant metastasis-free survival results from the randomized, phase 2 mRNA-4157-P201/KEYNOTE-942 trial. J Clin Oncol. (2023) 41(17):LBA9503. doi: 10.1200/JCO.2023.41.17_suppl.LBA9503
221. Weide B, Pascolo S, Scheel B, Derhovanessian E, Pflugfelder A, Eigentler TK, et al. Direct injection of protamine-protected mRNA: results of a phase 1/2 vaccination trial in metastatic melanoma patients. J Immunother. (2009) 32:498–507. doi: 10.1097/CJI.0b013e3181a00068
222. Heesen L, Jabulowsky R, Loquai C, Utikal J, Gebhardt C, Hassel J, et al. A first-in-human phase I/II clinical trial assessing novel mRNA-lipoplex nanoparticles encoding shared tumor antigens for potent melanoma immunotherapy. Ann Oncol. (2017) 28:xi14–5. doi: 10.1093/annonc/mdx711.030
223. Clifton GT, Litton JK, Arrington K, Ponniah S, Ibrahim NK, Gall V, et al. Results of a phase Ib trial of combination immunotherapy with a CD8+ T cell eliciting vaccine and trastuzumab in breast cancer patients. Ann Surg Oncol. (2017) 24:2161–7. doi: 10.1245/s10434-017-5844-0
224. Niemi JV, Sokolov AV, Schiöth HB. Neoantigen vaccines; clinical trials, classes, indications, adjuvants and combinatorial treatments. Cancers. (2022) 14:5163.
225. Crosby EJ, Acharya CR, Haddad AF, Rabiola CA, Lei G, Wei JP, et al. Stimulation of oncogene-specific tumor-infiltrating T cells through combined vaccine and αPD-1 enable sustained antitumor responses against established HER2 breast cancer. Clin Cancer Res. (2020) 26:4670–81. doi: 10.1158/1078-0432.CCR-20-0389
226. Jiang XT, Liu Q. mRNA vaccination in breast cancer: current progress and future direction. J Cancer Res Clin Oncol. (2023) 149:9435–50. doi: 10.1007/s00432-023-04805-z
227. Lin X, Chen H, Xie Y, Zhou X, Wang Y, Zhou J, et al. Combination of CTLA-4 blockade with MUC1 mRNA nanovaccine induces enhanced anti-tumor CTL activity by modulating tumor microenvironment of triple negative breast cancer. Trans Oncol. (2022) 15:101298. doi: 10.1016/j.tranon.2021.101298
228. Crosby EJ, Gwin W, Blackwell K, Marcom PK, Chang S, Maecker HT, et al. Vaccine-induced memory CD8+ T cells provide clinical benefit in HER2 expressing breast cancer: a mouse to human translational study. Clin Cancer Res. (2019) 25:2725–36. doi: 10.1158/1078-0432.CCR-18-3102
229. De Mey W, Esprit A, Thielemans K, Breckpot K, Franceschini L. RNA in cancer immunotherapy: unlocking the potential of the immune system. Clin Cancer Res. (2022) 28:3929–39. doi: 10.1158/1078-0432.CCR-21-3304
230. Mackensen A, Koenecke C, Haanen J, Alsdorf W, Desuki A, Wagner-Drouet E, et al. 958 BNT211: a phase I/II trial to evaluate safety and efficacy of CLDN6 CAR-T cells and vaccine-mediated in vivo expansion in patients with CLDN6-positive advanced solid tumors. J ImmunoTher Cancer. (2021) 9:A1008. doi: 10.1136/jitc-2021-SITC2021.958
231. Haanen JB, Mackensen A, Koenecke C, Alsdorf W, Desuki A, Wagner-Drouet E, et al. LBA1 BNT211: a phase I/II trial to evaluate safety and efficacy of CLDN6 CAR-T cells and CARVac-mediated in vivo expansion in patients with CLDN6+ advanced solid tumors. Ann Oncol. (2021) 32:S1392. doi: 10.1016/j.annonc.2021.10.216
232. Gwin WR, Hobeika A, Osada T, Hartman Z, Cheng Q, Broadwater G, et al. Effect of alphavirus vaccine encoding HER2 during concurrent anti-HER2 therapies on induction of oligoclonal T cell and antibody responses against HER2. J Clin Oncol. (2015) 33(15):3081. doi: 10.1200/jco.2015.33.15_suppl.3081
233. Tchou J, Zhao Y, Levine BL, Zhang PJ, Davis MM, Melenhorst JJ, et al. Safety and efficacy of intratumoral injections of chimeric antigen receptor (CAR) T cells in metastatic breast cancer. Cancer Immunol Res. (2017) 5:1152–61. doi: 10.1158/2326-6066.CIR-17-0189
234. Schmidt M, Vogler I, Derhovanessian E, Omokoko T, Godehardt E, Attig S, et al. 88MO T-cell responses induced by an individualized neoantigen specific immune therapy in post (neo) adjuvant patients with triple negative breast cancer. Ann Oncol. (2020) 31:S276. doi: 10.1016/j.annonc.2020.08.209
235. Rojas LA, Sethna Z, Soares KC, Olcese C, Pang N, Patterson E, et al. Personalized RNA neoantigen vaccines stimulate T cells in pancreatic cancer. Nature. (2023) 618:144–50. doi: 10.1038/s41586-023-06063-y
236. Kang N, Zhang S, Wang Y. A personalized mRNA vaccine has exhibited potential in the treatment of pancreatic cancer. Holist Integr Oncol. (2023) 2:18. doi: 10.1007/s44178-023-00042-z
237. Hong J, Guo G, Wu S, Lin S, Zhou Z, Chen S, et al. Altered MUC1 epitope-specific CTLs: A potential target for immunotherapy of pancreatic cancer. J Leukocyte Biol. (2022) 112:1577–90. doi: 10.1002/JLB.5MA0922-749R
238. Abou-Alfa GK, Chapman PB, Feilchenfeldt J, Brennan MF, Capanu M, Gansukh B, et al. Targeting mutated K-ras in pancreatic adenocarcinoma using an adjuvant vaccine. American journal of clinical oncology. (2011) 34:321–5.
239. Suso EM, Dueland S, Rasmussen AM, Vetrhus T, Aamdal S, Kvalheim G, et al. hTERT mRNA dendritic cell vaccination: complete response in a pancreatic cancer patient associated with response against several hTERT epitopes. Cancer Immunol Immunother. (2011) 60:809–18. doi: 10.1007/s00262-011-0991-9
240. Cowzer D, Zameer M, Conroy M, Kolch W, Duffy AG. Targeting KRAS in pancreatic cancer. J Personal Med. (2022) 12:1870. doi: 10.3390/jpm12111870
241. Nagasaka M, Potugari B, Nguyen A, Sukari A, Azmi AS, Ou SH. KRAS inhibitors–yes but what next? Direct targeting of KRAS–vaccines, adoptive T cell therapy and beyond. Cancer Treat Rev. (2021) 101:102309. doi: 10.1016/j.ctrv.2021.102309
242. O’Reilly EM, Wainberg ZA, Weekes CD, Furqan M, Kasi PM, Devoe CE, et al. AMPLIFY-201, a first-in-human safety and efficacy trial of adjuvant ELI-002 2P immunotherapy for patients with high-relapse risk with KRAS G12D-or G12R-mutated pancreatic and colorectal cancer. J Clin Oncol. (2023) 41(Suppl 16):2528. doi: 10.1200/JCO.2023.41.16_suppl.2528
243. Sebastian M, Papachristofilou A, Weiss C, Früh M, Cathomas R, Hilbe W, et al. Phase Ib study evaluating a self-adjuvanted mRNA cancer vaccine (RNActive®) combined with local radiation as consolidation and maintenance treatment for patients with stage IV non-small cell lung cancer. BMC cancer. (2014) 14:1–0. doi: 10.1186/1471-2407-14-748
244. Yang R, Yu S, Xu T, Zhang J, Wu S. Emerging role of RNA sensors in tumor microenvironment and immunotherapy. J Hematol Oncol. (2022) 15:43. doi: 10.1186/s13045-022-01261-z
245. Sebastian M, Schröder A, Scheel B, Hong HS, Muth A, von Boehmer L, et al. A phase I/IIa study of the mRNA-based cancer immunotherapy CV9201 in patients with stage IIIB/IV non-small cell lung cancer. Cancer immunol Immunother. (2019) 68:799–812. doi: 10.1007/s00262-019-02315-x
246. Papachristofilou A, Hipp MM, Klinkhardt U, Früh M, Sebastian M, Weiss C, et al. Phase Ib evaluation of a self-adjuvanted protamine formulated mRNA-based active cancer immunotherapy, BI1361849 (CV9202), combined with local radiation treatment in patients with stage IV non-small cell lung cancer. J immunother cancer. (2019) 7:1–4. doi: 10.1186/s40425-019-0520-5
247. Morse MA, Nair SK, Mosca PJ, Hobeika AC, Clay TM, Deng Y, et al. Immunotherapy with autologous, human dendritic cells transfected with carcinoembryonic antigen mRNA. Cancer Invest. (2003) 21:341–9. doi: 10.1081/CNV-120018224
248. Kiousi E, Lyraraki V, Mardiki GL, Stachika N, Damianou AK, Malainou CP, et al. Progress and Challenges of Messenger RNA Vaccines in the Therapeutics of NSCLC. Cancers. (2023) 15(23):5589.
249. Chen R, Liu W, Brown DM, Bong YS, He J, Shen D, et al. A pan-ras mRNA vaccine elicits specific immune responses and inhibits tumor growth in the mouse model of colon cancer. Cancer Res. (2023) 83:5738. doi: 10.1158/1538-7445.AM2023-5738
250. Hewitt SL, Bailey D, Zielinski J, Apte A, Musenge F, Karp R, et al. Intratumoral IL12 mRNA therapy promotes TH1 transformation of the tumor microenvironment. Clin Cancer Res. (2020) 26:6284–98. doi: 10.1158/1078-0432.CCR-20-0472
251. Li Y, Ma X, Yue Y, Zhang K, Cheng K, Feng Q, et al. Rapid surface display of mRNA antigens by bacteria-derived outer membrane vesicles for a personalized tumor vaccine. Adv Mater. (2022) 34:2109984. doi: 10.1002/adma.202109984
Keywords: mRNA, vaccine, immunotherapy, cancer, immune response
Citation: Laila UE, An W and Xu Z-X (2024) Emerging prospects of mRNA cancer vaccines: mechanisms, formulations, and challenges in cancer immunotherapy. Front. Immunol. 15:1448489. doi: 10.3389/fimmu.2024.1448489
Received: 13 June 2024; Accepted: 18 October 2024;
Published: 25 November 2024.
Edited by:
Jianxun J. Song, Texas A&M Health Science Center, United StatesReviewed by:
Yina Hsing Huang, Dartmouth College, United StatesCopyright © 2024 Laila, An and Xu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhi-Xiang Xu, emhpeGlhbmd4dTA4QGdtYWlsLmNvbQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.