- Department of Blood Transfusion, China-Japan Union Hospital of Jilin University, Changchun, China
Background: Platelets play a significant role in the innate and adaptive processes of immunity and inflammation. Inflammatory bowel disease (IBD) is an autoimmune disease that is widely understood to be caused by a combination of genetic predisposition, aberrant immune responses, etc.
Methods: To examine the relationships between genetically determined platelet indices and IBD, we conducted a Mendelian randomization (MR) study. Data associated with platelet count (PLT), mean platelet volume (MPV), platelet distribution width (PDW), plateletcrit (PCT) were used from the UK Biobank. The outcome data, including IBD, Crohn’s disease (CD), ulcerative colitis (UC), were from the FinnGen database. The inverse variance-weighted (IVW), MR-Egger, weighted median methods were used for MR analyses.
Results: The MR estimations from the IVW approach show a significant connection between PLT and IBD. Similarly, PCT and IBD have a relationship following the IVW and MR-Egger approaches. While PLT and PCT have strong relationships with CD, according to the findings of all three approaches respectively. Nevertheless, PDW was the only relevant indicator of UC. The only significant result was IVW’s.
Conclusion: Our findings suggest that the fluctuation of platelet indicators is of great significance in the development of IBD. PLT and PCT have a close association with IBD and CD, respectively; PDW only has a connection with UC. Platelets play an important role in the progression of IBD (UC, CD).
Introduction
Platelets are blood cells in plasma that are well recognized for their critical role in sustaining blood hemostasis (1). Megakaryocytes (MKs) create billions of them every day. MKs perceive and respond to inflammatory stress, and they engage in host immunological responses, according to emerging data (2). Platelet count (PLT), mean platelet volume (MPV), platelet width of distribution (PDW), and plateletcrit (PCT) are major platelet indicators in clinical practice that may be utilized to indicate platelet biochemical and functional changes (3). Platelets also play important roles in innate and adaptive immunity and inflammation, and they are the first blood cells to respond to wound-healing and tissue-repair mechanisms (1). Small platelets manage to maintain vascular integrity when faced with challenges of infection, sterile inflammation, and even malignancy, where they aid in hemostasis and serve as early responders to microbial threats (4). Because of their quick recruitment dynamics, these tiny, anucleate cell fragments are the first cells to form not just at sites of damage but also at sites of inflammation (5). Intravital imaging indicated that platelets are recruited and behave as individual cells rather than clots in the inflamed microvasculature, indicating that the hemostatic mechanism is unique to classical thrombosis and hemostasis. Unlike the well-defined processes of hemostasis following vascular trauma, inflammation-associated hemorrhage, also known as inflammatory bleeding, is a simplified summary of a phenomenon that occurs in a variety of disease settings, including sterile inflammation, microbial infection, and malignant tumors (6–8). Predilection sites include mucosal membranes, with epistaxis, gum bleeding, gastrointestinal bleeds, and hematuria being the most common bleeding episodes in thrombocytopenia patients. Platelet-mediated hemostasis without clot formation is critical to maintaining vascular integrity under these conditions (9, 10).
The autoimmune illness known as inflammatory bowel disease (IBD) is a chronic, relapsing condition that has caused significant health problems and is becoming more commonplace worldwide (11, 12). It is well accepted that genetic predisposition, environmental variables, and abnormal immune responses combine to cause IBD (13). The two main IBD subtypes, ulcerative colitis (UC) and Crohn’s disease (CD), can differ significantly in terms of their molecular, immunological, morphological, and clinical features (14). Rectal bleeding, diarrhea, stomach discomfort, fever, anemia, and weight loss are some of the symptoms of UC (15, 16). CD may impact any region of the digestive tract in addition to causing diarrhea and abdominal pain (17). Up to 29.3 percent of IBD patients have at least one extra intestinal manifestation (EIM), which can have an effect on many systems, according to a Swiss cohort study (18). As per the European Crohn’s and Colitis Organization, at least one EIM is experienced by up to 50% of people with IBD (19). Because of its great prevalence, IBD not only drastically lowers patients’ quality of life but also places a major financial and medical burden on society (20), additionally accompanied by a number of issues or EIM (21). The most common areas of the body affected by the various types of EIMs are the musculoskeletal system, mucocutaneous system, ocular system, hepatobiliary tract, and oral cavity. There’s a chance that other systems, including the pancreatic, pulmonary, cardiovascular, and urogenital systems are also at play (22, 23). Hematological EIMs haven’t been thoroughly acknowledged or verified yet. Although the exact pathogenesis of EIMs is still unknown, it often involves dysregulated immunological responses, environmental factors, genetic vulnerability, and microbiota dysbiosis (19). Therefore, in order to obtain better prevention and control, it is essential to investigate the pathophysiology and risk factors of IBD. Determining causative relationships and possible risk factors for IBD represents an emerging public health concern.
A recent research by Vallet et al., which was published in the Journal of Clinical Investigation (24), demonstrates how the locations of megakaryocytes and the quality of platelet production alter with illness. Considering the vital role platelets play in coagulation, wound healing, tissue damage repair, immunological response, and inflammatory infections. Thus, assessments of platelet indices that reflect platelet bioactivity may be extremely important for tracking the onset, course, management, and prognosis of IBD.
In conclusion, it has not been established that platelet indices and IBD (UC and CD) are causally related. However, conventional observational study designs are limited in their ability to establish causality regarding the function of platelets in the development of IBD because of significant methodological constraints like reverse causation and residual confounding. A different strategy is the Mendelian randomization (MR) design, which makes use of genetic variations as instrumental variables (IVs) for an exposure in order to establish the causal relationship between the exposure and the outcome (25–28). By employing genetic variation as an indicator of causation, MR can remove the confounding bias seen in observational research. As alleles follow the principle of random assignment, different genotypes result in different intermediate phenotypes. If this phenotype represents an individual’s exposure characteristic, the association effect between genotype and disease can describe the impact of exposure factors on illness. This effect is unaffected by confounding factors and reverse causal associations, as in traditional epidemiological studies (25, 29). The MR study concept is founded on Mendel’s rule and functions similarly to a randomized controlled trial (RCT) but without the high expense (30).
In the current investigation, we employed a two-sample MR analysis to ascertain the association between platelet indices (PLT, MPV, PDW, and PCT) and IBD (UC and CD). It suggests that an IV-induced modifiable exposure caused the result. Therefore, we think the single-nucleotide polymorphisms (SNPs) used as research instruments had a modifying impact on the platelet indices, proving a positive causal relationship between the SNPs and the probability of developing IBD. However, interventions aimed at targeting the exposure are unlikely to be effective if there is a non-causal link between the exposure and the outcome.
Materials and methods
Study design
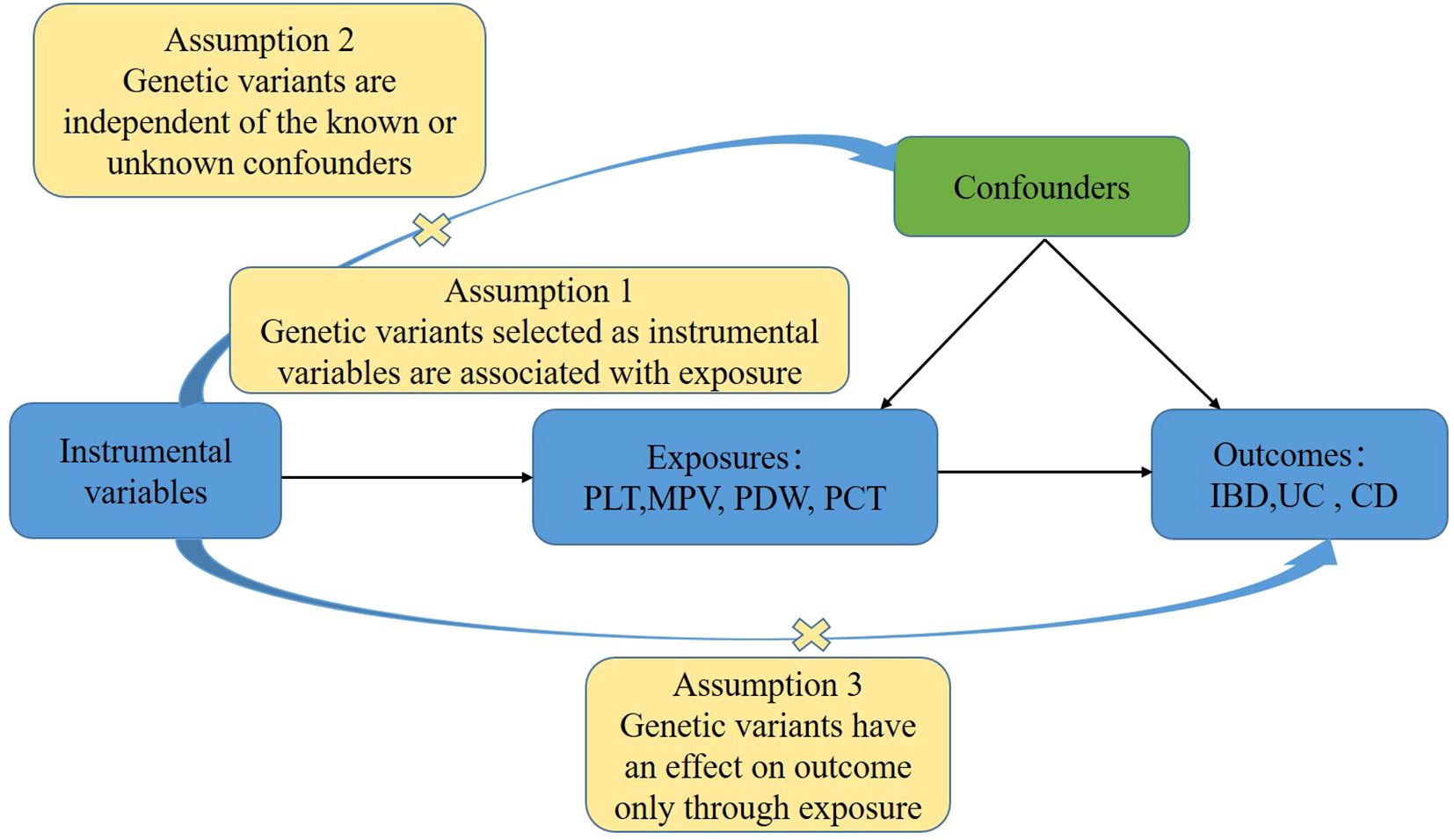
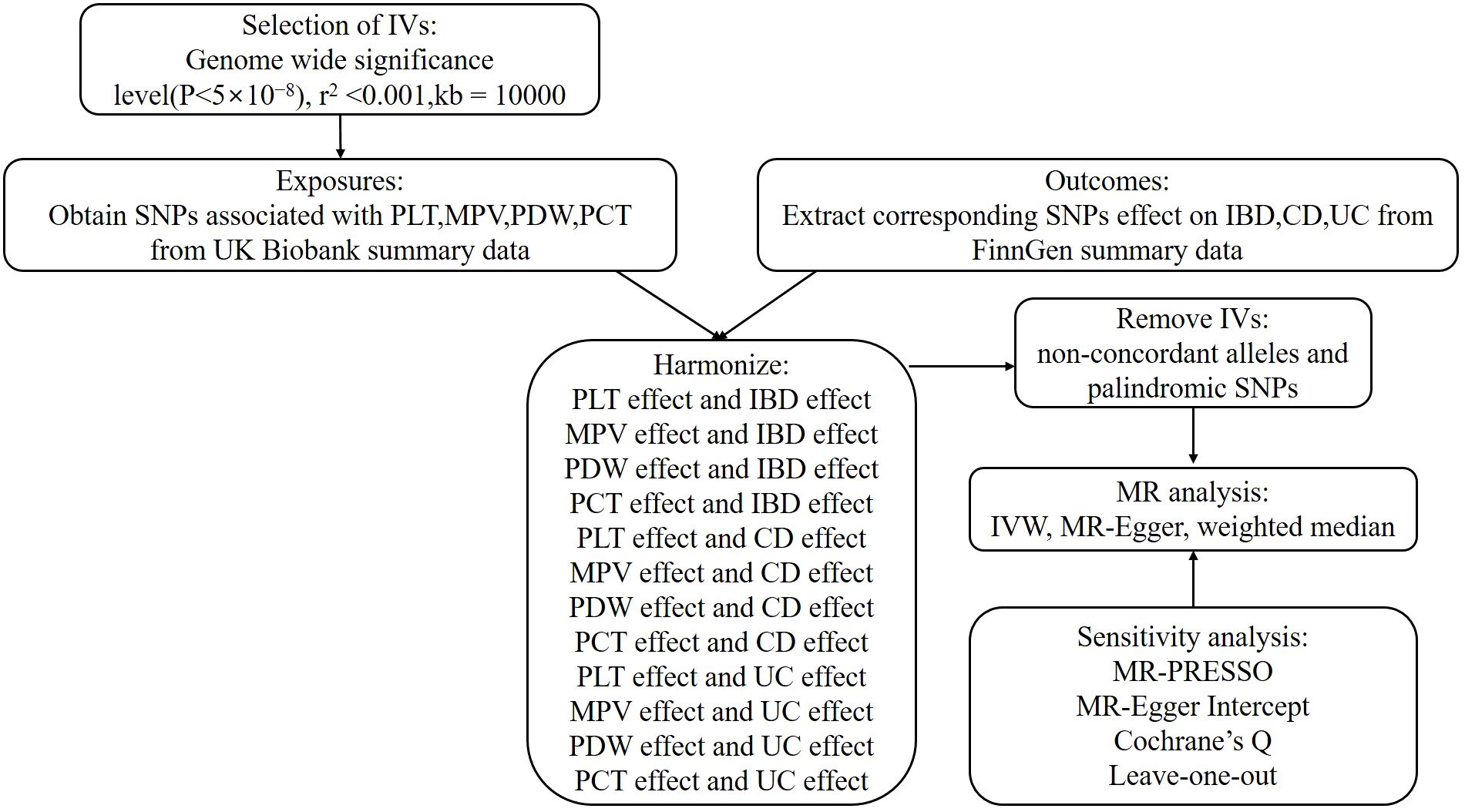
In order to investigate the associations between platelet indices (PLT, MPV, PDW, and PCT) and IBD (UC and CD), we used a two-sample MR design. Strengthening the Reporting of Observational Studies in Epidemiology Using Mendelian Randomization and the Fundamentals of MR were adhered to in the design of our study (31). Additionally, these selections underwent an MR analysis and satisfied three fundamental presumptions (Figure 1): Three things are relevant about the instrumental variables: (1) they are directly correlated with the exposure; (2) they are unaffected by confounders; and (3) genetic variations only influence outcomes through exposure (32). The purpose of the univariable MR study was to explore the relationship between platelet indices (PLT, MPV, PDW, and PCT) and IBD (UC and CD). The research design employed is shown in Figure 2.
Data source
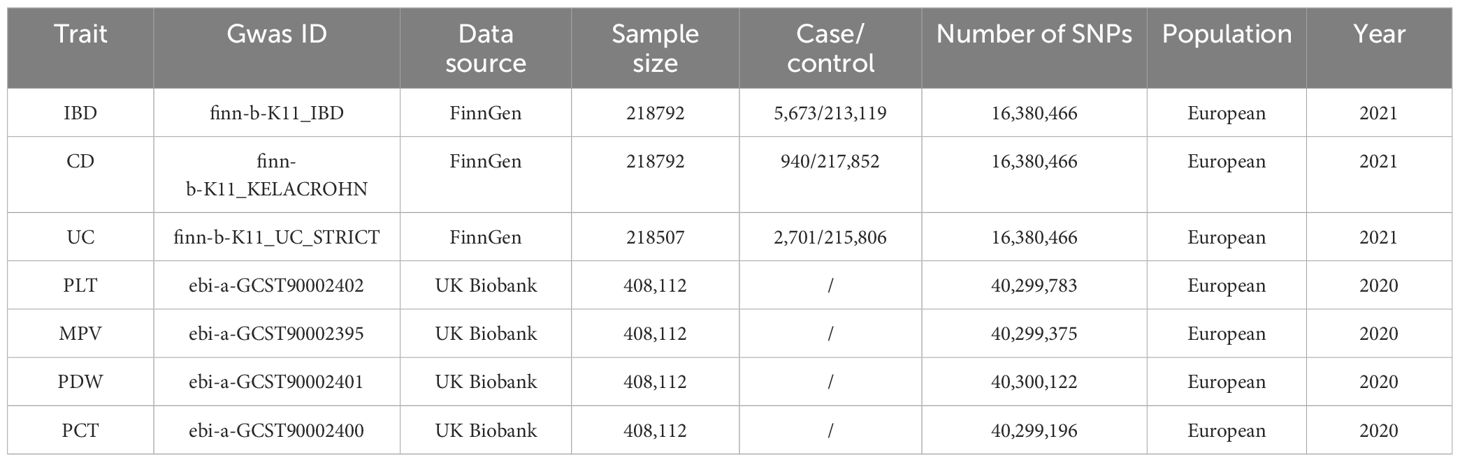
The genetic tools for the four platelet indices (PLT, MPV, PDW, and PCT) were chosen from a genome-wide association study (GWAS) that involved 408,112 participants in the UK Biobank (33). Every participant was descended from Europeans. Data from the FinnGen collaboration, which became publicly available in May 2021, was utilized to determine the outcomes. Which enrolled 218,792 European participants (cases/controls for IBD: 5,673/213,119; CD: 940/217,852); and 218,507 participants (cases/controls for UC: 2,701/215,806) (34). Since 2017, FinnGen has been a large-scale national effort that aims to improve human medicine by gathering genetic data and health record information from Finnish health registries and Biobanks, respectively. The detailed information on all traits involved was summarized in Table 1. Since all of the data are GWAS summary statistics that are available to the public, no further ethical approval or informed permission was needed.
Selection of instrumental variables
IVs were chosen as independent SNPs at genomewide significance (P<5×10−8) for every exposure taken into account in univariable MR analysis. Pairwise linkage disequilibrium, or independent SNPs, were found using criteria of (r2<0.001, clumping window=10,000 kb). To find and eliminate outlier instruments, MR pleiotropy residual sum and outlier (MR-PRESSO) analyses were carried out. The cumulative strength of the chosen SNPs was assessed using the F-statistic (F = beta2/se2), where beta denoted the exposure’s effect value and se denoted the exposure’s standard error. This helped to prevent weak instrument bias. F>10 is required to access the whole SNPs collection (35). The F-statistics used in the univariable MR analyses are provided in Supplementary Table 1.
Statistical analysis
Reverse causation can lead to an incorrect inference that the exposure and the outcome are causally connected if variations in the outcomes that exhibit greater relationships with outcomes than with exposures are employed in the MR analyses (36). Consequently, we must exclude the SNPs that have an outcome of P<5×10−8. And then, prior to analysis, we first harmonized exposure and outcome data to make alignments on effect alleles to the forward strand, if it is specified or could be inferred based on the allele frequency. Ambiguous SNPs with non-concordant alleles and palindromic SNPs that may create uncertainty regarding the identification of the effect allele in the exposure and outcome GWASs were excluded for further MR analyses (37, 38). After identifying the IV sets using the aforementioned selection criteria, we estimated the total effects using MR analysis. We performed a significance analysis using the IVW approach. Assuming that all SNPs are legitimate instrumental factors, this technique yields the maximum power estimate. When all IVs are genuine and horizontal pleiotropy is balanced, this method yields unbiased estimates of causal links even in the presence of variability across SNPs (39). The MR-Egger regression was used in secondary analyses to account for pleiotropy and assess the findings’ robustness. Although its power is limited, the MR-Egger method can identify and rectify directional pleiotropy. Even in the event that the second and third assumptions are false, it accounts for the directed pleiotropic effects of genetic instruments (40). The MR-Egger test produces a consistent causal estimate and a valid test of the null causal hypothesis, even in the case when all genetic variations are invalid (40). Nevertheless, MR-Egger shows poor statistical accuracy and is vulnerable to outlying genetic variations (41). The weighted median approach is the third method. It is substantially and continuously more accurate than the MR Egger approach and more resilient to violations of causal effects (42). It is predicated on the supposition that more than half of the IVs are believable. Furthermore, outliers and high-leverage genetic variants won’t have an impact on it (42). Otherwise, the IVW outcomes took precedence. The OR and accompanying 95% CI on the outcome risk of corresponding unit changes in exposure were used to represent the MR results. To evaluate the relative risk brought on by the existence of the illness of interest, the OR and 95% CI were shown. P<0.05 was used to indicate statistical significance in the univariable MR analysis for the findings of sensitivity analyses on the causal effects of exposures and outcomes. To depict the MR data, scatterplots, forest plots, and funnel plots were created in the interim.
We also assessed horizontal pleiotropy for significant estimates using the intercept tests of MR-Egger regression and MR-PRESSO. MR-Egger regression yielded an intercept, and intercept values that differ from zero indicate pleiotropy (here assessed using a p-value <0.05), which was suggestive of an overall directional pleiotropy (43). Using the global and SNP-specific observed residual sum of squares, the MR-PRESSO method screened for general horizontal pleiotropy (global test) and outliers (outlier test), with a significant threshold of 0.05 (44). Additionally, after eliminating outliers, it provided causal estimates and contrasted the raw values with the distortion. Additionally, 10,000 distribution points were allocated. By gradually eliminating each IV, leave-one-out analysis was performed in order to identify bias caused by a heterogeneous variation. In order to identify heterogeneity (p<0.05 shows heterogeneity), we also calculated the Cochrane’s Q value, which allowed us to identify the existence of pleiotropy (45). Each SNP’s heterogeneity in terms of causative effects was assessed using Cochran’s Q value (46). For the second and third assumptions to be satisfied, horizontal pleiotropy must be assessed (38). R statistical program (version 4.3.1, R Foundation for Statistical Computing, Vienna, Austria, 2023; https://www.R-project.org) was used for all statistical analyses, together with the Two-Sample MR and MR-PRESSO Packages (38).
Results
Selection of instrumental variables
Altogether, 477 index SNPs were shown to be possible genetic IVs for IBD, 482 SNPs for CD, and 479 SNPs for UC when PLT was taken into account as an exposure factor. In the presence of MPV as an exposure factor, 453 index SNPs were shown to be putative genetic IVs for IBD, 455 SNPs for CD, and 454 SNPs for UC, in that order. PDW as an exposure factor led to the identification of 379 index SNPs as putative genetic IVs for IBD, 378 SNPs for CD, and 375 SNPs for UC, in that order. In the case of PCT as an exposure factor, possible genetic IVs for IBD, CD, and UC were found to be 452 index SNPs, 454 SNPs, and 453 SNPs, respectively. Not only have all of these SNPs been harmonized and palindromic SNPs with intermediate allele frequencies removed, but they have also undergone the MR-PRESSO test, which was run in order to identify and eliminate outlier IVs. Once the outlier IVs were eliminated, MR estimations were reexamined. Thus, the SNPs listed above were taken into account for the MR analysis. Furthermore, each SNP’s F-value was greater than 10, which suggests that there is a minimal possibility of weak instrumental variable bias.
Mendelian randomization analysis
Overall, there was inconsistency in the results from the three approaches used to establish a causal relationship between platelet indicators (PLT, MPV, PDW, and PCT) and IBD (UC and CD). According to the IVW method’s MR estimations, there is a significant correlation between PLT and IBD (OR:1.11, 95%CI:1.02 to 1.21, P:0.013). However, IBD was not associated with the findings of the MR-Egger or weighted median techniques (OR:1.14, 1.11,95%CI:0.99 to 1.32,0.98 to 1.27, P:0.079,0.095), respectively. Likewise, there is a close link between PCT and IBD. IVW produced the following results: OR:1.10, 95%CI:1.01 to 1.20, P:0.034. OR:1.19, 95%CI:1.02 to 1.39, P:0.023 was the MR-Egger. However, there was no significant difference using the weighted median approach (OR:1.10, 95%CI:0.95 to 1.28, P:0.2). PLT and PCT were related to CD, whereas PDW was connected to UC, according to further study of the two subtypes. IVW (OR:1.35, 95%CI:1.15 to 1.59, P:0.0003), MR-Egger (OR:1.43, 95%CI:1.07 to 1.90, P:0.015), and weighted median (OR:1.41, 95%CI:1.06 to 1.86, P:0.017) were the values obtained from PLT to CD. PLT and CD have strong relationships, according to the findings of all three approaches. A comparison between PCT and CD revealed similarities in the IVW (OR:1.27, 95%CI:1.06 to 1.52, P:0.011), MR-Egger (OR:1.89, 95%CI:1.38 to 2.59, P:9.3×10-5), and weighted median (OR:1.36, 95%CI:1.01 to 1.85, P:0.046). PCT was closely associated to CD, according to the findings of all three methodologies. However, the only elevated factor with regard to UC was PDW. And only IVW’s finding (OR:1.14, 95%CI:1.01 to 1.29, P:0.032) was remarkable. We found no relationship between other platelet indices and IBD, CD, and UC; the detailed results and scatterplots are listed in Figures 3, 4. And the forest plots and funnel plots are shown in Supplementary Figures 1, 2.
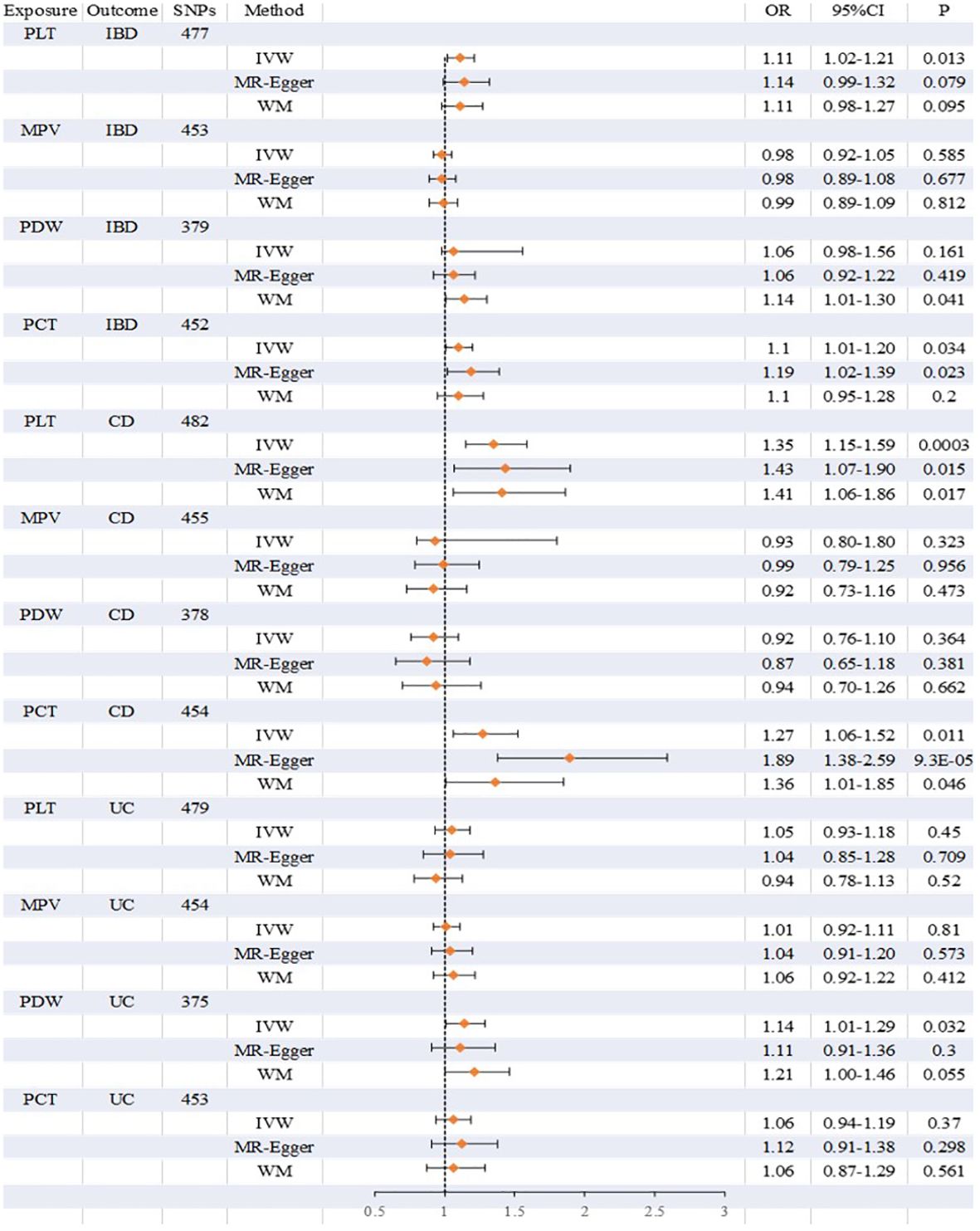
Figure 3 Detailed results on the association between platelet indices (PLT, MPV, PDW, PCT) and IBD, CD and UC.
Figure 4 Scatter plots of the MR analysis. (A) PLT on IBD. (B) MPV on IBD. (C) PDW on IBD. (D) PCT on IBD. (E) PLT on CD. (F) MPV on CD. (G) PDW on CD. (H) PCT on CD. (I) PLT on UC. (J) MPV on UC. (K) PDW on UC. (L) PCT on UC.
Sensitivity analysis
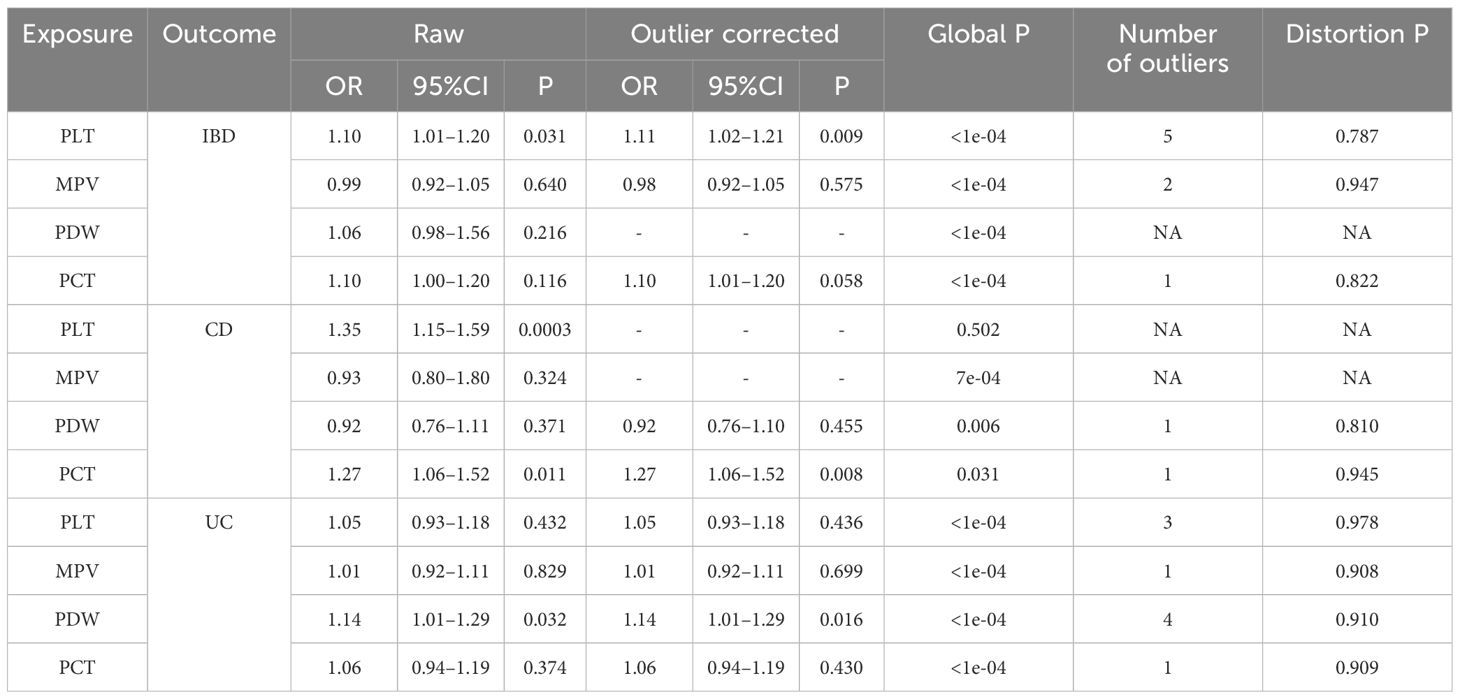
While some of the Cochran Q test findings showed heterogeneity, the major outcome of the random effects IVW analysis allowed for some heterogeneity. All except one of the p-values for the MR-Egger intercept were greater than 0.05. The results and details are provided in Supplementary Table 2. Furthermore, our results’ robustness was further validated by the fact that leave-one-out analysis failed to find any outlier IVs (Supplementary Figure 3). Additionally, following the global MR-PRESSO testing, we had to exclude a few SNPs. However, there were all significant SNPs after removing the outliers. The MR-PRESSO distortion test results showed the causal effect of genetically predicted platelet indices on IBD (CD, UC) after correction by removing outliers. On the other hand, genetically predicted platelet indices were shown to raise the risk of IBD (CD, UC) in both corrected and uncorrected data (Table 2).
Discussion
This is the first MR research that we are aware of that examines the relationship between platelet indices and IBD (UC and CD). The purpose of the current study was to investigate the relationship between IVs of the four platelet indices and IBD (CD, UC). We discovered in the univariable MR that a rise in IBD and CD was correlated with the amounts of PLT and PCT predicted by the provided genetics, while PDW was linked to UC. But there was no significant correlation between other platelet indicators and IBD (CD, UC). According to these results, PLT and PCT are the essential characteristics that generate favorable correlations between IBD and CD. PDW may only relevant to UC.
A two-sample MR analysis of the relationship between platelet indices and IBD was conducted for this investigation. There was shown to be a strong relationship between platelet indices and IBD. In order to better understand the association between platelet indices and IBD and to develop therapies for the disease, a greater study of the correlations between various platelet indices and IBD utilizing bigger and more diverse data sources is necessary. However, although they are categorized as IBD, CD, and UC, they are not the same in terms of pathophysiology, symptoms, complications, natural courses, and sequelae. In addition to severely impairing a patient’s quality of life, CD and UC both increase mortality and financial burden (12, 14, 47). Although the exact cause of IBD (CD, UC) is still unclear, genetic vulnerability, environmental factors, and the gut microbiome may all be significant (48). Further evidence of these two distinct situations was found in our research.
As is well known, PLT counts the number of platelets per unit volume of blood, PCT represents the proportion of blood volume occupied by platelets, and MPV indicates the average size of platelets. As a result, PCT is connected with the products of MPV and PLT, and may be thought of as a sort of analog of the total platelet volume. PDW, in comparison to PLT, PCT and MPV, is another significant metric. Thus, elevated indices may suggest that platelets play a part in understanding the IBD process (CD, UC). In our study, we have found there is a relevance between PLT, PDW, and PCT with IBD (CD, UC), so the platelet indices reflect this phenomenon and may be useful indicators for assessing the course of IBD (CD, UC). In clinical practice, it is important to highlight the independent and prominent roles that PLT, PCT, and PDW play among the four platelet indices.
Excessive clotting or unusual bleeding are the outcomes of elevated platelet levels (49). Because of the close involvement of their membrane receptors at different stages of the blood-coagulation cascade (50), a sequence of biochemical reactions that take place in the body in response to injury or damage to blood vessels, platelets play a critical role as the defenders of the integrity of the blood vasculature. The exterior membrane of platelets is extremely active and functional, expressing different integrin, glycoproteins, and antigens (1). These membrane constituents play a crucial role in coordinating the intricate interplay between platelets and sub endothelial structures that are exposed due to blood vessel wall damage. Additionally, proteins that make up fibrin clots and plasma coagulation factors and activators interact with biomolecules produced on platelet membranes. Membrane glycoproteins identify blood clotting factors and play a key role in platelet adherence and activation. Platelet membranes strongly express GPIIb/IIIa, GPIb-IX-V, GPVI, and P2Y12, all of which are essential in the hemostatic process that comes before the wound-healing phase (51). The immunological response of the body is improved by platelets. It has been demonstrated that platelet-derived CD40L may stimulate monocyte differentiation into dendritic cells (DC), DC maturation, and co-stimulatory molecule upregulation (52). This role of platelet-derived CD40L may be particularly important for autoimmune illnesses like systemic lupus erythematosus, where platelets stimulate B-cell secretion of antibodies via inducing DC differentiation and type-I interferon release (53). But IBD is an autoimmune disease that recurs frequently, causing intestinal bleeding, inflammatory responses, and EIMs such as cardiovascular problems. Furthermore, the precise aspects of its pathophysiology are yet unknown, but they appear to be linked to immune response problems and genetic predisposition. So combining the function of platelets and the MR results we obtained, platelet-related indices are indeed closely related to IBD and predict its occurrence and development.
We discovered the link between platelet indices and IBD (UC and CD), as previously mentioned. However, three presumptions relevance, independence, and exclusion-restriction are necessary for IVs to be valid in MR. The second and third assumptions, however, are dependent on every potential confounding factor of the exposure-outcome connection, both measurable and unmeasured, and only the first can be completely empirically evaluated. To provide a consistent estimate of the causative effect, all genetic variations included in the research as IVs must meet the MR assumptions for the IVW method (42). Both the weighted median and the MR-Egger methods were used to verify this. Even in cases where all genetic effects are null due to violations of the third assumption mentioned above, the MR-Egger approach reliably predicts the genuine causal impact under a lesser assumption (54). However, if all genetic variants have a comparable degree of connection with the exposure, then MR-Egger regression estimates become less accurate. On the other hand, if no single genetic variation accounts for more than 50% of the weight, the weighted median approach will yield a consistent estimate only if at least 50% of the weight originates from legitimate genetic variants. When it comes to faulty genetic variations, the weighted median method permits a more widespread violation of the MR assumptions than the MR-Egger method does (42). Therefore, we think that the remaining results suggest a causal relationship between platelet indices and IBD, even if an MR-Egger technique observation yielded a non-significant estimate.
Although we have identified a relationship between platelet indices and IBD through the MR study. There were a few more restrictions on this study. Firstly, it is probable that the putative gender-specific effects on the relationship were overlooked since we did not separate platelet indices and IBD (UC and CD) by gender. The UK Biobank sample was used for the GWAS of characteristics linked to platelet indices, while FinnGen provided data on IBD (CD, UC). As a result, bias and sample overlap are possible in relation to this fact (55). Furthermore, even though steps have been taken to identify and eliminate outlier SNPs, we cannot totally rule out the possibility that heterogeneity will have an impact on the results. Moreover, our work has demonstrated a causal association between platelet indices and IBD (UC and CD); nevertheless, additional research is necessary as the specific underlying processes are still unclear. Then, even with an MR research design, confounding cannot be totally minimized because the risk factors for IBD (CD, UC) comprise not just genetic variables but also other factors, such as environmental ones. Finally, the study only contained four platelet indices; more hematological indicators associated with platelets may exist, meaning that the relative importance of PLT, PCT, and PDW may need to be adjusted when considering other features.
Conclusions
Evidence supporting PLT, PCT, and PDW as distinct and predominant features explaining the relationship to IBD (CD, UC) may be found in the current MR investigation. Comprehending the function of platelets and their associated characteristics is beneficial for both public and clinical health. To strengthen the case for antiplatelet medication as the main preventive measure in IBD patients, stratified randomized controlled trials are also required. Our MR investigation showed that PLT and PCT had a connection to IBD and CD meanwhile that PDW had a relation to UC. To a certain extent, platelets and their associated characteristics influence the development of IBD (UC, CD). A possible preventative method for IBD might involve focusing on these characteristics. Further research is required to determine the precise mechanism and validate the therapeutic benefits of this kind of preventive therapy.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Ethics statement
Ethical approval was not required for the study involving humans in accordance with the local legislation and institutional requirements. Written informed consent to participate in this study was not required from the participants or the participants’ legal guardians/next of kin in accordance with the national legislation and the institutional requirements. Written informed consent was not obtained from the individual(s) for the publication of any potentially identifiable images or data included in this article because online public data does not require informed consent.
Author contributions
HL: Data curation, Formal analysis, Investigation, Software, Writing – original draft, Writing – review & editing. TL: Conceptualization, Funding acquisition, Writing – review & editing.
Funding
The authors declare financial support was received for the research, and/or publication of this article. This work was supported by the grants from National Key Research and Development Project of China (No. 2023YFC2413100).
Acknowledgments
The authors acknowledge and thank the investigators of the original GWAS studies for sharing the summary data used in this study.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2024.1377915/full#supplementary-material
Abbreviations
CD, Crohn’s disease; CI, Confidence intervals; EIM, Extra intestinal manifestation; IBD, Inflammatory bowel disease; IVs, Instrumental variables; IVW, Inverse variance-weighted; GWAS, Genome-wide association studies; MKs, Megakaryocytes; MPV, Mean platelet volume; MR, Mendelian randomization; MR-PRESSO, MR pleiotropy residual sum and outlier; OR, Odds ratios; PCT, Platelet crit; PDW, Platelet distribution width; PLT, Platelet count; SNPs, Single-nucleotide polymorphisms; UC, Ulcerative colitis; WM, Weighted Median.
References
1. Burnouf T, Chou ML, Lundy DJ, Chuang EY, Tseng CL, Goubran H. Expanding applications of allogeneic platelets, platelet lysates, and platelet extracellular vesicles in cell therapy, regenerative medicine, and targeted drug delivery. J BioMed Sci. (2023) 30:79. doi: 10.1186/s12929-023-00972-w
2. Khatib-Massalha E, Méndez-Ferrer S. Megakaryocyte diversity in ontogeny, functions and cell-cell interactions. Front Oncol. (2022) 12:840044. doi: 10.3389/fonc.2022.840044
3. Pogorzelska K, Krętowska A, Krawczuk-Rybak M, Sawicka-Żukowska M. Characteristics of platelet indices and their prognostic significance in selected medical condition - a systematic review. Adv Med Sci. (2020) 65:310–5. doi: 10.1016/j.advms.2020.05.002
4. Kaiser R, Escaig R, Nicolai L. Hemostasis without clot formation: how platelets guard the vasculature in inflammation, infection, and Malignancy. Blood. (2023) 142:1413–25. doi: 10.1182/blood.2023020535
5. Nicolai L, Massberg S. Platelets as key players in inflammation and infection. Curr Opin Hematol. (2020) 27:34–40. doi: 10.1097/MOH.0000000000000551
6. Ho-Tin-Noé B, Boulaftali Y, Camerer E. Platelets and vascular integrity: how platelets prevent bleeding in inflammation. Blood. (2018) 131:277–88. doi: 10.1182/blood-2017-06-742676
7. Ho-Tin-Noé B, Demers M, Wagner DD. How platelets safeguard vascular integrity. J Thromb Haemost. (2011) 9 Suppl 1:56–65. doi: 10.1111/j.1538-7836.2011.04317.x
8. Ho-Tin-Noé B, Goerge T, Wagner DD. Platelets: guardians of tumor vasculature. Cancer Res. (2009) 69:5623–6. doi: 10.1158/0008-5472.CAN-09-1370
9. Nachman RL, Rafii S. Platelets, petechiae, and preservation of the vascular wall. N Engl J Med. (2008) 359:1261–70. doi: 10.1056/NEJMra0800887
10. Wéra O, Lecut C, Servais L, Hego A, Delierneux C, Jiang Z, et al. P2X1 ion channel deficiency causes massive bleeding in inflamed intestine and increases thrombosis. J Thromb Haemost. (2020) 18:44–56. doi: 10.1111/jth.14620
11. Ng SC, Shi HY, Hamidi N, Underwood FE, Tang W, Benchimol EI, et al. Worldwide incidence and prevalence of inflammatory bowel disease in the 21st century: a systematic review of population-based studies. Lancet. (2017) 390:2769–78. doi: 10.1016/S0140-6736(17)32448-0
12. Kaplan GG. The global burden of IBD: from 2015 to 2025. Nat Rev Gastroenterol Hepatol. (2015) 12:720–7. doi: 10.1038/nrgastro.2015.150
13. Ramos GP, Papadakis KA. Mechanisms of disease: inflammatory bowel diseases. Mayo Clin Proc. (2019) 94:155–65. doi: 10.1016/j.mayocp.2018.09.013
14. Roda G, Chien Ng S, Kotze PG, Argollo M, Panaccione R, Spinelli A, et al. Crohn's disease. Nat Rev Dis Primers. (2020) 6:22. doi: 10.1038/s41572-020-0156-2
15. Bu F, Ding Y, Chen T, Wang Q, Wang R, Zhou JY, et al. Total flavone of Abelmoschus Manihot improves colitis by promoting the growth of Akkermansia in mice. Sci Rep. (2021) 11:20787. doi: 10.1038/s41598-021-00070-7
16. Kim HY, Cheon JH, Lee SH, Min JY, Back SY, Song JG, et al. Ternary nanocomposite carriers based on organic clay-lipid vesicles as an effective colon-targeted drug delivery system: preparation and in vitro/in vivo characterization. J Nanobiotechnology. (2020) 18:17. doi: 10.1186/s12951-020-0579-7
17. Levison SE, Fisher P, Hankinson J, Zeef L, Eyre S, Ollier WE, et al. Genetic analysis of the Trichuris muris-induced model of colitis reveals QTL overlap and a novel gene cluster for establishing colonic inflammation. BMC Genomics. (2013) 14:127. doi: 10.1186/1471-2164-14-127
18. Vavricka SR, Rogler G, Gantenbein C, Spoerri M, Prinz Vavricka M, Navarini AA, et al. Chronological order of appearance of extraintestinal manifestations relative to the time of IBD diagnosis in the swiss inflammatory bowel disease cohort. Inflammation Bowel Dis. (2015) 21:1794–800. doi: 10.1097/MIB.0000000000000429
19. Hedin CRH, Vavricka SR, Stagg AJ, Schoepfer A, Raine T, Puig L, et al. The pathogenesis of extraintestinal manifestations: implications for IBD research, diagnosis, and therapy. J Crohns Colitis. (2019) 13:541–54. doi: 10.1093/ecco-jcc/jjy191
20. Alatab S, Sepanlou SG, Ikuta K, Vahedi H, Bisignano C, Safiri S, et al. The global, regional, and national burden of inflammatory bowel disease in 195 countries and territories, 1990-2017: a systematic analysis for the Global Burden of Disease Study 2017. Lancet Gastroenterol Hepatol. (2020) 5:17–30. doi: 10.1016/s2468-1253(19)30333-4
21. Marotto D, Atzeni F, Ardizzone S, Monteleone G, Giorgi V, Sarzi-Puttini P. Extra-intestinal manifestations of inflammatory bowel diseases. Pharmacol Res. (2020) 161:105206. doi: 10.1016/j.phrs.2020.105206
22. Harbord M, Annese V, Vavricka SR, Allez M, Barreiro-de Acosta M, Boberg KM, et al. The first European evidence-based consensus on extra-intestinal manifestations in inflammatory bowel disease. J Crohns Colitis. (2016) 10:239–54. doi: 10.1093/ecco-jcc/jjv213
23. Rogler G, Singh A, Kavanaugh A, Rubin DT. Extraintestinal manifestations of inflammatory bowel disease: current concepts, treatment, and implications for disease management. Gastroenterology. (2021) 161:1118–32. doi: 10.1053/j.gastro.2021.07.042
24. Valet C, Magnen M, Qiu L, Cleary SJ, Wang KM, Ranucci S, et al. Sepsis promotes splenic production of a protective platelet pool with high CD40 ligand expression. J Clin Invest. (2022) 132:e153920. doi: 10.1172/JCI153920
25. Smith GD, Ebrahim S. 'Mendelian randomization': can genetic epidemiology contribute to understanding environmental determinants of disease? Int J Epidemiol. (2003) 32:1–22. doi: 10.1093/ije/dyg070
26. Sekula P, Del Greco MF, Pattaro C, Köttgen A. Mendelian randomization as an approach to assess causality using observational data. J Am Soc Nephrol. (2016) 27:3253–65. doi: 10.1681/ASN.2016010098
27. Davey Smith G, Hemani G. Mendelian randomization: genetic anchors for causal inference in epidemiological studies. Hum Mol Genet. (2014) 23:R89–98. doi: 10.1093/hmg/ddu328
28. Lawlor DA, Harbord RM, Sterne JA, Timpson N, Davey Smith G. Mendelian randomization: using genes as instruments for making causal inferences in epidemiology. Stat Med. (2008) 27:1133–63. doi: 10.1002/sim.3034
29. Burgess S, Butterworth A, Thompson SG. Mendelian randomization analysis with multiple genetic variants using summarized data. Genet Epidemiol. (2013) 37:658–65. doi: 10.1002/gepi.21758
30. Zhu X. Mendelian randomization and pleiotropy analysis. Quant Biol. (2021) 9:122–32. doi: 10.1007/s40484-020-0216-3
31. Skrivankova VW, Richmond RC, Woolf BAR, Davies NM, Swanson SA, VanderWeele TJ, et al. Strengthening the reporting of observational studies in epidemiology using mendelian randomisation (STROBE-MR): explanation and elaboration. BMJ. (2021) 375:n2233. doi: 10.1136/bmj.n2233
32. Davies NM, Holmes MV, Davey Smith G. Reading Mendelian randomisation studies: a guide, glossary, and checklist for clinicians. BMJ. (2018) 362:k601. doi: 10.1136/bmj.k601
33. Vuckovic D, Bao EL, Akbari P, Lareau CA, Mousas A, Jiang T, et al. The polygenic and monogenic basis of blood traits and diseases. Cell. (2020) 182:1214–1231.e1211. doi: 10.1016/j.cell.2020.08.008
34. Kurki MI, Karjalainen J, Palta P, Sipilä TP, Kristiansson K, Donner KM, et al. FinnGen provides genetic insights from a well-phenotyped isolated population. Nature. (2023) 613:508–18. doi: 10.1038/s41586-022-05473-8
35. Burgess S, Thompson SG. Avoiding bias from weak instruments in Mendelian randomization studies. Int J Epidemiol. (2011) 40:755–64. doi: 10.1093/ije/dyr036
36. Luo J, Xu Z, Noordam R, van Heemst D, Li-Gao R. Depression and inflammatory bowel disease: A bidirectional two-sample mendelian randomization study. J Crohns Colitis. (2022) 16:633–42. doi: 10.1093/ecco-jcc/jjab191
37. Hartwig FP, Davies NM, Hemani G, Davey Smith G. Two-sample Mendelian randomization: avoiding the downsides of a powerful, widely applicable but potentially fallible technique. Int J Epidemiol. (2016) 45:1717–26. doi: 10.1093/ije/dyx028
38. Hemani G, Zheng J, Elsworth B, Wade KH, Haberland V, Baird D, et al. The MR-Base platform supports systematic causal inference across the human phenome. ELife. (2018) 7:e34408. doi: 10.7554/eLife.34408
39. Bowden J, Del Greco MF, Minelli C, Davey Smith G, Sheehan N, Thompson J. A framework for the investigation of pleiotropy in two-sample summary data Mendelian randomization. Stat Med. (2017) 36:1783–802. doi: 10.1002/sim.7221
40. Bowden J, Davey Smith G, Burgess S. Mendelian randomization with invalid instruments: effect estimation and bias detection through Egger regression. Int J Epidemiol. (2015) 44:512–25. doi: 10.1093/ije/dyv080
41. Bowden J, Del Greco MF, Minelli C, Davey Smith G, Sheehan NA, Thompson JR. Assessing the suitability of summary data for two-sample Mendelian randomization analyses using MR-Egger regression: the role of the I2 statistic. Int J Epidemiol. (2016) 45:1961–74. doi: 10.1093/ije/dyw220
42. Bowden J, Davey Smith G, Haycock PC, Burgess S. Consistent estimation in mendelian randomization with some invalid instruments using a weighted median estimator. Genet Epidemiol. (2016) 40:304–14. doi: 10.1002/gepi.21965
43. Burgess S, Thompson SG. Interpreting findings from Mendelian randomization using the MR-Egger method. Eur J Epidemiol. (2017) 32:377–89. doi: 10.1007/s10654-017-0255-x
44. Verbanck M, Chen CY, Neale B, Do R. Detection of widespread horizontal pleiotropy in causal relationships inferred from Mendelian randomization between complex traits and diseases. Nat Genet. (2018) 50:693–8. doi: 10.1038/s41588-018-0099-7
45. Greco MF, Minelli C, Sheehan NA, Thompson JR. Detecting pleiotropy in Mendelian randomisation studies with summary data and a continuous outcome. Stat Med. (2015) 34:2926–40. doi: 10.1002/sim.6522
46. Bowden J, Del Greco MF, Minelli C, Zhao Q, Lawlor DA, Sheehan NA, et al. Improving the accuracy of two-sample summary-data Mendelian randomization: moving beyond the NOME assumption. Int J Epidemiol. (2019) 48:728–42. doi: 10.1093/ije/dyy258
47. Kobayashi T, Siegmund B, Le Berre C, Wei SC, Ferrante M, Shen B, et al. Ulcerative colitis. Nat Rev Dis Primers. (2020) 6:74. doi: 10.1038/s41572-020-0205-x
48. Ananthakrishnan AN, Bernstein CN, Iliopoulos D, Macpherson A, Neurath MF, Ali RAR, et al. Environmental triggers in IBD: a review of progress and evidence. Nat Rev Gastroenterol Hepatol. (2018) 15:39–49. doi: 10.1038/nrgastro.2017.136
49. Gregg D, Goldschmidt-Clermont PJ. Cardiology patient page. Platelets and cardiovascular disease. Circulation. (2003) 108:e88–90. doi: 10.1161/01.CIR.0000086897.15588.4B
50. Schenone M, Furie BC, Furie B. The blood coagulation cascade. Curr Opin Hematol. (2004) 11:272–7. doi: 10.1097/01.moh.0000130308.37353.d4
51. Gremmel T, Frelinger AL 3rd, Michelson AD. Platelet physiology. Semin Thromb Hemost. (2016) 42:191–204. doi: 10.1055/s-00000077
52. Kaneider NC, Kaser A, Tilg H, Ricevuti G, Wiedermann CJ. CD40 ligand-dependent maturation of human monocyte-derived dendritic cells by activated platelets. Int J Immunopathol Pharmacol. (2003) 16:225–31. doi: 10.1177/039463200301600307
53. Duffau P, Seneschal J, Nicco C, Richez C, Lazaro E, Douchet I, et al. Platelet CD154 potentiates interferon-alpha secretion by plasmacytoid dendritic cells in systemic lupus erythematosus. Sci Transl Med. (2010) 2:47ra63. doi: 10.1126/scitranslmed.3001001
54. Liu Z, Ye T, Sun B, Schooling M, Tchetgen ET. Mendelian randomization mixed-scale treatment effect robust identification and estimation for causal inference. Biometrics. (2023) 79:2208–19. doi: 10.1111/biom.13735
Keywords: Mendelian randomization, platelet indices, inflammatory bowel disease, Crohn’s disease, ulcerative colitis
Citation: Li H-y and Liu T-m (2024) Platelet indices and inflammatory bowel disease: a Mendelian randomization study. Front. Immunol. 15:1377915. doi: 10.3389/fimmu.2024.1377915
Received: 28 January 2024; Accepted: 25 June 2024;
Published: 09 July 2024.
Edited by:
Alex Tsoi, University of Michigan, United StatesReviewed by:
Shi Xue Dai, Guangdong Provincial People’s Hospital, ChinaShuai Wang, University of Texas Southwestern Medical Center, United States
Vincent Salvatore Gallicchio, Clemson University, United States
Copyright © 2024 Li and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Tie-mei Liu, bHRtQGpsdS5lZHUuY24=
 Hong-yang Li
Hong-yang Li Tie-mei Liu
Tie-mei Liu