
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Immunol., 04 April 2022
Sec. Cancer Immunity and Immunotherapy
Volume 13 - 2022 | https://doi.org/10.3389/fimmu.2022.833058
This article is part of the Research TopicRole of Non-Coding RNA in Gastrointestinal Cancer Tumor Immune Microenvironment and Anticancer TherapyView all 5 articles
 Chunyue Chen1,2†
Chunyue Chen1,2† Congcong Xia1,2†
Congcong Xia1,2† Hao Tang1,2
Hao Tang1,2 Yirun Jiang1,2
Yirun Jiang1,2 Shan Wang1,2,3
Shan Wang1,2,3 Xin Zhang1,2,3
Xin Zhang1,2,3 Tao Huang1,2,3
Tao Huang1,2,3 Xiaoqing Yuan4,5
Xiaoqing Yuan4,5 Junpu Wang1,2,3,6*
Junpu Wang1,2,3,6* Li Peng4,7*
Li Peng4,7*The immune system plays a complex role in tumor formation and development. On the one hand, immune surveillance can inhibit the growth of tumors; on the other hand, immune evasion of tumors can create conditions conducive for tumor development and growth. CircRNAs are endogenous non-coding RNAs with a covalently closed loop structure that are abundantly expressed in eukaryotic organisms. They are characterized by stable structure, rich diversity, and high evolutionary conservation. In particular, circRNAs play a vital role in the occurrence, development, and treatment of tumors through their unique functions. Recently, the incidence and mortality of digestive cancers, especially those of gastric cancer, colorectal cancer, and liver cancer, have remained high. However, the functions of circRNAs in digestive cancers immunity are less known. The relationship between circRNAs and digestive tumor immunity is systematically discussed in our paper for the first time. CircRNA can influence the immune microenvironment of gastrointestinal tumors to promote their occurrence and development by acting as a miRNA molecular sponge, interacting with proteins, and regulating selective splicing. The circRNA vaccine even provides a new idea for tumor immunotherapy. Future studies should be focused on the location, transportation, and degradation mechanisms of circRNA in living cells and the relationship between circRNA and tumor immunity. This paper provides a new idea for the diagnosis and treatment of gastrointestinal tumors.
In 1976, Sanger et al. (1) discovered a new single-stranded covalent circular RNA (circRNA) in plant viroids that is different from linear RNA. Subsequently, they proposed the concept of circRNA. CircRNA has been long regarded as a by-product of incorrect alternative splicing (2) and thus has not attracted the attention of the academic community. With the advancement in sequencing and bioinformatics technologies, our knowledge of circRNAs significantly increased. CircRNA is an endogenous non-coding RNA (ncRNA) molecule widely found in various biological cells. The most significant difference between circRNA and linear RNA is that the 3′ end and 5′ end of the circRNA molecule are connected to form a closed-loop structure. This particular structure provides circRNA with a high degree of abundance, conservation, stability, and distinctive expression pattern, which is tissue-specific and related to the developmental stage (3). At present, there are six main functions of circRNAs: (i) as a sponge of microRNAs (miRNAs), (ii) co-acting with proteins, (iii) regulation of gene transcription, (iv) selective splicing regulation, (v) as a translation template for proteins, and (vi) regulation of tumor immunity (3–6). More in-depth knowledge of circRNA can provide a new research direction to explore the occurrence, development mechanism, treatment, and prognosis of many diseases.
Digestive cancers mainly include oesophageal cancer, gastric cancer (GC), colorectal cancer (CRC), liver cancer, gallbladder cancer, cholangiocarcinoma, and pancreatic cancer. According to the Global Cancer Statistics 2020, the cancers with high mortality in 185 countries worldwide are lung cancer, liver cancer (8.3% of the cancer-related deaths), GC (7.7%), breast cancer, CRC (9.2%), oesophageal cancer (5.5%), pancreatic cancer (4.7%), and gallbladder cancer (0.9%) (7), with digestive cancers accounting for leading cancer deaths (36.3%). Similarly, the incidence of digestive cancers is estimated to reach as high as 5,091,327 new cases by 2040, which accounts for 26.4% of all 36 cancer types (7). These data indicate that cancers of the digestive system pose a serious health burden worldwide.
In the digestive system, immune tissue is widely distributed. The liver, digestive tract lymph node and mucosal immune tissue all perform different immune functions. The immune response plays a crucial role in the invasion of endogenous or exogenous substances. The role of immunity in tumors is similar, which is closely related to the occurrence and development of tumors (1–4). Immune surveillance and immune escape lead to different outcomes. Furthermore, circRNAs can regulate the expression of host genes. More principally, they serve as microRNA (miRNA) sponges to adjust the axis of miRNA/mRNA or regulate the activities of immune cells directly. Thus they play an essential role in the immune disorders of tumors (5–8). This paper summarizes the relationship between circRNA and digestive tumors, mainly focusing on tumor immunity.
In general, eukaryotic genes remove non-coding intron sequences through pre-mRNA splicing after transcription. And the exon sequences containing protein-coding information are sequentially joined together to form mature linear RNA molecules, thus achieving the transmission of genetic information. However, there is a method of reverse splicing, i.e., the reverse head-to-tail connection of the gene exon sequence, that forms circRNAs. The vast majority of circRNAs in organisms are processed and synthesized by precursor RNA (pre-mRNA) through various nonclassical reverse splicing methods.
Based on their composition, circRNAs can be divided into three categories as follows (Table 1): (i) exonic circRNAs (ecircRNAs), which are composed entirely of exons (9), (ii) exonic-intronic circRNAs (EIcircRNAs), which contain both exons and introns (10), and (iii) circular intronic RNAs (ciRNAs), which are composed entirely of introns (11). The splicing formation of circRNA can be classified into two categories: exon-cyclized and intron-cyclized. Jeck et al. (9) proposed three different formation models for ecircRNAs and EIcircRNAs. First is the Lariat-driven cyclization in which the 5′ donor site downstream of one exon is connected to the 3′ receptor site upstream of another exon to form a reverse covalently linked lasso structure. That is, the lasso-driven splicing donor and the splice acceptor are covalently combined to form an annular structure. After further splicing and removing the introns, the exons form ecircRNA through phosphodiester bonds. At this time, the synthesized circRNA is mainly located in the cytoplasm (12) (Figure 1A). The second model is RNA-binding protein (RBP)-dependent cyclization (13). Here, the upstream and downstream introns at both ends of the pre-mRNA exon bind specifically to RBP. Then, through a sequence that RBP can recognize, a dimer forms. Finally, the dimer cycles to form EIciRNA. At this time, the synthesized circRNA is mainly located in the nucleus (Figure 1B). The third model is cyclization driven by intron pairing. The introns on both sides of the pre-mRNA exons contain reverse complementary sequences, which form double-stranded RNA side by side at the splicing site in the duplex. Finally, ecircRNA containing exons or EIciRNA containing exons and introns are formed through differential splicing (14). Here, the synthesized circRNA is mainly located in the nucleus (Figure 1C). The ciRNA formed by the self-cyclization of introns is different from that of EcircRNA. It contains a unique 2′,5′ connection whose formation is related to the 7nt guanine and uracil-rich (GU-rich) sequence at both ends of the intron, the uracil-rich (U-rich) sequence, and the 11nt cytosine-rich (C-rich) sequence. The two ends of the intron GU-rich and C-rich elements are cyclized to form an exon-free lasso RNA structure with different tail lengths (15). After removing the tail branches, ciRNA is generated mainly in the nucleus (11) (Figure 1D).
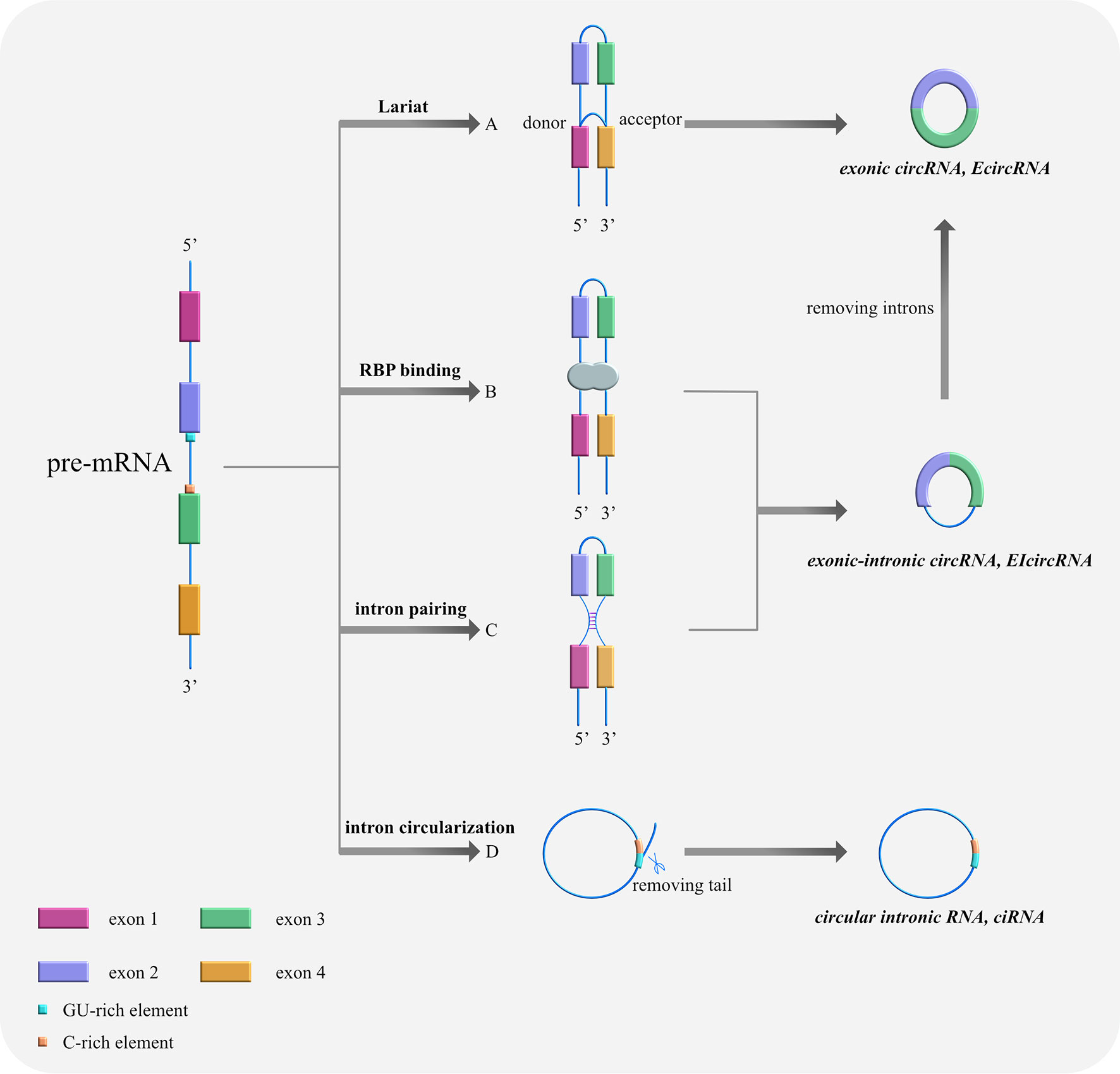
Figure 1 Cyclization models of circRNAs. (A) Lariat-driven cyclization. The 5′ donor splice site downstream of exon 4 is connected with the 3′ receptor splice site upstream of exon 1, forming a reverse covalently linked lariat structure. Then, the lasso containing skipped exons shears further and removes the introns, thus forming ecircRNA. (B) RNA-binding protein (RBP)-driven cyclization. RBPs bind to the splicing sites of both sides of exons 2 and 3 to assist cyclization and form ElciRNA. (C) Cyclization driven by intron pairing: complementary sequences at the flanks of exons 2 and 3 bring splicing sites together by base pairing, forming double-stranded RNA side by side. Afterwards, ecircRNA containing exons or EIciRNA containing exons and introns are produced through variable shearing. (D) Self-cyclization of introns: Pre-mRNA is spliced by spliceosome to remove intervening introns and there are only exons left, which are connected to form mature RNA. With the 7nt GU-rich sequence at both ends of the intron, the U-rich sequence, and the 11nt C-rich sequence, an exon-free lariat RNA structure is formed by 2′,5′ connection’s circularization.
Based on the gene location of the parents, circRNAs can be classified as intragenic and intergenic circRNAs. Each ecircRNA, EIcircRNA, and ciRNA molecule is derived from the splicing of different exons and/or introns within the same parent gene, so they are all considered intragenic circRNAs. Meanwhile, circRNA from the genome interval between different genes is called the intergenic circRNA.
CircRNAs are produced by special alternative splicing and largely exist in the cytoplasm of eukaryotic cells (2), although a small number of intron-derived circRNAs are found within the nucleus (16). Besides, circRNAs have solid specificity of cell, tissue, and development (17, 18). They are also widely found in human cells. In fact, more than 10% of the genes can produce circRNAs through reverse splicing and other pathways. The circRNA of some genes accumulates in the cells, whose number far exceeds that of linear mRNAs corresponding to the gene, and sometimes even up to ten times that of their linear isomers (19, 20). Furthermore, unlike the traditional linear RNA with 5′ and 3′ ends, the circRNA molecule has a covalently closed loop structure without a 5′- terminal cap and a 3′- terminal poly (A) tail; thus, it is not easily degraded by exonuclease RNase R and is more stable than linear RNA (19, 21, 22). Besides, many circRNAs are evolutionarily conserved among different species (19, 20). Jeck et al. (9) discovered that 457 of 2121 murine circles that readily mapped from human genomes could produce murine circular RNA, at a rate of 22%. The homology of mouse and human circRNAs varies from less than 5% to nearly 30% (23), whereas that of pig and mouse circRNAs varies from less than 15% to nearly 20% (24), and that of mouse and rat circRNAs varies ≥23% (25). In addition, some circRNAs exhibit rapid evolutionary changes (5, 16). Most circRNAs are derived from exons, and a small portion is formed by the direct circularisation of introns. Moreover, circRNA molecules are rich in miRNA response elements (MREs), which can act as miRNA sponges and competitive endogenous RNA (ceRNA) binding to miRNA. In this way, circRNAs can relieve the inhibition of miRNA on its target genes and up-regulate the expression level of target genes (14). Most circRNAs are non-coding RNAs (20). Jeck et al. (19) confirmed that there are more than 25 000 different RNAs in human fibroblasts that could produce circRNA. In addition, the same gene can produce different circRNAs through variable cyclization (16, 20); thus, circRNAs from the same gene source are diverse and enrich the species of circRNA. Other features of circRNAs remain to be elucidated. Compared with traditional linear RNA, circRNA has more quantity, higher stability and more MRE. It has great advantages in research, making it more likely to be used in the mechanism exploration and treatment of gastrointestinal tumors.
Studies have shown that circRNA is differentially expressed in cancer compared with normal tissues, which suggests that they serve a specific function in these cells (19). And exo-circRNA may distinguish patients with cancer from healthy controls, which illustrates its significant translational potential as a circulating biomarker for cancer diagnosis (26). These indirectly prove the role of circRNA in tumor development, and tumor immunity is an important factor determining the direction of tumor development. Therefore, the relationship between circRNA and tumor immunity is worth exploring.
To date, an increasing number of studies have confirmed that circRNAs play a critical role in immune response and the regulation of tumor immunity. For example, several circRNAs are associated with the infiltration of immune cells into tumors. Du et al. have reported that B and T cells infiltrate the tumor and the surrounding connective tissue in breast cancer and express circ-Foxo3. It indicates a host immune response to tumor xenografts, with circ-Foxo3 playing a tumor-suppressive role (27). Zou et al. also showed that circ-CDR1as is crucial for infiltrating immune cells in tumor tissues, especially CD8+ T cells (28). Furthermore, a ceRNA network involves hsa-miR-494, circ-UBAP2, and five central genes, particularly ZEB1 and CXCR4. The network can regulate pancreatic adenocarcinoma progression by controlling the infiltration of immune cells (16). CircRNA is also involved in tumor immunity by regulating intercellular cell adhesion molecule-1 (ICAM-1) molecules. In response to tumor cell invasion, hsa_circ_0007456 can regulate ICAM-1 expression via miR-6852-3p, thereby regulating natural killer (NK) cell activity to mediate tumor immune escape (17). Interestingly, in viral immunity, another circRasGEF1B molecule plays the opposite role. CircRasGEF1B regulates the level of ICAM-1 by modulating the stability of its mRNA, thus stimulating the innate immune response and protecting host cells from pathogens (18).
CircRNAs in tumors are also closely linked to immune-related molecules. For example, the nuclear factor kappa B (NF-κB) has been shown to play a unique role in the immunity of tumor cells (20–22). Its inhibition attenuates the ciRS-7-induced upregulation of MMP-2, thereby inhibiting ciRS-7-mediated oesophageal squamous cell carcinoma cell invasion (23). Wang et al. revealed that the circRNA-000911/miR-449a/Notch1/NF-κB network might be partly responsible for the oncogenic activity of breast cancer cells and maybe a new target for breast cancer therapy (24).
The primary known functions of circRNA include the following: (i) acts as a molecular sponge for miRNAs to silence the corresponding miRNAs and cause a series of alterations in downstream responses (25) (Figure 2A); (ii) interacts with a protein (protein decoy), thereby inhibiting its function (Figure 2B); (iii) regulation of gene transcription (Figure 2C); (iv) regulation of selective splicing (Figure 2D); and (v) translation into proteins (Figure 2E). Although current studies on the relationship between circRNAs and tumor immunity have mainly focused on their role as miRNA molecular sponges and protein decoys, studies on the regulation of selective splicing have also been conducted. Therefore, these three functions will be the main focus of our discussion.
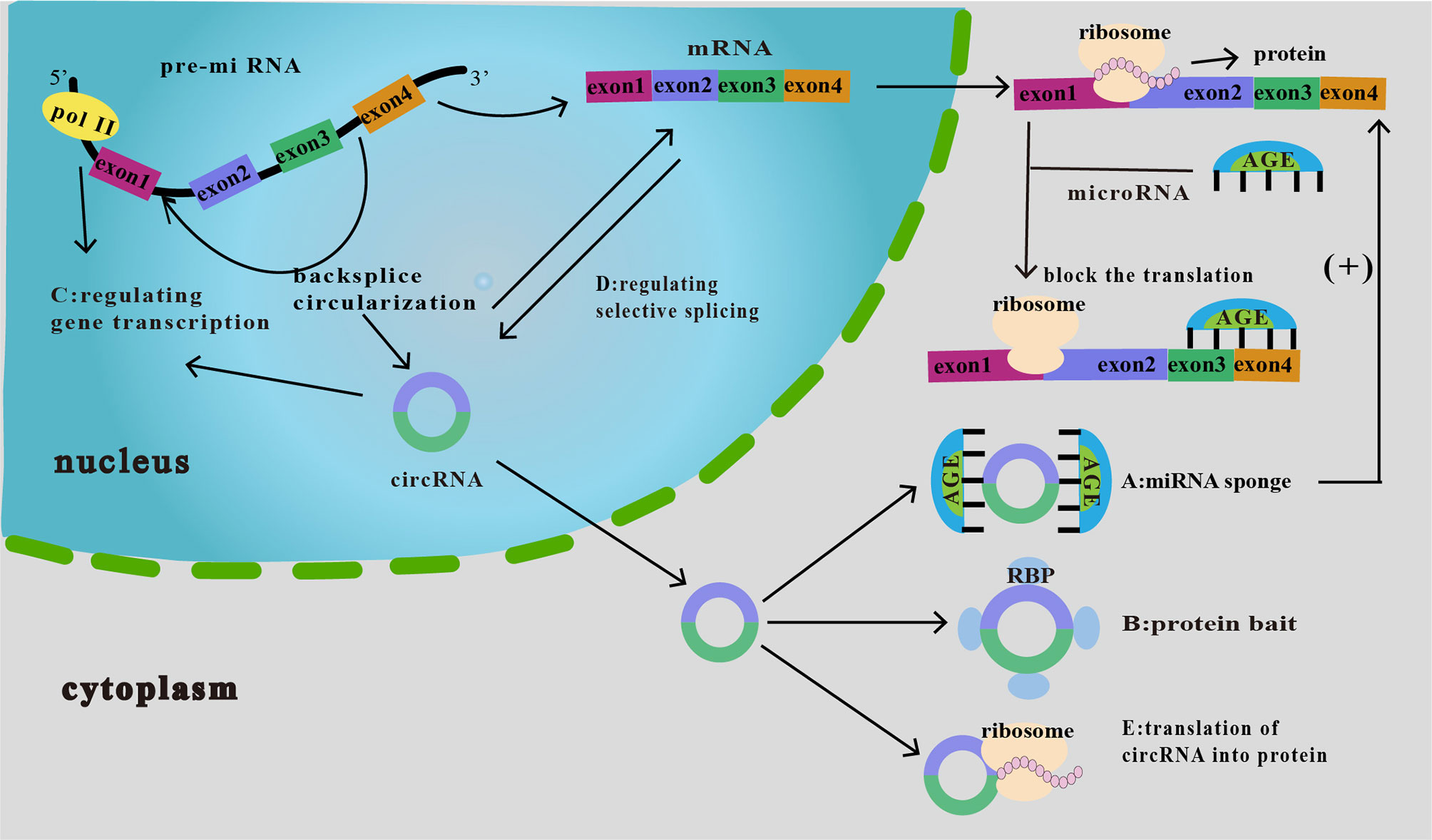
Figure 2 Functions of circRNAs. (A) As miRNA sponge. CircRNA binds miRNAs and inhibits their downstream responses, which then increases the expression of downstream target proteins, thereby affecting tumor proliferation, invasion, and migration. (B) As protein bait. CircRNAs act as protein decoys or antagonists to inhibit protein function, thus affecting the related progression of cancer. (C) Regulation of gene transcription. CircRNA interacts with RNA polymerase II to regulate the transcription and expression of parental genes. (D) Regulation of selective splicing. CircRNAs compete with linear RNAs; if the formation of circRNAs increases, linear splicing decreases. (E) Translation of circRNA into protein. CircRNAs can be translated into proteins in an unusual cap-independent manner.
MiRNAs are endogenous non-coding RNAs with regulatory functions in eukaryotic organisms and are roughly 19–25 nucleotides in length. They are involved in the formation of RNA-induced silencing complexes (RISC) and direct RISC to regulate post-transcriptional silencing (29, 30). However, circRNAs contain MREs that bind to miRNAs and act as miRNA sponges or ceRNAs, thus silencing the corresponding miRNAs and causing a series of alterations in downstream responses (25). Many of the genes with known ceRNA interactors identified so far have been implicated in human disease. Aberrant changes in ceRNA regulation may also contribute to disease initiation and progression (31). So ceRNA network interactions may have a role in determining the effectiveness of RNA-directed therapies (32). As a kind of Cerna, the role of circRNA in disease development is also worth studying.
CircRNAs regulate immunity in various types of cancer by acting as a miRNA sponge, and much attention has been paid to the regulation of immune escape. The interaction between programmed death receptor 1 (PD-1) and programmed cell death ligand 1 (PD-L1) can effectively suppress the activation of effector T lymphocytes, ultimately leading to tumor immune escape (Figure 3A). CircRNA-002178 has also been found to bind to miR-34a and suppress its expression, resulting in increased PD-L1 expression in cancer cells. CircRNA-002178 can also be transported from cancer cells to CD8+ T cells via exosomes. It suppresses miR-28-5p activity and promotes PD1 expression on CD8+ T cells (33). In addition, miR-138 activity is inhibited upon binding to hsa_circ_0020397, thus affecting its downstream responses and promoting the expression of PD-L1 and telomerase reverse transcriptase in CRC cells. Owing to the high expression level of has_circ_0020397 in CRC cells, the expression of PD-L1 is upregulated in turn (34, 35). Similarly, circRNA vimentin (circ-VIM) can bind to miR-124 and deregulate its downstream target PD-L1. Sevoflurane can synergistically inhibit immune escape from oesophageal cancer by circ-VIM silencing (36). The role of miRNAs in tumor immunity, especially in the regulation of PD-1/PD-L1 immune checkpoint expression, has been extensively investigated (37, 38). These results suggest that circRNAs promote tumor immune escape through the circRNA/miRNA/PD-1/PD-L1 axis.
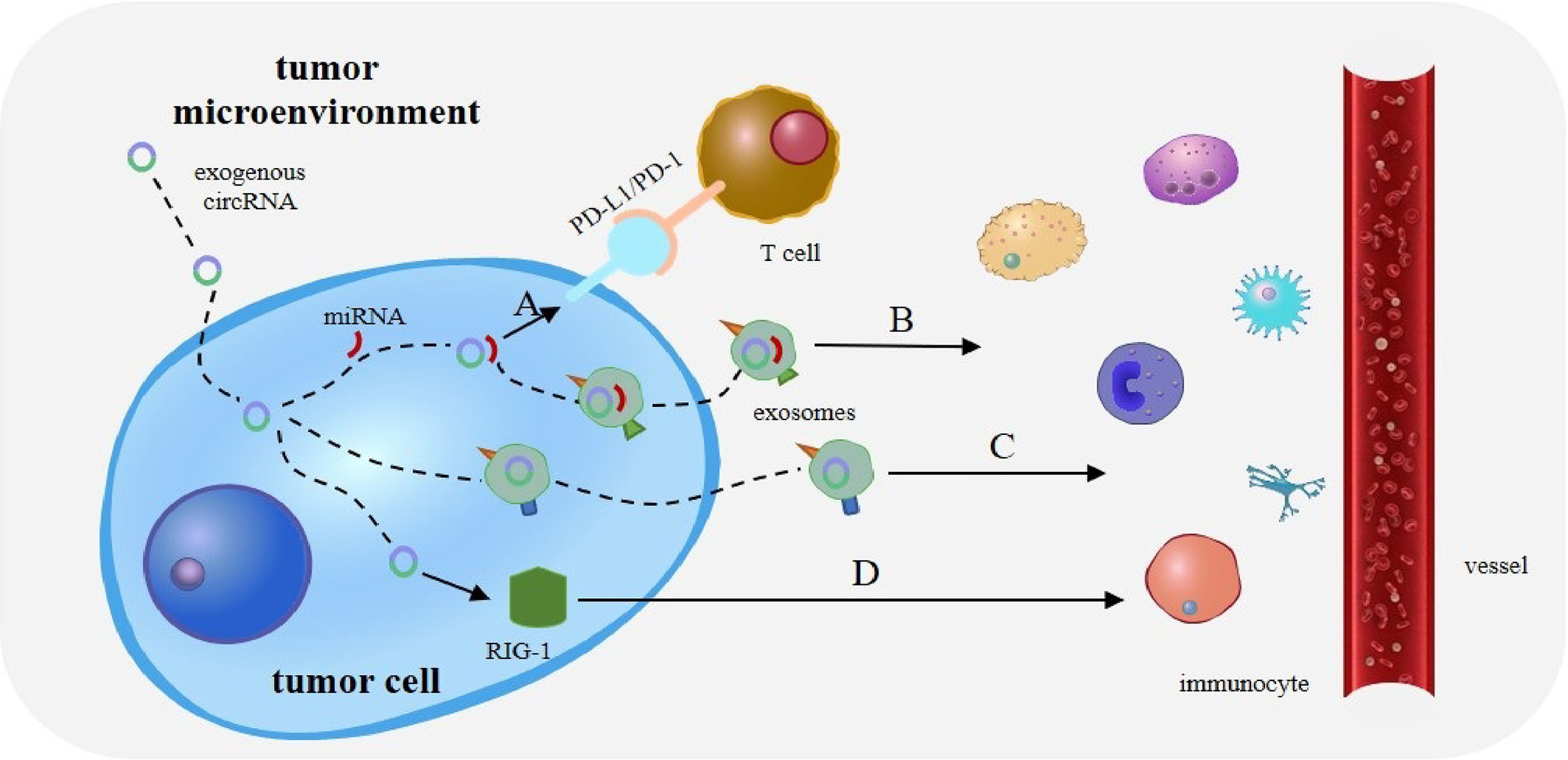
Figure 3 Role of circRNAs in tumor immunity. (A) The interaction between programmed death receptor 1 (PD-1) and programmed cell death ligand 1 (PD-L1) can effectively suppress the activation of effector T lymphocytes, ultimately leading to tumor immune escape. (B) CircRNA can also bind tumor-specific miRNAs and mRNAs through complementary sequences, to form a novel tumor antigen. CircRNA exists in exosomes to enhance the stability of these RNAs and is released upon reaching the target immune cells. (C) Exosomes and extracellular vesicles (EVs) can transport circRNA into immune cells. CircRNA can then act as potential tumor antigens to regulate immune responses in tumors. (D) Exogenous circRNA entering tumor cells may activate antitumor immunity by influencing RIG-I.
In breast cancer, circ_0000977 binds to miR-153 to counteract the inhibition of hypoxia-inducible factor 1-α (HIF1A) and a disintegrin and metalloproteinase domain-containing 10 (ADAM10) (39). However, hypoxia levels decrease the level of NKG2D on NK cells by inducing the upregulation of HIF1A, ADAM10, and soluble MICA, consequently tumor cells evade immune surveillance and cell lysis (40, 41). Thus, the circ_0000977/miR-153 axis may regulate the immune escape of breast cancer cells by increasing the levels of HIF1A and ADAM10 (39). In addition, circ-Amotl1 can indirectly inhibit the presence of miR-17-5p by upregulating DNMT3A. The process may lead to DNA methylation enrichment in the promoter region of miR-17 and result in subcellular translocation and STAT3 expression, thereby initiating immunosuppression in tumors (42, 43). These studies suggest that circRNAs can regulate the function of miRNAs related to cancer immunity. Further exploration of the relationship between circRNAs and miRNAs is needed to provide new ideas for tumor immunotherapy.
Interestingly, the binding of circRNA to miRNA has other effects besides miRNA silencing. CircRNA can also bind tumor-specific miRNAs and mRNAs through complementary sequences to form a novel tumor antigen. CircRNA also exists in exosomes to enhance the stability of these RNAs and is released upon reaching the target immune cells (35) (Figure 3B). This suggests that the role of circRNA as a miRNA sponge in tumor immunity needs to be explored further.
CircRNA can also act as a protein decoy or antagonist. CircRNA may serve as a decoy and bind to an intracellular protein to inhibit its function. Many endogenous circRNAs tend to form 16–26 bp double-stranded bodies (dsRNA) and inhibit the activity of the endogenous protein kinase (PKR) (44). In the model proposed by Zheng, this repression was achieved by the interaction between circRNA and PKR (45). However, after viral infection, endogenous circRNA can be degraded by RNase L to regulate dsRNA production, activate PKR, and promote innate immunity (44). In addition, circFoxo3 binds to p53-associated murine double minute2 to induce p53 degradation. P53 plays a crucial role in tumor development by increasing inducible nitric oxide synthase expression and regulating the immune response (46, 47). It is suggested that circRNA could regulate the immune response in tumors by regulating the stability of p53.
In non-small cell lung cancer (NSCLC), circNDUFB2 also inhibits NSCLC progression by disrupting insulin-like growth factor 2 mRNA-binding proteins and activating anti-tumor immunity. The inhibition can also be enhanced by N6-methyladenosine (m6A) modification of circNDUFB2. Therefore, circNDUFB2 may be involved in NSCLC progression as a potential tumor suppressor (48). However, not all circRNAs that bind to a protein can inhibit protein function. For instance, ectopic circ-Amotl1 binds to the nuclear oncogene c-myc and stabilizes it, thereby upregulating c-myc targets and promoting tumorigenesis (49).
Previously, we mentioned that circrna activates retinoic acid-induced gene I (RIG-I) and protects against viral infections through the immune pathway. In particular, circRNAs activate RIG-I via self-splicing introns, rather than the same circRNAs produced by endogenous introns and spliced by a cellular splicing mechanism. Thus, the activation of this immune response depends on the splicing mechanism of circRNAs (18). Furthermore, in preclinical models of hematological malignancies, the linear form of human plasmacytoma variant translocation 1 (PVT1) is overexpressed in myeloid-derived suppressor cells. The overexpression could induce immune tolerance. However, circPVT1 exhibits immunosuppressive properties in myeloid and lymphocyte subpopulations (50). Linear pvt1 and circpvt1 play two different roles in the immune regulation of hematological malignancies. The former involves cancer progression by promoting a more aggressive phenotype of malignant cells, and the latter presents a new potential therapy resistance mechanism to negatively regulate the immune system (50). This study provides new insights into the treatment of hematological malignancies and demonstrates the unique immunomodulatory role of circRNA.
Owing to their stability and specificity, circRNAs can act as tumor antigens and induce immune responses. Exosomes and extracellular vesicles (EVs) can transport circRNA to immune cells (51), thus acting as potential tumor antigens to regulate immune responses in tumors (Figure 3C). For example, CRC cells with KRAS mutations can transfer several circRNAs into exosomes (52). Many abnormal circRNAs are produced during tumorigenesis owing to gene mutations and chromosomal changes. In acute promyelocytic leukaemia, the chromosomal translocation of PML/RARα produces fused circRNAs (53). Newly formed abnormal circRNAs may then be transported to immune cells through exosomes and EVs secreted by tumor cells (35).
In addition, exogenous circRNAs can trigger immune responses in HeLa cells and stimulate the expression of innate immune genes, thus preventing viral infection (54), which may be achieved through their immunogenicity. Many circRNAs that regulate immune responses by inducing the expression of immune genes have been identified, including RIG-I (55, 56) and MDA5 (44, 54). RIG-I is a critical factor in the host antiviral defense system (57). It can isolate viral RNA from antiviral effector molecules, thus triggering an antiviral immune response (18). Subsequently, it can help the body better detect exogenous circRNA and form a positive feedback pathway, which can protect the body against viral infection (54). In addition, RIG-I agonists can activate anti-cancer immune responses against tumors (58, 59).
Exogenous circRNAs entering tumor cells may activate antitumor immunity by influencing RIG-I (Figure 3D). For example, circNDUFB2 can be recognized by RIG-I to activate the RIG-I MAVS signal cascade and recruit immune cells into the tumor microenvironment (TME) (48). Interestingly, exogenous circRNA without modification can avoid cellular RNA sensors. So it can avoid triggering an immune response in RIG-I and toll-like receptor cells and mice, resulting in reduced immunogenicity (60). In lung cancer, the overexpression of cirNDUFB2 leads to the overregulation of various immune genes and increased levels of CXCL10, CXCL11, CCL5, and IFNβ, thus enhancing the body’s immune defense (48).
Recently, the relationship between m6A modification and circRNA immunity has attracted much attention. Studies have shown that human circRNAs are labelled at birth by covalent m6A modification based on introns that program reverse splicing. The m6A modifier is marked as “SELF”, whereas the unmodified is marked as “FOREIGN”. RIG-I differentiates the unmodified and the m6A-modified circRNAs and is activated only by the former (61). CircSELF can evade innate immune surveillance through YTHDF2 (m6A reader protein)-mediated inhibition (62), which may inhibit RIG-I conformational conversion (activation) required for downstream signaling of immune genes in living cells (61).
Consistent with this, m6A-modified RNAs can be raised by the YTHDF protein and induced into disordered condensates of phase separation through its N-terminal domain (63). Furthermore, the human papillomavirus, which is closely related to the growth of CaSki cervical cancer cells in vitro and in vivo, can produce the oncogenic circRNA CircE7. In addition, m6A modification was found to be an essential motif of CircE7 protein-coding ability (64).
All in all, these studies show that the modification of m6A helps exogenous circRNAs evade immune surveillance. CircFOREIGN is also involved in immune signal transduction by forming a three-component signal transduction complex with RIG-I and K63-polyubiquitin chain (K63-UBN) (61).
Overall, circRNAs are involved in some critical processes of host immunity by maintaining internal homeostasis and providing adequate protection against microbial infection and malignancy. Thus, circRNA can be considered as a biomarker or therapeutic target. However, the underlying mechanisms remain largely unknown and should be further explored and clarified.
Although progress has been made in the treatment of digestive tract tumors, the tumor immune microenvironment limits the effect of therapy to a certain extent. As mentioned above, the incidence and mortality of digestive cancers remain high: CRC ranks 5th and 2nd, GC ranks 6th and 3rd, and liver cancer ranks 7th and 5th in new morbidities and cancer mortalities, respectively (65). Tumor immunity-related studies have focused on these three types of tumors. Therefore, we mainly discuss them in this review.
Immune tissue is widely distributed in the digestive system. The liver and digestive tract lymph nodes perform different immune functions. In addition, there is a mucosal immune system in the digestive tract, mainly gut-associated lymphatic tissue (GALT). The mucosa of the gastrointestinal tract can perform its innate immune function through recruitment of neutrophils and killing effect of IL-23/Th17 cell axis. In addition, the intestinal epithelium can produce protective sIgA due to the activation of adaptive immunity. Immune tolerance also exists: dendritic cells (DCs) recognize gut microbes, regulatory T cells are activated and begin to secrete interleukin-10 (IL-10) (66).
For the liver, it always maintains a low immune response state to avoid some unnecessary immune responses. But when it comes to tumor cells or other situations, the liver exerts its immune effect. The balance of immune tolerance and immune protection is important to maintain normal liver immune function. However, if immune tolerance is dominant, it may cause tumor metastasis and progression. In the liver microenvironment, Kupffer cells (KCs) show poor activation of adaptive immune response which leads to immune tolerance rather than immune protection (67, 68). And when NK cell function is impaired, it can result in limited tumor cell clearance. In addition, the innate immune cells, such as dendritic cells, innate lymphocytes cells, are all involved in the immune response of the liver (69).
During tumor development, tumor cells secrete cytokines, inflammatory mediators, and chemokines, leading to the reprogramming of the surrounding cells. The TME comprises tumor cells, stromal cells, immune cells, endothelial cells, and non-cellular components, playing an essential role in immune regulation (70). On one hand, tumor cells exhibit antigenic components that are different from normal cell tissues. Presenting tumor antigens to T cells can generate cellular immunity, humoral immunity, and cellular cytotoxic reactions of innate immune cells. These immune processes act as a defence in the early stages (71, 72). On the other hand, tumor cells can evade immune cells through complex signaling networks to achieve immune escape (70, 73). CircRNAs participate in many pathological processes, including cancer growth, metastasis, recurrence, and treatment resistance (74–80). These processes are realized primarily by sponging miRNA and interacting with proteins.
In the tumor immune microenvironment, circRNAs achieve immune escape through the following aspects:
i. Regulation of NK cell activity.
ii. Regulation of macrophage activity. In response to different stimuli, macrophages differentiate into M1 or M2 macrophages to perform their functions. CircRNAs may affect macrophage secretion.
iii. Regulation of lymphocyte activity. The proportion of tumor-infiltrating lymphocytes (TILs) in the TME is high and is closely related to prognosis. The regulation of circRNA on T lymphocyte PD-1 can improve tumor immune tolerance.
iv. Interaction with other immune-related factors (51, 73, 81).
We will introduce the role of circRNAs in the immunity of GC, CRC, and liver cancer in detail below (Table 2). The functions above are equally applicable to digestive cancers.
Although circRNAs have been reported to show many potential functions in tumorigenesis and tumor immune reactions, these functions are far from fully understood. As the relationship between miRNAs and tumor immunity is more evident than that between circRNAs presently, the role of circRNA in anti-tumor immunity is mainly understood through the circRNA/miRNA/mRNA axis. Peng et al. reported that circTMC5 targets RABL6 in GC by acting as a sponge for miR-361-3p to regulate the biological phenotype of GC cells (88). In addition, RABL6 participates in the immune regulation of GC through the production of IL-6 and IL-8, activation and transport of leukocytes, promotion of neuroinflammatory responses, and activation of macrophages during inflammation (89). The TISIDB database revealed the correlation between RABL6 and chemokines, including chemokine CCL14, CCL22, and CXCL3, in human GC samples using Spearman correlation (90). Among them, IL-6 and IL-8 can promote the genesis and development of tumors (96, 97), and CXCL3 may promote the malignant biological behavior of tumors (98, 99). In addition, a negative correlation between RABL6 expression and the infiltration of immune cells, including macrophages, B cells, CD8+T cells, neutrophils, and dendritic cells, has been observed (90). Thus, combining checkpoint blockade therapy with therapies that target this pathway may be a potential treatment strategy (100). Overall, the circTMC5/miR-361-3p/RABL6 axis plays a crucial role in the occurrence and development of GC and affects the tumor immune microenvironment.
The Epstein–Barr virus (EBV) is the earliest human tumor-associated virus (101). The EBV-associated gastric carcinoma is a group of GC in which EBV infects gastric epithelial cells during monoclonal growth (102, 103). One of its characteristics is the immune microenvironment in which viruses or virus-infected cells can grow normally. EBV BARTs are produced in large numbers in epithelial cells with EBV latent infection, and circRNAs have been identified in the BART loci; however, the function of circBARTs remains unclear. In addition, circRNA can be detected in the serum or plasma of patients because of its stability. EBV-specific circRNAs have great potential for future applications in this field.
As previously described, circRNA can be transported into immune cells by exosomes and EVs and act as potential tumor antigens to modulate immune reactions in tumors (51). For example, circRNA is downregulated and transferred by exosomes to KRAS-mutated colon cancer cells (52), demonstrating that it may act as a tumor antigen.
CircRNA can promote the immune escape of CRC cells through PD-L1. In CRC cells, circRNA CIRC-0020397 binds to miR-138 and inhibits the activity of miR-138, thereby facilitating downstream reactions, such as the production of telomerase reverse transcriptase and PD-L1. Since the expression of CIRC-0020397 is elevated in CRC cells, PD-L1 is upregulated. CIRC-0020397 interacts with PD-1 to induce T cell apoptosis, which causes immune escape. Studies have shown that PD-1/PD-L1 blockers provide better clinical benefits in patients with high PD-L1 levels (34). Similar studies have also reported that cirCCDR1-AS can significantly increase PD-L1 expression on the surface of colon cancer cells through CMTM4 and CMTM6, leading to a poor prognosis (92). Hsa_circ_0136666 promotes PD-L1 expression in CRC by inhibiting miR-497, which stimulates the production of Treg cells and causes immune escape (93). Circ-KRT6C can improve the presentation of PD-L1 by acting as a miR-485-3p sponge, thus promoting the malignant progression and immune escape of CRC cells (94). Therefore, targeting the relevant circRNAs to regulate PD-1/PD-L1 expression may provide a new direction for immune checkpoint therapy.
In addition, recent studies have proposed that the exosome CIRC-ABCC1 can bind to β-catenin to enter the nucleus and activate the Wnt pathway, which promotes the progression of CRC. Thus, circRNAs can promote malignant biological behavior and assist in the diagnosis and treatment of CRC.
The NKG2D ligand ULBP1 is upregulated by circARSP91 at mRNA and protein levels, thereby enhancing the responsiveness of hepatocellular carcinoma (HCC) cells to NK cells (82). However, the miRNA sponge does not achieve the above effect, which has been speculated to be related to tumor suppressors (104).
Compared with normal tissues, the level of circUHRF1 (hsa_circ_0048677) in HCC tissues increases. CircUHRF1 can enter NK cells via exosomes to achieve immune escape. Under the action of the circUHRF1/miR-449C-5P/T cell immunoglobulin domain and mucin domain protein-3 axis, NK cell infiltration in TME, and IFN-γ and TNF-α secretion decrease, leading to tumor immune escape. In addition, the above reactions regarding the circUHRF1 can increase HCC resistance to PD-1 therapy and the difficulty of treatment (83). The expression of circTRIM33-12 increases in HCC, leading to the inhibition of miR-191, in which the expression of TET1 is upregulated. Experiments showed a positive correlation between the expression of circTRIM33-12 and the number of NKG2D-positive cells. The decrease in NKG2D level is associated with the immune responses of NK cells, γδT cells, and CD8+ T cells to cancers (84, 105). In addition, hsa_circ_0007456 regulates NK cell activity via the hsa_circ_0007456/miR-6852-3p/ICAM-1 axis, which leads to immune escape (17).
Macrophages with different phenotypes can affect the progression of liver cancer. For example, hsa_circ_0074854 is upregulated in HCC cells, and its knockdown can inhibit the polarization of tumor-associated macrophages (TAM) to M2 (85). As M2 macrophages can contribute to the migration and invasion of tumor cells by secreting IL-10 (106, 107), the knockdown of hsa_circ_0074854 suppresses tumor growth. CircRNA regulation of macrophage infiltration and secretion is also involved in immune escape. The decreased expression of hsa-circ-0110102 suppresses that of PPARα via the action of miR-580-5p, thereby affecting the secretion of CCL2. CCL2 can inhibit the release of pro-inflammatory cytokines by regulating the COX-2/PGE2 axis to achieve immune escape (86). CircASAP1 regulates the infiltration of TAM by regulating the miR-326 or the miR-532-5p/CSF-1 pathway, thus affecting the anti-tumor ability of the TME (87).
The overexpression of circMET in HCC induces a tumor immunosuppressive microenvironment through the miR-30-5p/Snail/dipeptidyl peptidase 4/CXCL10 axis. The reduction in CXCL10 level leads to the decreased capacity for CD8 + lymphocyte transport, immune tolerance, and less sensitivity to PD-1 therapy (91). Studies have confirmed that hsa_circ_0064428 level is reduced in HCC patients with high TIL infiltration, and there is a negative relationship between hsa_circ_0064428 level and tumor size, metastasis rate, and survival rate. It is speculated that circRNA can regulate the tumor immune microenvironment by regulating TILs, and hsa-circ-0064428 can be used as a prognostic marker (95).
Blocking the tumor escape pathway associated with circRNA provides new ideas for future tumor therapy. In addition, circRNAs can be used as molecular markers for the diagnosis and prognosis of digestive cancers (108–111). The characteristics of circRNA make it more advantageous over other non-coding RNA. For example, the universality of circRNA make the therapeutic audiences wider and increase the sensitivity of tumor markers; diversity and richness provide the circRNA pathway with more alternative targets.
There are various treatment methods for gastrointestinal tumors, including surgery, endoscopic resection, radiotherapy and chemotherapy, immunotherapy, targeted therapy, and multiple combination therapies. Recently, the success of cancer immunotherapies, such as monoclonal antibodies, immune checkpoint inhibitors PD-1 and CTLA-4, chimeric antigen receptor T-cell, and other therapies, has attracted increasing attention (112). These therapies enhance the anti-tumor ability of the TME by activating or enhancing the body’s immune system, thus killing tumor cells.
Because of the obvious anti-tumor effect, blocking PD-1/PD-L1 pathway has a bright prospect in clinical tumor treatment. The interaction between PD-1 and PD-L1 transmits inhibitory signals to T cells, ultimately leading to a series of tumor immune escape reactions (113, 114). Recent studies have shown that this signaling pathway can block the G1 phase of the T cell cycle and induce iTregs to inhibit T cell activity (115, 116). Various types of circRNA can regulate the PD-1/PD-L1 pathway of digestive tumors through different signaling pathways. Therefore, future studies should investigate the regulation of the PD-1/PD-L1 pathway using circRNAs, which can contribute new ideas to immune checkpoint therapy for digestive tumors.
Cancer vaccines are similar to vaccines for common infectious diseases, including antigens and adjuvants. Adjuvants activate antigen-presenting cells and regulate surface molecules to initiate an adaptive immune response (117). Compared with vaccines for common infectious diseases, cancer vaccines are administered primarily for treatment rather than prevention. The application of circRNAs in tumor vaccines has recently entered the preliminary stage of exploration. Animal experiments showed that circFOREIGN decreased the tumor growth rate and improved the overall survival rate in OVA-B16 melanoma mouse model (61), demonstrating the potential of circRNA as an adjuvant for tumor vaccines. CircRNA can activate dendritic cells to promote antigenic cross-reactivity and activate CD4+ and CD8+T cells. CircRNA may also directly affect T cells and other immune cell types; however, this remains to be explored. Interestingly, the m6A modification of circRNA is recognized by the immune system as body tissue, thus weakening the immunogenicity of circRNA as an adjuvant (61).
To date, mRNA has been shown to be useful in developing cancer vaccines. Compared with traditional mRNA vaccines, the circRNA vaccine has a more stable circRNA ring structure (118); therefore, only a small amount of circRNA is required to initiate adaptive immunity. The CircRNA vaccine is highly expected due to its reduced dosage and improved efficiency. However, the immunogenicity of unmodified circRNA is lower than that of unmodified mRNA in vitro and depends on the purity of circRNA (60). Therefore, despite the promising prospects of the circRNA vaccine, its application as a tumor vaccine against gastrointestinal tumors remains challenging.
At present, there is no practical way to treat digestive tumors, except surgery, chemotherapy, and radiotherapy. Although significant progress has been made recently in immunotherapy, tumor cells still can lead to immune escape in various ways. That’s because the immunogenicity of tumor antigens is weak. Besides, some tumors have innate or acquired drug resistance, which hinders the progress of immunotherapy. Therefore, it is of great significance to explore the immune mechanisms of digestive tumors to improve the effect of immunotherapy.
Several studies have demonstrated the close relationship between circRNAs and tumor biological processes, such as proliferation, metastasis, and drug resistance. CircRNA can influence the immune microenvironment of gastrointestinal tumors by acting as a miRNA molecular sponge, interacting with proteins, and regulating selective splicing. In addition, by regulating the activities of NK cells, macrophages, lymphocytes, and other immune cells, circRNAs can lead to tumor immune escape. The circRNA vaccine, though just entering the stage of preliminary exploration at present, provides a new idea for tumor immunotherapy.
Since nowadays knowledge of circRNA remains insufficient, future studies should consider focusing on the following issues: (i) so far, most studies on circRNAs and tumors have been carried out on tumor cells and tissues. Studies on other cancer models, such as organoids and patient-derived xenografts, should be conducted; (ii) there are relatively a few circRNAs available, and the available loci are limited; (iii) the downstream reaction of circRNA is not well understood, so it is necessary to explore whether there are other effects; and (iv) as immunotherapy is expected to become more individualized, how to achieve this goal using circRNA will be a complex issue. Future studies should clarify the location, transport, and degradation mechanisms of circRNA in living cells, as well as the relationship between circRNA and tumor immunity. Safety studies should also be carried out to explore new paths for gastrointestinal tumor therapy.
CircRNAs have become a research hotspot worldwide. In this review, we summarized the relationship between circRNA and digestive tumors. Also, we found out the potential of circRNA as a vaccine against digestive tumors. With the progress of sequencing technology and bioinformatics analysis, knowledge of circRNA and gastrointestinal tumors is expected to be more in-depth. We believe that circRNA will provide a brand-new direction on immunotherapy, which will inevitably become an essential link in tumor therapy in the future.
JW, LP, and XY conceived and revised this manuscript. CC, CX, HT, and YJ wrote the manuscript. SW, XZ, and TH helped to draft the manuscript. All authors reviewed and approved the final manuscript.
This work was partially supported by the National Natural Science Foundation of China (Project No. 81602167 to JW, Project No. 81803636 to XY, Project No. 81972658 and 81802812 to LP), Hunan Provincial Natural Science Foundation of China (Project No. 2017JJ3494 and 2021JJ31100 to JW), the Science and Technology Program Foundation of Changsha City (Project No. kq2004085 to JW), and Guangdong Basic and Applied Basic Research Foundation (Project No. 2018A0303130329 to XY).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
ADAM10, a disintegrin and metalloproteinase domain-containing 10; ceRNA, competitive endogenous RNA; circRNAs, circular RNAs; ciRNAs, circular intronic RNAs; C-rich, cytosine-rich; CRC, colorectal cancer; dsRNA, double-stranded bodies; ecircRNAs, exonic circular RNAs; EIcircRNAs, exonic-intronic circular RNAs; EVs, extracellular vesicles; GC, gastric cancer; GU-rich, guanine and uracil rich; HIF1A, hypoxia-inducible factor 1-α; ICAM-1, intercellular cell adhesion molecule-1; MREs, miRNA response elements; NF-κB, nuclear factor kappa B; NSCLC, non-small cell lung cancer; PD-1, programmed death receptor 1; PD-L1, programmed cell death ligand 1; PKR, protein kinase; RBP, RNA-binding protein; RIG-I, retinoic acid-induced gene I; RISC, RNA-induced silencing complexes; TILs, tumor-infiltrating lymphocytes; TME, tumor microenvironment; U-rich, uracil-rich.
1. Wu C-Y, Lin J-T. The Changing Epidemiology of Asian Digestive Cancers: From Etiologies and Incidences to Preventive Strategies. Best Pract Res Clin Gastroenterol (2015) 29:843–53. doi: 10.1016/j.bpg.2015.09.016
2. IJsselsteijn ME, Sanz-Pamplona R, Hermitte F, de Miranda NFCC. Colorectal Cancer: A Paradigmatic Model for Cancer Immunology and Immunotherapy. Mol Aspects Med (2019) 69:123–9. doi: 10.1016/j.mam.2019.05.003
3. Li W, Deng Y, Chu Q, Zhang P. Gut Microbiome and Cancer Immunotherapy. Cancer Lett (2019) 447:41–7. doi: 10.1016/j.canlet.2019.01.015
4. Fu Y, Liu S, Zeng S, Shen H. From Bench to Bed: The Tumor Immune Microenvironment and Current Immunotherapeutic Strategies for Hepatocellular Carcinoma. J Exp Clin Cancer Res (2019) 38:396. doi: 10.1186/s13046-019-1396-4
5. Cui X, Wang J, Guo Z, Li M, Li M, Liu S, et al. Emerging Function and Potential Diagnostic Value of Circular RNAs in Cancer. Mol Cancer (2018) 17:123. doi: 10.1186/s12943-018-0877-y
6. Zhang L, Wang Y, Zhang Y, Zhao Y, Li P. Pathogenic Mechanisms and the Potential Clinical Value of Circfoxo3 in Cancers. Mol Ther Nucleic Acids (2021) 23:908–17. doi: 10.1016/j.omtn.2021.01.010
7. Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, et al. Natural RNA Circles Function as Efficient microRNA Sponges. Nature (2013) 495:384–8. doi: 10.1038/nature11993
8. Duan S, Wang S, Huang T, Wang J, Yuan X. circRNAs: Insight Into Their Role in Tumor-Associated Macrophages. Front Oncol (2021) 11:780744. doi: 10.3389/fonc.2021.780744
9. Jeck WR, Sorrentino JA, Wang K, Slevin MK, Burd CE, Liu J, et al. Circular RNAs are Abundant, Conserved, and Associated With ALU Repeats. RNA (2013) 19:141–57. doi: 10.1261/rna.035667.112
10. Li Z, Huang C, Bao C, Chen L, Lin M, Wang X, et al. Exon-Intron Circular RNAs Regulate Transcription in the Nucleus. Nat Struct Mol Biol (2015) 22:256–64. doi: 10.1038/nsmb.2959
11. Zhang Y, Zhang X-O, Chen T, Xiang J-F, Yin Q-F, Xing Y-H, et al. Circular Intronic Long Noncoding RNAs. Mol Cell (2013) 51:792–806. doi: 10.1016/j.molcel.2013.08.017
12. Kelly S, Greenman C, Cook PR, Papantonis A. Exon Skipping Is Correlated With Exon Circularization. J Mol Biol (2015) 427:2414–7. doi: 10.1016/j.jmb.2015.02.018
13. Conn SJ, Pillman KA, Toubia J, Conn VM, Salmanidis M, Phillips CA, et al. The RNA Binding Protein Quaking Regulates Formation of circRNAs. Cell (2015) 160:1125–34. doi: 10.1016/j.cell.2015.02.014
14. Zhang X-O, Wang H-B, Zhang Y, Lu X, Chen L-L, Yang L. Complementary Sequence-Mediated Exon Circularization. Cell (2014) 159:134–47. doi: 10.1016/j.cell.2014.09.001
15. Cocquerelle C, Mascrez B, Hétuin D, Bailleul B. Mis-Splicing Yields Circular RNA Molecules. FASEB J (1993) 7:155–60. doi: 10.1096/fasebj.7.1.7678559
16. Zhao R, Ni J, Lu S, Jiang S, You L, Liu H, et al. CircUBAP2-Mediated Competing Endogenous RNA Network Modulates Tumorigenesis in Pancreatic Adenocarcinoma. Aging (Albany NY) (2019) 11:8484–501. doi: 10.18632/aging.102334
17. Shi M, Li Z-Y, Zhang L-M, Wu X-Y, Xiang S-H, Wang Y-G, et al. Hsa_circ_0007456 Regulates the Natural Killer Cell-Mediated Cytotoxicity Toward Hepatocellular Carcinoma via the miR-6852-3p/ICAM-1 Axis. Cell Death Dis (2021) 12:94. doi: 10.1038/s41419-020-03334-8
18. Wang M, Yu F, Wu W, Zhang Y, Chang W, Ponnusamy M, et al. Circular RNAs: A Novel Type of non-Coding RNA and Their Potential Implications in Antiviral Immunity. Int J Biol Sci (2017) 13:1497–506. doi: 10.7150/ijbs.22531
19. Zheng Q, Bao C, Guo W, Li S, Chen J, Chen B, et al. Circular RNA Profiling Reveals an Abundant Circhipk3 That Regulates Cell Growth by Sponging Multiple miRNAs. Nat Commun (2016) 7:11215. doi: 10.1038/ncomms11215
20. Lowe JM, Menendez D, Bushel PR, Shatz M, Kirk EL, Troester MA, et al. P53 and NF-κb Coregulate Proinflammatory Gene Responses in Human Macrophages. Cancer Res (2014) 74:2182–92. doi: 10.1158/0008-5472.CAN-13-1070
21. Baratin M, Foray C, Demaria O, Habbeddine M, Pollet E, Maurizio J, et al. Homeostatic NF-κb Signaling in Steady-State Migratory Dendritic Cells Regulates Immune Homeostasis and Tolerance. Immunity (2015) 42:627–39. doi: 10.1016/j.immuni.2015.03.003
22. Köhler UA, Böhm F, Rolfs F, Egger M, Hornemann T, Pasparakis M, et al. NF-κb/RelA and Nrf2 Cooperate to Maintain Hepatocyte Integrity and to Prevent Development of Hepatocellular Adenoma. J Hepatol (2016) 64:94–102. doi: 10.1016/j.jhep.2015.08.033
23. Huang H, Wei L, Qin T, Yang N, Li Z, Xu Z. Circular RNA ciRS-7 Triggers the Migration and Invasion of Esophageal Squamous Cell Carcinoma via miR-7/KLF4 and NF-κb Signals. Cancer Biol Ther (2019) 20:73–80. doi: 10.1080/15384047.2018.1507254
24. Wang H, Xiao Y, Wu L, Ma D. Comprehensive Circular RNA Profiling Reveals the Regulatory Role of the circRNA-000911/miR-449a Pathway in Breast Carcinogenesis. Int J Oncol (2018) 52:743–54. doi: 10.3892/ijo.2018.4265
25. Bezzi M, Guarnerio J, Pandolfi PP. A Circular Twist on microRNA Regulation. Cell Res (2017) 27:1401–2. doi: 10.1038/cr.2017.136
26. Li Y, Zheng Q, Bao C, Li S, Guo W, Zhao J, et al. Circular RNA is Enriched and Stable in Exosomes: A Promising Biomarker for Cancer Diagnosis. Cell Res (2015) 25:981–4. doi: 10.1038/cr.2015.82
27. Du WW, Fang L, Yang W, Wu N, Awan FM, Yang Z, et al. Induction of Tumor Apoptosis Through a Circular RNA Enhancing Foxo3 Activity. Cell Death Differ (2017) 24:357–70. doi: 10.1038/cdd.2016.133
28. Zou Y, Zheng S, Deng X, Yang A, Xie X, Tang H, et al. The Role of Circular RNA CDR1as/ciRS-7 in Regulating Tumor Microenvironment: A Pan-Cancer Analysis. Biomolecules (2019) 9:E429. doi: 10.3390/biom9090429
29. Lu TX, Rothenberg ME. MicroRNA. J Allergy Clin Immunol (2018) 141:1202–7. doi: 10.1016/j.jaci.2017.08.034
30. Michlewski G, Cáceres JF. Post-Transcriptional Control of miRNA Biogenesis. RNA (2019) 25:1–16. doi: 10.1261/rna.068692.118
31. Tay Y, Rinn J, Pandolfi PP. The Multilayered Complexity of ceRNA Crosstalk and Competition. Nature (2014) 505:344–52. doi: 10.1038/nature12986
32. Almeida MI, Reis RM, Calin GA. Decoy Activity Through microRNAs: The Therapeutic Implications. Expert Opin Biol Ther (2012) 12:1153–9. doi: 10.1517/14712598.2012.693470
33. Wang J, Zhao X, Wang Y, Ren F, Sun D, Yan Y, et al. circRNA-002178 Act as a ceRNA to Promote PDL1/PD1 Expression in Lung Adenocarcinoma. Cell Death Dis (2020) 11:32. doi: 10.1038/s41419-020-2230-9
34. Zhang X-L, Xu L-L, Wang F. Hsa_circ_0020397 Regulates Colorectal Cancer Cell Viability, Apoptosis and Invasion by Promoting the Expression of the miR-138 Targets TERT and PD-L1. Cell Biol Int (2017) 41:1056–64. doi: 10.1002/cbin.10826
35. Xu Z, Li P, Fan L, Wu M. The Potential Role of circRNA in Tumor Immunity Regulation and Immunotherapy. Front Immunol (2018) 9:9. doi: 10.3389/fimmu.2018.00009
36. Gao C, Xu Y-J, Qi L, Bao Y-F, Zhang L, Zheng L. CircRNA VIM Silence Synergizes With Sevoflurane to Inhibit Immune Escape and Multiple Oncogenic Activities of Esophageal Cancer by Simultaneously Regulating miR-124/PD-L1 Axis. Cell Biol Toxicol (2021) 2021(1):1–21. doi: 10.1007/s10565-021-09613-0
37. Li D, Wang X, Yang M, Kan Q, Duan Z. Mir3609 Sensitizes Breast Cancer Cells to Adriamycin by Blocking the Programmed Death-Ligand 1 Immune Checkpoint. Exp Cell Res (2019) 380:20–8. doi: 10.1016/j.yexcr.2019.03.025
38. Xie W-B, Liang L-H, Wu K-G, Wang L-X, He X, Song C, et al. MiR-140 Expression Regulates Cell Proliferation and Targets PD-L1 in NSCLC. Cell Physiol Biochem (2018) 46:654–63. doi: 10.1159/000488634
39. Ou Z-L, Luo Z, Wei W, Liang S, Gao T-L, Lu Y-B. Hypoxia-Induced Shedding of MICA and HIF1A-Mediated Immune Escape of Pancreatic Cancer Cells From NK Cells: Role of Circ_0000977/miR-153 Axis. RNA Biol (2019) 16:1592–603. doi: 10.1080/15476286.2019.1649585
40. Barsoum IB, Hamilton TK, Li X, Cotechini T, Miles EA, Siemens DR, et al. Hypoxia Induces Escape From Innate Immunity in Cancer Cells via Increased Expression of ADAM10: Role of Nitric Oxide. Cancer Res (2011) 71:7433–41. doi: 10.1158/0008-5472.CAN-11-2104
41. Lu Y, Hu J, Sun W, Duan X, Chen X. Hypoxia-Mediated Immune Evasion of Pancreatic Carcinoma Cells. Mol Med Rep (2015) 11:3666–72. doi: 10.3892/mmr.2015.3144
42. Du WW, Zhang C, Yang W, Yong T, Awan FM, Yang BB. Identifying and Characterizing circRNA-Protein Interaction. Theranostics (2017) 7:4183–91. doi: 10.7150/thno.21299
43. Yang Z-G, Awan FM, Du WW, Zeng Y, Lyu J, Wu D, et al. The Circular RNA Interacts With STAT3, Increasing Its Nuclear Translocation and Wound Repair by Modulating Dnmt3a and miR-17 Function. Mol Ther (2017) 25:2062–74. doi: 10.1016/j.ymthe.2017.05.022
44. Liu C-X, Li X, Nan F, Jiang S, Gao X, Guo S-K, et al. Structure and Degradation of Circular RNAs Regulate PKR Activation in Innate Immunity. Cell (2019) 177:865–80.e21. doi: 10.1016/j.cell.2019.03.046
45. Zheng Z-M. Circular RNAs and RNase L in PKR Activation and Virus Infection. Cell Biosci (2019) 9:43. doi: 10.1186/s13578-019-0307-x
46. Du WW, Yang W, Liu E, Yang Z, Dhaliwal P, Yang BB. Foxo3 Circular RNA Retards Cell Cycle Progression via Forming Ternary Complexes With P21 and CDK2. Nucleic Acids Res (2016) 44:2846–58. doi: 10.1093/nar/gkw027
47. Huang Y, Yu P, Li W, Ren G, Roberts AI, Cao W, et al. P53 Regulates Mesenchymal Stem Cell-Mediated Tumor Suppression in a Tumor Microenvironment Through Immune Modulation. Oncogene (2014) 33:3830–8. doi: 10.1038/onc.2013.355
48. Li B, Zhu L, Lu C, Wang C, Wang H, Jin H, et al. Circndufb2 Inhibits non-Small Cell Lung Cancer Progression via Destabilizing IGF2BPs and Activating Anti-Tumor Immunity. Nat Commun (2021) 12:295. doi: 10.1038/s41467-020-20527-z
49. Yang Q, Du WW, Wu N, Yang W, Awan FM, Fang L, et al. A Circular RNA Promotes Tumorigenesis by Inducing C-Myc Nuclear Translocation. Cell Death Differ (2017) 24:1609–20. doi: 10.1038/cdd.2017.86
50. Ghetti M, Vannini I, Storlazzi CT, Martinelli G, Simonetti G. Linear and Circular PVT1 in Hematological Malignancies and Immune Response: Two Faces of the Same Coin. Mol Cancer (2020) 19:69. doi: 10.1186/s12943-020-01187-5
51. Zhang Q, Wang W, Zhou Q, Chen C, Yuan W, Liu J, et al. Roles of circRNAs in the Tumour Microenvironment. Mol Cancer (2020) 19:14. doi: 10.1186/s12943-019-1125-9
52. Dou Y, Cha DJ, Franklin JL, Higginbotham JN, Jeppesen DK, Weaver AM, et al. Circular RNAs Are Down-Regulated in KRAS Mutant Colon Cancer Cells and Can Be Transferred to Exosomes. Sci Rep (2016) 6:37982. doi: 10.1038/srep37982
53. Guarnerio J, Bezzi M, Jeong JC, Paffenholz SV, Berry K, Naldini MM, et al. Oncogenic Role of Fusion-circRNAs Derived From Cancer-Associated Chromosomal Translocations. Cell (2016) 165:289–302. doi: 10.1016/j.cell.2016.03.020
54. Chen YG, Kim MV, Chen X, Batista PJ, Aoyama S, Wilusz JE, et al. Sensing Self and Foreign Circular RNAs by Intron Identity. Mol Cell (2017) 67:228–238.e5. doi: 10.1016/j.molcel.2017.05.022
55. Barbalat R, Ewald SE, Mouchess ML, Barton GM. Nucleic Acid Recognition by the Innate Immune System. Annu Rev Immunol (2011) 29:185–214. doi: 10.1146/annurev-immunol-031210-101340
56. Wu J, Chen ZJ. Innate Immune Sensing and Signaling of Cytosolic Nucleic Acids. Annu Rev Immunol (2014) 32:461–88. doi: 10.1146/annurev-immunol-032713-120156
57. Cadena C, Hur S. Antiviral Immunity and Circular RNA: No End in Sight. Mol Cell (2017) 67:163–4. doi: 10.1016/j.molcel.2017.07.005
58. Iurescia S, Fioretti D, Rinaldi M. Nucleic Acid Sensing Machinery: Targeting Innate Immune System for Cancer Therapy. Recent Pat Anticancer Drug Discov (2018) 13:2–17. doi: 10.2174/1574892812666171030163804
59. Bourquin C, Pommier A, Hotz C. Harnessing the Immune System to Fight Cancer With Toll-Like Receptor and RIG-I-Like Receptor Agonists. Pharmacol Res (2020) 154:104192. doi: 10.1016/j.phrs.2019.03.001
60. Wesselhoeft RA, Kowalski PS, Parker-Hale FC, Huang Y, Bisaria N, Anderson DG. RNA Circularization Diminishes Immunogenicity and Can Extend Translation Duration In Vivo. Mol Cell (2019) 74:508–520.e4. doi: 10.1016/j.molcel.2019.02.015
61. Chen YG, Chen R, Ahmad S, Verma R, Kasturi SP, Amaya L, et al. N6-Methyladenosine Modification Controls Circular RNA Immunity. Mol Cell (2019) 76:96–109.e9. doi: 10.1016/j.molcel.2019.07.016
62. Zhang L, Hou C, Chen C, Guo Y, Yuan W, Yin D, et al. The Role of N6-Methyladenosine (M6a) Modification in the Regulation of circRNAs. Mol Cancer (2020) 19:105. doi: 10.1186/s12943-020-01224-3
63. Ries RJ, Zaccara S, Klein P, Olarerin-George A, Namkoong S, Pickering BF, et al. M6a Enhances the Phase Separation Potential of mRNA. Nature (2019) 571:424–8. doi: 10.1038/s41586-019-1374-1
64. Zhao J, Lee EE, Kim J, Yang R, Chamseddin B, Ni C, et al. Transforming Activity of an Oncoprotein-Encoding Circular RNA From Human Papillomavirus. Nat Commun (2019) 10:2300. doi: 10.1038/s41467-019-10246-5
65. Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin (2021) 71:209–49. doi: 10.3322/caac.21660
66. Perez-Lopez A, Behnsen J, Nuccio S-P, Raffatellu M. Mucosal Immunity to Pathogenic Intestinal Bacteria. Nat Rev Immunol (2016) 16:135–48. doi: 10.1038/nri.2015.17
67. Kubes P, Jenne C. Immune Responses in the Liver. Annu Rev Immunol (2018) 36:247–77. doi: 10.1146/annurev-immunol-051116-052415
68. Shetty S, Lalor PF, Adams DH. Liver Sinusoidal Endothelial Cells — Gatekeepers of Hepatic Immunity. Nat Rev Gastroenterol Hepatol (2018) 15:555–67. doi: 10.1038/s41575-018-0020-y
69. Doherty DG. Immunity, Tolerance and Autoimmunity in the Liver: A Comprehensive Review. J Autoimmun (2016) 66:60–75. doi: 10.1016/j.jaut.2015.08.020
70. Baghban R, Roshangar L, Jahanban-Esfahlan R, Seidi K, Ebrahimi-Kalan A, Jaymand M, et al. Tumor Microenvironment Complexity and Therapeutic Implications at a Glance. Cell Commun Signal (2020) 18:59. doi: 10.1186/s12964-020-0530-4
71. Zhang Q, Lou Y, Bai X-L, Liang T-B. Immunometabolism: A Novel Perspective of Liver Cancer Microenvironment and its Influence on Tumor Progression. World J Gastroenterol (2018) 24:3500–12. doi: 10.3748/wjg.v24.i31.3500
72. Liu X, Chen Y, Zhang S, Dong L. Gut Microbiota-Mediated Immunomodulation in Tumor. J Exp Clin Cancer Res (2021) 40:221. doi: 10.1186/s13046-021-01983-x
73. Fang Z, Jiang C, Li S. The Potential Regulatory Roles of Circular RNAs in Tumor Immunology and Immunotherapy. Front Immunol (2020) 11:617583. doi: 10.3389/fimmu.2020.617583
74. Mo WL, Jiang JT, Zhang L, Lu QC, Li J, Gu WD, et al. Circular RNA Hsa_Circ_0000467 Promotes the Development of Gastric Cancer by Competitively Binding to MicroRNA miR-326-3p. BioMed Res Int (2020) 2020:4030826. doi: 10.1155/2020/4030826
75. Wang N, Lu K, Qu H, Wang H, Chen Y, Shan T, et al. CircRBM33 Regulates IL-6 to Promote Gastric Cancer Progression Through Targeting miR-149. BioMed Pharmacother (2020) 125:109876. doi: 10.1016/j.biopha.2020.109876
76. Zhang X, Yang H, Jia Y, Xu Z, Zhang L, Sun M, et al. circRNA_0005529 Facilitates Growth and Metastasis of Gastric Cancer via Regulating miR-527/Sp1 Axis. BMC Mol Cell Biol (2021) 22:6. doi: 10.1186/s12860-020-00340-8
77. Niu Q, Dong Z, Liang M, Luo Y, Lin H, Lin M, et al. Circular RNA Hsa_Circ_0001829 Promotes Gastric Cancer Progression Through miR-155-5p/SMAD2 Axis. J Exp Clin Cancer Res (2020) 39:280. doi: 10.1186/s13046-020-01790-w
78. Bu X, Chen Z, Zhang A, Zhou X, Zhang X, Yuan H, et al. Circular RNA Circaff2 Accelerates Gastric Cancer Development by Activating miR-6894-5p and Regulating ANTXR 1 Expression. Clin Res Hepatol Gastroenterol (2021) 45:101671. doi: 10.1016/j.clinre.2021.101671
79. Yue F, Peng K, Zhang L, Zhang J. Circ_0004104 Accelerates the Progression of Gastric Cancer by Regulating the miR-539-3p/RNF2 Axis. Dig Dis Sci (2021) 66(12):4290–301. doi: 10.1007/s10620-020-06802-5
80. Lin J, Liao S, Li E, Liu Z, Zheng R, Wu X, et al. Circcyfip2 Acts as a Sponge of miR-1205 and Affects the Expression of Its Target Gene E2F1 to Regulate Gastric Cancer Metastasis. Mol Ther Nucleic Acids (2020) 21:121–32. doi: 10.1016/j.omtn.2020.05.007
81. Ma Z, Shuai Y, Gao X, Wen X, Ji J. Circular RNAs in the Tumour Microenvironment. Mol Cancer (2020) 19:8. doi: 10.1186/s12943-019-1113-0
82. Ma Y, Zhang C, Zhang B, Yu H, Yu Q. circRNA of AR-Suppressed PABPC1 91 Bp Enhances the Cytotoxicity of Natural Killer Cells Against Hepatocellular Carcinoma via Upregulating UL16 Binding Protein 1. Oncol Lett (2019) 17:388–97. doi: 10.3892/ol.2018.9606
83. Zhang P-F, Gao C, Huang X-Y, Lu J-C, Guo X-J, Shi G-M, et al. Cancer Cell-Derived Exosomal Circuhrf1 Induces Natural Killer Cell Exhaustion and may Cause Resistance to Anti-PD1 Therapy in Hepatocellular Carcinoma. Mol Cancer (2020) 19:110. doi: 10.1186/s12943-020-01222-5
84. Zhang P-F, Wei C-Y, Huang X-Y, Peng R, Yang X, Lu J-C, et al. Circular RNA Circtrim33-12 Acts as the Sponge of MicroRNA-191 to Suppress Hepatocellular Carcinoma Progression. Mol Cancer (2019) 18:105. doi: 10.1186/s12943-019-1031-1
85. Wang Y, Gao R, Li J, Tang S, Li S, Tong Q, et al. Downregulation of Hsa_Circ_0074854 Suppresses the Migration and Invasion in Hepatocellular Carcinoma via Interacting With HuR and via Suppressing Exosomes-Mediated Macrophage M2 Polarization. Int J Nanomed (2021) 16:2803–18. doi: 10.2147/IJN.S284560
86. Wang X, Sheng W, Xu T, Xu J, Gao R, Zhang Z. CircRNA Hsa_Circ_0110102 Inhibited Macrophage Activation and Hepatocellular Carcinoma Progression via miR-580-5p/Pparα/CCL2 Pathway. Aging (Albany NY) (2021) 13:11969–87. doi: 10.18632/aging.202900
87. Hu Z-Q, Zhou S-L, Li J, Zhou Z-J, Wang P-C, Xin H-Y, et al. Circular RNA Sequencing Identifies CircASAP1 as a Key Regulator in Hepatocellular Carcinoma Metastasis. Hepatology (2020) 72:906–22. doi: 10.1002/hep.31068
88. Xu P, Xu X, Wu X, Zhang L, Meng L, Chen Z, et al. CircTMC5 Promotes Gastric Cancer Progression and Metastasis by Targeting miR-361-3p/RABL6. Gastric Cancer (2021) 25(1):64–82. doi: 10.1007/s10120-021-01220-6
89. Vasaikar SV, Straub P, Wang J, Zhang B. LinkedOmics: Analyzing Multi-Omics Data Within and Across 32 Cancer Types. Nucleic Acids Res (2018) 46:D956–63. doi: 10.1093/nar/gkx1090
90. Ru B, Wong CN, Tong Y, Zhong JY, Zhong SSW, Wu WC, et al. TISIDB: An Integrated Repository Portal for Tumor-Immune System Interactions. Bioinformatics (2019) 35:4200–2. doi: 10.1093/bioinformatics/btz210
91. Huang X-Y, Zhang P-F, Wei C-Y, Peng R, Lu J-C, Gao C, et al. Circular RNA circMET Drives Immunosuppression and Anti-PD1 Therapy Resistance in Hepatocellular Carcinoma via the miR-30-5p/Snail/DPP4 Axis. Mol Cancer (2020) 19:92. doi: 10.1186/s12943-020-01213-6
92. Tanaka E, Miyakawa Y, Kishikawa T, Seimiya T, Iwata T, Funato K, et al. Expression of Circular RNA CDR1−AS in Colon Cancer Cells Increases Cell Surface PD−L1 Protein Levels. Oncol Rep (2019) 42:1459–66. doi: 10.3892/or.2019.7244
93. Xu Y-J, Zhao J-M, Gao C, Ni X-F, Wang W, Hu W-W, et al. Hsa_circ_0136666 Activates Treg-Mediated Immune Escape of Colorectal Cancer via miR-497/PD-L1 Pathway. Cell Signal (2021) 86:110095. doi: 10.1016/j.cellsig.2021.110095
94. Jiang Z, Hou Z, Liu W, Yu Z, Liang Z, Chen S. Circ-Keratin 6c Promotes Malignant Progression and Immune Evasion of Colorectal Cancer Through microRNA-485-3p/Programmed Cell Death Receptor Ligand 1 Axis. J Pharmacol Exp Ther (2021) 377:358–67. doi: 10.1124/jpet.121.000518
95. Weng Q, Chen M, Li M, Zheng Y-F, Shao G, Fan W, et al. Global Microarray Profiling Identified Hsa_Circ_0064428 as a Potential Immune-Associated Prognosis Biomarker for Hepatocellular Carcinoma. J Med Genet (2019) 56:32–8. doi: 10.1136/jmedgenet-2018-105440
96. Alfaro C, Sanmamed MF, Rodríguez-Ruiz ME, Teijeira Á, Oñate C, González Á, et al. Interleukin-8 in Cancer Pathogenesis, Treatment and Follow-Up. Cancer Treat Rev (2017) 60:24–31. doi: 10.1016/j.ctrv.2017.08.004
97. Masjedi A, Hashemi V, Hojjat-Farsangi M, Ghalamfarsa G, Azizi G, Yousefi M, et al. The Significant Role of Interleukin-6 and its Signaling Pathway in the Immunopathogenesis and Treatment of Breast Cancer. BioMed Pharmacother (2018) 108:1415–24. doi: 10.1016/j.biopha.2018.09.177
98. Sun X, He X, Zhang Y, Hosaka K, Andersson P, Wu J, et al. Inflammatory Cell-Derived CXCL3 Promotes Pancreatic Cancer Metastasis Through a Novel Myofibroblast-Hijacked Cancer Escape Mechanism. Gut [Internet] BMJ Publishing Group (2021) 71(1):129–47. doi: 10.1136/gutjnl-2020-322744
99. Korbecki J, Kojder K, Kapczuk P, Kupnicka P, Gawrońska-Szklarz B, Gutowska I, et al. The Effect of Hypoxia on the Expression of CXC Chemokines and CXC Chemokine Receptors—A Review of Literature. Int J Mol Sci (2021) 22:843. doi: 10.3390/ijms22020843
100. Wang W, Green M, Choi JE, Gijón M, Kennedy PD, Johnson JK, et al. CD8+ T Cells Regulate Tumor Ferroptosis During Cancer Immunotherapy. Nature (2019) 569:270–4. doi: 10.1038/s41586-019-1170-y
101. Epstein MA, Achong BG, Barr YM. Virus Particles in Cultured Lymphoblasts From Burkitt’s Lymphoma. Lancet (1964) 283:702–3. doi: 10.1016/S0140-6736(64)91524-7
102. Imai S, Koizumi S, Sugiura M, Tokunaga M, Uemura Y, Yamamoto N, et al. Gastric Carcinoma: Monoclonal Epithelial Malignant Cells Expressing Epstein-Barr Virus Latent Infection Protein. Proc Natl Acad Sci USA (1994) 91:9131–5. doi: 10.1073/pnas.91.19.9131
103. Fukayama M, Hayashi Y, Iwasaki Y, Chong J, Ooba T, Takizawa T, et al. Epstein-Barr Virus-Associated Gastric Carcinoma and Epstein-Barr Virus Infection of the Stomach. Lab Invest (1994) 71:73–81. doi: 10.1097/00002371-199407000-00007
104. Shi L, Yan P, Liang Y, Sun Y, Shen J, Zhou S, et al. Circular RNA Expression is Suppressed by Androgen Receptor (AR)-Regulated Adenosine Deaminase That Acts on RNA (ADAR1) in Human Hepatocellular Carcinoma. Cell Death Dis (2017) 8:e3171. doi: 10.1038/cddis.2017.556
105. Tang Y, Jiang M, Jiang H-M, Ye ZJ, Huang Y-S, Li X-S, et al. The Roles of circRNAs in Liver Cancer Immunity. Front Oncol (2020) 10:598464. doi: 10.3389/fonc.2020.598464
106. Qi L, Yu H, Zhang Y, Zhao D, Lv P, Zhong Y, et al. IL-10 Secreted by M2 Macrophage Promoted Tumorigenesis Through Interaction With JAK2 in Glioma. Oncotarget (2016) 7:71673–85. doi: 10.18632/oncotarget.12317
107. Liu Q, Yang C, Wang S, Shi D, Wei C, Song J, et al. Wnt5a-Induced M2 Polarization of Tumor-Associated Macrophages via IL-10 Promotes Colorectal Cancer Progression. Cell Commun Signal (2020) 18:51. doi: 10.1186/s12964-020-00557-2
108. Wu W, Zhen T, Yu J, Yang Q. Circular RNAs as New Regulators in Gastric Cancer: Diagnosis and Cancer Therapy. Front Oncol (2020) 10:1526. doi: 10.3389/fonc.2020.01526
109. Xu Y, Kong S, Qin X, Ju S. Comprehensive Assessment of Plasma Circ_0004771 as a Novel Diagnostic and Dynamic Monitoring Biomarker in Gastric Cancer. Onco Targets Ther (2020) 13:10063–74. doi: 10.2147/OTT.S263536
110. Wang X, Zhang Y, Huang L, Zhang J, Pan F, Li B, et al. Decreased Expression of Hsa_Circ_001988 in Colorectal Cancer and Its Clinical Significances. Int J Clin Exp Pathol (2015) 8:16020–5. doi: 10.1177/1010428317699125
111. Wang Y, Li Z, Xu S, Guo J. Novel Potential Tumor Biomarkers: Circular RNAs and Exosomal Circular RNAs in Gastrointestinal Malignancies. J Clin Lab Anal (2020) 34:e23359. doi: 10.1002/jcla.23359
112. Yang Y. Cancer Immunotherapy: Harnessing the Immune System to Battle Cancer. J Clin Invest (2015) 125:3335–7. doi: 10.1172/JCI83871
113. Li Z, Ruan Y, Zhang H, Shen Y, Li T, Xiao B. Tumor-Suppressive Circular RNAs: Mechanisms Underlying Their Suppression of Tumor Occurrence and Use as Therapeutic Targets. Cancer Sci (2019) 110:3630–8. doi: 10.1111/cas.14211
114. Xie F, Xu M, Lu J, Mao L, Wang S. The Role of Exosomal PD-L1 in Tumor Progression and Immunotherapy. Mol Cancer (2019) 18:146. doi: 10.1186/s12943-019-1074-3
115. Zhang S-A, Niyazi H-E-X-D, Hong W, Tuluwengjiang G-L-X, Zhang L, Zhang Y, et al. Effect of EBI3 on Radiation-Induced Immunosuppression of Cervical Cancer HeLa Cells by Regulating Treg Cells Through PD-1/PD-L1 Pathway. Tumour Biol (2017) 39:1010428317692237. doi: 10.1177/1010428317692237
116. Patsoukis N, Brown J, Petkova V, Liu F, Li L, Boussiotis VA. Selective Effects of PD-1 on Akt and Ras Pathways Regulate Molecular Components of the Cell Cycle and Inhibit T Cell Proliferation. Sci Signal (2012) 5:ra46. doi: 10.1126/scisignal.2002796
117. Mainini F, De Santis F, Fucà G, Di Nicola M, Rivoltini L, Eccles M. Nanobiotechnology and Immunotherapy: Two Powerful and Cooperative Allies Against Cancer. Cancers (Basel) (2021) 13:3765. doi: 10.3390/cancers13153765
Keywords: circular RNA, tumor immunity, tumor vaccine, digestive cancers, gastric cancer, colorectal cancer, liver cancer
Citation: Chen C, Xia C, Tang H, Jiang Y, Wang S, Zhang X, Huang T, Yuan X, Wang J and Peng L (2022) Circular RNAs Involve in Immunity of Digestive Cancers From Bench to Bedside: A Review. Front. Immunol. 13:833058. doi: 10.3389/fimmu.2022.833058
Received: 10 December 2021; Accepted: 01 March 2022;
Published: 04 April 2022.
Edited by:
Ximing Xu, Renmin Hospital of Wuhan University, ChinaReviewed by:
Lei Zhang, Qingdao University, ChinaCopyright © 2022 Chen, Xia, Tang, Jiang, Wang, Zhang, Huang, Yuan, Wang and Peng. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Li Peng, cGVuZ2xpOUBtYWlsLnN5c3UuZWR1LmNu; Junpu Wang, d2FuZy1qcDIwMTNAY3N1LmVkdS5jbg==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.