
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Immunol. , 26 January 2023
Sec. Inflammation
Volume 13 - 2022 | https://doi.org/10.3389/fimmu.2022.1089200
 Li-Ping Zhao
Li-Ping Zhao Jian Wu
Jian Wu Wei Quan
Wei Quan Yu Zhou
Yu Zhou Hui Hong
Hui Hong Gu-Yu Niu
Gu-Yu Niu Ting Li
Ting Li Shu-Bing Huang
Shu-Bing Huang Chen-Meng Qiao
Chen-Meng Qiao Wei-Jiang Zhao
Wei-Jiang Zhao Chun Cui
Chun Cui Yan-Qin Shen*
Yan-Qin Shen*Accumulative studies suggest that inflammatory bowel disease (IBD) may cause multiple central nervous system (CNS) pathologies. Studies have found that indoleamine-2,3-dioxygenase (IDO, rate-limiting enzyme of the kynurenine (Kyn) pathway) deficient mice were protected from endotoxin induced cognitive impairment, and Kyn administration induced cognitive memory deficits in both control and IDO-deficient mice. However, there is no investigation of the brain Kyn pathway in IBD, thus we investigated whether dextran sulfate sodium (DSS)-induced colitis could cause dysregulation of Kyn pathway in brain, and also in serum. C57BL/6J mice were given drinking water with 2% DSS for 10 consecutive days to induce colitis. In serum, we found significant increase in Kyn and kynurenic acid (Kyna) level, which was regulated by IDO-1 and KAT2 (rate-limiting enzymes of Trp-Kyn-Kyna pathway). Similarly, by analyzing GEO datasets, higher IDO-1 levels in peripheral blood monocytes and colon of UC patients was found. Furthermore, the Kyn pathway was significantly upregulated in the cerebral cortex under the action of IDO-1 after DSS treatment, which ultimately induced the neurotoxic phenotype of astrocytes. To investigate whether gut microbiota is involved in IBD-induced Kyn pathway dysregulation, we performed intestinal flora 16S rRNA sequencing and found that DSS-induced colitis significantly altered the composition and diversity of the gut microbiota. Metabolic function analysis also showed that Tryptophan metabolism, NOD-like receptor signaling pathway and MAPK signaling pathway were significantly up-regulated in the 2% DSS group. A significant association between intestinal flora and Trp metabolism (both in serum and brain) was found by correlation analysis. Overall, this study revealed that DSS-induced colitis causes dysregulation of the Kyn pathway in serum and brain by affecting rate-limiting enzymes and intestinal flora.
Inflammatory bowel disease (IBD) is characterized by chronic inflammation in gastrointestinal tract, including Crohn’s disease (CD) and ulcerative colitis (UC) (1). IBD basically own features of weight loss, diarrhea, abdominal pain and bloody stools (2). The etiology of IBD is still unknow, which is thought to be related to genetic susceptibility, immune dysfunction and microbial imbalance.
Recent years, increasing studies have focused on the influence of IBD on the central nervous system (CNS). Dextran sulfate sodium (DSS) treatment induced depressive-like behavior in mice (3–5), and depression/anxiety is more frequently in IBD patients compared with the general population (6). Furthermore, DSS-induced colitis impaired hippocampal neurogenesis and affected spatial and cognitive memory in mice (7, 8). However, the connection between gut and brain of IBD is still need to be explored.
Kynurenine (Kyn) pathway is the main pathway of tryptophan (Trp) metabolism and is involved in multiple CNS pathologies (9). The main rate-limiting enzyme of Kyn pathway is indoleamine-2,3-dioxygenase (IDO) and tryptophan-2,3-dioxygenase (TDO). IDO deficient mice were protected from cognitive impairment induced by endotoxin, and peripheral injection of Kyn caused recognition memory deficit in both Control and IDO null mice (10). In patients with Alzheimer’s disease, increased IDO-1/TDO expression was co-located with neurofibrillary tangles in hippocampus (11, 12). 1-Methyltryptophan (1-MT, an IDO-1 inhibitor) treatment improved neurotransmitter levels and behavioral parameters in 6-OHDA induced Parkinson’s disease mice while reducing the neuroinflammation marker (TNF-α, IFN-γ and IL-6), oxidative stress and neuronal apoptosis (Caspase-3) (13). These studies suggested that the Kyn pathway is an important mechanism involved in neuropathology, and it is necessary to clarify the change of Kyn pathway under the disease states. However, the effect and mechanism of IBD on brain Kyn pathway has not been explored.
Among pathogenic factors that affect both the gut and brain, the role of intestinal flora cannot be ignored. As an important medium of gut-brain communication, gut microbiota is involved in the pathogenesis of certain CNS diseases such as Parkinson’s disease (14), Alzheimer’s disease (15) and multiple sclerosis (16). Also, intestinal flora is essential for the development of DSS-induced colitis. Germ-free mice showed only minimal inflammation after the induction of colitis by DSS compared with conventional mice (17). Besides, studies have described the alterations in composition and function of the microbiota in IBD patients (18–21). And the alterations were also found in metabolite profiles including bile acids, short-chain fatty acids and Trp metabolites (22). In the intestinal tract, gut microbiota produces a variety of Trp metabolizing enzymes such as Trp decarboxylases, Trp monooxygenase and tryptophanase. These enzymes convert dietary Trp to tryptamine and other molecules, which altered host physiology by reducing available Trp and activated aryl hydrocarbon receptors (23–25). In serum and brain, the effect of gut microbiota on Trp metabolism and the Kyn pathway remain unclear.
Therefore, this study explored the effects of DSS-induced colitis on the Kyn pathway and whether intestinal flora is involved as the mechanism.
To verify the successful induction of IBD in mice, H&E staining of colon tissue was performed. Significant inflammatory infiltration was found in the mucosal layer after 2% DSS treatment, which lead to a higher histological score in the 2% DSS group compared with the Control group (Figures 1A, B). Besides, the expression level of inflammatory cytokines in colon confirmed the inflammation observed in colitis mice. Il-6 mRNA level (proinflammatory cytokine) was significantly increased 8.3 times (Figure 1C), and il-10 (anti-inflammatory cytokines) was significantly decreased by 44.2% in the 2% DSS group compared with the Control group (Figure 1D). Colonic inflammation was associated with epithelial integrity and mucus barrier disruption. Significant reduction of ZO-1 (72.1%) and muc2 (95.4%) was observed in the 2% DSS group (Figures 1E, F), which reflected the damage of mucosal barrier and mucous barrier. Furthermore, colon length and spleen weight were measured as macroscopic evaluations of colonic inflammation. In the 2% DSS group, a 29.4% reduction of colon length was found (Figure 1G), and spleen weight was elevated 3.0 times compared with Control group (Figure 1H). Disease activity index (DAI) values were significantly increased after DSS treatment due to the increased body weight loss, diarrhea and bleeding (Figures 1I, J). Overall, 2% DSS induced significant intestinal inflammation and intestinal barrier damage.
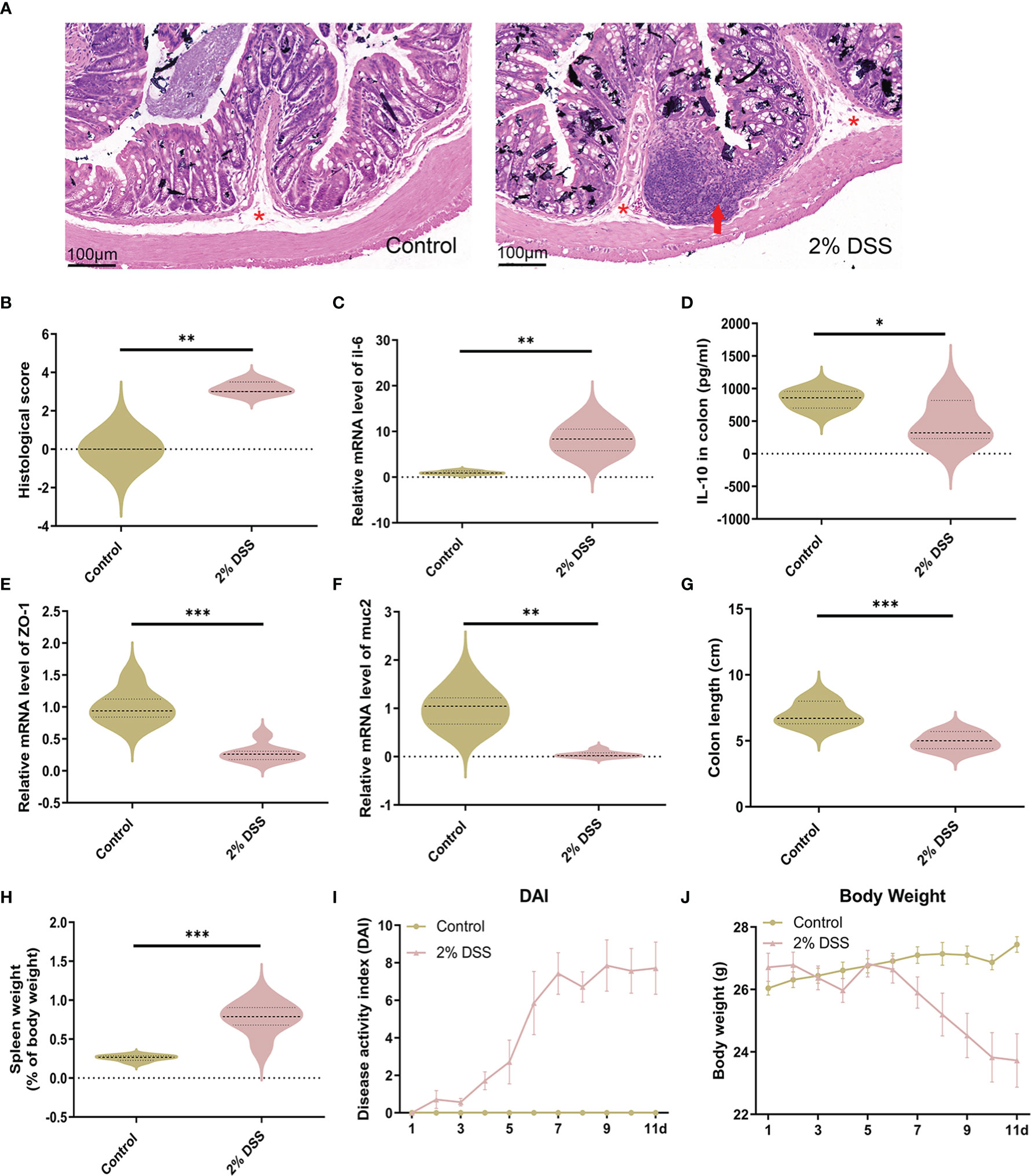
Figure 1 DSS treatment induces intestinal inflammation and intestinal barrier disruption. (A) Representative images of colon H&E staining (×25 magnification) showed significant inflammatory cell infiltration after 2% DSS treatment. Red asterisks indicate the location of submucosa, and red arrows indicate extensive infiltration of inflammatory cells in the mucosa. (B) Histological injury scores were calculated from inflammatory cell infiltration and tissue damage. (C) Il-6 (proinflammatory cytokine) mRNA levels in colon were detected by RT-qPCR. (D) Il-10 (anti-inflammatory cytokine) in colon were detected by ELISA. (E, F) ZO-1 (intestinal tight junction related molecules) and muc2 (mucins secreted by goblet cells) mRNA levels were detected by RT-qPCR. (G, H) Colon length and spleen weight were measured in two groups. (I) Evaluation of Disease Activity Index (DAI) during the experiment. (J) Change of body weight during the experiment. Data are presented as mean ± SEM, n=3 in (B), and n=5-7 in C–J. *P < 0.05, **P < 0.01, ***P < 0.001, P values were determined by the independent samples t test (B, C, E, G–J) or Mann-Whitney U test (D, F).
To investigate whether 2% DSS induced colitis affected the metabolism of serum Trp, we detected the levels of Trp and its metabolites (Kyn and Kyna) in serum. The level of serum Trp was decreased by 30.6% in 2% DSS group compared with Control group (Figure 2A). Kyn level in serum was increased 1.3 times (Figure 2B), while Kyna was decreased by 66.9% after DSS treatment (Figure 2C). Moreover, Kyn/Trp ratio and Kyna/Kyn ratio confirmed this effect. Kyn/Trp ratio was significantly elevated 1.8 times in 2% DSS group (Figure 2D), which reflected increased efficiency of Trp metabolism into Kyn. Kyna/Kyn ratio was decreased by 73.2% in 2% DSS treatment group (Figure 2E), indicated the reduction of Kyn metabolism into Kyna. These results indicated that DSS treatment significantly changed the levels of Trp and its metabolites in serum.
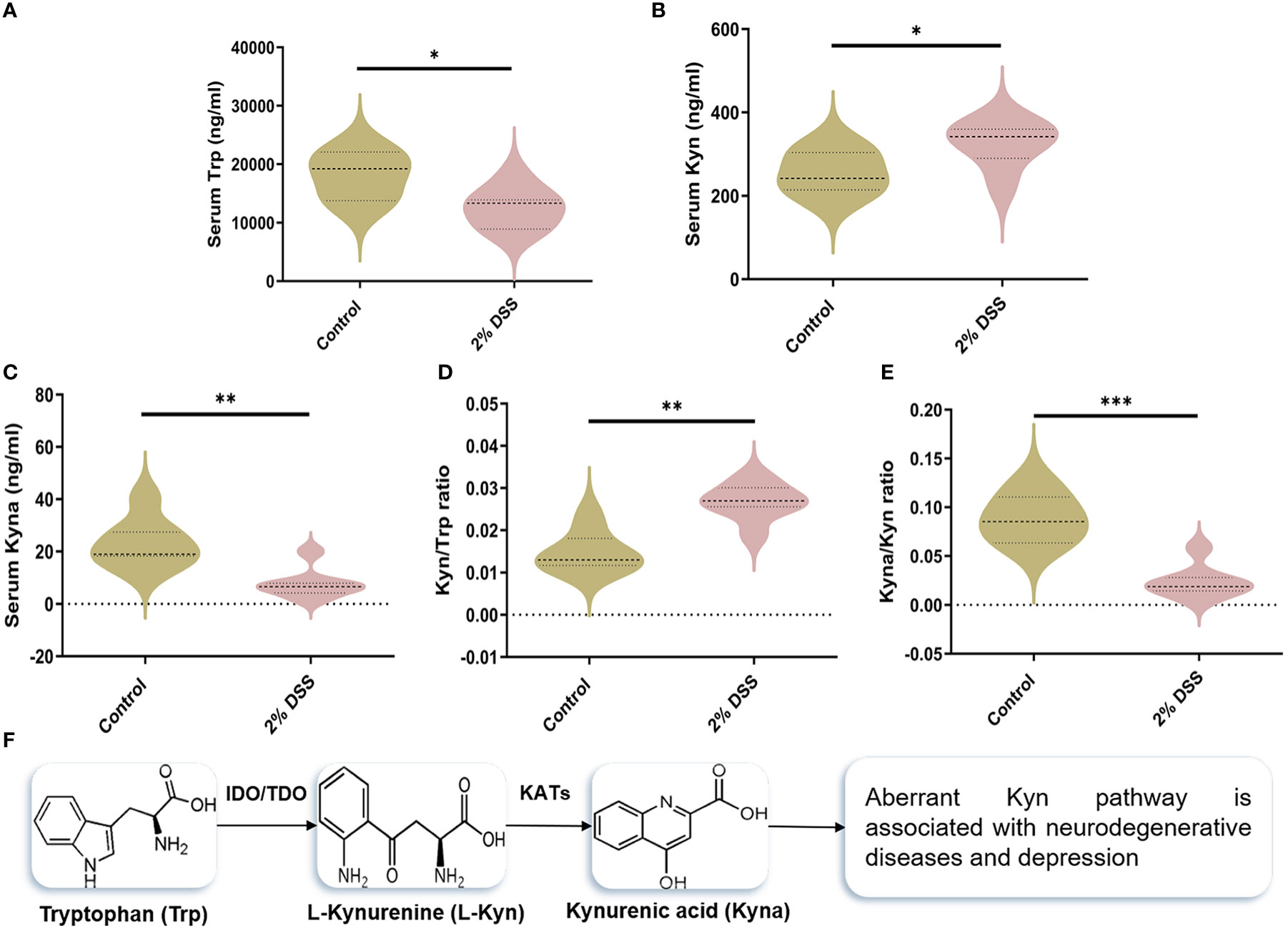
Figure 2 DSS-induced colitis alters tryptophan (Trp) metabolism in serum. (A–C) Serum levels of Trp, Kynurenine (Kyn) and kynurenic acid (Kyna) in each group was detected by liquid chromatography-mass spectrometry (LC-MS) assay. (D) Kyn/Trp ratio indicated elevated efficiency of Trp metabolism into Kyn. (E) Kyna/Kyn ratio indicated decreased efficiency of Kyn metabolism into Kyna. (F) Representation of Trp catabolism along the Kyn pathways. Data are presented as mean ± SEM, n=7. *P < 0.05, **P < 0.01, ***P < 0.001, P values were determined by the independent samples t test (A, B, E) or Mann-Whitney U test (C, D).
The levels of rate-limiting enzymes are the direct factor affecting Trp metabolites. IDO-1 and TDO-2 are the main rate-limiting enzymes that metabolize Trp into Kyn, and KAT2 is rate-limiting enzyme that metabolize Kyn into Kyna (Figure 2F). IDO-1, TDO-2 and KAT2 are all expressed in liver cells. Therefore, we detected the levels of IDO-1, TDO-2 and KAT2 in the liver to explore the causes of Trp metabolite changes. The protein level of IDO-1 in liver did not achieve significance between the two groups (Figures 3A, D), while a significant increase (5.9 times) of IDO-1 mRNA level was observed in 2% DSS group compared with Control group (Figure 3G). TDO-2 decreased by 64.1% and 41.2% respectively in protein level (Figures 3B, E) and mRNA level (Figure 3H) in 2% DSS group compared with Control group. This indicated that the increase in Kyn level was not due to elevated expression of IDO-1 and TDO2. Besides, KAT2 decreased by 28.8% and 67.0% respectively in protein level (Figures 3C, F) and mRNA level (Figure 3I) in 2% DSS group compared with Control group, which suggested that decreased Kyna level might be contributed to reduced KAT2 protein. In brief, the alteration of Trp-Kyn-Kyna pathway in serum was partly regulated by rate-limiting enzymes.
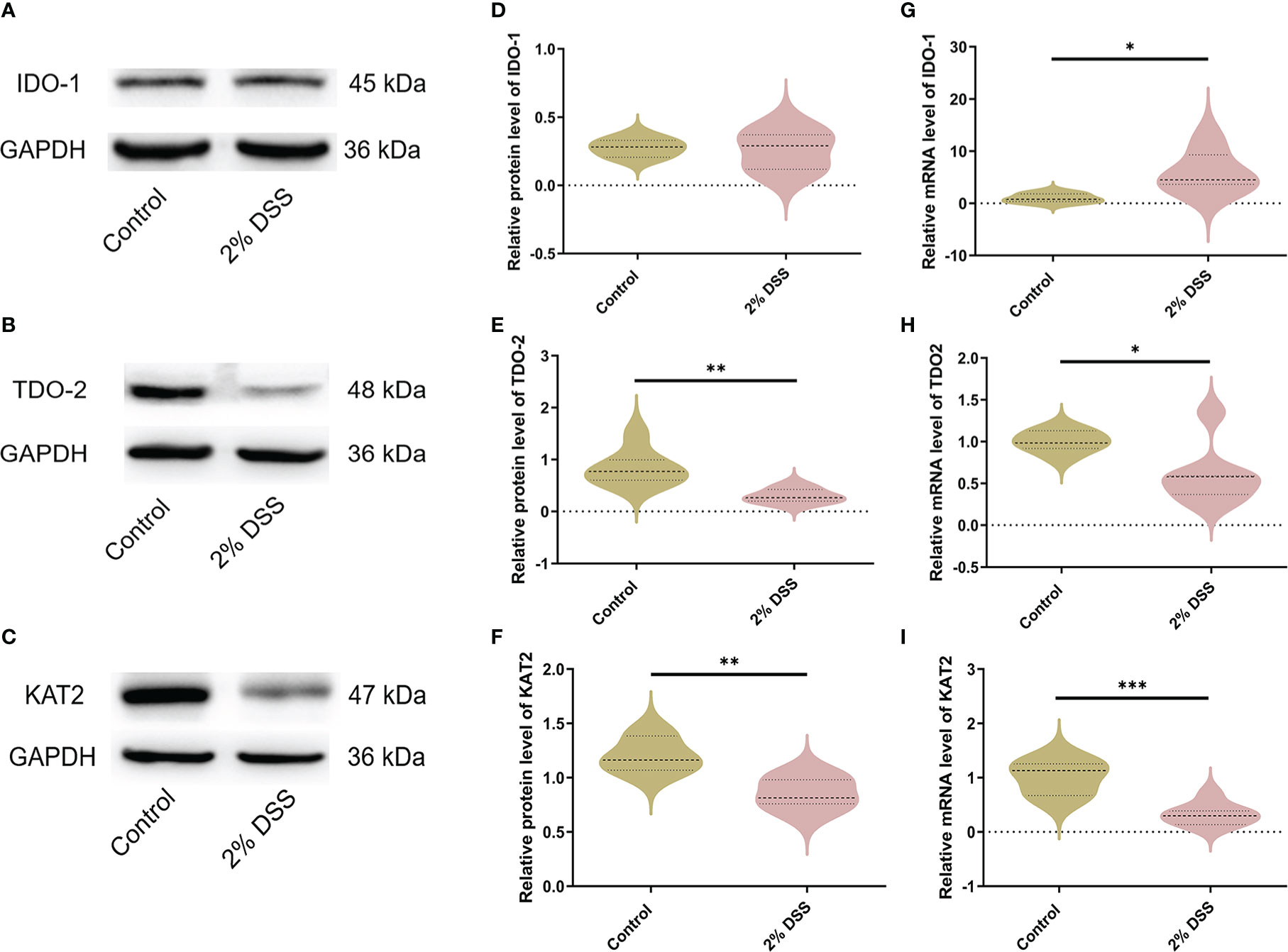
Figure 3 DSS-induced colitis alters metabolic enzyme levels of Kyn pathway in liver. (A–C) Representative western blot of IDO-1, TDO-2 and KAT2 expression in liver. (D–F) Intensity of bands was quantified with Image J software and quantitative data for IDO-1, TDO-2 and KAT2 following normalization to GAPDH. (G–I) IDO-1, TDO-2 and KAT2 mRNA levels in liver were detected by RT-qPCR. Data are presented as mean ± SEM, n=6-7. *P < 0.05, **P < 0.01, ***P < 0.001, P values were determined by the independent samples t test (D–F, G, I) or Mann-Whitney U test (H).
To further confirm the dysregulation of Kyn pathway in IBD patients, we analyzed four NCBI Gene Expression Omnibus (GEO) datasets (GDS1615, GDS4365, GDS3119, GDS3268) containing expression profiles of peripheral blood mononuclear cells and colonic mucosa from IBD patients, and one GEO dataset (GDS3859) containing expression profile of colon tissue from DSS-induced mouse colitis. We found that IDO-1 mRNA levels were significantly higher in peripheral blood mononuclear of UC patients but not CD patients compared with the normal group, which indicated that colitis induced the activation of IDO-1 in immune cells (Figure 4A). In colonic mucosa of UC patients, IDO-1 expression was significantly higher in UC-active/UC-inflamed patients compared with Control or UC-remission/UC-uninflamed patients, which reflected that colitis induced the activation of IDO-1 in colon (Figures 4B–E). Besides, colonic IDO-1 expression was significantly increased after 6 days treatment of DSS compared with normal group in C57BL/6J mice aged 12~14 weeks old (Figure 4F). Collectively, these results demonstrated the increase of IDO-1 in UC patients and colitis mice, indicated upregulation of Kyn pathway.
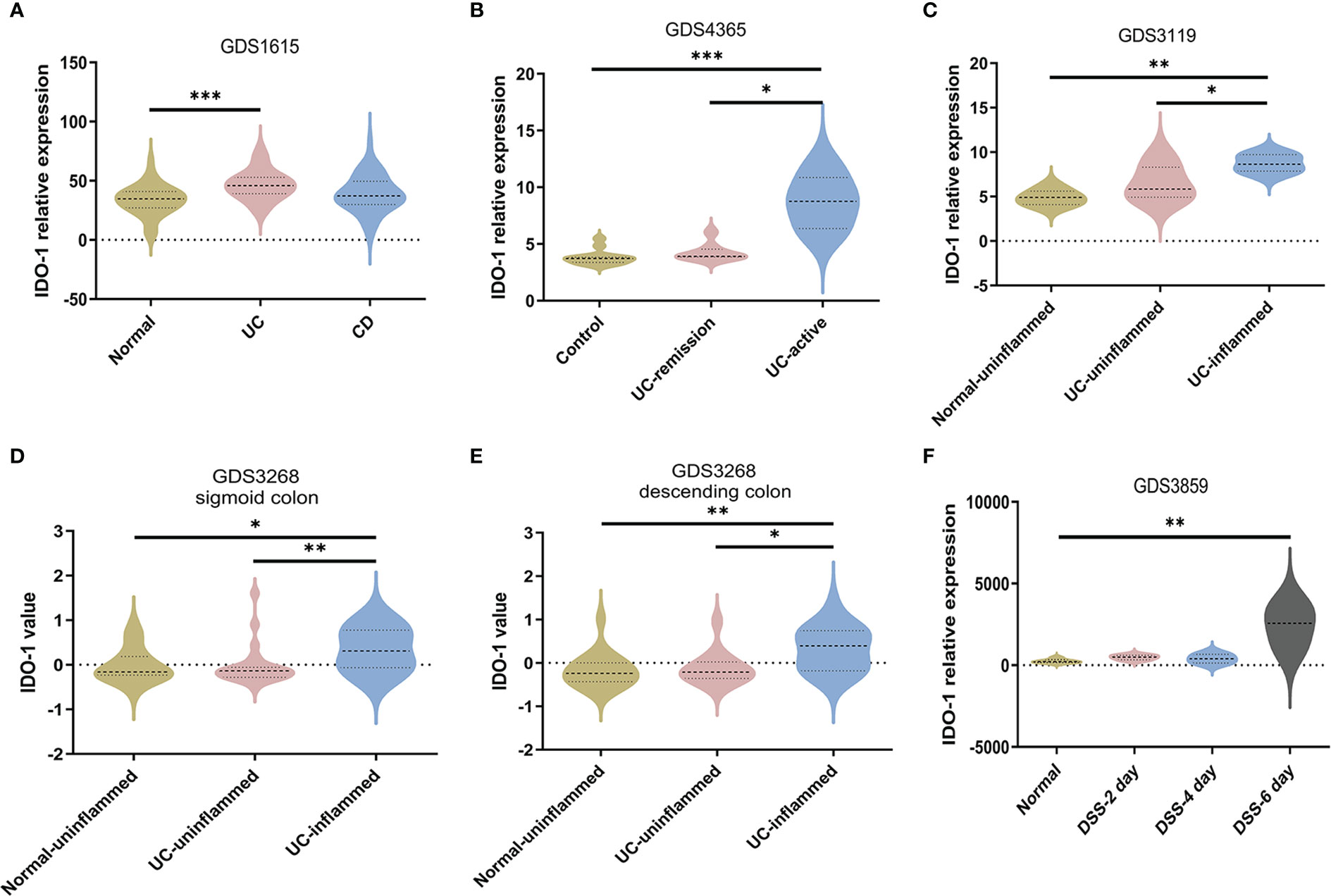
Figure 4 Upregulation of IDO-1 in peripheral blood mononuclear cells and colitis tissues. (A) IDO-1 expression in peripheral blood mononuclear cells from UC patients and CD patients were collected from GEO database (GDS1615, n=42 for normal group, n=26 for UC group and n=59 for CD group). (B) IDO-1 expression in control colon tissues and colonic tissues from UC patients (remission or active stage) were collected and analyzed from GDS4365 (n=13 for Control group, n=8 for UC-remission group and n=15 for UC-active group) (C–E) IDO-1 expression in uninflamed normal colonic tissues and colonic tissues from UC patients (uninflamed or inflamed colon) were collected and analyzed from GDS3119 (n=5 for Normal-uninflamed group, n=13 for UC-uninflamed group and n=8 for UC-inflamed group) and GDS3268 (sigmoid colon: n=24 for Normal-uninflamed group, n=25 for UC-uninflamed group and n=32 for UC-inflamed group; descending colon: n=22 for Normal-uninflamed group, n=15 for UC-uninflamed group and n=19 for UC-inflamed group). (F) IDO-1 expression in normal colonic tissues of C56BL/6J mice and 3% DSS-treated mice were collected and analyzed from GDS3859 (n=5 for Normal group, n=6 in DSS-2 day, DSS-4 day and DSS-6 day groups). Data are presented as mean ± SEM. *P < 0.05, **P < 0.01, ***P < 0.001, P values were determined by one-way ANOVA with post hoc comparisons of Bonferroni (A, C) or Kruskal-Wallis test (B, D–F).
Subsequently, we explored the effects of DSS-induced colitis on Trp metabolism in the cerebral cortex. Compared with Control group, IDO-1 mRNA level was significantly increased 1.8 times after 2% DSS treatment (Figure 5A), while the level of KAT2 was unchanged in 2% DSS group (Figure 5B). As a result, Kyn level was elevated in 2% DSS group compared with Control group, while Trp and Kyna did not achieve significance in the two groups (Figures 5C–E). This suggested that the level of Kyn and Kyna were under the regulation of IDO-1 and KAT2. Kyn/Trp ratio also showed that the efficiency of Trp metabolism to Kyn was improved (Figure 5F), which was consistent with elevated levels of IDO-1 and Kyn. The Kyna/Kyn ratio did not achieve significance between Control and 2% DSS group (Figure 5G), which reflected Kyn-Kyna pathway was not affected by DSS treatment. The transfer of Kyn from blood to brain is mediated by L-type amino acid transporter 1 (LAT1). We detected SLC3A2 (4F2hc heavy subunit) and SLC7A5 (CD98 light subunit) of LAT1 transporter subunits at the mRNA levels. There was no change in SLC3A2 between two groups (Figure 5H), while SLC7A5 was significantly decreased by 38.8% in 2% DSS group compared with Control group (Figure 5I). Therefore, we inferred that elevated Kyn in the cerebral cortex was mainly derived from IDO-1-mediated local synthesis rather than translocation from the blood.
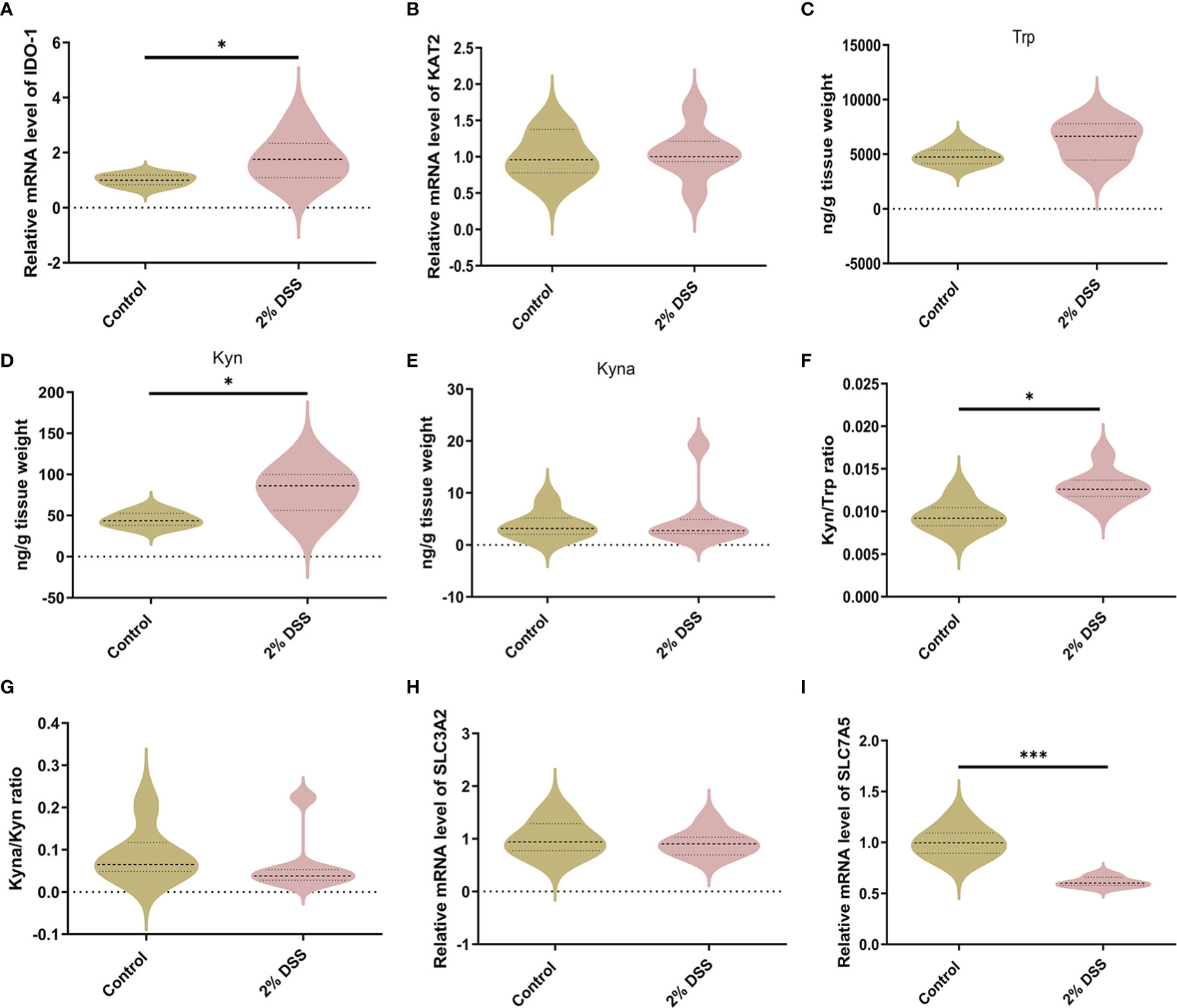
Figure 5 DSS-induced colitis alters Kyn pathway of Trp metabolism in the cortex. (A, B) Trp metabolic enzymes levels of IDO-1 and KAT2 in cortex were detected by RT-qPCR. (C–E) Trp metabolite levels including Trp, Kyn and Kyna in cortex were detected by LC-MS. (F) Kyn/Trp ratio indicated elevated efficiency of Trp metabolism into Kyn in cortex. (G) Kyna/Kyn ratio indicated unchanged efficiency of Kyn metabolism into Kyna in cortex. (H, I) SLC3A2 and SLC7A5 (L-type amino acid transporter LAT1 subunits) mRNA levels in cortex were detected by RT-qPCR. Data are presented as mean ± SEM, n=6-7. *P < 0.05, ***P < 0.001, P values were determined by the independent samples t test (A–D, F, H, I) or Mann-Whitney U test (E, G).
Previous studies have shown that DSS-induced colitis caused cortical inflammation, as demonstrated by significant elevation of proinflammation cytokines (il-6 and tnf-α) and increased microglial activation in cortical tissue (26). This suggested that the cerebral cortex is a brain target affected by colitis. Furthermore, Kyn administration also induced brain infiltration of proinflammatory monocytes and astrocytic activation (27, 28). Therefore, to further investigate whether Kyn elevation caused by colitis can alter astrocyte and microglia subtypes, we detected the expression levels of astrocyte and microglia markers. In A1-specific markers of astrocyte, H2-D1, H2-T23, GBP2 and CFB mRNA levels were significantly increased by 51.9%, 44.8%, 52.9% and 121.8% (Figures 6A–D), while SerpinG1 was decreased by 24.8% in 2% DSS group compared with Control group (Figure 6E), which indicated the expression of A1-specific markers of astrocyte was elevated in colitis group. In A2-specific markers of astrocyte, EMP1 was decreased by 35.7% in 2% DSS group compared with the Control group (Figure 6F), while there was no significant difference in S100a10 and PTX3 between the two groups (Figures 6G, H). These data suggested that astrocytes in colitis group were driven to an A1 phenotype.
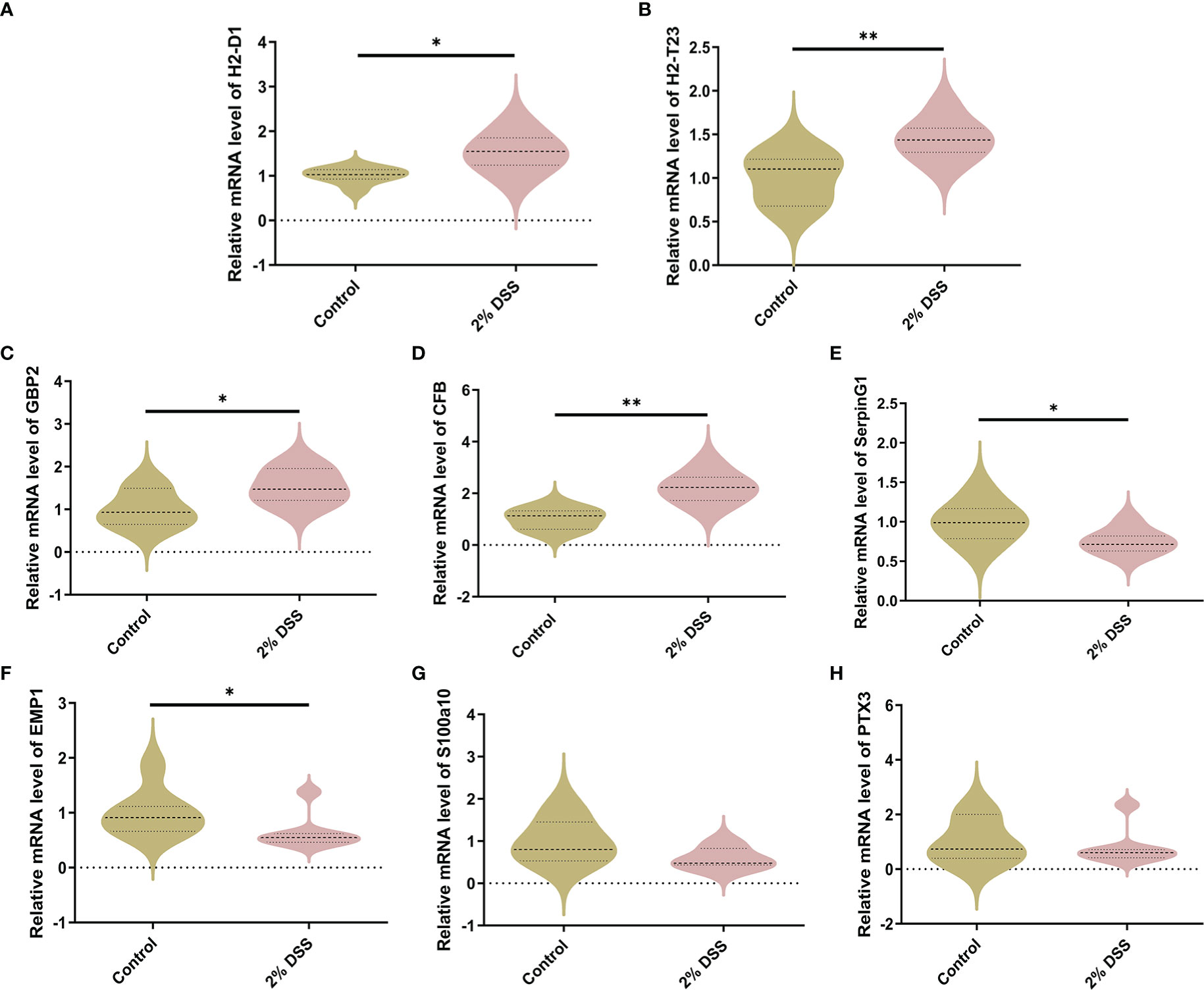
Figure 6 DSS-induced colitis induces neurotoxic phenotype (A1) of astrocyte. (A–E) A1-specific markers of astrocyte (H2-D1, H2-T23, GBP2, CFB and SerpingG1) were detected by RT-qPCR in cortex. (F–H) A2-specific markers of astrocyte (EMP1, S100a10 and PTX3) were detected by RT-qPCR in cortex. Data are presented as mean ± SEM, n=5-7. *P < 0.05, **P < 0.01, P values were determined by the independent samples t test (A–F) or Mann-Whitney U test (G, H).
For M1-specific markers of microglia, the mRNA levels of CD80 and CD32 were significantly elevated by 44.3% and 22.9% respectively in 2% DSS group (Figures 7A, B), while the levels of CD16 and CD86 did not achieve significance between 2% DSS group and Control group (Figures 7C, D). For M2-specific markers of microglia, the mRNA levels of TGF-β were significantly increased by 151.0% in 2% DSS group compared with Control group (Figure 7E), while there is no significant difference in YM-1 and CD206 between the two groups (Figures 7F, G). There was no significant change in M1/M2 phenotypes transition of microglia cells. Overall, these results suggested that DSS-induced colitis caused neurotoxic subtype of astrocytes, which may be attributed to increased levels of Kyn in the cerebral cortex.
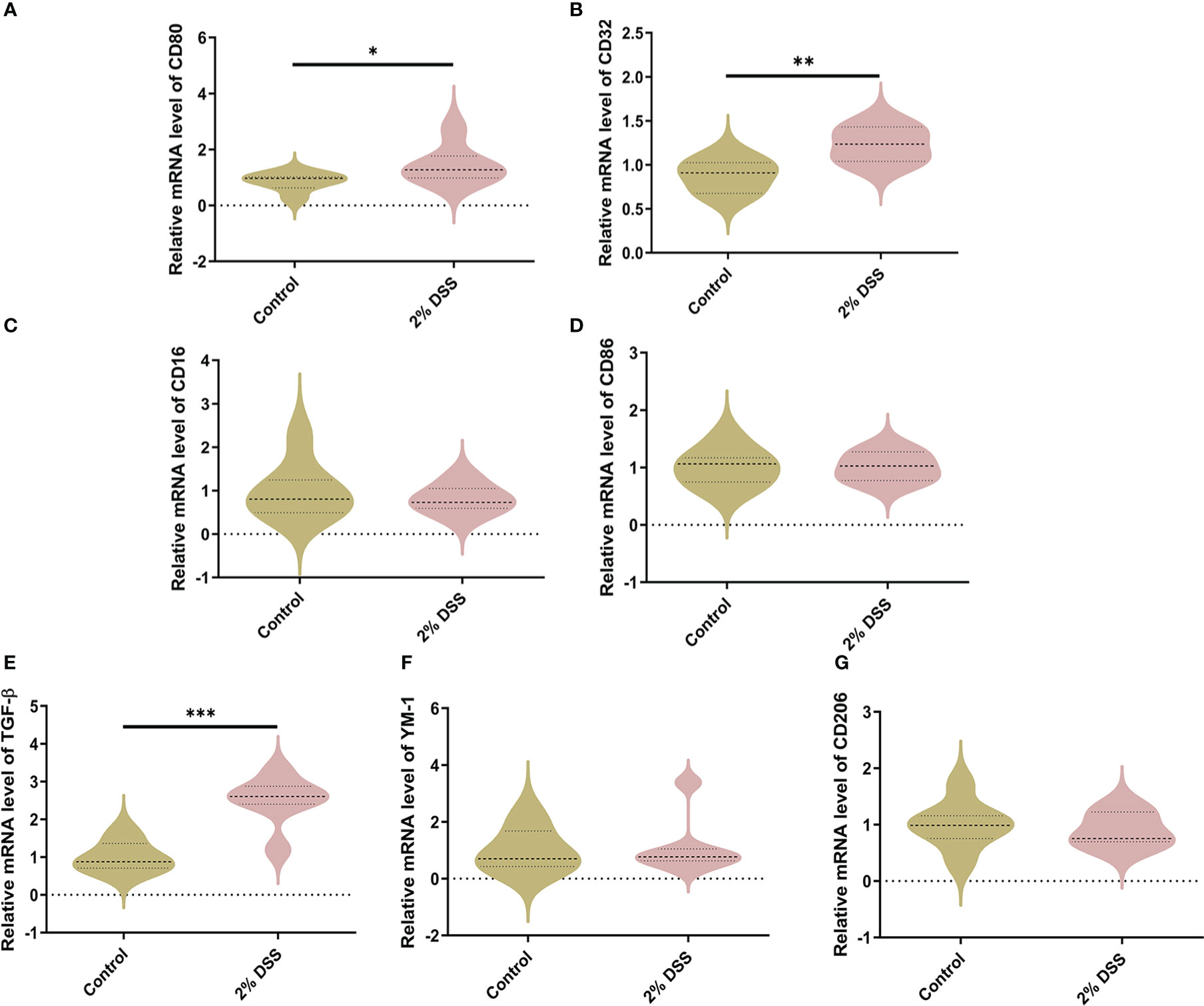
Figure 7 DSS-induced colitis alters microglial phenotypes related gene expression in cortex. (A–D) M1-specific markers of microglia (CD80, CD32, CD16, and CD86) were detected by RT-qPCR in cortex. (E–G) M2-specific markers of microglia (TGF-β, YM-1 and CD206) were detected by RT-qPCR in cortex. Data are presented as mean ± SEM, n=6-7. *P < 0.05, **P < 0.01, ***P < 0.001, P values were determined by the independent samples t test (A–D, G) or Mann-Whitney U test (E, F).
Several studies have shown that gut microbiota (such as Akkermansia muciniphila and Parabacteroides distasonis) is involved in the regulation of Trp metabolism (29–31). To investigate the characters of the gut flora after 2% DSS treatment, 16S rRNA sequencing was performed with the colon contents. Alpha diversity indices including Shannnon Index, Simpson Index and Pielou’s Evenness did not achieve significance between 2% DSS group and Control group (Figures 8A–C), which reflected unaffected species diversity and evenness. Principal coordinate analysis (PcoA) indicated a lower similarity of sample composition between the two groups (Figure 8D). Furthermore, Venn diagram showed 27 and 16 unique species respectively in Control group and 2% DSS group. And 31 common species were found in both groups (Figure 8E). The relative abundance of 31 common species was shown in the heatmap (Figure 8F), and only 8 common species achieved significance between Control group and 2% DSS group. The abundance of Bacteroides_acidifaciens, Shigella_sonnei, Parabacteroides_distasonis, Bacteroides_vulgatus, Ruminiclostridium_sp_KB18, Burkholderiales_bacterium_YL45 and Lachnospiraceae_bacterium_28-4 were significantly increased, and the abundance of Ileibacterium_valens was significantly decreased in 2% DSS group compared with Control group (Figure 9). These data reflected that DSS-induced colitis altered the diversity and composition of gut flora.
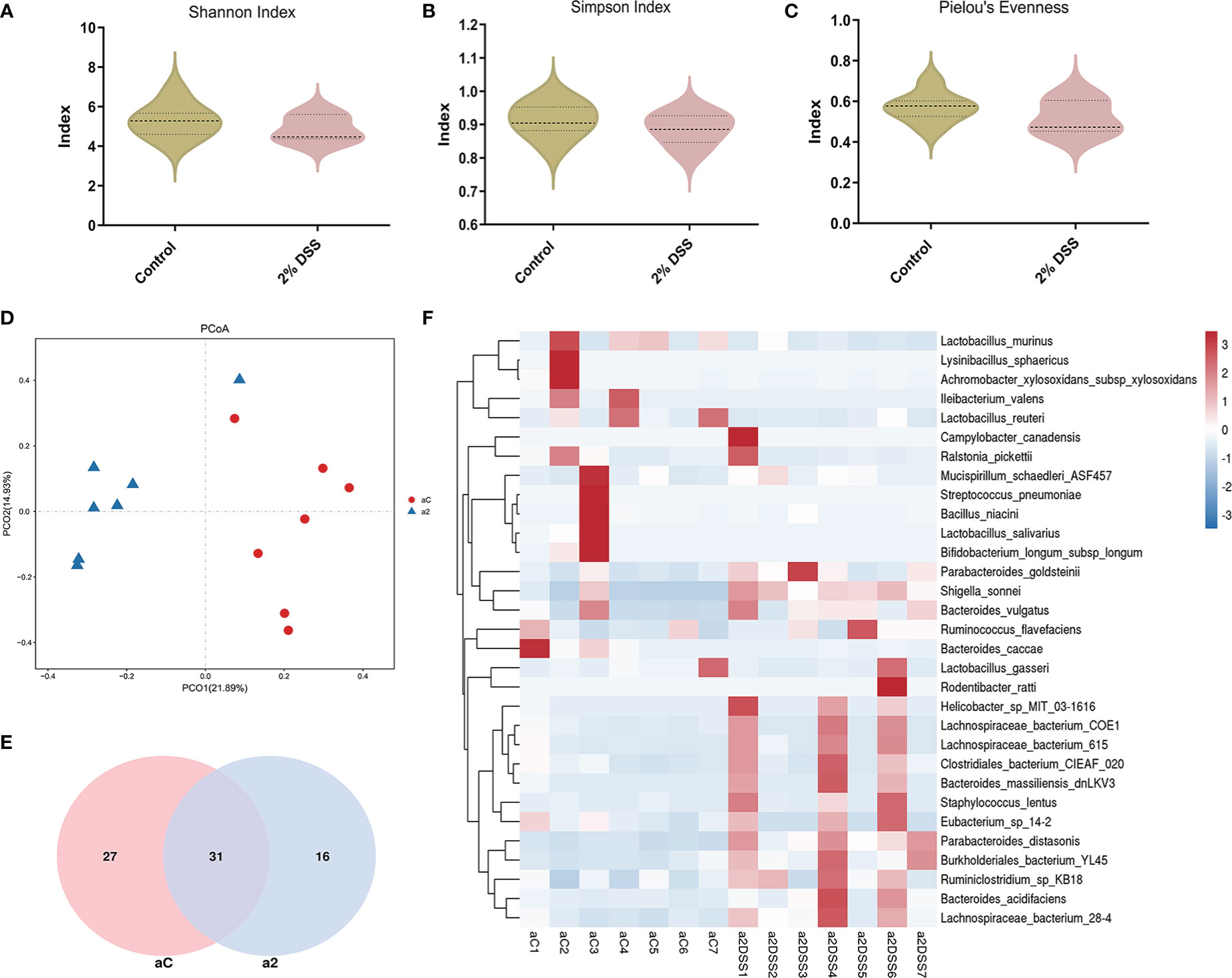
Figure 8 DSS-induced colitis significantly changed the beta diversity and species abundance of gut flora. (A–C) Analysis of alpha diversity showed no significant difference in Shannon index, Simpson index and Pielou’s evenness between Control group and 2% DSS group. (D) Principal coordinate analysis (PcoA, reflecting beta diversity) indicated lower sample composition similarity between two groups. (E) Venn diagram showed common and specific species of gut flora in Control group and 2% DSS group. (F) Heatmap showed the relative abundance of gut microbiota at the species level in Control group and 2% DSS group. Data are presented as mean ± SEM, n=7. P values were determined by the independent samples t test (A–C). aC, a2 at (D–F) represented Control group and 2% DSS group.
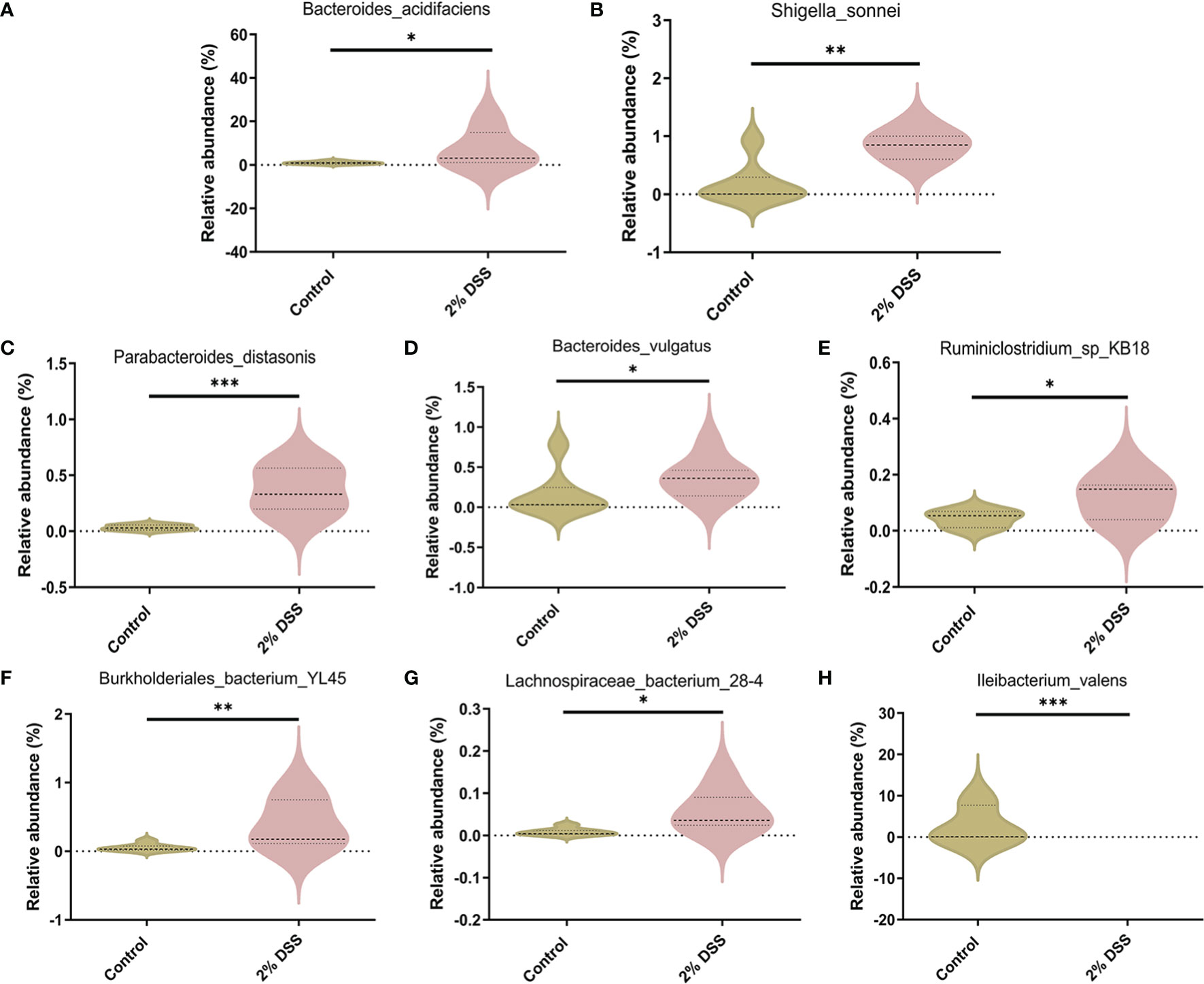
Figure 9 Species with significant abundance difference after 2% DSS treatment compared with the Control group. (A–H) At the species level, 2% DSS treatment significantly increased the abundance of Bacteroides_acidifaciens, Shigella_sonnei, Parabacteroides_distasonis, Bacteroides_vulgatus, Ruminiclostridium_sp_KB18, Burkholderiales_bacterium_YL45 and Lachnospiraceae_bacterium_28-4 (A–G), and decreased the abundance of Ileibacterium_valens (H). Data are presented as mean ± SEM, n=7. *P < 0.05, **P < 0.01, ***P < 0.001, P values were determined by the independent samples t test (C, E, G) or Mann-Whitney U test (A, B, D, F, H).
Given the change of Trp metabolism is related to intestinal flora, metabolic functions of the gut microbiota were assessed by PICRUSt2 based on 16S rRNA metagenomics data. Several functional pathways in the microbiome of 2% DSS group were found elevated, including Tryptophan metabolism, NOD-like receptor signaling pathway and MAPK signaling pathway (Figure 10). In addition, species with significant differences in abundance between the Control group and 2% DSS group were significantly associated with rate-limiting enzymes and metabolites of the Trp-Kyn-Kyna pathway (Figure 11), which indicates the regulatory effects of gut flora on Trp metabolism in the serum and cerebral cortex. Specifically, we found that several species were significantly correlated with the levels of IDO-1/TDO-2 in cortex and liver, but not with Kyn, suggested that gut microbiota may affect Kyn by regulating the level of IDO-1. The species involved in this process included: Bacteroides_acidifaciens, Shigella_sonnei, Parabacteroides_distasonis, Ileibacterium_valens, Bacteroides_vulgatus and Burkholderiales_bacterium_YL45. In addition, several species including Parabacteroides_distasonis, Ileibacterium_valens and Burkholderiales_bacterium_YL45 were significantly correlated with both KAT2 in liver and Kyna in serum, indicating that intestinal flora may regulate Kyna level in serum directly or by affecting KAT2.
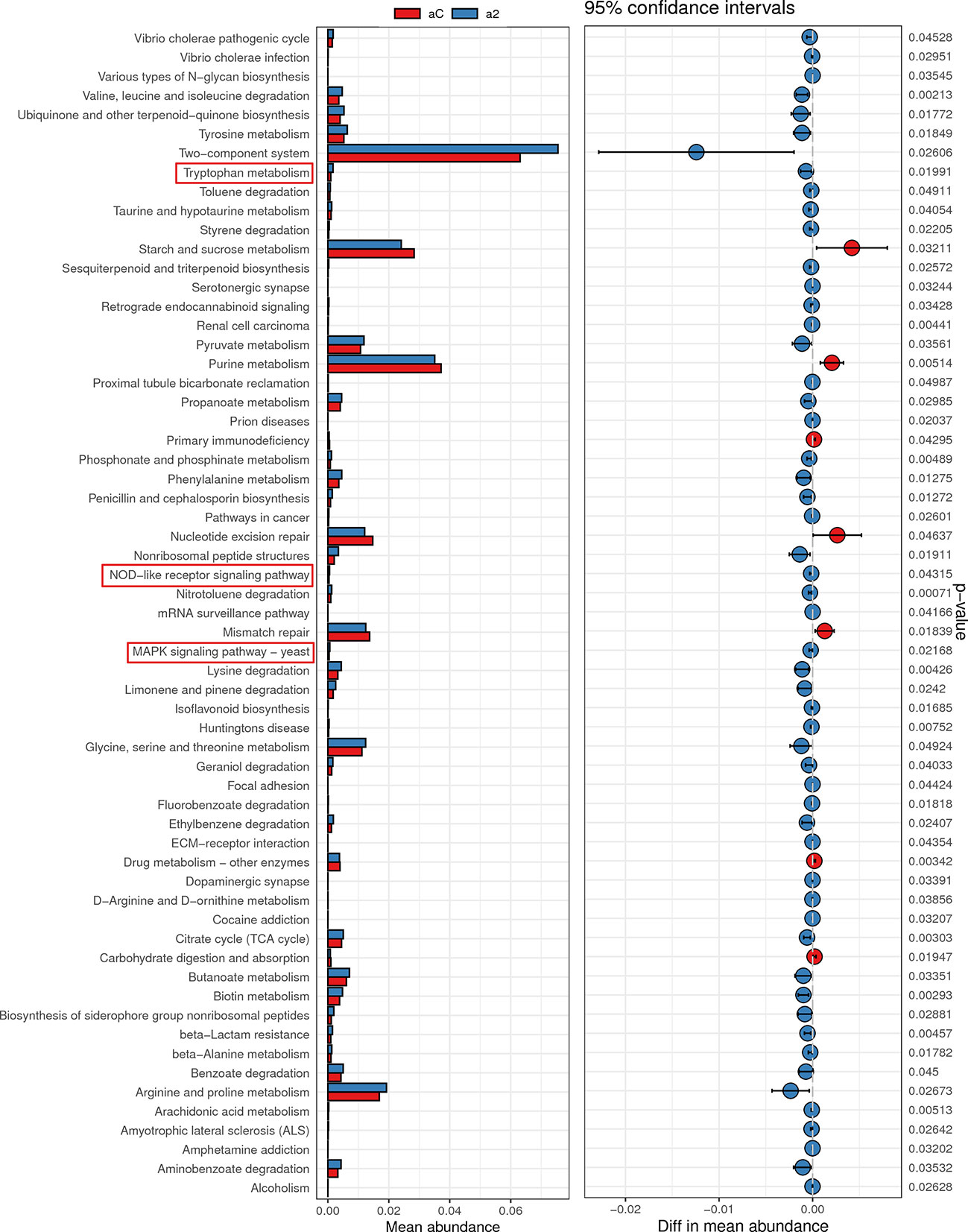
Figure 10 Metabolic function prediction of intestinal flora in Control group and 2% DSS group. Tax4Fun analysis was used to predict KEGG metabolic pathways. Relative abundance of Tryptophan metabolism, NOD-like receptor signaling pathway and MAPK signaling pathway were significantly increased in 2% DSS group compared with Control group.
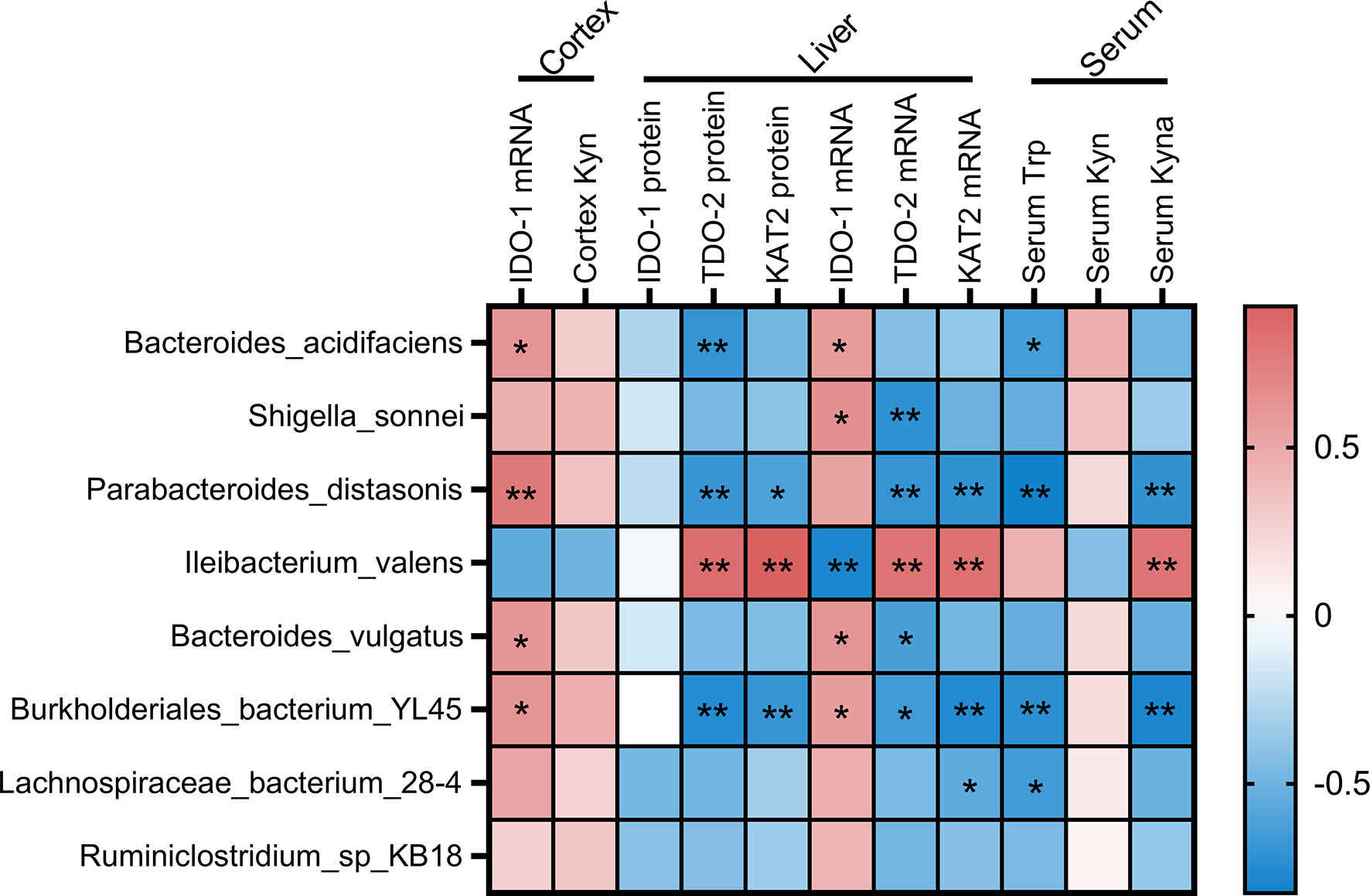
Figure 11 Associations of the intestinal flora and Trp metabolism. The intestinal flora listed in the figure were species with significant differences in abundance between Control and 2% DSS group. Pearson’s correlation coefficient was calculated for normally distributed data, and the Spearman’s correlation coefficient was calculated for non-normal data. Significant associations were displayed with asterisk (*P < 0.05, **P < 0.01).
This study investigated the effect of DSS-induced colitis on the Kyn pathway of Trp metabolism in the brain and serum, and the role of gut microbiota. In DSS-induced colitis mice, we found a significant increase in serum Kyn level and a significant decrease in serum Kyna level, which was mediated by the rate-limiting enzymes IDO-1 and KAT2. To verify this finding, we analyzed GEO datasets and found that IDO-1 levels were significantly upregulated in peripheral blood monocytes and colon of UC patients, indicated the dysregulation of Trp-Kyn pathway. Similarly, the Kyn pathway was significantly upregulated in the cerebral cortex under the action of IDO-1 after DSS treatment, which ultimately induced the neurotoxic phenotype of astrocytes. Furthermore, DSS-induced colitis altered gut microbiota composition and diversity. Metabolic functions analysis based on 16S rRNA metagenomics data found that Trp metabolism pathway was significantly elevated in 2% DSS group compared with the Control group. And significant correlation was found between gut microbiota and the Trp-Kyn pathway, implied the regulatory role of gut microbiota on Trp metabolism in serum and cerebral cortex.
In this study, we found that DSS-induced colitis caused a significant decrease in serum Trp content and increase in Kyn content, which was consistent with previous studies (32–34). The decrease in serum Trp content may be due to reduced food intake (the main source of Trp) in colitis mice (35). The elevation of Kyn content was mainly attributed to the decrease of its consumption. We detected the downstream product (Kyna) of Kyn and the rate-limiting enzyme (KAT2), and found that both of them were significantly reduced. In addition, the protein level of IDO-1, a rate-limiting enzyme for Kyn synthesis, did not achieve significance between Control and 2% DSS group, while the IDO-1 mRNA level was significantly increased after DSS treatment. The previous study found that only 40% of the protein variation could be explained by known mRNA abundance, and most part of the variation in proteins could be attributed to post-transcriptional regulation and measurement noise (36). We speculated that the inconsistent changes of IDO-1 mRNA and protein levels were probably due to post-transcriptional regulation.
The effect of colitis on the Kyn pathway in the brain has not been reported. In this study, we found that the Kyn pathway was significantly activated in the cerebral cortex after DSS-induced colitis, which was manifested by the significant elevation of Kyn and IDO-1. Approximately 60% of the Kyn in the brain comes from the periphery because it can cross the blood-brain barrier via the large neutral amino acid transporters (LATs) (37, 38). We detected two subunits of LAT1 and found that only SLC7A5 was significantly decreased in 2% DSS group, which implied that the increased Kyn in cerebral cortex is mainly due to IDO-1 mediated local elevation rather than from peripheral blood. Kyn in the brain can be metabolized by astrocytes and microglia to downstream products such as 3-hydroxykynurenine and Kyna, and can also affect cell status (39). Kyn (48 μM) treatment reduced LPS-induced expression of proinflammatory cytokines (IL-1β, IL-6 and TNF-α) in BV2 microglia (39). Besides, Kyn treatment resulted in up-regulation of NLRP2 protein expression in primary astrocytes in a concentration-dependent and time-dependent manner, which indicated the proinflammatory effect of Kyn (28). In our study, DSS-induced colitis causes neurotoxic phenotype of astrocytes but not microglia in the cerebral cortex, which may be attributed to elevated Kyn. However, the effect of DSS-induced elevation of Kyn on brain function remains unclear and needs to be further explored.
Some papers suggested gut microbiota is involved in Trp metabolism. For instance, fecal microbiota transplants from IDO-1-deficient mice directly attenuated the severity of DSS-induced colitis in IDO-1+/+ mice (40), which suggested that intestinal flora regulated the severity of colitis in Trp metabolism-dependent manner. In addition, fecal microbiota transplants from schizophrenic patients activated Kyn-Kyna pathway of Trp metabolism (41), which demonstrated the involvement of gut microbiota in Trp metabolism. In this study, we also found that DSS-induced colitis caused significant changes in the composition of gut microbiota compared with Control group, which is supportive to above-mentioned studies (42–44). In addition, we investigated the relationship between gut microbiota and Trp metabolism. PICRUSt analysis found that Trp metabolism-related microbial gene functions were higher in 2% DSS group. This is consistent with our findings in serum and brain, suggested that gut microbiota played an important role in Trp metabolism. Moreover, correlation analysis showed a significant association between intestinal flora and the Trp-Kyn-Kyna pathway of Trp metabolism. The previous study has shown that JAK-STAT signaling pathway and melatonin-JNK-FoxO1 pathway may be the regulators of IDO-1 expression (45). Our study found NOD-like receptor signaling pathway and MAPK signaling pathway of microbial gene functions were activated after DSS treatment, which implied that these two pathways may be the mechanism of gut microbiota affecting Trp metabolism in future studies.
Male C57BL/6 mice (aged 10 weeks, Vital River Laboratory Animal Technology, Pinghu, China) were used in this study. Animals were kept in specific pathogen-free environment (12h light/12h dark cycle) with regulated temperatures (24 ± 2.0°C) and humidity (55 ± 10%). Water and food were available ad libitum (Jiangsu Xietong Organism, China). Mice were acclimated for one week before the experiments. All animal experimental procedures were approved by the Animal Ethics Committee of Jiangnan University.
The mice were randomly allotted to two groups: Control group and 2% DSS group (n = 7 in each group). The Control group received sterile tap water as drinking water. Experimental colitis was induced by adding 2% DSS (36,000-50,000 Da, MP Biomedicals, Solon, USA) in drinking water for 10 days (from day 1 to day 10). Body weights, diarrhea (stool consistency) and rectal bleeding were daily recorded. All mice were sacrificed after being deeply anesthetized by isoflurane on day 11. Tissues (colon, liver and cerebral cortex), blood and colonic contents were collected. After centrifugation (4°C, 3000 rpm, 15 minutes), serum was separated from the blood.
Colonic tissue (proximal colon to the rectum) was collected and fixed in 4% paraformaldehyde at 4°C. Following embedded in paraffin, the colon was sectioned transversely (5 μm in thickness) and stained with H&E (C0105S, Beyotime, Shanghai, China). The histological changes were recorded under LuxScan 10K/B (CapitalBio, Beijing, China). As described previously, two parameters were used to calculate the histological damage score: tissue damage (score: 0-3) and infiltration of inflammatory cells (score: 0-3) in a double-blind fashion (46). Three sections obtained from each of three sites at 100 μm distance were evaluated, and mice were scored individually with each score representing the mean of nine sections (47).
According to the manufacturer’s instructions, total RNA from colon and cerebral cortex was extracted using Total RNA Extraction Reagent (R401-01, vazyme, Nanjing, China). Then, the total RNA was reverse transcribed into cDNA using HiScript III RT SuperMix for qPCR (R323-01, vazyme, Nanjing, China). PCR was performed using ChamQ Universal SYBR qPCR Master Mix (Q711-00, vazyme, Nanjing, China). Primers used are listed in Table 1. The 2-ΔΔCt method was used for mRNA quantification. The mRNA expression of target gene was normalized to GAPDH expression.
For extracting total protein, the liver was homogenized in a mixture of RIPA buffer (P0013C, Beyotime, Shanghai, China), 2% phosphatase inhibitor (P1081, Beyotime, Shanghai, China) and 1% phenylmethanesulfonyl fluoride (PMSF, ST506, Beyotime, Shanghai, China). Following centrifugation at 14,000 r/min for 15 minutes at 4°C, supernatants were collected. The level of il-10 (EK0417, Boster, Wuhan, China) in colon was quantified by using commercial ELISA kit according to the manufacturer’s instructions. The detection limit of the kit is 15.6-1000 pg/ml for il-10.
As previously described, body weight loss, stool consistency and bleeding were recorded each day to determine the Disease Activity Index (DAI) (48). The scores were quantified as follows: weight loss: 0 (no loss), 1 (1-5%), 2 (5-10%), 3 (10-20%), and 4 (>20%); stool consistency: 0 (normal), 2 (loose stool), and 4 (diarrhea); and bleeding: 0 (no blood), 1 (Hemoccult positive), 2 (Hemoccult positive and visual pellet bleeding), and 4 (gross bleeding, blood around anus). DAI for each mouse was calculated by adding together the scores for weight loss, stool consistency and fecal blood.
To detect the concentrations of Trp, Kyn and Kyna in serum and cerebral cortex, LC-MS/MS analysis was conducted according to Wang et al. with slight modifications (49). In brief, the analysis was performed using Orbitrap Q-Exactive Plus mass spectrometer (Thermo Fisher Scientific, Bremen, Germany) connected with Vanquish UHPLC System (Thermo Fisher Scientific, Bremen, Germany). Chromatographic separation was achieved on the ACQUITY UPLC BEH C18 column (100 mm × 2.1 mm, 1.7 μm, Waters Corporation, Milford, MA, USA). A linear gradient mobile phase containing 0.1% formic acid water (A) and acetonitrile (B) was programmed as follows: 0–0.5 min, 95% A; 0.5–5 min, 95–20% A; 5–6 min, 20% A; 6–6.1 min, 20–95% A and 6.1–8 min, 95% A (flow rate: 0.4 mL/min). For the electrospray ionization quantitative analysis, the positive ion mode with parallel reaction monitoring was utilized. The detailed MS ionization source conditions included spray voltage at 2800 V; Capillary temperature at 325°C; sheath gas flow rate at 35; aux gas flow rate at 15; sweep gas flow rate at 0; aux gas heater temperature at 350°C. Data were processed by using the Thermo Xcalibur software (Thermo Fisher Scientific, Waltham, MA, USA).
Total protein concentration in liver was determined with a BCA Protein Quantification Kit (E112-02, vazyme, Nanjing, China). After boiling denaturation, total protein (35 μg) was separated by 10%~12% SDS-PAGE, then transferred to PVDF membranes (ISEQ00010, Millipore, Billerica, MA, USA). The membranes were blocked with 5% skim milk for 2 h at room temperature and then incubated (overnight at 4°C) with the following primary antibodies: Rabbit anti-indoleamine 2,3-dioxygenase 1 (IDO-1, 1:1000, 13268-1-AP, Proteintech, Wuhan, China), Rabbit anti-TDO-2 (1:500, 15880-1-AP, Proteintech, Wuhan, China), Rabbit anti-AADAT (KAT2, 1:500, A13090, ABclonal, Wuhan, China) and Rabbit anti-GAPDH (1:8000, ET1601-4, Huabio, Hangzhou, China). Then the HRP conjugated goat anti-rabbit IgG was incubated for 2 h at room temperature. The protein bands were visualized by enhanced chemiluminescence (ECL) reagent (P90720, Millipore, USA) and Gel Image System (Bio-Rad, Hercules, CA, USA). Target signals were analyzed by using Image J software (NIH, Bethesda, MA, USA).
IDO-1 gene expression levels of patients with IBD (GDS1615, GDS4365, GDS3119 and GDS3268) and of mouse colitis models (GDS3859) were downloaded from the NCBI GEO database.
The genomic DNA was extracted from the fecal samples by using the PowerSoil DNA Isolation Kit (MOBIO, Carlsbad, CA, USA). The V3~V4 hypervariable regions of bacterial 16S rRNA gene were amplified with the primers 341F (5′-CCTACGGGNGGCWGCAG-3′) and 806R (5′-GGACTACHVGGGTATCTAAT-3′). The target fragments of PCR amplified products were recovered, and then the amplified products of PCR were quantified by fluorescence. The qualified sequencing library was used for Illumina MiSeq pyrosequencing analysis.
Using the SPSS 22.0 software (IBM SPSS Statistics, Armonk, New York, USA), normality was evaluated by the Shapiro-Wilk test. Differences between the two groups were analyzed by independent samples T test and Mann-Whitney U test respectively for parametric values and nonparametric values. Differences among three groups were analyzed by one-way ANOVA with post hoc comparisons of Bonferroni and Kruskal-Wallis test for parametric values and nonparametric values. Values were shown as mean ± SEM. P < 0.05 was considered statistically significant between groups (*P < 0.05, **P < 0.01, ***P < 0.001).
The data presented in the study are deposited in the SRA database repository, accession number PRJNA916349.
The animal study was reviewed and approved by Animal Ethics Committee of Jiangnan University.
Y-QS and L-PZ: conception and design of study. L-PZ, JW, WQ, YZ, HH, G-YN, TL and S-BH: acquisition of data. L-PZ, Y-QS, W-JZ, CC and C-MQ: analysis and interpretation of data. L-PZ and Y-QS: writing and review of the paper. All authors contributed to the article and approved the submitted version.
This study was supported by Grants from The National Natural Science Foundation of China (81771384 and 82171429 to Y-QS), Public Health Research Center at Jiangnan University (JUPH201801 to Y-QS), and National First-Class Discipline Program of Food Science and Technology (JUFSTR20180101 to Y-QS), Wuxi Municipal Health Commission (1286010241190480 to Y-QS).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
1. Xavier RJ, Podolsky DK. Unravelling the pathogenesis of inflammatory bowel disease. Nature (2007) 448:427–34. doi: 10.1038/nature06005
2. Ramos GP, Papadakis KA. Mechanisms of disease: Inflammatory bowel diseases. Mayo Clin Proc (2019) 94:155–65. doi: 10.1016/j.mayocp.2018.09.013
3. Takahashi K, Nakagawasai O, Nemoto W, Odaira T, Sakuma W, Onogi H, et al. Effect of enterococcus faecalis 2001 on colitis and depressive-like behavior in dextran sulfate sodium-treated mice: involvement of the brain-gut axis. J Neuroinflamm (2019) 16:201. doi: 10.1186/s12974-019-1580-7
4. Dong X, Lu K, Lin P, Che H, Li H, Song L, et al. Saccharina japonica ethanol extract ameliorates Depression/Anxiety-like behavior by inhibiting inflammation, oxidative stress, and apoptosis in dextran sodium sulfate induced ulcerative colitis mice. Front Nutr (2021) 8:784532. doi: 10.3389/fnut.2021.784532
5. Xia B, Liu X, Li X, Wang Y, Wang D, Kou R, et al. Sesamol ameliorates dextran sulfate sodium-induced depression-like and anxiety-like behaviors in colitis mice: the potential involvement of the gut-brain axis. Food Funct (2022) 13:2865–83. doi: 10.1039/D1FO03888E
6. Irving P, Barrett K, Nijher M, de Lusignan S. Prevalence of depression and anxiety in people with inflammatory bowel disease and associated healthcare use: population-based cohort study. Evid-Based Ment Heal (2021) 24:102–9. doi: 10.1136/ebmental-2020-300223
7. Gampierakis IA, Koutmani Y, Semitekolou M, Morianos I, Polissidis A, Katsouda A, et al. Hippocampal neural stem cells and microglia response to experimental inflammatory bowel disease (IBD). Mol Psychiatry (2021) 26:1248–63. doi: 10.1038/s41380-020-0651-6
8. He XF, Li LL, Xian WB, Li MY, Zhang LY, Xu JH, et al. Chronic colitis exacerbates NLRP3-dependent neuroinflammation and cognitive impairment in middle-aged brain. J Neuroinflamm (2021) 18:153. doi: 10.1186/s12974-021-02199-8
9. Platten M, Nollen EAA, Rohrig UF, Fallarino F, Opitz CA. Tryptophan metabolism as a common therapeutic target in cancer, neurodegeneration and beyond. Nat Rev Drug Discovery (2019) 18:379–401. doi: 10.1038/s41573-019-0016-5
10. Heisler JM, O’Connor JC. Indoleamine 2,3-dioxygenase-dependent neurotoxic kynurenine metabolism mediates inflammation-induced deficit in recognition memory. Brain Behav Immun (2015) 50:115–24. doi: 10.1016/j.bbi.2015.06.022
11. Bonda DJ, Mailankot M, Stone JG, Garrett MR, Staniszewska M, Castellani RJ, et al. Indoleamine 2,3-dioxygenase and 3-hydroxy-kynurenine modifications are found in the neuropathology of alzheimer’s disease. Redox Rep (2010) 15:161–8. doi: 10.1179/174329210X12650506623645
12. Wu W, Nicolazzo JA, Wen L, Chung R, Stankovic R, Bao SS, et al. Expression of tryptophan 2,3-dioxygenase and production of kynurenine pathway metabolites in triple transgenic mice and human alzheimer’s disease brain. PloS One (2013) 8:e59749. doi: 10.1371/journal.pone.0059749
13. Sodhi RK, Bansal Y, Singh R, Saroj P, Bhandari R, Kumar B, et al. IDO-1 inhibition protects against neuroinflammation, oxidative stress and mitochondrial dysfunction in 6-OHDA induced murine model of parkinson’s disease. Neurotoxicology (2021) 84:184–97. doi: 10.1016/j.neuro.2021.03.009
14. Sampson TR, Debelius JW, Thron T, Janssen S, Shastri GG, Ilhan ZE, et al. Gut microbiota regulate motor deficits and neuroinflammation in a model of parkinson’s disease. Cell (2016) 167:1469–80.e12. doi: 10.1016/j.cell.2016.11.018
15. Kim N, Jeon SH, Ju IG, Gee MS, Do J, Oh MS, et al. Transplantation of gut microbiota derived from alzheimer’s disease mouse model impairs memory function and neurogenesis in C57BL/6 mice. Brain Behav Immun (2021) 98:357–65. doi: 10.1016/j.bbi.2021.09.002
16. Berer K, Gerdes LA, Cekanaviciute E, Jia XM, Xiao L, Xia Z, et al. Gut microbiota from multiple sclerosis patients enables spontaneous autoimmune encephalomyelitis in mice. P Natl Acad Sci USA (2017) 114:10719–24. doi: 10.1073/pnas.1711233114
17. Hernandez-Chirlaque C, Aranda CJ, Ocon B, Capitan-Canadas F, Ortega-Gonzalez M, Carrero JJ, et al. Germ-free and antibiotic-treated mice are highly susceptible to epithelial injury in DSS colitis. J Crohns Colitis (2016) 10:1324–35. doi: 10.1093/ecco-jcc/jjw096
18. Ni J, Wu GD, Albenberg L, Tomov VT. Gut microbiota and IBD: causation or correlation? Nat Rev Gastroenterol Hepatol (2017) 14:573–84. doi: 10.1038/nrgastro.2017.88
19. Morgan XC, Tickle TL, Sokol H, Gevers D, Devaney KL, Ward DV, et al. Dysfunction of the intestinal microbiome in inflammatory bowel disease and treatment. Genome Biol (2012) 13:R79. doi: 10.1186/gb-2012-13-9-r79
20. Lane ER, Zisman TL, Suskind DL. The microbiota in inflammatory bowel disease: current and therapeutic insights. J Inflammation Res (2017) 10:63–73. doi: 10.2147/JIR.S116088
21. Blander JM, Longman RS, Iliev ID, Sonnenberg GF, Artis D. Regulation of inflammation by microbiota interactions with the host. Nat Immunol (2017) 18:851–60. doi: 10.1038/ni.3780
22. Lavelle A, Sokol H. Gut microbiota-derived metabolites as key actors in inflammatory bowel disease. Nat Rev Gastro Hepat (2020) 17:223–37. doi: 10.1038/s41575-019-0258-z
23. Gao K, Mu CL, Farzi A, Zhu WY. Tryptophan metabolism: A link between the gut microbiota and brain. Adv Nutr (2020) 11:709–23. doi: 10.1093/advances/nmz127
24. Williams BB, Van Benschoten AH, Cimermancic P, Donia MS, Zimmermann M, Taketani M, et al. Discovery and characterization of gut microbiota decarboxylases that can produce the neurotransmitter tryptamine. Cell Host Microbe (2014) 16:495–503. doi: 10.1016/j.chom.2014.09.001
25. Zelante T, Iannitti RG, Cunha C, De Luca A, Giovannini G, Pieraccini G, et al. Tryptophan catabolites from microbiota engage aryl hydrocarbon receptor and balance mucosal reactivity via interleukin-22. Immunity (2013) 39:372–85. doi: 10.1016/j.immuni.2013.08.003
26. Han Y, Zhao T, Cheng X, Zhao M, Gong SH, Zhao YQ, et al. Cortical inflammation is increased in a DSS-induced colitis mouse model. Neurosci Bull (2018) 34:1058–66. doi: 10.1007/s12264-018-0288-5
27. Zang X, Zheng X, Hou Y, Hu M, Wang H, Bao X, et al. Regulation of proinflammatory monocyte activation by the kynurenine-AhR axis underlies immunometabolic control of depressive behavior in mice. FASEB J (2018) 32:1944–56. doi: 10.1096/fj.201700853R
28. Zhang Q, Sun Y, He Z, Xu Y, Li X, Ding J, et al. Kynurenine regulates NLRP2 inflammasome in astrocytes and its implications in depression. Brain Behav Immun (2020) 88:471–81. doi: 10.1016/j.bbi.2020.04.016
29. Gu Z, Pei W, Shen Y, Wang L, Zhu J, Zhang Y, et al. Akkermansia muciniphila and its outer protein Amuc_1100 regulates tryptophan metabolism in colitis. Food Funct (2021) 12:10184–95. doi: 10.1039/D1FO02172A
30. Deng YY, Zhou MF, Wang JF, Yao JX, Yu J, Liu WW, et al. Involvement of the microbiota-gut-brain axis in chronic restraint stress: disturbances of the kynurenine metabolic pathway in both the gut and brain. Gut Microbes (2021) 13:1–16. doi: 10.1080/19490976.2020.1869501
31. Gimenez-Gomez P, Perez-Hernandez M, O’Shea E, Caso JR, Martin-Hernandez D, Cervera LA, et al. Changes in brain kynurenine levels via gut microbiota and gut-barrier disruption induced by chronic ethanol exposure in mice. FASEB J (2019) 33:12900–14. doi: 10.1096/fj.201900491RR
32. Zhang XJ, Yuan ZW, Qu C, Yu XT, Huang T, Chen PV, et al. Palmatine ameliorated murine colitis by suppressing tryptophan metabolism and regulating gut microbiota. Pharmacol Res (2018) 137:34–46. doi: 10.1016/j.phrs.2018.09.010
33. Qu C, Yuan ZW, Yu XT, Huang YF, Yang GH, Chen JN, et al. Patchouli alcohol ameliorates dextran sodium sulfate-induced experimental colitis and suppresses tryptophan catabolism. Pharmacol Res (2017) 121:70–82. doi: 10.1016/j.phrs.2017.04.017
34. Jing W, Dong S, Luo X, Liu J, Wei B, Du W, et al. Berberine improves colitis by triggering AhR activation by microbial tryptophan catabolites. Pharmacol Res (2021) 164:105358. doi: 10.1016/j.phrs.2020.105358
35. Wu Y, Li A, Liu H, Zhang Z, Zhang C, Ma C, et al. Lactobacillus plantarum HNU082 alleviates dextran sulfate sodium-induced ulcerative colitis in mice through regulating gut microbiome. Food Funct (2022) 13:10171–85. doi: 10.1039/D2FO02303B
36. Vogel C, Marcotte EM. Insights into the regulation of protein abundance from proteomic and transcriptomic analyses. Nat Rev Genet (2012) 13:227–32. doi: 10.1038/nrg3185
37. Gal EM, Sherman AD. L-kynurenine: its synthesis and possible regulatory function in brain. Neurochem Res (1980) 5:223–39. doi: 10.1007/BF00964611
38. Fukui S, Schwarcz R, Rapoport SI, Takada Y, Smith QR. Blood-brain barrier transport of kynurenines: implications for brain synthesis and metabolism. J Neurochem (1991) 56:2007–17. doi: 10.1111/j.1471-4159.1991.tb03460.x
39. Garrison AM, Parrott JM, Tunon A, Delgado J, Redus L, O’Connor JC. Kynurenine pathway metabolic balance influences microglia activity: Targeting kynurenine monooxygenase to dampen neuroinflammation. Psychoneuroendocrino (2018) 94:1–10. doi: 10.1016/j.psyneuen.2018.04.019
40. Shin JH, Lee YK, Shon WJ, Kim B, Jeon CO, Cho JY, et al. Gut microorganisms and their metabolites modulate the severity of acute colitis in a tryptophan metabolism-dependent manner. Eur J Nutr (2020) 59:3591–601. doi: 10.1007/s00394-020-02194-4
41. Zhu F, Guo R, Wang W, Ju Y, Wang Q, Ma Q, et al. Transplantation of microbiota from drug-free patients with schizophrenia causes schizophrenia-like abnormal behaviors and dysregulated kynurenine metabolism in mice. Mol Psychiatry (2020) 25:2905–18. doi: 10.1038/s41380-019-0475-4
42. Hu J, Huang H, Che Y, Ding C, Zhang L, Wang Y, et al. Qingchang huashi formula attenuates DSS-induced colitis in mice by restoring gut microbiota-metabolism homeostasis and goblet cell function. J Ethnopharmacol (2021) 266:113394. doi: 10.1016/j.jep.2020.113394
43. Cui L, Guan X, Ding W, Luo Y, Wang W, Bu W, et al. Scutellaria baicalensis georgi polysaccharide ameliorates DSS-induced ulcerative colitis by improving intestinal barrier function and modulating gut microbiota. Int J Biol Macromol (2021) 166:1035–45. doi: 10.1016/j.ijbiomac.2020.10.259
44. Zhang W, Cheng C, Han Q, Chen Y, Guo J, Wu Q, et al. Flos abelmoschus manihot extract attenuates DSS-induced colitis by regulating gut microbiota and Th17/Treg balance. BioMed Pharmacother (2019) 117:109162. doi: 10.1016/j.biopha.2019.109162
45. Li Y, Hu N, Yang D, Oxenkrug G, Yang Q. Regulating the balance between the kynurenine and serotonin pathways of tryptophan metabolism. FEBS J (2017) 284:948–66. doi: 10.1111/febs.14026
46. Wei J, Wei C, Wang M, Qiu X, Li Y, Yuan Y, et al. The GTPase-activating protein GIT2 protects against colitis by negatively regulating toll-like receptor signaling. Proc Natl Acad Sci U.S.A. (2014) 111:8883–8. doi: 10.1073/pnas.1309218111
47. Stopfer P, Obermeier F, Dunger N, Falk W, Farkas S, Janotta M, et al. Blocking lymphotoxin-beta receptor activation diminishes inflammation via reduced mucosal addressin cell adhesion molecule-1 (MAdCAM-1) expression and leucocyte margination in chronic DSS-induced colitis. Clin Exp Immunol (2004) 136:21–9. doi: 10.1111/j.1365-2249.2004.02402.x
48. Kim JJ, Shajib MS, Manocha MM, Khan WI. Investigating intestinal inflammation in DSS-induced model of IBD. J Vis Exp (2012) 60:3678. doi: 10.3791/3678
Keywords: DSS-induced colitis, tryptophan metabolism, kynurenine pathway (KP), indoleamine-2,3-dioxygenase (IDO), gut microbiota
Citation: Zhao L-P, Wu J, Quan W, Zhou Y, Hong H, Niu G-Y, Li T, Huang S-B, Qiao C-M, Zhao W-J, Cui C and Shen Y-Q (2023) DSS-induced colitis activates the kynurenine pathway in serum and brain by affecting IDO-1 and gut microbiota. Front. Immunol. 13:1089200. doi: 10.3389/fimmu.2022.1089200
Received: 04 November 2022; Accepted: 28 December 2022;
Published: 26 January 2023.
Edited by:
Teresa Zelante, University of Perugia, ItalyReviewed by:
Gerard M. Moloney, University College Cork, IrelandCopyright © 2023 Zhao, Wu, Quan, Zhou, Hong, Niu, Li, Huang, Qiao, Zhao, Cui and Shen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yan-Qin Shen, shenyanqin@jiangnan.edu.cn
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.