- 1Department of Pathogen Biology, School of Basic Medicine, Tongji Medical College and State Key Laboratory for Diagnosis and Treatment of Severe Zoonostic Infectious Disease, Huazhong University of Science and Technology, Wuhan, Hubei, China
- 2Cytek Biosciences, R&D Clinical Reagents, Fremont, CA, United States
- 3Department of Immunology, Institute of Biomedical Sciences, University of São Paulo (USP), São Paulo, SP, Brazil
- 4Centre for Immunobiology, Bizard Institute, Queen Mary University of London, London, United Kingdom
- 5Université de Paris, Institut de Recherche Saint-Louis, EMiLy, Paris, France
- 6Laboratory for Cytokine Regulation, Center for Integrative Medical Science (IMS), Rikagaku Kenkyusho, Institute of Physical and Chemical Research (RIKEN) Yokohama Institute, Yokohama, Japan
- 7Department of Ophthalmology, Rigshospitalet Glostrup, Copenhagen, Denmark
- 8Department of Clinical Medicine, University of Copenhagen, Copenhagen, Denmark
- 9Department of Paediatrics and Adolescent Medicine, Li Ka Shing Faculty of Medicine, The University of Hong Kong, Hong Kong, Hong Kong SAR, China
- 10Department of Immunology, Mayo Clinic, Rochester, MN, United States
- 11Division of Rheumatology, Department of Medicine, Mayo Clinic, Rochester, MN, United States
- 12State Key Laboratory of Quality Research in Chinese Medicine, Institute of Chinese Medical Sciences, University of Macau, Taipa, Macao SAR, China
- 13Department of Hematology, The Second Hospital of Anhui Medical University, Hefei, China
Raptor, a key component of mTORC1, is required for recruiting substrates to mTORC1 and contributing to its subcellular localization. Raptor has a highly conserved N-terminus domain and seven WD40 repeats, which interact with mTOR and other mTORC1-related proteins. mTORC1 participates in various cellular events and mediates differentiation and metabolism. Directly or indirectly, many factors mediate the differentiation and function of lymphocytes that is essential for immunity. In this review, we summarize the role of Raptor in lymphocytes differentiation and function, whereby Raptor mediates the secretion of cytokines to induce early lymphocyte metabolism, development, proliferation and migration. Additionally, Raptor regulates the function of lymphocytes by regulating their steady-state maintenance and activation.
Background
Mammalian target of rapamycin (mTOR) is a serine/threonine protein kinase that belongs to the PI3K-related kinase (PIKK) family. mTOR mediates phosphorylation of eukaryotic initiation factor 4E-binding protein 1 (4E-BP1) and p70 S6 kinase (p70S6k), which is essential in regulating daily metabolism, including mediating protein synthesis, cell differentiation, aging, and autophagy (1–4). The phosphatidylinositol-3-kinase (PI3K)-Akt-mTOR signaling pathway is a core pathway in many forms of human cancers (5), in which it is usually activated at abnormal levels. mTOR's role in cancer demonstrates its importance as a therapeutic target for cancer treatment (6–9). However, mTOR plays a vital role in normal physiological conditions as well, where it participates and mediates basic cell metabolism and differentiation (10).
The discovery of mTOR dates back to the 1960s. Its discovery came after that of rapamycin, which was identified as the mammalian target of Rapamycin (mTOR). Rapamycin, discovered in 1962, inhibits the autoimmune response and suppresses tumor growth (11). Rapamycin interacts with mTORC1 (12) and is an inhibitor of Raptor-mTORC1. Rapamycin can bind FKBP12 and form the complex, rapamycin–FKBP12, which is a specific inhibitor of Raptor- mTORC1 (13). The mTOR-FKBP12 complex, discovered in 1992, mediates mTOR anti-proliferative functions (14), and was identified in mammalian cells by genetic screening for Rapamycin-resistance in 1994 (15).
mTOR has two protein complexes and acts as a core component in the mTOR complex 1 (mTORC1) and mTOR complex 2 (mTORC2), which have different functions and are characterized by two different proteins: regulatory-associated protein of mTOR (Raptor) and rapamycin-insensitive companion of mTOR (Rictor) (1, 16). Since mTORC1 but not mTORC2 can be specifically inhibited by rapamycin, most studies have focused on mTORC1 and its role in metabolism. Previous studies have demonstrated mTORC1 as a central regulator of anabolism and catabolism, which features responding to nutrition condition (4). The function of mTORC1 is more focused on cell growth and survival, and mTORC2 is more inclined to cell proliferation and cytoskeletal remodeling. However, deciphering the mechanism of the function of mTORC2 is more challenging for lack of specific pharmacological inhibitors (17). mTORC1 is composed of three main components: mTOR (core component), Raptor (a regulatory protein), mLST8 (mammalian lethal with sec13 protein 8) (10, 18, 19) and two inhibitory subunits: PRAS40 (20), and DEPTOR (21) (Figure 1A).
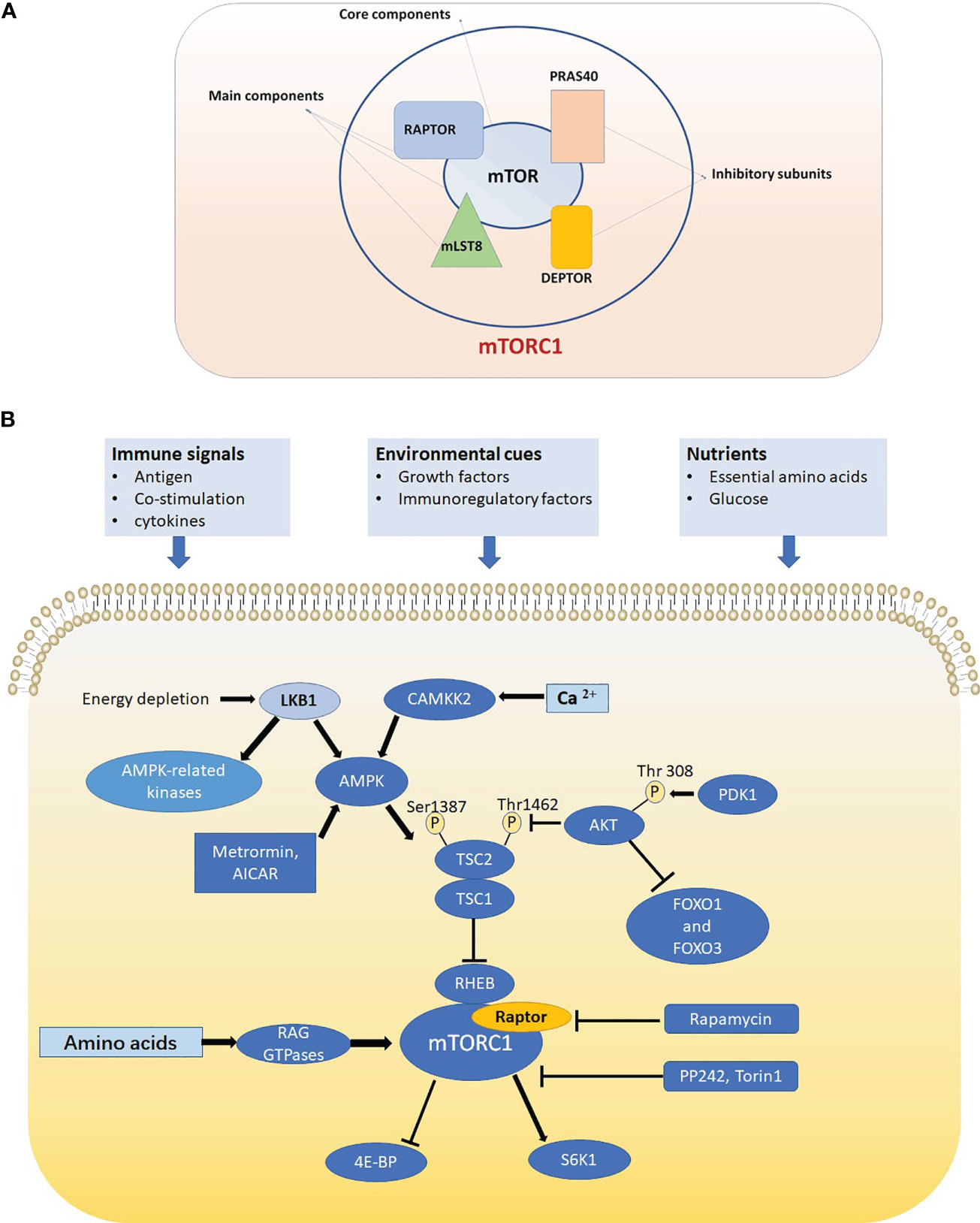
Figure 1 (A) mTORC1 is composed of mTOR, Raptor, mLST8, PRAS40 and DEPTOR. mTOR is the core component of mTORC1. Raptor and mLST8 are the two other main components and PRAS40 DEPTOR are two inhibitory subunits of mTORC1. (B) In T cell, mTORC1 is sensitive to three kinds of signals including immune signals, nutrients and environmental cues and mTORC1 can promote translation initiation and protein synthesis by directly phosphorylating ribosomal protein S6 kinases (S6Ks) and eIF4E-binding proteins (4E-BPs). Raptor-mTORC1 signaling pathway play crucial roles in cell signaling, metabolism and autophagy.
Raptor, a crucial part of mTORC1, recruits substrates to mTORC1 by binding to specific canonical substrates, which contributes to the subcellular localization of mTORC1 (22, 23). Raptor has a conserved domain in the N-terminus and seven WD40 repeats. This distinctive domain is expected to contribute to the interactions with mTOR and mTORC1-associated proteins (1, 19). The official name of Raptor gene is RPTOR, KOG1 as well as Mip1, and the gene ID is 57521. The human RPTOR gene locates in Chromosome 17 and starts from 80544838 to 80966368. The gene structure of mTORC1 and other related molecules has been clearly summarized by Tatebe, H. and Shiozaki, K (24). Growth factors, amino acids, lipids and cholesterol regulate the function of mTORC1 and lysosomes are the primary target (25, 26).
Growth factors such as epidermal growth factor (EGF) and insulin-like growth factor (IGF) bind to their receptors to mediate mTORC1-related signaling pathways like the PI3K-PDK1-Akt signaling pathway. As a core part of mTORC1, Raptor is also a target, which when phosphorylated, activates mTORC1 (27). For example, the activation of the mitogen-activated protein kinase (MAPK) pathway, ERK/RSK/ICK, leads to phosphorylation of Raptor, which activates mTORC1 (28–30). Raptor mediates mTORC1 functions such as cell metabolism, which consequently, affects cell growth and development. There are many studies demonstrating that Raptor/mTORC1 regulates lymphocyte function and differentiation through control of basic metabolism and finally influences the immunity (31, 32). The role of Raptor is different in various kinds of lymphocytes, and this review we systematically summarize the role of Raptor in them.
Raptor’s role in B cells
The developmental process of B cells is tightly regulated, which has many checkpoints during gene arrangement, positive selection and negative selection (31, 33). Looking at the expression of immunoglobulin chains and some cell surface proteins, the development and maturation process of B cells can be divided into the following stages: pre-pro B cell stage, pro B cell stage, early pre B cell stage, late pre B cell stage, immature and mature B cell stage, and the developing positions (BM or fetal liver) of B cell. If errors occur in the differentiation process, clonal deletion will happen (16, 34, 35). In order to avoid this interference when studying the function of Raptor-mTORC1, researchers knocked out the Raptor gene at specific stages. They found that Raptor-mTORC1 not only affects early B cell differentiation stages, but also B cells after the pre-B cell stage. Raptor was found to have many critical roles, including promoting B cells to produce Ig H chain (IgH), contributing to the maintenance of pre-B cell homeostasis and survival, and regulating the intracellular respiration and glycolysis of B cells to promote antibody production (16). In B-cells, Raptor regulates B cell differentiation and function, although it is more critical for regulating differentiation (36). Raptor impacts the differentiation process via cellular bioenergetics in B cells (16, 34). Interleukin-7 (IL–7) is an essential cytokine for B cell maturation (37, 38) and deletion of Raptor causes a decline in the production of IL–7, which causes a blockage in B cell development.
Raptor mediates the transition from pro-B cells to pre-B cells in an IL-7R signaling and Myc expression dependent way
By using Raptorfl/fl mice, and crossing Raptorfl/fl mice with transgenic Mb1-Cre mice to generate B cell lineage Raptor deletion mice, researchers examined the expression levels of mTORC1 pathway associated signaling proteins including Raptor, B220, CD43, HAS, BP1, IgH2, and Ig L chain (IgL) in KO and WT mouse bone marrow. They also aimed to determine the necessary expression conditions in the WT mouse group. The final result was that the expression level of Raptor was higher in large and small pre-B cells, while lower in small dormant immature and mature B cells. Not only the expression of proteins in mTORC1 related signaling pathway such as mTOR and Raptor is at higher levels, but also downstream proteins in the mTORC1 signaling pathway, p-S6R and p-4EBP1, are expressed at higher levels. These results suggest that in pro B and pre B cell stages, the mTORC1 signaling pathway and the expression of Raptor is more active. The reduced Raptor level in the late B cell stages corresponded with IgM and IgD expression (16). Looking further into the increased expression of Raptor in pro B to pre B stages by examining the expression of related proteins in WT and RaptorKO mice revealed that in B cell progenitors of RaptorKO mice, Raptor, mTOR, p-S6K1, p-S6R, and p-4EBP1 all had a significant decrease compared to the WT group. This strongly suggests that the lack of Raptor greatly disrupts the mTORC1 signaling pathway. The researchers also used flow cytometry and other techniques to detect the number of B cells in the RaptorKO group, and found that immature and mature B cells in the bone marrow of the RaptorKO group were largely absent relative to the WT group. Also, it was found that in peripheral tissues, there were decreased quantities of B220+ splenic B cells, B2 cells, and CD5+IgM+ B1a cells (16). Overall, Raptor plays an important role in B cell differentiation and the production of B cell surface markers; and the deficiency of Raptor causes a block in pro B to pre B stages. To explain this further, based on the fact that the proto-oncogene Myc is essential for B cell development (39–41), mTORC1 plays an irreplaceable role in IL-7 induced Myc expression in pro-B cells (42) (Figure 2A). Other studies also demonstrated that deficiency of Raptor inhibits early B cells’ ability to properly respond to early BCR stimulations or express BCR (16), thus, the deficiency of Raptor inhibits the B cell development process.
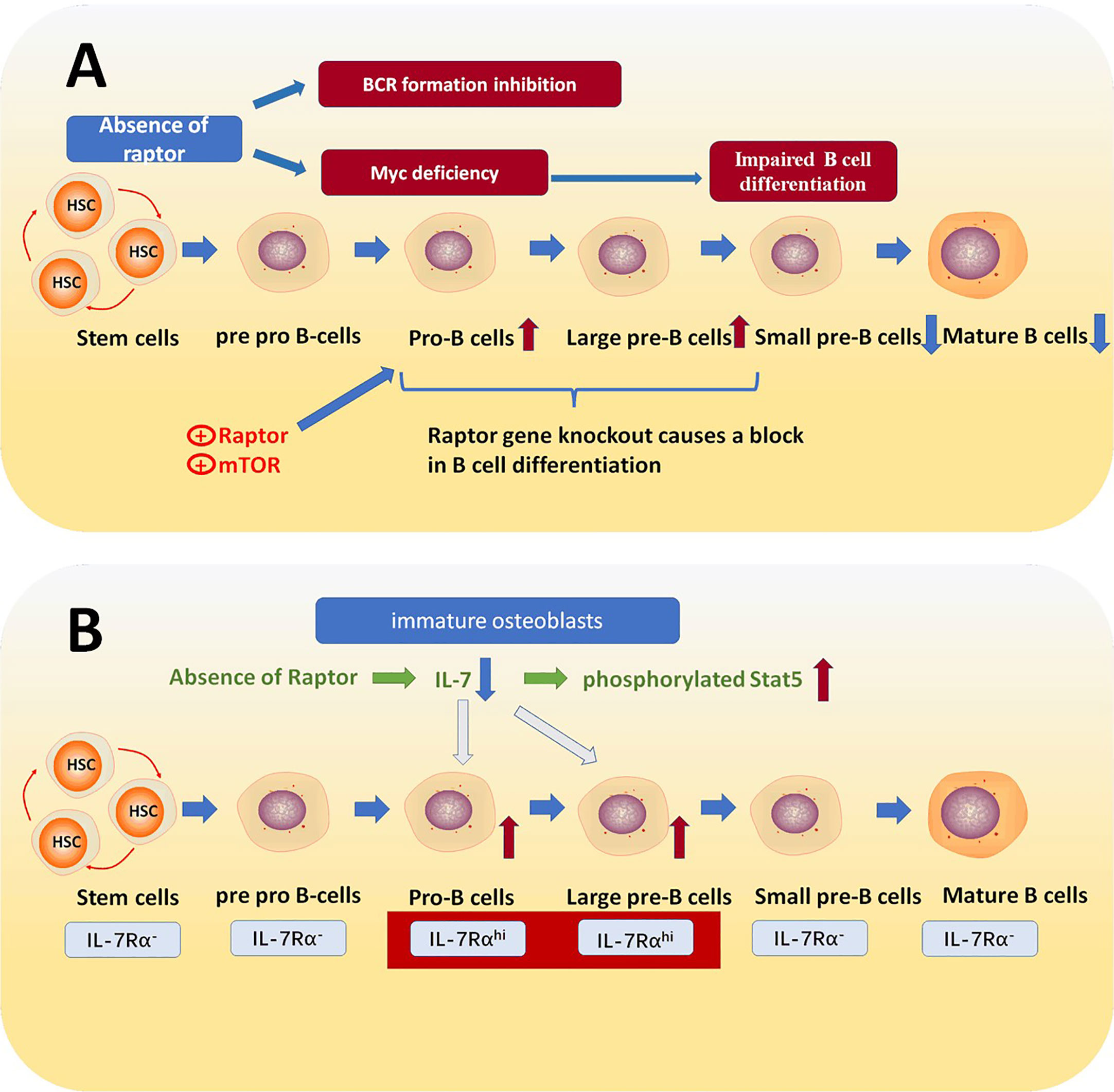
Figure 2 (A) Raptor mediates the transition from pro B cells to pre-B cells. The absence of raptor causes the inactivation of mTORC1, which is a downstream effector of IL-7R signaling and essential for Myc expression and consequently blocks the transition from pro-B cells to pre-B cells. (B) IL-7 receptors are increased in pro-B cell and large pre B cell stages, which is essential for B cell fate. IL-7 is a crucial cytokine in B cell differentiation process, and the absence of Raptor down-regulates the expression of IL-7 in immature osteoblasts, which leads to abnormal IL-7/Stat5 signaling and thereby prevents pro B cells transforming into pre B cells.
Raptor in immature osteoblasts blocks IL- 7 expression and then mediates B cell differentiation
B cells develop and mature in bone marrow (BM), so the bioactive components in the BM microenvironment impacts B cell differentiation (43). Furthermore, it has been already shown that mTORC1 regulates cell metabolism and proliferation as well as differentiation. The blockage of the mTORC1 signaling pathway in bone lineage cells causes reduced bone production in mice (44), and it has also been shown that Raptor deficiency in some osteolineage cells affects B cell differentiation (36). Moreover, early osteoblasts can produce C-X-C motif chemokine 12 (CXCL12) and IL-7, which are necessary in regulating B cell differentiation (45–47).
IL-7 is a crucial cytokine for various signaling pathways. For example, STAT5a/b, PI3 Kinase, STAT5 and SRC kinases can all be activated by IL-7, which is essential for immunity (48). Relevant studies have shown that IL-7 is necessary for the development and maturation of lymphocytes and that IL-7 mediates B cell development via activation of stromal derived factor1 (SDF1), and fms-related tyrosine kinase 3 ligand (FLT3LG) (49, 50). Osterix is a zinc finger-containing transcription factor proved to be essential for osteoblast differentiation (51). Studies have found that Raptor deficiency in Osterix-expressing cells leads to a decreased number of total B cells and suppresses pro-B cell transform into pre-B cells. Furthermore, it was also detected that the apoptosis rate of pre B cells is abnormally higher in Raptor deficiency mice than WT (36). In all, the deficiency of Raptor causes a block in the mTORC1 signaling pathway and inhibits IL-7 expression. Despite other pathways related to IL-7, what has been demonstrated is that Raptor deficiency in mice causes an increased level of phosphorylated Stat5 in pro B cells, which prevents development into pre B cells and then into immature B cells (36, 52, 53) (Figure 2B).
Interestingly, after providing RaptorKO mice with recombinant murine IL-7, the quantity of B cells in all developmental stages is close to normal (36, 54). Additionally, although CXCL12 is essential for B cell maturation and is expressed by osterix-expressing cells too, the quantity of CXCL12 is unaffected under the condition of Raptor gene specific knockout (36).
The role of raptor in T cells
Originating from the bone marrow and mature in the thymus, T cell is an essential part of the immune system to every Individual organism (55). In the thymus, T cells multiply and transit into helper, regulatory, cytotoxic, and memory T cells (56, 57). The transition process can be mediated by multiple factors and one of the factors is mTOR signaling pathway, which is sensitive to three kinds of signals including immune signals, nutrients and environmental cues (4, 58, 59) (Figure 1B). T cells are more dependent on Raptor than B cells. Both CD 4+, CD 8+ subsets and γδ, αβ subsets are regulated by Raptor (60). According to relevant studies, mTOR related signaling pathways regulate T cells’ differentiation and function by mediate their metabolic programing, which has intimate connections with membrane molecules expression and macromolecule synthesis (4, 61–63). CD4+ (helper) T cells play an irreplaceable role in immunity. T helper (Th) cells produce crucial cytokines that activate cytotoxic T cells in response to specific immunological challenges (64). Additionally, the cytokines produced by the Th lineage cells are crucial for immunity (65). As with other lymphocytes, mTORC1 can also regulates T helper cell metabolism, which including mediates the expression of glycolytic enzymes in these cells, thereby influencing their proliferation and function (66). It has been discovered that inhibiting mTOR with rapamycin influences naïve T cells to transform into Th1, Th2, and Th17 cells etc (67–69).
Raptor inhibits memory CD8 T cell differentiation via downstream effectors like S6K1 and eIF4E
CD8+ T cells, also known as cytotoxic T lymphocytes (CTLs), are the core component of cellular immunity. Thus, the activation of CD8 + T cells is the aim of improving many vaccines (70). Rapamycin can suppress interleukin-2-stimulated T cell proliferation. Thus, it is widely used to treat autoimmune diseases, particularly after organ transplantation (3, 12, 71). In a surprising study, after treating acute lymphocytic choriomeningitis virus infected mice with rapamycin, it was found that the amount of virus-specific CD8+ T cells increased. This phenomenon was undoubtedly controversial to previous perceptions. In this study, four biomarkers were used to testify the quantity and quality of memory CD 8 + T cells in rapamycin treated mice, including CD127, CD62L, KLRG1 and Bcl2, which are all relevant to long-lived protective immunity (72, 73). In order to explain this abnormal phenomenon, researchers specifically knocked down Raptor gene using a retrovirus based RNA interference (RNAi) system and demonstrated that rapamycin essentially inhibits memory CD8 T cell differentiation. While particularly in memory CD8 T cells, there is an unusual inhibition of the differentiation process caused by Raptor-mTORC1 signaling pathway intrinsically (60). Researchers finally found that the mechanism of Raptor-mTORC1 signaling pathway inhibition is via S6K1 and eIF4E, which are mTORC1 downstream effectors that down-regulate memory CD 8 + T cell differentiation (Figure 3) (60).
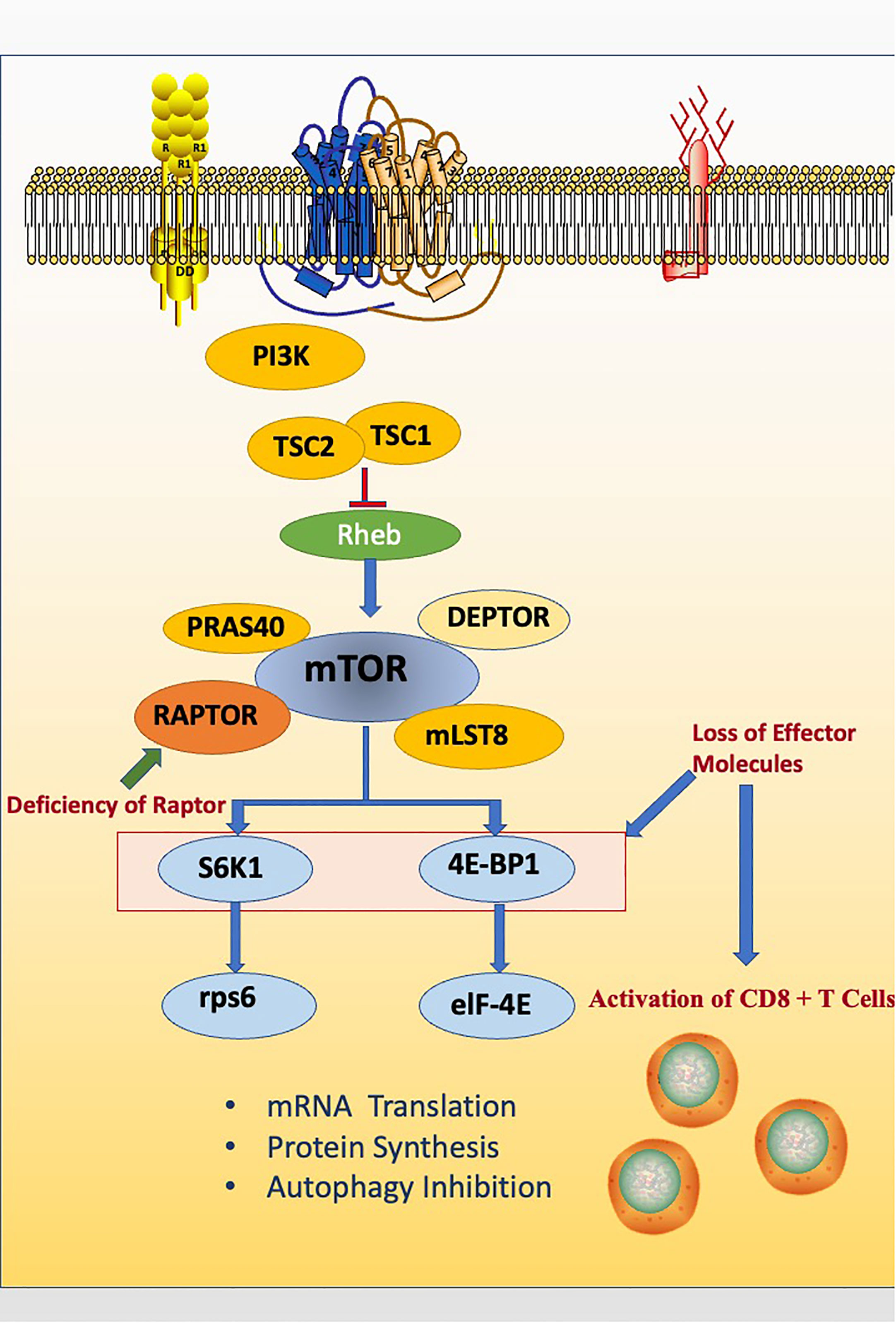
Figure 3 Raptor inhibits memory CD8 T cell differentiation via downstream effectors like S6K1 and eIF4E. The deficiency of Raptor leads to the abnormal activation of CD8 + T cells.
Additionally, other researchers reported that rapamycin can also suppress the proliferation of memory CD8 + T cells in intestinal and vaginal mucosa. Over all, these findings found both a new role of Raptor and rapamycin in lymphocyte functions (74).
WAVE2 maintains T cell homeostasis and function via binding to Raptor and mTOR
By regulating TCR-stimulated actin cytoskeletal dynamics, WASp family verprolin homologous protein 2 (WAVE2) can mediates T cell functions (75). According to some research, WAVE2 is expressed more in hematopoietic cells and the deficiency of WAVE2 causes abnormal activation of T cells. It has also been reported that WAVE2-related signaling pathways play crucial roles in the invasion process and metastasis keeping of cancer cells and may be a novel target for cancer therapy in future (76–78). Scientists have found that WAVE 2 KO mice have severe autoimmune disease and inflammatory response compared to the WT controls (79, 80). The WAVE KO mice suffered from splenomegaly, lymphadenopathy, multi-organ lymphocytic infiltration, and died early. It was also found that AKT-mTOR activation and metabolic reprogramming are dysregulated in WAVE 2 KO mice, which leads to overactivation of T cell and autoimmune disease. Furthermore, WAVE2 also acts as a signal coupling regulator between the antigens and cytokines of T cells. The compromised antigen-specific T cell immune response and greater apoptosis rate of T cells also occurs in WAVE 2 KO mice.
The mechanism is that WAVE2 colocalized with mTOR in the cell perinuclear region and can directly bind to mTOR which will coimmunoprecipitate mTOR in T cells and MYC-tagged mTOR in transfected cells. Furthermore, WAVE2 can also binds to Raptor, and then cause a coimmunoprecipitation, which leads to the reduction of the amount of mTOR and Raptor in T cells. The final result is that WAVE2 can reduce the amount of mTORC1 and consequently cause a reduction in mTORC1 activation. Moreover, in WAVE2 KO condition, it has been tested that there was an increased coprecipitation of mTOR with Raptor which means that WAVE2 actually suppresses the function of mTORC1. Above all, WAVE2 leads to a “mTOR equilibrium” which lays a solid foundation of T cell homeostasis and function (Figure 4) (80).
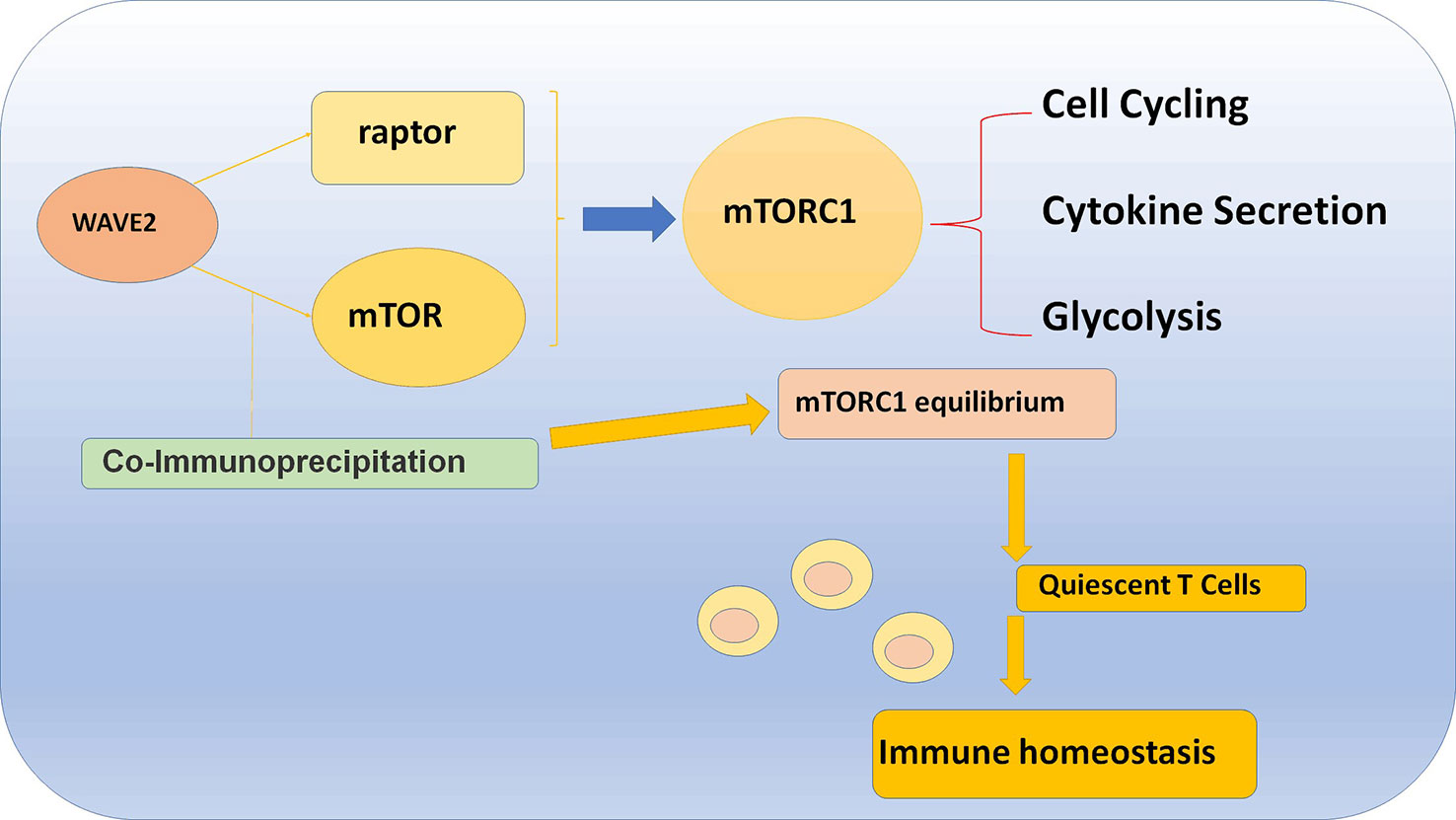
Figure 4 WAVE2 maintains T cell homeostasis and function via binding to Raptor and mTOR, which will cause a coimmunoprecipitation and lead to the reduction of the amount of mTOR and Raptor in T cell.
Raptor regulates T cell metabolism and promotes the differentiation into Th2 cells
According to studies, Raptor participates in naïve T cell-cycle priming and promotes the differentiation into Th2 cells (81). And the deficiency of Raptor inhibits naïve T cells from entering cell-cycle (82). The mechanism is that Raptor-mTORC1 mediates T cell glycolytic and lipogenic programs, which lays the foundation of the entry into the active cell cycle. Furthermore, the glycolytic and lipogenic condition and Raptor-mTORC1 pathway also intimately associated with cell fate decisions (83, 84). It is worth noting that Th2 cell differentiation have the highest rate of glycolysis among all effector T cells (85, 86).
T cell activation requires CD98 and CD71 (an iron intake receptor), which Raptor KO mice showed a decreased expression of, meaning Raptor mediates the cell-cycle required nutrient uptake. Further studies also demonstrated that the deficiency of Raptor inhibits the expressions of cell-cycle-related genes and the production of cell-cycle-related proteins such as cyclins D2 and E, CDK2, CDK4 and CDK6. Thus, Raptor regulates the cell-cycle machinery (81). Interestingly, it should be noted that Raptor is not essential for T cell proliferation when proliferation has already begun, which means that Raptor-mTORC1 signaling pathway plays a minor role in the subsequent cell division process.
Raptor can also mediate T cell metabolism and then indirectly regulates the proliferation of T cells. Hk2, Ldha, and Tpi1 are glycolytic enzymes, and the expression levels of their messenger RNA are decreased in Raptor KO T cells. What is more, the expression of c-Myc, which is an important transcription factor of T cell glycolysis (87) is decreased in Raptor KO T cells. Studies also concluded that Raptor can help T cells’ interactions with CD28 and TCR stimulation signals and then mediates the production of T cell proliferation-related cytokines including IL-4 and IL-2, which then finally affects the proliferation of T cells. Studies also concluded that the role of Raptor in promoting the differentiation into Th2 cells irrelevant to Rheb, mTORC2, or AKT, which are mTORC1 up-stream effector molecules (81, 88, 89).
Raptor mediates Th 1 and Th 17 cell differentiation via binding to mTOR
The mechanism of cell differentiation is very complex and often influenced by multiple signaling molecules in complex signal transduction pathways (90). In Th 1 cell differentiation process, IL-12 and IFN-γ stimulate naïve T cells to express T-bet and STAT4, which mediates naïve T cell transform to Th1 cells (91, 92). There are also many factors that regulate Th17 cell differentiation. What has been known clearly is that IL-6 and transforming growth factor (TGF)-β can upregulate the level of retinoic acid-related orphan receptor γt (RORγt), which subsequently promotes the conversion of naive T cells to Th17 cells and initiate CD4+ T cell differentiation process (93–95). Furthermore, phloretin is a dihydrochalcone structural flavonoid compound, which possesses many bioactive characteristics and mediates the AMPK signal pathway and consequently regulates the Th17 cell differentiation balance (96). Th17 cell differentiation is also influenced by glycolysis metabolic reprogramming. Pyruvate Kinase M2 (PKM2) is a glycolytic enzyme that translocates into the cell nucleus and activates STAT3 to promote Th17 cell differentiation (97). Moreover, in a STAT3-dependent way, Th17 cells are stabilized by IL-21 and IL-23 according to a related study (98).
Arctium lappa L (ALF) belongs to traditional Chinese medicine, which is used in treating inflammatory disorders, rheumatic pain and even fever. Arctigenin is a component of ALF that may be valid in treating ulcerative colitis via inhibiting Th 1 and 17 cell functions (99, 100). The mechanism of this traditional Chinese medicine is that it suppresses Th cell differentiation. In various CD4+ T cells, Arctigenin can selectively inhibit the activation of Th1 and Th17 cells (especially Th17 cells) by disturbing the stability of the mTOR1 complex. Arctigenin suppresses the formation of mTORC1 and the activation of the mTORC1 pathway (101) that further affects downstream molecules. In previous research, rapamycin blocks the combination of Raptor and mTOR (102, 103). It has been proven that Arctigenin does not interfere with the production of mTOR and Raptor, but also suppresses the binding of Raptor and mTOR, which consequently suppresses the phosphorylation of the mTORC1 signaling pathway downstream molecules such as p70S6K and RPS6 (101). Meanwhile, the overactivation of Raptor and mTOR can be relieved by the function of Arctigenin. This fully demonstrates the role of Raptor in Th17 cell development.
In addition, Raptor also regulates Th1 cell differentiation and function via other unknown mechanisms. Researchers have found that the decreased Th1 T cell response in the s.c. TME was observed in tumor-bearing Raptor conditional knockout (cKO) mice while the detailed mechanism still requires further research (104).
Ulcerative colitis (UC) is known to be a chronic nonspecific colonic mucosa inflammatory disorder (105). Although there is much debate about the cause of UC, more and more evidence demonstrates that the mechanism is an innate autoimmune response caused by Th1 and Th17 cells subsets and Treg cells (106, 107), which means that Raptor may be a new target of intestinal inflammation therapeutic strategy with strong clinic and therapeutic possibility (108–110).
Raptor regulates γδ T1 and γδ T17 cell differentiation and function
Like αβ T cells, γδ T cells are innate-like T cells that are essential for fighting infections and tumors (111–115). Though the number of them is far less than αβ T cells, they play an irreplaceable role in the immune function. Raptor not only mediates γδ T cell differentiation, but also regulates their immune functions. The regulatory effect of Raptor on the differentiation of γδ T cells is also mainly through the regulation of cytokine secretion. Interleukin-17 (IL-17)–producing γδ T (γδT17) and interferon-γ (IFN-γ)–producing γδ T (γδT1) cells have different metabolic requirements with being more dependent on glycolysis and phosphorylation respectively (116). Other researchers also found that CD27 is a unique regulator of γδ T1 and γδ T17 cell development. According to the studies, CD27+ γδ T cells will transform into γδ T1 cells, while the other type would differentiate into the γδ T17 cells (117). Further studies have demonstrated that Raptor-mTORC1 can even regulate T cell glycolysis to guide the direction of differentiation (118).
In a study, Raptor and rictor gene knockout mice were used to quantify γδ τ1 and γδ T17 cells. They found that the rate of γδ τ1 and γδ T17 cells were notably decreased in Raptor KO mice, while only the γδ T17 cell rate decreased in rictor KO cells. This phenomenon indicate that Raptor mediates both γδ T1 and γδ T17 cell differentiation, while rictor only participates in γδ T17 cell differentiation (118). Additionally, they further found that mTORC1 promotes IL-17 expression in γδ T cells (118).
Also, in a previous study, it was demonstrated that γδ T cells provide a cytokine environment, such as IFN-γ, NKG2D, to regulate immunity (112, 119). The researchers found that the deficiency of Raptor in γδ T cells leads to a decreased IFN-γ and NKG2D expression. Consequently, the immunological effector capacity of Raptor KO γδ T cells is significantly decreased (118).
The role of Raptor in NK cells
Natural killer cells (NK cells) are an integral part of the body’s immune system. Derived from hematopoietic stem cells, they mainly focused on antivirus-infection and tumor cell elimination. Moreover, NK cells are particularly involved in hypersensitivity reactions and autoimmune diseases (120). As for NK cells, Raptor can participate in regulating the early reprogramming process mediated by IL-15R indirectly (121). IL-15 has already been proved to be crucial for NK cell development and effector functions by binding with IL-15Rα (CD215) (122–124), and IL-15R can initiate the PI3K-p110δ/p85α in NK cells, which is a upstream signaling pathway and can trigger mTORC1 pathway (125). Studies have found that Lack of Raptor (mTORC1) resulted in decreased quantity of mature NK cell. Furthermore, Raptor-mTORC1 is required for the expression of effector molecules such as Eomesodermin (Eomes) and plays critical roles the transition from CD27 single-positive (SP) to double-positive (DP) NK stage. Additionally, reduced DP and CD11b SP NK cell populations can be observed in RaptorKO mice (126).
The differentiation process of NK cells includes many stages including common lymphoid progenitor stage (CLP stage), NK progenitor stage (NKp stage), immature NK stage (iNK stage), and mature NK stage (mNK stage) (127). Moreover, iNK cells can be grouped into three subsets by the expression of cell surface markers including CD27 and CD11b. The subsets are: CD27− CD11b− (DN, double negative, also refers as iNK stage), CD27+ CD11b−(single CD27 positive), CD27+ CD11b+ and CD27− CD11b+(double positive and single CD11b positive). The expression of CD11b marks the maturation of NK cells, so researchers put CD27+CD11b+ and CD27−CD11b+ subsets into one category (127). In some studies, it has been found that the deficiency of Raptor causes a blockage between the CD27+CD11b- stage and CD27+CD11b+ stage (128). Researchers also found that the development of NK cells are both regulated by mTORC1 and mTORC2. By generating Rptorfl/fl/CD122Cre/+and Rictorfl/fl/CD122Cre/+ mice to delete Raptor and Rictor, respectively, and comparing the phenotype of NK cells in Mtorfl/fl/CD122Cre/+ mice, researchers demonstrated that the absence of Raptor suppressed the differentiation process of immature NK cells (CD27+CD11b−) to mature NK cells while the absence of Rictor limited the differentiation of CD27−CD11b− early immature NK cells. Moreover, the specific deletion of Raptor can affect the process of NK cell terminal differentiation (129, 130).
Additionally, mediating factors such as E4 promoter-binding protein 4 (E4BP4), Eomesodermin (Eomes), T-bet as well as Tsc1 all mediate NK cell development and maturation (121, 131, 132). As have been demonstrated by other studies, E4BP4 and Eomes can maintain IL-15, which is of great importance to NK cell differentiation (133). The interaction between NK cells and IL-15 depends on the level of CD122 expressed on the NK cell surface (129, 134).
After examining and comparing the expression condition of E4BP4, Eomes, and T-bet in Rptorfl/fl/CD122Cre/+and Rictorfl/fl/CD122Cre/+ mice it was found that Eomes and T-bet were less expressed in Raptor KO NK cells, suggesting that Raptor plays an important role in inducing the production of E4BP4 and T-bet and consequently mediating NK cell early and late-stage differentiation (129) (Figure 5).
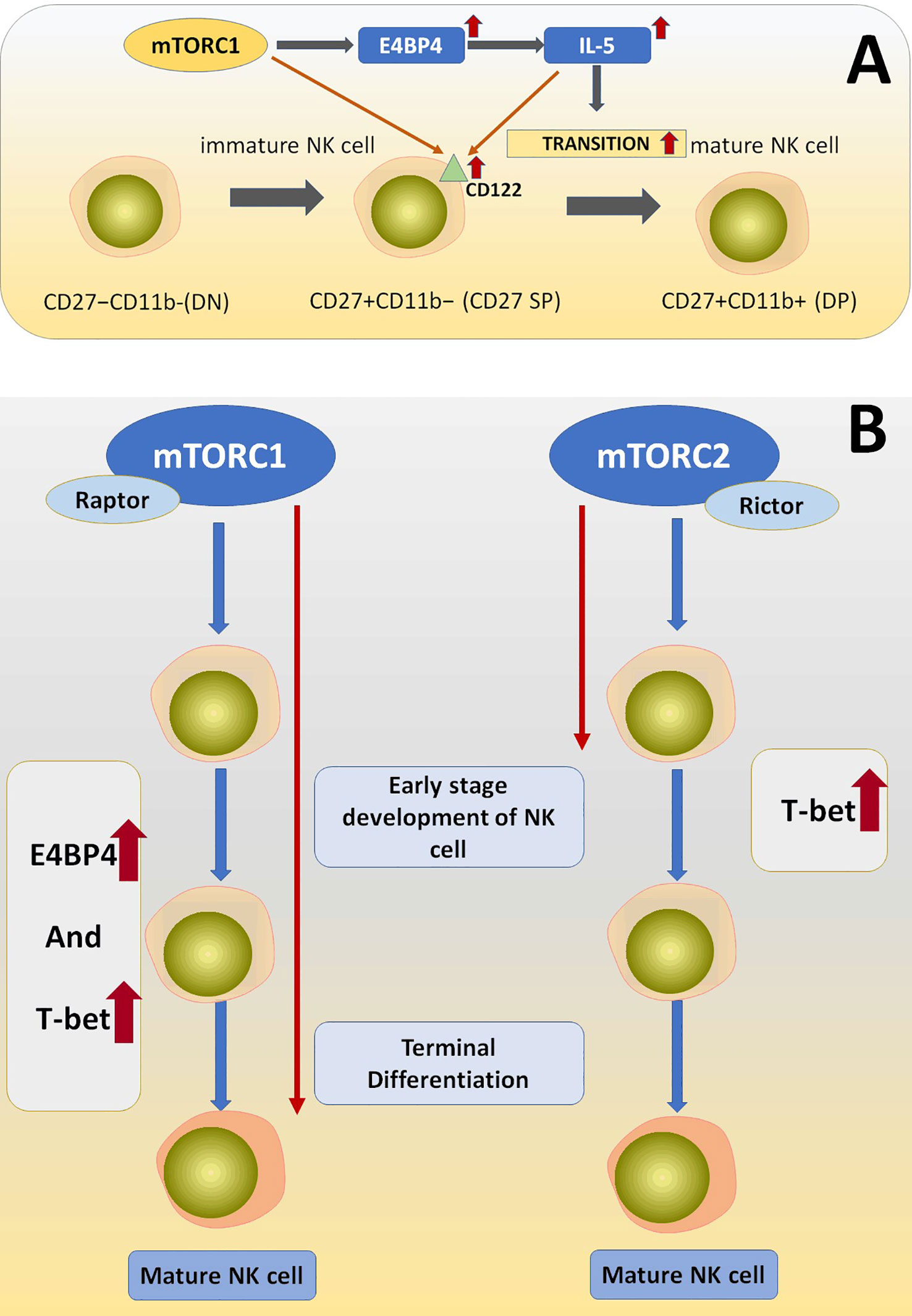
Figure 5 (A) The deficiency of Raptor downregulated the amount of CD122 on the NK cell surface. Raptor-mTORC1 signaling pathway play its role in NK cell differentiation via downstream effectors including E4BP4, T-bet, and IL-15, which is essentially needed during NK cell differentiation process. (B) mTORC1 is essential for early and late-stage development of NK cells via regulating E4BP4 and T-bet, while mTORC2 is only necessary for very early NK cell development through regulating T-bet.
As for the terminal maturation process of NK cells, researchers generated Rptorfl/fl/Ncr1-CreTg and Rictorfl/fl/Ncr1-CreTg mice. After comparing the amount and related subset phenotypes of NK cell (CD27+CD11b+ and CD27−CD11b+) between Mtorfl/fl/Ncr1-CreTg and Rptorfl/fl/Ncr1-CreTg and the conclusion is that mTORC1(Raptor) is a key molecule for the terminal differentiation stage of NK cells (129).
The role of Raptor in iNKT cells
Invariant Natural killer T cells (iNKT) are a unique lymphocyte lineage that are also regarded as CD1d-restricted cells. With the help of CD1d and TCR, iNKT cells can identify glycolipid antigens are not identified by other kinds of T cells (135–138). iNKT cells are regarded as typeINKT cells for their unique TCR α chain, which is encoded by Vα14Jα18 and Vα24Jα18 in mice and humans, respectively, while the β chain of the TCR is not invariant (139–141). Though the amount of iNKT cells is small compared to other types of T cells, iNKT cells are essential in cancer, autoimmunity, infection, allergy and even obesity (142–145). Mature iNKT cells can participate in innate immunity and are essential for adaptive immunity, which also have interactions with B cells (146). INKT cells play a part by producing cytokines such as IL-4, IL-17, IFN-γ and TNF-α after stimulation by the synthetic ligand α-galactosyl ceramide(α-GalCer) (147–149). α-GalCer is separated from the marine sponge Agelas mauritianus, and it can stimulate and activate the cytotoxic capacity of iNKT cells (150, 151). Raptor’s role in NKT cells is similar to that of B cells in that it controls development and function, and just like B cells, Raptor is more critical for cell differentiation than function. It affects both the level of CD molecules expressed on the NKT cell surface and the production of various cytokines, which play roles in signal transduction. Many scientists have been working in this field and this review summarizes their important findings.
iNKT cells can be grouped into categories according to differentiation stages represented by the expression levels of the surface markers CD24, CD44 and CD161 and they are grouped as follows: (CD24+CD44-CD161-), (CD24-CD44–), (CD24-CD44+CD161-), and (CD24-CD44+CD161+) named stage 0 to 3 in turn. Moreover, the expression of CD161 represents the maturation of iNKT cells (138, 140). The differentiation process and function of iNKT cells can be mediated by several transcription factors like RORγt, T-bet, Gata3, and promyelocytic leukemia zinc finger (PLZF) (138, 152–154), which have direct or indirect relationships with Raptor. The quantities of iNKT cells in different stages changed significantly in Raptor KO mice compared to the WT group. Specifically, the number of iNKT cells in stage 0 and 1 is within the normal range, while the number of iNKT cells in stage 2 and 3 is significantly decreased, indicating that Raptor-mTORC1 is needed during the stage 2 transformation and is essential in iNKT cell maturation (155, 156). Also, by further comparing the number of iNKT cells in stage 2 and 3, it can be concluded that Raptor also mediates the differentiation process of iNKT cells from stage 2 to 3.
As has been proven, PLZF is a crucial mediator of iNKT cells (157), and according to some studies, Raptor contributes to the localization of PLZF into the iNKT cell nucleus. The deficiency of Raptor suppresses the ability of PLZF to access differentiation-related gene promoters of iNKT cells (156), which causes a differentiation blockage between stage 1 and 2. This may be one of the reasons for the phenomenon above.
Furthermore, injecting mice with α-GalCer activates iNKT cells and leads to severe hepatitis caused by the overexpression of TNF-α (158, 159). However, in Raptor KO iNKT cells, this phenomenon is absent, which reveals that Raptor participates in regulating the production of TNF-α in iNKT cells. The expression levels of other cytokines such as IL-4 and IFN-γ decreased and the proliferation of iNKT cells was also abolished in Raptor KO mice after stimulating iNKT cells (156).
The deficiency of Raptor also causes reduced iNKT1, increased iNKT2, and normal iNKT17 cell ratios in mice. The explanation may be that Raptor can regulate the production of transcription factors of iNKT cells, although the deeper mechanism still needs to be explored (155, 156, 160).
Conclusion and future outlook
Raptor is a core component of mTORC1 and plays an essential and complex role in the differentiation processes and functions of lymphocytes. However, there are conflicting results on Raptor‘s role in regulating immune processes, which is specifically seen in studies using Raptor knockouts. With further technical and scientific advancements, researchers will be able to explore deeper into the specific roles of Raptor. The most current studies on Raptor are based on the findings of new phenomena, but none are linked to metabolic disease models such as gastric cancer, colon cancer, diabetes, etc. Since mTORC1 plays a role in the proliferation and metastasis of cancer cells, the regulation of Raptor in the proliferation and differentiation of cancer cells may be a direction worth exploring in the future. Finally, Raptor related studies are being conducted in mouse models, which have been limited to early stages of lymphocytes that only involve a few studies. In short, there is still a long way to go from basic medical theory to clinical application, and it is often very difficult to transition from mouse models to humans, thus if Raptor is to be used as clinical medicine, more efforts need to be made in its research. This review summarizes the current studies on the effects of Raptor on lymphocytes and aims to help researchers find new directions for clinical discovery.
Author contributions
JT wrote the article. LY, FG, HM, NC, LJ, KB, MK, SH, PL, JL, HZ, and CH revised the draft. ZZ and CL organized and revised the draft. All authors contributed to the article and approved the submitted version.
Funding
This project was supported by the Medjaden Academy & Research Foundation for Young Scientists (MJR20221030, MJR20221009).
Conflict of interest
Author HM was employed by company Cytek Biosciences.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Abbreviations
Raptor, regulatory-associated protein of mTOR; rictor, rapamycin-insensitive companion of mTOR; mTOR, Mammalian target of rapamycin; PIKK, PI3K-related kinase; 4E-BP1, 4E-binding protein 1; p70S6k, p70 S6 kinase; mTORC1, mTOR complex 1; mTORC2, mTOR complex 2; EGF, epidermal growth factor; IGF, insulin-like growth factor; MAPK, mitogen-activated protein kinase; BM, bone marrow; IgH, Ig H chain; IL, Interleukin; WT, Wild type; Th cell, T helper cell; NK cells, Natural killer cells; Eomes, Eomesodermin; SP, single-positive; DP, double-positive; iNKT, Invariant Natural killer T cells; CXCL12, C-X-C motif chemokine 12; WAVE2, WASp family verprolin homologous protein 2; ALF, Arctium lappa L; PLZF, promyelocytic leukemia zinc finger.
References
1. Hara K, Maruki Y, Long X, Yoshino K-I, Oshiro N, Hidayat S, et al. Raptor, a binding partner of target of rapamycin (TOR), mediates TOR action. Cell (2002) 110(2):177–89. doi: 10.1016/S0092-8674(02)00833-4
2. Sabatini DM. Twenty-five years of mTOR: uncovering the link from nutrients to growth. Proc Natl Acad Sci USA (2017) 114(45):11818–25. doi: 10.1073/pnas.1716173114
3. Arriola Apelo SI, Lamming DW. Rapamycin: an InhibiTOR of aging emerges from the soil of Easter island. J Gerontol Ser A: Biol Sci Med Sci (2016) 71(7):841–9. doi: 10.1093/gerona/glw090
4. Szwed A, Kim E, Jacinto E. Regulation and metabolic functions of mTORC1 and mTORC2. Physiol Rev (2021) 101(3):1371–426. doi: 10.1152/physrev.00026.2020
5. Mossmann D, Park S, Hall MN. mTOR signalling and cellular metabolism are mutual determinants in cancer. Nat Rev Cancer (2018) 18(12):744–57. doi: 10.1038/s41568-018-0074-8
6. Hua H, Kong Q, Zhang H, Wang J, Luo T, Jiang Y. Targeting mTOR for cancer therapy. J Hematol Oncol (2019) 12(1):71. doi: 10.1186/s13045-019-0754-1
7. Popova NV, Jücker M. The role of mTOR signaling as a therapeutic target in cancer. Int J Mol Sci (2021) 22(4):1743. doi: 10.3390/ijms22041743
8. Tan AC. Targeting the PI3K/Akt/mTOR pathway in non-small cell lung cancer (NSCLC). Thorac Cancer (2020) 11(3):511–8. doi: 10.1111/1759-7714.13328
9. Yang L, Shi P, Zhao G, Xu J, Peng W, Zhang J, et al. Targeting cancer stem cell pathways for cancer therapy. Signal Transduct Target Ther (2020) 5(1):8. doi: 10.1038/s41392-020-0110-5
10. Saxton RA, Sabatini DM. mTOR signaling in growth, metabolism, and disease. Cell (2017) 168(6):960–76. doi: 10.1016/j.cell.2017.02.004
11. Sehgal SN, Baker H, Vézina C. Rapamycin (AY-22,989), a new antifungal antibiotic. II. fermentation, isolation and characterization. J Antibiot (Tokyo) (1975) 28(10):727–32. doi: 10.7164/antibiotics.28.727
12. Benjamin D, Colombi M, Moroni C, Hall MN. Rapamycin passes the torch: a new generation of mTOR inhibitors. Nat Rev Drug Discov (2011) 10(11):868–80. doi: 10.1038/nrd3531
13. Rostamzadeh D, Yousefi M, Haghshenas MR, Ahmadi M, Dolati S, Babaloo Z. mTOR signaling pathway as a master regulator of memory CD8 + T-cells, Th17, and NK cells development and their functional properties. J Cell Physiol (2019) 234(8):12353–68. doi: 10.1002/jcp.28042
14. Chung J, Kuo CJ, Crabtree GR, Blenis J. Rapamycin-FKBP specifically blocks growth-dependent activation of and signaling by the 70 kd S6 protein kinases. Cell (1992) 69(7):1227–36. doi: 10.1016/0092-8674(92)90643-Q
15. Brown EJ, Albers MW, Shin TB, Ichikawa K, Keith CT, Lane WS, et al. A mammalian protein targeted by G1-arresting rapamycin-receptor complex. Nature (1994) 369(6483):756–8. doi: 10.1038/369756a0
16. Iwata TN, Ramirez JA, Tsang M, Park H, Margineantu DH, Hockenbery DM, et al. Conditional disruption of raptor reveals an essential role for mTORC1 in b cell development, survival, and metabolism. J Immunol (2016) 197(6):2250–60. doi: 10.4049/jimmunol.1600492
17. Rodrigues LF, Santos PHS, Pelozin BRA, Tobias GC. New insights into mTOR inhibition: understanding the mechanisms and their importance. J Physiol (2023) 601(1):3–4. doi: 10.1113/JP283999
18. Kim DH, Sarbassov DD, Ali SM, Latek RR, Guntur KV, Erdjument-Bromage H, et al. GbetaL, a positive regulator of the rapamycin-sensitive pathway required for the nutrient-sensitive interaction between raptor and mTOR. Mol Cell (2003) 11(4):895–904. doi: 10.1016/S1097-2765(03)00114-X
19. Kim DH, Sarbassov DD, Ali SM, King JE, Latek RR, Erdjument-Bromage H, et al. mTOR interacts with raptor to form a nutrient-sensitive complex that signals to the cell growth machinery. Cell (2002) 110(2):163–75. doi: 10.1016/S0092-8674(02)00808-5
20. Sancak Y, Thoreen CC, Peterson TR, Lindquist RA, Kang SA, Spooner E, et al. PRAS40 is an insulin-regulated inhibitor of the mTORC1 protein kinase. Mol Cell (2007) 25(6):903–15. doi: 10.1016/j.molcel.2007.03.003
21. Peterson TR, Laplante M, Thoreen CC, Sancak Y, Kang SA, Kuehl WM, et al. DEPTOR is an mTOR inhibitor frequently overexpressed in multiple myeloma cells and required for their survival. Cell (2009) 137(5):873–86. doi: 10.1016/j.cell.2009.03.046
22. Nojima H, Tokunaga C, Eguchi S, Oshiro N, Hidayat S, Yoshino K-I, et al. The mammalian target of rapamycin (mTOR) partner, raptor, binds the mTOR substrates p70 S6 kinase and 4E-BP1 through their TOR signaling (TOS) motif. J Biol Chem (2003) 278(18):15461–4. doi: 10.1074/jbc.C200665200
23. Schalm SS, Fingar DC, Sabatini DM, Blenis J. TOS motif-mediated raptor binding regulates 4E-BP1 multisite phosphorylation and function. Curr Biol (2003) 13(10):797–806. doi: 10.1016/S0960-9822(03)00329-4
24. Tatebe H, Shiozaki K. Evolutionary conservation of the components in the TOR signaling pathways. Biomolecules (2017) 7(4):77. doi: 10.3390/biom7040077
25. Deng L, Chen L, Zhao L, Xu Y, Peng X, Wang X, et al. Ubiquitination of rheb governs growth factor-induced mTORC1 activation. Cell Res (2019) 29(2):136–50. doi: 10.1038/s41422-018-0120-9
26. Loewith R, Jacinto E, Wullschleger S, Lorberg A, Crespo JL, Bonenfant D, et al. Two TOR complexes, only one of which is rapamycin sensitive, have distinct roles in cell growth control. Mol Cell (2002) 10(3):457–68. doi: 10.1016/S1097-2765(02)00636-6
27. Jewell JL, Fu V, Hong AW, Yu FX, Meng D, Melick CH, et al. GPCR signaling inhibits mTORC1 via PKA phosphorylation of raptor. Elife (2019) 8:e43038. doi: 10.7554/eLife.43038
28. Carriere A, Romeo Y, Acosta-Jaquez HA, Moreau J, Bonneil E, Thibault P, et al. ERK1/2 phosphorylate raptor to promote ras-dependent activation of mTOR complex 1 (mTORC1). J Biol Chem (2011) 286(1):567–77. doi: 10.1074/jbc.M110.159046
29. Wu D, Chapman JR, Wang L, Harris TE, Shabanowitz J, Hunt DF, et al. Intestinal cell kinase (ICK) promotes activation of mTOR complex 1 (mTORC1) through phosphorylation of raptor thr-908. J Biol Chem (2012) 287(15):12510–9. doi: 10.1074/jbc.M111.302117
30. Carrière A, Cargnello M, Julien LA, Gao H, Bonneil E, Thibault P, et al. Oncogenic MAPK signaling stimulates mTORC1 activity by promoting RSK-mediated raptor phosphorylation. Curr Biol (2008) 18(17):1269–77. doi: 10.1016/j.cub.2008.07.078
31. Herzog S, Reth M, Jumaa H. Regulation of b-cell proliferation and differentiation by pre-b-cell receptor signalling. Nat Rev Immunol (2009) 9(3):195–205. doi: 10.1038/nri2491
32. Ren A, Yin W, Miller H, Westerberg LS, Candotti F, Park CS, et al. Novel discoveries in immune dysregulation in inborn errors of immunity. Front Immunol (2021) 12:725587. doi: 10.3389/fimmu.2021.725587
33. Hardy RR, Hayakawa K. B cell development pathways. Annu Rev Immunol (2001) 19:595–621. doi: 10.1146/annurev.immunol.19.1.595
34. Reth M, Nielsen P. Signaling circuits in early b-cell development. Adv Immunol (2014) 122:129–75. doi: 10.1016/B978-0-12-800267-4.00004-3
35. Chen S, Guan F, Candotti F, Benlagha K, Camara NOS, Herrada AA, et al. The role of b cells in COVID-19 infection and vaccination. Front Immunol (2022) 13:988536. doi: 10.3389/fimmu.2022.988536
36. Wang Y, Xiao M, Tao C, Chen J, Wang Z, Yang J, et al. Inactivation of mTORC1 signaling in osterix-expressing cells impairs b-cell differentiation. J Bone Miner Res (2018) 33(4):732–42. doi: 10.1002/jbmr.3352
37. Corfe SA, Paige CJ. The many roles of IL-7 in b cell development; mediator of survival, proliferation and differentiation. Semin Immunol (2012) 24(3):198–208. doi: 10.1016/j.smim.2012.02.001
38. Almeida ARM, Neto JL, Cachucho A, Euzébio M, Meng X, Kim R, et al. Interleukin-7 receptor α mutational activation can initiate precursor b-cell acute lymphoblastic leukemia. Nat Commun (2021) 12(1):7268. doi: 10.1038/s41467-021-27197-5
39. Langdon WY, Harris AW, Cory S, Adams JM. The c-myc oncogene perturbs b lymphocyte development in e-mu-myc transgenic mice. Cell (1986) 47(1):11–8. doi: 10.1016/0092-8674(86)90361-2
40. Vallespinós M, Fernández D, Rodríguez L, Alvaro-Blanco J, Baena E, Ortiz M, et al. B lymphocyte commitment program is driven by the proto-oncogene c-myc. J Immunol (2011) 186(12):6726–36. doi: 10.4049/jimmunol.1002753
41. Sandoval GJ, Graham DB, Bhattacharya D, Sleckman BP, Xavier RJ, Swat W. Cutting edge: cell-autonomous control of IL-7 response revealed in a novel stage of precursor b cells. J Immunol (2013) 190(6):2485–9. doi: 10.4049/jimmunol.1203208
42. Zeng H, Yu M, Tan H, Li Y, Su W, Shi H, et al. Discrete roles and bifurcation of PTEN signaling and mTORC1-mediated anabolic metabolism underlie IL-7-driven b lymphopoiesis. Sci Adv (2018) 4(1):eaar5701. doi: 10.1126/sciadv.aar5701
43. Panaroni C, Wu JY. Interactions between b lymphocytes and the osteoblast lineage in bone marrow. Calcif Tissue Int (2013) 93(3):261–8. doi: 10.1007/s00223-013-9753-3
44. Chen J, Long F. mTORC1 signaling promotes osteoblast differentiation from preosteoblasts. PloS One (2015) 10(6):e0130627. doi: 10.1371/journal.pone.0130627
45. Greenbaum A, Hsu YM, Day RB, Schuettpelz LG, Christopher MJ, Borgerding JN, et al. CXCL12 in early mesenchymal progenitors is required for haematopoietic stem-cell maintenance. Nature (2013) 495(7440):227–30. doi: 10.1038/nature11926
46. Ding L, Morrison SJ. Haematopoietic stem cells and early lymphoid progenitors occupy distinct bone marrow niches. Nature (2013) 495(7440):231–5. doi: 10.1038/nature11885
47. Ren A, Sun J, Yin W, Westerberg LS, Miller H, Lee P, et al. Signaling networks in b cell development and related therapeutic strategies. J Leukoc Biol (2022) 111(4):877–91. doi: 10.1002/JLB.2RU0221-088RRR
48. Lin J, Zhu Z, Xiao H, Wakefield MR, Ding VA, Bai Q, et al. The role of IL-7 in immunity and cancer. Anticancer Res (2017) 37(3):963–7. doi: 10.21873/anticanres.11405
49. Hertz M, Nemazee D. BCR ligation induces receptor editing in IgM+IgD- bone marrow b cells in vitro. Immunity (1997) 6(4):429–36. doi: 10.1016/S1074-7613(00)80286-1
50. Rolink A, Grawunder U, Haasner D, Strasser A, Melchers F. Immature surface ig+ b cells can continue to rearrange kappa and lambda l chain gene loci. J Exp Med (1993) 178(4):1263–70. doi: 10.1084/jem.178.4.1263
51. Nakashima K, Zhou X, Kunkel G, Zhang Z, Deng JM, Behringer RR, et al. The novel zinc finger-containing transcription factor osterix is required for osteoblast differentiation and bone formation. Cell (2002) 108(1):17–29. doi: 10.1016/S0092-8674(01)00622-5
52. Malin S, McManus S, Cobaleda C, Novatchkova M, Delogu A, Bouillet P, et al. Role of STAT5 in controlling cell survival and immunoglobulin gene recombination during pro-b cell development. Nat Immunol (2010) 11(2):171–9. doi: 10.1038/ni.1827
53. Goetz CA, Harmon IR, O'Neil JJ, Burchill MA, Farrar MA. STAT5 activation underlies IL7 receptor-dependent b cell development. J Immunol (2004) 172(8):4770–8. doi: 10.4049/jimmunol.172.8.4770
54. Aguila HL, Mun SH, Kalinowski J, Adams DJ, Lorenzo JA, Lee SK. Osteoblast-specific overexpression of human interleukin-7 rescues the bone mass phenotype of interleukin-7-deficient female mice. J Bone Miner Res (2012) 27(5):1030–42. doi: 10.1002/jbmr.1553
55. Kumar BV, Connors TJ, Farber DL. Human T cell development, localization, and function throughout life. Immunity (2018) 48(2):202–13. doi: 10.1016/j.immuni.2018.01.007
56. Kishton RJ, Sukumar M, Restifo NP. Metabolic regulation of T cell longevity and function in tumor immunotherapy. Cell Metab (2017) 26(1):94–109. doi: 10.1016/j.cmet.2017.06.016
57. Zheng B, Zhang J, Chen H, Nie H, Miller H, Gong Q, et al. T Lymphocyte-mediated liver immunopathology of schistosomiasis. Front Immunol (2020) 11:61. doi: 10.3389/fimmu.2020.00061
58. Zoncu R, Efeyan A, Sabatini DM. mTOR: from growth signal integration to cancer, diabetes and ageing. Nat Rev Mol Cell Biol (2011) 12(1):21–35. doi: 10.1038/nrm3025
59. Iwasaki A, Medzhitov R. Control of adaptive immunity by the innate immune system. Nat Immunol (2015) 16(4):343–53. doi: 10.1038/ni.3123
60. Araki K, Turner AP, Shaffer VO, Gangappa S, Keller SA, Bachmann MF, et al. mTOR regulates memory CD8 T-cell differentiation. Nature (2009) 460(7251):108–12. doi: 10.1038/nature08155
61. Chapman NM, Chi H. mTOR links environmental signals to T cell fate decisions. Front Immunol (2014) 5:686. doi: 10.3389/fimmu.2014.00686
62. Delgoffe GM, Pollizzi KN, Waickman AT, Heikamp E, Meyers DJ, Horton MR, et al. The kinase mTOR regulates the differentiation of helper T cells through the selective activation of signaling by mTORC1 and mTORC2. Nat Immunol (2011) 12(4):295–303. doi: 10.1038/ni.2005
63. Chi H. Regulation and function of mTOR signalling in T cell fate decisions. Nat Rev Immunol (2012) 12(5):325–38. doi: 10.1038/nri3198
64. Stark JM, Tibbitt CA, Coquet JM. The metabolic requirements of Th2 cell differentiation. Front Immunol (2019) 10:2318. doi: 10.3389/fimmu.2019.02318
65. Walker JA, McKenzie ANJ. TH2 cell development and function. Nat Rev Immunol (2018) 18(2):121–33. doi: 10.1038/nri.2017.118
66. Pollizzi KN, Powell JD. Integrating canonical and metabolic signalling programmes in the regulation of T cell responses. Nat Rev Immunol (2014) 14(7):435–46. doi: 10.1038/nri3701
67. Lee K, Gudapati P, Dragovic S, Spencer C, Joyce S, Killeen N, et al. Mammalian target of rapamycin protein complex 2 regulates differentiation of Th1 and Th2 cell subsets via distinct signaling pathways. Immunity (2010) 32(6):743–53. doi: 10.1016/j.immuni.2010.06.002
68. Kurebayashi Y, Nagai S, Ikejiri A, Ohtani M, Ichiyama K, Baba Y, et al. PI3K-Akt-mTORC1-S6K1/2 axis controls Th17 differentiation by regulating Gfi1 expression and nuclear translocation of RORγ. Cell Rep (2012) 1(4):360–73. doi: 10.1016/j.celrep.2012.02.007
69. Sun IH, Oh MH, Zhao L, Patel CH, Arwood ML, Xu W, et al. mTOR complex 1 signaling regulates the generation and function of central and effector Foxp3(+) regulatory T cells. J Immunol (2018) 201(2):481–92. doi: 10.4049/jimmunol.1701477
70. Klebanoff CA, Gattinoni L, Restifo NP. CD8 + T-cell memory in tumor immunology and immunotherapy. Immunol Rev (2006) 211(1):214–24. doi: 10.1111/j.0105-2896.2006.00391.x
71. Wullschleger S, Loewith R, Hall MN. TOR signaling in growth and metabolism. Cell (2006) 124(3):471–84. doi: 10.1016/j.cell.2006.01.016
72. Li KP, Ladle BH, Kurtulus S, Sholl A, Shanmuganad S, Hildeman DA. T-Cell receptor signal strength and epigenetic control of bim predict memory CD8(+) T-cell fate. Cell Death Differ (2020) 27(4):1214–24. doi: 10.1038/s41418-019-0410-x
73. Sarkar S, Kalia V, Haining WN, Konieczny BT, Subramaniam S, Ahmed R. Functional and genomic profiling of effector CD8 T cell subsets with distinct memory fates. J Exp Med (2008) 205(3):625–40. doi: 10.1084/jem.20071641
74. Sowell RT, Rogozinska M, Nelson CE, Vezys V, Marzo AL. Cutting edge: generation of effector cells that localize to mucosal tissues and form resident memory CD8 T cells is controlled by mTOR. J Immunol (2014) 193(5):2067–71. doi: 10.4049/jimmunol.1400074
75. Nolz JC, Nacusi LP, Segovis CM, Medeiros RB, Mitchell JS, Shimizu Y, et al. The WAVE2 complex regulates T cell receptor signaling to integrins via abl- and CrkL–C3G-mediated activation of Rap1. J Cell Biol (2008) 182(6):1231–44. doi: 10.1083/jcb.200801121
76. Rana PS, Alkrekshi A, Wang W, Markovic V, Sossey-Alaoui K. The role of WAVE2 signaling in cancer. Biomedicines (2021) 9(9):1217. doi: 10.3390/biomedicines9091217
77. Kurisu S, Suetsugu S, Yamazaki D, Yamaguchi H, Takenawa T. Rac-WAVE2 signaling is involved in the invasive and metastatic phenotypes of murine melanoma cells. Oncogene (2005) 24(8):1309–19. doi: 10.1038/sj.onc.1208177
78. Kim JS, Kang CG, Kim SH, Lee EO. Rhapontigenin suppresses cell migration and invasion by inhibiting the PI3K-dependent Rac1 signaling pathway in MDA-MB-231 human breast cancer cells. J Nat Prod (2014) 77(5):1135–9. doi: 10.1021/np401078g
79. Yan C, Martinez-Quiles N, Eden S, Shibata T, Takeshima F, Shinkura R, et al. WAVE2 deficiency reveals distinct roles in embryogenesis and rac-mediated actin-based motility. EMBO J (2003) 22(14):3602–12. doi: 10.1093/emboj/cdg350
80. Liu M, Zhang J, Pinder BD, Liu Q, Wang D, Yao H, et al. WAVE2 suppresses mTOR activation to maintain T cell homeostasis and prevent autoimmunity. Science (2021) 371(6536):eaaz4544. doi: 10.1126/science.aaz4544
81. Yang K, Shrestha S, Zeng H, Karmaus PW, Neale G, Vogel P, et al. T Cell exit from quiescence and differentiation into Th2 cells depend on raptor-mTORC1-mediated metabolic reprogramming. Immunity (2013) 39(6):1043–56. doi: 10.1016/j.immuni.2013.09.015
82. Hoshii T, Kasada A, Hatakeyama T, Ohtani M, Tadokoro Y, Naka K, et al. Loss of mTOR complex 1 induces developmental blockage in early T-lymphopoiesis and eradicates T-cell acute lymphoblastic leukemia cells. Proc Natl Acad Sci USA (2014) 111(10):3805–10. doi: 10.1073/pnas.1320265111
83. Chapman NM, Boothby MR, Chi H. Metabolic coordination of T cell quiescence and activation. Nat Rev Immunol (2020) 20(1):55–70. doi: 10.1038/s41577-019-0203-y
84. Yang K, Chi H. mTOR and metabolic pathways in T cell quiescence and functional activation. Semin Immunol (2012) 24(6):421–8. doi: 10.1016/j.smim.2012.12.004
85. Michalek RD, Gerriets VA, Jacobs SR, Macintyre AN, MacIver NJ, Mason EF, et al. Cutting edge: distinct glycolytic and lipid oxidative metabolic programs are essential for effector and regulatory CD4+ T cell subsets. J Immunol (2011) 186(6):3299–303. doi: 10.4049/jimmunol.1003613
86. Macintyre AN, Gerriets VA, Nichols AG, Michalek RD, Rudolph MC, Deoliveira D, et al. The glucose transporter Glut1 is selectively essential for CD4 T cell activation and effector function. Cell Metab (2014) 20(1):61–72. doi: 10.1016/j.cmet.2014.05.004
87. Wang R, Dillon CP, Shi LZ, Milasta S, Carter R, Finkelstein D, et al. The transcription factor myc controls metabolic reprogramming upon T lymphocyte activation. Immunity (2011) 35(6):871–82. doi: 10.1016/j.immuni.2011.09.021
88. Cloëtta D, Thomanetz V, Baranek C, Lustenberger RM, Lin S, Oliveri F, et al. Inactivation of mTORC1 in the developing brain causes microcephaly and affects gliogenesis. J Neurosci (2013) 33(18):7799–810. doi: 10.1523/JNEUROSCI.3294-12.2013
89. Goorden SM, Hoogeveen-Westerveld M, Cheng C, van Woerden GM, Mozaffari M, Post L, et al. Rheb is essential for murine development. Mol Cell Biol (2011) 31(8):1672–8. doi: 10.1128/MCB.00985-10
90. Chen T, You Y, Jiang H, Wang ZZ. Epithelial-mesenchymal transition (EMT): a biological process in the development, stem cell differentiation, and tumorigenesis. J Cell Physiol (2017) 232(12):3261–72. doi: 10.1002/jcp.25797
91. Zhang Y, Zhang Y, Gu W, Sun B. TH1/TH2 cell differentiation and molecular signals. Adv Exp Med Biol (2014) 841:15–44. doi: 10.1007/978-94-017-9487-9_2
92. Johnson MO, Wolf MM, Madden MZ, Andrejeva G, Sugiura A, Contreras DC, et al. Distinct regulation of Th17 and Th1 cell differentiation by glutaminase-dependent metabolism. Cell (2018) 175(7):1780–1795.e19. doi: 10.1016/j.cell.2018.10.001
93. Li J, Wei B, Guo A, Liu C, Huang S, Du F, et al. Deficiency of beta-arrestin1 ameliorates collagen-induced arthritis with impaired TH17 cell differentiation. Proc Natl Acad Sci USA (2013) 110(18):7395–400. doi: 10.1073/pnas.1221608110
94. Xu J, Yang Y, Qiu G, Lal G, Yin N, Wu Z, et al. Stat4 is critical for the balance between Th17 cells and regulatory T cells in colitis. J Immunol (2011) 186(11):6597–606. doi: 10.4049/jimmunol.1004074
95. Kalim KW, Basler M, Kirk CJ, Groettrup M. Immunoproteasome subunit LMP7 deficiency and inhibition suppresses Th1 and Th17 but enhances regulatory T cell differentiation. J Immunol (2012) 189(8):4182–93. doi: 10.4049/jimmunol.1201183
96. Jiao A, Yang Z, Fu X, Hua X. Phloretin modulates human Th17/Treg cell differentiation In vitro via AMPK signaling. BioMed Res Int (2020) 2020:1–12. doi: 10.1155/2020/6267924
97. Damasceno LEA, Prado DS, Veras FP, Fonseca MM, Toller-Kawahisa JE, Rosa MH, et al. PKM2 promotes Th17 cell differentiation and autoimmune inflammation by fine-tuning STAT3 activation. J Exp Med (2020) 217(10):e20190613. doi: 10.1084/jem.20190613
98. McGeachy MJ, Chen Y, Tato CM, Laurence A, Joyce-Shaikh B, Blumenschein WM, et al. The interleukin 23 receptor is essential for the terminal differentiation of interleukin 17–producing effector T helper cells in vivo. Nat Immunol (2009) 10(3):314–24. doi: 10.1038/ni.1698
99. Yang M, Xu X, Xie C, Xie Z, Huang J, Yang D. Separation and purification of arctiin, arctigenin, matairesinol, and lappaol f fromFructus arctiiby high-speed counter-current chromatography. Separation Sci Technol (2013) 48(11):1738–44. doi: 10.1080/01496395.2012.753631
100. Park SY, Hong SS, Han XH, Hwang JS, Lee D, Ro JS, et al. Lignans from arctium lappa and their inhibition of LPS-induced nitric oxide production. Chem Pharm Bull (2007) 55(1):150–2. doi: 10.1248/cpb.55.150
101. Wu X, Dou Y, Yang Y, Bian D, Luo J, Tong B, et al. Arctigenin exerts anti-colitis efficacy through inhibiting the differentiation of Th1 and Th17 cells via an mTORC1-dependent pathway. Biochem Pharmacol (2015) 96(4):323–36. doi: 10.1016/j.bcp.2015.06.008
102. Zhang Y, Xu S, Lin J, Yao G, Han Z, Liang B, et al. mTORC1 is a target of nordihydroguaiaretic acid to prevent breast tumor growth in vitro and in vivo. Breast Cancer Res Treat (2012) 136(2):379–88. doi: 10.1007/s10549-012-2270-7
103. Beevers CS, Chen L, Liu L, Luo Y, Webster NJG, Huang S. Curcumin disrupts the mammalian target of rapamycin-raptor complex. Cancer Res (2009) 69(3):1000–8. doi: 10.1158/0008-5472.CAN-08-2367
104. Ding C, Sun X, Wu C, Hu X, Zhang H-G, Yan J. Tumor microenvironment modulates immunological outcomes of myeloid cells with mTORC1 disruption. J Immunol (2019) 202(5):1623–34. doi: 10.4049/jimmunol.1801112
105. Ordás I, Eckmann L, Talamini M, Baumgart DC, Sandborn WJ. Ulcerative colitis. Lancet (2012) 380(9853):1606–19. doi: 10.1016/S0140-6736(12)60150-0
106. Khanna PV, Shih DQ, Haritunians T, McGovern DP, Targan S. Use of animal models in elucidating disease pathogenesis in IBD. Semin Immunopathol (2014) 36(5):541–51. doi: 10.1007/s00281-014-0444-6
107. Boirivant M, Cossu A. Inflammatory bowel disease. Oral Dis (2012) 18(1):1–15. doi: 10.1111/j.1601-0825.2011.01811.x
108. Olsen T, Rismo R, Cui G, Goll R, Christiansen I, Florholmen J. TH1 and TH17 interactions in untreated inflamed mucosa of inflammatory bowel disease, and their potential to mediate the inflammation. Cytokine (2011) 56(3):633–40. doi: 10.1016/j.cyto.2011.08.036
109. Neurath MF. New targets for mucosal healing and therapy in inflammatory bowel diseases. Mucosal Immunol (2014) 7(1):6–19. doi: 10.1038/mi.2013.73
110. Monteleone G, Caruso R, Pallone F. Targets for new immunomodulation strategies in inflammatory bowel disease. Autoimmun Rev (2014) 13(1):11–4. doi: 10.1016/j.autrev.2013.06.003
111. Carding SR, Egan PJ. γδ T cells: functional plasticity and heterogeneity. Nat Rev Immunol (2002) 2(5):336–45. doi: 10.1038/nri797
112. Gao Y, Yang W, Pan M, Scully E, Girardi M, Augenlicht LH, et al. γδ T cells provide an early source of interferon γ in tumor immunity. J Exp Med (2003) 198(3):433–42. doi: 10.1084/jem.20030584
113. Born WK, Yin Z, Hahn YS, Sun D, O'Brien RL. Analysis of gamma delta T cell functions in the mouse. J Immunol (2010) 184(8):4055–61. doi: 10.4049/jimmunol.0903679
114. Silva-Santos B, Serre K, Norell H. γδ T cells in cancer. Nat Rev Immunol (2015) 15(11):683–91. doi: 10.1038/nri3904
115. Papotto PH, Ribot JC, Silva-Santos B. IL-17+ γδ T cells as kick-starters of inflammation. Nat Immunol (2017) 18(6):604–11. doi: 10.1038/ni.3726
116. Jensen KDC, Su X, Shin S, Li L, Youssef S, Yamasaki S, et al. Thymic selection determines γδ T cell effector fate: antigen-naive cells make interleukin-17 and antigen-experienced cells make interferon γ. Immunity (2008) 29(1):90–100. doi: 10.1016/j.immuni.2008.04.022
117. Ribot JC, Debarros A, Pang DJ, Neves JF, Peperzak V, Roberts SJ, et al. CD27 is a thymic determinant of the balance between interferon-γ- and interleukin 17–producing γδ T cell subsets. Nat Immunol (2009) 10(4):427–36. doi: 10.1038/ni.1717
118. Yang Q, Liu X, Liu Q, Guan Z, Luo J, Cao G, et al. Roles of mTORC1 and mTORC2 in controlling γδ T1 and γδ T17 differentiation and function. Cell Death Differ (2020) 27(7):2248–62. doi: 10.1038/s41418-020-0500-9
119. He W, Hao J, Dong S, Gao Y, Tao J, Chi H, et al. Naturally activated V gamma 4 gamma delta T cells play a protective role in tumor immunity through expression of eomesodermin. J Immunol (2010) 185(1):126–33. doi: 10.4049/jimmunol.0903767
120. Terren I, Orrantia A, Vitalle J, Zenarruzabeitia O, Borrego F. NK cell metabolism and tumor microenvironment. Front Immunol (2019) 10:2278. doi: 10.3389/fimmu.2019.02278
121. Yang C, Malarkannan S. Transcriptional regulation of NK cell development by mTOR complexes. Front Cell Dev Biol (2020) 8:566090. doi: 10.3389/fcell.2020.566090
122. Giri JG, Kumaki S, Ahdieh M, Friend DJ, Loomis A, Shanebeck K, et al. Identification and cloning of a novel IL-15 binding protein that is structurally related to the alpha chain of the IL-2 receptor. EMBO J (1995) 14(15):3654–63. doi: 10.1002/j.1460-2075.1995.tb00035.x
123. Becknell B, Caligiuri MA. Interleukin-2, interleukin-15, and their roles in human natural killer cells. Adv Immunol (2005) 86:209–39. doi: 10.1016/S0065-2776(04)86006-1
124. Marçais A, Viel S, Grau M, Henry T, Marvel J, Walzer T. Regulation of mouse NK cell development and function by cytokines. Front Immunol (2013) 4:450. doi: 10.3389/fimmu.2013.00450
125. Awasthi A, Samarakoon A, Dai X, Wen R, Wang D, Malarkannan S. Deletion of PI3K-p85alpha gene impairs lineage commitment, terminal maturation, cytokine generation and cytotoxicity of NK cells. Genes Immun (2008) 9(6):522–35. doi: 10.1038/gene.2008.45
126. Mayol K, Biajoux V, Marvel J, Balabanian K, Walzer T. Sequential desensitization of CXCR4 and S1P5 controls natural killer cell trafficking. Blood (2011) 118(18):4863–71. doi: 10.1182/blood-2011-06-362574
127. Narni-Mancinelli E, Chaix J, Fenis A, Kerdiles YM, Yessaad N, Reynders A, et al. Fate mapping analysis of lymphoid cells expressing the NKp46 cell surface receptor. Proc Natl Acad Sci USA (2011) 108(45):18324–9. doi: 10.1073/pnas.1112064108
128. Chiossone L, Chaix J, Fuseri N, Roth C, Vivier E, Walzer T. Maturation of mouse NK cells is a 4-stage developmental program. Blood (2009) 113(22):5488–96. doi: 10.1182/blood-2008-10-187179
129. Li D, Wang Y, Yang M, Dong Z. mTORC1 and mTORC2 coordinate early NK cell development by differentially inducing E4BP4 and T-bet. Cell Death Differ (2021) 28(6):1900–9. doi: 10.1038/s41418-020-00715-6
130. Yang C, Tsaih S-W, Lemke A, Flister MJ, Thakar MS, Malarkannan S. mTORC1 and mTORC2 differentially promote natural killer cell development. eLife (2018) 7:e35619. doi: 10.7554/eLife.35619
131. Goh W, Huntington ND. Regulation of murine natural killer cell development. Front Immunol (2017) 8:130. doi: 10.3389/fimmu.2017.00130
132. Held W, Jeevan-Raj B, Charmoy M. Transcriptional regulation of murine natural killer cell development, differentiation and maturation. Cell Mol Life Sci (2018) 75(18):3371–9. doi: 10.1007/s00018-018-2865-1
133. Kennedy MK, Glaccum M, Brown SN, Butz EA, Viney JL, Embers M, et al. Reversible defects in natural killer and memory CD8 T cell lineages in interleukin 15-deficient mice. J Exp Med (2000) 191(5):771–80. doi: 10.1084/jem.191.5.771
134. Chauvin J-M, Ka M, Pagliano O, Menna C, Ding Q, Deblasio R, et al. IL15 stimulation with TIGIT blockade reverses CD155-mediated NK-cell dysfunction in melanoma. Clin Cancer Res (2020) 26(20):5520–33. doi: 10.1158/1078-0432.CCR-20-0575
135. Mendiratta SK, Martin WD, Hong S, Boesteanu A, Joyce S, Van Kaer L. CD1d1 mutant mice are deficient in natural T cells that promptly produce IL-4. Immunity (1997) 6(4):469–77. doi: 10.1016/S1074-7613(00)80290-3
136. Kawano T, Cui J, Koezuka Y, Toura I, Kaneko Y, Motoki K, et al. CD1d-restricted and TCR-mediated activation of valpha14 NKT cells by glycosylceramides. Science (1997) 278(5343):1626–9. doi: 10.1126/science.278.5343.1626
137. Gapin L, Matsuda JL, Surh CD, Kronenberg M. NKT cells derive from double-positive thymocytes that are positively selected by CD1d. Nat Immunol (2001) 2(10):971–8. doi: 10.1038/ni710
138. Godfrey DI, Stankovic S, Baxter AG. Raising the NKT cell family. Nat Immunol (2010) 11(3):197–206. doi: 10.1038/ni.1841
139. Godfrey DI, Macdonald HR, Kronenberg M, Smyth MJ, Kaer LV. NKT cells: what's in a name? Nat Rev Immunol (2004) 4(3):231–7. doi: 10.1038/nri1309
140. Bendelac A, Savage PB, Teyton L. The biology of NKT cells. Annu Rev Immunol (2007) 25(1):297–336. doi: 10.1146/annurev.immunol.25.022106.141711
141. Taniguchi M, Harada M, Dashtsoodol N, Kojo S. Discovery of NKT cells and development of NKT cell-targeted anti-tumor immunotherapy. Proc Japan Academy Ser B (2015) 91(7):292–304. doi: 10.2183/pjab.91.292
142. Osman Y, Kawamura T, Naito T, Takeda K, Van Kaer L, Okumura K, et al. Activation of hepatic NKT cells and subsequent liver injury following administration of alpha-galactosylceramide. Eur J Immunol (2000) 30(7):1919–28. doi: 10.1002/1521-4141(200007)30:7<1919::AID-IMMU1919>3.0.CO;2-3
143. Terashima A, Watarai H, Inoue S, Sekine E, Nakagawa R, Hase K, et al. A novel subset of mouse NKT cells bearing the IL-17 receptor b responds to IL-25 and contributes to airway hyperreactivity. J Exp Med (2008) 205(12):2727–33. doi: 10.1084/jem.20080698
144. Berzins SP, Ritchie DS. Natural killer T cells: drivers or passengers in preventing human disease? Nat Rev Immunol (2014) 14(9):640–6. doi: 10.1038/nri3725
145. Van Kaer L, Parekh VV, Wu L. Invariant natural killer T cells as sensors and managers of inflammation. Trends Immunol (2013) 34(2):50–8. doi: 10.1016/j.it.2012.08.009
146. Zhu T, Wang R, Miller H, Westerberg LS, Yang L, Guan F, et al. The interaction between iNKT cells and b cells. J Leukoc Biol (2022) 111(3):711–23. doi: 10.1002/JLB.6RU0221-095RR
147. Coquet JM, Chakravarti S, Kyparissoudis K, McNab FW, Pitt LA, McKenzie BS, et al. Diverse cytokine production by NKT cell subsets and identification of an IL-17-producing CD4-NK1.1- NKT cell population. Proc Natl Acad Sci USA (2008) 105(32):11287–92. doi: 10.1073/pnas.0801631105
148. Milpied P, Massot B, Renand A, Diem S, Herbelin A, Leite-de-Moraes M, et al. IL-17-producing invariant NKT cells in lymphoid organs are recent thymic emigrants identified by neuropilin-1 expression. Blood (2011) 118(11):2993–3002. doi: 10.1182/blood-2011-01-329268
149. Brennan PJ, Brigl M, Brenner MB. Invariant natural killer T cells: an innate activation scheme linked to diverse effector functions. Nat Rev Immunol (2013) 13(2):101–17. doi: 10.1038/nri3369
150. Rudak PT, Haeryfar SMM. In Vivo Cytotoxicity by α-GalCer-transactivated NK cells. Methods Mol Biol (2021) 2388:157–74. doi: 10.1007/978-1-0716-1775-5_15
151. Zhang Y, Springfield R, Chen S, Li X, Feng X, Moshirian R, et al. α-GalCer and iNKT cell-based cancer immunotherapy: realizing the therapeutic potentials. Front Immunol (2019) 10:1126. doi: 10.3389/fimmu.2019.01126
152. Michel ML, Mendes-da-Cruz D, Keller AC, Lochner M, Schneider E, Dy M, et al. Critical role of ROR-γt in a new thymic pathway leading to IL-17-producing invariant NKT cell differentiation. Proc Natl Acad Sci USA (2008) 105(50):19845–50. doi: 10.1073/pnas.0806472105
153. Lee YJ, Holzapfel KL, Zhu J, Jameson SC, Hogquist KA. Steady-state production of IL-4 modulates immunity in mouse strains and is determined by lineage diversity of iNKT cells. Nat Immunol (2013) 14(11):1146–54. doi: 10.1038/ni.2731
154. Constantinides MG, Bendelac A. Transcriptional regulation of the NKT cell lineage. Curr Opin Immunol (2013) 25(2):161–7. doi: 10.1016/j.coi.2013.01.003
155. Zhang L, Tschumi BO, Corgnac S, Rüegg MA, Hall MN, Mach JP, et al. Mammalian target of rapamycin complex 1 orchestrates invariant NKT cell differentiation and effector function. J Immunol (2014) 193(4):1759–65. doi: 10.4049/jimmunol.1400769
156. Shin J, Wang S, Deng W, Wu J, Gao J, Zhong XP. Mechanistic target of rapamycin complex 1 is critical for invariant natural killer T-cell development and effector function. Proc Natl Acad Sci USA (2014) 111(8):E776–83. doi: 10.1073/pnas.1315435111
157. Savage AK, Constantinides MG, Han J, Picard D, Martin E, Li B, et al. The transcription factor PLZF directs the effector program of the NKT cell lineage. Immunity (2008) 29(3):391–403. doi: 10.1016/j.immuni.2008.07.011
158. Inui T, Nakashima H, Habu Y, Nakagawa R, Fukasawa M, Kinoshita M, et al. Neutralization of tumor necrosis factor abrogates hepatic failure induced by alpha-galactosylceramide without attenuating its antitumor effect in aged mice. J Hepatol (2005) 43(4):670–8. doi: 10.1016/j.jhep.2005.02.027
159. Biburger M, Tiegs G. Alpha-galactosylceramide-induced liver injury in mice is mediated by TNF-alpha but independent of kupffer cells. J Immunol (2005) 175(3):1540–50. doi: 10.4049/jimmunol.175.3.1540
Keywords: Raptor, mTORC1, T cell, B cell, immune
Citation: Tang J, Yang L, Guan F, Miller H, Camara NOS, James LK, Benlagha K, Kubo M, Heegaard S, Lee P, Lei J, Zeng H, He C, Zhai Z and Liu C (2023) The role of Raptor in lymphocytes differentiation and function. Front. Immunol. 14:1146628. doi: 10.3389/fimmu.2023.1146628
Received: 17 January 2023; Accepted: 28 April 2023;
Published: 22 May 2023.
Edited by:
Fredrick Rosario-Joseph, University of Colorado Denver, United StatesReviewed by:
Yifang Gao, The First Affiliated Hospital of Sun Yat-sen University, ChinaAndrea Pelosi, Bambino Gesù Children’s Hospital (IRCCS), Italy
Copyright © 2023 Tang, Yang, Guan, Miller, Camara, James, Benlagha, Kubo, Heegaard, Lee, Lei, Zeng, He, Zhai and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhimin Zhai, enp6bTg4OUAxNjMuY29t; Chaohong Liu, Y2hhb2hvbmdsaXU4MEAxMjYuY29t
 Jianing Tang
Jianing Tang Lu Yang1
Lu Yang1 Heather Miller
Heather Miller Louisa K. James
Louisa K. James Kamel Benlagha
Kamel Benlagha Masato Kubo
Masato Kubo Steffen Heegaard
Steffen Heegaard Pamela Lee
Pamela Lee Chengwei He
Chengwei He Chaohong Liu
Chaohong Liu