- 1Department of Microbiology, University of Chicago, Chicago, IL, United States
- 2Department of Immunobiology, Yale University, New Haven, CT, United States
- 3Committee on Microbiology, University of Chicago, Chicago, IL, United States
- 4Committee on Immunology, University of Chicago, Chicago, IL, United States
- 5Committee on Genetics, Genomics and System Biology, University of Chicago, Chicago, IL, United States
The vast diversity of mammalian adaptive antigen receptors allows for robust and efficient immune responses against a wide number of pathogens. The antigen receptor repertoire is built during the recombination of B and T cell receptor (BCR, TCR) loci and hypermutation of BCR loci. V(D)J recombination rearranges these antigen receptor loci, which are organized as an array of separate V, (D), and J gene segments. Transcription activation at the recombining locus leads to changes in the local three-dimensional architecture, which subsequently contributes to which gene segments are utilized for recombination. The endogenous retrovirus (ERV) mouse mammary tumor provirus 8 (Mtv8) resides on mouse chromosome 6 interposed within the large array of light chain kappa V gene segments. As ERVs contribute to changes in genomic architecture by driving high levels of transcription of neighboring genes, it was suggested that Mtv8 could influence the BCR repertoire. We generated Mtv8-deficient mice to determine if the ERV influences V(D)J recombination to test this possibility. We find that Mtv8 does not influence the BCR repertoire.
Introduction
The extensive diversity of the jawed vertebrate adaptive immune response depends on the programmed assembly and hypermutation of antigen receptor (AgR) genes (1). The first stage of AgR assembly is V(D)J recombination, initiated by lymphocyte-specific recombination activating genes 1 and 2 (RAG1 and RAG2), during which immunoglobulin (Ig) and T cell receptor (TCR) genes are recombined from discrete variable (V), diversity (D), and joining (J) gene segments (2).
AgR assembly is a sequential process during lymphocyte development. In B cells, the Ig heavy-chain (Igh) locus recombines in early and pro B cells prior to the kappa and lambda light chain (Igk, Igl) loci in pre B cells (2, 3). Igh recombines in two phases: first, DH-to-JH rearrangements occur in lymphoid progenitors; second, VH-to-DJH rearrangements occur in pro-B cells (2, 4). After successful (i.e. in-frame without premature stop codons) Igh recombination, Igk undergoes VK-to-JK recombination (3). If neither Igk allele successfully rearranges, the Igl locus recombines in a VL-to-JL fashion. During V(D)J recombination, RAG (a heterotetramer composed of RAG1 and RAG2 molecules) accumulates at recombination centers (RCs) that encompass J or DJ gene segments (for D-to-J/V-to-J and V-to-DJ recombination, respectively) (4). In RCs, RAG binds to a recombination signal sequences (RSS) that flanks the rearranging gene segment (4). Then, a partner RSS of the second gene segment is brought into the RC for synapsis and RAG-mediated cleavage.
V(D)J recombination determines the V, (D), and J gene segments used in a particular AgR gene and is dependent on three-dimensional chromosomal architecture. Specifically, V(D)J recombination is constrained to AgR loci by chromatin loops, the bases of which are defined by CCCTC-binding factor (CTCF)-bound CTCF-Binding Elements (CBEs) (5–9). The number of loops formed is dependent on the recombining locus (10). In addition to chromatin loops defining the AgR, chromosomal architecture also defines the mechanism by which recombining gene segments are brought into the RC (11).
The Igk locus is comprised of 92 functional (163 total) VK and four functional JK spread across 3.2 Mb (Figure 1A) and contracts into a recombination-competent chromosomal structure in developing pro- and pre-B cells (12, 13). Once contracted, the Igk locus forms a rosette-like structure with five VK-containing loops and one loop with JK and CK gene segments (10). Igk rearrangement occurs predominantly through the collision of, and subsequent recombination between, the JK-containing and a VK-containing loop (10). While the Igk loops are thought to be predominantly shaped by the CBEs throughout the locus, it is unknown whether other factors are involved.
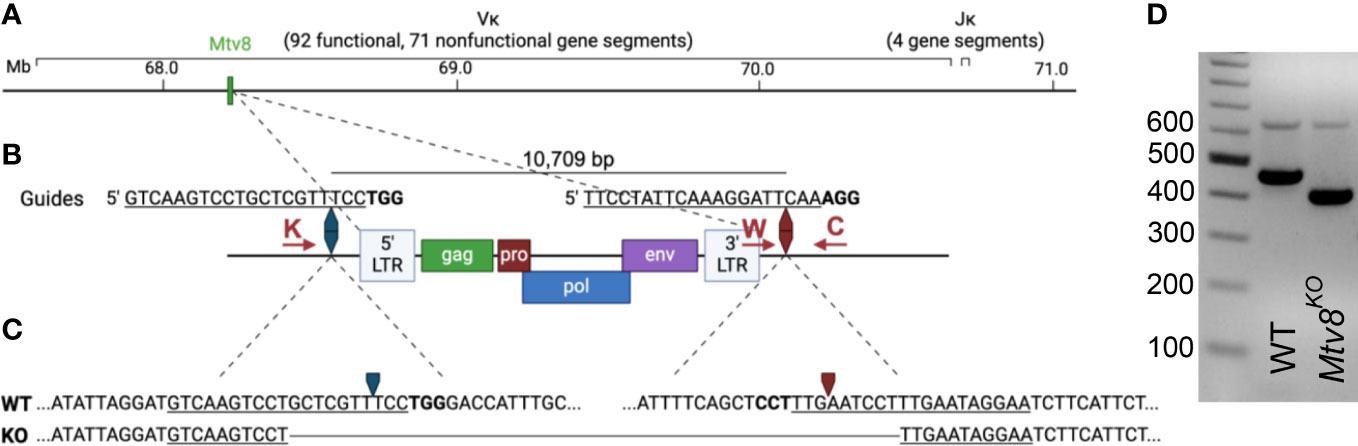
Figure 1 Generation of Mtv8KO mice. (A) Schematic of B6J murine Igk locus on chromosome 6. Location of Mtv8 is highlighted in green. (B) Schematic of Mtv8 and guides used for CRISPR/Cas9 targeting of Mtv8 to generate Mtv8KO mice. LTR, long-terminal repeat; gag, group-specific antigen; pro, protease; pol, polymerase; env, envelope. Red arrows: genotyping primers (K: KOF, W: WTF, C: CommonR). (C) Genomic sequence of WT and KO Mtv8 alleles. Only the flanking sequnces are shown for the WT allele. For (B, C), underlined text indicates guide sequence, bold text indicates PAM sequence, arrows indicate Cas9-cut site. (D) Gel image of genotyping representative WT and homozygous Mtv8KO mice. Mice were genotyped using a three-primer PCR with the KOF, WTF, and CommonR primers. WT product: 479 bp, KO product: 419 bp.
The endogenous retrovirus (ERV) mouse mammary tumor virus (MMTV), Mtv8 was mapped to the Igk locus on chromosome 6 (Figure 1A) (14–16). Mtv8 is a provirus with all open-reading frames (ORFs) intact, namely, gag, pro, pol, env, and superantigen (SAg) (Figure 1B). The provirus does not produce infectious virions and is silent in the mammary glands, the targeted tissue of all MMTVs, potentially due to hypermethylation of its promoter region (17–19). The Mtv8 SAg has a specificity for Vβ11 and Vβ12 (20). Mice strains harboring Mtv8 lack Vβ11+ and Vβ12+ T cells (21), indicating that Mtv8 is expressed in antigen presenting cells, such as dendritic cells and B cells which present SAg to cognate T cells (22). As Mtv8 is mapped within the Igk locus, it was hypothesized that it may contribute to Igk recombination by driving high levels of transcription in its vicinity (23). It was suggested that recombination to the first VK gene segment downstream of Mtv8 (VK14-111, formerly VK9M) could allow for enhancer activation of the Mtv8 promoter (23). To identify whether Mtv8 affects the Igk repertoire, the frequency of recombination of the JK gene segments to VK14-111 was analyzed in inbred mouse strains with [BALB/c, C58.C, A/J, and C57BL/6J (B6J)] and without (C58, C.C58, NZB, and PERA/Ei) Mtv8 (23). These experiments demonstrated that mice inheriting Mtv8 have higher recombination between VK14-111 and all JK gene segments compared to strains without Mtv8. While these analyses suggested differences in the usage of VK14-111, Ig repertoire comparisons between inbred mouse lines is not an ideal approach, as polymorphisms within the loci other than Mtv8 could also influence the repertoires.
To address whether Mtv8 shapes the repertoire of mice with this ERV, we used CRISPR/Cas9 technology to generate B6J mice lacking Mtv8 and compared their Ig repertoire to that of wild-type (WT) B6J mice. We found that the absence of Mtv8 had no significant effects on the Igh, Igk, and Igl gene segment usage. Thus, contribution of Mtv8 to the mouse Ig repertoires can be definitively ruled out.
Methods
Mice
Mice utilized in this study were bred and maintained at the animal facility of The University of Chicago. The studies described herein have been reviewed and approved by the Animal Care and Use Committee at the University of Chicago, which is accredited by the Association for Assessment and Accreditation of Laboratory Animal Care (AAALAC International).
C57BL/6J (B6J) mice were purchased from The Jackson Laboratory. Mtv8KO B6J mice were generated using CRISPR/Cas9 technology. Two guide RNAs targeting the flanking up- and downstream sequences of Mtv8 (5’ guide: 5’-GTCAAGTCCTGCTCGTTTCC; 3’ guide: 5’- TTCCTATTCAAAGGATTCAA) were co-injected into a single cell B6J embryos along with Cas9 (Figure 1B). Founder mice and subsequent offspring in which Mtv8 was eliminated were identified using PCR with primers flanking the guide cut sites (KOF: 5’-GAATTTGGGTGCTCTTGCAT; CommonR: 5’-AACACAAATGGAGGCAAAGC; KO product size: 419 bp). To identify mice with the WT allele, a separate PCR was used using the CommonR primer and a WT-specific reverse primer that lies in the excited region (WTF: 5’- CAGTCCTAACATTCACCTCT; WT product size: 479 bp). A founder line was established in which Mtv8 was eliminated with a 10,688 bp deletion (Figure 1C). The founder mouse was bred to a WT B6J mouse and the resulting F1 offspring were intercrossed to generate a homozygous KO line (Figure 1D). The deletion was confirmed at the DNA level by sequencing the KO allele using the KO-specific PCR primers.
RNA isolation from splenic B cells
CD19+ splenocytes were isolated from three WT and three Mtv8KO 10.5-week-old B6J female mice. Red blood cell lysed splenocytes were labeled with microbeads conjugated to monoclonal anti-mouse CD19 antibodies (Miltenyi Biotec, Bergisch Gladbach, Germany) and positively sorted as detailed by the manufacturer. RNA was isolated from sorted cells using guanidine thiocyanate extraction and CsCl gradient centrifugation (24).
Library preparation
Immunoglobulin libraries were generated from 1 μg of RNA using the NEBNext Immune Sequencing Kit (New England Biolabs, Ipswich, Massachusetts, USA) according to manufacturer’s instructions, specifically enriching for B cell receptor (BCR) chains during the first PCR step using the mouse NEBNext IS BCR primers included in the kit (Mus-IgGb: 5’-GATGGGGCTGTTGTTKTRGC; Mus-IgGa: 5’-GTGTCGTTTTGGCCTG; Mus-IgE: 5’-GGTTCCTGATAGAGGC; Mus-IgD: 5’-GTTCCTTTTTATCACC; Mus-IgM: 5’-TGACTCTCCTGMRGARAC; Mus-IgA: 5’-GTGGGTAGATGGTGGG; Mus-IgK: 5’-RCATCAGCCMGGTWT; Mus-IgL: 5’-ATGGHGWRGMCTTGGG). The libraries generated from the six individual mice were pooled in equimolar amounts and sequenced by paired-end 300 bp sequencing on an Illumina MiSeq by The University of Chicago Genomics Facility.
B cell receptor sequence processing and analysis
Preprocessing of BCR sequences was performed using the open-source workflow pRESTO NEBNext Immune Sequencing Kit Workflow (v3.2.0) on Galaxy (25). Reads with a Phred quality score <20 were removed for quality control. Reads that did not match to the constant region primer (maximum error rate 0.2) were removed. Reads that did not match to the template switch sequence (maximum error rate 0.5) were removed. The first 17 bp following the template switch site were a unique molecular identifier (UMI) on each read. Sequences with identical UMIs were collapsed into consensus sequences with sequences found in less than 60% of reads removed. Positions with more than 50% gap sequences were removed. Mate-pairs were assembled with a minimum of 8 bp overlap (maximum error rate of 0.3). Assembled reads were assigned isotype-constant region identities based on local alignment of the 3’ ends of the reads (maximum error rate of 0.3). Using the Mus musculus reference C57BL/6J genome (GRCm38/mm10), V, (D), and J gene segments were assigned using MiGMAP mapper (Galaxy Version 1.0.3+galaxy2). Subsequent analyses were done using R Studio. Statistically significance was calculated using unpaired Welch’s t test with Bonferroni correction. Graphing was done using GraphPad Prism version 10.0.1 for Mac (GraphPad Software, Boston, Massachusetts, USA).
Results
Generation of Mtv8-deficient B6J mice
Mtv8 knockout (KO) B6J mice were generated using a CRISPR/Cas9 approach. To target Mtv8 without disturbing the VK gene segments in its vicinity, we designed two guides to precisely delete the ERV: one 645 bp upstream of the 5’ LTR and one 79 bp downstream of the 3’ LTR (Figure 1B). A founder was identified using PCR with primers flanking the predicted deleted region and it was determined that a 10,688 bp deletion occurred, resulting in the elimination of the entire Mtv8 locus (Figures 1C, D). The founder was crossed to a wildtype (WT) B6J mouse and heterozygous, mutant allele-carrying progeny were interbred to generate a homozygous Mtv8-deficient line (Figure 1D).
Mtv8 does not contribute to light and heavy chain recombination
To investigate whether Mtv8 alters Ig gene segment usage, we analyzed the Ig repertoires of WT and Mtv8KO B6J mice (Figures 2-4, Supplementary Table 1). Accordingly, RNA isolated from CD19+ splenocytes was used to prepare Ig heavy and light chain-specific 5’-RACE libraries, which were analyzed to determine the BCR repertoire using the pRESTO toolkit using the Mus musculus reference C57BL/6J genome (GRCm38/mm10) (25). The repertoire of productive transcripts was analyzed. Productive transcripts were defined as in-frame without premature stop codons and are likely those transcripts that are translated into expressed Ig proteins.
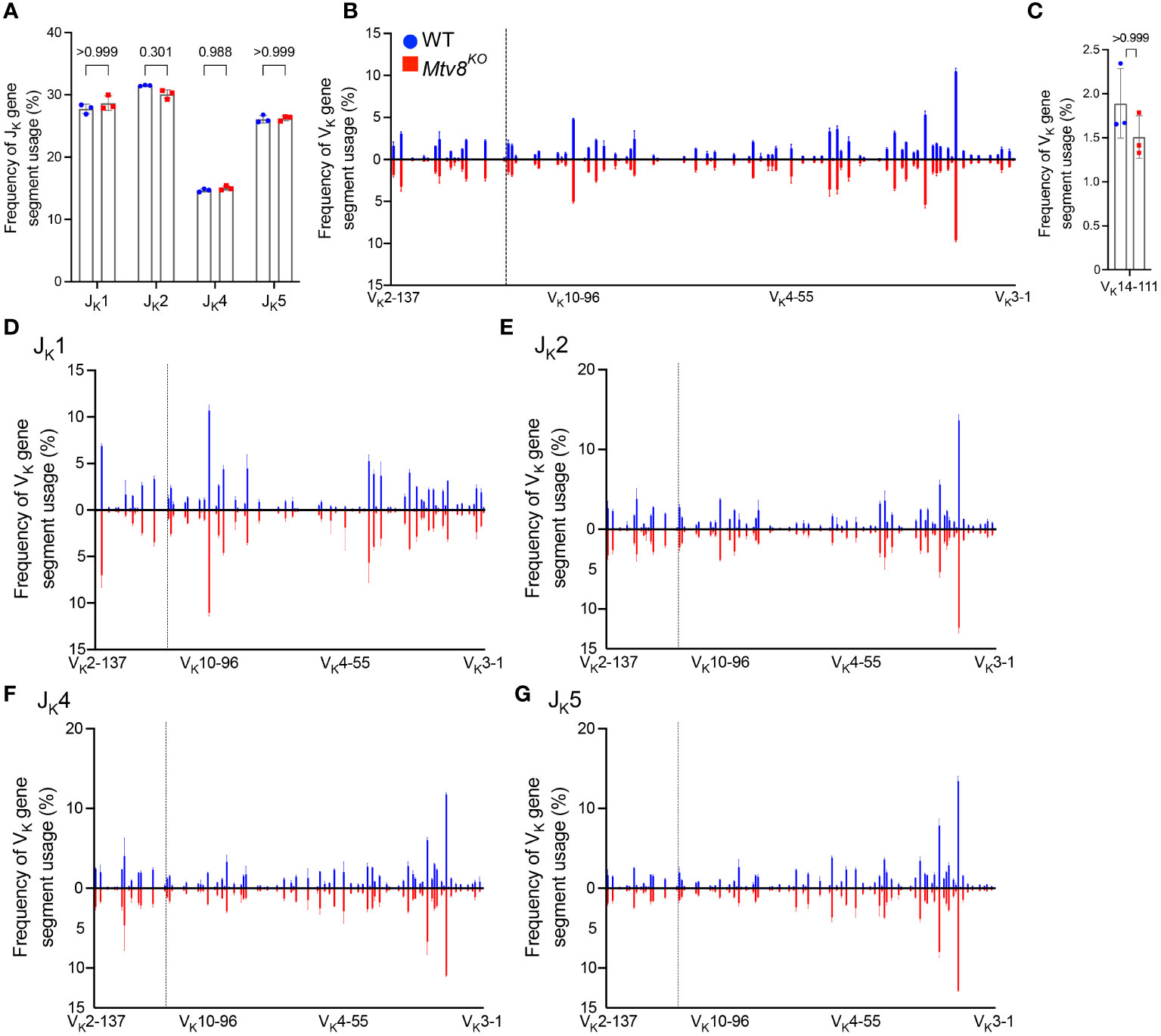
Figure 2 Igk repertoire of CD19+ splenocytes of Mtv8KO mice. Frequency of JK (A) and VK (B) gene segments in productive Igk transcripts from CD19+ splenocytes. Productive recombination events are in-frame without premature stop codons. (C) Frequency of VK14-111 (previously termed VK9M) in and productive Igk transcripts. (D-G) Frequency of VK gene segments in total Igk transcripts recombined to JK1 (D), JK2 (E), JK4 (F), and JK5 (G). (B, D-G) VK gene segments are arranged from 5’ distal to 3’ proximal to JK gene segments and the vertical dotted line represents chromosomal location of Mtv8. n=3 mice per group. Data presented as mean with error bars indicated SD. Statistical significance was determined by unpaired Welch’s t test with Bonferroni correction (numbers above bars indicate adjusted P value).
To determine whether Mtv8 influences Igk recombination, we examined the JK and VK usage in productive Igk transcripts from CD19+ splenocytes (Figure 2). There was no difference in JK usage (Figure 2A). Furthermore, the loss of Mtv8 led to no alterations in the total VK repertoire (Figure 2B). Notably, there was no change in usage of VK gene segments mapped in the close proximity of Mtv8 (dotted line; Figure 2B). An increased usage of the VK gene segment, VK14-111 (formerly called VK9M) directly downstream of Mtv8 in Mtv8+ mouse strains compared to Mtv8- strains was previously reported (23). We observed no difference in frequency of VK14-111 usage in Igk transcripts between WT and Mtv8KO B6J mice (Figure 2C). Thus, the previously observed disparities in VK14-111 usage between Mtv8+ and Mtv8- mice from distinct genetic backgrounds are independent of Mtv8.
The same Igk locus can undergo multiple rounds of recombination. While Mtv8 does not affect the overall frequency of VK usage, we wanted to test the possibility that it might affect either the first or subsequent recombination in distinct ways. Recombination of any VK to JK1 can only occur during a primary recombination event, as recombination to any other JK gene segment would remove JK1 from the Igk locus. Recombination to JK2, JK4, or JK5 can either occur during a primary or secondary recombination event. As such, we calculated the frequency of VK gene segment usage in total recombination events with each JK gene segment. We found that, in line with globally unaffected JK gene segment usage, frequency of any particular VK gene segment recombining to JK1, JK2, JK4, and JK5 was unchanged with the loss of Mtv8 (Figures 2D–G). Taken together, these data show that the Igk repertoire of Mtv8KO B6J mice does not differ from WT B6J and that Mtv8 does not influence the Igk recombination.
We also found no difference in the JL and VL usage among productive Igl transcripts between WT and Mtv8KO B6J mice (Figure 3). Similarly, we found no alterations in JH, DH, and VH gene segment usage between WT and Mtv8KO mice (Figure 4).
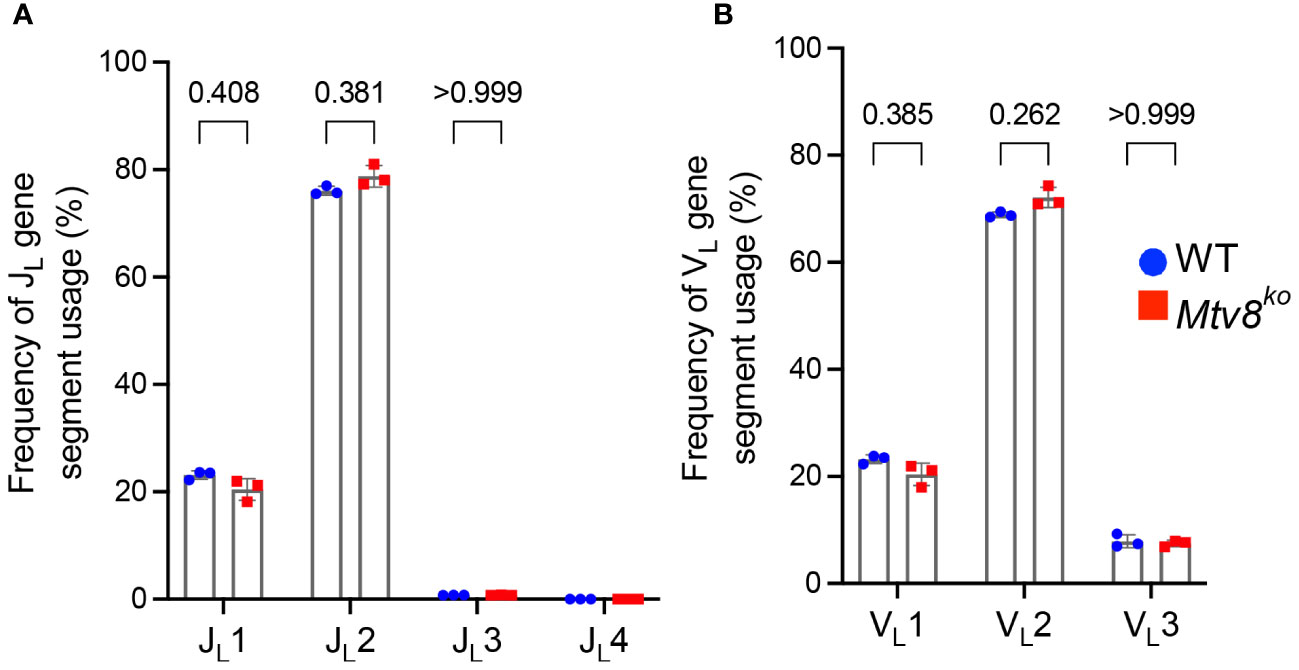
Figure 3 Igl repertoire of CD19+ splenocytes of Mtv8KO mice. Frequency of JL (A) and VL (B) gene segments in and productive Igl transcripts from CD19+ splenocytes. Productive recombination events are in-frame without premature stop codons. n=3 mice per group. Data presented as mean with error bars indicated SD. Statistical significance was determined by unpaired Welch’s t test with Bonferroni correction (numbers above bars indicate adjusted P value). No P value is indicated for JL4 as values for all samples are 0.
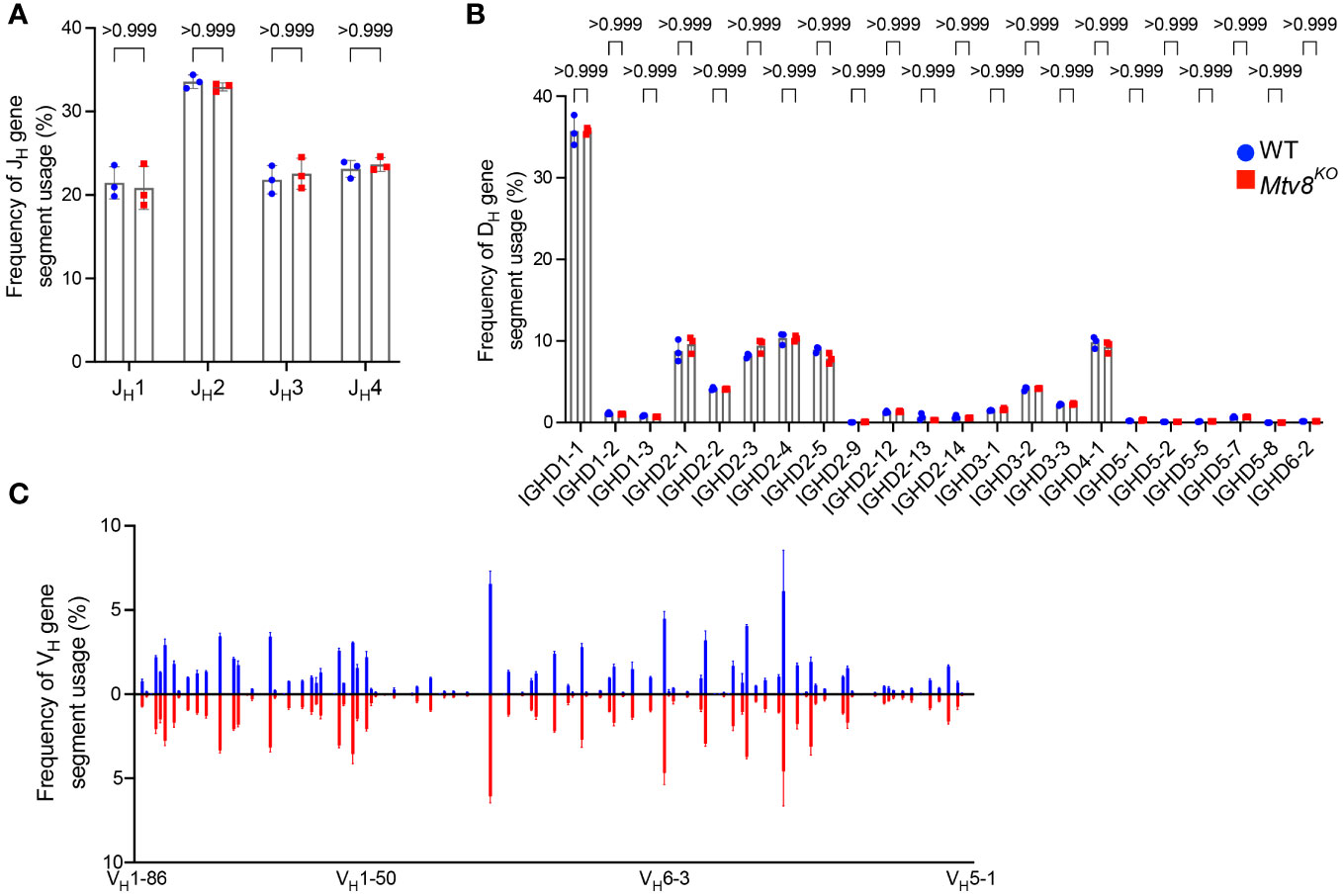
Figure 4 Igh repertoire of CD19+ splenocytes of Mtv8KO mice. Frequency of JH (A), DH (B), and VH (C) gene segments in productive Igh transcripts from CD19+ splenocytes. Productive recombination events are in-frame without premature stop codons. (B) DH gene segments are arranged from 5’ distal to 3’ proximal to JH gene segments. (C) Frequency of VH gene segments in productive Igh transcripts that are statistically different between WT and Mtv8KO mice. VH gene segments are arranged from 5’ distal to 3’ proximal to DH gene segments. n=3 mice per group. Data presented as mean with error bars indicated SD. Statistical significance was determined by unpaired Welch’s t test with Bonferroni correction (numbers above bars indicate adjusted P value).
Discussion
It is now accepted that the collisions of the rosette-like loops of the Igk locus are the major mechanism of VK-to-JK recombination (10). However, the identification of an ERV, Mtv8, in the middle of the VK array on the Igk locus led to the hypothesis that it may influence the Ig repertoire (14–16). Differences in VK usage in mouse strains with and without Mtv8 initially supported this hypothesis (23). Now, advancements in genome-editing technologies allow us to definitely address whether a genetic loss of Mtv8 can change the Ig repertoire within a mouse line. We show that Mtv8 has no effect on the Ig repertoire by analyzing the Igh, Igk, and Igl transcriptional repertoires of B cells isolated from WT and Mtv8KO B6J mice. These data demonstrate that while Mtv8 is an intact ERV at the Igk locus, it has no influence on Igk recombination.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE248410, GSE248410.
Ethics statement
The animal study was approved by University of Chicago Animal Resources Center. The study was conducted in accordance with the local legislation and institutional requirements.
Author contributions
HB: Conceptualization, Data curation, Formal analysis, Funding acquisition, Investigation, Methodology, Writing – original draft, Writing – review & editing. SE: Methodology, Writing – review & editing. TG: Funding acquisition, Methodology, Project administration, Resources, Supervision, Writing – review & editing.
Funding
The author(s) declare that no financial support was received for the research, authorship, and/or publication of this article.
Acknowledgments
We thank The University of Chicago Transgenic Mouse Facility (RRID:SCR_019171), especially Linda Degenstein, for the generation of the Mtv8-deficient B6J mouse line. We thank The University of Chicago Genomics Facility (RRID:SCR_019196), especially Dr. Pieter Faber, for their assistance with sequencing Ig libraries. We thank Dr. Alexander Chervonsky for helpful discussions. We also thank Dr. David G. Schatz for advice and guidance on the project. This work was supported by The University of Chicago fund to T. G. and by the Duchossois Family Institute at the University of Chicago to H. A. B.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2024.1345467/full#supplementary-material
References
1. Cooper MD, Alder MN. The evolution of adaptive immune systems. Cell. (2006) 124:815–22. doi: 10.1016/j.cell.2006.02.001
2. Schatz DG, Swanson PC. V(D)J recombination: mechanisms of initiation. Annu Rev Genet. (2011) 45:167–202. doi: 10.1146/annurev-genet-110410-132552
3. Alt FW, Zhang Y, Meng FL, Guo C, Schwer B. Mechanisms of programmed DNA lesions and genomic instability in the immune system. Cell. (2013) 152:417–29. doi: 10.1016/j.cell.2013.01.007
4. Schatz DG, Ji Y. Recombination centres and the orchestration of V(D)J recombination. Nat Rev Immunol. (2011) 11:251–63. doi: 10.1038/nri2941
5. Jhunjhunwala S, van Zelm MC, Peak MM, Murre C. Chromatin architecture and the generation of antigen receptor diversity. Cell. (2009) 138:435–48. doi: 10.1016/j.cell.2009.07.016
6. Zhang Y, Zhang X, Dai HQ, Hu H, Alt FW. The role of chromatin loop extrusion in antibody diversification. Nat Rev Immunol. (2022) 22:550–66. doi: 10.1038/s41577-022-00679-3
7. Davidson IF, Bauer B, Goetz D, Tang W, Wutz G, Peters JM. DNA loop extrusion by human cohesin. Science. (2019) 366:1338–45. doi: 10.1126/science.aaz3418
8. Kim Y, Shi Z, Zhang H, Finkelstein IJ, Yu H. Human cohesin compacts DNA by loop extrusion. Science. (2019) 366:1345–9. doi: 10.1126/science.aaz4475
9. Davidson IF, Peters JM. Genome folding through loop extrusion by SMC complexes. Nat Rev Mol Cell Biol. (2021) 22:445–64. doi: 10.1038/s41580-021-00349-7
10. Hill L, Wutz G, Jaritz M, Tagoh H, Calderón L, Peters JM, et al. Igh and Igk loci use different folding principles for V gene recombination due to distinct chromosomal architectures of pro-B and pre-B cells. Nat Commun. (2023) 14:2316. doi: 10.1038/s41467-023-37994-9
11. Zhang Y, Zhang X, Ba Z, Liang Z, Dring EW, Hu H, et al. The fundamental role of chromatin loop extrusion in physiological V(D)J recombination. Nature. (2019) 573:600–4. doi: 10.1038/s41586-019-1547-y
12. Stadhouders R, de Bruijn MJW, Rother MB, Yuvaraj S, de Almeida CR, Kolovos P, et al. Pre-B cell receptor signaling induces immunoglobulin kappa locus accessibility by functional redistribution of enhancer-mediated chromatin interactions. PloS Biol. (2014) 12:e1001791. doi: 10.1371/journal.pbio.1001791
13. Roldan E, Fuxa M, Chong W, Martinez D, Novatchkova M, Busslinger M, et al. Locus 'decontraction' and centromeric recruitment contribute to allelic exclusion of the immunoglobulin heavy-chain gene. Nat Immunol. (2005) 6:31–41. doi: 10.1038/ni1150
14. Robbins JM, Gallahan D, Hogg E, Kozak C, Callahan R. An endogenous mouse mammary tumor virus genome common in inbred mouse strains is located on chromosome 6. J Virol. (1986) 57:709–13. doi: 10.1128/jvi.57.2.709-713.1986
15. Yang JN, Boyd RT, Gottlieb PD, Dudley JP. The endogenous retrovirus Mtv-8 on mouse chromosome 6 maps near several kappa light chain markers. Immunogenetics. (1987) 25:222–7. doi: 10.1007/BF00404691
16. Yang JN, Dudley JP. The endogenous Mtv-8 provirus resides within the V kappa locus. J Virol. (1991) 65:3911–4. doi: 10.1128/jvi.65.7.3911-3914.1991
17. Golovkina TV. A novel mechanism of resistance to mouse mammary tumor virus infection. J Virol. (2000) 74:2752–9. doi: 10.1128/JVI.74.6.2752-2759.2000
18. Golovkina TV, Chervonsky A, Prescott JA, Janeway CA Jr., Ross SR. The mouse mammary tumor virus envelope gene product is required for superantigen presentation to T cells. J Exp Med. (1994) 179:439–46. doi: 10.1084/jem.179.2.439
19. Fanning TG, Morris DW, Cardiff RD, Bradshaw HD Jr. Characterization of an endogenous retrovirus-repetitive DNA chimera in the mouse genome. J Virol. (1985) 53:998–1000. doi: 10.1128/jvi.53.3.998-1000.1985
20. Holt MP, Shevach EM, Punkosdy GA. Endogenous mouse mammary tumor viruses (mtv): new roles for an old virus in cancer, infection, and immunity. Front Oncol. (2013) 3:287. doi: 10.3389/fonc.2013.00287
21. Dyson PJ, Knight AM, Fairchild S, Simpson E, Tomonari K. Genes encoding ligands for deletion of V beta 11 T cells cosegregate with mammary tumour virus genomes. Nature. (1991) 349:531–2. doi: 10.1038/349531a0
22. Acha-Orbea H, MacDonald HR. Superantigens of mouse mammary tumor virus. Annu Rev Immunol. (1995) 13:459–86. doi: 10.1146/annurev.iy.13.040195.002331
23. Yang JN, Dudley J. Endogenous Mtv-8 or a closely linked sequence stimulates rearrangement of the downstream V kappa 9 gene. J Immunol. (1992) 149:1242–51. doi: 10.4049/jimmunol.149.4.1242
24. Chirgwin JM, Przybyla AE, MacDonald RJ, Rutter WJ. Isolation of biologically active ribonucleic acid from sources enriched in ribonuclease. Biochemistry. (1979) 18:5294–9. doi: 10.1021/bi00591a005
Keywords: endogenous retrovirus, immunoglobulin repertoire, V(D)J recombination, MTV, B cell receptor (BCR) repertoire
Citation: Beilinson HA, Erickson SA and Golovkina T (2024) The endogenous Mtv8 locus and the immunoglobulin repertoire. Front. Immunol. 15:1345467. doi: 10.3389/fimmu.2024.1345467
Received: 27 November 2023; Accepted: 16 February 2024;
Published: 05 March 2024.
Edited by:
Duane R. Wesemann, Harvard Medical School, United StatesReviewed by:
Richard L. Frock, Stanford University, United StatesMing Tian, Harvard Medical School, United States
Copyright © 2024 Beilinson, Erickson and Golovkina. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Helen A. Beilinson, YmVpbGluc29uaEB1Y2hpY2Fnby5lZHU=
 Helen A. Beilinson
Helen A. Beilinson Steven A. Erickson2
Steven A. Erickson2 Tatyana Golovkina
Tatyana Golovkina