- 1Department of Oncology, The First Affiliated Hospital of Zhengzhou University, Zhengzhou, China
- 2Lymphoma Diagnosis and Treatment Centre of Henan Province, Zhengzhou, China
- 3Academy of Medical Sciences of Zhengzhou University, Zhengzhou, Henan, China
- 4State Key Laboratory of Esophageal Cancer Prevention and Treatment and Henan Key Laboratory for Esophageal Cancer Research, The First Affiliated Hospital of Zhengzhou University, Zhengzhou, Henan, China
Gut microbiota represents a hidden treasure vault encompassing trillions of microorganisms that inhabit the intestinal epithelial barrier of the host. In the past decade, numerous in-vitro, animal and clinical studies have revealed the profound roles of gut microbiota in maintaining the homeostasis of various physiological functions, especially immune modulation, and remarkable differences in the configuration of microbial communities between cancers and healthy individuals. In addition, although considerable efforts have been devoted to cancer treatments, there remain many patients succumb to their disease with the incremental cancer burden worldwide. Nevertheless, compared with the stability of human genome, the plasticity of gut microbiota renders it a promising opportunity for individualized treatment. Meanwhile, burgeoning findings indicate that gut microbiota is involved in close interactions with the outcomes of diverse cancer immunotherapy protocols, including immune checkpoint blockade therapy, allogeneic hematopoietic stem cell transplantation, and chimeric antigen receptor T cell therapy. Here, we reviewed the evidence for the capacity of gut microflora to modulate cancer immunotherapies, and highlighted the opportunities of microbiota-based prognostic prediction, as well as microbiotherapy by targeting the microflora to potentiate anticancer efficacy while attenuating toxicity, which will be pivotal to the development of personalized cancer treatment strategies.
Introduction
The human gut microbiota refers to the vast collection of various microbes living on the epithelial barrier surfaces of the gastrointestinal tract, including bacteria, fungi, viruses, archaea, and protozoa (1). With the advances of molecular tools and technologies such as 16S ribosomal RNA sequencing, metagenomic, metabolomic, and metatranscriptomic, as well as the use of gnotobiotic animal models, the intricate host-microbiota interactions are progressively being deciphered (2). For one thing, substantial researches have featured the key roles of gut microbiota in human pathophysiological processes (3–5), including immunity, metabolism, and inflammatory response. For another, albeit multiple factors are proposed to propel cancer progression, the deviation of gut microbiota, known as dysbiosis, is entertained as a harbinger, promoter or even cause of a variety of malignant conditions (6). Thereinto, a panel of potential pro-tumorigenesis or anti-tumorigenesis microbial species have been identified too (7), which lays the groundwork for the regulation of gut microbiota in cancer therapy.
Meanwhile, with the incremental cancer burden worldwide, it places greater demands on personalized cancer treatments with powerful efficacy (8), although substantial advancements have been made, especially cancer immunotherapy. Most notably, the limited efficacy and undesired toxicities still remain the major hurdles of current cancer therapies, which has been found to be heavily influenced by distinct gut microflora patterns (9). Of them, immunotherapy has been considered as a major revolution, which provides exciting hopes for patients in the fight against cancer, and the effects of certain gut species on immunotherapy have now become a topic of great scientificity (10, 11). In the light of these findings, there is emerging interest in microbiotherapy by the modulation of intestinal flora as one of the antitumor strategies in recent years.
In this review, we mainly discussed the interactions between gut microbiota and cancer immunotherapies, including immune checkpoint blockade (ICB) therapy, allogeneic hematopoietic stem cell transplantation (allo-HSCT), and chimeric antigen receptor T (CAR-T) cell therapy, and the opportunities of microbiota-based patient stratification strategies such as the prediction of response and the early recognition of toxic events, as well as the evidence for the ability of microbiotherapy in the management of cancer immunotherapy, including enhancing anticancer efficacy and alleviating toxicity, thus, to decipher the roadmap of gut microbiota in the exploitation of custom-fit therapeutic strategies for cancer care. A diagrammatic representation of the interactions between gut microbiota and cancer immunotherapy is described in Figure 1.
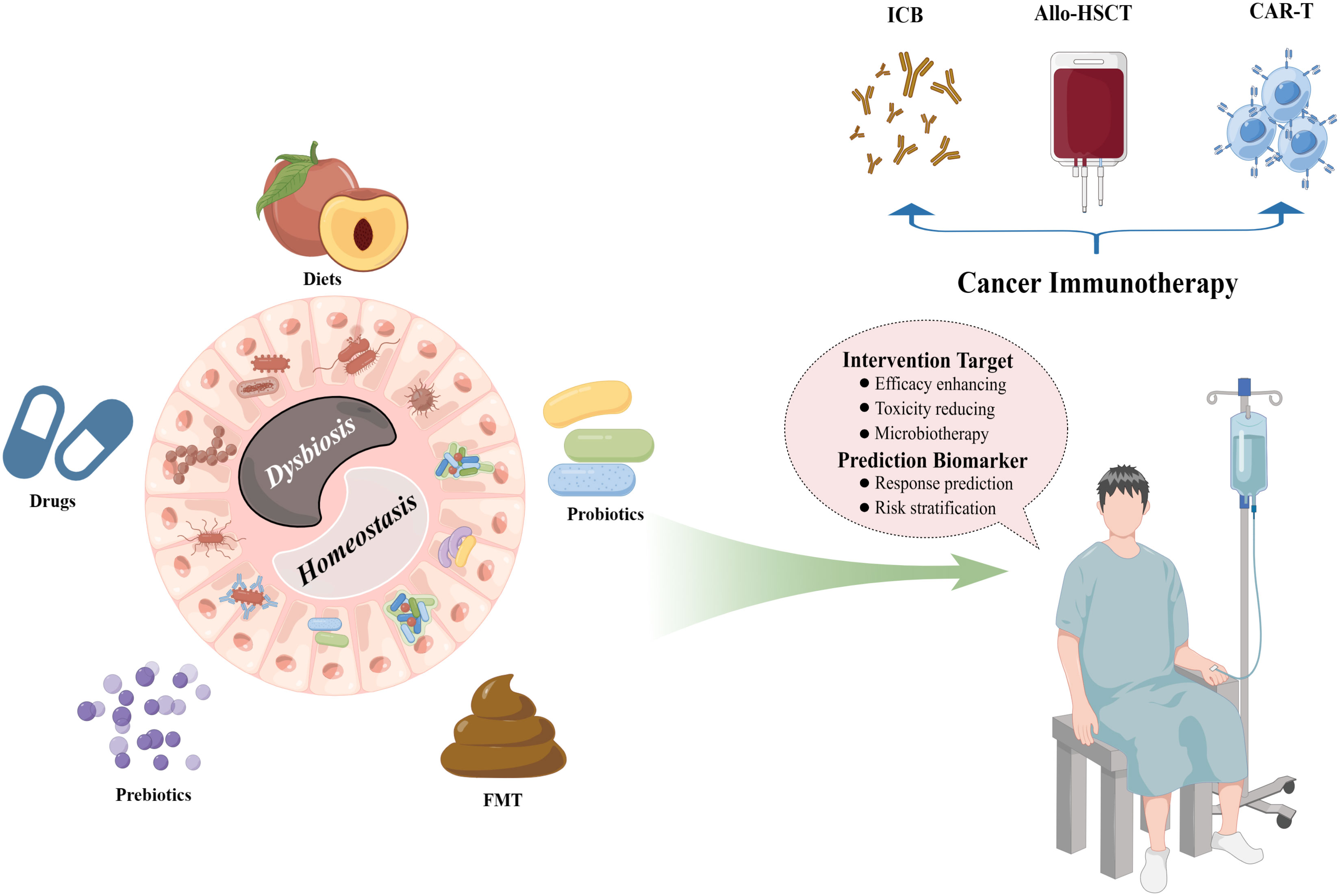
Figure 1 Interactions between the gut microbiota and cancer immunotherapy. The intestinal microbial ecosystem can be well modified by multiple patterns, including diets, drugs, prebiotics, probiotics, and FMT, which provide fascinating opportunities for the clinical managements of diverse cancer immunotherapy protocols such as ICB therapy, allo-HSCT, and CAR-T cell therapy. FMT, fecal microbiota transplantation; ICB, immune checkpoint blockade; allo-HSCT, allogeneic hematopoietic stem cell transplantation; CAR-T, chimeric antigen receptor T. (By Figdraw).
Immune checkpoint blockade therapy
Currently, one hotspot of cancer immunotherapy is the ICB therapy that inhibits programmed cell death protein 1/programmed cell death ligand 1 (PD-1/PD-L1) and cytotoxic T lymphocyte-associated antigen-4 (CTLA-4) signaling to reinvigorate CD8+ T cells in the tumor microenvironment (TME) to potentiate killing of tumor cells (12). Despite the remarkable effectiveness of ICB therapy in a subset of patients of several cancer types, including metastatic melanoma (13), classical Hodgkin lymphoma (14), non-small cell lung cancer (NSCLC) (15), and colon cancers (16), most patients were observed with primary or acquired resistance. Furthermore, a number of challenges such as the immune-related adverse events (irAEs) and biomarkers to predict response remain to be determined (17). However, accumulating data have pinpointed the indispensable roles of intestinal microbiota in ICB therapy.
The effects of antibiotics on the response of ICB therapy
Antibiotics-associated gut dysbiosis frequently confers deleterious effects on cancer patients treated with ICB (18). Derosa and colleagues (19) reported that antibiotics administration within 30 days of beginning ICB therapy was closely related to the inferior prognosis, including shorter progression free survival and overall survival (OS), in both advanced renal cell carcinoma and NSCLC patients. Similarly, the negative influences of antibiotics on the clinical outcomes of ICB have also be indicated in patients with melanoma (20), urothelial carcinoma (21), and bladder cancer (22). Nevertheless, Cheung et al. (23) and Fessas et al. (24) inversely revealed the detrimental and protective effects of antibiotics use on the survival of ICB treated hepatocellular carcinoma patients, respectively, which might be attributable to the difference of the antibiotic types, therapeutic regimens, baseline clinical characteristics and gut microbial features of patients. In addition, one caveat here is the antibiotics application might simply constitute a surrogate indicator of unsuited or immunodeficient cancer patients who were non-responsive for ICB therapy, which deserves further evaluation.
Influence of gut microbiota on the effectiveness of ICB therapy
The significance of commensal intestinal bacteria on the efficacy of ICB therapy has been well established in both pre-clinical models and patients. A plethora of microbial taxa, including Akkermansia muciniphila, Faecalibacterium spp., Bifidobacterium spp., and Bacteroides fragilis (25), have been reported to potentiate the antitumor efficacy of ICB therapy in both animal models and cancer patients. Of specific note, it has been well demonstrated that gut commensal bacteria such as Bifidobacterium and Bacteroidales could significantly improve tumor control of melanoma treated by anti-PD-L1 or anti-CTLA-4 via enhancing antitumor immunity response in mice models (26, 27). Further, this favorable role of commensal microbiome in ICB therapy was elucidated in melanoma patients (28, 29). Additionally, a higher diversity of gut microbiota at the starting point exhibited intimate relationships with the favorable responses to anti-PD-1 immunotherapy in patients with hepatocellular carcinoma and advanced NSCLC (30, 31), which might be involved in the enhanced memory T cell and natural killer cell signatures in the periphery in response to anti-PD-1 therapy. Interestingly, Helicobacter pylori seropositivity has been reported to be linked with an inferior NSCLC patient survival on anti-PD-1 therapy (32), and further be confirmed in in vitro co-culture assay and in H. pylori-infected mice with reduced number and activation status of tumor-specific CD8+ T cells in the tumors. Strikingly, apart from the linkages between individual bacterial taxa and ICB therapy outcome, the association of enteric microbiotypes (including diverse discrete ecologically balanced communities) with the response to melanoma patients treated by anti-PD-1 has also been proposed in a recent combination analysis (33). That is, four superclusters of a panel of microbial species, including two enriched in favorable taxa (Favorable 1: Bifidobacteriaceae, Eggerthelacea, Coriobacteriales, Akkermansia muciniphila, Fusobacteriaceae, Erysipelotrichaceae, Lachnospiraceae, Streptococcaceae, Lactobacillaceae, and Porphyromonadaceae; Favorable 2: Oscillospiraceae; by linear discriminant analyses) and two enriched in unfavorable taxa (Unfavorable 1: Prevotellaceae and Bacteroidales; Unfavorable 2: Rikenellaceae; by linear discriminant analyses), were defined, which comprised distinct microbiotypes with similar relationship between microbial composition and clinical outcome.
Various publications have now demonstrated a role for gut microbes in regulating responses to ICB therapy across several cancer types (Table 1). In a phase I clinical trial including ten patients with anti-PD-1-refractory metastatic melanoma (52), the researchers found that re-induction of anti-PD-1 combination with fecal microbiota transplantation (FMT) from complete response donors exhibited inspiring outcomes with clinical remission in three patients. Of them, FMT could remarkably increase the intra-tumoral immune activity, which supports the concept of overcoming resistance to immunotherapy through manipulating the intestinal microflora. Another multicenter retrospective study from Japan (53) revealed that probiotics administration was relevant to the survival and disease control in advanced or recurrent NSCLC patients that undergone anti-PD-1 monotherapy. Despite this, more thoughtful evaluations of the effects of current commercially available probiotic formulations on anticancer immunotherapy should be made, as they might be harmful in the setting of ICB therapy by impairing intra-tumoral IFN-γ T cell responses (40).
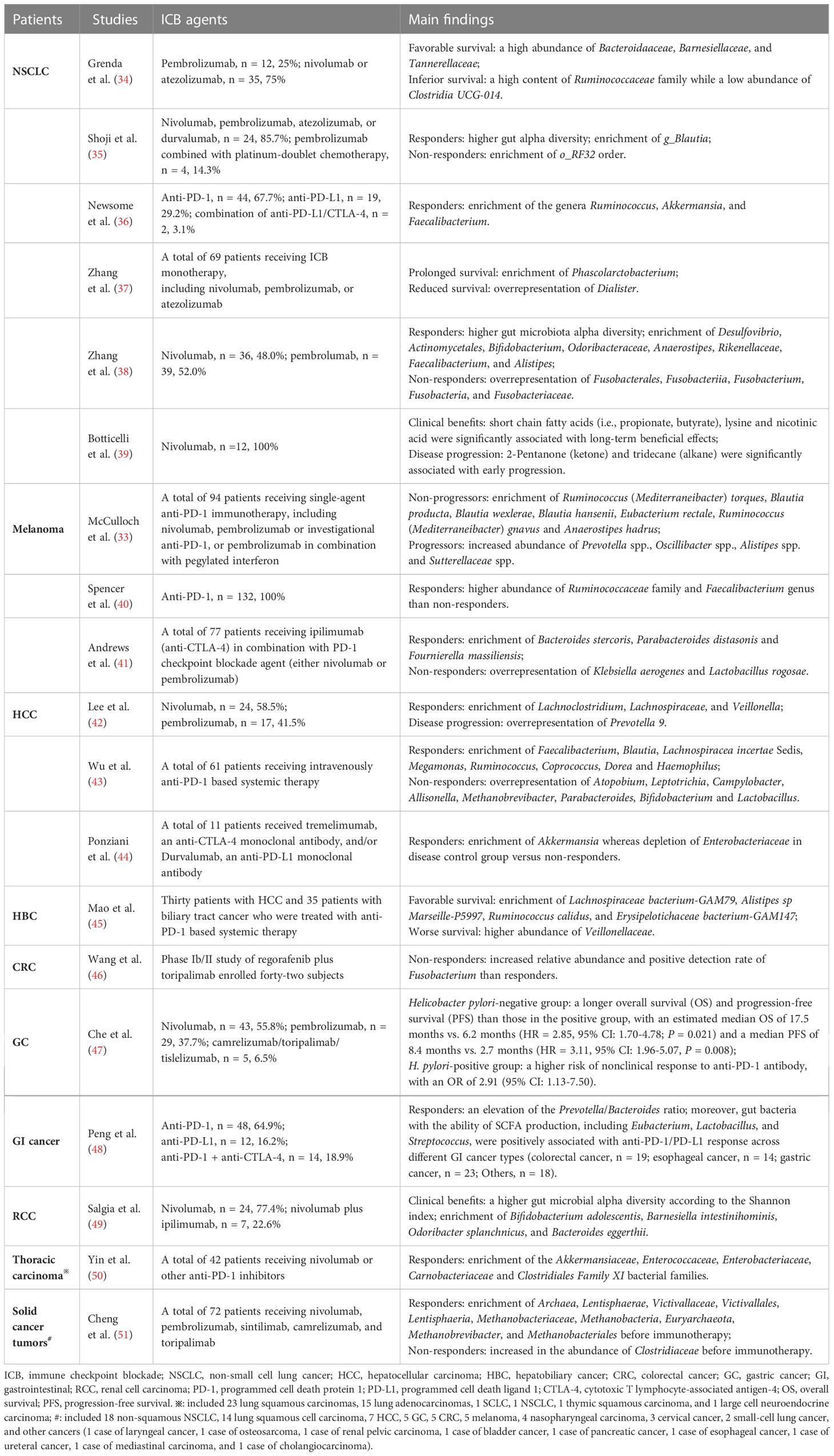
Table 1 Representative researches on the interactions between gut microbiota and the outcomes of ICB therapy across cancers in recent three years.
Interactions of gut microbiota with the toxicities related to ICB therapy
Evidence is accumulating that certain fecal microbiota composition is related to the development of several toxicities following ICB therapy such as irAEs, which result from off-tumor immune activation. McCulloch et al. (33) indicated that two microbial signatures, enriched for Streptococcaceae spp. and Lachnospiraceae spp., were involved in distinct irAEs, and melanoma patients with high Streptococcus spp. abundance in pretreatment microbiome samples tended to develop irAEs. Although higher rates of irAEs than anti-PD-1 or anti-CTLA-4 monotherapy, responders to combined ICB therapy targeting both CTLA-4 and PD-1 and responders to monotherapy exhibited similar compositional characteristics of gut microbiota with an enrichment of Ruminococcus/Ruminococcaceae consistently observed across diverse melanoma cohorts (41). Moreover, the researchers found a significantly higher abundance of Bacteroides intestinalis in patients developed ≥grade 3 irAEs versus those who did not, with upregulation of mucosal IL-1β in patient samples of colitis and in pre-clinical models.
Disturbances of intestinal homeostasis play a key role in driving ICB-associated toxicity. Stat3+/+ melanoma-bearing mice with acquired gastrointestinal impairment by Citrobacter rodentium infection and dextran sodium sulfate treatment displayed a predilection for anti-CTLA-4-mediated irAEs, with accumulation of neutrophils, cytotoxic and IFN-γ+ CD8+ and CD4+ T cells, and inflammatory cytokines such as IFN-γ and IL-6 in the colon (54). Furthermore, the pre-inflammation fecal microbiota of melanoma patients that presented a paucity of genetic pathways related to polyamine transport and B vitamin biosynthesis was linked with an increased risk of colitis (55). Remarkably, modulation of the gut microbiota can mitigate irAEs in cancers (56). Of them, ICB-related colitis could be successfully treated by FMT, with reconstitution of the intestinal microflora and increase in the proportion of regulatory T cells within the colonic mucosa (57). Additionally, microbial metabolites working at the interface between microorganisms and host immune system might abrogate ICB-induced colitis too. Renga et al. demonstrated that indole-3-carboxaldehyde, a microbial tryptophan catabolite, protected ICB-induced colitis mice from intestinal injury through a dual action on both the host and the microbes (58), which provides a new avenue in optimizing ICB therapy based on bacterial metabolome.
Prognostic utility of the gut microbiota-derived models for the outcome of ICB therapy
Gut microflora has emerged as a tumor-extrinsic predictive biomarker to the response of ICB therapy, and the machine learning models trained by microbial features provide a hopeful opportunity for outcome prediction. Recently, despite the heterogeneity across five melanoma cohorts, three modified leave-one-out cross-validation methods, including generalized linear model, random forest, and polynomial support-vector machine, based on batch-corrected intestinal microbiome data consistently predicted the outcomes to anti-PD-1 therapy in all cohorts (33). Of them, the Clostridium phylum was identified as a predictor of favorable outcome for a subset of cohorts, while the Bacteroidetes phylum was entertained as an unfavorable predictive indicator for the response of most melanoma cohorts. In addition, based on the bacterial signatures of five cancer cohorts, including melanoma, NSCLC, and renal cell carcinoma, treated with ICB, Shaikh et al. (59) constructed a non-responder “Integrated Microbiome Prediction Index” (calculated by assigning a weighted coefficient for the microbial species enriched in non-responders, including Bacteroides coprocola, Bacteroides fragilis, Bacteroides thetaiotaomicron, Bacteroides uniformis, Clostridium hathewayi, Clostridium hylemonae, Clostridium methylpentosum, Megasphaera micronuciformis, Oribacterium sinus, Parasutterella excrementihominis, Scardovia wiggsiae, and Veillonella parvula), rather than responder, that displayed the strongest and most consistent signal using a random effects model, which highlighted a novel avenue to recognize specific patients that probably benefit from microbiota-derived interventions to improve the outcomes of ICB therapy.
Gut microbiota-derived metabolites mediate the responses of ICB therapy
The gut microbial metabolites, a vast array of small molecules produced or transformed by intestinal microorganisms, represent one of the primary patterns by which the gut microbiota regulate antitumor immunity response, which are capable of conferring both local and systemic effects by spreading from their original location in the gastrointestinal tract to circulatory system (10, 60). Accordingly, it is necessary to dissect the underlying mechanistic pathways through which the specific bacterial metabolites impact on antitumor immunity and immunotherapeutic responses (Figure 2). Most notably, short−chain fatty acids (SCFAs, mainly including acetate, propionate, and butyrate), synthetized by the bacterial fermentation of dietary fiber, play a central role in the complicated gut microbial immune and metabolic networks (61). Of them, the gut microbial metabolite butyrate has been reported to engage in the enhanced anti-PD-1 therapeutic efficacy through increasing the CD4+ and CD8+ T cell infiltration in the TME in the tumor-bearing mice humanized with the intestinal microbes from colorectal cancer (CRC) patients (62). Moreover, replenishing butyrate prior to anti-PD-1 treatment was sufficient to recover the therapeutic efficacy in the non-responders. Similarly, He and colleagues indicated that the SCFAs butyrate could directly potentiate the antitumor CD8+ T cell response via ID2-dependent IL-12 signaling (63), suggesting the potential beneficial roles of butyrate supplementation in anticancer immunity therapy. However, the roles of SCFAs in ICB therapy might be ambiguous. In a study conducted by Coutzac and colleagues in both mice and melanoma patients treated with anti-CTLA-4 monoclonal antibody, the authors found that SCFAs limited the efficacy of anti-CTLA-4 treatment. Namely, high levels of blood butyrate and propionate were involved in the resistance to CTLA-4 blockade and higher frequency of Treg cells, and butyrate could impede the accumulation of tumor-specific T cells and memory T cells (64). Therefore, further evaluations are warranted to reveal a more nuanced illustration for the effects of SCFAs on antitumor immune and the responses to ICB therapy.
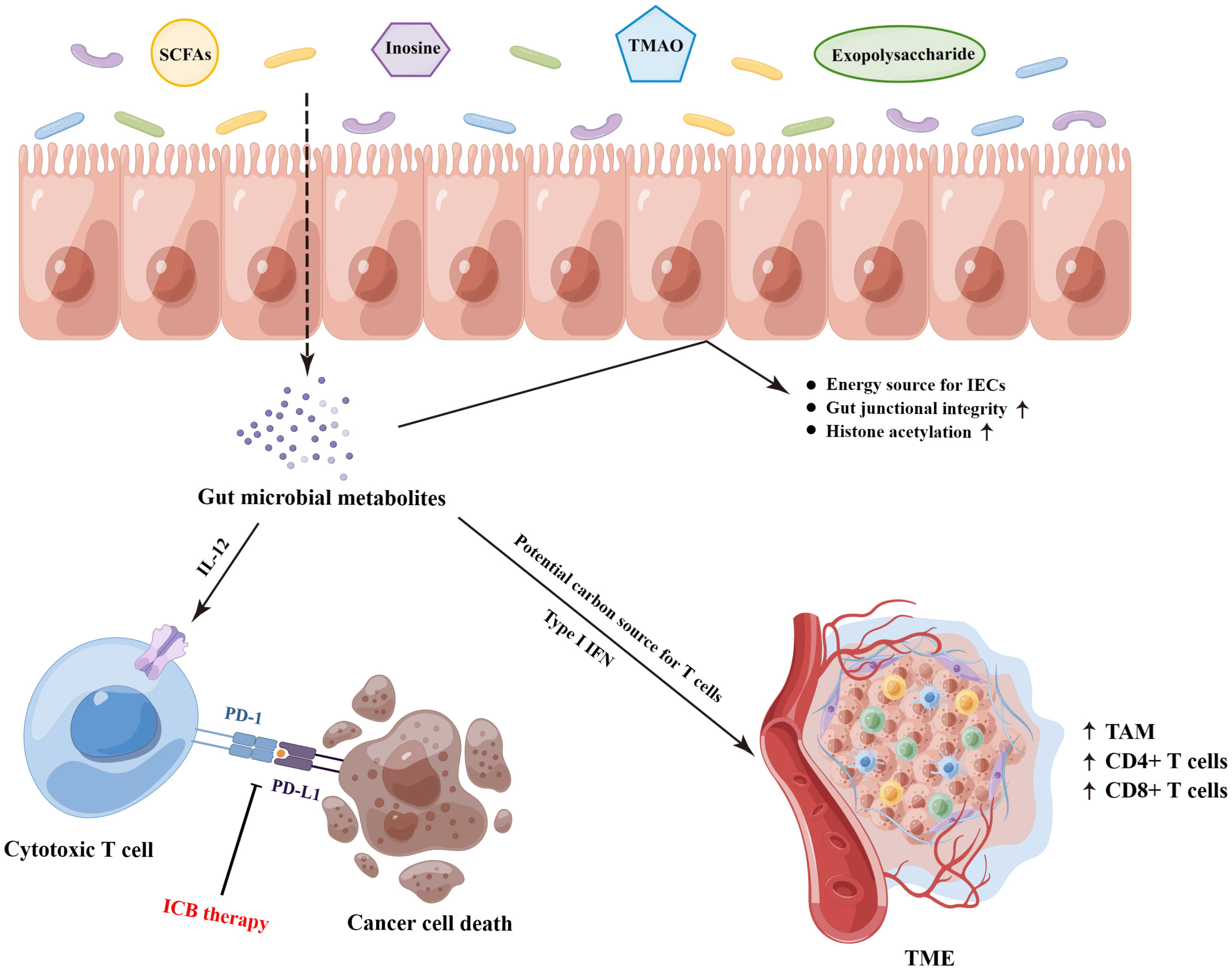
Figure 2 The underlying molecular mechanisms on the gut microbiota-derived metabolites that mediated the responses of ICB therapy. SCFAs, short−chain fatty acids; TMAO, trimethylamine N-oxide; IECs, intestinal epithelial cells; ICB, immune checkpoint blockade; TAM, tumor-associated macrophage; TME, tumor microenvironment. (By Figdraw).
In addition to SCFAs, other gut microbial metabolites also exhibit profound effects on the treatment of ICB. Strikingly, the bacterial metabolite inosine has been demonstrated to modulate enhanced ICB therapy response in mouse models of intestinal and epithelial tumors, which was dependent on T cell expression of the adenosine A2A receptor to promote Th1 cell activation (65). Furthermore, as a substitute carbon source for the metabolism of T lymphocyte in glucose-restricted environments such as TME, inosine can assist T cell proliferation and differentiation while fueling sensitivity to ICB therapy (66). Another gut microbiota-derived metabolite trimethylamine N-oxide, identified as a driver of antitumor immunity, exhibited the ability to boost the response to ICB therapy in pancreatic cancer-bearing mouse model (67). Of them, the administration of trimethylamine N-oxide was related to an immunostimulatory tumor-associated macrophage phenotype, and activated effector T cell response in the TME in a type I IFN-dependent manner. Interestingly, Kawanabe-Matsuda et al. (68) illustrated that oral consumption of Lactobacillus-derived exopolysaccharide could bolster the efficacy of ICB therapy against CCL20-expressing tumors via inducing CCR6+ CD8+ T cells in Peyer’s patches and improving the TME in experimental mouse tumor models, which provided compelling evidence on the dietary ingestion of exopolysaccharide for further clinical trials. Altogether, these studies lay the groundwork for the potential cancer immunotherapeutic strategies by targeting gut microbiota-derived metabolites.
Allogeneic hematopoietic stem cell transplantation
Allo-HSCT remains a curative approach for a range of hematological malignancies and might be recognized as one of the earliest effective modalities for cancer immunotherapy, but it is still hindered by high mortality rates, mainly because of graft-versus-host disease (GVHD) (69). Notably, at present the interactions between the intestinal microbiota and patient outcome after allo-HSCT have been well established (70). Particularly, ample evidence indicates an effect of gut microbial dysbiosis on GVHD. Furthermore, gut microbiota modulation through FMT also exhibits a promising revolution in the managements of allo-HSCT recipients, including ameliorating treatment-associated complications and improving patient outcomes.
Relationships between gut microbiota and allo-HSCT
Holler et al. (71) conducted a prospective research to collect stool specimens from 31 patients receiving allo-HSCT, and revealed that the loss of bacterial diversity and predominance of enterococci induced by systemic antibiotics might involve in the pronounced gastrointestinal GVHD for the first time. The patterns of microbial dysregulation during allo-HSCT were similar across diverse transplantation centers and geographic locations, and the depletion of gut diversity during allo-HSCT (accompanied by the domination of single taxa such as the genera enterococcus and streptococcus) has been observed to be linked with higher risks of transplantation-associated death in a large multi-center study (72). Recently, Andrlová and colleagues (73) indicated that a diverse gut microbiota early after allo-HSCT could produce more activating ligands for innate-like mucosal-associated invariant T cells and Vδ2 cells to maintain the immunological link between these populations, which contributed to improved OS and less acute GVHD. Furthermore, Enterococcal expansion after allo-HSCT as a remarkable risk factor for the occurrence of acute GVHD and reduced OS has been observed again in another multi-center study including 1325 recipients, and further be demonstrated in mouse models (74). Moreover, the researchers also found that posttransplant enterococcal enrichment was accompanied by the depletion of clostridia, with a significant reduction in fecal butyrate in both pre-clinical models and patients with GVHD. This result was consistent with a recent prospective single-center study that included 201 patients undergoing allo-HSCT and 28 healthy donors (75), indicating that butyrate-producing Clostridiales diminished early in the course of allo-HSCT, which was involved in the increased acute GVHD severity and transplantation associated mortality. In addition, it has also been illustrated that patients suffering chronic GVHD exhibited lower circulating concentrations of the butyrate and propionate in day 100 plasma samples (76). Most recently, Hino and colleagues (77) analyzed the gut microbial signatures of 59 long-term survivors (1-21.7 years; median, 6.4 years) after allo-HSCT, and found that intestinal dysbiosis with decreased abundance of the butyrate-producing bacteria was present over a 10-year lifetime after discharge following allo-HSCT. Of them, only limited chronic GVHD patients displayed no depletion of butyrate-producing Faecalibacterium. Similarly, a study including 541 patients admitted for allo-HSCT conducted by Peled et al. (78) indicated that patients with the dominance of another butyrate-producing Eubacterium limosum also displayed a close association with the reduced risk of relapse or progression of disease (hazard ratio [HR], 0.82 per 10-fold increase in abundance; 95% confidence interval (CI), 0.71 to 0.95; P = .009). Of note, as a major energy source for intestinal epithelial cells (IECs), evidence indicated that butyrate could improve IECs junctional integrity, decrease apoptosis and mitigate GVHD in mice, and the loss of butyrate led to reduced degree of histone acetylation in IECs (79).
Apart from the associations with GVHD, gut microbiota also has potential implications for a variety of other toxic effects, including γ-proteobacteria domination predicting pulmonary complications after engraftment (80), and gram-negative intestinal domination predicting subsequent bloodstream infection (81), while a more stable gut microbial configuration protecting febrile neutropenia (82), and three distinct bacterial taxa (Bacteroidetes, Lachnospiraceae, and Ruminococcaceae) protecting post-engraftment Clostridium difficile infection (83). In addition, it has to be noted the critical roles of intestinal microorganisms beyond bacteria played in allo-HSCT. Of them, Legoff and colleagues (84), characterizing the dynamic evolution of gut virome in 44 recipients during allo-HSCT by metagenomics, found that the overall proportion of vertebrate viral sequences in the guts of all recipients increased progressively during the weeks following transplantation, and the RNA viral reads from picobirnaviruses were predictive of later occurrence of severe enteric GVHD of stage 2 or higher (HR = 2.66; 95% CI = 1.46-4.86; P = 0.001) through a time-dependent Cox proportional-hazards model. In addition, Rolling et al. (85) reported the fungal dysbiosis in a cohort of 156 patients during allo-HSCT by both longitudinal amplicon-based and culture-dependent analyses in 1279 fecal samples. Notably, Candida parapsilosis complex species, including C. parapsilosis, C. orthopsilosis, and C. metapsilosis, were the most common cultured fungi. Compared with those without pre-engraftment domination by C. parapsilosis complex species, patients with C. parapsilosis complex domination pre-engraftment exhibited a higher transplant-related mortality and worse OS.
Effects of the gut microbial modulation on allo-HSCT recipients
On the basis of these findings, there has been tremendous interests in gut microbial modulation with the aim to improve the outcome of patients undergoing allo-HSCT (86–88), including antibiotics, diets, prebiotics, probiotics, and FMT.
Antibiotics
Recently, Severyn and colleagues (89) found that gut decontamination, by oral vancomycin-polymyxin B in patients undergoing allo-HSCT, might protect recipients against gut-derived bloodstream infection by reducing the prevalence of gut pathogens. Additionally, previous research has suggested that occurrence of GVHD after allo-HSCT in obesity mice could be mitigated by prophylactic antibiotic treatment (90). Despite this, great caution should be exercised when delineating the effects of antibiotics on allo-HSCT as increasing evidence has illustrated the detrimental roles of antibiotic administration in recipients. A retrospective research examined 857 allo-HSCT recipients from Shono and colleagues (91) reported that the use of antibiotics such as imipenem-cilastatin and piperacillin-tazobactam were linked with elevated GVHD-related mortality at 5 years. Through GVHD mice model, the authors further illustrated that imipenem-cilastatin treatment led to the loss of the protective mucus lining of the colon and intestinal barrier impairment, which might be explained by the enrichment of mucus-degrading Akkermansia muciniphila. Furthermore, an increased risk of patients occurring acute and intestinal GVHD by gut decontamination and prophylaxis has also been revealed in a meta-analysis of 18 references (92). Of note, the conflicting clinical results regarding the influence of antibiotics on the outcome of allo-HSCT might be explained by the different types of antibiotics and timing of treatment (87).
Diets, prebiotics, and probiotics
Dietary elements and nutritional strategies have been increasingly evaluated regarding the impact on allo-HSCT outcomes through modulating intestinal microorganisms (93). It has been revealed that mice with diet-induced obesity exhibited reduced survival associated with acute and severe gut GVHD, which was consistent with the inferior survival of allo-HSCT recipients with a high body mass index (BMI, >30) that presented decreased gut diversity and Clostridiaceae abundance (90). In addition, Li and colleagues (94) demonstrated the roles of tyrosine in acute GVHD murine models. Specifically, additional tyrosine supplementation could significantly prolong OS, alleviate symptoms at the early stage of acute GVHD by regulating the microbial composition and fecal metabolic phenotype. Likewise, prebiotic intake has also been observed to be an effective strategy for preventing acute GVHD in allo-HSCT in a prospective study (95). Namely, from pre-transplantation conditioning to day 28 after allo-HSCT, the combined administration of resistant starch and a commercially available prebiotics mixture (including glutamine, fiber, and oligosaccharide) decreased the incidence of all acute GVHD grades combined and of acute GVHD grades 2 to 4, and maintained the intestinal diversity and butyrate-producing bacterial population.
Compared with prebiotics, probiotics are viable microorganisms for healthy gut restoration. Importantly, the protective roles of the butyrate-producing Clostridia have been well demonstrated in preclinical allo-HSCT models (79, 96). Of them, Mathewson and colleagues (79) indicated that altering the indigenous microbiota, using the cocktail of 17 rationally selected Clostridial strains with the ability to produce high amounts of butyrate, could remarkably attenuate GVHD severity and improve survival. Furthermore, the safety and feasibility of another probiotic: Lactobacillus plantarum (LBP), have been also evaluated in children and adolescents undergoing allo-HSCT (97), and with no cases of LBP-bacteremia or LBP-associated severe adverse events recorded. Nevertheless, the safety and efficacy of probiotics in HSCT therapy remain elusive. For example, Lactobacillus acidophilus sepsis secondary to the excessive consumption of probiotic-enriched yogurt has been reported in a case with mantle cell lymphoma receiving HSCT (98).
Fecal microbiota transplantation
FMT refers to the transfer of fecal microbial content from a healthy donor to the intestine of a recipient (99), and represents a promising approach for the management of allo-HSCT patients (88), including alleviating infection of multidrug-resistant bacteria and GVHD, as well as promoting gut microbiota reconstitution. Bluestone and colleagues (100) reported that FMT displayed better safety and tolerance in three children developing recurrent Clostridium difficile infection after allo-HSCT, and one case did obtain successful clearance of C. difficile at follow-up 1 year 10 months after the FMT. Moreover, the safety and efficacy of FMT in the decolonization of multidrug-resistant bacteria, including vancomycin-resistant enterococci (n=2) or carbapenemase-producing bacteria (n=8), have also been presented in ten allo-HSCT patients with hematologic malignancies (101). Of them, seven of ten patients achieved decolonization and almost all patients without severe infectious events occurred during the first three months after FMT.
In a prospective, single-center, single-arm study enrolling 15 patients with steroid-refractory or steroid-dependent, acute or late-onset acute intestinal GVHD suffering allo-HSCT, van Lier et al. (102) found that ten of 15 subjects obtained a complete clinical response within 1 month after FMT, with a partial engraftment of donor microbial species, increased gut microbial α-diversity, and enrichment of butyrate-producing Clostridiales and Blautia species. As mentioned above, loss of intestinal diversity involves unfavorable allo-HSCT outcomes. Interestingly, FMT after allo-HSCT tends to be related to the improvement of recipients’ gut diversity that could be attributable to expansion of stool-donor taxa (103). In addition, it has been reported that autologous FMT (feces were provided by participants before the initiation of allo-HSCT), after microbiota-depleting antibiotic treatment, had the ability to boost microbial diversity and reestablished the commensal bacterial populations at the critical early immune reconstitution stage after allo-HSCT (104). Taken together, although FMT seemed safe and well-tolerated, further larger prospective studies are urgently required to deal with several safety concerns such as potential risks of infection upon FMT in these immunocompromised patients.
Chimeric antigen receptor T cell therapy
CAR-T cell therapy stands at the novel forefront of current cancer therapy, which has demonstrated unprecedented responses in patients with high-risk hematologic malignancies, including lymphoma, leukemia, and multiple myeloma (105–108). CAR-T cell therapy involves genetically modified T cells that express specific CAR, followed by in vitro cell amplification and reinfusion back into the patient to eradicate tumors (109). Given the intimate interactions of gut microflora with human T cell function and anti-tumor immunity (110, 111), it is not unexpected that the interactions and potential mechanisms of gut microbiota with CAR-T cell therapy have begun to be investigated in recent years (112, 113). Of them, Uribe-Herranz et al. (114) illustrated that gut microflora could modulate the anti-tumor efficacy of adoptive T cell therapy, mediated by CD8α+ dendritic cells and IL-12, in the tumor-bearing mice model.
Although the success of CAR-T cell therapy, several obstacles, including CAR-mediated toxicities, CAR-T cell dysfunction, antigen loss, tumor heterogeneity, and disease relapse, have impeded the utility of CAR-T cell therapy. Therefore, biomarkers for the favorable prognostic identification of patients receiving CAR-T cells are urgently needed. Inspiringly, in a multi-center retrospective study including patients with B-cell lymphoma and leukemia, Smith et al. (115) found that exposure to antibiotics prior to CD19 CAR-T cell infusion was involved in significantly inferior survival and increased immune effector cell-associated neurotoxicity syndrome. Moreover, enrichment of certain members within the class Clostridia, including Faecalibacterium, Ruminococcus, and Bacteroides, were linked with day 100 complete response to CAR-T cell therapy. Similarly, Hu and colleagues (116) also revealed the significant differences in the abundance of Bifidobacterium, Prevotella, Sutterella, and Collinsella between multiple myeloma patients in complete remission and those in partial remission, and observed a higher abundance of Bifidobacterium, Leuconostoc, Stenotrophomonas, and Staphylococcus in patients with severe cytokine release syndrome. Altogether, despite the research on the role of intestinal microbiota in CAR-T cell therapy is still at the very earliest stages, these findings suggest the tremendous potential of gut microbiota as a non-invasive prognostic marker for CAR-T cell therapy, and provide a novel reference to alleviate CAR-T cell therapy-induced toxic effects and to improve therapeutic outcome by modulating the gut microbiota.
Future perspectives and current challenges
Although major strides have been made toward the treatment of cancers, there remain many patients succumb to their disease (117). However, different from the stability of human genome, the modifiable nature of gut microbiota renders it a promising opportunity for cancer therapy (118). And meanwhile, there are emerging lines of evidence suggest the therapeutic potential of microbiotherapy by targeting the microbial flora. Among these, FMT appears central in the intervention options to restore microbial richness, as well as amend microbial dysbiosis and altered host-microbiota symbiosis related to cancer genesis and treatment (70). Moreover, utilizing bacteria strains or its proteins and peptides substances, including bacteriocins and toxins, as the anticancer agents on various cancers, termed bacteriotherapy, has also attracted salient attention, which can be employed alone or in conjunction with traditional therapies as an enhancer (119). Of interest, Montalban-Arques et al. (120) reported that oral supplementations of a mix of four Clostridiales species, namely Roseburia intestinalis, Eubacterium hallii, Faecalibacterium prausnitzii, and Anaerostipes caccae, outperformed anti-PD-1 therapy in mouse models of CRC and melanoma, which provided a strong preclinical foundation for exploring gut flora as novel stand-alone therapy against solid tumors. Additionally, despite the safety of probiotics in the management of cancer patients remains largely undefined, several gut next-generation probiotics such as Faecalibacterium prausnitzii, Akkermansia muciniphila, and Bacteroides fragilis exhibit their beneficial roles in supporting cancer therapy (121).
Most importantly, with mounting evidence of microorganisms colonizing tumors, synthetic biology approaches are being leveraged to improve the effectiveness of bacteriotherapy agents by repurposing bacteria. As an intelligent medicine, engineering bacteria are able to demonstrate autonomous control, sensing and responding to the internalization process, and subsequently releasing cargo (122). Furthermore, combinations of engineering bacteria with drug-loaded nanoparticles, monoclonal antibodies, oncolytic virus, and even CAR-T cells will also open charming options in oncology (123, 124).
Following the tremendous advances of cultivation-independent technologies and microbial analysis tools, the profiles of gut microbiota have been extensively revealed. While much attention has been given to gut bacteria, the contributions of other intestinal microorganisms such as viruses and fungi to cancer genesis and treatment also deserve further scrutiny. Furthermore, given the overlapped alterations of gut species across cancers (125, 126), future work is warranted to clarify the roles of gut microbiota-derived strategies, using machine learning algorithms, in precise risk stratification, prognostication, and therapeutic decision-making of cancer patients.
Although we believe that modulation of gut microflora will probably be the next vanguard in the management of cancer patients, however, several potential challenges should be mentioned. First, the exact mechanisms of action between gut dysbiosis and cancer genesis and therapy remain poorly characterized, and proofs of causation between them are still lacking. Therefore, continual efforts should be made to rationally select intestinal probiotics. On top of that, as a living body, the complexity of bacteria determines the hardships and risks such as biocontainment and safety concerns of transforming them into weapons to fight against cancers. Finally, considering the complex physiological conditions such as gastric acid and diverse enzymes that might digest or deactivate bacteriotherapy agents before they reach the action site, appropriate delivery route and dose of administration are also need to be investigated during clinical translations.
Conclusion
In recent years, overwhelming pre-clinical and patient-oriented evidence supports a critical role of gut microbiota in cancer immunotherapies such as improving efficacy and mitigating toxicity, and the manipulation of gut microbiota confers a promising therapeutic strategy for the clinical management of malignancies as well. Currently, microbiotherapy for cancers is still in its infancy. With the formidable challenges notwithstanding, it deserves further mechanistic dissection by cellular and animal studies as well as validation with larger longitudinal clinical cohorts.
Author contributions
MZ, ZL, and ZS conceived the study. ZS, HL, WS, and ZZ collected the related references and participated in the discussion. ZS and ZL drafted the manuscript and prepared the figures. MZ and ZS revised the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the Funds for Creative Research Groups of the First Affiliated Hospital of Zhengzhou university (QNCXTD2023012) and the National Natural Science Foundation of China (81970184; 82170183; U1904139; 82070209).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Abbreviations
ICB, immune checkpoint blockade; allo-HSCT, allogeneic hematopoietic stem cell transplantation; CAR-T, chimeric antigen receptor T; PD-1/PD-L1, programmed cell death protein 1/programmed cell death ligand 1; CTLA-4, cytotoxic T lymphocyte-associated antigen-4; TME, tumor microenvironment; NSCLC, non-small cell lung cancer; irAEs, immune-related adverse events; OS, overall survival; FMT, fecal microbiota transplantation; SCFAs, short−chain fatty acids; CRC, colorectal cancer; GVHD, graft-versus-host disease; HR, hazard ratio; CI, confidence interval; IECs, intestinal epithelial cells; LBP, Lactobacillus plantarum.
References
1. Aggarwal N, Kitano S, Puah GRY, Kittelmann S, Hwang IY, Chang MW. Microbiome and human health: Current understanding, engineering, and enabling technologies. Chem Rev (2023) 123(1):31–72. doi: 10.1021/acs.chemrev.2c00431
2. de Vos WM, Tilg H, Van Hul M, Cani PD. Gut microbiome and health: mechanistic insights. Gut (2022) 71(5):1020–32. doi: 10.1136/gutjnl-2021-326789
3. Scott AJ, Alexander JL, Merrifield CA, Cunningham D, Jobin C, Brown R, et al. International cancer microbiome consortium consensus statement on the role of the human microbiome in carcinogenesis. Gut (2019) 68(9):1624–32. doi: 10.1136/gutjnl-2019-318556
4. Fan Y, Pedersen O. Gut microbiota in human metabolic health and disease. Nat Rev Microbiol (2021) 19(1):55–71. doi: 10.1038/s41579-020-0433-9
5. Shi Z, Zhang M. Emerging roles for the gut microbiome in lymphoid neoplasms. Clin Med Insights Oncol (2021) 15:11795549211024197. doi: 10.1177/11795549211024197
6. Cullin N, Azevedo Antunes C, Straussman R, Stein-Thoeringer CK, Elinav E. Microbiome and cancer. Cancer Cell (2021) 39(10):1317–41. doi: 10.1016/j.ccell.2021.08.006
7. Bhatt AP, Redinbo MR, Bultman SJ. The role of the microbiome in cancer development and therapy. CA Cancer J Clin (2017) 67(4):326–44. doi: 10.3322/caac.21398
8. Kocarnik JM, Compton K, Dean FE, Fu W, Gaw BL, Harvey JD, et al. Cancer incidence, mortality, years of life lost, years lived with disability, and disability-adjusted life years for 29 cancer groups from 2010 to 2019: A systematic analysis for the global burden of disease study 2019. JAMA Oncol (2022) 8(3):420–44. doi: 10.1001/jamaoncol.2021.6987
9. Liu L, Shah K. The potential of the gut microbiome to reshape the cancer therapy paradigm: A review. JAMA Oncol (2022) 8(7):1059–67. doi: 10.1001/jamaoncol.2022.0494
10. Wang Y, Zhang H, Liu C, Wang Z, Wu W, Zhang N, et al. Immune checkpoint modulators in cancer immunotherapy: Recent advances and emerging concepts. J Hematol Oncol (2022) 15(1):111. doi: 10.1186/s13045-022-01225-3
11. Gopalakrishnan V, Helmink BA, Spencer CN, Reuben A, Wargo JA. The influence of the gut microbiome on cancer, immunity, and cancer immunotherapy. Cancer Cell (2018) 33(4):570–80. doi: 10.1016/j.ccell.2018.03.015
12. Ribas A, Wolchok JD. Cancer immunotherapy using checkpoint blockade. Science (2018) 359(6382):1350–5. doi: 10.1126/science.aar4060
13. Robert C, Thomas L, Bondarenko I, O’Day S, Weber J, Garbe C, et al. Ipilimumab plus dacarbazine for previously untreated metastatic melanoma. New Engl J Med (2011) 364(26):2517–26. doi: 10.1056/NEJMoa1104621
14. Ansell SM, Lesokhin AM, Borrello I, Halwani A, Scott EC, Gutierrez M, et al. PD-1 blockade with nivolumab in relapsed or refractory hodgkin’s lymphoma. New Engl J Med (2015) 372(4):311–9. doi: 10.1056/NEJMoa1411087
15. Reck M, Rodríguez-Abreu D, Robinson AG, Hui R, Csőszi T, Fülöp A, et al. Pembrolizumab versus chemotherapy for PD-L1-Positive non-Small-Cell lung cancer. New Engl J Med (2016) 375(19):1823–33. doi: 10.1056/NEJMoa1606774
16. Le DT, Uram JN, Wang H, Bartlett BR, Kemberling H, Eyring AD, et al. PD-1 blockade in tumors with mismatch-repair deficiency. New Engl J Med (2015) 372(26):2509–20. doi: 10.1056/NEJMoa1500596
17. Morad G, Helmink BA, Sharma P, Wargo JA. Hallmarks of response, resistance, and toxicity to immune checkpoint blockade. Cell (2021) 184(21):5309–37. doi: 10.1016/j.cell.2021.09.020
18. Elkrief A, Derosa L, Kroemer G, Zitvogel L, Routy B. The negative impact of antibiotics on outcomes in cancer patients treated with immunotherapy: a new independent prognostic factor? Ann Oncol (2019) 30(10):1572–9. doi: 10.1093/annonc/mdz206
19. Derosa L, Hellmann MD, Spaziano M, Halpenny D, Fidelle M, Rizvi H, et al. Negative association of antibiotics on clinical activity of immune checkpoint inhibitors in patients with advanced renal cell and non-small-cell lung cancer. Ann Oncol (2018) 29(6):1437–44. doi: 10.1093/annonc/mdy103
20. Mohiuddin JJ, Chu B, Facciabene A, Poirier K, Wang X, Doucette A, et al. Association of antibiotic exposure with survival and toxicity in patients with melanoma receiving immunotherapy. J Natl Cancer Inst (2021) 113(2):162–70. doi: 10.1093/jnci/djaa057
21. Hopkins AM, Kichenadasse G, Karapetis CS, Rowland A, Sorich MJ. Concomitant antibiotic use and survival in urothelial carcinoma treated with atezolizumab. Eur Urol (2020) 78(4):540–3. doi: 10.1016/j.eururo.2020.06.061
22. Pederzoli F, Bandini M, Raggi D, Marandino L, Basile G, Alfano M, et al. Is there a detrimental effect of antibiotic therapy in patients with muscle-invasive bladder cancer treated with neoadjuvant pembrolizumab? Eur Urol (2021) 80(3):319–22. doi: 10.1016/j.eururo.2021.05.018
23. Cheung KS, Lam LK, Seto WK, Leung WK. Use of antibiotics during immune checkpoint inhibitor treatment is associated with lower survival in hepatocellular carcinoma. Liver Cancer (2021) 10(6):606–14. doi: 10.1159/000518090
24. Fessas P, Naeem M, Pinter M, Marron TU, Szafron D, Balcar L, et al. Early antibiotic exposure is not detrimental to therapeutic effect from immunotherapy in hepatocellular carcinoma. Liver Cancer (2021) 10(6):583–92. doi: 10.1159/000519108
25. Routy B, Gopalakrishnan V, Daillere R, Zitvogel L, Wargo JA, Kroemer G. The gut microbiota influences anticancer immunosurveillance and general health. Nat Rev Clin Oncol (2018) 15(6):382–96. doi: 10.1038/s41571-018-0006-2
26. Sivan A, Corrales L, Hubert N, Williams JB, Aquino-Michaels K, Earley ZM, et al. Commensal bifidobacterium promotes antitumor immunity and facilitates anti-PD-L1 efficacy. Science (2015) 350(6264):1084–9. doi: 10.1126/science.aac4255
27. Vétizou M, Pitt JM, Daillère R, Lepage P, Waldschmitt N, Flament C, et al. Anticancer immunotherapy by CTLA-4 blockade relies on the gut microbiota. Science (2015) 350(6264):1079–84. doi: 10.1126/science.aad1329
28. Matson V, Fessler J, Bao R, Chongsuwat T, Zha Y, Alegre ML, et al. The commensal microbiome is associated with anti-PD-1 efficacy in metastatic melanoma patients. Science (2018) 359(6371):104–8. doi: 10.1126/science.aao3290
29. Gopalakrishnan V, Spencer CN, Nezi L, Reuben A, Andrews MC, Karpinets TV, et al. Gut microbiome modulates response to anti-PD-1 immunotherapy in melanoma patients. Science (2018) 359(6371):97–103. doi: 10.1126/science.aan4236
30. Zheng Y, Wang T, Tu X, Huang Y, Zhang H, Tan D, et al. Gut microbiome affects the response to anti-PD-1 immunotherapy in patients with hepatocellular carcinoma. J Immunother Cancer (2019) 7(1):193. doi: 10.1186/s40425-019-0650-9
31. Jin Y, Dong H, Xia L, Yang Y, Zhu Y, Shen Y, et al. The diversity of gut microbiome is associated with favorable responses to anti-programmed death 1 immunotherapy in Chinese patients with NSCLC. J Thorac Oncol (2019) 14(8):1378–89. doi: 10.1016/j.jtho.2019.04.007
32. Oster P, Vaillant L, Riva E, McMillan B, Begka C, Truntzer C, et al. Helicobacter pylori infection has a detrimental impact on the efficacy of cancer immunotherapies. Gut (2022) 71(3):457–66. doi: 10.1136/gutjnl-2020-323392
33. McCulloch JA, Davar D, Rodrigues RR, Badger JH, Fang JR, Cole AM, et al. Intestinal microbiota signatures of clinical response and immune-related adverse events in melanoma patients treated with anti-PD-1. Nat Med (2022) 28(3):545–56. doi: 10.1038/s41591-022-01698-2
34. Grenda A, Iwan E, Krawczyk P, Frąk M, Chmielewska I, Bomba A, et al. Attempting to identify bacterial allies in immunotherapy of NSCLC patients. Cancers (Basel) (2022) 14(24):6250. doi: 10.3390/cancers14246250
35. Shoji F, Yamaguchi M, Okamoto M, Takamori S, Yamazaki K, Okamoto T, et al. Gut microbiota diversity and specific composition during immunotherapy in responders with non-small cell lung cancer. Front Mol Biosci (2022) 9:1040424. doi: 10.3389/fmolb.2022.1040424
36. Newsome RC, Gharaibeh RZ, Pierce CM, da Silva WV, Paul S, Hogue SR, et al. Interaction of bacterial genera associated with therapeutic response to immune checkpoint PD-1 blockade in a united states cohort. Genome Med (2022) 14(1):35. doi: 10.1186/s13073-022-01037-7
37. Zhang F, Ferrero M, Dong N, D’Auria G, Reyes-Prieto M, Herreros-Pomares A, et al. Analysis of the gut microbiota: An emerging source of biomarkers for immune checkpoint blockade therapy in non-small cell lung cancer. Cancers (Basel) (2021) 13(11):2514. doi: 10.3390/cancers13112514
38. Zhang C, Wang J, Sun Z, Cao Y, Mu Z, Ji X. Commensal microbiota contributes to predicting the response to immune checkpoint inhibitors in non-small-cell lung cancer patients. Cancer Sci (2021) 112(8):3005–17. doi: 10.1111/cas.14979
39. Botticelli A, Vernocchi P, Marini F, Quagliariello A, Cerbelli B, Reddel S, et al. Gut metabolomics profiling of non-small cell lung cancer (NSCLC) patients under immunotherapy treatment. J Trans Med (2020) 18(1):49. doi: 10.1186/s12967-020-02231-0
40. Spencer CN, McQuade JL, Gopalakrishnan V, McCulloch JA, Vetizou M, Cogdill AP, et al. Dietary fiber and probiotics influence the gut microbiome and melanoma immunotherapy response. Science (2021) 374(6575):1632–40. doi: 10.1126/science.aaz7015
41. Andrews MC, Duong CPM, Gopalakrishnan V, Iebba V, Chen WS, Derosa L, et al. Gut microbiota signatures are associated with toxicity to combined CTLA-4 and PD-1 blockade. Nat Med (2021) 27(8):1432–41. doi: 10.1038/s41591-021-01406-6
42. Lee PC, Wu CJ, Hung YW, Lee CJ, Chi CT, Lee IC, et al. Gut microbiota and metabolites associate with outcomes of immune checkpoint inhibitor-treated unresectable hepatocellular carcinoma. J Immunother Cancer (2022) 10(6):e004779. doi: 10.1136/jitc-2022-004779
43. Wu H, Zheng X, Pan T, Yang X, Chen X, Zhang B, et al. Dynamic microbiome and metabolome analyses reveal the interaction between gut microbiota and anti-PD-1 based immunotherapy in hepatocellular carcinoma. Int J Cancer (2022) 151(8):1321–34. doi: 10.1002/ijc.34118
44. Ponziani FR, De Luca A, Picca A, Marzetti E, Petito V, Del Chierico F, et al. Gut dysbiosis and fecal calprotectin predict response to immune checkpoint inhibitors in patients with hepatocellular carcinoma. Hepatol Commun (2022) 6(6):1492–501. doi: 10.1002/hep4.1905
45. Mao J, Wang D, Long J, Yang X, Lin J, Song Y, et al. Gut microbiome is associated with the clinical response to anti-PD-1 based immunotherapy in hepatobiliary cancers. J Immunother Cancer (2021) 9(12):e003334. doi: 10.1136/jitc-2021-003334
46. Wang F, He MM, Yao YC, Zhao X, Wang ZQ, Jin Y, et al. Regorafenib plus toripalimab in patients with metastatic colorectal cancer: a phase Ib/II clinical trial and gut microbiome analysis. Cell Rep Med (2021) 2(9):100383. doi: 10.1016/j.xcrm.2021.100383
47. Che H, Xiong Q, Ma J, Chen S, Wu H, Xu H, et al. Association of helicobacter pylori infection with survival outcomes in advanced gastric cancer patients treated with immune checkpoint inhibitors. BMC Cancer (2022) 22(1):904. doi: 10.1186/s12885-022-10004-9
48. Peng Z, Cheng S, Kou Y, Wang Z, Jin R, Hu H, et al. The gut microbiome is associated with clinical response to anti-PD-1/PD-L1 immunotherapy in gastrointestinal cancer. Cancer Immunol Res (2020) 8(10):1251–61. doi: 10.1158/2326-6066.CIR-19-1014
49. Salgia NJ, Bergerot PG, Maia MC, Dizman N, Hsu J, Gillece JD, et al. Stool microbiome profiling of patients with metastatic renal cell carcinoma receiving anti-PD-1 immune checkpoint inhibitors. Eur Urol (2020) 78(4):498–502. doi: 10.1016/j.eururo.2020.07.011
50. Yin H, Yang L, Peng G, Yang K, Mi Y, Hu X, et al. The commensal consortium of the gut microbiome is associated with favorable responses to anti-programmed death protein 1 (PD-1) therapy in thoracic neoplasms. Cancer Biol Med (2021) 18(4):1040–52. doi: 10.20892/j.issn.2095-3941.2020.0450
51. Cheng X, Wang J, Gong L, Dong Y, Shou J, Pan H, et al. Composition of the gut microbiota associated with the response to immunotherapy in advanced cancer patients: A Chinese real-world pilot study. J Clin Med (2022) 11(18):5479. doi: 10.3390/jcm11185479
52. Baruch EN, Youngster I, Ben-Betzalel G, Ortenberg R, Lahat A, Katz L, et al. Fecal microbiota transplant promotes response in immunotherapy-refractory melanoma patients. Science (2021) 371(6529):602–9. doi: 10.1126/science.abb5920
53. Takada K, Shimokawa M, Takamori S, Shimamatsu S, Hirai F, Tagawa T, et al. Clinical impact of probiotics on the efficacy of anti-PD-1 monotherapy in patients with nonsmall cell lung cancer: A multicenter retrospective survival analysis study with inverse probability of treatment weighting. Int J Cancer (2021) 149(2):473–82. doi: 10.1002/ijc.33557
54. Zhou Y, Medik YB, Patel B, Zamler DB, Chen S, Chapman T, et al. Intestinal toxicity to CTLA-4 blockade driven by IL-6 and myeloid infiltration. J Exp Med (2023) 220(2):e20221333. doi: 10.1084/jem.20221333
55. Dubin K, Callahan MK, Ren B, Khanin R, Viale A, Ling L, et al. Intestinal microbiome analyses identify melanoma patients at risk for checkpoint-blockade-induced colitis. Nat Commun (2016) 7:10391. doi: 10.1038/ncomms10391
56. Chang AE, Golob JL, Schmidt TM, Peltier DC, Lao CD, Tewari M. Targeting the gut microbiome to mitigate immunotherapy-induced colitis in cancer. Trends Cancer (2021) 7(7):583–93. doi: 10.1016/j.trecan.2021.02.005
57. Wang Y, Wiesnoski DH, Helmink BA, Gopalakrishnan V, Choi K, DuPont HL, et al. Fecal microbiota transplantation for refractory immune checkpoint inhibitor-associated colitis. Nat Med (2018) 24(12):1804–8. doi: 10.1038/s41591-018-0238-9
58. Renga G, Nunzi E, Pariano M, Puccetti M, Bellet MM, Pieraccini G, et al. Optimizing therapeutic outcomes of immune checkpoint blockade by a microbial tryptophan metabolite. J Immunother Cancer (2022) 10(3):e003725. doi: 10.1136/jitc-2021-003725
59. Shaikh FY, White JR, Gills JJ, Hakozaki T, Richard C, Routy B, et al. A uniform computational approach improved on existing pipelines to reveal microbiome biomarkers of nonresponse to immune checkpoint inhibitors. Clin Cancer Res (2021) 27(9):2571–83. doi: 10.1158/1078-0432.CCR-20-4834
60. Krautkramer KA, Fan J, Backhed F. Gut microbial metabolites as multi-kingdom intermediates. Nat Rev Microbiol (2021) 19(2):77–94. doi: 10.1038/s41579-020-0438-4
61. Deleu S, Machiels K, Raes J, Verbeke K, Vermeire S. Short chain fatty acids and its producing organisms: An overlooked therapy for IBD? EBioMedicine (2021) 66:103293. doi: 10.1016/j.ebiom.2021.103293
62. Zhang SL, Mao YQ, Zhang ZY, Li ZM, Kong CY, Chen HL, et al. Pectin supplement significantly enhanced the anti-PD-1 efficacy in tumor-bearing mice humanized with gut microbiota from patients with colorectal cancer. Theranostics (2021) 11(9):4155–70. doi: 10.7150/thno.54476
63. He Y, Fu L, Li Y, Wang W, Gong M, Zhang J, et al. Gut microbial metabolites facilitate anticancer therapy efficacy by modulating cytotoxic CD8(+) T cell immunity. Cell Metab (2021) 33(5):988–1000 e7. doi: 10.1016/j.cmet.2021.03.002
64. Coutzac C, Jouniaux JM, Paci A, Schmidt J, Mallardo D, Seck A, et al. Systemic short chain fatty acids limit antitumor effect of CTLA-4 blockade in hosts with cancer. Nat Commun (2020) 11(1):2168. doi: 10.1038/s41467-020-16079-x
65. Mager LF, Burkhard R, Pett N, Cooke NCA, Brown K, Ramay H, et al. Microbiome-derived inosine modulates response to checkpoint inhibitor immunotherapy. Science (2020) 369(6510):1481–9. doi: 10.1126/science.abc3421
66. Samami E, Aleebrahim-Dehkordi E, Mohebalizadeh M, Yaribash S, Saghazadeh A, Rezaei N. Inosine, gut microbiota, and cancer immunometabolism. Am J Physiol Endocrinol Metab (2023) 324(1):E1–e8. doi: 10.1152/ajpendo.00207.2022
67. Mirji G, Worth A, Bhat SA, El Sayed M, Kannan T, Goldman AR, et al. The microbiome-derived metabolite TMAO drives immune activation and boosts responses to immune checkpoint blockade in pancreatic cancer. Sci Immunol (2022) 7(75):eabn0704. doi: 10.1126/sciimmunol.abn0704
68. Kawanabe-Matsuda H, Takeda K, Nakamura M, Makino S, Karasaki T, Kakimi K, et al. Dietary lactobacillus-derived exopolysaccharide enhances immune-checkpoint blockade therapy. Cancer Discovery (2022) 12(5):1336–55. doi: 10.1158/2159-8290.CD-21-0929
69. Chang YJ, Pei XY, Huang XJ. Haematopoietic stem-cell transplantation in China in the era of targeted therapies: current advances, challenges, and future directions. Lancet Haematol (2022) 9(12):e919–e29. doi: 10.1016/S2352-3026(22)00293-9
70. Malard F, Gaugler B, Mohty M. Faecal microbiota transplantation in patients with haematological malignancies undergoing cellular therapies: From translational research to routine clinical practice. Lancet Haematol (2022) 9(10):e776–e85. doi: 10.1016/S2352-3026(22)00223-X
71. Holler E, Butzhammer P, Schmid K, Hundsrucker C, Koestler J, Peter K, et al. Metagenomic analysis of the stool microbiome in patients receiving allogeneic stem cell transplantation: loss of diversity is associated with use of systemic antibiotics and more pronounced in gastrointestinal graft-versus-host disease. Biol Blood Marrow Transplant (2014) 20(5):640–5. doi: 10.1016/j.bbmt.2014.01.030
72. Peled JU, Gomes ALC, Devlin SM, Littmann ER, Taur Y, Sung AD, et al. Microbiota as predictor of mortality in allogeneic hematopoietic-cell transplantation. New Engl J Med (2020) 382(9):822–34. doi: 10.1056/NEJMoa1900623
73. Andrlová H, Miltiadous O, Kousa AI, Dai A, DeWolf S, Violante S, et al. MAIT and Vδ2 unconventional T cells are supported by a diverse intestinal microbiome and correlate with favorable patient outcome after allogeneic HCT. Sci Trans Med (2022) 14(646):eabj2829. doi: 10.1126/scitranslmed.abj2829
74. Stein-Thoeringer CK, Nichols KB, Lazrak A, Docampo MD, Slingerland AE, Slingerland JB, et al. Lactose drives enterococcus expansion to promote graft-versus-host disease. Science (2019) 366(6469):1143–9. doi: 10.1126/science.aax3760
75. Meedt E, Hiergeist A, Gessner A, Dettmer K, Liebisch G, Ghimire S, et al. Prolonged suppression of butyrate-producing bacteria is associated with acute gastrointestinal graft-vs-Host disease and transplantation-related mortality after allogeneic stem cell transplantation. Clin Infect Dis (2022) 74(4):614–21. doi: 10.1093/cid/ciab500
76. Markey KA, Schluter J, Gomes ALC, Littmann ER, Pickard AJ, Taylor BP, et al. The microbe-derived short-chain fatty acids butyrate and propionate are associated with protection from chronic GVHD. Blood (2020) 136(1):130–6. doi: 10.1182/blood.2019003369
77. Wang Y, Zhang Z, Liu B, Zhang C, Zhao J, Li X, et al. A study on the method and effect of the construction of a humanized mouse model of fecal microbiota transplantation. Front Microbiol (2022) 13:1031758. doi: 10.3389/fmicb.2022.1031758
78. Peled JU, Devlin SM, Staffas A, Lumish M, Khanin R, Littmann ER, et al. Intestinal microbiota and relapse after hematopoietic-cell transplantation. J Clin Oncol (2017) 35(15):1650–9. doi: 10.1200/JCO.2016.70.3348
79. Mathewson ND, Jenq R, Mathew AV, Koenigsknecht M, Hanash A, Toubai T, et al. Gut microbiome-derived metabolites modulate intestinal epithelial cell damage and mitigate graft-versus-host disease. Nat Immunol (2016) 17(5):505–13. doi: 10.1038/ni.3400
80. Harris B, Morjaria SM, Littmann ER, Geyer AI, Stover DE, Barker JN, et al. Gut microbiota predict pulmonary infiltrates after allogeneic hematopoietic cell transplantation. Am J Respir Crit Care Med (2016) 194(4):450–63. doi: 10.1164/rccm.201507-1491OC
81. Stoma I, Littmann ER, Peled JU, Giralt S, van den Brink MRM, Pamer EG, et al. Compositional flux within the intestinal microbiota and risk for bloodstream infection with gram-negative bacteria. Clin Infect Dis (2021) 73(11):e4627–e35. doi: 10.1093/cid/ciaa068
82. Masetti R, D’Amico F, Zama D, Leardini D, Muratore E, Ussowicz M, et al. Febrile neutropenia duration is associated with the severity of gut microbiota dysbiosis in pediatric allogeneic hematopoietic stem cell transplantation recipients. Cancers (Basel) (2022) 14(8):1932. doi: 10.3390/cancers14081932
83. Lee YJ, Arguello ES, Jenq RR, Littmann E, Kim GJ, Miller LC, et al. Protective factors in the intestinal microbiome against clostridium difficile infection in recipients of allogeneic hematopoietic stem cell transplantation. J Infect Dis (2017) 215(7):1117–23. doi: 10.1093/infdis/jix011
84. Legoff J, Resche-Rigon M, Bouquet J, Robin M, Naccache SN, Mercier-Delarue S, et al. The eukaryotic gut virome in hematopoietic stem cell transplantation: new clues in enteric graft-versus-host disease. Nat Med (2017) 23(9):1080–5. doi: 10.1038/nm.4380
85. Rolling T, Zhai B, Gjonbalaj M, Tosini N, Yasuma-Mitobe K, Fontana E, et al. Haematopoietic cell transplantation outcomes are linked to intestinal mycobiota dynamics and an expansion of candida parapsilosis complex species. Nat Microbiol (2021) 6(12):1505–15. doi: 10.1038/s41564-021-00989-7
86. Ciernikova S, Kasperova B, Drgona L, Smolkova B, Stevurkova V, Mego M. Targeting the gut microbiome: An emerging trend in hematopoietic stem cell transplantation. Blood Rev (2021) 48:100790. doi: 10.1016/j.blre.2020.100790
87. Khuat LT, Dave M, Murphy WJ. The emerging roles of the gut microbiome in allogeneic hematopoietic stem cell transplantation. Gut Microbes (2021) 13(1):1966262. doi: 10.1080/19490976.2021.1966262
88. Henig I, Yehudai-Ofir D, Zuckerman T. The clinical role of the gut microbiome and fecal microbiota transplantation in allogeneic stem cell transplantation. Haematologica (2021) 106(4):933–46. doi: 10.3324/haematol.2020.247395
89. Severyn CJ, Siranosian BA, Kong ST, Moreno A, Li MM, Chen N, et al. Microbiota dynamics in a randomized trial of gut decontamination during allogeneic hematopoietic cell transplantation. JCI Insight (2022) 7(7):e154344. doi: 10.1172/jci.insight.154344
90. Khuat LT, Le CT, Pai CS, Shields-Cutler RR, Holtan SG, Rashidi A, et al. Obesity induces gut microbiota alterations and augments acute graft-versus-host disease after allogeneic stem cell transplantation. Sci Trans Med (2020) 12(571):eaay7713. doi: 10.1126/scitranslmed.aay7713
91. Shono Y, Docampo MD, Peled JU, Perobelli SM, Velardi E, Tsai JJ, et al. Increased GVHD-related mortality with broad-spectrum antibiotic use after allogeneic hematopoietic stem cell transplantation in human patients and mice. Sci Trans Med (2016) 8(339):339ra71. doi: 10.1126/scitranslmed.aaf2311
92. Gavriilaki M, Sakellari I, Anagnostopoulos A, Gavriilaki E. The impact of antibiotic-mediated modification of the intestinal microbiome on outcomes of allogeneic hematopoietic cell transplantation: Systematic review and meta-analysis. Biol Blood Marrow Transplant (2020) 26(9):1738–46. doi: 10.1016/j.bbmt.2020.05.011
93. Muratore E, Leardini D, Baccelli F, Venturelli F, Prete A, Masetti R. Nutritional modulation of the gut microbiome in allogeneic hematopoietic stem cell transplantation recipients. Front Nutr (2022) 9:993668. doi: 10.3389/fnut.2022.993668
94. Li X, Lin Y, Li X, Xu X, Zhao Y, Xu L, et al. Tyrosine supplement ameliorates murine aGVHD by modulation of gut microbiome and metabolome. EBioMedicine (2020) 61:103048. doi: 10.1016/j.ebiom.2020.103048
95. Yoshifuji K, Inamoto K, Kiridoshi Y, Takeshita K, Sasajima S, Shiraishi Y, et al. Prebiotics protect against acute graft-versus-host disease and preserve the gut microbiota in stem cell transplantation. Blood Adv (2020) 4(19):4607–17. doi: 10.1182/bloodadvances.2020002604
96. Simms-Waldrip TR, Sunkersett G, Coughlin LA, Savani MR, Arana C, Kim J, et al. Antibiotic-induced depletion of anti-inflammatory clostridia is associated with the development of graft-versus-Host disease in pediatric stem cell transplantation patients. Biol Blood Marrow Transplant (2017) 23(5):820–9. doi: 10.1016/j.bbmt.2017.02.004
97. Ladas EJ, Bhatia M, Chen L, Sandler E, Petrovic A, Berman DM, et al. The safety and feasibility of probiotics in children and adolescents undergoing hematopoietic cell transplantation. Bone Marrow Transplant (2016) 51(2):262–6. doi: 10.1038/bmt.2015.275
98. Mehta A, Rangarajan S, Borate U. A cautionary tale for probiotic use in hematopoietic SCT patients-lactobacillus acidophilus sepsis in a patient with mantle cell lymphoma undergoing hematopoietic SCT. Bone Marrow Transplant (2013) 48(3):461–2. doi: 10.1038/bmt.2012.153
99. Keller JJ, Ooijevaar RE, Hvas CL, Terveer EM, Lieberknecht SC, Högenauer C, et al. A standardised model for stool banking for faecal microbiota transplantation: a consensus report from a multidisciplinary UEG working group. United Eur Gastroenterol J (2021) 9(2):229–47. doi: 10.1177/2050640620967898
100. Bluestone H, Kronman MP, Suskind DL. Fecal microbiota transplantation for recurrent clostridium difficile infections in pediatric hematopoietic stem cell transplant recipients. J Pediatr Infect Dis Soc (2018) 7(1):e6–8. doi: 10.1093/jpids/pix076
101. Battipaglia G, Malard F, Rubio MT, Ruggeri A, Mamez AC, Brissot E, et al. Fecal microbiota transplantation before or after allogeneic hematopoietic transplantation in patients with hematologic malignancies carrying multidrug-resistance bacteria. Haematologica (2019) 104(8):1682–8. doi: 10.3324/haematol.2018.198549
102. van Lier YF, Davids M, Haverkate NJE, de Groot PF, Donker ML, Meijer E, et al. Donor fecal microbiota transplantation ameliorates intestinal graft-versus-host disease in allogeneic hematopoietic cell transplant recipients. Sci Trans Med (2020) 12(556):eaaz8926. doi: 10.1126/scitranslmed.aaz8926
103. DeFilipp Z, Peled JU, Li S, Mahabamunuge J, Dagher Z, Slingerland AE, et al. Third-party fecal microbiota transplantation following allo-HCT reconstitutes microbiome diversity. Blood Adv (2018) 2(7):745–53. doi: 10.1182/bloodadvances.2018017731
104. Taur Y, Coyte K, Schluter J, Robilotti E, Figueroa C, Gjonbalaj M, et al. Reconstitution of the gut microbiota of antibiotic-treated patients by autologous fecal microbiota transplant. Sci Trans Med (2018) 10(460):eaap9489. doi: 10.1126/scitranslmed.aap9489
105. Raje N, Berdeja J, Lin Y, Siegel D, Jagannath S, Madduri D, et al. Anti-BCMA CAR T-cell therapy bb2121 in relapsed or refractory multiple myeloma. New Engl J Med (2019) 380(18):1726–37. doi: 10.1056/NEJMoa1817226
106. Jacobson CA, Chavez JC, Sehgal AR, William BM, Munoz J, Salles G, et al. Axicabtagene ciloleucel in relapsed or refractory indolent non-Hodgkin lymphoma (ZUMA-5): a single-arm, multicentre, phase 2 trial. Lancet Oncol (2022) 23(1):91–103. doi: 10.1016/S1470-2045(21)00591-X
107. Locke FL, Miklos DB, Jacobson CA, Perales MA, Kersten MJ, Oluwole OO, et al. Axicabtagene ciloleucel as second-line therapy for Large b-cell lymphoma. New Engl J Med (2022) 386(7):640–54. doi: 10.1056/NEJMoa2116133
108. Brentjens RJ, Rivière I, Park JH, Davila ML, Wang X, Stefanski J, et al. Safety and persistence of adoptively transferred autologous CD19-targeted T cells in patients with relapsed or chemotherapy refractory b-cell leukemias. Blood (2011) 118(18):4817–28. doi: 10.1182/blood-2011-04-348540
109. Lu J, Jiang G. The journey of CAR-T therapy in hematological malignancies. Mol Cancer (2022) 21(1):194. doi: 10.1186/s12943-022-01663-0
110. Tanoue T, Morita S, Plichta DR, Skelly AN, Suda W, Sugiura Y, et al. A defined commensal consortium elicits CD8 T cells and anti-cancer immunity. Nature (2019) 565(7741):600–5. doi: 10.1038/s41586-019-0878-z
111. Rangan P, Mondino A. Microbial short-chain fatty acids: a strategy to tune adoptive T cell therapy. J Immunother Cancer (2022) 10(7):e004147. doi: 10.1136/jitc-2021-004147c
112. Abid MB, Shah NN, Maatman TC, Hari PN. Gut microbiome and CAR-T therapy. Exp Hematol Oncol (2019) 8:31. doi: 10.1186/s40164-019-0155-8
113. Schubert ML, Rohrbach R, Schmitt M, Stein-Thoeringer CK. The potential role of the intestinal micromilieu and individual microbes in the immunobiology of chimeric antigen receptor T-cell therapy. Front Immunol (2021) 12:670286. doi: 10.3389/fimmu.2021.670286
114. Uribe-Herranz M, Bittinger K, Rafail S, Guedan S, Pierini S, Tanes C, et al. Gut microbiota modulates adoptive cell therapy via CD8α dendritic cells and IL-12. JCI Insight (2018) 3(4):e94952. doi: 10.1172/jci.insight.94952
115. Smith M, Dai A, Ghilardi G, Amelsberg KV, Devlin SM, Pajarillo R, et al. Gut microbiome correlates of response and toxicity following anti-CD19 CAR T cell therapy. Nat Med (2022) 28(4):713–23. doi: 10.1038/s41591-022-01702-9
116. Hu Y, Li J, Ni F, Yang Z, Gui X, Bao Z, et al. CAR-T cell therapy-related cytokine release syndrome and therapeutic response is modulated by the gut microbiome in hematologic malignancies. Nat Commun (2022) 13(1):5313. doi: 10.1038/s41467-022-32960-3
117. Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer statistics, 2022. CA Cancer J Clin (2022) 72(1):7–33. doi: 10.3322/caac.21708
118. Schupack DA, Mars RAT, Voelker DH, Abeykoon JP, Kashyap PC. The promise of the gut microbiome as part of individualized treatment strategies. Nat Rev Gastroenterol Hepatol (2022) 19(1):7–25. doi: 10.1038/s41575-021-00499-1
119. Soleimanpour S, Hasanian SM, Avan A, Yaghoubi A, Khazaei M. Bacteriotherapy in gastrointestinal cancer. Life Sci (2020) 254:117754. doi: 10.1016/j.lfs.2020.117754
120. Montalban-Arques A, Katkeviciute E, Busenhart P, Bircher A, Wirbel J, Zeller G, et al. Commensal clostridiales strains mediate effective anti-cancer immune response against solid tumors. Cell Host Microbe (2021) 29(10):1573–88 e7. doi: 10.1016/j.chom.2021.08.001
121. Kaźmierczak-Siedlecka K, Skonieczna-Żydecka K, Hupp T, Duchnowska R, Marek-Trzonkowska N, Połom K. Next-generation probiotics - do they open new therapeutic strategies for cancer patients? Gut Microbes (2022) 14(1):2035659. doi: 10.1080/19490976.2022.2035659
122. Gurbatri CR, Arpaia N, Danino T. Engineering bacteria as interactive cancer therapies. Science (2022) 378(6622):858–64. doi: 10.1126/science.add9667
123. Huang X, Pan J, Xu F, Shao B, Wang Y, Guo X, et al. Bacteria-based cancer immunotherapy. Adv Sci (Weinh) (2021) 8(7):2003572. doi: 10.1002/advs.202003572
124. Vincent RL, Gurbatri CR, Redenti A, Coker C, Arpaia N, Danino T. Probiotic-guided CAR-T cells for universal solid tumor targeting. bioRxiv (2021) 2021:10. doi: 10.1101/2021.10.10.463366
125. Su Q, Liu Q, Lau RI, Zhang J, Xu Z, Yeoh YK, et al. Faecal microbiome-based machine learning for multi-class disease diagnosis. Nat Commun (2022) 13(1):6818. doi: 10.1038/s41467-022-34405-3
Keywords: Gut microbiota, immunotherapy, immune checkpoint blockade, allogeneic hematopoietic stem cell transplantation, chimeric antigen receptor T cell therapy
Citation: Shi Z, Li H, Song W, Zhou Z, Li Z and Zhang M (2023) Emerging roles of the gut microbiota in cancer immunotherapy. Front. Immunol. 14:1139821. doi: 10.3389/fimmu.2023.1139821
Received: 07 January 2023; Accepted: 13 February 2023;
Published: 22 February 2023.
Edited by:
Yeonseok Chung, Seoul National University, Republic of KoreaReviewed by:
Hun Sik Kim, University of Ulsan, Republic of KoreaYoungjun Park, Jeju National University, Republic of Korea
Copyright © 2023 Shi, Li, Song, Zhou, Li and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Mingzhi Zhang, mingzhi_zhang1@163.com; Zhaoming Li, fcclizm@zzu.edu.cn
 Zhuangzhuang Shi1,2,3
Zhuangzhuang Shi1,2,3