- 1Seattle Children’s Research Institute, Center for Global Infectious Disease Research, Seattle Children’s, Seattle, WA, United States
- 2Department of Global Health, University of Washington, Seattle, WA, United States
- 3Department of Genetics, University of North Carolina at Chapel Hill, Chapel Hill, NC, United States
- 4Department of Immunology and Infectious Diseases, Harvard T. H. Chan School of Public Health, Boston, MA, United States
- 5Broad Institute of Massachusetts Institute of Technology (MIT), and Harvard, Cambridge, MA, United States
- 6Department of Immunology, University of Washington, Seattle, WA, United States
- 7Department of Pediatrics, University of Washington School of Medicine, Seattle, WA, United States
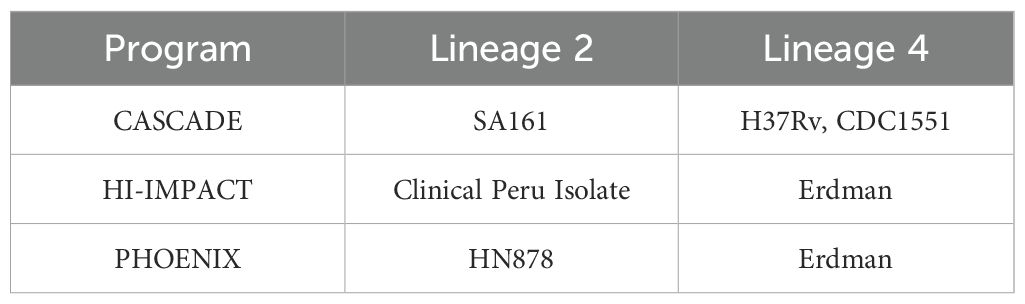
The three programs that make up the Immune Mechanisms of Protection Against Mycobacterium tuberculosis Centers (IMPAc-TB) had to prioritize and select strains to be leveraged for this work. The CASCADE team based at Seattle Children’s Research Institute are leveraging M.tb H37Rv, M.tb CDC1551, and M.tb SA161. The HI-IMPACT team based at Harvard T.H. Chan School of Public Health, Boston, have selected M.tb Erdman as well as a novel clinical isolate recently characterized during a longitudinal study in Peru. The PHOENIX team also based at Seattle Children’s Research Institute have selected M.tb HN878 and M.tb Erdman as their isolates of choice. Here, we describe original source isolation, genomic references, key virulence characteristics, and relevant tools that make these isolates attractive for use. The global context for M.tb lineage 2 and 4 selection is reviewed including what is known about their relative abundance and acquisition of drug resistance. Host–pathogen interactions seem driven by genomic differences on each side, and these play an important role in pathogenesis and immunity. The few M.tb strains chosen for this work do not reflect the vast genomic diversity within this species. They do, however, provide specific virulence, pathology, and growth kinetics of interest to the consortium. The strains selected should not be considered as “representative” of the growing available array of M.tb isolates, but rather tools that are being used to address key outstanding questions in the field.
1 Introduction
Across each of the Immune Mechanisms of Protection Against Mycobacterium tuberculosis Centers (IMPAc-TB), there are mechanistic questions of special focus. The CASCADE team at Seattle Children’s Research Institute are examining the stages of infection through detailed interrogation of pulmonary pathology and natural or vaccine-induced protection across this continuum in mice, non-human primates, and human studies. The HI-IMPACT team based at Harvard T.H. Chan School of Public Health, Boston, are leveraging key immune correlates of protection from their seminal intravenous (IV) bacillus Calmette–Guérin (BCG) vaccination strategy in rhesus macaques (1, 2) and comparing those with human cohort responses with computational modeling to better design vaccines against tuberculosis (TB). The PHOENIX team based at Seattle Children’s Research Institute are focused on a cross-species analysis of candidate vaccine-induced immunity and efficacy for prevention of disease or infection in preclinical mouse, guinea pig, and non-human primates as well as a human aerosol challenge model using BCG as a representative mycobacteria.
While these centers are focused on discrete but complementary work, each are using modeling, escalating from in vitro or in silico through in vivo preclinical and first-in-man experimental medicine studies. In the TB research field, we challenge in vitro or in vivo models with “representative” isolates of Mycobacterium tuberculosis (M.tb) that fit some categorical aspect of what we would like to examine or that provide continuity with previous work to continue a specific story. Recent data are challenging the use of “representative” to define our M.tb isolates of choice and instead suggest there is an underrepresentation of the breadth of biological diversity that exists in M.tb (3–5). Indeed, even preclinical drug therapy studies have demonstrated significant differences in in vivo sensitivity between M.tb strains (6). Next-generation tools and use of big data pipelines may allow us to better profile huge arrays of M.tb isolates or lineages in the near future, as groups within this consortium are already leveraging artificial intelligence and systems biology for complex data and predictive modeling.
In the absence of current or established tools that allow us to readily select challenge isolates spanning a vast array of M.tb lineages, it was necessary for each of the IMPAc-TB consortiums to choose the few they would study across their programs. The six strains used in the CASCADE (SCRI), HI-IMPACT (Harvard), or PHOENIX (SCRI) centers are described here (Table 1). We outline the source of each isolate, reference the sequenced genome if available, include specific tools in that background, give a summary of models using that strain, and provide seminal aspects of the isolate including virulence or kinetics if known and key historical work using that isolate. These basic features helped define the selection of isolates for specific consortium endpoints.
2 Materials and methods
2.1 M.tb genome sequencing
2.1.1 Genomic DNA extraction
In order to obtain high-quality genomic DNA (gDNA) from M.tb cultures of all of the isolates except the g2g clinical isolate described above, we followed the methodology described previously (7). In summary, mycobacterial cultures (M.tb SA161, H37Rv, CDC1551, Erdman, and HN878) were grown to an optical density 600 nm (O.D. 600) of 1.5. Subsequently, the cultures were harvested by centrifugation, washed in PBS, and dissolved in TE buffer. To lyse the mycobacteria, the bacterial suspension was subjected to 80°C for 20 min. After allowing the tubes to cool to room temperature, lysozyme solution (100 mg/ml) was added to each tube, followed by incubation at 37°C for 3 h with intermittent mixing. Next, a mixture of SDS (10%) and proteinase K (20 mg/ml) was added to each tube in an 88:12 ratio. The tubes were then incubated at 65°C for 2 h. Following this incubation period, 100 µl of 5 M NaCl was added to each tube and incubated at 65°C for an additional 10 min. Subsequently, an 80 µl aliquot of 10% cetyl trimethyl ammonium bromide (CTAB) solution (Sigma-Aldrich, St. Louis, MO) was added, mixed thoroughly, and incubated at 65°C for another 10 min. The DNA was then extracted first with an equal volume of phenol/chloroform/isoamyl alcohol (25:24:1 v/v/v) and then with chloroform/isoamyl alcohol (24:1 v/v). The DNA was precipitated by adding 0.6 volumes of ice-cold isopropanol. The precipitated DNA was pelleted by centrifugation at 10,000 rpm and 4°C for 15 min. The resulting DNA pellets were washed with cold 75% ethanol and dried using a SpeedVac for 5 min. Finally, the genomic DNA was resuspended in 50 µl of sterile distilled water. The quality of the gDNA was assessed using a NanoDrop spectrophotometer (Thermo Scientific, Wilmington, DE).
2.1.2 Library production
Starting with a minimum of 750 ng of DNA, samples are sheared in a 96-well format using a Covaris LE220 focused ultrasonicator targeting 380-bp inserts. This insert size improves overall library performance and allows the longer sequencing read lengths on Illumina sequencing platforms (150 bp) to be efficiently used without producing a significant number of overlapping reads. The resulting sheared DNA was cleaned with Agencourt AMPure XP beads to remove sample impurities prior to library construction. Shearing was followed by size selection performed using the KAPA Hyper Prep kit (KR0961 v1.14). End-repair, A-tailing, and ligation are performed as directed. Two final AMPure cleanups are performed after ligation to remove excess adapter dimers from the library. All library construction steps are automated on the Perkin Elmer Janus platform. Library yield was quantified using Quant-IT dsDNA High Sensitivity kit (Invitrogen Q33120). Libraries were validated in triplicate using the Bio-Rad CFX384 Real-Time System and KAPA Library Quantification Kit (KK4824).
2.1.3 Whole-genome sequencing
Massively parallel sequencing-by-synthesis with fluorescently labeled, reversibly terminating nucleotides was carried out using the Illumina NovaSeq 6000 platform (RTA 3.1.5) and was performed at the University of Washington Center for Rare Disease Research (UW-CRDR). Barcoded genome libraries were pooled with liquid handling robotics prior to loading. Raw sequence reads with an average read length of 150 bp were assembled against the sequence of the reference strain M.tb H37Rv (NCBI accession number: NC_018143.2) using SPADES v3.15.4 (8). Whole-genome sequences were deposited into GenBank BioProject: PRJNA1051793, as described with specific accession numbers stated.
2.2 In vivo mouse challenge and survival studies
In all studies, mice were housed in the Seattle Children’s Research Institute (SCRI) biosafety level 3 (BSL-3) animal facility under pathogen-free conditions and were handled in accordance with approved protocols from the SCRI Institutional Animal Care and Use Committee (IACUC). All methods were carried out in accordance with animal welfare guidelines and regulations. For survival studies, breeding pairs of C57BL/6 bg/bg (beige) mice were originally purchased through Jackson Laboratories. In-house bred male beige mice were used for experiments at 4–6 weeks of age and had not been previously used for breeding purposes. Mice were maintained at four animals per cage. Mice were challenged with a low-dose aerosol (target 25–100 CFU per mouse upon infection) of either M.tb SA161 or M.tb HN878 using a Glas-Col whole-body aerosol infection chamber. Bacterial burden was evaluated at 24 h and 4 weeks post challenge as previously described (9). Survival was monitored in 10 mice per group following M.tb infection. Animals with greater than 20% weight loss, or moribund condition, were euthanized. For ultra-low dose aerosol (targets 1–3 CFU per mouse upon infection) studies, female C57BL/6 mice purchased from Jackson Laboratories (age 8–27 weeks old) were challenged with either M.tb Erdman or M.tb H37Rv also using a Glas-Col whole-body aerosol infection chamber. Pulmonary bacterial burden was evaluated 35–42 days post challenge.
2.3 Statistical analysis
For ex vivo plating of organ homogenate to enumerate bacterial CFU in a single organ and timepoint (24 h, 4 weeks, or 6 weeks post-challenge), we used a two-tailed unpaired Student’s t test to compare between M.tb challenge strains. For probability of survival, cohorts were compared using the log-rank Mantel–Cox test. All statistical analysis was performed using GraphPad Prism version 10.2.2.
3 Results
3.1 M.tb isolates
As the consortium matures, we are reflecting on opportunities to cross-validate or harmonize so the discrete but complimentary data being generated can have the largest impact for the TB pathogenesis, immunology, and vaccine communities. Interestingly, and without prior discussions, each of the programs focused their M.tb isolates across two modern lineages, 2 and 4. Lineage 2 is notable in its expanding global distribution, “hypervirulence,” correlations with relapse, and enriched acquisition of antibiotic drug resistance (10–16). In vitro, lineage 2 Beijing isolates persist and grow normally within macrophages compared with other lineages that are relatively more inhibited once they are intracellular (17). These features make lineage 2 high-priority in preclinical studies for evaluation of disease progression and efficacy of vaccine candidates. M.tb isolates SA161 and HN878 and the clinical isolate from Peru are lineage 2 isolates represented in the IMPAc-TB consortiums. Lineage 4, the Euro-American lineage, is also well distributed globally and can be associated with unique features of disease pathology and treatment failure in humans (18–21). M.tb H37Rv, Erdman, and CDC1551 are each lineage 4 isolates being leveraged by one or more of the IMPAc-TB programs. Each isolates’ key features and rationale for selection are described below.
3.2 M.tb HN878 (lineage 2)
M.tb HN878, a W-Beijing lineage isolate, was sourced by the PHOENIX consortium directly from BEI Resources (NR-13647). This clinical isolate was derived from a patient sample in Houston, Texas, in a 1990s outbreak and has been well studied since (22) with the parental genome sequence available on GenBank: ADNF01000000.1. Many other M.tb-derived materials including M.tb HN878 whole-cell lysate (NR-14824), derived lipid fraction (NR-14839), and genomic DNA (NR-14867) are available within the BEI Resources Repository. Defined as hypervirulent (23), M.tb HN878 induces a waning host T helper 1 (Th1) response and rapid expansion of regulatory T cells in chronic preclinical settings (22), as well as progressive pulmonary pathology and morbidity in C57BL/6 mice (24, 25). Interestingly, this isolate can acquire large genetic duplications with serial in vitro passage dramatically reducing virulence in vivo (25, 26). For the PHOENIX consortium, we generated a large batch single passage culture in 2019 to avoid the duplication issue and ensure continuity across studies, and we confirmed virulence of our stock culture in mouse survival studies (25). The single passage PHOENIX batch of M.tb HN878 was sequenced, and that genome can be found at GenBank: SAMN38797941.
The M.tb HN878 strain has been evaluated extensively in vitro as well as in preclinical mouse and guinea pig models of aerosol challenge. This includes recent examinations of single-cell preparations and of the influence of detergents on the mycobacterial cell wall and their impacts on interactions with innate immune cells (27). In vivo, M.tb HN878 induces more granulomatous-like structures in preclinical models that better reflect human pathology than other well-used isolates (22, 23, 28). This includes induction of a blood-based TB transcriptional signature in C3HeB/FeJ mice by M.tb HN878 infection, whereas similar studies using less virulent M.tb H37Rv to infect C57BL/6 mice did not result in production of this human-derived biomarker of TB disease (29). Recent evidence suggests that while aerosol infection with a high dose of M.tb HN878 does induce bone-marrow-derived myelopoiesis, there is a concomitant reduction in trained innate immune responses in a mouse model (30). A key feature which led to the selection of this strain is that the high virulence of M.tb HN878 may be due to the preferential recruitment of pathology-inducing neutrophils and relatively damped adaptive responses (29). Despite the observed virulence and immune skewing from clinical M.tb HN878 isolate infection, prophylactic vaccination with BCG provides protection from bacterial burden and pulmonary pathology and enhances survival in several preclinical models (31–33). M.tb HN878 was selected by the PHOENIX consortium due to its well profiled virulence discussed above and its membership in lineage 2, which has high global prevalence. This M.tb isolate serves as a suitable strain to evaluate vaccine-specific efficacy in both prevention of infection (POI) and prevention of disease (POD) contexts, key components of the PHOENIX goals for the program. To our knowledge, this will also be the first time M.tb HN878 is used as a challenge strain in nonhuman primates.
3.3 M.tb SA161 (lineage 2)
M.tb SA161 is also a W-Beijing-type Lineage 2 isolate (34). SA161 is derived from a cluster of cases in Arkansas and was originally sourced by the CASCADE team from Dr. Ian Orme at Colorado State University (34). The M.tb SA161 being used by the Urdahl Lab in the CASCADE program was sequenced, and the genome sequence is deposited at GenBank: SAMN38797942.
Like M.tb HN878, SA161 is a highly virulent W-Beijing strain of M.tb (35). Historical data suggest that lineage 2 Beijing isolates may associate with TB infections in persons living with human immunodeficiency virus (PLWHIV) (35). As such, M.tb SA161 has been evaluated in several models with seminal features of HIV infection including mice transgenic for HIV transcription protein Tat (35). M.tb SA161 is frequently paired with highly susceptible mouse strains like C3HeB/FeJ to model high frequencies of pulmonary pathology and use survival as primary endpoints (32, 35). Despite the highly virulent M.tb—highly susceptible mouse strain pairing—prophylactic vaccination with BCG still affords protection from bacterial burden and pathology in both mouse and guinea pig infections at intermediate timepoints (32, 36). At later timepoints post challenge, W-Beijing strains M.tb HN878 and M.tb SA161 upregulate regulatory T-cell responses and the early protection wanes as mice succumb to high bacterial burden (> 6 Log10 CFU) and pathology in a more resistant C57BL/6 mouse strain (31), which was a key feature for its selection in this consortium.
At the start of the IMPAc-TB contract, the PHOENIX and CASCADE consortiums performed a head-to-head virulence survival study of M.tb SA161 and M.tb HN878 to both compare their relative virulence as lineage 2 members as well as infer any similarities or differences worth noting for future comparisons across programs. In this study, male Beige mice were challenged with a low-dose aerosol (25–100 CFU) of either lineage 2 isolate. Beige mice were used in this study because they are highly susceptible to mycobacterial infections and offered a compressed survival timeline (<200 days) for this exploratory head-to-head study. In addition, both in human epidemiology and in mouse preclinical studies, males are more sensitive to TB disease [well reviewed here (37)], which also afforded an accelerated assessment between M.tb isolates. Samples were collected 24 h post challenge to confirm deposition in lungs, and for HN878, we observed 23, 67, and 52 CFU per mouse (averaging 47 CFU) and 14, 31, and 59 CFU were observed for SA161 challenge (average 35 CFU; n=3 per group). At 4 weeks post challenge, lung and spleen bacterial burden were evaluated (n=6–7) with no significant difference in either site between the two challenge strains at this timepoint observed (Figures 1A, B). A cohort of mice (n=10 each) was followed for morbidity endpoints resulting in probability of survival over time (Figure 1C). The median survival for mice challenged with M.tb HN878 was 165 days, whereas the median survival for M.tb SA161 infected mice was 119.5 days.
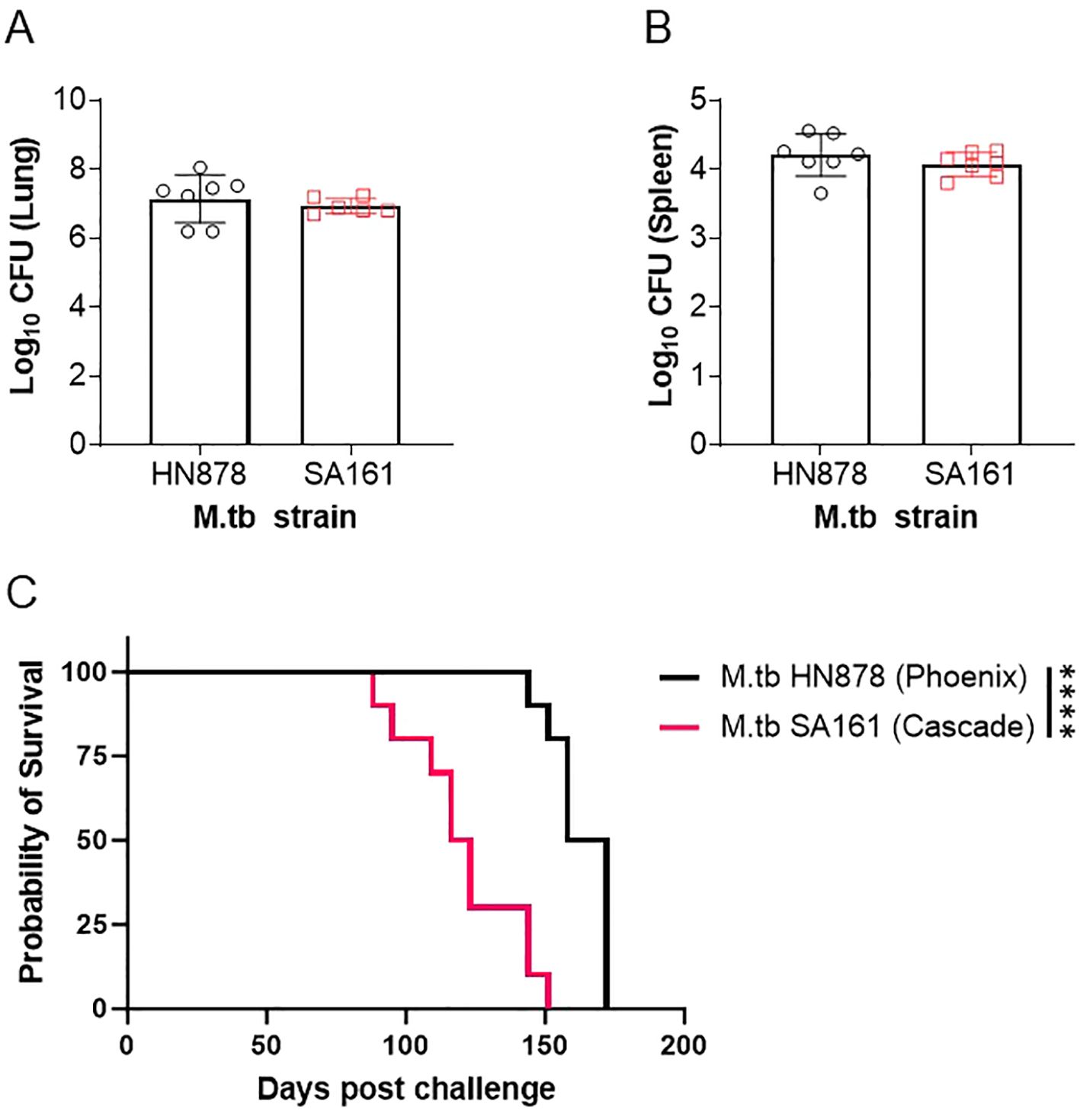
Figure 1. Survival of mice challenged with Beijing isolates of M.tb. Male Beige mice were challenged with a low-dose aerosol of either M.tb HN878 (open black circles) or M.tb SA161 (open red squares). At 4 weeks post-challenge, (A) lung and (B) spleen bacterial burden were evaluated in n=6-7. Two-tailed unpaired Student’s t test found no significant differences between lung (p = 0.4997) or spleen (p = 0.3365) groups. Mean Log10 CFU for lung = 7.1 HN878 and 6.9 for SA161. Mean Log10 CFU for spleen = 4.2 HN878 and 4.0 for SA161 (C) Cohorts of = 10 mice per challenge (M.tb HN878 = black line or M.tb SA161 = red line) isolate were followed over time with weekly or more frequent weighing as a measure of morbidity due to infection. When mice reached 20% weight loss, they were considered moribund and humanely euthanized. Asterisks represent significance (p < 0.0001) between the groups using a Log-rank Mantel–Cox test comparing survival curves. Data representative of one experiment.
In this comparative study, we observed a different survival profile in a Beige mouse background between M.tb HN878 and M.tb SA161 low-dose aerosol challenges, despite relatively equivalent bacterial burden at 4 week post challenge in both the lung and spleen. Interestingly, M.tb SA161 induces an earlier initiation of morbidity (day 88 to 151, 63 days of group decline) but the overall group mortality is more heterogeneous over time when compared with M.tb HN878 (day 144 to 172, 28 days of group decline), which seems to have a relatively delayed but more uniform morbidity of the total group. To our knowledge, this is the first report in male Beige mice for either M.tb isolate with these endpoints. This different pattern of morbidity between isolates in highly sensitive Beige mice mirrors that observed in the literature in more resistant C57BL/6 mice (31); however, the data reported here are less virulent in Beige mice comparatively by survival presumably due to the lower challenge dose at infection. In the context of the PHOENIX program where differences between vaccine groups are being evaluated, M.tb HN878 is a preferred isolate, but if, in contrast, interest is in the mechanism of disease progression and host variability in progressions to disease, then M.tb SA161 is a more reasonable strain to select. In the context of the CASCADE program, M.tb SA161 has been the primary isolate used to characterize pathological and immunological disease outcomes following vaccination due to its ability to generate large necrotic granulomas in the C3HeB/FeJ background. These profiles and specific endpoints of interest are well tailored to the overall objectives of the PHOENIX and CASCADE programs.
3.4 Clinical Peru isolate (lineage 2)
A distinct clinical isolate was recently discovered after the initiation of the IMPAc-TB program during a genome-to-genome (G2G) study conducted in Lima, Peru (38) (manuscript accepted, pending publication, SRA accession codes referenced there). This G2G study was designed to identify the genetic interaction between host genetic background and M.tb genetic background. The researchers discovered a single host variant (located in a non-coding region of the FLOT1 gene) that was associated with infection by a particular M.tb strain they called g2g-L2. They found the g2g-L2 strain locally emerged in Peru around 60 years ago but has since undergone rapid expansion. In a 2010 cohort, the g2g-L2 strain accounted for 6.6% of all M.tb strains circulating in Lima, Peru, but by 2020, the prevalence had almost doubled (12.6%). Additionally, it was shown that 87% of the g2g-L2 strains were in genomic clusters, whereas the clustering rates for the neighboring L2 clades were only 18% to 28%. These data indicate that the g2g-L2 strain has increased transmissibility in the local population. While the g2g-L2 strain can transmit successfully in Lima, Peru, it was not found elsewhere in the world. The authors also showed that g2g-L2 strains have a distinctive redox state compared with their nearest neighbor strains, manifested as a more oxidative cell state and resistance to reductive stress. The authors narrowed down a mutation (Thr2Asn) in trxB2 that was specific to the g2g-L2 strain, and they found that this trxB2 mutation can lead to higher activity of thioredoxin reductase and a significant shift in the NAD+/NADH ratio toward the oxidized state.
The authors delved into the underlying interaction between Mtb strains (g2g-L2 vs. nearest-neighbor strains, termed non-g2g-L2) and host immune cells with differing genetic backgrounds (FLOT1, AT vs. TT). RNA-seq analysis was employed to profile the innate immune response to M.tb infection, with the expression of 20 infection-responsive genes used to gauge the “infection score,” where higher expression indicated a stronger response. The study revealed that overall, AT host cells exhibited an elevated infection response compared with TT host cells. However, within the AT host cells, g2g-L2 infection blunted this response relative to non-g2g-L2 infection. Specifically, FLOT1-AT cells exhibited increased expression of genes associated with type 1 and type 2 interferon pro-inflammatory signaling, MHC-I antigen processing and presentation, IL-1B signaling, cytosolic DNA sensing, and zinc homeostasis. The authors posited that FLOT1-AT donors mount a robust transcriptional response to infection, potentially due to increased sensitivity in inducing innate responses. However, this response was skewed toward interferon following non-g2g-L2 infection, contrasting with IL-1B dominant responses observed after g2g-L2 infection. As the most recently isolated M.tb within the consortium, this g2g strain helps ground the clinical relevance of findings back to human epidemiologic data, a feature unique to the HI-IMPACT program.
3.5 M.tb Erdman (lineage 4)
M.tb Erdman K01 was sourced by the PHOENIX consortium directly from BEI Resources (NR-15404). This original clinical isolate was derived from human sputum at the Mayo Clinic in 1945 and is noted to be highly virulent (39). A low passage culture of M.tb Erdman was generated by the Coler Lab in the PHOENIX program. The stock material was sequenced and the assembled genome deposited at GenBank: SAMN38797939. A notable feature of M.tb Erdman was the development of a barcoded library (BEI Resources NR-50781) where each bacterium contains a unique and trackable sequence, enabling the profiling of a single cells’ longitudinal fate in vivo. This library has been leveraged in non-human primate models coupled with pathology and imaging to interrogate individual lesion-level disease progression and infection dynamics (40).
M.tb Erdman has been extensively used for aerosol challenge with diversity outbred mice to identify critical gene loci (41–44), innate cell influx, pulmonary necrosis (45), and age-related influences (46) that correlate with a spectrum of disease in this model. In mice, prophylactic BCG vaccination provides protection from early pulmonary and peripheral bacterial burden after challenge with M.tb Erdman (33). Studies in cynomolgus macaques infected with M.tb Erdman identified peripheral blood transcriptional changes occurring early (<6 months) post infection (47), which share similarities with human risk signatures predicting advancement to active TB disease. The similarities in risk signatures between M.tb Erdman-infected macaques and humans highlights the usefulness of this model to predict vaccine outcomes in humans.
3.6 M.tb H37Rv (lineage 4)
M.tb H37 was isolated from a human lung sample at the Trudeau Laboratory in 1905. M.tb H37Rv was derived from this isolate after selection for rough colony morphology and virulence in 1934 (48) and is one of the most widely used strains of M.tb available. Despite researchers identifying notable polymorphisms between M.tb H37Rv isolates maintained in different laboratories (49), the M.tb H37Rv genome is commonly used as the base reference. Indeed, antigens included in high-priority clinical vaccine candidates are sourced from the H37Rv genome, including ID93 (50), M72 (51), GamTBvac (52), and H107e (53). The M.tb H37Rv being leveraged by the CASCADE consortium was a gift from the lab of Dr. Joel Ernst. This isolate was derived from ATCC TMC 102 (catalog #27294), and the genome was partially sequenced and published in 1998. Despite this isolate serving as the de facto M.tb genome reference, the full genome sequence was only recently completed, comprising over 6,000 novel base pair regions, using a new paradigm of assembly called Bact-Builder (54). The CASCADE program M.tb H37Rv strain has been serially passaged through mice and has maintained virulence properties compared with other lab-adapted strains, perhaps through retention of the virulence factor PDIM, which is rapidly lost upon in vitro passage (55). The CASCADE M.tb H37Rv isolate was sequenced, and that assembled genome can be found at GenBank: SAMN38797937.
Like M.tb Erdman, M.tb H37Rv has been developed as a barcoded strain, allowing for the individual tracking and monitoring of deposited bacteria at the time of challenge and through chronic timepoints of infection and disease (56). This is an especially useful tool in the context of ultra-low-dose aerosol infections where as few as one bacterium per mouse is inhaled and monitored over time (56), better reflecting the conditions of human challenge. In general, M.tb H37Rv laboratory isolates are notably less virulent across models, with higher rates of survival and less necrotic lesions in the translational guinea pig preclinical model compared with other strains being used in this program including M.tb Erdman, M.tb HN878, or M.tb CDC1551 (23). Lower virulence of M.tb H37Rv compared with M.tb Erdman has also been observed in a New Zealand White rabbit model of aerosol challenge by 5 weeks post infection (57). Despite these reports, the CASCADE M.tb H37Rv isolate has similar virulence to M.tb Erdman and is more virulent than M.tb CDC1551, in studies by the group in C57BL/6 mice (Figure 2 and article by some of these same authors presented in this special issue (58), respectively). These patterns of virulence align with prior studies using the CASCADE programs’ M.tb H37Rv in C57BL/6 mice (56, 59). As such, the CASCADE program has used H37Rv as the primary lineage 4 strain to draw direct comparisons with M.tb SA161. Interestingly, in a BALB/c drug therapy model, different isolates of M.tb H37Rv were similarly sensitive to front-line drug treatment with rifampicin, isoniazid, and pyrazinamide as M.tb Erdman (6). However, M.tb H37Rv has been the strain of choice to date when assessing genetic components influencing host susceptibility in the context of the collaborative cross mice (60–63).
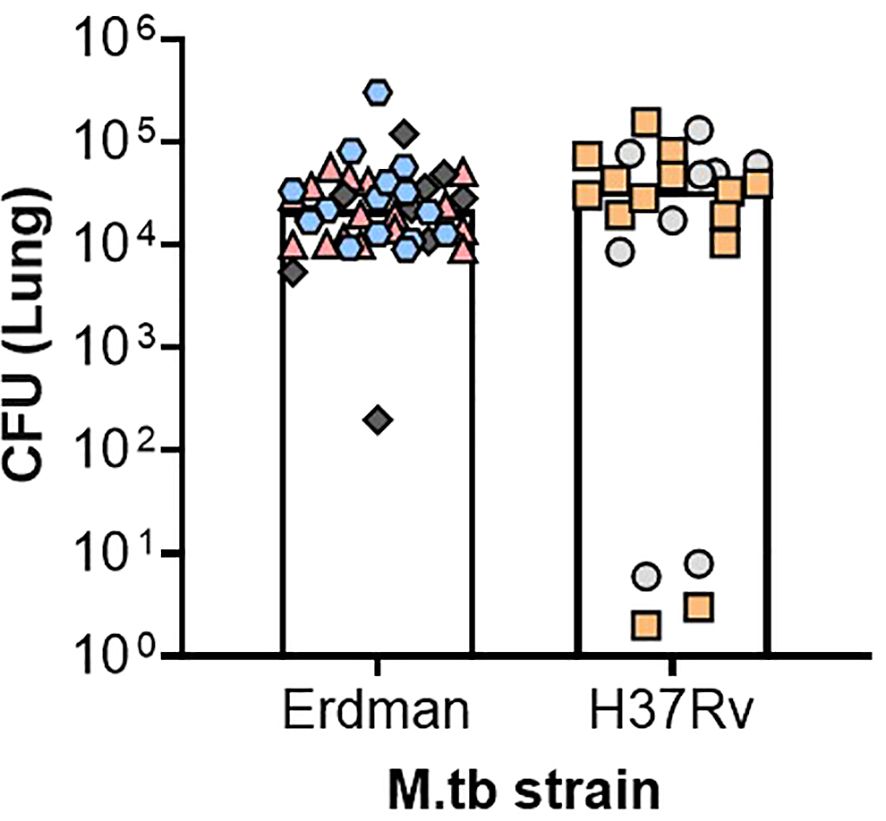
Figure 2. Comparable pulmonary bacterial burden between M.tb H37Rv and Erdman after ultra-low-dose challenge. Female C57BL/6 mice were challenged between 8 and 27 weeks of age with an ultra-low-dose challenge of either M.tb Erdman or M.tb H37Rv. At 35–42 days post-infection, lung bacterial burden was evaluated. Data depict six different challenge experiments, denoted in the figure as different-colored shapes, where each shape represents an individual mouse whose CFU was over 1. Two-tailed unpaired Student’s t test found no significant differences between the challenged groups (p = 0.5196) where the mean CFU for M.tb Erdman was 3.4 × 104 and for M.tb H37Rv was 4.2 × 104.
3.7 M.tb CDC1551 (lineage 4)
M.tb CDC1551 is a clinical isolate collected in the mid-1990s amid an outbreak with uncharacteristically highly positive tuberculin skin testing rates in rural Kentucky and Tennessee (64). The CASCADE M.tb CDC1551 isolate, originally a gift from W.R. Bishai, was sequenced, and that assembled genome can be found at GenBank: SAMN38797940. The BEI repository hosts numerous M.tb CDC1551-derived materials including cell lysate (NR-14835), polyclonal anti-sera from a guinea pig infection (NR-13818), genomic DNA (NR-48981), and a large assortment of transposon mutants. Live-attenuated vaccines are gaining traction in the TB research community, and one notable vaccine based in a parent CD1551 background is M.tbΔsigH (65).
M.tb H37Rv, M.tb Erdman, and M.tb CDC1551 contain a frame-shift deletion in a gene cluster responsible for phenolic glycolipid (PGL) production, resulting in a relatively reduced virulence compared with lineage 2 isolates (including M.tb HN878) which do not contain this deletion (66). Furthermore, prophylactic BCG vaccination provides better protection against these lineage 4 isolates compared with lineage 2 isolates in an intravenous infectious challenge of pooled barcoded M.tb in a C57BL/6 mouse model (67). Although the initial paper describing this strain in an in vivo mouse model found that, when compared with M.tb Erdman, M.tb CDC1551 expanded rapidly and notably reached two logs higher lung bacterial burden by day 20 (64), curiously, the M.tb Erdman peaked at ~104, which is ~2 logs lower than the typical CFU load following infection with virulent M.tb strains, such as M.tb Erdman or M.tb H37Rv. A subsequent study found that M.tb CDC1551 was actually less virulent than several other M.tb strains tested, including M.tb H37Rv, M.tb Erdman, and M.tb HN878, while inducing a more hyperinflammatory immune response (68). Consistent with the more recent study, the CASCADE M.tb CDC1551 isolate is less virulent in C57BL/6 mice than the CASCADE M.tb H37Rv isolate, with an approximately 1-log reduction in the peak lung bacterial burden.
4 Discussion
Aside from strain identity and genome sequence, there are many factors that influence how M.tb can behave both in vitro and in vivo. This includes culture conditions like the presence, absence, or concentration of Tween which directly changes the cell wall and some subsequent interactions with host cells (69–71) or growth medium albumin choice. Large first stock batches are made for each consortium where M.tb were grown in 10% OADC, 0.05% Tween, and in the absence of C02. For enumeration for mouse infection studies, the CASCADE program uses OD600nm of 1.0 and Phoenix uses direct CFU plating of aliquots and both programs combine these results with empirical titration experiments to determine nebulizer stock dilutions to attain the infection range per mouse desired. The Coler, Fortune, and Urdahl labs serve as distributor partners for each consortium where large low-passage batches (>900 1.0-ml aliquots in the case of the PHOENIX program) are made and stocks are distributed to contract partners to reduce the influence of strain passage on results obtained within each consortium. Annual CFU plating by all partners is a contract deliverable for some of the consortiums as a simple and direct check that stocks remain viable and at or near expected CFU. The status of specific lipids, like phthiocerol dimycocerosates (PDIMs) on the bacterial cell envelope, which have been observed to affect pathogenesis and virulence, is a similar factor important for understanding and comparing M.tb strain outcomes (72). The mouse-passaged M.tb H37Rv used by the CASCADE program recently confirmed PDIM status (73), and M.tb Erdman was similarly confirmed prior to DNA barcoding (40) and no subsequent colony purification occurred. PDIM status was not a planned screen throughout the program, although we acknowledge that this could affect results.
While each program is using different isolates, they are consistently from lineages 2 and 4, which provides some harmony of interest. Importantly, each isolate suits the needs of each consortium; for example, CASCADE is mechanistically interrogating the diversity of the immune response including pathology endpoints and the variability within an M.tb SA161 infection, possibly identifying key differences in disease progression. Conversely, PHOENIX is interested in vaccine efficacy endpoints and M.tb HN878 provides a more homologous response and disease progression that affords resolution between immunization strategies. The ability of HI-IMPACT to leverage a recent clinical isolate with a direct relationship to epidemiological data and human genetic variabilities make their work in preclinical models highly translational and informative. However, this collective work has excluded other lineages, including 1 and 3 which are found in Southeast Asian countries like Indonesia and the Philippines, which are among the top eight high-TB burden countries globally (74). Should these consortiums find critical disparities in relative immune responses or protection from vaccinations, it would be necessary to perform similar analyses using lineages 1 and 3. Other ex vivo tools like the mycobacterial growth inhibition assay (75–77), which allow for multiple challenge isolates to be used, are also being integrated into specific programs and may help address this need for an increased breadth of M.tb challenges to increase the scope of findings and areas of follow-up.
By identifying salient differences between strains being used across the three programs, we aim to transparently identify what may be common findings versus those that may be unique due to model organism selection. It is clear from the literature that whole-genome sequencing can reveal key genes involved in drug resistance, adaptations to stress conditions, and virulence of M.tb isolates (78–80). The sequences reported here aim to contribute to those efforts and provide pathogen genomic comparators for phenotypic outcomes across the consortium. Future work will align the sequences reported here to parent lineage-specific reference genomes available, clinical isolates, and an ancestral, non-lineage-specific, M.tb complex genome (termed MTBC0) (81). We expect that placing these isolates in the advanced phylogeny available for M.tb, through a number of platforms such as TB-Annotator (82) or MAGMA (83), will provide more opportunities to further link protection or pathogenic outcomes derived from IMPAc-TB with existing literature and clinical trends. In addition to M.tb isolate identity, the consortium is actively discussing challenge doses and how this directly influences outcomes in the preclinical models being studied. Furthermore, use of diversity outbred and collaborative cross mouse strains is an acknowledgement that host diversity plays a significant role in TB outcomes and may help uncover critical immune correlates of immunity.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/genbank/, BioProject: PRJNA1051793.
Ethics statement
The animal study was approved by Seattle Childrens’ Research Institute Animal Care and Use Committee. The study was conducted in accordance with the local legislation and institutional requirements.
Author contributions
SL: Conceptualization, Formal analysis, Investigation, Project administration, Resources, Visualization, Writing – original draft. HA: Data curation, Investigation, Software, Writing – original draft. CP: Resources, Writing – review & editing. SC: Writing – review & editing. HK: Investigation, Resources, Writing – review & editing. QL: Writing – original draft. MH: Investigation, Resources, Writing – review & editing. BB: Writing – review & editing. SB: Writing – review & editing. SF: Funding acquisition, Writing – review & editing. KU: Funding acquisition, Writing – review & editing. RC: Funding acquisition, Project administration, Supervision, Writing – review & editing. HB: Investigation, Visualization, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This project has been funded in whole or in part with Federal funds from the National Institute of Allergy and Infectious Diseases, National Institutes of Health, Department of Health and Human Services, under Contract Nos. 75N93021C00029 (to RC), 75N93019C00070 (to KU), and 75N93019C00071 (to SF). Additional support was from the National Institute of Allergy and Infectious Diseases, National Institutes of Health, under contract number P30AI168023 for the Seattle Tuberculosis Research Advancement Center - SEATRAC. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Acknowledgments
The authors would like to thank all consortium partner institutions for their collaboration and dedication to advancing our knowledge about protective mechanisms against M.tb. The following reagents were obtained through BEI Resources by the Coler Lab, NIAID, NIH: Mycobacterium tuberculosis, Strain HN878, NR-13647, and Strain Erdman K01, NR-15404. The authors acknowledge Research Scientific Computing at Seattle Children’s Research Institute for providing HPC resources that have contributed to the research results reported within this paper. The authors would also like to thank the SCRI Office of Animal Care (OAC) for their management of animal resources.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Darrah PA, Zeppa JJ, Maiello P, Hackney JA, Wadsworth MH 2nd, Hughes TK, et al. Prevention of tuberculosis in macaques after intravenous BCG immunization. Nature. (2020) 577(7788):95–102. doi: 10.1038/s41586-019-1817-8
2. Larson EC, Ellis-Connell AL, Rodgers MA, Gubernat AK, Gleim JL, Moriarty RV, et al. Intravenous bacille calmette-guérin vaccination protects simian immunodeficiency virus-infected macaques from tuberculosis. Nat Microbiol. (2023) 8(11):2080–92. doi: 10.1038/s41564-023-01503-x
3. O'Neill MB, Mortimer TD, Pepperell CS. Diversity of Mycobacterium tuberculosis across evolutionary scales. PloS Pathog. (2015) 11(11):e1005257. doi: 10.1371/journal.ppat.1005257
4. Stanley S, Spaulding CN, Liu Q, Chase MR, Ha DTM, Thai PVK, et al. High-throughput phenogenotyping of Mycobacteria tuberculosis clinical strains reveals bacterial determinants of treatment outcomes. bioRxiv. (2023). doi: 10.1101/2023.04.09.536166
5. Smith CM, Sassetti CM. Modeling diversity: Do homogeneous laboratory strains limit discovery? Trends Microbiol. (2018) 26(11):892–5. doi: 10.1016/j.tim.2018.08.002
6. De Groote MA, Gruppo V, Woolhiser LK, Orme IM, Gilliland JC, Lenaerts AJ. Importance of confirming data on the in vivo efficacy of novel antibacterial drug regimens against various strains of Mycobacterium tuberculosis. Antimicrob Agents Chemother. (2012) 56(2):731–8. doi: 10.1128/aac.05701-11
7. Abdelaal HFM, Spalink D, Amer A, Steinberg H, Hashish EA, Nasr EA, et al. Genomic polymorphism associated with the emergence of virulent isolates of mycobacterium bovis in the nile delta. Sci Rep. (2019) 9(1):11657. doi: 10.1038/s41598-019-48106-3
8. Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, et al. Spades: A new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. (2012) 19(5):455–77. doi: 10.1089/cmb.2012.0021
9. Larsen SE, Erasmus JH, Reese VA, Pecor T, Archer J, Kandahar A, et al. An RNA-based vaccine platform for use against Mycobacterium tuberculosis. Vaccines (Basel). (2023) 11(1):130. doi: 10.3390/vaccines11010130
10. Merker M, Blin C, Mona S, Duforet-Frebourg N, Lecher S, Willery E, et al. Evolutionary history and global spread of the Mycobacterium tuberculosis beijing lineage. Nat Genet. (2015) 47(3):242–9. doi: 10.1038/ng.3195
11. Glynn JR, Whiteley J, Bifani PJ, Kremer K, van Soolingen D. Worldwide occurrence of Beijing/W strains of Mycobacterium tuberculosis: A systematic review. Emerg Infect Dis. (2002) 8(8):843–9. doi: 10.3201/eid0805.020002
12. Kikuchi T, Nakamura M, Hachisu Y, Hirai S, Yokoyama E. Molecular epidemiological analysis of Mycobacterium tuberculosis modern beijing genotype strains isolated in chiba prefecture over 10 years. J Infect Chemother. (2022) 28(4):521–5. doi: 10.1016/j.jiac.2021.12.020
13. Shaik J, Pillay M, Moodley J, Jeena P. Predominance of the Mycobacterium tuberculosis beijing strain amongst children from a high tuberculosis burden township in South Africa. Tuberculosis (Edinb). (2022) 136:102250. doi: 10.1016/j.tube.2022.102250
14. Barbier M, Wirth T. The evolutionary history, demography, and spread of the Mycobacterium tuberculosis complex. Microbiol Spectr. (2016) 4(4). doi: 10.1128/microbiolspec.TBTB2-0008-2016
15. Tsolaki AG, Gagneux S, Pym AS, Goguet de la Salmoniere YO, Kreiswirth BN, Van Soolingen D, et al. Genomic deletions classify the Beijing/W strains as a distinct genetic lineage of Mycobacterium tuberculosis. J Clin Microbiol. (2005) 43(7):3185–91. doi: 10.1128/jcm.43.7.3185-3191.2005
16. He C, Cheng X, Kaisaier A, Wan J, Luo S, Ren J, et al. Effects of Mycobacterium tuberculosis lineages and regions of difference (Rd) virulence gene variation on tuberculosis recurrence. Ann Transl Med. (2022) 10(2):49. doi: 10.21037/atm-21-6863
17. Adams KN, Verma AK, Gopalaswamy R, Adikesavalu H, Singhal DK, Tripathy S, et al. Diverse clinical isolates of Mycobacterium tuberculosis develop macrophage-induced rifampin tolerance. J Infect Dis. (2019) 219(10):1554–8. doi: 10.1093/infdis/jiy710
18. Stucki D, Brites D, Jeljeli L, Coscolla M, Liu Q, Trauner A, et al. Mycobacterium tuberculosis lineage 4 comprises globally distributed and geographically restricted sublineages. Nat Genet. (2016) 48(12):1535–43. doi: 10.1038/ng.3704
19. Nahid P, Bliven EE, Kim EY, Mac Kenzie WR, Stout JE, Diem L, et al. Influence of M. tuberculosis lineage variability within a clinical trial for pulmonary tuberculosis. PloS One. (2010) 5(5):e10753. doi: 10.1371/journal.pone.0010753
20. He CJ, Wan JL, Luo SF, Guo RJ, Paerhati P, Cheng X, et al. Comparative study on tuberculosis drug resistance and molecular detection methods among different Mycobacterium tuberculosis lineages. Infect Drug Resist. (2023) 16:5941–51. doi: 10.2147/idr.S423390
21. Du DH, Geskus RB, Zhao Y, Codecasa LR, Cirillo DM, van Crevel R, et al. The effect of M. tuberculosis lineage on clinical phenotype. PloS Glob Public Health. (2023) 3(12):e0001788. doi: 10.1371/journal.pgph.0001788
22. Ordway D, Henao-Tamayo M, Harton M, Palanisamy G, Troudt J, Shanley C, et al. The hypervirulent Mycobacterium tuberculosis strain HN878 induces a potent TH1 response followed by rapid down-regulation. J Immunol. (2007) 179(1):522–31. doi: 10.4049/jimmunol.179.1.522
23. Palanisamy GS, Smith EE, Shanley CA, Ordway DJ, Orme IM, Basaraba RJ. Disseminated disease severity as a measure of virulence of Mycobacterium tuberculosis in the guinea pig model. Tuberculosis (Edinb). (2008) 88(4):295–306. doi: 10.1016/j.tube.2007.12.003
24. Baldwin SL, Reese VA, Huang PW, Beebe EA, Podell BK, Reed SG, et al. Protection and long-lived immunity induced by the Id93/Gla-se vaccine candidate against a clinical Mycobacterium tuberculosis isolate. Clin Vaccine Immunol. (2016) 23(2):137–47. doi: 10.1128/cvi.00458-15
25. Berube BJ, Larsen SE, McNeil MB, Reese VA, Pecor T, Kaur S, et al. Characterizing in vivo loss of virulence of an Hn878 Mycobacterium tuberculosis isolate from a genetic duplication event. Tuberculosis (Edinb). (2022) 137:102272. doi: 10.1016/j.tube.2022.102272
26. Domenech P, Rog A, Moolji JU, Radomski N, Fallow A, Leon-Solis L, et al. Origins of a 350-kilobase genomic duplication in Mycobacterium tuberculosis and its impact on virulence. Infect Immun. (2014) 82(7):2902–12. doi: 10.1128/iai.01791-14
27. Mittal E, Roth AT, Seth A, Singamaneni S, Beatty W, Philips JA. Single cell preparations of Mycobacterium tuberculosis damage the mycobacterial envelope and disrupt macrophage interactions. Elife. (2023) 12:e85416. doi: 10.7554/eLife.85416
28. Choreño-Parra JA, Bobba S, Rangel-Moreno J, Ahmed M, Mehra S, Rosa B, et al. Mycobacterium tuberculosis Hn878 infection induces human-like B-cell follicles in mice. J Infect Dis. (2020) 221(10):1636–46. doi: 10.1093/infdis/jiz663
29. Moreira-Teixeira L, Tabone O, Graham CM, Singhania A, Stavropoulos E, Redford PS, et al. Mouse transcriptome reveals potential signatures of protection and pathogenesis in human tuberculosis. Nat Immunol. (2020) 21(4):464–76. doi: 10.1038/s41590-020-0610-z
30. Maceiras AR, Silvério D, Gonçalves R, Cardoso MS, Saraiva M. Infection with hypervirulent Mycobacterium tuberculosis triggers emergency myelopoiesis but not trained immunity. Front Immunol. (2023) 14:1211404. doi: 10.3389/fimmu.2023.1211404
31. Ordway DJ, Shang S, Henao-Tamayo M, Obregon-Henao A, Nold L, Caraway M, et al. Mycobacterium bovis BCG-mediated protection against W-Beijing strains of Mycobacterium tuberculosis is diminished concomitant with the emergence of regulatory T cells. Clin Vaccine Immunol. (2011) 18(9):1527–35. doi: 10.1128/cvi.05127-11
32. Henao-Tamayo M, Obregón-Henao A, Creissen E, Shanley C, Orme I, Ordway DJ. Differential mycobacterium bovis BCG vaccine-derived efficacy in C3heb/Fej and C3h/Heouj mice exposed to a clinical strain of Mycobacterium tuberculosis. Clin Vaccine Immunol. (2015) 22(1):91–8. doi: 10.1128/cvi.00466-14
33. Jeon BY, Derrick SC, Lim J, Kolibab K, Dheenadhayalan V, Yang AL, et al. Mycobacterium bovis BCG immunization induces protective immunity against nine different Mycobacterium tuberculosis strains in mice. Infect Immun. (2008) 76(11):5173–80. doi: 10.1128/iai.00019-08
34. Palanisamy GS, DuTeau N, Eisenach KD, Cave DM, Theus SA, Kreiswirth BN, et al. Clinical strains of Mycobacterium tuberculosis display a wide range of virulence in guinea pigs. Tuberculosis (Edinb). (2009) 89(3):203–9. doi: 10.1016/j.tube.2009.01.005
35. Honda JR, Shang S, Shanley CA, Caraway ML, Henao-Tamayo M, Chan ED, et al. Immune responses of HIV-1 tat transgenic mice to Mycobacterium tuberculosis W-Beijing SA161. Open AIDS J. (2011) 5:86–95. doi: 10.2174/1874613601105010086
36. Obregón-Henao A, Shanley C, Bianco MV, Cataldi AA, Basaraba RJ, Orme IM, et al. Vaccination of guinea pigs using mce operon mutants of Mycobacterium tuberculosis. Vaccine. (2011) 29(26):4302–7. doi: 10.1016/j.vaccine.2011.04.027
37. Gupta M, Srikrishna G, Klein SL, Bishai WR. Genetic and hormonal mechanisms underlying sex-specific immune responses in tuberculosis. Trends Immunol. (2022) 43(8):640–56. doi: 10.1016/j.it.2022.06.004
38. Luo Y, Huang C-C, Liu Q, Howard N, Li X, Zhu J, et al. A Flot1 host regulatory allele is associated with a recently expanded mtb clade in patients with tuberculosis. medRxiv. (2022) 2022.02.07.22270622. doi: 10.1101/2022.02.07.22270622
39. Miyoshi-Akiyama T, Matsumura K, Iwai H, Funatogawa K, Kirikae T. Complete annotated genome sequence of Mycobacterium tuberculosis erdman. J Bacteriol. (2012) 194(10):2770. doi: 10.1128/jb.00353-12
40. Martin CJ, Cadena AM, Leung VW, Lin PL, Maiello P, Hicks N, et al. Digitally barcoding Mycobacterium tuberculosis reveals in vivo infection dynamics in the macaque model of tuberculosis. mBio. (2017) 8(3):e00312–17. doi: 10.1128/mBio.00312-17
41. Gatti DM, Tyler AL, Mahoney JM, Churchill GA, Yener B, Koyuncu D, et al. Systems genetics uncover new loci containing functional gene candidates in Mycobacterium tuberculosis-infected diversity outbred mice. bioRxiv. (2023). doi: 10.1101/2023.12.21.572738
42. Kurtz SL, Mittereder LR, Lehman CC, Khan H, Gould VA, Elkins KL. Intravenous BCG vaccination of diversity outbred mice results in moderately enhanced protection against challenge with Mycobacterium tuberculosis compared to intradermal vaccination. Infect Immun. (2023) 91(7):e0016823. doi: 10.1128/iai.00168-23
43. Koyuncu D, Niazi MKK, Tavolara T, Abeijon C, Ginese ML, Liao Y, et al. Cxcl1: A new diagnostic biomarker for human tuberculosis discovered using diversity outbred mice. PloS Pathog. (2021) 17(8):e1009773. doi: 10.1371/journal.ppat.1009773
44. Tavolara TE, Niazi MKK, Gower AC, Ginese M, Beamer G, Gurcan MN. Deep learning predicts gene expression as an intermediate data modality to identify susceptibility patterns in Mycobacterium tuberculosis infected diversity outbred mice. EBioMedicine. (2021) 67:103388. doi: 10.1016/j.ebiom.2021.103388
45. Niazi MK, Dhulekar N, Schmidt D, Major S, Cooper R, Abeijon C, et al. Lung necrosis and neutrophils reflect common pathways of susceptibility to Mycobacterium tuberculosis in genetically diverse, immune-competent mice. Dis Model Mech. (2015) 8(9):1141–53. doi: 10.1242/dmm.020867
46. Harrison DE, Astle CM, Niazi MKK, Major S, Beamer GL. Genetically diverse mice are novel and valuable models of age-associated susceptibility to Mycobacterium tuberculosis. Immun Ageing. (2014) 11(1):24. doi: 10.1186/s12979-014-0024-6
47. Gideon HP, Skinner JA, Baldwin N, Flynn JL, Lin PL. Early whole blood transcriptional signatures are associated with severity of lung inflammation in cynomolgus macaques with Mycobacterium tuberculosis infection. J Immunol (Baltimore Md 1950). (2016) 197(12):4817–28. doi: 10.4049/jimmunol.1601138
48. Steenken W, Oatway WH, Petroff SA. Biological studies of the tubercle bacillus : III. dissociation and pathogenicity of the R and S variants of the human tubercle bacillus (H(37)). J Exp Med. (1934) 60(4):515–40. doi: 10.1084/jem.60.4.515
49. Ioerger TR, Feng Y, Ganesula K, Chen X, Dobos KM, Fortune S, et al. Variation among genome sequences of H37Rv strains of Mycobacterium tuberculosis from multiple laboratories. J Bacteriol. (2010) 192(14):3645–53. doi: 10.1128/jb.00166-10
50. Baldwin SL, Reese V, Granger B, Orr MT, Ireton GC, Coler RN, et al. The Id93 tuberculosis vaccine candidate does not induce sensitivity to purified protein derivative. Clin Vaccine Immunol. (2014) 21(9):1309–13. doi: 10.1128/CVI.00372-14
51. Skeiky YA, Lodes MJ, Guderian JA, Mohamath R, Bement T, Alderson MR, et al. Cloning, expression, and immunological evaluation of two putative secreted serine protease antigens of Mycobacterium tuberculosis. Infect Immun. (1999) 67(8):3998–4007. doi: 10.1128/iai.67.8.3998-4007.1999
52. Tkachuk AP, Gushchin VA, Potapov VD, Demidenko AV, Lunin VG, Gintsburg AL. Multi-subunit BCG booster vaccine GamTBvac: Assessment of immunogenicity and protective efficacy in murine and guinea pig TB models. PloS One. (2017) 12(4):e0176784. doi: 10.1371/journal.pone.0176784
53. Woodworth JS, Clemmensen HS, Battey H, Dijkman K, Lindenstrøm T, Laureano RS, et al. A Mycobacterium tuberculosis-specific subunit vaccine that provides synergistic immunity upon co-administration with Bacillus Calmette-Guérin. Nat Commun. (2021) 12(1):6658. doi: 10.1038/s41467-021-26934-0
54. Chitale P, Lemenze AD, Fogarty EC, Shah A, Grady C, Odom-Mabey AR, et al. A comprehensive update to the Mycobacterium tuberculosis H37Rv reference genome. Nat Commun. (2022) 13(1):7068. doi: 10.1038/s41467-022-34853-x
55. Domenech P, Reed MB. Rapid and spontaneous loss of phthiocerol dimycocerosate (PDIM) from Mycobacterium tuberculosis grown in vitro: Implications for virulence studies. Microbiol (Reading). (2009) 155(Pt 11):3532–43. doi: 10.1099/mic.0.029199-0
56. Plumlee CR, Duffy FJ, Gern BH, Delahaye JL, Cohen SB, Stoltzfus CR, et al. Ultra-low dose aerosol infection of mice with Mycobacterium tuberculosis more closely models human tuberculosis. Cell Host Microbe. (2021) 29(1):68–82.e5. doi: 10.1016/j.chom.2020.10.003
57. Manabe YC, Dannenberg AM Jr., Tyagi SK, Hatem CL, Yoder M, Woolwine SC, et al. Different strains of Mycobacterium tuberculosis cause various spectrums of disease in the rabbit model of tuberculosis. Infect Immun. (2003) 71(10):6004–11. doi: 10.1128/iai.71.10.6004-6011.2003
58. Cohen SB, Plumlee CR, Engels L, Mai D, Murray TA, Jahn AN, et al. Host and pathogen genetic diversity shape vaccine-mediated protection to Mycobacterium tuberculosis. Front Immunol. (2024) 15:1427846. doi: 10.3389/fimmu.2024.1427846
59. Plumlee CR, Barrett HW, Shao DE, Lien KA, Cross LM, Cohen SB, et al. Assessing vaccine-mediated protection in an ultra-low dose Mycobacterium tuberculosis murine model. PloS Pathog. (2023) 19(11):e1011825. doi: 10.1371/journal.ppat.1011825
60. Smith CM, Proulx MK, Lai R, Kiritsy MC, Bell TA, Hock P, et al. Functionally overlapping variants control tuberculosis susceptibility in collaborative cross mice. mBio. (2019) 10(6):e02791–19. doi: 10.1128/mbio.02791-19
61. Smith CM, Proulx MK, Olive AJ, Laddy D, Mishra BB, Moss C, et al. Tuberculosis susceptibility and vaccine protection are independently controlled by host genotype. mBio. (2016) 7(5):01516–16. doi: 10.1128/mbio.01516-16
62. Lai R, Gong DN, Williams T, Ogunsola AF, Cavallo K, Lindestam Arlehamn CS, et al. Host genetic background is a barrier to broadly effective vaccine–mediated protection against tuberculosis. J Clin Invest. (2024) 133(13):e167762. doi: 10.1172/JCI167762
63. Smith CM, Baker RE, Proulx MK, Mishra BB, Long JE, Park SW, et al. Host-pathogen genetic interactions underlie tuberculosis susceptibility in genetically diverse mice. eLife. (2022) 11:e74419. doi: 10.7554/eLife.74419
64. Valway SE, Sanchez MP, Shinnick TF, Orme I, Agerton T, Hoy D, et al. An outbreak involving extensive transmission of a virulent strain of Mycobacterium tuberculosis. N Engl J Med. (1998) 338(10):633–9. doi: 10.1056/nejm199803053381001
65. Kaushal D, Foreman TW, Gautam US, Alvarez X, Adekambi T, Rangel-Moreno J, et al. Mucosal vaccination with attenuated Mycobacterium tuberculosis induces strong central memory responses and protects against tuberculosis. Nat Commun. (2015) 6:8533. doi: 10.1038/ncomms9533
66. Reed MB, Domenech P, Manca C, Su H, Barczak AK, Kreiswirth BN, et al. A glycolipid of hypervirulent tuberculosis strains that inhibits the innate immune response. Nature. (2004) 431(7004):84–7. doi: 10.1038/nature02837
67. Carey AF, Wang X, Cicchetti N, Spaulding CN, Liu Q, Hopkins F, et al. Multiplexed strain phenotyping defines consequences of genetic diversity in Mycobacterium tuberculosis for infection and vaccination outcomes. mSystems. (2022) 7(3):e0011022. doi: 10.1128/msystems.00110-22
68. Manca C, Tsenova L, Barry CE 3rd, Bergtold A, Freeman S, Haslett PA, et al. Mycobacterium tuberculosis CDC1551 induces a more vigorous host response in vivo and in vitro, but is not more virulent than other clinical isolates. J Immunol. (1999) 162(11):6740–6. doi: 10.4049/jimmunol.162.11.6740
69. Leisching G, Pietersen RD, Mpongoshe V, van Heerden C, van Helden P, Wiid I, et al. The host response to a clinical MDR mycobacterial strain cultured in a detergent-free environment: A global transcriptomics approach. PloS One. (2016) 11(4):e0153079. doi: 10.1371/journal.pone.0153079
70. Sani M, Houben EN, Geurtsen J, Pierson J, de Punder K, van Zon M, et al. Direct visualization by cryo-EM of the mycobacterial capsular layer: A labile structure containing ESX-1-Secreted proteins. PloS Pathog. (2010) 6(3):e1000794. doi: 10.1371/journal.ppat.1000794
71. Pietersen RD, du Preez I, Loots DT, van Reenen M, Beukes D, Leisching G, et al. Tween 80 induces a carbon flux rerouting in Mycobacterium tuberculosis. J Microbiol Methods. (2020) 170:105795. doi: 10.1016/j.mimet.2019.105795
72. Rens C, Chao JD, Sexton DL, Tocheva EI, Av-Gay Y. Roles for phthiocerol dimycocerosate lipids in Mycobacterium tuberculosis pathogenesis. Microbiol (Reading). (2021) 167(3). doi: 10.1099/mic.0.001042
73. Cohen SB, Gern BH, Delahaye JL, Adams KN, Plumlee CR, Winkler JK, et al. Alveolar macrophages provide an early Mycobacterium tuberculosis niche and initiate dissemination. Cell Host Microbe. (2018) 24(3):439–46.e4. doi: 10.1016/j.chom.2018.08.001
75. Painter H PS, Cia F, Stockdale L, Tanner R, Willcocks S, Reljic R, et al. Adaption of the ex vivo mycobacterial growth inhibition assay for use with murine lung cells. Sci Rep. (2020) 10:1–9. doi: 10.1038/s41598-020-60223-y
76. Painter H, Willcocks S, Zelmer A, Reljic R, Tanner R, Fletcher H. Demonstrating the utility of the ex vivo murine mycobacterial growth inhibition assay (MGIA) for high-throughput screening of tuberculosis vaccine candidates against multiple Mycobacterium tuberculosis complex strains. Tuberculosis (Edinb). (2024) 146:102494. doi: 10.1016/j.tube.2024.102494
77. Painter H, Harriss E, Fletcher HA, McShane H, Tanner R. Development and application of the direct mycobacterial growth inhibition assay: A systematic review. Front Immunol. (2024) 15:1355983. doi: 10.3389/fimmu.2024.1355983
78. Stanley S, Spaulding CN, Liu Q, Chase MR, Ha DTM, Thai PVK, et al. Identification of bacterial determinants of tuberculosis infection and treatment outcomes: A phenogenomic analysis of clinical strains. Lancet Microbe. (2024) 5(6):e570–e80. doi: 10.1016/s2666-5247(24)00022-3
79. Atavliyeva S, Auganova D, Tarlykov P. Genetic diversity, evolution and drug resistance of Mycobacterium tuberculosis lineage 2. Front Microbiol. (2024) 15:1384791. doi: 10.3389/fmicb.2024.1384791
80. Billows N, Phelan J, Xia D, Peng Y, Clark TG, Chang YM. Large-scale statistical analysis of Mycobacterium tuberculosis genome sequences identifies compensatory mutations associated with multi-drug resistance. Sci Rep. (2024) 14(1):12312. doi: 10.1038/s41598-024-62946-8
81. Harrison LB, Kapur V, Behr MA. An imputed ancestral reference genome for the Mycobacterium tuberculosis complex better captures structural genomic diversity for reference-based alignment workflows. Microb Genom. (2024) 10(1). doi: 10.1099/mgen.0.001165
82. Senelle G, Sahal MR, La K, Billard-Pomares T, Marin J, Mougari F, et al. Towards the reconstruction of a global TB history using a new pipeline "TB-Annotator". Tuberculosis (Edinb). (2023) 143s:102376. doi: 10.1016/j.tube.2023.102376
Keywords: Mycobacterium tuberculosis, tuberculosis, TB, lineages, diversity, isolates, IMPAc-TB
Citation: Larsen SE, Abdelaal HFM, Plumlee CR, Cohen SB, Kim HD, Barrett HW, Liu Q, Harband MH, Berube BJ, Baldwin SL, Fortune SM, Urdahl KB and Coler RN (2024) The chosen few: Mycobacterium tuberculosis isolates for IMPAc-TB. Front. Immunol. 15:1427510. doi: 10.3389/fimmu.2024.1427510
Received: 03 May 2024; Accepted: 06 September 2024;
Published: 28 October 2024.
Edited by:
Anna Kathleen Coussens, The University of Melbourne, AustraliaReviewed by:
Suraj B. Sable, Centers for Disease Control and Prevention (CDC), United StatesBen Gold, Weill Cornell Medicine, United States
Brian Weinrick, Trudeau Institute, United States
Copyright © 2024 Larsen, Abdelaal, Plumlee, Cohen, Kim, Barrett, Liu, Harband, Berube, Baldwin, Fortune, Urdahl and Coler. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Rhea N. Coler, UmhlYS5Db2xlckBTZWF0dGxlY2hpbGRyZW5zLm9yZw==
 Sasha E. Larsen
Sasha E. Larsen Hazem F. M. Abdelaal
Hazem F. M. Abdelaal Courtney R. Plumlee
Courtney R. Plumlee Sara B. Cohen
Sara B. Cohen Ho D. Kim1
Ho D. Kim1 Qingyun Liu
Qingyun Liu Susan L. Baldwin
Susan L. Baldwin Sarah M. Fortune
Sarah M. Fortune Rhea N. Coler
Rhea N. Coler