- 1Department of Dermatology and Venerology, Peking University First Hospital, Beijing, China
- 2Research Center for Medical Mycology, Peking University, Beijing, China
- 3National Clinical Research Center for Skin and Immune Diseases, Beijing, China
- 4Beijing Key Laboratory of Molecular Diagnosis on Dermatoses, Beijing, China
Dermatophytosis is a common superficial infection caused by dermatophytes, a group of pathogenic keratinophilic fungi. Apart from invasion against skin barrier, host immune responses to dermatophytes could also lead to pathologic inflammation and tissue damage to some extent. Therefore, it is of great help to understand the pathogenesis of dermatophytes, including fungal virulence factors and anti-pathogen immune responses. This review aims to summarize the recent advances in host-fungal interactions, focusing on the mechanisms of anti-fungal immunity and the relationship between immune deficiency and chronic dermatophytosis, in order to facilitate novel diagnostic and therapeutic approaches to improve the outcomes of these patients.
1 Introduction
Dermatophytes are the most common pathogenic filamentous fungi, with an infection rate of as high as 20%-25% worldwide (1). Dermatophytes usually infect the nails, skin, and hairs (2, 3), causing multiple superficial dermatophytoses, such as tinea capitis, onychomycosis, tinea corporis, and tinea pedis (4). Unfrequently, dermatophytes may also invade the dermal tissue and even deep organs, particularly in immunocompromised patients with congenital or acquired immunodeficiency (5), and these infections can progress to life-threatening conditions if appropriate treatment is not provided (6).
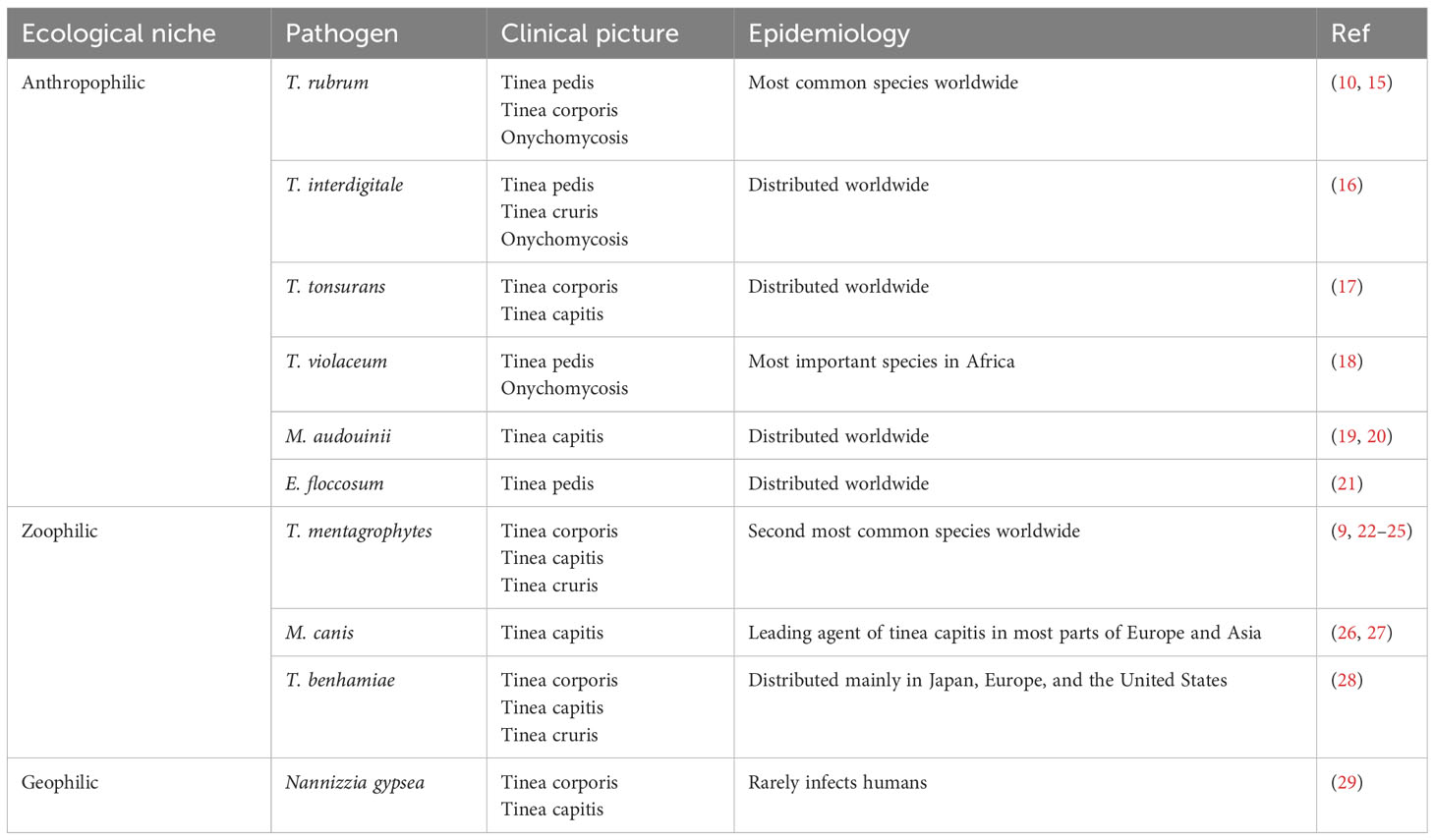
Dermatophytes can be categorized into three types based on host preferences and ecological niches (7): anthropophilic dermatophytes are mainly transmitted from person to person and usually result in chronic infections with moderate clinical symptoms; zoophilic dermatophytes prefer selective animal hosts but can normally infect other species, including humans, often causing inflammatory skin infections; and geophilic dermatophytes survive on keratinized waste present in the soil and are rarely pathogenic but can produce more severe inflammation than anthropophilic species (8). Trichophyton rubrum is among the most frequently detected species globally and is responsible for 50%-90% of dermatophytoses (9–13). However, a sudden shift in the most prevalent pathogen from T. rubrum to T. mentagrophytes complex in India has been observed in recent years, which might result from advances in molecular identification (14). Other important species include Microsporum canis, Epidermophyton floccosum, and T. tonsurans (Table 1) (22, 30, 31). In addition, a new emerging drug-resistant dermatophyte, T. indotineae, has caused a concurrent overwhelming circumstantial increase in reports on recalcitrance and drug resistance in India (32–34). This strain has also been isolated from several European, American, and Asian countries and has become a public health issue due to the number of individuals affected and the misery it causes (16, 35–42).
In recent years, the prevalence of dermatophytosis has continuously increased, especially in tropical or subtropical countries such as India (21, 43). Several risk factors may further increase the risk of dermatophyte infection, including type 2 diabetes, lack of physical activities, vascular disease, anemia, immunosuppression due to leukemia, organ transplant, acquired immunodeficiency syndrome (AIDS), and the use of immunosuppressants (44, 45). The elevated incidence of dermatophyte infections, especially chronic and recurrent dermatophytosis, has a large impact on patients’ quality of life and often requires extended treatments, causing psychological and economic burden (46–51). To provide a comprehensive understanding of the mechanism of dermatophytosis, this review summarizes recent findings on the pathogenesis of and the host’s immune responses to dermatophyte infections.
2 Virulence factors related to the pathogenicity of dermatophytes
When dermatophytes infect human skin, the first obstacles they need to overcome include physical, chemical, and morphological barriers of the skin. Abnormalities in the stratum corneum, such as macerations and occlusions, may promote fungal infection (52). Once the pathogen crosses the skin barriers mentioned above, colonization begins, and various other processes occur, including adhesion, germination, and invasion. The first stage of infection by arthroconidia includes adherence to the host epidermis via special fungal surface proteins (53). For example, T. rubrum binds epithelial cells through carbohydrate-specific adhesins on the microconidial surface, while T. mentagrophytes protrudes fibrillar projections when it requires adherence capabilities (54). In the next stage, arthroconidia identify a favorable environment and initiate metabolic reactivation and growth as hyphae (55). During the last stage, the epidermal cornified layer is invaded by the germinating tubes produced by hyphae that secrete various keratinases to digest keratin into smaller peptides and amino acids (56, 57), which are absorbed by dermatophytes as nutrients for growth and reproduction. The initial stage of native keratin biodegradation is sulfitolysis, during which the extensive keratin disulfide bridges are hydrolyzed (58, 59). Afterward, keratin can be further hydrolyzed by various proteases secreted by dermatophytes (60, 61). Endoproteases degrade keratin to release free peptides, on which exoproteases act to further decompose the peptides into smaller peptides and amino acids (62, 63) (Figure 1).
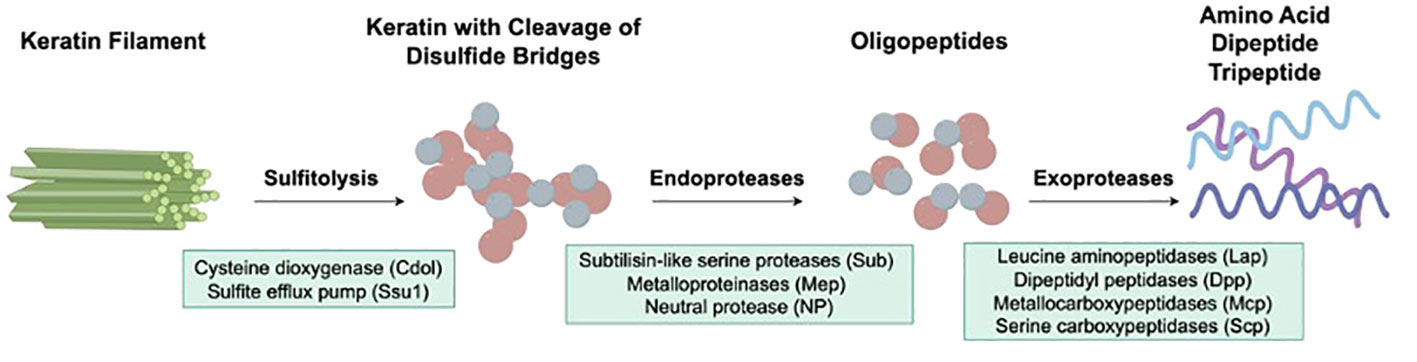
Figure 1 Virulence factors of dermatophytes involved in the keratinolysis process. The initial stage of native keratin biodegradation is sulfitolysis, the key enzymes of which are cysteine dioxygenase (Cdol) and sulfite efflux pump (Ssu1). Then, endoproteases degrade keratin to release free peptides, which are further cleaved into amino acids by exoproteases.
2.1 Virulence factors involved in the sulfitolysis process
Dermatophytes secrete a variety of proteases, most of which cannot directly hydrolyze natural keratins. Disulfide bonds within keratin molecules can cross-link within and between peptide chains, forming a network structure that is 3-dimensional, making natural keratin difficult to decompose (64). Therefore, cleavage of disulfide bridges must be the first step in keratinolysis. The key enzymes involved in the sulfitolysis process are cysteine dioxygenase (Cdol) and sulfite efflux pump (Ssu1) (65). Kunert and Truper (66) first reported the mechanisms of cysteine metabolism by N. gypseum hyphae in 1986: Cdol catalyzes the oxidation of cysteine to cysteine sulfinic acid, which is eventually metabolized to produce sulfites, sulfates and taurine; sulfites can be excreted by Ssu1. Subsequently, researchers found that L-cystine could induce the expression and activation of Cdol in T. mentagrophytes, and they successfully isolated recombinant Cdol (67, 68). Moreover, expression of the CDO1 gene in T. schoenleinii was significantly upregulated after coculture with keratin (69). In addition, CDO1 and SSU1 knockout strains of T. benhamiae showed obvious growth defects when cocultured with human hair and nails, suggesting that Cdo1 and Ssu1 are important virulence factors of dermatophytes (56).
2.2 Proteases of the main species of dermatophytes
Subtilisin-like serine protease 3 (Sub3) is the first keratinase found in M. canis (70, 71) and is essential for adhesion to the epidermis (72) (Figure 1). Subsequently, Brouta et al. (73, 74) revealed the expression of metalloproteinases (Mep) in M. canis in vitro and in vivo and discovered that metalloproteinases could mediate humoral and cellular immune responses (75). Subsequently, the virulence activities of Mep and Sub have been studied in various dermatophyte species. Mep4, Mep5 and Sub6 proved to be the predominant virulence factors of T. mentagrophytes (76, 77), while SUB6, SUB7, MEP1, MEP2 and MEP5 expression in T. tonsurans was associated with invasion (78). Leng et al. (79) demonstrated that Sub3, Sub4 and Mep4 were required for T. rubrum to invade skin. The expression of SUB1, SUB6-7, and MEP3 was upregulated significantly after culturing in a medium containing nail chips, illustrating that these genes could contribute to the pathogenicity of T. rubrum as well (80, 81). Recently, Baumbach et al. (82) observed SUB3 and SUB6 expression in T. benhamiae in vitro and in vivo using immunofluorescence methods, indicating that Sub3 and Sub6 might play a primary role in the adhesion and invasion of T. benhamiae. Apart from endoproteases, exoproteases also draw tremendous attention in research on the pathogenicity of dermatophytes (Figure 1). In 2005, Monod et al. (83) identified leucine aminopeptidase (Lap) 1 and Lap2 and dipeptidyl peptidase (Dpp) IV and DppV produced by coculturing T. rubrum with keratin, which were also found in T. violaceum and T. benhamiae (62, 84–86). In addition, the expression level of family of serine hydrolases 1 (FSH1) in M. canis cocultured with infant scalp was significantly increased (87), and FSH1 knock-out significantly decreased M. canis virulence (88), proving FSH1 as a potential virulence factor of M. canis.
2.3 Adaptation to skin pH changes
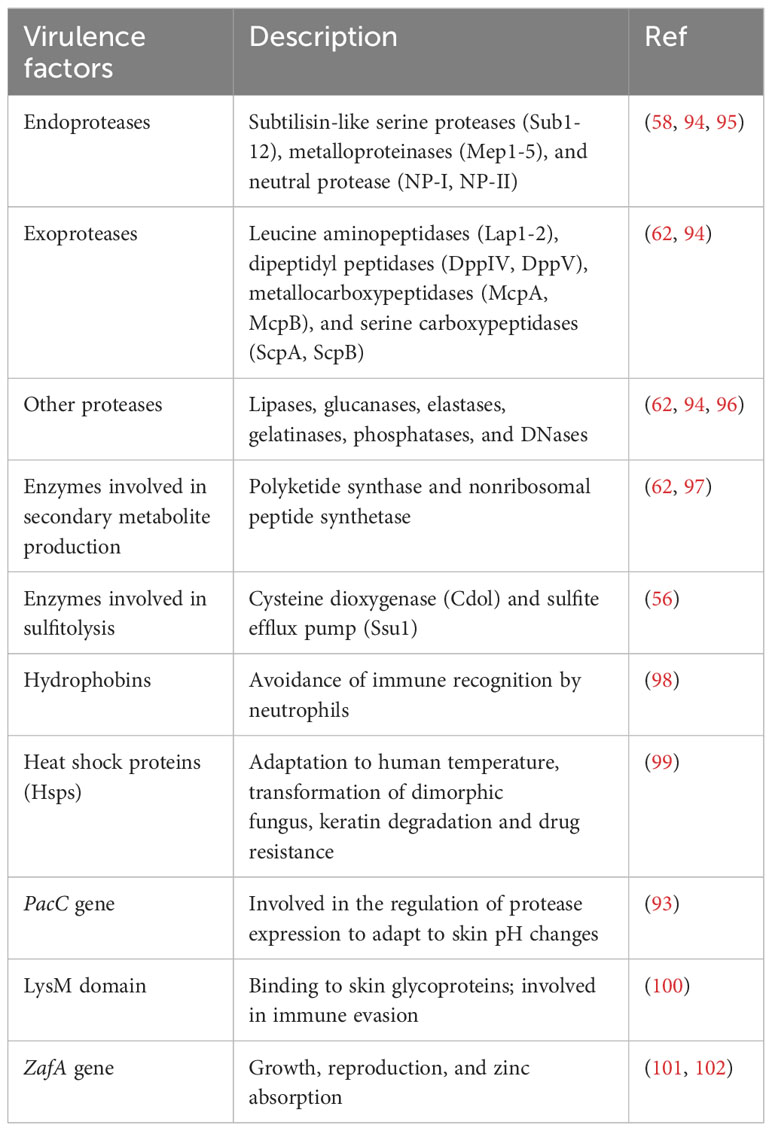
The pH of healthy skin and nails is slightly acidic; however, the amino acid metabolism during keratin breakdown causes an alkaline shift (89). Accordingly, the dermatophyte keratinases released during the early infection stages exhibit optimal activity at a slightly acidic pH, and other keratinases found in the later phases during keratin breakdown have maximal activity at higher pH values (90), which demonstrates that pathogenic fungi can sense and respond to the environmental pH. This adaptive response relies on the conserved PacC/Pal signal transduction pathway, which includes active PacC protein as a pH signaling transcription regulator (91, 92). PacC is essential for dermatophytes to grow on human tissues, as PacC gene disruption reduces keratinolytic protease secretion and the ability of mutant strains to invade the stratum corneum (93). Table 2 summarizes the main virulence factors of dermatophytes.
2.4 Mechanisms of antifungal resistance
There has been increasing concern about antifungal resistance during the last decade, especially after the emergence of T. indotineae, which can cause chronic or recurrent widespread superficial infections (103, 104). The frequency of terbinafine resistance in T. indotineae isolates is approximately 75% in India and more than 15% in other countries (33, 105). In addition, clinical failure of T. indotineae infection to azole treatments has also been reported globally (16, 35, 106, 107). Typically, antifungal resistance is acquired due to changes that directly or indirectly affect the drug–target interaction, including genetic changes to the target binding site (108), elevated drug efflux activity for intracellular drugs (103), or inhibition of prodrug activation (109). In terms of T. indotineae, terbinafine resistance has been associated to single nucleotide polymorphisms in the gene encoding the terbinafine binding protein squalene epoxidase (SQLE). In terms of T. indotineae, terbinafine resistance has been associated with single nucleotide polymorphisms in the gene encoding the terbinafine binding protein squalene epoxidase (SQLE). The Leu393Phe, Leu393Ser, and Phe397Leu substitutions in the enzyme have been the most frequently reported from resistant isolates (36). In addition, the overexpression of the ATP-binding cassette transporter (ABC) family gene MDR3 and the amplification of the C14-α-demethylase-encoding gene CYP51B are associated with azole resistance (40, 110, 111).
3 Host immune responses to dermatophytes
Apart from the direct damage caused by fungal virulence factors, host immune responses to dermatophyte infection also result in inflammatory damage to skin. Host immune responses, including those of the adaptive and innate immune systems, are determined by the interaction of pathogen-associated molecular patterns (PAMPs) and damage-associated molecular patterns (DAMPs) with pattern recognition receptors (PRRs) (1).
3.1 Innate immune responses
In shaping immune responses, dermatophytes can be detected by nonimmune and immune mediators via their cell wall components, secreted extracellular molecules, or intracellular content by multiple PRRs, which, when bound, cause transduction of intracellular signals that stimulate phagocyte lysis, cytokine and chemokine secretion, fungal phagocytosis, respiratory burst, etc. (112, 113). Usually, PRRs are classified according to their composition and activities: C-type lectin receptors (CLRs), retinoic acid inducible gene (RIG)-like receptors (RLRs), Toll-like receptors (TLRs), and nucleotide-binding and oligomerization domain (NOD)-like receptors (NLRs) (114, 115). Dermatophytes interact with the TLR, CLR, and NLR signaling pathways, which regulate host antifungal immunity.
CLRs, including dectins, Mincle, and mannose receptor (MR), recognize glycans, glycolipids, and glycoproteins contained in fungal cell walls that are not found in mammals (113, 114). Expressed on myeloid cells (neutrophils, dendritic cells [DCs], monocytes, and macrophages), keratinocytes, and human B-cell and T-cell subsets (116, 117), dectin-1 recognizes many pathogenic fungi by β-1,3-glucans present on their cell wall (112) and is thus one of the most important receptors in antifungal immune activities. The immunoreceptor tyrosine-based activation motif (ITAM)-comprising cytoplasmic domain is involved in dectin-1 signaling and is phosphorylated to recruit Syk kinase via a Src family kinase, activating mucosa-associated lymphoid tissue lymphoma translocation protein 1 (MALT1), caspase-associated recruitment domain 9 (CARD9), and a molecular scaffold composed of B-cell lymphoma/leukemia 10 (Bcl10) (118). This activation subsequently activates the nuclear factor κB (NF-κB) pathway, the canonical (NOD-like receptor thermal protein domain-associated protein 3 [NLRP3]/caspase 1) and noncanonical (MALT1/caspase 8) inflammasomes, and the Raf-1 kinase pathway (119) (Figure 2).
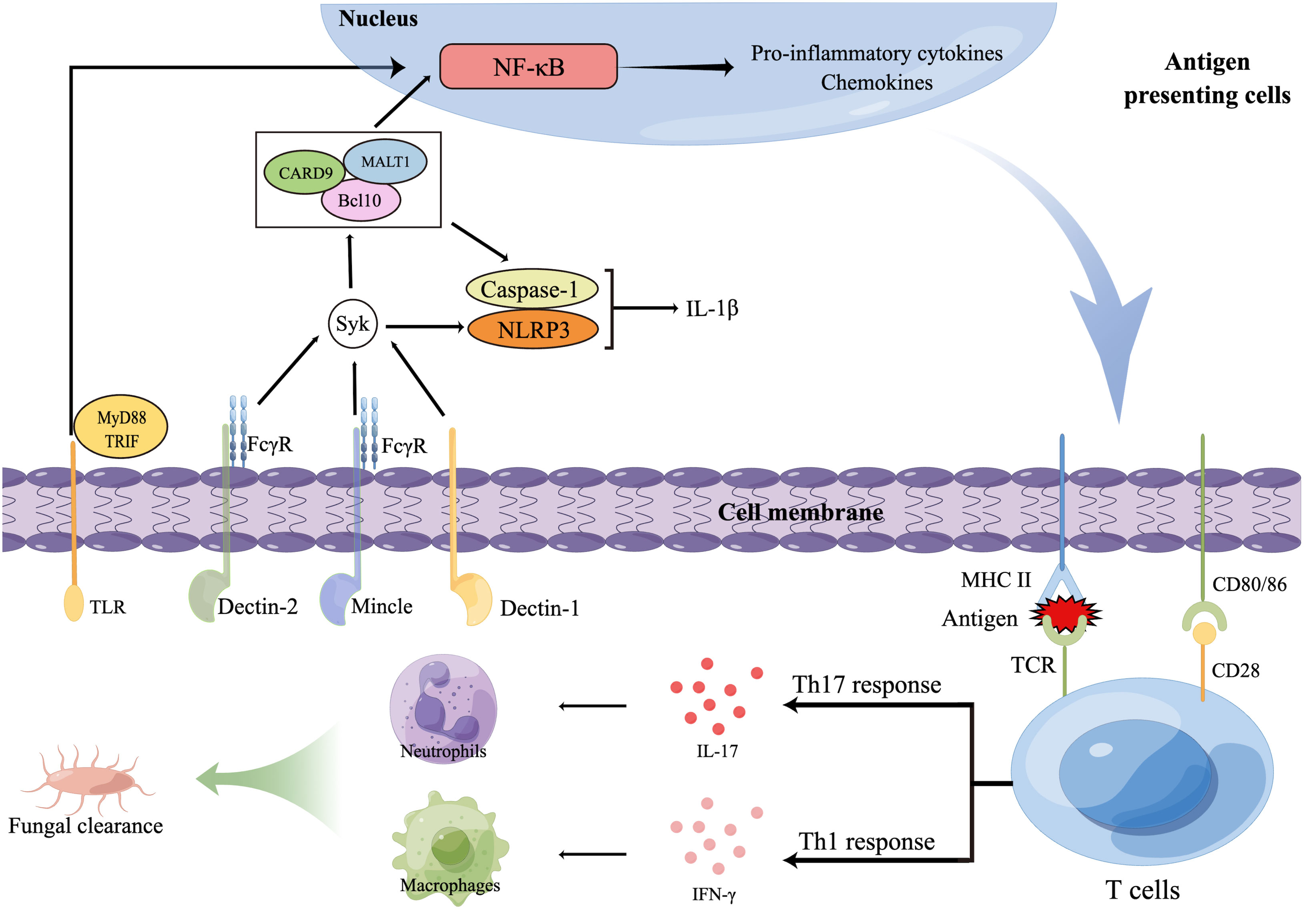
Figure 2 Innate and adaptive immune responses against dermatophytes. Several pattern recognition receptors (PRRs) on antigen presenting cells (APCs), including C-type lectin receptors (CLR) and TLR, are able to recognize dermatophytes, resulting in the release of transcription factors to nucleus that will regulate expression of inflammatory cytokinesThe foreign antigens APCs present and pro-inflammatory cytokines APCs secrete promote the polarization of T cells. T helper 17 (Th17) cells and other IL-17-secreting T cells (not shown) produce IL-17, promoting the production of neutrophil-recruiting chemokines. Interferon γ induced by Th1 cells activates macrophages. Then, macrophages and neutrophils together phagocytose and kill dermatophytes. Bcl10, B-cell lymphoma/leukemia 10; CARD9, Caspase-associated recruitment domain 9; FcγR, Fc receptor γ-chain; IFN-γ, Interferon γ; IL, Interleukin; MALT1, Mucosa-associated lymphoid tissue lymphoma translocation protein 1; MHC, Major histocompatibility complex; MyD88, Myeloid differentiation primary response 88; NF-κB, Nuclear factor κB; NLRP3, NOD-like receptor thermal protein domain-associated protein 3; TCR, T-cell receptor; Th cells, T helper cells; TLRs, Toll-like receptors; TRIF, Toll IL-1 receptor domain (TIR)-comprising adaptor inducer interferon-β.
Dectin-2 and Mincle belong to the dectin-2 cluster, which are predominantly expressed on myeloid cells and can identify O-linked mannoproteins and α-mannans from multiple pathogenic fungi (113). These receptors not only interact with the ITAM-comprising Fc receptor γ-chain (FcγR) to stimulate intracellular signaling through the Syk-CARD9 pathway (120) but also antagonize or synergize with other CLRs, such as TLRs, dectin-1, and inflammasomes (121).
Previous reports have described the function of dectin-1 and dectin-2 in protecting against dermatophyte infection. Both of these proteins can identify and bind to T. rubrum and M. audouinii, mediating innate immune responses (122). In addition, dectin-1 expression was markedly upregulated in T. rubrum cocultured with keratinocytes (123). Defects in these receptors severely impair the production of interferon γ (IFN-γ), tumor necrosis factor α (TNF-α), interleukin-1β (IL-1β), IL-10, and IL-6, culminating in deep dermatophytosis (124–126). These studies illustrated the major role of dectins in antidermatophyte immunity.
As a type-I transmembrane protein mainly expressed by DCs and macrophages, MR is responsible for L-fucose, D-mannose, or N-acetyl glucosamine recognition (113); however, its function in immunity to fungal pathogens seems controversial. MR activation led to Grb2 recruitment followed by Rac/Pak/Cdc-42 signaling cascade activation, which in turn limited phospho-inositide-3 kinase (PI3K) activity and phagosome-lysosome fusion (127). Nevertheless, blockade of MR using soluble antibodies inhibited macrophages from phagocytosing T. rubrum spores in vitro (128). Therefore, further investigations are required to understand the redundant roles of MR in the innate immune response.
TLRs are type-I integral membrane glycoproteins that can also recognize fungal PAMPs; however, the primary structures of fungal ligands are still only partially resolved to date. TLRs can hetero- or homodimerize with CLRs in the presence of ligands and then mediate intracellular signal transduction by Toll IL-1 receptor domain (TIR)-comprising adaptor inducer interferon-β (TRIF) and myeloid differentiation primary response 88 (MyD88), thereby initiating inflammatory responses.
TLR2 and TLR4 are representative TLRs in the recognition of dermatophytes. Upon coculture with dermatophytes, TLR2 and TLR4 expression in keratinocytes, neutrophils, myeloid cells, and fibroblasts was significantly increased (123, 129, 130). Similar results were obtained in patients with dermatophytosis (131). Interestingly, reduced expression of TLR4 in patients with disseminated dermatophytosis was found compared to that in healthy controls, indicating that the lack of TLR4 might contribute to the lack of infection resolution and the resulting chronic state of dermatophytosis (132). TLR2 blockade by neutralizing antibodies disrupts monocyte fungicidal activity against T. rubrum and monocyte TNF-α secretion, suggesting the importance of and requirement for TLR2 for effective conidium phagocytosis, and the absence of TLR2 in human monocytes may disrupt the successful inflammatory response (133). However, another study showed that TLR2-deficient and wild-type (WT) mice exhibited similar control of deep dermatophyte infection; nevertheless, the TLR2-deficient mice exhibited a notable elevation in IFN-γ, IL-10, and IL-17 production and an increased percentage of splenic regulatory T (Treg) cells (134). Therefore, how TLR2 exerts its immune activities during dermatophyte infection is still not completely clear, and more investigations are needed to elucidate its role in protection against dermatophytosis.
Similar to TLRs, NLRs are also associated with antifungal immunity primarily via recognition of other PRRs. NLRP3 is the most important NLR for antifungal immunity, and it is a part of the inflammasome, characterized by subsequent IL-18 and IL-1β generation owing to caspase-1 activation. Previous research has demonstrated the critical role of the NLRP3 inflammasome in host innate immunity against M. canis infection since M. canis-stimulated IL-1β secretion relies on NLRP3, whereas dectin-1, Syk, and CARD9 are linked with IL-1β secretion via pro-IL-1β transcription modulation (135). In addition, bone marrow-derived macrophages generate IL-1β in response to T. rubrum conidia in a caspase-1 and NLRP3-dependent manner, exerting protection against T. rubrum infection (136). A similar phenomenon was observed in monocytes during T. schoenleinii infection (137). In vivo experiments also showed that T. benhamiae, Arthroderma vanbreuseghemii and M. canis could promote IL-1β production in epithelial cells through the NLRP3-caspase1 pathway (138), which played a protective role against dermatophyte infection. Taken together, these results suggest that manipulating NLRP3 signaling can be a novel approach for the control of dermatophytosis.
Mast cells (MCs) also largely contribute to the immune responses in dermatophytosis infections as they are the first cells to encounter pathogens along with the other innate immune cells (Figure 3). MCs can participate in the direct killing of organisms by phagocytosis and the production of reactive oxygen species (ROS) and antimicrobial peptides (AMPs) (139). Another antifungal mechanism of MCs is the formation of extracellular traps composed of DNA, histones, and granule proteins (140). Mast cells can modulate host innate immune responses by secreting eicosanoid metabolites, chemokines, and cytokines (141). The release of histamine and other vasoactive mediators increases vascular permeability and local blood flow to increase the clearance of fungus (139). Chemotactic factors can enhance the recruitment of multiple inflammatory cells, including eosinophils (eotaxin), natural killer (NK) cells (IL-8), and neutrophils (IL-8 and TNF-α) (142).
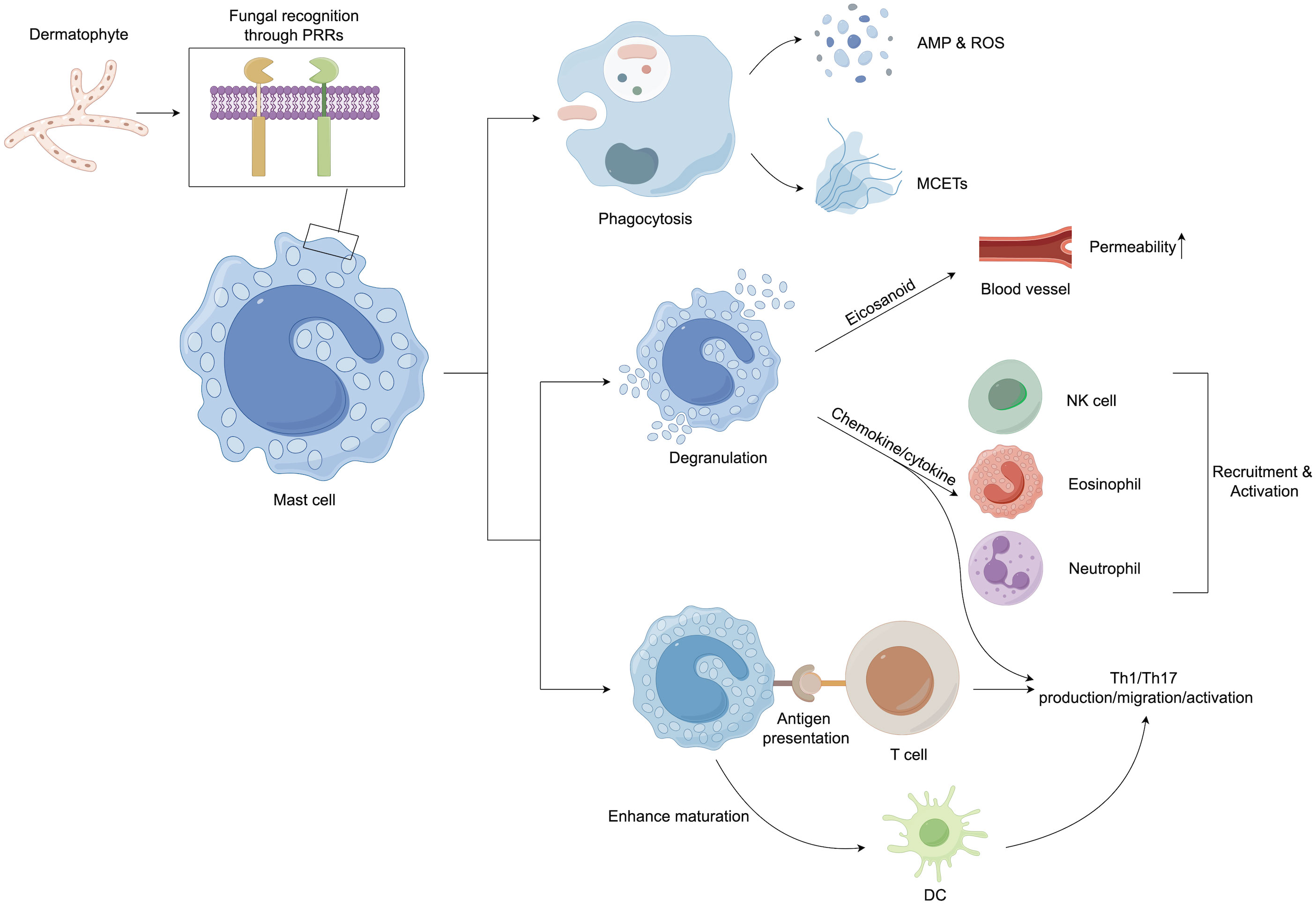
Figure 3 Mast cells play an important role in host defense against dermatophytes. After recognition of the pathogen, mast cells initiate both direct and indirect host defenses. They can kill pathogenic microorganisms by phagocytosis and the production of ROS, AMP, and extracellular traps. In addition, granulation mast cells can release several proinflammatory mediators, including histamine, chemokines, and cytokines, which can upregulate the innate immune response. Mast cell products have also been implicated in the regulation of adaptive immune responses, especially the promotion of Th17 reactions. In addition, mast cells can enhance the maturation of dendritic cells (DCs), priming DCs to facilitate Th17 and Th1 polarization. AMP, Antimicrobial peptide; DCs, Dendritic cells; MCETs, Extracellular traps of mast cells; NK cells, Natural killer cells; PRRs, Pattern recognition receptors; ROS, Reactive oxygen species; Th cells, T helper cells.
3.2 Adaptive immune responses
Adaptive and innate immunity are inextricably linked, and a successful adaptive immune response represents a cooperative effort that requires stimulation of antigen-presenting cells (APCs), tissue resident cells, and antigen-specific B and T cells (143). The T cell-mediated adaptive immune response is essential in antifungal defense (144, 145). PRR activation initiates a signal cascade that stimulates the MAPK and NF-kB pathways, which in turn induce naive T-cell transformation into T helper (Th) cells (146). Differentiation of Th cells is critical for antifungal immunity, as these cells generate proinflammatory cytokines, such as IL-17 and IFN-γ, which promote the killing of fungi by recruiting and activating phagocytes (146) (Figure 2).
The IL-17-mediated (type 17 or Th17) response was reported to be a critical pathway for host defense against fungal invasion (145). The IL-17 family consists of 6 six members (IL-17A to IL-17F) that exert their biological activities via interaction with IL-17 receptors (IL-17RA to IL-17RE). The most prevalent of these members are IL-17A and IL-17F, which exert their biological functions by interacting with IL-17RA and IL-17RC (147). In the skin, myeloid cell PRRs recognize fungi, fibroblasts and keratinocytes and stimulate IL-23, IL-1β, IL-6, and IL-21 synthesis (129, 148, 149); these cytokines interact with their lymphocytic receptors, alternatively inducing the phosphorylation and thus activation of cellular signal transducer and transcription 3 (STAT3), activation of retinoic-acid-receptor-related orphan nuclear receptor gamma (RORγt) transcription factor activity and subsequently stimulating type 17 cytokines (IL-22 and IL-17) or Th17 lineage production (150). Several studies have found that MCs may play an essential role in the functions of Th17 lymphocytes (Figure 3). MC-derived TNF is required to develop IL-17-secreting Th17 cells in a murine model (151). MCs can also induce Th17 differentiation in eosinophil-deficient mice, leading to neutrophil-dominant inflammation (152). In addition, MC-secreting chemokines might participate in Th17 infiltration (153). Interestingly, Th17 cells express the functional histamine H4 receptor; thus, MC-derived histamine may affect Th17 activity (154). Indeed, stimulation with histamine or an H4 receptor agonist increases the production of IL-17 by human Th17 cells (154). IL-17 can stimulate the generation of various cytokines (such as TNF and IL-6), vascular endothelial growth factor (VEGF), and chemokines (CXCL1 and CXCL8), increase AMP expression, and promote keratinocyte proliferation (145), which may essentially lead to the clearance of cutaneous fungal infection.
Mounting evidence has illustrated the potent antifungal immunity of AMPs induced by IL-17, which mainly involves S100 proteins, cathelicidin, and β-defensins. Cathelicidin is a synergistic agent for Th17 differentiation, and mice lacking cathelicidin are unable to produce adequate IL-17 levels in response to inflammation (155). β-defensins are key components of innate immunity that directly kill or inhibit the growth of pathogens (156). S-100 proteins can exert direct antifungal effects on fungal growth and indirect antifungal activity by regulating host immune responses (157). Previous studies have demonstrated that the expression of AMPs is increased in dermatophytosis (158) and that the reduced production of AMPs is related to chronic and widespread infection (159). Interestingly, some cytokines, such as IL-22 and TNF-α, synergize with IL-17 function by increasing AMPs production, thus playing an indispensable role in limiting the dissemination of pathogens (160, 161).
Several in vivo and in vitro studies have indicated the protective function of the Th17 immune response against dermatophyte infection. In guinea pigs infected with T. benhamiae and A. vanbreuseghemii, the in situ cytokine profile was characterized by the overexpression of transforming growth factor-β (TGF-β), IL-1β and IL-6 during infection, illustrating Th17 pathway involvement in the establishment of immunity (138). Increased levels of Th17 cells were also found in mice infected with T. mentagrophytes (162). Moreover, Il-17ra-/- or Il17a/f -/- mice suffered from a higher fungal load after inoculation of M. canis on the skin, which proved the role of the type 17 response in reducing the fungal burden (148). Consistent with this finding, patients with STAT3 mutations have enhanced dermatophytosis and candidiasis susceptibility because of a diminished type 17 response (163). In addition, a reduction in type 17 immune activity has been linked to dermatophytosis (invasive or chronic) susceptibility, as has been indicated in adult T-cell leukemia/lymphoma (ATLL) individuals (164), those with STAT3-related malfunction (163), patients with gain-of-function (GOF) mutations (165), and those receiving anti-IL-17 antibody-related therapy (i.e., secukinumab and ixekizumab) (166–168). Thus, the type 17 immune response is essential for protecting the host from dermatophyte invasion.
The function of IFN-γ-modulated (Th1 or type 1) activities related to anti-dermatophyte-relevant skin immunoprotection is not adequately understood compared to IL-17-induced immune activities. Baltazar et al. (169) established an Ifn-γ-/- mouse model of T. rubrum infection and found that the fungal load increased significantly compared with that of WT mice. Moreover, Ifn-γ-/- mouse macrophages were less effective at engulfing T. rubrum conidia and killing them by generating ROS and IL-1β (169). Sardana et al. (146) therefore concluded that the elimination of a fungal infection is mainly mediated by an IFN-γ-induced (type 1 or Th1) response, as Th1 cells can produce proinflammatory cytokines and stimulate phagocytes.
However, in an M. canis infection model, WT mice with dermatophyte infection did not exhibit an elevation of the number of antigen-related IFN-γ-secreting T cells in the lymph nodes draining the skin (148). However, mice in which IL-17 expression was downregulated exhibited an alternative pathway of Th1 activity, indicating the presence of IFN-γ-regulated compensation for controlling M. canis infection. Nevertheless, studies with IL-17RA KO mice revealed that IFN-γ neutralization enhanced the secretion of Th17 lineage cytokines (IL-22, IL-1β, IL-17, and IL-6) in the skin and markedly suppressed fungal growth (148). Thus, IFN-γ deregulation is a possibility when IL-17 signaling is absent and might cause superficial M. canis overgrowth by suppressing type 17-linked activities. Similarly, individuals with STAT1 GOF mutations that induce IFN gene transcription and alleviate IL-17-regulated immunity have an enhanced chance of developing chronic dermatophytosis and mucocutaneous candidiasis (165, 170). Regarding this finding, an elevated STAT1 response to Th1 cytokines (IL-27 and IFN-γ) inhibited IL-17-producing T-cell differentiation (171, 172). Enhanced STAT1 phosphorylation activities caused by IFN-γ can be inhibited after Janus kinase (JAK) inhibitor and ruxolitinib treatment (173). Interestingly, individuals with STAT1 GOF mutations who are managed with ruxolitinib suffer from mucocutaneous candidiasis remission (174). The M. canis model experimental results are supported by the clinical data, indicating that the cause of dermatophyte susceptibility is correlated with IL-17-induced immune-related deficiency and that type 1 and 17 immunities oppositely modulate each other. The impact of a dysregulated Th1 response on dermatophyte infection in the absence of a functional type 17 immune response needs to be addressed in the future (175).
Nevertheless, in mice infected with T. benhamiae, the type 1 and type 17 responses were both found to be involved in fungal clearance, and persistent superficial infection occurred only when the IL-17 and IFN-γ pathways were both defective (176), suggesting that Th1 and Th17 responses function in a complementary manner. The contrasting results regarding the type 1 response in T. rubrum, M. canis, and T. benhamiae infection reminded us that different pathogens might have distinct virulence factors or activate unique subsets of APCs, thus selectively inducing the activation of different immune pathways. Briefly, the Th1 response may impair the protection mediated by the Th17 response but may also complement with the Th17 response to clear dermatophyte infection.
Treg cells ensure a controlled immune response in the presence of microbes, thereby preventing pathological immune responses (145). However, pathogen clearance might be hindered, and persistent infection can be promoted if Tregs excessively inhibit the immune response (146). Thus, maintaining a balance between immunological disease prevention and protective immune responses against pathogens requires optimal Treg activity and activation. Treg cell marker expression on peripheral blood CD4+ T cells was found to be considerably higher in individuals with persistent dermatophytosis than in healthy controls (177). Similarly, Kaya et al. (178) discovered that onychomycosis patients had a greater expression of CD25+ CD4+ Treg cells than healthy controls. These findings demonstrated that elevated Treg cell levels might contribute to vulnerability to dermatophytosis and infection persistence.
4 CARD9 deficiencies in deep dermatophytosis
Invasions of dermatophytes into the deeper tissues and even the hypodermis and dermis define the severe, resistant, and, in rare cases, fatal illness known as deep dermatophytosis. Deep dermatophytosis is more common in those who have had a solid organ transplant, are using immunosuppressants topically, or have a CARD9 mutation (6).
CARD9 is a caspase recruitment domain-containing signaling protein essential for CLR downstream signaling and cross-signaling with other innate receptors (179). In addition, it is also involved in stimulating T-cell differentiation into different Th cells, initiating optimal adaptive immune responses (146). Therefore, CARD9 is vital for antifungal adaptive and innate immunity.
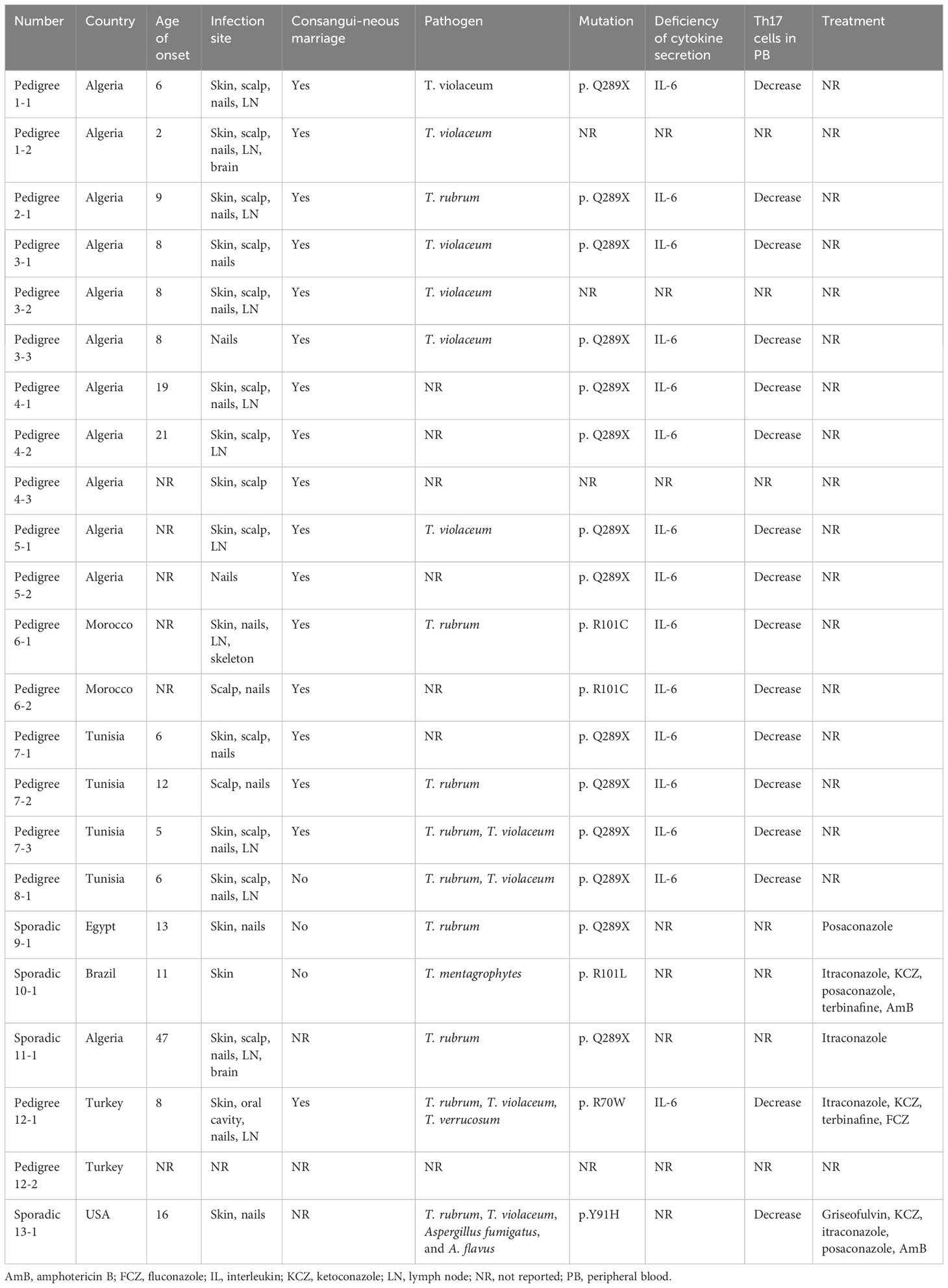
Patients with CARD9 deficiency have impaired cytokine and chemokine production in response to fungal infections (180). A homozygous premature stop codon mutation (Q289) could be detected in 15 individuals within seven unrelated Tunisian and Algerian families, and a homozygous missense mutation (R101C) could be detected in two Moroccan related families, providing the first evidence linking autosomal recessive (AR) CARD9 deficiency and T. violaceum- or T. rubrum-mediated deep dermatophytosis (181). Subsequently, a CARD9 Q289 mutation was documented in Egyptian individuals suffering from T. rubrum affecting the skin, nails, and other superficial tissues (182), as well as in an Algerian women with T. rubrum infection of her brain (183). Since then, novel AR CARD9 mutations, such as R101L and R70W, have been found in patients with deep and chronic dermatophytosis (184, 185). We summarized the CARD9 mutations found in dermatophyte infection patients (Table 3) (181–186), more than 50% of whom are from North Africa. Interestingly, CARD9 mutations are more common in Asia (187), however, African patients accounted for the majority of CARD9 mutation-related dermatophytosis cases. One possible explanation is that different CARD9 mutations causing different infections might have different geographic distributions. For example, homozygous (HMZ) p.Q289X, HMZ p.Q295X and HMZ p.D274fsX60 are the most commonly identified CARD9 mutations, among which the HMZ p.Q289X mutation is mainly found in Africa and is associated with a significant increase in the risk of developing deep dermatophytosis compared to other mutations (187). This could explain the high prevalence of dermatophyte infection in African CARD9-deficient patients, as mutations are specific in particular populations or geographic regions.
CARD9 deficiency is also related to an impaired type 17 immune response, since the Th17 cell proportion and IL-17A and IL-22 expression in CARD9-defective patients are significantly lower than that in healthy people (188). Interestingly, in individuals with both STAT3 and CARD9 mutations, high eosinophil and serum immunoglobulin E (IgE) levels were observed in addition to an increased susceptibility to fungal infections (189), which indicated an efficient biological mechanism that resulted in reduced Th17 pathway activity and enhanced Th2 pathway activity. Therefore, serum IgE and eosinophil levels could be utilized as indices in gene mutation tests for individuals with invasive dermatophytosis. In summary, the differentiation of Th cells (i.e., Th17) and the function of adaptive immune pathways were significantly impaired in CARD9 mutant patients due to a deficiency in the production of proinflammatory cytokines (i.e., IL-6, IL-1β, TNF-α), resulting in increased susceptibility to chronic or deep fungal infections.
5 Conclusion
Dermatophytes are the most common pathogenic fungi worldwide, causing superficial and even deep infections. In the past few decades, scientists have made tremendous breakthroughs in understanding the pathogenicity of dermatophytes and the host immune responses against pathogenic fungi through in vivo and in vitro studies. With the increasing incidence of dermatophytosis and the emergence of drug-resistant T. indotineae strains (190), there is an urgent need for a better understanding of the virulence factors of dermatophytes and pathogen−host interactions to identify new targets for clinical treatment.
Author contributions
RD: Conceptualization, Writing – original draft. XW: Writing – original draft. RL: Supervision, Writing – review & editing.
Funding
The author(s) declare that no financial support was received for the research, authorship, and/or publication of this article.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Glossary
References
1. Gupta C, Das S, Gaurav V, Singh PK, Rai G, Datt S, et al. Review on host-pathogen interaction in dermatophyte infections. J Mycol Med (2023) 33:101331. doi: 10.1016/j.mycmed.2022.101331
2. Faway E, Lambert de Rouvroit C, Poumay Y. In vitro models of dermatophyte infection to investigate epidermal barrier alterations. Exp Dermatol (2018) 27:915–22. doi: 10.1111/exd.13726
3. Morales EG, Iorizzo M, Lucchini GM, Mainetti C. Trichophyton violaceum: an emerging pathogen in Southern Switzerland. Dermatology (2019) 235:434–39. doi: 10.1159/000501271
4. Degreef H. Clinical forms of dermatophytosis (ringworm infection). Mycopathologia (2008) 166:257–65. doi: 10.1007/s11046-008-9101-8
5. Smith KJ, Welsh M, Skelton H. Trichophyton rubrum showing deep dermal invasion directly from the epidermis in immunosuppressed patients. Br J Dermatol (2001) 145:344–8. doi: 10.1046/j.1365-2133.2001.04331.x
6. Wang R, Huang C, Zhang Y, Li R. Invasive dermatophyte infection: a systematic review. Mycoses (2021) 64:340–8. doi: 10.1111/myc.13212
7. de Hoog S, Monod M, Dawson T, Boekhout T, Mayser P, Gräser Y. Skin fungi from colonization to infection. Microbiol Spectr (2017) 5. doi: 10.1128/microbiolspec.FUNK-0049-2016
8. Ma Y, Wang X, Li R. Cutaneous and subcutaneous fungal infections: recent developments on host-fungus interactions. Curr Opin Microbiol (2021) 62:93–102. doi: 10.1016/j.mib.2021.05.005
9. Zhan P, Liu W. The changing face of dermatophytic infections worldwide. Mycopathologia (2017) 182:77–86. doi: 10.1007/s11046-016-0082-8
10. Martinez-Rossi NM, Peres NTA, Bitencourt TA, Martins MP, Rossi A. State-of-the-art dermatophyte infections: epidemiology aspects, pathophysiology, and resistance mechanisms. J Fungi (Basel) (2021) 7:629. doi: 10.3390/jof7080629
11. Song G, Zhang M, Liu W, Liang G. Children onychomycosis, a neglected dermatophytosis: A retrospective study of epidemiology and treatment. Mycoses (2023) 66:448–54. doi: 10.1111/myc.13571
12. Landreau A, Simon L, Delaunay P, Pomares C, Hasseine L. Superficial fungal infections in the south of France-is fusariosis the next emergent onychopathy? Med Mycol (2023) 61. doi: 10.1093/mmy/myad015
13. Song G, Zhang M, Liu W, Liang G. Epidemiology of onychomycosis in chinese mainland: A 30-year retrospective study. Mycopathologia (2022) 187:323–31. doi: 10.1007/s11046-022-00647-4
14. Kumar P, Ramachandran S, Das S, Bhattacharya SN, Taneja B. Insights into changing dermatophyte spectrum in India through analysis of cumulative 161,245 cases between 1939 and 2021. Mycopathologia (2023) 188:183–202. doi: 10.1007/s11046-023-00720-6
15. Lipner SR, Scher RK. Onychomycosis: clinical overview and diagnosis. J Am Acad Dermatol (2019) 80:835–51. doi: 10.1016/j.jaad.2018.03.062
16. Klinger M, Theiler M, Bosshard PP. Epidemiological and clinical aspects of Trichophyton mentagrophytes/Trichophyton interdigitale infections in the Zurich area: a retrospective study using genotyping. J Eur Acad Dermatol Venereol (2021) 35:1017–25. doi: 10.1111/jdv.17106
17. Hiruma J, Ogawa Y, Hiruma M. Trichophyton tonsurans infection in Japan: epidemiology, clinical features, diagnosis and infection control. J Dermatol (2015) 42:245–9. doi: 10.1111/1346-8138.12678
18. Coulibaly O, L'Ollivier C, Piarroux R, Ranque S. Epidemiology of human dermatophytoses in Africa. Med Mycol (2018) 56:145–61. doi: 10.1093/mmy/myx048
19. Sacheli R, Adjetey C, Darfouf R, Harag S, Huynen P, Meex C, et al. A one-year survey of Microsporum audouinii infections in Belgium: epidemiological and genotypic characterization. Clin Microbiol Infect (2016) 22:285.e9–17. doi: 10.1016/j.cmi.2015.11.012
20. Aneke CI, Cmokova A, Hubka V, Rhimi W, Otranto D, Cafarchia C. Subtyping options for microsporum canis using microsatellites and MLST: a case study from Southern Italy. Pathogens (2021) 11:4. doi: 10.3390/pathogens11010004
21. Verma SB, Panda S, Nenoff P, Singal A, Rudramurthy SM, Uhrlass S, et al. The unprecedented epidemic-like scenario of dermatophytosis in India: I. Epidemiology, risk factors and clinical features. Indian J Dermatol Venereol Leprol (2021) 87:154–75. doi: 10.25259/ijdvl_301_20
22. Havlickova B, Czaika VA, Friedrich M. Epidemiological trends in skin mycoses worldwide. Mycoses (2008) 51 Suppl 4:2–15. doi: 10.1111/j.1439-0507.2008.01606.x
23. Rezaei-Matehkolaei A, Rafiei A, Makimura K, Graser Y, Gharghani M, Sadeghi-Nejad B. Epidemiological aspects of dermatophytosis in Khuzestan, southwestern Iran, an update. Mycopathologia (2016) 181:547–53. doi: 10.1007/s11046-016-9990-x
24. Lemus-Espinoza D, Teresa Maniscalchi M, Villarroel O, Bonoli SB, Wahab F, Garcia O. Superficial mycoses in patients from Anzoategui state, Venezuela, period 2002-2012. Invest Clin (2014) 55:311–20.
25. Miklić P, Skerlev M, Budimcić D, Lipozencić J. The frequency of superficial mycoses according to agents isolated during a ten-year period (1999-2008) in Zagreb area, Croatia. Acta Dermatovenerol Croat (2010) 18:92–8.
26. Chen XQ, Zheng DY, Xiao YY, Dong BL, Cao CW, Ma L, et al. Aetiology of tinea capitis in China: a multicentre prospective study. Br J Dermatol (2022) 186:705–12. doi: 10.1111/bjd.20875
27. Song G, Zhang M, Liu W, Liang G. Changing face of epidemiology of dermatophytoses in Chinese Mainland: A 30 years nationwide retrospective study from 1991 to 2020. Mycoses (2022) 65:440–8. doi: 10.1111/myc.13425
28. Sabou M, Denis J, Boulanger N, Forouzanfar F, Glatz I, Lipsker D, et al. Molecular identification of Trichophyton benhamiae in Strasbourg, France: a 9-year retrospective study. Med Mycol (2018) 56:723–34. doi: 10.1093/mmy/myx100
29. Dolenc-Voljc M, Gasparic J. Human infections with Microsporum gypseum complex (Nannizzia gypsea) in Slovenia. Mycopathologia (2017) 182:1069–75. doi: 10.1007/s11046-017-0194-9
30. Ziolkowska G, Nowakiewicz A, Gnat S, Troscianczyk A, Zieba P, Dziedzic BM. Molecular identification and classification of Trichophyton mentagrophytes complex strains isolated from humans and selected animal species. Mycoses (2015) 58:119–26. doi: 10.1111/myc.12284
31. Ozkutuk A, Ergon C, Yulug N. Species distribution and antifungal susceptibilities of dermatophytes during a one year period at a university hospital in Turkey. Mycoses (2007) 50:125–9. doi: 10.1111/j.1439-0507.2006.01333.x
32. Salehi Z, Shams-Ghahfarokhi M, Razzaghi-Abyaneh M. Molecular epidemiology, genetic diversity, and antifungal susceptibility of major pathogenic dermatophytes isolated from human dermatophytosis. Front Microbiol (2021) 12:643509. doi: 10.3389/fmicb.2021.643509
33. Jabet A, Normand AC, Brun S, Dannaoui E, Bachmeyer C, Piarroux R, et al. Trichophyton indotineae, from epidemiology to therapeutic. J Mycol Med (2023) 33:101383. doi: 10.1016/j.mycmed.2023.101383
34. Singh A, Masih A, Monroy-Nieto J, Singh PK, Bowers J, Travis J, et al. A unique multidrug-resistant clonal Trichophyton population distinct from Trichophyton mentagrophytes/Trichophyton interdigitale complex causing an ongoing alarming dermatophytosis outbreak in India: Genomic insights and resistance profile. Fungal Genet Biol (2019) 133:103266. doi: 10.1016/j.fgb.2019.103266
35. Nenoff P, Verma SB, Ebert A, Süß A, Fischer E, Auerswald E, et al. Spread of terbinafine-resistant trichophyton mentagrophytes type VIII (India) in Germany-"The tip of the iceberg?". J Fungi (Basel) (2020) 6. doi: 10.3390/jof6040207
36. Jabet A, Brun S, Normand AC, Imbert S, Akhoundi M, Dannaoui E, et al. Extensive dermatophytosis caused by terbinafine-resistant trichophyton indotineae, France. Emerg Infect Dis (2022) 28:229–33. doi: 10.3201/eid2801.210883
37. Sacheli R, Hayette MP. Antifungal resistance in dermatophytes: genetic considerations, clinical presentations and alternative therapies. J Fungi (Basel) (2021) 7. doi: 10.3390/jof7110983
38. Siopi M, Efstathiou I, Theodoropoulos K, Pournaras S, Meletiadis J. Molecular epidemiology and antifungal susceptibility of trichophyton isolates in Greece: emergence of terbinafine-resistant trichophytonmentagrophytes type VIII locally and globally. J Fungi (Basel) (2021) 7. doi: 10.3390/jof7060419
39. Astvad KMT, Hare RK, Jørgensen KM, Saunte DML, Thomsen PK, Arendrup MC. Increasing terbinafine resistance in danish trichophyton isolates 2019-2020. J Fungi (Basel) (2022) 8. doi: 10.3390/jof8020150
40. Kong X, Tang C, Singh A, Ahmed SA, Al-Hatmi AMS, Chowdhary A, et al. Antifungal susceptibility and mutations in the squalene epoxidase gene in dermatophytes of the trichophyton mentagrophytes species complex. Antimicrob Agents Chemother (2021) 65:e0005621. doi: 10.1128/aac.00056-21
41. Posso-De Los Rios CJ, Tadros E, Summerbell RC, Scott JA. Terbinafine resistant trichophyton indotineae isolated in patients with superficial dermatophyte infection in canadian patients. J Cutan Med Surg (2022) 26:371–6. doi: 10.1177/12034754221077891
42. Ngo TMC, Ton Nu PA, Le CC, Ha TNT, Do TBT, Tran Thi G. First detection of Trichophyton indotineae causing tinea corporis in Central Vietnam. Med Mycol Case Rep (2022) 36:37–41. doi: 10.1016/j.mmcr.2022.05.004
43. Sharma Y, Jain S, Chandra K, Khurana VK, Kudesia M. Clinico-mycological evaluation of dermatophytes and non-dermatophytes isolated from various clinical samples: a study from north India. J Res Med Sci (2012) 17:817–8.
44. John AM, Schwartz RA, Janniger CK. The kerion: an angry tinea capitis. Int J Dermatol (2018) 57:3–9. doi: 10.1111/ijd.13423
45. da Silva BC, Paula CR, Auler ME, Ruiz Lda S, Dos Santos JI, Yoshioka MC, et al. Dermatophytosis and immunovirological status of HIV-infected and AIDS patients from Sao Paulo city, Brazil. Mycoses (2014) 57:371–6. doi: 10.1111/myc.12169
46. Das A, Sil A, Fatima F, Podder I, Jafferany M. Impact of chronic and recurrent dermatophytosis on quality of life and psychologic morbidity-a cross-sectional study. J Cosmet Dermatol (2022) 21:3586–92. doi: 10.1111/jocd.14676
47. Achterman RR, White TC. A foot in the door for dermatophyte research. PloS Pathog (2012) 8:e1002564. doi: 10.1371/journal.ppat.1002564
48. Bongomin F, Olum R, Nsenga L, Namusobya M, Russell L, de Sousa E, et al. Estimation of the burden of tinea capitis among children in Africa. Mycoses (2021) 64:349–63. doi: 10.1111/myc.13221
49. Narang T, Bhattacharjee R, Singh S, Jha K, Kavita, Mahajan R, et al. Quality of life and psychological morbidity in patients with superficial cutaneous dermatophytosis. Mycoses (2019) 62:680–5. doi: 10.1111/myc.12930
50. Mushtaq S, Faizi N, Amin SS, Adil M, Mohtashim M. Impact on quality of life in patients with dermatophytosis. Australas J Dermatol (2020) 61:e184–8. doi: 10.1111/ajd.13191
51. Verma S, Vasani R, Reszke R, Matusiak L, Szepietowski JC. The influence of superficial dermatophytoses epidemic in India on patients' quality of life. Postepy Dermatol Alergol (2021) 38:102–5. doi: 10.5114/ada.2019.90088
52. Celestrino GA, Veasey JV, Benard G, Sousa MGT. Host immune responses in dermatophytes infection. Mycoses (2021) 64:477–83. doi: 10.1111/myc.13246
53. Esquenazi D, de Souza W, Alviano CS, Rozental S. The role of surface carbohydrates on the interaction of microconidia of Trichophyton mentagrophytes with epithelial cells. FEMS Immunol Med Microbiol (2003) 35:113–23. doi: 10.1016/s0928-8244(03)00007-5
54. Esquenazi D, Alviano CS, de Souza W, Rozental S. The influence of surface carbohydrates during in vitro infection of mammalian cells by the dermatophyte Trichophyton rubrum. Res Microbiol (2004) 155:144–53. doi: 10.1016/j.resmic.2003.12.002
55. Liu T, Zhang Q, Wang L, Yu L, Leng W, Yang J, et al. The use of global transcriptional analysis to reveal the biological and cellular events involved in distinct development phases of Trichophyton rubrum conidial germination. BMC Genomics (2007) 8:100. doi: 10.1186/1471-2164-8-100
56. Grumbt M, Monod M, Yamada T, Hertweck C, Kunert J, Staib P. Keratin degradation by dermatophytes relies on cysteine dioxygenase and a sulfite efflux pump. J Invest Dermatol (2013) 133:1550–5. doi: 10.1038/jid.2013.41
57. Gnat S, Nowakiewicz A, Lagowski D, Zieba P. Host- and pathogen-dependent susceptibility and predisposition to dermatophytosis. J Med Microbiol (2019) 68:823–36. doi: 10.1099/jmm.0.000982
58. Mercer DK, Stewart CS. Keratin hydrolysis by dermatophytes. Med Mycol (2019) 57:13–22. doi: 10.1093/mmy/myx160
59. Burstein VL, Beccacece I, Guasconi L, Mena CJ, Cervi L, Chiapello LS. Skin immunity to dermatophytes: from experimental infection models to human disease. Front Immunol (2020) 11:605644. doi: 10.3389/fimmu.2020.605644
60. Sharma B, Nonzom S. Superficial mycoses, a matter of concern: Global and Indian scenario-an updated analysis. Mycoses (2021) 64:890–908. doi: 10.1111/myc.13264
61. Ciesielska A, Kawa A, Kanarek K, Soboń A, Szewczyk R. Metabolomic analysis of Trichophyton rubrum and Microsporum canis during keratin degradation. Sci Rep (2021) 11:3959. doi: 10.1038/s41598-021-83632-z
62. Burmester A, Shelest E, Glockner G, Heddergott C, Schindler S, Staib P, et al. Comparative and functional genomics provide insights into the pathogenicity of dermatophytic fungi. Genome Biol (2011) 12:R7. doi: 10.1186/gb-2011-12-1-r7
63. Zhan P, Dukik K, Li D, Sun J, Stielow JB, Gerrits van den Ende B, et al. Phylogeny of dermatophytes with genomic character evaluation of clinically distinct Trichophyton rubrum and T. violaceum. Stud Mycol (2018) 89:153–75. doi: 10.1016/j.simyco.2018.02.004
64. Kasperova A, Kunert J, Raska M. The possible role of dermatophyte cysteine dioxygenase in keratin degradation. Med Mycol (2013) 51:449–54. doi: 10.3109/13693786.2013.794310
65. Nenoff P, Krüger C, Ginter-Hanselmayer G, Tietz HJ. Mycology - an update. Part 1: Dermatomycoses: causative agents, epidemiology and pathogenesis. J Dtsch Dermatol Ges (2014) 12:188–209; quiz 210, 188-211; quiz 212. doi: 10.1111/ddg.12245
66. Kunert J, Trüper HG. Cystine catabolism in mycelia of Microsporum gypseum, a dermatophytic fungus. Arch Microbiol (1986) 145:181–6. doi: 10.1007/bf00446777
67. Kasperova A, Kunert J, Horynova M, Weigl E, Sebela M, Lenobel R, et al. Isolation of recombinant cysteine dioxygenase protein from Trichophyton mentagrophytes. Mycoses (2011) 54:e456–62. doi: 10.1111/j.1439-0507.2010.01948.x
68. Kasperova A, Cahlikova R, Kunert J, Sebela M, Novak Z, Raska M. Exposition of dermatophyte Trichophyton mentagrophytes to L-cystine induces expression and activation of cysteine dioxygenase. Mycoses (2014) 57:672–8. doi: 10.1111/myc.12220
69. Ge LY, Liu J, Zheng HL, Mei H, Liang GZ, Liu WD. Comprehensive genome and transcriptome analysis of the dermatophyte Trichophyton schoenleinii reveals the candidate pathogenic genes. Mycoses (2021) 64:624–33. doi: 10.1111/myc.13257
70. Hamaguchi T, Morishita N, Usui R, Takiuchi I. Characterization of an extracellular keratinase from Microsporum canis. Nihon Ishinkin Gakkai Zasshi (2000) 41:257–62. doi: 10.3314/jjmm.41.257
71. Descamps F, Brouta F, Monod M, Zaugg C, Baar D, Losson B, et al. Isolation of a Microsporum canis gene family encoding three subtilisin-like proteases expressed in vivo. J Invest Dermatol (2002) 119:830–5. doi: 10.1046/j.1523-1747.2002.01784.x
72. Bagut ET, Baldo A, Mathy A, Cambier L, Antoine N, Cozma V, et al. Subtilisin Sub3 is involved in adherence of Microsporum canis to human and animal epidermis. Vet Microbiol (2012) 160:413–9. doi: 10.1016/j.vetmic.2012.06.011
73. Brouta F, Descamps F, Fett T, Losson B, Gerday C, Mignon B. Purification and characterization of a 43.5 kDa keratinolytic metalloprotease from Microsporum canis. Med Mycol (2001) 39:269–75. doi: 10.1080/mmy.39.3.269.275
74. Brouta F, Descamps F, Monod M, Vermout S, Losson B, Mignon B. Secreted metalloprotease gene family of Microsporum canis. Infect Immun (2002) 70:5676–83. doi: 10.1128/IAI.70.10.5676-5683.2002
75. Brouta F, Descamps F, Vermout S, Monod M, Losson B, Mignon B. Humoral and cellular immune response to a Microsporum canis recombinant keratinolytic metalloprotease (r-MEP3) in experimentally infected Guinea pigs. Med Mycol (2003) 41:495–501. doi: 10.1080/13693780310001615385
76. Zhang X, Wang Y, Chi W, Shi Y, Chen S, Lin D, et al. Metalloprotease genes of Trichophyton mentagrophytes are important for pathogenicity. Med Mycol (2014) 52:36–45. doi: 10.3109/13693786.2013.811552
77. Shi Y, Niu Q, Yu X, Jia X, Wang J, Lin D, et al. Assessment of the function of SUB6 in the pathogenic dermatophyte Trichophyton mentagrophytes. Med Mycol (2016) 54:59–71. doi: 10.1093/mmy/myv071
78. Preuett BL, Schuenemann E, Brown JT, Kovac ME, Krishnan SK, Abdel-Rahman SM. Comparative analysis of secreted enzymes between the anthropophilic-zoophilic sister species Trichophyton tonsurans and Trichophyton equinum. Fungal Biol (2010) 114:429–37. doi: 10.1016/j.funbio.2010.03.004
79. Leng W, Liu T, Wang J, Li R, Jin Q. Expression dynamics of secreted protease genes in Trichophyton rubrum induced by key host's proteinaceous components. Med Mycol (2009) 47:759–65. doi: 10.3109/13693780802524522
80. Chen J, Yi J, Liu L, Yin S, Chen R, Li M, et al. Substrate adaptation of Trichophyton rubrum secreted endoproteases. Microb Pathog (2010) 48:57–61. doi: 10.1016/j.micpath.2009.12.001
81. Bitencourt TA, Macedo C, Franco ME, Assis AF, Komoto TT, Stehling EG, et al. Transcription profile of Trichophyton rubrum conidia grown on keratin reveals the induction of an adhesin-like protein gene with a tandem repeat pattern. BMC Genomics (2016) 17:249. doi: 10.1186/s12864-016-2567-8
82. Baumbach CM, Michler JK, Nenoff P, Uhrlass S, Schrodl W. Visualising virulence factors: Trichophyton benhamiaes subtilisins demonstrated in a Guinea pig skin ex vivo model. Mycoses (2020) 63:970–8. doi: 10.1111/myc.13136
83. Monod M, Lechenne B, Jousson O, Grand D, Zaugg C, Stocklin R, et al. Aminopeptidases and dipeptidyl-peptidases secreted by the dermatophyte Trichophyton rubrum. Microbiol (Reading) (2005) 151:145–55. doi: 10.1099/mic.0.27484-0
84. Giddey K, Monod M, Barblan J, Potts A, Waridel P, Zaugg C, et al. Comprehensive analysis of proteins secreted by Trichophyton rubrum and Trichophyton violaceum under in vitro conditions. J Proteome Res (2007) 6:3081–92. doi: 10.1021/pr070153m
85. Staib P, Zaugg C, Mignon B, Weber J, Grumbt M, Pradervand S, et al. Differential gene expression in the pathogenic dermatophyte Arthroderma benhamiae in vitro versus during infection. Microbiol (Reading) (2010) 156:884–95. doi: 10.1099/mic.0.033464-0
86. Tran VD, De Coi N, Feuermann M, Schmid-Siegert E, Bagut ET, Mignon B, et al. RNA sequencing-based genome reannotation of the dermatophyte Arthroderma benhamiae and characterization of its secretome and whole gene expression profile during infection. mSystems (2016) 1:e00036–16. doi: 10.1128/mSystems.00036-16
87. Zhang FR, Zhang Y, Zhang ZY, Yang GL, Jing LJ, Bai YG. Analysis of the differentially expressed genes in Microsporum canis in inducing smooth skin and scalp tissue conditions. Clin Exp Dermatol (2011) 36:896–902. doi: 10.1111/j.1365-2230.2011.04157.x
88. Zhang F, Tan C, Xu Y, Yang G. FSH1 regulates the phenotype and pathogenicity of the pathogenic dermatophyte Microsporum canis. Int J Mol Med (2019) 44:2047–56. doi: 10.3892/ijmm.2019.4355
89. Lange L, Huang Y, Busk PK. Microbial decomposition of keratin in nature-a new hypothesis of industrial relevance. Appl Microbiol Biotechnol (2016) 100:2083–96. doi: 10.1007/s00253-015-7262-1
90. Peres NT, Sanches PR, Falcão JP, Silveira HC, Paião FG, Maranhão FC, et al. Transcriptional profiling reveals the expression of novel genes in response to various stimuli in the human dermatophyte Trichophyton rubrum. BMC Microbiol (2010) 10:39. doi: 10.1186/1471-2180-10-39
91. Silveira HC, Gras DE, Cazzaniga RA, Sanches PR, Rossi A, Martinez-Rossi NM. Transcriptional profiling reveals genes in the human pathogen Trichophyton rubrum that are expressed in response to pH signaling. Microb Pathog (2010) 48:91–6. doi: 10.1016/j.micpath.2009.10.006
92. Peres NT, Silva LG, Santos Rda S, Jacob TR, Persinoti GF, Rocha LB, et al. In vitro and ex vivo infection models help assess the molecular aspects of the interaction of Trichophyton rubrum with the host milieu. Med Mycol (2016) 54:420–7. doi: 10.1093/mmy/myv113
93. Ferreira-Nozawa MS, Silveira HC, Ono CJ, Fachin AL, Rossi A, Martinez-Rossi NM. The pH signaling transcription factor PacC mediates the growth of Trichophyton rubrum on human nail in vitro. Med Mycol (2006) 44:641–5. doi: 10.1080/13693780600876553
94. Eymann C, Wachlin G, Albrecht D, Tiede S, Krummrei U, Jünger M, et al. Exoproteome analysis of human pathogenic dermatophyte species and identification of immunoreactive proteins. Proteomics Clin Appl (2018) 12:e1800007. doi: 10.1002/prca.201800007
95. Méhul B, de Coi N, Grundt P, Genette A, Voegel JJ, Monod M. Detection of Trichophyton rubrum and Trichophyton interdigitale in onychomycosis using monoclonal antibodies against Sub6 (Tri r 2). Mycoses (2019) 62:32–40. doi: 10.1111/myc.12843
96. Martins MP, Silva LG, Rossi A, Sanches PR, Souza LDR, Martinez-Rossi NM. Global analysis of cell wall genes revealed putative virulence factors in the dermatophyte trichophyton rubrum. Front Microbiol (2019) 10:2168. doi: 10.3389/fmicb.2019.02168
97. Martinez DA, Oliver BG, Graser Y, Goldberg JM, Li W, Martinez-Rossi NM, et al. Comparative genome analysis of Trichophyton rubrum and related dermatophytes reveals candidate genes involved in infection. mBio (2012) 3:e00259–12. doi: 10.1128/mBio.00259-12
98. Heddergott C, Bruns S, Nietzsche S, Leonhardt I, Kurzai O, Kniemeyer O, et al. The Arthroderma benhamiae hydrophobin HypA mediates hydrophobicity and influences recognition by human immune effector cells. Eukaryot Cell (2012) 11:673–82. doi: 10.1128/ec.00037-12
99. Martinez-Rossi NM, Jacob TR, Sanches PR, Peres NT, Lang EA, Martins MP, et al. Heat shock proteins in dermatophytes: current advances and perspectives. Curr Genomics (2016) 17:99–111. doi: 10.2174/1389202917666151116212437
100. Kar B, Patel P, Free SJ. Trichophyton rubrum LysM proteins bind to fungal cell wall chitin and to the N-linked oligosaccharides present on human skin glycoproteins. PloS One (2019) 14:e0215034. doi: 10.1371/journal.pone.0215034
101. Dai P, Lv Y, Gao Y, Gong X, Zhang Y, Zhang X. ZafA gene is important for Trichophyton mentagrophytes growth and pathogenicity. Int J Mol Sci (2019) 20:848. doi: 10.3390/ijms20040848
102. Dai P, Lv Y, Gong X, Han J, Gao P, Xu H, et al. RNA-seq analysis of the effect of zinc deficiency on microsporum canis, zafA gene is important for growth and pathogenicity. Front Cell Infect Microbiol (2021) 11:727665. doi: 10.3389/fcimb.2021.727665
103. Fisher MC, Alastruey-Izquierdo A, Berman J, Bicanic T, Bignell EM, Bowyer P, et al. Tackling the emerging threat of antifungal resistance to human health. Nat Rev Microbiol (2022) 20:557–71. doi: 10.1038/s41579-022-00720-1
104. Logan A, Wolfe A, Williamson JC. Antifungal resistance and the role of new therapeutic agents. Curr Infect Dis Rep (2022) 24:105–16. doi: 10.1007/s11908-022-00782-5
105. Taghipour S, Shamsizadeh F, Pchelin IM, Rezaei-Matehhkolaei A, Zarei Mahmoudabadi A, Valadan R, et al. Emergence of terbinafine resistant trichophyton mentagrophytes in Iran, harboring mutations in the squalene epoxidase (SQLE) gene. Infect Drug Resist (2020) 13:845–50. doi: 10.2147/idr.S246025
106. Ebert A, Monod M, Salamin K, Burmester A, Uhrlaß S, Wiegand C, et al. Alarming India-wide phenomenon of antifungal resistance in dermatophytes: A multicentre study. Mycoses (2020) 63:717–28. doi: 10.1111/myc.13091
107. Brasch J, Gräser Y, Beck-Jendroscheck V, Voss K, Torz K, Walther G, et al. "Indian" strains of Trichophyton mentagrophytes with reduced itraconazole susceptibility in Germany. J Dtsch Dermatol Ges (2021) 19:1723–7. doi: 10.1111/ddg.14626
108. Robbins N, Caplan T, Cowen LE. Molecular evolution of antifungal drug resistance. Annu Rev Microbiol (2017) 71:753–75. doi: 10.1146/annurev-micro-030117-020345
109. Edlind TD, Katiyar SK. Mutational analysis of flucytosine resistance in Candida glabrata. Antimicrob Agents Chemother (2010) 54:4733–8. doi: 10.1128/aac.00605-10
110. Burmester A, Hipler UC, Uhrlaß S, Nenoff P, Singal A, Verma SB, et al. Indian Trichophyton mentagrophytes squalene epoxidase erg1 double mutants show high proportion of combined fluconazole and terbinafine resistance. Mycoses (2020). doi: 10.1111/myc.13150
111. Yamada T, Yaguchi T, Maeda M, Alshahni MM, Salamin K, Guenova E, et al. Gene amplification of CYP51B: a new mechanism of resistance to azole compounds in trichophyton indotineae. Antimicrob Agents Chemother (2022) 66:e0005922. doi: 10.1128/aac.00059-22
112. Salazar F, Brown GD. Antifungal innate immunity: a perspective from the last 10 years. J Innate Immun (2018) 10:373–97. doi: 10.1159/000488539
113. Lionakis MS, Levitz SM. Host control of fungal infections: lessons from basic studies and human cohorts. Annu Rev Immunol (2018) 36:157–91. doi: 10.1146/annurev-immunol-042617-053318
114. Goyal S, Castrillón-Betancur JC, Klaile E, Slevogt H. The interaction of human pathogenic fungi with C-type lectin receptors. Front Immunol (2018) 9:1261. doi: 10.3389/fimmu.2018.01261
115. Kasper L, König A, Koenig PA, Gresnigt MS, Westman J, Drummond RA, et al. The fungal peptide toxin Candidalysin activates the NLRP3 inflammasome and causes cytolysis in mononuclear phagocytes. Nat Commun (2018) 9:4260. doi: 10.1038/s41467-018-06607-1
116. Brown GD. Dectin-1: a signalling non-TLR pattern-recognition receptor. Nat Rev Immunol (2006) 6:33–43. doi: 10.1038/nri1745
117. van den Berg LM, Zijlstra-Willems EM, Richters CD, Ulrich MM, Geijtenbeek TB. Dectin-1 activation induces proliferation and migration of human keratinocytes enhancing wound re-epithelialization. Cell Immunol (2014) 289:49–54. doi: 10.1016/j.cellimm.2014.03.007
118. Gross O, Gewies A, Finger K, Schäfer M, Sparwasser T, Peschel C, et al. Card9 controls a non-TLR signalling pathway for innate anti-fungal immunity. Nature (2006) 442:651–6. doi: 10.1038/nature04926
119. Drummond RA, Swamydas M, Oikonomou V, Zhai B, Dambuza IM, Schaefer BC, et al. CARD9(+) microglia promote antifungal immunity via IL-1beta- and CXCL1-mediated neutrophil recruitment. Nat Immunol (2019) 20:559–70. doi: 10.1038/s41590-019-0377-2
120. Ostrop J, Lang R. Contact, collaboration, and conflict: signal integration of Syk-coupled C-type lectin receptors. J Immunol (2017) 198:1403–14. doi: 10.4049/jimmunol.1601665
121. Wevers BA, Kaptein TM, Zijlstra-Willems EM, Theelen B, Boekhout T, Geijtenbeek TB, et al. Fungal engagement of the C-type lectin mincle suppresses dectin-1-induced antifungal immunity. Cell Host Microbe (2014) 15:494–505. doi: 10.1016/j.chom.2014.03.008
122. Patin EC, Thompson A, Orr SJ. Pattern recognition receptors in fungal immunity. Semin Cell Dev Biol (2019) 89:24–33. doi: 10.1016/j.semcdb.2018.03.003
123. Huang XZ, Liang PP, Ma H, Yi JL, Yin SC, Chen ZR, et al. Effect of culture supernatant derived from trichophyton rubrum grown in the nail medium on the innate immunity-related molecules of haCaT. Chin Med J (Engl) (2015) 128:3094–100. doi: 10.4103/0366-6999.169106
124. Yoshikawa FS, Yabe R, Iwakura Y, de Almeida SR, Saijo S. Dectin-1 and Dectin-2 promote control of the fungal pathogen Trichophyton rubrum independently of IL-17 and adaptive immunity in experimental deep dermatophytosis. Innate Immun (2016) 22:316–24. doi: 10.1177/1753425916645392
125. Nakamura T, Nishibu A, Yoshida N, Yasoshima M, Anzawa K, Watanabe Y, et al. Glycyrrhetinic acid inhibits contact hypersensitivity induced by trichophytin via dectin-1. Exp Dermatol (2016) 25:299–304. doi: 10.1111/exd.12931
126. Nakamura T, Nishibu A, Yasoshima M, Tanoue C, Yoshida N, Hatta J, et al. Analysis of Trichophyton antigen-induced contact hypersensitivity in mouse. J Dermatol Sci (2012) 66:144–53. doi: 10.1016/j.jdermsci.2012.02.008
127. Rajaram MVS, Arnett E, Azad AK, Guirado E, Ni B, Gerberick AD, et al. M. tuberculosis-initiated human mannose receptor signaling regulates macrophage recognition and vesicle trafficking by FcRgamma-chain, Grb2, and SHP-1. Cell Rep (2017) 21:126–40. doi: 10.1016/j.celrep.2017.09.034
128. Santiago K, Bomfim GF, Criado PR, Almeida SR. Monocyte-derived dendritic cells from patients with dermatophytosis restrict the growth of Trichophyton rubrum and induce CD4-T cell activation. PloS One (2014) 9:e110879. doi: 10.1371/journal.pone.0110879
129. Hesse-Macabata J, Morgner B, Elsner P, Hipler UC, Wiegand C. Tryptanthrin promotes keratinocyte and fibroblast responses in vitro after infection with Trichophyton benhamiae DSM6916. Sci Rep (2020) 10:1863. doi: 10.1038/s41598-020-58773-2
130. Cambier LC, Heinen MP, Bagut ET, Antoine NA, Mignon BR. Overexpression of TLR-2 and TLR-4 mRNA in feline polymorphonuclear neutrophils exposed to Microsporum canis. Vet Dermatol (2016) 27:78–81e22. doi: 10.1111/vde.12295
131. Brasch J, Mörig A, Neumann B, Proksch E. Expression of antimicrobial peptides and toll-like receptors is increased in tinea and pityriasis versicolor. Mycoses (2014) 57:147–52. doi: 10.1111/myc.12118
132. Oliveira CB, Vasconcellos C, Sakai-Valente NY, Sotto MN, Luiz FG, Junior WB, et al. Toll-like receptors (TLR) 2 and 4 expression of keratinocytes from patients with localized and disseminated dermatophytosis. Rev Inst Med Trop Sao Paulo (2015) 57:57–61. doi: 10.1590/S0036-46652015000100008
133. Celestrino GA, Reis APC, Criado PR, Benard G, Sousa MGT. Trichophyton rubrum elicits phagocytic and pro-inflammatory responses in human monocytes through toll-like receptor 2. Front Microbiol (2019) 10:2589. doi: 10.3389/fmicb.2019.02589
134. Almeida DF, Fraga-Silva TF, Santos AR, Finato AC, Marchetti CM, Golim MA, et al. TLR2(-/-) mice display increased clearance of dermatophyte trichophyton mentagrophytes in the setting of hyperglycemia. Front Cell Infect Microbiol (2017) 7:8. doi: 10.3389/fcimb.2017.00008
135. Mao L, Zhang L, Li H, Chen W, Wang H, Wu S, et al. Pathogenic fungus Microsporum canis activates the NLRP3 inflammasome. Infect Immun (2014) 82:882–92. doi: 10.1128/iai.01097-13
136. Yoshikawa FS, Ferreira LG, de Almeida SR. IL-1 signaling inhibits Trichophyton rubrum conidia development and modulates the IL-17 response in vivo. Virulence (2015) 6:449–57. doi: 10.1080/21505594.2015.1020274
137. Li H, Wu S, Mao L, Lei G, Zhang L, Lu A, et al. Human pathogenic fungus Trichophyton schoenleinii activates the NLRP3 inflammasome. Protein Cell (2013) 4:529–38. doi: 10.1007/s13238-013-2127-9
138. Cambier L, Weatherspoon A, Defaweux V, Bagut ET, Heinen MP, Antoine N, et al. Assessment of the cutaneous immune response during Arthroderma benhamiae and A. vanbreuseghemii infection using an experimental mouse model. Br J Dermatol (2014) 170:625–33. doi: 10.1111/bjd.12673
139. Urb M, Sheppard DC. The role of mast cells in the defence against pathogens. PloS Pathog (2012) 8:e1002619. doi: 10.1371/journal.ppat.1002619
140. Elieh Ali Komi D, Kuebler WM. Significance of mast cell formed extracellular traps in microbial defense. Clin Rev Allergy Immunol (2022) 62:160–79. doi: 10.1007/s12016-021-08861-6
141. Żelechowska P, Pastwińska J, Brzezińska-Błaszczyk E, Agier J. Do mast cells contribute to the antifungal host defense? Cells (2021) 10. doi: 10.3390/cells10102510
142. Mukai K, Tsai M, Saito H, Galli SJ. Mast cells as sources of cytokines, chemokines, and growth factors. Immunol Rev (2018) 282:121–50. doi: 10.1111/imr.12634
143. Bartemes KR, Kita H. Innate and adaptive immune responses to fungi in the airway. J Allergy Clin Immunol (2018) 142:353–63. doi: 10.1016/j.jaci.2018.06.015
144. Griffiths JS, Camilli G, Kotowicz NK, Ho J, Richardson JP, Naglik JR. Role for IL-1 family cytokines in fungal infections. Front Microbiol (2021) 12:633047. doi: 10.3389/fmicb.2021.633047
145. Mills KHG. IL-17 and IL-17-producing cells in protection versus pathology. Nat Rev Immunol (2023) 23:38–54. doi: 10.1038/s41577-022-00746-9
146. Sardana K, Gupta A, Mathachan SR. Immunopathogenesis of dermatophytoses and factors leading to recalcitrant infections. Indian Dermatol Online J (2021) 12:389–99. doi: 10.4103/idoj.IDOJ_503_20
147. McGinley AM, Sutton CE, Edwards SC, Leane CM, DeCourcey J, Teijeiro A, et al. Interleukin-17A serves a priming role in autoimmunity by recruiting IL-1beta-producing myeloid cells that promote pathogenic T cells. Immunity (2020) 52:342–56.e6. doi: 10.1016/j.immuni.2020.01.002
148. Burstein VL, Guasconi L, Beccacece I, Theumer MG, Mena C, Prinz I, et al. IL-17-mediated immunity controls skin infection and T helper 1 response during experimental Microsporum canis dermatophytosis. J Invest Dermatol (2018) 138:1744–53. doi: 10.1016/j.jid.2018.02.042
149. Kashem SW, Kaplan DH. Skin immunity to Candida albicans. Trends Immunol (2016) 37:440–50. doi: 10.1016/j.it.2016.04.007
150. Chong WP, Mattapallil MJ, Raychaudhuri K, Bing SJ, Wu S, Zhong Y, et al. The cytokine IL-17A limits Th17 pathogenicity via a negative feedback loop driven by autocrine induction of IL-24. Immunity (2020) 53:384–97.e5. doi: 10.1016/j.immuni.2020.06.022
151. Nakae S, Suto H, Berry GJ, Galli SJ. Mast cell-derived TNF can promote Th17 cell-dependent neutrophil recruitment in ovalbumin-challenged OTII mice. Blood (2007) 109:3640–8. doi: 10.1182/blood-2006-09-046128
152. Cho KA, Suh JW, Sohn JH, Park JW, Lee H, Kang JL, et al. IL-33 induces Th17-mediated airway inflammation via mast cells in ovalbumin-challenged mice. Am J Physiol Lung Cell Mol Physiol (2012) 302:L429–40. doi: 10.1152/ajplung.00252.2011
153. Wang A, Wang Z, Cao Y, Cheng S, Chen H, Bunjhoo H, et al. CCL2/CCR2-dependent recruitment of Th17 cells but not Tc17 cells to the lung in a murine asthma model. Int Arch Allergy Immunol (2015) 166:52–62. doi: 10.1159/000371764
154. Mommert S, Gschwandtner M, Koether B, Gutzmer R, Werfel T. Human memory Th17 cells express a functional histamine H4 receptor. Am J Pathol (2012) 180:177–85. doi: 10.1016/j.ajpath.2011.09.028
155. Minns D, Smith KJ, Alessandrini V, Hardisty G, Melrose L, Jackson-Jones L, et al. The neutrophil antimicrobial peptide cathelicidin promotes Th17 differentiation. Nat Commun (2021) 12:1285. doi: 10.1038/s41467-021-21533-5
156. Jiang L, Fang M, Tao R, Yong X, Wu T. Recombinant human interleukin 17A enhances the anti-Candida effect of human oral mucosal epithelial cells by inhibiting Candida albicans growth and inducing antimicrobial peptides secretion. J Oral Pathol Med (2020) 49:320–7. doi: 10.1111/jop.12889
157. Rabeony H, Petit-Paris I, Garnier J, Barrault C, Pedretti N, Guilloteau K, et al. Inhibition of keratinocyte differentiation by the synergistic effect of IL-17A, IL-22, IL-1α, TNFα and oncostatin M. PloS One (2014) 9:e101937. doi: 10.1371/journal.pone.0101937
158. Pham CVA, Rademacher F, Hinrichs H, Beck-Jendroschek V, Harder M, Brasch J, et al. Expression of epidermal antimicrobial peptides is increased in tinea pedis. Mycoses (2021) 64:763–70. doi: 10.1111/myc.13279
159. Jensen JM, Pfeiffer S, Akaki T, Schröder JM, Kleine M, Neumann C, et al. Barrier function, epidermal differentiation, and human beta-defensin 2 expression in tinea corporis. J Invest Dermatol (2007) 127:1720–7. doi: 10.1038/sj.jid.5700788
160. Dixon BR, Radin JN, Piazuelo MB, Contreras DC, Algood HM. IL-17a and IL-22 Induce Expression of Antimicrobials in Gastrointestinal Epithelial Cells and May Contribute to Epithelial Cell Defense against Helicobacter pylori. PloS One (2016) 11:e0148514. doi: 10.1371/journal.pone.0148514
161. Guilloteau K, Paris I, Pedretti N, Boniface K, Juchaux F, Huguier V, et al. Skin inflammation induced by the synergistic action of IL-17A, IL-22, oncostatin M, IL-1{alpha}, and TNF-{alpha} Recapitulates some features of psoriasis. J Immunol (2010) 184:5263–70. doi: 10.4049/jimmunol.0902464
162. Hurabielle C, Link VM, Bouladoux N, Han SJ, Merrill ED, Lightfoot YL, et al. Immunity to commensal skin fungi promotes psoriasiform skin inflammation. Proc Natl Acad Sci U.S.A. (2020) 117:16465–74. doi: 10.1073/pnas.2003022117
163. Simpson JK, Frobel P, Seneviratne SL, Brown M, Lowe DM, Grimbacher B, et al. Invasive dermatophyte infection with Trichophyton interdigitale is associated with prurigo-induced pseudoperforation and a signal transducer and activator of transcription 3 mutation. Br J Dermatol (2018) 179:750–4. doi: 10.1111/bjd.15781
164. Sawada Y, Nakamura M, Kabashima-Kubo R, Shimauchi T, Kobayashi M, Tokura Y. Defective epidermal innate immunity and resultant superficial dermatophytosis in adult T-cell leukemia/lymphoma. Clin Cancer Res (2012) 18:3772–9. doi: 10.1158/1078-0432.Ccr-12-0292
165. Toubiana J, Okada S, Hiller J, Oleastro M, Lagos Gomez M, Aldave Becerra JC, et al. Heterozygous STAT1 gain-of-function mutations underlie an unexpectedly broad clinical phenotype. Blood (2016) 127:3154–64. doi: 10.1182/blood-2015-11-679902
166. Emelianov V, Feldmeyer L, Yawalkar N, Heidemeyer K. Tinesmall a, cyrillic corporis with Trichophyton Rubrum mimicking a flare-up of psoriasis undertreatment with IL17-inhibitor ixekizumab. Case Rep Dermatol (2021) 13:347–51. doi: 10.1159/000515702
167. Neema S, Singh S, Pathak N, Khan MA. Widespread superficial dermatophytosis in patient on secukinumab for treatment of chronic plaque psoriasis. Indian Dermatol Online J (2019) 10:76–8. doi: 10.4103/idoj.IDOJ_170_18
168. Quach OL, Hsu S. Perianal dermatophytosis during secukinumab therapy for plaque psoriasis. JAMA Dermatol (2016) 152:486–7. doi: 10.1001/jamadermatol.2015.4992
169. Baltazar Lde M, Santos PC, Paula TP, Rachid MA, Cisalpino PS, Souza DG, et al. IFN-gamma impairs Trichophyton rubrum proliferation in a murine model of dermatophytosis through the production of IL-1beta and reactive oxygen species. Med Mycol (2014) 52:293–302. doi: 10.1093/mmy/myt011
170. Okada S, Asano T, Moriya K, Boisson-Dupuis S, Kobayashi M, Casanova JL, et al. Human STAT1 gain-of-function heterozygous mutations: chronic mucocutaneous candidiasis and Type I interferonopathy. J Clin Immunol (2020) 40:1065–81. doi: 10.1007/s10875-020-00847-x
171. Depner M, Fuchs S, Raabe J, Frede N, Glocker C, Doffinger R, et al. The extended clinical phenotype of 26 patients with chronic mucocutaneous candidiasis due to gain-of-function mutations in STAT1. J Clin Immunol (2016) 36:73–84. doi: 10.1007/s10875-015-0214-9
172. Liu L, Okada S, Kong XF, Kreins AY, Cypowyj S, Abhyankar A, et al. Gain-of-function human STAT1 mutations impair IL-17 immunity and underlie chronic mucocutaneous candidiasis. J Exp Med (2011) 208:1635–48. doi: 10.1084/jem.20110958
173. Stark MA, Huo Y, Burcin TL, Morris MA, Olson TS, Ley K. Phagocytosis of apoptotic neutrophils regulates granulopoiesis via IL-23 and IL-17. Immunity (2005) 22:285–94. doi: 10.1016/j.immuni.2005.01.011
174. Higgins E, Al Shehri T, McAleer MA, Conlon N, Feighery C, Lilic D, et al. Use of ruxolitinib to successfully treat chronic mucocutaneous candidiasis caused by gain-of-function signal transducer and activator of transcription 1 (STAT1) mutation. J Allergy Clin Immunol (2015) 135:551–3. doi: 10.1016/j.jaci.2014.12.1867
175. Sparber F, LeibundGut-Landmann S. IL-17 takes center stage in dermatophytosis. J Invest Dermatol (2018) 138:1691–3. doi: 10.1016/j.jid.2018.03.1518
176. Heinen MP, Cambier L, Antoine N, Gabriel A, Gillet L, Bureau F, et al. Th1 and Th17 immune responses act complementarily to optimally control superficial dermatophytosis. J Invest Dermatol (2019) 139:626–37. doi: 10.1016/j.jid.2018.07.040
177. Rai G, Das S, Ansari MA, Singh PK, Pandhi D, Tigga RA, et al. The interplay among Th17 and T regulatory cells in the immune dysregulation of chronic dermatophytic infection. Microb Pathog (2020) 139:103921. doi: 10.1016/j.micpath.2019.103921
178. Kaya TI, Eskandari G, Guvenc U, Gunes G, Tursen U, Cimen MYB, et al. CD4+CD25+ Treg cells in patients with toenail onychomycosis. Arch Dermatol Res (2009) 301:725–9. doi: 10.1007/s00403-009-0941-y
179. Spesso MF, Nuncira CT, Burstein VL, Masih DT, Dib MD, Chiapello LS. Microsatellite-primed PCR and random primer amplification polymorphic DNA for the identification and epidemiology of dermatophytes. Eur J Clin Microbiol Infect Dis (2013) 32:1009–15. doi: 10.1007/s10096-013-1839-3
180. Corvilain E, Casanova JL, Puel A. Inherited CARD9 deficiency: invasive disease caused by ascomycete fungi in previously healthy children and adults. J Clin Immunol (2018) 38:656–93. doi: 10.1007/s10875-018-0539-2
181. Lanternier F, Pathan S, Vincent QB, Liu L, Cypowyj S, Prando C, et al. Deep dermatophytosis and inherited CARD9 deficiency. N Engl J Med (2013) 369:1704–14. doi: 10.1056/NEJMoa1208487
182. Jachiet M, Lanternier F, Rybojad M, Bagot M, Ibrahim L, Casanova JL, et al. Posaconazole treatment of extensive skin and nail dermatophytosis due to autosomal recessive deficiency of CARD9. JAMA Dermatol (2015) 151:192–4. doi: 10.1001/jamadermatol.2014.2154
183. Stambouli OB, Amrani N, Stambouli KB, Bouali F. Dermatophytic disease with deficit in CARD9: a new case with a brain impairment. J Mycol Med (2017) 27:250–3. doi: 10.1016/j.mycmed.2017.01.001
184. Grumach AS, de Queiroz-Telles F, Migaud M, Lanternier F, Filho NR, Palma SM, et al. A homozygous CARD9 mutation in a Brazilian patient with deep dermatophytosis. J Clin Immunol (2015) 35:486–90. doi: 10.1007/s10875-015-0170-4
185. de Medeiros AKA, Lodewick E, Bogaert DJ, Haerynck F, Van Daele S, Lambrecht B, et al. Chronic and invasive fungal infections in a family with CARD9 deficiency. J Clin Immunol (2016) 36:204–9. doi: 10.1007/s10875-016-0255-8
186. Nazarian RM, Lilly E, Gavino C, Hamilos DL, Felsenstein D, Vinh DC, et al. Novel CARD9 mutation in a patient with chronic invasive dermatophyte infection (tinea profunda). J Cutan Pathol (2020) 47:166–70. doi: 10.1111/cup.13574
187. Vaezi A, Fakhim H, Abtahian Z, Khodavaisy S, Geramishoar M, Alizadeh A, et al. Frequency and geographic distribution of CARD9 mutations in patients with severe fungal infections. Front Microbiol (2018) 9:2434. doi: 10.3389/fmicb.2018.02434
188. Zhang Y, Mijiti J, Huang C, Song Y, Wan Z, Li R, et al. Deep dermatophytosis caused by Microsporum ferrugineum in a patient with CARD9 mutations. Br J Dermatol (2019) 181:1093–5. doi: 10.1111/bjd.18146
189. Al-Shaikhly T, Ochs HD. Hyper IgE syndromes: clinical and molecular characteristics. Immunol Cell Biol (2019) 97:368–79. doi: 10.1111/imcb.12209
Keywords: dermatophyte, protease, innate immune response, pattern recognition receptors, caspase-associated recruitment domain 9, type 1 immune response, type 17 immune response
Citation: Deng R, Wang X and Li R (2023) Dermatophyte infection: from fungal pathogenicity to host immune responses. Front. Immunol. 14:1285887. doi: 10.3389/fimmu.2023.1285887
Received: 30 August 2023; Accepted: 23 October 2023;
Published: 02 November 2023.
Edited by:
Youcef Shahali, Centre Hospitalier Universitaire de Besançon, FranceReviewed by:
Francois Niyonsaba, Juntendo University, JapanTarun Narang, Postgraduate Institute of Medical Education and Research, India
Copyright © 2023 Deng, Wang and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ruoyu Li, ruoyuli_myco@163.com
 Ruixin Deng
Ruixin Deng