
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Immunol. , 19 January 2023
Sec. Comparative Immunology
Volume 13 - 2022 | https://doi.org/10.3389/fimmu.2022.1099637
 Yangyang Zhu1†
Yangyang Zhu1† Lei Liu1†
Lei Liu1† Chuchu Zhang1†
Chuchu Zhang1† Chao Zhang1†
Chao Zhang1† Tingting Han1
Tingting Han1 Renjie Duan1,2
Renjie Duan1,2 Yiheng Jin1
Yiheng Jin1 Huimin Guo3
Huimin Guo3 Kan She1
Kan She1 Yihua Xiao1
Yihua Xiao1 Akira Goto4
Akira Goto4 Qingshuang Cai1,5*
Qingshuang Cai1,5* Shanming Ji1*
Shanming Ji1*In Drosophila, the endoplasmic reticulum-associated protein degradation (ERAD) is engaged in regulating pleiotropic biological processes, with regard to retinal degeneration, intestinal homeostasis, and organismal development. The extent to which it functions in controlling the fly innate immune defense, however, remains largely unknown. Here, we show that blockade of the ERAD in fat bodies antagonizes the Toll but not the IMD innate immune defense in Drosophila. Genetic approaches further suggest a functional role of Me31B in the ERAD-mediated fly innate immunity. Moreover, we provide evidence that silence of Xbp1 other than PERK or Atf6 partially rescues the immune defects by the dysregulated ERAD in fat bodies. Collectively, our study uncovers an essential function of the ERAD in mediating the Toll innate immune reaction in Drosophila.
Innate immunity is the first line of the defense for the host against invasion with foreign pathogenic microorganisms (1, 2). Upon activation of the pattern recognition receptors (PRRs) through sensing distinct pathogen-associated molecular patterns (PAMPs), a series of innate immune signaling pathways entail complex downstream cascades to evoke the synthesis of multiple pro-inflammatory cytokines contributing to defeating the invading pathogens (1–4). Up to date, a large body of pioneering studies have highlighted the importance of the precise control of these signals, as defective or over-excessive innate immune response routinely trigger the occurrences of autoimmune disease, allergic reaction, and even cancer (5–9). Unraveling the molecular mechanisms of how innate immune signaling pathways are dynamically modulated has always been one of the hotspots in the basic immunology research.
Over the last decades, Drosophila melanogaster has served as one of the fundamental animal models for studying the regulatory mechanisms of the host innate immune reaction. In Drosophila, the innate immune defense is mainly dominated by two classical nuclear factor-kappa B (NF-κB)-related signaling pathways, namely the Toll and the immune deficiency (IMD) pathways, which rely on their productions of various antimicrobial peptides (AMPs) to function as anti-infective agents (10, 11). The Drosophila Toll pathway receptors are normally activated following infection of fungi or some types of Gram-positive bacteria. This activation further induces signal transductions involving factors including Myd88, pelle, as well as tube, and ultimately the phosphorylation of cactus, releasing the transcription factor Dif/dorsal into the nucleus to direct the production of a series of AMPs, such as Drosomycin (Drs) and Metchnikowin (Mtk) (12, 13). The IMD signaling pathway is typically activated post challenging of most Gram-negative bacteria or some species of Gram-positive bacteria, controlling the expression of an alternative set of AMPs such as Attacin (Att) and Diptericin (Dpt) (12, 14). In particular, both signaling pathways have been demonstrated to be precisely controlled by a large body of modulators. For instance, ubiquitination modification plays a pivotal role in Imd signaling transduction and leveling off (15–25), thus contributing to its homeostasis. While ubiquitin-involved cascade in the Toll pathway mediation was previously thought to be relatively rare, recent findings have been shedding light on this issue (26, 27).
In an effort to uncover novel regulators of the fly Toll pathway, we adopted the widely-used drosomycin promoter-luciferase (Drs-Luc) reporter system and performed an unbiased screening of various genes in Drosophila S2 cells. Of interest, we found that silence of Septin-interacting protein 3 (Sip3, also known as Hrd1) resulted in marked reduction in the Toll signaling activities (Figure S1A), implying that Sip3 is potentially a positive contributor of the Toll pathway. Previous literature has revealed that Sip3 is a multiplexed transmembrane protein in the endoplasmic reticulum (ER) behaving as an E3 ubiquitin ligase (28). The predominant function of Sip3 is to catalyze the degradation of misfolded/unfolded proteins in the ER via specifically accepting ubiquitin from the ER-associated E2 enzyme and transferring it to the substrate to facilitate its degradation (known as ER-associated degradation, ERAD) (28, 29). We therefore hypothesized that the Sip3-involved ERAD is probably essential for the Toll-mediated innate immune defense in Drosophila.
In this study, we demonstrate a critical role of the ERAD in regulating fly innate immunity. We show that blockade of the ERAD cascade in Drosophila fat bodies results in reduced inductions of the Toll downstream AMPs upon Gram-positive bacterial challenging. We further provide a series of evidence that the ERAD is required for the host resistance against various pathogenic microbes. Additionally, our proteomic and genetic results imply that the dysregulated ERAD leads to abnormal accumulation of Me31B, which may further negatively contribute to the Toll innate immune defense primarily in an Xbp1-dependent manner. Overall, our studies shed lights on an essential role of the ERAD in modulating the Toll innate immunity in Drosophila.
In order to investigate whether the E3 ligase Sip3 is involved in controlling Drosophila Toll innate immune response in vivo, we infected Sip3 loss-of-function (LOF) mutant flies (Sip31, isogenized with w1118) and the controls (w1118 as the wild-type control and MyD88KG03447 homozygote as the positive control) with Enterococcus faecalis (E. faecalis), a type of Gram-positive bacteria that has been widely utilized to activate the Toll signaling pathway in flies. The reverse transcription plus quantitative polymerase chain reaction (RT-qPCR) assays were performed to monitor the transcript levels of the Toll downstream AMPs, including Drs and Mtk. As illustrated in Figures 1A, B, loss of Sip3 resulted in marked decreases in the mRNA levels of both Drs (by ~42%) and Mtk (by ~51%) upon bacterial challenging, which is consistent with what we found in S2 cells (Figure S1A). Of note, similar results were obtained (Figures 1C, D) when flies were challenged with another type of Gram-positive bacteria Staphylococcus aureus (S. aureus).
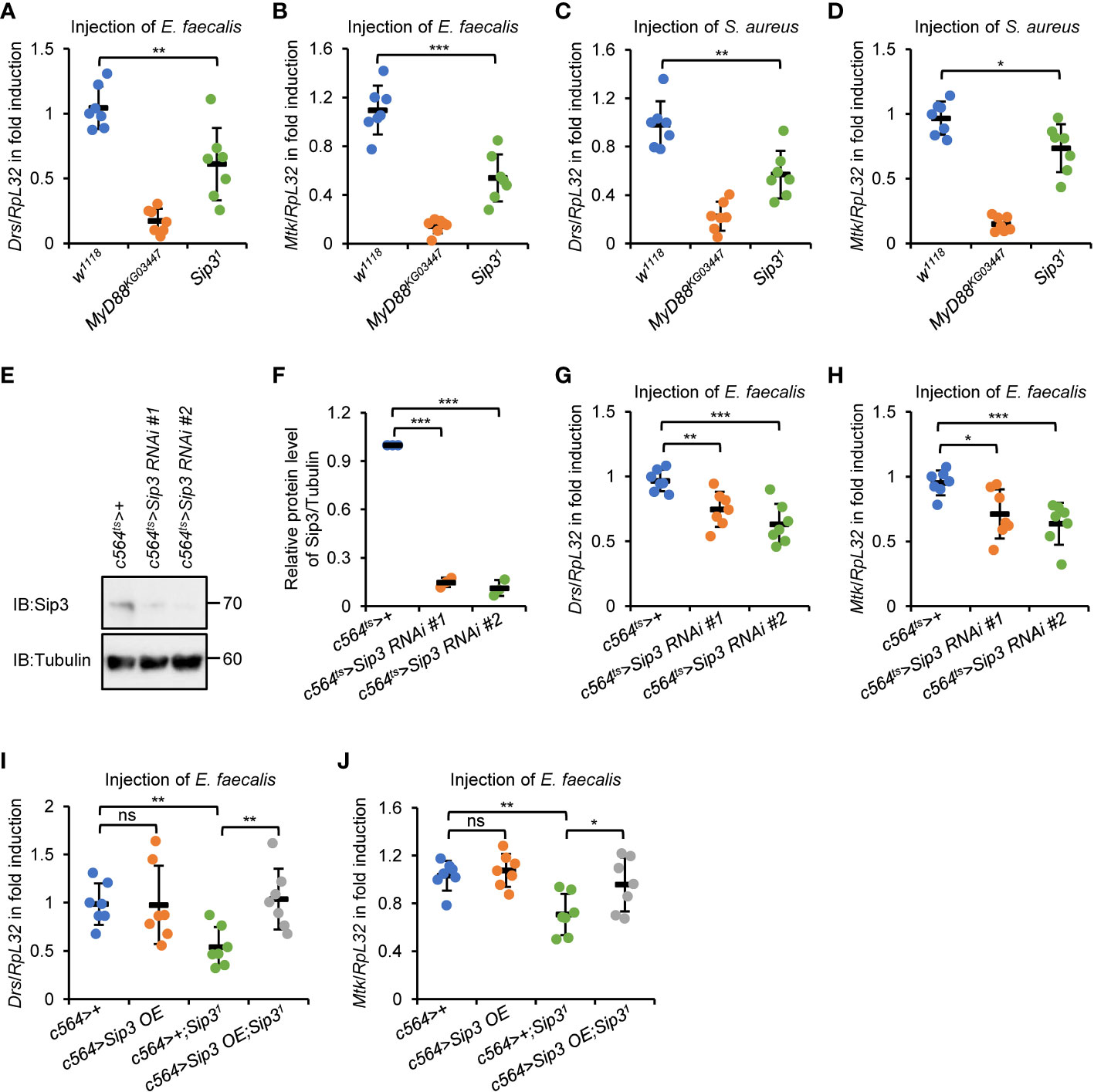
Figure 1 E3 ligase Sip3 positively regulates Toll signaling in Drosophila. (A–D) Wild-type (w1118), MyD88KG03447, and Sip31 mutant flies were injected with E. faecalis (A, B) or S. aureus (C, D) for 12 h, followed by RT-qPCR assays to monitor the expression levels of Drs (A, C) or Mtk (B, D). Each dot represents one independent replicate (10 flies for each replicate). (E, F) Fat bodies were dissected from flies (10 flies for each sample) including c564ts>+, c564ts>Sip3 RNAi #1, and c564ts>Sip3 RNAi #2, followed by Western blot assays (E). Tubulin was used as loading control. Densitometry analysis to quantify the protein level of samples in (E) is shown in (F). (G, H) Flies including c564ts>+, c564ts>Sip3 RNAi #1, and c564ts>Sip3 RNAi #2 were infected with E. faecalis. 12 h later, flies were harvested for RT-qPCR assays to detect the transcript levels of Drs (G) or Mtk (H). Each dot represents one independent replicate (10 flies for each replicate). (I) and (J) Flies including c564>+, c564>Sip3 OE, c564>+;Sip31, and c564>Sip3OE;Sip31 were infected with E. faecalis, followed by RT-qPCR assays as in (G) and (H). Each dot represents one independent replicate (10 flies for each replicate). In (A–D) and (F, J), data are shown as mean ± standard errors. *p < 0.05; **p < 0.01; ***p < 0.001; ns, not significant.
To test whether Sip3 modulates Toll signaling through its potential role in the fat body, which is the dominant immune organ/tissue of flies during systemic infection, we adopted two different Sip3 RNAi strains (see Materials and Methods) and crossed them with c564-gal4 to produce progenies with fat body-specific down-regulation of Sip3. The tub-gal80ts was utilized in our experimental system to carry out RNAi of Sip3 at adult stage. The RNAi efficiency in those Sip3 RNAi progenies (referred as c564ts>Sip3 RNAi #1 and c564ts>Sip3 RNAi #2) was tested by Western blot using anti-Sip3 antibody (Figures 1E, F). Further, these flies were infected with E. faecalis and subjected to RT-qPCR as described above. As illustrated in Figures 1G, H, silencing Sip3 reduced the transcript levels of both Drs (by ~23% to 35%) and Mtk (by ~25% to 28%) relative to those in the controls (c564ts>+). Moreover, ectopic expression of Sip3 in the fat body reversed the reductions in Drs and Mtk transcript levels by loss of Sip3 upon infection (Figures 1I, J), implying that the E3 ligase Sip3 controls the activation of Toll signaling post bacterial stimuli primarily through its essential role in the fat body. Notably, we failed to observe any apparent alterations in the context of pathogen-driven inductions of the Toll downstream AMPs in the c564>Sip3 flies, compared to those in the controls (Figures 1I, J).
A detailed understanding of the physiological function of Sip3 in regulating the host innate immune defense was achieved by analyzing the fly survival post infection of E. faecalis or S. aureus, as Sip3 LOF mutants were more susceptible to both pathogens than the wild-type flies (Figures 2A–C). Further, we quantified the amounts of bacteria present in the flies at various time points post infection (0, 6, and 12h, respectively). Much higher levels of colony-forming units (CFU) were observed in the samples of Sip3 LOF mutants (increased on average by more than 1-fold), relative to those in the control samples (Figures 2D, E), implying that Sip3 defective flies are less capable of scavenging pathogenic microorganisms. Similar results were obtained when we performed infection experiments with E. faecalis utilizing Sip3 RNAi and the control flies (Figures S2A, B).
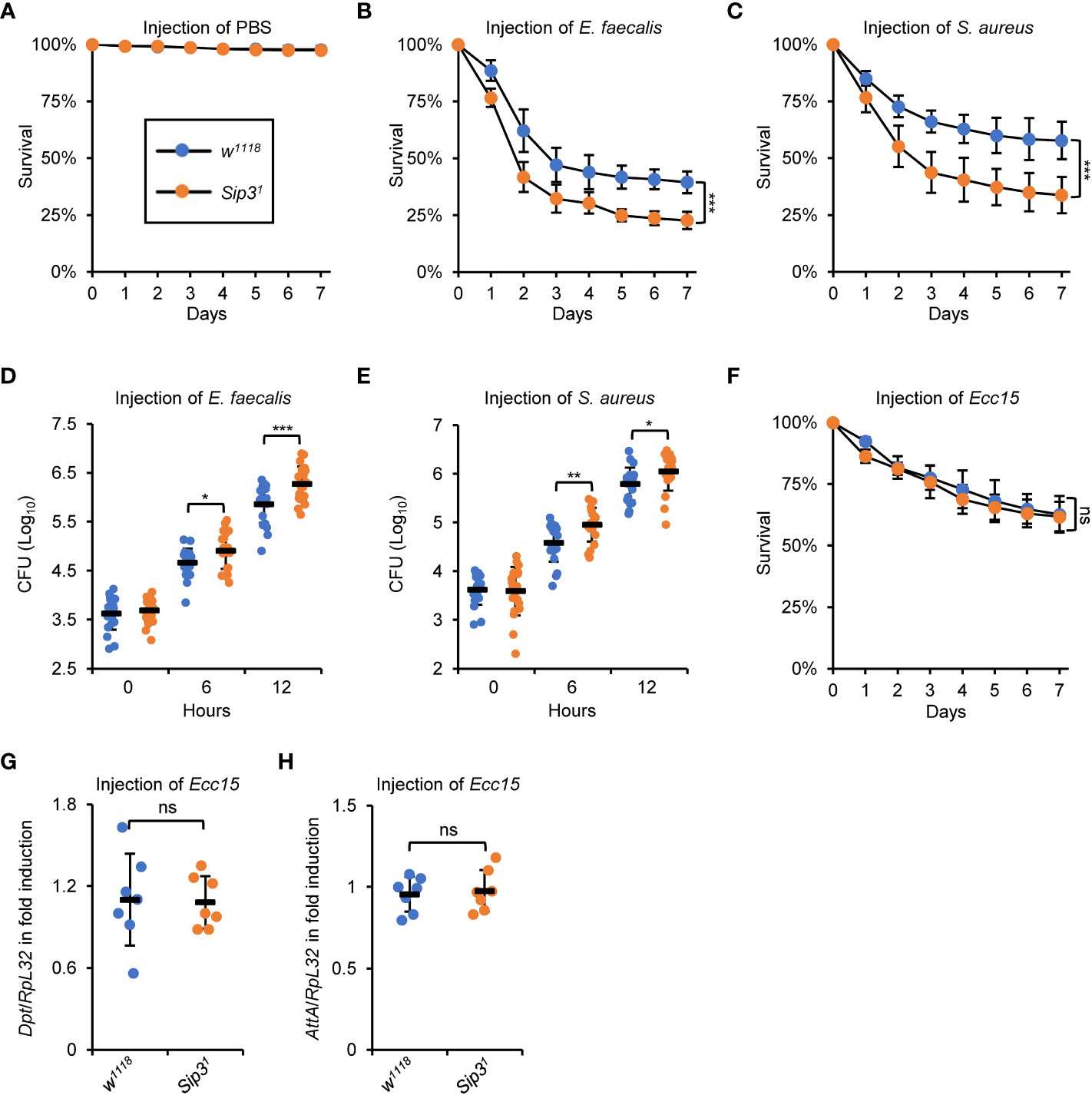
Figure 2 Sip3 contributes to the host resistance against injected bacteria. (A–C) Wild-type and Sip3 mutant flies were infected with sterile PBS (A), E. faecalis (B), or S. aureus (C). Flies were then counted for mortality every day. The numbers of flies analyzed in A, B and C were as follows. In (A), WT: 104, 105, 105; Sip3: 105, 102, 107. In (B), WT: 107, 106, 103; Sip3: 104, 107, 105. In (C), WT: 105, 107, 103; Sip3: 104, 102, 106. (D, E) Wild-type and Sip3 mutant flies were infected with E. faecalis (D), or S. aureus (E). At indicated time points (0, 6, and 12 h), flies were subjected to bacterial burden assays. Each dot represents one independent replicate (10 flies for each replicate). (F–H) Wild-type and Sip3 mutant flies were infected with Ecc15, followed by survival rate assays (F) or RT-qPCR assays (G, H). In (F), the numbers of flies were as follows. WT: 104, 107, 105; Sip3: 105, 102, 107. In (G) and (H), each dot represents one independent replicate (10 flies for each replicate). All the data were shown as mean ± standard errors. *p < 0.05; **p < 0.01; ***p < 0.001; ns, not significant.
In summary, our results indicate that E3 ligase Sip3 plays a crucial role in the Toll innate immune defense in fruit flies.
In Drosophila, the humoral immune defense is principally governed by two signaling pathways, namely the Toll and the IMD pathways (11). It is thus reasonable to explore whether Sip3 is as well involved in regulating the fly IMD innate immunity. To this end, an alternative set of infection experiment was conducted utilizing the Erwinia carotovora carotovora 15 (Ecc15), a type of broadly-used Gram-negative bacteria activating the IMD pathway in flies. Unfortunately, we hardly detected apparent alterations in the contexts of host survival (Figure 2F) and IMD downstream AMP inductions (Figures 2G, H), between those of the Sip3 LOF mutants and the wild-type controls upon Ecc15 injection. Collectively, our data endorse the notion that Sip3 is required for the Toll but not the IMD innate immune reaction in Drosophila.
As mentioned in the Introduction, Sip3 has been demonstrated to be responsible for the degradation of misfolded/unfolded proteins accumulated in the ER following a range of stresses (for instance metabolic dysfunction, excessive accumulation of reactive oxygen species, infection, etc.), a process known as ERAD (28, 29). We then sought to investigate whether other components of the ERAD also participate in affecting fly innate immunity, which is analogous to Sip3. To do this, we again debited the c564ts system to obtain flies with fat body-specific RNAi of Hrd3, Derlin-1, or Herp, which encode pivotal elements of the fly ERAD cascade (28, 30). As shown in Figures 3A, B, silence of Hrd3, Derlin-1, or Herp resulted in marked abatements (by ~34% to 58%) in the inductions of Drs and Mtk post bacterial stimuli. Accordingly, the c564ts>Hrd3 RNAi, c564ts>Derlin-1 RNAi, and c564ts>Herp RNAi flies were more sensitive to the injected E. faecalis and S. aureus (Figures 3C–E) but not Ecc15 (Figure 3F) with respective to the controls. Taken together, our results support the notion that the ERAD is responsible for the Drosophila Toll innate immune defense upon bacterial infection.
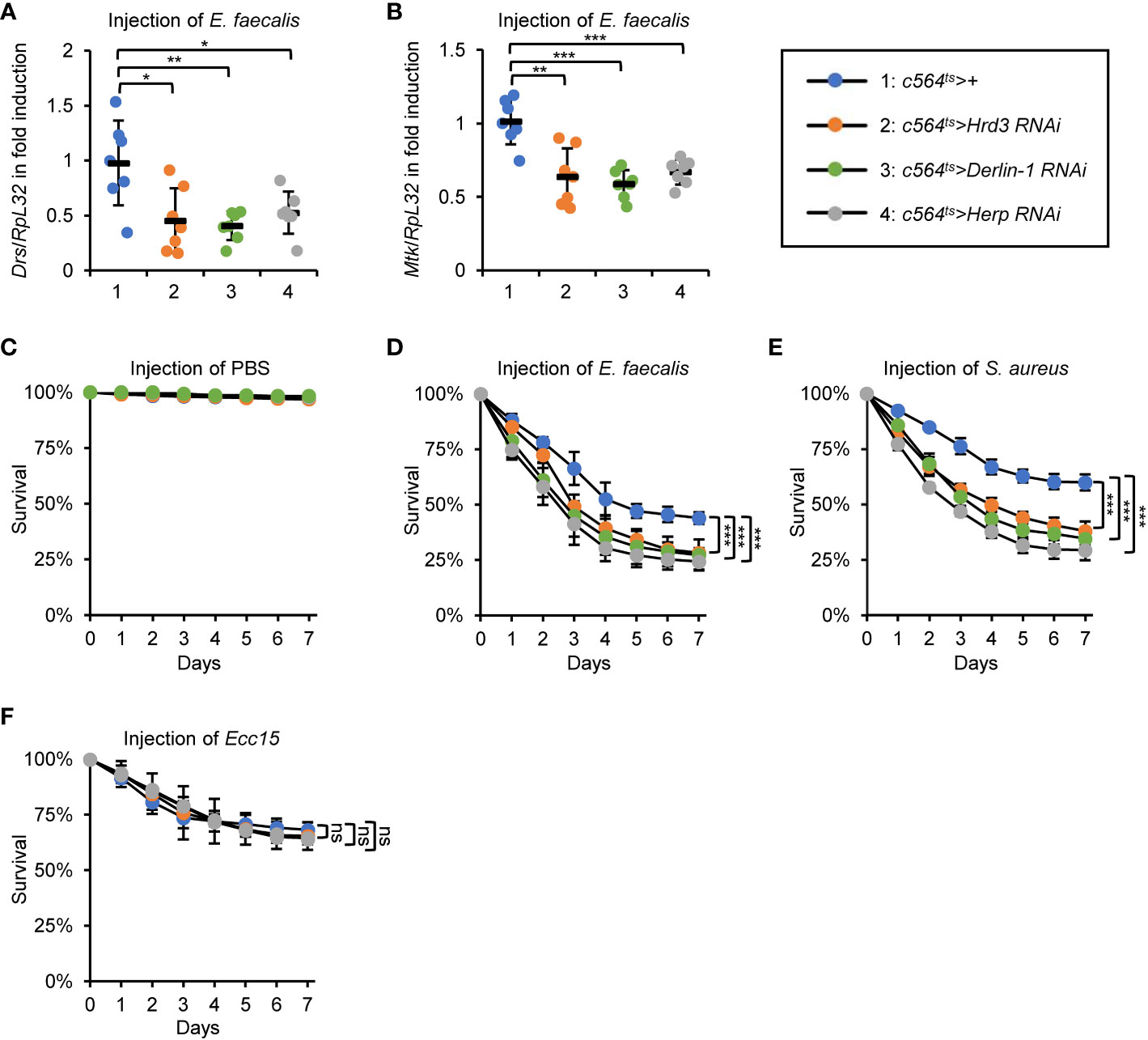
Figure 3 ERAD is essential for the fly Toll innate immune defense. (A, B) Flies including 1: c564ts>+, 2: c564ts>Hrd3 RNAi, 3: c564ts>Derlin-1 RNAi, and 4: c564ts>Herp RNAi were infected with E. faecalis. 12 h later, flies were harvested for RT-qPCR assays to detect the mRNA levels of Drs (A) or Mtk (B). Each dot represents one independent replicate (10 flies for each replicate). (C–F) Flies including 1: c564ts>+, 2: c564ts>Hrd3 RNAi, 3: c564ts>Derlin-1 RNAi, and 4: c564ts>Herp RNAi were injected with sterile PBS (C), E. faecalis (D), S. aureus (E), or Ecc15 (F), followed by survival rate assays. The numbers of flies were as follows. In (C), c564ts>+: 107, 106, 106; c564ts>Hrd3 RNAi: 102, 107, 105; c564ts>Derlin-1 RNAi: 103, 104, 107; c564ts>Herp RNAi: 103, 102, 102. In (D), c564ts>+: 104, 103, 105; c564ts>Hrd3 RNAi: 105, 106, 104; c564ts>Derlin-1 RNAi: 107, 105, 104; c564ts>Herp RNAi: 105, 102, 106. In (E), c564ts>+: 106, 104, 107; c564ts>Hrd3 RNAi: 105, 106, 105; c564ts>Derlin-1 RNAi: 102, 103, 105; c564ts>Herp RNAi: 104, 104, 102. In (F), c564ts>+: 104, 103, 104; c564ts>Hrd3 RNAi: 104, 105, 107; c564ts>Derlin-1 RNAi: 106, 102, 103; c564ts>Herp RNAi: 103, 105, 105. All the data were shown as mean ± standard errors. *p < 0.05; **p < 0.01; ***p < 0.001; ns, not significant.
We next sought to address how the ERAD is implicated in impacting the innate immune defense in Drosophila. A proteomic analysis was performed using dissected fat bodies from c564ts>Sip3 RNAi and the control flies following bacterial injection (Supplementary Table 1). Whereas over 1500 proteins/peptides were identified in this assay, the majority (more than 95%) of them overlapped in both samples (Figure 4A), implying that the E3 ligase Sip3 is of low potential for silencing or evoking gene expression. The fact that knockdown of Sip3 in S2 cells certainly leads to a reduction in Toll signaling activity (Figure S1A) makes us assume that when Sip3 is silenced, one (or possibly more) pivotal factors of the Toll pathway is dysregulated. However, we failed to observe any significant differences when we compared the appearances of protein/peptide of Myd88, pll, tube, cactus, dorsal, and Dif from both samples (Figure S3A), even though the Toll downstream effectors (e.g. BaraA2, Drs, Mtk, IM1) were markedly reduced by Sip3 RNAi (Figure S3B). Thus, we reasoned that the Sip3-mediated ERAD likely engage in an indirect way to modulate the fly Toll pathway.
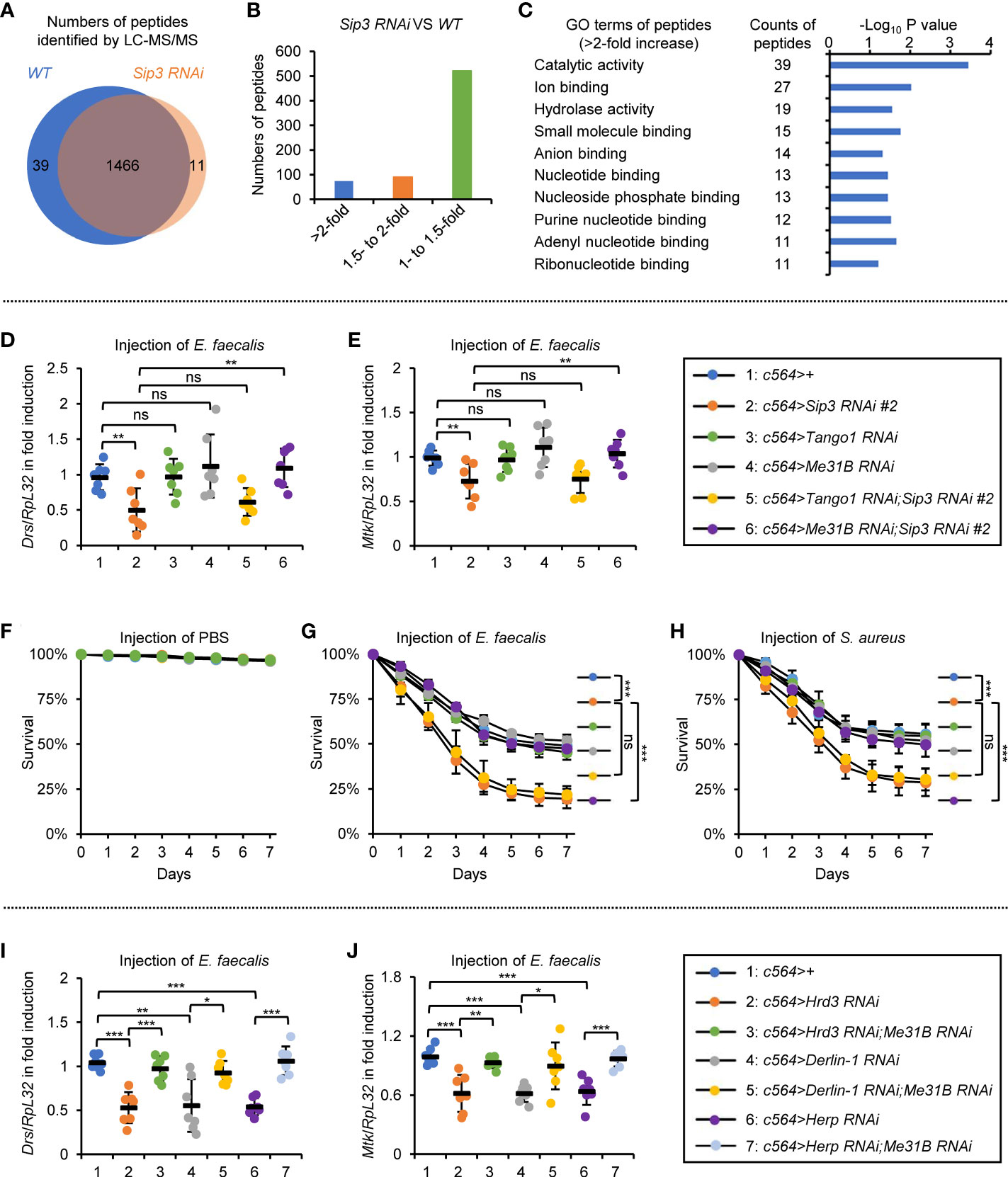
Figure 4 ERAD modulates Toll signaling in an Me31B-dependent manner. (A–C) Proteomic analysis of fat bodies dissected from wild-type (c564ts) and Sip3 RNAi (c564ts>Sip3 RNAi 2#) flies (100 flies for each sample, 3 biological replicate for each group). In (A), total numbers of peptides/proteins that were identified by the LC-MS/MS assay. In (B), numbers of peptides that were increased by various indicated folds (>2-fold, 1.5- to 2-fold, or 1- to 1.5-fold) in the Sip3 RNAi samples relative to the controls. In (C), gene ontology analysis of the increased (>2-fold) peptides/proteins. (D, E) Flies including 1: c564>+, 2: c564>Sip3 RNAi #2, 3: c564>Tango1 RNAi, 4: c564>Me31B RNAi, 5: c564>Tango1 RNAi;Sip3 RNAi #2, and 6: c564>Me31B RNAi;Sip3 RNAi #2 were infected with E. faecalis. 12 h later, flies were subjected to RT-qPCR assays. Each dot represents one independent replicate (10 flies for each replicate). (F–H) Flies with same genotypes as in (D) were injected with sterile PBS (F), E. faecalis (G), or S. aureus (H), followed by survival rate assays. The numbers of flies were as follows. In (F), c564>+:105, 103, 102; c564>Sip3 RNAi #2: 104, 105, 106; c564>Tango1 RNAi: 105, 106, 105; c564>Me31B RNAi: 102, 103, 105; c564>Tango1 RNAi;Sip3 RNAi #2: 106, 104, 107; c564>Me31B RNAi;Sip3 RNAi #2: 106, 104, 102. In (G), c564>+:103, 104, 107; c564>Sip3 RNAi #2: 103, 105, 103; c564>Tango1 RNAi: 105, 102, 103; c564>Me31B RNAi: 106, 106, 106; c564>Tango1 RNAi;Sip3 RNAi #2: 104, 106, 105; c564>Me31B RNAi;Sip3 RNAi #2: 106, 102, 104. In (H), c564>+:101, 105, 104; c564>Sip3 RNAi #2: 104, 104, 104; c564>Tango1 RNAi: 104, 102, 105; c564>Me31B RNAi: 103, 106, 102; c564>Tango1 RNAi;Sip3 RNAi #2: 106, 104, 103; c564>Me31B RNAi;Sip3 RNAi #2: 102, 107, 106. (I) and (J) Flies including 1: c564>+, 2: c564>Hrd3 RNAi, 3: c564>Hrd3 RNAi;Me31B RNAi, 4: c564>Derlin-1 RNAi, 5: c564>Derlin-1 RNAi;Me31B RNAi, 6: c564>Herp RNAi, and 7: c564>Herp RNAi;Me31B RNAi were infected with E. faecalis. 12 h later, flies were subjected to RT-qPCR assays. Each dot represents one independent replicate (10 flies for each replicate). All the data were shown as mean ± standard errors. *p < 0.05; **p < 0.01; ***p < 0.001; ns, not significant.
Since blockade of the ERAD leads to accumulation of misfolded/unfolded proteins, we thus prioritized higher existences of proteins/peptides present in the Sip3 RNAi flies, as we hypothesized that they are probably the key to the deregulated Toll signaling. Based upon the data from the proteomic analysis, only around 70 proteins/peptides out of the ~1500 were significantly increased (by more than 2-fold) by down-regulation of Sip3 (Figure 4B). Gene ontology (GO) analysis upon the increased proteins/peptides indicated that the top 3 categories regarding their molecular functions were 1) catalytical activity, 2) ion binding, and 3) hydrolase activity (Figure 4C).
We first focused on the top 10 increased proteins/peptides (including Cbp80, Kat60, Arts, Tango1, Cpb, LManV, Me31B, Vasa, CG7900, and CG31974). They were barely detectable in the control samples, whereas they enormously accumulated in the Sip3 RNAi fat bodies. Among these, Tango1 and Me31B are the only two candidates considered to be ER-related proteins (31, 32), allowing us to postulate that they are likely under the control of the ERAD and play potential roles in the ERAD-mediated innate immunity of Drosophila. To test this assumption, we used Tango1 RNAi and Me31B RNAi flies for genetic manipulations. We observed that the induction of Drs or Mtk post bacterial infection was not affected by knockdown of Tango1 or Me31B in fat bodies (Figures 4D, E). However, double knock down of Me31B and Sip3 restored the induction of the Toll pathway-regulated AMPs, and this was not observed in the case of Tango1 and Sip3 double RNAi (Figures 4D, E). Pioneering studies have demonstrated that Drosophila Me31B is a putative DEAD-box helicase, which is functionally involved in protein translation and mRNA decay through a set of associations with other adaptor proteins (33–37). Consistently, when we looked at the fly survival post infection of E. faecalis or S. aureus, we found that the low resistances to bacterial infection in Sip3 RNAi flies were profoundly elevated by Me31B RNAi (Figures 4F–H). Moreover, loss of Me31B also prevented the passive impacts on bacteria-driven AMP inductions by RNAi of Hrd3, Derlin-1, or Herp (Figures 4I, J). Taken together, our results indicate that the ERAD contributes to the Toll innate immune defense to a large extent in an Me31B-dependent manner in Drosophila.
Our next aim is to explore how accumulation of Me31B negatively impacts on Toll signaling. It is reported that the accumulated misfolded/unfolded proteins on ER can be recognized by diverse stress sensors, namely inositol acquisition enzyme-1 (IRE1), double-stranded RNA-activated protein kinase-like ER kinase (PERK), and/or activated transcription factor 6 (ATF6), in a manner that leads to the activation of specific signaling cascades (referred to as unfolded protein response to ER, UPRER) (29, 38–40). We thereby sought to determine whether the stockpiled Me31B by the ERAD dysfunction is responsible for the activation of UPRER, which passively impacts on the Toll innate immune pathway. Indeed, we observed an elevated mRNA occurrence of Xbp1RB (generated by splicing of Xbp1 mRNA) and PERK in the Sip3 RNAi fat bodies (Figures S4A, B), implicating that activation of UPRER in the fly fat bodies with the ERAD blockade. Notably, we detected decreased transcript levels of both Drs and Mtk in the Sip3 RNAi samples relative to those in the control (Figures S4C, D). In addition, we obtained flies with fat body-specific RNAi of Sip3 combined with Xbp1, PERK, or Atf6 RNAi and conducted infection experiments. As illustrated in Figures 5A, B, down regulation of PERK or Atf6 was dispensable for affecting the Sip3 RNAi-mediated decrease of Drs or Mtk induction following E. faecalis infection. However, loss of Xbp1 partially rescued the reduced transcript levels of Drs and Mtk in the Sip3 RNAi flies upon challenging. Regarding fly survival, we observed that the Xbp1 and Sip3 double RNAi flies were more resistant to both E. faecalis and S. aureus than flies with Sip3 RNAi alone (Figures 5C–E), implying an epistatic relationship between Xbp1 and Sip3 in modulating the fly innate immunity.
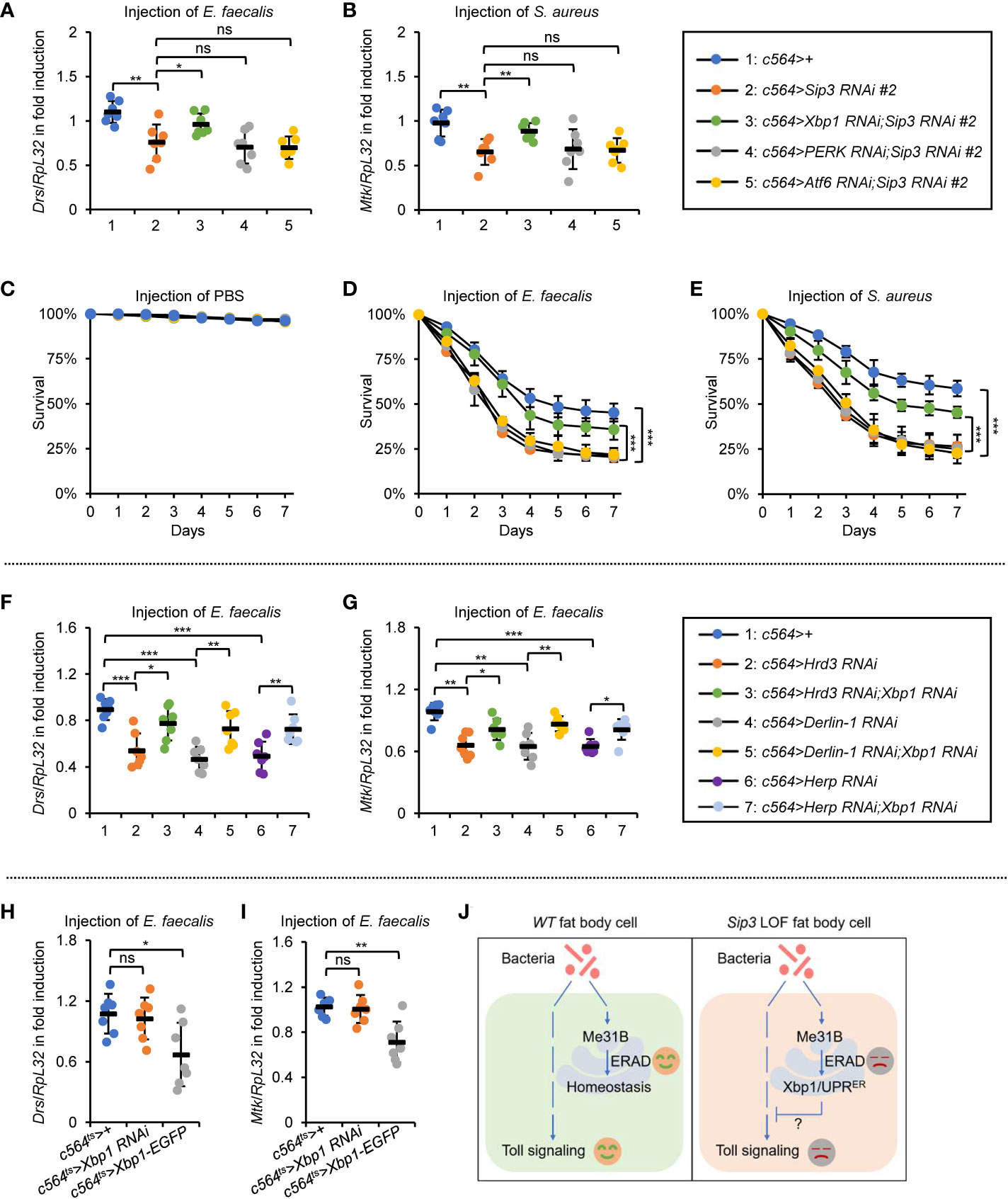
Figure 5 ERAD controls the Toll innate immunity partially via the Xbp1 axis of UPRER. (A, B) Flies including 1: c564>+, 2: c564>Sip3 RNAi #2, 3: c564>Xbp1 RNAi;Sip3 RNAi #2, 4: c564>PERK RNAi;Sip3 RNAi #2, and 5: c564>Atf6 RNAi;Sip3 RNAi #2 were infected with E. faecalis, followed by RT-qPCR assays. Each dot represents one independent replicate (10 flies for each replicate). (C–E) Flies with same genotypes as in (A) were injected with sterile PBS (C), E. faecalis (D), or S. aureus (E), followed by survival rate assays. The numbers of flies were as follows. In (C), c564>+: 106, 102, 104; c564>Sip3 RNAi #2: 104, 106, 103; c564>Xbp1 RNAi;Sip3 RNAi #2: 103, 105, 103; c564>PERK RNAi;Sip3 RNAi #2: 103, 105, 106; c564>Atf6 RNAi;Sip3 RNAi #2: 102, 107, 102. In (D), c564>+: 103, 105, 103; c564>Sip3 RNAi #2: 104, 106, 106; c564>Xbp1 RNAi;Sip3 RNAi #2: 107, 105, 105; c564>PERK RNAi;Sip3 RNAi #2: 104, 102, 103; c564>Atf6 RNAi;Sip3 RNAi #2: 105, 102, 102. In (E), c564>+: 102, 104, 103; c564>Sip3 RNAi #2: 106, 105, 107; c564>Xbp1 RNAi;Sip3 RNAi #2: 105, 102, 104; c564>PERK RNAi;Sip3 RNAi #2: 106, 106, 101; c564>Atf6 RNAi;Sip3 RNAi #2: 103, 104, 105. (F) and (G) Flies including 1: c564>+, 2: c564>Hrd3 RNAi #2, 3: c564>Hrd3 RNAi;Xbp1 RNAi, 4: c564>Derlin-1 RNAi, 5: c564>Derlin-1 RNAi;Xbp1 RNAi, 6: c564>Herp RNAi, and 7: c564>Herp RNAi;Xbp1 RNAi were infected with E. faecalis, followed by RT-qPCR assays. Each dot represents one independent replicate (10 flies for each replicate). (H) and (I) Flies including c564ts>+, c564ts>Xbp1 RNAi, and c564ts>Xbp1-EGFP were infected with E. faecalis. RT-qPCR assays were performed to monitor the transcript levels of Drs (H) and Mtk (I). Each dot represents one independent replicate (10 flies for each replicate). (J) Working model by which ERAD modulates Drosophila Toll innate immune response. In the wild-type (WT) fat bodies, the ERAD remains homeostasis without negatively impacting Toll signaling. When ERAD is blocked (e.g. in the Sip3 LOF fat bodies), accumulated Me31B activates the Xbp1 axis of the UPRER, thereby antagonizing the Toll pathway. In (A–I), data were shown as mean ± standard errors. *p < 0.05; **p < 0.01; ***p < 0.001; ns, not significant.
To obtain additional genetic evidence, we generated Hrd3, Derlin-1, or Herp RNAi flies together with Xbp1 RNAi. When these flies were challenged with E. faecalis, we obtained similar results with respect to the inductions of AMPs (Figures 5F, G). Further, we subjected flies with fat body specific down regulation or ectopic expression of Xbp1 to bacterial infection. We observed remarkable reduction in the inductions of Drs and Mtk in the c564ts>Xbp1-EGFP flies, although it was not the case in the c564ts>Xbp1 RNAi samples (Figures 5H, I). Since Xbp1 mainly functions as a transcription factor governing expression of downstream targets (29, 41–43), it is probably that one/some factor(s) of the Toll pathway is/are under the control of Xbp1. Ultimately, our data point to a working model in which the accumulated Me31B in the ERAD defective flies largely relies on the Xbp1-involved axis of the UPRER pathway to antagonize the Toll innate immune defense (Figure 5J).
ER has been well known for its fundamental importance in the proper post-translational modification and folding of proteins (44). Nevertheless, it is constantly challenged by diverse pathological insults and/or physiological defects, which may lead to ER dysfunction and the accumulation of some misfolded/unfolded proteins in ER (29, 38–40). As an E3 ligase, Drosophila Sip3 harbors a critical action in the ubiquitination and degradation of misfolded/unfolded ER proteins (also known as ERAD) (28), thus positively contributing to the maintenance of ER homeostasis to ensure its function. In flies, the Sip3-involved ERAD on retinal degeneration has been well established (28), yet whether it fulfils a role in innate immune modulation remains elusive. In the present study, we observed remarkably reduced induction of the Toll downstream AMPs and increased mortality in the Sip3 LOF mutants following bacterial infection. Importantly, ectopic expression of Sip3 in fat bodies almost fully rescued the Sip3 LOF mutant phenotypes in our experimental system. Further, we provided a series of genetic evidence suggesting that the ERAD is involved in governing the Toll innate immune defense. Our study provides a potential functional linkage between ER homeostasis and innate immunity in Drosophila.
How does the ERAD positively contribute to modulating the Toll innate immune defense? Despite our proteomic analyses revealing an increase of a series of proteins/peptides that are primarily involved in several pathways in the Sip3 RNAi fat body cells, it appears that none of the crucial factors of the canonical Toll pathway is altered. We therefore shifted our attention unbiasedly to the most increased proteins/peptides (top10) and focused on the proteins shown to be localized in ER, because Sip3 plays a critical role in the ERAD. Between the two potential candidates (Me31B and Tango1 in our study), we identified that Me31B is likely the major downstream effector in the ERAD-mediated Toll innate immune defense, as silencing Me31B nearly completely reverses the innate immune defects in the Sip3, Hrd3, Derlin-1, or Herp RNAi flies. Nonetheless, our current data cannot address whether the accumulated Me31B is misfolded/unfolded and if so, how the E3 ligase Sip3 promotes its ubiquitination and degradation. It would be worthwhile in the future to obtain a Me31B variant that could somehow mimic its misfolding/unfolding pattern and to inspect its effect in the fly innate immune regulation.
Several lines of evidence have indicated that ERAD and UPRER can intersect to control ER homeostasis and relevant cellular processes (30, 45, 46). Interestingly, in blocking the Xbp1 axis of the UPRER, we observed a striking impact on the ERAD-involved Toll innate immune reaction, albeit the immune dysfunction in the ERAD-deficient flies cannot be entirely rescued by Xbp1 RNAi. One possibility is that the three UPRER signaling cascades could be somehow reciprocally interrelated under the current experimental conditions. Nevertheless, in accordance with our results, we would like to hypothesize that while the ERAD is blocked, bacterial stimuli possibly lead to excessive accumulation of unfolded/misfolded Me31B protein in ER, where it activates the Xbp1-involved UPRER to antagonize the Toll innate immune defense in flies (Figure 5J). In agreement with this model, we further observed that ectopic expression of Xbp1 in fat bodies markedly prevented the pathogen-driven induction of the Toll downstream AMPs, through a way in which we currently have no knowledge. As a reminder, ERAD has been proposed to be deployed for the clearance of proteins not only misfolded/unfolded (“quality” control), but also excessively abundant (“quantity” control) (47). Indeed, several pioneering studies revealed that Me31B is abundant in variable types of ribonucleoprotein granules as a putative ATP-dependent RNA helicase for post-transcriptionally modulating specific target RNAs (36, 48). It would thus not be surprising if further studies demonstrate a functional utility of Me31B in correlating with and governing the fates of some mRNAs instrumental to the Drosophila Toll pathway.
All flies were maintained under normal condition (12h light/12h dark, 65% humidity) with medium (6.65% cornmeal, 7.15% dextrose, 5% yeast, 0.66% agar, 2.2% nipagin, and 3.4 ml/l propionic acid). The c564-gal4;tub-gal80ts (c564ts) was used to allow gene manipulation in fat bodies at adult stage. The following strains were obtained from Bloomington Drosophila Stock Center: Sip31 (#86518), MyD88KG03447 (#14091), Me31B RNAi (#38923), Tango1 RNAi (#67887), and UAS-Xbp1-EGFP (#60730). The following strains were purchased from Vienna Drosophila RNAi Center: Sip3 RNAi 2# (#6870), Hrd3 RNAi (#1161), Derlin-1 RNAi (#44210), Herp RNAi (#11724), Xbp1 RNAi (#109312), PERK RNAi (#16427), and Atf6 RNAi (#36504). The Sip3 RNAi 1# (#TH01506.N) was obtained from Tsinghua Stock Center. The UAS-Sip3 strain was described previously (49).
RT-qPCR assays were performed according to protocols described previously (50). Briefly, whole flies (male) or dissected fat bodies were homogenized in the RNA-easy Isolation Reagent (Vazyme) using glass beads. Total RNA was extracted, followed by cDNA synthesis using the first-strand cDNA synthesis kit (Transgen) according to the manufacturer’s instructions. Quantitative PCR assays were performed using SYBR Green Mix (Transgen) in triplicate on a Bio-Rad iCycler iQ5 PCR Thermal Cycler. All results were normalized to the endogenous reference RpL32, which encodes the ribosomal protein Rp49. Primers used in RT-qPCR assays were shown as follows. RpL32-s: GCTAAGCTGTCGCACAAATG; RpL32-as: GTTCGATCCGTAACCGATGT; Drs-s: CGTGAGAACCTTTTCCAATATGATG; Drs-as: TCCCAGGACCACCAGCAT; Mtk-s: CAGTGCTGGCAGAGCCTCAT; Mtk-as: ATAAATTGGACCCGGTCTTG; Dpt-s: TTTGCAGTCCAGGGTCACCA; Dpt-as: CACGAGCCTCCATTCAGTCCAATCTCGG; CecA1-s: ACGCGTTGGTCAGCACACT; CecA1-as: ACATTGGCGGCTTGTTGAG; Xbp1RB-s: CAACCTTGGATCTGCCGCAGGG; Xbp1RB-as: CGCTCCAGCGCCTGTTTCCAG; PERK-s: CTGCGCAGTCTTCGGGACGG; PERK-as: AGCTGCTGAAGGTGCGGCTG.
Overnight bacterial cultures were harvested and diluted in sterile 1×PBS at a concentration of OD600 = 1. Indicated male flies were injected with 4.6nl of diluted bacteria or the same volume of PBS as controls. After infection, flies were immediately transferred to fresh vials (30-40 flies per vial) every day. For survival assays, the numbers of death were counted every day. Flies that died within 2h (< 5% of the total) after bacterial challenge were not considered in the analyses.
For bacterial burden assays, 10 flies were harvested, dipped in 75% EtOH and subsequently volatilized with EtOH on the fly pad for several minutes. Flies were then homogenized in 200μl of sterile 1×PBS. The ground homogenate was serially diluted and 100μl of each diluent was placed on LB agar. Plates were incubated in a 30°C culture hood overnight before counting the numbers of bacterial colonies.
Western blot assays were performed as described previously (51). In brief, fat bodies were lysed in lysis buffer (150mM NaCl, 50mM Tris-HCl, pH=7.4, 10% glycerol, 0.5% TritonX-100, and 1mM PMSF). Lysates were then centrifuged at 12,000rpm for 10min at 4°C. The supernatant was subjected to Western blot analysis. The Sip3 polyclonal antibody was generated by immunizing mice with purified GST-tagged Sip3527-626. The β-Tubulin antibody was purchased from Cowin (1:2000, Cat#CW0098M).
Proteomic analysis by LC-MS/MS was performed according to previously described method (25). Briefly, Fat bodies were harvested from indicated flies in three independent biological replicates, followed by lysate preparation using lysis buffer (50mM Tris-HCl, pH=7.5, 150mM NaCl, 0.5% TritonX-100, 10% glycerol with 1mM PMSF). Samples were precipitated overnight with acetone on ice. Proteins/peptides were digested with Trypsin (Promega) and then desalted (PiecrceTMC-18 Spin Columns, Thermo) according to manufacturer’s instructions. Peptides were then subjected to LC-MS/MS assays. Resulting MS/MS data were processed using Thermo Proteome Discovery (version 1.4.1.14) and tandem mass spectra were searched against UniProt-Drosophila database.
All statistical analyses were performed by using GraphPad Prism 8. Data were shown as mean and standard errors. Statistical significance was determined by using the ANOVA or Mann-Whitney tests except for survival assays, in which the Log-Rank test (Kaplan-Meier method) was used for statistical analysis. The p value of less than 0.05 was considered statistically significant. *p < 0.05; **p < 0.01; ***p < 0.001; ns, not significant.
The data presented in the study are deposited in the "Mendeley" repository (https://data.mendeley.com/datasets/6xbrt3n75v/1).
YZ, LL, QC, and SJ conceived and designed the experiments. YZ, LL, ChuZ, ChaZ, TH, RD, YJ, KS, YX, and QC performed the experiments. YZ, LL, and QC analyzed the data. YZ and LL performed statistical analyses. YZ, AG, QC, and SJ wrote the manuscript. All authors contributed to the article and approved the submitted version.
This work was supported by grants from the National Natural Science Foundation of China (32100702) and the Anhui Provincial Natural Science Foundation (2008085J14).
We thank Bloomington Drosophila Stock Center, Vienna Drosophila RNAi Center, and Tsinghua Stock Center for the fly resources. We thank the staff members at Omics-Laboratory of the Biotechnology Centre of Anhui Agricultural University for providing technical supports in mass data collection.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2022.1099637/full#supplementary-material
1. Hoffmann JA. Innate immunity of insects. Curr Opin Immunol (1995) 7(1):4–10. doi: 10.1016/0952-7915(95)80022-0
2. Beutler B. Innate immunity: an overview. Mol Immunol (2004) 40(12):845–59. doi: 10.1016/j.molimm.2003.10.005
3. Nguyen TA, Pang KC, Masters SL. Intercellular communication for innate immunity. Mol Immunol (2017) 86:16–22. doi: 10.1016/j.molimm.2016.10.002
4. Wang Q, Ren M, Liu X, Xia H, Chen K. Peptidoglycan recognition proteins in insect immunity. Mol Immunol (2019) 106:69–76. doi: 10.1016/j.molimm.2018.12.021
5. Fitzgerald KA, Kagan JC. Toll-like receptors and the control of immunity. Cell. (2020) 180(6):1044–66. doi: 10.1016/j.cell.2020.02.041
6. Zhang Q, Cao X. Epigenetic remodeling in innate immunity and inflammation. Annu Rev Immunol (2021) 39:279–311. doi: 10.1146/annurev-immunol-093019-123619
7. Mantovani A. Wandering pathways in the regulation of innate immunity and inflammation. J Autoimmun (2017) 85:1–5. doi: 10.1016/j.jaut.2017.10.007
8. Brubaker SW, Bonham KS, Zanoni I, Kagan JC. Innate immune pattern recognition: a cell biological perspective. Annu Rev Immunol (2015) 33:257–90. doi: 10.1146/annurev-immunol-032414-112240
9. Zhang Y, Cao X. Long noncoding RNAs in innate immunity. Cell Mol Immunol (2016) 13(2):138–47. doi: 10.1038/cmi.2015.68
10. Hoffmann JA. The immune response of drosophila. Nature. (2003) 426(6962):33–8. doi: 10.1038/nature02021
11. Lemaitre B, Hoffmann J. The host defense of drosophila melanogaster. Annu Rev Immunol (2007) 25:697–743. doi: 10.1146/annurev.immunol.25.022106.141615
12. Ferrandon D, Imler JL, Hetru C, Hoffmann JA. The drosophila systemic immune response: sensing and signalling during bacterial and fungal infections. Nat Rev Immunol (2007) 7(11):862–74. doi: 10.1038/nri2194
13. Valanne S, Wang JH, Ramet M. The drosophila toll signaling pathway. J Immunol (2011) 186(2):649–56. doi: 10.4049/jimmunol.1002302
14. Myllymaki H, Valanne S, Ramet M. The drosophila imd signaling pathway. J Immunol (2014) 192(8):3455–62. doi: 10.4049/jimmunol.1303309
15. Meinander A, Runchel C, Tenev T, Chen L, Kim CH, Ribeiro PS, et al. Ubiquitylation of the initiator caspase DREDD is required for innate immune signalling. EMBO J (2012) 31(12):2770–83. doi: 10.1038/emboj.2012.121
16. Kleino A, Valanne S, Ulvila J, Kallio J, Myllymaki H, Enwald H, et al. Inhibitor of apoptosis 2 and TAK1-binding protein are components of the drosophila imd pathway. EMBO J (2005) 24(19):3423–34. doi: 10.1038/sj.emboj.7600807
17. Tsuda M, Langmann C, Harden N, Aigaki T. The RING-finger scaffold protein plenty of SH3s targets TAK1 to control immunity signalling in drosophila. EMBO Rep (2005) 6(11):1082–7. doi: 10.1038/sj.embor.7400537
18. Khush RS, Cornwell WD, Uram JN, Lemaitre B. A ubiquitin-proteasome pathway represses the drosophila immune deficiency signaling cascade. Curr Biol (2002) 12(20):1728–37. doi: 10.1016/S0960-9822(02)01214-9
19. Tsichritzis T, Gaentzsch PC, Kosmidis S, Brown AE, Skoulakis EM, Ligoxygakis P, et al. A drosophila ortholog of the human cylindromatosis tumor suppressor gene regulates triglyceride content and antibacterial defense. Development. (2007) 134(14):2605–14. doi: 10.1242/dev.02859
20. Guntermann S, Primrose DA, Foley E. Dnr1-dependent regulation of the drosophila immune deficiency signaling pathway. Dev Comp Immunol (2009) 33(1):127–34. doi: 10.1016/j.dci.2008.07.021
21. Thevenon D, Engel E, Avet-Rochex A, Gottar M, Bergeret E, Tricoire H, et al. The drosophila ubiquitin-specific protease dUSP36/Scny targets IMD to prevent constitutive immune signaling. Cell Host Microbe (2009) 6(4):309–20. doi: 10.1016/j.chom.2009.09.007
22. Yagi Y, Lim YM, Tsuda L, Nishida Y. Fat facets induces polyubiquitination of imd and inhibits the innate immune response in drosophila. Genes Cells (2013) 18(11):934–45. doi: 10.1111/gtc.12085
23. Engel E, Viargues P, Mortier M, Taillebourg E, Coute Y, Thevenon D, et al. Identifying USPs regulating immune signals in drosophila: USP2 deubiquitinates imd and promotes its degradation by interacting with the proteasome. Cell Commun Signal (2014) 12:41. doi: 10.1186/s12964-014-0041-2
24. Fernando MD, Kounatidis I, Ligoxygakis P. Loss of trabid, a new negative regulator of the drosophila immune-deficiency pathway at the level of TAK1, reduces life span. PLos Genet (2014) 10(2):e1004117. doi: 10.1371/journal.pgen.1004117
25. Ji S, Luo Y, Cai Q, Cao Z, Zhao Y, Mei J, et al. LC domain-mediated coalescence is essential for otu enzymatic activity to extend drosophila lifespan. Mol Cell (2019) 74(2):363–77.e5. doi: 10.1016/j.molcel.2019.02.004
26. Kanoh H, Tong LL, Kuraishi T, Suda Y, Momiuchi Y, Shishido F, et al. Genome-wide RNAi screening implicates the E3 ubiquitin ligase Sherpa in mediating innate immune signaling by toll in drosophila adults. Sci Signal (2015) 8(400):ra107. doi: 10.1126/scisignal.2005971
27. Ji S, Sun M, Zheng X, Li L, Sun L, Chen D, et al. Cell-surface localization of pellino antagonizes toll-mediated innate immune signalling by controlling MyD88 turnover in drosophila. Nat Commun (2014) 5:3458. doi: 10.1038/ncomms4458
28. Kang MJ, Ryoo HD. Suppression of retinal degeneration in drosophila by stimulation of ER-associated degradation. Proc Natl Acad Sci U S A. (2009) 106(40):17043–8. doi: 10.1073/pnas.0905566106
29. Mitra S, Ryoo HD. The unfolded protein response in metazoan development. J Cell Sci (2019) 132(5):1–9. doi: 10.1242/jcs.217216
30. Hwang J, Qi L. Quality control in the endoplasmic reticulum: Crosstalk between ERAD and UPR pathways. Trends Biochem Sci (2018) 43(8):593–605. doi: 10.1016/j.tibs.2018.06.005
31. Thomson T, Liu N, Arkov A, Lehmann R, Lasko P. Isolation of new polar granule components in drosophila reveals p body and ER associated proteins. Mech Dev (2008) 125(9-10):865–73. doi: 10.1016/j.mod.2008.06.005
32. Saito K, Chen M, Bard F, Chen S, Zhou H, Woodley D, et al. TANGO1 facilitates cargo loading at endoplasmic reticulum exit sites. Cell. (2009) 136(5):891–902. doi: 10.1016/j.cell.2008.12.025
33. Gotze M, Dufourt J, Ihling C, Rammelt C, Pierson S, Sambrani N, et al. Translational repression of the drosophila nanos mRNA involves the RNA helicase belle and RNA coating by Me31B and trailer hitch. RNA. (2017) 23(10):1552–68. doi: 10.1261/rna.062208.117
34. Jeske M, Moritz B, Anders A, Wahle E. Smaug assembles an ATP-dependent stable complex repressing nanos mRNA translation at multiple levels. EMBO J (2011) 30(1):90–103. doi: 10.1038/emboj.2010.283
35. Nakamura A, Amikura R, Hanyu K, Kobayashi S. Me31B silences translation of oocyte-localizing RNAs through the formation of cytoplasmic RNP complex during drosophila oogenesis. Development. (2001) 128(17):3233–42. doi: 10.1242/dev.128.17.3233
36. Jensen L, Venkei ZG, Watase GJ, Bisai B, Pletcher S, Lee CY, et al. me31B regulates stem cell homeostasis by preventing excess dedifferentiation in the drosophila male germline. J Cell Sci (2021) 134(14):1–10. doi: 10.1242/jcs.258757
37. Tritschler F, Eulalio A, Helms S, Schmidt S, Coles M, Weichenrieder O, et al. Similar modes of interaction enable trailer hitch and EDC3 to associate with DCP1 and Me31B in distinct protein complexes. Mol Cell Biol (2008) 28(21):6695–708. doi: 10.1128/MCB.00759-08
38. Han J, Kaufman RJ. Physiological/pathological ramifications of transcription factors in the unfolded protein response. Genes Dev (2017) 31(14):1417–38. doi: 10.1101/gad.297374.117
39. Hetz C, Papa FR. The unfolded protein response and cell fate control. Mol Cell (2018) 69(2):169–81. doi: 10.1016/j.molcel.2017.06.017
40. Walter P, Ron D. The unfolded protein response: From stress pathway to homeostatic regulation. Science. (2011) 334(6059):1081–6. doi: 10.1126/science.1209038
41. Hollien J, Weissman JS. Decay of endoplasmic reticulum-localized mRNAs during the unfolded protein response. Science. (2006) 313(5783):104–7. doi: 10.1126/science.1129631
42. Acosta-Alvear D, Zhou Y, Blais A, Tsikitis M, Lents NH, Arias C, et al. XBP1 controls diverse cell type- and condition-specific transcriptional regulatory networks. Mol Cell (2007) 27(1):53–66. doi: 10.1016/j.molcel.2007.06.011
43. Huang HW, Zeng X, Rhim T, Ron D, Ryoo HD. The requirement of IRE1 and XBP1 in resolving physiological stress during drosophila development. J Cell Sci (2017) 130(18):3040–9. doi: 10.1242/jcs.203612
44. Braakman I, Hebert DN. Protein folding in the endoplasmic reticulum. Cold Spring Harb Perspect Biol (2013) 5(5):a013201. doi: 10.1101/cshperspect.a013201
45. Ghosh R, Wang L, Wang ES, Perera BG, Igbaria A, Morita S, et al. Allosteric inhibition of the IRE1alpha RNase preserves cell viability and function during endoplasmic reticulum stress. Cell. (2014) 158(3):534–48. doi: 10.1016/j.cell.2014.07.002
46. Kyostila K, Cizinauskas S, Seppala EH, Suhonen E, Jeserevics J, Sukura A, et al. A SEL1L mutation links a canine progressive early-onset cerebellar ataxia to the endoplasmic reticulum-associated protein degradation (ERAD) machinery. PLos Genet (2012) 8(6):e1002759. doi: 10.1371/journal.pgen.1002759
47. Hegde RS, Ploegh HL. Quality and quantity control at the endoplasmic reticulum. Curr Opin Cell Biol (2010) 22(4):437–46. doi: 10.1016/j.ceb.2010.05.005
48. McCambridge A, Solanki D, Olchawa N, Govani N, Trinidad JC, Gao M. Comparative proteomics reveal Me31B's interactome dynamics, expression regulation, and assembly mechanism into germ granules during drosophila germline development. Sci Rep (2020) 10(1):564. doi: 10.1038/s41598-020-57492-y
49. Ryoo HD, Domingos PM, Kang MJ, Steller H. Unfolded protein response in a drosophila model for retinal degeneration. EMBO J (2007) 26(1):242–52. doi: 10.1038/sj.emboj.7601477
50. Hua Y, Zhu Y, Hu Y, Kong F, Duan R, Zhang C, et al. A feedback regulatory loop involving dTrbd/dTak1 in controlling IMD signaling in drosophila melanogaster. Front Immunol (2022) 13:932268. doi: 10.3389/fimmu.2022.932268
Keywords: Sip3, Me31B, Xbp1, ERAD, Toll innate immune defense, Drosophila melanogaster
Citation: Zhu Y, Liu L, Zhang C, Zhang C, Han T, Duan R, Jin Y, Guo H, She K, Xiao Y, Goto A, Cai Q and Ji S (2023) Endoplasmic reticulum-associated protein degradation contributes to Toll innate immune defense in Drosophila melanogaster. Front. Immunol. 13:1099637. doi: 10.3389/fimmu.2022.1099637
Received: 16 November 2022; Accepted: 22 December 2022;
Published: 19 January 2023.
Edited by:
Liang Jiang, Southwest University, ChinaReviewed by:
Ilias Kounatidis, The Open University, United KingdomCopyright © 2023 Zhu, Liu, Zhang, Zhang, Han, Duan, Jin, Guo, She, Xiao, Goto, Cai and Ji. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Qingshuang Cai, YnJpc2txaW5nc2h1YW5nQDE2My5jb20=; Shanming Ji, amlzbUBhaGF1LmVkdS5jbg==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.