
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
MINI REVIEW article
Front. Immunol. , 12 August 2022
Sec. Microbial Immunology
Volume 13 - 2022 | https://doi.org/10.3389/fimmu.2022.988708
This article is part of the Research Topic Immune Interactions with Pathogenic and Commensal Fungi View all 17 articles
 Liuyang Cai1,2†
Liuyang Cai1,2† Peigen Gao1,3†
Peigen Gao1,3† Zeyu Wang1,3†
Zeyu Wang1,3† Chenyang Dai1,3
Chenyang Dai1,3 Ye Ning1,3
Ye Ning1,3 Macit Ilkit4
Macit Ilkit4 Xiaochun Xue5*
Xiaochun Xue5* Jinzhou Xiao6*
Jinzhou Xiao6* Chang Chen1,3*
Chang Chen1,3*Species within the Aspergillus spp. cause a wide range of infections in humans, including invasive pulmonary aspergillosis, chronic pulmonary aspergillosis, and allergic bronchopulmonary aspergillosis, and are associated with high mortality rates. The incidence of pulmonary aspergillosis (PA) is on the rise, and the emergence of triazole-resistant Aspergillus spp. isolates, especially Aspergillus fumigatus, limits the efficacy of mold-active triazoles. Therefore, host-directed and novel adjunctive therapies are required to more effectively combat PA. In this review, we focus on PA from a microbiome perspective. We provide a general overview of the effects of the lung and gut microbiomes on the growth of Aspergillus spp. and host immunity. We highlight the potential of the microbiome as a therapeutic target for PA.
Pulmonary aspergillosis (PA) is an infection or allergic response caused by Aspergillus spp (1). Aspergillus spp. are widely present in the environment and are mainly transmitted via airborne conidia (2, 3) (Figure 1). Aspergillus fumigatus is one of the most common Aspergillus spp. It is responsible for the majority of PA incidences (6). Depending on host immunity, pulmonary ailments caused by Aspergillus spp. can be mainly classified as invasive pulmonary aspergillosis (IPA), chronic pulmonary aspergillosis (CPA), or allergic bronchopulmonary aspergillosis (ABPA) (6, 7). Recent global estimates revealed that 8,000,000 cases of PA occur annually (8). Mold-active triazole exerts cidal activity, and as such are the frontline antifungals used to treat aspergillosis, while echinocandins show static activity and amphotericin B prescription is limited owing to its cytotoxic activity (9). Unfortunately, the extensive use of fungicides in the environment as well as in the clinic, has resulted in the increasing emergence of triazole resistant aspergillosis, mostly owing to A. fumigatus (9–12). Additionally, expensive and/or toxic drugs, drug-drug interactions, and unequal clinical resources in different regions reduce the potential for survival and recovery (13, 14). Therefore, there is an unmet need to identify/design novel antifungal drugs as well as make use of host-directed strategies to combat PA and to improve the clinical outcomes of inflicted patients.
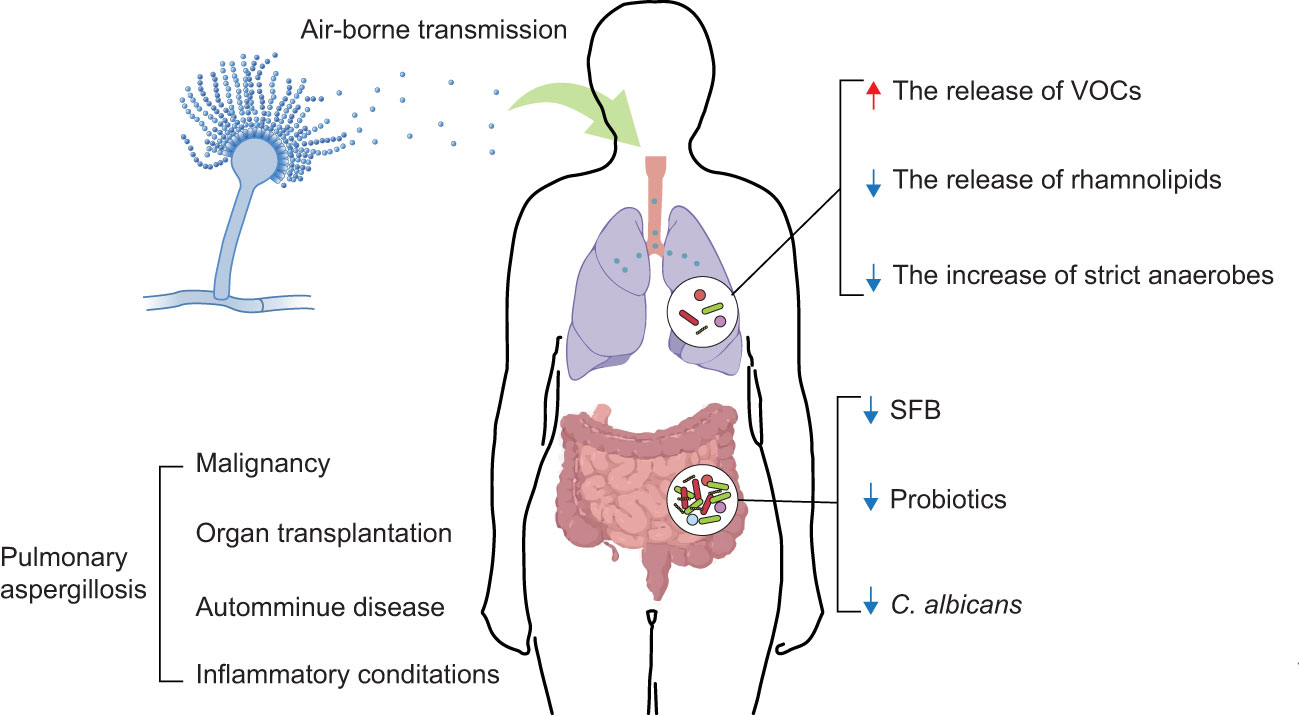
Figure 1 Interaction between the microbiome and PA. Aspergillus spp. conidia usually enter the human body through air-borne transmission. The colonization and infection of Aspergillus spp. have dramatically increased, considering the growing numbers of patients with an impaired immune state associated with the treatment of malignancy, organ transplantation, autoimmune diseases, and inflammatory conditions (4, 5). The development of PA is influenced by composition and metabolites of the lung and gut microbiomes (e.g., VOCs, rhamnolipids, strict anaerobes, SFB, probiotics, C. albicans). ↑: increase; ↓: decrease.
Despite being a novel field, the current paradigm suggests that communities of microbes living on various epithelial surfaces, known as microbiome, are linked to an array of complications in humans and dysregulation in the composition of such communities, known as microbiome dysbiosis, can have profound impact on predisposition to various infections and complications ranging from pulmonary infections and cancer to diabetes and neurological disorders (15–19). On the one hand, the application of fecal microbial transplantation from healthy donors to patients was found to be promising against a wide range of ailments. On the other hand, human complications are often accompanied by microbiome alterations, and as such, determination of the microbiome signature could potentially offer a robust diagnostic tool, and its leverage could subsequently aid in timely and effective treatment. For instance, Hérivaux et al. (20) reported that microbiome diversity was found to predict IPA onset and the mortality rate associated with this complication. Apart from scattered studies reported thus far, the association of the human microbiome with PA remains largely elusive. Determination of a clear picture of the healthy microbiome and dysbiosis in the context of PA could potentially enhance the therapeutic capacity; therefore, the current study thoroughly discusses and links the human microbiome to PA.
Although the microbiome composition of the lung remained elusive in early times, the development of quantitative molecular sequencing methods has identified a complicated microbial community inhabiting the lung, known as the lung microbiome (21). The lung microbiome is associated with immune activation and regulation (22); it is also known to diverge substantially between healthy (23) and diseased states (24, 25). Dysbiosis of the lung microbiome is related to the exacerbations of several respiratory diseases such as bronchiectasis, cystic fibrosis (CF), and chronic obstructive pulmonary disease (26–28). The components and metabolites of the gut microbiome can also influence immune responses (29). Intestinal dysbiosis has been linked to alterations in host immunity and disease development, including respiratory diseases (30). Moreover, numerous pieces of evidence support the key contribution of the microbiome in the prevention and treatment of respiratory diseases. Gram-negative bacilli (Pseudomonas aeruginosa, Acinetobacter baumannii, and Escherichia coli, etc.) that colonize the lungs usually cause nosocomial pneumonia. Antimicrobial therapy improves the outcome of nosocomial pneumonia (31). Gut commensal microbiome regulate immune responses in the respiratory mucosa and resist respiratory virus infections (32). Although the significance of the microbiome has already been established in respiratory diseases, the mechanisms by which the lung and gut microbiomes influence PA are relatively unknown (33, 34). Considering this, in this review, we aim to discuss the role of the lung and gut microbiomes in the growth of Aspergillus spp. and host immunity (Figure 1, Table 1). We hope it will serve as a vital foundation for the further analysis of the interactions between immunity, the microbiome, and PA. Moreover, our review will contribute to the development of a more reliable clinical treatment for PA.
Conventionally, a few members of the Aspergillus spp. have reached the alveoli and exposed the cell wall pathogen-associated molecular patterns, such as β-D-glucan (52, 53). In immunocompetent individuals, different pattern recognition receptors (PRRs) include Toll-like receptors, C-type lectin receptors (CLRs), and Nod-like receptors. PRRs can recognize Aspergillus spp. and initiate an early immune response (54). For instance, Dectin-1, a CLR, recognizes fungal β-glucan and modulates the inflammatory responses by inducing the expression of the anti-inflammatory cytokine interleukin (IL)-10 (55). Subsequently, innate immune cells (macrophages, neutrophils, etc.) actively participate in the cellular immune responses against Aspergillus spp. by engulfing and killing the conidia. As the main resident leukocytes in the lungs, alveolar macrophages can rapidly adhere to and take up conidia that enter the alveolar space (56). In contrast to delayed killing mediated by alveolar macrophages, neutrophilic granulocytes rapidly kill hyphae of Aspergillus spp. through an active oxygen-dependent mechanism at the cell surface (57). These innate immune responses constitute the first line of defense against pulmonary host defense and the natural and chemical barriers of the organism.
Nonetheless, for patients with chronic respiratory disease or impaired immune function (e.g., neutropenia), these innate immune responses do not function normally leading to Aspergillus spp. colonization and infection. In this case, the adaptive immune responses are activated. CD4 (including Th1, Th2, Th17, etc.) or CD8 T-cell responses play a critical role in PA. After infection with Aspergillus spp., Th1 cells enhance the antifungal activities of macrophages and neutrophils and express the pro-inflammatory cytokines TNF-α and IFN-γ (58). Conversely, Th2 cell activation inhibits Th1 cell responses. Allard et al. (59) reported a direct airway exposure to Aspergillus spp. Lysates boost the Th2 cell responses in the lungs of mice, resulting in symptoms similar to those of ABPA. Symptoms include eosinophilic inflammation, mucus hypersecretion, and increased airway resistance. In contrast, the role of Th17 cell responses in Aspergillus spp. infection is debatable. IL-17 and IL-23 produced by Th17 cells can suppress Th1-mediated protective immunity against fungi and increase susceptibility to Aspergillus spp. in mice (60). However, some studies have concluded that IL-17 is involved in protective responses against PA. For example, Werner et al. (61) observed that the neutralization of IL-17 significantly impaired A. fumigatus clearance. In summary, innate and adaptive immune responses help host resistance against PA.
Relevant advances have been made in devising immunotherapeutic strategies for PA. Among the innate immune responses, Bruton’s tyrosine kinase (BTK), a key molecule in multiple signaling pathways, activates fungal recognition immune responses. The clinical application of BTK inhibitors is to impair several immune functions of platelets in response to A. fumigatus and increases the risk of invasive aspergillosis in patients with chronic lymphocytic leukemia (62). Among the adaptive immune responses, from a neutropenia perspective, transfusable neutrophil progenitors serve as new cellular therapies for the prevention of IPA. This treatment produces unlimited numbers of homogenous granulocyte-macrophage progenitors, greatly improving survival in models of PA (63). Although an increasing number of new therapeutic strategies are being discovered, most of the studies are still limited to animal experiments. Whether these therapeutic strategies are applicable to humans remains uncertain and cannot be extrapolated directly. Thus, further studies are needed to determine how the immune system functions during Aspergillus spp. infection. Researchers should strive to translate these findings into valuable therapeutic tools for clinical settings.
Lung bacteria are vital in protecting against PA. Pseudomonas aeruginosa and A. fumigatus frequently coexist in the lungs. These two species have competitive interactions that can influence the growth of the microbiome and disease outcomes. Volatile bacterial organic compounds (VOCs) produced by P. aeruginosa or other gram-negative bacteria (e.g., E. coli and Burkholderia cepacia) can stimulate the growth of A. fumigatus without direct contact (35, 36) (Figure 1, Table 1). Li et al. (64) identified VOCs that can be used as biomarkers for differential diagnosis and therapeutic response prediction in patients with CPA. In contrast, A. fumigatus biofilm formation is inhibited by direct contact with P. aeruginosa (65). Pseudomonas aeruginosa showed a strong association with A. fumigatus hyphae. When P. aeruginosa is in direct contact with A. fumigatus, the diffusible extracellular molecules produced by P. aeruginosa disrupt its growth. Specifically, rhamnolipids secreted by P. aeruginosa block fungal β1,3 glucan synthase activity. Rhamnolipids inhibit the growth of A. fumigatus in in vitro experiments (37) (Figure 1, Table 1). Moreover, Hérivaux et al. (20) observed a loss of bacterial diversity and overgrowth of bacteria (e.g., Staphylococcus, Escherichia, Paraclostridium, and Finegoldia genera) in the lungs of patients with IPA. These changes in the lung microbiome were predictive of disease outcomes across IPA. In summary, there were complex reactions between lung bacteria and A. fumigatus. The growth of A. fumigatus may be regulated by lung bacteria, which, in turn, affects the severity of PA. Most of the studies were conducted in the context of CF, which has some similarities to the regulation of A. fumigatus growth by lung bacteria during PA; however, further validation is needed.
However, little is known about the direct regulation of PA by lung fungi. Several studies have focused on fungi that interact with lung bacteria and indirectly influence PA. Candida albicans colonization of the airway increases the prevalence of P. aeruginosa in rat lungs by inhibiting the production of reactive oxygen species by alveolar macrophages (66). An increase in the prevalence of P. aeruginosa in the lungs is likely to accelerate the growth of A. fumigatus and induce PA. Additionally, some studies have shown that changes in the composition of the lung microbiome can predict the survival of patients with IPA. On the one hand, the variety of lung bacteria declines, whereas lung fungi increase quickly. These changes worsen the prognosis of patients with IPA (20) (Table 1). On the other hand, the increase in strict anaerobes in the lungs reduces the risk of A. fumigatus infection by limiting the expansion of pathogenic Proteobacteria (38, 39) (Figure 1, Table 1). Notably, the lung microbiome has been shown to play a role in the regulation of A. fumigatus growth and even influence the progression of PA. However, studies on how the lung microbiome affects PA remain inadequate. As a potential treatment for PA, there is immense potential for future research on the lung microbiome.
The role of the lung microbiome in PA is probably largely underestimated because of non-specific and insensitive sampling and diagnostic tools. Compared with the gut microbiome, the lung microbiome is not easy to obtain and has low microbial biomass (67). Owing to the existence of physiological processes (e.g., aspiration), it is difficult to avoid the oral microbiome when trying to isolate the lung microbiome (23, 68). Deep sputum conjoint culture has been shown to distinguish oropharyngeal flora from lung fungi and diagnose lung fungal infections early (69). Pragman et al. (68) concluded that the lung lobectomy protocol utilized is well suited for obtaining reasonable non-invasive samples. Among the several methods used to obtain lung microbiome samples, bronchoscopy may cause sample contamination, but its effects are largely negligible (23). Micro-anatomical differences exist in the lung microbiome. Different parts of the lungs of the same individual have different microbiomes (70). In addition, individuals from different regions can also contain different lung microbiomes (71, 72). Rapid advances in technology have helped advance the study of the lung microbiome; however, several related problems exist owing to the lack of standardization. Diagnostic tools include high-throughput sequencing, phylogenetic microarray analysis, terminal restriction fragment length polymorphism, and amplicon length heterogeneity-polymerase chain reaction. The results obtained by different analytical methods vary (73, 74). In conclusion, many studies on the lung microbiome have been limited by small sample sizes, different sample collection techniques, different sampling sites, different regions of the subjects, and different analysis techniques. These limitations make it difficult to compare the results of the different studies. The impact of the lung microbiome on PA is an emerging area that needs further exploration and validation. Moreover, to better assess different studies and acquire reliable findings, continued research in this field is likely to establish a standardized method for obtaining and analysing the lung microbiome.
Bacteria colonizing the intestinal mucosa are involved in the maintenance of host immune homeostasis. Normal immune homeostasis further helps the host remove invasive fungi from the outside world. Ivanov et al. (40) found that segmented filamentous bacteria (SFB) can colonize the surface of the ileum in mice and induce intestinal CD4(+) T helper cells to produce IL-17 and IL-22 (Th17 cells). Furthermore, during fungal infections, SFB can induce lung autoimmunity by stimulating the systemic release of IL-1 receptor ligands and inducing gut-lung axis Th17 cells expressing dual TCR (41, 42) (Figure 1, Table 1). Mice infected with A. fumigatus show changes in the diversity of their gut bacteria, which affects intestinal immune tolerance and predisposes them to intestinal inflammation (75).
Probiotics are promising new targets for antifungal treatments. Probiotics can influence the constituents of the gut microbiome by directly affecting immune cells or releasing health-promoting metabolites, which, in turn, affects systemic immunity (43) (Figure 1, Table 1). For example, oral treatment with live Lactobacillus reuteri and Bifidobacterium longum reduces allergic airway reactions (e.g., ABPA) by increasing the number of Tregs in the lungs (44, 45). Oral administration of bacteria expressing high levels of α-Gal can protect turkeys against an infectious challenge with A. fumigatus by reducing the levels of lung anti-α-Gal IgA (46). Despite the lack of oral experiments, the E. coli DH5α strain also inhibited the development of A. fumigatus conidia in in vitro experiments (47). In summary, understanding the interaction between gut bacteria and host immunity provides a potential therapeutic strategy for the treatment of PA.
Studies have revealed the existence of cross-protective immunity between A. fumigatus and C. albicans. The gastrointestinal system of mice treated with C. albicans was protected against IPA and vice versa. In addition, cross-protection between A. fumigatus and C. albicans is mediated by Th1 immunity and dependent on IFN-γ. IFN-γ-deficient mice vaccinated with A. fumigatus or C. albicans show no reduced fungal growth in the lungs or the gastrointestinal system, respectively (76). Noverr et al. (48–50) demonstrated that antibiotic treatment changed the composition of the gut microbiome, causing overgrowth of intestinal bacteria and C. albicans in mice. As a result, Aspergillus-infected mice were more sensitive to CD4 T cell-mediated pulmonary allergic airway responses (e.g., ABPA). The reason for this may be that the overgrowth of C. albicans increases plasma concentrations of prostaglandin E2 (PGE2) and induces M2 macrophage polarization in the lungs (77). PGE2 is required for the Th17 response, and C. albicans is the major fungal inducer of human Th17 responses (51, 78). Global antifungal Th17 modulation by C. albicans promotes pathogenic airway inflammation triggered by A. fumigatus in susceptible patients via the selective recruitment of cross-reactive Th17 cells (51) (Figure 1, Table 1). Particularly, even if mice are exposed to Aspergillus spp., allergic reactions will not occur in the airways if the gut microbiome is not damaged by antibiotics (49, 50). Advances in the understanding of the relationship between the increase in intestinal C. albicans and the occurrence of ABPA have highlighted the importance of gut fungi in maintaining host immunity and resistance to Aspergillus spp. Nevertheless, there is still some confusion and defects in the mechanisms by which gut fungi regulate pulmonary immunity after infection with Aspergillus spp. This should be further explored in the future and should not be limited to C. albicans.
The microbiome plays an important role in the prevention of PA by inhibiting the growth of Aspergillus spp. or by increasing host immunity. However, this new research field poses several technical challenges and unanswered questions. Thus far, there is a lack of standardized sampling of the lungs and uniform sequencing techniques for the identification of the lung microbiome (79). Subsequently, many questions remain unanswered regarding the interaction of the lung microbiome with the gut microbiome during Aspergillus spp. infection (80). The influence of the lung and gut microbiomes on healthy and immunocompromised individuals during Aspergillus spp. infection remains unclear (81). Further studies focused on these issues will contribute to a better understanding of the effect of the microbiome and immune system on PA. The development of PA will lead to the development of novel treatment strategies.
XX, JX, and CC conceptualized the ideas. LC, PG, and ZW performed the literature search. LC wrote the original manuscript. CD, YN, and MI revised the manuscript. All authors contributed to the article and approved the submitted version.
This study was supported by the scientific and technological innovation action plan of Science and Technology Commission of Shanghai Municipality (No. 20DZ2253700) and Shanghai Hospital Development Center (SHDC2020CR5012).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
1. Kolwijck E, van de Veerdonk FL. The potential impact of the pulmonary microbiome on immunopathogenesis of aspergillus-related lung disease. Eur J Immunol (2014) 44(11):3156–65. doi: 10.1002/eji.201344404
2. Pfaller MA, Diekema DJ. Rare and emerging opportunistic fungal pathogens: concern for resistance beyond candida albicans and aspergillus fumigatus. J Clin Microbiol (2004) 42(10):4419–31. doi: 10.1128/jcm.42.10.4419-4431.2004
3. Kwon-Chung KJ, Sugui JA. Aspergillus fumigatus–what makes the species a ubiquitous human fungal pathogen? PloS Pathog (2013) 9(12):e1003743. doi: 10.1371/journal.ppat.1003743
4. Russo A, Tiseo G, Falcone M, Menichetti F. Pulmonary aspergillosis: an evolving challenge for diagnosis and treatment. Infect Dis Ther (2020) 9(3):511–24. doi: 10.1007/s40121-020-00315-4
5. Gao Y, Soubani A. Advances in the diagnosis and management of pulmonary aspergillosis. Adv Respir Med (2019) 87(6):231–43. doi: 10.5603/arm.2019.0061
6. Latgé JP. Aspergillus fumigatus and aspergillosis. Clin Microbiol Rev (1999) 12(2):310–50. doi: 10.1128/cmr.12.2.310
7. Kosmidis C, Denning DW. The clinical spectrum of pulmonary aspergillosis. Thorax (2015) 70(3):270–7. doi: 10.1136/thoraxjnl-2014-206291
8. Bongomin F, Gago S, Oladele RO, Denning DW. Global and multi-national prevalence of fungal diseases-estimate precision. J Fungi (Basel) (2017) 3(4):57. doi: 10.3390/jof3040057
9. Arastehfar A, Carvalho A, Houbraken J, Lombardi L, Garcia-Rubio R, Jenks JD, et al. Aspergillus fumigatus and aspergillosis: from basics to clinics. Stud Mycol (2021) 100:100115. doi: 10.1016/j.simyco.2021.100115
10. Echeverria-Esnal D, Martín-Ontiyuelo C, Navarrete-Rouco ME, Barcelo-Vidal J, Conde-Estévez D, Carballo N, et al. Pharmacological management of antifungal agents in pulmonary aspergillosis: an updated review. Expert Rev Anti Infect Ther (2022) 20(2):179–97. doi: 10.1080/14787210.2021.1962292
11. Denning DW, Cadranel J, Beigelman-Aubry C, Ader F, Chakrabarti A, Blot S, et al. Chronic pulmonary aspergillosis: rationale and clinical guidelines for diagnosis and management. Eur Respir J (2016) 47(1):45–68. doi: 10.1183/13993003.00583-2015
12. Fisher MC, Alastruey-Izquierdo A, Berman J, Bicanic T, Bignell EM, Bowyer P, et al. Tackling the emerging threat of antifungal resistance to human health. Nat Rev Microbiol (2022), 1–15. doi: 10.1038/s41579-022-00720-1
13. Moss RB. Treating allergic bronchopulmonary aspergillosis: the way forward. Eur Respir J (2016) 47(2):385–7. doi: 10.1183/13993003.01816-2015
14. Alastruey-Izquierdo A, Cadranel J, Flick H, Godet C, Hennequin C, Hoenigl M, et al. Treatment of chronic pulmonary aspergillosis: current standards and future perspectives. Respiration (2018) 96(2):159–70. doi: 10.1159/000489474
15. Choi HH, Cho YS. Fecal microbiota transplantation: current applications, effectiveness, and future perspectives. Clin Endosc (2016) 49(3):257–65. doi: 10.5946/ce.2015.117
16. Sencio V, Machado MG, Trottein F. The lung-gut axis during viral respiratory infections: the impact of gut dysbiosis on secondary disease outcomes. Mucosal Immunol (2021) 14(2):296–304. doi: 10.1038/s41385-020-00361-8
17. Sheflin AM, Whitney AK, Weir TL. Cancer-promoting effects of microbial dysbiosis. Curr Oncol Rep (2014) 16(10):406. doi: 10.1007/s11912-014-0406-0
18. Gurung M, Li Z, You H, Rodrigues R, Jump DB, Morgun A, et al. Role of gut microbiota in type 2 diabetes pathophysiology. EBioMedicine (2020) 51:102590. doi: 10.1016/j.ebiom.2019.11.051
19. Korf JM, Ganesh BP, McCullough LD. Gut dysbiosis and age-related neurological diseases in females. Neurobiol Dis (2022) 168:105695. doi: 10.1016/j.nbd.2022.105695
20. Hérivaux A, Willis JR, Mercier T, Lagrou K, Gonçalves SM, Gonçales RA, et al. Lung microbiota predict invasive pulmonary aspergillosis and its outcome in immunocompromised patients. Thorax (2022) 77(3):283–91. doi: 10.1136/thoraxjnl-2020-216179
21. Dickson RP, Erb-Downward JR, Martinez FJ, Huffnagle GB. The microbiome and the respiratory tract. Annu Rev Physiol (2016) 78:481–504. doi: 10.1146/annurev-physiol-021115-105238
22. Budden KF, Shukla SD, Rehman SF, Bowerman KL, Keely S, Hugenholtz P, et al. Functional effects of the microbiota in chronic respiratory disease. Lancet Respir Med (2019) 7(10):907–20. doi: 10.1016/s2213-2600(18)30510-1
23. Dickson RP, Erb-Downward JR, Freeman CM, McCloskey L, Falkowski NR, Huffnagle GB, et al. Bacterial topography of the healthy human lower respiratory tract. mBio (2017) 8(1):e02287–16. doi: 10.1128/mBio.02287-16
24. Jorth P, Ehsan Z, Rezayat A, Caldwell E, Pope C, Brewington JJ, et al. Direct lung sampling indicates that established pathogens dominate early infections in children with cystic fibrosis. Cell Rep (2019) 27(4):1190–204.e3. doi: 10.1016/j.celrep.2019.03.086
25. Pragman AA, Kim HB, Reilly CS, Wendt C, Isaacson RE. The lung microbiome in moderate and severe chronic obstructive pulmonary disease. PloS One (2012) 7(10):e47305. doi: 10.1371/journal.pone.0047305
26. Rogers GB, Zain NM, Bruce KD, Burr LD, Chen AC, Rivett DW, et al. A novel microbiota stratification system predicts future exacerbations in bronchiectasis. Ann Am Thorac Soc (2014) 11(4):496–503. doi: 10.1513/AnnalsATS.201310-335OC
27. Acosta N, Heirali A, Somayaji R, Surette MG, Workentine ML, Sibley CD, et al. Sputum microbiota is predictive of long-term clinical outcomes in young adults with cystic fibrosis. Thorax (2018) 73(11):1016–25. doi: 10.1136/thoraxjnl-2018-211510
28. Leitao Filho FS, Alotaibi NM, Ngan D, Tam S, Yang J, Hollander Z, et al. Sputum microbiome is associated with 1-year mortality after chronic obstructive pulmonary disease hospitalizations. Am J Respir Crit Care Med (2019) 199(10):1205–13. doi: 10.1164/rccm.201806-1135OC
29. Budden KF, Gellatly SL, Wood DL, Cooper MA, Morrison M, Hugenholtz P, et al. Emerging pathogenic links between microbiota and the gut-lung axis. Nat Rev Microbiol (2017) 15(1):55–63. doi: 10.1038/nrmicro.2016.142
30. McAleer JP, Kolls JK. Contributions of the intestinal microbiome in lung immunity. Eur J Immunol (2018) 48(1):39–49. doi: 10.1002/eji.201646721
31. Lee SC, Hua CC, Yu TJ, Shieh WB, See LC. Risk factors of mortality for nosocomial pneumonia: importance of initial anti-microbial therapy. Int J Clin Pract (2005) 59(1):39–45. doi: 10.1111/j.1742-1241.2005.00281.x
32. Ichinohe T, Pang IK, Kumamoto Y, Peaper DR, Ho JH, Murray TS, et al. Microbiota regulates immune defense against respiratory tract influenza a virus infection. Proc Natl Acad Sci USA (2011) 108(13):5354–9. doi: 10.1073/pnas.1019378108
33. Yagi K, Huffnagle GB, Lukacs NW, Asai N. The lung microbiome during health and disease. Int J Mol Sci (2021) 22(19):10872. doi: 10.3390/ijms221910872
34. Chunxi L, Haiyue L, Yanxia L, Jianbing P, Jin S. The gut microbiota and respiratory diseases: new evidence. J Immunol Res (2020) 2020:2340670. doi: 10.1155/2020/2340670
35. Briard B, Heddergott C, Latgé JP. Volatile compounds emitted by pseudomonas aeruginosa stimulate growth of the fungal pathogen aspergillus fumigatus. mBio (2016) 7(2):e00219. doi: 10.1128/mBio.00219-16
36. Latgé JP, Chamilos G. Aspergillus fumigatus and aspergillosis in 2019. Clin Microbiol Rev (2019) 33(1):e00140–18. doi: 10.1128/cmr.00140-18
37. Briard B, Rasoldier V, Bomme P, ElAouad N, Guerreiro C, Chassagne P, et al. Dirhamnolipids secreted from pseudomonas aeruginosa modify anjpegungal susceptibility of aspergillus fumigatus by inhibiting β1,3 glucan synthase activity. Isme J (2017) 11(7):1578–91. doi: 10.1038/ismej.2017.32
38. Nunzi E, Renga G, Palmieri M, Pieraccini G, Pariano M, Stincardini C, et al. A shifted composition of the lung microbiota conditions the antifungal response of immunodeficient mice. Int J Mol Sci (2021) 22(16):8474. doi: 10.3390/ijms22168474
39. Costantini C, Nunzi E, Spolzino A, Palmieri M, Renga G, Zelante T, et al. Pharyngeal microbial signatures are predictive of the risk of fungal pneumonia in hematologic patients. Infect Immun (2021) 89(8):e0010521. doi: 10.1128/iai.00105-21
40. Ivanov II, Atarashi K, Manel N, Brodie EL, Shima T, Karaoz U, et al. Induction of intestinal th17 cells by segmented filamentous bacteria. Cell (2009) 139(3):485–98. doi: 10.1016/j.cell.2009.09.033
41. McAleer JP, Nguyen NL, Chen K, Kumar P, Ricks DM, Binnie M, et al. Pulmonary th17 antifungal immunity is regulated by the gut microbiome. J Immunol (2016) 197(1):97–107. doi: 10.4049/jimmunol.1502566
42. Bradley CP, Teng F, Felix KM, Sano T, Naskar D, Block KE, et al. Segmented filamentous bacteria provoke lung autoimmunity by inducing gut-lung axis th17 cells expressing dual tcrs. Cell Host Microbe (2017) 22(5):697–704.e4. doi: 10.1016/j.chom.2017.10.007
43. Marsland BJ, Trompette A, Gollwitzer ES. The gut-lung axis in respiratory disease. Ann Am Thorac Soc (2015) 12 Suppl 2:S150–6. doi: 10.1513/AnnalsATS.201503-133AW
44. Karimi K, Inman MD, Bienenstock J, Forsythe P. Lactobacillus reuteri-induced regulatory t cells protect against an allergic airway response in mice. Am J Respir Crit Care Med (2009) 179(3):186–93. doi: 10.1164/rccm.200806-951OC
45. MacSharry J, O’Mahony C, Shalaby KH, Sheil B, Karmouty-Quintana H, Shanahan F, et al. Immunomodulatory effects of feeding with bifidobacterium longum on allergen-induced lung inflammation in the mouse. Pulm Pharmacol Ther (2012) 25(4):325–34. doi: 10.1016/j.pupt.2012.05.011
46. Mateos-Hernández L, Risco-Castillo V, Torres-Maravilla E, LG Bermúdez-Humarán, Alberdi P, Hodžić A, et al. Gut microbiota abrogates anti-α-Gal iga response in lungs and protects against experimental aspergillus infection in poultry. Vaccines (Basel) (2020) 8(2):285. doi: 10.3390/vaccines8020285
47. Balhara M, Ruhil S, Kumar M, Dhankhar S, Chhillar AK. Inhibition of conidiophore development in aspergillus fumigatus by an escherichia coli dh5α strain, a promising antifungal candidate against aspergillosis. J Mycol Med (2014) 24(1):1–12. doi: 10.1016/j.mycmed.2013.07.055
48. Noverr MC, Falkowski NR, McDonald RA, McKenzie AN, Huffnagle GB. Development of allergic airway disease in mice following antibiotic therapy and fungal microbiota increase: role of host genetics, antigen, and interleukin-13. Infect Immun (2005) 73(1):30–8. doi: 10.1128/iai.73.1.30-38.2005
49. Noverr MC, Huffnagle GB. Does the microbiota regulate immune responses outside the gut? Trends Microbiol (2004) 12(12):562–8. doi: 10.1016/j.tim.2004.10.008
50. Noverr MC, Noggle RM, Toews GB, Huffnagle GB. Role of antibiotics and fungal microbiota in driving pulmonary allergic responses. Infect Immun (2004) 72(9):4996–5003. doi: 10.1128/iai.72.9.4996-5003.2004
51. Bacher P, Hohnstein T, Beerbaum E, Röcker M, Blango MG, Kaufmann S, et al. Human anti-fungal th17 immunity and pathology rely on cross-reactivity against candida albicans. Cell (2019) 176(6):1340–55.e15. doi: 10.1016/j.cell.2019.01.041
52. van de Veerdonk FL, Gresnigt MS, Romani L, Netea MG, Latgé JP. Aspergillus fumigatus morphology and dynamic host interactions. Nat Rev Microbiol (2017) 15(11):661–74. doi: 10.1038/nrmicro.2017.90
53. Cunha C, Carvalho A, Esposito A, Bistoni F, Romani L. Damp signaling in fungal infections and diseases. Front Immunol (2012) 3:286. doi: 10.3389/fimmu.2012.00286
54. Patin EC, Thompson A, Orr SJ. Pattern recognition receptors in fungal immunity. Semin Cell Dev Biol (2019) 89:24–33. doi: 10.1016/j.semcdb.2018.03.003
55. Steele C, Rapaka RR, Metz A, Pop SM, Williams DL, Gordon S, et al. The beta-glucan receptor dectin-1 recognizes specific morphologies of aspergillus fumigatus. PloS Pathog (2005) 1(4):e42. doi: 10.1371/journal.ppat.0010042
56. Aderem A, Underhill DM. Mechanisms of phagocytosis in macrophages. Annu Rev Immunol (1999) 17:593–623. doi: 10.1146/annurev.immunol.17.1.593
57. Diamond RD, Clark RA. Damage to aspergillus fumigatus and rhizopus oryzae hyphae by oxidative and nonoxidative microbicidal products of human neutrophils in vitro. Infect Immun (1982) 38(2):487–95. doi: 10.1128/iai.38.2.487-495.1982
58. Sales-Campos H, Tonani L, Cardoso CR, Kress MR. The immune interplay between the host and the pathogen in aspergillus fumigatus lung infection. BioMed Res Int (2013) 2013:693023. doi: 10.1155/2013/693023
59. Allard JB, Rinaldi L, Wargo MJ, Allen G, Akira S, Uematsu S, et al. Th2 allergic immune response to inhaled fungal antigens is modulated by tlr-4-independent bacterial products. Eur J Immunol (2009) 39(3):776–88. doi: 10.1002/eji.200838932
60. Zelante T, De Luca A, Bonifazi P, Montagnoli C, Bozza S, Moretti S, et al. Il-23 and the th17 pathway promote inflammation and impair antifungal immune resistance. Eur J Immunol (2007) 37(10):2695–706. doi: 10.1002/eji.200737409
61. Werner JL, Metz AE, Horn D, Schoeb TR, Hewitt MM, Schwiebert LM, et al. Requisite role for the dectin-1 beta-glucan receptor in pulmonary defense against aspergillus fumigatus. J Immunol (2009) 182(8):4938–46. doi: 10.4049/jimmunol.0804250
62. Nasillo V, Lagreca I, Vallerini D, Barozzi P, Riva G, Maccaferri M, et al. Btk inhibitors impair platelet-mediated antifungal activity. Cells (2022) 11(6):1003. doi: 10.3390/cells11061003
63. Sykes DB, Martinelli MM, Negoro P, Xu S, Maxcy K, Timmer K, et al. Transfusable neutrophil progenitors as cellular therapy for the prevention of invasive fungal infections. J Leukoc Biol (2022) 111(6):1133–45. doi: 10.1002/jlb.4hi1221-722r
64. Li ZT, Zeng PY, Chen ZM, Guan WJ, Wang T, Lin Y, et al. Exhaled volatile organic compounds for identifying patients with chronic pulmonary aspergillosis. Front Med (Lausanne) (2021) 8:720119. doi: 10.3389/fmed.2021.720119
65. Mowat E, Rajendran R, Williams C, McCulloch E, Jones B, Lang S, et al. Pseudomonas aeruginosa and their small diffusible extracellular molecules inhibit aspergillus fumigatus biofilm formation. FEMS Microbiol Lett (2010) 313(2):96–102. doi: 10.1111/j.1574-6968.2010.02130.x
66. Roux D, Gaudry S, Dreyfuss D, El-Benna J, de Prost N, Denamur E, et al. Candida albicans impairs macrophage function and facilitates pseudomonas aeruginosa pneumonia in rat. Crit Care Med (2009) 37(3):1062–7. doi: 10.1097/CCM.0b013e31819629d2
67. Marsland BJ, Yadava K, Nicod LP. The airway microbiome and disease. Chest (2013) 144(2):632–7. doi: 10.1378/chest.12-2854
68. Pragman AA, Lyu T, Baller JA, Gould TJ, Kelly RF, Reilly CS, et al. The lung tissue microbiota of mild and moderate chronic obstructive pulmonary disease. Microbiome (2018) 6(1):7. doi: 10.1186/s40168-017-0381-4
69. Gitelman I. Saliva-controlled sputum culture (ssc) - a high value diagnostic tool for deep pulmonary infections. J Microbiol Methods (2020) 179:105986. doi: 10.1016/j.mimet.2020.105986
70. Erb-Downward JR, Thompson DL, Han MK, Freeman CM, McCloskey L, Schmidt LA, et al. Analysis of the lung microbiome in the “healthy” smoker and in copd. PloS One (2011) 6(2):e16384. doi: 10.1371/journal.pone.0016384
71. Lackner M, Coassin S, Haun M, Binder U, Kronenberg F, Haas H, et al. Geographically predominant genotypes of aspergillus terreus species complex in austria: s microsatellite typing study. Clin Microbiol Infect (2016) 22(3):270–6. doi: 10.1016/j.cmi.2015.10.021
72. Stressmann FA, Rogers GB, Klem ER, Lilley AK, Donaldson SH, Daniels TW, et al. Analysis of the bacterial communities present in lungs of patients with cystic fibrosis from american and british centers. J Clin Microbiol (2011) 49(1):281–91. doi: 10.1128/jcm.01650-10
73. Doud M, Zeng E, Schneper L, Narasimhan G, Mathee K. Approaches to analyse dynamic microbial communities such as those seen in cystic fibrosis lung. Hum Genomics (2009) 3(3):246–56. doi: 10.1186/1479-7364-3-3-246
74. Doud MS, Light M, Gonzalez G, Narasimhan G, Mathee K. Combination of 16s rrna variable regions provides a detailed analysis of bacterial community dynamics in the lungs of cystic fibrosis patients. Hum Genomics (2010) 4(3):147–69. doi: 10.1186/1479-7364-4-3-147
75. Kulas J, Mirkov I, Tucovic D, Zolotarevski L, Glamoclija J, Veljovic K, et al. Pulmonary aspergillus fumigatus infection in rats affects gastrointestinal homeostasis. Immunobiology (2019) 224(1):116–23. doi: 10.1016/j.imbio.2018.10.001
76. Stuehler C, Khanna N, Bozza S, Zelante T, Moretti S, Kruhm M, et al. Cross-protective th1 immunity against aspergillus fumigatus and candida albicans. Blood (2011) 117(22):5881–91. doi: 10.1182/blood-2010-12-325084
77. Kim YG, Udayanga KG, Totsuka N, Weinberg JB, Núñez G, Shibuya A. Gut dysbiosis promotes m2 macrophage polarization and allergic airway inflammation via fungi-induced pge₂. Cell Host Microbe (2014) 15(1):95–102. doi: 10.1016/j.chom.2013.12.010
78. Smeekens SP, van de Veerdonk FL, van der Meer JW, Kullberg BJ, Joosten LA, Netea MG. The candida th17 response is dependent on mannan- and beta-glucan-induced prostaglandin e2. Int Immunol (2010) 22(11):889–95. doi: 10.1093/intimm/dxq442
79. Carney SM, Clemente JC, Cox MJ, Dickson RP, Huang YJ, Kitsios GD, et al. Methods in lung microbiome research. Am J Respir Cell Mol Biol (2020) 62(3):283–99. doi: 10.1165/rcmb.2019-0273TR
80. Shukla SD, Budden KF, Neal R, Hansbro PM. Microbiome effects on immunity, health and disease in the lung. Clin Transl Immunol (2017) 6(3):e133. doi: 10.1038/cti.2017.6
Keywords: pulmonary aspergillosis, fungal diseases, Aspergillus, immunity, microbiome
Citation: Cai L, Gao P, Wang Z, Dai C, Ning Y, Ilkit M, Xue X, Xiao J and Chen C (2022) Lung and gut microbiomes in pulmonary aspergillosis: Exploring adjunctive therapies to combat the disease. Front. Immunol. 13:988708. doi: 10.3389/fimmu.2022.988708
Received: 07 July 2022; Accepted: 25 July 2022;
Published: 12 August 2022.
Edited by:
Wenjie Fang, Shanghai Changzheng Hospital, ChinaReviewed by:
Lei Zhang, Shaanxi Provincial People’s Hospital, ChinaCopyright © 2022 Cai, Gao, Wang, Dai, Ning, Ilkit, Xue, Xiao and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiaochun Xue, eHhjMjAyMUAxMjYuY29t; Jinzhou Xiao, c2lnbmFsNjExQDE2My5jb20=; Chang Chen, Y2hlbnRob3JhY2ljQDE2My5jb20=
†These authors have contributed equally to this work and share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.