- 1Department of Translational Medical Sciences, University of Naples Federico II, Naples, Italy
- 2World Allergy Organization (WAO), Center of Excellence (CoE), Naples, Italy
- 3Center for Basic and Clinical Immunology Research (CISI), University of Naples Federico II, Naples, Italy
- 4Institute of Experimental Endocrinology and Oncology “G. Salvatore”, National Research Council (CNR), Naples, Italy
- 5Division of Allergy and Clinical Immunology, The Johns Hopkins University School of Medicine, Baltimore, MD, United States
Basophils bind IgE via FcεRI-αβγ2, which they uniquely share only with mast cells. In doing so, they can rapidly release mediators that are hallmark of allergic disease. This fundamental similarity, along with some morphological features shared by the two cell types, has long brought into question the biological significance that basophils mediate beyond that of mast cells. Unlike mast cells, which mature and reside in tissues, basophils are released into circulation from the bone marrow (constituting 1% of leukocytes), only to infiltrate tissues under specific inflammatory conditions. Evidence is emerging that basophils mediate non-redundant roles in allergic disease and, unsuspectingly, are implicated in a variety of other pathologies [e.g., myocardial infarction, autoimmunity, chronic obstructive pulmonary disease, fibrosis, cancer, etc.]. Recent findings strengthen the notion that these cells mediate protection from parasitic infections, whereas related studies implicate basophils promoting wound healing. Central to these functions is the substantial evidence that human and mouse basophils are increasingly implicated as important sources of IL-4 and IL-13. Nonetheless, much remains unclear regarding the role of basophils in pathology vs. homeostasis. In this review, we discuss the dichotomous (protective and/or harmful) roles of basophils in a wide spectrum of non-allergic disorders.
1 Basic concepts of basophils
Basophils are rare blood cells, accounting for 1% or less of the circulating leukocytes-a feature evident both in humans and mice. Basophils share several morphological and functional characteristics with tissue-resident mast cells. Most recognized are the cytoplasmic granules that each cell possesses and that stain so predominantly with basic stains. Phenotypically, both cell types s uniquely express the αβγ2 structure of the high-affinity receptor (FcεRI) for IgE, which enables both cells to rapidly release pre-formed histamine and newly generated cysteinyl leukotriene C4 (LTC4), upon encountering relevant allergen (1, 2). Accordingly, basophils were initially viewed, incorrectly, as blood-circulating mast cells, which prompted the notion of using them as surrogates to study tissue mast cells, which proved far more difficult to obtain (2). However, it is now widely accepted that basophils and mast cells profoundly differ in several fundamental aspects (3). For example, the lifespan of basophils (~days) is much shorter than the months estimated for mast cells (4). Transcriptionally, basophils are more closely related to eosinophils than mast cells (5, 6). These differences (among many more discussed elsewhere (7) suggest that basophils have unique pathophysiological roles different from those of mast cells.
IL-3 is central to the growth, differentiation, priming, and overall activation of both human and mouse basophils (8, 9). It does so by binding, with high-affinity, to the α subunit of its receptor (IL-3Rα/CD123) highly expressed on basophils (10). Many cell types are implicated in producing the IL-3 that impacts basophil development and function, including T cells (11, 12), B cells (13), human eosinophils and neutrophils (14), but also mast cells and even basophils (15, 16). Although the IL-3 receptor is highly expressed on basophils (17–28), mice incapable of producing IL-3 and/or deficient in IL-3Rα/CD123 reportedly develop all blood lineages, including basophils and mast cells (29–31). In this regard, thymic stromal lymphopoietin (TSLP) is also reported to regulate mouse basophil development (32, 33) and activation (9) in vivo and may therefore represent an important early growth factor for these cells. In contrast, numerous studies show that IL-3 is quite sufficient in promoting the in vitro growth of functional human and mouse basophil-like cells from progenitors. TSLP is reported to activate human basophils from asthmatic subjects by promoting histamine release and cytokine secretion, along with inducing cell surface expression of CD203c and IL-3Rα (34). In contrast, several other investigators have since reported that TSLP does not activate human basophils isolated from healthy subjects or allergic patients (9, 10, 35). In light of the latter findings, TSLP may have very different effects on human versus mouse basophils (9). Finally, IL-3 is well known for its capacity to mediate synergistic (or priming) effects when combined with a diverse array of co-stimuli (9, 36–40).
It has been shown in mice that basophils originate from hematopoietic stem cells (HSCs) in the bone marrow (41, 42). So-called granulocyte-macrophage progenitors (GMPs), which develop later than the HSCs giving rise to most of the common myeloid progenitors, are thought to be the relevant basophil progenitors (BaPs (43). Common basophil-mast cell progenitors are also present in the spleen (43, 44). Single-cell transcriptomic analyses have highlighted the differentiation pathways of various cell lineages in mice (45–47). Single-cell culture of mouse bone marrow progenitors generated FcϵRI+ basophils and erythroid cells (48). The erythroid trajectory is close to that of basophils/mast cells, both in mice (49) and humans (50–53). Human CD131+ CMP progenitors in the bone marrow can differentiate into basophil/mast cell/eosinophil and erythroid/megakaryocyte populations (51). Likewise, studies of human bone marrow cells using single-cell transcriptome analysis found the basophil trajectory to be more linked with that of the megakaryocyte and erythroid lineages, rather than those of granulocytes/monocytes (52). It is likely that the differentiation pathways of basophils and mast cells are more closely linked to those of the erythroid/megakaryocyte lineages, rather than to granulocytes/monocytes, both in mice and humans.
Several analytical tools for the study of mouse basophil biology have been developed in recent years. In particular, the use of antibodies capable of depleting basophils in vivo (54, 55) as well as mice that are genetically altered to be deficient of basophils (56–61), which includes reporter mouse models (58, 61), and basophil-specific Cre-expressing mice (58, 62, 63). The results obtained with these different models have demonstrated non-redundant roles of basophils in experimental Th2-type inflammation, comprising certain aspects of various allergic responses (3, 64, 65). Likewise, these models have substantiated the long-held belief that basophils help mediate immunity against parasitic infections (66–69). Some of these analytical tools have been employed to evaluate the role of mouse basophils in myocardial infarction (MI) (70), renal fibrosis (71), cancer (72–75), autoimmune disorders (76, 77), and chronic obstructive pulmonary disease (COPD) (62). Table 1 lists the antibody-mediated and genetic models for analyzing the in vivo contribution of mouse basophils in various pathophysiological conditions.
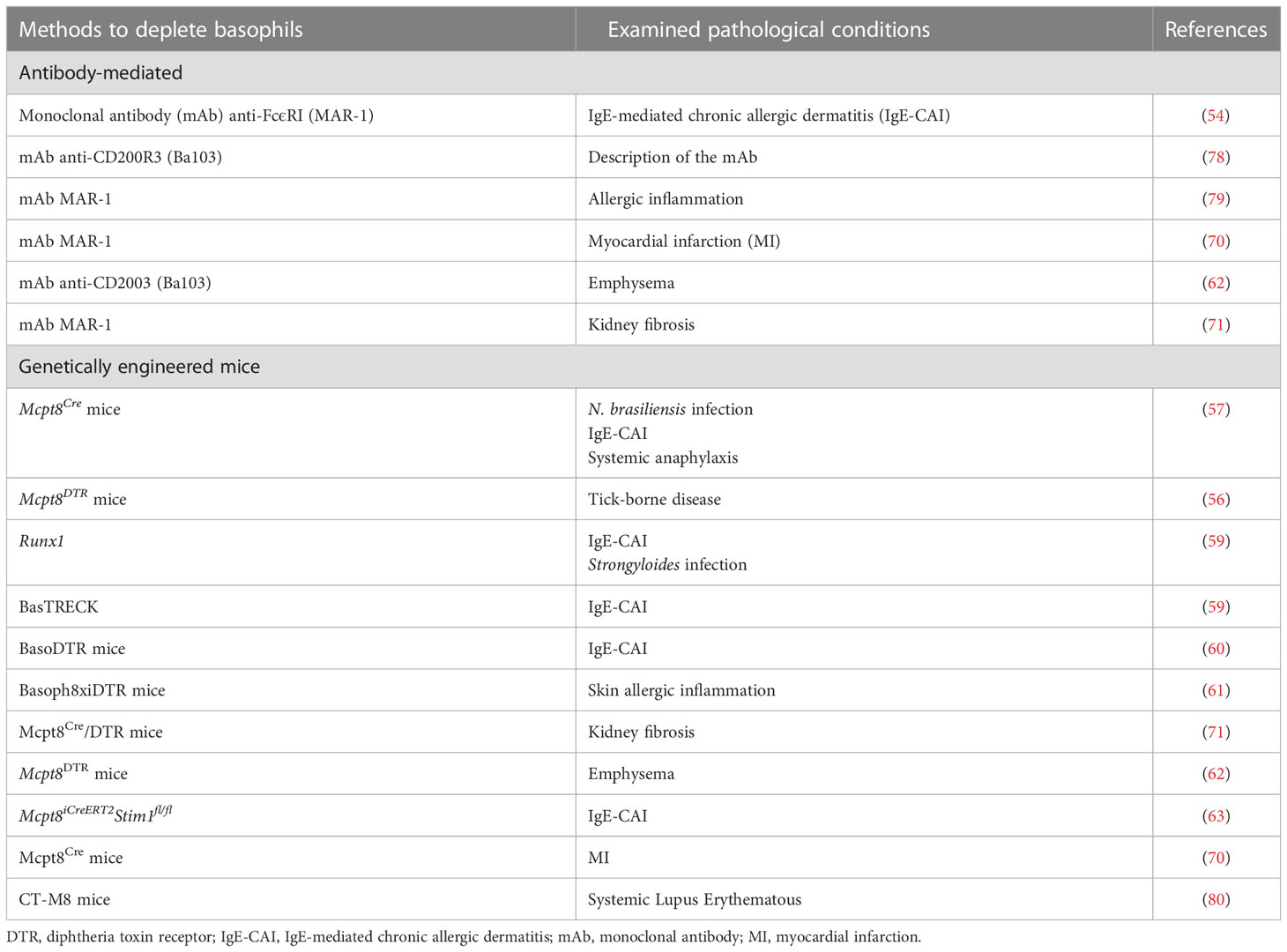
Table 1 Antibody-mediated and genetic depletion models for the in vivo study of basophils in different pathological conditions.
Several outstanding reviews have discussed the roles of mouse and human basophils in allergic disorders (1, 64, 74, 81, 82) and parasitic infections (66–68). Increasing evidences indicate that basophils also play relevant roles in several other types of responses, including autoimmunity (83, 84), myocardial infarction (70), fibrosis (70, 71, 85), cancer (86–88), and COVID-19 (89). In this review, we discuss the recent basophil contribution to the pathogenesis of several non-allergic inflammatory diseases.
2 Basophils in myocardial infarction
Myocardial infarction (MI) occurs when coronary arteries that supply oxygen and nutrients to the heart become obstructed by atherosclerotic arterial walls (90). The consequence is an ischemic injury that mobilizes a repertoire of innate and adaptive immune cells (91, 92). Shortly, after ischemic occurs, resident cardiac mast cells release their preformed mediators (93), resident macrophages and cardiomyocytes produce cytokines and chemokines (94, 95), fibroblasts release growth factors (96) and endothelial cells are activated. These events typically cause an influx of various immune cells, including neutrophils, monocytes, macrophages (92, 97), and mast cells (98, 99).
The inflammatory response following MI deeply affects subsequent cardiac remodeling and fibrosis (100, 101). The composition of immune cell types identified in the infarcted myocardium consists mostly of macrophages, monocytes, neutrophils, DCs, B and T cells, and NK cells (70, 97). Using a mouse model, Sicklinger and coworkers demonstrated that basophils infiltrate infarcted hearts, reaching a peak between days 3 and 7 and reverting to baseline on day 14 (70). The administration of the monoclonal antibody (mAb) anti-FcϵRI (MAR-1) depleted basophils in the heart, peripheral blood, and spleen. In contrast, mast cells and a subset of DCs expressing FcϵRI were not altered following MAR-1 administration. Depletion of basophils reduced left ventricular ejection fraction 4 weeks after MI and increased heart weight compared to control. Moreover, basophil-depleted mice showed reduced scar thickness.
Sicklinger et al. also studied the inflammatory response after MI in Mcpt8-Cre-transgenic (Baso-KO) mice constitutively deficient in basophils (57). In this model, the infarct size did not differ between Baso-KO compared to WT mice. However, 28 days after inducing the MI, the basophil-deficient mice developed cardiac dysfunction and increased heart weight compared to their WT littermates. Finally, Baso-KO mice showed increased scar thinning compared to controls. MI in genetic basophil ablation mice was associated with an altered cellular inflammatory response in infarcted hearts. Four days after MI, there was a change in the composition of monocyte subpopulations in the infarcted myocardium of the basophil-depleted mice, namely a shift from reparative Ly6Clo macrophages toward inflammatory Ly6Chi monocytes. This proinflammatory response could be reversed by the adoptive transfer of basophils into the basophil-deficient mice. The absence of basophils was associated with lower concentrations of cardiac IL-4 and IL-13, two cytokines typically released by mouse (9, 57, 102–105) and human basophils (9, 16, 36–38, 106–108). The authors concluded that the IL-4/IL-13 secreted by basophils infiltrating these lesions is critical in the transition from inflammatory monocytes to reparative macrophages (81, 109) Figure 1 illustrates the proposed mechanisms by which basophils influence the inflammatory response following myocardial infarction.
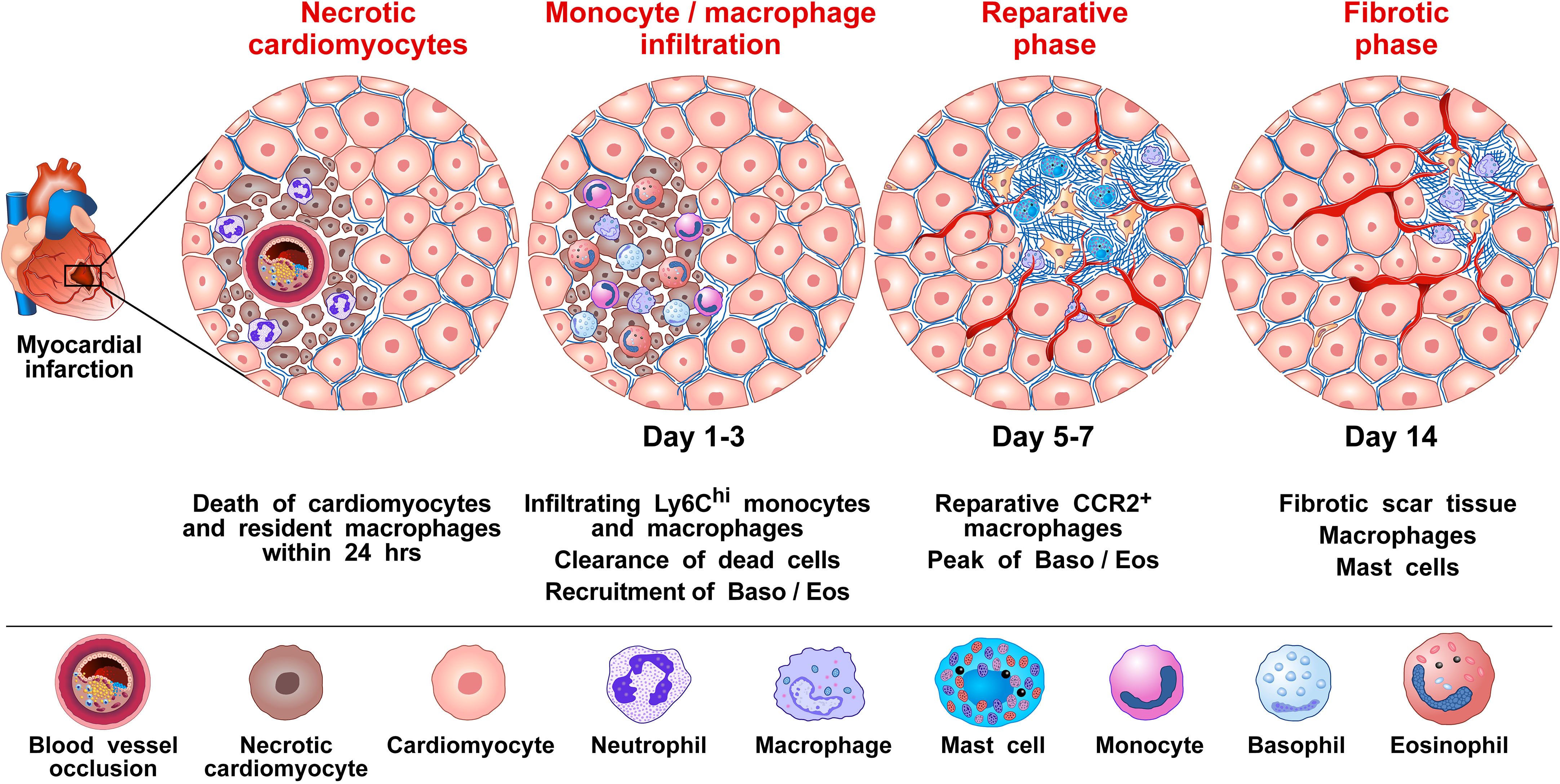
Figure 1 Proposed mechanism by which basophils influence the inflammatory response to promote wound healing and tissue repair following myocardial infarction (MI). MI is caused by the rupture of an atherosclerotic plaque causing the occlusion of a coronary artery, which then results in cardiac tissue damage due to ischemia (90). It has been shown in mice that several immune cells [e.g., monocytes/macrophages, neutrophils, dendritic cells (DCs), B and T cells, and natural killer (NK) cells, basophils and eosinophils infiltrate the heart after experimental MI (70, 97, 110). For basophils, this infiltration into the heart is evident 3 days following MI and peaks 7 days after the MI event (70). Monocytes/macrophages represent the most prevalent immune cells after MI. Cardiac resident macrophages contribute to the initial neutrophil infiltration into the ischemic area (111). Resident macrophages are reduced in murine models 1 day post-infarction (112). Within 1-3 days infiltrating bone marrow- and spleen-derived Ly6Chi monocytes are recruited into the injured cardiac tissue and differentiate to Ly6Clow macrophages facilitating clearance of necrotic cardiomyocytes. At approximately 5-7 days post MI, macrophages adopt a reparative phenotype, contributing to the resolution of inflammation and fibrotic tissue formation (70). By day 3, infiltrating basophils into the injured cardiac tissue release IL-4 and IL-13, which induce phenotypical and functional changes within macrophages expressing anti-inflammatory and tissue repair genes (70). Formation of neovessels in the healing infarct play an important role in repairing the infarcted myocardium (113). Basophils (114), macrophages (115–117), and cardiac mast cells (118, 119), are major sources of angiogenic factors. Collectively, results in mice models of MI indicate that basophils infiltrating infarcted heart promote resolution of cardiac inflammation and scar formation.
The authors also evaluated the cytokines produced in the heart 3 days after the MI event, both in the Baso-KO and WT mice. Among the cytokines commonly reported to be produced by mouse basophils (IL-4, IL-13, IL-6, TNF-α), there was a reduction only of IL-4 in the injured heart tissue of the basophil-deficient mice. Mice-deficient in IL-4/IL-13 showed a higher proportion of inflammatory Ly6Chi monocytes and worsened cardiac function following MI. In contrast, the increased release of IL-4 by basophils following the administration of the glycoprotein IPSE/α-1 (a known stimulus of these cytokines from basophils) resulted in enhanced post-MI healing. The authors concluded that myocardial basophils are activated to produce IL-4 following MI and that this response is critical in healing the damaged myocardium (70). What currently remains unknown, however, is the exact mode of stimulation in the myocardium responsible for inducing basophils to produce IL-4.
These experimental results were supported by observations that human subjects presented with decreased blood basophil numbers within the first week following an MI event, and that this basopenia associated with an increased scar size, as measured by late gadolinium enhancement cardiac MRI after one year of follow-up (70). Importantly, this correlation persisted after the adjustment of possible confounders (e.g., initial infarct size, systemic inflammation, cardiovascular risk factors). The authors suggested that basophils may also influence cardiac remodeling after MI in humans.
These studies, emphasizing the protective role of basophils following MI, might have translational relevance. For example, a growing number of allergic patients (e.g., asthma, atopic dermatitis) are being treated with biologics that block the IL-4/IL-13 axis (e.g., dupilumab, an anti-IL-4Rα mAb) (82, 120). Thus, the possible protective role of basophil-derived IL-4/IL-13 in MI should stimulate further mechanistic studies to investigate possible links between these therapies and whether they might impact myocardial healing following MI.
3 Basophils in kidney fibrosis
Chronic kidney disease (CKD) is a final manifestation of renal fibrosis and its incidence is increasing (121). Various inflammatory stimuli, including chronic infections, tissue injury, autoimmune disorders, chemical insults, and radiation result in kidney fibrosis (117, 122). Chronic low-grade inflammation is a crucial promoter of fibrosis (117, 123), but immune pathways orchestrating kidney fibrosis are largely unknown. Doke and collaborators investigated the interactions between altered renal tubules and basophils in a mouse model of kidney fibrosis by employing single-cell RNA-seq analysis (71). In this model of CKD, mice experienced either a sham operation or underwent unilateral ureter obstruction (UUO) surgery. Injured tubular cells (PTs) expressed several cytokines and chemokines known to induce the recruitment of basophils and other immune cells. PTs also released platelet-derived growth factor B (PDGFB), which upon binding to its receptor (PDGFBR) on fibroblasts induces these cells to release TGF-β. CXCL1, secreted by profibrotic tubules, recruited CXCR2+ basophils. The density of basophils (FcϵRI+CD200R3+CD49b+ cells) was markedly increased in UUO kidneys compared to sham operation. Using antibody-mediated and genetic approaches to delete basophils, the authors explored the role of these cells in this model. In the latter model, injection of diphtheria toxin (DT) into Mcpt8Cre/DTR mice induced depletion of basophils in the kidney and mitigated fibrosis in UUO kidney. Single-cell analysis and in situ hybridization demonstrated overexpression of Il6 by basophils in UUO kidneys, indicating that mouse basophils are a source of this cytokine in UUO kidneys. In the other model, basophil depletion was mediated by MAR-1 administration into WT mice, followed by UUO surgery and kidney examination 7 days later. MAR-1-treated mice showed a reduction of the fibrosis markers induced by the UUO surgery. These results from two complementary models of basophil depletion highlight the importance of these cells in the development of experimental kidney fibrosis.
There is evidence that TH17 cells contribute to renal fibrosis (124). For example, basophils were shown to directly interact with TH17 cells and macrophages (104, 125). Both TH17 cell number and IL-17A expression were increased in UUO, but they were lower in UUO kidneys of basophil-depleted mice. Single-cell RNA-seq analysis indicated a shift toward TH17 cells in fibrosis. Basophil-derived IL-6 contributed to enhanced TH17 cell differentiation from CD4+ T cells in UUO kidney (126). Moreover, the expression of Il17a and Tgfb1 were higher in UUO kidneys and were lower in UUO kidneys of basophil-depleted mice. Mice treated with an anti-IL-6R antibody were partially protected from renal fibrosis.
To evaluate the relevance of the above experimental findings to human kidney fibrosis, Doke and collaborators examined human kidneys, comparing those from healthy controls and CKD subjects using single-cell RNA-seq (71). They found that basophil numbers were increased in the kidney of patients with CKD, compared to healthy controls. Moreover, a correlation between renal fibrosis and basophil density was evident in the kidneys of CKD patients. There was also a positive correlation between IL6 expression and the severity of renal fibrosis, which further showed a negative correlation between IL6 and kidney function. Moreover, renal IL6 correlated with CKD severity. Collectively, the above results reveal several correlations between both basophil density and their function and renal fibrosis. Figure 2 schematically illustrates the contribution of basophil-derived cytokines and TH17 as downstream mediators in kidney fibrosis.
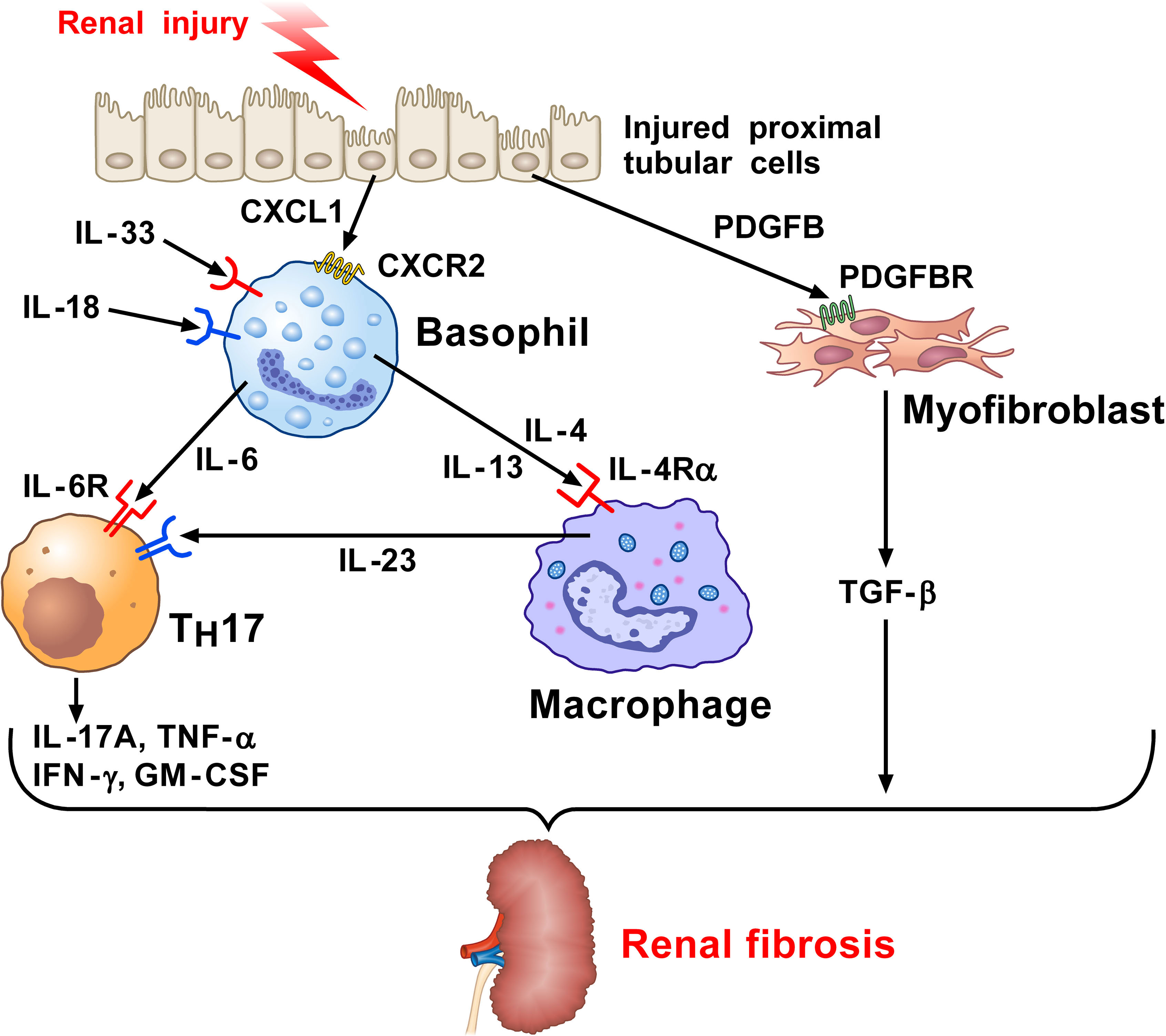
Figure 2 Kidney and wild-type mice subjected to unilateral ureter obstruction (UUO) surgery revealed the presence of neutrophils, monocytes/macrophages, dendritic cells (DCs), and basophils (71). Injured proximal tubular cells (PTs) in UUO kidney express Il34, Cxcl10, and the key profibrotic factor (71), platelet derived growth factor subunit B (PDGFB). PDGFB released by injured tubular activates the PDGFB receptor (PDGFBR) on fibroblasts to release TGF-β. Profibrotic PT cells participate in the recruitment of myeloid and lymphoid cells and the local fibroblast activation. CXCL1 released from PT cells induces the recruitment of basophils through the engagement of CXCR2. Basophils in UUO kidney can be activated by IL-33 and IL-18 released from the stroma to secrete IL-6. This cytokine favors TH17 differentiation from CD4+ T cells in UUO kidneys. IL-17A and TGF-β released from TH17 cells contribute to renal fibrosis. IL-4 and IL-13 released from activated basophils can contribute to macrophage activation (127–130). PDGF released from injured PT cells activates the PDGFR on myofibroblasts causing the release of TGF-β. Macrophages are also a major source of IL-6. Collectively, these findings indicate that basophils and their mediators contribute to kidney fibrosis.
4 Basophils in cancer
There is mounting evidence showing that basophils are an important component within the tumor microenvironment (TME) of several human (72, 88, 131, 132) and mouse experimental cancers (72, 73, 132, 133). Moreover, these studies indicate that basophils may play an active role in the onset and development of both solid and hematologic tumors (74, 86, 134). The results from these studies reveal that basophils can have both pro-tumor and antitumor effects depending on the context and type of tumor.
In particular, immune profiling studies show that basophils constitute a portion, albeit small, of the immune landscape in human non-small cell lung cancer (NSCLC) tumors (131) and in the immune infiltrate seen in the early stage of lung adenocarcinoma (132). Several studies additionally show that mouse and human basophils support the development and expansion of M2-like monocytes/macrophages (127–130), which are often prevalent in the TME favoring tumorigenesis. An in vivo study in mice points to the importance of IL-4/IL-13, promoting carcinogenesis by reducing Th1-like immunity (72). Likewise, basophils are known to secrete vascular endothelial growth factor-A (VEGF-A) (114) and cysteinyl leukotriene C4 (LTC4) (18, 19) with the latter more recently implicated in tumorigenesis and metastasis formation (135). In particular, both tumor growth and metastases were reduced in mice deficient in the cysteinyl leukotriene 2 receptor (CysLT2R). Moreover, administration of a CysLT2R antagonist reduced tumor growth and metastases in WT mice (135).
In exploring the immune cells involved in human pancreatic cancer (PC), IL4-expressing basophils were identified in the tumor-draining lymph nodes (TDLNs). Moreover, their presence was a negative prognostic marker of patient survival (72). To further investigate the underlying mechanisms of this association, the Mcpt8Cre basophil deficient mouse strain (57) and WT mice were implanted with PC cells. Strikingly, 80% of the WT mice developed PC-like cancer, but this was not observed in the basophil-deficient mice (72). The authors reported that TSLP released from basophils and cancer-associated fibroblasts (CAFs) within TDLNs activated CD4+ T cells to produce IL-3. CCL7, derived from DCs and monocytes, promoted basophil recruitment into TDLNs. IL-3-activated basophils exerted a pro-tumorigenic role by secreting IL-4, which induced the polarization of Th2 and M2 cells. Thus, these results not only confirmed/supported the notion that basophil-derived IL-4/IL-13 promote Th2 and M2-like cells, but also demonstrated that these cells actively participate in promoting PC.
With the concept that various basophil-derived products (e.g., IL-4, IL-13, VEGF-A, LTC4) promote tumorigenesis, an equally important issue pertains to the stimuli mediating their release. Schroeder and colleagues have shown that human basophils release copious amounts of histamine, IL-4 and IL-13 when co-cultured with the human lung adenocarcinoma cell line A549 (16). These responses were dependent on basophils expressing IgE, since removal/depletion of this immunoglobulin prevented basophil activation. Since pharmacologic inhibitors of FcϵRI signaling also suppressed these responses, it seemed clear that basophils were being activated via IgE/FcϵRI crosslinking to secrete these cytokines. Importantly, direct contact between basophils and A549 was necessary and occurred even if the adenocarcinoma cells were fixed with paraformaldehyde prior to co-culture. In a follow-up study, the IgE-binding lectin, galectin-3 (Gal-3) expressed on the A549 cells, proved crucial for basophil activation in these co-cultures, as A549 clones lacking Gal-3 failed to activate basophils (136). Gal-3 is widely implicated in various cancers and is a marker of chronic inflammation (137). These findings reveal a potentially new mechanism by which Gal-3 expressed by human lung adenocarcinoma cells can activate basophils to release cytokines and pro-inflammatory mediators that promote tumorigenesis. Additional investigations are required to fully understand all aspects of this mechanism and how it might be targeted for therapeutic intervention.
By utilizing a model whereby the skin of mice were topically exposed to the proinflammatory 12-0-tetradecanoylphorbol-13-acetate (TPA), Hayes et al. showed that serum IgE increased in these animals, which was accompanied by increased numbers of IgE-bearing basophils that promoted skin tumorigenesis (73). In a similar model of epithelial carcinogenesis involving the use of [7,12-dimyethylbenz(a)anthracene (DMBA) and subsequent exposure to TPA], mice lacking IgE (lgh7-/-) developed less tumors compared to WT mice. The influx of basophils into skin was promoted by CXCR4, TSLP and IL-3. IgE-signaling played a key role in basophil activation and infiltrating tissue basophils expressed Cxcr2, Cxcr4, and Ptgdr2 (CRTH2, the PGD2 receptor). Tumor development was markedly reduced when conducting the same experiment in Mcpt8Cre/+ mice, which were made deficient in basophils but retained normal mast cell numbers (57). Collectively, these in vivo results further indicate that FcεRI-signaling in basophils promotes inflammation-driven epithelial hyperplasia and tumor growth. While the role of galectin-3 in this tumorigenesis was not investigated, it seems worthy of future investigation, as mechanisms of this response are further elucidated.
In contrast to the belief that basophils contribute to tumorigenesis, association studies have shown evidence that higher expression of basophils (i.e., CD123+, CCR3+, FcεRI+) in tumors correlated with better overall survival (88). In particular, increased basophil numbers are associated with beneficial outcomes in several cancers, including sarcoma, lung, and breast. While several additional markers (e.g., CD63, CD203c) indicated that these tumor-associated basophils were, indeed, activated, relevant mediators commonly released by these cells (histamine, LTC4, IL-4, IL-13) were not investigated. Thus, the exact contribution of basophils in the increased survival rates remains challenging to interpret at this time. Likewise, the same group has reported evidence that the in vitro responses of peripheral blood basophils from cancer patients can predict survival rates. While such correlations are intriguing, the exact mechanisms by which basophils contribute to increased survival rates is an area requiring further elucidation.
In agreement with the concept that basophils mediate a beneficial role in cancer, evidence from a mouse melanoma model showed that basophils released CCL3 and CCL4, which induced CD8+ T cell recruitment and promoted tumor rejection (75). MAR-1 administration in these melanoma-bearing mice depleted basophils and prevented melanoma rejection. However, it is important to note that basophil depletion using the MAR-1 is also reported to deplete/activate other immune cells expressing FcϵRI, including mast cells, monocytes and DCs (138, 139). Whether these cells were also depleted and possibly involved in tumoricidal activity remains unclear.
IL-33 has been shown to promote tumoricidal activity mediated by eosinophils (140, 141), possibly by upregulating granzyme B (142). As noted, this cytokine also activates both human and mouse basophils (9, 36, 38, 143–145). Hence, IL-33-activated basophils co-cultured with B16.F10 melanoma cells were shown to inhibit tumor growth compared to melanoma cells co-cultured with un-stimulated basophils (142).
Overall, there are several studies indicating that basophils promote tumorigenesis (72, 74). In this instance, the tumor cells cause basophils to release cytokines/chemokines that may facilitate the development of protumorigenic TME (Figure 3). Interestingly, many of the same TME elements involved in this activity (e.g., IL-4, IL-13, galectin-3, VEGF-A, M2 and Th2 cells) are also implicated in promoting wound healing. Conversely, in certain tumors (e.g., melanoma), basophils mediate anti-tumor effects (75, 88, 154) (Figure 4). The mechanisms underlying the protective effects of basophils remain largely unknown. It has been suggested that certain mediators (e.g., TNF-α and granzyme B) released by basophils exert tumoricidal effect. In addition, other molecules (e.g., CCL3 and CCL4) can favor the recruitment of cytotoxic CD8+ T cells (74). Collectively, these findings highlight some apparently conflicting results regarding the role that basophils potentially exert in different models of tumorigenesis, and thus warrant further investigation.
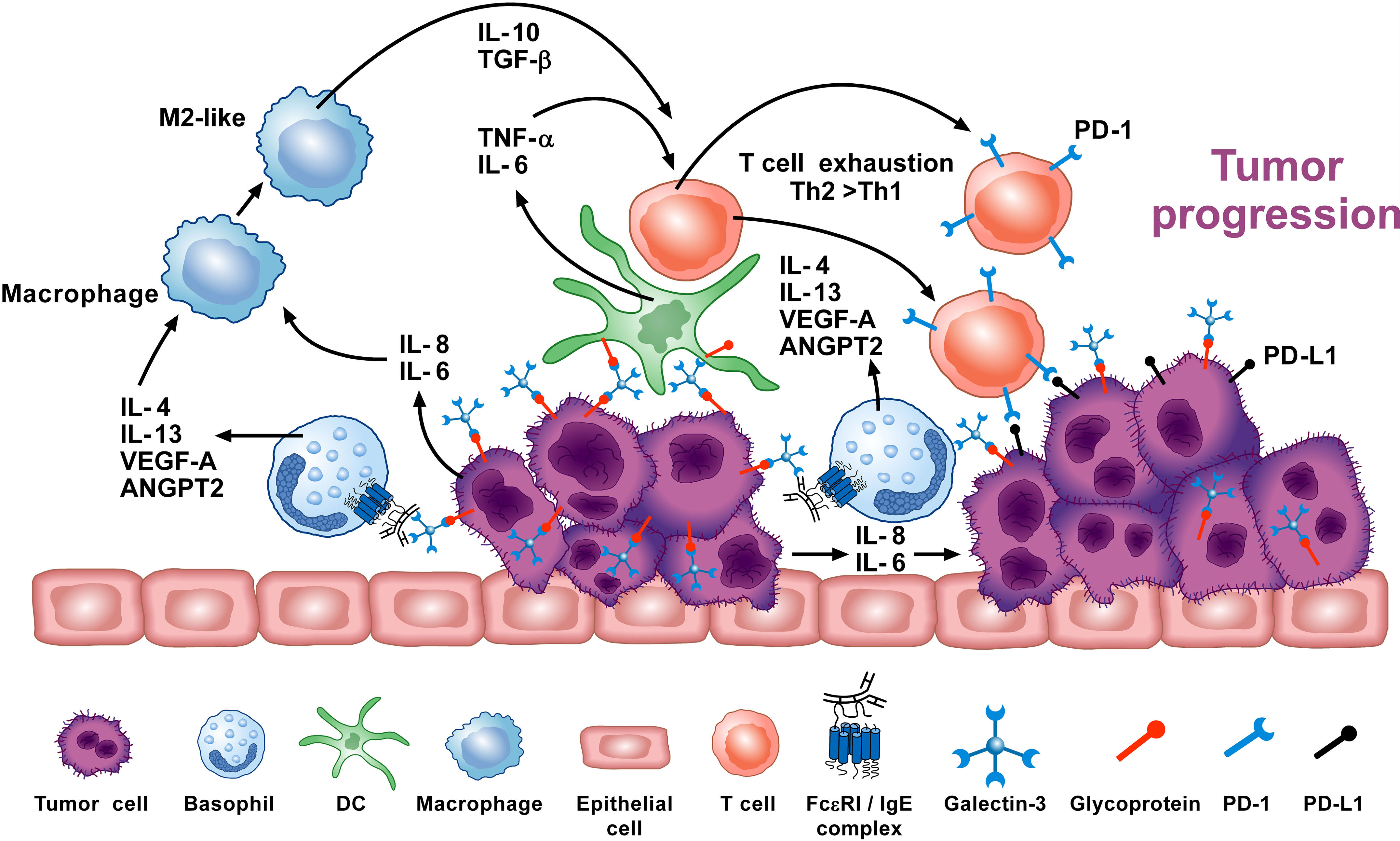
Figure 3 Basophils can promote tumor progression through different mechanisms. Galectin-3 (Gal-3) is a lectin expressed by several cancer cells (137), including the A549 adenocarcinoma cell line (EC-Gal-3). Gal-3 activates human basophils to release IL-4 and IL-13 (16, 136), which are widely known to promote M2-like macrophages, the major players in the TME (127–130). IL-4+ basophils have been found in the TME of human and experimental pancreatic cancer (72). Human and mouse basophils also secrete VEGF-A and angiopoietin 2 (ANGPT2) that can promote tumor angiogenesis (114, 146–148). Basophils can promote IL-6 and IL-8 release from epithelial cell lines through a mechanism requiring cell-to-cell contact (149) (JTS, unpublished). Tumor cell-derived IL-6/IL-8 play a critical role in metastasis formation (150). Dendritic cells and monocytes activated by EC-Gal-3 release TNF-α and IL-6 in vitro (151). These cytokines, combined with M2 cell-derived IL-10 and TGF-β induce T-cell exhaustion by up-regulating checkpoint inhibitors (i.e., PD-1), which interact with tumor cell-associated PD-L1 to decrease cytotoxic T cell activity (152, 153). These results suggest that basophils can promote tumorigenesis in certain experimental and clinical conditions. Adapted from Poto et al. (74).
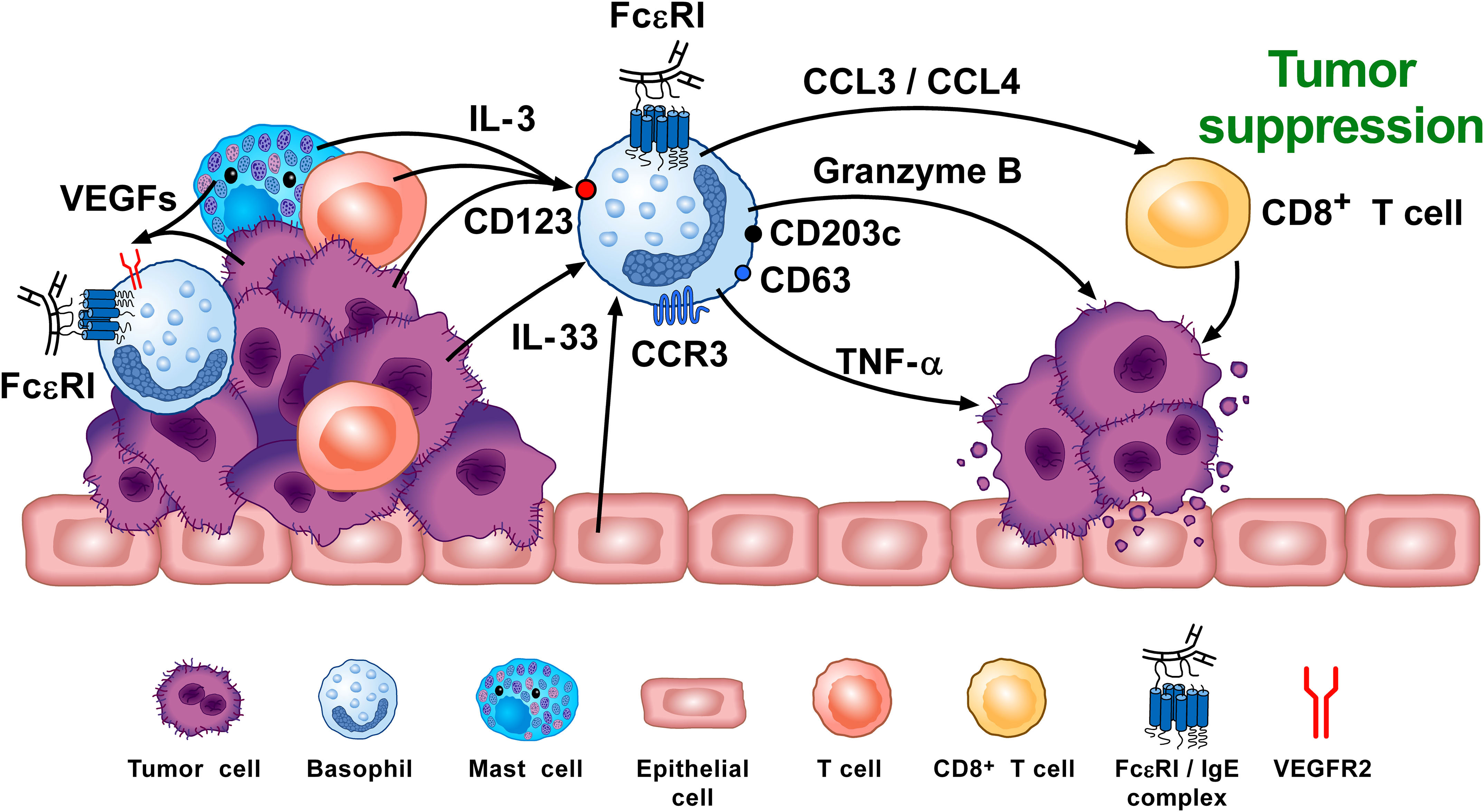
Figure 4 Basophils can promote tumor suppression through different mechanisms. Vascular endothelial growth factors (VEGFs) released by tumor and immune cells in the TME (e.g., macrophages, mast cells) (155–159) induce basophil recruitment via the activation of VEGFR2 on these cells (155). IL-3, released from intratumoral lymphocytes, mast cells and tumor cells (10, 160, 161), is the major growth, differentiation, priming and activating factor for both human and mouse basophils via the activation of the IL-3 receptor (IL-3Rα/CD123) (8–10). Intratumoral basophils secrete CCL3 and CCL4 which favor CD8+ T cell infiltration in TME, favoring melanoma rejection in mice (75). IL-33 produced by epithelial and tumor cells, plays a critical role in tumorigenesis (162) by upregulating granzyme B mRNA and the surface expression of CD63 in basophils. Mouse basophils activated by IL-33 cause melanoma cell death in vitro (142). Mouse (104, 163) and, in certain conditions, human basophils (164, 165) release TNF-α and granzyme B (142, 166), which exerts cytotoxic activity on cancer cells (102, 167). Tumor resident basophils overexpressing CD123, CCR3, CD63, CD203c mRNAs are associated with improved outcome in ovarian cancer (88, 154). These findings indicate that, under specific experimental and clinical circumstances, basophils can play an anti-tumorigenic role. Adapted from Poto et al. (74).
5 Basophils in autoimmune disorders
5.1 Systemic lupus erythematosus
With the discovery of IgE (168, 169), immunologists focused their attention on understanding its relevance for allergic disorders and host defense against parasitic infestations (2, 64, 81, 170). However, circulating IgE autoantibodies in rheumatoid arthritis and SLE patients had been reported as early as the late 70’s (171). While these early studies were conducted mostly using small cohorts of patients, they did confound the thought at the time that atopy was generally limited to patients suffering from allergic disease and/or parasitic infestations.
Systemic lupus erythematosus (SLE) is an autoimmune disorder associated with circulating self-reactive antibodies (172) (i.e., IgG anti-double-stranded DNA: anti-dsDNA). Several studies reported increased serum IgE in SLE, which correlated with severe disease manifestations (76, 173–175). A portion of the circulating IgE in these SLE patients was determined to be self-reactive, binding to nucleic acids, as was often the case for most IgG autoantibodies (176). In fact, several studies identified IgE against at least one autoantigen in SLE patients (171, 173, 177–182). Importantly, IgE anti-dsDNA antibodies are associated with disease activity and hypocomplementemia (177). Moreover, the levels of IgE anti-dsDNA proved to be an independent risk factor for SLE activity, even after excluding the levels of IgG anti-dsDNA (178). One study reported that IgE anti-dsDNA antibodies are found in ~ 70% of lupus patients, and are possibly linked to kidney damage (178). In a Franco-American cohort, IgE anti-dsDNA antibodies did associate with lupus nephritis, whereas IgE against other nucleic acid–containing autoantigens (Sm, SS-A/Ro, and SS-B/La) did not associate with disease (177). These findings suggested that IgE autoantibodies could play a role in the pathophysiologic mechanisms of lupus nephritis. The French-American collaborative study identified IgE autoantibodies against three new autoantigens: APEX nuclease 1, N-methylpurine DNA glycosylase and CAP-Gly domain-containing linker protein family member 4. These autoantigens specifically elicited IgE autoantibodies but not IgG autoantibodies (177). Collectively, these results indicate that IgE autoantibodies are prevalent in lupus nephritis patients and are associated with disease activity. Likewise, these findings provided the impetus for treating SLE patients in a randomized clinical trial using anti-IgE mAb (omalizumab) (NCT01716312).
Charles et al. first demonstrated mechanistic evidence that basophils are implicated in the pathobiology of lupus nephritis by using a spontaneous murine model of SLE (Lyn-/- mice) (76). This observation was subsequently confirmed using a model of pristane-induced lupus-like nephritis (183) as well as in a cohort of SLE patients (181). Basophils from SLE patients express significantly higher levels of the basophil activation marker, CD203c, compared to healthy controls (76). It was also found that the basophil density in both lymph nodes and spleen of SLE patients was higher than controls. Basophil-derived IL-4 reportedly induced B cell class switching toward IgE, and the autoreactive IgE produced was determined to be a relevant inducer of lupus (177, 178, 181, 184). Basophils from human patients with SLE and from two different lupus-like mouse models, overexpress both PGD2 receptors (PTGDR-1 and PTGDR-2) and CXCR4, the receptor for CXCL12 (185). Basophils seemingly contribute to SLE pathobiology by migrating to secondary lymphoid organs in a prostaglandin D2 (PGD2)- and CXCL12-dependent manner (185). These basophils can then support plasma cell functions by amplifying the production of autoantibodies and circulating immune complexes (76, 183, 185). Figure 5 schematically illustrates the mechanisms presumably linking IgE and basophils to SLE.
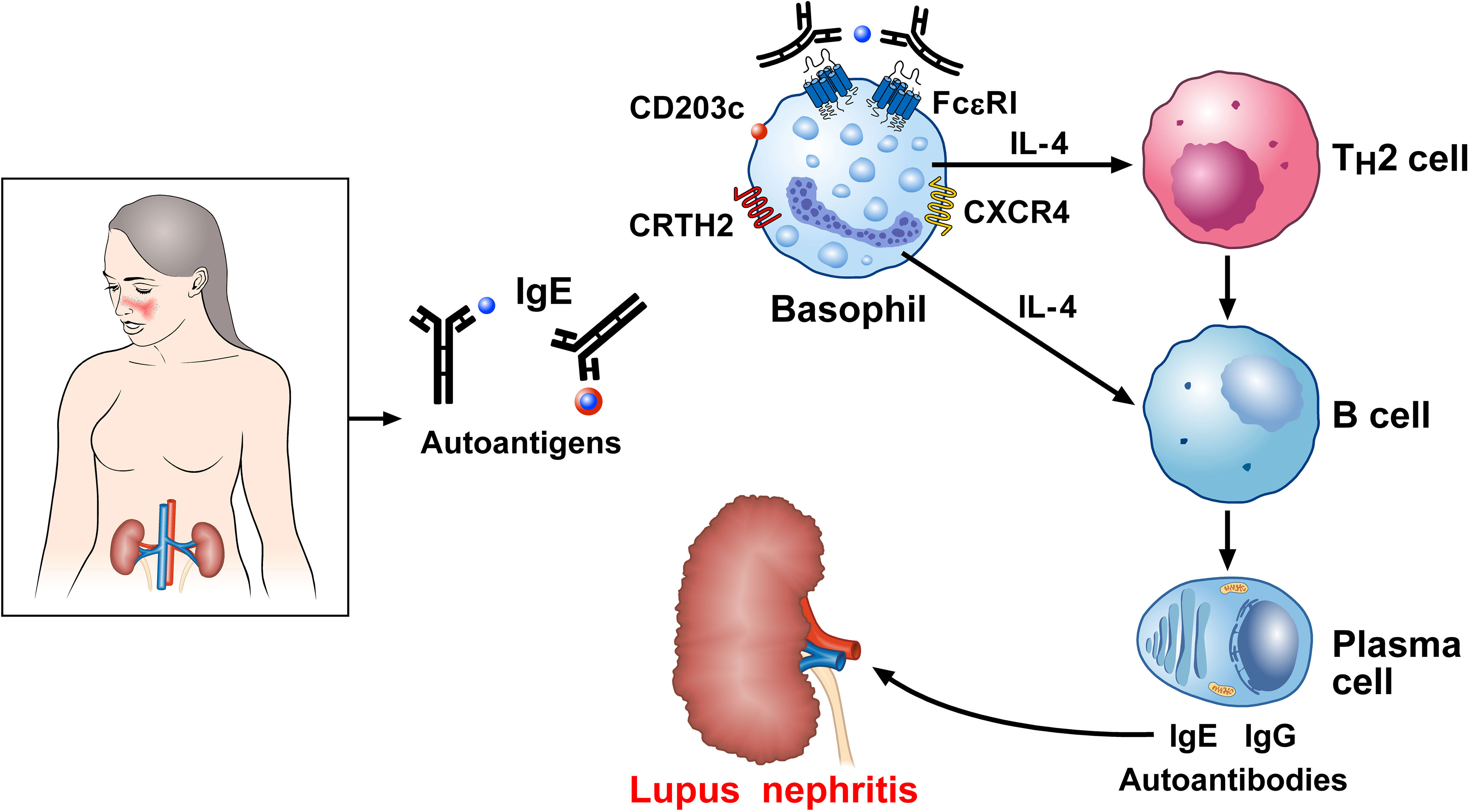
Figure 5 Proposed mechanism linking IgE basophils to autoimmunity in systemic lupus erythematosus (SLE). Serum IgE levels are increased in SLE and correlate with severe disease manifestations (76, 173–175). IgE against several autoantigens have been reported in SLE (171, 173, 177–182). Basophils from SLE patients show an activated phenotype in overexpressing CD203c (76), the prostaglandin D2 (PGD2) receptor [chemoattractant receptor-homologous molecule (CRTH2) expressed on Th2 cells], and CXCR4, the receptor for CXCL12 (185). Once recruited to the secondary lymphoid organs, activated basophils release IL-4, which drives B cell isotype switching toward IgE and autoreactive IgE (177, 181). Dendritic cells (DCs) in lymph nodes also act on B cells, triggering their differentiation into plasma cells and potentiating the formation of self-reactive autoantibodies (186). IgE immune complexes contribute to basophil activation. Deposits of IgG and IgE autoantibodies in the kidney play a major role in lupus nephritis.
5.2 Rheumatoid arthritis
Rheumatoid arthritis (RA) is a systemic autoimmune disease primarily involving inflammation of the joints (187). On a genetic background (i.e., HLA-DR4 found in ~ 70% of RA patients compared to 30% of controls), post-translational citrullination of several self-proteins generates altered self-antigens that activate CD4+ T cell responses in RA patients. Citrullination occurs via the conversion of arginine into citrulline by peptidyl arginine deiminases (PADs). Anti-citrullinated protein antibodies (ACPAs) are specific and predictive for RA and are implicated in the pathogenesis of RA (187).
IgE antibodies against citrullinated fibrinogen were detected in the serum of ~ 60% of ACPA+ RA patients (188). These authors reported that basophils from ACPA+ RA patients can be activated by citrullinated protein, whereas basophils from healthy controls were not activated. Serum from IgE-ACPA+ RA patients passively sensitized human FcϵRI+ expressing rat basophil cells (RBL) for activation by citrullinated proteins. These finding indicate that basophils from IgE-ACPA+ RA patients can be activated by citrullinated antigens. The results of this original study deserve to be extended using citrullinated proteins specific for RA patients.
5.3 Autoimmune encephalomyelitis
Experimental autoimmune encephalomyelitis (EAE) is an animal model widely used to investigate the mechanisms underlying multiple sclerosis (MS) (189). EAE differs from MS in needing to be induced rather than occurring spontaneously, although recent transgenic mouse models have indicated spontaneous development of EAE (189, 190). However, inoculation with central nervous system antigens and adjuvant or passive transfer of lymphocytes reactive with these antigens are often employed to induce EAE in many animal strains (189).
Yuk and collaborators have investigated the mechanisms by which basophils can contribute to TH17 differentiation and EAE pathogenesis (126). For example, IL-17 is highly expressed in MS lesions (191) and TH17 cells mediate blood-brain barrier disruption and the expression of IL-17 and IL-22 (192). TH17 differentiation requires IL-6 and TGF-β (193), yet whether basophils promote TH17 induction in EAE had remained unknown. To address this possibility, Yuk and coworkers demonstrated that IgE cross-linking, or the use of cholera toxin (CT), induced the release of IL-6 and IL-4 from bone marrow-derived basophils (126). Moreover, they found that basophils mediate TH17 differentiation through IL-6 secretion. The authors also examined whether basophils contribute to TH17 polarization in vivo. WT and IL-6-deficient mice were challenged with CT plus antigen. IL-17A producing CD4+ T cells were reduced in IL-6 deficient animals, suggesting that IL-6 is critical for the antigen-induced TH17 response. The role of basophils was also examined in basophil-deficient mice. The authors found that basophil-derived IL-6 cooperates with DCs to promote the differentiation of CD4 T cells into TH17 cells. TH17 responses were reduced in the absence of basophils or IL-6. Collectively, these findings suggest that basophil-derived mediators (e.g., IL-6) are involved in TH17 cell differentiation, allowing TH17 cells to migrate to the site of inflammation mediating pathogenic functions in EAE. These studies identify basophils and their mediators as candidates for investigating pathogenic mechanisms in MS patients. It should be noted that EAE pathology is not driven exclusively by TH17 and IL-17; other cells (e.g., CD8+, T cells, γδ T cells) and cytokines may also be involved (194).
5.4 Mixed connective tissue disease
Mixed connective tissue disease (MCTD) is a rare systemic autoimmune disease (incidence ~ 2 per 100,000 adults) affecting mainly women (~ 90%) (195). Its clinical manifestations often overlap with other connective tissue disorders, including SLE, systemic sclerosis, or myositis (196). The defining immunological feature of MCTD is the presence of autoantibodies recognizing the 70-kDa subunit of the U1 small nuclear ribonucleoprotein (U1-snRNP 70k) in the absence of IgG against dsDNA or to Sm, two SLE hallmarks (197). The pathophysiology underlying MCTD remains poorly understood, but posttranslational modifications of U1-snRNP are known to generate neoepitopes that may contribute to the disease (198). These neoepitopes can result in T cells recognizing U1-snRNP, which ultimately lead to the induction and proliferation of autoreactive B cells synthesizing autoantibodies (199). Immune complexes made of anti-U1snRNP antibodies and their antigen can activate endothelium and immune cell via a variety of receptors (e.g., Fc, complement, and Toll-like receptors, TLR), resulting in vascular disease and tissue injury (200–203). Pulmonary involvement characterizes more than 70% of MCTD patients (197). A mouse model has been described whereby mice immunized with human U1-snRNP develop a MCTD-like lung disorder (204).
Lamri and collaborators observed that basophils from patients with MCTD present an activated phenotype (77), sharing some features with basophils from SLE patients (i.e., overexpression of CD203c, CXCR4) (76, 185). In addition, basophils from MCTD expressed increased surface markers such as CCR3, yet unchanged expression levels of CD62L (77). A similar basophil phenotype was found in a MCTD-like mouse model in which activated basophils infiltrated in the lungs and lymph nodes. To study the contribution of basophils in the development of lung pathology in this model, basophils were depleted through the injection of DT in female Bcpt8DTR mice. Basophil depletion reduced the cellular infiltrates (e.g., CD4+ T cells) in the lungs. The authors also examined the MCTD-like lung disease in IgE-deficient mice (Igh7-/-). Similar to that seen with basophil depletion, IgE deficiency also protected mice from developing immune cell infiltration and lung fibrosis. These results indicate that basophils play a major effector role in inducing lung fibrosis via an IgE-dependent mechanism. The authors suggested that basophils, activated by the U1-snRNP antibodies complex, accumulate in the airways, where they release IL-4 contributing to lung fibrosis development. In this scenario, IgE-mediated basophil activation may play both immunoregulatory and effector roles in the development of MCTD lung disease. These mouse models identify basophils, and IgE as candidates for investigating pathogenic mechanisms in patients with MCTD.
6 Basophils in IgG4-related disease
IgG4-related disease (IgG4-RD) is a rare multi-organ disorder characterized by lympho-plasmacytic infiltration, fibrosis, and obliterative phlebitis (205, 206). This condition is characterized by IgG4+ plasma cell infiltration in different organs (e.g., biliary tree, pancreas, retroperitoneum, salivary and lacrimal glands, and lymph nodes) (207, 208). The disease was first described in 2003 in a cohort of seven patients with a diagnosis of autoimmune pancreatitis (AIP) associated with IgG4+ plasma cell infiltration (209). Although the pathogenic mechanisms underlying IgG4-RD remain elusive (206), an increased production of Th2 cytokines (IL-4, IL-5, IL-13) has been identified in IgG4-related cholangitis and pancreatitis (210). These cytokines favor IgE production and eosinophil recruitment. It has also been reported that in patients with IgG4-RD, there is an accumulation of T regulatory cells (Tregs) in the blood, along with evidence that these cells infiltrate affected tissues, showing overexpression of IL-10 and TGF-β (211, 212). TGF-β released from Tregs can stimulate fibroblasts to produce collagen. IL-10 produced by Tregs can also stimulate secretion of IgG4 from plasma cells. The involvement of IL-10 and TGF-β secreting basophils has been suggested in patients with IgG4-related submandibular gland disease (213). B cell activating factor (BAFF) and APRIL, in combination with IL-21, can promote the expansion of IgG4-committed B cells (214, 215).
Two studies performed by different investigators in Japan proposed a possible mechanism whereby basophils are stimulated via a TLR-dependent activation involving IgG4-RD (214, 216). When activated by TLR2/TLR4 agonists, basophils from healthy donors induced B cells to produce IgG4 and IgG1 (214). TLR4 activation of basophils induced the release of IL-13 and BAFF. Basophils from IgG4-RD patients, upon activation with TLR2 and TLR4 ligands, induced more IgG4 than IgG1 when co-cultured with B cells. The authors suggested that the activation of TLRs in basophils play a role in IgG4-RD development (214).
Another study examined the role of basophils from peripheral blood and pancreatic tissue in patients with autoimmune pancreatitis (AIP) (216). AIP is a manifestation of IgG4-RD (208). Basophil density in the pancreas of AIP patients was higher than in alcoholic pancreatitis (216). In some of these patients, peripheral blood and intrapancreatic basophils were TLR2 or TLR4 positive. The authors suggested that basophils activated by TLRs could play a role in AIP. At present, the possible involvement of basophils and their mediators in the pathogenesis of different localizations of IgG4-RD remains unknown.
7 Basophils in chronic obstructive pulmonary disease
Chronic obstructive pulmonary disease (COPD) is a primary cause of morbidity and mortality worldwide (217). COPD is characterized by chronic inflammation, progressive airflow limitation and emphysema. Relative to asthma, the cellular and molecular mechanisms of COPD remain ill defined (117). It also differs in being characterized by a non-reversible airway obstruction (82, 218).
Shibata and collaborators elegantly investigated the potential role of basophils and their mediators in an elastase-induced murine model of COPD (62). Intranasal elastase elicited the recruitment of monocytes to the lung, followed by differentiation into interstitial macrophages (IMs) rather than alveolar macrophages (AMs). Matrix metalloproteinase-12 (MMP-12) played a key role in developing elastase-induced emphysema and was mainly expressed by IMs. The expression of Il4, but not Il10, Il13, or Tgfb was upregulated in the lung after the instillation of elastase. Expression of Il4 mRNA was detected mainly in basophils, which accumulated in the lung. The authors used two complementary methods to deplete basophils in vivo, namely: diphtheria toxin (DT) treatment of Mcpt8DTR mice and anti-CD200R3 antibody treatment of WT mice. Using these models, they demonstrated impaired emphysema formation in basophil-depleted mice. They suggested that basophil-derived IL-4 promoted the differentiation of infiltrating monocytes into MMP-12–producing IMs that caused the alveolar wall destruction and emphysema formation. The authors concluded that the basophil-derived IL-4/monocyte–derived IM/MMP-12 axis plays a role in emphysema development. They also proposed that this novel cellular and humoral axis may be a potential target for COPD treatment.
In other findings, both eosinophils and basophils have been detected in several lung compartments of COPD patients, particularly in very severe COPD (219). Eosinophilic infiltration was patchy, and mainly confined eotaxin signatures with CCL11+ fibroblasts and CCL24+ macrophages. Basophils were preferentially localized in lymphoid tissue. These studies identify basophils and perhaps eosinophils as candidates for future investigations on their role in the pathogenic mechanisms of COPD.
8 Basophils in COVID-19
The current COVID-19 pandemic is caused by the novel severe acute respiratory syndrome Coronavirus 2 (SARS-CoV-2) (220). A dysregulated innate immune response is a key driver of clinical complications culminating in COVID-19 (221, 222). High levels of several cytokines (e.g., IL-1, IL-6, TNF-α, CXCL8) are detected early after viral infection, and many of these mediators are associated with granulocyte activation (223). The recombinant S1 subunit of the SARS-CoV-2 Spike protein activated in vitro human peripheral blood monocytes to release several cytokines (e.g., IL-6, IL-1β, TNF-α) and chemokines (e.g., CXCL10/IP-10, CCL3/MIP-1α, CCL4/MIP-1β) linked to COVID-19 (224). In this study, the S1 subunit did not induce any of these cytokines/chemokines from highly purified basophils (224). Another study reported that live SARS-CoV-2 virus induced IL-4 and IL-13 release in vitro from unprimed and IL-3-primed basophils (225). Although basophils have been implicated in the host response to other viruses (119, 226–229), the in vivo significance of basophil-derived cytokines/chemokines in the pathogenesis of COVID-19 remains unclear.
A detailed analysis at the single-cell resolution of granulocyte diversity in peripheral blood of COVID-19 patients demonstrated an increased level of both mature and immature neutrophils (230). By contrast, decreased basophils and eosinophils are often associated with severe COVID-19 (230, 231). Moreover, the emergence of PD-L1 expression on peripheral blood basophils (as defined as CD11b+SSlowCrTH2+ cells) has been associated with COVID-19 severity (232). It should be pointed out that in vitro incubation of live SARS-CoV-2 with basophils purified from normal donors did not induce the expression of PD-L1 (225), whereas INF-γ increased PD-L1 expression on IL-3-primed basophils (233). High basophil counts are associated with a lower risk of developing severe COVID-19 (234). Collectively, these interesting results potentially implicate that basophils and/or their mediators play a protective role in COVID-19.
9 Basophils in inflammatory bowel diseases
Crohn’s disease (CD) and ulcerative colitis (UC) are the most common chronic inflammatory bowel disorders (IBDs) (235, 236). The inflammatory infiltrate in IBDs is canonically characterized by activated T cells, macrophages, DCs, neutrophils, and TH17 cells (236). Basophils were identified in the inflamed mucosa of IBD patients that also expressed IL-33 (125). When activated by IL-3 and IL-33, basophils amplified TH17 cytokine expression in T cells. Basophils, but not mast cells, accumulated in inflamed CD and UC tissues compared to non-inflamed mucosa (237). No basophils were detected in colons of healthy control donors, indicating selective recruitment and/or survival of these cells at inflamed mucosal sites in patients with IBDs. The accumulation of basophils occurred in colons of untreated patients as well as in patients treated with 5-aminosalicylate acid or immunomodulators (e.g., glucocorticoids and/or immunosuppressive agents and/or biologics). Activated T cells infiltrate inflamed colons and are a major source of IL-3 (10) that may contribute to the infiltration and/or survival of basophils locally (238). Basophils increased IL-17 production and promoted the differentiation of IL-17+ cells. Collectively, these results demonstrate that basophils accumulate in the inflamed colon in patients with the two most frequent IBDs and may thus contribute to CD and UC pathogenesis. Figure 6 schematically illustrates the potential mechanisms by which basophils, together with other immune cells, contribute to IBD.
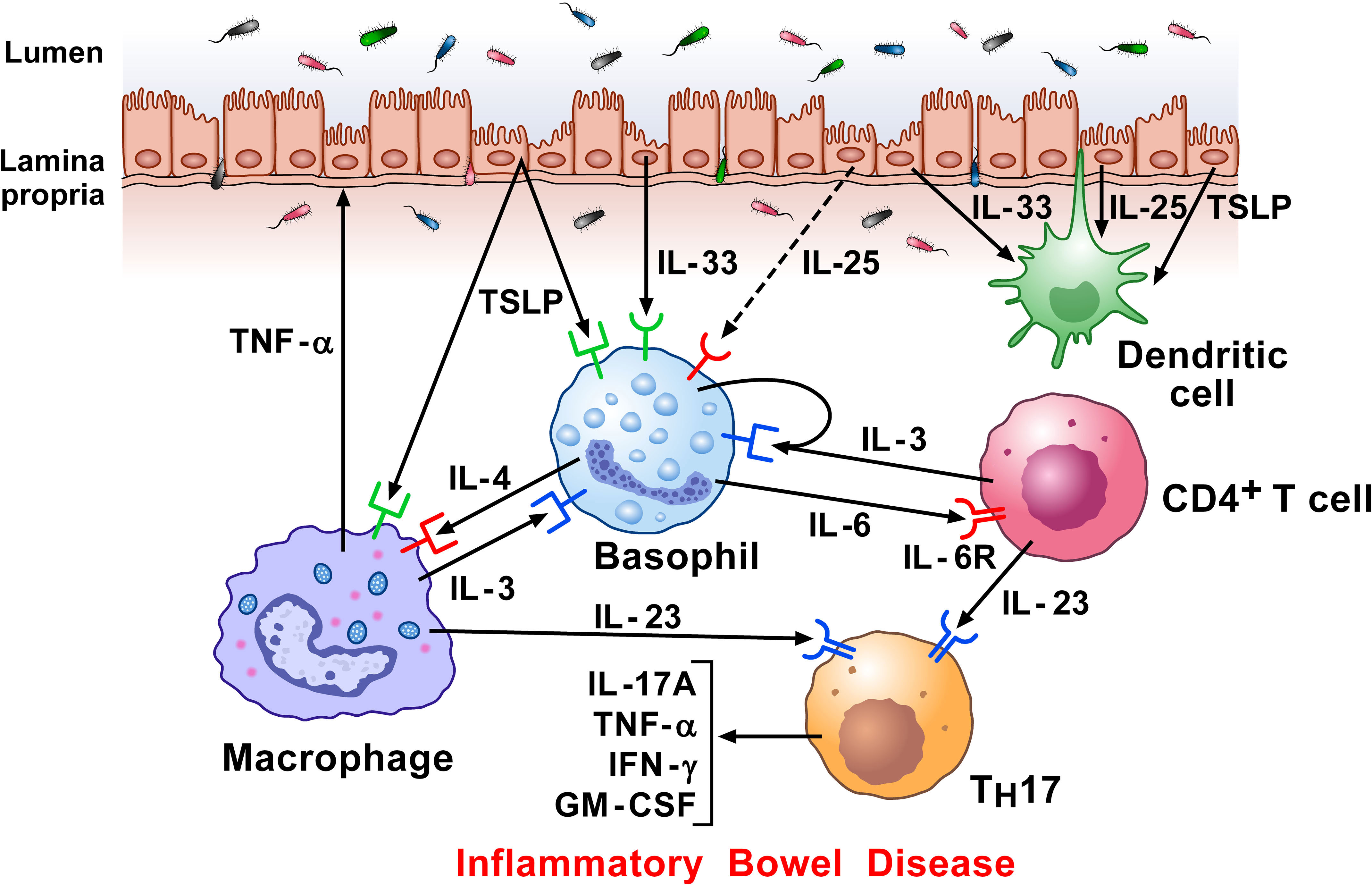
Figure 6 Hypothetical mechanisms by which dysregulated epithelial cells and inflammatory signaling by lamina propria immune cells in response to microbiota, contribute to inflammatory bowel disease (IBD) pathogenesis. Intestinal epithelial cells separate the lamina propria and deeper tissues from the luminal environment containing the intestinal microbiota (239). Increased intestinal permeability can potentiate immune-mediated systemic and intestinal inflammation in IBD (240). Damaged epithelial cells release alarmins (IL-33, TSLP, and IL-25) (115, 123, 241), which then regulate underlying immune cells (242), including basophils (9), macrophages (157), and DCs (243). Macrophages can damage epithelial cells directly by TNF-α secretion. Basophils accumulate in inflamed IBD compared to non-inflamed mucosa and to colon of healthy controls (125). Activated T cells infiltrate inflamed colons and release IL-3 which can contribute to the attraction and/or survival of basophils locally (238). Specific components of gut microbiota induce the emergence of intestinal TH17 cells. Basophils may also promote TH17 responses (125). Activated T cells release IL-23, which converts homeostatic TH17 cells to pathogenic TH17 cells, and play a major role in Crohn’s disease (244).
10 Basophils in eosinophilic granulomatosis with polyangiitis
Eosinophilic granulomatosis with polyangiitis (EGPA) is a rare systemic disease characterized by eosinophilic asthma, sinus and pulmonary infiltrates, and eosinophil vasculitis (245). Lung biopsies are rarely done in EGPA and adequate animal models are not currently available. Therefore, the lung immunopathology of this disorder has not been carefully examined. Basophils were detected in four of five EGPA open lung biopsies (246), whereas no basophils were identified in seven control lung biopsies. Mast cell density was increased in EGPA patients compared to the control lungs. These preliminary data show that EGPA lung immunopathology includes infiltrates of eosinophils, basophils, and mast cells. Further studies appear necessary to identify possible interlinks between basophils and IgE and delineate the protective versus rather harmful effects of these conditions in EGPA.
Therapeutic management of EGPA is based on glucocorticoids alone and often in combination with immunosuppressive agents (247). Several observational studies have evaluated the role of omalizumab on maintenance therapy in EGPA (247–249). The results of these studies suggest that omalizumab may be clinically beneficial for EGPA patients improving asthma symptoms, lung function, and may have a glucocorticoid-sparing effect (247–249). There is the possibility that the effects of omalizumab in EGPA patients may be related, at least in part, to its effects on human basophils (250).
11 Basophils in eosinophilic esophagitis
Eosinophilic esophagitis (EoE) is a chronic, food-driven allergic disease characterized by esophageal eosinophilia that affects children and adults (251–253). The histopathological and clinical features of EoE have been attributed to overproduction of the type 2 cytokines IL-4, IL-5 and IL-13, which mediate profound alterations in the esophageal epithelium (254–256). The esophageal epithelium likely has an important role in the initiation of EoE via production of the epithelium-derived cytokines thymic stromal lymphopoietin (TSLP) and IL-33 (257, 258). EoE is associated with polymorphism in the gene that encodes TSLP in children (259, 260). In a mouse model, EoE-like disease developed independently of IgE, but was dependent on TSLP and basophils (257). Targeting TSLP or basophil depletion during the sensitization phase limited disease and improved established EoE-like disease. Interestingly, increased TSLP expression and basophil responses were demonstrated in esophageal biopsies of patients with EoE (257). Collectively, these results suggest that the TSLP-basophil axis contributes to the pathogenesis of EoE.
In another model of EoE-like disease, mice were epicutaneously sensitized with ovalbumin (OVA), followed by intranasal OVA challenge (258). This procedure promoted eosinophilic esophagitis, upregulation of Th2-like cytokines and the IL-33 receptor (ST2). In vivo basophil depletion or disruption of the IL-33-ST2 axis mitigated these features. These results suggest that basophils mediate experimental EoE through IL-33-ST2 interaction. These authors also found that pediatric patients with EoE have increased expression of IL33 and IL1RL1 (encoding ST2) in esophageal biopsies (258).
Taken together, these studies endorse the paradigm that epithelium-derived cytokines (i.e., TSLP and IL-33) play a role in the pathogenesis of EoE through the activation of basophils and the development of type 2 inflammatory milieu.
12 Concluding remarks and perspectives
Basophils are extremely rare cells, accounting for 1% or less of the circulating blood leukocytes, both in humans and mice. As a result, there was limited capacity to investigate the biology of these immune cells for several decades following their discovery in 1879 (261). However, advances during the past ~30 years have increased interest with compelling new evidence that they represent important effector cells in allergic inflammation (1, 64, 81, 82) and exert a protective role in parasitic infections (66–68). The development of new murine genetic tools and different models of inflammation has also generated novel insight into the potential contribution of basophils to an increasing spectrum of diseases. In particular, basophils and their mediators are now implicated as important participants in pathophysiologic conditions never before considered, including MI (70), kidney fibrosis (71), several autoimmune disorders (76, 77, 126), different cancers (72, 73, 75), COPD (62), and COVID-19 (230–232, 234).
In several pathological conditions, such as kidney fibrosis (71), autoimmune disorders (76, 77, 125, 126), some cancers (72, 73), COPD (62), IgG4-RD (208), IBD (125, 237), and EoE (257, 258) basophils and their mediators play a harmful role. In other inflammatory disorders, such as MI (70), certain cancers (154) (75), and COVID-19 (230–232, 234), basophils appear to play a protective role. The dichotomous pathogenic role of basophils is intriguing and will undoubtedly be the subject of future investigations. There is the possibility that, like mast cells (262–266), macrophages (104, 132, 267, 268), neutrophils (269–272), and eosinophils (273, 274), subpopulations of basophils may also exist. In this regard, distinct phenotypic and functional basophil subpopulations have been described in human peripheral blood (275). Moreover, it has already been demonstrated that tissue-resident basophils differ from circulating basophils in mice (276) and possibly in humans. Finally, basophils might possess a high degree of plasticity and can modify their phenotype and functional characteristics when exposed to different local environments. Whatever the case, the possible existence of basophil subpopulations and the disease-specific heterogeneity of these cells need to be thoroughly and accurately explored in both humans and mice by novel analytical tools (e.g., single-cell RNA seq, CyTOF).
Finally, several biologics have been approved for the treatment of severe allergic disorders and are showing remarkable efficacy (218). Those designed primarily to target mast cells, eosinophils, and Th2 cells (e.g., omalizumab, mepolizumab, benralizumab and dupilumab) also target human basophils and/or their products (250, 277). Thus, there is the possibility that these biologics could prove efficacious in helping to combat other unsuspected conditions/diseases (e.g., cancer, autoimmunity, fibrosis) where basophils are recently implicated. In contrast, with mounting evidence that basophils and their mediators also play critical homeostatic and protective roles (70, 75, 226, 230–232, 234), caution may be warranted when these therapeutic interventions are used.
Author contributions
RP, GM, JS, GV drafted the manuscript and interpreted data; RP, SL, GM, AS, AP, JS, GV edited the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported in part by grants from the CISI-Lab Project (University of Naples Federico II), TIMING and Campania Bioscience Projects (Regione Campania) (GM and GV), and NIAID and NIH grants AI115703 and AI141486 (JS).
Acknowledgments
We would like to thank Dr. Gjada Criscuolo for her excellent managerial assistance in preparing this manuscript, the medical artist Fabrizio Fiorbianco for preparing the figures and the administrative staff (Dr. Roberto Bifulco, Dr. Anna Ferraro and Dr. Maria Cristina Fucci), without whom it would not be possible to work as a team. The authors apologize to the many researchers who have contributed importantly to the field and whose work was not cited due to space constraints.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Abbreviations
ACPA, anticitrullinated protein antibody; AIP, autoimmune pancreatitis; AM, alveolar macrophage; BAFF, B cell activating factor; BAP, basophil progenitor; CAF, cancer-associated fibroblast; CD, Crohn disease; CKD, chronic kidney disease; COPD, chronic obstructive pulmonary disease; CMP, common myeloid progenitor; CRTH2, chemoattractant receptor-homologous molecule expressed on Th2 cells; CT, cholera toxin; CXCR4, CX-C motif chemokine receptor 4; DC, dendritic cell; DMBA, 7,12-dimethylbenz[a]anthracene; dsDNA, double-stranded DNA; DT, diphtheria toxin; EAE, experimental autoimmune encephalomyelitis; EGPA, eosinophil granulomatosis with polyangitis; EoE, eosinophilic esophagitis; FcεRI, high affinity IgE receptor; GMP, granulocyte-macrophage progenitor; HSC, hematopoietic stem cell; IBD, inflammatory bowel disease; IFN-γ, interferon- γ; IgG4-RD, IgG4-related disease; IL, interleukin; IM, interstitial macrophages; LTC4, cysteinyl leukotriene C4; MI, myocardial infarction; MCTD, mixed connective tissue disease; MRI, magnetic resonance imaging; MS, multiple sclerosis; NK cell, natural killer cell; NSCLC, non-small cell lung cancer; OVA, ovalbumin; PAD, peptidyl arginine deiminase; PDAC, ductal adenocarcinoma; PDGFB, platelet derived growth factor subunit B; PDGFBR, platelet derived growth factor subunit B receptor; PGD2, prostaglandin D2; PT, proximal tubular cell; RA, rheumatoid arthritis; RBL, rat basophil cell; SLE, systemic lupus erythematosus; TDLN, tumor-draining lymph node; TGF-β, transforming growth factor-β; TME, tumormicroenvironment; TPA, tetradecanoylphorbol-13-acetate; Treg cell, T regulatory cell; TSLP, thymic stromal lymphopoietin; UC, ulcerative colitis; UUO, unilateral ureter obstruction; WT, wild type.
References
1. Schroeder JT. Basophils: emerging roles in the pathogenesis of allergic disease. Immunol Rev (2011) 242:144–60. doi: 10.1111/j.1600-065X.2011.01023.x
2. Marone G, Borriello F, Varricchi G, Genovese A, Granata F. Basophils: historical reflections and perspectives. Chem Immunol Allergy (2014) 100:172–92. doi: 10.1159/000358734
3. Varricchi G, Raap U, Rivellese F, Marone G, Gibbs BF. Human mast cells and basophils-how are they similar how are they different? Immunol Rev (2018) 282:8–34. doi: 10.1111/imr.12627
4. St John AL, Rathore APS, Ginhoux F. New perspectives on the origins and heterogeneity of mast cells. Nat Rev Immunol (2023) 23:55–68. doi: 10.1038/s41577-022-00731-2
5. Dwyer DF, Barrett NA, Austen KF. And immunological genome project, c., expression profiling of constitutive mast cells reveals a unique identity within the immune system. Nat Immunol (2016) 17:878–87. doi: 10.1038/ni.3445
6. Motakis E, Guhl S, Ishizu Y, Itoh M, Kawaji H, de Hoon M, et al. Redefinition of the human mast cell transcriptome by deep-CAGE sequencing. Blood (2014) 123:e58–67. doi: 10.1182/blood-2013-02-483792
7. Schroeder JT, Kagey-Sobotka A. And Lichtenstein, L.M., the role of the basophil in allergic inflammation. Allergy (1995) 50:463–72. doi: 10.1111/j.1398-9995.1995.tb01180.x
8. Kirshenbaum AS, Goff JP, Kessler SW, Mican JM, Zsebo KM, Metcalfe DD. Effect of IL-3 and stem cell factor on the appearance of human basophils and mast cells from CD34+ pluripotent progenitor cells. J Immunol (1992) 148:772–7.
9. Gambardella AR, Poto R, Tirelli V, Schroeder JT, Marone G, Mattei F, et al. Differential effects of alarmins on human and mouse basophils. Front Immunol (2022) 13:894163. doi: 10.3389/fimmu.2022.894163
10. Varricchi G, Poto R, Marone G, Schroeder JT. IL-3 in the development and function of basophils. Semin Immunol (2021) 54:101510. doi: 10.1016/j.smim.2021.101510
11. Dotsika EN, Sanderson CJ. Interleukin-3 production as a sensitive measure of T-lymphocyte activation in the mouse. Immunology (1987) 62:665–8.
12. Renner K, Hellerbrand S, Hermann F, Riedhammer C, Talke Y, Schiechl G, et al. IL-3 promotes the development of experimental autoimmune encephalitis. JCI Insight (2016) 1:e87157. doi: 10.1172/jci.insight.87157
13. Weber GF, Chousterman BG, He S, Fenn AM, Nairz M, Anzai A, et al. Interleukin-3 amplifies acute inflammation and is a potential therapeutic target in sepsis. Science (2015) 347:1260–5. doi: 10.1126/science.aaa4268
14. Kita H, Ohnishi T, Okubo Y, Weiler D, Abrams JS, Gleich GJ. Granulocyte/macrophage colony-stimulating factor and interleukin 3 release from human peripheral blood eosinophils and neutrophils. J Exp Med (1991) 174:745–8. doi: 10.1084/jem.174.3.745
15. Woolhiser MR, Brockow K. And metcalfe, D.D., activation of human mast cells by aggregated IgG through FcgammaRI: additive effects of C3a. Clin Immunol (2004) 110:172–80. doi: 10.1016/j.clim.2003.11.007
16. Schroeder JT, Bieneman AP. Activation of human basophils by A549 lung epithelial cells reveals a novel IgE-dependent response independent of allergen. J Immunol (2017) 199:855–65. doi: 10.4049/jimmunol.1700055
17. MacDonald SM, Schleimer RP, Kagey-Sobotka A, Gillis S, Lichtenstein LM. Recombinant IL-3 induces histamine release from human basophils. J Immunol (1989) 142:3527–32.
18. Ochensberger B, Tassera L, Bifrare D, Rihs S, Dahinden CA. Regulation of cytokine expression and leukotriene formation in human basophils by growth factors, chemokines and chemotactic agonists. Eur J Immunol (1999) 29:11–22. doi: 10.1002/(SICI)1521-4141(199901)29:01<11::AID-IMMU11>3.0.CO;2-B
19. Miura K, MacGlashan DW Jr. Dual phase priming by IL-3 for leukotriene C4 generation in human basophils: difference in characteristics between acute and late priming effects. J Immunol (2000) 164:3026–34. doi: 10.4049/jimmunol.164.6.3026
20. Hong L, Tang Y, Pan S, Xu M, Shi Y, Gao S, et al. Interleukin 3-induced GITR promotes the activation of human basophils. Cytokine (2020) 136:155268. doi: 10.1016/j.cyto.2020.155268
21. Rohner L, Reinhart R, Hagmann B, Odermatt A, Babirye A, Kaufmann T, et al. FcvarepsilonRI cross-linking and IL-3 protect human basophils from intrinsic apoptotic stress. J Allergy Clin Immunol (2018) 142:1647–1650 e3. doi: 10.1016/j.jaci.2018.06.040
22. Zellweger F, Buschor P, Hobi G, Brigger D, Dahinden CA, Villiger PM, et al. IL-3 but not monomeric IgE regulates FcepsilonRI levels and cell survival in primary human basophils. Cell Death Dis (2018) 9:510. doi: 10.1038/s41419-018-0526-9
23. Hagmann BR, Odermatt A, Kaufmann T, Dahinden CA, Fux M. Balance between IL-3 and type iinterferons and their interrelationship with FasL dictates lifespan and effector functions of human basophils. Clin Exp Allergy (2017) 47:71–84. doi: 10.1111/cea.12850
24. Kampfer SS, Odermatt A, Dahinden CA, Fux M. Late IL-3-induced phenotypic and functional alterations in human basophils require continuous IL-3 receptor signaling. J Leukoc Biol (2017) 101:227–38. doi: 10.1189/jlb.2A0715-292RR
25. Bischoff SC, de Weck AL, Dahinden CA. Interleukin 3 and granulocyte/macrophage-colony-stimulating factor render human basophils responsive to low concentrations of complement component C3a. Proc Natl Acad Sci USA (1990) 87:6813–7. doi: 10.1073/pnas.87.17.6813
26. Valent P, Schmidt G, Besemer J, Mayer P, Zenke G, Liehl E, et al. Interleukin-3 is a differentiation factor for human basophils. Blood (1989) 73:1763–9.
27. Qi Y, Operario DJ, Oberholzer CM, Kobie JJ, Looney RJ, Georas SN, et al. Human basophils express amphiregulin in response to T cell-derived IL-3. J Allergy Clin Immunol (2010) 126:1260–6.e4. doi: 10.1016/j.jaci.2010.08.040
28. Iype J, Odermatt A, Bachmann S, Coeudevez M. And fux, m., IL-1beta promotes immunoregulatory responses in human blood basophils. Allergy (2021) 76:2017–29. doi: 10.1111/all.14760
29. Mach N, Lantz CS, Galli SJ, Reznikoff G, Mihm M, Small C, et al. Involvement of interleukin-3 in delayed-type hypersensitivity. Blood (1998) 91:778–83.
30. Ichihara M, Hara T, Takagi M, Cho LC, Gorman DM. And miyajima, a., impaired interleukin-3 (IL-3) response of the A/J mouse is caused by a branch point deletion in the IL-3 receptor alpha subunit gene. EMBO J (1995) 14:939–50. doi: 10.1002/j.1460-2075.1995.tb07075.x
31. Lantz CS, Boesiger J, Song CH, Mach N, Kobayashi T, Mulligan RC, et al. Role for interleukin-3 in mast-cell and basophil development and in immunity to parasites. Nature (1998) 392:90–3. doi: 10.1038/32190
32. Siracusa MC, Saenz SA, Hill DA, Kim BS, Headley MB, Doering TA, et al. TSLP promotes interleukin-3-independent basophil haematopoiesis and type 2 inflammation. Nature (2011) 477:229–33. doi: 10.1038/nature10329
33. Giacomin PR, Siracusa MC, Walsh KP, Grencis RK, Kubo M, Comeau MR, et al. Thymic stromal lymphopoietin-dependent basophils promote Th2 cytokine responses following intestinal helminth infection. J Immunol (2012) 189:4371–8. doi: 10.4049/jimmunol.1200691
34. Salter BM, Oliveria JP, Nusca G, Smith SG, Watson RM, Comeau M, et al. Thymic stromal lymphopoietin activation of basophils in patients with allergic asthma is IL-3 dependent. J Allergy Clin Immunol (2015) 136:1636–44. doi: 10.1016/j.jaci.2015.03.039
35. Salabert-Le Guen N, Hemont C, Delbove A, Poli C, Braudeau C, Fantou A, et al. Thymic stromal lymphopoietin does not activate human basophils. J Allergy Clin Immunol (2018) 141:1476–1479 e6. doi: 10.1016/j.jaci.2017.11.012
36. Pecaric-Petkovic T, Didichenko SA, Kaempfer S, Spiegl N, Dahinden CA. Human basophils and eosinophils are the direct target leukocytes of the novel IL-1 family member IL-33. Blood (2009) 113:1526–34. doi: 10.1182/blood-2008-05-157818
37. Redrup AC, Howard BP, MacGlashan DW Jr., Kagey-Sobotka A, Lichtenstein LM, Schroeder JT. Differential regulation of IL-4 and IL-13 secretion by human basophils: their relationship to histamine release in mixed leukocyte cultures. J Immunol (1998) 160:1957–64.
38. Rivellese F, Suurmond J, de Paulis A, Marone G, Huizinga TW. And toes, R.E., IgE and IL-33-mediated triggering of human basophils inhibits TLR4-induced monocyte activation. Eur J Immunol (2014) 44:3045–55. doi: 10.1002/eji.201444731
39. Schroeder JT, Chichester KL, Bieneman AP. Human basophils secrete IL-3: evidence of autocrine priming for phenotypic and functional responses in allergic disease. J Immunol (2009) 182:2432–8. doi: 10.4049/jimmunol.0801782
40. Ochensberger B, Daepp GC, Rihs S, Dahinden CA. Human blood basophils produce interleukin-13 in response to IgE-receptor-dependent and -independent activation. Blood (1996) 88:3028–37.
41. Huang H, Li Y, Liu B. Transcriptional regulation of mast cell and basophil lineage commitment. Semin immunopathology (2016) 38:539–48. doi: 10.1007/s00281-016-0562-4
42. Sasaki H, Kurotaki D, Tamura T. Regulation of basophil and mast cell development by transcription factors. Allergology Int Off J Japanese Soc Allergology (2016) 65:127–34. doi: 10.1016/j.alit.2016.01.006
43. Arinobu Y, Iwasaki H, Gurish MF, Mizuno S, Shigematsu H, Ozawa H, et al. Developmental checkpoints of the basophil/mast cell lineages in adult murine hematopoiesis. Proc Natl Acad Sci USA (2005) 102:18105–10. doi: 10.1073/pnas.0509148102
44. Qi X, Hong J, Chaves L, Zhuang Y, Chen Y, Wang D, et al. Antagonistic regulation by the transcription factors C/EBPalpha and MITF specifies basophil and mast cell fates. Immunity (2013) 39:97–110. doi: 10.1016/j.immuni.2013.06.012
45. Paul F, Arkin Y, Giladi A, Jaitin DA, Kenigsberg E, Keren-Shaul H, et al. Transcriptional heterogeneity and lineage commitment in myeloid progenitors. Cell (2015) 163:1663–77. doi: 10.1016/j.cell.2015.11.013
46. Olsson A, Venkatasubramanian M, Chaudhri VK, Aronow BJ, Salomonis N, Singh H, et al. Single-cell analysis of mixed-lineage states leading to a binary cell fate choice. Nature (2016) 537:698–702. doi: 10.1038/nature19348
47. Busch K, Klapproth K, Barile M, Flossdorf M, Holland-Letz T, Schlenner SM, et al. Fundamental properties of unperturbed haematopoiesis from stem cells. vivo. Nat (2015) 518:542–6. doi: 10.1038/nature14242
48. Tusi BK, Wolock SL, Weinreb C, Hwang Y, Hidalgo D, Zilionis R, et al. Population snapshots predict early haematopoietic and erythroid hierarchies. Nature (2018) 555:54–60. doi: 10.1038/nature25741
49. Dahlin JS, Hamey FK, Pijuan-Sala B, Shepherd M, Lau WWY, Nestorowa S, et al. A single-cell hematopoietic landscape resolves 8 lineage trajectories and defects in kit mutant mice. Blood (2018) 131:e1–e11. doi: 10.1182/blood-2017-12-821413
50. Gorgens A, Radtke S, Mollmann M, Cross M, Durig J, Horn PA, et al. Revision of the human hematopoietic tree: granulocyte subtypes derive from distinct hematopoietic lineages. Cell Rep (2013) 3:1539–52. doi: 10.1016/j.celrep.2013.04.025
51. Drissen R, Thongjuea S, Theilgaard-Monch K, Nerlov C. Identification of two distinct pathways of human myelopoiesis. Sci Immunol (2019) 4:eaau7148. doi: 10.1126/sciimmunol.aau7148
52. Pellin D, Loperfido M, Baricordi C, Wolock SL, Montepeloso A, Weinberg OK, et al. A comprehensive single cell transcriptional landscape of human hematopoietic progenitors. Nat Commun (2019) 10:2395. doi: 10.1038/s41467-019-10291-0
53. Grootens J, Ungerstedt JS, Wu C, Hamberg Levedahl K, Nilsson G. And dahlin, J.S., CD203c distinguishes the erythroid and mast cell-basophil differentiation trajectories among human FcepsilonRI(+) bone marrow progenitors. Allergy (2020) 75:211–4. doi: 10.1111/all.13981
54. Obata K, Mukai K, Tsujimura Y, Ishiwata K, Kawano Y, Minegishi Y, et al. Basophils are essential initiators of a novel type of chronic allergic inflammation. Blood (2007) 110:913–20. doi: 10.1182/blood-2007-01-068718
55. Denzel A, Maus UA, Rodriguez Gomez M, Moll C, Niedermeier M, Winter C, et al. Basophils enhance immunological memory responses. Nat Immunol (2008) 9:733–42. doi: 10.1038/ni.1621
56. Wada T, Ishiwata K, Koseki H, Ishikura T, Ugajin T, Ohnuma N, et al. Selective ablation of basophils in mice reveals their nonredundant role in acquired immunity against ticks. J Clin Invest (2010) 120:2867–75. doi: 10.1172/JCI42680
57. Ohnmacht C, Schwartz C, Panzer M, Schiedewitz I, Naumann R, Voehringer D. Basophils orchestrate chronic allergic dermatitis and protective immunity against helminths. Immunity (2010) 33:364–74. doi: 10.1016/j.immuni.2010.08.011
58. Sullivan BM, Liang HE, Bando JK, Wu D, Cheng LE, McKerrow JK, et al. Genetic analysis of basophil function. vivo. Nat Immunol (2011) 12:527–35. doi: 10.1038/ni.2036
59. Sawaguchi M, Tanaka S, Nakatani Y, Harada Y, Mukai K, Matsunaga Y, et al. Role of mast cells and basophils in IgE responses and in allergic airway hyperresponsiveness. J Immunol (2012) 188:1809–18. doi: 10.4049/jimmunol.1101746
60. Matsuoka K, Shitara H, Taya C, Kohno K, Kikkawa Y. And yonekawa, h., novel basophil- or eosinophil-depleted mouse models for functional analyses of allergic inflammation. PloS One (2013) 8:e60958. doi: 10.1371/journal.pone.0060958
61. Pellefigues C, Mehta P, Prout MS, Naidoo K, Yumnam B, Chandler J, et al. The Basoph8 mice enable an unbiased detection and a conditional depletion of basophils. Front Immunol (2019) 10:2143. doi: 10.3389/fimmu.2019.02143
62. Shibata S, Miyake K, Tateishi T, Yoshikawa S, Yamanishi Y, Miyazaki Y, et al. Basophils trigger emphysema development in a murine model of COPD through IL-4-mediated generation of MMP-12-producing macrophages. Proc Natl Acad Sci U.S.A. (2018) 115:13057–62. doi: 10.1073/pnas.1813927115
63. Yoshikawa S, Oh-Hora M, Hashimoto R, Nagao T, Peters L, Egawa M, et al. Pivotal role of STIM2, but not STIM1, in IL-4 production by IL-3-stimulated murine basophils. Sci Signaling (2019) 12:eaav2060. doi: 10.1126/scisignal.aav2060
64. Miyake K, Shibata S, Yoshikawa S. And karasuyama, h., basophils and their effector molecules in allergic disorders. Allergy (2021) 76:1693–706. doi: 10.1111/all.14662
65. Nakashima C, Otsuka A, Kabashima K. Recent advancement in the mechanism of basophil activation. J Dermatol Sci (2018) 91:3–8. doi: 10.1016/j.jdermsci.2018.03.007
66. Eberle JU, Voehringer D. Role of basophils in protective immunity to parasitic infections. Semin immunopathology (2016) 38:605–13. doi: 10.1007/s00281-016-0563-3
67. Karasuyama H, Miyake K, Yoshikawa S. Immunobiology of acquired resistance to ticks. Front Immunol (2020) 11:601504. doi: 10.3389/fimmu.2020.601504
68. Peng J, Siracusa MC. Basophils in antihelminth immunity. Semin Immunol (2021) 53:101529. doi: 10.1016/j.smim.2021.101529
69. Donnelly EL, Cespedes N, Hansten G, Wagers D, Briggs AM, Lowder C, et al. Basophil depletion alters host immunity, intestinal permeability, and mammalian host-to-Mosquito transmission in malaria. Immunohorizons (2022) 6:581–99. doi: 10.4049/immunohorizons.2200055
70. Sicklinger F, Meyer IS, Li X, Radtke D, Dicks S, Kornadt MP, et al. Basophils balance healing after myocardial infarction via IL-4/IL-13. J Clin Invest (2021) 131:e136778. doi: 10.1172/JCI136778
71. Doke T, Abedini A, Aldridge DL, Yang YW, Park J, Hernandez CM, et al. Single-cell analysis identifies the interaction of altered renal tubules with basophils orchestrating kidney fibrosis. Nat Immunol (2022) 23:947–59. doi: 10.1038/s41590-022-01200-7
72. De Monte L, Wormann S, Brunetto E, Heltai S, Magliacane G, Reni M, et al. Basophil recruitment into tumor-draining lymph nodes correlates with Th2 inflammation and reduced survival in pancreatic cancer patients. Cancer Res (2016) 76:1792–803. doi: 10.1158/0008-5472.CAN-15-1801-T
73. Hayes MD, Ward S, Crawford G, Seoane RC, Jackson WD, Kipling D, et al. Inflammation-induced IgE promotes epithelial hyperplasia and tumour growth. Elife (2020) 9:e51862. doi: 10.7554/eLife.51862
74. Poto R, Gambardella AR, Marone G, Schroeder JT, Mattei F, Schiavoni G, et al. Basophils from allergy to cancer. Front Immunol (2022) 13:1056838. doi: 10.3389/fimmu.2022.1056838
75. Sektioglu IM, Carretero R, Bulbuc N, Bald T, Tuting T, Rudensky AY, et al. Basophils promote tumor rejection via chemotaxis and infiltration of CD8+ T cells. Cancer Res (2017) 77:291–302. doi: 10.1158/0008-5472.CAN-16-0993
76. Charles N, Hardwick D, Daugas E, Illei GG, Rivera J. Basophils and the T helper 2 environment can promote the development of lupus nephritis. Nat Med (2010) 16:701–7. doi: 10.1038/nm.2159
77. Lamri Y, Vibhushan S, Pacreau E, Boedec E, Saidoune F, Mailleux A, et al. Basophils and IgE contribute to mixed connective tissue disease development. J Allergy Clin Immunol (2021) 147:1478–1489 e11. doi: 10.1016/j.jaci.2020.12.622
78. Kojima T, Obata K, Mukai K, Sato S, Takai T, Minegishi Y, et al. Mast cells and basophils are selectively activated in vitro and in vivo through CD200R3 in an IgE-independent manner. J Immunol (2007) 179:7093–100. doi: 10.4049/jimmunol.179.10.7093
79. Sokol CL, Barton GM, Farr AG, Medzhitov R. A mechanism for the initiation of allergen-induced T helper type 2 responses. Nat Immunol (2008) 9:310–8. doi: 10.1038/ni1558
80. Tchen J, Simon Q, Chapart L, Pellefigues C, Karasuyama H, Miyake K, et al. CT-M8 mice: a new mouse model demonstrates that basophils have a nonredundant role in lupus-like disease development. Front Immunol (2022) 13:900532. doi: 10.3389/fimmu.2022.900532
81. Miyake K, Ito J, Karasuyama H. Role of basophils in a broad spectrum of disorders. Front Immunol (2022) 13:902494. doi: 10.3389/fimmu.2022.902494
82. Varricchi G, Ferri S, Pepys J, Poto R, Spadaro G, Nappi E, et al. Biologics and airway remodeling in severe asthma. Allergy (2022) 77:3538–52. doi: 10.1111/all.15473
83. Poddighe D, Dossybayeva K, Bexeitov Y, Mukusheva Z. Basophils in autoimmunity: systemic lupus erythematosus and more? Autoimmun Rev (2021) 20:102790. doi: 10.1016/j.autrev.2021.102790
84. Poto R, Quinti I, Marone G, Taglialatela M, de Paulis A, Casolaro V, et al. IgG autoantibodies against IgE from atopic dermatitis can induce the release of cytokines and proinflammatory mediators from basophils and mast cells. Front Immunol (2022) 13:880412. doi: 10.3389/fimmu.2022.880412
85. Schiechl G, Hermann FJ, Rodriguez Gomez M, Kutzi S, Schmidbauer K, Talke Y, et al. Basophils trigger fibroblast activation in cardiac allograft fibrosis development. Am J Transplant Off J Am Soc Transplant Am Soc Transplant Surgeons (2016) 16:2574–88. doi: 10.1111/ajt.13764
86. Marone G, Schroeder JT, Mattei F, Loffredo S, Gambardella AR, Poto R, et al. Is there a role for basophils in cancer? Front Immunol (2020) 11:2103. doi: 10.3389/fimmu.2020.02103
87. Mukaida N, Tanabe Y, Baba T. Cancer non-stem cells as a potent regulator of tumor microenvironment: a lesson from chronic myeloid leukemia. Mol biomedicine (2021) 2:7. doi: 10.1186/s43556-021-00030-7
88. Chauhan J, Stavraka C, Grandits M, Palhares L, Josephs DH, Lacy KE, et al. Clinical and translational significance of basophils in patients with cancer. Cells (2022) 11:438. doi: 10.3390/cells11030438
89. Murdaca G, Di Gioacchino M, Greco M, Borro M, Paladin F, Petrarca C, et al. Basophils and mast cells in COVID-19 pathogenesis. Cells (2021) 10:2754. doi: 10.3390/cells10102754
90. Anderson JL, Morrow DA. Acute myocardial infarction. N Engl J Med (2017) 376:2053–64. doi: 10.1056/NEJMra1606915
91. Swirski FK, Nahrendorf M. Cardioimmunology: the immune system in cardiac homeostasis and disease. Nat Rev Immunol (2018) 18:733–44. doi: 10.1038/s41577-018-0065-8
92. Kubota A, Frangogiannis NG. Macrophages in myocardial infarction. Am J Physiol Cell Physiol (2022) 323:C1304–24. doi: 10.1152/ajpcell.00230.2022
93. Frangogiannis NG, Lindsey ML, Michael LH, Youker KA, Bressler RB, Mendoza LH, et al. Resident cardiac mast cells degranulate and release preformed TNF-alpha, initiating the cytokine cascade in experimental canine myocardial ischemia/reperfusion. Circulation (1998) 98:699–710. doi: 10.1161/01.cir.98.7.699
94. Gwechenberger M, Mendoza LH, Youker KA, Frangogiannis NG, Smith CW, Michael LH, et al. Cardiac myocytes produce interleukin-6 in culture and in viable border zone of reperfused infarctions. Circulation (1999) 99:546–51. doi: 10.1161/01.cir.99.4.546
95. Frangogiannis NG, Dewald O, Xia Y, Ren G, Haudek S, Leucker T, et al. Critical role of monocyte chemoattractant protein-1/CC chemokine ligand 2 in the pathogenesis of ischemic cardiomyopathy. Circulation (2007) 115:584–92. doi: 10.1161/CIRCULATIONAHA.106.646091
96. Anzai A, Choi JL, He S, Fenn AM, Nairz M, Rattik S, et al. The infarcted myocardium solicits GM-CSF for the detrimental oversupply of inflammatory leukocytes. J Exp Med (2017) 214:3293–310. doi: 10.1084/jem.20170689
97. Swirski FK, Nahrendorf M. Leukocyte behavior in atherosclerosis, myocardial infarction, and heart failure. Science (2013) 339:161–6. doi: 10.1126/science.1230719
98. Varricchi G, Marone G, Kovanen PT. Cardiac mast cells: underappreciated immune cells in cardiovascular homeostasis and disease. Trends Immunol (2020) 41:734–46. doi: 10.1016/j.it.2020.06.006
99. Ngkelo A, Richart A, Kirk JA, Bonnin P, Vilar J, Lemitre M, et al. Mast cells regulate myofilament calcium sensitization and heart function after myocardial infarction. J Exp Med (2016) 213:1353–74. doi: 10.1084/jem.20160081
100. Frangogiannis NG. The inflammatory response in myocardial injury, repair, and remodelling. Nat Rev Cardiol (2014) 11:255–65. doi: 10.1038/nrcardio.2014.28
101. Libby P, Nahrendorf M, Swirski FK. Leukocytes link local and systemic inflammation in ischemic cardiovascular disease: an expanded "Cardiovascular continuum". J Am Coll Cardiol (2016) 67:1091–103. doi: 10.1016/j.jacc.2015.12.048
102. Motomura Y, Morita H, Moro K, Nakae S, Artis D, Endo TA, et al. Basophil-derived interleukin-4 controls the function of natural helper cells, a member of ILC2s, in lung inflammation. Immunity (2014) 40:758–71. doi: 10.1016/j.immuni.2014.04.013
103. Sokol CL, Chu NQ, Yu S, Nish SA, Laufer TM, Medzhitov R. Basophils function as antigen-presenting cells for an allergen-induced T helper type 2 response. Nat Immunol (2009) 10:713–20. doi: 10.1038/ni.1738
104. Cohen M, Giladi A, Gorki AD, Solodkin DG, Zada M, Hladik A, et al. Lung single-cell signaling interaction map reveals basophil role in macrophage imprinting. Cell (2018) 175:1031–1044 e18. doi: 10.1016/j.cell.2018.09.009
105. Schneider E, Petit-Bertron AF, Bricard R, Levasseur M, Ramadan A, Girard JP, et al. IL-33 activates unprimed murine basophils directly in vitro and induces their in vivo expansion indirectly by promoting hematopoietic growth factor production. J Immunol (2009) 183:3591–7. doi: 10.4049/jimmunol.0900328
106. MacGlashan D Jr., White JM, Huang SK, Ono SJ, Schroeder JT, Lichtenstein LM. Secretion of IL-4 from human basophils. the relationship between IL-4 mRNA and protein in resting and stimulated basophils. J Immunol (1994) 152:3006–16.
107. Genovese A, Borgia G, Bjorck L, Petraroli A, de Paulis A, Piazza M, et al. Immunoglobulin superantigen protein l induces IL-4 and IL-13 secretion from human fc epsilon RI+ cells through interaction with the kappa light chains of IgE. J Immunol (2003) 170:1854–61. doi: 10.4049/jimmunol.170.4.1854
108. Patella V, Giuliano A, Bouvet JP, Marone G. Endogenous superallergen protein fv induces IL-4 secretion from human fc epsilon RI+ cells through interaction with the VH3 region of IgE. J Immunol (1998) 161:5647–55.
109. Prabhu SD, Frangogiannis NG. The biological basis for cardiac repair after myocardial infarction: from inflammation to fibrosis. Circ Res (2016) 119:91–112. doi: 10.1161/CIRCRESAHA.116.303577
110. Toor IS, Ruckerl D, Mair I, Ainsworth R, Meloni M, Spiroski AM, et al. Eosinophil deficiency promotes aberrant repair and adverse remodeling following acute myocardial infarction. JACC Basic Transl Sci (2020) 5:665–81. doi: 10.1016/j.jacbts.2020.05.005
111. Li W, Hsiao HM, Higashikubo R, Saunders BT, Bharat A, Goldstein DR, et al. Heart-resident CCR2(+) macrophages promote neutrophil extravasation through TLR9/MyD88/CXCL5 signaling. JCI Insight (2016) 1:e87315. doi: 10.1172/jci.insight.87315
112. Heidt T, Courties G, Dutta P, Sager HB, Sebas M, Iwamoto Y, et al. Differential contribution of monocytes to heart macrophages in steady-state and after myocardial infarction. Circ Res (2014) 115:284–95. doi: 10.1161/CIRCRESAHA.115.303567
113. Wu X, Reboll MR, Korf-Klingebiel M, Wollert KC. Angiogenesis after acute myocardial infarction. Cardiovasc Res (2021) 117:1257–73. doi: 10.1093/cvr/cvaa287
114. de Paulis A, Prevete N, Fiorentino I, Rossi FW, Staibano S, Montuori N, et al. Expression and functions of the vascular endothelial growth factors and their receptors in human basophils. J Immunol (2006) 177:7322–31. doi: 10.4049/jimmunol.177.10.7322
115. Cristinziano L, Poto R, Criscuolo G, Ferrara AL, Galdiero MR, Modestino L, et al. IL-33 and superantigenic activation of human lung mast cells induce the release of angiogenic and lymphangiogenic factors. Cells (2021) 10:145. doi: 10.3390/cells10010145
116. Staiano RI, Loffredo S, Borriello F, Iannotti FA, Piscitelli F, Orlando P, et al. Human lung-resident macrophages express CB1 and CB2 receptors whose activation inhibits the release of angiogenic and lymphangiogenic factors. J Leukoc Biol (2016) 99:531–40. doi: 10.1189/jlb.3HI1214-584R
117. Poto R, Loffredo S, Palestra F, Marone G, Patella V. And varricchi, g., angiogenesis, lymphangiogenesis, and inflammation in chronic obstructive pulmonary disease (COPD): few certainties and many outstanding questions. Cells (2022) 11:1720. doi: 10.3390/cells11101720
118. Varricchi G, Loffredo S, Borriello F, Pecoraro A, Rivellese F, Genovese A, et al. Superantigenic activation of human cardiac mast cells. Int J Mol Sci (2019) 20:1828. doi: 10.3390/ijms20081828
119. Marone G, Rossi FW, Pecoraro A, Pucino V, Criscuolo G, Paulis A, et al. HIV gp120 induces the release of proinflammatory, angiogenic, and lymphangiogenic factors from human lung mast cells. Vaccines (Basel) (2020) 8:208. doi: 10.3390/vaccines8020208
120. Ferrando M, Bagnasco D, Varricchi G, Bernardi S, Bragantini A, Passalacqua G, et al. Personalized medicine in allergy. Allergy Asthma Immunol Res (2017) 9:15–24. doi: 10.4168/aair.2017.9.1.15
121. Jager KJ, Kovesdy C, Langham R, Rosenberg M, Jha V, Zoccali C. A single number for advocacy and communication-worldwide more than 850 million individuals have kidney diseases. Nephrology dialysis Transplant Off Publ Eur Dialysis Transplant Assoc - Eur Renal Assoc (2019) 34:1803–5. doi: 10.1093/ndt/gfz174
122. Wynn TA. Cellular and molecular mechanisms of fibrosis. J Pathol (2008) 214:199–210. doi: 10.1002/path.2277
123. Marone G, Spadaro G, Braile M, Poto R, Criscuolo G, Pahima H, et al. Tezepelumab: a novel biological therapy for the treatment of severe uncontrolled asthma. Expert Opin Investig Drugs (2019) 28:931–40. doi: 10.1080/13543784.2019.1672657
124. Coppock GM, Aronson LR, Park J, Qiu C, Park J, DeLong JH, et al. Loss of IL-27Ralpha results in enhanced tubulointerstitial fibrosis associated with elevated Th17 responses. J Immunol (2020) 205:377–86. doi: 10.4049/jimmunol.1901463
125. Wakahara K, Baba N, Van VQ, Begin P, Rubio M, Ferraro P, et al. Human basophils interact with memory T cells to augment Th17 responses. Blood (2012) 120:4761–71. doi: 10.1182/blood-2012-04-424226
126. Yuk CM, Park HJ, Kwon BI, Lah SJ, Chang J, Kim JY, et al. Basophil-derived IL-6 regulates T(H)17 cell differentiation and CD4 T cell immunity. Sci Rep (2017) 7:41744. doi: 10.1038/srep41744
127. Kuroda E, Ho V, Ruschmann J, Antignano F, Hamilton M, Rauh MJ, et al. SHIP represses the generation of IL-3-induced M2 macrophages by inhibiting IL-4 production from basophils. J Immunol (2009) 183:3652–60. doi: 10.4049/jimmunol.0900864
128. Egawa M, Mukai K, Yoshikawa S, Iki M, Mukaida N, Kawano Y, et al. Inflammatory monocytes recruited to allergic skin acquire an anti-inflammatory M2 phenotype via basophil-derived interleukin-4. Immunity (2013) 38:570–80. doi: 10.1016/j.immuni.2012.11.014
129. Borriello F, Longo M, Spinelli R, Pecoraro A, Granata F, Staiano RI, et al. IL-3 synergises with basophil-derived IL-4 and IL-13 to promote the alternative activation of human monocytes. Eur J Immunol (2015) 45:2042–51. doi: 10.1002/eji.201445303
130. Pernot S, Evrard S, Khatib AM. The give-and-Take interaction between the tumor microenvironment and immune cells regulating tumor progression and repression. Front Immunol (2022) 13:850856. doi: 10.3389/fimmu.2022.850856
131. Stankovic B, Bjorhovde HAK, Skarshaug R, Aamodt H, Frafjord A, Muller E, et al. Immune cell composition in human non-small cell lung cancer. Front Immunol (2018) 9:3101. doi: 10.3389/fimmu.2018.03101
132. Lavin Y, Kobayashi S, Leader A, Amir ED, Elefant N, Bigenwald C, et al. Innate immune landscape in early lung adenocarcinoma by paired single-cell analyses. Cell (2017) 169:750–765 e17. doi: 10.1016/j.cell.2017.04.014
133. Crawford G, Hayes MD, Seoane RC, Ward S, Dalessandri T, Lai C, et al. Epithelial damage and tissue gammadelta T cells promote a unique tumor-protective IgE response. Nat Immunol (2018) 19:859–70. doi: 10.1038/s41590-018-0161-8
134. Varricchi G, Ameri P, Cadeddu C, Ghigo A, Madonna R, Marone G, et al. Antineoplastic drug-induced cardiotoxicity: a redox perspective. Front Physiol (2018) 9:167. doi: 10.3389/fphys.2018.00167
135. Duah E, Teegala LR, Kondeti V, Adapala RK, Keshamouni VG, Kanaoka Y, et al. Cysteinyl leukotriene 2 receptor promotes endothelial permeability, tumor angiogenesis, and metastasis. Proc Natl Acad Sci U.S.A. (2019) 116:199–204. doi: 10.1073/pnas.1817325115
136. Schroeder JT, Adeosun AA, Do D, Bieneman AP. Galectin-3 is essential for IgE-dependent activation of human basophils by A549 lung epithelial cells. J Allergy Clin Immunol (2019) 144:312–315 e1. doi: 10.1016/j.jaci.2019.03.001
137. Sciacchitano S, Lavra L, Morgante A, Ulivieri A, Magi F, De Francesco GP, et al. Galectin-3: one molecule for an alphabet of diseases, from a to z. Int J Mol Sci (2018) 19:379. doi: 10.3390/ijms19020379
138. Hammad H, Plantinga M, Deswarte K, Pouliot P, Willart MA, Kool M, et al. Inflammatory dendritic cells–not basophils–are necessary and sufficient for induction of Th2 immunity to inhaled house dust mite allergen. J Exp Med (2010) 207:2097–111. doi: 10.1084/jem.20101563
139. Hubner MP, Larson D, Torrero MN, Mueller E, Shi Y, Killoran KE, et al. Anti-FcepsilonR1 antibody injections activate basophils and mast cells and delay type 1 diabetes onset in NOD mice. Clin Immunol (2011) 141:205–17. doi: 10.1016/j.clim.2011.08.004
140. Lucarini V, Ziccheddu G, Macchia I, La Sorsa V, Peschiaroli F, Buccione C, et al. IL-33 restricts tumor growth and inhibits pulmonary metastasis in melanoma-bearing mice through eosinophils. Oncoimmunology (2017) 6:e1317420. doi: 10.1080/2162402X.2017.1317420
141. Andreone S, Spadaro F, Buccione C, Mancini J, Tinari A, Sestili P, et al. IL-33 promotes CD11b/CD18-mediated adhesion of eosinophils to cancer cells and synapse-polarized degranulation leading to tumor cell killing. Cancers (Basel) (2019) 11:E1664. doi: 10.3390/cancers11111664
142. Marone G, Gambardella AR, Mattei F, Mancini J, Schiavoni G, Varricchi G. Basophils in tumor microenvironment and surroundings. Adv Exp Med Biol (2020) 1224:21–34. doi: 10.1007/978-3-030-35723-8_2
143. Silver MR, Margulis A, Wood N, Goldman SJ, Kasaian M. And chaudhary, d., IL-33 synergizes with IgE-dependent and IgE-independent agents to promote mast cell and basophil activation. Inflammation Res (2010) 59:207–18. doi: 10.1007/s00011-009-0088-5
144. Smithgall MD, Comeau MR, Yoon BR, Kaufman D, Armitage R. And smith, D.E., IL-33 amplifies both Th1- and Th2-type responses through its activity on human basophils, allergen-reactive Th2 cells, iNKT and NK cells. Int Immunol (2008) 20:1019–30. doi: 10.1093/intimm/dxn060
145. Suzukawa M, Iikura M, Koketsu R, Nagase H, Tamura C, Komiya A, et al. An IL-1 cytokine member, IL-33, induces human basophil activation via its ST2 receptor. J Immunol (2008) 181:5981–9. doi: 10.4049/jimmunol.181.9.5981
146. Cerny-Reiterer S, Ghanim V, Hoermann G, Aichberger KJ, Herrmann H, Muellauer L, et al. Identification of basophils as a major source of hepatocyte growth factor in chronic myeloid leukemia: a novel mechanism of BCR-ABL1-independent disease progression. Neoplasia (2012) 14:572–84. doi: 10.1593/neo.12724
147. Varricchi G, Poto R, Ferrara AL, Gambino G, Marone G, Rengo G, et al. Angiopoietins, vascular endothelial growth factors and secretory phospholipase A(2) in heart failure patients with preserved ejection fraction. Eur J Intern Med (2022) 106:111–9. doi: 10.1016/j.ejim.2022.10.014
148. Varricchi G, Loffredo S, Bencivenga L, Ferrara AL, Gambino G, Ferrara N, et al. Angiopoietins, vascular endothelial growth factors and secretory phospholipase A(2) in ischemic and non-ischemic heart failure. J Clin Med (2020) 9:1928. doi: 10.3390/jcm9061928
149. Wong CK, Cao J, Yin YB, Lam CW. Interleukin-17A activation on bronchial epithelium and basophils: a novel inflammatory mechanism. Eur Respir J (2010) 35:883–93. doi: 10.1183/09031936.00088309
150. Jayatilaka H, Tyle P, Chen JJ, Kwak M, Ju J, Kim HJ, et al. Synergistic IL-6 and IL-8 paracrine signalling pathway infers a strategy to inhibit tumour cell migration. Nat Commun (2017) 8:15584. doi: 10.1038/ncomms15584
151. Schroeder JT, Adeosun AA, Bieneman AP. Epithelial cell-associated galectin-3 activates human dendritic cell subtypes for pro-inflammatory cytokines. Front Immunol (2020) 11:524826. doi: 10.3389/fimmu.2020.524826
152. Todoric J, Antonucci L, Karin M. Targeting inflammation in cancer prevention and therapy. Cancer Prev Res (Phila) (2016) 9:895–905. doi: 10.1158/1940-6207.CAPR-16-0209
153. Poto R, Troiani T, Criscuolo G, Marone G, Ciardiello F, Tocchetti CG, et al. Holistic approach to immune checkpoint inhibitor-related adverse events. Front Immunol (2022) 13:804597. doi: 10.3389/fimmu.2022.804597
154. Bax HJ, Chauhan J, Stavraka C, Khiabany A, Nakamura M, Pellizzari G, et al. Basophils from cancer patients respond to immune stimuli and predict clinical outcome. Cells (2020) 9:E1631. doi: 10.3390/cells9071631
155. Detoraki A, Staiano RI, Granata F, Giannattasio G, Prevete N, de Paulis A, et al. Vascular endothelial growth factors synthesized by human lung mast cells exert angiogenic effects. J Allergy Clin Immunol (2009) 123:1142–9,1149.e1-5. doi: 10.1016/j.jaci.2009.01.044
156. Granata F, Frattini A, Loffredo S, Staiano RI, Petraroli A, Ribatti D, et al. Production of vascular endothelial growth factors from human lung macrophages induced by group IIA and group X secreted phospholipases A2. J Immunol (2010) 184:5232–41. doi: 10.4049/jimmunol.0902501
157. Braile M, Fiorelli A, Sorriento D, Di Crescenzo RM, Galdiero MR, Marone G, et al. Human lung-resident macrophages express and are targets of thymic stromal lymphopoietin in the tumor microenvironment. Cells (2021) 10:2012. doi: 10.3390/cells10082012
158. Varricchi G, Loffredo S, Galdiero MR, Marone G, Cristinziano L, Granata F. Innate effector cells in angiogenesis and lymphangiogenesis. Curr Opin Immunol (2018) 53:152–60. doi: 10.1016/j.coi.2018.05.002
159. Loffredo S, Borriello F, Iannone R, Ferrara AL, Galdiero MR, Gigantino V, et al. Group V secreted phospholipase A2 induces the release of proangiogenic and antiangiogenic factors by human neutrophils. Front Immunol (2017) 8:443. doi: 10.3389/fimmu.2017.00443
160. Sharma M, Das M, Stephen-Victor E, Galeotti C, Karnam A, Maddur MS, et al. Regulatory T cells induce activation rather than suppression of human basophils. Sci Immunol (2018) 3:eaan0829. doi: 10.1126/sciimmunol.aan0829
161. Plaut M, Pierce JH, Watson CJ, Hanley-Hyde J, Nordan RP, Paul WE. Mast cell lines produce lymphokines in response to cross-linkage of fc epsilon RI or to calcium ionophores. Nature (1989) 339:64–7. doi: 10.1038/339064a0
162. Afferni C, Buccione C, Andreone S, Galdiero MR, Varricchi G, Marone G, et al. The pleiotropic immunomodulatory functions of IL-33 and its implications in tumor immunity. Front Immunol (2018) 9:2601. doi: 10.3389/fimmu.2018.02601
163. Piliponsky AM, Shubin NJ, Lahiri AK, Truong P, Clauson M, Niino K, et al. Basophil-derived tumor necrosis factor can enhance survival in a sepsis model in mice. Nat Immunol (2019) 20:129–40. doi: 10.1038/s41590-018-0288-7
164. Chen K, Xu W, Wilson M, He B, Miller NW, Bengten E, et al. Immunoglobulin d enhances immune surveillance by activating antimicrobial, proinflammatory and b cell-stimulating programs in basophils. Nat Immunol (2009) 10:889–98. doi: 10.1038/ni.1748
165. Falkencrone S, Poulsen LK, Bindslev-Jensen C, Woetmann A, Odum N, Poulsen BC, et al. IgE-mediated basophil tumour necrosis factor alpha induces matrix metalloproteinase-9 from monocytes. Allergy (2013) 68:614–20. doi: 10.1111/all.12143
166. Tschopp CM, Spiegl N, Didichenko S, Lutmann W, Julius P, Virchow JC, et al. A novel mediator of allergic inflammation: its induction and release in blood basophils and human asthma. Blood (2006) 108:2290–9. doi: 10.1182/blood-2006-03-010348
167. Voskoboinik I, Whisstock JC, Trapani JA. Perforin and granzymes: function, dysfunction and human pathology. Nat Rev Immunol (2015) 15:388–400. doi: 10.1038/nri3839
168. Ishizaka K, Ishizaka T, Hornbrook MM. Physicochemical properties of reaginic antibody. v. correlation of reaginic activity wth gamma-e-globulin antibody. J Immunol (1966) 97:840–53.
169. Johansson SG. Raised levels of a new immunoglobulin class (IgND) in asthma. Lancet (1967) 2:951–3. doi: 10.1016/s0140-6736(67)90792-1
170. Marone G, Galdiero MR, Pecoraro A, Pucino V, Criscuolo G, Triassi M, et al. Prostaglandin D(2) receptor antagonists in allergic disorders: safety, efficacy, and future perspectives. Expert Opin Investig Drugs (2019) 28:73–84. doi: 10.1080/13543784.2019.1555237
171. Permin H, Wiik A. The prevalence of IgE antinuclear antibodies in rheumatoid arthritis and systemic lupus erythematosus. Acta pathologica et microbiologica Scandinavica. Section C. Immunology (1978) 86C:245–9. doi: 10.1111/j.1699-0463.1978.tb02587.x
172. La Cava A. Overview of the pathogenesis of systemic lupus erythematosus. In: Systemic lupus erythematosus (Second Edition)Basic, applied and clinical aspects. Academic Press, Cambridge (2021). p. 69–75. Chapter 9.
173. Atta AM, Sousa CP, Carvalho EM, Sousa-Atta ML. Immunoglobulin e and systemic lupus erythematosus. Braz J Med Biol Res = Rev Bras pesquisas medicas e biologicas (2004) 37:1497–501. doi: 10.1590/s0100-879x2004001000008
174. Liphaus BL, Jesus AA, Silva CA, Coutinho A, Carneiro-Sampaio M. Increased IgE serum levels are unrelated to allergic and parasitic diseases in patients with juvenile systemic lupus erythematosus. Clinics (2012) 67:1275–80. doi: 10.6061/clinics/2012(11)09
175. Rebhun J, Quismorio F Jr., Dubois E, Heiner DC. Systemic lupus erythematosus activity and IgE. Ann Allergy (1983) 50:34–6.
176. Zhu H, Luo H, Yan M, Zuo X, Li QZ. Autoantigen microarray for high-throughput autoantibody profiling in systemic lupus erythematosus. Genomics Proteomics Bioinf (2015) 13:210–8. doi: 10.1016/j.gpb.2015.09.001
177. Dema B, Pellefigues C, Hasni S, Gault N, Jiang C, Ricks TK, et al. Autoreactive IgE is prevalent in systemic lupus erythematosus and is associated with increased disease activity and nephritis. PloS One (2014) 9:e90424. doi: 10.1371/journal.pone.0090424
178. Henault J, Riggs JM, Karnell JL, Liarski VM, Li J, Shirinian L, et al. Self-reactive IgE exacerbates interferon responses associated with autoimmunity. Nat Immunol (2016) 17:196–203. doi: 10.1038/ni.3326
179. Rhyner C, Daigle I, Crameri R. Auto-reactive IgE responses to acidic ribosomal P(2) protein in systemic lupus erythematosus. Allergy (2011) 66:1127–9. doi: 10.1111/j.1398-9995.2011.02581.x
180. Atta AM, Santiago MB, Guerra FG, Pereira MM, Sousa Atta ML. Autoimmune response of IgE antibodies to cellular self-antigens in systemic lupus erythematosus. Int Arch Allergy Immunol (2010) 152:401–6. doi: 10.1159/000288293
181. Dema B, Charles N, Pellefigues C, Ricks TK, Suzuki R, Jiang C, et al. Immunoglobulin e plays an immunoregulatory role in lupus. J Exp Med (2014) 211:2159–68. doi: 10.1084/jem.20140066
182. Barone C, Bartoloni C, Gentiloni N, Grieco A, Flamini G. Systemic lupus erythematosus with only IgE-class antinuclear antibodies. Arthritis rheumatism (1981) 24:1441–3. doi: 10.1002/art.1780241120
183. Dema B, Lamri Y, Pellefigues C, Pacreau E, Saidoune F, Bidault C, et al. Basophils contribute to pristane-induced lupus-like nephritis model. Sci Rep (2017) 7:7969. doi: 10.1038/s41598-017-08516-7
184. Pellefigues C, Charles N. The deleterious role of basophils in systemic lupus erythematosus. Curr Opin Immunol (2013) 25:704–11. doi: 10.1016/j.coi.2013.10.003
185. Pellefigues C, Dema B, Lamri Y, Saidoune F, Chavarot N, Loheac C, et al. Prostaglandin D(2) amplifies lupus disease through basophil accumulation in lymphoid organs. Nat Commun (2018) 9:725. doi: 10.1038/s41467-018-03129-8
186. Sanjuan MA, Sagar D, Kolbeck R. Role of IgE in autoimmunity. J Allergy Clin Immunol (2016) 137:1651–61. doi: 10.1016/j.jaci.2016.04.007
187. Alivernini S, Firestein GS, McInnes IB. The pathogenesis of rheumatoid arthritis. Immunity (2022) 55:2255–70. doi: 10.1016/j.immuni.2022.11.009
188. Schuerwegh AJ, Ioan-Facsinay A, Dorjee AL, Roos J, Bajema IM, van der Voort EI, et al. Evidence for a functional role of IgE anticitrullinated protein antibodies in rheumatoid arthritis. Proc Natl Acad Sci USA (2010) 107:2586–91. doi: 10.1073/pnas.0913054107
189. McCombe PA, Greer JM. Effects of biological sex and pregnancy in experimental autoimmune encephalomyelitis: it's complicated. Front Immunol (2022) 13:1059833. doi: 10.3389/fimmu.2022.1059833
190. Pollinger B, Krishnamoorthy G, Berer K, Lassmann H, Bosl MR, Dunn R, et al. Spontaneous relapsing-remitting EAE in the SJL/J mouse: MOG-reactive transgenic T cells recruit endogenous MOG-specific b cells. J Exp Med (2009) 206:1303–16. doi: 10.1084/jem.20090299
191. Lock C, Hermans G, Pedotti R, Brendolan A, Schadt E, Garren H, et al. Gene-microarray analysis of multiple sclerosis lesions yields new targets validated in autoimmune encephalomyelitis. Nat Med (2002) 8:500–8. doi: 10.1038/nm0502-500
192. Kebir H, Kreymborg K, Ifergan I, Dodelet-Devillers A, Cayrol R, Bernard M, et al. Human TH17 lymphocytes promote blood-brain barrier disruption and central nervous system inflammation. Nat Med (2007) 13:1173–5. doi: 10.1038/nm1651
193. Stockinger B, Omenetti S. The dichotomous nature of T helper 17 cells. Nature reviews. Immunology (2017) 17:535–44. doi: 10.1038/nri.2017.50
194. Mills KHG. IL-17 and IL-17-producing cells in protection versus pathology. Nat Rev Immunol (2023) 23:38–54. doi: 10.1038/s41577-022-00746-9
195. Venables PJ. Mixed connective tissue disease. Lupus (2006) 15:132–7. doi: 10.1191/0961203306lu2283rr
196. Ungprasert P, Crowson CS, Chowdhary VR, Ernste FC, Moder KG, Matteson EL. Epidemiology of mixed connective tissue disease, 1985-2014: a population-based study. Arthritis Care Res (Hoboken) (2016) 68:1843–8. doi: 10.1002/acr.22872
197. Sapkota B, Al Khalili Y. Mixed connective tissue disease. Treasure Island, FL: StatPearls (2020).
198. Hof D, Cheung K, de Rooij DJ, van den Hoogen FH, Pruijn GJ, van Venrooij WJ, et al. Autoantibodies specific for apoptotic U1-70K are superior serological markers for mixed connective tissue disease. Arthritis Res Ther (2005) 7:R302–9. doi: 10.1186/ar1490
199. Greidinger EL, Zang YJ, Jaimes K, Martinez L, Nassiri M, Hoffman RW. CD4+ T cells target epitopes residing within the RNA-binding domain of the U1-70-kDa small nuclear ribonucleoprotein autoantigen and have restricted TCR diversity in an HLA-DR4-transgenic murine model of mixed connective tissue disease. J Immunol (2008) 180:8444–54. doi: 10.4049/jimmunol.180.12.8444
200. Enomoto K, Takada T, Suzuki E, Ishida T, Moriyama H, Ooi H, et al. Bronchoalveolar lavage fluid cells in mixed connective tissue disease. Respirology (2003) 8:149–56. doi: 10.1046/j.1440-1843.2003.00445.x
201. Paradowska-Gorycka A. U1-RNP and TLR receptors in the pathogenesis of mixed connective tissue diseasePart i. the U1-RNP complex and its biological significance in the pathogenesis of mixed connective tissue disease. Reumatologia (2015) 53:94–100. doi: 10.5114/reum.2015.51509
202. Greidinger EL, Foecking MF, Magee J, Wilson L, Ranatunga S, Ortmann RA, et al. A major b cell epitope present on the apoptotic but not the intact form of the U1-70-kDa ribonucleoprotein autoantigen. J Immunol (2004) 172:709–16. doi: 10.4049/jimmunol.172.1.709
203. Okawa-Takatsuji M, Aotsuka S, Uwatoko S, Takaono M, Iwasaki K, Kinoshita M, et al. Endothelial cell-binding activity of anti-U1-ribonucleoprotein antibodies in patients with connective tissue diseases. Clin Exp Immunol (2001) 126:345–54. doi: 10.1046/j.1365-2249.2001.01669.x
204. Greidinger EL, Zang Y, Jaimes K, Hogenmiller S, Nassiri M, Bejarano P, et al. A murine model of mixed connective tissue disease induced with U1 small nuclear RNP autoantigen. Arthritis rheumatism (2006) 54:661–9. doi: 10.1002/art.21566
205. Kamisawa T, Zen Y, Pillai S, Stone JH. IgG4-related disease. Lancet (2015) 385:1460–71. doi: 10.1016/S0140-6736(14)60720-0
206. Floreani A, Okazaki K, Uchida K, Gershwin ME. IgG4-related disease: changing epidemiology and new thoughts on a multisystem disease. J Trans Autoimmun (2021) 4:100074. doi: 10.1016/j.jtauto.2020.100074
207. Della-Torre E, Lanzillotta M, Doglioni C. Immunology of IgG4-related disease. Clin Exp Immunol (2015) 181:191–206. doi: 10.1111/cei.12641
208. Watanabe T, Minaga K, Kamata K, Kudo M, Strober W. Mechanistic insights into autoimmune pancreatitis and IgG4-related disease. Trends Immunol (2018) 39:874–89. doi: 10.1016/j.it.2018.09.005
209. Kamisawa T, Funata N, Hayashi Y, Eishi Y, Koike M, Tsuruta K, et al. A new clinicopathological entity of IgG4-related autoimmune disease. J Gastroenterol (2003) 38:982–4. doi: 10.1007/s00535-003-1175-y
210. Zen Y, Fujii T, Harada K, Kawano M, Yamada K, Takahira M, et al. Th2 and regulatory immune reactions are increased in immunoglobin G4-related sclerosing pancreatitis and cholangitis. Hepatology (2007) 45:1538–46. doi: 10.1002/hep.21697
211. Kusuda T, Uchida K, Miyoshi H, Koyabu M, Satoi S, Takaoka M, et al. Involvement of inducible costimulator- and interleukin 10-positive regulatory T cells in the development of IgG4-related autoimmune pancreatitis. Pancreas (2011) 40:1120–30. doi: 10.1097/MPA.0b013e31821fc796
212. Tsuboi H, Matsuo N, Iizuka M, Tsuzuki S, Kondo Y, Tanaka A, et al. Analysis of IgG4 class switch-related molecules in IgG4-related disease. Arthritis Res Ther (2012) 14:R171. doi: 10.1186/ar3924
213. Takeuchi M, Sato Y, Ohno K, Tanaka S, Takata K, Gion Y, et al. T Helper 2 and regulatory T-cell cytokine production by mast cells: a key factor in the pathogenesis of IgG4-related disease. Mod Pathol (2014) 27:1126–36. doi: 10.1038/modpathol.2013.236
214. Watanabe T, Yamashita K, Sakurai T, Kudo M, Shiokawa M, Uza N, et al. Toll-like receptor activation in basophils contributes to the development of IgG4-related disease. J Gastroenterol (2013) 48:247–53. doi: 10.1007/s00535-012-0626-8
215. Watanabe T, Yamashita K, Fujikawa S, Sakurai T, Kudo M, Shiokawa M, et al. Involvement of activation of toll-like receptors and nucleotide-binding oligomerization domain-like receptors in enhanced IgG4 responses in autoimmune pancreatitis. Arthritis rheumatism (2012) 64:914–24. doi: 10.1002/art.33386
216. Yanagawa M, Uchida K, Ando Y, Tomiyama T, Yamaguchi T, Ikeura T, et al. Basophils activated via TLR signaling may contribute to pathophysiology of type 1 autoimmune pancreatitis. J Gastroenterol (2018) 53:449–60. doi: 10.1007/s00535-017-1390-6
217. Adeloye D, Song P, Zhu Y, Campbell H, Sheikh A, Rudan I, et al. Global, regional, and national prevalence of, and risk factors for, chronic obstructive pulmonary disease (COPD) in 2019: a systematic review and modelling analysis. Lancet Respir Med (2022) 10:447–58. doi: 10.1016/S2213-2600(21)00511-7
218. Poto R, Criscuolo G, Marone G, Brightling CE, Varricchi G. Human lung mast cells: therapeutic implications in asthma. Int J Mol Sci (2022) 23:14466. doi: 10.3390/ijms232214466
219. Jogdand P, Siddhuraj P, Mori M, Sanden C, Jonsson J, Walls AF, et al. Eosinophils, basophils and type 2 immune microenvironments in COPD-affected lung tissue. Eur Respir J (2020) 55:1900110. doi: 10.1183/13993003.00110-2019
220. Berlin DA, Gulick RM, Martinez FJ. Severe covid-19. N Engl J Med (2020) 383:2451–60. doi: 10.1056/NEJMcp2009575
221. Bencivenga L, Rengo G, Varricchi G. Elderly at time of COronaVIrus disease 2019 (COVID-19): possible role of immunosenescence and malnutrition. GeroScience (2020) 42:1089–92. doi: 10.1007/s11357-020-00218-9
222. Palestra F, Poto R, Ciardi R, Opromolla G, Secondo A, Tedeschi V, et al. SARS-CoV-2 spike protein activates human lung macrophages. Int J Mol Sci (2023) 24:3036.
223. Mantovani A, Cassatella MA, Costantini C, Jaillon S. Neutrophils in the activation and regulation of innate and adaptive immunity. Nat Rev Immunol (2011) 11:519–31. doi: 10.1038/nri3024
224. Schroeder JT, Bieneman AP. The S1 subunit of the SARS-CoV-2 spike protein activates human monocytes to produce cytokines linked to COVID-19: relevance to galectin-3. Front Immunol (2022) 13:831763. doi: 10.3389/fimmu.2022.831763
225. Bonam SR, Chauvin C, Levillayer L, Mathew MJ, Sakuntabhai A, Bayry J. SARS-CoV-2 induces cytokine responses in human basophils. Front Immunol (2022) 13:838448. doi: 10.3389/fimmu.2022.838448
226. Karasuyama H, Miyake K, Yoshikawa S, Yamanishi Y. Multifaceted roles of basophils in health and disease. J Allergy Clin Immunol (2018) 142:370–80. doi: 10.1016/j.jaci.2017.10.042
227. Rosenberg HF, Dyer KD, Domachowske JB. Eosinophils and their interactions with respiratory virus pathogens. Immunol Res (2009) 43:128–37. doi: 10.1007/s12026-008-8058-5
228. Patella V, Florio G, Petraroli A, Marone G. HIV-1 gp120 induces IL-4 and IL-13 release from human fc epsilon RI+ cells through interaction with the VH3 region of IgE. J Immunol (2000) 164:589–95. doi: 10.4049/jimmunol.164.2.589
229. Hsieh JT, Rathore APS, Soundarajan G. And St John, A.L., Japanese encephalitis virus neuropenetrance is driven by mast cell chymase. Nat Commun (2019) 10:706. doi: 10.1038/s41467-019-08641-z
230. Lourda M, Dzidic M, Hertwig L, Bergsten H, Palma Medina LM, Sinha I, et al. High-dimensional profiling reveals phenotypic heterogeneity and disease-specific alterations of granulocytes in COVID-19. Proc Natl Acad Sci U.S.A. (2021) 118:e2109123118. doi: 10.1073/pnas.2109123118
231. Laing AG, Lorenc A, Del Molino Del Barrio I, Das A, Fish M, Monin L, et al. A dynamic COVID-19 immune signature includes associations with poor prognosis. Nat Med (2020) 26:1623–35. doi: 10.1038/s41591-020-1038-6
232. Vitte J, Diallo AB, Boumaza A, Lopez A, Michel M, Allardet-Servent J, et al. A granulocytic signature identifies COVID-19 and its severity. J Infect Dis (2020) 222:1985–96. doi: 10.1093/infdis/jiaa591
233. Bonam SR, Chauvin C, Mathew MJ, Bayry J. IFN-gamma induces PD-L1 expression in primed human basophils. Cells (2022) 11:801. doi: 10.3390/cells11050801
234. Sun Y, Zhou J, Ye K. White blood cells and severe COVID-19: a mendelian randomization study. J Pers Med (2021) 11:195. doi: 10.3390/jpm11030195
235. Kaser A, Zeissig S, Blumberg RS. Inflammatory bowel disease. Annu Rev Immunol (2010) 28:573–621. doi: 10.1146/annurev-immunol-030409-101225
236. Abraham C, Abreu MT, Turner JR. Pattern recognition receptor signaling and cytokine networks in microbial defenses and regulation of intestinal barriers: implications for inflammatory bowel disease. Gastroenterology (2022) 162:1602–1616 e6. doi: 10.1053/j.gastro.2021.12.288
237. Chapuy L, Bsat M, Mehta H, Rubio M, Wakahara K, Van VQ, et al. Basophils increase in crohn disease and ulcerative colitis and favor mesenteric lymph node memory TH17/TH1 response. J Allergy Clin Immunol (2014) 134:978–81.e1. doi: 10.1016/j.jaci.2014.05.025
238. Siracusa MC, Kim BS, Spergel JM, Artis D. Basophils and allergic inflammation. J Allergy Clin Immunol (2013) 132:789–801;quiz 788. doi: 10.1016/j.jaci.2013.07.046
239. Varricchi G, Poto R, Ianiro G, Punziano A, Marone G, Gasbarrini A, et al. Gut microbiome and common variable immunodeficiency: few certainties and many outstanding questions. Front Immunol (2021) 12:712915. doi: 10.3389/fimmu.2021.712915
240. Turpin W, Lee SH, Raygoza Garay JA, Madsen KL, Meddings JB, Bedrani L, et al. Increased intestinal permeability is associated with later development of crohn's disease. Gastroenterology (2020) 159:2092–2100 e5. doi: 10.1053/j.gastro.2020.08.005
241. Andreone S, Gambardella AR, Mancini J, Loffredo S, Marcella S, La Sorsa V, et al. Anti-tumorigenic activities of IL-33: a mechanistic insight. Front Immunol (2020) 11:571593. doi: 10.3389/fimmu.2020.571593
242. Schiering C, Krausgruber T, Chomka A, Frohlich A, Adelmann K, Wohlfert EA, et al. The alarmin IL-33 promotes regulatory T-cell function in the intestine. Nature (2014) 513:564–8. doi: 10.1038/nature13577
243. Ebina-Shibuya R, Leonard WJ. Role of thymic stromal lymphopoietin in allergy and beyond. Nat Rev Immunol (2023) 23:24–37. doi: 10.1038/s41577-022-00735-y
244. Feagan BG, Sandborn WJ, D'Haens G, Panes J, Kaser A, Ferrante M, et al. Induction therapy with the selective interleukin-23 inhibitor risankizumab in patients with moderate-to-severe crohn's disease: a randomised, double-blind, placebo-controlled phase 2 study. Lancet (2017) 389:1699–709. doi: 10.1016/S0140-6736(17)30570-6
245. White J, Dubey S. Eosinophilic granulomatosis with polyangiitis: a review. Autoimmun Rev (2023) 22:103219. doi: 10.1016/j.autrev.2022.103219
246. Dong ZM, Lin E, Wechsler ME, Weller PF, Klion AD, Bochner BS, et al. Pulmonary eosinophilic granulomatosis with polyangiitis has IgG4 plasma cells and immunoregulatory features. Am J Pathol (2020) 190:1438–48. doi: 10.1016/j.ajpath.2020.03.005
247. Jachiet M, Samson M, Cottin V, Kahn JE, Le Guenno G, Bonniaud P, et al. Anti-IgE monoclonal antibody (Omalizumab) in refractory and relapsing eosinophilic granulomatosis with polyangiitis (Churg-strauss): data on seventeen patients. Arthritis Rheumatol (2016) 68:2274–82. doi: 10.1002/art.39663
248. Detoraki A, Di Capua L, Varricchi G, Genovese A, Marone G, Spadaro G. Omalizumab in patients with eosinophilic granulomatosis with polyangiitis: a 36-month follow-up study. J Asthma (2016) 53:201–6. doi: 10.3109/02770903.2015.1081700
249. Basta F, Mazzuca C, Nucera E, Schiavino D, Afeltra A, Antonelli Incalzi R. Omalizumab in eosinophilic granulomatosis with polyangiitis: friend or foe? a systematic literature review. Clin Exp Rheumatol (2020) 38 Suppl 124:214–20.
250. MacGlashan D Jr., Saini S, Schroeder JT. Response of peripheral blood basophils in subjects with chronic spontaneous urticaria during treatment with omalizumab. J Allergy Clin Immunol (2021) 147:2295–2304 e12. doi: 10.1016/j.jaci.2021.02.039
251. Straumann A, Katzka DA. Diagnosis and treatment of eosinophilic esophagitis. Gastroenterology (2018) 154:346–59. doi: 10.1053/j.gastro.2017.05.066
252. Avlas S, Shani G, Rhone N, Itan M, Dolitzky A, Hazut I, et al. Epithelial cell-expressed type II IL-4 receptor mediates eosinophilic esophagitis. Allergy (2023) 78:464–76. doi: 10.1111/all.15510
253. Stern E, Taft T, Zalewski A, Gonsalves N, Hirano I. Prospective assessment of disease-specific quality of life in adults with eosinophilic esophagitis. Dis Esophagus (2018) 31. doi: 10.1093/dote/dox128
254. Muir A, Falk GW. Eosinophilic esophagitis: a review. JAMA (2021) 326:1310–8. doi: 10.1001/jama.2021.14920
255. Wen T, Aronow BJ, Rochman Y, Rochman M, Kc K, Dexheimer PJ, et al. Single-cell RNA sequencing identifies inflammatory tissue T cells in eosinophilic esophagitis. J Clin Invest (2019) 129:2014–28. doi: 10.1172/JCI125917
256. Khokhar D, Marella S, Idelman G, Chang JW, Chehade M, Hogan SP. Eosinophilic esophagitis: immune mechanisms and therapeutic targets. Clin Exp Allergy (2022) 52:1142–56. doi: 10.1111/cea.14196
257. Noti M, Wojno ED, Kim BS, Siracusa MC, Giacomin PR, Nair MG, et al. Thymic stromal lymphopoietin-elicited basophil responses promote eosinophilic esophagitis. Nat Med (2013) 19:1005–13. doi: 10.1038/nm.3281
258. Venturelli N, Lexmond WS, Ohsaki A, Nurko S, Karasuyama H, Fiebiger E, et al. Allergic skin sensitization promotes eosinophilic esophagitis through the IL-33-basophil axis in mice. J Allergy Clin Immunol (2016) 138:1367–1380 e5. doi: 10.1016/j.jaci.2016.02.034
259. Rothenberg ME, Spergel JM, Sherrill JD, Annaiah K, Martin LJ, Cianferoni A, et al. Common variants at 5q22 associate with pediatric eosinophilic esophagitis. Nat Genet (2010) 42:289–91. doi: 10.1038/ng.547
260. Sherrill JD, Gao PS, Stucke EM, Blanchard C, Collins MH, Putnam PE, et al. Variants of thymic stromal lymphopoietin and its receptor associate with eosinophilic esophagitis. J Allergy Clin Immunol (2010) 126:160–5 e3. doi: 10.1016/j.jaci.2010.04.037
261. Ehrlich P. Über die specifischen granulationen des blutes. Arch Anat Physiol Phys Abt (1879), 571–7.
262. Plum T, Wang X, Rettel M, Krijgsveld J, Feyerabend TB, Rodewald HR. Human mast cell proteome reveals unique lineage, putative functions, and structural basis for cell ablation. Immunity (2020) 52:404–416 e5. doi: 10.1016/j.immuni.2020.01.012
263. Varricchi G, de Paulis A, Marone G, Galli SJ. Future needs in mast cell biology. Int J Mol Sci (2019) 20:4397. doi: 10.3390/ijms20184397
264. Gentek R, Ghigo C, Hoeffel G, Bulle MJ, Msallam R, Gautier G, et al. Hemogenic endothelial fate mapping reveals dual developmental origin of mast cells. Immunity (2018) 48:1160–1171.e5. doi: 10.1016/j.immuni.2018.04.025
265. Li Z, Liu S, Xu J, Zhang X, Han D, Liu J, et al. Adult connective tissue-resident mast cells originate from late erythro-myeloid progenitors. Immunity (2018) 49:640–653.e5. doi: 10.1016/j.immuni.2018.09.023
266. Akula S, Paivandy A, Fu Z, Thorpe M, Pejler G, Hellman L. Quantitative in-depth analysis of the mouse mast cell transcriptome reveals organ-specific mast cell heterogeneity. Cells (2020) 9:211. doi: 10.3390/cells9010211
267. Chevrier S, Levine JH, Zanotelli VRT, Silina K, Schulz D, Bacac M, et al. An immune atlas of clear cell renal cell carcinoma. Cell (2017) 169:736–749.e18. doi: 10.1016/j.cell.2017.04.016
268. Balestrieri B, Granata F, Loffredo S, Petraroli A, Scalia G, Morabito P, et al. Phenotypic and functional heterogeneity of low-density and high-density human lung macrophages. Biomedicines (2021) 9:505. doi: 10.3390/biomedicines9050505
269. Zilionis R, Engblom C, Pfirschke C, Savova V, Zemmour D, Saatcioglu HD, et al. Single-cell transcriptomics of human and mouse lung cancers reveals conserved myeloid populations across individuals and species. Immunity (2019) 50:1317–1334 e10. doi: 10.1016/j.immuni.2019.03.009
270. Xie X, Shi Q, Wu P, Zhang X, Kambara H, Su J, et al. Single-cell transcriptome profiling reveals neutrophil heterogeneity in homeostasis and infection. Nat Immunol (2020) 21:1119–33. doi: 10.1038/s41590-020-0736-z
271. Veglia F, Hashimoto A, Dweep H, Sanseviero E, De Leo A, Tcyganov E, et al. Analysis of classical neutrophils and polymorphonuclear myeloid-derived suppressor cells in cancer patients and tumor-bearing mice. J Exp Med (2021) 218:e20201803. doi: 10.1084/jem.20201803
272. Fridlender ZG, Sun J, Kim S, Kapoor V, Cheng G, Ling L, et al. Polarization of tumor-associated neutrophil phenotype by TGF-beta: "N1" versus "N2" TAN. Cancer Cell (2009) 16:183–94. doi: 10.1016/j.ccr.2009.06.017
273. Mesnil C, Raulier S, Paulissen G, Xiao X, Birrell MA, Pirottin D, et al. Lung-resident eosinophils represent a distinct regulatory eosinophil subset. J Clin Invest (2016) 126:3279–95. doi: 10.1172/JCI85664
274. Gurtner A, Borrelli C, Gonzalez-Perez I, Bach K, Acar IE, Nunez NG, et al. Active eosinophils regulate host defence and immune responses in colitis. Nature (2023) 615:151–7. doi: 10.1038/s41586-022-05628-7
275. Vivanco Gonzalez N, Oliveria JP, Tebaykin D, Ivison GT, Mukai K, Tsai MM, et al. Mass cytometry phenotyping of human granulocytes reveals novel basophil functional heterogeneity. iScience (2020) 23:101724. doi: 10.1016/j.isci.2020.101724
276. Pellefigues C, Mehta P, Chappell S, Yumnam B, Old S, Camberis M, et al. Diverse innate stimuli activate basophils through pathways involving syk and IkappaB kinases. Proc Natl Acad Sci U.S.A. (2021) 118:e2019524118. doi: 10.1073/pnas.2019524118
Keywords: alarmins, allergy, autoimmunity, basophil, cancer, COVID-19, myocardial infarction
Citation: Poto R, Loffredo S, Marone G, Di Salvatore A, de Paulis A, Schroeder JT and Varricchi G (2023) Basophils beyond allergic and parasitic diseases. Front. Immunol. 14:1190034. doi: 10.3389/fimmu.2023.1190034
Received: 20 March 2023; Accepted: 14 April 2023;
Published: 02 May 2023.
Edited by:
Christophe Pellefigues, CNRS EMR8252 Centre de Recherche sur l’Inflammation, FranceReviewed by:
Jagadeesh Bayry, Indian Institute of Technology Palakkad, IndiaBernhard F. Gibbs, University of Oldenburg, Germany
Copyright © 2023 Poto, Loffredo, Marone, Di Salvatore, de Paulis, Schroeder and Varricchi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Gilda Varricchi, gildanet@gmail.com; John T. Schroeder, schray@jhmi.edu
†ORCID: Remo Poto, orcid.org/0000-0002-4723-0167
Stefania Loffredo, orcid.org/0000-0002-5871-1898
Gianni Marone, orcid.org/0000-0002-9849-4701
Antonio Di Salvatore, orcid.org/0000-0002-6434-3112
Amato de Paulis, orcid.org/0000-0003-0347-2540
John T. Schroeder, orcid.org/0000-0002-5263-7850
Gilda Varricchi, orcid.org/0000-0002-9285-4657
 Remo Poto
Remo Poto Stefania Loffredo
Stefania Loffredo Gianni Marone
Gianni Marone Antonio Di Salvatore1†
Antonio Di Salvatore1† Amato de Paulis
Amato de Paulis John T. Schroeder
John T. Schroeder Gilda Varricchi
Gilda Varricchi